-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaYY1 Regulates Melanocyte Development and Function by Cooperating with MITF
Studies of coat color mutants have greatly contributed to the discovery of genes that regulate melanocyte development and function. Here, we generated Yy1 conditional knockout mice in the melanocyte-lineage and observed profound melanocyte deficiency and premature gray hair, similar to the loss of melanocytes in human piebaldism and Waardenburg syndrome. Although YY1 is a ubiquitous transcription factor, YY1 interacts with M-MITF, the Waardenburg Syndrome IIA gene and a master transcriptional regulator of melanocytes. YY1 cooperates with M-MITF in regulating the expression of piebaldism gene KIT and multiple additional pigmentation genes. Moreover, ChIP–seq identified genome-wide YY1 targets in the melanocyte lineage. These studies mechanistically link genes implicated in human conditions of melanocyte deficiency and reveal how a ubiquitous factor (YY1) gains lineage-specific functions by co-regulating gene expression with a lineage-restricted factor (M-MITF)—a general mechanism which may confer tissue-specific gene expression in multiple lineages.
Published in the journal: . PLoS Genet 8(5): e32767. doi:10.1371/journal.pgen.1002688
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1002688Summary
Studies of coat color mutants have greatly contributed to the discovery of genes that regulate melanocyte development and function. Here, we generated Yy1 conditional knockout mice in the melanocyte-lineage and observed profound melanocyte deficiency and premature gray hair, similar to the loss of melanocytes in human piebaldism and Waardenburg syndrome. Although YY1 is a ubiquitous transcription factor, YY1 interacts with M-MITF, the Waardenburg Syndrome IIA gene and a master transcriptional regulator of melanocytes. YY1 cooperates with M-MITF in regulating the expression of piebaldism gene KIT and multiple additional pigmentation genes. Moreover, ChIP–seq identified genome-wide YY1 targets in the melanocyte lineage. These studies mechanistically link genes implicated in human conditions of melanocyte deficiency and reveal how a ubiquitous factor (YY1) gains lineage-specific functions by co-regulating gene expression with a lineage-restricted factor (M-MITF)—a general mechanism which may confer tissue-specific gene expression in multiple lineages.
Introduction
Waardenburg syndrome, Tietz syndrome and piebaldism represent disorders of melanocyte migration, proliferation, or survival during embryonic development and are characterized by stable congenital white patches of the skin and hair. They are caused by mutations in various genes, including PAX3 (paired-box 3), SOX10 (sex-determining region Y-box 10), MITF (microphthalmia-associated transcription factor), EDN3 (endothelin 3), EDNRB (endothelin receptor B) and KIT, resulting in hypopigmentation due to a lack of melanocytes rather than a lack of pigment in viable melanocytes, as occurs in albinism [1], [2], [3]. Among these genes, MITF is one of the earliest melanocyte-specific transcription factors and is a master regulator of melanocyte development and function. In humans, germline loss-of-function mutations of MITF are associated with Waardenburg Syndrome (WS) type IIA and Tietz syndrome, autosomal dominant conditions which exhibit melanocytic deficiencies and pigmentation abnormalities together with variable severity of sensorineural deafness [4], [5], [6]. M-MITF is the melanocyte-specific isoform of MITF. The importance of MITF in melanocyte differentiation is highlighted by its direct and lineage-specific transcription of essential pigmentation enzymes and melanosome components, e.g., tyrosinase (TYR), dopachrome tautomerase (DCT) and silver (SILV). MITF also regulates the receptor tyrosine kinase KIT [7], which is necessary for the survival and dispersal of melanocyte precursors from the migration staging area. Inactivating mutations or deletion of KIT lead to piebaldism in humans, with loss of melanocytes typically restricted to the hair and skin [3], [8], [9]. MITF thus has diverse functions in melanocyte differentiation, growth and survival pathways [10], [11], [12].
Yin Yang 1 (YY1) is a ubiquitously expressed zinc-finger transcription factor. It can act as transcriptional repressor or activator [13]. The essential role of YY1 in development is underscored by the fact that genetic ablation of Yy1 in mice resulted in peri-implantation lethality [14]. During B cell and oligodendrocyte lineage development, YY1 functions as a pro-differentiation factor [15], [16]. In mouse spermatogenesis, YY1 is required for maintaining heterochromatin structure integrity [17]. YY1 thus has important functions in several lineages, but given its ubiquitous expression in the majority of tissues, it is not known whether it is able to regulate select genes in a lineage-specific manner. This paper reports a key role for YY1 in melanocytic lineage development and describes how melanocyte-specific functions of YY1 may be directed by its interaction with M-MITF.
Results
YY1 is required for melanocyte development and survival in vivo and in vitro
To study the function of YY1 in melanocyte development, we generated melanocyte-specific Yy1 conditional knockout mice (TyrCre, yy1f/f) by crossing yy1flox/flox mice [18] with TyrCre mice in which Cre expression is constitutively driven by the melanocyte-specific tyrosinase (Tyr) promoter [19]. Cre-mediated genetic recombination starts from embryonic day 10.5. TyrCre, yy1f/f mice were born at the expected Mendelian ratio. Shortly after birth (P4), TyrCre, yy1f/f mice showed profoundly lighter skin pigmentation compared with littermate controls TyrCre, yy1f/+ (Figure 1A, P4). In the first hair cycle (P0–P28), ventral hairs of TyrCre, yy1f/f mice were essentially devoid of pigment (Figure 1A, P10). Hairs from dorsal skin of TyrCre, yy1f/f mice were much less pigmented than those in control mice (Figure 1A, P10), and H&E sections of dorsal skin revealed small amounts of residual hair follicle melanin (Figure 1B, P4, arrows). As indicated by immunofluorescence (Figure 1C–1H), the residual dorsal melanocytes (DCT positive, Figure 1D) continued to express YY1 (compare nuclear YY1 signal to cytoplasmic DCT, Figure 1G), indicating incomplete Cre-mediated deletion. MITF expression was not affected in the residual hair follicle melanocytes of P4 TyrCre, yy1f/f mice (Figure S1A). In the second hair cycle anagen phase (P28–P42), new dorsal hair follicles of TyrCre, yy1f/f mice completely lacked melanin pigment (Figure 1B, P38) as well as DCT positive melanocytes (Figure 1I and 1J) and corresponded to subsequent white dorsal fur (Figure 1A, P45), indicating an ongoing need for YY1 in post-developmental melanocytes. Further support for melanocyte absence, rather than absence of pigment within viable melanocytes, came from TyrCre, yy1f/f, Dct-lacZ mice, which carry a lacZ reporter under the control of the melanocytic-specific Dct promoter [20]. XGal staining of whole-mount skin sections confirmed the absence of melanocytes or pigment in hair follicles of TyrCre, yy1f/f, Dct-lacZ mice at the anagen phase of the second hair cycle (P38) (Figure 1K and 1L, Figure S1B). Collectively these data suggest that YY1 is required for melanocyte development and post-developmental survival in vivo.
