-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamadMyc Functions Downstream of Yorkie to Promote the Supercompetitive Behavior of Hippo Pathway Mutant Cells
Genetic analyses in Drosophila epithelia have suggested that the phenomenon of “cell competition” could participate in organ homeostasis. It has been speculated that competition between different cell populations within a growing organ might play a role as either tumor promoter or tumor suppressor, depending on the cellular context. The evolutionarily conserved Hippo (Hpo) signaling pathway regulates organ size and prevents hyperplastic disease from flies to humans by restricting the activity of the transcriptional cofactor Yorkie (yki). Recent data indicate also that mutations in several Hpo pathway members provide cells with a competitive advantage by unknown mechanisms. Here we provide insight into the mechanism by which the Hpo pathway is linked to cell competition, by identifying dMyc as a target gene of the Hpo pathway, transcriptionally upregulated by the activity of Yki with different binding partners. We show that the cell-autonomous upregulation of dMyc is required for the supercompetitive behavior of Yki-expressing cells and Hpo pathway mutant cells, whereas the relative levels of dMyc between Hpo pathway mutant cells and wild-type neighboring cells are critical for determining whether cell competition promotes a tumor-suppressing or tumor-inducing behavior. All together, these data provide a paradigmatic example of cooperation between tumor suppressor genes and oncogenes in tumorigenesis and suggest a dual role for cell competition during tumor progression depending on the output of the genetic interactions occurring between confronted cells.
Published in the journal: . PLoS Genet 6(9): e32767. doi:10.1371/journal.pgen.1001140
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1001140Summary
Genetic analyses in Drosophila epithelia have suggested that the phenomenon of “cell competition” could participate in organ homeostasis. It has been speculated that competition between different cell populations within a growing organ might play a role as either tumor promoter or tumor suppressor, depending on the cellular context. The evolutionarily conserved Hippo (Hpo) signaling pathway regulates organ size and prevents hyperplastic disease from flies to humans by restricting the activity of the transcriptional cofactor Yorkie (yki). Recent data indicate also that mutations in several Hpo pathway members provide cells with a competitive advantage by unknown mechanisms. Here we provide insight into the mechanism by which the Hpo pathway is linked to cell competition, by identifying dMyc as a target gene of the Hpo pathway, transcriptionally upregulated by the activity of Yki with different binding partners. We show that the cell-autonomous upregulation of dMyc is required for the supercompetitive behavior of Yki-expressing cells and Hpo pathway mutant cells, whereas the relative levels of dMyc between Hpo pathway mutant cells and wild-type neighboring cells are critical for determining whether cell competition promotes a tumor-suppressing or tumor-inducing behavior. All together, these data provide a paradigmatic example of cooperation between tumor suppressor genes and oncogenes in tumorigenesis and suggest a dual role for cell competition during tumor progression depending on the output of the genetic interactions occurring between confronted cells.
Introduction
Growth regulation requires the fine tuning between the rate of cell death and cell proliferation in developing organs. Studies in Drosophila have revealed that somatic cells within a growing epithelium compete with one another for contribution to the adult organ and this phenomenon, known as “cell competition” [1], is possibly conserved among organisms, for a review [2]. Cell competition was discovered several decades ago comparing the clonal growth parameters of Drosophila wild type cells (+/+) and slow-dividing Minute/+ cells [1]. From those analyses and recent data [3], it has been concluded that the contact between wild type and slow-growing cells, in genetic mosaics, favors the positive selection and clonal expansion of faster cells (winners) at the expense of slow-dividing ones (losers), although eventually the final number of cells in the organs is unaffected [3]. The biological function of cell competition remains unclear but it is thought to contribute to tissue homeostasis by coordinating the rate of cell proliferation and cell death [4], [5]. One of the best examples illustrating cell competition was obtained from the analysis of Drosophila myc [4], [5], opening to the speculation that this phenomenon might play a role in tumorigenesis [2], [6], however the basis of cell competition in tumorous situations has just begun to be investigated [7]. dmyc is an evolutionarily conserved proto-oncogene associated with different cellular processes, including cell cycle progression, cell growth and apoptosis [8]–[11]. The function of dMyc protein is both necessary and sufficient to control rRNA synthesis and ribosome biogenesis [12]. In Drosophila, cells carrying hypomorphic alleles of dmyc are viable in a homotypic context, but they are outcompeted and excluded from the epithelium when surrounded by wild type cells [5]. By contrast, dmyc overexpressing cells become “supercompetitors” able to kill wild type surrounding cells [4], [5]. Remarkably, dMyc upregulation is related with many types of human cancers [13] and it favors the clonal expansion of cells carrying additional oncogenic mutations [14], [15].
During the last years, the Hippo (Hpo) tumor suppressor pathway has emerged as a safeguard system restricting organ growth and preventing hyperplastic disease in metazoans [16], [17]. Mutations in several members of this pathway have been associated with tumor formation both in Drosophila and in humans [18]. It has also been reported that mutations in many members of the Hpo pathway can rescue the viability of heterozygous M/+ cells in genetic mosaics [19], suggesting that these mutant cells behave as “supercompetitors”. Therefore the detailed analysis of Hpo pathway members appears to be an attractive model in which to evaluate the relationship between cell competition and tumor growth, as well as the molecular mechanisms required for this crosstalk. Hpo, Salvador (Sav) and Warts (Wts) constitute the core of the Hpo pathway that regulates by phosphorylation the downstream transcriptional co-activator Yorkie (Yki) [18], [20]. The hyperphosphorylated form of Yki is retained in the cytoplasm [21], [22], thereby preventing the expression of several target genes involved in cell proliferation control (Cyclin E, E2F1, bantam miRNA) [16], [23]–[25], cell death (dIAP1) [16] and cell signaling regulation (dally and dally-like) [26]. It has been demonstrated that Yki regulates its target genes by binding to Scalloped (Sd), a TEAD/TEF family transcription factor [27]–[30]. In addition, recent data indicate that Yki is also able to bind to the homeoprotein Homothorax (Hth) forming a complex which regulates the transcription of bantam in the eye disc [31]. The atypical cadherins Fat (Ft) [26], [32]–[37] and Dachsous (Ds) [20], [26], [33], [38], as well as the FERM-domain proteins Expanded (Ex) and Merlin (Mer) [39], have also been implicated in the pathway as upstream components. Although their biochemical functions are still uncertain, it is assumed that they converge on Wts to regulate Yki activity [40], [41].
Here we provide a detailed analysis of the autonomous and non-autonomous effects on growth of yki-expressing cells and mutations of members of the Hpo pathway. In addition we show that dmyc is a transcriptional target of Yki, able to confer competitive properties to the Hpo pathway mutant cells in the Drosophila wing. Furthermore, dmyc upregulation is essential to sustain the high rate of cell proliferation of Hpo mutant cells and to protect them from being eliminated in a competitive background. Finally, we show that the relative levels of dMyc protein between neighboring cells are critical in order to define the role of cell competition during tumor progression.
Results
Hpo pathway mutant cells display supercompetitive properties
In order to analyze the competitive properties of Hpo pathway mutant cells, we used mosaic analysis to compare the size of yki overexpressing clones (hereafter referred to as ykiover) with their wild type twins. While clones and twins showed a comparable size in the wild type control (Figure 1A, 1F, and 1G, and Figure S1A, S1C, S1D), ykiover clones were notably larger than their wild type twins in wing discs dissected either 60h (Figure 1D, 1H, and 1I) or 48h (Figure S1B, S1E, S1F) after heat-shock induction. Furthermore, ykiover wild type twins were almost disappeared from the epithelium at 120h after egg laying (AEL) (Figure 1B and 1C). These differences in size were also prominent when discs were dissected at 96h AEL (Figure 1D and 1E). Interestingly, the clonal expansion of ykiover cells was also correlated with non-autonomous apoptosis, as revealed by active Caspase 3 immunoreactivity of a subset of surrounding wild type cells (Figure 1B–1E). The size advantage of ykiover clones and the induction of apoptosis in wild type cells is consistent with the broadly assumed definition of cell competition, which implies that the clonal expansion of the winner cells occurs at the expense of the juxtaposed losers, that are eliminated by apoptotic death [2], [42], [43]. The pattern of cell death in wild type and ykiover cells (Figure 1B–1E) was not confined to the interface between the two cell types; as can be seen in Figure 1E, cell death extends several cell diameters away and wild type cells tend to die massively when enclosed between nearby mutant clones (Figure 1E, yellow arrowhead). A similar pattern of non-autonomous cell death was observed in wild type cells nearby mutant clones for other members of the Hpo pathway, such as ft and ex (Figure S2A, S2B, S2C). Strikingly, ykiover clones and wts mutant clones grown for a longer period presented autonomous cell death (Figure 1C, see active Caspase 3 staining, and Figure S2D), despite the upregulation of anti-apoptotic molecules such as dIAP1 [16]; this might be possibly due to either developmental constraints compensating for excessive proliferation of the entire organ or toxicity caused by high and constant levels of Yki. Altogether, these results confirm the previously suggested supercompetitive properties of the Hpo pathway mutant clones [19] by revealing their ability to overgrow and eliminate surrounding wild type cells.
