-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaA Novel Actin mRNA Splice Variant Regulates ACTG1 Expression
Cytoplasmic actins are abundant, ubiquitous proteins in nucleated cells. However, actin expression is regulated in a tissue - and development-specific manner. We identified a novel cytoplasmic-γ-actin (Actg1) transcript that includes a previously unidentified exon (3a). Inclusion of this exon introduces an in-frame termination codon. We hypothesized this alternatively-spliced transcript down-regulates γ-actin production by targeting these transcripts for nonsense-mediated decay (NMD). To address this, we investigated conservation between mammals, tissue-specificity in mice, and developmental regulation using C2C12 cell culture. Exon 3a is 80% similar among mammals and varies in length from 41 nucleotides in humans to 45 in mice. Though the predicted amino acid sequences are not similar between all species, inclusion of exon 3a consistently results in the in the introduction of a premature termination codon within the alternative Actg1 transcript. Of twelve tissues examined, exon 3a is predominantly expressed in skeletal muscle, cardiac muscle, and diaphragm. Splicing to include exon 3a is concomitant with previously described down-regulation of Actg1 in differentiating C2C12 cells. Treatment of differentiated C2C12 cells with an inhibitor of NMD results in a 7-fold increase in exon 3a-containing transcripts. Therefore, splicing to generate exon 3a-containing transcripts may be one component of Actg1 regulation. We propose that this post-transcriptional regulation occurs via NMD, in a process previously described as “regulated unproductive splicing and translation” (RUST).
Published in the journal: . PLoS Genet 9(10): e32767. doi:10.1371/journal.pgen.1003743
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1003743Summary
Cytoplasmic actins are abundant, ubiquitous proteins in nucleated cells. However, actin expression is regulated in a tissue - and development-specific manner. We identified a novel cytoplasmic-γ-actin (Actg1) transcript that includes a previously unidentified exon (3a). Inclusion of this exon introduces an in-frame termination codon. We hypothesized this alternatively-spliced transcript down-regulates γ-actin production by targeting these transcripts for nonsense-mediated decay (NMD). To address this, we investigated conservation between mammals, tissue-specificity in mice, and developmental regulation using C2C12 cell culture. Exon 3a is 80% similar among mammals and varies in length from 41 nucleotides in humans to 45 in mice. Though the predicted amino acid sequences are not similar between all species, inclusion of exon 3a consistently results in the in the introduction of a premature termination codon within the alternative Actg1 transcript. Of twelve tissues examined, exon 3a is predominantly expressed in skeletal muscle, cardiac muscle, and diaphragm. Splicing to include exon 3a is concomitant with previously described down-regulation of Actg1 in differentiating C2C12 cells. Treatment of differentiated C2C12 cells with an inhibitor of NMD results in a 7-fold increase in exon 3a-containing transcripts. Therefore, splicing to generate exon 3a-containing transcripts may be one component of Actg1 regulation. We propose that this post-transcriptional regulation occurs via NMD, in a process previously described as “regulated unproductive splicing and translation” (RUST).
Introduction
All mammals express six isoforms of actin: α-cardiac (Actc1, NM_009608), α-skeletal (Acta1, NM_009606), α-aortic (Acta2, NM_007392), γ-enteric (Actg2, NM_009610), β-cytoplasmic (Actb, NM_007393), and γ-cytoplasmic (Actg1, NM_009609). Each actin is encoded on a separate chromosome but the coding sequence of the actins are 71% identical and there is 92% amino acid sequence identity between actin proteins. This degree of conservation is indicative of intolerance of these proteins to changes in amino acid composition, presumably because of the large number of proteins that interact directly with actin. Although the coding sequences are similar between actin isoforms, the genomic architecture of actin isoforms differs between the cytoplasmic (six exons), smooth muscle (nine exons), and cardiac and skeletal isoforms (seven exons). The genomic sequence of Actg1 was first described in 1986 by Erba and colleagues [1] and no splice variants of this gene have been reported.
In most dividing cells the two cytoplasmic actins are expressed at high levels. For example, mature skeletal and cardiac muscle derive from myoblasts, which express high levels of β - and γ-actin in their undifferentiated form. However, during differentiation, and in mature skeletal and cardiac muscle, the cytoplasmic actins are down-regulated to comprise only a small fraction of the total actin content, and α-skeletal and α-cardiac actins, respectively, become the predominant isoforms [2]–[4]. Nevertheless Actg1-null mice demonstrate that γ-actin is crucial for the normal function of mature skeletal muscle, as its complete absence results in a progressive myopathy in adult mice [5], [6].
C2C12 mouse myoblast cell culture is widely used to study the expression and regulation of genes during skeletal muscle development. In this system, myoblasts proliferate until induced to differentiate either via serum-starvation or substitution of horse serum into the growth medium. Differentiation of myoblasts involves exit from the cell cycle, fusion with neighboring cells, elongation into myotubes, and movement of the nuclei to the periphery of the myotubes. Subsequent maturation is characterized by bundling of α-actin thin filaments to form myofibrils. Down-regulation of Actb in differentiated C2C12 cells was previously attributed to a 40 nucleotide long conserved element in the 3′ UTR of the Actb transcript [7]. In contrast, Actg1 down-regulation was proposed to involve inhibition of splicing of intron 3 from the primary Actg1 transcript, thus preventing the production of a mature RNA [2].
A potentially relevant mechanism for post-transcriptional down-regulation is Regulated Unproductive Splicing and Translation [8]. RUST occurs by alternative splicing to include a regulatory exon which either contains, or creates via frameshift, a premature termination codon (PTC). Introduction of a PTC results in subsequent degradation of the mRNA by nonsense-mediated decay (NMD). Recent evidence suggests that as many as 4% of transcripts include alternatively spliced exons with PTCs, though there is some debate as to whether this splicing is functional or an artifact of highly active splicing factors in cells and therefore should be considered transcriptional noise [9], [10]. To help resolve this, Zhang and colleagues proposed criteria for deciding whether a splicing event is functional or noise. Briefly, they have suggested that alternative splicing is likely to be functional if the exon is: 1) conserved among species, 2) developmentally regulated, and 3) tissue specific [11].
We identified an alternatively spliced exon in Actg1 transcripts from mouse skeletal muscle cDNA. This alternative transcript includes a novel 45 bp exon (exon 3a) located in the middle of Actg1 intron 3 that introduces a PTC. A similar alternative Actg1 transcript is found in skeletal muscle from multiple other mammals. In mouse, splicing to include this exon is regulated in a tissue and development specific manner. Here we demonstrate that inclusion of exon 3a results in post-transcriptional down-regulation of Actg1 by RUST.
Results
Identification of a novel Actg1 transcript
While investigating γ-actin expression in a knock-in mouse model harboring a targeted mutation in exon 4 of Actg1, we identified a novel, alternatively spliced Actg1 transcript in wild-type animals. PCR amplification of skeletal muscle cDNA designed to amplify mouse Actg1 exon 3 to 4 (Figure 1A, Table 1), but to not allow amplification of Actb and Acta1, yielded an unexpected product. In addition to the predicted 102 bp product, an amplicon of 147 bp was observed in skeletal muscle, heart and diaphragm (Figure 1B). Sequencing of the 147 bp product from skeletal muscle revealed an alternatively spliced transcript that includes a 45 bp exon located in intron 3, which we designate exon 3a (Figure 2A,B). It should be noted that in addition to the PCR products corresponding to the two alternatively spliced transcripts, larger amplicons that were not present in the no-RT controls were observed. Gel extraction and Sanger sequencing of these products revealed these were partially spliced products which included regions of intron 3 upstream and downstream of exon 3a (Figure 2C). In mouse, exon 3a is flanked by canonical splice acceptor and donor sites, and inclusion predicts introduction of an in-frame termination codon (Figure 2B,D). A BLAST search of the NCBI mouse EST database revealed transcripts that included exon 3a (http://blast.ncbi.nlm.nih.gov/Blast.cgi).
