-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaDistinct SUMO Ligases Cooperate with Esc2 and Slx5 to Suppress Duplication-Mediated Genome Rearrangements
Suppression of duplication-mediated gross chromosomal rearrangements (GCRs) is essential to maintain genome integrity in eukaryotes. Here we report that SUMO ligase Mms21 has a strong role in suppressing GCRs in Saccharomyces cerevisiae, while Siz1 and Siz2 have weaker and partially redundant roles. Understanding the functions of these enzymes has been hampered by a paucity of knowledge of their substrate specificity in vivo. Using a new quantitative SUMO-proteomics technology, we found that Siz1 and Siz2 redundantly control the abundances of most sumoylated substrates, while Mms21 more specifically regulates sumoylation of RNA polymerase-I and the SMC-family proteins. Interestingly, Esc2, a SUMO-like domain-containing protein, specifically promotes the accumulation of sumoylated Mms21-specific substrates and functions with Mms21 to suppress GCRs. On the other hand, the Slx5-Slx8 complex, a SUMO-targeted ubiquitin ligase, suppresses the accumulation of sumoylated Mms21-specific substrates. Thus, distinct SUMO ligases work in concert with Esc2 and Slx5-Slx8 to control substrate specificity and sumoylation homeostasis to prevent GCRs.
Published in the journal: . PLoS Genet 9(8): e32767. doi:10.1371/journal.pgen.1003670
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1003670Summary
Suppression of duplication-mediated gross chromosomal rearrangements (GCRs) is essential to maintain genome integrity in eukaryotes. Here we report that SUMO ligase Mms21 has a strong role in suppressing GCRs in Saccharomyces cerevisiae, while Siz1 and Siz2 have weaker and partially redundant roles. Understanding the functions of these enzymes has been hampered by a paucity of knowledge of their substrate specificity in vivo. Using a new quantitative SUMO-proteomics technology, we found that Siz1 and Siz2 redundantly control the abundances of most sumoylated substrates, while Mms21 more specifically regulates sumoylation of RNA polymerase-I and the SMC-family proteins. Interestingly, Esc2, a SUMO-like domain-containing protein, specifically promotes the accumulation of sumoylated Mms21-specific substrates and functions with Mms21 to suppress GCRs. On the other hand, the Slx5-Slx8 complex, a SUMO-targeted ubiquitin ligase, suppresses the accumulation of sumoylated Mms21-specific substrates. Thus, distinct SUMO ligases work in concert with Esc2 and Slx5-Slx8 to control substrate specificity and sumoylation homeostasis to prevent GCRs.
Introduction
The human genome contains many “at-risk” sequences that are prone to mutations including diverse repeated sequences, segmental duplications and regions of copy number variations [1], [2]. Such repetitive sequence elements can cause genome rearrangements through non-allelic homologous recombination (NAHR) and many human diseases are caused by chromosomal rearrangements mediated by NAHR [3]. Moreover, many cancers exhibit ongoing genome rearrangements, stimulated by numerous “at-risk” sequences in the genome. The yeast Saccharomyces cerevisiae provides a powerful model system to study genome rearrangements [4], and many genes in S. cerevisiae have been found to suppress gross chromosomal rearrangements (GCRs), including those mediated by single-copy sequences and those mediated by segmental duplications [5]. Interestingly, a number of these genes are involved in protein sumoylation [5]: Esc2 contains tandem SUMO-like domains [6]–[8], and Slx5 is a subunit of the Slx5-Slx8 complex, which was shown to be a SUMO-targeted ubiquitin ligase [9],[10]. In contrast, Siz1, a SUMO E3 ligase [11], does not have a significant role in suppressing duplication-mediated GCRs [12]. However, deletion of SIZ1 suppresses the GCR defect of asf1 mutant mediated by single-copy sequences to essentially wild-type levels [13]. Taken together, these observations raise the possibility that protein sumoylation may have a role in regulating and in some cases suppressing GCRs.
SUMO (small ubiquitin-like modifier) is a member of the ubiquitin-like protein family, and, like ubiquitin, is covalently attached by an isopeptide bond through its C-terminus to lysine residues of the target proteins via an enzymatic cascade [14]. The SMT3 gene encodes SUMO in S. cerevisiae, and the Smt3 protein is activated by the Aos1-Uba2 complex, which is the S. cerevisiae SUMO E1 activating enzyme [15]. Smt3 is then transferred to Ubc9, the sole SUMO E2 enzyme in S. cerevisiae [16], which catalyzes sumoylation of target proteins. Although Ubc9 is capable of sumoylation by itself in vitro, substrate selectivity in vivo is generally controlled by SUMO E3 ligases. The three known S. cerevisiae SUMO E3 ligases in mitotic cells are Siz1, Siz2, and Mms21 [11], [17]. These SUMO E3 ligases are members of the PIAS (protein inhibitor of activated STAT) family and contain an SP-RING (Siz/PIAS RING) domain, which is similar to the RING domain in ubiquitin E3 ligases [14]. While Siz1 and Siz2 are single-polypeptide enzymes, Mms21 is a subunit of the multi-subunit Smc5-Smc6 complex [17]. Two SUMO-specific isopeptidases, Ulp1 and Ulp2, cleave SUMO from modified proteins [18]–[20]. Ulp2 is present throughout the nucleus, whereas Ulp1 is localized to the nuclear envelope through interactions with the nuclear pores [21], [22]. The non-redundant genes in the sumoylation pathway, SMT3, AOS1, UBA2, and UBC9, are all essential for viability in S. cerevisiae. In addition, ULP1 and MMS21 are also essential; however, the SUMO E3 ligase domain of MMS21 is not required for cell viability [17]. Moreover, the SUMO-ligase deficient mms21 mutation is lethal when it is combined with both siz1 and siz2 mutations, indicating a requirement of SUMO E3 ligases for sumoylation in vivo [23]. Protein sumoylation regulates numerous cellular processes, including protein transport, gene transcription, chromosome segregation, DNA repair, and meiosis [14], [24], [25]. The structure and enzymology of these sumoylation enzymes are relatively well understood; however, the identities of sumoylated substrates and specificity of the SUMO E3 ligases in vivo have been more poorly characterized, leaving a gap in our knowledge of the roles of sumoylation in vivo.
Several approaches have been taken to identify proteins sumoylated in vivo. A number of sumoylated proteins have been identified via a candidate-protein approach [23], [26]–[30]; however, this approach is difficult to apply to a proteome-wide study. On the other hand, mass spectrometry (MS) based methods have been used to more broadly identify sumoylated proteins in yeast [31]–[35]. In these studies, epitope-tagged SUMO was used for affinity-based purification of sumoylated proteins, which were either subjected to SDS-PAGE and in-gel digestion or liquid chromatography-based method for MS analysis. SDS-PAGE separation helps to remove unconjugated monomeric SUMO prior to MS analysis; however, this process limits the throughput and sensitivity of MS analysis [31]–[33]. In cases where liquid chromatography was used, the presence of the highly abundant Smt3 could compromise the detection of lower abundant sumoylated proteins [34], [35]. Importantly, varying levels of contaminant proteins are unavoidable during purification of sumoylated proteins. As a consequence, the lack of quantitative MS analysis to examine the levels of contaminant proteins in these previous studies limits the confidence that all of the proteins identified were actually sumoylated. Here we describe the development of a new proteomics technology for a quantitative analysis of protein sumoylation. Using stable isotope labeling method, we were able to exclude contaminant proteins and provide quantitative MS evidence for sumoylated proteins identified here. We then applied this new technology to determine the substrate specificity of SUMO E3 ligases Siz1, Siz2, and Mms21 on a proteome-wide scale for the first time. We further discovered a novel function of Esc2 in regulating the substrate selectivity of Mms21 and showed that the Slx5-Slx8 complex works in concert with Siz1, Siz2 and Mms21 to regulate substrate-specific sumoylation homeostasis in vivo. Together, these findings provide mechanistic insights into the functions of Esc2 and Slx5-Slx8 in suppressing GCRs and maintaining sumoylation homeostasis in conjunction with distinct SUMO ligases.
Results
Distinct SUMO ligases cooperate with Esc2 and Slx5-Slx8 to suppress duplication-mediated GCRs
To explore the roles of the SUMO E3 ligases in genome stability, we introduced deletions of SIZ1 and SIZ2 and two alleles of the essential MMS21 gene, mms21-11 and mms21-CH, into strains to measure the rates of accumulating GCRs. We tested the mutations in two strain backgrounds; strains containing the yel068c::CAN1/URA3 assay monitor GCRs formed using single-copy sequences, whereas strains containing the yel072w::CAN1/URA3 assay monitor GCRs formed both by a segmental duplication and by single-copy sequences (Table 1 and Figure S1) [5]. Neither the siz1Δ mutation nor the siz2Δ mutation caused an appreciable defect in suppressing GCR formed in either assay. In contrast, the siz1Δ siz2Δ double mutant strain had a synergistic increase in the rate of GCR formation specific to the duplication-mediated GCR assay. This is consistent with the increase in mitotic recombination (loss of heterozygosity) seen in the siz1Δ siz2Δ double mutant [36]. The mms21-11 allele, which encodes a truncated Mms21 that lacks the SUMO ligase domain at the C-terminus [17], caused substantially increased GCR rates both in the yel068c::CAN1/URA3 assay (∼47-fold), consistent with increases seen in a related GCR assay of the smc6 mutant [37], and in the yel072w::CAN1/URA3 assay (∼603-fold; Table 1). Combining the mms21-11 allele with the siz1Δ mutation caused a 3-fold increase in the yel068c::CAN1/URA3 assay and no increase in the yel072w::CAN1/URA3 assay. In contrast, the mms21-11 siz2Δ double mutant strain had a ∼1,800-fold increase in the yel072w::CAN1/URA3 assay relative to wild-type and a ∼3-fold increase relative to the mms21-11 strain, which is the highest GCR rate yet reported with this assay [5], [12]. To verify that the increased GCR rates in mms21-11 allele were due to loss of SUMO ligase activity, we also introduced the mms21-CH allele into the two GCR assay strains. The mms21-CH allele encodes an inactive SUMO ligase domain caused by alanine substitutions at the conserved Cys200 and His202 of the SP-RING domain [38]. The increases in GCR rates caused by the mms21-CH allele were very similar to mms21-11: (i) the single mutant strain caused a substantially increased GCR rate in both assays (∼9-fold and ∼215-fold for non-duplication mediated and duplication mediated assays, respectively), and (ii) combinations with deletions of SIZ1 and SIZ2 caused increased GCRs, with the largest effects being observed in the yel072w::CAN1/URA3 assay and caused by additional deletion of SIZ2 (Table 1). Taken together, these data indicate that the SUMO ligase activity of Mms21 suppresses GCRs mediated by single-copy sequences and plays an even more important role in suppressing GCRs mediated by segmental duplications. In contrast, Siz1 and Siz2 are partially redundant, consistent with previous studies [27], and relatively less important than Mms21 in suppressing GCRs. Additionally, Siz2 plays a relatively stronger role in suppressing GCRs than Siz1 in the absence of Mms21 SUMO ligase activity.
