-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaThe Same but Different: Worms Reveal the Pervasiveness of Developmental System Drift
article has not abstract
Published in the journal: . PLoS Genet 10(2): e32767. doi:10.1371/journal.pgen.1004150
Category: Perspective
doi: https://doi.org/10.1371/journal.pgen.1004150Summary
article has not abstract
For at least 50 years, biologists have appreciated that some genes are universally conserved, some are restricted to particular clades, and some are found only in one species. It would be reasonable to expect that orthologous genes persist because they retain a conserved function, and in cell and developmental genetics, “function” has been defined practically by the consequences of inactivation or inappropriate activation. This simple but powerful paradigm revolutionized many fields [e.g.], [ 1]–[4] and has produced a huge body of gene-function relationships from a few genetic model organisms. Monogenic human diseases can provide a similar sort of information about our own species when the nature of the causative mutations is known. However, the phylogenetic sparseness of research models means that even when orthologous gene perturbations have been performed, it is often difficult to know what the “same” phenotype even means.
The ideal approach to characterizing the conservation of gene function would compare the effects of perturbing ortholog activity in organisms with similar anatomy, physiology, and laboratory tractability. Though we might expect that under these circumstances essentially all ortholog perturbations would give identical results, a small but growing literature suggests this is not necessarily the case. For example, a survey of over 100 transcription factor knockouts in Candida albicans, a distant relative of Saccharomyces cerevisiae, allowed the phenotypes of orthologs to be compared [5]. Most were similar, but a small number of striking shifts in the “wiring” of otherwise conserved metabolic circuits were observed (one previously described [6]). Similarly, both fushi tarazu and oskar, first identified for their roles in patterning the Drosophila melangaster embryo, may have initially functioned in the central nervous system and were only later co-opted into early development [7], [8]. These studies confirm that even when a trait is under strong stabilizing selection, with enough time, the orthologous genes that produce it can evolve surprisingly distinct roles—a phenomenon called developmental system drift (DSD) [9]. In a recent study in PLOS Genetics, Verster and colleagues [10] have undertaken a systematic search for DSD in more closely related animals.
The experimental system used by Verster et al. [10], Caenorhabditis nematodes, is especially advantageous for large-scale comparisons of gene function. Across the over 20 easily cultured species in the genus, anatomy and the cell lineages that produce it are essentially invariant, so there is no doubt of homology [11], [12]. RNAi knockdown is broadly applicable, though only a few species are susceptible to the simplest, food-borne method of introducing dsRNA [13]–[16]. For self-fertile species, such as C. elegans [2] and C. briggsae [17]–[19], screens for recessive mutations impacting development are also simple to conduct. These approaches have shown that, as expected, many genes do have highly similar knockdown/knockout phenotypes. However, surprising functional divergence has also been reported for orthologs regulating processes as diverse as sex determination [20]–[22], early embryonic patterning [23], vulval development [19], [24], and excretory physiology [25]. These differences indicate that even organs that are identical at the cellular level across a range of species can experience rapid DSD. However, these cases were not necessarily representative of the genome as a whole, and for genes involved in sex determination interpretation is complicated by the convergent evolution of hermaphroditism in C. elegans, C. briggsae, and C. sp. 11 [26], [27].
To give an unbiased estimate of the extent of DSD, Verster et al. [10] performed what may be the largest comparative analysis of gene function ever. Starting with over 1,300 genes both with strong RNAi phenotypes in C. elegans and with C. briggsae orthologs, they systematically compared the effects of knockdown in both species. After imposing several control filters, they found 91 cases of likely functional divergence (as defined by qualitatively different phenotypes), or about 7%. Though some of these may be false positives due to differential knockdown efficacy, careful quantitation for a sample of genes suggests that the fraction attributable to such artifacts is quite small. The set of genes with different phenotypes includes a disproportionately large number of transcription factors and of genes restricted to the nematode phylum, but few genes related to universal processes, such as protein synthesis factors. This makes some sense—knockdown of genes essential to cell viability will generally be lethal across the board—while transcription factors and new genes are likely to have more restricted roles and thus more potential for deviation.