Fig. 1. Splicing to include exon 3a into the Actg1 transcript is tissue specific. 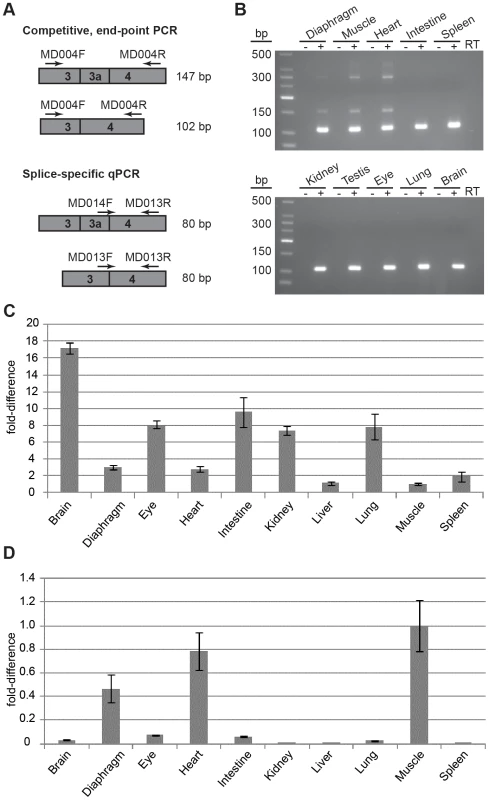
(A) Two PCR-based assays were employed to screen for the presence of Actg1 – competitive end-point PCR and splice-specific qPCR. Actg1-specific primers were designed to amplify either both the normal and alternative transcripts in a single reaction (competitive end-point PCR), or specifically the normal or alternative transcripts in separate reactions (splice-specific qPCR). (B) The competitive end-point PCR assay was used to amplify all Actg1 transcripts in various tissues from adult mouse. The larger PCR products at ∼390 bp and ∼290 bp are intermediate spliceforms of Actg1. The 147 bp product represents alternatively spliced, exon 3a-containing Actg1 transcripts, while the 102 bp product represents normally spliced Actg1 transcripts. (C) Expression data from qPCR to amplify normal Actg1 (exon 3 – exon 4) shows a high degree of variability of γ-actin expression between tissues. (D) Splicing to include exon 3a, as measured by qPCR is primarily limited to skeletal and cardiac muscle, with very low levels in the brain, eye, and intestine. All qPCR data is normalized to Rplp0 and Rrn18s expression and presented as a fold-difference to skeletal muscle. Fig. 2. Exon 3a and flanking sequence display a high degree of conservation among mammals. 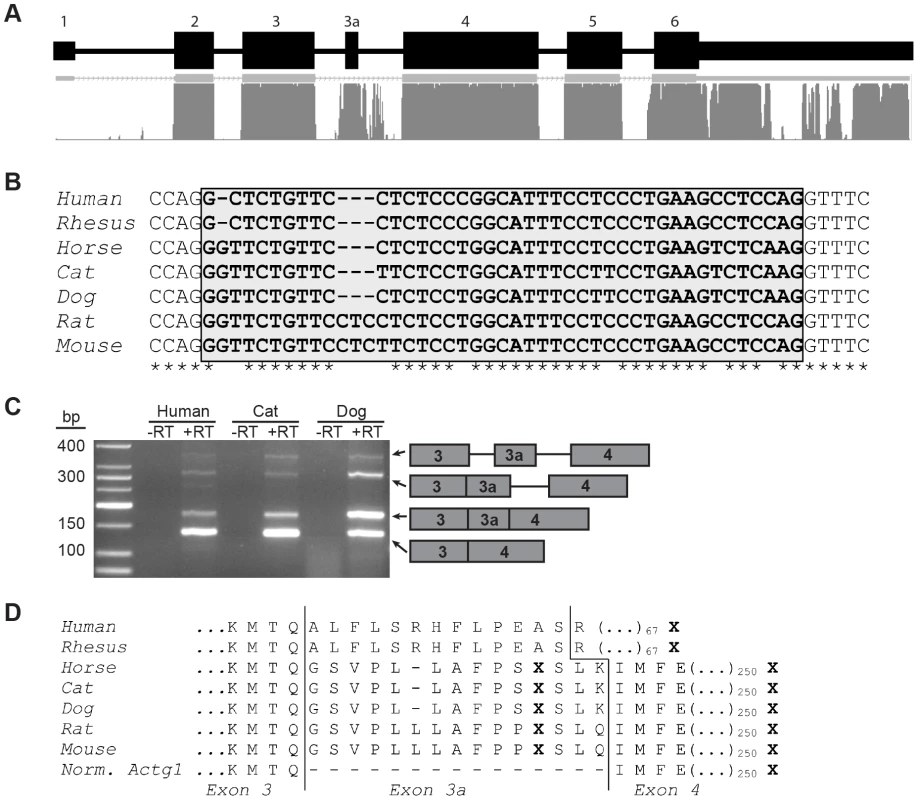
(A) Actg1 gene structure and mammalian conservation based on UCSC Genome Browser. (B) Alignment of the nucleotide sequence of exon 3a (shaded box) in seven mammals shows that exon 3a is flanked by canonical splice sites in all species and is 80% conserved. (C) Species- and isoform- specific primers were used to amplify across the exon 3 – exon 4 junction of Actg1 in humans, cat, and dog skeletal muscle cDNA. Similar to the competitive end-point PCR assay described in Figure 1, larger products are present corresponding to intermediate spliceforms. The primary Actg1 transcript corresponds to a 135 bp product, whereas inclusion of exon 3a results in a larger, 177 bp product (176 bp in human), as visualized on a 3% agarose gel. (D) Predicted translated products resulting from inclusion of exon 3a creates an in-frame stop codon (X) and is nearly perfectly conserved in all species with the exception of primates. In primates, an additional 1 bp deletion generates a frameshift resulting in degeneration of amino acid conservation of exon 3a and introduction of a premature stop codon (X) 68 amino acids into exon 4. Tab. 1. The sequence of primers used in this study. 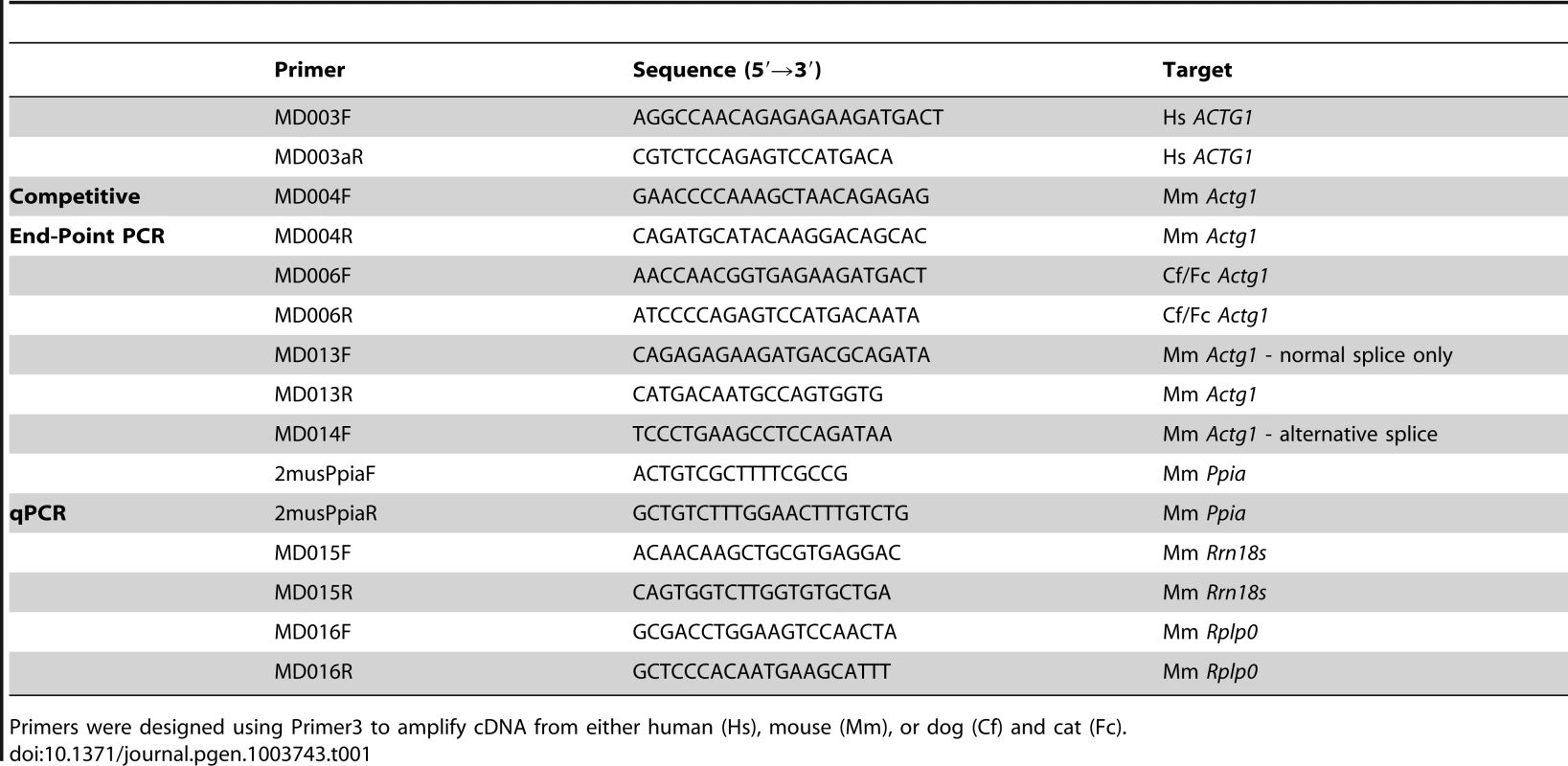
Primers were designed using Primer3 to amplify cDNA from either human (Hs), mouse (Mm), or dog (Cf) and cat (Fc). Exon 3a-containing transcripts are enriched in muscle
To determine if this transcript was present in other tissues of the adult mouse, cDNA was prepared from diaphragm, skeletal muscle, heart, intestine, spleen, kidney, testis, eye, lung, and brain from adult mice. Only skeletal and cardiac muscle were positive for the presence of alternatively spliced transcripts in addition to the normal Actg1 transcript, which was the major product amplified (Figure 1B).
