-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaInsertional Mutagenesis and Deep Profiling Reveals Gene Hierarchies and a -Dependent Bottleneck in Lymphomagenesis
Retroviral insertional mutagenesis (RIM) is a powerful tool for cancer genomics that was combined in this study with deep sequencing (RIM/DS) to facilitate a comprehensive analysis of lymphoma progression. Transgenic mice expressing two potent collaborating oncogenes in the germ line (CD2-MYC, -Runx2) develop rapid onset tumours that can be accelerated and rendered polyclonal by neonatal Moloney murine leukaemia virus (MoMLV) infection. RIM/DS analysis of 28 polyclonal lymphomas identified 771 common insertion sites (CISs) defining a ‘progression network’ that encompassed a remarkably large fraction of known MoMLV target genes, with further strong indications of oncogenic selection above the background of MoMLV integration preference. Progression driven by RIM was characterised as a Darwinian process of clonal competition engaging proliferation control networks downstream of cytokine and T-cell receptor signalling. Enhancer mode activation accounted for the most efficiently selected CIS target genes, including Ccr7 as the most prominent of a set of chemokine receptors driving paracrine growth stimulation and lymphoma dissemination. Another large target gene subset including candidate tumour suppressors was disrupted by intragenic insertions. A second RIM/DS screen comparing lymphomas of wild-type and parental transgenics showed that CD2-MYC tumours are virtually dependent on activation of Runx family genes in strong preference to other potent Myc collaborating genes (Gfi1, Notch1). Ikzf1 was identified as a novel collaborating gene for Runx2 and illustrated the interface between integration preference and oncogenic selection. Lymphoma target genes for MoMLV can be classified into (a) a small set of master regulators that confer self-renewal; overcoming p53 and other failsafe pathways and (b) a large group of progression genes that control autonomous proliferation in transformed cells. These findings provide insights into retroviral biology, human cancer genetics and the safety of vector-mediated gene therapy.
Published in the journal: . PLoS Genet 10(2): e32767. doi:10.1371/journal.pgen.1004167
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1004167Summary
Retroviral insertional mutagenesis (RIM) is a powerful tool for cancer genomics that was combined in this study with deep sequencing (RIM/DS) to facilitate a comprehensive analysis of lymphoma progression. Transgenic mice expressing two potent collaborating oncogenes in the germ line (CD2-MYC, -Runx2) develop rapid onset tumours that can be accelerated and rendered polyclonal by neonatal Moloney murine leukaemia virus (MoMLV) infection. RIM/DS analysis of 28 polyclonal lymphomas identified 771 common insertion sites (CISs) defining a ‘progression network’ that encompassed a remarkably large fraction of known MoMLV target genes, with further strong indications of oncogenic selection above the background of MoMLV integration preference. Progression driven by RIM was characterised as a Darwinian process of clonal competition engaging proliferation control networks downstream of cytokine and T-cell receptor signalling. Enhancer mode activation accounted for the most efficiently selected CIS target genes, including Ccr7 as the most prominent of a set of chemokine receptors driving paracrine growth stimulation and lymphoma dissemination. Another large target gene subset including candidate tumour suppressors was disrupted by intragenic insertions. A second RIM/DS screen comparing lymphomas of wild-type and parental transgenics showed that CD2-MYC tumours are virtually dependent on activation of Runx family genes in strong preference to other potent Myc collaborating genes (Gfi1, Notch1). Ikzf1 was identified as a novel collaborating gene for Runx2 and illustrated the interface between integration preference and oncogenic selection. Lymphoma target genes for MoMLV can be classified into (a) a small set of master regulators that confer self-renewal; overcoming p53 and other failsafe pathways and (b) a large group of progression genes that control autonomous proliferation in transformed cells. These findings provide insights into retroviral biology, human cancer genetics and the safety of vector-mediated gene therapy.
Introduction
The oncogenic potential of murine γ-retroviruses (MLVs) stems from proviral integration into host DNA, a mutagenic process which can result in activation or disruption of critical host cell genes [1]. Moreover, by sequential integrations in the nascent tumour cell, MLVs can drive multiple steps in the oncogenic process. These features have led to the use of MLVs as screening tools for genes relevant to cancer, particularly haematopoietic malignancies. The reach of this approach has grown considerably with the development of high throughput methods for cloning and sequencing analysis of host-virus junctions at insertion sites, facilitating screens of large tumour panels and identifying hundreds of genes of potential relevance to cancer. Importantly, genes identified by this method frequently map to orthologous sites of mutation in human cancer [2], [3]. Moreover, retroviral insertional mutagenesis (RIM) provides a complementary approach to whole genome sequencing and copy number analysis in cancer, as RIM has the potential to uncover genes that are rarely mutated but more commonly subject to indirect processes including epigenetic modification [4]. Furthermore, large scale analyses of co-occurrence of target genes can identify patterns indicating collaborative or redundant relationships between cancer genes [5], [6]. Despite the wealth of information provided by these studies, it is not yet known whether two events are sufficient for lymphoid transformation or whether higher order collaborations between more than two target genes are required. Target gene interactions can be explored functionally when combined with manipulation of the mouse genome and mice with an activated oncogene or mutant tumour suppressor gene in the germ-line often show accelerated tumour onset [7], [8]. RIM tagging in this context reveals preferential targeting of specific collaborating genes, which can be confirmed by analysis of compound transgenic mice [1].
Moloney murine leukaemia virus (MoMLV) is an oncogenic γ-retrovirus that has been widely used in RIM studies [3], [9], [10] and owes its potency to a duplicated enhancer element in the proviral long terminal repeats (LTRs) [11]. Notably, the LTRs and backbone of this virus formed the basis of retroviral vector systems used in early trials of human gene therapy, where leukaemia resulting from insertional activation of host genes has been a significant adverse outcome [12]. In mice, the target genes for MoMLV that have been identified to date show a predominance of oncogene activation events over tumour suppressor disruption, consistent with the observed low rate of loss of heterozygosity in MoMLV lymphomas [13]. However, these findings presented a long-standing puzzle in light of the effect of germ-line inactivation of the major tumour suppressor p53, which confers rapid onset T-cell lymphomas with a similar broad phenotypic spectrum to MoMLV but shows relatively weak cooperation with MoMLV [14]. We hypothesised previously that the MoMLV oncogenic programme must neutralise the tumour suppressor activity of p53, circumventing the need for direct mutations in the pathway [14], [15]. In support of this proposal we showed that the potent combination of two MoMLV target genes, Myc and Runx2, could overcome the need for genetic inactivation of the p53 pathway, despite the fact that both oncogenes evoke p53 growth suppression and collaborate strongly with p53 deficiency [16]. Nevertheless, this combination still appears to be insufficient for full transformation, as double transgenic tumours emerge as clonal outgrowths from a polyclonal premalignant phase [17]. We showed previously that tumour onset could be accelerated by retroviral infection and a RIM screen identified a number of candidate third hit genes, including Pim1, a gene that accelerates tumour onset when combined with MYC/Runx2 in the germ-line [9], [18].
In this study we have conducted a further screen on the same progressing lymphomas, using a deep sequencing method (splinkerette/454) which is orders of magnitude more sensitive than previous shotgun cloning methods. Sequencing at this depth raises another potential concern, as γ-retroviruses including MoMLV display preferential integration at transcriptional start sites and other chromatin feature that may also entail a bias towards proto-oncogenes [19]–[21]. However, we present multiple lines of evidence for post-integration selection as the dominant force shaping the progression ‘integrome’. Moreover, we find that a surprisingly large fraction of the known MoMLV target gene spectrum is detectable in the integrome, indicating that any one among hundreds of genes can contribute to driving clonal outgrowth. However, there is a clear hierarchy of target genes that are selected from a large gene pool generated by the intrinsic preferences of γ-retrovirus integration. Another striking finding is the genetic bottleneck to transformation imposed by transgenic CD2-MYC, which is highly dependent on Runx gene activation. Comparison with other transgenic models of Myc over-expression shows that these each display potent selection from a small pool of master collaborating genes. These genes share the capacity to suppress the p53 pathway but are differentially recruited according to lymphoid lineage and developmental stage. The identification of a small gene set that confers the lymphoma initiating cell phenotype and is conserved in human disease has significant implications for targeted interventions.
Results
Deep sequencing of progressing lymphomas reveals a Darwinian clonal selection process involving many target genes
Relevant features of the CD2-MYC and CD2-Runx2 transgenic mice are displayed in Figures 1 and S1. The disease-free survival of most parental transgenic mice has been attributed to variegated expression under CD2 locus control region (LCR) control [22] along with counter-selection by failsafe processes [22], [23]. As previously described [9], [17], [23], co-expression of both transgenes results in rapid onset lymphomas in 100% of mice, but the tumours typically display a single predominant clone as illustrated by T-cell receptor gene rearrangement (Figure S1). Neonatal infection with MoMLV leads to accelerated lymphoma onset, increased clonal complexity and lymphoid dissemination, although the tumours retain the characteristic bimodal phenotype seen in the absence of infection (CD8+,CD4+/−,TCRhi) [16].
