-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaInvestigating the Host Binding Signature on the PfEMP1 Protein Family
The Plasmodium falciparum erythrocyte membrane protein 1
(PfEMP1) family plays a central role in antigenic variation and cytoadhesion of
P. falciparum infected erythrocytes. PfEMP1
proteins/var genes are classified into three main
subfamilies (UpsA, UpsB, and UpsC) that are hypothesized to have different roles
in binding and disease. To investigate whether these subfamilies have diverged
in binding specificity and test if binding could be predicted by adhesion domain
classification, we generated a panel of 19 parasite lines that primarily
expressed a single dominant var transcript and assayed binding
against 12 known host receptors. By limited dilution cloning, only UpsB and UpsC
var genes were isolated, indicating that UpsA
var gene expression is rare under in vitro
culture conditions. Consequently, three UpsA variants were obtained by rosette
purification and selection with specific monoclonal antibodies to create a more
representative panel. Binding assays showed that CD36 was the most common
adhesion partner of the parasite panel, followed by ICAM-1 and TSP-1, and that
CD36 and ICAM-1 binding variants were highly predicted by adhesion domain
sequence classification. Binding to other host receptors, including CSA, VCAM-1,
HABP1, CD31/PECAM, E-selectin, Endoglin, CHO receptor “X”, and
Fractalkine, was rare or absent. Our findings identify a category of larger
PfEMP1 proteins that are under dual selection for ICAM-1 and CD36 binding. They
also support that the UpsA group, in contrast to UpsB and UpsC
var genes, has diverged from binding to the major
microvasculature receptor CD36 and likely uses other mechanisms to sequester in
the microvasculature. These results demonstrate that CD36 and ICAM-1 have left
strong signatures of selection on the PfEMP1 family that can be detected by
adhesion domain sequence classification and have implications for how this
family of proteins is specializing to exploit hosts with varying levels of
anti-malaria immunity.
Published in the journal: . PLoS Pathog 7(5): e32767. doi:10.1371/journal.ppat.1002032
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1002032Summary
The Plasmodium falciparum erythrocyte membrane protein 1
(PfEMP1) family plays a central role in antigenic variation and cytoadhesion of
P. falciparum infected erythrocytes. PfEMP1
proteins/var genes are classified into three main
subfamilies (UpsA, UpsB, and UpsC) that are hypothesized to have different roles
in binding and disease. To investigate whether these subfamilies have diverged
in binding specificity and test if binding could be predicted by adhesion domain
classification, we generated a panel of 19 parasite lines that primarily
expressed a single dominant var transcript and assayed binding
against 12 known host receptors. By limited dilution cloning, only UpsB and UpsC
var genes were isolated, indicating that UpsA
var gene expression is rare under in vitro
culture conditions. Consequently, three UpsA variants were obtained by rosette
purification and selection with specific monoclonal antibodies to create a more
representative panel. Binding assays showed that CD36 was the most common
adhesion partner of the parasite panel, followed by ICAM-1 and TSP-1, and that
CD36 and ICAM-1 binding variants were highly predicted by adhesion domain
sequence classification. Binding to other host receptors, including CSA, VCAM-1,
HABP1, CD31/PECAM, E-selectin, Endoglin, CHO receptor “X”, and
Fractalkine, was rare or absent. Our findings identify a category of larger
PfEMP1 proteins that are under dual selection for ICAM-1 and CD36 binding. They
also support that the UpsA group, in contrast to UpsB and UpsC
var genes, has diverged from binding to the major
microvasculature receptor CD36 and likely uses other mechanisms to sequester in
the microvasculature. These results demonstrate that CD36 and ICAM-1 have left
strong signatures of selection on the PfEMP1 family that can be detected by
adhesion domain sequence classification and have implications for how this
family of proteins is specializing to exploit hosts with varying levels of
anti-malaria immunity.Introduction
Plasmodium falciparum erythrocyte membrane protein 1 (PfEMP1) is a clonally variant adhesion protein that mediates binding of infected erythrocytes (IE) to blood microvasculature and other host cells [1]. Adherence of IEs to microvascular endothelium is a major virulence factor and, in conjunction with the related phenomenon of rosetting with uninfected erythrocytes, prevents parasitized erythrocyte circulation to the spleen where parasites may be destroyed [2]. Each parasite strain encodes ∼60 PfEMP1 proteins, or var genes, which are expressed in a mutually exclusive fashion [3], [4]. Switches in var gene expression enable infected erythrocytes to evade host immunity and may modify disease manifestations by changing parasite binding tropism [5]–[7].
Efforts to unravel the role of PfEMP1 proteins in disease are complicated by the vast diversity of var genes. Each parasite has a diverse repertoire of genes, and there is limited overlap of repertoires between parasite genomes [8]–[10]. However, genes can be classified into three main subfamilies denoted Groups A, B, and C [11], plus three unusual strain-transcendent variants (var1csa, var2csa, and type 3 var) [12]–[15]. The var gene subfamilies possess distinctive upstream flanking regions termed UpsA, UpsB, and UpsC and are found in characteristic locations in the subtelomeric or central regions of chromosomes [4], [9], [11], [12]. It has been hypothesized that var gene organization may contribute to a gene recombination hierarchy that influences gene function and evolution [1].
A number of studies have sought to correlate specific parasite adhesion traits with disease outcome [16]–[19]. To date, at least 12 host receptors have been reported to mediate P. falciparum IE binding [20]. CD36 binding is the most common adhesion trait in the parasite population, followed by intercellular adhesion molecule 1 (ICAM-1) [17], [19]. These two receptors can synergize under flow conditions to mediate infected erythrocyte binding to microvasculature endothelium [21]–[23]. Most other binding properties appear to be rarer or have not been studied in more than one or a few parasite isolates. ICAM-1 binding has been associated with cerebral malaria in some studies [17], [24], but not in others [19], [25]. In addition, infected erythrocyte rosetting, or binding of parasitized red blood cells to uninfected red blood cells, has been associated with disease severity in African children [26]–[28]. The clearest disease association is placental malaria, in which parasites express the unusually strain-transcendent VAR2CSA PfEMP1 protein and adhere to chondroitin sulfate A (CSA) in the placenta [14], [29]. VAR2CSA is a leading candidate for a pregnancy malaria vaccine and a paradigm for syndrome-specific anti-disease vaccine efforts.
Although the molecular basis for other adhesion-based complications of P. falciparum is less established than for pregnancy malaria, several observations suggest the antigenic diversity of severe malaria isolates may also be limited. For instance, immunity to severe malaria appears to be acquired after relatively few infections [30], [31]. In addition, isolates from severe malaria cases appear to express a relatively restricted variant antigen surface repertoire [32]–[34]. Furthermore, seroepidemiological and var transcriptional profiling studies suggest that UpsA variants are more commonly expressed in young African children with limited immunity and in severe malaria infections [35]–[39]. Therefore it is possible the UpsA group has become specialized to exploit individuals with limited anti-malaria immunity, and it is important to understand what may account for this expression profile.
To gain insight into PfEMP1 binding properties, sequence classification has been performed [40]. The extracellular binding region of PfEMP1 proteins is comprised of 2–7 receptor-like domains called Duffy Binding-Like (DBL) and Cysteine Rich Interdomain Region (CIDR) [41], [42]. DBL and CIDR domains are classified into different major types (α to ε) and sub-types by sequence criteria [10], [40]. PfEMP1 proteins can be further subdivided by protein architecture into small proteins with a four-domain extracellular binding region (DBL-CIDR-DBL-CIDR) and large proteins with a more complex domain composition [43]. By comparison to other groups, nearly all of the UpsA proteins are in the large protein category [9], [10]. The best characterized binding interactions are between CIDR::CD36 and DBLβ::ICAM-1 [44]–[46]. In a repertoire-wide binding comparison with CIDR recombinant proteins, the majority of proteins encoded CD36 binding function, except for the UpsA group, which had different CIDR sequence types than the UpsB and UpsC groups [11], [12], [46]. UpsA proteins may also be under less selection to bind ICAM-1, as 7 of 23 DBLβ domains from the IT4 parasite strain bound ICAM-1, but none of the 9 DBLβ domains tested from the UpsA group were ICAM-1 binders [44]. However, using a different binding analysis in a BioPlex system, only a single DBLβ recombinant protein from the 3D7 parasite strain bound ICAM-1, and it was from an UpsA protein [45]. UpsA proteins have also been reported to bind ICAM-1 (PFD1235w) and PECAM-1 (PF11_0008) [47]. Taken together, sequence and binding analysis suggest the UpsA group forms a preferential gene recombination group that is under less selection to bind the primary microvasculature receptor CD36. Furthermore, it is possible UpsA genes may have evolved specialized binding properties that contribute to their preferential expression in the malaria non-immune.
While sequence and binding analysis of isolated domains have provided significant insights into PfEMP1 function, few binding predictions have been confirmed for native proteins at the IE surface, and it is not yet established whether binding differences truly exist between var gene subfamilies. Furthermore, it is possible that recombinant protein binding properties may be modified by adjacent domains [48] or may not extrapolate to the native PfEMP1 molecule [49]. Thus, there remain significant uncertainties in our ability to predict IE binding, and there is still limited understanding of how host selection is shaping the PfEMP1 variant antigen repertoire for parasite survival and transmission. For this study, we generated a large panel of cloned parasite lines from the cytoadhesive IT4/FCR3 parasite strain and selected three highly enriched UpsA parasite lines with specific monoclonal antibodies. This panel was employed to both investigate the major host selection binding pressures operating on the protein family and to evaluate binding predictions based on sequence information and isolated domain binding assays.