To quantify expression of Actg1 isoforms, we designed primers compatible with qPCR to specifically amplify transcripts resulting from either an exon 3 - exon 4 (normal transcript) or an exon 3a - exon 4 splice (alternative transcript) (Figure 1A). Unlike competitive end-point PCR, the qPCR assay permits detection of low levels of the alternatively spliced Actg1 3a transcript. Using this method, we were able to obtain the relative abundance of normal Actg1 and alternative Actg1 transcript levels across tissues (Figure 1C,D). We found that brain exhibited the highest level of normal Actg1, whereas skeletal muscle and liver had the least. As expected from the results of the end-point PCR, skeletal muscle had the highest level of alternative Actg1.
The relative level of the alternative transcript was compared to that of the normal transcript using combined data from three qPCR experiments. The efficiencies of the PCR reactions for the normal and alternative transcript were within <1% of each other and the threshold was set at 0.2 for both reactions. This analysis revealed average ΔCt values of 20.8 for the normal and 22.7 for the alternative transcripts in skeletal muscle corresponding to a 3.5∶1 ratio of normal to alternatively spliced Actg1 (Table 2).
Tab. 2. Ct values for a skeletal muscle cDNA control demonstrate no plate-to-plate variability. 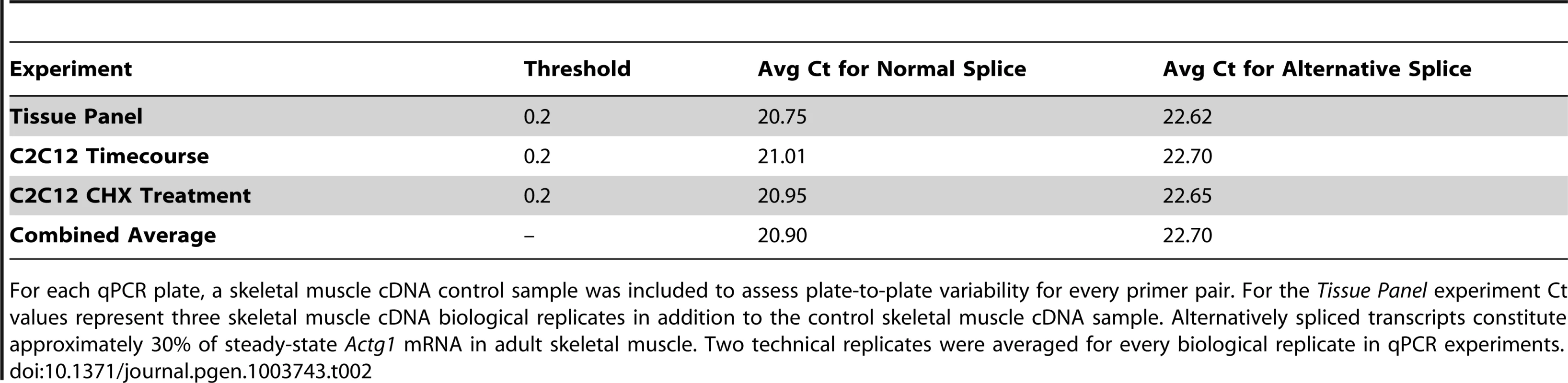
For each qPCR plate, a skeletal muscle cDNA control sample was included to assess plate-to-plate variability for every primer pair. For the Tissue Panel experiment Ct values represent three skeletal muscle cDNA biological replicates in addition to the control skeletal muscle cDNA sample. Alternatively spliced transcripts constitute approximately 30% of steady-state Actg1 mRNA in adult skeletal muscle. Two technical replicates were averaged for every biological replicate in qPCR experiments. Exon 3a is highly conserved among mammals
Evolutionary conservation of nucleotide sequence is typically indicative of functional significance. While no conservation of intron 3 is detected in fish or chicken cytoplasmic actin, the nucleotide sequence of Actg1 intron 3 is highly conserved among mammals. Specifically, the region containing the 45 bp alternatively spliced exon and flanking splice sites are 80% identical between humans and mice (Figure 2A,B). To determine if splicing of the Actg1 transcript is an evolutionarily conserved event in vivo, we prepared cDNA from human, dog, and cat skeletal muscle total RNA. Species - and isoform-specific primers were designed similar to the competitive end-point PCR assay for mouse described above and outlined in Figure 1A. Splicing to include exon 3a was observed in skeletal muscle cDNA from the species assayed (Figure 2C). All PCR products were sequenced to confirm the imputed exon 3a sequence (Figure 2B, human, cat, dog). Alignment of sequences obtained from the UCSC genome browser (http://genome.ucsc.edu) indicates that inclusion of exon 3a is predicted to introduce an in-frame PTC in non-primate mammals. In humans and rhesus, exon 3a is 41 nt in length and results in a frameshift of the ACTG1 coding sequence, thus creating a PTC in exon 4. While the amino acid sequence of the predicted polypeptide generated by inclusion of exon 3a is highly conserved in non-primate mammals, the frameshift generated by the 4 nt deletion in primates results in a complete loss of this conservation (Figure 2D).
Developmental regulation of Actg1 alternative splicing
To investigate the potential function of the alternative Actg1 transcript in a relevant tissue, we utilized the well-characterized C2C12 mouse myoblast cell line as a proxy for skeletal muscle development. C2C12 cells are frequently used to study transcriptional and proteome changes during the differentiation of myoblasts into myotubes (shown in Figure 3A) [12], [13]. Using these cells, we first asked if Actg1 alternative splicing is developmentally regulated. Total RNA was isolated from myoblasts prior to addition of differentiation medium and at 2 day intervals after addition of differentiation media. qPCR was used to determine expression levels of normal and alternatively spliced Actg1. In agreement with previous studies by Lloyd and Gunning [2], our results demonstrate that normal Actg1 is down-regulated during myoblast differentiation (Figure 3B). These data also reveal that concurrent with the decrease in normal Actg1 expression, alternatively spliced Actg1 transcripts increase during differentiation (Figure 3B).
Fig. 3. Splicing to include exon 3a is a developmentally regulated event in skeletal muscle. 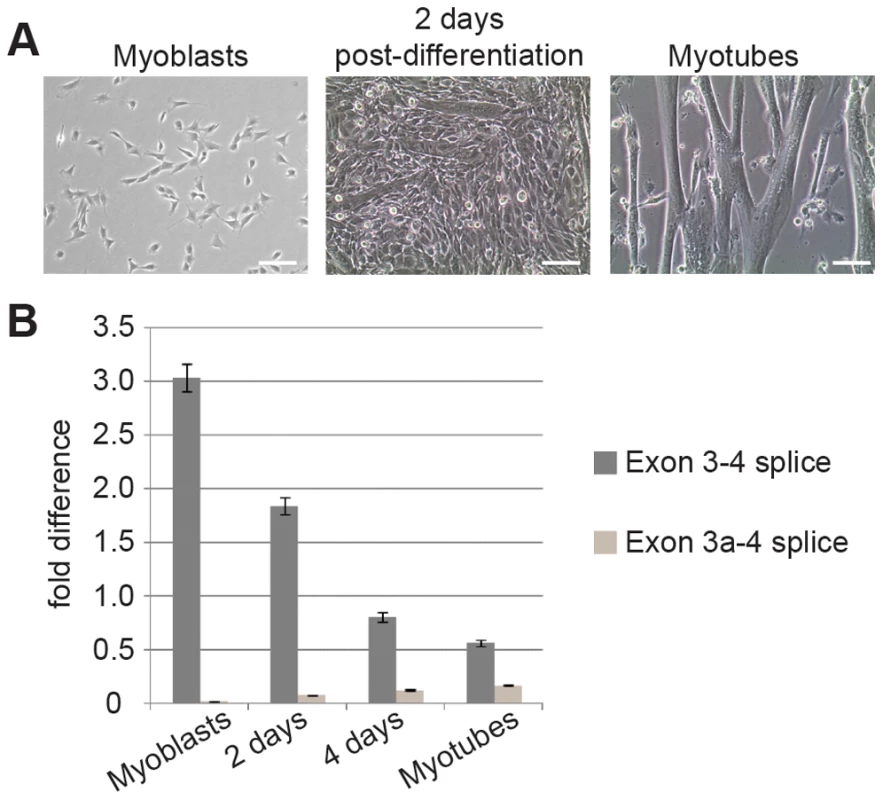
(A) Microscopy of cell cultures before, during and after differentiation. C2C12 myoblasts were grown to 70% confluence and induced to differentiate in DMEM+10% horse serum. Partially differentiated cultures containing both myoblasts and myotubes were observed by 2 days post-differentiation. After 48 hours, medium was replaced with DMEM+2% horse serum and 10 µM Ara-C and cultured for an additional 4 days. (B) qPCR of RNA harvested in Trizol showed that concurrent with a decrease in normal Actg1, splicing to generate alternative Actg1 increases during differentiation into myotubes. Expression of both the normal and alternative transcripts was normalized to Ppia and is presented as fold-difference compared to skeletal muscle. A two-tailed type 2 Student's T-test was used to compare expression differences between time points. For all time points compared, p<0.0001. Exon 3a-containing transcripts are exported to the cytoplasm, but no corresponding protein product is present
Given the lack of conservation in the amino acid sequence generated by inclusion of exon 3a (Figure 2D), we hypothesized that a protein product is not produced from the alternative Actg1 transcript. To address this, we first sought to determine if the alternative transcript is exported to the cytoplasm and therefore available for translation. Total RNA was isolated from both cytoplasmic and nuclear fractions of mature myotube cultures. Using competitive end-point PCR, we found that the nuclear fraction contained all splice products including putative splicing intermediates, whereas only the normal and alternatively spliced Actg1 transcripts were present in the cytoplasmic fraction (Figure 4A,B).
