-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaExtent, Causes, and Consequences of Small RNA Expression Variation in Human Adipose Tissue
Small RNAs are functional molecules that modulate mRNA transcripts and have been implicated in the aetiology of several common diseases. However, little is known about the extent of their variability within the human population. Here, we characterise the extent, causes, and effects of naturally occurring variation in expression and sequence of small RNAs from adipose tissue in relation to genotype, gene expression, and metabolic traits in the MuTHER reference cohort. We profiled the expression of 15 to 30 base pair RNA molecules in subcutaneous adipose tissue from 131 individuals using high-throughput sequencing, and quantified levels of 591 microRNAs and small nucleolar RNAs. We identified three genetic variants and three RNA editing events. Highly expressed small RNAs are more conserved within mammals than average, as are those with highly variable expression. We identified 14 genetic loci significantly associated with nearby small RNA expression levels, seven of which also regulate an mRNA transcript level in the same region. In addition, these loci are enriched for variants significant in genome-wide association studies for body mass index. Contrary to expectation, we found no evidence for negative correlation between expression level of a microRNA and its target mRNAs. Trunk fat mass, body mass index, and fasting insulin were associated with more than twenty small RNA expression levels each, while fasting glucose had no significant associations. This study highlights the similar genetic complexity and shared genetic control of small RNA and mRNA transcripts, and gives a quantitative picture of small RNA expression variation in the human population.
Published in the journal: . PLoS Genet 8(5): e32767. doi:10.1371/journal.pgen.1002704
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1002704Summary
Small RNAs are functional molecules that modulate mRNA transcripts and have been implicated in the aetiology of several common diseases. However, little is known about the extent of their variability within the human population. Here, we characterise the extent, causes, and effects of naturally occurring variation in expression and sequence of small RNAs from adipose tissue in relation to genotype, gene expression, and metabolic traits in the MuTHER reference cohort. We profiled the expression of 15 to 30 base pair RNA molecules in subcutaneous adipose tissue from 131 individuals using high-throughput sequencing, and quantified levels of 591 microRNAs and small nucleolar RNAs. We identified three genetic variants and three RNA editing events. Highly expressed small RNAs are more conserved within mammals than average, as are those with highly variable expression. We identified 14 genetic loci significantly associated with nearby small RNA expression levels, seven of which also regulate an mRNA transcript level in the same region. In addition, these loci are enriched for variants significant in genome-wide association studies for body mass index. Contrary to expectation, we found no evidence for negative correlation between expression level of a microRNA and its target mRNAs. Trunk fat mass, body mass index, and fasting insulin were associated with more than twenty small RNA expression levels each, while fasting glucose had no significant associations. This study highlights the similar genetic complexity and shared genetic control of small RNA and mRNA transcripts, and gives a quantitative picture of small RNA expression variation in the human population.
Introduction
A world of noncoding RNA molecules has been uncovered in the last decades, expanding our understanding of functional elements in the genome [1]. After it was found that the small (∼15–30 nt) noncoding RNAs can directly modulate protein levels [2], [3], and via that, almost any cellular process [4], they have been subject to vigorous study, leading to the recognition that several different types of small RNAs can act as posttranscriptional regulators [5].
MicroRNA genes (miRNAs) were the first animal small RNA genes to be discovered [6], and over 1,500 examples have been found in humans to date [7]. The primary miRNA transcript has a stem loop structure that is recognised and cleaved via RNA processing enzymes to produce a double stranded duplex [8]. The mature miRNA strand is loaded into a complex containing Argonaute family proteins and guided to targeting, while the other strand is assumed to be degraded. miRNAs target mRNA transcripts via base pair complementarity, typically in the 3′ untranslated region [8], [9], but also coding sequence [10]. This targeting can induce transcript cleavage, degradation, destabilisation, or repression of translation, thus modulating protein levels. Small nucleolar RNAs (snoRNAs) are typically longer genes (60–300 nt) that facilitate RNA editing within ribosomal or spliceosomal RNAs [11]. However, their full sequences can also be processed into snoRNA derived RNAs that exert a similar mode of action as miRNAs [12], [13], [14].
The recent ability to quantify levels of small RNA expression invites questions about the extent and causes of their variability in the human population. Importantly, the quantity and quality of transcripts are the only way genetic variation can influence phenotype. Thus, the genetic contribution to small RNA expression trait variability has to be assessed for accurate understanding of transmission of heritable information. Such questions have already been successfully addressed for mRNA expression levels, where variability between tissues [15], populations [16], and diseased and healthy individuals [17], as well as the contribution of genotype [16], [18], [19], [20] have been thoroughly characterised. Previous studies have found genetic contribution to miRNA levels in both human fibroblasts [21] as well as adipose tissue [22] using miRNA microarrays. However, other types of small RNAs have not been assayed, and a full account of small RNA sequence and transcriptome variability in a reference cohort is missing.
Small RNA expression can be viewed as a primary genetic trait to be mapped in isolation, but also as a quantitative trait with downstream influences on gene expression and other phenotypes. Recent studies have been successful in combining information about genotype and intermediate phenotypes (such as mRNA levels [17], [23], [24] or inferred cellular activations [25]) to understand how the genetic signal is mediated. In this light, it is especially interesting to analyse small RNA transcript levels as intermediate traits potentially causative for downstream effects, as both miRNAs and snoRNAs have already been implicated in many human disease phenotypes ranging from obesity and autism to cancer [26], [27], [28], [29], [30], [31], [32].
The MuTHER (Multi-Tissue Heritability Resource) cohort was established with the aim of analysing the genetics of gene expression in multiple human tissues in over 800 individuals [19], [20], [33]. This cohort is a subset of the UK Twins [34], and has extensive information on genotype and gene expression, as well as a plethora of clinical phenotypes. We set out to characterise small RNA variability in 131 abdominal fat samples from MuTHER resource using high throughput sequencing technology. We quantified the content of the small RNA transcriptome, the extent of sequence and transcript level variation, the relative levels of miRNA expression from both arms of the molecule, as well as coexpression of miRNAs from the same cluster. Since high density genotype data, mRNA levels from the same RNA sample as well as obesity-related phenotypes were available for these individuals, we associated these measurements with the small RNA levels to find out about the extent of genetic control, mRNA and miRNA expression correlates, and relation of small RNAs and global metabolic traits.
Results
Largest small RNA sequencing dataset to date
We sequenced subcutaneous adipose tissue small RNAs of 131 females from the UK TWINS cohort [34] included in the MuTHER study [19] on the Illumina GAII platform (Materials and Methods, data available at the EGA, submission ID EGAS00001000212). After filtering, quality control, and mapping, we obtained 331 million total reads, with a median of 2.3 million reads per sample aligning to the genome (Materials and Methods, Figure S1). The majority of the reads (93%) mapped to annotated mature miRNA sequences (mirBase v17 [7]), with the rest divided between tRNAs (2%), snoRNAs (0.6%), lincRNAs (0.3%), and other noncoding RNA features annotated in Ensembl v63 [35] (Table S1). This distribution is expected, as we size-selected for 15–30 base pair fragments, which excludes other functional RNA species except for degradation products. In addition, we found reads mapped to loci previously unannotated for noncoding RNA transcription. We identified 12 novel miRNA gene candidates using MapMi ([36], Materials and Methods, Dataset S1), and 701 short (<100 bp) regions with at least 1000 total mapped reads across all samples (2% of all mapped reads, Table S2). These regions were significantly enriched in DNAse hypersensitivity sites (237/701, one-tailed binomial p<10−10, Materials and Methods), which often harbour enhancer elements that are known to give rise to short transcripts [37]. The rest overlapped exons (149/701, one-tailed binomial p<10−10) and introns (190/701, not significant), with 233 regions arising from intergenic sequence.
Highly expressed miRNAs are implicated in adipose and blood cell development
We quantified expression levels of 418 known miRNA gene products, 239 tRNAs, 173 snoRNAs, 111 lincRNAs and 107 other RNAs that had at least 1000 total sequencing reads (Figure 1A, Table S1). For further analyses, we focused on miRNAs and snoRNA derived sequences as the only known functional molecules in our selected size range. The adipose tissue small RNA transcriptome is of medium complexity, with a median of 17 species of molecules required to account for 75% of the mapped reads (Figure 1B). The most highly expressed small RNAs (Figure 2A) have previously been associated with adipose development (mir-143-3p [38], mir-21-5p [12]), angiogenesis (mir-126-3p [39], mir-378a-3p [40]), and erythropoiesis (mir-24-3p [41], mir-451a [42]). We compared the average expression levels in adipose tissue to public human small RNA sequencing data from B-cells [43], liver [44], pigment cells [45], pooled thymocytes, bone marrow, CD34+ progenitor cells [46], lung, kidney, skeletal muscle, heart, pancreas, frontal orbital gyrus, spleen, and liver tissue [47] after processing them with our pipelines (Materials and Methods, Table S3). While seven of the ten most highly expressed small RNA genes and gene families were highly expressed in all tissues (let7 family, mir-24-3p, mir-378a-3p, mir-21-5p) other highly expressed small RNAs (mir-143-3p, mir-126-3p) were specific to adipose tissue (average q-value of pairwise comparisons <0.1, Materials and Methods). In total, there were 12 miRNAs with significantly higher expression (q<0.1) compared to mean of every other tissue, and no such snoRNAs, with mir-126-3p, mir-340-3p, mir-190a, and mir-335-3p showing the strongest specificity signal (Figure 2B).