Fig. 1. YY1 is required for melanocyte development in vivo. 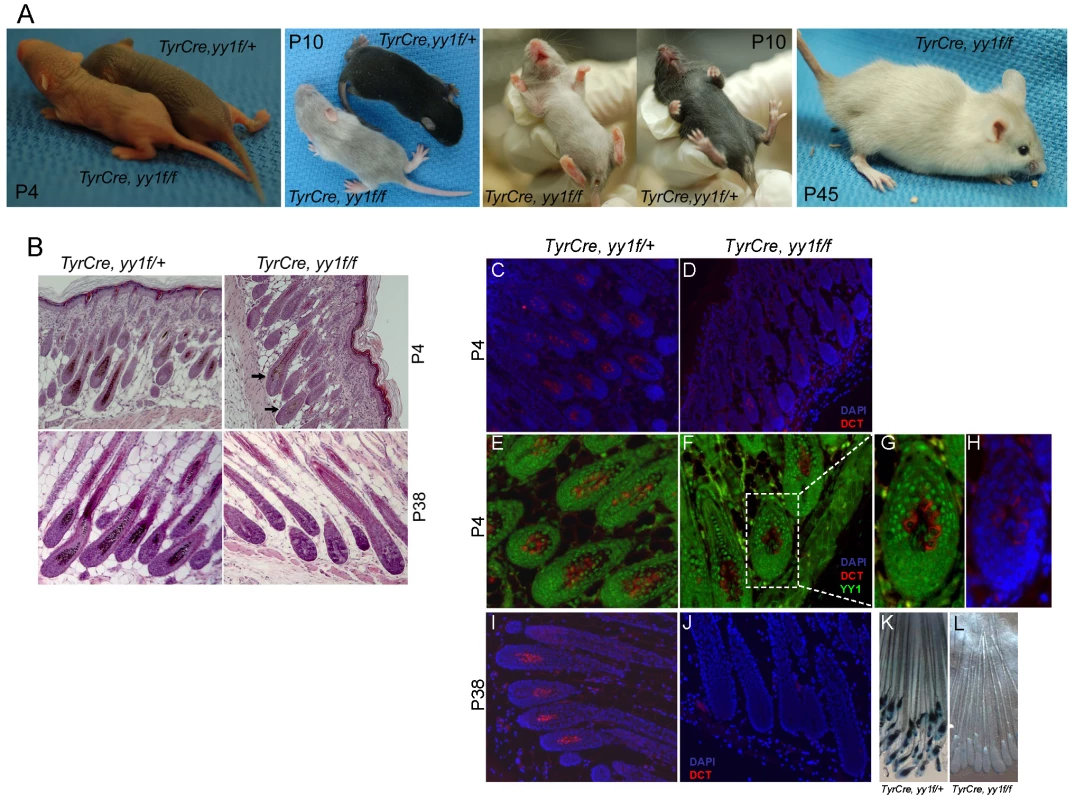
(A) Skin and hair pigmentation phenotype of TyrCre, yy1f/fl in the first (P4, P10) and second (P45) hair cycles. (B) H&E staining of hair follicles from skin sections of TyrCre, yy1f/+ and TyrCre, yy1f/f mice in the first (P4) and second (P38) hair cycles. Arrows point to the hair follicles still containing pigment. (C,D) Immunofluorescence staining of DCT (red) in P4 hair follicles of TyrCre, yy1f/+ (C) and TyrCre, yy1f/f (D) mice. Skin sections were stained with goat anti-DCT primary antibody and donkey anti-goat Alexa 594 secondary antibody. Nuclei were counterstained with DAPI (blue). (E,F) Immunofluorescence staining of DCT (red) and YY1 (green) in P4 hair follicles of TyrCre, yy1f/+ (E) and TyrCre, yy1f/f (F) mice. Skin sections were double-stained with goat anti-DCT and rabbit anti-YY1 primary antibodies and then donkey anti-goat Alexa 594 and goat anti-rabbit Alexa 488 secondary antibodies. (G,H) Zoom-in view of the dashed box area in (F). DAPI stain is in blue in (H). (I,J) Immunofluorescence staining of DCT (red) in P38 hair follicles of TyrCre, yy1f/+ (I) and TyrCre, yy1f/f (J) mice. Nuclei were stained with DAPI (blue). (K&L) XGal stain of whole-mount skin sections from P38 TyrCre, yy1f/+, Dct-LacZ (K) and TyrCre, yy1f/f, Dct-LacZ (L) mice. To determine whether YY1 is also required for melanocytic cell survival in vitro, we stably knocked down endogenous YY1 using two lentiviral shRNAs (yy1shR#1) (Figure 2A). Prolonged depletion of YY1 led to decreased cell numbers in both YY1 knockdown MALME-3M melanoma cells and human foreskin primary melanocytes (HFM), similar to MITF knockdown (Figure 2B and 2C, Figure S2A). While a decrease in cell numbers clearly indicates cell death, it is plausible that there could also be anti-proliferative effects in some (surviving) cells, a possibility that will require additional study.
Fig. 2. YY1 is required for melanocyte survival and the expression of melanocyte differentiation markers in vitro. 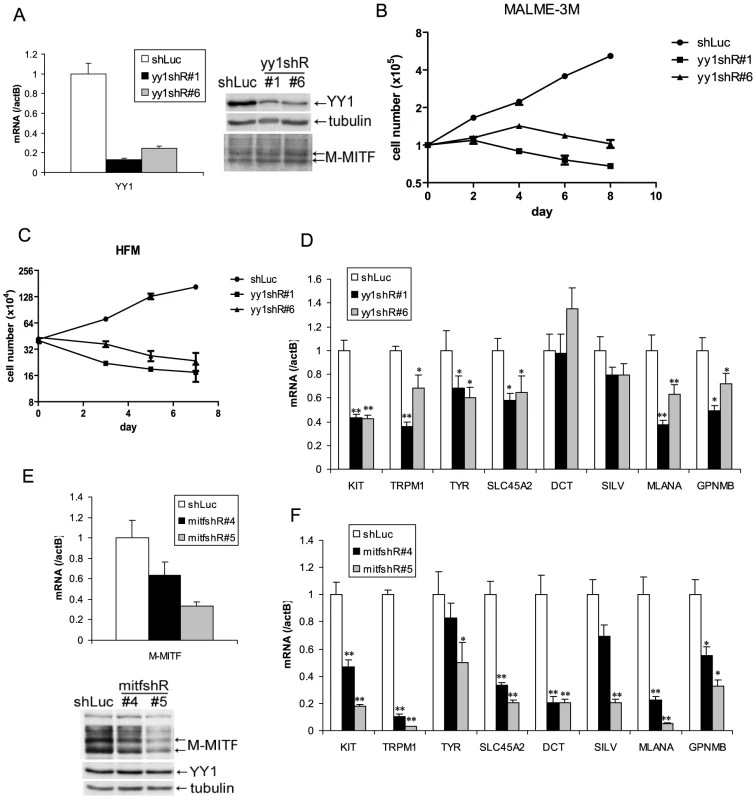
(A,E) Knockdown of endogenous YY1 (A) and MITF (E) in MALME-3M cells. MALME-3M cells were infected with lentivirus carrying YY1 shRNA (yy1shR#1 & #6), MITF shRNA (mitfshR#4 & #5) or control shRNA (shLuc). After overnight infection, cells were selected with puromycin for 3 days. Total RNA and cell lysates were harvested for mRNA and protein measurements by RT-qPCR and western blotting. mRNA levels were normalized to β-actin (actB). (B) Growth curve of YY1 knockdown MALME-3M cells. After 3 days of puromycin selection of YY1 knockdown and control cells as in (A), cells were re-seeded at 1×105 per 6 cm plate. Cell numbers were counted every other day. (C) Growth curve of YY1 knockdown HFM cells. HFM were infected and selected with puromycin as in (A). After puromycin selection, cell numbers were counted on days 0, 3, 5, 7. (D,F) mRNA levels were quantitated for KIT and multiple melanocyte differentiation genes in YY1 (D) and MITF (F) knockdown cells. Error bars represent s.d. of triplicates. *, p<0.05; **, p<0.01 (Student t-test). YY1 is required for the expression of melanocyte survival and differentiation genes
Melanocyte deficiency and premature gray hair in melanocyte-specific Yy1 conditional knockout mice are reminiscent of the melanocyte deficiency phenotype in human Waardenburg syndrome and related disorders and suggest that YY1 may transcriptionally regulate genes important for lineage development or survival. We therefore performed expression profiling analysis using MALME-3M melanoma cells with or without YY1 knockdown. Although the Waardenburg syndrome genes PAX3, SOX10, MITF, EDN3 and EDNRB were not dramatically affected at the mRNA level (Table S1), expression of the piebaldism gene KIT was significantly down-regulated upon loss of YY1 (Table S1 and Figure 2D). It is currently unclear whether the diminished MALME-3M viability upon loss of YY1 is mediated by suppression of KIT expression, or the other survival factors. As demonstrated by immunofluorescence, KIT protein was significantly reduced in the residual hair follicle melanocytes (DCT positive) in P4 TyrCre, yy1f/f mice (Figure S2B). The mRNA levels of multiple melanocyte differentiation factors TYR, SLC45A2, MLANA, TRPM1, and GPNMB, were also down-regulated in YY1 knockdown cells (Table S1 and Figure 2D). These melanocyte differentiation genes are well-known direct targets of MITF [11]. Indeed, MITF knockdown also strongly down-regulated these and other pigmentation genes (e.g. DCT and SILV, whose MITF binding sites have been previously reported [21], [22]) (Figure 2E and 2F). KIT has been shown to be a target of MITF in mast cells [7]. MITF knockdown also reduced the expression of KIT in MALME-3M melanoma cells (Figure 2F). BCL2, another known target of MITF [23], was modestly down-regulated in both MITF and YY1 knockdown MALME-3M cells (Figure S2C).