Results
Generation of a panel of cloned parasite lines from a cytoadhesive parasite line
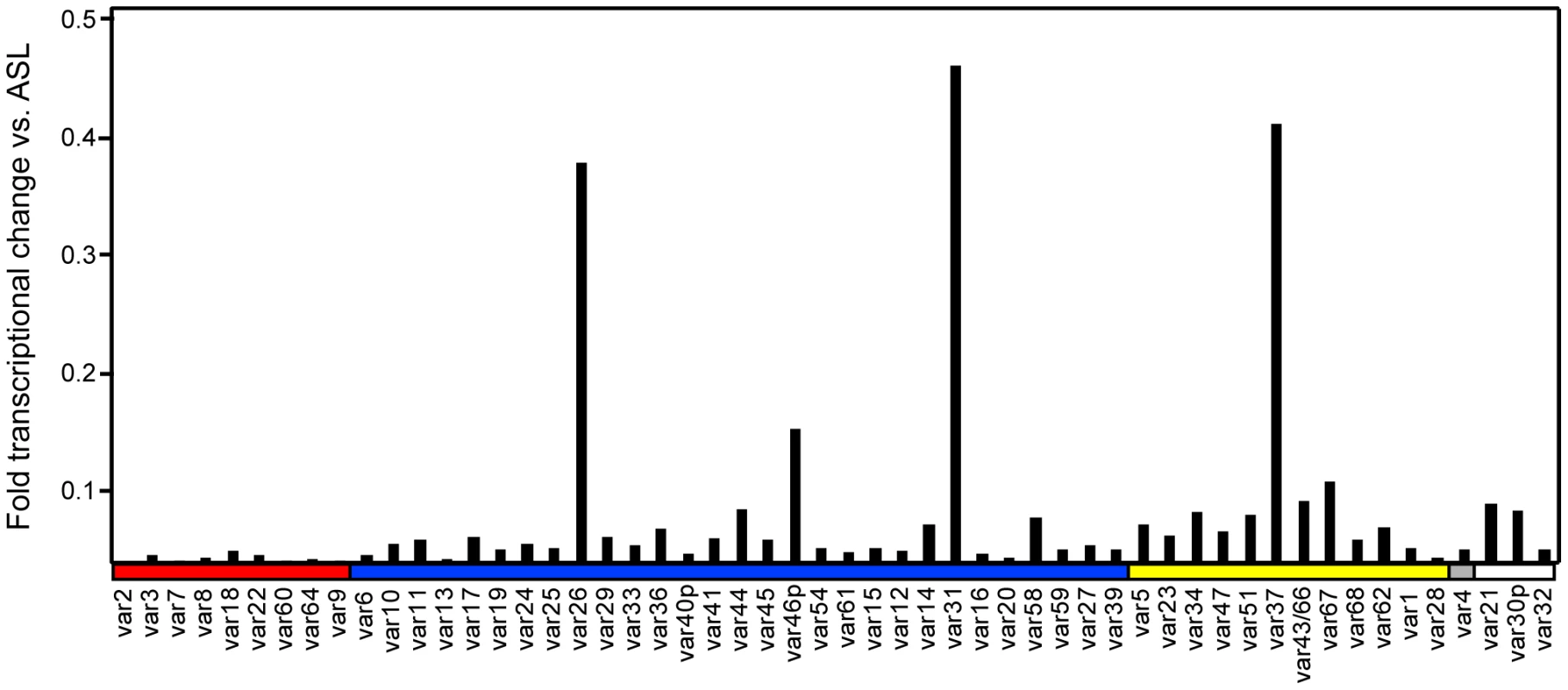
To create a panel of parasites for phenotypic analysis, parasites were cloned from a long-term, continuous culture of the IT4/25/5 clone A4 (Figure 1) [6]. The IT4/25/5 (IT4) parasite genotype is unusual because the parasite maintained its cytoadhesion capabilities after in vitro adaptation [50], [51], making it a primary model for this virulence determinant. A set of 54 var genes has been reported from the IT4 parasite genotype [9], [10]. The A4 cloned parasite line expresses a var gene (A4var/IT4var14) that has an unusually high switch frequency (∼1–2% per generation) [6], [52], resulting in PfEMP1 heterogeneity at the population level in the long term culture. After 70 parasite divisions in continuous culture, the long-term A4 culture had completely switched away from the A4var gene (IT4var14) and expressed a mixture of different var genes at low levels with IT4var26, IT4var31, and IT4var37 predominating (Figure 2). Both IT4var31 (previously referred to as C18var) and IT4var37 (previously referred to as AFBR6) were also found to be common switch events in two previous studies of var gene switching within the A4 parasite lineage [7], [52], suggesting that these particular genes may have high “on” rates in unselected cultures.
Fig. 1. Derivation of parasite lines for phenotypic analysis. 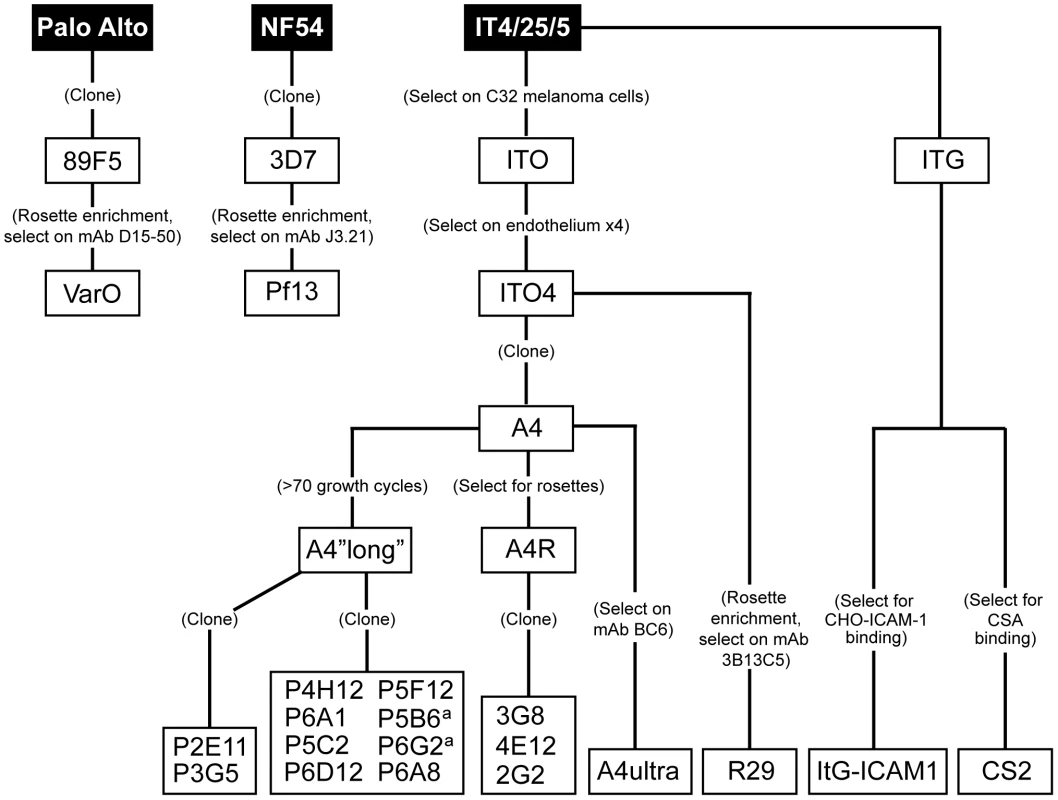
Initially, 17 subclones were isolated from the long-term A4 parasite culture by limited dilution cloning (Figure 1). From var transcription profiling, 6 of the subclones transcribed IT4var31 as either the primary or secondary var transcript, 8 transcribed dominant var gene transcripts distinct from each other, and a dominant var transcript (present at greater than 50% of the total var transcripts) could not be identified in 3 of the subclones by qRT-PCR analysis (Table 1, and data not shown). Ten subclones that primarily expressed single dominant var transcripts, including two that expressed IT4var31, were selected for phenotypic analysis (Figure 3).
Of interest, there was negligible UpsA transcription in the long-term A4 culture (Figure 2), and none of the isolated subclones expressed an UpsA var gene (Figure 3). To attempt to enrich for UpsA variants, the long-term A4 culture was panned on CD36 receptor protein and non-adherent parasites were selected. Although the var transcriptional profile was modified after CD36 negative selection, this approach did not enrich for UpsA variants. Instead, the frequent switch variant IT4var31 was the resulting major transcript (data not shown). This again indicates that UpsA genes are rare switch events in long-term A4 cultures.
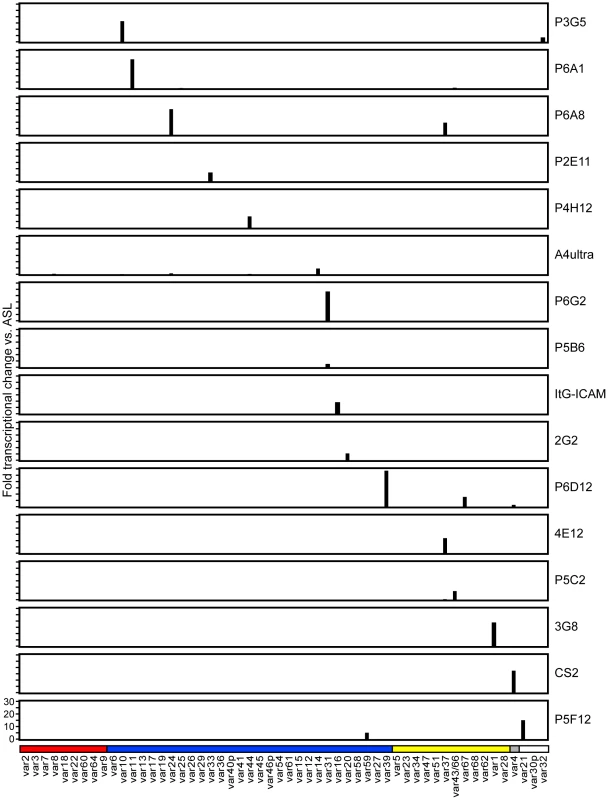
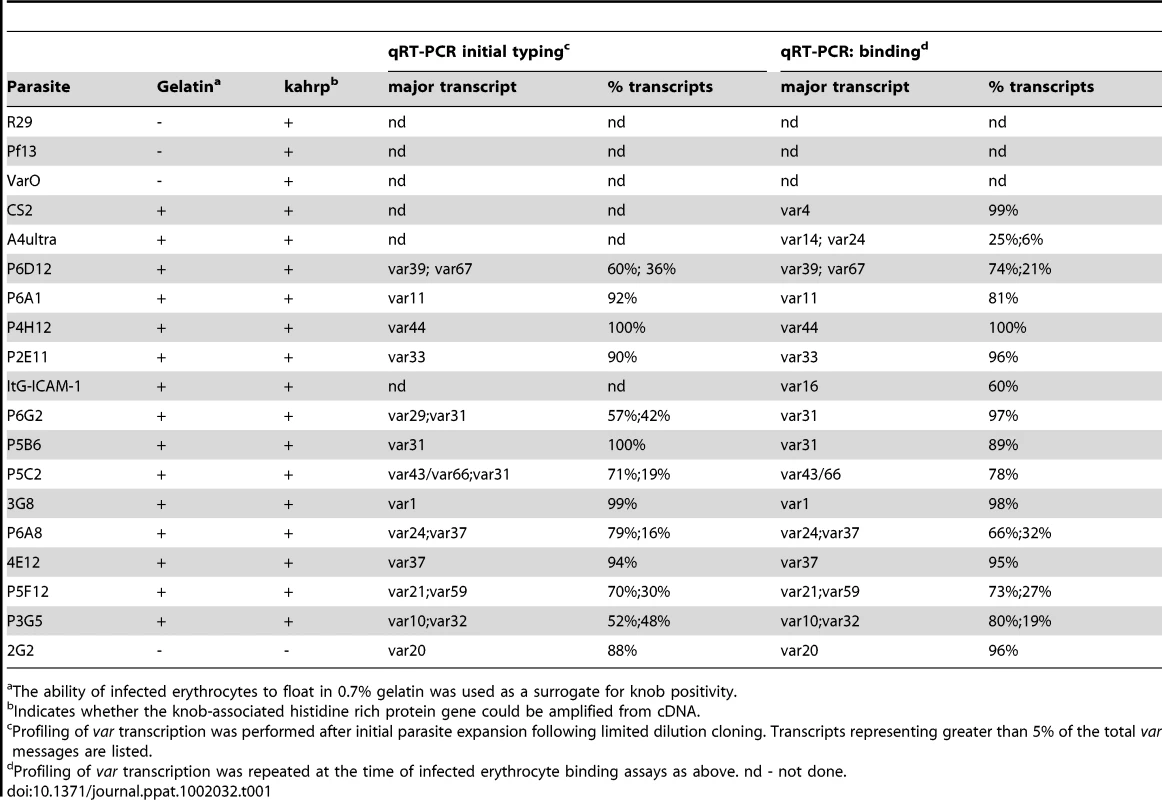
To create a more representative panel for phenotypic analysis, six previously isolated parasite lines from the IT4/FCR3 strain and three UpsA parasite lines from different parasite strains (IT4/FCR3, Palo Alto 89F5, and 3D7) (Figure 1) were included in the binding studies. The three UpsA parasite lines (R29, VarO, and Pf13) were isolated by rosette enrichment and selected for high purity using specific monoclonal antibodies to the respective PfEMP1 proteins [53]. Altogether, 19 parasite lines were examined representing all three major var gene groups. Three of the parasites in the panel expressed an UpsA protein as the dominant var transcript, ten expressed an UpsB var gene, three expressed an UpsC var, one expressed the unique UpsE linked transcript (IT4var4, var2CSA), and for one parasite, the Ups category of the dominant var transcript has yet to be determined (Figure 4). The remaining parasite in the panel, 2G2, is knobless and was employed as a negative binding control (Table 1) [54]. Most parasites in the panel expressed distinct dominant var transcripts, except two subclones (P6G2 and P5B6) expressed IT4var31, and two others (P6A8 and 4E12) expressed IT4var37/AFBR6 as either the dominant or secondary var transcript (Figure 3 and Table 1).