Fig. 1. Summaries of sequenced small RNAs. 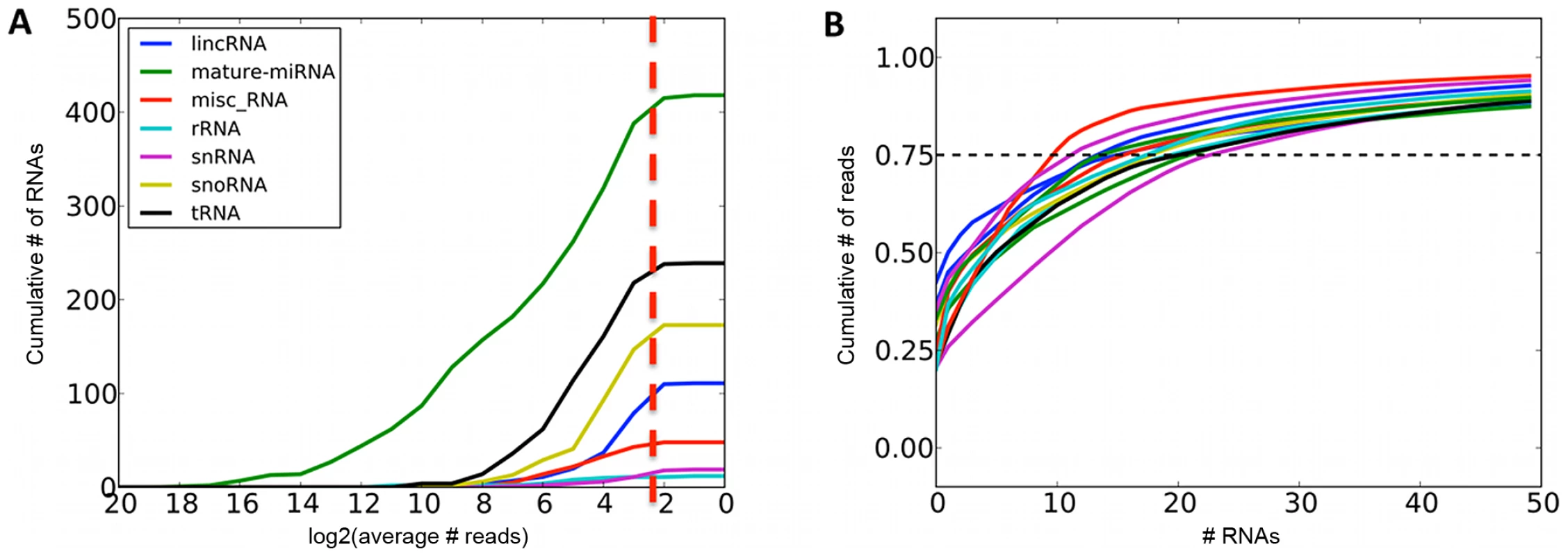
A. Cumulative number of different small RNA species recovered at required minimum coverage of 1000 total mapped reads (approximately 8 reads per sample, red line). B. Cumulative fraction of mapped reads accounted for by the most highly expressed small RNAs for twelve randomly chosen samples, with 75th percentile marked with the dashed line. Fig. 2. Most highly expressed and most adipose-specific small RNA genes. 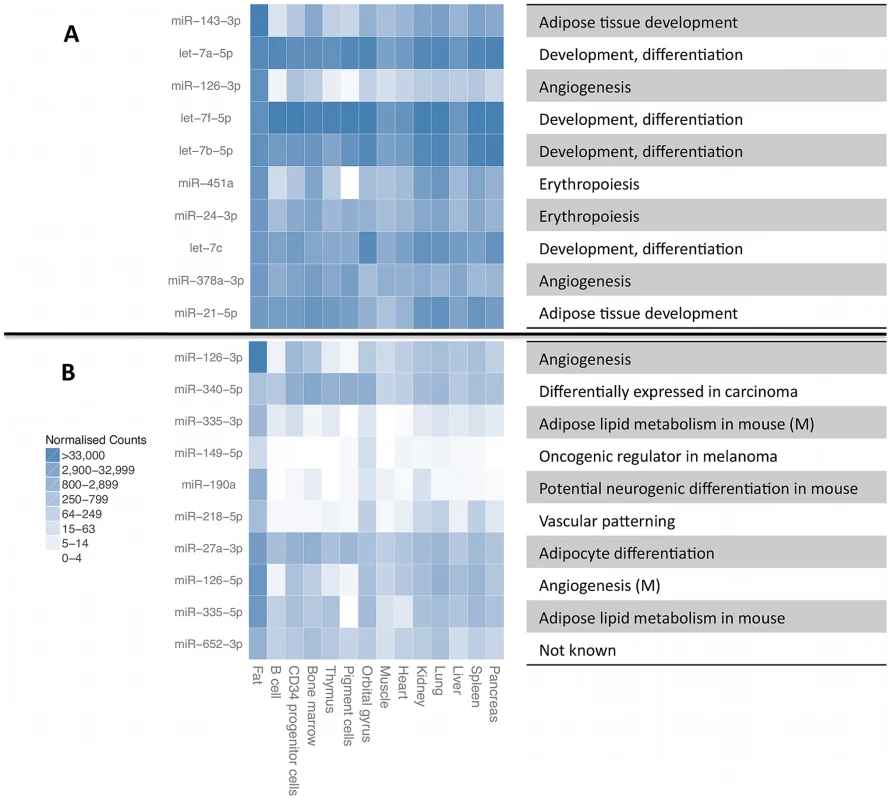
Most highly expressed (A) and most adipose-specific (B) small RNA genes, their relative expression level in B-cells [43], liver [44], pigment cells [45], pooled thymocytes, bone marrow, CD34+ progenitor cells [46], lung, kidney, skeletal muscle, heart, pancreas, frontal orbital gyrus, spleen and liver [47]. The annotated function of the miRNAs is given on the right, with information for mature miRNAs (M) given for some of the sequences from the other miRNA arm that did not have independent annotation. Expressed miRNA and snoRNA genes have reduced genetic variability
Next, we called variants from the RNA sequence data (Materials and Methods), and found one mature miRNA and two snoRNA polymorphisms, all with independent evidence from whole genome sequencing of the UK10K cohort (personal communication, UK10K Consortium) (Table S4). All three found variants had relatively low (<11%) minor allele frequency (MAF). Assuming Hardy-Weinberg equilibrium, and equal expression from both gene copies, the miRNA sequence variant represents a fraction of 7×10−5 of the 14,005 mature miRNA and star sequence sites that could pass our filters, consistent with previous reports of strong purifying selection in the functional small RNA regions [48]. The same regions in the UK10K project harboured 13 called polymorphic sites, 9 of which had MAF<1%. We detected one of these sites using small RNA sequencing (MAF = 11%), and did not find the rest. Based on the MAF of each UK10K DNA variant, and expression levels of the small RNAs, we expected to recover one additional site (Materials and Methods). While it is possible that other polymorphisms are present in sequences coding for miRNA and snoRNA products, the derived alleles were not observed on at least 10 reads in our data, and could thus not be reliably detected. In addition to genetic variants, we found three A to I RNA editing events in the mature miRNA regions (Materials and Methods, Table S4). These sites were the 7th, 8th, and 9th bases of the mature product, and edited in 25, 18, and 11 percent of the reads, indicating that additional variability is tolerated in the functionally important seed region. We also observed bases at the ends of mapped reads not matching the genome in line with previous reports ([49], Table S5), but as similar discrepancies were not observed at comparable frequency in the data from other tissues, we considered them more likely to be sequencing or library preparation artefacts than true RNA modifications.
Genetically variable small RNAs have low expression level and variation
As mature miRNA sequences and analogous snoRNA products function via base pair complementarity, there is selective pressure against accumulating variants in their regions. Previous reports from DNA sequence data have confirmed increased conservation of miRNA sequence compared to intronic and intergenic background, but also a more pronounced effect for more highly expressed genes. We also observed a lack of miRNAs with at least 1000 reads on average and UCSC primate conservation score of less than 0 (Figure 3A, p<3×10−5, chi-squared test, Materials and Methods). Moreover, we assessed if the variability in the expression levels is under similar influence. Indeed, we observed a lack of small RNAs with expression variance of at least 5, and a conservation score below 0 (Figure 3B, nominal p<3×10−4, chi-squared test), suggesting that selection acts on not just average expression, but also expression variation.
Fig. 3. Lack of small RNAs. 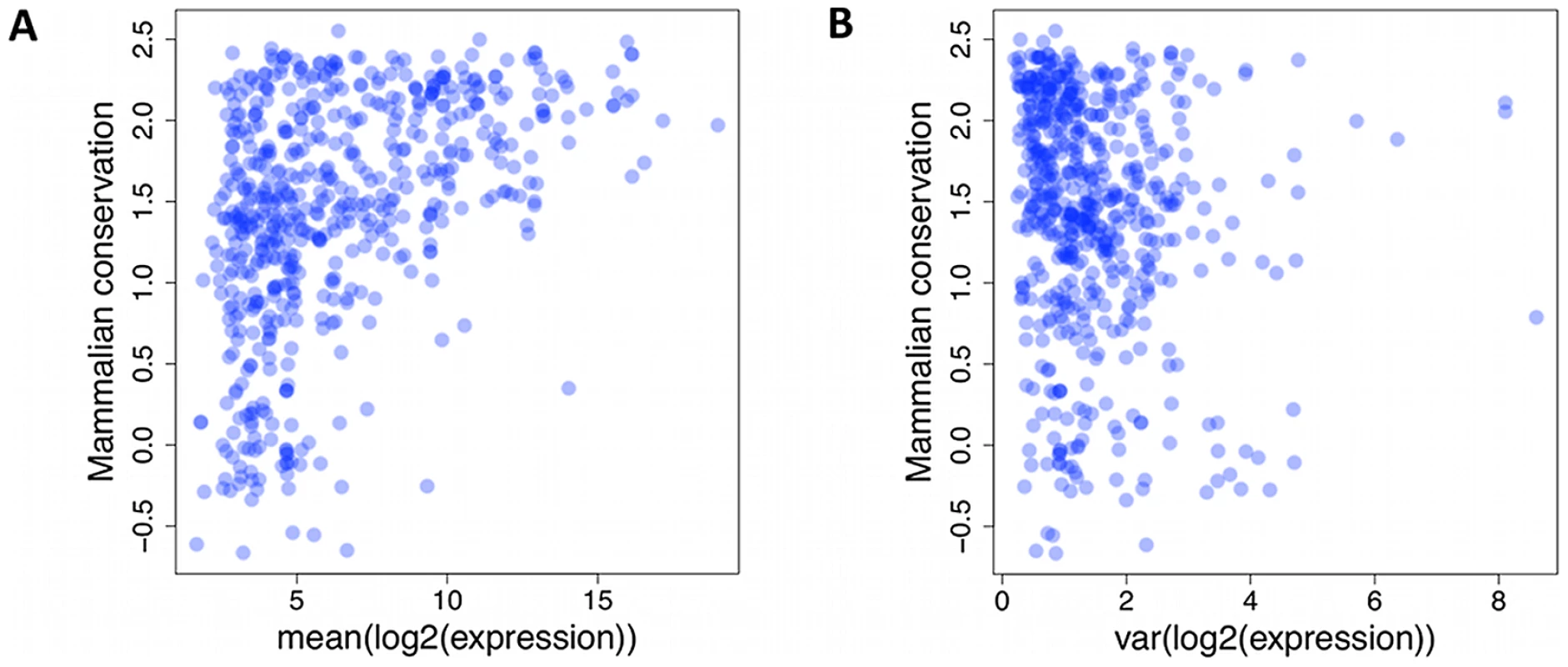
Lack of small RNAs with low mammalian conservation score (UCSC Genome Browser [50], y-axis) and high average (A) or variance (B) of expression level (x-axis). Each dot represents a single miRNA or snoRNA gene. Batch effects and covariates have a strong effect on expression levels
After analysing the variability of RNA expression levels within and between tissues, we next addressed inter-individual variation. First, we tested whether experimental confounders influenced small RNA expression variability between samples. To this end, we performed principal components analysis of log-transformed, normalised read counts (Materials and Methods, Tables S6 and S7), and associated first twenty components (PCs) to known covariates of sample multiplexing tag, library batch, sequencing flow cell, RNA integrity score (RIN, [51]), and RNA concentration (Materials and Methods). We found significant associations (Bonferroni-corrected p<0.05) for RIN (PC1), library batches (PC1, 2, 3, 6, 10, 11, 12, 13, 18), and two multiplexing tags (PC5, 14). As these components capture major directions of variation in the data, we included the associated covariates measured for all samples (age, library batch and multiplexing tag) in eventual analyses. Since it has been demonstrated that unmeasured confounders similarly have an influence on expression levels [52], [53], we tested whether applying the Bayesian factor analysis package PEER [54], to account for these confounders, increases the number of discoveries. As we already corrected for 30 known covariates, including the additional inferred factors did not increase findings in downstream analyses, and were not used.
Small RNA levels are associated with genotype
To identify miRNA and snoRNA genes whose expression is driven by cis-acting genetic variation, we performed association tests between their transcript levels and SNPs within 100 kb of the transcript (Materials and Methods). We found significant cis-eQTLs (nominal p<2.4×10−4, FDR<5%) for eight of 418 miRNAs and six of 173 snoRNAs (Table 1). In comparison, 462 eQTLs were found for 27,499 mRNA probes in the same tissue and cohort with comparable sample size and FDR in a previous study [19], suggesting a similar level of genetic control for mRNA and small RNA transcript levels.
