-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaHow to Choose the Right Mate
article has not abstract
Published in the journal: . PLoS Genet 9(10): e32767. doi:10.1371/journal.pgen.1003881
Category: Perspective
doi: https://doi.org/10.1371/journal.pgen.1003881Summary
article has not abstract
Genetically programmed recombination plays an important role in differentiation, antigenic variation, and evolution in many systems. Mating-type switching is an example of programmed recombination, and studies of both the budding yeast [1] and fission yeast [2] have provided a wealth of knowledge on how epigenetic and genetic machineries interact with each other to control this process.
Fission yeast has evolved a potent mating-type switching process that rapidly establishes a mixed population, containing roughly the same proportion of P (for plus) and M (for minus) cells, thus allowing sexual reproduction of individuals in a clonal cell population. The sexual region is located on the right arm of chromosome 2 and contains three cassettes—the expressed mat1 and the two silent mat2-P and mat3-M loci. Each locus is flanked by the H1 and H2 sequences. The expressed mat1 locus contains the P genes (Pi and Pc) or the M genes (Mi and Mc) that determine the mating type of the cell [3]. The silent loci located 17 kb from mat1 are embedded in a 20 kb heterochromatic domain delimited by two boundary elements and enriched in Swi6/HP1 chromodomain protein [4], [5]. The allele present at mat1 is converted by copying the genetic information from mat2-P or mat3-M silent donor loci by a non-reciprocal homologous recombination process. Mating-type switching is initiated by a DNA strand-specific imprinting located at mat1 at the junction between H1 and the specific mating-type allele that is transformed into a polar double strand DNA break (DSB) during DNA replication [6]. The broken DNA does not use the intact sister chromatid, but instead engages recombinational repair by copying the genetic information located between the H1 and H2 sequence of one of the silent donors [7]. This process allows the DNA replication fork to restart, in order to maintain cellular viability and to switch mating-type [8], [9]. Pedigree analysis of switching indicated that this process is very efficient, such that the mat2-P donor is favored in M cells and mat3-M donor is favored in P cells, with 80% efficiency (reviewed in 2). This intriguing property raises the question of donor preference (i.e., of the directionality of the switching event), which is the topic of a new study by Jakočiūnas et al. [10], published in the current issue of PLOS Genetics.
In an initial study, Thon and Klar [11] exchanged the alleles present at mat2 and mat3 silent donors (mat2-M, mat3-P) and found a strong reduction of switching (20%). In the absence of Swi6 or the machinery for histone H3 lysine 9 methylation [12], [13], the switching efficiency becomes random regardless of donor configuration. Collectively, these results demonstrated that the location of the donors, rather than their content, regulates the choice and indicates that the heterochromatic status of the silent donors is critical for the search of the broken mat1 DNA strand, a process influenced by the mating-type allele present or expressed at mat1. Grewal's lab [14] identified a switching recombination enhancer (SRE3) adjacent to mat3-M that strongly biases the usage of mat3 as a donor in P cells and not in M cells. Jakočiūnas et al. have now identified a second enhancer (SRE2) adjacent to mat2-P. By swapping the silent cassettes, with or without their cognate enhancers, they showed that these enhancers compete with each other. However, both enhancers are not equivalent with respect to Swi6. Two other switching factors, Swi2 and Swi5 are known to work in the same switching step as Swi6 [15]. Swi2 and Swi5 form a recombination mediator complex. Swi5 is required for general recombination, and Swi2 is specific for mating-type switching, and interacts with itself, Swi5, Swi6, and Rad51, a central protein essential for homologous recombination [16]. In P cells, Swi2/5 is bound to the switching recombination enhancer (SRE3) element, located next to the silent mat3-M locus, independently of Swi6. In M cells, Swi2/5 covers the entire silent region and reaches mat2-P in a Swi6-dependent fashion [14]. The Grewal lab proposed a spreading model, whereby Swi2/5 complex anchored at SRE3 in P cells will slide onto the Swi6 protein to reach mat2-P in M cells. More recently, Grewal's [17] and Klar's [18] laboratories found that the mat1-Mc transcription factor, together with the CENP-B homolog, Abp1 [19], bind the swi2 gene and regulate its expression. Their results support the notion that the differential distribution of Swi2/5 complex is controlled, at least in part, by mat1-Mc cell type-specific transcription factor. However, it is not clear how the distribution of Swi2/5 over the silent region restricts the usage of mat2-P as a donor in M cells. The discovery of the second enhancer thus simplifies the model for directionality (summarized in Figure 1) without the assumption of spreading. Moreover, by using a careful and laborious statistical approach, Jakočiūnas et al. observed a large fluctuation of P/M colonies in swi2Δ and swi5Δ mutant strains, indicative of inefficient switching, rather than solely a defect in directionality.
