-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaTeaching Fido New ModiFICation Tricks
article has not abstract
Published in the journal: . PLoS Pathog 10(9): e32767. doi:10.1371/journal.ppat.1004349
Category: Pearls
doi: https://doi.org/10.1371/journal.ppat.1004349Summary
article has not abstract
The Fic (filamentation induced by cAMP) family of proteins comprises several thousand members that are found in all domains of life [1], [2]. In bacteria, these proteins are often expressed by pathogens as virulence factors that disrupt signaling in the mammalian host cell through inhibition of one or more GTPases or kinases involved in cell signaling. By integrating surprising new insights, this Pearl highlights the spectrum of potent posttranslational modifications catalyzed by the Fic family of proteins in bacterial pathogens. We will also discuss how Fic proteins are predicted to endow bacterial pathogens with the ability to perturb essential signaling pathways in their host cells (for secreted Fic proteins) or enable the pathogen to endure stresses such as antibiotic exposure (for intracellular Fic proteins derived from toxin-antitoxin modules).
All Fic family proteins (also referred to as “Fido” because the family includes Doc toxins [3]) contain a Fic domain of 100–140 amino acids that exhibits two hallmarks, striking structural similarity and a highly conserved, nine amino acid active site (Figure 1). At the structural level, Fic domains have an α-helical core consisting of six or eight α-helices (reviewed in [3], [4]). Yet, there is no significant sequence similarity among all Fic domain proteins beyond the 9-residue active site, the Fic motif. These conserved features alone did not provide clues to the function of Fic domain proteins. Only upon molecular analysis of the Vibrio parahemolyticus virulence factor VopS by the Orth group [5] was a precise enzymatic activity of Fic domain proteins revealed. This discovery set the stage for a series of unanticipated twists demonstrating that Fic domains can influence pathogenesis by inactivating target proteins involved in cell signaling by one of four posttranslational modifications: AMP, UMP, phosphocholine, or phosphate (summarized in Figure 2).
