-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaA Long-Chain Flavodoxin Protects from Oxidative Stress and Host Bacterial Clearance
Long-chain flavodoxins, ubiquitous electron shuttles containing flavin mononucleotide (FMN) as prosthetic group, play an important protective role against reactive oxygen species (ROS) in various microorganisms. Pseudomonas aeruginosa is an opportunistic pathogen which frequently has to face ROS toxicity in the environment as well as within the host. We identified a single ORF, hereafter referred to as fldP (for flavodoxin from P. aeruginosa), displaying the highest similarity in length, sequence identity and predicted secondary structure with typical long-chain flavodoxins. The gene was cloned and expressed in Escherichia coli. The recombinant product (FldP) could bind FMN and exhibited flavodoxin activity in vitro. Expression of fldP in P. aeruginosa was induced by oxidative stress conditions through an OxyR-independent mechanism, and an fldP-null mutant accumulated higher intracellular ROS levels and exhibited decreased tolerance to H2O2 toxicity compared to wild-type siblings. The mutant phenotype could be complemented by expression of a cyanobacterial flavodoxin. Overexpression of FldP in a mutT-deficient P. aeruginosa strain decreased H2O2-induced cell death and the hypermutability caused by DNA oxidative damage. FldP contributed to the survival of P. aeruginosa within cultured mammalian macrophages and in infected Drosophila melanogaster, which led in turn to accelerated death of the flies. Interestingly, the fldP gene is present in some but not all P. aeruginosa strains, constituting a component of the P. aeruginosa accessory genome. It is located in a genomic island as part of a self-regulated polycistronic operon containing a suite of stress-associated genes. The collected results indicate that the fldP gene encodes a long-chain flavodoxin, which protects the cell from oxidative stress, thereby expanding the capabilities of P. aeruginosa to thrive in hostile environments.
Published in the journal: . PLoS Genet 10(2): e32767. doi:10.1371/journal.pgen.1004163
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1004163Summary
Long-chain flavodoxins, ubiquitous electron shuttles containing flavin mononucleotide (FMN) as prosthetic group, play an important protective role against reactive oxygen species (ROS) in various microorganisms. Pseudomonas aeruginosa is an opportunistic pathogen which frequently has to face ROS toxicity in the environment as well as within the host. We identified a single ORF, hereafter referred to as fldP (for flavodoxin from P. aeruginosa), displaying the highest similarity in length, sequence identity and predicted secondary structure with typical long-chain flavodoxins. The gene was cloned and expressed in Escherichia coli. The recombinant product (FldP) could bind FMN and exhibited flavodoxin activity in vitro. Expression of fldP in P. aeruginosa was induced by oxidative stress conditions through an OxyR-independent mechanism, and an fldP-null mutant accumulated higher intracellular ROS levels and exhibited decreased tolerance to H2O2 toxicity compared to wild-type siblings. The mutant phenotype could be complemented by expression of a cyanobacterial flavodoxin. Overexpression of FldP in a mutT-deficient P. aeruginosa strain decreased H2O2-induced cell death and the hypermutability caused by DNA oxidative damage. FldP contributed to the survival of P. aeruginosa within cultured mammalian macrophages and in infected Drosophila melanogaster, which led in turn to accelerated death of the flies. Interestingly, the fldP gene is present in some but not all P. aeruginosa strains, constituting a component of the P. aeruginosa accessory genome. It is located in a genomic island as part of a self-regulated polycistronic operon containing a suite of stress-associated genes. The collected results indicate that the fldP gene encodes a long-chain flavodoxin, which protects the cell from oxidative stress, thereby expanding the capabilities of P. aeruginosa to thrive in hostile environments.
Introduction
Microorganisms living in aerobic environments are constantly exposed to the harmful effects of reactive oxygen species (ROS), including H2O2 and the superoxide radical, which are generated as unavoidable by-products of oxygen utilization [1]. In addition, commensal and pathogenic bacteria have to face the host oxidative response, such as H2O2 production from phagocytes [1], [2]. Aerobic organisms have evolved multigenic responses to prevent and/or repair the cellular damage potentially inflicted by these toxic compounds. Whenever defenses are overcome by the amounts of ROS produced, cells are afflicted by a condition called oxidative stress [1], [2]. Protective mechanisms deployed by stressed organisms include regulation of membrane permeability, antioxidant and repair systems, and replacement of ROS-sensitive targets by resistant isofunctional versions. In microorganisms as distantly related as enterobacteria and cyanobacteria, induction of the mobile electron shuttle flavodoxin (Fld) appears to be a common feature of the antioxidant response [3], [4]. Flds contain flavin mononucleotide (FMN) as prosthetic group and are largely isofunctional with the ubiquitous electron carrier ferredoxin (Fd), exchanging reducing equivalents with a promiscuous lot of donors and acceptors. Fld induction is assumed to act as a backup for Fd, which harbors a ROS-sensitive iron-sulfur cluster as redox-active cofactor and whose levels are down-regulated under conditions of environmental stress or iron starvation [5], [6]. Accordingly, Fld overexpression has been shown to confer augmented tolerance toward various sources of oxidative stress in organisms with very different lifestyles, such as Escherichia coli [7], rhizobia [8] and plants [9]. Unlike Fds, which are present in all major kingdoms, Flds are restricted to various groups of prokaryotes and some oceanic algae [10]. From sequence alignments and structural considerations, they can be divided into two classes, short-chain and long-chain Flds, which differ by the presence of a 20-amino acid loop of a so far unknown function [11]. Phylogenetic analyses indicate that the two lineages have diverged only once [12].
Pseudomonas aeruginosa is a free-living bacterium commonly found in soil, water, moist locations, and most man-made environments throughout the Earth. P. aeruginosa has a wide metabolic versatility as the reflection of a large and flexible genome with a substantial number of genes, which facilitates its adaptability to thrive in different habitats, and allows a quick response to diverse environmental stimuli and challenges [13]. This remarkable versatility enables P. aeruginosa to infect damaged animal tissues or immunocompromised individuals, where it constitutes an important opportunistic pathogen highly prevalent in nosocomial infections [14], [15]. Indeed, this bacterium is of particular concern to patients with cystic fibrosis (CF) who are highly susceptible to P. aeruginosa and suffer severe and often fatal chronic airway infections [16].
The present research seeks to identify and characterize putative long-chain Flds in P. aeruginosa which could play a protective role under environmental stress conditions. Both P. aeruginosa and its relative Pseudomonas putida contain a single-copy gene encoding a short-chain Fld [17], [18], annotated as mioC due to the homology of the product with the homonymous Fld from E. coli, but no long-chain Fld orthologs have been so far reported in pseudomonads. We identified a gene, PA14_22540 (hereafter referred to as fldP, for flavodoxin from P. aeruginosa), which displays low but significant sequence homology to the Anabaena and E. coli Fld genes (named isiB and fldA, respectively), and whose recombinant product was able to display Fld activity in vitro. Expression of the fldP gene in P. aeruginosa was induced by H2O2 treatment via an OxyR-independent pathway, whereas its disruption increased H2O2-induced killing and accumulation of intracellular ROS. The mutant phenotype could be complemented by transformation with the isiB gene from Anabaena. Overexpression of fldP mitigated H2O2-induced cell death in a mutT-deficient P. aeruginosa strain as well as the hypermutability caused by DNA oxidative damage. The presence of a functional fldP gene contributed to P. aeruginosa survival in two model systems of infection: cultured mammalian macrophages and Drosophila melanogaster. Improved bacterial endurance in Drosophila resulted in higher death tolls of the infected flies. In line with its presumptive adaptive role, the fldP gene was found to be part of a self-regulated operon belonging to the P. aeruginosa accessory genome, a collection of strain-specific gene clusters which are acquired en bloc and expand the genomic repertoire to fit the needs for survival in adverse environments. Then, the collected results indicate that the fldP gene encodes a long-chain flavodoxin which is induced when P. aeruginosa is under oxidative stress to exert a protective role against the physiological and mutational damage caused by ROS.
Results
Search and structural analysis of a long-chain flavodoxin in the P. aeruginosa genome
We performed an in silico survey of putative Flds in the genome of P. aeruginosa PA14 and compared the retrieved sequences with those reported for Anabaena (IsiB) and E. coli (FldA) long-chain Flds, whose protective roles against oxidative stress have been extensively documented [12]. To find orthologs, the IsiB and FldA sequences were compared to the P. aeruginosa PA14 genome using the Domain Enhanced Lookup Time Accelerated Basic Local Alignment Search Tool (DELTA-BLAST), which is sensitive in detecting remote protein homologs [19]. Four unique open reading frames (ORFs) displaying both sequence homology and similar domain organization were retrieved. They were a putative oxidoreductase with a covalently-linked Fld-like domain (PA14_58560), the repressor binding protein WrbA (PA14_51990), a short-chain Fld similar to E. coli MioC (PA14_19660), and an ORF (PA14_22540, tentatively referred to as FldP), which displayed the highest similarity in length and sequence with the long-chain Flds used as baits. Analysis of ORF PA14_22540 indicated that it would encode a 184-amino acid protein with a molecular mass of ∼20 kDa, which is in the range of those typically observed for long-chain Flds (170–185 amino acids). Multiple-sequence alignment between FldA, IsiB and FldP showed that while FldA and IsiB display 47% identity and 67% similarity, FldP exhibits 23% identity with both flavodoxins, and 50% and 41% similarity with IsiB and FldA, respectively.
By using the JPred3 software we next performed a prediction of the secondary structure of FldP and compared it with those of IsiB (Accession number P0A3E0) and FldA (Accession number P61949). According to this prediction, FldP displays a significant similarity, at the level of the secondary structures, with both FldA and IsiB (Figure 1). The three proteins fit the common scheme of five β-sheets intercalated with five α-helices, as well as the loosely structured region dividing β5 into β5a and β5b (between residues 130 and 150 of FldP), which is typical of the long-chain class of Flds [11], [12].
Fig. 1. Primary and secondary structures of FldP from P. aeruginosa. 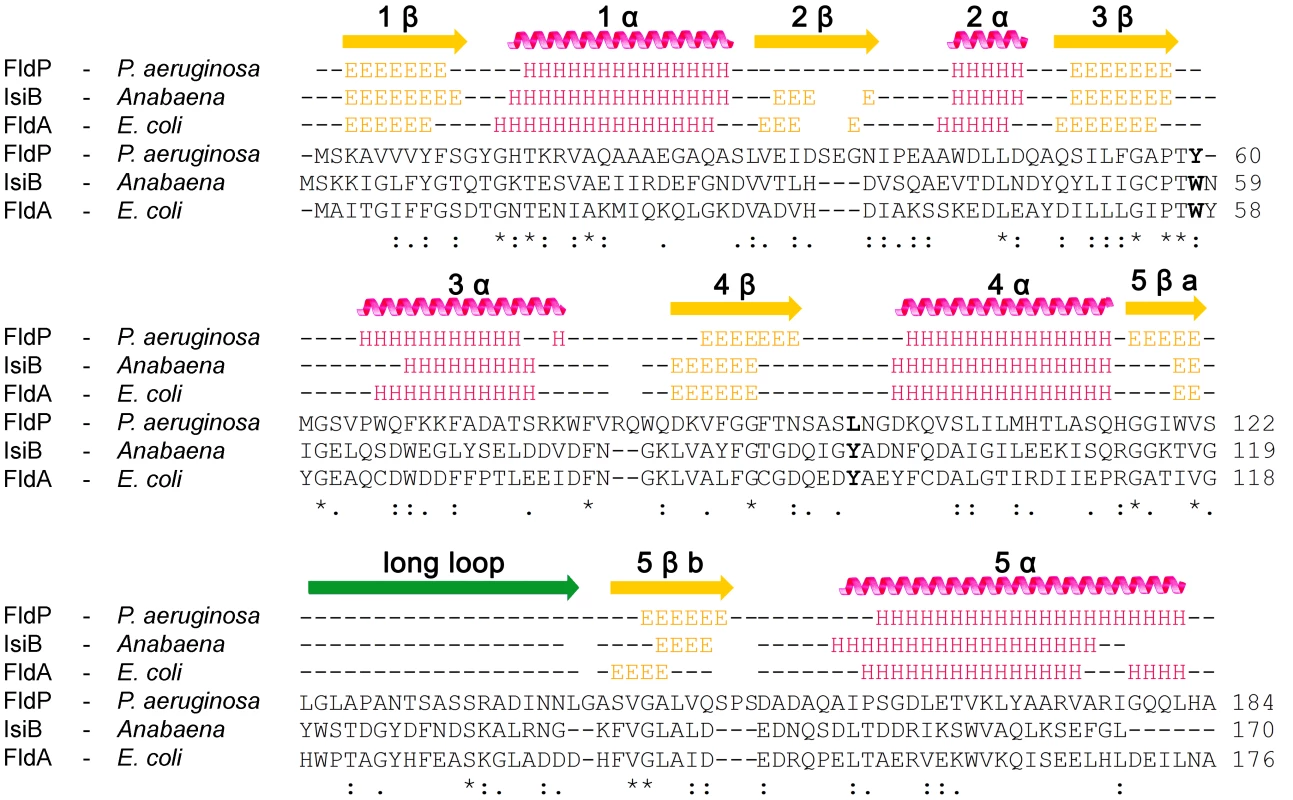
FldP sequence was compared with those of well-characterized flavodoxins from Anabaena (IsiB) and E. coli (FldA). Secondary structure predictions are represented above the sequence alignments. “E” identifies amino acids involved in β-sheets (yellow arrows), and “H”, those which are part of α-helices (pink helices). The green arrow corresponds to the extra loop that characterizes long-chain Flds. Amino acids in bold (corresponding to W58 and Y95 in Anabaena IsiB) indicate the aromatic residues stacking onto the isoalloxazine ring system in IsiB and FldA, and their equivalents in FldP. The fldP gene encodes a functional flavodoxin
To determine if the product of the fldP gene is a functional Fld, the coding sequence was expressed in E. coli under the control of an inducible promoter (see Materials and Methods). The recombinant protein accumulated to high levels in the bacterial host, but unlike IsiB, which was readily soluble and assembled its prosthetic group in the E. coli cytosol [20], the P. aeruginosa protein was recovered largely as insoluble inclusion bodies (Figure S1A). Only after the simultaneous expression of a suite of molecular chaperones, a minor but significant amount of the protein could be solubilized and purified by affinity chromatography on Ni-NTA columns (Figure S1B).