The above quantification of DSD is a major contribution, but Verster et al. went further to explain how divergent knockdown phenotypes evolved. They considered three possibilities, including (1) a change in expression pattern, (2) changes in protein sequence that alter the molecular function, and (3) changes in interacting genes that alter the role or required expression level of orthologs. These were then tested using reporter constructs (to infer expression patterns) and gene chimeras that mixed and matched the regulatory and coding sequences from the two species. For some orthologs, different expression patterns were observed and use of the endogenous promoter was required for cross-species rescue. This supports the first hypothesis above. Evidence for the third hypothesis was found as well. If a contextual change alone is responsible for the distinct phenotypes, then the genes themselves should be interchangeable between species. Indeed, several cases were found in which orthologs with distinct phenotypes exhibited complete cross-species transgene rescue. Recent studies of the Lef/TCF homolog pop-1 [23] and of genes regulating germline sex determination [20], [21] and vulval development [28] have also revealed important roles for genetic context in determining the outcome of ortholog inactivation. Taken together, it appears that gene regulatory networks are constantly being reconfigured even when phenotypes are not (Figure 1), the essence of DSD.
Fig. 1. Two mechanisms for the functional divergence of orthologous genes in Caenorhabiditis. 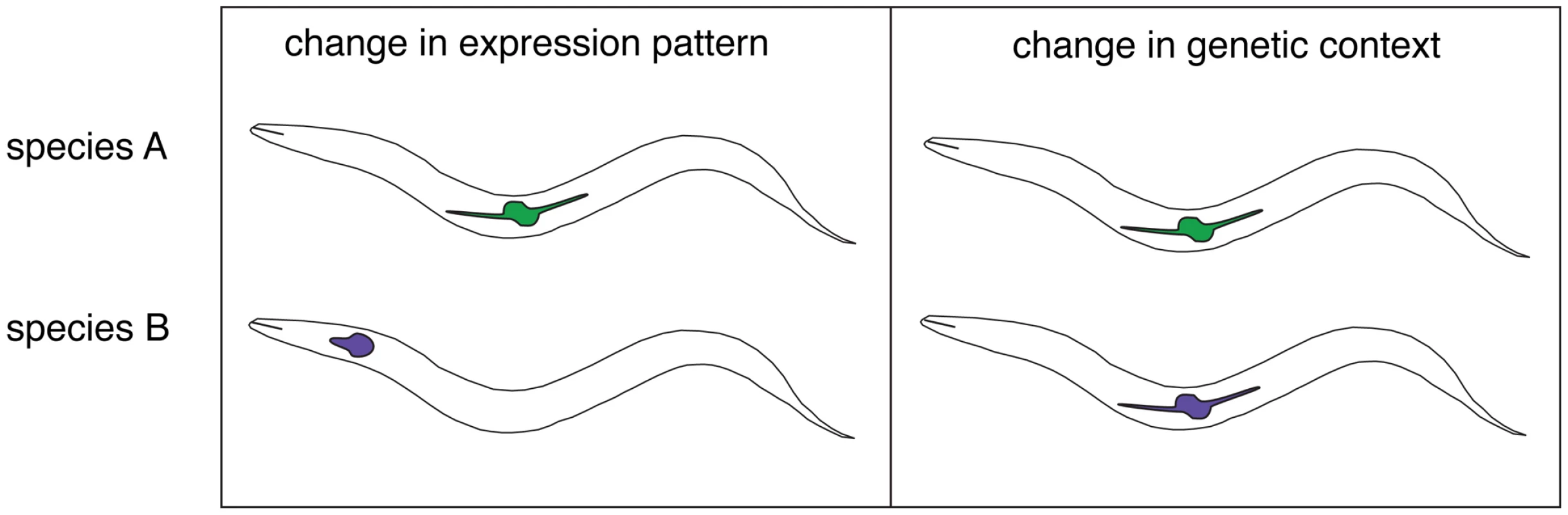
Left: The site of expression of orthologous genes (depicted by colored shapes) has diverged in Species A and Species B and is essential for the normal development and/or function of their respective tissues. A promoter swap is necessary and sufficient for cross-species rescue of a null mutation. Right: The expression of the orthologs has not changed, yet the consequence of inactivation produces a distinct phenotype in each. Cross-species rescue is effective without alteration of the ortholog, indicating that the locus of functional evolution is not the gene itself, but in the context for the gene's activity. Interestingly, in six attempts, Verster et al. failed to identify a clear case where genes with different knockdown phenotypes required the conspecific coding sequence to rescue a mutant. This is a small set of data, but the result is interesting given the different phenotypes and the substantial amino acid divergence between orthologs tested. This suggests that the main engines of DSD may be ongoing fluctuations in the regulation of gene expression and the shifting molecular context that such regulatory changes impart in a given cell type. For example, a novel expression domain for a transcriptional regulator may make another such regulator partially or completely redundant, with the excess capacity now capable of being shifted to either restore conservation (by a reversal) or to create DSD [29]. The need to simultaneously accommodate directional selection in one aspect of a pleiotropic regulator while retaining function of another under strong purifying selection may further accelerate DSD [30]. It will thus be of great interest to determine whether there is a correlation between a gene's tendency to exhibit DSD and the extent of its pleiotropy.
