-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaA Minimal Nitrogen Fixation Gene Cluster from sp. WLY78 Enables Expression of Active Nitrogenase in
Most biological nitrogen fixation is catalyzed by molybdenum-dependent nitrogenase, an enzyme complex comprising two component proteins that contains three different metalloclusters. Diazotrophs contain a common core of nitrogen fixation nif genes that encode the structural subunits of the enzyme and components required to synthesize the metalloclusters. However, the complement of nif genes required to enable diazotrophic growth varies significantly amongst nitrogen fixing bacteria and archaea. In this study, we identified a minimal nif gene cluster consisting of nine nif genes in the genome of Paenibacillus sp. WLY78, a gram-positive, facultative anaerobe isolated from the rhizosphere of bamboo. We demonstrate that the nif genes in this organism are organized as an operon comprising nifB, nifH, nifD, nifK, nifE, nifN, nifX, hesA and nifV and that the nif cluster is under the control of a σ70 (σA)-dependent promoter located upstream of nifB. To investigate genetic requirements for diazotrophy, we transferred the Paenibacillus nif cluster to Escherichia coli. The minimal nif gene cluster enables synthesis of catalytically active nitrogenase in this host, when expressed either from the native nifB promoter or from the T7 promoter. Deletion analysis indicates that in addition to the core nif genes, hesA plays an important role in nitrogen fixation and is responsive to the availability of molybdenum. Whereas nif transcription in Paenibacillus is regulated in response to nitrogen availability and by the external oxygen concentration, transcription from the nifB promoter is constitutive in E. coli, indicating that negative regulation of nif transcription is bypassed in the heterologous host. This study demonstrates the potential for engineering nitrogen fixation in a non-nitrogen fixing organism with a minimum set of nine nif genes.
Published in the journal: . PLoS Genet 9(10): e32767. doi:10.1371/journal.pgen.1003865
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1003865Summary
Most biological nitrogen fixation is catalyzed by molybdenum-dependent nitrogenase, an enzyme complex comprising two component proteins that contains three different metalloclusters. Diazotrophs contain a common core of nitrogen fixation nif genes that encode the structural subunits of the enzyme and components required to synthesize the metalloclusters. However, the complement of nif genes required to enable diazotrophic growth varies significantly amongst nitrogen fixing bacteria and archaea. In this study, we identified a minimal nif gene cluster consisting of nine nif genes in the genome of Paenibacillus sp. WLY78, a gram-positive, facultative anaerobe isolated from the rhizosphere of bamboo. We demonstrate that the nif genes in this organism are organized as an operon comprising nifB, nifH, nifD, nifK, nifE, nifN, nifX, hesA and nifV and that the nif cluster is under the control of a σ70 (σA)-dependent promoter located upstream of nifB. To investigate genetic requirements for diazotrophy, we transferred the Paenibacillus nif cluster to Escherichia coli. The minimal nif gene cluster enables synthesis of catalytically active nitrogenase in this host, when expressed either from the native nifB promoter or from the T7 promoter. Deletion analysis indicates that in addition to the core nif genes, hesA plays an important role in nitrogen fixation and is responsive to the availability of molybdenum. Whereas nif transcription in Paenibacillus is regulated in response to nitrogen availability and by the external oxygen concentration, transcription from the nifB promoter is constitutive in E. coli, indicating that negative regulation of nif transcription is bypassed in the heterologous host. This study demonstrates the potential for engineering nitrogen fixation in a non-nitrogen fixing organism with a minimum set of nine nif genes.
Introduction
Although fixed nitrogen plays a critical role in the global food supply, overuse of chemical nitrogen fertilizers has led to increased costs for farmers and harmful consequences for the environment and human health. Biological nitrogen fixation, the conversion of atmospheric N2 to NH3, offers a natural means of providing nitrogen for plants [1]. There has been a long-standing interest in reducing dependence on fertilizers through engineering non-legume crops that “fix” nitrogen but maintain growth yields [2], [3]. Achieving this goal will require elucidating the minimal number of genes required to sustain biological nitrogen fixation.
Most biological nitrogen fixation is catalyzed by molybdenum-dependent nitrogenase, which is distributed within bacteria and archaea. This enzyme is composed of two component proteins, MoFe protein and Fe protein. The MoFe protein component is an α2β2 heterotetramer (encoded by nifD and nifK) that contains two metalloclusters; FeMo-co, a [Mo-7Fe-9S-C-homocitrate] cluster which serves as the active site of substrate binding and reduction and the P-cluster, a [8Fe-7S] cluster which shuttles electrons to FeMo-co. The Fe protein (encoded by nifH) is a homodimer bridged by an intersubunit [4Fe-4S] cluster that serves as the obligate electron donor to the MoFe protein. The assembly pathway for the biosynthesis of nitrogenase is complex. Apart from the structural subunits encoded by nifH, nifD and nifK, several genes are required for the biosynthesis of the metalloclusters, in addition to other gene products necessary to produce a fully functional enzyme. It is now well established from genetic and biochemical analysis that nifE nifN, nifX nifB, nifQ, nifV, nifY and nifH contribute to the synthesis and insertion of FeMo-co into nitrogenase, that nifU nifS and nifZ play an important role in synthesis of metalloclusters and that nifM is required for proper folder of nitrogenase Fe protein [4]–[7].
The inventory of genes required for diazotrophy varies greatly amongst species, dependent upon the environmental niche and physiology of the host. For example, in Klebsiella oxytoca, twenty nif genes are co-located within a ∼24 kb cluster [8], whereas in Azotobacter vinelandii the nif genes are more dispersed and distributed as two clusters in the genome [9] (Figure 1). However, in contrast to these paradigm diazotrophs, other nitrogen fixing organisms possess a more restricted nif gene set, for example the archeon, Methanococcus maripaludis, contains only 9 nif genes (Figure 1), two of which nifI1 and nifI2, are not essential for nitrogen fixation, but serve a regulatory function [10]. Analysis of the distribution of nif gene sequences within microbial genomes indicates that nearly all diazotrophs have a minimal gene set consisting of six conserved genes nifH, nifD, nifK, nifE, nifN, and nifB [11]. This concurs with the minimal catalytic core required to assemble FeMo-co in vitro [12].
Fig. 1. Comparison of the Paenibacillus sp. WLY78 nif gene cluster with representative clusters from diverse diazotrophic bacteria and archaea. 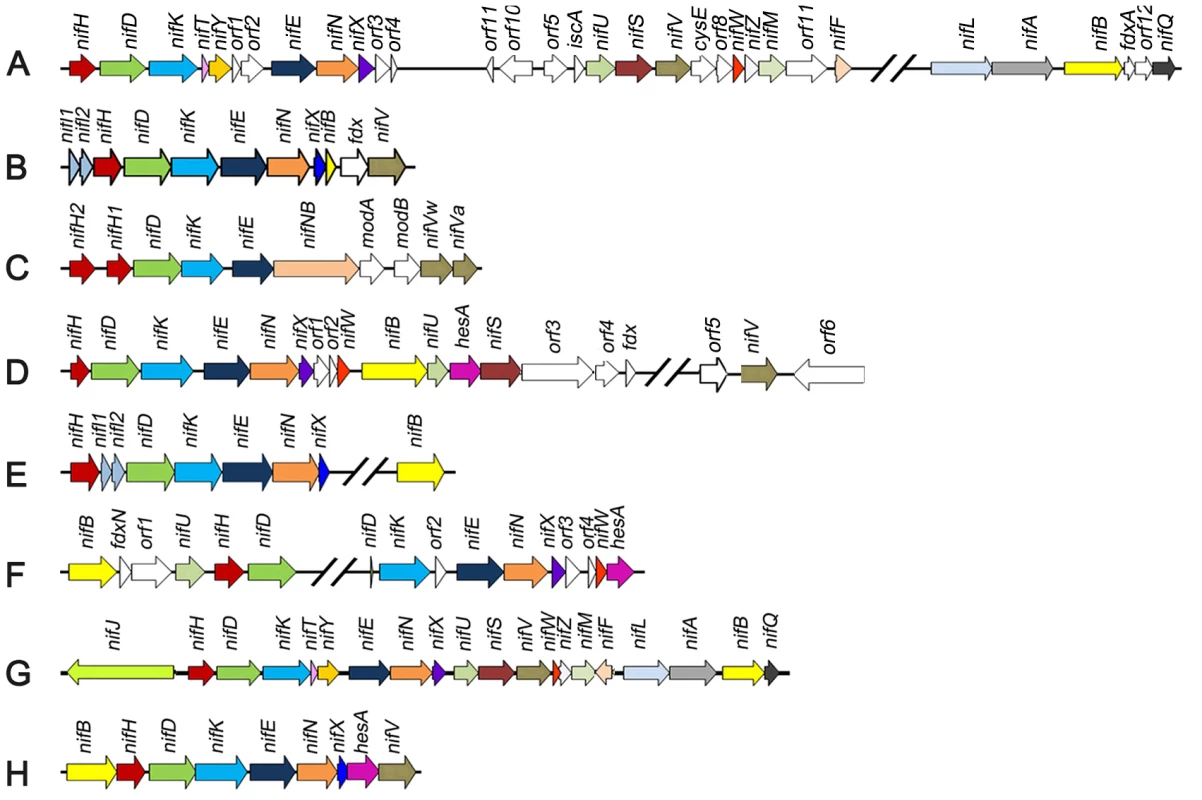
(A) Azotobacter vinelandii, (B) Heliobacterium chlorum, (C) Clostridium acetobutylicum W5, (D) Frankia sp. EAN1pec, (E) Methanococus maripaludis, (F) Anabaena variabilis ATCC 29413, (G) Klebsiella oxytoca M5al, (H) Paenibacillus sp. WLY78. One of the difficulties in determining the precise genetic requirements for nitrogen fixation in diazotrophs arises from the presence of “housekeeping” counterparts in the genome that may substitute for the function of known nif genes. This may be particularly important in the case of diazotrophs that possess minimal nif gene clusters. One approach to investigate the inventory of genes required for diazotrophy in such cases is to transfer the nif cluster to a distantly related organism that does not have the capacity to fix nitrogen. Escherichia coli provides an important model organism for such studies as physiology and gene function is extremely well understood. Since transfer of the complete cluster of 20 nif genes from K. oxytoca to E. coli confers the ability to fix nitrogen [13], we were interested to determine whether a more evolutionary distant nif gene cluster would also enable nitrogenase activity in E. coli. In this study, we identified a minimal nif cluster consisting of nine genes, in the genome of Paenibacillus sp. WLY78 (Figure 1). The cluster is apparently transcribed from a single σA (σ70)-like promoter that functions in E. coli to express active nitrogenase. Our results may have important implications for future engineering of nitrogen fixation in non-diazotrophs.
