-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaSix Novel Susceptibility Loci for Early-Onset Androgenetic Alopecia and Their Unexpected Association with Common Diseases
Androgenetic alopecia (AGA) is a highly heritable condition and the most common form of hair loss in humans. Susceptibility loci have been described on the X chromosome and chromosome 20, but these loci explain a minority of its heritable variance. We conducted a large-scale meta-analysis of seven genome-wide association studies for early-onset AGA in 12,806 individuals of European ancestry. While replicating the two AGA loci on the X chromosome and chromosome 20, six novel susceptibility loci reached genome-wide significance (p = 2.62×10−9–1.01×10−12). Unexpectedly, we identified a risk allele at 17q21.31 that was recently associated with Parkinson's disease (PD) at a genome-wide significant level. We then tested the association between early-onset AGA and the risk of PD in a cross-sectional analysis of 568 PD cases and 7,664 controls. Early-onset AGA cases had significantly increased odds of subsequent PD (OR = 1.28, 95% confidence interval: 1.06–1.55, p = 8.9×10−3). Further, the AGA susceptibility alleles at the 17q21.31 locus are on the H1 haplotype, which is under negative selection in Europeans and has been linked to decreased fertility. Combining the risk alleles of six novel and two established susceptibility loci, we created a genotype risk score and tested its association with AGA in an additional sample. Individuals in the highest risk quartile of a genotype score had an approximately six-fold increased risk of early-onset AGA [odds ratio (OR) = 5.78, p = 1.4×10−88]. Our results highlight unexpected associations between early-onset AGA, Parkinson's disease, and decreased fertility, providing important insights into the pathophysiology of these conditions.
Published in the journal: . PLoS Genet 8(5): e32767. doi:10.1371/journal.pgen.1002746
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1002746Summary
Androgenetic alopecia (AGA) is a highly heritable condition and the most common form of hair loss in humans. Susceptibility loci have been described on the X chromosome and chromosome 20, but these loci explain a minority of its heritable variance. We conducted a large-scale meta-analysis of seven genome-wide association studies for early-onset AGA in 12,806 individuals of European ancestry. While replicating the two AGA loci on the X chromosome and chromosome 20, six novel susceptibility loci reached genome-wide significance (p = 2.62×10−9–1.01×10−12). Unexpectedly, we identified a risk allele at 17q21.31 that was recently associated with Parkinson's disease (PD) at a genome-wide significant level. We then tested the association between early-onset AGA and the risk of PD in a cross-sectional analysis of 568 PD cases and 7,664 controls. Early-onset AGA cases had significantly increased odds of subsequent PD (OR = 1.28, 95% confidence interval: 1.06–1.55, p = 8.9×10−3). Further, the AGA susceptibility alleles at the 17q21.31 locus are on the H1 haplotype, which is under negative selection in Europeans and has been linked to decreased fertility. Combining the risk alleles of six novel and two established susceptibility loci, we created a genotype risk score and tested its association with AGA in an additional sample. Individuals in the highest risk quartile of a genotype score had an approximately six-fold increased risk of early-onset AGA [odds ratio (OR) = 5.78, p = 1.4×10−88]. Our results highlight unexpected associations between early-onset AGA, Parkinson's disease, and decreased fertility, providing important insights into the pathophysiology of these conditions.
Introduction
A main advantage of genome-wide association (GWA) studies is that their hypothesis-free scan of the genome for genes associated with common disease enables identification of novel associations between different diseases and pathways. For example, a pioneering GWA study demonstrated that susceptibility alleles for age-related macular degeneration were found in the complement pathway [1], giving rise to novel insights into the etiology and treatment of this condition [2].
Through previous GWA studies, we have identified genetic determinants of androgenetic alopecia (AGA, male-pattern baldness) that highlight the importance of the androgen pathway in this condition [3], [4]. However, these studies did not identify readily apparent novel pathways that link AGA to other conditions.
AGA is the most common form of hair loss in humans, affecting 80% of men by age 80 [5]. Its etiologic factors are androgen dependency and genetic predisposition [5]. While largely a cosmetic condition, the mechanisms influencing its etiology may also impact upon important medical conditions such as coronary heart disease, metabolic syndrome, and prostate cancer [6], [7].
Prostate cancer is the most frequently diagnosed cancer and ranks second as a cancer killer among men in the United States [8]. Androgens play a key role in stimulation of normal prostate growth and are essential in prostate cancer initiation and progression [9]. The main genetic determinant of AGA is the androgen receptor (AR) [3], [4] and prostate cancer susceptibility loci identified through recent GWA studies overlap with androgen receptor binding sites [10], [11] demonstrating a shared etiologic factor in these two conditions. However, while evidence suggests that AGA and prostate cancer are strongly influenced by androgen sensitivity [12], [13], attempts to examine the relationship between AGA and prostate cancer through case-control studies have yielded inconsistent results [7] [14]–[18].
Since previous GWA studies for AGA explained only 13.7% of the variance in this condition [3] we aimed to identify novel determinants of AGA and test their association with common diseases, by undertaking a large-scale meta-analysis of GWA studies involving 12,806 Europeans from seven cohorts included in the Meta-Analysis for Androgenetic Alopecia Novel Determinants (MAAN) consortium.
Results/Discussion
The current analysis comprised 3,891 cases and 8,915 controls of European ancestry from seven independent studies: Bonn, CoLaus, TwinsUK, Nijmegen Biomedical, 23andMe, Icelandic, and an Australian population based twin study (Table 1). Briefly, we used an extreme discordant case-control design to contrast individuals with early-onset AGA to older individuals without alopecia as assessed by questionnaire, clinical visit or photographs evaluated by a dermatologist, where available. Genome-wide genotyping, using standard platforms, and imputation using the CEU panel of Phase II HapMap were performed. After the quality control criteria were applied, 2,391,230 SNPs remained for genotype-phenotype association analysis.
