-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaOrigin and Functional Diversification of an Amphibian Defense Peptide Arsenal
The skin secretion of many amphibians contains an arsenal of bioactive molecules, including hormone-like peptides (HLPs) acting as defense toxins against predators, and antimicrobial peptides (AMPs) providing protection against infectious microorganisms. Several amphibian taxa seem to have independently acquired the genes to produce skin-secreted peptide arsenals, but it remains unknown how these originated from a non-defensive ancestral gene and evolved diverse defense functions against predators and pathogens. We conducted transcriptome, genome, peptidome and phylogenetic analyses to chart the full gene repertoire underlying the defense peptide arsenal of the frog Silurana tropicalis and reconstruct its evolutionary history. Our study uncovers a cluster of 13 transcriptionally active genes, together encoding up to 19 peptides, including diverse HLP homologues and AMPs. This gene cluster arose from a duplicated gastrointestinal hormone gene that attained a HLP-like defense function after major remodeling of its promoter region. Instead, new defense functions, including antimicrobial activity, arose by mutation of the precursor proteins, resulting in the proteolytic processing of secondary peptides alongside the original ones.
Although gene duplication did not trigger functional innovation, it may have subsequently facilitated the convergent loss of the original function in multiple gene lineages (subfunctionalization), completing their transformation from HLP gene to AMP gene. The processing of multiple peptides from a single precursor entails a mechanism through which peptide-encoding genes may establish new functions without the need for gene duplication to avoid adaptive conflicts with older ones.
Published in the journal: . PLoS Genet 9(8): e32767. doi:10.1371/journal.pgen.1003662
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1003662Summary
The skin secretion of many amphibians contains an arsenal of bioactive molecules, including hormone-like peptides (HLPs) acting as defense toxins against predators, and antimicrobial peptides (AMPs) providing protection against infectious microorganisms. Several amphibian taxa seem to have independently acquired the genes to produce skin-secreted peptide arsenals, but it remains unknown how these originated from a non-defensive ancestral gene and evolved diverse defense functions against predators and pathogens. We conducted transcriptome, genome, peptidome and phylogenetic analyses to chart the full gene repertoire underlying the defense peptide arsenal of the frog Silurana tropicalis and reconstruct its evolutionary history. Our study uncovers a cluster of 13 transcriptionally active genes, together encoding up to 19 peptides, including diverse HLP homologues and AMPs. This gene cluster arose from a duplicated gastrointestinal hormone gene that attained a HLP-like defense function after major remodeling of its promoter region. Instead, new defense functions, including antimicrobial activity, arose by mutation of the precursor proteins, resulting in the proteolytic processing of secondary peptides alongside the original ones.
Although gene duplication did not trigger functional innovation, it may have subsequently facilitated the convergent loss of the original function in multiple gene lineages (subfunctionalization), completing their transformation from HLP gene to AMP gene. The processing of multiple peptides from a single precursor entails a mechanism through which peptide-encoding genes may establish new functions without the need for gene duplication to avoid adaptive conflicts with older ones.Introduction
In response to stress or injury, many amphibians release a viscous secretion through granular glands in their skin. In several frog families, the most abundant class of secreted molecules consists of peptides and proteins. Since the 1960s, several of these peptides have been identified as structural analogues of neurohormones that are evolutionarily conserved among vertebrates and play key roles in gastrointestinal functioning. These skin-secreted hormone-like peptides (hereafter abbreviated as HLPs) have been hypothesized to provide passive defense against predation, by disturbing gastrointestinal processes upon ingestion [1], [2], [3]. Other peptides however, were found to show little similarity to any vertebrate hormone, and their function was unclear at the time of their discovery [4], [5], [6].
In 1987, two 23-AA-long peptides in the skin secretion of the African clawed frog Xenopus laevis were shown capable to kill a broad range of microorganisms [7]. Both peptides, called magainins, were cleaved from a larger precursor protein as predicted from cloned cDNA sequences [7], [8] and are thus encoded by a single gene. The discovery of similar gene-encoded antimicrobial peptides (AMPs) in other amphibians fueled the perception that these animals possess a genetically controlled arsenal of antimicrobials in their skin that provides first-line protection against infectious microorganisms in their environment. AMPs are now considered key effectors of the innate immune system of many organisms, but amphibian skin secretions continue to be explored as promising sources of potential lead compounds for the development of new antibiotics. Amphibian species in which skin AMPS have been found typically secrete 5–20 different peptides [9], [10] although in Odorrana species, the number may exceptionally exceed even 100 [11]. A recent study suggested that AMPs in distantly related anuran lineages represent independently evolved defense arsenals [12]. However, the genetic mechanisms and processes that underlay the evolutionary origin and functional diversification of any single defense arsenal remains unknown.
Frogs of the family Pipidae (including the genera Xenopus and Silurana) possess some of the best-studied skin secretions of all amphibians [13]–[23]. The model species X. laevis for example, secretes the HLPs caerulein [24], levitide [25], and xenopsin [26] and apart from the two magainins, confirmed AMPs include PGLa [4], and pGQ [27], caerulein precursor factor (CPF), and two xenopsin precursor factors (XPF). cDNA sequences have shown that HLPs and AMPs in X. laevis are posttranslationally cleaved from strikingly similar precursor proteins [8], [28], and in some cases, both types of peptide are even processed from the very same precursor [e.g. the HLP xenopsin and the AMP xenopsin precursor fragment, the HLP caerulein and the AMP caerulein precursor fragment [5], [25], [29]. Consequently, these peptides are not only subject to the same gene-regulatory mechanisms, they also share interdependent evolutionary histories.
Previoulsy, we identified a precursor gene in the species Silurana tropicalis with homology to the caerulein genes of X. laevis [30]. Phylogenetic analyses showed that they represented a single gene lineage, which evolved from the cholecystokinin (cck) hormone gene in a pipid ancestor. In the present study, we combined transcriptome, genome, and peptidome data to obtain a comprehensive overview of the entire defense peptide arsenal that evolved from this gene lineage, from the underlying genes to the active peptides. Besides identifying new peptides with potential therapeutic applications, our analyses disclose the full gene repertoire that encodes the AMP/HLP arsenal of S. tropicalis, elucidates the birth-and-death process by which it diversified, and illustrate how peptides with new functions arose in the expanding arsenal. Throughout this study, we apply the nomenclature introduced by Conlon et al. [13]–[23] to describe new peptides, and extend its use to their precursor proteins and genes.
Results
1. Charting the AMP/HLP gene repertoire of Silurana tropicalis
To chart the full repertoire of AMP genes in the S. tropicalis genome, we obtained new transcriptome data by preparing a cDNA library of skin samples of two S. tropicalis frogs. Comparative alignments and BLAST analyses of 384 randomly sequenced clones yielded 113 query sequences with homology to X. laevis mRNA transcripts and genes encoding known AMPs and HLPs (Table 1). This number corresponds to a remarkably high proportion (∼29%) of the recovered skin transcriptome. Together, they represent nine different mRNA transcripts, one of which encodes the recently described Xt6LP precursor [30] (Following the nomenclature of Conlon et al., this gene is hereafter called cpf-St7, where ‘cpf’ stands for ‘caerulein precursor fragment’, ‘St’ stands for S. tropicalis and ‘7’ indicates that it encodes the seventh CPF peptide known for this species). BLAST-screening of the S. tropicalis genome using the recovered transcripts identifies a single cluster of 15 homologous genes spanning a ∼380-kb region over the scaffolds 665 and 811 (Fig. 1A). They occur in both orientations, are composed of three (cpf-St4), four (cpf-St5, cpf-St6, and cpf-St7) or five exons (magainin-St1, xpf-St4, xpf-St5, xpf-St6, xpf-St7, xpf-St8, pgla-St2, and pgla-St3) and show considerable variation in length, ranging from ∼3.6 kb (cpf-St6) to over 23.5 kb (xpf-St5). One gene (xpf-St8p) shows signs of pseudogenization, with a degraded 5′-UTR and a premature stop-codon, while another (xpf-St7p) seems to lack a 5′-UTR and start codon altogether. The thirteen others are transcriptionally active, as evidenced by the nine query transcripts and by non-annotated EST sequences available in GenBank (Table 1; Text S1). One of these genes, cpf-St4, lacks a terminal exon but 17 GenBank EST sequences show that it is involved in the production of alternative splicing variants, by combining the cpf-St4 exons with the terminal exon of the adjacent cpf-St5 (Fig. 1A). No related genes were found on any other scaffold, suggesting that the entire AMP gene repertoire of S. tropicalis is organized in a single cluster. This cluster is flanked by the genes trak-1, ulk-4 and ctnnb-1 on scaffold 665, which in other vertebrates lie adjacent to cck (Fig. 1B), delivering solid genomic support for the hypothesis that the pipid AMP/HLP gene family evolved from an ancestral cck gene [30].
Fig. 1. Genomic organization of the Silurana tropicalis AMP gene repertoire. 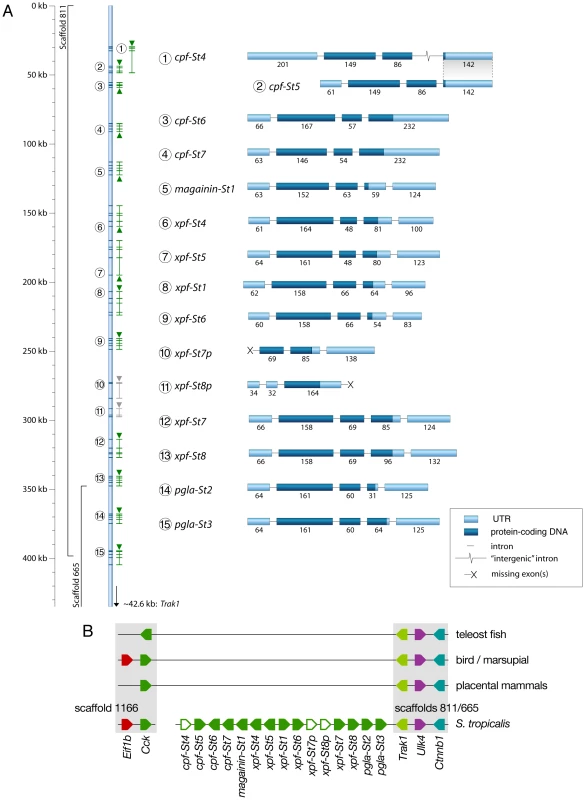
(A) Gene cluster map showing gene and transcript positions (indicated by numbers 1–15 on the left) and gene orientation (indicated by upward or downward pointing triangles). Genes with incomplete coding sequences are colored grey. Exon organization of each gene/transcript is shown on the right (labeled by the numbers of the gene map), with exon lengths indicated as numbers below bars, untranslated regions colored light blue and coding sequences colored dark-blue. (B) Schematic representation of the S. tropicalis AMP gene cluster and adjacent genes showing preserved synteny with the cck gene in other vertebrates. Tab. 1. Transcript abundance of <i>S. tropicalis</i> AMP genes in our cDNA library and in the EST division of GenBank. 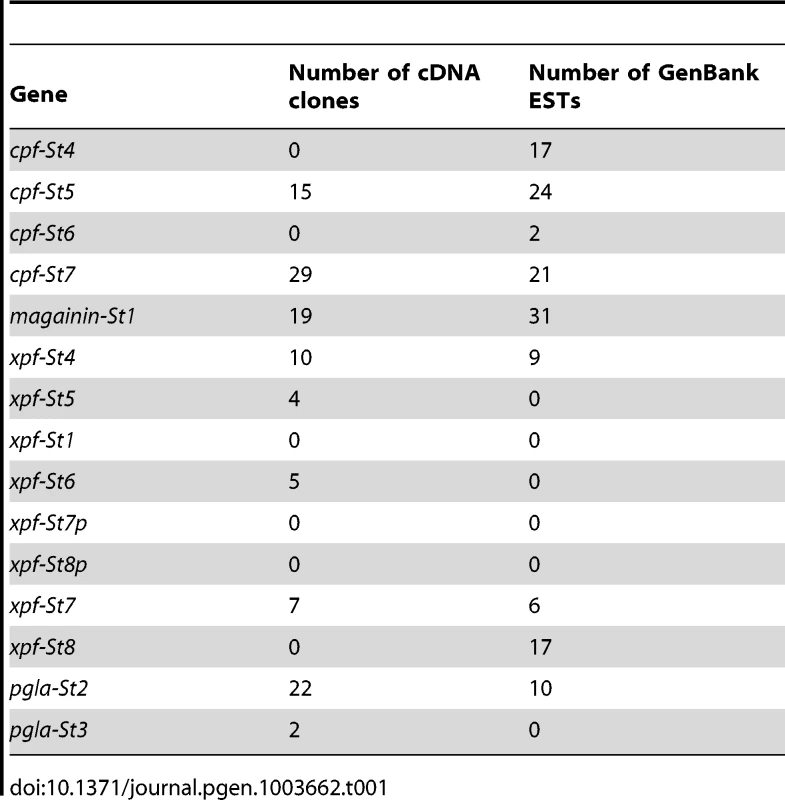
2. The AMP/HLP arsenal of S. tropicalis
All S. tropicalis AMP genes encode precursor proteins of 75 to 96 amino acids (AA) spanning exons 2–4 and containing an N-terminal signal peptide as predicted by the SignalP server [31]. Comparison of the inferred precursor proteins with previously isolated peptides from Silurana and Xenopus species allows us to predict the position and cleavage of functional peptides (Fig. 2). All precursor sequences share a region divided over exons 2 and 3, with sequence homology to previously reported AMPs. In nearly all cases, this region is flanked by an N-terminal -RXXR - motif and a C-terminal -RXXR-, -KR-, or -RR - motif, corresponding to common cleavage sites in other vertebrate peptide precursors. In addition, several of the S. tropicalis genes, similar to those of X. laevis, seem to encode secondary peptides, some of which show homology to known HLPs. The C-terminal region of prepro-CPF-St6 shows sequence similarity to the HLP caerulein, as previously reported for prepro-CPF-St7 [30]. Prepro-XPF-St4 and prepro-XPF-St5 share a C-terminal region with similarity to the X. laevis peptide levitide [25], and prepro-XPF-St7, prepro-XPF-St7p and prepro-XPF-St8 show similarity to peptide phenylalanine-glutamine-amide (pFQa), a 14-AA-long peptide of Silurana epitropicalis [16].
