-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaCOP9 Signalosome Integrity Plays Major Roles for Hyphal Growth, Conidial Development, and Circadian Function
The COP9 signalosome (CSN) is a highly conserved multifunctional complex that has two major biochemical roles: cleaving NEDD8 from cullin proteins and maintaining the stability of CRL components. We used mutation analysis to confirm that the JAMM domain of the CSN-5 subunit is responsible for NEDD8 cleavage from cullin proteins in Neurospora crassa. Point mutations of key residues in the metal-binding motif (EXnHXHX10D) of the CSN-5 JAMM domain disrupted CSN deneddylation activity without interfering with assembly of the CSN complex or interactions between CSN and cullin proteins. Surprisingly, CSN-5 with a mutated JAMM domain partially rescued the phenotypic defects observed in a csn-5 mutant. We found that, even without its deneddylation activity, the CSN can partially maintain the stability of the SCFFWD-1 complex and partially restore the degradation of the circadian clock protein FREQUENCY (FRQ) in vivo. Furthermore, we showed that CSN containing mutant CSN-5 efficiently prevents degradation of the substrate receptors of CRLs. Finally, we found that deletion of the CAND1 ortholog in N. crassa had little effect on the conidiation circadian rhythm. Our results suggest that CSN integrity plays major roles in hyphal growth, conidial development, and circadian function in N. crassa.
Published in the journal: . PLoS Genet 8(5): e32767. doi:10.1371/journal.pgen.1002712
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1002712Summary
The COP9 signalosome (CSN) is a highly conserved multifunctional complex that has two major biochemical roles: cleaving NEDD8 from cullin proteins and maintaining the stability of CRL components. We used mutation analysis to confirm that the JAMM domain of the CSN-5 subunit is responsible for NEDD8 cleavage from cullin proteins in Neurospora crassa. Point mutations of key residues in the metal-binding motif (EXnHXHX10D) of the CSN-5 JAMM domain disrupted CSN deneddylation activity without interfering with assembly of the CSN complex or interactions between CSN and cullin proteins. Surprisingly, CSN-5 with a mutated JAMM domain partially rescued the phenotypic defects observed in a csn-5 mutant. We found that, even without its deneddylation activity, the CSN can partially maintain the stability of the SCFFWD-1 complex and partially restore the degradation of the circadian clock protein FREQUENCY (FRQ) in vivo. Furthermore, we showed that CSN containing mutant CSN-5 efficiently prevents degradation of the substrate receptors of CRLs. Finally, we found that deletion of the CAND1 ortholog in N. crassa had little effect on the conidiation circadian rhythm. Our results suggest that CSN integrity plays major roles in hyphal growth, conidial development, and circadian function in N. crassa.
Introduction
The COP9 signalosome (CSN) is an evolutionarily conserved multifunctional complex in eukaryotes; it is composed of eight subunits (CSN1–CSN8) in plants and mammals [1]. The CSN was initially discovered to be an important regulator of photomorphogenesis in Arabidopsis thaliana [2] and was later found to participate in a wide range of processes [1], [3]. The CSN potentially influences these cellular pathways by regulating the activity of cullin-RING ubiquitin ligases (CRLs, e.g., CRL1, CRL3, and CRL4 complexes in most eukaryotes) [3], [4], [5]. CRLs are a big family of ubiquitin ligases that share a common cullin/RING-E2 module [6], [7], [8]. They are necessary for substrate ubiquitination in a cascade of enzymatic reactions involving E1, E2, and E3 [9]. Under the control of the CSN-regulated ubiquitin-proteasome pathway, cells coordinate the expression of an array of genes involved in the regulation of growth and development in order to respond to environmental signals, such as light, temperature, and changes in nutrient conditions [1], [10]. Loss-of-function mutations in CSN subunits result in dysfunction of hundreds of CRLs [3], which explains the pleiotropic phenotypes observed in CSN mutants [1], [3], [7].
In 2002, Deshaies and his colleagues first described that the CSN-5 metalloprotease (JAMM) motif is required for removing the ubiquitin-like protein NEDD8 from Cul1 [11]. Later studies confirmed that the isopeptidase activity of the CSN complex is responsible for cullin deneddylation in eukaryotes [1], [3], [4], [5], [12]. In this process, the CSN binds to CRL E3 ligase and cleaves NEDD8 from cullins via the catalytic activity of its CSN-5 subunit, and then inhibits CRL activity [5], [13], [14], [15]. Thus, the deneddylation activity of CSN requires the metalloprotease motif located in the CSN-5 subunit and the functional core subunits of the CSN [11], [16]. However, CSN-5–dependent metalloprotease activity is not essential in Schizosaccharomyces pombe, as no obvious phenotype was detected in csn-5 deletion strains [17], [18].
The physiological importance of CSN deneddylation activity in development and cell differentiation was examined in Drosophila melanogaster, in which the lethality of csn-5Δ/Δ animals was rescued by expression of a CSN-5 transgene but no adult flies were recovered upon equivalent expression of CSN-5 (D148N) (loss of deneddylation activity) [11], [19]. In CSN-5-downregulated HeLa cells, however, the accelerated degradation of c-Jun was rescued equally by over-expression of either the JAMM domain mutant CSN-5D151N or wild-type CSN-5 [20]. These results suggest that the requirement for neddylation/deneddylation cycle of cullins is not absolutely necessary during normal growth and certain developmental stages. In plants, genetic studies suggest that although neddylation/deneddylation cycle is not absolutely necessary during early embryonic development and germination, it is required during seedling establishment and the later developmental stages [12], [21]. In Aspergillus nidulans, deletion of csnE/csn-5 or mutation in JAMM domain results in a block in fruiting body formation at the primordial stage, with a few other observed phenotypic changes, such as light-dependent signaling [22], [23]. Although deneddylation is a major activity of the CSN, it alone cannot explain all of the phenomena described above. These observations raise the possibility that the CSN may have other functional activities in addition to its deneddylation activity.
Recent genetic evidence suggest that the CSN has one additional major function: it controls the stability of CRL ubiquitin ligases in vivo by mediating assembly/disassembly of CRL complexes and by protecting substrate receptors in CRLs from degradation [3], [24], [25], [26], [27], [28]. A recent structural and biochemical study showed that the protective effect of the CSN on DDB2 and CSA autoubiquitination in CRL4 complexes does not require CSN-5–mediated deneddylation activity [29]. However, both of the CSN activities occur when CSN associates with cullins in CRL E3 complexes. Furthermore, there is also a tight correlation between CSN deneddylation activity and the ability of the complex to modulate the stability of CRLs [3]. Thus, it is difficult to determine which function is more important for growth and development through regulation of CRL activity, or how these two functions cooperate with each other in regulating CRL dynamicity in eukaryotes. In A. thaliana, the MPN (Mpr-Pad1-N-terminal domain) subunits CSN-5 and CSN-6 are essential for the structural integrity of the CSN holocomplex [12]. Several studies have shown that point mutations in the JAMM metal-binding site of CSN-5 do not interfere with the proper assembly of CSN complexes in S. pombe, A. thaliana, and A. nidulans [11], [21], [23]. In N. crassa, CSN-5 is not an essential gene; the deletion mutant can survive, and displays obvious growth and developmental defects, making it an excellent model system for investigating the distinctions between the deneddylation and CRL complex assembly/disassembly functions of the CSN [16].
The CSN takes part in a wide range of cellular and developmental processes in N. crassa, including hyphal growth, conidial formation, light and temperature responses, and circadian clock function [16], [24]. To further investigate its biological function in vivo, we created a series of point mutations in the JAMM metal-binding motif of the CSN-5 subunit to disrupt the deneddylation activity of the CSN complex. In those mutant strains, the integrity of the CSN and its interactions with Cul1 and Cul4 were not affected. Surprisingly, mutated CSN-5 almost retained the ability to restore the phenotypic defects of a csn-5KO strain and partially maintained the stability of the SCFFWD-1 complex, which was able to carry out degradation of the clock protein FREQUENCY (FRQ) in vivo. Moreover, the stability of four other substrate receptors of CRLs can be efficiently restored by the CSN containing mutant CSN-5. However, deletion of the CAND1ortholog in N. crassa had little effect on conidiation circadian rhythm and the degradation of FRQ. Our results suggest that the integrity of CSN plays major roles in hyphal growth, conidial development, and circadian function in N. crassa.
Results
Mutations within the CSN-5 JAMM metal-binding site abolish CSN–mediated deneddylation of cullins
The N. crassa genome encodes seven COP9 signalosome subunits (CSN-1–CSN-7) [16], [24]. Several studies have shown that the JAMM metal-binding sites in the MPN domain of CSN-5 are required for metalloprotease activity in the CSN [11], [21], [23]. When the CSN-5 protein sequence was used in a BLAST search against protein databases, a highly conserved MPN domain in the N. crassa CSN-5 subunit was identified. As shown in Figure 1A, three conserved residues corresponding to His127, His129, and Asp140 lie within the putative metal-binding motif (EXnHXHX10D) of the N. crassa CSN-5 JAMM domain. To determine whether these conserved residues form the metalloprotease-like active site of JAMM, we used the JAMM domain of Archaeoglobus fulgidus as a template to generate the tertiary structure of N. crassa CSN-5 [30]. Because of the low similarity between these two JAMM domains, the generated structure was poor. Thus, we instructed SWISS-MODEL to automatically select a template protein for generating the structure of N. crassa CSN-5 [31]. SWISS-MODEL selected the pre-mRNA splicing factor Prp8 as template (Protein Data Bank [PDB] accession number 2P8R) for N. crassa CSN-5. The functional sites were mapped into predicted structure according to the structural alignment with AfJAMM (PDB accession number 1R5X). As shown in Figure 1B, His127, His129, and Asp140 within EXnHXHX10D of the N. crassa CSN-5 JAMM corresponded to the putative metal-binding motif as metalloprotease-like active site in AfJAMM [30], [32].