As knockdown of YY1 did not change MITF protein levels (Figure 2A), the fact that it significantly affected the levels of many MITF target genes suggested that YY1 might functionally cooperate with MITF. To explore this possibility, we performed expression profiling analysis with MITF knockdown melanoma cells (Figure 2E). We found that 1241 RefSeq genes showed significantly reduced expression after MITF knockdown (Table S1). YY1's DNA binding sequence was one of the top five motifs showing the greatest enrichment in open chromatin regions of the MITF-responsive gene promoters (Materials and Methods, and Table S2), further strengthening the possibility that YY1 might cooperate with MITF.
YY1 interacts with MITF
To test whether YY1 and MITF physically interact in a protein complex, we first co-transfected expression constructs of Flag-tagged YY1 and HA-tagged M-MITF or a close MiT family member TFE3 into 293T cells. Flag-tagged YY1 co-immunoprecipitated with M-MITF as well as with TFE3 (Figure 3A). Interaction of TFE3 with E2F3b but not with E2F2 served as positive and negative controls respectively, as previously described [24]. Endogenous YY1 also co-immunoprecipitated with endogenous MITF in MALME-3M cells (Figure 3B). YY1 contains an N-terminal histidine-rich region (His) flanked by acidic amino acids (Acidic), a central glycine/lysine-rich region (GK) and C-terminal zinc fingers [25] (Figure 3C). MITF contains a central basic helix-loop-helix leucine-zipper domain (b-HLH-Zip) and two transcription activation domains (TAD) (Figure 3C and reference therein [10]). Expression constructs encoding full-length or truncated mutants of YY1 and M-MITF revealed required binding domains for YY1-MITF complex formation: M-MITF interacted with full-length as well as the C-terminal domain (amino acid 251–320) of YY1 (Figure 3D, Figure S3A); YY1 interacted with full-length, N-terminal TAD (amino acid 1–180) and central b-HLH-Zip (amino acid 181–300) of M-MITF, but not with C-terminal TAD (amino acid 301–419) (Figure 3E, Figure S3B). A smaller b-HLH-Zip domain (amino acid 181–264) of MITF showed a weaker interaction with YY1 compared with the full length central domain (Figure S3B), suggesting that the whole b-HLH-Zip domain is important in mediating the strongest interaction
Fig. 3. YY1 interacts with MITF. 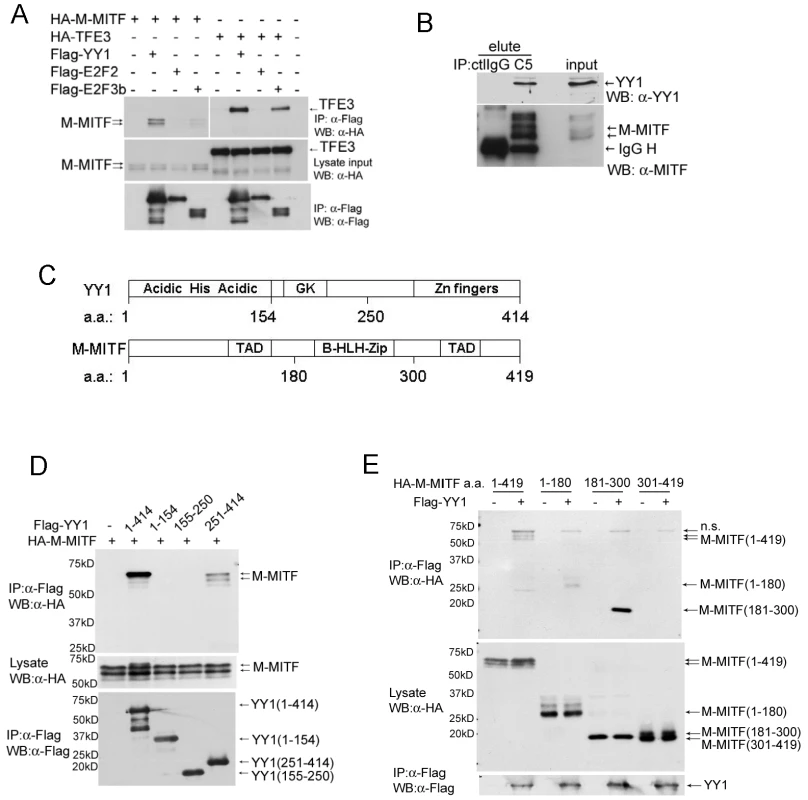
(A) Interaction between Flag-tagged YY1 and HA-tagged M-MITF in 293T cells. Total cell lysate of 293T cells transfected with indicated plasmids was immunoprecipitated with anti-Flag M2 agarose beads. Immunocomplex and lysate input were analyzed by western blotting with anti-Flag and anti-HA antibodies. (B) Endogenous protein interaction between YY1 and MITF. Total cell lysate of MALME-3M was immunoprecipitated with mouse control IgG (ctlIgG) or mouse anti-MITF monoclonal antibody (C5). Immunocomplex (elute) and lysate input were analyzed by western blotting with anti-MITF and anti-YY1 antibodies. (C) Schematic diagrams of YY1 and M-MITF proteins. GK, glycine and lysine rich region; TAD, transcription activation domain; b-HLH-Zip, basic helix-loop-helix leucine zipper. (D) M-MITF interacts with the C-terminal domain (a.a. 251–414) of YY1. (E) YY1 interacts with the N-terminal TAD (a.a. 1–180) and the central b-HLH-Zip (a.a. 181–300) of M-MITF. YY1 cooperates with MITF in gene transcription
Using the expression profiling data, we further analyzed the cooperation between YY1 and MITF. Figure 4A shows the scatter plot of log fold-change of gene expression in MITF and YY1 knockdown cells relative to that in control cells. The bagged regression curve (red line) shows a significant association between the sets of genes co-activated by MITF and YY1, as well as genes co-repressed by MITF and YY1. We also compared the overlap between the top ranking differentially expressed genes after MITF knockdown or YY1 knockdown. Statistical significance is observed for genes that are either co-repressed or co-activated by MITF and YY1, but not for genes on which MITF and YY1 have antagonistic effects (Figure 4B). The statistical significance is seen to be robust and independent of the number of top ranking differentially expressed genes (Figure 4B). Of note, MITF and YY1 knockdown effects correlated for 131 pigmentation-related genes (Figure 4C).