Tab. 1. Demographic properties of the study subjects in participant studies. 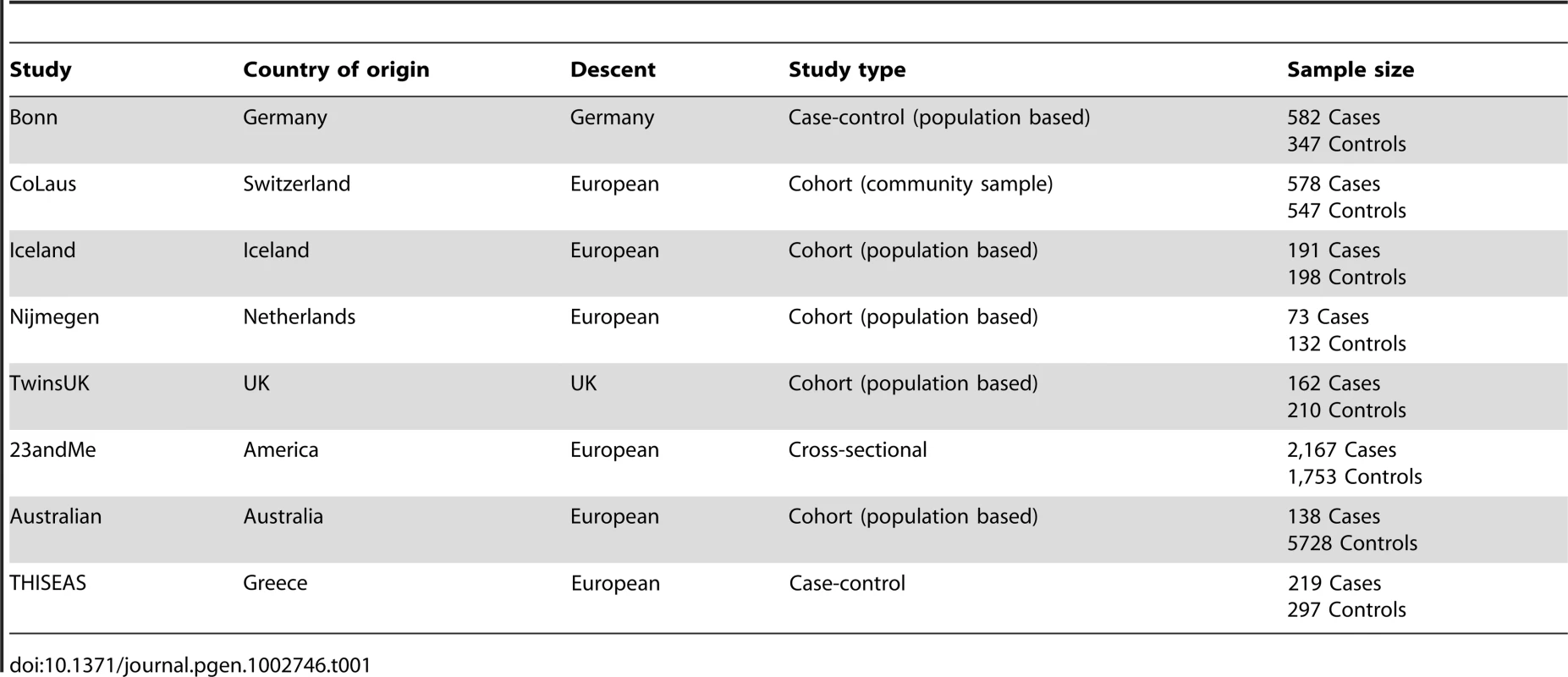
The sample size of the current meta-analysis (n = 12,806) was more than fourfold that of the earlier GWA studies of AGA [3], [4]. The genomic inflation factors of the individual studies (Table S1) and overall meta-analysis (λgc = 1.02) were low, indicating that the observed GWA results were not due to population stratification. The fixed-effect meta-analytic data from all seven cohorts demonstrated a substantial excess of significant associations with AGA at the tail of the QQ plot (Figure S1). We identified a total of 645 SNPs that achieved genome-wide significance (p<5×10−8, Table 2, Table S2). The fixed-effect meta-analytic results showed no evidence of substantial heterogeneity across populations (Tables S1 and S2).
Tab. 2. Summary result for the lead SNP from the genome-wide significant loci. 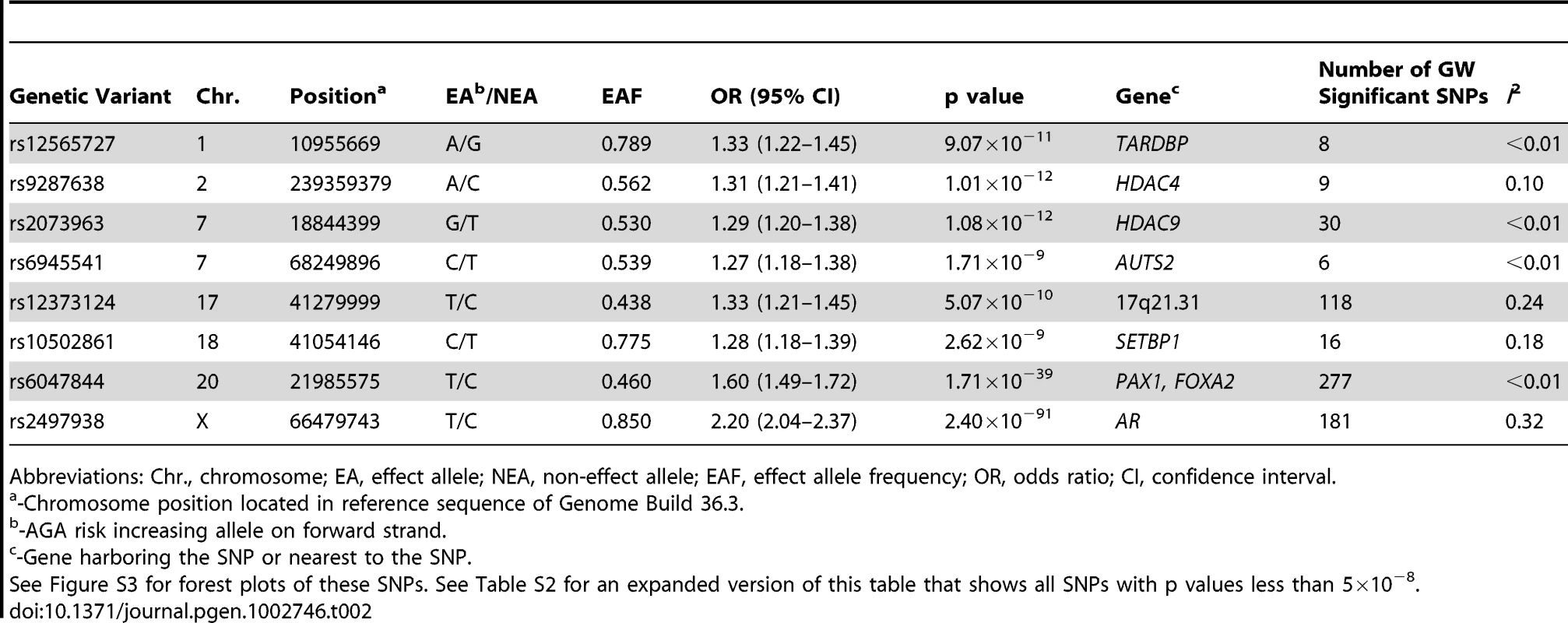
Abbreviations: Chr., chromosome; EA, effect allele; NEA, non-effect allele; EAF, effect allele frequency; OR, odds ratio; CI, confidence interval. Consistent with our previous reports [3], [4], a susceptibility locus for AGA on chromosome 20p11 was confirmed over a ∼253 kb interval with the strongest signal arising at rs6047844 (p = 1.71×10−39, odds ratio (OR) = 1.60, 95% confidence interval (CI) = 1.49–1.72) (Figure 1). We also showed confirmed association with AGA at AR gene by highly significant signal spanning the gene region (OR = 2.20, 95% CI = 2.04–2.37, p = 2.40×10−91 for risk allele T of top SNP rs2497938). Even after removing SNPs in these two established AGA loci, the QQ-plot demonstrated an excess of SNPs associated with AGA (Figure S2).