Results
Genome sequencing of Paenibacillus sp. WLY78 identifies a minimal nitrogen fixation (nif) gene cluster
Paenibacillus sp. WLY78 is a gram-positive, facultative anaerobic, endospore-forming bacterium isolated from the rhizosphere of bamboo [14]. This bacterium has potential use in agriculture, since it is able to fix nitrogen and also produces antimicrobial substances. We therefore determined the genome sequence of this organism and identified a nitrogen fixation gene cluster consisting of nine genes arranged within a 10.5 kb region in the order, nifB, nifH, nifD, nifK, nifE, nifN, nifX, hesA and nifV (Figure 1). The nif cluster is flanked by genes coding for a hypothetical protein upstream and an ABC transporter downstream. The G+C content of this nif cluster was higher than the average of the entire genome (52.8% vs. 45.1%), suggesting that it may have been acquired by horizontal gene transfer. The Paenibacillus sp. WLY78 nif cluster is one of the most compact compared with other dizaotrophs described to date (Figure 1). Similar nif gene arrangements and neighborhoods are observed in other Paenibacillus strains, including Paenibacillus terrae HPL-003. Multiple alignments revealed that the predicted protein products of the Paenibacillus nif genes showed 67–80% identity to the corresponding nif gene products of their gram-positive counterparts [15], but showed only 35–69% identity to the corresponding nif genes of K. oxytoca and A. vinelandii (Table S1). The gene designated as hesA, which is located between nifX and nifV is found in other nif clusters (Figure 1) and the predicted product shares ∼45% identity with the putative molybdenum cofactor biosynthesis protein HesA of Frankia alni ACN14a [16] and Cyanothece sp. ATCC 51142 [17]. HesA is a member of the ThiF-MoeB-HesA family and contains an N-terminal nucleotide binding domain and a C-terminal MoeZ/MoeB-like domain.
RT-PCR experiments using primers designed to span across intergenic regions indicated that the nine genes within the nif cluster are organized in a single operon (Figure 2). Single operon nif clusters have been reported in gram-positive prokaryotes and in the archaea, e.g. Heliobacterium chlorum [18] and Methanococcus maripaludis [19]. However, in contrast to these nif clusters Paenibacillus sp. WLY78 does not contain the negative regulatory genes nifI1 and nifI2 (homologues of glnB), which are involved in post-translational regulation of nitrogenase activity in response to fixed nitrogen [10].
Fig. 2. The nif genes of Paenibacillus sp. WLY78 are organized in an operon as determined by RT-PCR. 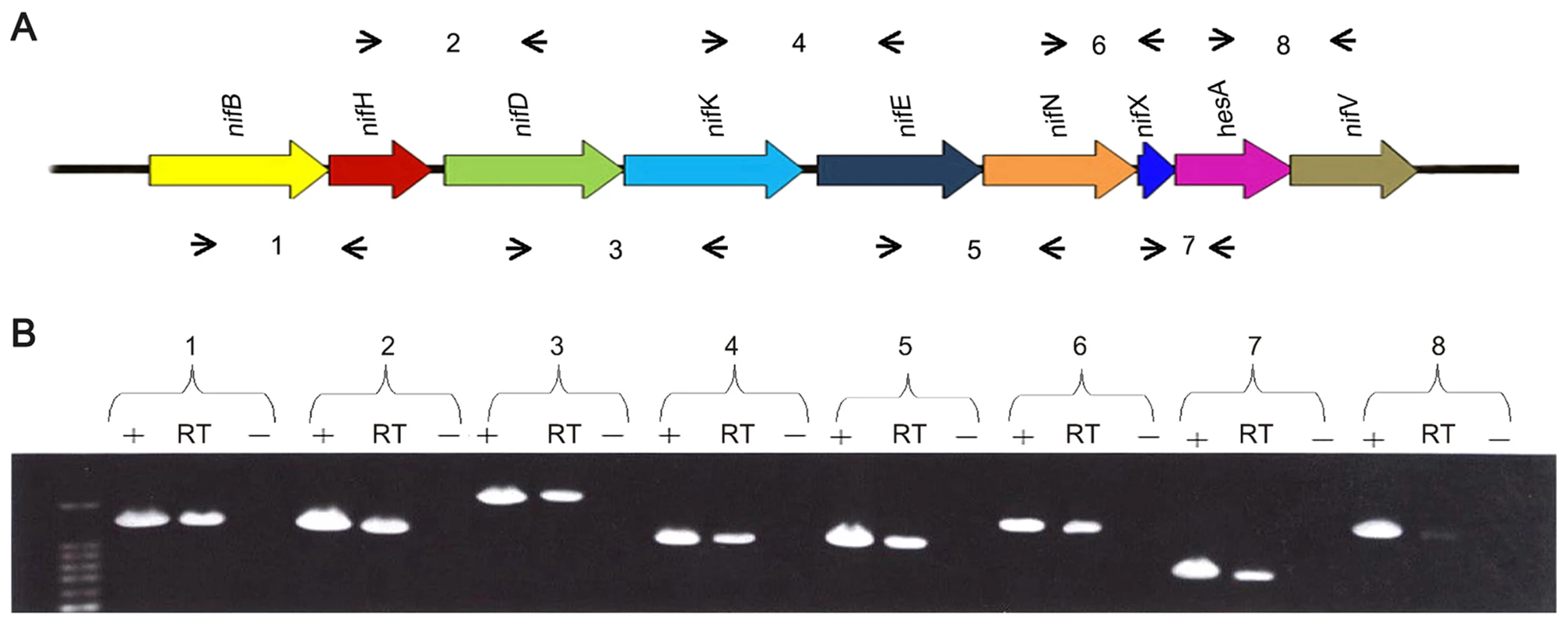
(A) Outline of the strategy. Primers used and amplified products (numbered) are given below the schematic representation of the genes. (B) Result of RT-PCR reactions with RNA from Paenibacillus sp. WLY78 grown under N2-fixing conditions. The numbering on the top of the gels corresponds to the product numbers drawn schematically in the outline given above. RT, standard RT-PCR reaction; (−), negative control in which no reverse transcriptase was added to the RT reaction; (+), positive control in which genomic DNA was used as template in the RT-PCR. Characterization of the Paenibacillus sp. WLY78 nif promoter and transcription unit
The transcriptional start site (TSS) of the nif gene cluster in Paenibacillus sp. WLY78 was determined by using the 5′-RACE (Rapid Amplification of cDNA Ends) method. The TSS was located 59 bp upstream of the translational start site of nifB and a putative promoter was identified 6 nucleotides preceding the TSS (Figure 3). The −35 (TTGACT) and −10 (TAAGAT) sequences in the nifB promoter were similar to the corresponding consensus sequences (TTGACA and TATAAT respectively) of E. coli σ70-dependent promoters. Unlike other members of the Bacillales, the Paenibacillus sp. WLY78 genome does not contain a homolog of rpoN and consequently σ54-dependent −24/−12 promoter sequences were not observed either upstream of the nif cluster or in the 5′ regions of other genes in the Paenibacillus sp. WLY78 genome (data not shown). Downstream of nifV, a potential transcriptional termination site was identified, containing two potential stem loops followed by a T-rich region (Figure 3B). These findings indicate that the nif genes in Paenibacillus sp. WLY78 are organized as a single operon containing 9 genes, which is transcribed from an rpoD-dependent promoter.
Fig. 3. Characterization of the nif promoter of Paenibacillus sp. WLY78. 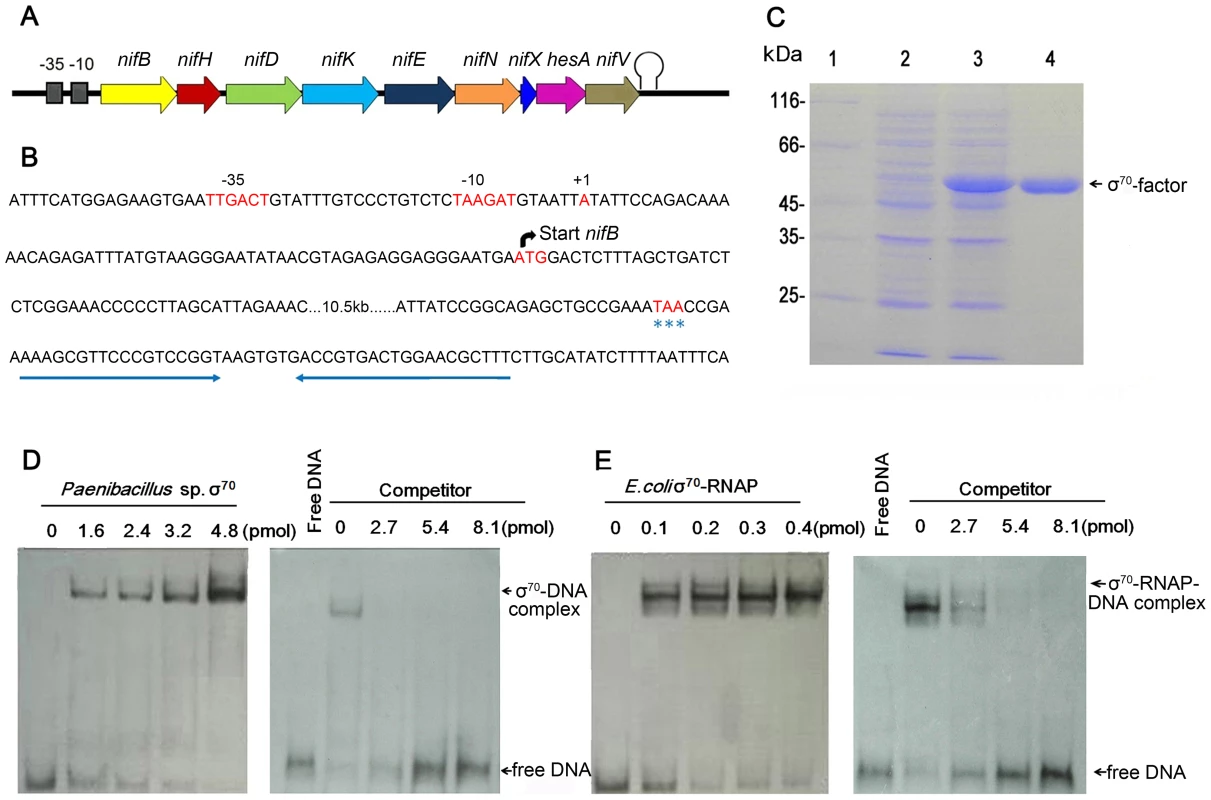
(A) Schematic representation of the Paenibacillus sp. WLY78 nif operon. (B) Nucleotide sequence of the nifB promoter and the putative terminator sequence flanking the 3′ end of nifV. The asterisks below TAA indicate the nifV stop codon. (C) Overexpression and purification of σ70 from Paenibacillus sp. WLY78. Lane 1: protein marker; lane 2: uninduced protein; lane 3: induced protein; lanes 4: purified σ70 factor. (D) Electrophoretic mobility shift assays (EMSA) demonstrating binding of Paenibacillus σ70 to the 50 bp nifB promoter DNA fragment (final concentration 0.03 pmol). The protein concentration is indicated in pmol above each lane (left hand panel). In the right hand panel, the protein concentration was maintained at 2.4 pmol and unlabeled nifB promoter fragment was added as competitor (concentration indicated above each lane). (E) EMSA experiments demonstrating binding of E. coli σ70-RNAP to the 50 bp nifB promoter DNA fragment (final concentration 0.03 pmol). The protein concentration is indicated in pmol above each lane (left hand panel). In the right hand panel, the protein concentration was maintained at 0.2 pmol and unlabeled nifB promoter fragment was added as competitor (concentration indicated above each lane). To analyze the σ70-dependentcy of the nifB promoter, electrophoretic mobility shift assays (EMSA) were carried out using either E. coli σ70-RNAP (RNA polymerase) or σ70 from Paenibacillus sp. WLY78, which was overexpressed and purified from E. coli (Figure 3C). EMSA experiments revealed that both purified σ70 from Paenibacillus sp. WLY78 and E. coli σ70-RNAP holoenzyme bind to the 50 bp nifB promoter fragment. Competition experiments with non-labelled nifB DNA indicated that the E. coli RNAP holoenzyme binds more tightly to this DNA fragment, since higher concentrations of competitor were apparently required to dissociate the E. coli σ70-RNAP (Figure 3, panels D and E). EMSA experiments with a scrambled double-stranded oligonucleotide did not reveal binding of either protein (data not shown). These results are consistent with the ability of σA (σ70) of Bacillus subtilis to bind to promoters independent of core RNAP [20], [21].