Fig. 1. Overview of Fic domain proteins, their targets, and their roles in bacterial virulence. 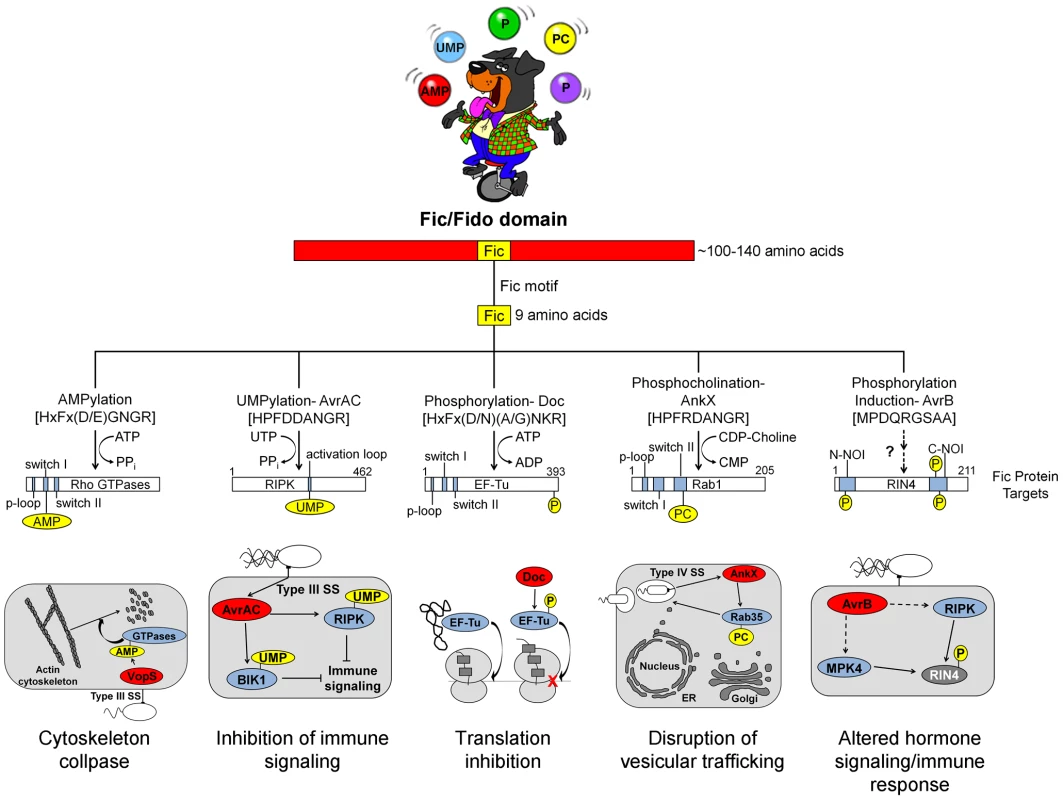
The Fic domain is defined by a conserved structural fold composed of six to eight α-helices (red bar). A nine amino acid Fic motif (yellow box) within this core fold comprises the key residues for catalysis. Both the structure of the Fic domain and the sequence of the Fic motif informs the activity of the Fic domain protein. Important domains of the target proteins are shown as blue bars; modifications as yellow circles (not to scale). Known physiological manifestations of each class of Fic domain protein are illustrated below their target. The exact role of the NOI domains of RIN4 is not known, though they are exclusively represented in plant proteins and have been hypothesized to play a role in host response to pathogens [32]. Abbreviations on juggled balls: AMP, adenosine monophosphate; UMP, uridine monophosphate; P, phosphate; PC, phosphocholine. Fig. 2. Features and functions of effector proteins that contain a Fic domain. 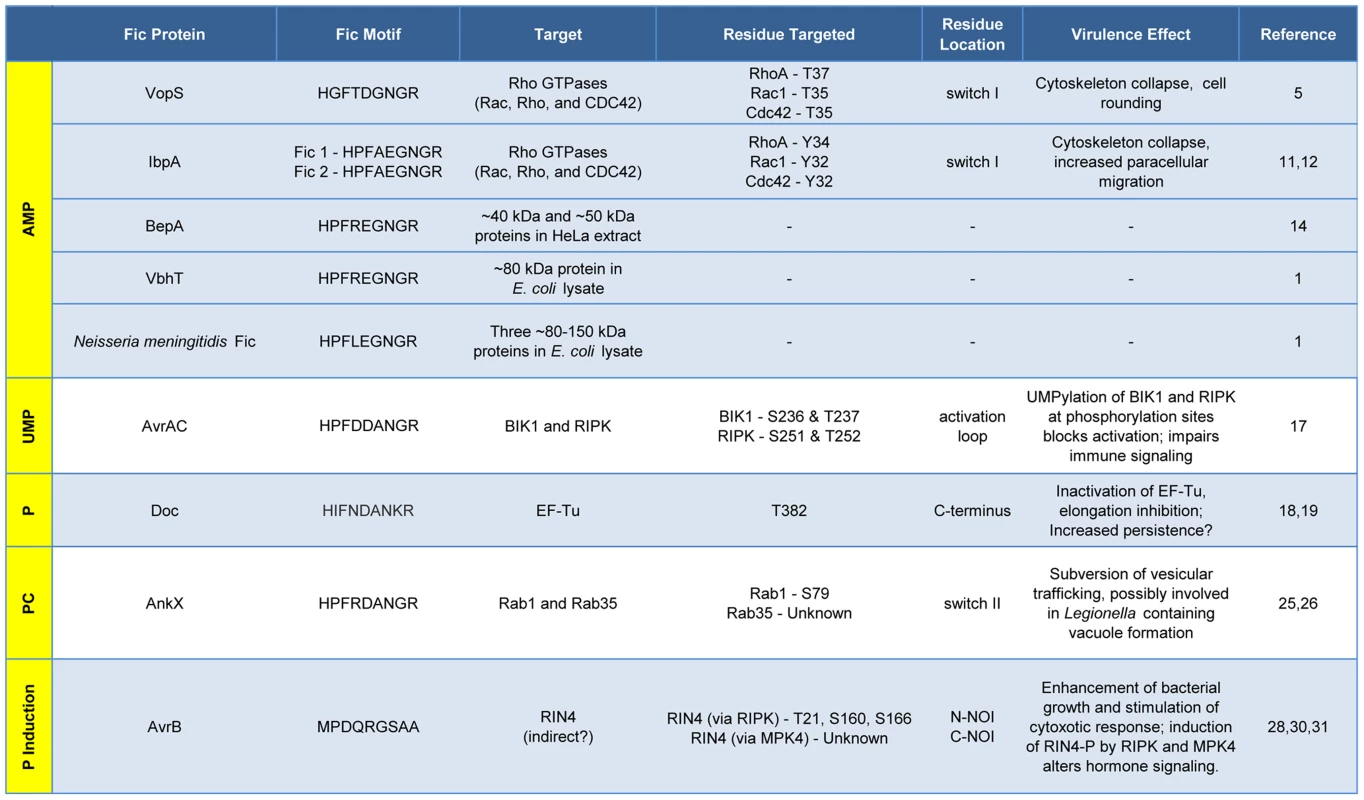
Fic proteins, their targets, and the residue(s) modified are shown. The far left column, running vertically, shows the modification type added by each Fic protein. Dashes, data not known. Inactivation of Host Cell GTPases by Addition of AMP
VopS is an effector protein secreted by the foodborne, gastrointestinal pathogen V. parahemolyticus.
The Fic domain of VopS catalyzes the stable transfer of a single adenylyl group derived from ATP [5], a process referred to as adenylylation [6] or AMPylation [5], [7]. Although modulation of protein activity by AMPylation had been documented earlier for glutamine synthetase [8], VopS represented the first example of the use of AMPylation in the context of bacterial virulence. The type III secretion system in V. parahemolyticus secretes VopS into the mammalian host cell, inactivates Rho family GTPases by AMPylation at a single site in the functionally essential switch I region, and results in collapse of the actin cytoskeleton [5]. VopS-mediated AMPylation