Fig. 2. Precursors encoded by the Silurana tropicalis AMP gene repertoire. 
Peptides are predicted based on sequence homology to previously identified peptides (both highlighted in black) and putative cleavage sites (highlighted in gray). Underlined sequences represent predicted signal peptides. Asterisks represent translation stops. Nano-liquid chromatography tandem mass spectrometry data (nanoLC-MS/MS) from skin extracts confirm that S. tropicalis synthesizes a peptide arsenal that parallels the AMP/HLP diversity of X. laevis. Screening of these spectra against a database composed of AMP/HLP precursor proteins predicted from cDNA and gene sequences confirms the cleavage and posttranslational processing of 11 predicted peptides, while providing support (albeit with insignificant identity scores) for two more (Table 2, Text S2). These include nine of the predicted AMP homologues, two levitide-related peptides (Levitide-St1 and Levitide-St2) and a single homologue of pFQa (pFQa-St1). Our analysis additionally indicates the processing of a secondary peptide (PLD-St1) from prepro-PGLa-St3 without any apparent homology to known peptides. Visual inspection of this precursor confirms the proximity of an N-terminal -KR - site (Fig. 2). This cleavage site is separated from PLD-St1 by the dipeptide Met-Ala, indicating that a cleaved 16-AA-long peptide may be further processed by exopeptidases to obtain PLD-St1. Posttranslational modifications of individual residues are observed in six of the cleaved peptides, all of which undergo C-terminal amidation, and two of which undergo N-terminal pyroglutamate formation (Table 2).
Tab. 2. Overview of transcriptionally active S. tropicalis AMP genes, predicted peptides and their molecular weights. 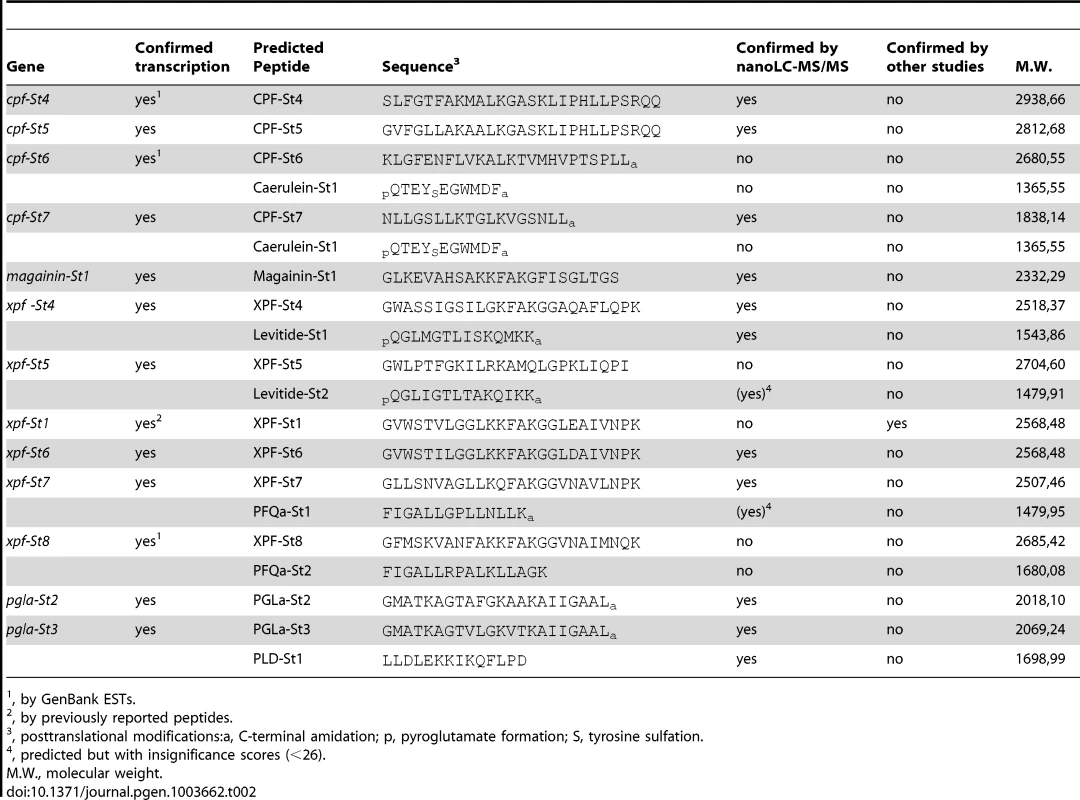
, by GenBank ESTs. 3. Structural and functional analysis of S. tropicalis AMPs
The majority of the predicted peptides show the typical structural features of amphibian AMPs (Table 3). Most of them have the potential to form an alpha-helix according to secondary structure predictions [32], are cationic at a pH of 7.0, and show an alternated sequence of hydrophylic/cationic and hydrophobic AAs, which in an alpha-helical configuration results in a strong amphipathic structure. Upon contact with cell membranes, the cationic side of the helix allows interaction with negatively charged phospholipid heads on the membrane surfaces, while the hydrophobic side facilitates alignment with, or intrusion into the intermembrane region, potentially inducing membrane pore formation and eventually, cell lysis [33], [34].
Tab. 3. Physicochemical properties of S. tropicalis AMPs and HLPs. 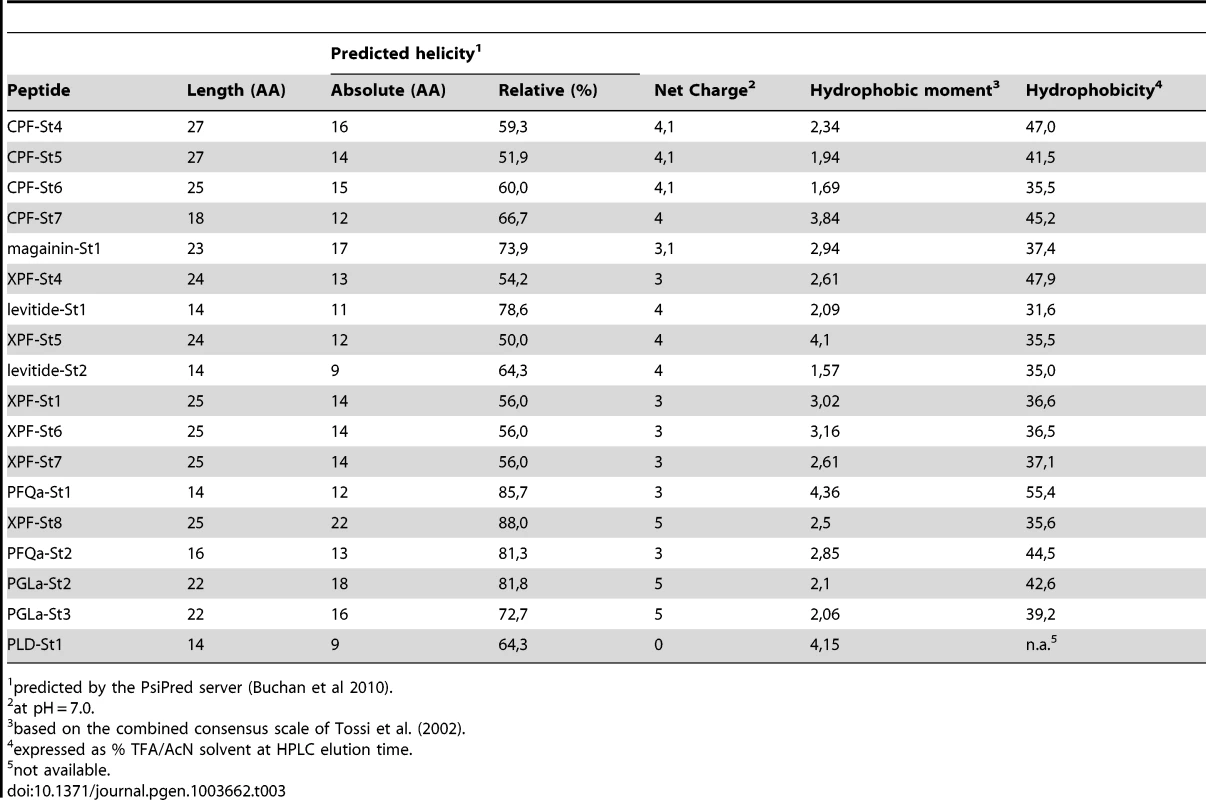
predicted by the PsiPred server (Buchan et al 2010). It was recently argued that AMPs, besides being effectors of innate immune response, may play a role in antipredator defense through their cytolytic effects [35]. We therefore investigated the activity of 17 peptides by concentration-dependent assays in liquid media against both microorganisms and vertebrate cells. Target cells included two gram-negative bacteria (Escherichia coli and Pseudomonas aeruginosa), two gram-positive bacteria (Staphylococcus aureus and Micrococcus luteus), a fungus (Saccharomyces cerevisiae), a protozoan parasite (Trypanosoma brucei), mouse (Mus musculus) red blood cells, and mouse spleen cultures (containing a mixture of T-lymphocytes, macrophages and myeloid cells). The variable nature of these cells required the use of different measures of peptide activity (see Materials and Methods) but combined, their results support a number of distinct patterns.
First, 13 of the 17 synthesized peptides show antimicrobial activity as evidenced by minimum inhibitory concentrations (MIC). These include 12 peptides homologous to known AMPs, and PFQa-St2 (Table 4). In all cases examined, subculturing from MIC solutions to agar media lacking peptides did not yield any colonies, confirming that growth of the population was inhibited by effectively killing the cells.
Tab. 4. Activity of S. tropicalis peptides against a broad range of cell types. 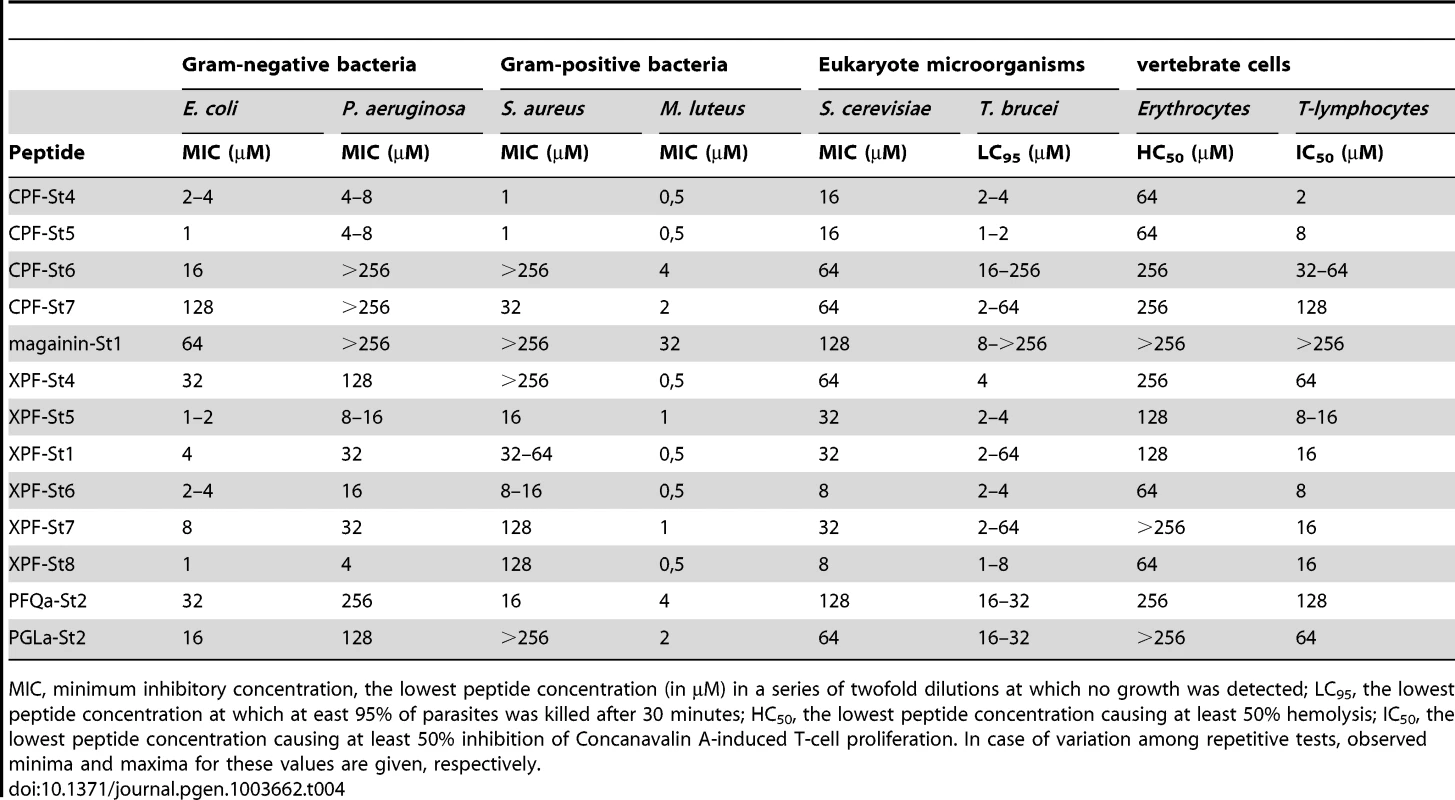
MIC, minimum inhibitory concentration, the lowest peptide concentration (in µM) in a series of twofold dilutions at which no growth was detected; LC95, the lowest peptide concentration at which at east 95% of parasites was killed after 30 minutes; HC50, the lowest peptide concentration causing at least 50% hemolysis; IC50, the lowest peptide concentration causing at least 50% inhibition of Concanavalin A-induced T-cell proliferation. In case of variation among repetitive tests, observed minima and maxima for these values are given, respectively. Second, ten of the peptides are to some extent capable of inducing lysis of red blood cells. Observed hemolytic activity is generally relatively low, with peptide concentrations inducing 50% hemolysis (HC50) ranging down to 64 µM. In addition, 12 peptides are capable to inhibit the Concanavalin A-stimulated proliferation of T-lymphocytes in spleen cell cultures. Peptide concentrations inducing 50% inhibition of T-cell proliferation (IC50) range down to 2 µM (CPF-St4) while higher concentrations typically eliminate any proliferation. Concanavalin A induces T-cell proliferation indirectly, by stimulating macrophages to activate T-cells. Consequently, the peptides may inhibit proliferation directly (by suppression or lysis of T-cells), indirectly (by inhibition or lysis of macrophages), or both. These results confirm that amphibian AMPs may indeed contribute to antipredator defense through cytolytic and/or immunosuppressive effects.