Fig. 1. (A) Features of the system and experimental design of the RIM/DS progression screen (see also Figure S1). 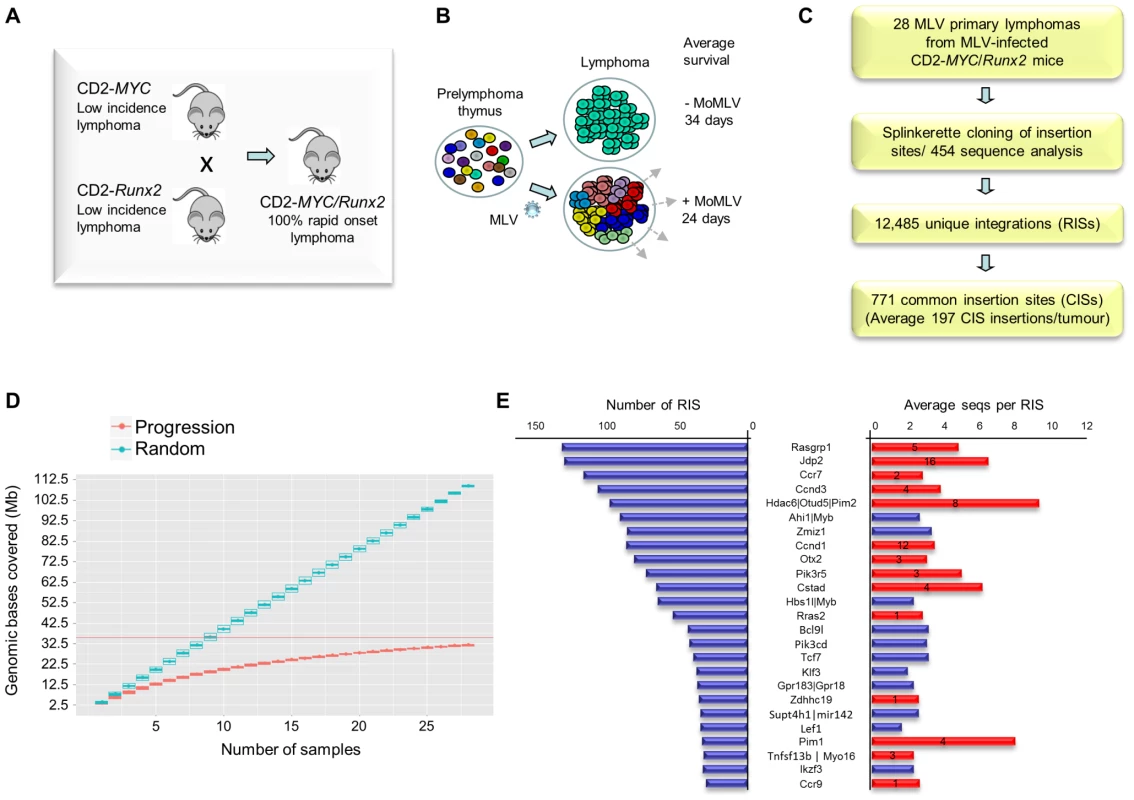
Mice carrying CD2-MYC or CD2-Runx2 transgenes each develop a low incidence of lymphoma, while double transgenics develop lymphomas early with 100% penetrance. (B) Infection of double transgenic mice with MoMLV increases the rate of lymphoma development and the clonal complexity of the resulting tumours. (C) Flowchart of sequencing analysis: Splinkerette clones from 28 double transgenic tumours were sequenced by Roche 454 to identify 12,485 unique retroviral insertion sites (RISs). Gaussian kernel convolution statistical analysis identified 771 common insertion sites (CISs). (D) Saturation analysis of common insertion sites from the 28 MoMLV-accelerated thymic lymphomas. The number of genomic bases covered by predicted CISs increases as the number of samples used increases. The increase is linear if RIS are randomly distributed (upper) but approaches saturation in our real dataset (lower), indicating that 28 samples is sufficient to identify almost all positively selected CISs in this experimental system. (E) The 25 most frequently targeted CISs ranked by number of individual RISs. The right-hand panel shows the average number of reads for RIS. Red bars denote those detected in a previous shotgun cloning screen, with numbers denoting the number of clones detected [9]. A positive correlation (R = 0.56) was noted between with the number of reads/RIS and likelihood of detection by the lower-powered shotgun cloning methodology. Here, a panel of 28 lymphomas was analysed by RIM/DS (splinkerette/454). Processing of reads as described in Methods yielded 12,485 unique retroviral insertion sites (RISs), compared to 272 by previous manual cloning and sequencing methods [9]. Common insertion sites (CISs) were identified using a multi-scale Gaussian Kernel Convolution approach [24] yielding 771 significant CISs compared to 0–3 expected from simulations of random integration (Table S1). A list of all RIS is provided as a .bed file for visualisation in genome browsers, version mm9 (Table S2). Notably, analysis of CIS accrual by number of tumours indicated that this system is approaching saturation and that virtually all the retrievable CISs have been detected (Figure 1D). Target genes affected by integration at CISs were identified by computational methods [25] followed by manual curation.
All 14 target genes identified by shotgun cloning methods [26] featured prominently (Figure 1E; Table S3). There was a positive correlation between the number of clones previously detected by shot-gun cloning and the number of 454 reads (linear regression analysis; R = 0.56) showing that earlier lower powered methods detect only the “tip of the iceberg” of clonal expansion. While splinkerette/454 analysis is only semi-quantitative due to restriction enzyme site distribution and primary sequence constraints on PCR efficiency, we noted that the most abundant RIS corresponding to Pim-1 insertions were also detectable as rearrangements by Southern blot analysis (Figure S2). Moreover, the top 40 RISs (by number of reads) show few apparent passenger insertions, defined as isolated RIS far from any known target gene (5/40), although these predominate (85%) in the total population of 12,485 RISs. The possibility that most of these clones have acquired two separate driver insertions without any passenger insertions appears unlikely, suggesting that most highly proliferative clones contained only a single provirus.
Comparison with CISs from end-stage MoMLV lymphomas reveals major overlap
If the progression network consists of target genes that can complete the oncogenic transformation process, they would be expected to feature strongly in the dominant clones found in end-stage MoMLV-induced lymphomas. To test this assumption, we examined the overlap between the 771 progression CISs in this study with a meta-analysis by Kool and co-workers involving CISs identified by shotgun cloning of 19,923 unique RIS from 977 MoMLV-induced lymphomas of wild-type or tumour suppressor deficient mice [3]. Due to the lower sensitivity of the approach, these CISs should be enriched for major expanded tumour clones. A remarkable 346 CISs (45%) were found in common between the Kool CISs and the progression CISs, indicating that a significant proportion of the target genes involved have been implicated previously as drivers of lymphoma development (Figure 2A, Table S4).
Fig. 2. (A) Comparison between CISs detected in 19,900 MoMLV insertions derived from 937 lymphomas by shotgun cloning [25] and the progression CISs defined in this study. ![(A) Comparison between CISs detected in 19,900 MoMLV insertions derived from 937 lymphomas by shotgun cloning <em class="ref">[25]</em> and the progression CISs defined in this study.](https://www.prolekarniky.cz/media/cache/resolve/media_object_image_small/media/image/b4466f9de390ba7e103e0b61c721a205.png)
There is substantial overlap in the CISs detected. (B) Peak distance of RISs from the nearest transcription start site (TSS). RISs that fall outside CISs (bottom panel) display a distribution similar to that reported for unselected MoMLV insertions [19], [21], and strongly cluster around the TSS. RISs that comprise the ‘biased CIS’ set (top panel) display a relaxed clustering at TSS, while ‘non-biased CIS’ set present an intermediate picture. (C) Orientation bias analysis of 771 progression CISs. Bias is plotted against the number of RISs in each CIS (after Benjamini-Hochberg correction for multiple testing). Those with a p-value <0.05 define the ‘biased CIS’ set, and the others the ‘non-biased CIS’ set. (D) Examples of orientation bias of RISs targeting Ccr7, Ccnd1 and Ahi1/Myb. Each vertical bar represents an individual RIS, coloured to depict orientation (green forward, red reverse) relative to the DNA+strand. Positions of exons and introns are abstracted from the UCSC genome browser (NCBI37/mm9). Percentages refer to predominant orientation at each CIS. Notably, this analysis implicates Myb as the target of long-range insertions from both 5′ and 3′ ends. Further evidence of oncogenic selection: orientation bias and network analysis
Preferential integration of γ-retroviruses around transcriptional initiation sites is an established phenomenon [19] and on the basis of this and further evidence of non-random behaviour it has been argued that the observation of a CIS is insufficient evidence that post-integration selection for growth has occurred, particularly in large scale analyses [27]. While the ideal comparison with the progression CISs identified here would be normal thymocytes immediately after infection, there are significant technical challenges in obtaining a reliable in vivo baseline measurement due to the kinetics of infection and ongoing replication. We therefore chose to compare some aspects of our data to a published large-scale study of human CD34+ cells obtained after in vitro infection with a non-replicating MLV vector. This study by Cattoglio et al. is described as ‘near-baseline’, as analysis was not carried out until 10 days post-transduction [21].
Notably, preference for transcriptional start sites was relaxed in the CISs observed in our study and this trend was more evident still in CISs with an orientation bias, consistent with the increasing importance of post-integration oncogenic selection in this subset (Figure 2B). Moreover, we noted that most of the highly targeted CISs displayed the pronounced orientation bias that is classically associated with enhancer-mode gene activation [1]. As orientation bias does not arise at the level of integration [28], this feature provides direct evidence of post-integration clonal selection. Stringent filtering of CISs for orientation bias yielded 17 examples which we will refer to as biased CISs (Figure 2C; Table S5). We applied the same approach to the Cattoglio ‘near-baseline’ dataset [21] and found no clusters with significant orientation bias after correction for multiple testing. CIS target genes displaying strong orientation bias were also the most frequently targeted and often displayed the greatest levels of clonal expansion, suggesting that enhancer mode activation is the most efficient process by which MoMLV drives lymphoma progression.
An interesting outcome of this analysis shown in Figure 2D is that it provides strong support for the Myb gene as the target of long-range activation by insertions both 5′ and 3′, including the CIS annotated as Ahi1, in accord with hypotheses based on gene expression studies in lymphoma cell lines [29], [30]. Further examples of genes subject to enhancer mode insertions are shown in Figures 2D and S3.
Evidence that the biased CIS targets form part of a larger progression network under selection was provided by KEGG pathway analysis which showed that some of the most frequent CIS targets (e.g. Ccnd3, Ccr7, Pik3cd, Pik3r5, Rasgrp1) map to metanodes that include many of the less frequent targets (Figure S4). Furthermore, KEGG pathway enrichment analysis showed that statistically significant over-representation of specific signalling pathways (T-cell receptor, chemokine, JAK-STAT) was evident even when the top 50–100 target genes were excluded from the analysis (P = <1×10E-5), arguing that oncogenic selection may also be occurring at sites that harbour only a few insertions (Figure S5).
While orientation bias is useful to identify oncogenic selection on a background of preferential integration, we noted that there was a second frequent CIS group defined by intragenic insertions that displayed no statistical bias in orientation. Evidence that these are also under oncogenic selection is provided by the fact that 17 of the 20 most frequent targets have been observed in end-stage lymphomas (Table S6) and by the fact that a significant subset have annotation suggestive of tumour suppressor or oncogene function (Ikzf3, Mad1l1, Als2, Ppp1r16b, Prex1, Ttc28 and Ptprc). The typical pattern of insertions distributed across the target genes is suggestive of a tumour suppressor role, although a role for oncogenic truncated isoforms is also plausible [1], [9], [31]
The progression network provides strong evidence of complementation
Although the majority of top ranking MLV target genes were shared between our progression dataset and the Kool meta-analysis of end-stage lymphomas, there were also notable differences. This was evident from comparison of CIS peak heights and relative rank order of CISs between the datasets where the most discordant examples are listed (Table S7). Oncogenic complementation was evident, with greatly reduced targeting of Myc/Pvt1, Mycn and Runx family genes in the progression set. However, there was also a marked loss of selection for some major targets recorded by Kool et al. including Gfi1 and Notch1. It appears that the combination of MYC and Runx2 in this context also renders these insertions redundant, which is intriguing as insertions at Gfi1 have been shown to be positively selected in some CD2-Runx2 lymphomas [18].
Also of interest was the large number of novel CISs in the progression set (Table S8, examples shown in Figure S3). The most frequently targeted CIS targets displaying strong evidence of enhancer mode activation included Otx2, a homeobox transcription factor which plays a major oncogenic role in medulloblastoma [32] but has not previously been observed in haematopoietic cancers and Myo16, an atypical nuclear myosin with links to survival, cell cycle progression and PI3K signalling [33]. Moreover, a number of prominent targets for potentially disruptive intragenic insertions were unique to the progression set. These included Endou (Pp11), a placental poly-U endonuclease over-expressed in ovarian adenocarcinomas [34], Xrra1, which has been shown to modulate the response to X-ray irradiation [35], and Ttc28 (Tprbk), encoding a large tetratricopeptide domain protein that is regulated by p53, complexes with BRCA1 and suppresses the growth of Ras-transformed cells [36].
