-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaLoss of a Conserved tRNA Anticodon Modification Perturbs Cellular Signaling
Transfer RNA (tRNA) modifications enhance the efficiency, specificity and fidelity of translation in all organisms. The anticodon modification mcm5s2U34 is required for normal growth and stress resistance in yeast; mutants lacking this modification have numerous phenotypes. Mutations in the homologous human genes are linked to neurological disease. The yeast phenotypes can be ameliorated by overexpression of specific tRNAs, suggesting that the modifications are necessary for efficient translation of specific codons. We determined the in vivo ribosome distributions at single codon resolution in yeast strains lacking mcm5s2U. We found accumulations at AAA, CAA, and GAA codons, suggesting that translation is slow when these codons are in the ribosomal A site, but these changes appeared too small to affect protein output. Instead, we observed activation of the GCN4-mediated stress response by a non-canonical pathway. Thus, loss of mcm5s2U causes global effects on gene expression due to perturbation of cellular signaling.
Published in the journal: . PLoS Genet 9(8): e32767. doi:10.1371/journal.pgen.1003675
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1003675Summary
Transfer RNA (tRNA) modifications enhance the efficiency, specificity and fidelity of translation in all organisms. The anticodon modification mcm5s2U34 is required for normal growth and stress resistance in yeast; mutants lacking this modification have numerous phenotypes. Mutations in the homologous human genes are linked to neurological disease. The yeast phenotypes can be ameliorated by overexpression of specific tRNAs, suggesting that the modifications are necessary for efficient translation of specific codons. We determined the in vivo ribosome distributions at single codon resolution in yeast strains lacking mcm5s2U. We found accumulations at AAA, CAA, and GAA codons, suggesting that translation is slow when these codons are in the ribosomal A site, but these changes appeared too small to affect protein output. Instead, we observed activation of the GCN4-mediated stress response by a non-canonical pathway. Thus, loss of mcm5s2U causes global effects on gene expression due to perturbation of cellular signaling.
Introduction
Transfer RNAs (tRNAs) from all domains of life contain numerous post-transcriptional modifications, many of which are highly conserved. These modifications enhance the efficiency, specificity and fidelity of translation [1]–[3]. In the budding yeast Saccharomyces cerevisiae, three tRNAs are modified by addition of 5-methoxycarbonylmethyl (mcm5) and 2-thio (s2) groups to uridine at the 5′ nucleotide of the tRNA anticodon (U34), resulting in an mcm5s2U nucleotide. The mcm5s2U modification (MSUM) and many of the responsible modifying enzymes are conserved across eukaryotes, having been identified in fungi [4], [5], plants [6], worms [7] and mammals [8]. Despite widespread conservation, and extensive biochemical characterization, the physiological role of MSUM is unknown.
Genes required for MSUM are unusual among tRNA modification genes in the number and severity of their mutant phenotypes. Most yeast strains lacking tRNA modifications are viable and show no growth impairment [2], [3], but S. cerevisiae and C. elegans double mutants lacking both mcm5 and s2 are not viable [7], [9]. In yeast, single mutants lacking either mcm5 or s2 have numerous phenotypes including temperature sensitivity, various chemical stress sensitivities, exocytosis defects, and transcriptional defects [10], [11]. In C. elegans, mutants of the Elongator complex (comprised of elp1 through elp6), which is required to produce the mcm5 modification, display neurological defects [7]. In humans, mutations in IBKAP, the elp1 homolog, cause familial dysautonomia (FD) [12], and mutations in elp4 are associated with Rolandic epilepsy [13].
The molecular connection between these cellular/organismal phenotypes and the lack of specific tRNA anticodon modifications is currently unknown. Loss of either mcm5 or s2 impairs reading of both Watson-Crick (VAA) and wobble (VAG) cognate codons by the modified tRNAs [14], [15], and chemical removal or modification of the s2 moiety leads to a reduction in the rate of tRNA charging in vitro [16], [17]. The MSUM phenotypes were originally attributed to a proposed role of the Elongator complex in transcriptional elongation [18] before its function in tRNA modification was discovered [4]. However, the phenotypes of yeast MSUM mutants, including the lethality in mutants lacking both mcm5 and s2, can be suppressed by overexpression of unmodified versions of two tRNAs that normally contain mcm5s2U – and [11]. These observations indicate that at least a subset of the yeast cellular phenotypes are tied to tRNA function. It has been argued that loss of MSUM leads to codon-specific translation defects leading to insufficient protein production, either from many genes, or from a few genes required to carry out particular cellular processes or stress responses, but this hypothesis has not been directly tested.
In this study, we examined codon level ribosome distributions genome-wide using ribosome footprint profiling (Ribo-seq). We found that loss of mcm5 or s2 leads to slow translation elongation specifically at codons that Watson-Crick pair with MSUM tRNAs, but the magnitude of these changes appeared insufficient to affect protein output. Surprisingly, all of the MSUM strains showed gene expression signatures consistent with activation of the Gcn4p-mediated stress response pathway. We demonstrate that disruption of this pathway suppresses the MSUM mutant phenotypes independently of tRNA concentration.
Results
Ribosome Footprint Profiling Reveals Features of Translation for Specific Codons
We set out to determine whether MSUM mutants display codon-specific translation defects. Translational activity genome-wide was determined using Ribo-seq, which consists of isolating and sequencing ribosome-protected mRNA fragments from RNase-treated whole-cell lysates [19]. This method reveals ribosome positions at single nucleotide resolution, and thus has the potential to identify translational defects affecting single codons [19], [20]. Wild type (WT) yeast, as well as strains lacking the s2 moiety (ncs2Δ, ncs6Δ, and uba4Δ), or mcm5 (elp3Δ) (Figure 1A), were profiled by Ribo-seq, as well as RNA-seq. To assess the impact of these modifications on translation, the ribosome dwell time at specific codons was determined as follows. The positions of the A, P and E site codons within ribosome footprints of various lengths (25–31 nt) were determined by examining the 5′ ends of footprints mapping to start codons, where initiating ribosomes are expected to contain start codons in their P sites (Figure 1B) [21]. Next, to determine the genome-wide average ribosome dwell time for a given codon (Figure 1C, left), all instances of that codon in the genome were aligned, and 5′ ends of reads mapping to the surrounding positions (Figure 1C) were summed (see Materials and Methods). The resulting metacodon plots show the relative number of ribosome footprints, and thus the relative amount of time the ribosome spends at each position, as the codon moves through the A, P and E sites. Codon identity is not expected to affect translation from the outer sites (±1, ±2), so the entire plot was normalized to the height of these peaks. The height of each peak is the bulk occupancy for that codon in that ribosomal site, similar to a previously described metric [20]. The metacodon distributions for ATG and stop codons indicated that the reads were properly assigned to the ribosomal sites (Figure 1C, right). We observed distinct and reproducible patterns of ribosome density for different codons in WT yeast (Figure 1C, S1A,B), consistent with the single-nucleotide resolution of this technique.
Fig. 1. Genetic ablation of mcm5 or s2 leads to ribosome accumulation at specific codons. 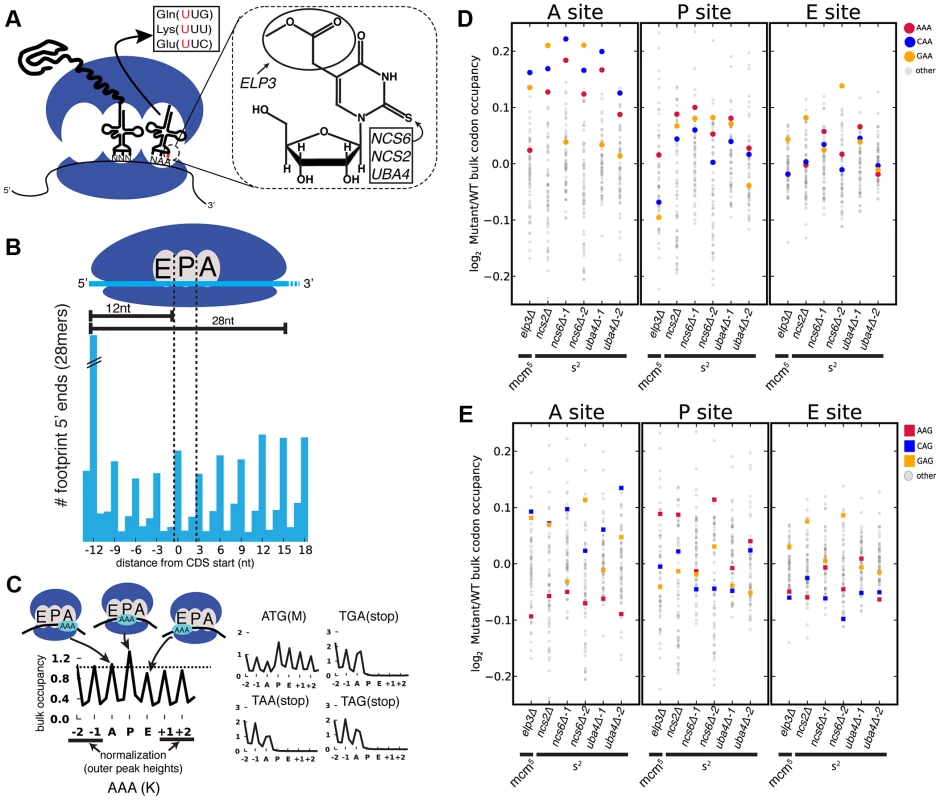
(A) (left) mcm5s2U is found at the 5′ nucleotide of the anticodon in three yeast tRNAs. (right) The structure of mcm5s2U, and the subset of modification genes whose mutants were profiled in this study are indicated. (B) (top) Anatomy of a ribosome footprint, with P-site offset for 28 mer reads indicated. (bottom) Metaplot of WT ribosome footprint reads summed across all start codons. The peak of upstream reads corresponds to ribosomes with start codons in their P site. The location of this peak is used to determine the location of A, P and E sites for each read length. (C) (left) Explanation of metacodon plots. Similar to panel B, all in-frame instances of a given codon in the genome are aligned, and the reads mapping around those positions are summed. The resulting plot is then offset by the P-site distance, and normalized to the average peak height of the outer sites (±1, ±2). The peak heights for each site are the bulk codon occupancies, a proxy for the amount of time the ribosome spends with a given codon in each site, compared to its neighbors. (right) ATG codons and stop codons display the expected distributions with this metric. All plots are from WT yeast. (D and E) Changes in bulk codon occupancy in MSUM mutants. Both plots are the same, with different codons highlighted. Independent biological replicates were done for ncs6Δ and uba4Δ. All mutants are compared to a WT sample prepared and processed simultaneously. The metacodon plots of WT yeast provided insights into the determinants of translation rate for specific codons. Notably, all four proline codons spent over 2-fold more time in the P site than the average codon, while glycine codons spent ∼40–50% more time in the A site (Figure S1C). This effect was additive for Pro-Gly pairs in the P and A sites, but not if the codon order was reversed (Figure S1D), indicating that the effects of Pro and Gly were specific to the P and A sites, respectively. This proline effect is reminiscent of the proline/glycine pausing recently discovered in bacteria lacking elongation factor P [22]–[24]. The observed effects were consistent with in vitro data which showed that peptidyl transfer can be rate limiting for A-site glycine and proline codon translation at physiological pH [25], and that proline induces particularly slow peptide bond formation when it is at the carboxyl terminus of the growing peptide chain [26] (Figure S1E). These results suggest that peptidyl transfer is rate limiting for certain Pro and Gly codons in yeast cells as well.