of Rho GTPases inhibits NLRC4 inflammasome activation [9] but indirectly activates the Pyrin inflammasome [10]. This counteracting effect of Pyrin stems from its newly identified role as a host cell immune sensor that specifically detects the downstream effects of inactivating modifications to Rho GTPases (i.e., AMPylation, deamidation, glucosylation, and ADP-ribosylation) to restore an inflammatory response [10].
Subsequent work uncovered several more AMPylating Fic domain proteins (Figure 2). IbpA from Histophilus somni, an opportunistic pathogen that infects the mucosa of cattle and sheep, has two Fic domains [11]. As with VopS, IbpA AMPylates Rho GTPases at a single site in switch I and leads to cell rounding and cytoskeleton collapse. IbpA is a major virulence factor; H. somni strains lacking it are avirulent as defined by their lack of cytotoxicity and the inability to transmigrate across a cultured cell monolayer [12]. Treatment of cultured bovine cells with an antibody to one of the two Fic domains in IbpA also blocks virulence; i.e., it prevents cytotoxicity and cell migration [12]. BepA (Bartonella effector protein) from Bartonella henselae, which causes cat scratch fever, is one of seven effector proteins translocated by the VirB type IV secretion system [13]. In vitro, BepA AMPylates two proteins in a HeLa cell extract [14]. Another pathogen with a characterized Fic domain protein, Bartonella schoenbuchensis, is harbored by a louse fly that typically bites and infects ruminants but may also incidentally infect humans [15]. B. schoenbuchensis Fic domain protein VbhT is the toxin component of the VbhA-VbhT toxin-antitoxin (TA) system [1]. VbhT has a carboxy terminal BID (Bartonella intracellular delivery) domain that mediates delivery to its mammalian host cell via type IV secretion system [1]. In vitro experiments demonstrated that pure VbhT exhibits AMPylation activity against a single protein in an Escherichia coli cell extract [1]. Finally, Neisseria meningitides, which most commonly causes meningitis in children and young adults, has a Fic domain protein that appears to AMPylate a few proteins in an E. coli cell extract [1].
