-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaA New Family of Secreted Toxins in Pathogenic Neisseria Species
Many bacteria are able to secrete toxins targeted against neighboring cells. In order to protect themselves against their own toxin, they also express an “immunity” protein. In silico analysis of bacterial genomes predicts that numerous genes could encode potential new toxin-immunity systems. The recently described CDI system is involved in contact-dependent inhibition of growth and confers to its host strain a significant advantage in competitive ecosystems such as the gastro-intestinal tract. Indeed, an Escherichia coli CDI+ strain is able to outcompete CDI - strains and to become predominant. Here, we show that a large family of genes called “maf”, found in pathogenic Neisseria species, encodes a toxin-immunity system. We demonstrate that a toxin named MafBMGI-1NEM8013 inhibits the growth of E. coli by degrading RNA and show that the immunity protein MafIMGI-1NEM8013 is able to abolish the toxicity. MafB toxins exhibit highly variable toxic domains. This variability of secreted toxins could be important to compete against bacteria of different species sharing the same reservoir. Since a strain may contain numerous toxin-immunity systems that can all play a role in interbacterial competition, deciphering interactions between these systems will allow a better understanding of complex bacterial communities.
Published in the journal: . PLoS Pathog 11(1): e32767. doi:10.1371/journal.ppat.1004592
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1004592Summary
Many bacteria are able to secrete toxins targeted against neighboring cells. In order to protect themselves against their own toxin, they also express an “immunity” protein. In silico analysis of bacterial genomes predicts that numerous genes could encode potential new toxin-immunity systems. The recently described CDI system is involved in contact-dependent inhibition of growth and confers to its host strain a significant advantage in competitive ecosystems such as the gastro-intestinal tract. Indeed, an Escherichia coli CDI+ strain is able to outcompete CDI - strains and to become predominant. Here, we show that a large family of genes called “maf”, found in pathogenic Neisseria species, encodes a toxin-immunity system. We demonstrate that a toxin named MafBMGI-1NEM8013 inhibits the growth of E. coli by degrading RNA and show that the immunity protein MafIMGI-1NEM8013 is able to abolish the toxicity. MafB toxins exhibit highly variable toxic domains. This variability of secreted toxins could be important to compete against bacteria of different species sharing the same reservoir. Since a strain may contain numerous toxin-immunity systems that can all play a role in interbacterial competition, deciphering interactions between these systems will allow a better understanding of complex bacterial communities.
Introduction
The growing number of sequenced bacterial genomes has led to the computer-based prediction of numerous novel bacterial factors possibly involved in virulence. As a result, many novel putative bacterial toxins have been identified by sequence-homology criteria. However, very few of these bacterial proteins have been tested for their toxic activity. Using in silico analysis, Aravind and colleagues have recently described widespread genes encoding putative secreted multi-domain toxins grouped under the name of bacterial polymorphic toxin systems (or polymorphic toxin-immunity systems) [1]–[3]. In silico analysis identified over 150 distinct toxin domains in these systems including many putative peptidase, nuclease or deaminase domains. Immunity genes found immediately downstream of the toxin genes encode highly variable proteins that protect bacteria from their own toxins or from toxins secreted by neighboring cells [4]–[6]. Immunity genes are a characteristic of polymorphic toxin systems that distinguishes them from host-directed toxins (i.e. cholera toxin or pertussis toxin) [7]. The polymorphic toxin systems are typically encoded on hypervariable chromosomal islands with characteristics of horizontal gene transfer [1]. These systems are found in both Gram - negative and positive bacteria [8]. The dominant hypothesis is that polymorphic toxin systems are primarily involved in conflict between related bacterial strains. The N-terminal domain of the toxin is typically related to trafficking mode whereas the C-terminal domain carries the toxic activity [1]. In a defined family of polymorphic toxins, the N-terminal domains are similar, while the C-terminal domains are highly variable. Toxins are potentially secreted by Type II, V, VI, or VII (ESX) secretion systems [1]. Toxins are encoded in loci that also contain standalone cassettes and immunity genes. Cassettes encoding alternative C-termini could promote diversity of toxic activities through genetic recombination [1], [9], [10].
The recently described contact-dependent growth inhibition (CDI) systems are a subgroup of polymorphic toxin systems [1], [11]. Toxins encoded by CDI systems are large filamentous proteins that exhibit RHS (rearrangement hotspot) or filamentous haemagglutinin repeats in their central region [4], [6], [8]. Rhs proteins are likely to be exported through the Type VI secretion machinery [6], whereas toxins with filamentous haemagglutinin repeats are exported through the Type 5 Secretion System (T5SS) [8], [12]. The first CDI toxin secreted by a T5SS was reported in Escherichia coli EC93 (CdiAEC93) [11]. E. coli EC93 was found to inhibit the growth of other E. coli strains (i.e. E. coli K-12) in co-culture experiments. The growth inhibition mediated by E. coli EC93 required a direct contact between toxic and target cells. In CDI systems, the toxin CdiA is secreted by an outer membrane transporter named CdiB. CdiA and CdiB are part of a two-partner secretion protein family (type Vb). Subsequently, several studies have demonstrated that CDI systems are present in many species including Neisseria meningitidis [4], [5], [13]. Moreover, it has been recently demonstrated that the two-partner system TpsAB is indeed a functional CDI system in N. meningitidis strain B16B6 [14].
In addition to non-pathogenic commensal species, the genus Neisseria includes two human pathogens: N. gonorrhoeae (the gonococcus) and N. meningitidis (the meningococcus). N. gonorrhoeae colonizes the uro-genital tract and is a common cause of sexually transmitted infections [15]. N. meningitidis is commonly found in the nasopharynx of healthy individuals, where it can cross the mucosal epithelium and cause sepsis or meningitis [16]. For yet unknown reasons, some meningococcal strains belonging to a limited number of clonal complexes, known as hyper invasive clonal complexes, are much more likely to cause disease than others [17].
Comparison of the genomes of related bacteria that exhibit distinct pathogenic phenotypes can identify relevant genetic variations linked to virulence. The availability of complete genome sequences for several strains of both pathogenic and non-pathogenic species of Neisseria genus enabled their in silico comparison [18]–[23]. Genes involved in adherence to epithelial cells, in capsule biosynthesis, or in iron uptake are well known to be crucial for pathogenicity [16], [21], [24]. Nevertheless, their presence is not sufficient to explain the invasiveness of pathogenic strains compared to non-pathogenic strains. Thus, to date, genomic comparisons between pathogenic and non-pathogenic Neisseria species have failed to identify genes sufficient and necessary to cause disease [18], [21], [25]. The accessory genome, which is composed of genes found only in some strains, confers strain-specific traits and is commonly acquired through horizontal transfer [18]. The accessory genome may be linked to virulence as illustrated by pathogenicity islands (PAI) that are present in pathogenic strains, and absent in non-pathogenic strains of one species. PAIs are genomic islands (GIs) encoding virulence factors such as toxins, adhesins or invasins. Identification of GIs is primarily based on a different G+C content from the rest of the genome and on their association with insertion sequence (IS) elements or tRNA genes at their boundaries [26], [27].
There are several identified islands and prophages in meningococci and gonococci [23], [28]–[34]. It has been recently suggested that an island composed of 22 genes in the N. meningitidis isolate 053442 genome (NMCC_0592 to NMCC_0613) and called IHT-G (Island of Horizontally Transferred DNA-G) could be a “meningococcal pathogenicity island-like region” [20]. This island, which is adjacent to a tRNA-Pro gene, contains genes belonging to the multiple adhesin family (maf). Maf proteins were first described in the gonococcal strain MS11 as ligands interacting with a specific glycolipid (GgO4) [35]. Indeed, the heterologous expression of the neisserial protein in E. coli allows bacterial adhesion to GgO4 [35]. Since multiple genes in the gonococcus chromosome encode these proteins, they were subsequently termed “MafA adhesins”. The gene immediately downstream of mafA, the function of which was unknown, was termed mafB because both genes are organized in a putative operon.
In this study, we analyzed loci containing maf genes in several strains of N. meningitidis and N. gonorrhoeae. We propose here a novel uniform nomenclature of these loci. We demonstrated experimentally that mafB genes encode polymorphic toxins and that genes immediately downstream of mafB encode a specific immunity protein (MafI). Furthermore, we demonstrated that overexpression of one of the four MafB toxins of strain NEM8013 provides an advantage in competition assays.
Results
General features of maf genomic islands in pathogenic Neisseria species
A uniform nomenclature for loci containing maf genes
Our in silico analysis of the genome of 12 strains of N. meningitidis (Z2491, MC58, FAM18, H44/76, M04-240196, M01-240355, NZ-05/33, WUE2594, 053442, G2136, M6190 and NEM8013) and 3 strains of N. gonorrhoeae (FA1090, MS11 and NCCP 11945) revealed that all strains contain several loci in which genes previously designated mafA and mafB are present. To facilitate comparison of these loci, we propose the following uniform nomenclature.
Typical maf loci contain at least one module of two genes: mafB and a small ORF that we designated mafI (Fig. 1A). If a locus contains this module, the locus is thereafter named MGI for maf genomic island. MGIs have conserved chromosomal locations (Fig. 1B, Fig. 2) and exhibit hallmarks of horizontal gene transfer. They are indeed located near a tRNA gene or flanked by a transposable element (IS1016 element), and exhibit a nucleotide composition different from the rest of the genome. The average GC% of a MGI is close to 40% whereas the average GC% of a pathogenic Neisseria genome is close to 52%.