Fig. 1. Fission yeast cells switch mating-type in a directional manner, by gene conversions of the mat1 locus. 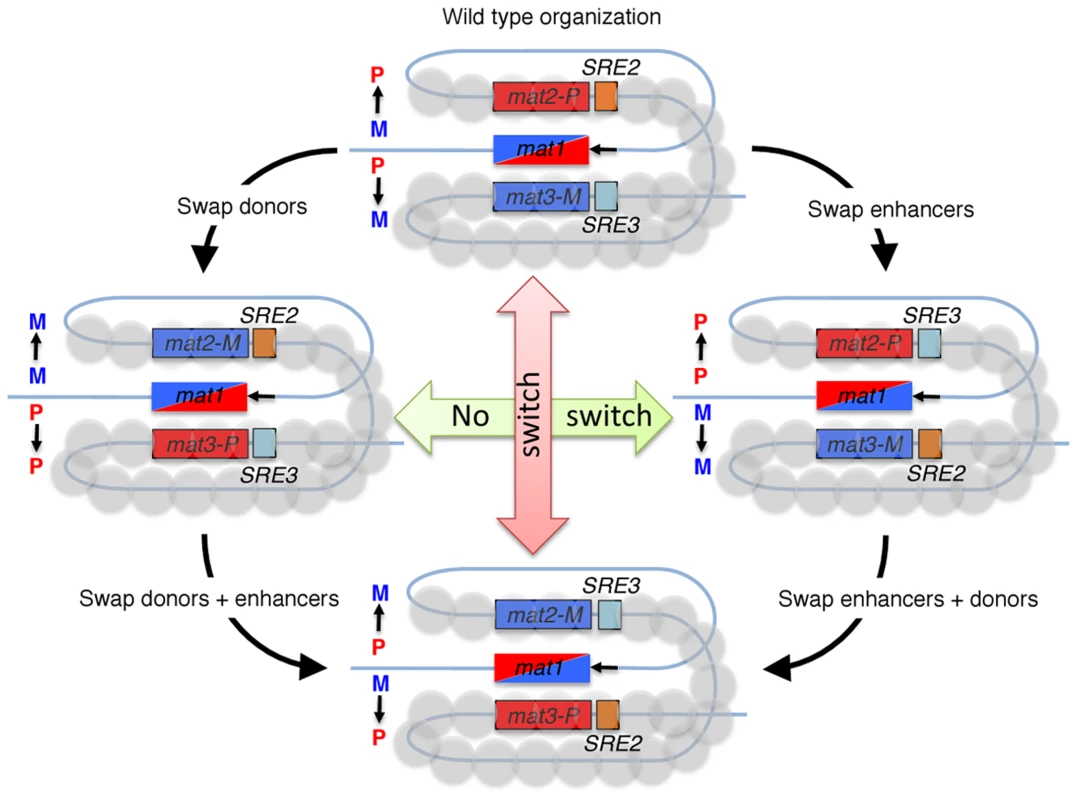
A recombinogenic DNA end is formed at mat1 during DNA replication (black arrow) and the broken DNA invades a donor whose genetic information is copied into mat1. Wild-type M cells (mat1-M allele) use the mat2-P donor, while P cells (mat1-P allele) use the mat3-M donor, as depicted in the top drawing. The recombination enhancers SRE2 and SRE3 are central to these choices. Experiments in which donors and enhancers are swapped, alone or in combination, show that SRE2 and SRE3 are recognized in a cell-type specific manner to promote use of their adjacent donor. The heterochromatic structure of the mat2-P–mat3-M region is required for this differential recognition. It is important to recall that the Schizosaccharomyces pombe mutant strain containing a deletion of the mat2-P and mat3-M region remains fully viable, since the sister chromatid provides the template for mat1 DSB repair [20]. Thus, the break at mat1 in the absence of Swi2 (or Swi5) is more likely repaired off the sister chromatid than off the silent donors. As a consequence, the initial allele present at mat1 of the seeding cells will bias the overall mating-type of the colony, possibly imposing the fluctuation shown by Jakočiūnas et al. Another relevant observation is that the second recombination mediator complex, Rad55/Rad57, also participates in the mating-type switching process [9].
At this stage, it is becoming increasingly clear that several challenging experiments will be necessary to further investigate the mechanism of directionality in fission yeast. State-of-the-art chromosome conformation-capture and classical functional approaches will have to be designed. Moreover, since the heterochromatin association of Swi6/HP1 is regulated during the cell cycle and mating-type switching is triggered only in S-phase, it is important to determine when and how the differential distribution of Swi2/5 (and Rad55/57) complexes is achieved and becomes functional.