Fig. 1. Genome-wide meta-analysis results for AGA in MAAN. 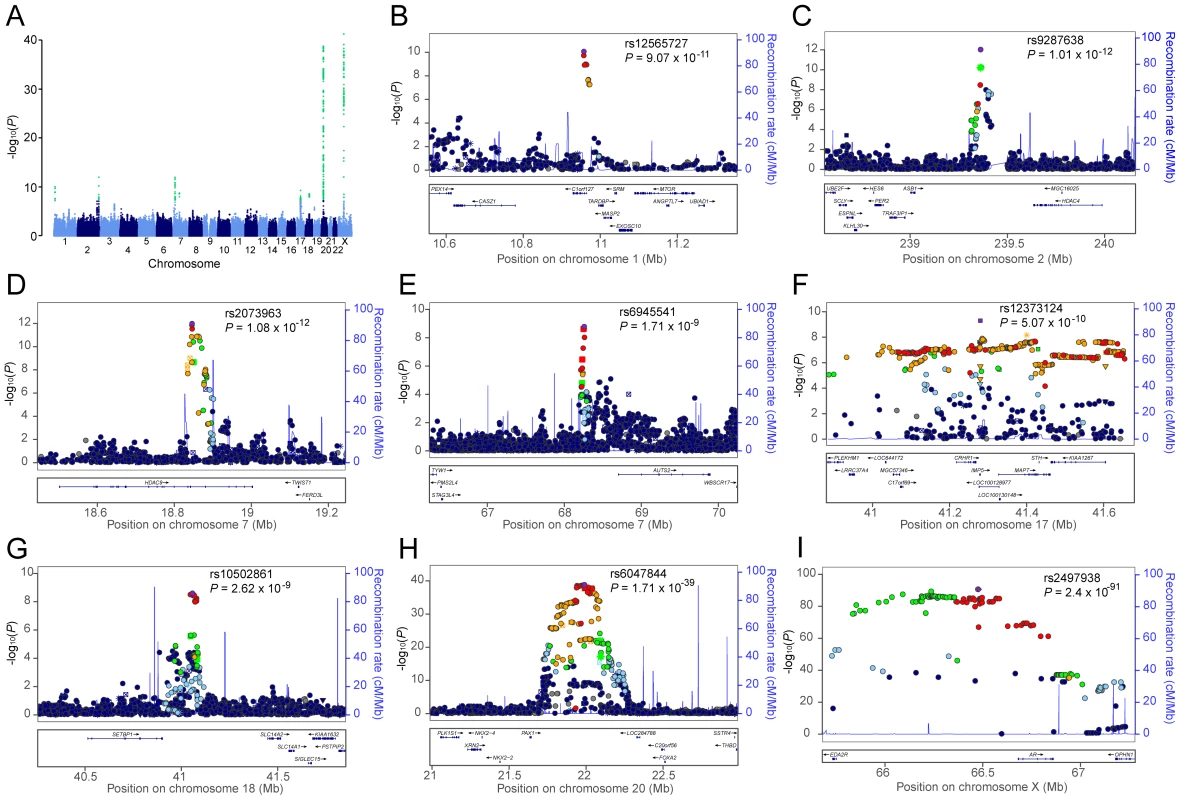
(A) Manhattan plot showing the −log10 p value of SNPs against their chromosomal positions. The genome-wide significant SNPs are green (p value<5×10−8). The points with p value <1×10−40 were truncated; the smallest p value was 2.4×10−91 at AR gene. (B–I) Regional association plots for eight loci associated with AGA. In each panel, the lead SNP is denoted in purple with its rs ID and association p value. The color of other SNPs indicates the LD with the lead SNP as red (0.8≤r2≤1), orange (0.6≤r2<0.8), green (0.4≤r2<0.6), light blue (0.2≤r2<0.4), and dark blue (r2<0.2). Estimated recombination rates are in light blue. FOXA2 (forkhead box A2) is worth noting among the flanking genes at the chromosome 20 locus. Foxa2, the transcription factor encoded by FOXA2 in mouse, interacts with AR, especially through DNA binding domain, to regulate gene expression [19]. It is expressed in prostate tissue and plays a pivotal role in neuroendocrine prostate tumors, a form of metastatic prostate tumors, by inducing development from androgen-dependent tumors to androgen-independent tumors [20]–[22]. Its expression was associated with the invasive phenotype in the primary prostate cancer [22]. Furthermore, the association of metastatic prostate cancer and AGA through AR polymorphisms was previously described in a well-defined case-control study [14].
Multiple SNPs in strong linkage disequilibrium (LD) (r2>0·6) revealed a newly discovered association interval encompassing several genes on chromosome 17q21.31. The most significant signal of this region was detected for the synonymous (His649His) SNP rs12373124 (P = 5.07×10−10, OR = 1.33, 95%CI = 1.21–1.45 for risk allele T) at the gene IMP5 (intramembrane protease 5). However, as the genome-wide significant SNPs are highly-correlated with each other in this region, the most proximal gene is not necessarily the gene functionally affected by causal SNPs. The MAPT (microtubule-associated protein tau) gene, which encodes the tau protein, is of particular interest in this region because its expression was detected in hair follicles (p = 9.22×10−3) but not in the other tissues examined in our tissue expression analysis (Table S3).
The SNP with the smallest p value, apart from the AGA locus on chromosome 20, (rs9287638 [A]: OR = 1.31, 95% CI = 1.21–1.41, P = 1.01×10−12), is located on chromosome 2q37, with seven other genome-wide significant SNPs in the same region. These SNPs lie 558 kb downstream of HDAC4 (histone deacetylase 4). The next locus mapped within HDAC9 (histone deacetylase 9) on chromosome 7p21.1 (rs2073963 [G]: OR = 1.29, 95% CI = 1.20–1.38, P = 1.08×10−12). Our expression analysis revealed that HDAC9 and HDAC4 were well expressed in hair follicles (p = 6.59×10−3 and 2.64×10−3 respectively), and HDAC9 was not expressed in skin or scalp tissues (Table S3).
Thus, two independent signals arose from HDACs (HDAC4 and HDAC9) which act as transcriptional corepressors by deacetylating nucleosomal histones [23]. By interaction with transcription factors ARR19 and CRIF1, HDAC4 plays a critical role in inhibition of AR transactivation [24], [25] and its accumulation coincides with loss of androgen sensitivity in prostate cancer [26]. Furthermore, HDAC9 and HDAC4 share conserved residues and their tissue specific expression pattern overlaps [27]. Very recently, ZNF652 which has been shown to be involved in transcriptional repression effect of HDACs [28] was identified to be a prostate cancer susceptibility locus [29]. All together, our findings give rise to the possibility that HDAC4 and HDAC9 might influence pathogenesis of AGA through dysregulation of the androgen pathway, highlighting a shared etiologic factor in this condition and prostate cancer.
The AUTS2 (autism susceptibility candidate 2) gene was also identified (lead SNP rs6945541[C]: OR = 1.27, 95%CI = 1.18–1.38, p = 1.71×10−9) at the chromosome 7q11.22 locus. The mechanism underlying the association between the AUTS2 locus and AGA is currently unknown, but its expression profile reveals abundance of transcript in hair, skin, and scalp (p = 2.64×10−3, <1.00×10−3, and <1.00×10−3 respectively, Table S3), but not blood. Additionally, variants at AUTS2 were recently associated with the regulation of alcohol consumption through a GWA study [30]. The function of AUTS2 is currently unknown. It was previously associated with autism and mental retardation [30]. How the AUTS2 variants affect AGA outcomes deserves further investigation.
Remaining novel findings include one locus on chromosome 1 which is near the genes TARDBP (TAR DNA binding protein), PEX14 (peroxisomal biogenesis factor 14), MASP2 (mannan-binding lectin serine peptidase 2), and SRM (spermidine synthase) (lowest p = 9.07×10−11 for SNP rs12565727 [A], OR = 1.33, 95%CI = 1.22–1.45) and another locus on chromosome 18q21.1, near the 3′ end of SETBP1 (SET binding protein 1), of which the most significant was rs10502861 [C] (P = 2.62×10−9, OR = 1.28, 95%CI = 1.18–1.39). SETBP1 again demonstrated expression in hair, skin, and scalp (detection p values are 2.64×10−3, <1.00×10−3, and <1.00×10−3 respectively, Table S3), but not blood.