Fig. 1. yki overexpression confers cells a supercompetitive behavior. 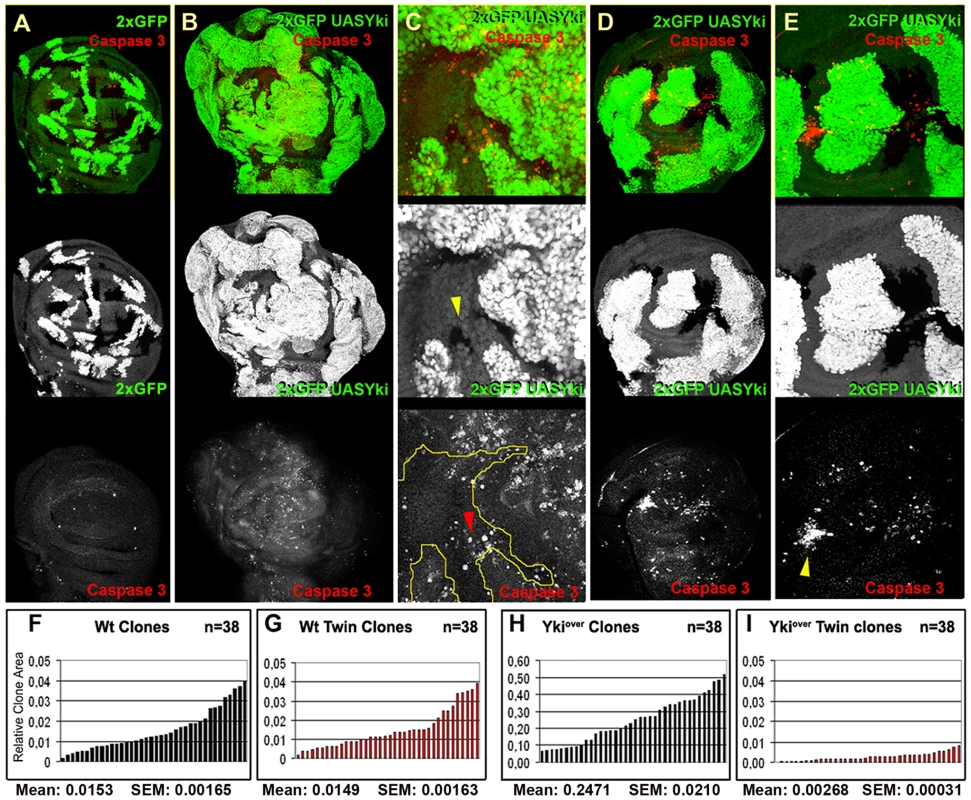
(A) yw, hs-Flp, tub-Gal4, UAS-GFP; FRT42D, tub-Gal80/FRT42D, Ubi-GFP clones induced at 48–72 h AEL. Twin clones are marked by the lack of GFP (0xGFP); imaginal discs were dissected at 120h AEL. (B–E) yw, hs-Flp,tub-Gal4, UAS-GFP; FRT42D, tub-Gal80/FRT42D, Ubi-GFP; UAS-yki/+ clones (2xGFP = 2XUbi-GFP+tub-GFP) sorrounded by wild type cells. (B,D) Clones were induced at 24–48 h AEL and dissected at 120 h AEL (B) or 96h AEL (D). C and E are magnifications of B and D respectively. Cell death assayed by Caspase 3 staining is shown in red (A–E). Yellow arrowhead indicates a wild type twin in C (0xGFP), whereas the red arrowhead indicates wild type cells dying far from ykiover clones. Note that wild type cells preferentially die when enclosed by ykiover cells (yellow arrowhead in E). (F–I) Histograms showing the surface area of wild type and ykiover clones and respective twins induced at 48–72 h AEL and allowed to grow until 120h AEL. (F, G) Wild type clones (F) and their twins (G) display the same size profile. (H) The size profile of ykiover clones indicates that they are larger either than wild type controls (F, G) or than their wild type twins (I). Note also that wild type twins (I) of ykiover clones (H) are smaller than wild type clones induced at the same stage of development (F, G). SEM = Standard Error of the Mean. P<0.0001. dmyc is a Hpo pathway target gene regulated by the activity of Yki
It is well documented that the confrontation of different levels of dMyc protein between two populations of cells either in vivo [4], [5] or in cell culture [44] can trigger cell competition, however the molecular mechanism by which this occurs is unknown. In addition, myc family oncogenes are frequently overexpressed in human cancers and it contributes to tumor progression of YAP-expressing cells (mammalian orthologue of yki) [17]. We have previously shown that a transcriptional activation of dmyc occurs in ft mutant tissues and that ft clones fail to grow in a dmyc hypomorphic background [45], indicating a possible regulation of this oncogene by the Hpo pathway. Moreover, the expression pattern of dMyc is complementary to that of Ds in the wing imaginal disc (Figure 2A), suggesting a possible functional interaction. To validate this hypothesis, we analyzed dMyc expression in mutant clones for several members of the Hpo pathway and in ykiover cells by immunofluorescence. Noticeably, we found that dMyc was upregulated in a cell-autonomous manner in ykiover clones throughout the wing disc (Figure 2B and Figure S3), with the weakest activation in the lateral regions, and in a subset of clones mutant for several Hpo pathway members (Figure 2C–2F). These differences in dMyc activation between ykiover clones and clones mutant for other members of the Hpo signaling pathway might be due to additional levels of regulation of the Hpo cascade operating on upstream members. According to our previous observations, we would predict a repression of dMyc upon Hpo pathway hyperactivation. To investigate this hypothesis, we expressed Hpo in the spalt expression domain of the developing wing disc. Since Hpo overexpressing cells die massively by apoptosis during development [25], we coexpressed the anti-apoptotic factor p35. As expected, cells coexpressing Hpo and p35 show reduced levels of dMyc with respect to the control (Figure S4A) in both late (Figure S4B) and early (Figure S4C) wing discs. Thus dMyc levels can be regulated by the Hpo pathway activity.
Fig. 2. dmyc oncogene is regulated by the Hpo pathway. 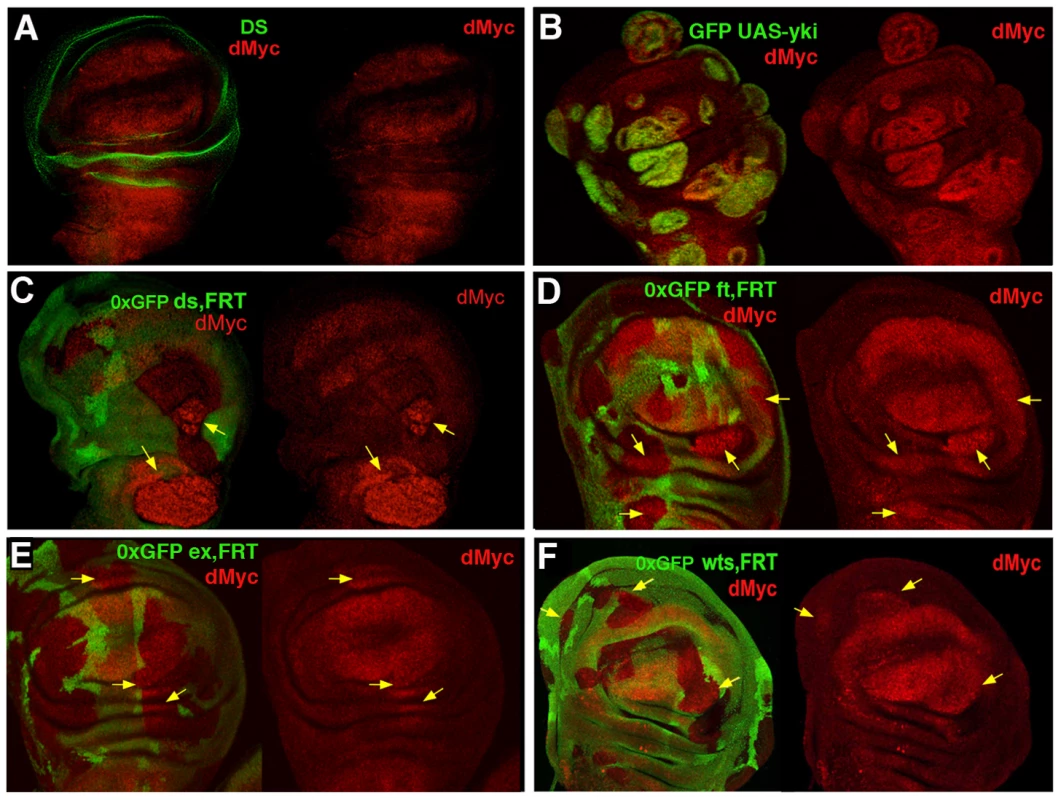
(A) Double staining with anti-dMyc (red) and anti-Ds (green) reveals that dMyc and Ds proteins show a complementary pattern of expression in the wing disc; dMyc is highly expressed in the wing pouch and less in the notum, while Ds localizes mainly in the hinge and pleura, where dMyc expression is the lowest. (B) dMyc staining of yw, hs-Flp; actFRTy+FRTGal4, UAS-GFP/UAS-yki Flp-Out clones (GFP+) show increased dMyc protein level. (C–F) dMyc protein levels are increased in dsd36, ftG-rv, exE1 and wtsX1 LOF clones (arrows). Clones were induced by the Flp/FRT system and are marked by the lack of GFP (0xGFP). Clones were induced at 42–54 h AEL (C–F) or at 54–66 h AEL (B). dmyc is transcriptionally regulated by Yki
dmyc was observed upregulated in RT-PCRs performed on ft mutant imaginal discs [45], suggesting that it could be a transcriptional target of the Hpo pathway. In order to investigate this, we first performed an in situ hybridization in Drosophila wing discs expressing yki under the control of the decapentaplegic (dpp) promoter. As expected, dmyc transcript is detectable in the dpp domain both in yki and control dmyc-expressing discs (Figure 3A). No signal within the dpp domain was detected in dpp>GFP control discs (not shown). We were able to reproduce these data using a dmyc>lacZ line [46] which recapitulates accurately the dmyc pattern throughout the wing disc during development [7], [47]. As can be seen in Figure 3B, the ßGal expression is increased in the dpp domain upon yki expression, indicating that Yki acts upon dmyc transcription. This result was supported using clonal analysis, both in ykiover cells, as shown in Figure 3C, and in cells mutant for ft (Figure S5). Altogether, these data demonstrate the ability of the Hpo pathway to regulate dmyc transcription in the imaginal wing disc.
