-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaUDP-Galactose 4′-Epimerase Activities toward UDP-Gal and UDP-GalNAc Play Different Roles in the Development of
In both humans and Drosophila melanogaster, UDP-galactose 4′-epimerase (GALE) catalyzes two distinct reactions, interconverting UDP-galactose (UDP-gal) and UDP-glucose (UDP-glc) in the final step of the Leloir pathway of galactose metabolism, and also interconverting UDP-N-acetylgalactosamine (UDP-galNAc) and UDP-N-acetylglucosamine (UDP-glcNAc). All four of these UDP-sugars serve as vital substrates for glycosylation in metazoans. Partial loss of GALE in humans results in the spectrum disorder epimerase deficiency galactosemia; partial loss of GALE in Drosophila melanogaster also results in galactose-sensitivity, and complete loss in Drosophila is embryonic lethal. However, whether these outcomes in both humans and flies result from loss of one GALE activity, the other, or both has remained unknown. To address this question, we uncoupled the two activities in a Drosophila model, effectively replacing the endogenous dGALE with prokaryotic transgenes, one of which (Escherichia coli GALE) efficiently interconverts only UDP-gal/UDP-glc, and the other of which (Plesiomonas shigelloides wbgU) efficiently interconverts only UDP-galNAc/UDP-glcNAc. Our results demonstrate that both UDP-gal and UDP-galNAc activities of dGALE are required for Drosophila survival, although distinct roles for each activity can be seen in specific windows of developmental time or in response to a galactose challenge. By extension, these data also suggest that both activities might play distinct and essential roles in humans.
Published in the journal: . PLoS Genet 8(5): e32767. doi:10.1371/journal.pgen.1002721
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1002721Summary
In both humans and Drosophila melanogaster, UDP-galactose 4′-epimerase (GALE) catalyzes two distinct reactions, interconverting UDP-galactose (UDP-gal) and UDP-glucose (UDP-glc) in the final step of the Leloir pathway of galactose metabolism, and also interconverting UDP-N-acetylgalactosamine (UDP-galNAc) and UDP-N-acetylglucosamine (UDP-glcNAc). All four of these UDP-sugars serve as vital substrates for glycosylation in metazoans. Partial loss of GALE in humans results in the spectrum disorder epimerase deficiency galactosemia; partial loss of GALE in Drosophila melanogaster also results in galactose-sensitivity, and complete loss in Drosophila is embryonic lethal. However, whether these outcomes in both humans and flies result from loss of one GALE activity, the other, or both has remained unknown. To address this question, we uncoupled the two activities in a Drosophila model, effectively replacing the endogenous dGALE with prokaryotic transgenes, one of which (Escherichia coli GALE) efficiently interconverts only UDP-gal/UDP-glc, and the other of which (Plesiomonas shigelloides wbgU) efficiently interconverts only UDP-galNAc/UDP-glcNAc. Our results demonstrate that both UDP-gal and UDP-galNAc activities of dGALE are required for Drosophila survival, although distinct roles for each activity can be seen in specific windows of developmental time or in response to a galactose challenge. By extension, these data also suggest that both activities might play distinct and essential roles in humans.
Introduction
Galactose is an essential component of glycoproteins and glycolipids in metazoans, and as a constituent monosaccharide of the milk sugar, lactose, also serves as a key nutrient for mammalian infants. Galactose is also found in notable quantities in some fruits, vegetables, and legumes. Galactose is both synthesized and catabolized in all species via the Leloir pathway, which is highly conserved across branches of the evolutionary tree [1].
The reactions of the Leloir pathway are catalyzed by the sequential activities of three enzymes: (1) galactokinase (GALK) which phosphorylates alpha-D-galactose to form galactose-1-phosphate (gal-1P), (2) galactose-1-phosphate uridylyltransferase (GALT), which transfers uridine monophosphate (UMP) from uridine diphosphoglucose (UDP-glc) to gal-1P, forming UDP-galactose (UDP-gal) and releasing glucose-1-phosphate (glc-1P), which can proceed to phosphoglucomutase and the glycolytic pathway, and (3) UDP-galactose 4′-epimerase (GALE) which interconverts UDP-gal and UDP-glc [1]. In addition to a role in the Leloir pathway, metazoan GALE enzymes also interconvert UDP-N-acetylgalactosamine (UDP-galNAc) and UDP-N-acetylglucosamine (UDP-glcNAc) (Figure 1). Because it catalyzes reversible reactions, GALE therefore not only contributes to the catabolism of dietary galactose, but also enables the endogenous biosynthesis of both UDP-gal and UDP-galNAc [2], [3] when exogenous sources are limited.
Fig. 1. The Leloir pathway of galactose metabolism. 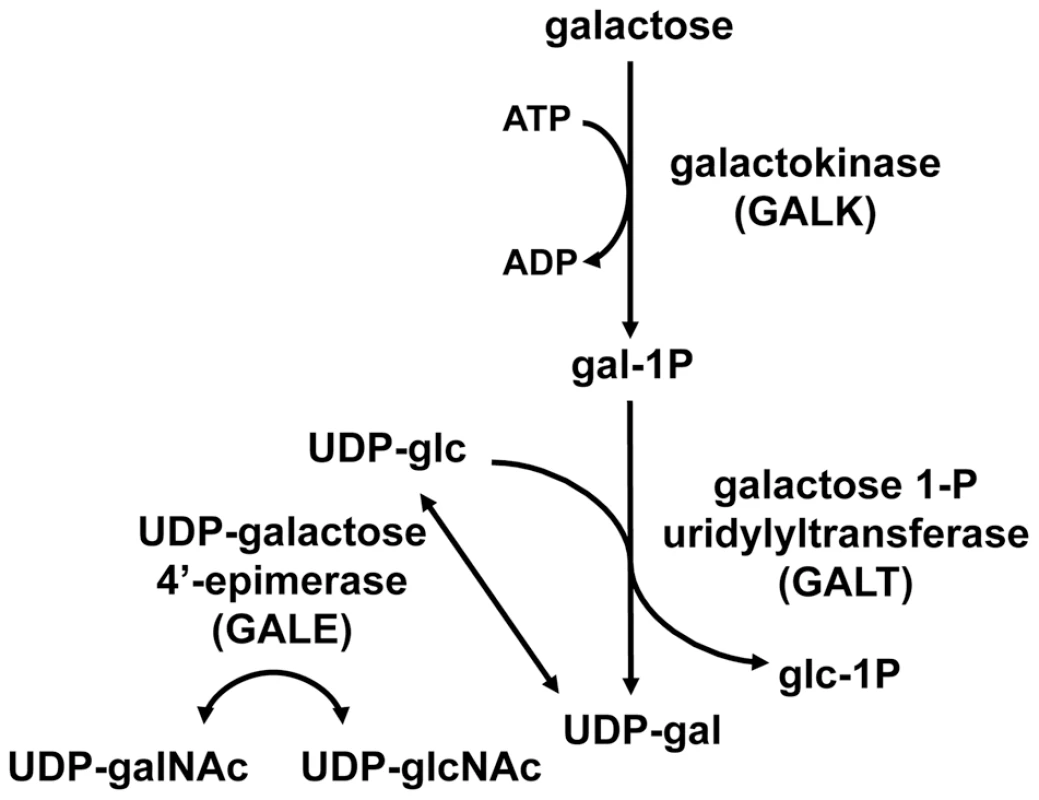
UDP-galactose 4′-epimerase, the third enzyme in the pathway, also interconverts UDP-N-acetylgalactosamine (UDP-galNAc) and UDP-N-acetylglucosamine (UDP-glcNAc) in humans, Drosophila, and other metazoans tested. Deficiency in any of the three Leloir enzymes in humans results in a form of the metabolic disorder galactosemia, although the symptoms and clinical severity differ according to which enzyme is impaired and the extent of the impairment. Profound loss of hGALE results in generalized epimerase-deficiency galactosemia, an autosomal recessive and potentially severe disorder. To date, however, no patient has been reported with complete loss of GALE, and even the most severely affected demonstrate at least 5% residual enzyme activity [4]. Previous studies have indicated that different patient mutations impair hGALE to different extents [5]–[9]. Further, while some mutations impair both GALE activities similarly, others do not. For example, the hGALE allele V94M, which leads to severe epimerase-deficiency galactosemia in the homozygous state, encodes an enzyme that retains ∼5% residual activity toward UDP-gal but ∼25% residual activity toward UDP-galNAc [8], [9]. Disparities such as this have raised the question of whether the pathophysiology of epimerase deficiency galactosemia results from the loss of GALE activity toward UDP-gal/UDP-glc, or toward UDP-galNAc/UDP-glcNAc, or both.
