-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaStructural Analysis of HIV-1 Maturation Using Cryo-Electron Tomography
HIV-1 buds form infected cells in an immature, non-infectious form. Maturation into an infectious virion requires proteolytic cleavage of the Gag polyprotein at five positions, leading to a dramatic change in virus morphology. Immature virions contain an incomplete spherical shell where Gag is arranged with the N-terminal MA domain adjacent to the membrane, the CA domain adopting a hexameric lattice below the membrane, and beneath this, the NC domain and viral RNA forming a disordered layer. After maturation, NC and RNA are condensed within the particle surrounded by a conical CA core. Little is known about the sequence of structural changes that take place during maturation, however. Here we have used cryo-electron tomography and subtomogram averaging to resolve the structure of the Gag lattice in a panel of viruses containing point mutations abolishing cleavage at individual or multiple Gag cleavage sites. These studies describe the structural intermediates correlating with the ordered processing events that occur during the HIV-1 maturation process. After the first cleavage between SP1 and NC, the condensed NC-RNA may retain a link to the remaining Gag lattice. Initiation of disassembly of the immature Gag lattice requires cleavage to occur on both sides of CA-SP1, while assembly of the mature core also requires cleavage of SP1 from CA.
Published in the journal: . PLoS Pathog 6(11): e32767. doi:10.1371/journal.ppat.1001215
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1001215Summary
HIV-1 buds form infected cells in an immature, non-infectious form. Maturation into an infectious virion requires proteolytic cleavage of the Gag polyprotein at five positions, leading to a dramatic change in virus morphology. Immature virions contain an incomplete spherical shell where Gag is arranged with the N-terminal MA domain adjacent to the membrane, the CA domain adopting a hexameric lattice below the membrane, and beneath this, the NC domain and viral RNA forming a disordered layer. After maturation, NC and RNA are condensed within the particle surrounded by a conical CA core. Little is known about the sequence of structural changes that take place during maturation, however. Here we have used cryo-electron tomography and subtomogram averaging to resolve the structure of the Gag lattice in a panel of viruses containing point mutations abolishing cleavage at individual or multiple Gag cleavage sites. These studies describe the structural intermediates correlating with the ordered processing events that occur during the HIV-1 maturation process. After the first cleavage between SP1 and NC, the condensed NC-RNA may retain a link to the remaining Gag lattice. Initiation of disassembly of the immature Gag lattice requires cleavage to occur on both sides of CA-SP1, while assembly of the mature core also requires cleavage of SP1 from CA.
Introduction
The assembly of HIV-1 occurs at the plasma membrane of infected cells. The primary structural component of the assembling virus is the 55-kDa polyprotein Gag. Multiple copies of Gag assemble to form an incomplete sphere underneath the plasma membrane, which recruits components of the cellular ESCRT machinery to mediate membrane scission and release of the budding virus from the cell [1], [2], [3]. Gag consists of three major structural components: MA, the membrane binding domain, CA, the capsid domain, and NC, the nucleocapsid domain, which interacts with the viral RNA. CA and NC are separated by a short linker peptide SP1, and downstream of NC are two further peptide domains, SP2 and p6 (Figure 1A) [4]. p6 contains the short linear motifs which are responsible for ESCRT recruitment [2], [5], [6]. The viral genome is recruited to the assembling particle from the pool of cellular mRNA via interactions between a packaging signal (Ψ) present at the 5′ end of the genome [7] and zinc-fingers present in NC [4].
Fig. 1. Steps in HIV-1 proteolytic maturation, and variants analysed. 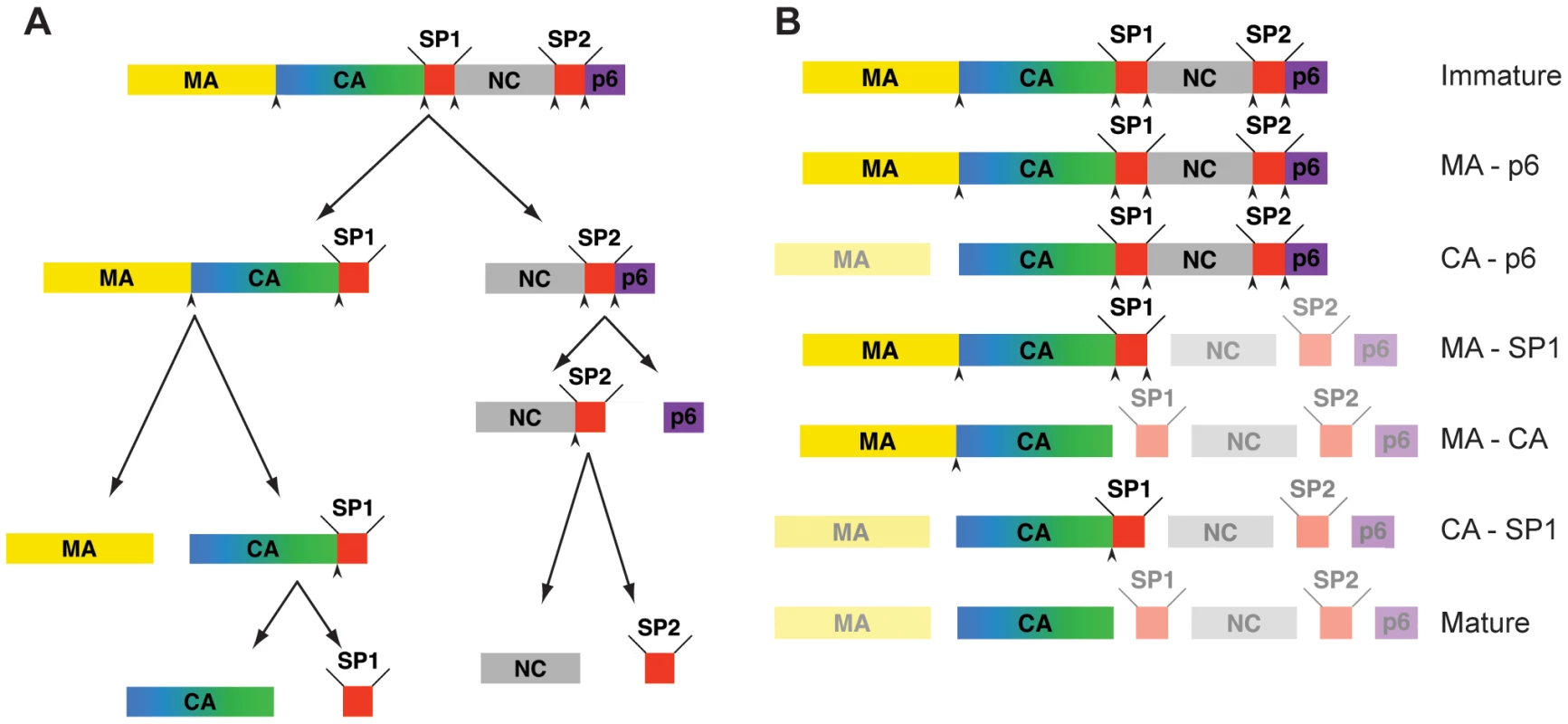
A) Schematic outline of the proteolytic cleavages which take place in Gag during the HIV-1 maturation process. Arrowheads indicate proteolytic sites before cleavage. The order of cleavage events shown is based on the rates of cleavage in vitro as described in [22]. B) Schematic representation of Gag after completion of cleavage for each variant analyzed. The non-cleaved products due to the inactivation of the proteolytic sites are highlighted. Mutated, and therefore uncleaved processing sites are indicated by an arrowhead. During or concomitant with budding, the viral protease (PR) cleaves Gag in five positions into its component domains. These cleavages lead to the dramatic morphological changes required for creation of a mature, infectious virion. Proteolytic maturation is essential for infectivity, and PR inhibitors are a key element of current antiretroviral therapies [8]. Maturation appears to be a rapid process, and viruses imaged by electron microscopy (EM) in the vicinity of infected cells are predominantly mature, with only occasional immature particles. Intermediate maturation states have not been detected so far. For this reason, the majority of morphological and structural studies have focused either on the mature virus, or on the immature virus that can be produced in the presence of a PR inhibitor or by mutation of the PR active site.