Third, the tested peptides show notable variation in activity against any single cell type. For S. aureus for example, MIC values range from >512 µM (e.g., magainin-St1) down to 1 µM (e.g., CPF-St5). For T. brucei, a single peptide may additionally show variation in activity across repeated assays. This pattern may be related to the use of pleomorphic parasite cultures, representing a variable mixture of two parasite forms (“long-slender” and “short-stumpy”) with potentially different susceptibility to membrane permeabilization.
Fourth, our assays do not provide evidence of target cell specificity or complementary target spectra among different peptides as observed in other frogs (e.g., [36]). Although there is substantial variation in the sensitivity of cell types to the AMPs, (e.g, the gram-positive M. luteus is highly sensitive to most peptides), the measured activities of peptides across cell types are correlated. In other words, peptides either seem effective against the broad range of cell types (e.g., CPF-St5) or hardly effective at all (e.g., magainin-St1). Previous studies have demonstrated that relatively weak AMPs from the same amphibian species may yield an increased antimicrobial effect when applied in combination [11], [36], [37], [38]. We searched for the existence of such synergistic effects among the S. tropicalis AMPs by MIC assays against S. aureus in which peptides were pairwise combined in a 1∶1 stochiometry. Our analyses reveal a single case of synergistic activity: combination of two of the weakest AMPs, magainin-St1 and PGLa-St1, yields a combined MIC of 64 µM (32 µM for each peptide), while individually, magainin-St1 had no apparent effect on S. aureus (MIC>512 µM) and PGLa-St1 had a very weak effect (MIC = 512 µM).
4. Evolutionary diversification of the pipid AMP gene repertoire
Origin of the pipid AMP gene promoter
Similar to CCK, AMPs have been found in the gastronintestinal tract of X. laevis [27]. However, the transition of the neurohormone function of CCK to the defense function of these AMPs implies that changes in gene regulatory mechanisms entailed a shift in gene expression to the skin glands. The transcriptional regulation of CCK is well-studied and comparative analyses of its promoter region have shown the evolutionary conservation of several cis-regulatory elements (CREs) across vertebrates [39], [40]. These include a TATA-box, GC-box, one or several E-box elements (consensus sequence CANNTG), and a combined cAMP response element/tetradecanoylphorbol-13-acetate response element (CRE/TRE; consensus sequence TGCGTCAG). Binding of basic Leucine-zipper (bZIP) transcription factors like CREB and Jun/Fos (AP-1) heterodimers to the CRE/TRE responsive element has been shown to increase cck promoter activity. Instead, binding of basic-helix-loop-helix (bHLH) transcription factors like the c-Myc/Max heterodimer to the E-box responsive element inhibits CRE/TRE activation, indicating a mechanism of negative cooperativity between the juxtaposed elements [40], [41].
Comparative alignment of the promoter regions of the pipid AMP genes with those of tetrapod cck genes shows a major renovation of otherwise evolutionary conserved sequence motifs in the promoter region (Fig. 3). The TATA-box is retained in all AMP genes, but the E-box and CRE/TRE elements, conserved at a fixed positions 10–20 bp apart in cck gene promoters, are absent, implying loss of the abovementioned negative cooperativity mechanism. These binding sites became evolutionarily degraded to give way to high sequence variation across AMP genes (Fig. 3). Instead, our analyses identify two motifs that show strong evolutionary conservation across AMP gene promoters. The first motif (CRE1) is 36–41 bp long and starts approximately 120–155 bp upstream of the transcription initiation site. Motif discovery searches using MEME [42] show that CRE1 occurs twice within the promoter region of the X. laevis caerulein genes. In addition, the same analyses recognize homologous sequences in the cck core promoters of both Lithobates catesbeianus (American bullfrog) and S. tropicalis as variants of the same motif, suggesting that the evolution of CRE1 reached an identifiable stage at the time of divergence of ranid and xenopodine frogs, during early anuran radiation. The second motif (CRE2) is located approximately 50 bp upstream of the TATA-box and contains the conserved octamer TGCAAANA in 13 out of 16 investigated AMP genes in S. tropicalis and X. laevis. This motif evolved to an inverted orientation in the X. laevis caerulein gene promoters, and originated secondarily in a more upstream position in the promoter regions of xpf-St7 through pgla-St2. The TGCAAANA octamer is present at the same position in the cck gene promoters of Anolis carolinensis (anole lizard), L. catesbeianus, and S. tropicalis, indicating that it may represent a much older, previously overlooked CRE retained in the AMP gene promoters. The level of sequence conservation of the two motifs, and the fact that they occur (sometimes twice) in the majority of AMP gene promoters is a strong indication that they are subject to purifying selection, and play a fundamental role in the genetic regulation of the AMP/HLP arsenal. In addition, the presence of both motifs in cck genes of various taxa implies that their evolution predated the loss of cck-specific CREs and that their evolution was well underway before the diversification of pipid AMP genes.
Fig. 3. Comparative alignment of the promoter regions of vertebrate cck genes and pipid AMP genes. 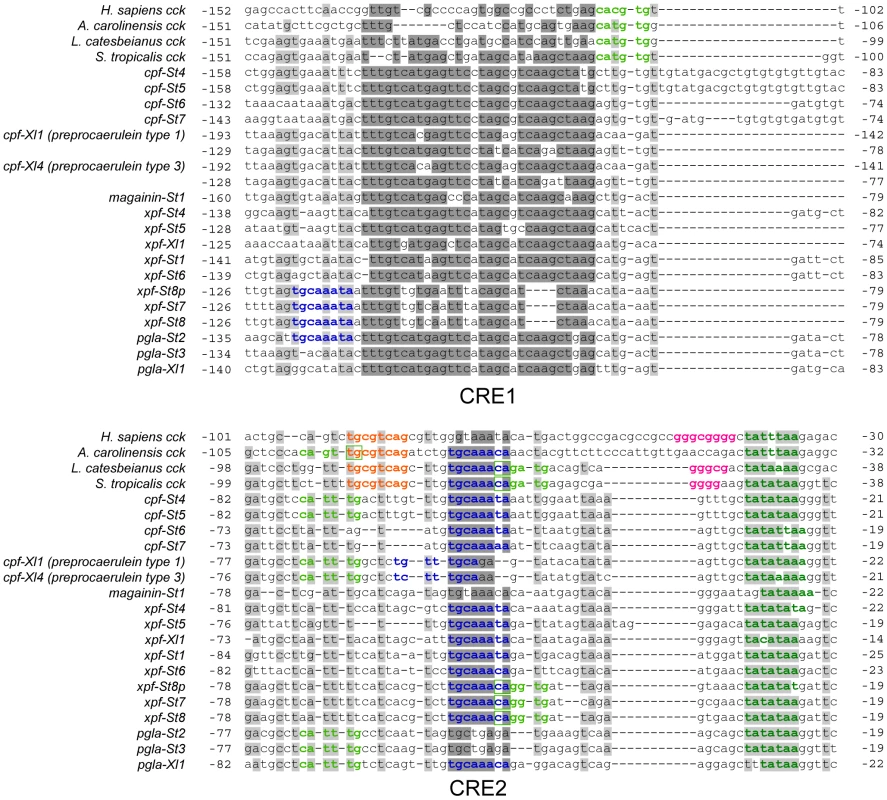
Sites with >75% sequence conservation across AMP gene promoters are indicated in grey; those additionally discussed as conserved sequence motifs in the text are indicated in dark grey. Sequence motifs corresponding to known or putative regulatory elements are colored as follows: dark green, TATA-box; light green, E-box; Orange, CRE/TRE element; pink, GC-box; dark blue, CRE2-conserved octamer. Screening of the two newly identified promoter motifs against the TRANSFAC public database [43] predicts the potential binding of several other transcription factors known to be involved in innate immunity. The CRE1 motif in the majority of AMP genes contains two sites with sequence similarity to known binding elements of the bZIP transcription factors C-Fos, C-Jun, Fra-1, JunB, JunD, and XBP1. Several of those have been shown to play a role in various immune responses [44], including the expression of AMP genes in other vertebrates [45]. In addition, CRE2 shows strong similarity to known binding elements of CCAAT/Enhancer binding proteins (C/EBP) and POU class 2 homeobox factor 1 (POU2F1), again transcription factors associated with the synthesis of antimicrobial peptides and proteins in mammals [46]. Binding of these transcription factors to overlapping sites has been observed before in the promoter of interleukin-8, where C/EBP causes activation of the gene while POU2F1 causes repression [46].
The pipid AMP gene promoters show little similarity to promoters characterized in skin-expressed AMP genes of other amphibians. Analysis of the gaegurein 4 gene in Rana rugosa revealed the lack of a TATA box, and the presence of Dl and NF-IL6 transcription factor binding sites, known to regulate acute phase immune response in mammals and insects [47]. The promoter regions of the bombinin-like peptide 3 (blp3) and bombinin-like peptide 7 (blp7) genes in Bombina orientalis [48], besides NF-IL6 elements, contain target sites for NF-kB, another key regulator of inflammatory and immune responses. While the NF-IL6 binding motif in blp3 shows resemblance to CRE2 in the AMP gene promoters (NF-IL6 is a member of the C/EBP protein family), dl and NF-kB binding sites are absent in the AMP gene promoters. These differences in core promoter motifs indicate that AMP arsenals in distantly related frog lineages are not only encoded by independently evolved gene families [12], but are also differently “wired” into the innate immune response pathway.
Phylogeny of the pipid AMP gene repertoire
We reconstructed the evolution of the pipid AMP arsenal by phylogenetic analyses of the genes of S. tropicalis and X. laevis using amphibian cck gene sequences as outgroups. Bayesian analyses based on fixed alignments [49] or via direct optimization (integrating sequence alignment and tree estimation in a single analysis [50]) yield a well-resolved gene tree elucidating the rise of the AMP arsenal with respect to pipid evolution (Fig. 4). First, expansion of the gene cluster was well underway prior to the divergence between Silurana and Xenopus, which has been estimated by molecular clock analyses to have taken place approximately 42 (30–59) million years (Myr) ago [51]. At least five gene duplication events (grey nodes in Fig. 4) followed the basal gene duplication of the cck gene (black node) and predated the Silurana-Xenopus split, implying that the most recent ancestor of both genera had at least six AMP genes (X-marks in Fig. 4). Second, five well-supported branches in the tree identify the orthologous peptides in S. tropicalis of all known X. laevis AMPs and HLPs. For example, the two synergistic AMPs magainin-St1 and PGLa-St2 are confirmed to be orthologous to magainin-Xl2 and PGLa-Xl1, respectively, two X. laevis peptides with a similar synergistic effect [37], [38]. Levitide-St1 and levitide-St2 are confirmed to be orthologues of X. laevis levitide. Unexpectedly, the structurally distinct X. laevis xenopsin is recovered as closer related to X. laevis levitide than the S. tropicalis levitides, suggesting that this HLP descended from a levitide-like ancestral peptide after the Xenopus-Silurana split. Third, the tree indicates that several Silurana genes, after the divergence from Xenopus, underwent parallel gene duplication, yielding multiple pairs of closely related sister genes. Nine gene duplication events occurred in the Silurana lineage after it divergence from Xenopus (blue nodes in Fig. 4). In contrast, only three gene duplication events are observed for Xenopus since its divergence from Silurana (red nodes in Fig. 4).