Fig. 1. Organization and location of maf genomic islands in pathogenic Neisseria species. 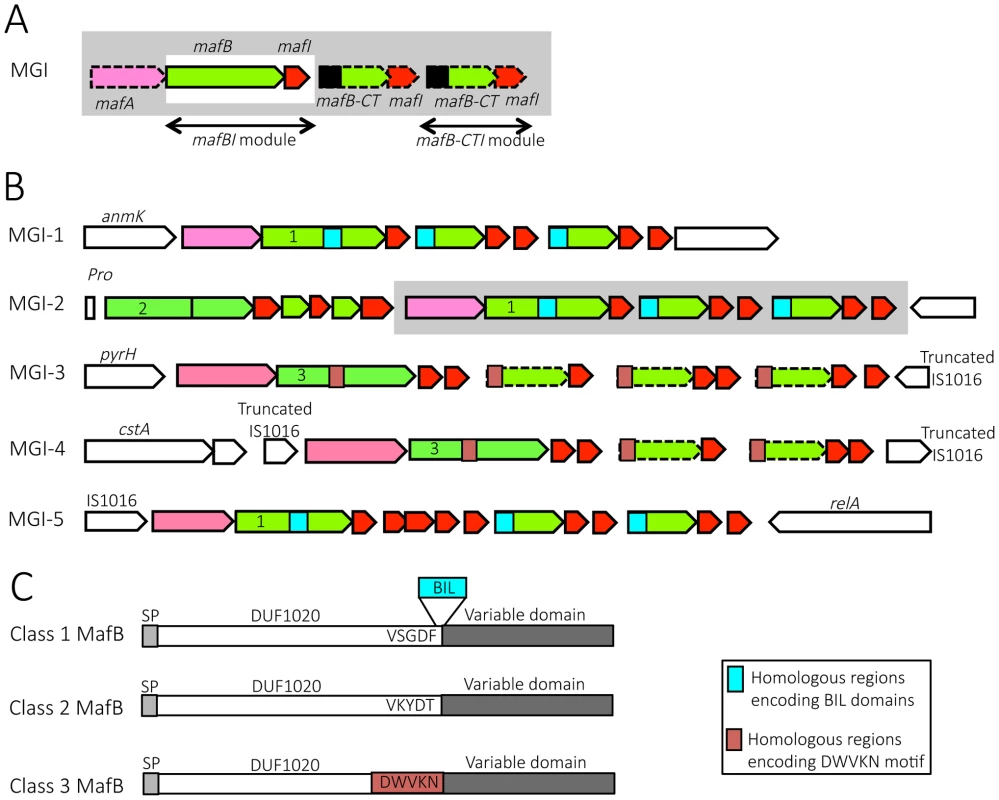
A) Schematic depiction of maf genomic island (MGI). By definition, a GI containing a mafBI module is a MGI. Each mafBI module is composed of two genes: mafB (green) and mafI (red). Additional genes (on a grey background) are mafA (pink), ORFs encoding alternative C-terminal domains of MafB (mafB-CT, green) associated with their cognate mafI gene (red). Black box at the 5′ end of mafB-CT indicates a region potentially involved in antigenic variation of MafB. In class 1 MafBs, this region encodes a bacterial intein-like (BIL) domain whereas in class 3 MafBs, this region encodes a DWVKN motif. B) Simplified genomic organizations and flanking genes of the 5 MGIs found in pathogenic Neisseria. MGIs 1, 2 and 3 are found in N. meningitidis and N. gonorrhoeae while MGIs 4 and 5 are only found in N. gonorrhoeae. MGI-1 flanking genes encode an Anhydro-N-acetylmuramic acid kinase (AnmK) on the 5′end of the island and a hypothetical periplasmic protein on the 3′end. MGI-2 flanking genes encode a Proline tRNA on the 5′end of the island and Trk system potassium uptake protein on the 3′end of the island. A cluster of genes, which is only present in some strains, is represented on a grey background. MGI-3 flanking genes encode an uridylate kinase (PyrH) and a truncated IS1016 element. MGI-4 is flanked by two truncated IS1016 elements. MGI-5 flanking genes encode an IS1016 and a ppGpp synthase (RelA). Color code: conserved flanking genes (white), mafA (pink), mafB and mafB-CT cassettes (green), mafI (red), location of sequence encoding BIL domain (blue box), location of sequence encoding DWVKN motif (salmon rose box). The number (1, 2 or 3) inside the 5′ end of mafB genes indicates the corresponding class of MafB (Class 1, 2 or 3 respectively). The dotted outline indicates a mafB-CT gene without initiation codon. C) A schematic representation of the 3 classes of MafB proteins. All MafBs contain a signal peptide (SP, light grey), a N-terminal conserved domain named DUF1020 (white) and a C-terminal variable region (dark grey). Class 1 MafBs contain a VSGDF motif at the end of the N-terminal conserved domain, and between the conserved and variable regions a bacterial intein-like (BIL) domain can be inserted (blue box). Class 2 MafBs contain a VKYDT motif at the end of the N-terminal conserved domain. Class 3 MafBs contain a DWVKN motif (salmon rose box) at the end of the N-terminal conserved domain. Fig. 2. Pairwise comparison of the genetic organization of MGI-1, MGI-2 and MGI-3 loci in 6 representative Neisseria genomes. 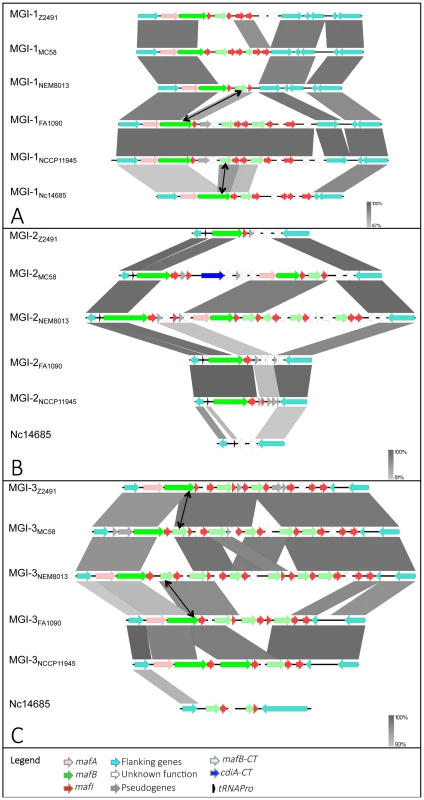
Nucleotide comparison of MGI-1 (A), MGI-2 (B) and MGI-3 (C) from meningococcal strains Z2491, MC58 and NEM8013, gonococcal strains FA1090 and NCCP11945 and N. cinerea strain ATCC 14685. Genome comparisons were generated using BLASTn implemented in Easyfig 2.1 with a cutoff value of 80%. Grey vertical blocks indicate regions of shared similarity shaded according to BLASTn identity. The level of nucleotide identity is shown in the gradient scale for each MGI. Genes are indicated with arrows colored according to their predicted functions with the color code illustrated in the legend. Double-headed arrows connect mafB-CT genes that are identical to the 3′ region of a full-length mafB gene. The orientation of genes is as in the published genomic sequences, except for MGI-1NEM8013, MGI-1MC58, MGI-2NEM8013, MGI-3Z2491 and MGI-3FA1090, which have been reversed for clarity purpose. Genomic regions found in N. cinerea at the location of MGI-2 and MGI-3 are not considered as MGIs because they do not encode a full-length MafB toxin. We propose to give the same name (MGI-1, MGI-2…) to identify different MGIs that are located at the same location across multiple strains. Since each sequenced genome exhibits a unique gene composition of MGIs, the name of the strain is mentioned as a lower index (i.e. MGI-1Z2491). We identified 5 different types of MGIs (MGI-1 to MGI-5) (Fig. 1B).
MGI-1 is located at the 3′ end of anmK (encoding an anhydro-N-acetylmuramic acid kinase) and at the 5′ end of a conserved gene encoding a hypothetical periplasmic protein (Fig. 1B, Fig. 2A). MGI-2 is located at the 3′ end of a tRNA-Pro gene and at the 3′ end of a conserved gene encoding a putative Trk system potassium uptake protein (Fig. 1B, Fig. 2B). MGI-3 is located at the 3′ end of pyrH (encoding an uridylate kinase) and at the 5′ end of a truncated IS1016 element (Fig. 1B, Fig. 2C). MGI-1, MGI-2 and MGI-3 are present in N. meningitidis and in N. gonorrhoeae. N. gonorrhoeae genomes contain two additional MGIs: i) MGI-4 is flanked by two truncated IS1016 elements that are located between a small ORF encoding a putative protein and a nalP pseudogene (Fig. 1B), ii) MGI-5 is located at the 3′ end of an IS1016 and at the 3′ end of relA (encoding a ppGpp synthase) (Fig. 1B).
To designate a gene located in a MGI, we propose to abandon the nomenclature from the initial genome annotation which was solely based on the order of appearance of maf genes in the genome (i.e. in NEM8013: mafB1 = NMV_0410, mafB2 = NMV_1757 and mafB3 = NMV_2312), and to name a gene by its nature mafA, mafB or mafI followed in a lower index by the name of the genomic island where it is found and the name of the strain. For example, NMV_0410 is referred as MafBMGI-1NEM8013 and NMV_2312 is referred as MafBMGI-3NEM8013. Correspondence between locus tags, old and new nomenclature is summarized in S1 Table. If there are several mafB genes in a MGI, a number that refers to the position in the locus is added. This is especially the case for MGI-2 that has two mafB genes, the first mafB gene on the 5′end of the island is numbered 1. For example in MGI-2NEM8013, NMV_1766 is now referred as MafB1MGI-2NEM8013 and NMV_1757 is referred as MafB2MGI-2NEM8013. The small ORF found immediately downstream of a mafB gene is termed mafI, i.e. mafI1MGI-2NEM8013 designates the gene found immediately downstream of mafB1MGI-2NEM8013.
Strikingly, there are few or no MGIs in commensal species, in contrast to strains of pathogenic species where multiple MGIs are present. MGI-1 is found in N. lactamica (strain 020-06), N. cinerea (ATCC 14685) and N. polysaccharea (ATCC 43768). N. cinerea (ATCC 14685) and N. polysaccharea (ATCC 43768) do not contain additional MGI. N. lactamica is the non-pathogenic species that is the most closely related to the two pathogenic Neisseria species. In N. lactamica 020-06 a MGI-1 and a MGI-3 are present. A MGI-2 and a MGI-5 are also present but the corresponding mafB exhibit a frameshift in both of these MGIs. In the available unassembled genomes of commensal species N. subflava (NJ9703), N. mucosa (ATCC 25996 and C102 strains) or N. elongata subsp. glycolytica (ATCC 29315) we did not find a mafA gene or a full-length mafB gene. In N. flavescens (NRL 30031/H210), there is an incomplete MGI-5 with a mafA gene (NEIFLAOT_01129) and genes encoding C-terminal regions of mafB but without a full-length mafB gene.
Classification of MafB proteins based on their N-terminal domain
Sequence alignment of MafB proteins revealed that MafB has a N-terminal conserved domain of unknown function named DUF1020 (or PF06255 in PFAM Database) and a C-terminal (CT) variable region (Fig. 1C). DUF1020 is restricted to Neisseria genus and is preceded by a signal peptide domain (approximately 25 residues long). Amino acids alignment of 150 sequences of MafB proteins (see details in Materials and Methods section) revealed 3 classes of MafB with 3 conserved N-terminal regions (Fig. 1C). The percentage of sequence identity within N-terminal conserved regions of MafB proteins belonging to the same class is over 85% (90%, 97% and 89% for class 1, 2, 3, respectively) whereas the percentage of sequence identity between the conserved regions of different classes is approximately 35%.
Class 1 MafB proteins exhibit a conserved motif (VSGDF) located approximately 300 amino acids from the N-terminus (Fig. 1C and S1 Fig.). This motif is located at the transition between the conserved N-termini and the variable CT sequences. On the other hand, in class 2 and 3 MafB proteins, the conserved motifs found at the transition between the conserved and the variable regions are VKYDT and DWVKN, respectively (Fig. 1C, S2-S3 Fig.).
Some class 1 MafB proteins have a striking feature at the end of their N-terminal constant domain. Indeed, an A-type Bacterial Intein-Like (BIL) domain of approximately 140 amino acids is present at the junction between the conserved N-terminal region and the variable C-terminal region (i.e. in MGI-1FA1090) (Fig. 1C and S1 Fig.). This BIL domain is located immediately after the conserved motif (VSGDF) (S1 Fig.).
All the MafB sequences, that we analyzed using the SignalP 4.1 program [36] showed a signal peptide (SP) recognized by type 1 signal peptidase. For example in NEM8013, MafBMGI-1NEM8013 and MafB2MGI-2NEM8013 harbor a predicted cleavage site between amino acid residues 27 and 28, MafB1MGI-2NEM8013 between amino acid residues 24 and 25 and MafBMGI-3NEM8013 between amino acid residues 26 and 27. Despite the presence of two consecutive arginine residues in the signal-peptide sequence of class 1 MafBs, PRED-TAT [37], TatP 1.0 [38] or TATFIND 1.4 [39] programs did not predict a putative tat (twin arginine translocation) signal. Thus, the translocation of the 3 classes of MafB proteins through the inner membrane is likely to occur via the Sec pathway.
In most cases, the variable domain of MafB shares no homology with protein of known function. Only in few cases a homology with CT extremities of polymorphic Cdi and Rhs toxins family can be noticed. However, in contrast with Cdi and Rhs toxins no repeat domains are found in the N-terminal region of MafB.
Gene content of the different maf genomic islands
In addition to the mafB-mafI module, a mafA gene is frequently found immediately upstream of genes encoding class 1 and class 3 MafB but not upstream of genes encoding class 2 MafB (Fig. 1A, 1B). A BLASTp search using MafA sequence as a query evidenced that mafA is also specific of the Neisseria genus. The LipoP 1.0 server [40] predicts that MafA exhibits a lipoprotein signal peptide without an aspartic acid in position +2 after the cleavage site and thus could be a lipoprotein attached to the outer membrane. It has been shown by immunoelectronic microscopy [35] that MafA of gonococcal strain MS11 is surface exposed and is able to bind to glycolipids [35]. The location of MafA in the outer membrane and in outer membrane vesicles (OMV) has been confirmed by several proteomic studies [41]–[43].
Genes encoding class 1 MafB are present in MGI-1, MGI-5 and some MGI-2, whereas genes encoding class 3 MafB are present in MGI-3 and MGI-4 (Fig. 1B). Genes encoding class 2 MafB are only found in MGI-2 (Fig. 1B). Class 1 MafB encoded in the MGI-2NEM8013 (MafB2MGI-2NEM8013) and MGI-2MC58 (MafB2MGI-2MC58) differs from the class 1 MafB encoded in the MGI-1NEM8013 and MGI-1MC58 by a deletion of 50 amino acids in their N-terminal region.
Examination of maf clusters downstream of the full-length mafB gene shows many genes encoding alternative MafB-C-terminal cassettes. All these mafB-CT genes are followed by at least one mafI gene (Fig. 1A, 1B). There are two types of mafB-CT genes: i) mafB-CTs starting with an initiation codon, which are potentially translated, and ii) mafB-CTs devoid of initiation codon (ATG, GTG, TTG, ATT or CTG), which are potentially silent cassettes.
Intriguingly, potentially expressed cassettes are only found in MGIs encoding class 1 MafB whereas silent cassettes are only found in MGIs encoding class 3 MafB (Fig. 1B). In MGIs encoding class 1 MafB, the mafB-CTs that have an initiation codon encode an A-type BIL (Fig. 1B) and have no homologous region with the conserved region of the full-length class 1 MafB (Fig. 1B) that could allow a recombination. The initiation codon for the translation of the BILs is a TTG and is the initiation codon of the mafB-CT. It should be pointed out that BILs of the MafB-CTs lack the first 7 amino acids compared to complete BIL sequence. On the other hand, the full-length mafB genes located upstream of these mafB-CTs may contain a complete BIL sequence (i.e. in MGI-1FAM18).
In the MGI containing class 3 MafB, the mafB-CTs are gene fragments that usually encode a conserved WDWVKN motif present in the full-length class 3 MafB, but as mentioned above these mafB-CTs do not have an initiation codon (Fig. 1B).