Tab. 1. Small RNA <i>cis</i>-eQTLs - significant (nominal p<2.4×10<sup>−4</sup>, FDR<5%) associations of miRNA and snoRNA expression levels to SNPs within 100 kb from the transcript. 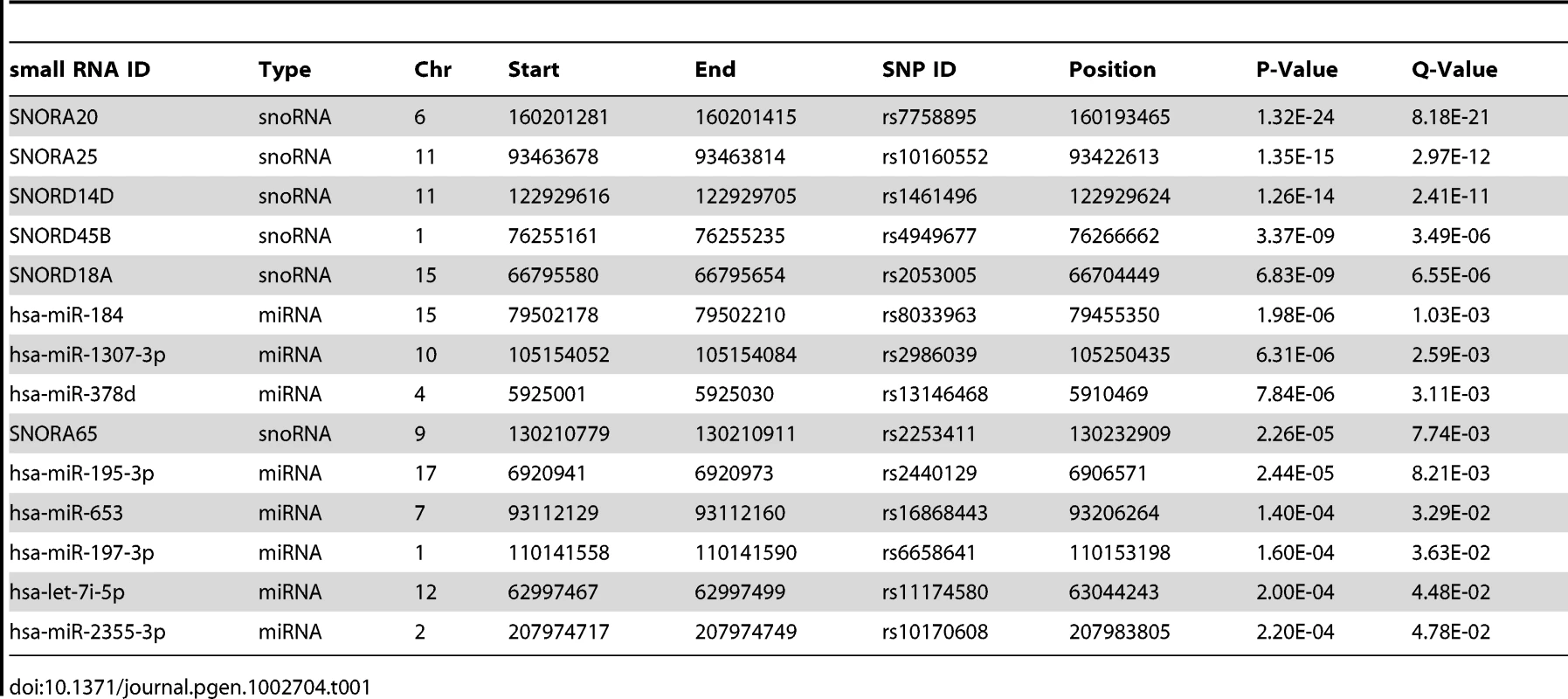
We validated our eQTL findings in an independent cohort of 70 human samples with array-based miRNA expression data from abdominal adipose tissue [22]. Five of the eight miRNAs with an eQTL in our study were assayed in this study, with three of them replicating (nominal p<0.05, Table S8) and p-values across the full set of eQTLs tested in each study concordant (Spearman rank correlation p<8×10−4, Figure S2). However, we found no overlap between our significant cis-eQTL results and 12 significant (p<0.05 from 10,000 permutations) miRNA cis-eQTLs reported in human fibroblasts [21], likely due to a different set of expressed genes and lack of replication power. As mRNA studies in larger cohorts have found only up to a third of genetic associations to be tissue-specific [19], [55], we also expect many of the small RNA eQTLs to have an effect in other tissues in better powered studies.
The full MuTHER cohort of 776 individuals was profiled for mRNA levels from adipose tissue using the same RNA sample for the individuals in our study, as well as skin and lymphoblastoid cell lines from a separate RNA sample. Thus, we could directly assess any overlap in genetic control of transcripts of different type and across multiple tissues. We found seven of our small RNA eQTL SNPs to also be significantly associated with a nearby mRNA probe (Table 2). The mRNA transcripts were the nearest annotated transcript to the two miRNAs and two snoRNAs, but at least one and up to four annotated mRNA transcripts away from the rest of the snoRNAs. Further, in three of the eight cases, the mRNA and small RNA did not share the direction of the SNP effect. This suggests nontrivial shared genetic control, either via enhancer or promoter, or a single transcript that is spliced to form multiple genes.
Tab. 2. Overlap of small RNA cis-eQTLs and mRNA cis-eQTLs from the MuTHER study. 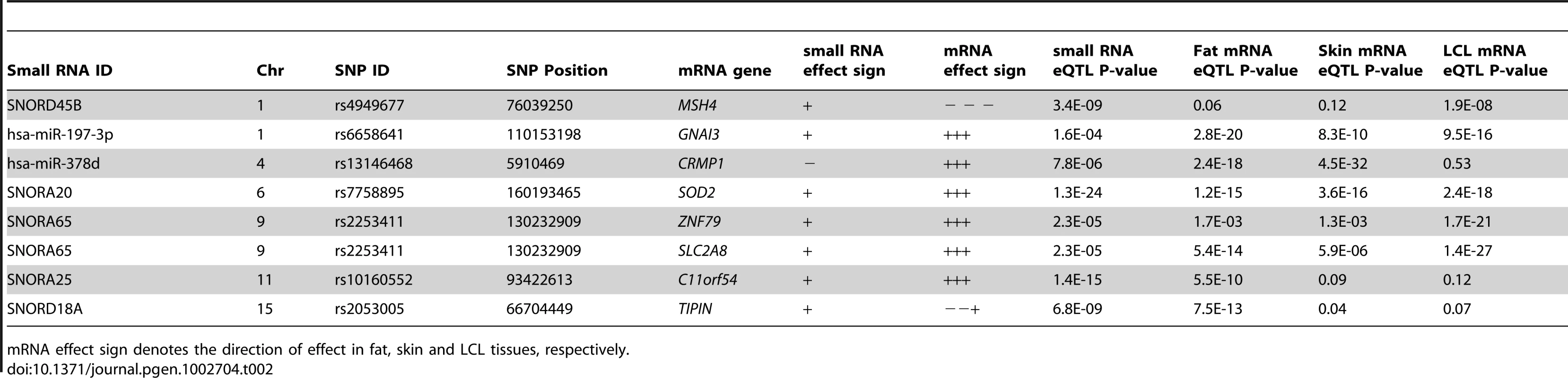
mRNA effect sign denotes the direction of effect in fat, skin and LCL tissues, respectively. Small RNA expression levels are associated with obesity-related phenotypes
As our cohort has been phenotyped for DEXA-derived measurements of percentage trunk fat mass (PTFM), BMI, fasting insulin, and fasting glucose (summaries in Table S9), we examined the association between small RNA expression and these obesity-related phenotypes (Materials and Methods). We found 47, 41 and 23 out of the 591 tested small RNAs to be associated with PTFM, BMI and fasting insulin respectively (per-trait FDR<5%). As these traits are highly correlated (Pearson's r>0.45 for all pairwise comparisons), there is also considerable overlap in the associated small RNAs between the traits (Table 3).
Tab. 3. Top 14 associations between small RNA expression levels and percent trunk fat mass (PTFM), body mass index (BMI), and fasting insulin (FI), ordered by lowest q-value. 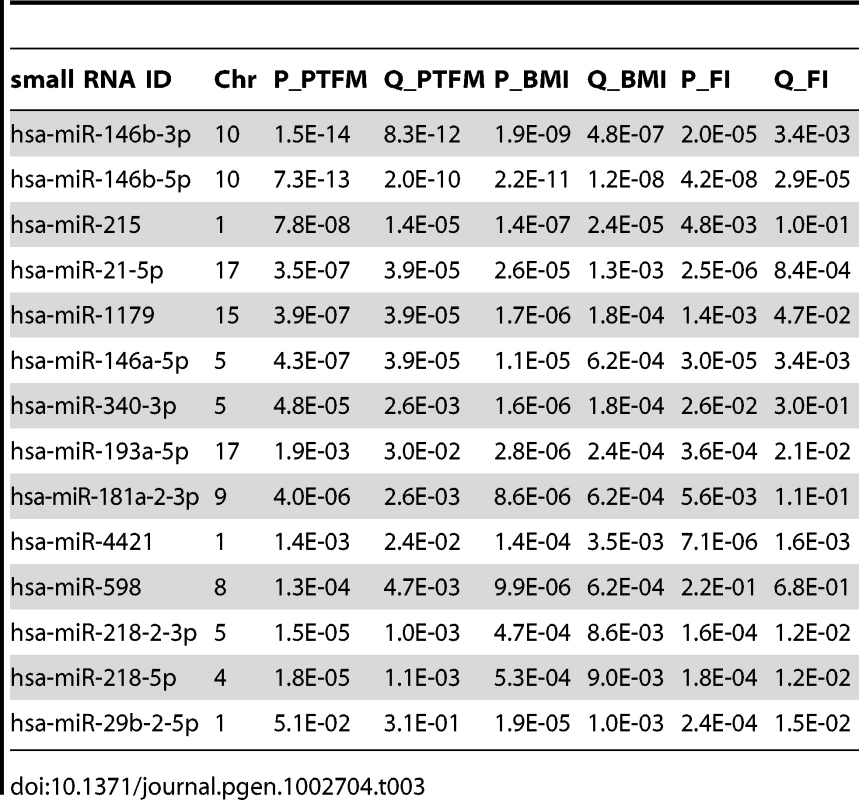
Fourteen small RNAs were highly significantly associated with at least one of the phenotypes (FDR<0.1%, Table 3, miRNA targets and functional enrichment analysis [56] in Table S10, Figure S3, S4). As a complement to the association analysis, we also contrasted small RNA gene expression levels between lean (BMI<25; n = 45) and obese (BMI>30; n = 36) subjects, and found that 43 small RNAs showed significant differences between the two groups (FDR<0.05, p<5.7×10−3, Table S11), including all significant hits from Table 3.
Four of the phenotype-associated small RNAs have previously been associated with metabolic phenotypes and/or adipogenesis. In a recent study, mir-1179 was found to be significantly associated (FDR<5%) with metabolic syndrome case control status, with lower expression levels in cases [22]. Here, we report similar associations between mir-1179 and obesity phenotypes with lower expression levels associated with increasing BMI, PTFM and FI, all of which are major components of metabolic syndrome. Mir-21, here significantly associated with obesity phenotypes (Table 3), has been reported to be involved in regulation of adipogenesis and lipid metabolism through its gene targets TGFBR2 and PPARalpha respectively [57]. Furthermore, mir-21 as well as mir-146b have been reported to be expressed at higher levels in skin tissue from diabetic mice [58], and in response to glucose stimulation in mouse adipocytes [59], [60]. Overexpression of mir-29 isoforms in mouse adipocytes resulted in an insulin resistant phenotype [59]. In a recent study, carried out in mouse islets, isoforms of mir-29 were found to contribute to the beta-cell-specific silencing of MCT1 (SLC16A1) expression required for appropriate insulin secretion [61]. In our study, mir-29b-2-5p was significantly associated BMI and fasting insulin, but not PTFM. Immune processes have previously been found to be enriched among mRNAs associated with metabolic phenotypes [17], [62], and mir-146a involved in inflammatory processes [63] and innate immunity [64] was here found to be associated with PTFM, BMI and insulin.