To address this question, we applied a Drosophila melanogaster model of GALE deficiency [10]. Using this model, we have previously established that GALE is essential in Drosophila; animals completely lacking endogenous dGALE succumb as embryos, and conditional loss of dGALE in larvae results in death within two to four days of knockdown. Finally, partial loss of dGALE leads to galactose sensitivity in larvae, and transgenic expression of human GALE (hGALE) rescues each of these negative outcomes [7].
Here we have applied our transgenic Drosophila model to uncouple and examine the individual roles of GALE separately. Toward that end, we generated flies that lacked endogenous dGALE and expressed either of two prokaryotic transgenes, one encoding E. coli GALE (eGALE) which exhibits an approximately 8,000-fold substrate preference for UDP-gal/UDP-glc over UDP-galNAc/UDP-glcNAc [11], and the other encoding P. shigelloides wbgU, which exhibits an approximately 2,000-fold substrate preference for UDP-galNAc/UDP-glcNAc over UDP-gal/UDP-glc [12]. By expressing these prokaryotic transgenes individually or in combination in dGALE-deficient Drosophila we determined that both GALE activities are required for survival of embryos and larvae. We also found that restoration of one activity or the other in later development rescued some phenotypes. Combined, these results provide insight into the varied roles of dGALE in Drosophila development and homeostasis, and by extension, suggest that hGALE may play similarly complex and essential roles in humans.
Results
The Drosophila GALE enzyme efficiently interconverts both UDP-gal/UDP-glc and UDP-galNAc/UDP-glcNAc
Human and other mammalian GALE enzymes efficiently interconvert both UDP-gal/UDP-glc and UDP-galNAc/UDP-glcNAc (e.g. [13]–[15]). Previously, we reported that Drosophila GALE interconverts the first of these substrate pairs (UDP-gal/UDP-glc) [10], but did not address whether dGALE could also interconvert the second. Here we demonstrate that dGALE from wild-type adult flies efficiently interconverts both substrate sets (left most bar, Figure 2). Of note, while purified human GALE [15] and dGALE each interconvert both UDP-gal/UDP-glc and UDP-galNAc/UDP-glcNAc, the apparent specific activity of both human and fly enzymes toward UDP-gal is significantly higher than toward UDP-galNAc.
Fig. 2. Enzyme activities of flies expressing different GALE 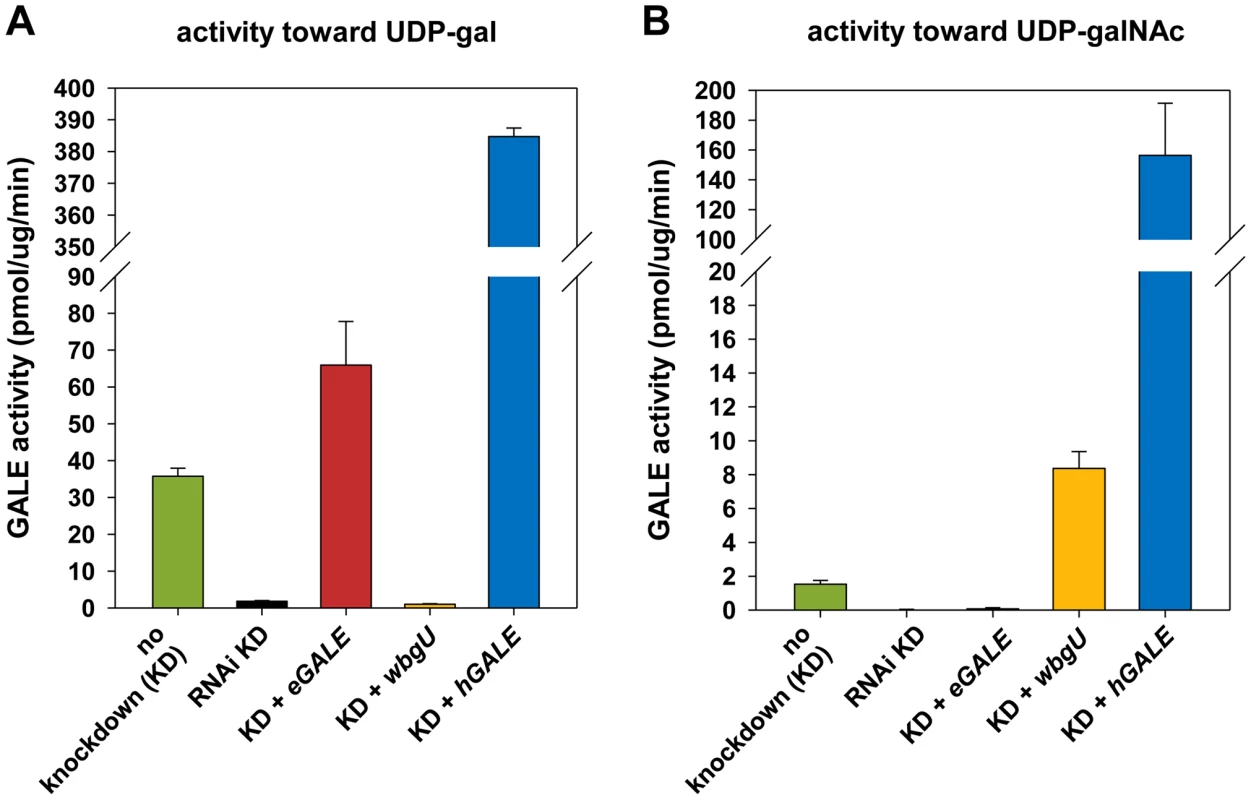
transgenes. Assays for all genotypes were performed on flies with dGALE knockdown (KD) driven by the Act5C-GAL4 driver with the exception of flies labeled “no knockdown”; those flies carried the same UAS-RNAidGALE and GAL80ts alleles, but were balanced over TSTL, and thus lacked the driver. In addition to RNAi knockdown of dGALE, Act5C-GAL4 also drives expression of the specified transgenes in these animals. Panel A: GALE activity using UDP-gal as substrate. Panel B: GALE activity using UDP-galNAc as substrate. eGALE and wbgU transgenes enable the expression of individual GALE activities in Drosophila
To generate flies with epimerase activity toward only UDP-gal/UDP-glc or only UDP-galNAc/UDP-glcNAc, we created transgenic lines expressing eGALE (UAS-eGALE) or wbgU (UAS-wbgU), respectively, each in a conditionally dGALE-impaired background. Each of these prokaryotic GALE genes has been demonstrated previously to encode epimerase activity toward only one of the two sets of epimer pairs (e.g. [11], [12]). To minimize background, activities of the encoded eGALE and WbgU enzymes toward UDP-gal and UDP-galNAc were assayed in flies knocked down for endogenous dGALE; results for the transgenes that demonstrated activities closest to those seen in wild-type Drosophila, eGALE62A and wbgU19A, are presented in Figure 2. As expected, lysates from dGALE knockdown flies expressing the eGALE transgene demonstrated strong activity toward UDP-gal, but not UDP-galNAc, and lysates from dGALE knockdown flies expressing the wbgU transgene demonstrated strong activity toward UDP-galNAc, but not UDP-gal. As a control we also tested lysates from dGALE knockdown flies expressing a human GALE transgene; as expected, those samples demonstrated very strong activity toward both substrates.
