-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaThe Histone H3K36 Methyltransferase MES-4 Acts Epigenetically to Transmit the Memory of Germline Gene Expression to Progeny
Methylation of histone H3K36 in higher eukaryotes is mediated by multiple methyltransferases. Set2-related H3K36 methyltransferases are targeted to genes by association with RNA Polymerase II and are involved in preventing aberrant transcription initiation within the body of genes. The targeting and roles of the NSD family of mammalian H3K36 methyltransferases, known to be involved in human developmental disorders and oncogenesis, are not known. We used genome-wide chromatin immunoprecipitation (ChIP) to investigate the targeting and roles of the Caenorhabditis elegans NSD homolog MES-4, which is maternally provided to progeny and is required for the survival of nascent germ cells. ChIP analysis in early C. elegans embryos revealed that, consistent with immunostaining results, MES-4 binding sites are concentrated on the autosomes and the leftmost ∼2% (300 kb) of the X chromosome. MES-4 overlies the coding regions of approximately 5,000 genes, with a modest elevation in the 5′ regions of gene bodies. Although MES-4 is generally found over Pol II-bound genes, analysis of gene sets with different temporal-spatial patterns of expression revealed that Pol II association with genes is neither necessary nor sufficient to recruit MES-4. In early embryos, MES-4 associates with genes that were previously expressed in the maternal germ line, an interaction that does not require continued association of Pol II with those loci. Conversely, Pol II association with genes newly expressed in embryos does not lead to recruitment of MES-4 to those genes. These and other findings suggest that MES-4, and perhaps the related mammalian NSD proteins, provide an epigenetic function for H3K36 methylation that is novel and likely to be unrelated to ongoing transcription. We propose that MES-4 transmits the memory of gene expression in the parental germ line to offspring and that this memory role is critical for the PGCs to execute a proper germline program.
Published in the journal: . PLoS Genet 6(9): e32767. doi:10.1371/journal.pgen.1001091
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1001091Summary
Methylation of histone H3K36 in higher eukaryotes is mediated by multiple methyltransferases. Set2-related H3K36 methyltransferases are targeted to genes by association with RNA Polymerase II and are involved in preventing aberrant transcription initiation within the body of genes. The targeting and roles of the NSD family of mammalian H3K36 methyltransferases, known to be involved in human developmental disorders and oncogenesis, are not known. We used genome-wide chromatin immunoprecipitation (ChIP) to investigate the targeting and roles of the Caenorhabditis elegans NSD homolog MES-4, which is maternally provided to progeny and is required for the survival of nascent germ cells. ChIP analysis in early C. elegans embryos revealed that, consistent with immunostaining results, MES-4 binding sites are concentrated on the autosomes and the leftmost ∼2% (300 kb) of the X chromosome. MES-4 overlies the coding regions of approximately 5,000 genes, with a modest elevation in the 5′ regions of gene bodies. Although MES-4 is generally found over Pol II-bound genes, analysis of gene sets with different temporal-spatial patterns of expression revealed that Pol II association with genes is neither necessary nor sufficient to recruit MES-4. In early embryos, MES-4 associates with genes that were previously expressed in the maternal germ line, an interaction that does not require continued association of Pol II with those loci. Conversely, Pol II association with genes newly expressed in embryos does not lead to recruitment of MES-4 to those genes. These and other findings suggest that MES-4, and perhaps the related mammalian NSD proteins, provide an epigenetic function for H3K36 methylation that is novel and likely to be unrelated to ongoing transcription. We propose that MES-4 transmits the memory of gene expression in the parental germ line to offspring and that this memory role is critical for the PGCs to execute a proper germline program.
Introduction
Regulation of gene expression through DNA packaging into chromatin has emerged as an important theme in development (reviewed in [1]). Chromatin consists of nucleosomal units of 147 bp segments of DNA wrapped around histone octamers composed of H2A, H2B, H3, and H4 [1]–[3]. Post-translational modification of histones via methylation, acetylation and addition of other covalent marks constitutes an important mechanism to regulate chromatin structure, DNA accessibility, and recruitment of regulatory factors during transcription [4], [5]. Some modifications, such as H3K27 methylation, additionally serve epigenetic roles by propagating particular chromatin states and gene expression patterns from mother to daughter cells [e.g. 6], [7].
This paper focuses on MES-4, a C. elegans enzyme that methylates histone H3 on lysine 36 (H3K36me) [8]. In yeast, all H3K36 methylation is carried out by the SET domain-containing protein Set2 [9]. In more complex eukaryotes, H3K36 methylation is carried out by two groups of enzymes. The first group includes the Set2-related proteins, called MET-1 in C. elegans, Hypb/Set2 in fruit flies, and HYPB/Setd2 in mammals, [10]–[13]. The second group includes MES-4-related proteins called MES-4 in C. elegans, dMES-4 in fruit flies, and NSD1 (for nuclear receptor-binding SET domain protein 1), NSD2/WHSC1/MMSET, and NSD3/WHSC1L1 in mammals [11], [14]–[16]. Mutations in the human MES-4/NSD genes underlie developmental disorders and various human malignancies [15]–[17].
In C. elegans, the MES-4 histone methyltransferase (HMT) is essential for germ cell viability. The genetics of mes-4 demonstrate that MES-4 serves a transgenerational (mother to offspring) role. Maternal mes-4(+) enables even homozygous mes-4/mes-4 progeny to form a fully functional germ line, while absence of maternal mes-4(+) severely impairs the ability of the primordial germ cells (PGCs) in progeny to proliferate and causes them to deteriorate early [18]. Based on immunostaining of germ lines and embryos, MES-4 and the H3K36me2 marks generated by MES-4 are dramatically concentrated on the autosomes and absent from all but the left tip of the X chromosomes [8], [19]. Surprisingly, microarray analysis of adult germ lines from fertile mes-4/mes-4 mothers revealed up-regulation of numerous (345) X-linked genes, along with mis-regulation of autosomal genes (155 up - and 115 down-regulated) (data from [8]; our unpublished results) suggesting that MES-4 participates in silencing the X chromosomes in wild-type adult germ lines [8], [20], [21]. Learning how MES-4 and H3K36 methylation promote PGC survival hinges on identifying the targets of MES-4 binding at the gene level and elucidating the role of MES-4-generated H3K36 methyl marks.
Studies of yeast Set2 have established one paradigm for binding and function of H3K36 HMTs. Yeast Set2 associates via its “SRI domain” with the C-terminal domain of RNA Polymerase II (Pol II) during the elongation phase of transcription and methylates H3K36 within the body of genes, predominantly in the 3′ coding region [22]–[26]. Set2-catalyzed H3K36me marks are recognized by the chromodomain of Eaf3 and the plant homeo domain (PHD) of Rco1, both in a complex with the Rpd3 enzyme that deacetylates histones. Delivery of Rpd3 to the body of genes suppresses spurious transcription initiation within the coding region [27]–[29]. Studies of X-chromosome dosage compensation in Drosophila have revealed another role for H3K36 methylation in mediating spreading of the dosage compensation complex from sites of initial recruitment [12], [30].
To investigate the targeting and roles of MES-4 in C. elegans, we employed ChIP-chip to determine the genome-wide distribution at high resolution of MES-4, H3K36me2, H3K36me3, and Pol II in extracts from early-stage embryos. Our analysis revealed that MES-4 accumulates over the bodies of ∼20% of genes, and that the majority of MES-4-bound genes are on the five autosomes and the left tip of the X. Comparing MES-4 and Pol II association with classes of genes with different temporal-spatial patterns of expression revealed that MES-4 resides on genes expressed in the maternal germ line and not genes newly expressed in early embryos. These findings, especially the observation that genes specifically expressed in the adult germ line are bound by MES-4 but not Pol II in early embryos, point to a mode of MES-4 recruitment that is different than Set2. MES-4 associates with and maintains methylation of germline-expressed genes through cell division and in a transcription-independent manner, establishing a novel epigenetic role for H3K36 methylation.
Results
MES-4, but not MET-1, is required for early-embryo maintenance of H3K36 trimethylation in the absence of RNA Polymerase II
Previous studies demonstrated that MES-4 generates H3K36me2 in adult germ lines and embryos [8], and the Set2 ortholog MET-1 generates H3K36me3 in embryos [10]. Our immunostaining experiments demonstrate that MES-4 contributes significantly to H3K36me3 as well, since H3K36me3 signal is abolished only in double mutant germ lines and embryos lacking both MES-4 and MET-1 (Figure 1A–1D; germline data not shown). To investigate the roles served by these two H3K36me3 HMTs, we tested the dependence of each HMT on RNA Pol II.
Fig. 1. Two C. elegans HMTs, MES-4 and MET-1, perform H3K36 trimethylation and display different dependence on Pol II. 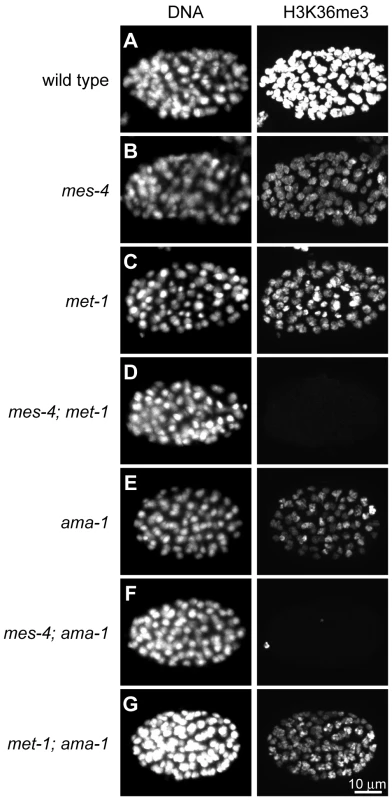
H3K36me3 levels are reduced relative to wild type (A) in mes-4 mutant (B), met-1 mutant (C), and ama-1(RNAi) (E) ∼80–110-cell embryos. H3K36me3 is undetectable in mes-4; met-1 (D) and mes-4; ama-1(RNAi) (F), but is still present in met-1; ama-1(RNAi) (G) embryos. Embryos were stained in parallel and images acquired and processed identically. See Figure S1 for specificity of H3K36me3 antibody. Previous studies suggested a Pol II-independent component of MES-4 function. MES-4-generated H3K36me2 immunofluorescence signal in embryos persists after RNAi depletion of AMA-1, the large subunit of Pol II [8]. In this study we tested the transcription dependence of both MES-4 and MET-1 in embryos by treating wild-type, mes-4 mutant, and met-1 mutant worms with RNAi to ama-1 and analyzing 80–110-cell embryos for H3K36me3. The persistence of some H3K36me3 after depletion of AMA-1 from wild-type suggests that H3K36me3 is at least partly Pol II-independent (Figure 1E). H3K36me3 staining was undetectable after simultaneous loss of MES-4 and AMA-1, but was easily detected after simultaneous loss of MET-1 and AMA-1 (Figure 1F and 1G). These results are consistent with MET-1 mediating Pol II-dependent H3K36me3, and indicate that MES-4 mediates at least some Pol II-independent H3K36me3. There are no precedents for Pol II-uncoupled H3K36me3. To further investigate the Pol II-independent binding and methylation of targets by MES-4, and to help understand how this activity promotes the viability of PGCs, we analyzed MES-4 binding across the genome at high resolution using ChIP-chip.