Zdroje
1. HaberJE (2012) Mating-type genes and MAT switching in Saccharomyces cerevisiae. Genetics 191 : 33–64.
2. KlarAJ (2007) Lessons learned from studies of fission yeast mating-type switching and silencing. Annu Rev Genet 41 : 213–236.
3. KellyM, BurkeJ, SmithM, KlarA, BeachD (1988) Four mating-type genes control sexual differentiation in the fission yeast. EMBO J 7 : 1537–1547.
4. LorentzA, OstermannK, FleckO, SchmidtH (1994) Switching gene swi6, involved in repression of silent mating-type loci in fission yeast, encodes a homologue of chromatin-associated proteins from Drosophila and mammals. Gene 143 : 139–43.
5. EkwallK, JaverzatJP, LorentzA, SchmidtH, CranstonG, AllshireR (1995) The chromodomain protein Swi6: a key component at fission yeast centromeres. Science 269 : 1429–31.
6. ArcangioliB, de LahondesR (2000) Fission yeast switches mating type by a replication-recombination coupled process. EMBO J. 2000 Mar 15;19(6):1389–96. EMBO J 17 : 4503–4510.
7. HolmesAM, KaykovA, ArcangioliB (2005) Molecular and cellular dissection of mating-type switching steps in Schizosaccharomyces pombe. Mol Cell Biol 25 : 303–311.
8. ArcangioliB (2000) Fate of mat1 DNA strands during mating-type switching in fission yeast. EMBO Rep 1 : 145–150.
9. RoseaulinL, YamadaY, TsutsuiY, RussellP, IwasakiH, ArcangioliB (2008) Mus81 is essential for sister chromatid recombination at broken replication forks. EMBO J 27 : 1378–87.
10. JakočiūnasT, HolmLR, Verhein-HansenJ, TrusinaA, ThonG (2013) Two portable recombination enhancers direct donor choice in fission yeast heterochromatin. PLoS Genet 9: e1003762 doi:10.1371/journal.pgen.1003762
11. ThonG, KlarAJ (1993) Directionality of fission yeast mating-type interconversion is controlled by the location of the donor loci. Genetics 134 : 1045–1054.
