-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaA Component of the Xanthomonadaceae Type IV Secretion System Combines a VirB7 Motif with a N0 Domain Found in Outer Membrane Transport Proteins
Type IV secretion systems (T4SS) are used by Gram-negative bacteria to translocate protein and DNA substrates across the cell envelope and into target cells. Translocation across the outer membrane is achieved via a ringed tetradecameric outer membrane complex made up of a small VirB7 lipoprotein (normally 30 to 45 residues in the mature form) and the C-terminal domains of the VirB9 and VirB10 subunits. Several species from the genera of Xanthomonas phytopathogens possess an uncharacterized type IV secretion system with some distinguishing features, one of which is an unusually large VirB7 subunit (118 residues in the mature form). Here, we report the NMR and 1.0 Å X-ray structures of the VirB7 subunit from Xanthomonas citri subsp. citri (VirB7XAC2622) and its interaction with VirB9. NMR solution studies show that residues 27–41 of the disordered flexible N-terminal region of VirB7XAC2622 interact specifically with the VirB9 C-terminal domain, resulting in a significant reduction in the conformational freedom of both regions. VirB7XAC2622 has a unique C-terminal domain whose topology is strikingly similar to that of N0 domains found in proteins from different systems involved in transport across the bacterial outer membrane. We show that VirB7XAC2622 oligomerizes through interactions involving conserved residues in the N0 domain and residues 42–49 within the flexible N-terminal region and that these homotropic interactions can persist in the presence of heterotropic interactions with VirB9. Finally, we propose that VirB7XAC2622 oligomerization is compatible with the core complex structure in a manner such that the N0 domains form an extra layer on the perimeter of the tetradecameric ring.
Published in the journal: . PLoS Pathog 7(5): e32767. doi:10.1371/journal.ppat.1002031
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1002031Summary
Type IV secretion systems (T4SS) are used by Gram-negative bacteria to translocate protein and DNA substrates across the cell envelope and into target cells. Translocation across the outer membrane is achieved via a ringed tetradecameric outer membrane complex made up of a small VirB7 lipoprotein (normally 30 to 45 residues in the mature form) and the C-terminal domains of the VirB9 and VirB10 subunits. Several species from the genera of Xanthomonas phytopathogens possess an uncharacterized type IV secretion system with some distinguishing features, one of which is an unusually large VirB7 subunit (118 residues in the mature form). Here, we report the NMR and 1.0 Å X-ray structures of the VirB7 subunit from Xanthomonas citri subsp. citri (VirB7XAC2622) and its interaction with VirB9. NMR solution studies show that residues 27–41 of the disordered flexible N-terminal region of VirB7XAC2622 interact specifically with the VirB9 C-terminal domain, resulting in a significant reduction in the conformational freedom of both regions. VirB7XAC2622 has a unique C-terminal domain whose topology is strikingly similar to that of N0 domains found in proteins from different systems involved in transport across the bacterial outer membrane. We show that VirB7XAC2622 oligomerizes through interactions involving conserved residues in the N0 domain and residues 42–49 within the flexible N-terminal region and that these homotropic interactions can persist in the presence of heterotropic interactions with VirB9. Finally, we propose that VirB7XAC2622 oligomerization is compatible with the core complex structure in a manner such that the N0 domains form an extra layer on the perimeter of the tetradecameric ring.
Introduction
Bacteria employ multiprotein secretion systems to translocate effector proteins or nucleoprotein complexes across the cell envelope where they modulate the bacterium's interactions with the environment. In Gram-negative bacteria, the secretion systems have been classified into 6 different types [1], [2]. The type IV secretion systems (T4SSs) are ancestrally related to bacterial conjugation machines [3] and are able to translocate proteins and/or protein-DNA complexes to the extracellular milieu or the host interior, in many cases contributing to the ability of the bacterial pathogen to colonize the host and evade its immune system [4]. T4SSs are important in the pathogenic mechanism of many microbes, including the animal pathogens Legionella pneumophila (Legionnaires' disease; [5]), Bordetella pertussis (whooping cough; [6]), Coxiella burnetii (Q fever; [7]), Bartonella henselae (cat scratch disease, [8]), Brucella spp. (brucellosis; [9]) and Helicobacter pylori (gastritis, ulcers, stomach cancer; [10]), as well as the plant pathogen Agrobacterium tumefaciens (crown gall disease; [11]) which has the prototype T4SS, composed of 12 proteins, VirB1-VirB11 and VirD4 [12].
Xanthomonas citri subsp. citri (formerly Xanthomonas axonopodis pv. citri) (Xac) is a gammaproteobacterial phytopathogen which causes citrus canker, a disease that affects all citrus plants and has significant economic impact worldwide [13]. The Xac genome was sequenced and many potential virulence-related genes were identified [14], including an 18 kb chromosomal vir locus that codes for a T4SS [13], [14], [15]. Orthologous T4SSs have been identified in the closely related species Xanthomonas campestris pv. campestris (strains ATCC 33913 [14], 8004 [16] and B100 [17]), Xanthomonas albilineans [18], Xanthomonas campestris pv. vasculorum NCPPB702 [19], Xanthomonas campestris pv. musacearum NCPPB4381 [19] and Stenotrophomonas maltophilia (strains K279A [20] and R551-3 [21]). The vir locus is incomplete in Xanthomonas campestris pv. vesicatoria [22] and is absent in four Xanthomonas oryzae strains (KACC10331 [23], MAFF311018 [24], PXO99A [25] and BLS256 (GenBank database AAQN00000000)) and in two Xanthomonas fuscans subsp. aurantifolii lineages (10535 and 11122 [26]). The unrelated Gram-negative bacteria Neisseria flavescens SK114 (GenBank database ACQV00000000) and Neisseria sp. oral taxon 014 str. F0314 (GenBank database ADEA00000000) also present a locus highly similar to the T4SS loci found in the Xanthomonadaceae family. Some Xanthomonas genomes, including Xac, carry megaplasmids that code for a second T4SS probably involved in plasmid mobilization and whose structural components exhibit only very low sequence identity to their counterparts in the chromosomally encoded T4SS under study [15], [27].
One particularly distinctive feature of the Xanthomonad T4SSs is related to the VirB7 component. In well-characterized T4SSs, VirB7 is a lipoprotein attached to the periplasmic side of the outer membrane [28]. It has been shown to interact with several T4SS subunits [29], including itself [30] and the VirB9 C-terminal region [31], [32]. The structure of a VirB7-VirB9-VirB10 complex (TraN-TraO-TraF) from the T4SS of the conjugative plasmid pKM101 demonstrated that these proteins form a hetero-tetradecameric outer membrane channel through which the substrates pass [33], [34]. Structures of the complex formed between the TraO C-terminal domain and TraN revealed that the 33-residue TraN forms an extended structure that winds around the TraO β-sandwich [34], [35].
Members of the VirB7 family are typically 45–65 residues long [36], becoming 15–20 residues shorter after removal of the N-terminal signal sequence and covalent attachment to lipid molecules. In Xac, several lines of evidence indicate that the gene xac2622 codes for a VirB7 equivalent; specifically, the gene position, the presence of signals for outer membrane localization and lipidation and the interaction of its product with VirB9 [15]. However, XAC2622 does not exhibit any sequence similarity with the VirB7 family and it is much larger than a typical VirB7 protein (139 and 118 residues before and after signal sequence removal, respectively).
In order to understand the unique structural features of Xanthomonad VirB7, we have solved the solution nuclear magnetic resonance (NMR) and X-ray crystal structures of XAC2622, which we now call VirB7XAC2622, and studied its interaction with VirB9. VirB7XAC2622 presents all of the characteristics of VirB7: predicted signal peptide and lipobox sequences followed by a short and extended region that contains the VirB9 binding site. However, unlike other VirB7 proteins, VirB7XAC2622 has an extra domain whose topology is strikingly similar to that of N0 domains found in proteins from different multi-subunit complexes involved in transport across the bacterial outer membrane. We show that VirB7XAC2622 oligomerizes through interactions involving conserved residues in the folded domain and the unfolded N-terminus and that oligomerization and the formation of the VirB7XAC2622-VirB9 complex can occur simultaneously. We propose that these interactions are compatible with the tetradecameric core complex structure, resulting in the formation of an extra ring layer.
Results
Solution Structure of Xac VirB7 (VirB7XAC2622_24–139)
The construct used for NMR structure determination corresponds to residues 24–139 of VirB7XAC2622 preceded by a Gly-Ser-His-Met sequence. Bioinformatics analysis predicts that residues 1–21 would be removed and Cys22 lipidated, leading to anchoring in the bacterial outer membrane [15]. Approximately 98% of main chain and aliphatic side chain and 93% of aromatic side chain 1H, 13C and 15N resonances were assigned (Table 1 and Figure S1). A total of 2176 experimental NMR restraints, consisting of 2056 interproton distances and 120 torsion angles, were used in the structure calculation (Table 1). A small set of nuclear Overhauser effect (NOE) cross-peaks that were not assigned by CYANA [37] were subsequently identified as resulting from intermolecular contacts (see below).
Tab. 1. Assignment and structural statistics for the solution structure of VirB7XAC2622_24–139. 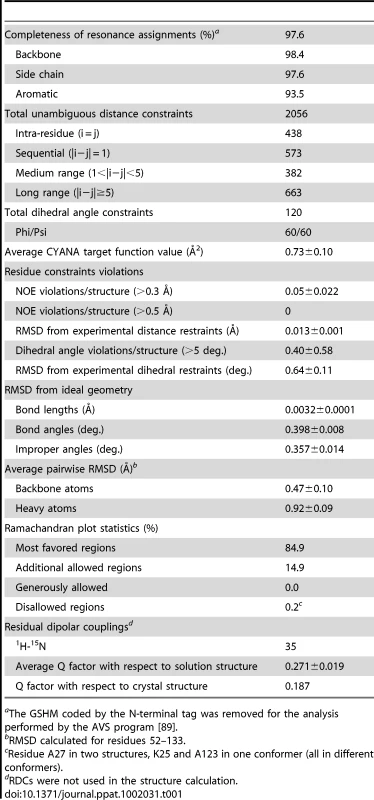
aThe GSHM coded by the N-terminal tag was removed for the analysis performed by the AVS program [89]. The solution structure of VirB7XAC2622_24–139 consists of a globular domain (residues 52–133), flanked by a long disordered N-terminus (amino acids 24–51) and a short flexible C-terminus (residues 134–139) (Figure 1A). The globular domain is composed of two α-helices sandwiched between a mixed three stranded β-sheet on one side and an antiparallel two-stranded β-sheet plus a short 310 helix on the other (Figure 1B). The NMR structures were independently validated by 1H-15N residual dipolar couplings (RDCs). The average Q factor obtained by fitting RDCs from secondary structure elements to the whole NMR ensemble is 0.271±0.019, consistent with a good quality structure [38], [39], [40].
Fig. 1. NMR and X-ray models of VirB7XAC2622. 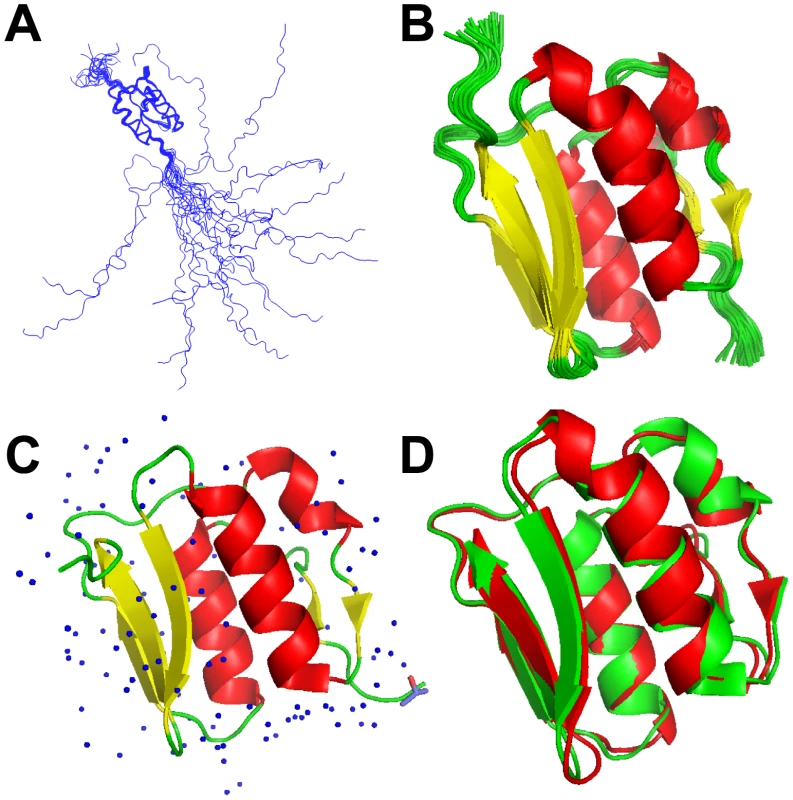
Superposition of 20 lowest energy VirB7XAC2622_24–139 solution structures: (A) backbone traces of the full-length protein (residues 24–139) and (B) ribbon representation of the folded domain (residues 51–134) in the NMR ensemble. (C) Ribbon representation of the X-ray crystal structure of VirB7XAC2622_51–134. Water molecules and one isopropanol ligand are depicted. (D) Superposition of the X-ray (green) and NMR (lowest energy model; red) structures of the VirB7XAC2622 globular domain. High Resolution X-ray Crystal Structure of the VirB7XAC2622 Globular Domain
To better characterize the globular domain, we cloned and purified a recombinant fragment encompassing residues 51–134 (VirB7XAC2622_51–134) which was submitted to crystallization trials that resulted in large plates (Figure S2) which belong to space group C2221 and diffracted up to 1.0 Å. Molecular replacement was performed using the solution structure of the VirB7XAC2622 globular domain as the search model. Details of the data collection and structure refinement are listed in Table 2 and the model is presented in Figure 1C. There was clear electron density for all residues, with the exception of the first amino acid (threonine 51) and residues 122 and 123 in the loop between β4 and β5. As expected, the crystal structure is very similar to the NMR ensemble (Figure 1D), displaying an average root mean square deviation (RMSD) for the backbone heavy atoms and all heavy atoms of 1.05±0.05 Å and 1.61±0.06 Å for residues 53–130, and excellent agreement with the backbone 1H-15N RDCs [Q factor of 0.187 (regular secondary structures only) and 0.272 (all residues)].
