-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaMolecular Insights Into the Evolutionary Pathway of O1 Atypical El Tor Variants
In this report, we suggest a genetic mechanism of how the V. cholerae atypical El Tor variants were generated from classical and prototype El Tor biotype strains. An intermediary strain, containing the CTX-1 and CTX-2 prophages, was identified among the clinical isolates that were collected in 1991, when the atypical strains emerged. This strain can be converted into various Wave 2 atypical El Tor strains by eliminating prototype components, CTX-1 and RS1. Further, new types of the CTX phage genome can be generated from the intermediary strain by inter-chromosomal recombination between CTX phages and recombination between the CTX phage and RS1. These new CTX phages can be transduced into other El Tor strains, transforming them into Wave 3 atypical strains. This is a demonstrated instance of how a single-segment-genome CTX phage re-organizes its genome through recombination between different types of phage, leading to generation of new phage variants and atypical El Tor strains.
Published in the journal: . PLoS Pathog 10(9): e32767. doi:10.1371/journal.ppat.1004384
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1004384Summary
In this report, we suggest a genetic mechanism of how the V. cholerae atypical El Tor variants were generated from classical and prototype El Tor biotype strains. An intermediary strain, containing the CTX-1 and CTX-2 prophages, was identified among the clinical isolates that were collected in 1991, when the atypical strains emerged. This strain can be converted into various Wave 2 atypical El Tor strains by eliminating prototype components, CTX-1 and RS1. Further, new types of the CTX phage genome can be generated from the intermediary strain by inter-chromosomal recombination between CTX phages and recombination between the CTX phage and RS1. These new CTX phages can be transduced into other El Tor strains, transforming them into Wave 3 atypical strains. This is a demonstrated instance of how a single-segment-genome CTX phage re-organizes its genome through recombination between different types of phage, leading to generation of new phage variants and atypical El Tor strains.
Introduction
Vibrio cholerae O1 serogroup strains have been categorized into 2 biotypes - classical and El Tor -based on microbiological properties and the CTX prophage that they harbor [1], [2]. Classical biotype strains contain the classical CTX prophage (CTXcla), and El Tor strains are believed to contain the El Tor CTX prophage (CTXEl Tor, or CTX-1).
The CTX phage comprises 10 genes (rstR, rstA, rstB, psh, cep, orfU, ace, zot, ctxA, and ctxB). Whereas rstR is phage type-specific, other genes differ between phages by several SNPs, except for ctxA, which is identical in the 2 phages [3]. The binding subunit of cholera toxin (CTB) is encoded by ctxB, and the CTBs between phages differ by 2 of their 125 amino acids (residues 39 and 68) [1]. An evolutionary model of pathogenic V. cholerae O1 strains, in which El Tor biotype strains acquired only the El Tor CTX phage, whereas the classical strains obtained the classical phage, has been widely accepted [4].
Atypical El Tor variants, defined as El Tor biotype strains that produce classical cholera toxin, were first recognized in 2006, and several atypical El Tor variants have since been reported [5]–[7]. Two atypical CTX phages that contain ctxBcla have been reported among atypical El Tor strains [8], the first of which was discovered in V. cholerae clinical isolates of cholera outbreaks in Mozambique in 2004 and was later found to have existed in South Asian countries since the early 1990s [9]. This CTX phage contains the classical biotype-specific rstR (rstRcla) and ctxB (ctxBcla), and was thus considered CTXcla. However, it was later found to contain other genes of CTX-1 and was renamed CTX-2 [10]. V. cholerae strains that contain CTX-2 harbor a tandem repeat of CTX-2 on chromosome 2. Based on the analysis of SNPs in the genome and the CTX phage they harbor, the El Tor strains have been categorized into 3 Waves [10]. The prototype El Tor strains that contain CTX-1 are considered Wave 1 strains. The strains contain CTX-2 constitute a phylogenetic subgroup among the seventh cholera pandemic strains by genome analysis and are therefore categorized as Wave 2 strains [10].
The second atypical CTX phage (CTX-3) was first reported in Vietnam in 2007 and has the same genetic structure and sequence as CTX-1, with the exception of SNPs in rstA and ctxB (Figure 1) [11]. Strains that harbor CTX-3 typically contain a satellite phage, RS1, followed by a CTX-3 on chromosome 1 [6]. These strains have existed since the early 1990s on the Indian subcontinent [6]. A variant of CTX-3 that contains a new type of ctxB (ctxB genotype 7) emerged from India in 2006 (Figure 1). In addition to 2 SNPs in ctxBcla of CTX-3 compared with ctxBEl Tor of CTX-1, CTX-3b has an additional SNP (nucleotide position 58) at amino acid residue 20 [12]. Most current global clinical isolates of V. cholerae are atypical El Tor variants that harbor CTX-3 or CTX-3b [6]. A recent surveillance study in India has shown that strains containing CTX-3b have been gradually replacing strains with CTX-3, as isolates that contain CTX-3b constitute 93.3% of all isolates that were collected in 2011 [13]. V. cholerae strains that contain CTX-3 or CTX-3b are distinguished from Wave 2 strains, based on the CTX phage and genomic variations, and are thus categorized as Wave 3 strains of the seventh cholera pandemic [10].
Fig. 1. Genetic map of CTX phages of Vibrio cholerae O1 El Tor Wave 3 atypical strains aligned with Wave 1 strain N16961 (genomes not shown to scale). 
ctxAB of N16961 (ctxB genotype 3) is shown in blue, and ctxABs (classical ctxB, or ctxB genotype 1) of CTX-3, CTX-4, CTX-5, and CTX-6 are shown in red. ctxB of genotype 7 (Haiti strain type) of CTX-3b and CTX-6b is shown in purple. SNPs of rstA and rstB are shown as superscripts (position 927, 933, and 942 of rstA and positions 74–76, 87, 93, 105, and 189 of rstB). Del indicates 3-nucleotide (74–76 of rstB) deletion. The catastrophic cholera outbreak in Haiti in 2010 was caused by a Wave 3 strain that contained ctxB genotype 7 [10], [14], [15]. We noted the CTX phage of Haitian strains differed from CTX-3b by several SNPs. We surveyed Wave 3 V. cholerae strains that were collected in Kolkata, India from 2003–2007 to verify that the Haitian-type CTX phage had existed earlier. We identified a CTX phage that contains identical SNPs as the Haitian-type CTX phage (except for ctxB) and other variant CTX phages. The SNPs of these variant CTX phages appeared to originate from the RS1 satellite phage, implying that the variant CTX phages were generated by recombination of homologous genes (rstR, rstA, and rstB) between RS1 and CTX.
V212-1, a strain that bears CTX-1 and CTX-2, was collected when atypical strains first appeared (Table 1) and was considered to be an intermediary between prototype and atypical El Tor strains [16]. In this study, we demonstrate that Wave 2 strains can be generated directly from V212-1 by progressive elimination of CTX-1 and RS1 through intra-strand recombination. Further, novel mosaic CTX phages can be produced by an inter-chromosomal recombination between 2 different CTX prophages in V212-1, and the mosaic phages can be transmitted to new host bacteria, converting them into Wave 3 strains. These results indicate that the intra-strand and inter-strand recombination between CTXs and RS1 element played an important role in the evolution of CTX phage and V. cholerae, as previously suggested [17].