Few animals are as amenable to reverse genetics as Caenorhabditis nematodes, the study of which has now created an important insight into the divergence of gene function among close relatives. However, the recent application of custom site-specific nucleases [e.g., 31] promises to greatly accelerate this kind of work in a range of systems. The main constraint seems to be the ability to introduce the effectors into a single-cell stage of development (i.e., an egg or zygote) and raise the progeny long enough to identify new mutants. This may exclude many species, but it opens up hundreds or thousands to rigorous genetic manipulation for the first time. It will be exciting to see the results of these studies in the near future.
Zdroje
1. HartwellLH, CulottiJ, PringleJR, ReidBJ (1974) Genetic control of the cell division cycle in yeast. Science 183 : 46–51.
2. BrennerS (1974) The genetics of Caenorhabditis elegans. Genetics 77 : 71–94.
3. Nusslein-VolhardC, WieschausE (1980) Mutations affecting segment number and polarity in Drosophila. Nature 287 : 795–801.
4. BowmanJL, SmythDR, MeyerowitzEM (1989) Genes directing flower development in Arabidopsis. Plant Cell 1 : 37–52.
5. HomannOR, DeaJ, NobleSM, JohnsonAD (2009) A phenotypic profile of the Candida albicans regulatory network. PLOS Genet 5: e1000783 doi:10.1371/journal.pgen.1000783
6. MartchenkoM, LevitinA, HoguesH, NantelA, WhitewayM (2007) Transcriptional rewiring of fungal galactose-metabolism circuitry. Curr Biol 17 : 1007–1013.
7. HefferA, XiangJ, PickL (2013) Variation and constraint in Hox gene evolution. Proc Natl Acad Sci U S A 110 : 2211–2216.
8. Ewen-CampenB, SroujiJR, SchwagerEE, ExtavourCG (2012) Oskar predates the evolution of germ plasm in insects. Curr Biol 22 : 2278–2283.
9. TrueJR, HaagES (2001) Developmental system drift and flexibility in evolutionary trajectories. Evol Dev 3 : 109–119.
10. VersterA, RamaniA, McKayS, FraserA (2014) Comparative RNAi screens in C. elegans and C. briggsae reveal the impact of Developmental System Drift on gene function. PLOS Genet 10: e1004077 doi:10.1371/journal.pgen.1004077
11. FelixMA (2007) Cryptic quantitative evolution of the vulva intercellular signaling network in Caenorhabditis. Curr Biol 17 : 103–114.
12. ZhaoZ, BoyleTJ, BaoZ, MurrayJI, MericleB, et al. (2008) Comparative analysis of embryonic cell lineage between Caenorhabditis briggsae and Caenorhabditis elegans. Dev Biol 314 : 93–99.
13. HaagES, KimbleJ (2000) Regulatory elements required for development of Caenorhabditis elegans hermaphrodites are conserved in the tra-2 homologue of C. remanei, a male/female sister species. Genetics 155 : 105–116.
14. KuwabaraPE (1996) Interspecies comparison reveals evolution of control regions in the nematode sex-determining gene tra-2. Genetics 144 : 597–607.
15. WinstonWM, SutherlinM, WrightAJ, FeinbergEH, HunterCP (2007) Caenorhabditis elegans SID-2 is required for environmental RNA interference. Proc Natl Acad Sci U S A 104 : 10565–10570.