Tab. 2. Crystallographic data collection and refinement statistics of VirB7XAC2622_51–134. 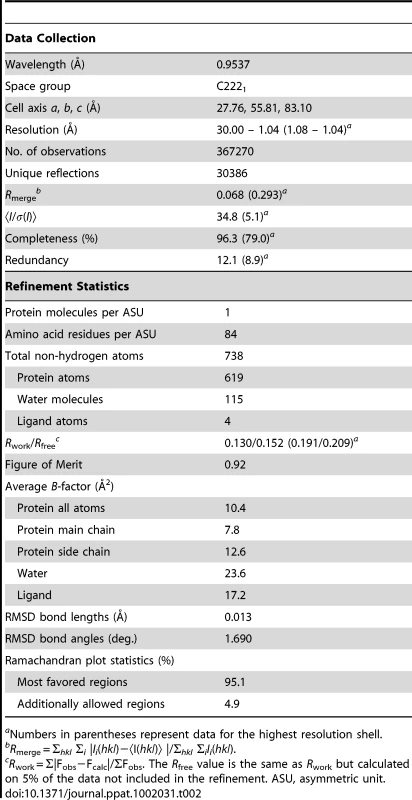
aNumbers in parentheses represent data for the highest resolution shell. VirB7 Oligomerization
Changes in 15N heteronuclear single-quantum coherence (15N-HSQC) cross-peak positions as a function of protein concentration indicated that VirB7XAC2622 oligomerizes in fast exchange on the NMR time scale (Figure 2A). Consistent with this observation, alterations in 15N T1 and T2 relaxation times indicated that the overall tumbling slows significantly at higher protein concentrations (see below). Chemical shift perturbation data showed that two regions are involved in VirB7XAC2622 self-interactions: a region in the unfolded N-terminus (residues 42–49) and a patch on the surface of the globular domain made up of residues 63–65, 85–93, 111–119 and 131 (Figure 2B and C).
Fig. 2. VirB7XAC2622_24–139 oligomerization. 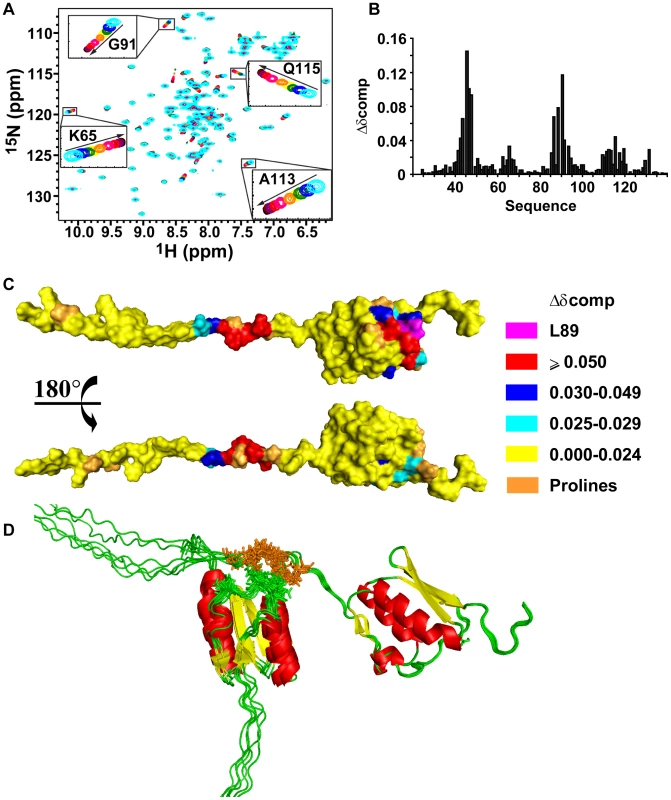
(A) Superposition of 15N-HSQC spectra of 15N-VirB7XAC2622_24–139 at different concentrations. Cyan: 850 µM; blue: 600 µM; green: 400 µM; yellow: 300 µM; orange: 200 µM; pink: 100 µM; red: 50 µM; purple: 25 µM; brown: 13 µM and black: 7 µM. The inserts are amplifications of selected spectral regions showing peak movements as the protein concentration varies from 850 to 7 µM in the direction of the arrows. (B) Weighted chemical shift changes (Δδcomp) observed upon dilution from 100 µM to 7 µM. (C) Surface representation of VirB7XAC2622_24–139 colored according to the weighted chemical shift changes. Residues involved in oligomerization are located in the folded domain and in the unfolded N-terminus. 1H-15N correlation of Leucine 89 (L89) was not detected in any 15N-HSQC spectrum probably due to chemical exchange. (D) Ribbon representations of docking models of the VirB7XAC2622 – VirB7XAC2622 interaction. Helices, β-strands and coil regions are colored red, yellow and green, respectively. Residues involved in intermolecular NOEs are shown as stick models colored brown (residues A43, T45, E46, I47, L49) and green (T63, S85, D86, Y87, T88, I90). Six models are shown. Two possible interaction schemes can be envisaged that would be compatible with the chemical shift perturbation data: i) a side-by-side arrangement in which the N-terminal unfolded segment and the globular domain of one molecule interact, respectively, with the same regions of a second molecule, or ii) a head-to-tail complex arrangement where the N-terminus of one molecule recognizes the folded domain of the other. In order to test the hypothesis of the formation of a head to tail complex, we performed a titration of the 15N-VirB7XAC2622_51–134 globular domain with an unlabeled peptide encompassing residues 38–52 from the N-terminal region (VirB7XAC2622_38–52). During the titration, we observed changes in the same cross-peaks which were perturbed in the entire protein in a concentration dependent manner (Figure S3). This hypothesis was corroborated by the analysis of a set of 13 NOEs not assigned by CYANA [37], which showed that they could be accounted for by the formation of head-to-tail dimers. Indeed, those NOEs were observed between proton pairs derived from amino acids whose main-chain 1H-15N chemical shifts are perturbed in a concentration dependent manner (the NOEs are listed in the Materials and Methods).
We then used the set of 13 intermolecular NOEs as geometric restraints to drive computational docking simulations of the VirB7XAC2622 dimer using HADDOCK2.0 [41]. In one simulation, a fragment corresponding to the disordered N-terminal region (residues 24–50) was docked with a fragment corresponding to the globular domain (residues 51–139) while the second simulation involved the docking between two full length proteins (residues 24–139). The resulting models from both simulations were highly similar. In the second simulation three of the thirteen NOEs were violated in all solutions by approximately 1.4 to 3 Å. They correspond to weak peaks and could have contributions of spin diffusion or chemical exchange. These docking simulations were therefore able to determine the general nature of the oligomerization interface whose resolution is necessarily limited by the small number of restraints and by the absence of structural information for the N-terminal region. One cluster of docking solutions using full length VirB7XAC2622 is shown in Figure 2D.
To investigate whether VirB7XAC2622_24–139 forms dimers or higher-order oligomers, glutaraldehyde cross-linking experiments were performed. These assays showed that VirB7XAC2622_24–139 forms dimers, trimers and higher order oligomers (Figure S4A). When the cross-linking experiment was performed in the presence of 1% SDS, no covalently cross-linked oligomers were observed, indicating that oligomerization requires the presence of a correctly folded protein (Figure S4A). As expected, no cross-links were observed when the experiment was performed using VirB7XAC2622_51–134 which lacks the disordered N-terminal region (Figure S4B).
VirB7XAC2622 Interaction with VirB9XAC2620
To analyze this interaction, VirB7XAC2622_24–139_His was titrated into a solution of VirB9XAC2620_34–255 (full-length VirB9XAC2620 minus the first 33 residues including a predicted signal peptide) and changes in intrinsic fluorescence were monitored. A monotonic increase in fluorescence emission was detected until saturation at a 1∶1 VirB7∶VirB9 stoichiometry (Figure S5). A dissociation constant of 4×10−8 M for the complex was estimated by fitting the observed fluorescence changes to a binding isotherm (equation 2, Materials and Methods).
To study this interaction by NMR, the 15N-HSQC spectrum of 15N-VirB7XAC2622_24–139 was acquired in the presence of unlabeled VirB9XAC2620_34–255. At a 1∶1 molar ratio, essentially no cross peaks were detected, except for those derived from the flexible N - and C-terminal residues (amino acids 24–25 and 134–139; data not shown). This observation is consistent with the formation of a tight complex with long correlation time, with the majority of cross peaks beyond detection.
VirB7XAC2622 was previously shown to interact with the C-terminal domain of VirB9XAC2620 [15] as has been shown for other VirB7-VirB9 homologs [32], [35]. We therefore produced a fragment, VirB9XAC2620_154–255, corresponding to the C-terminal domain of VirB9XAC2620. Changes in the 15N-HSQC spectrum of VirB7XAC2622_24–139 upon adding VirB9XAC2620_154–255 showed that the binding occurs in slow exchange on the NMR time scale, consistent with the sub-micromolar affinity detected by fluorescence spectroscopy for the interaction with full length VirB9 (VirB9XAC2620_34–255). In order to map the VirB9 binding site on the structure of VirB7, we assigned the backbone resonances of VirB7XAC2622_24–139 in complex with VirB9XAC2620_154–255 and analyzed the chemical shift differences (Figure 3A and B). This analysis showed that only residues 27–41, within the disordered VirB7 N-terminus, undergo significant chemical shift perturbations (Figure 3C and D). This region is adjacent to, but does not overlap, the N-terminal region involved in VirB7XAC2622 oligomerization (residues 42–49). 1H-15N RDCs and backbone chemical shifts for VirB7XAC2622_24–139 alone and in the presence of VirB9XAC2620_154–255 are essentially identical, with the exception of the N-terminal region that is involved in binding to VirB9 (Figure 3 and Figure S6). Thus, the folded domain does not participate in the recognition of the VirB9XAC2620 C-terminal domain and the linker between the VirB9 interaction site and the VirB7 globular domain is flexible enough to permit the two regions to align independently in the alignment medium. Interestingly, VirB7XAC2622_24–139 is able to simultaneously oligomerize and interact with VirB9XAC2620_154–255, as the cross-peaks related to the regions involved in VirB7 oligomerization shift even in the presence of VirB9XAC2620_154–255 (Figure S7).
Fig. 3. The VirB7XAC2622 N-terminus recognizes the VirB9XAC2620 C-terminal domain. 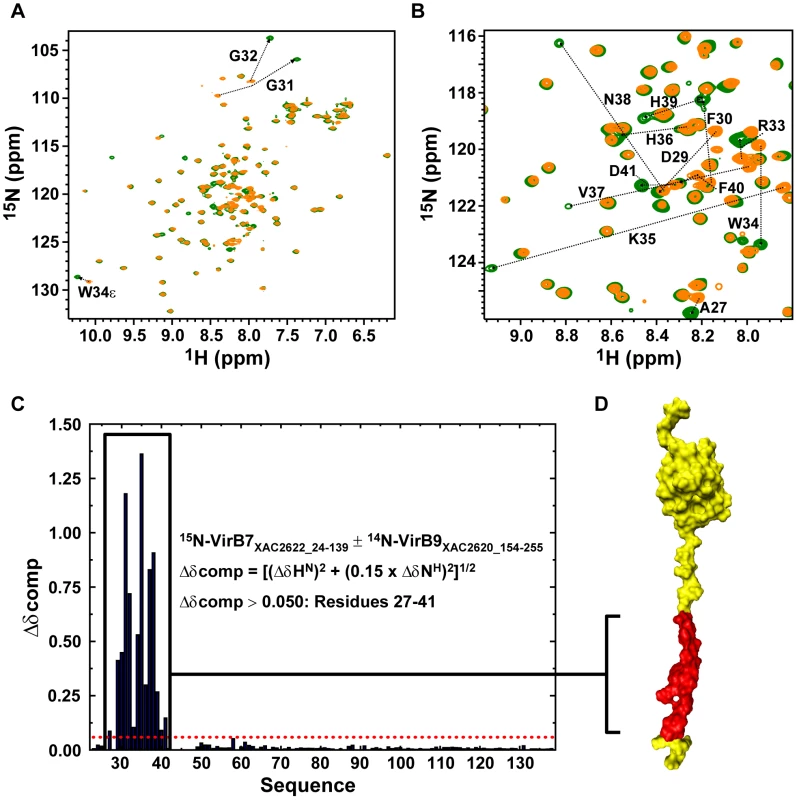
(A) Superposition of the 15N-HSQC spectra of 15N-VirB7XAC2622_24–139 alone (orange) and in the presence of 14N-VirB9XAC2620_154–255 (green). (B) Same as A, but with a close-up view of the central spectral region. The residues for which significant changes in peak positions were observed upon complex formation are indicated. (C) Weighted chemical shift changes (Δδcomp) of VirB7XAC2622 upon binding to VirB9XAC2620. (D) Residues affected by interaction with VirB9XAC2620_154–255 (residues 27–41; red) are color coded on the structure of VirB7XAC2622_24–139. VirB7XAC2622-Induced Changes in the VirB9XAC2620 C-terminal Domain
The 15N-HSQC spectrum of VirB9XAC2620_154–255 showed characteristics of poor line shape and chemical shift dispersion (Figure 4; red) suggestive of conformational disorder and a probable lack of stable tertiary structure. However, when unlabeled VirB7XAC2622_24–139 was added to the solution containing 15N-VirB9XAC2620_154–255, the 1H-15N cross peaks became more uniform in shape and more highly resolved (Figure 4; green). These observations suggest that VirB9XAC2620_154–255 on its own is in a dynamic conformational equilibrium or not properly folded in the absence of VirB7XAC2622_24–139. Nevertheless, it undergoes a significant conformational change which greatly reduces its conformational flexibility when bound to the latter protein.