12. NomaK, AllisCD, GrewalSI (2001) Transitions in distinct histone H3 methylation patterns at the heterochromatin domain boundaries. Science 293 : 1150–1155.
13. ThonG, HansenKR, AltesSP, SidhuD, SinghG, Verhein-HansenJ, BonaduceMJ, KlarAJ (2005) The Clr7 and Clr8 directionality factors and the Puc4 cullin mediate heterochromatin formation in fission yeast Schizosaccharomyces pombe. Genetics 171 : 1583–95.
14. JiaS, YamadaT, GrewalSI (2004) Heterochromatin regulates cell type-specific long-range chromatin interactions essential for directed recombination. Cell 119 : 469–480.
15. EgelR, BeachDH, KlarAJ (1984) Genes required for initiation and resolution steps of mating-type switching in fission yeast. Proc Natl Acad Sci USA 81 : 3481–3485.
16. AkamatsuY, TsutsuiY, MorishitaT, SiddiqueMS, KurokawaY, et al. (2007) Fission yeast Swi5/Sfr1 and Rhp55/Rhp57 differentially regulate Rhp51-dependent recombination outcomes. EMBO J 26 : 1352–1362.
17. MatsudaE, Sugioka-SugiyamaR, MizuguchiT, MehtaS, CuiB, et al. (2011) A homolog of male sex-determining factor SRY cooperates with a transposon-derived CENP-B protein to control sex-specific directed recombination. Proc Natl Acad Sci USA 108 : 18754–18759.
18. YuC, BonaduceMJ, KlarAJ (2012) Going in the right direction: mating-type switching of Schizosaccharomyces pombe is controlled by judicious expression of two different swi2 transcripts. Genetics 190 : 977–987.
19. Aguilar-ArnalL, MarsellachFX, AzorínF (2008) The fission yeast homologue of CENP - B, Abp1, regulates directionality of mating-type switching. EMBO J 27 : 1029–1038.
20. KlarAJ, MiglioLM (1986) Initiation of meiotic recombination by double-strand DNA breaks in S. pombe. Cell 46 : 725–731.
Štítky
Genetika Reprodukční medicína
Článek Defending Sperm FunctionČlánek Conserved Translatome Remodeling in Nematode Species Executing a Shared Developmental TransitionČlánek Genome-Wide and Cell-Specific Epigenetic Analysis Challenges the Role of Polycomb in SpermatogenesisČlánek The Integrator Complex Subunit 6 (Ints6) Confines the Dorsal Organizer in Vertebrate EmbryogenesisČlánek Multiple bHLH Proteins form Heterodimers to Mediate CRY2-Dependent Regulation of Flowering-Time inČlánek Playing the Field: Sox10 Recruits Different Partners to Drive Central and Peripheral MyelinationČlánek A Minimal Nitrogen Fixation Gene Cluster from sp. WLY78 Enables Expression of Active Nitrogenase inČlánek Evolutionary Tuning of Protein Expression Levels of a Positively Autoregulated Two-Component System
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2013 Číslo 10
-
Všechny články tohoto čísla
- Defending Sperm Function
- How to Choose the Right Mate
- A Mutation in the Gene in Labrador Retrievers with Hereditary Nasal Parakeratosis (HNPK) Provides Insights into the Epigenetics of Keratinocyte Differentiation
- Conserved Translatome Remodeling in Nematode Species Executing a Shared Developmental Transition
- A Novel Actin mRNA Splice Variant Regulates ACTG1 Expression
- Tracking Proliferative History in Lymphocyte Development with Cre-Mediated Sister Chromatid Recombination
- Correlated Occurrence and Bypass of Frame-Shifting Insertion-Deletions (InDels) to Give Functional Proteins
- Chimeric Protein Complexes in Hybrid Species Generate Novel Phenotypes
- Loss of miR-10a Activates and Collaborates with Activated Wnt Signaling in Inducing Intestinal Neoplasia in Female Mice
- Both Rare and Copy Number Variants Are Prevalent in Agenesis of the Corpus Callosum but Not in Cerebellar Hypoplasia or Polymicrogyria
