-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaUnravelling the Multiple Functions of the Architecturally Intricate β-galactosidase, BgaA
The adherence of bacteria to host cells is a critical step in most bacterial infections; yet, mechanisms are poorly understood for many bacteria, including Streptococcus pneumoniae (pneumococcus), a human pathogen of global relevance. The surface of this bacterium is decorated with a landscape of large and structurally sophisticated proteins that mediate contact with the host. Here we show that the sugar-degrading β-galactosidase BgaA, can bind and cleave sugars through separate portions of this protein, which is one of the largest pneumococcal surface proteins and a model for architecturally intricate carbohydrate-active surface proteins. Non-enzymatic carbohydrate-binding modules in BgaA mediate adherence to specific host-cell surface carbohydrates. The identification of the first adhesin-carbohydrate receptor pair in S. pneumoniae provides critical molecular-level support for the long-held hypothesis that pneumococci bind carbohydrates on host cells and extends the paradigm of carbohydrate-binding module function. The enzymatically active portion of BgaA enables the bacterium to grow on host-derived glycans and evade the immune system, aspects of the host-pathogen interaction we show can be modulated by a specific inhibitor of enzymatic activity. Our work advances the concept that large bacterial surface proteins mediate complex host-bacterial interactions through specific functions of the varied regions comprising these proteins.
Published in the journal: . PLoS Pathog 10(9): e32767. doi:10.1371/journal.ppat.1004364
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1004364Summary
The adherence of bacteria to host cells is a critical step in most bacterial infections; yet, mechanisms are poorly understood for many bacteria, including Streptococcus pneumoniae (pneumococcus), a human pathogen of global relevance. The surface of this bacterium is decorated with a landscape of large and structurally sophisticated proteins that mediate contact with the host. Here we show that the sugar-degrading β-galactosidase BgaA, can bind and cleave sugars through separate portions of this protein, which is one of the largest pneumococcal surface proteins and a model for architecturally intricate carbohydrate-active surface proteins. Non-enzymatic carbohydrate-binding modules in BgaA mediate adherence to specific host-cell surface carbohydrates. The identification of the first adhesin-carbohydrate receptor pair in S. pneumoniae provides critical molecular-level support for the long-held hypothesis that pneumococci bind carbohydrates on host cells and extends the paradigm of carbohydrate-binding module function. The enzymatically active portion of BgaA enables the bacterium to grow on host-derived glycans and evade the immune system, aspects of the host-pathogen interaction we show can be modulated by a specific inhibitor of enzymatic activity. Our work advances the concept that large bacterial surface proteins mediate complex host-bacterial interactions through specific functions of the varied regions comprising these proteins.
Introduction
The cell surfaces of bacterial pathogens are complex landscapes of molecules that create an elaborate interface between the host and the bacterium. Integral to this landscape are cell-surface presented proteins that provide a variety of functions from cellular maintenance to communicating with the external environment to interaction with host tissues. A common feature of these proteins, particularly in Gram-positive bacteria, is their very large size and structural sophistication. These architecturally intricate proteins are also often functionally complex and thereby contribute to different aspects of pathogenesis.
Carbohydrate-active enzymes (CAZymes), particularly those that break glycosidic bonds joining sugar residues, are frequently found on the surface of bacterial species and are commonly architecturally intricate. By definition these enzymes contain a catalytic domain that confers the ability to break glycosidic bonds; the most common super-family is the glycoside hydrolases (GH), which are further organized into families based on sequence similarity [1]. GHs often contain numerous ancillary modules, the most common of which are the carbohydrate-binding modules (CBM) that non-catalytically mediate enzyme-carbohydrate interactions [2], [3]. The paradigm of CBM function has been that these modules concentrate enzymes onto carbohydrate substrates and, through this local concentration effect, enhance catalytic activity. This, however, has been based largely on non-surface attached enzyme systems.
CAZymes have been a focus of study for the opportunistic pathogen Streptococcus pneumoniae (pneumococcus), a leading cause of pneumonia, bacteremia and meningitis. Pneumococci express eight known surface CAZymes that have distinct specificities and that together can modify a wide range of host glycans including N-linked glycans, O-linked glycans and glycosaminoglycans [4]–[6]. S. pneumoniae expresses several exoglycosidases that cleave terminal carbohydrates. Neuraminidase NanA cleaves terminal α-2,3 and α-2,6 linked sialic acid, while pneumococci express two β-galactosidases BgaA, specific for terminal galactose (Gal) β-1,4 linked to N-acetylglucosamine (GlcNAc) or glucose and BgaC, specific for terminal galactose β-1,3 linked to GlcNAc [7]–[9]. N-acetylglucosaminidase StrH, contains two GH20 catalytic modules that both recognize terminal GlcNAc residues that are β-1,2 linked to mannose within complex N-linked glycans, but have subtle differences in enzymatic activity [10], [11]. S. pneumoniae also expresses endoglycosidases EndoD, an Endo-β-N-acetylglucosaminidase, which cleaves the chitobiose core of N-linked glycans and Eng which cleaves the core-1 (Galβ-1,3 N-acetylgalactosamine) structure of O-linked glycans [12], [13]. S. pneumoniae also expresses α-glucanase SpuA, which has specificity for α-1,6 linkages of glucose in the context of stretches of α-1,4 linked glucose, such as those found in glycogen [14], [15]. Finally, pneumococci express a hyaluronate lyase (Hyl) that cleaves the β-1,4 linkage of hyaluronic acid by β-elimination [4], [16], [17]. Several of these enzymes contribute to the ability of pneumococci to colonize or cause disease in vivo [6], [18]–[21]. Furthermore, many of these CAZymes have been shown, using in vitro assays, to contribute to specific steps in pathogenesis including growth, avoidance of clearance by the immune system, adherence and biofilm formation [6], [15], [19], [20], [22]–[31].
Pneumococcal surface-associated glycosidases are multimodular suggesting that they have complex interactions with soluble glyconjugates, mucin layers, and/or the glycocalyx layer that coats mammalian cells. At over 2200 amino acids and with at least 17 modules/domains of 7 different types the β-galactosidase BgaA, is among the largest cell surface attached proteins expressed by S. pneumoniae [7], [32] (Figure 1A). At present, none of the individual modules of BgaA have been ascribed functions and the functions of similar modules in other proteins, except the predicted catalytic module, remain ambiguous. The gene encoding BgaA is present in all sequenced pneumococcal strains and all strains tested possess β-galactosidase activity [32]–[35]. BgaA is specific for galactose β-1,4-linked to glucose or GlcNAc [lactose or N-acetyllactosamine (LacNAc) motifs, respectively] found in glycoconjugates. This activity is required for the release of galactose from N-linked glycans and for efficient growth on glycoconjugates having these modifications [5], [7], [22], [36], [37]. BgaA is also linked to pneumococcal resistance against complement deposition and the resulting phagocytic killing [23] and strongly involved with adherence to epithelial cells [31]. At present, deeper insight into the complex biological roles that BgaA plays is hindered by an absence of studies that relate the complex architecture of this enzyme to its varied functions.
Fig. 1. The structural features of BgaA from S. pneumoniae. 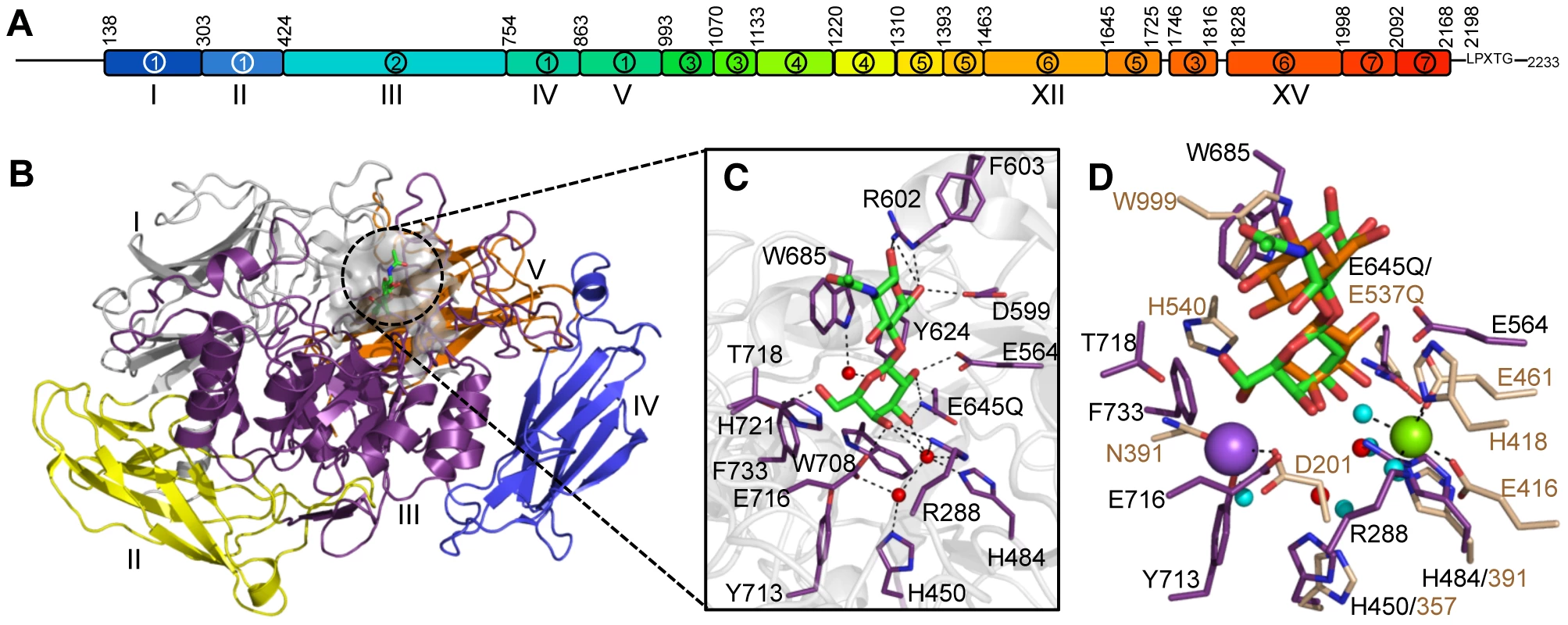
(A) The architecture of BgaA showing the 17 modules/domains defined on the basis of fold recognition using the Phyre2 server [39]. The 7 different modules/domains are labeled with Arabic numbers: 1, sequence similarity to GH2-associated Ig-like; 2, sequence similarity to GH2 (β/α)8-barrel; 3, fold similarity to PDB ID 2LY7 (>98% confidence); 4, fold similarity to a fragment of a bacterial invasin (95% confidence); 5, fold similarity to bacterial Ig-like modules (98% confidence); 6, predicted β-sandwich fold similar to that of family 32 CBMs (99.5% confidence); 7, fold similarity to pneumococcal G5 modules (>98% confidence). The LPXTG cell wall anchoring motif is shown. The modules/domains that are the focus of this study are labeled beneath the schematic with Roman numerals. Amino acid numbering for the module/domain boundaries is given above the schematic. (B) Cartoon representation of the structure of the catalytic region comprising domains I–V (colored sequentially as gray, yellow, purple, blue, and orange). The bound LacNAc molecule is shown as green sticks and the surface of the active site in transparent gray. (C) Specific interactions of the BgaA active site with LacNAc (green). Water molecules are shown as red spheres and hydrogen bonds as dashed lines. (D) Overlap of the BgaA active site (purple stick representation for side chains, green sticks for LacNAc, and red spheres for waters) with the active site of E. coli LacZ in complex with lactose (tan stick representation for side chains, orange sticks for lactose, and blue spheres for waters, green sphere for Mg2+, and purple sphere for Na+; PDB ID 1JYN). In this study, the varied biological functions of BgaA are deconvoluted from the complex architecture of this enzyme. Through detailed structural and functional analyses the molecular basis for the catalytic specificity of BgaA is defined and this activity is demonstrated as critical for the ability of pneumococci to utilize complex N-linked glycans as a carbon source and protect the bacterium from opsonophagocytic killing. Further analyses also revealed the presence of non-catalytic CBMs within the C-terminal region of BgaA that mediate adherence to host cell surface LacNAc and/or lactose. Notably, this is the first demonstration that a CBM within a CAZyme can mediate adherence of a pathogen to host cells, thus extending the paradigm of CBM function. As CBM containing CAZymes are on the surface of many other bacterial species, we hypothesize that BgaA is a member of a novel class of adhesins. Furthermore, we show that these functions can be specifically modulated with small molecule inhibitors or competitors. Together these data highlight that understanding the architectural sophistication of surface-attached proteins can increase our understanding of the different mechanisms by which these proteins can contribute to bacterial pathogenesis and potentially aid in the development of strategies to inhibit these pathogenic mechanisms.