Fig. 3. Yki regulates dmyc transcription. 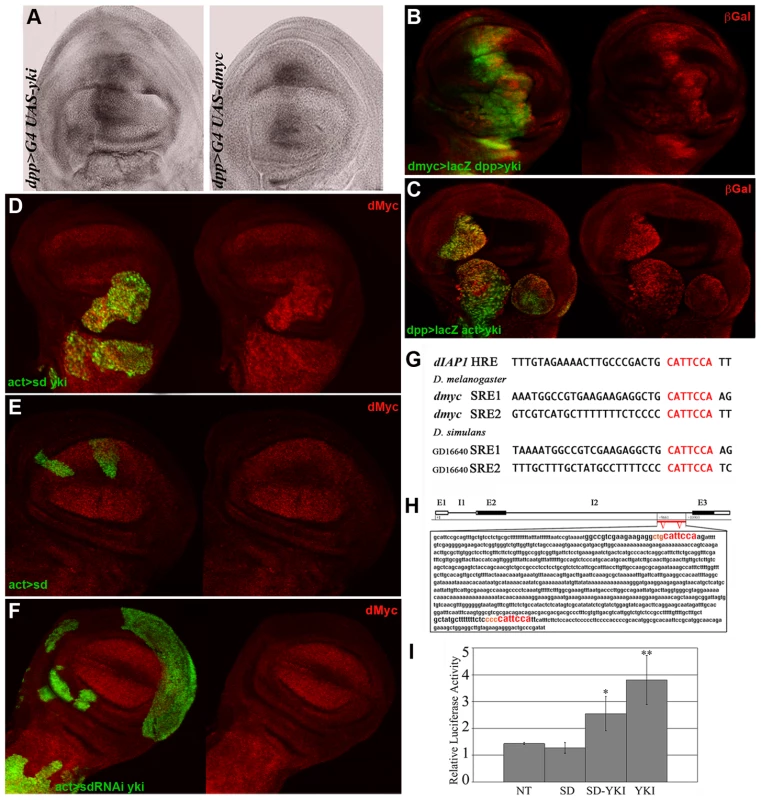
(A) In situ hybridization on imaginal wing discs with a full-length dmyc RNA probe. The dmyc transcript is robustly upregulated in the dpp>yki disc on the left. A dpp>dmyc disc is shown on the right as a control. (B) ßGal staining (red) of dmyc>lacZG0354/+; dpp>Gal4, UAS-GFP/UAS-yki imaginal wing discs. dmyc expression is upregulated in the dpp domain (GFP+). (C) ßGal staining (red) of dmyc>lacZG0354/hs-Flp; actFRTy+FRTGal4, UAS-GFP/UAS-yki imaginal wing discs. ykiover clones (GFP+) were induced at 42–54h AEL. As can be observed, a robust activation of dmyc regulatory sequences is visible within the mutant clones. (D,E) dMyc staining of yw, hs-Flp/+; UAS-sd/+; actFRTy+FRTGal4, UAS-GFP/UAS-yki and yw, hs-Flp/+; UAS-sd/+; actFRTy+FRTGal4, UAS-GFP/+ imaginal wing discs respectively. Clones (GFP+) were induced at 42–54 h AEL. As can be observed in E, dMyc levels are comparable inside and outside the mutant clones. (F) dMyc staining of yw, hs-Flp/+; UAS-sd-RNAi/+; actFRTy+FRTGal4, UAS-GFP/UAS-yki imaginal wing discs. Clones (GFP+) were induced at 42–54 h AEL. No clones in the wing pouch region were recovered and those outside the wing pouch never took the tumorous shape typical of ykiover clones. All larvae were dissected at 120h AEL. (G) dIAP1 Hpo Responsive Elements (HRE) for Yki/Sd complexes are indicated in red [28]. The same consensus sites are found twice in the second intron of dmyc both in D. melanogaster and D. simulans, indicating a possible functional conservation. (H) Scheme of the dmyc locus showing the exons (E) and introns (I). The red bar indicates the region of the second intron containing the putative Yki/Sd consensus sites. (I) Luciferase assay on S2 Drosophila cells. NT: cells transfected with dmyc-firefly alone. P value is significative for Sd/Yki (P<0,05) and Yki (P<0,01). Error bars represent standard deviation (triplicate wells). Yki transcriptional activity depends on the formation of tissue-specific complexes with different partners such as Scalloped and Homothorax [27]–[31]. In order to study the contribution of Sd to dmyc upregulation by Yki in the wing disc, we generated ykiover clones coexpressing either a UAS-sd or a UAS-sd-RNAi construct (see Figure S6A for validation). As can be seen in Figure 3D, sdover; ykiover clones overgrew relative to ykiover clones (compare with Figure 2B, 68% increase on average, n = 27, P<0,005) confirming previous data [29], but we were not able to detect significant differences in dMyc protein levels compared to ykiover clones (n = 22, P = 0,43). As expected, control sdover clones did not overgrow and did not deregulate dMyc (Figure 3E), demonstrating that Yki is required for dMyc upregulation. We were not able to recover sd-RNAi; ykiover clones in the wing pouch region, but clones generated in other territories of the wing disc, although large, did not upregulate dMyc (Figure 3F), nor showed the same degree of hyperplasia as Yki expression alone (Figure 1B–1E). sd-RNAi control clones were very small and did not deregulate dMyc (not shown). These data indicate a key role for Sd in vivo in upregulating dMyc in ykiover clones, and in contributing to the ykiover tumorous phenotype.
Interestingly, examination of dmyc locus revealed the existence of several CATTCCA repeats in non-coding regions of the gene, which perfectly match the mammalian [48], [49] and Drosophila [28], [29] TEAD/TEF family transcription factor consensus binding motifs (mammaliam orthologues of Scalloped). In addition, these putative binding motifs for Yki/Sd complexes are evolutionarily conserved in D. simulans (Figure 3G) and relatively close to the insertion point of P elements that recapitulate the endogenous expression of the gene (dmPL35 LacZ [50], [51] and dmBG02383 Gal4 insertions - http://flybase.org/reports/FBti0018138.html).
To test the significance of these sequences in dmyc regulation, we generated a dmyc-firefly reporter containing the putative responsive elements for Yki/Sd complexes (Figure 3H) and performed a transient dual luciferase assay in S2 cells. As can be seen in Figure 3I, the reporter was specifically activated upon Sd and Yki cotransfection but, unexpectedly, the transfection of Yki alone was able to activate the reporter as efficiently as the cotransfection Yki/Sd (Figure 3I). This result suggests that in presence of high levels of Yki alone, additional partners such as Hth [31] could bind it and co-regulate dmyc expression.
Indeed, complementarily to Yki/Sd complexes, Yki/Hth complexes seemed to play the same role in the presumptive thoracic region of the wing disc. Supporting this conclusion, hth-RNAi; ykiover clones down-regulated dMyc in the notum (30% reduction on average, n = 15, P<0,05, Figure S6B, yellow arrows) and did not grow as tumors in that region. By contrast, they were undistinguishable from ykiover clones in the wing pouch region (Figure S6B, white arrowhead), where Hth expression is almost undetectable (Figure S6C). Altogether, these latter results indicate that Sd and Hth play a role in Yki-induced tumorigenesis by regulating dmyc expression in the wing disc, with Sd playing a more critical role in the pouch and Hth acting in the presumptive thorax.
dMyc upregulation enhances cell proliferation of the Hpo pathway mutant cells in an autonomous manner
With the aim to investigate the cell-autonomous contribution of dMyc overexpression to ykiover phenotypes, we first compared the size of ykiover clones with that of ykiover; dmyc-RNAi clones (Figure 4, see also Figure S7A, S7A′, and [7] for RNAi construct validation ). As expected, dmyc-RNAi clones showed a reduced number of cells with respect to that observed in wild type clones (21% reduction on average, compare Figure 4B and 4B′ with Figure 4A and 4A′, P<0.01). The reduction in cell number displayed by the ykiover; dmyc-RNAi clones with respect to the ykiover clones was even more evident (43% reduction on average, compare Figure 4D and 4D′ with Figure 4C and 4C′, P<0.01), and this percentage raised up to 65% (n = 87, P<0,001) when these clones were induced earlier in development (42–54h AEL), indicating a strong cell-autonomous requirement of dMyc protein for the expansion of ykiover clones. We also observed that the non-autonomous apoptosis induced by yki overexpression was reduced upon dmyc deprivation (32% on average, n = 28, P<0,01, Figure S7B). These data suggest that dMyc upregulation promotes cell proliferation of ykiover clones in an autonomous manner, and also promotes their competitive behavior.