Regional association plots for seven loci associated with AGA are shown in Figure 1. Forest plots of the top SNPs are shown in Figure S3. All these loci were associated with AGA in random-effect results except the locus on chromosome 18 (Table S4).
While any of the above susceptibility loci may impart a small risk, examining the combined effect of these loci in individuals harboring more than one risk allele may improve the ability to identify individuals at high risk of AGA. Using the top SNPs identified in this study from both the 6 novel loci and the 2 confirmed regions on chromosome 20p11 and AR gene, we constructed a genotype risk score based on the weighted number of susceptibility alleles in an independent replication sample from 23andMe study. As shown in Table 3, there was an increased risk for AGA across each quartile of the genotypic risk score. For the individuals with a genotype score in the highest quartile, we observed substantially increased odds for AGA (OR = 5.78, 95% CI = 4.86–6.87, p = 1.4×10−88), compared to individuals at the lowest quartile.
Tab. 3. Genotype score associated with the risk of androgenetic alopecia. 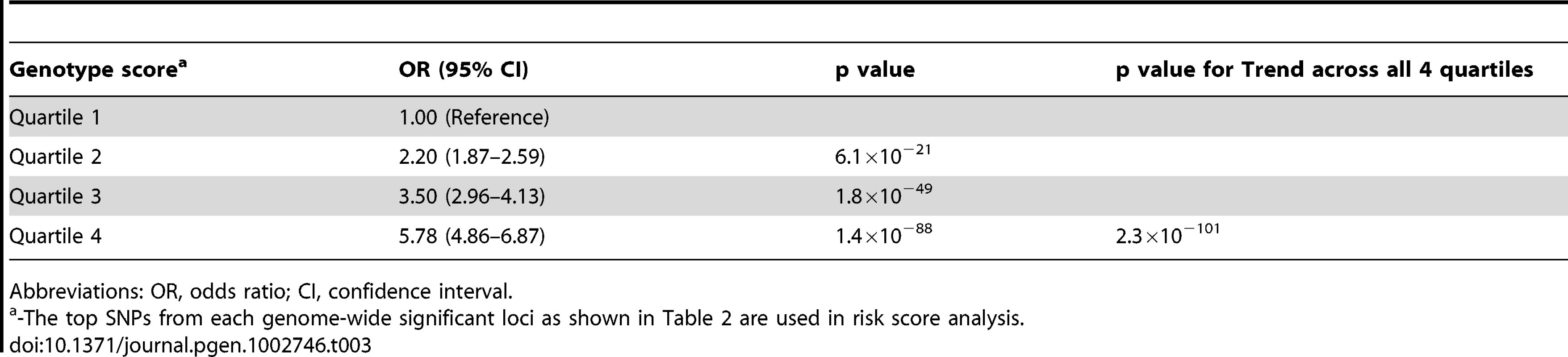
Abbreviations: OR, odds ratio; CI, confidence interval. Reasoning that AR gene is replicated as the most substantial genetic component of AGA, we explored the relationship between AR gene and other AGA susceptibility loci by several complimentary approaches. First, we examined the effect of AR gene on other AGA loci by adding an interaction term in the logistic regression model. We found no evidence for interaction (Table S5), which is consistent with our previous reports [4], [31]. Second, we sought clues of interaction between AR gene and other AGA loci mediated by AR binding sites since AR functions as a transcription factor. Although a large number of genes were found to be targets of AR, no AR binding sites have been identified in any of the candidate genes at AGA loci [32], [33]. However, as aforementioned, among these candidate genes, interaction with AR and consequent regulation on an epididymis-specific gene has been shown on FOXA2 (chromosome 20 locus), and the candidate gene at chromosome 2 locus, HDAC4, was reported to act as an inhibitor of AR, especially in prostate cancer cells, suggesting that these two loci may have a role in androgen-dependent pathway.
We identified that the 17q21.31 locus overlaps with a widely replicated locus that is strongly associated with Parkinson's disease [34]–[38]. Interestingly, MAPT on this locus contained identical genome-wide significant risk alleles that were shared between AGA (rs2942168 [G], [OR = 1.25, 95% CI = 1.15–1.36, p = 1.95×10−7]) and Parkinson's disease (OR = 1.32, 95%CI = 1.23–1.39, p = 1.62×10−18) reported in a recently published large GWA study [38]. The lead AGA SNP (rs12373124) at the 17q21.31 locus is in high linkage disequilibrium (r2 = 0·87) with SNPs that have been associated with Parkinson's disease in recent GWA studies [34], [35].
To explore this unexpected relationship between Parkinson's disease and early-onset AGA we next sought to understand whether AGA itself was a risk factor for Parkinson's disease. From among a superset of the early-onset AGA cases and controls using the same definition for AGA as in the meta-analysis, we identified 568 self-reported physician diagnosed Parkinson's disease cases, and 7,664 population controls [39]. This definition of Parkinson's disease has been used previously to replicate genetic loci for this condition in the 23andMe cohort [39]. We found that AGA cases had significantly higher odds of Parkinson's disease (OR = 1.28, 95% CI = 1.06–1.55, p = 8.9×10−3). Restricting the analysis to 714 individuals with current age 70 or higher, the association was stronger (OR = 1.94, 95% CI = 1.31–2.88, p = 6.5×10−4).
To investigate whether the shared genetic association at 17q21.31 explained the association between Parkinson's disease and AGA, we evaluated a regression model with an additional term for an individual's genotype at rs12185268, which is the variant in this region most strongly associated with Parkinson's disease in the 23andMe study. The OR for association with early-onset AGA and its significance were essentially unchanged (OR = 1.96, 95% CI = 1.32–2.90, p = 6.0×10−4). In the individuals in the age 70 or higher group, we also tested the association between Parkinson's disease and AGA stratified by rs12185268 genotypes. The AG and GG genotypes are pooled due to the small proportion of GG homozygotes. There is no essential difference between the odds ratios for individuals with rs12185268 AA genotypes (OR = 1.93, 95% CI = 1.16–3.21, p = 8.7×10−3) and AG+GG genotypes (OR = 2.07, 95% CI = 1.09–3.93, p = 2.2×10−2), indicating that rs12185268 does not modify the association between Parkinson's disease and early-onset AGA.
We next looked for evidence that any other loci associated with Parkinson's disease were also associated with early-onset AGA, or vice versa. We identified the lead SNPs from 27 loci with p<1.0×10−5 for association with AGA, and 31 loci with p<1.0×10−5 for association with Parkinson's disease. We tested the AGA loci for association with Parkinson's disease, and the Parkinson's disease loci for association with AGA. The 17q21.31 locus was the only locus demonstrating convincing evidence for association across both phenotypes (Tables S6 and S7).
Parkinson's disease is the second most common neurodegenerative disorder with a prevalence of one percent in individuals that are over 60 years old [40]. Despite the often-reported higher prevalence of Parkinson's disease in men, as compared to women [40], there are no previous reports investigating the relationship between AGA and Parkinson's disease. This novel association between Parkinson's disease and early-onset AGA indicated that there could be a shared genetic or environmental cause for both conditions.