To confirm the presence of knobs on the IE surface, which are known to be important in PfEMP1 anchoring and infected erythrocyte binding [41], [54], [55], parasites were tested for transcription of the knob associated histidine rich protein (kahrp+) by RT-PCR and floated by gelatin sedimentation (gelatin+). All parasites in the panel were positive in both assays, except for the negative control 2G2 parasite line, which lacks kahrp and therefore sedimented in gelatin. In addition, the three rosette-forming UpsA parasites all transcribed kahrp but sedimented in gelatin because they were originally isolated on the basis of their property to sediment in Ficoll (Table 1). To confirm the identity of var gene transcription at the time of binding assays, RNA was harvested within the same growth cycle that binding assays were performed. For these assays, thawed parasite stabilates were grown for 4 to 5 cycles to generate sufficient parasite material, and parasites were generally analyzed a total of 18–20 cycles from initial parasite cloning. In general, the dominant var transcript did not change between the initial qRT-PCR characterization performed after limited dilution cloning and the second round of -typing done at the binding assay (Table 1). In only one parasite line, P6G2, the previous dominant transcript was replaced by the secondary var transcript that was present before freezing (Table 1). At the time of the binding analysis, the average fold transcription of dominant var transcripts relative to the asl housekeeping gene was 14.2 (range 2.8–28.1). Furthermore, most parasite lines were significantly enriched for a single predominant var transcript (Figure 3), and only 8 parasite lines contained a secondary var transcript at greater than 5% of the total var transcripts (Table 1). In most cases, the secondary transcript was present at much lower levels than the dominant var transcript. Thus, var gene transcription was stable over the short-term culture period used to perform these assays. For the three UpsA variants, PfEMP1 expression was established by flow cytometry with specific monoclonal antibodies to be 79% or higher using conservative gating criteria (Figure S1). Furthermore, all three lines formed rosettes in O-type RBCs: R29 (rosetting rate = 37%, 89% mAb reactivity R29), VarO (rosetting rate = 73%, 79% mAb reactivity VarO), Pf13 (rosetting rate = 52%, 94% mAb reactivity Pf13_0003). Therefore, all of the parasites in the panel were highly homogenous for one or two var transcripts, and UpsA parasite lines were highly pure for a single expressed PfEMP1 variant.
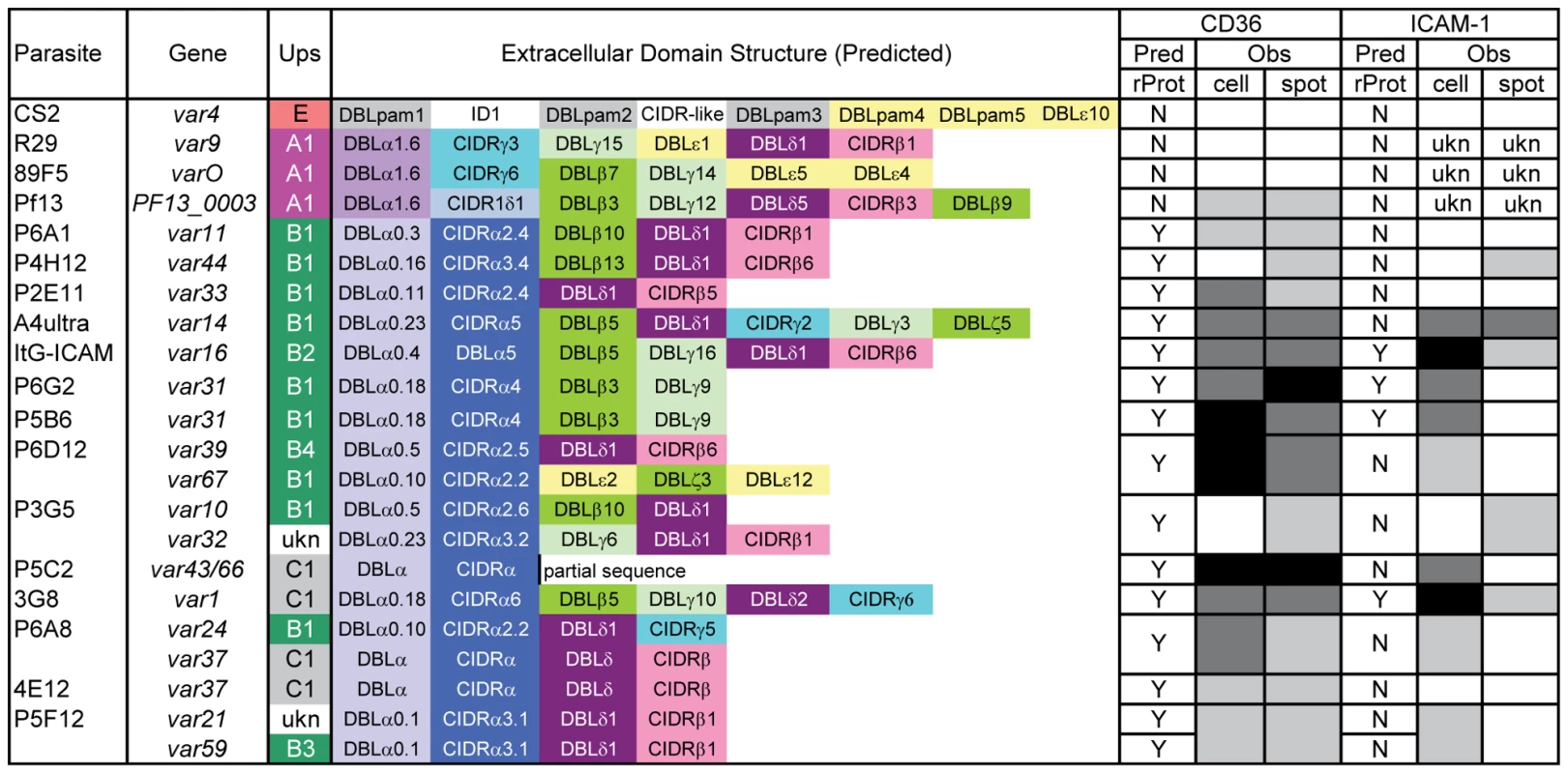
CD36 binding of infected erythrocytes is highly predicted by the type of CIDR domain in PfEMP1 proteins
To investigate whether infected erythrocyte binding to CD36 could be predicted from sequence information and binding studies of isolated CIDR domains [46], the complete panel of parasite lines was analyzed for binding to both CHO745-CD36 and immobilized CD36 recombinant protein. Because rosettes of uninfected red blood cells can interfere with binding or make bound IEs more susceptible to disruption during washing stages, the rosettes of the three UpsA variants were first disrupted using heparin sulfate prior to binding analysis. Previous work has shown that sulfated glycoconjugates can enhance binding to CD36 on cell surfaces [56]. Therefore, as a control for the three rosetting parasite lines, all of the parasites in the panel were treated with heparin sulfate and tested for binding to immobilized CD36 recombinant protein. Heparin sulfate treatment greatly diminished rosette formation in the three UpsA parasite lines (<10%) (Figure S2), but had minimal effect on infected erythrocyte binding to immobilized CD36 recombinant protein. Overall, parasites had comparable binding levels in the presence or absence of heparin sulfate (Figure S2). In addition, two non-rosetting, CD36 binding parasite lines (A4ultra and ItG-ICAM-1) were tested for binding to CHO745-CD36 cells in the presence or absence of heparin sulfate. Similar to what has been reported previously [56], sulfated glycoconjugates increased IE binding to CHO745-CD36 (Figure S3). Because heparin sulfate may slightly enhance IE adhesion to CHO-CD36 and did not modify IE adhesion to immobilized CD36, the binding assay was then repeated for all of the non-UpsA parasites in the absence of sulfated glycoconjugates. In contrast, binding of the three UpsA lines to CHO-CD36 and immobilized CD36 was repeated in the presence of sulfated glyconjugates to prevent infected erythrocyte rosetting interfering with the binding results.
Overall, there was a significant correlation between CHO745-CD36 and spotted CD36 protein formats (Figure 5, Spearman's Rho = 0.75, p<0.001). Although the level of CD36 binding varied between parasite lines, most of the parasites bound CD36, with the exception of UpsA/E groups (Figure 6). The three UpsA parasites were at the lower spectrum of CD36 binding in both cell and recombinant protein binding assays, and were basically indistinguishable from the negative control, knobless parasite line, and the UpsE parasite line that does not bind CD36 (Figure 6). Furthermore, CD36 binding was highly predicted by the type of CIDR1 domain in the PfEMP1 head structure (Figure 4). Indeed, only two parasites in the panel that were predicted to bind CD36 did not bind to CHO-CD36 cells. However, both exceptions (P4H12 and P3G5) bound at a low level to 50 µg/mL rCD36, but not to 5 µg/mL rCD36 (Figure 5), and therefore may be lower affinity CD36 binders. In group-wide comparisons, UpsB and UpsC had a higher mean CD36 binding level than UpsA. This difference was significantly different in the immobilized CD36 binding assay and between the UpsC and UpsA groups in the CHO-CD36 assay, and just missed significance between the UpsB and UpsA groups in the CHO-CD36 assay (Figure 6). Taken together, infected erythrocyte binding was highly predictable based on the type of CIDR domain (Figure 4), and the UpsA group appears to be under less selection to bind CD36.