Tab. 1. Genetic variations in RS1 and CTX prophages aligned with CTX-1 of N16961. 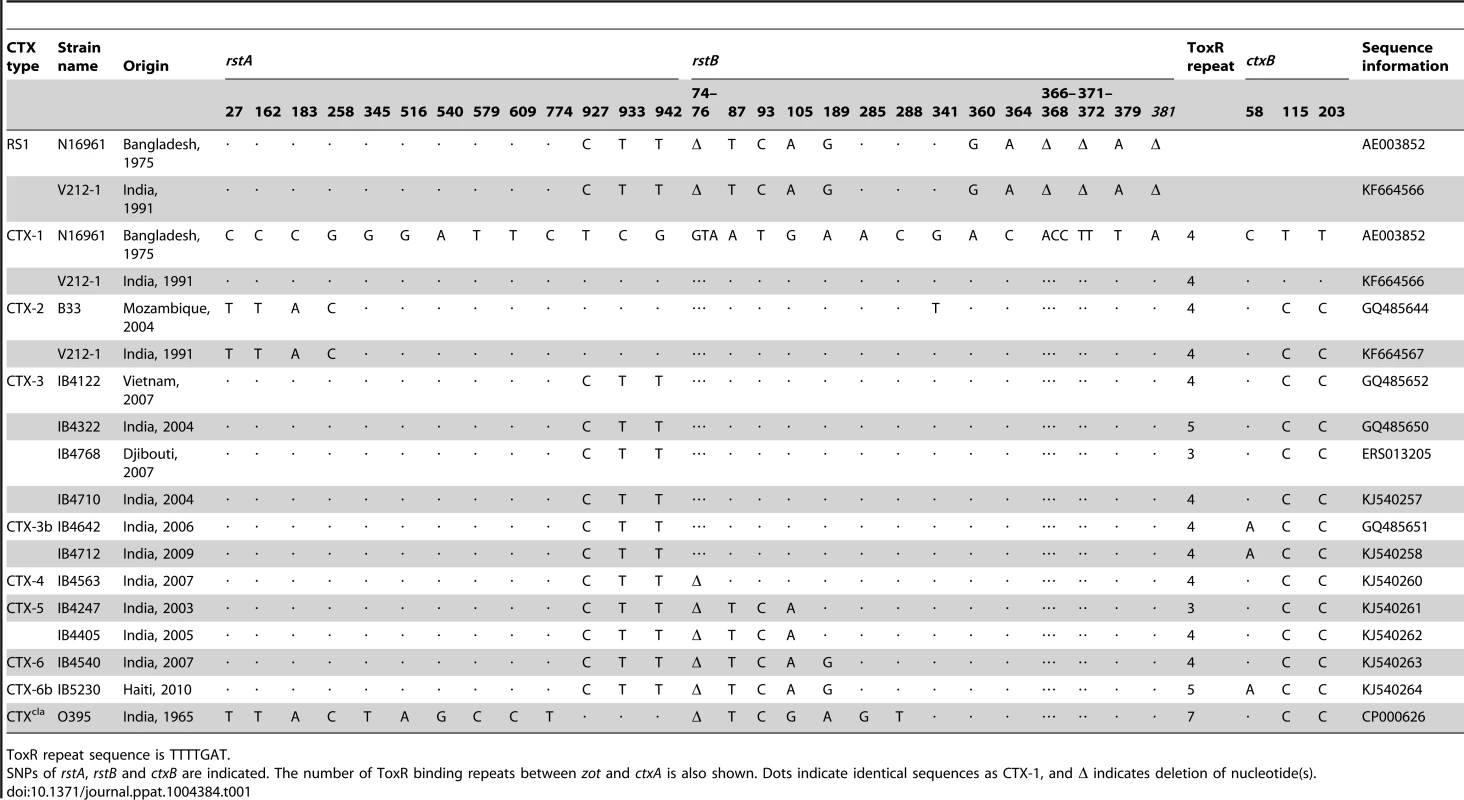
ToxR repeat sequence is TTTTGAT. Results
Variants of mosaic CTX phages in V. cholerae O1 El Tor Wave 3 atypical strains
The genome sequences of CTX prophages indicated that the CTX phage genome can be classified into several types, based on SNPs of rstA, rstB, and ctxB, as suggested by Mutreja et al. [10]. The CTX-3 prophages in V. cholerae isolates belonging to Wave 3 strains that were collected in Vietnam and India contained 3 SNPs (nucleotides 927, 933, and 942) in rstA compared with CTX-1 of N16961 and V212-1, as shown in Table 1 [8], [18]. The CTX prophage in Haitian isolates that were collected during the 2010 cholera outbreak was believed to be CTX-3b, which contains ctxB genotype 7. However, SNPs (3-nucleotide [GTA] deletion of positions 74–76 and SNPs at positions 87, 93, 105, and 189) in rstB, in addition to the 3 SNPs of rstA, were identified by sequencing the CTX genome of Haitian strains (Table 1). The difference in SNPs between CTX-3 and Haitian-type CTX suggests that more CTX variants exist among Wave 3 V. cholerae strains and that the CTX phages in Wave 3 strains can be classified further.
Because the GTA deletion in rstB could be used as a marker for identifying the Haitian-type CTX phage genome, a PCR strategy was designed to detect the presence and absence of this deletion (Figure S1). rstB PCR and DMAMA-PCR for ctxB were performed on 365 clinical isolates that were collected in Kolkata, India between 2003 and 2007 [13], [19]. Most clinical isolates contained CTX-3 or CTX-3b, whereas 11 had GTA-deleted CTX prophage (Table S1).
CTX-3 was the most prevalent type; CTX-3b emerged in 2006, as previously reported [12]; and CTX-4, CTX-5, and CTX-6 were identified in 2004, 2003–2005, and 2007, respectively. The 20 clinical isolates from the cholera outbreak in Haiti in 2010 contained the same CTX prophage that could be categorized as CTX-6b, according to this scheme. However, strains that contained CTX-6b had not been identified in India by 2007.
Notably, the location of the SNPs in rstA and rstB in these CTX phages appear to be of RS1, implying that the CTX phage genomes are mosaics of CTX-1 and RS1 (Table 1).
V212-1, an intermediary strain between prototype and atypical El Tor strains
DNA sequence analysis of CTX-2 in Wave 2 strains and various CTX phages in Wave 3 suggested that these phages in atypical V. cholerae El Tor strains are mosaics of CTX-1, CTXcla, and RS1 (Table 1) [18], [20]. It is unlikely that genetic exchange occurred between viral genomes independently of a host bacterial cell. Thus, we examined whether an intermediate strain that contained various CTX types was the origin of atypical strains. The infection of a classical strain with CTX-1, resulting in a single strain that harbors both CTX types is possible in vitro but, no clinical isolates or reference strains that contained both types of CTX in a single bacterial cell are available [17], [21]_ENREF_15. Instead, an intermediary strain, V212-1, that contains CTX-1 and CTX-2 has been reported [16].
We determined and verified the exact array of CTX and RS1 in V212-1—TLC:RS1:CTX-1:RS1 on chromosome 1 and CTX-2:CTX-2 on chromosome 2—by sequencing (Table 2). CTX-1 on chromosome 1 was identified as an authentic CTX-1, as the SNPs in rstA and rstB were identical to those of CTX-1 in N16961.