The maf clusters harbor numerous small ORFs, designated mafI, immediately downstream of mafB. Similarly to the high variability of MafB-CT, MafI sequences are also highly variable, suggesting that the encoded proteins could specifically interact with cognate MafB-CTs. In support of this hypothesis, when two mafB-CTs are almost identical their associated mafI gene are also identical. For instance, the C-terminal region of MafBMGI-3Z2491 (NMA0324) and a cassette (NMB2107) found in MGI-3 of MC58 exhibit 100% amino acid identity (Fig. 2C). The corresponding immunity proteins MafIMGI-3Z2491 (NMA0323) and NMB2108 are also 100% identical (Fig. 2C). Similarly, MafBMGI-3FA1090 (NGO1971) and a cassette (NMV_2314) found in MGI-3 of NEM8013 exhibit 90% amino acid identity (Fig. 2C). The corresponding immunity proteins MafIMGI-3FA1090 (NGO1970) and NMV_2315 are 100% identical (Fig. 2C).
A search in the Conserved Domain Database (CDD) [44] of the NCBI revealed that many mafI genes encode proteins containing domains typically found in putative immunity proteins of bacterial polymorphic toxin systems [1], [2]. These recently described domains include Imm17, Imm21, Imm22, Imm47, SUFU and SMI1 domains. However, almost half of the small ORFs in MGIs do not contain any known domains. Small ORFs are considered immunity genes if they are located immediately downstream of a mafB-CT gene or if the corresponding amino acids sequence contain a predicted immunity domain in CD search database.
mafB-mafI modules encode a new family of toxin-immunity
Analysis of the amino acid sequences of several MafB-CT regions using the CDD server revealed homologies with putative or known toxic domains. For instance, proteins encoded by mafBMGI-1NEM8013 (NMV_0410) contains a domain belonging to the RNase EndoU-fold, mafBMGI-1FA19 (NGEG_01276) contains a domain belonging to a nucleotide deaminase superfamily and mafBMGI-5FA1090 (NGO1392) contains a domain belonging to the DNase HNH/EndoVII-fold. In this study, we decided to focus on the four putative MafB toxins encoded in meningococcal strain NEM8013 which are MafBMGI-1NEM8013, MafB2MGI-2NEM8013, MafBMGI-3NEM8013 and MafB1MGI-2NEM8013 (formerly MafB1, MafB2, MafB3 and MafB-related respectively).
MafBMGI-1NEM8013 and MafB MGI-3NEM8013 proteins of strain NEM8013 inhibit cell growth when expressed in E. coli and expression of cognate MafI counteracts MafB toxicity
To assess the putative function of mafB-mafI modules, we expressed the four predicted MafB toxins of strain NEM8013 in E. coli. We individually cloned the four mafB genes of strain NEM8013 into tightly controllable expression vector pBAD33 [45]. Induction of the expression of mafBMGI-1NEM8013 and mafBMGI-3NEM8013 was highly toxic for E. coli both on agar Luria-Bertani (LB) plates and in liquid LB culture (Fig. 3). Induction of the expression of mafBMGI-1NEM8013 and mafBMGI-3NEM8013 resulted in growth inhibition. On the other hand, induction of mafB1MGI-2NEM8013 and mafB2MGI-2NEM8013 genes did not alter growth of E. coli (Fig. 3A). Because some toxins targets are only present in the periplasmic space, we cloned mafB1MGI-2NEM8013 and mafB2MGI-2NEM8013 devoid of their own signal sequence in pET-22 and added pelB-signal sequence at the N-termini of both proteins to direct them to E. coli periplasm. MafB1MGI-2NEM8013 and MafB2MGI-2NEM8013 remained non-toxic even when exported to E. coli periplasm (S4A Fig.). These results demonstrate that a toxic activity could be detected only for MafBMGI-1NEM8013 and MafBMGI-3NEM8013.
Fig. 3. Toxic effect of MafB proteins of NEM8013 strain on E. coli growth. 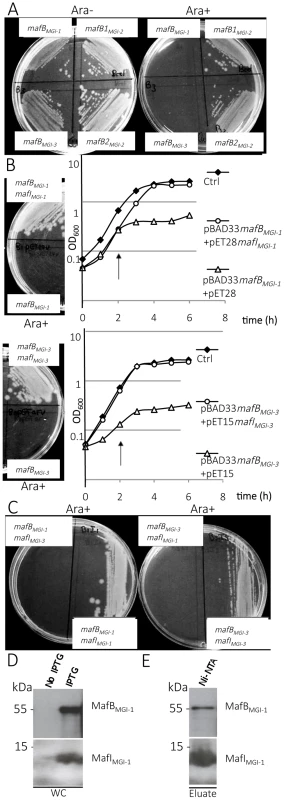
A) Effect of NEM8013 MafB putative toxins over-expression on E. coli grown in the presence of 0.2% L-arabinose (Ara+) or without arabinose (Ara−). BL21(DE3) cells were transformed with vector pBAD33 carrying mafB genes found in NEM8013 strain (mafBMGI-1NEM8013, mafB1MGI-2NEM8013, mafB2MGI-2NEM8013 and mafBMGI-3NEM8013). B) Inhibition of growth due to the toxin is counteracted by cognate immunity protein co-expression on LB agar plates (0.2% L-arabinose) and in LB broth. LB agar plates and LB broth contain 0.01 mM IPTG to induce expression of mafI. Toxin expression was induced by adding 0.2% L-arabinose in LB broth 2 h after inoculation (arrow). mafBMGI-1NEM8013 and mafBMGI-3NEM8013 are cloned in pBAD33, mafIMGI-1NEM8013 is cloned in pET28 and mafIMGI-3NEM8013 is cloned in pET15. Control strains (Ctrl) contain empty vectors. C) Co-expression of a non-cognate immunity protein does not confer protection against MafB toxicity. LB agar plates contain 0.01 mM IPTG and 0.2% L-arabinose. The results shown are from one of three independent experiments. D) MafBMGI-1NEM8013 and MafIMGI-1NEM8013 copurify. mafB and mafI from MGI-1NEM8013 were cloned under the control of two independent T7 promoters in plasmid pcolaDUET. Upon induction with IPTG, E. coli BL21(DE3) carrying pcolaDuet-mafBmafI expressed His6- MafIMGI-1NEM8013 and MafBMGI-1NEM8013, as evidenced by immunoblotting of whole-cell lysates (WC). E) His6-tagged MafIMGI-1NEM8013 and MafBMGI-1NEM8013 were recovered from Ni-NTA-agarose column eluate (Eluate). Copurified proteins were analysed by immunoblotting. Antibodies used for immunoblotting of WC and Ni-NTA eluate were Anti- MafBMGI-1NEM8013 and Anti- MafIMGI-1NEM8013. Ni-NTA, Nickel-nitrilotriacetic acid. In order to test whether the toxicity of MafB resides in its C-terminal domain, we cloned only the DNA region encoding the C-terminal domain or the-N terminal domain of MafBMGI-1NEM8013 in E. coli using pBAD33. As expected, expression of the C-terminal domain of MafBMGI-1NEM8013 was toxic for E. coli but expression of the N-terminal domain had no effect on E. coli growth (S4B–S4C Fig.).
To verify if the small ORF immediately downstream of the mafB gene was indeed an immunity gene, we cloned mafIMGI-1NEM8013 and mafIMGI-3NEM8013 under the control of the IPTG inducible promoter of pET-28 and pET-15 respectively. While the expression of MafBMGI-1NEM8013 or MafBMGI-3NEM8013 inhibited growth of E. coli, the co-expression of MafBMGI-1NEM8013 and MafIMGI-1NEM8013 or the co-expression of MafBMGI-3NEM8013 and MafIMGI-3NEM8013 did not impede growth of E. coli (Fig. 3B). There was no cross-protection conferred by MafIMGI-1NEM8013 against MafBMGI-3NEM8013 or by MafIMGI-1NEM8013 against MafBMGI-3NEM8013 (Fig. 3C). Thus, the small ORF adjacent to the toxin mafB gene encodes a protein providing immunity to the cognate toxin and preventing self-intoxication.
To explore the mechanism of toxin inactivation we investigated a potential direct toxin-antitoxin binding. For this, we cloned in the same vector (pcolaDUET) under two different IPTG promoters, mafBMGI-1NEM8013 and mafIMGI-1NEM8013 in order to co-express in E. coli both proteins. Only MafIMGI-1NEM8013 harbored a His6-tag. We tested whether MafBMGI-1NEM8013 co-purified with His6-tagged MafIMGI-1NEM8013 using Ni2+-affinity chromatography. Despite the very low expression of the MafBMGI-1NEM8013 toxin in E. coli, we could co-purify MafBMGI-1NEM8013 with His6-tagged MafIMGI-1NEM8013 (Fig. 3D–E). Thus MafIMGI-1NEM8013 is likely to inhibit MafBMGI-1NEM8013 toxicity by a direct interaction.
MafBMGI-1NEM8013, MafBMGI-3NEM8013 and MafB1MGI-2NEM8013 proteins are toxic in their original NEM8013 Neisseria strain
In a first attempt to assess the toxicity of MafB proteins in meningococcus, we tried to generate mutants of the immunity gene located downstream of each of the four mafB genes in NEM8013. It was impossible to delete the immunity gene of mafBMGI-1NEM8013, mafB1MGI-2NEM8013 and mafBMGI-3NEM8013 (respectively mafIMGI-1NEM8013, mafI1MGI-2NEM8013 and mafIMGI-3NEM8013). Every attempt of mutagenesis led to duplication of the gene. On the other hand, the replacement of mafI2MGI-2NEM8013 immunity gene by a chloramphenicol resistance cassette was possible. We next tried to insert an ectopic copy of the four mafB genes in the NEM8013 genome using pGCC4 constructs (see Materials and Methods section) [46]. Similar results as above were obtained and no strain carrying an additional copy of mafBMGI-1NEM8013, mafB1MGI-2NEM8013 and mafBMGI-3NEM8013 could be obtained. On the other hand, it was possible to insert an additional copy of mafB2MGI-2NEM8013 in the wild-type (WT) strain and in a mafI2MGI-2NEM8013 deleted mutant. On the other hand, it was possible to insert an additional ectopic copy of the four whole modules (mafB with mafI) using pGCC4 constructs. These results evidenced that all MafB proteins of strain 8013 except MafB2MGI-2NEM8013 have a toxic effect in NEM8013, unlike what was observed in E. coli where MafB1MGI-2NEM8013 and MafB2MGI-2NEM8013 were non-toxic.
Overexpression of the toxin MafB1MGI-2NEM8013 gives a competitive advantage
Since Cdi and Rhs toxins have been involved in inter-bacterial competition, we searched for a role of MafB in competition assays. We used strain NEM8013 and tested the impact of the overexpression of the four MafB proteins in competition assays. Constructions overexpressing each of the four MafB toxins with their cognate MafI proteins, in strain NEM8013, were used as inhibitor cells. As it has been shown previously that expression of a capsule could block CdiA mediated toxicity [47], we used an unencapsulated derivative of meningococcal strain NEM8013 as target cell. Competition assays were performed overnight on a solid media at an inhibitor to target ratio of 10∶1 (see Materials and Methods). Under these experimental conditions, we evidenced a significant competitive advantage only for the strain overexpressing MafB1MGI-2NEM8013 (Fig. 4). The advantage was lost when target cells overexpressed the cognate immunity MafI1MGI-2NEM8013 (Fig. 4). These data suggest that MafB1MGI-2NEM8013 could be employed by strain NEM8013 to outcompete strains that do not possess or express at a sufficient level the cognate immunity. Besides, the fact that the introduction of an additional copy of mafB1MGI-2NEM8013 in wild-type NEM8013 is impossible without the simultaneous introduction of an additional copy of the cognate immunity gene, suggests that the toxicity resulting of an overexpression of the toxin cannot be counteracted by the basal expression of the cognate immunity. Altogether these data demonstrate that MafB1MGI-2NEM8013 has a toxic effect on a non-capsulated derivative of strain NEM8013.
