-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaDistinct DNA Binding Sites Contribute to the TCF Transcriptional Switch in and
Regulation of gene expression by signaling pathways often occurs through a transcriptional switch, where the transcription factor responsible for signal-dependent gene activation represses the same targets in the absence of signaling. T-cell factors (TCFs) are transcription factors in the Wnt/ß-catenin pathway, which control numerous cell fate specification events in metazoans. The TCF transcriptional switch is mediated by many co-regulators that contribute to repression or activation of Wnt target genes. It is typically assumed that DNA recognition by TCFs is important for target gene location, but plays no role in the actual switch. TCF/Pangolin (the fly TCF) and some vertebrate TCF isoforms bind DNA through two distinct domains, a High Mobility Group (HMG) domain and a C-clamp, which recognize DNA motifs known as HMG and Helper sites, respectively. Here, we demonstrate that POP-1 (the C. elegans TCF) also activates target genes through HMG and Helper site interactions. Helper sites enhanced the ability of a synthetic enhancer to detect Wnt/ß-catenin signaling in several tissues and revealed an unsuspected role for POP-1 in regulating the C. elegans defecation cycle. Searching for HMG-Helper site clusters allowed the identification of a new POP-1 target gene active in the head muscles and gut. While Helper sites and the C-clamp are essential for activation of worm and fly Wnt targets, they are dispensable for TCF-dependent repression of targets in the absence of Wnt signaling. These data suggest that a fundamental change in TCF-DNA binding contributes to the transcriptional switch that occurs upon Wnt stimulation.
Published in the journal: . PLoS Genet 10(2): e32767. doi:10.1371/journal.pgen.1004133
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1004133Summary
Regulation of gene expression by signaling pathways often occurs through a transcriptional switch, where the transcription factor responsible for signal-dependent gene activation represses the same targets in the absence of signaling. T-cell factors (TCFs) are transcription factors in the Wnt/ß-catenin pathway, which control numerous cell fate specification events in metazoans. The TCF transcriptional switch is mediated by many co-regulators that contribute to repression or activation of Wnt target genes. It is typically assumed that DNA recognition by TCFs is important for target gene location, but plays no role in the actual switch. TCF/Pangolin (the fly TCF) and some vertebrate TCF isoforms bind DNA through two distinct domains, a High Mobility Group (HMG) domain and a C-clamp, which recognize DNA motifs known as HMG and Helper sites, respectively. Here, we demonstrate that POP-1 (the C. elegans TCF) also activates target genes through HMG and Helper site interactions. Helper sites enhanced the ability of a synthetic enhancer to detect Wnt/ß-catenin signaling in several tissues and revealed an unsuspected role for POP-1 in regulating the C. elegans defecation cycle. Searching for HMG-Helper site clusters allowed the identification of a new POP-1 target gene active in the head muscles and gut. While Helper sites and the C-clamp are essential for activation of worm and fly Wnt targets, they are dispensable for TCF-dependent repression of targets in the absence of Wnt signaling. These data suggest that a fundamental change in TCF-DNA binding contributes to the transcriptional switch that occurs upon Wnt stimulation.
Introduction
Transcriptional switches are common in the regulation of gene expression by cell-cell signaling pathways [1]. This mechanism is typified by active repression of transcription under basal conditions, which is converted to activation by the respective signaling pathway. Examples include the Notch and Wnt/ß-catenin signaling pathways [1], [2], as well as class II nuclear hormone receptors [3]. These switches are important to ensure the proper pattern of expression in development [4], [5] and physiology [6].
The T-cell factor (TCF) family of transcription factors (TFs) offers a prominent example of a transcriptional switch [7]. TCFs are major nuclear mediators of Wnt/ß-catenin signaling, which controls numerous cell fate decisions during development and whose misregulation has been linked to cancer and other human pathologies [8]–[11]. Wnt signaling promotes the stabilization and nuclear accumulation of β-catenin [12], [13]. In the nucleus, ß-catenin is recruited to Wnt response elements (WREs), the cis-regulatory modules that control Wnt target gene transcription, through direct binding to TCFs [7], [14]. TCFs recognize DNA through their High Mobility Group (HMG) domain and act as repressors of gene transcription in the absence of ß-catenin. However, when bound by ß-catenin, they become transcriptional activators [7], [9], [14].
Drosophila and C. elegans each possess a single TCF gene, TCF/Pangolin (TCF/Pan) in flies and pop-1 in nematodes. Genetic evidence indicates that both these TCFs operate as transcriptional switches. For example, in C. elegans embryos, Wnt signaling activates transcription of end-1 and other endoderm-specific genes in the E blastomere [15]–[18]. The adjacent MS blastomere, which does not receive Wnt signal, doesn't express endoderm genes and develops into mesoderm [15]–[20]. In pop-1 mutants, there is a reduction of end-1 expression in E cells [16], [18] while MS cells now express end-1 and other endoderm markers [15]–[18], [20], leading to both blastomeres adopting an endoderm-like fate [19]–[23]. Similarly, mutation of a single HMG binding site in a end-1 WRE reporter results in a similar pattern of GFP expression as seen in POP-1 mutants [16]. Similar data are found in flies, where loss of TCF/Pan results in both loss of activation and expansion of Wnt targets [24], [25] and mutation of HMG sites in WRE reporters display loss of activation as well as derepression of expression [26]–[28]. Thus POP-1 and TCF/Pan repress Wnt targets in the absence of signaling and activate the same genes upon Wnt stimulation.
The current model for the TCF regulatory switch is that ß-catenin promotes a dramatic change in transcriptional co-regulators associated with TCFs. Transducin-like enhancer of Split (TLE) co-repressors such as Groucho in flies [24] and UNC-37 in nematodes [29] bind to TCFs in the absence of signaling and recruit histone deacetylases, promoting gene silencing [7], [14]. ß-catenin binding to TCFs displaces these TLE proteins, somehow antagonizes other co-repressors, and in turn recruits several co-activators [7], [14].
In addition to this classic switch, many Wnt dependent targets in C. elegans utilize an additional mechanism, often referred to as the “Wnt/ß-catenin asymmetry” (WßA) pathway. WβA signaling promotes nuclear influx of the ß-catenin homolog SYS-1, while also promoting nuclear efflux of POP-1 [30]–[32]. This efflux of POP-1 requires the transforming-growth-factor - ß-activated kinase MOM-4, the Nemo-like kinase LIT-1 and the ß-catenin homolog WRM-1 [33]–[35]. WRM-1 acts with LIT-1 to phosphorylate POP-1, causing its nuclear export [36], which shifts the balance from POP-1 mediated repression to activation [30]–[32].
All TCFs contain a HMG domain, which recognizes a 9 bp consensus of SCTTTGATS (S = G/C) with high affinity [11], [37], [38]. In addition, most invertebrate TCFs and E-tail isoforms of vertebrate TCF1 and TCF4 contain a second DNA binding domain just downstream of the HMG domain called the C-clamp [11], [39]. This domain enables TCFs to recognize a second DNA motif, termed the Helper site, which is essential for the Wnt responsiveness of WREs in Drosophila and mammalian cell culture [39]–[41]. Functional Helper sites are often found near (<10 bp) functional HMG sites [39]–[41]. This supports a model where C-clamp containing TCFs recognize WREs through HMG domain-HMG site and C-clamp-Helper site interactions [9], [11].
In this study, we extended our analysis of Helper sites to three WREs directly regulated by the WßA pathway, from the ceh-22, psa-3 and end-1 loci. We found that all three had Helper site motifs near the functional HMG sites, and these Helper sites were crucial for expression of WRE reporters in transgenic worms. The presence of Helper sites dramatically increased binding of POP-1 to HMG sites in vitro. In some cases, multiple Helper sites functioned with a single HMG site to enhance POP-1 binding and mediate expression in vivo. An in silico search of the C. elegans genome for similar Helper sites-HMG site clusters uncovered a new WRE upstream of the K08D12.3 gene, which is expressed in head muscles and the gut. In addition, a synthetic reporter containing concatamerized HMG and Helper sites displayed a novel POP-1 dependent pattern in the int9 cells of the larval intestine, leading us to discover that pop-1 regulates the C. elegans defecation cycle. As previously reported [16], mutation of a single HMG site in the end-1 WRE reporter results in derepression in MS blastomere, but this was not observed upon mutation of two nearby Helper sites, although these motifs were required for expression of the reporter in the E blastomere. This differential requirement of Helper sites in the transcriptional switch was also observed in a fly WRE. Consistent with the cis-regulatory mutagenesis data, we also found that the C-clamp is not required for TCF-mediated repression in the developing fly wing, but is essential for activation. These data support a model where basal repression of WREs occurs through HMG-HMG site interactions, whereas conversion of TCF to a transcriptional activator requires HMG-HMG site and C-clamp-Helper site recognition of DNA. Our data indicate that the TCF binding site is not passive during the transcriptional switch, but rather plays a more active role than previously suspected.
Results
Helper Sites Are Essential for POP-1 Regulation of the ceh-22b and psa-3 WREs
Like TCF/Pan and vertebrate TCF1E and TCF4E isoforms, POP-1 contains a C-clamp downstream of its HMG domain [9], [11]. Given the importance of Helper sites in fly and mammalian WREs that are regulated by C-clamp containing TCFs [39]–[41], we wanted to test the possibility that similar DNA motifs were present in C. elegans WREs. Therefore, we examined three targets that have previously been shown to be directly regulated by the WßA pathway, ceh-22 [42], psa-3 [43] and end-1 [16] (Figure 1).
Fig. 1. Schematics of the ceh-22, psa-3 and end-1 loci. 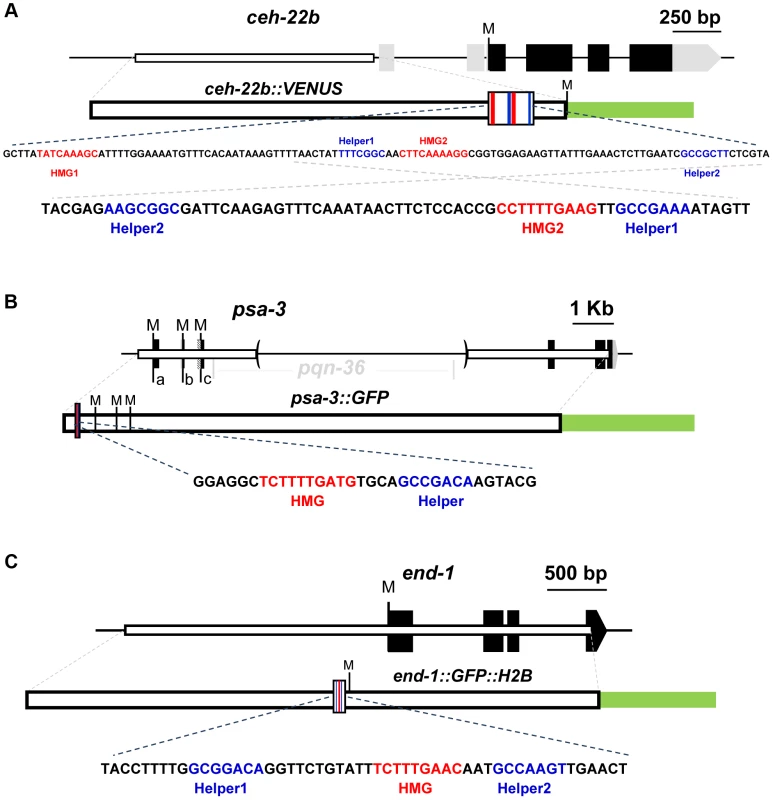
For each locus, black boxes represent exons and gray boxes untranslated regions (UTRs). Start codons representing the Translation Start Site (TlSS) for each isoform are marked by ‘M’. White boxes represent the genomic region used to construct the WRE reporters and the green box the GFP variant used. The larger white boxes in the WRE reporter show the location of the HMG (red lines) and Helper sites (blue lines). Below each schematic are the genomic sequences highlighting the putative Helper sites (blue) and functional HMG sites (red) that were targeted for mutagenesis. (A) For the ceh-22 gene (Gene ID: 179485), a transcriptional fusion of the ceh-22b isoform called ceh-22b::VENUS [42] was used for reporter analysis (nucleotides −1853 to −633 with the first nucleotide of the ceh-22b TlSS representing +1). (B) For psa-3 (Gene ID: 181631), a translational fusion (psa-3::GFP) including promoter sequences (starting at -382) and the first exons of the a, b & c isoforms was used, where the pqn-36 gene, located in the third intron was deleted, as indicated by the parentheses [43]. (C) For end-1 (Gene ID: 179893), a translational fusion containing ∼2.2 kb of promoter sequence, known as end-1::GFP::H2B was used [16]. ceh-22 encodes a homeodomain TF that is required for specification of the distal tip cells (DTCs) [42]. The DTCs play an essential role in gonadal arm elongation during development and in maintenance of the gonadal stem cell niche during adulthood [42], [44]. The ceh-22 locus produces three isoforms, but the ceh-22b/c isoforms (termed ceh-22b) (Figure 1A) are sufficient to rescue gonadal defects in ceh-22 mutants [42]. In hermaphrodites, a ∼1.2 kb transcriptional fusion upstream of the ceh-22b isoform (ceh-22b::VENUS) is expressed in the somatic gonadal precursors (SGPs) and their descendants Z1.a and Z4.p, the distal daughters of which become the DTCs [42]. Maintenance of this expression is dependent on sys-1 and pop-1, and two HMG sites in the ceh-22b::VENUS reporter [42]. An examination of the sequences surrounding these functional HMG sites (termed HMG1 and HMG2) revealed the presence of two motifs (Helper1 and Helper2) similar to the Helper sites found in fly and mammalian systems (Figure 1A).
Mutagenesis of the HMG and Helper sites in the ceh-22b::VENUS reporter revealed that they all contribute to expression in SGP descendants. Transgenic worms expressing stably integrated versions of the wild type (WT) or mutant ceh-22b::VENUS reporters were generated (see Table S1 for details of the altered sequences). In addition to the SGP descendants during specific larval stages, ceh-22b::VENUS is also expressed in the pharynx (Figure 2A′) and this latter pattern is not dependent on sys-1 and pop-1 [42] . Therefore, we used pharyngeal expression as an internal control for transgene copy number, selecting lines with similar expression (Figures 2A′–2G′) to test the functionality of HMG and Helper sites. The WT reporter recapitulated the previously reported pattern of this WRE [42] in the Z1.a and Z4.p descendants in the late L1 hermaphrodites and subsequent stages (Figure 2A; data not shown). Mutation of HMG2 or Helper1 abolished the gonadal expression of this reporter (Figure 2C, 2D, 2H). Mutation of HMG1 or Helper2 resulted in a slightly less severe reduction (Figure 2H) with some animals expressing VENUS in the Z1.a or Z4.p daughters (Figure 2B, 2E). Simultaneous mutation of the two HMG or both Helper sites abolished gonadal expression (Figure 2F, 2G, 2H). These results indicated that Helper sites are required for activation of the ceh-22b::VENUS reporter in the Z1.a and Z4.p daughters.