In the immature virus, Gag is arranged in a radial fashion, with the N-terminal MA domain associated with the viral membrane, and the C-terminus of the protein pointed towards the centre of the particle. Gag adopts a curved hexameric lattice with an 8 nm unit cell [9], which closes through the incorporation of heterogeneously shaped defects [10]. After proteolytic cleavage, the MA layer is thought to remain associated with the viral membrane, whereas NC and the RNA are condensed into a ribonucleoprotein complex layer surrounded by the viral CA capsid core. The core is most often cone-shaped, though other shapes including capped tubes are seen. The core is thought to have a fullerene cone geometry formed from a hexameric lattice with twelve pentamers incorporated to allow closure [11], five at the narrow end of the cone and seven at the broad end of the cone shaped structure. CA adopts a larger 10 nm unit cell in the mature core [12], [13].
The structure of the immature lattice has been explored by cryo-electron tomography combined with sub-tomogram averaging [10], [14]. The N-terminal CA domain forms hexameric rings surrounding central holes. Below these rings, 2-fold symmetric densities link adjacent hexamers. These densities have been assigned to the dimeric C-terminal CA domain. Six such dimers converge below the holes in the N-terminal CA domain into a rod-like structure suggested to represent a six-helix bundle formed by SP1 [14]. This rod-like structure descends towards the centre of the virus into the NC-RNA layer, which is not hexagonally ordered [10].
Studies of the arrangement of CA in the mature core are much more advanced through the use of in vitro assembly of purified proteins to form regular arrays appropriate for high-resolution structural study [15], [16]. In the mature core, the N-terminal residues of CA are folded back into the structure to form a β-hairpin [17]. The formation of this structure appears to be conserved in all retroviruses with the exception of the spumaviruses [17], [18], [19], [20]. Mutagenesis studies suggest that the formation of the β-hairpin is important for the formation of the mature core [17], [21]. This structure cannot be present in the immature virus since the N-terminal residues are covalently linked to the MA domain.
The five proteolytic cleavage sites in Gag are cleaved at very different rates in vitro. The fastest cleavage is that between SP1 and NC. Relative to this cleavage, SP2-p6 is cleaved 9x slower, MA-CA 14x slower, NC-SP2 350x slower, and CA-SP1 400x slower [22]. These differences, together with the observation of distinct and reproducible processing intermediates in lysates from infected cells or upon partial inhibition of PR [23], suggest that cleavage is a stepwise process in vivo. The order of cleavage based on these rates determined in vitro is illustrated in Figure 1A with initial processing separating NC-p6 from the membrane-bound N-terminal part of Gag, secondary cleavages separating MA from CA-SP1 and p6 from NC-SP2 and final processing removing the two spacer peptides from the C-termini of CA and NC, respectively. Mutational studies revealed that the efficacy and order of these cleavage events is highly relevant for viral infectivity: blocking cleavage at any individual processing site - with the exception of NC-SP2 - severely diminished or abolished virion infectivity [24], [25], [26], [27], mostly affecting fusion or early post-fusion events [24], [25], [28].
Due to the fast time scale of maturation and the absence of any system for synchronizing the process, studies of maturation intermediates have relied upon viruses in which individual cleavage sites have been mutated [22], [27], [29], [30]. These studies showed that mutation of the cleavage site between MA and CA led to normal condensation of the NC-RNA complex without formation of the cone shaped capsid [27], [29]. Inhibition of both cleavages flanking SP1 led to viruses with immature-like morphology, whereas inhibition of the CA-SP1 cleavage allowed NC-RNA condensation, but prevented formation of a mature-like core [27]. A thin electron-dense layer separated from the virion membrane was observed in this case and suggested to correspond to CA. A similar phenotype was observed when virus was produced in the presence of bevirimat [31], which specifically inhibits cleavage between CA and SP1. Although the inhibition of CA-SP1 processing by bevirimat is incomplete [31], [32], the infectivity of virions raised in its presence is severely diminished. Similarly, partial inhibition of polyprotein processing by co-expression and co-assembly of wild-type (wt) and cleavage-site mutated Gag polyprotein showed a strong trans-dominant effect of the mutated proteins, which appeared strongest for mutations affecting the CA-NC border [25], [26], [33]. Mutations spanning the CA-SP1 regions also had a trans-dominant negative effect on processing of the respective wt protein when co-assembled into particles, suggesting that the CA-SP1 region forms a stable multimeric structure [26].
Here we set out to elucidate and describe the stepwise transitions occurring during HIV-1 maturation on a structural level. By doing so we also aimed to shed light on the structure and integrity of the Gag lattice by understanding structural changes associated with specific cleavage events. To do this we have applied cryo-electron tomography and sub-tomogram averaging to describe the morphology and structure of an extended pool of cleavage site mutants.
Results
Virus production and cleavage efficiency
We analyzed a panel of viruses derived from HIV-1 NL43, which carry point mutations at single or multiple of the cleavage sites in Gag [24], [27], [28] to prevent proteolytic cleavage at the targeted sites. In parallel we transfected a wt proviral plasmid in the presence or absence of the PR inhibitor Lopinavir to generate immature and mature viruses, respectively. The constructs used are shown in Figure 1B. Based on the scheme shown in Figure 1A [22], MA-p6 (together with PR inhibited virus), MA-SP1, CA-SP1 (CA5 in [27]) and the wt virus represent true maturation products. The other constructs CA-p6, and MA-CA were selected to shed light on the importance of individual cleavage sites upstream and downstream of CA.
HIV-1 wt and all variant viruses were produced by transfection of 293T cells, and purified by gradient centrifugation as described previously [10]. We assessed the degree of proteolytic processing via SDS PAGE followed by silver staining or immunoblotting (Figure 2A and data not shown). In most cases all predicted cleavage products were seen, indicating that proteolytic cleavage at non-mutated sites proceeded to completion, while mutated sites were blocked. The exception was MA-CA carrying a single mutation at the cleavage site between MA and CA, but exhibiting strongly reduced cleavage efficiency at the CA-SP1 boundary of only 20%–50% depending on the virus preparation. Failure to cleave at MA-CA therefore leads to a reduction in the efficiency of cleavage at the CA-SP1 boundary.