Sites of MES-4 binding are distributed along the five autosomes and are under-represented on the X chromosome
To determine the distribution of MES-4 across the genome by ChIP-chip, we used rabbit anti-MES-4 antibody to immunoprecipitate endogenous MES-4 (Figure 2A and Figure S2A), and as an alternative strategy used a mouse anti-FLAG antibody to immunoprecipitate FLAG-tagged MES-4 (Figure S2C). Tagged MES-4 was judged to be functional by two criteria: the autosomal concentration of MES-4::GFP::FLAG resembles that of endogenous MES-4 (Figure S2), and the transgene rescues mes-4(bn73) mutant worms (see Text S1). ChIP was performed on extracts from early embryos for several reasons. First, MES-4 levels appear relatively low in the maternal germ line and dramatically increase after fertilization (Y. Fong and S. Strome, unpublished result), suggesting that early embryogenesis is a period important for MES-4 function and that ChIP signals may be more robust from early embryos than from isolated germline tissue. Second, MES-4 binding is concentrated on autosomes at all stages analyzed, including early embryos, suggesting that the mechanisms that control selective recruitment to the autosomes are operational in early embryos [19]. Third, early embryos can be harvested in quantities sufficient for ChIP, while pure germline tissue currently cannot. ChIP was performed in triplicate from sheared chromatin prepared from formaldehyde-fixed early embryos (see Materials and Methods). As controls, we performed ChIP using Protein A and Protein G beads and also beads coated with antibodies that were not specific to C. elegans proteins. Endogenous MES-4 and tagged MES-4 displayed remarkably concordant ChIP distributions, which differed significantly from those of controls (Figure 3 and Figure S3). Here, we focus on ChIP analysis of endogenous MES-4.
Fig. 2. MES-4 is concentrated on the autosomes and the left tip of the X chromosome. 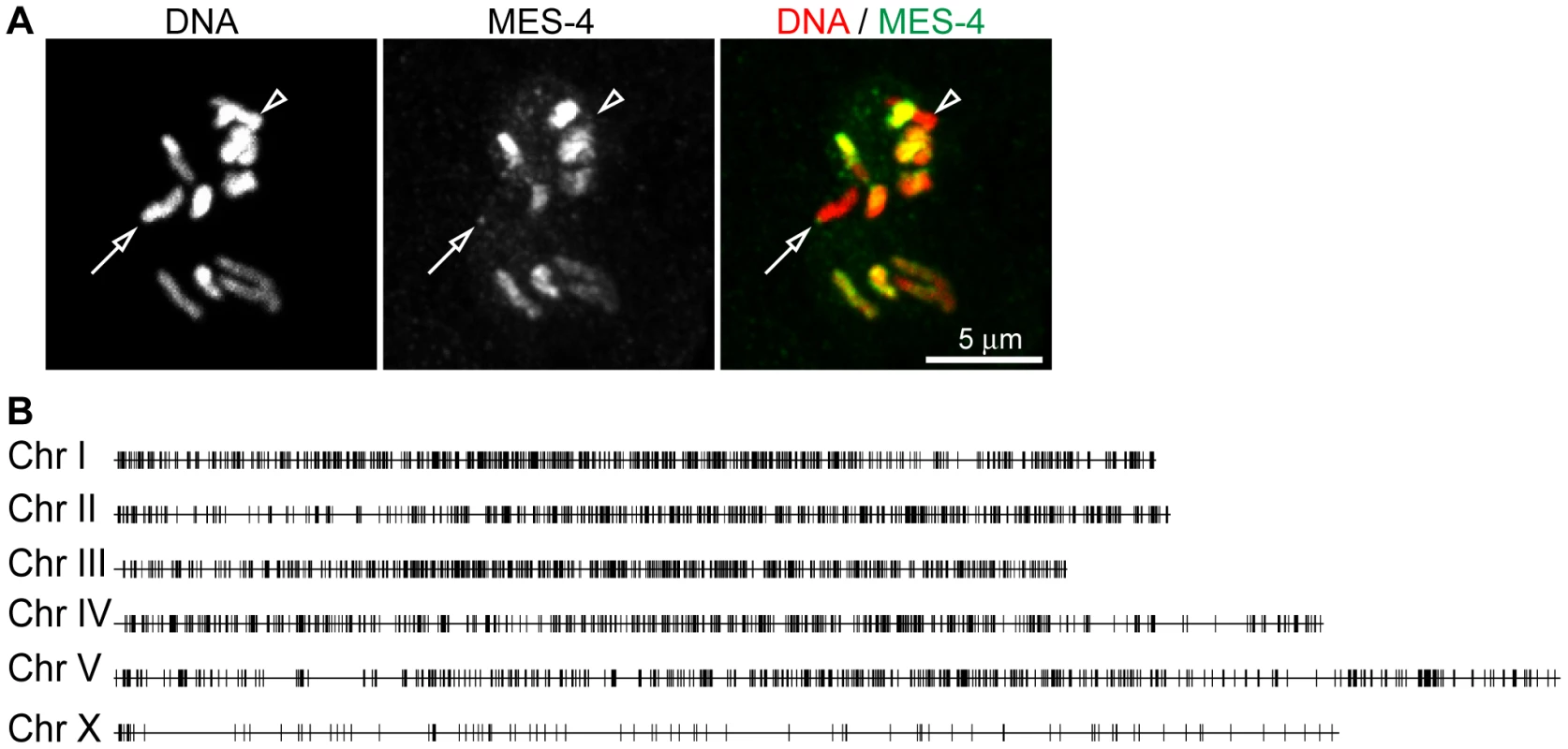
(A) The anti-MES-4 antibody used for ChIP stains the 10 autosomes and the left tip of the X (arrow; the other X chromosome is marked with an arrowhead), similar to previously characterized antibodies [8], [19]. The MES-4 signal was over-exposed to show the dot of MES-4 on the left end of X and the absence of detectable MES-4 along the rest of the X. Additional staining, including absence of MES-4 signal in a mes-4 mutant, is shown in Figure S2B. (B) 5291 of the 5391 MES-4 binding sites are distributed along the five autosomes. Of the 100 MES-4 peaks on the X chromosome, 25 are in the leftmost 300 kb. The expected and observed numbers of MES-4 peaks on each chromosome are in Figure S2D. All of the ChIPs shown in Figure 2, Figure 3, Figure 4, Figure 5, and Figure 6 were performed in early embryo extracts. Fig. 3. MES-4 is mostly concentrated on Pol II–bound genes. 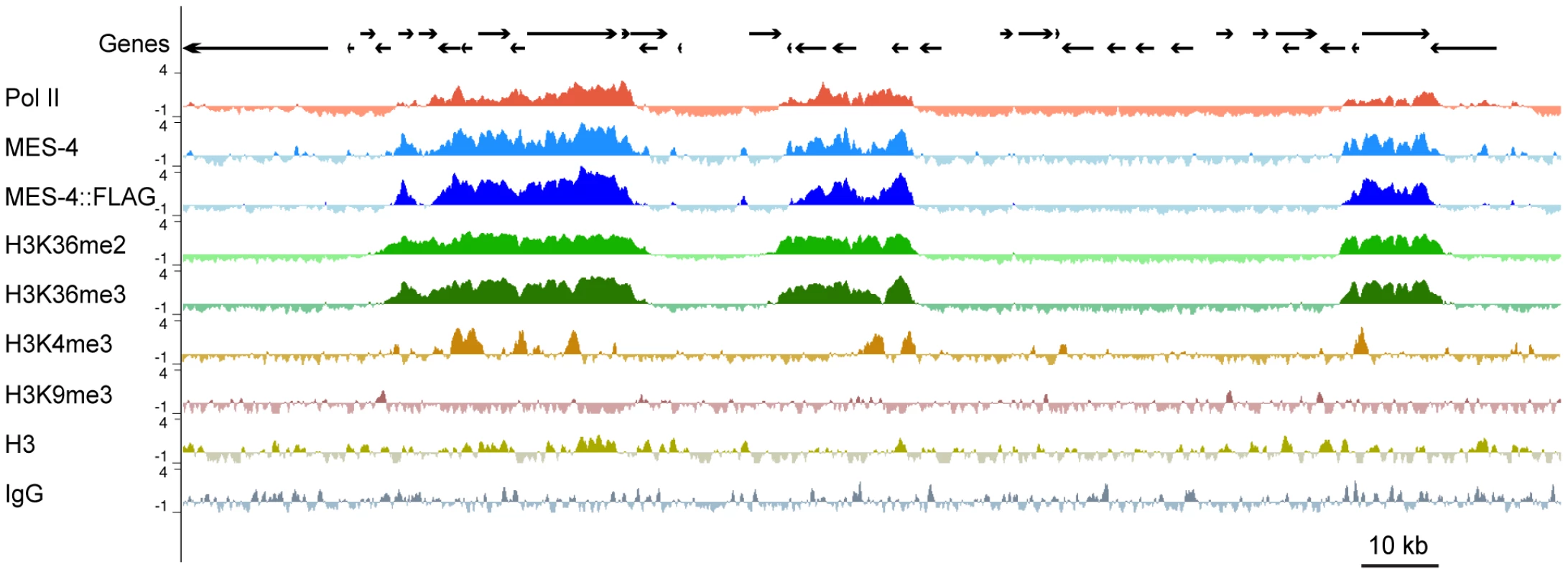
For a representative ∼180 kb autosomal (ChrIV) region of the genome, annotated genes (arrows; for genes encoding multiple transcript variants, the longest ones are shown) and ChIP z-scores (standardized log2 ratios of ChIP/Input signals) for Pol II, MES-4, MES-4::FLAG, H3K36me2, H3K36me3, H3K4me3, H3K9me3, H3, and a control ChIP (Protein A beads + rabbit IgG) are shown. Pol II, MES-4, H3K36me2 and H3K36me3 generally colocalize over bodies of transcribed genes. H3K4me3 is mainly found in the promoter and 5′ coding region of transcribed genes. H3K9me3 is absent from expressed genes. See Figure S3 for analysis of reproducibility and concordance among ChIPs. As observed by immunofluorescence staining of whole chromosomes, MES-4 ChIP peaks are concentrated on the autosomes and the left tip of the X. The peak-finding algorithm ChIPOTLe [31] (http://sourceforge.net/projects/chipotle-2/) was used to identify 5391 genomic regions with significant MES-4 enrichment. 5291 of the 5391 MES-4 binding sites are distributed along the five autosomes. Only 100 of the 5391 MES-4 sites (1.9%) are located on the X chromosome, in contrast to the 953 (17.7%) expected on the X if MES-4 were distributed uniformly across chromosomes (p-value<10−131) (Figure 2B and Figure S2D). 25% of MES-4 binding sites on the X are located within the leftmost 300 kb (1.7%) of the X. The density of binding sites on the left tip of the X (0.08 sites/kb) is similar to that on the autosomes (0.06 sites/kb). The remaining ∼17.4 Mb of the X chromosome have an extremely low density of MES-4 sites (0.004 sites/kb), which is apparently too low to detect by immunofluorescence. The far left tip of X, which resembles the autosomes in MES-4 binding density, is indeed marked by a dot of MES-4 immunostaining [8] (Figure 2).