Genetic control of small RNA expression associated with obesity-related traits
We overlapped the SNPs for our 14 significant cis-eQTLs (cis-SNPs), with SNPs that are directly associated with obesity-related phenotypes in published genome wide association study (GWAS) data [65]. Four of the cis-SNPs were associated (nominal p<0.05) with body mass index (BMI) [65], one each with waist-hip-ratio adjusted for BMI (WHRadjBMI) [65], low density lipoprotein (LDL) high density lipoprotein (HDL), and total cholesterol (TC), and none with triglycerides (TG) [66] (Table 4).
Tab. 4. Overlap of <i>cis</i>-eQTLs (FDR<5%, p<2.4×10<sup>−4</sup>) and GWAS SNPs nominally significant (p<0.05) for body mass index (BMI), waist-hip-ratio adjusted for BMI (WHR(adjBMI)), low density lipoprotein (LDL), high density lipoprotein (HDL), and total cholesterol (TC). 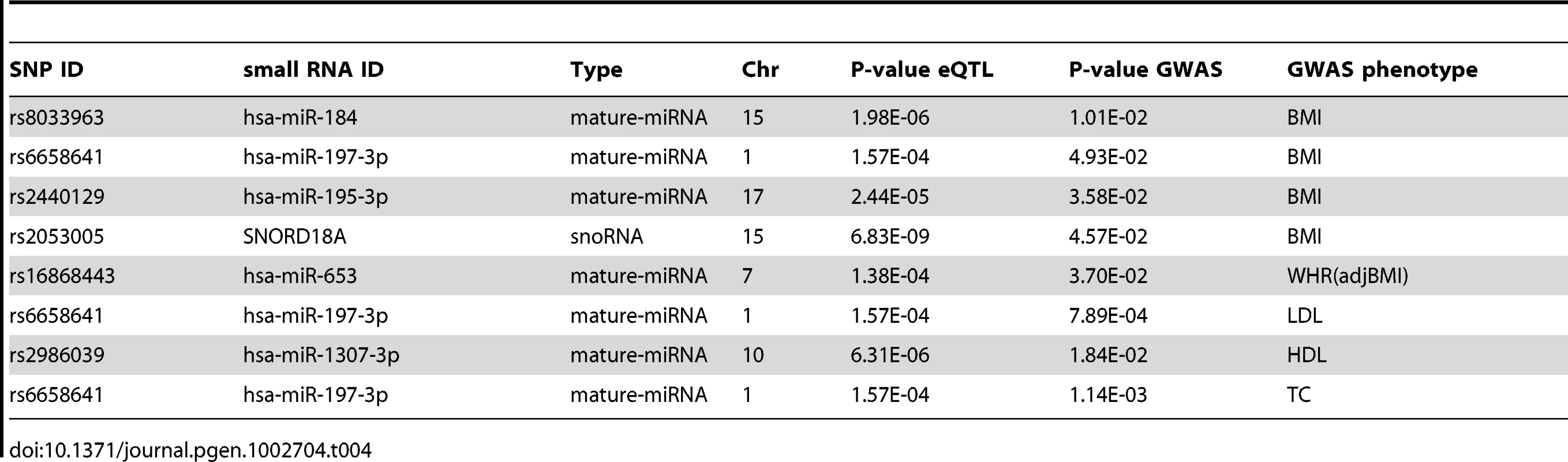
While on the whole, none of the cis-SNPs were genome-wide significant in the GWAS data, they were significantly enriched for nominally significant (p<0.05) SNPs in the BMI GWAS results ([65], binomial p = 0.007), indicating either their pleiotropic effect, or metabolic trait regulation through small RNA expression levels. rs2440129 was nominally significant in the BMI GWAS lookup [65], while mir-195-3p was significantly associated with both rs2440129 in cis (FDR<5%, p<2.4×10−5), as well as BMI (FDR<5%, p<3.9×10−3) and PTFM (FDR<5%, p<4.1×10−3), suggesting a mechanism for the rs2440129 association. Rs6658641 has a significant (FDR<5%, p<1.6×10−4) cis association with mir-197-3p in our data (Table 1), GNAI3 mRNA in three tissues (Table 2), as well as nominally significant associations to metabolic traits in GWAS. As mir-197 has been reported to regulate the expression of tumour suppressor gene FUS1 [67] and to be upregulated in type two diabetes patients [32], it is plausible that the effect of rs6658641 genotype on downstream expression and metabolic traits is mediated via the miRNA expression level.
miRNAs from the same cluster are correlated in expression, while miRNAs and mRNAs from the same transcript are not
miRNA genes are either processed from intronic mRNA sequence, or transcribed from endogenous promoters [68]. A single miRNA promoter can give rise to a transcript that includes a cluster of miRNAs that are then individually cleaved [69]. We tested whether pairwise correlations between expression levels of miRNAs in the same cluster (defined by Saini et al. [68] to be within a 10 kb block) are larger than those between random miRNAs, and found significant enrichment of positive correlation (Materials and Methods, Figure S5). The median of median pairwise correlations between cluster member expression levels was 0.37, compared to 0.03 of random miRNA sets of same size (p<10−8, Mann-Whitney U test). On the other hand, we found little evidence for relation between miRNA expression level and expression of its nearest mRNA probe. The distribution of correlation coefficients was centered on zero, without a heavy tail of positive correlation (Figure S6), a statistically significant difference to distribution of random small RNA-mRNA pairs (p>0.37, Mann-Whitney U test), or a trend for higher correlation for less distant probes. This shows that mRNA transcript levels are not good predictors of intronic miRNA levels in our dataset, and suggests that more miRNAs are expressed from an endogenous promoter than commonly appreciated, in line with recent findings [69], [70], [71].
There is little evidence for abundance of negative correlation between mRNA and miRNA levels
One of the two modes of miRNA action is directly regulating the transcript level via influencing the stability of the transcript, or direct cleavage [72]. To test whether variability in the miRNA expression levels is related to variability in its target mRNA expression, we calculated correlations between miRNA expression levels and their validated mRNA targets from miRecords [73] or predicted mRNA targets from tarBase v5 [74] both with and without accounting for experimental confounders in mRNA and miRNA data sets (Materials and Methods). To our surprise, we found that the average correlation between miRNA expression levels and their 522 validated targets was −0.012, and their 194,205 predicted targets −0.004. While these averages are statistically significantly less than 0 (one-sample t-test p<0.05 and 10−5 respectively), they indicate no strong enrichment of extreme negative correlations compared to random miRNA-mRNA pairs (Figure S7). We also tested whether the miRNA seed sequences are overrepresented in the 3′ UTR regions of the mRNA expression levels most negatively correlated to the miRNA using Sylamer ([75], Methods). Again, we found no evidence for significant enrichment (all q-values>0.5). This suggests that at a genome-wide level inter-individual variation of small RNA expression levels in our reference cohort does not have a detectably large effect on mRNA expression.
Expression ratio between miRNA arms varies across genes and individuals
The mature miRNA is processed from a double-stranded RNA hairpin by the Dicer RNAse [72], with the other arm assumed to be degraded [76]. The basis for choosing one of the hairpin arms as a mature product, and the extent to which the alternate arm (the less commonly observed product, previously also referred to as the star sequence) is functional, are not well understood [77], [78], [79]. To assess the extent of expression of both arms, and the variability of the relative expression ratio, we quantified the expression level of the alternate arm for 63 miRNAs. Other miRNA genes had only one arm detectably expressed, and only eight out of the 63 alternate arms were expressed at average level of at least 250 reads per sample. For seven miRNA genes, the alternate arm was on average more highly expressed compared to the mature product according to miRBase (Figure S8). Looking at variation between individuals, we found 12 mature sequence expression levels to be significantly correlated with their alternate arm sequence expression level (|Spearman's rho|>0.4, nominal p<2×10−5). For mir-186 and mir-29a, high abundance of the alternate arm sequence was indicative of low mature sequence levels, suggesting mutually exclusive selection of the arms. As the arm choice is suggested to be influenced by the nearby RNA context [78], [80], we tested for whether DNA variants in the region are correlated with the relative abundance of sequence from the two arms. We found SNP rs13174179 to be associated with the expression difference of miR-378 arms (nominal p = 5.3×10−4, FDR<10%).
Discussion
We have presented the largest small RNA sequencing dataset in a human reference cohort to date, and demonstrated the extent, causes, and consequences of the variability in small RNA expression.
Extent of variability
In spite of the medium complexity of the small RNA transcriptome, we quantified close to 1,000 different small RNA species. The highly expressed small RNAs fell into two categories in terms of inter-tissue variability - adipose-specific, and ubiquitously expressed microRNAs, corroborating previous observations [81], [82]. We confirmed that small RNA sequences have low genetic variability. This finding was especially pronounced for small RNAs highly expressed in the tissue we assayed, as only three derived alleles and three editing events were found. Additional genetic variants have been seen using DNA sequencing methods, but their potential functional impact remains to be assessed in other tissues where the genes are expressed above background level. Purifying selection acting on highly expressed as well as highly variable small RNAs was evident from their high conservation throughout the mammalian lineage, reiterating the importance of these functional molecules.
Causes of variability
Unexpectedly, some of the largest sources of variability in our data were due to the experimental protocol. The barcoding method used in this study, whereby the indexing tag and the unknown RNA are sequenced in the same read, caused a bias in terms of the profile of small RNAs that were captured. This could be addressed by using a generic 5′ adaptor and one that incorporates the indexing tag via PCR, such that the RNA sequence and the indexing tag are determined in separate reads, or performing the reverse transcription step directly on the flow cell [83]. Similar issues with tag bias have been observed and addressed in recent work published after the experiments reported here were carried out [84], [85]. Additional limitations for the library preparation were the quality and quantity of the starting material. Although not always feasible in a clinical situation, every attempt should be made to ensure that the quality of the total RNA is of a very high standard (minimum RIN of 8), and it is subject to minimal handling and freeze/thaw cycles prior to library construction. These considerations forced us to employ statistical methods to account for batch effects due to multiplexing tags, and to drop 37 samples from our initial design due to poor RNA quality.
Differences in sample preparation and sequencing platform introduce technical variation that biases and reduces the power of direct comparisons between small RNA sequencing studies [86]. We limited such confounding effects on our assessment of small RNA expression tissue specificity by using only Illumina short read data from other studies, and treating their raw reads in an identical manner to our samples. While we do not expect this to fully mitigate the problem, we do not expect that the residual bias produces the reported large differences between tissues. These considerations do not affect the rest of our analysis, for which the small RNA and mRNA data were collected from the same RNA samples, and genotyping and phenotyping were performed on the same individuals.
Another important issue for comparing RNA levels between samples and finding genetic associations was mapping bias due to sequence variants. Previously uncharacterised polymorphisms resulted in fewer reads mapped to samples with derived alleles, which also created a significant eQTL at a known linked SNP. We recommend projects using small RNA sequencing to employ our technique of including known genetic variation in the reference sequence, and to use an ambiguity aware aligner, such as NovoAlign, to avoid such pitfalls.
Correcting for these technical issues, we were able to explore the biological causes of small RNA expression variation. We found genetic associations at a rate comparable with mRNA transcripts, and replicated them in an independent cohort. Unexpectedly, we found eight cases of a locus genotype influencing expression levels of a nearby mRNA and a nearby small RNA, where in four of these cases the two were unlikely to share a transcript as they were separated by at least one additional transcribed region. This highlights that cis, or proximal signal does not have to be contained to the near vicinity of the transcript, and that distal regulatory sites are shared between multiple genes.