Fig. 4. Evolutionary diversification of pipid AMP genes. 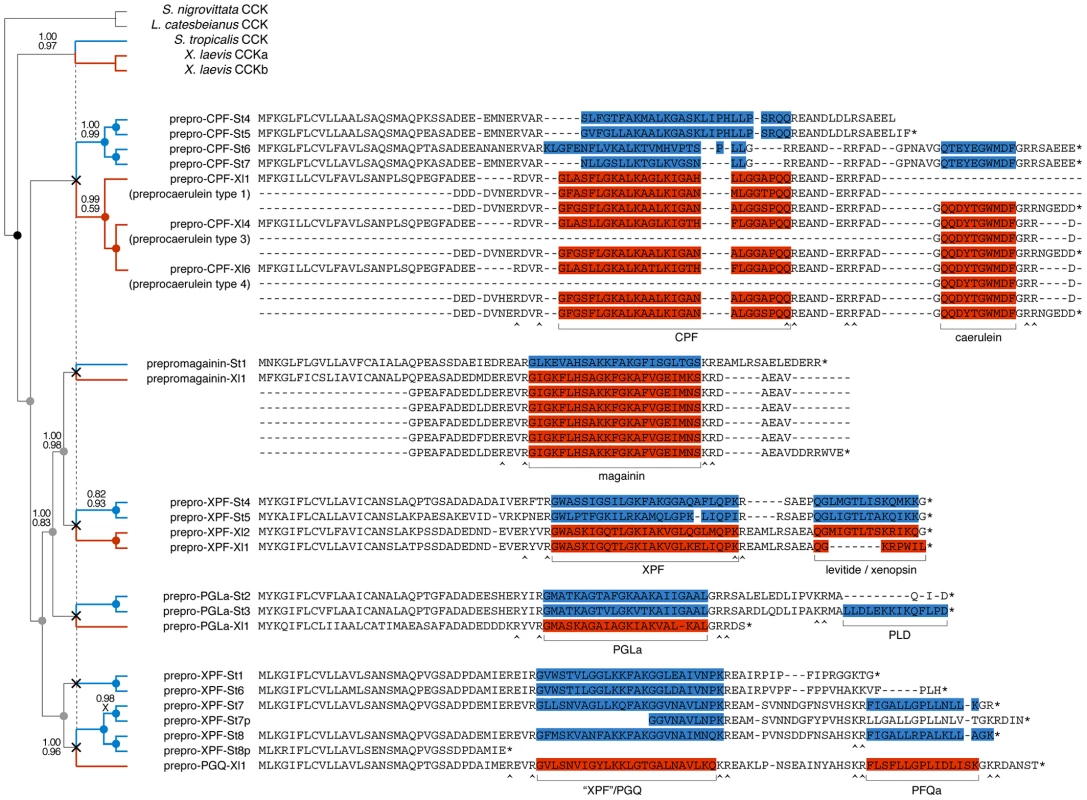
Phylogenetic relationships of S. tropicalis and X. laevis AMP genes are shown in a consensus tree obtained by Bayesian analyses using the traditional two-step approach of phylogeny inference (separate alignment and Bayesian phylogeny inference) and using direct optimization (integrated Bayesian alignment and phylogeny inference). Each gene is represented here by its precursor protein sequence aligned to visualize similarity with its closely related homologues. Note that several of the X. laevis sequences occupy multiple lines because duplicated exons were aligned to each other. Unlabeled branches in the tree are supported by maximum posterior probabilities (1.00) under both methods; branches that received less support by one or both methods are labeled by their posterior probability under the two-step method (top) and under direct optimization (bottom). Nodes representing gene duplication events are labeled by circles and color-coded as follows: black, the split between cck and the ancestral pipid AMP gene; grey, gene duplication in an ancestor of Silurana and Xenopus; blue, gene duplication in Silurana; and red, gene duplication in Xenopus. Crosses linked by a vertical dashed line mark the divergence of Silurana and Xenopus, the resulting orthologous gene lineages are marked by blue and red branches respectively. Peptides in the precursor proteins are color-coded accordingly. Selection on the AMP precursor proteins
Alignment of the AMP/HLP precursor proteins indicates variation in amino acid substitution rates along their length (Fig. 4) in a pattern that is consistent with those observed in the AMP precursors of other amphibians [9], [10]. Codon-based likelihood analyses comparing sitewise nonsynonymous and synonymous substitution rates confirm that this rate variation reflects mixed patterns of diversifying (positive) and purifying (negative) selection (Fig. 5). High site-specific ratios of nonsynonymous over synonymous substitution rates are mostly observed for the spacer - and AMP-encoding regions, and all sites that are identified as showing significant evidence of diversifying selection lie in the these regions. This pattern may be consistent with selective pressures for a large structural diversity in amphibian AMP arsenals in adaptation to target a diverse spectrum of microorganisms [9], [10]. Conversely, sites with low site-specific ratios of nonsynonymous over synonymous substitution rates and significant evidence of purifying selection are concentrated in the regions encoding the signal peptide, peptide cleavage sites and the CCK-like bioactive site preserved in the X. laevis caerulein genes, cpf-St6, and cpf-St7 (Fig. 5). Hence, similar to the evolutionarily conserved sequence motifs in the AMP gene promoter regions, parts of the coding sequences are subject to purifying selection, most likely to retain proper processing of the defense peptides.
Fig. 5. Patterns of diversifying and purifying selection in pipid AMP/HLP precursor proteins. 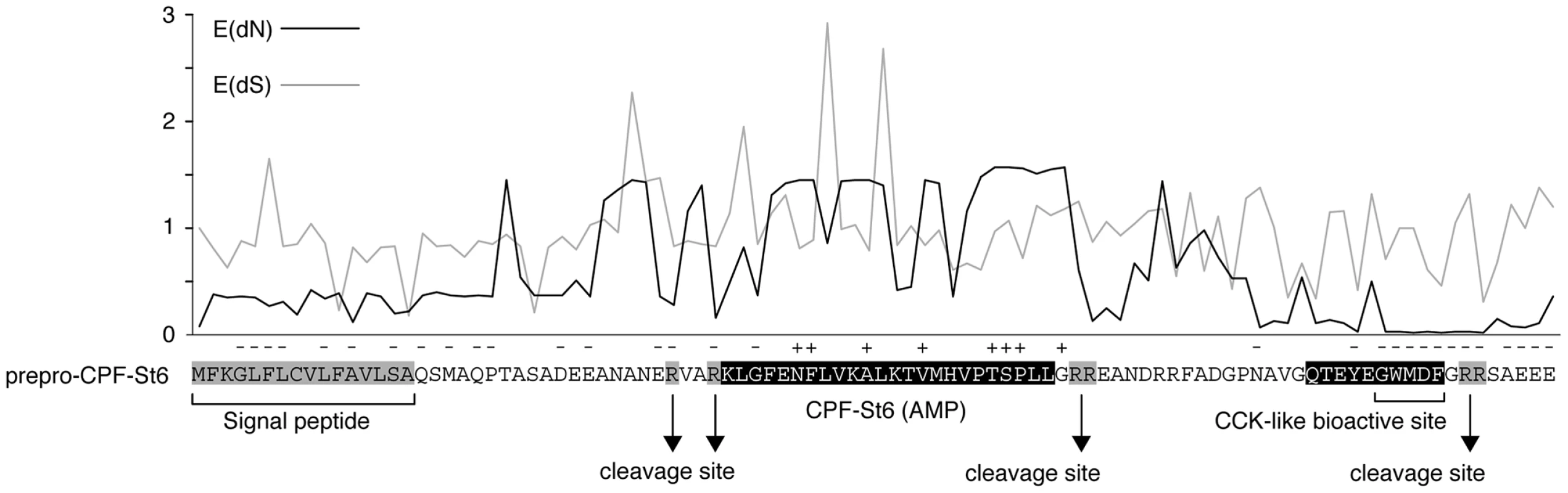
Site-specific mean posteriors of nonsynonymous (E(dN), black line) and synonymous (E(dS), grey line) codon substitution rates are plotted along an exemplary precursor protein sequence (prepro-CPF-St6). Sites for which the black plot line rises above grey one have dN/dS ratios >1 (suggesting diversifying selection); those for which the black plot stays below the grey have dN/dS ratios <1 (suggesting purifying selection). Sites showing significant evidence of diversifying or purifying selection (at Bayes factor >50) are labeled with ‘+’ and ‘−’, respectively. Discussion
By combining transcriptome and genome data with peptidome analyses, we present the first overview of the defensive peptide arsenal of an amphibian and the gene repertoire that underlies its synthesis. Together, the 13 charted genes encode up to 19 different peptides, a diversity comparable to, or even larger than those currently known from other pipid frogs, but far less extensive than the peptide arsenals reported from ranid frogs of the genus Odorrana, where transcripts of a single species encode for a diversity of 107 AMPs (55 AMPs in a single individual) [11], [52]. Our cDNA library further suggests variation in the level of transcription of individual genes, as indicated by respective numbers of randomly sequenced clones (Table 1). This pattern suggests that all AMP genes may not contribute equally to the arsenal of a single frog at any time, and may show both temporal and/or individual variation in expression.
Although our assays indicate that several of the S. tropicalis peptides are capable to kill a broad range of microorganisms at micromolar concentrations, the level of protection provided by the entire AMP arsenal against typical amphibian pathogens remains unknown. For example, unlike X. laevis, S. tropicalis has been suggested to be susceptible to the widespread chytrid fungus Batrachochytrium dendrobatidis [53], [54]. Microarray studies have shown that skin infection with this fungus hardly induces an immunogenetic response in S. tropicalis, and does not stimulate the expression of any AMP gene [55], [56]. This finding may indicate that the S. tropicalis AMP arsenal failed to protect against this fungus because its gene repertoire was not stimulated to an effective level in the first place.
Several authors have recently questioned the singular role of amphibian AMPs as first-line antimicrobials [12]–[16]. The weak activity of some AMPs compared to others has been interpreted as an indication that they evolved another, yet unknown function [17], [20]. On the one hand, there are indications that AMPs play a far more integrated role in the innate immune system, e.g. by acting as chemoattractants for immune cells or as modulators of cytokine release [11]. On the other hand, the cytolytic effect of AMPs on vertebrate cells, and their frequent co-expression with HLPs in various frog families may indicate that they serve a role in antipredator defense [35]. Our analyses confirm that they show activity against vertebrate red blood cells as well as T-cells at micromolar concentrations. At lower concentrations however, these AMPs may be ineffective in killing vertebrate cells, and instead, may serve a more subtle function. Some AMPs for example, are capable to stimulate insulin release by beta cells at nanomolar concentrations [57]. Although findings like these have a clear pharmaceutical potential, it remains uncertain whether such activity provides an adaptive benefit to amphibians when being attacked by a predator. Regardless of other potential adaptive functions, it seems unlikely that the broad-scale antimicrobial activity of some AMPs would not provide any adaptive benefit to the host species, even if it fails to provide protection to some pathogens.
1. Differential evolution of the AMP/HLP arsenal in Silurana and Xenopus
The phylogeny of the pipid AMP/HLP arsenal is consistent with a birth-and-death model of gene evolution, similar to those implicated for vertebrate immune gene families [58] and reptile toxin genes [59], [60], in which some gene lineages survive for a prolonged time, and may give rise to additional genes through subsequent duplication, while others are eventually lost. Gene loss is confirmed by the presence of several incomplete genes in the S. tropicalis cluster, one of which shows clear signs of pseudogenisation (cpf-St8b), and the apparent absence of orthologues of xpf-St1 and xpf-St6 in X. laevis. Comparative peptidome analyses of several Silurana and Xenopus species have recently indicated that ancient genome duplication events did not result in an increased AMP diversity in allopolyploid species [16], [17], [20]. This observation led to the conclusion that polyploidization events were compensated by subsequent gene losses, counterbalancing the expected doubling of the AMP arsenal. Our analyses confirm similar levels of AMP diversity in S. tropicalis and X. laevis but also reveal that the comparable arsenals in both taxa arose under contrasting patterns of gene evolution. The diploid S. tropicalis has the largest AMP/HLP gene repertoire (due to higher rates of tandem gene duplication or lower rates of gene loss) but each gene encodes a single AMP and/or a single HLP. Instead, X. laevis, despite being tetraploid, has fewer genes (due to lower rates of gene duplication or increased gene loss), but several of them (e.g. the magainin and caerulein genes) are characterized by multiple tandem-repeated peptide–encoding exons. The difference between both patterns may have been maintained by the self-sustaining nature of tandem repeats (whether genes or exons), because as their number increases, so does the probability of unequal crossing-over among neighboring copies, potentially creating additional duplicates of the same type. Exon duplication is considered an important mechanism of transcriptional economy by which peptide diversity can be increased, either through alternatives splicing [61] or by differential cleavage of tandem-encoded peptides [62]. However, duplicated exons in the caerulein and magainin genes show quasi-zero sequence divergence (Fig. 4), possibly reflecting very recent duplication events, extremely low evolutionary rates, or gene conversion. Alternative splicing therefore adds little to peptide diversity, but in the X. laevis genes encoding multiple tandem-repeated exons may provide a way to boost the synthesis of peptides at a low transcriptional cost.
2. Dynamic structural and functional evolution of the pipid AMP gene repertoire
Our phylogenetic analyses allow us to formulate an evolutionary scenario for the timing of origin and loss of specific peptide types and their corresponding functions in light of major structural changes in the pipid AMP gene repertoire (Fig. 6). The first major step in the evolution of the peptide arsenal entails the transition of a neurohormone (CCK) to a basal defense peptide (Figs. 6A, 6B). The HLP caerulein, encoded by three genes in X. laevis, contains the same bioactive site as CCK and shares its capacity to bind vertebrate CCK receptors, thereby inducing pancreatitis, vomiting, diarrhea, hypotension, and inhibition of exploratory and feeding behavior [63]. In addition, C-terminal sequences homologous to caerulein are retained in the S. tropicalis precursors prepro-CPF-St6 and prepro-CPF-St7. Together, these Xenopus and Silurana genes represent an early-diverged lineage in the pipid AMP gene tree. It is therefore likely that caerulein retained the basal defense function of the cck-derived gene family. Its origin was accompanied by a gene duplication event (gene duplication 1 in Fig. 6B) and a shift in expression from the gastrointestinal tract and brain to the granular skin glands. The taxonomic distribution of newly identified promoter elements (Fig. 3) provide an indication that at least part of the changes required for this expression shift may have happened long before the cck gene duplication, implying that the ancestral cck gene was “preconditioned” to acquire a skin-secretory function.