Results
Structure and inhibition of the BgaA catalytic region
The N-terminal region of BgaA comprising amino acid residues 138–993 has amino acid sequence identity with GH family 2 enzymes. The X-ray crystal structures of a catalytically active fragment of BgaA was determined to 2.7 Å resolution (data not shown) and an inactive Glu645Gln nucleophile mutant in complex with unhydrolyzed LacNAc to 2.2 Å resolution (Figure 1B and Figure S1A). This polypeptide had five distinct domains, four with immunoglobulin (Ig)-like folds that are arranged to create a nest in which the central (α/β)8-barrel domain III sits. All other structurally characterized GH2 enzymes with known β-galactosidase activity have a LacZ-type architecture where Ig-like domain V is replaced by a super β-sandwich domain (Figure S1B). The catalytic site of BgaA resides in a pocket located at the center of domain III (Figure 1B) and makes a series of direct and water-mediated hydrogen bonds with both residues of the disaccharide while the a-face of the GlcNAc residues lies parallel to Trp685 in a classical carbohydrate ring-aromatic amino acid sidechain interaction (Figure 1C). The LacNAc in this complex does not fully engage the catalytic residues: neither Glu564, the acid base, nor Gln645, mutated from the glutamic acid that would normally act as the nucleophile, are appropriately positioned to perform a catalytic function. This “shallow” mode of substrate binding representing an active site loading step is the same as that observed for Escherichia coli LacZ Glu537Gln mutant in complex with lactose (Figure 1D). The positions of the nucleophile and catalytic acid, Glu645 and Glu564, respectively, in BgaA are conserved with the analogous residues Glu537 and Glu461 of LacZ. Notably, however, there was no evidence of bound metals in the active site of BgaA. Indeed, the side chain of Arg288 occupies the space where a Mg2+ atom is bound in LacZ while Tyr713 and Glu716 fill the region occupied by a Na+ atom. Consistent with this, the activity of our catalytic region construct displayed no sensitivity to the presence or absence of metal ions (data not shown).
Given the shallow loading mode of LacNAc binding we also examined the binding of BgaA to the galactoisofagomine (GIF) and galactonojirimycin (GNJ), which are known potent galactosidase inhibitors [38] to provide additional insight into sugar recognition. GIF (Figure 2A) had a Ki of 25.0 (±4.4) nM (Figure 2B), displayed a competitive mode of inhibition (Figure 2C), and isothermal titration calorimetry (ITC; Figure S2A) confirmed the tight binding [Kd of 26.0 (±3.2) nM] and 1∶1 stoichiometry. GNJ (Figure 2D) had a more moderate Ki at 33.9 (±1.6) µM (Figure 2E) and also displayed a competitive mode of inhibition (Figure 2F). Despite the different chemical structures of the inhibitors they bound with very similar sets of interactions with Glu645 positioned beneath the atom equivalent to C1 at a distance of ∼3.5 Å, consistent with the role of this residue as a nucleophile (Figure 2G). GIF binding results in only subtle structural changes compared with LacNAc binding, despite the deeper binding mode of GIF (Figure 2H). These two complexes appear to represent a trajectory that progresses through a substrate-loading mode to a mode where the −1 catalytic subsite is fully engaged. The catalytically non-productive loading mode appears to provide BgaA with its substrate specificity through a pre-(−1)-subsite that recognizes terminal galactose residues and a preceding pre-(+1)-subsite that accommodates the β-1,4-linked GlcNAc residue through primary interactions with Trp685 and a series of hydrogen bonds between O6 of this sugar residue and a tailored pocket (Figure 2H). The steric constraints imposed by this architecture legislate against β-1,6-linked GlcNAc, with its longer overall length, and β-1,3-linked GlcNAc, where the 2-acetamido group would clash with the O6-specific pocket in the active site. Indeed, BgaA has insignificant activity on these sugars. This substrate-loading mode does not, however, suggest a mechanism for discrimination between lactose and LacNAc, where the latter is preferred by a factor of ∼10-fold [37]. It is possible that additional specificity for the 2-acetamido group of the GlcNAc is provided in the transition from the loading mode to fully involving the catalytic site and formation of the Michaelis complex where a deeper binding mode and/or distortion of the substrate might result in the engagement of this chemical group.
Fig. 2. Characterization of BgaA inhibitors. 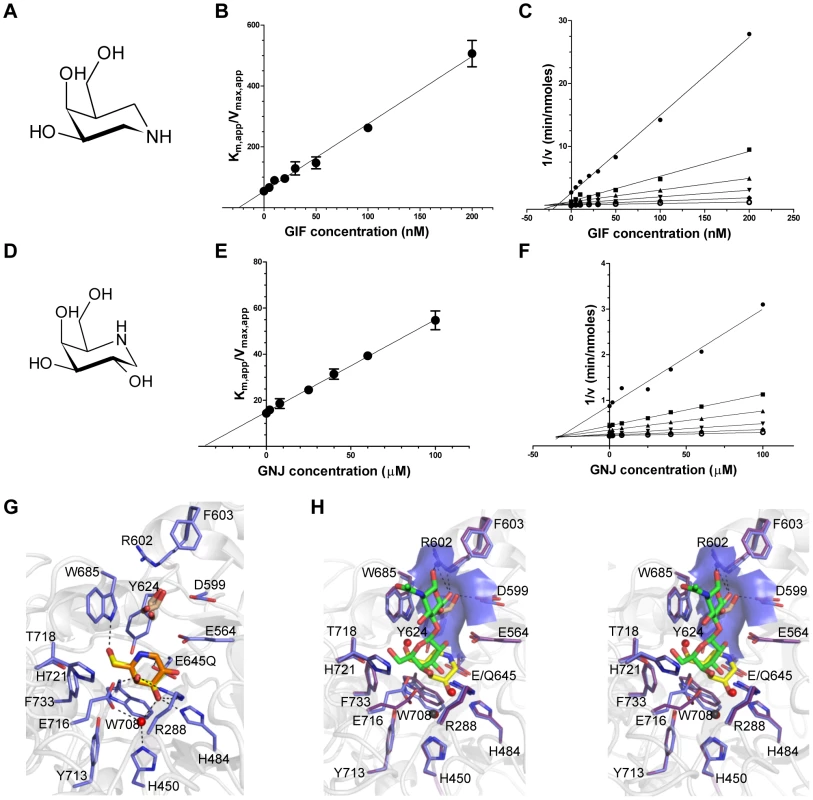
(A) and (D) The chemical structures of GIF and GNJ, respectively. (B) and (E) Plots of the apparent Km against GIF and GNJ concentration, respectively. Solid lines represent the best fits from linear regression analysis. (C) and (F) Dixon plots of the data shown in panels (B) and (E) for GIF and GNJ, respectively. (G) Specific interactions of the BgaA active site with GIF (yellow sticks) and GNJ (orange sticks) with the protein from the GIF complex shown. Water molecules are shown as red spheres, ethylene glycol molecules as brown sticks, and hydrogen bonds as dashed lines. (H) Divergent stereo view of an overlap of the BgaA LacNAc complex (purple stick representation for side chains, green sticks for LacNAc, and red spheres for waters) with the BgaA GIF complex (blue stick representation for side chains, yellow sticks for LacNAc, brown sticks for a bound ethylene glycol, and black spheres for waters). The surface of the pocket accommodating the O6 of the GlcNAc residue is shown as transparent blue. BgaA catalytic activity is required for efficient growth on glycoconjugates and immune evasion, but not adherence
As previously observed, deletion of bgaA resulted in significantly reduced growth on N-linked glycans decorating glycoproteins [22] (Figure 3A). The addition of 1 µM GIF reduced the growth of TIGR4 on aisalofetuin to approximately that of the bgaA mutant (Figure 3A). The reduction of growth by GIF was dose dependent with an inhibitor concentration ∼75 nM giving half the maximum reduction in growth, which is consistent with the measured Ki and Kd values (Figure 3B).