Fig. 4. YY1 cooperates with MITF in gene transcription. 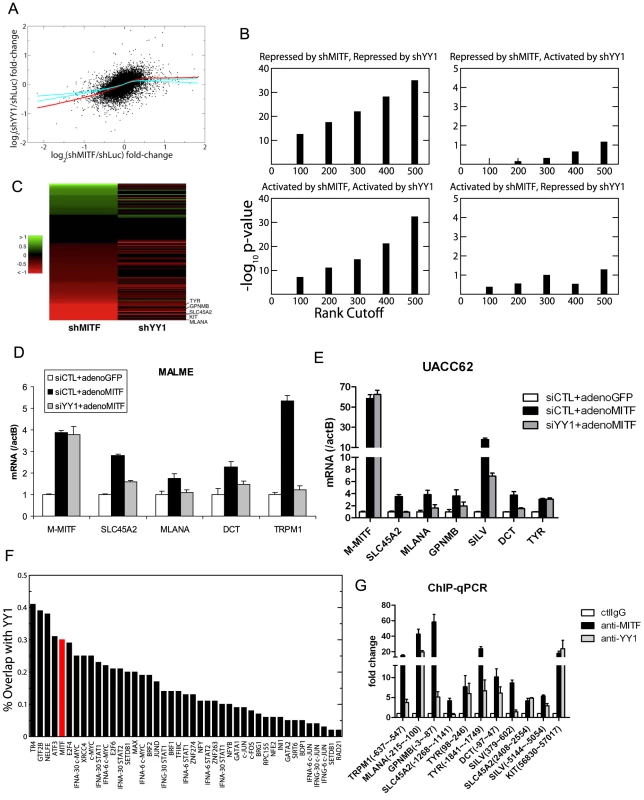
(A) Scatter plot of mean log fold-changes in MALME-3M and UACC62 cells treated with MITF shRNA (shMITF) and YY1 shRNA (shYY1) relative to the control (shLuc). The red line is the average of 100 lowess regression curves fitted to 100 bootstrap simulations. The cyan curves represent the upper and lower bounds of 1000 lowess curves fitted to the data obtained by randomizing the x-coordinates of differentially expressed genes after YY1-shRNA. The fact that the observed fit lies well outside the error bounds of randomized data in the lower left quadrant shows that YY1 and MITF co-activate a statistically significant number of common target genes. (B) Fisher's exact test p-values are plotted for the significance of the overlap between the top ranking differentially expressed genes after shMITF and shYY1, where the differential expression was ranked by RSA analysis p-values. Statistical significance is observed for genes that are either co-repressed or co-activated by MITF and YY1, but not for genes on which MITF and YY1 have antagonistic effects. (C) Gene Ontology analysis found 131 genes to play a role in melanosome function and pigmentation regulation. The heatmap shows the log expression fold-change of those 131 genes after MITF and YY1 knockdown. (D,E) Loss of YY1 inhibits M-MITF-dependent transcriptional up-regulation of multiple melanocytic markers. MALME-3M cells (D) or UACC62 cells (E) were transfected with 10 nM of YY1 siRNA (siYY1) or control siRNA (siCTL). After 24 h, cells were infected with adenovirus encoding cDNA of M-MITF or GFP at MOI 500 (D) or 100 (E). Cells were harvested 48 hours post infection. mRNA levels of the different genes were measured by RT-qPCR and normalized to β-actin (actB). Error bars represent s.d. of triplicates. (F) The fraction of transcription factor (TF) binding sites overlapping with YY1 was computed for 42 TFs mapped by the ENCODE consortium in the K562 cell line (black bar). The overlap between MITF binding sites in 501MEL and YY1 binding sites in MALME-3M (red bar) was 30%. (G) Co-localization of YY1 and MITF at the proximal promoter of multiple melanocyte differentiation markers in MALME-3M cells. ChIP-qPCR primer position relative to the transcription start site is indicated in brackets. Data are normalized to control IgG ChIP. Error bar represents s.d. of triplicates. Although knockdown of endogenous YY1 affected the basal transcription of multiple melanocyte differentiation genes (Figure 2D), overexpression of YY1 alone did not affect the expression of these genes (data not shown). In MALME-3M (high endogenous M-MITF level) and UACC62 cells (low endogenous M-MITF level), overexpression of M-MITF by adenovirus infection was sufficient to induce the transcription of multiple melanocyte differentiation genes (Figure 4D and 4E, Figure S4). However knockdown of YY1 inhibited the M-MITF-dependent transcriptional upregulation of most of these genes except TYR. These results suggest a dependency of M-MITF-induced transcription on YY1. Moreover this dependency is target specific: M-MITF depends on YY1 to upregulate the transcription of many target genes, like SLC45A2, MLANA, GPNMB, SILV, DCT and TRPM1; for other targets like TYR, although YY1 cooperates with MITF for its basal transcription (Figure 2D), knockdown of YY1 did not affect M-MITF-dependent transcriptional upregulation (Figure 4E), suggesting that M-MITF is a dominant factor in controlling their induction. The mechanistic basis for these variable dependencies will require further analysis.