MES-4 associates with the coding regions of expressed genes
MES-4-bound regions generally overlie the coding regions of single genes and sets of adjacent genes (Figure 3). Of the 5391 MES-4 binding sites, 5217 overlap the coding region of at least 1 gene, 143 overlap the 1 kb upstream (64) or downstream (79) of at least 1 gene, and 31 reside in intergenic regions (Table S1A). Conversely, of the 22,241 annotated genes in WormBase, 4400 overlap a MES-4-bound site and an additional 98 overlap in their 1 kb upstream or downstream region. Thus, the large majority (>96%) of MES-4 overlies genic regions, and approximately 1 in 5 genes is associated with MES-4.
To investigate whether MES-4 generally accumulates on expressed or silent genes, we compared the distribution of MES-4 in early embryos to that of Pol II across the genome. ChIP analysis of Pol II was performed using the 8WG16 monoclonal antibody, which detects both unphosphorylated and phosphoryated forms of Pol II [32]. Most genes bound by MES-4 are also associated with Pol II. This is illustrated both by viewing individual genes (Figure 3) and by plotting MES-4 ChIP signal versus Pol II ChIP signal for all genes (Figure S4C). However, as discussed below, certain gene classes with specific temporal or spatial expression patterns have high MES-4 but no Pol II or high Pol II and no MES-4, indicating that Pol II is neither necessary nor sufficient to recruit MES-4.
Viewing the distribution of MES-4 and Pol II across gene bodies stratified by transcript accumulation in early embryos revealed that MES-4 levels are highest on the most highly transcribed genes and that MES-4 overlies gene bodies (Figure 4). Genes aligned at their transcript start sites (TSS) and end sites (TES) were binned into 5 equally sized groups based on microarray measurements of transcript levels in the extracts used for ChIP. Average MES-4 and Pol II profiles for genes within each bin were plotted starting from 1 kb upstream to 1.5 kb downstream of the TSS and from 1.5 kb upstream to 1 kb downstream of the TES (Figure 4). Genes with highest RNA level show the highest MES-4 binding, followed by progressively lower levels of MES-4 on genes with lower RNA levels. In the most highly expressed genes, MES-4 levels rise across the TSS, reaching a maximum value ∼500 bp into the gene body, gradually decline beyond that and drop at the TES. Pol II shows a different profile than MES-4. Pol II levels peak at the TSS and again 3′ of the TES (Figure 4). Although MES-4 and Pol II accumulate on mostly the same genes, the 3′ peak of Pol II compared to the 3′ drop of MES-4 suggests that MES-4 does not track with Pol II or that it tracks only with a subpopulation of Pol II.
Fig. 4. MES-4 and H3K36 methyl marks are concentrated over gene bodies of highly expressed genes. 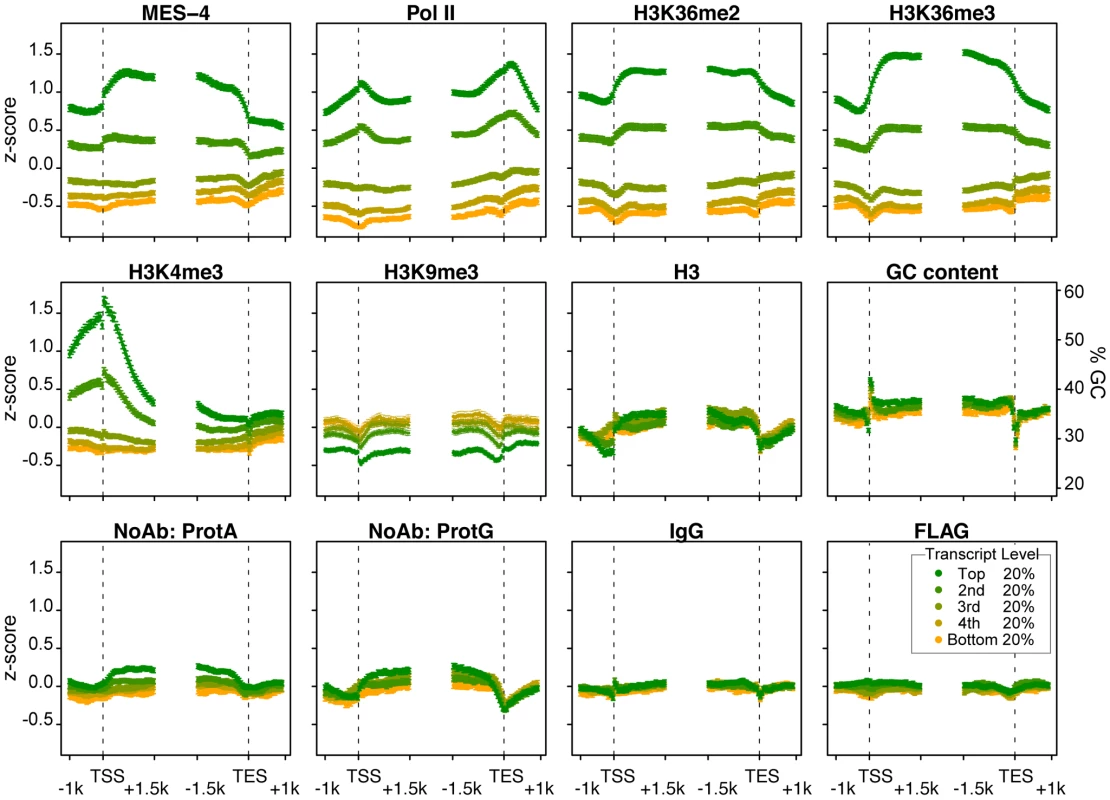
Genes >2 kb long (9932 genes) were grouped into 5 equal sized bins based on expression in early embryos. Colors display bins with highest (green) to lowest (orange) transcript level in early embryo extracts generated in parallel with ChIP extracts. Average z-score profiles for each ChIP target are shown in 50 bp steps in the 2.5 kb around the transcript start site (TSS) and the transcript end site (TES). Error bars indicate 95% confidence intervals for the means. The bottom row shows four different control ChIPs: Protein A beads, Protein G beads, Protein A beads coated with rabbit IgG, and sheep anti-mouse beads coated with mouse anti-FLAG. GC content variation is also shown. The methylation of H3K36 generally mirrors MES-4 accumulation
To compare the distribution of MES-4 to the distributions of H3K36 methyl marks in early embryos, we performed ChIP-chip using antibodies to H3K36me2 and H3K36me3. We also analyzed H3K4me3 and H3K9me3. All histone mark ChIPs were performed using highly specific monoclonal antibodies [33] (Figure S1).
The H3K36me2 and H3K36me3 ChIP profiles generally mirror those of MES-4. MES-4 and H3K36me2 and me3 ChIP signals on all genes are highly correlated (Figure 3 and ). In the bodies of actively expressed genes, H3K36me2 and me3 levels rise across the TSS and drop across the TES (Figure 4). The relatively flat profiles of H3K36me2 and me3 across gene bodies contrast with the 3′ enriched profiles of H3K36 methylation generated by Set2 homologs in yeast and other systems [e.g. 11], [25]. The distinctive gene-body profiles of H3K36me2 and me3 are not shared by other histone methyl marks. In keeping with H3K4me3 being a hallmark of active chromatin and specifically marking transcription initiation, H3K4me3 is distributed around the TSS, and declines sharply within gene bodies (Figure 4). H3K9me3, which is mostly associated with transcriptionally silent chromatin, indeed shows an inverse relationship to gene expression (Figure 4). The histone mark patterns described above can be compared to histone H3, which reflects nucleosome occupancy. We find that H3 is modestly elevated in gene bodies and shows a pronounced dip in the promoter region of highly expressed genes, as has been reported in other organisms [e.g. 25].
Genes bound by MES-4 in early embryos are those that had been expressed previously in the maternal germ line
MES-4 is essential for germline development and survival, and knowing which genes are bound by MES-4 is critical to understanding its mechanism of action. Gene Ontology (GO) analysis of all MES-4-bound genes revealed that MES-4 preferentially associates with genes that participate in reproduction, growth and development (Table S1C). To further characterize MES-4 target genes, we analyzed RNA levels in conjunction with MES-4, Pol II, and H3K36me2 and me3 ChIP signals on genes that exhibit different temporal-spatial expression patterns (Figure 5A). We generated six gene categories, based on previous studies (Table S2): (1) 4693 “germline-expressed” genes whose transcripts are present in dissected adult hermaphrodite germ lines according to SAGE analysis [34]; (2) 169 “germline-specific” genes whose transcripts are expressed exclusively in the maternal germ line (transcripts were detected in dissected hermaphrodite germ lines by SAGE, are enriched in the germ line based on whole-worm microarrays, accumulate in embryos strictly by maternal contribution as determined by microarray analysis of staged embryos, and are not represented in muscle, gut, or neuron SAGE sets) [21], [34]–[36]; (3) 797 “embryo-expressed” genes whose transcripts are not maternally provided and increase in level during embryogenesis [35]; (4) 323 “soma-specific” genes whose transcripts are detected in muscle, gut, and neuron SAGE sets but not in the adult germ line by SAGE or microarray analysis [21], [34], [36]; (5) 2580 “ubiquitous” or housekeeping genes whose transcripts are shared by muscle, gut, neuron and adult germline SAGE sets [34], [36]; and (6) 415 “silent” genes, serpentine receptor genes that are expressed in a few mature neurons and are not expected to be expressed in cells of early embryos [37]. The distinctive patterns of MES-4, Pol II, and H3K36 methylation on genes in different temporal-spatial classes provided key insights into MES-4 recruitment and function.