Fig. 4. The conformation of the VirB9XAC2620 C-terminal domain changes significantly upon interacting with VirB7XAC2622. 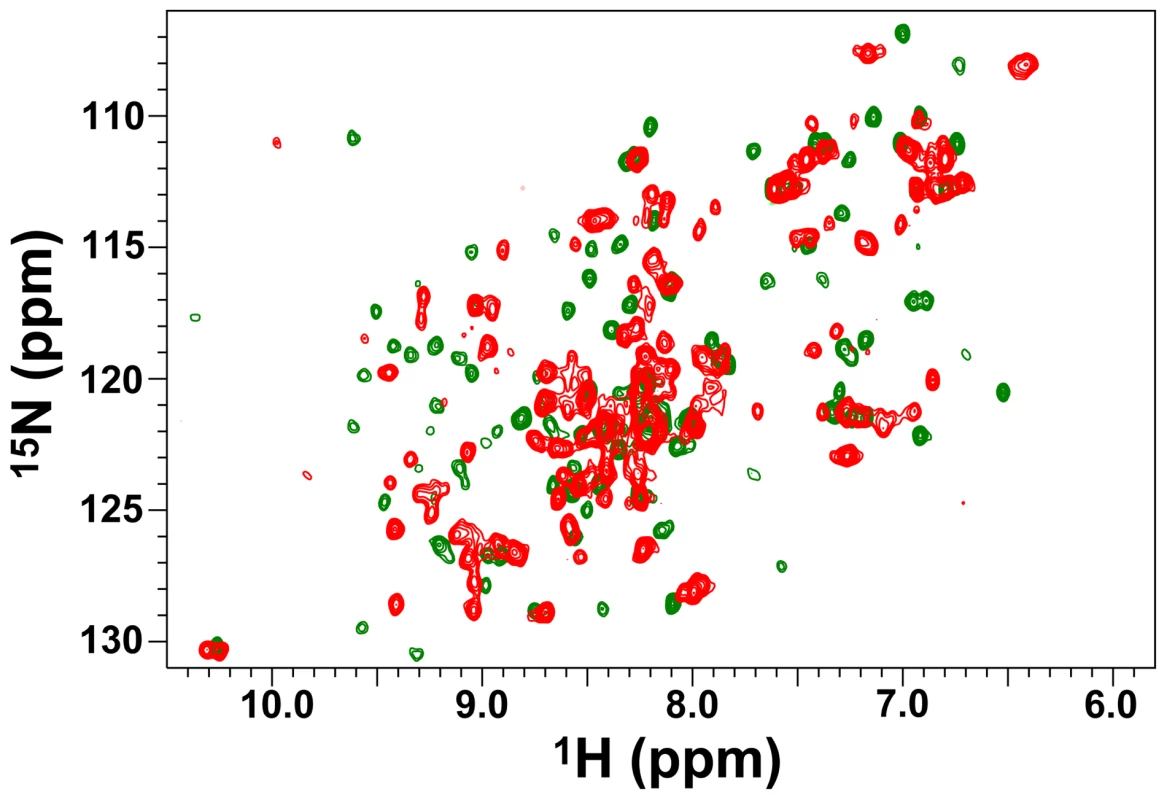
15N-HSQC spectra of 15N-VirB9XAC2620_154–255 in the absence (red) and in the presence (green) of 14N-VirB7XAC2622_24–139. Spectra were collected at 30°C on a 500 MHz spectrometer equipped with a room temperature probe. In order to further investigate whether the interaction of VirB9XAC2620_154–255 with the N-terminal domain of VirB7XAC2622 alone is sufficient to drive the formation of a specific VirB7-VirB9 complex, we studied the interaction of 15N-labeled VirB9 with an unlabeled peptide derived from the VirB7XAC2622 N-terminus (residues 24–46), which includes the VirB7XAC2622 N-terminal segment that interacts with VirB9XAC2620 (amino acids 27–41). Indeed, the perturbations observed in the 15N-HSQC spectra of VirB9XAC2620_154–255 in the presence of the full-length VirB7XAC2622_24–139 or the VirB7XAC2622_24–46 peptide are essentially the same (Figure 4 and Figure S8), demonstrating that the VirB7 N-terminal region is sufficient to interact with the VirB9XAC2620 C-terminal domain.
To investigate whether aggregation could be responsible for the line broadening observed in the 15N-HSQC spectrum of VirB9 C-terminal domain, we calculated the approximate overall rotational correlation time (τc) from estimates of 15N T1 and T2 obtained using 1D versions of the 15N relaxation experiments. The values of τc of VirB9XAC2610_154–255 alone or in complex with the VirB7XAC2622_24–46 peptide are approximately 6.5 ns (data not shown), which suggest that the VirB9XAC2620 C-terminal domain is monomeric in both conditions.
Relaxation Studies of VirB7XAC2622_24–139
Measurements of 15N relaxation times (T1 and T2) and heteronuclear {1H}-15N NOE were performed for 15N-VirB7XAC2622_24–139 alone (at 100 and 800 µM) and in complex with unlabeled VirB9XAC2620_154–255. The relaxation data are consistent with the presence of a flexible N-terminal tail and a more rigid globular domain (Figure 5). It is worth noting that the VirB9 binding surface of VirB7XAC2622_24–139 (residues 27–41) becomes less flexible upon binding to VirB9 (Figure 5A and C). In the absence of VirB9, residues 29–42 display {1H}-15N heteronuclear NOE values close to zero, indicating that this segment is not fully flexible and may present transient structures poised to interact with VirB9. Changes in 15N-T1 and 15N-T2 observed upon raising the VirB7XAC2622_24–139 concentration from 100 to 800 µM indicate a significant decrease in the overall rotational correlation rate, consistent with the formation of oligomers (Figure 5B and C). Relaxation parameters for residues involved in oligomerization (43–47 and some between positions 86 and 93) were not obtained because they were not detectable in the 15N-HSQC spectrum at high protein concentration (800 µM). Although these cross peaks become visible upon diluting the sample, they display too low amplitude to allow precise measurements. From the 15N T1 and T2 data, the overall rotational correlation times of VirB7XAC2622 at the concentrations of 100 and 800 µM were estimated to be 5.8 and 12.6 ns, respectively. These values are consistent with the predominance of a monomer at lower concentration and a dimer at higher concentration. This observation, however, does not exclude the possibility of the existence of an equilibrium which includes higher-order oligomers, as shown by glutaraldehyde cross-linking data.
Fig. 5. 15N relaxation data for VirB7XAC2622_24–139. 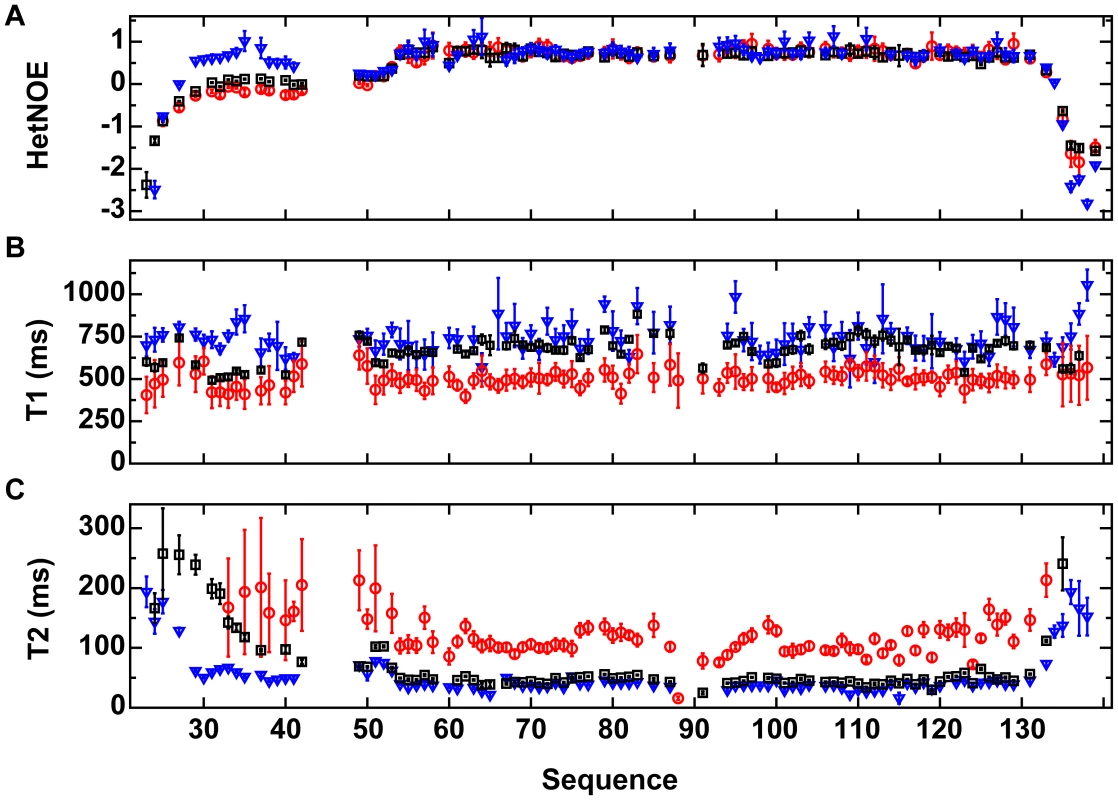
(A) Heteronuclear {1H}-15N NOE (HetNOE), (B) 15N-T1 and (C) 15N-T2 relaxation rates as a function of the protein sequence. Black squares: 15N-VirB7XAC2622_24–139 at 800 µM; red circles: 15N-VirB7XAC2622_24–139 at 100 µM; blue inverted triangles: 15N-VirB7XAC2622_24–139 – 14N-VirB9XAC2620_154–255 complex at 400 µM. Data are presented as mean ± uncertainty of the fitted parameter. Relaxation parameters for residues involved in oligomerization (43–47 and some between positions 86 and 93) were not obtained because they were not detectable in the 15N HSQC spectrum at high protein concentration (800 µM) and display amplitudes too low to allow precise measurements at lower concentrations. VirB7XAC2622 Contributes to VirB9XAC2620 and VirB10XAC2619 Stability in vivo
In Agrobacterium tumefaciens cells, the VirB7 protein contributes to VirB9 and VirB10 stability [42]. We therefore produced a xac2622 gene knockout strain and performed immunoblot experiments to assess the expression of VirB7XAC2622, VirB9XAC2620 and VirB10XAC2619. The production of all three proteins could be detected in wild type cells but none could be detected in the ΔvirB7XAC2622 strain (Figure 6A). To evaluate if the absence of VirB9XAC2620 and VirB10XAC2619 in the mutant strain is due to the lack of the VirB7XAC2622 production or to a polar effect in the T4SS operon, ΔvirB7XAC2622 cells were complemented with a plasmid encoding the VirB7XAC2622 protein in trans. The expression of VirB7XAC2622 from the pUFR-VirB7 plasmid restored VirB9XAC2620 and VirB10XAC2619 protein levels (Figure 6A). Furthermore, no significant differences in virB9XAC2620 transcript levels were detected between the wild-type, ΔvirB7XAC2622 and ΔvirB7XAC2622+pUFR-VirB7 strains in quantitative RT-PCR experiments (data not shown). These results show that VirB7XAC2622 is necessary for the stability of VirB9XAC2620 and VirB10XAC2619 proteins in vivo.
Fig. 6. A complex between VirB7XAC2622, VirB9XAC2620 and VirB10XAC2619 is formed in vivo in Xac cells. 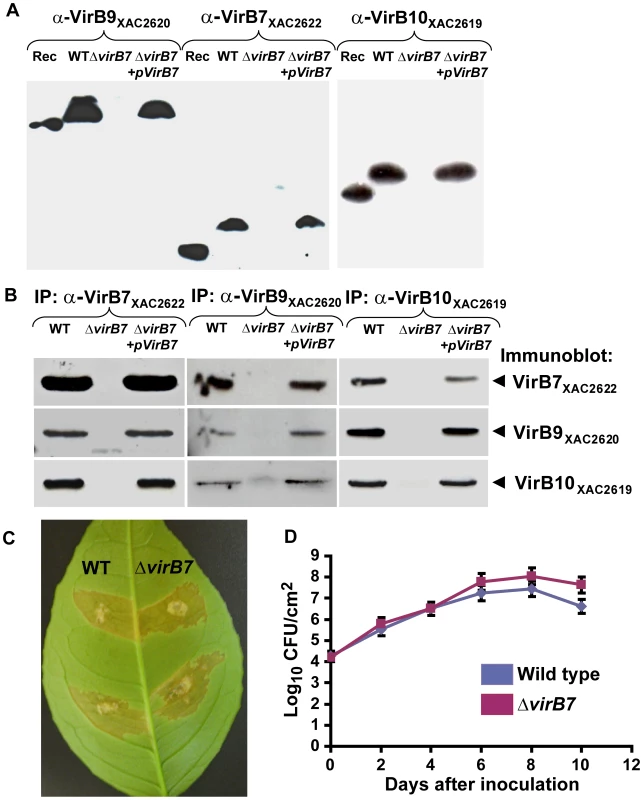
(A) Immunoblot analysis of VirB9XAC2620, VirB7XAC2622 and VirB10XAC2619 protein levels in bacterial total protein extracts. Polyclonal antibodies raised against VirB9XAC2620 (α-VirB9XAC2620), VirB7XAC2622 (α-VirB7XAC2622) and VirB10XAC2619 (α-VirB10XAC2619) were used. Rec: purified recombinant proteins used as positive controls; WT: Wild type Xac strain; ΔvirB7: ΔvirB7XAC2622 knockout strain and ΔvirB7+pVirB7: ΔvirB7XAC2622 knockout cells complemented with the pUFR-VirB7 plasmid. Recombinant VirB7XAC2622_24–139 has a slightly greater electrophoretic mobility than native VirB7XAC2622, possibly due to the presence of a covalently attached lipid moiety or retention of the signal sequence of the native protein. VirB9XAC2620_34–255 (26 kDa) and VirB10XAC2619_85–389_His (34 kDa) are also smaller than the respective native proteins. (B) Coimmunoprecipitation assays detecting reciprocal interactions between VirB proteins. Xac total cell lysates were immunoprecipitated (IP) with anti-VirB7XAC2622, anti-VirB9XAC2620 or anti-VirB10XAC2619 serum and the presence of VirB7, VirB9 or VirB10 was detected by immunoblot analysis. Lane labels are the same as in part (A). No VirB proteins were detected in control experiments using pre-immune sera (data not shown). (C) Characterization of the ΔvirB7XAC2622 gene knockout in Citrus sinensis infections. Macroscopic symptoms on abaxial surface of orange leaves 12 days post inoculation with wild type (left) and ΔvirB7 (right) strains of Xac. (D) Growth curves of Xac strains on orange leaves. Blue: wild type; red: ΔvirB7. Data are presented as mean ± standard deviation. The VirB7, VirB9 and VirB10 orthologs TraN, TraO and TraF form a stable trimeric complex [33], [34]. In order to determine whether the VirB7XAC2622, VirB9XAC2620 and VirB10XAC2619 form a stable complex in Xac cells, we performed immunoprecipitation experiments using antisera for each of the three proteins. Results described in Figure 6B show that VirB7XAC2622, VirB9XAC2620 and VirB10XAC2619 are present in the material immunoprecipitated by each of the three antisera in wild-type cells. Furthermore, none of the three proteins were immunoprecipitated from ΔvirB7XAC2622 cells while all three proteins were detected in immunoprecipitation experiments using ΔvirB7XAC2622+pUFR-VirB7 cells (Figure 6B). Altogether, these results show that a complex between VirB7XAC2622, VirB9XAC2620 and VirB10XAC2619 is formed in vivo in Xac cells.