The transcriptome of prelymphoma MYC/Runx2 thymus provides insights into progression gene selection and chemokine-receptor interplay in lymphoma dissemination
A previously published analysis of preferential integration targets in early passage CD34+ cells showed a good correlation between basal transcriptional levels and integration frequency [21]. To test whether progression RIS targets were also selected by their high transcription rates in premalignant cells, we compared the transcriptomes of Runx2/MYC and control thymus at 10 days of age, several weeks before clonal tumours emerge. Figure 3 shows expression scatter plots for all gene probes. Basal expression of the most prominent progression targets was widely variable, and only Ccnd1 showed significant up-regulation compared to control thymus. Moreover, the frequent MoMLV targets that were not enriched in the progression network showed a similarly wide distribution with regard to expression levels. The exquisite selection by RIM of specific members of multigene families (e.g. Jdp2, D cyclins) also appeared to be poorly correlated with expression level, strengthening evidence for post-integration selection as the predominant force shaping the progression network.
Fig. 3. Global gene expression analysis in 10 day old (prelymphoma) MYC/Runx2 thymus compared to wild type controls as determined by Affymetrix microarray. 
Scatter plots showing relative expression of genes in transgenic vs control mouse thymus with particular gene sets highlighted as indicated. Target genes positively selected in the progression network are denoted by red dots, others by blue dots (A) Biased CIS gene set, with annotation of novel targets (B) Common MoMLV target genes absent from, or under-represented in, progression CISs. (C) Jun family, noting the prominent target Jdp2. (D) Cyclin genes, noting prominent D-cyclin targets (E), CC chemokine receptors, noting prominent targets Ccr7 and Ccr9, and (F) CC chemokine ligands showing significantly down-regulated Ccl19,21,25. Frequent targeting of Ccr7, and to a lesser extent Ccr9, is interesting in view of their central roles in mediating T-cell progenitor homing to thymus [37], [38]. Moreover, Ccr7 has been reported as a mediator of progression and homing to lymph nodes in multiple tumour types, and to stimulate survival pathways by autocrine or paracrine mechanisms [39]. The cognate ligands for Ccr7 and Ccr9 (Ccl21a, Ccl19, Ccl25), are highly expressed in normal thymus, but intriguingly were significantly down-regulated genes in premalignant organs (validation shown in Figure S6). The respective chemokine genes are normally expressed only in non-lymphoid elements of the thymus including epithelial cells [40]. The possibility that these genes were aberrantly activated to drive autocrine growth in the lymphoma cells was tested by direct analysis of isolated lymphoma cells (Figure S7). However, expression of the ligand genes was below detectable levels in Runx2/MYC or CD2-MYC/p53 null lymphoma cells suggesting that activation of Ccr7/9 provides a growth advantage by a paracrine mechanism that is dependent on thymic stroma. Falling expression of ligand genes in 10-day Runx2/MYC thymus may be due to down-regulation or simple occlusion of non-lymphoid cells by nascent lymphoma cells, which is virtually complete at later stages (Figure S1).
Analysis of single transgenic tumours reveals a Myc-directed bottleneck and collaboration between Runx2 and Ikzf1
To compare the progression network with genes selected during earlier events in tumorigenesis, a second RIM/DS barcode screen was conducted, including MoMLV-infected end-stage lymphomas from parental CD2-MYC, -Runx2 and wild-type mice with a subset of Runx2/MYC progressing tumours (Figure 4, Table S9). All insertions, sorted by genotype are provided as a .bed file for visualisation in genome browsers, version mm9 (Table S10). MYC and Runx2 transgenes each cooperate with MoMLV to accelerate lymphoma onset to around 60 days post-infection [8], [17], [18]. Compared to MYC/Runx2, the other three tumour sets yielded many more reads, but from a much smaller number of unique RISs, reflecting the presence of highly expanded tumour clones (Figure 4A, B). The massive number of RISs per tumour (221–276) shows that in MoMLV lymphomas the predominant end-stage clones co-exist with a polyclonal background of minor populations.
Fig. 4. (A) Features and design of the RIM/DS complementation screen. Average lifespan (days) of wildtype (WT), CD2-MYC, CD2-Runx2 and CD2-MYC/Runx2 double transgenic mice, without and with MoMLV infection. 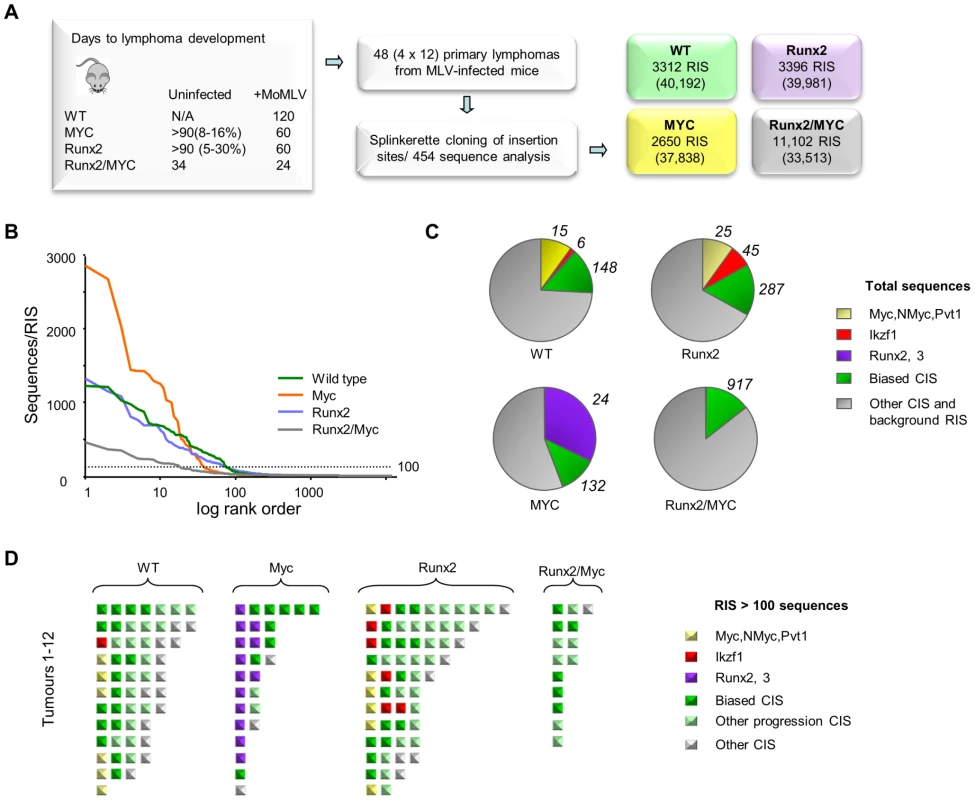
Percentage value indicates lifetime lymphoma incidence. 12 lymphomas from each genotype were analysed by RIM/DS, identifying the indicated number of RISs (total reads in parentheses). (B) Individual RIS in log rank order according to number of reads. Horizontal dotted line represents 100 reads per RIS and was used as a threshold for expanded CISs shown in (d). (C) Total reads in each genotype cohort comprising the CISs around Myc/Mycn/Pvt1 (yellow), Ikzf1 (red), Runx2/Runx3 (purple), around other CISs from the biased CIS set (green) or other RISs (grey). The total numbers of RISs that contribute to the overall read count are indicated outside the pie charts (D) Schematic representation of all RISs with at least 100 reads detected by DS. Each square represents a single RIS, with colour coding as in (c). Expanded RISs not falling within a CIS (presumptive passenger RIS) are not depicted. These analyses illustrate the reduced complexity and greater clonal expansion in MoMLV accelerated CD2-MYC tumours. Application of an abundance threshold of 100 copies (Figure 4B) yielded a RIS number close to that expected from Southern blot analyses of end-stage MoMLV tumours that estimated 4–6 RISs in each dominant clone [41]. In most cases this cut-off correlated well with previous direct analyses for gene rearrangement [18], [42], although rearrangements of Myc detected by Southern blot in two of the CD2-Runx2 tumours analysed here failed to register in the splinkerette/454 analysis. Occasionally ‘missing’ clones might be explained by technical limitation e.g. due to sequence drift in primer sequences. In this regard, it is noted that the bias towards Myc family insertions was less marked here than in a Southern blot-based analysis of a larger CD2-Runx2 tumour cohort [18]. Nevertheless, there were clear and profound differences between cohorts, as MYC transgenic tumours resolved into fewer clones with substantially greater clonal enrichment compared to other genotypes while the double transgenics showed greater complexity as expected (Fig. 4B, C). This apparent difference in the mode of tumour acceleration is interesting as CD2-Runx2 mice harbour an expanded population of transformation-prone thymocytes, which has no parallel in CD2-MYC mice, most of which remain healthy with no obvious abnormality [8], [16], [43].
The most striking features of the single transgenic tumours were evident when the most abundant RISs were sorted according to gene family (Figure 4C, D). High copy RIS mapping to Runx2 or Runx3 were almost ubiquitous in, but exclusive to, CD2-MYC tumours (P = 0.0001, Fisher's Exact Test). A number of high abundance RIS mapped far upstream of Runx2, adding this gene to the list of those subject to long-range activation. Only two tumours displayed no detectable Runx insertion.
Another salient observation from analysis of the end-stage lymphomas was that the low abundance RIS left after subtraction of the major clones frequently correspond to progression network genes (Tables S11, S12). It is conceivable that these represent tumour subclones that have acquired a further hit of proviral insertion, although the alternative possibility that these represent insertions in prelymphoma cells cannot be excluded. The possibility that this background reflects preferential integration in untransformed cells appears unlikely, as such cells form only a tiny fraction of the thymic mass and the hallmark orientation bias at major targets (Figure 1C) is also evident in these minor populations. Moreover, expanded RIS indicative of third hit genes in CD2-MYC, CD2-Runx2 and wild-type mice appeared to be selected from a broad cross-section of the progression network, with the ‘winners’ of the progression race largely recapitulating the expansion rate measured by earlier analysis of the progression network (Figure 1D).
Second hit genes represent a narrow genetic bottleneck to transformation
We reasoned that specific ‘second hit’ collaborating genes would be distinguishable from progression genes on the basis of (a) positive selection in lymphomas of single transgenic mice compared to wild-type and (b) loss of selection or reduction to background levels in double transgenics. As expected, the Runx genes (Runx2 and Runx3) and Myc family targets (Myc, Mycn) conformed to this pattern, being selected in CD2-MYC and CD2-Runx2 respectively and effectively disappearing from the double transgenic tumours (Figure 4C). Surprisingly, inspection of the entire CIS list revealed only one other target gene with statistically significant correspondence to this pattern: intragenic insertions in Ikzf1 were significantly more abundant in CD2-Runx2 transgenic tumours than in the other three genotypes and showed more frequent representation in dominant clones (Figure 5). Intriguingly, analysis of the CD34+ ‘random integration’ vector dataset [21] shows two hotspots for integration in the human IKZF1 gene that correspond to active chromatin marks. The murine Ikzf1 gene showed a similar background pattern, although 3–4 clusters of insertions could be discerned in the murine gene. These observations suggest a two-step model for targeting of Ikzf1 by MoMLV, with preferential integration at sensitive sites within the gene leading to sustained clonal expansion only in the presence of a collaborating lesion such as deregulated Runx expression (Figure 5B).