16. NuezI, FelixMA (2012) Evolution of susceptibility to ingested double-stranded RNAs in Caenorhabditis nematodes. PLOS One 7: e29811 doi:10.1371/journal.pone.0029811
17. GuoY, LangS, EllisR (2009) Independent recruitment of F-box genes to regulate hermaphrodite development during nematode evolution. Curr Biol 19 : 1853–1860.
18. KelleherDF, de CarvalhoCE, DotyAV, LaytonM, ChengAT, et al. (2008) Comparative genetics of sex determination: masculinizing mutations in Caenorhabditis briggsae. Genetics 178 : 1415–1429.
19. SharanyaD, ThillainathanB, MarriS, BojanalaN, TaylorJ, et al. (2012) Genetic control of vulval development in Caenorhabditis briggsae. G3 (Bethesda) 2 : 1625–1641.
20. BeadellAV, LiuQ, JohnsonDM, HaagES (2011) Independent recruitments of a translational regulator in the evolution of self-fertile nematodes. Proc Natl Acad Sci U S A 108 : 19672–19677.
21. HillRC, de CarvalhoCE, SalogiannisJ, SchlagerB, PilgrimD, et al. (2006) Genetic flexibility in the convergent evolution of hermaphroditism in Caenorhabditis nematodes. Dev Cell 10 : 531–538.
22. NayakS, GoreeJ, SchedlT (2005) fog-2 and the evolution of self-fertile hermaphroditism in Caenorhabditis. PLOS Biology 3: e6 doi:10.1371/journal.pbio.0030006
23. LinKT, Broitman-MaduroG, HungWW, CervantesS, MaduroMF (2009) Knockdown of SKN-1 and the Wnt effector TCF/POP-1 reveals differences in endomesoderm specification in C. briggsae as compared with C. elegans. Dev Biol 325 : 296–306.
24. RudelD, KimbleJ (2001) Conservation of glp-1 regulation and function in nematodes. Genetics 157 : 639–654.
25. WangX, ChamberlinHM (2004) Evolutionary innovation of the excretory system in Caenorhabditis elegans. Nat Genet 36 : 231–232.
26. KiontkeK, FélixM-A, AilionM, RockmanM, BraendleC, et al. (2011) A phylogeny and molecular barcodes for Caenorhabditis, with numerous new species from rotting fruits. BMC Evol Biol 11 : 339.
27. KiontkeK, GavinNP, RaynesY, RoehrigC, PianoF, et al. (2004) Caenorhabditis phylogeny predicts convergence of hermaphroditism and extensive intron loss. Proc Natl Acad Sci U S A 101 : 9003–9008.
28. SommerRJ, EizingerA, LeeKZ, JungblutB, BubeckA, et al. (1998) The Pristionchus HOX gene Ppa-lin-39 inhibits programmed cell death to specify the vulva equivalence group and is not required during vulval induction. Development 125 : 3865–3873.
29. HaagE (2007) Compensatory vs. pseudocompensatory evolution in molecular and developmental interactions. Genetica 129 : 45–55.
30. JohnsonN, PorterA (2007) Evolution of branched regulatory genetic pathways: directional selection on pleiotropic loci accelerates developmental system drift. Genetica 129 : 57–70.
31. SungYH, KimJM, KimHT, LeeJ, JeonJ, et al. (2013) Highly efficient gene knockout in mice and zebrafish with RNA-guided endonucleases. Genome Res 24 : 125–131.