Fig. 1. Mutations within the JAMM motif of CSN-5 abolish CSN–mediated deneddylation activity for Cul1, Cul3, and Cul4. 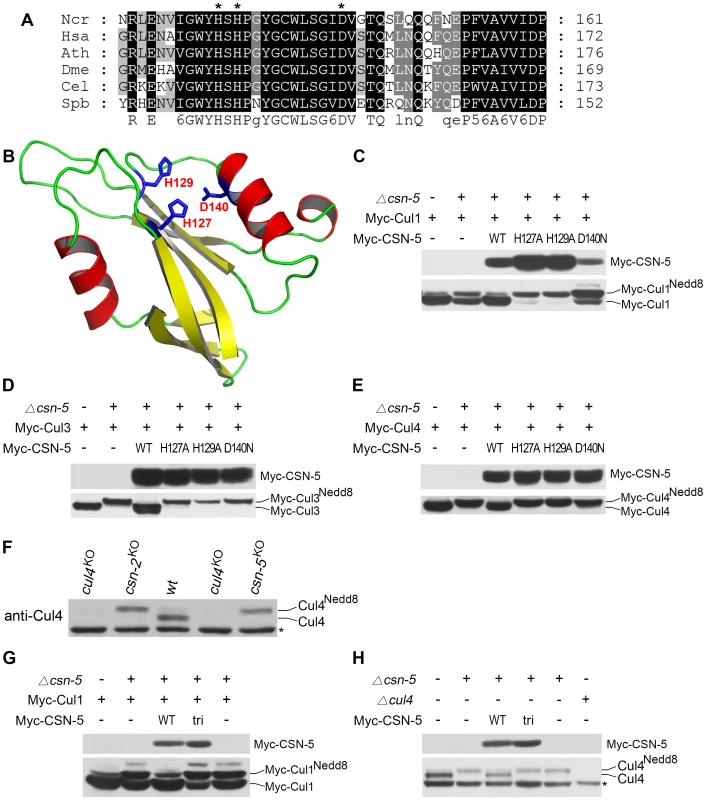
(A) Amino acid alignment of conserved JAMM motifs of CSN-5 homologs from Neurospora crassa (Ncr), Homo sapiens (Hsa), Arabidopsis thaliana (Ath), Drosophila melanogaster (Dme), Caenorhabditis elegans (Cel), and Schizosaccharomyces pombe (Spb). (B) Predicted structure of the N. crassa CSN-5 JAMM domain. The structure was generated by the SWISS-MODEL using the structure of the pre-mRNA splicing factor Prp8 as template (PDB accession number 2P8R), and the functional sites (His127, His129, and Asp140) were mapped according to the structure alignment with the AfJAMM structure (PDB accession number 1R5X). (C–E) Western blot analyses with c-Myc antibody of expression profiles of Myc-Cul1 (C), Myc-Cul3 (D), and Myc-Cul4 (E) in the wild-type strain, csn-5KO, and CSN-5 complementation strains. (F) Western blot analysis showing the expression of Cul4 in the wild-type, cul4KO, and csn-5KO strains. (G) Western blot analyses with c-Myc antibody of expression profiles of Myc-Cul1 in the wild-type strain, csn-5KO, and csn-5KO, pcsn-5-Myc-CSN-5 or csn-5KO, pcsn-5-Myc-CSN-5tri complementation strains. (H) Western blot analyses showing the expression of endogenous Cul4 in the wild-type strain, csn-5KO, csn-5KO, pcsn-5-Myc-CSN-5 or csn-5KO, pcsn-5-Myc-CSN-5tri complementation strains. The asterisk indicates a nonspecific cross-reacted protein band recognized by our Cul4 antiserum (in F and H). To confirm the contribution of CSN-5 to CSN-mediated deneddylation of cullins, we mutated these three highly conserved amino acids (H127A, H129A or D140N) using site-directed mutagenesis. We then introduced quinic acid (QA)–inducible Myc-tagged wild-type CSN-5 or one of the three mutant CSN-5 constructs into a csn-5KO strain expressing Myc-Cul1 protein. As shown in Figure 1C, Myc-CSN-5, Myc-CSN-5H127A, Myc-CSN-5H129A, and Myc-CSN-5D140N were expressed in the csn-5KO strains in the presence of QA. Expression of Myc-tagged wild-type CSN-5 in the csn-5KO strain resulted in a decrease in hyperneddylated Cul1 to the level of the wild-type strain (Figure 1C), indicating that the Myc-tagged CSN-5 protein was functional for CSN deneddylation activity. In contrast, expression of mutant CSN-5 (H127A, H129A, or D140N) failed to decrease the hyperneddylated Cul1 in the csn-5KO strain (Figure 1C). Similarly, hyperneddylation of Cul3 (Figure 1D) and Cul4 (Figure 1E) in the csn-5KO strain was rescued by expressing the Myc-tagged wild-type CSN-5, but not by any of the mutated Myc-CSN-5s. This indicates that the metal-binding motif of JAMM is essential for CSN-mediated deneddylation of cullins.
Because all of the Cul3 and Cul4 was neddylated while not all of the Cul1 was neddylated in the csn-5KO strain and csn-5KO strains complemented by JAMM-domain mutant CSN-5, we rechecked Cul1 modification in the csn mutants. As shown in Figure S1, c-Myc antibody detected three specific protein bands in first generation of csn-5KO or csn-6KO transformants and two specific bands in the csn-1KO transformants. In most positive transformants, there was slightly less unneddylated Cul1 than neddylated Cul1, but the signal remained strong. This is different from the studies in yeast, plants, and fruit fly in which deletion of csn-5 results in hyperneddylation of Cul1 [11], [21], [33]. Possible explanations are that N. crassa genome codes for another deneddylase in addition to CSN complex or there is large amount of newly synthesized Cul1 proteins. We next examined the neddylation of Cul4 using a polyclonal antibody that recognizes the N terminus of N. crassa Cul4. As shown in Figure 1F, only the neddylated Cul4 was detected in csn-5KO strain, while in the wild-type strain, most of the detected Cul4 was unneddylated. Next, we transferred endogenous csn-5 promoter-driven constructs of either wild-type CSN-5 or CSN-5 with JAMM triple point mutations (H127A, H129A, and D140N) (hereafter referred to as CSN-5tri) into a csn-5KO strain expressing Myc-Cul1 protein. Myc-CSN-5 and Myc-CSN-5tri were expressed in the csn-5KO strains (Figure 1G). Similar to what we observed in csn-5KO expressing CSN-5 with a single point mutation (Figure 1C), expression of the CSN-5tri failed to decrease the hyperneddylation of Cul1 in the csn-5KO strain (Figure 1G) as well. Interestingly, the amount of unneddylated Cul1 in csn-5KO strains expressing either single (Figure 1C) or triple (Figure 1G) point mutant CSN-5 was less than that in a csn-5KO strain. Furthermore, expression of CSN-5tri in the csn-5KO strain also failed to decrease hyperneddylated Cul4 to the levels observed in the wild-type or csn-5KO strain complemented with wild-type CSN-5 (Figure 1H). Taken together, these data confirm that the JAMM domain metal-binding motif of N. crassa CSN-5 is essential for the deneddylation activity of the CSN.
The rescue of growth and developmental defects in the csn-5KO strain by CSN-5 with JAMM domain mutations
To examine whether the JAMM metal-binding site of CSN-5 functions in growth and development, we analyzed the phenotypes of the csn-5KO strain expressing either Myc-tagged wild-type or mutant CSN-5. On minimal slants with QA, the csn-5KO strain produced fewer aerial hyphae and conidia than the wild-type strain (Figure 2A). Expression of wild-type CSN-5 in the csn-5KO strain restored aerial hyphal growth and conidial formation to levels similar to those in the wild-type strain (Figure 2A). Surprisingly, when csn-5KO, Myc-CSN-5H127A; csn-5KO, Myc-CSN-5H129A; and csn-5KO, Myc-CSN-5D140N strains (hereafter referred to as csn-5H127A, csn-5H129A, and csn-5D140N, respectively) were grown in minimal slants containing QA, the transformants exhibited hyphal formation and conidiation that were the same as the wild-type strain and the csn-5KO, Myc-CSN-5 strain (Figure 2A). We next measured the growth rates of the wild-type strain, the csn-5KO strain, and the transformants by race tube assay in constant darkness. Interestingly, the growth of csn-5H127A, csn-5H129A, and csn-5D140N strains was slightly faster than that of the wild-type strain (approximately 4.2 cm per day vs. 3.7 cm per day, respectively) and the csn-5KO, Myc-CSN-5 strain (Figure 2B). These results suggest that these CSN-5s with a point mutation within the JAMM metal-binding motif function similarly as the wild-type CSN-5 on N. crassa growth and conidiation. In QA-containing race tubes, the conidiation rhythms of the csn-5H127A, csn-5H129A, and csn-5D140N strains (a period of about 22.5 h) were pretty much (only slightly longer) to those of the wild-type and csn-5KO, Myc-CSN-5 strains (a period about 22.2 h) (Figure 2C) in constant darkness after light entrainment. To characterize the effect on light response of each CSN-5 point mutation, we further examined the light-entrained conidiation rhythm of each csn-5KO transformant during light–dark (LD) cycles (12 h light/12 h dark). As shown in Figure 2D, although the LD cycles entrained the conidiation rhythm of the csn-5KO strains expressing wild-type CSN-5 or mutant CSN-5, however, the conidiation bands of the csn-5H127A, csn-5H129A, and csn-5D140N strains were broader than those of the wild-type and csn-5KO, Myc-CSN-5 strains. Similarly, 12 h 27°C/12 h 22°C temperature cycles entrained the conidiation rhythm of the csn-5H127A, csn-5H129A, and csn-5D140N strains, but not the patterns of conidiation bands (Figure 2E). Taken together, these results suggest that point mutations within CSN-5 are functional in growth and conidiation, and partially functional in circadian rhythm, light response, and temperature-entrained clock process.