Tab. 1. Effect of mutation of SUMO E3 ligases, Esc2 and Slx5 on duplication-mediated GCRs. 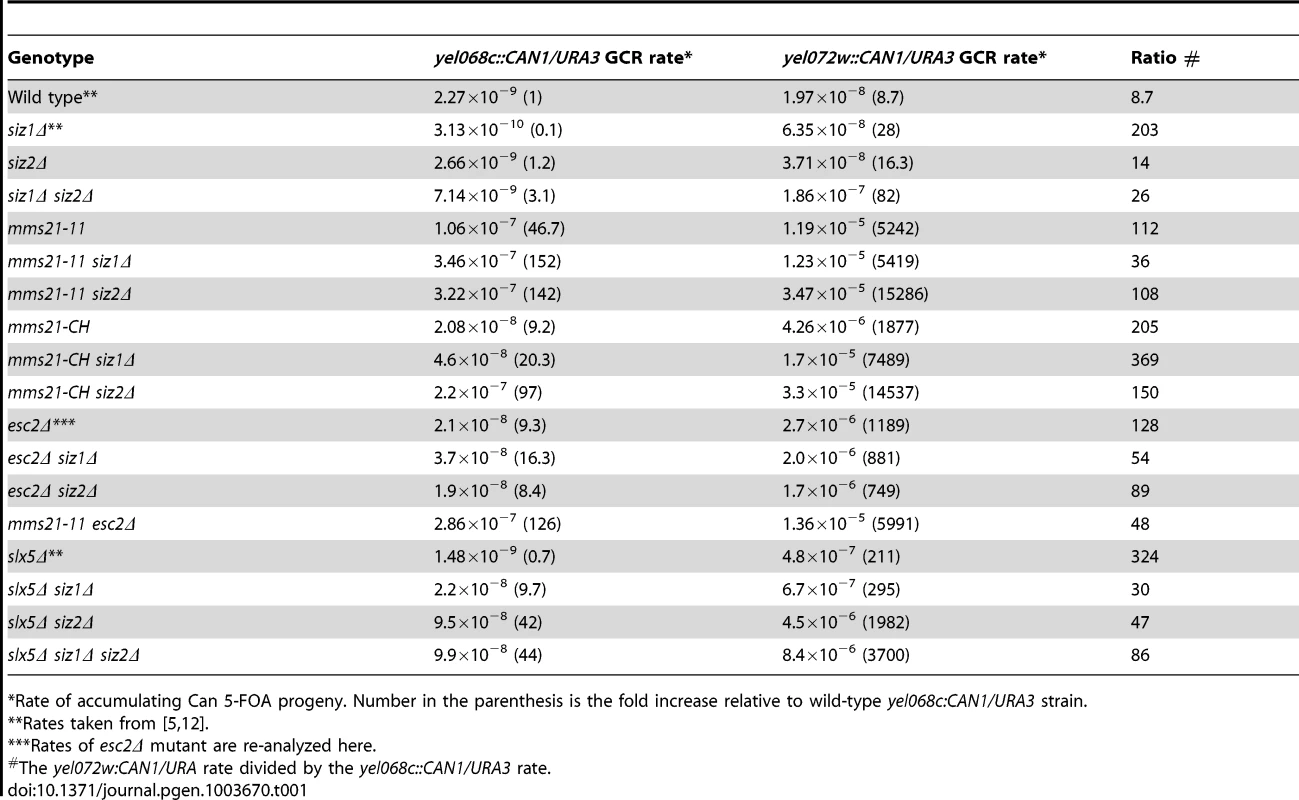
Rate of accumulating Can 5-FOA progeny. Number in the parenthesis is the fold increase relative to wild-type yel068c:CAN1/URA3 strain. ESC2 encodes a protein containing two SUMO-like domains [6]–[8] and plays a role in sister chromatid cohesion in conjunction with Smc5-Smc6 in the repair of damage during DNA replication [39], [40]. Deletion of ESC2 also causes a strong defect in suppressing GCRs, particularly duplication-mediated GCRs, though less than that caused by the mms21-11 allele (Table 1) [5]. Combining esc2Δ with siz1Δ or siz2Δ did not cause further increases in the GCR rates from either assay relative to the esc2Δ. The esc2Δ mms21-11 double mutant strain had a similar GCR rate to the mms21-11 rates in both GCR assays. Together these data suggest epistasis between esc2Δ and mms21-11, but cannot distinguish between an epistatic or additive effect of combining esc2Δ with siz1Δ and siz2Δ given the weak effects of the siz1Δ and siz2Δ mutations in the rate of accumulating GCRs.
Combining a deletion of SLX5 with mutations of either ESC2 or MMS21 causes lethality [7], [29], suggesting SLX5 functions in pathways that are redundant to ESC2 and MMS21, and is potentially redundant with SIZ1 and SIZ2. We therefore examined the genetic interactions between SLX5, SIZ1 and SIZ2. The slx5Δ single mutation did not cause an increase in the yel068c::CAN1/URA3 assay but did in the duplication-mediated yel072w::CAN1/URA3 assay. Combining slx5Δ with siz1Δ or siz2Δ caused a synergistic increase in GCR rates in the yel068c::CAN1/URA3 assay, and the slx5Δ siz1Δ siz2Δ triple mutant was not appreciably different than the slx5Δ siz2Δ double mutant. All of the slx5Δ, siz1Δ, and siz2Δ single mutations had stronger effects on the accumulation of GCRs in the duplication-mediated yel072w::CAN1/URA3 assay (Table 1). Similarly, the combination of slx5Δ with siz1Δ and/or siz2Δ also caused a much greater effect on the accumulation of GCRs in the yel072w::CAN1/URA3 assay than the yel068c::CAN1/URA3 assay; however, the relative effects of the mutations was the same as in the yel068c::CAN1/URA3 assay. Taken together the data indicate that SLX5 functions redundantly with SIZ1 and SIZ2 to suppress GCRs, with SIZ2 playing a more important role than SIZ1 in the absence of SLX5.
Quantitative and proteome-wide analysis identifies SUMO targets
In order to monitor sumoylation on a proteome-wide scale, we developed a quantitative proteomics method to identify and quantify sumoylated proteins (Figure 1A). We first integrated a 6×HIS-3×FLAG (HF) tag at the 5′-end of the SMT3 gene at the chromosomal locus so that HF-SUMO and proteins covalently modified by HF-SUMO could be isolated by a tandem-affinity purification using Ni-NTA and anti-Flag columns. Since contaminant proteins are unavoidable and they could vary between independent purifications, we chose to metabolically label HF-SUMO cells and untagged cells using the Stable Isotope Labeling via the Amino acid in Cell culture (SILAC) method [41], [42]. An HF-SUMO tagged strain and an untagged strain that are both unable to synthesize lysine or arginine were grown in media that contained either 14N/12C-incorporated lysine and arginine or 15N/13C-incorporated lysine and arginine. Equal numbers of cells from both cultures were combined prior to cell lysis and purification of sumoylated proteins as one sample. This double labeling method then allowed us to monitor contaminant proteins during the purification of sumoylated proteins.
Fig. 1. A new quantitative proteomics technology to characterize the SUMO-proteome. 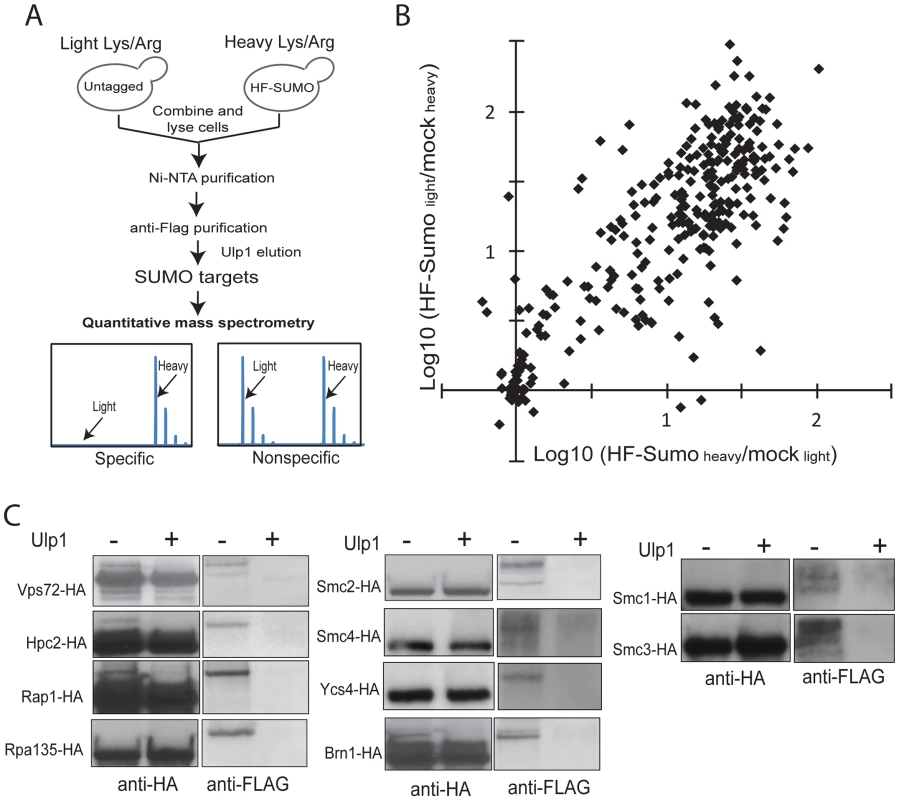
A) Method to identify and quantify sumoylated proteins using SILAC and MS. Untagged cells and HF-SUMO cells were combined for purification of sumoylated proteins. Ulp1 was used to elute sumoylated proteins for MS analysis. B) Scatter plot of the identified proteins based on two replicate experiments. The majority of the proteins show a large abundance ratio between HF-SUMO purification versus mock purification, indicating a highly purified sample. Candidate SUMO targets are identified based on at least 10-fold abundance ratio between HF-SUMO and mock sample in both replicate experiments. C) Western blot analysis of several SUMO targets confirmed them being sumoylated. Ulp1 treatment was used to treat half of the anti-HA immunoprecipitated sample. To remove the highly abundant unconjugated SUMO, which is a major barrier in the use of liquid chromatography-based proteomics technology for studying sumoylated proteins, we eluted the sumoylated proteins from the anti-Flag antibody column by specifically cleaving the isopeptide bonds between these proteins and HF-SUMO with Ulp1 [18]. The Ulp1-eluted proteins were then digested by trypsin and directly analyzed by multi-dimensional liquid chromatography and MS [43]. Proteins enriched in the HF-SUMO cells relative to the untagged cells, which are readily distinguished and quantified based on their isotope signatures, are candidate sumoylated proteins whereas those proteins with both isotope signatures are contaminant proteins that can be excluded from further analysis. We performed two replicate experiments with the HF-SUMO cells labeled by light lysine and arginine in the first experiment and by heavy lysine and arginine in the second experiment. We identified 653 proteins in the first and 695 proteins in the second experiment, and 306 proteins were identified in both replicates (Figure 1B). Among them, 176 proteins were strongly enriched (>10-fold) from HF-SUMO cells in both experiments (Table 2, Tables S1 and S2). Sites of modification could not be identified as Ulp1 treatment removes the SUMO modification; however, most targets were identified with fewer than 10 peptides and relatively low protein sequence coverage that would not be sufficient for SUMO site identification anyway (Tables S1 and S2). Our use of SILAC labeling, however, allows identification of sumoylated proteins at much higher sensitivities than methods dependent upon identification of sumoylation sites. We also compared the previously proteomic data sets and found a general lack of concordance between them (Figure S2A) [32]–[35]. Among them, the findings of Wohlschlegel et al and Denison et al share the most overlap with the targets identified here (Figure S2B), which were generally identified with more peptides per protein and likely represent the more abundant SUMO targets.