Distinct from bacterial pathogen virulence factors, there are currently only two eukaryotic Fic domain proteins known to possess AMPylation activity. Humans and Drosophila each have a single Fic domain protein. Drosophila Fic (also known as CG9523 [3]) plays an essential role in visual neural transmission; flies lacking this protein are viable and fertile, but blind [16]. The human Fic protein, FicD/HYPE, AMPylates Rho GTPases Rac, Rho, and Cdc42 in vitro, but the physiological effects of FicD/HYPE activity are not known [11].
Inactivation of Kinases by Addition of UMP
There is currently one example of a Fic domain protein that inhibits the activity of its target by covalent addition of a UMP moiety instead of AMP (Figures 1 and 2). AvrAC from the bacterial plant pathogen Xanthomonas campestris infects members of the plant family Brassicaceae, including important food crops such as radishes, cauliflower, and cabbage, as well as the model organism Arabidopsis thaliana. In A. thaliana, AvrAC catalyzes the addition of UMP to the activation loop of kinase targets BIK1 and RIPK, precluding phosphorylation required for their activation [17]. Consequently, the immune response normally mediated by these two kinases is not triggered [17].
Doc Toxins and Phosphorylation
Although the Doc family of proteins belongs to the Fic/Fido family, they exhibit notable differences in their structural core and catalytic motif. Doc proteins possess only six α-helices in their core folds compared to eight in AMPylating Fic domain proteins [3], and their consensus motif, HxFx(D/N)(A/G)NKR, differs slightly from that found in canonical Fic proteins, HxFx(D/E)GNGR [18], [19] (Figure 1). All Doc family members are toxin components of Phd-Doc TA systems. TA systems are small operons that encode a stable toxin and a labile antitoxin (reviewed in [20]). Under normal conditions, the antitoxin physically interacts with the toxin to inhibit its activity. Specific stress conditions result in the degradation of the antitoxin, freeing the toxin to act on its target, typically leading to bacterial cell growth arrest [20]. In relation to pathogenesis and treatment of bacterial infections, expression of TA systems is associated with an increase in the formation of persister cells [21]. Persister cells exploit slow growth, sometimes to the point of dormancy, to avoid antibiotic-mediated killing and may contribute to the tenacity of certain bacterial pathogens that cause characteristically chronic or recurring infections.
The Doc toxin from the bacteriophage P1 TA system has served as a model for this family and is the only member of this family studied in detail. P1 Doc phosphorylates the essential elongation factor and GTPase EF-Tu, resulting in translation arrest [18], [19]. EF-Tu inactivation through a single phosphorylation event represents a highly effective conduit for regulation of growth and the formation of persister cells.
The slight variations in the conserved Fic motif of Doc, highlighted above, combined with structural differences account for its ability to catalyze a reaction distinct from transfer of an adenylyl group. Canonical Fic proteins possess a glycine at position 8 in their 9-residue Fic motif; instead, Doc family members contain a lysine at this position in the active site. This change forces ATP to bind Doc in an inverted orientation [18], leading to addition of a phosphate moiety to the target protein rather than an adenylyl moiety. Therefore, all Doc family members containing this altered signature motif should also act as kinases. These family members include Doc toxins in the chromosomes of several important pathogens including Streptococcus pneumoniae, Vibrio cholera, Clostridium tetani, and Salmonella enterica Typhimurium [22].
Phosphocholination by AnkX
The Fic domain protein AnkX is a virulence factor from Legionella pneumophila, an intracellular pathogen and the causative agent of Legionnaires' disease. Upon inhalation, L. pneumophila is taken up by alveolar macrophages and evades killing by altering phagosome–lysosome fusion [23]. The AnkX virulence factor is released into the macrophages by a type IV secretion system [24] and catalyzes the transfer of phosphocholine from CDP-choline to the Rab1 and Rab35 GTPases [25]. Since these GTPases are master regulators of vesicular trafficking, their inactivation results in the disruption of the host cell secretory pathway [26], presumably in a manner that favors establishment and maintenance of Legionella-containing vacuoles. Structural studies revealed that AnkX binds to its CDP-choline substrate in the opposite orientation of that for substrates binding canonical Fic proteins, similar to the manner in which Doc binds ATP [27]. This inverted binding is mediated by residues within two distinct domains of AnkX, the Fic domain and a domain unique to AnkX called the CMP domain. AnkX then transfers a phosphocholine moiety onto the target rather than a CMP; however, it does so using the same chemical reaction and catalytic residues as AMPylating Fic proteins [27].