Fig. 4. MafB1MGI-2NEM8013 provides an advantage in competition assay. 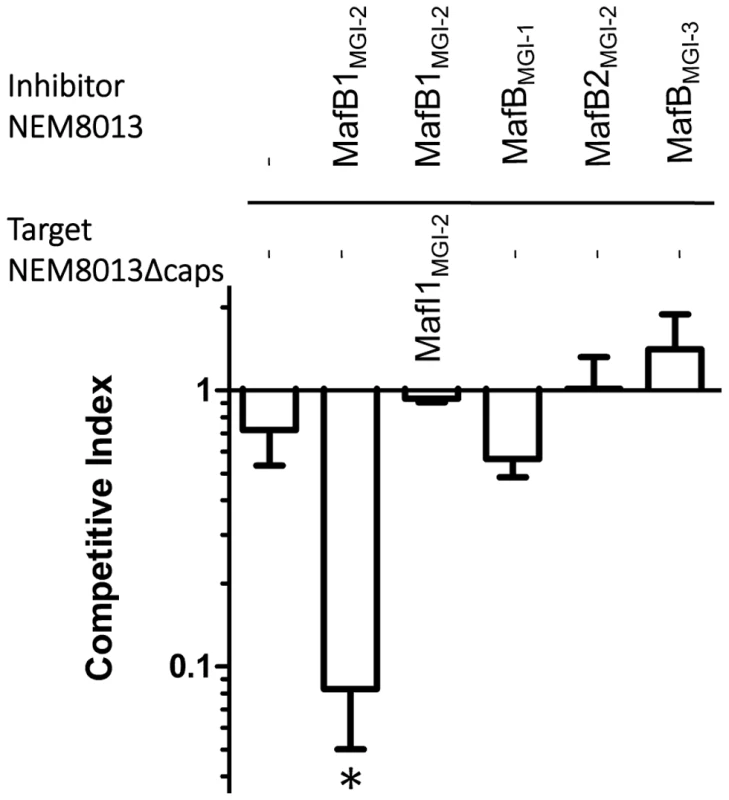
Competition assays were performed with an initial inhibitor to target cell ratio of 10 to 1. Putative inhibitory cells were NEM8013 overexpressing each of the four mafB toxins and their cognate mafI immunity genes using pGCC4 constructs. Putative target cells were an unencapsulated derivative of NEM8013 with a transposon insertion in ctrA gene (NEM8013Δcaps) or this unencapsulated derivative overexpressing MafI1MGI-2NEM8013. The overexpressed toxin of the inhibitor cells is indicated (otherwise a – indicates that there is no overexpressed toxin) and the overexpressed immunity of the target cells is indicated (otherwise a – indicates that there is no overexpressed immunity protein). Mixed cultures were spotted on a membrane filter placed on GCB agar plate containing 1 mM IPTG and incubated overnight. Filters recovered after overnight incubation were used to perform viable counts and the competitive index was calculated as the inhibitor/target ratio in the output divided by the initial inhibitor/target ratio. The data from three independent experiments were examined for significance using a two-tailed Student's t-test. * p-value p<0.05. MafB toxins are secreted in a signal-peptide dependent manner
We focused on MafBMGI-1NEM8013 (Class 1 MafB) and MafBMGI-3NEM8013 (Class 3 MafB) of strain NEM8013 and we engineered NEM8013 strains chromosomally expressing a FLAG-tagged version of these proteins (Fig. 5A). Using Western blotting, we were able to detect MafBMGI-1NEM8013 (Fig. 5B) in the culture supernatant. MafBMGI-1NEM8013 was no longer detected in the supernatant of a strain expressing MafBMGI-1NEM8013 devoid of its signal peptide (Fig. 5B). Similar results were obtained after overexpression of the wild type sequence of MafBMGI-1NEM8013 instead of the FLAG-tagged version using anti MafBMGI-1NEM8013 antibodies (S5 Fig.). Thus, the first step of MafB secretion across the inner membrane involves signal peptide recognized by the general secretory pathway. To gain access to the culture medium MafB has to cross the outer membrane, potentially through an outer membrane secretin. As PilQ is the only characterized secretin in Neisseria sp, we overexpressed MafBMGI-1NEM8013 construction in a pilQ mutant of strain NEM8013. The lack of PilQ secretin had no impact on MafBMGI-1NEM8013 level in the culture supernatant (S5 Fig.). We also aimed at studying the impact of MafA on MafB secretion. We used N. cinerea ATCC 14685 that possesses only one copy of mafA. Deletion of the unique copy of mafA in N. cinerea did not prevent secretion of MafB-FLAG when the strain was transformed with pGCC4-mafBMGI-1NEM8013FLAG (S6A–S6B Fig.). Besides, we could not evidence the presence of MafBMGI-1NEM8013 in E. coli supernatant when the protein was produced from a pET vector (S6C Fig.). Taken together, these data suggest that secretion of MafB requires a Neisseria-specific factor that is neither PilQ nor MafA.
Fig. 5. MafBMGI-1NEM8013 is a secreted toxin. 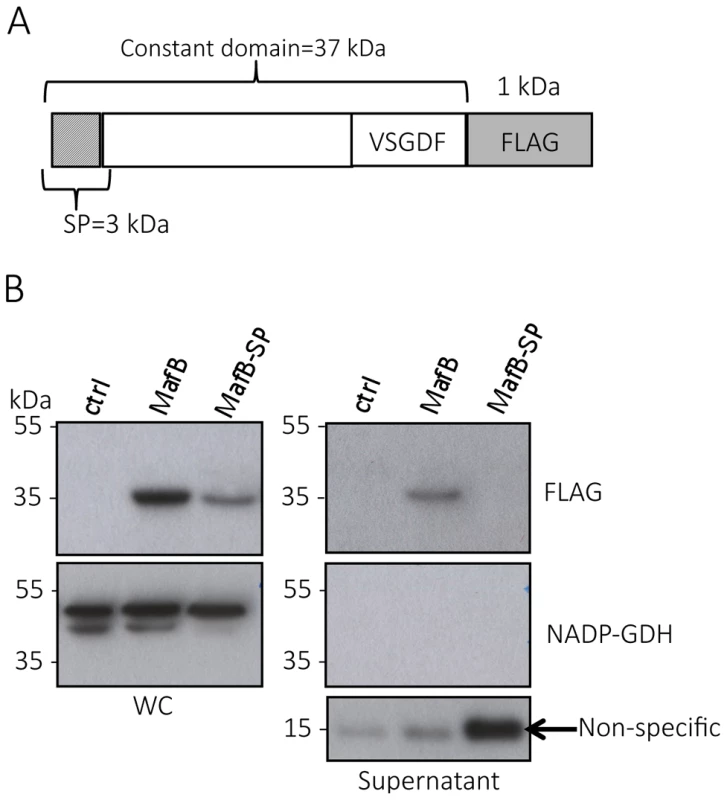
A) Schematic representation of the construct expressed under IPTG inducible promoter in NEM8013. This construction has been inserted in an intergenic region of NEM8013 chromosome using pGCC4 vector. B) C-terminal FLAG-tagged MafBMGI-1NEM8013 (MafB) with or without its signal sequence (MafB-SP) are detected in the whole-cell lysates (WC) of NEM8013 strain expressing MafB or MafB-SP under an IPTG inducible promoter. NEM8013 parental strain is used as a control (ctrl). MafB is detected in the supernatant only when its signal sequence is present. As the production of MafB-SP was less efficient than the production of MafB, a larger quantity of supernatant was loaded as shown by the intensity of the nonspecific band (arrow) detected with Anti-FLAG antibody. Antibodies used for immunoblotting of WC and supernatants were Anti-NADP-GDH (NADP-dependent glutamate dehydrogenase, as a cytoplasmic marker protein) and Anti-FLAG to detect C-terminal, FLAG-tagged MafB. MafBMGI-1NEM8013 is a bacterial EndoU nuclease that can degrade multiple sources of RNA
Previous bioinformatics studies conducted by Zhang et coll. (in Supplementary material of [2]) suggested that MafB2MGI-2Nm053442 (encoded by gene NMCC_0602 GI: 161869586) could contain a nucleasic toxin domain of the EndoU fold. Amino acid sequences comparison between MafBMGI-1NEM8013 and MafB2MGI-2Nm053442 showed 95% identity over the entire length and 100% identity over the CT region. A search in the GenBank database with the BLASTp program using the CT domain of MafBMGI-1NEM8013 protein as a query confirmed the presence of a putative EndoU catalytic domain with two conserved histidine residues (Fig. 6A).
Fig. 6. MafBMGI-1NEM8013 is a bacterial EndoU nuclease. 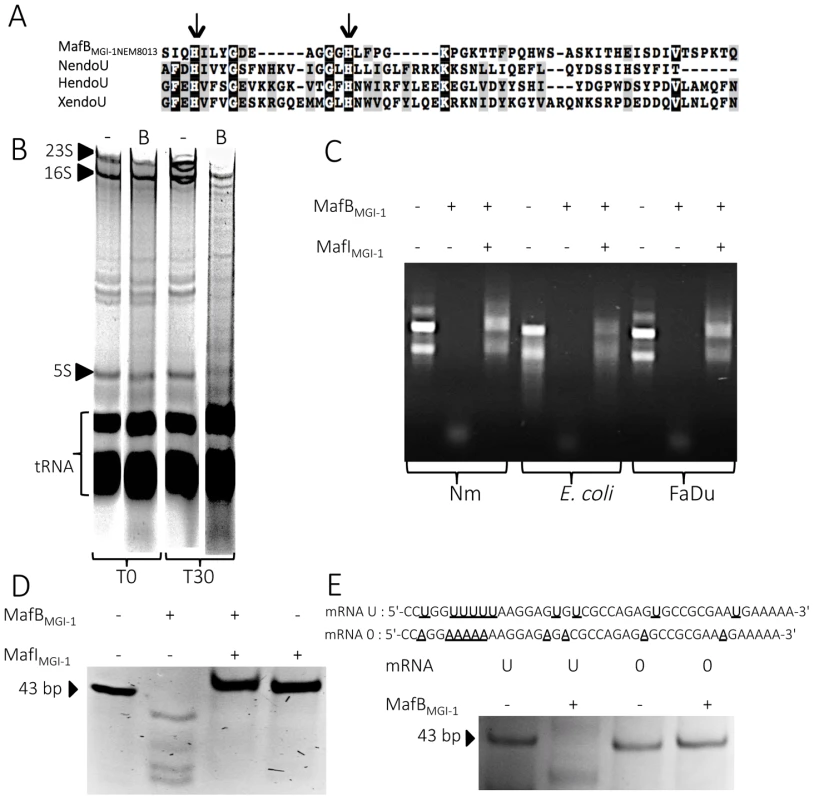
A) Partial sequence alignment of EndoU nuclease domain of Nidovirus Nsp15 protein (NendoU; NP_740619), Xenopus laevis XendoU (Q8JFY9), human placental protein PP11 (HendoU P21128) and MafBMGI-1 from NEM8013 strain (C9X2Z7). The arrows indicate two conserved histidine residues, which are part of the catalytic site of previously characterized EndoU nucleases. Multiple alignment was performed using MUSCLE and shaded using the BoxShade server. Residues that are identical or similar in at least three of the four sequences are shaded with black or grey background respectively. B) Analysis of in vivo impact of MafBMGI-1NEM8013 expression in E. coli. Total RNA from E. coli expressing MafBMGI-1NEM8013 (pBAD33-mafBMGI-1NEM8013) was isolated before induction (T0) and 30 min after addition of L-arabinose (T30). Samples were run on 5% denaturing polyacrylamide gels and stained with ethidium bromide. Positions of 23S, 16S, and 5S rRNAs and tRNAs are shown. -, empty vector control; B, E. coli expressing MafBMGI-1NEM8013 from pBAD33 C) RNase activity of purified recombinant MafBMGI-1NEM8013 was assessed by incubating MafBMGI-1NEM8013 -His6 alone or with MafIMGI-1NEM8013 -His6 with total RNA isolated from different sources (N. meningitidis NEM8013, E. coli TOP10 and human epithelial cells FaDu). Each reaction was performed for 30 min at 37°C with 4 µg of RNA in Tris-EDTA buffer. D) Synthetic mRNA of 43 bp was incubated with purified MafBMGI-1NEM8013-His6 alone or with MafIMGI-1NEM8013 -His6 in Tris-EDTA buffer for 15 min at 37°C. The cleavage products were separated by electrophoresis in a 14% polyacrylamide/8M urea gel and were visualized by ethidium bromide staining. E) Synthetic oligoribonucleotides containing several uridylates (U) or none (0) were incubated with purified MafBMGI-1NEM8013-His6 in Tris-EDTA buffer for 20 min at 37°C. The reaction products were analyzed by 14% polyacrylamide/8 M urea gel. Sequences of synthetic oligonucleotides used in this experiment are shown. First, we examined total RNA from E. coli cells expressing MafBMGI-1NEM8013 for evidence of RNA degradation. Gel analysis of total RNA isolated from cells before and after induction of MafBMGI-1NEM8013 production revealed that expression of MafBMGI-1NEM8013 led to the non-specific cleavage of most cellular RNAs (Fig. 6B). To further characterize MafBMGI-1NEM8013 ribonuclease activity, we purified a recombinant His6-tagged - MafBMGI-1NEM8013 protein and incubated it with various nucleic acids substrates. Recombinant MafBMGI-1NEM8013 was able to degrade purified total RNA from E. coli, N. meningitidis and human epithelial cells (Fig. 6C). Thus, MafBMGI-1NEM8013 is a ribonuclease. Incubation of MafBMGI-1NEM8013 with a synthetic RNA confirmed the RNAse activity (Fig. 6D). Addition of MafIMGI-1NEM8013 blocked the toxic activity of MafBMGI-1NEM8013 towards RNA (Fig. 6D). In order to determine whether MafBMGI-1NEM8013 ribonuclease had an uridylate specific activity, characteristic of the EndoU fold, we obtained a similar synthetic RNA where all the Us were replaced by As. This synthetic RNA was no longer cleaved even when incubation was extended up to 1 hour (Fig. 6E). Our results evidenced that MafBMGI-1NEM8013 preferred cleavage sites contain uridylates. Thus, with MafBMGI-1NEM8013, we characterized the first bacterial EndoU ribonuclease.
Discussion
In this study, we have demonstrated that mafB encodes a functional polymorphic toxin and mafI, the downstream gene, a specific immunity protein. Focusing on MafBMGI-1NEM8013, we were able to characterize its EndoU ribonuclease activity. Toxins carrying EndoU activities are predicted to be widespread among diverse polymorphic toxin systems, however no EndoU activity carrying toxins had been previously experimentally confirmed in bacteria.
Polymorphic toxins encompass numerous families distributed in all bacterial lineages. Nevertheless, very few of these systems have been experimentally characterized except the Cdi and the Rhs families in Gram-negative bacteria [6], [8] and the PF04740 family from Gram-positive bacteria [48]. CdiA and Rhs toxins are the best characterized and can be found in many species including E. coli, Yersinia pestis, Dickeya dadantii or Burkholderia pseudomallei [8]. CdiA and Rhs toxins are large filamentous proteins (over 1000 amino acids) with multiple repetitive elements. In contrast, MafB toxins are predicted to have a globular structure. Organization of the loci encoding CdiA and Rhs toxins shares similarities with MGIs organization. In particular, the presence of genes encoding CT cassettes/immunity modules downstream of the toxin gene is a common feature [8]. Interestingly, mafB genes are restricted to the genus Neisseria, which is very unusual in polymorphic toxin systems [1]. A mafA gene, encoding a surface exposed outer membrane lipoprotein, is frequently found immediately upstream of mafB. mafA is also specific of the Neisseria genus. It has been shown that gonococcal MafA binds to glycolipids but its biological function remains elusive [35]. Unlike CDI two-partner secretion system in which CdiB mediates the secretion of CdiA [49], the mafA gene located 5′ of mafB shares no homology with known secretion systems. Furthermore, the sequence of MafA offers no other clues to its function.
As observed for maf genes, there are several loci bearing haemagglutinin related genes in meningococcal genomes. Haemagglutinin related genes are termed tpsA and tpsB genes. tpsA encodes a secreted filamentous haemagglutinin and tpsB encodes its dedicated transporter. Of note, haemagglutinin related genes are only present as pseudogenes in gonococcal genomes [50]. Analysis of the sequences of the TpsA proteins encoded in N. meningitidis genomes revealed the presence of three distinct groups [51]. Group 1 is found in all meningococcal genomes whereas tpsA genes of group 2 and 3 are overrepresented in disease isolates compared to carriage isolates [51]. In meningococcus, the number of tpsA genes range from 1 to 5 [52]. Genomes of strains FAM18, B16B6 and Z2491 contain only one tpsA gene of group 1 system while strain MC58 contains five tpsA genes (NMB0493, NMB0497, NMB1214, NMB1768 and NMB1779) belonging to the three different groups [51].
The tps locus of meningococcal strain FAM18 is referred as a cdi locus in the comparative genomic study conducted by Poole et al. in 2011 [4]. Indeed, the FAM18 tpsA gene (NMC0444 also termed cdiA by Poole et al.) encodes a protein that exhibits a filamentous haemagglutinin family N-terminal domain and a Pre-toxin domain with a VENN motif, which is located before the putative C-terminal toxin domain. Since this study, several recent works have been published on the meningococcal tps loci. It has been shown that meningococcal strain B16B6 had an advantage in competition assay against a B16B6 deletion mutant lacking the cognate immunity gene of tpsA [14]. The tpsA gene of strain B16B6 (HQ420265) encodes a protein with >99% sequence identity with that of FAM18 [14] confirming the prediction of Poole et al. [4] that meningococcal TpsA proteins from group 1 constitute functional CDI systems. It remains to be investigated whether TpsA proteins from group 2 and 3 are also able to mediate growth inhibition.
The six secretion pathways identified in Gram-negative bacteria can be classified in two categories, either the pathways transporting proteins in a single-step across both inner and outer membranes (i.e. type I, III, IV and VI) or the two-step secretion pathways (i.e. type II and V), where proteins are first targeted to a machinery that recognizes their N-terminal signal peptide [53]–[55]. Type V secretion systems encompass auto-transporter proteins (type Va) and two-partner secretion systems (type Vb) [54]. Cdi toxins belong to the type Vb subclass. Only type I, Va and Vb secretion pathways are found in N. meningitidis, whereas only type IV and Va secretion pathways are found in N. gonorrhoeae [56]. Thus, the only common secretion pathway shared by pathogenic Neisseria species is autotransporters pathway. Autotransporters are single polypeptides that consist of a surface-exposed variable N-terminal domain (“passenger domain”) that can be released from the cell surface by a proteolytic cleavage and a C-terminal domain (“translocator domain”) folded into a β-barrel structure in the outer membrane. The β-barrel of translocator domains is in most cases composed of 14 β-strands [57]. The IgA protease of N. gonorrhoeae was the first described autotransporter [58]. Since MafB toxins possess an N-terminal signal peptide and are found in N. meningitidis and N. gonorrhoeae, they are likely to be secreted by a two-step secretion pathway found in both species and not yet identified. Indeed, according to their domains organization and the lack of β-barrel structure prediction (using TBBpred server [59] and HMM-TM server [60]), MafB toxins do not belong to the autotransporters family. Besides, secreted proteins known to be targeted to a TpsB transporter of two-partners systems exhibit a TPS secretion domain adjacent to the signal peptide sequence [61]. There is no such TPS domain in the N-terminal region of MafBs that could target the toxin to a TpsB transporter. Thus, in contrast to Cdi toxins, MafB toxins are unlikely to be secreted via type V secretion systems. Moreover, in contrast to Rhs toxins that can be secreted through type VI secretion system, there is no type VI secretion system in pathogenic Neisseria species.
In addition to these six types of secretion systems, Gram-negative bacteria, including Neisseria pathogenic species, constitutively produce outer membrane vesicles (OMV) during their normal growth [62], [63]. OMVs are mainly composed of outer membrane and periplasmic components. OMVs enable the secretion of virulence factors to the surrounding environment or directly to neighboring bacteria [64], [65] or to eukaryotic cells [66], [67]. For instance, OMVs derived from P. aeruginosa is able to kill other bacterial species [64] by the release of murein hydrolases capable of degrading the peptidoglycan of other species. P. aeruginosa is also able to deliver multiple virulence factors directly into the host cell cytoplasm by fusion of OMV with host cell membrane lipid raft [67]. Thus, OMVs can interact both with competing bacteria and with host cell to promote bacterial colonization of the host or pathogenesis. Pathogenic Neisseria are well known for their release of OMVs [56], [62]. Natural and engineered OMVs have recently gained interest for use as vaccine or adjuvants. The OMV vaccine strategy has been successfully used during several clonal outbreaks of serogroup B meningococcal strains in Cuba, Norway, and New Zealand [68]. As a consequence, several recent proteomic studies analyzed the Neisseria OMVs content. A proteomic study of naturally released OMVs isolated from four gonococcal strains (FA1090, F62, MS11, and 1291) revealed the presence of MafA and MafBMGI-2 (corresponding to ORFs NGO0225, NGNG_00563, NGFG_00362 and NGAG_00430) in OMVs of 4 strains [43]. In addition, supplemental data published by Zielke et al. in the same proteomic study suggest that four other MafB toxins produced by these gonococcal strains could also be present in OMVs. Since MafA has been previously described as an adhesin able to bind cellular glycolipids [35], the presence of MafA in OMVs could mediate attachment of OMVs to eukaryotic cells. This suggests that OMV-mediated release could be a mean for delivery of MafB toxins to neighboring bacteria or to eukaryotic cells. The presence of MafB toxins in OMVs and its potential implications for Neisseria pathogenesis have to be further explored.
A hallmark of MGIs is the presence of numerous mafB-CT genes. Genetic recombination, resulting from the replacement of the 5′ end of a full-length mafB gene by an alternative CT cassette is supported by genome comparison both for MGI encoding class 1 or class 3 MafBs. For example in MGI-1s, the mafB-CT cassette found in MGI-1NEM8013 (NVM_0408) encodes the same CT region that mafBMGI-1FA1090 (Fig. 2A). In MGI-3s, the mafB-CT cassette found in MGI-3NEM8013 (NVM_2314) encodes the same CT region that mafBMGI-3FA1090 (Fig. 2C). Genetic recombination of polymorphic toxins has been recently demonstrated for CdiA in N. meningitidis [14] and for Rhs in Salmonella enterica serovar Typhimurium [10].
In this study, we demonstrated a functional role for one of the four MafB toxins of NEM8013. Indeed, we showed that an unencapsulated derivative of NEM8013 was outcompeted by a strain overexpressing MafB1MGI-2NEM8013. Several studies have evidenced that meningococci frequently become acapsulated in the nasopharynx as a result of phase variation [69], [70] or by down-regulation of the genes involved in capsule biosynthesis [71], [72]. Thus, both capsulated and unencapsulated strains are likely to compete with each other for colonization of the nasopharyngeal niche. The function of the three other mafB genes of this strain remains unknown. Given the diversity of the CT extremities of MafBs, it is also plausible that they exhibit various biological targets such as other bacterial species or eukaryotic cells. Thus, biological function of maf genes remains a challenging question. Since these genes represent 2% of the genome of pathogenic Neisseria, but are virtually absent from non-pathogenic species, it is likely that they play important biological roles, including in pathogenesis. Regulation of the expression of maf genes could give clues on their physiological role. It has been recently demonstrated by Fagnocchi et colleagues that NadR is a regulator of maf operons [73]. The NadR regulon in meningococcal MC58 strain comprises nadA (encoding an adhesin), and the operons NMB0375-374 (encoding MafA and MafB in MGI-1MC58) and NMB0652-654 (encoding MafA, MafB and MafI in MGI-2MC58). In the presence of human saliva (or the small metabolite 4HPA that is secreted in human saliva) Fagnocchi et colleagues showed that maf genes are repressed, while nadA is induced in a NadR-dependent manner. This coordinate regulation indicates an adaptation of the bacteria in response to the signal molecules present in saliva, and suggests a role in colonization of the port-of-entry.
It has been suggested that, in addition to growth-inhibiting function, Rhs and Cdi might have a broader role in interbacterial communication. CdiA - or Rhs-CTs could serve as signal molecules when translocated in neighboring cells protected by the cognate immunity protein, in a manner similar to quorum sensing. Furthermore, recent studies showed that CDI plays a role in biofilm formation in Burkholderia thailandensis [13], [74], [75]. Indeed, CDI systems might be a mechanism allowing bacteria to discern kin versus non-kin within a complex population (e.g. in a polymicrobial biofilm). Only the cells expressing the same set of toxins and of immunity proteins will be able to live in close proximity. However, it is difficult to predict the evolution of a complex community since a strain may contain numerous toxin systems (such as Maf, Rhs, Cdi, bacteriocins or recently described type VI secretion systems effectors) that can all play a role in interbacterial competition. Deciphering interactions between these systems is a challenging question for future studies.
Materials and Methods
Bioinformatic analysis
We used 6 fully sequenced and annotated genomes of N. meningitidis (Z2491, MC58, FAM18 and NEM8013) and N. gonorrhoeae (FA1090 and NCCP11945) present in the MicroScope database with curated annotations from the NeMeSys project [19], [76]–[80]. We also used N. gonorrhoeae MS11 genome sequence available from the Broad Institute, N. cinerea ATCC 14685 genome sequenced by Washington University and the following meningococcal strains: H44/76 [81], M04-240196 [82], M01-240355 [82], G2136 [82], M6190 [82], NZ-05/33 [82], WUE2594 [83] and 053442 [20]. We used BLASTp with default parameters to search for orthologs of the Maf proteins in other Neisserial strains present in the NCBI non-redundant protein database.
To identify the different classes of MafB proteins, we downloaded 150 sequences stored on the PFAM database server [84] that have been used to build the DUF1020 (PF06255) family. The alignment was performed with ClustalW (http://www.genome.jp/tools/clustalw/) [85], [86] using default parameters and a rooted phylogenic tree (UPGMA) was generated.
Multiple alignments were also performed using MUSCLE [87] or Clustal Omega [88] with default parameters and shaded using the BoxShade server (http://www.ch.embnet.org/software/BOX_form.html). The LipoP 1.0 server [40] and SignalP server 4.1 [36] was used to predict the presence of a signal sequence with default options for Gram-negative bacteria. PRED-TAT [37], TatP 1.0 [38] or TATFIND 1.4 [39] servers were used to identify putative tat (twin arginine translocation) signal. β-barrel structure prediction were performed with TBBpred server [59] and HMM-TM server [60] and conserved domains were identified with the Conserved Domain Database (CDD) [44] of the NCBI server. Pairwise genome comparisons were visualized using Easyfig [89].
Accession numbers of proteins mentioned in the text with corresponding locus tags are listed in the supplementary information (S1 Table).
Bacterial strains and growth conditions
All strains used in this study can be found in S2 Table.
Meningococci NEM8013 and N. cinerea were grown at 37°C in a moist atmosphere containing 5% CO2 on GCB (« Gonococcal Broth »; Difco) agar plates containing Kellog's supplements and appropriate antibiotics (100 µg/ml kanamycin, 6 µg/ml chloramphenicol and/or 4.5 µg/ml erythromycin for NEM8013 or 9 µg/ml erythromycin for N. cinerea). E. coli TOP10 (Life technologies) or BL21(DE3) (Life technologies) were grown at 37°C in liquid or solid Luria-Bertani (LB) medium (Difco), which contained appropriate antibiotics (50 µg/ml ticarcillin, 10 µg/ml chloramphenicol and/or 50 µg/ml kanamycin).