The purified protein showed a typical flavoprotein spectrum with absorption maxima at 374 and 446 nm, close to those of free FMN in aqueous solution (Figure 2A). The 446-nm peak, which corresponds to transition I of the flavin, is strongly red-shifted in the flavodoxins from Anabaena and E. coli (Figure 2A), indicating that the environment of the prosthetic group is more hydrophobic and/or less solvent-exposed in these flavoproteins. The flavin moiety of IsiB is sandwiched between the aromatic side-chains of Trp58 and Tyr95, in a coplanar conformation, and the π–π interaction thus established has been regarded as the major cause for the spectral red shift [12], [21]. The two amino acids are conserved in FldA, but not in FldP, where the tryptophan is replaced by a tyrosine and the tyrosine by a leucine (Figure 1). The substitutions will prevent aromatic stacking of the isoalloxazine ring system, and introduce conformational changes in its immediate environment, since both residues are located in flexible loops between 3β-3α and 4β-4α (Figure 1). Then, the absence of red-shifted peaks in the visible absorption spectrum of FldP likely results from a combination of higher solvent exposure and decreased aromatic stacking on the flavin.
Fig. 2. FldP from P. aeruginosa is a functional flavodoxin. 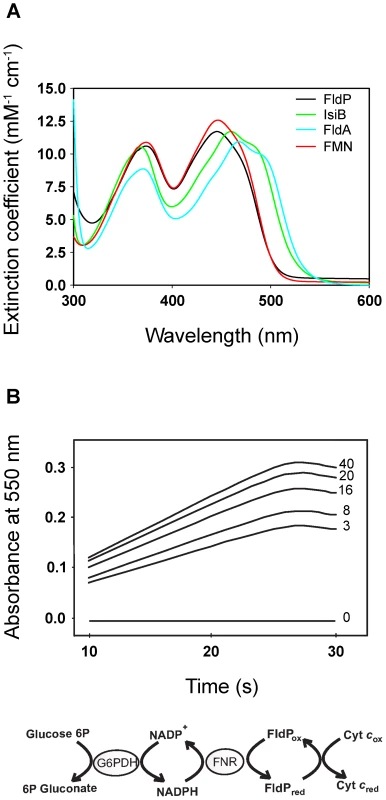
The FldP holoprotein was expressed in E. coli and purified by affinity chromatography as described in Materials and Methods. (A) Visible absorption spectra of purified FldP (black), FldA (blue), IsiB (green) and FMN (red) in 50 mM Tris-HCl pH 8.5. (B) Kinetics of cytochrome c reductase activity as determined by the increase in absorbance at 550 nm. Numerals on the side of each curve indicate FldP concentration (in µM) in the reaction medium. The Fld-driven reaction sequence is indicated at the bottom. Experimental conditions are given in the text. FldP was assayed in vitro as a substrate of cyanobacterial ferredoxin-NADP(H) reductase (FNR). Figure 2B shows that purified FldP was able to mediate FNR-driven cytochrome c reduction in a concentration-dependent manner, with an apparent KM of 1.3±0.2 µM and a kcat of 22.0±1.8 min−1. Under similar conditions, IsiB displayed a kcat of 48.5±3.8 min−1 (data not shown). The collected results indicate that the product of the fldP gene displayed the structural and functional properties of a bona fide flavodoxin.
Inactivation of fldP decreases survival of P. aeruginosa and increases intracellular ROS levels upon exposure to H2O2
In order to determine the functional role of FldP in P. aeruginosa, we tested the tolerance exhibited by a fldP-deficient mutant strain to H2O2 toxicity. In the absence of stress, the viabilities of the wt and fldP mutant strains were similar (Figure S2), but fldP cells were ∼5-fold more sensitive than their wt siblings to the H2O2 treatment (P = 0.024, Figure 3A). Complementation of this mutant with the fldP gene cloned in plasmid p2 (p2-fldP) led to an approximately 1.4-fold increase in the percentage of surviving cells respect to that observed with the wt strain, although the difference was not statistically significant (P = 0.3929, Figure 3A). Noteworthy, expression of IsiB from the p2-isiB plasmid provided even higher levels of protection against the deleterious effects of H2O2, with a rise of 2.2-fold relative to the wt strain (P = 0.125, Figure 3A).
Fig. 3. Role of FldP in cell survival and ROS accumulation upon exposure to H2O2. 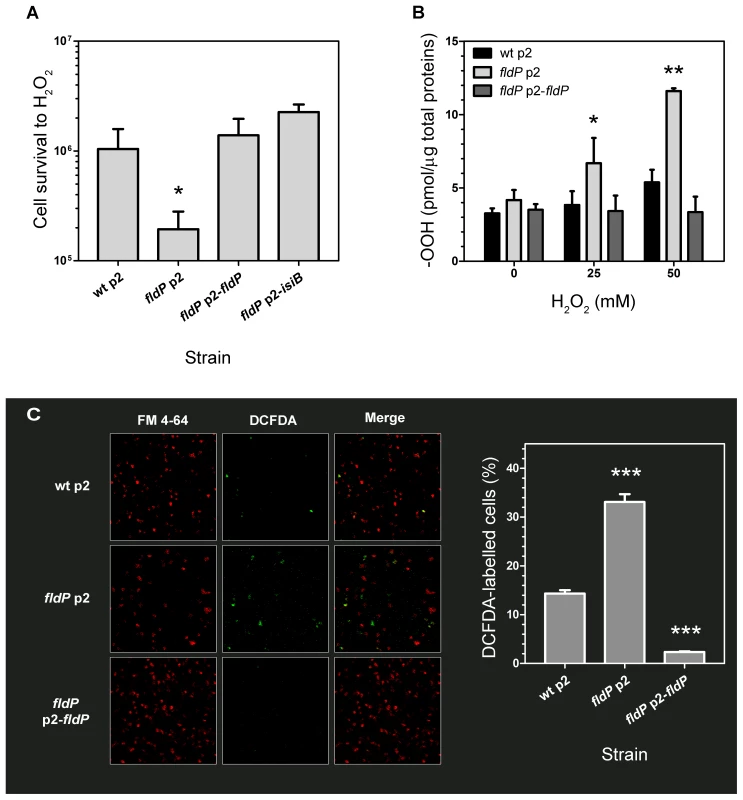
(A) Flavodoxin increases oxidative stress tolerance of P. aeruginosa exposed to H2O2. Cells from wt P. aeruginosa PA14 transformed with the empty plasmid p2, and from the isogenic fldP mutant strain harboring vectors p2, p2-fldP or p2-isiB were normalized to ∼108 cells and then incubated with 50 mM H2O2 for 30 min at 37°C. Viability was estimated as described in Materials and Methods. (B) Expression of FldP prevents ROS build-up in stressed P. aeruginosa cells. Bacteria were exposed to the indicated concentrations of H2O2 for 30 min, and total peroxides (-OOH) were determined in cleared extracts using the FOX II assay as described in the experimental section. Measurements in (A) and (B) were carried out in triplicate for two independent experiments, and the results are expressed as means with their SEM. Statistically significant differences P<0.005 and P<0.05 are identified by ** and *, respectively (one-tailed Mann-Whitney test). (C) Fluorescent detection of ROS formation in cells from wt, mutant and fldP-complemented strains challenged with 25 mM H2O2 for 15 min, using a ROS-dependent fluorescent probe (DCFDA). Panels on the left show membrane fluorescence after staining with FM4-64; those on the middle depict ROS-dependent fluorescence, with the merges displayed in the right panels. Bars show percentages of ROS-stained cells relative to membrane-stained cells. Values presented are the means ± SEM of 8–10 independent fields. Statistically significant differences respect to wt (P<0.0005) are identified by *** (one-tailed Mann-Whitney test). Taking into account these observations, we further tested whether FldP may be involved in the control of intracellular ROS build-up, presumably the cause of H2O2-induced killing. ROS accumulation was quantified in bacterial extracts from the wt, the fldP and the complemented mutant strains after exposure to H2O2. Cells from the wt strain displayed a small increase of their ROS (-OOH) levels as the H2O2 concentration was raised (Figure 3B). Although this increase was not statistically significant, it was consistently observed in a number of experiments. On the other hand, lack of a functional FldP led to ROS build-up in the fldP mutant, whereas expression of FldP from a plasmid in the complemented cells decreased the total -OOH levels to those observed in untreated wt bacteria (Figure 3B).
ROS accumulation was also detected in whole P. aeruginosa cells by using the fluorogenic dye 2′,7′-dichlorofluorescein diacetate (DCFDA), and visualized by confocal microscopy. Figure 3C shows that the fraction of labelled cells above the detection threshold was significantly higher in the fldP mutant compared to their wt siblings (33% vs. 14%), but decreased to 2% after complementation with the p2-fldP plasmid, presumably due to the effect of increased genic doses provided by the plasmid.
Overexpression of fldP and isiB increases resistance to H2O2 in a mutT-deficient strain of P. aeruginosa
It has been recently reported that P. aeruginosa strains deficient in the 8-oxodeoxiguanine system (GO) are particularly vulnerable to oxidative stress [22], [23]. Specifically, mutT-deficient cells showed to be the most susceptible to oxidants such as H2O2 or methyl viologen. We therefore used this strain to further characterize the protective activity of FldP against ROS. A mutT P. aeruginosa strain was transformed with either p2-fldP or p2-isiB, and the resulting transformants were tested for their susceptibility to H2O2. Parallel controls were carried out by using the wt and mutT strains harboring an empty p2 plasmid.
As shown in Figure 4, the mutT strain showed a 20-fold decrease in survival after exposure to H2O2, being significantly more susceptible than the wt strain (P = 0.0119). However, when fldP or isiB were overexpressed in the mutT mutant, cell survival increased dramatically (8 - and 16-fold, respectively, P = 0.0119), relative to mutT transformed with the empty p2 vector (Figure 4). The results suggest that the antioxidant role of FldP and IsiB can partially compensate the increased susceptibility of mutT-deficient cells to oxidative stress.