Fig. 5. Infected erythrocyte binding to CD36 and ICAM-1. 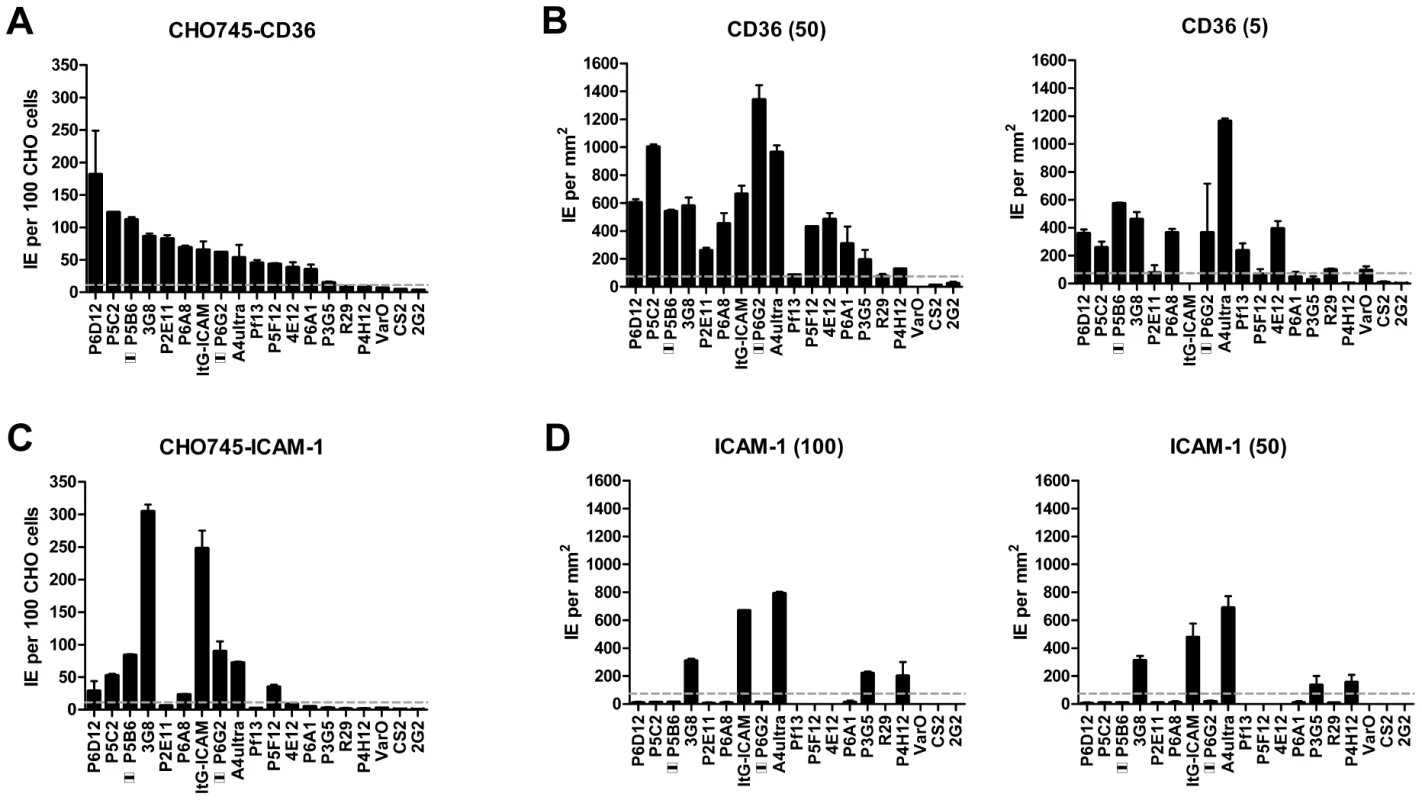
Fig. 6. Infected erythrocyte binding to CD36 and ICAM-1. 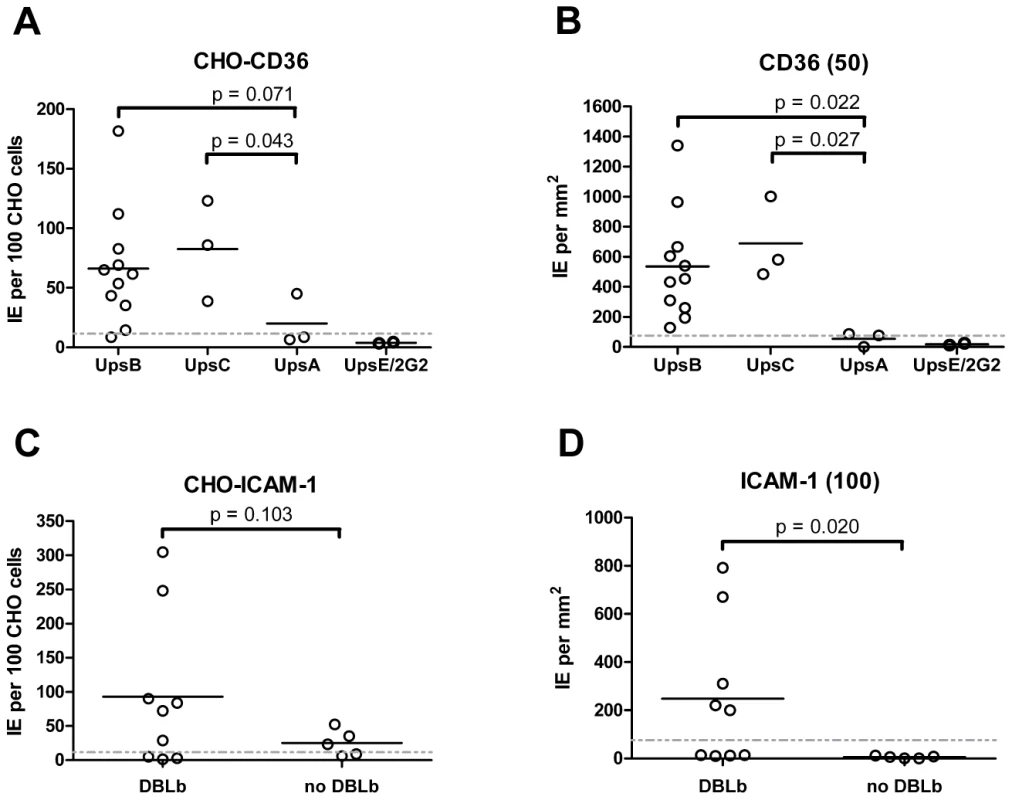
ICAM-1 binding was strongly associated with larger, DBLβ containing PfEMP1 proteins
To test whether ICAM-1 binding was associated with larger PfEMP1 proteins containing DBLβ domains [44], the parasite panel was analyzed for binding to CHO745-ICAM-1 and recombinant ICAM-1 protein. Again, to prevent rosettes from interfering with the binding analysis, the three UpsA parasite lines were treated with sulfated glycoconjugates prior to binding analysis, and as a control, two non-rosetting, ICAM-1 binding parasite lines (A4ultra and ItG-ICAM-1) were tested for ICAM-1 binding in the presence or absence of sulfated glycoconjugates. Sulfated glyconjugates reduced binding of A4ultra in the CHO745-ICAM-1 assay and binding of both parasite lines to spotted ICAM-1 recombinant protein (Figure S3), similar to what has been reported before [56]. Because of the potential for sulfated glyconjugates to interfere with ICAM-1 binding in the cell and recombinant protein assays, the three UpsA parasite lines were not considered in the ICAM-1 binding analysis.
In the cell binding assay, two parasite lines bound at a high level (>2 IEs/CHO745-ICAM-1), three bound at moderate level (0.5–2 IEs/CHO745-ICAM-1), and the remaining parasite lines bound at a low level or did not bind ICAM-1 (Figure 5). While there was good consistency between the cell and recombinant protein assays for the two high level ICAM-1 binders, there was more discordance for weaker ICAM-1 binders (Figure 5). Only three parasite lines bound ICAM-1 in both platforms (3G8, ItG-ICAM-1, and A4ultra), and two parasite lines that bound at a moderate level to CHO745-ICAM-1 did not bind to immobilized ICAM-1 protein (Figure 5). Notably, both parasite lines express the IT4var31 transcript, which has been suggested to be a weaker ICAM-1 binding variant that is trypsin-resistant [57], [58]. To confirm whether binding was trypsin-resistant, P5B6-infected erythrocytes expressing IT4var31 were treated with 1 mg/mL trypsin prior to ICAM-1 binding analysis. Trypsin treatment reduced CD36 binding and increased binding to recombinant ICAM-1 (Figure S4), and therefore may have cleaved or truncated the PfEMP1 head structure. The increase in ICAM-1 binding could be blocked by anti-ICAM-1 antibody (mAb 15.2) and not by anti-CD36 isotype control antibody (FA6-152) (Figure S4). In contrast, identical trypsin treatment of 3G8 (IT4var1) and ItG-ICAM-1 parasite lines (IT4var16) abolished binding to both CD36 and ICAM-1 (data not shown). Thus, as predicted from binding of the isolated DBLβ domain [58], IT4var31 was associated with ICAM-1 binding, but the cell binding assay was more sensitive than immobilized protein in detecting this interaction. Two of the parasite lines also bound at a low level to immobilized ICAM-1 recombinant protein but did not bind CHO745-ICAM-1. Thus, there may be differences in the sensitivity of the two platforms to detect lower affinity ICAM-1 interactions, or some of the low level binding interactions may not have been specific.
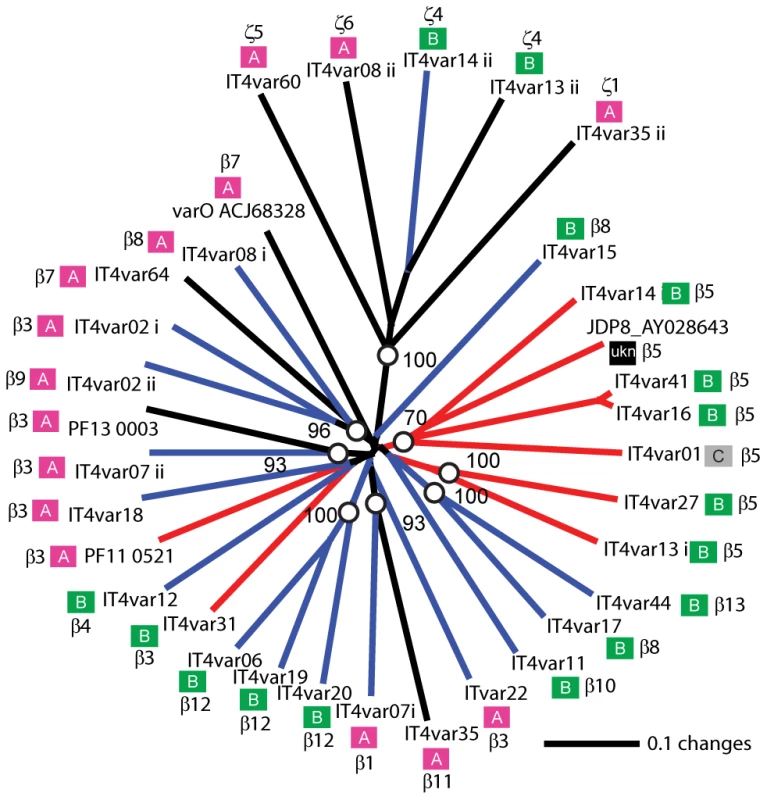
Overall, ICAM-1 binding was strongly associated with larger PfEMP1 proteins that contained a DBLβ domain. Seven of the ten parasites lines that expressed a dominant var transcript containing a DBLβ domain bound to ICAM-1 in either the cell or recombinant protein platform (Figure 4), and parasite lines without a DBLβ either bound extremely weakly or did not bind ICAM-1 (Figure 6). This difference was significant in the immobilized ICAM-1 assays (1-tailed t-test, p = 0.020) and just missed significance in the CHO745-ICAM-1 assay (1-tailed t-test, p = 0.103). Recently, there has been a reclassification of DBL and CIDR domains into additional subtypes based on a comparison of 7 parasite genomes in which DBLβ domains were subclassified into 13 sub-types [10]. Of interest, all three parasites that bound in both the CHO-ICAM-1 and immobilized ICAM-1 assays expressed a DBLβ5 domain (Figure 4). To investigate if DBLβ5 could be a marker for ICAM-1 binding, we reanalyzed the recombinant DBLβ-ICAM-1 binding data [44]. In the IT4 parasite genotype, 7 of 23 DBLβ domains bound ICAM-1. Of the 7 ICAM-1 binders, 6 were DBLβ5 sequences, and there were no DBLβ5 domains that did not bind ICAM-1 (Figure 7). Significantly, an ICAM-1 binding parasite from India (JDP8-ICAM-1, AY028643) [59] also uses a DBLβ5 domain to bind ICAM-1 (Figure 7). The fact that ICAM-1 binding was 100% predictable in the IT4 parasite genotype, and that a different parasite isolate from India also uses DBLβ5 for binding, strongly supports this domain as a marker for ICAM-1 binding. There are also two DBLβ3 sequences that bound ICAM-1, one from the IT4 parasite genotype and one from the 3D7 parasite genotype [45], but several other DBLβ3 sequences did not bind ICAM-1 as recombinant proteins (Figure 7). Taken together, ICAM-1 binding was strongly associated with the DBLβ domain, and the DBLβ5 marks a category of larger PfEMP1 variants that encode this adhesion property.