In order to test the role of VirB7XAC2622 in Xac virulence, sweet orange leaves were infiltrated with the wild type and ΔvirB7XAC2622 strains. Both strains presented essentially the same infection phenotypes, including water-soaking, hyperplasia and necrosis (Figure 6C) [13]. In planta growth curves of the wild type and ΔvirB7XAC2622 cells revealed that the two strains replicate at a similar rate (Figure 6D). These data indicate that the VirB7XAC2622 protein does not participate in the Xac's ability to induce canker symptoms in orange leaves under the experimental conditions tested. Similarly, a T4SS knockout failed to affect the pathogenicity of X. campestris pv. campestris on several host plants [43]. Xac pathogenicity in citrus and canker symptoms are strongly dependent on the action of virulence factors secreted by the Type III secretion system [13]. The T4SS is probably involved in other, as yet unidentified, cellular functions. As noted in the Introduction, the Xac pXAC64 megaplasmid codes for a second T4SS probably involved in plasmid mobilization [15], [27] and whose structural components exhibit only very low sequence identity to their counterparts in the chromosomally encoded T4SS under study. For example, its VirB9 and VirB10 homologs (XACb0039 and XACb0038) are only 23–24% identical to VirB9XAC2620 and VirB10XAC2619 [15]. Furthermore, the pXAC64 plasmid does not code for any proteins with similarity to VirB7XAC2622 or any other known VirB7 proteins [15]. Therefore, it is unlikely that the two Xac T4SSs exercise redundant functions.
Discussion
The structure of the outer membrane complex of the T4SS coded by the conjugative E. coli plasmid pKM101 has been determined by X-ray crystallography and consists of 14 repeats of the TraN – TraOCT – TraFCT trimer [34], homologs of VirB7, the C-terminal domain of VirB9 and the C-terminal domain of VirB10, respectively. This structure revealed that the inner lining of the channel is formed by the C-terminal domains of the 14 TraF subunits. The TraN-TraO subcomplexes surround the internal ring formed by TraF. The 33 residue long lipidated TraN subunit is the outermost subunit of the complex whose external diameter is 172 Å [34].
Xanthomonas VirB7 Structure and Interactions in the Context of the T4SS Core Complex
VirB7XAC2622 is structurally distinct from all VirB7 homologs characterized to date. Its structural motifs are illustrated in Figure 7A. Like all other VirB7 proteins, it has a signal peptide, a conserved cysteine residue (Cys22) within a lipobox for covalent attachment to outer membrane lipids and a short extended region involved in interaction with VirB9. These motifs are all contained within the first 41 residues, which corresponds relatively well with the size of the majority of unprocessed VirB7 proteins (45–65 amino acids) [36].
Fig. 7. VirB7XAC2622 structure and interactions in the context of the T4SS outer membrane complex. 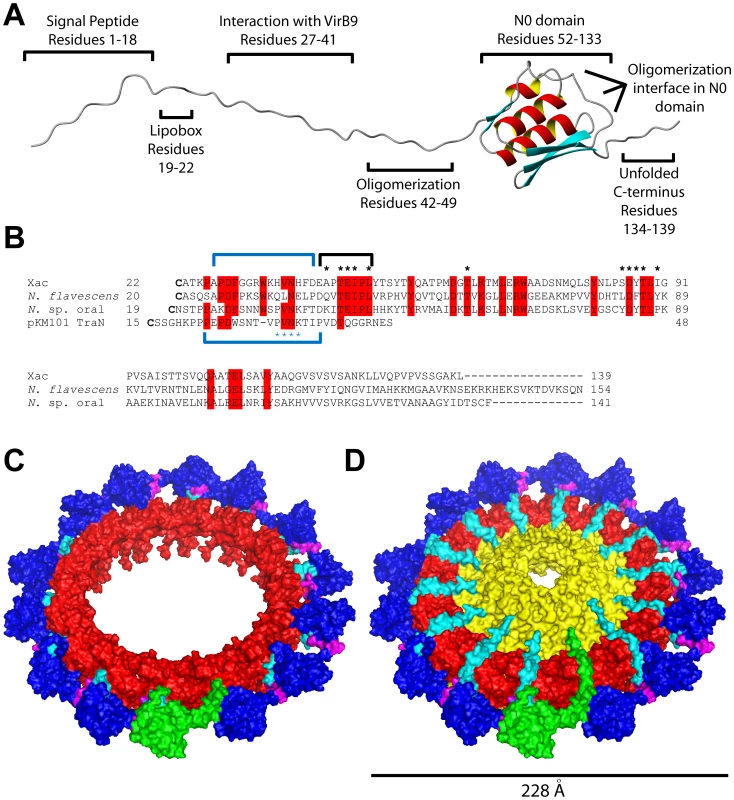
(A) Schematic organization of the VirB7XAC2622 protein, based on bioinformatics and experimental data. (B) Sequence alignment of the VirB7 proteins from Xanthomonas citri subsp. citri (GenBank code AAM37471), Neisseria flavescens (EER57212), Neisseria sp. oral taxon 014 str. F0314 (EFI23373) and TraN from pKM101 (AAA86456). Sequences begin at the cysteine residues of the predicted lipidation sites. Black asterisks indicate residues involved in observed intermolecular NOEs. Blue asterisks indicate the PVNK motif found in the pKM101 TraN protein. Blue brackets above and below the alignment indicate the VirB9XAC2620 and TraO binding sites in VirB7XAC2622 and TraN, respectively. The black bracket indicates the N-terminal oligomerization site. Residues outlined in red are conserved in at least three of the four sequences. Note that the codon for V55 of the protein from Neisseria flavescens is incorrectly assigned as a start codon in the GenBank database. (C) Final molecular dynamics configuration of the (VirB7XAC2622_37–139-TraOCT)14 complex. TraOCT is represented in red, VirB7XAC2622 is in cyan (residues 37–41), magenta (residues 42–49) and dark blue (residues 50–139). One VirB7XAC2622_37–139 unit is shown in green. (D) Putative model for the VirB7XAC2622-VirB9XAC2620-VirB10XAC2619 outer membrane complex produced by adding TraN residues 19–32 (representing VirB7XAC2622 residues 22–36, see alignment in part B) and TraFCT (representing VirB10XAC2619) to the structure shown in part C. The color scheme is the same as in part C except that cyan represents VirB7XAC2622 residues 22–41 and TraF/VirB10 is shown in yellow. One complete VirB7XAC2622 molecule is shown in green. VirB7 proteins are very poorly conserved though it has been observed that many proteins of this family have in common a PVNK motif that is involved in the interaction with VirB9 [35]. The VirB9 binding site in VirB7XAC2622 includes a HVNH sequence (residues 36–39) found in almost all Xanthomonadaceae orthologs (PVNR in X. albilineans and Stenotrophomonas maltophilia) which aligns with a perfect PVNK motif in the Neisseria sp. oral taxon 014 str. F0314 ortholog (Figure 7B). The side chains of the two most conserved residues of this motif, Val and Asn, make intimate contacts with TraO/VirB9 in the pKM101 outer membrane complex structure [35]: the Val side chain is inserted between the two TraO β-sheets while the Asn amide Oδ1 and Nδ2 atoms makes a set of three H-bonds with the TraO main chain. Furthermore, the PVNK motif in TraN is preceded by an approximately 10 amino acid segment that makes contacts with TraO, including a 3 residue β-strand that adds to the edge of one of the TraO β-sheets [34]. The corresponding regions in VirB7XAC2622 are precisely those that demonstrate the greatest chemical shift perturbations upon binding to VirB9XAC2620 (Figure 3). This suggests that interactions between VirB7XAC2622 and VirB9XAC2620 are very similar to that observed in the TraN-TraO complex.
The VirB7-VirB9 interactions described are most likely responsible for the loss of conformational flexibility observed for the N-terminal tail of VirB7XAC2622 and the C-terminal domain of VirB9XAC2620. Furthermore, interaction with VirB7XAC2622 is likely to be accompanied by a significant change in the structure of VirB9XAC2620_154–255, as revealed by the very large chemical shift perturbations observed for the majority of the 1H-15N cross-peaks (Figure 4). These changes in VirB9XAC2620 structure and dynamics may be related to its instability in vivo in the absence of VirB7XAC2622. Our immunoblot and coimmunoprecipitation data suggest that VirB7XAC2622, VirB9XAC2620 and VirB10XAC2619 form a stable trimeric complex in vivo and that the stabilities of VirB9XAC2620 and VirB10XAC2619 are dependent on VirB7XAC2622 (Figure 6). This may be a physiologically relevant mechanism by which excess VirB9 and VirB10 are degraded to maintain proper VirB7-VirB9-VirB10 stoichiometry.
VirB7XAC2622 has a mosaic structure (Figure 7A). In addition to the canonical VirB7-like N-terminal region, VirB7XAC2622 has a C-terminal region that includes a globular domain (residues 52–133). This globular domain does not interact with VirB9XAC2620_154–255 but does interact with an extended region (residues 42–49) that immediately precedes the globular domain in another VirB7XAC2622 molecule (Figure 2D), leading to the formation of homo-oligomers. The unique structural features of VirB7XAC2622 raise the question of how they may be incorporated into the model for the T4SS outer membrane complex tetradecamer previously solved for the pKM101 conjugation machine [34]. Specifically: can VirB7XAC2622 oligomerization occur while maintaining VirB7-VirB9 interactions in the context of the pore? And how are the VirB7XAC2622 globular domains oriented with respect to the pore? In order to investigate these questions, we used the VirB7XAC2622-TraN alignment depicted in Figure 7B to manually place 14 different NMR models of residues 37–139 of VirB7XAC2622 (VirB7XAC2622_37–139) around a tetradecameric ring made up of TraOCT subunits derived from the pKM101 TraN-TraOCT-TraFCT structure [34]. The VirB7XAC2622_37–139 subunits were placed so that Val37 and Asn38 occupied positions similar to Val33 and Asn34 of TraN. This (VirB7XAC2622_37–139-TraOCT)14 complex was used as a starting model in a molecular dynamics simulation in which the following restraints were used: i) The 14 TraOCT subunits were fixed in space, ii) VirB7XAC2622 residues 37 and 38 were fixed in space and iii) intermolecular NOE derived distance restraints were placed on 5 hydrogen pairs between neighboring VirB7XAC2622 subunits. During the initial cycles of the molecular dynamics simulation, the distance observed between the 13 intermolecular VirB7XAC2622 hydrogen pairs for which NOEs were measured (Figure 2D and Results above) decreases from the 12–25 Å range observed in the starting model to a 4–7 Å range, consistent with observation of NOE signals. These short contact distances are then observed throughout the simulation for all 14 VirB7XAC2622 pair interfaces made up of residues A43, T45, E46, I47, L49 in one VirB7XAC2622 molecule and residues T63, S85, D86, Y87, T88, I90 in a neighboring VirB7XAC2622. This is a consequence of the strong distance restraints included in the potential energy function. The final molecular dynamics configuration is shown in Figure 7C. Adding models of TraN residues 19–32 to represent VirB7XAC2622 residues 22–36 (see alignment in Figure 7B) and TraFCT (to represent VirB10XAC2619) results in the structure shown in Figure 7D made up of 14 repeats of VirB7XAC2622_37–139, TraN19–32, TraOCT and TraFCT. This can be seen as putative model for the VirB7XAC2622-VirB9XAC2620-VirB10XAC2619 outer membrane complex. Note that in Figures 7C and 7D, the VirB7 N0 domains (dark blue) adopt a variety of orientations with respect to the central ring and the membrane normal. This flexibility is derived from the conformational freedom of the VirB7XAC2622 regions immediately before and after residues 42–49 (magenta) involved in VirB7-VirB7 interactions.
Our proposal of a physiological role for VirB7XAC2622 oligomerization is supported by an analysis of conserved residues in the small group of full-length VirB7XAC2622 homologs found in Xanthomonadaceae and the two Neisseria species Neisseria flavescens SK114 and Neisseria sp. oral taxon 014 str. F0314. An alignment of VirB7XAC2622 and the two Neisseria proteins is shown in Figure 7B. The three proteins have only 21% sequence identity. However, 6 out of 11 of the residues involved in intermolecular NOEs at the B7-B7 interface (E46, I47, L49, T63, D86 and T88) are absolutely conserved in all Xanthomonadaceae, Neisseria str. F0314 and Neisseria flavescens and is either Phe or Tyr at position 87. Thus, the oligomerization interface is well conserved in an otherwise highly diverse protein family.