Fig. 3. Inhibition of S. pneumoniae BgaA. 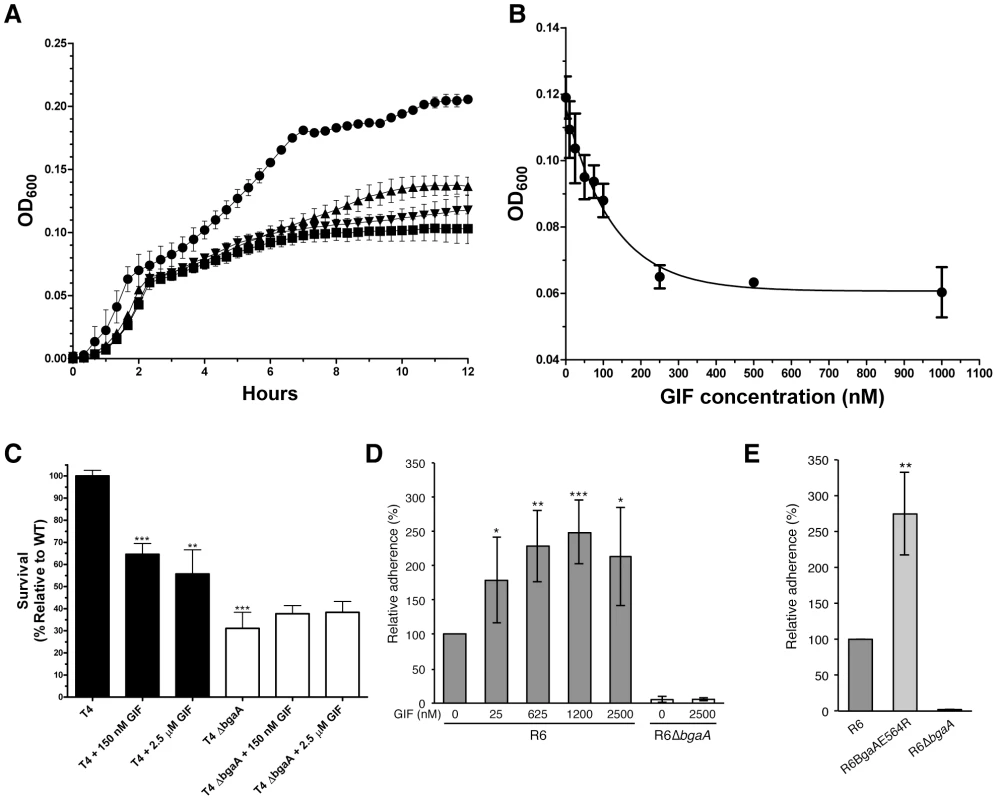
(A) Growth curves of S. pneumoniae (TIGR4) performed using a semi-defined medium supplemented with bovine asialofetuin. Circles represent growth of the TIGR4 strain, triangles the growth of TIGR4 strain in the presence of 1000 nM GIF, and inverted triangles the growth of the ΔbgaA strain. Also shown as squares is the growth of the TIGR4 strain in the absence of a carbon source. Error bars represent the standard deviation of triplicate experiments run in parallel. The experiment was performed three times with highly similar results. (B) Inhibition of growth on asialofetuin as a function of GIF concentration. Results represent the mean measurements of three independent experiments where culture densities were taken at 6 hr of culture growth. The error bars represent the standard deviations of the independent measurements. (C) Survival of S. pneumoniae TIGR4 in neutrophil killing assays, showing comparisons of wild-type (filled bars) and ΔbgaA strain (open bars) in the presence and absence of GIF. Asterisks above sample bars represent statistical comparison of that sample with the reference, which is the TIGR4 strain with no inhibitor. Data are mean values compiled from two independent experiments performed in duplicate ± standard deviation. The ΔbgaA samples with inhibitors were compared with ΔbgaA in the absence of inhibitors and were found to have p values>0.1 and thus were not significantly different. (D) Addition of GIF (25–2500 nM) significantly increases adherence of S. pneumoniae R6 to D562 cells. Data are the mean ± SD of four independent experiments performed in triplicate. Asterisks above sample bars represent statistical comparison of R6 and R6 + GIF. (E) Adherence of an S. pneumoniae strain expressing enzymatically inactive BgaA (R6BgaAE564R) to D562 cells is significantly higher than the adherence of parental strain (R6). Data are the mean ± SD of three independent experiments performed in triplicate. Asterisks above sample bars represent statistical comparison between R6 and R6BgaAE564R. Statistically significant differences were assessed using a two-tailed Student's t-tests. * p≤0.05, ** p≤0.007, *** p≤0.0007. As previously reported, the survival of the bgaA mutant in an opsonophagocytic killing assay was reduced to ∼30% of that of the parental strain (Figure 3C) [23]. The addition of 150 nM or 2.5 µM GIF to the assays significantly reduced the survival of TIGR4 to ∼60%; GIF had no significant influence on the survival of the bgaA mutant (Figure 3C).
It has previously been reported that bgaA mutants in some genetic backgrounds including R6, but not TIGR4, were significantly reduced in adherence [31]. Consistent with the published data we observed a significant reduction in adherence of an R6 bgaA mutant to epithelial cells (Figure 3D). GIF did not reduce adherence of the R6 strain and indeed caused a significant increase in adherence in a dose dependent manner (Figure 3D). The concentration of GIF giving an approximately 50% increase in adherence was ∼25 nM, again consistent with the Ki determined for this inhibitor. GIF treatment resulted in a decrease in β-galactosidase activity associated with the bacterium indicating effective inhibition of BgaA catalytic activity (Figure S2D). This observation of increased adherence by inactivation of BgaA β-galactosidase activity was further supported by a similar increase in adherence of a mutant where substitution of the catalytic acid base residue, Glu564, by a bulky arginine residue to block the −1 subsite destroyed the catalytic activity of the enzyme (Figure 3E and Figure S2E). Thus, BgaA requires neither β-galactosidase activity nor an accessible active site to mediate adherence. Remarkably, the catalytic activity is in fact antagonistic to adherence.
Together these results show that the catalytic activity of BgaA is required for nutrient acquisition by this enzyme and protection from the innate immune system. Furthermore, these biological roles can be specifically inhibited by targeting the catalytic activity with an inhibitor. The mechanism by which the catalytic activity of BgaA provides protection from complement-mediated killing is presently unknown; however, it appears to be related to an effect of glycan modification, likely on complement components, that reduces complement deposition. In contrast, the catalytic activity of BgaA appears to inhibit adherence, suggesting that the portion of BgaA that mediates adherence is distinct from the catalytic site and, further, that the receptor may be a substrate for the BgaA catalytic region, and therefore a carbohydrate.
The C-terminal region of BgaA is sufficient to facilitate pneumococcal adherence
To test the hypothesis that the C-terminal region of BgaA mediates adherence, pneumococcal strains expressing either a surface-associated BgaA C-terminal region (BgaAC) or a surface-associated BgaA N-terminal enzymatic module (BgaAN) were constructed in strains previously used to demonstrate a role for BgaA in adherence, R6 and a low passage clinical isolate C06_18 (Figure 4A). For both strain backgrounds significantly higher adherence of the BgaAC strain as compared to the bgaA mutant to normal human bronchial epithelial (NHBE) cells and the pharyngeal cell line Detroit 562 (D562) was observed (Figures 4B, 4C, S3A and S3Bcbm). In contrast, BgaAN strains showed no significant difference in adherence from that of the bgaA mutant. An immunoblot was used to confirm that the N-terminal construct was properly expressed and localized (Figure S3C). Despite appropriate expression and localization, R6BgaAN had reduced β-galactosidase activity (Figure S3D); however, reduced adherence of R6BgaAN could not be attributed to reduced enzyme activity as catalytically inactive BgaA still facilitates efficient adherence (Figure 3E) [31].
Fig. 4. The C-terminal region of BgaA is sufficient to facilitate BgaA mediated pneumococcal adherence. 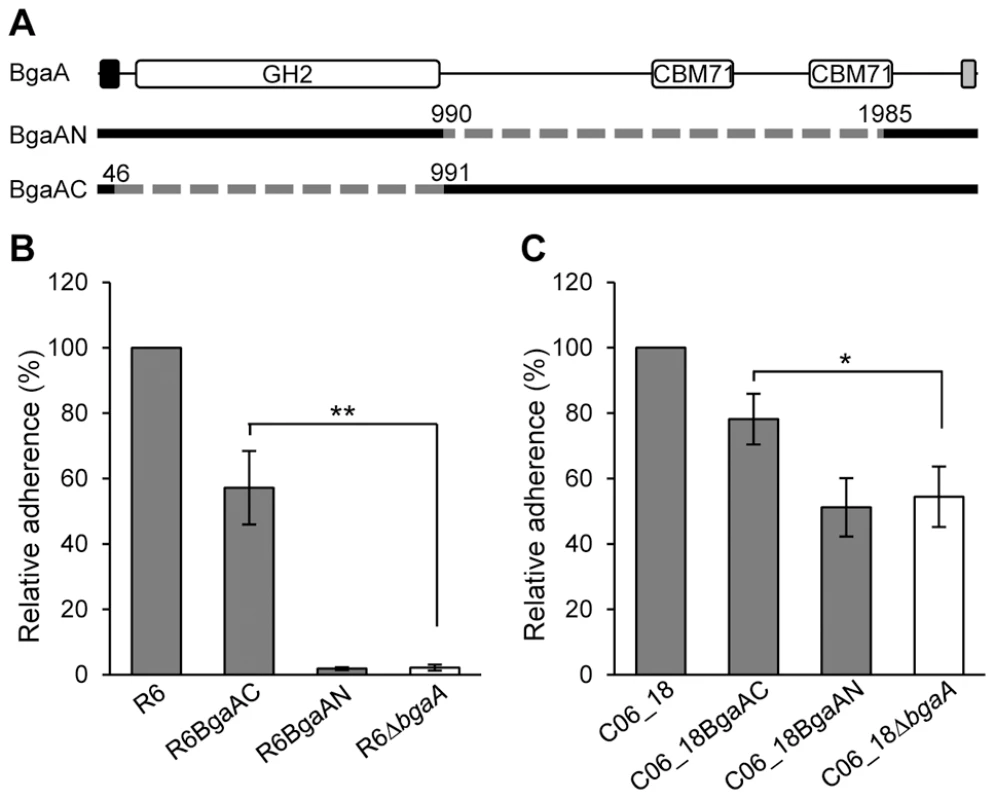
(A) Schematic indicating regions of BgaA deleted to generate BgaAN and BgaAC. Dashed lines indicate regions deleted and numbers indicate amino acid boundaries of the deletions. The signal sequence, shown as the black box, and the Gram positive anchor domain, shown as a grey boxβ, were retained in both constructs for proper localization of expressed proteins. (B) R6BgaAC has significantly higher adherence to NHBE cells as compared to R6ΔbgaA. Adherence of R6BgaAN is not significantly different when compared to R6ΔbgaA. (C) CO6_18BgaAC has significantly higher adherence to NHBE cells as compared to CO6_18ΔbgaA. Adherence of CO6_18BgaAN is not significantly different when compared to CO6_18ΔbgaA. Adherence data are the means ± SD of three independent experiments performed in triplicate. Statistically significant differences were assessed using a two-tailed Student's t-tests. * p≤0.03, ** p≤0.002. The significant increase in adherence previously discerned in the absence of BgaA enzymatic activity was not observed for pneumococci expressing only the C-terminal region of BgaA, while the reason for this is unclear it may be that the large deletion affects surface presentation or stability of the protein. Nevertheless, these data indicate that the C-terminal region of BgaA mediates adherence to receptors on the epithelial cell surface. Furthermore, the observation that the catalytic activity of BgaA is antagonistic to adherence suggests that the receptor for BgaA adherence is the carbohydrate substrate of the catalytic domain.