Fig. 4. Exon 3a containing alternatively spliced Actg1 is exported to the cytoplasm, but does not produce a stable protein. 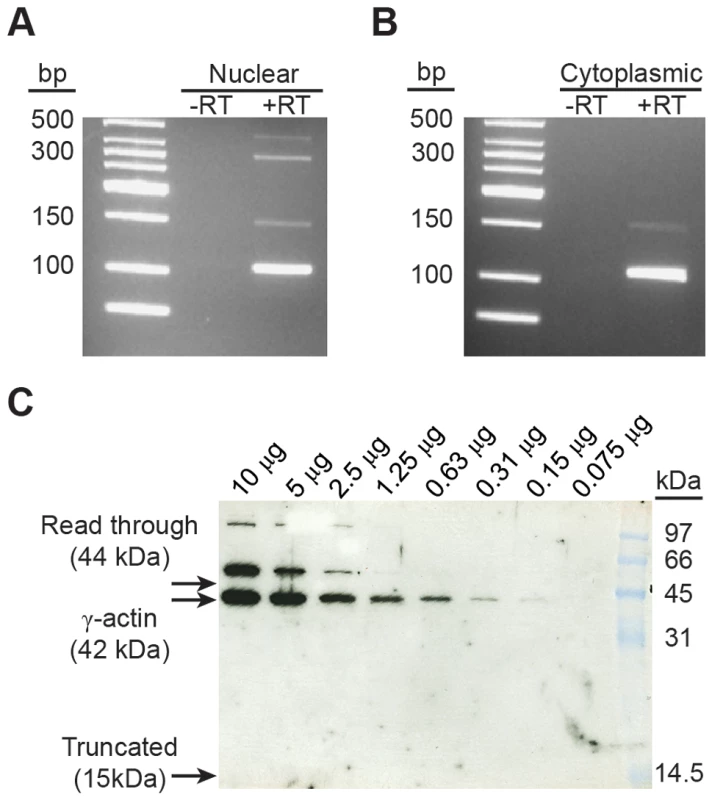
Nuclear (A) and cytoplasmic (B) RNA fractions were harvested from C2C12 myotubes and evaluated for the presence of alternative Actg1 using competitive, end-point PCR. All Actg1 spliceforms, including partially spliced transcripts, were present in the nuclear fraction, however, only the normal and exon 3a transcripts were observed in the cytoplasmic fraction. (C) Western blot using an anti-γ-actin specific antibody was used to probe mouse skeletal muscle lysate for the presence of a protein product corresponding to the inclusion of exon 3a. Usage of the in-frame stop codon in exon 3a would generate a 15 kDa protein. Alternatively, read-through of the stop codon would increase the size of the ACTG1 protein by 2 kDa, resulting in a 44 kDa protein. Neither of these protein products is present. A γ-actin protein at 52 kDa is observed and likely represents a post-translational modification. Molecular weight marker is prestained Low Range (BioRad). Knowing that the alternative transcript is exported to the cytoplasm, we used western blotting to detect a protein product corresponding to either the use of the premature stop codon or a read-through of the stop codon. Because the levels of alternative transcript in the skeletal muscle samples exceed that of the myotubes in culture, we used skeletal muscle lysate for this experiment. Using an anti-γ-actin specific antibody directed against the N-terminus of the polypeptide, we probed for the presence of a protein product from alternatively spliced Actg1 transcripts. Based on the qPCR data, there should be a 3∶1 ratio of normal ACTG1 to alternative ACTG1 (Table 2). We loaded 1∶2 serial dilutions to determine our lower-limit of detection by western blot, beginning with 10 µg of total protein. No band corresponding either to usage of the termination codon (15 kDa) or a read-through of the termination codon in exon 3a (45 kDa) was detected (Figure 4C). We did observe a larger, 52 kDa protein in these samples which is consistent in size with previously described modifications of cytoplasmic and skeletal muscle actins, specifically mono-sumoylation [14] or mono-ubiquitination [15].
Inhibition of nonsense-mediated decay results in an increase of exon 3a-containing transcripts
We reasoned that exon 3a is alternatively spliced to post-transcriptionally down-regulate expression of Actg1. To address the hypothesis that exon 3a represses translation of Actg1 by targeting the transcript for NMD, we treated cells with cycloheximide to block translation. Cycloheximide targets the small ribosomal subunit and can be used to inhibit translation-dependent NMD of PTC-containing transcripts [16]. Cultures of proliferating myoblasts and mature myotubes were treated with either 40 µg/mL cycloheximide in ethanol or an ethanol-only control for three hours in otherwise standard growth conditions. A three-hour treatment with cycloheximide resulted in an approximately 7-fold increase of exon 3a transcripts as measured by qPCR (Figure 5). These data strongly suggest that exon 3a targets the transcript for translation-dependent NMD. Furthermore, they indicate that splicing to include exon 3a is a frequent event in mature myotubes, given the rapid increase in the relative abundance of the alternatively spliced product.
Fig. 5. Cycloheximide (CHX) treatment of myotubes results in a 7-fold increase in alternative Actg1. 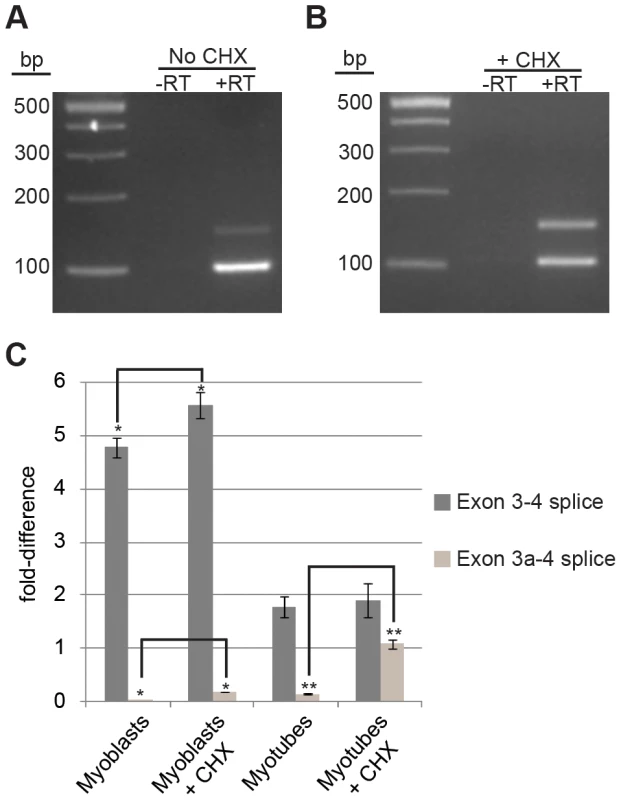
(A,B) Competitive end-point PCR and (C) qPCR were used to evaluate relative abundance in untreated (A,C) and CHX treated cells (B,C). Expression of both the normal and alternative transcripts were normalized to Ppia and are presented as fold-difference compared to skeletal muscle. A two-tailed type 2 Student's T-test was used to compare expression differences between samples. * p<0.05, ** p<0.001. Discussion
In this study, we identified a novel Actg1 splice variant enriched in cardiac and skeletal muscle. We propose that production of this alternative transcript is regulated and functional. Despite the fact that cytoplasmic actins are well-studied and widely used as reference genes, this is the first report of alternative splicing for any actin transcript. Why might exon 3a containing transcripts have been overlooked? The most divergent sequence for distinguishing between actin transcripts reside in the 5′ and 3′ UTRs. PCR primers designed to specifically amplify one isoform are typically in the UTRs and inclusion of a small exon, such as the 41–45 nt exon 3a, is likely to be overlooked in a large, >1 kb PCR product. Furthermore, short PCR products in which a 41–45 nt difference could be resolved are unlikely to involve exons 3–5 of the actin transcript because the high degree of nucleotide similarity between actin isoforms in these exons. Given that the alternative exon appears to be specific to γ-actin, a product corresponding to the alternative transcript would be lost among the abundance of the other actin cDNAs amplified.