Fig. 4. FldP mitigates H2O2-induced cell death in a mutT-deficient P. aeruginosa. 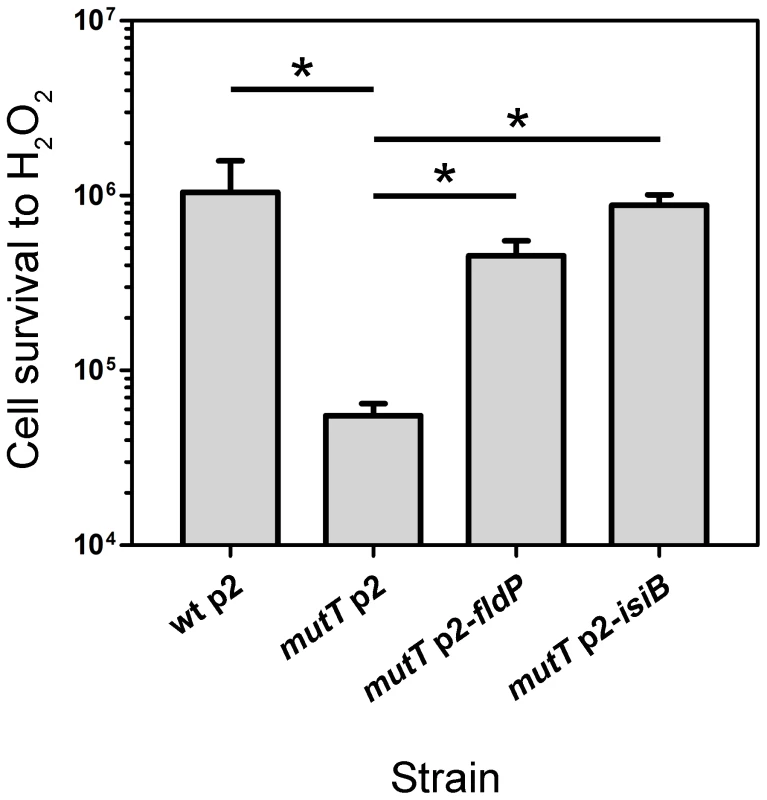
The mutT mutant strain (transformed with the empty plasmid p2) was tested for resistance to H2O2, and compared with its wt parental strain (harboring p2), and with mutT mutants transformed with p2-fldP or p2-isiB. Bacterial cultures were normalized to ∼108 cells and then incubated with 50 mM H2O2 for 30 min at 37°C. Viability was estimated as described in Materials and Methods. Measurements were carried out in triplicate for two independent experiments, and the results were expressed as means with their ± SEM. Statistically significant differences (P<0.05) are identified by * (one-tailed Mann-Whitney test). FldP and IsiB decrease the mutation frequency induced by H2O2 in a mutT-deficient strain of P. aeruginosa
DNA damage produced by ROS can lead to increased mutation frequencies [24] and emergence of adaptive phenotypes [25], [26]. Particularly in mutT-deficient mutants, in which the damage produced by oxidative stress cannot be avoided, mutation frequencies can increase 100 - to 1000-fold, leading to hypermutator phenotypes [22]. We therefore investigated whether fldP and isiB could display an antimutator effect and alleviate the hypermutability that is typically observed in a mutT-deficient background. Thus, we exposed a mutT strain, which overexpressed fldP or isiB to H2O2 and measured the mutation frequency by determining the emergence of mutants resistant to streptomycin. The mutT strain showed a ∼1200-fold increase in the H2O2-induced mutation frequency (3.44×10−6), relative to the wt (2.83×10−9). Expression of either fldP or isiB in the mutT strain diminished the mutation frequencies to 4.49×10−7 and 2.63×10−7, which represent a 13% (P = 0.05) and 8% (P = 0.0286), respectively, of the mutation frequency showed by mutT cells transformed with p2 (Figure 5). This last result indicates that overexpression of a functional Fld could avoid the majority (∼90%), but not all H2O2-induced lesions produced as a consequence of mutT deficiency. The protective effect presumably results from the antioxidant properties of these flavoproteins. Accordingly, the antimutator effect conferred by both fldP and isiB was not observed when the spontaneous mutation frequency was tested (data not shown).
Fig. 5. FldP decreases the H2O2-induced mutation frequency in mutT-deficient P. aeruginosa. 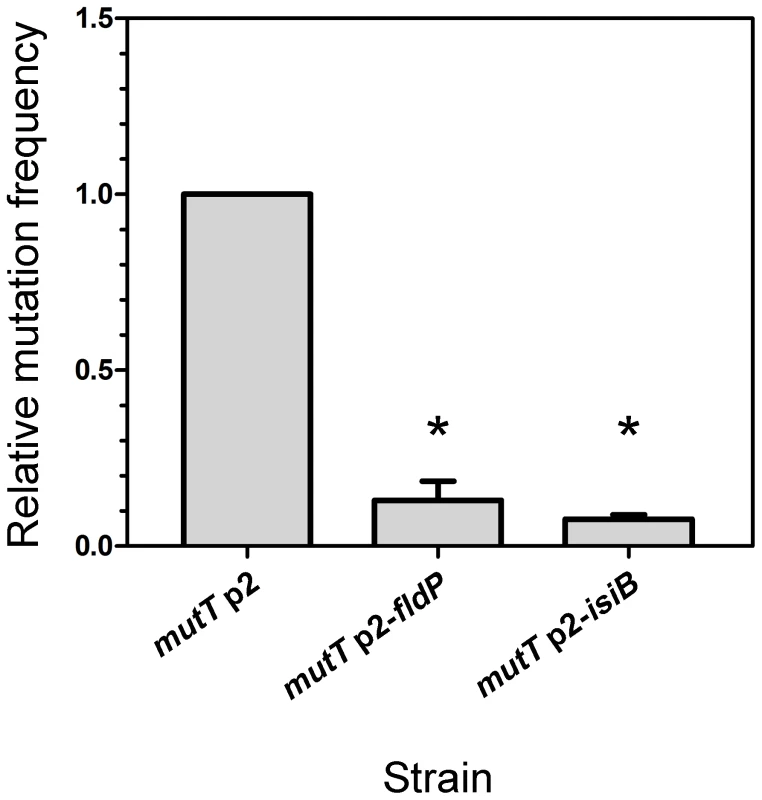
The mutT mutant strain transformed with the empty vector p2 was tested for its frequency of mutations induced by H2O2 (50 mM, 30 min), and compared with mutT mutants expressing fldP or isiB. The H2O2-induced mutation frequency was calculated as the ratio between the number of streptomycin-resistant cells and the number of viable cells and normalized to 1 in the mutT strain. The experiments were carried out in quintuplicate for three independent replicas, and the results were expressed as means ± SEM. Statistically significant differences (P<0.05) are identified by * (one-tailed Mann-Whitney test). Expression of the fldP gene is induced by H2O2 treatment via an OxyR-independent pathway
To gain further insight into the role played by FldP in oxidative stress tolerance, we studied the transcriptional induction of the fldP gene by H2O2 treatment. The expression of fldP was monitored by semi-quantitative, two-step, reverse transcription-PCR (RT-PCR), using expression of the constitutive housekeeping rpoD gene as control for equal amounts of cDNA in each reaction. Figure 6 shows that expression of fldP in the wt strain was clearly induced (3.9±0.7-fold relative to untreated cultures, P = 0.004), after H2O2 treatment.
Fig. 6. Induction and regulation of fldP expression. 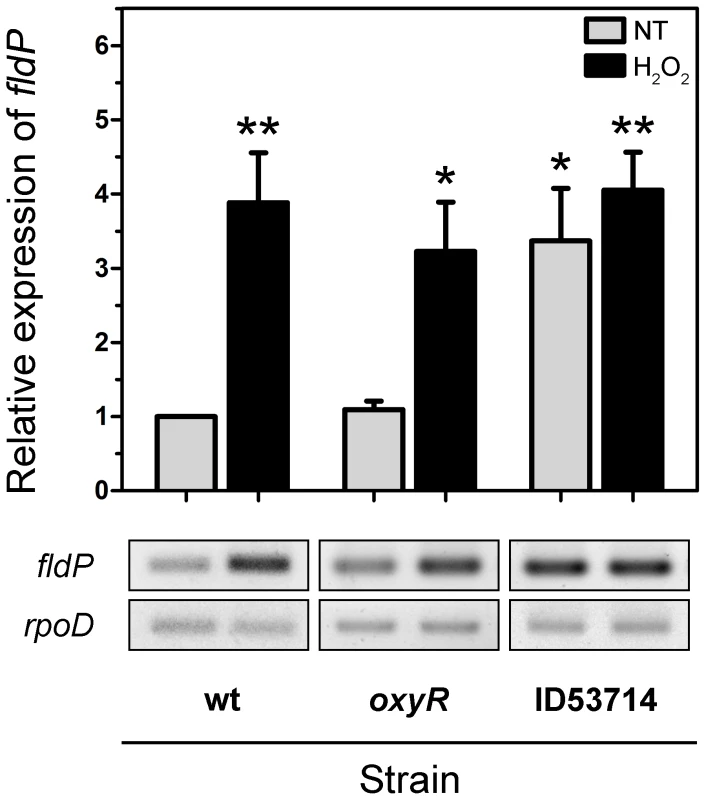
Total RNA was extracted from cultures (OD600 = 0.8) of wt, oxyR and ID53714 (PA14_22550 mutant) strains of P. aeruginosa which had been treated with 50 mM H2O2 for 15 min or left untreated (NT). Semi-quantitative RT-PCR was performed with specific primers for fldP and the amplification products resolved by agarose gel electrophoresis. The rpoD transcripts were used as housekeeping controls. The upper panel shows relative expression levels averaged over at least 4 experiments (means ± SEM), with NT expression of the wt strain normalized to 1. Statistically significant differences at P<0.005 and P<0.05 are identified by ** and *, respectively (two-tailed Student's t test). The lower panels illustrate typical results obtained with fldP and rpoD. We further investigated whether this increased expression of fldP was dependent on OxyR, the main oxidative stress response regulator of P. aeruginosa [27]. An oxyR deletion mutant strain of P. aeruginosa was constructed and tested for fldP expression in response to H2O2. Interestingly, no differences were observed in the oxyR strain compared to its isogenic wt, showing a 3.0±0.7-fold induction in the expression of fldP relative to untreated cultures (P = 0.0045, Figure 6). Importantly, we also evaluated expression of fldP in a second oxyR strain (ID54029) from the PA14 insertion mutant library [28], which yielded equivalent results (data not shown). These observations indicate that although fldP is a stress-responsive gene in P. aeruginosa, as in other bacterial species [3], [4], this response is triggered by an OxyR-independent mechanism.
FldP enhances P. aeruginosa survival within mammalian macrophages and during in vivo infection of Drosophila melanogaster
The use of ROS to kill bacterial pathogens, such as H2O2 production from phagocytes, is a common feature of the innate immune response of eukaryotic organisms. Considering that FldP sheltered P. aeruginosa from oxidative stress under in vitro conditions, we tried to determine if this protective role of FldP could also provide an advantage to cope with the host immune defenses.
As a first approach, we evaluated the capacity of the different P. aeruginosa strains to survive in the intracellular milieu of phagocytes by using monolayers of the macrophagic cell line RAW 264.7, which were inoculated with wt P. aeruginosa or its isogenic fldP-deficient strain, complemented or not with p2-fldP. Then, with the addition of antibiotics to kill extracellular bacteria we were able to compare the proportion of bacterial cells of each strain that survived during a 3-h period inside the phagocytes (see Materials and Methods) by lysing the cell monolayer and plating the lysates on LB agar. Figure 7A shows that inactivation of fldP produced a moderate but significant decrease of ∼24% in the intracellular survival of P. aeruginosa in phagocytic cells (P = 0.0103). Importantly, this decrease could be reverted by complementation with p2-fldP, even surpassing the wt values. This result indicates that FldP is contributing to the intracellular survival of P. aeruginosa in macrophagic cells, probably by enhancing bacterial resistance to the ROS produced by phagocytes.
Fig. 7. FldP enhances P. aeruginosa survival within mammalian macrophages and during in vivo infection of Drosophila melanogaster. 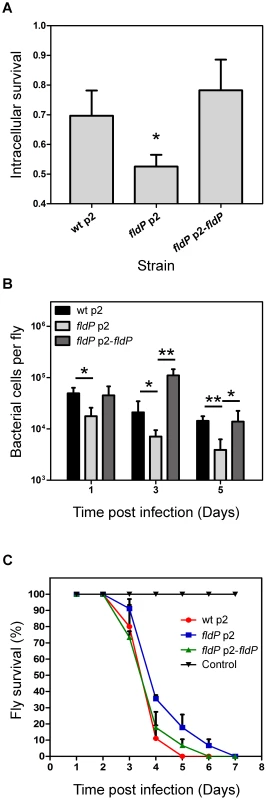
Comparison between the P. aeruginosa wt strain and its isogenic fldP mutant complemented or not with a functional copy of the fldP gene (p2-fldP), with respect to: (A) the fraction of bacteria that survived after 3 h in the intracellular milieu of RAW 264.7 macrophagic cells. Three wells of cells were used for each strain in each experiment, and all experiments were repeated three times. Statistically significant differences relative to the wt strain (P<0.05) are identified by * (one-tailed Student's t test); (B) survival of bacterial cells within D. melanogaster flies after 1, 3 and 5 days of infection. Assays were carried out in quadruplicate for two independent experiments and the results are expressed as means ± SEM. Statistically significant differences at P<0.005 and P<0.05 are identified by ** and *, respectively (one-tailed Student's t test). (C) Percentage of flies that survived at each time point after being fed with the different P. aeruginosa strains. Each point of the survival curve is the average of triplicate experiments expressed as means ± SEM. A representative experiment out of 3 is shown in the figure. To evaluate if this protective role of FldP could also be observed in the context of a whole organism infection, which is a more complex system than cultured cells, we used the insect-infection model of D. melanogaster, as previously validated to evaluate virulence traits of P. aeruginosa [29], [30]. Thus, flies were fed with the same strains of P. aeruginosa mentioned above and left for periods of 1, 3 or 5 days, after which the ability of each P. aeruginosa strain to survive within the host was scored by counting colony forming units (CFU). Interestingly, the capacity of the fldP mutant to survive within the host dropped to 28 to 36% (P<0.05) at the three time-periods assayed, which could recover to wt values after complementation with p2-fldP (Figure 7B).