The C-terminal region of BgaA contains two CBMs that bind lactose and LacNAc
Amino acid sequence similarity searches failed to identify candidate CBMs in the C-terminal portion of BgaA. However, fold prediction using the Phyre2 server [39], which does not rely on amino acid sequence similarity, distinguished two regions (XII and XV, Figure 1A) with a high probability of adopting the β-sandwich fold common to many CBMs found in CAZymes. These two ∼175 amino acid residue modules, which we refer to as CBM71-1 and CBM71-2, share ∼35% amino acid identity with one another, but have no identity with known CBMs. The two predicted CBMs were recombinantly produced and the polypeptides screened for binding to all commonly occurring monosaccharides by UV difference spectroscopy; only D-galactose gave a signature UV difference spectrum consistent with sugar binding (Figures S4A and S4B). Subsequently, this approach was expanded to the relevant galactose-containing sugars LacNAc, lactose, galactopyranosyl-β-1,3-N-acetyl-D-glucosamine (lacto-N-biose), and galactopyranosyl-β-1,3-N-acetyl-D-galactosamine [Thomsen-Freidenreich (TF) epitope] and binding was only observed to LacNAc and lactose. For both CBMs, the binding to galactose was too weak to quantify. The dissociation constants (Kds) determined for CBM71-1 by ITC were 251 (±29) µM and 368 (±52) µM for LacNAc and lactose, respectively (Figures S4C and S4D). Similar values of 247 (±37) µM and 378 (±30) µM for LacNAc and lactose, respectively, were obtained for CBM71-2 (Figures S4E and S4F). Significantly, the CBMs only bound with significant affinity to sugars that are substrates for the catalytic domain. Though relatively weak, these affinities are consistent with those determined for other CBMs with similar binding specificities [40].
The structure of CBM71-1 solved by X-ray crystallography in complex with LacNAc revealed its β-sandwich fold comprising opposing sheets of 4 - and 5-anti-parallel β-strands (Figure 5A). A single bound metal ion was modeled as Ca2+ on the basis of coordination geometry and B-factor analysis. The shallow LacNAc binding site sits at the apex of the β-fold opposite the N - and C-termini (Figure 5A). The structure of CBM71-2 is highly similar to that of CBM71-1 with the most obvious difference being an extended loop adjacent to the binding site (Figure 5B). Though a bound complex of CBM71-2 was not obtained the binding sites of the two CBMs are very well conserved, consistent with the shared specificity of the CBMs and similar binding affinities (Figure 5C).
Fig. 5. Structures of the CBM71 modules in BgaA. 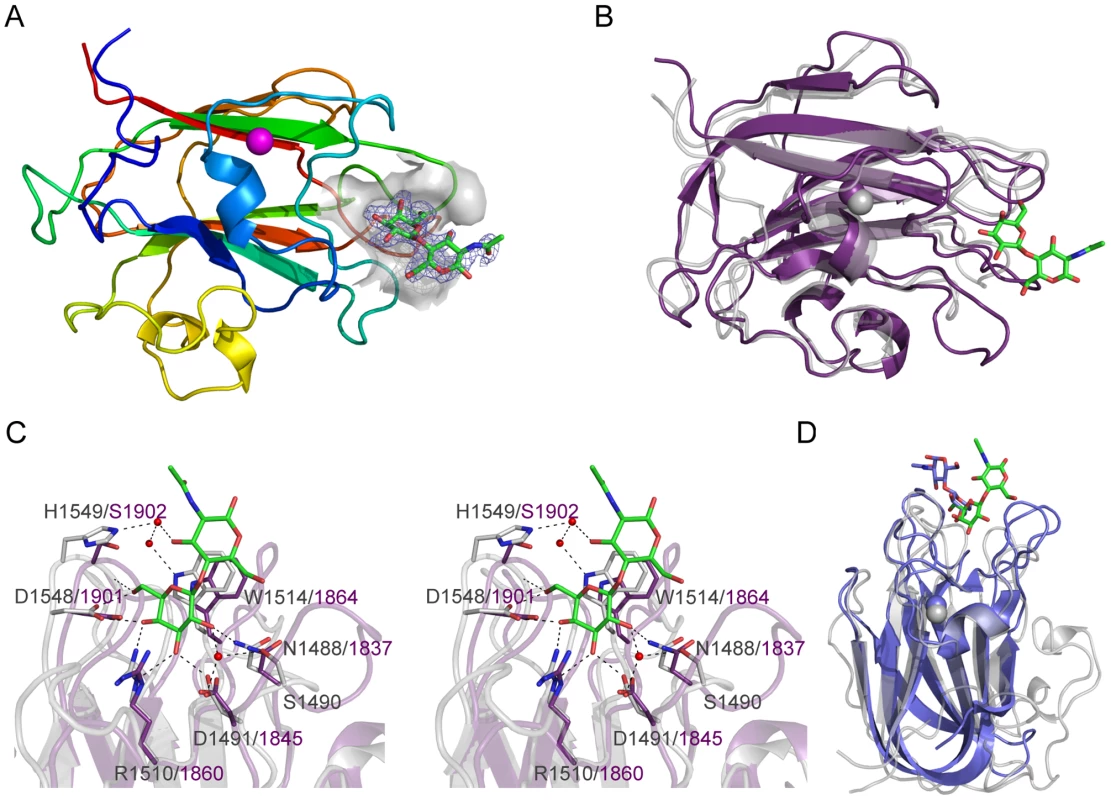
(A) A cartoon representation of CBM71-1 in complex with LacNAc. The protein is color ramped red to blue from the N-terminus to the C-terminus. A bound calcium atom is shown in magenta. The binding site of the CBM is shown as a grey transparent surface with the bound LacNAc molecule shown as green stick. The electron density for the LacNAc is shown as a blue-mesh Fo-Fc maximum-likelihood/σA-weighted map contoured at 3σ (0.33 e−/Å3). (B) An overlap of the structure of CBM71-2 (purple) with CBM71-1 (grey). The LacNAc molecule bound to CBM71-1 is shown as green sticks. (C) Expanded view of the CBM71 binding site shown in divergent stereo. CBM71-1 is shown in grey with the LacNAc shown as green sticks, residues involved in binding the LacNAc as grey sticks, and the interacting water network as red spheres. Black dashed lines represent potential hydrogen bonds. CBM71-2 is shown in purple; residues conserved with CBM71-2 are shown as purple sticks. (D) An overlap of the CBM71-1 LacNAc complex (grey with LacNAc in green) with the CBM32 from C. perfringens NagJ (blue with bound LacNAc shown as blue sticks). The base of the CBM71-1 active site provides amino acid sidechains that provide specificity for a terminal galacto-configured sugar but prevent accommodation of a 2-acetamido group, providing an explanation for the lack of binding to N-acetylgalactosamine (Figure 5C). Tryptophan 1514 lies directly beneath the glycoside bond and coplanar with the disaccharide thus providing CH-π interactions with both pyranose rings and a higher affinity for β-linked disaccharides than for galactose alone. This binding site architecture accommodates lactose and LacNAc, but would limit the recognition of other sugars terminating in β-linked galactose. Given their carbohydrate binding activity, but lack of amino acid sequence identity between the BgaA CBMs and known CBM families, CBM71-1 and CBM71-2 constitute the founding members of a new CBM family, CBM71, which is most similar in three-dimensional structure to CBM family 32 (Figure 5D).
CBMs in BgaA mediate pneumococcal adherence
The ability of these CBMs to mediate adherence to host cells was explored using the free carbohydrates galactose, lactose and LacNAc as well as soluble recombinant CBMs as specific competitors of adherence. The addition of 250 µM CBM71-1 or CBM71-2 significantly reduced adherence of R6 and C06_18 to both NHBE and D562 cells (Figures 6A, 6B, S5A and S5B). The CBM71-1.2 tandem construct that comprises both CBMs and the two intervening modules reduced adherence more than either CBM alone; although, this difference was not significant for CO6_18. Importantly, recombinant CBMs had no significant effect on adherence of a bgaA mutant, demonstrating that the effect of CBMs on adherence was BgaA specific (Figures 6A, 6B, S5A and S5B).
Fig. 6. Recombinant CBMs and LacNAc and/or lactose reduce pneumococcal adherence in a BgaA-dependent manner. 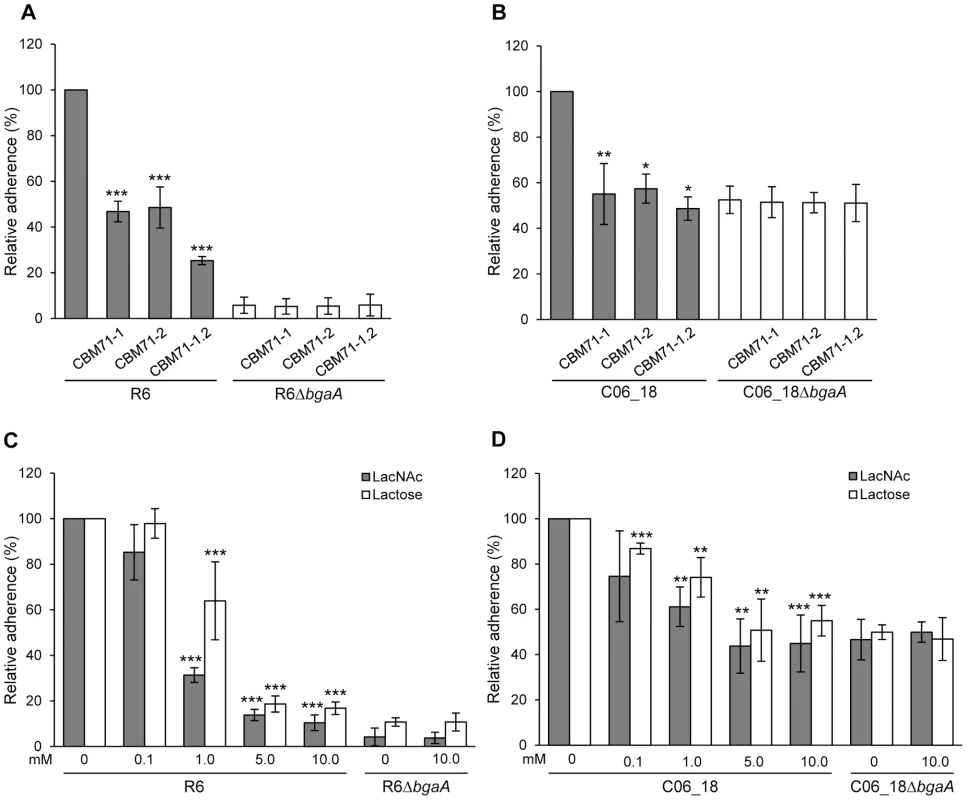
(A) Adherence of S. pneumoniae strain R6 and R6ΔbgaA to NHBE cells in the presence of CBM71-1, CBM71-2 or CBM71-1.2 (250 µM). Asterisks indicate significant differences in adherence in the presence or absence of CBM. (B) Adherence of S. pneumoniae strain C06_18 and C06_18ΔbgaA to NHBE cells in the presence of CBM71-1, CBM71-2 or CBM71-1.2 (250 µM). (C) Adherence of S. pneumoniae strain R6 and R6ΔbgaA to NHBE cells in the presence of LacNAc and lactose (0–10 mM). Asterisks indicate significant differences in adherence in the presence or absence of disaccharide. (D) Adherence of S. pneumoniae strain C06_18 and C06_18ΔbgaA to NHBE cells in the presence of LacNAc and lactose (0–10 mM). As in (C) except using pneumococcal strain C06_18. Adherence assays are mean ± SD of three independent experiments each performed in triplicate. Statistically significant differences were assessed using a two-tailed Student's t-test. * p≤0.04, ** p≤0.007, *** p≤8×10−4. Lactose, LacNAc and galactose significantly reduced adherence to NHBE and D562 cells, though, consistent with the low affinity of these CBMs for galactose, this monosaccharide reduced adherence (Figure S5E and data not shown) significantly less than the same concentration of disaccharides (Figure 6C, 6D, S5C and S5D). The effect of lactose and LacNAc was BgaA-specific and dose-dependent.