To globally identify YY1 target genes in the melanocytic lineage, we performed a ChIP-seq analysis in MALME-3M melanoma cells. We obtained 15,940 peaks by using the Skellam statistic at a p-value cutoff of 10−6 (Materials and Methods, and Table S3). The overlap between recently published MITF binding sites [12] and our YY1 binding sites is 30% (Figure 4F), higher than the overlap for c-Myc, which has been previously documented to interact with YY1 [26]. Figure 4F shows that apart from some general transcription factors, MITF is the top differentiation factor whose binding sites significantly overlap with those of YY1. Gene ontology analysis found that MITF and YY1 co-regulate pathways involving mitochondria biogenesis, cytoskeleton, mitosis, as well as pigment granule and melanosome synthesis (Table S4).
We designed ChIP-qPCR primers at multiple reported MITF binding sites of the pigmentation genes [11], [12], and confirmed that YY1 co-localized with MITF on the proximal promoter of TRPM1, MLANA, GPNMB, TYR and DCT (Figure 4G). For SLC45A2 and SILV, YY1 co-localized with MITF at the 2nd nearest MITF binding sites [12] (Figure 4G). We obtained 8,899 melanocyte-specific YY1 binding sites by removing the sites that are found within 5 kb of YY1 ChIP-seq sites in GM12878, K562, and NT2D cell lines profiled by the ENCODE Consortium. We found that the nearest YY1 binding site on the KIT gene is localized in the 7th intron (peak at 56899 bp from the transcription start site). This YY1 binding site is melanocyte-specific and has an MITF binding site [12] within 50 bp, as confirmed by ChIP-qPCR (Figure 4G).
To validate the cooperativity of MITF and YY1 on a target gene promoter, we utilized a 700 bp upstream promoter region of TRPM1 fused to a firefly luciferase reporter [27]. There are three MITF consensus binding E-box sequences (E1, catgtg; E2, catgtg; E3, cacatg) and one YY1 consensus binding sequence (Y1, gccatc) within this promoter region, as shown in Figure S5. We mutated E-box sequences (mut2) or YY1 binding sequence (mut1) and found that mutation of the YY1 binding sequence modestly, but significantly reduced reporter activity by 22%, while mutation of MITF binding sequences dramatically reduced the reporter activity (Figure S5). While TRPM1 appears to be a melanocyte-specific gene (also called “melastatin” [27]), these experiments demonstrated a measurable contribution to its expression by the ubiquitous factor YY1.
Discussion
In summary, our findings reveal a critical role of YY1 in melanocytic lineage development and function, as melanocyte-specific Yy1 conditional knockout mice display complete loss of hair follicle melanocytes and premature gray hair after the first hair cycle. For a ubiquitous protein such as YY1 to gain cell type-specific functions, one of the most frequently adopted strategies is to interact with additional factors [25]. YY1 has been shown to directly interact with other ubiquitous factors, Smads, to activate cardiac development genes [28]. Here, we show that YY1 interacts with a lineage-specific transcription factor, M-MITF, to activate critical survival and pigmentation genes in melanocytic cells and that the transcriptional activity of M-MITF is dependent on YY1 for many target genes. Importantly, we demonstrate that despite its ubiquitous expression, YY1 acquires cell type-specific functions by interacting with a lineage-specific transcription factor. Since YY1 most strongly interacts with the intact b-HLH-Zip region of M-MITF (Figure 3E), which is present in other non-melanocytic isoforms of MITF, other isoforms of MITF might also interact with YY1 and potentially undergo functional effects within different lineages on the basis of similar biochemical forms of cooperativity. Novel mechanisms through which MITF and YY1 cooperate to confer survival to the melanocyte lineage are interesting subjects for future investigation.
Materials and Methods
Reagents
Antibodies used were anti-DCT (D-18), anti-YY1 (H-414) and anti-c-Kit (H-300) from Santa Cruz Biotechnology, anti-MITF (C5 mouse monoclonal and rabbit polyclonal) [22], anti-Flag (F3165, Sigma) and anti-HA (HA.11, Covance). Anti-Flag M2 agarose beads were obtained from Sigma-Aldrich.
Generation and genotyping of TyrCre, yy1f/f mice
All animal work has been conducted according to MGH and national guidelines. Male TyrCre mouse (Cre transgene is on the X chromosome) [19] was crossed with yy1f/f female mouse to obtain TyrCre, yy1f/+ female (F1). TyrCre, yy1f/+ F1 female was then crossed with yy1f/f male to obtain TyrCre, yy1f/f male (F2). Genotyping primers are listed in Table S5.
Histology and immunofluorescence staining
Mouse skin was fixed in 10% formalin after dissection and submitted to our histopathology core (Massachusetts General Hospital) for paraffin embedding, tissue section and H&E staining. For immunofluorescence staining, paraffin-embedded sections were washed twice in Xylene and then passed through 100%, 100%, 95% and 80% ethanol and H2O. Antigen retrieval was performed in EDTA buffer (5 mM Tris-HCl, 1 mM EDTA, PH 8.0) at 98°C for 20 min followed by cooling for 1 h. After PBS washing, tissue sections were blocked in 3%BSA/PBS for 1 h and then incubated with primary antibodies (1∶100 dilution) at 4°C overnight. After PBS washing three times, tissue sections were incubated with fluorescent secondary antibody (1∶1000 dilution) at room temperature for 1 h. Slides were mounted using VECTASHIELD mounting media with DAPI (Vector Laboratories). Fluorescent images were taken under Zeiss Axio Observer A1 microscope with AxioVision software.
Whole-mount LacZ staining
Skin samples from Dct-LacZ mice were washed sequentially with PBS and LacZ wash buffer (2 mM MgCl2, 0.01% sodium deoxycholate, 0.02% Nonidet P40 in PBS PH 7.4). Samples were then incubated with LacZ solution (0.5 mg/ml XGal, 5 mM potassium ferricyanide, 5 mM potassium ferrocyanide in LacZ wash buffer) at 37°C for 24 h. After PBS washing, samples were fixed with fresh 4% paraformaldehyde/PBS for 10 min at room temperature. Stained (and fixed) samples were transferred onto microscope slides and mounted in Fluoromount-G mounting media (SouthernBiotech).
Construction of YY1 and M-MITF expression plasmids
A plasmid containing cDNA encoding full-length human YY1 was obtained from the American Type Culture Collection (IMAGE clone ID, 5815774). The cDNA fragments encoding full-length or truncated YY1 were PCR amplified from IMAGE clone and inserted into p3XFLAG-CMV vector at EcoRI/BamHI sites. The cDNA fragments encoding truncated M-MITF were PCR amplified from pCDNA3-HA-hMi plasmid and inserted into pCDNA3-HA vector at EcoRI/NotI sites.
MITF and YY1 RNAi
Lentiviral shRNA vector (pLKO.1) containing MITF and YY1 RNAi sequences were obtained from the Broad Institute RNAi Consortium (Cambridge, MA). Lentivirus was generated in 293T cells 72 h post transfection. Cells were infected with lentivirus for 24 h and selected in puromycin for 3 days. The shRNA sequences are: MITFshRNA#4, CGTGGACTATATCCGAAAGTT (sense); MITFshRNA#5, CGGGAAACTTGATTGATCTTT (sense); YY1shRNA#1, GCCTCTCCTTTGTATATTATT (sense) and YY1shRNA#6, GGGAGCAGAAGCAGGTGCAGAT (sense). Synthetic siRNA oligos targeting YY1 (siGENOME SMARTpool) and non-targeting control oligos were obtained from Thermo Fisher Scientific. Cells were transfected with siRNA using lipidoid reagent C12-113 and assayed 72 h post transfection.