Previous studies suggested that the phenomenon of retention of intron 3 in the primary Actg1 RNA would be responsible for the down-regulation of γ-actin during differentiation of myoblasts [2], [17]. Here we add support for this hypothesis and provide evidence for regulated splicing to incorporate an additional exon situated in the middle of intron 3 which is sufficient for down-regulation of Actg1 via a mechanism not previously shown for an actin transcript. Cycloheximide inhibition of translation results in an elevation in the level of exon 3a-containing transcripts, a finding that is consistent with translation-dependent NMD. Cycloheximide is commonly used as an inhibitor of NMD [16], [18], [19], however there exists the possibility that the increase in exon 3a-containing transcripts is the result of another effect of the cycloheximide treatment rather than a direct correlation with inhibition of the NMD pathway. Therefore, assuming no additional effects of treatment with cycloheximide, we hypothesize that γ-actin is down-regulated via alternative splicing to introduce a PTC, leading to the degradation of Actg1 transcripts via NMD [8], [20].
We considered the possibility that the presence of the alternatively spliced Actg1 transcript is the result of ‘noisy splicing’. Large-scale analyses indicate that the majority of alternatively spliced transcripts are likely generated in error because of their low abundance across multiple tissues and lack of correlation with expression differences in the genes examined [9], [21], [22]. However, several examples of RUST as a mechanism of post-translational down-regulation exist [23]–[26]. Using the guidelines established by Zhang and colleagues to distinguish between noisy and functional splicing, we evaluated whether inclusion of exon 3a in Actg1 transcripts is likely to be spurious [11], [22]. A primary feature of ‘noisy splicing’ is lack of conservation of the alternative splice form in other species. However, we find that, similar to the normal coding exons of Actg1, exon 3a is highly conserved in mammals. Furthermore, in agreement with criteria for functional splice variants, we demonstrated that the alternative splice form is abundant only in skeletal and cardiac muscle, and that this splicing event is differentially regulated in developing myotubes.
Splicing to include exon 3a in Actg1 transcripts maintains the proper reading frame in most species, however in primates exon 3a is only 41 nucleotides, thereby creating a frameshift. This subtle difference in the sequence of exon 3a between primates and lower mammals further supports the regulatory hypothesis in that it is the generation of a PTC and not a translated product that is evolutionarily conserved. Previous studies of RUST indicate that this type of regulation is not only conserved across species, but is typically found as a regulatory mechanism for members of the same gene family. We believe RUST may post-transcriptionally regulate both cytoplasmic actins, γ and β. Both cytoplasmic actins have a similar genomic structure and similar to γ -actin, β-actin has high degree of conservation in a portion of intron 3 [27]. In addition, it was recently demonstrated that an expression construct utilizing the β-actin promoter in combination with the 3′ UTR resulted in high level expression in skeletal muscle [28], [29]. It is possible the conserved region of intron 3 of β-actin may confer an additional level of regulation in addition to the previously reported conserved 40 nt element in the 3′UTR [7]. The genomic structure of the non-cytoplasmic actins is markedly different from the cytoplasmic actins, and lack the high degree conservation observed in Actg1 and Actb intron 3. Therefore, RUST is an unlikely mechanism of regulation for the non-cytoplasmic actin isoforms.
The premise of RUST seems counter-intuitive as a regulatory mechanism, since the most efficient means of down-regulation would be at the transcriptional level. However, Soergel et al note that the production of large transcripts in any instance can be an inherently wasteful endeavor, as introns can constitute up to 95% of a primary RNA transcript [30], [31]. It is possible that RUST can serve as a mechanism to quickly modulate expression of genes that are typically highly expressed, form very stable transcripts, or are essential in the cell. When a particular environmental or physiologic change dictates that only moderate levels of protein are required, production can be down-regulated without entirely switching-off transcription. In such an instance, post-transcriptional degradation of a portion of excess transcripts produced via NMD may be more energy efficient for the cell than either switching on or off transcription or post-translational degradation of unnecessary proteins. This model is suitable for γ-actin in muscle, as it is expressed at high levels in proliferating myoblasts, but is also necessary at lower levels in differentiated myotubes and developed skeletal muscle. RUST due to inclusion of exon 3a is unlikely to be the only mechanism of regulation for cytoplasmic actin transcripts. Though oftentimes used as a constitutive promoter in in vitro systems, the Actb and Actg1 promoters may also supply further control to the down-regulation of actin at a transcriptional level. Furthermore, retention of the entire intron 3 of the immature Actg1 message may contribute to delayed processing of mature transcripts [2]. Indeed, the increased abundance of incompletely spliced heterogeneous RNA observed when exon 3a-containing transcripts were present (figures 1B, 2C, 4a) may suggest a role for intron retention in down-regulation.
While the alternative transcript is enriched in skeletal and cardiac muscle, we were also able to detect very low levels in brain, eye, and intestine; tissues which also expressed normal Actg1 at high levels. Regulation of β-actin synthesis was previously shown to be influenced by the levels of actin monomers in cells treated with an actin depolymerizing agent, latrunculin A [32]. We speculate that γ-actin expression may also involve an auto-regulatory mechanism by which an excess of free actin monomers in the cell induces splicing to include exon 3a of the Actg1 transcript, thus temporarily halting production in cell types which normally require high levels of actin. In such a situation, very low levels of exon 3a-containing transcipts would be expected in tissues with a high cytoplasmic actin content.
In skeletal and cardiac muscle, exon 3a splicing may be regulated by splicing enhancer or splicing repressor elements. The intronic region immediately adjacent to the 5′ donor site of exon 3a is well conserved among mammals (Figure 6). During differentiation of myoblasts, multiple transcription and splicing factors are required to coordinate changes in gene expression crucial for differentiation. As such, the intronic conservation immediately 3′ of exon 3a may contain recognition motifs for splicing enhancers or repressors. Indeed, an exon splicing enhancer prediction software, ESE Finder v3.0, identified several splicing enhancer recognition motifs (Figure 6). Of particular interest are the recognition motifs for SF2/ASF and SC35, which were shown previously to effect alternative splicing of β-tropomyosin in a tissue-specific manner [33].