To further examine the specificity of binding of E. coli σ70-RNAP to the Paenibacillus sp. WLY78 nifB promoter, we made substitutions in the −35 (TTGACT to GCTACT) and −10 (TAAGAT to GCAGAC) regions of the promoter (Figure 4A). Binding of E. coli σ70-RNAP to the nifB promoter fragment was weakened considerably by the presence of the −35 and −10 substitutions (compare Figure 4, panels B and C), suggesting that E. coli σ70-RNAP specifically interacts with the nifB promoter from Paenibacillus sp. WLY78. In order to confirm this, we performed DNAse I footprinting with a fluorescently labeled 319 bp DNA target carrying the nifB promoter and analyzed the digested DNA fragments using a capillary sequencer. As expected, the region protected from DNAse I digestion corresponded to the nifB promoter, confirming that E. coli σ70-RNAP specifically binds to the −35 and −10 regions upstream of the transcription start site. (Figure 4D). Our studies thus demonstrate that the nifB promoter of Paenibacillus sp. WLY78 is σ70-dependent and thus distinct from the typical σ54-dependent −24/−12 promoters found upstream of nif genes in gram-negative diazotrophs.
Fig. 4. E. coli σ70-RNAP binds preferentially to the −35 region and −10 region of the nifB promoter of Paenibacillus sp. WLY78. 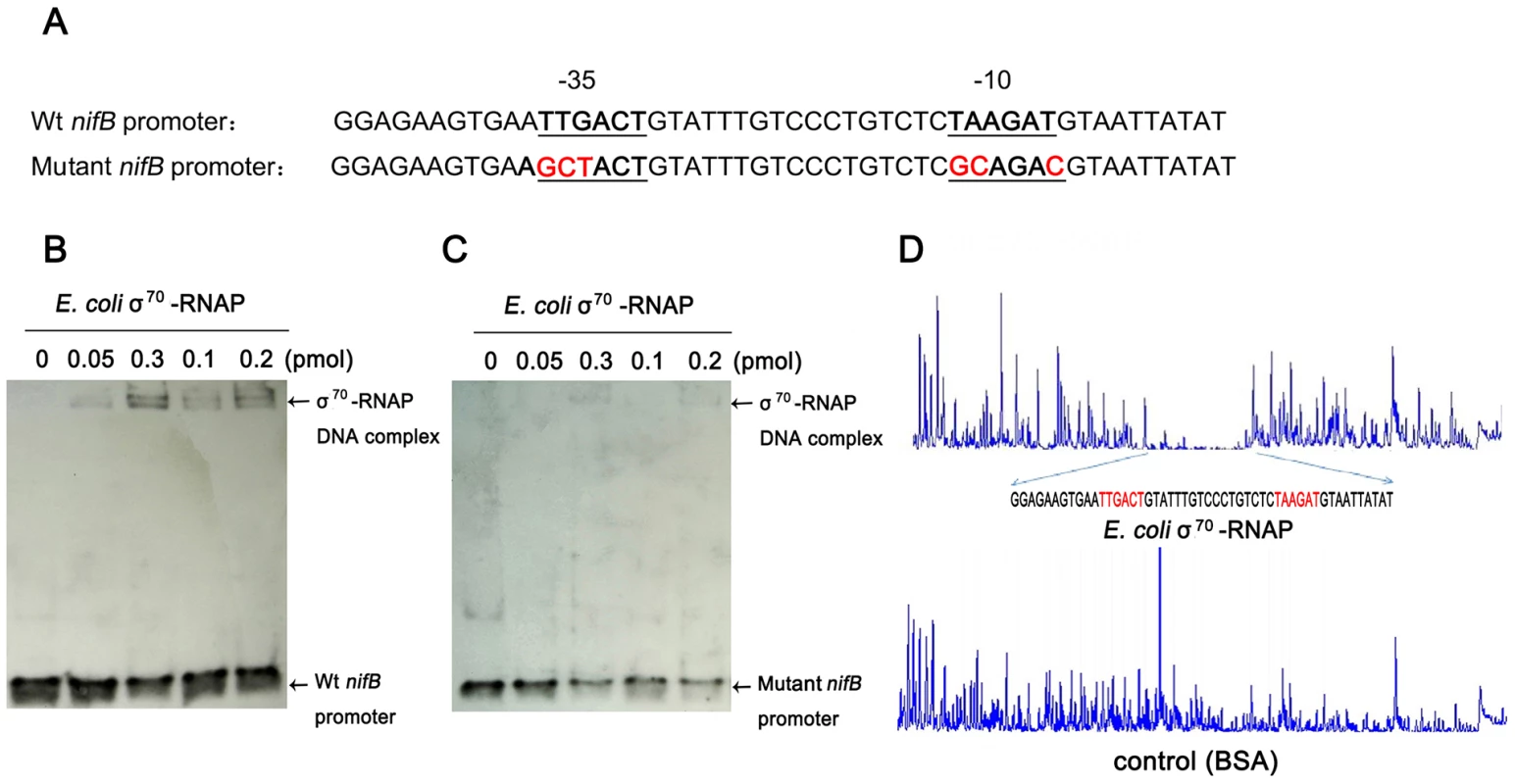
(A) Substitutions introduced in the nifB promoter sequence. The sequences of the −35 and −10 regions of the nifB promoter are underlined (Wt indicates the wild-type sequence). Base substitutions in the mutant promoter are indicated in red. (B) and (C) EMSA experiments comparing the binding of E. coli σ70-RNAP to the wild-type nifB promoter fragment (panel B) with the mutant promoter fragment (panel C). The protein concentration is indicated above each lane. (D) DNase I footprinting of the interaction of E. coli σ70-RNAP with the nifB promoter using an automated capillary sequencer. The top lane is an electropherogram obtained in the presence of σ70-RNAP with the sequence protected from cleavage shown below. A control electropherogram obtained from a reaction containing BSA is shown in the bottom lane. To verify if the nifB promoter of Paenibacillus sp. WLY78 is functional in E. coli, it was fused to the lacZ reporter gene. The level of β-galactosidase activity expressed from the PnifB::lacZ fusion in E. coli strain JM109 was not influenced either by the concentration of fixed nitrogen in the culture medium or by the external oxygen concentration (Figure 5). Hence, the Paenibacillus sp. WLY78 nifB promoter is apparently recognized by E. coli σ70 RNA polymerase in vivo. These data concur with previous studies where promoters of gram-positive bacteria, for example, Bacillus stereothermophilus [22] and Corynebacterium glutamicum [23], were shown to be functional in E. coli.