AvrB and Phosphorylation
The fifth member of the Fic/Fido family is the AvrB effector protein from Pseudomonas syringae, a pathogen of plants. This domain has an α-helical core fold that is similar to both Fic and Doc proteins, but contains a drastically different nine amino acid sequence in place of the catalytic Fic motif ([3], Figure 1). After transport into plant cells via a type III secretion system [28], AvrB enhances P. syringae growth in susceptible plants [29] and causes cytotoxicity [28]. AvrB interacts with the immune regulator RIN4 and the signaling kinases MPK4 and RIPK; it also induces phosphorylation of RIN4 and MPK4 [28], [30], [31]. However, direct phosphorylation of RIN4 or MPK4 by AvrB was not observed in vitro. Therefore, the mechanism of AvrB-induced phosphorylation is still unclear.
Common Themes among Bacterial Fic Proteins
This Pearl serves as a snapshot of an evolving field that has uncovered provocative new roles for Fic proteins as modulators of bacterial virulence or persistence. Some notable themes have emerged from family members whose enzymatic activity and cellular targets are known. First, Fic proteins post-translationally modify a functionally critical amino acid through stable, covalent addition of adenosine monophosphate, uridine monophosphate, phosphocholine, or phosphate. With the exception of AvrAC, a single residue in the target protein(s) is modified; AvrAC adds UMP to a contiguous serine-threonine pair [17]. Third, the modification disrupts signaling by, or the function of, the target protein. Fic proteins never enhance the function of their target proteins, in contrast to kinases that are typically activated by modification (phosphorylation). Finally, GTPases are common targets of Fic proteins, a logical choice since they typically have pivotal roles in cell signaling and physiology. In fact, to date, GTPases are the exclusive target of Doc, AnkX, and all of the Fic proteins that AMPylate. With thousands of members of the Fic domain family, this summary undoubtedly represents the tip of the iceberg. Coupled with the complementary body of elegant structural and biophysical studies [4], future advances will surely be accelerated now that a general framework for the function of Fic proteins has been established.
Zdroje
1. EngelP, GoepfertA, StangerFV, HarmsA, SchmidtA, et al. (2012) Adenylylation control by intra - or intermolecular active-site obstruction in Fic proteins. Nature 482 : 107–110.
2. MattooS, DurrantE, ChenMJ, XiaoJ, LazarCS, et al. (2011) Comparative analysis of Histophilus somni immunoglobulin-binding protein A (IbpA) with other fic domain-containing enzymes reveals differences in substrate and nucleotide specificities. J Biol Chem 286 : 32834–32842.
3. KinchLN, YarbroughML, OrthK, GrishinNV (2009) Fido, a novel AMPylation domain common to fic, doc, and AvrB. PLoS ONE 4: e5818.
4. Garcia-PinoA, ZenkinN, LorisR (2014) The many faces of Fic: structural and functional aspects of Fic enzymes. Trends Biochem Sci 39 : 121–129.
5. YarbroughML, LiY, KinchLN, GrishinNV, BallHL, et al. (2009) AMPylation of Rho GTPases by Vibrio VopS disrupts effector binding and downstream signaling. Science 323 : 269–272.
6. ItzenA, BlankenfeldtW, GoodyRS (2011) Adenylylation: renaissance of a forgotten post-translational modification. Trends Biochem Sci 36 : 221–228.
7. WooleryAR, LuongP, BrobergCA, OrthK (2010) AMPylation: Something Old is New Again. Front Microbiol 1 : 113.
8. StadtmanER (2001) The story of glutamine synthetase regulation. J Biol Chem 276 : 44357–44364.
9. HigaN, TomaC, KoizumiY, NakasoneN, NoharaT, et al. (2013) Vibrio parahaemolyticus effector proteins suppress inflammasome activation by interfering with host autophagy signaling. PLoS Pathog 9: e1003142.