Vectors construction for toxicity assays in E. coli BL21(DE3)
All vectors and primers used in this study can be found in S3 Table and S4 Table.
Full-length mafB genes including the putative signal peptide sequence or only the 5′ or 3′ regions of mafB genes from NEM8013 were cloned in pBAD33. Briefly, PCR products of mafB1MGI-2NEM8013, mafBMGI-1NEM8013, the 5′ region of mafBMGI-1NEM8013 (the first 1020 nucleotides), the 3′ region of mafBMGI-1NEM8013 (the last 477 nucleotides) and the 3′ region of mafBMGI-3NEM8013 (the last 339 nucleotides) were digested by SacI and XbaI, whereas PCR products of mafB2MGI-2NEM8013, mafBMGI-3NEM8013 and the 5′ region of mafBMGI-3NEM8013 (the first 1107 nucleotides) were digested by SmaI and XbaI. The digested PCR products were ligated to pBAD33 under the control of arabinose-inducible PBAD promoter. mafB2MGI-2NEM8013 and mafB1MGI-2NEM8013 amplified without their putative signal peptide sequence (without the first 93 nucleotides for mafB2MGI-2NEM8013 and the first 120 nucleotides for mafB1MGI-2NEM8013) were also cloned in pET22 using BamHI and XhoI to obtain proteins containing PelB peptide signal. mafIMGI-1NEM8013 was cloned in pET28 using NcoI and XhoI and mafIMGI-3NEM8013 was cloned in pET15 using XhoI and BamHI. pBAD33-mafB or pET22-mafB constructs were transformed in E. coli BL21(DE3) with or without pET-mafI constructs to perform toxicity assays.
All vectors constructed were verified by PCR and sequencing.
Vectors construction for protein production in E. coli BL21(DE3)
In order to product and purify MafBMGI-1NEM8013, the operon mafBIMGI-1NEM8013 (NMV_0410-NMV_0409) without the sequence of the signal peptide of MafBMGI-1NEM8013 was cloned downstream of an IPTG inducible promoter in pET15 using XhoI and BamHI. The resulting MafB protein harbors hexahistidine N-terminal tag.
In order to product and purify MafIMGI-1NEM8013, NMV_0409 was cloned downstream of an IPTG inducible promoter in pET28 using NcoI and XhoI. The resulting MafI protein harbors hexahistidine C-terminal tag.
In order to assess the presence of MafB in the supernatant of E. coli, the operon mafBIMGI-1NEM8013 (NMV_0410-NMV_0409) was cloned downstream of an IPTG inducible promoter in pET28 using NcoI and XhoI.
To assess the potential co-purification of MafBMGI-1NEM8013 and MafIMGI-1NEM8013, mafBMGI-1NEM8013 and mafIMGI-1NEM8013 were cloned in two different multiple cloning site (MCS) under two IPTG inducible promoters in pcolaDUET. mafBMGI-1NEM8013 was cloned in MCS2 using BglII and KpnI and mafIMGI-1NEM8013 was cloned in MCS1 using BamHI and HindIII.
All vectors constructed were verified by PCR and sequencing.
Vector construction for mafABI deletion in N. cinerea
PCR reactions were used to amplify 400 bp upstream mafA (NEICIv1_50108) and 770 bp downstream mafI (NEICIv1_50110). The resulting products were digested by EcoRI/BamHI and BamHI/HindIII respectively and cloned into pUC19 digested by EcoRI/HindIII. A kanamycin resistance cassette apha-3 was then inserted in the BamHI site. The final construct containing kanamycin resistance cassette apha-3 flanked by homologous regions for recombination was amplified by PCR using primers with DNA uptake sequence. Amplicon was introduced in N. cinerea by transformation and transformants were verified by PCR and sequencing.
Vectors construction for maf genes expression in NEM8013 or N. cinerea
mafB genes, mafI genes or mafB-mafI operons of NEM8013 were cloned in pGCC4 under the control of an IPTG inducible promoter using PacI and ScaI. pGCC4-mafB or pGCC4-mafB-mafI were transformed in NEM8013, pGCC4-mafI1MGI-2NEM8013 was transformed in NEM8013ctrA [80] to insert mafB, mafI or mafB-mafI in the meningococcal intergenic region between lctP and aspC. NEM8013 strains harbouring IPTG inducible mafB-mafI operons have been used in competition assays. pGCC4-mafBIMGI-1NEM8013 was also transformed in NEM8013pilQ- [90] to assess the role of PilQ on MafB secretion.
In order to express a FLAG-tagged MafBMGI-1NEM8013 protein in NEM8013, the 5′ region of mafBMGI-1NEM8013, with or without the sequence of its signal peptide, was amplified with a reverse primer encoding the FLAG epitope (DYKDDDDK). The resulting PCR product was cloned in pGCC4 using PacI and ScaI. pGCC4-mafBMGI-1NEM8013FLAG or pGCC4-mafBMGI-1NEM8013FLAG-SP was transformed in NEM8013. pGCC4-mafBMGI-1NEM8013FLAG was also transformed in N. cinerea and in N. cinerea ΔmafABI to assess the role of MafA on MafB secretion.
All strains constructed were verified by PCR and sequencing.
Toxicity assays in E. coli
Toxin activity was assessed by growing E. coli BL21(DE3) carrying pBAD33-mafBMGI-1NEM8013, pBAD33-mafB1MGI-2NEM8013, pBAD33-mafB2MGI-2NEM8013 or pBAD33-mafBMGI-3NEM8013 at 37°C on LB agar plates with or without 0.2% L-arabinose to induce gene expression. Growth curves were performed in LB broth at 37°C (200 rpm) and toxin expression was induced either by addition of L-arabinose to a final concentration of 0.2% (pBAD33-mafB) or by 1 mM IPTG (pET22 - mafB).
To assess the protective role of mafI, E. coli BL21(DE3) were co-transformed with pBAD33-mafB and either cognate or non-cognate pET-mafI. Growth curves were performed with 0.01 mM IPTG to induce the antitoxin production throughout the experiment whereas L-arabinose (0.2%) was added 2 h post-inoculation to induce the toxin production. Growth was monitored every hour with OD 600 nm. Viability was assessed in parallel of the growth curves by spotting 5 microliters of bacterial cultures onto LB agar plates containing D-glucose (0.2%) before and after the induction of toxin expression.
Purification of proteins
Toxin MafBMGI-1NEM8013 and cognate immunity protein MafIMGI-1NEM8013 were expressed in E. coli BL21 (DE3) using plasmid pET15 resulting in the production of N-terminal hexahistidine-tagged toxin and untagged immunity protein. Protein expression was induced for 2 h at 37°C with 1 mM IPTG. Proteins were purified using NiNTA metal affinity resin (Qiagen) in denaturing conditions. Bacterial pellets were lysed by sonication in lysis buffer (100 mM NaH2PO4, 10 mM TrisHCl, 8 M urea, pH 8) and centrifuged for 20 mn at 10 000 g. Supernatants were incubated with Ni-NTA resin and loaded onto columns. The resin was washed with denaturing wash buffer (100 mM NaH2PO4, 10 mM TrisHCl, 8 M urea, pH 6.3) to first remove untagged immunity protein and then, MafBMGI-1NEM8013 was eluted using the same buffers by lowering pH (pH 5.9 and pH 4.5). Renaturation of MafBMGI-1NEM8013 was achieved by serial dialysis against buffers containing decreasing concentrations of urea.
Immunity MafIMGI-1NEM8013 was expressed in E. coli BL21(DE3) using plasmid pET28 resulting in the production of a C-terminal hexahistidine-tagged immunity protein. Protein expression was induced for 1 h at 37°C with 1 mM IPTG. Proteins were purified using NiNTA resin in native conditions. Bacterial pellets were lysed by sonication in lysis buffer (50 mM NaH2PO4, 300 mM NaCl, 10 mM imidazole, pH 8) and centrifuged for 20 mn at 10 000 g. Supernatants were loaded on NiNTA resin columns. The resin was washed (50 mM NaH2PO4, 300 mM NaCl, 20 mM and 50 mM imidazole, pH 8) and MafIMGI-1NEM8013 was eluted using a buffer containing 250 mM imidazole (pH 8).
MafBMGI-1NEM8013 and MafIMGI-1NEM8013 were co-expressed in E. coli BL21(DE3) using pcolaDUET. Protein expression was induced for 2 h at 37°C with 1 mM IPTG. Proteins were purified using NiNTA resin in native conditions as described above for MafIMGI-1NEM8013 alone. Bound complexes were eluted with native elution buffer containing 250 mM imidazole (pH 8) for SDS-PAGE analysis and immunoblotting.
In vivo RNase activity
E. coli Top10 carrying pBAD33 or pBAD33-mafBMGI-1NEM8013 were grown in LB containing chloramphenicol at 37°C, 200 rpm, until OD600 reached 0.2, then L-Arabinose was added to a final concentration of 0.2%. Total RNA from E. coli was isolated before induction (T0) and 30 min after addition of L-arabinose using TRIzol reagent (Life Technologies) according to manufacturer's instructions. Total RNAs were analyzed by denaturing gel electrophoresis (5% polyacrylamide/8 M urea) and visualized by staining with ethidium bromide.
In vitro RNase activity
Purified MafBMGI-1NEM8013-His6 alone (3 µM) and/or MafIMGI-1NEM8013-His6 (100 µM) were incubated with 4 µg of total RNA isolated from different sources with TRIzol reagent method (N. meningitidis NEM8013, E. coli TOP10 and human epithelial cells FaDu). Each reaction was performed for 30 min at 37°C in Tris-EDTA buffer and run on native 1% agarose gels containing ethidium bromide. To assess the role of divalent cations, buffers containing Mg2+ (10 mM Tris-HCl, 2.5 mM MgCl2) or Mn2+ (25 mM HEPES pH 7.4, 50 mM NaCl, 5 mM MnCl2, 1 mM DTT) have been used instead of Tris-EDTA buffer.
To assess the ability of MafBMGI-1NEM8013 to cleave synthetic RNA in vitro, purified MafBMGI-1NEM8013-His6 alone (3 µM) and/or MafIMGI-1NEM8013-His6 (100 µM) were incubated with 3 µM of a synthetic oligoribonucleotide in Tris-EDTA buffer for 15 min at 37°C. Two synthetic oligoribonucleotides (synthetized by Integrated DNA Technologies) were used: 5′-CCUGGUUUUUAAGGAGUGUCGCCAGAGUGCCGCGAAUGAAAAA -3′ (mRNA U) and 5′ - CCAGGAAAAAAAGGAGAGACGCCAGAGAGCCGCGAAAGAAAAA-3′ (mRNA 0). The reactions were stopped by the addition of an equal volume of Gel Loading Buffer II (95% formamide, 18 mM EDTA, 0.025% SDS; Ambion) and incubation for 5 min at 95°C. The reaction products were separated by electrophoresis in 14% polyacrylamide/8 M urea and were visualized by ethidium bromide staining.
Immunoblotting
Preparation of protein samples, SDS-PAGE separation, transfer to membranes and immunoblotting were performed using standard molecular biology techniques. Proteins were quantified using NanoDrop, following manufacturer's instructions.
We raised polyclonal antibodies in rabbits against purified recombinant protein MafIMGI-1NEM8013 and against a synthetic peptide (KNSNIHEKNYGRD) of the COOH-terminal region of MafBMGI-1NEM8013 protein (Proteogenix). We used rabbit polyclonal anti-FLAG antibody directed against DYKDDDDK epitope (Cell Signaling Technology) and mouse monoclonal antibody directed against NADP-dependent glutamate dehydrogenase. Bound primary antibodies were detected by goat Anti-Rabbit or Anti-Mouse HRP-linked antibodies (Cell Signaling Technology) using ECL Plus detection reagents (Pierce).
Preparation of supernatants from meningococcal and E. coli cultures
Overnight cultures of N. meningitidis grown on GCB agar plates were used to inoculate RPMI 1640 medium (PAA) containing Kellog's supplements and 1 mM IPTG. Overnight cultures of BL21(DE3) grown on LB agar plates were used to inoculate LB medium containing appropriate antibiotic. When OD 600 reached 0.2, IPTG was added to a final concentration of 1 mM. When OD 600 reached 0.5, the cells were harvested by centrifugation (3 000× g for 30 min), and supernatants were passed through a 0.22-µm pore size filter unit. Supernatant proteins were concentrated by ultrafiltration (Amicon, Ultra-15, 3 kDa cutoff) according to the manufacturer's instructions. The pellets and the concentrated supernatants were used for immunoblotting analysis.
Competition assays
Overnight cultures of putative target cells and putative inhibitory cells grown on GCB agar plates were used to inoculate Ham's F12 (PAA) containing Kellog's supplements and 1 mM IPTG. Putative target cells were an unencapsulated derivative of NEM8013 with a transposon insertion in the ctrA gene [80] or this unencapsulated derivative overexpressing MafI1MGI-2NEM8013. Putative inhibitory cells were NEM8013 overexpressing each of the four mafB toxins and their cognate mafI immunity genes. When OD 600 reached 0.8, cultures were mixed at an inhibitor to target cell ratio of 10 to 1 (∼8×109 inhibitor cells and ∼8×108 target cells) and centrifuged at 4 000 rpm for 5 min. 10 µl of the mixed culture pellet were spotted on a membrane filter (pore size, 0.45 µm) placed on GCB agar plate containing 1 mM IPTG and incubated overnight. 10 µl of mixed cultures were also used for determination of the inhibitor/target initial ratio by plating on GCB with appropriate antibiotics. Filters recovered after overnight incubation were used to perform viable counts and the competitive index (CI) was calculated as the inhibitor/target ratio in the output divided by the initial inhibitor/target ratio. The data from three independent experiments were examined for significance using a two-tailed Student's t-test. A p-value p<0.05 was considered significant.