Fig. 2. Rescue of growth and developmental defects in the csn-5KO strain by the expression of JAMM-motif mutant CSN-5. 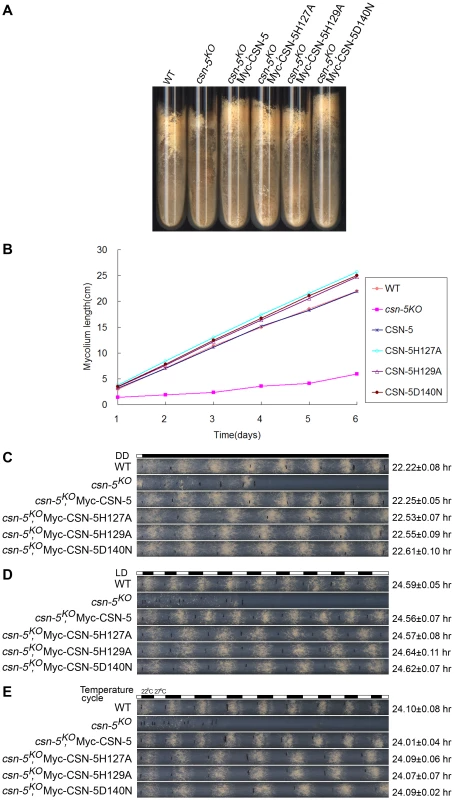
(A) Wild-type, csn-5KO, and the different CSN-5 complementation strains growing on slants containing QA. csn-5KO strains produced significantly less conidia and aerial hyphae than wild-type or CSN-5 complementation strains. (B) Growth rates of the wild-type, csn-5KO, and CSN-5 complementation strains, measured at 25°C by race tube assays in constant darkness after 1 d of light treatment. (C–E) Rescue of conidiation rhythms in the different CSN-5 complementation strains, measured by race tube assay in dark–dark (C), light–dark (D), and temperature cycles (E). At least four replicates were tested under each condition. Black lines indicate the growth fronts every 24 h. JAMM domain mutations do not disrupt CSN complex integrity or its interactions with Cul1 and Cul4
The loss of deneddylation activity of the JAMM domain mutations may be due to the disruption of the CSN complex. To examine this, we tested the interactions between the CSN-6 subunit and wild-type or mutant CSN-5s. Myc-tagged CSN-6 was co-expressed with Flag-tagged CSN-5 or mutant CSN-5 proteins in csn-5KO strains. As shown in Figure 3A, the Flag-tagged CSN-5 strongly interacted with Myc-tagged CSN-6 in an immunoprecipitation reaction, suggesting that they were both in the intact CSN complexes. As expected, the Flag antibody pulled down the Myc-tagged CSN-6 protein in the csn-5KO strain co-expressing Myc-CSN-6 and each of the mutant Flag-CSN-5 proteins (Figure 3A), similar to what was observed in the csn-5KO strain co-expressing CSN-6 and wild-type CSN-5. This result indicates that the point mutations within the CSN-5 JAMM metal-binding motif did not affect the interactions between the CSN-5 and CSN-6 subunits and those two MPN proteins within PCI (Proteasome, COP9, eukaryotic Initiation factor 3) complexes may form dimers. To further examine whether Myc-His-tagged CSN-5 point mutants are incorporated into a larger molecular mass CSN complex, we performed gel filtration and followed by Western blot analysis. As shown in Figure 3B, like wild-type CSN-5, CSN-5H127A, CSN-5H129A, and CSN-5D140N fusion proteins were eluted in larger molecular mass fractions, suggesting that each of the Myc-tagged CSN-5 point mutants can be incorporated into the intact CSN complex.
Fig. 3. Point mutations do not disrupt integrity of the CSN or interactions of the CSN with Cul1 and Cul4. 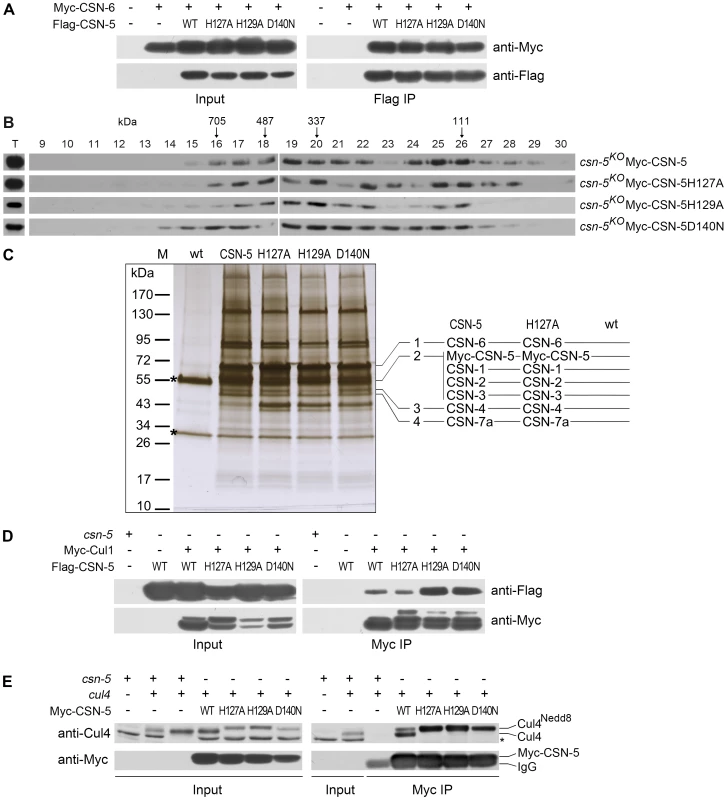
(A) Immunoprecipitation assays confirming interactions between different versions of Flag-CSN-5 and Myc-CSN-6. Wild-type strain and wild-type strain expressing Myc-CSN-6 were used as negative controls. (B) The Myc-CSN-5, Myc-CSN-5H127A, Myc-CSN-5H129A, or Myc-CSN-5D140N in csn-5KO properly incorporates into CSN complex. (C) Silver-stained SDS-PAGE showing the two-step purification of Myc-His-CSN-5, Myc-His-CSN-5H127A, Myc-His-CSN-5H129A, or Myc-His-CSN-5D140N in the csn-5KO strains. A wild-type strain was used as the negative control. CSN subunits identified by mass spectrometry analysis in products of Myc-His-CSN-5 or Myc- His-CSN-5H127A are indicated. Asterisks indicate the two IgG bands. (D) Immunoprecipitation assays confirming the interaction between different versions of Flag-CSN-5 and Myc-Cul1. (E) Immunoprecipitation assays showing the interaction between different versions of Myc-CSN-5 and endogenous Cul4. The asterisk indicates a nonspecific cross-reacted protein band recognized by our Cul4 antiserum. Using protein affinity purification followed by Mass Spectrometry analysis, we further examined whether the CSN complex is properly assembled with CSN-5 point mutants. Myc-His-tagged CSN-5H127A, CSN-5H129A, CSN-5D140N, or wild-type CSN-5 was purified on a nickel column followed by immunoprecipitation with a c-Myc monoclonal antibody. As shown in Figure 3C, similar immunoprecipitated protein profiles were detected in the Myc-His-CSN-5H127A, Myc-His-CSN-5H129A, Myc-His-CSN-5D140N, and Myc-His-CSN-5 (wild-type CSN-5) samples, but not in the wild-type strain (negative control). Liquid chromatography–mass spectrometry/mass spectrometry (LC-MS/MS) analysis of excised gel bands led to the identification of seven subunits, from CSN-1 to CSN-7a, in the Myc-His-CSN-5 purified products and in the Myc-His-CSN-5H127A purified products (Figure 3C). Taken together, these results confirm that the integrity of the CSN complex is not affected by mutations within the JAMM motif of CSN-5 in N. crassa.
Next, we examined whether CSN complexes with mutant CSN-5 subunits can still interact with Cul1 protein. As shown in Figure 3D, both wild-type CSN-5 and each of the mutant CSN-5 proteins co-immunoprecipitated with Cul1 protein. We further examined whether CSN complexes with mutant CSN-5 subunits can also interact with Cul4 protein in vivo by IP/western blotting experiments. As shown in Figure 3E, the Myc-tagged wild-type CSN-5 co-immunoprecipitated with the neddylated and unneddylated Cul4, indicating that the N. crassa CSN complex can interact with all species of Cul4 in vivo. Similarly, the Myc-tagged mutant CSN-5s also co-precipitated with Cul4 (Figure 3E), further confirming that mutations within the JAMM metal-binding motif of CSN-5 do not interfere with interaction between CSN and cullins. These results strongly suggest that the point mutations within the JAMM metal-binding motif abolish NEDD8 isopeptidase activity but have no effect on the integrity of the CSN or on its interactions with cullins.
CSN-5 with mutations in the metal-binding motif of JAMM domain can partially restore SCF-mediated FRQ degradation in the csn-5KO strain
In N. crassa, the clock protein FREQUENCY (FRQ) is a negative regulator in the negative feedback loop that controls the circadian clock under constant conditions [34], [35]. Impaired FRQ degradation in csn-2 mutants results in the loss of circadian rhythm [24]. To investigate whether the mutant CSN-5s can rescue circadian rhythm defects in the csn-5KO strain, we examined the degradation of FRQ protein in the wild-type and csn-5KO strains expressing wild-type CSN-5 or mutant CSN-5s after addition of the protein synthesis inhibitor cycloheximide (CHX). In the wild-type strain, the FRQ was gradually degraded after CHX treatment, with a half-life of about 2.5 h (Figure 4A and 4B). However, in the csn-5KO strain, the degradation of FRQ was mostly blocked (Figure 4A and 4B), similar to what was observed in the csn-2KO strain and the fwd1RIP mutant [24], [36]. As shown in Figure 4A and 4B, the expression of Myc-tagged wild-type CSN-5 in the csn-5KO strain restored the degradation of FRQ to wild-type levels, so that the conidiation period on race tubes was similar to that of the wild-type strain (Figure 2C). We next checked FRQ degradation in the csn-5KO strain expressing CSN-5 proteins with mutations in the JAMM metal-binding site. As shown in Figure 4A and 4B, the expression of Myc-tagged CSN-5H127A, CSN-5H129A, or CSN-5D140N in the csn-5KO strain partially rescued the degradation of FRQ in the csn-5KO strain. FRQ was degraded slightly slower in the mutants than the wild-type strain or the csn-5KO strain complemented by wild-type CSN-5, with a half-life of ∼5 h, consistent with the prolonged period of the conidiation rhythms in the csn-5KO strains expressing the mutant CSN-5, indicating that both deneddylation activity and integrity of CSN are needed in this process. Taken together, these results demonstrate that CSN-5 with point mutations in the JAMM metal-binding site partially restore the SCF-mediated FRQ degradation.