Fig. 2. Helper sites are required for regulation of ceh-22b and psa-3 WRE reporters. 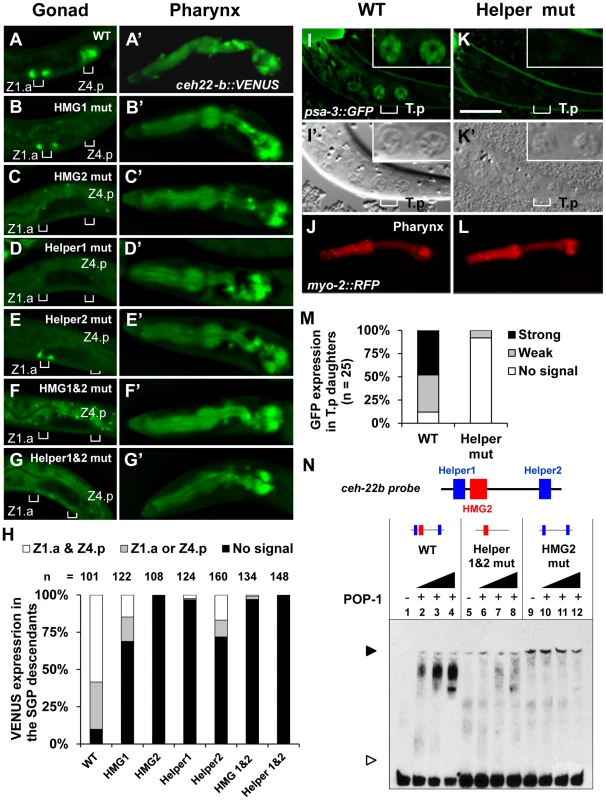
Deconvolved images of fixed L1 larvae showing the expression of stably integrated ceh-22b::VENUS post-division of distal SGP daughters Z1.a and Z4.p (A–G) and expression in the pharynx (A′–G′). VENUS expression in (A) Wildtype (WT), (B) HMG1 mutant, (C) HMG2 mutant, (D) Helper1 mutant, (E) Helper2 mutant, (F) HMG 1 & HMG2 mutant and (G) Helper1& Helper2 mutant were analyzed. Positions of the Z1.a & Z4.p daughter cells are indicated with brackets. (H) Semi-quantification of expression patterns of the different transgenic reporters, grouped three ways (expression in at least one daughter of both Z1.a & Z4.p cells; expression in at least one daughter in either Z1.a or Z4.p cells; no expression). Indicated individuals from three independent lines were examined for each construct except for the HMG1 & HMG2 double mutant construct where two lines were examined. Deconvolved (I & K) and Nomarski images (I′ & K′) of live L1 larvae with (I) Wildtype (WT) and (K) Helper mutant psa-3::GFP reporter. GFP expression was scored in the T cell granddaughters T.pa and T.pp which have the granular nuclear morphology of a neuroblast (I′ and K′, inset) [107]. myo-2::RFP was used as a coinjection marker. WT (J) and Helper mutant (L) animals with a similar RFP expression level in the pharynx were analyzed. (M) Semi-quantification of expression patterns in the T.p daughters of L1 larvae extrachromosomally expressing psa-3::GFP WT or Helper mutant reporter. (N) DNA binding of recombinant POP-1 to a 69 bp ceh-22b WRE probe (see Figure 1A for complete sequence) containing the HMG2 site and both Helper sequences was determined with EMSA. Wildtype probe (WT) shows a POP-1 dependent shift (lanes 1–4), while mutation of both Helper sites (lanes 5–8) or the HMG2 site (lanes 9–12) results in almost no detectable binding. The black arrowhead represents the DNA-protein complex and the white arrowhead represents unbound probe. The data shown are representative of more than three separate binding assays. To extend our analysis of Helper site function to another WßA pathway target, the cis-regulatory region of the Meis-related factor psa-3 was examined. In hermaphrodites, POP-1 regulates the expression of psa-3 in the posterior T-cell descendants, which give rise to the phasmid socket cells [43]. A translational psa-3 fusion (Figure 1B), which can rescue the psa-3 mutant phenotype [43], was used as the starting point to examine the functionality of a Helper site located near a HMG site (Figure 1B). This HMG site, located upstream of the psa-3 Translational Start Site (TlSS; Figure 1B) was previously shown to be essential for expression of the translational reporter in the posterior T cell granddaughters T.pa and T.pp during the mid-L1 stage [43](Figure 2I). Wild-type (WT) and Helper mutant reporters (see Table S1 for base pair substitutions) were co-injected with the myo-2::RFP reporter, the pharyngeal expression of which was used as an internal control in the transgenic lines that were established (Figure 2J, 2L). Mutation of the Helper site abolished detectable expression of the psa-3::GFP reporter in the posterior granddaughters T.pa and T.pp in majority of transgenic larvae examined (Figure 2K, 2M). These data demonstrate that a Helper site near the functional HMG site plays a crucial role in activation of the WßA pathway target psa-3.
In analogy with fly and mammalian WREs [39]–[41], the Helper sites in the ceh-22b and psa-3 reporters may enhance WßA signaling by increasing POP-1 binding. To test this, we performed Electrophoretic Mobility Shift Assays (EMSAs) with recombinant POP-1 and DNA probes containing Helper 1, HMG2 and Helper 2 from the ceh-22b reporter (Figure 1A) and the functional HMG and Helper sites from psa-3::GFP (Figure 1B). Both probes showed robust binding when incubated with POP-1 (Figure 2N; Figure S1). Mutation of the HMG2 site or Helper 1 & 2 sites dramatically reduced POP-1 binding to the ceh-22b probe (Figure 2N). Competition assays with unlabeled oligonucleotides demonstrated that Helper 1 & 2 sites are both significant contributors to POP-1 binding (Figure S1A). Similarly, the HMG and Helper site were both required to compete with labeled psa-3 probe binding to POP-1 (Figure S1B). These results are consistent with a model where both HMG and Helper sites are required for high affinity binding of POP-1 to the ceh-22b and psa-3 WREs.
In Silico Identification of a cis-regulatory Element Upstream of the K08D12.3/ZNF9 Locus Containing a Functional HMG-Helper Site Cluster
We next wanted to test whether a computational search for HMG and Helper site clusters could identify new POP-1 targets in the C. elegans genome. We utilized the open source algorithm Target Explorer [45], which we have used previously to detect novel WREs in Drosophila [40]. A position weight matrix for each motif was created based on the sequence of the functional HMG or Helper sites from ceh-22b, psa-3 and end-1 reporters plus the optimal HMG and Helper sites used in the synthetic reporter described below (Table S2). Because many cis-regulatory elements are located just upstream of nematode genes [46]–[49], we restricted the search to 500 bp regions upstream of every annotated TlSS or Transcriptional Start Site (TSS) in the C. elegans genome (24841 in total). Based on the organization of functional Helper and HMG sites in ceh-22b, psa-3 and end-1 (Figure 1), we utilized a search criterion where a 50 bp stretch had to contain at least two Helper sites and one HMG site to score positive. This generated a list of 115 clusters. 19 of these had the HMG site between the two Helper sites as found in the ceh-22b and end-1 WREs (list provided in Table S3). Of this subgroup, three regions were selected for EMSA analysis with POP-1, based on the presence of more than two Helpers sites within 50 bp of the HMG site (K08D12.3) or the quality of the HMG and Helper sites (RO5G6.5 and Y66d12A.9).
Of the three Helper-HMG clusters tested, only the region upstream of the annotated K08D12.3/ZNF9 gene had significant binding to POP-1 (Figure S2). The original K08D12.3 probe contained three Helper sites and one HMG site. A shorter probe missing one of the Helper sites was still bound by POP-1, but to a lesser degree (Figure S2), suggesting that Helper 1 was required for maximal POP-1 binding. Indeed, competition experiments indicated that the HMG site and all three Helper sites were required for POP-1 binding, with the HMG and Helper3 sites having the greatest contribution (Figure 3F). Therefore, all four sites were considered in our in vivo analysis.
Fig. 3. Identification of a new POP-1 target using a computational search for Helper site-HMG site clusters. 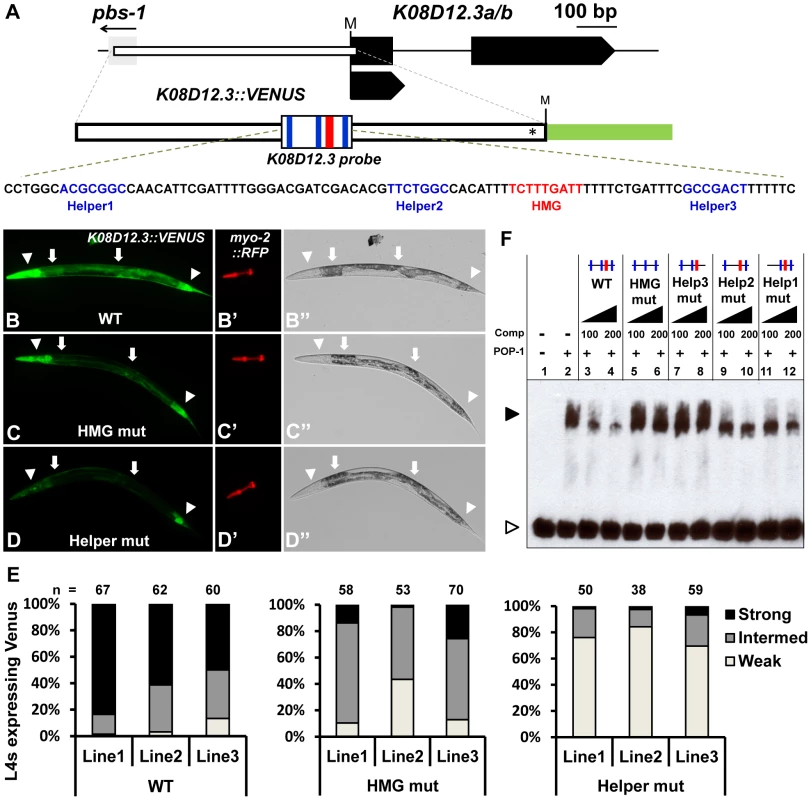
(A) Schematic depicting the K08D12.3 locus (Gene ID: 176979) with black boxes representing exons and the gray box the flanking gene pbs-1. The start codon is marked by ‘M’. The white box indicates the genomic region used to construct the GFP transcriptional reporter (nucleotides −579 to +14; first nucleotide of TlSS represents +1), with the asterisk indicating where the K08D12.3 start codon was mutated to allow GFP to be read in the correct frame. The location of the HMG and Helper sites are indicated in red and blue respectively. Fluorescence (B–D; B′–D′) and Brightfield (B″–D″) images of live late L4 larvae extrachromosomally expressing the K08D12.3::VENUS reporter. Strong expression was seen in the head muscles, pharyngeal muscles, posterior intestine and hindgut (arrowheads) and moderate expression in the midgut (arrows). (B) Wildtype, (C) HMG mutant and (D) Helper mutant worms were scored based on the VENUS expression in the head muscles, pharyngeal muscles and intestine. (E) Histogram showing the expression analysis of late L4 larvae from three independent lines carrying either the WT, HMG mutant or Helper mutant K08D12.3::VENUS reporters, grouped into strong, intermediate or weak expressers, represented by the images in panels B, C & D, respectively. (F) Competition analysis using EMSA with POP-1 protein with a 90 bp probe (sequence shown in panel A) containing the three functional Helper sites and the functional HMG site from the K08D12.3 WRE. The POP-1 dependent shift (lane 2) is competed by an excess of unlabeled WT probe (lanes 3, 4), while unlabeled HMG mutant probe (lanes 5, 6) or the Helper3 mutant probe (lanes 7, 8) does not compete even at 200 fold excess competitor levels. Unlabeled Helper1 mutant (lanes 9, 10) and Helper2 mutant (lanes 11, 12) probes displayed a moderate level of competition. The black arrowhead represents the DNA-protein complex and the white arrowhead represents unbound probe. The data is representative of three independent experiments. To test whether the region upstream of K08D12.3/ZNF9 could drive expression of a reporter gene, a 585 bp stretch upstream of the TlSS was cloned behind a VENUS reporter (Figure 3A), and tested for expression in vivo. Strong reporter expression was observed in the foregut, anterior body wall musculature (dorsal and ventral head muscles), posterior intestine and hindgut during larval stages till adulthood (Figure 3B, 3B″; data not shown). Weaker expression was also seen in the midgut, mid-body wall musculature and the posterior body wall muscles during these stages (Figure 3B, 3B″; data not shown). This expression pattern was similar (but less intense) than a previously reported pattern of a ∼2.3 kb K08D12.3::GFP transcriptional fusion [47], [48]. No decrease in the K08D12.3 reporter was observed in pop-1 (q645) mutants or animals depleted of POP-1 via RNAi feeding (data not shown). However, these conditions only partially reduce pop-1 gene activity [50], [51] and are therefore inconclusive.
To provide a more definitive test of POP-1 regulation of the K08D12.3 reporter, late L4 hermaphrodites containing a wild-type, a HMG mutant or a triple Helper mutant reporter were scored based on VENUS expression in the hindgut and midgut, as well as the head and pharyngeal muscles, and characterized as having strong, intermediate or weak expression (Figure 3B–E). As with the psa-3 reporter, myo-2::RFP was used as an internal control (Figure 3B′–3D′). Mutation of the HMG site and Helper sites significantly affected VENUS expression in the head muscles, midgut, and foregut with less reduction in hindgut expression (Figure 3B–E). The Helper site mutant lines had less expression than the HMG site mutants, most notably in the pharyngeal muscles (Figure 3C–E). These data suggest that the K08D12.3 reporter is a direct target of POP-1.
The C-Clamp of POP-1 Is Required for Enhanced Binding to HMG-Helper Site Clusters
The finding that Helper sites are important for high affinity POP-1 binding (Figure 2N) suggests that the C-clamp of POP-1 is required for this binding. To test this, recombinant POP-1 containing two substitutions in the C-clamp (K365A & R367E) was expressed and purified. The corresponding mutations in TCF/Pan abolish DNA binding to Helper site DNA (A. Ravindranath and K. M. Cadigan, unpublished). This C-clamp mutant displayed a dramatic reduction in binding to a ceh-22b HMG-Helper site probe (Figure 4A). Mutation of the C-clamp also dramatically reduced affinity of POP-1 for HMG-Helper site containing probes from the psa-3 and K08D12.3 loci (Figure 4B, 4C). These data demonstrate that high affinity binding of POP-1 to functionally important WRE DNA requires the C-clamp domain.
