-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaSlowing Replication in Preparation for Reduction
article has not abstract
Published in the journal: . PLoS Genet 8(5): e32767. doi:10.1371/journal.pgen.1002715
Category: Perspective
doi: https://doi.org/10.1371/journal.pgen.1002715Summary
article has not abstract
Meiosis reduces the ploidy of the genome to generate haploid gametes for sexual reproduction. As gametes are portals for the generational transfer of genetic material, it is imperative that the genome is copied accurately and that chromosomes segregate equally into each haploid gamete. Proper chromosome segregation requires the formation of specialized chromosome axes to create and maintain an environment competent for double-strand break (DSB) formation and homologous recombination. Although the fundamental copying mechanism appears to be identical in mitosis and meiosis, the S phase that precedes meiosis (meiS) is at least twice as long as mitotic S phase (mitS) [1]–[4]. The underlying basis for an extended S phase prior to meiosis has, until now, been mysterious. While it is postulated that meiS length contributes to the dramatic chromosome reorganization that occurs during meiotic prophase (Figure 1), there is conflicting data concerning the interdependencies of meiS, chromosome morphogenesis, and DSB formation [5]–[9]. In this issue of PLoS Genetics, Blitzblau et al. [10] use innovative genome-wide approaches in yeast to elucidate mechanisms underlying meiS length and provide insight into the relationship between DNA replication and meiotic prophase events.
Fig. 1. Meiotic DNA replication, chromosome axes, and DSB formation. 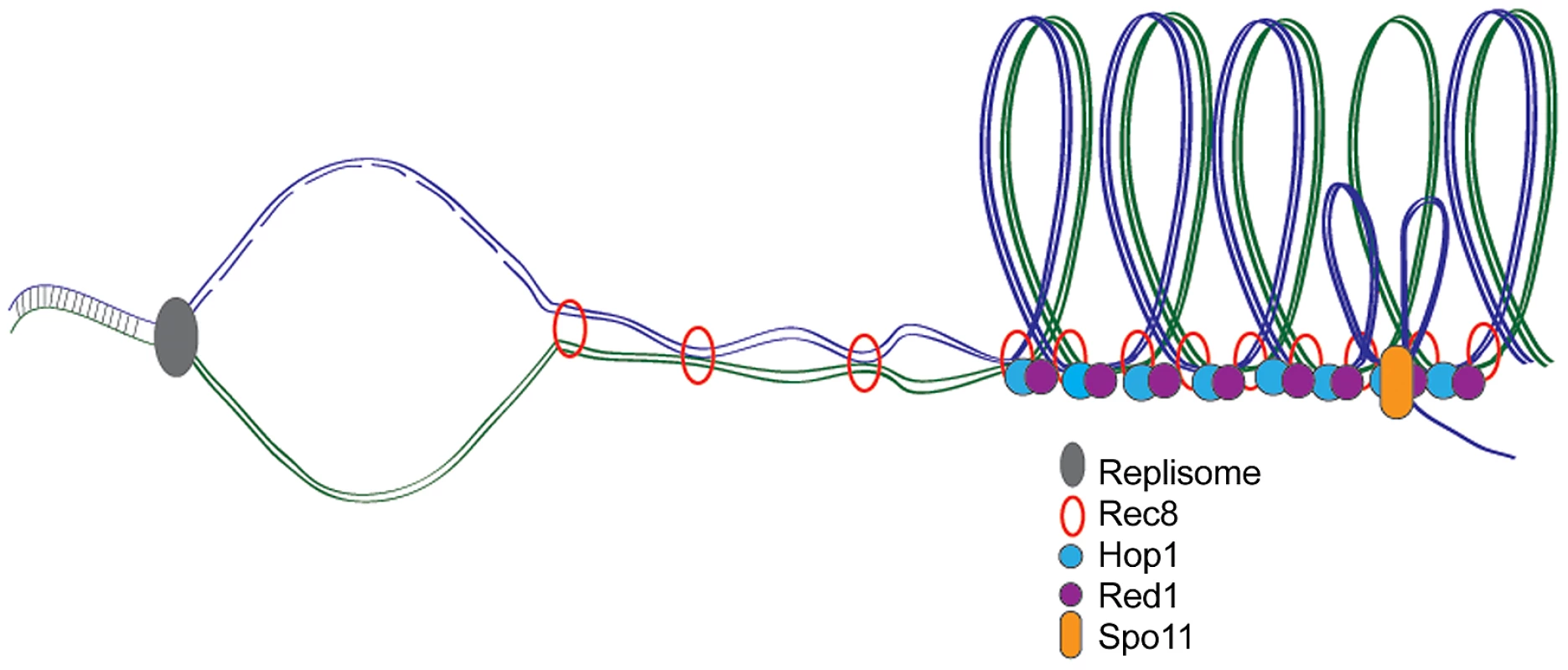
During meiosis, DNA is replicated and meiosis-specific cohesin Rec8 holds sister chromatids together while axis proteins Red1 and Hop1 associate to form the loop axis structure. Endonuclease Spo11 creates DSBs required for homologous recombination and crossover formation at the axis where single-stranded DNA is exposed to facilitate homology search. Delayed Origin Firing Slows Meiotic S Phase
To compare mitS and meiS, Blitzblau et al. performed genome-wide chromatin immunoprecipitation (ChIP) to identify Mcm replicative helicase binding sites in conjunction with microarray analysis to monitor actively replicating origins. Importantly, synchrony was achieved by taking advantage of an ATP analog–sensitive mutant of Ime2, a kinase essential for DNA replication and meiotic entry, allowing for conditional inactivation. The authors found that Mcm bound to a significant fraction of the same origins in both mitosis and meiosis; the few that differed were located near cycle-specific, actively transcribed genes, consistent with studies suggesting competition between replication and transcription machinery [11]. Thus, the small differences in Mcm occupancy are unlikely to account for the timing differences between mitS and meiS. In contrast, while origins fired in the same relative order, as reported previously for a single chromosome [12], replication initiation at a significant fraction of origins was delayed in meiS compared to mitS.