Fig. 2. Characterisation of HIV-1 variants. 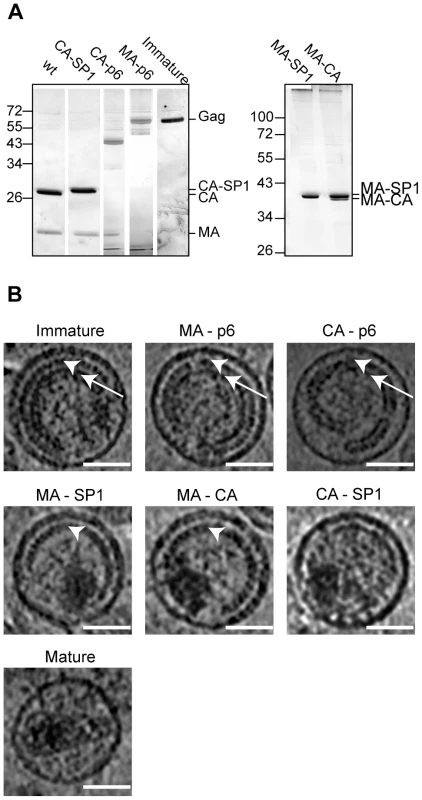
A) Partially processed Gag-derived products detected by SDS-PAGE. Iodixanol gradient purified particle preparations of the indicated HIV-1 variants were separated on 12.5% SDS gels. Virion-associated proteins were visualized by silver staining according to standard procedures, showing MA and CA containing Gag derivatives as the most prominent bands. Lanes in the left panel were combined from two independent gels and were size adjusted for comparison. The right panel illustrates incomplete processing at the CA-SP1 boundary for variant MA-CA. Positions of molecular mass standards (in kDa) are indicated to the left. B) Central sections of 0.8 nm thickness from 6 µm defocus tomographic reconstructions of the HIV-1 variants analyzed. White arrowheads point to the immature CA layer, whereas white arrows point to the NC-RNA layer. Immature, MA-p6 and CA-p6 variants show both CA and NC-RNA layers. In CA-p6 the distance between the CA layer and the membrane is more variable. In MA-SP1 and MA-CA, the CA layer is seen, but the NC-RNA has condensed. CA-SP1 and the mature virus do not show the characteristic striated CA layer. The scale bar is 50 nm. 3D morphology of the viruses
In order to characterize the effect of cleavage site mutations on structure and morphology of the viruses, purified viruses were plunge-frozen in liquid ethane and imaged in the electron microscope. To eliminate morphological variability resulting from transfection or purification, a minimum of 3 independent preparations of each virus were imaged. With the exception of the CA-SP1 construct (see below), independent preparations of the same variant showed no significant differences in morphology. In order to obtain three-dimensional structural information, cryo-electron tomograms were acquired. A minimum of 46 virus particles were reconstructed in 3D for each variant, in each case distributed across all 3 preparations.
Figure 2B shows central sections through representative tomograms of viruses from each of the variants (see also Figure S1). Both the PR inactivated wt virus and the MA-p6 variant show the characteristic morphology, which has previously been described for immature HIV imaged by cryo-electron microscopy and cryo-electron tomography [10], [14], [34]. These virus particles are approximately spherical and display an incomplete protein lattice underneath the viral membrane. The membrane appears thicker in regions where Gag is present, due to the presence of the membrane associated MA domain in these regions [10]. The protein lattice shows a striated pattern in the outer ring of density, previously assigned to the CA domain (Figure 2B, arrowheads), and a smoother inner ring of density, previously assigned to the NC-RNA complex (Figure 2B, arrows) [34]. The similarity of the PR-inhibited virus and the MA-p6 variant indicates that there are no substantial morphological differences between a virus which has an active PR, but mutations in all proteolytic cleavage sites in Gag, and a virus with intact cleavage sites but an inhibited PR.
The characteristic two layers of density corresponding to the CA and NC-RNA domains were also seen in the CA-p6 virus; however, the distance between these layers and the membrane was significantly more variable than in the immature virus (compare CA-p6 with immature in Figure 2B), leading to greater variability in the curvature of the layer.
MA-SP1 and MA-CA showed a thinner Gag layer. The striated CA layer of density was present, but the inner NC-RNA layer of density that is seen in the immature virus was missing. Cleavage between CA and NC therefore leads to loss of the NC-RNA layer. Instead we observed a globular dense structure within the virus that is likely to represent condensed NC-RNA. The volume of the condensed NC-RNA was measured by visual assessment of the boundaries of the condensed region. In MA-SP1 it was 4.4±1.1 * 104 nm3 (n = 46), and in MA-CA was 4.3±1.2 * 104 nm3 (n = 46), corresponding to a ratio between the volume of the virus particle and the volume of the condensed NC-RNA of 26.9±8.5 for MA-SP1 and 28.0±8.2 for MA-CA and thus indicating no detectable difference in the degree of condensation.
The CA-SP1 viruses presented high variability in the morphology depending on the preparation. Of the 5 independent preparations, 2 showed no internal structures, whereas the other 3 showed features, and were further analysed. In contrast to what was observed for the other mutated variants, the striated CA layer was not present in any of the particles, while a discontinuous and thin layer was visible in ∼45% of the viruses (29 of 64 particles). The NC-RNA was condensed to the same degree as in MA-SP1 and MA-CA (volume 4.3±1.0 * 104 nm3 (n = 46), ratio of virus volume to NC-RNA volume 26.4±7.2). The membrane appears thinner, similar to the regions of the immature virus where Gag is absent.
The mature virus showed the same morphology as previously described by tomography of equivalent preparations [35]. Virus particles contained predominantly cone shaped cores consisting of a thin CA lattice containing density which we attribute to the NC-RNA. The NC-RNA volume was 3.5±0.8 * 104 nm3 (n = 46), corresponding to a ratio between the volume of the virus particle and the volume of the condensed NC-RNA of 35.0±8.9. The slightly lower NC-RNA volume in the mature virus when compared to MA-SP1, MA-CA and CA-SP1 probably reflects the better definition of the NC-RNA edges when confined within the core, rather than a change in the degree of condensation. The membrane thickness appears similar to that in the CA-SP1 viruses.
Global arrangement of the Gag lattice
To describe the changes in the global arrangement of the Gag lattices in greater detail, we applied subtomogram averaging to the cryo-electron tomography data for those variants which showed a striated lattice (the thinner CA layer in CA-SP1 and wt proved insufficiently featured for subtomogram averaging). By plotting in 3D the position of the subtomograms after the alignments, we were able to define the position and the orientation of the centre of each identified hexagon of the lattice. All the lattice maps showed the same general arrangement of a continuous hexameric lattice with irregularly shaped defects, partially covering the inner surface of the viral membrane, but leaving one large gap (Figure 3). There was no substantial difference in the degree of shell completeness between the different variants.
Fig. 3. The global arrangement of the Gag layer. 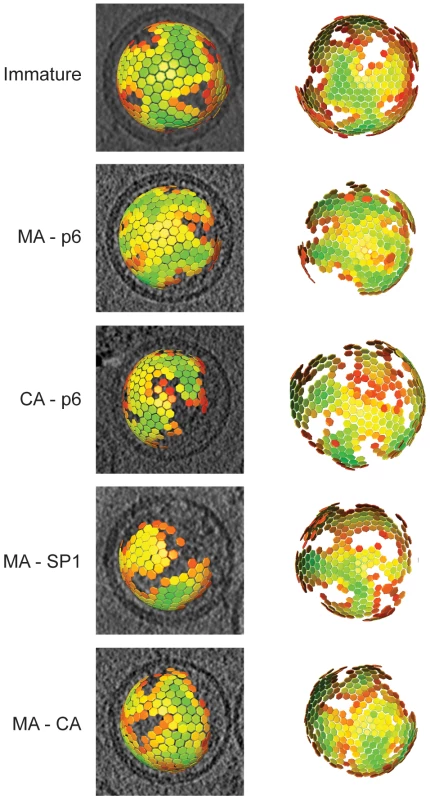
Global lattice maps for each variant superimposed on central sections of the tomographic reconstruction of the virus (left panels), or viewed from the direction of the largest gap in the lattice (right panels). The centres of each hexameric unit cell are marked with hexamers, which are coloured according to cross correlation on a scale from low (red) to high (green). Higher cross correlation values indicate that the subtomogram is more similar to the average structure. The cross-correlation range in each map has been set between the minimum and the maximum cross-correlation value present in the map. Maps are shown in perspective such that hexamers on the rear surface of the particle appear smaller. Structure of the Gag lattice by sub-tomogram averaging
To describe the changes in the Gag lattice structures during the maturation process in greater detail, we analysed the average structures generated during subtomogram averaging. These represent a 3D reconstruction of the local structure of the Gag lattice in each variant.