Fig. 4. dMyc boosts the proliferative abilities of ykiover cells. 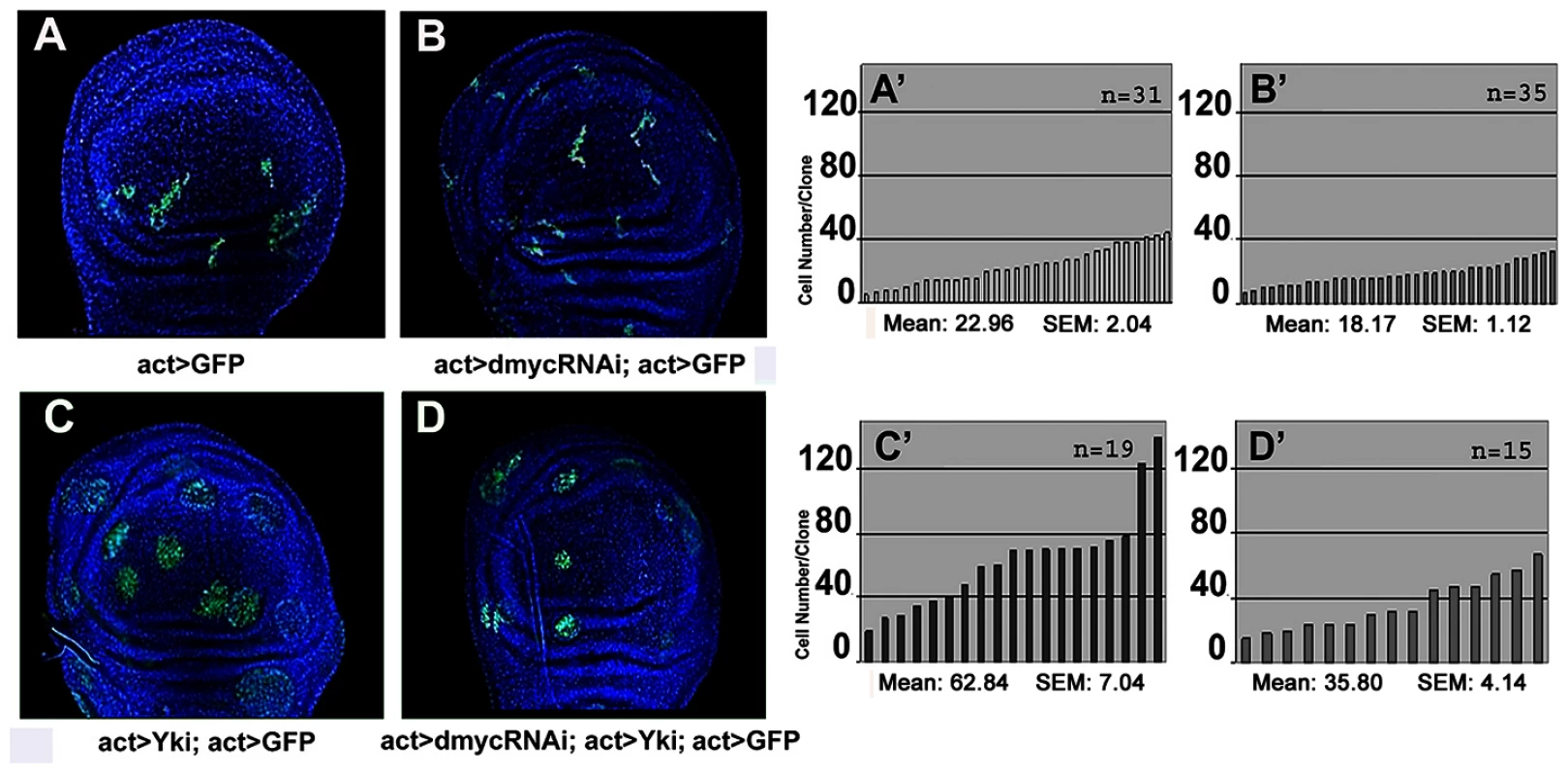
Four types of clones were simultaneously induced through the Flp-Out system at 48–72h AEL and allowed to grow for a comparable period (until 120h AEL). Clones are GFP+, nuclei are counterstained with DAPI. (A–A′) act>GFP control clones. (B–B′) act>dmyc-RNAi clones. (C–C′) act>ykiover clones proliferate faster than wild type clones. (D–D′) act>dmyc-RNAi; ykiover clones: proliferation is reduced relative to act>ykiover clones. (A′–D′) Histograms showing the number of cells/clone of the genotypes indicated in A–D. SEM = Standard Error of the Mean. P<0.01. To further characterize this proliferation-promoting effect of dMyc, we compared the clonal behavior of various mutations in members of the Hpo pathway grown in two different genetic backgrounds: a wild type context and a genetic background overexpressing dmyc under the control of a hedgehog promoter in the posterior (P) compartment of the wing disc. We found that ft, ex and ds mutant clones were consistently larger in those territories expressing uniform levels of dMyc than in the wild-type background (Figure 5 and Figure S8). It is however described that the overexpression of dMyc is able to autonomously increase apoptosis [8]–[11]. In fact, the wild type tissue expressing high amounts of dMyc tends to die and does not overgrow (see active Caspase 3 stainings in Figure 5A and 5D). Noticeably, the apoptosis mediated by dMyc overexpression seems to be extremely reduced inside ft and ex clones (Figure 5A and 5D) with respect to the wild type surrounding territories, likely due to the upregulation of antiapoptotic genes such as dIAP1, a target of the Hpo pthway [20]. In addition, the dying cells in this genetic background might induce morphogens to promote compensatory proliferation [52] that may contribute to the extra-growth of ft - or ex-UAS-dmyc expressing clones. To circumvent this problem, we repeated the same experiment coexpressing dmyc and dIAP1. As can be seen in Figure S9, both ft (Figure S9A) and ex (Figure S9D) mutant clones grown in the P compartment were still consistently larger than those originated in the A compartment, thus confirming a specific cooperation of dmyc and Hpo pathway mutants in clonal expansion.
Fig. 5. dMyc overexpression enhances the proliferation of Hpo pathway mutant cells. 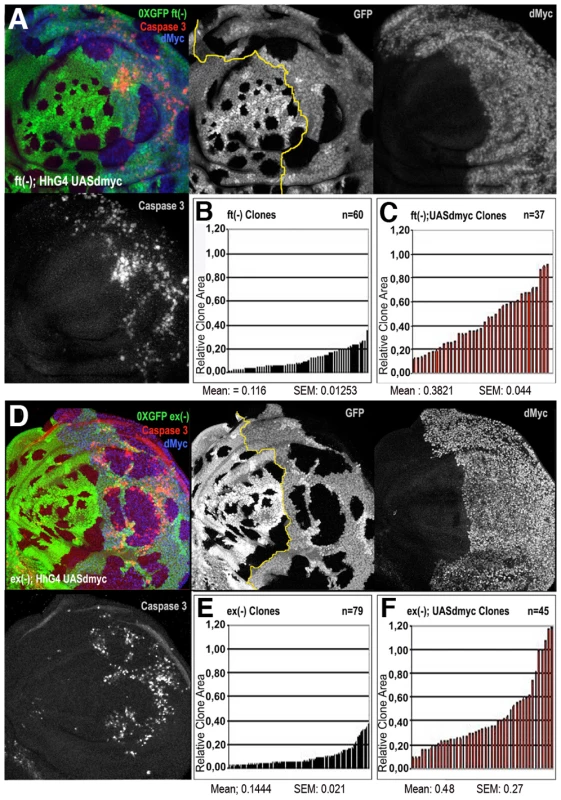
ft (A–C) and ex (D–F) LOF clones (0xGFP) generated in a background where posterior (P) cells ectopically express dmyc under the control of hh-Gal4 (A and P compartments are separated by a yellow line in A and D; P is on the right). dMyc overexpression strongly enhances the proliferative activity of ft (A–C) and ex (D–F) mutant cells; mutant clones are larger in dMyc-expressing territories (P compartment in histograms C and F) than in a wild type background (A compartment in histograms B and E). SEM = Standard Error of the Mean. P<0.001. High levels of active Caspase 3 signal are evident in dMyc-expressing cells outside ft and ex mutant clones in the posterior compartment. Hpo mutant cells therefore seem to show the ability to take advantage of the cell mass accumulation boosted by dMyc overexpression to proliferate faster.
dMyc upregulation prevents the Hpo pathway mutant clones from being restrained in a competitive background
To address the non-autonomous relevance of dmyc upregulation in providing ykiover cells with a supercompetitive behavior, we compared the size of ykiover clones generated in a wild type background to that of ykiover clones generated in a background ubiquitously overexpressing dmyc under the control of a tubulin (tub) promoter (cell competition assay, [4], [5]). In this assay cells express the endogenous dmyc gene plus an extra copy of the gene under the control of a tub promoter that ensures two-to-threefold increase of dmyc transcript [5]. This extra copy of dmyc is located in a removable cassette between the tub promoter and a Gal4 cDNA. Upon dmyc cassette excision, the tub promoter drives Gal4 expression in the clones and, as a result, those cells express lower levels of dmyc relative to the background and are rapidly eliminated from the tissue by cell competition. Only few genes have so far been found whose overexpression rescues cell viability in this context [5]. The relative difference in dMyc levels between yki-expressing cells and the surrounding tub>dmyc cells was minimized in a competitive background compared to a wild type context (compare Figure S7C and S7C′ with Figure 2B). In this competitive background, ykiover clones showed a diminished ability to overgrow compared to a wild type background (44% reduction on average, compare Figure 6C and 6C′ and Figure 6B and 6B′; P<0,01). Besides the reduction in size, ykiover clones showed an important reduction in clone number both in discs (Figure 6C) and adult wings (compare Figure S7E to Figure S7D). Moreover, ykiover clones induced earlier in development (42–54h AEL) were never recovered at the end of larval development (not shown). These data indicate that the competitive properties of ykiover cells are extremely reduced when they are surrounded by cells expressing very high amounts of dMyc.