Infected erythrocyte binding to additional receptors was rare
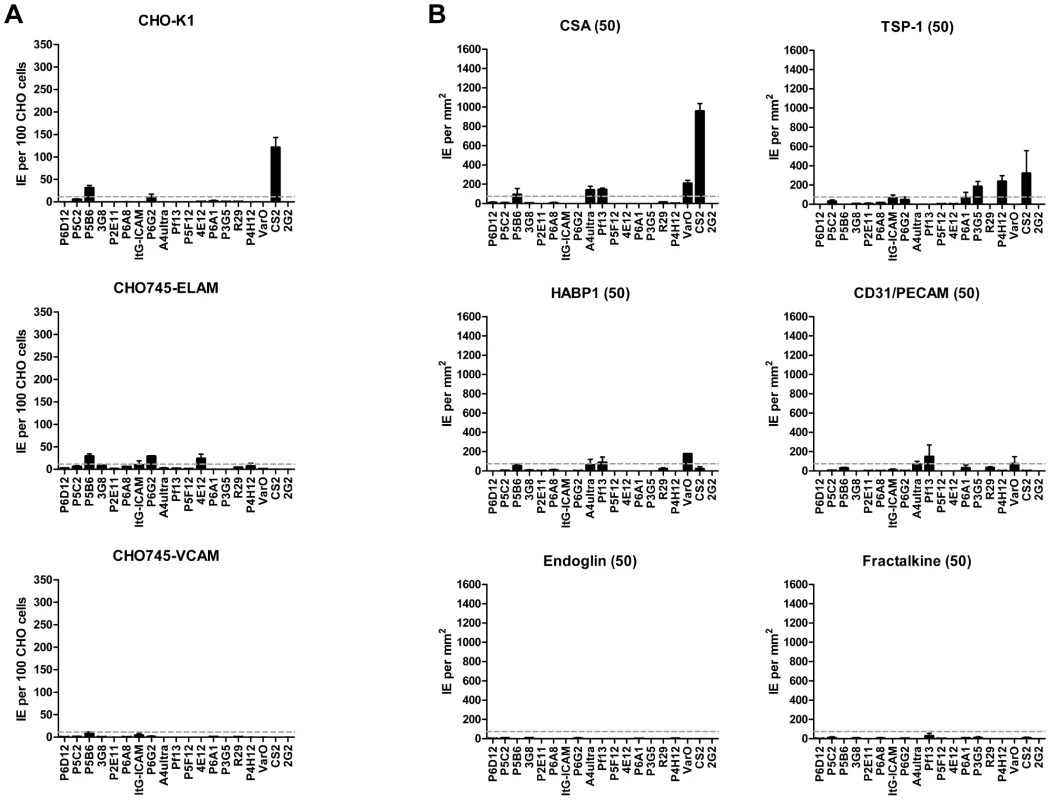
Infected erythrocytes have been reported to bind a number of host receptors [20], but for the most part binding has only been tested on one or a few parasite lines. Using transfected cells or recombinant proteins, the 19 parasite lines were assayed against 8 additional receptors: Endothelial Leukocyte Adhesion Molecule 1 (E-selectin), Vascular Cell Adhesion Molecule 1 (VCAM-1), CHO receptor “X”, Hyaluronan Binding Protein 1 (HABP1), Platelet Endothelial Cell Adhesion Molecule-1 (CD31/PECAM-1), Thrombospondin-1 (TSP-1), CSA, and Fractalkine. Whereas a few parasite lines bound at a low level to TSP-1 and CHO-ELAM-1, there was negligible binding to most receptors tested (Figure 8). Two of the UpsA parasites (Pf13 and VarO) bound at a low level to HABP1, CD31, and CSA. However, binding of UpsA parasites was performed in the presence of sulfated glycoconjugates to disrupt rosettes, and they also had higher background binding to bovine serum albumin (BSA) employed as a blocking agent for binding assays (Figure 8, and data not shown). As expected, the strongest CSA-binder in the panel was the CS2 parasite line in both the CHO-K1 cell and CSA spot formats (Figure 8). CS2 expresses the VAR2CSA PfEMP1 protein that has been shown to be the primary PfEMP1 variant associated with CSA binding [60], [61]. Most of the other receptors tested did not support strong adhesion of infected erythrocytes in these binding assays and it is questionable whether all of the observed weak interactions are physiologically relevant.
Discussion
PfEMP1 proteins/var genes are classified into three main subfamilies (UpsA, UpsB, and UpsC) that have different host expression profiles [35]–[37], [39]. Both binding strength and specificity of IEs are likely to influence disease severity during an infection; therefore, it is important to understand whether PfEMP1 subfamilies have evolved specialized properties for distinct host/biological niches. Studies of malaria during pregnancy have demonstrated how a specific PfEMP1 variant can precipitate severe disease in otherwise immune women by altering IE tropism for the placenta [14], [29], [62]. Although VAR2CSA appears to be unique in its ability to confer high-affinity binding to CSA in the placenta [60], [61], [63], it offers a paradigm for the role of specific PfEMP1s in disease. UpsA classified PfEMP1 proteins are frequently observed in young children with limited anti-malaria immunity or experiencing severe malaria [35]–[39]. Unlike VAR2CSA, the adherence characteristics of UpsA proteins are poorly understood and limited largely to predictions of binding based on studies of isolated adhesion domains [44]–[46]. To investigate a correlation of PfEMP1 binding specificities with disease outcome, the binding characteristics of at least a representative sample of the three main subgroups (UpsA, UpsB and UpsC) have to be known. In this study, we employed a panel of different PfEMP1 types to test binding predictions based upon studies of single PfEMP1 domains.
While UpsA variants appear to be commonly expressed in early childhood infections and non-immune individuals [35]–[39], very little is known about what may account for this preferential expression in the malaria naïve. Investigation is hampered because most P. falciparum infections contain a mixture of PfEMP1 variants and even minor parasite subsets may obscure binding analysis. In addition, gene silencing of UpsA variants has been observed upon in vitro adaptation [64]. In long term in vitro adapted parasite cultures grown without selection for specific var gene expression, UpsA variants were expressed at a low level, and an UpsB (IT4var31/C18var) and an UpsC (IT4var37/AFBR6) var gene appeared to be the most common switch events. Both were also found to be frequently activated in previous clonal analyses in this strain background [7], [52] and thus may have a higher “on” rate under in vitro culture conditions. One study found that var genes in central chromosome regions had lower switch rates than those in telomeric regions [65], but inherent differences were not consistently observed in a different parasite line [52]. The chromosome positions of IT4var31 (UpsB) and IT4var37 (UpsC) have not been mapped and therefore we cannot comment on whether this observation held true in our study or not. However, our findings indicate that promoter type is not the main determinant of var gene “on” rate as far as UpsB and UpsC type var genes are concerned. In the case of UpsA variants, the promoter type did seem to determine var gene expression rate by significantly reducing it. To overcome these problems, we used specific monoclonal antibodies to generate three distinct UpsA parasite lines of high purity for the parasite panel.
In epidemiological studies, CD36 and ICAM-1 binding are the most common adhesion traits in the parasite population [17], [19], but their distribution among different members of the PfEMP1 family is only partially understood [44]–[46], [58], [66]. In the parasite panel, CD36 was by far the most common binding partner, followed by ICAM-1 and TSP-1. CD36-binding was nearly 100% predictable and was always associated with a CIDRα type domain in the protein head structure, while the three UpsA variants had different sequence types (CIDRγ and CIDRδ) and did not bind CD36 or only bound at a low level. Thus, in the absence of a CIDRα domain, other potential CD36 ligands [67], [68] were unable to compensate for infected erythrocyte binding. Moreover, the level of CD36 binding differed between isogenic parasites expressing different PfEMP1 variants, suggesting that PfEMP1 sequence variability or surface expression levels have an important role in influencing the overall binding affinity of infected erythrocytes.
The UpsA group contains three different types of CIDR1 sequences (α1, γ, or δ) [10], [12], [40], [46]. Although the three UpsA parasites in the panel were all selected for rosetting, “rosetting” and “non-CD36 binding” can exist as independent phenotypes. For instance, the non-CD36 binding CIDR domains identified in this study may potentially be found in non-rosetting group A genes, and there is evidence that CD36 is able to act as a host receptor for rosetting in the Malayan Camp parasite strain and some field isolates [69]. This parasite panel did not contain any representation of the CIDRα1 subtype, which is found in approximately half of UpsA proteins [10]. However, it has previously been shown that recombinant CIDRα1 subtype domains do not bind CD36 [46], and CD36 selection led to loss of expression of an UpsA gene in a mixed parasite culture that expressed a CIDRα1 subtype [70]. Taken together, the results suggest the UpsA group is not under strong selection for CD36 binding, and it will be interesting to determine if the UpsA protein head structure is selected for specific binding properties that support microvasculature sequestration by a mechanism different from CD36 binding. Part of this selection may be for infected erythrocyte rosetting [71], [72], but the UpsA group may encode other adhesion properties [47].
After CD36, ICAM-1 is one of the most common adhesion properties, and the two receptors synergize to mediate infected erythrocyte binding under flow [22], [23]. ICAM-1 is upregulated on brain endothelium during malaria infections and has been proposed to be a potential cerebral sequestration receptor [24]. ICAM-1 binding has previously been mapped to the DBLβ domain [44], [45], [58], [59], [73]. Our study confirms this association as the DBLβ5 domain was 100% associated with ICAM-1 binding in both parasite lines and recombinant proteins. It also shows that not all DBLβ domains bind to ICAM-1. In future work using patient samples it may be interesting to investigate how well transcription of var genes containing a DBLβ5 domain can predict ICAM-1 binding. Overall, this study identifies a category of large UpsB and UpsC PfEMP1 containing CIDRα and DBLβ5 subtype domains that were 100% associated with CD36 and ICAM-1 binding. In a comparison of var gene repertoires from 7 parasite strains, the CIDRα and DBLβ5 domains were always found together in tandem arrangement (27 of 399 full or partial length var genes), and the DBLβ5 domain was never associated with a predicted “non-CD36 binding” CIDR domain. This suggests the association has not evolved by chance and that the CIDRα-DBLβ5 domain combination may be under dual selection for binding to CD36 and ICAM-1. Both receptors are co-displayed on many of the same cell types (endothelial, monocyte, and dendritic cells) and may provide the parasite opportunities to manipulate host cells [74], [75], thus contributing to their strong selection in the PfEMP1 repertoire. There were also a few DBLβ3 domains that bound to ICAM-1, but these were found in association with both CD36 binding and non-CD36 binding CIDR domains. Thus, CD36 and ICAM-1 have left strong signatures of selection detectable by PfEMP1 adhesion domain sequence classification, despite the extensive sequence diversity in the family.
Other PfEMP1 adhesion properties examined appear to be much rarer or may only play an additive role in overall binding affinity. Nearly all PfEMP1 proteins have four or more extracellular domains. In addition to undefined binding properties, other PfEMP1 domains may also function as “spacers” to position the PfEMP1 head structure and adjacent DBLβ away from the IE surface in order to engage CD36 and ICAM-1 [76]. A potential caveat is that binding was performed under static adhesion conditions, and individual host recombinant proteins were employed in the protein binding assays. However, all host receptors examined were originally defined under similar static adhesion conditions. Furthermore, static adhesion assays are capable of detecting host receptor interactions that support both rolling (ICAM-1, TSP-1) and stationary (CD36) cytoadhesion of infected erythrocytes under flow conditions [21]. Cooperative binding is likely necessary to mediate firm adhesion under flow [21]–[23], but from this analysis CD36 binding is under greatest selection and contributes the greatest binding avidity in different PfEMP1 proteins.
These results reveal a fundamental difference in CD36 binding between Ups groups that has important implications for how parasites establish infections in individuals of varying levels of immunity. UpsA proteins are more commonly expressed in children with low immunity [35], [36], [39]. Later, as malaria immunity develops, it may be significant that the proportion of non-UpsA types and CD36 binding variants increases. It is interesting to speculate that non-CD36-binding parasites may experience a selective advantage over their CD36-binding counterparts in patients with limited exposure to malaria. CD36-binding parasites are thought to manipulate both host innate and adaptive immune responses by interacting with monocytes and dendritic cells [74], [75], [77], [78]. In the malaria naïve, these interactions may be less important, or UpsA variants may possess other advantages or means of host manipulation. While UpsA variants have not been clearly associated with disease in all studies [79], they are more abundant in patients with severe malaria [80], [81] and have been associated with cerebral malaria infections in children in Mali [38]. A greater proportion of UpsA variants in early infections could potentially contribute to why CD36 binding levels are very low in children with severe malaria anemia [17], [19], or these variants could alter the pattern of sequestration to microvascular beds, such as brain endothelium, where CD36 binding levels are extremely low [24]. Therefore, it will be important to learn more about this group of proteins.