The structure of the Gag lattice in the PR inhibited virus (Figure 4) was consistent with that previously published for immature HIV using these methods [10]. The lattice had a six-fold symmetric structure with an inter-hexagon distance of 8 nm measured at the C-terminal CA domain. The outermost density, previously assigned to the N-terminal domain of CA, formed hexameric rings around large holes. The C-terminal domain of CA sat at a lower radius at the 2-fold symmetrical position between the hexameric rings. These rings were linked at the 6-fold positions, beneath the holes in the N-terminal CA layer, where they joined a rod-like density, previously proposed to represent SP1 descending into the NC-RNA layer, which was not hexagonally ordered. At this resolution, the structure of MA–p6 was indistinguishable from the PR-inhibited wt virus.
Fig. 4. The local structure of the Gag layer. 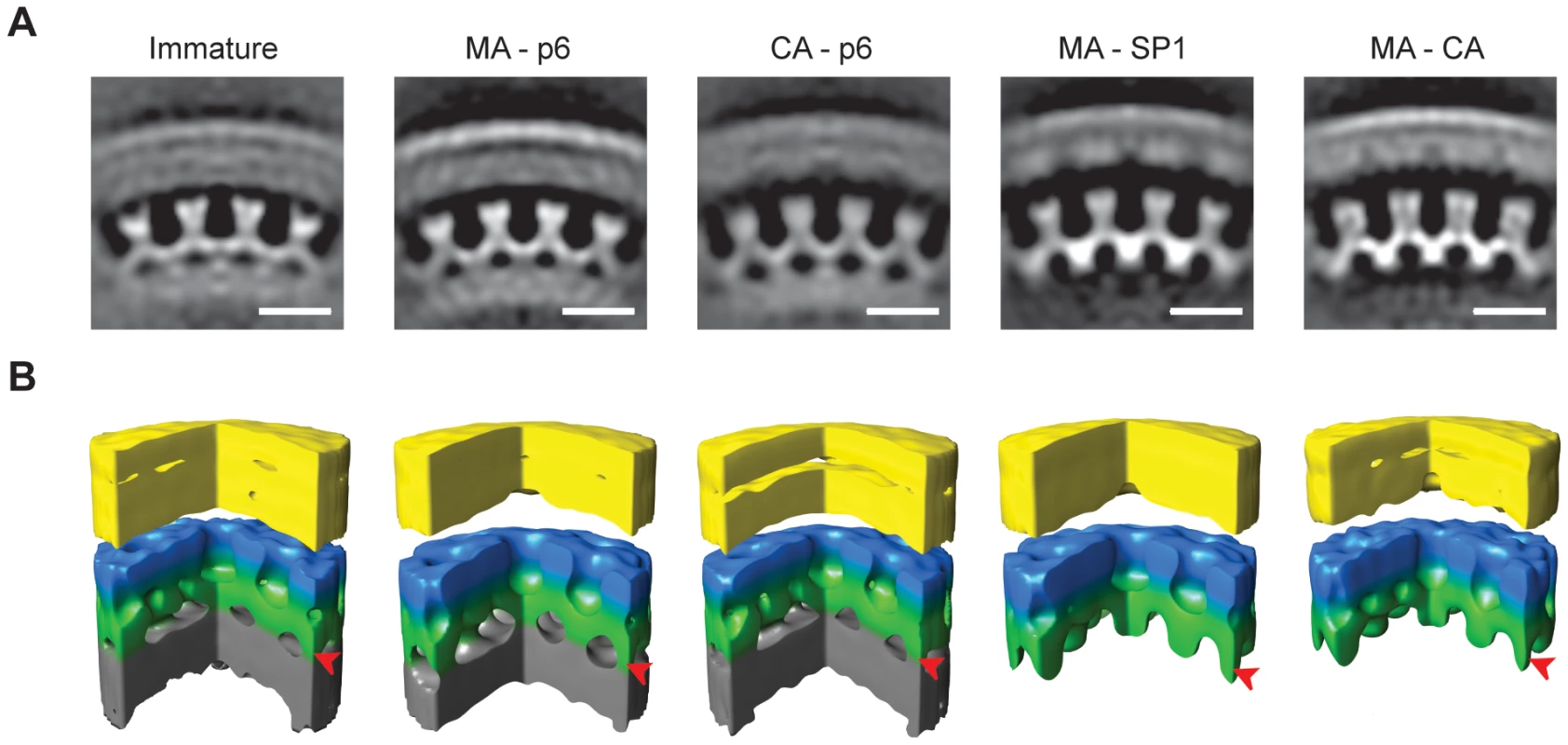
A) Radial sections from the subtomogram average reconstructions coming from each variant. Density is white. The scale bar is 10 nm. B) Surface rendering of the subtomogram average reconstructions. A sector of 90° has been cut out to better show the organization of the lattice. The colours on the surfaces are for illustrative purposes. The membrane and the MA layer, which cannot be delineated at this resolution, are yellow. N-terminal CA domain is blue. C-terminal CA domain and SP1 spacer are green. NC-RNA layer is grey. In MA-SP1 and MA-CA the grey NC-RNA layer is missing because condensation has already taken place, and rod-like protrusions are seen descending from the C-terminal CA domain towards the centre of the virus particle. Arrowheads mark the radial position of the rod-like densities. The structure of the CA–p6 lattice showed that the outer hexameric rings corresponding to N-CA, the 2-fold densities corresponding to C-CA and the rod-like densities were all unchanged. The inter-hexagon distance remained 8 nm. These observations imply that the CA, SP1 and NC-RNA layers in the CA-p6 variant adopt a structure that is similar to that in the immature virus.
The N-CA and C-CA features of the MA-SP1 structure were similar to the PR-inhibited virus. The inter-hexagon distance remained 8 nm. The rod-like densities that descend from the CA layer towards the NC-RNA could also still be seen in this structure. Below the rod-like densities, the reconstruction of the lattice of the MA-SP1 variant lacked the inner disordered layer that in the immature virus has been assigned to the NC-RNA. The structure of the lattice in MA-CA was identical to the one in MA-SP1, despite the partial cleavage of the SP1 region.
A link between partially cleaved Gag and the NC-RNA
All preparations contained some partially broken virus particles. Condensed NC-RNA, and sections of Gag lattice, which were not part of intact virus particles, could be visualised in the tomograms. We noticed that sections of MA-SP1 lattice from broken particles were commonly associated with condensed NC-RNA (data not shown), suggesting the possibility of an interaction between the condensed NC-RNA and the MA-SP1 lattice.