Fig. 4. Mutations in the JAMM motif of CSN-5 partially restore SCF-mediated FRQ degradation in the csn-5KO strain. 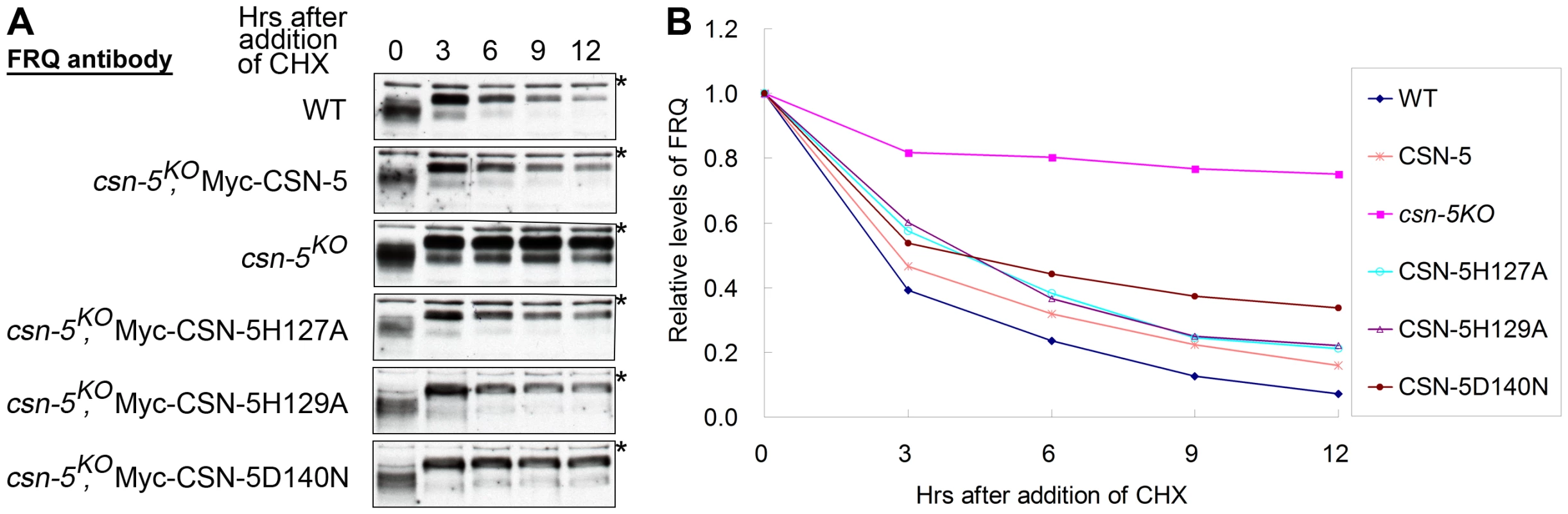
(A) Western blot analyses showing degradation of FRQ protein in csn-5 mutant and the different CSN-5 complementation strains after addition of cycloheximide (10 mg/mL). Asterisks indicate nonspecific bands detected by FRQ antibody. (B) Densitometric analyses from four independent experiments showing the degradation of FRQ in different strains. Catalytically dead CSN-5 partially stabilizes the SCFFWD-1 complex
Previous studies showed that FRQ ubiquitination and degradation is mediated by the SCFFWD-1 E3 ligase complex [24], [36], and that the stability of E3 ligase components is controlled by CSN in vivo [3], [7], [16], [24]. Because the ectopic expression of mutated CSN-5 partially rescued both the circadian conidiation rhythm and FRQ degradation in the csn-5KO strain, we decided to check whether CSN with mutant CSN-5 can prevent the degradation of components of the SCFFWD-1complex. As shown in Figure 5A, Myc-Cul1 was stable after induced expression of Myc-CSN-5 in the csn-5KO strain, with a half-life of >9 h in the presence of CHX, similar to that of the wild-type strain. In the csn-5 mutant, however, both the neddylated and unneddylated Myc-Cul1 became very unstable, with a half-life about 1.5 h (Figure 5A and 5D) [16]. Interestingly, the expression of JAMM mutant CSN-5 had a differential effect on the neddylated and unneddylated Cul1. In mutant CSN-5 transformants, the stability of neddylated Cul1 was only partially rescued, with a half-life of >3 h in the presence of CHX (Figure 5A and 5D), whereas the stability of unneddylated Cul1 was almost rescued, with a half-life of >12 h (Figure 5A and 5D). These data indicate that although CSN containing JAMM mutated CSN-5 fails to cleave NEDD8 from neddylated Cul1, it still functions to protect hyperneddylated and unneddylated Cul1 from degradation to a certain extent.
Fig. 5. CSN-5–mutated CSN efficiently prevents degradation of components of the SCFFWD-1 complex. 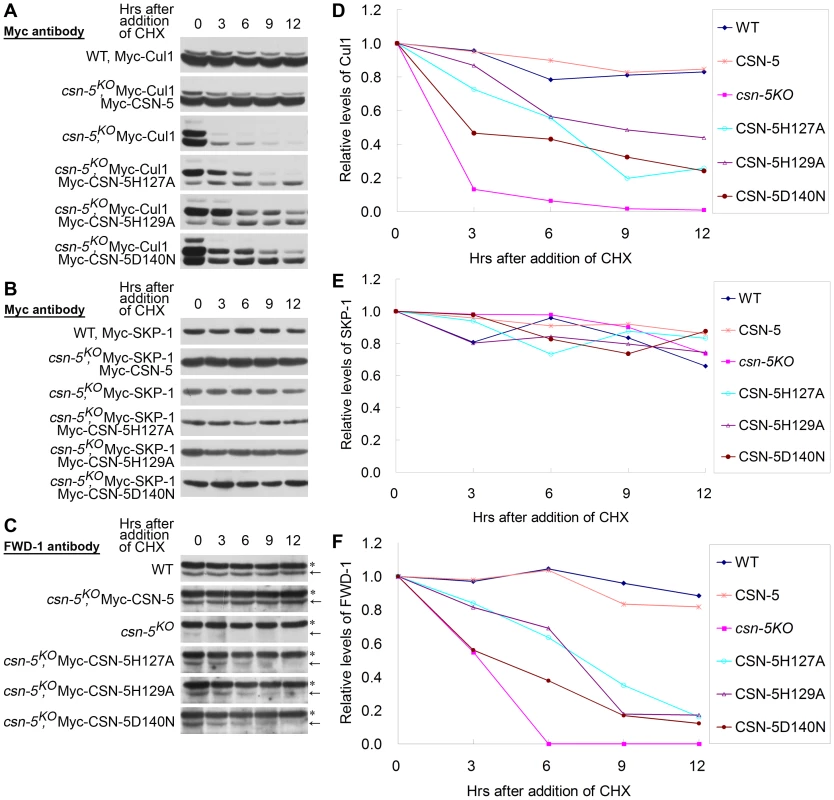
(A–C) Western blot analyses with labeled antibodies showing degradation of Myc-Cul1 (A), Myc-SKP-1 (B), and FWD-1 (C) after addition of cycloheximide (10 mg/mL) in the wild-type, csn-5KO, and different CSN-5 complementation strains. Arrows point out FWD-1 protein bands. Asterisks indicate nonspecific bands detected by FWD-1 antibody. (D–F) Densitometric analyses from four independent experiments showing the degradation of Myc-Cul1 (D), Myc-SKP-1 (E), and FWD-1 (F). In N. crassa, deletion of csn-5 or csn-3 has no effect on the stability of SKP-1 protein in the SCFFWD-1 complex [16]. As expected, Myc-SKP-1 were very stable in the wild-type strain and the csn-5KO strain and in the complementation strains with mutant CSN-5, with a half-life of >12 h (Figure 5B and 5E).
FWD-1, the substrate-recruiting subunit of the SCFFWD-1 complex, was quite stable in the wild-type strain, whereas it became undetectable after only 3 h of CHX treatment in csn-5KO strain (Figure 5C and 5F). In the csn-5H127A, csn-5H129A, and csn-5D140N strains, however, FWD-1 signals could still be detected after 6 h of CHX treatment (Figure 5C and 5F), indicating that CSN with mutated CSN-5 partially functions to protect F-box proteins from degradation. This finding further confirms that regulation of SCF-mediated FRQ degradation by the CSN is a key step in the N. crassa circadian clock. Therefore, both the deneddylation activity and the integrity of the CSN are important for preventing the degradation of components of the SCFFWD-1 complex.
CSN complex containing mutant CSN-5 efficiently prevents degradation of substrate receptors of CRLs
We next asked whether CSN with mutated CSN-5 still functions to protect other CRL substrate receptors from degradation. N. crassa SCON-2, an F-box protein involved in regulating sulfur metabolism, was previously shown to interact with SKP-1 and is very unstable in a csn-2KO strain [24], [37]. We compared the stability of Myc-SCON-2 in wild-type, csn-5KO and csn-5KO expressing wild-type CSN-5 or mutant CSN-5H127A strains. The half-life of Myc-SCON-2 was approximately 12 h in the wild-type and csn-5KO expressing wild-type CSN-5 strains in the presence of CHX. Myc-SCON-2 was very unstable in the csn-5 mutant and became undetectable after 3 h of CHX treatment (Figure 6A and 6B). In the csn-5H127A strain, the detection of Myc-SCON-2 signal extended to 6 h after CHX treatment (Figure 6A and 6B). FBP94 (NCU04785), another F-box–containing protein in N. crassa, can also interact with SKP-1 (data not shown). As shown in Figure 6C and 6D, FBP94 was quite stable in the wild-type strain and csn-5KO strain complemented with Myc-CSN-5, whereas in the csn-5KO strain it became undetectable after only 6 h of CHX treatment. In the csn-5H127A strain, detection of FBP94 signal extended to 12 h after CHX treatment (Figure 6C and 6D). Therefore, CSN complex with mutated JAMM domain can partially function in maintaining the stability of other F-box–containing adaptor proteins.
Fig. 6. CSN containing mutant CSN-5 efficiently prevents degradation of substrate receptors of CRLs. 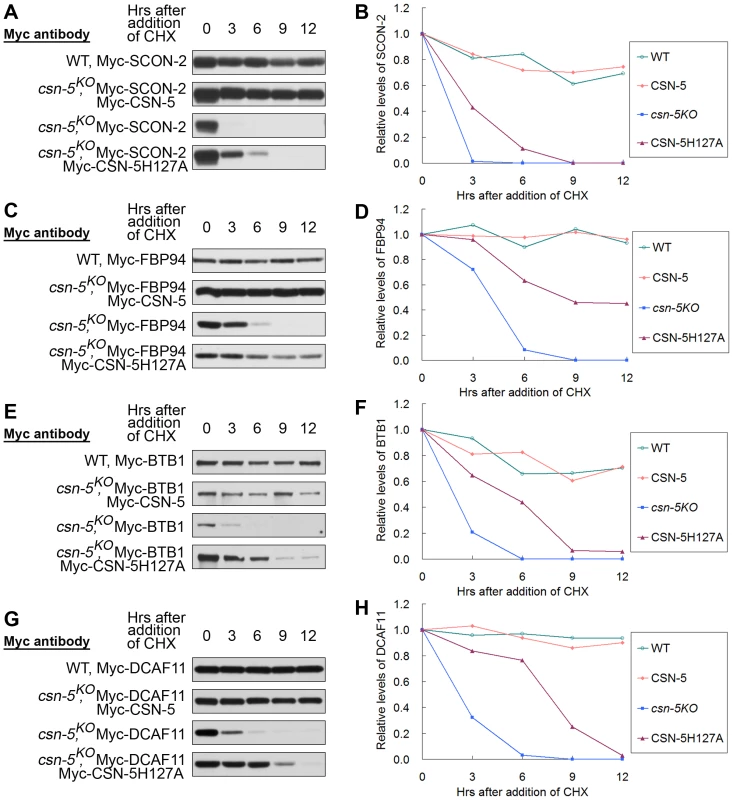
Western blot analyses with labeled antibody showing degradation of Myc-SCON-2 (A), Myc-FBP94 (C), Myc-BTB1 (E), and Myc-DCAF11 (G) after addition of cycloheximide (10 mg/mL) in the wild-type, csn-5KO, csn-5KO complementation with CSN-5 and CSN-5H127A strains. Densitometric analyses from four independent experiments showing the degradation of Myc-SCON-2 (B), Myc-FBP94 (D), Myc-BTB1 (F), and Myc-DCAF11 (H). In a previous study, we determined that the N. crassa Cul3 protein interacts with BTB1 protein, and both proteins become unstable in the csn-5KO strain [16]. The half-life of Myc-BTB1 was >12 h in the wild-type and csn-5KO expressing wild-type CSN-5 strains in the presence of CHX, whereas in the csn-5KO strain it became undetectable after 6 h of CHX treatment (Figure 6E and 6F). As expected, in the csn-5H127A strain, BTB1 signals were detectable at 12 h after CHX treatment (Figure 6E and 6F), indicating that CSN with the JAMM mutated CSN-5 still partially functions to protect the substrate adaptor proteins of CRL3 from degradation. We also investigated whether CSN with the JAMM mutated CSN-5 regulates the substrate receptor protein of CRL4 in a similar manner. N. crassa Cul4 was previously shown to interact with DCAF11, a putative substrate receptor of CRL4DCAF11 [16], [38]. As shown in Figure 6G and 6H, the half-life of Myc-DCAF11 was >12 h in the wild-type and csn-5KO expressing wild-type CSN-5 strains in the presence of CHX, whereas in the csn-5KO strain it became undetectable after 6 h of CHX treatment [16]. As expected, in the csn-5H127A strain, the detection of DCAF11 signal was extended to 9 h after CHX treatment (Figure 6G and 6H). Taken together, these in vivo results indicate that the CSN complex containing mutant CSN-5 efficiently prevents degradation of substrate receptors of CRLs.