Fig. 5. Distribution of RNA, Pol II, MES-4, H3K36me2, and H3K36me3 levels for various gene sets. 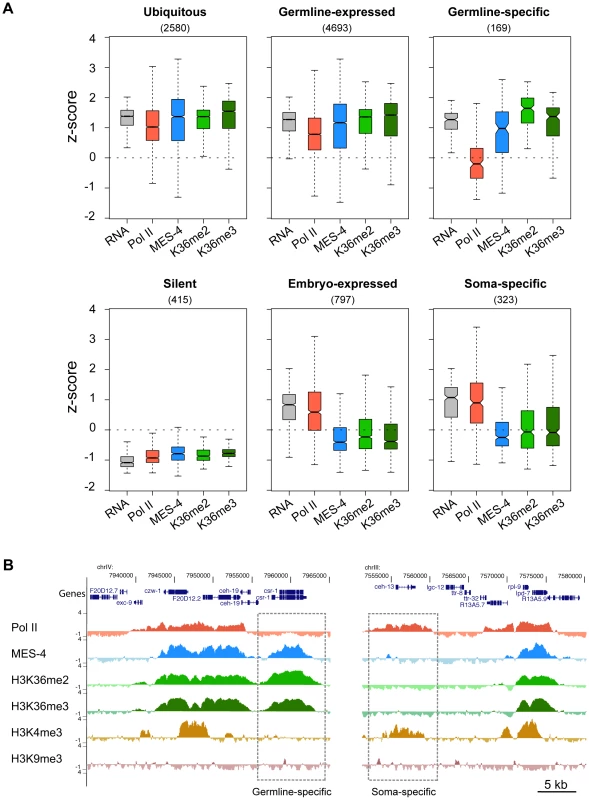
(A) Gene classes were defined as described in Results. The number of genes in each set is shown in parentheses. For each gene set, boxplots show the distribution of RNA level (standardized to z-score calculated from our microarray analysis) and Pol II, MES-4, H3K36me2 and H3K36me3 average z-scores from early embryo ChIP-chip analysis. Each box extends from the 25th to 75th percentile of the z-scores in the set, and the whiskers extending from a box indicate the 2.5th and 97.5th percentile. Wedges around the median indicate 95% confidence interval for the medians. See Figure S4C for scatter plots of the ubiquitous, germline-expressed, germline-specific and embryo-expressed categories. (B) Examples of Pol II- MES-4+ H3K36me+ and Pol II+ MES-4− H3K36me− genes. The germline-expressed gene csr-1 (left dashed box) contains high levels of MES-4, H3K36me2 and H3K36me3 but lacks Pol II and H3K4me3. The soma-expressed gene ceh-13 (right dashed box) contains Pol II and H3K4me3 but lacks MES-4, H3K36me2 and H3K36me3. Both csr-1 and ceh-13 are flanked by genes that show the typical high concordance between Pol II, MES-4 and H3K36me marks. See Table S2 for lists of genes in different temporal-spatial classes and Figure 6C for more examples of individual genes. First, the two largest classes of genes, germline-expressed genes and ubiquitously-expressed genes, share a pattern of high RNA accumulation and high Pol II, MES-4 and H3K36me2/me3 association (Figure 5A and Figure S4C). The common theme among these genes is that they are expressed in the maternal germ line. Thus, in early embryo extracts, genes expressed in the maternal germ line typically remain bound by Pol II, MES-4, and H3K36me2/me3. As an interesting sidelight, the clustering of Pol II and MES-4-bound genes displayed in Figure 3 and observed across all autosomes is consistent with the finding that ∼40% of genes with germline-enriched expression are organized in operons, and that those operons further cluster with monocistronic germline-expressed genes [38].
Second, silent genes exhibit low RNA accumulation and low Pol II, MES-4, and H3K36me2/me3 (Figure 5A). This result fits with seeing lowest levels of Pol II, MES-4, and H3K36me2/me3 association with genes in the bottom quantiles of expression (Figure 4).
Third and most importantly, the germline-specific class shows a distinct pattern: high RNA accumulation, high MES-4 and H3K36me2/me3 signals, but low Pol II (Figure 5A and Figure S4C). To explore this deviation from the usual high correlation between MES-4 and Pol II, we analyzed individual germline-specific genes in the UCSC genome browser. Numerous genes displayed robust MES-4 and H3K36me2/me3 ChIP signal but no detectable Pol II ChIP signal (Figure 5B and Figure 6C). We identified 214 genes as having high MES-4 and high H3K36me3 (z-scores>1) and low Pol II (z-score<0) (Table S3). Notably, 180 of the 214 genes (84%) show evidence of germline expression based on microarray and/or SAGE analysis [21], [34], and many of those genes are known to be involved in germline-specific processes (e.g. the meiosis genes him-8, htp-1, htp-2, htp-3, rec-8, syp-2, zim-2, zim-3, and the P-granule genes pgl-1, pgl-3, glh-2, glh-4). The germline-specific class of genes indicates that MES-4 can persist on genes that were previously expressed in the maternal germ line but are no longer being actively transcribed in early embryos, revealing that MES-4 can associate with genes independently of Pol II. This class probably contributes to the MES-4 and H3K36me2 and me3 staining observed after depletion of Pol II (Figure 1E) [8].
Fig. 6. RNAi depletion of MES-4 substantially reduces H3K36me3 on germline-expressed genes. 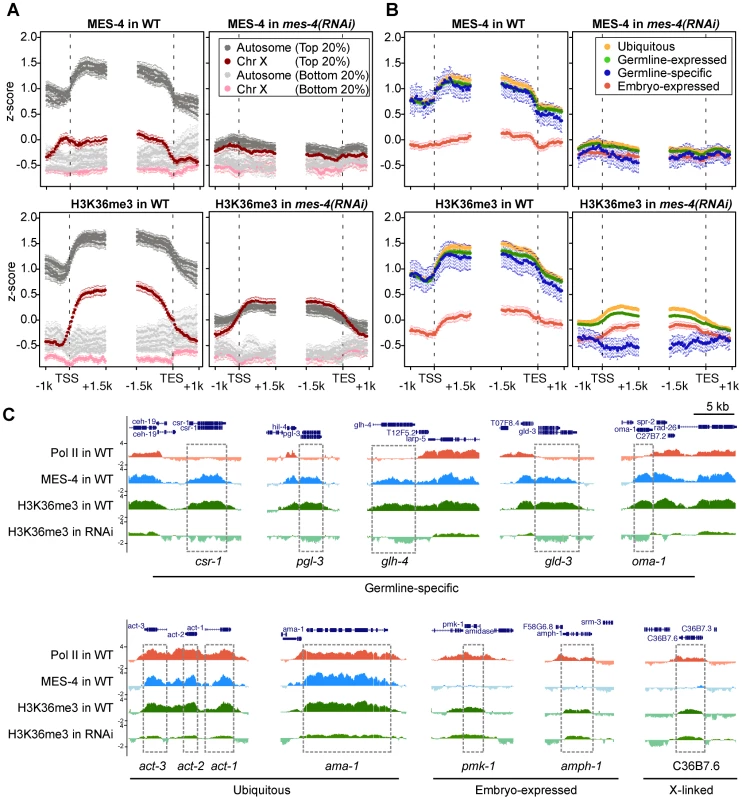
(A) MES-4 and H3K36me3 ChIP-chip signal on the autosomes (gray) and the X chromosome (red), specifically on genes with the highest expression in wild type (top 20% in dark color) and genes with the lowest expression in wild type (lowest 20% in light color). MES-4 is dramatically reduced genome-wide in mes-4(RNAi). H3K36me3 on autosomes is reduced to the lower level observed on the X, from which MES-4 is largely absent in wild type. The remaining low level of H3K36me3 on transcribed genes in mes-4(RNAi) is most likely due to transcription-coupled methylation by MET-1. (B) MES-4 and H3K36me3 ChIP-chip signal on ubiquitously-expressed (orange), germline-expressed (green), germline-specific (blue), and embryo-expressed (red) gene sets in wild type and mes-4(RNAi). In mes-4(RNAi), H3K36me3 is dramatically reduced on genes expressed in the germline (ubiquitous and germline-expressed) and undetectable on germline-specific genes. The embryo-expressed class (and soma-specific class, not shown) lack MES-4 in wild type and show little change in mes-4(RNAi). (C) Examples of individual genes illustrating the patterns observed in (B). Fourth, embryonically-expressed genes and soma-specific genes are silent in the maternal germ line but must be activated during embryogenesis. These genes display the unusual pattern of high RNA accumulation and high Pol II but low MES-4 and H3K36me2/me3 (Figure 5A and Figure S4C). In fact, inspection of individual genes in this class identified examples of robust Pol II ChIP signal but low or no detectable MES-4 or H3K36me2/me3 (Figure 5B, Figure 6C, and Figure S4C). These findings suggest that although MES-4 is found mostly on Pol II-bound genes (the first two large classes discussed above) (Figure 3, Figure S3B, and Figure S4C), Pol II accumulation is not sufficient to recruit MES-4.
In addition to clues about MES-4, some members of the embryonically-expressed and soma-specific genes provide the first natural examples of transcription elongation in the absence of detectable H3K36 methylation (Figure 5B and Figure S4C), a phenomenon observed only in yeast set2 mutants previously [9]. The C. elegans Set2 ortholog MET-1, which participates in transcription-dependent H3K36 methylation in embryos (Figure 1), is expected to methylate H3K36 on genes newly expressed in embryos. By immunofluorescence, MET-1-mediated H3K36me3 is not detected in 2 - to 40-cell embryos and gradually rises from the 40-cell stage onward (Furuhashi et al., unpublished and our unpublished results). In our ChIP experiments, genes bound by Pol II but lacking MES-4 display little or no H3K36me2/me3 (Figure S4C). Thus, in our early embryo samples, MET-1 appears to be a relatively minor contributor to H3K36 methylation.
MES-4 is responsible for H3K36 methylation of germline-specific genes
The above results strongly suggest that MES-4 mediates the majority of H3K36 methylation in early embryos. To investigate MES-4's contribution, we depleted MES-4 by RNAi and performed ChIP-chip analysis of MES-4 and H3K36me3. Evidence that RNAi was effective was provided by the following observations: the mes-4(RNAi) embryos used for ChIP lacked detectable MES-4 immunostaining, and MES-4 and H3K36me3 ChIP signal on autosomes, where MES-4 overwhelmingly resides in wild type, was reduced in mes-4(RNAi) extract to the low level observed on the X chromosome (Figure 6A). Importantly, H3K36me3 ChIP signal in mes-4(RNAi) was completely lost from germline-specific genes (Figure 6B and 6C), supporting our hypothesis that these genes are H3K36 methylated solely by MES-4 and in the absence of active transcription. H3K36me3 signal on ubiquitously-expressed and germline-expressed genes, the majority of which are actively transcribed in early embryos, was reduced but not eliminated (Figure 6B and 6C). The remaining H3K36 methylation of these genes is probably due to (low) MET-1 activity and possibly incomplete knock-down of MES-4. Embryo-expressed and soma-specific genes showed little change in H3K36me3 levels in mes-4(RNAi), many retaining the low levels of H3K36me3 seen in wild type (Figure 6B and 6C). This again suggests that detection of H3K36me3 is due to low MET-1 activity on this set of actively transcribed genes. Taken together, our RNAi results demonstrate that during embryogenesis MES-4 is responsible for H3K36 trimethylation of germline-specific genes and contributes the majority of H3K36 trimethylation on other germline-expressed genes as well.
The RNA polymerase-independent epigenetic function of MES-4 is reflected in the distribution of H3K36me3 on target genes
The above results reveal that in embryos MES-4 can associate with genes and maintain H3K36 methylation in a Pol II-independent manner. If MET-1 resembles Set2 in being recruited to genes by elongating Pol II, then MET-1-generated H3K36me3 should be enriched in the 3′ coding region of genes, similar to H3K36me3 generated by other known Set2 homologs [11], [23], [25], [39]. Indeed, soma-specific genes, which generally lack MES-4 binding and are presumed to acquire H3K36 methylation from MET-1, display such an enrichment in the 3′ gene body (Figure 7A in red). This pattern is relatively low-level in early embryos in which MET-1 activity is low (see discussion in previous section) and more pronounced in later stages (L3 larvae; data from [37]) in which somatic cells are actively engaged in transcription and MET-1 level and activity are expected to be elevated. In contrast, germline-specific genes display a distribution of H3K36me3 that is slightly 5′ enriched, similar to the distribution of MES-4 (Figure 7A in blue). The difference in H3K36me3 profiles on germline - versus soma-specific genes strengthens the notion that MES-4 and MET-1 are recruited to genes by different mechanisms and serve different roles on their targets. We hypothesize that MES-4 serves an epigenetic role and transmits the memory of gene expression in the adult germ line to cells of the embryo (see Discussion).