Fig. 5. Insertions at Ikzf1 display dual features of preferential integration and oncogenic selection. 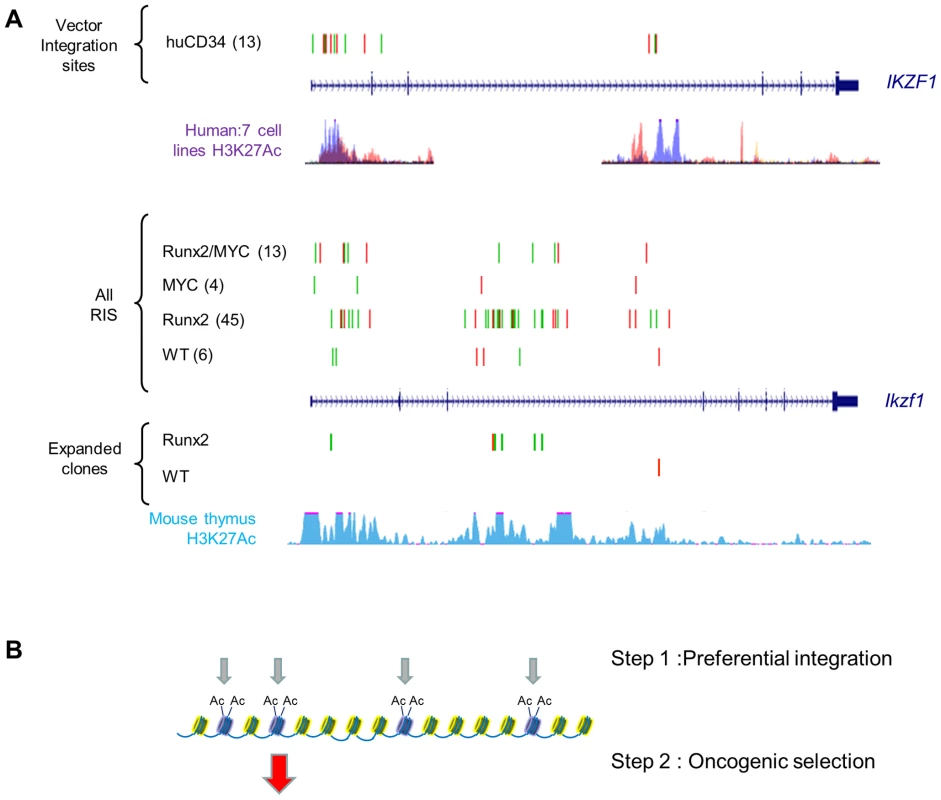
A) Upper panel : Inspection of >32,000 MLV vector integrations in early passage human CD34+ cells (Cattoglio set [21]) shows two clusters of integration in the IKZF1 gene which map to sites of active chromatin marks (H3K27 acetylation, ENCODE data for 7 cell lines). Lower panel: A pattern of low abundance integrations within Ikzf1 is present in all 4 genotypes in our study, suggesting a conserved process of preferential integration at the murine gene corresponding again to chromatin features (H3K27 acetylation in C57/BL thymus). However, many more insertions are evident in the CD2-Runx2 background, and substantial expansions (>100 reads) in end-stage lymphomas show a similar genotype bias. (B) Diagrammatic model of a two-stage process of oncogenic selection on a background of MoMLV preferential integration. Table S13 summarises the genes showing strongest evidence of complementation in parental transgenic mice. In addition, there is evidence of reduced selection for Gfi1 and Notch1 insertions on the CD2-MYC background which directly mirrors findings on the progression set compared to common MoMLV targets (Table S7) suggesting that this bias is conferred by the CD2-MYC transgene. Targeting of both genes in wild-type controls and CD2-Runx2 in this study rules out mouse strain differences as the basis of this phenomenon. Notably, Notch1 has been shown to block p53-dependent apoptosis due to Myc over-expression [44], while Gfi1 has recently been shown to modulate p53 responses indirectly by altering protein methylation [45]. The latter finding illuminates early RIM screens of Eμ-Myc mice which suggested that Gfi1 and Bmi1 belong to the same complementation group [7], and Bmi1 is known to control p53 responses by transcriptional suppression of Arf [46]. As we have shown that Runx2 also inhibits Myc-induced apoptosis in vivo and that the Runx2/MYC combination neutralises selection for loss of p53 [16], we propose the model in Figure 6 to account for the respective gene interactions in different transgenic backgrounds in a three-hit model of MoMLV lymphomagenesis.
Fig. 6. Hierarchical model of MoMLV-induced T-cell lymphomagenesis and preferred target genes in Myc transgenic systems. 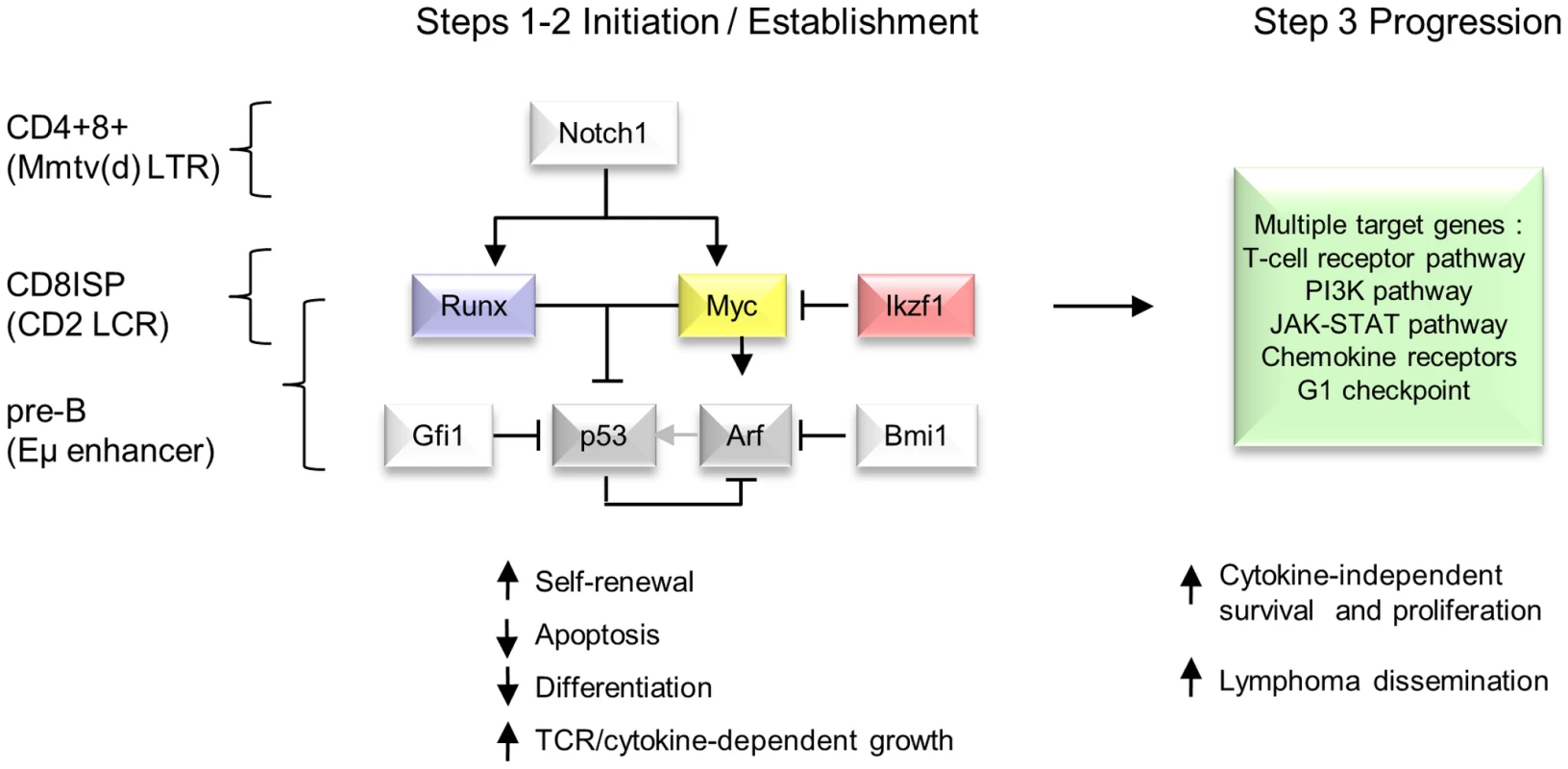
In this model, critical genes in lymphoma initiation and establishment converge on Myc and p53, while Ikzf1 insertions are postulated to de-repress c-myc [31], [67]. Lymphoma phenotype and preferred target genes vary according to expression control element and lineage [7], [53], [56]. A much larger set of target genes and downstream pathways is capable of driving clonal expansion at the tumour progression stage. Comparison with murine and human T-cell lymphomas reveals extensive overlap in common insertion sites and domains of copy number alteration
The extent to which the pathways targeted in retrovirus-induced lymphomas overlap with similar tumours of non-viral origin, including human cancers, is also of considerable interest. We compared the comprehensive CIS database with regions of chromosomal gain and loss described in a previous study of T-cell lymphomas arising in mice defective in telomerase, p53 and ATM (‘TKO’) mice [47], where a strong overlap was noted with human T-ALL. Remarkably, 16/18 regions of syntenic deletion or amplification contained CISs, corresponding to 43/771 CISs (for this overlap P = <0.0001; Table S14). Notably, no known cancer genes could be found at the majority of these domains [47], suggesting that the genes targeted at these CISs represent candidates for gain or loss of function that is conserved between human and mouse cancers. Significantly, many of the target genes display intragenic insertions, particularly for the deleted regions (13/22). An interesting example is Gpr132, located on chromosome 12, which encodes a G-protein coupled receptor with apparent tumour suppressor activity [48].
Discussion
In this study we examined an established system of oncogene cooperation and retroviral acceleration using a deep sequencing (DS) platform. RIM/DS increases sensitivity of RIS detection by almost two orders of magnitude over earlier methodologies [9] and when applied to a lymphoma progression model shows evidence of saturation, indicating that all relevant major CISs have been obtained. The remarkable observation that much of the large repertoire of MoMLV target genes from almost one thousand end-stage T-cell lymphomas can be found in the progression network from only 28 lymphomas shows the enormous potential of RIM/DS when applied to polyclonal populations under strong selection. While statistical and pathway analyses provide useful tools to discriminate genes under oncogenic selection from preferential integration, our findings suggest that the phenomena may not be completely separable. The example of Ikzf1 illustrates the principle whereby a gene may be selectively targeted by γ-retroviral integration but leads to clonal expansion in the presence of a complementary oncogenic programme provided in this case by Runx gene activation. It has been demonstrated recently that γ-retroviral integration at transcriptional start sites is a consequence of interaction with BET chromatin regulators that direct the process towards genomic regions rich in acetylated histones [49], [50]. The integration specificity of γ-retroviruses is clearly fundamental to their efficient replication and transmission in nature. In wild-type mice, the rate of oncogenic transformation due to successive integration events is reduced by retroviral interference, but the process is accelerated in oncogene transgenic mice where fewer hits are required.