We also looked for coordinated transcription by direct correlation of nearby transcripts. Small RNAs are known to be expressed in clusters from a shared promoter, as well as cleaved from intronic RNA sequence [68]. While we found support for increased correlation between miRNAs from the same cluster, we did not see a global signal for correlation between intronic miRNAs and their nearest mRNA probe expression. Previous results have shown a strong relationship between average tissue mRNA expression level and the intronic miRNA expression [81], but our results suggest the additional variability around the average level is not as tightly linked, possibly due to an independent promoter of the miRNA, or additional postprocessing regulation of the spliced mRNA transcript.
Finally, phenotypic and environmental differences can and do elicit changes in the transcriptome. To this end, we found 51 small RNA genes whose expression level is significantly associated with metabolic phenotypes available for our cohort. Given the strength of the observed signal, it is not possible without additional information to distinguish between causal, reactive, and common cause models for the relationship between the expression and phenotype traits. Studies in mouse models and human cohorts have shown that environmental factors, such as diet, can influence the expression of both mRNA [87] as well as small RNA [88] in adipose tissue. We used fat biopsies taken from individuals who had been instructed to fast the day of the biopsy to control for potential confounding effect of the daily food consumption, but long term dietary behaviour was not available for these samples and thus could not be analysed. Modelling potential hidden causes of variation in the expression data did not increase the number of discoveries, suggesting that even if the environmental factors were observed, they could not be accounted for in a simple linear manner. Despite this, we can not infer in general that the phenotypic variability is due to changes in small RNA expression. In some cases however, previous findings suggest a plausible regulatory effect of small RNAs on phenotypes as highlighted in the results.
The MuTHER cohort was set up with the aim to assess heritability of gene expression in different tissues using twins. However, as using highly related subjects reduces the power to map eQTLs using association, we focused our resources on unrelated individuals in the clinically relevant adipose tissue for which related phenotype data and an eQTL replication cohort were available. Analysing multiple tissues, or employing a co-twin design to provide heritability estimates and immediate replication of the results could be followed up from this pilot study.
Consequences of variability
A major goal of this study was to assess the effect of naturally occurring variation in miRNA expression levels on the mRNA levels. However, we found no evidence for miRNA expression variation to be correlated with target mRNA variation. This negative result cannot be due to the amount of noise in our data alone, as we could successfully detect genetic effects and phenotype correlations. Thus, the strength of association between natural variation of miRNA expression and variation in their target mRNA expression is limited to a smaller scale than that of genetic control or downstream effects of global metabolic phenotypes. This lack of tight target regulation supports the growing body of evidence [22] that quantitative variation of small RNA expression within a tissue does not have even a moderately sized effect on its target mRNA levels, and is consistent with a primary role of miRNAs being to buffer mRNA levels, for example to a random fluctuations of transcriptional regulators.
The small effect size of drastic miRNA level perturbation via knockdown, transfection, or overexpression of a single miRNA on its target mRNA expression levels has already been shown in several recent studies in human cell lines. For example, the median log2 expression level change of the top 150 TargetScan conserved targets was 0.096 (6.9%) for mir-29 knockdown in fetal lung fibroblasts [89], 0.131 (9.5%) for mir-145 transfection of MB-231 breast cancer cells [90], 0.173 (12.7%) for mir-30 overexpression in melanoma cell lines [91], and 0.465 (38.0%) for mir-7 overexpression in A549 cancer cells [92]. Thus, even for these extreme perturbations of miRNA levels, the observed effects on the target mRNAs are not pronounced. It is therefore not surprising that the naturally occurring inter-individual variation also does not have a large effect.
For the first time, we were able to assess the expression variation of both microRNA arms. We found that while the alternative arms (star sequences) are not highly expressed in general, there are several of them that are not degraded, and are expressed at appreciable levels. We also observed examples of high mature miRNA expression being correlated with low expression of the alternate arm, and a relatively strong genetic signal for arm choice of one miRNA.
Materials and Methods
Samples and phenotypes
The unrelated individuals included in this study are part of the MuTHER study of Caucasian females (median age 58) recruited from the UK Adult Twin Registry (TwinsUK, [34]). Punch biopsies (8 mm) were taken from a relatively photo-protected area adjacent and inferior to the umbilicus, subcutaneous adipose tissue was dissected followed by DNA and RNA extraction as described in [20]. For inclusion in this study the requirements were that the individuals were not under hormone replacement therapy, and did not have confirmed Diabetes Mellitus Type 2. Subjects were instructed to fast on the day of the biopsy to avoid potential biases due to food consumption. We used genotypes obtained, filtered and imputed to HapMap2 as described in [20]. The previously published gene expression values [20] were obtained using the Illumina Human HT-12 V3 BeadChips, followed by filtering and normalisation, and are available at the ArrayExpress [93] (www.ebi.ac.uk/arrayexpress) under accession number E-TABM-1140. Metabolic phenotypes were measured at the same time point as the biopsies and were collected as previously described, including BMI [94], DEXA measurements of percentage trunk fat mass, fasting glucose [95] and fasting insulin [96].
Library preparation and sequencing
Only samples with good quality total RNA (no visible degradation in BioAnalyzer profile and RIN scores in excess of 6.7) were selected for small RNA isolation. Low molecular weight RNA (<40 nucleotides) was size-selected from between 0.5 to 1.0 µg total RNA using a flashPAGE Fractionator (Ambion, Austin, TX, USA). The recovered small RNAs were first ligated to the Illumina v1.5 small RNA 3′ adaptor (Illumina, Inc., San Diego, CA, USA) using T4 RNA ligase 2 - truncated (New England Biolabs, Ipswich, MA, USA). This was followed by a second ligation, using T4 RNA ligase 1, to one of twelve modified Illumina SRA 5′ adaptors, each with a six-base index tag at the 3′ end (Sigma-Aldrich, Haverhill, UK). Both ligation steps were performed according to the Illumina v1.5 protocol. The 5′ and 3′ adaptor-ligated small RNAs were immediately reverse transcribed, amplified and size-selected as described in the Illumina v1.5 protocol. The completed cDNA libraries were pooled (12 libraries per pool) in equimolar amounts and were sequenced using 37 base reads on the Illumina GAII platform.
Raw data analysis
Raw sequencing data was obtained in FASTQ format, and processed with R [97](Bioconductor [98], [99], Biostrings and ShortRead [98], [99] packages) and python scripts. We first assigned the raw reads to their corresponding multiplexing tags. For this, we calculated the edit distance of the first six bases to all 12 index tag sequences used in the study, considering 0.25 as the distance between N and any other base. Reads with edit distances of at least 2.75 to all tags were discarded as well as those with the same minimum edit distance to more than one tag. The remaining reads were assigned to the library corresponding to the shortest edit distance, and their first six bases were removed before proceeding. The next step consisted of locating and trimming sequences matching the small RNA 3′ adaptor using the trimLRPatterns function and allowing for mismatches of up to 20% of the alignment length. The first 12 bases of the 3′ adaptor sequence were allowed to align to any location within the short reads, and if no alignment was found, a shortened adaptor sequence was realigned iteratively by removing one base from the 3′ end and anchoring the alignment to the 3′ end of the short reads. To further clean up the short reads to help avoid ambiguous mappings, any window of five bases with at least three Ns was located and the read was trimmed starting at the position of the first N. Any occurrence of an N within two bases of the 3′ end of the read was also trimmed. The reads were then low-complexity filtered to remove those with > = 90% of a single base. After all filtering steps, reads with less than 16 bases were discarded. All remaining read sequences should correspond to short RNA molecules present in the samples, and length histograms were produced to confirm the enrichment of a miRNA peak around 22 bases.
Mapping and quantification
Accurate quantification of small RNA molecule counts from read data is challenging due to genetic variation in the sequence, ambiguities in read mapping, and frequent contamination by large numbers of adaptor dimers. To solve these problems, we used a multi-stage mapping approach to exclude contaminating molecules that could be due to the library preparation kit, prioritise alignments to known small RNAs, and take genetic variation into account.
First, we aligned the known small RNA molecule sequences against the human reference genome (NCBI build 37), retrieved all known variants in the mapped regions from the UK10K sequencing data (July 2011 release, personal communication, UK10K Consortium), and created an individual sequence of each small RNA, with variable bases denoted with the corresponding IUPAC ambiguity codes. We included all mapped regions for RNAs that mapped to more than one genomic location. We then created five synthetic reference genomes (all with the ambiguous bases at variable sites) corresponding to:
-
Contaminating sequences of adaptors, linkers, adaptor-linker, and adaptor-tag combinations with all possible single nucleotide alterations (mutation, insertion, deletion).
-
Human mature miRNA and miRNA* sequences from miRBase v17 [7], but using the hairpin sequence to extend them by up to 3 bp from the 5′ end and 5 bp from the 3′ end
-
All known and predicted human non-coding RNA sequences from Ensembl version 63 together with full-length miRBase hairpin sequences [7], [35]
-
All known human non-coding RNA pseudogenes from Ensembl version 63 [35]
-
Human reference genome (NCBI Build 37)
We mapped reads to these references using BWA [100] (bwa aln -n 2 -o 1) and with novoalign v. 2.07.11 (http://www.novocraft.com, parameters -h 60 60 -t30 -s -m -l 16 -R 0 -r A 30). The latter is aware of sequence ambiguities, but the former was more sensitive at detecting reads aligning to the contaminating sequences. We also tested Bowtie [101], but did not use it due to the inability of the tested version to handle indels.
For the mapping calls, we excluded reads mapping to contaminating sequence with either method. For both aligners, we then took all the alignments in the highest stratum of references (miRNA>ncRNA>pseudogenes>genome), and picked the ones with the smallest edit distance. Conservatively, we only retained alignments of a read if both aligners agreed on all the aligned locations. For reads mapping genome-wide, but not any known ncRNAs, we created the set of uncharacterised RNA loci covered by at least one read in at least one sample without gaps, and assigned reads to their corresponding uncharacterised loci.
Finally, we quantified the expression level of each ncRNA and unannotated locus by counting the number of reads aligning to it. If a read mapped between k alternative sequences or loci in one reference, we added 1/k to the count of each. We trimmed the data matrix to contain only RNAs that were observed at least 1000 times across all individuals, or at least 100 times in a single individual. We discarded individuals with less than 500,000 mapped reads. This retained 131 individuals, including 129 individuals with at least 800,000 reads, and 119 individuals with at least 1,500,000 reads.
This multi-stage approach excludes mapping contaminating sequences to the reference, avoids allelic imbalance due to ability to map by incorporating information on genetic variation, and resolves potential mapping ambiguities to reflect our belief of how small RNA molecules are generated.
Normalisation
To use the read counts quantitatively, we normalised the data to have a comparable total number of reads for each individual. We estimated a size factor s for library j as the median inflation factor across all genes: sj = mediang (ngj/GMj(ngj)), where GM stands for the geometric mean, and ngj is the read count of gene g for individual j as recommended by Anders and Huber [102]. For further analyses, we used the log2-transformed corrected values log2(ngj/sj) to account for heteroskedasticity in the data.
Unannotated read overlaps
We downloaded UCSC genome browser [50] tracks for DNASE hypersensitivity sites, and ENSEMBL gene structures for human genome version 37, and calculated their total length, as well as overlap with the loci giving rise to unannotated small RNA molecules. We calculated the significance of the enrichment of unannotated regions in the track from the probability of observing at least as many overlaps of the 701 unannotated regions given the frequency of bases covered by each track using a standard binomial test.
