-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaMediator Directs Co-transcriptional Heterochromatin Assembly by RNA Interference-Dependent and -Independent Pathways
Heterochromatin at the pericentromeric repeats in fission yeast is assembled and spread by an RNAi-dependent mechanism, which is coupled with the transcription of non-coding RNA from the repeats by RNA polymerase II. In addition, Rrp6, a component of the nuclear exosome, also contributes to heterochromatin assembly and is coupled with non-coding RNA transcription. The multi-subunit complex Mediator, which directs initiation of RNA polymerase II-dependent transcription, has recently been suggested to function after initiation in processes such as elongation of transcription and splicing. However, the role of Mediator in the regulation of chromatin structure is not well understood. We investigated the role of Mediator in pericentromeric heterochromatin formation and found that deletion of specific subunits of the head domain of Mediator compromised heterochromatin structure. The Mediator head domain was required for Rrp6-dependent heterochromatin nucleation at the pericentromere and for RNAi-dependent spreading of heterochromatin into the neighboring region. In the latter process, Mediator appeared to contribute to efficient processing of siRNA from transcribed non-coding RNA, which was required for efficient spreading of heterochromatin. Furthermore, the head domain directed efficient transcription in heterochromatin. These results reveal a pivotal role for Mediator in multiple steps of transcription-coupled formation of pericentromeric heterochromatin. This observation further extends the role of Mediator to co-transcriptional chromatin regulation.
Published in the journal: . PLoS Genet 9(8): e32767. doi:10.1371/journal.pgen.1003677
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1003677Summary
Heterochromatin at the pericentromeric repeats in fission yeast is assembled and spread by an RNAi-dependent mechanism, which is coupled with the transcription of non-coding RNA from the repeats by RNA polymerase II. In addition, Rrp6, a component of the nuclear exosome, also contributes to heterochromatin assembly and is coupled with non-coding RNA transcription. The multi-subunit complex Mediator, which directs initiation of RNA polymerase II-dependent transcription, has recently been suggested to function after initiation in processes such as elongation of transcription and splicing. However, the role of Mediator in the regulation of chromatin structure is not well understood. We investigated the role of Mediator in pericentromeric heterochromatin formation and found that deletion of specific subunits of the head domain of Mediator compromised heterochromatin structure. The Mediator head domain was required for Rrp6-dependent heterochromatin nucleation at the pericentromere and for RNAi-dependent spreading of heterochromatin into the neighboring region. In the latter process, Mediator appeared to contribute to efficient processing of siRNA from transcribed non-coding RNA, which was required for efficient spreading of heterochromatin. Furthermore, the head domain directed efficient transcription in heterochromatin. These results reveal a pivotal role for Mediator in multiple steps of transcription-coupled formation of pericentromeric heterochromatin. This observation further extends the role of Mediator to co-transcriptional chromatin regulation.
Introduction
Heterochromatin is a silent higher-order chromatin structure that is associated with various genome functions such as transcriptional regulation, chromosomal segregation, suppression of recombination and repression of selfish elements. The fission yeast Schizosaccharomyces pombe provides a good model system for investigating heterochromatin formation. In fission yeast, heterochromatin is preferentially enriched across large chromosomal domains at the pericentromeres, subtelomeres and the mating-type locus. These regions are rich in methylation of histone H3K9 (H3K9me), which is catalyzed by the histone methyltransferase Clr4, a homolog of mammalian SUV39h [1], [2]. The modification of H3K9me is critical for the binding of HP1 proteins [2], [3], which recruit various factors for the assembly of repressive chromatin and associated various functions [4], [5].
Several distinct pathways promote heterochromatin assembly in fission yeast. At the pericentromere, RNAi machinery plays essential roles in heterochromatin formation [6], [7]. Pericentromeric heterochromatin is assembled on the outer repeat (otr) region (containing of dg and dh repeats), and the outer portion of the innermost repeats (imr), which surround the central core (cnt) domain, the site of kinetochore assembly [8]. The repeats are transcribed by RNA polymerase II (RNAPII) to produce non-coding RNAs (ncRNAs) during S-phase [9], [10], [11]. Transcribed ncRNAs give rise to double-strand RNA via the RNA-dependent RNA polymerase complex (RDRC), comprised of Rdp1, Cid12 and Hrr1, and are processed into small interfering RNAs (siRNAs) by the RNase III helicase Dicer (Dcr1). The siRNAs are then loaded into an RNA-induced transcriptional silencing (RITS) complex composed of Ago1, Tas3 and Chp1 [7]. siRNAs target the RITS complex to cognate nascent transcripts, resulting in the recruitment of additional factors, including RDRC and ultimately Clr4, to methylate histone H3K9. Generation of siRNAs and heterochromatin assembly are interdependent processes that form a self-enforcing loop [12], [13]. Importantly, RNAPII appears to couple transcription at the target loci with the generation of siRNAs. This was shown by the fact that a specific mutation in RNAPII results in a decrease in heterochromatic histone modifications, accumulation of pericentromeric transcripts, and accompanying loss of siRNAs, which are effects that were observed previously in RNAi mutants [10].
Heterochromatin, once established, spreads into neighboring region, which is typically shown by the heterochromatin formation and silencing of the genes inserted into heterochromatin. This process depends on RNAi system and probably couples with transcription [14], [15].
Nuclear RNA is monitored by a nuclear RNA surveillance system involving exosomes with 3′-5′ exonuclease activity, and a portion of the ncRNA at the pericentromere has been shown to be degraded by the nuclear exosome [16], [17]. In addition to RNA degradation, 3′-5′ exonuclease Rrp6, a component of the nuclear exosome, was shown to mediate heterochromatin formation in parallel with RNAi, which is demonstrated by the cumulative increase and decrease of H3K9me at the pericentromere in the double null-mutant of ago1 and rrp6 [18]. Since the amount of siRNA is not affected by depletion of Rrp6, Rrp6-dependent heterochromatin formation occurs via a pathway that is distinct from that of RNAi-dependent siRNA generation [16]. The molecular basis of the Rrp6-dependent pathway is not yet clear.
The cenH sequence, which shows 96% homology to centromeric dg and dh repeats, is present at the silent mating-type (mat2/3) locus and serves as an RNAi-dependent heterochromatin nucleation center [6], [19]. In parallel with the RNAi-dependent pathway, the ATF/CREB family DNA-binding proteins, Atf1 and Pcr1, participate in heterochromatin nucleation with a histone deacetylase, Clr3 [20].
Mediator, which is a well-conserved protein complex consisting of at least 20 subunits, was first identified as a factor that mediates DNA transcription factors binding at regulatory sequneces and RNAPII at promoters for the efficient start of transcription [21], [22] and has been shown to be required for transcription of almost all protein-coding genes in vivo [23], [24], [25]. Structural analysis indicates that this complex consists of four distinct structural domains: head, middle, tail and kinase. The head domain is responsible for extensive interaction with RNAPII, and the Med18/Pmc6-Med20 heterodimer, which is a portion of the head domain, binds to the core head domain through the C-terminal helix of Med8 [26]. The head domain stabilizes the connection between RNAPII and TFIIH, which facilitates the transition from initiation complex to elongation complex [27]. In addition to the promotion of general transcription from protein-coding genes, recent studies have revealed a new function of Mediator. In Arabidopsis thaliana, Mediator directs the transcription of ncRNA genes by recruiting RNAPII to their promoters [28]. In mammalian cells, a specific subunit of Mediator functions as an interaction site for alternative mRNA splicing or transcription elongation factors [29], [30]. These data suggest that Mediator might play roles in both transcription elongation and the subsequent processing of transcripts as a platform for the recruitment of various factors.
Since both Rrp6-dependent heterochromatin formation and RNAi-dependent heterochromatin formation are coupled with transcription, we assumed that the factor(s) that interacts with RNAPII directs the coupling. Therefore, we assessed the role of several RNAPII-interacting factors in pericentromeric heterochromatin assembly. We found that the disruption of Med18 and Med20, non-essential subunits of the Mediator head domain (MHD), compromised both RNAi-dependent and Rrp6-dependent heterochromatin assembly at the pericentromere. In addition, the head domain is required for transcriptional activation in heterochromatin. Therefore, we propose that Mediator links transcription of ncRNA and its processing by RNAi and exosomes for the formation of centromeric heterochromatin.
Results
Mediator is required for heterochromatic silencing at the pericentromeres
To investigate whether Mediator is involved in heterochromatin assembly, each gene encoding a non-essential subunit of mediator was disrupted in a strain possessing marker genes in the pericentromeric heterochromatin (otr1R::ade6+ and imr1L::ura4+) to monitor heterochromatic silencing (Figure 1A) [31]. Since the otr1R::ade6+ and imr1L::ura4+ genes are repressed by heterochromatin, the wild-type strain formed red colonies on a solid medium containing a limiting amount of adenine (Low Ade) and was resistant to 5-fluoroorotic acid (5-FOA), a counter-selective drug for ura4+ expression. By contrast, heterochromatin mutants, such as clr4Δ, formed white or pink colonies on the Low Ade plate and showed sensitivity to 5-FOA (Figure 1B). Among the eight non-essential subunits of mediator tested (med1/pmc2, med27/pmc3, med18/pmc6, med20, med19/rox3, med12/srb8, med13/srb9 and cdk8/srb10), only disruption of med18 (also known as pmc6) and med20 resulted in the formation of pink colonies and increased sensitivity to 5-FOA (Figure 1B), which is in accordance with growth on plates lacking uracil or adenine (Figure S1A). Closer examination revealed that both med18Δ med20Δ cells formed a mixture of white and pink colonies on Low Ade plates. In addition, point mutants of med8 (med8-K9) and med31 (med31-H1), which were isolated by the screening of heterochromatic mutants (Figure S1B; Kato et al. submitted), also formed a mixture of pink and white colonies (Figure 1C, Figure S1C).