Fig. 6. The intronic sequence immediately 3′ of exon 3a is highly conserved. 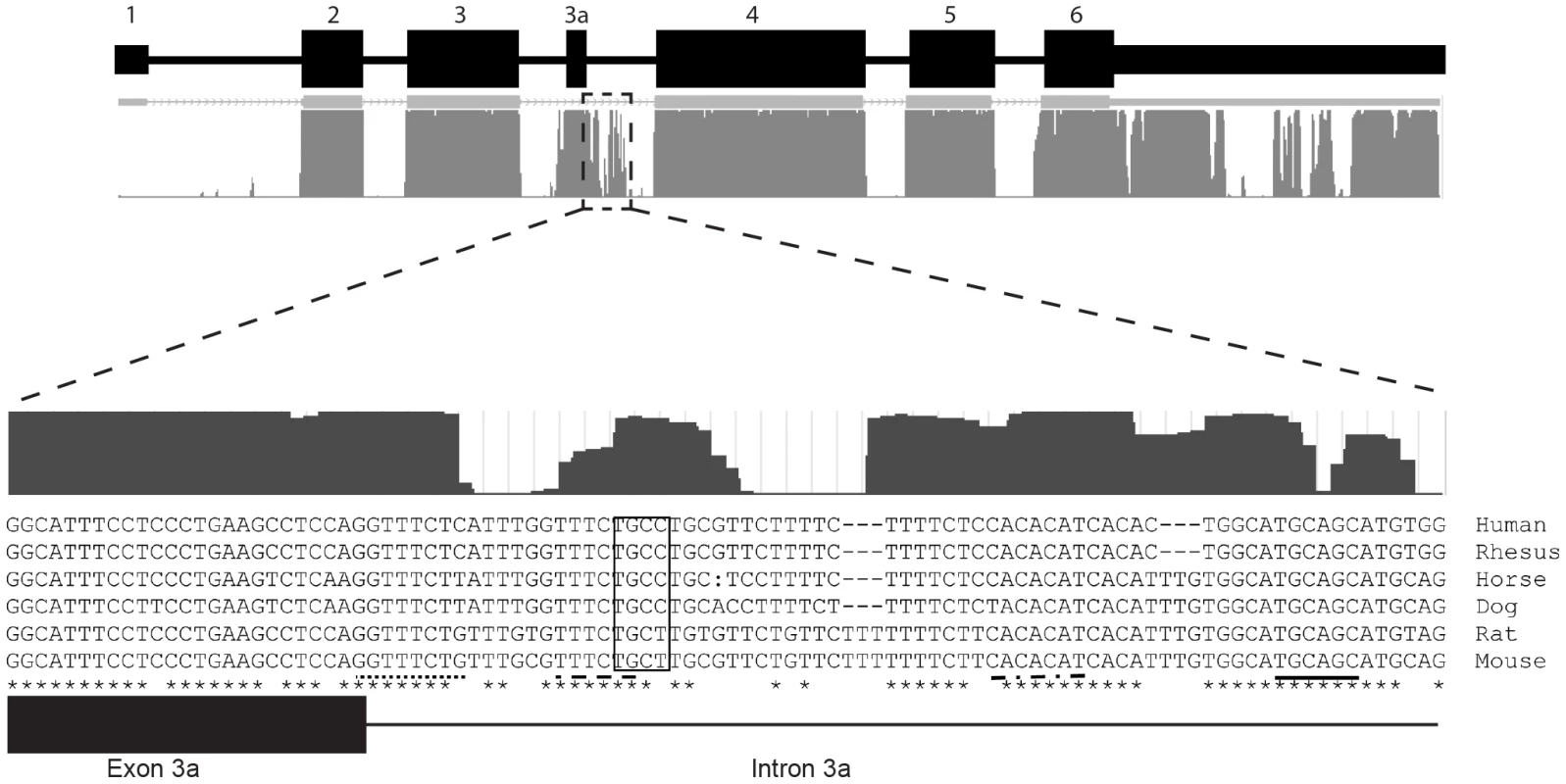
The UCSC mammalian conservation track shows a high degree of conservation in the 3′ flanking sequence. ESE Finder v3.0 was used to identify consensus sequences for multiple splicing enhancer elements in human and mouse, including SRSF1 (dot-dash line), SRSF2 (dotted line), SRSF5 (dashed line), SRSF6 (solid line). All predicted ESEs shown in this figure are considered “high-score motifs” because the threshold is greater than the baseline threshold for each motif: SRSF1 = 3.658 (minimum 1.956); SRSF2 = 4.893 (minimum 2.383); SRSF5 = 3.877 (minimum 2.670); SRSF6 = 4.713 (minimum 2.676). In addition, a muscleblind-like 1 YCGY motif (box) is found 3′ of the exon 3a [34]. In closing, this report documents the first identification and characterization of an alternatively spliced actin transcript. These data provide evidence for the dynamic regulation of Actg1 and further functional evidence for RUST.
Materials and Methods
Bioinformatics
Sequence ascertainment and analysis was performed using UCSC Genome Browser/BLAT (http://www.genome.ucsc.edu/), Ensembl! (http://www.ensembl.org), Sequencher 6.0 (Gene Codes Corp.), and Clustal X2 (http://www.clustal.org). Primers were designed using Primer3 software (http://frodo.wi.mit.edu/). Potential exon splicing enhancers were identified using ESEFinder v3.0 (http://rulai.cshl.edu/cgi-bin/tools/ESE3/esefinder.cgi)
Animals and tissue preparation
All animals were maintained according to Michigan State University IACUC and NIH guidelines. Tissue samples were harvested from three 1 year old C57Bl/6J mice, snap frozen on dry ice, and stored at −80°C. Prior to use for RNA or protein isolation, samples were chopped into 100–200 mg pieces. Cat and dog skeletal muscle samples were provided courtesy of Dr. John Fyfe (Michigan State University). Human skeletal muscle total RNA was purchased from Ambion (catalog AM7982; Austin, TX).
Cell culture
All cell culture media and supplements were purchased from Invitrogen (Carlsbad, CA), unless noted otherwise. C2C12 myoblast cells were purchased from ATCC (Manassas, VA; CRL-1772) and propagated in DMEM containing 10% heat-inactivated fetal bovine serum and 2 mM L-glutamine. Differentiation of myoblasts into myotubes was achieved by culturing cells >70% confluent in DMEM supplemented with 10% horse serum in place of fetal bovine serum. Forty-eight hours post differentiation, DMEM with 2% horse serum and 10 µM Ara-C (Sigma, St. Louis, MO) was used to maintain differentiated myotubes and inhibit the proliferation of myoblasts. Cells were maintained at 37°C in 5% CO2.
RNA isolation and cDNA synthesis
Total RNA isolation from tissue and cell culture samples was achieved using TRIzol (Invitrogen, Carlsbad, CA) purification followed by a DNaseI treatment using RNeasy mini-columns (Qiagen, Hilden, Germany). Total RNA was quantified using a NanoDrop (Thermo, Wilmington, DE). cDNA was synthesized using SuperScriptIII reverse transcriptase (Invitrogen, Carlsbad, CA) according to manufacturer's instructions. Reaction volume was 20 µl; for qPCR, 300 ng of RNA was used per reaction, and for endpoint PCR, 1 µg of RNA was used. Following incubation at 55°C for 1 hour, samples were heat inactivated at 75°C for 20 minutes then stored at −20°C until used.
Nuclear and cytoplasmic RNA isolation
Myotube cultures were lysed rapidly with 1% Triton X-100 in PBS, cell debris and nuclei were gently scraped from the culture dishes and evaluated by brightfield microscopy for the presence of intact nuclei. Samples were centrifuged at 1,000×g for 5 minutes to pellet nuclei. The cytosol-containing supernatant was examined under the microscope to assure the absence of nuclei in the cytosolic fraction prior to the addition of TRIzol LS to the supernatant and TRIzol to the pellet (Invitrogen, Carlsbad, CA). RNA purification of both the cytosolic and nuclear fractions was done according to manufacturer's instructions.
Competitive end-point PCR amplification
Actg1 cDNA was amplified using Promega GoTaq polymerase per manufacturer's instructions (Madison, WI) with 5 µM each of species - and isoform-specific primers located in exons 3 and 4 of Actg1 (Table 1). For most reactions, 28 cycles were sufficient to amplify within the perceived linear range. Products were evaluated on 3% agarose gels containing 0.3 µg/mL ethidium bromide and visualized using BioRad GelDoc System (Hercules, CA). Pixel intensity of the PCR products was quantitated using GelDoc software (BioRad, Hercules, CA) for semi-quantitative analysis.
qPCR
Quantitative PCR (qPCR) was performed on an ABI7000, using Power SYBR Green (Invitrogen, Carlsbad, CA) as the reporter dye, and data were collected using StepOne Plus software (Applied BioSystems, Carlsbad, CA). Data were analyzed using qbasePLUS software (Biogazelle, Zwijnaarde, Belgium). Actg1 transcripts were normalized to Ppia for experiments using C2C12 cells and to Rplp0 and Rrn18s for experiments using mouse tissues. See Table 1 for primer sequences. Averaged data represent two technical replicates for each of three biological replicates. A two-tailed type 2 Student's T-test was used to compare expression differences between samples.
Sequencing
Sequence data for exon 3a were obtained by Sanger sequencing on an ABI Prism 3700 DNA Analyzer at the Research Technology Support Facility (Michigan State University) using competitive, end-point PCR (Table 1).
Cycloheximide treatment
Cycloheximide (Sigma, St. Louis, MO) was dissolved in 100% ethanol at a stock concentration of 40 mg/mL and added to growth medium at a final concentration of 40 µg/mL. Cells were incubated in cycloheximide containing medium for 3 hours and immediately harvested in TRIzol for RNA isolation and cDNA synthesis as described above. Each experiment was repeated three times.
Protein isolation
Approximately 100 mg of skeletal muscle from a 1 year old mouse was lysed using a Polytron rotor homogenizer in lysis buffer containing 100 mM KCl, 10 mM PIPES, 5 mM EGTA, 1% Triton X-100 and Complete Protease Inhibitors (Roche, Basel, Switzerland), and incubated on ice for 1 hour to allow further lysis. Protein content in the total lysate was determined using a Bradford assay (BioRad, Hercules, CA).
Western blotting
Proteins were separated via SDS-PAGE on discontinuous 12% bis-acrylamide gels [10]. Proteins were transferred in 10 mM Tris pH 7.4, 100 mM glycine, 15% methanol (transfer buffer) at 4°C overnight at a constant current of 5 mAmp onto polyvinylidene difluoride (PVDF) membranes (BioRad, Hercules, CA). Membranes were incubated in PBS (pH 7.4) containing 5% non-fat milk and 0.025% Tween-20 (blocking buffer) for one hour at room temperature. A previously validated rabbit polyclonal anti-γ-actin specific antiserum raised against the first 15 amino acids of the polypeptide [5] was diluted 1∶10,000 in blocking buffer. Membranes were incubated with primary antiserum for 2 hours at room temperature. Goat polyclonal anti-rabbit IgG-HRP conjugated secondary antibody (Sigma, St. Louis, MO) was used at 1∶3,000 in blocking buffer for one hour at room temperature. Proteins were detected using an ECL Detection Kit (GE Healthcare, Waukesha, WI) with Amersham Hyperfilm MP autoradiography film (GE healthcare, Waukesha, WI).
Zdroje
1. ErbaHP, GunningP, KedesL (1986) Nucleotide sequence of the human gamma cytoskeletal actin mRNA: anomalous evolution of vertebrate non-muscle actin genes. Nucleic Acids Res 14 : 5275–5294.
2. LloydC, GunningP (2002) beta - and gamma-actin genes differ in their mechanisms of down-regulation during myogenesis. J Cell Biochem 84 : 335–342.
3. RubensteinPA, SpudichJA (1977) Actin microheterogeneity in chick embryo fibroblasts. Proc Natl Acad Sci U S A 74 : 120–123.
4. StortiRV, CoenDM, RichA (1976) Tissue-specific forms of actin in the developing chick. Cell 8 : 521–527.
5. BelyantsevaIA, PerrinBJ, SonnemannKJ, ZhuM, StepanyanR, et al. (2009) Gamma-actin is required for cytoskeletal maintenance but not development. Proc Natl Acad Sci U S A 106 : 9703–9708.
6. SonnemannKJ, FitzsimonsDP, PatelJR, LiuY, SchneiderMF, et al. (2006) Cytoplasmic gamma-actin is not required for skeletal muscle development but its absence leads to a progressive myopathy. Dev Cell 11 : 387–397.
7. DePonti-ZilliL, Seiler-TuynsA, PatersonBM (1988) A 40-base-pair sequence in the 3′ end of the beta-actin gene regulates beta-actin mRNA transcription during myogenesis. Proc Natl Acad Sci U S A 85 : 1389–1393.
8. LewisBP, GreenRE, BrennerSE (2003) Evidence for the widespread coupling of alternative splicing and nonsense-mediated mRNA decay in humans. Proc Natl Acad Sci U S A 100 : 189–192.
9. McGlincyNJ, SmithCW (2008) Alternative splicing resulting in nonsense-mediated mRNA decay: what is the meaning of nonsense? Trends Biochem Sci 33 : 385–393.
10. LareauLF, GreenRE, BhatnagarRS, BrennerSE (2004) The evolving roles of alternative splicing. Curr Opin Struct Biol 14 : 273–282.
11. ZhangZ, XinD, WangP, ZhouL, HuL, et al. (2009) Noisy splicing, more than expression regulation, explains why some exons are subject to nonsense-mediated mRNA decay. BMC Biol 7 : 23.
12. CasadeiL, ValloraniL, GioacchiniAM, GuesciniM, BurattiniS, et al. (2009) Proteomics-based investigation in C2C12 myoblast differentiation. Eur J Histochem 53 : 261–268.
13. KislingerT, GramoliniAO, PanY, RahmanK, MacLennanDH, et al. (2005) Proteome dynamics during C2C12 myoblast differentiation. Mol Cell Proteomics 4 : 887–901.
14. HofmannWA, ArduiniA, NicolSM, CamachoCJ, LessardJL, et al. (2009) SUMOylation of nuclear actin. J Cell Biol 186 : 193–200.
15. KudryashovaE, KudryashovD, KramerovaI, SpencerMJ (2005) Trim32 is a ubiquitin ligase mutated in limb girdle muscular dystrophy type 2H that binds to skeletal muscle myosin and ubiquitinates actin. Journal of Molecular Biology 354 : 413–424.
16. CarterMS, DoskowJ, MorrisP, LiS, NhimRP, et al. (1995) A regulatory mechanism that detects premature nonsense codons in T-cell receptor transcripts in vivo is reversed by protein synthesis inhibitors in vitro. J Biol Chem 270 : 28995–29003.
17. LloydC, GunningP (1993) Noncoding regions of the gamma-actin gene influence the impact of the gene on myoblast morphology. J Cell Biol 121 : 73–82.
18. LambaJK, AdachiM, SunD, TammurJ, SchuetzEG, et al. (2003) Nonsense mediated decay downregulates conserved alternatively spliced ABCC4 transcripts bearing nonsense codons. Hum Mol Genet 12 : 99–109.
19. NoensieEN, DietzHC (2001) A strategy for disease gene identification through nonsense-mediated mRNA decay inhibition. Nat Biotechnol 19 : 434–439.
20. GreenRE, LewisBP, HillmanRT, BlanchetteM, LareauLF, et al. (2003) Widespread predicted nonsense-mediated mRNA decay of alternatively-spliced transcripts of human normal and disease genes. Bioinformatics 19 Suppl 1: i118–121.
21. PanQ, SaltzmanAL, KimYK, MisquittaC, ShaiO, et al. (2006) Quantitative microarray profiling provides evidence against widespread coupling of alternative splicing with nonsense-mediated mRNA decay to control gene expression. Genes Dev 20 : 153–158.
22. MelamudE, MoultJ (2009) Stochastic noise in splicing machinery. Nucleic Acids Res 37 : 4873–4886.
23. HyvonenMT, UimariA, VepsalainenJ, KhomutovAR, KeinanenTA, et al. (2012) Tissue-specific alternative splicing of spermidine/spermine N1-acetyltransferase. Amino Acids 42 : 485–493.
24. De RosaM, MorelliG, CesaroE, DuraturoF, TuranoM, et al. (2007) Alternative splicing and nonsense-mediated mRNA decay in the regulation of a new adenomatous polyposis coli transcript. Gene 395 : 8–14.
25. UranoE, MiyauchiK, IchikawaR, FutahashiY, KomanoJ (2012) Regulation of cyclin T1 expression and function by an alternative splice variant that skips exon 7 and contains a premature termination codon. Gene 505 : 1–8.
26. OchsMJ, SorgBL, PufahlL, GrezM, SuessB, et al. (2012) Post-transcriptional regulation of 5-lipoxygenase mRNA expression via alternative splicing and nonsense-mediated mRNA decay. PLoS One 7: e31363.
27. NgSY, GunningP, EddyR, PonteP, LeavittJ, ShowsT, KedesL (1985) Evolution of the functional human beta-actin gene and its multi-pseudogene family: conservation of noncoding regions and chromosomal dispersion of pseudogenes. Mol Cell Biol 5 (10) 2720–2732.
28. KeeAJ, SchevzovG, Nair-ShallikerV, RobinsonCF, Vrhovski, et al. (2004) Sorting of a nonmuscle tropomyosin to a novel cytoskeletal compartment in skeletal muscle results in muscular dystrophy. J Cell Biol 166 : 685–696.
29. SchevzovG, FathT, VrhovskiB, VlahovichN, RajanS, et al. (2008) Divergent Regulation of the Sarcomere and the Cytoskeleton. J Biol Chem 283 : 275–283.
30. Soergel DAW, Lareau LF, Brenner SE (2006) Regulation of Gene Expression by Coupling of Alternative Splicing and NMD. In: Maquat LE, editor. Nonsense-Mediated mRNA Decay. Austin, Tx: Landes Bioscience.
31. LanderES, LintonLM, BirrenB, NusbaumC, ZodyMC, et al. (2001) Initial sequencing and analysis of the human genome. Nature 409 : 860–921.
32. LyubimovaA, BershadskyAD, Ben-Ze'evA (2000) Autoregulation of actin synthesis requires the 3 ′-UTR of actin mRNA and protects cells from actin overproduction. J Cell Biochem 76 : 1–12.
33. GallegoME, GattoniR, SteveninJ, MarieJ, Expert-BezanconA (1997) The SR splicing factors ASF/SF2 and SC35 have antagonistic effects on intronic enhancer-dependent splicing of the beta-tropomyosin alternative exon 6A. EMBO J 16 : 1772–1784.
34. GoersES, PurcellJ, VoelkerRB, GatesDP, BerglundJA (2010) MBNL1 binds GC motifs embedded in pyrimidines to regulate alternative splicing. Nucleic Acids Res 38 : 2467–2484.
Štítky
Genetika Reprodukční medicína
Článek Defending Sperm FunctionČlánek How to Choose the Right MateČlánek Conserved Translatome Remodeling in Nematode Species Executing a Shared Developmental TransitionČlánek Genome-Wide and Cell-Specific Epigenetic Analysis Challenges the Role of Polycomb in SpermatogenesisČlánek The Integrator Complex Subunit 6 (Ints6) Confines the Dorsal Organizer in Vertebrate EmbryogenesisČlánek Multiple bHLH Proteins form Heterodimers to Mediate CRY2-Dependent Regulation of Flowering-Time inČlánek Playing the Field: Sox10 Recruits Different Partners to Drive Central and Peripheral MyelinationČlánek A Minimal Nitrogen Fixation Gene Cluster from sp. WLY78 Enables Expression of Active Nitrogenase inČlánek Evolutionary Tuning of Protein Expression Levels of a Positively Autoregulated Two-Component System
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2013 Číslo 10
-
Všechny články tohoto čísla
- Defending Sperm Function
- How to Choose the Right Mate
- A Mutation in the Gene in Labrador Retrievers with Hereditary Nasal Parakeratosis (HNPK) Provides Insights into the Epigenetics of Keratinocyte Differentiation
- Conserved Translatome Remodeling in Nematode Species Executing a Shared Developmental Transition
- A Novel Actin mRNA Splice Variant Regulates ACTG1 Expression
- Tracking Proliferative History in Lymphocyte Development with Cre-Mediated Sister Chromatid Recombination
- Correlated Occurrence and Bypass of Frame-Shifting Insertion-Deletions (InDels) to Give Functional Proteins
- Chimeric Protein Complexes in Hybrid Species Generate Novel Phenotypes
- Loss of miR-10a Activates and Collaborates with Activated Wnt Signaling in Inducing Intestinal Neoplasia in Female Mice
- Both Rare and Copy Number Variants Are Prevalent in Agenesis of the Corpus Callosum but Not in Cerebellar Hypoplasia or Polymicrogyria
- Reverse PCA, a Systematic Approach for Identifying Genes Important for the Physical Interaction between Protein Pairs
- Partial Deletion of Chromosome 8 β-defensin Cluster Confers Sperm Dysfunction and Infertility in Male Mice
- Genome-Wide and Cell-Specific Epigenetic Analysis Challenges the Role of Polycomb in Spermatogenesis
- Coordinate Regulation of Mature Dopaminergic Axon Morphology by Macroautophagy and the PTEN Signaling Pathway
- Cooperation between RUNX1-ETO9a and Novel Transcriptional Partner KLF6 in Upregulation of in Acute Myeloid Leukemia
- Mobility of the Native Conjugative Plasmid pLS20 Is Regulated by Intercellular Signaling
- FliZ Is a Global Regulatory Protein Affecting the Expression of Flagellar and Virulence Genes in Individual Bacterial Cells
- Specific Tandem Repeats Are Sufficient for Paramutation-Induced Trans-Generational Silencing
- Condensin II Subunit dCAP-D3 Restricts Retrotransposon Mobilization in Somatic Cells
- Dominant Mutations in Identify the Mlh1-Pms1 Endonuclease Active Site and an Exonuclease 1-Independent Mismatch Repair Pathway
- The Insulator Homie Promotes Expression and Protects the Adjacent Gene from Repression by Polycomb Spreading
- Human Intellectual Disability Genes Form Conserved Functional Modules in
- Coordination of Cell Proliferation and Cell Fate Determination by CES-1 Snail
- ORFs in Drosophila Are Important to Organismal Fitness and Evolved Rapidly from Previously Non-coding Sequences
- Different Roles of Eukaryotic MutS and MutL Complexes in Repair of Small Insertion and Deletion Loops in Yeast
- The Spore Differentiation Pathway in the Enteric Pathogen
- Acceleration of the Glycolytic Flux by Steroid Receptor Coactivator-2 Is Essential for Endometrial Decidualization
- The Human Nuclear Poly(A)-Binding Protein Promotes RNA Hyperadenylation and Decay
- Genome Wide Analysis Reveals Zic3 Interaction with Distal Regulatory Elements of Stage Specific Developmental Genes in Zebrafish
- Xbp1 Directs Global Repression of Budding Yeast Transcription during the Transition to Quiescence and Is Important for the Longevity and Reversibility of the Quiescent State
- The Integrator Complex Subunit 6 (Ints6) Confines the Dorsal Organizer in Vertebrate Embryogenesis
- Incorporating Motif Analysis into Gene Co-expression Networks Reveals Novel Modular Expression Pattern and New Signaling Pathways
- The Bacterial Response Regulator ArcA Uses a Diverse Binding Site Architecture to Regulate Carbon Oxidation Globally
- Direct Monitoring of the Strand Passage Reaction of DNA Topoisomerase II Triggers Checkpoint Activation
- Multiple bHLH Proteins form Heterodimers to Mediate CRY2-Dependent Regulation of Flowering-Time in
- A Reversible Histone H3 Acetylation Cooperates with Mismatch Repair and Replicative Polymerases in Maintaining Genome Stability
- ALS-Associated Mutations Result in Compromised Alternative Splicing and Autoregulation
- Robust Demographic Inference from Genomic and SNP Data
- Preferential Binding to Elk-1 by SLE-Associated Risk Allele Upregulates Expression
- Rad52 Sumoylation Prevents the Toxicity of Unproductive Rad51 Filaments Independently of the Anti-Recombinase Srs2
- The Serum Resistome of a Globally Disseminated Multidrug Resistant Uropathogenic Clone
- Identification of 526 Conserved Metazoan Genetic Innovations Exposes a New Role for Cofactor E-like in Neuronal Microtubule Homeostasis
- SUMO Localizes to the Central Element of Synaptonemal Complex and Is Required for the Full Synapsis of Meiotic Chromosomes in Budding Yeast
- Integrated Enrichment Analysis of Variants and Pathways in Genome-Wide Association Studies Indicates Central Role for IL-2 Signaling Genes in Type 1 Diabetes, and Cytokine Signaling Genes in Crohn's Disease
- Genome-Wide High-Resolution Mapping of UV-Induced Mitotic Recombination Events in
- Genome-Wide Analysis of Cell Type-Specific Gene Transcription during Spore Formation in
- Playing the Field: Sox10 Recruits Different Partners to Drive Central and Peripheral Myelination
- Two Portable Recombination Enhancers Direct Donor Choice in Fission Yeast Heterochromatin
- Mining the Human Phenome Using Allelic Scores That Index Biological Intermediates
- Yeast Tdh3 (Glyceraldehyde 3-Phosphate Dehydrogenase) Is a Sir2-Interacting Factor That Regulates Transcriptional Silencing and rDNA Recombination
- A Minimal Nitrogen Fixation Gene Cluster from sp. WLY78 Enables Expression of Active Nitrogenase in
- A Review of Bacteria-Animal Lateral Gene Transfer May Inform Our Understanding of Diseases like Cancer
- High Throughput Sequencing Reveals Alterations in the Recombination Signatures with Diminishing Spo11 Activity
- Partitioning the Heritability of Tourette Syndrome and Obsessive Compulsive Disorder Reveals Differences in Genetic Architecture
- Eleven Candidate Susceptibility Genes for Common Familial Colorectal Cancer
- A GDF5 Point Mutation Strikes Twice - Causing BDA1 and SYNS2
- Systematic Unraveling of the Unsolved Pathway of Nicotine Degradation in
- Natural Genetic Variation of Integrin Alpha L () Modulates Ischemic Brain Injury in Stroke
- Evolutionary Tuning of Protein Expression Levels of a Positively Autoregulated Two-Component System
- Evolutionary Change within a Bipotential Switch Shaped the Sperm/Oocyte Decision in Hermaphroditic Nematodes
- Limiting of the Innate Immune Response by SF3A-Dependent Control of MyD88 Alternative mRNA Splicing
- Multiple Signaling Pathways Coordinate to Induce a Threshold Response in a Chordate Embryo
- Distinct Regulatory Mechanisms Act to Establish and Maintain Pax3 Expression in the Developing Neural Tube
- Genome Wide Analysis of Narcolepsy in China Implicates Novel Immune Loci and Reveals Changes in Association Prior to Versus After the 2009 H1N1 Influenza Pandemic
- Mismatch Repair Genes and Modify CAG Instability in Huntington's Disease Mice: Genome-Wide and Candidate Approaches
- The Histone H3 K27 Methyltransferase KMT6 Regulates Development and Expression of Secondary Metabolite Gene Clusters
- Hsp70-Hsp40 Chaperone Complex Functions in Controlling Polarized Growth by Repressing Hsf1-Driven Heat Stress-Associated Transcription
- Function and Evolution of DNA Methylation in
- Stimulation of mTORC1 with L-leucine Rescues Defects Associated with Roberts Syndrome
- Transcription Termination and Chimeric RNA Formation Controlled by FPA
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Dominant Mutations in Identify the Mlh1-Pms1 Endonuclease Active Site and an Exonuclease 1-Independent Mismatch Repair Pathway
- Eleven Candidate Susceptibility Genes for Common Familial Colorectal Cancer
- The Histone H3 K27 Methyltransferase KMT6 Regulates Development and Expression of Secondary Metabolite Gene Clusters
- A Mutation in the Gene in Labrador Retrievers with Hereditary Nasal Parakeratosis (HNPK) Provides Insights into the Epigenetics of Keratinocyte Differentiation
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