Blitzblau et al. identified early meiS replication sites by analyzing replication in the presence of the ribonucleotide reductase inhibitor hydroxyurea (HU), which depletes nucleotide pools and prevents late origins from firing [13]. While all early replication meiS sites were shared in mitS, only 38% of early mitS sites initiated replication in meiS, consistent with the delay in origin firing and extended meiS length.
In the course of these experiments, the authors found that meiS is more sensitive to HU and has more robust checkpoint signaling than mitS. To explore whether this is a consequence of reduced nucleotide pools, Blitzblau et al. treated meiotic cells with HU in the absence of ribonucleotide reductase inhibitor SML1 [14]. The number of early firing origins increased, but not to the level utilized in mitotic cells, suggesting nucleotide depletion contributes to delayed origin firing and meiS timing.
MeiS, Chromosomal Axis Formation, and DSB Competency Can Be Uncoupled
It has been proposed that meiS is slowed to facilitate the elaboration of meiosis-specific chromosomal structures and DSB formation (Figure 1) [5]. Blitzblau et al. probed the relationship between axis and DSB formation and meiS timing by examining early origin firing in the presence of HU in axis mutants (rec8Δ) or in cells that do not induce DSBs (spo11Δ). No significant differences between the replication profiles were observed, suggesting that loading of meiosis-specific proteins and break formation do not regulate meiotic replication timing.
To test whether DNA synthesis, in turn, affected axis and DSB formation, the authors examined the association of meiosis-specific axis components (Red1 and Hop1) following replication arrest by HU, depletion of Mcm loading factor Cdc6, or in the absence of B-type cyclins. In all situations, axis proteins were loaded onto chromosomes, suggesting replication is not an absolute prerequisite for axis formation. Blitzblau et al. also found that the cdc6 mutant was competent for DSB formation, indicating that breaks can occur on unreplicated chromosomes. This is in contrast to a previous study that found that delaying replication delayed break formation [8]. The discrepancy is most likely due to shared regulatory components between DNA replication and DSB formation that are disrupted in cdc6 mutants.