Fig. 7. The distribution of and requirement for MES-4-catalyzed H3K36 methyl marks. 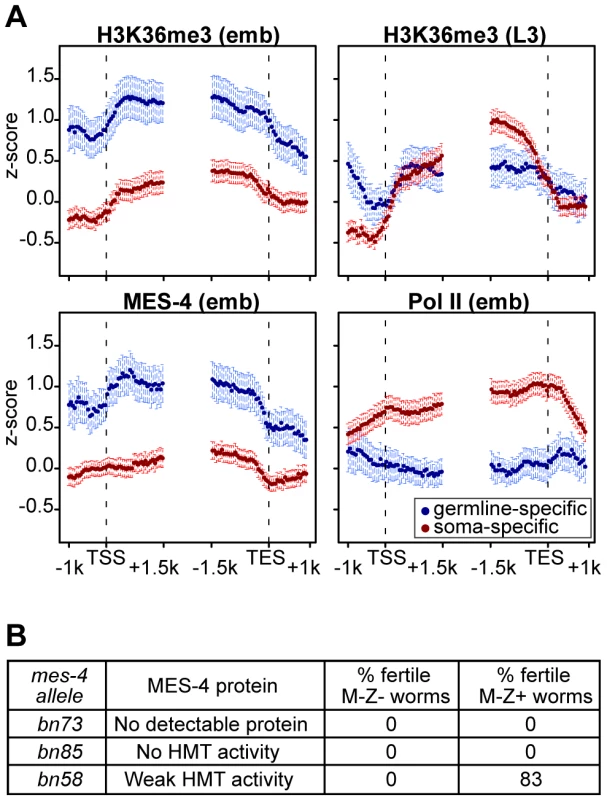
(A) Germline-specific and soma-specific genes display different profiles of H3K36me3, MES-4, and Pol II in early embryos (emb) and H3K36me3 in L3 larvae (L3 data are from [37]). Gene profiles are as described for Figure 4, using the germline- and soma-specific genes described in Results. This figure highlights the reciprocal relationship between Pol II levels and MES-4/H3K36me3 levels on germline- and soma-specific genes in early embryos (also see Figure 5). On germline-specific genes, H3K36me3, like MES-4, is slightly more elevated in 5′ than 3′ gene bodies, whereas on soma-specific genes, H3K36me3 is more elevated in 3′ than 5′ gene bodies, especially in L3s, probably due to transcription-dependent methylation by MET-1. (B) Embryonic expression of MES-4(+) rescues fertility of mes-4 mutants only when maternal MES-4 retains some HMT activity. Analysis of mutant MES-4 proteins is from [8]. M-Z− (lacking both maternal and zygotic expression of mes-4+) worms were mes-4/mes-4 hermaphrodites produced by mes-4/mes-4 mothers. M-Z+ worms were mes-4/+ hermaphrodites produced by mating mes-4/mes-4 mothers with wild-type males. At least 24 M-Z+ hermaphrodites were assessed for fertility. 75% of bn58/+ hermaphrodites produced viable progeny; 8% produced embryos that failed to hatch. The H3K36 methyltransferase activity of maternal MES-4 is crucial for germline development in progeny
Embryos from mes-4/mes-4 mothers develop into sterile adults, and embryonic expression of MES-4(+) is not sufficient to restore fertility [18]. Based on our findings that in embryos MES-4 persists on germline-expressed genes even in the absence of ongoing transcription, we reasoned that the failure of embryonic expression of MES-4(+) to rescue fertility was likely due to loss of H3K36 methylation generated by maternal MES-4(+) and an inability of embryonically expressed MES-4(+) to newly generate the patterns of H3K36 methylation needed for normal PGC development. Previous tests for rescue of fertility by embryonic expression of MES-4(+) were performed using mes-4/mes-4 mutant mothers that lack detectable H3K36 HMT activity (bn23, bn50, and bn67 alleles of mes-4 [8], [18]). We tested several additional alleles with the goal of assessing whether mes-4/mes-4 mutant mothers with weak HMT activity would provide sufficient H3K36 marking to enable embryonic expression of MES-4(+) to rescue fertility. The bn73 allele does not produce detectable MES-4 protein, the bn85 allele produces MES-4 lacking part of the catalytic SET domain, and the bn58 allele produces MES-4 with weak HMT activity [8]. Embryonic expression of MES-4(+) did not restore fertility to animals from MES-4(−) mothers (bn73 allele) or from mothers that produce MES-4 lacking HMT activity (bn85 allele) (Figure 7B). Importantly, embryonic expression of MES-4(+) restored fertility to animals whose mothers produced mutant MES-4 with weak HMT activity (bn58 allele) (Figure 7B). This demonstrates that the HMT activity of MES-4 is critical for maternal MES-4 function, and strengthens the view that MES-4 acts epigenetically and transmits developmentally important H3K36 marks from the parents' germ line to embryos, to ensure normal development of PGCs.
Discussion
MES-4 has emerged as a critical germline regulator in several studies. Its maternal-effect sterile (mes) phenotype demonstrates that the action of MES-4(+) in the maternal germ line and/or a maternal supply of MES-4(+) is required for the PGCs in offspring to thrive and survive [18]. Germline functions that require MES-4 are repression of genes on the X chromosome, transgene silencing, cosuppression, and RNAi [8], [40]–[42]. In addition to all of these repressive roles, MES-4 is required to promote ectopic expression of germline genes in somatic cells of synMuv B mutant larvae [43], [44]. The current study identified MES-4 binding targets as being genes expressed in the adult germ line. The targeting and function of MES-4 appear to be distinct from the well-studied Set2 family of H3K36 HMTs. We speculate that MES-4 transmits the memory of germline gene expression patterns from the parental germ line to the PGCs in offspring, and that loss of this function causes PGCs to degenerate in young larvae.
By both immunofluorescence and ChIP-chip analysis, MES-4 is concentrated on the five autosomes and the very left tip of the X ([8], [19] and this study). The strong autosomal bias of MES-4 ChIP signal is not due to an X chromosome structure that is recalcitrant to ChIP, as ChIP analysis of the dosage compensation complex preferentially targets the X [45], [46]. Instead, the autosomal enrichment of MES-4 is likely explained by the association of MES-4 with germline-expressed genes, which are significantly under-represented on the X [21]. The left end of X has several distinquishing properties, including a relatively high concentration of H3K9me3 and low density of periodic AA/TT clusters compared to the rest of the X [47] and the presence of the X chromosome “pairing center” for meiosis [48]. The autosome-like higher density of MES-4 binding sites on the left end of X is consistent with its being “pseudo-autosomal” in nature. Perhaps it arose by translocation of an autosomal segment to the end of the X. In fact, comparison of the genomes of C. elegans and Pristionchus pacificus suggests that they evolved from a common ancestor in which chromosomes X and V were fused ([49] and R. Sommer, personnal communication). The autosomal concentration of MES-4 is regulated by the other three MES proteins, MES-2, MES-3, and MES-6, which form the C. elegans Polycomb Repressive Complex 2 and like PRC2 catalyze methylation of H3K27 [50]–[52]. Loss of any component of the MES-2/3/6 complex leads to loss of silencing of the X chromosomes in the germ line and significant MES-4 immunofluorescence signal along the full length of the X in oocytes and embryos [8], [19]. Our model predicts that in mes-2, mes-3, and mes-6 mutant embryos, elevated MES-4 signal will be observed on X-linked genes whose expression was up-regulated in the maternal germ line.
Our studies demonstrate that in early embryos MES-4 associates with genes that were expressed in the maternal germ line. Genes expressed exclusively in the maternal germ line have high MES-4 and low Pol II. Some individual genes are MES-4+ Pol II−. These genes strongly argue that Pol II is not required to maintain MES-4 association with genes. Conversely, genes newly expressed in embryos and genes expressed specifically in somatic cells have high Pol II and low MES-4. Indeed, numerous individual genes are Pol II+ MES-4−. This is consistent with Pol II not being sufficient to recruit MES-4 to expressed genes.
Additional evidence supports the notion that MES-4 is capable of associating with gene bodies and methylating H3K36 independently of Pol II. First, RNAi depletion of the large subunit of RNA Pol II does not impair MES-4 binding to chromosomes or H3K36 di − or trimethylation, as detected by immunofluorescence ([8] and this study). Second, the slightly 5′ enriched distribution of MES-4 across genes bodies is quite different from the 3′ enriched distribution of Set2 homologs, the latter driven by association of Set2 with elongating Pol II [11], [23], [25], [39]. Recent studies in yeast have revealed that Set2 can be recruited to genes independently of Pol II, but is impaired in H3K36 di − and trimethylation [53]. In contrast, MES-4 appears capable of di − and trimethylating H3K36 in the absence of Pol II. Taken together, our findings suggest that MES-4 and perhaps the related mammalian NSD proteins provide another layer of function for H3K36 methylation that is novel and likely to be unrelated to ongoing transcription.
An attractive model is that MES-4 serves as a maintenance HMT to mark germline-expressed genes and pass the memory of gene expression from one generation of germ cells to the next. In this model, the H3K36 HMT MET-1 serves a Set2-like role and tracks with Pol II to methylate H3K36 during gene expression in the adult germ line; during embryogenesis, MES-4 maintains H3K36 methylation on those genes independently of Pol II. We think that MES-4 can serve this maintenance role even in transcriptionally repressed cells (e.g. in the germline blastomeres) and potentially for generations (e.g. in met-1 mutant worms). In support of this model, MES-4 does not appear to be capable of de novo H3K36 methylation in the soma and embryonic PGCs: embryonic expression of MES-4 in embryos that lack maternal H3K36me3 (mes-4; met-1 double mutant mothers) does not generate detectable H3K36me3 signal (Furuhashi et al., unpublished). In contrast, embryonic expression of MET-1 generates robust H3K36me3. Thus, our model posits that MES-4 is a specialized maintenance HMT that in embryos can only methylate chromatin with pre-existing H3K36 methyl marks. Consistent with this, embryonic expression of MES-4(+) can rescue the fertility of embryos from mothers that produce mutant MES-4 with weak HMT activity, but not embryos from mothers that lack MES-4 or that produce MES-4 that lacks HMT activity. The maintenance HMT activity of MES-4 enables MES-4 to serve a truly epigenetic role, propagation of a particular chromatin state through meiosis and mitosis. Recent studies demonstrate that the Polycomb Repressive Complex 2 both initiates and maintains a repressed chromatin state, the latter by binding the chromatin marks that it generates [6].