Lipidoid synthesis
Lipidoid delivery agent C12-113 was synthesized and characterized as previously described [29]. Briefly, 3 equivalents of 1,2-epoxydodecane was combined with 1 equivalent of 2,2′-diamino-N-methyldiethylamine (TCI America) in a glass scintillation vial. Reaction was stirred for 3 days at 90°C. Following synthesis, reaction mixture was characterized by MALDI-TOF mass spectroscopy to confirm mass of expected products. Synthesized material is used in in vitro biological assays without further purification. Lipidoid was dissolved in 25 mM NaOAc buffer (pH∼5.2) and added to solution of siRNA for complexation.
Real-time quantitative PCR
The total volume of each reaction was 25 µl, including 12.5 µl 2× SYBR Green master mix (Bio-Rad), 0.25 µl reverse transcriptase (Qiagen), 1 µl of each primer (10 µM stock) and 100 ng of total RNA. Reverse transcription was carried out at 48°C for 30 min. Then 40 cycles of PCR reaction were carried out at 95°C for 15 s and 60°C for 30 s using 7500 Fast Real Time PCR system (Applied Biosystems). Data were acquired and analyzed with 7500 Fast System SDS software. qPCR primer sequences are listed in Table S5.
Chromatin immunoprecipitation (ChIP)
Cells were fixed by adding formaldehyde to the culture media to a final concentration of 1% and incubated for 20 min at room temperature. Cells were harvested by scraping with ice cold PBS containing protease inhibitor (Roche). Cell pellets were re-suspended in SDS lysis buffer (1%SDS, 10 mM EDTA, 50 mM Tris-HCl, pH 8.0), incubated for 10 min on ice, and then sonicated to reduce DNA length to ∼500 base pairs. Samples were centrifuged to remove debris and then diluted 10 fold in IP buffer (0.01% SDS, 1.1% TritonX-100, 1.2 mM EDTA, 16.7 mM Tris-HCl, pH 8.0, 167 mM NaCl and protease inhibitors). To reduce nonspecific background, chromatin solution was pre-cleared with 80 µl of 50% protein A/G slurry containing 0.25 mg/ml sonicated salmon sperm DNA and 1 mg/ml BSA in TE (10 mM Tris, 1 mM EDTA, pH 8.0) for 1 h at 4°C. Antibodies were then added to pre-cleared chromatin solution and incubated overnight at 4°C. Protein A/G slurry (as used in the pre-clear step) were added to the sample and incubated for another 1 h at 4°C. The immuno-complexes were washed sequentially with buffer I (0.1% SDS, 1% TritonX-100, 2 mM EDTA, 20 mM Tris-HCl, pH 8.0, 150 mM NaCl), buffer II (0.1% SDS, 1% TritonX-100, 2 mM EDTA, 20 mM Tris-HCl, pH 8.0, 500 mM NaCl), buffer III (0.25M LiCl, 1% NP40, 1% sodium deoxycholate, 1 mM EDTA, 10 mM Tris-HCl, pH 8.0) and twice with TE. The immuno-complexes were eluted from beads with 1% SDS in 0.1M NaHCO3 twice for 15 min at room temperature. Crosslinks were reversed overnight at 65°C. Protein was digested with proteinase K for 1 h at 55°C. DNA was purified with QIAquick PCR purification kit (Qiagen). qPCR primers are listed in Table S5.
Adenovirus infection
Adenoviruses encoding wild-type M-MITF or GFP were generated as previously described [27]. 2×105 cells were plated in 6-well plates for 24 h. Cells were infected overnight with concentrated adenoviruses in complete media at MOI 500 for MALME-3M cells or MOI 100 for UACC62 cells and HFM. Media was changed the next day and total RNA was harvest with RNeasy plus mini kit (Qiagen) 48 h post infection.
Expression profiling
mRNA expression in the melanoma cell lines MALME-3M and UACC62 were profiled using Affymetrix U133P2 microarrays. Each cell line had four samples, corresponding to two experiments treated with two independent shRNA constructs against MITF and two controls transfected with shRNA against luciferase. We also obtained two additional paired samples before and after siMITF (GSE16249, Gene Expression Omnibus). The data were normalized using RMA and the latest RefSeq probe mapping to the reference human genome [30], [31]. For each gene g, let xM1,g and xM2,g denote the fold-changes in shMITF-treated MALME-3M relative to the average control expression values in MALME-3M. Similarly denote the fold-changes in UACC62 and GSE16249 as xU1,g, xU2,g and xG1,g, xG2,g, respectively. In order to assess the significance of differential expression after knocking down MITF, we pooled together these 6 fold-changes for all genes and ranked them in an increasing order, and we then computed the minimum p-value for the rank distribution of each gene's fold-changes by using the hypergeometric test, as was done in the Redundant siRNA Activity (RSA) analysis [32]. The p-values were adjusted for multiple hypothesis testing and turned into q-values, as described in [33]. At a q-value cutoff of 10−2, 1241 RefSeq genes showed significantly reduced expression after MITF knockdown. A similar RSA analysis was performed for YY1 shRNA.
Motif analysis
We found 814 open chromatin regions within the 1241 MITF-responsive RefSeq promoters in melanocytes, as recently mapped by the ENCODE Consortium. We also generated 814 matching random regions in the human genome. We then scanned the open chromatin and random regions with TRANSFAC and JASPAR position-specific scoring matrices (PSSM); the cutoff PSSM scores were chosen separately for each motif to minimize the binomial p-value for the over-representation or under-representation of each motif in the open chromatin regions of MITF-responsive promoters relative to random sequences.
YY1 ChIP–seq
YY1 chromatin-immunoprecipitated DNA was sequenced by using Illumina Genome Analyzer II, yielding ∼15 million mappable reads. As a control, ∼9 million DNA fragments from randomly sonicated chromatin were also sequenced. Peaks were detected by using the Skellam distribution as a null model, similar to the Poisson null model used by MACS [34]. We used a p-value cutoff of 10−6; see Text S1 for further discussion. Regions containing satellites and rRNAs (ENCODE Duke Excluded Regions) were filtered out. Normalization of ChIP-seq data is described in detail in Text S1.
Pathway analysis
We considered the genes down-regulated by both MITF and YY1 KD at a q-value cutoff of 0.01 and also bound by YY1 within 5 kb of transcriptional start site or in intron (Table S1). Gene ontology of those genes was analyzed by using DAVID (http://david.abcc.ncifcrf.gov).
GSK cancer cell line expression analysis
954 CEL files were normalized together by using RMA [30] and the RefSeq CDF [31]. Replicate samples were averaged to yield summary expression levels for each cell line. Cell lines were then grouped into cancer subtypes, and the t-test was applied between each cancer subtype and the rest.