Fig. 1. Mediator is required for heterochromatic silencing at the pericentromere. 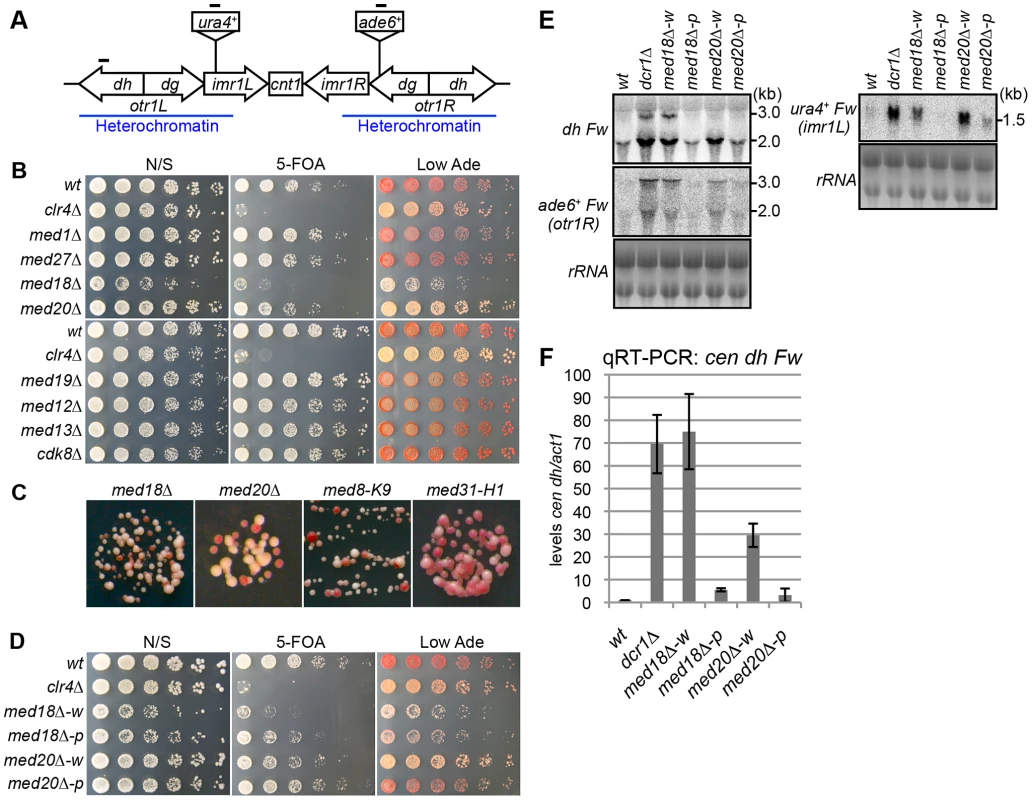
(A) Schematic of fission yeast centromere 1. Locations of ura4 and ade6 reporter genes inserted within the pericentromeric region are shown (imr1L::ura4+ and otr1R::ade6). Black bars indicate the location of primers or probes used for ChIP, RT-PCR and northern analysis. (B) Silencing assay at the pericentromere. Shown are the results of serial dilutions of the indicated strains spotted onto non-selective media (N/S), medium with 5-fluoroorotic acid (5-FOA), and medium with a limited amount of adenine (Low Ade) to assay ura4+ and ade6+ expression. (C) Spots on Low Ade medium using Mediator mutants (med18Δ, med20Δ, med8-K9 and med31-H1), which are defective in heterochromatic silencing at the pericentromere. (D) Silencing assay at the pericentromere. Shown are the results of serial dilutions of the indicated strains spotted onto N/S, 5-FOA and Low Ade media to assay ura4+ and ade6+ expression. w indicates white epiclones (med18Δ-w and med20Δ-w), and p indicates pink epiclones (med18Δ-p and med20Δ-p). (E) Northern Analysis of dh, otr1R::ade6 and imr1L::ura4 forward strand transcripts in wild-type (wt) and mutant cells using oligonucleotide probes. rRNA was used as a loading control. (F) Quantitative RT-PCR analysis of cen dh forward transcript levels relative to a control act1+, normalized to the wild type in the indicated strains. Error bars show the standard error of the mean (n = 3). The variegation in the color of colonies by the mutation of Mediator subunits suggested that silencing of the otr1R::ade6+ gene was variegated in the mutant cells and that distinct levels of otr1R::ade6+ silencing were epigenetically inherited. To test the stability of the pink and white phenotype, pink and white colonies of each mutant were selected, cultured in YES media overnight and re-spotted onto Low Ade and 5-FOA plates. Re-spotting of the cells from white colonies (med18Δ-w, med20Δ-w and med8-K9-w) and pink colonies (med18Δ-p med20Δ-p and med8-K9-p) produced predominantly white colonies and pink colonies, respectively (Figure 1D, Figure S1C). This indicated that the white and pink phenotypes were epigenetically inherited through generation but exchangeable, which is further confirmed by the measurements of the conversion rates between white and pink epiclones (Figure S2). The conversion rates are different in each mutants, but in all mutants, conversion rates from pink to white is higher than those of white to pink, showing that white-epiclones, in which heterochromatin is compromised, are more stable. Hereafter, we designate the epigenetic clones derived from white colonies and red colonies as white and pink “epiclones”, respectively. med18Δ-w and med20Δ-w showed greater sensitivity to 5-FOA than med18Δ-p and med20Δ-p (Figure 1D), indicating that silencing at imr1L::ura4+ was also compromised more severely in the white epiclones and that the silencing defect at otr1R::ade6+ is connected with that at imr1L::ura4+. This suggested that the white phenotype reflected silencing defects of the entire pericentromeric heterochromatin. It should be noted that it was difficult to separate the white epiclones from the pink epiclones of med31-H1 cells (med31-H1-w and med31-H1-p in Figure S1C) because of frequent variegation between the two (Figure S2).
The loss of heterochromatic gene silencing was confirmed in Mediator mutants by measuring the accumulation of transcripts from the pericentromeric repeats (dg and dh) and inserted marker genes. Strand-specific northern analysis showed a large increase in those transcripts in both med18Δ- and med20Δ-w cells (Figure 1E and Figure S3), which was consistent with the observed silencing defects (Figure 1D), while only marginal accumulation was observed in med18Δ-p and med20Δ-p cells. Both point mutants of the other Mediator subunits (med8-K9 and med31-H1) also showed accumulation of the transcripts (Figure S3). Accumulation of heterochromatic transcripts from dh repeats was also demonstrated by strand-specific RT-PCR (Figure 1F). These results showed that the Mediator subunits Med8, Med18, Med20 and Med31 were involved in silencing of pericentromeric heterochromatin. Med18 and Med20 form a heterodimer that associates with head domain core complex through the C-terminal linker region of Med8 [26], and the connection between the Med18/Med20 heterodimer and head domain appears to be lost in the med8-K9 mutant (Figure S1B). In addition, Med31, a component of the middle domain, is located close to the head domain. Because these findings indicate that Mediator functions in pericentromeric heterochromatin via the head domain, Med18 and Med20 were selected for closer examination.
Mediator localizes with RNAPII at the pericentromeric repeats
Both RNAi - and Rrp6-dependent heterochromatin formation, which occur in parallel at the pericentromere, appear to be coupled with the transcription of ncRNA at the pericentromeric repeats [10], [18]. We, hence, assume that Mediator contributed to pericentromeric heterochromatin formation directly through the transcription of ncRNA and/or processing of ncRNA. If this assumption was true, Mediator should localize to the transcribed region in heterochromatic repeats. To test this possibility, the localization of Med20-5Flag and RNAPII to the transcribed regions of dh repeats was examined by Chromatin immunoprecipitation (ChIP) assay. Since heterochromatic ncRNA is mainly transcribed during G1/S-phase [11], cell cycle was synchronized using the cdc25 temperature-sensitive mutation. The results show that RNAPII accumulated during G1 to early S-phase, followed by the accumulation of transcripts (, C, D). Med20-5Flag showed a similar oscillating pattern, but the peak disappeared slightly earlier than the Pol2 peak (Figure S4B). This is consistent with the speculation that Mediator is involved in the heterochromatic ncRNA transcription.
Loss of Med18 or Med20 causes defects in heterochromatin structure at the pericentromeres
To gain further insight into the roles of Mediator during heterochromatin organization, the occupancy of H3K9me2, Swi6 and RNAPII at the centromeric heterochromatin was assessed by ChIP assay. At the inserted marker gene (otr1R::ade6+), the levels of histone H3K9me and Swi6 were decreased and RNAPII occupancy was increased in the white epiclones of med18Δ and med20Δ (Figure 2A–C). This indicated that heterochromatin structure at the marker gene was disrupted in the white epiclones. By contrast, in the pink epiclones of med18Δ and med20Δ, the decrease in H3K9me/Swi6 and increase in RNAPII were less prominent than those in white epiclones. This reflected the difference in silencing defects in each epiclone (Figure 1D). At the heterochromatic repeats, dh, H3K9me/Swi6 and RNAPII were also decreased and increased in the Mediator mutants, respectively, but the differences between the white and pink epiclones were less prominent than at otr1R::ade6+. These results showed that the accumulation of transcripts from heterochromatic repeats and marker genes is, at least in part, due to an increase in transcription induced by the disruption of heterochromatin structure. These results also confirm that Mediator is required for heterochromatin formation at the pericentromere.