To further explore the possibility of a direct link between the condensed NC-RNA and the remaining Gag lattice, we gently disrupted MA-SP1 and MA-CA virus particles by vortexing for 15 s in the presence of 10 nm gold beads. Cryo-electron micrographs of these preparations showed large numbers of virus particles with disrupted membranes. All of the broken particles had condensed densities associated with the remaining MA-SP1 lattice (Figure 5). In some cases the presence of neighbouring particles or the orientation made interpretation difficult, but in most of the cases, the size and shape of the density suggested that it represented the condensed NC-RNA.
Fig. 5. Analysis of disrupted virus particles. 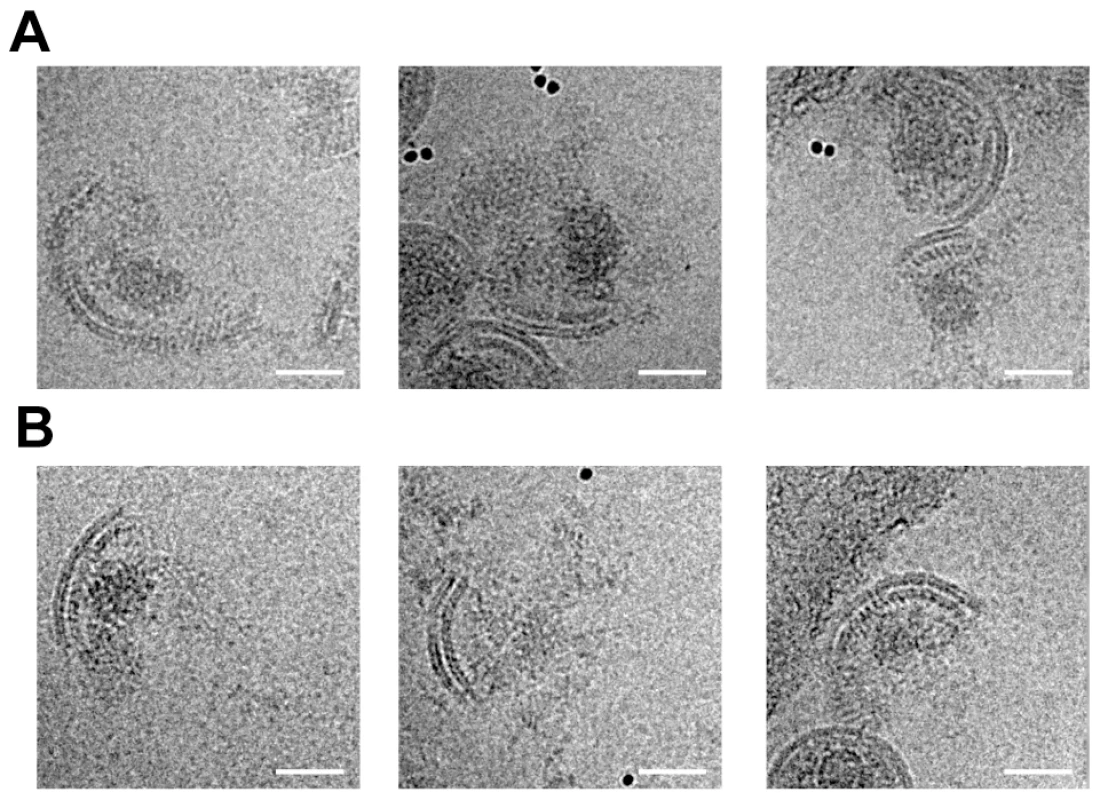
Micrographs of MA – SP1 (A) and MA – CA (B) virus particles after disruption by vortexing in the presence of gold beads show patches of viral membrane with underlying CA lattice. The condensed NC – RNA is visible as an irregular, globular density similar to that seen within intact virus particles (compare with figure 2B). Rather than diffusing away from the broken particles, the NC-RNA appears to remain associated with the CA lattice. The scale bar is 50 nm. Discussion
Here we have studied HIV-1 maturation by using cryo-electron tomography of a panel of viruses with mutations in proteolytic cleavage sites to describe the structures of potential maturation intermediates at macromolecular resolution. Analysis of these viruses reveals how individual cleavage events influence the structure and disassembly of the immature lattice as well as the formation of the mature lattice.
MA-p6 viruses lacking functional cleavage sites in Gag, but exhibiting wt PR activity and regular processing of Pol, present a lattice structure that is indistinguishable from immature HIV-1 produced in the presence of a PR inhibitor. The completeness of the Gag shell and the size of the viruses is also the same as for protease inhibited wt viruses. This suggests that (i) processing of the Pol domain does not detectably influence the lattice structure if Gag remains uncleaved, (ii) the alteration of cleavage sites did not have a significant effect on the structure and assembly of the Gag layer. Further analysis of variants with mutated cleavage sites revealed that processing at both sides of CA-SP1 is needed for dissolution of the immature lattice, while release of SP1 is needed, in addition, for formation of the regular mature core.
MA-SP1 forms a structure which is stable in the absence of NC
The structure of the MA-SP1 variant shows that cleavage between SP1 and NC leads to removal of the NC-RNA layer from Gag, and to condensation of the NC-RNA core. On the other hand, the structure of the MA-SP1 region of the lattice remained unchanged compared to the immature virus. NC-RNA interactions or other non-specific interaction domains downstream of SP1 have been shown to be essential for virus-like particle assembly both in vitro and in tissue culture, however [36]. The stability of the lattice in MA-SP1 indicates that these interactions concentrate and align Gag at the membrane during assembly, but are dispensable for maintaining the structure or stability of the Gag lattice once it is formed.
The same structure as is observed here for MA-CA and MA-SP1 was also seen in a subset of cellular budding sites and released particles in vivo, (see accompanying manuscript by Carlson et al). This structure can therefore also be formed by wt Gag if premature SP1-NC cleavage occurs and leads to loss of PR from the budding particle. Observing the same structure also in particles carrying wt Gag polyproteins provides further support for it representing a true maturation intermediate.
In the MA-CA construct, all cleavage sites downstream of CA remain in their wt form. Nevertheless, cleavage efficiency for the CA-SP1 site is reduced to between 20% and 50%, due to the failure of cleavage between MA and CA. These observations suggest that failure to cleave upstream of CA inhibits access of PR to the CA-SP1 cleavage site as has also been reported for mutation of the downstream SP1-NC cleavage site [37]. It has been shown that cleavage at the wt CA-SP1 site is also inhibited in a trans-dominant way if small amounts of MA-SP1 are incorporated into HIV-1 particles [26]. Our data indicate that in all of these situations there is a failure to sufficiently disassemble the immature CA lattice (see below), and that disassembly of the immature CA layer is required for efficient access of PR to the CA-SP1 cleavage site.
Both MA-CA and MA-SP1 variants showed rod-like structures descending from the CA layer towards the centre of the virus. These rod-like structures were not disrupted by cleavage between SP1 and NC, or by subsequent partial cleavage between CA and SP1. This argues that SP1-SP1 interactions together with CA-SP1 interactions are sufficient to maintain the integrity of the rod-like structures even in the presence of partial CA-SP1 cleavage. SP1 contains important assembly determinants, and this observation suggests that these direct formation of a specific structure, which does not require NC tethering for its stability.
CA-NC forms a structure which is stable in the absence of MA
In solution, cleavage between MA and CA has been demonstrated to cause rearrangement in the N-terminal domain of CA, leading to part of the MA-CA flexible-linker folding back into the N-terminal domain of CA to form a β-hairpin structure [20]. However in the CA-p6 variant, where this cleavage has occurred, the CA lattice was identical to the one present in the immature virus, at the resolution of the reconstructions. It also had the same overall completeness. This suggests that formation of the β-hairpin structure either requires downstream processing in addition, or that its formation is insufficient to cause structural changes in the immature lattice.