Fig. 5. Expression of the Paenibacillus sp. WLY78 PnifB::lacZ promoter fusion is constitutive in E. coli. 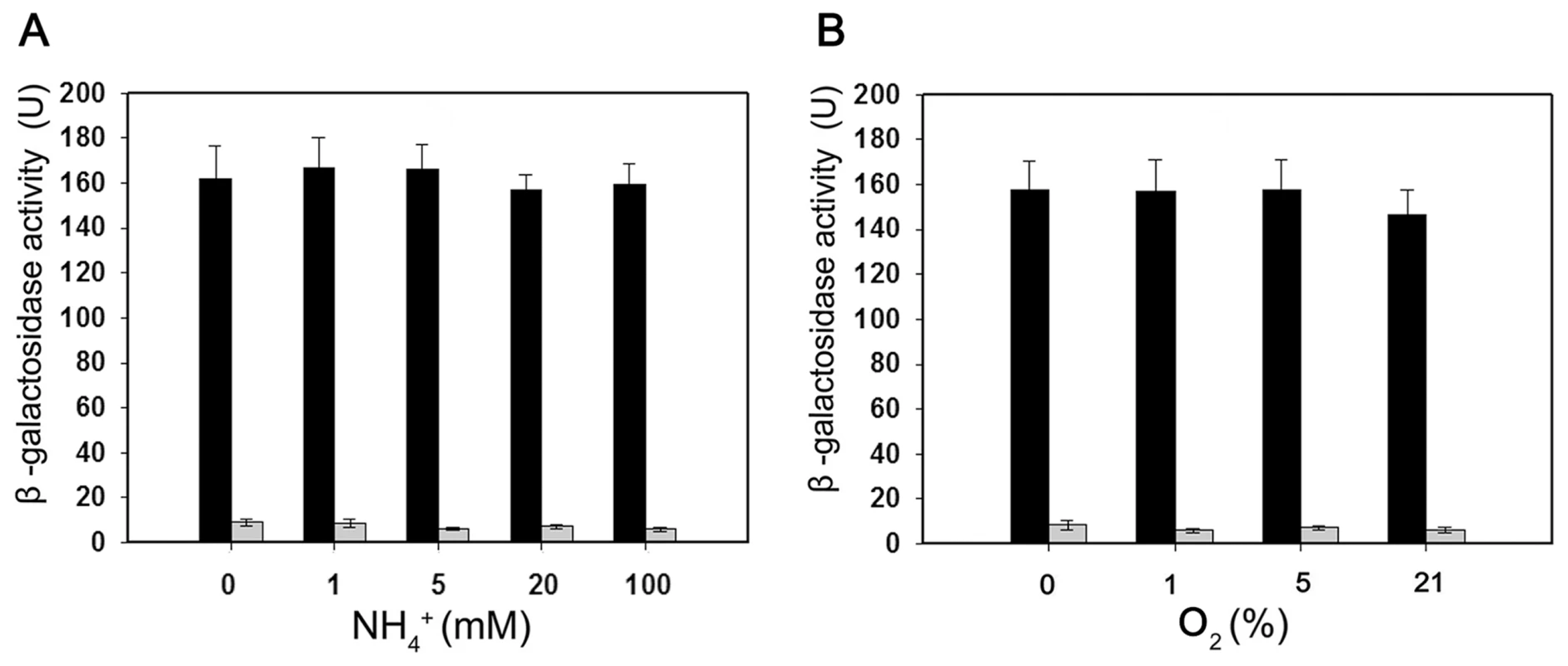
Black bars indicate expression of ß-galactosidase driven by the nifB promoter; grey bars indicate the level of ß-galactosidase activity exhibited by the vector plasmid (pPR9TT) alone. Cultures were grown in nitrogen deficient medium, with 2 mM glutamate as nitrogen source, either anaerobically with the indicated concentrations of NH4Cl (left panel) or with the indicated initial oxygen concentrations shown in the right-hand panel. Error bars indicate the standard deviation observed from at least two independent experiments. The Paenibacillus nif gene cluster enables nitrogen fixation by E. coli
To transfer the Paenibacillus nif gene cluster to E. coli, we cloned a 10.5-kb DNA fragment (containing the sequence from the ATG start codon of nifB to the TAA stop codon of nifV) in the expression vector pET-28b bringing the nif genes under control of the T7 promoter. This construct was then transformed into E. coli BL21 (DE3), yielding the engineered E. coli strain 78-32. We further cloned the 11-kb full-length nif cluster containing its own nif promoter and the contiguous nine genes nifBHDKENXhesAnifV into the multicopy plasmid pHY300PLK and transformed this into E. coli JM109, yielding the engineered E. coli strain 78-7 (Figure 6A). To determine whether the Paenibacillus nif cluster functions in E. coli, we employed two independent methods to assess nitrogenase activity; firstly, reduction of the alternative substrate acetylene to ethylene, which can be readily quantified by gas chromatography [24], [25] and secondly, a 15N2 enrichment assay to directly measure the incorporation of this tracer into organic nitrogen [26]. When grown anaerobically in nitrogen-deficient medium, Paenibacillus sp. WLY78 exhibits both acetylene reduction and 15N2 incorporation (Figure 6, panels B and C). The engineered E. coli strain 78-7, which expresses the nif genes from the native promoter showed approximately 10% of the specific activity for acetylene reduction when compared with Paenibacillus and was competent to assimilate 15N2. In contrast, when expressed from the T7 promoter and induced with 2 mM IPTG the Paenibacillus nif cluster exhibited relatively low levels of nitrogenase activity in the recombinant E. coli strain 78-32 (Figure 6). Therefore, the engineered E. coli strain 78-7 was used for most of the studies reported here. When compared with the recipient E. coli strain JM109, the engineered strain 78-7 had an identical cellular phenotype when analyzed by Biolog phenotypic microarrays [27] (data not shown). In comparison with the Paenibacillus sp. WLY78 strain, which is capable of diazotrophic growth, the engineered E. coli strain 78-7 grew poorly in liquid media with dinitrogen as the sole nitrogen source (data not shown). Therefore, although the recombinant strain expresses active nitrogenase and assimilates 15N2, this does not enable the engineered E. coli strain to grow as a diazotroph.
Fig. 6. Engineered E. coli strains and their nitrogen fixation abilities. 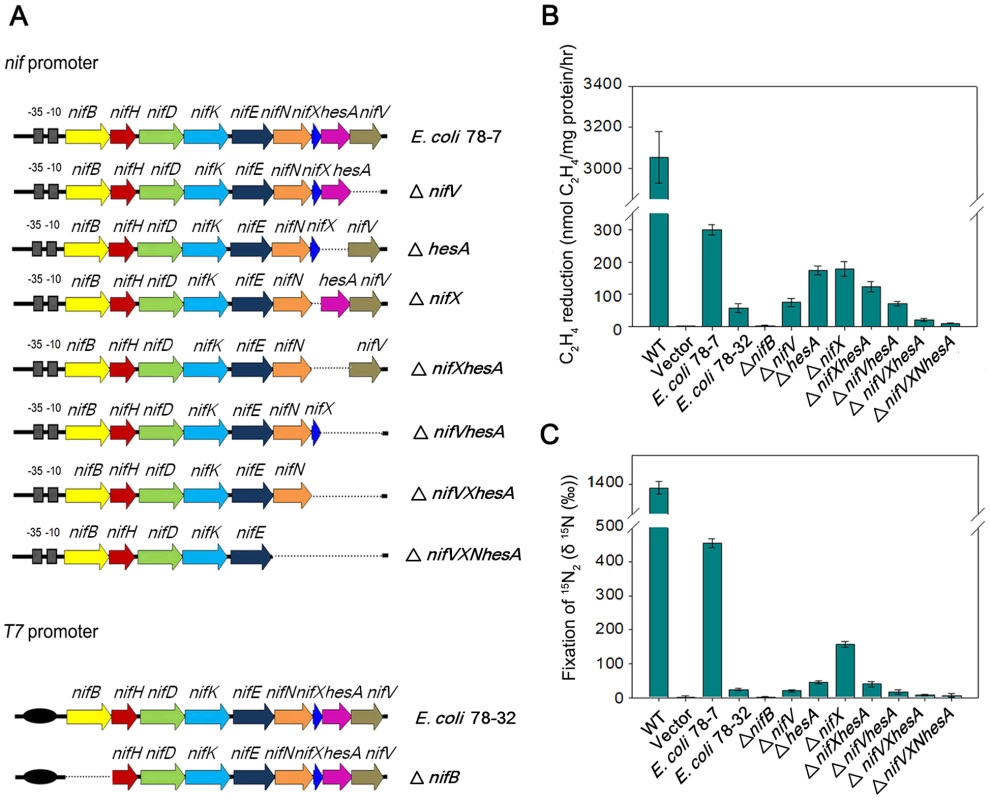
(A) Scheme showing the genetic organization of the engineered E. coli strains. (B) and (C) Nitrogenase activities of engineered strains and their deletion variants compared with Paenibacillus sp. WLY78 (bars marked as “WT”) and E. coli JM109 carrying the empty vector plasmid pHY300PLK (bars marked as “vector”). Strains were grown anaerobically in nitrogen-deficient conditions and the cultures were assayed either for acetylene reduction (panel B) or for 15N2 incorporation (panel C). Error bars indicate the standard deviation observed from at least two independent experiments. Minimal Paenibacillus nif genes required for nitrogenase activity
To further determine the minimal nif genes required for nitrogen fixation, we constructed a series of nif gene deletions (Figure 6). Neither acetylene nor 15N2 incorporation was detectable in the nifB deletion, supporting the original observation that nifB is essential for synthesis of nitrogenase [5]. When nifV was deleted, 15N2 assimilation decreased more significantly than acetylene reduction, in agreement with the substrate reduction properties of nifV mutants [28], which are unable to synthesize the homocitrate moiety of FeMo-co [29]. Deletion of hesA also influenced 15N2 incorporation more significantly than acetylene reduction, suggesting that hesA is required for nitrogen fixation. In contrast, deleting nifX gave rise to a similar decrease (∼50%) in the reduction of both substrates. In the ΔnifXhesA double deletion, nitrogenase activity was similar to that in the single hesA mutant, whereas in the double ΔhesAnifV deletion, activities were similar to those exhibited by the single nifV mutant. Deletion of three (nifXhesAnifV) or four genes (nifNXhesAnifV) ablated nitrogenase activity. In all cases the phenotypic defects exhibited by the deletions could be reversed by complementation with plasmids bearing the missing genes (data not shown). These results suggest that all nine Paenibacillus genes (nifBHDKENXhesAnifV) are necessary for optimal nitrogenase activity in E. coli.
Effects of fixed nitrogen and oxygen on nif transcription
In many diazotrophs such as K. oxytoca and A. vinelandii, expression of the nif genes is tightly controlled at the transcriptional level in response to the concentration of fixed nitrogen and the oxygen [30]. In addition, the activity of nitrogenase itself can be regulated at the post-translational level in response to environmental effectors [31]. To examine whether the Paenibacllus nif cluster is subject to similar regulation, we compared the effects of NH4+ and O2 on in vivo nitrogenase activity and nif gene transcription in the native Paenibacillus sp. WLY78 strain with that of engineered E. coli 78-7 (Figure 7). Both Paenibacillus sp. WLY78 and the engineered E. coli 78-7 strain did not exhibit nitrogenase activity at O2 concentrations above 5% (Figure 7A). In addition, acetylene reduction by Paenibacillus sp. WLY78 was not observed at NH4+ concentrations above 1 mM. In contrast, the engineered E. coli strain 78-7 exhibited nitrogenase activity even in the presence of 200 mM NH4Cl (Figure 7B). The latter observation suggests that the Paenibacillus nif cluster is not subject to regulation by fixed nitrogen in E. coli. In agreement with the acetylene reduction data, the α and ß subunits of the MoFe protein and the Fe protein component of nitrogenase were only detectable by Western blotting in Paenibacillus sp. WLY78 grown under nitrogen fixation conditions, whereas nitrogenase components were detectable in the engineered E. coli strain even in the presence of oxygen (Figure S1).