Sialidase treated human epithelial cells showed significantly increased adherence to immobilized CBM71-1 and CBM71-2, as compared to immobilized BSA, indicating that the CBMs within BgaA directly interact with the host cell (Figure 7A and 7B). Furthermore, adherence to CBMs was reduced if epithelial cells were treated with both sialidase and the catalytic domain of BgaA, indicating that the receptor mediating adherence is a substrate of BgaA: terminal β-1,4-linked galactose. To ensure that this interaction was relevant in the context of intact bacteria we constructed a strain designed to abrogate CBM binding through point mutations in bgaA that target critical binding residues in the CBMs. W1514 and W1864 in the structures of CBM71-1 and CBM71-2, respectively, make classical aromatic amino acid side chain – carbohydrate ring interactions, which are typically critical to CBM binding [2]. Thus, these residues were chosen for alanine substitutions. As predicted, the strain expressing the S. pneumoniae mutant carrying the BgaAW1514A,W1864A variant showed dramatically reduced adherence that was not significantly different from the bgaA mutant (Figure 7C). This reduction in adherence was not due to differences in expression, localization or activity of BgaA (Figure S6A and S6B). Together these data strongly support the hypothesis that CBMs in BgaA contribute to pneumococcal adherence by binding to LacNAc and lactose containing cell surface glycoconjugates.
Fig. 7. CBMs within BgaA directly bind epithelial cells. 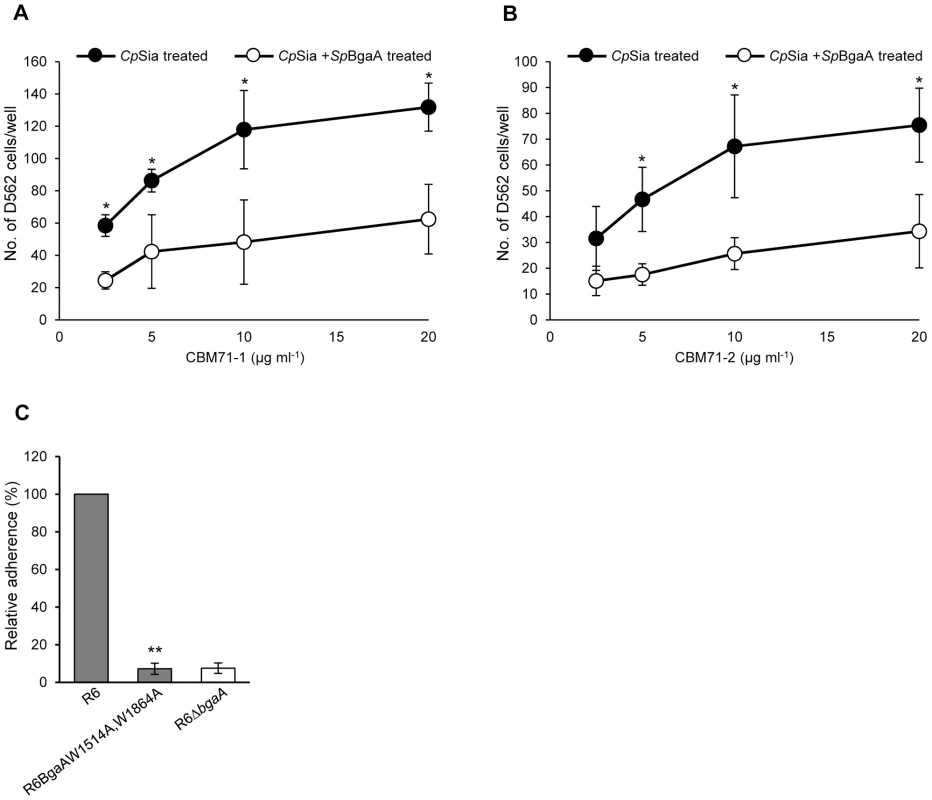
(A) Binding of D562 cells to immobilized recombinant CBM71-1. A range of concentrations (2.5–20 µg ml−1) of CBM71-1 were immobilized on the bottom of a 96 well plate. D562 cells pretreated with 0.002 Unit ml−1 C. perfringens sialidase (CpSia) with or without 0.054 µM SpBgaA146-990 (SpBgaA) were allowed to adhere to the plate. Adherence to control wells coated with 1 mg ml−1 of BSA was subtracted from the data (maximum 22 cells per well). Asterisks indicate significant differences in the number of adherent D562 cells following pretreatment with sialidase and sialidase plus SpBgaA146-990. (B) Binding of D562 cells to immobilized recombinant CBM71-2. As in (A) except using CBM71-2. (C) Adherence of S. pneumoniae strain R6 and R6BgaAW1514A,W1864A to D562 cells. Asterisks indicate significant differences in adherence of R6 and R6BgaAW1514A,W1864A. Data are the mean ± SD of three independent experiments each performed in triplicate. Statistically significant differences were assessed using a two-tailed Student's t-test. * p≤0.04, ** p≤2.00×10−7. Heterologous complementation of S. pneumoniae bgaA mutant by Streptococcus gordonii bgaA
Although the majority of β-galactosidases lack the large C-terminal region found within BgaA (Figure 4A), a relatively large number of host-adapted streptococci, including S. gordonii, encode similar β-galactosidases [41] (Figure S7). In order to test if BgaA orthologs may represent a previously uncharacterized class of bacterial adhesins, we tested adherence of an S. pneumoniae bgaA mutant expressing S. gordonii BgaA (R6ΔbgaA SgbgaA+) at the same locus and under control of the native promoter. Adherence and enzymatic activity of the pneumococcal strain expressing the S. gordonii BgaA was not significantly different from that of the parental strain (Figure 8). These data indicate that other BgaA orthologs including S. gordonii BgaA have the potential to act as bacterial adhesins.
Fig. 8. S. gordonii bgaA can restore the β-galactosidase activity and adherence of a S. pneumoniae bgaA mutant. 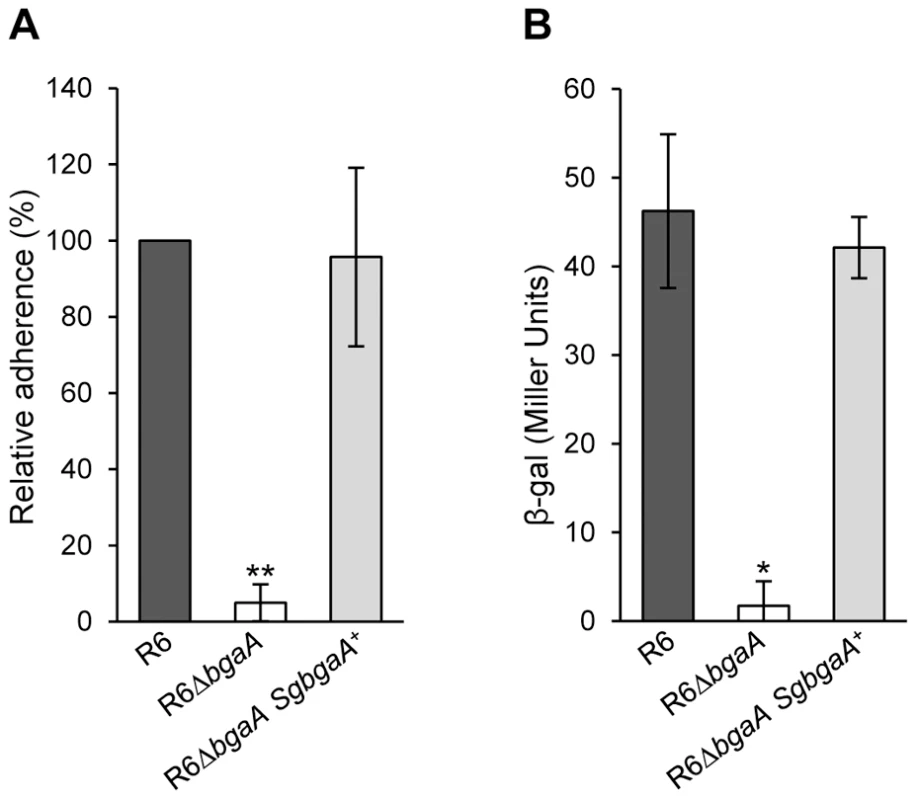
(A) S. gordonii bgaA can restore the adherence of a S. pneumoniae bgaA mutant. (B) S. gordonii bgaA can restore the β-galactosidase activity of a S. pneumoniae bgaA mutant. Data presented here are mean ± SD of three independent experiments each performed in triplicate. Asterisks indicate statistically significant differences between R6ΔbgaA and both R6ΔbgaA SgbgaA+ and R6 calculated using a two-tailed Student's t-test. * p≤0.002, ** p≤5×10−6. Discussion
The catalytic specificity of BgaA is reported to be for LacNAc and lactose, carbohydrate motifs found on a wide variety of glycoconjugates, though the activity on lactose is lower [37]. This catalytic specificity appears to be initially provided by an unusual pre-active or substrate loading complex in the active site that is similar to what has been observed for E. coli LacZ and selects for the β-1,4-linkage in these sugars. The overall architecture of the active site, however, which generally accommodates only a disaccharide, suggests that the enzyme would be quite tolerant of sugar residues preceding a LacNAc or lactose motif, consistent with the ability of BgaA to release galactose from a wide variety of glycoconjugates terminating in LacNAc or lactose motifs [42]. Through the use of a S. pneumoniae mutant lacking bgaA the ability to process these sugar motifs has been linked to growth on a glycoconjugate and protection from opsonophagocytosis [5], [22], [23]. Here, the use of a tight binding inhibitor that specifically targets the active site of BgaA conclusively links the necessity of having an available catalytic site with these biological outcomes and indicates that glycan processing is responsible for the protective effect of BgaA against opsonophagocytosis.