Fig. 2. Loss of Med18/20 causes defects in heterochromatin structure at the pericentromere. 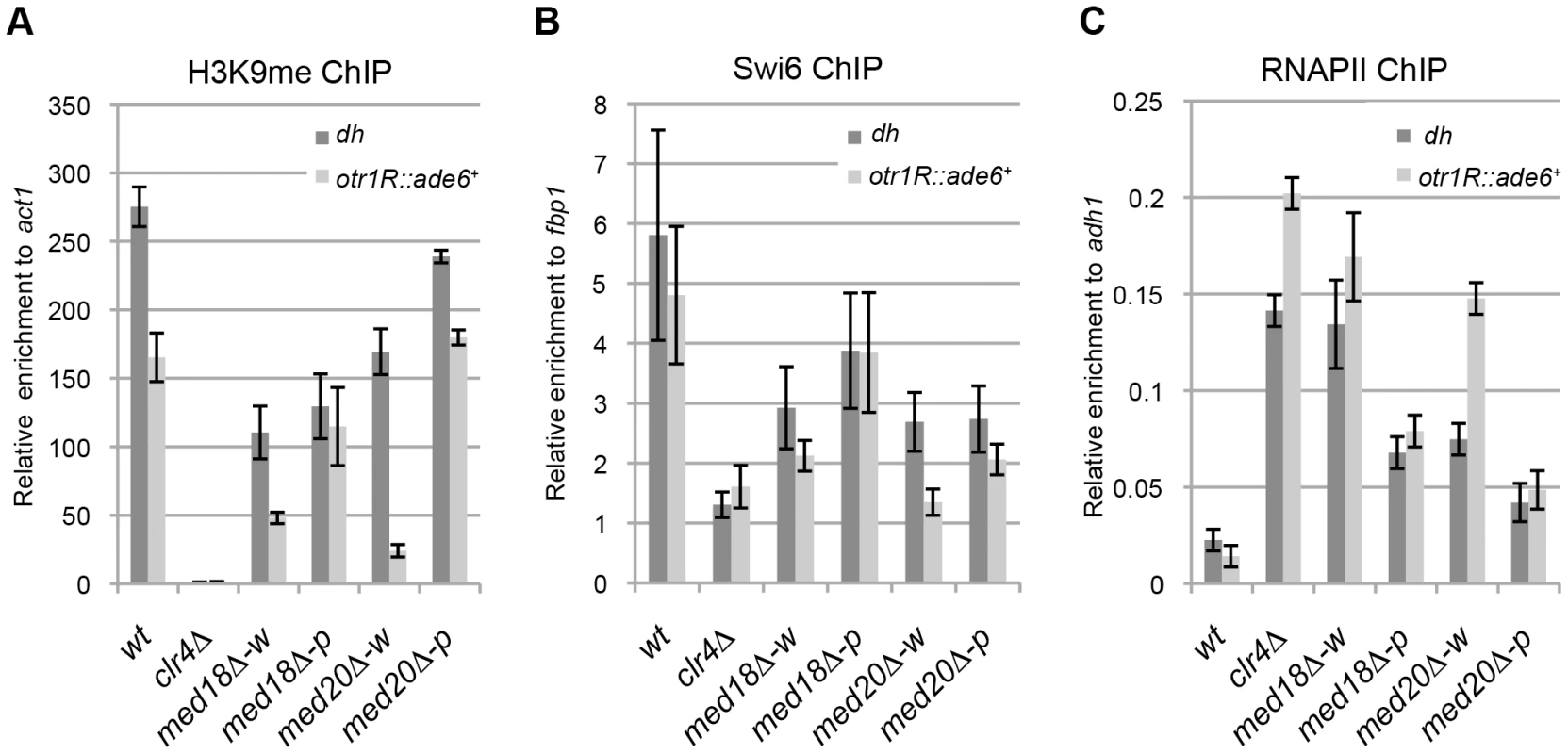
ChIP analysis of H3K9me (A), Swi6 (B) and RNAPII (C) at dh repeats or otr1R::ade6+ relative to act1, fbp1 or adh1, respectively. Error bars show the standard error of the mean (n = 3). Med18 is required for Rrp6-dependent H3K9 methylation at the pericentromere
There are two distinct pathways for heterochromatin formation at the pericentromeric repeats: RNAi-dependent and Rrp6-dependent pathways. In RNAi mutants such as dcr1Δ, H3K9me is diminished at the inserted marker genes but substantially retained at the pericentromeric repeats, while disruption of rrp6 did not affect H3K9 me at the marker genes [18]. The distribution of H3K9me in the white epiclones of the Mediator mutants resembled that observed in dcr1Δ cells; the level of H3K9me at the marker genes was lower than that at heterochromatic repeats. Thus, We speculated that Mediator is involved in the RNAi-dependent pathway. To confirm this, a med18Δ dcr1Δ double mutant was established and used to examine heterochromatin silencing and the amount of H3K9me and Swi6 at dh repeats and imr1:: ura4+ (Figure 3A, B). Note that since med18Δ Δdcr1Δ cells did not exhibit the variegated phenotype observed in the med18Δ single mutant (Figure 3A), white and pink epiclones of med18Δ cells were not separated in the following experiments (Figure 1B, C and D). If Mediator functions in the RNAi-dependent pathway, the med18Δ dcr1Δ double mutants would retain H3K9me and Swi6 to the level similar to those in single mutant. However, in the double mutant, the retained H3K9me/Swi6 at the dh repeats was significantly decreased compared to each single mutant (Figure 3B), suggesting that, contrary to our speculation, Med18 functions in a pathway distinct from the RNAi-dependent pathway.
Fig. 3. Med18/Mediator is required for Rrp6/Exosome-dependent H3K9 methylation at the pericentromere. 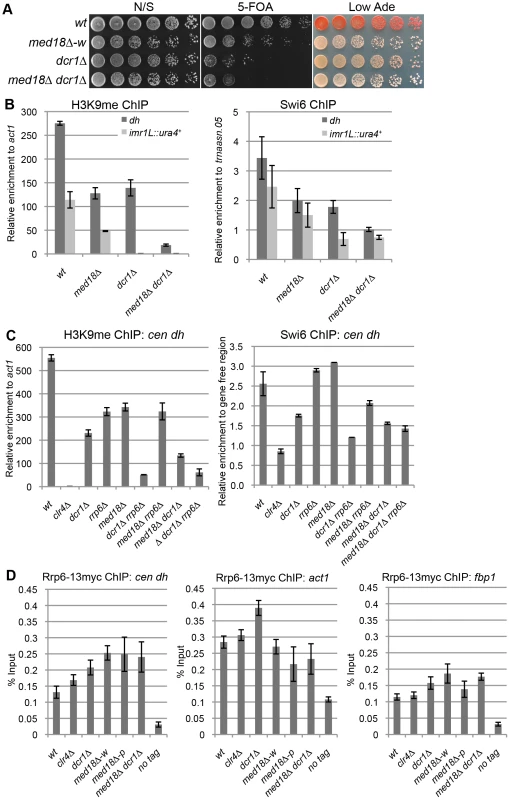
(A) Silencing assay at the pericentromere. Shown are the results of serial dilutions of the indicated strains spotted onto N/S, 5-FOA and Low Ade media to assay ura4+ and ade6 expression. (B) ChIP analysis of H3K9me (left panel) and Swi6 (right panel) at dh repeats or imr1L::ura4+ relative to act1 or a tRNA genes (trnaasn.05), respectively. Error bars show the standard error of the mean (n = 3). (C) ChIP analysis of H3K9me (left panel) and Swi6 (right panel) at the dh repeats relative to act1 or a gene-free region, respectively. Error bars show the standard error of the mean (n = 3). (D) ChIP analysis of Rrp6-13myc at dh repeats (left panel), act1 (middle panel) and fbp1 (right panel). Enrichment relative to the input whole cell extract (WCE) in the indicated strains are shown. Error bars represent the standard error of the mean (n = 3). Because the results in Figure 3B were reminiscent of the results reported by Reyes-Trucu et al., in which the amount of H3K9me retained at the centromeric repeats in ago1Δ cells was significantly decreased by further disruption of rrp6 [18], we speculated that Mediator functioned in an Rrp6-dependent heterochromatin formation pathway. To confirm this, single, double and triple mutant cells of dcr1, med18 and rrp6 were used to measure the amount of H3K9me at the dh repeats (Figure 3C, left panel). Each single mutant, as well as the med18Δ rrp6Δ double mutants, retained similar amounts of H3K9me. By contrast, combination of dcr1Δ with rrp6Δ caused a substantial decrease in H3K9me, which was consistent with the previous proposal that both RNAi - and Rrp6-dependent pathways contribute to heterochromatin formation at the pericentromeres [18]. Similarly, the combination of dcr1Δ with med18Δ also caused a significant decrease in H3K9me, while med18Δ rrp6Δ cells maintained a level of H3K9me comparable to each single disruptant. The H3K9me retained in med18Δ rrp6Δ cells was also decreased by the introduction of dcr1Δ. The amount of Swi6 in each mutant reflects the amount of H3K9me (Figure 3C, left panel). These results clearly indicate that Med18 functions in the same heterochromatin formation pathway as Rrp6 at the dh repeats. Therefore, H3K9me in dcr1Δ cells was retained by the Rrp6/Med18-dependent pathway, while H3K9me in each of the med18Δ and rrp6Δ cells was maintained by the RNAi-dependent pathway.
Details of the Rrp6-dependent pathway are not clear yet; even the localization of Rrp6 at heterochromatin has not been examined. We, thereby, analyzed the localization of Rrp6 tagged with myc epitope at heterochromatin (dh) as well as euchromatin (act1 and fbp1) (Figure. 3D). Compered with no-tag control, Rrp6-myc was enriched at both dh and euchromatic genes to the same extent. Depletion of clr4 did not affect the localization of Rrp6-myc, while deletion of dcr1 caused a slight increase at all loci. The enrichment of Rrp6 at dh increased in med18Δ-w and med18Δ-p epiclones, and also in the med18Δ dcr1Δ double mutant, while the enrichment at euchromatic genes was marginally affected in those mutant cells. This suggests that Mediator functions in a step after association of Rrp6 on chromatin for heterochromatin formation.
Mediator is required for the generation of siRNA from pericentromeric ncRNA
Both dcr1Δ and rrp6Δ cells retained similar levels of H3K9me at centromeric repeats (Figure 3C). By contrast, deletion of rrp6 does not affect H3K9me at the marker genes inserted in centromeric repeats [18], whereas deletion of dcr1 caused the loss of H3K9me on the marker genes (Figure 3B), indicating that the spreading of H3K9me into the inserted marker genes occurs via an RNAi-dependent mechanism [14], [15]. While H3K9me at otr1R::ade6+ was severely decreased in the white epiclones of med18Δ and med20Δ, it was substantially retained in the pink epiclones (Figure 2A), indicating that the spreading of H3K9me in the marker genes was variegated in the Mediator mutants. In addition, introduction of, dcr1Δ to med18Δ cells abolished a variegated phenotype (Figure 3A). These data confirm that the loss of Med18/Mediator results in the variegation of RNAi-dependent heterochromatin spreading. In other words, Med18/Mediator is also involved in the RNAi-dependent heterochromatin pathway.
To examine the involvement of Mediator in RNAi, siRNA derived from dg and dh repeats was analyzed in the Mediator mutants. siRNAs corresponding to the pericentromeric repeats were not detected in dcr1Δ cells (Figure 4A). In the white epiclones of the Mediator mutants, a marginal amount of siRNA from the dg and dh repeats was detected (Figure 4A and Figure S5A, B). The marginal amount of siRNA was diminished by introduction of dcr1Δ (Figure S5B), showing that the siRNA observed in med18Δ cells are produced through RNAi pathway. In the pink epiclones, reduced but significant amounts of siRNAs (approximately 10–50% of that of wild-type cells) were detectable. Note that the amount was varied because of the state of variegation. Since the structure of heterochromatin (H3K9me and Swi6) at the repeats was substantially maintained in both white epiclones (Figure 2A, B) and the maintenance was dependent upon RNAi-pathway as shown above (Figure 3C), the small amount of siRNA synthesized in the white epiclones appears to be sufficient to maintain heterochromatin structure at the repeats. A similar reduction of siRNA was observed in the white and pink epiclones of med8-K9 cells and med31-H1 cells (Figure S5). Note that when siRNA derived from dg and dh repeats was analyzed separately, each siRNA was found to be reduced in med18Δ cells (Figure S5). These data indicate that Mediator is required for siRNA generation at pericentromeric heterochromatin and the defect of the MHD causes variegation of the spreading of H3K9me into the marker genes.