Fig. 7. Effects of O2 and NH4+ on nitrogenase activity and nif gene transcription. 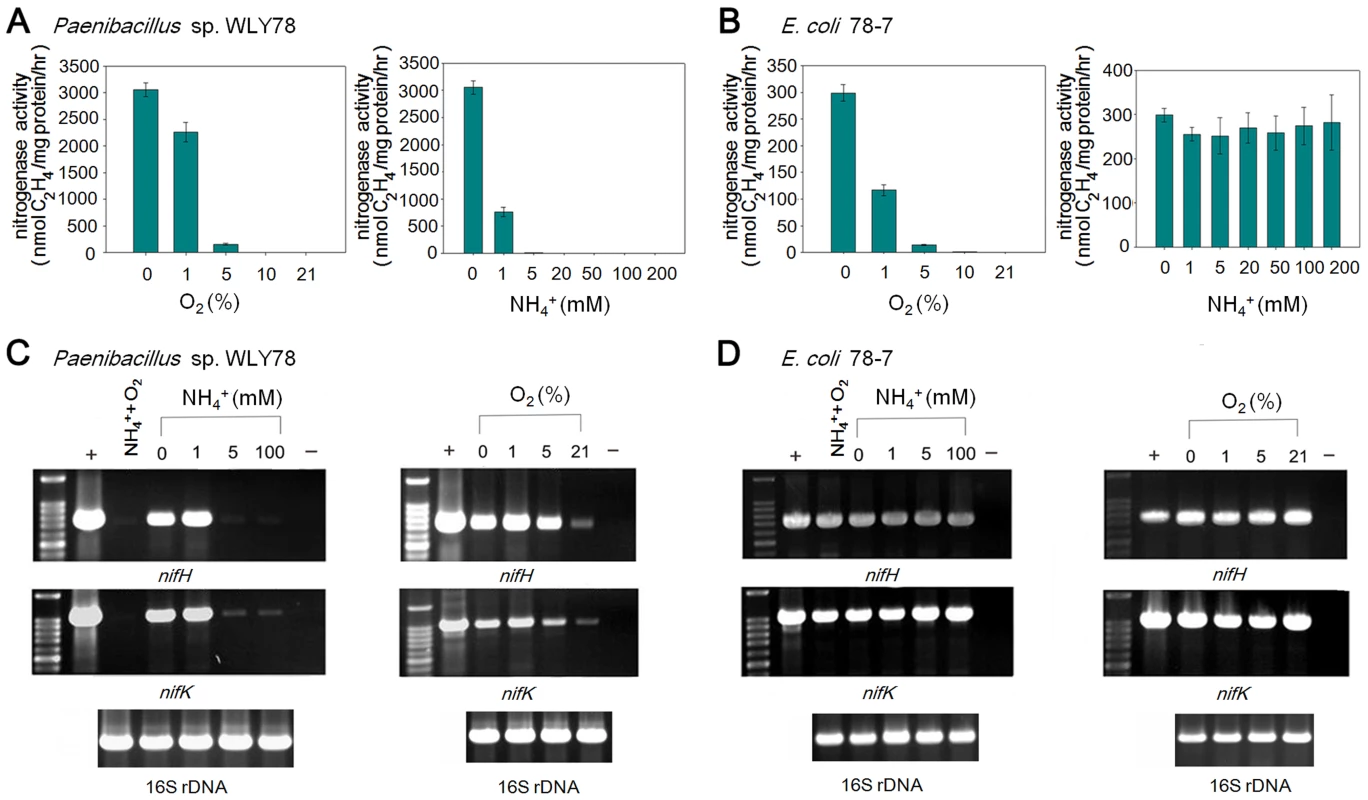
(A) and (B) Comparison of the acetylene reduction activities of Paenibacillus sp. WLY78 (panel A) and the engineered E. coli 78-7 strain (panel B), when cultures are grown in the presence of either oxygen or ammonium (at the initial concentrations shown on the y axis). Error bars indicate the standard deviation observed from at least two independent experiments. (C) and (D) Comparison of transcription of nifH and nifK as determined by RT-PCR in Paenibacillus sp. WLY78 (panel C) and E. coli 78-7 (panel D). Initial concentrations of ammonium and oxygen are indicated above relevant lanes. Lanes labeled “NH4+,O2” indicate that both 2 mM ammonium and 21% oxygen were present. Lanes labeled “+” indicate positive controls in which genomic DNA was used as template in the RT-PCR. Lanes labeled “−” indicate negative controls in which no reverse transcriptase was added to the RT-PCR reaction. In each case a parallel RT-PCR reaction was performed to detect the level of 16S rRNA, to provide a loading control (shown beneath relevant lanes). The influence of oxygen and fixed nitrogen on transcription was assessed by RT-PCR using nifH and nifK probes. Conversant with the acetylene reduction data, nif transcription in Paenibacillus sp. WLY78 was inhibited by NH4+ concentrations above 1 mM and by the presence of 21% oxygen (Figure 7C). In contrast, nifH and nifK transcription in E. coli 78-7 was insensitive to the presence of oxygen and fixed nitrogen (Figure 7D). Thus the Paenibacillus nif genes are constitutively transcribed in the engineered strain indicating that the transcriptional regulation observed in the native host does not occur in E. coli.
Discussion
Although the biochemical properties and structure of molybdenum nitrogenases are remarkably similar when purified from diverse bacteria and archaea, genetic requirements for the synthesis and assembly of the enzyme and maintenance of its activity differ widely amongst diazotrophs [11], [32], [33]. Some of this diversity is undoubtedly determined by the environmental lifestyle of each diazotroph, the need to protect the enzyme from damage by oxygen and the requirement to provide sufficient ATP and reductant to support enzyme activity under different physiological conditions. Although the conserved nature of the structural genes and the assembly pathway for FeMoco biosynthesis dictates the presence of a common core of nif genes, other functions may be provided by protein counterparts encoded elsewhere in the genome. Alternatively, the large nif gene clusters found primarily in Proteobacteria may have evolved from more simple clusters in which assembly, processing and maintenance of nitrogenase activity is less well optimized.
In contrast with earlier studies in which transfer of the complete complement of 20 nif genes from K. oxytoca enabled E. coli to fix nitrogen [12], our results with Paenibacillus sp. WLY78 demonstrate that only nine nif genes are needed to synthesize active nitrogenase in E. coli. The specific activity of the enzyme expressed in E. coli was approximately 10% of that observed in Paenibacillus, but nevertheless sufficient to provide 15N2 assimilation. However, synthesis of active nitrogenase in the recombinant E. coli strain did not enable diazotrophic growth. This implies that this level of enzyme activity is insufficient to support growth on dinitrogen as sole nitrogen source. However, we cannot rule out the possibility that other physiological factors in E. coli, for example the ability to synthesis high levels of nitrogenase proteins under conditions of nitrogen starvation, limit the capacity for diazotrophic growth. Considering the physiological background of E. coli, one of the notable absences in the minimal Paenibacillus nif gene cluster is the presence of nifF and nifJ, which provide the electron transport chain to nitrogenase in some diazotrophs [34], [35]. The activity of Paenibacillus nitrogenase is therefore likely to be reductant limited in E. coli in the absence of this electron transport chain. Another notable absence is nifM, which encodes a cis-trans peptidyl prolyl isomerase required for proper folding of nitrogenase Fe protein in diazotrophic proteobacteria [6]. Potentially this function is provided by a counterpart enzyme encoded elsewhere in the genome in other diazotrophs such as Paenibacillus. However, a functional equivalent of nifM is not present in E. coli, since assembly of active K. oxytoca Fe protein in this background requires the presence of both nifH and nifM [36]. The Paenibacillus NifH sequence contains the seven conserved proline residues identified in other NifH sequences that are considered to be potential substrates for NifM [6]. However, it is possible that other amino acid substitutions in NifH may enable assembly of Fe protein in the absence of NifM. The Paenibacillus sp. WLY78 nif gene operon does not contain homologs of the nitrogen fixation-specific iron-sulphur cluster assembly pathway encoded by nifU and nifS. As in the case of other diazotrophs, this function may be provided by the Suf system, encoded elsewhere in the Paenibacillus genome. When nifH and nifM are expressed in E. coli, assembly of the 4Fe-4S cluster in the K. oxytoca Fe protein does not require nifU and nifS [36], [37]. This function is probably provided by the general Isc, Csd or Suf machineries for iron-sulphur cluster biosynthesis in E. coli. However, K. oxytoca nifS is apparently required for the biosynthesis of the P cluster in the MoFe protein, when Nif polypetides are expressed in E. coli [38]. Although nifU and nifS also participate in FeMoco biosynthesis [37], the requirement for these genes is not absolute, particularly if nifB is strongly expressed [38].
Systematic deletion of genes in the Paenibacillus nif gene cluster suggests they have functions similar to those of other diazotrophs. As anticipated, nifB is essential for nitrogen fixation in E. coli and the substrate reduction profile of the nifV deletion is expected for a mutant lacking homocitrate synthase and therefore unable to make the homocitrate moiety of FeMo-co [39]. The co-localisation of hesA within the nif operon is an interesting feature of Paenibacillus and other minimal nif clusters such as those of cyanobacteria and Frankia (Figure 1). Our deletion analysis demonstrates that hesA is important for nitrogenase activity, but the function of hesA in nitrogen fixation has not so far been determined. Well-characterized homologs belonging to the ThiF-MoeB-HesA family engage in an ATP-dependent process that activates the C-terminus of partner ubiquitin-like proteins by forming an acyl adenylate complex that facilitates sulfur transfer [40], [41]. Ubiquitin-like proteins contain a conserved C-terminal Gly-Gly motif that is the target for adenylylation by the activating enzyme [42]. Intriguingly, both NifB and NifN from Paenibacillus contain C-terminal Gly-Gly motifs and therefore are potential targets for adenylylation by HesA. Given the potential role of HesA as an activating enzyme for sulphur transfer, it is tempting to speculate that HesA may perform a role in metallocluster biosynthesis.
In the Proteobacteria, nif genes are generally transcribed from σ54-dependent promoters that are subject to transcriptional activation by the enhancer binding protein NifA and are regulated in response to fixed nitrogen and oxygen [30]. However, much less in known about nif gene regulation in other diazotrophs where this paradigm is absent. Our results demonstrate that the nif cluster of Paenibacillus sp. WLY78 is transcribed from a σ70-dependent promoter, most likely as a single operon, and that transcription of the nif genes is subject to regulation in response to the extracellular concentration of oxygen and fixed nitrogen in Paenibacillus. As no transcriptional regulation by either oxygen or fixed nitrogen was detectable when the Paenibacillus sp. WLY78 nif cluster was expressed from the native nifB promoter in E. coli, it seems likely that the transcriptional regulation of the nif system in Paenibacillus involves repression mechanisms. Potential candidates for repression of transcription in response to the nitrogen source are the global nitrogen regulators GlnR and TnrA, which are present in Paenibacillus [43].
In summary our results demonstrate that a minimal nif gene cluster derived from a gram-positive bacterium can function to synthesize active nitrogenase when expressed in the very different host environment of E. coli. This raises various questions concerning the repertoire of genes required for nitrogen fixation and may have important biotechnological implications for engineering diazotophic eukaryotes.