Our data specifically identify genetic variation in the 17q21.31 region as shared genetic risk factors for these two conditions. To date, only drug-induced hair loss has been described in patients having Parkinson's disease after use of dopamine agonist [41], although most of the patients affected by drug-induced hair loss are females [41]. As noted above, a greater incidence of Parkinson's disease has been reported in elderly men than in women [40] and androgen mediated neurotoxicity has been proposed to contribute to the gender bias in Parkinson's disease [42]. Since this association is entirely novel, it is unlikely that our results have arisen due to recall bias. In addition, the AGA cases in 23andMe study had an age of onset less than 40 years old. The mean age at diagnosis of Parkinson's disease is 70.5 years [43]. Therefore it is highly unlikely that Parkinson's disease occurred before AGA, as defined in the 23andMe study. At present we are unaware of any prospective Parkinson's disease cohorts having collected AGA data to further explore this relationship, and our evidence provides rationale to undertake such studies.
Furthermore, the 17q21.31 locus harbors an inversion polymorphism that has previously been described to be under negative selection pressure and has been demonstrated to be associated with decreased fertility in women [44]. We found a genome-wide significant SNP for early-onset AGA, whose risk allele (rs1800547 [A], p = 2.85×10−8, OR = 1.27, 95%CI = 1.16–1.39) represents the H1 haplotype of the 17q21.31 inversion. This H1 haplotype is under negative selection pressure in Europeans [45] and Icelandic female carriers of the H1 lineage have fewer children than non-carriers, while men sharing this haplotype have a trend towards decreased fertility [44]. We note that previous studies have identified an association between polycystic ovarian syndrome (PCOS), which is the most common cause of anovulatory infertility in women [46], and early-onset AGA in their male relatives [47], [48]. And increased androgen levels strongly affect both traits. Our findings, therefore, provide rationale to explore the androgen pathway as a possible explanation for the decreased fertility associated with the H1 haplotype in women.
Even though the definition of hair loss and the method of sampling differed between the eight study groups, the associations of five genome-wide significant loci were essentially identical in fixed - and random-effects analysis, suggesting that our results are unlikely to be influenced by heterogeneity.
We are aware that some other diseases, including progressive supranuclear palsy, corticobasal degeneration, frontotemporal dementia and Pick disease are also strongly associated with 17q21.31 region [49], [50]. However it is difficult to test the association between these diseases and AGA. In this study, we focused on the early-onset AGA. The AGA cases we recruited are young. In addition, due to the low prevalence (between 6.4 and 15 per 100,000 [51]–[53]) of these diseases compared to Parkinson's disease, we do not have samples to test the association.
Since 17q21.31 locus has been recognized as a risk factor for Parkinson's disease, it is plausible that an individual's status of Parkinson's disease may have an effect on the association between 17q21.31 locus and early-onset AGA. We undertook an association analysis conditioned on Parkinson's disease in 8,232 individuals for whom both phenotype are available and confirmed that status of Parkinson's disease does not affect the strength of association between AGA and rs12185268, the top variant at 17q21.31 locus in this sample (unconditioned: OR = 0.76, 95% CI = 0.70–0.82, p = 7.9×10−13; conditioned: OR = 0.76, 95% CI = 0.70–0.82, p = 7.9×10−13). This result was verified within strata of Parkinson's disease: OR is 0.69 in the cases (95% CI = 0.51–0.94, p = 1.8×10−2) and 0.76 for individuals without Parkinson's disease (95% CI = 0.71–0.83, p = 1.1×10−11). Further, the relationship between the 17q21.31 locus and Parkinson's disease was not affected by conditioning on AGA status although the association with rs12185268 was not significant (unconditioned: OR = 0.99, 95% CI = 0.84–1.15, p = 0.86; conditioned: OR = 1.00, 95% CI = 0.86–1.17, p = 0.97).
Our data demonstrated several aspects of the relationship between AGA and Parkinson's disease: First, variants at 17q21.31 locus including the MAPT gene are associated with both risk of Parkinson's disease (p = 2.8×10−12) and early onset AGA (p = 9.3×10−8) as shown in Table S6 and S7. Second, early onset AGA (age of onset <40 years), is a risk factor for Parkinson's disease (p = 8.9×10−3). Third, controlling for variation at rs12185268, the top SNP at 17q21.31 locus, does not eliminate the relationship between AGA and Parkinson's disease. And fourth, the other genetic determinants of AGA, as described through GWAS, do not influence risk of Parkinson's disease and vice versa. Given that the 17q21.31 locus does not fully explain the association between AGA and Parkinson's disease, and that we do not see more broad overlap between susceptibility loci for AGA and Parkinson's disease, it could be that the association between these phenotypes is mediated by an unobserved, shared environmental or genetic risk factor.
The identification of the new associations in this report was driven primarily by augmented power arising from the expanded sample size, which was more than fourfold that of previous GWA analyses [3], [4]. Similar to previous GWA studies [54], this increase in power was associated with a decrease in effect sizes from newly identified loci (top ORs range from 1·27 to 1·33 for six new loci versus OR = 1·60 for the lead SNP on chromosome 20). In this regard, it seems likely that more variants with smaller effect sizes could be uncovered by future larger studies to full describe the allelic architecture of early-onset AGA.
In conclusion, our findings provide fresh insights into the pathogenesis of early-onset AGA. As these newly identified susceptibility loci are also implicated in Parkinson's disease, prostate cancer and fertility, our results highlight the importance of hypothesis-free genetic studies, which allow unexpected genetic relationships between conditions to uncover shared etiologies.
Materials and Methods
Ethics Statement
All seven studies were approved by institutional ethics review committees at the relevant organizations, and written informed consent was provided by all participating individuals.
Study Participants
All the participants for this genome-wide meta-analysis were drawn from seven studies: Bonn (582 cases and 347 controls), CoLasu (578 cases and 547 controls), TwinsUK cohort (162 cases and 210 controls), Nigmegen Biomedical Study (73 cases and 132 controls), 23andMe (2,167 cases and 1,753 controls), Iceland (191 cases and 198 controls), Australian population based twin study (138 unrelated cases and 5728 unrelated controls). A detailed description of all these studies and phenotype definitions used in current study is provided in Text S1.
Genotyping and Quality Controls
The genotyping platforms, imputation methods and genome wide association methods used in participant studies are provided in Table S1. Extensive quality control thresholds were applied to include common SNPs (minor allele frequency ≥1%) with a high call rate (≥95%) for genotyped SNPs, and imputed SNPs with high quality metrics (variance ratio ≥0.3 for MACH and proper info statistic ≥0.4 for IMPUTE) [55], [56]. In addition, SNPs demonstrating deviation from Hardy Weinberg Equilibrium (p>10−6) were excluded. The test statistics for each cohort at each SNP were corrected for their respective genomic inflation factors to avoid inflation of results due to population stratification.
Meta-Analysis
We carried out a meta-analysis under both fixed - and random-effects models using the inverse-variance method to combine results from each study using an additive genetic model, while correcting for the genomic inflation factor for each study and the overall meta-analysis. To implement this strategy GWAMA software was used for SNPs on autosomes [57]. Based on the data available, association results using pre-imputation SNPs from Bonn, CoLaus, TwinsUK cohort, Nigmegen Biomedical Study, 23andMe and Australian population based twin study were used for meta-analysis on X chromosome through YAMAS program (http://yamas.meb.uni-bonn.de/index.html). Proxy association was applied to gain a higher power. We also tested for evidence of heterogeneity of effects between SNPs and AGA across studies using the Cochran's Q statistics and I2 measurement. SNPs with low heterogeneity (Q p value>0·10 and I2<50%) and present in at least three individual studies are reported. In order to test for an inflation of test statistics and the presence of a signal arising from the data for the variants influencing AGA, we constructed quantile-quantile (QQ) plots [58]. Genome-wide significance was set at a p value of 5×10−8 [59].