Tab. 2. Genetic information on <i>V. cholerae</i> strains and pCTXs generated in this study. 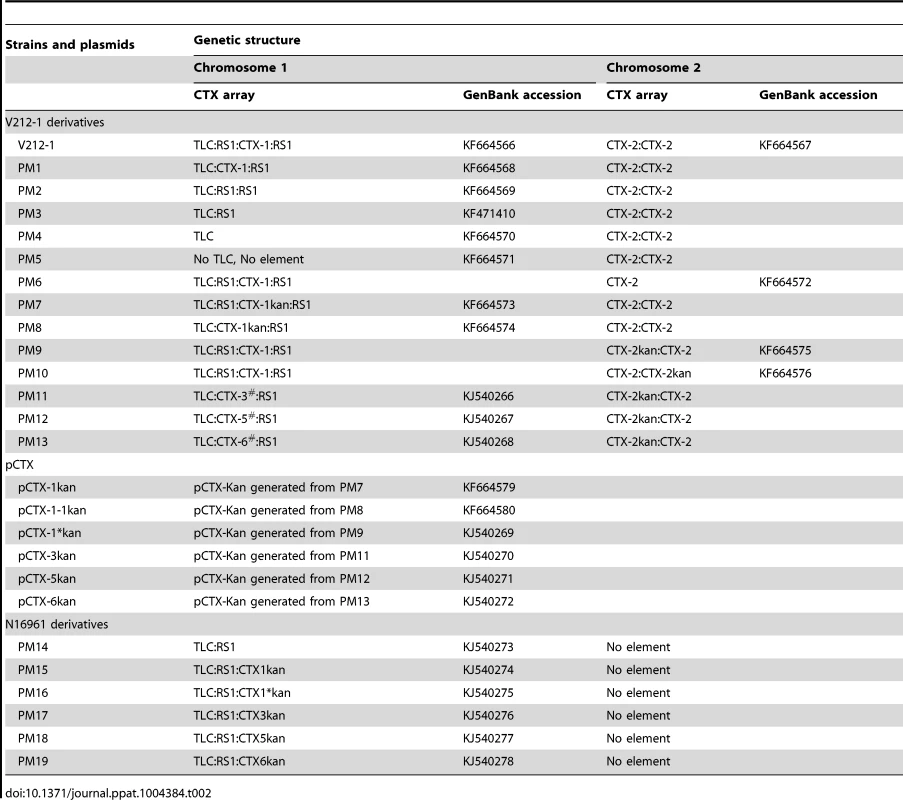
Generation of Wave 2 strains from V212-1
Whole-genome sequence analysis and the presence of a tandem repeat of CTX-2 on chromosome 2 indicate that V212-1 belongs to the Wave 2 strains [10]. However, V212-1 could be considered as an intermediary strain between Wave 1 and Wave 2 strains, because it also contained CTX-1 on chromosome 1. The generation of previously reported Wave 2 strains from V212-1 by the excision of CTX-1 and RS1 and, ultimately, excision of the entire RS1:CTX-1:RS1 array was determined as follows.
The removal of CTX-1 and RS1 was monitored in a recombinant strain V212-1CVD that was constructed by inserting a pCVD442-derived plasmid, pCVDrstRET, into rstR of CTX-1 of V212-1 (Figure S2). Three derivatives of V212-1 (PM1–PM3) were generated by excision of the first RS1, CTX-1, and CTX-1:RS1, respectively from chromosome 1 (Figure 2 and Figure S2). The overall excision rate was estimated to be 1/(4.4×103), and the efficiency of generation of each array varied (Table 3). The most frequently obtained derivative was a strain that contained a solitary RS1.
Fig. 2. Generation of Wave 2 strains by excision of CTX-1 and RS1 from V212-1. 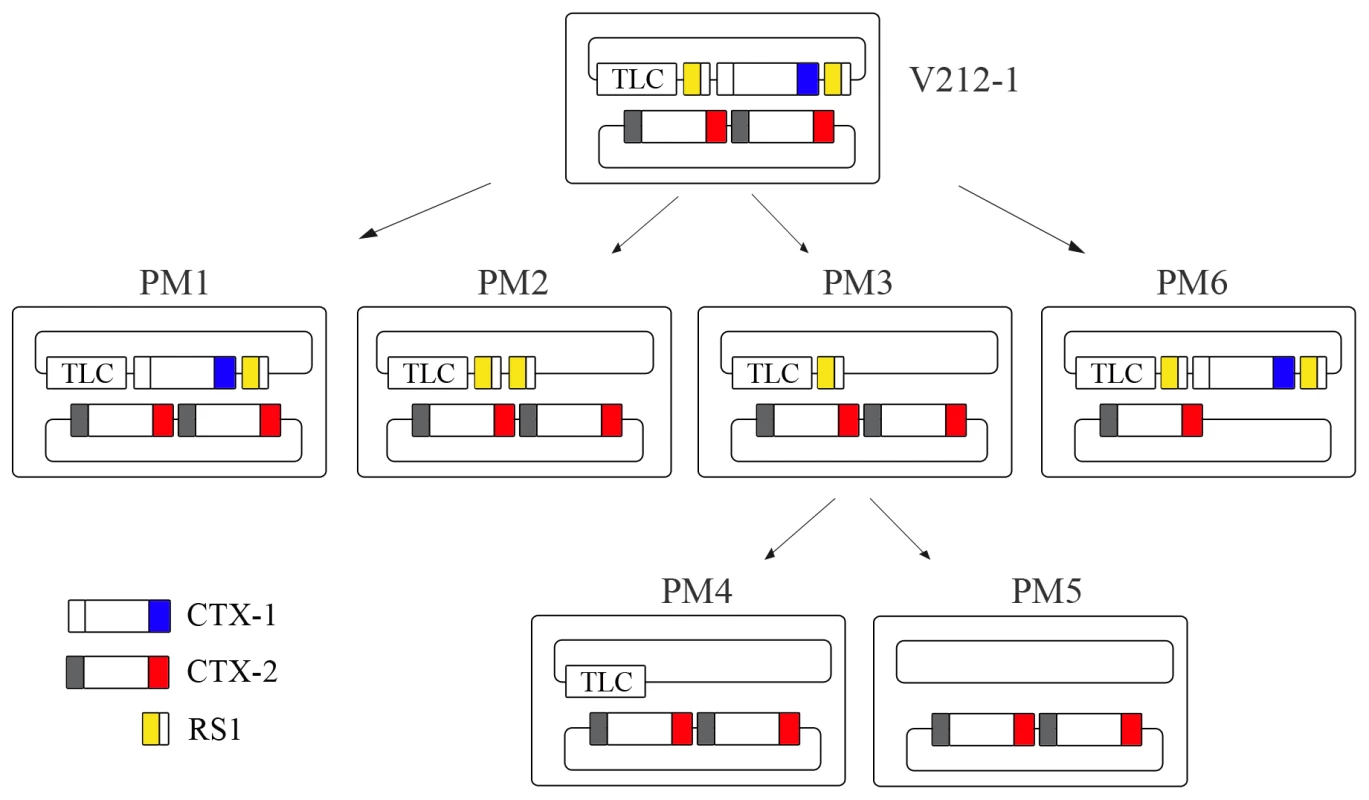
V212-1 can be converted into several variants that belong to Wave 2 strains by excision of RS1s, CTX-1, and TLC (toxin-linked cryptic) from chromosome 1. Six variants of V212-1—PM1, PM2, PM3, PM4, PM5, and PM6—were generated in this report. Although PM4 and PM5 were generated from PM3 in this report, they could be generated directly from V212-1. RS1 is shown as a rectangle divided into 2 parts: RS2 (yellow) and rstC (white). CTX is shown as a rectangle divided into 3 parts: RS2 (white for CTX-1, grey for CTX-2), core, and ctxB (El Tor type ctxB is shown in blue and classical type ctxB is shown in red). Tab. 3. Frequency of generation of each array by excision of CTX-1, RS1, CTX-1:RS1 from chromosome 1 of V212-1CVD and V212-1CVD recA−. 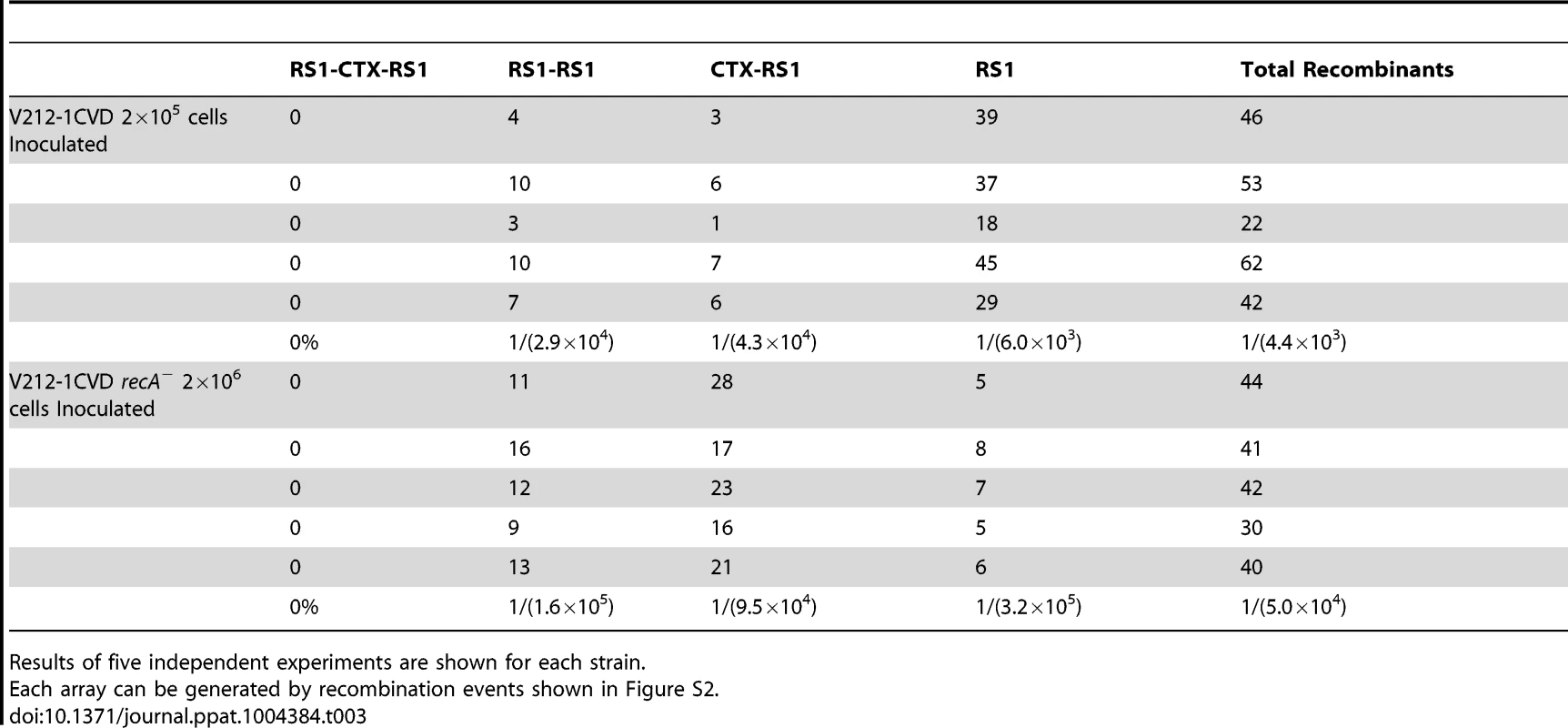
Results of five independent experiments are shown for each strain. The excision of RS1 and/or CTX-1 from chromosome 1 seemed to be mainly by a homologous recombination, since the recombination frequency was reduced 2-fold, 5-fold, and 50-fold in recA− background depending on the recombination position (Table 3). However, the frequency of recombination was not reduced evenly, indicating the deletion of RS1 and/or CTX might be mediated also by other mechanism.
Whereas the CTX prophage of strain PM1 was identified to be the authentic CTX-1 by DNA sequencing, 3 other variant CTX phages were obtained among the derivatives that contain the CTX:RS1 array (Figure 3 and Table 2). These CTX variants can be generated by various recombination events between the RS2 region (rstR, rstA, and rstB) of CTX-1 and RS1 (Figure 3). The SNPs in rstA and rstB of these CTX variants were identical to the CTX-3, CTX-5, and CTX-6 prophages of clinical isolates, respectively, and were thus designated CTX-3#, CTX-5#, and CTX-6# to clarify that they contained ctxBEl Tor (Table 2). These variant strains were used later to produce pCTX-3kan, pCTX-5kan, and pCTX-6kan.
Fig. 3. The generation of new mosaic CTX phages from V212-1 by inter-strand recombination between CTX phages and intra-strand recombination between CTX-1 and RS1 on chromosome 1. 