Fig. 4. The C-clamp of POP-1 facilitates binding to DNA containing Helper sites. 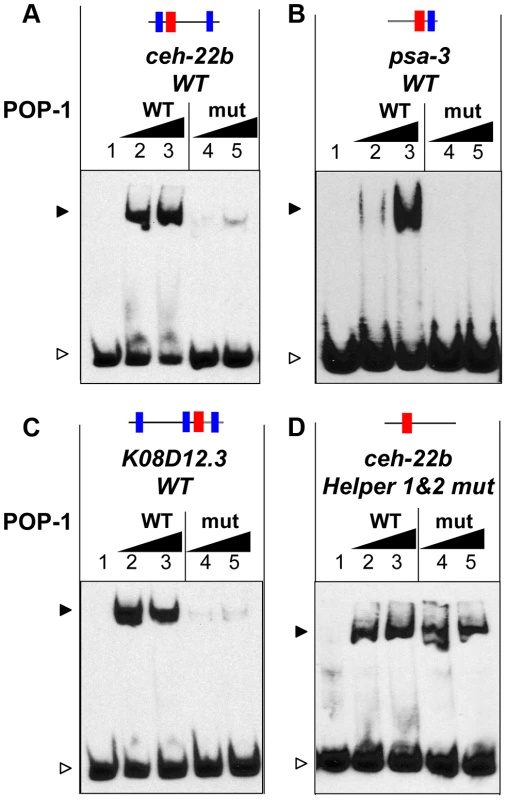
(A–D) EMSAs showing binding of wild-type recombinant POP-1 and a POP-1 C-clamp mutant to the ceh-22b WRE probe (1.5 femtomoles/reaction) described in Figure 1A and 2N (A), the psa-3 probe (3 femtomoles/reaction) described in Figure 1B and S1B (B), the K08D12.3 probe (4 femtomoles/reaction) described in Figure 3A and 3F (C) and the ceh-22b probe (3 femtomoles/reaction) with both Helper sites mutated (D). The ceh-22b, psa-3 and K08D12.3 WT probes show strong binding with increasing amounts (0.4 and 0.8 µg/reaction) of POP-1 WT protein (lanes 2 and lane 3 respectively) but not with the POP-1 C-clamp mutant (lane 4 and lane 5 respectively). Under conditions designed to detect lower affinity binding (0.75 and 1.5 µg of POP-1; 3 femtomoles of probe and longer exposure times), binding to the ceh-22b probe lacking Helper sites (containing only the HMG2 site) was similar with WT and mutant POP-1. The data are representative of three independent experiments. As described above, Helper sites are required for high affinity binding to DNA by POP-1 (Figure 2N, Figure 3F, Figure S1). This requirement is not altered by mutation of the C-clamp (Figure S3). However, POP-1 can still recognize HMG site DNA in the absence of Helper motifs or a functional C-clamp, albeit at lower affinity. Under conditions allowing the detection of weaker binding, i.e., increased probe concentration and longer exposure times, wild-type and C-clamp mutant POP-1 showed comparable binding to a ceh-22b probe where the Helper sites are mutated (Figure 4D; see also Figure S3, which uses conditions that only detect higher affinity binding). These data demonstrate that the HMG domain of POP-1 can recognize HMG site DNA, but higher affinity binding required a bipartite mechanism similar to what we previously reported for TCF/Pan [40].
A Synthetic Reporter Containing HMG-Helper Sequences Reveals a POP-1 Dependent Pattern in int9 Intestinal Cells
Synthetic reporters containing concatemerized high-affinity HMG sites have been used as Wnt/ß-catenin signaling readouts in several systems [52], e.g, TOPFLASH in mammalian cell culture [53] and TOPGAL in transgenic mice [54]. Similar HMG site reporters do not work well in Drosophila [40], but multiple HMG-Helper site pairs provide a much more sensitive indicator of Wnt/ß-catenin signaling [40]. In C. elegans, a reporter known as POPTOP (POP-1and TCF Optimal Promoter) contains seven copies of a high affinity HMG site, which displays a wide range of expression including several cells where Wnt/POP-1 signaling is known to occur [55]. To test whether Helper sites can improve the sensitivity or selectivity of HMG sites in this synthetic context, we constructed a reporter containing six HMG-Helper site pairs (see Materials and Methods for sequence) called POPHHOP (POP-1 and HMG-Helper Optimal Promoter) and tested it for expression in C. elegans.
Similar to POPTOP, stably integrated POPHHOP was expressed in several cells where Wnt/ß-catenin signaling is known to be active (Figure 5D; Figure S4). The POPHHOP reporter was active in cells not previously known to receive Wnt signals, e.g., unidentified tail neurons and posterior cells in the ventral nerve cord during the early L1 stage (Figure 5A; Figure S4A). Expression was also seen in seam cell nuclei, muscle nuclei along the anterior/posterior axis and the QL.d nuclei (Figure S4C–G). The most prominent novel pattern observed with POPHHOP was in the posterior most intestinal cells known as ‘int9 cells’, during the early L1 stage (Figure 5A, 5C, 5G; Figure S4E, S4F) onward to adults (data not shown). This pattern was lost in a genetic background homozygous for the hypomorphic allele pop-1(hu-9) (Figure 5H). In sum, the inclusion of Helper sites in a HMG site synthetic reporter altered the specificity of reporter expression, and should be a useful tool to study POP-1 readouts in C. elegans.
Fig. 5. A synthetic HMG-Helper site reporter reveals a novel POP-1 function in rhythmic defecation behaviour. 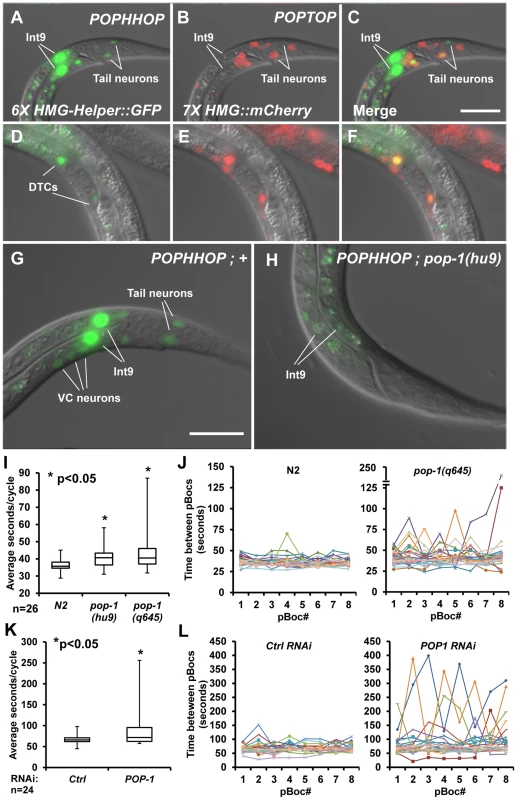
(A–F) Nomarski images of animals with stably integrated POPHHOP (6× HMG-Helper::GFP) and POPTOP (7× HMG::mCherry) reporters showing GFP (A, D) and mCherry (B, E) fluorescence. Live L1 larvae have overlapping expression of GFP and mCherry in some tail neurons (A–C) and live L3 larvae display overlapping DTC expression (D–F). In addition, POPHHOP displayed strong expression in the int9 intestinal cells of early L1 Larvae (A) onward through adulthood (not shown). (G–H) Stably integrated POPHHOP animals in a wild-type (G) or pop-1(hu9) background (H). The reporter expression seen in the int9 cells, tail neurons, and occasionally in the VC neurons is low or undetectable in the pop-1 mutants. Scale bars = 10 µm. (I) Box-whisker plot showing the median (line inside the box), third quartile (upper box), first quartile (lower box), longest pBoc cycle time (upper whisker limit) and shortest pBoc cycle time (lower whisker limit) for N2 controls and two pop-1 alleles at the L2 stage. A statistically significant increase was seen in the pBoc cycle time based on a Student's two-tailed t test (see Table 1). (J) 8 individual pBocs (X-axis) were monitored (n = 26, each color representing one larva) for each genotype and plotted against time between each pBoc (y-axis). pop-1(q645) mutants have greater variability between pBocs than the wild-type N2 control. (K) Box-whisker plot showing the pBoc period of pop-1 depleted worms compared to ctrl RNAi worms using the OLB11 strain, which allows intestine-specific RNAi [65], [66]. Animals were assayed at the young adult stage. A statistically significant increase was seen in the pBoc cycle time based on a Student's two-tailed t test (see Table 1). (L) 8 individual pBocs (X-axis) were monitored in young adults (n = 24, each color representing one adult) for each genotype and plotted against time between each pBoc (y-axis). pop-1 RNAi leads to a high variability in the cycle time in pop-1 depleted adults compared to controls. The Role of POP-1 in the C. elegans Defecation Cycle
The expression pattern of POPHHOP in int9 cells was intriguing, given the central role that these cells are known to play in regulating the defecation cycle in C. elegans [56]. The cycle starts with contractions in the posterior body wall muscles (pBoc) at regular (45–50 sec) second intervals in adult worms [57]. Each pBoc is followed by anterior body wall contractions (aBoc) and expulsion (Exp) of the fecal content completing a defecation cycle [57]. The intestine acts as a pacemaker where calcium oscillations set the period of each cycle [56], [58]–[60]. Calcium spikes originate in the int9 cells, which initiate the pBoc step [56]. POP-1 is expressed in these cells [61] and is required for POPHHOP expression in int9 cells (Figure 5H). This suggested that POP-1 could play a role in regulating rhythmic defecation in C. elegans.
Strong alleles of pop-1 are early larval or embryonic lethal [19], [50], [62]. Therefore, two hypomorphic alleles of pop-1, both containing single amino acid substitutions in the ß-catenin binding domain, were used to examine the role of POP-1 in the defecation cycle. pop-1(hu9) homozygotes survive to adulthood [63], [64], while a fraction of hermaphrodites homozygous for the stronger allele pop-1(q645) make it to adulthood [50]. Since POPHHOP was regulated by POP-1 at the early L1 stage and onward (Figure 5H; data not shown), we examined pBoc cycle lengths and expulsion events in control N2 animals and the hypomorphic pop-1 mutants in early L2 larvae, well before any lethality was apparent in the pop-1(q645) animals.
We found that control N2 animals have a highly regular defecation cycle at the L2 stage, displaying a mean periodicity of 36.4 seconds between pBocs with a standard deviation of 3.5 seconds. L2 larvae homozygous for the pop-1(hu9) and pop-1(q645) alleles had a 11–18% increase in mean cycle time and greater arrhythmia than N2 controls (Table 1). The increased arrhythmia can be observed when individual cycle times are plotted for each background (Figure 5J) and in a box-whisker plot (Figure 5I). In addition, the expulsion of fecal content, which is tightly coupled to the pBoc in each cycle, failed to occur 6.7% of the time in the pop-1(q645) mutants, while such failures were never seen in control or pop-1(hu9) animals (Table 1). Taken together, our analysis of the expression pattern of the POPHHOP reporter led to the discovery of a previously unsuspected requirement for POP-1 in regulating the cycle time and rhythmicity of C. elegans defecation.
Tab. 1. Reduction of pop-1 gene activity results in a prolonged defecation cycle. 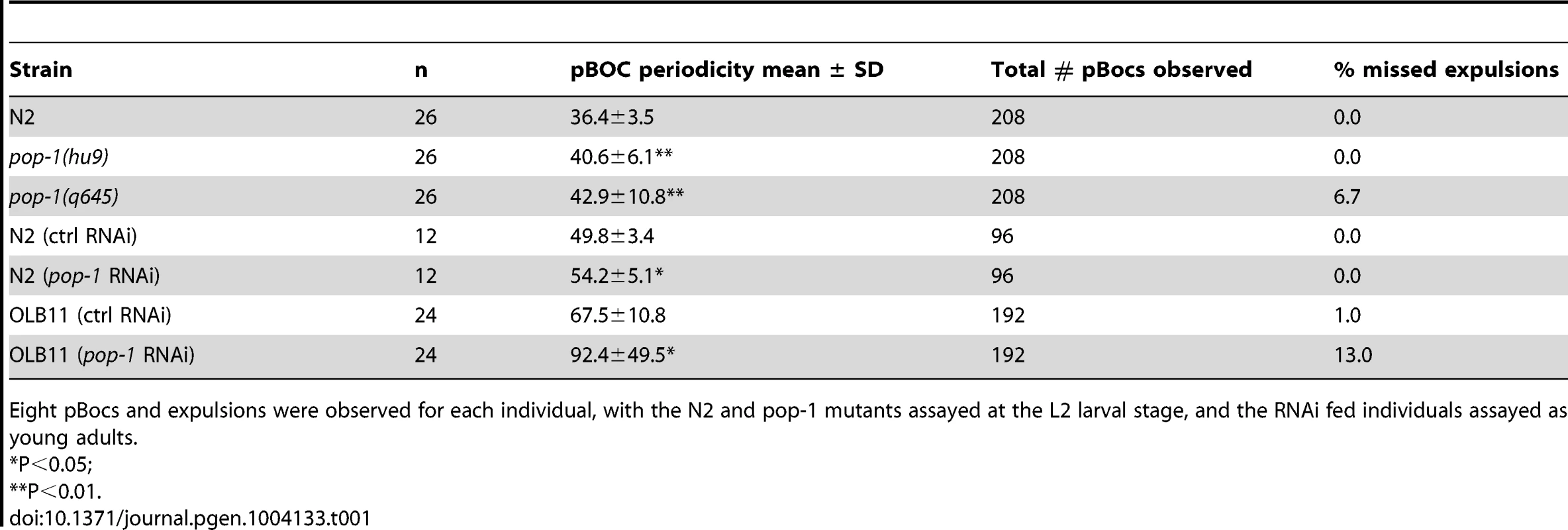
Eight pBocs and expulsions were observed for each individual, with the N2 and pop-1 mutants assayed at the L2 larval stage, and the RNAi fed individuals assayed as young adults. Although the pop-1 alleles used above have no obvious anatomical defects at the L2 stage and survive to adulthood, the possibility remains that the effect on defecation in these mutants was indirectly due to abnormal embryonic development. To address this, RNAi feeding was performed with L1 larvae, with defecation measured in young adults (0–12 hours). RNAi depletion of pop-1 resulted in a significant increase (8.8% in the mean period) in the defecation cycle and greater variability compared with control fed animals (Table 1 and data not shown). When pop-1 depletion was limited to intestinal cells, using strain OLB11, which is mutant for the Argonaute gene rde-1 and contains a transgene expressing rde-1 under the control of the elt-2 intestinal promoter [65], [66], a more dramatic effect on defecation was observed (36.9% increase in the mean period) with a concomitant increase in variation in cycle length (Figure 5K, 5L). The frequency of missed expulsions following pBoc was 13 times higher than controls (Table 1). These results indicate that the requirement for POP-1 in regulating the defecation cycle resides in intestinal cells.