Genome-wide suggestive SNPs (5×10−8<p<5×10−6, n = 397) that were imputable using the MetaboChip platform were followed-up in an expanded meta-analysis including the THISEAS study [60] (Text S1, 297 controls and 219 cases) and the original seven cohorts. This additional cohort joined the consortium after completion of the main meta-analysis. No additional loci achieved genome-wide significance after inclusion of this cohort.
Expression Methods
To understand whether the identified SNPs are in close proximity to genes that show differential expression in hair follicles, we performed tissue expression analysis. In brief, total RNA extracted from human hair follicles, skin from temple, scalp, and whole blood were used for array-based gene expression analysis. The differential expression of genes in these tissues was determined by the average signal of identical probes and detection p values (which is significant, if a gene is reliably expressed). Further details are provided in Text S1.
Genotype Risk Score
In genotype risk score analysis, the association between a genotype risk score based on the weighted number of susceptibility alleles and AGA status was determined. Six novel susceptibility loci, chromosome 20p11 locus and AR gene are included. The weights were established using β coefficients for AGA susceptibility from meta-analysis. The resultant genotypic risk score was divided into quartiles and the risk of AGA for each quartile was tested in an additional set of subjects in 23andMe study, which were not included in the original meta-analysis, using the lowest risk quartile as the reference group. We identified 1582 controls and 1765 cases for this analysis, using the same phenotypic definition used in the meta-analysis. The genotype data is complete without missing values based on phasing and imputation. The trend for risk across the quartiles was tested using the non-parametric trend test [61].
Interaction Analysis
To detect the potential modification effect of AR gene on any other AGA loci, an interaction analysis was deployed using top SNPs identified from meta-analysis. Each SNP was coded by the number of AGA risk-increasing allele. The test of interaction is based on the coefficient b3 of the interaction term in the logistic regression model Y = b0+b1 * A+b2 * B+b3 * AB+e. The Bonferroni correction was applied to address the multiple comparisons problem and the significance level was set to p value<7×10−3.
Association with Other Common Diseases
In order to better understand whether the identified risk loci from the meta-analysis were associated with common diseases, we searched the AGA susceptibility loci using the GWAS Integrator embedded in HuGE Navigator website [62]. This online reference tool collects data from all published genetic studies in humans to facilitate the identification of shared etiologic pathways between diseases. Due to the frequent publication of new GWAS, only the GWAS searched out before June 2011 were used.
To examine the relationship between Parkinson's disease and AGA, we performed a logistic regression of Parkinson's disease status against AGA status, evaluating significance of the AGA term by analysis of deviance using a likelihood ratio test. To control for confounding factors, we included age, age2, and the top five principal components derived from genotype data as covariates.
Supporting Information
Zdroje
1. KleinRJZeissCChewEYTsaiJYSacklerRS 2005 Complement factor H polymorphism in age-related macular degeneration. Science 308 385 389
2. DonosoLAVrabecTKuivaniemiH 2010 The role of complement Factor H in age-related macular degeneration: a review. Surv Ophthalmol 55 227 246
3. RichardsJBYuanXGellerFWaterworthDBatailleV 2008 Male-pattern baldness susceptibility locus at 20p11. Nat Genet 40 1282 1284
4. HillmerAMBrockschmidtFFHannekenSEigelshovenSSteffensM 2008 Susceptibility variants for male-pattern baldness on chromosome 20p11. Nat Genet 40 1279 1281
5. HamiltonJB 1951 Patterned loss of hair in man; types and incidence. Ann N Y Acad Sci 53 708 728
6. LoJCFeigenbaumSLYangJPressmanARSelbyJV 2006 Epidemiology and adverse cardiovascular risk profile of diagnosed polycystic ovary syndrome. J Clin Endocrinol Metab 91 1357 1363
7. GilesGGSeveriGSinclairREnglishDRMcCredieMR 2002 Androgenetic alopecia and prostate cancer: findings from an Australian case-control study. Cancer Epidemiol Biomarkers Prev 11 549 553
8. JemalATiwariRCMurrayTGhafoorASamuelsA 2004 Cancer statistics, 2004. CA Cancer J Clin 54 8 29
9. HeinleinCAChangC 2004 Androgen receptor in prostate cancer. Endocr Rev 25 276 308
10. JiaLLandanGPomerantzMJaschekRHermanP 2009 Functional enhancers at the gene-poor 8q24 cancer-linked locus. PLoS Genet 5 e1000597 doi:10.1371/journal.pgen.1000597
11. LuYZhangZYuHZhengSLIsaacsWB 2011 Functional annotation of risk loci identified through genome-wide association studies for prostate cancer. Prostate 71 955 963
12. EllisJASinclairRHarrapSB 2002 Androgenetic alopecia: pathogenesis and potential for therapy. Expert Rev Mol Med 4 1 11
13. SuzukiHUedaTIchikawaTItoH 2003 Androgen receptor involvement in the progression of prostate cancer. Endocr Relat Cancer 10 209 216
14. HayesVMSeveriGEggletonSAPadillaEJSoutheyMC 2005 The E211 G>A androgen receptor polymorphism is associated with a decreased risk of metastatic prostate cancer and androgenetic alopecia. Cancer Epidemiol Biomarkers Prev 14 993 996
15. HayesVMSeveriGPadillaEJMorrisHATilleyWD 2007 5alpha-Reductase type 2 gene variant associations with prostate cancer risk, circulating hormone levels and androgenetic alopecia. Int J Cancer 120 776 780
16. CremersRGAbenKKVermeulenSHden HeijerMvan OortIM 2010 Androgenic alopecia is not useful as an indicator of men at high risk of prostate cancer. Eur J Cancer 46 3294 3299
17. WrightJLPageSTLinDWStanfordJL 2010 Male pattern baldness and prostate cancer risk in a population-based case-control study. Cancer Epidemiol 34 131 135
18. YassaMSaliouMDe RyckeYHemeryCHenniM 2011 Male pattern baldness and the risk of prostate cancer. Ann Oncol
19. YuXGuptaAWangYSuzukiKMirosevichJ 2005 Foxa1 and Foxa2 interact with the androgen receptor to regulate prostate and epididymal genes differentially. Ann N Y Acad Sci 1061 77 93
20. MirosevichJGaoNGuptaAShappellSBJoveR 2006 Expression and role of Foxa proteins in prostate cancer. Prostate 66 1013 1028
21. QiJNakayamaKCardiffRDBorowskyADKaulK 2010 Siah2-dependent concerted activity of HIF and FoxA2 regulates formation of neuroendocrine phenotype and neuroendocrine prostate tumors. Cancer Cell 18 23 38
22. YuXWangYDegraffDJWillsMLMatusikRJ 2011 Wnt/beta-Catenin activation promotes prostate tumor progression in a mouse model. Oncogene 30 1868 1879