Materials and Methods
Strains and media
Paenibacillus sp. WLY78 was isolated from the rhizosphere of bamboo in Beijing, China by enrichment in nitrogen-free medium after heating at 100°C for 10 min [14]. Strain WLY78 is similar to P. polymyxa based on 16S rDNA phylogeny and whole genome sequencing. E. coli strains JM109 and BL21 were used as the recipient strains for constructing the engineered E. coli strains carrying nitrogen fixation genes.
Paenibacillus sp. WLY78 and the engineered E. coli strains were routinely grown in LB or LD medium (per liter contains: 2.5 g NaCl, 5 g yeast and 10 g tryptone) at 30°C with shaking. When appropriate, antibiotics were added in the following concentrations: 40 µg/ml chloramphenicol, 100 µg/ml ampicillin, and 20 µg/ml tetracycline for maintenance of plasmids.
Nitrogen-free, nitrogen-deficient and nitrogen-excess media were used in this study. Nitrogen-free medium contained (per liter) 10.4 g Na2HPO4, 3.4 g KH2PO4, 26 mg CaCl2• 2H2O, 30 mg MgSO4, 0.3 mg MnSO4, 36 mg Ferric citrate, 7.6 mg Na2MoO4·2H2O, 10 µg p-aminobenzoic acid, 5 µg biotin and 4 g glucose as carbon source. Nitrogen-deficient medium contained 2 mM glutamate as nitrogen source in nitrogen-free medium. Nitrogen-excess medium contained 100 mM NH4Cl in nitrogen-free medium [14].
Acetylene reduction assays
For nitrogenase activity assays, Paenibacillus sp.WLY78 and the engineered E. coli strains were grown in 5 ml of LD media (supplemented with antibiotics) in 50 ml flasks shaken at 250 rpm for 16 h at 30°C. The cultures were collected by centrifugation, washed three times with sterilized water and then resuspended in nitrogen-deficient medium containing 2 mM glutamate as nitrogen source (supplemented with antibiotics for the engineered E. coli strains and IPTG when necessary) to a final OD600 of 0.2–0.4. Then, 1 ml of the culture was transferred to a 25-ml test tube and the test tube was sealed with robber stopper. The headspace in the tube was then evacuated and replaced with argon gas [14]. After incubating the cultures for 6–8 h at 30°C with shaking at 250 rpm, C2H2 (10% of the headspace volume) was injected into the test tubes. After incubating the cultures for a further 3 h, 100 µl of culture headspace was withdrawn through the rubber stopper with a gas tight syringe and manually injected into a HP6890 gas chromatograph to quantify ethylene production. All treatments were in three replicates and all the experiments were repeated three or more times.
For measuring the effect of ammonium on nitrogenase activity, nitrogen-deficient medium was supplemented with NH4Cl at the concentrations indicated and the cultures were also grown under anaerobic conditions. For measuring the effect of oxygen on nitrogenase activity, nitrogen-deficient medium containing 2 mM glutamate as nitrogen source was used, and oxygen was adjusted to the initial concentration indicated at the start of the incubation.
15N2 incorporation assay
Paenibacillus sp.WLY78 and the engineered E. coli strains were grown overnight in LD medium. The cultures were collected and resuspended in 70 ml nitrogen-deficient medium containing 2 mM glutamate as nitrogen source, to an OD600 of 0.4 in a 120 ml serum bottle. The serum bottles were filled with N2 gas, and then 8 ml gas was removed and 5 ml 15N2 (99%+, Shanghai Engineering Research Center for Stable Isotope) gas was injected. After 72 h of incubation at 30°C, the cultures were collected, and were freeze dried, ground, weighed and sealed into tin capsules. Isotope ratios are expressed as δ15N whose values are a linear transform of the isotope ratios 15N/14N, representing the per mille difference between the isotope ratios in a sample and in atmospheric N2 [26].
Genome sequencing, assembly and annotation
Total DNA was extracted from Paenibacillus sp. WLY78. DNA sequencing was performed using Illumina technologies. A total length of 600,000,120 base pairs of reads was obtained, to enable the assembly of all tags using SOAP denovov. 1.04 assembler [44]. Finally, 87 scaffolds were assembled, giving 101.3-fold coverage of the genome. Glimmer 3 (version 3.0.2) was used for gene finding [45]. Transfer RNA genes were identified by the program tRNAscan-SE [46]. Genes coding for proteins with known functions were annotated by searches against KEGG Genes, Pfam, and SWISSPROT. The complete genome sequence of Paenibacillus sp. WLY78 has been deposited at DDBJ/EMBL/Genbank under the accession ALJV00000000. The version described in this paper is version ALJV01000000.
Construction of recombinant plasmids and recombinant E. coli strains
Genomic DNA of Paenibacillus sp. WLY78 was used as template for cloning nif genes. Primers used for construction of the engineered E. coli strains are listed in Table S2. Recombinant plasmids and strains are listed in Table S3.
Transcription start site identification
The 5′-RACE method was used to determine the transcription start site (TSS) using the SMARTer RACE cDNA Amplification Kit (Clontech). Gene-specific primers are listed in Table S2. The PCR product was cloned into the pMD18-T Vector and then sequenced.
Overexpression and purification of σ70 from Paenibacillus sp. WLY78 in E. coli
A 1134 bp DNA fragment carrying the rpoD gene (encoding σ70 of Paenibacillus sp. WLY78) was PCR amplified with primers sigma A-F and sigma A-R (Table S2). The PCR product was ligated to the pET-28b expression vector, yielding plasmid pET28-σ70. E. coli strain BL21 (DE3) was transformed with expression plasmid pET28-σ70 and utilized for protein expression. The bacterial cells were grown in LB medium to the end of log phase and then a final concentration of 1 mM IPTG (isopropyl-β-D-thiogalactopyranoside) was added to the culture and the cells were harvested after incubation for another 4 h at 16°C. The cells were then harvested and disrupted by sonication on ice. The protein was purified from the supernatant with Ni2+-NTA agarose (Qiagen) according to the manufacturer's instructions.
Electrophoretic mobility shift assay (EMSA)
For the electrophoretic mobility shift assay (EMSA), a 50 bp nif promoter fragment (from −47 to +3 relative to the transcription start site of nifB in Paenibacillus sp. WLY78) was synthesized by Sangon Biotech Co., Ltd (Shanghai). To do this, two DNA fragments corresponding to the sequences of the first strand (5′ - GGAGAAGTGAATTGACTGTATTTGTCCCTGTCTCTAAGATGTAATTATAT-3′) and the complementary DNA strand (5′ - ATATAATTACATCTTAGAGACAGGGACAAATACAGTCAATTCACTTCTCC-3′) were synthesized. The two strands were annealed and then labeled with digoxin using the DIG Gel Shift Kit (Roche). The binding of E. coli σ70-RNAP (RNA polymerase) (Epicentre) or σ70 of Paenibaciillus sp. WLY78 to the nif promoter was carried out using a gel shift kit (Roche). A scrambled 39 bp DNA fragment formed by annealing the following complementary oligonucleotides (5′ - GTACGGAGTATCCAGCTCCGTAGCATGCAAATCCTCTGG-3′) and (5′-CCAGAGGATTTGCATGCTACGGAGCTGGATACTCCGTAC -3′) was used to assay non-specific binding.
To examine the specificity of binding to the promoter sequence per se, primers designed with substitutions in the −35 (TTGACT to GCTACT) and −10 (TAAGAT to GCAGAC) regions of the nifB promoter were utilized and were annealed and labeled as described above.
DNAse I footprinting
The DNase I footprinting assay was performed as described by Zianni et al. [47]. A 365 bp nif promoter fragment (from −315 to +50 relative to the transcription start site) was PCR amplified from Paenibacillus sp. WLY78 with primer pfoot-up whose terminal base was fluorescent 6-carboxyfluorescein (FAM)-labeled and primer pfoot-down (Table S2). The 5′-FAM-labeled DNA fragment (400 ng) was incubated with the E. coli σ70-RNAP (10 pmol) for 30 min at 25°C. Bovine serum albumin (BSA) was used for the control experiment. After incubation, the mixtures were digested with DNase I for 40 seconds at 37°C and then the reactions were stopped by adding 0.2 M EDTA (pH 8.0). The digested DNA fragments were extracted with phenol-chloroform, precipitated with ethanol, and the pellets dissolved in Mini-Q water. The samples were sequenced with the ABI 3730 DNA analyzer by Genolab Co. and the data were analyzed with GeneMarker software.
Construction of a nifB promoter::lacZ fusion
A 100 bp DNA fragment (Pnif) (from −97 to +3 relative to the nifB transcription start codon) containing the nifB promoter was amplified from total DNA of Paenibacillus sp. WLY78 using primers (Table S2). The fragment was cloned into the promoterless plasmid pPR9TT, yielding plasmid pPR9TT-Pnif. The plasmid was then transformed into E. coli JM109, yielding E. coli/PnifB::lacZ.
For β-galactosidase activity assays, E. coli JM109/pPR9TT and E. coli/PnifB::lacZ were grown overnight in LB medium at 30°C with shaking. The cultures were collected by centrifugation, washed three times with sterilized water and then resuspended in nitrogen-deficient medium containing 2 mM glutamate as nitrogen source to a final OD600 of 0.2–0.4. For measuring the effect of ammonium on nitrogenase activity, 1 ml culture was transferred to a 25 ml test tube supplemented with the concentration of NH4Cl indicated and the culture was incubated for 20 h at 30°C with shaking under anaerobic conditions. For measuring the effect of oxygen on nitrogenase activity, the test tubes were capped and filled with argon, and the oxygen concentration was adjusted to the initial concentration indicated and cultures were then incubated for 20 h at 30°C with shaking.
β-galactosidase activity was assayed according to the method described by Miller [48]. A 100 µl sample was taken and then mixed with 900 µl Z buffer containing β-mercaptoethanol, 40 µl chloroform and 20 µl 10% SDS and then shaken for 20 sec. Then 200 µl o-nitrophenyl-β-D-galactopyranoside (ONPG) (4 mg/ml) was added to the mixture and incubated in a water bath for 20 min at 28°C. The reaction was stopped with 500 µl 1M Na2CO3 solution. The mixture was then centrifuged for 15 min at 12000 rpm and the supernatant was used to measure the OD420 and OD550 values. 1 unit of β-galactosidase = [1000×(OD420−1.7 OD550)]/[Time (min)×vol (ml)×OD600].