Both eGALE and wbgU transgene activities are required, in combination, to rescue viability of dGALE–deficient Drosophila
Previously, we created and characterized two dGALE-deficient alleles, dGALEf00624.4 and dGALEΔy, which allowed us to demonstrate that GALE is essential for survival in Drosophila [10]. To examine the requirement for the two different epimerase activities separately, we set up crosses which allowed for the expression of eGALE or wbgU, individually or in combination, driven by Act5C-GAL4 in an otherwise dGALE-deficient background (dGALEf00624.4/dGALEΔy). Table 1 shows the observed to expected ratios of surviving transgenic offspring that eclosed from these crosses. As presented in Table 1, neither eGALE alone nor wbgU alone was sufficient to rescue survival of the dGALE-deficient animals; however, expression of both eGALE and wbgU, in combination, was sufficient. These results demonstrate that GALE activities toward both UDP-gal and UDP-galNAc are essential for survival of D. melanogaster. To rule out the possibility that rescue with eGALE plus wbgU in combination occurred not because both GALE activities are essential but rather because neither individual transgene expressed sufficient enzyme, we also tested additional eGALE and wbgU transgenes that individually demonstrated higher levels of expression; none was sufficient to rescue (data not shown). Of note, there also was no apparent over-expression phenotype; for example, animals expressing either eGALE or wbgU in addition to endogenous dGALE, and animals dramatically over-expressing human GALE, remained viable, fertile, and appeared morphologically normal (data not shown).
Tab. 1. Crosses to test rescue of <i>wbgU</i> and <i>eGALE</i> transgenes individually and in combination. 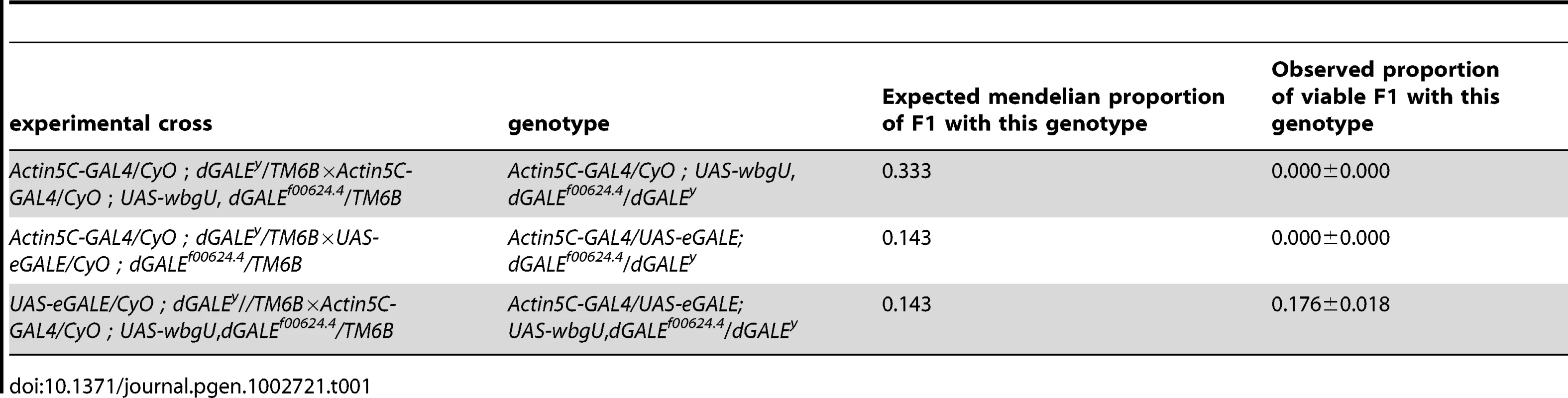
Different requirements for GALE activities at different stages of Drosophila development
Previously, we described an approach that achieves conditional knockdown of dGALE in Drosophila using a UAS-RNAidGALE transgene (12030-R2, National Institute of Genetics Fly Stock Center, Mishima, Shizuoka, Japan) in combination with a temperature sensitive allele of yeast GAL80 (GAL80ts) ([10] and Figure 3A). Using this system, we found that dGALE is required from embryogenesis through pupation, and that loss of dGALE during pupation leads to defects in fecundity and perhaps also a shortened life span [10].
Fig. 3. Differentially impaired fecundity of flies lacking different GALE activities. 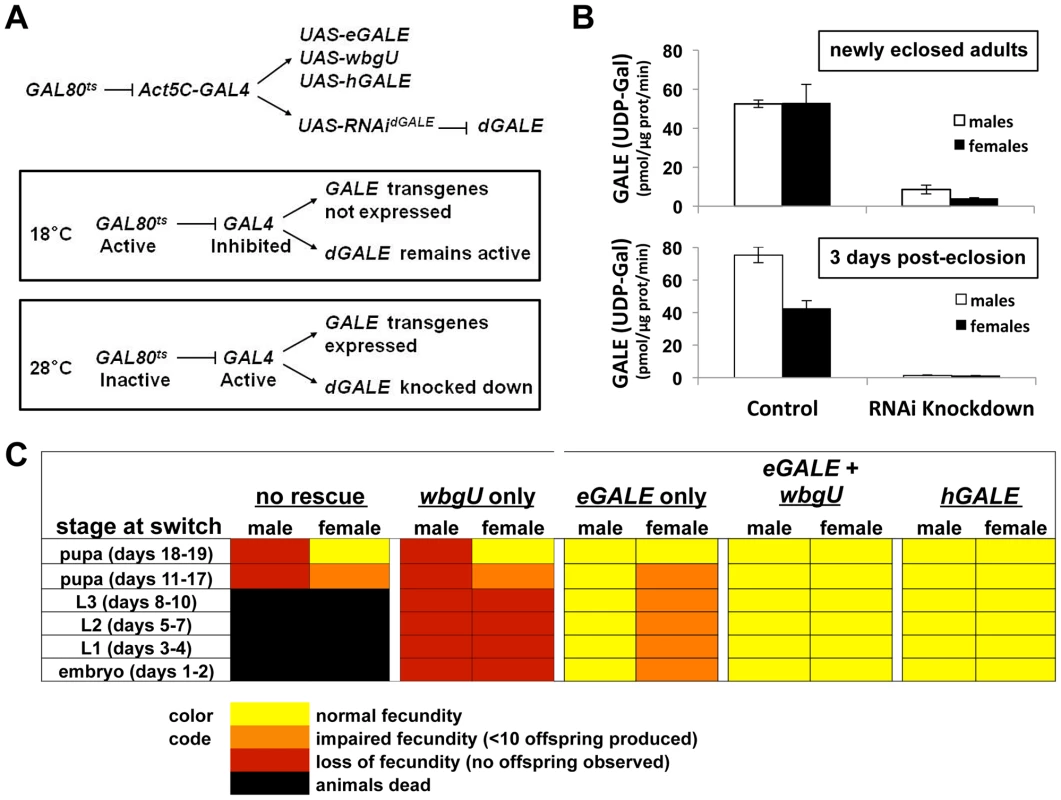
(A) Diagram of the method used to achieve expression of different GALE transgenes in the background of dGALE knockdown animals. The timing of knockdown and concurrent transgene expression was controlled by switching flies from the permissive temperature (18°C) to the restrictive temperature (28–29°C), as indicated. (B) Knockdown efficiency in male and female animals switched to the restrictive temperature as early to mid-stage pupa and harvested for biochemical analysis as newly eclosed adults or three days after eclosion. Of note, GALT activity was completely normal in all samples tested and apparently unaffected by the dGALE knockdown (data not shown). (C) Each box represents the outcome of flies switched from 18°C to 28°C at the stage indicated in the column on the left. The number of days the flies developed at 18°C to reach each stage is shown in parentheses. Here we have expanded the GAL80tsconditional dGALE knockdown system to include different GAL4-dependent GALE transgenes and have applied this expanded system to test the ability of each transgene, or pair of transgenes, to compensate for the loss of endogenous dGALE. By using age-synchronized cohorts of animals and shifting from the permissive (18°C) to the restrictive temperature (28–29°C) at different times we also were able to test the ability of each GALE transgene, or pair of transgenes, to sustain survival and fecundity at different stages of development. At 18°C these animals expressed endogenous dGALE, but not their transgenes, and at 28–29°C these animals expressed their transgenes but not dGALE (Figure 3A). Specifically, we tested Drosophila that carried no GALE transgene, an eGALE transgene, a wbgU transgene, both eGALE and wbgU transgenes, or an hGALE transgene.