In conclusion, the PfEMP1 protein family has diversified under dual selection to evade host immunity and mediate infected erythrocyte binding. The development of a parasite panel enriched for distinct PfEMP1 expression from the major Ups groups has facilitated the testing of binding predictions, and may have potential applications for investigating immune acquisition to the family of proteins. This comparative analysis demonstrates the predictability of P. falciparum-IE binding to the two major cytoadhesion receptors CD36 and ICAM-1 and provides new insight into how natural selection may be shaping the PfEMP1 binding repertoire to exploit distinct host niches of varying anti-malaria immunity.
Materials and Methods
Ethics statement
Human blood was used for P. falciparum culture in this study. Donor blood was obtained from healthy volunteers under a minimal risk, standardized, Institute protocol (protocol number HS013) that was approved by the Western Institutional Review Board. Written informed consent was obtained from all blood donor study participants.
P. falciparum culture conditions
The three UpsA variants were isolated by gelatin sedimentation followed by positive selection with specific monoclonal antibodies against the respective NTS-DBLα domain. The VarO parasite clone was generated from the Palo Alto strain as described by rosette enrichment and selection with monoclonal antibody D15–50 [82]. The R29 parasite (IT4 parasite strain) has been described previously [6], [7], [83]. Highly enriched parasite cultures expressing the R29 PfEMP1 protein and Pf13 (3D7 strain) were isolated by similar methodologies to the VarO parasite line using rosette enrichment and specific monoclonal antibodies against the R29-DBLα domain (3B13C5) or the Pf13_0003-DBLα domain (J3.21) [53]. The ItG-ICAM-1 parasite line was derived by ICAM-1 selection [18], CS2 by CSA selection [84], and the 3G8, 4E12, and 2G2 parasite lines by limited dilution cloning [52]. The remaining parasite lines were derived from IT4/25/5 clone A4 [6] by limited dilution cloning. Infected erythrocytes were cultured under standard conditions using human O red blood cells (RBCs) in RPMI-1640 medium (Invitrogen) supplemented with 10% pooled human A+ serum and an atmosphere of 5% CO2, 5% O2, and 95% N2 at 37°C. Synchronization of parasite growth was achieved by treatment with 5% sorbitol in PBS. Gelatin sedimentation assays were performed in RPMI-1640 medium containing 0.7% porcine gelatin (Sigma) for 45 minutes at 37°C. Enrichment of infected erythrocytes (IE) in the gelatin supernatant was determined by counting >300 methanol-fixed, Giemsa-stained RBCs under 1000X magnification. Rosette formation was visualized after infected red blood cell nuclei were stained by ethidium bromide. The rosetting rate was calculated by determining the percentage of rosette-forming infected cells in the mature parasite population.
Chinese hamster ovary cell culture conditions
CHO-K1, CHO745, and CHO745 transfectants expressing CD36, ICAM-1, E-selectin, or VCAM-1 were cultured in F-12 Kaighn's medium supplemented with 10% fetal calf serum and 0.5 mg/mL geneticin (Gibco). The CHO745 transfectants were described in Buffet et al. [85]. Recombinant protein surface expression was monitored by flow cytometry on a monthly basis using receptor-specific monoclonal antibodies (R&D Systems), and cells were replaced if the percentage of transfected cells or mean fluorescence intensity diminished by greater than 20%.
Limited dilution cloning of parasite lines
An A4 parasite clonal line [6] was grown continuously under standard conditions for more than 70 growth cycles in the absence of overt selection. IEs were periodically enriched for knob expression by floatation in 0.7% porcine gelatin (Sigma) dissolved in RPMI-1640 (Invitrogen) at 37°C. Prior to limited dilution cloning, RNA was collected and a profile of var transcription was determined by quantitative real-time polymerase chain reaction (qRT-PCR) using a primer set designed to amplify unique sequence tags within the repertoire of IT4 var genes [86]. Individual infected erythrocytes were obtained on two separate occasions by limited dilution cloning after more than 78 and 84 cycles of continuous parasite growth, respectively, at a seeding rate of 0.5 infected erythrocytes per well. Initial frozen stabilates were collected after approximately 14–15 cycles of growth and parasite lines were typed for var gene expression by qRT-PCR.
Determination of var transcription by qRT-PCR
The determination of var gene transcription profiles was performed using primers and PCR conditions as previously described [86]. In brief, RNA was extracted in Trizol LS (Invitrogen) from ring stage parasites at ∼6–12 hours post-invasion and purified on RNeasy Micro columns with on-column DNaseI treatment (QIAGEN) according to manufacturer's protocols. cDNA was synthesized from 4 µg total RNA using Multi-Scribe reverse transcriptase (Applied Biosystems) and one half of this material was used for each real-time reaction against the complete set of primers. Real-time reactions were performed on an ABI Prism 7500 thermocycler at optimized final primer concentrations of 0.05 µM-0.5 µM using Power-SYBR Green Master Mix in 20 µL reaction volumes under the following PCR conditions: 50°C for 1 min, 95°C for 10 min, then 40 cycles of dissociation, annealing, and extension at 95°C for 15 sec, 52°C for 15 sec, and 60°C for 45 sec, respectively. Relative transcription was determined by normalization to the adenylosuccinate lyase (ASL, PFB0295w) control housekeeping gene. After optimizing primer efficiencies, residual primer bias was corrected by calculating the average difference in CT values between each optimized IT4 var primer pair and ASL using genomic DNA as template to provide a final normalized correction.
Infected erythrocyte binding assays
Parasite RNA was collected and binding assays performed within the same growth cycle to accurately assess var transcription at the time of the binding assay. For binding assays, individual CHO cell lines were grown to subconfluent levels on 60-mm tissue culture-treated dishes (BD Falcon) and recombinant proteins were immobilized by overnight incubation onto 60-mm polystyrene dishes (Corning). The following proteins were analyzed: CD36-Fc (R&D Systems), ICAM-1-Fc (R&D Systems), HABP1/gC1qR-6x HIS (R&D Systems), Fractalkine-6x-HIS (R&D Systems), CD31/PECAM-1 (R&D Systems), TSP-1-10x HIS (R&D Systems), and CSA (Sigma). All proteins and CSA were applied at 50 µg/mL except for CD36 and ICAM-1, which were additionally applied at 5 µg/mL and 100 µg/mL. On the day of the assay, dishes containing CHO cells were washed twice with pre-warmed cell binding medium (BMcell: RPMI-1640 medium containing 0.1% bovine serum albumin, pH 7.2) and protein spots were blocked with 2% bovine serum albumin for 45 min at 37°C, then washed twice with pre-warmed protein binding medium (BMprotein: RPMI-1640 medium containing 0.1% bovine serum albumin, pH 6.8). Infected erythrocytes (3-8% parasitemia) were washed and resuspended to 1% hematocrit in either BMcell or BMprotein then overlayed onto CHO cells or spotted onto immobilized proteins, respectively, and incubated for 1 hr at 37°C. Prior to binding assays, rosettes in the three UpsA parasite lines were disrupted in binding medium containing 100 Units/mL heparin sulfate (Sigma). The same conditions were used when testing the effect of heparin sulfate on all of the parasites in the panel. In additional assays to test the effect of sulfated glycoconjugates on IE binding, either 10 µg/mL dextran sulfate (MW >500,000; Sigma) or 100 Units/mL heparin sulfate were included during binding assays. Non-binding erythrocytes were removed by gently flooding each dish with warm binding medium, rocking the dish back and forth several times to resuspend non-binding erythrocytes, then pouring off and replacing the medium. The initial washing procedure was performed on CHO745 cells and 2% BSA spots and was repeated until non-binding erythrocytes were sufficiently removed by observation under 400X magnification. The remaining cells and spots then received the same number of washes. For quantification, dishes were fixed in 1% glutaraldehyde for 20 min at room temperature, then stained with 1X Giemsa for 15 minutes. Binding was quantified by determining the number of IE adhering to at least 300 cells under 1000X magnification or the number of IE per mm2 in 4 random fields under 400X magnification. All binding assays were repeated in duplicate.
Flow cytometry analysis
Trophozoite stage infected RBCs were incubated for one hour at room temperature with specific monoclonal mouse antibodies against R29var NTS-DBLα (mAb 3B13C5, 1∶500) Pf13_0003 NTS-DBLα (mAb J3.21, 1∶20), or VarO NTS-DBLα (mAb D15-50, 1∶20). Antibody labeling was detected with goat anti-mouse IgG-R-Phycoerythrin (Sigma) (1∶20) for 30 minutes. Infected erythrocyte nuclei were detected with SYTO 61 DNA dye (Invitrogen) (1∶1000) added with the secondary antibody. Stained cells were washed in PBS and analyzed on an LSRII FACS machine (BD Biosciences). Analysis was performed using FlowJo 8 (Tree Star, Inc).
Supporting Information
Zdroje
1. KraemerSMSmithJD
2006
A family affair: var genes, PfEMP1 binding, and malaria
disease.
Curr Opin Micro
9
374
380
2. MillerLHBaruchDIMarshKDoumboOK
2002
The pathogenic basis of malaria.
Nature
415
673
679
3. FrankMDeitschK
2006
Activation, silencing and mutually exclusive expression within
the var gene family of Plasmodium falciparum.
Int J Parasitol
36
975
985
4. GardnerMJHallNFungEWhiteOBerrimanM
2002
Genome sequence of the human malaria parasite Plasmodium
falciparum.
Nature
419
498
511
5. BiggsBAAndersRFDillonHEDavernKMMartinM
1992
Adherence of infected erythrocytes to venular endothelium selects
for antigenic variants of Plasmodium falciparum.
J Immunol
149
2047
2054
6. RobertsDJCraigAGBerendtARPinchesRNashG
1992
Rapid switching to multiple antigenic and adhesive phenotypes in
malaria.
Nature
357
689
92
7. SmithJDChitnisCECraigAGRobertsDJHudson-TaylorDE
1995
Switches in expression of Plasmodium falciparum var genes
correlate with changes in antigenic and cytoadherent phenotypes of infected
erythrocytes.
Cell
82
101
110
8. Freitas-JuniorLHBottiusEPirritLADeitschKWScheidigC
2000
Frequent ectopic recombination of virulence factor genes in
telomeric chromosome clusters of P. falciparum.
Nature
407
1018
22
9. KraemerSMKyesSAAggarwalGSpringerALNelsonSO
2007
Patterns of gene recombination shape var gene repertoires in
Plasmodium falciparum: comparisons of geographically diverse
isolates.
BMC Genomics
8
45
10. RaskTSHansenDATheanderTGGormPALavstsenT
2010
Plasmodium falciparum Erythrocyte Membrane Protein 1 Diversity in
Seven Genomes - Divide and Conquer.
PLoS Comput Biol
6
e1000933
11. LavstsenTSalantiAJensenATArnotDETheanderTG
2003
Sub-grouping of Plasmodium falciparum 3D7 var genes based on
sequence analysis of coding and non-coding regions.
Malar J
2
27
12. KraemerSMSmithJD
2003
Evidence for the importance of genetic structuring to the
structural and functional specialization of the Plasmodium falciparum var
gene family.
Mol Microbiol
50
1527
38
13. RoweJAKyesSARogersonSJBabikerHARazaA
2002
Identification of a conserved Plasmodium falciparum var gene
implicated in malaria in pregnancy.