These results suggest that increased bacterial loads could have an impact on host survival. To assess this, D. melanogaster flies were starved for 3 h and then continuously fed with P. aeruginosa. The number of surviving flies was monitored every day until all flies died. The results showed that the flies that had been fed with the wt strain of P. aeruginosa died faster than those fed with the fldP mutant (Figure 7C), which is in agreement with the better capacity of the wt P. aeruginosa strain to survive within the host. Complementation of the mutant bacteria with p2-fldP increased the mortality rate to wt values (Figure 7C).
Taken together, these findings suggest that FldP could be playing a role in P. aeruginosa pathogenesis by increasing its resistance to the ROS-dependent clearance carried out by the host immune system, which in turn would result in a more lethal infection due to a higher load of viable bacterial cells in the host.
The fldP gene is a component of the P. aeruginosa accessory genome
The P. aeruginosa genome is made up by a mosaic of “core” and “accessory and variable” gene clusters [31], [32]. While the gene composition of the core genome is conserved in almost every strain of P. aeruginosa, genes belonging to the accessory genome are found in discrete patches, referred to as regions of genome plasticity (RGP), which can vary in occurrence and location among strains [33]. By using genome sequence information available online, we evaluated the occurrence of the fldP gene in thirteen P. aeruginosa strains (namely PAO1, PA14, 2192, 39016, C3719, LESB58, PACS2, M18, NCGM2.S1, B136-33, RP73, DK2 and PA7), whose genomes have been completely sequenced and made available at the Pseudomonas Genome Database (www.pseudomonas.com) [34]. The genome of the environmental strain Hex1T [35], which has been recently sequenced (Feliziani et al., unpublished), was also included in the analysis. The survey showed that only six of the fourteen strains (PA14, 39016, NCGM2.S1, B136-33, PA7 and Hex1T) contained the fldP gene, suggesting that it is a component of the P. aeruginosa accessory genome. Indeed, fldP is located in the region of genome plasticity RGP32 [33]. With the exception of the taxonomic outlier PA7, all other strains showed a conserved synteny of RGP32, with fldP (PA14_22540) being the fifth gene in a six-gene cluster (PA14_22500 to PA14_22550 in the PA14 genome) (Figure 8). The DNA sequence of RGP32 is highly conserved among strains PA14, 39016, NCGM2.S1, B136-33 and Hex1T, displaying up to 97% identity. The five ORFs accompanying fldP in RGP32 have been assigned putative functions, based on similarity with known genes (Table S1). Interestingly, RGP32 is flanked by two palindromic sequences (inverted sequence repeats) which could have played a role in the acquisition of this gene cluster. On the other hand, the genome of PA7 only showed orthologs for fldP (PSPA7_2449) with 62% identity, and for the two flanking genes of RGP32 (PSPA7_2618 and PSPA7_2620), both with 61% identity respect to their PA14 orthologs. Importantly, none of these genes share the genomic location observed for RGP32 in the other five strains.
Fig. 8. fldP is a component of the P. aeruginosa variable genome. 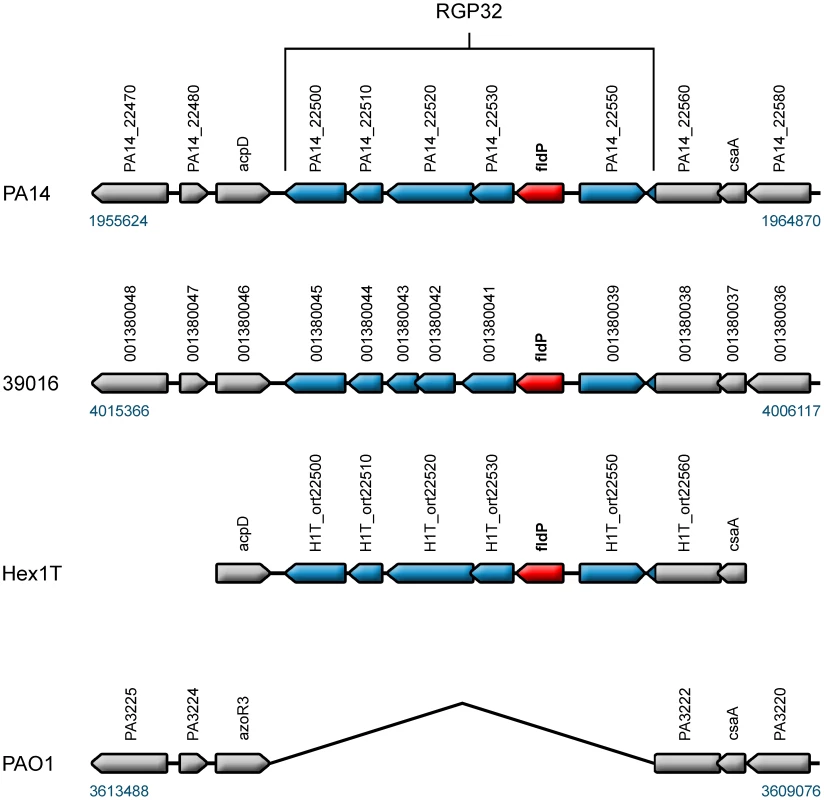
The fldP gene (in red) is located in a cluster of six genes which is referred to as RGP32 [33], here represented in blue in strains PA14, 39016 and Hex1T. The genomic context of RGP32 was outlined in gray in the three strains and also in the prototypic strain PAO1, which does not harbor RGP32. To further investigate the prevalence of fldP, we carried out PCR analysis using conserved primers (Table S2) on a collection of clinical and environmental isolates of P. aeruginosa, which were selected on the basis of being clonally different [36]. While highly divergent fldP versions could have been overlooked by this experimental approach, the high degree of sequence conservation of fldP observed among the characterized P. aeruginosa genomes makes this possibility rather improbable. The analysis showed that ∼23% of clones amplified a fragment corresponding to the fldP gene (Figure S3). Considering this low prevalence, the origin of RGP32 in P. aeruginosa is better explained as a consequence of DNA acquisition through one or more horizontal gene transfer processes rather than a genetic loss in those isolates which do not harbor RGP32.
fldP is the first gene of an operon regulated by a LysR-type transcriptional regulator
We subsequently analyzed the transcriptional organization of RGP32 in order to elucidate whether the fldP gene behaved as a single transcriptional unit or showed co-expression with other genes of the same variable region, thereby constituting an operon. We carried out PCRs using cDNA as template and primers designed to amplify fragments containing regions of two neighboring genes. We measured co-expression of those RGP32 genes which shared the same orientation and were located downstream from fldP (fldP to PA14_22500). As shown in Figure 9A, we were able to amplify fragments between fldP and PA14_22530, PA14_22530 and PA14_22520, and PA14_22520 and PA14_22510. In contrast, no PCR fragments could be amplified between PA14_22510 and PA14_22500. Importantly, amplicons were obtained among all adjacent genes, even between PA14_22510 and PA14_22500, when genomic DNA templates were used as positive controls (Figure 9A). These results depict a transcriptional organization of RGP32 structured in one polycistronic operon containing fldP, PA14_22530, PA14_22520 and PA14_22510, and two monocistronic transcriptional units for genes PA14_22500 and PA14_22550. To strengthen further these observations, we performed RT-PCRs to measure transcripts of PA14_22530 and PA14_22500 in cultures which were treated or not with H2O2. Interestingly, expression of PA14_22530 exhibited a 3-fold H2O2-dependent induction, similar to that previously observed for fldP, whereas transcripts of PA14_22500 showed only a small increase (Figure 9B). This result is in line with the transcriptional organization of RGP32 described above, and raises the question as to how this fldP-containing operon could be regulated. It has been previously observed that genomic islands often possess their own regulatory elements. In this sense, gene PA14_22550, which is predicted to encode a LysR family transcriptional regulator, appears as a promising candidate to fulfill this role. We investigated this possibility by using a PA14_22550 null mutant strain (ID53714) [28], and compared the expression of fldP and its response to H2O2 with those previously observed for the isogenic wt strain. Surprisingly, RT-PCR analyses showed that inactivation of PA14_22550 produced a 3.4-fold increase in the expression of fldP (P = 0.0285), even in untreated cells (Figure 6). In fact, treatment with H2O2 only raised this difference to 4.1-fold (P = 0.004). Thus, following the increase observed due to PA14_22550 inactivation, exposure to H2O2 did not produce any further rise of fldP expression (P = 0.4558). Then, the collected results indicate that fldP is part of a polycistronic operon which is repressed by the LysR-like transcriptional regulator present in the same RGP.
Fig. 9. Transcriptional organization of RGP32. 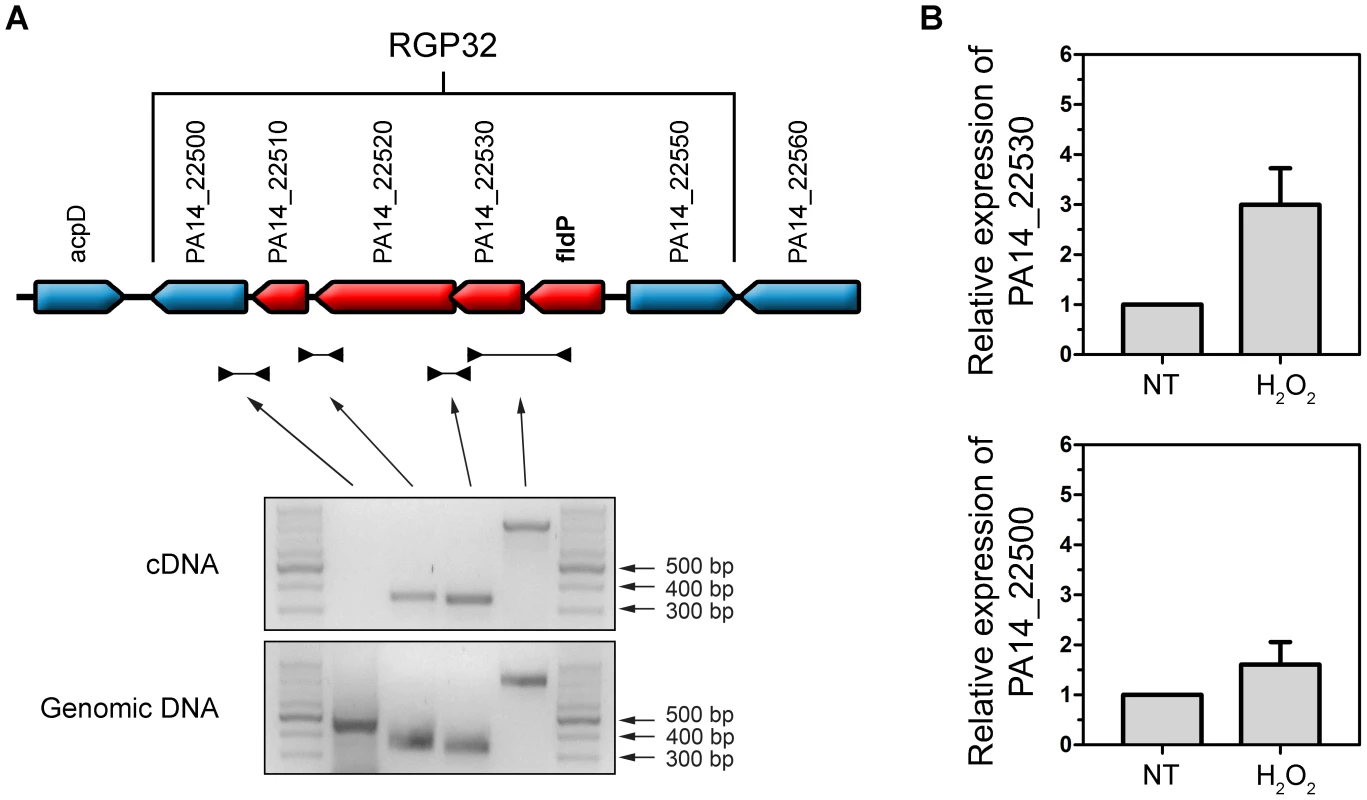
(A) The fldP gene is co-expressed with genes PA14_22530, PA14_22520 and PA14_22510 as a transcriptional unit, thereby constituting an operon (in red). Genes which are not co-transcribed with fldP are shown in blue. The gels below represent typical PCR amplifications obtained using cDNA or genomic DNA as templates. (B) Semi-quantitative RT-PCR was performed with specific primers for PA14_22530, which shows a H2O2-induced expression similar to that of fldP. This induction was not observed in PA14_22500, indicating that expression of this gene is independent from that of fldP. The rpoD transcripts were used as housekeeping controls. Bars represent relative expression levels averaged over two independent experiments (means ± SEM), with expression of the untreated (NT) wt strain normalized to 1. Discussion
Even under optimal growth conditions a low percentage of the electrons involved in cellular redox pathways are diverted to oxygen with concomitant ROS generation. This fraction can be dramatically increased under adverse environmental situations, leading to a condition known as oxidative stress. Moreover, cellular auto-oxidations are not the only source of ROS and oxidative stress; they can also be generated by external redox processes, such as phagocytes and other eukaryotic cells, which douse invading pathogens with H2O2 as a strategy to prevent infection (reviewed in [1], [2]).