10. XuH, YangJ, GaoW, LiL, LiP, et al. (2014) Innate immune sensing of bacterial modifications of Rho GTPases by the Pyrin inflammasome. Nature E-pub ahead of print. doi:10.1038/nature13449
11. WorbyCA, MattooS, KrugerRP, CorbeilLB, KollerA, et al. (2009) The fic domain: regulation of cell signaling by adenylylation. Mol Cell 34 : 93–103.
12. ZekariasB, MattooS, WorbyC, LehmannJ, RosenbuschRF, et al. (2010) Histophilus somni IbpA DR2/Fic in virulence and immunoprotection at the natural host alveolar epithelial barrier. Infect Immun 78 : 1850–1858.
13. PulliainenAT, DehioC (2009) Bartonella henselae: subversion of vascular endothelial cell functions by translocated bacterial effector proteins. Int J Biochem Cell Biol 41 : 507–510.
14. PalaniveluDV, GoepfertA, MeuryM, GuyeP, DehioC, et al. (2011) Fic domain-catalyzed adenylylation: insight provided by the structural analysis of the type IV secretion system effector BepA. Protein Sci 20 : 492–499.
15. DehioC, SauderU, HiestandR (2004) Isolation of Bartonella schoenbuchensis from Lipoptena cervi, a blood-sucking arthropod causing deer ked dermatitis. J Clin Microbiol 42 : 5320–5323.
16. RahmanM, HamH, LiuX, SugiuraY, OrthK, et al. (2012) Visual neurotransmission in Drosophila requires expression of Fic in glial capitate projections. Nat Neurosci 15 : 871–875.
17. FengF, YangF, RongW, WuX, ZhangJ, et al. (2012) A Xanthomonas uridine 5′-monophosphate transferase inhibits plant immune kinases. Nature 485 : 114–118.
18. Castro-RoaD, Garcia-PinoA, De GieterS, van NulandNA, LorisR, et al. (2013) The Fic protein Doc uses an inverted substrate to phosphorylate and inactivate EF-Tu. Nat Chem Biol 9 : 811–817.
19. CruzJW, RothenbacherFP, MaehigashiT, LaneWS, DunhamCM, et al. (2014) Doc toxin is a kinase that inactivates elongation factor Tu. J Biol Chem 289 : 7788–7798.
20. YamaguchiY, InouyeM (2011) Regulation of growth and death in Escherichia coli by toxin-antitoxin systems. Nat Rev Microbiol 9 : 779–790.
21. MaisonneuveE, GerdesK (2014) Molecular mechanisms underlying bacterial persisters. Cell 157 : 539–548.
22. PandeyDP, GerdesK (2005) Toxin-antitoxin loci are highly abundant in free-living but lost from host-associated prokaryotes. Nucleic Acids Res 33 : 966–976.
23. FieldsBS, BensonRF, BesserRE (2002) Legionella and Legionnaires' disease: 25 years of investigation. Clin Microbiol Rev 15 : 506–526.
24. NinioS, RoyCR (2007) Effector proteins translocated by Legionella pneumophila: strength in numbers. Trends Microbiol 15 : 372–380.
25. MukherjeeS, LiuX, ArasakiK, McDonoughJ, GalanJE, et al. (2011) Modulation of Rab GTPase function by a protein phosphocholine transferase. Nature 477 : 103–106.
26. GoodyRS, ItzenA (2013) Modulation of small GTPases by legionella. Curr Top Microbiol Immunol 376 : 117–133.
27. CampanacciV, MukherjeeS, RoyCR, CherfilsJ (2013) Structure of the Legionella effector AnkX reveals the mechanism of phosphocholine transfer by the FIC domain. EMBO J 32 : 1469–1477.
28. DesveauxD, SingerAU, WuAJ, McNultyBC, MusselwhiteL, et al. (2007) Type III effector activation via nucleotide binding, phosphorylation, and host target interaction. PLoS Pathog 3: e48.
29. AshfieldT, KeenNT, BuzzellRI, InnesRW (1995) Soybean resistance genes specific for different Pseudomonas syringae avirulence genes are allelic, or closely linked, at the RPG1 locus. Genetics 141 : 1597–1604.