J Infect Dis
185
1207
11
14. SalantiAStaalsoeTLavstsenTJensenATSowaMP
2003
Selective upregulation of a single distinctly structured var gene
in chondroitin sulphate A-adhering Plasmodium falciparum involved in
pregnancy-associated malaria.
Mol Microbiol
49
179
91
15. TrimnellARKraemerSMMukherjeeSPhippardDJJanesJH
2006
Global genetic diversity and evolution of var genes associated
with placental and severe childhood malaria.
Mol Biochem Parasitol
148
169
180
16. HoMSinghBLooareesuwanSDavisTMBunnagD
1991
Clinical correlates of in vitro Plasmodium falciparum
cytoadherence.
Infect Immun
59
873
878
17. NewboldCWarnPBlackGBerendtACraigA
1997
Receptor-specific adhesion and clinical disease in Plasmodium
falciparum.
Am J Trop Med Hyg
57
389
98
18. OckenhouseCFHoMTandonNNVan SeventerGAShawS
1991
Molecular basis of sequestration in severe and uncomplicated
Plasmodium falciparum malaria: differential adhesion of infected
erythrocytes to CD36 and ICAM-1.
J Infect Dis
164
163
9
19. RogersonSJTembenuRDobanoCPlittSTaylorTE
1999
Cytoadherence characteristics of Plasmodium falciparum-infected
erythrocytes from Malawian children with severe and uncomplicated
malaria.
Am J Trop Med Hyg
61
467
72
20. RoweJClaessensACorriganRArmanM
2010
Adhesion of Plasmodium falciparum-infected erythrocytes to human
cells: molecular mechanisms and therapeutic implications.
Expert Rev Mol Med
11
e16
21. CookeBMBerendtARCraigAGMacGregorJNewboldCI
1994
Rolling and stationary cytoadhesion of red blood cells
parasitized by Plasmodium falciparum: separate roles for ICAM-1, CD36 and
thrombospondin.
Br J Haematol
87
162
70
22. GrayCMcCormickCTurnerGCraigA
2003
ICAM-1 can play a major role in mediating P. falciparum adhesion
to endothelium under flow.
Mol Biochem Parasitol
128
187
193
23. HoMHickeyMJMurrayAGAndoneguiGKubesP
2000
Visualization of Plasmodium falciparum-endothelium interactions
in human microvasculature: mimicry of leukocyte recruitment.
J Exp Med
192
1205
1211
24. TurnerGDMorrisonHJonesMDavisTMLooareesuwanS
1994
An immunohistochemical study of the pathology of fatal malaria.
Evidence for widespread endothelial activation and a potential role for
intercellular adhesion molecule-1 in cerebral sequestration.
Am J Pathol
145
1057
69
25. FryAEAuburnSDiakiteMGreenARichardsonA
2008
Variation in the ICAM1 gene is not associated with severe malaria
phenotypes.
Genes Immun
9
462
469
26. CarlsonJHelmbyHHillAVBrewsterDGreenwoodBM
1990
Human cerebral malaria: association with erythrocyte rosetting
and lack of anti-rosetting antibodies.
Lancet
336
1457
60
27. RoweAObeiroJNewboldCIMarshK
1995
Plasmodium falciparum rosetting is associated with malaria
severity in Kenya.
Infect Immun
63
2323
6
28. TreutigerCJHedlundIHelmbyHCarlsonJJepsonA
1992
Rosette formation in Plasmodium falciparum isolates and
anti-rosette activity of sera from Gambians with cerebral or uncomplicated
malaria.
Am J Trop Med Hyg
46
503
510
29. FriedMDuffyPE
1996
Adherence of Plasmodium falciparum to chondroitin sulfate A in
the human placenta.
Science
272
1502
4
30. GuptaSSnowRWDonnellyCAMarshKNewboldC
1999
Immunity to non-cerebral severe malaria is acquired after one or
two infections.
Nat Med
5
340
3
31. MarshKSnowRW
1997
Host-parasite interaction and morbidity in malaria endemic
areas.
Philos Trans R Soc Lond B Biol Sci
352
1385
1394
32. BullPCLoweBSKortokMMarshK
1999
Antibody recognition of Plasmodium falciparum erythrocyte surface
antigens in Kenya: evidence for rare and prevalent variants.
Infect Immun
67
733
9
33. BullPCKortokMKaiONdunguFRossA
2000
Plasmodium falciparum-infected erythrocytes: agglutination by
diverse Kenyan plasma is associated with severe disease and young host
age.
J Infect Dis
182
252
9
34. NielsenMAStaalsoeTKurtzhalsJAGokaBQDodooD
2002
Plasmodium falciparum variant surface antigen expression varies
between isolates causing severe and nonsevere malaria and is modified by
acquired immunity.
J Immunol
168
3444
50
35. ChamGKTurnerLLusinguJVestergaardLMmbandoBP
2009
Sequential, ordered acquisition of antibodies to Plasmodium
falciparum erythrocyte membrane protein 1 domains.
J Immunol
183
3356
3363
36. ChamGKTurnerLKurtisJDMutabingwaTFriedM
2010
Hierarchical, domain type-specific acquisition of antibodies to
Plasmodium falciparum erythrocyte membrane protein 1 in Tanzanian
children.
Infect Immun
78
4653
4659
37. JensenATMagistradoPSharpSJoergensenLLavstsenT
2004
Plasmodium falciparum associated with severe childhood malaria
preferentially expresses PfEMP1 encoded by group A var
genes.
J Exp Med
199
1179
90
38. KyriacouHMStoneGNChallisRJRazaALykeKE
2006
Differential var gene transcription in Plasmodium falciparum
isolates from patients with cerebral malaria compared to
hyperparasitaemia.
Mol Biochem Parasitol
150
211
218
39. WarimweGMKeaneTMFeganGMusyokiJNNewtonCR
2009
Plasmodium falciparum var gene expression is modified by host
immunity.
Proc Natl Acad Sci U S A
106
21801
21806
40. SmithJDSubramanianGGamainBBaruchDIMillerLH
2000
Classification of adhesive domains in the Plasmodium falciparum
erythrocyte membrane protein 1 family.
Mol Biochem Parasitol
110
293
310
41. BaruchDIPasloskeBLSinghHBBiXMaXC
1995
Cloning the P. falciparum gene encoding PfEMP1, a malarial
variant antigen and adherence receptor on the surface of parasitized human
erythrocytes.
Cell
82
77
87
42. SuXZHeatwoleVMWertheimerSPGuinetFHerrfeldtJA
1995
The large diverse gene family var encodes proteins involved in
cytoadherence and antigenic variation of Plasmodium falciparum-infected
erythrocytes.
Cell
82
89
100
43. SmithJDGamainBBaruchDIKyesS
2001
Decoding the language of var genes and Plasmodium falciparum
sequestration.
Trends Parasitol
17
538
45
44. HowellDPLevinEASpringerALKraemerSMPhippardDJ
2008
Mapping a common interaction site used by Plasmodium falciparum
Duffy binding-like domains to bind diverse host receptors.
Mol Microbiol
67
78
87
45. OleinikovAVAmosEFryeITRossnagleEMutabingwaTK
2009
High throughput functional assays of the variant antigen PfEMP1
reveal a single domain in the 3D7 Plasmodium falciparum genome that binds
ICAM1 with high affinity and is targeted by naturally acquired neutralizing
antibodies.
PLoS Pathog
5
e1000386
46. RobinsonBAWelchTLSmithJD
2003
Widespread functional specialization of Plasmodium falciparum
erythrocyte membrane protein 1 family members to bind CD36 analysed across a
parasite genome.
Mol Microbiol
47
1265
78
47. JoergensenLBengtssonDCBengtssonARonanderEBergerSS
2010
Surface co-expression of two different PfEMP1 antigens on single
Plasmodium falciparum-infected erythrocytes facilitates binding to ICAM1 and
PECAM1.
PLoS Pathog
6
e1001083
48. GamainBGratepancheSMillerLHBaruchDI
2002
Molecular basis for the dichotomy in Plasmodium falciparum
adhesion to CD36 and chondroitin sulfate A.
Proc Natl Acad Sci U S A
99
10020
10024
49. DahlbackMNielsenMASalantiA
2010
Can any lessons be learned from the ambiguous glycan binding of
PfEMP1 domains?
Trends Parasitol
26
230
235
50. BourkePFHoltDCSutherlandCJKempDJ
1996
Disruption of a novel open reading frame of Plasmodium falciparum
chromosome 9 by subtelomeric and internal deletions can lead to loss or
maintenance of cytoadherence.
Mol Biochem Parasitol
82
25
36
51. UdeinyaIJGravesPMCarterRAikawaMMillerLH
1983
Plasmodium falciparum: effect of time in continuous culture on
binding to human endothelial cells and amelanotic melanoma
cells.
Exp Parasitol
56
207
214
52. HorrocksPPinchesRChristodoulouZKyesSANewboldCI
2004
Variable var transition rates underlie antigenic variation in
malaria.
Proc Natl Acad Sci U S A
101
11129
34
53. Vigan-WomasIGuillotteMJuilleratAVallieresCLewit-BentleyA
2011
Allelic diversity of the Plasmodium falciparum erythrocyte
membrane protein 1 entails variant-specific red cell surface
epitopes.
PLoS One
6
e16544
54. CrabbBSCookeBMReederJCWallerRFCaruanaSR
1997
Targeted gene disruption shows that knobs enable malaria-infected
red cells to cytoadhere under physiological shear stress.
Cell
89
287
296
55. RugMPrescottSWFernandezKMCookeBMCowmanAF
2006
The role of KAHRP domains in knob formation and cytoadherence of
P falciparum-infected human erythrocytes.
Blood
108
370
378
56. McCormickCJNewboldCIBerendtAR
2000
Sulfated glycoconjugates enhance CD36-dependent adhesion of
Plasmodium falciparum-infected erythrocytes to human microvascular
endothelial cells.
Blood
96
327
333
57. GardnerJPPinchesRARobertsDJNewboldCI
1996
Variant antigens and endothelial receptor adhesion in Plasmodium
falciparum.
Proc Natl Acad Sci U S A
93
3503
8
58. SmithJDCraigAGKriekNHudson-TaylorDKyesS
2000
Identification of a Plasmodium falciparum intercellular adhesion
molecule-1 binding domain: a parasite adhesion trait implicated in cerebral
malaria.
Proc Natl Acad Sci U S A
97
1766
71
59. ChattopadhyayRTanejaTChakrabartiKPillaiCRChitnisCE
2004
Molecular analysis of the cytoadherence phenotype of a Plasmodium
falciparum field isolate that binds intercellular adhesion
molecule-1.
Mol Biochem Parasitol
133
255
65
60. DuffyMFMaierAGByrneTJMartyAJElliottSR
2006
VAR2CSA is the principal ligand for chondroitin sulfate A in two
allogeneic isolates of Plasmodium falciparum.
Mol Biochem Parasitol
148
117
124
61. ViebigNKGamainBScheidigCLepolardCPrzyborskiJ
2005
A single member of the Plasmodium falciparum var multigene family
determines cytoadhesion to the placental receptor chondroitin sulphate
A.
EMBO Rep
6
775
781
62. SmithJDDeitschKW
2004
Pregnancy-associated malaria and the prospects for
syndrome-specific antimalaria vaccines.
J Exp Med
200
1093
7
63. MagistradoPSalantiATuikue NdamNGMwakalingaSBResendeM
2008
VAR2CSA expression on the surface of placenta-derived Plasmodium
falciparum-infected erythrocytes.
J Infect Dis
198
1071
1074
64. PetersJMFowlerEVKrauseDRChengQGattonML
2007
Differential Changes in Plasmodium falciparum var Transcription
during Adaptation to Culture.
The J Infect Dis
195
748
755
65. FrankMDzikowskiRAmulicBDeitschK
2007
Variable switching rates of malaria virulence genes are
associated with chromosomal position.