Although the structure between CA and p6 in the CA-p6 variant was the same as that in the immature virus, the distance between CA and the membrane became more variable, indicating the absence of a MA-CA link. The curvature of the CA layer was more variable than in the immature virus, suggesting that interactions with the membrane contribute to the uniformity of the lattice curvature.
Cleavage upstream and downstream of CA-SP1 is required for immature lattice disassembly: cleavage within CA-SP1 for mature lattice assembly
As discussed above, both MA-SP1 and CA-p6 show CA lattices with an immature structure. We can therefore conclude that cleavage either only upstream or only downstream of the CA-SP1 domain does not affect the organization of the immature lattice. The immature lattice is disassembled, however, when cleavage has occurred both upstream of CA and downstream of SP1 in the case of the CA-SP1 variant, which contains no characteristic, striated CA lattice. In thin section electron micrographs, an irregular electron-dense structure separated from the virion membrane was commonly observed for this virus [27]. Such irregular layers were occasionally seen in our analyses as well, but were too thin to represent an immature CA lattice. They are more comparable in thickness to the mature CA core, which shows a thinner CA layer than the immature virus due to the closer proximity of the N - and C - terminal CA domains. Inhibition of cleavage at the CA-SP1 cleavage site therefore does not appear to prevent disassembly of the immature lattice, but to affect formation of the regular mature core.
CA-SP1 and even CA-NC constructs are able to assemble in vitro into mature-like lattices [13], [21], [36], indicating that there is no absolute requirement for cleavage at the CA-SP1 junction for mature CA lattice assembly. Indeed the thin layers sometimes seen in CA-SP1 particles may represent patches of mature CA lattice, but formation of the closed mature core around the NC-RNA complex does not occur unless SP1 is removed from CA. It is currently not clear why SP1 cleavage is needed for formation of the mature core inside the virion, while being dispensable for in vitro assembly of the mature-like lattice. We speculate that this may be due to a defect in nucleating formation of the cone-shaped mature core in the virion if SP1 is not cleaved. It is interesting, in this context, that in vitro assembly of CA or C-terminally extended versions of CA generally leads to hollow tubes, and formation of a closed cone is observed only very rarely. The in vitro system may thus not completely mimic this aspect of virion maturation. Cleavage between CA and SP1 is the site of action of the maturation inhibitor bevirimat, suggesting that this drug has no effect on disassembly of the immature lattice but acts by preventing formation of the closed cone-shaped core of the mature virion.
A link between the condensed NC-RNA and the immature CA lattice
In the immature virus the NC-RNA is organized in a disordered but compact layer below the CA lattice. When Gag is cleaved downstream of SP1, the NC region condenses. Approximately the same degree of condensation was seen in the MA-SP1, MA-CA, CA-SP1 and wt variants, suggesting that there is no role for free CA in NC-RNA condensation. The high variability in shape and volume (if related to the size of the virus) indicates that the condensed NC-RNA does not form a well-defined structure.
MA-SP1 and MA-CA exhibited a condensed NC-RNA, as well as an intact immature CA layer. Surprisingly, broken MA-SP1 and MA-CA particles exhibited associated density consistent with NC-RNA. The size and shape of this density and the absence of any other strong candidates in the purified virus preparation suggests to us that this density is indeed NC-RNA. We expect that diffusion of the NC-RNA away from the protein shell would have occurred during the vortexing procedure used to disrupt the particles, or within the time taken to transfer the sample to the grid and prepare it for electron microscopy, if no physical linkage was present. These observations therefore suggest that the NC-RNA remains attached to the CA lattice, suggesting the presence of an interaction or link between the partially cleaved lattice and the condensed NC-RNA in these variants.
If a non-covalent interaction is present, it could represent a non-specific interaction between the NC-RNA complex and the CA lattice. The interaction could also be specific: a cellular or viral protein present within the CA lattice may expose a defined binding site for a component of the NC-RNA complex. A more attractive hypothesis is that there is a direct link between NC-RNA and CA in the form of uncleaved Gag at this position. Such a link could be locally maintained if, for example, binding of the RNA packaging signal to NC protects the SP1-NC cleavage site such that it is not accessible to the viral PR. It is tempting to speculate that maintenance of such a link during maturation may allow the NC-RNA to function as a template for formation of the conical core, and/or to ensure that the NC-RNA is packaged within the core.
In summary, our data suggest that cleavages at both sides of the CA-SP1 module are required to disassemble the immature Gag lattice with removal of the NC-p6 region initiating the independent condensation of the NC-RNA core, still connected with the membrane bound lattice. Subsequent cleavage between MA and CA causes dissociation of the immature lattice, but the resulting CA-SP1 module requires final cleavage of the SP1 peptide to permit assembly of the mature cone.
Methods
Constructs and virus preparation
Derivatives of proviral plasmid pNL4-3 carrying mutations at specific PR cleavage sites within Gag, as well as the PR defective variant NL4-3D25A have been described previously [26], [38].
293T cells were maintained in DMEM with 10% fetal calf serum and antibiotics. Transfections with the indicated proviral derivatives were performed using the calcium phosphate method. Culture media were harvested at 42 h post transfection cleared by low speed centrifugation (5 min, 1500 g) followed by filtration through 0.45 µM nitrocellulose filters. Particles were purified by centrifugation through a 20% (w/w) sucrose cushion and subsequent centrifugation on an Iodixanol gradient as described [39]. Purified virus was inactivated with 1% paraformaldehyde for 1 h on ice. Successful inactivation was confirmed by infection of C8166 cells and scoring for syncytia formation up to 10 days after inoculation. At least 3 independent particle preparations were analyzed for each variant shown with no variability in the structures with the exception of the CA-SP1.
Sample preparation and data acquisition
Purified viruses were mixed with 10 nm colloidal gold particles, deposited on C-flat holey Carbon grids, and vitrified by plunge-freezing in liquid ethane. Tilt series were collected on an FEI Tecnai F30 “Polara” transmission electron microscope with Gatan GIF 2002 post column energy filter and 2 k×2 k Multiscan CCD camera. Data collection was performed at 300 kV using the SerialEM Software. Tilt series were collected between 60° and −60° with 3° angular increment; the total electron dose applied to the tomograms was approximately 90 e/Å2. Tomograms were acquired at defocuses between 2.6 and 6.0 µm, with a magnification of 34000X resulting in a pixel size at the specimen level of 4.0 Å.
Image processing
Tomograms were reconstructed using IMOD [40]. Subtomogram averaging was carried out as described in [10] and below using MATLAB (Mathworks). Lattice map representations were generated using Amira (Visage Imaging), together with the EM Package [41]. A hexagon is placed at the final aligned position of each tomogram, and coloured according to the cross-correlation value between the sub-tomogram and the average. Tomograms aligned to an inappropriate radial position were excluded based on the radii distribution of the set of 20 nearest subtomograms surrounding each subtomogram. If the radius of the selected subtomogram was in the first or in the fourth quartile of the distribution the subtomogram was excluded. Only hexagons with a cross-correlation over a defined threshold are displayed. The threshold was appropriately set such that hexagons were not displayed where density corresponding to Gag was absent in the tomogram. Density maps were displayed using Amira, surface rendering was done using UCSF Chimera [42].