Helper Sites Are Required for Activation of the end-1 WRE, but Not Basal Repression by POP-1
Although the TCF transcriptional switch is the prevailing model for Wnt/β-catenin gene regulation, for many WREs, there is little evidence for repression in the absence of signaling [9]. For example, there is a dramatic loss of activation when HMG sites are mutated in the ceh-22b, psa-3 and K08D12.3 reporters discussed thus far, but no detectable derepression. This is likely due to a lack of local activators in these elements that could drive expression in the absence of POP-1 mediated repression [1], [9]. To address a possible role for Helper sites in POP-1 basal repression, we examined the WRE regulating the GATA factor end-1 (Figure 1C). In early embryogenesis, the WßA pathway is required for maximal expression of end-1 in the endodermal descendant of EMS, known as E cells [15], [16], [18]. In the other EMS derived daughter cell, the mesodermal MS, POP-1 represses end-1 expression [15]–[20]. Consistent with this, mutation of a single HMG site in an end-1 translational fusion (end-1::GFP::H2B) causes a reduction in expression in E cells, accompanied by a dramatic elevation of expression in MS cells [16]. Two putative Helper sites are located near this functional HMG site (Figure 1C). Thus, the bimodal regulation of end-1 by POP-1 provides an ideal system in which to test the role of Helper sites in a target where both sides of the transcriptional switch are robustly apparent.
Transgenic worms expressing stably integrated WT or mutant end-1::GFP::H2B reporter fusions were generated (Figure 1C; Table S1) and examined at the 2E stage. As previously reported [16], the WT reporter had a strong GFP expression in the two E cell daughters while mutation of the HMG site led to a strong decrease of expression in these cells, and ectopic expression in the MS descendants with high penetrance (Figure 6A, 6B, 6D). Mutation of the two putative Helper sites resulted in the same dramatic decrease in E cell expression as seen in the HMG site mutant (Figure 6C, 6D). However, mutation of the Helper sites resulted in almost no detectable derepression in the MS cells (Figure 6C, 6D). These data indicated that the Helper sites are largely dispensable for repression of end-1 in the MS cells, while they are required for its maximal activation in the E cells.
Fig. 6. HMG and Helper sites contribute differentially to the regulation of end-1 during early embryogenesis. 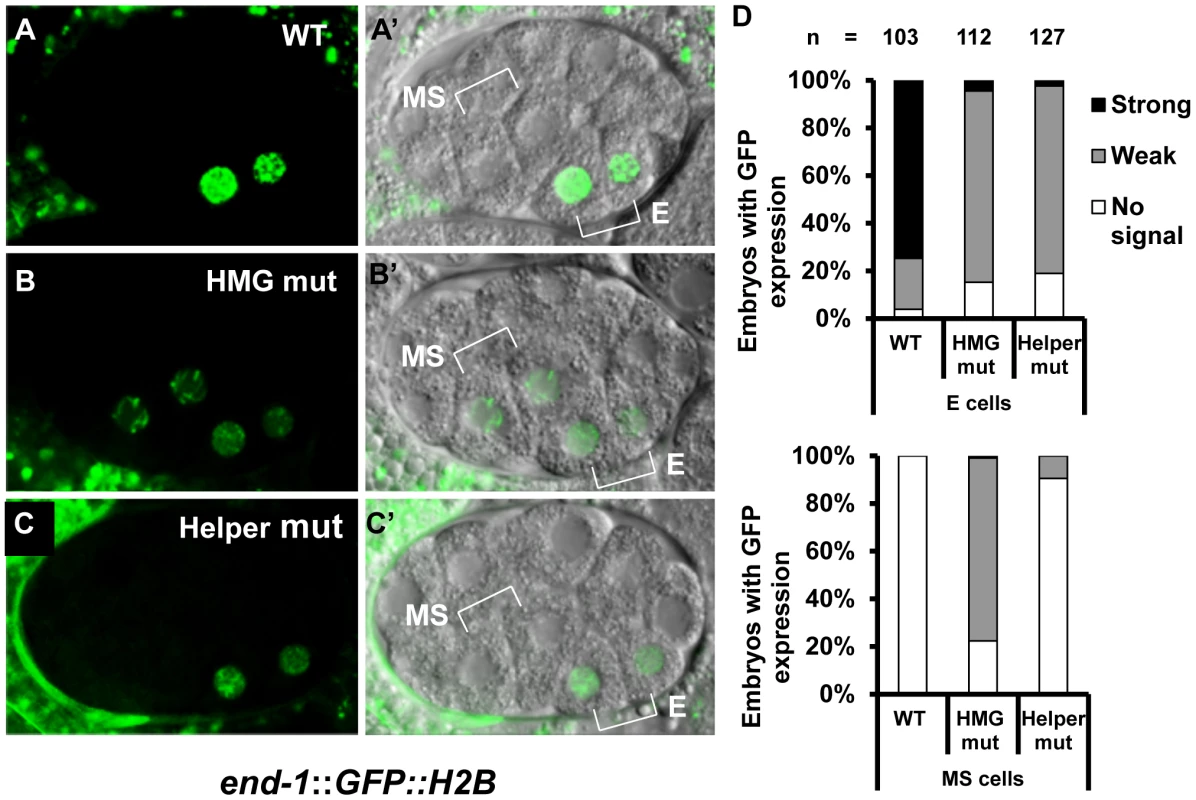
Deconvolved (A–C) and Nomarski (A′–C′) images showing expression of a stably integrated end-1::GFP::H2B reporter in the endodermal (E) and/or mesodermal (MS) daughters of live embryos at the 2E stage. The wild type (WT) reporter shows strong GFP expression in the E cell daughters (A, A′). Mutation of the HMG site leads to a significant reduction of GFP expression in the E daughters and a significant derepression of end-1::GFP::H2B in the MS daughters (B, B′). Mutation of two Helper sites leads to a significant reduction of GFP in the E daughters (C, C′), but little or no depression in the MS daughters (C, C′). (D) Histograms summarizing the results from over 100 embryos from three independent lines for each construct, grouped by strong, weak or no expression in the E (upper graph) and MS (lower graph) cells. The Differential Requirement of HMG and Helper sites in the Transcriptional TCF Switch Is Conserved in Flies
In our previous report on the role of Helper sites in Drosophila, the four WREs tested in transgenic reporter assays were similar to the ceh-22b and psa-3 reporters, i.e., they are strongly activated by Wnt/ß-catenin signaling but have little detectable derepression when their HMG sites are removed [40]. However, we have found a WRE upstream of the pxb gene (Gene ID: 41899), originally identified through a computational search for HMG-Helper site clusters in the fly genome [40], that has a significant basal repression component. When this WRE was placed upstream of a minimal promoter driving lacZ and inserted into the fly genome through P-element transgenesis, expression was observed in a pattern that overlapped with Wingless (Wg, a fly Wnt) in the second constriction of the embryonic midgut (Figure 7A–C; see arrow). The pxb-WRE reporter was also expressed at low levels in the anterior midgut (Figure 7B, first arrowhead) and in the hindgut where there is no detectable Wg expression (Figures 7A–C). Mutation of two HMG sites in the pxb-WRE resulted in a strong derepression throughout the midgut (Figure 7E, arrow and arrowheads) and no change in the hindgut expression (Figure 7E). These results indicate that in the embryonic midgut, there is a large degree of basal repression of the pxb-WRE by TCF/Pan.
Fig. 7. Helper sites and the C-clamp are not required for basal repression of Wg targets in Drosophila. 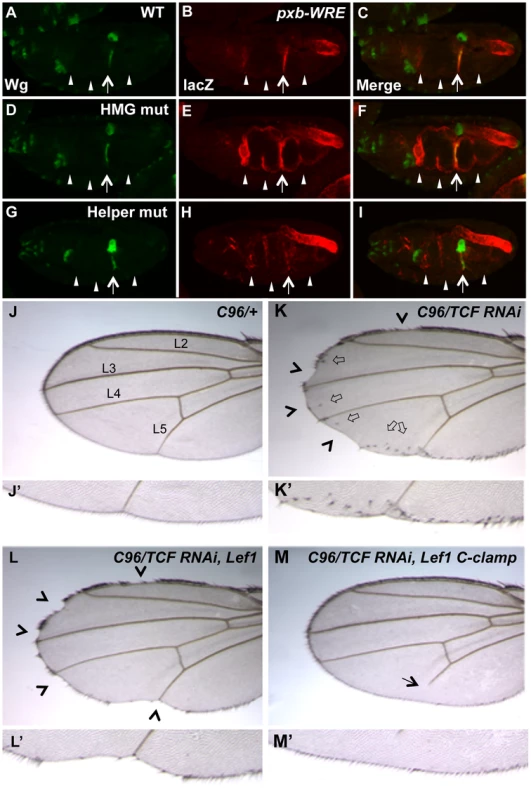
(A–I) Confocal images of stage 16–17 embryos containing a pxb::lacZ WRE reporter immunostained for Wg (green) (A, D & G), lacZ (red) (B, E & H) or merged (C, F & I). The wild-type reporter shows a pattern overlapping with Wg in the second constriction of the midgut, and a non-overlapping pattern in the hindgut (A–C). Mutation of two HMG sites leads to a strong depression through the entire midgut (arrowheads), without affecting lacZ expression in the second constriction (arrow) (D–F). Mutation of two Helper sites leads to a significant decrease in the lacZ expression in the second constriction (arrow) with weak ectopic expression (arrowheads)(G–I). The hindgut expression did not vary in the different constructs and was used as an internal control. All images are representative of at least 20 embryos. (J–M) Images of adult wings containing the wing driver C96-Gal4 crossed to wildtype (WT) (J, J′), UAS-TCF/Pan RNAi (K, K′) or UAS-TCF/Pan RNAi plus UAS-LEF1 (L, L′) or UAS-LEF1 plus the C-clamp of TCF/Pan (M, M′). Knockdown of TCF/Pan leads to notches (arrowheads) and ectopic wing margin bristles (block arrows) along the periphery of the wing (where C96-Gal4 is active; K, K′). Expression of the human LEF1 transgene significantly rescues the ectopic bristle expression, but not the notches (L, L′). Expression of a LEF1-C-clamp chimera rescues the wing margin defects and prevents ectopic bristle formation, and causes a L5 vein defect (arrow). Details about the penetrance of these phenotypes are listed in Table 1. In contrast to the HMG sites, mutation of two nearby Helper sites caused a moderate decrease of pxb-WRE expression in the second midgut constriction (Figure 7H, arrow), with only mild ectopic expression in the midgut (Figure 7H, arrowheads). This derepression was extremely weak compared to that seen in the HMG mutant embryos (Figure 7E, 7H). In both mutant constructs, expression in the hindgut was not significantly affected, serving as an internal control (Figure 7B, 7E, 7H). These data indicate that the Helper sites are required for activation of pxb-WRE by Wg signaling but are largely dispensable for repression of this reporter in cells with no detectable Wg expression.
If Helper sites are primarily required for TCF/ß-catenin activation of gene expression and not basal repression by TCF, is the C-clamp domain of TCFs only required for the activation side of the transcriptional switch? To test this idea, a TCF/Pan rescue assay was established in the developing fly wing. Wg signaling is required for specification of the wing margin and adjacent sensory bristles, with loss of signaling resulting in notches in the wing blade [67], [68] and ectopic signaling causing ectopic sensory bristles [69], [70]. When TCF/Pan was depleted in flies containing the wing margin specific Gal4 driver C96 [71] and a UAS-TCF/Pan RNAi construct [72], notches along most of the distal margin were observed with 100% penetrance (Figure 7K, 7K′; Table 2). In addition, a large number of ectopic wing margin bristles (∼22/wing) were seen (Figure 7K, 7K′; Table 2), likely due to derepression of the Wg targets specifying sensory bristles.
Tab. 2. The C-clamp is required for Wg activation but not basal repression in a TCF/Pan rescue assay. 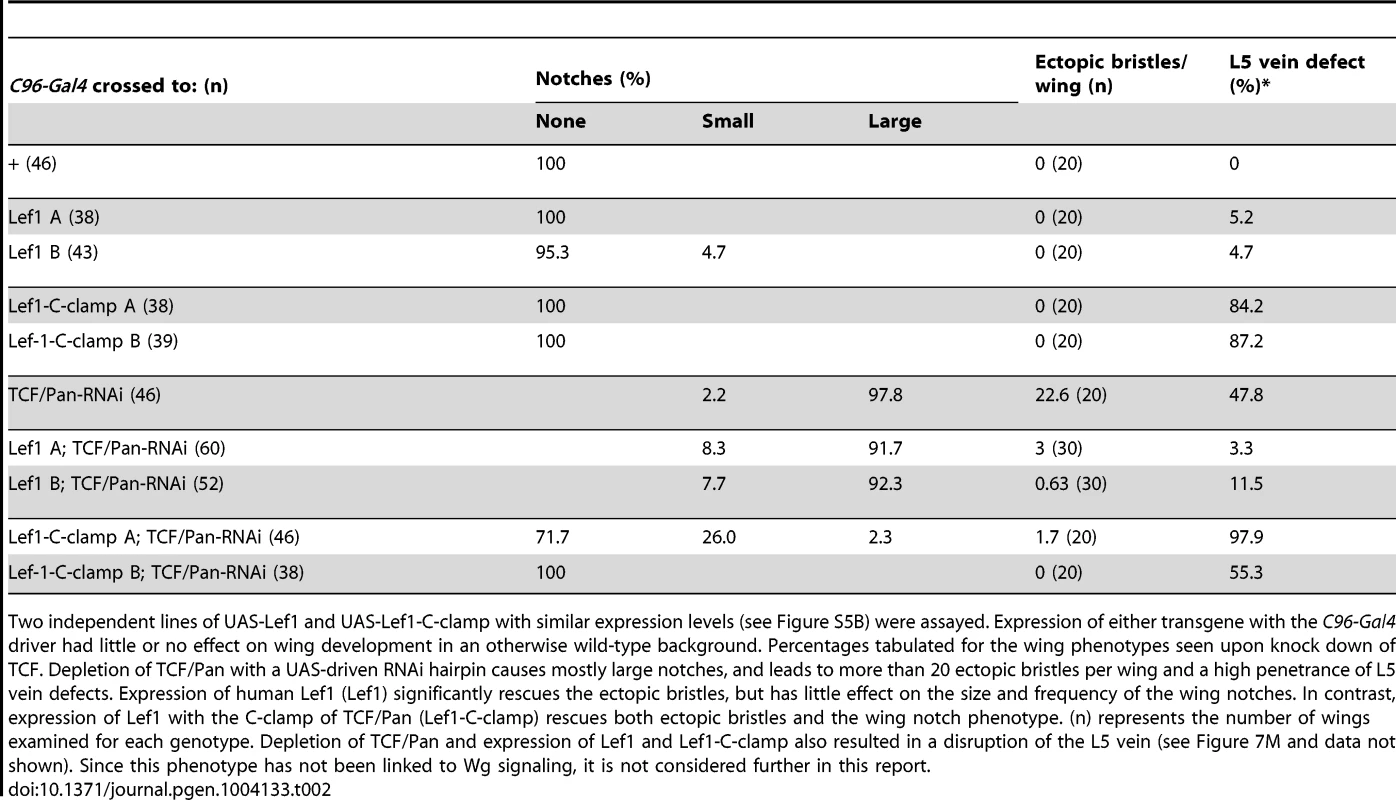
Two independent lines of UAS-Lef1 and UAS-Lef1-C-clamp with similar expression levels (see Figure S5B) were assayed. Expression of either transgene with the C96-Gal4 driver had little or no effect on wing development in an otherwise wild-type background. Percentages tabulated for the wing phenotypes seen upon knock down of TCF. Depletion of TCF/Pan with a UAS-driven RNAi hairpin causes mostly large notches, and leads to more than 20 ectopic bristles per wing and a high penetrance of L5 vein defects. Expression of human Lef1 (Lef1) significantly rescues the ectopic bristles, but has little effect on the size and frequency of the wing notches. In contrast, expression of Lef1 with the C-clamp of TCF/Pan (Lef1-C-clamp) rescues both ectopic bristles and the wing notch phenotype. (n) represents the number of wings examined for each genotype. Depletion of TCF/Pan and expression of Lef1 and Lef1-C-clamp also resulted in a disruption of the L5 vein (see Figure 7M and data not shown). Since this phenotype has not been linked to Wg signaling, it is not considered further in this report. The TCF/Pan RNAi phenotypes were used to assay the ability of UAS transgenes expressing the human TCF family member LEF1 (which contains a HMG domain but no C-clamp), or LEF1 with the C-clamp of TCF/Pan (LEF1-C-clamp; see Figure S5A for description of LEF1 proteins), to rescue the derepression and loss of activation phenotypes in C96::TCF/Pan RNAi wings. A human TCF was used in this assay because it is insensitive to the UAS-TCF/Pan RNA hairpin, which targets the ORF of endogenous TCF/Pan mRNA.