The implications of our analyses are also interesting for retroviral vector-based gene therapy. As the most potently selected insertions mediate enhancer-mode gene activation, the removal of enhancer elements in self-inactivating vectors [51] is likely to improve safety margins. However, failure to deal with the targeting apparatus will leave a residual risk, particularly for gene disruption events which, from their lack of obvious orientation bias, may not require strong enhancer function (e.g. at Ikzf1).
While intrinsic preference for integration at transcriptional start sites and other chromatin features [19]–[21] creates the platform on which oncogenic selection operates, it is clear that post-integration selection events play a decisive role in shaping the genetic profile of end-stage tumours. The progression network is highly adapted to the T-cell environment but is not simply a cross-section of highly expressed and therefore available target genes. This principle is illustrated by the strong selection for specific members of multigene families (e.g. Jdp2, D cyclins) that show no correlation with basal transcription levels. Similarly the targeting of novel genes that were not seen in previous large-scale screens of MoMLV-induced T-cell lymphomas (e.g. Otx2, Myo16) is not merely due to their up-regulation in the background of the Runx2/MYC model. These findings suggest that it will be of value to employ RIM/DS to probe the growth checkpoint networks in tissues and cell lineages that have been less well explored to date.
While most of the functionally annotated progression network genes are predicted to confer autonomous proliferation, an exception to this rule was provided by the frequent activation of Ccr7 and Ccr9, which in their normal developmental roles promote T-cell homing to thymus and ligand-dependent survival and proliferation [37]. Moreover, Ccr7 is stimulated by Notch signalling [52], and we would predict that retroviral activation bypasses this requirement. It appears that the result of Ccr7/9 activation in Runx2/MYC lymphomas is likely to be paracrine growth stimulation, as expression of the cognate ligands (Ccl19, 21, 25) is restricted to thymic stromal cells. Moreover, declining levels of ligand transcripts in Runx2/MYC thymus offers a rationale for the accelerated dissemination of lymphoma cells towards highly expressing peripheral lymphoid tissues [9]. Export of lymphoma cells with Ccr7 insertions is also in accord with the relatively low read/RIS ratio in primary thymic lymphomas. Identification of Ccr7 as a major target highlights the complementary value of RIM screening, as this gene does not appear to be subject to mutation or amplification in human cancer, yet is required for CNS metastasis of human leukaemia cells [52].
Comparison of the progression network with a large scale meta-analysis of MoMLV targets in T-cell lymphomas from various genetic backgrounds [3] showed that the principles of complementation apply where the two germ-line oncogenes are present, as insertions at Myc and Runx family members were massively under-represented in the progression set. Moreover, while most major targets overlapped strongly, a few prominent targets including Gfi1 and Notch1 were also greatly diminished in the progression network. Our second RIM/DS of parental transgenic mice shed further light on this observation, as the CD2-MYC parental transgenic system in particular did not select for these targets but instead showed virtual dependence on activation of a Runx family gene with the order Runx2>Runx3>Runx1 in targeting frequency in accord with previous observations [53]–[55]. Comparison of several Myc transgenic model systems (CD2-MYC, Eμ-Myc, Mmtv(d)-Myc) shows that these have massively divergent preferences for collaborating genes detected by RIM, presumably reflecting the lineage and stage-specificity of Myc expression control [7], [42], [56]. However, it is notable that all of these potently selected collaborating genes share the ability to suppress the p53 response in the context of activated Myc [16], [44], [46], [57]. There is an obvious parallel with the observation that the combination of CD2-Runx2/MYC overcomes the requirement for genetic inactivation of the p53 pathway [16], providing a rationale for the reduced selection for Notch and Gfi1 on this background.
The foregoing observations invite the model presented in Figure 6, where the interaction of this small gene set is presented as a bottleneck to transformation in contrast to the broad range of progression genes that can be recruited at later stages. In addition to the simple outline shown here, it appears that the MoMLV ‘core’ gene programme can also neutralise p53-independent failsafe pathways, as p53 deficiency has relatively modest effects on MoMLV-induced tumour onset and target gene spectrum [5], [14], [15]. It should also be noted that at least some of the genes in the progression network can also serve as initiators when expressed as transgenes, showing that the mutational order may not be fixed [58]–[60].
Why do the major collaborating gene targets vary so markedly between Myc transgenic models? The most obvious rationale is presented by the lineage and stage-specificity of Myc expression. RIM targeting of Bmi1 is largely a feature of B-cell lymphomas in the mouse [7], while Notch targeting predominates in the CD4+CD8+ lymphomas of Mmtv(d)-Myc mice [56]. The CD2 LCR confers strong T-cell specificity but is also active in B-cells [61], implying that its developmental activation may occur at the level of committed lymphoid progenitors. High level Myc expression in this niche appears to lead to cell death, unless combined with loss of p53 or an activated Runx allele [17], [62], [63]. We hypothesise that Notch1 or Gfi1 pathways are not available for RIM targeting at this stage and that Runx2, the ‘bone-specific’ family member, which is also transcriptionally active in early haematopoietic development [64], becomes the primary target for activation in this niche. As mounting evidence indicates that Runx family members are downstream of Notch signalling in expression control and effector functions [65], it is tempting to suggest that dual activation of Runx and Myc supplants the need for activation of Notch. The model we propose has implications for therapeutic targeting of Notch signalling with γ-secretase inhibitors [66], as up-regulation of Runx and Myc may represent another pathway to resistance.
Although CD2-Runx2 selects strongly for activation of Myc family genes by RIM [18] it appears less critically dependent, possibly due to the survival of Runx2 expressing thymocytes as a premalignant, slowly proliferating population blocked at the DN/CD8ISP stage [43]. This study shows that Ikzf1 is also favoured as a collaborating target on this background. Notably, Ikzf1 is a haplo-insufficient tumour suppressor that has been reported to act as a transcriptional suppressor of Myc [67], while intragenic retroviral insertions lead to expression of truncated isoforms with dominant negative potential [31]. We therefore suggest that de-repression of Myc may be one of the consequences of Ikzf1 targeting that leads to its co-selection with Runx2. It would interesting in this regard to test whether lymphomas of Runx2 transgenic mice with reduced Ikzf1 function [68] would show reduced RIM targeting of both Myc family genes and Ikzf1.
This analysis has wider implications for the genetics of human lymphomas and other cancers. It appears that the final step in lymphoid transformation by MLV can be accomplished by a wide range of genes with the common functional end-point of growth factor-independent proliferation. As the progression network also includes numerous genes that are mutated, amplified or deleted in human cancer (Table S14), it is tempting to suggest that many of the acquired mutations in human cancer are also late embellishments. Another important insight is provided by the evidence of a small network of genes (Myc, Runx, Ikzf1, Gfi1, Notch1, and Bmi1) that act in pairwise combinations to confer lymphoma self-renewal and overcome failsafe responses via the p53 pathway. It seems likely that this network operates under normal physiological conditions to licence cell growth and is co-ordinately subverted in cells carrying mutations in the pathways. The recent description of Gfi1 as an ‘oncorequisite’ factor that is rarely directly mutated but nevertheless required for growth of ALL cells [45] highlights the potential for targeting this network. The Runx genes are heavily implicated in human leukaemia but show paradoxical features of either gain or loss of function in disease subsets [69]. The demonstration here that Runx activation is virtually essential for MYC transformation of early murine T-cell lymphoma suggests that it may be fruitful to examine the requirement for RUNX function in human leukaemia/lymphomas driven by amplified MYC or NOTCH/IKZF1 mutations.
Methods
Ethics statement
Animals were routinely monitored and sacrificed when showing signs of ill health in line with the UK Animals (Scientific Procedures) Act, 1986.
Animals
CD2-MYC, CD2-Runx2, and CD2-MYC/CD2-Runx2 transgenic animals and maintenance were described previously [9]. Neonates were infected within 24 hours of birth with ∼105 infectious units of MoMLV as previously described [42]. Littermate-matched genotype controls were used to control for mouse strain.
DNA extraction
DNA was extracted from approximately 20 mg of frozen enlarged lymphoid/tumour tissue using Gentra Puregene Genomic DNA Purification Kit (Qiagen, UK) according to the manufacturer's instructions.
Isolation of retroviral insertion sites
Isolation of the retroviral insertion sites from the tissues was performed using splinkerette PCR to produce barcoded PCR products that were pooled and sequenced on 454 GS-FLX sequencers (Roche Diagnostics platform) as described previously [70], [71]. The restriction enzymes used to digest the genomic DNA were Sau3AI and Tsp509I, and the enzyme used to digest MoMLV DNA was EcoRV.
Bioinformatic analysis of 454 sequencing results
Processing of 454 reads, identification of insertion sites, and Gaussian kernel convolution (GKC) statistical methods used to identify common insertion sites (CISs) have been described previously [6], [24], [71], [72]. In summary, 454 reads were mapped to the mouse mm9 genome assembly, where the only modification to the previous alignment procedure was the removal of the stringency check as to whether an alignment was located neighbouring a TA dinucleotide site (the insertion locations preferred by Sleeping Beauty transposons on which the bioinformatics processing method was developed). Reads from the same sample whose start genomic locations aligned within three nucleotides of each other were merged together. Reads from the same sample that were more than three nucleotides apart were considered independent integration events. CISs were identified using the multi-scale GKC approach [6], [24]
Analysis of sample saturation
In order to determine whether the MLV screen had reached some level of saturation, the Gaussian Kernel Convolution (GKC) CIS calls from all 28 samples were analysed using the ACT software package [73].
ACT considers genomic locations generated by multiple samples for specific biological phenomenon under study (e.g. ChIP-seq peaks) to determine the saturation of a screen. The program considers the various combinations in which samples can be added so that the increase in base pair coverage is a range of values based on all the samples. The results can be depicted as a series of boxplots showing the increase in base pair coverage, where the boxplot at each position n on the x-axis shows the coverage values of all combinations of n samples. Boxplots that approach a horizontal asymptote indicate that the coverage has reached saturation.
For the GKC CISs generated by all 28 samples, the insertion sites that contributed to CISs were extracted, resulting in a set of 7,485 sites. The insertion sites were then selected per sample and pseudo-kernels of 7.5k nucleotides either side of each insertion were applied to mimic GKC kernels of 15k nucleotides. Overlapping kernels within each sample were merged into continuous genomic regions. These 28 modified insertions files were then analysed using ACT. For each combination of samples the median values, and 25th and 75th percentiles were plotted using ggplot2 [74].
As a control, the 28 samples were re-analysed where the same number of insertion sites per sample were selected at random across the mouse genome. The pseudo-15k nucleotide kernels were applied.
While the analysis does not produce a clear-cut asymptote this is to be expected due to the type of data under consideration. ACT was designed to analyse such data as ChIP-seq arrays for predicting transcription factor binding sites. In these scenarios ChIP-seq replicates should ideally report the same key binding sites/genomic locations. Hence across multiple samples the same locations should be reported.
For MLV screens however, while insertions in the same gene will be found from different samples, the locations of the insertion sites will not overlap perfectly, even with the addition of the 15k nucleotide pseudo kernels. Hence each sample will introduce novel regions, such that the overall coverage will continue to increase even if the screen has truly reached a ‘saturation’ point. Also not all samples will contribute to all CISs. Different combinations of samples will thereby result in varying coverages, causing the coverage profile not to asymptote perfectly.