Batch effects
We tested for significance of the correlation coefficient r between covariates and principal components of the raw read count data as well as log-transformed and normalised data by calculating a statistic t = r ((1−r2)/129)−0.5, and calculating the (two-tailed) probability of observing at least as extreme a value, and Bonferroni correcting for 131 tests (one for each PC). We called the correlation significant, if the corrected p-value was less than 0.05, corresponding to |r|>0.31.
Sequence variation calling
For each sample, we created sorted BAM files from the alignment output, and called segregating sites using Samtools v.0.1.12 (samtools pileup –vcf ) [103]. We then combined the list of all called variable sites across all samples with sites from the UK10K project (June 2011 release, personal communication, UK10K Consortium), created pileup files at them for each sample (samtools pileup –l sites.tab –f ref.fa), and combined all the information into a single table giving the number of times each nucleotide was observed in every sample for each site. The sites were filtered to have information from at least 20 samples, have at least one sample with at least 10 observed alleles, and have at least one sample with at least 20% non-reference allele frequency. We further discarded three sites as likely false positives – one had 23 observed non-reference alleles, with 16 in one sample and no DNA evidence (see below), and the other two were variants in the last base of the mature miRNA, consistent with a modified degradation product. For validating the genotypes using genome sequencing data, we constructed DNA read pileup files at same sites for 40 of our samples sequenced in the UK10K cohort. We called a site to be a DNA polymorphism, if it had at least five DNA sequencing reads supporting the non-reference allele. An A to I edit was called if there were no more than two DNA sequencing reads with a G allele, and both A and G alleles were observed at least 90% of the samples, implying extreme deviation from Hardy-Weinberg equilibrium.
Novel miRNA gene calling
We applied the MapMi pipeline [36] to find potential novel miRNA loci. We retrieved the sequences of unannotated genomic regions, calculated their corresponding RNA secondary structure using RNAfold [104], and applied the MapMi classifier to obtain a structure score s. We calculated the self-containment score c of hairpins with s>35 as described in [105], and retained hairpins satisfying s*c>35. We then mapped all reads to the filtered candidate hairpins using bowtie, allowing for zero mismatches, and manually assessed the structural characteristics, genomic context, and alignment pileup shape for each candidate hairpin.
Evolutionary analyses
Mammalian conservation scores were downloaded from the UCSC genome browser [50]. A chi-square test with one degree of freedom was used to test the deviation of the fraction of highly expressed (average log-scale expression>10) unconserved (conservation score <0) genes from expectation. Similar test was used for highly variable (log scale variability>5) unconserved genes.
RNA correlation analyses
We subtracted off the linear fit of sample covariates (library batch and multiplexing tag), from the log-transformed, normalised data, and calculated the Pearson correlation coefficient between the residual expression levels and other small RNAs, miRNA star expression levels, and mRNA probe expression levels from [20]. For mRNA levels, we used both raw measurements, as well as residuals after correcting for global variance components using PEER. We also tested for correlation with uncorrected expression levels, and using linear models as described below, but found no additional enrichment of statistical signal.
miRNA seed enrichment
miRNA binding specificity is controlled through binding of its seed region (bases 1–8 of the mature miRNA) with seed complementary regions (SCRs) in the 3′ UTR of mRNAs. Binding is enhanced if a SCR matches the first seed nucleotide with an adenosine, irrespective of the seed nucleotide [72]. As the strongest statistical associations have been reported for regions of length 6, 7 and 8 (the full region), we combined analyses for these seed lengths.
For each miRNA, we ordered probes and their associated 3′ UTRs by correlation of probe expression values to the miRNA, with correlations calculated in four different ways as described above. For each possible 8-nucleotide sequence s8 ending in an adenosine, we considered its middle 6-mer s6, the two constituent 7-mers s7,1 and s7,2 and the 8-mer s8 itself as the seeds. For each of these four seeds s and given n, we used Sylamer 08-185 [75] to calculate a hypergeometric p-value pn(s) to assess the extent to which the number of their SCR incidences in the top n of the ordered 3′ UTRs deviated from the expected. Potential nucleotide composition biases were accounted for using third order Markov correction (flag -m 4). The seed enrichment score for s8 was calculated as maxn(−log10 pn(s6)−log10 pn(s7,1)−log10 pn(s7,2)−log10 pn(s8)) using a grid of values for n.
For each miRNA that produced a ranked list of probes, the null distribution of observed scores was estimated by fitting an extreme value distribution for all calculated adenosine-ending 8-mer scores using the R function fgev from the evd package. For the miRNA used to generate the list, the significance of its influence on the mRNA expression was evaluated by testing its seed enrichment score against the estimated null. q-values were calculated for the ordered list of miRNA p-values.
Reanalysis of miRNA perturbation experiments
We selected series GSE18651, GSE19737, GSE27718, GSE14507 from the Gene Expression Omnibus [106] in which a particular miRNA was directly perturbed, either by knockdown or overexpression. We downloaded the normalized expression data using Bioconductor package GEOquery [107], and performed differential expression analysis using limma [108] to sort the genes according to fold-change in response to the perturbation. We first validated that the mRNA expression changes actually represent the direct effect of a miRNA on its targets. To do so we used Sylamer [75] to search for enrichment of seed-matches in the 3′UTR sequences in the appropriate portion of the genelist, i.e. in knockdown experiments targets should be up-regulated, upon overexpression targets should be down-regulated. We then obtained the targets of each miRNA according to TargetScan v5 [109], and calculated their median fold-change in the corresponding experiment. We tested different sets of targets, prioritizing by evolutionary conservation (PCT) or by context-score, and selecting the 150 targets with the best scores. In all cases the median fold-change of these target sets was quite low, representing changes of 5–38%. Selecting more targets led to a reduction in the median fold-change. We also calculated the median fold-change of all possible targets, taking the full set of transcripts with at least a 7mer seed-match in their 3′UTR. These larger sets had the lowest median fold-changes, representing a 2–8% change in expression. All this confirms the notion that miRNAs do not act as on-off switches on the majority of their targets. Even in experiments that dramatically alter miRNA abundance, the average effect upon targets is modest.
Genotype and phenotype associations
Associations between snoRNA and miRNA expression and mean genotypes (expected minor allele count under IMPUTE posterior probabilities, MAF>5%, IMPUTE info value>0.8) or phenotypes were tested using a linear model implemented in R [97]. Cis-eQTL analysis was limited to SNPs located within 100 kB either side of the transcript. The linear model was adjusted for age, multiplex tag and library batch. The significance of the genotype or phenotype effect was calculated from the Chi-square distribution with 1 degree of freedom using −2log(likelihood ratio) as the test statistic. False discovery rate (FDR) was calculated using the qvalue package implemented in R 2.11 [97]. Corrections for multiple testing were done using q-values to control the false discovery rate (FDR) at 5%. To calculate the FDR, the associations between the 591 small RNAs and all the cis-located SNPs for each small RNA, were considered.
To test for difference in small RNA expression between obese (BMI>30) and lean (BMI<25) individuals we treated BMI, for subjects falling into one of the two BMI groups, as a binary categorical variable. Linear models where fitted with small RNA expression level as response variable and the lean/obese categorical variable as the predictor while adjusting for relevant covariates (age, library batch, multiplex tag). Significance of the effect size estimates of the lean/obese predictor was determined by a likelihood ratio test, and FDR was calculated using the qvalue package.
Supporting Information
Zdroje
1. MattickJSMakuninIV 2006 Non-coding RNA. Hum Mol Genet 15 Spec No 1 R17 29
2. LauNCLimLPWeinsteinEGBartelDP 2001 An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science 294 858 862
3. Lagos-QuintanaMRauhutRLendeckelWTuschlT 2001 Identification of novel genes coding for small expressed RNAs. Science 294 853 858
4. HeLHannonGJ 2004 MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet 5 522 531
5. CzechBHannonGJ 2011 Small RNA sorting: matchmaking for Argonautes. Nat Rev Genet 12 19 31
6. LeeRCFeinbaumRLAmbrosV 1993 The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75 843 854
7. KozomaraAGriffiths-JonesS 2011 miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res 39 D152 157
8. WinterJJungSKellerSGregoryRIDiederichsS 2009 Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat Cell Biol 11 228 234
9. KimVNHanJSiomiMC 2009 Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol 10 126 139
10. FangZRajewskyN 2011 The impact of miRNA target sites in coding sequences and in 3′UTRs. PLoS ONE 6 e18067 doi:10.1371/journal.pone.0018067
11. KissT 2002 Small nucleolar RNAs: an abundant group of noncoding RNAs with diverse cellular functions. Cell 109 145 148
12. BrameierMHerwigAReinhardtRWalterLGruberJ 2011 Human box C/D snoRNAs with miRNA like functions: expanding the range of regulatory RNAs. Nucleic Acids Res 39 675 686
13. MarzMGruberARHoner Zu SiederdissenCAmmanFBadeltS 2011 Animal snoRNAs and scaRNAs with exceptional structures. RNA Biol 8
14. TaftRJGlazovEALassmannTHayashizakiYCarninciP 2009 Small RNAs derived from snoRNAs. RNA 15 1233 1240
15. LukkMKapusheskyMNikkilaJParkinsonHGoncalvesA 2010 A global map of human gene expression. Nat Biotechnol 28 322 324
16. StrangerBENicaACForrestMSDimasABirdCP 2007 Population genomics of human gene expression. Nat Genet 39 1217 1224
17. EmilssonVThorleifssonGZhangBLeonardsonASZinkF 2008 Genetics of gene expression and its effect on disease. Nature 452 423 428