Slow S, Replication Fidelity, and Metazoans
The current study provides strong evidence that in S. cerevisiae there is reduced replication capacity during meiS, at least in part due to limiting nucleotide pools. This manifests as delayed firing of a significant fraction of origins and extended S phase; a similar pattern of origin firing has been observed in S. pombe [2]. In both of these organisms, meiosis is initiated by starvation conditions, which presumably alters the activity of cell cycle components as well as decreases nucleotide pools. The applicability of this finding to metazoans, where meiS is also extended [1], [3] is unknown as there is no evidence that germ cells in multicellular organisms experience limiting nutrients. The authors suggest that extended S phase increases replication fidelity; however, in metazoans, germ cells can undergo multiple rounds of DNA replication prior to meiotic entry; these replications must also occur accurately. While no direct comparison of error rate between mitS and meiS has been performed, DNA polymerase mutants that have an inherently lower misincorporation frequency also have reduced processivity [15], suggesting that slowed replication could increase fidelity. However, this would manifest in reduced fork rates, something not directly addressed in this study. Perhaps the enhanced checkpoint signaling observed in meiS reflects more robust surveillance mechanisms that promote fidelity irrespective of nucleotide pools. Future work examining nucleotide pools, replication fidelity, and checkpoints may shed light on the significance of extended meiotic S phase in both single-celled and multicellular organisms.
While the authors conclusively demonstrate that chromosomal axis and DSB formation can occur in the absence of DNA replication and do not directly impinge on replication timing, these processes are nonetheless linked and occur successively in wild-type cells (Figure 1). Thus there is still much to be learned about how DNA replication is modified in meiosis to ensure the transfer of genetic material from one generation to the next.
Zdroje
1. Jaramillo-LambertAEllefsonMVilleneuveAMEngebrechtJ 2007 Differential timing of S phases, X chromosome replication, and meiotic prophase in the C. elegans germ line. Dev Biol 308 206 221
2. HeichingerCPenkettCJBahlerJNurseP 2006 Genome-wide characterization of fission yeast DNA replication origins. EMBO J 25 5171 5179
3. HottaYSternH 1971 Analysis of DNA synthesis during meiotic prophase in Lilium. J Mol Biol 55 337 355
4. WilliamsonDHJohnstonLHFennellDJSimchenG 1983 The timing of the S phase and other nuclear events in yeast meiosis. Exp Cell Res 145 209 217
5. ChaRSWeinerBMKeeneySDekkerJKlecknerN 2000 Progression of meiotic DNA replication is modulated by interchromosomal interaction proteins, negatively by Spo11p and positively by Rec8p. Genes Dev 14 493 503
6. DollEMolnarMCuanoudGOctobreGLatypovV 2008 Cohesin and recombination proteins influence the G1-to-S transition in azygotic meiosis in Schizosaccharomyces pombe. Genetics 180 727 740
7. MaloneREHaringSJForemanKEPansegrauMLSmithSM 2004 The signal from the initiation of meiotic recombination to the first division of meiosis. Eukaryot Cell 3 598 609
8. BordeVGoldmanASLichtenM 2000 Direct coupling between meiotic DNA replication and recombination initiation. Science 290 806 809
9. MurakamiHNurseP 2001 Regulation of premeiotic S phase and recombination-related double-strand DNA breaks during meiosis in fission yeast. Nat Genet 28 290 293
10. BlitzblauHGChanCSHochwagenABellSP 2012 Separation of DNA replication from the assembly of break-competent meiotic chromosomes. PLoS Genet 6 e1002643 doi:10.1371/journal.pgen.1002643
11. SnyderMSapolskyRJDavisRW 1988 Transcription interferes with elements important for chromosome maintenance in Saccharomyces cerevisiae. Mol Cell Biol 8 2184 2194