RT-PCR
For RT-PCR, Paenibacillus sp. WLY78 and the recombinant E. coli strains were grown in N2-fixing conditions (without NH4Cl and O2), non-N2-fixing conditions (100 mM NH4Cl and 21% O2) or at different concentrations of NH4Cl in the absence of O2 or at different concentration of O2 in the absence of NH4Cl. The cultures were harvested by centrifugation at 4°C, and total RNA was isolated using the PrimeScript RT reagent Kit with gDNA Eraser (Takara Bio) according to the manufacturer's instructions. The possibility of contamination of genomic DNA was eliminated by digestion with RNase-free DNase I (Takara Bio). The integrity and size distribution of the RNA was verified by agarose gel electrophoresis, and the concentration was determined spectrophotometrically. Synthesis of cDNA was carried out using RT Prime Mix according to the manufacturer's specifications (Takara Bio). 0.8 µg of cDNA was used for RT-PCR. The nifH and nifK transcripts were detected by using an RT-PCR Kit with 16S rDNA as a control. Primers for nifH, nifK and 16S rDNA used for PCR are listed in Table S2.
Western blot assays for NifH and NifDK expression
For Western blotting, cultures of Paenibacillus sp. WLY78 and the engineered E. coli strains were grown either in non-N2-fixing conditions (LD medium and 21% O2) and harvested after 6–8 h of incubation or grown in N2-fixing conditions (2 mM glutamate and without O2) and harvested after 20 h of incubation, respectively. The cell pellet collected from 4 ml cultures at OD600 = 1 was dissolved in 200 µl sodium dodecyl sulfate (SDS) gel-loading buffer, boiled for 5 min and then 20 µl was loaded onto the stacking gel. Proteins were separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) with an acrylamide∶bis-acrylamide ratio of 172∶1. Antisera raised against MoFe protein and Fe protein of K. oxytoca M5al were used as probes for Western blotting. The MoFe protein and Fe protein components of nitrogenase were purified from K. oxytoca M5al under anaerobic conditions and then used to make rabbit antiserum.
Supporting Information
Zdroje
1. FalkowskiPG (1997) Evolution of the nitrogen cycle and its influence on the biological sequestration of CO2 in the ocean. Nature 387 : 272–275.
2. MerrickM, DixonR (1984) Why don't plants fix nitrogen? Trends Biotechnol 2 : 162–166.
3. BeattyPH, GoodAG (2011) Future prospects for cereals that fix nitrogen. Science 333 : 416–417.
4. RubioLM, LuddenPW (2008) Biosynthesis of the iron-molybdenum cofactor of nitrogenase. Annu Rev Microbiol 62 : 93–111.
5. RobertsCP, MacnellT, MacnellD, BrillWJ (1978) Regulation and characterization of protein products coded by the nif (nitrogen fixation) genes of Klebsiella pneumoniae. J Bacteriol 136 : 267–279.
6. GaviniN, TungturS, PulakatL (2006) Peptidyl-prolylcis/trans isomerase-independent functional NifH mutant of Azotobacter vinelandii. J Bacteriol 188 : 6020–6025.
7. HuY, RibbeMW (2011) Biosynthesis of nitrogenase FeMoco. Coord Chem Rev 255 : 1218–1224.
8. ArnoldW, RumpA, KlippW, PrieferUB, PühlerA (1988) Nucleotide sequence of a 24,206-base-pair DNA fragment carrying the entire nitrogen fixation gene cluster of Klebsiella pneumoniae. J Mol Biol 203 : 715–738.
9. SetubalJC, dos SantosP, GoldmanBS, ErtesvåH, EspinG (2009) Genome Sequence of Azotobacter vinelandii, an obligate aerobe specialized to support diverse anaerobic metabolic processes. J Bacteriol 191 : 4534–4545.
10. DodsworthJA, LeighJA (2006) Regulation of nitrogenase by 2-oxoglutarate-reversible, direct binding of a PII-like nitrogen sensor protein to dinitrogenase. Proc Natl Acad Sci USA 103 : 9779–9784.
11. Dos SantosPC, FangZ, MasonSW, Setubal JC. DixonR (2012) Distribution of nitrogen fixation and nitrogenase-like sequences amongst microbial genomes. BMC Genomics 13 : 162.
12. CurattiL, HernandezJA, IgarashiRY, SobohB, ZhaoD (2007) In vitro synthesis of the iron–molybdenum cofactor of nitrogenase from iron, sulfur, molybdenum, and homocitrate using purified proteins. Proc Natl Acad Sci USA 104 : 17626–17631.
13. DixonR, PostgateJ (1972) Genetic transfer of nitrogen fixation from Klebsiella pneumoniae to Escherichia coli. Nature 237 : 102–103.
14. XieJB, BaiLQ, WangLY, ChenSF (2012) Phylogeny of 16S rRNA and nifH genes and regulation of nitrogenase by oxygen and ammonium in the genus Paenibacillus. Microbiology 81 : 5–6.
15. ZhaoH, XieB, ChenSF (2006) Cloning and sequencing of nifBHDKENX genes of Paenibacillus massiliensis T7 and its nif promoter analysis. China C Life Sci 49 : 115–122.
16. OhCJ, KimHB, KimJ, KimWJ, LeeH, et al. (2012) Organization of nif gene cluster in Frankia sp. EuIK1 strain, a symbiont of Elaeagnus umbellate. Arch Microbiol 194 : 29–34.
17. WelshEA, LibertonM, StockelJ, SatoH, LohT (2008) The genome of Cyanothece 51142, a unicellular diazotrophic cyanobacterium important in the marine nitrogen cycle. Proc Natl Acad Sci USA 105 : 15094–15099.
18. Enkh-AmgalanJ, KawasakiH, Oh-okaH, SekiT (2006) Cloning and characterization of a novel gene involved in nitrogen fixation in Heliobacterium chlorum, a possible regulatory gene. Arch Microbiol 186 : 327–337.
19. KesslerPS, BlankC, LeighJA (1998) The nif gene operon of the methanogenic archaeon Methanococcus maripaludis. J Bacteriol 180 : 1504–1511.
20. YehHY, ChenTC, LiouKM, HsuHT, ChungKM (2011) The core-independent promoter-specific interaction of primary sigma factor. Nucleic Acids Res 39 : 913–925.
21. JarmerH, LarsenTS, KroghA, SaxildHH, BrunakS (2001) Sigma A recognition sites in the Bacillus subtilisgenome. Microbiology 147 : 2417–2424.
22. YamadaM, KuboM, MiyakeT, SakaguchiR, HigoY, et al. (1991) Promoter sequence analysis in Bacillus and Escherichia, construction of strong promoters in E. coli. Gene 99 : 109–114.
23. Barriuso-IglesiasM, BarreiroC, FlechosoF, MartínJF (2006) Transcriptional analysis of the F0F1 ATPase operon of Corynebacterium glutamicum ATCC 13032 reveals strong induction by alkaline pH. Microbiology 152 : 11–21.
24. DilworthMJ (1966) Acetylene reduction by nitrogen fixing preparations from Clostridium pasteurianum. Biochem Biophys Acta 127 : 285–294.
25. SchöllhornR, BurrisRH (1967) Acetylene as a competitive inhibitor of N-2 fixation. Proc Natl Acad Sci USA 58 : 213–216.
26. MontoyaJP, VossM, KahlerP, CaponeDG (1996) A simple, high-precision, high-sensitivity tracer assay for N2 fixation. Appl Environ Microbiol 62 : 986–993.
27. BochnerBR, GadzinskiP, PanomitrosE (2011) Phenotype microarrays for high-throughput phenotypic yesting and assay of gene function. Genome Res 11 : 1246–1255.
28. McLeanP, DixonR (1981) Requirement of nifV gene for production of wild-type nitrogenase enzyme in Klebsiella pneumoniae. Nature 292 : 655–656.
29. HooverTR, ImperialJ, LuddenPW, ShahVK (1988) Homocitrate cures the NifV-phenotype in Klebsiella pneumoniae. J Bacteriol 170 : 1978–1989.
30. DixonR, KahnD (2004) Genetic regulation of biological nitrogen fixation. Nat Rev Microbiol 2 : 621–631.
31. HuergoLF, PedrosaFO, Muller-SantosM, ChubatsuLS, MonteiroRA, et al. (2012) PII signal transduction proteins: pivotal players in post-translational control of nitrogenase activity. Microbiology 158 : 176–190.
32. Masson-BoivinC, GiraudE, PerretX, BatutJ (2009) Establishing nitrogen-fixing symbiosis with legumes: how many rhizobium recipes? Trends Microbiol 17 : 458–66.
33. BoydES, AnbarAD, MillerS, HamiltonTL, LavinM, et al. (2011) A late methanogen origin for molybdenum-dependent nitrogenase. Geobiology 9 : 221–232.
34. Nieva-GómezD, RobertsGP, KlevickisS, BrillWJ (1980) Electron transport to nitrogenase in Klebsiella pneumoniae. Proc Natl Acad Sci USA 77 : 2555–5558.
35. HillS, KavanaghEP (1980) Roles of nifF and nifJ gene products in electron transport to nitrogenase in Klebsiella pneumoniae. J Bacteriol 141 : 470–475.
36. HowardKS, McLeanPA, HansenFB, LemleyPV, KoblanKS, et al. (1986) Klebsiella pneumoniae nifM gene product is required for stabilization and activation of nitrogenase iron protein in Escherichia coli. J Biol Chem 261 : 772–778.
37. ZhaoD, CurattiL, RubioLM (2007) Evidence for nifU and nifS participation in the biosynthesis of the iron-molybdenum cofactor of nitrogenase. J Bio Chem 282 : 37016–37025.
38. HarrisGS, WhiteTC, FloryJE, Orme-JohnsonWH (1990) Genes required for formation of the apoMoFe protein of Klebsiella pneumoniae nitrogenase in Escherichia coli. J Biol Chem 265 : 15909–15919.
39. ImperialJ, HooverTR, MaddenMS, LuddenPW, ShahVK (1989) Substrate reduction properties of dinitrogenase activated in vitro are dependent upon the presence of homocitrate or its analogs during iron-molybdenum cofactor synthesis. Biochemistry 28 : 7796–7799.
40. LakeMW, WuebbensMM, RajagopalanKV, SchindelinH (2001) Mechanism of ubiquitin activation revealed by the structure of a bacterial MoeB-MoaD complex. Nature 414 : 325–329.
41. LehmannC, BegleyTP, EalickSE (2006) Structure of the Escherichia coli ThiS-ThiF complex, a key component of the sulfur transfer system in thiamin biosynthesis. Biochemistry 45 : 9–11.