Fig. 4. Mediator is required for siRNA formation at the pericentromeric repeats. 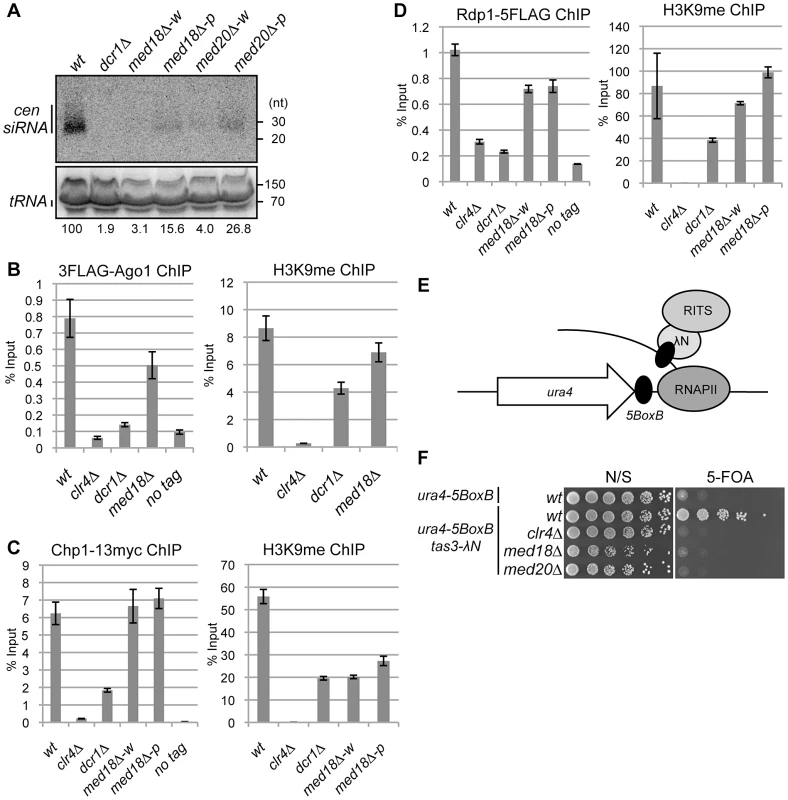
(A) Northern analysis of siRNA isolated from the indicated strains using oligonucleotide probes specific for dg and dh centromeric repeats. tRNA was used as a loading control. (B) ChIP analysis of 3Flag-Ago1 and H3K9me at dh repeats relative to input WCE in the indicated strains. Error bars show the standard error of the mean (n = 3). (C) ChIP analysis of Chp1-13myc and H3K9me at dh repeats relative to input WCE in the indicated strains. Error bars show the standard error of the mean (n = 3). (D) ChIP analysis of Rdp1- 5Flag and H3K9me at dh repeats relative to input WCE in the indicated strains. Error bars show the standard error of the mean (n = 3). (E) Schematic representation of artificial heterochromatin formation by RNA-induced transcriptional silencing (RITS) tethering to the ura4 mRNA. In this system, RITS was tethered artificially to ura4 RNA via binding of the λN protein fused to Tas3 (a subunit of RITS) to its recognition sequence, BoxB, five copies of which are inserted into the 3′ UTR of the ura4 mRNA. This induced siRNA generation and subsequent heterochromatin formation at the ura4 locus in an RNAi-dependent manner. (F) Gene silencing of ura4 via tethering of RITS. Serial dilution of strains harboring the RITS-tethering system (ura4-5boxB, tas3λN) or ura4-5boxB alone were spotted onto N/S and 5-FOA media for evaluating the silencing state of ura4 gene. A strain harboring clr4Δ was also included as a control. Mediator promotes efficient siRNA production from RITS-bound ncRNA
Since RNAi machinery localizes on heterochromatin for processing of ncRNA into siRNA [3], [6], [12], [32], the requirement of Med18 for the recruitment of RNAi factors to heterochromatin was investigated. Binding of the components of the RITS complex (3Flag-Ago1 and Chp1-13myc) and of RDRC (Rdp1-5Flag) to pericentromeric repeats was examined by ChIP assay (Figure 4 B–D). As reported, 3Flag-Ago1 bound to dh repeats in a heterochromatin - and/or RNAi-dependent manner [3], [12], [32], as evidenced by the finding that the binding of Ago1 was reduced to a level comparable to that of the no-Flag-tag control in clr4Δ and dcr1Δ cells (Figure 4B left panel). By contrast, a substantial amount of 3Flag-Ago1 was retained in med18Δ cells that formed a mixture of pink and white epiclones (Figure 4B left panel). Binding of Chp1-13myc to dh repeats was abolished by deletion of clr4, while reduced but significant Chp1-13myc localization was observed at the dh repeats in dcr1Δ cells, representing the binding of the chromo-domain of Chp1 to H3K9me that was retained at the pericentromeric repeats in these cells (Figure 4C, right panel). By contrast, the binding of Chp1-13myc was not affected by the deletion of med18. Even in white epiclones, in which H3K9me is reduced to the same level as in dcr1Δ cells (Figure 4C, right panel), Chp1-13myc binds to dh repeats at the same level as in wild-type cells (Figure 4C, left panel). Association of Rdp1-5Flag in each mutant was similar to that of 3Flag-Ago1 in that it was almost abolished in clr4Δ and dcr1Δ cells, but significantly retained in both med18Δ-w and med18Δ-p cells (Figure 4D, left panel). All together, the RITS complex and RDRC associated with heterochromatin even in med18Δ-w cells, probably because the small amount of siRNA synthesized in med18Δ cells was sufficient for the association of RITS with heterochromatin. Together with the data on the accumulation of ncRNA and reduction of siRNA in Mediator mutants, these data indicate that Mediator is not required for the association of the RITS complex and RDRC to heterochromatin but is required for efficient siRNA production from the ncRNA by heterochromatin-bound RNAi machinery.
It has been previously reported that the tethering RITS to ura4 RNA induces RNAi - and heterochromatin-dependent gene silencing of the ura4 gene, indicating that binding of the RITS complex to ncRNA is a key step in the RNAi-directed formation of heterochromatin by inducing H3K9 methylation and conversion of ncRNA to siRNA [33]. Tethering of RITS is achieved by the fusion of Tas3, a subunit of the RITS complex, to the λN protein, which binds to the 5BoxB sequence inserted at the 3′ UTR region of ura4 RNA (Figure 4E). To determine whether Med18 or Med20 is required for Tas3-λN-induced silencing of the ura4-5boxB gene, the effect of the deletion of these subunits on silencing induced by artificial tethering of the RITS complex was examined. Disruption of med18 or med20 resulted in the loss of ura4-5BoxB silencing (Figure 4F), similar to the effect of clr4 disruption. This result showed that Med18 and Med20 are required for Tas3-λN-induced silencing of the ura4-5BoxB locus and that Mediator plays a role in the step following the binding of the RITS complex to target RNA.
Mediator is required for efficient transcription in heterochromatin
Since Mediator regulates general transcription in euchromatin, it is possible that it also regulates the transcription of heterochromatic non-coding RNA. Indeed, recent reports suggest a negative role of MHD subunits (Med18 and Med20) in heterochromatic transcription, based on the observation of an increase in the transcription of pericentromeric ncRNA in Mediator mutants [34], [35]. However, it is difficult to state conclusively whether the observed increase is due to the direct effects of the absence of Mediator because it is also possible that the deletion of Mediator subunits causes disruption of the heterochromatin, which secondarily induces an increase in transcription. To avoid this dilemma, heterochromatin at the mating-type locus was selected for examination (Figure 5A) because the RNAi-dependent pathway is dispensable for the maintenance of heterochromatin here due to the existence of another pathway mediated by the DNA-binding proteins Atf1and Pcr1 [20]. Thus, mutation of Mediator would not be expected to affect the mating-type locus heterochromatin, making it possible to directly measure the effect of the mutation on transcription activity in heterochromatin.
Fig. 5. Mediator is required for transcriptional activation in heterochromatin. 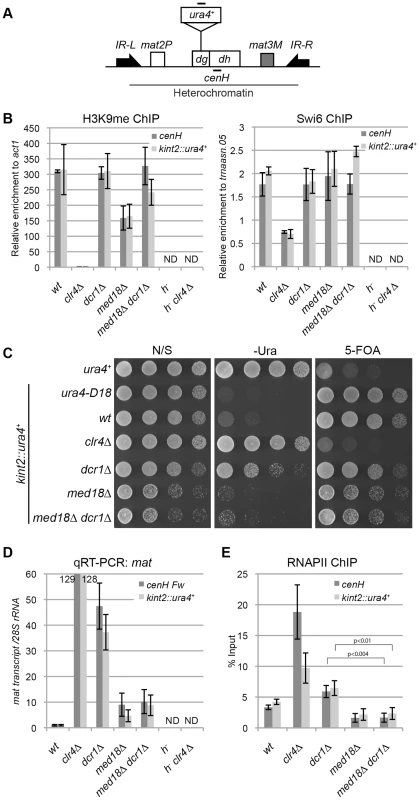
(A) Schematic of the fission yeast mating-type locus. Location of the ura4 reporter inserted within the cenH is shown (kint2::ura4+). Black bars indicate the location of primers or probes used for ChIP, RT-PCR and northern analysis. (B) ChIP analysis of H3K9me and Swi6 at cenH dh repeats or kint2::ura4+, each relative to act1 or trnaasn.05. Error bars show the standard error of the mean (n = 3). (C) Silencing assay at the mating-type locus. The results of serial dilutions of the indicated strains spotted onto N/S, 5-FOA media and medium without uracil (-URA) for the silencing of ura4 are shown. Note that PMGS (PM medium containing L-glutamic acid as a nitrogen source instead of ammonium chloride) plates were used as N/S plates. (D) Quantitative RT-PCR analysis of cenH dh repeats or kint2::ura4+ forward transcript levels relative to a control SPRRNA.48, whose levels were normalized to that of the wild-type in the indicated strains. Error bars show the standard error of the mean (n = 3). (E) ChIP analysis of RNAPII at cenH dh repeats or kint2::ura4+ relative to SPRRNA.48. Error bars show the standard error of the mean (n = 3). P values were determined using a two-sided Student's t-test. Note that in Figures B, D and E, h− strains (h−, h−clr4Δ), which do not have cenH sequence, were included to show the primers used in the experiments only detect cenH and do not detect the pericentromeric repeats. First, a ChIP assay was performed for H3K9me and Swi6 to examine heterochromatin structure at the mating-type locus in various mutants using specific primers for the cenH sequence (Figure 5B). As expected, high levels of H3K9me2 and Swi6 were maintained at the cenH sequence at the mating-type locus and the inserted ura4+ gene (kint2::ura4+) in dcr1Δ and dcr1Δ med18Δ mutants. In med18Δ cells, the level of H3K9me was decreased to half of that of wild-type cells for an unknown reason, but the level of Swi6, which is essential for transcriptional gene silencing in heterochromatin, was maintained, consistent with the observation that silencing at this locus was not affected by med18Δ (Figure 5B). Hence, the Atf1/Pcr1-dependent pathway probably retains heterochromatin structure and silencing without Med18 function.
Next, the effect of deletion of dcr1 and med18 on the silencing of kint2::ura4+ was examined (Figure 5C). While the wild-type strain was able to grow on a 5-FOA-containing plate but not on a uracil-lacking (-Ura) plate, the clr4Δ strain was hypersensitive to 5-FOA but grew well on an –Ura plate (Figure 5C), showing that kint2::ura4+ was silenced and expressed, respectively, in each strain. By contrast, dcr1Δ cells, like the wild-type cells, hardly grew on 5-FOA containing media, while some cells were able to grow on an –Ura plate, suggesting that silencing is only weakly compromised in dcr1Δ cells. However, med18Δ cells showed a phenotype similar to that of wild-type cells, showing no silencing defect at the mating-type locus. Introduction of med18Δ to dcr1Δ cells suppressed the silencing defect detected on the –Ura plate. RT-PCR analysis of transcripts from cenH and kint2::ura4+ was consistent with the silencing assay; more than 100-fold, approximately 40-fold, and approximately 10-fold accumulation of transcripts from cenH and kint2::ura4 were observed in clr4Δ, dcr1Δ and med18Δ cells, respectively (Figure 5D). In addition, introduction of med18Δ into dcr1Δ cells caused a decrease in transcripts to a level similar to that of med18Δ cells.