As expected from prior results ([10] and Table 1), animals expressing no GALE transgene succumbed when shifted to the restrictive temperature as larvae, while animals expressing either human GALE or both eGALE plus wbgU remained viable and fertile (Figure 3C). Surprisingly, expression of either eGALE or wbgU alone was also sufficient to rescue survival, albeit to a lesser extent. The fact that the individual prokaryotic transgenes were sufficient to rescue dGALE knockdown animals, but not animals genetically null for dGALE (Table 1), suggests that trace residual dGALE expression in the knockdown animals lowered the threshold of transgene function required for rescue.
Of note, while dGALE knockdown animals encoding either eGALE or wbgU remained viable following a shift to the restrictive temperature in early to mid-development (Figure 3C), these survivors were not entirely healthy. Specifically, these animals demonstrated either partial or complete loss of fecundity as adults. To test whether the degree of dGALE knockdown was comparable between males and females, and therefore not a confounding factor in differential outcome, we performed GALE and GALT enzyme assays on newly eclosed and three day old male and female knockdown adults that carried no GALE transgene and that had been switched to the restrictive temperature as early to mid-stage pupa. The degree of GALE knockdown in both males and females was profound and comparable (Figure 3B). As expected, the level of GALE activity was even lower in the older animals, presumably because any GALE synthesized prior to the temperature switch had three additional days to decay. Also as expected, GALT activity was normal and apparently unaffected by the dGALE knockdown in all samples tested (data not shown).
To examine fecundity, we collected and sequestered newly eclosed virgin female and male flies from each surviving cohort, crossed them to an equal number of wild-type flies of the opposite sex, and counted the numbers of viable offspring resulting from each cross. Crosses resulting in large numbers of viable offspring (>50) were scored as “normal fecundity”. Crosses resulting in fewer than 10 viable offspring were scored as “reduced fecundity,” and crosses resulting in no viable offspring were scored as “loss of fecundity” (Figure 3C). For example, when dGALE knockdown was initiated during early to mid-stage pupal development, animals of both sexes displayed diminished fecundity. Expression of eGALE alone, but not wbgU alone, rescued the male defect, whereas expression of both prokaryotic transgenes in combination, or hGALE alone, was required to rescue the female defect. These results indicate that GALE activity toward UDP-gal is both necessary and sufficient for male fecundity, but that GALE activities toward both UDP-gal and UDP-galNAc are required for female fecundity.
Galactose exposure of transgenic flies with late-onset dGALE knockdown reveals differential roles of GALE activities toward UDP-gal and UDP-galNAc
We have previously demonstrated that Drosophila expressing a hypomorphic allele of dGALE are viable but sensitive to galactose exposure [10]. To assess the roles of the two GALE activities in coping with environmental galactose, we collected adult flies in which dGALE knockdown coupled with hGALE, eGALE, wbgU, or eGALE plus wbgU transgene expression was initiated using the GAL80ts system during late larval or early-to-mid-pupal development. These animals were allowed to develop on a standard molasses-based food, and were then transferred as newly eclosed adults to food containing either 555 mM glucose as the sole sugar, or 555 mM glucose plus 175 mM galactose.
We assessed the lifespan of each cohort of animals on both foods; as a control, knockdown animals expressing no GALE transgene were also monitored (Figure 4). In the absence of galactose, all cohorts showed similar longevity profiles, although females (Figure 4C) showed greater variability than males (Figure 4A). In the presence of galactose, however, both males and females expressing either no GALE transgene, or only the wbgU transgene, demonstrated a dramatic reduction in life span (p<0.0001, Figure 4B and 4D). Females expressing eGALE alone exhibited a slight decrease in life span that was independent of diet. Animals expressing hGALE or eGALE+wbgU had lifespans comparable to control animals expressing endogenous dGALE, regardless of diet. These data implicate loss of UDP-gal activity as responsible for the galactose-dependent early demise of adult dGALE-impaired Drosophila.