VirB7XAC2622 oligomerization is different from the dimerization observed in the canonical VirB7 protein from A. tumefaciens [30] which occurs via the formation of disulfide bonds involving residue 24. The structure of the outer membrane complex [34] however, is not compatible with the maintenance of these disulfide bonds in a fully assembled T4SS. Furthermore, VirB7XAC2622 does not have any cysteine residues other than at the N-terminal lipidation site.
The VirB7XAC2622 Globular Domain is Structurally Similar to Proteins Associated with Transport Across Bacterial Outer Membranes
The VirB7XAC2622 globular domain has no significant sequence similarity to proteins of known structure. We therefore used the Dali Server [44] to search for proteins with similar topology to this domain. This analysis revealed that the VirB7XAC2622 folded region resembles domains found in the following proteins (Table 3 and Figure 8): i) the TonB-dependent receptors (periplasmic signaling domain; Figure 8A) [45], [46], ii) the outer membrane secretin channel GspD from the type II secretion system (T2SS; Figure 8B) [47], iii) the secretin EscC from the Type III secretion system (T3SS; Figure 8C) [48], iv) the needle-like cell-puncturing device components gp27 and gp44 from long-tailed phages like T4 and Mu (Figure 8D) [49], [50] and v) the Type VI secretion system (T6SS) protein VgrG (superposition not shown) [51]. These domains are found at the N-termini of T2SS and T3SS secretins and have been denominated N0 domains which have also been identified in the Type IV pilus secretin PilQ [47] and the filamentous phage secretin pIV [52]. It is striking that in spite of its small and compact nature, domains with the VirB7XAC2622 topology are found only in a restricted number of proteins, all of which are involved in the transport of molecules across bacterial outer membranes. These observations suggest that all of these proteins may be distantly related and have evolved in the periplasm or outer membrane to adopt a variety of functions, from structural modules in outer membrane pores (secretins from type II and type III secretion systems, type IV pili and filamentous phages) to membrane-penetrating devices in T6SS and long-tailed bacteriophages, and signal-transduction modules in TonB-dependent receptors.
Fig. 8. Superposition of the structure of the VirB7XAC2622 N0 domain with structural homologs. 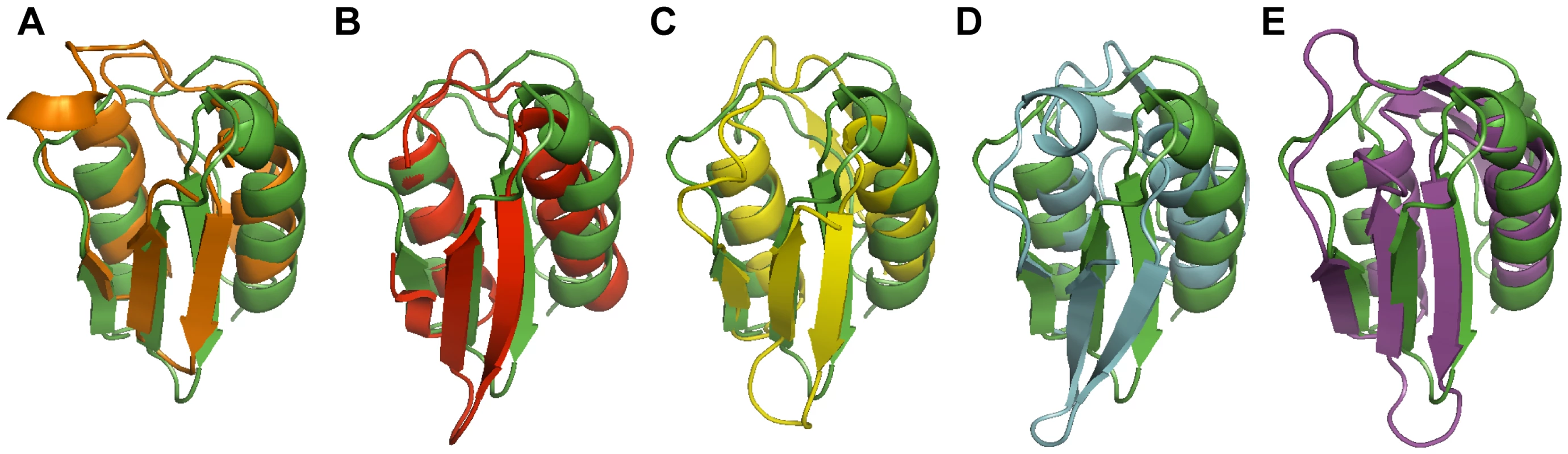
The VirB7XAC2622 globular domain (residues 51–134; green) is superposed with (A) the signaling domain of PupA (orange, residues 3–76, PDB: 2A02); (B) the N0 domain of GspD (red, amino acids 3–79, PDB: 3EZJ); (C) the N0 domain of EscC (yellow, amino acids 31–103, PDB: 3GR5), (D) residues 97–176 of gene product 44 from bacteriophage Mu (light blue, PDB: 1WRU) and (E) residues 81–161 of DotD from L. pneumophila (purple, PDB: 3ADY). Tab. 3. Structural similarity of the VirB7XAC2622 globular domaina. 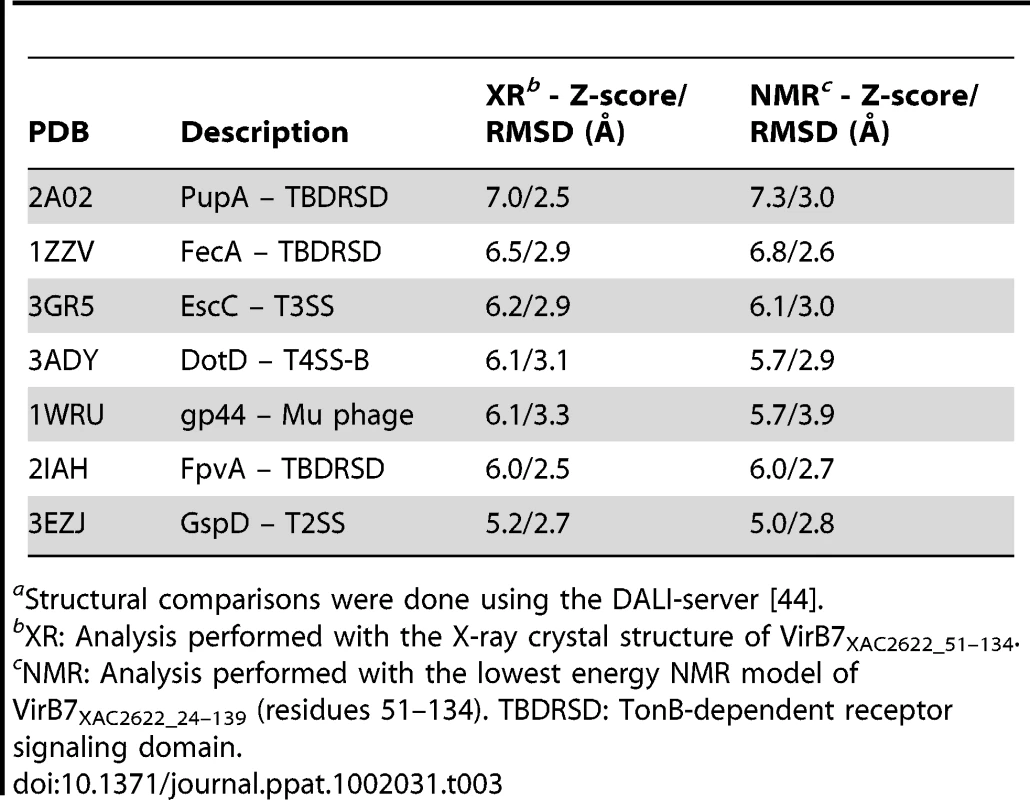
aStructural comparisons were done using the DALI-server [44]. Nakano et al. [53] have recently described the crystal structure of the DotD lipoprotein of the Type IV-B secretion system from Legionella pneumophila. Type IV-B secretion systems have weak or no sequence similarity with the larger group of Type IV-A secretion systems [54] to which the Xanthomonad T4SS pertains. Interestingly, the DotD crystal structure presents a C-terminal region with an N0 domain topology similar to that of VirB7XAC2622 structure (Figure 8E), though the proteins present no significant sequence similarity. An interesting difference between the two structures is that a 6 amino acid sequence (called the “lid”) of the otherwise disordered N-terminal region is visible in the crystal structure and makes β-strand addition to β1. This interaction is different from that observed in the VirB7XAC2622 oligomer described here. Since no electron density for the 23 residues between the lid and the N0 domain were visible, it was not clear whether they come from the same molecule in the crystal lattice. We therefore predict that DotD will eventually be shown to exhibit the same characteristics as VirB7XAC2622, namely to interact with both a VirB9-like protein and to form head-to-tail polymeric rings based on interactions between the flexible N-terminal region and globular N0 domains of neighboring subunits. This analysis points to previously unrecognized common structural features between the outer membrane complexes of Type IV-A and Type IV-B secretion systems.
The novel VirB7XAC2622 structure, its oligomerization and its interaction with VirB9 described here point to a possible structural variation in the Xanthomonas T4SS that could result in the formation of an extra ring layer in the core complex. While the VirB7-VirB7 interactions observed in the NMR experiments were of relatively low affinity (fast exchange chemical shift changes were observed in the 7–850 µM range), these interactions would be strengthened by the increased effective concentration afforded by the fourteen neighboring VirB7XAC2622 binding sites on the outer surface of the VirB9/VirB10 ring. Several hypotheses regarding the physiological role of this VirB7XAC2622 ring can be proposed. One possibility is that the weak B7-B7 interactions break and reform again to accommodate different conformational states of the outer pore during substrate translocation or to permit allosteric communication between the outer and inner membrane core complexes. Another possibility is that the VirB7XAC2622 N0 domains could act as a conduit for signal transmission between substrates or signaling molecules in the periplasm and the VirB9 and VirB10 subunits. Whether the VirB7XAC2622 N0 domain carries out a purely structural function or is involved in substrate recognition or signal transduction will have to be tested in the future.
Materials and Methods
Cloning, Protein Expression and Purification
Plasmids, oligonucleotides and bacterial strains used in this study are listed in Table S1. The DNA fragments encoding residues 24–139 of VirB7XAC2622 (GenBank AAM37471), 154–255 of VirB9XAC2620 (GenBank AAM37469) and 85–389 of VirB10XAC2619 (GenBank AAM37468) were amplified by PCR from Xanthomonas citri subsp. citri strain 306 genomic DNA and inserted into the pET28a expression vector (Novagen) using the NdeI and XhoI (VirB7XAC2622) or NdeI and BamHI (VirB9XAC2620 and VirB10XAC2619) cloning sites, to express the recombinant proteins fused with an N-terminal His-tag. DNA fragments encoding residues 51–134 of VirB7XAC2622 and 34–255 of VirB9XAC2620 were cloned into the pET11a vector (Novagen) using NdeI and BamHI sites. All constructs were confirmed by DNA sequencing. E. coli BL21(DE3)RP and BL21(DE3)Star strains (Novagen) were transformed with the recombinant plasmids for expression of the protein products which are referred to as VirB7XAC2622_24–139_His, VirB9XAC2620_154–255_His, VirB7XAC2622_51–134, VirB9XAC2620_34–255 and VirB10XAC2619_85–389_His. Cells were grown in 2XTY for expression of unlabeled proteins or in M9 minimal media containing 15NH4Cl and 12C-glucose or 13C-glucose (Cambridge Isotope Laboratories) for production of 15N or 15N and 13C isotopically labeled proteins. Recombinant protein expression was induced at midlog phase by the addition of 0.5 mM isopropyl β-D-1-thiogalactopyranoside and the cells were grown for four hours at 37°C. Cells were then harvested and lysed by French Press. In the case of VirB7XAC2622_24–139_His and VirB9XAC2620_154–255_His, the lysis supernatants were separated by passage through Q-Sepharose (VirB7XAC2622_24–139_His) or SP-Sepharose (VirB9XAC2620_154–255_His) ion exchange columns (GE Healthcare) followed by affinity chromatography using a nickel affinity column. The N-terminal His-tags were removed using the Thrombin CleanCleave kit (Sigma), creating the proteins VirB7XAC2622_24–139 and VirB9XAC2620_154–255. A final purification step consisted of passage through a Superdex 75 26/60 size exclusion column (GE Healthcare). VirB7XAC2622_51–134 was purified using Q-sepharose anion exchange and Superdex 75 gel filtration chromatography. VirB9XAC2620_34–255 and VirB10XAC2619_85–389_His are expressed as insoluble proteins present in inclusion bodies. After lysis, VirB9XAC2620_34–255 was solubilized in the presence of 8 M urea and purified by SP-Sepharose cation exchange and Superdex 75 size exclusion chromatographies using buffers containing 8 M urea. VirB10XAC2619_85–389_His was purified by affinity chromatography using buffers containing 8 M urea. Purified proteins were refolded by successive dialysis against decreasing concentrations of urea in 10 mM sodium acetate (pH 5.0) for VirB9XAC2620_34–255 or 10 mM Tris-HCl (pH 8.0), 150 mM NaCl for VirB10XAC2619_85–389_His. Synthetic peptides corresponding to VirB7XAC2622_24–46 and VirB7XAC2622_38–52 were purchased from Biomatik Corporation (Cambridge, Ontario, Canada) and EZBiolab (Carmel, Indiana, USA), respectively.