A common strategy for coping with ROS damage and oxidative stress is the use of electron shuttles to relieve the excess of reducing equivalents and redox-active compounds. One of the most conspicuous among them is the electron carrier flavoprotein Fld, which has been consistently associated with stress protection in a number of organisms. The long-chain Flds from Anabaena and E. coli (IsiB and FldA, respectively) are encoded by oxidant-inducible genes, and confer increased tolerance to environmental and nutritional hardships [3], [4].
Despite the physiological importance of Fld as adaptive resource, little had been described about these flavoproteins in the versatile opportunistic human pathogen P. aeruginosa. In this work, we report the presence of a long-chain Fld (FldP) in P. aeruginosa and describe its role in the defense of the bacterial cell against oxidative conditions. FldP is encoded by the PA14_22540 gene as a 184-amino acid product sharing overall sequence similarity and common structural signatures with well-characterized flavodoxins such as FldA and IsiB (Figure 1). The fldP gene was cloned, expressed in E. coli and the resulting product purified to homogeneity, displaying the spectral properties and activity of a functional Fld (Figure 2).
We deemed it necessary to confirm that FldP was a functional flavodoxin because short-chain Flds have been already identified in both P. aeruginosa and P. putida [17],[18]. These genes have been annotated as mioC by comparison with their ortholog in E. coli. Pseudomonas MioC behaves as a functional Fld, mediating FNR-catalyzed electron transfer to cytochrome c with a kcat of about 6 min−1 [18], comparable to that displayed by FldP (22 min−1, Figure 2B), but its physiological role remains yet to be determined. Null mutants in mioC did not display growth phenotypes in E. coli [37], [38] or P. aeruginosa [17]. In the latter organism, however, mioC mutations exhibited other pleiotropic effects, including altered response to iron stress, increased production of the extracellular pigments pyocyanin and pyoverdine, and modified resistance to antibiotics [17]. In contrast to the enhanced sensitivity of fldP mutants toward H2O2 toxicity (Figure 3A), P. aeruginosa cells deficient in mioC were more tolerant than the wt to oxidative stress caused by methyl viologen or H2O2 [17].
The presence of different non-redundant Flds in a single organism is not uncommon. The E. coli genome, for instance, contains at least four genes predicted to encode flavodoxins: fldA, fldB, mioC, and yqcA [38]. E. coli Flds engage in different cellular pathways, and in general they cannot be functionally exchanged [38], [39]. Moreover, the fldA gene is specifically induced by H2O2 and redox-cycling oxidants [40], [41], whereas mioC is not [37]. Likewise, FldP and MioC, while displaying essentially the same activity in vitro, appear to play different roles in P. aeruginosa, with FldP contributing to the protection against oxidative challenges, and MioC in the response to iron stress [17].
A search of fldP orthologs in the Pseudomonas genomes available in the Pseudomonas Genome Project (http://www.pseudomonas.com) [34] and the environmental Hex1T strain (Feliziani et al., unpublished) revealed their presence in just six out of fourteen strains. The P. aeruginosa genome is made up of a conserved core component disrupted by numerous strain-specific RGPs, the sum of which conform the accessory and variable genome [31], [33]. The fldP gene is part of the region of genome plasticity RGP32, belonging to the accessory genome (Figure 8). Synteny and genome location are highly conserved in the PA14, PA39016, NCGM2.S1, B136-33 and Hex1T strains, with flanking genes being present in PAO1, suggesting that RGP32 is the result of a common insertion event. In this sense, we identified conserved palindromic inverted repeats flanking RGP32 in all these genomes, which provide circumstantial evidences on the origin and acquisition of this genome block.
While the conserved repertoire of the core genome codes for central functions required for survival and reproduction in any habitat [31], [42], the variable regions may play customized roles in adaptation to particular environments [33], [43]. This singular genome architecture, combining conserved and variable components, bestows P. aeruginosa pangenome upon its capability to handle a broad metabolic potential in order to adapt to the widest range of environmental niches. In this context, fldP, as part of the accessory genome, may provide specialized oxidative stress-related functions that benefit survival under stressful conditions, conferring RGP32 a role as adaptive island.
Indeed, several lines of evidence indicate that FldP is involved in protection against oxidative stress, including (i) the strong induction of the fldP gene in response to H2O2 (Figure 6), (ii) the enhanced ROS build-up and lower survival of fldP null mutants exposed to H2O2 (Figure 3), and (iii) the partial protection conferred by FldP overexpression to P. aeruginosa cells deficient in mutT against the deleterious effects (Figure 4) and increased mutational burden (Figure 5) caused by H2O2 treatment.
P. aeruginosa houses a multifaceted antioxidant response, comprising scavenging enzymes such as catalases and superoxide dismutases, thioredoxins and glutaredoxins, as well as small antioxidant molecules such as glutathione and melanine [44]. OxyR can be considered the master P. aeruginosa oxidative stress adaptive response [27]. Furthermore, recent studies have extended the implication of OxyR in other P. aeruginosa important responses, such as quorum sensing regulation, iron homeostasis and oxidative phosphorylation, also identifying a large number of target genes and revealing a far more complex cellular response than previously envisaged [45]. It was therefore tempting to speculate that fldP could be just another effector gene of this transcriptional regulator. Analysis of fldP expression in two oxyR loss-of-function mutants ruled out this possibility (Figure 6), indicating that induction of fldP in response to oxidative stress proceeds through an OxyR-independent pathway.
Inspection of the gene content and organization of RGP32 suggested another attractive possibility. The fldP gene is the first in a row of five ORFs displaying the same transcriptional orientation and extending up to the left end of RGP32 (Figure 8). This accessory genomic region contains still another ORF (PA14_22550), which is divergently transcribed and encodes a putative protein related to the LysR family of transcriptional regulators (Figure 8). LysR is the prototype for the most extended family of transcription factors in the bacterial world. Although originally described as activators of divergently transcribed genes, subsequent research placed LysR proteins as global transcriptional regulators, acting as either activators or repressors of single or operonic genes (reviewed in [46]). We found that four of the five divergent ORFs (including fldP) were co-transcribed in response to oxidative stress (Figure 9A), therefore constituting an operon. Moreover, inactivation of PA14_22550 led to full induction of fldP in the absence of oxidants, and to H2O2 insensitivity (Figure 6), indicating that the LysR homologue acts as a repressor of fldP and presumably the entire operon. The sixth ORF of RGP32, whose presumptive product is homologous to protein-disulfide isomerases (Table S1), is not part of the operon.
Stress-associated traits are often encoded by loci adjacent to those of other defensive products, especially when they are co-regulated [31], [42]. In RGP32, the ORF immediately downstream from fldP encodes a putative glutathione S-transferase (Table S1). Involvement of this superfamily of conjugative enzymes in the protection against oxidative stress has been extensively documented in all types of organisms (see, for instance [47]). On the other hand, nucleoside-disphosphate-sugar epimerases as that presumably encoded by PA14_22520 have been consistently associated with stress responses in plants [48], fungi [49] and bacteria [50]. Finally, PA14_22510 encodes a putative H protein from the glycine cleavage system (Table S1). These are lipoate-containing carrier proteins which provide reducing power (in the form of thiols) to the multi-enzymatic complex involved in glycine decarboxylation. The gene encoding this protein has been shown to be strongly up-regulated by H2O2 in streptococci [51], and lipoic acid is known to participate in the cellular response against oxidative stress in different organisms [52]. Then, all members of the operon have the potential to contribute to the defense against oxidative stress at different levels. We therefore propose that RGP32 represents a stress-inducible, self-regulated genetic element which confers increased tolerance to oxidative challenges. Under normal growth conditions, expression of RGP32 genes should be repressed by the LysR-type regulator encoded by PA14_22550. Oxidants somehow inactivate this transcription factor allowing induction of fldP and other components of the operon. The mechanism by which oxidants modulate the activity of the LysR-like protein of RGP32 is at present unknown and deserves further investigation.
The protective effect displayed in vitro by FldP against oxidative stress prompted us to evaluate whether this tolerance could be advantageous to P. aeruginosa cells exposed to the oxidative assault of cells belonging to the mammalian immune system. Indeed, wt and complemented P. aeruginosa strains expressing FldP from the chromosome or a plasmid exhibited better survival within infected macrophages relative to an isogenic mutant lacking this flavoprotein (Figure 7A). Cifani et al. [53] have recently shown that the oxidative burst produced by P. aeruginosa-infected macrophages plays a key role in the short-term killing of intracellular bacteria following invasion, strongly suggesting that the protective effect of FldP stems from its antioxidant function as it occurs in vitro.
We also used the insect model system of D. melanogaster (in which the PA14 strains has been shown to be particularly aggressive [29]), to further investigate the importance of this protective effect on a real infection process. In good agreement with the differential sensitivity observed in macrophages, fldP mutant bacteria accumulated to lower levels in D. melanogaster, as compared to the wt or the complemented strains (Figure 7B). Noteworthy, the mortality kinetics was delayed for ∼24 h in the mutant-infected flies, whereas the death rate of the flies infected with the fldP-deficient bacteria complemented with p2-fldP was equivalent to that of the wt strain (Figure 7C). Thus, the oxidative killing of P. aeruginosa within Drosophila hemolymph may involve mechanisms similar to those utilized by mammalian hosts. While the FldP effect suggests that the electron shuttle could be involved in P. aeruginosa virulence, it is more likely that the different death rates observed in Figure 7C are a consequence of longer persistence of FldP-containing bacteria in the fly due to the adaptive advantages conferred by the flavoprotein. In line with this proposal, mutation of the P. aeruginosa oxyR gene had similar effects on bacterial survival and Drosophila killing as those reported here [54].
Then, our data identify an oxidant-responsive long-chain flavodoxin in P. aeruginosa, which participates in the defense against ROS and contributes to the bacterial tolerance to the oxidative onslaught elicited by the host immune system. We anticipate that studies on this direction will lead to a more comprehensive panorama of the mechanisms allowing this opportunistic pathogen to adapt and persist in stressful and dynamic environments.
Materials and Methods
Bacterial strains, plasmids and growth conditions
P. aeruginosa PA14 and its isogenic strains ID38939 (PA14_22540 mutant, fldP), ID53714 (PA14_22550 mutant) and ID54029 (PA14_70560 mutant, oxyR) were kindly provided by Dr Eliana Drenkard and Dr Jonathan Urbach from the Massachusetts General Hospital, Boston, USA [28], whereas P. aeruginosa MPAO1 and its isogenic mutT strain were provided by Dr Michael Jacobs from the University of Washington Genome Center, USA [55]. Insertion of the MAR2xT7 mini-transposon in mutant ID38939 is unlikely to have polar effects on the expression of downstream genes since the consecutive aacC1 promoter is oriented in the same direction. To prepare inocula, bacteria were routinely cultured on Luria-Bertani (LB) agar plates from frozen stocks and subcultured overnight in LB liquid medium at 37°C with shaking at 220 r.p.m. Antibiotics were used at the following concentrations: 30 µg ml−1 gentamicin (Gm); 250 µg ml−1 kanamycin (Km).
Bioinformatics analysis
To find Fld homologs in P. aeruginosa PA14, the Anabaena IsiB and E. coli FldA sequences were compared against the entire PA14 genome using the DELTA-BLAST Search Tool [19]. Multiple DNA sequence alignments were performed by using ClustalW (http://www.clustal.org). Secondary structures were predicted using the Jpred3 software provided by the Dundee Scotland University (http://www.compbio.dundee.ac.uk/www-jpred).