Fig. 4. Flies lacking GALE activity toward UDP-gal/UDP-glc have a shortened life span when exposed to galactose as adults. 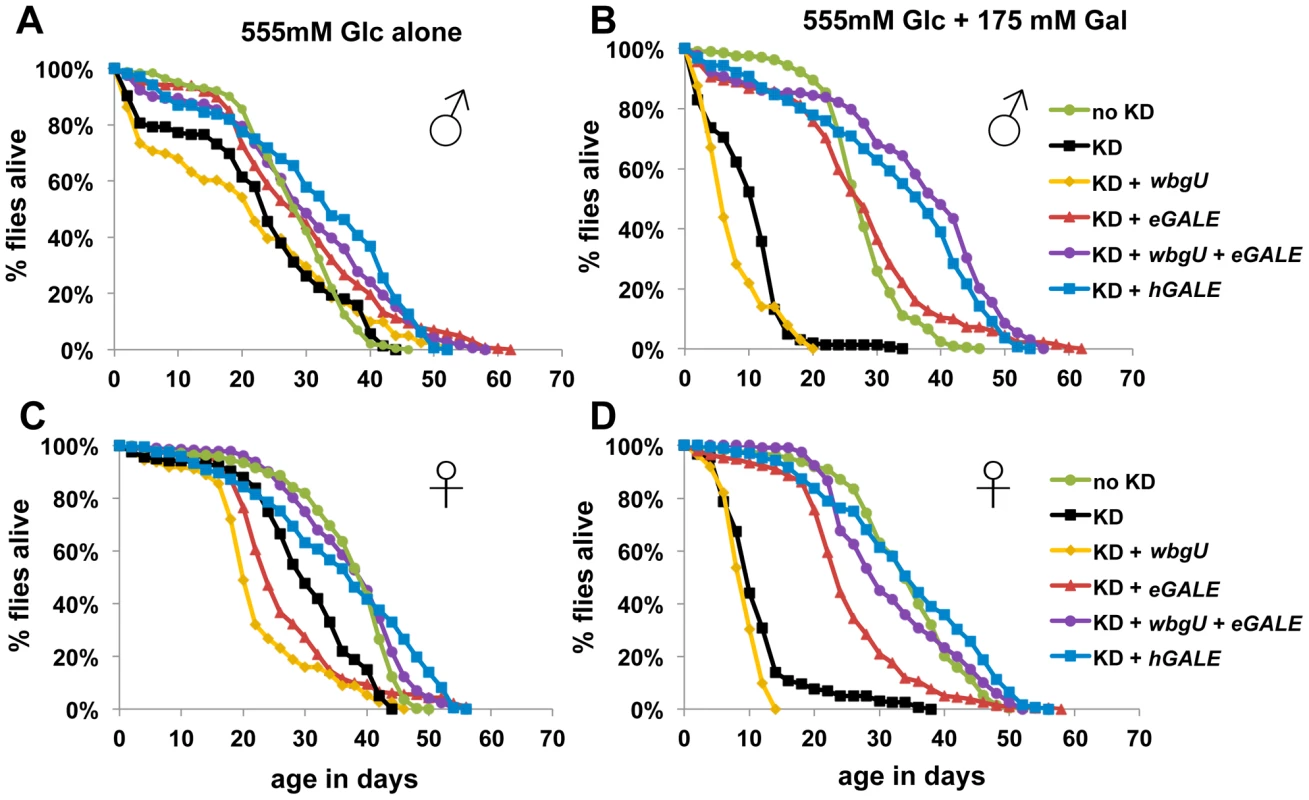
The life spans of male (A and B) and female (C and D) flies reared on molasses food and then tapped as newly eclosed adults to food containing either 555 mM glucose only (A and C), or 555 mM glucose plus 175 mM galactose (B and D), is illustrated. As indicated by the key, these cohorts of flies included controls expressing endogenous dGALE as well as animals that expressed endogenous dGALE early in development but then were subjected late in development to dGALE knockdown coupled with induced expression of either no GALE transgene, or wbgU, eGALE, hGALE, or both wbgU and eGALE in combination. Based on Log rank and Wilcoxon tests for significance, the life spans of knockdown animals expressing either no transgene or expressing only wbgU were significantly decreased on food containing galactose compared with food containing only glucose (p<0.0001). Differentially GALE–impaired flies exposed to galactose demonstrate different metabolic abnormalities
As one approach to explore the pathophysiology underlying the different galactose-dependent outcomes observed in Drosophila deficient in GALE activity toward UDP-gal or UDP-galNAc we measured the levels of gal-1P, UDP-gal, and UDP-galNAc in lysates prepared from galactose-exposed third instar larvae expressing different GALE transgenes. As illustrated in Figure 5, galactose exposed animals deficient in both GALE activities (bars marked “KD” for knockdown) accumulated abnormally high levels of gal-1P (Figure 5A and 5D) and UDP-gal (Figure 5B and 5E). Animals deficient only in GALE activity toward UDP-gal (bars marked “wbgU” in Figure 5) also demonstrated elevated gal-1P (Figure 5A and 5D) and UDP-gal (Figure 5B and 5E). In contrast, galactose exposed larvae deficient only in GALE activity toward UDP-galNAc (bars marked “eGALE” in Figure 5C and 5F) demonstrated no extraordinary metabolic abnormalities, although, as expected, the absolute level of UDP-galNAc was diminished in these animals independent of diet relative to the “no knockdown” control (Figure 5C). Also as expected, animals expressing either hGALE or both eGALE plus wbgU demonstrated no clear metabolic abnormalities (Figure 5).
Fig. 5. Metabolite profiles of Drosophila exposed to galactose. 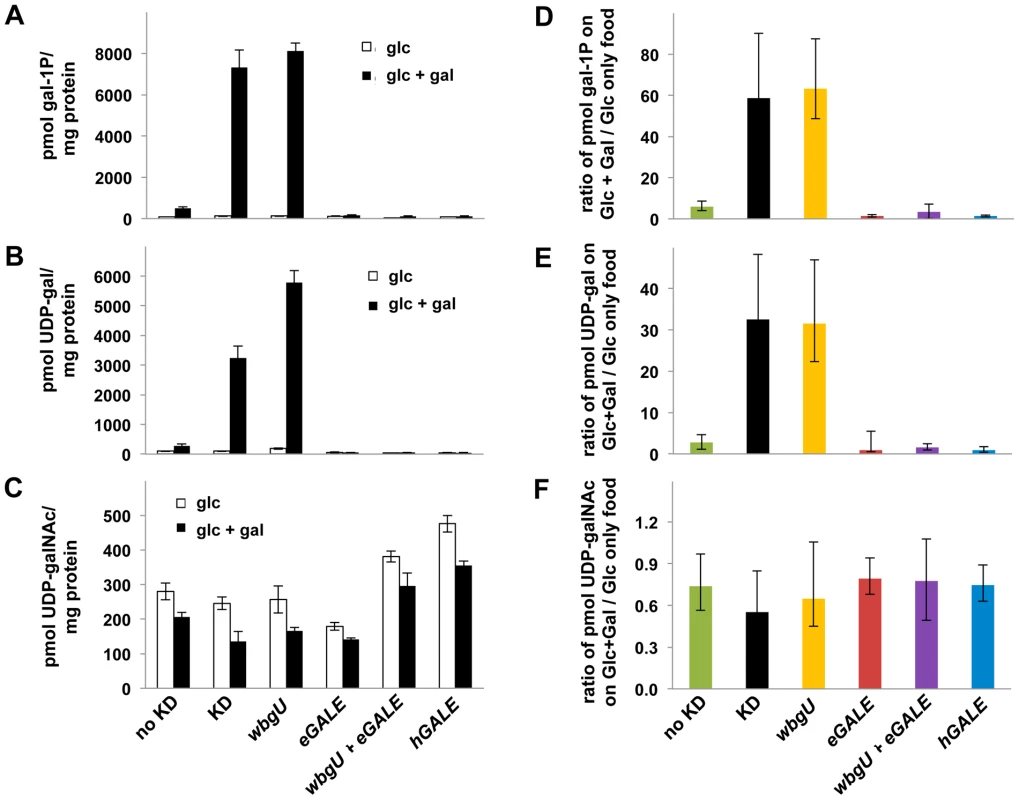
Metabolites were extracted from cohorts of larvae raised on food containing either 555 mM glucose or 555 mM glucose+175 mM galactose. Animals were shifted from the permissive temperature (18°C) to the restrictive temperature (28°C) as first instar larvae and allowed to develop for four days before harvest. Accumulated metabolite values for gal-1P (A), UDP-gal (B), and UDP-galNAc (C) are shown on food containing glucose and glucose+galactose. To demonstrate the impact of diet on metabolite levels, values for gal-1P (D), UDP-gal (E), and UDP-galNAc (F) are shown as ratios of the amount of each metabolite accumulated by animals on food containing galactose over that accumulated by animals of the same genotype on food containing only glucose. Error bars show the 95% confidence interval for each ratio. Discussion
UDP-galactose 4′-epimerase (GALE) is an essential enzyme in Drosophila [10] and in humans [16], but until now the relative contributions of the two distinct GALE activities to development and galactose-tolerance has remained unclear. Understanding these roles has important implications regarding mechanism of galactose sensitivity, and may be applicable to diagnosis and prognosis in humans with epimerase-deficiency galactosemia. Our experiments described here exploit the genetic and biochemical facility of Drosophila melanogaster to test the consequences of losing each of the two GALE activities individually at different stages of development, or under different conditions of galactose exposure.