Experiments in recombinant systems have led to the strong expectation that translation times for codons should be inversely proportional to the concentrations of their cognate tRNAs [27], [28]. To investigate potential sources of the distinctive metacodon distributions we observed, we performed unsupervised hierarchical clustering on them (Figure S2A). This analysis clustered many codons together based on their encoded amino acid or the first two nucleotides of the codon. Notably, codons did not cluster by tRNA adaptation index (tAI), a proxy for cognate tRNA abundance [27]. More directly, the bulk occupancies did not show a negative correlation with tAI in the A site (Figure S2B). There was also no correlation of codon occupancy with tRNA abundance measurements, genomic copy number, or a more recent codon usage metric which accounts for tRNA competition [29] (data not shown). These results demonstrate that translation rates for particular yeast codons are not determined by the cellular concentrations of their cognate tRNAs, consistent with findings from Ribo-seq experiments in mice and bacteria [30], [31] and from protein synthesis reporters (containing codon repeats) in yeast [32].
Loss of MSUM Genes Reduces Translation Rate at AAA, CAA, GAA Codons
Having established the ability to detect differences in the translation of different codons, we next examined changes in codon-specific translation in the MSUM strains. Bulk occupancy for each codon in each ribosomal site (the height of the peaks in the metacodon plots) was determined for each mutant. All of the strains lacking the s2 modification showed increases in ribosome density corresponding to CAA and AAA in the A site, while the elp3Δ strain showed an increase in the CAA and GAA codons (Figure 1D). The magnitude of the changes was largest when the affected codon was found in the ribosomal A-site. The magnitude and direction of change for the GAA codon was variable between mutants lacking the same modification, and even between biological replicates (Figure 1D), indicative of some underlying biological or technical noise in this measurement. Nonetheless, in all but one replicate, the largest increases in each mutant were for codons decoded by Watson-Crick pairing with MSUM tRNAs.
mcm5s2U Is Not Required for Wobble Decoding of AAG, CAG, and GAG Codons In Vivo
MSUM is necessary for wobble decoding of G-ending codons in strains that lack other cognate tRNAs [14], but it is not clear whether the modified tRNAs contribute to decoding in the WT state where these other tRNAs are present. In our datasets AAG, CAG, and GAG codons showed smaller increases in bulk occupancy (and some net decreases) compared to their A-ending counterparts, suggesting that MSUM is mainly required for translation of VAA codons (Figure 1E). In order to assess the statistical significance of these changes, a metric for ribosome dwell time at individual codons was developed (Figure 2A). This metric normalizes the read counts at a particular codon by the mean read density of the open reading frame that contains it. The genome-wide distributions for all instances of each codon were compared between mutant and WT strains using the K-S test (Figure 2B, C). Due to the noise inherent in read sampling, many codons showed statistically significant changes. However, the VAA codons had p values many orders of magnitude smaller than all other codons, particularly in the ncs6Δ and uba4Δ datasets, which were from pooled biological replicates (Figure 2C). The pooled datasets provided data for approximately twice as many codons and may have averaged out biological and technical noise. Consistent with our analysis of bulk codon occupancy, the effect of MSUM loss was strongest in the A site for all 3 VAA codons. We did not see a corresponding statistical significance for the VAG codons (Figure 2C), indicating that mcm5s2U does not significantly contribute to the decoding of these codons in vivo. This result does not contradict previous evidence that the modifications are required for translation of VAG codons by wobble pairing [14], but indicates that tRNAsUUB contribute minimally to the translation of VAG codons in vivo, where tRNAsCUB with Watson-Crick complementarity are available.
Fig. 2. A single-codon occupancy metric shows that ribosome footprint accumulations at AAA, CAA, and GAA are statistically significant. 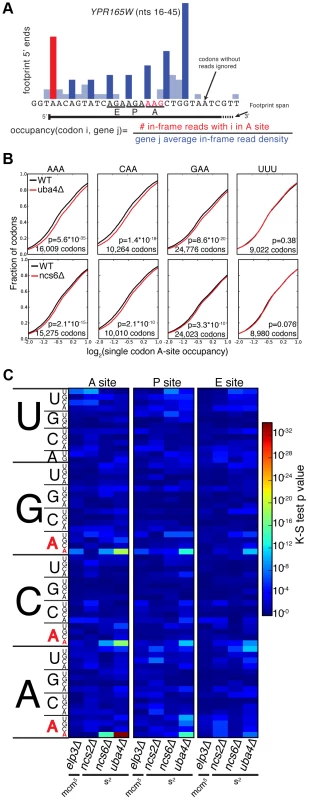
(A) Description of the single codon occupancy metric. The occupancy for a given codon in a given site is the number of in-frame reads for that codon in that site, compared to the average in-frame read density for the parent gene. (B) Cumulative distributions of single-codon occupancy for select codons in ncs6Δ and uba4Δ. (C) Heatmap of K-S test p-values for all sense codons in all mutants. For ncs6Δ and uba4Δ, mutant and WT replicates were pooled to improve the accuracy of the metric. The Elongation Defects in MSUM Strains Appear Insufficient to Affect Protein Levels
Despite the statistical significance of the increased ribosome dwell times at VAA codons in MSUM mutants, the magnitude of the changes does not seem to be large enough to generally affect protein output. Initiation, not elongation, is the rate-limiting step of eukaryotic translation in most circumstances [33], [34], and the mean ribosome density is only 1 per 164 nts [35]. Given this sparse spacing of ribosomes on yeast mRNAs, transcripts with mean ribosome density would require an elongation delay greater than the average translation time of 50 codons in order for an MSUM mutation to make elongation rate limiting. The most densely populated messages would require a 20-fold elongation delay. The average bulk increase observed for VAA codons was less than 17% (Figure 1D), and the largest confidently assigned (≥32 reads) single-codon change was less than 5-fold (Figure 3A, S3A). In the event of an elongation delay long enough to affect protein output, ribosome queuing should occur behind AAA and CAA codons with increased occupancy. However, no queuing was observed (Figure 3B, S3B). Codons with more read coverage display smaller changes than codons with low read coverage, indicating that the range of this metric is not being limited by sequencing depth (Figure 3A, S3A). We also did not observe increased ribosome density at stretches of 2 or more VAA codons (data not shown). These results were consistent with the polysome gradient profiles of the MSUM strains, which were indistinguishable from WT (data not shown), indicating that translation elongation in bulk was unaffected.