- Reverse PCA, a Systematic Approach for Identifying Genes Important for the Physical Interaction between Protein Pairs
- Partial Deletion of Chromosome 8 β-defensin Cluster Confers Sperm Dysfunction and Infertility in Male Mice
- Genome-Wide and Cell-Specific Epigenetic Analysis Challenges the Role of Polycomb in Spermatogenesis
- Coordinate Regulation of Mature Dopaminergic Axon Morphology by Macroautophagy and the PTEN Signaling Pathway
- Cooperation between RUNX1-ETO9a and Novel Transcriptional Partner KLF6 in Upregulation of in Acute Myeloid Leukemia
- Mobility of the Native Conjugative Plasmid pLS20 Is Regulated by Intercellular Signaling
- FliZ Is a Global Regulatory Protein Affecting the Expression of Flagellar and Virulence Genes in Individual Bacterial Cells
- Specific Tandem Repeats Are Sufficient for Paramutation-Induced Trans-Generational Silencing
- Condensin II Subunit dCAP-D3 Restricts Retrotransposon Mobilization in Somatic Cells
- Dominant Mutations in Identify the Mlh1-Pms1 Endonuclease Active Site and an Exonuclease 1-Independent Mismatch Repair Pathway
- The Insulator Homie Promotes Expression and Protects the Adjacent Gene from Repression by Polycomb Spreading
- Human Intellectual Disability Genes Form Conserved Functional Modules in
- Coordination of Cell Proliferation and Cell Fate Determination by CES-1 Snail
- ORFs in Drosophila Are Important to Organismal Fitness and Evolved Rapidly from Previously Non-coding Sequences
- Different Roles of Eukaryotic MutS and MutL Complexes in Repair of Small Insertion and Deletion Loops in Yeast
- The Spore Differentiation Pathway in the Enteric Pathogen
- Acceleration of the Glycolytic Flux by Steroid Receptor Coactivator-2 Is Essential for Endometrial Decidualization
- The Human Nuclear Poly(A)-Binding Protein Promotes RNA Hyperadenylation and Decay
- Genome Wide Analysis Reveals Zic3 Interaction with Distal Regulatory Elements of Stage Specific Developmental Genes in Zebrafish
- Xbp1 Directs Global Repression of Budding Yeast Transcription during the Transition to Quiescence and Is Important for the Longevity and Reversibility of the Quiescent State
- The Integrator Complex Subunit 6 (Ints6) Confines the Dorsal Organizer in Vertebrate Embryogenesis
- Incorporating Motif Analysis into Gene Co-expression Networks Reveals Novel Modular Expression Pattern and New Signaling Pathways
- The Bacterial Response Regulator ArcA Uses a Diverse Binding Site Architecture to Regulate Carbon Oxidation Globally
- Direct Monitoring of the Strand Passage Reaction of DNA Topoisomerase II Triggers Checkpoint Activation
- Multiple bHLH Proteins form Heterodimers to Mediate CRY2-Dependent Regulation of Flowering-Time in
- A Reversible Histone H3 Acetylation Cooperates with Mismatch Repair and Replicative Polymerases in Maintaining Genome Stability
- ALS-Associated Mutations Result in Compromised Alternative Splicing and Autoregulation
- Robust Demographic Inference from Genomic and SNP Data
- Preferential Binding to Elk-1 by SLE-Associated Risk Allele Upregulates Expression
- Rad52 Sumoylation Prevents the Toxicity of Unproductive Rad51 Filaments Independently of the Anti-Recombinase Srs2
- The Serum Resistome of a Globally Disseminated Multidrug Resistant Uropathogenic Clone
- Identification of 526 Conserved Metazoan Genetic Innovations Exposes a New Role for Cofactor E-like in Neuronal Microtubule Homeostasis