Supporting Information
Zdroje
1. ZhangD, de SouzaRF, AnantharamanV, IyerLM, AravindL (2012) Polymorphic toxin systems: Comprehensive characterization of trafficking modes, processing, mechanisms of action, immunity and ecology using comparative genomics. Biol Direct 7 : 18.
2. ZhangD, IyerLM, AravindL (2011) A novel immunity system for bacterial nucleic acid degrading toxins and its recruitment in various eukaryotic and DNA viral systems. Nucleic Acids Res 39 : 4532–4552.
3. IyerLM, ZhangD, RogozinIB, AravindL (2011) Evolution of the deaminase fold and multiple origins of eukaryotic editing and mutagenic nucleic acid deaminases from bacterial toxin systems. Nucleic Acids Res 39 : 9473–9497.
4. PooleSJ, DinerEJ, AokiSK, BraatenBA, t'Kint de RoodenbekeC, et al. (2011) Identification of functional toxin/immunity genes linked to contact-dependent growth inhibition (CDI) and rearrangement hotspot (Rhs) systems. PLoS Genet 7: e1002217.
5. AokiSK, DinerEJ, de RoodenbekeCT, BurgessBR, PooleSJ, et al. (2010) A widespread family of polymorphic contact-dependent toxin delivery systems in bacteria. Nature 468 : 439–442.
6. KoskiniemiS, LamoureuxJG, NikolakakisKC, t'Kint de RoodenbekeC, KaplanMD, et al. (2013) Rhs proteins from diverse bacteria mediate intercellular competition. Proc Natl Acad Sci U S A 110 : 7032–7037.
7. BeddoeT, PatonAW, Le NoursJ, RossjohnJ, PatonJC (2010) Structure, biological functions and applications of the AB5 toxins. Trends Biochem Sci 35 : 411–418.
8. HayesCS, KoskiniemiS, RuheZC, PooleSJ, LowDA (2014) Mechanisms and biological roles of contact-dependent growth inhibition systems. Cold Spring Harb Persp Med 4 (2).
9. AokiSK, PooleSJ, HayesCS, LowDA (2011) Toxin on a stick: modular CDI toxin delivery systems play roles in bacterial competition. Virulence 2 : 356–359.
10. KoskiniemiS, Garza-SanchezF, SandegrenL, WebbJS, BraatenBA, et al. (2014) Selection of orphan Rhs toxin expression in evolved Salmonella enterica serovar Typhimurium. PLoS Genet 10: e1004255.
11. AokiSK, PammaR, HerndayAD, BickhamJE, BraatenBA, et al. (2005) Contact-dependent inhibition of growth in Escherichia coli. Science 309 : 1245–1248.
12. ur RahmanS, van UlsenP (2013) System specificity of the TpsB transporters of coexpressed two-partner secretion systems of Neisseria meningitidis. J Bacteriol 195 : 788–797.
13. AndersonMS, GarciaEC, CotterPA (2012) The Burkholderia bcpAIOB genes define unique classes of two-partner secretion and contact dependent growth inhibition systems. PLoS Genet 8: e1002877.
14. ArenasJ, SchipperK, van UlsenP, van der EndeA, TommassenJ (2013) Domain exchange at the 3′ end of the gene encoding the fratricide meningococcal two-partner secretion protein A. BMC Genomics 14 : 622.
15. UnemoM, ShipitsynaE, DomeikaM, Eastern EuropeanS (2011) Reproductive Health Network Antimicrobial Resistance G (2011) Gonorrhoea surveillance, laboratory diagnosis and antimicrobial susceptibility testing of Neisseria gonorrhoeae in 11 countries of the eastern part of the WHO European region. APMIS 119 : 643–649.
16. CarbonnelleE, HillDJ, MorandP, GriffithsNJ, BourdoulousS, et al. (2009) Meningococcal interactions with the host. Vaccine 27 Suppl 2: B78–89.
17. CaugantDA, TzanakakiG, KrizP (2007) Lessons from meningococcal carriage studies. FEMS Microbiol Rev 31 : 52–63.
18. BennettJS, BentleySD, VernikosGS, QuailMA, CherevachI, et al. (2010) Independent evolution of the core and accessory gene sets in the genus Neisseria: insights gained from the genome of Neisseria lactamica isolate 020-06. BMC Genomics 11 : 652.
19. BentleySD, VernikosGS, SnyderLA, ChurcherC, ArrowsmithC, et al. (2007) Meningococcal genetic variation mechanisms viewed through comparative analysis of serogroup C strain FAM18. PLoS Genet 3: e23.
20. PengJ, YangL, YangF, YangJ, YanY, et al. (2008) Characterization of ST-4821 complex, a unique Neisseria meningitidis clone. Genomics 91 : 78–87.
21. SchoenC, BlomJ, ClausH, Schramm-GluckA, BrandtP, et al. (2008) Whole-genome comparison of disease and carriage strains provides insights into virulence evolution in Neisseria meningitidis. Proc Natl Acad Sci U S A 105 : 3473–3478.
22. SnyderLA, ButcherSA, SaundersNJ (2001) Comparative whole-genome analyses reveal over 100 putative phase-variable genes in the pathogenic Neisseria spp. Microbiology 147 : 2321–2332.
23. Dunning HotoppJC, GrifantiniR, KumarN, TzengYL, FoutsD, et al. (2006) Comparative genomics of Neisseria meningitidis: core genome, islands of horizontal transfer and pathogen-specific genes. Microbiology 152 : 3733–3749.
24. SunYH, BakshiS, ChalmersR, TangCM (2000) Functional genomics of Neisseria meningitidis pathogenesis. Nat Med 6 : 1269–1273.
25. SnyderLA, SaundersNJ (2006) The majority of genes in the pathogenic Neisseria species are present in non-pathogenic Neisseria lactamica, including those designated as ‘virulence genes’. BMC Genomics 7 : 128.
26. DobrindtU, HochhutB, HentschelU, HackerJ (2004) Genomic islands in pathogenic and environmental microorganisms. Nat Rev Microbiol 2 : 414–424.
27. JuhasM, van der MeerJR, GaillardM, HardingRM, HoodDW, et al. (2009) Genomic islands: tools of bacterial horizontal gene transfer and evolution. FEMS Microbiol Rev 33 : 376–393.
28. BilleE, ZaharJR, PerrinA, MorelleS, KrizP, et al. (2005) A chromosomally integrated bacteriophage in invasive meningococci. J Exp Med 201 : 1905–1913.
29. KleeSR, NassifX, KusecekB, MerkerP, BerettiJL, et al. (2000) Molecular and biological analysis of eight genetic islands that distinguish Neisseria meningitidis from the closely related pathogen Neisseria gonorrhoeae. Infect Immun 68 : 2082–2095.
30. PerrinA, BonacorsiS, CarbonnelleE, TalibiD, DessenP, et al. (2002) Comparative genomics identifies the genetic islands that distinguish Neisseria meningitidis, the agent of cerebrospinal meningitis, from other Neisseria species. Infect Immun 70 : 7063–7072.
31. SnyderLA, DaviesJK, RyanCS, SaundersNJ (2005) Comparative overview of the genomic and genetic differences between the pathogenic Neisseria strains and species. Plasmid 54 : 191–218.
32. PiekarowiczA, KlyzA, MajchrzakM, Adamczyk-PoplawskaM, MaugelTK, et al. (2007) Characterization of the dsDNA prophage sequences in the genome of Neisseria gonorrhoeae and visualization of productive bacteriophage. BMC Microbiol 7 : 66.
33. DillardJP, SeifertHS (2001) A variable genetic island specific for Neisseria gonorrhoeae is involved in providing DNA for natural transformation and is found more often in disseminated infection isolates. Mol Microbiol 41 : 263–277.
34. SnyderLA, JarvisSA, SaundersNJ (2005) Complete and variant forms of the ‘gonococcal genetic island’ in Neisseria meningitidis. Microbiology 151 : 4005–4013.
35. ParuchuriDK, SeifertHS, AjiokaRS, KarlssonKA, SoM (1990) Identification and characterization of a Neisseria gonorrhoeae gene encoding a glycolipid-binding adhesin. Proc Natl Acad Sci U S A 87 : 333–337.
36. PetersenTN, BrunakS, von HeijneG, NielsenH (2011) SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods 8 : 785–786.
37. BagosPG, NikolaouEP, LiakopoulosTD, TsirigosKD (2010) Combined prediction of Tat and Sec signal peptides with hidden Markov models. Bioinformatics 26 : 2811–2817.
38. BendtsenJD, KiemerL, FausbollA, BrunakS (2005) Non-classical protein secretion in bacteria. BMC Microbiol 5 : 58.
39. RoseRW, BruserT, KissingerJC, PohlschroderM (2002) Adaptation of protein secretion to extremely high-salt conditions by extensive use of the twin-arginine translocation pathway. Mol Microbiol 45 : 943–950.
40. JunckerAS, WillenbrockH, Von HeijneG, BrunakS, NielsenH, et al. (2003) Prediction of lipoprotein signal peptides in Gram-negative bacteria. Protein Sci 12 : 1652–1662.
41. VipondC, WheelerJX, JonesC, FeaversIM, SukerJ (2005) Characterization of the protein content of a meningococcal outer membrane vesicle vaccine by polyacrylamide gel electrophoresis and mass spectrometry. Hum Vaccin 1 : 80–84.
42. WuHJ, SeibKL, SrikhantaYN, EdwardsJ, KiddSP, et al. (2010) Manganese regulation of virulence factors and oxidative stress resistance in Neisseria gonorrhoeae. J Proteomics 73 : 899–916.
43. ZielkeRA, WierzbickiIH, WeberJV, GafkenPR, SikoraAE (2014) Quantitative proteomics of the Neisseria gonorrhoeae cell envelope and membrane vesicles for the discovery of potential therapeutic targets. Mol Cell Proteomics 13 : 1299–1317.
44. Marchler-BauerA, LuS, AndersonJB, ChitsazF, DerbyshireMK, et al. (2011) CDD: a Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res 39: D225–229.
45. GuzmanLM, BelinD, CarsonMJ, BeckwithJ (1995) Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol 177 : 4121–4130.
46. MehrIJ, LongCD, SerkinCD, SeifertHS (2000) A homologue of the recombination-dependent growth gene, rdgC, is involved in gonococcal pilin antigenic variation. Genetics 154 : 523–532.
47. AokiSK, MalinverniJC, JacobyK, ThomasB, PammaR, et al. (2008) Contact-dependent growth inhibition requires the essential outer membrane protein BamA (YaeT) as the receptor and the inner membrane transport protein AcrB. Mol Microbiol 70 : 323–340.
48. HolbergerLE, Garza-SanchezF, LamoureuxJ, LowDA, HayesCS (2012) A novel family of toxin/antitoxin proteins in Bacillus species. FEBS Lett 586 : 132–136.
49. WebbJS, NikolakakisKC, WillettJL, AokiSK, HayesCS, et al. (2013) Delivery of CdiA nuclease toxins into target cells during contact-dependent growth inhibition. PLoS One 8: e57609.
50. SchielkeS, FroschM, KurzaiO (2010) Virulence determinants involved in differential host niche adaptation of Neisseria meningitidis and Neisseria gonorrhoeae. Med Microbiol Immunol 199 : 185–196.
51. van UlsenP, RuttenL, FellerM, TommassenJ, van der EndeA (2008) Two-partner secretion systems of Neisseria meningitidis associated with invasive clonal complexes. Infect Immun 76 : 4649–4658.
52. ur RahmanS, ArenasJ, OzturkH, DekkerN, van UlsenP (2014) The polypeptide transport-associated (POTRA) domains of TpsB transporters determine the system specificity of two-partner secretion systems. J Biol Chem 289 : 19799–19809.
53. RemautH, WaksmanG (2004) Structural biology of bacterial pathogenesis. Curr Opin Struct Biol 14 : 161–170.
54. DesvauxM, HebraudM, TalonR, HendersonIR (2009) Secretion and subcellular localizations of bacterial proteins: a semantic awareness issue. Trends Microbiol 17 : 139–145.
55. SilvermanJM, BrunetYR, CascalesE, MougousJD (2012) Structure and regulation of the type VI secretion system. Annu Rev Microbiol 66 : 453–472.
56. van UlsenP, TommassenJ (2006) Protein secretion and secreted proteins in pathogenic Neisseriaceae. FEMS Microbiol Rev 30 : 292–319.
57. DesvauxM, ParhamNJ, HendersonIR (2004) Type V protein secretion: simplicity gone awry? Curr Issues Mol Biol 6 : 111–124.