A double crossover recombination event between 2 prophages on each chromosome of V212-1 (indicated by arrows) results in the generation of the CTX-1* prophage which contains ctxBcla on chromosome 1. Intra-stand recombination between CTX-1* and RS1 generates a mosaic CTX prophage. Depending on the recombination position (shown as R1, R2, and R3), CTX-3, -5, and -6 can be generated (CTX-4 can also be generated, but not shown in this figure). The infectious CTX-3, -5, or -6 virions can be transduced to a new host to give rise to Wave 3 strains. Two additional recombinant strains of V212-1, in which pCVDrstRET was inserted into rstR in each RS1, were constructed to examine the excision of CTX-1 and RS1 further. From a recombinant strain that contained pCVDrstRET in the first RS1, the overall excision rate of RS1 and/or CTX-1 was 1/(4.4×104). Of the resulting 3 arrays (RS1:CTX-1:RS1, CTX-1:RS1, and RS1) from this strain, CTX-1:RS1 array was the most frequently produced. In a strain that contained the pCVDrstRET in the second RS1, the overall excision rate was 1/(4.7×105) and the solitary RS1 array was primarily generated.
No strain that had lost the entire array of chromosome 1 was produced directly from V212-1; therefore, stepwise excision of the entire array was tested in strain PM3. pCVDrstRET was inserted into the rstR of RS1 in PM3 to construct PM3CVD. A strain that had lost RS1 (PM4) and 4 strains (one of them was analyzed as PM5) that had lost RS1 and the TLC (toxin-linked cryptic) were obtained among 110 recombinants that were screened (Figure S3 and Table S2). After the removal of RS1 (PM4) and TLC:RS1 (PM5), the authentic dif-1 sequence (CCTTAATTTAACATAACATACATAATGCGCACTAGGAACATTT) and the defective dif-like sequence (CCTTAATTTAACATAACATACATAATATGCACTGAGGTATTTT) were restored, respectively, indicating that the excision of RS1 and TLC:RS1 was by a precise process. The exact mechanism of the excision of entire array remains unknown, although site-specific recombination through att sequences that flank the RS1s and CTX was proposed to explain the generation of Wave 2 strains [22]. The excision of RS1 and/or TLC:RS1 was also investigated in the recA− strain, and excision efficiencies were significantly reduced in the recA− strain, indicating that the excision of entire array from chromosome 1 might be mediated by an additional mechanism.
These results suggest that the entire array of chromosome 1 in V212-1 can be excised either by a stepwise manner, or by a single step process. A CTX-2 on chromosome 2 can also be excised, resulting in a strain that harbors a single CTX-2 on chromosome 2 (PM6).
The CTX arrays of some V212-1variants that we generated were identical to those in the clinical isolates of Wave 2 strains. PM2 has the TLC:RS1:RS1 array on chromosome 1 and CTX-2:CTX-2 on chromosome 2 and has the same structure as MG116926 [8]. PM5 has the same structure as B33 and MJ1236 [23]. Our results suggest that the intermediary strains such as V212-1 might be the immediate ancestor of Wave 2 strains.
Infectious CTX-1 virions can be generated from chromosome 1 of V212-1
CTX virions can be generated from chromosome 1 of V212-1 and PM1, based on the replication mechanism of CTX phage [24]. ctxAB of CTX-1 of V212-1 and PM1 was replaced by a kanamycin cassette to construct strains PM7 and PM8, respectively, and the generation of kanamycin-resistant CTX virions from PM7 and PM8 was confirmed by generation of the kanamycin-resistant O395 transductant. The titer of CTX virions was approximately 104 virions/ml and 105 virions/ml in the culture supernatant of PM7 and PM8, respectively, in the presence of mitomycin C. pCTX-1kan generated from PM7 and pCTX-1-1kan generated from PM8 were extracted from the recipients and analyzed by sequencing. pCTX-1kan and pCTX-1-1kan had identical DNA sequences, because they were replication products from the same CTX-1 prophage genome of chromosome 1. No CTX-2 virions were generated from the tandem repeat of CTX-2 on chromosome 2 of PM9 and PM10, as no virions were produced from B33 [22].