Štítky
Genetika Reprodukční medicína
Článek Natural Polymorphisms in Influence Negative Selection and CD4∶CD8 Lineage Commitment in the RatČlánek MicroRNAs Located in the Hox Gene Clusters Are Implicated in Huntington's Disease PathogenesisČlánek Comparative RNAi Screens in and Reveal the Impact of Developmental System Drift on Gene FunctionČlánek Mutation of SLC35D3 Causes Metabolic Syndrome by Impairing Dopamine Signaling in Striatal D1 Neurons
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2014 Číslo 2
-
Všechny články tohoto čísla
- Fifteen Years Later: Hard and Soft Selection Sweeps Confirm a Large Population Number for HIV In Vivo
- The Same but Different: Worms Reveal the Pervasiveness of Developmental System Drift
- Serine Carboxypeptidase SCPEP1 and Cathepsin A Play Complementary Roles in Regulation of Vasoconstriction via Inactivation of Endothelin-1
- Coherent Functional Modules Improve Transcription Factor Target Identification, Cooperativity Prediction, and Disease Association
- A Long-Chain Flavodoxin Protects from Oxidative Stress and Host Bacterial Clearance
- Mammalian E-type Cyclins Control Chromosome Pairing, Telomere Stability and CDK2 Localization in Male Meiosis
- Influenza Virus Drug Resistance: A Time-Sampled Population Genetics Perspective
- Transcriptome-Wide Analyses of 5′-Ends in RNase J Mutants of a Gram-Positive Pathogen Reveal a Role in RNA Maturation, Regulation and Degradation
- Selective Disruption of Aurora C Kinase Reveals Distinct Functions from Aurora B Kinase during Meiosis in Mouse Oocytes
- X Chromosome Control of Meiotic Chromosome Synapsis in Mouse Inter-Subspecific Hybrids
- A Cohesin-Independent Role for NIPBL at Promoters Provides Insights in CdLS
- Extreme Population Differences in the Human Zinc Transporter ZIP4 (SLC39A4) Are Explained by Positive Selection in Sub-Saharan Africa
- Classic Selective Sweeps Revealed by Massive Sequencing in Cattle
- Genomic Networks of Hybrid Sterility
- Natural Polymorphisms in Influence Negative Selection and CD4∶CD8 Lineage Commitment in the Rat
- Oxidative Stress Is Not a Major Contributor to Somatic Mitochondrial DNA Mutations
- Molecular Identification of Collagen 17a1 as a Major Genetic Modifier of Laminin Gamma 2 Mutation-Induced Junctional Epidermolysis Bullosa in Mice
- Uncoupling of Molecular Maturation from Peripheral Target Innervation in Nociceptors Expressing a Chimeric TrkA/TrkC Receptor
- MicroRNAs Located in the Hox Gene Clusters Are Implicated in Huntington's Disease Pathogenesis
- Loss of Trabid, a New Negative Regulator of the Immune-Deficiency Pathway at the Level of TAK1, Reduces Life Span
- Targeted Ablation of Nesprin 1 and Nesprin 2 from Murine Myocardium Results in Cardiomyopathy, Altered Nuclear Morphology and Inhibition of the Biomechanical Gene Response
- Identification of Novel Genetic Loci Associated with Thyroid Peroxidase Antibodies and Clinical Thyroid Disease
- CEP-1, the p53 Homolog, Mediates Opposing Longevity Outcomes in Mitochondrial Electron Transport Chain Mutants
- Transcriptomics and Functional Genomics of ROS-Induced Cell Death Regulation by
- Quantitative Genome-Wide Genetic Interaction Screens Reveal Global Epistatic Relationships of Protein Complexes in
- Cascades of Genetic Instability Resulting from Compromised Break-Induced Replication
- Serine- and Threonine/Valine-Dependent Activation of PDK and Tor Orthologs Converge on Sch9 to Promote Aging
- Zfp322a Regulates Mouse ES Cell Pluripotency and Enhances Reprogramming Efficiency
- Insertional Mutagenesis and Deep Profiling Reveals Gene Hierarchies and a -Dependent Bottleneck in Lymphomagenesis
- DAAM Is Required for Thin Filament Formation and Sarcomerogenesis during Muscle Development in Drosophila
- Plasma Cholesterol–Induced Lesion Networks Activated before Regression of Early, Mature, and Advanced Atherosclerosis
- High-Resolution Profiling of Stationary-Phase Survival Reveals Yeast Longevity Factors and Their Genetic Interactions
- Comparative RNAi Screens in and Reveal the Impact of Developmental System Drift on Gene Function