How is MES-4 initially recruited to germline-expressed genes? Our findings that MES-4 can associate with genes and generate H3K36 methylation independently of Pol II do not rule out the possibility that MES-4 has a Pol II-dependent mode as well. In the adult germ line, MES-4 may be initially recruited to expressed genes in a Pol II-dependent manner. Another possibility is that MES-4, perhaps via its PHD fingers, binds H3K36 methyl marks generated by MET-1 and/or MES-4. Yet a third possibility is that the chromodomain protein MRG-1, like its counterparts in other systems [12], [27], [28], [30], [54], associates with methylated H3K36 and helps recruit MES-4. In fact, MRG-1, like MES-4, displays autosomal enrichment by immunofluorescence, and mrg-1 mutants display a suite of mes-4-like defects, including maternal-effect death of PGCs [55]. MES-4 association with autosomes is not lost in mrg-1 mutants and vice versa [55]. Determining whether MRG-1 participates in recruiting MES-4 and/or is a downstream effector of MES-4-mediated H3K36 methylation is in progress.
An important question for future investigation is why the PGCs in embryos from mes-4 mutant mothers die. An attractive scenario is that MES-4 marking of genes expressed in the maternal germ line identifies genes to be expressed in the progeny's germ line. A related scenario, which might temporally precede the first, is that MES-4 marking of genes helps keep those genes repressed in the PGCs during embryogenesis. Indeed, wild-type PGCs generally do not acquire marks of active transcription until after embryos hatch into L1s, while mes-4 mutant PGCs acquire such marks prematurely, during mid-embryogenesis ([56] and Furuhashi et al., unpublished). In addition to a potential role in up - or down-regulating transcription in the PGCs, MES-4 may influence gene expression at the level of regulation of splicing. Recent papers report that H3K36me3 is enriched in expressed exons relative to introns [37] and that the level of H3K36me3 influences alternative splicing [57]. Like H3K36me3, MES-4 appears to be exon-enriched ( and Figure S5), raising the exciting possibility that MES-4 preferentially associates with exons and methylates them in a manner that facilitates or regulates splicing. Experiments are planned to isolate PGCs from wild-type and mes-4 mutant embryos and compare their RNA accumulation and splicing patterns. In the meantime, we analyzed the overlap between MES-4-bound genes in embryos and genes mis-regulated in mes-4 mutant adult germ lines, and found it to be small (Figure S6). The small overlap may be due to the stage difference (i.e. embryo versus adult germ line) or to technical and biological effects that cause even well documented transcription regulators to display a low overlap between factor-bound genes and genes mis-regulated when the factor is absent [e.g. 58], [59]. Despite the absence of gene expression data in PGCs, the strong dependence of early PGC development on maternal MES-4 demonstrates the functional importance of MES-4.
MES-4 and its HMT activity are detected in all cells of early embryos and become restricted to the PGCs during mid to late embryogenesis ([19] and our unpublished results). Our model that MES-4 propagates the memory of germline gene expression through embryogenesis raises the question how somatic cells in the embryo deal with that signal. Our current view is that the synMuv B chromatin regulators antagonize germline fate in somatic cells. Loss of synMuv B proteins causes somatic cells to express germline-specific genes, and concomitant loss of maternal MES-4 suppresses the germline potential of somatic cells [43], [44]. Further tests of our model and of the interplay between MES-4, MET-1, MRG-1 and the synMuv B chromatin regulators will shed light on how germline identity is passed from generation to generation and how germline gene expression patterns are controlled.
Materials and Methods
RNAi depletion of RNA polymerase II
We depleted AMA-1 by feeding N2 worms bacteria expressing dsRNA against ama-1 (from the Ahringer RNAi feeding library [60]). In brief, L3-L4 hermaphrodites were fed for 24 hours at 24°C, then transferred to new plates to score the embryonic lethality of F1 progeny (>98%) or dissected to obtain embryos for staining.
Generation of MES-4 ChIP reagents
Affinity purified rabbit antibodies directed against amino acids 729–828 of MES-4 were generated by Strategic Diagnostic Inc. (SDI, Newark, DE). mes-4 animals expressing a MES-4::GFP::FLAG transgene were generated as described in Supplement.
Preparation of N2 and mes-4(RNAi) early embryo extracts and chromatin
For preparation of embryo extracts, N2 adult worms were grown from synchronized L1s in standard S-basal medium with shaking at 230 rpm. N2 worms were fed HB101 for 58–60 hr at 20°C, or fed bacteria expressing dsRNA against mes-4 (from the Ahringer RNAi feeding library [60]) for 48 hr at 24°C. To ensure that a majority of embryos used for extract preparation were early stages, worms were processed for extract only if the majority of worms contained 10 or fewer embryos. Embryos were obtained by dissolving adult worms with bleach and analyzed by DAPI staining. Embryo preparations contained an average of 37% <28-cell embryos, 30% 28–100-cell embryos, and 33% ∼100–300-cell embryos. 50 µl of packed embryos were set aside for RNA extraction and expression profiling. The remaining embryos were washed and cross-linked with 1.85% formaldehyde in M9 buffer for 30 min at room temperature, then washed for 5 min in each of the following buffers: twice with M9, once with 100 mM Tris-HCl pH7.5, once with 10 ml FA buffer (50 mM HEPES/KOH pH7.5, 1 mM EDTA, 1% Triton X-100, 0.1% sodium deoxycholate, 150 mM NaCl). Embryos were frozen at −80°C. 2 ml of embryos, thawed and resuspended in 4 ml of FA buffer containing protease inhibitors (Roche protease inhibitor cocktail tablet), were dounce-homogenized 30 times using a tight pestle at 4°C. Chromatin was sheared to an approximate size range of 200–800 bp using a Branson sonicator or a Diagenode bioruptor. To remove cellular debris, samples were centrifuged at 13,000 rpm, 4°C for 15 min. To remove lipids, samples were filtered through Millipore Ultrafree-MC 0.45 µm filter units at 13,000 rpm, 4°C for 1 min. Protein concentration was determined and samples were stored at −80°C.
Chromatin immunoprecipitation (ChIP) from early embryos
ChIPs were performed using 1–6 mg of worm embryo protein and 2–5 µg antibody per IP. 5–10% of starting material was reserved as input. Rabbit antibodies used for IP were anti-histone H3 serum (Active Motif AR-0144) and affinity-purified anti-MES-4 (SDI). Mouse monoclonal antibodies used for IP were M2 anti-FLAG (Sigma F3165), 8WG16 anti-Pol II (Abcam ab817), anti-H3K4me3 (Wako 305-34819), anti-H3K9me3 (from Hiroshi Kimura), anti-H3K36me2 (from Hiroshi Kimura), and anti-H3K36me3 (from Hiroshi Kimura).
MES-4 and Pol II were ChIPed using 3 mg embryo protein and 5 µg antibody. For each ChIP reaction, antibody was incubated overnight at 4°C with embryo extract. Protein A - or Protein G-coupled Dynabeads (Invitrogen) equilibrated in 50 µl FA buffer were added, nutated for 2 hr at 4°C, concentrated using a Dynal Magnetic Particle Concentrator (MPC) (Invitrogen) and washed at room temperature for 5 min in 1 ml of each of the following buffers: FA buffer, 2 washes; FA buffer with 1M NaCl, 1 wash. Beads were transferred to a new tube, washed once for 10 min at room temperature in 1 ml FA buffer with 500 mM NaCl, once with TEL buffer (0.25 M LiCl, 1% NP-40, 1% sodium deoxycholate, 1 mM EDTA, 10 mM Tris-HCl, pH 8.0) and twice for 5 min at room temperature in 1 ml TE. To elute complexes, beads were incubated twice at 65°C in 150 µl of elution buffer (1% SDS in TE with 250 mM NaCl) for 15 min. Input samples were brought to a volume of 300 µl and incubated with 20 µg of RNAse A for 30 minutes at 37°C. Both input and IP samples were incubated with 20 µg of proteinase K for 1 hr at 55°C. Crosslinks were reversed overnight at 65°C. DNA was purified using a Zymo DNA purification column.
Histone H3 and methyl marks were ChIPed using 1 mg embryo protein and 2 µg antibody and the same method as above, except IP beads were Dynabeads coupled to sheep anti-mouse IgG (Invitrogen) equilibrated in 50 µl of 5 mg/ml IgG-free BSA (Sigma) in FA buffer, and both input and IP samples were incubated with RNAse A before proteinase K.
Amplification by LM–PCR
Input and IPed DNAs were blunted with T4 DNA polymerase, purified using a Zymo DNA purification column, and ligated to a unidirectional linker prepared from the following HPLC-purified oligos: Long: 5′-AGAAGC TTGAATTCGAGCAGTCAG-3′ and Short: 5′-CTGCTCGAATTCAAGCTTCT-3′. DNAs were purified using a Zymo DNA purification column and amplified with the Short oligo in an 80 µl volume using the following cycling conditions: 55°C for 4 min, 72°C for 5 min, 94°C for 5 min followed by 25 cycles of: 94°C for 1 min, 55°C for 1 min, 72°C for 1 min. Samples were incubated for an additional 5 min at 72°C. DNA was purified using a Qiagen PCR purification kit and eluted in 30 or 50 µl dH2O.
Microarray hybridizations and data analysis and display
NimbleGen 2.1M probe tiling arrays, with 50 bp probes, designed against WS170 (ce4) were used. 2–3 independent ChIPs were performed with each antibody and for each no antibody control (NoAb). Amplified samples were labeled and hybridized by the Roche NimbleGen Service Laboratory. Most ChIP samples were labeled with Cy5 and their input reference with Cy3; for most targets, one ChIP per antibody was dye swapped. For each probe, the intensity from the sample channel was divided by the reference channel and transformed to log2 space. The enrichment scores for each replicate were calculated by standardizing the log ratios to mean zero and standard deviation one (z-score). The mes-4(RNAi) ChIP samples were normalized with respect to the X chromosome in wild type. MES-4 was found to be largely absent from the X in wild type, and therefore was expected to be affected the least by RNAi. The log2 ratios for MES-4 and H3K36me3 in mes-4(RNAi) were normalized so that the X chromosome had the same mean and standard deviation as the X chromosome in the respective wild-type ChIP samples (after the wild-type ChIPs had been z-score normalized). Genome-wide Pearson correlations were calculated between all ChIP targets and replicates using all probes' z-scores after smoothing over 250 bp. The average z-score across replicates was used for all analyses.
Scatter plots and boxplots were generated by first obtaining an average z-score per gene. Z-scores of probes located completely within the transcript start site (TSS) and end site (TES) were averaged for each gene.
Gene body profile plots for the various ChIP targets were generated by aligning genes of length greater than 2 kb at their TSS and TES. The genomic regions 1.5 kb upstream to 1 kb downstream from the TSS and 1.5 kb upstream to 1 kb downstream from the TES were divided into 50 50-bp bins each. Probes in those genomic regions were assigned to the nearest bin. A profile for a group of genes was generated by averaging probes' z-scores within each bin across genes in the group. Error bars indicate 95% confidence intervals for the mean.