30. CuiH, WangY, XueL, ChuJ, YanC, et al. (2010) Pseudomonas syringae effector protein AvrB perturbs Arabidopsis hormone signaling by activating MAP kinase 4. Cell Host Microbe 7 : 164–175.
31. LiuJ, ElmoreJM, LinZJ, CoakerG (2011) A receptor-like cytoplasmic kinase phosphorylates the host target RIN4, leading to the activation of a plant innate immune receptor. Cell Host Microbe 9 : 137–146.
32. AfzalAJ, KimJH, MackeyD (2013) The role of NOI-domain containing proteins in plant immune signaling. BMC Genomics 14 : 327.
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek Out-of-Sequence Signal 3 as a Mechanism for Virus-Induced Immune Suppression of CD8 T Cell ResponsesČlánek RNF26 Temporally Regulates Virus-Triggered Type I Interferon Induction by Two Distinct MechanismsČlánek Mouse, but Not Human, ApoB-100 Lipoprotein Cholesterol Is a Potent Innate Inhibitor of Pneumolysin
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2014 Číslo 9- Jak souvisí postcovidový syndrom s poškozením mozku?
- Měli bychom postcovidový syndrom léčit antidepresivy?
- Farmakovigilanční studie perorálních antivirotik indikovaných v léčbě COVID-19
- 10 bodů k očkování proti COVID-19: stanovisko České společnosti alergologie a klinické imunologie ČLS JEP
-
Všechny články tohoto čísla
- Virus Control Goes Epigenetic
- The Role of Iron in Prion Disease and Other Neurodegenerative Diseases
- The Ins and Outs of Rust Haustoria
- Prion Strains and Amyloid Polymorphism Influence Phenotypic Variation
- Teaching Fido New ModiFICation Tricks
- Can Enhance Infection in Mosquitoes: Implications for Malaria Control?
- MIF Contributes to Associated Immunopathogenicity Development
- Persistence of Virus Reservoirs in ART-Treated SHIV-Infected Rhesus Macaques after Autologous Hematopoietic Stem Cell Transplant
- Bacillus Calmette-Guerin Infection in NADPH Oxidase Deficiency: Defective Mycobacterial Sequestration and Granuloma Formation
- EhCoactosin Stabilizes Actin Filaments in the Protist Parasite
- Molecular Insights Into the Evolutionary Pathway of O1 Atypical El Tor Variants
- LprG-Mediated Surface Expression of Lipoarabinomannan Is Essential for Virulence of
- Structural Correlates of Rotavirus Cell Entry
- Multivalent Adhesion Molecule 7 Clusters Act as Signaling Platform for Host Cellular GTPase Activation and Facilitate Epithelial Barrier Dysfunction
- The Effects of Vaccination and Immunity on Bacterial Infection Dynamics
- Myeloid Derived Hypoxia Inducible Factor 1-alpha Is Required for Protection against Pulmonary Infection
- Functional Characterisation of Germinant Receptors in and Presents Novel Insights into Spore Germination Systems
- Global Analysis of Neutrophil Responses to Reveals a Self-Propagating Inflammatory Program
- Host Cell Invasion by Apicomplexan Parasites: The Junction Conundrum
- Comparative Phenotypic Analysis of the Major Fungal Pathogens and
- Unravelling the Multiple Functions of the Architecturally Intricate β-galactosidase, BgaA
- Sialylation of Prion Protein Controls the Rate of Prion Amplification, the Cross-Species Barrier, the Ratio of PrP Glycoform and Prion Infectivity
- Symbionts Commonly Provide Broad Spectrum Resistance to Viruses in Insects: A Comparative Analysis of Strains
- Ontogeny of Recognition Specificity and Functionality for the Broadly Neutralizing Anti-HIV Antibody 4E10
- Identification and Characterisation of a Hyper-Variable Apoplastic Effector Gene Family of the Potato Cyst Nematodes
- Crimean-Congo Hemorrhagic Fever Virus Entry into Host Cells Occurs through the Multivesicular Body and Requires ESCRT Regulators
- Age-Dependent Enterocyte Invasion and Microcolony Formation by
- CD160-Associated CD8 T-Cell Functional Impairment Is Independent of PD-1 Expression