- Accurate and Robust Genomic Prediction of Celiac Disease Using Statistical Learning
- Sex-Specific Embryonic Gene Expression in Species with Newly Evolved Sex Chromosomes
- Chromosome X-Wide Association Study Identifies Loci for Fasting Insulin and Height and Evidence for Incomplete Dosage Compensation
- Negative Feedback and Transcriptional Overshooting in a Regulatory Network for Horizontal Gene Transfer
- DNA Sequence Explains Seemingly Disordered Methylation Levels in Partially Methylated Domains of Mammalian Genomes
- Insights into the Genomic Landscape: Comparative Genomics Reveals Variations in Ploidy and Nutrient Utilisation Potential amongst Wine Isolates
- Molecular Evidence for the Inverse Comorbidity between Central Nervous System Disorders and Cancers Detected by Transcriptomic Meta-analyses
- The Centriolar Satellite Protein AZI1 Interacts with BBS4 and Regulates Ciliary Trafficking of the BBSome
- Fine-Mapping the Region Detects Common Variants Tagging a Rare Coding Allele: Evidence for Synthetic Association in Prostate Cancer
- Transmission Distortion Affecting Human Noncrossover but Not Crossover Recombination: A Hidden Source of Meiotic Drive
- A Variant in the Neuropeptide Receptor is a Major Determinant of Growth and Physiology
- Mutation of SLC35D3 Causes Metabolic Syndrome by Impairing Dopamine Signaling in Striatal D1 Neurons
- NSUN4 Is a Dual Function Mitochondrial Protein Required for Both Methylation of 12S rRNA and Coordination of Mitoribosomal Assembly
- MicroRNA-133 Inhibits Behavioral Aggregation by Controlling Dopamine Synthesis in Locusts
- Convergence of Light and ABA Signaling on the Promoter
- Arf4 Is Required for Mammalian Development but Dispensable for Ciliary Assembly
- Distinct Requirements for Cranial Ectoderm and Mesenchyme-Derived Wnts in Specification and Differentiation of Osteoblast and Dermal Progenitors
- Chk2 and P53 Regulate the Transmission of Healed Chromosomes in the Male Germline
- Ddc2 Mediates Mec1 Activation through a Ddc1- or Dpb11-Independent Mechanism
- Mapping the Fitness Landscape of Gene Expression Uncovers the Cause of Antagonism and Sign Epistasis between Adaptive Mutations
- Euchromatic Transposon Insertions Trigger Production of Novel Pi- and Endo-siRNAs at the Target Sites in the Germline
- miR-100 Induces Epithelial-Mesenchymal Transition but Suppresses Tumorigenesis, Migration and Invasion
- Canine Hereditary Ataxia in Old English Sheepdogs and Gordon Setters Is Associated with a Defect in the Autophagy Gene Encoding
- Within-Host Spatiotemporal Dynamics of Plant Virus Infection at the Cellular Level
- Analysis of Meiosis in SUN1 Deficient Mice Reveals a Distinct Role of SUN2 in Mammalian Meiotic LINC Complex Formation and Function
- Genome-Wide Association Study of Metabolic Traits Reveals Novel Gene-Metabolite-Disease Links
- Mechanistically Distinct Mouse Models for -Associated Retinopathy
- DAF-16/FoxO Directly Regulates an Atypical AMP-Activated Protein Kinase Gamma Isoform to Mediate the Effects of Insulin/IGF-1 Signaling on Aging in
- Chromosome I Controls Chromosome II Replication in
- Integrated Genomic Characterization Reveals Novel, Therapeutically Relevant Drug Targets in FGFR and EGFR Pathways in Sporadic Intrahepatic Cholangiocarcinoma
- The Iodotyrosine Deiodinase Ortholog SUP-18 Functions through a Conserved Channel SC-Box to Regulate the Muscle Two-Pore Domain Potassium Channel SUP-9
- The Genome of Highlights a Fish Pathogen Adapted to Fluctuating Environments
- Distinct DNA Binding Sites Contribute to the TCF Transcriptional Switch in and
- The Streamlined Genome of spp. Relative to Human Pathogenic Kinetoplastids Reveals a Parasite Tailored for Plants
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Genome-Wide Association Study of Metabolic Traits Reveals Novel Gene-Metabolite-Disease Links
- A Cohesin-Independent Role for NIPBL at Promoters Provides Insights in CdLS
- Classic Selective Sweeps Revealed by Massive Sequencing in Cattle
- Arf4 Is Required for Mammalian Development but Dispensable for Ciliary Assembly
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