Integration site location mapping relative to transcription start sites (TSS)
The genomic coordinates of the ‘UCSC Genes’ set was downloaded via the UCSC genome browser for mouse assembly mm9. Each of the 12,485 MoMLV integration sites was then mapped relative to the transcription start site (TSS) of its closest UCSC-defined ‘known’ gene.
Bioinformatic analysis of Kool et al. 2012 insertion sites
The Kool set of 19,923 mouse retroviral insertions sites was downloaded from the Mutapedia website (http://mutapedia.nki.nl/) [3]. In the original paper, 596 CISs were identified using the GKC statistical framework with a fixed kernel width of 30k nucleotides. The insertion sites were re-analysed using the same multi-scale kernel approach that was applied to the MoMLV insertion sites. As a result of the multi-scale kernels and a less stringent cut-off value, 977 CISs were identified.
Defining the width of a CIS as spanning the minimum and maximum genomic coordinates of insertion sites that contribute to a CIS, CISs were compared between the progression set and the re-analysed Kool set for overlaps. CISs were called overlapping if at least one nucleotide was overlapping between the two CIS sets.
Integration site orientation bias analysis
MLV CISs from this study
For each MoMLV CIS, the integration sites that contributed to it were collated, divided into forward - and reverse-orientation sites, and their frequencies counted. A one-tailed Fisher's exact test was then performed using the frequencies of the CIS-specific integrations versus the frequencies of remaining integration sites not present in the current CIS. Multiple test correction was performed using the Benjamini-Hochberg procedure [75].
MLV vector CISs in CD34+ cells
A set of 32,592 human MLV-based vector integration sites was kindly provided by Cattoglio and co-workers as previously published [21]. In the original study genomic regions were considered as significant if three or more integration sites were found clustered within regions of 12,587 nucleotides. This threshold was applied to the 32,592 integrations sites resulting in the identification of 3,453 clusters. Taking the integration sites within the clusters, a similar Fisher's exact test method was used to assess the orientation bias of the integration sites as for the MoMLV CISs. Following multiple test correction no clusters exhibited any orientation bias.
Microarray analysis
RNA was isolated and purified from the thymuses of 10 day old wild type and CD2-MYC/Runx2 double transgenic mice using an RNeasy Mini Kit as per the manufacturer's instructions (Qiagen, UK) with mechanical lysis using a pellet pestle in a microfuge tube (Sigma). RNA purity was assessed using a Nanodrop 2000 Spectrophotometer (Thermo Scientific), and integrity verified using the Agilent 2100 Bioanalyser with RNA 6000 Nano Reagents kit (Agilent Biotechnologies) as per the manufacturer's protocol. Whole genome expression profiling was performed using Affymetrix mouse GeneChip microarrays (MoGene-1) in triplicate as per the manufacturer's protocol (Affymetrix, UK). Data analysis was carried out using the Partek Genomic Suite (Partek Inc., St. Louis, MO, USA). Briefly, after Robust Multichip Average normalisation [76] with GC content pre-background adjustment, the differentially expression analysis was performed using ANOVA. Multiple testing correction was done using the ‘q value’ cut-off [77] with gene changes of p<0.05 considered significant. Graphical representations of data were prepared using CLC Genomics Workbench 4.
Supporting Information
Zdroje
1. UrenAG, KoolJ, BernsA, vanLM (2005) Retroviral insertional mutagenesis: past, present and future. Oncogene 24 : 7656–7672 1209043 [pii];10.1038/sj.onc.1209043 [doi].
2. MattisonJ, KoolJ, UrenAG, de RidderJ, WesselsL, et al. (2010) Novel candidate cancer genes identified by a large-scale cross-species comparative oncogenomics approach. Cancer Res 70 : 883–895 0008-5472.CAN-09-1737 [pii];10.1158/0008-5472.CAN-09-1737 [doi].
3. KoolJ, UrenAG, MartinsCP, SieD, de RidderJ, et al. (2010) Insertional mutagenesis in mice deficient for p15Ink4b, p16Ink4a, p21Cip1, and p27Kip1 reveals cancer gene interactions and correlations with tumor phenotypes. Cancer Res 70 : 520–531 0008-5472.CAN-09-2736 [pii];10.1158/0008-5472.CAN-09-2736 [doi].
4. AlbihnA, JohnsenJI, HenrikssonMA (2010) MYC in oncogenesis and as a target for cancer therapies. Adv Cancer Res 107 : 163–224 S0065-230X(10)07006-5 [pii];10.1016/S0065-230X(10)07006-5 [doi].
5. UrenAG, KoolJ, MatentzogluK, de RidderJ, MattisonJ, et al. (2008) Large-scale mutagenesis in p19(ARF) - and p53-deficient mice identifies cancer genes and their collaborative networks. Cell 133 : 727–741 S0092-8674(08)00436-4 [pii];10.1016/j.cell.2008.03.021 [doi].
6. de RidderJ, UrenA, KoolJ, ReindersM, WesselsL (2006) Detecting statistically significant common insertion sites in retroviral insertional mutagenesis screens. PLoS Comput Biol 2: e166 06-PLCB-RA-0052R3 [pii];10.1371/journal.pcbi.0020166 [doi].
7. van LohuizenM, VerbeekS, ScheijenB, WientjensE, van der GuldenH, et al. (1991) Identification of cooperating oncogenes in E mu-myc transgenic mice by provirus tagging. Cell 65 : 737–752 0092-8674(91)90382-9 [pii].
8. StewartM, CameronE, CampbellM, McFarlaneR, TothS, et al. (1993) Conditional expression and oncogenicity of c-myc linked to a CD2 gene dominant control region. Int J Cancer 53 : 1023–1030.
9. StewartM, MackayN, HanlonL, BlythK, ScobieL, et al. (2007) Insertional mutagenesis reveals progression genes and checkpoints in MYC/Runx2 lymphomas. Cancer Res 67 : 5126–5133 67/11/5126 [pii];10.1158/0008-5472.CAN-07-0433 [doi].
10. HwangHC, MartinsCP, BronkhorstY, RandelE, BernsA, et al. (2002) Identification of oncogenes collaborating with p27Kip1 loss by insertional mutagenesis and high-throughput insertion site analysis. Proc Natl Acad Sci U S A 99 : 11293–11298 10.1073/pnas.162356099 [doi];162356099 [pii].
11. LiY, GolemisE, HartleyJW, HopkinsN (1987) Disease specificity of nondefective Friend and Moloney murine leukemia viruses is controlled by a small number of nucleotides. J Virol 61 : 693–700.
12. Hacein-Bey-AbinaS, GarrigueA, WangGP, SoulierJ, LimA, et al. (2008) Insertional oncogenesis in 4 patients after retrovirus-mediated gene therapy of SCID-X1. J Clin Invest 118 : 3132–3142 10.1172/JCI35700 [doi].
13. LanderJK, FanH (1997) Low-frequency loss of heterozygosity in Moloney murine leukemia virus-induced tumors in BRAKF1/J mice. J Virol 71 : 3940–3952.
14. BaxterEW, BlythK, DonehowerLA, CameronER, OnionsDE, et al. (1996) Moloney murine leukemia virus-induced lymphomas in p53-deficient mice: overlapping pathways in tumor development? J Virol 70 : 2095–2100.
15. BaxterEW, BlythK, CameronER, NeilJC (2001) Selection for loss of p53 function in T-cell lymphomagenesis is alleviated by Moloney murine leukemia virus infection in myc transgenic mice. J Virol 75 : 9790–9798 10.1128/JVI.75.20.9790-9798.2001 [doi].
16. BlythK, VaillantF, HanlonL, MackayN, BellM, et al. (2006) Runx2 and MYC collaborate in lymphoma development by suppressing apoptotic and growth arrest pathways in vivo. Cancer Res 66 : 2195–2201 66/4/2195 [pii];10.1158/0008-5472.CAN-05-3558 [doi].
17. VaillantF, BlythK, TerryA, BellM, CameronER, et al. (1999) A full-length Cbfa1 gene product perturbs T-cell development and promotes lymphomagenesis in synergy with myc. Oncogene 18 : 7124–7134 10.1038/sj.onc.1203202 [doi].
18. BlythK, TerryA, MackayN, VaillantF, BellM, et al. (2001) Runx2: a novel oncogenic effector revealed by in vivo complementation and retroviral tagging. Oncogene 20 : 295–302 10.1038/sj.onc.1204090 [doi].
19. WuX, LiY, CriseB, BurgessSM (2003) Transcription start regions in the human genome are favored targets for MLV integration. Science 300 : 1749–1751 10.1126/science.1083413 [doi];300/5626/1749 [pii].
20. RothSL, MalaniN, BushmanFD (2011) Gammaretroviral integration into nucleosomal target DNA in vivo. J Virol 85 : 7393–7401 JVI.00635-11 [pii];10.1128/JVI.00635-11 [doi].
21. CattoglioC, PellinD, RizziE, MaruggiG, CortiG, et al. (2010) High-definition mapping of retroviral integration sites identifies active regulatory elements in human multipotent hematopoietic progenitors. Blood 116 : 5507–5517 blood-2010-05-283523 [pii];10.1182/blood-2010-05-283523 [doi].
22. WilliamsA, HarkerN, KtistakiE, Veiga-FernandesH, RoderickK, et al. (2008) Position effect variegation and imprinting of transgenes in lymphocytes. Nucleic Acids Res 36 : 2320–2329 gkn085 [pii];10.1093/nar/gkn085 [doi].
23. BlythK, VaillantF, HanlonL, MackayN, BellM, et al. (2006) Runx2 and MYC collaborate in lymphoma development by suppressing apoptotic and growth arrest pathways in vivo. Cancer Res 66 : 2195–2201 66/4/2195 [pii];10.1158/0008-5472.CAN-05-3558 [doi].
24. Perez-ManceraPA, RustAG, van der WeydenL, KristiansenG, LiA, et al. (2012) The deubiquitinase USP9X suppresses pancreatic ductal adenocarcinoma. Nature 486 : 266–270 nature11114 [pii];10.1038/nature11114 [doi].
25. de JongJ, de RidderJ, van der WeydenL, SunN, vanUM, et al. (2011) Computational identification of insertional mutagenesis targets for cancer gene discovery. Nucleic Acids Res 39: e105 gkr447 [pii];10.1093/nar/gkr447 [doi].
26. StewartM, MackayN, HanlonL, BlythK, ScobieL, et al. (2007) Insertional mutagenesis reveals progression genes and checkpoints in MYC/Runx2 lymphomas. Cancer Res 67 : 5126–5133 67/11/5126 [pii];10.1158/0008-5472.CAN-07-0433 [doi].
27. WuX, LukeBT, BurgessSM (2006) Redefining the common insertion site. Virology 344 : 292–295 S0042-6822(05)00621-5 [pii];10.1016/j.virol.2005.08.047 [doi].
28. BushmanFD (2003) Targeting survival: integration site selection by retroviruses and LTR-retrotransposons. Cell 115 : 135–138 S0092867403007608 [pii].
29. HanlonL, BarrNI, BlythK, StewartM, HaviernikP, et al. (2003) Long-range effects of retroviral insertion on c-myb: overexpression may be obscured by silencing during tumor growth in vitro. J Virol 77 : 1059–1068.