Generation of new mosaic CTX phages from V212-1
An inter-chromosomal recombination event between ctxABEl Tor of CTX-1 on chromosome 1 and ctxABcla of CTX-2 on chromosome 2 in V212-1, generating new mosaic phage genomes, was verified by the generation of CTX-1*kan virions from strain PM9 (Table 2 and Figure 3). These CTX-1*kan virions were transduced into a recipient strain, from which pCTX-1*kan was obtained. The CTX-1*kan virion titer was 50 virions/ml in the presence of mitomycin C in PM9 culture supernatant (results of five independent experiments). The total number of CTX-1 virions and CTX-1*kan virions that were generated in PM9 should equal the number of CTX-1kan virions that were generated in PM7 (104 virions/ml); thus, the efficiency of generation of CTX-1* virions from V212-1 was estimated to be 1 per 200 progeny virions. CTX-1* can be defined as a CTX phage that is identical to authentic CTX-1, except that it contains ctxBcla.
The inter-strand recombination frequency was reduced to 1/10 in the PM9 recA− strain (6 virions/ml in culture supernatant of PM9 recA−). This suggests that the inter-strand recombination is mainly mediated by homologous recombination; however, further detailed analysis is necessary to reveal the nature of the inter-strand recombination.
pCTX-1*kan was generated only from strain PM9, which has a kanamycin cassette in the first CTX-2 on chromosome 2. No CTX-1*kan was obtained from PM10, which contains the kanamycin cassette in the second CTX-2, in over 5 transduction experiments, possibly because the homologous region downstream of ctxB of the second CTX-2 is not long enough for recombination. However, under natural conditions without the kanamycin cassette, recombination between CTX-1 and the second CTX-2 should be possible. It needs to be elucidated if the inter-strand recombination event happens between two prophages (CTX-1 and CTX-2) or, between the replicative pCTX generated from chromosome 1 and the CTX-2 prophage. Perhaps, the recombination occurs more frequently between pCTX and the CTX-2 prophage than between two prophages.
PM11, PM12, and PM13 were generated similarly from strains that contained CTX-3#:RS1, CTX-5#:RS1, and CTX-6#:RS1, respectively, by replacing the ctxABcla of CTX-2 with a kanamycin resistance cassette (Table 2). pCTX-3kan, pCTX-5kan, and pCTX-6kan were generated from PM11, PM12, and PM13, respectively, by inter-strand recombination like pCTX-1*kan (Table 2 and Figure 3).
These results demonstrate that new mosaic phages are generated through combination of inter-chromosomal recombination event between 2 phages and intra-strand recombination event between the resulting recombinant CTX phage genome and RS1 in an intermediary strain.
Generation of Wave 3 strains
Because new mosaic CTX phages were generated from V212-1, we examined their transmission to novel El Tor strains to generate Wave 3 strains. Construction of the Wave 3 strains required a strain that contained only RS1 on chromosome 1, but no such strains were available among clinical isolates. PM14, which contains only RS1 on chromosome 1, was generated by the excision of CTX-1 from N16961. PM14 was transformed with various pCTXs that were extracted from transductants. pCTXs could be maintained in PM14 in plasmid form under selective antibiotic pressure or as an integrated prophage form. PM15, PM16, PM17, PM18, and PM19, which contained only the integrated pCTX-1kan, pCTX-1*kan, pCTX-3kan, pCTX-5kan, and pCTX-6kan next to the RS1 on chromosome 1, respectively, were screened in the transformants (Table 2).
These results demonstrate that the mosaic CTX phages that are produced from an intermediary strain such as V212-1 can be transmitted to an El Tor strain that contains an RS1 and can be integrated into the genome of a new host bacterial cell, enabling it to develop into a Wave 3 strain.
Discussion
The emergence of atypical El Tor strains traces back to 1991 on the Indian subcontinent, where prototype and atypical El Tor strains began to co-existed for several years [6]. It is assumed that prototype El Tor strains have been extinguished, because only atypical El Tor strains have since been isolated from cholera patients globally [6]. Phylogenetic analyses that are based on SNPs in the V. cholerae genome have indicated that prototype and atypical El Tor strains originated from a common ancestor in the 1950's [10], [25]. Although the genome of El Tor V. cholerae strains has modulated gradually, the changes of their CTX phage have occurred stepwise manner. In addition, strains that harbor various CTX phages constitute phylogenetically distinct subgroups, implying that the acquisition of a new CTX phage is independent of evolution of the bacterial genome [10], [26].
Although several studies have proposed models for the generation of atypical El Tor strains, suggesting that the atypical variants originated through recombination events and lateral gene transfer, the exact process by which the mosaic CTX phages and atypical El Tor strains were generated remains unknown [16], [17], [22], [27]. In this study, we propose a genetic mechanism by which atypical El Tor variants are generated. We demonstrated how a single-segmented CTX virus reorganized its genome through recombination of 2 prophages and/or between a CTX prophage and RS1 in a host bacterial cell, resulting in a novel mosaic CTX virus, which can be transduced to a new host.
Homologous recombination between the common genes (rstRET, rstA, and rstB) between RS1 and CTX-1 or between any gene, except rstR, of the CTX-1 and CTX-2 (or, CTXcla), mediates generation of mosaic CTX phage genomes. Although the creation of CTX-2 was not shown directly—because no V. cholerae strains that have both CTX-1 and CTXcla were available—our results support the existence of an ancestral strain that harbors CTX-1 and CTXcla in nature. Further, V. cholerae O141 strains have been shown to be a reservoir of CTXcla, and an El Tor strain has been shown to take up CTXcla from them [17].
Intra-strand recombination between CTX-1 and RS1 is essential not only for the generation of mosaic CTX phages but also for the emergence of Wave 3 V. cholerae strains that harbor these mosaic phages. CTX and/or RS1 can be excised from chromosome 1 if the array contains a CTX repeat or a CTX:RS1 repeat. When the array ends with RS1, the ultimate product of the excision is a strain that contains a lone RS1. Depending on the structure of the array, the excision rate can reach 1/(4.4×103), indicating that a substantial proportion of the V. cholerae population tends to shed the CTX phage they harbor and is competent to acquire a new CTX phage. Thus, the Wave 3 atypical strains perhaps arose from prototype strains that had lost CTX-1 and obtained mosaic CTX phages that were generated from an intermediary strain.
Based on these results, we propose a pathway by which atypical El Tor variants were generated from prototype El Tor strains (Figure 4). A prototype El Tor strain was infected by a CTX-2 virion, and the CTX-2 genome was integrated as a tandem repeat in the small chromosome—thus, the prototype strain turned into an intermediary strain. The prototype components were removed from the intermediary strain stepwise, and the resulting strains from each step developed into a group of Wave 2 strains. The intermediate strain was also a source of new mosaic phages of Wave 3 strains. New mosaic CTX phages were generated from the intermediary strain, and the progeny mosaic CTX phages were transduced to progenitor strains of Wave 3 strains.
Fig. 4. A model of the generation of Wave 2 and Wave 3 strains. 