Mol Microbiol
64
1486
1498
66. BaruchDIMaXCSinghHBBiXPasloskeBL
1997
Identification of a region of PfEMP1 that mediates adherence of
Plasmodium falciparum infected erythrocytes to CD36: conserved function with
variant sequence.
Blood
90
3766
75
67. CrandallILandKMShermanIW
1994
Plasmodium falciparum: pfalhesin and CD36 form an
adhesin/receptor pair that is responsible for the pH-dependent portion of
cytoadherence/sequestration.
Exp Parasitol
78
203
9
68. OckenhouseCFKlotzFWTandonNNJamiesonGA
1991
Sequestrin, a CD36 recognition protein on Plasmodium falciparum
malaria - infected erythrocytes identified by anti-idiotype
antibodies.
Proc Natl Acad Sci U S A
88
3175
9
69. HandunnettiSMvan SchravendijkMRHaslerTBarnwellJWGreenwaltDEHowardRJ
1992
Involvement of CD36 on erythrocytes as a rosetting receptor for
Plasmodium falciparum-infected erythrocytes.
Blood
80
2097
104
70. MagistradoPAStaalsoeTTheanderTGHviidLJensenAT
2008
CD36 selection of 3D7 Plasmodium falciparum associated with
severe childhood malaria results in reduced VAR4 expression.
Malar J
7
204
71. HBehrCMercereau-PuijalonOMichelJ
2000
Plasmodium falciparum in the squirrel monkey (Saimiri sciureus):
infection of non-splenectomised animals as a model for exploring clinical
manifestations of malaria.
Microbes Infect
2
945
954
72. KaulDKRothEFJrNagelRLHowardRJHandunnettiSM
1991
Rosetting of Plasmodium falciparum-infected red blood cells with
uninfected red blood cells enhances microvascular obstruction under flow
conditions.
Blood
78
812
819
73. SpringerALSmithLMMackayDQNelsonSOSmithJD
2004
Functional interdependence of the DBLbeta domain and c2 region
for binding of the Plasmodium falciparum variant antigen to
ICAM-1.
Mol Biochem Parasitol
137
55
64
74. UrbanBCFergusonDJPainAWillcoxNPlebanskiM
1999
Plasmodium falciparum-infected erythrocytes modulate the
maturation of dendritic cells.
Nature
400
73
7
75. UrbanBCRobertsDJ
2002
Malaria, monocytes, macrophages and myeloid dendritic cells:
sticking of infected erythrocytes switches off host cells.
Curr Opin Immunol
14
458
465
76. MelcherMMuhleRAHenrichPPKraemerSMAvrilM
2010
Identification of a role for the PfEMP1 semi-conserved head
structure in protein trafficking to the surface of Plasmodium falciparum
infected red blood cells.
Cell Microbiol
12
1446
62
77. PatelSNLuZAyiKSerghidesLGowdaDC
2007
Disruption of CD36 impairs cytokine response to Plasmodium
falciparum glycosylphosphatidylinositol and confers susceptibility to severe
and fatal malaria in vivo.
J Immunol
178
3954
3961
78. SerghidesLSmithTGPatelSNKainKC
2003
CD36 and malaria: friends or foes?
Trends Parasitol
19
461
469
79. KalmbachYRottmannMKombilaMKremsnerPGBeckHP
2010
Differential var gene expression in children with malaria and
antidromic effects on host gene expression.
J Infect Dis
202
313
317
80. KirchgatterKPortillo HdelA
2002
Association of severe noncerebral Plasmodium falciparum malaria
in Brazil with expressed PfEMP1 DBL1 alpha sequences lacking cysteine
residues.
Mol Med
8
16
23
81. RottmannMLavstsenTMugasaJPKaestliMJensenATR
2006
Differential Expression of var Gene Groups Is Associated with
Morbidity Caused by Plasmodium falciparum Infection in Tanzanian
Children.
Infect Immun
74
3904
3911
82. Vigan-WomasIGuillotteMLeSCIgonetSPetresS
2008
An in vivo and in vitro model of Plasmodium falciparum rosetting
and autoagglutination mediated by varO, a group A var gene encoding a
frequent serotype.
Infect Immun
76
5565
5580
83. RoweJAMouldsJMNewboldCIMillerLH
1997
P. falciparum rosetting mediated by a parasite-variant
erythrocyte membrane protein and complement-receptor 1.
Nature
388
292
5
84. ReederJCCowmanAFDavernKMBeesonJGThompsonJK
1999
The adhesion of Plasmodium falciparum-infected erythrocytes to
chondroitin sulfate A is mediated by P. falciparum erythrocyte membrane
protein 1.
Proc Natl Acad Sci U S A
96
5198
202
85. BuffetPAGamainBScheidigCBaruchDSmithJD
1999
Plasmodium falciparum domain mediating adhesion to chondroitin
sulfate A: a receptor for human placental infection.
Proc Natl Acad Sci U S A
96
12743
8
86. ViebigNKLevinEDechavanneSRogersonSJGysinJ
2007
Disruption of var2csa gene impairs placental malaria associated
adhesion phenotype.
PLoS One
2
e910
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek Distribution of the Phenotypic Effects of Random Homologous Recombination between Two Virus SpeciesČlánek SIV Nef Proteins Recruit the AP-2 Complex to Antagonize Tetherin and Facilitate Virion ReleaseČlánek Dual Function of the NK Cell Receptor 2B4 (CD244) in the Regulation of HCV-Specific CD8+ T CellsČlánek A Large and Intact Viral Particle Penetrates the Endoplasmic Reticulum Membrane to Reach the CytosolČlánek Interleukin-13 Promotes Susceptibility to Chlamydial Infection of the Respiratory and Genital Tracts
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2011 Číslo 5- Jak souvisí postcovidový syndrom s poškozením mozku?
- Měli bychom postcovidový syndrom léčit antidepresivy?
- Farmakovigilanční studie perorálních antivirotik indikovaných v léčbě COVID-19
- 10 bodů k očkování proti COVID-19: stanovisko České společnosti alergologie a klinické imunologie ČLS JEP
-
Všechny články tohoto čísla
- Lymphoadenopathy during Lyme Borreliosis Is Caused by Spirochete Migration-Induced Specific B Cell Activation
- Infections Are Virulent and Inhibit the Human Malaria Parasite in
- A Gamma Interferon Independent Mechanism of CD4 T Cell Mediated Control of Infection
- MDA5 and TLR3 Initiate Pro-Inflammatory Signaling Pathways Leading to Rhinovirus-Induced Airways Inflammation and Hyperresponsiveness
- The OXI1 Kinase Pathway Mediates -Induced Growth Promotion in Arabidopsis
- An E2F1-Mediated DNA Damage Response Contributes to the Replication of Human Cytomegalovirus
- Quantitative Subcellular Proteome and Secretome Profiling of Influenza A Virus-Infected Human Primary Macrophages
- Distribution of the Phenotypic Effects of Random Homologous Recombination between Two Virus Species
- Inhibition of Both HIV-1 Reverse Transcription and Gene Expression by a Cyclic Peptide that Binds the Tat-Transactivating Response Element (TAR) RNA
- A Viral Satellite RNA Induces Yellow Symptoms on Tobacco by Targeting a Gene Involved in Chlorophyll Biosynthesis using the RNA Silencing Machinery
- Misregulation of Underlies the Developmental Abnormalities Caused by Three Distinct Viral Silencing Suppressors in Arabidopsis
- Investigating the Host Binding Signature on the PfEMP1 Protein Family
- Human Neutrophil Clearance of Bacterial Pathogens Triggers Anti-Microbial γδ T Cell Responses in Early Infection
- Septation of Infectious Hyphae Is Critical for Appressoria Formation and Virulence in the Smut Fungus
- A Family of Helminth Molecules that Modulate Innate Cell Responses via Molecular Mimicry of Host Antimicrobial Peptides
- Phospholipids Trigger Capsular Enlargement during Interactions with Amoebae and Macrophages
- CTL Escape Mediated by Proteasomal Destruction of an HIV-1 Cryptic Epitope
- Evolution of Th2 Immunity: A Rapid Repair Response to Tissue Destructive Pathogens
- Extensive Genome-Wide Variability of Human Cytomegalovirus in Congenitally Infected Infants
- The Antiviral Efficacy of HIV-Specific CD8 T-Cells to a Conserved Epitope Is Heavily Dependent on the Infecting HIV-1 Isolate
- Epstein-Barr Virus Infection of Polarized Epithelial Cells via the Basolateral Surface by Memory B Cell-Mediated Transfer Infection
- Reactive Oxygen Species Hydrogen Peroxide Mediates Kaposi's Sarcoma-Associated Herpesvirus Reactivation from Latency
- Crystal Structure and Functional Analysis of the SARS-Coronavirus RNA Cap 2′-O-Methyltransferase nsp10/nsp16 Complex
- The Dot/Icm System Delivers a Unique Repertoire of Type IV Effectors into Host Cells and Is Required for Intracellular Replication
- AAV Exploits Subcellular Stress Associated with Inflammation, Endoplasmic Reticulum Expansion, and Misfolded Proteins in Models of Cystic Fibrosis
- Suboptimal Activation of Antigen-Specific CD4 Effector Cells Enables Persistence of In Vivo
- SIV Nef Proteins Recruit the AP-2 Complex to Antagonize Tetherin and Facilitate Virion Release
- Dual Function of the NK Cell Receptor 2B4 (CD244) in the Regulation of HCV-Specific CD8+ T Cells
- Transition of Sporozoites into Liver Stage-Like Forms Is Regulated by the RNA Binding Protein Pumilio
- A Large and Intact Viral Particle Penetrates the Endoplasmic Reticulum Membrane to Reach the Cytosol
- Transcriptome Analysis of in Human Whole Blood and Mutagenesis Studies Identify Virulence Factors Involved in Blood Survival
- Interleukin-13 Promotes Susceptibility to Chlamydial Infection of the Respiratory and Genital Tracts
- Structural Insights into Viral Determinants of Nematode Mediated Transmission
- Protective Efficacy of Serially Up-Ranked Subdominant CD8 T Cell Epitopes against Virus Challenges
- Viral CTL Escape Mutants Are Generated in Lymph Nodes and Subsequently Become Fixed in Plasma and Rectal Mucosa during Acute SIV Infection of Macaques
- Taking Some of the Mystery out of Host∶Virus Interactions
- Viral Small Interfering RNAs Target Host Genes to Mediate Disease Symptoms in Plants
- : An Emerging Cause of Sexually Transmitted Disease in Women
- Mitochondrial Ubiquitin Ligase MARCH5 Promotes TLR7 Signaling by Attenuating TANK Action
- The Hexamer Structure of the Rift Valley Fever Virus Nucleoprotein Suggests a Mechanism for its Assembly into Ribonucleoprotein Complexes
- Acquisition of Human-Type Receptor Binding Specificity by New H5N1 Influenza Virus Sublineages during Their Emergence in Birds in Egypt
- Stromal Down-Regulation of Macrophage CD4/CCR5 Expression and NF-κB Activation Mediates HIV-1 Non-Permissiveness in Intestinal Macrophages
- A Component of the Xanthomonadaceae Type IV Secretion System Combines a VirB7 Motif with a N0 Domain Found in Outer Membrane Transport Proteins
- Perturbs Iron Trafficking in the Epithelium to Grow on the Cell Surface
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Crystal Structure and Functional Analysis of the SARS-Coronavirus RNA Cap 2′-O-Methyltransferase nsp10/nsp16 Complex
- Lymphoadenopathy during Lyme Borreliosis Is Caused by Spirochete Migration-Induced Specific B Cell Activation
- The OXI1 Kinase Pathway Mediates -Induced Growth Promotion in Arabidopsis
- : An Emerging Cause of Sexually Transmitted Disease in Women
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání