-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaDominant Mutations in Identify the Mlh1-Pms1 Endonuclease Active Site and an Exonuclease 1-Independent Mismatch Repair Pathway
Lynch syndrome (hereditary nonpolypsis colorectal cancer or HNPCC) is a common cancer predisposition syndrome. Predisposition to cancer in this syndrome results from increased accumulation of mutations due to defective mismatch repair (MMR) caused by a mutation in one of the mismatch repair genes MLH1, MSH2, MSH6 or PMS2/scPMS1. To better understand the function of Mlh1-Pms1 in MMR, we used Saccharomyces cerevisiae to identify six pms1 mutations (pms1-G683E, pms1-C817R, pms1-C848S, pms1-H850R, pms1-H703A and pms1-E707A) that were weakly dominant in wild-type cells, which surprisingly caused a strong MMR defect when present on low copy plasmids in an exo1Δ mutant. Molecular modeling showed these mutations caused amino acid substitutions in the metal coordination pocket of the Pms1 endonuclease active site and biochemical studies showed that they inactivated the endonuclease activity. This model of Mlh1-Pms1 suggested that the Mlh1-FERC motif contributes to the endonuclease active site. Consistent with this, the mlh1-E767stp mutation caused both MMR and endonuclease defects similar to those caused by the dominant pms1 mutations whereas mutations affecting the predicted metal coordinating residue Mlh1-C769 had no effect. These studies establish that the Mlh1-Pms1 endonuclease is required for MMR in a previously uncharacterized Exo1-independent MMR pathway.
Published in the journal: . PLoS Genet 9(10): e32767. doi:10.1371/journal.pgen.1003869
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1003869Summary
Lynch syndrome (hereditary nonpolypsis colorectal cancer or HNPCC) is a common cancer predisposition syndrome. Predisposition to cancer in this syndrome results from increased accumulation of mutations due to defective mismatch repair (MMR) caused by a mutation in one of the mismatch repair genes MLH1, MSH2, MSH6 or PMS2/scPMS1. To better understand the function of Mlh1-Pms1 in MMR, we used Saccharomyces cerevisiae to identify six pms1 mutations (pms1-G683E, pms1-C817R, pms1-C848S, pms1-H850R, pms1-H703A and pms1-E707A) that were weakly dominant in wild-type cells, which surprisingly caused a strong MMR defect when present on low copy plasmids in an exo1Δ mutant. Molecular modeling showed these mutations caused amino acid substitutions in the metal coordination pocket of the Pms1 endonuclease active site and biochemical studies showed that they inactivated the endonuclease activity. This model of Mlh1-Pms1 suggested that the Mlh1-FERC motif contributes to the endonuclease active site. Consistent with this, the mlh1-E767stp mutation caused both MMR and endonuclease defects similar to those caused by the dominant pms1 mutations whereas mutations affecting the predicted metal coordinating residue Mlh1-C769 had no effect. These studies establish that the Mlh1-Pms1 endonuclease is required for MMR in a previously uncharacterized Exo1-independent MMR pathway.
Introduction
DNA mismatch repair (MMR) acts to repair the potentially mutagenic misincorporation errors that occur during normal DNA replication and the absence of MMR results in increased rates of accumulating mutations. Consequently, defects in human MMR genes cause the hereditary cancer susceptibility syndrome HNPCC (hereditary nonpolypsis colorectal cancer, otherwise known as Lynch syndrome) [1], [2], and loss of MMR function also appears to underlie the development of some sporadic cancers [3]–[7]. MMR also repairs mispaired bases that occur in recombination intermediates as well as prevents inappropriate recombination between DNAs with imperfect homology where recombination could result in genome rearrangements [8]–[10].
The mechanism of MMR has been extensively characterized in both E. coli and different eukaryotic systems, with E. coli MMR being the best characterized [11]–[14]. In E. coli MMR, mismatches are recognized by the MutS homodimer [15], [16]. Mispair bound MutS then recruits the MutL homodimer [17]. This recruitment leads to activation of the MutH endonuclease, which introduces single strand breaks, called nicks, at unmethylated GATC sites in the newly replicated and hemimethylated DNA strand [18]. Next, a combination of the UvrD helicase and one of four single stranded DNA specific exonucleases excise the nicked strand past the mispair and the resulting singled-stranded gap is filled in by DNA polymerase III, single strand DNA binding protein and DNA ligase [14], [19].
In eukaryotes mispairs are recognized by either Msh2-Msh6 or Msh2-Msh3, two partially redundant heterodimers of MutS family member proteins [12], [20], [21]. Mispair bound Msh2-Msh6 and Msh2-Msh3 recruit the MutL related complex, called Mlh1-Pms1 in S. cerevisiae and Mlh1-Pms2 in human and mouse [11], [12], [22]–[24]. The Pms1/Pms2 subunit of the Mlh1-Pms1/Pms2 complex is known to contain an endonuclease active site, suggesting that Mlh1-Pms1/Pms2 may be analogous to a combination of both E. coli MutL and MutH [25], [26]. Exo1, a DNA exonuclease from the Rad2/XPG family, has been implicated in the excision step of eukaryotic MMR; however, mutations in S. cerevisiae and mouse EXO1 only result in partial MMR defects, suggesting the existence of additional excision mechanisms [27]–[29]. Genetic and biochemical studies have also implicated DNA polymerase δ, RPA, RFC and PCNA in MMR [12], [30]–[37] and have suggested that several of these proteins including PCNA and RFC may function both prior to excision and in the resynthesis steps of MMR [25], [26], [33], [38].
MMR is spatially and temporally coupled to replication in vivo [38], [39], providing a mechanism to bring MMR proteins into the proximity of newly formed mispairs. DNA replication generates nicks in the nascent DNA strands that may be involved in MMR [30], [34], [40], [41], consistent with the observation that discontinuous lagging strand MMR is more dependent on excision catalyzed by Exo1 than leading strand MMR [38], [42]. Furthermore, preexisting nicks in DNA target the Mlh1-Pms1/Pms2 endonuclease to the nicked strand in vitro [43]. However, these results raise two unresolved questions: Why is it necessary to target additional nicks to an already nicked DNA strand, and if preexisting nicks can support MMR in vitro then why is Mlh1-Pms1 absolutely required for MMR in vivo? Part of the answer to the apparent contradictions implied by these experimental results could be the presence of multiple MMR pathways in which the same MMR proteins have differing roles. Consistent with this, biochemical studies have identified two types of excision mechanisms that may function in MMR, excision by Exo1 [44] and strand displacement synthesis toward the mispair by DNA polymerase δ potentially coupled with flap cleavage [45]. Both mechanisms could act at either a pre-existing 5′ nick or a 5′ nick introduced by the Mlh1-Pms1/Pms2 endonuclease. Genetic studies have also identified Exo1-dependent and -independent MMR pathways [27]. The Exo1-independent pathway requires the PCNA-Msh2-Msh6 interaction and the Pol32 subunit of DNA polymerase δ and is inactivated by separation-of-function mutations affecting Mlh1, Pms1, Msh2, Msh3, and PCNA that do not affect Exo1-dependent MMR [27], [38]. How these mutations specifically affect in the Exo1-independent MMR pathway and how this pathway excises the nascent DNA strand is unclear.
To better understand the role of both Mlh1-Pms1 and Exo1 in MMR, we performed a genetic screen for dominant mutations in the PMS1 gene. We identified pms1 null missense mutations that caused weakly dominant MMR defects when present in a wild-type S. cerevisiae strain on a single-copy plasmid. Interestingly, these mutations caused much stronger MMR defects when present on a single-copy plasmid in an exo1Δ mutant. Analysis of these mutations using the structure of the C-terminal domains of Mlh1-Pms1 [46] predicted that three amino acids altered by these mutations were metal ligands in the Mlh1-Pms1 nuclease active site and the fourth was a residue adjacent to the metal binding site. Biochemical analysis of mutant proteins confirmed that both pms1 and mlh1 mutations affecting the predicted active site eliminated or significantly reduced the RFC-PCNA dependent nuclease activity of Mlh1-Pms1 and in vivo imaging showed that the same mutations resulted in accumulation of Mlh1-Pms1-4GFP foci consistent with failure to execute a downstream step in MMR. These results both define the nuclease active site of the Mlh1-Pms1 endonuclease and thoroughly characterize the role of this endonuclease activity in a previously uncharacterized Exo1-independent MMR sub-pathway.
Results
Identification of dominant pms1 mutations
To gain insight into the role of PMS1 in mismatch repair, we sought to generate novel pms1 mutations that cause a dominant MMR defect. First we mutagenized the PMS1 gene by PCR amplification and gap-repaired the resulting DNA fragments into the low copy number ARS CEN pRS316 plasmid by co-transformation into a S. cerevisiae strain with a wild-type PMS1 gene. A total of 38,000 transformants were screened for increased reversion of the lys2-10A frameshift mutation. This screen identified 211 transformants potentially containing dominant pms1 mutations. Rescreening these 211 transformants for increased reversion of the hom3-10 frameshift mutation identified 8 transformants with mutator phenotypes. The pms1 mutation-bearing plasmids were isolated from these 8 transformants, and the PMS1 gene from each plasmid was sequenced. Site directed mutagenesis and sub-cloning were used to construct PMS1 plasmids containing single point mutations. These mutant plasmids were retested in the three mutator assays confirming four dominant pms1 mutations resulting from amino acid substitutions in the C-terminal domain of Pms1: G683E, C817R, C848S, and H850R.
Three dominant mutations eliminate metal binding ligands in the Pms1 endonuclease domain
The four amino acid substitutions resulting from the dominant mutations altered highly conserved amino acids (Figure 1A). Initial analysis of these amino acid substitutions using a homology model based on the C-terminal domains of endonuclease-proficient and zinc-binding Neisseria gonorrhoeae and Bacillus subtilis MutL homologs [47], [48] indicated that they affect the active site of the Pms1 endonuclease. Mapping of the amino acid substitutions onto the newly available structure of the C-terminal domains of Mlh1-Pms1 (Figure 1B,C) confirmed that the C817R, C848S, and H850R amino acid substitutions each eliminated one of the 5 metal ligands in the Pms1 endonuclease active site; all 5 ligands are conserved in eukaryotic ScPms1/HsPMS2 proteins and in the N. gonorrhoeae and B. subtilis MutL homologs [47], [48]. The fourth amino acid substitution, G683E, mapped to a conserved position adjacent to sites of metal coordination and could sterically disrupt the site or locally perturb the structure. We also constructed mutations resulting in the amino acid substitutions H703A and E707A to eliminate the remaining two predicted metal ligands not identified in our screen.
Fig. 1. Conserved amino acid residues comprise the Mlh1-Pms1 endonuclease active site. 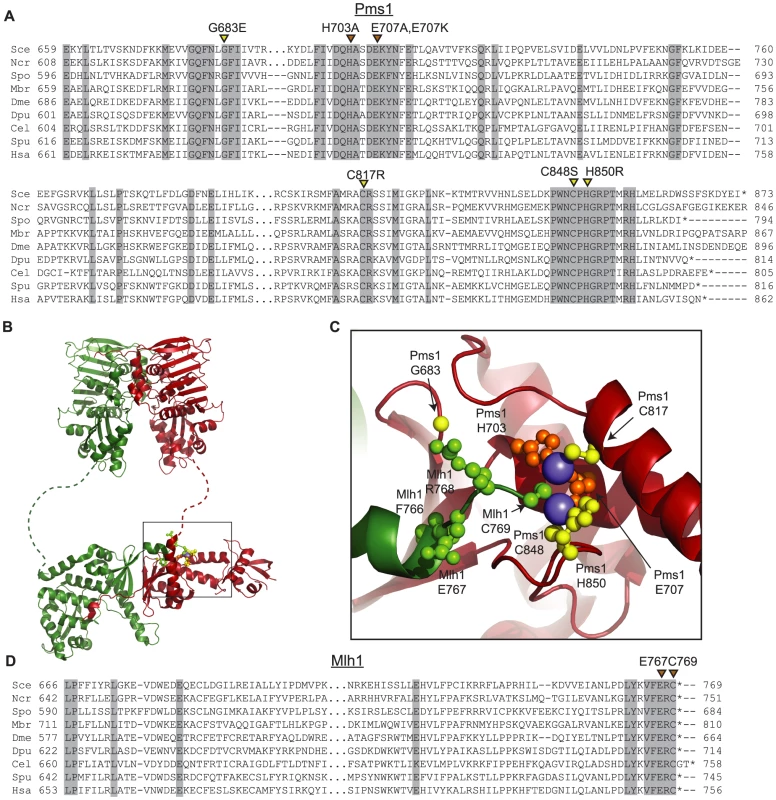
A. Alignments of fungal Pms1 and animal Pms2 sequences from Sce (Saccharomyces cerevisiae NP_0141317.2), Ncr (Neurospora crassa XP_962690.2), Spo (Schizosaccharomyces pombe NP_594417.1), Mbr (Monosigna brevicollis XP_001747286.1), Dme (Drosophila melanogaster NP_477023.1), Dpu (Daphnia pulex EFX87911.1), Cel (Caenorhabitis elegans NP_505933.1), Spu (Strongylocentrotus purpuratus XP_786592.2), Hsa (Homo sapiens NP_000526.1) using Clustal Omega [72]. Asterisks indicate stop codons. Highlighted columns indicate perfectly conserved residues in the nine aligned species. Yellow triangles indicate the positions of mutations isolated in the screen. Orange triangles indicate the positions of engineered mutations generated in this study. B. Homology model of S. cerevisiae Mlh1-Pms1 with Mlh1 in green and Pms1 in red. The unstructured linker between the N- and C-terminal domains depicted as a dashed line. The position of the C-terminal active site is boxed. C. Modeled Pms1 endonuclease active site is displayed with the positions of amino acid substitutions isolated in the genetic screen shown with yellow ball-and-stick side chains and the position of the other metal ligands in orange. Position of the Mlh1 C-terminal FERC motif displayed as green ball-and-stick side chains. D. Alignments of fungal and animal Mlh1 sequences as in panel A from Sce (Saccharomyces cerevisiae NP_013890.1), Ncr (Neurospora crassa XP_962522.1), Spo (Schizosaccharomyces pombe NP_596199.1), Mbr (Monosigna brevicollis, reannotated starting from XP_001745742.1), Dme (Drosophila melanogaster, NP_477022.1), Dpu (Daphnia pulex, EFX86130.1), Cel (Caenorhabitis elegans NP_499796.2), Spu (Strongylocentrotus purpuratus XP_793318.2) and Hsa (Homo sapiens NP_000240.1). Fluctuation analysis was performed to evaluate the mutator effects of the dominant pms1 mutants. Mutation rates were measured using the CanR forward mutation assay and the hom3-10 and lys2-10A frameshift reversion assays [21], [27] when the pms1 dominant mutations were present on low copy plasmids in a strain with a wild-type PMS1 gene (Table 1). None of the mutant plasmids caused more than a 2-fold increased mutation rate in the CanR assay, which has a relatively high background mutation rate in wild-type cells and a low sensitivity for detecting MMR defects. In contrast, the mutations on the low copy plasmid caused between a 2 - and 102-fold increase in mutation rate in the hom3-10 and lys2-10A assays compared to introduction of a wild-type copy of PMS1 on a low copy plasmid. Introduction of these mutations onto a high copy 2-micron plasmid resulted in much higher rates (Table S1), but still did not cause mutation rates that were as high as caused by deletion of PMS1 (Compare Table S1 to Table S2). The pms1 mutant plasmids were also unable complement a pms1Δ strain above that seen for the vector control (Table S2). Taken together, the fluctuation analysis indicated that the dominant pms1 mutations were null PMS1 alleles that caused a copy-number dependent dominant mutator phenotype. This dominant mutator phenotype was stronger for mutations causing amino acid substitutions of metal ligands as compared to the pms1-G683E mutation.
Tab. 1. Mutation rates of pms1 metal coordination mutants on a low-copy plasmid in wild-type and exo1Δ strains. 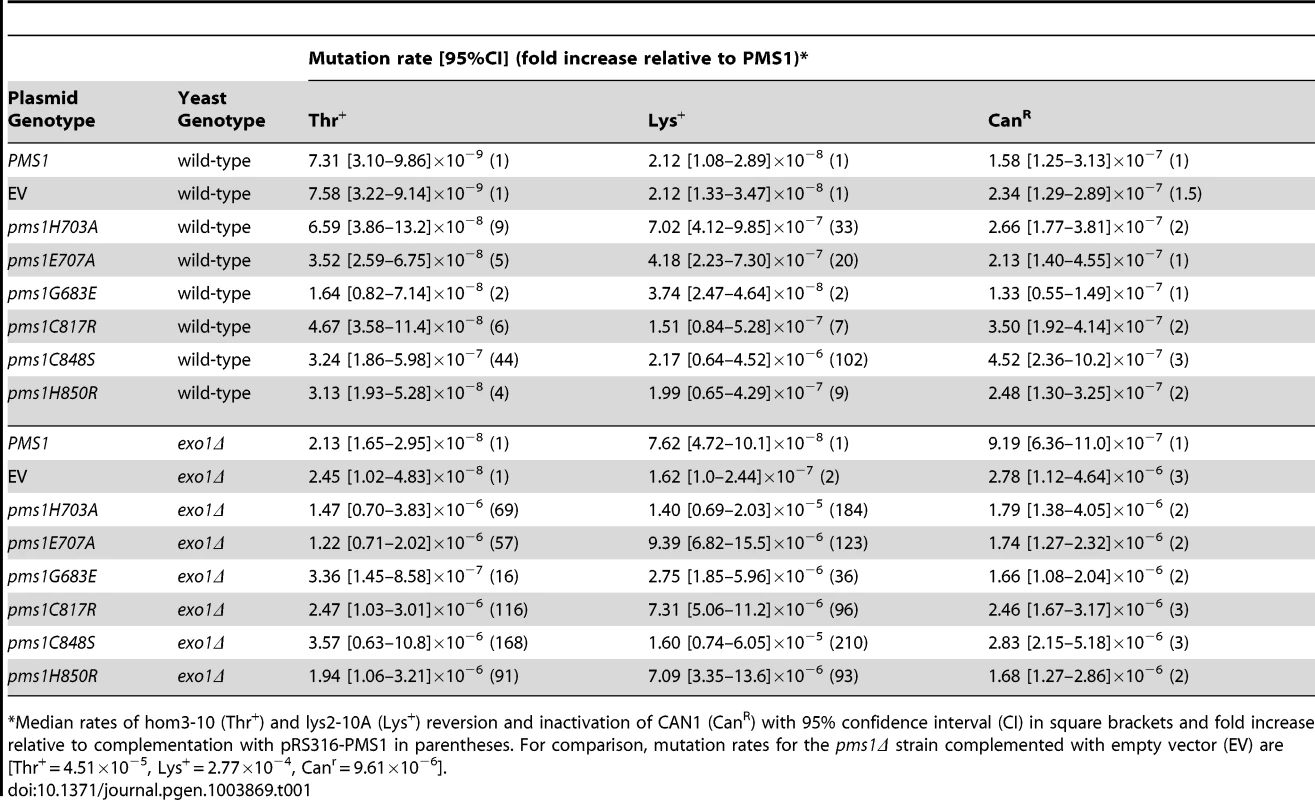
Median rates of hom3-10 (Thr+) and lys2-10A (Lys+) reversion and inactivation of CAN1 (CanR) with 95% confidence interval (CI) in square brackets and fold increase relative to complementation with pRS316-PMS1 in parentheses. For comparison, mutation rates for the pms1Δ strain complemented with empty vector (EV) are [Thr+ = 4.51×10−5, Lys+ = 2.77×10−4, Canr = 9.61×10−6]. Disruption of metal coordination inhibits endonuclease function
Previous studies have shown that S. cerevisiae Mlh1-Pms1 has a metal-dependent endonuclease activity that can be stimulated by RFC and PCNA [26]. To determine if the dominant pms1 mutations affecting metal ligating amino acids disrupt the endonuclease function of Mlh1-Pms1, we expressed and purified the S. cerevisiae wild-type Mlh1-Pms1 complex and the mutant Mlh1-Pms1-G683E, Mlh1-Pms1-C817R, Mlh1-Pms1-C848S, and Mlh1-Pms1-H850R complexes and assayed the ability of these complexes to nick supercoiled pRS425 plasmid DNA with or without accessory factors RFC-Δ1N and PCNA (Figure 2A). Wild-type Mlh1-Pms1 alone showed little endonuclease activity. However, addition of PCNA and RFC-Δ1N to reactions containing wild-type Mlh1-Pms1 resulted in a 20-fold increase in endonuclease activity resulting in cleavage of nearly half of the original substrate DNA. The newly identified Mlh1-Pms1 mutant proteins and the previously studied Mlh1-Pms1-E707K mutant protein did not exhibit any PCNA and RFC-Δ1N stimulated endonuclease activity, with the exception of the Mlh1-Pms1-H850R mutant protein (Figure 2A). An explanation of the ability of the Mlh1-Pms1-H850R mutant protein to nick supercoiled DNA is provided in the “Discussion”. These results support the idea that loss of metal coordination by Pms1 inhibits the endonuclease activity of Mlh1-Pms1.
Fig. 2. Metal coordination mutations eliminate the ability of Mlh1-Pms1 to nick closed circular DNA. 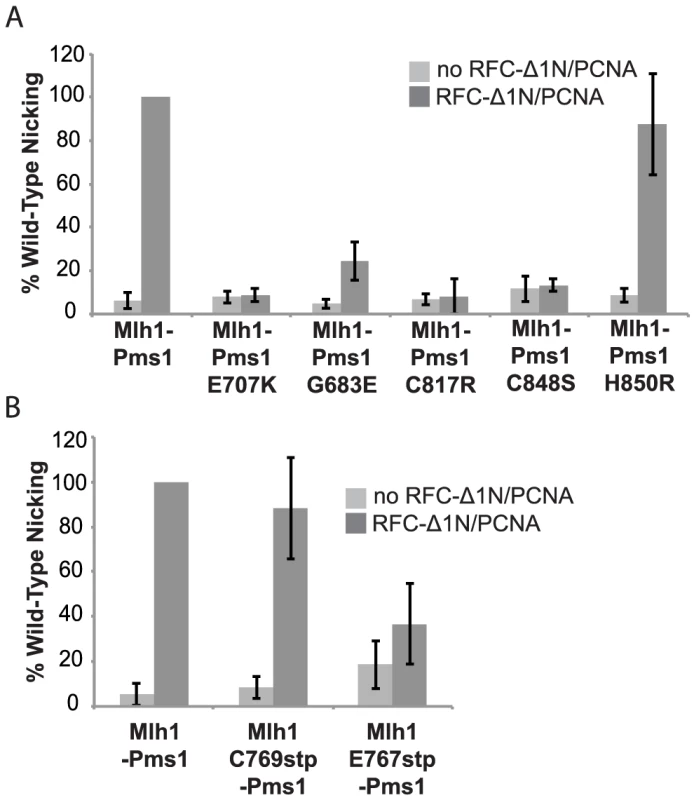
Endonuclease reactions to nick closed circular DNA were performed with Mlh1-Pms1 alone or with Mlh1-Pms1, PCNA and RFC-Δ1N. A. Percentage of wild-type cleaved product formed by wild-type Mlh1-Pms1 and mutant Mlh1-Pms1 complexes containing the indicated Pms1 amino acid changes. B. Percentage of wild-type cleaved product formed by wild-type Mlh1-Pms1 and mutant Mlh1-Pms1 complexes containing the indicated Mlh1 amino acid changes. 100% cleavage of the 100 ng of pRS425 in the assay is 2.2 fmoles of cleavage events. PMS1 metal coordination mutations cause a synthetic mutator phenotype when combined with an exo1Δ mutation
Exo1 is a 5′-3′ nuclease that functions in MMR in vitro by resecting DNA from a preexisting nick to a point past the mispair [28], [44], [49]. Loss of Exo1 function in vivo by deletion of EXO1 or missense mutations inactivating the Exo1 active site only results in a weak mutator phenotype and hence only partial loss of MMR (Table 1) [27]–[29], [50]. To determine the consequences of eliminating the two known nucleases involved in S. cerevisiae MMR, we tested the effect of introducing plasmids containing the dominant pms1 mutations into an exo1Δ mutant strain containing a wild-type PMS1 gene. Relative to the effects in a wild-type strain, introduction of the dominant pms1 mutations on a low-copy plasmid into the exo1Δ mutant strain increased the mutation rate to a much greater extent (Table 1). The pms1-G683E mutation again caused the weakest mutator phenotype, whereas the other five pms1 mutations that affect the metal ligands caused relatively high mutation rates ranging from a 57 - to a 210-fold increased mutation rate depending on the mutation and the assay. These results show that the dominant pms1 mutations cause a greater MMR defect in an exo1Δ mutant strain than in a wild-type strain, suggesting that Exo1 and the Mlh1-Pms1 nuclease have a redundant function in MMR.
Role of the conserved Mlh1 C-terminus in endonuclease activity and MMR
The four C-terminal residues of Mlh1 are almost completely conserved as the amino acids FERC in fungal and animal species (Figure 1D). Furthermore, Mlh1 has no C-terminal extension beyond the FERC residues in almost all sequenced eukaryotic organisms except nematodes (FERCG[T/S]) whereas the ScPms1/HsPMS2 family of proteins have variable length C-terminal extensions (Figure 1D). Consistent with a special role for the C-terminus of Mlh1, C-terminal fusions to the S. cerevisiae Mlh1 are non-functional for MMR in vivo, whereas C-terminal fusions to S. cerevisiae Pms1 do not affect function [38]. A structure of this highly conserved Mlh1 C-terminus (Figure 1B,C) revealed that this region might be appropriately positioned to play a role at the endonuclease active site of Pms1, with the C-terminal Mlh1 cysteine potentially acting as a metal ligand. We therefore generated the mlh1-E767stp, mlh1-C769stp, mlh1-C769A and mlh1-C769S mutations to probe the role of these highly conserved residues in endonuclease activity in vitro and MMR in vivo.
Unlike the pms1 mutations affecting metal ligands, none of the mutations affecting the C of the conserved FERC motif of Mlh1 that is predicted to be a metal ligand including the mlh1-C769A, mlh1-C769S, and mlh1-C769stp mutations caused a dominant mutator phenotype when present on an ARS CEN plasmid in a wild-type strain or an exo1Δ strain (Table 2). The mlh1-C769A, mlh1-C769S, and mlh1-C769stp mutants fully complimented the MMR defect of an mlh1Δ strain (Table S2) consistent with previously published results for the mlh1-C769A mutation but not the mlh1-C769S mutation [51] or the mlh1-C769stp mutation [46]. In this regard, it should be noted that our studies used a broader series of mutator assays, including more sensitive assays, than previous studies of mutations affecting Mlh1-C767. In contrast, the mlh1-E767stp mutant plasmid failed to complement the MMR defect of an mlh1Δ strain and resulted in a null phenotype (Table S2). Furthermore, the mlh1-E767stp mutation on an ARS CEN plasmid caused a weak dominant mutator phenotype when present in a wild-type strain and a stronger dominant mutator phenotype when present in an exo1Δ strain (Table 2), although not to the extent as that caused by the PMS1 metal ligand mutations.
Tab. 2. Mutations rates of MLH1-FERC motif mutations on a low-copy plasmid in wild-type and exo1Δ strains. 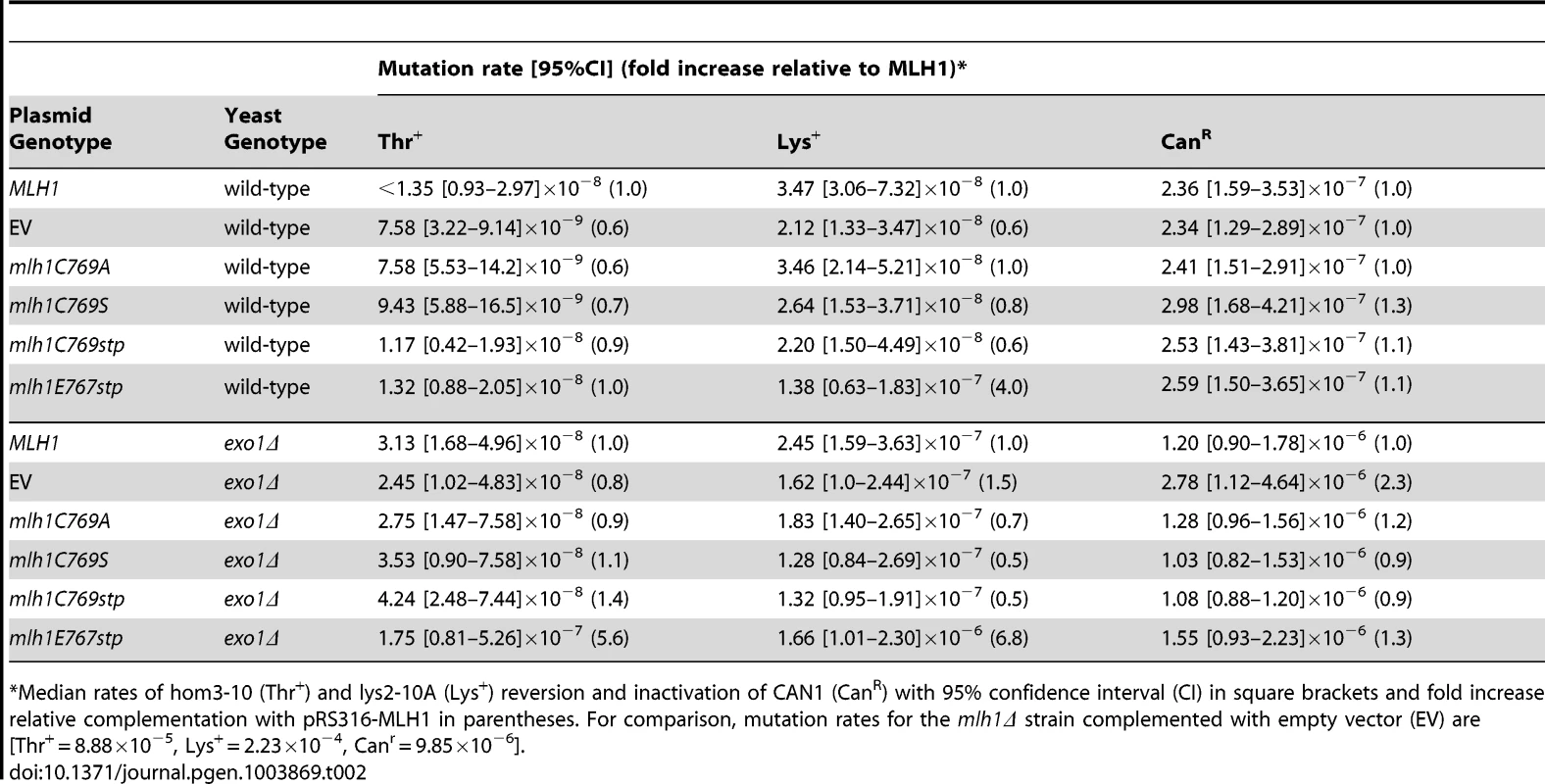
Median rates of hom3-10 (Thr+) and lys2-10A (Lys+) reversion and inactivation of CAN1 (CanR) with 95% confidence interval (CI) in square brackets and fold increase relative complementation with pRS316-MLH1 in parentheses. For comparison, mutation rates for the mlh1Δ strain complemented with empty vector (EV) are [Thr+ = 8.88×10−5, Lys+ = 2.23×10−4, Canr = 9.85×10−6]. We also tested the effect of the mlh1-C769stp and mlh1-E767stp mutations in the endonuclease assay (Figure 2B). The mutant Mlh1-Pms1 protein lacking only the Mlh1 C-terminal cysteine that did not cause an MMR defect in vivo nicked supercoiled DNA to the same extent as the wild-type Mlh1-Pms1 protein. In contrast, the mutant Mlh1-Pms1 protein resulting from the mlh1-E767stp mutation was significantly defective for nicking supercoiled plasmid DNA, which parallels the effect of this mutation on MMR in vivo. These results support the idea that the C-terminus of Mlh1 functions in the endonuclease active site although if the terminal cysteine coordinates bound metal then this role is not required for MMR or endonuclease activity (Figure 1C).
Mlh1-Pms1 metal coordination mutants do not preferentially affect MMR on the leading or lagging DNA strands
Mutations affecting the active sites of leading and lagging strand DNA polymerases Pol ε, pol2-M644G, and Pol δ, pol3-L612M, have been identified that preferentially introduce misincorporation errors in their respective strand during DNA replication [52], [53]. In a wild-type background, these lesions are then efficiently corrected by MMR. This strand-biased misincorporation can be used to determine strand preferences for MMR [52], [54], [55]. Here we probed mutants containing these polymerase active site mutations with ARS CEN plasmids encoding endonuclease defective pms1 and mlh1 mutations to investigate whether the Mlh1-Pms1 endonuclease preferentially functions in leading or lagging strand MMR. We found that ARS CEN PMS1 plasmids containing the pms1-C817R, pms1-C848S or pms1-H850R mutations caused a statistically similar synergistic increase in mutation rate when present in strains containing mutations affecting either DNA polymerase (Table 3). Of nine pairwise comparisons between pol2-M644G and pol3-L612M mutants containing the same pms1 mutation on a low copy plasmid using three different mutation rate assays, seven were not different (p-value>0.05, Mann-Whitney test). For the two comparisons that showed a difference, one showed a modestly higher rate in the pol2-M644G strain while the other showed a modestly higher rate in the pol3-L612M strain (p-value<0.05, Mann-Whitney test). This degree of similarity between the pol2-M644G and pol3-L612M mutants in these comparisons is in marked contrast to the effect of an exo1Δ mutation that caused a 9-fold higher increase in mutation rate in a pol3-L612M mutant compared to a pol2-M644G mutant [38]. Overall, these results suggest that Mlh1-Pms1 functions similarly on both the leading and lagging strands during MMR.
Tab. 3. Mutations rates of pms1 metal coordination mutants on a low-copy plasmid in polymerase mutant strains pol2M644G and pol3L612M. 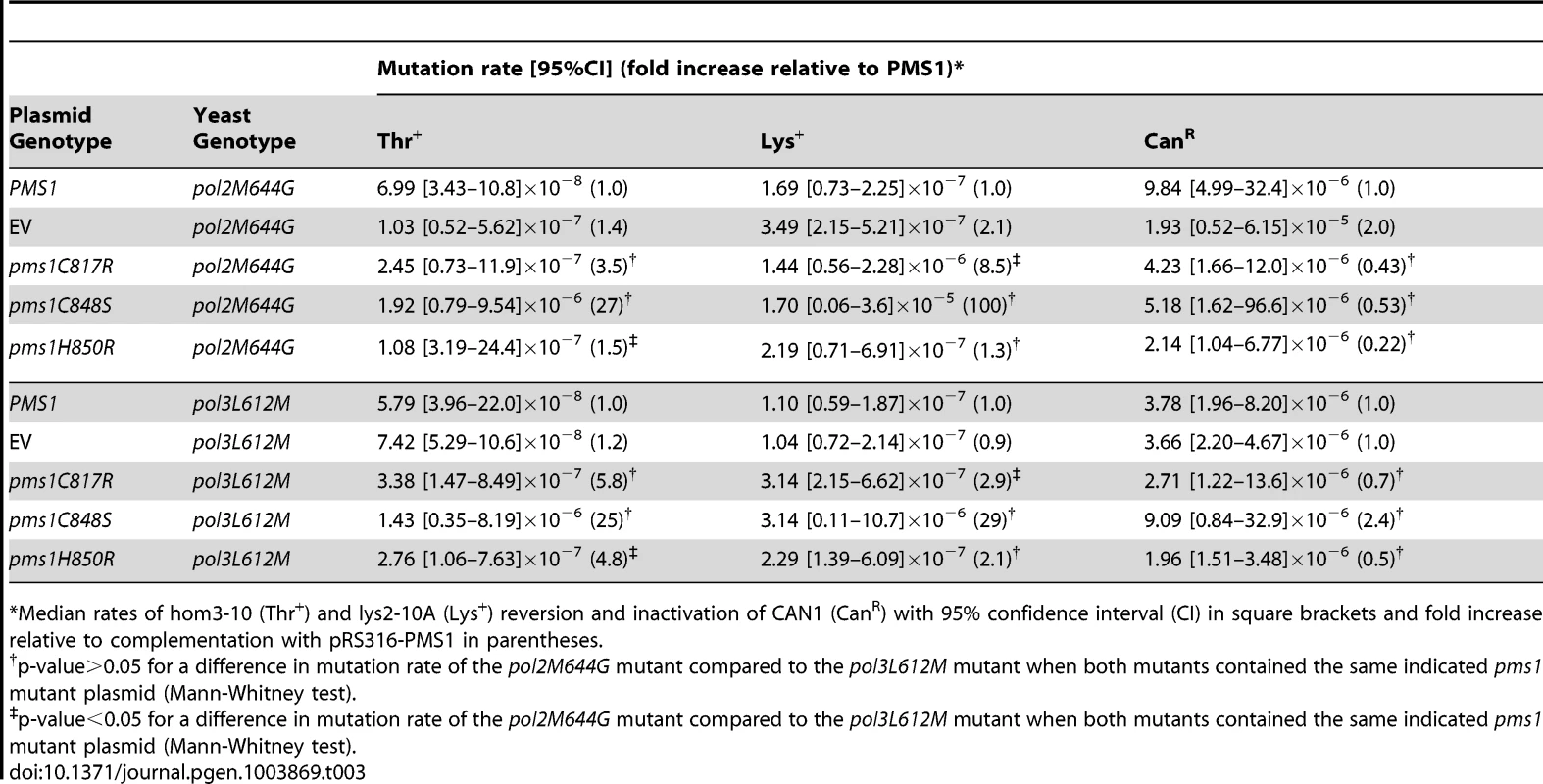
Median rates of hom3-10 (Thr+) and lys2-10A (Lys+) reversion and inactivation of CAN1 (CanR) with 95% confidence interval (CI) in square brackets and fold increase relative to complementation with pRS316-PMS1 in parentheses. Mutations that affect Mlh1-Pms1 endonuclease function exhibit increased levels of Pms1 foci
We have previously demonstrated that Mlh1-Pms1 foci are an intermediate in MMR and that blocking MMR downstream of Mlh1-Pms1 recruitment results in increased levels of Mlh1-Pms1 foci [38]. To test the effect of the Mlh1-Pms1 active site mutations on the levels of Pms1 foci, different mlh1 and pms1 mutations were introduced into the relevant endogenous locus in a strain in which the single wild-type copy of PMS1 was functionally tagged with four tandem copies of GFP. Normally, Mlh1-Pms1-4GFP foci are present in approximately 10% of logarithmically growing wild-type cells whereas all of the mutations that affected endonuclease function in vitro that were tested caused increased Mlh1-Pms1-4GFP foci formation (Figure 3). These included the metal coordination pms1 mutations pms1-E707K, which was previously tested [38], as well as pms1-C817R, pms1-C848S and pms1-H850R, the pms1-G683E mutation and the mlh1-E767stp endonuclease active site mutation, which increased the proportion of cells containing Mlh1-Pms1-4GFP foci to between 64 and 96%. In contrast, the endonuclease and MMR proficient mlh1-C769stp mutation did not alter the levels of Mlh1-Pms1-4GFP foci compared to wild-type strain. These results are consistent with the idea that mispairs are recognized normally in these Mlh1-Pms1 endonuclease active site mutants and that there is proper loading of the mutant Mlh1-Pms1 on DNA but instead there is a mispair processing defect resulting in decreased turnover of the mutant Mlh1-Pms1 from the DNA [38].
Fig. 3. Zinc Metal coordination mutations cause increased levels of Mlh1-Pms1-4GFP foci. 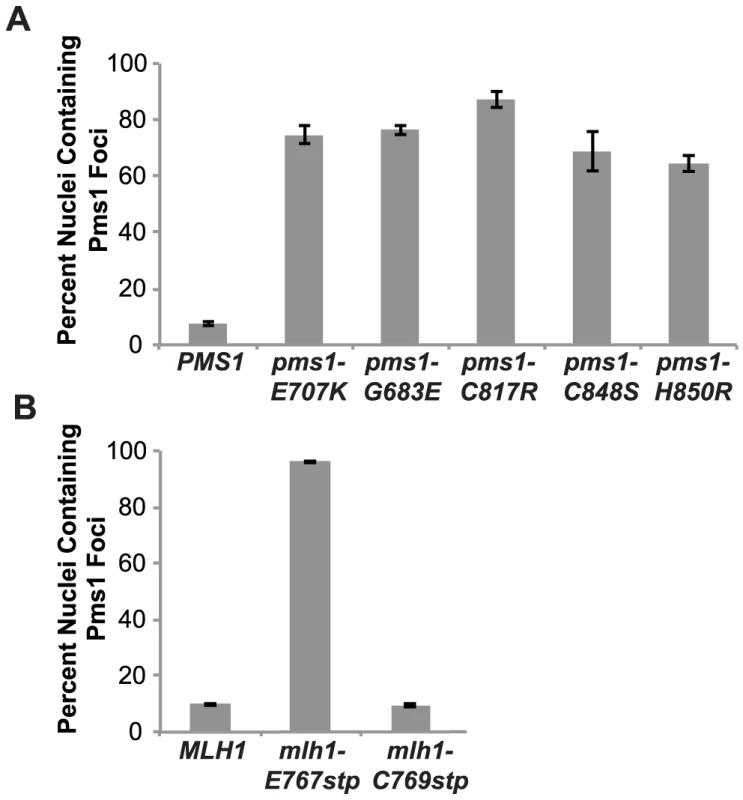
A. C-terminally tagged Pms1-GFP was imaged in logarithmically growing asynchronous cultures of the wild-type and the indicated pms1 mutant strains. The quantitative data presented here, is the result of at least two independent experiments, each performed using two independent strain isolates. The total number of cells/nuclei (n) analyzed for each strain is indicated, and bars indicate the error of the mean (SEM). B. C-terminally tagged Pms1-GFP was imaged in logarithmically growing asynchronous cultures of wild-type and the indicated mlh1-FERC mutant strains as described under A. Discussion
In this study, we used the highly sensitive lys2-10A frameshift mutation reversion assay to screen mutagenized low copy PMS1 plasmids for pms1 mutations that caused a dominant mutator phenotype in the presence of a single-copy of the wild-type PMS1 gene. We identified four null mutations that caused a weak dominant phenotype under these conditions. These mutations all caused amino acid substitutions in or near the region predicted to contain the Pms1 endonuclease active site and three of the amino acid substitutions including Pms1-C817R, Pms1-C848S, and Pms1-H850R affected predicted metal binding motifs while Pms1-G683 was in close proximity to the metal coordination site. The weak effect of these mutations explains why a prior study of a predicted Pms1 active site mutation did not observe a dominant effect [56]. A model of the endonuclease active site predicted Pms1 amino acids H703 and E707 as well as Mlh1-C769 to also be a part of the Mlh1-Pms1 active site [46]. Consistent with this model, the pms1-H703A and pms1-H707A mutations were found to cause the same phenotypes as the other PMS1 metal ligand mutations. In contrast, no mutation affecting Mlh1-C769 caused either a dominant mutator phenotype or affected Mlh1 function whereas the mlh1-E767stp mutation caused phenotypes that were similar to those caused by the PMS1 metal ligand mutations. Remarkably, all of the pms1 mutations caused a stronger dominant mutator phenotype when present in an exo1Δ strain on a low copy plasmid. The mlh1-E767stp also caused an increased dominant mutator affect in the exo1Δ strain, although not to the extent seen with the pms1 mutations. The phenotype of these mutants is similar to previously described separation-of-function mutations in MSH2, MSH3, MSH6, MLH1, PMS1, POL30 and POL32 that cause strong defects in Exo1-independent MMR but little if any defect in MMR when Exo1 is functional [27].
The asymmetry of the endonuclease active site is consistent with proposed roles of Mlh1-Pms1 in nicking double-stranded DNA during MMR; however, this asymmetry is not present in homodimeric bacterial MutL homologs with endonuclease function [47], [48]. The similarity of the eukaryotic Mlh1-Pms1 and bacterial MutL homologs suggests that MutL-DNA complexes may be functionally asymmetric so that only one active site is positioned to cleave DNA, analogous to the functional asymmetry during mispair recognition by bacterial MutS homodimers [15], [16]. The asymmetry of the eukaryotic MutL complexes, however, allows specialization of each subunit. The highly conserved C-terminus of Mlh1 is positioned in the Mlh1-Pms1 structure in a way that suggests that the Mlh1-ScPms1/HsPMS2 and Mlh1-Mlh3 complexes have a composite endonuclease active site. Potential roles for residues in the Mlh1 C-terminus include coordinating DNA phosphates, promoting nucleophilic attack by a water molecule or stabilization of the Pms1 active site. Consistent with this, the mlh1-E767stp mutant plasmid did not complement the mutator phenotype caused by a deletion of MLH1, and the mlh1-E767stp mutation resulted in the accumulation of Mlh1-Pms1-4GFP foci and reduced endonuclease activity similar to mutations in PMS1 affecting the endonuclease active site. It was surprising that mutation or deletion of the highly conserved C-terminal cysteine did not cause a MMR defect in vivo or reduce endonuclease activity in vitro given the possibility that this residue might coordinate with metals in the endonuclease active site. It is possible that conservation of the C-terminal cysteine may reflect other roles for Mlh1, potentially including crossover resolution during meiosis [57]–[59].
Analysis of the genetically identified and structure-based mutations in the MLH1 and PMS1 genes revealed that disruption of the metal binding sites leads to disruption of the Mlh1-Pms1 endonuclease activity and arrest of MMR repair at a step following recruitment of Mlh1-Pms1 into microscopically-observable foci. In a Mn2+-, RFC-, and PCNA-dependent endonuclease assay, amino acid substitutions affecting all of the metal ligands caused defects in the Mlh1-Pms1 endonuclease activity, which confirms and extends previous studies of the human Mlh1-Pms2 E705K amino acid substitution [25], [26]. The pms1-H850R and the mlh1-E767stp mutations resulted in proteins with partial defects in the in vitro endonuclease assay, but caused complete MMR defects in vivo. The partial endonuclease defect caused by these mutations may reflect the fact that the in vivo metal ion in eukaryotic and bacterial homologs is Zn2+, which has tetrahedral coordination geometry, whereas the metal that promotes the in vitro assay is Mn2+, which prefers an octahedral coordination geometry [25], [48], [60]–[63]. All of the pms1 mutations that affect predicted metal binding ligands, as well as mlh1-E767stp, caused complete MMR defects in vivo, consistent with the observation that metal ligand defects in human Mlh1-Pms2 inactivate MMR in vitro [26], [63]. These mutations also caused an accumulation of Mlh1-Pms1-4GFP foci indicating the step at which these mutations disrupt MMR is after loading of Mlh1-Pms1 by Msh2-Msh6, suggesting that loss of endonuclease activity leads to a turnover defect of Mlh1-Pms1 during MMR.
A striking property of the dominant pms1 and mlh1 mutations is that when present on a low copy plasmid they cause a much greater defect in Exo1-independent MMR compared to MMR when Exo1 is functional even though Mlh1-Pms1 appears to be absolutely required for all MMR. This phenotype is similar to the phenotype caused by the previously described separation-of-function mutations in genes like MSH2, MSH3, MSH6, MLH1, PMS1, POL30 and POL32 that result in strong defects in Exo1-independent MMR but little if any defect in MMR when Exo1 is functional [27]. A hypothesis that could explain these observations is that Mlh1-Pms1 has two roles in MMR, one involving activation of the Mlh1-Pms1 endonuclease and one where Mlh1-Pms1 plays a role in the recruitment of downstream MMR factors. It was previously shown that Exo1 interacts with Mlh1 and that this interaction is required for Exo1 to function in MMR [50]. This suggests the possibility that mispair recognition by Msh2-Msh6 or Msh2-Msh3 recruits Mlh1-Pms1 which then targets Exo1 to DNA where it could promote excision at pre-existing nicks in the DNA, consistent with the observation that lagging strand MMR is more Exo1 dependent than leading strand MMR [38], [42]. Such a reaction would be expected to be relatively insensitive to inhibition by competition with an endonuclease inactive but structurally normal form of Mlh1-Pms1 that would still bind Exo1 and target it to the site of MMR. In contrast, MMR in the absence of Exo1 might be completely dependent on the Mlh1-Pms1 endonuclease activity. This reaction would be expected to be competed for and interfered with by the presence of endonuclease inactive but structurally normal form of Mlh1-Pms1. A model that summarizes these concepts is presented in Figure 4.
Fig. 4. Model of Exo1-dependent and Exo1-independent resection of mispaired DNA. 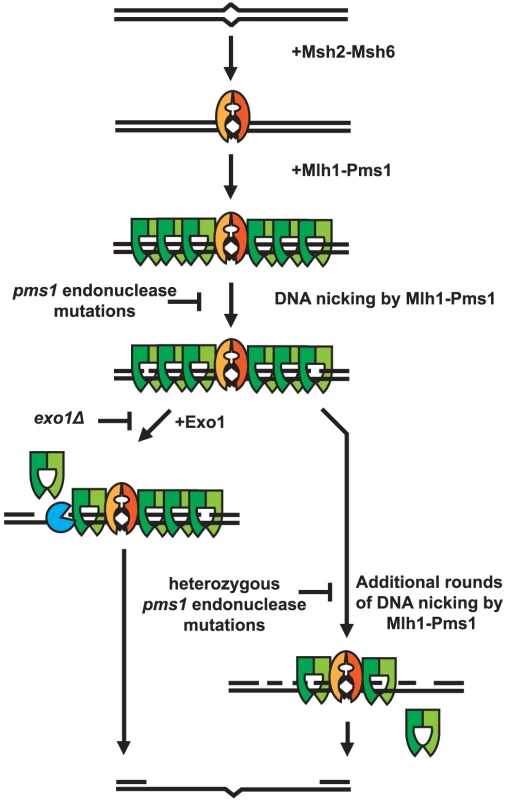
Mispaired DNA is recognized by the Msh2-Msh6 heterodimer and subsequently recruits super-stoichiometric amounts of Mlh1-Pms1. In the absence of endonuclease function, Mlh1-Pms1 has slow or no turnover and persistently remains on damaged DNA. When the Mlh1-Pms1 endonuclease is functional, it can introduce nicks into the DNA and either work in conjunction with Exo1 or function alone to resect past damaged DNA allowing for repair of DNA damage. Materials and Methods
Strains and media
S. cerevisiae cells were grown in YEPD (1% yeast extract, 2% Bacto peptone and 2% dextrose with or without 2% Bacto agar) or SD (0.67% yeast nitrogen base and 2% dextrose with or without 2% Bacto agar) medium. SD medium was supplemented with the appropriate dropout mix of amino acids (USA Biological). The S. cerevisiae strains used in genetic experiments were derived from an S288c parental strain and the strains used for protein purification were derived from RDKY1293 or RDKY8053 (listed in Table S3). All strains were constructed using standard gene disruption and transformation procedures. E. coli strains were propagated in LB media (0.5% yeast extract, 1% tryptone, 0.5% NaCl, 50 µg/ml thymine with or without 2% Bacto agar) containing 100 µg/ml ampicillin as required.
Plasmids and plasmid construction
All plasmids (listed in Table S4) were maintained in E. coli TOP 10F′. A pRS316 Ampr URA3 ARS-CEN PMS1 plasmid pRDK1667 was constructed by recombination in vivo. Briefly, PMS1 was amplified from S. cerevisiae S288c chromosomal DNA using the primers 5′ACGACGGCCAGTGAATTGTAATACGACTCACTATAGGGCGAATTGGAGCTattgccaaacaggcaaagac that contains 50 bp of homology to pRS316 upstream of the multiple cloning site followed by 20 bp of homology to the PMS1 genes 707 bp upstream of the promoter starting at chromosome XIV coordinate 472684 and 5′TTAACCCTCACTAAAGGGAACAAAAGCTGGGTACCGGGCCCCCCCTCGAGgcatacaagaaacaacgcga that contains 50 bp of homology to pRS316 encompassing the XhoI, DraII, ApaI and KpnI sites of the multiple cloning site followed by 20 bp at the 3′ end with homology to the PMS1 genes 302 bp downstream of the stop codon from chromosome XIV coordinate 476314. The PCR product was mixed with an equimolar amount of pRS316 that had been linearized by digestion with SmaI and co-transformed into the wild-type S. cerevisiae strain RDKY3590. The transformants were selected on SC-uracil drop out plates, DNA was isolated from individual transformants, rescued by transformation into E. coli and sequenced. The plasmid selected for further use has a silent C3055T mutation. A pRS426 Ampr URA3 2-micron PMS1 plasmid, pRDK1689, was constructed by subcloning the XhoI to StuI (StuI cuts in the URA3 gene) PMS1 fragment from pRDK1667 into the XhoI to StuI backbone of pRS426. The pRS316 Ampr URA3 ARS-CEN MLH1 plasmid pRDK1338 was from our laboratory collection and contains a SacI to XhoI MLH1 fragment inserted between the SacI and XhoI sites of pRS316. The MLH1 fragment starts at the native SacI site 2281 bp upstream of the MLH1 ATG and ends at an XhoI site inserted by PCR 121 bp downstream of the MLH1 stop codon. The 2-micron Mlh1 and Pms1 over-expression plasmids pRDK573 TRP1 GAL10-MLH1 and pRDK1099 LEU2 GAL10-PMS1-FLAG have been described previously [64]. Mutations were made in these plasmids using standard site-directed mutagenesis methods or by subcloning from a mutant plasmid and the resulting plasmids were verified by DNA sequencing. The PMS1 mutant alleles E707K, H850R, C848S, G683E and C817R were introduced at the chromosomal locus using standard pop in/out techniques employing the integrative plasmids listed in Table S4. These integration plasmids were generated by subcloning the XhoI-StuI fragment containing the pms1 mutant sequence from their respective pRS316-pms1 mutant series plasmids into the XhoI-StuI sites of the pRS306 backbone and were linearized with BlpI prior to transformation for integration into the strains of interest. The MLH1 mutant alleles E767stp and C769stp were introduced at the chromosomal locus using standard gene disruption employing an HPH disruption cassette generated by PCR such the upstream homology targeted the C-terminus of MLH1 and contained the mutations needed to introduce the E767stp and C769stp alleles. All of the chromosomal pms1 and mlh1 mutations were verified by sequencing the entire PMS1 or MLH1 gene as relevant, which also ensured that no additional mutations were introduced during strain construction.
Isolation of random mutations in the PMS1 gene
Mutagenesis of the PMS1 gene by PCR was performed essentially as previously described [65] with the following modifications. The primers used for PCR were those described above for amplification of PMS1. Ten PCR reactions were performed using Klentaq DNA polymerase and a PMS1 gene containing plasmid pRDK433 from our laboratory collection as a template. The PCR reactions were pooled, aliquots of DNA were mixed with an equimolar amount of pRS316 that had been linearized by digestion with SmaI and co-transformed into the wild-type S. cerevisiae strain RDKY3590. The transformants were plated on SC-uracil drop out plates to select for transformants, which were then replica plated onto SC-uracil-lysine drop out plates to screen for colonies that had increased rates of reversion of the lys2-A10 frameshift mutation. Candidate mutator mutants were retrieved from the uracil drop out plates, restreaked on SC-uracil drop out plates, patched in duplicate onto uracil drop out plates and replica plated onto threonine-uracil drop out plates to screen for patches that had increased rates of reversion of the hom3-10 frameshift mutation. Plasmid DNA was isolated from each mutator mutant, transformed into E. coli TOP10F′ and sequenced. Individual mutations identified were then transferred to a new pRS316 PMS1 plasmid pRDK1667 by either sub-cloning using appropriate restriction endonuclease cleavage sites or by site-directed mutagenesis and retested essentially as described for the initial screen above.
Protein purification
S. cerevisiae Mlh1-Pms1 was purified from 2.2 L of culture of the overproduction strain RDKY7608 (RDKY1293 containing the 2-micron plasmids pRDK573 TRP1 GAL10-MLH1 and pRDK1099 LEU2 GAL10pr-PMS1-FLAG) (Table S3) according to a previously published procedure [64], except with the following 6 modifications: (1) Cell growth and induction of Mlh1-Pms1 expression utilized a published lactate to galactose shift protocol according to previously published methods [66]; (2) 2 mM β-mercaptoethanol was substituted for the 1 mM DTT in the buffers used to run the Heparin and FLAG antibody columns whereas all other buffers contained 1 mM DTT; (3) After washing the Heparin column with Buffer A containing 200 mM NaCl, the proteins were eluted using a single step of 1 M NaCl in Buffer A; (4) The pooled Heparin column fractions were diluted with Buffer A to obtain a final NaCl concentration of 500 mM prior to being subjected to 3 cycles of binding and elution from the FLAG antibody column; (5) The SP Sepharose column fractions were diluted with Buffer A to a final NaCl concentration of 200 mM prior to being loaded onto a 1 ml HiTrap Q column (GE Healthcare) followed by elution with a 100 mM to 1 M linear NaCl gradient run in Buffer A; and (6) The HiTrap Q column fractions containing the Mlh1-Pms1 were concentrated and desalted using a Centraprep (Ultracel 30K) spin column. The resulting Mlh1-Pms1 was contained in 0.5 ml of Buffer A +100 mM NaCl, and was frozen in liquid nitrogen and stored at −80 C. The Mlh1-Pms1-E707K, Mlh1-Pms1-C817R, Mlh1-Pms1-C848S and Mlh1-Pms1-H850R proteins were purified using the overproduction strains RDKY7696, RDKY7756, RDKY7759 and RDKY7793 (Table S3). The Mlh1-C769stp-Pms1 and Mlh1-E767stp-Pms1 proteins were purified using the overproduction strains RDKY8055 and RDKY8057 for which the RDKY8053 host strain (Table S3) was a derivative of RDKY1293 containing a deletion of the MLH1 gene. Because mlh1-E767stp allele does not compliment the MLH1 deletion in the host, after the protein expression period, DNA was isolated from the culture, 20 independent MLH1 and PMS1 plasmids were rescued by transformation into E. coli and sequenced to ensure that no mutations had occurred in the expression plasmids. S. cerevisiae PCNA and RFC-Δ1N were purified exactly as described in published procedures [66]–[68]. All of the protein preparations used in these studies were greater than 98% pure as analyzed by SDS-PAGE.
Mlh1-Pms1 endonuclease assay
Mismatch-independent endonuclease assays were performed as a modification of one used previously [26]. 40 µL reactions containing 1 mM MnSO4, 20 mM Tris pH 7.5, 0.5 mM ATP 0.2 mg/mL bovine serum albumin (BSA), 2 mM DTT and 100 ng pRS425 were incubated at 30°C for 30 minutes. Reactions were terminated by incubation at 55°C following introduction of SDS, EDTA, glycerol and proteinase K at concentrations of 0.1%, 14 mM, 8% and 0.5 ug/ml respectively. Mlh1-Pms1, PCNA, or RFC-Δ1N were diluted to the appropriate working concentrations with a buffer comprised of 10% glycerol, 200 mM NaCl, 2 mM DTT and 20 mM Tris pH 7.5. Following termination of the reaction the samples were electrophoresed on a 0.8% agarose gel, the gel was stained with ethidium bromide, extensively destained and then the bands were quantified using a BioRad ChemiDoc XP imaging system. Serial dilutions of XhoI linearized pRS425 ranging from 10–100 ng were used as a concentration standard for quantification.
Determination of mutation rates
Mutation rates were determined by fluctuation analysis. A single colony was used to inoculate a culture that was then diluted and used for transformation with a selectable plasmid carrying the desired allele and transformed colonies were selected by growth for 3 days at 30°C on SC-uracil dropout plates. 7 independent colonies were used to inoculate individual overnight cultures containing 10 ml of SC-uracil dropout media. Following cell growth, appropriate dilutions of the cultures were plated onto SC –uracil, –uracil-lysine, -uracil-threonine, and -uracil-arginine+canavanine dropout plates. The resulting colonies counted after growth at 30°C for 3 days and the average mutation rate was calculated for each strain as described previously [21], [27]. Each experiment was performed independently up to 4 times.
Molecular modeling
Site-directed mutagenesis was guided by a molecular model of the C-terminal domains of Mlh1-Pms1. The C-terminus of S. cerevisiae Mlh1 was modeled using Phyre, xfit, and CNS [69]–[71] starting from the crystal structure of the human Mlh1 C-terminal domain (PDB id 3rbn). The C-terminus of S. cerevisiae Pms1 was similarly modeled using the crystal structures of N. gonorrhoeae [PDB id 3ncv; [47]] and B. subtilis [PDB ids 3gab, 3kdg, 3kdk; [48]] MutL homologs. Subsequently, the amino acid substitutions studied were mapped onto the newly available structure of the C-terminal domains of S. cerevisiae Mlh1-Pms1 [PDB id 4e4w; [46]].
Live-cell imaging and image analysis
For microscopy studies, the C-terminus of each PMS1 protein of interest was fluorescently tagged by targeting a 4GFP tag to the chromosomal locus so that the native promoter was intact and expression remained unaffected. Previous analysis of the tagged PMS1 gene demonstrated that the 4GFP tag did not affect the biological activity of Pms1 [38]. Exponentially growing cultures were washed and resuspended in water, placed on minimal media agar pads, covered with a coverslip, and imaged on a Deltavision (Applied Precision) microscope with an Olympus 100× 1.35NA objective. Fourteen 0.5 µm z sections were acquired and deconvolved with softWoRx software. Further image processing, including maximum intensity projections and intensity measurements were performed using ImageJ.
Supporting Information
Zdroje
1. de la ChapelleA (2004) Genetic predisposition to colorectal cancer. Nat Rev Cancer 4 : 769–780.
2. PeltomakiP, VasenHF (1997) Mutations predisposing to hereditary nonpolyposis colorectal cancer: database and results of a collaborative study. The International Collaborative Group on Hereditary Nonpolyposis Colorectal Cancer. Gastroenterology 113 : 1146–1158.
3. BorresenAL, LotheRA, MelingGI, LystadS, MorrisonP, et al. (1995) Somatic mutations in the hMSH2 gene in microsatellite unstable colorectal carcinomas. Hum Mol Genet 4 : 2065–2072.
4. KaneMF, LodaM, GaidaGM, LipmanJ, MishraR, et al. (1997) Methylation of the hMLH1 promoter correlates with lack of expression of hMLH1 in sporadic colon tumors and mismatch repair-defective human tumor cell lines. Cancer Res 57 : 808–811.
5. Network TCGA (2012) Comprehensive molecular characterization of human colon and rectal cancer. Nature 487 : 330–337.
6. Network TCGA (2013) Integrated genomic characterization of endometrial carcinoma. Nature 497 : 67–73.
7. PeltomakiP (2003) Role of DNA mismatch repair defects in the pathogenesis of human cancer. J Clin Oncol 21 : 1174–1179.
8. DattaA, AdjiriA, NewL, CrouseGF, Jinks RobertsonS (1996) Mitotic crossovers between diverged sequences are regulated by mismatch repair proteins in Saccaromyces cerevisiae. Mol Cell Biol 16 : 1085–1093.
9. MaticI, RayssiguierC, RadmanM (1995) Interspecies gene exchange in bacteria: the role of SOS and mismatch repair systems in evolution of species. Cell 80 : 507–515.
10. PutnamCD, HayesTK, KolodnerRD (2009) Specific pathways prevent duplication-mediated genome rearrangements. Nature 460 : 984–989.
11. IyerRR, PluciennikA, BurdettV, ModrichPL (2006) DNA mismatch repair: functions and mechanisms. Chem Rev 106 : 302–323.
12. KolodnerRD, MarsischkyGT (1999) Eukaryotic DNA mismatch repair. Curr Opin Genet Dev 9 : 89–96.
13. KunkelTA, ErieDA (2005) DNA mismatch repair. Annu Rev Biochem 74 : 681–710.
14. LahueRS, AuKG, ModrichP (1989) DNA mismatch correction in a defined system. Science 245 : 160–164.
15. LamersMH, PerrakisA, EnzlinJH, WinterwerpHH, de WindN, et al. (2000) The crystal structure of DNA mismatch repair protein MutS binding to a G×T mismatch. Nature 407 : 711–717.
16. ObmolovaG, BanC, HsiehP, YangW (2000) Crystal structures of mismatch repair protein MutS and its complex with a substrate DNA. Nature 407 : 703–710.
17. AcharyaS, FosterPL, BrooksP, FishelR (2003) The coordinated functions of the E. coli MutS and MutL proteins in mismatch repair. Mol Cell 12 : 233–246.
18. WelshKM, LuAL, ClarkS, ModrichP (1987) Isolation and characterization of the Escherichia coli mutH gene product. J Biol Chem 262 : 15624–15629.
19. BurdettV, BaitingerC, ViswanathanM, LovettST, ModrichP (2001) In vivo requirement for RecJ, ExoVII, ExoI, and ExoX in methyl-directed mismatch repair. Proc Natl Acad Sci U S A 98 : 6765–6770.
20. AcharyaS, WilsonT, GradiaS, KaneMF, GuerretteS, et al. (1996) hMSH2 forms specific mispair-binding complexes with hMSH3 and hMSH6. Proc Natl Acad Sci U S A 93 : 13629–13634.
21. MarsischkyGT, FilosiN, KaneMF, KolodnerR (1996) Redundancy of Saccharomyces cerevisiae MSH3 and MSH6 in MSH2-dependent mismatch repair. Genes Dev 10 : 407–420.
22. ProllaTA, PangQ, AlaniE, KolodnerRD, LiskayRM (1994) MLH1, PMS1, and MSH2 interactions during the initiation of DNA mismatch repair in yeast. Science 265 : 1091–1093.
23. MendilloML, MazurDJ, KolodnerRD (2005) Analysis of the interaction between the Saccharomyces cerevisiae MSH2-MSH6 and MLH1-PMS1 complexes with DNA using a reversible DNA end-blocking system. J Biol Chem 280 : 22245–22257.
24. BlackwellLJ, WangS, ModrichP (2001) DNA chain length dependence of formation and dynamics of hMutSalpha.hMutLalpha.heteroduplex complexes. J Biol Chem 276 : 33233–33240.
25. KadyrovFA, DzantievL, ConstantinN, ModrichP (2006) Endonucleolytic function of MutLalpha in human mismatch repair. Cell 126 : 297–308.
26. KadyrovFA, HolmesSF, AranaME, LukianovaOA, O'DonnellM, et al. (2007) Saccharomyces cerevisiae MutLalpha is a mismatch repair endonuclease. J Biol Chem 282 : 37181–37190.
27. AminNS, NguyenMN, OhS, KolodnerRD (2001) exo1-Dependent mutator mutations: model system for studying functional interactions in mismatch repair. Mol Cell Biol 21 : 5142–5155.
28. TishkoffDX, BoergerAL, BertrandP, FilosiN, GaidaGM, et al. (1997) Identification and characterization of Saccharomyces cerevisiae EXO1, a gene encoding an exonuclease that interacts with MSH2. Proc Natl Acad Sci U S A 94 : 7487–7492.
29. WeiK, ClarkAB, WongE, KaneMF, MazurDJ, et al. (2003) Inactivation of Exonuclease 1 in mice results in DNA mismatch repair defects, increased cancer susceptibility, and male and female sterility. Genes Dev 17 : 603–614.
30. ConstantinN, DzantievL, KadyrovFA, ModrichP (2005) Human mismatch repair: reconstitution of a nick-directed bidirectional reaction. J Biol Chem 280 : 39752–39761.
31. Flores-RozasH, ClarkD, KolodnerRD (2000) Proliferating cell nuclear antigen and Msh2p-Msh6p interact to form an active mispair recognition complex. Nat Genet 26 : 375–378.
32. LinYL, ShivjiMK, ChenC, KolodnerR, WoodRD, et al. (1998) The evolutionarily conserved zinc finger motif in the largest subunit of human replication protein A is required for DNA replication and mismatch repair but not for nucleotide excision repair. J Biol Chem 273 : 1453–1461.
33. UmarA, BuermeyerAB, SimonJA, ThomasDC, ClarkAB, et al. (1996) Requirement for PCNA in DNA mismatch repair at a step preceding DNA resynthesis. Cell 87 : 65–73.
34. ZhangY, YuanF, PresnellSR, TianK, GaoY, et al. (2005) Reconstitution of 5′-directed human mismatch repair in a purified system. Cell 122 : 693–705.
35. XieY, CounterC, AlaniE (1999) Characterization of the repeat-tract instability and mutator phenotypes conferred by a Tn3 insertion in RFC1, the large subunit of the yeast clamp loader. Genetics 151 : 499–509.
36. DzantievL, ConstantinN, GenschelJ, IyerRR, BurgersPM, et al. (2004) A defined human system that supports bidirectional mismatch-provoked excision. Mol Cell 15 : 31–41.
37. LongleyMJ, PierceAJ, ModrichP (1997) DNA polymerase delta is required for human mismatch repair in vitro. J Biol Chem 272 : 10917–10921.
38. HombauerH, CampbellCS, SmithCE, DesaiA, KolodnerRD (2011) Visualization of eukaryotic DNA mismatch repair reveals distinct recognition and repair intermediates. Cell 147 : 1040–1053.
39. HombauerH, SrivatsanA, PutnamCD, KolodnerRD (2011) Mismatch repair, but not heteroduplex rejection, is temporally coupled to DNA replication. Science 334 : 1713–1716.
40. LangstonLD, O'DonnellM (2006) DNA replication: keep moving and don't mind the gap. Mol Cell 23 : 155–160.
41. BenkovicSJ, ValentineAM, SalinasF (2001) Replisome-mediated DNA replication. Annu Rev Biochem 70 : 181–208.
42. LibertiSE, LarreaAA, KunkelTA (2013) Exonuclease 1 preferentially repairs mismatches generated by DNA polymerase alpha. DNA Repair (Amst) 12 : 92–96.
43. PluciennikA, DzantievL, IyerRR, ConstantinN, KadyrovFA, et al. (2010) PCNA function in the activation and strand direction of MutLalpha endonuclease in mismatch repair. Proc Natl Acad Sci U S A 107 : 16066–16071.
44. GenschelJ, ModrichP (2003) Mechanism of 5′-directed excision in human mismatch repair. Mol Cell 12 : 1077–1086.
45. KadyrovFA, GenschelJ, FangY, PenlandE, EdelmannW, et al. (2009) A possible mechanism for exonuclease 1-independent eukaryotic mismatch repair. Proc Natl Acad Sci U S A 106 : 8495–8500.
46. GueneauE, DherinC, LegrandP, Tellier-LebegueC, GilquinB, et al. (2013) Structure of the MutLalpha C-terminal domain reveals how Mlh1 contributes to Pms1 endonuclease site. Nat Struct Mol Biol 20 : 461–468.
47. NamaduraiS, JainD, KulkarniDS, TabibCR, FriedhoffP, et al. (2010) The C-terminal domain of the MutL homolog from Neisseria gonorrhoeae forms an inverted homodimer. PLoS One 5: e13726.
48. PillonMC, LorenowiczJJ, UckelmannM, KlockoAD, MitchellRR, et al. (2010) Structure of the endonuclease domain of MutL: unlicensed to cut. Mol Cell 39 : 145–151.
49. TranPT, ErdenizN, SymingtonLS, LiskayRM (2004) EXO1-A multi-tasking eukaryotic nuclease. DNA Repair (Amst) 3 : 1549–1559.
50. TranPT, ErdenizN, DudleyS, LiskayRM (2002) Characterization of nuclease-dependent functions of Exo1p in Saccharomyces cerevisiae. DNA Repair (Amst) 1 : 895–912.
51. PangQ, ProllaTA, LiskayRM (1997) Functional domains of the Saccharomyces cerevisiae Mlh1p and Pms1p DNA mismatch repair proteins and their relevance to human hereditary nonpolyposis colorectal cancer-associated mutations. Mol Cell Biol 17 : 4465–4473.
52. Nick McElhinnySA, StithCM, BurgersPM, KunkelTA (2007) Inefficient proofreading and biased error rates during inaccurate DNA synthesis by a mutant derivative of Saccharomyces cerevisiae DNA polymerase delta. J Biol Chem 282 : 2324–2332.
53. Nick McElhinnySA, KumarD, ClarkAB, WattDL, WattsBE, et al. (2010) Genome instability due to ribonucleotide incorporation into DNA. Nat Chem Biol 6 : 774–781.
54. Nick McElhinnySA, WattsBE, KumarD, WattDL, LundstromEB, et al. (2010) Abundant ribonucleotide incorporation into DNA by yeast replicative polymerases. Proc Natl Acad Sci U S A 107 : 4949–4954.
55. PursellZF, IsozI, LundstromEB, JohanssonE, KunkelTA (2007) Regulation of B family DNA polymerase fidelity by a conserved active site residue: characterization of M644W, M644L and M644F mutants of yeast DNA polymerase epsilon. Nucleic Acids Res 35 : 3076–3086.
56. DeschenesSM, TomerG, NguyenM, ErdenizN, JubaNC, et al. (2007) The E705K mutation in hPMS2 exerts recessive, not dominant, effects on mismatch repair. Cancer Lett 249 : 148–156.
57. ZakharyevichK, TangS, MaY, HunterN (2012) Delineation of joint molecule resolution pathways in meiosis identifies a crossover-specific resolvase. Cell 149 : 334–347.
58. BrownMS, LimE, ChenC, NishantKT, AlaniE (2013) Genetic Analysis of mlh3 Mutations Reveals Interactions Between Crossover Promoting Factors During Meiosis in Baker's Yeast. G3 (Bethesda) 3 : 9–22.
59. HunterN, BortsRH (1997) Mlh1 is unique among mismatch repair proteins in its ability to promote crossing-over during meiosis. Genes Dev 11 : 1573–1582.
60. DuppatlaV, BoddaC, UrbankeC, FriedhoffP, RaoDN (2009) The C-terminal domain is sufficient for endonuclease activity of Neisseria gonorrhoeae MutL. Biochem J 423 : 265–277.
61. FukuiK, NishidaM, NakagawaN, MasuiR, KuramitsuS (2008) Bound nucleotide controls the endonuclease activity of mismatch repair enzyme MutL. J Biol Chem 283 : 12136–12145.
62. IinoH, KimK, ShimadaA, MasuiR, KuramitsuS, et al. (2011) Characterization of C - and N-terminal domains of Aquifex aeolicus MutL endonuclease: N-terminal domain stimulates the endonuclease activity of C-terminal domain in a zinc-dependent manner. Biosci Rep 31 : 309–322.
63. KosinskiJ, PlotzG, GuarneA, BujnickiJM, FriedhoffP (2008) The PMS2 subunit of human MutLalpha contains a metal ion binding domain of the iron-dependent repressor protein family. J Mol Biol 382 : 610–627.
64. HargreavesVV, ShellSS, MazurDJ, HessMT, KolodnerRD (2010) Interaction between the Msh2 and Msh6 nucleotide-binding sites in the Saccharomyces cerevisiae Msh2-Msh6 complex. J Biol Chem 285 : 9301–9310.
65. UmezuK, SugawaraN, ChenC, HaberJE, KolodnerRD (1998) Genetic analysis of yeast RPA1 reveals its multiple functions in DNA metabolism. Genetics 148 : 989–1005.
66. GerikKJ, GarySL, BurgersPM (1997) Overproduction and affinity purification of Saccharomyces cerevisiae replication factor C. J Biol Chem 272 : 1256–1262.
67. AyyagariR, ImpellizzeriKJ, YoderBL, GarySL, BurgersPM (1995) A mutational analysis of the yeast proliferating cell nuclear antigen indicates distinct roles in DNA replication and DNA repair. Mol Cell Biol 15 : 4420–4429.
68. FienK, StillmanB (1992) Identification of replication factor C from Saccharomyces cerevisiae: a component of the leading-strand DNA replication complex. Mol Cell Biol 12 : 155–163.
69. BrungerAT, AdamsPD, CloreGM, DeLanoWL, GrosP, et al. (1998) Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr 54 : 905–921.
70. McReeDE (1999) XtalView/Xfit–A versatile program for manipulating atomic coordinates and electron density. J Struct Biol 125 : 156–165.
71. Bennett-LovseyRM, HerbertAD, SternbergMJ, KelleyLA (2008) Exploring the extremes of sequence/structure space with ensemble fold recognition in the program Phyre. Proteins 70 : 611–625.
72. SieversF, WilmA, DineenD, GibsonTJ, KarplusK, et al. (2011) Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7 : 539.
Štítky
Genetika Reprodukční medicína
Článek Defending Sperm FunctionČlánek How to Choose the Right MateČlánek Conserved Translatome Remodeling in Nematode Species Executing a Shared Developmental TransitionČlánek Genome-Wide and Cell-Specific Epigenetic Analysis Challenges the Role of Polycomb in SpermatogenesisČlánek The Integrator Complex Subunit 6 (Ints6) Confines the Dorsal Organizer in Vertebrate EmbryogenesisČlánek Multiple bHLH Proteins form Heterodimers to Mediate CRY2-Dependent Regulation of Flowering-Time inČlánek Playing the Field: Sox10 Recruits Different Partners to Drive Central and Peripheral MyelinationČlánek A Minimal Nitrogen Fixation Gene Cluster from sp. WLY78 Enables Expression of Active Nitrogenase inČlánek Evolutionary Tuning of Protein Expression Levels of a Positively Autoregulated Two-Component System
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2013 Číslo 10
-
Všechny články tohoto čísla
- Defending Sperm Function
- How to Choose the Right Mate
- A Mutation in the Gene in Labrador Retrievers with Hereditary Nasal Parakeratosis (HNPK) Provides Insights into the Epigenetics of Keratinocyte Differentiation
- Conserved Translatome Remodeling in Nematode Species Executing a Shared Developmental Transition
- A Novel Actin mRNA Splice Variant Regulates ACTG1 Expression
- Tracking Proliferative History in Lymphocyte Development with Cre-Mediated Sister Chromatid Recombination
- Correlated Occurrence and Bypass of Frame-Shifting Insertion-Deletions (InDels) to Give Functional Proteins
- Chimeric Protein Complexes in Hybrid Species Generate Novel Phenotypes
- Loss of miR-10a Activates and Collaborates with Activated Wnt Signaling in Inducing Intestinal Neoplasia in Female Mice
- Both Rare and Copy Number Variants Are Prevalent in Agenesis of the Corpus Callosum but Not in Cerebellar Hypoplasia or Polymicrogyria
- Reverse PCA, a Systematic Approach for Identifying Genes Important for the Physical Interaction between Protein Pairs
- Partial Deletion of Chromosome 8 β-defensin Cluster Confers Sperm Dysfunction and Infertility in Male Mice
- Genome-Wide and Cell-Specific Epigenetic Analysis Challenges the Role of Polycomb in Spermatogenesis
- Coordinate Regulation of Mature Dopaminergic Axon Morphology by Macroautophagy and the PTEN Signaling Pathway
- Cooperation between RUNX1-ETO9a and Novel Transcriptional Partner KLF6 in Upregulation of in Acute Myeloid Leukemia
- Mobility of the Native Conjugative Plasmid pLS20 Is Regulated by Intercellular Signaling
- FliZ Is a Global Regulatory Protein Affecting the Expression of Flagellar and Virulence Genes in Individual Bacterial Cells
- Specific Tandem Repeats Are Sufficient for Paramutation-Induced Trans-Generational Silencing
- Condensin II Subunit dCAP-D3 Restricts Retrotransposon Mobilization in Somatic Cells
- Dominant Mutations in Identify the Mlh1-Pms1 Endonuclease Active Site and an Exonuclease 1-Independent Mismatch Repair Pathway
- The Insulator Homie Promotes Expression and Protects the Adjacent Gene from Repression by Polycomb Spreading
- Human Intellectual Disability Genes Form Conserved Functional Modules in
- Coordination of Cell Proliferation and Cell Fate Determination by CES-1 Snail
- ORFs in Drosophila Are Important to Organismal Fitness and Evolved Rapidly from Previously Non-coding Sequences
- Different Roles of Eukaryotic MutS and MutL Complexes in Repair of Small Insertion and Deletion Loops in Yeast
- The Spore Differentiation Pathway in the Enteric Pathogen
- Acceleration of the Glycolytic Flux by Steroid Receptor Coactivator-2 Is Essential for Endometrial Decidualization
- The Human Nuclear Poly(A)-Binding Protein Promotes RNA Hyperadenylation and Decay
- Genome Wide Analysis Reveals Zic3 Interaction with Distal Regulatory Elements of Stage Specific Developmental Genes in Zebrafish
- Xbp1 Directs Global Repression of Budding Yeast Transcription during the Transition to Quiescence and Is Important for the Longevity and Reversibility of the Quiescent State
- The Integrator Complex Subunit 6 (Ints6) Confines the Dorsal Organizer in Vertebrate Embryogenesis
- Incorporating Motif Analysis into Gene Co-expression Networks Reveals Novel Modular Expression Pattern and New Signaling Pathways
- The Bacterial Response Regulator ArcA Uses a Diverse Binding Site Architecture to Regulate Carbon Oxidation Globally
- Direct Monitoring of the Strand Passage Reaction of DNA Topoisomerase II Triggers Checkpoint Activation
- Multiple bHLH Proteins form Heterodimers to Mediate CRY2-Dependent Regulation of Flowering-Time in
- A Reversible Histone H3 Acetylation Cooperates with Mismatch Repair and Replicative Polymerases in Maintaining Genome Stability
- ALS-Associated Mutations Result in Compromised Alternative Splicing and Autoregulation
- Robust Demographic Inference from Genomic and SNP Data
- Preferential Binding to Elk-1 by SLE-Associated Risk Allele Upregulates Expression
- Rad52 Sumoylation Prevents the Toxicity of Unproductive Rad51 Filaments Independently of the Anti-Recombinase Srs2
- The Serum Resistome of a Globally Disseminated Multidrug Resistant Uropathogenic Clone
- Identification of 526 Conserved Metazoan Genetic Innovations Exposes a New Role for Cofactor E-like in Neuronal Microtubule Homeostasis
- SUMO Localizes to the Central Element of Synaptonemal Complex and Is Required for the Full Synapsis of Meiotic Chromosomes in Budding Yeast
- Integrated Enrichment Analysis of Variants and Pathways in Genome-Wide Association Studies Indicates Central Role for IL-2 Signaling Genes in Type 1 Diabetes, and Cytokine Signaling Genes in Crohn's Disease
- Genome-Wide High-Resolution Mapping of UV-Induced Mitotic Recombination Events in
- Genome-Wide Analysis of Cell Type-Specific Gene Transcription during Spore Formation in
- Playing the Field: Sox10 Recruits Different Partners to Drive Central and Peripheral Myelination
- Two Portable Recombination Enhancers Direct Donor Choice in Fission Yeast Heterochromatin
- Mining the Human Phenome Using Allelic Scores That Index Biological Intermediates
- Yeast Tdh3 (Glyceraldehyde 3-Phosphate Dehydrogenase) Is a Sir2-Interacting Factor That Regulates Transcriptional Silencing and rDNA Recombination
- A Minimal Nitrogen Fixation Gene Cluster from sp. WLY78 Enables Expression of Active Nitrogenase in
- A Review of Bacteria-Animal Lateral Gene Transfer May Inform Our Understanding of Diseases like Cancer
- High Throughput Sequencing Reveals Alterations in the Recombination Signatures with Diminishing Spo11 Activity
- Partitioning the Heritability of Tourette Syndrome and Obsessive Compulsive Disorder Reveals Differences in Genetic Architecture
- Eleven Candidate Susceptibility Genes for Common Familial Colorectal Cancer
- A GDF5 Point Mutation Strikes Twice - Causing BDA1 and SYNS2
- Systematic Unraveling of the Unsolved Pathway of Nicotine Degradation in
- Natural Genetic Variation of Integrin Alpha L () Modulates Ischemic Brain Injury in Stroke
- Evolutionary Tuning of Protein Expression Levels of a Positively Autoregulated Two-Component System
- Evolutionary Change within a Bipotential Switch Shaped the Sperm/Oocyte Decision in Hermaphroditic Nematodes
- Limiting of the Innate Immune Response by SF3A-Dependent Control of MyD88 Alternative mRNA Splicing
- Multiple Signaling Pathways Coordinate to Induce a Threshold Response in a Chordate Embryo
- Distinct Regulatory Mechanisms Act to Establish and Maintain Pax3 Expression in the Developing Neural Tube
- Genome Wide Analysis of Narcolepsy in China Implicates Novel Immune Loci and Reveals Changes in Association Prior to Versus After the 2009 H1N1 Influenza Pandemic
- Mismatch Repair Genes and Modify CAG Instability in Huntington's Disease Mice: Genome-Wide and Candidate Approaches
- The Histone H3 K27 Methyltransferase KMT6 Regulates Development and Expression of Secondary Metabolite Gene Clusters
- Hsp70-Hsp40 Chaperone Complex Functions in Controlling Polarized Growth by Repressing Hsf1-Driven Heat Stress-Associated Transcription
- Function and Evolution of DNA Methylation in
- Stimulation of mTORC1 with L-leucine Rescues Defects Associated with Roberts Syndrome
- Transcription Termination and Chimeric RNA Formation Controlled by FPA
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Dominant Mutations in Identify the Mlh1-Pms1 Endonuclease Active Site and an Exonuclease 1-Independent Mismatch Repair Pathway
- Eleven Candidate Susceptibility Genes for Common Familial Colorectal Cancer
- The Histone H3 K27 Methyltransferase KMT6 Regulates Development and Expression of Secondary Metabolite Gene Clusters
- A Mutation in the Gene in Labrador Retrievers with Hereditary Nasal Parakeratosis (HNPK) Provides Insights into the Epigenetics of Keratinocyte Differentiation
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