The accumulation of RNA in dcr1Δ cells and med18Δ cells could be explained by defects in RNA degradation by RNAi and/or the exosome (Noma et al., 2004, Buhler et al., 2007), or by an increase in heterochromatic transcription. To examine the latter possibility, localization of RNAPII at cenH and kint2::ura4+ was examined by ChIP assay (Figure 5E). Unexpectedly, RNAPII was significantly increased in dcr1Δ cells at both loci in spite of the maintenance of heterochromatin in this strain, suggesting that Dcr1 negatively regulates heterochromatin transcription. Note that RNAi machinery has been shown to interact with RNAPII and modulate transcription in other organisms [36], [37]. Thus, an increase in transcription and prevention of processing of RNA into siRNA could cause the observed accumulation of transcripts in dcr1Δ cells (Figure 5D). By contrast, the level of RNAPII in med18Δ cells was comparable to that of wild-type cells (Figure 5E). Similar results of RNAPII localization were obtained with ChIP assay using the antibody against RNAPII-C-terminal repeats phosphorylated at the second serine, which represents elongating RNAPII (Figure S6). These indicated that Med18 does not repress transcription in heterochromatin. The approximately 10-fold accumulation of cenH and kint2::ura4+ RNA observed in med18Δ cells might be due to a defect in exosome-dependent degradation of RNA [16], which would indicate that significant transcription took place in the absence of Med18. Importantly, introduction of med18Δ into dcr1Δ cells caused a decrease in RNAPII to the level of wild-type cells, suggesting that Mediator is required for efficient transcription in heterochromatin in dcr1Δ cells.
To analyze the role of Med18 on the transcription in the absence of heterochromatin, we compared RNAPII occupancy at centromeric repeats of dcr1Δ rrp6Δ cells with those of dcr1Δ rrp6Δ med18Δcells (Figure S7). Note that both strains showed similarly low levels of H3K9me at dh repeats (Figure 3C). Introduction of med18Δ caused the moderate increase of RNAPII. This suggested that Med18 negatively regulates transcription in the compromised heterochromatin.
From these data, we suggest that in the fully assembled heterochromatin, Med18/Mediator does not negatively regulate pericentromeric transcription; rather, it might be required for efficient transcription in heterochromatin.
Effect of Mediator disruption on euchromatic genes
The effect of med18Δ and med20Δ on euchromatic gene expression was further examined using microarray. Pink and white epiclones of med18Δ and med20Δ cells were separated and the expression pattern of each epiclone was compared. Analysis of the genes that showed ≥1.5-fold increase (Up) or decrease (Down) in expression between the epiclones revealed that a common set of genes were affected in both med18Δ cells and med20Δ cells, irrespective of the state of heterochromatic silencing (Figure 6A). Therefore, a clear difference between the white and pink epiclones was observed at pericentromeric ncRNA expression. Indeed, in the white epiclones, no euchromatic genes showed stronger induction than centromeric ncRNA; the most strongly increased euchromatic gene showed an approximately 14-fold and 26-fold increase in med18Δ-w and med20Δ-w cells, respectively, which is much weaker than the increase in centromeric ncRNA (which increased by more than 100-fold). Comparison of med18Δ-w and med20Δ-w, or med18Δ-p and med20Δ-p, showed that both subunits shared a common set of targets (Figure S8). This is consistent with the fact that both Med18 and Med20 formes heterodimers submodule in the MHD.
Fig. 6. Effect of Mediator disruption on euchromatic genes. 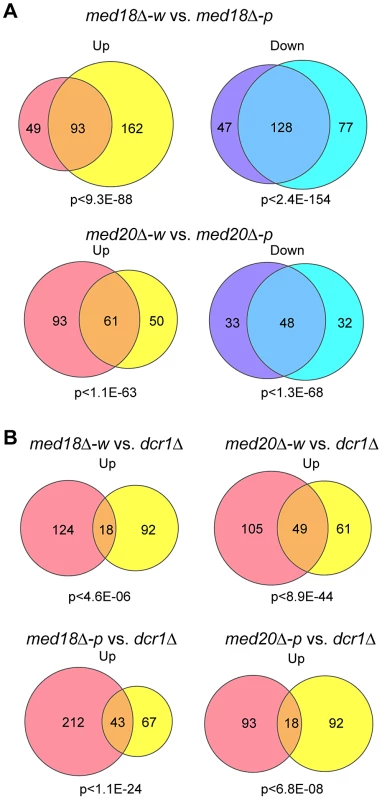
Venn diagram showing the number of transcripts whose expression levels are increased (up) or decreased (down) >1.5-fold in mutants compared to the wild-type. P-values were calculated using Fisher's exact test. (A) Transcripts of med18Δ-w (left circles) vs. med18Δ-p (right circles) mutants (top) and med20Δ-w (left circles) vs. med20Δ-p (right circles) mutants (bottom). (B) Transcripts of med18Δ-w (left circles) vs. dcr1Δ (right circles) mutants (upper left), med20Δ-w (left circles) vs. dcr1Δ (right circles) mutants (upper right), med18Δ-p (left circles) vs. dcr1Δ (right circles) mutants (lower left), and med20Δ-p (left circles) vs. dcr1Δ (right circles) mutants (lower right). When the expression pattern in the white epiclones of the Mediator mutants (med18Δ-w and med20Δ-w) was compared with that in dcr1Δ cells, it was evident that the expression of a common set of genes was upregulated (Figure 6B, upper panels). Similar sharing of target genes was observed between the pink epiclones of the Mediator mutants (med18Δ-p and med20Δ-p) and dcr1Δ cells (Figure 6B, lower panel). Interestingly, gene ontology analysis showed significant enrichment of terms pertaining to stress responses in the shared target genes. For example, the top GO terms included “cellular response to stimulus” (GO: 0033554, med18Δ-w vs. dcr1Δ P = 5.35×10−3, med18Δ-p vs. dcr1Δ P = 2.2×10−6, med20Δ-w vs. dcr1Δ P = 2.19×10−6, med20Δ-p vs. dcr1Δ P = 1.27×10−4). These results suggest that some euchromatic genes, including stress response genes, are repressed by the RNAi/Mediator system, which may function via a mechanism partly similar to that of RNAi-mediated heterochromatin.
Discussion
In this study, we showed that the specific subunits of Mediator, Med18, Med20, Med8 and Med31 were involved in pericentromeric heterochromatin formation. The Med18-Med20 heterodimer is a component of the head domain of Mediator [26]. Because the med8-K9 mutation causes truncation of the C-terminal domain that interacts with the Med18/Med20 heterodimer (Figure S1B), it resulted in the loss of the heterodimer. Importantly, Med31 belongs to the middle domain but is located close to the head domain [38], suggesting that the head domain does not function alone in heterochromatin formation, but rather as a part of Mediator. Therefore, we suggest that Mediator specifically plays multiple roles in the formation of pericentromeric heterochromatin via the MHD (Fig. 7).
Fig. 7. Model of the functions of Mediator in pericentromeric heterochromatin assembly in fission yeast. 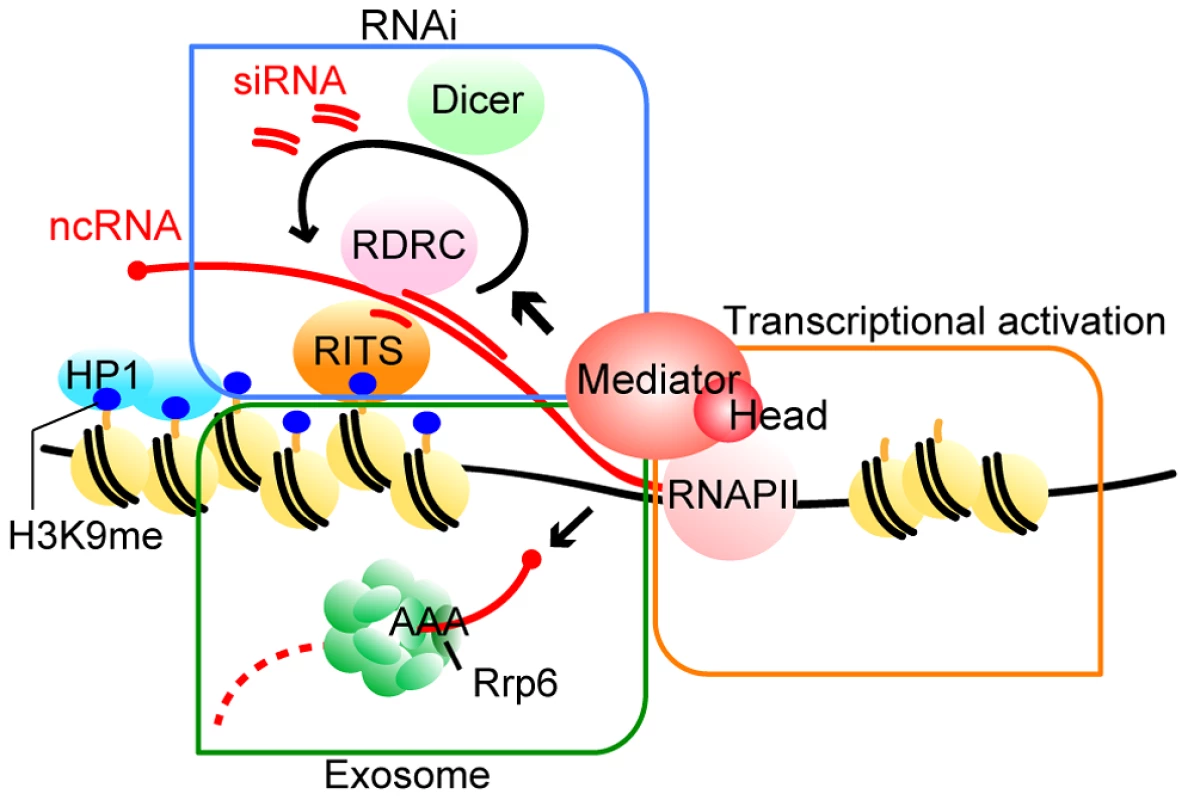
Mediator localizes to the pericentromeres to recruit RNAPII for efficient transcription. Furthermore, Mediator regulates processing of transcribed ncRNA by RNAi machineries, which directs heterochromatin formation. In addition, Mediator is also required for Rrp6-dependent heterochromatin formation. For details, see Discussion. Two distinct mechanisms, the RNAi-dependent and Rrp6-dependent pathways, function in heterochromatin formation at pericentromeric repeats, while the spreading of H3K9me onto marker genes mainly depends on the RNAi pathway [14], [15]. We found that at the pericentromeric heterochromatin, in the absence of MHD, the Rrp6-dependent pathway is compromised and H3K9me is largely maintained by the RNAi-dependent pathway (Fig. 3). The finding that the amount of siRNA produced in the white epiclones of MHD mutants decreased to 3–20% of that in wild-type cells (Fig. 4A and Figure S5) indicates that only a small amount of siRNA is necessary to maintain heterochromatin at the pericentromeric repeats. The remaining H3K9me at the repeats in MHD mutants spreads onto the marker genes by an RNAi-dependent mechanism. This process was also compromised by the decrease of siRNA caused by the absence of MHD, resulting in variegation of the level of H3K9me at the marker genes, which ultimately caused the appearance of white and pink epiclones. The spreading process appears to require more efficient siRNA production than the maintenance of heterochromatin at the pericentromeric repeats because med18Δ-p cells that produced more siRNA showed more efficient spreading of H3K9me and silencing of marker genes than med18Δ-w cells. In contrast, dcr1Δ cells did not show the variegation of silencing because of loss of siRNA production. It is noteworthy that the variegated phenotype was metastable, which suggests that once heterochromatin was spread into the marker gene, it could be maintained in an MHD-independent manner, probably through the small amount of siRNA produced in MHD mutants.