NMR Spectroscopy
NMR samples containing 15N or 15N/13C labeled protein were prepared in 10 mM 2H-sodium acetate (pH 5.0), containing 50 mM sodium chloride, 0.05% (w/v) sodium azide, 7% (v/v) 2H2O and 1 mM trimethylsilyl-2,2,3,3-tetradeuteropropionic acid as internal standard for 1H chemical shift referencing. NMR spectra were acquired on a 600 MHz Varian Unity-Inova spectrometer equipped with inverse triple resonance (1H, 15N, 13C) cold probe, or on BRUKER 500 MHz DRX or 800 MHz Avance III spectrometers equipped with inverse triple resonance (1H, 15N, 13C) room temperature probes. Unless otherwise mentioned, all NMR experiments were performed at 40°C. The backbone resonance assignment of 15N/13C-VirB7XAC2622_24–139 was achieved by analyzing the following three-dimensional spectra: HNCA, HN(CO)CA, HNCO, HN(CA)CO, HNCACB and CBCA(CO)NH [55]. Side-chain resonance assignments were determined by analyzing the following spectra: 2D 13C-HSQC, 2D TOCSY, 2D NOESY, 3D HNHA, 3D HBHA(CBCACO)NH, 3D 15N-TOCSY-HSQC, 3D H(CCO)NH-TOCSY, 3D (H)CC(CO)NH-TOCSY, 3D 15N-NOESY-HSQC, 3D H(C)CH-TOCSY and 3D 13C-NOESY-HSQC (specific for the aliphatic or the aromatic regions). The 3D H(C)CH-TOCSY and 3D 13C-NOESY-HSQC spectra were recorded in >99% (v/v) 2H2O. NOESY experiments were carried out using 0.2–0.3 mM protein samples and all NOESY mixing times were 100 ms. Backbone resonance assignments for 15N/13C-VirB7XAC2622_24–139 in complex with unlabeled VirB9XAC2620_154–255 were obtained from the analysis of three-dimensional HNCA, HN(CO)CA, HNCO, HNCACB, CBCA(CO)NH, HBHA(CBCACO)NH and 15N-NOESY-HSQC experiments recorded on a sample of 15N/13C-VirB7XAC2622_24–139 in the presence of a 20% molar excess of unlabeled VirB9XAC2620_154–255. The backbone resonance assignment (15N and 1H) for VirB7XAC2622_51–134 was performed by comparison with the 15N-HSQC spectrum of the larger construct, VirB7XAC2622_24–139. Residual 1DNH dipolar couplings for VirB7XAC2622_24–139 (0.25 mM), alone or in complex with VirB9XAC2620_154–255, were determined from the differences in 1H-15N splittings obtained in isotropic and anisotropic media. The 1H-15N splittings were extracted from a series of J-modulated 2D 1H-15N-HSQC experiments acquired at 30°C on the 500 MHz spectrometer, and analyzed essentially as described [56]. Residual alignment was induced by a liquid crystalline medium consisting of 5% penta-ethyleneglycol monododecyl ether (C12E5)/n-hexanol (r = 0.96) [57]. Fittings of RDCs to the NMR and X-ray structures were carried out using PALES [58] or MODULE [59]. All NMR spectra were processed with NMRPipe [60] and analyzed with CCPN Analysis [61]. Resonance assignments were deposited in the Biological Magnetic Resonance Data Bank (BMRB accession code 17257).
NMR Structure Calculation
Structure calculation and automated NOE assignment were performed using CYANA 2.1 [37]. Chemical shifts of 1Hα, 13Cα, 13Cβ, 13C′ and 15NH were used as input for TALOS [62] to predict φ and ψ dihedral angles that were subsequently used as restraints in the structure calculation. A total of 3471 NOE cross peaks were manually picked in the 15N-NOESY-HSQC and 13C-NOESY-HSQC (acquired specifically for the aliphatic or the aromatic regions) spectra and used as input for CYANA. The CYANA protocol consisted of 7 cycles of simulated annealing in torsion angle space with 10000 integration steps. In each cycle 300 conformers were calculated. The intra-residue, sequential and 10 unambiguous long range NOE cross peaks were assigned manually and imposed during all stages of the protocol. Chemical shift tolerances for automated NOE assignment were set to ±0.025 ppm in the direct 1H dimension, ±0.030 ppm in the indirect 1H dimension and ±0.25 ppm for the heteronuclei. The best 50 CYANA solutions were selected based on a target function criteria, and further refined in explicit water using CNS2.1 and HADDOCK2.0 [41]. The 1616 NOEs assigned by CYANA, the manually assigned NOEs, and the TALOS dihedral angle restraints were used as input during water refinement in HADDOCK. The best 20 lowest-energy conformers after water refinement were deposited in the Protein Data Bank (PDB code 2L4W). The quality of the final NMR structures was investigated by PROCHECK-NMR [63] and PSVS [64]. The structures were visualized using MOLMOL [65] or PYMOL (http://www.pymol.org).
15N Relaxation Measurements
Experiments for measuring backbone 15N longitudinal (T1) and transverse (T2) relaxation times, and the heteronuclear {1H}-15N NOE, were recorded at 40°C on a Varian Inova 600 MHz spectrometer using standard Biopack pulse sequences (Varian, Inc.). The inversion recovery delays used for sampling T1 relaxation were 50, 250, 450, 650, 850, 1050, 1250, 1450 and 1650 ms. The CPMG delays used for sampling T2 relaxation were 10, 30, 50, 70, 90, 110, 130, 150, 170, 190 and 210 ms. The 1H saturation period in the heteronuclear {1H}-15N NOE experiment was 3 s. All 2D spectra were acquired sequentially as matrices of 512(1H)×128(15N) complex points, and inter scan delay of 3 s. Relaxation rates were obtained by fitting the time decay of peak intensities to an exponential decay function with three fitted parameters, the relaxation rate, the intensity at time zero and an offset, using the CCPN Analysis software [61]. Overall rotational correlation times (τc) were obtained either from the mean ratios of 15N T1/T2 relaxation rates of 1H-15N bond vectors located in regions of secondary structure using TENSOR2 [66], or from estimates of 15N T1 and T2 relaxation times obtained from 1D 15N-edited relaxation experiments, as previously described [67], [68].
NMR Titration Experiments
Unlabeled VirB9XAC2620_154–255 or VirB9XAC2620_34–255 were titrated into a solution of 15N-labeled VirB7XAC2622_24–139 and a 15N-HSQC spectrum was acquired after the addition of each aliquot until a molar excess of 1.5 of unlabeled protein with respect to 15N-VirB7XAC2622_24–139. In analogous experiments 15N-labeled VirB9XAC2620_154–255 was titrated with unlabeled VirB7XAC2622_24–139 or VirB7XAC2622_24–46. VirB7XAC2622_24–139 oligomerization was studied by monitoring changes in the 15N-HSQC spectra as a function of protein concentration in the range of 7 to 850 µM. 15N-labeled VirB7XAC2622_51–134 was also titrated with unlabeled VirB7XAC2622_38–52. Chemical shift perturbations due to sample dilution or protein-protein interaction were obtained from the weighted chemical shift changes in the 15N-HSQC spectra calculated according to Eq. 1 [69]:(1)where ΔδHN and ΔδNH are the chemical shift changes (in ppm) of the amide proton and nitrogen resonances, respectively.
Protein-Protein Docking
Docking simulations were used to build a docking model for the dimer of VirB7XAC2622_24–139. The calculations were performed with HADDOCK2.0 using CNS2.1 [41], and the first model of the NMR ensemble as starting structure. The docking was driven by unambiguous intermolecular NOEs and by ambiguous interaction restraints (AIR) between active residues and active plus passive residues. The following 13 NOE restraints were used: Ala43Hβ - Ile90Hδ1; Thr45Hα - T88Hγ2; Thr45Hβ - T88Hγ2; Glu46Hβ - Tyr87Hε; Ile47Hδ1 - Thr63Hβ; Ile47Hδ1 - Thr63Hγ2; Ile47Hδ1 - T88Hα; Ile47Hδ1 - T88Hγ2; Leu49Hγ - Ser85Hα; Leu49 Hδ1 - Asp86Hα; Leu49Hδ1 - Asp86Hβ1; Leu49Hδ1 - Asp86Hβ2 and Leu49Hδ2 - Asp86Hα. Active residues were identified as those showing chemical shift perturbations of at least one standard deviation above the mean, for 15N-VirB7XAC2622_24–139 as a function of protein concentration (Figure 2). Active residues were E42, A43, T45, E46 and I47 at the N-terminal tail of the first subunit and D86, T88, I90, G91, Q115 of the C-terminal globular domain of the second subunit. Passive residues were F40, D41, P44, P48 and L49 in the N-terminal segment, and P84, S85, Y87, P92, A113 and A114 in the C-terminal domain. The N-terminal tails (residues 24–50) were kept fully flexible during all stages of the docking, while C-terminal domain residues at the interface, 84–92, 61–63 and 113–115, were maintained semi-flexible, being allowed to move only during semi-flexible refinement. The rigid body stage consisted of the calculation of 1000 solutions, from which 100 with the lowest HADDOCK scores were selected for semi-rigid simulated annealing in torsion angle space including backbone and side chain flexibility, and final refinement in Cartesian space with explicit water. The refined solutions were then clustered based on the backbone RMSD at the interface. An analogous protocol was used for docking the N-terminal (residues 24–50) and C-terminal (residues 51–139) fragments of VirB7, except that in this calculation NOE restraints were not included and flexibility was introduced only at the interface.
Model Building and Molecular Dynamics Simulations
Models of residues 37–139 of VirB7XAC2622 (VirB7XAC2622_37–139) were positioned around a tetradecameric ring made up of TraOCT subunits derived from the pKM101 T4SS TraN-TraOCT-TraFCT structure [34]. The VirB7XAC2622_37–139 subunits were placed so that Val37 and Asn38 occupied positions similar to Val33 and Asn34 of TraN. Molecular mechanics optimization and dynamics were then carried out from this initial model. The OPLS/AA force-field [70] for the protein and an implicit solvent representation of the generalized Born-formalism [71] were used. In addition to the molecular force-field, distance restraints with a flat-bottom harmonic functional form were included such that an energy penalty was added to the potential when the distance between specified pairs of atoms was less than a minimum threshold value of 3 Å or exceeded a maximum of 6 Å. A harmonic force constant of 5000 kJ⋅mol−1⋅Å−2 was used. The following 5 NOE proton pairs were specified for each VirB7XAC2622 pair interface: Ala43Hβ - Ile90Hδ1, Thr45Hβ - T88Hγ2, Ile47Hδ1 - Thr63Hβ; Leu49Hγ - Ser85Hα; and Leu49Hδ2 - Asp86Hα. These pairs correspond to a subset of the pairs for which intermolecular NOEs were detected. Thus, a total of 70 (14×5) distance restrains were included in the potential. All ionizable residues besides histidine were treated in their charged forms. Molecular dynamics were carried out at a temperature of 300 K for a total time of 2 ns. All computations were carried out with the GROMACS 4.5.3 software [72].
Crystallization, Data Collection and Processing
A construct corresponding to the VirB7 C-terminal region, VirB7XAC2622_51–134, at 14 mg/mL in 5 mM Tris-HCl pH 7.5 and 25 mM sodium chloride, was submitted to vapor diffusion sitting-drop crystallization trials at 18°C. Large plates appeared after one day over a reservoir solution comprising 1.4 M ammonium sulfate and 4% (v/v) isopropyl alcohol. Reservoir solution supplemented with 25% (v/v) glycerol was used as cryoprotectant. Crystals were flash frozen at 100 K and submitted to X-ray diffraction at beam line W01B-MX2 of the Brazilian Synchrotron Light Laboratory (LNLS) coupled to a Marmosaic-225 CCD detector (Mar, USA). The diffraction data were indexed, integrated and scaled using HKL2000 [73].
X-ray Structure Determination and Refinement
The crystal structure of VirB7XAC2622_51–134 was determined by molecular replacement by Phaser [74] using residues 51–134 of the lowest energy NMR structure as the search model. The model was iteratively refined using the graphics program Coot [75] with rounds of restrained refinement in Refmac5 [76] with individual anisotropic B-factor for all non-hydrogen atoms. All programs used for refinement were from the CCP4 suite [77]. The quality of the structure was analyzed by the programs Coot, Refmac5, Procheck [78], Rampage [79] and MolProbity [80]. The coordinates of the crystal structure were deposited in the PDB (PDB code 3OV5).
Fluorescence Spectroscopy
Fluorescence experiments were conducted in an AVIV ATF-105 spectrofluorimeter (AVIV Instruments). Fluorescence emission was collected after successive addition of 0.1 µM VirB7XAC2622_24–139_His aliquots into a sample containing 1 µM VirB9XAC2620_34–255 in 5 mM sodium acetate (pH 5.0). The final VirB7XAC2622_24–139_His concentration in the titration was 2.2 µM. Samples were pre-equilibrated for 2 min at 25°C, excited at 280 nm (2 nm bandwidth) and fluorescence emission was detected from 334–342 nm (7 nm bandwidth) at 2 nm intervals. The dissociation constant was calculated from a nonlinear regression fit of fluorescence titration data to Eq. 2 [81], [82], using the SigmaPlot program (SPSS, Inc.):(2)where F is the fluorescence intensity at any given point of the titration curve, v is the initial volume, V is volume at any given point of the titration, [X] and [Y] are the VirB9XAC2620_34–255 and VirB7XAC2622_24–139_His concentrations at any given point of the titration, respectively, Kd is the dissociation constant and α is the ratio between the maximum fluorescence intensity and the initial fluorescence intensity.
Glutaraldehyde Cross-Linking Experiments
Samples with different concentrations of VirB7XAC2622_24–139 or VirB7XAC2622_51–134 were pre-equilibrated for 15 min, and then incubated with 0.01% (v/v) glutaraldehyde. After 20 min, the cross-linking reaction was stopped by the addition of SDS-PAGE sample buffer (100 mM Tris-HCl pH 6.8, 3.7% (w/v) SDS, 18.7% (v/v) glycerol, 140 mM 2-mercaptoethanol and 0.01% (w/v) bromophenol blue). The cross-linking products were analyzed by 16% Tricine SDS-PAGE [83].
virB7XAC2622 Knockout and Complementation
Approximately 1 kb of the upstream and downstream regions of the virB7XAC2622 gene were amplified by PCR from Xac genomic DNA and the two fragments were ligated to produce an in frame deletion, leaving only the region coding for the first three and last five codons. This sequence was then cloned into the BamHI restriction site of the pNPTS138 suicide vector (M. R. Alley, unpublished), thereby generating the plasmid pNPTS-Δxac2622. This vector was introduced into Xac by electroporation and replacement of the wild-type copy by the deleted version was obtained after two recombination events as described [84]. In order to complement the virB7XAC2622 knockout, a fragment including the virB7XAC2622 gene plus 1000 pb of upstream sequence was amplified by PCR from Xac genomic DNA and inserted into the pUFR047 vector [85] at the BamHI restriction site, creating the plasmid pUFR-VirB7. This plasmid was then transferred to the ΔvirB7XAC2622 strain by electroporation and selection of gentamicin resistance.