Cloning of the fldP gene from P. aeruginosa PA14 and the isiB gene from Anabaena PCC7119
A DNA fragment containing the entire coding region of the fldP gene (PA14_22540) was amplified by PCR from PA14 genomic DNA using oligonucleotides FldP-F and FldP-R (Table S2), containing BamHI and HindIII restriction sites, respectively. The PCR product was ligated to the pGem-T Easy vector (Promega) and subsequently cloned into the broad-host-range plasmid pBBR1MCS2 (p2), which harbors a Km resistance marker [56], to generate p2-fldP. A similar strategy was employed to prepare p2-isiB containing the Fld-encoding gene from Anabaena PCC7119, originally cloned in pEMBL8-isiB [20], using oligonucleotides IsiB-F and IsiB-R as forward and reverse primers, respectively (Table S2). The resulting plasmids (p2-fldP and p2-isiB) and the empty p2 vector (as control), were introduced in the different P. aeruginosa strains via electroporation [57].
Construction of an oxyR deletion-mutant strain
Complete deletion of the oxyR gene was carried out as previously described [57]. All primer sequences are described in Table S2. Briefly, a first round of three PCR reactions was performed in which the 5′ and 3′ flanking regions of oxyR, as well as a Gm resistance cassette were amplified from plasmid pPS856 [58] using four gene-specific primers (Oxy-UpF-GWL, Oxy-UpR-Gm, Oxy-DnF-Gm and Oxy-DnR-GWR) and the common Gm-specific primers (Gm-F and Gm-R). This generated three fragments with partial overlaps either to each other or the attB1 and attB2 recombination sites. The purified fragments were then assembled in vitro by overlap extension during the second round PCR using the common primers GW-attB1 and GW-attB2. This resulted in an oxyR-deletion-mutant PCR fragment which was subsequently cloned into pDONR221 (Invitrogen) via the BP clonase reaction to create pDONR221-oxyR::Gm. This construct served as the substrate for LR clonase-mediated recombination into the destination vector pEX18ApGW. The resulting suicide vector pEX18ApGW-oxyR::Gm was then transferred to P. aeruginosa and the plasmid-borne oxyR-deletion mutation was exchanged with the chromosome via homologous recombination to generate the chromosomal deletion mutant.
Expression and purification of recombinant FldP and IsiB
For expression in E. coli, a 568-bp fragment encoding the complete sequence of the fldP gene was obtained by PCR amplification, using p2-fldP as template, and primers Rec-FldP-F and Rec-FldP-R, which contain restriction sites for NdeI and HindIII, respectively (Table S2). The amplified fragment was digested with the corresponding enzymes, cloned into compatible sites of pET-TEV (Novagen) under the control of the T7 promoter, and fused in-frame to an N-terminal His-tag. To improve solubility, BL21 E. coli cells were co-transformed with this vector and plasmid pG-Tf2 (Takara Bio Inc) expressing E. coli molecular chaperones (GroEL, GroES and Trigger Factor). After induction with 0.2 mM isopropyl-β-D-thiogalactoside (IPTG), the soluble flavoprotein was purified from cleared lysates in a Ni-NTA column by elution with 500 mM imidazole. Expression and purification of IsiB were carried out according to Fillat et al. [20]. UV-visible spectra of the purified recombinant proteins and FMN were recorded in 50 mM Tris-HCl pH 8.5.
Functional characterization of FldP
The ability of both electron shuttles, FldP and IsiB, to mediate the cytochrome c reductase activity of Anabaena FNR was assayed according to Shin [59]. The reaction mixture contained 3 mM glucose 6-phosphate, 0.3 mM NADP+ and 1 unit ml−1 glucose 6-phosphate dehydrogenase (G6PDH, to generate NADPH), 0.5 µM FNR, 50 µM equine heart cytochrome c and various amounts of Fld in 50 mM Tris-HCl pH 8.5. Cytochrome c reduction was followed at 30°C by the increase in absorbance at 550 nm (ε550 = 19 mM−1 cm−1).
ROS detection
A modified version of the FOX II assay [60] was used to quantify the presence of peroxides in bacterial extracts. Cultures of the parental PA14 and the fldP mutant strains transformed with either p2 or p2-fldP were grown aerobically in LB broth at 37°C for 5 h with the appropriate antibiotics. Then, H2O2 was added to final concentrations of 0, 25 and 50 mM, and bacterial suspensions were incubated for 30 min with vigorous shaking. Cultures were split into two equal portions; one of them was used to measure protein concentration, while the other was centrifuged, washed with 0.9% (w/v) NaCl and finally resuspended in 1 ml of an 80∶20 ethanol/water solution containing 0.01% (w/v) butylated hydroxytoluene (BHT). Samples were disrupted by sonic oscillation (10 times for 10 sec each, 30% amplitude), centrifuged at 10,000 g for 10 min, and 250 µl of the supernatants were combined with 250 µl of 10 mM Tris-phenyl phosphine in methanol (TPP, a -OOH reducing agent), or with 250 µl of methanol, to measure total oxidants. Mixtures were incubated for 30 min to allow complete -OOH reduction by TPP. Five hundred µl of FOX reagent (100 µM xylenol orange, 4 mM BHT, 250 µM ferrous ammonium sulphate and 25 mM H2SO4 in 90% (v/v) methanol) were then added to each sample, and the absorbance at 560 nm was recorded 10 min after reagent addition. The absorbance differences between equivalent samples with and without TPP indicate the amounts of -OOH, which were calculated using a 0–20 µM H2O2 standard curve. Protein concentrations were estimated in cleared lysates in 50 mM Tris-HCl pH 8.0 by a dye binding method [61], using bovine serum albumin as standard.
For observation at the confocal microscope, DCFDA was introduced into P. aeruginosa cells by electroporation. Briefly, 10-ml overnight cultures of the various strains were collected by centrifugation at 12,000 g for 2 min, washed three times with 1 ml of 0.3 M sucrose and finally resuspended in 100 µl of the same solution. DCFDA (in dimethyl sulfoxide) was added to a final concentration of 500 µM and the suspension transferred to electroporation cuvettes of 1-mm width. Cells were electroporated in an Electro Cell Manipulator 600, BTX electroporation System at 25 µF, 2.5 kV and 200 Ω for 5 ms, diluted with 300 µl of 0.3 M sucrose and divided into two equal fractions. One of them was incubated with 25 mM H2O2 at 37°C for 15 min and the other kept under the same conditions without the oxidant. Seven µl of the suspensions were mixed with 3 µl of the membrane-staining dye FM 4-64 (0.1 mg ml−1) on the surface of a glass plate and the lid was sealed with colorless nail paint. Cells were observed at excitation/emission maxima of ∼515/640 nm in a Nikon TE-2000-E2 confocal microscope. The number of fluorescent spots was determined for each dye by using the Image J 1.46 software.
H2O2 susceptibility measurements
For H2O2 susceptibility assays, cells were grown to mid-exponential phase (OD600∼0.8), collected by centrifugation and washed with 0.9% (w/v) NaCl solution. Then, bacteria were normalized to ∼108 cells and treated with 50 mM H2O2 for 30 min at 37°C. Cells were centrifuged again, washed twice with 0.9% (w/v) NaCl and finally resuspended in fresh LB medium. Serial dilutions were spotted on LB agar plates and incubated overnight at 37°C to determine viability. Non-treated controls were included. Bacterial susceptibility was expressed as the ratio between the survivals of treated to untreated cells.
Estimation of spontaneous and H2O2-induced mutant frequencies
For estimation of spontaneous-mutation frequencies, five bacterial colonies of the different P. aeruginosa strains were cultured overnight in 10 ml LB medium at 37°C with shaking at 220 r.p.m. Appropriate dilutions of the cultures were plated on LB agar to determine the total number of viable cells, or on LB agar supplemented with 500 µg ml−1 streptomycin to count the number of streptomycin-resistant cells, after overnight incubation at 37°C. Spontaneous-mutation frequency was determined as the ratio between the number of streptomycin–resistant cells and the number of viable cells.
Determination of H2O2-induced mutant frequency was carried out according to previously described protocols [62], with some modifications. Briefly, five independent bacterial cultures for each strain were grown in LB to mid-exponential phase, collected by centrifugation and washed with 0.9% (w/v) NaCl. Cells were normalized to ∼108 and subsequently treated with 50 mM H2O2 for 30 min at 37°C, centrifuged again and washed twice with 1 ml of 0.9% (w/v) NaCl. An aliquot of treated cells (0.5 ml) was diluted 10-fold in fresh LB broth and cultured overnight at 37°C with shaking at 220 r.p.m. Then, 100 µl of each overnight culture were plated onto LB agar supplemented with 500 µg ml−1 streptomycin, whereas aliquots from appropriate dilutions were plated on LB agar without antibiotic to measure cell viability. The H2O2-induced mutation frequency was calculated as above.
Semi-quantitative RT-PCR
P. aeruginosa PA14 mid-exponential phase cultures (OD600∼0.8) were treated with 50 mM H2O2 for 15 min, centrifuged and subsequently washed twice with 0.9% (w/v) NaCl. Treated and untreated cultures were used to extract total RNA using the RNA Purification Kit (Fermentas). RNA was quantified by UV spectrophotometry, and its integrity was checked by electrophoresis in 1.5% (w/v) agarose gels. Then, 1 µg of total RNA was reverse-transcribed using the QuantiTect Reverse Transcription Kit (QIAGEN). PCR primers were manually designed with the assistance of the Netprimer software (PREMIER Biosoft International, Palo Alto, CA) and evaluated for their specificity with the BLAST program at the NCBI Web site. Specific transcripts were semi-quantitatively measured by RT-PCR using primers FldP-RT-F and FldP-RT-R (for fldP), 22530-RT-F and 22530-RT-R (for PA14_22530), and 22500-RT-F and 22500-RT-R (for PA14_22500). Transcripts of the rpoD gene were amplified with primers RpoD-RT-F and RpoD-RT-R and served as housekeeping controls. All primer sequences are described in Table S2. The optimal number of cycles was determined in advance to evaluate expression in the exponential phase of amplification. Final cycling conditions included a hot start at 95°C for 10 min, followed by 32 cycles at 94°C for 30 sec, 60°C for 30 sec and 72°C for 30 sec, and a final extension at 72°C for 5 min. Specificity was verified by agarose gel electrophoresis. Fold change in gene expression was calculated by measuring band intensities with the Gel-Pro Analyzer Software. No amplification was observed in PCR reactions containing water or non-reverse transcribed RNA as template.
Characterization of RGP32 transcriptional organization
Primers were designed in order to amplify PCR products containing regions of two neighboring genes for those genes of RGP32 which share the same orientation and are located downstream from fldP (Figure 9A). The cDNA to be used as a template for PCR was obtained by reverse transcription of purified total RNA as described above. Thus, primers FldP-F and FldP-30-R were employed to determine the co-expression of genes fldP and PA14_22530 (Table S2). In the same way, co-expression of downstream contiguous genes was analyzed with the following primers: a) 30-20-F and 30-20-R to analyze PA14_22530 and PA14_22520 co-expression; b) 20-10-F and 20-10-R for PA14_22520 and PA14_22510; and c) 10-00-F and 10-00-R for PA14_22510 and PA14_22500 (Table S2). Genomic DNA was used as template for positive controls of the PCR reactions. No amplification was observed in PCR reactions containing water or non-reverse transcribed RNA as template.
Intracelullar survival in macrophagic cells
Cells of the macrophagic-derived line RAW 264.7 were grown on 12-mm-diameter wells until they reached 95–100% confluence. To prepare inocula, overnight cultures of P. aeruginosa were washed with phosphate buffered saline (PBS) and suspended in Dulbecco's Modified Eagle's Medium (DMEM) at a final concentration of 107 cells ml−1. Bacterial antibiotic protection assays were conducted as previously described with few modifications [63]. Briefly, macrophagic cells were inoculated with the various P. aeruginosa strains by the addition of 500 µl of the bacterial suspension, corresponding to a multiplicity of infection (MOI) of about 20, followed by centrifugation for 10 min at 1000 r.p.m. to facilitate cell contact. After a 2-h incubation period at 37°C, the supernatant was removed and the cells were washed three times with PBS to remove non-associated bacteria. To count intracellular P. aeruginosa, fresh DMEM with streptomycin (1 mg ml−1) and carbenicillin (400 µg ml−1) was added to the cell monolayer and incubated for 2 h to kill extracellular bacteria. Antibiotic bactericidal activity was confirmed by plating 100-µl aliquots from the wells directly on LB-agar. Cells were again washed to remove antibiotics, then lysed by incubation with 0.1% (v/v) Triton X-100 in PBS. At this initial stage (Ti) intracellular bacteria were enumerated by plating serial dilutions of cell lysates on LB agar and counting CFU. To estimate P. aeruginosa intracellular survival, an equivalent set of wells were incubated with antibiotics for an additional 3-h period (Tf), cells were lysed and surviving bacteria enumerated as described above. Intracellular survival was calculated as the ratio between the final and the initial (Tf/Ti) CFU counts. Three wells of cells were used for each strain in each experiment, and all experiments were repeated three times. To rule out the pre-existence of resistant bacteria in the overnight cultures, 10× inocula (5×107 cells) were plated on LB agar containing streptomycin (1 mg ml−1) and carbenicillin (400 µg ml−1), with not a single CFU observed even after 72 h of incubation.
Phagocytes viability was monitored at every time point of the experiment by measuring release of LDH (CellTiter 96 Aqueous Non-Radioactive Cell Proliferation Assay, Promega), which showed an initial decrease of ∼50% in the first 2 h, but remained unaltered after the antibiotic treatment.
Bacterial survival in Drosophila melanogaster
P. aeruginosa cells of the various strains were grown overnight, washed and diluted in PBS, 10% (w/v) sucrose, 250 µg ml−1 Km to OD600 = 0.025. D. melanogaster strain W1118 flies (4–5 days old) were starved for 3 h and then fed continuously at 25°C on cottons plugs, which had been previously embedded in the bacterial solution. For each time point, 3 groups of 5 flies were pestle homogenized in 200 µl of PBS and the homogenate was serially diluted in PBS and plated on LB agar containing 250 µg ml−1 Km, to quantify the number of bacterial colonies.
Fly survival experiments
Overnight cultures of each P. aeruginosa strain were diluted in fresh LB to an OD600 = 0.25. This solution was then diluted 10-fold in PBS, 10% (w/v) sucrose, 250 µg ml−1 Km. Groups of 15 D. melanogaster W1118 male flies were starved during 3 h and then placed in vials with sterile cotton plugs, which had been previously embedded in 3 ml of the bacterial suspension. Flies were kept at 25°C and survival was monitored daily. Three groups of 15 flies were used for each condition in three independent experiments.
Statistical analysis
Statistical analyses were performed using one-tailed Mann-Whitney test appropriate for nonparametric adjustment. When appropriate, one - or two-tailed Student's t test was applied. In all cases, P values less than or equal to 0.05 were considered statistically significant.
Ethics statements
The collection of P. aeruginosa clinical isolates used in this work has already been published by Feliziani et al. [36]. In this previous study, the informed consent as well as the approval from an Institutional Research Committee were appropriately evaluated and fulfilled the PLOS ethical standards.
Supporting Information
Zdroje
1. ImlayJA (2008) Cellular defenses against superoxide and hydrogen peroxide. Annu Rev Biochem 77 : 755–776.
2. MishraS, ImlayJ (2012) Why do bacteria use so many enzymes to scavenge hydrogen peroxide? Arch Biochem Biophys 525 : 145–160.
3. ZhengM, DoanB, SchneiderTD, StorzG (1999) OxyR and SoxRS regulation of fur. J Bacteriol 181 : 4639–4643.
4. FuldaS, HagemannM (1995) Salt treatment induces accumulation of flavodoxin in the cyanobacterium Synechocystis sp. PCC6803. J Plant Physiol 146 : 520–526.
5. MazouniK, DomainF, ChauvatF, Cassier-ChauvatC (2003) Expression and regulation of the crucial plant-like ferredoxin of cyanobacteria. Mol Microbiol 49 : 1019–1029.
6. SinghAK, LiH, ShermanLA (2004) Microarray analysis and redox control of gene expression in the cyanobacterium Synechocystis sp. PCC 6803. Physiol Plant 120 : 27–35.
7. KrappAR, HumbertMV, CarrilloN (2011) The soxRS response of Escherichia coli can be induced in the absence of oxidative stress and oxygen by modulation of NADPH content. Microbiology 157 : 957–965.
8. RedondoFJ, de la PenaTC, MorcilloCN, LucasMM, PueyoJJ (2009) Overexpression of flavodoxin in bacteroids induces changes in antioxidant metabolism leading to delayed senescence and starch accumulation in alfalfa root nodules. Plant Physiol 149 : 1166–1178.
9. TognettiVB, PalatnikJF, FillatMF, MelzerM, HajirezaeiMR, et al. (2006) Functional replacement of ferredoxin by a cyanobacterial flavodoxin in tobacco confers broad-range stress tolerance. Plant Cell 18 : 2035–2050.
10. LodeyroAF, CeccoliRD, Pierella KarlusichJJ, CarrilloN (2012) The importance of flavodoxin for environmental stress tolerance in photosynthetic microorganisms and transgenic plants. Mechanism, evolution and biotechnological potential. FEBS Lett 586 : 2917–2924.
11. López-LlanoJ, MaldonadoS, JainS, LostaoA, Godoy-RuizR, et al. (2004) The long and short flavodoxins: II. The role of the differentiating loop in apoflavodoxin stability and folding mechanism. J Biol Chem 279 : 47184–47191.
12. SanchoJ (2006) Flavodoxins: sequence, folding, binding, function and beyond. Cell Mol Life Sci 63 : 855–864.
13. StoverCK, PhamXQ, ErwinAL, MizoguchiSD, WarrenerP, et al. (2000) Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 406 : 959–964.
14. BodeyGP, BolivarR, FainsteinV, JadejaL (1983) Infections caused by Pseudomonas aeruginosa. Rev Infect Dis 5 : 279–313.
15. MorrisonAJJr, WenzelRP (1984) Epidemiology of infections due to Pseudomonas aeruginosa. Rev Infect Dis 6 Suppl 3: S627–642.
16. LyczakJB, CannonCL, PierGB (2002) Lung infections associated with cystic fibrosis. Clin Microbiol Rev 15 : 194–222.
17. YeomJ, ParkW (2012) Pleiotropic effects of the mioC mutation on the physiology of Pseudomonas aeruginosa PAO1. FEMS Microbiol Lett 335 : 47–57.
18. YeomJ, ParkW (2012) Biochemical characterization of ferredoxin-NADP(+) reductase interaction with flavodoxin in Pseudomonas putida. BMB Rep 45 : 476–481.
19. BoratynGM, SchafferAA, AgarwalaR, AltschulSF, LipmanDJ, et al. (2012) Domain enhanced lookup time accelerated BLAST. Biol Direct 7 : 12.
20. FillatMF, BorriasWE, WeisbeekPJ (1991) Isolation and overexpression in Escherichia coli of the flavodoxin gene from Anabaena PCC 7119. Biochem J 280 (Pt 1) 187–191.
21. PellettJD, BeckerDF, SaengerAK, FuchsJA, StankovichMT (2001) Role of aromatic stacking interactions in the modulation of the two-electron reduction potentials of flavin and substrate/product in Megasphaera elsdenii short-chain acyl-coenzyme A dehydrogenase. Biochemistry 40 : 7720–7728.
22. MoreroNR, ArgarañaCE (2009) Pseudomonas aeruginosa deficient in 8-oxodeoxyguanine repair system shows a high frequency of resistance to ciprofloxacin. FEMS Microbiol Lett 290 : 217–226.
23. SandersLH, SudhakaranJ, SuttonMD (2009) The GO system prevents ROS-induced mutagenesis and killing in Pseudomonas aeruginosa. FEMS Microbiol Lett 294 : 89–96.
24. CookeMS, EvansMD, DizdarogluM, LunecJ (2003) Oxidative DNA damage: mechanisms, mutation, and disease. FASEB J 17 : 1195–1214.
25. MoyanoAJ, LujánAM, ArgarañaCE, SmaniaAM (2007) MutS deficiency and activity of the error-prone DNA polymerase IV are crucial for determining mucA as the main target for mucoid conversion in Pseudomonas aeruginosa. Mol Microbiol 64 : 547–559.
26. MatheeK, CiofuO, SternbergC, LindumPW, CampbellJI, et al. (1999) Mucoid conversion of Pseudomonas aeruginosa by hydrogen peroxide: a mechanism for virulence activation in the cystic fibrosis lung. Microbiology 145 (Pt 6) 1349–1357.
27. OchsnerUA, VasilML, AlsabbaghE, ParvatiyarK, HassettDJ (2000) Role of the Pseudomonas aeruginosa oxyR-recG operon in oxidative stress defense and DNA repair: OxyR-dependent regulation of katB-ankB, ahpB, and ahpC-ahpF. J Bacteriol 182 : 4533–4544.
28. LiberatiNT, UrbachJM, MiyataS, LeeDG, DrenkardE, et al. (2006) An ordered, nonredundant library of Pseudomonas aeruginosa strain PA14 transposon insertion mutants. Proc Natl Acad Sci U S A 103 : 2833–2838.
29. LutterEI, FariaMM, RabinHR, StoreyDG (2008) Pseudomonas aeruginosa cystic fibrosis isolates from individual patients demonstrate a range of levels of lethality in two Drosophila melanogaster infection models. Infect Immun 76 : 1877–1888.
30. Limmer S, Haller S, Drenkard E, Lee J, Yu S, et al. Pseudomonas aeruginosa RhlR is required to neutralize the cellular immune response in a Drosophila melanogaster oral infection model. Proc Natl Acad Sci U S A 108 : 17378–17383.
31. KiewitzC, TümmlerB (2000) Sequence diversity of Pseudomonas aeruginosa: impact on population structure and genome evolution. J Bacteriol 182 : 3125–3135.
32. SpencerDH, KasA, SmithEE, RaymondCK, SimsEH, et al. (2003) Whole-genome sequence variation among multiple isolates of Pseudomonas aeruginosa. J Bacteriol 185 : 1316–1325.
33. MatheeK, NarasimhanG, ValdesC, QiuX, MatewishJM, et al. (2008) Dynamics of Pseudomonas aeruginosa genome evolution. Proc Natl Acad Sci U S A 105 : 3100–3105.
34. WinsorGL, LamDK, FlemingL, LoR, WhitesideMD, et al. (2011) Pseudomonas Genome Database: improved comparative analysis and population genomics capability for Pseudomonas genomes. Nucleic Acids Res 39: D596–600.
35. SmaniaAM, SeguraI, PezzaRJ, BecerraC, AlbesaI, et al. (2004) Emergence of phenotypic variants upon mismatch repair disruption in Pseudomonas aeruginosa. Microbiology 150 : 1327–1338.
36. FelizianiS, LujánAM, MoyanoAJ, SolaC, BoccoJL, et al. (2010) Mucoidy, quorum sensing, mismatch repair and antibiotic resistance in Pseudomonas aeruginosa from cystic fibrosis chronic airways infections. PLOS One 5: e12669.
37. Løbner-OlesenA, BoyeE (1992) Different effects of mioC transcription on initiation of chromosomal and minichromosomal replication in Escherichia coli. Nucleic Acids Research 20 : 3029–3036.
38. BirchOM, HewitsonKS, FuhrmannM, BurgdorfK, BaldwinJE, et al. (2000) MioC is an FMN-binding protein that is essential for Escherichia coli biotin synthase activity in vitro. J Biol Chem 275 : 32277–32280.
39. PuanKJ, WangH, DairiT, KuzuyamaT, MoritaCT (2005) fldA is an essential gene required in the 2-C-methyl-D-erythritol 4-phosphate pathway for isoprenoid biosynthesis. FEBS Lett 579 : 3802–3806.
40. PomposielloPJ, BennikMH, DempleB (2001) Genome-wide transcriptional profiling of the Escherichia coli responses to superoxide stress and sodium salicylate. J Bacteriol 183 : 3890–3902.
41. ZhengM, WangX, TempletonLJ, SmulskiDR, LaRossaRA, et al. (2001) DNA microarray-mediated transcriptional profiling of the Escherichia coli response to hydrogen peroxide. J Bacteriol 183 : 4562–4570.
42. SchmidtKD, TümmlerB, RömlingU (1996) Comparative genome mapping of Pseudomonas aeruginosa PAO with P. aeruginosa C, which belongs to a major clone in cystic fibrosis patients and aquatic habitats. J Bacteriol 178 : 85–93.
43. SilbyMW, WinstanleyC, GodfreySA, LevySB, JacksonRW (2011) Pseudomonas genomes: diverse and adaptable. FEMS Microbiol Rev 35 : 652–680.
44. HassettDJ, CohenMS (1989) Bacterial adaptation to oxidative stress: implications for pathogenesis and interaction with phagocytic cells. FASEB J 3 : 2574–2582.