Many processes involved in RNA processing, such as RNA splicing and RNA transport, are coupled to transcription by RNAPII. siRNA production is also coupled with RNAPII-dependent transcription [10]. Our results showed that the mutation of MHD resulted in a large decrease in siRNA. By contrast, rrp6Δ cells, which have levels of H3K9me and Swi6 at the pericentromeric repeats similar to med18Δ cells (Fig. 3), produced the same amount of siRNA as wild-type cells [16]. These results indicate that MHD is somehow involved in siRNA production after transcription of ncRNA. MHD is localized at the transcribed region in pericentromeric repeats (Figure S4; [34], [35] and is required for transcription in heterochromatin, suggesting that it directly functions in the coupling of transcription of heterochromatic ncRNA by RNAPII and processing of the siRNA by RNAi machinery. Retention of RNAi factors at the pericentromeric repeats in med18Δ cells and the requirement for MHD in heterochromatin formation via the artificial tethering of the RITS complex to RNA suggest that MHD functions after RITS associates with heterochromatic repeats and/or target RNA. Recently, we showed that RNAi factors are assembled into an siRNA amplification compartment that includes transcriptionally active heterochromatin [39]. Thus, MHD might be involved in the formation of this compartment. Although we were not able to detect a stable interaction between MHD components and RNAi factors, such as Ago1, by co-immunoprecipitation experiments (data not shown), it is still possible that MHD recruits factors required for siRNA generation to transcriptionally active heterochromatin through direct or indirect interactions. In any case, further experiments are necessary to clarify the molecular function of Mediator in the RNAi pathway.
In contrast to the RNAi pathway, little is known about the Rrp6-dependent heterochromatin formation pathway. As deletion of rrp6 marginally affects H3K9me and silencing at the inserted marker genes [16], [18], the Rrp6-dependent pathway mainly functions at the pericentromeric repeats. Our genetic experiments showed that rrp6 and med18 were epistatic in the formation of pericentromeric heterochromatin (Fig. 3), indicating that MHD functions in Rrp6-dependent heterochromatin formation. Rrp6 is an exonuclease that is a subunit of the nuclear exosome involved in RNA-quality control [40]. A functional relationship between Mediator and the nuclear exosome has not been reported. We found that Rrp6 associates with both heterochromatin (dh) as well as euchromatin (act1 and fbp1) and deletion of med18 did not affect the localization, suggesting Mediator acts in a step after the association of Rrp6/exsosome with chromatin. This is analogous to the function of Mediator in the RNAi-dependent pathway; MHD functions after recruitment of RITS complex and RDRC to chromatin. Considering the co-transcriptional nature of RNA-quality control [41], [42] and recruitment of RNA-splicing factors to transcripts by Mediator [29], we speculate that MHD plays a role in the co-transcriptional function of chromatin-associated Rrp6 and/or other co-factors to promote heterochromatin formation. Alternatively, given that the RNAi-independent heterochromatin nucleation pathway and Mediator functionally interact with RNAPII processivity factors [18], [30], Mediator may promote Rrp6-dependent heterochromatin formation by affecting elongation by RNAPII through interaction with these processivity factors. It is also possible that the same mechanism is also involved in RNAi-dependent heterochromatin formation through MHD.
Recently, two reports showed that MHD was important for heterochromatin formation at pericentromeres [34], [35]. However, there are several discrepancies between their data and ours. Firstly, the decrease in H3K9me and Swi6 in med20Δ cells was much more severe than that in ours. Secondly, Carlsten et al. claimed that siRNA from the dh repeat in med20Δ cells was diminished but that siRNA from the dg repeats was comparable to that in wild-type cells. Thirdly, both papers assert that Mediator negatively regulates heterochromatic transcription. The first two discrepancies could be caused by the variegated phenotype of MHD mutants. If this variegated phenotype was overlooked or disregarded, the results would be affected by which epiclones were used in the experiments. In addition, since the amount of H3K9me/Swi6 varies depending on the position in the repeats, the discrepancies between their results and ours might reflect a difference in the sites used for ChIP analysis. The third discrepancy could be explained by the use of pericentromeric transcription for their analysis. As described in the Results section, it is hard to argue definitively for the direct influence of MHD mutants on transcription in pericentromeric heterochromatin because it is difficult to determine whether the observed increase is due to the direct effect of depletion of MHD or a secondary effect resulting from the disruption of heterochromatin. Our data using the mating-type locus heterochromatin showed that disruption of Mediator did not cause increased transcription in heterochromatin, rather it caused a decrease in transcription enhanced by deletion of dcr1 (Fig. 6). Interestingly, when heterochromatin was compromised, Mediator appears to negatively regulate transcription, which might also explain the discrepancy.
Recently, Dcr1 was shown to repress a set of genes, including stress response genes, through the degradation of target RNA [43]. We identified a similar set of euchromatic genes that were up-regulated in med20Δ and dcr1Δ cells, suggesting that MHD functions in co-transcriptional degradation of euchromatic RNAs in collaboration with Dcr1. Note that previous transcriptome analysis of the mediator mutants also showed that a similar set of genes were up regulated in med18Δ and med20Δ cells but not in med12Δ cells, supporting the collaborative function of MHD and Dcr1 [44]. In addition, Rrp6-dependent heterochromatin formation was observed at several meiotic genes [45]. Moreover, the exosome and RNAi are shown to regulate a set of genes, including retrotransposons and developmental genes [46]. Therefore, it is also possible that Mediator functions at these loci to silence genes by regulating both RNAi and exosomal machineries.
Emerging evidence shows that Mediator works as a platform for various factors that function in transcription and RNA processing, using a distinct subunit for particular interactions with the factors [29], [30]. Our results further extend the range of Mediator function to include regulation of higher-order chromatin structure in the genome. It is now widely accepted that RNAPII transcribes almost all of the genome. Mediator might not only mediate transcription factors and RNAPII at each gene, but also mediate RNAPII and genome-wide regulation of higher-order chromatin structure.
Materials and Methods
Strains and culture media
The S. pombe strains used in this study are described in Table S1. The media and genetic methods used in the study were essentially as described previously [47]. Yeast cells were cultured in YES at 30°C. For deletion or epitope-tagging of the target genes, the PCR-based module method [48] was used.
Silencing assays
Silencing assays were conducted from overnight unsaturated cultures grown in 10 ml YES. A 5-fold dilution series of cells was spotted on N/S plates (YES in all spot figures, except Figure 5B), 5-FOA plates (N/S plates with the addition of 1 g/l 5-fluoroorotic acid), and Low Ade plates (N/S plates including limited amount of adenine). The plates were then incubated at 30°C for 3 days.
Chromatin immunoprecipitation (ChIP) analysis
ChIP was performed as described previously [39] with the following changes: crosslinking with formaldehyde was performed for 30 min and digestion with Proteinase K was carried out for 1 h at 42°C. The following antibodies were used: anti-H3-K9-me2 monoclonal (a gift from T. Urano, Shimane University), anti-Swi6 (produced in-house), anti-RNA polymerase II (8WG16, Abcam), anti-FLAG (M2, Sigma), or anti-myc (4A6, Millipore). Primer sequences are shown in Table S2.
RNA analysis
Total RNA was isolated from logarithmically-growing S. pombe (in YES media) using the hot phenol method [49]. For northern blotting of centromeric and mat RNA, 50 µg of total RNA was electrophoresed on a 1% agarose gel containing 1× MOPS and 1% formaldehyde. RNA was transferred to positively-charged nylon membranes (Amersham Biosciences) in 10× SSC by standard capillary blotting. Following UV crosslinking of the RNA to the nylon filter, prehybridization and hybridization were carried out at 42°C in UltraHyb-Oligo buffer (Ambion). For hybridization, 50 pmol oligos were end-labeled with [γ32P]dATP (3000 Ci/mmol) using T4 Polynucleotide Kinase (TOYOBO). After hybridization for 24 h, membranes were washed four times in 2× SSC/0.1% SDS for 10 min at 42°C before exposure to an imaging plate for 1–2 days. For re-probing, probes on the membrane were stripped by boiling in 200 ml of 0.5× SSC/0.1% with shaking. Detection of siRNA was performed as described previously [39]. Oligonucleotides used as probes are shown in Table S2.
RT-PCR
For RT-PCR analysis, total RNA was cleaned up and treated with Recombinant DNase I (RNase-free) (TaKaRa) according to the manufacturer's instructions. RT-PCR was performed using PrimeScript Reverse Transcriptase (TaKaRa) according to the manufacturer's instructions. Primer sequences are shown in Table S2.
qPCR
qPCR was performed using SYBR premix Ex-Taq (TaKaRa) and the Thermal Cycler Dice Real time system TP800 (TaKaRa). Primer sequences are shown in Table S2.
ChIP-qPCR and RT-PCR using synchronized cdc25-22 cells
cdc25-22 cells were grown at 25°C to a concentration of 2×106 cells/ml and then shifted to 36°C for 4 hr and 15 min to stop the cell cycle at the G2/M phase. Samples for ChIP assay were collected every 30 min for 300 min after shifting the cells back to 25°C to release cell cycle block. ChIP assay was performed as described in the Experimental Procedures. To prepare RNA for RT-PCR, the input fractions of ChIP were adjusted to 0.25% SDS and 0.25 mg/ml proteinase K and incubated for 45 min at 45°C and then at 65°C for more than 4 hours to reverse crosslinking. Samples were extracted once with phenol-chloroform. After ethanol precipitation, the samples were resuspended in a suitable volume of DEPC-treated distilled water. RT-PCR was performed as described in the Experimental Procedures.