12. CollinsINewlonCS 1994 Chromosomal DNA replication initiates at the same origins in meiosis and mitosis. Mol Cell Biol 14 3524 3534
13. YabukiNTerashimaHKitadaK 2002 Mapping of early firing origins on a replication profile of budding yeast. Genes Cells 7 781 789
14. ZhaoXMullerEGRothsteinR 1998 A suppressor of two essential checkpoint genes identifies a novel protein that negatively affects dNTP pools. Mol Cell 2 329 340
15. HerrAJWilliamsLNPrestonBD 2011 Antimutator variants of DNA polymerases. Crit Rev Biochem Mol Biol 46 548 570
Štítky
Genetika Reprodukční medicína
Článek Functional Centromeres Determine the Activation Time of Pericentric Origins of DNA Replication inČlánek Dynamic Deposition of Histone Variant H3.3 Accompanies Developmental Remodeling of the TranscriptomeČlánek Integrin α PAT-2/CDC-42 Signaling Is Required for Muscle-Mediated Clearance of Apoptotic Cells inČlánek Prdm5 Regulates Collagen Gene Transcription by Association with RNA Polymerase II in Developing BoneČlánek Acquisition Order of Ras and p53 Gene Alterations Defines Distinct Adrenocortical Tumor Phenotypes
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2012 Číslo 5
-
Všechny články tohoto čísla
- Slowing Replication in Preparation for Reduction
- Chromosome Pairing: A Hidden Treasure No More
- Loss of Imprinting Differentially Affects REM/NREM Sleep and Cognition in Mice
- Six Novel Susceptibility Loci for Early-Onset Androgenetic Alopecia and Their Unexpected Association with Common Diseases
- Regulation by the Noncoding RNA
- UDP-Galactose 4′-Epimerase Activities toward UDP-Gal and UDP-GalNAc Play Different Roles in the Development of
- Deletion of PTH Rescues Skeletal Abnormalities and High Osteopontin Levels in Mice
- Karyotypic Determinants of Chromosome Instability in Aneuploid Budding Yeast
- Genome-Wide Copy Number Analysis Uncovers a New HSCR Gene:
- MicroRNA-277 Modulates the Neurodegeneration Caused by Fragile X Premutation rCGG Repeats
- Functional Centromeres Determine the Activation Time of Pericentric Origins of DNA Replication in
- Dynamic Deposition of Histone Variant H3.3 Accompanies Developmental Remodeling of the Transcriptome
- Scientist Citizen: An Interview with Bruce Alberts
- YY1 Regulates Melanocyte Development and Function by Cooperating with MITF
- Congenital Heart Disease–Causing Gata4 Mutation Displays Functional Deficits
- Recombination Drives Vertebrate Genome Contraction
- KATNAL1 Regulation of Sertoli Cell Microtubule Dynamics Is Essential for Spermiogenesis and Male Fertility
- Re-Patterning Sleep Architecture in through Gustatory Perception and Nutritional Quality
- Using Whole-Genome Sequence Data to Predict Quantitative Trait Phenotypes in
- Genome-Wide Analysis of GLD-1–Mediated mRNA Regulation Suggests a Role in mRNA Storage
- Meiotic Chromosome Pairing Is Promoted by Telomere-Led Chromosome Movements Independent of Bouquet Formation
- LINT, a Novel dL(3)mbt-Containing Complex, Represses Malignant Brain Tumour Signature Genes
- The H3K27 Demethylase UTX-1 Is Essential for Normal Development, Independent of Its Enzymatic Activity
- Suppresses Senescence Programs and Thereby Accelerates and Maintains Mutant -Induced Lung Tumorigenesis
- Genome-Wide Association of Pericardial Fat Identifies a Unique Locus for Ectopic Fat
- An Essential Role for Katanin p80 and Microtubule Severing in Male Gamete Production
- Identification of Genes That Promote or Antagonize Somatic Homolog Pairing Using a High-Throughput FISH–Based Screen
- Principles of Carbon Catabolite Repression in the Rice Blast Fungus: Tps1, Nmr1-3, and a MATE–Family Pump Regulate Glucose Metabolism during Infection
- Integrin α PAT-2/CDC-42 Signaling Is Required for Muscle-Mediated Clearance of Apoptotic Cells in
- Histone H3 Localizes to the Centromeric DNA in Budding Yeast
- Collapse of Telomere Homeostasis in Hematopoietic Cells Caused by Heterozygous Mutations in Telomerase Genes
- Hypersensitive to Red and Blue 1 and Its Modification by Protein Phosphatase 7 Are Implicated in the Control of Arabidopsis Stomatal Aperture
- Extent, Causes, and Consequences of Small RNA Expression Variation in Human Adipose Tissue
- TBC-8, a Putative RAB-2 GAP, Regulates Dense Core Vesicle Maturation in
- Regulating Repression: Roles for the Sir4 N-Terminus in Linker DNA Protection and Stabilization of Epigenetic States
- Common Genetic Determinants of Intraocular Pressure and Primary Open-Angle Glaucoma
- Prdm5 Regulates Collagen Gene Transcription by Association with RNA Polymerase II in Developing Bone
- Fitness Landscape Transformation through a Single Amino Acid Change in the Rho Terminator
- Repeated, Selection-Driven Genome Reduction of Accessory Genes in Experimental Populations
- Allelic Variation and Differential Expression of the mSIN3A Histone Deacetylase Complex Gene Promote Mammary Tumor Growth and Metastasis
- DNA Demethylation and USF Regulate the Meiosis-Specific Expression of the Mouse
- Knowledge-Driven Analysis Identifies a Gene–Gene Interaction Affecting High-Density Lipoprotein Cholesterol Levels in Multi-Ethnic Populations
- A Duplication CNV That Conveys Traits Reciprocal to Metabolic Syndrome and Protects against Diet-Induced Obesity in Mice and Men
- EMT Inducers Catalyze Malignant Transformation of Mammary Epithelial Cells and Drive Tumorigenesis towards Claudin-Low Tumors in Transgenic Mice
- Inactivation of a Novel FGF23 Regulator, FAM20C, Leads to Hypophosphatemic Rickets in Mice
- Genome-Wide Association for Abdominal Subcutaneous and Visceral Adipose Reveals a Novel Locus for Visceral Fat in Women
- Stratifying Type 2 Diabetes Cases by BMI Identifies Genetic Risk Variants in and Enrichment for Risk Variants in Lean Compared to Obese Cases
- New Insight into the History of Domesticated Apple: Secondary Contribution of the European Wild Apple to the Genome of Cultivated Varieties
- Activated Cdc42 Kinase Has an Anti-Apoptotic Function
- The Region Is Critical for Birth Defects and Electrocardiographic Dysfunctions Observed in a Down Syndrome Mouse Model
- COP9 Signalosome Integrity Plays Major Roles for Hyphal Growth, Conidial Development, and Circadian Function
- Bmps and Id2a Act Upstream of Twist1 To Restrict Ectomesenchyme Potential of the Cranial Neural Crest
- Psip1/Ledgf p52 Binds Methylated Histone H3K36 and Splicing Factors and Contributes to the Regulation of Alternative Splicing
- The Number of X Chromosomes Causes Sex Differences in Adiposity in Mice
- Target Gene Analysis by Microarrays and Chromatin Immunoprecipitation Identifies HEY Proteins as Highly Redundant bHLH Repressors
- Acquisition Order of Ras and p53 Gene Alterations Defines Distinct Adrenocortical Tumor Phenotypes
- ELK1 Uses Different DNA Binding Modes to Regulate Functionally Distinct Classes of Target Genes
- Histone H1 Depletion Impairs Embryonic Stem Cell Differentiation
- IDN2 and Its Paralogs Form a Complex Required for RNA–Directed DNA Methylation
- Separation of DNA Replication from the Assembly of Break-Competent Meiotic Chromosomes
- Genomic Hypomethylation in the Human Germline Associates with Selective Structural Mutability in the Human Genome
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Inactivation of a Novel FGF23 Regulator, FAM20C, Leads to Hypophosphatemic Rickets in Mice
- Genome-Wide Association of Pericardial Fat Identifies a Unique Locus for Ectopic Fat
- Slowing Replication in Preparation for Reduction
- An Essential Role for Katanin p80 and Microtubule Severing in Male Gamete Production
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