We also identified ancillary CBMs that mediate adherence of the bacterium. Many bacterial species bind host tissues through protein-carbohydrate interactions, which is achieved through a potential myriad of proteins from single, dedicated surface proteins to components of complex flagellar structures [43]–[46]. This is, however, the first demonstration of a CBM mediating adherence of a pathogen to host cells. CBMs typically function to maintain CAZymes in proximity of substrate, thereby enhancing catalytic activity. This may indeed be also be the case with the CBMs in BgaA; however, the overall role in adhering the bacterium to a host cell is a new function for CBMs, not only expanding the repertoire of bacterial adhesins but altering the paradigm of CBM function. It may seem counterintuitive that adherence can be mediated by interactions of CBMs with host glycans that are cleaved by an enzymatic domain within the same protein. However, we propose a dynamic interaction between common host cell surface glycans and multiple copies of a bacterial surface protein. Multiple adhesion events also increase the avidity of the interaction and may provide an explanation of how CBMs with relatively weak affinity for glycans mediate adherence.
Though the CBMs in BgaA clearly mediate an interaction with carbohydrate motifs, namely LacNAc and lactose, the exact nature of the glycoconjugate receptor(s) remains unknown. LacNAc is very common in the N - and O-linked glycans that decorate glycproteins on the surface of epithelial cells while both LacNAc and lactose are frequent motifs in glycosphingolipids. BgaA is active on both LacNAc and lactose, albeit with approximately 10-fold higher activity on LacNAc [37], while the CBMs within BgaA show a minor preference for LacNAc, suggesting that LacNAc is the most likely receptor. Given that it has previously been reported that BgaA may bind a non-proteinacious receptor [31] the members of the neolactoceramide subfamily of glycosphingolipids, which contain LacNAc motifs, are possible candidates as glycoconjugate receptors for BgaA.
As other bacterial species adept at modifying carbohydrates encode surface-associated CAZymes predicted to contain CBMs [41], [47]–[49], we propose that BgaA may be a member of a novel class of bacterial adhesins. This hypothesis is supported by our data demonstrating that bgaA from S. gordonii can complement a S. pneumoniae bgaA mutant. In addition to BgaA, pneumococcal surface-associated glycoside hydrolases NanA, EndoD, Eng and SpuA, contain, or are predicted to contain, CBMs [1], [12], [50], [51]. Two of these enzymes, NanA and Eng, have been demonstrated to contribute to pneumococcal adherence [6], [26], [27]. Although NanA acts to reveal a receptor for BgaA-mediated adherence to epithelial cells, enzymatic activity is not required for adherence to endothelial cells [27]. In fact, an N-terminal region including a putative CBM is required for adherence to endothelial cells. The role of Eng in adherence remains to be defined.
It is likely that CBM-mediated adherence affects the pathogenesis of multiple bacterial species, but these data are of particular significance to the study of pneumococcal biology. It has long been proposed that initial adherence of pneumococci to host tissue occurs via binding carbohydrates on the epithelial cell surface but the identification of specific adhesin-receptor pairs has been lacking [52], [53]. This study elucidates the first carbohydrate-mediated pneumococcal adherence mechanism. It was previously reported that mutation of BgaA does not reduce adherence of all pneumococcal strains, but this mechanism of adherence is very likely relevant to pneumococcal pathogenesis as it affects adherence of multiple strains, including low-passage clinical isolates to all human airway epithelial cell lines tested and primary airway epithelial cells [31]. Pneumococci are a very diverse species and variances in the contribution of different adherence mechanisms between strains has previously been reported [54]–[56]. Differential expression of the five CBM containing CAZymes encoded by pneumococci could explain the differential role of BgaA to adherence, especially in light of the published evidence that some of these CAZymes contribute to adherence [6], [27].
Understanding the specific contributions of different domains/modules of complex proteins to bacterial pathogenesis provides the opportunity to identify inhibitors of these mechanisms. We significantly reduced pneumococcal adherence by the addition of recombinant CBM or free carbohydrate-receptor (i.e. lactose or LacNAc). Additionally, the tight binding β-galactosidase inhibitor GIF inhibited enzymatic activity on the surface of the bacteria to reduce growth on glycoconjugates and resistance to opsonophagocytosis. The demonstrated capacity to modulate the multiple functions of architecturally complex bacterial surface-associated CAZymes with simple molecules may provide a framework for developing approaches to targeting pathogens utilizing such proteins in the host-pathogen interaction.
Materials and Methods
Bacterial strains, plasmids and culture media
Bacterial strains and plasmids used in this study are described in Table S1. S. pneumoniae S. gordonii, and E. coli strains were grown using routine conditions for these organisms and where appropriate media was supplemented with antibiotics. For details see Supporting Information.
Cloning, protein expression and purification
All cloning was performed using standard molecular biology procedures. Protein production in E. coli was done using pET 28-based expression vectors and purification of the polypeptides using procedures described previously and primers detailed in Table S2 [10]. Protein concentrations were determined by measuring the absorbance at 280 nm and using calculated molar extinction coefficient of 174070 cm−1.M−1 for GH2 and GH2-E645Q, 29540 cm−1.M−1 for CBM71-1, 33920 cm−1.M−1 for CBM71-2, and 76320 cm−1.M−1 for CBM71-1.2, [57]. For details see Supporting Information.
Crystallography procedures
All crystallization experiments were performed using sitting-drop vapor diffusion for screening and hanging drop vapor diffusion for optimization, all at 18°C. Diffraction data were collected on cryo-protected crystals at 100 K and data was processed using MOSFLM and SCALA [58], [59]. All data collection and processing statistics are shown in Table S3. The structure of CBM71-1 was determined a by single-anomalous dispersion experiment optimized for selenium using the program ShelXC/D/E [60]. All other structures were solved by molecular replacement using standard procedures. For details see Supporting Information. All data collection, processing, and structure refinement statistics are given in Table S3.
Enzyme inhibition and binding studies
All steady state kinetic studies were performed in triplicate at 37°C in a Cary/Varian 300 Bio UV-Visible Spectrophotometer as previously described [10]. The Ki values for GNJ and GIF were determined from plots of the apparent Km/Vmax against inhibitor concentration. Qualitative UV difference scan and ITC were performed using methods already described [10], [40], [61], [62]. All experiments were performed at 25°C in triplicate. For details see Supporting Information.
Generation of S. pneumoniae mutants
S. pneumoniae TIGR4ΔbgaA strain, was obtained by a PCR ligation technique to replace bgaA with a chloramphenicol cassette [15], [63]. S. pneumoniae R6 and C06_18 strains expressing the surface attached N-terminal (BgaAN) or C-terminal region of BgaA (BgaAC), R6 expressing enzymatically inactive BgaA (R6BgaAE564R), R6 expressing BgaA with point mutants in the CBMs that abrogate carbohydrate binding (R6BgaAW1514A,W1864A), and the S. pneumoniae bgaA mutant expressing S. gordonii BgaA (R6ΔbgaA SgbgaA+) were generated using the Janus cassette selection method using primers described in Table S2 [64]. For details see Supporting Information.
Growth assays and opsonophagocytic killing assays
The protocol for the growth assays of wild-type and ΔbgaA S. pneumoniae TIGR4 strains on bovine asialofetuin was adapted from Battig et al. [65] and performed as described previously [10]. Neutrophil killing assays were performed essentially as previously described with S. pneumoniae TIGR4 wild type strain with or without inhibitors and ΔbgaA strain in the presence of inhibitors or a vehicle control (+++ buffer) [10], [23], [66]. For details see Supporting Information.
Adherence assays
Adherence of S. pneumoniae to monolayer of D562 cells (ATCC CCL-138) and primary NHBE cells (Lonza), grown in 24 well tissue culture plates was determined essentially as previously described [31], [67]. For details see Supporting Information.
Cell adhesion assay to immobilized CBM
Ninety-six well plates coated in a range of concentrations of CBM71-1, CBM71-2 or BSA (control) were blocked with 1% BSA (w/v) before addition of D562 cells treated with Clostridium perfringens sialidase (CpSia) or sialidase and S. pneumoniae BgaA146-990 (SpBgaA). Following incubation for 1 h at 37°C unbound cells were removed by washing and cells were fixed, stained and counted using an inverted light microscope. The average number of cells bound to BSA coated wells was subtracted from the number of cells attached to CBM coated wells. For details see Supporting Information.
Statistical analysis
Data from opsonophagocytic, adherence assays and cell-binding assays were assessed for statistically significant differences using a two tailed Student's t-test and data points with p value≤0.05 were considered significant.
Accession codes
Protein Data Bank. Coordinates and structure factors have been deposited with the following accession codes: native BgaA catalytic domain, 4cu6; BgaA catalytic domain in complex with GIF, 4cu7; BgaA catalytic domain in complex with GNJ, 4cu8; BgaA catalytic domain E645Q complex with LacNAc, 4cuc; CBM71-1 Se-met, 4cua; CBM71-1 in complex with LacNAc, 4cub; CBM71-2, 4cu9.
Supporting Information
Zdroje
1. CantarelBL, CoutinhoPM, RancurelC, BernardT, LombardV, et al. (2009) The Carbohydrate-Active EnZymes database (CAZy): an expert resource for Glycogenomics. Nucleic Acids Res 37: D233–238.
2. BorastonAB, BolamDN, GilbertHJ, DaviesGJ (2004) Carbohydrate-binding modules: fine-tuning polysaccharide recognition. Biochem J 382 : 769–781.
3. Ficko-BleanE, BorastonAB (2012) Insights into the recognition of the human glycome by microbial carbohydrate-binding modules. Curr Opin Struct Biol 22 : 570–577.
4. BerryAM, LockRA, ThomasSM, RajanDP, HansmanD, et al. (1994) Cloning and nucleotide sequence of the Streptococcus pneumoniae hyaluronidase gene and purification of the enzyme from recombinant Escherichia coli. Infect Immun 62 : 1101–1118.
5. KingSJ, HippeKR, WeiserJN (2006) Deglycosylation of human glycoconjugates by the sequential activities of exoglycosidases expressed by Streptococcus pneumoniae. Mol Microbiol 59 : 961–974.
