-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaDynamic Rewiring of the Retinal Determination Network Switches Its Function from Selector to Differentiation
Organ development is directed by selector gene networks. Eye development in the fruit fly Drosophila melanogaster is driven by the highly conserved selector gene network referred to as the “retinal determination gene network,” composed of approximately 20 factors, whose core comprises twin of eyeless (toy), eyeless (ey), sine oculis (so), dachshund (dac), and eyes absent (eya). These genes encode transcriptional regulators that are each necessary for normal eye development, and sufficient to direct ectopic eye development when misexpressed. While it is well documented that the downstream genes so, eya, and dac are necessary not only during early growth and determination stages but also during the differentiation phase of retinal development, it remains unknown how the retinal determination gene network terminates its functions in determination and begins to promote differentiation. Here, we identify a switch in the regulation of ey by the downstream retinal determination genes, which is essential for the transition from determination to differentiation. We found that central to the transition is a switch from positive regulation of ey transcription to negative regulation and that both types of regulation require so. Our results suggest a model in which the retinal determination gene network is rewired to end the growth and determination stage of eye development and trigger terminal differentiation. We conclude that changes in the regulatory relationships among members of the retinal determination gene network are a driving force for key transitions in retinal development.
Published in the journal: . PLoS Genet 9(8): e32767. doi:10.1371/journal.pgen.1003731
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1003731Summary
Organ development is directed by selector gene networks. Eye development in the fruit fly Drosophila melanogaster is driven by the highly conserved selector gene network referred to as the “retinal determination gene network,” composed of approximately 20 factors, whose core comprises twin of eyeless (toy), eyeless (ey), sine oculis (so), dachshund (dac), and eyes absent (eya). These genes encode transcriptional regulators that are each necessary for normal eye development, and sufficient to direct ectopic eye development when misexpressed. While it is well documented that the downstream genes so, eya, and dac are necessary not only during early growth and determination stages but also during the differentiation phase of retinal development, it remains unknown how the retinal determination gene network terminates its functions in determination and begins to promote differentiation. Here, we identify a switch in the regulation of ey by the downstream retinal determination genes, which is essential for the transition from determination to differentiation. We found that central to the transition is a switch from positive regulation of ey transcription to negative regulation and that both types of regulation require so. Our results suggest a model in which the retinal determination gene network is rewired to end the growth and determination stage of eye development and trigger terminal differentiation. We conclude that changes in the regulatory relationships among members of the retinal determination gene network are a driving force for key transitions in retinal development.
Introduction
During organogenesis, cells undergo progressive cell fate restriction coupled with a loss of pluripotency. This process is hallmarked by the stages of specification, proliferation, and differentiation [1]. The transitions between each of these states mark major changes in developmental competence and plasticity during tissue and organ development.
The adult fly eye develops from a larval structure called the eye imaginal disc [2], [3]. Following specification and growth during early larval development, the retinal field begins to differentiate during the third larval stage, or instar [4]. Drosophila eye differentiation occurs progressively, proceeding from the posterior to the anterior margins of the disc; its progress is marked by a morphologically and molecularly detectable event called the morphogenetic furrow [5]–[7]. Anterior to the morphogenetic furrow, cells are determined and proliferating, while posterior to it cells exit the cell cycle and differentiate. Within the morphogenetic furrow, cells transition from proliferation to differentiation. Thus, the developing Drosophila eye is an ideal system to study how cells regulate the transition from pluripotency to terminal differentiation.
Selector genes direct the development of many organs from their primordia [8]. The development of the eye imaginal disc into the adult eye is directed by a conserved network of transcriptional regulators called the retinal determination (RD) gene network. The core members of this network, twin of eyeless (toy), eyeless (ey), sine oculis (so), eyes absent (eya), and dachshund (dac), are each necessary for normal eye development and are sufficient to drive ectopic eye development in other imaginal discs [9]–[17]. During normal development, Toy activates ey expression in the first instar [17]. Initially, Ey is expressed throughout the disc and activates the expression of eya, so, and dac [18]–[21]. Once established, So maintains its own expression, as well as that of dac and ey [19], [22]. Such positive feedback mechanisms within the network are well characterized [17]–[19], [23]–[25]. The downstream RD network members Eya, So, and Dac are expressed and necessary in cells posterior to the morphogenetic furrow (Figure 1B–D) [9]–[13], [22], [26]. In contrast, at the morphogenetic furrow, ey expression is sharply down-regulated (Figure 1A), but how the positive feedback loops are terminated remains unknown [14], [18].
Fig. 1. Ey repression at the morphogenetic furrow is necessary for differentiation. 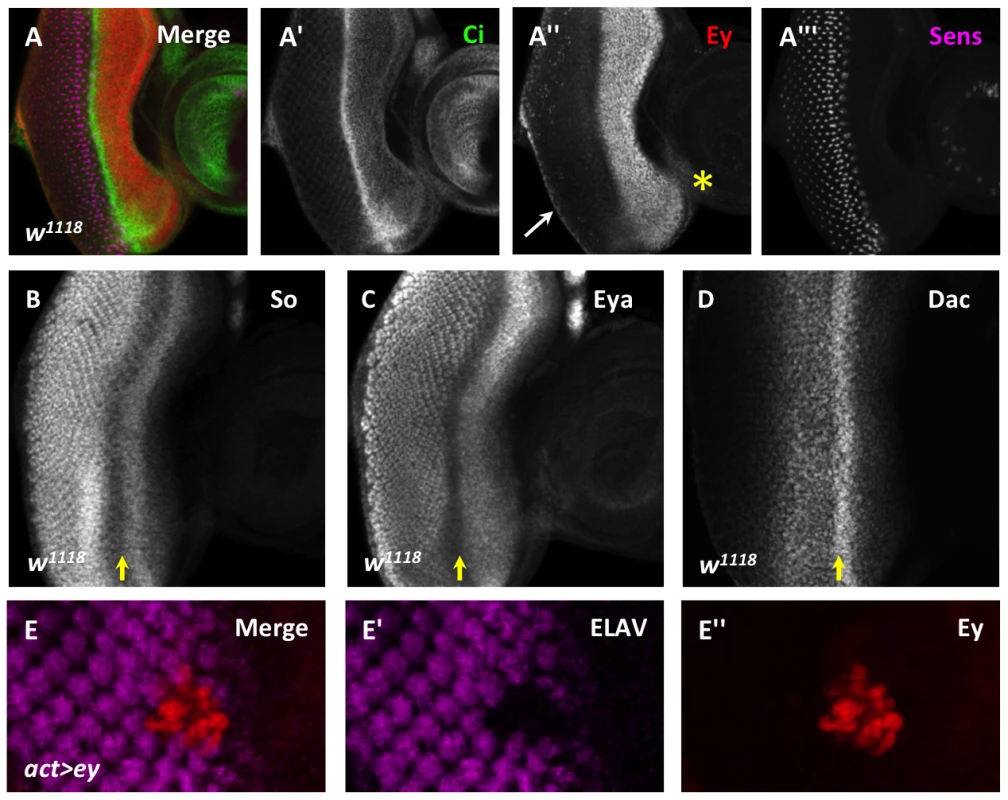
(A) Cubitus interruptus (Ci), Eyeless (Ey), and Senseless (Sens) expression in a w1118 third instar eye-antennal imaginal disc. (A′) Strong Ci accumulation marks the morphogenetic furrow. (A″) Ey expression; white arrow marks cuboidal margin cells, yellow asterisk marks ventral head capsule. (A′″) Sens expression shows R8 differentiation. (B–D) Yellow arrow marks the morphogenetic furrow: (B) Sine oculis (So) expression, (C) Eyes absent (Eya) expression, (D) Dachshund (Dac) expression. (E) Overexpression of Ey posterior to the morphogenetic furrow using Flipout-Gal4 inhibits photoreceptor differentiation. (E′) ELAV from panel E showing differentiation. (E″) Eyeless expression from panel E. In the region just anterior to the morphogenetic furrow where Dac, Eya, So, and Ey overlap, these proteins cooperate to initiate the expression of low levels of the proneural gene atonal (ato), which is required for the onset of photoreceptor differentiation [27]–[29]. However, without further amplification and refinement by Notch signaling in the morphogenetic furrow, the low level of Ato expression induced in this region of the eye is not sufficient to induce photoreceptor differentiation, and Ey expression persists [30]–[32]. Thus, while RD gene activity is required to initially activate one of the most upstream genes required for the onset of differentiation, this is not sufficient to fully trigger differentiation.
In this work, we show that maintaining expression of ey posterior to the morphogenetic furrow blocks photoreceptor differentiation. In addition, we identify a key regulatory switch in the RD gene network required for the repression of ey. Specifically, So directly regulates ey anterior to the furrow to promote high levels of expression, and via the same enhancer binding site blocks high levels of ey expression posterior to the furrow. Our results support a model that ey expression posterior to the furrow is regulated indirectly by eya and dac expression, and is triggered by signaling events in the morphogenetic furrow. These results suggest a model in which rewiring of the RD gene network is a key driving force during retinal organogenesis.
Results
Ey repression is necessary for the onset of differentiation
During the third instar, Eyeless (Ey) is strongly expressed anterior to the morphogenetic furrow. However, its expression sharply decreases at the morphogenetic furrow, and is detected only weakly in the differentiating eye field (Figure 1A). In contrast, the downstream RD gene network members are expressed not only in undifferentiated cells anterior to the morphogenetic furrow, but also in differentiating cells posterior to the morphogenetic furrow (Figure 1B–D). To determine if reducing Ey expression at the morphogenetic furrow is important for normal eye development, we overexpressed Ey posterior to the furrow using two methods. First, using the Flipout-Gal4 system we generated clones of cells that maintained Ey expression beyond the passage of the furrow [12], [33]. This caused cells to fail to differentiate, as assayed by expression of the pan-neuronal marker ELAV (Figure 1E). Second, we reactivated Ey expression in cells posterior to the furrow using the GMR-Gal4 and lz-Gal4 drivers [34], [35]. GMR-Gal4 eventually drives expression in all cells posterior to the furrow, while lz-Gal4 drives expression in cells that generate the future photoreceptors R1, 6, and 7 as well as in the cone and pigment cell precursors. ELAV expression is not affected in these genotypes, suggesting that Ey is not sufficient to block differentiation once differentiation has begun (Figure S1A–C). However, adult eyes of lz-Gal4; UAS-ey show defects in ommatidial shape and pigment when compared to wild-type (Figure S1D,E). Sections through lz-Gal4; UAS-ey eyes showed that photoreceptors survive, but that rhabdomere morphogenesis and ommatidial rotation are abnormal, suggesting that terminal differentiation events are disrupted by ectopic Ey expression (Figure S1F,G). From these results we conclude that down-regulation of Ey expression is necessary for normal photoreceptor differentiation.