- Functional Fluorescent Protein Insertions in Herpes Simplex Virus gB Report on gB Conformation before and after Execution of Membrane Fusion
- The Tudor Domain Protein Spindlin1 Is Involved in Intrinsic Antiviral Defense against Incoming Hepatitis B Virus and Herpes Simplex Virus Type 1
- Transgenic Analysis of the MAP Kinase MPK10 Reveals an Auto-inhibitory Mechanism Crucial for Stage-Regulated Activity and Parasite Viability
- Evidence for a Transketolase-Mediated Metabolic Checkpoint Governing Biotrophic Growth in Rice Cells by the Blast Fungus
- Incomplete Deletion of IL-4Rα by LysM Reveals Distinct Subsets of M2 Macrophages Controlling Inflammation and Fibrosis in Chronic Schistosomiasis
- Identification and Functional Expression of a Glutamate- and Avermectin-Gated Chloride Channel from , a Southern Hemisphere Sea Louse Affecting Farmed Fish
- Out-of-Sequence Signal 3 as a Mechanism for Virus-Induced Immune Suppression of CD8 T Cell Responses
- Strong Epistatic Selection on the RNA Secondary Structure of HIV
- Hematopoietic but Not Endothelial Cell MyD88 Contributes to Host Defense during Gram-negative Pneumonia Derived Sepsis
- Delineation of Interfaces on Human Alpha-Defensins Critical for Human Adenovirus and Human Papillomavirus Inhibition
- Exploitation of Reporter Strains to Probe the Impact of Vaccination at Sites of Infection
- RNF26 Temporally Regulates Virus-Triggered Type I Interferon Induction by Two Distinct Mechanisms
- Helminth Infections Coincident with Active Pulmonary Tuberculosis Inhibit Mono- and Multifunctional CD4 and CD8 T Cell Responses in a Process Dependent on IL-10
- MHC Class II Restricted Innate-Like Double Negative T Cells Contribute to Optimal Primary and Secondary Immunity to
- Reactive Oxygen Species Regulate Caspase-11 Expression and Activation of the Non-canonical NLRP3 Inflammasome during Enteric Pathogen Infection
- Evolution of Plastic Transmission Strategies in Avian Malaria
- A New Human 3D-Liver Model Unravels the Role of Galectins in Liver Infection by the Parasite
- Translocates into the Myocardium and Forms Unique Microlesions That Disrupt Cardiac Function
- Mouse, but Not Human, ApoB-100 Lipoprotein Cholesterol Is a Potent Innate Inhibitor of Pneumolysin
- The Cofilin Phosphatase Slingshot Homolog 1 (SSH1) Links NOD1 Signaling to Actin Remodeling
- Kaposi's Sarcoma Herpesvirus MicroRNAs Induce Metabolic Transformation of Infected Cells
- Reorganization of the Endosomal System in -Infected Cells: The Ultrastructure of -Induced Tubular Compartments
- Distinct Dictation of Japanese Encephalitis Virus-Induced Neuroinflammation and Lethality via Triggering TLR3 and TLR4 Signal Pathways
- Exploitation of the Complement System by Oncogenic Kaposi's Sarcoma-Associated Herpesvirus for Cell Survival and Persistent Infection
- The Secreted Peptide PIP1 Amplifies Immunity through Receptor-Like Kinase 7
- Structural Insight into Host Recognition by Aggregative Adherence Fimbriae of Enteroaggregative
- The CD14CD16 Inflammatory Monocyte Subset Displays Increased Mitochondrial Activity and Effector Function During Acute Malaria
- Infection Induces Expression of a Mosquito Salivary Protein (Agaphelin) That Targets Neutrophil Function and Inhibits Thrombosis without Impairing Hemostasis
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- The Secreted Peptide PIP1 Amplifies Immunity through Receptor-Like Kinase 7
- Symbionts Commonly Provide Broad Spectrum Resistance to Viruses in Insects: A Comparative Analysis of Strains
- MIF Contributes to Associated Immunopathogenicity Development
- The Ins and Outs of Rust Haustoria
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