Our results demonstrate that developing animals require at least some GALE activity toward both epimer pairs, even in the absence of dietary galactose. Complete loss of either activity in embryos is lethal (Table 1). In animals with trace dGALE activity left by knockdown rather than genetic deletion or disruption, however, transgenic expression of either GALE activity alone is sufficient for rescue (Figure 3C). Further, in animals that expressed both GALE activities as larvae, knockdown of both activities during pupation is not lethal.
However, knockdown of either GALE activity in early development, or knockdown of both activities in later development has consequences. For example, loss of activity toward UDP-gal in larvae results in impaired fecundity of both males and females, while loss of activity toward UDP-galNAc in larvae results in impaired fecundity of females but not males. Individual loss of one activity or the other later in development also results in differential sensitivity to galactose. Specifically, both male and female flies deficient in GALE activity toward UDP-gal exhibit a markedly reduced lifespan when exposed to galactose; this effect is not seen in wild-type flies or in flies uniquely deficient in GALE activity toward UDP-galNAc.
These results support two important conclusions. First, the essential role of GALE in development and homeostasis of Drosophila extends beyond the Leloir pathway. Whether GALE activity toward UDP-galNAc is essential because of its presumed role in establishing and maintaining substrate pools for glycosylation, or for some other reason, remains unknown. Prior studies in GALE-deficient mammalian cells [17] showed that uridine supplementation could rescue growth and some metabolic abnormalities caused by galactose exposure, raising the possibility that depleted pools of uridine or uridine-derivatives might also be contributing factors. In the current study it is also unclear whether animals subjected to knockdown of one or both GALE activities later in development demonstrate a less severe outcome than those knocked down earlier in development because the products of GALE function, namely UDP-gal, UDP-glc, UDP-galNAc, and UDP-glcNAc, are less essential later in development, or rather because these UDP sugars have already accumulated to sufficient levels and can be recycled for use. Similarly, the differential sensitivities of male and female fecundity to loss of GALE activity later in development may reflect fundamental differences in male and female development, or alternatively may reflect differential sensitivity to loss; for example, eggs may require a more substantial pool of specific UDP-sugar substrates than sperm to give rise to a viable embryo.
Implications for mechanism
The disparate metabolic profiles observed in GALE-impaired flies exposed to galactose provide a window of insight into potential mechanisms behind the outcomes observed. For example, gal-1P accumulates to abnormal levels in animals missing GALE activity toward UDP-gal but not UDP-galNAc, and only those animals demonstrate substantially reduced lifespan when exposed to galactose as adults. This metabolic result is expected, since only GALE activity toward UDP-gal should impact the Leloir pathway, and this outcome result implies that gal-1P might contribute to the early demise of these animals. However, the gal-1P result also implies that the negative outcomes observed in Drosophila deficient in GALE activity toward UDP-galNAc, e.g. compromised survival in embryos and compromised fecundity in adult females, do not result from gal-1P accumulation. This is an important point because it challenges the common supposition that gal-1P underlies pathophysiology in both classic and epimerase deficiency galactosemias. Clearly there must be another basis for the negative outcomes observed in these animals. It is also interesting to note that while loss of GALE activity toward UDP-galNAc in developing animals has phenotypic consequences, at least for female fecundity, it does not appear to negatively impact the “global” level of UDP-galNAc in animals exposed to galactose. The explanation for this apparent disparity might involve subtle or tissue-specific differences below the threshold of detection of our experimental approach.
Implications for patients
The implications of this work for patients with epimerase deficiency galactosemia are two-fold. First, these results demonstrate that both GALE activities are essential for health of flies, and possibly also people. To our knowledge clinical laboratories that test patient samples for GALE activity only test activity toward UDP-gal. While this practice is certainly understandable, given that mutations may impact the two GALE activities differently [18]–[20], the results presented here raise the possibility that rare patients with GALE deficiency limited to UDP-galNAc activity could be missed. Second, given the impact of GALE-loss on both male and female fecundity in flies, these results suggest that long-term studies of both male and female reproductive issues in epimerase-deficiency galactosemia patients might be warranted.
Materials and Methods
Drosophila stocks and maintenance
The Drosophila stocks used in this study are listed in Table S1. Stocks were maintained at 25°C on a molasses-based food that contained 43.5 g/l cornmeal, 17.5 g/l yeast extract, 8.75 g/l agar, 54.7 ml/l molasses, 10 mls propionic acid and 14.4 ml/l tegosept mold inhibitor (10% w/v in ethanol). For experiments in which the levels and types of sugar were to be varied, we used a glucose-based food [5.5 g/l agar, 40 g/l yeast, 90 g/l cornmeal, 100 g/l glucose, 10 ml/l propionic acid and 14.4 ml/l tegosept mold inhibitor (10% w/v in ethanol)] [21] supplemented with galactose, as indicated.
Generation of transgenic lines
UAS-eGALE and UAS-wbgU transgenes were generated by subcloning the eGALE and wbgU coding sequences, respectively, as EcoRI/XhoI fragments, into pUAST [22] using the EcoRI and XhoI sites in the pUAST polylinker region. The wbgU sequence was amplified from a plasmid generously provided by Peng George Wang (Ohio State University). Resulting plasmids were confirmed by sequence analysis. UAS-eGALE stocks were generated using standard transgenic techniques following injection of the transgene into embryos by the fly core of the Massachusetts General Hospital, Charlestown, MA. UAS-wbgU stocks were generated using standard transgenic techniques following injection of the transgene into embryos by Genetic Services, Inc., Cambridge, MA. Transformants were selected by the presence of the white gene within pUAST. Expression of functional eGALE or wbgU was confirmed by enzymatic assay of lysates from transformants.
GALK, GALT, and GALE (UDP-gal) enzyme assays
Lysates were prepared and assays for GALK, GALT and GALE with UDP-gal as the substrate were performed (n≥3) as described previously [10].
GALE assay conditions for activity toward UDP-galNAc
Activity was calculated from the conversion of UDP-galNAc to UDP-glcNAc. The initial reaction mixture concentrations were: 100 mM glycine pH 8.7, 1.6 mM UDP-galNAc and 0.5 mM NAD. Enzyme assays were performed as described in Sanders et al. [10] except for the following changes: To start each reaction, 7.5 µl of diluted protein and 5 µl of a cocktail of substrates and cofactors were combined. Reaction mixtures were incubated at 25°C for 30 minutes and then quenched by the addition of 112.5 µl of ice-cold high-performance liquid chromatography (HPLC)-grade water (Fisher). Lysates were diluted 1∶4, except for those prepared from animals with RNAi knockdown, which were undiluted, and those prepared from animals overexpressing hGALE or wbgU transgenes, which were diluted to a greater extent. Lysates from Act5C>hGALE22C animals were diluted 1∶60. Lysates from Act5C>wbgU19A animals were diluted 1∶20.