Supporting Information
Zdroje
1. DessiniotiCStratigosAJRigopoulosDKatsambasAD 2009 A review of genetic disorders of hypopigmentation: lessons learned from the biology of melanocytes. Exp Dermatol 18 741 749
2. PingaultVEnteDDastot-Le MoalFGoossensMMarlinS 2010 Review and update of mutations causing Waardenburg syndrome. Hum Mutat 31 391 406
3. ThomasIKihiczakGGFoxMDJannigerCKSchwartzRA 2004 Piebaldism: an update. Int J Dermatol 43 716 719
4. SmithSDKelleyPMKenyonJBHooverD 2000 Tietz syndrome (hypopigmentation/deafness) caused by mutation of MITF. J Med Genet 37 446 448
5. HughesAENewtonVELiuXZReadAP 1994 A gene for Waardenburg syndrome type 2 maps close to the human homologue of the microphthalmia gene at chromosome 3p12–p14.1. Nat Genet 7 509 512
6. TassabehjiMNewtonVEReadAP 1994 Waardenburg syndrome type 2 caused by mutations in the human microphthalmia (MITF) gene. Nat Genet 8 251 255
7. TsujimuraTMoriiENozakiMHashimotoKMoriyamaY 1996 Involvement of transcription factor encoded by the mi locus in the expression of c-kit receptor tyrosine kinase in cultured mast cells of mice. Blood 88 1225 1233
8. GiebelLBSpritzRA 1991 Mutation of the KIT (mast/stem cell growth factor receptor) protooncogene in human piebaldism. Proc Natl Acad Sci U S A 88 8696 8699
9. FleischmanRASaltmanDLStastnyVZneimerS 1991 Deletion of the c-kit protooncogene in the human developmental defect piebald trait. Proc Natl Acad Sci U S A 88 10885 10889
10. LevyCKhaledMFisherDE 2006 MITF: master regulator of melanocyte development and melanoma oncogene. Trends Mol Med 12 406 414
11. HoekKSSchlegelNCEichhoffOMWidmerDSPraetoriusC 2008 Novel MITF targets identified using a two-step DNA microarray strategy. Pigment Cell Melanoma Res 21 665 676
12. StrubTGiulianoSYeTBonetCKeimeC 2011 Essential role of microphthalmia transcription factor for DNA replication, mitosis and genomic stability in melanoma. Oncogene
13. DengZCaoPWanMSuiG 2010 Yin Yang 1: A multifaceted protein beyond a transcription factor. Transcr 1 81 84
14. DonohoeMEZhangXMcGinnisLBiggersJLiE 1999 Targeted disruption of mouse Yin Yang 1 transcription factor results in peri-implantation lethality. Mol Cell Biol 19 7237 7244
15. LiuHSchmidt-SupprianMShiYHobeikaEBartenevaN 2007 Yin Yang 1 is a critical regulator of B-cell development. Genes Dev 21 1179 1189
16. HeYDupreeJWangJSandovalJLiJ 2007 The transcription factor Yin Yang 1 is essential for oligodendrocyte progenitor differentiation. Neuron 55 217 230
17. WuSHuYCLiuHShiY 2009 Loss of YY1 impacts the heterochromatic state and meiotic double-strand breaks during mouse spermatogenesis. Mol Cell Biol 29 6245 6256
18. Affar elBGayFShiYLiuHHuarteM 2006 Essential dosage-dependent functions of the transcription factor yin yang 1 in late embryonic development and cell cycle progression. Mol Cell Biol 26 3565 3581
19. DelmasVMartinozziSBourgeoisYHolzenbergerMLarueL 2003 Cre-mediated recombination in the skin melanocyte lineage. Genesis 36 73 80
20. MackenzieMAJordanSABuddPSJacksonIJ 1997 Activation of the receptor tyrosine kinase Kit is required for the proliferation of melanoblasts in the mouse embryo. Dev Biol 192 99 107
21. YasumotoKTakedaKSaitoHWatanabeKTakahashiK 2002 Microphthalmia-associated transcription factor interacts with LEF-1, a mediator of Wnt signaling. EMBO J 21 2703 2714
22. DuJWidlundHRHorstmannMARamaswamySRossK 2004 Critical role of CDK2 for melanoma growth linked to its melanocyte-specific transcriptional regulation by MITF. Cancer Cell 6 565 576
23. McGillGGHorstmannMWidlundHRDuJMotyckovaG 2002 Bcl2 regulation by the melanocyte master regulator Mitf modulates lineage survival and melanoma cell viability. Cell 109 707 718
24. GiangrandePHHallstromTCTunyaplinCCalameKNevinsJR 2003 Identification of E-box factor TFE3 as a functional partner for the E2F3 transcription factor. Mol Cell Biol 23 3707 3720
25. HeYCasaccia-BonnefilP 2008 The Yin and Yang of YY1 in the nervous system. J Neurochem 106 1493 1502
26. ShrivastavaAYuJArtandiSCalameK 1996 YY1 and c-Myc associate in vivo in a manner that depends on c-Myc levels. Proc Natl Acad Sci U S A 93 10638 10641
27. MillerAJDuJRowanSHersheyCLWidlundHR 2004 Transcriptional regulation of the melanoma prognostic marker melastatin (TRPM1) by MITF in melanocytes and melanoma. Cancer Res 64 509 516
28. LeeKHEvansSRuanTYLassarAB 2004 SMAD-mediated modulation of YY1 activity regulates the BMP response and cardiac-specific expression of a GATA4/5/6-dependent chick Nkx2.5 enhancer. Development 131 4709 4723
29. LoveKTMahonKPLevinsCGWhiteheadKAQuerbesW 2010 Lipid-like materials for low-dose, in vivo gene silencing. Proc Natl Acad Sci U S A 107 1864 1869
30. IrizarryRAHobbsBCollinFBeazer-BarclayYDAntonellisKJ 2003 Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4 249 264
31. DaiMWangPBoydADKostovGAtheyB 2005 Evolving gene/transcript definitions significantly alter the interpretation of GeneChip data. Nucleic Acids Res 33 e175
32. KonigRChiangCYTuBPYanSFDeJesusPD 2007 A probability-based approach for the analysis of large-scale RNAi screens. Nat Methods 4 847 849
33. StoreyJD 2002 A direct approach to false discovery rates. Journal of the Royal Statistical Society Series B-Statistical Methodology 64 479 498
34. ZhangYLiuTMeyerCAEeckhouteJJohnsonDS 2008 Model-based analysis of ChIP-Seq (MACS). Genome Biol 9 R137
Štítky
Genetika Reprodukční medicína
Článek Functional Centromeres Determine the Activation Time of Pericentric Origins of DNA Replication inČlánek Dynamic Deposition of Histone Variant H3.3 Accompanies Developmental Remodeling of the TranscriptomeČlánek Integrin α PAT-2/CDC-42 Signaling Is Required for Muscle-Mediated Clearance of Apoptotic Cells inČlánek Prdm5 Regulates Collagen Gene Transcription by Association with RNA Polymerase II in Developing BoneČlánek Acquisition Order of Ras and p53 Gene Alterations Defines Distinct Adrenocortical Tumor Phenotypes
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2012 Číslo 5
-
Všechny články tohoto čísla
- Slowing Replication in Preparation for Reduction
- Chromosome Pairing: A Hidden Treasure No More
- Loss of Imprinting Differentially Affects REM/NREM Sleep and Cognition in Mice