Subtomogram averaging
For subtomogram averaging, tomograms collected at defocuses between 2.6 and 2.9 µm were used. Sub-volumes of (38.3 nm)3 were extracted from tomograms along the surface of a sphere centred in the centre of the virus and with a radius equal to the mean radius at CA level. The sub-volumes were iteratively aligned. The initial reference used for the alignment was the average of the subtomograms in the extraction position. All variants subjected to sub-tomogram averaging show 6-fold symmetry, as evidenced using radius-angle-frequency plots (see Text S1 and Figure S2). 6-fold symmetry was therefore applied to the average after all iterations. The threshold for the subtomograms to be averaged was set to the mean cross-correlation value between all subtomograms and the reference. The final reconstructions had a resolution according to the Fourier shell correlation with a 0.5 criterion of approximately 26.5 Å in the CA region (Figure S3A) and are filtered to this resolution. The resolution varies with radius, with the highest resolution in the CA region (Figure S3B).
Supporting Information
Zdroje
1. DemirovDG
FreedEO
2004 Retrovirus budding. Virus Res 106 87 102
2. MoritaE
SundquistWI
2004 Retrovirus budding. Annu Rev Cell Dev Biol 20 395 425
3. Ganser-PornillosBK
YeagerM
SundquistW
2008 The structural biology of HIV assembly. Curr Opin Struct Biol 18 203 217
4. CoffinJM
HughesSH
VarmusHE
1998 Retroviruses
5. HurleyJH
2008 ESCRT complexes and the biogenesis of multivesicular bodies. Curr Opin Cell Biol 20 4 11
6. WilliamsRL
UrbeS
2007 The emerging shape of the ESCRT machinery. Nat Rev Mol Cell Biol 8 355 368
7. AldoviniA
YoungRA
1990 Mutations of RNA and protein sequences involved in human immunodeficiency virus type 1 packaging result in production of noninfectious virus. J Virol 64 1920 1926
8. AndersonJ
SchifferC
LeeSK
SwanstromR
2009 Viral protease inhibitors. Handb Exp Pharmacol 85 110
9. BriggsJAG
SimonM
GrossI
KrausslichHG
FullerS
2004 The stoichiometry of Gag protein in HIV-1. Nat Struct Mol Biol 11 672 675
10. BriggsJAG
RichesJD
GlassB
BartonovaV
ZanettiG
2009 Structure and assembly of immature HIV. Proc Natl Acad Sci USA 106 11090 11095
11. GanserBK
LiS
KlishkoV
FinchJ
SundquistW
1999 Assembly and analysis of conical models for the HIV-1 core. Science 283 80 83
12. BriggsJAG
WilkT
WelkerR
KrausslichHG
FullerSD
2003 Structural organization of authentic, mature HIV-1 virions and cores. EMBO J 22 1707 1715
13. LiS
HillC
SundquistW
FinchJ
2000 Image reconstructions of helical assemblies of the HIV-1 CA protein. Nature 407 409 413
14. WrightE
SchoolerJ
DingH
KiefferC
FillmoreC
2007 Electron cryotomography of immature HIV-1 virions reveals the structure of the CA and SP1 Gag shells. EMBO J 26 2218 2226
15. Ganser-PornillosBK
ChengA
YeagerM
2007 Structure of full-length HIV-1 CA: a model for the mature capsid lattice. Cell 131 70 79
16. PornillosO
Ganser-PornillosBK
KellyBN
HuaY
WhitbyFG
2009 X-ray structures of the hexameric building block of the HIV capsid. Cell 137 1282 1292
17. von SchwedlerUK
StemmlerTL
KlishkoVY
LiS
AlbertineKH
1998 Proteolytic refolding of the HIV-1 capsid protein amino-terminus facilitates viral core assembly. EMBO J 17 1555 1568
18. KingstonRL
Fitzon-OstendorpT
EisenmesserEZ
SchatzGW
VogtVM
2000 Structure and self-association of the Rous sarcoma virus capsid protein. Structure 8 617 628
19. CornilescuCC
BouamrF
YaoX
CarterC
TjandraN
2001 Structural analysis of the N-terminal domain of the human T-cell leukemia virus capsid protein. J Mol Biol 306 783 797
20. TangC
NdassaY
SummersMF
2002 Structure of the N-terminal 283-residue fragment of the immature HIV-1 Gag polyprotein. Nat Struct Biol 9 537 543
21. GrossI
HohenbergH
HuckhagelC
KräusslichHG
1998 N-Terminal extension of human immunodeficiency virus capsid protein converts the in vitro assembly phenotype from tubular to spherical particles. J Virol 72 4798 4810
22. PettitSC
MoodyMD
WehbieRS
KaplanAH
NantermetPV
1994 The p2 domain of human immunodeficiency virus type 1 Gag regulates sequential proteolytic processing and is required to produce fully infectious virions. J Virol 68 8017 8027
23. KaplanAH
ManchesterM
SwanstromR
1994 The activity of the protease of human immunodeficiency virus type 1 is initiated at the membrane of infected cells before the release of viral proteins and is required for release to occur with maximum efficiency. J Virol 68 6782 6786
24. CorenLV
ThomasJA
ChertovaE
SowderRC2nd
GagliardiTD
2007 Mutational analysis of the C-terminal gag cleavage sites in human immunodeficiency virus type 1. J Virol 81 10047 10054
25. LeeSK
HarrisJ
SwanstromR
2009 A strongly transdominant mutation in the human immunodeficiency virus type 1 gag gene defines an Achilles heel in the virus life cycle. J Virol 83 8536 8543
26. MüllerB
AndersM
AkiyamaH
WelschS
GlassB
2009 HIV-1 Gag processing intermediates trans-dominantly interfere with HIV-1 infectivity. J Biol Chem 284 29692 29703
27. WiegersK
RutterG
KottlerH
TessmerU
HohenbergH
1998 Sequential steps in human immunodeficiency virus particle maturation revealed by alterations of individual Gag polyprotein cleavage sites. J Virol 72 2846 2854
28. WymaDJ
JiangJ
ShiJ
ZhouJ
LinebergerJE
2004 Coupling of human immunodeficiency virus type 1 fusion to virion maturation: a novel role of the gp41 cytoplasmic tail. J Virol 78 3429 3435
29. GottlingerHG
SodroskiJG
HaseltineWA
1989 Role of capsid precursor processing and myristoylation in morphogenesis and infectivity of human immunodeficiency virus type 1. Proc Natl Acad Sci U S A 86 5781 5785
30. KräusslichHG
FäckeM
HeuserAM
KonvalinkaJ
ZentgrafH
1995 The spacer peptide between human immunodeficiency virus capsid and nucleocapsid proteins is essential for ordered assembly and viral infectivity. J Virol 69 3407 3419
31. LiF
Goila-GaurR
SalzwedelK
KilgoreNR
ReddickM
2003 PA-457: a potent HIV inhibitor that disrupts core condensation by targeting a late step in Gag processing. Proc Natl Acad Sci USA 100 13555 13560
32. ZhouJ
YuanX
DismukeD
ForsheyBM
LundquistC
2004 Small-molecule inhibition of human immunodeficiency virus type 1 replication by specific targeting of the final step of virion maturation. J Virol 78 922 929
33. CheckleyMA
LuttgeBG
SoheilianF
NagashimaK
FreedEO
The capsid-spacer peptide 1 Gag processing intermediate is a dominant-negative inhibitor of HIV-1 maturation. Virology 400 137 144
34. WilkT
GrossI
GowenB
RuttenT
de HaasF
2001 Organization of immature human immunodeficiency virus type 1. J Virol 75 759 771
35. BriggsJAG
GrunewaldK
GlassB
ForsterF
KrausslichHG
2006 The mechanism of HIV-1 core assembly: insights from three-dimensional reconstructions of authentic virions. Structure 14 15 20