Both the LEF1 and LEF1-C-clamp transgenes had biological activity in the fly wing, but with dramatically different specificities. LEF1 was unable to rescue the wing notch phenotype (i.e., activation) but strongly suppressed the formation of ectopic bristles (basal repression) (Figure 7L, 7L′; Table 2). In contrast, the LEF1-C-clamp chimera was able to rescue both the notch and bristle phenotypes (Figure 7M, 7M′; Table 2). More than a dozen independent UAS-LEF1 and UAS-LEF1-C-clamp lines were generated, and the ones at the lower end of the expression spectrum were used in this rescue experiment, because higher expression of either transgene caused wing notches in an otherwise wild-type background (data not shown). We suspect that too much of either LEF1 protein inhibits Wg signaling by titrating out fly ß-catenin in the nucleus, in analogy to high nuclear POP-1 levels in the WßA pathway [18], [73]. Western blot analysis revealed that the LEF1 and LEF1-C-clamp transgenes used for the rescue were expressed at similar levels (Figure S5B). These data fit with the cis-regulatory mutagenesis of the end-1 and pxb WREs (Figure 6; Figure 7A–7I), supporting a model where basal repression by TCFs involves HMG domain-HMG site interactions, while ß-catenin dependent activation requires HMG domain-HMG site and C-clamp-Helper site binding.
Discussion
Helper Sites and POP-1 Recognition of C. elegans WREs
In this report, we demonstrate that Helper sites enhance POP-1's ability to bind to DNA with high affinity and are critical for the expression of four distinct C. elegans WREs expressed in a variety of tissues and developmental stages (Figure 2; Figure 3; Figure 6). In this respect, C. elegans is similar to Drosophila, where we have previously shown that Helper sites are just as important as HMG sites for WRE activity in vivo [40]. HMG and Helper sites are also equally important for activation of specific WREs in mammalian cell culture by TCF1 and TCF4 isoforms containing a C-clamp [41]. Since we have found functional Helper sites in every fly and worm WRE that we have rigorously characterized (a total of ten), it is tempting to suggest that many more WREs in these organisms utilize a similar HMG-Helper site mechanism. We also suggest that in other invertebrate phyla, such as porifera, cnidarians and echinoderms, which possess one TCF gene with a C-clamp [74]–[76], TCFs likely recognize WREs through a similar bipartite mechanism to perform many of their essential functions.
While Helper sites are clearly important for several fly, nematode and mammalian WREs [40], [41; this report], it is also true that multimerized HMG sites are sufficient for Wnt responsiveness in reporters such as TOPFLASH and POPTOP [52]–[54]. However, such high density clusters (typically 4–7 are used) of perfect consensus HMG sites are not found in endogenous WREs [9], [11], [52]. While these synthetic reporters are useful tools for studying Wnt signaling in many systems, they do not reflect the situation in naturally occurring WREs, where the lower density and degeneracy of HMG sites requires additional mechanisms to increase the DNA binding specificity of TCFs. For TCF family members containing a C-clamp, the presence of Helper sites near HMG sites is one such mechanism.
There are several similarities between the DNA binding sites for POP-1, TCF/Pan and vertebrate TCFs, but there are also some important differences. While the five functional HMG sites examined in this report have a consensus that is similar to other TCFs (Figure 8A) [11], [38], there are some differences, most notably HMG sites in the ceh-22b and psa-3 WREs with CTTTTG instead of the traditional CTTTG (Figure 1A, 1B). POP-1 can bind to “classic” HMG sites used in POPTOP [77], but can also tolerate an extra T in the core of the HMG site. Whether this property is unique to POP-1 requires further study.
Fig. 8. POP-1 consensus HMG and Helper sites and models for the TCF transcriptional switch. 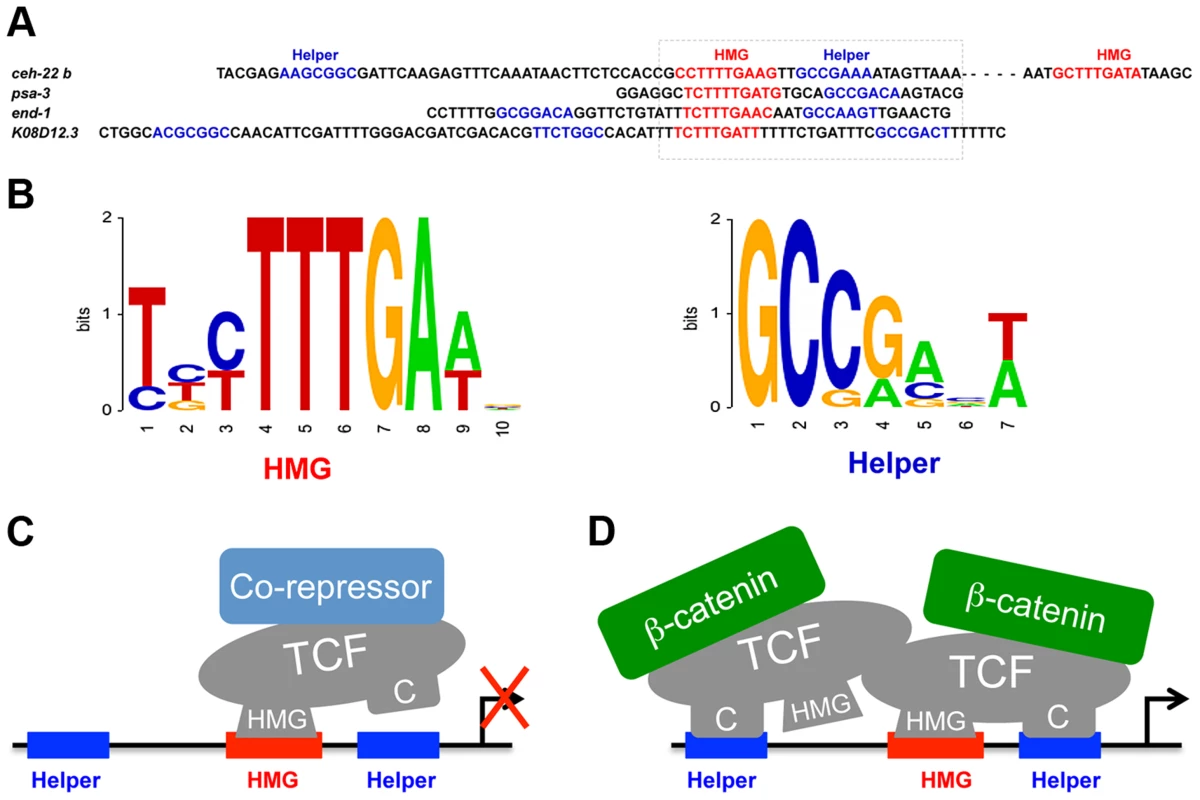
(A) Genomic sequences of the functional HMG and Helper sites, with the box indicating a HMG-Helper site pair with similar orientation in each WRE. (B) Sequence logos showing the consensus of HMG and Helper sites, based on the functional sites used in this study. (C, D) Model to explain the differential requirement of HMG and Helper sites in the TCF transcriptional switch, without (C) and with (D) Wnt/ß-catenin signaling . We propose that the DNA binding properties of TCF are influenced by co-regulators, with ß-catenin stabilizing the HMG-Helper site interaction. It is suggested that POP-1 may recognize HMG sites surrounded by two Helper sites as a dimer. This model does not preclude the existence of addition DNA-binding co-factors for POP-1 in either the absence or presence of signaling. The functional Helper sites identified in this report are also similar but different from those in other systems. The C. elegans Helper consensus shown in Figure 7B, derived from the eight functional sites, is GCCRAnW (R = A/G; W = A/T). This is slightly different from the fly (GCCGCCR) and vertebrate (GCSGS) consensus sites [11]. The C-clamp domain of POP-1 is the most diverged of all the sequenced metazoan TCFs [9]. For example, the C-clamps of POP-1 and TCF/Pan are 59% identical/72% conserved, while TCF/Pan and human TCF4E are 83% identical/90% conserved. A more systematic comparison of Helper site binding from different TCF family members is required to determine if the above differences are due to intrinsic differences in their C-clamp domains.
Functional Helper sites are usually found near functional HMG sites in WREs [40], [41]. Mutation of individual HMG and Helper sites in fly WREs indicates that they act in pairs (Chang and Cadigan, unpublished observations) and this view is supported by studies of TCF binding in vitro, where the TCFs containing a C-clamp bind to adjacent HMG and Helper sites with high affinity [39]–[41]. However, the analysis of POP-1 binding clearly demonstrates that multiple Helper sites can augment binding of POP-1 to DNA containing a HMG site (Figure 3F; Figure S1A). An alignment of the four Helper-HMG clusters is shown in Figure 8A. All four WREs have a HMG-Helper pair in similar orientation, while three have additional Helper sites that contribute to in vivo WRE activity (Figure 2, 3 & 6). The mechanism by which POP-1 binds to multiple Helper sites and a single HMG site is not clear, though it seems likely that POP-1 homo-oligomerization is involved (Figure 8C; Bhambhani and Cadigan, unpublished results). While additional experiments are needed to resolve this issue, it seems clear that these results should be taken into account in designing computational searches for additional WREs.
To test whether knowledge of the importance of Helper sites can enhance our ability to detect WREs in silico, we conducted a genome-wide search for Helper-HMG site clusters. After secondary screening (see Results and Table S3) three candidates were tested for POP-1 binding (Figure 3F; Figure S2) and the one with strong binding (K08D12.3/ZNF9) was tested in a transgenic GFP reporter assay (Figure 3A). The reporter was active in several muscle cell types and the gut (Figure 3B), a pattern that has considerable overlap with a POP-1 transcriptional fusion [78]. Indeed, mutation of the HMG and Helper sites in the K08D12.3 reporter caused a dramatic reduction in expression (Figure 3C, 3D). These results provide a proof of principle that novel POP-1 targets can be identified through HMG-Helper site directed computational searches.
There are some well-known targets of Wnt/ß-catenin signaling in C. elegans where WREs have not been identified, e.g., mab-5 and lin-39 [77], [79]–[82]. It is possible that these are indirect targets of the pathway, but an alternative is that the bona fide POP-1 binding sites have been missed because they contain sub-optimal HMG sites combined with multiple Helper sites. For example, one of the HMG-Helper site clusters identified in our computational search (among the initial 115 hits) is upstream of the pha-4 gene [83], [84] (Figure S6A), whose expression is POP-1 dependent [85]. egl-18 was also recently reported to be a direct target of POP-1, based on a partial reduction in reporter expression when a single HMG site was mutated [86]. Examination of the region near this site revealed another HMG-Helper site cluster nearby (Figure S7). While these observations require experimental validation, our work strongly suggests that the presence of Helper sites will in many cases facilitate the identification of functional POP-1 binding sites in Wnt targets.
The TCF Transcriptional Switch
In this study, we have shown that four worm WREs require Helper sites for maximal expression (Figure 2; Figure 3; Figure 6). For one of these, plus a fly WRE where mutation of HMG sites resulted in substantial derepression of expression (Figure 6B; Figure 7E), Helper sites made little or no contribution to TCF basal repression (Figure 6C; Figure 7H). In addition, a TCF rescue assay in the developing fly wing demonstrated that the C-clamp was required for activation of Wnt readouts (Figure 7; Table 2) but was dispensable for repression of ectopic bristles (Figure 7L, 7L′; Table 2). Taken together, these results support a model where TCFs repress Wnt target gene expression in the absence of signaling through HMG domain-HMG site binding, but require HMG domain-HMG site and C-clamp-Helper site interactions to mediate activation (Figure 8C). Interestingly, a vertebrate TCF (TCF3) which is associated with repression of Wnt targets in the absence of signaling [87], [88] does not have a C-clamp, perhaps supporting the dispensability of C-clamp-Helper sites in basal repression.
It should be noted that a prior report has implicated the C-clamp as being required for the ability of POP-1 to repress endoderm genes in MS blastomeres and their descendants [23]. However, the deletion constructs used removed additional portions of POP-1 besides the C-clamp, and the precise region required for endoderm repression was not identified [23]. To resolve the discrepancy between this work and our results, more surgical mutations in the C-clamp should be employed to test its role in basal repression of Wnt targets.
One model to explain our data is depicted in Figure 8C. In this scenario, TCF DNA binding is allosterically regulated by associated co-regulators. Co-repressors would promote HMG domain-HMG site binding while ß-catenin would stimulate binding to both DNA motifs (Figure 8C). It is known that the C-clamp increases the affinity of TCFs for DNA containing HMG and Helper sites [39], [40], so this model predicts that TCFs would bind DNA more efficiently in the presence of high levels of nuclear ß-catenin. Indeed, in fly cells and tissues, Wg signaling causes a robust increase in TCF binding to WRE chromatin, as measured by Chromatin Immuno-Precipitation (ChIP) [89]–[92]. This increase in TCF binding is dependent on Armadillo, the fly ß-catenin [92]. This is in contrast to human LEF1, which lacks a C-clamp, where Wnt/ß-catenin signaling has no effect on the level of LEF-1 binding to a c-myc WRE [93]. Further biochemical analysis is required to determine the exact mechanisms involved in the differential requirement of Helper sites in the TCF transcriptional switch.