In planta Studies
Virulence assays were performed by infiltration of the Xac strains into sweet orange leaves (Citrus sinensis L. Osbeck). The cell cultures were adjusted to an optical density of 0.1 at 600 nm and leaves were inoculated by syringe infiltration with needle. The plants were maintained at 28°C with a 12 h photoperiod and the development of the symptoms was regularly observed. Bacterial growth was measured by harvesting 10 mm2 citrus leaf discs for each inoculated Xac strain, and the leaf discs were macerated in 150 mM sodium chloride. The solution was serially diluted and spread onto LBON plates [1% (w/v) tryptone, 0.5% (w/v) yeast extract and 1.5% (w/v) agar] containing ampicillin. The mean number of colony forming units per square centimeter (CFU/cm2) was calculated by counting individual colonies obtained from each dilution.
Antibody Production and Immunoblot
Rabbit polyclonal antibodies were raised against VirB7XAC2622_24–139, VirB9XAC2620_34–255 and VirB10XAC2619_85–389_His. To analyze protein levels in Xac, cells were grown in XVM2 medium [86] to an optical density (600 nm) of 1.0. Recombinant proteins and Xac cellular extracts were separated by 18% SDS-PAGE followed by immunoblot analysis. The rabbit sera were used at 1∶1000 dilutions and the antibodies were detected with staphylococcal protein A conjugated to horseradish peroxidase (Sigma). Immunoblots were developed with the ECL Advance Western Blotting system and exposed to Amersham Hyperfilm ECL film (GE Healthcare).
Immunoprecipitation
Strains of Xac were grown in XVM2 to an optical density (600 nm) of 1.0 and equal numbers of cells were harvested by centrifugation. Cells were resuspended in 500 µl of 50 mM Hepes (pH 8.0) containing 5 mM EDTA. Coimmunoprecipitation experiments were performed after solubilization of cells with deoxycholate (DOC) and N,N–Dimethyldodecylamine N-oxide (LDAO) detergents, as described [87]. Bacterial suspensions were treated with 200 µg/ml lysozyme for 1 h at 4°C. Cells were lysed by sonication in the presence of 100 mM NaCl, 1% (w/v) DOC and protease inhibitor cocktail (Sigma Aldrich). Volumes were adjusted to 1 mL with 50 mM Hepes (pH 8.0) and 2% (w/v) LDAO and lysates were solubilized by incubation for 16 h at 4°C with agitation. Solubilized material was isolated by centrifugation at 15,000×g for 20 min and incubated with Protein G agarose beads (Millipore) for 4 h at 4°C, removing nonspecifically bound proteins. For immunoprecipitation, supernatants from this pre-clearing step were incubated with protein G agarose beads in the presence of pre-immune serum (controls) or anti-VirB7, anti-VirB9 or anti-VirB10 antibody, for 16 h at 4°C, with agitation. The unbound material was discarded after centrifugation (400×g, 5 min) and beads were washed three times with decreasing concentrations of LDAO and DOC (final wash contained 0.1% DOC and 0.1% LDAO), each time for 10 min at 25°C, with agitation. Immunoprecipitates were eluted by addition of SDS sample buffer and boiling for 10 min. Samples were analyzed by SDS-PAGE followed by immunoblot.
Quantitative RT-PCR
RNAs from Xac cultures grown in XVM2 medium to an optical density (600 nm) of 1.0 were extracted using Illustra RNAspin Mini kit, according to manufacturer's instructions (GE Healthcare). Purified RNA was treated with 1 U/µg DNaseI, RNase-free (Fermentas) and successful removal of contaminating DNA was confirmed by PCR. Reverse transcription was performed using 1 µg of DNaseI-treated RNA and RevertAid H Minus First Strand cDNA Synthesis Kit, following manufacturer's protocol (Fermentas). Quantitative amplification of the resulting cDNA (40 ng) was performed using 0.3 µM of each primer (F_virB9_RT and R_virB9_RT; Table S1) and SYBR Green/ROX qPCR Master Mix in the ABI7300 Real-Time System (Applied Biosystems). Relative quantification of gene expression was performed using gene xac1631 (that codes for subunit A of DNA gyrase) as an endogenous control and the 2−ΔΔCT method [88]. Primers were designed using the PrimerExpress Software (Applied Biosystems). Triplicates of two independent biological samples were used.
Accession Numbers
1H, 13C and 15N resonance assignments of VirB7XAC2622 were deposited in the BMRB: entry 17257. NMR and X-ray crystallography coordinates have been deposited in the PDB with accession codes 2L4W and 3OV5, respectively.
Supporting Information
Zdroje
1. TsengTTTylerBMSetubalJC 2009 Protein secretion systems in bacterial-host associations, and their description in the Gene Ontology. BMC Microbiol 9 Suppl 1 S2
2. EconomouAChristiePJFernandezRCPalmerTPlanoGV 2006 Secretion by numbers: Protein traffic in prokaryotes. Mol Microbiol 62 308 319
3. JuhasMCrookDWHoodDW 2008 Type IV secretion systems: tools of bacterial horizontal gene transfer and virulence. Cell Microbiol 10 2377 2386
4. BackertSMeyerTF 2006 Type IV secretion systems and their effectors in bacterial pathogenesis. Curr Opin Microbiol 9 207 217
5. NinioSRoyCR 2007 Effector proteins translocated by Legionella pneumophila: strength in numbers. Trends Microbiol 15 372 380
6. ShrivastavaRMillerJF 2009 Virulence factor secretion and translocation by Bordetella species. Curr Opin Microbiol 12 88 93
7. VothDEHeinzenRA 2009 Coxiella type IV secretion and cellular microbiology. Curr Opin Microbiol 12 74 80
8. DehioC 2008 Infection-associated type IV secretion systems of Bartonella and their diverse roles in host cell interaction. Cell Microbiol 10 1591 1598
9. CelliJ 2006 Surviving inside a macrophage: the many ways of Brucella. Res Microbiol 157 93 98
10. BackertSSelbachM 2008 Role of type IV secretion in Helicobacter pylori pathogenesis. Cell Microbiol 10 1573 1581
11. McCullenCABinnsAN 2006 Agrobacterium tumefaciens and plant cell interactions and activities required for interkingdom macromolecular transfer. Annu Rev Cell Dev Biol 22 101 127
12. ChristiePJAtmakuriKKrishnamoorthyVJakubowskiSCascalesE 2005 Biogenesis, architecture, and function of bacterial type IV secretion systems. Annu Rev Microbiol 59 451 485
13. BruningsAMGabrielDW 2003 Xanthomonas citri: breaking the surface. Mol Plant Pathol 4 141 157
14. da SilvaACFerroJAReinachFCFarahCSFurlanLR 2002 Comparison of the genomes of two Xanthomonas pathogens with differing host specificities. Nature 417 459 463
15. AlegriaMCSouzaDPAndradeMODocenaCKhaterL 2005 Identification of new protein-protein interactions involving the products of the chromosome - and plasmid-encoded type IV secretion loci of the phytopathogen Xanthomonas axonopodis pv. citri. J Bacteriol 187 2315 2325
16. QianWJiaYRenSXHeYQFengJX 2005 Comparative and functional genomic analyses of the pathogenicity of phytopathogen Xanthomonas campestris pv. campestris. Genome Res 15 757 767
17. VorholterFJSchneikerSGoesmannAKrauseLBekelT 2008 The genome of Xanthomonas campestris pv. campestris B100 and its use for the reconstruction of metabolic pathways involved in xanthan biosynthesis. J Biotechnol 134 33 45
18. PierettiIRoyerMBarbeVCarrereSKoebnikR 2009 The complete genome sequence of Xanthomonas albilineans provides new insights into the reductive genome evolution of the xylem-limited Xanthomonadaceae. BMC Genomics 10 616
19. StudholmeDJKemenEMacleanDSchornackSArituaV 2010 Genome-wide sequencing data reveals virulence factors implicated in banana Xanthomonas wilt. FEMS Microbiol Lett 310 182 192
20. CrossmanLCGouldVCDowJMVernikosGSOkazakiA 2008 The complete genome, comparative and functional analysis of Stenotrophomonas maltophilia reveals an organism heavily shielded by drug resistance determinants. Genome Biol 9 R74
21. RoccoFDe GregorioEColonnaBDi NoceraPP 2009 Stenotrophomonas maltophilia genomes: a start-up comparison. Int J Med Microbiol 299 535 546
22. ThiemeFKoebnikRBekelTBergerCBochJ 2005 Insights into genome plasticity and pathogenicity of the plant pathogenic bacterium Xanthomonas campestris pv. vesicatoria revealed by the complete genome sequence. J Bacteriol 187 7254 7266
23. LeeBMParkYJParkDSKangHWKimJG 2005 The genome sequence of Xanthomonas oryzae pathovar oryzae KACC10331, the bacterial blight pathogen of rice. Nucleic Acids Res 33 577 586
24. OchiaiHInoueVTakeyaMSasakiAKakuH 2005 Genome sequence of Xanthomonas oryzae pv. oryzae suggests contribution of large numbers of effector genes and insertion sequences to its race diversity. Jpn Agric Res Q 39 275 287
25. SalzbergSLSommerDDSchatzMCPhillippyAMRabinowiczPD 2008 Genome sequence and rapid evolution of the rice pathogen Xanthomonas oryzae pv. oryzae PXO99A. BMC Genomics 9 204
26. MoreiraLMAlmeidaNFJrPotnisNDigiampietriLAAdiSS 2010 Novel insights into the genomic basis of citrus canker based on the genome sequences of two strains of Xanthomonas fuscans subsp. aurantifolii. BMC Genomics 11 238
27. El YacoubiBBruningsAMYuanQShankarSGabrielDW 2007 In planta horizontal transfer of a major pathogenicity effector gene. Appl Environ Microbiol 73 1612 1621
28. FernandezDDangTASpudichGMZhouXRBergerBR 1996 The Agrobacterium tumefaciens virB7 gene product, a proposed component of the T-complex transport apparatus, is a membrane-associated lipoprotein exposed at the periplasmic surface. J Bacteriol 178 3156 3167
29. SchroderGLankaE 2005 The mating pair formation system of conjugative plasmids-A versatile secretion machinery for transfer of proteins and DNA. Plasmid 54 1 25
30. SpudichGMFernandezDZhouXRChristiePJ 1996 Intermolecular disulfide bonds stabilize VirB7 homodimers and VirB7/VirB9 heterodimers during biogenesis of the Agrobacterium tumefaciens T-complex transport apparatus. Proc Natl Acad Sci U S A 93 7512 7517
31. DasAAndersonLBXieYH 1997 Delineation of the interaction domains of Agrobacterium tumefaciens VirB7 and VirB9 by use of the yeast two-hybrid assay. J Bacteriol 179 3404 3409
32. WardDVDraperOZupanJRZambryskiPC 2002 Peptide linkage mapping of the Agrobacterium tumefaciens vir-encoded type IV secretion system reveals protein subassemblies. Proc Natl Acad Sci U S A 99 11493 11500
33. FronzesRSchaferEWangLSaibilHROrlovaEV 2009 Structure of a type IV secretion system core complex. Science 323 266 268
34. ChandranVFronzesRDuquerroySCroninNNavazaJ 2009 Structure of the outer membrane complex of a type IV secretion system. Nature 462 1011 1015
35. BaylissRHarrisRCoutteLMonierAFronzesR 2007 NMR structure of a complex between the VirB9/VirB7 interaction domains of the pKM101 type IV secretion system. Proc Natl Acad Sci U S A 104 1673 1678
36. CaoTBSaierMHJr 2001 Conjugal type IV macromolecular transfer systems of Gram-negative bacteria: organismal distribution, structural constraints and evolutionary conclusions. Microbiology 147 3201 3214
37. HerrmannTGuntertPWuthrichK 2002 Protein NMR structure determination with automated NOE assignment using the new software CANDID and the torsion angle dynamics algorithm DYANA. J Mol Biol 319 209 227
38. BaxA 2003 Weak alignment offers new NMR opportunities to study protein structure and dynamics. Protein Sci 12 1 16
39. CornilescuGMarquardtJLOttigerMBaxA 1998 Validation of protein structure from anisotropic carbonyl chemical shifts in a dilute liquid crystalline phase. J Am Chem Soc 120 6836 6837
40. BaxAGrishaevA 2005 Weak alignment NMR: a hawk-eyed view of biomolecular structure. Curr Opin Struct Biol 15 563 570
41. DominguezCBoelensRBonvinAM 2003 HADDOCK: a protein-protein docking approach based on biochemical or biophysical information. J Am Chem Soc 125 1731 1737
42. FernandezDSpudichGMZhouXRChristiePJ 1996 The Agrobacterium tumefaciens VirB7 lipoprotein is required for stabilization of VirB proteins during assembly of the T-complex transport apparatus. J Bacteriol 178 3168 3176
43. HeYQZhangLJiangBLZhangZCXuRQ 2007 Comparative and functional genomics reveals genetic diversity and determinants of host specificity among reference strains and a large collection of Chinese isolates of the phytopathogen Xanthomonas campestris pv. campestris. Genome Biol 8 R218
44. HolmLSanderC 1996 Mapping the protein universe. Science 273 595 603
45. FergusonADAmezcuaCAHalabiNMChelliahYRosenMK 2007 Signal transduction pathway of TonB-dependent transporters. Proc Natl Acad Sci U S A 104 513 518
46. Garcia-HerreroAVogelHJ 2005 Nuclear magnetic resonance solution structure of the periplasmic signalling domain of the TonB-dependent outer membrane transporter FecA from Escherichia coli. Mol Microbiol 58 1226 1237
47. KorotkovKVPardonESteyaertJHolWG 2009 Crystal structure of the N-terminal domain of the secretin GspD from ETEC determined with the assistance of a nanobody. Structure 17 255 265