Fig. 6. ykiover clonal expansion is restrained by dmyc-induced cell competition. 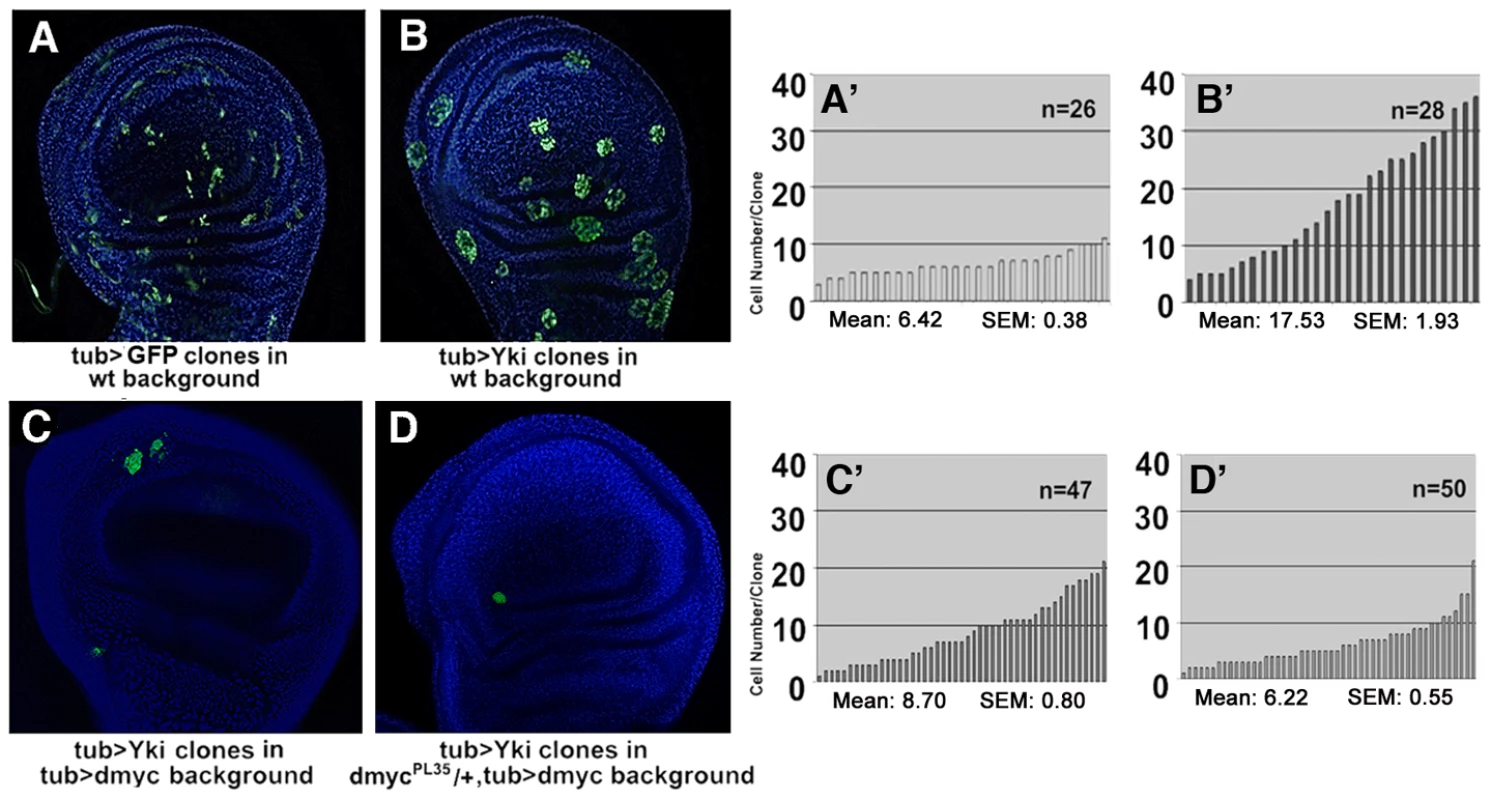
(A–D′) Cell competition assay shows that, while wild type clones are outcompeted in this genetic background [5], the clonal expansion of ykiover cells is partially restrained. Clones were induced at 60–84 h AEL and allowed to grow until 120h AEL. Clones are GFP+, nuclei are counterstained with DAPI. (A–A′) wild type control clones generated through a tubulin (tub) Flp-OUT system. (B–B′) ykiover clones generated with the same system as the wild type control. (C–C′) ykiover clones in a tub>dmyc background are smaller than in a wild type background. P<0.01. (D–D′) dmycPL35/+; tub>ykiover clones in a tub>dmyc background are smaller than in C (P<0.01), confirming that the relative dMyc levels outside vs inside the clones affect the competitive ability of ykiover cells. (A′–D′) Histograms showing the number of cells/clone of the genotypes indicated in A–D. In C and D discs are larger due to the overall increase in body size of tub>dmyc individuals [59] SEM = Standard Error of the Mean. We then performed the same competition assay as before while reducing dmyc activity inside the clones. We used the pupal lethal dmycPL35 allele [49] and, taking advantage of dmyc locus association to chromosome X, we were able to analyze both female (heterozygous condition, the expression of dmyc is halved) and male (hemizygous condition, the expression of dmyc is completely removed) larvae. In dmycPL35/+; tub>dmyc females, ykiover clones were smaller than those described in the previous assay (28% reduction on average, compare Figure 6D and 6D′ to Figure 6C and 6C′, P<0,05), whereas they were completely outcompeted by 48h after the heat shock in males (not shown). Since it has been observed that a dmycPL35 heterozygous condition does not impair cell growth or proliferation rate [49], our results reveal an important role for dmyc-induced cell competition in controlling the clonal expansion of ykiover cells, which may occur via their non-autonomous capabilities to compete with neighboring wild type cells.
dMyc expression alone is not sufficient to prevent the elimination of yki mutant cells
yki LOF clones generated in a wild type background are not able to grow [16], [25] and the ectopic expression of the antiapoptotic proteins dIAP1 [25] or p35 (Figure S10A) poorly rescues their viability, whereas a Minute background [53] or bantam overexpression within yki clones has been shown to partially rescue their growth [25]. Since our results have indicated that dmyc participates in tumor growth of the Hpo pathway mutant cells, we therefore analyzed if the expression of dMyc was sufficient to prevent the death of yki mutant cells. The overexpression of dMyc failed to rescue the viability of yki−/− cells (Figure S10B). Since yki mutant cells express low levels of the apoptosis inhibitor dIAP1 (not shown), this result is not surprising, considering the autonomous cell death described for cells overexpressing dMyc [11]. However, yki mutant cells coexpressing dMyc and p35 also failed to grow (Figure S10C). The lack of expression of additional antiapoptotic genes and cell cycle regulators [18] possibly impedes the clonal growth of yki mutant cells even though they overexpress dMyc. This result suggests that dmyc expression is able to enhance the ability of Hpo pathway mutant cells to grow, but it is not sufficient to rescue tissue growth of yki−/− clones.
Discussion
Cells within a tissue coordinate and execute complex genetic programs in order to succeed in completing a variety of processes during development. In this context, the phenomenon of cell competition may be part of the developmental plan that ensures removal and replacement of defective cells in growing organs, thus keeping their size invariant. In this work, we have evaluated in details the relationships between the phenomenon of cell competition and the clonal expansion of tumorous cells, using for that purpose mutants in components of the evolutionarily conserved Hpo pathway. From our studies we reveal that the Hpo pathway regulates dMyc expression, and show that this is critical for the tissue growth and competitive behavior of Hpo pathway mutant clones.
dMyc is a Hpo pathway transcriptional target
dmyc upregulation has been demonstrated in many studies to provide cells with supercompetitive properties [4], [5], [7]. The model explaining how dMyc can confer competitive properties to cells is based on the relative levels of this protein in neighboring cell populations, transforming those cells expressing higher levels of dMyc into supercompetitors [4], [5]. dmyc overexpression is nevertheless insufficient to drive tumorous growth; dmycover clones fail to overproliferate and show strong autonomous apoptosis [9]. Interestingly, we found that dMyc protein is overexpressed in Hpo pathway mutant clones, indicating an involvement for this cascade in dmyc regulation (Figure 2). Furthermore, the upregulation of dMyc in Yki-expressing cells correlates with an increase in the amount of mRNA, observed by in situ hybridization (Figure 3A) and using a dmyc>lacZ line (Figure 3B and 3C). Finally, we have identified a regulatory region in the second intron of dmyc that is sensitive to Yki abundance; importantly, this regulatory region includes predicted consensus-binding motifs for Sd (Figure 3H). Clonal experiments in the wing disc indicate that Sd is necessary for Yki function in vivo, since upon Sd downregulation Yki is no longer able to induce tumorous growth and does not upregulate dMyc (Figure 3F). All these findings support the notion that there is a transcriptional regulation of dMyc mediated by Yki/Sd complexes in the wing pouch. Importantly, similar results were observed for dMyc regulation in the notum by Yki/Hth complexes, suggesting that tumor growth and dmyc regulation are tissue-specific.