18. CheungVGNayakRRWangIXElwynSCousinsSM 2010 Polymorphic cis - and trans-regulation of human gene expression. PLoS Biol 8 e1000480 doi:10.1371/journal.pbio.1000480
19. NicaACPartsLGlassDNisbetJBarrettA 2011 The architecture of gene regulatory variation across multiple human tissues: the MuTHER study. PLoS Genet 7 e1002003 doi:10.1371/journal.pgen.1002003
20. SmallKSHedmanAKGrundbergENicaACThorleifssonG 2011 Identification of an imprinted master trans regulator at the KLF14 locus related to multiple metabolic phenotypes. Nat Genet 43 561 564
21. BorelCDeutschSLetourneauAMigliavaccaEMontgomerySB 2011 Identification of cis - and trans-regulatory variation modulating microRNA expression levels in human fibroblasts. Genome Res 21 68 73
22. RantalainenMHerreraBMNicholsonGBowdenRWillsQF 2011 MicroRNA Expression in Abdominal and Gluteal Adipose Tissue Is Associated with mRNA Expression Levels and Partly Genetically Driven. PLoS ONE 6 e27338 doi:10.1371/journal.pone.0027338
23. NicaACMontgomerySBDimasASStrangerBEBeazleyC 2010 Candidate causal regulatory effects by integration of expression QTLs with complex trait genetic associations. PLoS Genet 6 e1000895 doi:10.1371/journal.pgen.1000895
24. FehrmannRSJansenRCVeldinkJHWestraHJArendsD 2011 Trans-eQTLs reveal that independent genetic variants associated with a complex phenotype converge on intermediate genes, with a major role for the HLA. PLoS Genet 7 e1002197 doi:10.1371/journal.pgen.1002197
25. PartsLStegleOWinnJDurbinR 2011 Joint genetic analysis of gene expression data with inferred cellular phenotypes. PLoS Genet 7 e1001276 doi:10.1371/journal.pgen.1001276
26. TakanabeROnoKAbeYTakayaTHorieT 2008 Up-regulated expression of microRNA-143 in association with obesity in adipose tissue of mice fed high-fat diet. Biochem Biophys Res Commun 376 728 732
27. BrestPLapaquettePSouidiMLebrigandKCesaroA 2011 A synonymous variant in IRGM alters a binding site for miR-196 and causes deregulation of IRGM-dependent xenophagy in Crohn's disease. Nat Genet 43 242 245
28. GehrkeSImaiYSokolNLuB 2010 Pathogenic LRRK2 negatively regulates microRNA-mediated translational repression. Nature 466 637 641
29. KumarMSLuJMercerKLGolubTRJacksT 2007 Impaired microRNA processing enhances cellular transformation and tumorigenesis. Nat Genet 39 673 677
30. SahooTdel GaudioDGermanJRShinawiMPetersSU 2008 Prader-Willi phenotype caused by paternal deficiency for the HBII-85 C/D box small nucleolar RNA cluster. Nat Genet 40 719 721
31. EstellerM 2011 Non-coding RNAs in human disease. Nat Rev Genet 12 861 874
32. KlotingNBertholdSKovacsPSchonMRFasshauerM 2009 MicroRNA expression in human omental and subcutaneous adipose tissue. PLoS ONE 4 e4699 doi:10.1371/journal.pone.0004699
33. Grundberg ESKSHedmanÅKNicaACBuilAKeildsonSBellJT 2012 Mapping cis and trans regulatory effects across multiple tissues in twins: the MuTHER study. submitted
34. SpectorTDWilliamsFM 2006 The UK Adult Twin Registry (TwinsUK). Twin Res Hum Genet 9 899 906
35. FlicekPAmodeMRBarrellDBealKBrentS 2011 Ensembl 2011. Nucleic Acids Res 39 D800 806
36. Guerra-AssuncaoJAEnrightAJ 2010 MapMi: automated mapping of microRNA loci. BMC Bioinformatics 11 133
37. KimTKHembergMGrayJMCostaAMBearDM 2010 Widespread transcription at neuronal activity-regulated enhancers. Nature 465 182 187
38. EsauCKangXPeraltaEHansonEMarcussonEG 2004 MicroRNA-143 regulates adipocyte differentiation. J Biol Chem 279 52361 52365
39. van SolingenCSeghersLBijkerkRDuijsJMRoetenMK 2009 Antagomir-mediated silencing of endothelial cell specific microRNA-126 impairs ischemia-induced angiogenesis. J Cell Mol Med 13 1577 1585
40. LeeDYDengZWangCHYangBB 2007 MicroRNA-378 promotes cell survival, tumor growth, and angiogenesis by targeting SuFu and Fus-1 expression. Proc Natl Acad Sci U S A 104 20350 20355
41. WangQHuangZXueHJinCJuXL 2008 MicroRNA miR-24 inhibits erythropoiesis by targeting activin type I receptor ALK4. Blood 111 588 595
42. Bruchova-VotavovaHYoonDPrchalJT 2010 miR-451 enhances erythroid differentiation in K562 cells. Leuk Lymphoma 51 686 693
43. KuchenSReschWYamaneAKuoNLiZ 2010 Regulation of microRNA expression and abundance during lymphopoiesis. Immunity 32 828 839
44. HouJLinLZhouWWangZDingG 2011 Identification of miRNomes in human liver and hepatocellular carcinoma reveals miR-199a/b-3p as therapeutic target for hepatocellular carcinoma. Cancer Cell 19 232 243
45. StarkMSTyagiSNancarrowDJBoyleGMCookAL 2010 Characterization of the Melanoma miRNAome by Deep Sequencing. PLoS ONE 5 e9685 doi:10.1371/journal.pone.0009685
46. SchotteDMoqadamFALange-TurenhoutEAChenCvan IjckenWF 2011 Discovery of new microRNAs by small RNAome deep sequencing in childhood acute lymphoblastic leukemia. Leukemia 25 1389 1399
47. FaghihiMAZhangMHuangJModarresiFVan der BrugMP 2010 Evidence for natural antisense transcript-mediated inhibition of microRNA function. Genome Biol 11 R56
48. SaundersMALiangHLiWH 2007 Human polymorphism at microRNAs and microRNA target sites. Proc Natl Acad Sci U S A 104 3300 3305
49. WymanSKKnoufECParkinRKFritzBRLinDW 2011 Post-transcriptional generation of miRNA variants by multiple nucleotidyl transferases contributes to miRNA transcriptome complexity. Genome Res 21 1450 1461
50. FujitaPARheadBZweigASHinrichsASKarolchikD 2011 The UCSC Genome Browser database: update 2011. Nucleic Acids Res 39 D876 882
51. SchroederAMuellerOStockerSSalowskyRLeiberM 2006 The RIN: an RNA integrity number for assigning integrity values to RNA measurements. BMC Mol Biol 7 3
52. StegleOPartsLDurbinRWinnJ 2010 A Bayesian framework to account for complex non-genetic factors in gene expression levels greatly increases power in eQTL studies. PLoS Comput Biol 6 e1000770 doi:10.1371/journal.pcbi.1000770
53. LeekJTStoreyJD 2007 Capturing heterogeneity in gene expression studies by surrogate variable analysis. PLoS Genet 3 e161 doi:10.1371/journal.pgen.0030161
54. StegleOPartsLPiipariMWinnJDurbinR 2012 Using probabilistic estimation of expression residuals (PEER) to obtain increased power and interpretability of gene expression analyses. Nat Protoc 7 500 507
55. FuJWolfsMGDeelenPWestraHJFehrmannRS 2012 Unraveling the regulatory mechanisms underlying tissue-dependent genetic variation of gene expression. PLoS Genet 8 e1002431 doi:10.1371/journal.pgen.1002431
56. ReimandJArakTViloJ 2011 g:Profiler–a web server for functional interpretation of gene lists (2011 update). Nucleic Acids Res 39 W307 315
57. KidaKNakajimaMMohriTOdaYTakagiS 2011 PPARalpha is regulated by miR-21 and miR-27b in human liver. Pharm Res 28 2467 2476
58. MadhyasthaRMadhyasthaHNakajimaYOmuraSMaruyamaM 2011 MicroRNA signature in diabetic wound healing: promotive role of miR-21 in fibroblast migration. Int Wound J
59. HeAZhuLGuptaNChangYFangF 2007 Overexpression of micro ribonucleic acid 29, highly up-regulated in diabetic rats, leads to insulin resistance in 3T3-L1 adipocytes. Mol Endocrinol 21 2785 2794
60. HerreraBMLockstoneHETaylorJMRiaMBarrettA 2010 Global microRNA expression profiles in insulin target tissues in a spontaneous rat model of type 2 diabetes. Diabetologia 53 1099 1109
61. PullenTJda Silva XavierGKelseyGRutterGA 2011 miR-29a and miR-29b contribute to pancreatic beta-cell-specific silencing of monocarboxylate transporter 1 (Mct1). Mol Cell Biol 31 3182 3194
62. ChenYZhuJLumPYYangXPintoS 2008 Variations in DNA elucidate molecular networks that cause disease. Nature 452 429 435
63. PerryMMMoschosSAWilliamsAEShepherdNJLarner-SvenssonHM 2008 Rapid changes in microRNA-146a expression negatively regulate the IL-1beta-induced inflammatory response in human lung alveolar epithelial cells. J Immunol 180 5689 5698
64. WilliamsAEPerryMMMoschosSALarner-SvenssonHMLindsayMA 2008 Role of miRNA-146a in the regulation of the innate immune response and cancer. Biochem Soc Trans 36 1211 1215
65. LindgrenCMHeidIMRandallJCLaminaCSteinthorsdottirV 2009 Genome-wide association scan meta-analysis identifies three Loci influencing adiposity and fat distribution. PLoS Genet 5 e1000508 doi:10.1371/journal.pgen.1000508
66. TeslovichTMMusunuruKSmithAVEdmondsonACStylianouIM 2010 Biological, clinical and population relevance of 95 loci for blood lipids. Nature 466 707 713
67. DuLSchagemanJJSubausteMCSaberBHammondSM 2009 miR-93, miR-98, and miR-197 regulate expression of tumor suppressor gene FUS1. Mol Cancer Res 7 1234 1243
68. SainiHKGriffiths-JonesSEnrightAJ 2007 Genomic analysis of human microRNA transcripts. Proc Natl Acad Sci U S A 104 17719 17724
69. IsikMKorswagenHCBerezikovE 2010 Expression patterns of intronic microRNAs in Caenorhabditis elegans. Silence 1 5
70. MartinezNJOwMCReece-HoyesJSBarrasaMIAmbrosVR 2008 Genome-scale spatiotemporal analysis of Caenorhabditis elegans microRNA promoter activity. Genome Res 18 2005 2015
71. OzsolakFPolingLLWangZLiuHLiuXS 2008 Chromatin structure analyses identify miRNA promoters. Genes Dev 22 3172 3183
72. BartelDP 2009 MicroRNAs: target recognition and regulatory functions. Cell 136 215 233
73. XiaoFZuoZCaiGKangSGaoX 2009 miRecords: an integrated resource for microRNA-target interactions. Nucleic Acids Res 37 D105 110
74. PapadopoulosGLReczkoMSimossisVASethupathyPHatzigeorgiouAG 2009 The database of experimentally supported targets: a functional update of TarBase. Nucleic Acids Res 37 D155 158
75. van DongenSAbreu-GoodgerCEnrightAJ 2008 Detecting microRNA binding and siRNA off-target effects from expression data. Nat Methods 5 1023 1025
76. RubyJGStarkAJohnstonWKKellisMBartelDP 2007 Evolution, biogenesis, expression, and target predictions of a substantially expanded set of Drosophila microRNAs. Genome Res 17 1850 1864
77. WarfMBJohnsonWEBassBL 2011 Improved annotation of C. elegans microRNAs by deep sequencing reveals structures associated with processing by Drosha and Dicer. RNA 17 563 577
78. Griffiths-JonesSHuiJHMarcoARonshaugenM 2011 MicroRNA evolution by arm switching. EMBO Rep 12 172 177
79. de WitELinsenSECuppenEBerezikovE 2009 Repertoire and evolution of miRNA genes in four divergent nematode species. Genome Res 19 2064 2074
80. OkamuraKPhillipsMDTylerDMDuanHChouYT 2008 The regulatory activity of microRNA* species has substantial influence on microRNA and 3′ UTR evolution. Nat Struct Mol Biol 15 354 363
81. LiangYRidzonDWongLChenC 2007 Characterization of microRNA expression profiles in normal human tissues. BMC Genomics 8 166
82. LandgrafPRusuMSheridanRSewerAIovinoN 2007 A mammalian microRNA expression atlas based on small RNA library sequencing. Cell 129 1401 1414
83. MamanovaLTurnerDJ 2011 Low-bias, strand-specific transcriptome Illumina sequencing by on-flowcell reverse transcription (FRT-seq). Nat Protoc 6 1736 1747
84. AlonSVigneaultFEminagaSChristodoulouDCSeidmanJG 2011 Barcoding bias in high-throughput multiplex sequencing of miRNA. Genome Res 21 1506 1511
85. Van NieuwerburghFSoetaertSPodshivalovaKAy-Lin WangESchafferL 2011 Quantitative Bias in Illumina TruSeq and a Novel Post Amplification Barcoding Strategy for Multiplexed DNA and Small RNA Deep Sequencing. PLoS ONE 6 e26969 doi:10.1371/journal.pone.0026969