58. KoomeyJM, GillRE, FalkowS (1982) Genetic and biochemical analysis of gonococcal IgA1 protease: cloning in Escherichia coli and construction of mutants of gonococci that fail to produce the activity. Proc Natl Acad Sci U S A 79 : 7881–7885.
59. NattNK, KaurH, RaghavaGP (2004) Prediction of transmembrane regions of beta-barrel proteins using ANN - and SVM-based methods. Proteins 56 : 11–18.
60. BagosPG, LiakopoulosTD, HamodrakasSJ (2006) Algorithms for incorporating prior topological information in HMMs: application to transmembrane proteins. BMC Bioinformatics 7 : 189.
61. HodakH, Jacob-DubuissonF (2007) Current challenges in autotransport and two-partner protein secretion pathways. Res Microbiol 158 : 631–637.
62. DevoeIW, GilchristJE (1973) Release of endotoxin in the form of cell wall blebs during in vitro growth of Neisseria meningitidis. J Exp Med 138 : 1156–1167.
63. LloubesR, BernadacA, HouotL, PommierS (2013) Non classical secretion systems. Res Microbiol 164 : 655–663.
64. KadurugamuwaJL, BeveridgeTJ (1996) Bacteriolytic effect of membrane vesicles from Pseudomonas aeruginosa on other bacteria including pathogens: conceptually new antibiotics. J Bacteriol 178 : 2767–2774.
65. MacDonaldIA, KuehnMJ (2012) Offense and defense: microbial membrane vesicles play both ways. Res Microbiol 163 : 607–618.
66. KestyNC, MasonKM, ReedyM, MillerSE, KuehnMJ (2004) Enterotoxigenic Escherichia coli vesicles target toxin delivery into mammalian cells. EMBO J 23 : 4538–4549.
67. BombergerJM, MaceachranDP, CoutermarshBA, YeS, O'TooleGA, et al. (2009) Long-distance delivery of bacterial virulence factors by Pseudomonas aeruginosa outer membrane vesicles. PLoS Pathog 5: e1000382.
68. HolstJ, OsterP, ArnoldR, TatleyMV, NaessLM, et al. (2013) Vaccines against meningococcal serogroup B disease containing outer membrane vesicles (OMV): lessons from past programs and implications for the future. Hum Vaccin Immunother 9 : 1241–1253.
69. HammerschmidtS, HilseR, van PuttenJP, Gerardy-SchahnR, UnkmeirA, et al. (1996) Modulation of cell surface sialic acid expression in Neisseria meningitidis via a transposable genetic element. EMBO J 15 : 192–198.
70. HammerschmidtS, MullerA, SillmannH, MuhlenhoffM, BorrowR, et al. (1996) Capsule phase variation in Neisseria meningitidis serogroup B by slipped-strand mispairing in the polysialyltransferase gene (siaD): correlation with bacterial invasion and the outbreak of meningococcal disease. Mol Microbiol 20 : 1211–1220.
71. HeyA, LiMS, HudsonMJ, LangfordPR, KrollJS (2013) Transcriptional profiling of Neisseria meningitidis interacting with human epithelial cells in a long-term in vitro colonization model. Infect Immun 81 : 4149–4159.
72. DeghmaneAE, GiorginiD, LarribeM, AlonsoJM, TahaMK (2002) Down-regulation of pili and capsule of Neisseria meningitidis upon contact with epithelial cells is mediated by CrgA regulatory protein. Mol Microbiol 43 : 1555–1564.
73. FagnocchiL, PigozziE, ScarlatoV, DelanyI (2012) In the NadR regulon, adhesins and diverse meningococcal functions are regulated in response to signals in human saliva. J Bacteriol 194 : 460–474.
74. GarciaEC, AndersonMS, HagarJA, CotterPA (2013) Burkholderia BcpA mediates biofilm formation independently of interbacterial contact-dependent growth inhibition. Mol Microbiol 89 : 1213–1225.
75. AndersonMS, GarciaEC, CotterPA (2014) Kind discrimination and competitive exclusion mediated by contact-dependent growth inhibition systems shape biofilm community structure. PLoS Pathog 10: e1004076.
76. ParkhillJ, AchtmanM, JamesKD, BentleySD, ChurcherC, et al. (2000) Complete DNA sequence of a serogroup A strain of Neisseria meningitidis Z2491. Nature 404 : 502–506.
77. TettelinH, SaundersNJ, HeidelbergJ, JeffriesAC, NelsonKE, et al. (2000) Complete genome sequence of Neisseria meningitidis serogroup B strain MC58. Science 287 : 1809–1815.
78. VallenetD, LabarreL, RouyZ, BarbeV, BocsS, et al. (2006) MaGe: a microbial genome annotation system supported by synteny results. Nucleic Acids Res 34 : 53–65.
79. ChungGT, YooJS, OhHB, LeeYS, ChaSH, et al. (2008) Complete genome sequence of Neisseria gonorrhoeae NCCP11945. J Bacteriol 190 : 6035–6036.
80. RusniokC, VallenetD, FloquetS, EwlesH, Mouze-SoulamaC, et al. (2009) NeMeSys: a biological resource for narrowing the gap between sequence and function in the human pathogen Neisseria meningitidis. Genome Biol 10: R110.
81. PietJR, Huis in 't VeldRA, van SchaikBD, van KampenAH, BaasF, et al. (2011) Genome sequence of Neisseria meningitidis serogroup B strain H44/76. J Bacteriol 193 : 2371–2372.
82. BudroniS, SienaE, Dunning HotoppJC, SeibKL, SerrutoD, et al. (2011) Neisseria meningitidis is structured in clades associated with restriction modification systems that modulate homologous recombination. Proc Natl Acad Sci U S A 108 : 4494–4499.
83. SchoenC, Weber-LehmannJ, BlomJ, JosephB, GoesmannA, et al. (2011) Whole-genome sequence of the transformable Neisseria meningitidis serogroup A strain WUE2594. J Bacteriol 193 : 2064–2065.
84. FinnRD, BatemanA, ClementsJ, CoggillP, EberhardtRY, et al. (2014) Pfam: the protein families database. Nucleic Acids Res 42: D222–230.
85. KanehisaM (1997) Linking databases and organisms: GenomeNet resources in Japan. Trends Biochem Sci 22 : 442–444.
86. LarkinMA, BlackshieldsG, BrownNP, ChennaR, McGettiganPA, et al. (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23 : 2947–2948.
87. EdgarRC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32 : 1792–1797.
88. SieversF, HigginsDG (2014) Clustal Omega, accurate alignment of very large numbers of sequences. Methods Mol Biol 1079 : 105–116.
89. SullivanMJ, PettyNK, BeatsonSA (2011) Easyfig: a genome comparison visualizer. Bioinformatics 27 : 1009–1010.
90. HelaineS, CarbonnelleE, ProuvensierL, BerettiJL, NassifX, et al. (2005) PilX, a pilus-associated protein essential for bacterial aggregation, is a key to pilus-facilitated attachment of Neisseria meningitidis to human cells. Mol Microbiol 55 : 65–77.
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek Differential Reliance on Autophagy for Protection from HSV Encephalitis between Newborns and AdultsČlánek The Molecular Basis for Control of ETEC Enterotoxin Expression in Response to Environment and HostČlánek Different Infectivity of HIV-1 Strains Is Linked to Number of Envelope Trimers Required for EntryČlánek Preferential Use of Central Metabolism Reveals a Nutritional Basis for Polymicrobial Infection
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2015 Číslo 1- Jak souvisí postcovidový syndrom s poškozením mozku?
- Měli bychom postcovidový syndrom léčit antidepresivy?
- Farmakovigilanční studie perorálních antivirotik indikovaných v léčbě COVID-19
- 10 bodů k očkování proti COVID-19: stanovisko České společnosti alergologie a klinické imunologie ČLS JEP
-
Všechny články tohoto čísla
- The Importance of Pathogen Load
- Implication of Gut Microbiota in Nonalcoholic Fatty Liver Disease
- Infections in Humans and Animals: Pathophysiology, Detection, and Treatment
- Helminth-Induced Immune Regulation: Implications for Immune Responses to Tuberculosis
- The M3 Muscarinic Receptor Is Required for Optimal Adaptive Immunity to Helminth and Bacterial Infection
- An Iron-Mimicking, Trojan Horse-Entering Fungi—Has the Time Come for Molecular Imaging of Fungal Infections?
- Modulates the Unfolded Protein Response in during Infection
- Differential Reliance on Autophagy for Protection from HSV Encephalitis between Newborns and Adults
- Identification of HNRNPK as Regulator of Hepatitis C Virus Particle Production
- Parasite Biomass-Related Inflammation, Endothelial Activation, Microvascular Dysfunction and Disease Severity in Vivax Malaria
- : Trypanosomatids Adapted to Plant Environments
- Early Virus-Host Interactions Dictate the Course of a Persistent Infection
- TLR3 Signaling in Macrophages Is Indispensable for the Protective Immunity of Invariant Natural Killer T Cells against Enterovirus 71 Infection
- The Epstein-Barr Virus Encoded BART miRNAs Potentiate Tumor Growth
- Macrophage-Derived Human Resistin Is Induced in Multiple Helminth Infections and Promotes Inflammatory Monocytes and Increased Parasite Burden
- Dissemination of a Highly Virulent Pathogen: Tracking The Early Events That Define Infection
- Variability in Tuberculosis Granuloma T Cell Responses Exists, but a Balance of Pro- and Anti-inflammatory Cytokines Is Associated with Sterilization
- The Shear Stress of Host Cell Invasion: Exploring the Role of Biomolecular Complexes
- The Molecular Basis for Control of ETEC Enterotoxin Expression in Response to Environment and Host
- Different Infectivity of HIV-1 Strains Is Linked to Number of Envelope Trimers Required for Entry
- Secreted Herpes Simplex Virus-2 Glycoprotein G Modifies NGF-TrkA Signaling to Attract Free Nerve Endings to the Site of Infection
- Preferential Use of Central Metabolism Reveals a Nutritional Basis for Polymicrobial Infection
- A New Family of Secreted Toxins in Pathogenic Neisseria Species
- A Human Type 5 Adenovirus-Based Therapeutic Vaccine Re-programs Immune Response and Reverses Chronic Cardiomyopathy
- Regulation of Oncogene Expression in T-DNA-Transformed Host Plant Cells
- GITR Intrinsically Sustains Early Type 1 and Late Follicular Helper CD4 T Cell Accumulation to Control a Chronic Viral Infection
- Cell Cycle-Independent Phospho-Regulation of Fkh2 during Hyphal Growth Regulates Pathogenesis
- Virus-Induced NETs – Critical Component of Host Defense or Pathogenic Mediator?
- Environmental Drivers of the Spatiotemporal Dynamics of Respiratory Syncytial Virus in the United States
- Protective Efficacy of Centralized and Polyvalent Envelope Immunogens in an Attenuated Equine Lentivirus Vaccine
- Transmitted Virus Fitness and Host T Cell Responses Collectively Define Divergent Infection Outcomes in Two HIV-1 Recipients
- Systemic Expression of Kaposi Sarcoma Herpesvirus (KSHV) Vflip in Endothelial Cells Leads to a Profound Proinflammatory Phenotype and Myeloid Lineage Remodeling
- Dengue Virus RNA Structure Specialization Facilitates Host Adaptation
- DNA Is an Antimicrobial Component of Neutrophil Extracellular Traps
- Uropathogenic Superinfection Enhances the Severity of Mouse Bladder Infection
- Well-Ordered Trimeric HIV-1 Subtype B and C Soluble Spike Mimetics Generated by Negative Selection Display Native-like Properties
- The Phylogenetically-Related Pattern Recognition Receptors EFR and XA21 Recruit Similar Immune Signaling Components in Monocots and Dicots
- Reprogramming of from Virulent to Persistent Mode Revealed by Complex RNA-seq Analysis
- Compartment-Specific and Sequential Role of MyD88 and CARD9 in Chemokine Induction and Innate Defense during Respiratory Fungal Infection
- Bacterial Flagella: Twist and Stick, or Dodge across the Kingdoms
- Elucidation of the RamA Regulon in Reveals a Role in LPS Regulation
- IL-1α Signaling Is Critical for Leukocyte Recruitment after Pulmonary Challenge
- Chronic Filarial Infection Provides Protection against Bacterial Sepsis by Functionally Reprogramming Macrophages
- Specificity and Dynamics of Effector and Memory CD8 T Cell Responses in Human Tick-Borne Encephalitis Virus Infection
- Promiscuous RNA Binding Ensures Effective Encapsidation of APOBEC3 Proteins by HIV-1
- Viral Activation of MK2-hsp27-p115RhoGEF-RhoA Signaling Axis Causes Cytoskeletal Rearrangements, P-body Disruption and ARE-mRNA Stabilization
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Infections in Humans and Animals: Pathophysiology, Detection, and Treatment
- : Trypanosomatids Adapted to Plant Environments
- Environmental Drivers of the Spatiotemporal Dynamics of Respiratory Syncytial Virus in the United States
- Dengue Virus RNA Structure Specialization Facilitates Host Adaptation
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