30. ZhangJ, MarkusJ, BiesJ, PaulT, WolffL (2012) Three murine leukemia virus integration regions within 100 kilobases upstream of c-myb are proximal to the 5′ regulatory region of the gene through DNA looping. J Virol 86 : 10524–10532 JVI.01077-12 [pii];10.1128/JVI.01077-12 [doi].
31. BeverlyLJ, CapobiancoAJ (2003) Perturbation of Ikaros isoform selection by MLV integration is a cooperative event in Notch(IC)-induced T cell leukemogenesis. Cancer Cell 3 : 551–564 S1535610803001375 [pii].
32. BuntJ, HasseltNE, ZwijnenburgDA, HamdiM, KosterJ, et al. (2012) OTX2 directly activates cell cycle genes and inhibits differentiation in medulloblastoma cells. Int J Cancer 131: E21–E32 10.1002/ijc.26474 [doi].
33. CameronRS, LiuC, MixonAS, PihkalaJP, RahnRJ, et al. (2007) Myosin16b: The COOH-tail region directs localization to the nucleus and overexpression delays S-phase progression. Cell Motil Cytoskeleton 64 : 19–48 10.1002/cm.20162 [doi].
34. InabaN, IshigeH, IjichiM, SatohN, OhkawaR, et al. (1982) Immunohistochemical detection of pregnancy-specific protein (SP1) and placenta-specific tissue proteins (PP5, PP10, PP11 and PP12) in ovarian adenocarcinomas. Oncodev Biol Med 3 : 379–389.
35. MesakFM, OsadaN, HashimotoK, LiuQY, NgCE (2003) Molecular cloning, genomic characterization and over-expression of a novel gene, XRRA1, identified from human colorectal cancer cell HCT116Clone2_XRR and macaque testis. BMC Genomics 4 : 32 10.1186/1471-2164-4-32 [doi].
36. BradyCA, JiangD, MelloSS, JohnsonTM, JarvisLA, et al. (2011) Distinct p53 transcriptional programs dictate acute DNA-damage responses and tumor suppression. Cell 145 : 571–583 S0092-8674(11)00312-6 [pii];10.1016/j.cell.2011.03.035 [doi].
37. ZlotoffDA, SambandamA, LoganTD, BellJJ, SchwarzBA, et al. (2010) CCR7 and CCR9 together recruit hematopoietic progenitors to the adult thymus. Blood 115 : 1897–1905 blood-2009-08-237784 [pii];10.1182/blood-2009-08-237784 [doi].
38. CalderonL, BoehmT (2011) Three chemokine receptors cooperatively regulate homing of hematopoietic progenitors to the embryonic mouse thymus. Proc Natl Acad Sci U S A 108 : 7517–7522 1016428108 [pii];10.1073/pnas.1016428108 [doi].
39. MburuYK, WangJ, WoodMA, WalkerWH, FerrisRL (2006) CCR7 mediates inflammation-associated tumor progression. Immunol Res 36 : 61–72 IR:36 : 1:61 [pii];10.1385/IR:36 : 1:61 [doi].
40. GrayDH, TullD, UenoT, SeachN, ClassonBJ, et al. (2007) A unique thymic fibroblast population revealed by the monoclonal antibody MTS-15. J Immunol 178 : 4956–4965 178/8/4956 [pii].
41. SuzukiT, ShenH, AkagiK, MorseHC, MalleyJD, et al. (2002) New genes involved in cancer identified by retroviral tagging. Nat Genet 32 : 166–174 10.1038/ng949 [doi];ng949 [pii].
42. StewartM, TerryA, O'HaraM, CameronE, OnionsD, et al. (1996) til-1: a novel proviral insertion locus for Moloney murine leukaemia virus in lymphomas of CD2-myc transgenic mice. J Gen Virol 77 (Pt 3) 443–446.
43. VaillantF, BlythK, AndrewL, NeilJC, CameronER (2002) Enforced expression of Runx2 perturbs T cell development at a stage coincident with beta-selection. J Immunol 169 : 2866–2874.
44. DemarestRM, DahmaneN, CapobiancoAJ (2011) Notch is oncogenic dominant in T-cell acute lymphoblastic leukemia. Blood 117 : 2901–2909 blood-2010-05-286351 [pii];10.1182/blood-2010-05-286351 [doi].
45. KhandanpourC, MoroyT (2013) Growth factor independence 1 (Gfi1) as a regulator of p53 activity and a new therapeutical target for ALL. Oncotarget 4 : 374–375 933 [pii].
46. JacobsJJ, ScheijenB, VonckenJW, KieboomK, BernsA, et al. (1999) Bmi-1 collaborates with c-Myc in tumorigenesis by inhibiting c-Myc-induced apoptosis via INK4a/ARF. Genes Dev 13 : 2678–2690.
47. MaserRS, ChoudhuryB, CampbellPJ, FengB, WongKK, et al. (2007) Chromosomally unstable mouse tumours have genomic alterations similar to diverse human cancers. Nature 447 : 966–971 nature05886 [pii];10.1038/nature05886 [doi].
48. LeLQ, KabarowskiJH, WongS, NguyenK, GambhirSS, et al. (2002) Positron emission tomography imaging analysis of G2A as a negative modifier of lymphoid leukemogenesis initiated by the BCR-ABL oncogene. Cancer Cell 1 : 381–391 S1535610802000582 [pii].
49. SharmaA, LarueRC, PlumbMR, MalaniN, MaleF, et al. (2013) BET proteins promote efficient murine leukemia virus integration at transcription start sites. Proc Natl Acad Sci U S A 110 : 12036–12041 1307157110 [pii];10.1073/pnas.1307157110 [doi].
50. GuptaSS, MaetzigT, MaertensGN, SharifA, RotheM, et al. (2013) Bromo and ET domain (BET) chromatin regulators serve as co-factors for murine leukemia virus integration. J Virol JVI.01942-13 [pii];10.1128/JVI.01942-13 [doi].
51. ThornhillSI, SchambachA, HoweSJ, UlaganathanM, GrassmanE, et al. (2008) Self-inactivating gammaretroviral vectors for gene therapy of X-linked severe combined immunodeficiency. Mol Ther 16 : 590–598 6300393 [pii];10.1038/sj.mt.6300393 [doi].
52. BuonamiciS, TrimarchiT, RuoccoMG, ReavieL, CathelinS, et al. (2009) CCR7 signalling as an essential regulator of CNS infiltration in T-cell leukaemia. Nature 459 : 1000–1004 nature08020 [pii];10.1038/nature08020 [doi].
53. StewartM, TerryA, HuM, O'HaraM, BlythK, et al. (1997) Proviral insertions induce the expression of bone-specific isoforms of PEBP2alphaA (CBFA1): evidence for a new myc collaborating oncogene. Proc Natl Acad Sci U S A 94 : 8646–8651.
54. StewartM, MackayN, CameronER, NeilJC (2002) The common retroviral insertion locus Dsi1 maps 30 kilobases upstream of the P1 promoter of the murine Runx3/Cbfa3/Aml2 gene. J Virol 76 : 4364–4369.
55. WottonS, StewartM, BlythK, VaillantF, KilbeyA, et al. (2002) Proviral insertion indicates a dominant oncogenic role for Runx1/AML-1 in T-cell lymphoma. Cancer Res 62 : 7181–7185.
56. GirardL, HannaZ, BeaulieuN, HoemannCD, SimardC, et al. (1996) Frequent provirus insertional mutagenesis of Notch1 in thymomas of MMTVD/myc transgenic mice suggests a collaboration of c-myc and Notch1 for oncogenesis. Genes Dev 10 : 1930–1944.
57. KhandanpourC, PhelanJD, VassenL, SchutteJ, ChenR, et al. (2013) Growth factor independence 1 antagonizes a p53-induced DNA damage response pathway in lymphoblastic leukemia. Cancer Cell 23 : 200–214 S1535-6108(13)00036-6 [pii];10.1016/j.ccr.2013.01.011 [doi].
58. van LohuizenM, VerbeekS, KrimpenfortP, DomenJ, SarisC, et al. (1989) Predisposition to lymphomagenesis in pim-1 transgenic mice: cooperation with c-myc and N-myc in murine leukemia virus-induced tumors. Cell 56 : 673–682 0092-8674(89)90589-8 [pii].
59. BodrugSE, WarnerBJ, BathML, LindemanGJ, HarrisAW, et al. (1994) Cyclin D1 transgene impedes lymphocyte maturation and collaborates in lymphomagenesis with the myc gene. EMBO J 13 : 2124–2130.
60. KlingerMB, GuilbaultB, GouldingRE, KayRJ (2005) Deregulated expression of RasGRP1 initiates thymic lymphomagenesis independently of T-cell receptors. Oncogene 24 : 2695–2704 1208334 [pii];10.1038/sj.onc.1208334 [doi].
61. ScobieL, HectorRD, GrantL, BellM, NielsenAA, et al. (2009) A novel model of SCID-X1 reconstitution reveals predisposition to retrovirus-induced lymphoma but no evidence of gammaC gene oncogenicity. Mol Ther 17 : 1031–1038 mt200959 [pii];10.1038/mt.2009.59 [doi].
62. BlythK, TerryA, O'HaraM, BaxterEW, CampbellM, et al. (1995) Synergy between a human c-myc transgene and p53 null genotype in murine thymic lymphomas: contrasting effects of homozygous and heterozygous p53 loss. Oncogene 10 : 1717–1723.
63. BlythK, SlaterN, HanlonL, BellM, MackayN, et al. (2009) Runx1 promotes B-cell survival and lymphoma development. Blood Cells Mol Dis 43 : 12–19 S1079-9796(09)00046-1 [pii];10.1016/j.bcmd.2009.01.013 [doi].
64. KuoYH, ZaidiSK, GornostaevaS, KomoriT, SteinGS, et al. (2009) Runx2 induces acute myeloid leukemia in cooperation with Cbfbeta-SMMHC in mice. Blood 113 : 3323–3332 blood-2008-06-162248 [pii];10.1182/blood-2008-06-162248 [doi].
65. NakagawaM, IchikawaM, KumanoK, GoyamaS, KawazuM, et al. (2006) AML1/Runx1 rescues Notch1-null mutation-induced deficiency of para-aortic splanchnopleural hematopoiesis. Blood 108 : 3329–3334 blood-2006-04-019570 [pii];10.1182/blood-2006-04-019570 [doi].
66. O'NeilJ, GrimJ, StrackP, RaoS, TibbittsD, et al. (2007) FBW7 mutations in leukemic cells mediate NOTCH pathway activation and resistance to gamma-secretase inhibitors. J Exp Med 204 : 1813–1824 jem.20070876 [pii];10.1084/jem.20070876 [doi].
67. MaS, PathakS, MandalM, TrinhL, ClarkMR, et al. (2010) Ikaros and Aiolos inhibit pre-B-cell proliferation by directly suppressing c-Myc expression. Mol Cell Biol 30 : 4149–4158 MCB.00224-10 [pii];10.1128/MCB.00224-10 [doi].