Prototype El Tor strains transform into an intermediary strain by CTX-2 phage infection. It is unknown how the CTX-2 phage was generated, but we hypothesize that CTX-2 is generated in a strain containing CTX-1 and CTXcla. The intermediary stain is converted into Wave 2 strains by stepwise removal of CTX-1 and RS1. Mosaic CTX phages, such as CTX-3, CTX-4, are generated from the intermediary strain, and transduced into a new host bacterial strain that harbors only RS1 on chromosome 1, resulting in Wave 3 strains. ctxB type 1 of some Wave 3 strains is further converted into ctxB type 7 by a point mutation. We are interested in examining the dynamics of the emergence of Wave 2 and Wave 3 atypical strains. Classical biotype and prototype El Tor biotype strains coexisted for approximately 2 decades after the prototype El Tor strains first appeared in 1962 in South Asian countries [6]. Yet, inexplicably, the initial appearance of atypical strains was not documented until 1991—30 years after prototype El Tor strains originated. Mosaic CTX phages were most likely generated during the early seventh cholera pandemic period. Whether they were generated in the 1960s—when prototype El Tor strains and classical strains coexisted—and took 30 years to spread, or in the 1990s, shortly before dissemination into the human population, has not been determined. Further study is required to establish when the atypical strains were generated in their natural habitat and disseminated throughout the human population. Coincidentally, had the first atypical strains appeared around the early 1990s, their emergence would have concurred with that of the new O139 serogroup, which is also responsible for epidemic cholera.
The whole-genome analysis of the Haitian strains and a group of Nepalese strains showed that their genomes differ by 1 or 2 base pairs, indicating that the Haitian V. cholerae strains originated from Nepal [28]. The strain that was isolated from the 2010 Haiti cholera outbreak contained CTX-6b and the proportion of strains that contain ctxB genotype 7 has been increasing rapidly in Kolkata, India, since it first emerged in 2006, [13]. Strains that harbored CTX-6b had not been found until 2007 in India, whereas a strain with CTX-6 was isolated in 2007. We hypothesize that mosaic CTX phages that contain ctxB genotype 7 emerged through 2 possible mechanisms: ctxB of the intermediary strain changed first, and progeny mosaic viruses that harbored ctxB genotype 7 were produced; alternatively, CTX-3b and CTX-6b independently arose from CTX-3 and CTX-6 by point mutation of ctxB. CTX-3b and CTX-6b are the CTXs that harbor ctxB genotype 7 that have been identified, but the existence of CTX-4b or CTX-5b or new types of CTXs should be examined. A surveillance study of the generation and dissemination of strains that contain ctxB genotype 7 will provide evidence on how the V. cholerae population has changed and aid in tracking strains with specific CTX prophage.
A link between the increase in severe cholera cases and atypical V. cholerae strains has been proposed, but the precise mechanism by which the clinical symptoms and genetic changes in the bacterial strains correlate remains unknown [29]. Although our results demonstrate the mechanism of the genesis of atypical strains, further studies are required to explain how new variants differ in pathology and how they dominate earlier prevalent strains. It is unclear whether population changes in clinically isolated V. cholerae strains reflect those of V. cholerae in the natural habitat. In addition to bacterial and environmental factors, human activity and, perhaps, the human immune response against temporarily prevalent strains influence population changes in clinically isolated V. cholerae strains.
Materials and Methods
Bacterial strains and genetic structure analysis
The bacterial strains and pCTX are shown in Tables 1 and 2. The CTX and RS1 arrays of the V. cholerae strains were determined and analyzed by sequencing. The DNA sequences of the CTX arrays of bacterial strains and pCTXs that were generated in this study were deposited into GenBank (Table 2).
Discrimination PCR of SNPs of rstB
A PCR primer set was designed for detection of the trinucleotide GTA (nt position 74–76) in rstB, similar to MAMA PCR for distinguishing classical, El Tor, and Haitian type ctxB [13]. For the Wave 3 strains that contained the RS1:CTX array, the forward primer, rstBFw (CTC ATT CTG AAG GGG TGA GTA A) was designed to anneal to rstB of RS1 (Figure S1), and the reverse primer, GTARev (GGT GCA CCA GTC TTA CAA C) was designed to detect GTA with rstBFw (annealing temperature at 63°C). The primer DelRev (GCA CCA GTC TTA CGT AC) was designed to verify the absence of GTA with rstBFw (annealing temperature 58°C).
Surveillance of various CTX prophage genomes in Wave 3 strains
The GTA trinucleotide was examined in rstB in 365 clinical isolates of Wave 3 strains that were collected from 2004 to 2007 in Kolkata, India and 20 isolates that were gathered in Haiti in 2010 [14], [19]. DMAMA (Double-Mismatch-Amplification Mutation Assay)-PCR for ctxB was also performed with these isolates. CTX phages that contained GTA-deleted rstB were confirmed by DNA sequencing of the entire RS1:CTX array.
Excision of the CTX and RS1 from chromosome 1 and chromosome 2 of V212-1
A DNA fragment (690 bp) that contained rstRET (339 bp) and the first 226 bp of rstA was inserted into pCVD442 to generate a recombinant plasmid, pCVDrstRET. The recombinant plasmid was inserted into rstRET of CTX-1 and RS1s of V212-1 to construct V212-1CVD [30]. The excision of CTX-1and/or RS1 was verified by analyzing the genetic structure of strains that were selected on LB plates with 15% sucrose. In addition, pCVDrstRET was inserted into rstRET of PM3 to construct PM3CVD, and excision of the RS1 element was confirmed as described. Excision of CTX-2 from the tandem repeat of CTX-2 on chromosome 2 was examined similarly by inserting rstRcla fragment-containing pCVD442 into each rstRcla of CTX-2s.
Replacement of ctxAB in V212 with a kanamycin cassette
The entire ctxA gene and the first 166 bp (of 375 bp) of ctxB of CTX-1 on chromosome 1 and CTX-2s on chromosome 2 of V212-1 were individually replaced by a kanamycin resistance cassette by allele exchange method [21]. The residual ctxB fragment on each recombinant strain contained the second SNP (nucleotide 203) of ctxBEl Tor (T) and ctxBcla (C) on chromosomes 1 and 2, respectively. This SNP was used to distinguish pCTX-1kan and pCTX-1*kan. Similarly, a kanamycin cassette was introduced into CTX-1 on chromosome 1 of PM1, resulting in strain PM8. Strains PM11, PM12, and PM13 were constructed similarly by replacing the ctxABcla of the first CTX-2 on chromosome 2 with a kanamycin cassette.
Production of CTX-1 phage from chromosome 1 of V212-1
Transduction of kanamycin-resistant CTX virions was monitored as described [21], [31]. Briefly, PM7 culture supernatant (LB containing 20 ng/ml of mitomycin C) was mixed with a classical strain, O395, as the recipient. The mixture was incubated for 30 min for phage infection and plated on LB plates that contained kanamycin. pCTX-1kan, a replicative form of the CTX genome in the transductants, was extracted and analyzed by sequencing. CTX-1-1kan virions were generated similarly from PM8 and were determined to have the same DNA sequence as CTX-1kan (Table 2).
Generation of Wave 3 strains
PM14 was generated from N16961 by excision of CTX-1. The recombinant plasmid pCVDrstRET was inserted into CTX-1 of N16961, and strain PM14 was screened as described above. PM14 was transformed by the replicative form of CTXs that were generated in this study (pCTX-1kan, pCTX-1*kan, pCTX-3kan, pCTX5kan, and pCTX-6kan). Integration of pCTXs next to RS1 on chromosome 1 of PM14 was confirmed by DNA sequencing (Table 1) [8].
recA− mutants construction
The internal 600 nt DNA fragment (nucleotide position 119–720) of recA was amplified with primers recASacIF: CGC GCG GAG CTC CGA CCC GAT TTC GAC ACA and recAEcoRIR: CCG GGC GAA TTC CTT GAG GTT ACC CGT CAG by using PCR. This fragment was inserted into a suicide plasmid pSW23.oriT to construct a recombinant plasmid pSWrecA [32]. The recombinant plasmid was individually transferred by conjugation to V212-1CVD, PM3CVD, and PM9. The recA of each strain was disrupted by insertion of pSWrecA.