Fig. 3. Single codon occupancy changes may be insufficient to affect protein output. 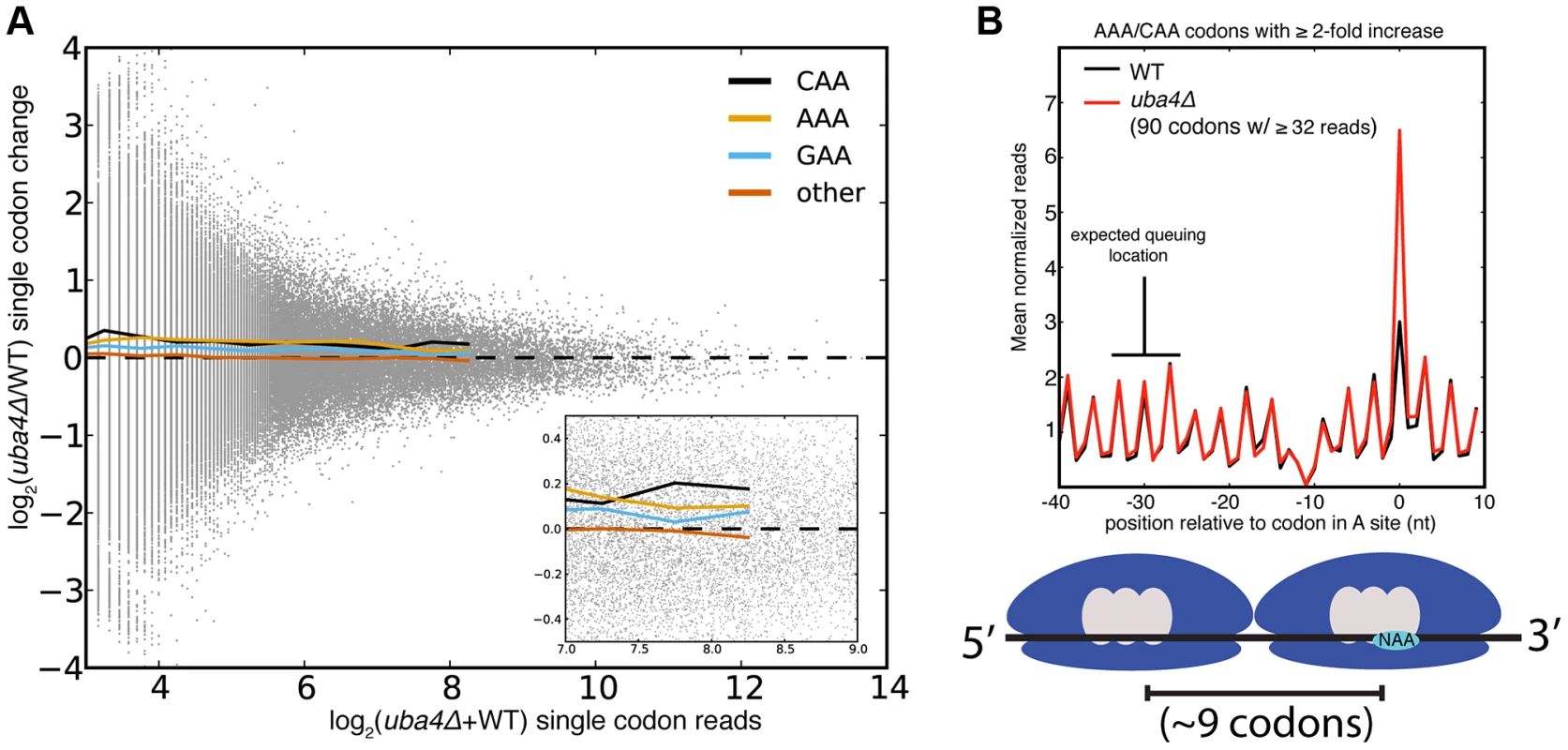
(A) Fold changes for all single codons in uba4Δ are plotted against their read density in grey. Colored lines are the mean fold changes for the specified codons over read-coverage bins of width 0.2 (log2 scaled). “Other” is a pool of all non-VAA codons. (B) Metaplot of ribosome footprint density around all AAA and CAA codons with ≥2-fold change in uba4Δ, and ≥32 reads in both datasets. Reads at each position were normalized by the total number of reads for the parent gene, and averaged across all host genes that overlap that position. The plot is offset such that 0 corresponds to having the codon in the A site. The expected location of a ribosome queuing event is indicated, and a diagram of such an event is shown below. The dip in ribosome footprint density at −10 is a computational artifact, due to an inability to determine read lengths of poly-adenylated fragments when they end in one or more adenosines. The GCN4-Mediated Stress Response Is Activated in MSUM Strains
In search of an alternative explanation for MSUM mutant phenotypes, we examined global ribosome footprint densities and transcript levels for perturbations in the MSUM mutant strains. Consistent with previous reports [19], [36], gene expression values from Ribo-seq were highly reproducible (Figure S4A). Furthermore, all of the mutant strains showed similar RNA-seq and Ribo-seq changes when compared to WT strains (Figure S4B,C), indicating that these gene expression changes are likely to be downstream of a common defect. Replicate data for ncs6Δ and uba4Δ enabled us to assess the significance of particular changes using counting statistics [37]. This analysis identified a set of genes with significant changes in ribosome footprint density, which were largely shared between ncs6Δ and uba4Δ (Figure 4A,4B,S4D). The changes in ribosome footprint density were correlated with changes in transcript levels (r = 0.59 for ncs6Δ, 0.64 for uba4Δ), indicating that these gene expression changes were largely due to changes in the mRNA pool (Figure 4A, 4D). Intriguingly, a significant fraction (24/68) of the affected genes are known targets of the GCN4 transcription factor [38] (Figure 4A,4B,S4D). To investigate the specificity of the observed induction of GCN4 targets in MSUM mutants, we examined the behavior of GCN4 targets in 1,924 yeast microarray studies using data from the SPELL curated yeast microarray compendium. This compendium includes experiments sampling a broad range of environmental and genetic perturbations [39]. We determined the significance of overlap between GCN4 targets and the set of upregulated (≥2-fold) genes in each of these 1,924 datasets. Notably, the overlap between GCN4 targets and induced genes in MSUM strains was more statistically significant than the overlap between GCN4 targets and induced genes in 82% of the SPELL datasets. The datasets with a higher degree of overlap consisted mostly (at least 276/343) of gene deletions and stress conditions in which GCN4 is known to play a role (e.g. heat, nutritional perturbation, osmotic stress and DNA damage) (Table S4, data not shown). Furthermore, GCN4 targets as a whole showed increased ribosome footprint density in all MSUM strains (Figure 4C, data not shown). We further confirmed this enrichment for functional GCN4 targets by examining the predicted Gcn4p binding affinity of the promoters for the affected genes [40]. The promoter regions of the upregulated genes were enriched for Gcn4p binding motifs (Figure 4D). Using the same sets of upregulated genes from the SPELL compendium as above, less than 6% of these upregulated gene sets had a mean predicted Gcn4p occupancy greater than the genes upregulated in the MSUM strains (Table S4). Thus, GCN4 target genes are transcriptionally upregulated in all MSUM strains.
Fig. 4. MSUM strains show the gene-expression signatures of GCN4 activation. 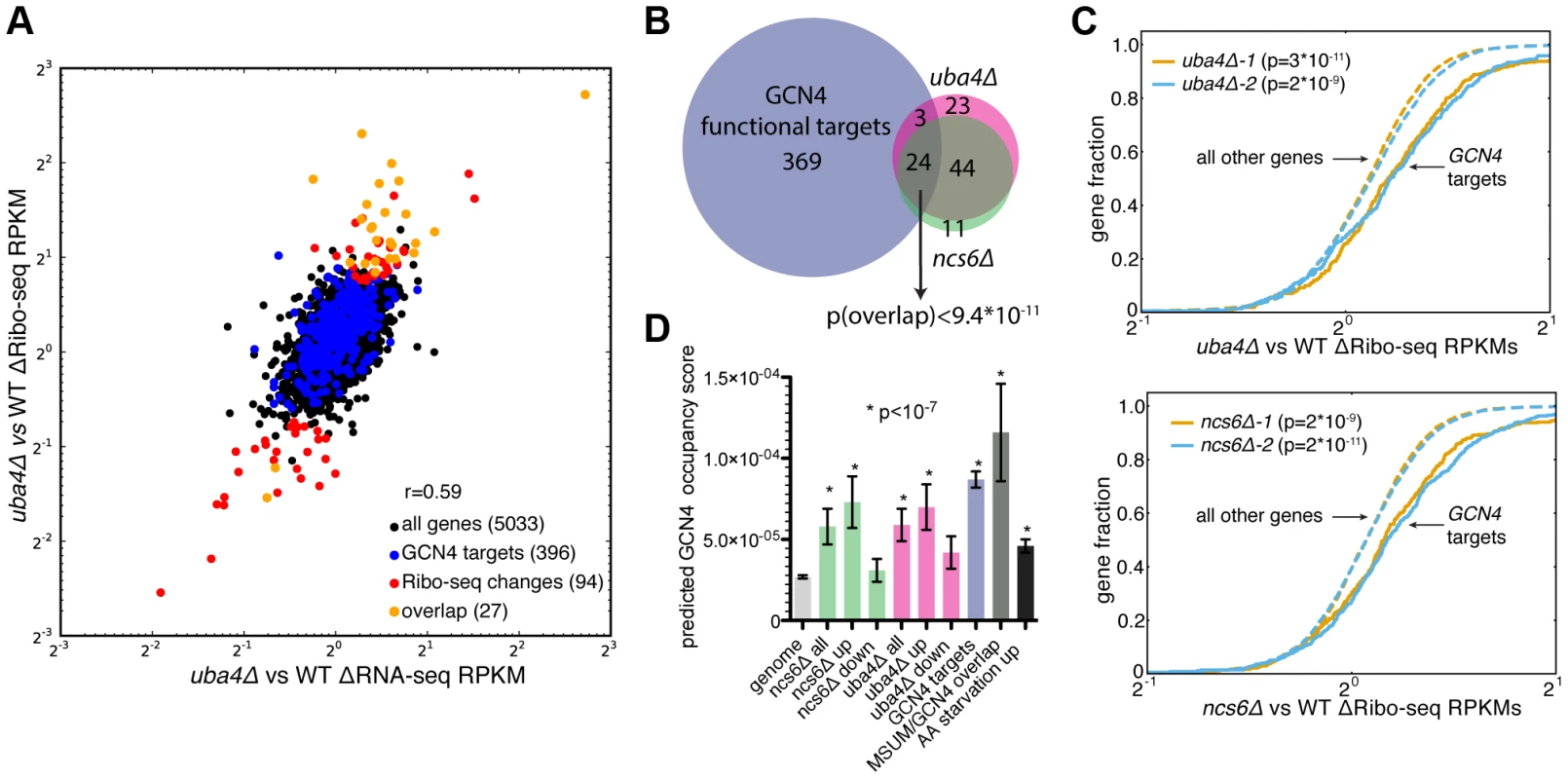
(A) Comparison of RNA-seq and Ribo-seq RPKM changes in uba4Δ. GCN4 targets and statistically significant Ribo-seq changes are indicated. Values are the means of 2 biological replicates. (B) Venn diagram of overlap between GCN4 functional targets (blue) and significant Ribo-seq RPKM changes in uba4Δ (pink) and ncs6Δ (green). The significance of the overlap was computed using the hypergeometric distribution. (C) Cumulative distribution plots of fold Ribo-seq changes for GCN4 targets (solid lines) compared to all other genes (dashed lines) in uba4Δ (top) and ncs6Δ. P values are from a KS test of GCN4 targets against the rest of the genome. (D) Mean±SEM of predicted Gcn4p occupancy for groups of genes from panel B and figure S5, as determined by high-throughput in vitro binding assays [40]. Bars are colored to match groups in panel B. P values are from t-tests comparing the indicated gene set against all genes in the genome. To provide context for these gene expression changes, the same analyses were performed on Ribo-seq data from yeast subjected to amino acid (AA) starvation, a well-characterized GCN4-inducing condition [19]. 20 minutes of amino acid starvation leads to a 4-fold increase in ribosome footprints on the GCN4 ORF (data not shown). A larger number of genes displayed changes in AA starvation compared to MSUM ablation, and GCN4 targets as a group had larger fold changes (median 2.0-fold induction vs. 1.2 and 1.1-fold for uba4Δ and ncs6Δ respectively). (Figure S5A, S5B). However, a smaller fraction of the significantly changing genes are GCN4 targets (13% in AA-starved cells, vs 29% and 30% for uba4Δ and ncs6Δ respectively) (Figure 5B, S5C). Furthermore, the starvation-induced genes had a smaller enrichment for predicted Gcn4p occupancy in their promoters compared to genes upregulated in the MSUM strains (Figure 5D). The limited induction of high-affinity Gcn4p targets in MSUM mutants is consistent with a weak but specific activation of the GCN4 pathway.