- Six Novel Susceptibility Loci for Early-Onset Androgenetic Alopecia and Their Unexpected Association with Common Diseases
- Regulation by the Noncoding RNA
- UDP-Galactose 4′-Epimerase Activities toward UDP-Gal and UDP-GalNAc Play Different Roles in the Development of
- Deletion of PTH Rescues Skeletal Abnormalities and High Osteopontin Levels in Mice
- Karyotypic Determinants of Chromosome Instability in Aneuploid Budding Yeast
- Genome-Wide Copy Number Analysis Uncovers a New HSCR Gene:
- MicroRNA-277 Modulates the Neurodegeneration Caused by Fragile X Premutation rCGG Repeats
- Functional Centromeres Determine the Activation Time of Pericentric Origins of DNA Replication in
- Dynamic Deposition of Histone Variant H3.3 Accompanies Developmental Remodeling of the Transcriptome
- Scientist Citizen: An Interview with Bruce Alberts
- YY1 Regulates Melanocyte Development and Function by Cooperating with MITF
- Congenital Heart Disease–Causing Gata4 Mutation Displays Functional Deficits
- Recombination Drives Vertebrate Genome Contraction
- KATNAL1 Regulation of Sertoli Cell Microtubule Dynamics Is Essential for Spermiogenesis and Male Fertility
- Re-Patterning Sleep Architecture in through Gustatory Perception and Nutritional Quality
- Using Whole-Genome Sequence Data to Predict Quantitative Trait Phenotypes in
- Genome-Wide Analysis of GLD-1–Mediated mRNA Regulation Suggests a Role in mRNA Storage
- Meiotic Chromosome Pairing Is Promoted by Telomere-Led Chromosome Movements Independent of Bouquet Formation
- LINT, a Novel dL(3)mbt-Containing Complex, Represses Malignant Brain Tumour Signature Genes
- The H3K27 Demethylase UTX-1 Is Essential for Normal Development, Independent of Its Enzymatic Activity
- Suppresses Senescence Programs and Thereby Accelerates and Maintains Mutant -Induced Lung Tumorigenesis
- Genome-Wide Association of Pericardial Fat Identifies a Unique Locus for Ectopic Fat
- An Essential Role for Katanin p80 and Microtubule Severing in Male Gamete Production
- Identification of Genes That Promote or Antagonize Somatic Homolog Pairing Using a High-Throughput FISH–Based Screen
- Principles of Carbon Catabolite Repression in the Rice Blast Fungus: Tps1, Nmr1-3, and a MATE–Family Pump Regulate Glucose Metabolism during Infection
- Integrin α PAT-2/CDC-42 Signaling Is Required for Muscle-Mediated Clearance of Apoptotic Cells in
- Histone H3 Localizes to the Centromeric DNA in Budding Yeast
- Collapse of Telomere Homeostasis in Hematopoietic Cells Caused by Heterozygous Mutations in Telomerase Genes
- Hypersensitive to Red and Blue 1 and Its Modification by Protein Phosphatase 7 Are Implicated in the Control of Arabidopsis Stomatal Aperture
- Extent, Causes, and Consequences of Small RNA Expression Variation in Human Adipose Tissue
- TBC-8, a Putative RAB-2 GAP, Regulates Dense Core Vesicle Maturation in
- Regulating Repression: Roles for the Sir4 N-Terminus in Linker DNA Protection and Stabilization of Epigenetic States
- Common Genetic Determinants of Intraocular Pressure and Primary Open-Angle Glaucoma
- Prdm5 Regulates Collagen Gene Transcription by Association with RNA Polymerase II in Developing Bone
- Fitness Landscape Transformation through a Single Amino Acid Change in the Rho Terminator
- Repeated, Selection-Driven Genome Reduction of Accessory Genes in Experimental Populations
- Allelic Variation and Differential Expression of the mSIN3A Histone Deacetylase Complex Gene Promote Mammary Tumor Growth and Metastasis
- DNA Demethylation and USF Regulate the Meiosis-Specific Expression of the Mouse
- Knowledge-Driven Analysis Identifies a Gene–Gene Interaction Affecting High-Density Lipoprotein Cholesterol Levels in Multi-Ethnic Populations
- A Duplication CNV That Conveys Traits Reciprocal to Metabolic Syndrome and Protects against Diet-Induced Obesity in Mice and Men
- EMT Inducers Catalyze Malignant Transformation of Mammary Epithelial Cells and Drive Tumorigenesis towards Claudin-Low Tumors in Transgenic Mice
- Inactivation of a Novel FGF23 Regulator, FAM20C, Leads to Hypophosphatemic Rickets in Mice
- Genome-Wide Association for Abdominal Subcutaneous and Visceral Adipose Reveals a Novel Locus for Visceral Fat in Women
- Stratifying Type 2 Diabetes Cases by BMI Identifies Genetic Risk Variants in and Enrichment for Risk Variants in Lean Compared to Obese Cases
- New Insight into the History of Domesticated Apple: Secondary Contribution of the European Wild Apple to the Genome of Cultivated Varieties
- Activated Cdc42 Kinase Has an Anti-Apoptotic Function
- The Region Is Critical for Birth Defects and Electrocardiographic Dysfunctions Observed in a Down Syndrome Mouse Model
- COP9 Signalosome Integrity Plays Major Roles for Hyphal Growth, Conidial Development, and Circadian Function
- Bmps and Id2a Act Upstream of Twist1 To Restrict Ectomesenchyme Potential of the Cranial Neural Crest
- Psip1/Ledgf p52 Binds Methylated Histone H3K36 and Splicing Factors and Contributes to the Regulation of Alternative Splicing
- The Number of X Chromosomes Causes Sex Differences in Adiposity in Mice
- Target Gene Analysis by Microarrays and Chromatin Immunoprecipitation Identifies HEY Proteins as Highly Redundant bHLH Repressors
- Acquisition Order of Ras and p53 Gene Alterations Defines Distinct Adrenocortical Tumor Phenotypes
- ELK1 Uses Different DNA Binding Modes to Regulate Functionally Distinct Classes of Target Genes
- Histone H1 Depletion Impairs Embryonic Stem Cell Differentiation
- IDN2 and Its Paralogs Form a Complex Required for RNA–Directed DNA Methylation
- Separation of DNA Replication from the Assembly of Break-Competent Meiotic Chromosomes
- Genomic Hypomethylation in the Human Germline Associates with Selective Structural Mutability in the Human Genome
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Inactivation of a Novel FGF23 Regulator, FAM20C, Leads to Hypophosphatemic Rickets in Mice
- Genome-Wide Association of Pericardial Fat Identifies a Unique Locus for Ectopic Fat
- Slowing Replication in Preparation for Reduction
- An Essential Role for Katanin p80 and Microtubule Severing in Male Gamete Production
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