CAND1 is not required for regulation of the circadian rhythm and SCFFWD-1 ubiquitin ligase
Current models suggest that the activity and assembly of CRLs are controlled by cycles of CRL deneddylation and CAND1 binding of deneddylated cullins [39], [40], [41], [42], [43], [44], [45], [46]. In plants and worms, CAND1 mutants exhibit defects consistent with a positive role in regulating the function of a subset of CRLs [40], [47], [48], [49]. However, in yeast and human cells, loss of CAND1 has little effect on the abundance of neddylated cullins, suggesting that the neddylation/deneddylation cycle may function independently of CAND1 [50], [51]. To test whether CAND1 is involved in maintaining the function of CRLs in N. crassa, we examined the role of CAND1 in the regulation of circadian conidiation rhythm and proper functioning of the SCFFWD-1 complex. We first measured the growth rates of the wild-type and cand1KO strains by race tube assay under constant darkness. The growth of the cand1KO strain (about 3.0 cm per day) was slightly slower than that of the wild-type strain (about 3.7 cm per day), suggesting that CAND1 is involved in regulating hyphal growth. After entrainment by light, like the wild-type strain, the cand1KO strain exhibited a robust circadian conidiation rhythm with a period of about 22 h at 25°C in constant darkness (Figure 7A), suggesting that CAND1 is not required for circadian rhythms in N. crassa. To test whether CAND1 functions in a manner similar to the CSN, we examined the conidiation rhythms of the cand1 mutant in LD cycles (12 h light/12 h dark). As shown in Figure 7B, the conidiation rhythms of the cand1KO strain were entrained by LD cycles, indicating that unlike CSN, CAND1 is not required in light regulation of the circadian clock. We also examined the responses of the cand1 mutant to temperature entrainment using race tube assays. As expected, in 12 h 27°C/12 h 22°C temperature cycles, as shown in Figure 7C, like the wild-type strain, the conidiation rhythm of the cand1KO strain was synchronized by the temperature cycles, indicating that CAND1 is not required for the temperature-entrained conidiation process. These results suggest that CAND1 does not play a significant role in the regulation of circadian rhythm in N. crassa.
Fig. 7. CAND1 is not required for regulation of the circadian rhythm. 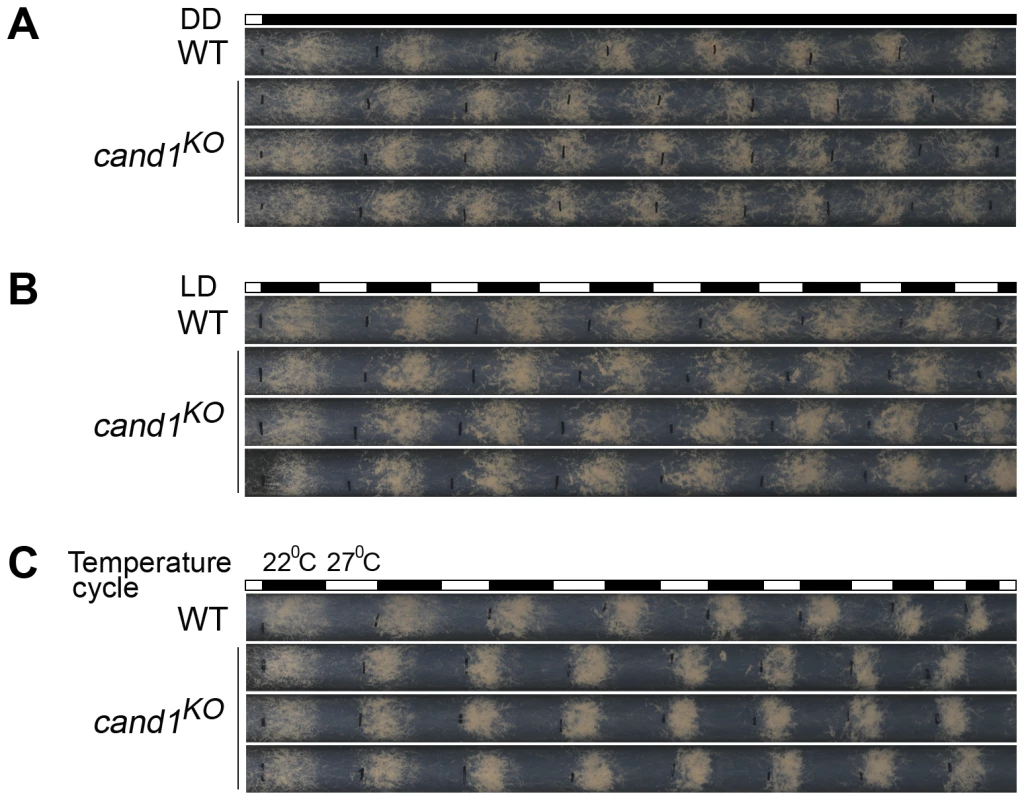
(A–C) Normal conidiation rhythms of cand1 mutants in dark–dark (A), light–dark (B), and temperature cycles (C) measured by race tube assays. At least four replicates were tested under each condition. Black lines indicate the growth fronts every 24 h. Deletion of cand1 also had no effect on degradation of the clock protein FRQ, which is the substrate of the SCFFWD-1 ubiquitin ligase complex in N. crassa (Figure 8A and 8B). We also examined the stability of FWD-1 of the SCFFWD-1 complex in the cand1KO strain. As shown in Figure 8C and 8D, FWD-1 was very stable in the cand1 mutant, as in the wild-type strain, with a half-life of >12 h. Together, these results suggest that CAND1 is not required for regulation of the circadian rhythm and for maintaining the proper function of the SCFFWD-1 complex in N. crassa.
Fig. 8. CAND1 is not required for regulation of the SCFFWD-1 ubiquitin ligase. 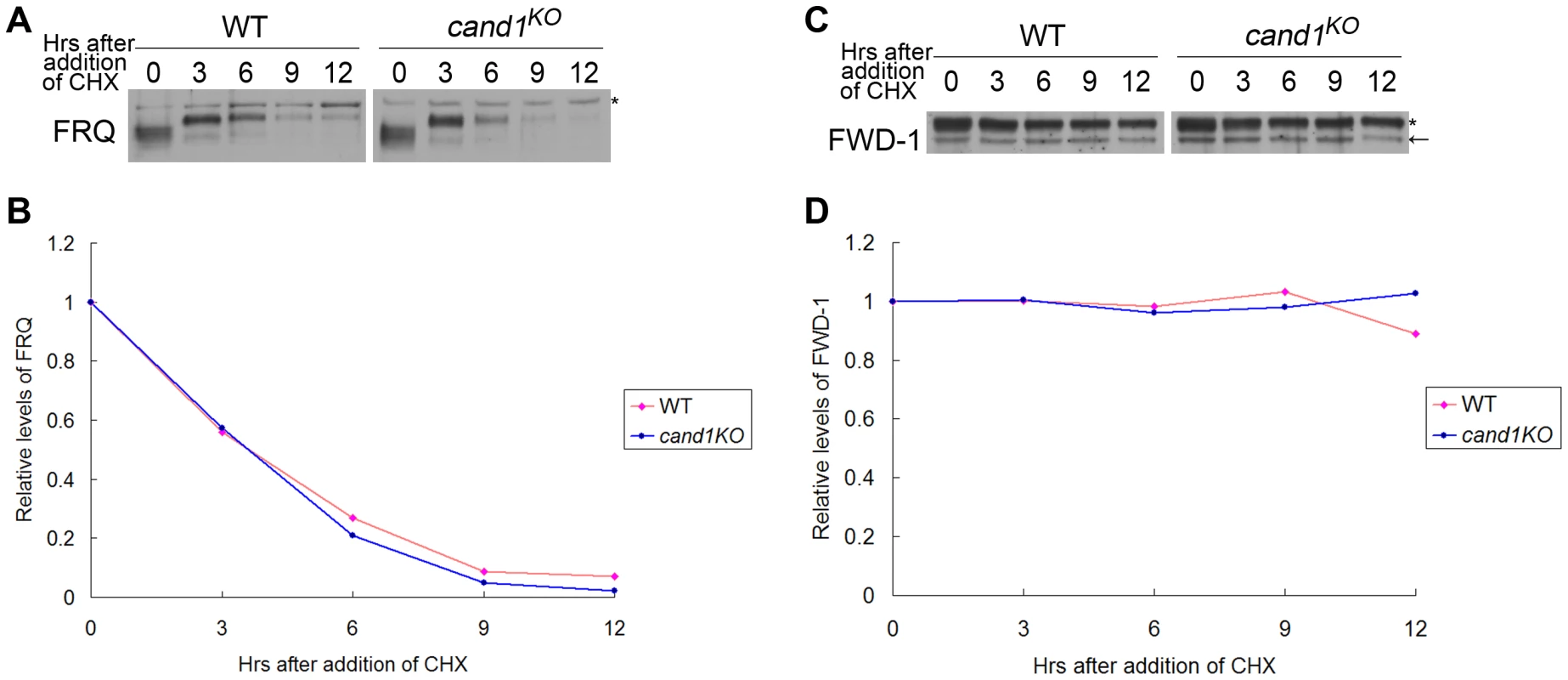
Western blot analyses showing degradation of FRQ (A) and FWD-1 (C) in wild-type and cand1KO strains after addition of cycloheximide (10 mg/mL). Densitometric analyses from four independent experiments showing the degradation of FRQ (B) and FWD-1 (D). Arrows point out FWD-1 protein bands. Asterisks indicate nonspecific bands detected by FRQ antibody (A) or FWD-1 antibody (C). Discussion
In eukaryotes, the COP9 signalosome (CSN) is a highly conserved multifunctional complex that has two major biochemical roles: cleaving NEDD8 from cullin proteins [1], [3], [7], [11] and maintaining the stability of the CRL components [7], [24]. In this study, we used mutation analysis to confirm that the JAMM metal-binding motif of the CSN-5 subunit is responsible for NEDD8 cleavage from cullin proteins in N. crassa. Point mutations of the key residues in the metal-binding motif (EXnHXHX10D) of the CSN-5 disrupted CSN deneddylation activity without interfering with the CSN assembly. We demonstrated that those mutant CSN-5s could almost restore the growth and conidiation defects of the csn-5KO strain. Furthermore, even without the deneddylation activity, the CSN partially maintained the stability of the SCFFWD-1 complex and partially restored the degradation of clock protein FRQ in vivo. Finally, we also showed that CSN containing mutant CSN-5 could efficiently prevent the degradation of the substrate receptors of CRLs. In addition, deletion of the CAND1 ortholog in N. crassa had little effect on the circadian rhythm of conidiation. Thus, our results suggest that maintenance of CRL stability by the CSN integrity is even more crucial in hyphal growth, conidial development, and circadian function in N. crassa.