Accession numbers
The nucleotide sequences of CTX prophages and pCTXs were deposited in GenBank under accession numbers KF471410, KF664566–KF664576, KF664579, KF664580, and KJ540257–KJ540278.
Supporting Information
Zdroje
1. KaperJB, MorrisJGJr, LevineMM (1995) Cholera. Clin Microbiol Rev 8 : 48–86.
2. SackDA, SackRB, NairGB, SiddiqueAK (2004) Cholera. Lancet 363 : 223–233.
3. ChoiSY, LeeJH, KimEJ, LeeHR, JeonYS, et al. (2010) Classical RS1 and environmental RS1 elements in Vibrio cholerae O1 El Tor strains harbouring a tandem repeat of CTX prophage: revisiting Mozambique in 2005. J Med Microbiol 59 : 302–308.
4. BoydEF, HeilpernAJ, WaldorMK (2000) Molecular analyses of a putative CTXΦ precursor and evidence for independent acquisition of distinct CTXΦs by toxigenic Vibrio cholerae. J Bacteriol 182 : 5530–5538.
5. NairGB, QadriF, HolmgrenJ, SvennerholmAM, SafaA, et al. (2006) Cholera due to altered El Tor strains of Vibrio cholerae O1 in Bangladesh. J Clin Microbiol 44 : 4211–4213.
6. SafaA, NairGB, KongRY (2010) Evolution of new variants of Vibrio cholerae O1. Trends Microbiol 18 : 46–54.
7. MoritaM, OhnishiM, ArakawaE, YamamotoS, NairGB, et al. (2010) Emergence and genetic diversity of El Tor Vibrio cholerae O1 that possess classical biotype ctxB among travel-associated cases of cholera in Japan. J Med Microbiol 59 : 708–712.
8. LeeJH, ChoiSY, JeonYS, LeeHR, KimEJ, et al. (2009) Classification of hybrid and altered Vibrio cholerae strains by CTX prophage and RS1 element structure. J Microbiol 47 : 783–788.
9. AnsaruzzamanM, BhuiyanNA, NairBG, SackDA, LucasM, et al. (2004) Cholera in Mozambique, variant of Vibrio cholerae. Emerg Infect Dis 10 : 2057–2059.
10. MutrejaA, KimDW, ThomsonNR, ConnorTR, LeeJH, et al. (2011) Evidence for several waves of global transmission in the seventh cholera pandemic. Nature 477 : 462–465.
11. NguyenBM, LeeJH, CuongNT, ChoiSY, HienNT, et al. (2009) Cholera outbreaks caused by an altered Vibrio cholerae O1 El Tor biotype strain producing classical cholera toxin B in Vietnam in 2007 to 2008. J Clin Microbiol 47 : 1568–1571.
12. ChoiSY, LeeJH, JeonYS, LeeHR, KimEJ, et al. (2010) Multilocus variable-number tandem repeat analysis of Vibrio cholerae O1 El Tor strains harbouring classical toxin B. J Med Microbiol 59 : 763–769.
13. NahaA, PazhaniGP, GangulyM, GhoshS, RamamurthyT, et al. (2012) Development and evaluation of a PCR assay for tracking the emergence and dissemination of Haitian variant ctxB in Vibrio cholerae O1 strains isolated from Kolkata, India. J Clin Microbiol 50 : 1733–1736.
14. ChinCS, SorensonJ, HarrisJB, RobinsWP, CharlesRC, et al. (2011) The origin of the Haitian cholera outbreak strain. N Engl J Med 364 : 33–42.
15. ShakyaG, KimDW, ClemensJD, MallaS, UpadhyayaBP, et al. (2012) Phenotypic and genetic characterization of Vibrio cholerae O1 clinical isolates collected through national antimicrobial resistance surveillance network in Nepal. World J Microbiol Biotechnol 28 : 2671–2678.
16. PatraT, ChatterjeeS, RaychoudhuriA, MukhopadhyayAK, RamamurthyT, et al. (2011) Emergence and progression of Vibrio cholerae O1 El Tor variants and progenitor strains of Mozambique variants in Kolkata, India. Int J Med Microbiol 301 : 310–317.
17. UddenSM, ZahidMS, BiswasK, AhmadQS, CraviotoA, et al. (2008) Acquisition of classical CTX prophage from Vibrio cholerae O141 by El Tor strains aided by lytic phages and chitin-induced competence. Proc Natl Acad Sci U S A 105 : 11951–11956.
18. KimEJ, LeeD, MoonSH, LeeCH, KimDW (2014) CTX prophages in Vibrio cholerae O1 strains. J Microbiol Biotechnol 24 : 725–731.
19. SurD, LopezAL, KanungoS, PaisleyA, MannaB, et al. (2009) Efficacy and safety of a modified killed-whole-cell oral cholera vaccine in India: an interim analysis of a cluster-randomised, double-blind, placebo-controlled trial. Lancet 374 : 1694–1702.
20. LeeJH, HanKH, ChoiSY, LucasME, MondlaneC, et al. (2006) Multilocus sequence typing (MLST) analysis of Vibrio cholerae O1 El Tor isolates from Mozambique that harbour the classical CTX prophage. J Med Microbiol 55 : 165–170.
21. WaldorMK, MekalanosJJ (1996) Lysogenic conversion by a filamentous phage encoding cholera toxin. Science 272 : 1910–1914.
22. FaruqueSM, TamVC, ChowdhuryN, DiraphatP, DziejmanM, et al. (2007) Genomic analysis of the Mozambique strain of Vibrio cholerae O1 reveals the origin of El Tor strains carrying classical CTX prophage. Proc Natl Acad Sci U S A 104 : 5151–5156.
23. GrimCJ, HasanNA, TavianiE, HaleyB, ChunJ, et al. (2010) Genome sequence of hybrid Vibrio cholerae O1 MJ-1236, B-33, and CIRS101 and comparative genomics with V. cholerae. J Bacteriol 192 : 3524–3533.
24. DavisBM, WaldorMK (2000) CTXΦ contains a hybrid genome derived from tandemly integrated elements. Proc Natl Acad Sci U S A 97 : 8572–8577.
25. ChunJ, GrimCJ, HasanNA, LeeJH, ChoiSY, et al. (2009) Comparative genomics reveals mechanism for short-term and long-term clonal transitions in pandemic Vibrio cholerae. Proc Natl Acad Sci U S A 106 : 15442–15447.
26. ChoYJ, YiH, LeeJH, KimDW, ChunJ (2010) Genomic evolution of Vibrio cholerae. Curr Opin Microbiol 13 : 646–651.
27. HalderK, DasB, NairGB, BhadraRK (2010) Molecular evidence favouring step-wise evolution of Mozambique Vibrio cholerae O1 El Tor hybrid strain. Microbiology 156 : 99–107.
28. HendriksenRS, PriceLB, SchuppJM, GilleceJD, KaasRS, et al. (2011) Population genetics of Vibrio cholerae from Nepal in 2010: evidence on the origin of the Haitian outbreak. mBio 2: e00157–11.
29. SiddiqueAK, NairGB, AlamM, SackDA, HuqA, et al. (2010) El Tor cholera with severe disease: a new threat to Asia and beyond. Epidemiol Infect 138 : 347–352.
30. DonnenbergMS, KaperJB (1991) Construction of an eae deletion mutant of enteropathogenic Escherichia coli by using a positive-selection suicide vector. Infect Immun 59 : 4310–4317.
31. DasB, BischerourJ, BarreFX (2011) VGJΦ integration and excision mechanisms contribute to the genetic diversity of Vibrio cholerae epidemic strains. Proc Natl Acad Sci U S A 108 : 2516–2521.