What is the contribution of dMyc to the Hpo pathway mutant phenotypes?
We found that dMyc upregulation is a common feature of Hpo pathway mutant cells. Since dmyc has been repeatedly associated with tumor progression and cell competition, we analyzed its role in the clonal expansion of Hpo pathway mutant cells. We observed that the reduction of dMyc expression restricts the ability of Hpo pathway mutant cells to proliferate (Figure 4), whereas its uniform overexpression strongly promotes their proliferation (Figure 5). Furhermore, while dMyc-expressing wild type cells surrounding mutant clones are rapidly eliminated by autonomous apoptosis, Hpo pathway mutant cells are able to take advantage of dMyc role in protein biosynthesis and cellular growth to divide rapidly. This is a clear example of functional cooperation between different genes in order to favor tumor progression, but it also indicates a specific role of dMyc in promoting the clonal expansion of Hpo pathway mutant cells. According to these data, we conclude that dMyc behaves as a growth-promoting factor which sustains the hyperplastic phenotype of Hpo pathway mutant cells. Importantly, this specific cooperation might be evolutionarily conserved, since c-myc appears to be upregulated in a murine model of YAP-induced carcinoma [17].
Relative levels of dMyc in neighboring cells restrict/promote clonal expansion of hyperplastic cells, likely through cell competition
It has been suggested that cell competition may be a mechanism potentially restricting the clonal expansion of tumorous cells [7], but it might also help faster proliferation of transformed cells. Our data indicate that Hpo pathway mutant cells are able to use high levels of dMyc to proliferate rapidly (Figure 5), but in a competitive context, where neighboring cells express high levels of dMyc, clonal expansion of ykiover cells is restrained (Figure 6), therefore suggesting a tumor suppressor role for cell competition. Conversely, dMyc upregulation in ykiover clones grown in a wild type background favors their clonal expansion promoting cell autonomous proliferation and also conferring the ability to outcompete sourrounding cells in a non-autonomous manner. These findings suggest that the phenomenon of cell competition may play a dual role in tumor progression depending on the output of the genetic interactions occurring between adjacent cells.
In summary, we have shown a tumor-braking gene network in Drosophila epithelia which tightly controls cell proliferation, apoptosis and cell competition via the Hpo pathway and dMyc expression. Importantly, YAP deregulation has been reported in several types of human cancers [54]–[56], therefore the mechanism of clonal expansion of Hpo pathway mutant cells in Drosophila might be relevant to understand tumor progression in mammals.
Materials and Methods
Genotypes and clonal analysis
The fly strains used in the present work were obtained by the Bloomington Stock Center and are described at http://flybase.bio.indiana.edu. The following strains were instead obtained by: w; UAS-yki (D Pan); yw, tubFRTdmycFRTGal4 and yw, dmycPL35, actFRTy+FRTGal4 (P Gallant); w, hs-FLP; actFRTy+FRTGal4, UAS-GFP (B Edgar); w; FRT40A, dsD36 (I Rodríguez). The UAS-RNAi constructs for dmyc, sd and hth were obtained from the VDRC.
All experiments were carried out at 25°C unless otherwise indicated.
MARCM UAS-yki twin-spot clones were induced at different stages of development by a 35-minutes heat shock at 37°C and larvae of the following genotype were dissected at either 84-100h AEL or 120h AEL: yw, hs-Flp, tub-Gal4, UAS-GFP; FRT42D, tub-Gal80/FRT42D, Ubi-GFP; UAS-yki/+. Clones of the same genotype were induced 54–66 h AEL and dissected 48h after a 20-minutes heat shock (Figure S1). For FRT-Flp twin analysis, the following hypomorphic or null alleles were used: dsD36, ftG-rv, exE1, wtsX1, ykiB5. Loss-of-function clones of ds, ft, ex and wts in either wild-type or mutant backgrounds overexpressing different transgenes in the posterior compartment were induced at 48–72h AEL by 1 hour heat shock at 37°C. Larvae of the following genotype were dissected at 120h AEL:
yw, hs-Flp; FRT40A, Ubi-GFP/FRT40A, dsD36 or ftG-rv or exE1
yw, hs-Flp; FRT82B, Ubi-GFP/FRT82B, wtsX1
yw, hs-Flp; FRT40A, Ubi-GFP/FRT40A, dsD36 or ftG-rv or exE1; hh-Gal4/UAS-dmyc
yw, hs-Flp; FRT40A, Ubi-GFP/FRT40A, ftG-rv or exE1; hh-Gal4/UAS-dmyc, UAS-dIAP1
The size of non-confluent clones was measured drawing each Z-stack of the confocal images using ImageJ software (http://rsbweb.nih.gov/ij). Afterwards the area of the clones was normalized dividing by the area of the wing pouch, considered as the territory encircled by the first outer folding of the wing. In Figure S1, the narrower window of clonal induction allowed us to compare clonal size without size normalization respect to the wing pouch. Statistical analysis was performed with Microsoft Excel and R (www.r-project.org). Statistical significance was determined by two tailed Student's t test and reported as the associated probability value (P).
Flp-Out clones were induced at 60h AEL by a 8-minutes heat shock at 37°C; imaginal discs of the following genotype were dissected at 120h AEL:
yw, hs-Flp; actFRTy+FRTGal4, UAS-GFP
yw, hs-Flp; UAS-dmycRNAi/+; actFRTy+FRTGal4, UAS-GFP/+
yw, hs-Flp; actFRTy+FRTGal4, UAS-GFP/UAS-yki
yw, hs-Flp; UAS-dmycRNAi/+; actFRTy+FRTGal4, UAS-GFP/UAS-yki.
yw, hs-Flp/w, dmyc>lacZG0354; actFRTy+FRTGal4, UAS-GFP/UAS-yki.
Cell competition assays were performed at 72h AEL inducing a 40-minutes heat shock at 36°C. Larvae of the following genotype were dissected at 120h AEL:
yw, tubFRTy+FRTGal4/hs-Flp; UAS-GFP/+
yw, tubFRTy+FRTGal4/hs-Flp; UAS-GFP/+; UAS-yki/+
yw, tubFRTdmycFRTGal4/hs-Flp; UAS-GFP/+; UAS-yki/+
yw, dmycPL35, hs-Flp, tubFRTdmycFRTGal4/+-Y; UAS-GFP/+; UAS-yki/+.
MARCM yki clones overexpressing p35, dMyc or both were generated at 48–72h AEL by a 45-minutes heat shock at 37°C and larvae were dissected 48h later.
Immunofluorescence
Immunostainings were performed using standard protocols. The following primary antibodies were used: mouse anti-dMyc (1∶5, P Gallant), mouse anti-En (1∶50, DSHB), rabbit anti-active Caspase 3 (1∶100, Cell Signaling Technology), rabbit anti-p35 (1∶1000, Stratagene), rabbit anti-Ds (1∶100, D Strutt), rabbit anti-Hth (1∶400, A Salzberg, [57]), mouse anti-dIAP1 (1∶100, B Hay) and rabbit anti-ßGal (1∶400, F Graziani). Anti-mouse and anti-rabbit Alexa Fluor 555 (1∶200) (Molecular Probes) and anti-mouse Cy5 (1∶200) (Jackson Laboratories) against corresponding primary antibodies were used as secondary antibodies. Imaginal discs were mounted in Vectashield (Vector Laboratories) for confocal imaging. Single Z stacks were acquired with Leica SP2 and SP5 confocal microscopes. Images for Figure 4 and Figure 6 were captured with an epifluorescence Nikon 90i microscope. Entire images were elaborated with Photoshop CS2 (Adobe) and the projections along the Z axis were rebuilt starting from 35–55 Z stacks using the ImageJ public software (NIH). For measurements of dMyc abundance, fluorescence intensity was calculated using the ImageJ public software (NIH) as the average gray value within selectioned portions of confocal Z stacks. For measurement of active Caspase 3 signal outside UAS-dmyc-RNAi; UAS-yki and UAS-yki clones, staged wing discs were chosen containing as few clones as possible and single cells positive to active Caspase 3 observed at a maximum distance of five nuclei (counterstained with DAPI) from the border of the clone were counted on confocal Z stacks. In situ hybridization was performed with a full length dmyc probe [9] on wing imaginal discs of L3 larvae expressing UAS-GFP, UAS-dmyc or UAS-yki under the control of dpp-Gal4. RNA in situ hybridization was carried out using digoxigenin-labeled RNA probes [58].
Luciferase transient expression assays
Drosophila S2 cells were grown at 25°C in Schneider medium (GIBCO) supplemented with 10% heat-inactivated FCS and 100 units of penicillin.
1189 base pairs located in the second intron of the dmyc sequence (Figure 3H) were subcloned into a pGL3-firefly vector (Promega) and co-transfected with Sd and/or Yki-expressing pAc5.1/V5-HisB plasmids [28] using Effectene Qiagen Transfection Kit. The primers used for that purpose were:
5′ CAGCGGTACCAGTTTGCTGTCCTCTGC 3′
5′GCACTCTAGAGCCATGCGGAATTGTGCG 3′.