Both deneddylation activity and integrity of CSN are required for maintenance of CRL stability
As the key regulator of CRLs, both deneddylation and maintenance of CRL stability by the CSN occurs when the CSN binds to CRLs. Thus, it is difficult to distinguish which function is more important for maintaining the proper function of CRLs in eukaryotes. To precisely determine the function of the CSN in maintaining the stability of CRLs, we sought to separate the two functional aspects of CSN from one other in N. crassa. In those csn-5KO strains expressing CSN-5 proteins with different point mutations in the JAMM metal-binding motif, the deneddylation activity was disrupted, while the assembly of the CSN complex and interactions between CSN and cullin proteins were not affected. Therefore, this system has great potential as a model for distinguishing between the two activities of the CSN.
A recent study suggests that neddylated Cul1 and Cul3 are unstable in D. melanogaster csn mutant cells due to a defect in CSN deneddylation activity, whereas unneddylated cullins are stable in csn-5 mutant cells [33]. The results presented here show that unstable forms of Cul1 in the csn-5 mutant were partially restored by expression of mutant CSN-5 protein without deneddylation activity. Like the stability of unneddylated Cul1 and Cul3 in D. melanogaster CSN-5–defective cells [33], unneddylated Cul1 remained stable in the csn-5H127A, csn-5H129A, and csn-5D140N strains, similar to that in the wild-type strain (Figure 5A), indicating that CSN integrity with catalytically dead CSN-5 effectively maintains the stability of cullins. Studies in D. melanogaster and A. nidulans CSN-5 mutants indicated that the CSN deneddylation activity is essential for cell differentiation and developmental initiation [11], [19], [22], [33]. However, in the A. thaliana fus6/C231 mutant (a CSN1 N-terminal deletion mutant), although the Cul1 neddylation still works in a wild-type pattern, it was lethal and exhibited severe gene expression defects [52]. This genetic evidence raises questions concerning whether the CSN has other important functions aside from its deneddylation activity. The accelerated degradation of c-Jun in HeLa cells in which CSN-5 is downregulated is rescued equally by over-expression of the deneddylation mutant CSN-5D151N or wild-type CSN-5 [20]. These data suggest that two activities of CSN may function parallel for regulating the activity of CRLs.
Bennett et al. found that Cul1K720R (a constitutively unneddylated Cul1 mutant) assembles with CSN, SKP-1, and most F-box proteins to the same extent as wild-type Cul1 [51]. Our IP experiments also show that wild-type CSN interacts with both neddylated and unneddylated Cul4. These findings suggest that the CSN can interact with CRLs independent of the prior neddylation of cullins. In plants, genetic results also suggest that during early embryo development and germination, neddylation/deneddylation cycling is not absolutely required, although it becomes more important during seedling establishment and later in development [21], suggesting that the CSN has distinct biochemical functions that orchestrate development in the appropriate spatial and temporal setting.
Protection of substrate receptors by the CSN has been described for the CRLs in vivo [24], [25], [26], [27], [28]. We found that CSN with mutated CSN-5 had a contribution to the stabilities of five receptor proteins of CRLs in vivo. These results provide evidence for the idea that the abundance of adaptor modules (rather than cycles of neddylation/deneddylation and CAND1 binding) drives CRL network organization [51]. This possibility is supported by our genetic observations that the csn-5H127A, csn-5H129A, and csn-5D140N strains exhibited normal growth and conidiation phenotypes. The integrity of CSN was maintained in these JAMM mutation complementation strains, thus it can serve as a platform to recruit other proteins for regulating the activities of CRLs, such as the recruitment of UBP12 in yeast, as well as USP15 in human [14], [53]. In addition, a non-catalytic CSN itself may stabilize the substrate receptors of CRLs. A very recent study has shown that the protective effect of the CSN on DDB2 and CSA autoubiquitination is independent of CSN-5 mediated deneddylation in vitro [29]. These results suggest that the partial rescue of stability of substrate receptors by the catalytically dead CSN is mainly dependent on its protective effect. Therefore, the stability of cullins and some substrate receptors of CRLs are dependent on both deneddylation activity and integrity of the CSN in N. crassa.
Maintaining stability of the SCFFWD-1 complex is the key process in circadian rhythm regulation
The csn-5KO strain exhibits abnormal conidiation rhythms in DD, which cannot be entrained by either LD or temperature cycles, indicating that light and temperature regulation of the conidiation process is impaired in this mutant [16]. We found that degradation of the clock protein FRQ is impaired in the csn-5KO strain, especially when protein synthesis is completely blocked. To further characterize the molecular mechanism of how the CSN regulates the conidiation rhythm, we focused on the SCFFWD-1 ubiquitin ligase, which controls the N. crassa circadian rhythm by ubiquitinating FRQ [36]. Our results demonstrated that defective FRQ degradation in the csn-5KO strain is due to the drastically reduced stability and levels of FWD-1 and Cul1 proteins in the SCFFWD-1 complex. Ectopic expression of mutant CSN-5 without deneddylation activity restored the defects of growth and conidiation in the csn-5KO strain, and almost restored the defects of the circadian conidiation rhythm in DD and FRQ degradation in the csn-5KO strain. Our data further showed that the low levels of FWD-1 in the csn-5KO strain were dramatically increased after expression of each of the CSN-5 proteins with point mutations in the JAMM metal-binding site, however, the increased stability and levels of the components in the SCFFWD-1 ubiquitin ligase are not enough to fully restore the degradation of FRQ to wild-type level, indicating that regulation of FRQ degradation plays a key role in maintaining the precise period length of conidiation rhythm in N. crassa. This is further supported by the finding that accelerated degradation of c-Jun in HeLa cells in which CSN-5 is downregulated can be rescued equally by over-expression of the deneddylation mutant CSN-5D151N or wild-type CSN-5; however, accelerated c-Jun degradation is not rescued in CSN-1 – or CSN-3–downregulated cells by over-expression of wild-type CSN-5 [20]. Furthermore, the degradation of EB1 (microtubule-end-binding protein 1) is accelerated by over-expression of wild-type CSN-5 or CSN-5D151N in HeLa cells [20]. These results suggest that the integrity of CSN might contribute more to regulating the stability of some substrates of CRLs. Current models suggest that the CRL complex is controlled by cycles of CRL deneddylation and CAND1 binding [7]. Our experiments further suggested that CAND1, a putative regulator of CRLs, is not required for maintenance of SCFFWD-1 ubiquitin ligase activity and circadian rhythm in N. crassa. These data provide additional evidence that the CSN is an important regulator of the circadian clock in N. crassa through maintenance of SCFFWD-1 ubiquitin ligase stability.
In conclusion, the results of our experiments indicate that even without deneddylation activity, the N. crassa CSN can still regulate hyphal growth, conidial development, and circadian function by regulating the activities of E3 ubiquitin ligases. Because the function of the CSN in the regulation of CRL activities is conserved in higher eukaryotes, we propose that the CSN may have a similar role in plants and animals.
Materials and Methods
Strains and culture conditions
The N. crassa strain 87-3 (bd, a) was used as the wild-type strain in this study. The bd ku70RIP strain, which was generated previously [54], was used as the host strain for creating the cand1 knockout mutants. We also used csn-5KO, csn-2KO and csn-5KO, his-3 strains that were generated previously [16]. The 301-6 (bd, his-3, A) strain and the csn-5KO, his-3 strain were used as the host strains for the his-3 targeting construct transformation [24]. Liquid culture conditions were the same as described previously [34]. For QA-induced protein expression, 0.01 M QA (pH 5.8) was added to liquid medium containing 1× Vogel's medium, 0.1% glucose, and 0.17% arginine. The medium for the race tube assay contained 1× Vogel's, 0.1% glucose, 0.17% arginine, 50 ng/mL biotin, and 1.5% agar [55]. For race tubes containing QA (10−3 M), glucose was excluded from the medium.
Plasmids
All three JAMM point mutations of CSN-5 were generated using the Quikchange Site-Directed Mutagenesis Kit (Stratagene). pUC19-CSN-5 was used as the template for mutagenesis. Afterwards, the mutated CSN-5 DNA fragments were subcloned into either the pqa-5Myc-6His or pqa-3Flag vectors. The triple point mutant CSN-5 (H127A, H129A and D140N) generated from pUC19-CSN-5 was subcloned into the endogenous csn-5 promoter-driven vector pcsn-5-Myc-His-CSN-5, resulting in pcsn-5-Myc-His-CSN-5tri. The previously constructed plasmids pqa-Myc-Cul1, pqa-Myc-His-Cul3, pqa-Myc-His-Cul4, pqa-Myc-His-CSN-6, pqa-Myc-His-SCON-2, pqa-Myc-His-FBP94, pqa-Myc-His-BTB1, and pqa-Myc-His-DCAF11 were also used for his-3 targeting transformation in the csn-5KO, his-3 and 301-6 (bd, his-3, A) strains [16] and cotransformation in the csn-5H127A, csn-5H129A, and csn-5D140N strains.
Generation of antiserum against Cul4
GST-Cul4 (containing Cul4 amino acids 1–113) fusion protein was expressed in RIL cells and the recombinant protein was purified and used as the antigen to generate rabbit polyclonal antiserum, as described previously [56].