Determining requirement for GALE activities in development and homeostasis
Generation of animals in which GALE knockdown was initiated at 24-hour intervals throughout development was achieved as described previously [10]. A stock homozygous for both P{tubP-GAL80ts}10 and 12030R-2 was used in all crosses. These flies were then crossed to the appropriate genotypes to obtain offspring expressing various transgenes; for: no transgene, P{Act5C-GAL4}25FO1 ; +/T(2;3)TSTL, Tb, Hu; eGALE only, P{Act5C-GAL4}25FO1, UAS-eGALE62A/CyO; wbgU only, P{Act5C-GAL4}25FO1/CyO; P{Act5C-GAL4}25FO1/CyO ; UAS-wbgU19A/TM6B; eGALE plus wbgU, P{Act5C-GAL4}25FO1, UAS-eGALE62A/CyO ; UAS-wbgU19A/TM6B; hGALE, P{Act5C-GAL4}25FO1/CyO ; UAS-hGALE22C/TM6B. Adult flies eclosing from the vials were scored for the presence or absence of humeral and/or curly, as appropriate for each cross.
Measurement of life span
Animals in which dGALE knockdown with concurrent transgene expression was achieved throughout development were obtained as described above. These animals were maintained on standard molasses medium until eclosion. Within 24 hours after eclosion, approximately 20 virgin male or female flies were placed in fresh vials of food containing 555 mM glucose only or 555 mM glucose plus 175 mM galactose. Flies were transferred to fresh food every 2–3 days, and the number of dead flies in each vial was recorded every other day. Log rank and Wilcoxon tests were used for statistical analysis using the program JMP (http://www.jmp.com/).
Measuring metabolite accumulation in GALE–deficient larvae
Cohorts of newly hatched larvae raised at 18°C were transferred to vials of food containing either 555 mM glucose only or 555 mM glucose plus 175 mM galactose. The larvae were maintained at 18°C for one additional day, then transferred to 28°–29°C and allowed to develop for another four days prior to harvest. Metabolites were extracted and quantified as described previously [10], and were separated and quantified using a Dionex HPLC, as described previously [23] with the following changes: UDP-gal and UDP-galNAc were separated using a high salt isocratic procedure with a flow rate of 0.5 mL/min and buffer concentrations of 45% A and 55% B (0–61 min), followed by washing with a linear increase of B to 95% (61–80 min). For all samples, 20 µl were injected into a 25 µl injection loop. Ratios of the level of each metabolite on food containing galactose over the level on food containing glucose only were calculated, and 95% confidence intervals were determined using Fieller's theorem.
Supporting Information
Zdroje
1. HoldenHMRaymentIThodenJB 2003 Structure and function of enzymes of the Leloir pathway for galactose metabolism. J Biol Chem 278 43885 43888
2. KingsleyDKozarskyKFHobbieLKriegerM 1986 Reversible defects in O-linked glycosylation and LDL receptor expression in a UDP-Gal/UDP-GalNAc 4-epimerase deficient mutant. Cell 44 749 759
3. BerryGMoatePReynoldsRYagerCNingC 2004 The rate of de novo galactose synthesis in patients with galactose-1-phosphate uridyltransferase deficiency. Mol Genet Metab 81 22 30
4. HoltonJB 2000 8 ed: McGraw Hill 1553 1587
5. MaceratesiPDallapicollaBNovelliGOkanoYIsshikiGReichardtJ 1996 American Journal of Human Genetics. 59 A204
6. MaceratesiPDaudeNDallapiccolaBNovelliGAllenR 1998 Human UDP-galactose 4′ epimerase (GALE) gene and identification of five missense mutations in patients with epimerase-deficiency galactosemia. Molecular Genetics & Metabolism 63 26 30
7. QuimbyBBAlanoAAlmashanuSDeSandroAMCowanTM 1997 Characterization of two mutations associated with epimerase-deficiency galactosemia, by use of a yeast expression system for human UDP-galactose-4-epimerase. American Journal of Human Genetics 61 590 598
8. WohlersTMChristacosNCHarremanMTFridovich-KeilJL 1999 Identification and Characterization of a Mutation, in the Human UDP-Galactose - 4-Epimerase Gene, Associated with Generalized Epimerase-Deficiency Galactosemia. American Journal of Human Genetics: American Society for Human Genetics
9. WohlersTMFridovich-KeilJL 2000 Studies of the V94M-substituted human UDP-galactose-4-epimerase enzyme associated with generalized epimerase-deficiency galactosaemia. Journal of Inherited Metabolic Disease 23 713 729
10. SandersRDSeftonJMIMobergKHFridovich-KeilJL 2010 UDP-galactose 4′ epimerase (GALE) is essential for development of Drosophila melanogaster. Disease Models & Mechanisms 3 628 638
11. ThodenJHendersonJFridovich-KeilJHoldenH 2002 Structural analysis of the Y299C mutant of Escherichia coli UDP-galactose 4-epimerase. Teaching an old dog new tricks. J Biol Chem 277 27528 27534
12. KowalPWangP 2002 New UDP-GlcNAc C4 epimerase involved in the biosynthesis of 2-acetamino-2-deoxy-L-altruronic acid in the O-antigen repeating units of Plesiomonas shigelloides O17. Biochemistry 41 15410 15414
13. MaleyFMaleyGF 1959 The enzymatic conversion of glucosamine to galactosamine. Biochim Biophys Acta 31 577 578
14. PillerFHanlonMHHillRL 1983 Co-purification and characterization of UDP-glucose 4-epimerase and UDP-N-acetylglucosamine 4-epimerase from porcine submaxillary glands. J Biol Chem 258 10774 10778
15. SchulzJWatsonASandersRRossKThodenJ 2004 Determinants of function and substrate specificity in human UDP-galactose 4′-epimerase. J Biol Chem 279 32796 32803
16. Fridovich-KeilJBeanLHeMSchroerR 2011 Epimerase Deficiency Galactosemia. PagonRBirdTDolanCStephensK GeneReviews Seattle, (WA) University of Washington, Seattle
17. SchulzJRossKMalmstromKKriegerMFridovich-KeilJ 2005 Mediators of galactose sensitivity in UDP-galactose 4′-epimerase-impaired mammalian cells. J Biol Chem 280 13493 13502
18. OpenoKSchulzJVargasCOrtonCEpsteinM 2006 Epimerase-deficiency galactosemia is not a binary condition. Am J Hum Genet 78 89 102
19. WohlersTFridovich-KeilJL 2000 Studies of the V94M-substituted human UDP-galactose-4-epimerase enzyme associated with generalized epimerase-deficiency galactosemia. J Inher Metab Dis 23 713 729
20. WohlersTMChristacosNCHarremanMTFridovich-KeilJL 1999 Identification and Characterization of a Mutation, in the Human UDP Galactose-4-Epimerase Gene, Associated with Generalized Epimerase-Deficiency Galactosemia. Am J Hum Gen 64 462 470
21. HonjoKFTK 2005 Induction of cAMP response element-binding protein-dependent medium-term memory by appetitive gustatory reinforcement in Drosophila larvae. Journal of Neuroscience 25 7905 7913
22. BrandAHPerrimonN 1993 Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118 401 415
23. RossKLDavisCNFridovich-KeilJL 2004 Differential roles of the Leloir pathway enzymes and metabolites in defining galactose sensitivity in yeast. Mol Gen Metab 83 103 116
Štítky
Genetika Reprodukční medicína