Fig. 6. Structural and functional evolution of the pipid defense peptide arsenal. 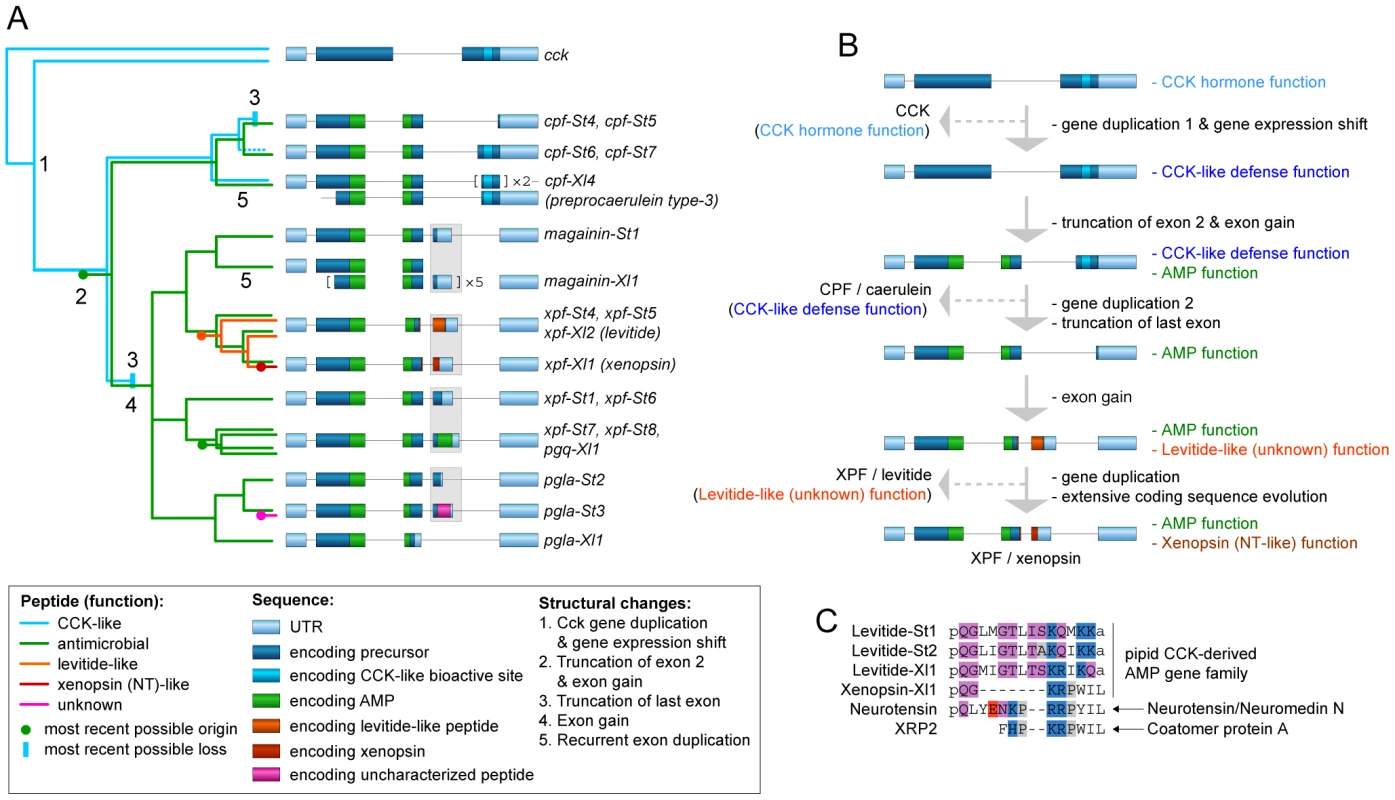
(A) Evolutionary scenario for the origin and loss of defense peptides and their function along a summarized phylogenetic tree. Numbers along tree branches represent structural changes as mentioned in the legend. (B) Evolutionary trajectory showing structural changes and functional transitions from the ancestral cck gene to the present-day X. laevis xpf-Xl1 gene, encoding the AMP XPF and the HLP xenopsin. Only gene duplication events relevant to functional changes (see text) are indicated. (C) Comparative alignment of levitide-like peptides, X. laevis xenopsin and the vertebrate neurohormones with which it shows evolutionary convergence. The origins of the different peptides are indicated on the right. Amino acids are color-coded as follows: white, hydrophobic; grey, near-neutral; purple, uncharged polar; blue, cationic (basic); red; anionic (acidic). A second major step involves the origin of a cationic alpha-helical AMP in the central region of the ancestral precursor protein, leading to a bifunctional AMP/HLP gene product with an architecture similar to, e.g. the present day prepro-CPF-St6 (Figs. 6A, 6B). This step is marked by truncation of the N-terminal spacer region, and the origin of an additional exon, encoding the C-terminal half of pipid AMPs. In contrast, an evolutionary conserved -RXXR - site, inducing the N-terminal cleavage of the longest functional CCK-isoforms in vertebrates (e.g. CCK-58 in humans) has been preserved in the majority of pipid AMP precursors, where it induces cleavage of the functional AMPs.
Third, after establishment of an antimicrobial function, the basal CCK-like defense function was lost twice independently (Fig. 6A), yielding monofunctional AMP precursors with an architecture similar to e.g., the present-day prepro-CPF-St5 precursor (Figs. 6A, 6B). One loss occurred in the gene ancestral to cpf-St4 and cpf-St5, while a second loss happened in the gene ancestral to magainin-St1 through pgla-St3. In both cases, the loss of the CCK-like function followed gene duplication events and were accompanied by the 5′-truncation of the genes' last exon, eliminating the sequence encoding the CCK-like bioactive site.
Fourth, after loss of the basal CCK-like defense function, new AMPs and HLP functions (pFQa-like peptides, levitide-like peptides, PLD-St1), arose alongside the antimicrobial function, again yielding a bifunctional gene. All of these secondary peptides are encoded by an extra exon (exon 4 in xpf-St4, -St5, -St-6, -St7, and -St8, and pgla-St2, and -St3), but due to their lack of structural similarity, it is unclear whether they originated independently, or share a common origin. The latter would imply extraordinary structural diversification of the peptides due to extremely high substitution rates in exon 4.
On at least two occasions, evolution of the pipid AMP genes gave rise to striking cases of evolutionary convergence in peptide sequence. One case is represented by caerulein, which independently evolved to an identical structure in Litoria frogs, starting from a different neurohormone (gastrin) [30]. A second example is represented by xenopsin, which according to our phylogenetic analyses evolved from a levitide-like peptide (Figs. 4, 6A, 6B). Apart from AA substitutions, this evolutionary process involved a 7-AA deletion in the peptide and loss of C-terminal amidation (Fig. 6C). Xenopsin shows strong structural and functional similarities to the hormones neurotensin (NT) and xenopsin-related peptide 2 (XRP-2), two universal NT receptor agonists in vertebrates [26], [64], [65]. These hormones share five and six C-terminal AAs with xenopsin, respectively, all of which are involved in neurotensin receptor binding [65]. Effects of NT and XRP-2 include stimulation of exocrine pancreatic secretion, suppression of food intake, increase of vascular permeability (enhancing inflammation responses and hypotension), and stimulation of histamin release by mast cells (enhancing an allergenic reaction) [66], [67]. While NT is cleaved from its own precursor along with the hormone neuromedin N [68], XRP-2 is cleaved from the N-terminal region of Coatomer Subunit alpha (CopA), a 138-kDa intracellular protein [69]. Because xenopsin is unrelated to either, its evolution is likely to reflect adaptive convergence to neurotensin receptor binding.
3. The pipid AMP arsenal as a model system of functional innovation
Toxin gene families showing complex functional diversity among their members in venomous animal taxa have been used as model systems to investigate the role of gene duplication and diversifying selection in functional innovation, and lended support to different theoretical models of neofunctionalisation [59], [70], [71], [72]. The pipid AMP/HLP arsenal provides an opportunity to extend the same theoretical framework to amphibian defense peptide arsenals and presents an empiricial illustration of how peptide-encoding genes may acquire new functions independent of gene duplication.
Current theoretical models invoke gene duplication as a key factor of functional innovation. Under the original model of neofunctionalisation (“mutation during nonfunctionality”, [73]) gene duplication delivers a functionally redundant gene, which, released of selective constraints to maintain the ancestral function, accumulates mutations that lead to a new function. Alternative models instead invoke subfunctionalisation following gene duplication to divide multiple functions of the parental gene over the duplicates in a nonadaptive way (“duplication-degeneration-complementation” ; [74]), or to resolve adaptive conflicts between different ancestral functions (“escape from adaptive conflict” [75], [76]). Although gene duplication of the cck gene allowed the origin of a CCK-like defense function in a pipid ancestor, it did not trigger subsequent functional innovation. Instead, new functions recurrently arose by mutation of a precursor protein, resulting in the posttranslational processing of a secondary peptide (an AMP) alongside the original one (e.g. a CCK-like HLP). The resulting bifunctional gene is consistent with the starting condition in several theoretical models invoking subsequent gene duplication [74], [76], [77]. However, pipid AMP/HLP genes deviate from these models (or represent a special case) by attaining multifunctionality through the production of multiple peptides, rather than a single peptide or protein with multiple functions.
One implication of this mechanism is that genes in the AMP/HLP arsenal acquired new functions while being under purifying selection to retain an older one. This could have been possible by mutations in a gene region that is subject to only marginal purifying selection [10]. Selection on a gene to retain its original function is likely to affect only regions of the gene that are crucial for the proper expression, processing and functioning of the peptide [10], [78]. In the case of the Pipid AMP/HLP gene repertoire, this is confirmed by purifying selection on gene regulatory elements, and exon regions encoding signal peptide, cleavage sites and the CCK-like bioactive site (Figs. 3 and 5). The evolution of a new functional peptide in precursor regions under relaxed selection thereby not avoids adaptive conflicts with the original peptide but may even rely on the gene regions under purifying selection to obtain an adaptive value.
Another implication is that selection on a gene to retain its original function is likely to affect the probability of possible new functions to arise. In pipid frogs, purifying selection to retain a gene with all the necessary features to produce a skin-secretory peptide, increases the probability that any new function will be related to skin secretion as well. Therefore, while representing a red line through the evolutionary history of the pipid AMP gene repertoire, skin gland expression acted as a key determinant of functional innovation.
Although gene duplication did not act a the direct trigger of functional innovation, it may have subsequently still led to subfunctionalisation [74], [75], [76], by relaxing selective pressures to retain both functions in each daughter gene. Subfunctionalisation may explain the convergent loss of the original CCK-like defense function in multiple gene lineages after duplication, completing the final step in their functional transformation from HLP gene to AMP gene (Fig. 6B).
The pipid defense peptide arsenal illustrates an evolutionary mechansim independent of gene duplication that could explain the presence of precursor proteins yielding multiple peptides with distinct structures and bioactivities in the poison or venom glands of various animals. Transcripts of a single gene isolated from Bombina toad skins encodes both bradykinin peptides and a structurally unrelated bradykinin inhibitor [79], while natriuretic peptide precursors produced in the venom glands of vipers and Heloderma lizards are further cleaved to obtain angiotensin-converting enzyme (ACE) inhibitors, and bradykinin receptor antagonists, respectively [80], [81]. Furthermore, the evolution of multiple peptides within a single precursor, when followed by gene duplication and subfunctionalisation, provides an explanation for the existence of closely related genes encoding structurally and functionally distinct peptides. Homologous genes encoding fundamentally different peptides, like AMPs, bradykinin-like HLPs and opioid peptides, have been identified in ranid and hylid frogs [3], [10], [82], [83]. A recent evolutionary study of snake venom metalloproteinases (SVMPs) showed how differential protein domain loss following gene duplication gave rise to proteins with distinct toxin functions [70]. This study implied that the ancestral SVMP toxin was composed of multiple domains, each with a distinct function, but did not explain how these domains were combined in a single protein.
Materials and Methods
1. Ethics statement
The experiments, maintenance and care of mice complied with the guidelines of the European Convention for the Protection of Vertebrate Animals used for Experimental and other Scientific Purposes (CETS n° 123). The experiments for this study were approved by the Ethical Committee for Animal Experiments of the Vrije Universiteit Brussel, VUB, Brussels, Belgium (Permit Number: 08-220-8).
2. Charting the S. tropicalis AMP gene repertoire
Freshly dissected skin tissue of S. tropicalis was snap-frozen in liquid nitrogen, ground to powder with mortar and pestle, and stored at −80°C. mRNA was extracted using the Qiagen Oligotex mRNA midi kit and cDNA libraries were made using the Clontech Creator SMART cDNA library construction kit. Plasmids with cDNA inserts were cloned in Oneshot Electrocompetent GeneHog E. coli cells (Invitrogen Corp.) and 384 randomly selected clones were sequenced by the Australian Genome Research Facility. Clones corresponding to AMP-encoding transcripts were identified by BLAST searches based on sequence homology to mRNA and gene sequences encoding known peptides in X. laevis. Subsequent online translation of these transcripts yielded the corresponding precursor sequences. Functional peptides in the precursors were predicted by comparison with previously described peptides from various Silurana and Xenopus species.
The S. tropicalis AMP gene repertoire was charted by BLAST-searches against the Xenopus tropicalis 4.1 genome (DOE Joint Genome Institute; http://genome.jgi-psf.org/Xentr4/Xentr4.home.html; X. tropicalis is the previous name of S. tropicalis) using the newly obtained transcripts as queries. Based on a large sequence overlap including pgla-St2 and pgla-St3, we concatenated genomic scaffolds 665 and 811. We assume that these scaffolds were kept separate during contig assembly due to sequence gaps whose lengths were improperly estimated, resulting in apparent sequence conflicts. Intron/exon boundaries of identified AMP genes were checked by verifying the presence of GT/AG intron borders flanking the inferred exons.