Purification of Myc-His-CSN-5 and mutant Myc-His-CSN-5 proteins from N. crassa
The csn-5KO, Myc-His-CSN-5H127A, csn-5KO, Myc-His-CSN-5H129A, or csn-5KO, Myc-His-CSN-5D140N strain, wild-type strain (negative control), and csn-5KO, Myc-His-CSN-5 strain (positive control) were cultured for approximately 24 h in constant light (LL) in liquid medium containing QA (0.01 M QA, 1× Vogel's medium, 0.1% glucose, and 0.17% arginine). Approximately 10 g of tissue from each strain grown in LL was harvested. The purification procedure was the same as described previously [16]. Fractions containing purified Myc-tagged CSN proteins were immunoprecipitated by adding 25 µL of c-Myc monoclonal antibody-coupled agarose beads (9E10AC, Santa Cruz Biotechnology). The precipitates of each sample were analyzed by SDS-PAGE (4%–20% acrylamide), which was subsequently silver stained following the manufacturer's instructions (ProteoSilver Plus, Sigma). Specific bands in the Myc-His-CSN-5 purified products or in the Myc-His-CSN-5H127A purified products were excised and subjected to tryptic digestion and LC-MS/MS.
Gel filtration chromatography of Myc-His-CSN-5 or mutant CSN-5s in csn-5 mutant
The protocol of gel filtration chromatography was the same as described previously [16], [21]. Briefly, purified proteins (400 µg) were loaded onto a Superdex™ 200 (GE) gel filtration column that was equilibrated with 25 mL (150 mM NaCl, 20 mM Tris Cl pH 7.4). The proteins were eluted in the same buffer at a flow rate of 0.3 mL/min. Fractions of 0.4 mL were collected starting from the onset of the column void volume (8.0 mL) and finishing at 18 mL (25 fractions). 20 µL of each fraction were prepared in 20 µL of 2× SDS loading buffer, separated by 7.5% SDS-PAGE, and then examined by Western blot analysis using c-Myc antibody (9E10, Santa Cruz Biotechnology).
Protein analyses
Protein extraction, quantification, western blot analysis, protein degradation assays, and immunoprecipitation assays were performed as described previously [24], [56]. Western blot analyses using a monoclonal c-Myc antibody (9E10, Santa Cruz Biotechnology) or Flag antibody (F3165-5MG, Sigma) were performed to identify the positive transformants. Immunoprecipitates or equal amounts of total protein (40 µg) were loaded into each protein lane for SDS-PAGE. After electrophoresis, proteins were transferred onto a PVDF membrane, and western blot analysis was performed using c-Myc antibody, Flag antibody, FWD-1 antiserum, FRQ antiserum, or Cul4 antiserum.
Supporting Information
Zdroje
1. WeiNDengXW 2003 The COP9 signalosome. Annual review of cell and developmental biology 19 261 286
2. WeiNDengXW 1992 COP9: A new genetic locus involved in light-regulated development and gene expression in Arabidopsis. The Plant Cell Online 4 1507
3. CopeGADeshaiesRJ 2003 COP9 signalosome: a multifunctional regulator of SCF and other cullin-based ubiquitin ligases. Cell 114 663 671
4. SchwechheimerCSerinoGCallisJCrosbyWLLyapinaS 2001 Interactions of the COP9 signalosome with the E3 ubiquitin ligase SCFTIRI in mediating auxin response. Science 292 1379 1382
5. LyapinaSCopeGShevchenkoASerinoGTsugeT 2001 Promotion of NEDD-CUL1 conjugate cleavage by COP9 signalosome. Science 292 1382 1385
6. BosuDRKipreosET 2008 Cullin-RING ubiquitin ligases: global regulation and activation cycles. Cell Div 3 7
7. PetroskiMDDeshaiesRJ 2005 Function and regulation of cullin-RING ubiquitin ligases. Nat Rev Mol Cell Biol 6 9 20
8. HottonSKCallisJ 2008 Regulation of cullin RING ligases. Annu Rev Plant Biol 59 467 489
9. PickartCM 2001 Mechanisms underlying ubiquitination. Annu Rev Biochem 70 503 533
10. von ArnimAGSchwechheimerC 2006 Life Is Degrading–Thanks to Some Zomes. Molecular Cell 23 621 629
11. CopeGASuhGSAravindLSchwarzSEZipurskySL 2002 Role of predicted metalloprotease motif of Jab1/Csn5 in cleavage of Nedd8 from Cul1. Science 298 608 611
12. GusmaroliGFigueroaPSerinoGDengXW 2007 Role of the MPN subunits in COP9 signalosome assembly and activity, and their regulatory interaction with Arabidopsis Cullin3-based E3 ligases. Plant Cell 19 564 581
13. YangXMenonSLykke-AndersenKTsugeT 2002 The COP9 signalosome inhibits p27kip1 degradation and impedes G1-S phase progression via deneddylation of SCF Cul1. Current Biology 12 667 672
14. ZhouCWeeSRheeENaumannMDubielW 2003 Fission yeast COP9/signalosome suppresses cullin activity through recruitment of the deubiquitylating enzyme Ubp12p. Molecular Cell 11 927 938
15. GroismanRPolanowskaJKuraokaISawadaJSaijoM 2003 The ubiquitin ligase activity in the DDB2 and CSA complexes is differentially regulated by the COP9 signalosome in response to DNA damage. Cell 113 357 367
16. WangJHuQChenHZhouZLiW 2010 Role of individual subunits of the Neurospora crassa CSN complex in regulation of deneddylation and stability of cullin proteins. PLoS Genet 6 e1001232 doi:10.1371/journal.pgen.1001232
17. MundtKELiuCCarrAM 2002 Deletion mutants in COP9/signalosome subunits in fission yeast Schizosaccharomyces pombe display distinct phenotypes. Molecular biology of the cell 13 493
18. ZhouCSeibertVGeyerRRheeELyapinaS 2001 The fission yeast COP9/signalosome is involved in cullin modification by ubiquitin-related Ned8p. BMC biochemistry 2 7
19. SuhGSPoeckBChouardTOronESegalD 2002 Drosophila JAB1/CSN5 acts in photoreceptor cells to induce glial cells. Neuron 33 35 46
20. PethABerndtCHenkeWDubielW 2007 Downregulation of COP9 signalosome subunits differentially affects the CSN complex and target protein stability. BMC biochemistry 8 27
21. GusmaroliGFengSDengXW 2004 The Arabidopsis CSN5A and CSN5B subunits are present in distinct COP9 signalosome complexes, and mutations in their JAMM domains exhibit differential dominant negative effects on development. The Plant Cell Online 16 2984
22. BuschSEckertSEKrappmannSBrausGH 2003 The COP9 signalosome is an essential regulator of development in the filamentous fungus Aspergillus nidulans. Molecular microbiology 49 717 730
23. BuschSSchwierEUNahlikKBayramHelmstaedtK 2007 An eight-subunit COP9 signalosome with an intact JAMM motif is required for fungal fruit body formation. Proceedings of the National Academy of Sciences 104 8089
24. HeQChengPLiuY 2005 The COP9 signalosome regulates the Neurospora circadian clock by controlling the stability of the SCFFWD-1 complex. Genes Dev 19 1518 1531
25. WeeSGeyerRKTodaTWolfDA 2005 CSN facilitates Cullin-RING ubiquitin ligase function by counteracting autocatalytic adapter instability. Nat Cell Biol 7 387 391
26. CopeGADeshaiesRJ 2006 Targeted silencing of Jab1/Csn5 in human cells downregulates SCF activity through reduction of F-box protein levels. BMC biochemistry 7 1
27. DentiSFernandez-SanchezMERoggeLBianchiE 2006 The COP9 signalosome regulates Skp2 levels and proliferation of human cells. J Biol Chem 281 32188 32196
28. Luke-GlaserSRoyMLarsenBLe BihanTMetalnikovP 2007 CIF-1, a shared subunit of the COP9/signalosome and eukaryotic initiation factor 3 complexes, regulates MEL-26 levels in the Caenorhabditis elegans embryo. Mol Cell Biol 27 4526 4540
29. FischerESScrimaABohmKMatsumotoSLingarajuGM 2011 The molecular basis of CRL4DDB2/CSA ubiquitin ligase architecture, targeting, and activation. Cell 147 1024 1039
30. AmbroggioXIReesDCDeshaiesRJ 2004 JAMM: a metalloprotease-like zinc site in the proteasome and signalosome. PLoS Biol 2 e2 doi:10.1371/journal.pbio.0020002
31. KieferFArnoldKKunzliMBordoliLSchwedeT 2009 The SWISS-MODEL Repository and associated resources. Nucleic Acids Res 37 D387 392
32. TranHJAllenMDLoweJBycroftM 2003 Structure of the Jab1/MPN domain and its implications for proteasome function. Biochemistry 42 11460 11465
33. WuJTLinHCHuYCChienCT 2005 Neddylation and deneddylation regulate Cul1 and Cul3 protein accumulation. Nature cell biology 7 1014 1020
34. AronsonBDJohnsonKALorosJJDunlapJC 1994 Negative feedback defining a circadian clock: autoregulation of the clock gene frequency. Science 263 1578 1584
35. LiuYBell-PedersenD 2006 Circadian rhythms in Neurospora crassa and other filamentous fungi. Eukaryotic Cell 5 1184
36. HeQChengPYangYYuHLiuY 2003 FWD1-mediated degradation of FREQUENCY in Neurospora establishes a conserved mechanism for circadian clock regulation. The EMBO journal 22 4421 4430
37. SizemoreSTPaiettaJV 2002 Cloning and characterization of scon-3+, a new member of the Neurospora crassa sulfur regulatory system. Eukaryot Cell 1 875 883
38. XuHWangJHuQQuanYChenH 2010 DCAF26, an adaptor protein of Cul4-based E3, is essential for DNA methylation in Neurospora crassa. PLoS Genet 6 e1001132 doi:10.1371/journal.pgen.1001132
39. ZhengJYangXHarrellJMRyzhikovSShimEH 2002 CAND1 binds to unneddylated CUL1 and regulates the formation of SCF ubiquitin E3 ligase complex. Molecular Cell 10 1519 1526
40. FengSShenYSullivanJARubioVXiongY 2004 Arabidopsis CAND1, an unmodified CUL1-interacting protein, is involved in multiple developmental pathways controlled by ubiquitin/proteasome-mediated protein degradation. The Plant Cell Online 16 1870
41. SchmidtMWMcQuaryPRWeeSHofmannKWolfDA 2009 F-box-directed CRL complex assembly and regulation by the CSN and CAND1. Molecular Cell 35 586 597
42. GoldenbergSJCascioTCShumwaySDGarbuttKCLiuJ 2004 Structure of the Cand1-Cul1-Roc1 complex reveals regulatory mechanisms for the assembly of the multisubunit cullin-dependent ubiquitin ligases. Cell 119 517 528