48. SpreterTYipCKSanowarSAndreIKimbroughTG 2009 A conserved structural motif mediates formation of the periplasmic rings in the type III secretion system. Nat Struct Mol Biol 16 468 476
49. KanamaruSLeimanPGKostyuchenkoVAChipmanPRMesyanzhinovVV 2002 Structure of the cell-puncturing device of bacteriophage T4. Nature 415 553 557
50. KondouYKitazawaDTakedaSTsuchiyaYYamashitaE 2005 Structure of the central hub of bacteriophage Mu baseplate determined by X-ray crystallography of gp44. J Mol Biol 352 976 985
51. LeimanPGBaslerMRamagopalUABonannoJBSauderJM 2009 Type VI secretion apparatus and phage tail-associated protein complexes share a common evolutionary origin. Proc Natl Acad Sci U S A 106 4154 4159
52. SpagnuoloJOpalkaNWenWXGagicDChabaudE 2010 Identification of the gate regions in the primary structure of the secretin pIV. Mol Microbiol 76 133 150
53. NakanoNKuboriTKinoshitaMImadaKNagaiH 2010 Crystal Structure of Legionella DotD: Insights into the Relationship between Type IVB and Type II/III Secretion Systems. PLoS Pathog 6 e1001129
54. ChristiePJVogelJP 2000 Bacterial type IV secretion: conjugation systems adapted to deliver effector molecules to host cells. Trends Microbiol 8 354 360
55. SattlerMSchleucherJGriesingerC 1999 Heteronuclear multidimensional NMR experiments for the structure determination of proteins in solution employing pulsed field gradients. Prog NMR Spectrosc 34 93 158
56. TjandraNGrzesiekSBaxA 1996 Magnetic field dependence of nitrogen-proton J splittings in N-15-enriched human ubiquitin resulting from relaxation interference and residual dipolar coupling. J Am Chem Soc 118 6264 6272
57. RuckertMOttingG 2000 Alignment of biological macromolecules in novel nonionic liquid crystalline media for NMR experiments. J Am Chem Soc 122 7793 7797
58. ZweckstetterMBaxA 2000 Prediction of sterically induced alignment in a dilute liquid crystalline phase: Aid to protein structure determination by NMR. J Am Chem Soc 122 3791 3792
59. DossetPHusJCMarionDBlackledgeM 2001 A novel interactive tool for rigid-body modeling of multi-domain macromolecules using residual dipolar couplings. J Biomol NMR 20 223 231
60. DelaglioFGrzesiekSVuisterGWZhuGPfeiferJ 1995 NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR 6 277 293
61. VrankenWFBoucherWStevensTJFoghRHPajonA 2005 The CCPN data model for NMR spectroscopy: development of a software pipeline. Proteins 59 687 696
62. CornilescuGDelaglioFBaxA 1999 Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J Biomol NMR 13 289 302
63. LaskowskiRARullmannnJAMacArthurMWKapteinRThorntonJM 1996 AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J Biomol NMR 8 477 486
64. BhattacharyaATejeroRMontelioneGT 2007 Evaluating protein structures determined by structural genomics consortia. Proteins 66 778 795
65. KoradiRBilleterMWuthrichK 1996 MOLMOL: a program for display and analysis of macromolecular structures. J Mol Graph 14 51 55, 29–32
66. DossetPHusJCBlackledgeMMarionD 2000 Efficient analysis of macromolecular rotational diffusion from heteronuclear relaxation data. J Biomol NMR 16 23 28
67. FarrowNAMuhandiramRSingerAUPascalSMKayCM 1994 Backbone dynamics of a free and phosphopeptide-complexed Src homology 2 domain studied by 15N NMR relaxation. Biochemistry 33 5984 6003
68. RossiPSwapnaGVHuangYJAraminiJMAnklinC 2010 A microscale protein NMR sample screening pipeline. J Biomol NMR 46 11 22
69. MulderFASchipperDBottRBoelensR 1999 Altered flexibility in the substrate-binding site of related native and engineered high-alkaline Bacillus subtilisins. J Mol Biol 292 111 123
70. JorgensenWLMaxwellDSTirado-RivesJ 1996 Development and testing of the OPLS all-atom force field on conformational energetics and properties of organic liquids. J Am Chem Soc 118 11225 11236
71. RouxBSimonsonT 1999 Implicit solvent models. Biophys Chem 78 1 20
72. HessBKutznerCvan der SpoelDLindahlE 2008 Gromacs 4: Algorithms for highly efficient, load-balanced, and scalable molecular simulation. J Chem Theory Comput 4 435 447
73. OtwinowskiZMinorW 1997 Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol 276 307 326
74. McCoyAJGrosse-KunstleveRWAdamsPDWinnMDStoroniLC 2007 Phaser crystallographic software. J Appl Crystallogr 40 658 674
75. EmsleyPCowtanK 2004 Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60 2126 2132
76. MurshudovGNVaginAADodsonEJ 1997 Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr 53 240 255
77. Collaborative Computational Project Number 4 1994 The CCP4 Suite - Programs for protein crystallography. Acta Crystallogr D Biol Crystallogr 50 760 763
78. LaskowskiRAMacarthurMWMossDSThorntonJM 1993 Procheck - a program to check the stereochemical quality of protein structures. J Appl Crystallogr 26 283 291
79. LovellSCDavisIWArendallWB3rdde BakkerPIWordJM 2003 Structure validation by Calpha geometry: phi,psi and Cbeta deviation. Proteins 50 437 450
80. DavisIWLeaver-FayAChenVBBlockJNKapralGJ 2007 MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res 35 W375 383
81. CorreaFSalinasRKBonvinAMFarahCS 2008 Deciphering the role of the electrostatic interactions in the alpha-tropomyosin head-to-tail complex. Proteins 73 902 917
82. CorreaFFarahCS 2007 Different effects of trifluoroethanol and glycerol on the stability of tropomyosin helices and the head-to-tail complex. Biophys J 92 2463 2475
83. SchaggerH 2006 Tricine-SDS-PAGE. Nat Protoc 1 16 22
84. GuzzoCRSalinasRKAndradeMOFarahCS 2009 PILZ protein structure and interactions with PILB and the FIMX EAL domain: implications for control of type IV pilus biogenesis. J Mol Biol 393 848 866
85. De FeyterRYangYGabrielDW 1993 Gene-for-genes interactions between cotton R genes and Xanthomonas campestris pv. malvacearum avr genes. Mol Plant Microbe Interact 6 225 237
86. WengelnikKMarieCRusselMBonasU 1996 Expression and localization of HrpA1, a protein of Xanthomonas campestris pv. vesicatoria essential for pathogenicity and induction of the hypersensitive reaction. J Bacteriol 178 1061 1069
87. CascalesEChristiePJ 2004 Agrobacterium VirB10, an ATP energy sensor required for type IV secretion. Proc Natl Acad Sci U S A 101 17228 17233
88. LivakKJSchmittgenTD 2001 Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods 25 402 408
89. MoseleyHNSahotaGMontelioneGT 2004 Assignment validation software suite for the evaluation and presentation of protein resonance assignment data. J Biomol NMR 28 341 355
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek Distribution of the Phenotypic Effects of Random Homologous Recombination between Two Virus SpeciesČlánek SIV Nef Proteins Recruit the AP-2 Complex to Antagonize Tetherin and Facilitate Virion ReleaseČlánek Dual Function of the NK Cell Receptor 2B4 (CD244) in the Regulation of HCV-Specific CD8+ T CellsČlánek A Large and Intact Viral Particle Penetrates the Endoplasmic Reticulum Membrane to Reach the CytosolČlánek Interleukin-13 Promotes Susceptibility to Chlamydial Infection of the Respiratory and Genital Tracts
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2011 Číslo 5- Jak souvisí postcovidový syndrom s poškozením mozku?
- Měli bychom postcovidový syndrom léčit antidepresivy?
- Farmakovigilanční studie perorálních antivirotik indikovaných v léčbě COVID-19
- 10 bodů k očkování proti COVID-19: stanovisko České společnosti alergologie a klinické imunologie ČLS JEP
-
Všechny články tohoto čísla
- Lymphoadenopathy during Lyme Borreliosis Is Caused by Spirochete Migration-Induced Specific B Cell Activation
- Infections Are Virulent and Inhibit the Human Malaria Parasite in
- A Gamma Interferon Independent Mechanism of CD4 T Cell Mediated Control of Infection
- MDA5 and TLR3 Initiate Pro-Inflammatory Signaling Pathways Leading to Rhinovirus-Induced Airways Inflammation and Hyperresponsiveness
- The OXI1 Kinase Pathway Mediates -Induced Growth Promotion in Arabidopsis
- An E2F1-Mediated DNA Damage Response Contributes to the Replication of Human Cytomegalovirus
- Quantitative Subcellular Proteome and Secretome Profiling of Influenza A Virus-Infected Human Primary Macrophages
- Distribution of the Phenotypic Effects of Random Homologous Recombination between Two Virus Species
- Inhibition of Both HIV-1 Reverse Transcription and Gene Expression by a Cyclic Peptide that Binds the Tat-Transactivating Response Element (TAR) RNA
- A Viral Satellite RNA Induces Yellow Symptoms on Tobacco by Targeting a Gene Involved in Chlorophyll Biosynthesis using the RNA Silencing Machinery
- Misregulation of Underlies the Developmental Abnormalities Caused by Three Distinct Viral Silencing Suppressors in Arabidopsis
- Investigating the Host Binding Signature on the PfEMP1 Protein Family
- Human Neutrophil Clearance of Bacterial Pathogens Triggers Anti-Microbial γδ T Cell Responses in Early Infection
- Septation of Infectious Hyphae Is Critical for Appressoria Formation and Virulence in the Smut Fungus
- A Family of Helminth Molecules that Modulate Innate Cell Responses via Molecular Mimicry of Host Antimicrobial Peptides
- Phospholipids Trigger Capsular Enlargement during Interactions with Amoebae and Macrophages
- CTL Escape Mediated by Proteasomal Destruction of an HIV-1 Cryptic Epitope
- Evolution of Th2 Immunity: A Rapid Repair Response to Tissue Destructive Pathogens
- Extensive Genome-Wide Variability of Human Cytomegalovirus in Congenitally Infected Infants
- The Antiviral Efficacy of HIV-Specific CD8 T-Cells to a Conserved Epitope Is Heavily Dependent on the Infecting HIV-1 Isolate
- Epstein-Barr Virus Infection of Polarized Epithelial Cells via the Basolateral Surface by Memory B Cell-Mediated Transfer Infection
- Reactive Oxygen Species Hydrogen Peroxide Mediates Kaposi's Sarcoma-Associated Herpesvirus Reactivation from Latency
- Crystal Structure and Functional Analysis of the SARS-Coronavirus RNA Cap 2′-O-Methyltransferase nsp10/nsp16 Complex
- The Dot/Icm System Delivers a Unique Repertoire of Type IV Effectors into Host Cells and Is Required for Intracellular Replication
- AAV Exploits Subcellular Stress Associated with Inflammation, Endoplasmic Reticulum Expansion, and Misfolded Proteins in Models of Cystic Fibrosis
- Suboptimal Activation of Antigen-Specific CD4 Effector Cells Enables Persistence of In Vivo
- SIV Nef Proteins Recruit the AP-2 Complex to Antagonize Tetherin and Facilitate Virion Release
- Dual Function of the NK Cell Receptor 2B4 (CD244) in the Regulation of HCV-Specific CD8+ T Cells
- Transition of Sporozoites into Liver Stage-Like Forms Is Regulated by the RNA Binding Protein Pumilio
- A Large and Intact Viral Particle Penetrates the Endoplasmic Reticulum Membrane to Reach the Cytosol
- Transcriptome Analysis of in Human Whole Blood and Mutagenesis Studies Identify Virulence Factors Involved in Blood Survival
- Interleukin-13 Promotes Susceptibility to Chlamydial Infection of the Respiratory and Genital Tracts
- Structural Insights into Viral Determinants of Nematode Mediated Transmission
- Protective Efficacy of Serially Up-Ranked Subdominant CD8 T Cell Epitopes against Virus Challenges
- Viral CTL Escape Mutants Are Generated in Lymph Nodes and Subsequently Become Fixed in Plasma and Rectal Mucosa during Acute SIV Infection of Macaques
- Taking Some of the Mystery out of Host∶Virus Interactions
- Viral Small Interfering RNAs Target Host Genes to Mediate Disease Symptoms in Plants
- : An Emerging Cause of Sexually Transmitted Disease in Women
- Mitochondrial Ubiquitin Ligase MARCH5 Promotes TLR7 Signaling by Attenuating TANK Action
- The Hexamer Structure of the Rift Valley Fever Virus Nucleoprotein Suggests a Mechanism for its Assembly into Ribonucleoprotein Complexes
- Acquisition of Human-Type Receptor Binding Specificity by New H5N1 Influenza Virus Sublineages during Their Emergence in Birds in Egypt
- Stromal Down-Regulation of Macrophage CD4/CCR5 Expression and NF-κB Activation Mediates HIV-1 Non-Permissiveness in Intestinal Macrophages
- A Component of the Xanthomonadaceae Type IV Secretion System Combines a VirB7 Motif with a N0 Domain Found in Outer Membrane Transport Proteins
- Perturbs Iron Trafficking in the Epithelium to Grow on the Cell Surface
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Crystal Structure and Functional Analysis of the SARS-Coronavirus RNA Cap 2′-O-Methyltransferase nsp10/nsp16 Complex
- Lymphoadenopathy during Lyme Borreliosis Is Caused by Spirochete Migration-Induced Specific B Cell Activation
- The OXI1 Kinase Pathway Mediates -Induced Growth Promotion in Arabidopsis
- : An Emerging Cause of Sexually Transmitted Disease in Women
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