3. Tracing predicted AMPs and HLPs in the S. tropicalis skin peptidome
Freshly dissected dorsal skin tissue of S. tropicalis frogs were placed in a 90∶9∶1 (v∶v∶v) CH3OH/H20/HCOOH solution and stored at −20°C. After sonication (5 times 2 min) and centrifugation (10 min at 9000 g), the supernatant was lyophilized (SpeedVac Concentrator). Dry peptide extracts were reconstituted in 100 µL of 0.1% aqueous HCOOH and lipids were removed by re-extraction with equal volumes of ethyl acetate and n-hexane respectively. The sample was filtered through a 0.22 µm spin-down filter (Ultrafree-MC; Millipore) and 1/10th of the sample was analyzed by nanoLC-MS/MS on an Ultimate 3000 Nano and Capillary LC System (Dionex) coupled to a microTOF-Q mass spectrometer (Bruker Daltonics GmbH). The peptides were separated on a Pepmap C18 column (3 µm, 75 µm×150 mm, LC Packings) using a 50 min linear gradient from 95% solvent A and 5% solvent B to 50% solvent A and 50% solvent B at a flow rate of 200 nl/min (solvent A: deionised water containing 0.1% HCOOH; and solvent B: CH3CN containing 0.1% HCOOH). In the mass spectrometer, doubly or triply charged ions of sufficient abundance are selected for fragmentation by the software (MS/MS). [84]. MS/MS peak list files were submitted to an in-house version of MASCOT server (Matrix Science, USA) and screened against a database of predicted AMP/HLP precursor protein sequences. This way, the actual cleavage and posttranslational processing of predicted peptides from the precursors was verified. The resulting spectra and corresponding tables are provided in Text S2.
4. Structural and functional analyses of S. tropicalis AMPs
Secondary structure of AMPs was predicted using a neural network algorithm as implemented on the Psipred server [32]. Peptides predicted to have an alpha-helical structure were projected in a wheel projection to visualize their amphipathic nature. Hydrophobic moments were calculated using the combined consensus scale implemented in HydroMCalc [85].
Seventeen predicted and recovered peptides were de novo synthesized using solid-phase technology by CASLO Laboratory ApS (Lyngby, Denmark) and delivered as HPLC-purified (>95%) trifluoracetate (TFA) salts. The peptide salts were stored as 5.12 mM stock solutions in 0.01% (V/V) acetic acid/0.2% (m/V) BSA. Peptides were tested against Escherichia coli (ATCC 25922) and Pseudomonas aeruginosa (ATCC 15692), Staphylococcus aureus (ATCC 25923), Micrococcus luteus, Saccharomyces cerevisiae, a protozoan parasite (Trypanosoma brucei brucei), mouse (Mus musculus) red blood cells and mouse spleen tissue cultures (containing 50–60% B-cells, ∼25–30% T-cells, and 5% myeloid cells). All assays were performed in duplicate or triplicate.
Activity against the bacteria and fungus was measured by assessing the lowest peptide concentration in a series of twofold dilutions at which no growth was detected (known as the minimum inhibitory concentration, MIC). Bacterial and yeast cultures (5×105 colony forming units/ml) were prepared in Müller-Hinton (MH) broth and Luria-Bertani (LB) broth respectively and transferred to serial dilutions of peptides ranging from 512 or 256 µM down to 1 or 0.5 µM in 96-well polypropylene plates. After incubation at 37°C (bacteria) or 30°C (S. cerevisiae) for 18 hours, growth of the cultures was checked by eye. In addition to MIC, we determined minimum microbicidal concentrations (MMC) by inoculating samples of the MIC assays on agar plates lacking peptides and checking for overnight growth. In all cases, MIC and MMC were identical.
Activity against T. brucei parasites was assessed as the minimum concentration in a series of twofold dilutions required to kill 95% of parasites in 30 minutes (LC95). Frozen stabilates of Trypanosoma brucei brucei AnTat1.1 bloodstream parasites were expanded by infection of C57Black/6 mice (Janvier). Mice with systemic parasitaemia (typically 4–5 days post infection) were exsanguinated and parasites were purified from heparinized blood by DEAE-cellulose (DE52, Whatman) chromatography. Collected parasites were washed twice with Phosphate-Saline-Glucose buffer (PSG-buffer) and enumerated microscopically using a Bürker hematocytometer. The DEAE52-purified parasites (stock: 106 parasites/ml PSG buffer containing 5% FCS) were added to polypropylene 96-well plates containing the serial peptide dilutions to obtain total volumes of 200 µl. After incubation for 30 minutes, trypanolytic activity was assessed using a light microscope by calculating the percentage of dead parasites in a minimum of 200 counted cells.
Activity against mouse red blood cells was examined by assessing the lowest peptide concentration in a series of twofold dilutions causing at least 50% hemolysis (HC50). Briefly, heparinized blood was obtained by cardiac puncture of mice euthanized via CO2. The blood was diluted 1/100 in RPMI and added to polypropylene 96-well plates containing the serial peptide dilutions to obtain total volumes of 100 µl. The plates were incubated at 37°C in a humidified atmosphere for 30 minutes and centrifuged at 1400 rpm for 2 minutes to pellet intact red blood cells allowing visual observation of hemolysis. Supernatants were subsequently transferred to 96-well flat-bottom plates to allow spectrophotometry using an ELISA reader (OD measured at 550 nm). The percentage of hemolyis for each peptide concentration was calculated as 100×(ODobs−OD0%)/(OD100%−OD0%), where ODobs is the OD measured for the peptide concentration, OD0% is the average OD in the absence of peptides (0% hemolysis), and OD100% is the average OD in the presence of 1% Tween-20 (100% hemolysis).
Activity against T-lymphocytes was assessed as the lowest peptide concentration in a series of twofold dilutions causing at least 50% inhibition of Concanavalin A-induced T-cell proliferation (IC50). Solutions containin 2×105 naive C57Black/6 splenocytes in the presence of 2.5 µg Concanavalin A were added to polypropylene 96-well plates containing the serial peptide dilutions to obtain total volumes of 100 µl. After incubation for 24 hours at 37°C in a humidified atmosphere, the cells were pulsed with [3H]thymidine and incubated for an additional 18 hours. The amount of [3H]thymidine incorporation was assessed by a β-counter as a measure of cell proliferation and the percentage of proliferation inhibition for each peptide concentration was calculated as 100×(1−(Tobs/T100%)), where Tobs is the [3H]thymidine level measured for the peptide concentration, and T100% is the [3H]thymidine level measured for Concanavalin-A-induced cell proliferation in the absence of peptide (representing 100% proliferation).
5. Analysis of gene promoter regions
Intergenic regions in the cluster were screened for the presence of evolutionary conserved elements (phylogenetic footprints) using the program Tracker [86]. Tracker identified the gene promoter regions as only conserved non-repeat regions in between adjacent AMP genes. The 500-bp upstream regions of all S. tropicalis AMP genes, four X. laevis AMP genes and the cck genes of five vertebrates were aligned using the EINSI algorithm in MAFFT 6.704 [87]. Candidate regulatory elements in the gene promoter regions were identified by: (1) determining aligned sites with more than 75% sequence conservation across all AMP genes, and (2) by searching for significantly overrepresented sequence motifs in the promoter regions using MEME 4.8.0 [42].
6. Phylogenetic reconstruction of pipid AMP gene diversification
AMP gene sequences retrieved from the S. tropicalis genome were combined in a single dataset with related gene and mRNA sequences of X. laevis retrieved from GenBank. Five amphibian cck gene or mRNA sequences were added to serve as outgroups. Phylogenetic relationships were estimated using the conventional ‘two-step’ approach, involving sequence alignment and tree reconstruction as separate steps of phylogeny inference, and a ‘direct optimization’ approach that accounts for alignment uncertainty by integrating sequence alignment and tree construction in a single algorithm. For the two-step approach, sequences were aligned using MAFFT. To avoid the erroneous alignment of nonhomologous sequences, exons were aligned separately and subsequently concatenated. Phylogenetic analyses were performed with MrBayes 3.1.2 [49] using a GTR+G+I model of DNA substitution. Two parallel runs of four incrementally heated (temperature parameter = 0.2) Markov chain Monte Carlo (MCMC) chains were performed, with a length of 10,000,000 generations, a sampling frequency of 1 per 1,000 generations, and a burn-in corresponding to the first 2,000,000 generations. Convergence of the parallel runs was confirmed by split frequency standard deviations (<0.01) and potential scale reduction factors (approximating 1.0) for all model parameters, as reported by MrBayes. Adequate posterior sampling was verified using Tracer 1.5 [88], by checking if the runs had reached effective sampling sizes >200 for all model parameters.
Bayesian analyses under direct optimization were conducted with BAli-Phy 2.0.2 [50]. Alignment constraints were imposed to maintain gene alignments that respect exon boundaries. The data set was analyzed under a GTR+G+I model of DNA substitution combined with a RS07 model of DNA insertion/deletion. Four independent analyses of a single MCMC chain each were run for ten million generations, and alignments and trees were sampled every 1000 generations. Again, convergence of the runs, and effective sampling size of the log-likelihood values and model parameters was checked using Tracer.
7. Selection analyses
We investigated whether diversifying (positive) or purifying (negative) selection affected the evolution of coding sequences in the expanding gene cluster using the random effects likelihood (REL) method, as implemented in the HYPHY software package [89] Kosakovsky Pond et al., 2005]. This method is considered the most suitable for datasets <50 sequences, draws sitewise synonymous and nonsynonymous codon substitution rates from separate rate heterogeneity distributions, and implements an empirical Bayes approach to test for significant selection at any specific site [90]. As such, REL represents an extension of the methods implementend in the benchmark program PAML [91]. Analyses were conducted using a MG94 codon substitution model ‘crossed’ with a GTR nucleotide substitution model, and codon sites were identified as subject to significant diversifying or purifying selection at Bayes factors >50. Due to the gain, loss and truncation of exons in the history of pipid AMP genes, the selection analyses were conducted on separate data sets for exon 2 (combining CCK and AMP gene sequences), exon 3 (composed of AMP gene sequences only) and the last exon (composed of CCK gene sequences and the homologous sequences of four AMPs that encode a CCK-like bioactive site).
Supporting Information
Zdroje
1. BevinsCL, ZasloffM (1990) Peptides from frog skin. Annu Rev Biochem 59 : 395–414.
2. BasirYJ, KnoopFC, DulkaJ, ConlonJM (2000) Multiple antimicrobial peptides and peptides related to bradykinin and neuromedin N isolated from skin secretions of the pickerel frog, Rana palustris. Biochim Biophys Acta 1543 : 95–105.
3. LiL, BjoursonAJ, HeJ, CaiG, RaoP, et al. (2003) Bradykinins and their cDNA from piebald odorous frog, Odorrana schmackeri, skin. Peptides 24 : 863–872.
4. HoffmannW, RichterK, KreilG (1983) A novel peptide designated PYLa and its precursor as predicted from cloned mRNA of Xenopus laevis skin. EMBO J 2 : 711–714.
5. GibsonBW, PoulterL, WilliamsDH, MaggioJE (1986) Novel peptide fragments originating from PGLa and the caerulein and xenopsin precursors from Xenopus laevis. J Biol Chem 261 : 5341–5349.
6. GiovanniniMG, PoulterL, GibsonBW, WilliamsDH (1987) Biosynthesis and degradation of peptides derived from Xenopus laevis prohormones. Biochem J 243 : 113–120.
7. ZasloffM (1987) Magainins, a class of antimicrobial peptides from Xenopus skin: Isolation, characterization of two active forms, and partial cDNA sequence of a precursor. Proc Natl Acad Sci USA 84 : 5449–5453.
8. TerryAS, PoulterL, WilliamsDH, NutkinsJC, GiovanniniMG, et al. (1988) The cDNA sequence coding for Prepro-PGS (Prepro-magainins) and aspects of the processing of this prepro-polypeptide. J Biol Chem 263 : 5745–5751.
9. DudaTF, VanhoyeD, NicolasP (2002) Roles of diversifying selection and coordinated evolution in the evolution of amphibian antimicrobial peptides. Mol Biol Evol 19 : 858–864.
10. VanhoyeD, BrustonF, NicolasP, AmicheM (2003) Antimicrobial peptides from hylid and ranin frogs originated from a 150-million-year-old ancestral precursor with a conserved signal peptide but a hypermutable antimicrobial domain. Eur J Biochem 270 : 2068–2081.
11. LiJ, XuX, XuC, ZhouW, ZhangK, et al. (2007) Anti-infection peptidomics of amphibian skin. Mol Cell Prot 6 : 882–894.
12. KönigE, Bininda-EmondsOR (2011) Evidence for convergent evolution in the antimicrobial peptide system in anuran amphibians. Peptides 32 : 20–25.
13. AliMF, SotoA, KnoopFC, ConlonJM (2001) Antimicrobial peptides isolated from skin secretions of the diploid frog, Xenopus tropicalis (Pipidae). Biochim Biophys Acta 1550 : 81–89.
14. ConlonJM, Al-GhaferiN, AhmedE, MeetaniMA, LeprinceJ, et al. (2010) Orthologs of magainin, PGLa, procaerulein-derived, and proxenopsin-derived peptides from skin secretions of the octoploid frog Xenopus amieti (Pipidae). Peptides 31 : 989–994.
15. ConlonJM, MechkarskaM, AhmedE, LeprinceJ, VaudryH, et al. (2011) Purification and properties of antimicrobial peptides from skin secretions of the Eritrea clawed frog Xenopus clivii (Pipidae). Comp Biochem Physiol C Toxicol Pharmacol 153 : 350–354.