86. ToedlingJServantNCiaudoCFarinelliLVoinnetO 2012 Deep-Sequencing Protocols Influence the Results Obtained in Small-RNA Sequencing. PLoS ONE 7 e32724 doi:10.1371/journal.pone.0032724
87. DahlmanILinderKArvidsson NordstromEAnderssonILidenJ 2005 Changes in adipose tissue gene expression with energy-restricted diets in obese women. Am J Clin Nutr 81 1275 1285
88. ParraPSerraFPalouA 2010 Expression of adipose microRNAs is sensitive to dietary conjugated linoleic acid treatment in mice. PLoS ONE 5 e13005 doi:10.1371/journal.pone.0013005
89. CushingLKuangPPQianJShaoFWuJ 2011 miR-29 is a major regulator of genes associated with pulmonary fibrosis. Am J Respir Cell Mol Biol 45 287 294
90. GotteMMohrCKooCYStockCVaskeAK 2010 miR-145-dependent targeting of junctional adhesion molecule A and modulation of fascin expression are associated with reduced breast cancer cell motility and invasiveness. Oncogene 29 6569 6580
91. Gaziel-SovranASeguraMFDi MiccoRCollinsMKHannifordD 2011 miR-30b/30d regulation of GalNAc transferases enhances invasion and immunosuppression during metastasis. Cancer Cell 20 104 118
92. WebsterRJGilesKMPriceKJZhangPMMattickJS 2009 Regulation of epidermal growth factor receptor signaling in human cancer cells by microRNA-7. J Biol Chem 284 5731 5741
93. ParkinsonHSarkansUKolesnikovNAbeygunawardenaNBurdettT 2011 ArrayExpress update–an archive of microarray and high-throughput sequencing-based functional genomics experiments. Nucleic Acids Res 39 D1002 1004
94. SkidmorePMCassidyASwaminathanRRichardsJBSpectorTD 2008 Relation of birth weight, body mass index, and change in size from birth to adulthood to insulin resistance in a female twin cohort. J Clin Endocrinol Metab 93 516 520
95. ProkopenkoILangenbergCFlorezJCSaxenaRSoranzoN 2009 Variants in MTNR1B influence fasting glucose levels. Nat Genet 41 77 81
96. FalchiMWilsonSGPaximadasDSwaminathanRSpectorTD 2008 Quantitative linkage analysis for pancreatic B-cell function and insulin resistance in a large twin cohort. Diabetes 57 1120 1124
97. R Core Development Team 2010 R: A language and environment for statistical computing: R Foundation for Statistical Computing, Vienna, Austria
98. GentlemanRCCareyVJBatesDMBolstadBDettlingM 2004 Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 5 R80
99. MorganMAndersSLawrenceMAboyounPPagesH 2009 ShortRead: a bioconductor package for input, quality assessment and exploration of high-throughput sequence data. Bioinformatics 25 2607 2608
100. LiHDurbinR 2009 Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25 1754 1760
101. LangmeadBTrapnellCPopMSalzbergSL 2009 Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10 R25
102. AndersSHuberW 2010 Differential expression analysis for sequence count data. Genome Biol 11 R106
103. LiHHandsakerBWysokerAFennellTRuanJ 2009 The Sequence Alignment/Map format and SAMtools. Bioinformatics 25 2078 2079
104. IvoLHofackerWFStadlerPeterFBonhoefferSebastianTackerManfredSchusterPeter 1994 Fast folding and comparison of RNA secondary structures. Mh Chemie 125 167 188
105. LeeMTKimJ 2008 Self containment, a property of modular RNA structures, distinguishes microRNAs. PLoS Comput Biol 4 e1000150 doi:10.1371/journal.pcbi.1000150
106. BarrettTTroupDBWilhiteSELedouxPEvangelistaC 2011 NCBI GEO: archive for functional genomics data sets–10 years on. Nucleic Acids Res 39 D1005 1010
107. SeanDMeltzerPS 2007 GEOquery: a bridge between the Gene Expression Omnibus (GEO) and BioConductor. Bioinformatics 23 1846 1847
108. SmythGK 2005 Limma: linear models for microarray data. Gentleman VCRDudoitSIrizarryRHuberW Bioinformatics and Computational Biology Solutions using R and Bioconductor New York Springer 397 420
109. FriedmanRCFarhKKBurgeCBBartelDP 2009 Most mammalian mRNAs are conserved targets of microRNAs. Genome Res 19 92 105
Štítky
Genetika Reprodukční medicína
Článek Functional Centromeres Determine the Activation Time of Pericentric Origins of DNA Replication inČlánek Dynamic Deposition of Histone Variant H3.3 Accompanies Developmental Remodeling of the TranscriptomeČlánek Integrin α PAT-2/CDC-42 Signaling Is Required for Muscle-Mediated Clearance of Apoptotic Cells inČlánek Prdm5 Regulates Collagen Gene Transcription by Association with RNA Polymerase II in Developing BoneČlánek Acquisition Order of Ras and p53 Gene Alterations Defines Distinct Adrenocortical Tumor Phenotypes
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2012 Číslo 5
-
Všechny články tohoto čísla
- Slowing Replication in Preparation for Reduction
- Chromosome Pairing: A Hidden Treasure No More
- Loss of Imprinting Differentially Affects REM/NREM Sleep and Cognition in Mice
- Six Novel Susceptibility Loci for Early-Onset Androgenetic Alopecia and Their Unexpected Association with Common Diseases
- Regulation by the Noncoding RNA
- UDP-Galactose 4′-Epimerase Activities toward UDP-Gal and UDP-GalNAc Play Different Roles in the Development of
- Deletion of PTH Rescues Skeletal Abnormalities and High Osteopontin Levels in Mice
- Karyotypic Determinants of Chromosome Instability in Aneuploid Budding Yeast
- Genome-Wide Copy Number Analysis Uncovers a New HSCR Gene:
- MicroRNA-277 Modulates the Neurodegeneration Caused by Fragile X Premutation rCGG Repeats
- Functional Centromeres Determine the Activation Time of Pericentric Origins of DNA Replication in
- Dynamic Deposition of Histone Variant H3.3 Accompanies Developmental Remodeling of the Transcriptome
- Scientist Citizen: An Interview with Bruce Alberts
- YY1 Regulates Melanocyte Development and Function by Cooperating with MITF
- Congenital Heart Disease–Causing Gata4 Mutation Displays Functional Deficits
- Recombination Drives Vertebrate Genome Contraction
- KATNAL1 Regulation of Sertoli Cell Microtubule Dynamics Is Essential for Spermiogenesis and Male Fertility
- Re-Patterning Sleep Architecture in through Gustatory Perception and Nutritional Quality
- Using Whole-Genome Sequence Data to Predict Quantitative Trait Phenotypes in
- Genome-Wide Analysis of GLD-1–Mediated mRNA Regulation Suggests a Role in mRNA Storage
- Meiotic Chromosome Pairing Is Promoted by Telomere-Led Chromosome Movements Independent of Bouquet Formation
- LINT, a Novel dL(3)mbt-Containing Complex, Represses Malignant Brain Tumour Signature Genes
- The H3K27 Demethylase UTX-1 Is Essential for Normal Development, Independent of Its Enzymatic Activity
- Suppresses Senescence Programs and Thereby Accelerates and Maintains Mutant -Induced Lung Tumorigenesis
- Genome-Wide Association of Pericardial Fat Identifies a Unique Locus for Ectopic Fat
- An Essential Role for Katanin p80 and Microtubule Severing in Male Gamete Production
- Identification of Genes That Promote or Antagonize Somatic Homolog Pairing Using a High-Throughput FISH–Based Screen
- Principles of Carbon Catabolite Repression in the Rice Blast Fungus: Tps1, Nmr1-3, and a MATE–Family Pump Regulate Glucose Metabolism during Infection
- Integrin α PAT-2/CDC-42 Signaling Is Required for Muscle-Mediated Clearance of Apoptotic Cells in
- Histone H3 Localizes to the Centromeric DNA in Budding Yeast
- Collapse of Telomere Homeostasis in Hematopoietic Cells Caused by Heterozygous Mutations in Telomerase Genes
- Hypersensitive to Red and Blue 1 and Its Modification by Protein Phosphatase 7 Are Implicated in the Control of Arabidopsis Stomatal Aperture
- Extent, Causes, and Consequences of Small RNA Expression Variation in Human Adipose Tissue
- TBC-8, a Putative RAB-2 GAP, Regulates Dense Core Vesicle Maturation in
- Regulating Repression: Roles for the Sir4 N-Terminus in Linker DNA Protection and Stabilization of Epigenetic States
- Common Genetic Determinants of Intraocular Pressure and Primary Open-Angle Glaucoma
- Prdm5 Regulates Collagen Gene Transcription by Association with RNA Polymerase II in Developing Bone
- Fitness Landscape Transformation through a Single Amino Acid Change in the Rho Terminator
- Repeated, Selection-Driven Genome Reduction of Accessory Genes in Experimental Populations
- Allelic Variation and Differential Expression of the mSIN3A Histone Deacetylase Complex Gene Promote Mammary Tumor Growth and Metastasis
- DNA Demethylation and USF Regulate the Meiosis-Specific Expression of the Mouse
- Knowledge-Driven Analysis Identifies a Gene–Gene Interaction Affecting High-Density Lipoprotein Cholesterol Levels in Multi-Ethnic Populations
- A Duplication CNV That Conveys Traits Reciprocal to Metabolic Syndrome and Protects against Diet-Induced Obesity in Mice and Men
- EMT Inducers Catalyze Malignant Transformation of Mammary Epithelial Cells and Drive Tumorigenesis towards Claudin-Low Tumors in Transgenic Mice
- Inactivation of a Novel FGF23 Regulator, FAM20C, Leads to Hypophosphatemic Rickets in Mice
- Genome-Wide Association for Abdominal Subcutaneous and Visceral Adipose Reveals a Novel Locus for Visceral Fat in Women
- Stratifying Type 2 Diabetes Cases by BMI Identifies Genetic Risk Variants in and Enrichment for Risk Variants in Lean Compared to Obese Cases
- New Insight into the History of Domesticated Apple: Secondary Contribution of the European Wild Apple to the Genome of Cultivated Varieties
- Activated Cdc42 Kinase Has an Anti-Apoptotic Function
- The Region Is Critical for Birth Defects and Electrocardiographic Dysfunctions Observed in a Down Syndrome Mouse Model
- COP9 Signalosome Integrity Plays Major Roles for Hyphal Growth, Conidial Development, and Circadian Function
- Bmps and Id2a Act Upstream of Twist1 To Restrict Ectomesenchyme Potential of the Cranial Neural Crest
- Psip1/Ledgf p52 Binds Methylated Histone H3K36 and Splicing Factors and Contributes to the Regulation of Alternative Splicing
- The Number of X Chromosomes Causes Sex Differences in Adiposity in Mice
- Target Gene Analysis by Microarrays and Chromatin Immunoprecipitation Identifies HEY Proteins as Highly Redundant bHLH Repressors
- Acquisition Order of Ras and p53 Gene Alterations Defines Distinct Adrenocortical Tumor Phenotypes
- ELK1 Uses Different DNA Binding Modes to Regulate Functionally Distinct Classes of Target Genes
- Histone H1 Depletion Impairs Embryonic Stem Cell Differentiation
- IDN2 and Its Paralogs Form a Complex Required for RNA–Directed DNA Methylation
- Separation of DNA Replication from the Assembly of Break-Competent Meiotic Chromosomes
- Genomic Hypomethylation in the Human Germline Associates with Selective Structural Mutability in the Human Genome
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Inactivation of a Novel FGF23 Regulator, FAM20C, Leads to Hypophosphatemic Rickets in Mice
- Genome-Wide Association of Pericardial Fat Identifies a Unique Locus for Ectopic Fat
- Slowing Replication in Preparation for Reduction
- An Essential Role for Katanin p80 and Microtubule Severing in Male Gamete Production
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