Fig. 5. GCN4 is induced independently of GCN2 in MSUM strains. 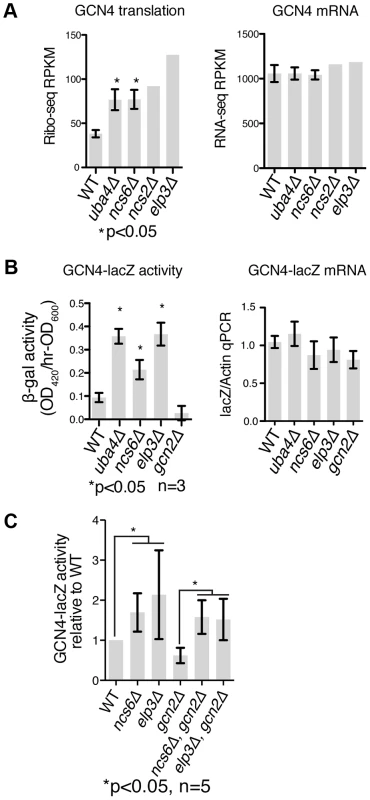
(A) Ribo-seq and RNA-seq RPKMs for the GCN4 open reading frame. Standard deviations are indicated for strains with replicate data. (B) The indicated strains were transformed with a reporter containing the promoter and transcript leader of GCN4 fused to lacZ. LacZ activity and mRNA levels were measured in log phase after overnight growth in YPD. (C) LacZ assays were performed as in panel B, with the addition of double mutant strains. P values are for t-test against WT unless otherwise indicated. Induction of GCN4 Occurs Independently of GCN2
We next sought to identify the mechanism of GCN4 pathway induction in MSUM strains. GCN4 is known to be translationally regulated in response to a variety of insults, most notably by amino acid starvation [41]. Translational repression of GCN4 is mediated by four upstream open reading frames (uORFs), which prevent ribosomes from initiating on the protein-coding ORF. Conditions that decrease the efficiency of re-initiation allow some ribosomes to scan through the uORFs and initiate at the GCN4 ORF. All four MSUM mutants showed ∼2-fold translational upregulation of GCN4, as evidenced by increased ribosome footprint density in the ORF with no increase in mRNA levels (Figure 5A).
A reporter construct containing the transcript leader of GCN4 fused to lacZ verified that the uORF-containing leader was sufficient to recapitulate the translational induction observed in MSUM strains (Figure 5B). The magnitude of this induction (2–4 fold) is consistent with a weak activation of the GCN pathway, as a 3 hr shift to SC-Ura, and a constitutive GCN2 allele [42] induced GCN4-lacz 7-fold and 50-fold, respectively (data not shown). The best-characterized pathway of inducing GCN4 involves the activation of the Gcn2p kinase by uncharged tRNA, leading to phosphorylation of eukaryotic initiation factor 2α (eIF2α) and reduced efficiency of initiation and re-initiation. We therefore tested the effect of gcn2Δ on GCN4 induction by MSUM mutants. Surprisingly, GCN4-lacZ was still induced in MSUM strains lacking GCN2 (Figure 5C). In addition, basal eIF2α phosphorylation levels were not increased in the MSUM strains, consistent with a GCN2-independent mechanism (data not shown). Thus, GCN4 translational induction in MSUM strains occurs by a non-canonical pathway.
In addition to the canonical GCN2-dependent response, some tRNA charging and modification defects have been shown to cause induction of GCN4 by a GCN2-independent mechanism [43]–[45]. MSUM mutations may affect charging. In vitro experiments have shown that loss of the s2 moiety of MSUM tRNAs reduces the efficiency of tRNA charging [16], [17], although steady state tRNA charging levels are unaltered in MSUM mutants [14]. We reasoned that a kinetic defect in tRNA charging could lead to compensatory increases in tRNA synthetase gene expression [46], which could suppress steady-state charging defects. We examined synthetase expression by unsupervised hierarchical clustering of mRNA abundance changes in all of the mutant strains. GlnRS, LysRS, GluRS and AspRS formed a cluster of increased expression in the MSUM mutants (Figure S6). Three of these synthetases (Gln, Lys, and Glu) have MSUM tRNAs as substrates. The specific upregulation of this set of tRNA synthetases, along with the global activation of GCN4 targets, suggests that MSUM mutants have adjusted their cellular state to cope with the loss of the mcm5s2U modification (see Discussion).
Disruption of the GCN Pathway Partially Suppresses Some MSUM Phenotypes
To investigate the functional significance of GCN4 misregulation in MSUM mutants, double mutants were constructed between gcn2Δ or gcn4Δ and ncs6Δ or elp3Δ, and tested for growth under conditions where MSUM mutants grow poorly. Under heat (40°C), caffeine and diamide stress, gcnΔ/MSUM double mutants showed some increase in growth compared to the single MSUM mutants (Figure 6A,S7A). On rapamycin, the suppression by gcn deletion was similar in magnitude to the suppression by high-copy (hc)-tRNA (Figure 6A). We did not observe any rescue of slow growth on YPD at 30°C with either GCN deletion or hc-tRNA expression (Figure 6A,S7B). Expressing hc-tRNA in the double mutant strains conferred additional resistance in all stress conditions, indicating that the GCN pathway contributes to the MSUM phenotypes independently of the pathway affected by hc-tRNA expression (Figure 6B).
Fig. 6. Disruption of the GCN pathway partially suppresses the stress sensitivity of MSUM strains, independently of tRNA overexpression. 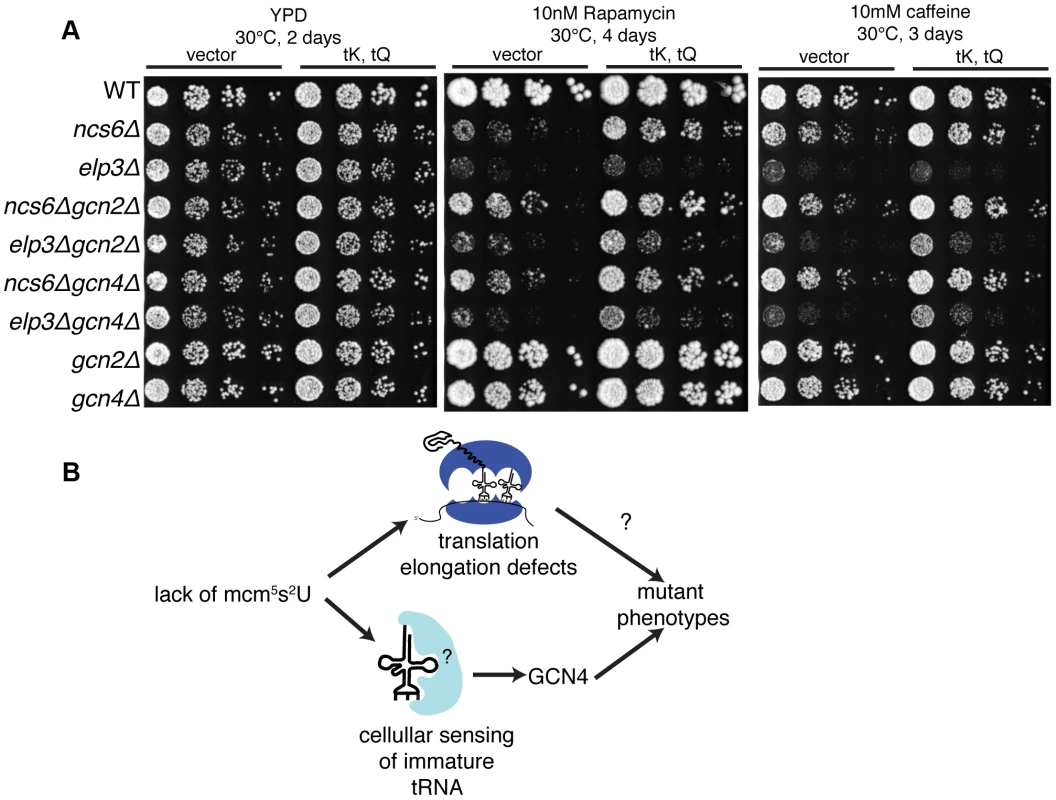
(A) Yeast was grown to saturation in selective media. 5-fold serial dilutions were spotted onto YPD containing the indicated drug, and grown at the indicated temperature. (B) The independent rescue of MSUM phenotypes by gcnΔ and hc-tRNA suggests that two independent pathways contribute to the mutant phenotypes. Discussion
MSUM tRNA modifications are conserved throughout eukarya and are required for organismal fitness in yeast, C. elegans, and humans. Due to the striking phenotypes of MSUM mutants, as well as the reported suppression by hc-tRNA [11], we expected to find large increases in ribosome density at codons decoded by MSUM tRNAs. We did detect increased ribosome density at VAA codons, and the largest effects of MSUM ablation occurred in the ribosomal A-site, the only site where tRNA binding, and thus concentration, is expected to play a role [21]. Thus, our analysis was capable of detecting codon-level translation defects in these mutants. However, the small magnitude of the observed effect makes it unlikely that protein output is generally affected. Additionally, suppression by hc-tRNA was incomplete in our hands, and the extent of both phenotypes and suppression varied between elp3Δ and ncs6Δ mutants when they were directly compared, as opposed to examined separately as in previous studies [11]. This suggests that MSUM genes may play additional roles in the cell, or create tRNA defects that are not suppressible by tRNA overexpression.