32. KimDW, LenzenG, PageAL, LegrainP, SansonettiPJ, et al. (2005) The Shigella flexneri effector OspG interferes with innate immune responses by targeting ubiquitin-conjugating enzymes. Proc Natl Acad Sci U S A 102 : 14046–14051.
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek Out-of-Sequence Signal 3 as a Mechanism for Virus-Induced Immune Suppression of CD8 T Cell ResponsesČlánek RNF26 Temporally Regulates Virus-Triggered Type I Interferon Induction by Two Distinct MechanismsČlánek Mouse, but Not Human, ApoB-100 Lipoprotein Cholesterol Is a Potent Innate Inhibitor of Pneumolysin
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2014 Číslo 9- Jak souvisí postcovidový syndrom s poškozením mozku?
- Měli bychom postcovidový syndrom léčit antidepresivy?
- Farmakovigilanční studie perorálních antivirotik indikovaných v léčbě COVID-19
- 10 bodů k očkování proti COVID-19: stanovisko České společnosti alergologie a klinické imunologie ČLS JEP
-
Všechny články tohoto čísla
- Virus Control Goes Epigenetic
- The Role of Iron in Prion Disease and Other Neurodegenerative Diseases
- The Ins and Outs of Rust Haustoria
- Prion Strains and Amyloid Polymorphism Influence Phenotypic Variation
- Teaching Fido New ModiFICation Tricks
- Can Enhance Infection in Mosquitoes: Implications for Malaria Control?
- MIF Contributes to Associated Immunopathogenicity Development
- Persistence of Virus Reservoirs in ART-Treated SHIV-Infected Rhesus Macaques after Autologous Hematopoietic Stem Cell Transplant
- Bacillus Calmette-Guerin Infection in NADPH Oxidase Deficiency: Defective Mycobacterial Sequestration and Granuloma Formation
- EhCoactosin Stabilizes Actin Filaments in the Protist Parasite
- Molecular Insights Into the Evolutionary Pathway of O1 Atypical El Tor Variants
- LprG-Mediated Surface Expression of Lipoarabinomannan Is Essential for Virulence of
- Structural Correlates of Rotavirus Cell Entry
- Multivalent Adhesion Molecule 7 Clusters Act as Signaling Platform for Host Cellular GTPase Activation and Facilitate Epithelial Barrier Dysfunction
- The Effects of Vaccination and Immunity on Bacterial Infection Dynamics
- Myeloid Derived Hypoxia Inducible Factor 1-alpha Is Required for Protection against Pulmonary Infection
- Functional Characterisation of Germinant Receptors in and Presents Novel Insights into Spore Germination Systems
- Global Analysis of Neutrophil Responses to Reveals a Self-Propagating Inflammatory Program
- Host Cell Invasion by Apicomplexan Parasites: The Junction Conundrum
- Comparative Phenotypic Analysis of the Major Fungal Pathogens and
- Unravelling the Multiple Functions of the Architecturally Intricate β-galactosidase, BgaA
- Sialylation of Prion Protein Controls the Rate of Prion Amplification, the Cross-Species Barrier, the Ratio of PrP Glycoform and Prion Infectivity
- Symbionts Commonly Provide Broad Spectrum Resistance to Viruses in Insects: A Comparative Analysis of Strains
- Ontogeny of Recognition Specificity and Functionality for the Broadly Neutralizing Anti-HIV Antibody 4E10
- Identification and Characterisation of a Hyper-Variable Apoplastic Effector Gene Family of the Potato Cyst Nematodes
- Crimean-Congo Hemorrhagic Fever Virus Entry into Host Cells Occurs through the Multivesicular Body and Requires ESCRT Regulators
- Age-Dependent Enterocyte Invasion and Microcolony Formation by
- CD160-Associated CD8 T-Cell Functional Impairment Is Independent of PD-1 Expression
- Functional Fluorescent Protein Insertions in Herpes Simplex Virus gB Report on gB Conformation before and after Execution of Membrane Fusion
- The Tudor Domain Protein Spindlin1 Is Involved in Intrinsic Antiviral Defense against Incoming Hepatitis B Virus and Herpes Simplex Virus Type 1
- Transgenic Analysis of the MAP Kinase MPK10 Reveals an Auto-inhibitory Mechanism Crucial for Stage-Regulated Activity and Parasite Viability
- Evidence for a Transketolase-Mediated Metabolic Checkpoint Governing Biotrophic Growth in Rice Cells by the Blast Fungus
- Incomplete Deletion of IL-4Rα by LysM Reveals Distinct Subsets of M2 Macrophages Controlling Inflammation and Fibrosis in Chronic Schistosomiasis
- Identification and Functional Expression of a Glutamate- and Avermectin-Gated Chloride Channel from , a Southern Hemisphere Sea Louse Affecting Farmed Fish
- Out-of-Sequence Signal 3 as a Mechanism for Virus-Induced Immune Suppression of CD8 T Cell Responses
- Strong Epistatic Selection on the RNA Secondary Structure of HIV
- Hematopoietic but Not Endothelial Cell MyD88 Contributes to Host Defense during Gram-negative Pneumonia Derived Sepsis
- Delineation of Interfaces on Human Alpha-Defensins Critical for Human Adenovirus and Human Papillomavirus Inhibition
- Exploitation of Reporter Strains to Probe the Impact of Vaccination at Sites of Infection
- RNF26 Temporally Regulates Virus-Triggered Type I Interferon Induction by Two Distinct Mechanisms
- Helminth Infections Coincident with Active Pulmonary Tuberculosis Inhibit Mono- and Multifunctional CD4 and CD8 T Cell Responses in a Process Dependent on IL-10
- MHC Class II Restricted Innate-Like Double Negative T Cells Contribute to Optimal Primary and Secondary Immunity to
- Reactive Oxygen Species Regulate Caspase-11 Expression and Activation of the Non-canonical NLRP3 Inflammasome during Enteric Pathogen Infection
- Evolution of Plastic Transmission Strategies in Avian Malaria
- A New Human 3D-Liver Model Unravels the Role of Galectins in Liver Infection by the Parasite
- Translocates into the Myocardium and Forms Unique Microlesions That Disrupt Cardiac Function
- Mouse, but Not Human, ApoB-100 Lipoprotein Cholesterol Is a Potent Innate Inhibitor of Pneumolysin
- The Cofilin Phosphatase Slingshot Homolog 1 (SSH1) Links NOD1 Signaling to Actin Remodeling
- Kaposi's Sarcoma Herpesvirus MicroRNAs Induce Metabolic Transformation of Infected Cells
- Reorganization of the Endosomal System in -Infected Cells: The Ultrastructure of -Induced Tubular Compartments
- Distinct Dictation of Japanese Encephalitis Virus-Induced Neuroinflammation and Lethality via Triggering TLR3 and TLR4 Signal Pathways
- Exploitation of the Complement System by Oncogenic Kaposi's Sarcoma-Associated Herpesvirus for Cell Survival and Persistent Infection
- The Secreted Peptide PIP1 Amplifies Immunity through Receptor-Like Kinase 7
- Structural Insight into Host Recognition by Aggregative Adherence Fimbriae of Enteroaggregative
- The CD14CD16 Inflammatory Monocyte Subset Displays Increased Mitochondrial Activity and Effector Function During Acute Malaria
- Infection Induces Expression of a Mosquito Salivary Protein (Agaphelin) That Targets Neutrophil Function and Inhibits Thrombosis without Impairing Hemostasis
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- The Secreted Peptide PIP1 Amplifies Immunity through Receptor-Like Kinase 7
- Symbionts Commonly Provide Broad Spectrum Resistance to Viruses in Insects: A Comparative Analysis of Strains
- MIF Contributes to Associated Immunopathogenicity Development
- The Ins and Outs of Rust Haustoria
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