43. LiuJFurukawaMMatsumotoTXiongY 2002 NEDD8 modification of CUL1 dissociates p120(CAND1), an inhibitor of CUL1-SKP1 binding and SCF ligases. Mol Cell 10 1511 1518
44. MinKWHwangJWLeeJSParkYTamuraTA 2003 TIP120A associates with cullins and modulates ubiquitin ligase activity. J Biol Chem 278 15905 15910
45. HwangJWMinKWTamuraTAYoonJB 2003 TIP120A associates with unneddylated cullin 1 and regulates its neddylation. FEBS Lett 541 102 108
46. OshikawaKMatsumotoMYadaMKamuraTHatakeyamaS 2003 Preferential interaction of TIP120A with Cul1 that is not modified by NEDD8 and not associated with Skp1. Biochem Biophys Res Commun 303 1209 1216
47. BosuDKipreosE 2008 Cullin-RING ubiquitin ligases: global regulation and activation cycles. Cell division 3 7
48. HottonSKCallisJ 2008 Regulation of cullin RING ligases. Annu Rev Plant Biol 59 467 489
49. ChuangHWZhangWGrayWM 2004 Arabidopsis ETA2, an apparent ortholog of the human cullin-interacting protein CAND1, is required for auxin responses mediated by the SCF(TIR1) ubiquitin ligase. Plant Cell 16 1883 1897
50. LeeJESweredoskiMJGrahamRLJKolawaNJSmithGT 2011 The steady-state repertoire of human SCF Ubiquitin ligase complexes does not require ongoing Nedd8 conjugation. Molecular & Cellular Proteomics 10
51. BennettEJRushJGygiSPHarperJW 2010 Dynamics of cullin-RING ubiquitin ligase network revealed by systematic quantitative proteomics. Cell 143 951 965
52. WangXKangDFengSSerinoGSchwechheimerC 2002 CSN1 N-terminal-dependent activity is required for Arabidopsis development but not for Rub1/Nedd8 deconjugation of cullins: A structure-function study of CSN1 subunit of COP9 signalosome. Molecular biology of the cell 13 646
53. HetfeldBKHelfrichAKapelariBScheelHHofmannK 2005 The zinc finger of the CSN-associated deubiquitinating enzyme USP15 is essential to rescue the E3 ligase Rbx1. Curr Biol 15 1217 1221
54. HeQChaJLeeHCYangYLiuY 2006 CKI and CKII mediate the FREQUENCY-dependent phosphorylation of the WHITE COLLAR complex to close the Neurospora circadian negative feedback loop. Genes Dev 20 2552 2565
55. ChengPYangYLiuY 2001 Interlocked feedback loops contribute to the robustness of the Neurospora circadian clock. Proceedings of the National Academy of Sciences 98 7408
56. ZhaoYShenYYangSWangJHuQ 2010 Ubiquitin ligase components Cullin4 and DDB1 are essential for DNA methylation in Neurospora crassa. J Biol Chem 285 4355 4365
Štítky
Genetika Reprodukční medicína
Článek Functional Centromeres Determine the Activation Time of Pericentric Origins of DNA Replication inČlánek Dynamic Deposition of Histone Variant H3.3 Accompanies Developmental Remodeling of the TranscriptomeČlánek Integrin α PAT-2/CDC-42 Signaling Is Required for Muscle-Mediated Clearance of Apoptotic Cells inČlánek Prdm5 Regulates Collagen Gene Transcription by Association with RNA Polymerase II in Developing BoneČlánek Acquisition Order of Ras and p53 Gene Alterations Defines Distinct Adrenocortical Tumor Phenotypes
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2012 Číslo 5
-
Všechny články tohoto čísla
- Slowing Replication in Preparation for Reduction
- Chromosome Pairing: A Hidden Treasure No More
- Loss of Imprinting Differentially Affects REM/NREM Sleep and Cognition in Mice
- Six Novel Susceptibility Loci for Early-Onset Androgenetic Alopecia and Their Unexpected Association with Common Diseases
- Regulation by the Noncoding RNA
- UDP-Galactose 4′-Epimerase Activities toward UDP-Gal and UDP-GalNAc Play Different Roles in the Development of
- Deletion of PTH Rescues Skeletal Abnormalities and High Osteopontin Levels in Mice
- Karyotypic Determinants of Chromosome Instability in Aneuploid Budding Yeast
- Genome-Wide Copy Number Analysis Uncovers a New HSCR Gene:
- MicroRNA-277 Modulates the Neurodegeneration Caused by Fragile X Premutation rCGG Repeats
- Functional Centromeres Determine the Activation Time of Pericentric Origins of DNA Replication in
- Dynamic Deposition of Histone Variant H3.3 Accompanies Developmental Remodeling of the Transcriptome
- Scientist Citizen: An Interview with Bruce Alberts
- YY1 Regulates Melanocyte Development and Function by Cooperating with MITF
- Congenital Heart Disease–Causing Gata4 Mutation Displays Functional Deficits
- Recombination Drives Vertebrate Genome Contraction
- KATNAL1 Regulation of Sertoli Cell Microtubule Dynamics Is Essential for Spermiogenesis and Male Fertility
- Re-Patterning Sleep Architecture in through Gustatory Perception and Nutritional Quality
- Using Whole-Genome Sequence Data to Predict Quantitative Trait Phenotypes in
- Genome-Wide Analysis of GLD-1–Mediated mRNA Regulation Suggests a Role in mRNA Storage
- Meiotic Chromosome Pairing Is Promoted by Telomere-Led Chromosome Movements Independent of Bouquet Formation
- LINT, a Novel dL(3)mbt-Containing Complex, Represses Malignant Brain Tumour Signature Genes
- The H3K27 Demethylase UTX-1 Is Essential for Normal Development, Independent of Its Enzymatic Activity
- Suppresses Senescence Programs and Thereby Accelerates and Maintains Mutant -Induced Lung Tumorigenesis
- Genome-Wide Association of Pericardial Fat Identifies a Unique Locus for Ectopic Fat
- An Essential Role for Katanin p80 and Microtubule Severing in Male Gamete Production
- Identification of Genes That Promote or Antagonize Somatic Homolog Pairing Using a High-Throughput FISH–Based Screen
- Principles of Carbon Catabolite Repression in the Rice Blast Fungus: Tps1, Nmr1-3, and a MATE–Family Pump Regulate Glucose Metabolism during Infection
- Integrin α PAT-2/CDC-42 Signaling Is Required for Muscle-Mediated Clearance of Apoptotic Cells in
- Histone H3 Localizes to the Centromeric DNA in Budding Yeast
- Collapse of Telomere Homeostasis in Hematopoietic Cells Caused by Heterozygous Mutations in Telomerase Genes
- Hypersensitive to Red and Blue 1 and Its Modification by Protein Phosphatase 7 Are Implicated in the Control of Arabidopsis Stomatal Aperture
- Extent, Causes, and Consequences of Small RNA Expression Variation in Human Adipose Tissue
- TBC-8, a Putative RAB-2 GAP, Regulates Dense Core Vesicle Maturation in
- Regulating Repression: Roles for the Sir4 N-Terminus in Linker DNA Protection and Stabilization of Epigenetic States
- Common Genetic Determinants of Intraocular Pressure and Primary Open-Angle Glaucoma
- Prdm5 Regulates Collagen Gene Transcription by Association with RNA Polymerase II in Developing Bone
- Fitness Landscape Transformation through a Single Amino Acid Change in the Rho Terminator
- Repeated, Selection-Driven Genome Reduction of Accessory Genes in Experimental Populations
- Allelic Variation and Differential Expression of the mSIN3A Histone Deacetylase Complex Gene Promote Mammary Tumor Growth and Metastasis
- DNA Demethylation and USF Regulate the Meiosis-Specific Expression of the Mouse
- Knowledge-Driven Analysis Identifies a Gene–Gene Interaction Affecting High-Density Lipoprotein Cholesterol Levels in Multi-Ethnic Populations
- A Duplication CNV That Conveys Traits Reciprocal to Metabolic Syndrome and Protects against Diet-Induced Obesity in Mice and Men
- EMT Inducers Catalyze Malignant Transformation of Mammary Epithelial Cells and Drive Tumorigenesis towards Claudin-Low Tumors in Transgenic Mice
- Inactivation of a Novel FGF23 Regulator, FAM20C, Leads to Hypophosphatemic Rickets in Mice
- Genome-Wide Association for Abdominal Subcutaneous and Visceral Adipose Reveals a Novel Locus for Visceral Fat in Women
- Stratifying Type 2 Diabetes Cases by BMI Identifies Genetic Risk Variants in and Enrichment for Risk Variants in Lean Compared to Obese Cases
- New Insight into the History of Domesticated Apple: Secondary Contribution of the European Wild Apple to the Genome of Cultivated Varieties
- Activated Cdc42 Kinase Has an Anti-Apoptotic Function
- The Region Is Critical for Birth Defects and Electrocardiographic Dysfunctions Observed in a Down Syndrome Mouse Model
- COP9 Signalosome Integrity Plays Major Roles for Hyphal Growth, Conidial Development, and Circadian Function
- Bmps and Id2a Act Upstream of Twist1 To Restrict Ectomesenchyme Potential of the Cranial Neural Crest
- Psip1/Ledgf p52 Binds Methylated Histone H3K36 and Splicing Factors and Contributes to the Regulation of Alternative Splicing
- The Number of X Chromosomes Causes Sex Differences in Adiposity in Mice
- Target Gene Analysis by Microarrays and Chromatin Immunoprecipitation Identifies HEY Proteins as Highly Redundant bHLH Repressors
- Acquisition Order of Ras and p53 Gene Alterations Defines Distinct Adrenocortical Tumor Phenotypes
- ELK1 Uses Different DNA Binding Modes to Regulate Functionally Distinct Classes of Target Genes
- Histone H1 Depletion Impairs Embryonic Stem Cell Differentiation
- IDN2 and Its Paralogs Form a Complex Required for RNA–Directed DNA Methylation
- Separation of DNA Replication from the Assembly of Break-Competent Meiotic Chromosomes
- Genomic Hypomethylation in the Human Germline Associates with Selective Structural Mutability in the Human Genome
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Inactivation of a Novel FGF23 Regulator, FAM20C, Leads to Hypophosphatemic Rickets in Mice
- Genome-Wide Association of Pericardial Fat Identifies a Unique Locus for Ectopic Fat
- Slowing Replication in Preparation for Reduction
- An Essential Role for Katanin p80 and Microtubule Severing in Male Gamete Production
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