68. DumortierA, JeannetR, KirstetterP, KleinmannE, SellarsM, et al. (2006) Notch activation is an early and critical event during T-Cell leukemogenesis in Ikaros-deficient mice. Mol Cell Biol 26 : 209–220 26/1/209 [pii];10.1128/MCB.26.1.209-220.2006 [doi].
69. BlythK, CameronER, NeilJC (2005) The RUNX genes: gain or loss of function in cancer. Nat Rev Cancer 5 : 376–387 nrc1607 [pii];10.1038/nrc1607 [doi].
70. UrenAG, MikkersH, KoolJ, van der WeydenL, LundAH, et al. (2009) A high-throughput splinkerette-PCR method for the isolation and sequencing of retroviral insertion sites. Nat Protoc 4 : 789–798 nprot.2009.64 [pii];10.1038/nprot.2009.64 [doi].
71. MarchHN, RustAG, WrightNA, ten HoeveJ, de RidderJ, et al. (2011) Insertional mutagenesis identifies multiple networks of cooperating genes driving intestinal tumorigenesis. Nat Genet 43 : 1202–1209 ng.990 [pii];10.1038/ng.990 [doi].
72. de RidderJ, UrenA, KoolJ, ReindersM, WesselsL (2006) Detecting statistically significant common insertion sites in retroviral insertional mutagenesis screens. PLoS Comput Biol 2: e166 06-PLCB-RA-0052R3 [pii];10.1371/journal.pcbi.0020166 [doi].
73. JeeJ, RozowskyJ, YipKY, LochovskyL, BjornsonR, et al. (2011) ACT: aggregation and correlation toolbox for analyses of genome tracks. Bioinformatics 27 : 1152–1154 btr092 [pii];10.1093/bioinformatics/btr092 [doi].
74. Wickham H. (2009) ggplot2: elegant graphics for data analysis. Springer New York.
75. BenjaminiY, HochbergY (1995) Controlling the false discovery rate: a practical and powerful approach to mutiple testing. J Roy Statist Soc Ser B 57 : 289–300.
76. IrizarryRA, HobbsB, CollinF, Beazer-BarclayYD, AntonellisKJ, et al. (2003) Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4 : 249–264 10.1093/biostatistics/4.2.249 [doi];4/2/249 [pii].
77. StoreyJD (2002) A direct approach to false discovery rates. Journal of the Royal Statistical Society, Series B 64 : 479–498.
Štítky
Genetika Reprodukční medicína
Článek Natural Polymorphisms in Influence Negative Selection and CD4∶CD8 Lineage Commitment in the RatČlánek MicroRNAs Located in the Hox Gene Clusters Are Implicated in Huntington's Disease PathogenesisČlánek Comparative RNAi Screens in and Reveal the Impact of Developmental System Drift on Gene FunctionČlánek Mutation of SLC35D3 Causes Metabolic Syndrome by Impairing Dopamine Signaling in Striatal D1 Neurons
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2014 Číslo 2
-
Všechny články tohoto čísla
- Fifteen Years Later: Hard and Soft Selection Sweeps Confirm a Large Population Number for HIV In Vivo
- The Same but Different: Worms Reveal the Pervasiveness of Developmental System Drift
- Serine Carboxypeptidase SCPEP1 and Cathepsin A Play Complementary Roles in Regulation of Vasoconstriction via Inactivation of Endothelin-1
- Coherent Functional Modules Improve Transcription Factor Target Identification, Cooperativity Prediction, and Disease Association
- A Long-Chain Flavodoxin Protects from Oxidative Stress and Host Bacterial Clearance
- Mammalian E-type Cyclins Control Chromosome Pairing, Telomere Stability and CDK2 Localization in Male Meiosis
- Influenza Virus Drug Resistance: A Time-Sampled Population Genetics Perspective
- Transcriptome-Wide Analyses of 5′-Ends in RNase J Mutants of a Gram-Positive Pathogen Reveal a Role in RNA Maturation, Regulation and Degradation
- Selective Disruption of Aurora C Kinase Reveals Distinct Functions from Aurora B Kinase during Meiosis in Mouse Oocytes
- X Chromosome Control of Meiotic Chromosome Synapsis in Mouse Inter-Subspecific Hybrids
- A Cohesin-Independent Role for NIPBL at Promoters Provides Insights in CdLS
- Extreme Population Differences in the Human Zinc Transporter ZIP4 (SLC39A4) Are Explained by Positive Selection in Sub-Saharan Africa
- Classic Selective Sweeps Revealed by Massive Sequencing in Cattle
- Genomic Networks of Hybrid Sterility
- Natural Polymorphisms in Influence Negative Selection and CD4∶CD8 Lineage Commitment in the Rat
- Oxidative Stress Is Not a Major Contributor to Somatic Mitochondrial DNA Mutations
- Molecular Identification of Collagen 17a1 as a Major Genetic Modifier of Laminin Gamma 2 Mutation-Induced Junctional Epidermolysis Bullosa in Mice
- Uncoupling of Molecular Maturation from Peripheral Target Innervation in Nociceptors Expressing a Chimeric TrkA/TrkC Receptor
- MicroRNAs Located in the Hox Gene Clusters Are Implicated in Huntington's Disease Pathogenesis
- Loss of Trabid, a New Negative Regulator of the Immune-Deficiency Pathway at the Level of TAK1, Reduces Life Span
- Targeted Ablation of Nesprin 1 and Nesprin 2 from Murine Myocardium Results in Cardiomyopathy, Altered Nuclear Morphology and Inhibition of the Biomechanical Gene Response
- Identification of Novel Genetic Loci Associated with Thyroid Peroxidase Antibodies and Clinical Thyroid Disease
- CEP-1, the p53 Homolog, Mediates Opposing Longevity Outcomes in Mitochondrial Electron Transport Chain Mutants
- Transcriptomics and Functional Genomics of ROS-Induced Cell Death Regulation by
- Quantitative Genome-Wide Genetic Interaction Screens Reveal Global Epistatic Relationships of Protein Complexes in
- Cascades of Genetic Instability Resulting from Compromised Break-Induced Replication
- Serine- and Threonine/Valine-Dependent Activation of PDK and Tor Orthologs Converge on Sch9 to Promote Aging
- Zfp322a Regulates Mouse ES Cell Pluripotency and Enhances Reprogramming Efficiency
- Insertional Mutagenesis and Deep Profiling Reveals Gene Hierarchies and a -Dependent Bottleneck in Lymphomagenesis
- DAAM Is Required for Thin Filament Formation and Sarcomerogenesis during Muscle Development in Drosophila
- Plasma Cholesterol–Induced Lesion Networks Activated before Regression of Early, Mature, and Advanced Atherosclerosis
- High-Resolution Profiling of Stationary-Phase Survival Reveals Yeast Longevity Factors and Their Genetic Interactions
- Comparative RNAi Screens in and Reveal the Impact of Developmental System Drift on Gene Function
- Accurate and Robust Genomic Prediction of Celiac Disease Using Statistical Learning
- Sex-Specific Embryonic Gene Expression in Species with Newly Evolved Sex Chromosomes
- Chromosome X-Wide Association Study Identifies Loci for Fasting Insulin and Height and Evidence for Incomplete Dosage Compensation
- Negative Feedback and Transcriptional Overshooting in a Regulatory Network for Horizontal Gene Transfer
- DNA Sequence Explains Seemingly Disordered Methylation Levels in Partially Methylated Domains of Mammalian Genomes
- Insights into the Genomic Landscape: Comparative Genomics Reveals Variations in Ploidy and Nutrient Utilisation Potential amongst Wine Isolates
- Molecular Evidence for the Inverse Comorbidity between Central Nervous System Disorders and Cancers Detected by Transcriptomic Meta-analyses
- The Centriolar Satellite Protein AZI1 Interacts with BBS4 and Regulates Ciliary Trafficking of the BBSome
- Fine-Mapping the Region Detects Common Variants Tagging a Rare Coding Allele: Evidence for Synthetic Association in Prostate Cancer
- Transmission Distortion Affecting Human Noncrossover but Not Crossover Recombination: A Hidden Source of Meiotic Drive
- A Variant in the Neuropeptide Receptor is a Major Determinant of Growth and Physiology
- Mutation of SLC35D3 Causes Metabolic Syndrome by Impairing Dopamine Signaling in Striatal D1 Neurons
- NSUN4 Is a Dual Function Mitochondrial Protein Required for Both Methylation of 12S rRNA and Coordination of Mitoribosomal Assembly
- MicroRNA-133 Inhibits Behavioral Aggregation by Controlling Dopamine Synthesis in Locusts
- Convergence of Light and ABA Signaling on the Promoter
- Arf4 Is Required for Mammalian Development but Dispensable for Ciliary Assembly
- Distinct Requirements for Cranial Ectoderm and Mesenchyme-Derived Wnts in Specification and Differentiation of Osteoblast and Dermal Progenitors
- Chk2 and P53 Regulate the Transmission of Healed Chromosomes in the Male Germline
- Ddc2 Mediates Mec1 Activation through a Ddc1- or Dpb11-Independent Mechanism
- Mapping the Fitness Landscape of Gene Expression Uncovers the Cause of Antagonism and Sign Epistasis between Adaptive Mutations
- Euchromatic Transposon Insertions Trigger Production of Novel Pi- and Endo-siRNAs at the Target Sites in the Germline
- miR-100 Induces Epithelial-Mesenchymal Transition but Suppresses Tumorigenesis, Migration and Invasion
- Canine Hereditary Ataxia in Old English Sheepdogs and Gordon Setters Is Associated with a Defect in the Autophagy Gene Encoding
- Within-Host Spatiotemporal Dynamics of Plant Virus Infection at the Cellular Level
- Analysis of Meiosis in SUN1 Deficient Mice Reveals a Distinct Role of SUN2 in Mammalian Meiotic LINC Complex Formation and Function
- Genome-Wide Association Study of Metabolic Traits Reveals Novel Gene-Metabolite-Disease Links
- Mechanistically Distinct Mouse Models for -Associated Retinopathy
- DAF-16/FoxO Directly Regulates an Atypical AMP-Activated Protein Kinase Gamma Isoform to Mediate the Effects of Insulin/IGF-1 Signaling on Aging in
- Chromosome I Controls Chromosome II Replication in
- Integrated Genomic Characterization Reveals Novel, Therapeutically Relevant Drug Targets in FGFR and EGFR Pathways in Sporadic Intrahepatic Cholangiocarcinoma
- The Iodotyrosine Deiodinase Ortholog SUP-18 Functions through a Conserved Channel SC-Box to Regulate the Muscle Two-Pore Domain Potassium Channel SUP-9
- The Genome of Highlights a Fish Pathogen Adapted to Fluctuating Environments
- Distinct DNA Binding Sites Contribute to the TCF Transcriptional Switch in and
- The Streamlined Genome of spp. Relative to Human Pathogenic Kinetoplastids Reveals a Parasite Tailored for Plants
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Genome-Wide Association Study of Metabolic Traits Reveals Novel Gene-Metabolite-Disease Links
- A Cohesin-Independent Role for NIPBL at Promoters Provides Insights in CdLS
- Classic Selective Sweeps Revealed by Massive Sequencing in Cattle
- Arf4 Is Required for Mammalian Development but Dispensable for Ciliary Assembly
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