Microarray analysis of Mediator mutants
Microarray analysis for gene expression was performed as described previously [50] using FY2002 as a parental strain. White and pink epiclones of med18Δ and med20Δ were analyzed separately. The sequences of the probes and original data from the microarray experiments were deposited to GEO (http://www.ncbi.nlm.nih.gov/geo) with accession number GSE43543.
Methods in the supplemental information
Methods used in the supplemental information are described in Text S1.
Supporting Information
Zdroje
1. ReaS, EisenhaberF, O'CarrollD, StrahlBD, SunZW, et al. (2000) Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature 406 : 593–599.
2. NakayamaJ, RiceJC, StrahlBD, AllisCD, GrewalSI (2001) Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science 292 : 110–113.
3. CamHP, SugiyamaT, ChenES, ChenX, FitzGeraldPC, et al. (2005) Comprehensive analysis of heterochromatin - and RNAi-mediated epigenetic control of the fission yeast genome. Nat Genet 37 : 809–819.
4. GrewalSI, JiaS (2007) Heterochromatin revisited. Nat Rev Genet 8 : 35–46.
5. ShimadaA, MurakamiY (2010) Dynamic regulation of heterochromatin function via phosphorylation of HP1-family proteins. Epigenetics 5 : 30–33.
6. VolpeTA, KidnerC, HallIM, TengG, GrewalSI, et al. (2002) Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science 297 : 1833–1837.
7. MotamediMR, VerdelA, ColmenaresSU, GerberSA, GygiSP, et al. (2004) Two RNAi complexes, RITS and RDRC, physically interact and localize to noncoding centromeric RNAs. Cell 119 : 789–802.
8. ChikashigeY, KinoshitaN, NakasekoY, MatsumotoT, MurakamiS, et al. (1989) Composite motifs and repeat symmetry in S. pombe centromeres: direct analysis by integration of NotI restriction sites. Cell 57 : 739–751.
9. DjupedalI, PortosoM, SpahrH, BonillaC, GustafssonCM, et al. (2005) RNA Pol II subunit Rpb7 promotes centromeric transcription and RNAi-directed chromatin silencing. Genes Dev 19 : 2301–2306.
10. KatoH, GotoDB, MartienssenRA, UranoT, FurukawaK, et al. (2005) RNA polymerase II is required for RNAi-dependent heterochromatin assembly. Science 309 : 467–469.
11. ChenES, ZhangK, NicolasE, CamHP, ZofallM, et al. (2008) Cell cycle control of centromeric repeat transcription and heterochromatin assembly. Nature 451 : 734–737.
12. NomaK, SugiyamaT, CamH, VerdelA, ZofallM, et al. (2004) RITS acts in cis to promote RNA interference-mediated transcriptional and post-transcriptional silencing. Nat Genet 36 : 1174–1180.
13. SugiyamaT, CamH, VerdelA, MoazedD, GrewalSI (2005) RNA-dependent RNA polymerase is an essential component of a self-enforcing loop coupling heterochromatin assembly to siRNA production. Proc Natl Acad Sci U S A 102 : 152–157.
14. IrvineDV, ZaratieguiM, ToliaNH, GotoDB, ChitwoodDH, et al. (2006) Argonaute slicing is required for heterochromatic silencing and spreading. Science 313 : 1134–1137.
15. LiH, MotamediMR, YipCK, WangZ, WalzT, et al. (2009) An alpha motif at Tas3 C terminus mediates RITS cis spreading and promotes heterochromatic gene silencing. Mol Cell 34 : 155–167.
16. BühlerM, HaasW, GygiSP, MoazedD (2007) RNAi-dependent and -independent RNA turnover mechanisms contribute to heterochromatic gene silencing. Cell 129 : 707–721.
17. BühlerM, SpiesN, BartelDP, MoazedD (2008) TRAMP-mediated RNA surveillance prevents spurious entry of RNAs into the Schizosaccharomyces pombe siRNA pathway. Nat Struct Mol Biol 15 : 1015–1023.
18. Reyes-TurcuFE, ZhangK, ZofallM, ChenE, GrewalSI (2011) Defects in RNA quality control factors reveal RNAi-independent nucleation of heterochromatin. Nat Struct Mol Biol 18 : 1132–1138.
19. GrewalSI, KlarAJ (1997) A recombinationally repressed region between mat2 and mat3 loci shares homology to centromeric repeats and regulates directionality of mating-type switching in fission yeast. Genetics 146 : 1221–1238.
20. JiaS, NomaK, GrewalSI (2004) RNAi-independent heterochromatin nucleation by the stress-activated ATF/CREB family proteins. Science 304 : 1971–1976.
21. KelleherRJ3rd, FlanaganPM, KornbergRD (1990) A novel mediator between activator proteins and the RNA polymerase II transcription apparatus. Cell 61 : 1209–1215.
22. KimYJ, BjorklundS, LiY, SayreMH, KornbergRD (1994) A multiprotein mediator of transcriptional activation and its interaction with the C-terminal repeat domain of RNA polymerase II. Cell 77 : 599–608.
23. SoutourinaJ, WydauS, AmbroiseY, BoschieroC, WernerM (2011) Direct interaction of RNA polymerase II and mediator required for transcription in vivo. Science 331 : 1451–1454.
24. LariviereL, SeizlM, CramerP (2012) A structural perspective on Mediator function. Curr Opin Cell Biol 24 : 305–313.
25. BourbonHM (2008) Comparative genomics supports a deep evolutionary origin for the large, four-module transcriptional mediator complex. Nucleic Acids Res 36 : 3993–4008.
26. LariviereL, GeigerS, HoeppnerS, RotherS, StrasserK, et al. (2006) Structure and TBP binding of the Mediator head subcomplex Med8-Med18-Med20. Nat Struct Mol Biol 13 : 895–901.
27. ImasakiT, CaleroG, CaiG, TsaiKL, YamadaK, et al. (2011) Architecture of the Mediator head module. Nature 475 : 240–243.
28. KimYJ, ZhengB, YuY, WonSY, MoB, et al. (2011) The role of Mediator in small and long noncoding RNA production in Arabidopsis thaliana. EMBO J 30 : 814–822.
29. HuangY, LiW, YaoX, LinQJ, YinJW, et al. (2012) Mediator complex regulates alternative mRNA processing via the MED23 subunit. Mol Cell 45 : 459–469.
30. TakahashiH, ParmelyTJ, SatoS, Tomomori-SatoC, BanksCA, et al. (2011) Human mediator subunit MED26 functions as a docking site for transcription elongation factors. Cell 146 : 92–104.
31. AllshireRC, NimmoER, EkwallK, JaverzatJP, CranstonG (1995) Mutations derepressing silent centromeric domains in fission yeast disrupt chromosome segregation. Genes Dev 9 : 218–233.
32. VerdelA, JiaS, GerberS, SugiyamaT, GygiS, et al. (2004) RNAi-mediated targeting of heterochromatin by the RITS complex. Science 303 : 672–676.
33. BühlerM, VerdelA, MoazedD (2006) Tethering RITS to a nascent transcript initiates RNAi - and heterochromatin-dependent gene silencing. Cell 125 : 873–886.
34. CarlstenJO, SzilagyiZ, LiuB, Davila LopezM, SzasziE, et al. (2012) Mediator Promotes CENP-A Incorporation at Fission Yeast Centromeres. Mol Cell Biol 32 : 4035–43.
35. ThorsenM, HansenH, VenturiM, HolmbergS, ThonG (2012) Mediator regulates non-coding RNA transcription at fission yeast centromeres. Epigenetics Chromatin 5 : 19.
36. KaviHH, BirchlerJA (2009) Interaction of RNA polymerase II and the small RNA machinery affects heterochromatic silencing in Drosophila. Epigenetics Chromatin 2 : 15.
37. GuangS, BochnerAF, BurkhartKB, BurtonN, PavelecDM, et al. (2010) Small regulatory RNAs inhibit RNA polymerase II during the elongation phase of transcription. Nature 465 : 1097–1101.
38. CaiG, ImasakiT, YamadaK, CardelliF, TakagiY, et al. (2010) Mediator head module structure and functional interactions. Nat Struct Mol Biol 17 : 273–279.
39. KawakamiK, HayashiA, NakayamaJ, MurakamiY (2012) A novel RNAi protein, Dsh1, assembles RNAi machinery on chromatin to amplify heterochromatic siRNA. Genes Dev 26 : 1811–1824.
40. HouseleyJ, LaCavaJ, TollerveyD (2006) RNA-quality control by the exosome. Nat Rev Mol Cell Biol 7 : 529–539.
41. HuertasP, AguileraA (2003) Cotranscriptionally formed DNA:RNA hybrids mediate transcription elongation impairment and transcription-associated recombination. Mol Cell 12 : 711–721.
42. SuganumaN, NakamuraY, YamamotoM, OhtaT, KoiwaH, et al. (2003) The Lotus japonicus Sen1 gene controls rhizobial differentiation into nitrogen-fixing bacteroids in nodules. Mol Genet Genomics 269 : 312–320.
43. WoolcockKJ, StunnenbergR, GaidatzisD, HotzHR, EmmerthS, et al. (2012) RNAi keeps Atf1-bound stress response genes in check at nuclear pores. Genes Dev 26 : 683–692.
44. LinderT, RasmussenNN, SamuelsenCO, ChatzidakiE, BaraznenokV, et al. (2008) Two conserved modules of Schizosaccharomyces pombe Mediator regulate distinct cellular pathways. Nucleic Acids Res 36 : 2489–2504.
45. ZofallM, YamanakaS, Reyes-TurcuFE, ZhangK, RubinC, et al. (2012) RNA elimination machinery targeting meiotic mRNAs promotes facultative heterochromatin formation. Science 335 : 96–100.
46. YamanakaS, MehtaS, Reyes-TurcuFE, ZhuangF, FuchsRT, et al. (2012) RNAi triggered by specialized machinery silences developmental genes and retrotransposons. Nature 493 : 557.
47. XhemalceB, KouzaridesT (2010) A chromodomain switch mediated by histone H3 Lys 4 acetylation regulates heterochromatin assembly. Genes Dev 24 : 647–652.
48. BahlerJ, WuJQ, LongtineMS, ShahNG, McKenzieA3rd, et al. (1998) Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 14 : 943–951.
49. MotamediMR, HongEJ, LiX, GerberS, DenisonC, et al. (2008) HP1 proteins form distinct complexes and mediate heterochromatic gene silencing by nonoverlapping mechanisms. Mol Cell 32 : 778–790.
50. TangeY, KurabayashiA, GotoB, HoeKL, KimDU, et al. (2012) The CCR4-NOT complex is implicated in the viability of aneuploid yeasts. PLoS Genet 8: e1002776.
51. GuglielmiB, van BerkumNL, KlapholzB, BijmaT, BoubeM, et al. (2004) A high resolution protein interaction map of the yeast Mediator complex. Nucleic acids research 32 : 5379–5391.