45. WeiQ, MinhPN, DotschA, HildebrandF, PanmaneeW, et al. (2012) Global regulation of gene expression by OxyR in an important human opportunistic pathogen. Nucleic Acids Res 40 : 4320–4333.
46. MaddocksSE, OystonPC (2008) Structure and function of the LysR-type transcriptional regulator (LTTR) family proteins. Microbiology 154 : 3609–3623.
47. HayesJD, FlanaganJU, JowseyIR (2005) Glutathione transferases. Annu Rev Pharmacol Toxicol 45 : 51–88.
48. RostiJ, BartonCJ, AlbrechtS, DupreeP, PaulyM, et al. (2007) UDP-glucose 4-epimerase isoforms UGE2 and UGE4 cooperate in providing UDP-galactose for cell wall biosynthesis and growth of Arabidopsis thaliana. Plant Cell 19 : 1565–1579.
49. CresnarB, PlaperA, BreskvarK, Hudnik-PlevnikT (1998) cDNA sequence and deduced amino acid sequence of a fungal stress protein induced in Rhizopus nigricans by steroids. Biochem Biophys Res Commun 250 : 664–667.
50. FanousA, HeckerM, GorgA, ParlarH, JacobF (2010) Corynebacterium glutamicum as an indicator for environmental cobalt and silver stress–a proteome analysis. J Environ Sci Health B 45 : 666–675.
51. GrifantiniR, ToukokiC, ColapricoA, GryllosI (2011) Peroxide stimulon and role of PerR in group A Streptococcus. J Bacteriol 193 : 6539–6551.
52. BrykR, LimaCD, Erdjument-BromageH, TempstP, NathanC (2002) Metabolic enzymes of mycobacteria linked to antioxidant defense by a thioredoxin-like protein. Science 295 : 1073–1077.
53. CifaniN, PompiliB, AnileM, PatellaM, DisoD, et al. (2013) Reactive-oxygen-species-mediated P. aeruginosa killing is functional in human cystic fibrosis macrophages. PLOS One 8: e71717.
54. LauGW, BritiganBE, HassettDJ (2005) Pseudomonas aeruginosa OxyR is required for full virulence in rodent and insect models of infection and for resistance to human neutrophils. Infect Immun 73 : 2550–2553.
55. JacobsMA, AlwoodA, ThaipisuttikulI, SpencerD, HaugenE, et al. (2003) Comprehensive transposon mutant library of Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 100 : 14339–14344.
56. KovachME, ElzerPH, HillDS, RobertsonGT, FarrisMA, et al. (1995) Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166 : 175–176.
57. ChoiKH, SchweizerHP (2006) mini-Tn7 insertion in bacteria with single attTn7 sites: example Pseudomonas aeruginosa. Nat Protoc 1 : 153–161.
58. HoangTT, Karkhoff-SchweizerRR, KutchmaAJ, SchweizerHP (1998) A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212 : 77–86.
59. ShinM (1971) Ferredoxin-NADP reductase from spinach. Methods Enzymol 23 : 440–447.
60. DeLongJM, PrangeRK, HodgesDM, ForneyCF, BishopMC, et al. (2002) Using a modified ferrous oxidation-xylenol orange (FOX) assay for detection of lipid hydroperoxides in plant tissue. J Agric Food Chem 50 : 248–254.
61. SedmakJJ, GrossbergSE (1977) A rapid, sensitive, and versatile assay for protein using Coomassie brilliant blue G250. Anal Biochem 79 : 544–552.
62. Rodríguez-RojasA, BlázquezJ (2009) The Pseudomonas aeruginosa pfpI gene plays an antimutator role and provides general stress protection. J Bacteriol 191 : 844–850.
63. FleiszigSM, Wiener-KronishJP, MiyazakiH, VallasV, MostovKE, et al. (1997) Pseudomonas aeruginosa-mediated cytotoxicity and invasion correlate with distinct genotypes at the loci encoding exoenzyme S. Infect Immun 65 : 579–586.
Štítky
Genetika Reprodukční medicína
Článek Natural Polymorphisms in Influence Negative Selection and CD4∶CD8 Lineage Commitment in the RatČlánek MicroRNAs Located in the Hox Gene Clusters Are Implicated in Huntington's Disease PathogenesisČlánek Comparative RNAi Screens in and Reveal the Impact of Developmental System Drift on Gene FunctionČlánek Mutation of SLC35D3 Causes Metabolic Syndrome by Impairing Dopamine Signaling in Striatal D1 Neurons
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2014 Číslo 2
-
Všechny články tohoto čísla
- Fifteen Years Later: Hard and Soft Selection Sweeps Confirm a Large Population Number for HIV In Vivo
- The Same but Different: Worms Reveal the Pervasiveness of Developmental System Drift
- Serine Carboxypeptidase SCPEP1 and Cathepsin A Play Complementary Roles in Regulation of Vasoconstriction via Inactivation of Endothelin-1
- Coherent Functional Modules Improve Transcription Factor Target Identification, Cooperativity Prediction, and Disease Association
- A Long-Chain Flavodoxin Protects from Oxidative Stress and Host Bacterial Clearance
- Mammalian E-type Cyclins Control Chromosome Pairing, Telomere Stability and CDK2 Localization in Male Meiosis
- Influenza Virus Drug Resistance: A Time-Sampled Population Genetics Perspective
- Transcriptome-Wide Analyses of 5′-Ends in RNase J Mutants of a Gram-Positive Pathogen Reveal a Role in RNA Maturation, Regulation and Degradation
- Selective Disruption of Aurora C Kinase Reveals Distinct Functions from Aurora B Kinase during Meiosis in Mouse Oocytes
- X Chromosome Control of Meiotic Chromosome Synapsis in Mouse Inter-Subspecific Hybrids
- A Cohesin-Independent Role for NIPBL at Promoters Provides Insights in CdLS
- Extreme Population Differences in the Human Zinc Transporter ZIP4 (SLC39A4) Are Explained by Positive Selection in Sub-Saharan Africa
- Classic Selective Sweeps Revealed by Massive Sequencing in Cattle
- Genomic Networks of Hybrid Sterility
- Natural Polymorphisms in Influence Negative Selection and CD4∶CD8 Lineage Commitment in the Rat
- Oxidative Stress Is Not a Major Contributor to Somatic Mitochondrial DNA Mutations
- Molecular Identification of Collagen 17a1 as a Major Genetic Modifier of Laminin Gamma 2 Mutation-Induced Junctional Epidermolysis Bullosa in Mice
- Uncoupling of Molecular Maturation from Peripheral Target Innervation in Nociceptors Expressing a Chimeric TrkA/TrkC Receptor
- MicroRNAs Located in the Hox Gene Clusters Are Implicated in Huntington's Disease Pathogenesis
- Loss of Trabid, a New Negative Regulator of the Immune-Deficiency Pathway at the Level of TAK1, Reduces Life Span
- Targeted Ablation of Nesprin 1 and Nesprin 2 from Murine Myocardium Results in Cardiomyopathy, Altered Nuclear Morphology and Inhibition of the Biomechanical Gene Response
- Identification of Novel Genetic Loci Associated with Thyroid Peroxidase Antibodies and Clinical Thyroid Disease
- CEP-1, the p53 Homolog, Mediates Opposing Longevity Outcomes in Mitochondrial Electron Transport Chain Mutants
- Transcriptomics and Functional Genomics of ROS-Induced Cell Death Regulation by
- Quantitative Genome-Wide Genetic Interaction Screens Reveal Global Epistatic Relationships of Protein Complexes in
- Cascades of Genetic Instability Resulting from Compromised Break-Induced Replication
- Serine- and Threonine/Valine-Dependent Activation of PDK and Tor Orthologs Converge on Sch9 to Promote Aging
- Zfp322a Regulates Mouse ES Cell Pluripotency and Enhances Reprogramming Efficiency
- Insertional Mutagenesis and Deep Profiling Reveals Gene Hierarchies and a -Dependent Bottleneck in Lymphomagenesis
- DAAM Is Required for Thin Filament Formation and Sarcomerogenesis during Muscle Development in Drosophila
- Plasma Cholesterol–Induced Lesion Networks Activated before Regression of Early, Mature, and Advanced Atherosclerosis
- High-Resolution Profiling of Stationary-Phase Survival Reveals Yeast Longevity Factors and Their Genetic Interactions
- Comparative RNAi Screens in and Reveal the Impact of Developmental System Drift on Gene Function
- Accurate and Robust Genomic Prediction of Celiac Disease Using Statistical Learning
- Sex-Specific Embryonic Gene Expression in Species with Newly Evolved Sex Chromosomes
- Chromosome X-Wide Association Study Identifies Loci for Fasting Insulin and Height and Evidence for Incomplete Dosage Compensation
- Negative Feedback and Transcriptional Overshooting in a Regulatory Network for Horizontal Gene Transfer
- DNA Sequence Explains Seemingly Disordered Methylation Levels in Partially Methylated Domains of Mammalian Genomes
- Insights into the Genomic Landscape: Comparative Genomics Reveals Variations in Ploidy and Nutrient Utilisation Potential amongst Wine Isolates
- Molecular Evidence for the Inverse Comorbidity between Central Nervous System Disorders and Cancers Detected by Transcriptomic Meta-analyses
- The Centriolar Satellite Protein AZI1 Interacts with BBS4 and Regulates Ciliary Trafficking of the BBSome
- Fine-Mapping the Region Detects Common Variants Tagging a Rare Coding Allele: Evidence for Synthetic Association in Prostate Cancer
- Transmission Distortion Affecting Human Noncrossover but Not Crossover Recombination: A Hidden Source of Meiotic Drive
- A Variant in the Neuropeptide Receptor is a Major Determinant of Growth and Physiology
- Mutation of SLC35D3 Causes Metabolic Syndrome by Impairing Dopamine Signaling in Striatal D1 Neurons
- NSUN4 Is a Dual Function Mitochondrial Protein Required for Both Methylation of 12S rRNA and Coordination of Mitoribosomal Assembly
- MicroRNA-133 Inhibits Behavioral Aggregation by Controlling Dopamine Synthesis in Locusts
- Convergence of Light and ABA Signaling on the Promoter
- Arf4 Is Required for Mammalian Development but Dispensable for Ciliary Assembly
- Distinct Requirements for Cranial Ectoderm and Mesenchyme-Derived Wnts in Specification and Differentiation of Osteoblast and Dermal Progenitors
- Chk2 and P53 Regulate the Transmission of Healed Chromosomes in the Male Germline
- Ddc2 Mediates Mec1 Activation through a Ddc1- or Dpb11-Independent Mechanism
- Mapping the Fitness Landscape of Gene Expression Uncovers the Cause of Antagonism and Sign Epistasis between Adaptive Mutations
- Euchromatic Transposon Insertions Trigger Production of Novel Pi- and Endo-siRNAs at the Target Sites in the Germline
- miR-100 Induces Epithelial-Mesenchymal Transition but Suppresses Tumorigenesis, Migration and Invasion
- Canine Hereditary Ataxia in Old English Sheepdogs and Gordon Setters Is Associated with a Defect in the Autophagy Gene Encoding
- Within-Host Spatiotemporal Dynamics of Plant Virus Infection at the Cellular Level
- Analysis of Meiosis in SUN1 Deficient Mice Reveals a Distinct Role of SUN2 in Mammalian Meiotic LINC Complex Formation and Function
- Genome-Wide Association Study of Metabolic Traits Reveals Novel Gene-Metabolite-Disease Links
- Mechanistically Distinct Mouse Models for -Associated Retinopathy
- DAF-16/FoxO Directly Regulates an Atypical AMP-Activated Protein Kinase Gamma Isoform to Mediate the Effects of Insulin/IGF-1 Signaling on Aging in
- Chromosome I Controls Chromosome II Replication in
- Integrated Genomic Characterization Reveals Novel, Therapeutically Relevant Drug Targets in FGFR and EGFR Pathways in Sporadic Intrahepatic Cholangiocarcinoma
- The Iodotyrosine Deiodinase Ortholog SUP-18 Functions through a Conserved Channel SC-Box to Regulate the Muscle Two-Pore Domain Potassium Channel SUP-9
- The Genome of Highlights a Fish Pathogen Adapted to Fluctuating Environments
- Distinct DNA Binding Sites Contribute to the TCF Transcriptional Switch in and
- The Streamlined Genome of spp. Relative to Human Pathogenic Kinetoplastids Reveals a Parasite Tailored for Plants
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Genome-Wide Association Study of Metabolic Traits Reveals Novel Gene-Metabolite-Disease Links
- A Cohesin-Independent Role for NIPBL at Promoters Provides Insights in CdLS
- Classic Selective Sweeps Revealed by Massive Sequencing in Cattle
- Arf4 Is Required for Mammalian Development but Dispensable for Ciliary Assembly
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