Tab. 2. A summary of 176 sumoylated proteins identified from two replicate MS experiments (see text for details). 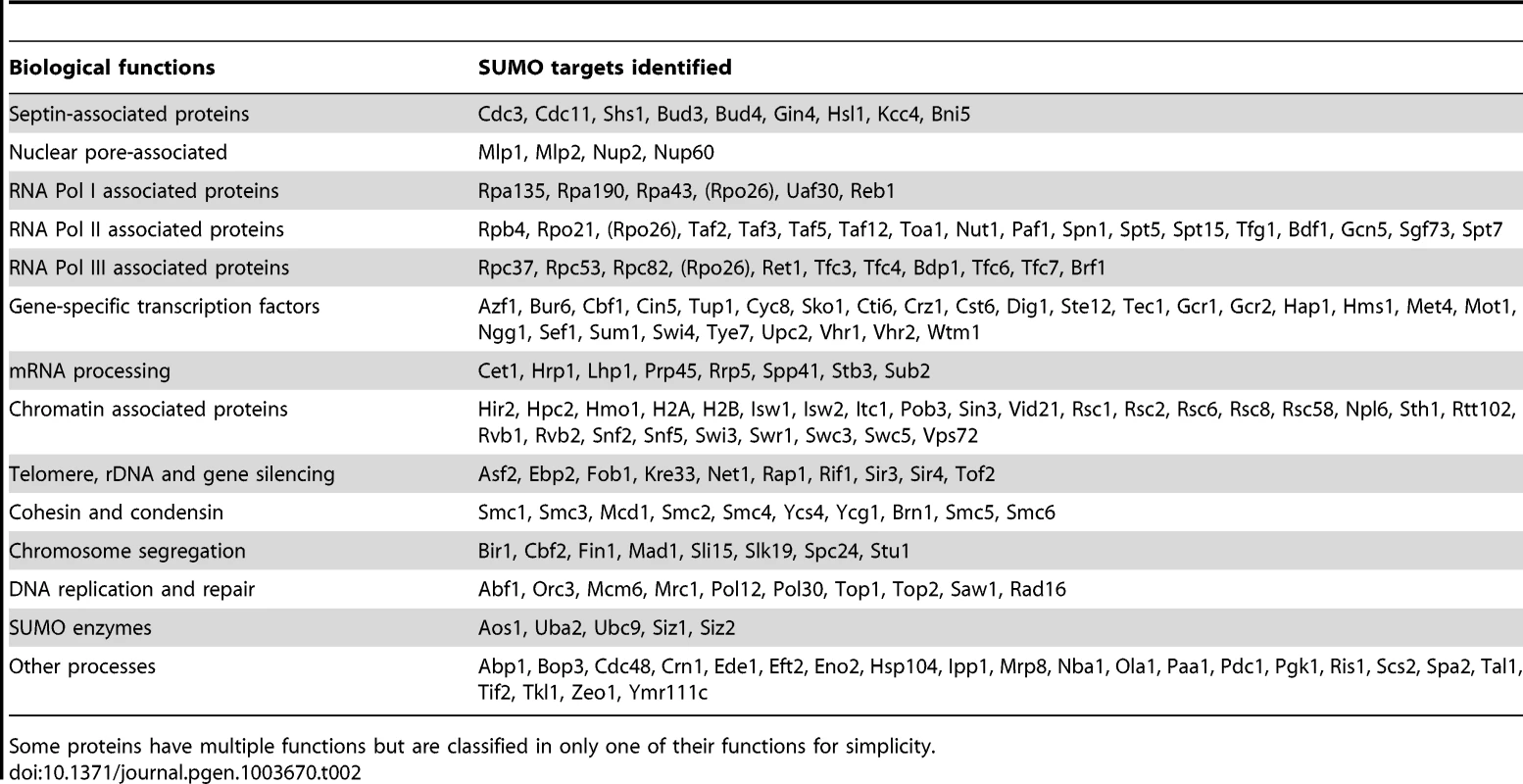
Some proteins have multiple functions but are classified in only one of their functions for simplicity. We selected 10 candidate proteins identified above to verify the presence of the SUMO modification in unperturbed cells, including Vps72, Hpc2, Rap1, Rpa135, subunits of cohesin (Smc1 and Smc3), and subunits of condensin (Smc2, Smc4, Ycs4, and Brn1). For each protein of interest, we integrated a 3×HA tag at 3′-end of the encoding gene at the endogenous locus in the HF-SUMO strain. We then lysed cultures and purified the tagged protein via anti-HA immunoprecipitation. Part of the immunoprecipitate was treated by recombinant Ulp1, and treated and untreated immunopreciptates were analyzed by anti-HA and anti-FLAG Western blots (Figure 1C). The anti-FLAG antibody, which recognizes HF-SUMO, detected a species that was eliminated by Ulp1 treatment. This anti-FLAG reactive species had a higher molecular weight than the species recognized by the anti-HA antibody, which primarily detects the unmodified protein. Thus this analysis indicated both that all of these candidate proteins are sumoylated, and that only a fraction of the endogenous protein is sumoylated under non-perturbed conditions.
We classified the SUMO targets identified by MS according to their known functions (Table 2). Consistent with previous findings [23], [26], [30], [44]–[47], Septins, PCNA and many known sumoylated proteins are identified. Despite the differences between previously proteomic studies and our study here [32]–[35], which could be attributed to the methods used, considerable overlaps in the identified proteins are found (Figure S2). By far, most SUMO targets identified are involved in gene expression, including the core subunits of RNA polymerases, their associated proteins, chromatin remodelers, numerous gene-specific transcription factors and proteins involved in mRNA processing and transport. Of particular interests here are the proteins involved in DNA replication and repair. We observed fewer DNA repair proteins than have been reported [26], likely due to the fact that the cells studied here were not exposed to DNA damaging treatments. It is also interesting to note that many proteins that function in chromosomal organization were found as sumoylated substrates, including cohesin (Smc1, Smc3, Mcd1), condensin (Smc2, Smc4, Brn1, Ycs4, Ycg1) and the Smc5-Smc6 complex. In summary, the majority of the SUMO targets identified here function in the nucleus; consistent with a diverse role of protein sumoylation in nuclear processes.
We also often identified multiple subunits of large protein complexes (Table 2). This is likely to be due to multiple subunits being sumoylated and unlikely to be due to co-purification. We used strong denaturing conditions during the purification (see Experimental procedures). Additionally, combining untagged cells and HF-SUMO cells prior to their lysis for subsequent purification would exclude subunits undergoing rapid exchange, as peptides from these subunits would be identified with both light and heavy isotope signatures. In the case of septin sumoylation, we identified the known sumoylation targets Cdc3, Cdc11, and Shs1, but not the Cdc10 or Cdc12 subunits that are not known to be modified [44]. Similarly, for most of the complexes for which we identified multiple subunits, we consistently identified the same subsets of the complex in multiple subsequent experiments. Indeed, sumoylation of multiple subunits of a protein complex often occurs [26], [44], [48]. This could be due to either promiscuous sumoylation in vivo, or alternatively redundancy and/or cooperativity during signaling in downstream events such as the recruitment of proteins via SUMO-interaction motifs [49], [50].
Specificity of the Siz1, Siz2 and Mms21 E3 SUMO ligases
We next sought to determine the specificity of the SUMO E3 ligases Siz1, Siz2 and Mms21. We therefore compared the relative abundance of each SUMO target in wild-type strains and various strains with defects in specific E3 SUMO ligases using quantitative MS. A wild-type strain expressing HF-SUMO and a siz1Δ, siz2Δ, mms21-11, or siz1Δ siz2Δ strain expressing HF-SUMO were grown in media that labeled proteins with arginines and lysines containing 15N/13C or 14N/12C, respectively. Equal numbers of cells from the wild-type strain and a mutant strain were then mixed and lysed, and the SUMO-modified proteins were isolated by tandem-affinity purification and Ulp1-elution. The eluent was then digested by trypsin, and peptides identified by tandem MS. Changes in the levels of sumoylation of targets were determined by calculating the ratio of intensities of individual peptides from the mutant strains relative to the intensities from the wild-type strain (Tables S3, S4, S5, S6).
Remarkably, sumoylation of only a few proteins was entirely or almost entirely dependent upon Siz1 (Figure 2). These included the septins Cdc3, Cdc11 and Shs1, as well as their associated proteins Bud3 and Bud4, in agreement with previous findings [11]. Other Siz1-specific targets were also identified, including the pre-mRNA splicing factor Prp45, the transcription factors Sum1 and Vhr1, and the proteins of unknown function Bop3 and Mrp8 (Table S3). In contrast, none of the SUMO targets was specific to Siz2 exclusively. Unexpectedly, the major effect of the siz2Δ mutation was a modest increase in the abundance of many SUMO targets compared to wild-type cells, which could possibly be caused by up-regulation of Siz1 and/or Mms21 activity in the siz2Δ strain. This up-regulation is unlikely to be due to delays in S-phase as siz2Δ was not identified in screen of mutation altering the S. cerevisiae replication timing program [51]. For the siz1Δ siz2Δ double mutant strain, we observed a strong reduction of sumoylation of the majority of SUMO targets, indicating that Siz1 and Siz2 mediate sumoylation of many of the same targets and that biochemical redundancy explains the genetic interactions between SIZ1 and SIZ2 in suppressing genomic instability (Table 1). Sumoylated proteins primarily dependent upon either Siz1 or Siz2 for sumoylation included Pol30/PCNA, which had undetectable sumoylation in the siz1Δ siz2Δ double mutant strain as previously reported [52]. In addition, we identified proteins involved in chromosome segregation (Bir1, Cbf2, and Spc24), DNA replication (Abf1 and Orc3), DNA repair (Saw1 and Rad16), RNA polymerase II transcription (Rpo21, Rpo26, Rpb4, Reb1, Toa1, and subunits of TFIIA, TFIID, TFIIG, and SAGA), and RNA polymerase III transcription (Rpc37, Rpc53, Rpc82, Rpo26, and subunits of TFIIIB and TFIIIC). In contrast, the amounts of RNA polymerase I specific subunits Rpa135 and Rpa190 were higher in the siz1Δ siz2Δ double mutant strain compared to the wild-type strain. Thus, Siz1 and Siz2 have a highly redundant role in the sumoylation of proteins involved in gene transcription, chromatin remodeling and others with the notable exception of RNA polymerase I subunits such as Rpa135.
Fig. 2. A summary of the effect of SUMO ligase-null mutations on the abundance of SUMO targets as measured by quantitative MS. 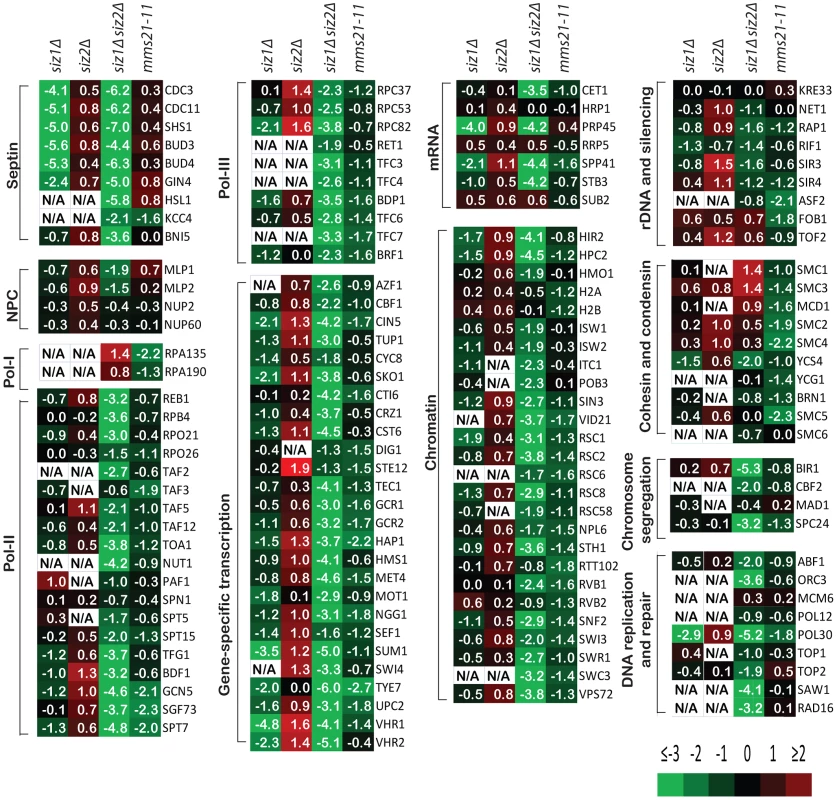
Data are shown in the same order of siz1Δ, siz2Δ, siz1Δ siz2Δ and mms21-11 mutants from left to right for each SUMO target. Log2 scale was used to calculate the relative abundance ratios. Occasionally, some SUMO targets were not identified and are indicated as not available (N/A). Additional information can be found in Tables S3, S4, S5, S6. We next examined the effect of mms21-11 mutation on the abundance of sumoylated proteins (Figure 2). In contrast to the siz1Δ siz2Δ mutant strain, most SUMO targets only underwent a modest decrease in sumoylation, including some of the RNA polymerase II and III related proteins and proteins involved in chromatin maintenance and remodeling. Sumoylated targets that were decreased primarily in the mms21-11 strain but not the siz1Δ siz2Δ double mutant strain included the RNA polymerase I subunit Rpa135 and the structural maintenance of chromosome (SMC) family of proteins, Smc1, Smc2, Smc3, Smc4 and Smc5. In summary, the siz1Δ, siz2Δ and mms21-11 mutations cause distinct perturbations to a subset of SUMO targets with Siz1 and Siz2 having redundant roles for the majority of proteins sumoylated in unperturbed cells.
Sumoylation of RNA polymerase I and the SMC-family proteins requires Mms21
Since the defects in suppression of GCRs caused by the tested mms21 hypomorphic alleles were stronger than that caused by the siz1Δ siz2Δ double mutation (Table 1), we sought to gain insight into Mms21-specific substrates. First, we calculated the ratio between Mms21-dependent ratio (mms21-11 versus wild-type) and Siz1/Siz2 dependent ratio (siz1Δ siz2Δ versus wild-type) for each SUMO target, which is equivalent to the abundance ratio between mms21-11 and siz1Δ siz2Δ mutants. Most SUMO targets were more specific to Siz1/Siz2 than Mms21 (Figure 3A, grey bars). For example, the amounts of septin-associated proteins are much higher in mms21-11 mutant compared to siz1Δ siz2Δ mutant. Similarly, numerous transcription-related proteins are more specific to Siz1/Siz2 than to Mms21. Interestingly, we observed a small subset of proteins that were less sumoylated in the mms21-11 mutant strain compared to the siz1Δ siz2Δ double mutant strain. These proteins included RNA polymerase I, the ribosomal DNA-associated proteins Fob1 and Tof2, and the SMC-family of cohesin and condensin subunits. Second, we confirmed these findings experimentally by using quantitative MS to compare the abundance of sumoylated proteins in the mms21-11 mutant strain with the siz1Δ siz2Δ double mutant strain (Figure 3A, black bars; Table S7). The results were essentially identical to the previously calculated ratio of ratios, which indicated the accuracy and quality of the MS data in three independent MS experiments (siz1Δ siz2Δ versus wild-type, mms21-11 versus wild-type, and mms21-11 versus siz1Δ siz2Δ). The robustness is due, in part, to the use of multiple peptides for protein quantification that allow changes as small as two-folds to be accurately measured.
Fig. 3. Identification of SUMO targets that are more specific to Mms21 than Siz1 and Siz2. 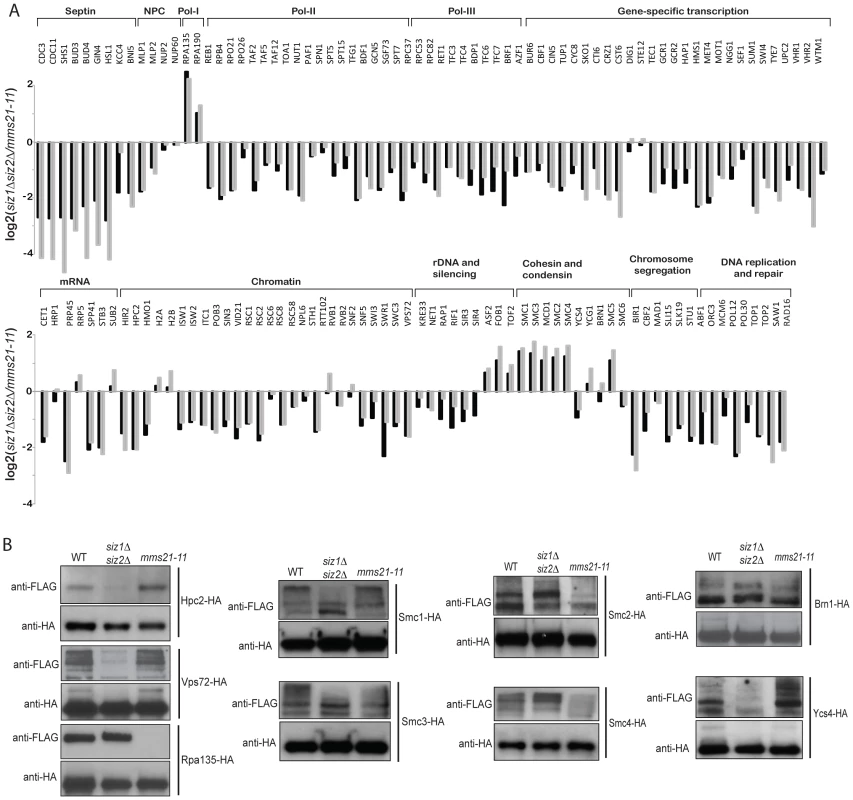
A) Grey bar shows the calculated ratio between Mms21-dependent ratio and Siz1/Siz2 dependent ratio, using the data shown in Figure 2. Black bar shows a direct comparison between mms21-11 and siz1Δ siz2Δ mutants using quantitative MS. B) Effects of mm21-11 and siz1Δ siz2Δ on the sumoylation of selected SUMO targets. Each SUMO target was purified by anti-HA antibody and probed for its sumoylation using anti-FLAG Western blot. To further substantiate the MS findings, we examined sumoylation of several SUMO targets using Western blot analysis. Among Siz1/Siz2-specific targets, sumoylation of Hpc2 and Vps72 were drastically reduced in the siz1Δ siz2Δ double mutant strain but not the mms21-11 mutant strain (Figure 3B), in agreement with the MS findings. Also consistent with the MS results, sumoylation of Rpa135 was not detected in the mms21-11 mutant strain but was unaffected in the siz1Δ siz2Δ double mutant strain (Figure 3B). On the other hand, sumoylation of the cohesin and condensin subunits had a more complex dependency on Siz1, Siz2 and Mms21. In contrast to a previous report [47], sumoylation of Smc1 and Smc3 of the cohesin complex was modestly reduced by the mms21-11 mutation (Figure 3B), consistent with our MS results (Figure 2). The condensin subunits Smc2 and Smc4 had a stronger reduction of sumoylation levels in the mms21-11 mutant strain than the siz1Δ siz2Δ double mutant strain (Figure 3B); however, sumoylation of the condensin subunit Ycs4 was more strongly reduced in the siz1Δ siz2Δ mutant strain, while sumoylation of the condensin subunit Brn1 was not significantly affected in either the mms21-11 or the siz1Δ siz2Δ mutant strains. These observations indicate that Mms21 has a relatively stronger role in the sumoylation of the SMC subunits of cohesin and condensin than Siz1 and Siz2. Moreover, sumoylation of RNA polymerase I subunit Rpa135 is almost entirely dependent on Mms21.
Esc2 regulates sumoylation of Mms21-preferred substrates
Esc2 has been shown to interact with Ubc9 and SUMO itself [7], and the ESC2 deletion was found to be epistatic to the mms21-11 mutation (Table 1). Thus, Esc2 might play a role in promoting the sumoylation of Mms21 substrates. To test this, we first used quantitative MS to determine the abundance ratios of sumoylated proteins between the wild-type strain and the esc2Δ mutant strain (Figure 4A, black bars; Table S8). Comparison with the abundance ratios of sumoylated proteins between the wild-type strain and the mms21-11 mutant strain (Figure 4A, grey bars) revealed an overall similarity between the effects of esc2Δ and mms21-11 on the abundances of most SUMO targets. For example, sumoylation of septins was elevated in both esc2Δ and mms21-11 mutant strains compared to the wild-type strain, while that of Rpa135, Smc2 and Smc4 was reduced in both esc2Δ and mms21-11 mutants. When we verified this similarity through direct comparison of the abundance of sumoylated proteins in the esc2Δ and the mms21-11 mutant strains by quantitative MS, we found little change in the abundances of most SUMO targets between esc2Δ and mms21-11 mutants (Figure 4B, Table S9). Together, these data indicate that Esc2 and Mms21 have similar roles on the sumoylated proteins in cells and are consistent with previously observed genetic epistasis in maintaining genome stability (Table 1).
Fig. 4. Effect of esc2Δ on the sumoylation of SUMO targets. 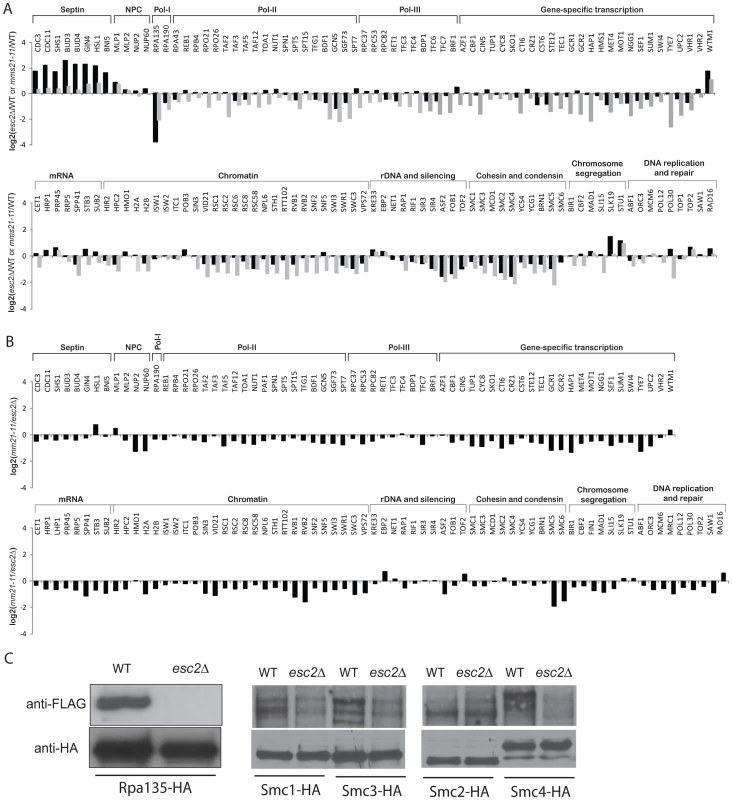
A) Bar graph shows the relative abundance of SUMO targets between esc2Δ and wild-type cells (black bars). As a comparison, the results of mms21-11 versus wild-type cells are shown in grey bars. B) Quantification of the relative abundance of SUMO targets between esc2Δ and mms21-11 mutants. C) Western blot analysis of the effect of esc2Δ on the sumoylation of SMC proteins and Rpa135. Each SUMO target was purified by anti-HA antibody and probed for its sumoylation by anti-FLAG Western blot. We also determined the relative specificity between Esc2 and Siz1/Siz2 for sumoylation of targets by calculating the ratio of the esc2Δ/wild-type (Figure 4A) and siz1Δ siz2Δ/wild-type (Figure 2) relative abundance ratios (Figure S3). Most SUMO targets were more strongly dependent on Siz1/Siz2 than Esc2; however, a small subset of SUMO targets was more specific to Esc2, including Rpa135 and the SMC subunits of cohesin and condensin, similar to the comparison between the mms21-11 and siz1Δ siz2Δ strains (Figure 3A). We confirmed these MS findings by Western blot analysis and found that sumoylation of Rpa135 was not detectable in the esc2Δ mutant (Figure 4C), similar to mms21-11 (Figure 3B). Sumoylation of Smc1, Smc2 and Smc3 were modestly reduced by esc2Δ, and sumoylation of Smc4 was more strongly reduced by esc2Δ. In summary, Esc2 and Mms21 had a similar, although not identical, effect on the sumoylation levels of most SUMO targets in vivo. Both Esc2 and Mms21 exhibited stronger specificity towards sumoylation of Rpa135 and the SMC-family subunits of cohesin and condensin such as Smc4.
Slx5 antagonizes sumoylation of Mms21-specific targets
Slx5 and its mammalian ortholog Rnf4 bind to sumoylated proteins and promote ubiquitination events [10], [53], [54]. To determine the role of Slx5-Slx8 complex in regulating the SUMO proteome, we compared the abundances of sumoylated proteins in the wild-type strain and the slx5Δ mutant strain using quantitative MS (Table S10). The net effect of deleting SLX5 was to increase the overall abundance of most of the sumoylated proteins (Figure 5A, black bars). Siz1-specific targets were only weakly affected; sumoylation of septins was modestly increased, whereas sumoylation of Pol30/PCNA, Sum1 and Prp45 was not appreciably altered. In contrast, sumoylation of Mms21-specific targets were elevated, including cohesin and condensin subunits; sumoylation of Smc4 increased by 4-fold. Together, these data suggested that Slx5 more specifically suppressed the accumulation of sumoylated Mms21 targets. Since mms21-11 had a broad effect on many SUMO targets (Figure 2), we compared the abundance ratios of slx5Δ/wild-type and mms21-11/wild-type and observed an inverse correlation between them with the exceptions of septins and RNA polymerase I (Figure 5A, compare black and grey bars). The sumoylation events down-regulated in the mms21-11 mutant strain, such as cohesin and condensin subunits, were up-regulated in the slx5Δ mutant strain, indicating an antagonistic relationship between Slx5 and Mms21.
Fig. 5. Effect of slx5Δ on the sumoylation of SUMO targets. 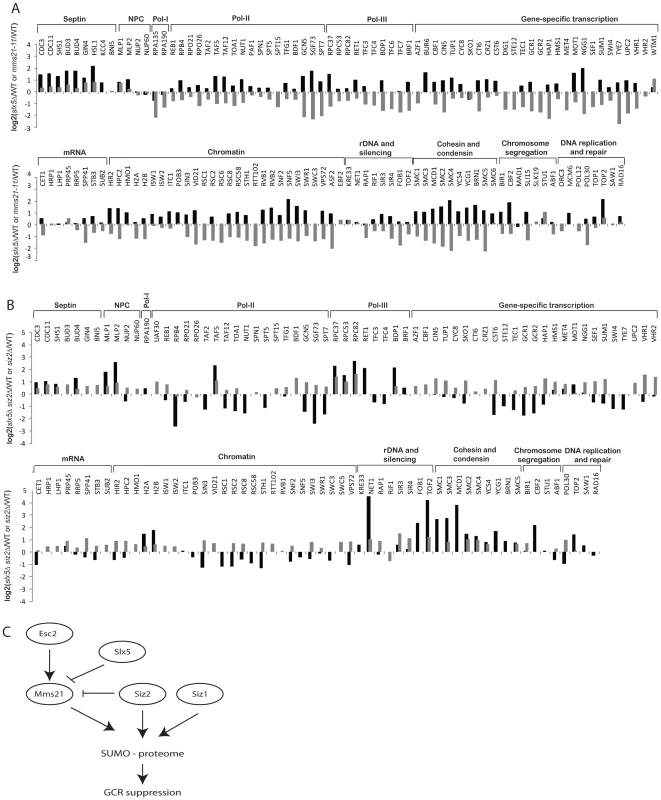
A) Bar graph shows the relative abundance of SUMO targets between slx5Δ and wild-type cells (black bars). Results of mms21-11 versus wild-type are included for comparison (grey bars). B) Bar graph shows the relative abundance of SUMO targets between slx5Δ siz2Δ and wild-type cells (black bars). Results of siz2Δ versus wild-type are included for comparison (grey bars). C) A proposed model of the regulation of sumoylation homeostasis in cells. Mms21-specific sumoylation is positively regulated by Esc2 and negatively regulated by Slx5 to achieve a balance of sumoylation in cells, which is critical to suppress GCRs. Next, we examined the effect of deleting SIZ1 on the SUMO proteome in slx5Δ cells. As expected, there was a drastic reduction of sumoylation of Siz1-specific targets including septins and Pol30 (Figure S4, Table S11). Overall, the effect of deleting SIZ1 on the SUMO proteome of the slx5Δ mutant strain was similar to that of deleting SIZ1 in the wild-type strain, indicating a lack of functional interaction between SIZ1 and SLX5. In contrast, deletion of SIZ2, which alone caused a modest increase in sumoylated proteins (Figure 2), caused a further increase in sumoylation of cohesion and condensin subunits when combined with deletion of SLX5 (Figure 5B, Table S12). As expected, Siz1-specific targets such as septins and Pol30 were not appreciably affected in the siz2Δ slx5Δ mutant. Thus, the deletion of SLX5 increases sumoylation of Mms21-specific targets such as cohesin and condensin subunits, which is further exacerbated by deletion of SIZ2.
Discussion
Mms21, Siz1, and Siz2 have overlapping and distinct substrate specificities
Protein sumoylation is an important post-translational modification that regulates many cellular processes including, protein transport, gene transcription, and DNA repair [14], [25], [55]. Genetic studies have suggested that the three E3 SUMO ligases in S. cerevisiae, Mms21, Siz1, and Siz2, have distinct as well as overlapping functions [17], [56], [57]. Although the E3 SUMO ligases for a few SUMO substrates were known [23], [45], [47], here we have comprehensively analyzed the proteome-wide substrate specificities of Siz1, Siz2, and Mms21 in S. cerevisiae using a novel quantitative proteomics strategy. Several key findings are summarized here.
First, Siz1 and Mms21 specifically sumoylate a subset of substrates, whereas we could not detect any Siz2-specific SUMO targets in unperturbed cells. Siz1 plays a major role in the sumoylation of septin subunits and septin-associated proteins as well as some factors involved in RNA polymerase II transcription. In contrast, Mms21 plays an important role in sumoylation of proteins in the nucleolus, including RNA polymerase I, Fob1, and Tof2, as well as cohesin and condensin subunits that play roles both in the nucleolus and the nucleus. Lack of Siz2-specific targets may be due to the lower abundance of Siz2-specific targets in unperturbed cells, to a specific role of Siz2 during certain stresses [48], or to redundancy between SIZ2 and other E3 SUMO ligases. In the siz1Δ siz2Δ double mutant strain, sumoylation of the majority of SUMO targets was down regulated. This result indicates that there is a strong overlap in the substrate specificity between Siz1 and Siz2, which is likely due to the fact that the SIZ1 and SIZ2 genes are paralogs arising from the whole genome duplication event that occurred in the ancestor of S. cerevisiae and related yeast species [58].
Second, multiple subunits of the same protein complex are often sumoylated. As an example, Smc2, Smc4, Ycs4 and Brn1 of condensin are found to be sumoylated. Interestingly, although Mms21 preferentially regulates sumoylation of Smc2 and Smc4, Siz1 and Siz2 are more important for the sumoylation of Ycs4. Moreover, both Siz1/Siz2 and Mms21 control sumoylation of Brn1. This observation argues against promiscuous sumoylation catalyzed by any single SUMO ligase in vivo. Instead, it raises the possibility that distinct sumoylation of different subunits of the same protein complex may lead to different biological outcomes.
Third, our MS data indicate that a balance of activities from Siz1, Siz2 and Mms21 controls the abundances of SUMO targets in cells. For example, siz2Δ causes increased sumoylation of many SUMO targets (Figure 2), which may be due to elevated activities of Siz1 and/or Mms21. Similarly; siz1Δ siz2Δ causes increased sumoylation of several SMC-family of proteins, consistent with up regulation of Mms21 activity. It should also be noted here that despite the importance of Mms21 for sumoylating the SMC-family proteins, Siz1 and Siz2 still contribute to their sumoylation to a lesser extent (Figure 3). Since MS only detects the levels of sumoylated proteins between WT and SUMO ligase mutants, we cannot exclude the possibility that the observed change of sumoylation could be due to its protein abundance change caused by mutations of SUMO ligases, which should be evaluated by alternative methods during further studies.
Esc2 is a regulator of Mms21 activity in vivo
Mms21 is part of the multi-subunit Smc5-Smc6 complex [17]. Little was known about how Mms21 achieves its substrate selectivity. Genetic analyses suggest that Esc2 may function with Mms21. Both MMS21 and ESC2 suppress duplication-mediated GCRs (Table 1), are epistatic with each other (Table 1), and prevent the formation of X-shaped DNA replication intermediates [7], [39], [40]. Esc2 contains SUMO-like domains and no known enzymatic activity [6], but physically interacts with components of the SUMO pathway, including Ubc9 and Smt3 itself [7]. Previous studies of Rad60, an ortholog of Esc2 in fission yeast, identified an interaction between Rad60 and the Smc5-Smc6 complex and showed that Rad60 functions in the same pathway as Nse2, the fission ortholog of Mms21 [59], [60]. Here we found that esc2Δ has a similar but not identical effect as mms21-11 on the sumoylation of most SUMO targets (Figure 4), indicating that Esc2 positively regulates Mms21 activity in vivo (Figure 5C), which is consistent with their epistatic relationship and the physical interactions between S. pombe Rad60 with Smc5-Smc6 and Ubc9 (Table 1) [59], [60]. Taken together, these data would be consistent with a role of Esc2 in promoting the formation of an active complex between Ubc9 and the Smc5-Smc6-Mms21 complex.
Slx5 and Siz2 suppress sumoylation of Mms21-specific substrates
The Slx5-Slx8 complex has been implicated in protein sumoylation pathway [29]. Studies of Slx5 in yeasts and its ortholog Rnf4 in mammalian cells have shown that its role to the SUMO pathway is related to the SUMO-interacting motifs (SIMs) of Slx5/Rnf4, which bind to sumoylated proteins to catalyze ubiquitination events [10], [53], [54], and these data have been used to suggest that Slx5-Slx8 regulates the abundance of sumoylated proteins in cells. In addition, poly-sumoylation was suggested to cause an slx8Δ defect in S. pombe [59]; paradoxically poly-sumoylation could also suppress slx5Δ phenotype in S. cerevisiae [61]. While we do not have information on the effect of slx5Δ on poly-sumoylation, we found that Slx5 has a specific and novel role in suppressing sumoylation of Mms21-specific substrates. Our quantitative MS results cannot distinguish between mechanisms in which Slx5-Slx8 antagonizes Mms21 by ubiquitination and degradation of its sumoylated targets, which is consistent with its known ubiquitin ligase and SUMO-binding activities, from mechanisms by which Slx5-Slx8 antagonizes by directly or indirectly preventing Mms21 from sumoylating its targets. Deletion of SIZ2 causes a modestly higher sumoylation of Mms21-specific targets, such as SMC-family proteins, and this increase in the accumulation of sumoylated targets is further elevated in the siz2Δ slx5Δ mutant strain (Figure 5B). As Siz2 mediates sumoylation, the effects of the siz2Δ mutation are likely indirect in which loss of Siz2 activity causes increased activity by Mms21 independently of the effects of the slx5Δ mutation. These findings thus reveal an intricate relationship between Slx5, Esc2 and various SUMO ligases towards sumoylation homeostasis in cells.
Protein sumoylation has an important role in suppressing duplication-mediated GCRs
Many genes suppress GCRs, and a number of pathways are specific to or have more important roles in suppressing GCRs formed by NAHR between repetitive sequences [4], [5], [12], [62]. Previously several genes involved in sumoylation, ESC2, SLX5, and SLX8 (Putnam et al. 2009), were shown to play important roles in suppression of duplication-mediated GCRs, whereas SIZ1 was shown to play little or no role [12], [13]. Here we have extensively examined interactions between genes involved in SUMO homeostasis, which in combination with identification of sumoylated targets specific to each E3 SUMO ligase provides some insight into roles of proteins sumoylation in maintenance of genome stability.
Mutations in ESC2 and MMS21 give rise to substantially greater defects in the duplication-mediated GCR assay than in the assay in which GCRs are formed by single copy sequences. Epistasis between SUMO E3 ligase domain mutations in MMS21 and ESC2 indicate that both genes acting the same pathway, although deletion of ESC2 appears to have a weaker phenotype in most assays than mms21-11 or mms21-CH. This epistasis is consistent with the fact that both genes suppress X-shaped DNA replication intermediates [7], [39], [40] and both promote sumoylation of the same targets (Figure 4), including the SMC-family of proteins and proteins in the nucleolus. In contrast, deletion of SIZ1 or SIZ2 causes much smaller effects, with the deletion of SIZ2 causing a greater defect in genome stability when combined with mutations of either MMS21 or SLX5. Together, these results suggest that an Mms21 - and Esc2 - specific sumoylation event plays a crucial role in suppressing duplication-mediated GCR formation.
Increases in the rate of duplication-mediated GCRs could arise through increased levels of DNA damage, increases in the overall levels of HR, and/or defects in suppressing NAHR relative to HR. Physical restraints between sister chromatids via cohesin or the damage-induced Smc5-Smc6 complex allows for appropriate repair. Consistent with this view, a single double-stranded break is sufficient to establish genome-wide sister chromatid cohesion even on undamaged chromosomes independent of DNA replication [63], [64]; cohesin mutants are sensitive to ionizing radiation [65]; and the DNA damage checkpoint also mediate the stability of the securin Pds1 to prevent sister chromatid separation [66]–[69]. Moreover, sumoylation of the cohesin subunit Mcd1 by Mms21 is induced by a double strand break and is required for establishing cohesion between sister chromatids both at the double-strand break and in the genome-wide response [28]. Consistent with the remarkable defects observed by mutation of MMS21 or ESC2 but not SIZ1 or SIZ2, we hypothesize that modification of Mms21-specific targets, most likely SMC-family proteins and/or associated proteins observed here, are required to maintain sister chromatid cohesion and prevent NAHR.
Remarkably, deletion of SLX5 and SLX8 also cause increased rates of duplication-mediated rearrangements (Table 1, Putnam et al., 2009), despite the fact that deletion of SLX5 causes an increase in the sumoylation of Mms21-targets. Moreover, the deletion of SIZ2, which also increases the sumoylation of Mms21-targets, causes a greater defect in suppressing duplication-mediated rearrangements alone and in combination with esc2Δ, slx5Δ, mms21-11, and mms21-CH than deletion of SIZ1, which does not affect Mms21-targets (Table 1). Together these data would suggest that careful maintenance of the sumoylation levels of Mms21 targets is required to maintain genomic stability or that deletion of SLX5, SLX8, and SIZ2 cause other defects leading to NAHR independent of how MMS21 suppresses genome stability.
Analysis of the synthetic interactions between mutations affecting viability, target sumoylation levels, or changes in genome stability suggests a mechanism whereby sumoylation by MMS21 functions in parallel to a pathway comprised of the partially redundant SIZ1 and SIZ2 genes (Figure 5C). ESC2 promotes the accumulation of sumoylated Mms21 targets, likely by recruiting Ubc9 or stabilizing the Mms21-Ubc9 complex, and the inability to generate the esc2Δ siz1Δ siz2Δ strain likely corresponds to a substantial loss of the sumoylated proteome. ESC2 and SLX5 act in opposing manners to regulate Mms21 activity towards a proper balance of protein sumoylation in cells, which is critical to prevent aberrant genome rearrangements driven by homeologous DNA sequences in the complex genomes of higher eukaryotes.
Materials and Methods
Yeast genetic methods
Standard yeast genetics technique was used to generate mutants listed in Table S13. Although no effect on GCRs was observed, 2-micron plasmid was eliminated in SUMO ligase-null mutants using a method described previously [56]. Method for GCR analysis was described previously [70].
Biochemical methods
To purify sumoylated proteins from HF-SUMO cells, equal amounts of cells from two difference cells, i.e., untagged versus HF-SUMO or wild-type versus SUMO ligase-null mutant, were first combined and lysed together. To identify sumoylated proteins, two replicate experiments were performed to compare untagged cells and HF-SUMO cells. The stable isotopes used were switched in these two replicate experiments. For all other comparisons between wild-type and SUMO ligase-null mutant, wild-type cells were always grown in the light Lys/Arg containing media, while the ligase-null mutant was grown in the heavy Lys/Arg containing media. The combined cells were broken using glass beads in a solution containing 0.2 M sodium hydroxide and 2% SDS. Following cell lysis, 0.2 M hydrochloric acid and 0.1 M Tris pH8.0 buffer was added to adjust the pH of sample to be 8.0. The sample was then heated to 100°C in the presence of 20 mM DTT to reduce proteins, cooled to room temperature and then 50 mM iodoacetamide was added to alkylate free cysteines. The purification of HF-SUMO was done using first Ni-NTA resins and then anti-FLAG resins essentially as described previously [31], [44]. To elute sumoylated proteins from anti-FLAG affinity column, 1 microgram of recombinant Ulp1 catalytic domain was used to cleave the isopeptide bond between sumoylated proteins and HF-SUMO [20]. The Ulp1-eluted sample was then subjected to trypsin digestion for MS analysis [43].
For Western blot analysis to evaluate sumoylation of 3×HA-tagged proteins, each tagged yeast strain was lysed using the same method as above. Cell lysate was then incubated with anti-HA resins, washed and probed by either anti-HA antibody for loading control or anti-FLAG antibody to detect the sumoylated form of the protein.
MS and data analysis methods
The proteomic method used was described previously [43]. Briefly, tryptic peptides were first fractionated using a HILIC column to generate 10 fractions. Each fraction was then analyzed by LC-MS/MS on an Orbitrap-LTQ MS system. MS data was searched using SEQUEST on a Sorcerer system and quantified using XPRESS as described previously [42]. For database searching, differential modifications of lysine and arginine were included. Each protein was quantified using at least three unique peptides, allowing us to calculate its abundance ratio based on the median and average of these ratios. The identification of SUMO targets further requires them being identified from two replicate experiments using a 10-fold ratio between mock and HF-SUMO as a cutoff (Tables S1 and S2). In cases where a peptide was not identified in the mock sample, the noise level (minimal ion intensity is set to be 1×103 if noise peaks were not detected) in the MS range of the expected peptide was used to calculate the abundance ratio. We then queried these SUMO targets against the results of comparisons between wild-type and each SUMO ligase-null mutant to obtain the abundance ratio for each SUMO target. For protein quantification, the relative abundances of multiple peptides of each SUMO target were used to calculate its abundance ratio based on the median of all ratios. The use of multiple peptides per protein allows an accurate measurement of its abundance change. Occasionally, some SUMO targets were not identified in all the MS experiments, which are indicated in Figure 2 and Supplementary Tables. The same methods were used to analyze the relative abundance of SUMO targets between wild-type and various mutants (see text for details).
Supporting Information
Zdroje
1. GordeninDA, ResnickMA (1998) Yeast ARMs (DNA at-risk motifs) can reveal sources of genome instability. Mutat Res 400 : 45–58.
2. DeiningerPL, BatzerMA (1999) Alu repeats and human disease. Mol Genet Metab 67 : 183–193 doi:10.1006/mgme.1999.2864
3. JiY, EichlerEE, SchwartzS, NichollsRD (2000) Structure of chromosomal duplicons and their role in mediating human genomic disorders. Genome Res 10 : 597–610.
4. KolodnerRD, PutnamCD, MyungK (2002) Maintenance of genome stability in Saccharomyces cerevisiae. Science 297 : 552–557 doi:10.1126/science.1075277
5. PutnamCD, HayesTK, KolodnerRD (2009) Specific pathways prevent duplication-mediated genome rearrangements. Nature 460 : 984–989 doi:10.1038/nature08217
6. OhyaT, AraiH, KubotaY, ShinagawaH, HishidaT (2008) A SUMO-like domain protein, Esc2, is required for genome integrity and sister chromatid cohesion in Saccharomyces cerevisiae. Genetics 180 : 41–50 doi:10.1534/genetics.107.086249
7. SollierJ, DriscollR, CastellucciF, FoianiM, JacksonSP, et al. (2009) The Saccharomyces cerevisiae Esc2 and Smc5-6 proteins promote sister chromatid junction-mediated intra-S repair. Mol Biol Cell 20 : 1671–1682 doi:10.1091/mbc.E08-08-0875
8. NovatchkovaM, BachmairA, EisenhaberB, EisenhaberF (2005) Proteins with two SUMO-like domains in chromatin-associated complexes: the RENi (Rad60-Esc2-NIP45) family. BMC Bioinformatics 6 : 22 doi:10.1186/1471-2105-6-22
9. XieY, KerscherO, KroetzMB, McConchieHF, SungP, et al. (2007) The yeast Hex3.Slx8 heterodimer is a ubiquitin ligase stimulated by substrate sumoylation. J Biol Chem 282 : 34176–34184 doi:10.1074/jbc.M706025200
10. PruddenJ, PebernardS, RaffaG, SlavinDA, PerryJJP, et al. (2007) SUMO-targeted ubiquitin ligases in genome stability. EMBO J 26 : 4089–4101 doi:10.1038/sj.emboj.7601838
11. JohnsonES, GuptaAA (2001) An E3-like factor that promotes SUMO conjugation to the yeast septins. Cell 106 : 735–744.
12. PutnamCD, HayesTK, KolodnerRD (2010) Post-replication repair suppresses duplication-mediated genome instability. PLoS Genet 6: e1000933 doi:10.1371/journal.pgen.1000933
13. KatsES, EnserinkJM, MartinezS, KolodnerRD (2009) The Saccharomyces cerevisiae Rad6 postreplication repair and Siz1/Srs2 homologous recombination-inhibiting pathways process DNA damage that arises in asf1 mutants. Mol Cell Biol 29 : 5226–5237 doi:10.1128/MCB.00894-09
14. HochstrasserM (2001) SP-RING for SUMO: new functions bloom for a ubiquitin-like protein. Cell 107 : 5–8.
15. JohnsonES, SchwienhorstI, DohmenRJ, BlobelG (1997) The ubiquitin-like protein Smt3p is activated for conjugation to other proteins by an Aos1p/Uba2p heterodimer. EMBO J 16 : 5509–5519 doi:10.1093/emboj/16.18.5509
16. JohnsonES, BlobelG (1997) Ubc9p is the conjugating enzyme for the ubiquitin-like protein Smt3p. J Biol Chem 272 : 26799–26802.
17. ZhaoX, BlobelG (2005) A SUMO ligase is part of a nuclear multiprotein complex that affects DNA repair and chromosomal organization. Proc Natl Acad Sci USA 102 : 4777–4782 doi:10.1073/pnas.0500537102
18. LiSJ, HochstrasserM (1999) A new protease required for cell-cycle progression in yeast. Nature 398 : 246–251 doi:10.1038/18457
19. LiSJ, HochstrasserM (2000) The yeast ULP2 (SMT4) gene encodes a novel protease specific for the ubiquitin-like Smt3 protein. Mol Cell Biol 20 : 2367–2377.
20. MossessovaE, LimaCD (2000) Ulp1-SUMO crystal structure and genetic analysis reveal conserved interactions and a regulatory element essential for cell growth in yeast. Mol Cell 5 : 865–876.
21. StrunnikovAV, HoganE, KoshlandD (1995) SMC2, a Saccharomyces cerevisiae gene essential for chromosome segregation and condensation, defines a subgroup within the SMC family. Genes Dev 9 : 587–599.
22. LiS-J, HochstrasserM (2003) The Ulp1 SUMO isopeptidase: distinct domains required for viability, nuclear envelope localization, and substrate specificity. J Cell Biol 160 : 1069–1081 doi:10.1083/jcb.200212052
23. ReindleA, BelichenkoI, BylebylGR, ChenXL, GandhiN, et al. (2006) Multiple domains in Siz SUMO ligases contribute to substrate selectivity. J Cell Sci 119 : 4749–4757 doi:10.1242/jcs.03243
24. MatunisMJ, PickartCM (2005) Beginning at the end with SUMO. Nat Struct Mol Biol 12 : 565–566 doi:10.1038/nsmb0705-565
25. BerginkS, JentschS (2009) Principles of ubiquitin and SUMO modifications in DNA repair. Nature 458 : 461–467 doi:10.1038/nature07963
26. CremonaCA, SarangiP, YangY, HangLE, RahmanS, et al. (2012) Extensive DNA damage-induced sumoylation contributes to replication and repair and acts in addition to the mec1 checkpoint. Mol Cell 45 : 422–432 doi:10.1016/j.molcel.2011.11.028
27. TakahashiY, Yong-GonzalezV, KikuchiY, StrunnikovA (2006) SIZ1/SIZ2 control of chromosome transmission fidelity is mediated by the sumoylation of topoisomerase II. Genetics 172 : 783–794 doi:10.1534/genetics.105.047167
28. McAleenanA, Cordón-PreciadoV, Clemente-BlancoA, LiuI-C, SenN, et al. (2012) SUMOylation of the α-Kleisin Subunit of Cohesin Is Required for DNA Damage-Induced Cohesion. Curr Biol 22 : 1564–1575 doi:10.1016/j.cub.2012.06.045
29. BurgessRC, RahmanS, LisbyM, RothsteinR, ZhaoX (2007) The Slx5-Slx8 complex affects sumoylation of DNA repair proteins and negatively regulates recombination. Mol Cell Biol 27 : 6153–6162 doi:10.1128/MCB.00787-07
30. MontpetitB, HazbunTR, FieldsS, HieterP (2006) Sumoylation of the budding yeast kinetochore protein Ndc10 is required for Ndc10 spindle localization and regulation of anaphase spindle elongation. J Cell Biol 174 : 653–663 doi:10.1083/jcb.200605019
31. ZhouW, RyanJJ, ZhouH (2004) Global analyses of sumoylated proteins in Saccharomyces cerevisiae. Induction of protein sumoylation by cellular stresses. J Biol Chem 279 : 32262–32268 doi:10.1074/jbc.M404173200
32. DenisonC, RudnerAD, GerberSA, BakalarskiCE, MoazedD, et al. (2005) A proteomic strategy for gaining insights into protein sumoylation in yeast. Mol Cell Proteomics 4 : 246–254 doi:10.1074/mcp.M400154-MCP200
33. PanseVG, HardelandU, WernerT, KusterB, HurtE (2004) A proteome-wide approach identifies sumoylated substrate proteins in yeast. J Biol Chem 279 : 41346–41351 doi:10.1074/jbc.M407950200
34. HannichJT, LewisA, KroetzMB, LiS-J, HeideH, et al. (2005) Defining the SUMO-modified proteome by multiple approaches in Saccharomyces cerevisiae. J Biol Chem 280 : 4102–4110 doi:10.1074/jbc.M413209200
35. WohlschlegelJA, JohnsonES, ReedSI, YatesJR (2004) Global analysis of protein sumoylation in Saccharomyces cerevisiae. J Biol Chem 279 : 45662–45668 doi:10.1074/jbc.M409203200
36. ChenXL, SilverHR, XiongL, BelichenkoI, AdegiteC, et al. (2007) Topoisomerase I-dependent viability loss in saccharomyces cerevisiae mutants defective in both SUMO conjugation and DNA repair. Genetics 177 : 17–30 doi:10.1534/genetics.107.074708
37. HwangJ-Y, SmithS, CeschiaA, Torres-RosellJ, AragónL, et al. (2008) Smc5-Smc6 complex suppresses gross chromosomal rearrangements mediated by break-induced replications. DNA Repair (Amst) 7 : 1426–1436 doi:10.1016/j.dnarep.2008.05.006
38. DuanX, SarangiP, LiuX, RangiGK, ZhaoX, et al. (2009) Structural and functional insights into the roles of the Mms21 subunit of the Smc5/6 complex. Mol Cell 35 : 657–668 doi:10.1016/j.molcel.2009.06.032
39. MankouriHW, NgoH-P, HicksonID (2009) Esc2 and Sgs1 act in functionally distinct branches of the homologous recombination repair pathway in Saccharomyces cerevisiae. Mol Biol Cell 20 : 1683–1694 doi:10.1091/mbc.E08-08-0877
40. ChoiK, SzakalB, ChenY-H, BranzeiD, ZhaoX (2010) The Smc5/6 complex and Esc2 influence multiple replication-associated recombination processes in Saccharomyces cerevisiae. Mol Biol Cell 21 : 2306–2314 doi:10.1091/mbc.E10-01-0050
41. OngS-E, BlagoevB, KratchmarovaI, KristensenDB, SteenH, et al. (2002) Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteomics 1 : 376–386.
42. ChenS-H, AlbuquerqueCP, LiangJ, SuhandynataRT, ZhouH (2010) A proteome-wide analysis of kinase-substrate network in the DNA damage response. J Biol Chem 285 : 12803–12812 doi:10.1074/jbc.M110.106989
43. AlbuquerqueCP, SmolkaMB, PayneSH, BafnaV, EngJ, et al. (2008) A multidimensional chromatography technology for in-depth phosphoproteome analysis. Mol Cell Proteomics 7 : 1389–1396 doi:10.1074/mcp.M700468-MCP200
44. JohnsonES, BlobelG (1999) Cell cycle-regulated attachment of the ubiquitin-related protein SUMO to the yeast septins. J Cell Biol 147 : 981–994.
45. HoegeC, PfanderB, MoldovanG-L, PyrowolakisG, JentschS (2002) RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature 419 : 135–141 doi:10.1038/nature00991
46. SilverHR, NissleyJA, ReedSH, HouY-M, JohnsonES (2011) A role for SUMO in nucleotide excision repair. DNA Repair (Amst) 10 : 1243–1251 doi:10.1016/j.dnarep.2011.09.013
47. TakahashiY, DulevS, LiuX, HillerNJ, ZhaoX, et al. (2008) Cooperation of sumoylated chromosomal proteins in rDNA maintenance. PLoS Genet 4: e1000215 doi:10.1371/journal.pgen.1000215
48. PsakhyeI, JentschS (2012) Protein Group Modification and Synergy in the SUMO Pathway as Exemplified in DNA Repair. Cell 151 : 807–820 doi:10.1016/j.cell.2012.10.021
49. SongJ, DurrinLK, WilkinsonTA, KrontirisTG, ChenY (2004) Identification of a SUMO-binding motif that recognizes SUMO-modified proteins. Proc Natl Acad Sci USA 101 : 14373–14378 doi:10.1073/pnas.0403498101
50. ArmstrongAA, MohideenF, LimaCD (2012) Recognition of SUMO-modified PCNA requires tandem receptor motifs in Srs2. Nature 483 : 59–63 doi:10.1038/nature10883
51. KorenA, SoiferI, BarkaiN (2010) MRC1-dependent scaling of the budding yeast DNA replication timing program. Genome Res 20 : 781–790 doi:10.1101/gr.102764.109
52. ParkerJL, BucceriA, DaviesAA, HeidrichK, WindeckerH, et al. (2008) SUMO modification of PCNA is controlled by DNA. EMBO J 27 : 2422–2431 doi:10.1038/emboj.2008.162
53. XieY, RubensteinEM, MattT, HochstrasserM (2010) SUMO-independent in vivo activity of a SUMO-targeted ubiquitin ligase toward a short-lived transcription factor. Genes Dev 24 : 893–903 doi:10.1101/gad.1906510
54. GalantyY, BelotserkovskayaR, CoatesJ, JacksonSP (2012) RNF4, a SUMO-targeted ubiquitin E3 ligase, promotes DNA double-strand break repair. Genes Dev 26 : 1179–1195 doi:10.1101/gad.188284.112
55. JohnsonES (2004) Protein modification by SUMO. Annu Rev Biochem 73 : 355–382 doi:10.1146/annurev.biochem.73.011303.074118
56. ChenXL, ReindleA, JohnsonES (2005) Misregulation of 2 microm circle copy number in a SUMO pathway mutant. Mol Cell Biol 25 : 4311–4320 doi:10.1128/MCB.25.10.4311-4320.2005
57. BranzeiD, SollierJ, LiberiG, ZhaoX, MaedaD, et al. (2006) Ubc9 - and mms21-mediated sumoylation counteracts recombinogenic events at damaged replication forks. Cell 127 : 509–522 doi:10.1016/j.cell.2006.08.050
58. ByrneKP, WolfeKH (2005) The Yeast Gene Order Browser: combining curated homology and syntenic context reveals gene fate in polyploid species. Genome Res 15 : 1456–1461 doi:10.1101/gr.3672305
59. PruddenJ, PerryJJP, NieM, VashishtAA, ArvaiAS, et al. (2011) DNA repair and global sumoylation are regulated by distinct Ubc9 noncovalent complexes. Mol Cell Biol 31 : 2299–2310 doi:10.1128/MCB.05188-11
60. BoddyMN, ShanahanP, McDonaldWH, Lopez-GironaA, NoguchiE, et al. (2003) Replication checkpoint kinase Cds1 regulates recombinational repair protein Rad60. Mol Cell Biol 23 : 5939–5946.
61. MullenJR, DasM, BrillSJ (2011) Genetic evidence that polysumoylation bypasses the need for a SUMO-targeted Ub ligase. Genetics 187 : 73–87 doi:10.1534/genetics.110.124347
62. ChanJE, KolodnerRD (2011) A genetic and structural study of genome rearrangements mediated by high copy repeat Ty1 elements. PLoS Genet 7: e1002089 doi:10.1371/journal.pgen.1002089
63. StrömL, KarlssonC, LindroosHB, WedahlS, KatouY, et al. (2007) Postreplicative formation of cohesion is required for repair and induced by a single DNA break. Science 317 : 242–245 doi:10.1126/science.1140649
64. UnalE, Heidinger-PauliJM, KoshlandD (2007) DNA double-strand breaks trigger genome-wide sister-chromatid cohesion through Eco1 (Ctf7). Science 317 : 245–248 doi:10.1126/science.1140637
65. SjögrenC, NasmythK (2001) Sister chromatid cohesion is required for postreplicative double-strand break repair in Saccharomyces cerevisiae. Curr Biol 11 : 991–995.
66. YamamotoA, GuacciV, KoshlandD (1996) Pds1p, an inhibitor of anaphase in budding yeast, plays a critical role in the APC and checkpoint pathway(s). J Cell Biol 133 : 99–110.
67. Cohen-FixO, KoshlandD (1997) The anaphase inhibitor of Saccharomyces cerevisiae Pds1p is a target of the DNA damage checkpoint pathway. Proc Natl Acad Sci USA 94 : 14361–14366.
68. GardnerR, PutnamCW, WeinertT (1999) RAD53, DUN1 and PDS1 define two parallel G2/M checkpoint pathways in budding yeast. EMBO J 18 : 3173–3185 doi:10.1093/emboj/18.11.3173
69. SanchezY, BachantJ, WangH, HuF, LiuD, et al. (1999) Control of the DNA damage checkpoint by chk1 and rad53 protein kinases through distinct mechanisms. Science 286 : 1166–1171.
70. PutnamCD, KolodnerRD (2010) Determination of gross chromosomal rearrangement rates. Cold Spring Harb Protoc 2010: pdb.prot5492.
Štítky
Genetika Reprodukční medicína
Článek Re-Ranking Sequencing Variants in the Post-GWAS Era for Accurate Causal Variant IdentificationČlánek Bypass of 8-oxodGČlánek Integrated Model of and Inherited Genetic Variants Yields Greater Power to Identify Risk GenesČlánek Comparative Genomic and Functional Analysis of 100 Strains and Their Comparison with Strain GGČlánek A Nuclear Calcium-Sensing Pathway Is Critical for Gene Regulation and Salt Stress Tolerance inČlánek Computational Identification of Diverse Mechanisms Underlying Transcription Factor-DNA OccupancyČlánek Reversible and Rapid Transfer-RNA Deactivation as a Mechanism of Translational Repression in StressČlánek Genome-Wide Association of Body Fat Distribution in African Ancestry Populations Suggests New Loci
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2013 Číslo 8
-
Všechny články tohoto čísla
- Genome-Wide DNA Methylation Analysis of Systemic Lupus Erythematosus Reveals Persistent Hypomethylation of Interferon Genes and Compositional Changes to CD4+ T-cell Populations
- Re-Ranking Sequencing Variants in the Post-GWAS Era for Accurate Causal Variant Identification
- Histone Variant HTZ1 Shows Extensive Epistasis with, but Does Not Increase Robustness to, New Mutations
- Past Visits Present: TCF/LEFs Partner with ATFs for β-Catenin–Independent Activity
- Functional Characterisation of Alpha-Galactosidase A Mutations as a Basis for a New Classification System in Fabry Disease
- A Flexible Approach for the Analysis of Rare Variants Allowing for a Mixture of Effects on Binary or Quantitative Traits
- Masculinization of Gene Expression Is Associated with Exaggeration of Male Sexual Dimorphism
- Genic Intolerance to Functional Variation and the Interpretation of Personal Genomes
- Endogenous Stress Caused by Faulty Oxidation Reactions Fosters Evolution of 2,4-Dinitrotoluene-Degrading Bacteria
- Transposon Domestication versus Mutualism in Ciliate Genome Rearrangements
- Comparative Anatomy of Chromosomal Domains with Imprinted and Non-Imprinted Allele-Specific DNA Methylation
- An Essential Function for the ATR-Activation-Domain (AAD) of TopBP1 in Mouse Development and Cellular Senescence
- Depletion of Retinoic Acid Receptors Initiates a Novel Positive Feedback Mechanism that Promotes Teratogenic Increases in Retinoic Acid
- Bypass of 8-oxodG
- Calpain-6 Deficiency Promotes Skeletal Muscle Development and Regeneration
- ATM Release at Resected Double-Strand Breaks Provides Heterochromatin Reconstitution to Facilitate Homologous Recombination
- Generation of Tandem Direct Duplications by Reversed-Ends Transposition of Maize Elements
- Loss of a Conserved tRNA Anticodon Modification Perturbs Cellular Signaling
- Integrated Model of and Inherited Genetic Variants Yields Greater Power to Identify Risk Genes
- High-Throughput Genetic and Gene Expression Analysis of the RNAPII-CTD Reveals Unexpected Connections to SRB10/CDK8
- Dynamic Rewiring of the Retinal Determination Network Switches Its Function from Selector to Differentiation
- β-Catenin-Independent Activation of TCF1/LEF1 in Human Hematopoietic Tumor Cells through Interaction with ATF2 Transcription Factors
- Genetic Mapping of Specific Interactions between Mosquitoes and Dengue Viruses
- A Highly Redundant Gene Network Controls Assembly of the Outer Spore Wall in
- Origin and Functional Diversification of an Amphibian Defense Peptide Arsenal
- Myc-Driven Overgrowth Requires Unfolded Protein Response-Mediated Induction of Autophagy and Antioxidant Responses in
- Integrative Modeling of eQTLs and Cis-Regulatory Elements Suggests Mechanisms Underlying Cell Type Specificity of eQTLs
- Species and Population Level Molecular Profiling Reveals Cryptic Recombination and Emergent Asymmetry in the Dimorphic Mating Locus of
- Ras-Induced Changes in H3K27me3 Occur after Those in Transcriptional Activity
- Characterization of the p53 Cistrome – DNA Binding Cooperativity Dissects p53's Tumor Suppressor Functions
- Global Analysis of Fission Yeast Mating Genes Reveals New Autophagy Factors
- Deficiency Suppresses Intestinal Tumorigenesis
- Introns Regulate Gene Expression in in a Pab2p Dependent Pathway
- Meiotic Recombination Initiation in and around Retrotransposable Elements in
- Comparative Oncogenomic Analysis of Copy Number Alterations in Human and Zebrafish Tumors Enables Cancer Driver Discovery
- Comparative Genomic and Functional Analysis of 100 Strains and Their Comparison with Strain GG
- A Model-Based Analysis of GC-Biased Gene Conversion in the Human and Chimpanzee Genomes
- Masculinization of the X Chromosome in the Pea Aphid
- The Architecture of a Prototypical Bacterial Signaling Circuit Enables a Single Point Mutation to Confer Novel Network Properties
- Distinct SUMO Ligases Cooperate with Esc2 and Slx5 to Suppress Duplication-Mediated Genome Rearrangements
- The Yeast Environmental Stress Response Regulates Mutagenesis Induced by Proteotoxic Stress
- Mediator Directs Co-transcriptional Heterochromatin Assembly by RNA Interference-Dependent and -Independent Pathways
- The Genome of and the Basis of Host-Microsporidian Interactions
- Regulation of Sister Chromosome Cohesion by the Replication Fork Tracking Protein SeqA
- Neuronal Reprograming of Protein Homeostasis by Calcium-Dependent Regulation of the Heat Shock Response
- A Nuclear Calcium-Sensing Pathway Is Critical for Gene Regulation and Salt Stress Tolerance in
- Cross-Species Array Comparative Genomic Hybridization Identifies Novel Oncogenic Events in Zebrafish and Human Embryonal Rhabdomyosarcoma
- : A Mouse Strain with an Ift140 Mutation That Results in a Skeletal Ciliopathy Modelling Jeune Syndrome
- The Relative Contribution of Proximal 5′ Flanking Sequence and Microsatellite Variation on Brain Vasopressin 1a Receptor () Gene Expression and Behavior
- Combining Quantitative Genetic Footprinting and Trait Enrichment Analysis to Identify Fitness Determinants of a Bacterial Pathogen
- The Innocence Project at Twenty: An Interview with Barry Scheck
- Computational Identification of Diverse Mechanisms Underlying Transcription Factor-DNA Occupancy
- GUESS-ing Polygenic Associations with Multiple Phenotypes Using a GPU-Based Evolutionary Stochastic Search Algorithm
- H2A.Z Acidic Patch Couples Chromatin Dynamics to Regulation of Gene Expression Programs during ESC Differentiation
- Identification of DSB-1, a Protein Required for Initiation of Meiotic Recombination in , Illuminates a Crossover Assurance Checkpoint
- Binding of TFIIIC to SINE Elements Controls the Relocation of Activity-Dependent Neuronal Genes to Transcription Factories
- Global Analysis of the Sporulation Pathway of
- Genetic Circuits that Govern Bisexual and Unisexual Reproduction in
- Deletion of microRNA-80 Activates Dietary Restriction to Extend Healthspan and Lifespan
- Fifty Years On: GWAS Confirms the Role of a Rare Variant in Lung Disease
- The Enhancer Landscape during Early Neocortical Development Reveals Patterns of Dense Regulation and Co-option
- Gene Expression Regulation by Upstream Open Reading Frames and Human Disease
- Sociogenomics of Cooperation and Conflict during Colony Founding in the Fire Ant
- The Intronic Long Noncoding RNA Recruits PRC2 to the Promoter, Reducing the Expression of and Increasing Cell Proliferation
- The , p.E318G Variant Increases the Risk of Alzheimer's Disease in -ε4 Carriers
- The Wilms Tumor Gene, , Is Critical for Mouse Spermatogenesis via Regulation of Sertoli Cell Polarity and Is Associated with Non-Obstructive Azoospermia in Humans
- Reversible and Rapid Transfer-RNA Deactivation as a Mechanism of Translational Repression in Stress
- QTL Analysis of High Thermotolerance with Superior and Downgraded Parental Yeast Strains Reveals New Minor QTLs and Converges on Novel Causative Alleles Involved in RNA Processing
- Genome Wide Association Identifies Novel Loci Involved in Fungal Communication
- Chromatin Sampling—An Emerging Perspective on Targeting Polycomb Repressor Proteins
- A Recessive Founder Mutation in Regulator of Telomere Elongation Helicase 1, , Underlies Severe Immunodeficiency and Features of Hoyeraal Hreidarsson Syndrome
- Genome-Wide Association of Body Fat Distribution in African Ancestry Populations Suggests New Loci
- Causal and Synthetic Associations of Variants in the Gene Cluster with Alpha1-antitrypsin Serum Levels
- Hard Selective Sweep and Ectopic Gene Conversion in a Gene Cluster Affording Environmental Adaptation
- Brittle Culm1, a COBRA-Like Protein, Functions in Cellulose Assembly through Binding Cellulose Microfibrils
- Chromosomal Copy Number Variation, Selection and Uneven Rates of Recombination Reveal Cryptic Genome Diversity Linked to Pathogenicity
- The Ribosomal Protein Rpl22 Controls Ribosome Composition by Directly Repressing Expression of Its Own Paralog, Rpl22l1
- Ras1 Acts through Duplicated Cdc42 and Rac Proteins to Regulate Morphogenesis and Pathogenesis in the Human Fungal Pathogen
- The DSB-2 Protein Reveals a Regulatory Network that Controls Competence for Meiotic DSB Formation and Promotes Crossover Assurance
- Recurrent Modification of a Conserved -Regulatory Element Underlies Fruit Fly Pigmentation Diversity
- Associations of Mitochondrial Haplogroups B4 and E with Biliary Atresia and Differential Susceptibility to Hydrophobic Bile Acid
- The Conditional Nature of Genetic Interactions: The Consequences of Wild-Type Backgrounds on Mutational Interactions in a Genome-Wide Modifier Screen
- A Critical Function of Mad2l2 in Primordial Germ Cell Development of Mice
- A Role for CF1A 3′ End Processing Complex in Promoter-Associated Transcription
- Vitellogenin Underwent Subfunctionalization to Acquire Caste and Behavioral Specific Expression in the Harvester Ant
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Chromosomal Copy Number Variation, Selection and Uneven Rates of Recombination Reveal Cryptic Genome Diversity Linked to Pathogenicity
- Genome-Wide DNA Methylation Analysis of Systemic Lupus Erythematosus Reveals Persistent Hypomethylation of Interferon Genes and Compositional Changes to CD4+ T-cell Populations
- Associations of Mitochondrial Haplogroups B4 and E with Biliary Atresia and Differential Susceptibility to Hydrophobic Bile Acid
- A Role for CF1A 3′ End Processing Complex in Promoter-Associated Transcription
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