Štítky
Genetika Reprodukční medicína
Článek Re-Ranking Sequencing Variants in the Post-GWAS Era for Accurate Causal Variant IdentificationČlánek Bypass of 8-oxodGČlánek Integrated Model of and Inherited Genetic Variants Yields Greater Power to Identify Risk GenesČlánek Comparative Genomic and Functional Analysis of 100 Strains and Their Comparison with Strain GGČlánek A Nuclear Calcium-Sensing Pathway Is Critical for Gene Regulation and Salt Stress Tolerance inČlánek Computational Identification of Diverse Mechanisms Underlying Transcription Factor-DNA OccupancyČlánek Reversible and Rapid Transfer-RNA Deactivation as a Mechanism of Translational Repression in StressČlánek Genome-Wide Association of Body Fat Distribution in African Ancestry Populations Suggests New Loci
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2013 Číslo 8
-
Všechny články tohoto čísla
- Genome-Wide DNA Methylation Analysis of Systemic Lupus Erythematosus Reveals Persistent Hypomethylation of Interferon Genes and Compositional Changes to CD4+ T-cell Populations
- Re-Ranking Sequencing Variants in the Post-GWAS Era for Accurate Causal Variant Identification
- Histone Variant HTZ1 Shows Extensive Epistasis with, but Does Not Increase Robustness to, New Mutations
- Past Visits Present: TCF/LEFs Partner with ATFs for β-Catenin–Independent Activity
- Functional Characterisation of Alpha-Galactosidase A Mutations as a Basis for a New Classification System in Fabry Disease
- A Flexible Approach for the Analysis of Rare Variants Allowing for a Mixture of Effects on Binary or Quantitative Traits
- Masculinization of Gene Expression Is Associated with Exaggeration of Male Sexual Dimorphism
- Genic Intolerance to Functional Variation and the Interpretation of Personal Genomes
- Endogenous Stress Caused by Faulty Oxidation Reactions Fosters Evolution of 2,4-Dinitrotoluene-Degrading Bacteria
- Transposon Domestication versus Mutualism in Ciliate Genome Rearrangements
- Comparative Anatomy of Chromosomal Domains with Imprinted and Non-Imprinted Allele-Specific DNA Methylation
- An Essential Function for the ATR-Activation-Domain (AAD) of TopBP1 in Mouse Development and Cellular Senescence
- Depletion of Retinoic Acid Receptors Initiates a Novel Positive Feedback Mechanism that Promotes Teratogenic Increases in Retinoic Acid
- Bypass of 8-oxodG
- Calpain-6 Deficiency Promotes Skeletal Muscle Development and Regeneration
- ATM Release at Resected Double-Strand Breaks Provides Heterochromatin Reconstitution to Facilitate Homologous Recombination
- Generation of Tandem Direct Duplications by Reversed-Ends Transposition of Maize Elements
- Loss of a Conserved tRNA Anticodon Modification Perturbs Cellular Signaling
- Integrated Model of and Inherited Genetic Variants Yields Greater Power to Identify Risk Genes
- High-Throughput Genetic and Gene Expression Analysis of the RNAPII-CTD Reveals Unexpected Connections to SRB10/CDK8
- Dynamic Rewiring of the Retinal Determination Network Switches Its Function from Selector to Differentiation
- β-Catenin-Independent Activation of TCF1/LEF1 in Human Hematopoietic Tumor Cells through Interaction with ATF2 Transcription Factors
- Genetic Mapping of Specific Interactions between Mosquitoes and Dengue Viruses
- A Highly Redundant Gene Network Controls Assembly of the Outer Spore Wall in
- Origin and Functional Diversification of an Amphibian Defense Peptide Arsenal
- Myc-Driven Overgrowth Requires Unfolded Protein Response-Mediated Induction of Autophagy and Antioxidant Responses in
- Integrative Modeling of eQTLs and Cis-Regulatory Elements Suggests Mechanisms Underlying Cell Type Specificity of eQTLs
- Species and Population Level Molecular Profiling Reveals Cryptic Recombination and Emergent Asymmetry in the Dimorphic Mating Locus of
- Ras-Induced Changes in H3K27me3 Occur after Those in Transcriptional Activity
- Characterization of the p53 Cistrome – DNA Binding Cooperativity Dissects p53's Tumor Suppressor Functions
- Global Analysis of Fission Yeast Mating Genes Reveals New Autophagy Factors
- Deficiency Suppresses Intestinal Tumorigenesis
- Introns Regulate Gene Expression in in a Pab2p Dependent Pathway
- Meiotic Recombination Initiation in and around Retrotransposable Elements in
- Comparative Oncogenomic Analysis of Copy Number Alterations in Human and Zebrafish Tumors Enables Cancer Driver Discovery
- Comparative Genomic and Functional Analysis of 100 Strains and Their Comparison with Strain GG
- A Model-Based Analysis of GC-Biased Gene Conversion in the Human and Chimpanzee Genomes
- Masculinization of the X Chromosome in the Pea Aphid
- The Architecture of a Prototypical Bacterial Signaling Circuit Enables a Single Point Mutation to Confer Novel Network Properties
- Distinct SUMO Ligases Cooperate with Esc2 and Slx5 to Suppress Duplication-Mediated Genome Rearrangements
- The Yeast Environmental Stress Response Regulates Mutagenesis Induced by Proteotoxic Stress
- Mediator Directs Co-transcriptional Heterochromatin Assembly by RNA Interference-Dependent and -Independent Pathways
- The Genome of and the Basis of Host-Microsporidian Interactions
- Regulation of Sister Chromosome Cohesion by the Replication Fork Tracking Protein SeqA
- Neuronal Reprograming of Protein Homeostasis by Calcium-Dependent Regulation of the Heat Shock Response
- A Nuclear Calcium-Sensing Pathway Is Critical for Gene Regulation and Salt Stress Tolerance in
- Cross-Species Array Comparative Genomic Hybridization Identifies Novel Oncogenic Events in Zebrafish and Human Embryonal Rhabdomyosarcoma
- : A Mouse Strain with an Ift140 Mutation That Results in a Skeletal Ciliopathy Modelling Jeune Syndrome
- The Relative Contribution of Proximal 5′ Flanking Sequence and Microsatellite Variation on Brain Vasopressin 1a Receptor () Gene Expression and Behavior
- Combining Quantitative Genetic Footprinting and Trait Enrichment Analysis to Identify Fitness Determinants of a Bacterial Pathogen
- The Innocence Project at Twenty: An Interview with Barry Scheck
- Computational Identification of Diverse Mechanisms Underlying Transcription Factor-DNA Occupancy
- GUESS-ing Polygenic Associations with Multiple Phenotypes Using a GPU-Based Evolutionary Stochastic Search Algorithm
- H2A.Z Acidic Patch Couples Chromatin Dynamics to Regulation of Gene Expression Programs during ESC Differentiation
- Identification of DSB-1, a Protein Required for Initiation of Meiotic Recombination in , Illuminates a Crossover Assurance Checkpoint
- Binding of TFIIIC to SINE Elements Controls the Relocation of Activity-Dependent Neuronal Genes to Transcription Factories
- Global Analysis of the Sporulation Pathway of
- Genetic Circuits that Govern Bisexual and Unisexual Reproduction in
- Deletion of microRNA-80 Activates Dietary Restriction to Extend Healthspan and Lifespan
- Fifty Years On: GWAS Confirms the Role of a Rare Variant in Lung Disease
- The Enhancer Landscape during Early Neocortical Development Reveals Patterns of Dense Regulation and Co-option
- Gene Expression Regulation by Upstream Open Reading Frames and Human Disease
- Sociogenomics of Cooperation and Conflict during Colony Founding in the Fire Ant
- The Intronic Long Noncoding RNA Recruits PRC2 to the Promoter, Reducing the Expression of and Increasing Cell Proliferation
- The , p.E318G Variant Increases the Risk of Alzheimer's Disease in -ε4 Carriers
- The Wilms Tumor Gene, , Is Critical for Mouse Spermatogenesis via Regulation of Sertoli Cell Polarity and Is Associated with Non-Obstructive Azoospermia in Humans
- Reversible and Rapid Transfer-RNA Deactivation as a Mechanism of Translational Repression in Stress
- QTL Analysis of High Thermotolerance with Superior and Downgraded Parental Yeast Strains Reveals New Minor QTLs and Converges on Novel Causative Alleles Involved in RNA Processing
- Genome Wide Association Identifies Novel Loci Involved in Fungal Communication
- Chromatin Sampling—An Emerging Perspective on Targeting Polycomb Repressor Proteins
- A Recessive Founder Mutation in Regulator of Telomere Elongation Helicase 1, , Underlies Severe Immunodeficiency and Features of Hoyeraal Hreidarsson Syndrome
- Genome-Wide Association of Body Fat Distribution in African Ancestry Populations Suggests New Loci
- Causal and Synthetic Associations of Variants in the Gene Cluster with Alpha1-antitrypsin Serum Levels
- Hard Selective Sweep and Ectopic Gene Conversion in a Gene Cluster Affording Environmental Adaptation
- Brittle Culm1, a COBRA-Like Protein, Functions in Cellulose Assembly through Binding Cellulose Microfibrils
- Chromosomal Copy Number Variation, Selection and Uneven Rates of Recombination Reveal Cryptic Genome Diversity Linked to Pathogenicity
- The Ribosomal Protein Rpl22 Controls Ribosome Composition by Directly Repressing Expression of Its Own Paralog, Rpl22l1
- Ras1 Acts through Duplicated Cdc42 and Rac Proteins to Regulate Morphogenesis and Pathogenesis in the Human Fungal Pathogen
- The DSB-2 Protein Reveals a Regulatory Network that Controls Competence for Meiotic DSB Formation and Promotes Crossover Assurance
- Recurrent Modification of a Conserved -Regulatory Element Underlies Fruit Fly Pigmentation Diversity
- Associations of Mitochondrial Haplogroups B4 and E with Biliary Atresia and Differential Susceptibility to Hydrophobic Bile Acid
- The Conditional Nature of Genetic Interactions: The Consequences of Wild-Type Backgrounds on Mutational Interactions in a Genome-Wide Modifier Screen
- A Critical Function of Mad2l2 in Primordial Germ Cell Development of Mice
- A Role for CF1A 3′ End Processing Complex in Promoter-Associated Transcription
- Vitellogenin Underwent Subfunctionalization to Acquire Caste and Behavioral Specific Expression in the Harvester Ant
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Chromosomal Copy Number Variation, Selection and Uneven Rates of Recombination Reveal Cryptic Genome Diversity Linked to Pathogenicity
- Genome-Wide DNA Methylation Analysis of Systemic Lupus Erythematosus Reveals Persistent Hypomethylation of Interferon Genes and Compositional Changes to CD4+ T-cell Populations
- Associations of Mitochondrial Haplogroups B4 and E with Biliary Atresia and Differential Susceptibility to Hydrophobic Bile Acid
- A Role for CF1A 3′ End Processing Complex in Promoter-Associated Transcription
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