Článek Functional Centromeres Determine the Activation Time of Pericentric Origins of DNA Replication inČlánek Dynamic Deposition of Histone Variant H3.3 Accompanies Developmental Remodeling of the TranscriptomeČlánek Integrin α PAT-2/CDC-42 Signaling Is Required for Muscle-Mediated Clearance of Apoptotic Cells inČlánek Prdm5 Regulates Collagen Gene Transcription by Association with RNA Polymerase II in Developing BoneČlánek Acquisition Order of Ras and p53 Gene Alterations Defines Distinct Adrenocortical Tumor Phenotypes
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2012 Číslo 5
-
Všechny články tohoto čísla
- Slowing Replication in Preparation for Reduction
- Chromosome Pairing: A Hidden Treasure No More
- Loss of Imprinting Differentially Affects REM/NREM Sleep and Cognition in Mice
- Six Novel Susceptibility Loci for Early-Onset Androgenetic Alopecia and Their Unexpected Association with Common Diseases
- Regulation by the Noncoding RNA
- UDP-Galactose 4′-Epimerase Activities toward UDP-Gal and UDP-GalNAc Play Different Roles in the Development of
- Deletion of PTH Rescues Skeletal Abnormalities and High Osteopontin Levels in Mice
- Karyotypic Determinants of Chromosome Instability in Aneuploid Budding Yeast
- Genome-Wide Copy Number Analysis Uncovers a New HSCR Gene:
- MicroRNA-277 Modulates the Neurodegeneration Caused by Fragile X Premutation rCGG Repeats
- Functional Centromeres Determine the Activation Time of Pericentric Origins of DNA Replication in
- Dynamic Deposition of Histone Variant H3.3 Accompanies Developmental Remodeling of the Transcriptome
- Scientist Citizen: An Interview with Bruce Alberts
- YY1 Regulates Melanocyte Development and Function by Cooperating with MITF
- Congenital Heart Disease–Causing Gata4 Mutation Displays Functional Deficits
- Recombination Drives Vertebrate Genome Contraction
- KATNAL1 Regulation of Sertoli Cell Microtubule Dynamics Is Essential for Spermiogenesis and Male Fertility
- Re-Patterning Sleep Architecture in through Gustatory Perception and Nutritional Quality
- Using Whole-Genome Sequence Data to Predict Quantitative Trait Phenotypes in
- Genome-Wide Analysis of GLD-1–Mediated mRNA Regulation Suggests a Role in mRNA Storage
- Meiotic Chromosome Pairing Is Promoted by Telomere-Led Chromosome Movements Independent of Bouquet Formation
- LINT, a Novel dL(3)mbt-Containing Complex, Represses Malignant Brain Tumour Signature Genes
- The H3K27 Demethylase UTX-1 Is Essential for Normal Development, Independent of Its Enzymatic Activity
- Suppresses Senescence Programs and Thereby Accelerates and Maintains Mutant -Induced Lung Tumorigenesis
- Genome-Wide Association of Pericardial Fat Identifies a Unique Locus for Ectopic Fat
- An Essential Role for Katanin p80 and Microtubule Severing in Male Gamete Production
- Identification of Genes That Promote or Antagonize Somatic Homolog Pairing Using a High-Throughput FISH–Based Screen
- Principles of Carbon Catabolite Repression in the Rice Blast Fungus: Tps1, Nmr1-3, and a MATE–Family Pump Regulate Glucose Metabolism during Infection
- Integrin α PAT-2/CDC-42 Signaling Is Required for Muscle-Mediated Clearance of Apoptotic Cells in
- Histone H3 Localizes to the Centromeric DNA in Budding Yeast
- Collapse of Telomere Homeostasis in Hematopoietic Cells Caused by Heterozygous Mutations in Telomerase Genes
- Hypersensitive to Red and Blue 1 and Its Modification by Protein Phosphatase 7 Are Implicated in the Control of Arabidopsis Stomatal Aperture
- Extent, Causes, and Consequences of Small RNA Expression Variation in Human Adipose Tissue
- TBC-8, a Putative RAB-2 GAP, Regulates Dense Core Vesicle Maturation in
- Regulating Repression: Roles for the Sir4 N-Terminus in Linker DNA Protection and Stabilization of Epigenetic States
- Common Genetic Determinants of Intraocular Pressure and Primary Open-Angle Glaucoma
- Prdm5 Regulates Collagen Gene Transcription by Association with RNA Polymerase II in Developing Bone
- Fitness Landscape Transformation through a Single Amino Acid Change in the Rho Terminator
- Repeated, Selection-Driven Genome Reduction of Accessory Genes in Experimental Populations
- Allelic Variation and Differential Expression of the mSIN3A Histone Deacetylase Complex Gene Promote Mammary Tumor Growth and Metastasis
- DNA Demethylation and USF Regulate the Meiosis-Specific Expression of the Mouse
- Knowledge-Driven Analysis Identifies a Gene–Gene Interaction Affecting High-Density Lipoprotein Cholesterol Levels in Multi-Ethnic Populations
- A Duplication CNV That Conveys Traits Reciprocal to Metabolic Syndrome and Protects against Diet-Induced Obesity in Mice and Men
- EMT Inducers Catalyze Malignant Transformation of Mammary Epithelial Cells and Drive Tumorigenesis towards Claudin-Low Tumors in Transgenic Mice
- Inactivation of a Novel FGF23 Regulator, FAM20C, Leads to Hypophosphatemic Rickets in Mice
- Genome-Wide Association for Abdominal Subcutaneous and Visceral Adipose Reveals a Novel Locus for Visceral Fat in Women
- Stratifying Type 2 Diabetes Cases by BMI Identifies Genetic Risk Variants in and Enrichment for Risk Variants in Lean Compared to Obese Cases
- New Insight into the History of Domesticated Apple: Secondary Contribution of the European Wild Apple to the Genome of Cultivated Varieties
- Activated Cdc42 Kinase Has an Anti-Apoptotic Function
- The Region Is Critical for Birth Defects and Electrocardiographic Dysfunctions Observed in a Down Syndrome Mouse Model
- COP9 Signalosome Integrity Plays Major Roles for Hyphal Growth, Conidial Development, and Circadian Function
- Bmps and Id2a Act Upstream of Twist1 To Restrict Ectomesenchyme Potential of the Cranial Neural Crest
- Psip1/Ledgf p52 Binds Methylated Histone H3K36 and Splicing Factors and Contributes to the Regulation of Alternative Splicing
- The Number of X Chromosomes Causes Sex Differences in Adiposity in Mice
- Target Gene Analysis by Microarrays and Chromatin Immunoprecipitation Identifies HEY Proteins as Highly Redundant bHLH Repressors
- Acquisition Order of Ras and p53 Gene Alterations Defines Distinct Adrenocortical Tumor Phenotypes
- ELK1 Uses Different DNA Binding Modes to Regulate Functionally Distinct Classes of Target Genes
- Histone H1 Depletion Impairs Embryonic Stem Cell Differentiation
- IDN2 and Its Paralogs Form a Complex Required for RNA–Directed DNA Methylation
- Separation of DNA Replication from the Assembly of Break-Competent Meiotic Chromosomes
- Genomic Hypomethylation in the Human Germline Associates with Selective Structural Mutability in the Human Genome
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Inactivation of a Novel FGF23 Regulator, FAM20C, Leads to Hypophosphatemic Rickets in Mice
- Genome-Wide Association of Pericardial Fat Identifies a Unique Locus for Ectopic Fat
- Slowing Replication in Preparation for Reduction
- An Essential Role for Katanin p80 and Microtubule Severing in Male Gamete Production
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