36. CampbellS
VogtVM
1995 Self-assembly in vitro of purified CA-NC proteins from Rous sarcoma virus and human immunodeficiency virus type 1. J Virol 69 6487 6497
37. PettitSC
HendersonGJ
SchifferCA
SwanstromR
2002 Replacement of the P1 amino acid of human immunodeficiency virus type 1 Gag processing sites can inhibit or enhance the rate of cleavage by the viral protease. J Virol 76 10226 10233
38. KonvalinkaJ
LitterstMA
WelkerR
KottlerH
RippmannF
1995 An active-site mutation in the human immunodeficiency virus type 1 proteinase (PR) causes reduced PR activity and loss of PR-mediated cytotoxicity without apparent effect on virus maturation and infectivity. J Virol 69 7180 7186
39. DettenhoferM
YuXF
1999 Highly purified human immunodeficiency virus type 1 reveals a virtual absence of Vif in virions. J Virol 73 1460 1467
40. KremerJR
MastronardeDN
McIntoshJR
1996 Computer visualization of three-dimensional image data using IMOD. J Struct Biol 116 71 76
41. PruggnallerS
MayrM
FrangakisAS
2008 A visualization and segmentation toolbox for electron microscopy. Journal of Struct Biol 164 161 165
42. PettersenEF
GoddardTD
HuangCC
CouchGS
GreenblattDM
2004 UCSF chimera - A visualization system for exploratory research and analysis. J Comp Chem 25 1605 1612
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek Tick Histamine Release Factor Is Critical for Engorgement and Transmission of the Lyme Disease AgentČlánek TGF-b2 Induction Regulates Invasiveness of -Transformed Leukocytes and Disease SusceptibilityČlánek The Origin of Intraspecific Variation of Virulence in an Eukaryotic Immune Suppressive ParasiteČlánek Glycosylation Focuses Sequence Variation in the Influenza A Virus H1 Hemagglutinin Globular Domain
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2010 Číslo 11- Jak souvisí postcovidový syndrom s poškozením mozku?
- Měli bychom postcovidový syndrom léčit antidepresivy?
- Farmakovigilanční studie perorálních antivirotik indikovaných v léčbě COVID-19
- 10 bodů k očkování proti COVID-19: stanovisko České společnosti alergologie a klinické imunologie ČLS JEP
-
Všechny články tohoto čísla
- Patients with Discordant Responses to Antiretroviral Therapy Have Impaired Killing of HIV-Infected T Cells
- A Molecular Mechanism for Eflornithine Resistance in African Trypanosomes
- Tyrosine Sulfation of the Amino Terminus of PSGL-1 Is Critical for Enterovirus 71 Infection
- Autoimmunity as a Predisposition for Infectious Diseases
- The Subtilisin-Like Protease AprV2 Is Required for Virulence and Uses a Novel Disulphide-Tethered Exosite to Bind Substrates
- Structural Analysis of HIV-1 Maturation Using Cryo-Electron Tomography
- Tick Histamine Release Factor Is Critical for Engorgement and Transmission of the Lyme Disease Agent
- Interferon-Inducible CXC Chemokines Directly Contribute to Host Defense against Inhalational Anthrax in a Murine Model of Infection
- TGF-b2 Induction Regulates Invasiveness of -Transformed Leukocytes and Disease Susceptibility
- The Origin of Intraspecific Variation of Virulence in an Eukaryotic Immune Suppressive Parasite
- CO Acts as a Signalling Molecule in Populations of the Fungal Pathogen
- SV2 Mediates Entry of Tetanus Neurotoxin into Central Neurons
- MAP Kinase Phosphatase-2 Plays a Critical Role in Response to Infection by
- Glycosylation Focuses Sequence Variation in the Influenza A Virus H1 Hemagglutinin Globular Domain
- Potentiation of Epithelial Innate Host Responses by Intercellular Communication
- Fcγ Receptor I Alpha Chain (CD64) Expression in Macrophages Is Critical for the Onset of Meningitis by K1
- ANK, a Host Cytoplasmic Receptor for the Cell-to-Cell Movement Protein, Facilitates Intercellular Transport through Plasmodesmata
- Analysis of the Initiating Events in HIV-1 Particle Assembly and Genome Packaging
- Evolution of Linked Avirulence Effectors in Is Affected by Genomic Environment and Exposure to Resistance Genes in Host Plants
- Structural Basis of HIV-1 Neutralization by Affinity Matured Fabs Directed against the Internal Trimeric Coiled-Coil of gp41
- Hepatitis C Virus (HCV) Evades NKG2D-Dependent NK Cell Responses through NS5A-Mediated Imbalance of Inflammatory Cytokines
- Host Cell Invasion and Virulence Mediated by Ssa1
- Global Gene Expression in Urine from Women with Urinary Tract Infection
- Should the Human Microbiome Be Considered When Developing Vaccines?
- HapX Positively and Negatively Regulates the Transcriptional Response to Iron Deprivation in
- Enhancing Oral Vaccine Potency by Targeting Intestinal M Cells
- Herpes Simplex Virus Reorganizes the Cellular DNA Repair and Protein Quality Control Machinery
- The Female Lower Genital Tract Is a Privileged Compartment with IL-10 Producing Dendritic Cells and Poor Th1 Immunity following Infection
- Zn Inhibits Coronavirus and Arterivirus RNA Polymerase Activity and Zinc Ionophores Block the Replication of These Viruses in Cell Culture
- Cryo Electron Tomography of Native HIV-1 Budding Sites
- Crystal Structure and Size-Dependent Neutralization Properties of HK20, a Human Monoclonal Antibody Binding to the Highly Conserved Heptad Repeat 1 of gp41
- Modelling the Evolution and Spread of HIV Immune Escape Mutants
- The Arabidopsis Resistance-Like Gene Is Activated by Mutations in and Contributes to Resistance to the Bacterial Effector AvrRps4
- Platelet-Activating Factor Receptor Plays a Role in Lung Injury and Death Caused by Influenza A in Mice
- Genetic and Structural Basis for Selection of a Ubiquitous T Cell Receptor Deployed in Epstein-Barr Virus Infection
- Ubiquitin-Regulated Nuclear-Cytoplasmic Trafficking of the Nipah Virus Matrix Protein Is Important for Viral Budding
- Pneumolysin Activates the NLRP3 Inflammasome and Promotes Proinflammatory Cytokines Independently of TLR4
- Immune Evasion by : Differential Targeting of Dendritic Cell Subpopulations
- Survival in Selective Sand Fly Vector Requires a Specific -Encoded Lipophosphoglycan Galactosylation Pattern
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Zn Inhibits Coronavirus and Arterivirus RNA Polymerase Activity and Zinc Ionophores Block the Replication of These Viruses in Cell Culture
- The Female Lower Genital Tract Is a Privileged Compartment with IL-10 Producing Dendritic Cells and Poor Th1 Immunity following Infection
- Crystal Structure and Size-Dependent Neutralization Properties of HK20, a Human Monoclonal Antibody Binding to the Highly Conserved Heptad Repeat 1 of gp41
- The Arabidopsis Resistance-Like Gene Is Activated by Mutations in and Contributes to Resistance to the Bacterial Effector AvrRps4
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