The PCR product was first cloned in pCR 2.1 TOPO-TA (Sigma) and then subcloned in KpnI/XhoI sites of pGL3 Promoter vector. For luciferase transient expression assays, 2×104 cells were plated in 96-well dishes. Cells were harvested at 48 hours after transfection and luciferase activity was measured using the Dual-Luciferase reporter assay system (Promega). Dual-Luciferase measurements were performed using a FLUOstar Optima luminometer (BMG Labtech) and normalized to the Renilla luciferase activity using pAct5C-seapansy as an internal control. All transient expression data reported in this paper represent the means from three parallel experiments, each performed in triplicate. Average relative luciferase activity was graphed and statistically analyzed by the Student's t-test.
Supporting Information
Zdroje
1. MorataG
RipollP
1975 Minutes: mutants of Drosophila autonomously affecting cell division rate. Dev Biol 42 211 221
2. MorenoE
2008 Is cell competition relevant to cancer? Nat Rev Cancer 8 141 147
3. MartínFA
HerreraFC
MorataG
2009 Cell competition, growth and size control in the Drosophila wing imaginal disc. Development 136 3747 3756
4. de la CovaC
AbrilM
BellostaP
GallantP
JohnstonLA
2004 Drosophila myc regulates organ size by inducing cell competition. Cell 117 107 116
5. MorenoE
BaslerK
2004 dMyc transforms cells into super-competitors. Cell 117 117 129
6. BakerNE
2008 Cell competition and its possible relation to cancer. Cancer Res 68 50505 50507
7. FroldiF
ZiosiM
GaroiaF
PessionA
GrzeschikNA
2010 The lethal giant larvae tumour suppressor mutation requires dMyc oncoprotein to promote clonal malignancy. BMC Biol 8 33
8. OsterSK
HoCS
SoucieEL
PennLZ
2002 The myc oncogene: Marvelous Complex. Adv Cancer Res 84 81 154
9. JohnstonLA
ProberDA
EdgarBA
EisenmanRN
GallantP
1999 Drosophila myc regulates cellular growth during development. Cell 98 779 790
10. MeyerN
KimSS
PennLZ
2006 The Oscar-worthy role of Myc in apoptosis. Semin Cancer Biol 16 275 287
11. MonteroL
MüllerN
GallantP
2008 Induction of Apoptosis by Drosophila Myc. Genesis 46 104 111
12. GrewalSS
LiL
OrianA
EisenmanRN
EdgarBA
2005 Myc-dependent regulation of ribosomal RNA synthesis during Drosophila development. Nat Cell Biol 7 295 302
13. VitaM
HenrikssonM
2006 The Myc oncoprotein as a therapeutic target for human cancer. Semin Cancer Biol 16 318 330
14. LandH
ParadaLF
WeinbergRA
1983 Cellular oncogenes and multistep carcinogenesis. Science 222 771 778
15. ZhanL
RosenbergA
BergamiKC
YuM
XuanZ
2008 Deregulation of Scribble Promotes Mammary Tumorigenesis and Reveals a Role for Cell Polarity in Carcinoma. Cell 135 865 878
16. HuangJ
WuS
BarreraJ
MatthewsK
PanD
2005 The Hippo signaling pathway coordinately regulates cell proliferation and apoptosis by inactivating Yorkie, the Drosophila homolog of YAP. Cell 122 421 434
17. DongJ
FeldmannG
HuangJ
WuS
ZhangN
2007 Elucidation of a universal size-control mechanism in Drosophila and mammals. Cell 130 1120 1133
18. SaucedoLJ
EdgarB
2007 Filling out the Hippo pathway. Nat Rev Mol Cell Biol 8 613 621
19. TylerDM
LiW
ZhuoN
PellockB
BakerNE
2006 Genes affecting cell competition in Drosophila. Genetics 175 643 657
20. HarveyK
TaponN
2007 The Salvador-Warts-Hippo pathway - an emerging tumour-suppressor network. Nat Rev Cancer 3 182 191
21. OhH
IrvineKD
2009 In vivo analysis of Yorkie phosphorilation sites. Oncogene 28 1916 1927
22. RenF
ZhangL
JiangJ
2009 Hippo signaling regulates Yorkie nuclear localization and activity through 14-3-3 dependent and independent mechanisms. Dev Biol 337 303 312
23. NicolayBN
FrolovMV
2008 Context-dependent requirement for dE2F during oncogenic proliferation. PLoS Genet 4 e1000205
24. NoloR
MorrisonCM
TaoC
ZhangX
HalderG
2006 The bantam MicroRNA is a target of the Hippo tumor-suppressor pathway. Curr Biol 16 1895 1904
25. ThompsonBJ
CohenSM
2006 The Hippo pathway regulates the bantam microRNA to control cell proliferation and apoptosis in Drosophila. Cell 126 767 774
26. Baena-LopezLA
RodriguezI
BaonzaA
2008 The tumor suppressor genes dachsous and fat modulate different signalling pathways by regulating dally and dally-like. Proc Natl Acad Sci USA 105 9645 9650
27. GoulevY
FaunyJD
Gonzalez-MartiB
FlagielloD
SilberJ
2008 SCALLOPED Interacts with YORKIE, the Nuclear Effector of the Hippo Tumor-Suppressor Pathway in Drosophila. Curr Biol 18 435 441
28. WuS
LiuY
ZhengQ
DongJ
PanD
2008 The TEAD/TEF family protein Scalopped mediates transcriptional output of the Hippo growth-regulatory pathway. Dev Cell 14 388 398
29. ZhangL
RenF
ZhangQ
ChenY
WangB
2008 The TEAD/TEF family of transcription factor Scalopped mediates Hippo signaling in organ size control. Dev Cell 14 377 387
30. ZhaoB
YeX
YuJ
LiL
LiW
2008 TEAD mediates YAP-dependent gene induction and growth control. Genes Dev 22 1962 1971
31. PengHW
SlatteryM
MannRS
2009 Transcription factor choice in the Hippo signaling pathway: homothorax and yorkie regulation of the microRNA bantam, in the progenitor domain of the Drosophila eye imaginal disc. Genes Dev 1 2307 2319
32. WilleckeM
HamaratogluF
Kango-SinghM
UdanR
ChenC
2006 The Fat Cadherin Acts through the Hippo Tumor-Suppressor Pathway to Regulate Tissue Size. Curr Biol 16 1 11
33. SilvaE
TsatskisY
GardanoL
TaponN
McNeillH
2006 The Tumor-Suppressor Gene fat Controls Tissue Growth Upstream of Expanded in the Hippo Signaling Pathway. Curr Biol 16 2081 2089
34. ChoE
FengY
RauskolbC
MaitraS
FehonR
2006 Delineation of a Fat tumor suppressor pathway. Nat Genet 38 1142 1150
35. BennettFC
HarveyKF
2006 Fat Cadherin Modulates Organ Size in Drosophila via the Salvador/Warts/Hippo Signaling Pathway. Curr Biol 16 2101 2110
36. ChoE
IrvineKD
2004 Action of fat, four-jointed, dachsous and dachs in distal-to-proximal wing signaling. Development 131 4489 4500
37. FengY
IrvineKD
2007 Fat and expanded act in parallel to regulate growth through warts. Proc Natl Acad Sci USA 104 20362 20367
38. WilleckeM
HamaratogluF
Sansores-GarciaL
TaoC
HalderG
2008 Boundaries of Dachsous Cadherin activity modulate the Hippo signaling pathway to induce cell proliferation. Proc Natl Acad Sci USA 105 14897 14902
39. HamaratogluF
WilleckeM
Kango-SinghM
NoloR
HyunE
2006 The tumour suppressor genes NF2/Merlin and Expanded act through Hippo signalling to regulate cell proliferation and apoptosis. Nat Cell Biol 8 27 36
40. WuS
HuangJ
DongJ
PanD
2003 hippo encodes a Ste-20 family protein kinase that restricts cell proliferation and promotes apoptosis in conjunction with salvador and warts. Cell 114 445 456
41. ReddyBVVG
IrvineKD
2008 The Fat and warts signaling pathways: new insights into their regulation, mechanism and conservation. Development 135 2827 2838
42. MorenoE
BaslerK
MorataG
2002 Cells compete for decapentaplegic survival factor to prevent apoptosis in Drosophila wing development. Nature 416 755 759
43. LiW
BakerNE
2007 Engulfment is required for cell competition. Cell 15 1215 1225
44. Senoo-MatsudaN
JohnstonLA
2007 Soluble factors mediate competitive and cooperative interactions between cells expressing different levels of Drosophila Myc. Proc Natl Acad Sci USA 104 18543 18548
45. GaroiaF
GrifoniD
TrottaV
GuerraD
PezzoliMC
2005 The tumor suppressor gene fat modulates the EGFR-mediated proliferation control in the imaginal tissues of Drosophila melanogaster. Mech Dev 122 175 187
46. PeterA
SchöttlerP
WernerM
BeinertN
DoweG
2002 Mapping and identification of essential gene functions on the X chromosome of Drosophila. EMBO Rep 31 34 38
47. CrannaN
QuinnL
2009 Impact of steroid hormone signals on Drosophila cell cycle during development. Cell Div 20 4 : 3
48. XiaoJH
DavidsonI
MatthesH
GarnierJM
ChambonP
1991 Cloning, expression, and transcriptional properties of the human enhancer factor TEF-1. Cell 65 551 568
49. LarkinSB
FarranceIK
OrdahlCP
1996 Flanking sequences modulate the cell specificity of M-CAT elements. Mol Cell Biol 16 3742 3755
50. BourbonHM
Gonzy-TreboulG
PeronnetF
AlinMF
ArdourelC
2002 A P-insertion screen identifying novel X-linked essential genes in Drosophila. Mech Dev 110 71 83
51. BenassayagC
MonteroL
ColombieN
GallantP