ChiPOTle peak finding and analysis
ChiPOTle 2.0 (http://sourceforge.net/projects/chipotle-2) [31] was run using a p-value cut-off of E-20 for the average z-score of MES-4 and the Protein A NoAb control (window size 500 bp, step size 100 bp, Bonferroni p-value correction). 5408 peaks were identified for MES-4 and 38 for the NoAb control. 17 MES-4 peaks that were within 1 step (100 bp) of a NoAb peak were eliminated, producing the final set of 5391 MES-4 peaks. Expected values for number of peaks on different chromosomes were calculated by dividing the total number of peaks by chromosome length (Figure S2D).
Peak-gene annotation analysis
The list of gene coordinates (transcript start-end) was downloaded from WormBase (http://www.wormbase.org/). We analyzed MES-4 peaks with respect to genomic annotations in three ways. First, MES-4 peaks that overlap with a gene by at least 1 bp in the gene body or within 1 kb of the gene's 5′ or 3′ were identified (Table S1A). Second, since one MES-4 peak could overlap with multiple genes, genes that overlap with a MES-4 peak by at least 1 bp within the gene body were identified, producing a set of “MES-4-bound” genes. Third, we determined the distribution of MES-4 peaks among different genomic annotations using Cis-regulatory Element Annotation System (CEAS) [61]. In CEAS, each peak is assigned to a single coordinate and using WormBase version WS170 the location of this coordinate with respect to the annotation classes exon, intron, 5′, 3′, and other (in our case >1 kb away) is determined. We chose the probe with the maximum enrichment value as a peak's coordinate. One coordinate can be assigned to multiple genomic annotations. Therefore, to assign a peak to a single region, we imposed the following hierarchy: exon, intron, 3′, 5′, 1 kb away. For comparison, all probes within the microarray were run through CEAS with the same parameters. Table S1B reports the results of MES-4 peak assignments to exons and introns.
Transcription profiling from early embryos
RNA was isolated from four of the early embryo preparations used for ChIP using Trizol and purified using the RNeasy kit (Qiagen, catalog 74104). Samples were analyzed on an Agilent Bioanalyzer to ensure that rRNAs were not degraded and that RNA was free of protein and DNA contamination. Two of the samples were treated with DNase for 30 min at room temperature. 20 µg of each RNA were hybridized to a single color 4-plex NimbleGen expression array with 72,000 probes (three 60-mer oligo probes per gene). Quantile normalization [62] and the Robust Multichip Average (RMA) algorithm [63] were used to normalize and summarize the multiple probe values per gene to obtain one expression value per gene and sample. The expression values per gene were averaged across the four samples.
Assessment of rescue by embryonic expression of MES-4(+)
mes-4(bn58, bn73, or bn85) dpy-11(e224)/DnT1[unc(n754)let] (IV;V) mothers were allowed to produce mes-4 dpy-11 (Dpy) hermaphrodites. These mes-4 dpy-11 (Dpy) hermaphrodites were mated to wild-type males. The Dpy progeny, the result of self-fertilization, were M-Z − (lacking both maternal and zygotic mes-4(+) product). The non-Dpy progeny, the result of cross-fertilization, were M-Z+. At least 24 of each category of progeny were scored for fertility (the production of embryos).
Supporting Information
Zdroje
1. AllisCD
JenuweinT
ReinbergD
2007 Epigenetics - Overview and concepts. Epigenetics Cold Spring Harbor Laboratory Press. Allis, Jenuwein, Reinberg, eds 23 61
2. FelsenfeldG
GroudineM
2003 Controlling the double helix. Nature 421 448 453
3. LugerK
MaderAW
RichmondRK
SargentDF
RichmondTJ
1997 Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 389 251 260
4. KouzaridesT
2007 Chromatin modifications and their function. Cell 128 693 705
5. StrahlBD
AllisCD
2000 The language of covalent histone modifications. Nature 403 41 45
6. MargueronR
JustinN
OhnoK
SharpeML
SonJ
2009 Role of the polycomb protein EED in the propagation of repressive histone marks. Nature 461 762 767
7. MullerJ
VerrijzerP
2009 Biochemical mechanisms of gene regulation by polycomb group protein complexes. Curr Opin Gen Dev 19 1 9
8. BenderLB
SuhJ
CarrollCR
FongY
FingermanIM
2006 MES-4: an autosome-associated histone methyltransferase that participates in silencing the X chromosomes in the C. elegans germ line. Development 133 3907 3917
9. StrahlBD
GrantPA
BriggsSD
SunZW
BoneJR
2002 Set2 is a nucleosomal histone H3-selective methyltransferase that mediates transcriptional repression. Mol Cell Biol 22 1298 1306
10. AndersenEC
HorvitzHR
2007 Two C. elegans histone methyltransferases repress lin-3 EGF transcription to inhibit vulval development. Development 134 2991 2999
11. BellO
WirbelauerC
HildM
ScharfAN
SchwaigerM
2007 Localized H3K36 methylation states define histone H4K16 acetylation during transcriptional elongation in Drosophila. EMBO J 26 4974 4984
12. LarschanE
AlekseyenkoAA
GortchakovAA
PengS
LiB
2007 MSL complex is attracted to genes marked by H3K36 trimethylation using a sequence-independent mechanism. Mol Cell 28 121 133
13. SunXJ
WeiJ
WuXY
HuM
WangL
2005 Identification and characterization of a novel human histone H3 lysine 36-specific methyltransferase. J Biol Chem 280 35261 35271
14. LiY
TrojerP
XuCF
CheungP
KuoA
2009 The target of the NSD family of histone lysine methyltransferases depends on the nature of the substrate. J Biol Chem 284 34283 34295
15. NimuraK
UraK
ShiratoriH
IkawaM
OkabeM
2009 A histone H3 lysine 36 trimethyltransferase links Nkx2-5 to Wolf-Hirschhorn syndrome. Nature 460 287 291
16. RayasamGV
WendlingO
AngrandPO
MarkM
NiederreitherK
2003 NSD1 is essential for early post-implantation development and has a catalytically active SET domain. EMBO J 22 3153 3163
17. WangGG
CaiL
PasillasMP
KampsMP
2007 NUP98-NSD1 links H3K36 methylation to Hox-A gene activation and leukaemogenesis. Nat Cell Biol 9 804 812
18. CapowskiEE
MartinP
GarvinC
StromeS
1991 Identification of grandchildless loci whose products are required for normal germline development in the nematode Caenorhabditis elegans. Genetics 129 1061 1072
19. FongY
BenderL
WangW
StromeS
2002 Regulation of the different chromatin states of autosomes and X chromosomes in the germ line of C. elegans. Science 296 2235 2238
20. KellyWG
SchanerCE
DernburgAF
LeeMH
KimSK
2002 X-chromosome silencing in the germline of C. elegans. Development 129 479 492
21. ReinkeV
GilIS
WardS
KazmerK
2004 Genome-wide germline-enriched and sex-biased expression profiles in Caenorhabditis elegans. Development 131 311 323
22. KizerKO
PhatnaniHP
ShibataY
HallH
GreenleafAL
2005 A novel domain in Set2 mediates RNA polymerase II interaction and couples histone H3 K36 methylation with transcript elongation. Mol Cell Biol 25 3305 3316
23. KroganNJ
KimM
TongA
GolshaniA
CagneyG
2003 Methylation of histone H3 by Set2 in Saccharomyces cerevisiae is linked to transcriptional elongation by RNA polymerase II. Mol Cell Biol 23 4207 4218
24. LiB
HoweL
AndersonS
YatesJR3rd
WorkmanJL
2003 The Set2 histone methyltransferase functions through the phosphorylated carboxyl-terminal domain of RNA polymerase II. J Biol Chem 278 8897 8903
25. PokholokDK
HarbisonCT
LevineS
ColeM
HannettNM
2005 Genome-wide map of nucleosome acetylation and methylation in yeast. Cell 122 517 527
26. RaoB
ShibataY
StrahlBD
LiebJD
2005 Dimethylation of histone H3 at lysine 36 demarcates regulatory and nonregulatory chromatin genome-wide. Mol Cell Biol 25 9447 9459
27. CarrozzaMJ
LiB
FlorensL
SuganumaT
SwansonSK
2005 Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell 123 581 592
28. JoshiAA
StruhlK
2005 Eaf3 chromodomain interaction with methylated H3-K36 links histone deacetylation to Pol II elongation. Mol Cell 20 971 978
29. LiB
GogolM
CareyM
LeeD
SeidelC
2007 Combined action of PHD and chromo domains directs the Rpd3S HDAC to transcribed chromatin. Science 316 1050 1054
30. SuralTH
PengS
LiB
WorkmanJL
ParkPJ
2008 The MSL3 chromodomain directs a key targeting step for dosage compensation of the Drosophila melanogaster X chromosome. Nat Struct Mol Biol 15 1318 1325
31. BuckMJ
NobelAB
LiebJD
2005 ChIPOTle: a user-friendly tool for the analysis of ChIP-chip data. Genome Biol 6 R97
32. ChoEJ
KoborMS
KimM
GreenblattJ
BuratowskiS
2001 Opposing effects of Ctk1 kinase and Fcp1 phosphatase at Ser 2 of the RNA polymerase II C-terminal domain. Genes Dev 15 3319 3329
33. KimuraH
Hayashi-TakanakaY
GotoY
TakizawaN
NozakiN
2008 The organization of histone H3 modifications as revealed by a panel of specific monoclonal antibodies. Cell Struct Funct 33 61 73
34. WangX
ZhaoY
WongK
EhlersP
KoharaY
2009 Identification of genes expressed in the hermaphrodite germ line of C. elegans using SAGE. BMC Genomics 10 213
35. BaughLR
HillAA
SlonimDK
BrownEL
HunterCP
2003 Composition and dynamics of the Caenorhabditis elegans early embryonic transcriptome. Development 130 889 900
36. MeissnerB
WarnerA
WongK
DubeN
LorchA
2009 An integrated strategy to study muscle development and myofilament structure in Caenorhabditis elegans. PLoS Genet 5 e1000537 doi:10.1371/journal.pgen.1000537
37. Kolasinska-ZwierzP
DownT
LatorreI
LiuT
LiuXS
2009 Differential chromatin marking of introns and expressed exons by H3K36me3. Nat Genet 41 376 381
38. ReinkeV
CutterAD
2009 Germline expression influences operon organization in the Caenorhabditis elegans genome. Genetics 181 1219 1228
39. BannisterAJ
SchneiderR
MyersFA
ThorneAW
Crane-RobinsonC
2005 Spatial distribution of di - and tri-methyl lysine 36 of histone H3 at active genes. J Biol Chem 280 17732 17736
40. DudleyNR
LabbeJC
GoldsteinB
2002 Using RNA interference to identify genes required for RNA interference. Proc Natl Acad Sci U S A 99 4191 4196
41. KellyWG
FireA
1998 Chromatin silencing and the maintenance of a functional germline in Caenorhabditis elegans. Development 125 2451 2456
42. RobertVJ
SijenT
van WolfswinkelJ
PlasterkRH
2005 Chromatin and RNAi factors protect the C. elegans germline against repetitive sequences. Genes Dev 19 782 787
43. UnhavaithayaY
ShinTH
MiliarasN
LeeJ
OyamaT