42. SchmitzJ, WuebbensMM, RajagopalanKV, LeimkühlerS (2007) Role of the C-terminal Gly-Gly motif of Escherichia coli MoaD, a molybdenum cofactor biosynthesis protein with a ubiquitin fold. Biochemistry 46 : 909–916.
43. Groot KormelinkT, KoendersE, HagemeijerY, OvermarsL, SiezenRJ, et al. (2012) Comparative genome analysis of central nitrogen metabolism and its control by GlnR in the class Bacilli. BMC Genomics 13 : 191.
44. LiR, LiY, KristiansenK, WangJ (2008) SOAP, short oligonucleotide alignment program. Bioinformatics 24 : 713–714.
45. DelcherAL, BratkeKA, PowersEC, SalzbergSL (2007) Identifying bacterial genes and endosymbiont DNA with Glimmer. Bioinformatics 23 : 673–679.
46. LoweTM, EddySR (1997) tRNAscan-SE, a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res 25 : 0955–0964.
47. ZianniM, TessanneK, MerighiM, LagunaR, TabitaFR (2006) Identification of the DNA bases of a DNase I footprint by the use of dye primer sequencing on an automated capillary DNA analysis instrument. J Biomol Tech 17 : 103–13.
48. Miller JH (1972) Experiments in Molecular Genetics. New York: Cold Spring Harbor Laboratory Press.
Štítky
Genetika Reprodukční medicína
Článek Defending Sperm FunctionČlánek How to Choose the Right MateČlánek Conserved Translatome Remodeling in Nematode Species Executing a Shared Developmental TransitionČlánek Genome-Wide and Cell-Specific Epigenetic Analysis Challenges the Role of Polycomb in SpermatogenesisČlánek The Integrator Complex Subunit 6 (Ints6) Confines the Dorsal Organizer in Vertebrate EmbryogenesisČlánek Multiple bHLH Proteins form Heterodimers to Mediate CRY2-Dependent Regulation of Flowering-Time inČlánek Playing the Field: Sox10 Recruits Different Partners to Drive Central and Peripheral MyelinationČlánek Evolutionary Tuning of Protein Expression Levels of a Positively Autoregulated Two-Component System
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2013 Číslo 10
-
Všechny články tohoto čísla
- Defending Sperm Function
- How to Choose the Right Mate
- A Mutation in the Gene in Labrador Retrievers with Hereditary Nasal Parakeratosis (HNPK) Provides Insights into the Epigenetics of Keratinocyte Differentiation
- Conserved Translatome Remodeling in Nematode Species Executing a Shared Developmental Transition
- A Novel Actin mRNA Splice Variant Regulates ACTG1 Expression
- Tracking Proliferative History in Lymphocyte Development with Cre-Mediated Sister Chromatid Recombination
- Correlated Occurrence and Bypass of Frame-Shifting Insertion-Deletions (InDels) to Give Functional Proteins
- Chimeric Protein Complexes in Hybrid Species Generate Novel Phenotypes
- Loss of miR-10a Activates and Collaborates with Activated Wnt Signaling in Inducing Intestinal Neoplasia in Female Mice
- Both Rare and Copy Number Variants Are Prevalent in Agenesis of the Corpus Callosum but Not in Cerebellar Hypoplasia or Polymicrogyria
- Reverse PCA, a Systematic Approach for Identifying Genes Important for the Physical Interaction between Protein Pairs
- Partial Deletion of Chromosome 8 β-defensin Cluster Confers Sperm Dysfunction and Infertility in Male Mice
- Genome-Wide and Cell-Specific Epigenetic Analysis Challenges the Role of Polycomb in Spermatogenesis
- Coordinate Regulation of Mature Dopaminergic Axon Morphology by Macroautophagy and the PTEN Signaling Pathway
- Cooperation between RUNX1-ETO9a and Novel Transcriptional Partner KLF6 in Upregulation of in Acute Myeloid Leukemia
- Mobility of the Native Conjugative Plasmid pLS20 Is Regulated by Intercellular Signaling
- FliZ Is a Global Regulatory Protein Affecting the Expression of Flagellar and Virulence Genes in Individual Bacterial Cells
- Specific Tandem Repeats Are Sufficient for Paramutation-Induced Trans-Generational Silencing
- Condensin II Subunit dCAP-D3 Restricts Retrotransposon Mobilization in Somatic Cells
- Dominant Mutations in Identify the Mlh1-Pms1 Endonuclease Active Site and an Exonuclease 1-Independent Mismatch Repair Pathway
- The Insulator Homie Promotes Expression and Protects the Adjacent Gene from Repression by Polycomb Spreading
- Human Intellectual Disability Genes Form Conserved Functional Modules in
- Coordination of Cell Proliferation and Cell Fate Determination by CES-1 Snail
- ORFs in Drosophila Are Important to Organismal Fitness and Evolved Rapidly from Previously Non-coding Sequences
- Different Roles of Eukaryotic MutS and MutL Complexes in Repair of Small Insertion and Deletion Loops in Yeast
- The Spore Differentiation Pathway in the Enteric Pathogen
- Acceleration of the Glycolytic Flux by Steroid Receptor Coactivator-2 Is Essential for Endometrial Decidualization
- The Human Nuclear Poly(A)-Binding Protein Promotes RNA Hyperadenylation and Decay
- Genome Wide Analysis Reveals Zic3 Interaction with Distal Regulatory Elements of Stage Specific Developmental Genes in Zebrafish
- Xbp1 Directs Global Repression of Budding Yeast Transcription during the Transition to Quiescence and Is Important for the Longevity and Reversibility of the Quiescent State
- The Integrator Complex Subunit 6 (Ints6) Confines the Dorsal Organizer in Vertebrate Embryogenesis
- Incorporating Motif Analysis into Gene Co-expression Networks Reveals Novel Modular Expression Pattern and New Signaling Pathways
- The Bacterial Response Regulator ArcA Uses a Diverse Binding Site Architecture to Regulate Carbon Oxidation Globally
- Direct Monitoring of the Strand Passage Reaction of DNA Topoisomerase II Triggers Checkpoint Activation
- Multiple bHLH Proteins form Heterodimers to Mediate CRY2-Dependent Regulation of Flowering-Time in
- A Reversible Histone H3 Acetylation Cooperates with Mismatch Repair and Replicative Polymerases in Maintaining Genome Stability
- ALS-Associated Mutations Result in Compromised Alternative Splicing and Autoregulation
- Robust Demographic Inference from Genomic and SNP Data
- Preferential Binding to Elk-1 by SLE-Associated Risk Allele Upregulates Expression
- Rad52 Sumoylation Prevents the Toxicity of Unproductive Rad51 Filaments Independently of the Anti-Recombinase Srs2
- The Serum Resistome of a Globally Disseminated Multidrug Resistant Uropathogenic Clone
- Identification of 526 Conserved Metazoan Genetic Innovations Exposes a New Role for Cofactor E-like in Neuronal Microtubule Homeostasis
- SUMO Localizes to the Central Element of Synaptonemal Complex and Is Required for the Full Synapsis of Meiotic Chromosomes in Budding Yeast
- Integrated Enrichment Analysis of Variants and Pathways in Genome-Wide Association Studies Indicates Central Role for IL-2 Signaling Genes in Type 1 Diabetes, and Cytokine Signaling Genes in Crohn's Disease
- Genome-Wide High-Resolution Mapping of UV-Induced Mitotic Recombination Events in
- Genome-Wide Analysis of Cell Type-Specific Gene Transcription during Spore Formation in
- Playing the Field: Sox10 Recruits Different Partners to Drive Central and Peripheral Myelination
- Two Portable Recombination Enhancers Direct Donor Choice in Fission Yeast Heterochromatin
- Mining the Human Phenome Using Allelic Scores That Index Biological Intermediates
- Yeast Tdh3 (Glyceraldehyde 3-Phosphate Dehydrogenase) Is a Sir2-Interacting Factor That Regulates Transcriptional Silencing and rDNA Recombination
- A Minimal Nitrogen Fixation Gene Cluster from sp. WLY78 Enables Expression of Active Nitrogenase in
- A Review of Bacteria-Animal Lateral Gene Transfer May Inform Our Understanding of Diseases like Cancer
- High Throughput Sequencing Reveals Alterations in the Recombination Signatures with Diminishing Spo11 Activity
- Partitioning the Heritability of Tourette Syndrome and Obsessive Compulsive Disorder Reveals Differences in Genetic Architecture
- Eleven Candidate Susceptibility Genes for Common Familial Colorectal Cancer
- A GDF5 Point Mutation Strikes Twice - Causing BDA1 and SYNS2
- Systematic Unraveling of the Unsolved Pathway of Nicotine Degradation in
- Natural Genetic Variation of Integrin Alpha L () Modulates Ischemic Brain Injury in Stroke
- Evolutionary Tuning of Protein Expression Levels of a Positively Autoregulated Two-Component System
- Evolutionary Change within a Bipotential Switch Shaped the Sperm/Oocyte Decision in Hermaphroditic Nematodes
- Limiting of the Innate Immune Response by SF3A-Dependent Control of MyD88 Alternative mRNA Splicing
- Multiple Signaling Pathways Coordinate to Induce a Threshold Response in a Chordate Embryo
- Distinct Regulatory Mechanisms Act to Establish and Maintain Pax3 Expression in the Developing Neural Tube
- Genome Wide Analysis of Narcolepsy in China Implicates Novel Immune Loci and Reveals Changes in Association Prior to Versus After the 2009 H1N1 Influenza Pandemic
- Mismatch Repair Genes and Modify CAG Instability in Huntington's Disease Mice: Genome-Wide and Candidate Approaches
- The Histone H3 K27 Methyltransferase KMT6 Regulates Development and Expression of Secondary Metabolite Gene Clusters
- Hsp70-Hsp40 Chaperone Complex Functions in Controlling Polarized Growth by Repressing Hsf1-Driven Heat Stress-Associated Transcription
- Function and Evolution of DNA Methylation in
- Stimulation of mTORC1 with L-leucine Rescues Defects Associated with Roberts Syndrome
- Transcription Termination and Chimeric RNA Formation Controlled by FPA
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Dominant Mutations in Identify the Mlh1-Pms1 Endonuclease Active Site and an Exonuclease 1-Independent Mismatch Repair Pathway
- Eleven Candidate Susceptibility Genes for Common Familial Colorectal Cancer
- The Histone H3 K27 Methyltransferase KMT6 Regulates Development and Expression of Secondary Metabolite Gene Clusters
- A Mutation in the Gene in Labrador Retrievers with Hereditary Nasal Parakeratosis (HNPK) Provides Insights into the Epigenetics of Keratinocyte Differentiation
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