23. MartinMKettmannRDequiedtF 2007 Class IIa histone deacetylases: regulating the regulators. Oncogene 26 5450 5467
24. JeongBCHongCYChattopadhyaySParkJHGongEY 2004 Androgen receptor corepressor-19 kDa (ARR19), a leucine-rich protein that represses the transcriptional activity of androgen receptor through recruitment of histone deacetylase. Mol Endocrinol 18 13 25
25. SuhJHShongMChoiHSLeeK 2008 CR6-interacting factor 1 represses the transactivation of androgen receptor by direct interaction. Mol Endocrinol 22 33 46
26. HalkidouKCookSLeungHYNealDERobsonCN 2004 Nuclear accumulation of histone deacetylase 4 (HDAC4) coincides with the loss of androgen sensitivity in hormone refractory cancer of the prostate. European urology 45 382 389; author reply 389
27. PetrieKGuidezFHowellLHealyLWaxmanS 2003 The histone deacetylase 9 gene encodes multiple protein isoforms. J Biol Chem 278 16059 16072
28. AmannJMNipJStromDKLutterbachBHaradaH 2001 ETO, a target of t(8;21) in acute leukemia, makes distinct contacts with multiple histone deacetylases and binds mSin3A through its oligomerization domain. Mol Cell Biol 21 6470 6483
29. HaimanCAChenGKBlotWJStromSSBerndtSI 2011 Genome-wide association study of prostate cancer in men of African ancestry identifies a susceptibility locus at 17q21. Nat Genet 43 570 573
30. SchumannGCoinLJLourdusamyACharoenPBergerKH 2011 Genome-wide association and genetic functional studies identify autism susceptibility candidate 2 gene (AUTS2) in the regulation of alcohol consumption. Proc Natl Acad Sci U S A 108 7119 7124
31. HeroldCSteffensMBrockschmidtFFBaurMPBeckerT 2009 INTERSNP: genome-wide interaction analysis guided by a priori information. Bioinformatics 25 3275 3281
32. MassieCEAdryanBBarbosa-MoraisNLLynchAGTranMG 2007 New androgen receptor genomic targets show an interaction with the ETS1 transcription factor. EMBO Rep 8 871 878
33. HuSYaoGGuanXNiZMaW 2010 Research resource: Genome-wide mapping of in vivo androgen receptor binding sites in mouse epididymis. Mol Endocrinol 24 2392 2405
34. EdwardsTLScottWKAlmonteCBurtAPowellEH 2010 Genome-wide association study confirms SNPs in SNCA and the MAPT region as common risk factors for Parkinson disease. Ann Hum Genet 74 97 109
35. Simon-SanchezJSchulteCBrasJMSharmaMGibbsJR 2009 Genome-wide association study reveals genetic risk underlying Parkinson's disease. Nat Genet 41 1308 1312
36. HamzaTHZabetianCPTenesaALaederachAMontimurroJ 2010 Common genetic variation in the HLA region is associated with late-onset sporadic Parkinson's disease. Nat Genet 42 781 785
37. SpencerCCPlagnolVStrangeAGardnerMPaisan-RuizC 2011 Dissection of the genetics of Parkinson's disease identifies an additional association 5′ of SNCA and multiple associated haplotypes at 17q21. Hum Mol Genet 20 345 353
38. NallsMAPlagnolVHernandezDGSharmaMSheerinUM 2011 Imputation of sequence variants for identification of genetic risks for Parkinson's disease: a meta-analysis of genome-wide association studies. Lancet 377 641 649
39. Do CBTJDorfmanEKieferAKDrabantEM 2011 Web-based genome-wide association study identifies two novel loci and a substantial genetic component for Parkinson's disease. PLoS Genet 7 e1002141 doi:10.1371/journal.pgen.1002141
40. de LauLMBretelerMM 2006 Epidemiology of Parkinson's disease. Lancet Neurol 5 525 535
41. MiwaHKondoT 2003 Hair loss induced by dopamine agonist: case report and review of the literature. Parkinsonism Relat Disord 10 51 52
42. CunninghamRLGiuffridaARobertsJL 2009 Androgens induce dopaminergic neurotoxicity via caspase-3-dependent activation of protein kinase Cdelta. Endocrinology 150 5539 5548
43. Van Den EedenSKTannerCMBernsteinALFrossRDLeimpeterA 2003 Incidence of Parkinson's disease: variation by age, gender, and race/ethnicity. Am J Epidemiol 157 1015 1022
44. StefanssonHHelgasonAThorleifssonGSteinthorsdottirVMassonG 2005 A common inversion under selection in Europeans. Nat Genet 37 129 137
45. VoightBFKudaravalliSWenXPritchardJK 2006 A map of recent positive selection in the human genome. PLoS Biol 4 e72 doi:10.1371/journal.pbio.0040072
46. NestlerJE 2008 Metformin for the treatment of the polycystic ovary syndrome. N Engl J Med 358 47 54
47. CareyAHWaterworthDPatelKWhiteDLittleJ 1994 Polycystic ovaries and premature male pattern baldness are associated with one allele of the steroid metabolism gene CYP17. Hum Mol Genet 3 1873 1876
48. GovindAObhraiMSClaytonRN 1999 Polycystic ovaries are inherited as an autosomal dominant trait: analysis of 29 polycystic ovary syndrome and 10 control families. J Clin Endocrinol Metab 84 38 43
49. PastorPEzquerraMPerezJCChakravertySNortonJ 2004 Novel haplotypes in 17q21 are associated with progressive supranuclear palsy. Ann Neurol 56 249 258
50. RademakersRCrutsMvan BroeckhovenC 2004 The role of tau (MAPT) in frontotemporal dementia and related tauopathies. Hum Mutat 24 277 295
51. SchragABen-ShlomoYQuinnNP 1999 Prevalence of progressive supranuclear palsy and multiple system atrophy: a cross-sectional study. Lancet 354 1771 1775
52. TogasakiDMTannerCM 2000 Epidemiologic aspects. Adv Neurol 82 53 59
53. RatnavalliEBrayneCDawsonKHodgesJR 2002 The prevalence of frontotemporal dementia. Neurology 58 1615 1621
54. ParkJHWacholderSGailMHPetersUJacobsKB 2010 Estimation of effect size distribution from genome-wide association studies and implications for future discoveries. Nat Genet 42 570 575
55. LiYWillerCJDingJScheetPAbecasisGR 2010 MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet Epidemiol 34 816 834
56. MarchiniJHowieBMyersSMcVeanGDonnellyP 2007 A new multipoint method for genome-wide association studies by imputation of genotypes. Nat Genet 39 906 913
57. MagiRMorrisAP 2010 GWAMA: software for genome-wide association meta-analysis. BMC Bioinformatics 11 288
58. BaldingDJ 2006 A tutorial on statistical methods for population association studies. Nat Rev Genet 7 781 791
59. FrazerKABallingerDGCoxDRHindsDAStuveLL 2007 A second generation human haplotype map of over 3.1 million SNPs. Nature 449 851 861
60. 2011 A genome-wide association study in Europeans and South Asians identifies five new loci for coronary artery disease. Nat Genet 43 339 344
61. CuzickJ 1985 A Wilcoxon-type test for trend. Stat Med 4 87 90
62. YuWYesupriyaAWulfAHindorffLADowlingN 2011 GWAS Integrator: a bioinformatics tool to explore human genetic associations reported in published genome-wide association studies. Eur J Hum Genet 19 1095 1099
Štítky
Genetika Reprodukční medicína
Článek Functional Centromeres Determine the Activation Time of Pericentric Origins of DNA Replication inČlánek Dynamic Deposition of Histone Variant H3.3 Accompanies Developmental Remodeling of the TranscriptomeČlánek Integrin α PAT-2/CDC-42 Signaling Is Required for Muscle-Mediated Clearance of Apoptotic Cells inČlánek Prdm5 Regulates Collagen Gene Transcription by Association with RNA Polymerase II in Developing BoneČlánek Acquisition Order of Ras and p53 Gene Alterations Defines Distinct Adrenocortical Tumor Phenotypes