2002 MEP-1 and a homolog of the NURD complex component Mi-2 act together to maintain germline-soma distinctions in C. elegans. Cell 111 991 1002
44. WangD
KennedyS
ConteDJr
KimJK
GabelHW
2005 Somatic misexpression of germline P granules and enhanced RNA interference in retinoblastoma pathway mutants. Nature 436 593 597
45. ErcanS
GiresiPG
WhittleCM
ZhangX
GreenRD
2007 X chromosome repression by localization of the C. elegans dosage compensation machinery to sites of transcription initiation. Nat Genet 39 403 408
46. JansJ
GladdenJM
RalstonEJ
PickleCS
MichelAH
2009 A condensin-like dosage compensation complex acts at a distance to control expression throughout the genome. Genes Dev 23 602 618
47. GuSG
FireA
2010 Partitioning the C. elegans genome by nucleosome modification, occupancy, and positioning. Chromosoma 119 73 87
48. PhillipsCM
MengX
ZhangL
ChretienJH
UrnovFD
2009 Identification of chromosome sequence motifs that mediate meiotic pairing and synapsis in C. elegans. Nat Cell Biol 11 934 942
49. HongRL
SommerRJ
2006 Pristionchus pacificus: a well-rounded nematode. Bioessays 28 651 659
50. BenderLB
CaoR
ZhangY
StromeS
2004 The MES-2/MES-3/MES-6 complex and regulation of histone H3 methylation in C. elegans. Curr Biol 14 1639 1643
51. KetelCS
AndersenEF
VargasML
SuhJ
StromeS
2005 Subunit contributions to histone methyltransferase activities of fly and worm polycomb group complexes. Mol Cell Biol 25 6857 6868
52. XuL
FongY
StromeS
2001 The Caenorhabditis elegans maternal-effect sterile proteins, MES-2, MES-3, and MES-6, are associated in a complex in embryos. Proc Natl Acad Sci U S A 98 5061 5066
53. DuHN
FingermanIM
BriggsSD
2008 Histone H3 K36 methylation is mediated by a trans-histone methylation pathway involving an interaction between Set2 and histone H4. Genes Dev 22 2786 2798
54. ZhangP
DuJ
SunB
DongX
XuG
2006 Structure of human MRG15 chromo domain and its binding to Lys36-methylated histone H3. Nucleic Acids Res 34 6621 6628
55. TakasakiT
LiuZ
HabaraY
NishiwakiK
NakayamaJI
2007 MRG-1, an autosome-associated protein, silences X-linked genes and protects germline immortality in Caenorhabditis elegans. Development 134 757 767
56. SchanerCE
DeshpandeG
SchedlPD
KellyWG
2003 A conserved chromatin architecture marks and maintains the restricted germ cell lineage in worms and flies. Dev Cell 5 747 757
57. LucoRF
PanQ
TominagaK
BlencoweBJ
Pereira-SmithOM
2010 Regulation of alternative splicing by histone modifications. Science 327 996 1000
58. ChuaG
MorrisQD
SopkoR
RobinsonMD
RyanO
2006 Identifying transcription factor functions and targets by phenotypic activation. Proc Natl Acad Sci U S A 103 12045 12050
59. HuZ
KillionPJ
IyerVR
2007 Genetic reconstruction of a functional transcriptional regulatory network. Nat Genet 39 683 687
60. KamathRS
AhringerJ
2003 Genome-wide RNAi screening in Caenorhabditis elegans. Methods 30 313 321
61. JiX
LiW
SongJ
WeiL
LiuXS
2006 CEAS: cis-regulatory element annotation system. Nucleic Acids Res 34 W551 554
62. BolstadBM
IrizarryRA
AstrandM
SpeedTP
2003 A comparison of normalization methods for high density oligonucleotide array data based on bias and variance. Bioinformatics 19 185 193
63. IrizarryRA
HobbsB
CollinF
Beazer-BarclayYD
AntonellisKJ
2003 Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4 249 264
Štítky
Genetika Reprodukční medicína
Článek Allelic Variation at the 8q23.3 Colorectal Cancer Risk Locus Functions as a Cis-Acting Regulator ofČlánek Allelic Selection of Amplicons in Glioblastoma Revealed by Combining Somatic and Germline AnalysisČlánek Lactic Acidosis Triggers Starvation Response with Paradoxical Induction of TXNIP through MondoAČlánek Rice a Cinnamoyl-CoA Reductase-Like Gene Family Member, Is Required for NH1-Mediated Immunity to pv.Článek Differentiation of Zebrafish Melanophores Depends on Transcription Factors AP2 Alpha and AP2 Epsilon
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2010 Číslo 9
-
Všechny články tohoto čísla
- Optimal Strategy for Competence Differentiation in Bacteria
- Mutational Patterns Cannot Explain Genome Composition: Are There Any Neutral Sites in the Genomes of Bacteria?
- Frail Hypotheses in Evolutionary Biology
- Genetic Architecture of Complex Traits and Accuracy of Genomic Prediction: Coat Colour, Milk-Fat Percentage, and Type in Holstein Cattle as Contrasting Model Traits
- Allelic Variation at the 8q23.3 Colorectal Cancer Risk Locus Functions as a Cis-Acting Regulator of
- Allelic Selection of Amplicons in Glioblastoma Revealed by Combining Somatic and Germline Analysis
- Germline Variation Controls the Architecture of Somatic Alterations in Tumors
- Mice Doubly-Deficient in Lysosomal Hexosaminidase A and Neuraminidase 4 Show Epileptic Crises and Rapid Neuronal Loss
- Analysis of Population Structure: A Unifying Framework and Novel Methods Based on Sparse Factor Analysis
- FliO Regulation of FliP in the Formation of the Flagellum
- Cdc20 Is Critical for Meiosis I and Fertility of Female Mice
- dMyc Functions Downstream of Yorkie to Promote the Supercompetitive Behavior of Hippo Pathway Mutant Cells
- DCAF26, an Adaptor Protein of Cul4-Based E3, Is Essential for DNA Methylation in
- Genome-Wide Double-Stranded RNA Sequencing Reveals the Functional Significance of Base-Paired RNAs in
- An Immune Response Network Associated with Blood Lipid Levels
- Genetic Variants and Their Interactions in the Prediction of Increased Pre-Clinical Carotid Atherosclerosis: The Cardiovascular Risk in Young Finns Study
- The Histone H3K36 Methyltransferase MES-4 Acts Epigenetically to Transmit the Memory of Germline Gene Expression to Progeny
- Long- and Short-Term Selective Forces on Malaria Parasite Genomes
- Lactic Acidosis Triggers Starvation Response with Paradoxical Induction of TXNIP through MondoA
- Identification of Early Requirements for Preplacodal Ectoderm and Sensory Organ Development
- Orphan CpG Islands Identify Numerous Conserved Promoters in the Mammalian Genome
- Analysis of the Basidiomycete Reveals Conservation of the Core Meiotic Expression Program over Half a Billion Years of Evolution
- ETS-4 Is a Transcriptional Regulator of Life Span in
- The SR Protein B52/SRp55 Is Required for DNA Topoisomerase I Recruitment to Chromatin, mRNA Release and Transcription Shutdown
- The Baker's Yeast Diploid Genome Is Remarkably Stable in Vegetative Growth and Meiosis
- Chromatin Landscape Dictates HSF Binding to Target DNA Elements
- The APETALA-2-Like Transcription Factor OsAP2-39 Controls Key Interactions between Abscisic Acid and Gibberellin in Rice
- Accurately Assessing the Risk of Schizophrenia Conferred by Rare Copy-Number Variation Affecting Genes with Brain Function
- Widespread Over-Expression of the X Chromosome in Sterile F Hybrid Mice
- The Characterization of Twenty Sequenced Human Genomes
- The Genome of a Pathogenic : Cooptive Virulence Underpinned by Key Gene Acquisitions
- A Single Element Maintains Repression of the Key Developmental Regulator
- Identification of New Genetic Risk Variants for Type 2 Diabetes
- Effect of Correlated tRNA Abundances on Translation Errors and Evolution of Codon Usage Bias
- Evidence of Selection upon Genomic GC-Content in Bacteria
- Proteomic Changes Resulting from Gene Copy Number Variations in Cancer Cells
- Rice a Cinnamoyl-CoA Reductase-Like Gene Family Member, Is Required for NH1-Mediated Immunity to pv.
- Longitudinal Genome-Wide Association of Cardiovascular Disease Risk Factors in the Bogalusa Heart Study
- Response to Mechanical Stress Is Mediated by the TRPA Channel Painless in the Heart
- DNMT3L Modulates Significant and Distinct Flanking Sequence Preference for DNA Methylation by DNMT3A and DNMT3B
- Identifying Signatures of Natural Selection in Tibetan and Andean Populations Using Dense Genome Scan Data
- Incremental Genetic Perturbations to MCM2-7 Expression and Subcellular Distribution Reveal Exquisite Sensitivity of Mice to DNA Replication Stress
- Loss of Maternal ATRX Results in Centromere Instability and Aneuploidy in the Mammalian Oocyte and Pre-Implantation Embryo
- Comparative Genomic Hybridization (CGH) Reveals a Neo-X Chromosome and Biased Gene Movement in Stalk-Eyed Flies (Genus )
- Differentiation of Zebrafish Melanophores Depends on Transcription Factors AP2 Alpha and AP2 Epsilon
- Gene–Environment Interactions at Nucleotide Resolution
- Dementia Revealed: Novel Chromosome 6 Locus for Late-Onset Alzheimer Disease Provides Genetic Evidence for Folate-Pathway Abnormalities
- Critical Functions of Rpa3/Ssb3 in S-Phase DNA Damage Responses in Fission Yeast
- Preferential Re-Replication of Heterochromatin in the Absence of Geminin
- The Potential for Enhancing the Power of Genetic Association Studies in African Americans through the Reuse of Existing Genotype Data
- Evidence That Mutation Is Universally Biased towards AT in Bacteria
- Perturbation Analysis of Heterochromatin-Mediated Gene Silencing and Somatic Inheritance
- Diversity of Eukaryotic DNA Replication Origins Revealed by Genome-Wide Analysis of Chromatin Structure
- Genetic Deletion of the Desmosomal Component Promotes Tumor Microinvasion in a Mouse Model of Pancreatic Neuroendocrine Carcinogenesis
- The Metabolic Enzyme ManA Reveals a Link between Cell Wall Integrity and Chromosome Morphology
- SNPs Associated with Cerebrospinal Fluid Phospho-Tau Levels Influence Rate of Decline in Alzheimer's Disease
- Synthesizing and Salvaging NAD: Lessons Learned from
- A Central Regulatory System Largely Controls Transcriptional Activation and Repression Responses to Phosphate Starvation in Arabidopsis
- An Insect Herbivore Microbiome with High Plant Biomass-Degrading Capacity
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Synthesizing and Salvaging NAD: Lessons Learned from
- Optimal Strategy for Competence Differentiation in Bacteria
- Long- and Short-Term Selective Forces on Malaria Parasite Genomes
- Identifying Signatures of Natural Selection in Tibetan and Andean Populations Using Dense Genome Scan Data
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