16. ConlonJM, MechkarskaM, PrajeepM, SonnevendA, CoquetL, et al. (2012) Host-defense peptides in skin secretions of the tetraploid frog Silurana epitropicalis with potent activity against methicillin-resistant Staphylococcus aureus (MRSA). Peptides 37 : 113–119.
17. ConlonJM, MechkarskaM, KingJD (2012) Host-defense peptides in skin secretions of African clawed frogs (Xenopodinae, Pipidae). Gen Comp Endocrinol 176 : 513–518.
18. KingJD, MechkarskaM, CoquetL, LeprinceJ, JouenneT, et al. (2012) Host-defense peptides from skin secretions of the tetraploid frogs Xenopus petersii and Xenopus pygmaeus, and the octoploid frog Xenopus lenduensis (Pipidae). Peptides 33 : 35–43.
19. MechkarskaM, AhmedE, CoquetL, LeprinceJ, JouenneT, et al. (2010) Antimicrobial peptides with therapeutic potential from skin secretions of the Marsabit clawed frog Xenopus borealis (Pipidae). Comp Biochem Physiol C Toxicol Pharmacol 152 : 467–472.
20. MechkarskaM, EmanA, CoquetL, JérômeL, JouenneT, et al. (2011) Genome duplications within the Xenopodinae do not increase the multiplicity of antimicrobial peptides in Silurana paratropicalis and Xenopus andrei skin secretions. Comp Biochem Physiol Part D Genomics Proteomics 6 : 206–212.
21. MechkarskaM, AhmedE, CoquetL, LeprinceJ, JouenneT, et al. (2011) Peptidomic analysis of skin secretions demonstrates that the allopatric populations of Xenopus muelleri (Pipidae) are not conspecific. Peptides 32 : 1502–1508.
22. MechkarskaM, MeetaniM, MichalakP, VaksmanZ, TakadaK, et al. (2012) Hybridization between the African clawed frogs Xenopus laevis and Xenopus muelleri (Pipidae) increases the multiplicity of antimicrobial peptides in skin secretions of female offspring. Comp Biochem Physiol Part D Genomics Proteomics 7 : 285–291.
23. ZahidOK, MechkarskaM, OjoOO, Abdel-WahabYH, FlattPR, et al. (2011) Caerulein-and xenopsin-related peptides with insulin-releasing activities from skin secretions of the clawed frogs, Xenopus borealis and Xenopus amieti (Pipidae). Gen Comp Endocrinol 172 : 314–320.
24. AnastasiA, BertacciniG, CeiJM, de CaroG, ErspamerV, et al. (1970) Presence of caerulein in extracts of the skin of Leptodactylus pentadactylus labyrinthicus and of Xenopus laevis. Br J Pharmacol 38 : 221–228.
25. PoulterL, TerryAS, WilliamsDH, GiovanniniMG, MooreCH, et al. (1988) Levitide, a neurohormone-like peptide from the skin of Xenopus laevis. J Biol Chem 263 : 3279–3283.
26. ArakiK, TachibanaS, UchiyamaM, NakajimaT, YasuharaT (1973) Isolation and structure of a new active peptide “Xenopsin” on the smooth muscle, especially on a strip of fundus from a rat stomach, from the skin of Xenopus laevis. Chem Pharm Bull 23 : 2801–2804.
27. MooreKS, BevinsCL, BrasseurMM, TomassiniN, TurnerK, et al. (1991) Antimicrobial peptides in the stomach of Xenopus laevis. J Biol Chem 266 : 19851–19857.
28. KuchlerK, KreilG, SuresI (1989) The genes for the frog skin peptides GLa, xenopsin, levitide and caerulein contain a homologous export exon encoding a signal peptide sequence and part of an amphiphilic peptide. Eur J Biochem 179 : 281–285.
29. RichterK, AschauerH, KreilG (1985) Biosynthesis of peptides in the skin of Xenopus laevis: isolation of novel peptides predicted from the sequence of cloned cDNAs. Peptides 6 : 17–21.
30. RoelantsK, FryBG, NormanJA, ClynenE, SchoofsL, et al. (2010) Identical skin toxins by convergent molecular adaptation in frogs. Curr Biol 20 : 125–130.
31. PetersenTN, BrunakS, von HeijneG, NielsenH (2011) SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods 8 : 785–786.
32. BuchanDW, WardSM, LobleyAE, NugentTC, BrysonK, et al. (2010) Protein annotation and modelling servers at University College London. Nucleic Acids Res 38: W563–568.
33. ZasloffM (2002) Antimicrobial peptides of multicellular organisms. Nature 415 : 389–395.
34. BrogdenKA (2005) Antimicrobial peptides: pore formers or metabolic inhibitors in bacteria? Nat Rev Microbiol 3 : 238–250.
35. KönigE, ZhouM, WangL, ChenT, Bininda-EmondsOR, et al. (2012) Antimicrobial peptides and alytesin are co-secreted from the venom of the Midwife toad, Alytes maurus (Alytidae, Anura): Implications for the evolution of frog skin defensive secretions. Toxicon 60 : 967–981.
36. MorA, HaniK, NicolasP (1994) The vertebrate peptide antibiotics dermaseptins have overlapping structural features but target specific microorganisms. J Biol Chem 269 : 31635–31641.
37. WesterhoffHV, ZasloffM, RosnerJL, HendlerRW, De WaalA, et al. (1995) Functional synergism of the magainins PGLa and magainin-2 in Escherichia coli, tumor cells and liposomes. Eur J Biochem 228 : 257–264.
38. MatsuzakiK, MitaniY, AkadaKY, MursaeO, YoneyamaS, et al. (1998) Mechanism of synergism between antimicrobial peptides magainin 2 and PGLa. Biochemistry 37 : 15144–15153.
39. RourkeIJ, RehfeldJF, MøllerM, JohnsenAH (1997) Characterization of the cholecystokinin and gastrin genes from the bullfrog, Rana catesbeiana: evolutionary conservation of primary and secondary sites of gene expression. Endocrinology 138 : 1719–1727.
40. HansenTV (2001) Cholecystokinin gene transcription: promoter elements, transcription factors and signaling pathways. Peptides 22 : 1201–1211.
41. RourkeIJ, HansenTV, NerlovC, RehfeldJF, NielsenFC (1999) Negative cooperativity between juxtaposed E-box and cAMP/TPA responsive elements in the cholecystokinin gene promoter. FEBS Lett 448 : 15–18.
42. BaileyTL, BodenM, BuskeFA, FrithM, GrantCE, et al. (2009) MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res 37: W202–208.
43. MatysV, FrickeE, GeffersR, GösslingE, HaubrockM, et al. (2003) TRANSFAC: Transcriptional regulation, from patterns to profiles. Nucleic Acids Res 31 : 374–378.
44. FolettaVC, SegalDH, CohenDR (1998) Transcriptional regulation in the immune system: all roads lead to AP-1. J Leukoc Biol 63 : 139–152.
45. BrahmacharyM, SchonbachC, YangL, HuangE, TanS, et al. (2006) Computational promoter analysis of mouse, rat and human antimicrobial peptide-coding genes. BMC Bioinformatics 7 Suppl 5: S8.
46. WuGD, LaiEJ, HuangN, WenX (1997) Oct-1 and CCAAT/enhancer-binding protein (C/EBP) bind to overlapping elements within the interleukin-8 promoter. The role of Oct-1 as a transcriptional repressor. J Biol Chem 272 : 2396–2403.
47. KwonSY, CarlsonBA, ParkJM, LeeBJ (2000) Structural organization and expression of the gaegurin 4 gene of Rana rugosa. Biochim Biophys Acta 1492 : 185–190.
48. MieleR, BjörklundG, BarraD, SimmacoM, EngströmY (2001) Involvement of Rel factors in the expression of antimicrobial peptide genes in amphibia. Eur J Biochem 268 : 443–449.
49. RonquistF, HuelsenbeckJP (2003) MrBayes version 3.0: Bayesian phylogenetic inference under mixed models. Bioinformatics 19 : 1572–1574.
50. SuchardMA, RedelingsBD (2006) BAli-Phy: simultaneous Bayesian inference of alignment and phylogeny. Bioinformatics 22 : 2047–2048.
51. RoelantsK, GowerDJ, WilkinsonM, LoaderSP, BijuSD, et al. (2007) Global patterns of diversification in the history of modern amphibians. Proc Natl Acad Sci USA 104 : 887–892.
52. YangX, LeeWH, ZhangY (2012) Extremely abundant antimicrobial peptides existed in the skins of nine kinds of Chinese odorous frogs. J Proteome Res 11 : 306–319.
53. ParkerJM, MikaelianI, HahnN, DiggsHE (2002) Clinical diagnosis and treatment of epidermal chytridiomycosis in African clawed frogs (Xenopus tropicalis). Comp Med 52 : 265–268.
54. RamseyJP, ReinertLK, HarperLK, WoodhamsDC, Rollins-SmithLA (2010) Immune defenses against Batrachochytrium dendrobatidis, a fungus linked to global amphibian declines, in the South African clawed frog, Xenopus laevis. Infect Immun 78 : 3981–3992.
55. RosenblumEB, PoortenTJ, SettlesM, MurdochGK, RobertJ, et al. (2009) Genome-Wide Transcriptional Response of Silurana (Xenopus) tropicalis to Infection with the Deadly Chytrid Fungus. PLoS ONE 4: e6494.
56. RibasL, LiM-S, DoddingtonBJ, RobertJ, SeidelJA, et al. (2009) Expression Profiling the Temperature-Dependent Amphibian Response to Infection by Batrachochytrium dendrobatidis. PLoS ONE 4: e8408.
57. SrinivasanD, MechkarskaM, Abdel-WahabYH, FlattPR, ConlonJM (2013) Caerulein precursor fragment (CPF) peptides from the skin secretions of Xenopus laevis and Silurana epitropicalis are potent insulin-releasing agents. Biochimie 95 : 429–435.
58. NeiM, RooneyAP (2005) Concerted and Birth-and-Death Evolution of Multigene Families. Annu Rev Genet 39 : 121–152.
59. LynchVJ (2007) Inventing an arsenal: adaptive evolution and neofunctionalization of snake venom phospholipase A2 genes. BMC Evol Biol 7 : 2.
60. FryBG, ScheibH, van der WeerdL, YoungB, McNaughtanJ, RamjanSFR, VidalN, PoelmannRE, NormanJA (2008) Evolution of an Arsenal. Structural and functional diversification of the venom system in the advanced snakes (Caenophidia). Mol Cell Proteomics 7 : 215–246.
61. KondrashovFA, KooninEV (2001) Origin of alternative splicing by tandem exon duplication. Hum Mol Genet 10 : 2661–2669.
62. McCruddenCM, ZhouM, ChenT, O'RourkeM, WalkerB, et al. (2007) The complex array of bradykinin-related peptides (BRPs) in the peptidome of pickerel frog (Rana palustris) skin secretion is the product of transcriptional economy. Peptides 28 : 1275–1281.
63. Bowie JH, Tyler MJ (2006) Host defense peptides from Australian amphibians: Caerulein and other neuropeptides. In: Kastin AJ, editor. Handbook of biologically active peptides. San Diego: Academic Press. pp. 283–289.
64. CheclerF, LabbéC, GranierC, van RietschotenJ, KitabgiP, VincentJP (1982) [TRP11]-neurotensin and xenopsin discriminate between rat and guinea-pig neurotensin receptors. Life Sci 31 : 1145–1150.
65. ClemensA, KatsoulisS, NustedeR, SeebeckJ, SeyfarthK, Morys-WortmannC, FeurleGE, FolschUR, SchmidtWE (1997) Relaxant effect of xenin on rat ileum is mediated by apamin-sensitive neurotensin-type receptors. Am J Physiol 272: G190–G196.
66. FeurleGE (1998) Xenin - – a review. Peptides 19 : 609–615.
67. KalafatakisK, TriantafyllouK (2011) Contribution of neurotensin in the immune and neuroendocrine modulation of normal and abnormal enteric function. Regul Pept 170 : 7–17.
68. KitabgiP (2006) Differential processing of pro-neurotensin/neuromedin N and relationship to pro-hormone convertases. Peptides 27 : 2508–2514.
69. ChowVTK, QuekHH (1997) Alpha coat protein COPA (HEP-COP): presence of an Alu repeat in cDNA and identity of the amino terminus to xenin. Ann Hum Genet 61 : 369–373.
70. CasewellNR, WagstaffSC, HarrisonRA, RenjifoC, WüsterW (2011) Domain Loss Facilitates Accelerated Evolution and Neofunctionalization of Duplicate Snake Venom Metalloproteinase Toxin Genes. Mol Biol Evol 28 : 2637–2649.
71. ChangD, DudaTFJr (2012) Extensive and continuous duplication facilitates rapid evolution and diversification of gene families. Mol Biol Evol 29 : 2019–2029.
72. WongES, BelovK (2012) Venom evolution through gene duplications. Gene 496 : 1–7.
73. Ohno S (1970) Evolution by Gene Duplication. Berlin: Springer.
74. ForceA, LynchM, Bryan PickettF, AmoresA, YanY, et al. (1999) Preservation of duplicate genes by complementary, degenerative mutations. Genetics 151 : 1531–1545.
75. HittingerCT, CarrollSB (2007) Gene duplication and the adaptive evolution of a classic genetic switch. Nature 449 : 677–681.
76. SikosekT, ChanHS, Bornberg-BauerE (2012) Escape from Adaptive Conflict follows from weak functional trade-offs and mutational robustness. Proc Natl Acad Sci USA 109 : 14888–14893.
77. BergthorssonU, AnderssonDI, RothJR (2007) Ohno's dilemma: evolution of new genes under continuous selection. Proc Natl Acad Sci U S A 104 : 17004–17009.
78. Kozminsky-AtiasA, Bar-ShalomA, MishmarD, ZilberbergN (2008) Assembling an arsenal, the scorpion way. BMC Evol Biol 8 : 333.