6. MarionC, LimoliDH, BobulskyGS, AbrahamJL, BurnaughAM, et al. (2009) Identification of a pneumococcal glycosidase that modifies O-linked glycans. Infect Immun 77 : 1389–1396.
7. ZahnerD, HakenbeckR (2000) The Streptococcus pneumoniae beta-galactosidase is a surface protein. J Bacteriol 182 : 5919–5921.
8. JeongJK, KwonO, LeeYM, OhDB, LeeJM, et al. (2009) Characterization of the Streptococcus pneumoniae BgaC protein as a novel surface {beta}-galactosidase with specific hydrolysis activity for the Gal{beta}-1-3GlcNAc moiety of oligosaccharide. J Bacteriol 191 : 3011–3023.
9. CamaraM, BoulnoisGJ, AndrewPW, MitchellTJ (1994) A neuraminidase from Streptococcus pneumoniae has the features of a surface protein. Infect Immun 62 : 3688–3695.
10. PluvinageB, HigginsMA, AbbottDW, RobbC, DaliaAB, et al. (2011) Inhibition of the pneumococcal virulence factor StrH and molecular insights into N-glycan recognition and hydrolysis. Structure 19 : 1603–1614.
11. ClarkeVA, PlattN, ButtersTD (1995) Cloning and expression of the beta-N-acetylglucosaminidase gene from Streptococcus pneumoniae. Generation of truncated enzymes with modified aglycon specificity. J Biol Chem 270 : 8805–8814.
12. CainesME, ZhuH, VuckovicM, WillisLM, WithersSG, et al. (2008) The structural basis for T-antigen hydrolysis by Streptococcus pneumoniae: a target for structure-based vaccine design. J Biol Chem 283 : 31279–31283.
13. MuramatsuH, TachikuiH, UshidaH, SongX, QiuY, et al. (2001) Molecular cloning and expression of endo-beta-N-acetylglucosaminidase D, which acts on the core structure of complex type asparagine-linked oligosaccharides. J Biochem (Tokyo) 129 : 923–928.
14. BongaertsRJ, HeinzHP, HaddingU, ZyskG (2000) Antigenicity, expression, and molecular characterization of surface-located pullulanase of Streptococcus pneumoniae. Infect Immun 68 : 7141–7143.
15. AbbottDW, HigginsMA, HyrnuikS, PluvinageB, Lammerts van BuerenA, et al. (2010) The molecular basis of glycogen breakdown and transport in Streptococcus pneumoniae. Mol Microbiol 77 : 183–199.
16. BerryAM, PatonJC (2000) Additive attenuation of virulence of Streptococcus pneumoniae by mutation of the genes encoding pneumolysin and other putative pneumococcal virulence proteins. Infect Immun 68 : 133–140.
17. LiS, KellySJ, LamaniE, FerraroniM, JedrzejasMJ (2000) Structural basis of hyaluronan degradation by Streptococcus pneumoniae hyaluronate lyase. Embo J 19 : 1228–1240.
18. MancoS, HernonF, YesilkayaH, PatonJC, AndrewPW, et al. (2006) Pneumococcal neuraminidases A and B both have essential roles during infection of the respiratory tract and sepsis. Infect Immun 74 : 4014–4020.
19. MarionC, StewartJM, TaziMF, BurnaughAM, LinkeCM, et al. (2012) Streptococcus pneumoniae can utilize multiple sources of hyaluronic acid for growth. Infect Immun 80 : 1390–1398.
20. TerraVS, HomerKA, RaoSG, AndrewPW, YesilkayaH (2010) Characterization of novel beta-galactosidase activity that contributes to glycoprotein degradation and virulence in Streptococcus pneumoniae. Infect Immun 78 : 348–357.
21. ZwijnenburgPJ, van der PollT, FlorquinS, van DeventerSJ, RoordJJ, et al. (2001) Experimental pneumococcal meningitis in mice: a model of intranasal infection. J Infect Dis 183 : 1143–1146.
22. BurnaughAM, FrantzLJ, KingSJ (2008) Growth of Streptococcus pneumoniae on human glycoconjugates is dependent upon the sequential activity of bacterial exoglycosidases. J Bacteriol 190 : 221–230.
23. DaliaAB, StandishAJ, WeiserJN (2010) Three surface exoglycosidases from Streptococcus pneumoniae, NanA, BgaA, and StrH, promote resistance to opsonophagocytic killing by human neutrophils. Infect Immun 78 : 2108–2116.
24. YesilkayaH, MancoS, KadiogluA, TerraVS, AndrewPW (2008) The ability to utilize mucin affects the regulation of virulence gene expression in Streptococcus pneumoniae. FEMS Microbiol Lett 278 : 231–235.
25. ParkerD, SoongG, PlanetP, BrowerJ, RatnerAJ, et al. (2009) The NanA neuraminidase of Streptococcus pneumoniae is involved in biofilm formation. Infect Immun 77 : 3722–3730.
26. BrittanJL, BuckeridgeTJ, FinnA, KadiogluA, JenkinsonHF (2012) Pneumococcal neuraminidase A: an essential upper airway colonization factor for Streptococcus pneumoniae. Mol Oral Microbiol 27 : 270–283.
27. UchiyamaS, CarlinAF, KhosraviA, WeimanS, BanerjeeA, et al. (2009) The surface-anchored NanA protein promotes pneumococcal brain endothelial cell invasion. J Exp Med 206 : 1845–1852.
28. HavaDL, CamilliA (2002) Large-scale identification of serotype 4 Streptococcus pneumoniae virulence factors. Mol Microbiol 45 : 1389–1406.
29. PolissiA, PontiggiaA, FegerG, AltieriM, MottlH, et al. (1998) Large-scale identification of virulence genes from Streptococcus pneumoniae. Infect Immun 66 : 5620–5629.
30. KingSJ (2010) Pneumococcal modification of host sugars: a major contributor to colonization of the human airway? Mol Oral Microbiol 25 : 15–24.
31. LimoliDH, SladekJA, FullerLA, SinghAK, KingSJ (2011) BgaA acts as an adhesin to mediate attachment of some pneumococcal strains to human epithelial cells. Microbiology 157 : 2369–2381.
32. TettelinH, NelsonKE, PaulsenIT, EisenJA, ReadTD, et al. (2001) Complete genome sequence of a virulent isolate of Streptococcus pneumoniae. Science 293 : 498–506.
33. GiefingC, MeinkeAL, HannerM, HenicsT, Bui MinhD, et al. (2008) Discovery of a novel class of highly conserved vaccine antigens using genomic scale antigenic fingerprinting of pneumococcus with human antibodies. J Exp Med 205 : 117–131.
34. LanieJA, NgWL, KazmierczakKM, AndrzejewskiTM, DavidsenTM, et al. (2007) Genome sequence of Avery's virulent serotype 2 strain D39 of Streptococcus pneumoniae and comparison with that of unencapsulated laboratory strain R6. J Bacteriol 189 : 38–51.
35. HoskinsJ, AlbornWEJr, ArnoldJ, BlaszczakLC, BurgettS, et al. (2001) Genome of the bacterium Streptococcus pneumoniae strain R6. J Bacteriol 183 : 5709–5717.
36. KharatAS, TomaszA (2003) Inactivation of the srtA gene affects localization of surface proteins and decreases adhesion of Streptococcus pneumoniae to human pharyngeal cells in vitro. Infect Immun 71 : 2758–2765.
37. ZelenyR, AltmannF, PraznikW (1997) A capillary electrophoretic study on the specificity of beta-galactosidases from Aspergillus oryzae, Escherichia coli, Streptococcus pneumoniae, and Canavalia ensiformis (jack bean). Anal Biochem 246 : 96–101.
38. IchikawaH, IgarashiY, IchikawaM, SuharaY (1998) 1-N-Iminosugars: potent and selective inhibitors of β-glycosidases. J Am Chem Soc 120 : 3007–3018.
39. KelleyLA, SternbergMJ (2009) Protein structure prediction on the Web: a case study using the Phyre server. Nat Protoc 4 : 363–371.
40. Ficko-BleanE, BorastonAB (2006) The interaction of a carbohydrate-binding module from a Clostridium perfringens N-acetyl-beta-hexosaminidase with its carbohydrate receptor. J Biol Chem 281 : 37748–37757.
41. VickermanMM, IobstS, JesionowskiAM, GillSR (2007) Genome-wide transcriptional changes in Streptococcus gordonii in response to competence signaling peptide. J Bacteriol 189 : 7799–7807.
42. KojimaK, IwamoriM, TakasakiS, KubushiroK, NozawaS, et al. (1987) Diplococcal beta-galactosidase with a specificity reacting to beta 1–4 linkage but not to beta 1–3 linkage as a useful exoglycosidase for the structural elucidation of glycolipids. Anal Biochem 165 : 465–469.
43. BuciorI, PielageJF, EngelJN (2012) Pseudomonas aeruginosa pili and flagella mediate distinct binding and signaling events at the apical and basolateral surface of airway epithelium. PLoS Pathog 8: e1002616.
44. KrogfeltKA, BergmansH, KlemmP (1990) Direct evidence that the FimH protein is the mannose-specific adhesin of Escherichia coli type 1 fimbriae. Infect Immun 58 : 1995–1998.
45. ChessaD, WinterMG, JakominM, BaumlerAJ (2009) Salmonella enterica serotype Typhimurium Std fimbriae bind terminal alpha(1,2)fucose residues in the cecal mucosa. Mol Microbiol 71 : 864–875.
46. PetheK, AumercierM, FortE, GatotC, LochtC, et al. (2000) Characterization of the heparin-binding site of the mycobacterial heparin-binding hemagglutinin adhesin. J Biol Chem 275 : 14273–14280.
47. ReichmannP, NuhnM, DenapaiteD, BrucknerR, HenrichB, et al. (2011) Genome of Streptococcus oralis strain Uo5. J Bacteriol 193 : 2888–2889.
48. SchellMA, KarmirantzouM, SnelB, VilanovaD, BergerB, et al. (2002) The genome sequence of Bifidobacterium longum reflects its adaptation to the human gastrointestinal tract. Proc Natl Acad Sci USA 99 : 14422–14427.
49. TurroniF, BottaciniF, ForoniE, MulderI, KimJH, et al. (2010) Genome analysis of Bifidobacterium bifidum PRL2010 reveals metabolic pathways for host-derived glycan foraging. Proc Natl Acad Sci U S A 107 : 19514–19519.
50. van BuerenAL, HigginsM, WangD, BurkeRD, BorastonAB (2007) Identification and structural basis of binding to host lung glycogen by streptococcal virulence factors. Nat Struct Mol Biol 14 : 76–84.
51. AbbottDW, BorastonA (2011) Structural analysis of a putative family 32 carbohydrate-binding module from the Streptococcus pneumoniae enzyme EndoD. Acta Crystallogr Sect F Struct Biol Cryst Commun 67 : 429–433.