Transcriptional switches are a common mechanism for transcription factors (TFs) working in cell-cell signaling pathways [1]–[3]. There are hints that the DNA-binding properties of some of these TFs may be controlled by signaling in a manner analogous with TCFs. For example, the genome-wide distribution of retinoic acid receptor (RAR) is dramatically altered by ligand in differentiating mouse embryonic stem cells [94]. In another case, fly CSL (also known as Suppressor of Hairless), the TF that mediates Notch signaling, displays a transient increase in binding to Notch regulatory elements upon signal stimulation [95]. Interestingly, co-activator binding to mammalian CSL has been shown to stabilize binding to dimeric CSL binding sites [96]. Whether different binding sites contribute to the RAR or CSL transcriptional switches is not known. Given that fly and worm TCFs have two distinct DNA binding domains and DNA binding sites, their differential requirements in the transcriptional switch may be more readily discernable. But it is possible that other TFs operating as transcriptional switches have similar differential binding requirements in the absence or presence of signaling.
Role of POP-1 in Post-embryonic Gut Physiology
While POP-1 is known to play important roles in gut development during embryogenesis [15], [18], [22], [73], its role in post-embryonic gut development is poorly understood. Our synthetic POPHHOP fluorescent reporter displays strong expression in ‘int9’ cells, the posterior most intestinal cells in the larval and adult stages (Figures 5A, 5G). This expression pattern is pop-1 dependent (Figure 5H) and it has been reported that POP-1 is expressed in these cells during larval development [61]. Since these cells have been known to play an important role in the defecation behavior of C. elegans [56], we tested if POP-1 had any role in post-embryonic gut physiology and found that POP-1 regulates the frequency and regularity of the defecation cycle (Figures 5I, 5J, 5K).
The defecation behavior is comprised of rhythmic contractions of the posterior body wall muscles, followed by the anterior body wall muscle and eventually the enteric muscles expelling the fecal matter. Under conditions of constant feeding, these steps are highly regular [57] and are controlled by neuronal and non-neuronal signals [97]. The intestine is thought to be the pacemaker for this process [56], [58]–[60] with int9 cells exhibiting rhythmic calcium fluxes which initiate each cycle [56]. Hypomorphic pop-1 mutants and animals with intestine-specific RNAi depletion of pop-1 have a prolonged cycle and display significant arrhythmia (Figures 5I–5L). These data, along with the expression of POPHHOP in int9 cells suggests that POP-1 is required in these cells for pacemaker function. Since both the pop-1(hu9) [63], [64] and pop-1(q645) [50] alleles are point mutations in the ß-catenin binding domain, it is likely that POP-1 is working with Wnt/ß-catenin signaling to control this rhythmic behavior. The Wnt genes egl-20, cwn-1 and lin-44 are expressed close to or overlapping the int9 cells [98], [99] and SYS-1 is important for attachment of the posterior intestine to the rectum [18]. Hence they could be contributing to the non-neuronal pacemaker function of these posterior intestinal cells to regulate the pBoc step of the defecation cycle.
What are the downstream targets of Wnt/ß-catenin signaling in int9 cells? Based on our behavioral analysis, pop-1 is in a class of mutants that show defecation cycle length and expulsion defects (Figure 5I-5L and Table 1). Of particular interest, loss of function mutations in a phospholipase Cβ (egl-8; Gene ID: 178537) lead to a dramatic increase in the cycle length with mutants displaying severe arrhythmia and expulsion defects [57], [59], [100]. pop-1 mutants show a subtle phenotype compared to egl-8, which could be attributed to the hypomorphic alleles used in our study. Interestingly, egl-8 is expressed in the posterior intestine [100] and we found two HMG-Helper site clusters in the intronic regions of egl-8 that could contribute to this pattern (Figure S6B). It will be interesting to see if the intestinal pattern of egl-8 is dependent on POP-1 and whether the HMG-Helper sites we identified are functional.
There is clearly more work to be done in elucidating the role of Wnt/ß-catenin signaling in the C. elegans defecation cycle. For example, are the calcium fluxes originating from the posterior intestinal region [56], [58], [59] affected in pop-1 mutants? While this and other questions remain, this report provides a powerful example of how knowledge of Helper sites working with HMG sites can not only lead to finding novel POP-1 targets, but also a better understanding of Wnt biology in C.elegans.
Materials and Methods
Plasmids
For C. elegans WRE reporters, specific mutations introduced into each HMG site and Helper site are listed in Supplemental Table S1. Site directed mutagenesis for all constructs was performed using the Quickchange II Kit (Stratagene). The WT sequences were replaced by subcloning the HMG or Helper mutant fragment into the PstI and BamHI site of ceh-22b::VENUS plasmid [42] (pJK1082, kindly provided by Dr. Judith Kimble), the PstI sites of psa-3::GFP plasmid [43] (kindly provided by Dr. Hitoshi Sawa) or the NaeI and AvrII sites of end-1::GFP::H2B plasmid [16] (kindly provided by Dr. Rueyling Lin). To generate the K08D12.3::VENUS plasmid, ceh-22b WRE was replaced by a 585 bp region spanning the TlSS and sequence upstream of the K08D12.3 gene. PCR based cloning was used to insert this fragment at the SphI and SmaI site of the ceh-22b::VENUS plasmid. The fragment was amplified using the reverse primer 5′GCCAATCCCGGGGATCCTTTCTGTCCGAGATTACTGCAA3′ with the start codon mutated (underlined) [48] and forward primer 5′TCGAAGCATGCCTGCAGCCGATTGCCGGAATGGCTTTGCGC3′. The POPHHOP plasmid was generated by cloning 6×HMG-Helper (6× GGAAGATCAAAGGGGGTAGCCGCCAGT) [40] upstream of a NLS-GFP in pPD107.94 (also known as L3135) vector using the NheI sites. 6×Histagged full length POP-1 plasmid (pRSETA-POP-1) was kindly provided by Dr. David M. Eisenmann and the C-clamp mutant used in Figure 4 was generated from this plasmid. The pxb-lacZ plasmid was generated by subcloning the pxb cluster3 WRE into the pH-Pelican vector as described previously [40] . pUAS LEF1V5 and pUAS LEF1C-clamp V5 were generated by PCR based cloning into pUAST vector. pUAS LEF1-V5 was generated by subcloning a human LEF1 fragment from an expression plasmid (kindly provided by Dr. Marian L. Waterman) into pActin5.1 to introduce a V5 tag. PCR based amplification of LEF1 was carried out using forward primer 5′CCCCGGTACCATGCCCCAACTCTCCGGA3′ and reverse primer 5′CAGTGAATTCTGCGATGTAGGCAGCTGTCATTCTTGG3′ and inserted into the KpnI and EcoRI sites of the pActin5.1 vector, with the stop codon mutated (underlined). Lef1V5 was digested using the KpnI and PmeI sites and inserted into the KpnI and XbaI sites of pUAST. Prior to insertion, pUAST vector was restricted with XbaI and sticky ends filled in by Klenow to create blunt ends, followed by a restriction with KpnI. pUAS-Lef1C-clamp V5 plasmid was generated by PCR based amplification of the C-clamp (i.e., KKCRARFGLDQQSQWCKPCRRKKKCIRYMEAL) from fly TCF/Pan and insertion into the EcoRI site of pUAS Lef1V5 by non-directional cloning.
C. elegans and D. melanogaster Transgenics and Genetics
Worm strains were derived from the wild-type C.elegans N2 Bristol strain and cultured using standard protocols. Transgenic strains with extrachromosomal arrays were generated by injecting WT or mutant versions of ceh-22b::VENUS (100 ng/µl), psa-3::GFP (50 ng/µl), end-1::GFP::H2B (100 ng/µl) or K08D12.3::VENUS (150 ng/µl) plasmid into N2 worms, along with coinjection marker myo-2::RFP plasmid (3 ng/µl) and pActin5.1 (up to a total of 200 ng/µl). Stable integrants were generated by UV irradiation using a Stratalinker (Stratagene) at power 325. POPHHOP plasmid (1 ng/µl) was injected along with a dpy-20(+) plasmid (50 ng/µl) and pBluescript (100 ng/µl) and stable integrants generated by gamma-irradiation. Animals with integrated transgenes were outcrossed at least three times. Transgenic POPTOP (7× HMG::mCherry) strain was kindly provided by Dr. Paul W. Sternberg [55]. It should be noted that the proximal promoter and 3′ UTR of POPHHOP and the POPTOP constructs are different, which could account for some of the differences in expression of these two synthetic reporters. POPHHOP was crossed into a pop1(hu9) [64] background and analyzed at different stages. All reporter strains were maintained at 25°C except for psa-3::GFP transgenic analysis during which synchronized L1s were grown at 20°C for 9 hours only for the reporter analysis. pop-1(q645) strain (JK2944) was obtained from Caenorhabatitis Genetics Center (CGC).
Transgenic pxb-lacz, Lef1 and Lef1 C-clamp flies were generated by P-element transgenesis (performed by BestGene Inc.). w1118 was obtained from Bloomington Stock Center. C96::Gal4 was kindly provided by Dr. Rolf Bodmer [71]. The TCF/Pan RNAi line (#25940) was obtained from Vienna Drosophila RNAi Center. All fly crosses were performed at 25°C.
Imaging
Methods for mounting and viewing C.elegans larvae and embryos by Nomarski optics have been described previously [101], [102]. ceh-22b::VENUS, psa-3::GFP, end-1::GFP::H2B, K08D12.3::VENUS and myo-2::RFP reporter expression was analyzed by fluorescence on a Olympus BX61 motorized X-drive microscope. Images were taken using a Hamamatsu ORGA-ERCA-CCD camera, with a specific exposure time for each WRE reporter and multiple focal planes were merged to obtain the representative image. Deconvolution was performed using slidebook 5.0 software and the nearest neighbors method. POPHHOP and POPTOP reporter expression was analyzed using a Zeiss Axioscope microscope equipped with a Zeiss Axiocam digital camera. pxb-lacZ WRE images were obtained using a Leica triple channel confocal microscope DM6000B-CS and multiple focal planes were merged to obtain the representative image. All images were processed using Adobe Photoshop 8.0.
EMSA
Full-length 6×His-POP-1 was expressed in E.Coli and purified using nickel beads (Sigma). ESMA was performed as described previously [40] using 6% native gels. POP-1 (300–900 ng unless otherwise indicated) in 10% glycerol and the biotin labeled DNA probes (4–8 femtomoles unless otherwise indicated) were incubated with 50 ug/ml poly (dI-dC), 0.05% NP-40, 5 mMMgCl2 and 2 µl of 50% glycerol in the presence of binding buffer (10 mM Tris–HCl, pH 7.5, 50 mM KCl, 1 mM DTT) in a final volume of 20 µl for 5 min on ice and 25 min at room temperature. For the competition assays, unlabeled probes were incubated with the reaction mixture containing POP-1 for 10 min prior to adding the labeled probe.
Fixation, Immunostains and Immunoblots
For ceh-22b::VENUS fluorescence analysis, worm larvae were fixed in 4% paraformaldehyde (in M9) for 15 min, using a protocol adapted from the whole-mount freeze-cracking method [103], without any immunostaining. For pxb-lacZ WRE analysis, fly embryos were fixed and immunostained using anti-lacZ and anti-Wg as described previously [40]. For comparing Lef1 and Lef1 C-clamp expression, wing imaginal discs from third instar larvae were dissected, immediately suspended in boiling 5×-Laemmli buffer, homogenized and boiled for 5 min. Immunoblots were performed using anti-V5 and anti-tubulin as described previously [104].
Behavioral Analysis
N2 worms and pop-1 mutants were maintained at 20°C, and scored for defecation at room temperature (∼23°C) using a Leica MZ16 F stereoscope at a 115× magnification. The protocol was adapted from a previous report [57]. Briefly, ten larvae (from synchronized L1s grown at 20°C for 15 hours) were transferred to a seeded NGM plate each time and allowed to settle down at 20°C for one hour followed by at least twenty minutes at room temp before assaying them for defecation at early L2. Pharyngeal pumping was observed for at least one minute in each worm before starting the defecation assay, to ensure the overall health of the animal and to confirm that they were not in the L1/L2 lethargus stage. Defecation cycle length was defined as the time between two consecutive pBocs [105]. Each larva was scored for eight consecutive cycles (nine consecutive pBocs) and the mean was calculated. The mean cycle lengths were used to calculate the mean, median and standard deviations for each genotype. Expulsion was observed at ∼three seconds after a pBoc. pop-1(hu9) worms were maintained as a homozygotes [64] and scored for defecation. The pop-1(q645) (JK2944; CGC) strain segregates as WT - green-fluorescing heterozygotes, and non-fluorescing homozygotes. pop-1(q645) hermaphrodites are sterile due to severe gonadal development defects [50]. Non-fluorescing pop-1(q645) larvae were scored for defecation cycles, transferred to individual plates and allowed to grow to adulthood to confirm that they were sterile and had a protruding vulva [50]. Four homozygotes for each allele were genotyped to confirm the point mutations for pop-1(hu9) (G to A)(E47 to K) [64] and pop-1(q645) (T to A)(D9 to E) [50] in the N-terminal β-catenin binding domain using forward primer 5′CTCCATGGCCTAACTTCCGCGGACC3′ and reverse primer 5′GTCGAAAGGCAATTGAGGTGGTCC 3′.
RNAi feeding of N2 and OLB11 strains were performed as described [106] using the pop-1 dsRNA expressing strain from the Ahringer RNAi library [51].
Wing Mounting
Adult flies were stored in 70% ethanol and soaked in 100% ethanol for two hours before dissection in 100% ethanol. Dissected wings were transferred into Xylene (Fisher) and mounted on a slide with Permount (Fisher). Wing notch and bristle images were obtained using the Nikon Eclipse 800 microscope. All images were processed using Adobe Photoshop 8.0.
Supporting Information
Zdroje
1. BaroloS, PosakonyJW (2002) Three habits of highly effective signaling pathways: principles of transcriptional control by developmental cell signaling. Genes Dev 16 : 1167–1181.
2. Bray S, Bernard F (2010) Notch Targets and Their Regulation. In: Raphael K, editor. Current Topics in Developmental Biology: Academic Press. pp. 253–275.
3. BaniahmadA (2005) Nuclear hormone receptor co-repressors. J Steroid Biochem Mol Biol 93 : 89–97.
4. MorelV, LecourtoisM, MassianiO, MaierD, PreissA, et al. (2001) Transcriptional repression by suppressor of hairless involves the binding of a hairless-dCtBP complex in Drosophila. Curr Biol 11 : 789–792.
5. HalfonMS, CarmenaA, GisselbrechtS, SackersonCM, JiménezF, et al. (2000) Ras Pathway Specificity Is Determined by the Integration of Multiple Signal-Activated and Tissue-Restricted Transcription Factors. Cell 103 : 63–74.
6. GhislettiS, HuangW, JepsenK, BennerC, HardimanG, et al. (2009) Cooperative NCoR/SMRT interactions establish a corepressor-based strategy for integration of inflammatory and anti-inflammatory signaling pathways. Genes Dev 23 : 681–693.