79. WangL, ChenY, YangM, ZhouM, ChenT, et al. (2010) Peptide DV-28 amide: An inhibitor of bradykinin-induced arterial smooth muscle relaxation encoded by Bombina orientalis skin kininogen-2. Peptides 31 : 979–982.
80. MurayamaN, HayashiMA, OhiH, FerreiraLA, HermannVV, et al. (1997) Cloning and sequence analysis of a Bothrops jararaca cDNA encoding a precursor of seven bradykinin-potentiating peptides and a C-type natriuretic peptide. Proc Natl Acad Sci USA 94 : 1189–1193.
81. FryBG, RoelantsK, WinterK, HodgsonWC, GriesmanL, et al. (2010) Novel venom proteins produced by differential domain-expression strategies in beaded lizards and gila monsters (genus Heloderma). Mol Biol Evol 27 : 395–407.
82. MorA, DelfaourA, NicolasP (1991) Identification of a D-alanine-containing polypeptide precursor for the peptide opioid, dermorphin. J Biol Chem 266 : 6264–6270.
83. ChenM, CheQ, WangX, LiJ, YangH, et al. (2010) Cloning and characterization of the first amphibian bradykinin gene. Biochimie 92 : 226–231.
84. PerkinsDN, PappinDJ, CreasyDM, CottrellJS (1999) Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20 : 3551–3567.
85. Tossi A, Sandri L, Giangaspero A (2002) New consensus hydrophobicity scale extended to non-proteinogenic amino acids. In: Peptides 2002: Proceedings of the twenty-seventh European peptide symposium. Napoli: Edizioni Ziino. pp. 416–417.
86. ProhaskaSJ, FriedC, FlammC, WagnerGP, StadlerPF (2004) Surveying phylogenetic footprints in large gene clusters: applications to Hox cluster duplications. Mol Phylogenet Evol 31 : 581–604.
87. KatohK, KumaK, TohH, MiyataT (2005) MAFFT version 5: Improvement in accuracy of multiple sequence alignment. Nucleic Acids Res 33 : 511–518.
88. Rambaut A, Drummond AJ (2007) Tracer v1.5. Available: http://beast.bio.ed.ac.uk/Tracer. Accessed: 01 February, 2010.
89. Kosakovsky PondSL, FrostSDW, MuseSV (2005) HyPhy: hypothesis testing using phylogenies. Bioinformatics 21 : 676–679.
90. Kosakovsky PondSL, FrostSDW (2005) Not So Different After All: A Comparison of Methods for Detecting Amino Acid Sites Under Selection. Mol Biol Evol 22 : 1208–1222.
91. YangZ (2007) PAML4: Phylogenetic Analysis by Maximum Likelihood. Mol Biol Evol 24 : 1586–1591.
Štítky
Genetika Reprodukční medicína
Článek Re-Ranking Sequencing Variants in the Post-GWAS Era for Accurate Causal Variant IdentificationČlánek Bypass of 8-oxodGČlánek Integrated Model of and Inherited Genetic Variants Yields Greater Power to Identify Risk GenesČlánek Comparative Genomic and Functional Analysis of 100 Strains and Their Comparison with Strain GGČlánek A Nuclear Calcium-Sensing Pathway Is Critical for Gene Regulation and Salt Stress Tolerance inČlánek Computational Identification of Diverse Mechanisms Underlying Transcription Factor-DNA OccupancyČlánek Reversible and Rapid Transfer-RNA Deactivation as a Mechanism of Translational Repression in StressČlánek Genome-Wide Association of Body Fat Distribution in African Ancestry Populations Suggests New Loci
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2013 Číslo 8
-
Všechny články tohoto čísla
- Genome-Wide DNA Methylation Analysis of Systemic Lupus Erythematosus Reveals Persistent Hypomethylation of Interferon Genes and Compositional Changes to CD4+ T-cell Populations
- Re-Ranking Sequencing Variants in the Post-GWAS Era for Accurate Causal Variant Identification
- Histone Variant HTZ1 Shows Extensive Epistasis with, but Does Not Increase Robustness to, New Mutations
- Past Visits Present: TCF/LEFs Partner with ATFs for β-Catenin–Independent Activity
- Functional Characterisation of Alpha-Galactosidase A Mutations as a Basis for a New Classification System in Fabry Disease
- A Flexible Approach for the Analysis of Rare Variants Allowing for a Mixture of Effects on Binary or Quantitative Traits
- Masculinization of Gene Expression Is Associated with Exaggeration of Male Sexual Dimorphism
- Genic Intolerance to Functional Variation and the Interpretation of Personal Genomes
- Endogenous Stress Caused by Faulty Oxidation Reactions Fosters Evolution of 2,4-Dinitrotoluene-Degrading Bacteria
- Transposon Domestication versus Mutualism in Ciliate Genome Rearrangements
- Comparative Anatomy of Chromosomal Domains with Imprinted and Non-Imprinted Allele-Specific DNA Methylation
- An Essential Function for the ATR-Activation-Domain (AAD) of TopBP1 in Mouse Development and Cellular Senescence
- Depletion of Retinoic Acid Receptors Initiates a Novel Positive Feedback Mechanism that Promotes Teratogenic Increases in Retinoic Acid
- Bypass of 8-oxodG
- Calpain-6 Deficiency Promotes Skeletal Muscle Development and Regeneration
- ATM Release at Resected Double-Strand Breaks Provides Heterochromatin Reconstitution to Facilitate Homologous Recombination
- Generation of Tandem Direct Duplications by Reversed-Ends Transposition of Maize Elements
- Loss of a Conserved tRNA Anticodon Modification Perturbs Cellular Signaling
- Integrated Model of and Inherited Genetic Variants Yields Greater Power to Identify Risk Genes
- High-Throughput Genetic and Gene Expression Analysis of the RNAPII-CTD Reveals Unexpected Connections to SRB10/CDK8
- Dynamic Rewiring of the Retinal Determination Network Switches Its Function from Selector to Differentiation
- β-Catenin-Independent Activation of TCF1/LEF1 in Human Hematopoietic Tumor Cells through Interaction with ATF2 Transcription Factors
- Genetic Mapping of Specific Interactions between Mosquitoes and Dengue Viruses
- A Highly Redundant Gene Network Controls Assembly of the Outer Spore Wall in
- Origin and Functional Diversification of an Amphibian Defense Peptide Arsenal
- Myc-Driven Overgrowth Requires Unfolded Protein Response-Mediated Induction of Autophagy and Antioxidant Responses in
- Integrative Modeling of eQTLs and Cis-Regulatory Elements Suggests Mechanisms Underlying Cell Type Specificity of eQTLs
- Species and Population Level Molecular Profiling Reveals Cryptic Recombination and Emergent Asymmetry in the Dimorphic Mating Locus of
- Ras-Induced Changes in H3K27me3 Occur after Those in Transcriptional Activity
- Characterization of the p53 Cistrome – DNA Binding Cooperativity Dissects p53's Tumor Suppressor Functions
- Global Analysis of Fission Yeast Mating Genes Reveals New Autophagy Factors
- Deficiency Suppresses Intestinal Tumorigenesis
- Introns Regulate Gene Expression in in a Pab2p Dependent Pathway
- Meiotic Recombination Initiation in and around Retrotransposable Elements in
- Comparative Oncogenomic Analysis of Copy Number Alterations in Human and Zebrafish Tumors Enables Cancer Driver Discovery
- Comparative Genomic and Functional Analysis of 100 Strains and Their Comparison with Strain GG
- A Model-Based Analysis of GC-Biased Gene Conversion in the Human and Chimpanzee Genomes
- Masculinization of the X Chromosome in the Pea Aphid
- The Architecture of a Prototypical Bacterial Signaling Circuit Enables a Single Point Mutation to Confer Novel Network Properties
- Distinct SUMO Ligases Cooperate with Esc2 and Slx5 to Suppress Duplication-Mediated Genome Rearrangements
- The Yeast Environmental Stress Response Regulates Mutagenesis Induced by Proteotoxic Stress
- Mediator Directs Co-transcriptional Heterochromatin Assembly by RNA Interference-Dependent and -Independent Pathways
- The Genome of and the Basis of Host-Microsporidian Interactions
- Regulation of Sister Chromosome Cohesion by the Replication Fork Tracking Protein SeqA
- Neuronal Reprograming of Protein Homeostasis by Calcium-Dependent Regulation of the Heat Shock Response
- A Nuclear Calcium-Sensing Pathway Is Critical for Gene Regulation and Salt Stress Tolerance in
- Cross-Species Array Comparative Genomic Hybridization Identifies Novel Oncogenic Events in Zebrafish and Human Embryonal Rhabdomyosarcoma
- : A Mouse Strain with an Ift140 Mutation That Results in a Skeletal Ciliopathy Modelling Jeune Syndrome
- The Relative Contribution of Proximal 5′ Flanking Sequence and Microsatellite Variation on Brain Vasopressin 1a Receptor () Gene Expression and Behavior
- Combining Quantitative Genetic Footprinting and Trait Enrichment Analysis to Identify Fitness Determinants of a Bacterial Pathogen
- The Innocence Project at Twenty: An Interview with Barry Scheck
- Computational Identification of Diverse Mechanisms Underlying Transcription Factor-DNA Occupancy
- GUESS-ing Polygenic Associations with Multiple Phenotypes Using a GPU-Based Evolutionary Stochastic Search Algorithm
- H2A.Z Acidic Patch Couples Chromatin Dynamics to Regulation of Gene Expression Programs during ESC Differentiation
- Identification of DSB-1, a Protein Required for Initiation of Meiotic Recombination in , Illuminates a Crossover Assurance Checkpoint
- Binding of TFIIIC to SINE Elements Controls the Relocation of Activity-Dependent Neuronal Genes to Transcription Factories
- Global Analysis of the Sporulation Pathway of
- Genetic Circuits that Govern Bisexual and Unisexual Reproduction in
- Deletion of microRNA-80 Activates Dietary Restriction to Extend Healthspan and Lifespan
- Fifty Years On: GWAS Confirms the Role of a Rare Variant in Lung Disease
- The Enhancer Landscape during Early Neocortical Development Reveals Patterns of Dense Regulation and Co-option
- Gene Expression Regulation by Upstream Open Reading Frames and Human Disease
- Sociogenomics of Cooperation and Conflict during Colony Founding in the Fire Ant
- The Intronic Long Noncoding RNA Recruits PRC2 to the Promoter, Reducing the Expression of and Increasing Cell Proliferation
- The , p.E318G Variant Increases the Risk of Alzheimer's Disease in -ε4 Carriers
- The Wilms Tumor Gene, , Is Critical for Mouse Spermatogenesis via Regulation of Sertoli Cell Polarity and Is Associated with Non-Obstructive Azoospermia in Humans
- Reversible and Rapid Transfer-RNA Deactivation as a Mechanism of Translational Repression in Stress
- QTL Analysis of High Thermotolerance with Superior and Downgraded Parental Yeast Strains Reveals New Minor QTLs and Converges on Novel Causative Alleles Involved in RNA Processing
- Genome Wide Association Identifies Novel Loci Involved in Fungal Communication
- Chromatin Sampling—An Emerging Perspective on Targeting Polycomb Repressor Proteins
- A Recessive Founder Mutation in Regulator of Telomere Elongation Helicase 1, , Underlies Severe Immunodeficiency and Features of Hoyeraal Hreidarsson Syndrome
- Genome-Wide Association of Body Fat Distribution in African Ancestry Populations Suggests New Loci
- Causal and Synthetic Associations of Variants in the Gene Cluster with Alpha1-antitrypsin Serum Levels
- Hard Selective Sweep and Ectopic Gene Conversion in a Gene Cluster Affording Environmental Adaptation
- Brittle Culm1, a COBRA-Like Protein, Functions in Cellulose Assembly through Binding Cellulose Microfibrils
- Chromosomal Copy Number Variation, Selection and Uneven Rates of Recombination Reveal Cryptic Genome Diversity Linked to Pathogenicity
- The Ribosomal Protein Rpl22 Controls Ribosome Composition by Directly Repressing Expression of Its Own Paralog, Rpl22l1
- Ras1 Acts through Duplicated Cdc42 and Rac Proteins to Regulate Morphogenesis and Pathogenesis in the Human Fungal Pathogen
- The DSB-2 Protein Reveals a Regulatory Network that Controls Competence for Meiotic DSB Formation and Promotes Crossover Assurance
- Recurrent Modification of a Conserved -Regulatory Element Underlies Fruit Fly Pigmentation Diversity
- Associations of Mitochondrial Haplogroups B4 and E with Biliary Atresia and Differential Susceptibility to Hydrophobic Bile Acid
- The Conditional Nature of Genetic Interactions: The Consequences of Wild-Type Backgrounds on Mutational Interactions in a Genome-Wide Modifier Screen
- A Critical Function of Mad2l2 in Primordial Germ Cell Development of Mice
- A Role for CF1A 3′ End Processing Complex in Promoter-Associated Transcription
- Vitellogenin Underwent Subfunctionalization to Acquire Caste and Behavioral Specific Expression in the Harvester Ant
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Chromosomal Copy Number Variation, Selection and Uneven Rates of Recombination Reveal Cryptic Genome Diversity Linked to Pathogenicity
- Genome-Wide DNA Methylation Analysis of Systemic Lupus Erythematosus Reveals Persistent Hypomethylation of Interferon Genes and Compositional Changes to CD4+ T-cell Populations
- Associations of Mitochondrial Haplogroups B4 and E with Biliary Atresia and Differential Susceptibility to Hydrophobic Bile Acid
- A Role for CF1A 3′ End Processing Complex in Promoter-Associated Transcription
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