CribbsD
2005 Human c-Myc isoforms differentially regulate cell growth and apoptosis in Drosophila melanogaster. Mol Cell Biol 25 9897 9909
52. MartínFA
Peréz-GarijoA
MorataG
2009 Apoptosis in Drosophila: compensatory proliferation and undead cells. Int J Dev Biol 53 1341 1347
53. OhH
IrvineKD
2008 In vivo regulation of Yorkie phosphorylation and localization. Development 135 1081 1088
54. Lam-HimlinDM
DanielsJA
GayyedMF
DongJ
MaitraA
2006 The hippo pathway in human upper gastrointestinal dysplasia and carcinoma: a novel oncogenic pathway. Int J Gastrointest Cancer 37 103 109
55. SteinhardtAA
GayyedMF
KleinAP
DongJ
MaitraA
2008 Expression of Yes-associated protein in common solid tumors. Hum Pathol 39 1582 1589
56. OverholtzerM
ZhangJ
SmolenGA
MuirB
LiW
2006 Transforming properties of YAP, a candidate oncogene on the chromosome 11q22 amplicon. Proc Natl Acad Sci USA 103 12405 12410
57. KurantE
PaiCY
SharfR
HalachmiN
SunYH
1998 Dorsotonals/homothorax, the Drosophila homologue of meis1, interacts with extradenticle in patterning of the embryonic PNS. Development 125 1037 1048
58. JohnstonLA
EdgarBA
1998 Wingless and Notch regulate cell-cycle arrest in the developing Drosophila wing. Nature 394 82 84
59. De la CovaC
JohnstonLA
2006 Myc in model organisms: a view from the fly room. Sem Cancer Biol 16 303 312
Štítky
Genetika Reprodukční medicína
Článek Allelic Variation at the 8q23.3 Colorectal Cancer Risk Locus Functions as a Cis-Acting Regulator ofČlánek Allelic Selection of Amplicons in Glioblastoma Revealed by Combining Somatic and Germline AnalysisČlánek Lactic Acidosis Triggers Starvation Response with Paradoxical Induction of TXNIP through MondoAČlánek Rice a Cinnamoyl-CoA Reductase-Like Gene Family Member, Is Required for NH1-Mediated Immunity to pv.Článek Differentiation of Zebrafish Melanophores Depends on Transcription Factors AP2 Alpha and AP2 Epsilon
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2010 Číslo 9
-
Všechny články tohoto čísla
- Optimal Strategy for Competence Differentiation in Bacteria
- Mutational Patterns Cannot Explain Genome Composition: Are There Any Neutral Sites in the Genomes of Bacteria?
- Frail Hypotheses in Evolutionary Biology
- Genetic Architecture of Complex Traits and Accuracy of Genomic Prediction: Coat Colour, Milk-Fat Percentage, and Type in Holstein Cattle as Contrasting Model Traits
- Allelic Variation at the 8q23.3 Colorectal Cancer Risk Locus Functions as a Cis-Acting Regulator of
- Allelic Selection of Amplicons in Glioblastoma Revealed by Combining Somatic and Germline Analysis
- Germline Variation Controls the Architecture of Somatic Alterations in Tumors
- Mice Doubly-Deficient in Lysosomal Hexosaminidase A and Neuraminidase 4 Show Epileptic Crises and Rapid Neuronal Loss
- Analysis of Population Structure: A Unifying Framework and Novel Methods Based on Sparse Factor Analysis
- FliO Regulation of FliP in the Formation of the Flagellum
- Cdc20 Is Critical for Meiosis I and Fertility of Female Mice
- dMyc Functions Downstream of Yorkie to Promote the Supercompetitive Behavior of Hippo Pathway Mutant Cells
- DCAF26, an Adaptor Protein of Cul4-Based E3, Is Essential for DNA Methylation in
- Genome-Wide Double-Stranded RNA Sequencing Reveals the Functional Significance of Base-Paired RNAs in
- An Immune Response Network Associated with Blood Lipid Levels
- Genetic Variants and Their Interactions in the Prediction of Increased Pre-Clinical Carotid Atherosclerosis: The Cardiovascular Risk in Young Finns Study
- The Histone H3K36 Methyltransferase MES-4 Acts Epigenetically to Transmit the Memory of Germline Gene Expression to Progeny
- Long- and Short-Term Selective Forces on Malaria Parasite Genomes
- Lactic Acidosis Triggers Starvation Response with Paradoxical Induction of TXNIP through MondoA
- Identification of Early Requirements for Preplacodal Ectoderm and Sensory Organ Development
- Orphan CpG Islands Identify Numerous Conserved Promoters in the Mammalian Genome
- Analysis of the Basidiomycete Reveals Conservation of the Core Meiotic Expression Program over Half a Billion Years of Evolution
- ETS-4 Is a Transcriptional Regulator of Life Span in
- The SR Protein B52/SRp55 Is Required for DNA Topoisomerase I Recruitment to Chromatin, mRNA Release and Transcription Shutdown
- The Baker's Yeast Diploid Genome Is Remarkably Stable in Vegetative Growth and Meiosis
- Chromatin Landscape Dictates HSF Binding to Target DNA Elements
- The APETALA-2-Like Transcription Factor OsAP2-39 Controls Key Interactions between Abscisic Acid and Gibberellin in Rice
- Accurately Assessing the Risk of Schizophrenia Conferred by Rare Copy-Number Variation Affecting Genes with Brain Function
- Widespread Over-Expression of the X Chromosome in Sterile F Hybrid Mice
- The Characterization of Twenty Sequenced Human Genomes
- The Genome of a Pathogenic : Cooptive Virulence Underpinned by Key Gene Acquisitions
- A Single Element Maintains Repression of the Key Developmental Regulator
- Identification of New Genetic Risk Variants for Type 2 Diabetes
- Effect of Correlated tRNA Abundances on Translation Errors and Evolution of Codon Usage Bias
- Evidence of Selection upon Genomic GC-Content in Bacteria
- Proteomic Changes Resulting from Gene Copy Number Variations in Cancer Cells
- Rice a Cinnamoyl-CoA Reductase-Like Gene Family Member, Is Required for NH1-Mediated Immunity to pv.
- Longitudinal Genome-Wide Association of Cardiovascular Disease Risk Factors in the Bogalusa Heart Study
- Response to Mechanical Stress Is Mediated by the TRPA Channel Painless in the Heart
- DNMT3L Modulates Significant and Distinct Flanking Sequence Preference for DNA Methylation by DNMT3A and DNMT3B
- Identifying Signatures of Natural Selection in Tibetan and Andean Populations Using Dense Genome Scan Data
- Incremental Genetic Perturbations to MCM2-7 Expression and Subcellular Distribution Reveal Exquisite Sensitivity of Mice to DNA Replication Stress
- Loss of Maternal ATRX Results in Centromere Instability and Aneuploidy in the Mammalian Oocyte and Pre-Implantation Embryo
- Comparative Genomic Hybridization (CGH) Reveals a Neo-X Chromosome and Biased Gene Movement in Stalk-Eyed Flies (Genus )
- Differentiation of Zebrafish Melanophores Depends on Transcription Factors AP2 Alpha and AP2 Epsilon
- Gene–Environment Interactions at Nucleotide Resolution
- Dementia Revealed: Novel Chromosome 6 Locus for Late-Onset Alzheimer Disease Provides Genetic Evidence for Folate-Pathway Abnormalities
- Critical Functions of Rpa3/Ssb3 in S-Phase DNA Damage Responses in Fission Yeast
- Preferential Re-Replication of Heterochromatin in the Absence of Geminin
- The Potential for Enhancing the Power of Genetic Association Studies in African Americans through the Reuse of Existing Genotype Data
- Evidence That Mutation Is Universally Biased towards AT in Bacteria
- Perturbation Analysis of Heterochromatin-Mediated Gene Silencing and Somatic Inheritance
- Diversity of Eukaryotic DNA Replication Origins Revealed by Genome-Wide Analysis of Chromatin Structure
- Genetic Deletion of the Desmosomal Component Promotes Tumor Microinvasion in a Mouse Model of Pancreatic Neuroendocrine Carcinogenesis
- The Metabolic Enzyme ManA Reveals a Link between Cell Wall Integrity and Chromosome Morphology
- SNPs Associated with Cerebrospinal Fluid Phospho-Tau Levels Influence Rate of Decline in Alzheimer's Disease
- Synthesizing and Salvaging NAD: Lessons Learned from
- A Central Regulatory System Largely Controls Transcriptional Activation and Repression Responses to Phosphate Starvation in Arabidopsis
- An Insect Herbivore Microbiome with High Plant Biomass-Degrading Capacity
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Synthesizing and Salvaging NAD: Lessons Learned from
- Optimal Strategy for Competence Differentiation in Bacteria
- Long- and Short-Term Selective Forces on Malaria Parasite Genomes
- Identifying Signatures of Natural Selection in Tibetan and Andean Populations Using Dense Genome Scan Data
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