Overall, we found complex and varied patterns of ribosome density surrounding the different codons of the genetic code. These patterns appear to be determined not by cognate tRNA concentrations, but by intrinsic properties of aminoacyl tRNAs or peptidyl transfer kinetics, consistent with previous data showing that synonymous codon usage had little effect on protein output when mRNAs were expressed at physiological levels [28], [47]. This overall result is also consistent with the results of a systematic study of protein output from codon-repeat reporters [32]. Our data do not recapitulate all of the findings of that study, most likely because the reporters contained unnaturally long stretches of rare codons and were expressed at levels high enough to deplete the native tRNA pool. Furthermore, unlike reporter gene assays, Ribo-seq is able to detect changes in translation rate that are too small to be detected in an assay for protein output.
Since tRNA concentrations vary over an order of magnitude [27], yet had little effect on ribosome distributions at different codons, it is hard to understand how a ∼2–3 fold overexpression of hypomodified tRNA [48] could strongly affect the rate of ribosome movement. Our data do not rule out the possibility that one or more lowly expressed genes have elongation defects in MSUM mutants that are sufficient to reduce protein output. If so, there must be additional features that make codons in those genes unusually sensitive to the lack of the mcm5s2U modification. Indeed, loss of MSUM has been shown to cause a reduction in protein output in artificially sensitized conditions, such as the readthrough of stop codons by a suppressor tRNA [4], [49]. It is also possible that larger codon-specific translation defects were not manifest in our growth conditions, which would be consistent with the inability of hc-tRNA to rescue the slow growth of MSUM mutants on YPD. Our data also do not rule out the possibility that a slight increase in ribosome dwell time could lead to amino acid misincorporation [50], misfolding of the protein product [51], or degradation of the mRNA and/or protein by the mRNA surveillance machinery [52]. Further experiments are needed to understand the mechanism(s) of phenotypic suppression by hc-tRNAs.
The largest changes detected in the MSUM mutants were transcriptional effects consistent with activation of the GCN4 pathway. The gene expression signature of GCN4 induction was noticed previously in elpΔ mutants [10], and was attributed to the presumed role of Elongator in transcription. However, the similarity of the elp3Δ gene expression changes to those of ncs6Δ, ncs2Δ and uba4Δ, which have clear roles in an independent tRNA modification pathway [5], [53], [54], argues against this explanation. Instead, it appears that improperly modified tRNAs elicit a cellular stress response.
There is precedent for GCN2-independent activation of the GCN4 pathway by perturbations of tRNAs. Nuclear aminoacylation of tRNAs facilitates export to the cytoplasm in yeast and Xenopus oocytes [55], [56], and disruption of this process can lead to nuclear accumulation of tRNA, as well as GCN2-independent GCN4 induction [43], [44]. Loss of the s2 modification has been previously shown to reduce the rate of in vitro aminoacylation reactions for MSUM tRNAs [16], [17]. This charging defect could lead to nuclear accumulation of tRNA and the observed GCN2-independent induction of GCN4, despite the normal steady-state levels of charged tRNA in MSUM strains [14]. The apparent transcriptional upregulation of all three synthetases that recognize MSUM tRNAs may reflect a cellular response to such a defect in tRNA charging. Consistent with a role for the GCN pathway in mediating physiologically relevant signaling in response to loss of MSUM, deletion of GCN2 or GCN4 partially suppressed the phenotypes of MSUM strains.
The observation that GCN deletion suppresses MSUM phenotypes independently of the phenotypic suppression conferred by hc-tRNA suggests that there are at least two independent pathways contributing to the MSUM phenotypes. This may have implications for Elongator complex mutants in higher eukaryotes. In C. elegans, rescue of MSUM phenotypes by hc-tRNA has not been demonstrated. Furthermore, the translational effects reported in C. elegans MSUM strains [7] are more consistent with a global decrease in translation initiation, as might be expected in conditions leading to GCN4 activation, than with codon-specific elongation defects. Such secondary effects on gene expression may also play a role in the neurological symptoms of patients with mutations in elp genes. Indeed, induced pluripotent stem cells from FD patients with hypomorphic alleles of elp1 display numerous transcriptional changes during differentiation compared to controls [57]. It will be important to determine the extent to which tRNA-responsive signaling and transcriptional changes, in addition to codon-specific translation defects, contributes to the phenotypes of MSUM mutants in higher eukaryotes, and the severe and varied symptoms of FD patients.
Materials and Methods
Yeast Strains and Culture Conditions
All strains (Table S1) were in the s288c BY4742 background (MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0). MSUM and GCN deletions strains were constructed by PCR-mediated gene replacement as previously described [58]. All strains were grown in YPD (1% Yeast extract, 2% Peptone, 0.01% Adenine hemisulfate, 2% Dextrose) unless otherwise indicated. For growth assays with hc-tRNA plasmids, strains were grown in SC-Leu to maintain selection. Strains were then plated onto YPD.
Ribo-seq and RNA-seq
Yeast strains were grown from an OD600 of ∼0.001–0.004 in aerated flasks at 30°C to mid-log phase (OD ∼0.7), treated with 0.1 mg/ml cycloheximide for 2 minutes, and harvested by centrifugation. Cells were lysed by vortexing with glass beads, and libraries were prepared essentially as described [19], [36]. For the WT-2, elp3Δ, ncs2Δ, uba4Δ-2, ncs6Δ-2 libraries, triton was omitted until after lysis. For any analysis in which only 2 libraries are compared, the mutant was always compared to the WT sample processed identically. Sequencing data were deposited in the GEO database with the accession number GSE45366.
Read Mapping and Positional Assignment
Data analysis was performed using custom Python and Bash scripts developed in-house, unless otherwise indicated. Reads were mapped based on their 5′ 21 nt using Bowtie [59]. Reads were first mapped to S. cerevisiae rRNA, allowing up to 3 mismatches, and any mapping multiplicity. Any reads mapping to rRNA were discarded. Reads were then mapped to the S. cerevisiae genome downloaded from the saccharomyces genome database (SGD) on 5/26/2010, allowing up to 3 mismatches and requiring unique mapping. Read lengths were determined by comparing the original read sequence to the genomic sequence. Reads for which the beginning of the in vitro poly-A tail coincides with a genomic A have ambiguous length, and were excluded from length-specific analyses. Open reading frame (ORF) annotations downloaded from SGD were used to produce mappings of reads relative to the start codon for each ORF, which were used for all downstream calculations. For all codon-level analyses, reads of each length were processed separately, and 5′ end mapping locations were subsequently pooled, and shifted 5′ with the appropriate offsets (25 : 0, 26 : 0, 27 : 0, 28 : 0, 29:-1, 30:-1, 31:-2, negative numbers imply a 3′ shift) to put them in frame with 28 mer reads. When computing RPKMs (reads per kilobase of ORF sequence per million ORF reads) and read counts for each ORF, an unsplit pool of reads was used. The ORF positions are defined from 12 nt upstream of the start codon to 14 nt upstream of the stop codon. The first 8 codons of each ORF were excluded from all gene expression calculations to exclude possible artifacts from cycloheximide incubation.
Metacodon Plots and Bulk Occupancy Calculations
The value of position i in the metacodon vector for codon NNN is computed as follows:
Where the 21 nt offset is the 28 mer P-site offset (12 nt) plus the distance from the p-site to the first nt in the metacodon plot. The normalized metacodon vector is computed by normalizing to the peak heights of the outer sites: The mapping of metacodon peaks to ribosomal sites is: (0:-2, 3:-1, 6:A, 9:P, 12:E,15:+1,18:+2). For Figure S1D, the summation is performed over all codon positions for the given amino-acid pair, using the position of the first nucleotide of the first codon in the pair.Single Codon Occupancy Metric
The single codon occupancy for codon i in gene j in ribosomal site k is computed as:
For both the numerator and denominator, only in-frame reads (those whose 5′ ends fall a multiple of 3 from the first nt of the site) were counted, and the first 4 codons, as well as codons with no in-frame reads were excluded.Hierarchical Clustering
For Figure S2, the normalized metacodon vectors for each codon were used as inputs for cluster 3.0 [60]. Codons were clustered using spearman correlation and single linkage. Heatmaps were generated using Java Treeview [61]. The tAI column was not used for clustering, and was only added afterwards for comparison. For Figure S5, centroid linkage was used for clustering.
Queuing Analysis
For each AAA and CAA codon with ≥2-fold increase, the reads at each surrounding position were normalized by the mean read density for the entire ORF. These values were summed relative to all of the codons analyzed, offset so that the 0 position corresponds to the codon in the A site, and the value at each position was divided by the total number of codons whose host gene overlapped the given position. A secondary ribosome pileup is expected to occur approximately one ribosome footprint width (∼28 nt) upstream of the slow codon. Due to the use of polyadenylation in library preparation, any read ending in an adenosine cannot be assigned a length, and is not included in this analysis. Because of this, there is a depletion of read density at ∼−10 nts, corresponding to reads that end with 1 or more adenosines.
Gene Expression Analysis
Significant Ribo-seq changes were called using edgeR [37]. Significance was assessed using a Bonferroni-corrected p-value cutoff of 0.05. The significance of overlap with GCN4 targets was assessed using the hypergeometric test, and the definition of target genes derived from Natarajan et al [38]. The background for the hypergeometric test was defined as the set of genes with confident expression values for all datasets (5034 genes for MSUM datasets, 2780 for amino acid starvation).
β-galactosidase Assays
Starter cultures containing the GCN4-lacZ reporter plasmid (Table S2) were grown to saturation in SC-URA, then diluted into YPAD and grown in conditions identical to the Ribo-seq samples. At an OD600 of 0.7–0.8, 1 ml aliquots each were taken for qPCR and β-galactosidase assays, spun down, media aspirated, and frozen. Pellets were resuspended in Z buffer and permeabilized as previously described [62]. Cell suspensions were transferred in triplicate to a transparent 96-well plate, and 1/5 volume of 4 mg/ml ONPG was added. OD420 was measured every minute for 1 hour in a Bio-Tek synergy HT plate reader. β-galactosidase activity was defined as the slope of the linear portion of the OD420 vs. time graph, normalized by the OD600 of the culture at harvest.
Quantitative RNA Analysis
RNA was purified from yeast pellets as described [63]. Reverse transcription and quantitative PCR was performed using Avian Myeloblastosis Virus Reverse Trancriptase (AMV-RT; Promega) and real-time reagents (Invitrogen) according to manufacturer's instructions using a Roche Lightcycler 480. See Table S3 for gene-specific primer sequences.
Automated Liquid Growth Assays
Liquid growth assays were carried out as previously described [64], except that saturated selective media starter cultures were diluted to an OD of 0.01 in YPD, then diluted 20-fold in YPD to a final volume of 100 µl.
Supporting Information
Zdroje
1. AgrisPF, VendeixFAP, GrahamWD (2007) tRNA's wobble decoding of the genome: 40 years of modification. Journal of Molecular Biology 366 : 1–13 doi:10.1016/j.jmb.2006.11.046
2. PhizickyEM, HopperAK (2010) tRNA biology charges to the front. Genes & Development 24 : 1832–1860 doi:10.1101/gad.1956510
3. JohanssonM, ByströmA (2005) Transfer RNA modifications and modifying enzymes in Saccharomyces cerevisiae. Fine-tuning of RNA functions by modification and editing 12 : 87–120 doi:10.1007/b105814
4. HuangB, JohanssonMJO, ByströmAS (2005) An early step in wobble uridine tRNA modification requires the Elongator complex. RNA 11 : 424–436 doi:10.1261/rna.7247705
5. LeidelS, PedrioliPGA, BucherT, BrostR, CostanzoM, et al. (2009) Ubiquitin-related modifier Urm1 acts as a sulphur carrier in thiolation of eukaryotic transfer RNA. Nature 458 : 228–232 doi:10.1038/nature07643
6. MehlgartenC, JablonowskiD, WrackmeyerU, TschitschmannS, SondermannD, et al. (2010) Elongator function in tRNA wobble uridine modification is conserved between yeast and plants. Mol Microbiol 76 : 1082–1094 doi:10.1111/j.1365-2958.2010.07163.x
7. ChenC, TuckS, ByströmAS (2009) Defects in tRNA modification associated with neurological and developmental dysfunctions in Caenorhabditis elegans elongator mutants. PLoS Genet 5: e1000561 doi:10.1371/journal.pgen.1000561
8. ChanJC, YangJA, DunnMJ, AgrisPF, WongTW (1982) The nucleotide sequence of a glutamine tRNA from rat liver. Nucleic Acids Research 10 : 3755–3758.
9. BjörkGR, HuangB, PerssonOP, ByströmAS (2007) A conserved modified wobble nucleoside (mcm5s2U) in lysyl-tRNA is required for viability in yeast. RNA 13 : 1245–1255 doi:10.1261/rna.558707
10. KroganNJ, GreenblattJF (2001) Characterization of a Six-Subunit Holo-Elongator Complex Required for the Regulated Expression of a Group of Genes in Saccharomyces cerevisiae. Mol Cell Biol 21 : 8203–8212 doi:10.1128/MCB.21.23.8203-8212.2001
11. EsbergA, HuangB, JohanssonMJO, ByströmAS (2006) Elevated Levels of Two tRNA Species Bypass the Requirement for Elongator Complex in Transcription and Exocytosis. Molecular Cell 24 : 139–148 doi:10.1016/j.molcel.2006.07.031
12. SlaugenhauptSA, BlumenfeldA, GillSP, LeyneM, MullJ, et al. (2001) Tissue-Specific Expression of a Splicing Mutation in the IKBKAP Gene Causes Familial Dysautonomia. The American Journal of Human Genetics 68 : 598–605 doi:10.1086/318810
13. StrugLJ, ClarkeT, ChiangT, ChienM, BaskurtZ, et al. (2009) Centrotemporal sharp wave EEG trait in rolandic epilepsy maps to Elongator Protein Complex 4 (ELP4). European Journal of Human Genetics 17 : 1171–1181 doi:10.1038/ejhg.2008.267
14. JohanssonMJO, EsbergA, HuangB, BjörkGR, ByströmAS (2008) Eukaryotic wobble uridine modifications promote a functionally redundant decoding system. Mol Cell Biol 28 : 3301–3312 doi:10.1128/MCB.01542-07
15. KrügerMK, PedersenS, HagervallTG, SørensenMA (1998) The modification of the wobble base of tRNAGlu modulates the translation rate of glutamic acid codons in vivo. Journal of Molecular Biology 284 : 621–631 doi:10.1006/jmbi.1998.2196
16. SenGC, GhoshHP (1976) Role of modified nucleosides in tRNA: effect of modification of the 2-thiouridine derivative located at the 5′-end of the anticodon of yeast transfer RNA Lys2. Nucleic Acids Research 3 : 523–535.
17. SenoT, AgrisPF, SöllD (1974) Involvement of the anticodon region of Escherichia coli tRNAGln and tRNAGlu in the specific interaction with cognate aminoacyl-tRNA synthetase. Alteration of the 2-thiouridine derivatives located in the anticodon of the tRNAs by BrCN or sulfur deprivation. Biochim Biophys Acta 349 : 328–338.
18. OteroG, FellowsJ, LiY, de BizemontT, DiracAM, et al. (1999) Elongator, a multisubunit component of a novel RNA polymerase II holoenzyme for transcriptional elongation. Molecular Cell 3 : 109–118.
19. IngoliaNT, GhaemmaghamiS, NewmanJRS, WeissmanJS (2009) Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 324 : 218–223.
20. StadlerM, FireA (2011) Wobble base-pairing slows in vivo translation elongation in metazoans. RNA 17 : 2063–2073 doi:10.1261/rna.02890211
21. KappLD, LorschJR (2004) The molecular mechanics of eukaryotic translation. Annu Rev Biochem 73 : 657–704 doi:10.1146/annurev.biochem.73.030403.080419
22. DoerfelLK, WohlgemuthI, KotheC, PeskeF, UrlaubH, et al. (2013) EF-P is essential for rapid synthesis of proteins containing consecutive proline residues. Science 339 : 85–88 doi:10.1126/science.1229017
23. UdeS, LassakJ, StarostaAL, KraxenbergerT, WilsonDN, et al. (2013) Translation elongation factor EF-P alleviates ribosome stalling at polyproline stretches. Science 339 : 82–85 doi:10.1126/science.1228985
24. WoolstenhulmeCJ, ParajuliS, HealeyDW, ValverdeDP, PetersenEN, et al. (2013) Nascent peptides that block protein synthesis in bacteria. Proceedings of the National Academy of Sciences 110: E878–E887 doi:10.1073/pnas.1219536110
25. JohanssonM, IeongK-W, TrobroS, StrazewskiP, ÅqvistJ, et al. (2011) pH-sensitivity of the ribosomal peptidyl transfer reaction dependent on the identity of the A-site aminoacyl-tRNA. Proceedings of the National Academy of Sciences 108 : 79–84 doi:10.1073/pnas.1012612107
26. PavlovMY, WattsRE, TanZ, CornishVW, EhrenbergM, et al. (2009) Slow peptide bond formation by proline and other N-alkylamino acids in translation. Proceedings of the National Academy of Sciences 106 : 50–54 doi:10.1073/pnas.0809211106
27. TullerT, CarmiA, VestsigianK, NavonS, DorfanY, et al. (2010) An evolutionarily conserved mechanism for controlling the efficiency of protein translation. Cell 141 : 344–354.
28. PedersenS (1984) Escherichia coli ribosomes translate in vivo with variable rate. EMBO J 3 : 2895–2898.
29. PechmannS, FrydmanJ (2012) Evolutionary conservation of codon optimality reveals hidden signatures of cotranslational folding. Nature Structural & Molecular Biology 20 : 237–243 doi:10.1038/nsmb.2466
30. IngoliaNT, LareauLF, WeissmanJS (2011) Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell 147 : 789–802 doi:10.1016/j.cell.2011.10.002
31. LiG-W, OhE, WeissmanJS (2012) The anti-Shine-Dalgarno sequence drives translational pausing and codon choice in bacteria. Nature 484 : 538–541 doi:10.1038/nature10965
32. LetzringDP, DeanKM, GrayhackEJ (2010) Control of translation efficiency in yeast by codon-anticodon interactions. RNA 16 : 2516–2528 doi:10.1261/rna.2411710
33. LodishHF, JacobsenM (1972) Regulation of hemoglobin synthesis. Equal rates of translation and termination of - and -globin chains. J Biol Chem 247 : 3622–3629.
34. WaldenWE, Godefroy-ColburnT, ThachRE (1981) The role of mRNA competition in regulating translation. I. Demonstration of competition in vivo. J Biol Chem 256 : 11739–11746.
35. AravaY, WangY, StoreyJD, LiuCL, BrownPO, et al. (2003) Genome-wide analysis of mRNA translation profiles in Saccharomyces cerevisiae. Proc Natl Acad Sci USA 100 : 3889–3894 doi:10.1073/pnas.0635171100
36. BrarGA, YassourM, FriedmanN, RegevA, IngoliaNT, et al. (2012) High-Resolution View of the Yeast Meiotic Program Revealed by Ribosome Profiling. Science 335 : 552–557 doi:10.1126/science.1215110
37. RobinsonMD, McCarthyDJ, SmythGK (2009) edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26 : 139–140 doi:10.1093/bioinformatics/btp616
38. NatarajanK, MeyerMR, JacksonBM, SladeD, RobertsC, et al. (2001) Transcriptional profiling shows that Gcn4p is a master regulator of gene expression during amino acid starvation in yeast. Mol Cell Biol 21 : 4347–4368.
39. HibbsMA, HessDC, MyersCL, HuttenhowerC, LiK, et al. (2007) Exploring the functional landscape of gene expression: directed search of large microarray compendia. Bioinformatics 23 : 2692–2699 doi:10.1093/bioinformatics/btm403
40. NutiuR, FriedmanRC, LuoS, KhrebtukovaI, SilvaD, et al. (2011) Direct measurement of DNA affinity landscapes on a high-throughput sequencing instrument. Nat Biotechnol 29 : 659–664 doi:10.1038/nbt.1882
41. HinnebuschAG (2005) Translational regulation of GCN4 and the general amino acid control of yeast. Annu Rev Microbiol 59 : 407–450 doi:10.1146/annurev.micro.59.031805.133833
42. RamirezM, WekRC, Vazquez de AldanaCR, JacksonBM, FreemanB, et al. (1992) Mutations activating the yeast eIF-2 alpha kinase GCN2: isolation of alleles altering the domain related to histidyl-tRNA synthetases. Mol Cell Biol 12 : 5801–5815 doi:10.1128/MCB.12.12.5801
43. de AldanaCRV, WekRC, SegundoPS, TruesdellAG, HinnebuschAG (1994) Multicopy tRNA genes functionally suppress mutations in yeast eIF-2 alpha kinase GCN2: evidence for separate pathways coupling GCN4 expression to unchanged tRNA. Mol Cell Biol 14 : 7920–7932 doi:10.1128/MCB.14.12.7920
44. QiuH, HuC, AndersonJ, BjorkGR, SarkarS, et al. (2000) Defects in tRNA processing and nuclear export induce GCN4 translation independently of phosphorylation of the alpha subunit of eukaryotic translation initiation factor 2. Mol Cell Biol 20 : 2505–2516.
45. DaugeronMC, LenstraTL, FrizzarinM, Yacoubi ElB, LiuX, et al. (2011) Gcn4 misregulation reveals a direct role for the evolutionary conserved EKC/KEOPS in the t6A modification of tRNAs. Nucleic Acids Research 39 : 6148–6160 doi:10.1093/nar/gkr178
46. FrugierM, RyckelynckM, GiegéR (2005) tRNA-balanced expression of a eukaryal aminoacyl-tRNA synthetase by an mRNA-mediated pathway. EMBO Rep 6 : 860–865 doi:10.1038/sj.embor.7400481
47. KudlaG, MurrayAW, TollerveyD, PlotkinJB (2009) Coding-Sequence Determinants of Gene Expression in Escherichia coli. Science 324 : 255–258 doi:10.1126/science.1170160
48. BjorkGR, HuangB, PerssonOP, BystromAS (2007) A conserved modified wobble nucleoside (mcm5s2U) in lysyl-tRNA is required for viability in yeast. RNA 13 : 1245–1255 doi:10.1261/rna.558707
49. ChenC, HuangB, EliassonM, RydénP, ByströmAS (2011) Elongator Complex Influences Telomeric Gene Silencing and DNA Damage Response by Its Role in Wobble Uridine tRNA Modification. PLoS Genet 7: e1002258 doi:10.1371/journal.pgen.1002258.t001
50. PatilA, ChanCTY, DyavaiahM, RooneyJP, DedonPC, et al. (2012) Translational infidelity-induced protein stress results from a deficiency in Trm9-catalyzed tRNA modifications. rnabiology 9 : 990–1001 doi:10.4161/rna.20531
51. ZhangG, HubalewskaM, IgnatovaZ (2009) Transient ribosomal attenuation coordinates protein synthesis and co-translational folding. Nature Structural & Molecular Biology 16 : 274–280 doi:10.1038/nsmb.1554
52. ShoemakerCJ, GreenR (2012) Translation drives mRNA quality control. Nature Structural & Molecular Biology 19 : 594–601 doi:10.1038/nsmb.2301
53. SchliekerCD, Van der VeenAG, DamonJR, SpoonerE, PloeghHL (2008) A functional proteomics approach links the ubiquitin-related modifier Urm1 to a tRNA modification pathway. Proceedings of the National Academy of Sciences 105 : 18255–18260 doi:10.1073/pnas.0808756105
54. NomaA, SakaguchiY, SuzukiT (2009) Mechanistic characterization of the sulfur-relay system for eukaryotic 2-thiouridine biogenesis at tRNA wobble positions. Nucleic Acids Research 37 : 1335–1352 doi:10.1093/nar/gkn1023
55. GrosshansH, HurtE, SimosG (2000) An aminoacylation-dependent nuclear tRNA export pathway in yeast. Genes & Development 14 : 830–840.
56. LundE, DahlbergJE (1998) Proofreading and Aminoacylation of tRNAs Before Export from the Nucleus. Science 282 : 2082–2085 doi:10.1126/science.282.5396.2082
57. LeeG, PapapetrouEP, KimH, ChambersSM, TomishimaMJ, et al. (2009) Modelling pathogenesis and treatment of familial dysautonomia using patient-specific iPSCs. Nature 461 : 402–406 doi:10.1038/nature08320
58. LongtineMS, McKenzieA, DemariniDJ, ShahNG, WachA, et al. (1998) AID-YEA293>3.0.CO;2-U.
59. LangmeadB, TrapnellC, PopM, SalzbergSL (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biology 10: R25 doi:10.1186/gb-2009-10-3-r25
60. EisenMB, SpellmanPT, BrownPO, BotsteinD (1998) Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA 95 : 14863–14868.