- SUMO Localizes to the Central Element of Synaptonemal Complex and Is Required for the Full Synapsis of Meiotic Chromosomes in Budding Yeast
- Integrated Enrichment Analysis of Variants and Pathways in Genome-Wide Association Studies Indicates Central Role for IL-2 Signaling Genes in Type 1 Diabetes, and Cytokine Signaling Genes in Crohn's Disease
- Genome-Wide High-Resolution Mapping of UV-Induced Mitotic Recombination Events in
- Genome-Wide Analysis of Cell Type-Specific Gene Transcription during Spore Formation in
- Playing the Field: Sox10 Recruits Different Partners to Drive Central and Peripheral Myelination
- Two Portable Recombination Enhancers Direct Donor Choice in Fission Yeast Heterochromatin
- Mining the Human Phenome Using Allelic Scores That Index Biological Intermediates
- Yeast Tdh3 (Glyceraldehyde 3-Phosphate Dehydrogenase) Is a Sir2-Interacting Factor That Regulates Transcriptional Silencing and rDNA Recombination
- A Minimal Nitrogen Fixation Gene Cluster from sp. WLY78 Enables Expression of Active Nitrogenase in
- A Review of Bacteria-Animal Lateral Gene Transfer May Inform Our Understanding of Diseases like Cancer
- High Throughput Sequencing Reveals Alterations in the Recombination Signatures with Diminishing Spo11 Activity
- Partitioning the Heritability of Tourette Syndrome and Obsessive Compulsive Disorder Reveals Differences in Genetic Architecture
- Eleven Candidate Susceptibility Genes for Common Familial Colorectal Cancer
- A GDF5 Point Mutation Strikes Twice - Causing BDA1 and SYNS2
- Systematic Unraveling of the Unsolved Pathway of Nicotine Degradation in
- Natural Genetic Variation of Integrin Alpha L () Modulates Ischemic Brain Injury in Stroke
- Evolutionary Tuning of Protein Expression Levels of a Positively Autoregulated Two-Component System
- Evolutionary Change within a Bipotential Switch Shaped the Sperm/Oocyte Decision in Hermaphroditic Nematodes
- Limiting of the Innate Immune Response by SF3A-Dependent Control of MyD88 Alternative mRNA Splicing
- Multiple Signaling Pathways Coordinate to Induce a Threshold Response in a Chordate Embryo
- Distinct Regulatory Mechanisms Act to Establish and Maintain Pax3 Expression in the Developing Neural Tube
- Genome Wide Analysis of Narcolepsy in China Implicates Novel Immune Loci and Reveals Changes in Association Prior to Versus After the 2009 H1N1 Influenza Pandemic
- Mismatch Repair Genes and Modify CAG Instability in Huntington's Disease Mice: Genome-Wide and Candidate Approaches
- The Histone H3 K27 Methyltransferase KMT6 Regulates Development and Expression of Secondary Metabolite Gene Clusters
- Hsp70-Hsp40 Chaperone Complex Functions in Controlling Polarized Growth by Repressing Hsf1-Driven Heat Stress-Associated Transcription
- Function and Evolution of DNA Methylation in
- Stimulation of mTORC1 with L-leucine Rescues Defects Associated with Roberts Syndrome
- Transcription Termination and Chimeric RNA Formation Controlled by FPA
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Dominant Mutations in Identify the Mlh1-Pms1 Endonuclease Active Site and an Exonuclease 1-Independent Mismatch Repair Pathway
- Eleven Candidate Susceptibility Genes for Common Familial Colorectal Cancer
- The Histone H3 K27 Methyltransferase KMT6 Regulates Development and Expression of Secondary Metabolite Gene Clusters
- A Mutation in the Gene in Labrador Retrievers with Hereditary Nasal Parakeratosis (HNPK) Provides Insights into the Epigenetics of Keratinocyte Differentiation
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