7. CadiganKM (2012) TCFs and Wnt/β-catenin signaling: more than one way ot throw the switch. Transcriptional Switches during Development 98 : 1–34.
8. GrigoryanT, WendP, KlausA, BirchmeierW (2008) Deciphering the function of canonical Wnt signals in development and disease: conditional loss - and gain-of-function mutations of beta-catenin in mice. Genes Dev 22 : 2308–2341.
9. ArchboldHC, YangYX, ChenL, CadiganKM (2012) How do they do Wnt they do?: regulation of transcription by the Wnt/β-catenin pathway. Acta Physiologica 204 : 74–109.
10. PolakisP (2012) Wnt signaling in cancer. Cold Spring Harb Perspect Biol 4: a008052.
11. CadiganKM, WatermanML (2012) TCF/LEFs and Wnt signaling in the nucleus. Cold Spring Harb Perspect Biol 4: a007906.
12. CadiganKM, PeiferM (2009) Wnt signaling from development to disease: insights from model systems. Cold Spring Harb Perspect Biol 1: a002881.
13. MacDonaldBT, TamaiK, HeX (2009) Wnt/beta-catenin signaling: components, mechanisms, and diseases. Dev Cell 17 : 9–26.
14. ValentaT, HausmannG, BaslerK (2012) The many faces and functions of β-catenin. EMBO J 31 : 2714–2736.
15. MaduroMF, KasmirJJ, ZhuJ, RothmanJH (2005) The Wnt effector POP-1 and the PAL-1/Caudal homeoprotein collaborate with SKN-1 to activate C. elegans endoderm development. Dev Biol 285 : 510–523.
16. ShettyP, LoMC, RobertsonSM, LinRL (2005) C-elegans TCF protein, POP-1, converts from repressor to activator as a result of Wnt-induced lowering of nuclear levels. Developmental Biology 285 : 584–592.
17. MaduroMF, Broitman-MaduroG, MengarelliI, RothmanJH (2007) Maternal deployment of the embryonic SKN-1→MED-1,2 cell specification pathway in C. elegans. Dev Biol 301 : 590–601.
18. HuangSY, ShettyP, RobertsonSM, LinR (2007) Binary cell fate specification during C-elegans embryogenesis driven by reiterated reciprocal asymmetry of TCF POP-1 and its coactivator beta-catenin SYS-1. Development 134 : 2685–2695.
19. LinR, ThompsonS, PriessJR (1995) pop-1 encodes an HMG box protein required for the specification of a mesoderm precursor in early C. elegans embryos. Cell 83 : 599–609.
20. MaduroMF, LinR, RothmanJH (2002) Dynamics of a developmental switch: recursive intracellular and intranuclear redistribution of Caenorhabditis elegans POP-1 parallels Wnt-inhibited transcriptional repression. Dev Biol 248 : 128–142.
21. LinKT, Broitman-MaduroG, HungWW, CervantesS, MaduroMF (2009) Knockdown of SKN-1 and the Wnt effector TCF/POP-1 reveals differences in endomesoderm specification in C. briggsae as compared with C. elegans. Dev Biol 325 : 296–306.
22. OwraghiM, Broitman-MaduroG, LuuT, RobersonH, MaduroMF (2010) Roles of the Wnt effector POP-1/TCF in the C. elegans endomesoderm specification gene network. Dev Biol 340 : 209–221.
23. RobertsonSM, LoMC, OdomR, YangXD, MedinaJ, et al. (2011) Functional analyses of vertebrate TCF proteins in C. elegans embryos. Dev Biol 355 : 115–123.
24. CavalloRA, CoxRT, MolineMM, RooseJ, PolevoyGA, et al. (1998) Drosophila Tcf and Groucho interact to repress Wingless signalling activity. Nature 395 : 604–608.
25. SchweizerL, NellenD, BaslerK (2003) Requirement for Pangolin/dTCF in Drosophila Wingless signaling. Proc Natl Acad Sci U S A 100 : 5846–5851.
26. RieseJ, YuX, MunnerlynA, EreshS, HsuSC, et al. (1997) LEF-1, a nuclear factor coordinating signaling inputs from wingless and decapentaplegic. Cell 88 : 777–787.
27. KnirrS, FraschM (2001) Molecular integration of inductive and mesoderm-intrinsic inputs governs even-skipped enhancer activity in a subset of pericardial and dorsal muscle progenitors. Dev Biol 238 : 13–26.
28. YangX, van BeestM, CleversH, JonesT, HurshDA, et al. (2000) decapentaplegic is a direct target of dTcf repression in the Drosophila visceral mesoderm. Development 127 : 3695–3702.
29. CalvoD, VictorM, GayF, SuiG, LukeMP, et al. (2001) A POP-1 repressor complex restricts inappropriate cell type-specific gene transcription during Caenorhabditis elegans embryogenesis. EMBO J 20 : 7197–7208.
30. PhillipsBT, KimbleJ (2009) A New Look at TCF and beta-Catenin through the Lens of a Divergent C-elegans Wnt Pathway. Developmental Cell 17 : 27–34.
31. JacksonBM, EisenmannDM (2012) beta-catenin-dependent Wnt signaling in C. elegans: teaching an old dog a new trick. Cold Spring Harb Perspect Biol 4: a007948.
32. SawaH (2012) Control of cell polarity and asymmetric division in C. elegans. Transcriptional Switches during Development 101 : 55–76.
33. MeneghiniMD, IshitaniT, CarterJC, HisamotoN, Ninomiya-TsujiJ, et al. (1999) MAP kinase and Wnt pathways converge to downregulate an HMG-domain repressor in Caenorhabditis elegans. Nature 399 : 793–797.
34. RocheleauCE, YasudaJ, ShinTH, LinRL, SawaH, et al. (1999) WRM-1 activates the LIT-1 protein kinase to transduce anterior posterior polarity signals in C-elegans. Cell 97 : 717–726.
35. ShinTH, YasudaJ, RocheleauCE, LinRL, SotoM, et al. (1999) MOM-4, a MAP kinase kinase kinase-related protein, activates WRM-1/LIT-1 kinase to transduce anterior/posterior polarity signals in C-elegans. Molecular Cell 4 : 275–280.
36. LoMC, GayF, OdomR, ShiY, LinF (2004) Phosphorylation by the beta-catenin/MAPK complex promotes 14-3-3-mediated nuclear export of TCF/POP-1 in signal-responsive cells in C-elegans. Cell 117 : 95–106.
37. van de WeteringM, CavalloR, DooijesD, van BeestM, van EsJ, et al. (1997) Armadillo coactivates transcription driven by the product of the Drosophila segment polarity gene dTCF. Cell 88 : 789–799.
38. HallikasO, PalinK, SinjushinaN, RautiainenR, PartanenJ, et al. (2006) Genome-wide prediction of mammalian enhancers based on analysis of transcription-factor binding affinity. Cell 124 : 47–59.
39. AtchaFA, SyedA, WuB, HoverterNP, YokoyamaNN, et al. (2007) A unique DNA binding domain converts T-cell factors into strong Wnt effectors. Mol Cell Biol 27 : 8352–8363.
40. ChangMV, ChangJL, GangopadhyayA, ShearerA, CadiganKM (2008) Activation of wingless targets requires bipartite recognition of DNA by TCF. Curr Biol 18 : 1877–1881.
41. HoverterNP, TingJH, SundareshS, BaldiP, WatermanML (2012) A WNT/p21 circuit directed by the C-clamp, a sequence-specific DNA binding domain in TCFs. Mol Cell Biol 32 : 3648–3662.
42. LamN, ChesneyMA, KimbleJ (2006) Wnt signaling and CEH-22/tinman/Nkx2.5 specify a stem cell niche in C-elegans. Current Biology 16 : 287–295.
43. ArataY, KouikeH, ZhangYP, HermanMA, OkanoH, et al. (2006) Wnt signaling and a Hox protein cooperatively regulate PSA-3/Meis to determine daughter cell fate after asymmetric cell division in C-elegans. Developmental Cell 11 : 105–115.
44. KimbleJE, WhiteJG (1981) On the control of germ cell development in Caenorhabditis elegans. Developmental Biology 81 : 208–219.
45. SosinskyA, BoninCP, MannRS, HonigB (2003) Target Explorer: an automated tool for the identification of new target genes for a specified set of transcription factors. Nucleic Acids Research 31 : 3589–3592.
46. GaudetJ, MuttumuS, HornerM, MangoSE (2004) Whole-genome analysis of temporal gene expression during foregut development. Plos Biology 2: e352.
47. Hunt-NewburyR, ViveirosR, JohnsenR, MahA, AnastasD, et al. (2007) High-throughput in vivo analysis of gene expression in Caenorhabditis elegans. Plos Biology 5 : 1981–1997.
48. DupuyD, BertinN, HidalgoCA, VenkatesanK, TuD, et al. (2007) Genome-scale analysis of in vivo spatiotemporal promoter activity in Caenorhabditis elegans. Nature Biotechnology 25 : 663–668.
49. SleumerMC, BilenkyM, HeA, RobertsonG, ThiessenN, et al. (2009) Caenorhabditis elegans cisRED: a catalogue of conserved genomic elements. Nucleic Acids Research 37 : 1323–1334.
50. SiegfriedKR, KimbleJ (2002) POP-1 controls axis formation during early gonadogenesis in C-elegans. Development 129 : 443–453.
51. KamathRS, AhringerJ (2003) Genome-wide RNAi screening in Caenorhabditis elegans. Methods 30 : 313–321.
52. BaroloS (2006) Transgenic Wnt/TCF pathway reporters: all you need is Lef? Oncogene 25 : 7505–7511.
53. KorinekV, BarkerN, MorinPJ, van WichenD, de WegerR, et al. (1997) Constitutive Transcriptional Activation by a β-Catenin-Tcf Complex in APC−/ − Colon Carcinoma. Science 275 : 1784–1787.
54. DasGuptaR, FuchsE (1999) Multiple roles for activated LEF/TCF transcription complexes during hair follicle development and differentiation. Development 126 : 4557–4568.
55. GreenJL, InoueT, SternbergPW (2008) Opposing Wnt pathways orient cell polarity during organogenesis. Cell 134 : 646–656.
56. TeramotoT, IwasakiK (2006) Intestinal calcium waves coordinate a behavioral motor program in C. elegans. Cell Calcium 40 : 319–327.
57. ThomasJH (1990) Genetic analysis of defecation in Caenorhabditis elegans. Genetics 124 : 855–872.
58. Dal SantoP, LoganMA, ChisholmAD, JorgensenEM (1999) The inositol trisphosphate receptor regulates a 50-second behavioral rhythm in C. elegans. Cell 98 : 757–767.
59. EspeltMV, EstevezAY, YinX, StrangeK (2005) Oscillatory Ca2+ signaling in the isolated Caenorhabditis elegans intestine: role of the inositol-1,4,5-trisphosphate receptor and phospholipases C beta and gamma. J Gen Physiol 126 : 379–392.
60. WangH, GirskisK, JanssenT, ChanJP, DasguptaK, et al. (2013) Neuropeptide secreted from a pacemaker activates neurons to control a rhythmic behavior. Curr Biol 23 : 746–754.
61. LinRL, HillRJ, PriessJR (1998) POP-1 and anterior-posterior fate decisions in C-elegans embryos. Cell 92 : 229–239.
62. KingRS, MaidenSL, HawkinsNC, KiddAR, KimbleJ, et al. (2009) The N - or C-terminal domains of DSH-2 can activate the C. elegans Wnt/beta-catenin asymmetry pathway. Developmental Biology 328 : 234–244.
63. GleasonJE, KorswagenHC, EisenmannDM (2002) Activation of Wnt signaling bypasses the requirement for RTK/Ras signaling during C-elegans vulval induction. Genes & Development 16 : 1281–1290.
64. KorswagenHC, CoudreuseDYM, BetistMC, van de WaterS, ZivkovicD, et al. (2002) The Axin-like protein PRY-1 is a negative regulator of a canonical Wnt pathway in C-elegans. Genes & Development 16 : 1291–1302.
65. McGheeJD, FukushigeT, KrauseMW, MinnemaSE, GoszczynskiB, et al. (2009) ELT-2 is the predominant transcription factor controlling differentiation and function of the C. elegans intestine, from embryo to adult. Developmental Biology 327 : 551–565.
66. PilipiukJ, LefebvreC, WiesenfahrtT, LegouisR, BossingerO (2009) Increased IP3/Ca2+ signaling compensates depletion of LET-413/DLG-1 in C. elegans epithelial junction assembly. Developmental Biology 327 : 34–47.
67. PhillipsRG, WhittleJR (1993) wingless expression mediates determination of peripheral nervous system elements in late stages of Drosophila wing disc development. Development 118 : 427–438.
68. CousoJP, BishopSA, Martinez AriasA (1994) The wingless signalling pathway and the patterning of the wing margin in Drosophila. Development 120 : 621–636.
69. BlairSS (1992) Shaggy (zeste-white 3) and the formation of supernumerary bristle precursors in the developing wing blade of Drosophila. Dev Biol 152 : 263–278.
70. CadiganKM, FishMP, RulifsonEJ, NusseR (1998) Wingless repression of Drosophila frizzled 2 expression shapes the Wingless morphogen gradient in the wing. Cell 93 : 767–777.
71. KruppJJ, YaichLE, WessellsRJ, BodmerR (2005) Identification of genetic loci that interact with cut during Drosophila wing-margin development. Genetics 170 : 1775–1795.
72. DietzlG, ChenD, SchnorrerF, SuKC, BarinovaY, et al. (2007) A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature 448 : 151–156.
73. PhillipsBT, KiddAR, KingR, HardinJ, KimbleJ (2007) Reciprocal asymmetry of SYS-1/beta-catenin and POP-1/TCF controls asymmetric divisions in Caenorhabditis elegans. Proceedings of the National Academy of Sciences of the United States of America 104 : 3231–3236.
74. HuangL, LiX, El-HodiriHM, DayalS, WikramanayakeAH, et al. (2000) Involvement of Tcf/Lef in establishing cell types along the animal-vegetal axis of sea urchins. Dev Genes Evol 210 : 73–81.
75. AdamskaM, LarrouxC, AdamskiM, GreenK, LovasE, et al. (2010) Structure and expression of conserved Wnt pathway components in the demosponge Amphimedon queenslandica. Evol Dev 12 : 494–518.
76. DuffyDJ, PlickertG, KuenzelT, TilmannW, FrankU (2010) Wnt signaling promotes oral but suppresses aboral structures in Hydractinia metamorphosis and regeneration. Development 137 : 3057–3066.
77. KorswagenHC, HermanMA, CleversHC (2000) Distinct beta-catenins mediate adhesion and signalling functions in C-elegans. Nature 406 : 527–532.
78. Reece-HoyesJS, ShinglesJ, DupuyD, GroveCA, WalhoutAJM, et al. (2007) Insight into transcription factor gene duplication from Caenorhabditis elegans promoterome-driven expression patterns. BMC Genomics 8 : 27.
79. HunterCP, HarrisJM, MaloofJN, KenyonC (1999) Hox gene expression in a single Caenorhabditis elegans cell is regulated by a caudal homolog and intercellular signals that inhibit Wnt signaling. Development 126 : 805–814.
80. MaloofJN, WhangboJ, HarrisJM, JongewardGD, KenyonC (1999) A Wnt signaling pathway controls Hox gene expression and neuroblast migration in C-elegans. Development 126 : 37–49.
81. EisenmannDM, MaloofJN, SimskeJS, KenyonC, KimSK (1998) The beta-catenin homolog BAR-1 and LET-60 Ras coordinately regulate the Hox gene lin-39 during Caenorhabditis elegans vulval development. Development 125 : 3667–3680.
82. HermanMA (2001) C-elegans POP-1/TCF functions in a canonical Wnt pathway that controls cell migration acid in a noncanonical Wnt pathway that controls cell polarity. Development 128 : 581–590.
83. HornerMA, QuintinS, DomeierME, KimbleJ, LabouesseM, et al. (1998) pha-4, an HNF-3 homolog, specifies pharyngeal organ identity in Caenorhabditis elegans. Genes & Development 12 : 1947–1952.
84. MangoSE, LambieEJ, KimbleJ (1994) The Pha-4 Gene Is Required to Generate the Pharyngeal Primordium of Caenorhabditis-Elegans. Development 120 : 3019–3031.
85. MurrayJI, BoyleTJ, PrestonE, VafeadosD, MericleB, et al. (2012) Multidimensional regulation of gene expression in the C. elegans embryo. Genome Research 22 : 1282–1294.
86. GorrepatiL, ThompsonKW, EisenmannDM (2013) C. elegans GATA factors EGL-18 and ELT-6 function downstream of Wnt signaling to maintain the progenitor fate during larval asymmetric divisions of the seam cells. Development 140 : 2093–2102.
87. SokolSY (2011) Wnt signaling through T-cell factor phosphorylation. Cell Research 21 : 1002–1012.
88. WuC-I, HoffmanJA, ShyBR, FordEM, FuchsE, et al. (2012) Function of Wnt/β-catenin in counteracting Tcf3 repression through the Tcf3–β-catenin interaction. Development 139 : 2118–2129.
89. ChangJL, LinHV, BlauwkampTA, CadiganKM (2008) Spenito and Split ends act redundantly to promote Wingless signaling. Developmental Biology 314 : 100–111.
90. FangM, LiJ, BlauwkampT, BhambhaniC, CampbellN, et al. (2006) C-terminal-binding protein directly activates and represses Wnt transcriptional targets in Drosophila. Embo Journal 25 : 2735–2745.
91. LiJ, SutterC, ParkerDS, BlauwkampT, FangM, et al. (2007) CBP/p300 are bimodal regulators of Wnt signaling. Embo Journal 26 : 2284–2294.
92. ParkerDS, NiYYY, ChangJHL, LiJ, CadiganKM (2008) Wingless signaling induces widespread chromatin remodeling of target loci. Molecular and Cellular Biology 28 : 1815–1828.
93. SierraJ, YoshidaT, JoazeiroCA, JonesKA (2006) The APC tumor suppressor counteracts beta-catenin activation and H3K4 methylation at Wnt target genes. Genes & Development 20 : 586–600.
94. MahonyS, MazzoniEO, McCuineS, YoungRA, WichterleH, et al. (2011) Ligand-dependent dynamics of retinoic acid receptor binding during early neurogenesis. Genome Biology 12: R2.
95. KrejciA, BrayS (2007) Notch activation stimulates transient and selective binding of Su(H)/CSL to target enhancers. Genes & Development 21 : 1322–1327.
96. ArnettKL, HassM, McArthurDG, IlaganMXG, AsterJC, et al. (2010) Structural and mechanistic insights into cooperative assembly of dimeric Notch transcription complexes. Nature Structural & Molecular Biology 17 : 1312–1317.
97. BranickyR, HekimiS (2006) What keeps C-elegans regular: the genetics of defecation. Trends in Genetics 22 : 571–579.
98. HarterinkM, KimDH, MiddelkoopTC, DoanTD, van OudenaardenA, et al. (2011) Neuroblast migration along the anteroposterior axis of C. elegans is controlled by opposing gradients of Wnts and a secreted Frizzled-related protein. Development 138 : 2915–2924.
99. CoudreuseDY, RoelG, BetistMC, DestreeO, KorswagenHC (2006) Wnt gradient formation requires retromer function in Wnt-producing cells. Science 312 : 921–924.
100. LacknerMR, NurrishSJ, KaplanJM (1999) Facilitation of synaptic transmission by EGL-30 G(q)alpha and EGL-8 PLC beta: DAG binding to UNC-13 is required to stimulate acetylcholine release. Neuron 24 : 335–346.
101. SulstonJE, HorvitzHR (1977) Post-embryonic cell lineages of the nematode, Caenorhabditis elegans. Dev Biol 56 : 110–156.
102. SulstonJE, SchierenbergE, WhiteJG, ThomsonJN (1983) The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev Biol 100 : 64–119.
103. Crittenden S, Kimble J (2009) Preparation and immunolabeling of Caenorhabditis elegans. Cold Spring Harb Protoc 2009: pdb prot5216.
104. BhambhaniC, ChangJL, AkeyDL, CadiganKM (2011) The oligomeric state of CtBP determines its role as a transcriptional co-activator and co-repressor of Wingless targets. Embo Journal 30 : 2031–2043.
105. BranickyR, ShibataY, FengJL, HekimiS (2001) Phenotypic and suppressor analysis of defecation in clk-1 mutants reveals that reaction to changes in temperature is an active process in Caenorhabditis elegans. Genetics 159 : 997–1006.
106. ColletteKS, PettyEL, GolenbergN, BembenekJN, CsanskovszkiG (2011) Different roles for Aurora B in condensin targeting during mitosis and meiosis. Journal Cell Science 124 : 3684–3694.
107. HermanMA, HorvitzHR (1994) The Caenorhabditis elegans gene lin-44 controls the polarity of asymmetric cell divisions. Development 120 : 1035–1047.
Štítky
Genetika Reprodukční medicína
Článek Natural Polymorphisms in Influence Negative Selection and CD4∶CD8 Lineage Commitment in the RatČlánek MicroRNAs Located in the Hox Gene Clusters Are Implicated in Huntington's Disease PathogenesisČlánek Comparative RNAi Screens in and Reveal the Impact of Developmental System Drift on Gene FunctionČlánek Mutation of SLC35D3 Causes Metabolic Syndrome by Impairing Dopamine Signaling in Striatal D1 Neurons
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2014 Číslo 2
-
Všechny články tohoto čísla
- Fifteen Years Later: Hard and Soft Selection Sweeps Confirm a Large Population Number for HIV In Vivo
- The Same but Different: Worms Reveal the Pervasiveness of Developmental System Drift
- Serine Carboxypeptidase SCPEP1 and Cathepsin A Play Complementary Roles in Regulation of Vasoconstriction via Inactivation of Endothelin-1
- Coherent Functional Modules Improve Transcription Factor Target Identification, Cooperativity Prediction, and Disease Association
- A Long-Chain Flavodoxin Protects from Oxidative Stress and Host Bacterial Clearance
- Mammalian E-type Cyclins Control Chromosome Pairing, Telomere Stability and CDK2 Localization in Male Meiosis
- Influenza Virus Drug Resistance: A Time-Sampled Population Genetics Perspective
- Transcriptome-Wide Analyses of 5′-Ends in RNase J Mutants of a Gram-Positive Pathogen Reveal a Role in RNA Maturation, Regulation and Degradation
- Selective Disruption of Aurora C Kinase Reveals Distinct Functions from Aurora B Kinase during Meiosis in Mouse Oocytes
- X Chromosome Control of Meiotic Chromosome Synapsis in Mouse Inter-Subspecific Hybrids
- A Cohesin-Independent Role for NIPBL at Promoters Provides Insights in CdLS
- Extreme Population Differences in the Human Zinc Transporter ZIP4 (SLC39A4) Are Explained by Positive Selection in Sub-Saharan Africa
- Classic Selective Sweeps Revealed by Massive Sequencing in Cattle
- Genomic Networks of Hybrid Sterility
- Natural Polymorphisms in Influence Negative Selection and CD4∶CD8 Lineage Commitment in the Rat
- Oxidative Stress Is Not a Major Contributor to Somatic Mitochondrial DNA Mutations
- Molecular Identification of Collagen 17a1 as a Major Genetic Modifier of Laminin Gamma 2 Mutation-Induced Junctional Epidermolysis Bullosa in Mice
- Uncoupling of Molecular Maturation from Peripheral Target Innervation in Nociceptors Expressing a Chimeric TrkA/TrkC Receptor
- MicroRNAs Located in the Hox Gene Clusters Are Implicated in Huntington's Disease Pathogenesis
- Loss of Trabid, a New Negative Regulator of the Immune-Deficiency Pathway at the Level of TAK1, Reduces Life Span
- Targeted Ablation of Nesprin 1 and Nesprin 2 from Murine Myocardium Results in Cardiomyopathy, Altered Nuclear Morphology and Inhibition of the Biomechanical Gene Response
- Identification of Novel Genetic Loci Associated with Thyroid Peroxidase Antibodies and Clinical Thyroid Disease
- CEP-1, the p53 Homolog, Mediates Opposing Longevity Outcomes in Mitochondrial Electron Transport Chain Mutants
- Transcriptomics and Functional Genomics of ROS-Induced Cell Death Regulation by
- Quantitative Genome-Wide Genetic Interaction Screens Reveal Global Epistatic Relationships of Protein Complexes in
- Cascades of Genetic Instability Resulting from Compromised Break-Induced Replication
- Serine- and Threonine/Valine-Dependent Activation of PDK and Tor Orthologs Converge on Sch9 to Promote Aging
- Zfp322a Regulates Mouse ES Cell Pluripotency and Enhances Reprogramming Efficiency
- Insertional Mutagenesis and Deep Profiling Reveals Gene Hierarchies and a -Dependent Bottleneck in Lymphomagenesis
- DAAM Is Required for Thin Filament Formation and Sarcomerogenesis during Muscle Development in Drosophila
- Plasma Cholesterol–Induced Lesion Networks Activated before Regression of Early, Mature, and Advanced Atherosclerosis
- High-Resolution Profiling of Stationary-Phase Survival Reveals Yeast Longevity Factors and Their Genetic Interactions
- Comparative RNAi Screens in and Reveal the Impact of Developmental System Drift on Gene Function
- Accurate and Robust Genomic Prediction of Celiac Disease Using Statistical Learning
- Sex-Specific Embryonic Gene Expression in Species with Newly Evolved Sex Chromosomes
- Chromosome X-Wide Association Study Identifies Loci for Fasting Insulin and Height and Evidence for Incomplete Dosage Compensation
- Negative Feedback and Transcriptional Overshooting in a Regulatory Network for Horizontal Gene Transfer
- DNA Sequence Explains Seemingly Disordered Methylation Levels in Partially Methylated Domains of Mammalian Genomes
- Insights into the Genomic Landscape: Comparative Genomics Reveals Variations in Ploidy and Nutrient Utilisation Potential amongst Wine Isolates
- Molecular Evidence for the Inverse Comorbidity between Central Nervous System Disorders and Cancers Detected by Transcriptomic Meta-analyses
- The Centriolar Satellite Protein AZI1 Interacts with BBS4 and Regulates Ciliary Trafficking of the BBSome
- Fine-Mapping the Region Detects Common Variants Tagging a Rare Coding Allele: Evidence for Synthetic Association in Prostate Cancer
- Transmission Distortion Affecting Human Noncrossover but Not Crossover Recombination: A Hidden Source of Meiotic Drive
- A Variant in the Neuropeptide Receptor is a Major Determinant of Growth and Physiology
- Mutation of SLC35D3 Causes Metabolic Syndrome by Impairing Dopamine Signaling in Striatal D1 Neurons
- NSUN4 Is a Dual Function Mitochondrial Protein Required for Both Methylation of 12S rRNA and Coordination of Mitoribosomal Assembly
- MicroRNA-133 Inhibits Behavioral Aggregation by Controlling Dopamine Synthesis in Locusts
- Convergence of Light and ABA Signaling on the Promoter
- Arf4 Is Required for Mammalian Development but Dispensable for Ciliary Assembly
- Distinct Requirements for Cranial Ectoderm and Mesenchyme-Derived Wnts in Specification and Differentiation of Osteoblast and Dermal Progenitors
- Chk2 and P53 Regulate the Transmission of Healed Chromosomes in the Male Germline
- Ddc2 Mediates Mec1 Activation through a Ddc1- or Dpb11-Independent Mechanism
- Mapping the Fitness Landscape of Gene Expression Uncovers the Cause of Antagonism and Sign Epistasis between Adaptive Mutations
- Euchromatic Transposon Insertions Trigger Production of Novel Pi- and Endo-siRNAs at the Target Sites in the Germline
- miR-100 Induces Epithelial-Mesenchymal Transition but Suppresses Tumorigenesis, Migration and Invasion
- Canine Hereditary Ataxia in Old English Sheepdogs and Gordon Setters Is Associated with a Defect in the Autophagy Gene Encoding
- Within-Host Spatiotemporal Dynamics of Plant Virus Infection at the Cellular Level
- Analysis of Meiosis in SUN1 Deficient Mice Reveals a Distinct Role of SUN2 in Mammalian Meiotic LINC Complex Formation and Function
- Genome-Wide Association Study of Metabolic Traits Reveals Novel Gene-Metabolite-Disease Links
- Mechanistically Distinct Mouse Models for -Associated Retinopathy
- DAF-16/FoxO Directly Regulates an Atypical AMP-Activated Protein Kinase Gamma Isoform to Mediate the Effects of Insulin/IGF-1 Signaling on Aging in
- Chromosome I Controls Chromosome II Replication in
- Integrated Genomic Characterization Reveals Novel, Therapeutically Relevant Drug Targets in FGFR and EGFR Pathways in Sporadic Intrahepatic Cholangiocarcinoma
- The Iodotyrosine Deiodinase Ortholog SUP-18 Functions through a Conserved Channel SC-Box to Regulate the Muscle Two-Pore Domain Potassium Channel SUP-9
- The Genome of Highlights a Fish Pathogen Adapted to Fluctuating Environments
- Distinct DNA Binding Sites Contribute to the TCF Transcriptional Switch in and
- The Streamlined Genome of spp. Relative to Human Pathogenic Kinetoplastids Reveals a Parasite Tailored for Plants
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Genome-Wide Association Study of Metabolic Traits Reveals Novel Gene-Metabolite-Disease Links
- A Cohesin-Independent Role for NIPBL at Promoters Provides Insights in CdLS
- Classic Selective Sweeps Revealed by Massive Sequencing in Cattle
- Arf4 Is Required for Mammalian Development but Dispensable for Ciliary Assembly
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