52. VossS, GamezG, HammerschmidtS (2012) Impact of pneumococcal microbial surface components recognizing adhesive matrix molecules on colonization. Mol Oral Microbiol 27 : 246–256.
53. HammerschmidtS (2006) Adherence molecules of pathogenic pneumococci. Curr Opin Microbiol 9 : 12–20.
54. BagnoliF, MoschioniM, DonatiC, DimitrovskaV, FerlenghiI, et al. (2008) A second pilus type in Streptococcus pneumoniae is prevalent in emerging serotypes and mediates adhesion to host cells. J Bacteriol 190 : 5480–5492.
55. CronLE, BootsmaHJ, NoskeN, BurghoutP, HammerschmidtS, et al. (2009) Surface-associated lipoprotein PpmA of Streptococcus pneumoniae is involved in colonization in a strain-specific manner. Microbiology 155 : 2401–2410.
56. BrockSC, McGrawPA, WrightPF, CroweJEJr (2002) The human polymeric immunoglobulin receptor facilitates invasion of epithelial cells by Streptococcus pneumoniae in a strain-specific and cell type-specific manner. Infect Immun 70 : 5091–5095.
57. GasteigerJ (2003) Physicochemical effects in the representation of molecular structures for drug designing. Mini Rev Med Chem 3 : 789–796.
58. PowellHR (1999) The Rossmann Fourier autoindexing algorithm in MOSFLM. Acta Crystallogr D Biol Crystallogr 55 : 1690–1695.
59. WinnMD, BallardCC, CowtanKD, DodsonEJ, EmsleyP, et al. (2011) Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr 67 : 235–242.
60. SheldrickGM (2008) A short history of SHELX. Acta Crystallogr A 64 : 112–122.
61. Ficko-BleanE, BorastonAB (2009) N-acetylglucosamine recognition by a family 32 carbohydrate-binding module from Clostridium perfringens NagH. J Mol Biol 390 : 208–220.
62. Ficko-BleanE, StubbsKA, NemirovskyO, VocadloDJ, BorastonAB (2008) Structural and mechanistic insight into the basis of mucopolysaccharidosis IIIB. Proc Natl Acad Sci U S A 105 : 6560–6565.
63. LauPC, SungCK, LeeJH, MorrisonDA, CvitkovitchDG (2002) PCR ligation mutagenesis in transformable streptococci: application and efficiency. J Microbiol Methods 49 : 193–205.
64. SungCK, LiH, ClaverysJP, MorrisonDA (2001) An rpsL cassette, janus, for gene replacement through negative selection in Streptococcus pneumoniae. Appl Environ Microbiol 67 : 5190–5196.
65. BattigP, HathawayLJ, HoferS, MuhlemannK (2006) Serotype-specific invasiveness and colonization prevalence in Streptococcus pneumoniae correlate with the lag phase during in vitro growth. Microbes Infect 8 : 2612–2617.
66. DavisKM, AkinbiHT, StandishAJ, WeiserJN (2008) Resistance to mucosal lysozyme compensates for the fitness deficit of peptidoglycan modifications by Streptococcus pneumoniae. PLoS Pathog 4: e1000241.
67. GouldJM, WeiserJN (2002) The inhibitory effect of C-reactive protein on bacterial phosphorylcholine platelet-activating factor receptor-mediated adherence is blocked by surfactant. J Infect Dis 186 : 361–371.
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek Out-of-Sequence Signal 3 as a Mechanism for Virus-Induced Immune Suppression of CD8 T Cell ResponsesČlánek RNF26 Temporally Regulates Virus-Triggered Type I Interferon Induction by Two Distinct MechanismsČlánek Mouse, but Not Human, ApoB-100 Lipoprotein Cholesterol Is a Potent Innate Inhibitor of Pneumolysin
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2014 Číslo 9- Jak souvisí postcovidový syndrom s poškozením mozku?
- Měli bychom postcovidový syndrom léčit antidepresivy?
- Farmakovigilanční studie perorálních antivirotik indikovaných v léčbě COVID-19
- 10 bodů k očkování proti COVID-19: stanovisko České společnosti alergologie a klinické imunologie ČLS JEP
-
Všechny články tohoto čísla
- Virus Control Goes Epigenetic
- The Role of Iron in Prion Disease and Other Neurodegenerative Diseases
- The Ins and Outs of Rust Haustoria
- Prion Strains and Amyloid Polymorphism Influence Phenotypic Variation
- Teaching Fido New ModiFICation Tricks
- Can Enhance Infection in Mosquitoes: Implications for Malaria Control?
- MIF Contributes to Associated Immunopathogenicity Development
- Persistence of Virus Reservoirs in ART-Treated SHIV-Infected Rhesus Macaques after Autologous Hematopoietic Stem Cell Transplant
- Bacillus Calmette-Guerin Infection in NADPH Oxidase Deficiency: Defective Mycobacterial Sequestration and Granuloma Formation
- EhCoactosin Stabilizes Actin Filaments in the Protist Parasite
- Molecular Insights Into the Evolutionary Pathway of O1 Atypical El Tor Variants
- LprG-Mediated Surface Expression of Lipoarabinomannan Is Essential for Virulence of
- Structural Correlates of Rotavirus Cell Entry
- Multivalent Adhesion Molecule 7 Clusters Act as Signaling Platform for Host Cellular GTPase Activation and Facilitate Epithelial Barrier Dysfunction
- The Effects of Vaccination and Immunity on Bacterial Infection Dynamics
- Myeloid Derived Hypoxia Inducible Factor 1-alpha Is Required for Protection against Pulmonary Infection
- Functional Characterisation of Germinant Receptors in and Presents Novel Insights into Spore Germination Systems
- Global Analysis of Neutrophil Responses to Reveals a Self-Propagating Inflammatory Program
- Host Cell Invasion by Apicomplexan Parasites: The Junction Conundrum
- Comparative Phenotypic Analysis of the Major Fungal Pathogens and
- Unravelling the Multiple Functions of the Architecturally Intricate β-galactosidase, BgaA
- Sialylation of Prion Protein Controls the Rate of Prion Amplification, the Cross-Species Barrier, the Ratio of PrP Glycoform and Prion Infectivity
- Symbionts Commonly Provide Broad Spectrum Resistance to Viruses in Insects: A Comparative Analysis of Strains
- Ontogeny of Recognition Specificity and Functionality for the Broadly Neutralizing Anti-HIV Antibody 4E10
- Identification and Characterisation of a Hyper-Variable Apoplastic Effector Gene Family of the Potato Cyst Nematodes
- Crimean-Congo Hemorrhagic Fever Virus Entry into Host Cells Occurs through the Multivesicular Body and Requires ESCRT Regulators
- Age-Dependent Enterocyte Invasion and Microcolony Formation by
- CD160-Associated CD8 T-Cell Functional Impairment Is Independent of PD-1 Expression
- Functional Fluorescent Protein Insertions in Herpes Simplex Virus gB Report on gB Conformation before and after Execution of Membrane Fusion
- The Tudor Domain Protein Spindlin1 Is Involved in Intrinsic Antiviral Defense against Incoming Hepatitis B Virus and Herpes Simplex Virus Type 1
- Transgenic Analysis of the MAP Kinase MPK10 Reveals an Auto-inhibitory Mechanism Crucial for Stage-Regulated Activity and Parasite Viability
- Evidence for a Transketolase-Mediated Metabolic Checkpoint Governing Biotrophic Growth in Rice Cells by the Blast Fungus
- Incomplete Deletion of IL-4Rα by LysM Reveals Distinct Subsets of M2 Macrophages Controlling Inflammation and Fibrosis in Chronic Schistosomiasis
- Identification and Functional Expression of a Glutamate- and Avermectin-Gated Chloride Channel from , a Southern Hemisphere Sea Louse Affecting Farmed Fish
- Out-of-Sequence Signal 3 as a Mechanism for Virus-Induced Immune Suppression of CD8 T Cell Responses
- Strong Epistatic Selection on the RNA Secondary Structure of HIV
- Hematopoietic but Not Endothelial Cell MyD88 Contributes to Host Defense during Gram-negative Pneumonia Derived Sepsis
- Delineation of Interfaces on Human Alpha-Defensins Critical for Human Adenovirus and Human Papillomavirus Inhibition
- Exploitation of Reporter Strains to Probe the Impact of Vaccination at Sites of Infection
- RNF26 Temporally Regulates Virus-Triggered Type I Interferon Induction by Two Distinct Mechanisms
- Helminth Infections Coincident with Active Pulmonary Tuberculosis Inhibit Mono- and Multifunctional CD4 and CD8 T Cell Responses in a Process Dependent on IL-10
- MHC Class II Restricted Innate-Like Double Negative T Cells Contribute to Optimal Primary and Secondary Immunity to
- Reactive Oxygen Species Regulate Caspase-11 Expression and Activation of the Non-canonical NLRP3 Inflammasome during Enteric Pathogen Infection
- Evolution of Plastic Transmission Strategies in Avian Malaria
- A New Human 3D-Liver Model Unravels the Role of Galectins in Liver Infection by the Parasite
- Translocates into the Myocardium and Forms Unique Microlesions That Disrupt Cardiac Function
- Mouse, but Not Human, ApoB-100 Lipoprotein Cholesterol Is a Potent Innate Inhibitor of Pneumolysin
- The Cofilin Phosphatase Slingshot Homolog 1 (SSH1) Links NOD1 Signaling to Actin Remodeling
- Kaposi's Sarcoma Herpesvirus MicroRNAs Induce Metabolic Transformation of Infected Cells
- Reorganization of the Endosomal System in -Infected Cells: The Ultrastructure of -Induced Tubular Compartments
- Distinct Dictation of Japanese Encephalitis Virus-Induced Neuroinflammation and Lethality via Triggering TLR3 and TLR4 Signal Pathways
- Exploitation of the Complement System by Oncogenic Kaposi's Sarcoma-Associated Herpesvirus for Cell Survival and Persistent Infection
- The Secreted Peptide PIP1 Amplifies Immunity through Receptor-Like Kinase 7
- Structural Insight into Host Recognition by Aggregative Adherence Fimbriae of Enteroaggregative
- The CD14CD16 Inflammatory Monocyte Subset Displays Increased Mitochondrial Activity and Effector Function During Acute Malaria
- Infection Induces Expression of a Mosquito Salivary Protein (Agaphelin) That Targets Neutrophil Function and Inhibits Thrombosis without Impairing Hemostasis
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- The Secreted Peptide PIP1 Amplifies Immunity through Receptor-Like Kinase 7
- Symbionts Commonly Provide Broad Spectrum Resistance to Viruses in Insects: A Comparative Analysis of Strains
- MIF Contributes to Associated Immunopathogenicity Development
- The Ins and Outs of Rust Haustoria
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