So maintains Ey in determined cells and represses Ey in differentiating cells
To identify how the change in Ey expression is regulated, we undertook a candidate gene approach based on the literature. Previous studies of the RD gene network member Sine oculis (So) indicate that So activates ey expression during the third instar; however, so loss-of-function clones posterior to the morphogenetic furrow contained Ey expression, suggesting either that So is also required to suppress Ey expression or alternatively that these cells are trapped in an earlier developmental state [18], [22], [36]. This apparent paradox in the literature led us to examine Ey expression in so3 null clones in different positions of the eye disc during the third instar. In so3 clones anterior to the morphogenetic furrow, Ey expression was reduced, supportive of the model that So positively regulates ey expression anterior to the furrow (Figure 2A, arrow) [22]. Posterior to the morphogenetic furrow, we observed strong Ey expression in so3 clones (Figure 2A) [36]. We conclude that So promotes Ey expression anterior to the furrow and suppresses Ey expression posterior to the furrow.
Fig. 2. So regulates Ey expression anterior and posterior to the morphogenetic furrow. 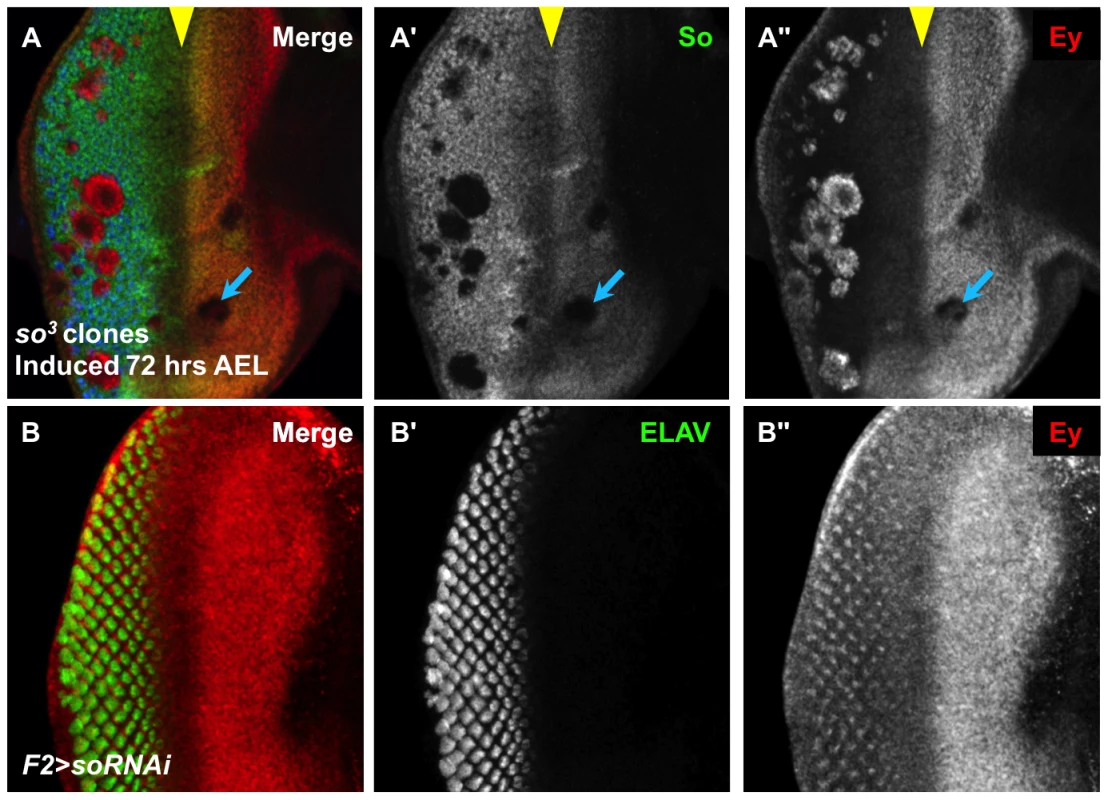
(A) so3 null clones, induced by hs-flp 72 hrs AEL showing So (green), Ey (red), and ELAV expression (blue); the yellow arrowhead marks the morphogenetic furrow, and the blue arrow indicates an anterior clone. (A′) Grayscale image of So expression, green in A; loss of So expression marks the clones. (A″) Grayscale image of Ey, red in A. (B) F2-Gal4 drives expression of soRNAi (VDRC transformant KK108128). (B′) Grayscale image of ELAV expression, green in B, marks differentiating photoreceptors. (B″) Grayscale image of Ey expression, red in B. Ey derepression posterior to the furrow matches previously described pattern of F2-Gal4 expression. We investigated the non-uniform appearance of Ey expression in posterior so3 clones, and observed that it is due to the morphology of the clones (Figure 2A). Specifically, orthogonal sections through clones displayed a spherical shape, with Ey expression being restricted to the so mutant tissue (Figure S2A,B). To determine if these cells lie in the interior of the clones that express low levels of Ey or no Ey, we co-labeled so3 clones for both Ey and Lamin, a marker of the nuclear membrane. We observed spaces within the clones that lack nuclei, and these spaces lack Ey (Figure S2C). Therefore, we conclude that Ey is robustly expressed cell autonomously in all so mutant cells posterior to the furrow. Our clonal analyses suggest that So cell autonomously promotes Ey expression anterior to the morphogenetic furrow, and suppresses Ey expression posterior to the morphogenetic furrow.
The presence of ey transcript or protein in so loss-of-function clones posterior to the morphogenetic furrow has been interpreted previously as a secondary consequence of failed furrow progression and/or differentiation [26], [36]. However, it may be that so expression is required posterior to the morphogenetic furrow to negatively regulate Ey. To distinguish between these models, we let Ey undergo normal regulation anterior to and within the morphogenetic furrow and then knocked down so expression specifically in differentiating cells posterior to the morphogenetic furrow. The F2-Gal4 driver, generated by our group with a characterized enhancer of the sens gene [37], initiates expression in the intermediate clusters within the furrow, posterior to Ey negative regulation, and is ultimately refined to drive expression most strongly in the R8 photoreceptor (Figure S2D,F). This driver permits analysis of the role of so in Ey regulation specifically in differentiating cells. Additionally, changes in expression are easily detectable because normal cells surround the knockdown cells. In F2-Gal4>UAS-so-RNAi discs, we observed Ey expression posterior to the morphogenetic furrow in an R8-like pattern (Figure 2B). Knockdown of So in F2-Gal4>UAS-so-RNAi discs is supported by So staining (Figure S2E) and results in a mildly disorganized adult eye (Figure S2G). Based on these results, we conclude that so is required to suppress Ey expression posterior to the morphogenetic furrow and that such suppression is required for normal eye development.
A single So binding site is required for ey maintenance and suppression
So is a homeodomain transcription factor, leading us to ask if So suppresses ey expression at the transcriptional level. To test this, we required a reporter that recapitulates ey regulation anterior and posterior to the morphogenetic furrow. Published ey enhancer reporters [22], [38], unlike Ey expression, persist posterior to the morphogenetic furrow, possibly due to perdurance of beta-galactosidase. We therefore constructed a new destabilized GFP (dGFP) reporter. To compare wild-type and mutant constructs while avoiding position effects, we utilized a vector that could integrate only at specific sites in our analysis [39]–[41]. We cloned a previously characterized full-length eye enhancer from the ey locus into this new dGFP vector, “ey-dGFP” [38], [39]. We detected robust expression with ey-dGFP throughout larval development (Figure 3A–C, Figure S3A–C). Similar to ey expression, ey-dGFP is expressed throughout the eye disc in first instar (not shown) and is maintained throughout the eye disc until furrow initiation (Figure 3A). During the third instar ey-dGFP is maintained anterior to the morphogenetic furrow and suppressed at the morphogenetic furrow, similar to Ey expression (Figure 3B). This expression pattern is maintained throughout the third instar (Figure 3C). Therefore, this enhancer recapitulates the Ey expression pattern in the eye field.
Fig. 3. So regulates ey expression through a binding site in an ey eye enhancer. 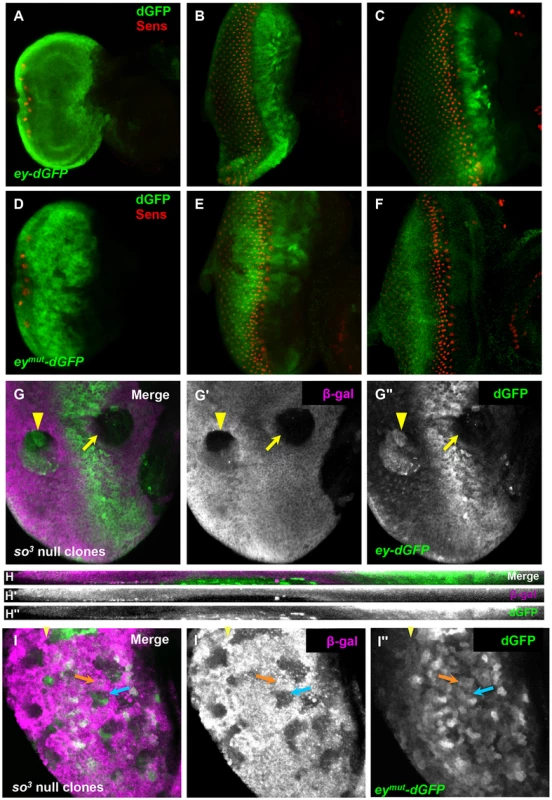
(A–F) reporter expression in third instar discs; columns of Sens positive cells were counted to compare furrow progression at different times. (A–C) expression of ey-dGFP (green) and Sens (red) in early (one column of photoreceptors) (A), mid (12 columns of photoreceptors) (B) and late (20 columns of photoreceptors) (C) third instar eye imaginal discs; individual channels shown in Figure S3. (D–F) expression of eymut-dGFP (green) and Sens (red) in third instar eye imaginal discs; individual channels shown in Figure S3. (D) one column of photoreceptors, (E) 11 columns of photoreceptors, (F) 18 columns of photoreceptors. (G) ey-dGFP expression in so3 null clone anterior (yellow arrow) and posterior (yellow arrowhead) to the morphogenetic furrow. (G′) Grayscale image of β-Galactosidase expression, magenta in G; loss of β-Galactosidase marks the clone (G″) Grayscale image of ey-dGFP expression, green in G (H–H″) Maximum projection of orthogonal sections through the posterior clone indicated by a yellow arrowhead in G–G″. (I) eymut-dGFP (green) expression in so3 null clone marked by loss of β-Galactosidase expression (magenta) in a disc aged between panels D and E. The yellow arrowhead marks the furrow; the orange arrow indicates non-clone tissue, blue arrow indicates anterior clone; similar expression detected in and out of clone (I′) Grayscale image of β-Galactosidase expression, magenta in I. (I″) Grayscale image of ey-dGFP expression, green in I. To determine if ey-dGFP can be regulated by So, we generated so3 clones and assayed reporter expression in clones anterior and posterior to the furrow. As with Ey, ey-dGFP reporter expression was reduced in anterior so3 clones, while it was induced in posterior clones (Figure 3G–H). Based on these results, we conclude that So regulates ey expression at the transcriptional level both anterior and posterior to the morphogenetic furrow.
To determine if So can regulate the expression of ey-dGFP directly, we mutated a previously well-characterized So binding site in the ey enhancer to generate eymut-dGFP [22]. From early development through initiation of the morphogenetic furrow eymut-dGFP is indistinguishable from ey-dGFP, consistent with published data that early ey expression is independent of So (Figure 3D, Figure S3D) [18]. However, during furrow progression, the expression pattern of eymut-dGFP is dynamic. The expression of eymut-dGFP anterior to the morphogenetic furrow is initially strong but weakens throughout the third instar, and eventually becomes barely detectable (Figure 3E,F, Figure S3E,F). This may indicate that additional positive regulators of ey are initially expressed in this domain, consistent with findings that Tsh promotes Ey expression in the same region [42], [43]. This is also consistent with our observation that Ey expression is diminished but not lost in the anterior so3 clones we observed (Figure 2A). By the time the furrow has progressed 7–8 columns, eymut-dGFP expression is detected posterior to the onset of Sens expression in the furrow. By 14 columns of photoreceptor recruitment, eymut-dGFP is expressed in most cells posterior to the morphogenetic furrow (Figure 3E, Figure S3E shows a disc at 11 columns). Posterior expression is detected weakly even in very late discs where anterior expression is lost (Figure 3F, Figure S3F shows a disc of 18 columns) suggesting that the So binding site is required posterior to the furrow to suppress activation of ey by another activator. We conclude that a So binding site is required to suppress expression of the ey enhancer reporter posterior to the furrow and to maintain reporter expression anterior to the furrow.
To determine if So can regulate eymut-dGFP expression, we examined eymut-dGFP expression in so3 clones. If mutation of the binding site is sufficient to make the reporter unresponsive to regulation by So, then we should not observe changes in the reporter expression pattern when we compare tissue within versus outside of clones. We chose to assay a time point early in furrow progression when the reporter is still expressed anterior to the furrow and is beginning to express posterior to it. We observed areas of identical reporter brightness both inside and outside of the clones, leading us to conclude that mutation of the binding site makes the reporter unresponsive to regulation by So (Figure 3I). Together with the fact that this binding site has been demonstrated to be bound by So in vitro [22], our analyses of ey-dGFP and eymut-dGFP lead us to conclude that So directly regulates the expression of Ey both anterior and posterior to the morphogenetic furrow through the same binding site.
The So cofactor Eya is necessary for Ey repression
We next wanted to investigate the mechanism by which So represses Ey posterior to the furrow. Sine oculis interacts with multiple cofactors that affect its function as a transcriptional regulator, including the transcriptional coactivator Eyes absent (Eya) and the TLE family corepressor Groucho (Gro) [25], [36], [44]–[46]. As both cofactors are expressed in the eye disc, we set out to determine which of them, if either, cooperates with So to regulate Ey. We performed loss-of-function analyses for each cofactor and assayed the effects on Ey expression in clones. Our primary candidate was Gro, which cooperates with So in the repression of targets in the eye [25], [46]. Surprisingly, null loss-of-function clones of gro had no effect on Ey expression anterior or posterior to the morphogenetic furrow (Figure S4A). We conclude that Gro is not necessary for the normal regulation of Ey expression during the third instar, and unlikely to cooperate with So in this process.
We next wanted to determine if Eya cooperates with So to regulate ey. Previous studies found that So and Eya physically interact to promote the activation of target genes [28], [36], [45], [47], [48]. Based on these studies, we predicted that eya would be necessary for the maintenance of Ey expression by So anterior to the morphogenetic furrow. To test this, we generated eya null clones and examined Ey expression. We observed, surprisingly, that Ey expression was normal in eya anterior clones (Figure 4A). As these clones were small and rare, we also used RNAi to knockdown eya expression using the Flipout-Gal4 technique. Even in large knock-down clones we observed that Ey expression was normal in clones anterior to the morphogenetic furrow (Figure 4B). These results indicate that Eya is not required to maintain Ey expression anterior to the furrow. Posterior to the furrow, both null and RNAi knockdown clones of eya expressed Ey strongly (Figure 4B,C). We also observed similar morphology changes in eya clones as in so clones posterior to the furrow (compare Figure 4C to Figure 2D). Based on these results, we conclude that eya expression is required for Ey suppression posterior to the furrow.
Fig. 4. Eya is necessary for ey repression posterior to the morphogenetic furrow. 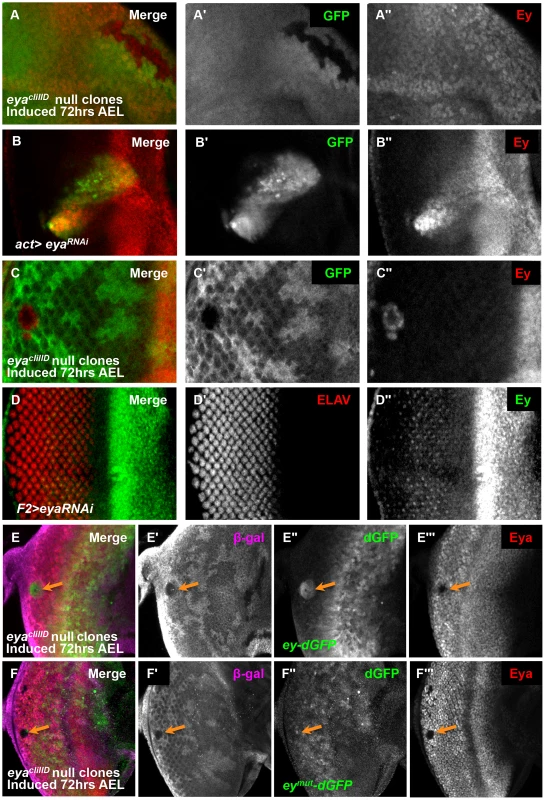
(A) eyacliIID null clones anterior to the morphogenetic furrow, induced by hs-flp 72 hrs AEL showing GFP and Ey expression (A′) Grayscale image of GFP expression, shown as green in A; loss of GFP expression marks the clones. (A″) Grayscale image of Ey channel alone, red in A. (B) Flipout-Gal4 drives expression of eyaRNAi (VDRC transformant KK108071). Merge of GFP and Ey expression shown. (B′) Grayscale image of GFP expression, shown as green in B; GFP expression marks the clones. (B″) Grayscale image of Ey channel alone, red in B. (C) eyacliIID null clones posterior to the morphogenetic furrow, induced by hs-flp 72 hours after egg lay (AEL) showing GFP and Ey expression (C′) Grayscale image of GFP expression, shown as green in C; loss of GFP expression marks the clones. (C″) Grayscale image of Ey channel alone, red in C. (D) F2-Gal4 drives expression of eyaRNAi (VDRC transformant KK108071). Merge of ELAV and Ey expression shown. (D′) Grayscale image of ELAV expression, shown as green in D, marks differentiating photoreceptors. (D″) Grayscale image of Ey expression, shown as red in D. (E) ey-dGFP (green) expression in eyacliIID null clone posterior to the morphogenetic furrow (indicated by orange arrow) marked by loss of β-Galactosidase expression (magenta) and Eya (red). (E′) Grayscale image of β-Galactosidase expression in E. (E″) Grayscale image of ey-dGFP expression in E as revealed by anti-GFP. (E′″) Grayscale image of Eya expression in E. (F) eymut-dGFP (green) expression in eyacliIID null clone posterior to the morphogenetic furrow (indicated by orange arrow) marked by loss of β-Galactosidase expression (magenta) and Eya (red). (F′) Grayscale image of β-Galactosidase expression in F. (F″) Grayscale image of ey-dGFP expression in F as revealed by anti-GFP. (F′″) Grayscale image of Eya expression in F. Eya is necessary for furrow progression and differentiation; therefore, failure of morphogenetic furrow progression through eya clones could result in the maintenance of Ey in these clones [26], [36], [49]–[51]. To test if Ey expression in posterior eya clones is an indirect effect of failed furrow progression, we used the F2-Gal4 driver to knock down eya expression specifically posterior to the furrow. We observed Ey expression in eya knockdown cells (Figure 4D). Staining for Eya indicates that the RNAi effectively knocks down eya expression (Figure S5A). Adults of F2-Gal4>eyaRNAi have disorganized eyes (Figure S5B). We conclude that Eya is required for Ey suppression posterior to the furrow.
To determine if eya is required for Ey suppression at the transcriptional level and dependent upon the So binding site, we examined ey-dGFP and eymut-dGFP expression in posterior eya clones. In clones posterior to the furrow, ey-dGFP was expressed, similar to so clone phenotypes, suggesting that eya is required for the negative regulation of ey at the transcriptional level (Figure 4E). In contrast to ey-dGFP, the expression of eymut-dGFP is not induced in posterior eya clones, suggesting that it no longer requires eya for its regulation (Figure 4F). From these results we conclude that Eya regulation of ey requires the So binding site.
High levels of eya and so are sufficient to repress endogenous Ey
Eya and So each overlap Ey expression just anterior to the morphogenetic furrow, but do not negatively regulate Ey expression in this zone. Therefore, we re-examined the expression of Eya and So in the eye imaginal disc to determine if their expression patterns could suggest how Eya and So could be required to suppress ey expression posterior to the furrow. Quantification of Eya and So expression in orthogonal sections revealed that expression of both factors is increased posterior to the morphogenetic furrow (Figure 5A,B). To test if the increased level is sufficient to repress Ey, we overexpressed both eya and so within the Ey domain using the Flipout-Gal4 strategy. Co-misexpression of eya and so was sufficient to repress Ey expression to background levels within the eye field, while ectopic Ey expression was detected in clones in other discs (Figure 5C, and data not shown). These data suggest that, within the developing retinal field, increased so and eya expression is sufficient to repress Ey expression anterior to the morphogenetic furrow. When we utilized the temperature sensitivity of the Gal4-UAS system to overexpress eya+so at 18°C, which results in lower expression of eya+so than at 25°C, they failed to repress Ey expression in the eye field, but were still sufficient to ectopically activate Ey expression in the antennal disc (Figure 5D, Figure S6A, white arrow).
Fig. 5. Eya and So can cooperate to negatively regulate Ey expression in vivo. 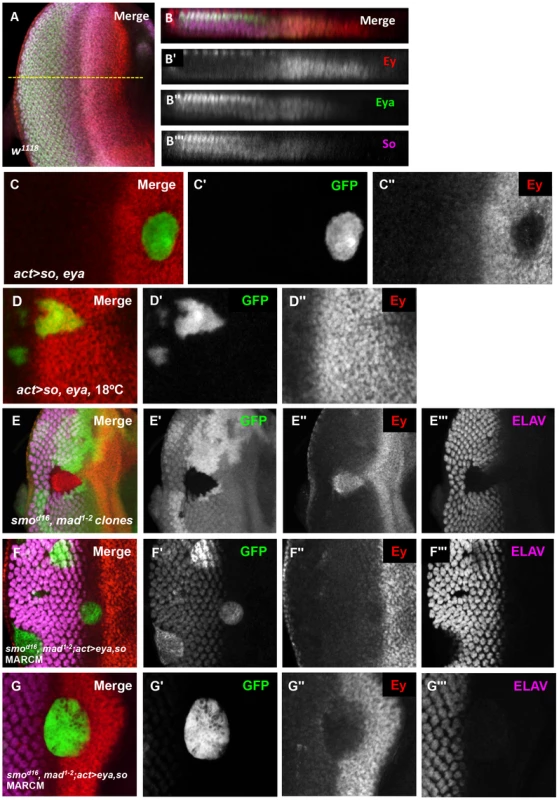
(A) Expression patterns of Ey, Eya, and So in a w1118 third instar eye imaginal disc. The yellow dashed line indicates the approximate location of the orthogonal section in B–B′″. (B) Orthogonal section of A. (B′) Grayscale image of Ey expression, red in B, apical Ey expression is detected in the peripodium, sections excluded in normal Z projection. (B″) Grayscale image of Eya expression, green in B. (B′″) Grayscale image of So expression, magenta in B. (C–G) Expression of UAS-GFP marks the clone(C, D, F, G); lack of GFP marks the clone in E. Crosses were raised at 25°C, except D, raised at 18°C. (C) UAS-so and UAS-eya were co-overexpressed anterior to the furrow. (C′) Grayscale image of GFP expression in C. (C″) Grayscale image of Ey expression in C. (D) Flipout-Gal4 was used to co-express UAS-so, UAS-eya, and UAS-GFP. (D′) Grayscale image of GFP expression, green in D. (D″) Grayscale image of Ey expression, red in D. (E) Double loss of function clones for smod16 null allele and mad1–2 hypomorphic allele were generated by inducing hs-flp expression at 48 hrs AEL. (E′) Grayscale image of GFP expression, green in E. (E″) Grayscale image of Ey expression, red in E. (E′″) Grayscale image of ELAV expression, magenta in E, shows differentiating photoreceptors. (F) MARCM clones that are mutant for smod16 and mad1–2 while overexpressing so and eya. (F′) Grayscale image of GFP expression in F; the ELAV-like pattern is due to non-specific antibody interaction. (F″) Grayscale image of Ey expression in F. (F′″) Grayscale image of ELAV expression in F shows differentiating photoreceptors. (G–G′″) Same as F showing a clone extending anterior to the furrow. The levels of So and Eya expression increase posterior to the morphogenetic furrow in response to activation of the Hedgehog (Hh) and Decapentaplegic (Dpp) signaling pathways [26]. Next, we asked if upregulation of Eya and So is sufficient to suppress ey even without the signaling pathways normally required for morphogenetic furrow movement. To test this, we made use of the MARCM system. We overexpressed eya and so simultaneously in smo3, mad1–2 double mutant clones, which cannot respond to either Hh or Dpp signaling. Clones doubly mutant for these two signaling effectors are known to lack furrow progression: they do not activate Notch signaling, they lack differentiation, and they retain Ey expression [5], [26], [49], [52]–[54] (Figure 5E). We observed that Ey is strongly repressed in clones anterior to the morphogenetic furrow, and is not expressed in clones posterior to the morphogenetic furrow (Figure 5F,G). Therefore, high levels of eya and so are sufficient to repress Ey in the absence of normal morphogenetic furrow signaling. Together, these data suggest that the increased levels of Eya and So induced by signals in the morphogenetic furrow are important for Ey repression.
Excess So can block ey transcription
To gain a better understanding of how Eya and So cooperate to regulate ey expression, we tested the response of the ey enhancer in vitro to So and/or Eya. In Drosophila S2 cells, when the ey enhancer is used to drive luciferase expression (ey-luc), reporter expression was induced by co-expression of So with Eya, but not by either factor alone (Figure 6A, “WT”). This suggests that the ey enhancer can be activated by Eya and So, and is consistent with previously published results that they cooperate to activate targets [45], [47]. Mutation or deletion of the So binding site (Mut or Short, respectively) within the reporter strongly reduced its induction by Eya/So (Figure 6A,B). This suggests that the activation of the construct in our assay depends primarily upon the So binding site.
Fig. 6. Excess So is sufficient to block enhancer activation in vitro. 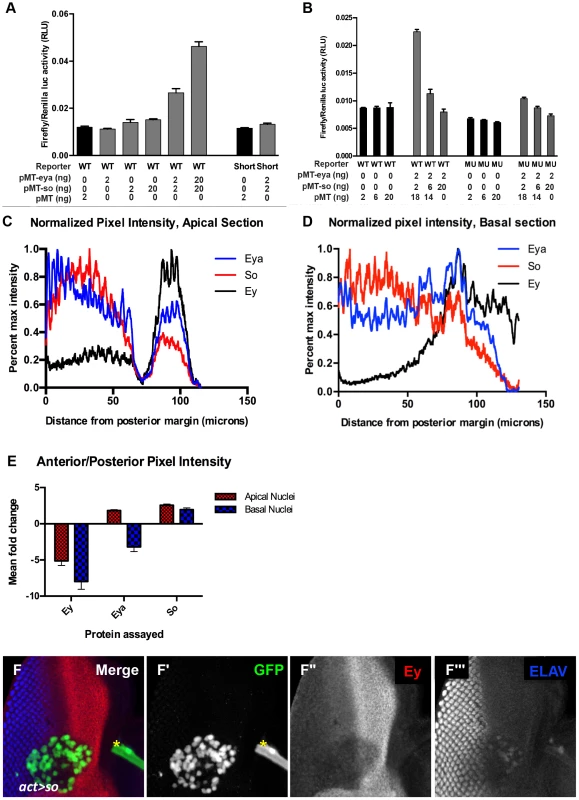
(A,B) Luciferase assay performed in transiently transfected S2 cells, reported as a ratio between Firefly and Renilla luciferase levels. Reporter constructs as indicated: WT = ey-Luc, Short = eyshort-Luc that deletes the So binding site, MU = eymut-Luc. Controls are graphed in black; manipulations are in gray. For each condition the nanograms transfected of pMT-eya, pMT-so, or pMT empty vector are indicated. (C,D) Normalized pixel intensity plots for fluorescent immunohistochemistry to assay Eya (blue), So (red), and Ey (black) expression in a w1118 disc (see Materials and Methods). (C) Staining intensity in apical nuclei. (D) Staining intensity in basal nuclei. (E) Mean fold change for each channel was calculated (n = 5), and plotted. Error bars indicate S.E.M. (F) Flipout-Gal4 driving UAS-GFP and UAS-So expression showing GFP, Ey, and ELAV. Yellow asterisk denotes a piece of trachea that is not part of the disc. (F′–F″) individual channels from panel F. (F′) GFP marks the clone, (F″) Ey, (F′″) ELAV shows differentiating photoreceptors. Our in vivo results indicate that high levels of Eya and So expression can repress Ey expression. However, even a 10 fold increase of both transfected plasmids did not repress; rather, the reporter was activated more strongly (Figure 6A). To generate additional hypotheses we re-examined the in vivo expression of Ey, So, and Eya. We quantified pixel intensity values for Eya, So, and Ey in orthogonal sections (as in Figure 5B) across multiple imaginal discs (n = 5) as a proxy to examine expression levels across the third instar disc. Values were normalized and plotted for each protein to generate a line graph that visually depicts staining intensity across the section (as shown in Figure 6C,D). We observed that So undergoes a greater average positive fold change (Posterior Max/Anterior max) than Eya in both apical and basal sections (Figure 6E). While this analysis is only semi-quantitative, it was highly reproducible, and could indicate that So is in excess to Eya in posterior cells. At a minimum it suggests that their relative levels of expression are different in anterior and posterior cells. To test the simple model that excess So can prevent ey expression, we increased the ratio of transfected so plasmid to eya plasmid in our in vitro system. In response, we observed a dramatic decrease of reporter expression (Figure 6B), leading us to conclude that excess So suppresses activation of ey-dGFP by the Eya/So complex in vitro. To test this model in vivo we overexpressed So anterior to the morphogenetic furrow. We observed that in some clones Ey expression was mildly repressed by overexpression of So (Figure 6F). Based on our in vivo and in vitro observations, we conclude that excess So expression can be sufficient to suppress ey expression.
Dac contributes to Ey repression within the morphogenetic furrow
Within the morphogenetic furrow, we observed that Eya and So levels are not increased until after the initial decrease of Ey expression, indicating that there must be an additional mechanism that contributes to Ey negative regulation in this domain. The Ski/Sno family member Dachshund (Dac) physically interacts with Eya [20], [55], and may cooperate to regulate targets of So and Eya [28]. In mammals, the ortholog Dach interacts with the Eya and So orthologs to repress targets [56], though this interaction has not been confirmed in Drosophila. To test if Dac is involved in Ey repression, we generated dac null clones. Anterior to the furrow, Ey expression was not affected in dac clones, suggesting that dac is not required for Ey expression anterior to the morphogenetic furrow (Figure 7A). As previously reported for clones posterior to the morphogenetic furrow, we observed increased Ey expression in dac clones near the furrow, but not clones distant from it (Figure 7A,B) [26]. This overlaps the highest levels of Dac expression posterior to the furrow (Figure S7A,B). This shows that dac is required for negative regulation of Ey specifically in the domain near the morphogenetic furrow.
Fig. 7. Dachshund is required for ey repression near the morphogenetic furrow. 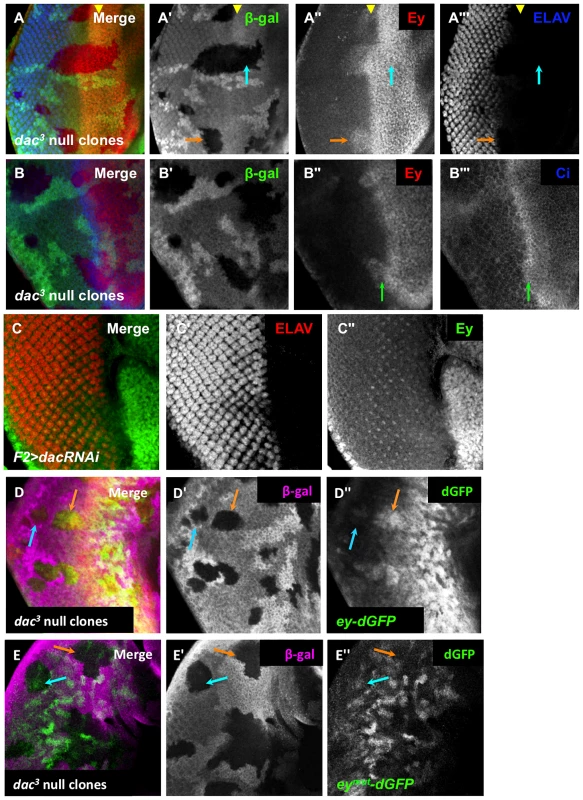
(A) dac3 null clones, induced by hs-flp 48 hours after egg lay (AEL) showing β-Galactosidase, ELAV and Ey expression. The yellow arrowhead indicates the furrow. The blue arrow indicates an anterior clone. The orange arrow indicates a posterior clone. (A′) Grayscale image of β-Galactosidase expression, shown as green in A; loss of β-Galactosidase expression marks the clones. (A″) Grayscale image of Ey channel alone, red in A. (A′″) Grayscale image of ELAV channel alone showing photoreceptor differentiation, blue in A. (B) dac3 null clones, induced by hs-flp 48 hours after egg lay (AEL) showing β-Galactosidase, Ci and Ey expression. The green arrow indicates the boundary between high and low levels of Ci. (B′) Grayscale image of β-Galactosidase expression, shown as green in B; loss of β-Galactosidase expression marks the clones. (B″) Grayscale image of Ey channel alone, red in B. (B′″) Grayscale image of Ci channel alone, blue in B. (C) F2-Gal4 drives expression of dacRNAi (TRiP collection transformant ID HMS01435). Merge of ELAV and Ey expression shown. (C′) Grayscale image of ELAV expression, shown as red in C, marks differentiating photoreceptors. (C″) Grayscale image of Ey expression, shown as green in C. (D) ey-dGFP (green) expression in dac3 null clone posterior to the morphogenetic furrow marked by loss of β-Galactosidase expression (magenta). The blue arrow indicates a clone far posterior to the furrow. The orange arrow indicates a clone posterior to but near the furrow. (D′) Grayscale image of β-Galactosidase expression in D. (D″) Grayscale image of ey-dGFP expression in D as revealed by anti-GFP. (E) eymut-dGFP (green) expression in dac3 null clone posterior to the morphogenetic furrow marked by loss of β-Galactosidase expression (magenta). The blue arrow indicates a clone far posterior to the furrow. The orange arrow indicates a clone near the furrow. (E′) Grayscale image of β-Galactosidase expression in E. (E″) Grayscale image of eymut-dGFP expression in E as revealed by anti-GFP. It is known that large dac clones can have delayed morphogenetic furrow progression, making it possible that Ey expression within these clones could be a secondary consequence of a delayed furrow [13]. To address this, we assayed furrow progression through small dac clones and compared this to the Ey expression boundary. Cubitus interruptus (Ci), the effector of Hedgehog signaling, normally accumulates to high levels in a tight band within the morphogenetic furrow, just posterior to the onset of Ey negative regulation (Figure 1A, Figure S7C). In dac clones spanning the furrow, Ci accumulation was not delayed, but Ey overlapped high levels of Ci, which was not observed in wild-type cells (Figure 7B, compare to Figure 1A). This result suggests that the leading edge of the morphogenetic furrow, normally correlating with Ey suppression, moves into and through these dac clones. As Ey suppression is delayed in these clones, it indicates that Dac is required for suppression of Ey near the furrow independent of its role in furrow progression. To further test if dac represses Ey posterior to the furrow, we used F2-Gal4 to drive multiple independent dac RNAi transgenes, and observed that Ey expression is detected in knockdown cells posterior to the furrow (Figure 7C and data not shown). This result shows that Dac is necessary to suppress Ey expression posterior to the furrow.
We used the reporter ey-dGFP to determine if Dac suppresses ey at the transcriptional level. Like Ey, ey-dGFP is expressed in dac clones near the furrow (Figure 7D, orange arrow), but not clones far posterior to the furrow (Figure 7D, blue arrow). This indicates that Dac is required to suppress ey transcription near the morphogenetic furrow, consistent with the expression pattern of Dac. We also examined eymut-dGFP in dac clones. First, near the morphogenetic furrow, we did not observe expression of eymut-dGFP in dac clones as we had observed with ey-dGFP (Figure 7E, orange arrow). This result indicates that the elevated levels of wild-type reporter expression observed in dac clones require the So binding site. By extrapolation, this result suggests that So still activates ey expression in dac clones near the MF; this places repression by Dac earlier than suppression by So during development. In clones far posterior to the morphogenetic furrow we observed that eymut-dGFP is expressed in dac clones (Figure 7E, blue arrow), suggesting the repression of the wild-type reporter observed in dac clones requires the So binding site. We conclude that the phenotypes of ey reporter expression in dac clones reflect regulation by So in these domains. Furthermore, we conclude that Dac suppression of ey expression is an earlier developmental event than repression by So.
We next overexpressed Dac with Eya or So to see if they were sufficient to suppress Ey expression anterior to the furrow. Overexpression of dac or eya alone did not alter Ey expression (data not shown). Co-overexpression of eya and dac also had no effect on Ey expression (data not shown). However, co-overexpression of so with dac was sufficient to repress Ey expression to modest levels (Figure 8A). We conclude that Dac and So can cooperate to reduce Ey expression in vivo.
Fig. 8. So and Eya cooperate with Dac in vivo to complete Ey repression. 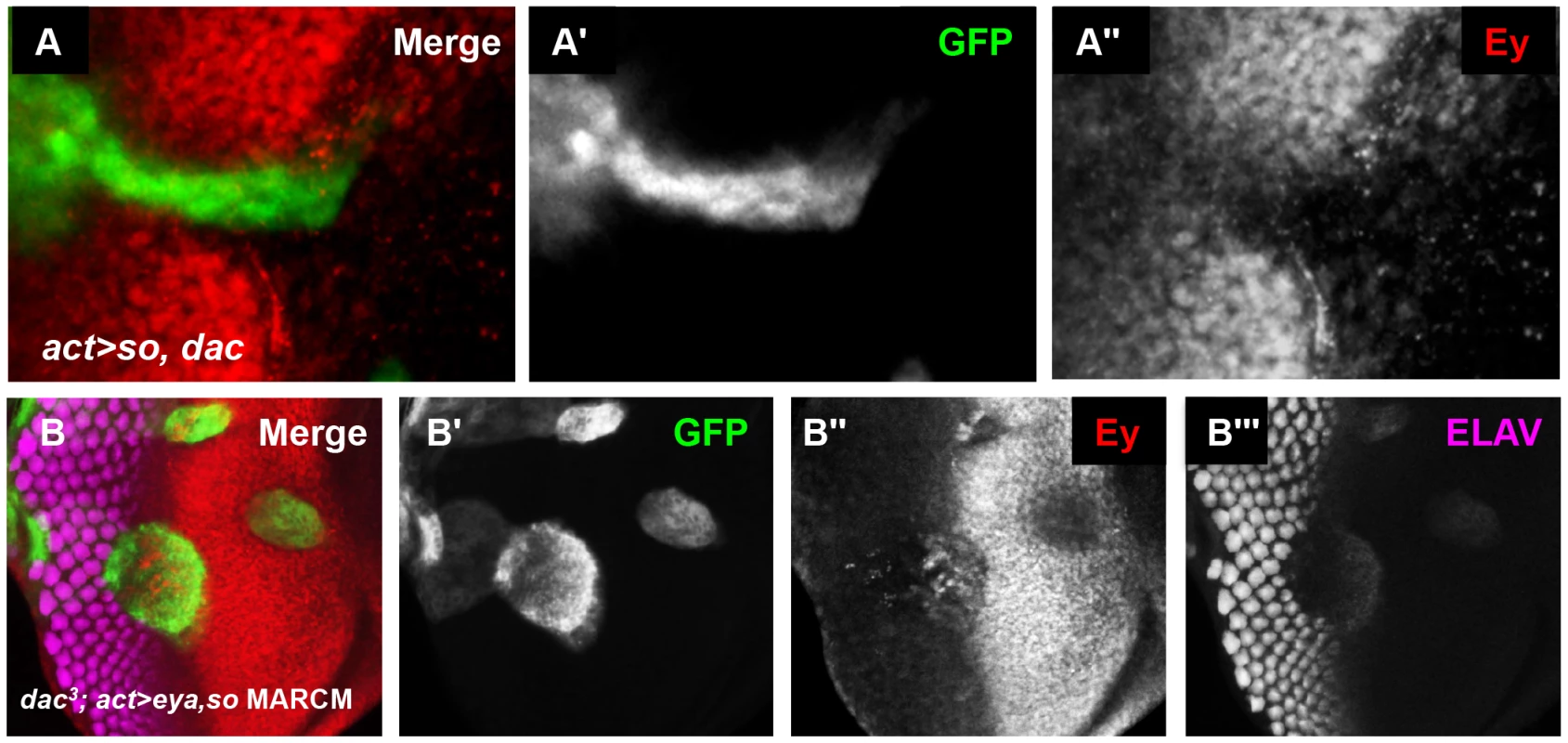
(A) UAS-so and UAS-dac7c4 were co-overexpressed anterior to the furrow. (A′) Grayscale image of GFP expression in A; GFP marks the clone. (A″) Grayscale image of Ey expression in A. (B) MARCM clones that are null for dac while overexpressing so and eya. (B′) Grayscale image of GFP expression in B; GFP marks the clone. (B″) Grayscale image of Ey expression in B. (B″) Grayscale image of ELAV expression in B shows differentiating photoreceptors. dac is a downstream target of the So/Eya complex in the eye [19], [25], [49]. Therefore, we wanted to determine if Ey repression anterior to the furrow by co-overexpression of Eya and So (Eya+So) requires the activation of dac by these genes. To test this, we generated Eya+So overexpression clones that were also null for dac using the MARCM technique [57]. So and Eya reduced Ey expression anterior to the furrow, though less effectively than in cells that can still express Dac (Figure 8B vs. Figure 5C). This suggests that So and Eya can repress Ey expression without Dac, but that full repression anterior to the furrow requires Dac. In MARCM clones spanning the furrow, the phenotype resembles dac null clones and Ey is not repressed, suggesting that Dac is specifically required in this domain (Figure 8B). Finally, in posterior clones distant from the furrow, Ey is not expressed (Figure 8B). This indicates that Eya and So are sufficient to completely suppress Ey in this domain. Together, these results indicate that Dac is required near the morphogenetic furrow to negatively regulate Ey expression, but that So and Eya can cooperate to repress Ey independent of Dac further posteriorly.
Discussion
In this work, we have found that a switch from high to low levels of Ey expression is required for normal differentiation during retinal development. We also present a mechanism of Ey regulation by the RD gene network members Eya, So, and Dac. Specifically, we report that So switches from being an activator to a suppressor of ey expression, both depending on a So binding site within an ey eye-specific enhancer. We additionally report that the So cofactors Eya and Dac are required for ey repression posterior to the furrow but not for its maintenance ahead of the furrow, and are sufficient to cooperate with So to mediate Ey repression within the normal Ey expression domain.
Our results support a Gro-independent mechanism for the suppression of target gene expression by the transcription factor Sine oculis (So). An independent study has also shown that So can repress the selector gene cut in the antenna in a Gro-independent process though the mechanism was not determined [46]. We observe that Ey is expressed at low levels posterior to the morphogenetic furrow. However, when so expression is lost in clones posterior to the furrow, Ey expression and ey-dGFP expression are strongly activated. We show that this is not simply a default response of ey to So loss, as removing So from developmentally earlier anterior cells results in reduced ey expression. We also observe that knockdown of So specifically in differentiating cells using RNAi causes a similar phenotype, suggesting that an activator of Ey expression is expressed in differentiating photoreceptors. Mutation of a known So binding site in ey-dGFP results in activation of the reporter posterior to the furrow, supporting a model that binding of So to the enhancer prevents inappropriate activation of ey expression posterior to the furrow. Finally, in vitro we observe that an excess of So is sufficient to prevent activation of the enhancer and observe that in vivo overexpression of So can suppress normal Ey expression. Our observations are consistent with what in vitro studies have indicated about So function: when So binds DNA without Eya, it can only weakly activate transcription [45, and this work]. However, our work introduces a novel mechanism of regulation for So targets, in which So occupancy of an enhancer prevents other transcription factors from inducing high levels of target gene expression. Our results also indicate that suppression of robust ey expression is an important developmental event. It is not yet clear if maintaining basal expression of ey, rather than completely repressing it, is developmentally important; however, it is possible that the ultimate outcome of a basal level of ey transcription may be necessary for the completion of retinal development [58].
Our results also show that eya is required for Ey suppression in vivo. However, consistent with its characterization as a transcriptional coactivator, our in vitro analysis does not indicate a direct role for Eya in repression. Previous studies, and our observations, indicate that Eya is required for the expression of So posterior to the furrow in the third instar [18], [24], [25], [36], and Figure S5. Additionally, our reporter analysis shows that Eya regulation of ey requires the So binding site. We propose that the simplest model for Eya function in the suppression of ey is through its established function as a positive regulator of So expression, as we observe that overexpression of So alone is sufficient to weakly repress Ey expression and to block reporter activation in vitro. This model could also account for the results reported by us and others regarding the inability of this UAS-so construct to induce ectopic eye formation [16], [36], [46], [59]. Briefly, the primary function of So in ectopic eye formation is to repress the non-eye program [46]. Overexpressing the So construct used in this study alone is not sufficient to induce this program, possibly because the transgene expression level is not sufficient; however, co-expression of the so positive regulator Eya is sufficient to induce robust ectopic eye formation [16], [36]. In light of our findings, we propose that Eya co-expression is necessary to induce So expression to sufficient levels to block transcriptional activation of non-eye targets to permit the induction of the ectopic eye program; however we cannot rule out that other functions of Eya may play a role.
We further demonstrate that dac expression is required specifically near the furrow for Ey repression. In addition, we show that the So binding site is required for strong ey expression in dac clones near the furrow, suggesting that So activates ey in these clones. This suggests that repression by Dac occurs before the transition to repression by So, making Dac the first repressor of ey expression at the furrow, and identifying how the initiation of repression occurs before So levels increase. We further show that Eya and So are sufficient to repress ey expression in dac mutant clones anterior to the furrow, though not as completely as in cells that express Dac. This result indicates that Dac is not an obligate partner with Eya and So in ey repression, but is required for the full suppression of ey. One model would be that Dac and So can cooperate in a complex to modestly repress eyeless directly. This would be consistent with our loss-of-function and reporter data as well as the observation that Dac and So misexpression can weakly cooperate to repress Ey anterior to the furrow. However, while a similar complex has been described in mammalian systems, previous studies have been unable to detect this physical interaction in Drosophila [44], [45], [55], [60]. An alternative model is that Dac suppresses ey expression indirectly and in parallel to Eya and So. A previous study has shown that dac expression is necessary and sufficient near the furrow to inhibit the expression of the zinc finger transcription factor Teashirt (Tsh) [26]. Tsh overlaps Ey expression anterior to the furrow, and can induce Ey expression when misexpressed [42], [43]. Furthermore, tsh repression is required for morphogenetic furrow progression and differentiation [42], [43]. In light of these previously published findings, we propose that a simpler model based on current knowledge is that Dac repression of tsh at the morphogenetic furrow reduces Ey expression indirectly. Future studies may distinguish between these mechanisms.
In addition to the role of the RD gene network in ey modulation, we identify that signaling events within the morphogenetic furrow indirectly regulate the switch to low levels of ey expression. It has been shown that signaling pathways activated in the morphogenetic furrow increase levels of Eya, So and Dac; furthermore, it is proposed that this upregulation alters their targets, creating an embedded loop within the circuitry governing retinal development and allowing signaling events to indirectly regulate targets through the RD network [26], [28], [61]. The identification of ey regulation by So posterior to the morphogenetic furrow represents a direct target consistent with this model.
In conclusion, we present a model that rewiring of the RD network activates different dominant sub-circuits to drive key transitions in development (Figure 9). To the interactions previously identified by others, we add that strong upregulation of So, dependent on Eya, results in minimal levels of ey transcription [18], [25]. We propose that the identification of this novel sub-circuit of the RD network provides a mechanism for terminating the self-perpetuating loop of determination associated with high levels of Ey, permitting the onset of differentiation and the completion of development. Together, these results give us a new view into how temporal rewiring within the RD network directs distinct developmental events.
Fig. 9. A model for dynamic RD gene network interactions during the third instar. 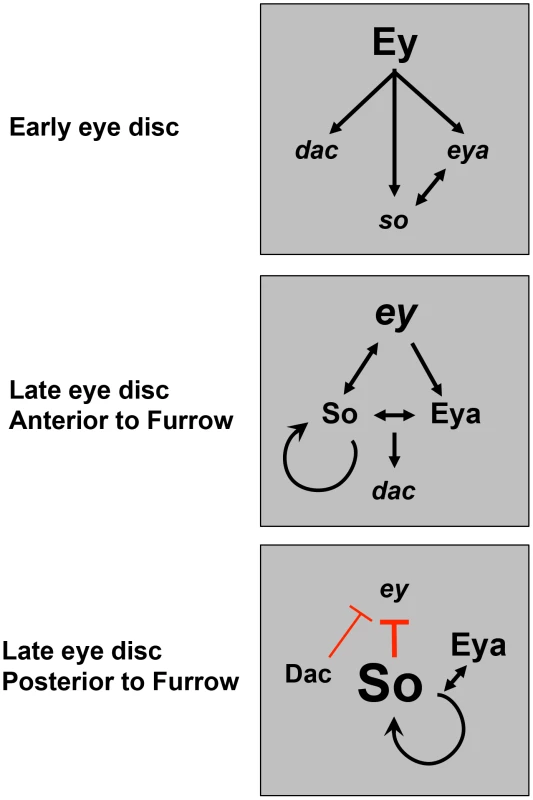
Previous studies have shown that prior to the third instar, Ey activates expression of downstream RD genes. This work shows that anterior to the furrow during third instar, positive feedback loops are maintained among the RD network members, with So feeding back to help promote and maintain ey expression. Just posterior to the morphogenetic furrow, Dac represses ey transcription. Finally, posterior to the furrow, high levels of So, induced by Eya, are sufficient to prevent activation of high levels of ey transcription. Materials and Methods
Generation of destabilized GFP (dGFP) constructs for in vivo experiments
pH-dGFP-attB
A 285 bp φC31 attB fragment was PCR-amplified from p[ACMAN] [39], cut with AatII, and cloned into pH-Stinger [41], resulting in the construct pH-Stinger-attB. dGFP encodes a destabilized variant of enhanced green fluorescent protein, amplified from 10XSTAT92E-GFP [62] with 5′ AgeI and 3′ NotI tails and cloned into pH-Stinger-attB, generating pH-dGFP-attB.
ey-dGFP and eymut-dGFP
ey-dGFP was generated by PCR on genomic DNA of wild-type flies by using the following primer sets: 5′-CGGAATTCCAAGTACAAACTGACTTCTTG-3′; 5′-CGCGGATCCGAATTCGAGAAATATCACATGGCC-3′. 5′ EcoRI and 3′ BamHI sites were added and used for subcloning into pH-dGFP-attB. The So site was mutated by changing GAG to CCC and introduced by two-step PCR to generate eymut-dGFP [22].
UAS-dGFP
To generate the UAS-dGFP construct, dGFP was first amplified from the 10XSTAT92E-GFP construct with XbaI and XhoI tails. PCR product was then digested and ligated into pUAST-attB vector (a gift from Konrad Basler). Positive clones were sequenced to confirm sequence integrity and orientation.
For transgenic fly generation, each construct was injected into a docking site at 68A (P2). Correct integration events were identified by genomic PCR by standard methods [22], [39].
Generation of ey-Luc, eyshort-Luc, and eymut-Luc
The enhancer sequences were amplified from ey-dGFP or eymut-dGFP with XhoI and NheI tails. PCR fragments were digested and ligated per the manufacturer's instructions (NEB, Takara) directionally into pGL3-Basic (Promega). Correct ligation events were identified by sequencing to generate ey-Luc and eymut-Luc, respectively. eyshort-Luc was amplified from ey-Luc and generates a truncated enhancer that ends 8 bp upstream of the So binding site.
S2 cell culture, transfection and luciferase assays
Drosophila S2 cells were cultured in Schneider's medium containing 10% fetal bovine serum and antibiotics. Cells were transiently transfected in 48-well plates using Cellfectin (Invitrogen) according to the manufacturer's protocol. Cells were transfected with ey-Luc, eyshort-Luc, or eymut-Luc, in the presence or absence of Eya and So in pMT vector (Invitrogen, a gift from Ilaria Rebay), along with tub-Renilla luciferase in pRL vector (a gift from K Basler). 24 hrs after transfection, cells were induced with CuSO4 at a final concentration of 500 µM. Luciferase activity was assayed 2 days after induction using the Dual-Glo kit (Promega) according to the manufacturer's protocol. Data were graphed in GraphPad Prism and labeled using Adobe Illustrator.
Crosses and fly husbandry
For a list of the genotypes used, please reference Table S1. All crosses were performed on standard cornmeal agar at 25°C unless otherwise noted. Heat shocks were performed at 37°C. Flipout-Gal4 [63] crosses were heat shocked for 8 min, 48 hrs after egg laying (AEL). For loss-of-function clones or MARCM clones [57], heat shocks were performed for 1 hr at 48 hrs AEL, or, for so3 and eyacliIID clones, 72 hrs AEL. Wandering third instar larvae were collected and dissected using standard methods as previously described [37].
Immunohistochemistry
Staining was performed as previously described [64]. For antibodies used, please reference Table S2.
Microscopy and Image processing
Imaginal disc images were captured using a Zeiss LSM confocal microscope. LSMs were stacked using ImageJ software and stacks were merged in ImageJ and prepared for figures using Adobe Photoshop. Staining quantification for Eya, Ey and So: orthogonal sections were generated using ImageJ and represent approximately 10 micron wide slices through the full depth of the disc (n = 5); sections were resliced in ImageJ to generate XZ stacks which were summed. The apical ROI was measured based on the width of the Eya signal in photoreceptors. The basal ROI was the same ROI, shifted basally to exclude the apical Eya signal. Pixel intensity was calculated using the plot profile function, and values were normalized. Pixel intensity plots and bar graph of average fold change were generated in GraphPad Prism. For adult images, adults were frozen at −80°C for 30 minutes. Light microscopy images of adult heads were captured on a Zeiss Axioplan microscope, and were processed with Adobe Photoshop software.
Supporting Information
Zdroje
1. Gilbert SF (2003) Developmental Biology. Sunderland: Sinauer Associates, Inc. 838 p.
2. Bodentstein D (1950) The Postembryonic development of Drosophila melanogaster. In: Demerec M, editor. Biology of Drosophila. New York: John Wiley and Sons, Inc. pp. 275–367.
3. Ferris GF (1950) External Morphology of the Adult. In: Demerec M, editor. Biology of Drosophila. New York: John Wiley and Sons, Inc. . pp. 368–419.
4. Cohen SM (1993) Imaginal Disc Development. In: Bate M, Martinez-Arias A, editors. The Development of Drosophila melanogaster: Cold Spring Harbor Laboratory Press. pp. 747–841.
5. HeberleinU, WolffT, RubinGM (1993) The TGF beta homolog dpp and the segment polarity gene hedgehog are required for propagation of a morphogenetic wave in the Drosophila retina. Cell 75 : 913–926.
6. MaC, ZhouY, BeachyPA, MosesK (1993) The segment polarity gene hedgehog is required for progression of the morphogenetic furrow in the developing Drosophila eye. Cell 75 : 927–938.
7. WolffT, ReadyDF (1991) The beginning of pattern formation in the Drosophila compound eye: the morphogenetic furrow and the second mitotic wave. Development 113 : 841–850.
8. MannRS, CarrollSB (2002) Molecular mechanisms of selector gene function and evolution. Curr Opin Genet Dev 12 : 592–600.
9. BoniniNM, BuiQT, Gray-BoardGL, WarrickJM (1997) The Drosophila eyes absent gene directs ectopic eye formation in a pathway conserved between flies and vertebrates. Development 124 : 4819–4826.
10. BoniniNM, LeisersonWM, BenzerS (1993) The eyes absent gene: genetic control of cell survival and differentiation in the developing Drosophila eye. Cell 72 : 379–395.
11. CheyetteBN, GreenPJ, MartinK, GarrenH, HartensteinV, et al. (1994) The Drosophila sine oculis locus encodes a homeodomain-containing protein required for the development of the entire visual system. Neuron 12 : 977–996.
12. HalderG, CallaertsP, GehringWJ (1995) Induction of ectopic eyes by targeted expression of the eyeless gene in Drosophila. Science 267 : 1788–1792.
13. MardonG, SolomonNM, RubinGM (1994) dachshund encodes a nuclear protein required for normal eye and leg development in Drosophila. Development 120 : 3473–3486.
14. QuiringR, WalldorfU, KloterU, GehringWJ (1994) Homology of the eyeless gene of Drosophila to the Small eye gene in mice and Aniridia in humans. Science 265 : 785–789.
15. ShenW, MardonG (1997) Ectopic eye development in Drosophila induced by directed dachshund expression. Development 124 : 45–52.
16. WeasnerB, SalzerC, KumarJP (2007) Sine oculis, a member of the SIX family of transcription factors, directs eye formation. Dev Biol 303 : 756–771.
17. CzernyT, HalderG, KloterU, SouabniA, GehringWJ, et al. (1999) twin of eyeless, a second Pax-6 gene of Drosophila, acts upstream of eyeless in the control of eye development. Mol Cell 3 : 297–307.
18. HalderG, CallaertsP, FlisterS, WalldorfU, KloterU, et al. (1998) Eyeless initiates the expression of both sine oculis and eyes absent during Drosophila compound eye development. Development 125 : 2181–2191.
19. PappuKS, OstrinEJ, MiddlebrooksBW, SiliBT, ChenR, et al. (2005) Dual regulation and redundant function of two eye-specific enhancers of the Drosophila retinal determination gene dachshund. Development 132 : 2895–2905.
20. BuiQT, ZimmermanJE, LiuH, BoniniNM (2000) Molecular analysis of Drosophila eyes absent mutants reveals features of the conserved Eya domain. Genetics 155 : 709–720.
21. NiimiT, SeimiyaM, KloterU, FlisterS, GehringWJ (1999) Direct regulatory interaction of the eyeless protein with an eye-specific enhancer in the sine oculis gene during eye induction in Drosophila. Development 126 : 2253–2260.
22. PauliT, SeimiyaM, BlancoJ, GehringWJ (2005) Identification of functional sine oculis motifs in the autoregulatory element of its own gene, in the eyeless enhancer and in the signalling gene hedgehog. Development 132 : 2771–2782.
23. OstrinEJ, LiY, HoffmanK, LiuJ, WangK, et al. (2006) Genome-wide identification of direct targets of the Drosophila retinal determination protein Eyeless. Genome Res 16 : 466–476.
24. PunzoC, SeimiyaM, FlisterS, GehringWJ, PlazaS (2002) Differential interactions of eyeless and twin of eyeless with the sine oculis enhancer. Development 129 : 625–634.
25. SalzerCL, KumarJP (2009) Position dependent responses to discontinuities in the retinal determination network. Dev Biol 326 : 121–130.
26. FirthLC, BakerNE (2009) Retinal determination genes as targets and possible effectors of extracellular signals. Dev Biol 327 : 366–375.
27. ZhangT, RanadeS, CaiCQ, ClouserC, PignoniF (2006) Direct control of neurogenesis by selector factors in the fly eye: regulation of atonal by Ey and So. Development 133 : 4881–4889.
28. Tanaka-MatakatsuM, DuW (2008) Direct control of the proneural gene atonal by retinal determination factors during Drosophila eye development. Dev Biol 313 : 787–801.
29. JarmanAP, GrellEH, AckermanL, JanLY, JanYN (1994) Atonal is the proneural gene for Drosophila photoreceptors. Nature 369 : 398–400.
30. KenyonKL, RanadeSS, CurtissJ, MlodzikM, PignoniF (2003) Coordinating proliferation and tissue specification to promote regional identity in the Drosophila head. Dev Cell 5 : 403–414.
31. LiY, BakerNE (2001) Proneural enhancement by Notch overcomes Suppressor-of-Hairless repressor function in the developing Drosophila eye. Curr Biol 11 : 330–338.
32. BakerNE, YuSY (1997) Proneural function of neurogenic genes in the developing Drosophila eye. Curr Biol 7 : 122–132.
33. PignoniF, ZipurskySL (1997) Induction of Drosophila eye development by decapentaplegic. Development 124 : 271–278.
34. CrewJR, BatterhamP, PollockJA (1997) Developing compound eye in lozenge mutants of Drosophila: lozenge expression in the R7 equivalence group. Development Genes and Evolution 206 : 481–493.
35. FreemanM (1996) Reiterative use of the EGF receptor triggers differentiation of all cell types in the Drosophila eye. Cell 87 : 651–660.
36. PignoniF, HuB, ZavitzKH, XiaoJ, GarrityPA, et al. (1997) The eye-specification proteins So and Eya form a complex and regulate multiple steps in Drosophila eye development. Cell 91 : 881–891.
37. PeppleKL, AtkinsM, VenkenK, WellnitzK, HardingM, et al. (2008) Two-step selection of a single R8 photoreceptor: a bistable loop between senseless and rough locks in R8 fate. Development 135 : 4071–4079.
38. HauckB, GehringWJ, WalldorfU (1999) Functional analysis of an eye specific enhancer of the eyeless gene in Drosophila. Proc Natl Acad Sci U S A 96 : 564–569.
39. VenkenKJ, HeY, HoskinsRA, BellenHJ (2006) P[acman]: a BAC transgenic platform for targeted insertion of large DNA fragments in D. melanogaster. Science 314 : 1747–1751.
40. EkasLA, BaegGH, FlahertyMS, Ayala-CamargoA, BachEA (2006) JAK/STAT signaling promotes regional specification by negatively regulating wingless expression in Drosophila. Development 133 : 4721–4729.
41. BaroloS, CarverLA, PosakonyJW (2000) GFP and beta-galactosidase transformation vectors for promoter/enhancer analysis in Drosophila. Biotechniques 29 : 726, 728, 730, 732.
42. BessaJ, GebeleinB, PichaudF, CasaresF, MannRS (2002) Combinatorial control of Drosophila eye development by eyeless, homothorax, and teashirt. Genes Dev 16 : 2415–2427.
43. PanD, RubinGM (1998) Targeted expression of teashirt induces ectopic eyes in Drosophila. Proc Natl Acad Sci U S A 95 : 15508–15512.
44. KenyonKL, LiDJ, ClouserC, TranS, PignoniF (2005) Fly SIX-type homeodomain proteins Sine oculis and Optix partner with different cofactors during eye development. Dev Dyn 234 : 497–504.
45. SilverSJ, DaviesEL, DoyonL, RebayI (2003) Functional dissection of eyes absent reveals new modes of regulation within the retinal determination gene network. Mol Cell Biol 23 : 5989–5999.
46. AndersonAM, WeasnerBM, WeasnerBP, KumarJP (2012) Dual transcriptional activities of SIX proteins define their roles in normal and ectopic eye development. Development 139 : 991–1000.
47. JemcJ, RebayI (2007) Identification of transcriptional targets of the dual-function transcription factor/phosphatase eyes absent. Dev Biol 310 : 416–429.
48. SuzukiT, SaigoK (2000) Transcriptional regulation of atonal required for Drosophila larval eye development by concerted action of eyes absent, sine oculis and hedgehog signaling independent of fused kinase and cubitus interruptus. Development 127 : 1531–1540.
49. CurtissJ, MlodzikM (2000) Morphogenetic furrow initiation and progression during eye development in Drosophila: the roles of decapentaplegic, hedgehog and eyes absent. Development 127 : 1325–1336.
50. PappuKS, ChenR, MiddlebrooksBW, WooC, HeberleinU, et al. (2003) Mechanism of hedgehog signaling during Drosophila eye development. Development 130 : 3053–3062.
51. HazelettDJ, BourouisM, WalldorfU, TreismanJE (1998) decapentaplegic and wingless are regulated by eyes absent and eyegone and interact to direct the pattern of retinal differentiation in the eye disc. Development 125 : 3741–3751.
52. DominguezM, HafenE (1997) Hedgehog directly controls initiation and propagation of retinal differentiation in the Drosophila eye. Genes Dev 11 : 3254–3264.
53. FuW, BakerNE (2003) Deciphering synergistic and redundant roles of Hedgehog, Decapentaplegic and Delta that drive the wave of differentiation in Drosophila eye development. Development 130 : 5229–5239.
54. GreenwoodS, StruhlG (1999) Progression of the morphogenetic furrow in the Drosophila eye: the roles of Hedgehog, Decapentaplegic and the Raf pathway. Development 126 : 5795–5808.
55. ChenR, AmouiM, ZhangZ, MardonG (1997) Dachshund and eyes absent proteins form a complex and function synergistically to induce ectopic eye development in Drosophila. Cell 91 : 893–903.
56. IkedaK, WatanabeY, OhtoH, KawakamiK (2002) Molecular interaction and synergistic activation of a promoter by Six, Eya, and Dach proteins mediated through CREB binding protein. Mol Cell Biol 22 : 6759–6766.
57. LeeT, LuoL (2001) Mosaic analysis with a repressible cell marker (MARCM) for Drosophila neural development. Trends Neurosci 24 : 251–254.
58. ShengG, ThouvenotE, SchmuckerD, WilsonDS, DesplanC (1997) Direct regulation of rhodopsin 1 by Pax-6/eyeless in Drosophila: evidence for a conserved function in photoreceptors. Genes Dev 11 : 1122–1131.
59. SalzerCL, KumarJP (2010) Identification of retinal transformation hot spots in developing Drosophila epithelia. PLoS One 5: e8510.
60. LiX, OghiKA, ZhangJ, KronesA, BushKT, et al. (2003) Eya protein phosphatase activity regulates Six1-Dach-Eya transcriptional effects in mammalian organogenesis. Nature 426 : 247–254.
61. BakerNE, FirthLC (2011) Retinal determination genes function along with cell-cell signals to regulate Drosophila eye development: examples of multi-layered regulation by master regulators. Bioessays 33 : 538–546.
62. BachEA, EkasLA, Ayala-CamargoA, FlahertyMS, LeeH, et al. (2007) GFP reporters detect the activation of the Drosophila JAK/STAT pathway in vivo. Gene Expr Patterns 7 : 323–331.
63. XuT, RubinGM (1993) Analysis of genetic mosaics in developing and adult Drosophila tissues. Development 117 : 1223–1237.
64. JusiakB, AbulimitiA, HaeltermanN, ChenR, MardonG (2012) MAPK target sites of eyes absent are not required for eye development or survival in Drosophila. PLoS One 7: e50776.
Štítky
Genetika Reprodukční medicína
Článek Re-Ranking Sequencing Variants in the Post-GWAS Era for Accurate Causal Variant IdentificationČlánek Bypass of 8-oxodGČlánek Integrated Model of and Inherited Genetic Variants Yields Greater Power to Identify Risk GenesČlánek Comparative Genomic and Functional Analysis of 100 Strains and Their Comparison with Strain GGČlánek A Nuclear Calcium-Sensing Pathway Is Critical for Gene Regulation and Salt Stress Tolerance inČlánek Computational Identification of Diverse Mechanisms Underlying Transcription Factor-DNA OccupancyČlánek Reversible and Rapid Transfer-RNA Deactivation as a Mechanism of Translational Repression in StressČlánek Genome-Wide Association of Body Fat Distribution in African Ancestry Populations Suggests New Loci
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2013 Číslo 8
-
Všechny články tohoto čísla
- Genome-Wide DNA Methylation Analysis of Systemic Lupus Erythematosus Reveals Persistent Hypomethylation of Interferon Genes and Compositional Changes to CD4+ T-cell Populations
- Re-Ranking Sequencing Variants in the Post-GWAS Era for Accurate Causal Variant Identification
- Histone Variant HTZ1 Shows Extensive Epistasis with, but Does Not Increase Robustness to, New Mutations
- Past Visits Present: TCF/LEFs Partner with ATFs for β-Catenin–Independent Activity
- Functional Characterisation of Alpha-Galactosidase A Mutations as a Basis for a New Classification System in Fabry Disease
- A Flexible Approach for the Analysis of Rare Variants Allowing for a Mixture of Effects on Binary or Quantitative Traits
- Masculinization of Gene Expression Is Associated with Exaggeration of Male Sexual Dimorphism
- Genic Intolerance to Functional Variation and the Interpretation of Personal Genomes
- Endogenous Stress Caused by Faulty Oxidation Reactions Fosters Evolution of 2,4-Dinitrotoluene-Degrading Bacteria
- Transposon Domestication versus Mutualism in Ciliate Genome Rearrangements
- Comparative Anatomy of Chromosomal Domains with Imprinted and Non-Imprinted Allele-Specific DNA Methylation
- An Essential Function for the ATR-Activation-Domain (AAD) of TopBP1 in Mouse Development and Cellular Senescence
- Depletion of Retinoic Acid Receptors Initiates a Novel Positive Feedback Mechanism that Promotes Teratogenic Increases in Retinoic Acid
- Bypass of 8-oxodG
- Calpain-6 Deficiency Promotes Skeletal Muscle Development and Regeneration
- ATM Release at Resected Double-Strand Breaks Provides Heterochromatin Reconstitution to Facilitate Homologous Recombination
- Generation of Tandem Direct Duplications by Reversed-Ends Transposition of Maize Elements
- Loss of a Conserved tRNA Anticodon Modification Perturbs Cellular Signaling
- Integrated Model of and Inherited Genetic Variants Yields Greater Power to Identify Risk Genes
- High-Throughput Genetic and Gene Expression Analysis of the RNAPII-CTD Reveals Unexpected Connections to SRB10/CDK8
- Dynamic Rewiring of the Retinal Determination Network Switches Its Function from Selector to Differentiation
- β-Catenin-Independent Activation of TCF1/LEF1 in Human Hematopoietic Tumor Cells through Interaction with ATF2 Transcription Factors
- Genetic Mapping of Specific Interactions between Mosquitoes and Dengue Viruses
- A Highly Redundant Gene Network Controls Assembly of the Outer Spore Wall in
- Origin and Functional Diversification of an Amphibian Defense Peptide Arsenal
- Myc-Driven Overgrowth Requires Unfolded Protein Response-Mediated Induction of Autophagy and Antioxidant Responses in
- Integrative Modeling of eQTLs and Cis-Regulatory Elements Suggests Mechanisms Underlying Cell Type Specificity of eQTLs
- Species and Population Level Molecular Profiling Reveals Cryptic Recombination and Emergent Asymmetry in the Dimorphic Mating Locus of
- Ras-Induced Changes in H3K27me3 Occur after Those in Transcriptional Activity
- Characterization of the p53 Cistrome – DNA Binding Cooperativity Dissects p53's Tumor Suppressor Functions
- Global Analysis of Fission Yeast Mating Genes Reveals New Autophagy Factors
- Deficiency Suppresses Intestinal Tumorigenesis
- Introns Regulate Gene Expression in in a Pab2p Dependent Pathway
- Meiotic Recombination Initiation in and around Retrotransposable Elements in
- Comparative Oncogenomic Analysis of Copy Number Alterations in Human and Zebrafish Tumors Enables Cancer Driver Discovery
- Comparative Genomic and Functional Analysis of 100 Strains and Their Comparison with Strain GG
- A Model-Based Analysis of GC-Biased Gene Conversion in the Human and Chimpanzee Genomes
- Masculinization of the X Chromosome in the Pea Aphid
- The Architecture of a Prototypical Bacterial Signaling Circuit Enables a Single Point Mutation to Confer Novel Network Properties
- Distinct SUMO Ligases Cooperate with Esc2 and Slx5 to Suppress Duplication-Mediated Genome Rearrangements
- The Yeast Environmental Stress Response Regulates Mutagenesis Induced by Proteotoxic Stress
- Mediator Directs Co-transcriptional Heterochromatin Assembly by RNA Interference-Dependent and -Independent Pathways
- The Genome of and the Basis of Host-Microsporidian Interactions
- Regulation of Sister Chromosome Cohesion by the Replication Fork Tracking Protein SeqA
- Neuronal Reprograming of Protein Homeostasis by Calcium-Dependent Regulation of the Heat Shock Response
- A Nuclear Calcium-Sensing Pathway Is Critical for Gene Regulation and Salt Stress Tolerance in
- Cross-Species Array Comparative Genomic Hybridization Identifies Novel Oncogenic Events in Zebrafish and Human Embryonal Rhabdomyosarcoma
- : A Mouse Strain with an Ift140 Mutation That Results in a Skeletal Ciliopathy Modelling Jeune Syndrome
- The Relative Contribution of Proximal 5′ Flanking Sequence and Microsatellite Variation on Brain Vasopressin 1a Receptor () Gene Expression and Behavior
- Combining Quantitative Genetic Footprinting and Trait Enrichment Analysis to Identify Fitness Determinants of a Bacterial Pathogen
- The Innocence Project at Twenty: An Interview with Barry Scheck
- Computational Identification of Diverse Mechanisms Underlying Transcription Factor-DNA Occupancy
- GUESS-ing Polygenic Associations with Multiple Phenotypes Using a GPU-Based Evolutionary Stochastic Search Algorithm
- H2A.Z Acidic Patch Couples Chromatin Dynamics to Regulation of Gene Expression Programs during ESC Differentiation
- Identification of DSB-1, a Protein Required for Initiation of Meiotic Recombination in , Illuminates a Crossover Assurance Checkpoint
- Binding of TFIIIC to SINE Elements Controls the Relocation of Activity-Dependent Neuronal Genes to Transcription Factories
- Global Analysis of the Sporulation Pathway of
- Genetic Circuits that Govern Bisexual and Unisexual Reproduction in
- Deletion of microRNA-80 Activates Dietary Restriction to Extend Healthspan and Lifespan
- Fifty Years On: GWAS Confirms the Role of a Rare Variant in Lung Disease
- The Enhancer Landscape during Early Neocortical Development Reveals Patterns of Dense Regulation and Co-option
- Gene Expression Regulation by Upstream Open Reading Frames and Human Disease
- Sociogenomics of Cooperation and Conflict during Colony Founding in the Fire Ant
- The Intronic Long Noncoding RNA Recruits PRC2 to the Promoter, Reducing the Expression of and Increasing Cell Proliferation
- The , p.E318G Variant Increases the Risk of Alzheimer's Disease in -ε4 Carriers
- The Wilms Tumor Gene, , Is Critical for Mouse Spermatogenesis via Regulation of Sertoli Cell Polarity and Is Associated with Non-Obstructive Azoospermia in Humans
- Reversible and Rapid Transfer-RNA Deactivation as a Mechanism of Translational Repression in Stress
- QTL Analysis of High Thermotolerance with Superior and Downgraded Parental Yeast Strains Reveals New Minor QTLs and Converges on Novel Causative Alleles Involved in RNA Processing
- Genome Wide Association Identifies Novel Loci Involved in Fungal Communication
- Chromatin Sampling—An Emerging Perspective on Targeting Polycomb Repressor Proteins
- A Recessive Founder Mutation in Regulator of Telomere Elongation Helicase 1, , Underlies Severe Immunodeficiency and Features of Hoyeraal Hreidarsson Syndrome
- Genome-Wide Association of Body Fat Distribution in African Ancestry Populations Suggests New Loci
- Causal and Synthetic Associations of Variants in the Gene Cluster with Alpha1-antitrypsin Serum Levels
- Hard Selective Sweep and Ectopic Gene Conversion in a Gene Cluster Affording Environmental Adaptation
- Brittle Culm1, a COBRA-Like Protein, Functions in Cellulose Assembly through Binding Cellulose Microfibrils
- Chromosomal Copy Number Variation, Selection and Uneven Rates of Recombination Reveal Cryptic Genome Diversity Linked to Pathogenicity
- The Ribosomal Protein Rpl22 Controls Ribosome Composition by Directly Repressing Expression of Its Own Paralog, Rpl22l1
- Ras1 Acts through Duplicated Cdc42 and Rac Proteins to Regulate Morphogenesis and Pathogenesis in the Human Fungal Pathogen
- The DSB-2 Protein Reveals a Regulatory Network that Controls Competence for Meiotic DSB Formation and Promotes Crossover Assurance
- Recurrent Modification of a Conserved -Regulatory Element Underlies Fruit Fly Pigmentation Diversity
- Associations of Mitochondrial Haplogroups B4 and E with Biliary Atresia and Differential Susceptibility to Hydrophobic Bile Acid
- The Conditional Nature of Genetic Interactions: The Consequences of Wild-Type Backgrounds on Mutational Interactions in a Genome-Wide Modifier Screen
- A Critical Function of Mad2l2 in Primordial Germ Cell Development of Mice
- A Role for CF1A 3′ End Processing Complex in Promoter-Associated Transcription
- Vitellogenin Underwent Subfunctionalization to Acquire Caste and Behavioral Specific Expression in the Harvester Ant
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Chromosomal Copy Number Variation, Selection and Uneven Rates of Recombination Reveal Cryptic Genome Diversity Linked to Pathogenicity
- Genome-Wide DNA Methylation Analysis of Systemic Lupus Erythematosus Reveals Persistent Hypomethylation of Interferon Genes and Compositional Changes to CD4+ T-cell Populations
- Associations of Mitochondrial Haplogroups B4 and E with Biliary Atresia and Differential Susceptibility to Hydrophobic Bile Acid
- A Role for CF1A 3′ End Processing Complex in Promoter-Associated Transcription
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