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2012 Číslo 5
-
Všechny články tohoto čísla
- Slowing Replication in Preparation for Reduction
- Chromosome Pairing: A Hidden Treasure No More
- Loss of Imprinting Differentially Affects REM/NREM Sleep and Cognition in Mice
- Six Novel Susceptibility Loci for Early-Onset Androgenetic Alopecia and Their Unexpected Association with Common Diseases
- Regulation by the Noncoding RNA
- UDP-Galactose 4′-Epimerase Activities toward UDP-Gal and UDP-GalNAc Play Different Roles in the Development of
- Deletion of PTH Rescues Skeletal Abnormalities and High Osteopontin Levels in Mice
- Karyotypic Determinants of Chromosome Instability in Aneuploid Budding Yeast
- Genome-Wide Copy Number Analysis Uncovers a New HSCR Gene:
- MicroRNA-277 Modulates the Neurodegeneration Caused by Fragile X Premutation rCGG Repeats
- Functional Centromeres Determine the Activation Time of Pericentric Origins of DNA Replication in
- Dynamic Deposition of Histone Variant H3.3 Accompanies Developmental Remodeling of the Transcriptome
- Scientist Citizen: An Interview with Bruce Alberts
- YY1 Regulates Melanocyte Development and Function by Cooperating with MITF
- Congenital Heart Disease–Causing Gata4 Mutation Displays Functional Deficits
- Recombination Drives Vertebrate Genome Contraction
- KATNAL1 Regulation of Sertoli Cell Microtubule Dynamics Is Essential for Spermiogenesis and Male Fertility
- Re-Patterning Sleep Architecture in through Gustatory Perception and Nutritional Quality
- Using Whole-Genome Sequence Data to Predict Quantitative Trait Phenotypes in
- Genome-Wide Analysis of GLD-1–Mediated mRNA Regulation Suggests a Role in mRNA Storage
- Meiotic Chromosome Pairing Is Promoted by Telomere-Led Chromosome Movements Independent of Bouquet Formation
- LINT, a Novel dL(3)mbt-Containing Complex, Represses Malignant Brain Tumour Signature Genes
- The H3K27 Demethylase UTX-1 Is Essential for Normal Development, Independent of Its Enzymatic Activity
- Suppresses Senescence Programs and Thereby Accelerates and Maintains Mutant -Induced Lung Tumorigenesis
- Genome-Wide Association of Pericardial Fat Identifies a Unique Locus for Ectopic Fat
- An Essential Role for Katanin p80 and Microtubule Severing in Male Gamete Production
- Identification of Genes That Promote or Antagonize Somatic Homolog Pairing Using a High-Throughput FISH–Based Screen
- Principles of Carbon Catabolite Repression in the Rice Blast Fungus: Tps1, Nmr1-3, and a MATE–Family Pump Regulate Glucose Metabolism during Infection
- Integrin α PAT-2/CDC-42 Signaling Is Required for Muscle-Mediated Clearance of Apoptotic Cells in
- Histone H3 Localizes to the Centromeric DNA in Budding Yeast
- Collapse of Telomere Homeostasis in Hematopoietic Cells Caused by Heterozygous Mutations in Telomerase Genes
- Hypersensitive to Red and Blue 1 and Its Modification by Protein Phosphatase 7 Are Implicated in the Control of Arabidopsis Stomatal Aperture
- Extent, Causes, and Consequences of Small RNA Expression Variation in Human Adipose Tissue
- TBC-8, a Putative RAB-2 GAP, Regulates Dense Core Vesicle Maturation in
- Regulating Repression: Roles for the Sir4 N-Terminus in Linker DNA Protection and Stabilization of Epigenetic States
- Common Genetic Determinants of Intraocular Pressure and Primary Open-Angle Glaucoma
- Prdm5 Regulates Collagen Gene Transcription by Association with RNA Polymerase II in Developing Bone
- Fitness Landscape Transformation through a Single Amino Acid Change in the Rho Terminator
- Repeated, Selection-Driven Genome Reduction of Accessory Genes in Experimental Populations
- Allelic Variation and Differential Expression of the mSIN3A Histone Deacetylase Complex Gene Promote Mammary Tumor Growth and Metastasis
- DNA Demethylation and USF Regulate the Meiosis-Specific Expression of the Mouse
- Knowledge-Driven Analysis Identifies a Gene–Gene Interaction Affecting High-Density Lipoprotein Cholesterol Levels in Multi-Ethnic Populations
- A Duplication CNV That Conveys Traits Reciprocal to Metabolic Syndrome and Protects against Diet-Induced Obesity in Mice and Men
- EMT Inducers Catalyze Malignant Transformation of Mammary Epithelial Cells and Drive Tumorigenesis towards Claudin-Low Tumors in Transgenic Mice
- Inactivation of a Novel FGF23 Regulator, FAM20C, Leads to Hypophosphatemic Rickets in Mice
- Genome-Wide Association for Abdominal Subcutaneous and Visceral Adipose Reveals a Novel Locus for Visceral Fat in Women
- Stratifying Type 2 Diabetes Cases by BMI Identifies Genetic Risk Variants in and Enrichment for Risk Variants in Lean Compared to Obese Cases
- New Insight into the History of Domesticated Apple: Secondary Contribution of the European Wild Apple to the Genome of Cultivated Varieties
- Activated Cdc42 Kinase Has an Anti-Apoptotic Function
- The Region Is Critical for Birth Defects and Electrocardiographic Dysfunctions Observed in a Down Syndrome Mouse Model
- COP9 Signalosome Integrity Plays Major Roles for Hyphal Growth, Conidial Development, and Circadian Function
- Bmps and Id2a Act Upstream of Twist1 To Restrict Ectomesenchyme Potential of the Cranial Neural Crest
- Psip1/Ledgf p52 Binds Methylated Histone H3K36 and Splicing Factors and Contributes to the Regulation of Alternative Splicing
- The Number of X Chromosomes Causes Sex Differences in Adiposity in Mice
- Target Gene Analysis by Microarrays and Chromatin Immunoprecipitation Identifies HEY Proteins as Highly Redundant bHLH Repressors
- Acquisition Order of Ras and p53 Gene Alterations Defines Distinct Adrenocortical Tumor Phenotypes
- ELK1 Uses Different DNA Binding Modes to Regulate Functionally Distinct Classes of Target Genes
- Histone H1 Depletion Impairs Embryonic Stem Cell Differentiation
- IDN2 and Its Paralogs Form a Complex Required for RNA–Directed DNA Methylation
- Separation of DNA Replication from the Assembly of Break-Competent Meiotic Chromosomes
- Genomic Hypomethylation in the Human Germline Associates with Selective Structural Mutability in the Human Genome
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Inactivation of a Novel FGF23 Regulator, FAM20C, Leads to Hypophosphatemic Rickets in Mice
- Genome-Wide Association of Pericardial Fat Identifies a Unique Locus for Ectopic Fat
- Slowing Replication in Preparation for Reduction
- An Essential Role for Katanin p80 and Microtubule Severing in Male Gamete Production
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