61. SaldanhaAJ (2004) Java Treeview–extensible visualization of microarray data. Bioinformatics 20 : 3246–3248 doi:10.1093/bioinformatics/bth349
62. AmbergDC, BurkeDJ, StrathernJN (2006) Assay of β-Galactosidase in Yeast: Permeabilized Cell Assay. Cold Spring Harbor Protocols 2006 : 4158 doi:10.1101/pdb.prot4158
63. CollartMA, OlivieroS (2001) Preparation of yeast RNA. Curr Protoc Mol Biol Chapter 13: Unit13.12 doi:10.1002/0471142727.mb1312s23
64. ToussaintM, ConconiA (2006) High-throughput and sensitive assay to measure yeast cell growth: a bench protocol for testing genotoxic agents. Nat Protoc 1 : 1922–1928 doi:10.1038/nprot.2006.304
65. FrugierM, GiegéR (2003) Yeast Aspartyl-tRNA Synthetase Binds Specifically its Own mRNA. Journal of Molecular Biology 331 : 375–383 doi:10.1016/S0022-2836(03)00767-8
Štítky
Genetika Reprodukční medicína
Článek Re-Ranking Sequencing Variants in the Post-GWAS Era for Accurate Causal Variant IdentificationČlánek Bypass of 8-oxodGČlánek Integrated Model of and Inherited Genetic Variants Yields Greater Power to Identify Risk GenesČlánek Comparative Genomic and Functional Analysis of 100 Strains and Their Comparison with Strain GGČlánek A Nuclear Calcium-Sensing Pathway Is Critical for Gene Regulation and Salt Stress Tolerance inČlánek Computational Identification of Diverse Mechanisms Underlying Transcription Factor-DNA OccupancyČlánek Reversible and Rapid Transfer-RNA Deactivation as a Mechanism of Translational Repression in StressČlánek Genome-Wide Association of Body Fat Distribution in African Ancestry Populations Suggests New Loci
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2013 Číslo 8
-
Všechny články tohoto čísla
- Genome-Wide DNA Methylation Analysis of Systemic Lupus Erythematosus Reveals Persistent Hypomethylation of Interferon Genes and Compositional Changes to CD4+ T-cell Populations
- Re-Ranking Sequencing Variants in the Post-GWAS Era for Accurate Causal Variant Identification
- Histone Variant HTZ1 Shows Extensive Epistasis with, but Does Not Increase Robustness to, New Mutations
- Past Visits Present: TCF/LEFs Partner with ATFs for β-Catenin–Independent Activity
- Functional Characterisation of Alpha-Galactosidase A Mutations as a Basis for a New Classification System in Fabry Disease
- A Flexible Approach for the Analysis of Rare Variants Allowing for a Mixture of Effects on Binary or Quantitative Traits
- Masculinization of Gene Expression Is Associated with Exaggeration of Male Sexual Dimorphism
- Genic Intolerance to Functional Variation and the Interpretation of Personal Genomes
- Endogenous Stress Caused by Faulty Oxidation Reactions Fosters Evolution of 2,4-Dinitrotoluene-Degrading Bacteria
- Transposon Domestication versus Mutualism in Ciliate Genome Rearrangements
- Comparative Anatomy of Chromosomal Domains with Imprinted and Non-Imprinted Allele-Specific DNA Methylation
- An Essential Function for the ATR-Activation-Domain (AAD) of TopBP1 in Mouse Development and Cellular Senescence
- Depletion of Retinoic Acid Receptors Initiates a Novel Positive Feedback Mechanism that Promotes Teratogenic Increases in Retinoic Acid
- Bypass of 8-oxodG
- Calpain-6 Deficiency Promotes Skeletal Muscle Development and Regeneration
- ATM Release at Resected Double-Strand Breaks Provides Heterochromatin Reconstitution to Facilitate Homologous Recombination
- Generation of Tandem Direct Duplications by Reversed-Ends Transposition of Maize Elements
- Loss of a Conserved tRNA Anticodon Modification Perturbs Cellular Signaling
- Integrated Model of and Inherited Genetic Variants Yields Greater Power to Identify Risk Genes
- High-Throughput Genetic and Gene Expression Analysis of the RNAPII-CTD Reveals Unexpected Connections to SRB10/CDK8
- Dynamic Rewiring of the Retinal Determination Network Switches Its Function from Selector to Differentiation
- β-Catenin-Independent Activation of TCF1/LEF1 in Human Hematopoietic Tumor Cells through Interaction with ATF2 Transcription Factors
- Genetic Mapping of Specific Interactions between Mosquitoes and Dengue Viruses
- A Highly Redundant Gene Network Controls Assembly of the Outer Spore Wall in
- Origin and Functional Diversification of an Amphibian Defense Peptide Arsenal
- Myc-Driven Overgrowth Requires Unfolded Protein Response-Mediated Induction of Autophagy and Antioxidant Responses in
- Integrative Modeling of eQTLs and Cis-Regulatory Elements Suggests Mechanisms Underlying Cell Type Specificity of eQTLs
- Species and Population Level Molecular Profiling Reveals Cryptic Recombination and Emergent Asymmetry in the Dimorphic Mating Locus of
- Ras-Induced Changes in H3K27me3 Occur after Those in Transcriptional Activity
- Characterization of the p53 Cistrome – DNA Binding Cooperativity Dissects p53's Tumor Suppressor Functions
- Global Analysis of Fission Yeast Mating Genes Reveals New Autophagy Factors
- Deficiency Suppresses Intestinal Tumorigenesis
- Introns Regulate Gene Expression in in a Pab2p Dependent Pathway
- Meiotic Recombination Initiation in and around Retrotransposable Elements in
- Comparative Oncogenomic Analysis of Copy Number Alterations in Human and Zebrafish Tumors Enables Cancer Driver Discovery
- Comparative Genomic and Functional Analysis of 100 Strains and Their Comparison with Strain GG
- A Model-Based Analysis of GC-Biased Gene Conversion in the Human and Chimpanzee Genomes
- Masculinization of the X Chromosome in the Pea Aphid
- The Architecture of a Prototypical Bacterial Signaling Circuit Enables a Single Point Mutation to Confer Novel Network Properties
- Distinct SUMO Ligases Cooperate with Esc2 and Slx5 to Suppress Duplication-Mediated Genome Rearrangements
- The Yeast Environmental Stress Response Regulates Mutagenesis Induced by Proteotoxic Stress
- Mediator Directs Co-transcriptional Heterochromatin Assembly by RNA Interference-Dependent and -Independent Pathways
- The Genome of and the Basis of Host-Microsporidian Interactions
- Regulation of Sister Chromosome Cohesion by the Replication Fork Tracking Protein SeqA
- Neuronal Reprograming of Protein Homeostasis by Calcium-Dependent Regulation of the Heat Shock Response
- A Nuclear Calcium-Sensing Pathway Is Critical for Gene Regulation and Salt Stress Tolerance in
- Cross-Species Array Comparative Genomic Hybridization Identifies Novel Oncogenic Events in Zebrafish and Human Embryonal Rhabdomyosarcoma
- : A Mouse Strain with an Ift140 Mutation That Results in a Skeletal Ciliopathy Modelling Jeune Syndrome
- The Relative Contribution of Proximal 5′ Flanking Sequence and Microsatellite Variation on Brain Vasopressin 1a Receptor () Gene Expression and Behavior
- Combining Quantitative Genetic Footprinting and Trait Enrichment Analysis to Identify Fitness Determinants of a Bacterial Pathogen
- The Innocence Project at Twenty: An Interview with Barry Scheck
- Computational Identification of Diverse Mechanisms Underlying Transcription Factor-DNA Occupancy
- GUESS-ing Polygenic Associations with Multiple Phenotypes Using a GPU-Based Evolutionary Stochastic Search Algorithm
- H2A.Z Acidic Patch Couples Chromatin Dynamics to Regulation of Gene Expression Programs during ESC Differentiation
- Identification of DSB-1, a Protein Required for Initiation of Meiotic Recombination in , Illuminates a Crossover Assurance Checkpoint
- Binding of TFIIIC to SINE Elements Controls the Relocation of Activity-Dependent Neuronal Genes to Transcription Factories
- Global Analysis of the Sporulation Pathway of
- Genetic Circuits that Govern Bisexual and Unisexual Reproduction in
- Deletion of microRNA-80 Activates Dietary Restriction to Extend Healthspan and Lifespan
- Fifty Years On: GWAS Confirms the Role of a Rare Variant in Lung Disease
- The Enhancer Landscape during Early Neocortical Development Reveals Patterns of Dense Regulation and Co-option
- Gene Expression Regulation by Upstream Open Reading Frames and Human Disease
- Sociogenomics of Cooperation and Conflict during Colony Founding in the Fire Ant
- The Intronic Long Noncoding RNA Recruits PRC2 to the Promoter, Reducing the Expression of and Increasing Cell Proliferation
- The , p.E318G Variant Increases the Risk of Alzheimer's Disease in -ε4 Carriers
- The Wilms Tumor Gene, , Is Critical for Mouse Spermatogenesis via Regulation of Sertoli Cell Polarity and Is Associated with Non-Obstructive Azoospermia in Humans
- Reversible and Rapid Transfer-RNA Deactivation as a Mechanism of Translational Repression in Stress
- QTL Analysis of High Thermotolerance with Superior and Downgraded Parental Yeast Strains Reveals New Minor QTLs and Converges on Novel Causative Alleles Involved in RNA Processing
- Genome Wide Association Identifies Novel Loci Involved in Fungal Communication
- Chromatin Sampling—An Emerging Perspective on Targeting Polycomb Repressor Proteins
- A Recessive Founder Mutation in Regulator of Telomere Elongation Helicase 1, , Underlies Severe Immunodeficiency and Features of Hoyeraal Hreidarsson Syndrome
- Genome-Wide Association of Body Fat Distribution in African Ancestry Populations Suggests New Loci
- Causal and Synthetic Associations of Variants in the Gene Cluster with Alpha1-antitrypsin Serum Levels
- Hard Selective Sweep and Ectopic Gene Conversion in a Gene Cluster Affording Environmental Adaptation
- Brittle Culm1, a COBRA-Like Protein, Functions in Cellulose Assembly through Binding Cellulose Microfibrils
- Chromosomal Copy Number Variation, Selection and Uneven Rates of Recombination Reveal Cryptic Genome Diversity Linked to Pathogenicity
- The Ribosomal Protein Rpl22 Controls Ribosome Composition by Directly Repressing Expression of Its Own Paralog, Rpl22l1
- Ras1 Acts through Duplicated Cdc42 and Rac Proteins to Regulate Morphogenesis and Pathogenesis in the Human Fungal Pathogen
- The DSB-2 Protein Reveals a Regulatory Network that Controls Competence for Meiotic DSB Formation and Promotes Crossover Assurance
- Recurrent Modification of a Conserved -Regulatory Element Underlies Fruit Fly Pigmentation Diversity
- Associations of Mitochondrial Haplogroups B4 and E with Biliary Atresia and Differential Susceptibility to Hydrophobic Bile Acid
- The Conditional Nature of Genetic Interactions: The Consequences of Wild-Type Backgrounds on Mutational Interactions in a Genome-Wide Modifier Screen
- A Critical Function of Mad2l2 in Primordial Germ Cell Development of Mice
- A Role for CF1A 3′ End Processing Complex in Promoter-Associated Transcription
- Vitellogenin Underwent Subfunctionalization to Acquire Caste and Behavioral Specific Expression in the Harvester Ant
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Chromosomal Copy Number Variation, Selection and Uneven Rates of Recombination Reveal Cryptic Genome Diversity Linked to Pathogenicity
- Genome-Wide DNA Methylation Analysis of Systemic Lupus Erythematosus Reveals Persistent Hypomethylation of Interferon Genes and Compositional Changes to CD4+ T-cell Populations
- Associations of Mitochondrial Haplogroups B4 and E with Biliary Atresia and Differential Susceptibility to Hydrophobic Bile Acid
- A Role for CF1A 3′ End Processing Complex in Promoter-Associated Transcription
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání







