-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaZn Inhibits Coronavirus and Arterivirus RNA Polymerase Activity and Zinc Ionophores Block the Replication of These Viruses in Cell Culture
Increasing the intracellular Zn2+ concentration with zinc-ionophores like pyrithione (PT) can efficiently impair the replication of a variety of RNA viruses, including poliovirus and influenza virus. For some viruses this effect has been attributed to interference with viral polyprotein processing. In this study we demonstrate that the combination of Zn2+ and PT at low concentrations (2 µM Zn2+ and 2 µM PT) inhibits the replication of SARS-coronavirus (SARS-CoV) and equine arteritis virus (EAV) in cell culture. The RNA synthesis of these two distantly related nidoviruses is catalyzed by an RNA-dependent RNA polymerase (RdRp), which is the core enzyme of their multiprotein replication and transcription complex (RTC). Using an activity assay for RTCs isolated from cells infected with SARS-CoV or EAV—thus eliminating the need for PT to transport Zn2+ across the plasma membrane—we show that Zn2+ efficiently inhibits the RNA-synthesizing activity of the RTCs of both viruses. Enzymatic studies using recombinant RdRps (SARS-CoV nsp12 and EAV nsp9) purified from E. coli subsequently revealed that Zn2+ directly inhibited the in vitro activity of both nidovirus polymerases. More specifically, Zn2+ was found to block the initiation step of EAV RNA synthesis, whereas in the case of the SARS-CoV RdRp elongation was inhibited and template binding reduced. By chelating Zn2+ with MgEDTA, the inhibitory effect of the divalent cation could be reversed, which provides a novel experimental tool for in vitro studies of the molecular details of nidovirus replication and transcription.
Published in the journal: . PLoS Pathog 6(11): e32767. doi:10.1371/journal.ppat.1001176
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1001176Summary
Increasing the intracellular Zn2+ concentration with zinc-ionophores like pyrithione (PT) can efficiently impair the replication of a variety of RNA viruses, including poliovirus and influenza virus. For some viruses this effect has been attributed to interference with viral polyprotein processing. In this study we demonstrate that the combination of Zn2+ and PT at low concentrations (2 µM Zn2+ and 2 µM PT) inhibits the replication of SARS-coronavirus (SARS-CoV) and equine arteritis virus (EAV) in cell culture. The RNA synthesis of these two distantly related nidoviruses is catalyzed by an RNA-dependent RNA polymerase (RdRp), which is the core enzyme of their multiprotein replication and transcription complex (RTC). Using an activity assay for RTCs isolated from cells infected with SARS-CoV or EAV—thus eliminating the need for PT to transport Zn2+ across the plasma membrane—we show that Zn2+ efficiently inhibits the RNA-synthesizing activity of the RTCs of both viruses. Enzymatic studies using recombinant RdRps (SARS-CoV nsp12 and EAV nsp9) purified from E. coli subsequently revealed that Zn2+ directly inhibited the in vitro activity of both nidovirus polymerases. More specifically, Zn2+ was found to block the initiation step of EAV RNA synthesis, whereas in the case of the SARS-CoV RdRp elongation was inhibited and template binding reduced. By chelating Zn2+ with MgEDTA, the inhibitory effect of the divalent cation could be reversed, which provides a novel experimental tool for in vitro studies of the molecular details of nidovirus replication and transcription.
Introduction
Zinc ions are involved in many different cellular processes and have proven crucial for the proper folding and activity of various cellular enzymes and transcription factors. Zn2+ is probably an important cofactor for numerous viral proteins as well. Nevertheless, the intracellular concentration of free Zn2+ is maintained at a relatively low level by metallothioneins, likely due to the fact that Zn2+ can serve as intracellular second messenger and may trigger apoptosis or a decrease in protein synthesis at elevated concentrations [1], [2], [3]. Interestingly, in cell culture studies, high Zn2+ concentrations and the addition of compounds that stimulate cellular import of Zn2+, such as hinokitol (HK), pyrrolidine dithiocarbamate (PDTC) and pyrithione (PT), were found to inhibit the replication of various RNA viruses, including influenza virus [4], respiratory syncytial virus [5] and several picornaviruses [6], [7], [8], [9], [10], [11]. Although these previous studies provided limited mechanistic information, this suggests that intracellular Zn2+ levels affect a common step in the replicative cycle of these viruses.
In cell culture, PT stimulates Zn2+ uptake within minutes and inhibits RNA virus replication through a mechanism that has only been studied in reasonable detail for picornaviruses [11], [12]. In vitro studies with purified rhinovirus and poliovirus 3C proteases revealed that protease activity was inhibited by Zn2+ [13], [14], which is in line with the inhibition of polyprotein processing by zinc ions that was observed in cells infected with human rhinovirus and coxsackievirus B3 [11]. The replication of segmented negative-strand RNA viruses such as influenza virus, however, does not depend on polyprotein processing and the effect of PDTC-mediated Zn2+ import was therefore hypothesized to result from inhibition of the viral RNA-dependent RNA polymerase (RdRp) and cellular cofactors [4]. Moreover, an inhibitory effect of Zn2+ on the activity of purified RdRps from rhinoviruses and hepatitis C virus was noted, but not investigated in any detail [15], [16].
Details on the effect of zinc ions are currently largely unknown for nidoviruses. This large group of positive-strand RNA (+RNA) viruses includes major pathogens of humans and livestock, such as severe acute respiratory syndrome coronavirus (SARS-CoV), other human coronaviruses, the arteriviruses equine arteritis virus (EAV), and porcine reproductive and respiratory syndrome virus (PRRSV) [17], [18]. The common ancestry of nidoviruses is reflected in their similar genome organization and expression strategy, and in the conservation of a number of key enzymatic functions in their large replicase polyproteins [19]. A hallmark of the corona - and arterivirus replicative cycle is the transcription of a 5′ - and 3′-coterminal nested set of subgenomic (sg) mRNAs from which the viral structural and accessory protein genes are expressed [20], [21].
Analogous to picornaviruses [13], [22], zinc ions were demonstrated to inhibit certain proteolytic cleavages in the processing of the coronavirus replicase polyproteins in infected cells and cell-free systems [23], [24]. In this study we report that the zinc-ionophore pyrithione (PT) in combination with Zn2+ is a potent inhibitor of the replication of SARS-coronavirus (SARS-CoV) and equine arteritis virus (EAV) in cell culture. To assess whether - besides a possible effect on proteolytic processing - nidovirus RTC subunits and RNA synthesis are directly affected by Zn2+, we employed in vitro systems for SARS-CoV and EAV RNA synthesis that are based on membrane-associated RTCs isolated from infected cells (from here on referred to as RTC assays) [25], [26]. In addition, we used in vitro recombinant RdRp assays to directly study the effect of zinc ions on the RdRps of SARS-CoV and EAV [27], [28].
Using these independent in vitro approaches, we were able to demonstrate that Zn2+ directly impairs nidovirus RNA synthesis, since it had a strong inhibitory effect in both RTC and RdRp assays. Interestingly, the Zn2+-mediated inhibition could be reversed through the addition of a Zn2+ chelator (MgEDTA). We therefore applied this compound to stop and restart the in vitro RNA-synthesizing activity at will. This convenient tool allowed us to study various mechanistic aspects of arteri - and coronavirus RNA synthesis in more detail. Additionally, the zinc-mediated inhibition of nidovirus RNA synthesis described here may provide an interesting basis to further explore the use of zinc-ionophores in antiviral therapy.
Results
Zinc and pyrithione inhibit nidovirus replication in vivo
Zinc ions are involved in many different cellular processes, but the concentration of free Zn2+ is maintained at a relatively low level by metallothioneins [1]. Zn2+ and compounds that stimulate import of Zn2+ into cells, such as PT, were previously found to inhibit replication of several picornaviruses, including rhinoviruses, foot-and-mouth disease virus, coxsackievirus, and mengovirus in cell culture [6], [7], [8], [9], [10], [11]. To determine whether Zn2+ has a similar effect on nidoviruses, we investigated the effect of PT and Zn2+ on the replication of EAV and SARS-CoV in Vero-E6 cells, using reporter viruses that express green fluorescent proteins (GFP), i.e., EAV-GFP [29] and SARS-CoV-GFP [30]. EAV-GFP encodes an N-terminal fusion of GFP to the viral nonstructural protein 2 (nsp2), one of the cleavage products of the replicase polyproteins, and thus provides a direct readout for translation of the replicase gene. In SARS-CoV-GFP, reporter expression occurs from sg mRNA 7, following the replacement of two accessory protein-coding genes (ORFs 7a and 7b) that are dispensable for replication in cell culture.
We first assessed the cytotoxicity of a range of PT concentrations (0–32 µM) in the presence of 0 to 8 µM ZnOAc2. Treatment with PT of concentrations up to 32 µM in combination with <4 µM ZnOAc2 did not reduce the viability of mock-infected cells after 18 h (Fig. 1A), as measured by the colorimetric MTS (3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium) viability assay. As elevated Zn2+ concentrations are known to inhibit cellular translation, we also used metabolic labeling with 35S-methionine to assess the effect of PT and Zn2+ on cellular protein synthesis. Incubation of Vero-E6 cells for 18 h with the combinations of PT and Zn2+ mentioned above, followed by a 2-h metabolic labeling, revealed no change in overall cellular protein synthesis when the concentration of ZnOAc2 was <4 µM (data not shown).
Fig. 1. The zinc ionophore pyrithione inhibits nidovirus replication in cell culture. 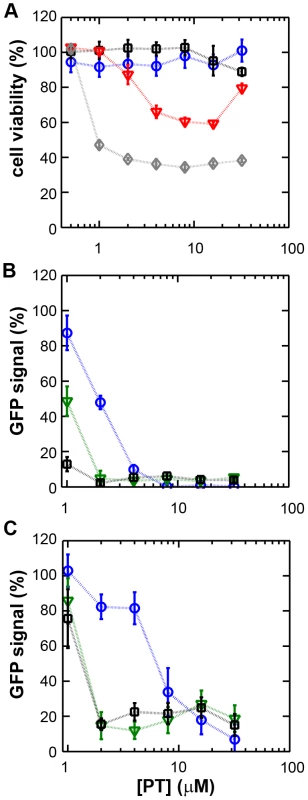
(A) Cytotoxicity of PT in Vero-E6 cells in the absence (blue circles) or presence of 2 (black squares), 4 (red triangles), or 8 µM (gray diamonds) ZnOAc2 as determined by the MTS assay after 18 hours of exposure. (B) Dose-response curves showing the effect of PT and Zn2+ on the GFP fluorescence in Vero-E6 cells infected with a GFP-expressing EAV reporter strain at 17 h p.i. Data were normalized to GFP expression in infected, untreated control cultures (100%). The different Zn2+ concentrations added to the medium were 0 (blue circles), 1 (green triangles), or 2 µM ZnOAc2 (black squares). (C) Effect of PT and Zn2+ on the GFP fluorescence in Vero-E6 cells infected with a GFP-expressing SARS-CoV reporter strain at 17 h p.i. Data were normalized to GFP expression in infected untreated control cells (100%). Colors for different Zn2+ concentrations as in Fig. 1B. Error bars indicate the standard deviation (n = 4). Using these non-cytotoxic conditions we subsequently tested the effect of PT and ZnOAc2 on EAV-GFP and SARS-CoV-GFP replication. To this end, Vero-E6 cells in 96-well plates were infected with a multiplicity of infection (m.o.i.) of 4. One hour post infection (h p.i.), between 0 and 32 µM of PT and 0, 1, or 2 µM ZnOAc2 were added to the culture medium. At 17 h p.i., a time point at which GFP expression in untreated infected cells reaches its maximum for both viruses, cells were fixed, and GFP fluorescence was quantified.
The reporter gene expression of both SARS-CoV-GFP and EAV-GFP was already significantly inhibited in a dose-dependent manner by the addition of PT alone (Fig. 1B and C). This effect was significantly enhanced when 2 µM of Zn2+ was added to the medium. We found that addition of ZnOAc2 alone also reduced virus replication, but only at levels that were close to the 50% cytotoxicity concentration (CC50) of ZnOAc2 in Vero-E6 cells (∼70 µM, data not shown). This is likely due to the poor solubility of Zn2+ in phosphate-containing medium and the inefficient uptake of Zn2+ by cells in the absence of zinc-ionophores. The combination of 2 µM PT and 2 µM ZnOAc2 resulted in a 98±1% and 85±3% reduction of the GFP signal for EAV-GFP and SARS-CoV-GFP, respectively. No cytotoxicity was observed for this combination of PT and ZnOAc2 concentrations. From the dose-response curves in Fig. 1, a CC50 value of 82 µM was calculated for PT in the presence of 2 µM zinc. Half maximal inhibitory concentrations (IC50) of 1.4 µM and 0.5 µM and selectivity indices of 59 and 164 were calculated for SARS-CoV and EAV, respectively.
Zn2+ reversibly inhibits the RNA-synthesizing activity of isolated nidovirus RTCs
We previously developed assays to study the in vitro RNA-synthesizing activity of RTCs isolated from cells infected with SARS-CoV or EAV [25], [26]. In these RTC assays [α-32P]CMP is incorporated into both genomic (replication) and sg mRNA (transcription) (Fig. 2). This allowed us to monitor the synthesis of the same viral RNA molecules that can be detected by hybridization of RNA from nidovirus-infected cells. A benefit of these assays is that the activity does not depend on continued protein synthesis and that it allows us to study viral RNA synthesis independent of other aspects of the viral replicative cycle [26]. To investigate whether the inhibitory effect of PT and zinc ions on nidovirus replication in cell culture is reflected in a direct effect of Zn2+ on viral RNA synthesis, we tested the effect of Zn2+ addition on RTC activity. For both EAV (Fig. 2A) and SARS-CoV (Fig. 2B), a dose-dependent decrease in the amount of RNA synthesized was observed when ZnOAc2 was present. For both viruses, a more than 50% reduction of overall RNA-synthesis was observed at a Zn2+ concentration of 50 µM, while less than 5% activity remained at a Zn2+ concentration of 500 µM. Both genome synthesis and sg mRNA production were equally affected.
Fig. 2. Inhibition of the in vitro RNA-synthesizing activity of isolated RTCs by Zn2+. 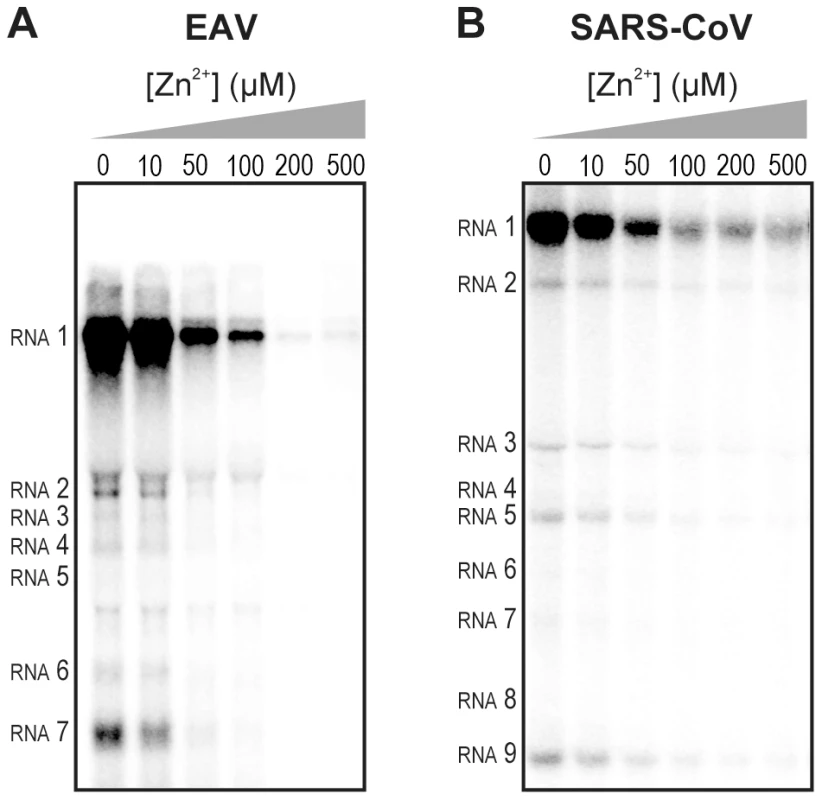
Incorporation of [α-32P]CMP into viral RNA by EAV (A) and SARS-CoV (B) in RTC assays in the presence of various Zn2+ concentrations, as indicated above each lane. To test whether the inhibition of RTC activity by Zn2+ was reversible, RTC reactions were started in the presence or absence of 500 µM Zn2+. After 30 min, these reactions were split into two aliquots and magnesium-saturated EDTA (MgEDTA) was added to one of the tubes to a final concentration of 1 mM (Fig. 3A). We used MgEDTA as Zn2+ chelator in these in vitro assays, because it specifically chelates Zn2+ while releasing Mg2+, due to the higher stability constant of the ZnEDTA complex. Uncomplexed EDTA inhibited RTC activity in all reactions (data not shown), most likely by chelating the Mg2+ that is crucial for RdRp activity [27], [28], whereas MgEDTA had no effects on control reactions without Zn2+ (Fig. 3B, compare lane 1 and 2). As shown in Fig. 2, the EAV RTC activity that was inhibited by Zn2+ (Fig. 3B&C, lane 3) could be restored by the addition of MgEDTA (Fig. 3B, lane 4) to a level observed for control reactions without Zn2+ (Fig. 3B, lane 1). Compared to the untreated control, the EAV RTC assay produced approximately 30% less RNA, which was consistent with the 30% shorter reaction time after the addition of the MgEDTA (100 versus 70 min for lanes 1 and 4, respectively). Surprisingly, SARS-CoV RTC assays that were consecutively supplemented with Zn2+ and MgEDTA incorporated slightly more [α-32P]CMP compared to untreated control reactions (Fig. 3C; compare lane 1 and 4). This effect was not due to chelation of the Zn2+ already present in the post-nuclear supernatant (PNS) of SARS-CoV-infected cells, as this increase was not observed when MgEDTA was added to a control reaction without additional Zn2+ (Fig. 3C, lane 2).
Fig. 3. Inhibition of nidovirus RTC activity by Zn2+ can be reversed by chelation. 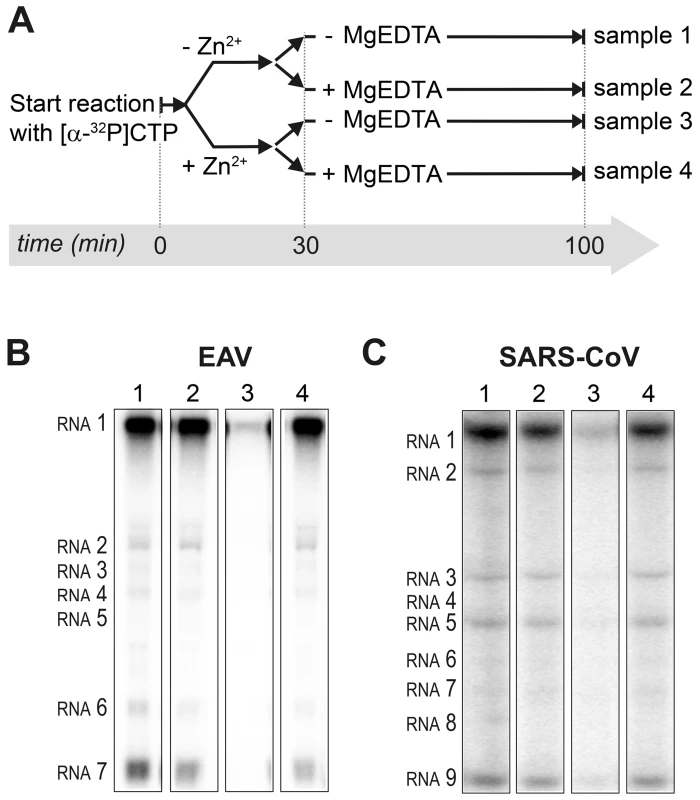
(A) Schematic representation of the in vitro assays with isolated RTCs, which were initiated with [α-32P]CTP, either in the absence (sample 1 and 2) or presence of 500 µM Zn2+. After a 30-min incubation at 30°C, both the untreated and Zn2+-treated samples were split into two aliquots, and 1 mM of the Zn2+ chelator MgEDTA was added to samples 2 and 4. All reactions were subsequently incubated for another 70 min before termination. (B) Analysis of RNA products synthesized in assays with EAV RTCs. Numbers above the lanes refer to the sample numbers described under (A). (C) In vitro activity assay with SARS-CoV RTCs. Zinc ions affect the in vitro activity of recombinant nidovirus RdRps
To establish whether inhibition of RTC activity might be due to a direct effect of Zn2+ on nidovirus RdRp activity, we tested the effect of Zn2+ on the activity of the purified recombinant RdRps of SARS-CoV (nsp12) and EAV (nsp9) using previously developed RdRp assays [27], [28]. Using an 18-mer polyU template, the EAV RdRp incorporated [α-32P]AMP into RNA products of up to 18 nt in length (Fig. 4A). Initiation was de novo, which is in line with previous observations and the presence of a conserved priming loop in the nsp9 sequence [28]. Unlike the EAV RdRp nsp9, the in vitro activity of the SARS-CoV RdRp nsp12 - which lacks a priming loop - was shown to be strictly primer-dependent [27]. Thus, to study the RdRp activity of SARS-CoV nsp12, a primed polyU template was used (Fig. 4B), thereby allowing us to sample [α-32P]AMP incorporation as described previously [27]. As specificity controls, we used the previously described SARS-CoV nsp12 mutant D618A [27], which contains an aspartate to alanine substitution in motif A of the RdRp active site, and EAV nsp9-D445A, in which we engineered an aspartate to alanine substitution at the corresponding site of EAV nsp9 [28], [31]. Both mutant RdRps showed greatly reduced [α-32P]AMP incorporation in our assays (Fig. 4), confirming once again that the radiolabeled RNA products derived from nidovirus RdRp activity.
Fig. 4. EAV and SARS-CoV RdRp assays with wild-type enzyme and active-site mutants. 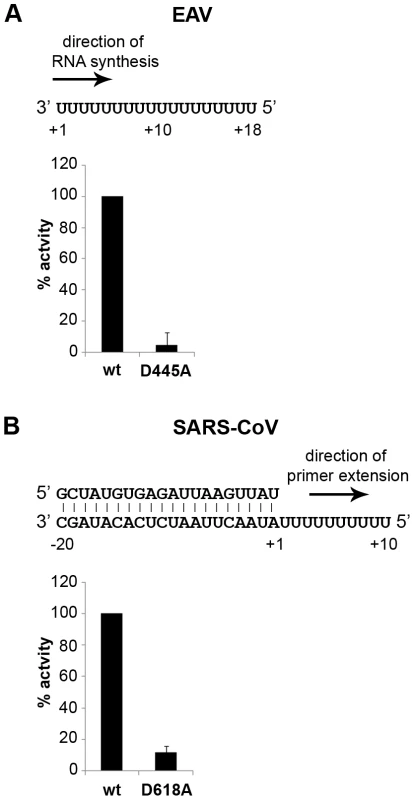
(A) The EAV polymerase was incapable of primer extension and required a free 3′ end and poly(U) residues to initiate. Nucleotide incorporating activity of the wild-type enzyme and D445A mutant of nsp9 on an 18-mer poly(U) template confirmed the specificity of our assay. (B) SARS-CoV nsp12 RdRp assays were performed with an RNA duplex with a 5′ U10 overhang as template. The bar graph shows the nucleotide incorporating activities of wild-type and D618A nsp12. Error bars represent standard error of the mean (n = 3). Addition of ZnOAc2 to RdRp assays resulted in a strong, dose-dependent inhibition of enzymatic activity for both the EAV and SARS-CoV enzyme (Fig. 5A and B, respectively), similar to what was observed in RTC assays. In fact, compared to other divalent metal ions such Co2+ and Ca2+, which typically bind to amino acid side chains containing oxygen atoms rather than sulfur groups, Zn2+ was the most efficient inhibitor of SARS-CoV nsp12 RdRp activity (Supplemental Fig. S1). To test whether, as in the RTC assay, the RdRp inhibition by zinc ions was reversible, RdRp assays were pre-incubated with 6 mM Zn2+, a concentration that consistently gave >95% inhibition. After 30 min, 8 mM MgEDTA was added to both a control reaction and the reaction inhibited with ZnOAc2, and samples were incubated for another 30 min (Fig. 5C). As shown in Fig. 5D, the inhibition of EAV RdRp activity by Zn2+ could be reversed by chelation of Zn2+ (Fig. 5D; compare lanes 3 and 4). The amount of product synthesized was consistently 60±5% of that synthesized in a 60-min control reaction (Fig. 5D; compare lanes 1 and 4), which was within the expected range given the shorter reaction time. The inhibition of the SARS-CoV RdRp was reversible as well. During the 30-min incubation after the addition of MgEDTA, SARS-CoV nsp12 incorporated 40±5% of the label incorporated during a standard 60-min reaction (Fig. 5E). This was slightly lower than the expected yield and may be caused by the elevated Mg2+ concentration, which was shown to be suboptimal for nsp12 activity [27] and results from the release of Mg2+ from MgEDTA upon chelation of Zn2+.
Fig. 5. The activity of the RdRps of EAV and SARS-CoV is reversibly inhibited by Zn2+. 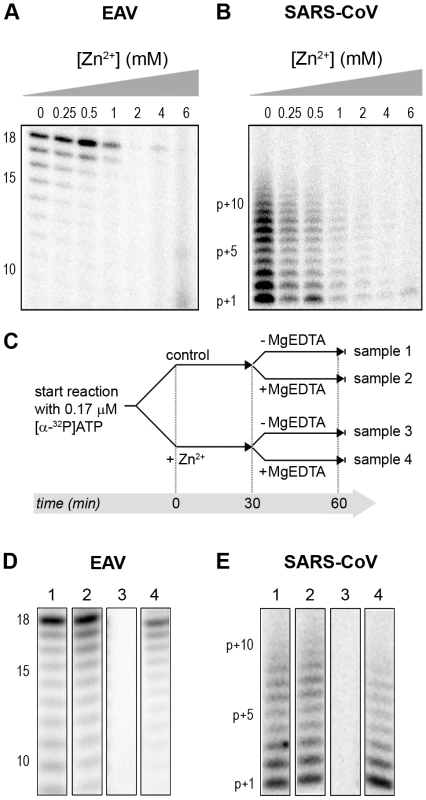
RdRp activity of purified EAV nsp9 (A) and SARS-CoV nsp12 (B) in the presence of various Zn2+ concentrations, as indicated above the lanes. (C) Schematic representation of the experiment to test whether Zn2+-mediated inhibition of RdRp activity could be reversed with MgEDTA. RdRp reactions, either untreated controls (sample 1 and 2) or reactions containing 6 mM Zn2+ (samples 3 and 4) were incubated for 30 min. Both Zn2+-containing and control samples were split into two aliquots and 6 mM MgEDTA was added to sample 2 and 4. All reactions were incubated for an additional 30 min and then terminated. Reaction products of the RdRp assays with EAV nsp9 and SARS-CoV nsp12 are shown in (D) and (E), respectively. Numbers above the lanes refer to the sample numbers described under (C). Differential effect of Zn2+ on the initiation and elongation phase of nidovirus RNA synthesis
For EAV, close inspection of the RdRp assays revealed a less pronounced effect of Zn2+ on the generation of full-length 18-nt products than on the synthesis of smaller reaction intermediates (Fig. 5A). This suggested that Zn2+ specifically inhibited the initiation step of EAV RNA synthesis. To test this hypothesis, an RTC assay was incubated for 30 min with unlabeled CTP (initiation), after which the reaction was split in two. Then, [α-32P]CTP was added to both tubes (pulse), 500 µM Zn2+ was added to one of the tubes, and samples were taken at different time points during the reaction (Fig. 6A). Fig. 6B shows that in the presence of Zn2+ [α-32P]CMP was predominantly incorporated into nascent RNA molecules that were already past the initiation phase at the moment that Zn2+ was added to the reaction. No new initiation occurred, as was indicated by the smear of short radiolabeled products that progressively shifted up towards the position of full-length genomic RNA. This suggested that Zn2+ does not affect the elongation phase of EAV RNA synthesis and that it specifically inhibits initiation. This also explains the relatively weak signal intensity of the smaller sg mRNA bands (e.g., compare the relative change in signal of RNA2 to RNA7) produced in the presence of Zn2+, since multiple initiation events are required on these short molecules to obtain signal intensities similar to those resulting from a single initiation event on the long genomic RNA, e.g., 16 times more in the case of RNA7. In contrast to EAV, the effect of Zn2+ on RNA synthesis by SARS-CoV RTCs was not limited to initiation, but appeared to impair the elongation phase as well, given that the addition of Zn2+ completely blocked further incorporation of [α-32P]CMP when added 40 min after the start of the reaction (Fig. 6C).
Fig. 6. Effect of Zn2+ on initiation and elongation in in vitro assays with isolated EAV and SARS-CoV RTCs. 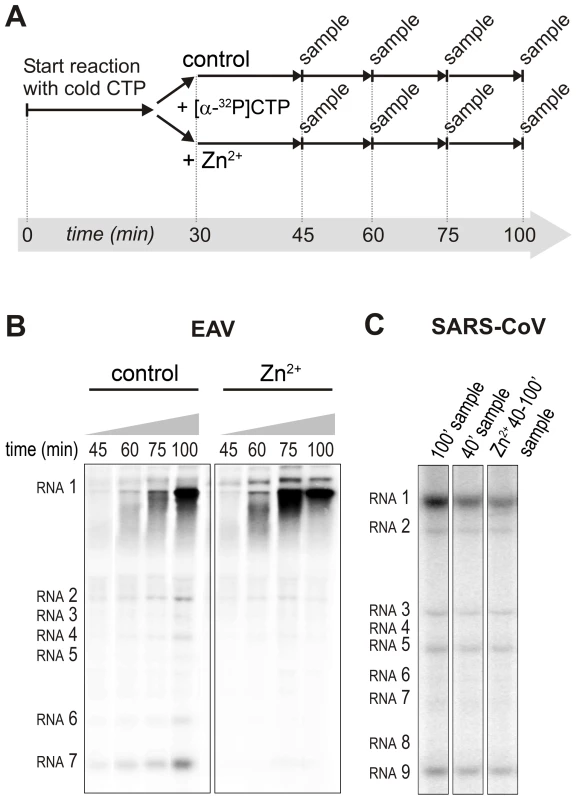
(A) An in vitro RTC assay with isolated EAV RTCs was allowed to initiate with unlabeled NTPs (initiation). After 30 min, [α-32P]CTP was added (pulse), the reaction was split into two equal volumes, and Zn2+ was added to a final concentration of 0.5 mM to one of the tubes. At the time points indicated, samples were taken and incorporation of [α-32P]CMP into viral RNA was analyzed. (B) Radiolabeled EAV RNA synthesized at the time points indicated above the lanes in the presence and absence of Zn2+. (C) Radiolabeled RNA synthesized by isolated SARS-CoV RTCs in reactions terminated after 100 (lane 1) and 40 (lane 2) min. Reaction products of a reaction to which 500 µM Zn2+ was added after 40 min, and that was terminated at t = 100 are shown in lane 3. In the RdRp assays, the short templates used made it technically impossible to do experiments similar to those performed with complete RTCs. However, we previously noticed that at low concentrations of [α-32P]ATP (∼0.17 µM) SARS-CoV nsp12 RdRp activity was restricted to the addition of only a single nucleotide to the primer [27]. EAV nsp9 mainly produced very short (2–3 nt long) abortive RNA products and only a fraction of full length products, as is common for de novo initiating RdRps [28]. This allowed us to separately study the effect of Zn2+ on initiation and elongation by performing an experiment in which a pulse with a low concentration of [α-32P]ATP was followed by a chase in the presence of 50 µM of unlabeled ATP, which increased processivity and allowed us to study elongation (Fig. 7A and C) as described previously [27]. The results of these experiments were in agreement with those obtained with isolated RTCs. For EAV, with initiation and dinucleotide synthesis completely inhibited by the presence of 6 mM Zn2+ (Supplemental Fig. S2A), the amount of reaction intermediates shorter than 18 nt diminished with time, while products from templates on which the RdRp had already initiated were elongated to full-length 18-nt molecules (Fig. 7B, right panel). This was consistent with the observation that the EAV RdRp remained capable of extending the synthetic dinucleotide ApA to trinucleotides in the presence of Zn2+ (Supplemental Fig. S2B). Likely due to the absence of reinitiation in the reactions shown in Fig. 7B, the low processivity of the EAV RdRp, and the substrate competition between the remaining [α-32P]ATP and the >200 fold excess of unlabeled ATP, the differences between the 5 - and 30-min time points were small. In the absence of Zn2+, the RdRp continued to initiate as indicated by the ladder of smaller-sized RNA molecules below the full-length product (Fig. 7B, left panel) and the time course shown in Supplemental Fig. S2A. In contrast, the addition of Zn2+ to a SARS-CoV RdRp reaction also blocked elongation, since extension of the radiolabeled primer as observed in the absence of Zn2+ (Fig. 7D, left panel) no longer occurred (Fig. 7D, right panel).
Fig. 7. The effect of Zn2+ on initiation and elongation activity of purified EAV and SARS-CoV RdRps. 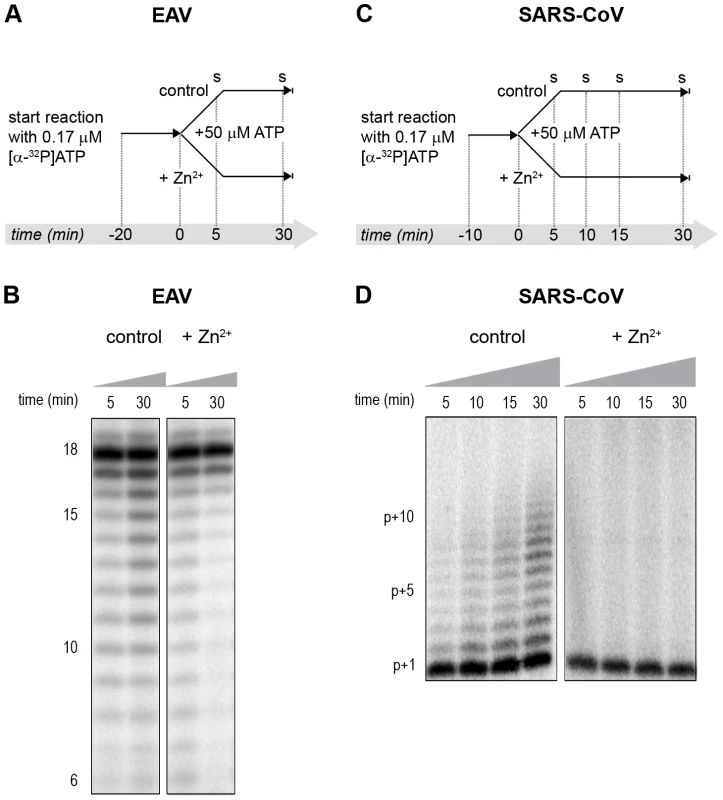
(A) An EAV RdRp reaction was initiated in the presence of [α-32P]ATP under conditions that do not allow elongation, i.e., low ATP concentration. After 20 min, the reaction was split into two equal volumes, and Zn2+ was added to one of the tubes. A chase with 50 µM unlabeled ATP, which allows elongation, was performed on both reactions and samples were taken after 5 and 30 min. (B) EAV RdRp reaction products that accumulated in the presence and absence of Zn2+ (indicated above the lanes) after a 5- and 30-min chase with unlabeled ATP. The length of the reaction products in nt is indicated next to the gel. (C) A SARS-CoV RdRp reaction was initiated in the presence of 0.17 µM [α-32P]ATP, which limits elongation. After 10 min, the reaction was split into two equal volumes, and Zn2+ was added to one of the tubes. A chase with 50 µM unlabeled ATP was performed on both reactions and samples were taken after 5, 10, 15, and 30 min. (D) SARS-CoV RdRp reaction products formed at the chase times indicated above the lanes in the presence and absence of Zn2+. The length of the reaction products in nt is indicated next to the gel (p is the primer length). Zinc affects SARS-CoV RdRp template binding
To assess whether Zn2+ affects the interaction of recombinant SARS-CoV nsp12 with the template used in our assays, we performed electromobility shift assays (EMSA) in the presence and absence of Zn2+ (Fig. 8A). To measure the binding affinity of the RdRp for the template, we determined the fraction of bound template at various protein concentrations and observed a 3–4 fold reduction in RNA binding when Zn2+ was present in the assay (Fig. 8B). We also assessed whether pre-incubation of the RdRp or RNA with Zn2+ was a requirement for this drop in binding affinity, but found no significant difference with experiments not involving such a preincubation (data not shown).
Fig. 8. The effect of Zn2+ on SARS-CoV nsp12 template binding. 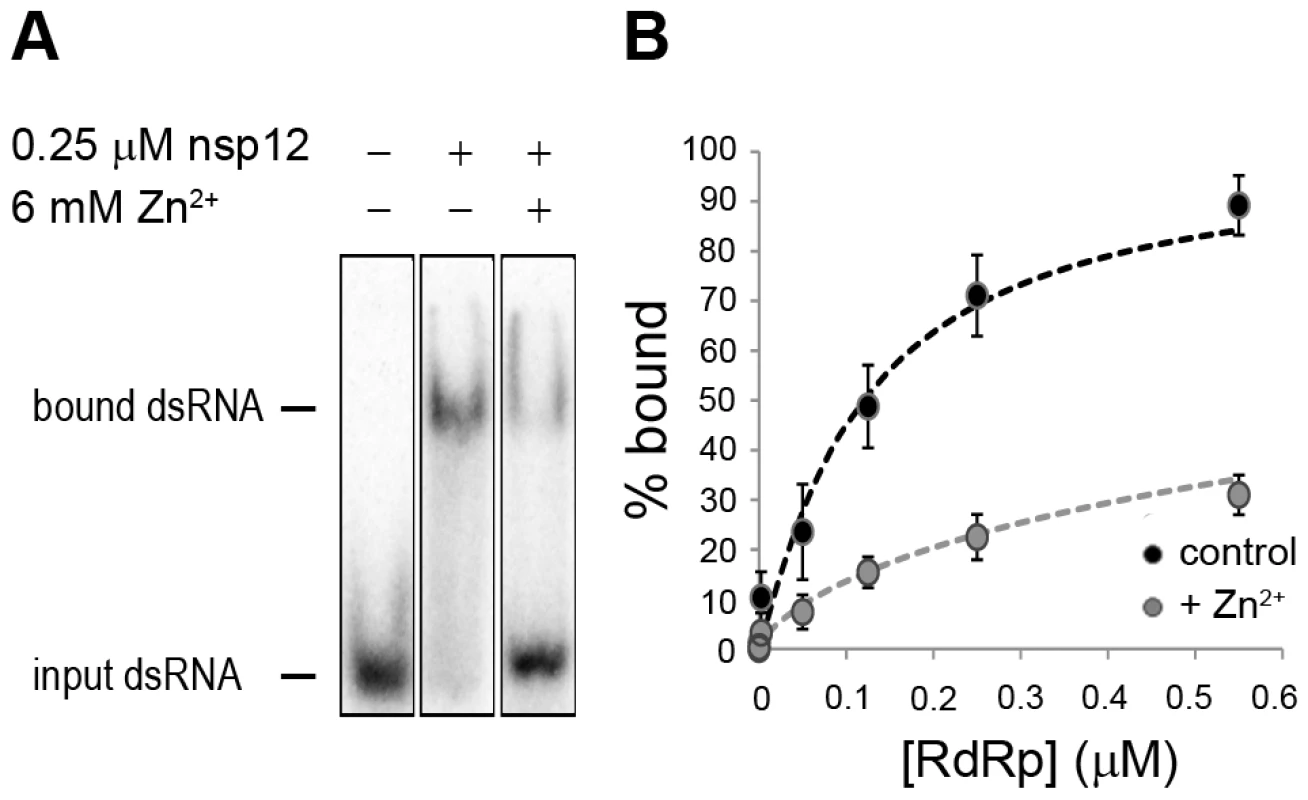
(A) Electrophoretic mobility shift assay with radiolabeled dsRNA and nsp12 in the presence and absence of Zn2+ (indicated above the lanes). The position of unbound and nsp12-bound RNA in the gel is marked on the left of the panel. (B) Determination of the nsp12 affinity for RNA in the presence and absence of Zn2+. A fixed amount of RNA was incubated with an increasing amount of nsp12. This revealed a 3–4 fold reduction in the percentage of bound RNA in the presence of zinc ions (grey) relative to the percentage of bound RNA in the absence of zinc ions (black). Error bars represent standard error of the mean (n = 3). No binding was observed when a similar RNA binding assay was performed with purified EAV RdRp. Likewise, nsp9 did not bind RNA in pull-down experiments with Talon-beads, His6-tagged nsp9, and radiolabeled EAV genomic RNA or various short RNA templates including polyU, whereas we were able to detect binding of a control protein (SARS-CoV nsp8, which has demonstrated RNA and DNA binding activity [32]) using this assay. It presently remains unclear why we are not able to detect the binding of recombinant EAV nsp9 to an RNA template.
Discussion
Although a variety of compounds have been studied, registered antivirals are currently still lacking for the effective treatment of SARS and other nidovirus-related diseases [33]. RdRps are suitable targets for antiviral drug development as their activity is strictly virus-specific and may be blocked without severely affecting key cellular functions. Several inhibitors developed against the polymerases of e.g. human immunodeficiency virus (HIV) and hepatitis C virus are currently being used in antiviral therapy or clinical trials [34], [35], [36]. Therefore, advancing our molecular knowledge of nidovirus RdRps and the larger enzyme complexes that they are part of, and utilizing the potential of recently developed in vitro RdRp assays [25], [26], [27], [28] could ultimately aid in the development of effective antiviral strategies.
Zinc ions and zinc-ionophores, such as PT and PDTC, have previously been described as potent inhibitors of various RNA viruses. We therefore investigated whether PT-stimulated import of zinc ions into cells also inhibited the replication of nidoviruses in cell culture. Using GFP-expressing EAV and SARS-CoV [29], [30], we found that the combination of 2 µM PT and 2 µM Zn2+ efficiently inhibited their replication, while not causing detectable cytoxicity (Fig. 1). Inhibition of replication by PT and Zn2+ at similar concentrations (2–10 µM) was previously observed for several picornaviruses such as rhinoviruses, foot-and-mouth disease virus, coxsackievirus, and mengovirus [6], [7], [8], [9], [10], [11].
The inhibitory effect of Zn2+ on the replication of picornaviruses appeared to be due to interference with viral polyprotein processing. In infections with the coronavirus mouse hepatitis virus (MHV), Zn2+ also interfered with some of the replicase polyproteins cleavages [24], albeit at a much higher concentration (100 µM Zn2+) than used in our studies. Since impaired replicase processing will indirectly affect viral RNA synthesis in the infected cell, we used two recently developed in vitro assays to investigate whether Zn2+ also affects nidovirus RNA synthesis directly. Our in vitro studies revealed a strong inhibitory effect of zinc ions on the RNA-synthesizing activity of isolated EAV and SARS-CoV RTCs. Assays with recombinant enzymes subsequently demonstrated that this was likely due to direct inhibition of RdRp function. The inhibitory effect could be reversed by chelating the zinc ions, which provides an interesting experimental (on/off) approach to study nidovirus RNA synthesis. Addition of Zn2+ following initiation of EAV RNA synthesis had little or no effect on NTP incorporation in molecules whose synthesis had already been initiated in the absence of Zn2+ (Fig. 6 and 7), indicating that Zn2+ does not affect elongation and does not increase the termination frequency, as was previously found for Mn2+ [25]. Therefore, Zn2+ appears to be a specific inhibitor of the initiation phase of EAV RNA synthesis. In contrast, Zn2+ inhibited SARS-CoV RdRp activity also during the elongation phase of RNA synthesis, probably by directly affecting template binding (Fig. 8). In coronaviruses, zinc ions thus appear to inhibit both the proper proteolytic processing of replicase polyproteins [23], [24] and RdRp activity (this study). Contrary to the RTC assays, millimolar instead of micromolar concentrations of ZnOAc2 were required for a nearly complete inhibition of nucleotide incorporation in RdRp assays.
It has been well established that DNA and RNA polymerases use a triad of conserved aspartate residues in motifs A and C to bind divalent metal ions like Mg2+, which subsequently coordinate incoming nucleotides during the polymerization reaction [37], [38]. Mg2+ is also the divalent metal ion that is required for the in vitro activity of isolated SARS-CoV and EAV RTCs and recombinant RdRps [25], [26], [27], [28], although de novo initiation of EAV nsp9 is primarily Mn2+-dependent. Zn2+ could not substitute for Mg2+ or Mn2+ as cofactor as it was incapable of supporting the polymerase activity of nidovirus RTCs and RdRps in the absence of Mg2+ (data not shown), as was also reported for the poliovirus RdRp [39]. Moreover, inhibition of nidovirus RdRp activity by Zn2+ was even observed at low concentrations and in the presence of a more than 25-fold excess of Mg2+, suggesting that either the affinity of the active site for Zn2+ is much higher or that Zn2+ does not compete for Mg2+-binding and binds to another zinc(-specific) binding site in the protein.
Specific protein domains or pockets that contain zinc ions may be involved in protein-protein interactions, protein-RNA/DNA interactions, or conformational changes in enzyme structures. Zinc-binding domains commonly consist of at least three conserved cysteine and/or histidine residues within a stretch of ∼10–30 amino acids, such as in zinc-finger motifs and metalloproteases [2], [40], [41]. However, in RdRps there are only few precedents for the presence of zinc-binding pockets, such as those identified in the crystal structure of the Dengue RdRp [42]. Sequence analysis of the EAV nsp9 amino acid sequence revealed that it lacks patches rich in conserved cysteines and/or histidines. In contrast, inspection of the SARS-CoV nsp12 amino acid sequence revealed two such patches, namely H295-C301-C306-H309-C310 and C799-H810-C813-H816. A crystal structure for nsp12 is presently unavailable, but a predicted structure that represents the C-terminal two-thirds of the enzyme has been published [31]. Interestingly, in this model, C799, H810, C813 and H816 are in a spatial arrangement resembling that of the Zn2+ coordinating residues in the Zn2 zinc-binding pocket found in motif E of the Dengue virus RdRp (see Supplemental Fig. S3). Clearly, an in-depth analysis of nidovirus RdRps, e.g. through structural analysis and subsequent mutational studies targeting aforementioned cysteines and histidines, is required to provide further insight into and a structural basis for the Zn2+-induced inhibitory effects on RdRp activity documented in this study. Such studies may, however, be complicated when Zn2+ binding proves to be very transient in nature and not detectable with currently available methods.
In summary, the combination of zinc ions and the zinc-ionophore PT efficiently inhibits nidovirus replication in cell culture. This provides an interesting basis for further studies into the use of zinc-ionophores as antiviral compounds, although systemic effects have to be considered [43], [44] and a water-soluble zinc-ionophore may be better suited, given the apparent lack of systemic toxicity of such a compound at concentrations that were effective against tumors in a mouse xenograft model [45]. In vitro, the reversible inhibition of the RdRp by Zn2+ has also provided us with a convenient research tool to gain more insight into the molecular details of (nido)viral RNA synthesis, and revealed novel mechanistic differences between the RdRps of SARS-CoV and EAV.
Materials and Methods
Cells and viruses
Vero-E6 cells were cultured and infected with SARS-CoV (strain Frankfurt-1; accession nr. AY291315) or SARS-CoV-GFP as described previously [46]. All procedures involving live SARS-CoV were performed in the biosafety level 3 facility at Leiden University Medical Center. BHK-21 or Vero-E6 cells were cultured and infected with EAV (Bucyrus strain; accession nr. NC_002532) or EAV-GFP [29] as described elsewhere [25].
Effect of zinc ions on nidovirus replication in cell culture
One day prior to infection, Vero-E6 cells were seeded in transparent or black (low fluorescence) 96-well clusters at 10,000 cells per well. The next day, cells were infected with SARS-CoV-GFP or EAV-GFP with an m.o.i. of 4, and 1 h p.i. the inoculum was removed and 100 µl of medium containing 2% fetal calf serum (FCS) was added to each well. In some experiments 0–32 µM of pyrithione (Sigma) was added in addition to 0–2 µM ZnOAc2. Infected cells were fixed at 17 h p.i. by aspirating the medium and adding 3% paraformaldehyde in PBS. After washing with PBS, GFP expression was quantified by measuring fluorescence with a LB940 Mithras plate reader (Berthold) at 485 nm. To determine toxicity of ZnOAc2 and PT, cells were exposed to 0–32 µM PT and 0–8 µM ZnOAc2. After 18 h incubation, cell viability was determined with the Cell Titer 96 AQ MTS assay (Promega). EC50 and CC50 values were calculated with Graphpad Prism 5 using the nonlinear regression model.
RNA templates and oligonucleotides
RNA oligonucleotides SAV557R (5′-GCUAUGUGAGAUUAAGUUAU-3′), SAV481R (5′-UUUUUUUUUUAUAACUUAAUCUCACAUAGC-3′) and poly(U)18 (5′-UUUUUUUUUUUUUUUUUU-3′) were purchased from Eurogentec, purified from 7 M Urea/15% PAGE gels and desalted through NAP-10 columns (GE healthcare). To anneal the RNA duplex SAV557R/SAV481R, oligonucleotides were mixed at equimolar ratios in annealing buffer (20 mM Tris-HCl pH 8.0, 50 mM NaCl and 5 mM EDTA), denatured by heating to 90°C and allowed to slowly cool to room temperature after which they were purified from 15% non-denaturing PAGE gels.
In vitro viral RNA synthesis assay with isolated RTCs
SARS-CoV and EAV RTCs were isolated from infected cells and assayed for activity in vitro as described previously [25], [26]. To assess the effect of Zn2+, 1 µl of a ZnOAc2 stock solution was added to standard 28-µl reactions, resulting in final Zn2+ concentrations of 10–500 µM. When Zn2+ had to be chelated in the course of the reaction, magnesium-saturated EDTA (MgEDTA) was added to a final concentration of 1 mM. After RNA isolation, the 32P-labeled reaction products were separated on denaturing 1% (SARS-CoV) or 1.5% (EAV) agarose formaldehyde gels. The incorporation of [α-32P]CMP into viral RNA was quantified by phosphorimaging of the dried gels using a Typhoon scanner (GE Healthcare) and the ImageQuant TL 7 software (GE Healthcare).
Expression and purification of nidovirus RdRps
SARS-CoV nsp12 and EAV nsp9 were purified essentially as described elsewhere [27], [28], but with modifications for nsp9. In short, E. coli BL21(DE3) with plasmid pDEST14-nsp9-CH was grown in auto-induction medium ZYM-5052 [47] for 6 hours at 37°C and a further 16 hours at 20°C. After lysis in buffer A (20 mM HEPES pH 7.4, 200 mM NaCl, 20 mM imidazole, and 0.05% Tween-20) the supernatant was applied to a HisTrap column (GE Healthcare). Elution was performed with a gradient of 20–250 mM imidazole in buffer A. The nsp9-containing fraction was further purified by gel filtration in 20 mM HEPES, 300 mM NaCl and 0.1% Tween-20 on a Superdex 200 column (GE Healthcare). The fractions containing nsp9-CH were pooled, dialyzed against 1000 volumes of buffer B (20 mM HEPES, 100 mM NaCl, 1 mM DTT and 50% glycerol) and stored at −20°C. RdRps with a D618A (SARS-CoV) or D445A (EAV) mutation were obtained by site-directed mutagenesis of the wild-type (wt) plasmid pDEST14-nsp9-CH [28] with oligonucleotides 5′-TACTGCCTTGAAACAGCCCTGGAGAGTTGTGAT-3′ and 5′-ATCACAACTCTCCAGGGCTGTTTCAAGGCAGTA-3′, and plasmid pASK3-Ub-nsp12-CHis6 with oligonucleotides 5′-CCTTATGGGTTGGGCTTATCCAAAATGTG-3′ and 5′-CACATTTTGGATAAGCCCAACCCATAAGGA-3′, as described elsewhere [27]. Mutant proteins were purified parallel to the wt enzymes.
RdRp assays with purified enzymes
Standard reaction conditions for the RdRp assay with 0.1 µM of purified SARS-CoV nsp12 are described elsewhere [27]. To study the effect of Zn2+ in this assay, 0.5 µl of a dilution series of 0–80 mM ZnOAc2 was added to the 5 µl reaction mixture, yielding final Zn2+ concentrations of 0–8 mM. The EAV RdRp assay contained 1 µM nsp9, 1 µM RNA template poly(U)18, 0.17 µM [α-32P]ATP (0.5 µCi/µl; Perkin-Elmer), 50 µM ATP, 20 mM Tris-HCl (pH 8.0), 10 mM NaCl, 10 mM KCl, 1 mM MnCl2, 4 mM MgOAc2, 5% glycerol, 0.1% Triton-X100, 1 mM DTT and 0.5 units RNaseOUT. ZnOAc2 was added to the reaction to give a final concentration of 0–6 mM. To chelate Zn2+ during reactions, MgEDTA was added to a final concentration of 8 mM. Reactions were terminated after 1 hour and analyzed as described [27].
SARS-CoV nsp12 electrophoretic mobility shift assay
SARS-CoV RdRp was incubated with 0.2 nM 5′ 32P-labeled SAV557R/SAV481R RNA duplex, for 10 minutes at 30°C either in presence or absence of 6 mM ZnOAc2. Reactions were analyzed as described previously [27].
Supporting Information
Zdroje
1. LazarczykM
FavreM
2008 Role of Zn2+ ions in host-virus interactions. J Virol 82 11486 11494
2. FredericksonCJ
KohJY
BushAI
2005 Neurobiology of zinc in health and disease. Nat Rev Neurosci 6 449 462
3. AlirezaeiM
NairnAC
GlowinskiJ
PremontJ
MarinP
1999 Zinc inhibits protein synthesis in neurons: potential rol of phosphorylation of translation initiation factor-2a. J Biol Chem 274 32433 32438
4. UchideN
OhyamaK
BesshoT
YuanB
YamakawaT
2002 Effect of antioxidants on apoptosis induced by influenza virus infection: inhibition of viral gene replication and transcription with pyrrolidine dithiocarbamate. Antiviral Res 56 207 217
5. SuaraRO
CroweJEJ
2004 Effect of zinc salts on respiratory syncytial virus replication. Antimicrob Agents Chemother 48 783 790
6. GaudernakE
SeipeltJ
TriendlA
GrassauerA
KuechlerE
2002 Antiviral Effects of Pyrrolidine Dithiocarbamate on Human Rhinoviruses. J Virol 76 6004 6015
7. SiX
McManusBM
ZhangJ
YuanJ
CheungC
2005 Pyrrolidine Dithiocarbamate Reduces Coxsackievirus B3 Replication through Inhibition of the Ubiquitin-Proteasome Pathway. J Virol 79 8014 8023
8. KorantBD
KauerJC
ButterworthBE
1974 Zinc ions inhibit replication of rhinoviruses. Nature 248 588 590
9. PolatnickJ
BachrachHL
1978 Effect of zinc and other chemical agents on foot-and-mouth-disease virus replication. Antimicrob Agents Chemother 13 731 734
10. LankeK
KrennBM
MelchersWJG
SeipeltJ
van KuppeveldFJM
2007 PDTC inhibits picornavirus polyprotein processing and RNA replication by transporting zinc ions into cells. J Gen Virol 88 1206 1217
11. KrennBM
GaudernakE
HolzerB
LankeK
Van KuppeveldFJM
2009 Antiviral Activity of the Zinc Ionophores Pyrithione and Hinokitiol against Picornavirus Infections. J Virol 83 58 64
12. ZalewskiPD
ForbesIJ
BettsWH
1993 Correlation of apoptosis with change in intracellular labile Zn(II) using Zinquin [(2-methyl-8-p-toluenesulphonamide-6-quinolyloxy)acetic acid], a new specific fluorescent probe for Zn(II). Biochem J 296 403 408
13. BaumEZ
BebernitzGA
PalantO
MuellerT
PlotchSJ
1991 Purification, properties, and mutagenesis of poliovirus 3C protease. Virology 165 140 150
14. CordingleyMG
RegisterRB
CallahanPL
GarskyVM
ColonnoRJ
1989 Cleavage of small peptides in vitro by human rhinovirus 14 3C protease expressed in Escherichia coli. J Virol 63 5037 5045
15. FerrariE
Wright-MinogueJ
FangJW
BaroudyBM
LauJY
1999 Characterization of soluble hepatitis C virus RNA-dependent RNA polymerase expressed in Escherichia coli. J Virol 73 1649 1654
16. HungM
GibbsCS
TsiangM
2002 Biochemical characterization of rhinovirus RNA-dependent RNA polymerase. Antiviral Res 56 99 114
17. PerlmanS
NetlandJ
2009 Coronaviruses post-SARS: update on replication and pathogenesis. Nat Rev Micro 7 439 450
18. GorbalenyaAE
EnjuanesL
ZiebuhrJ
SnijderEJ
2006 Nidovirales: evolving the largest RNA virus genome. Virus Res 117 17 37
19. SnijderEJ
BredenbeekPJ
DobbeJC
ThielV
ZiebuhrJ
2003 Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage. J Mol Biol 331 991 1004
20. PasternakAO
SpaanWJ
SnijderEJ
2006 Nidovirus transcription: how to make sense…? J Gen Virol 80 1403 1421
21. SawickiSG
SawickiDL
SiddellSG
2007 A Contemporary View of Coronavirus Transcription. J Virol 81 20 29
22. ButterworthBE
KorantBD
1974 Characterization of the large picornaviral polypeptides produced in the presence of zinc ion. J Virol 14 282 291
23. DenisonMR
PerlmanS
1986 Translation and processing of mouse hepatitis virus virion RNA in a cell-free system. J Virol 60 12 18
24. DenisonMR
ZoltickPW
HughesSA
GiangrecoB
OlsonAL
1992 Intracellular processing of the N-terminal ORF 1a proteins of the coronavirus MHV-A59 requires multiple proteolytic events. Virology 189 274 284
25. van HemertMJ
de WildeAH
GorbalenyaAE
SnijderEJ
2008 The in Vitro RNA Synthesizing Activity of the Isolated Arterivirus Replication/Transcription Complex Is Dependent on a Host Factor. J Biol Chem 283 16525 16536
26. van HemertMJ
van den WormSHE
KnoopsK
MommaasAM
GorbalenyaAE
2008 SARS-Coronavirus Replication/Transcription Complexes Are Membrane-Protected and Need a Host Factor for Activity In Vitro. PLoS Pathog 4 e1000054
27. te VelthuisAJ
ArnoldJJ
CameronCE
van den WormSH
SnijderEJ
2009 The RNA polymerase activity of SARS-coronavirus nsp12 is primer dependent. Nucleic Acids Res 38 203 214
28. BeerensN
SeliskoB
RicagnoS
ImbertI
van der ZandenL
2007 De Novo Initiation of RNA Synthesis by the Arterivirus RNA-Dependent RNA Polymerase. J Virol 81 8384 8395
29. van den BornE
PosthumaCC
KnoopsK
SnijderEJ
2007 An infectious recombinant equine arteritis virus expressing green fluorescent protein from its replicase gene. J Gen Virol 88 1196 1205
30. SimsAC
BurkettSE
YountB
PicklesRJ
2008 SARS-CoV replication and pathogenesis in an in vitro model of the human conducting airway epithelium. Virus Res 133 33 44
31. XuX
LiuY
WeissS
ArnoldE
SarafianosSG
2003 Molecular model of SARS coronavirus polymerase: implications for biochemical functions and drug design. Nucleic Acids Res 31 7117 7130
32. ZhaiY
SunF
LiX
PangH
XuX
2005 Insights into SARS-CoV transcription and replication from the structure of the nsp7-nsp8 hexadecamer. Nat Struct Mol Biol 12 980 986
33. StockmanLJ
BellamyR
GarnerP
2006 SARS: Systematic Review of Treatment Effects. PLoS Med 3 e343
34. ThompsonA
PatelK
TillmanH
McHutchisonJG
2009 Directly acting antivirals for the treatment of patients with hepatitis C infection: A clinical development update addressing key future challenges. J Hepatol 50 184 194
35. De ClercqE
2004 Antivirals and antiviral strategies. Nat Rev Microbiol 2 704 720
36. ThompsonAJV
McHutchisonJG
2009 Antiviral resistance and specifically targeted therapy for HCV (STAT-C). J Viral Hepat 16 377 387
37. YangW
LeeJY
NowotnyM
2006 Making and Breaking Nucleic Acids: Two-Mg2+-Ion Catalysis and Substrate Specificity. Mol Cell 22 5 13
38. CastroC
SmidanskyE
MaksimchukKR
ArnoldJJ
KorneevaVS
2007 Two proton transfers in the transition state for nucleotidyl transfer catalyzed by RNA - and DNA-dependent RNA and DNA polymerases. Proc Natl Acad Sci USA 104 4267 4272
39. ArnoldJJ
GhoshSK
CameronCE
1999 Poliovirus RNA-dependent RNA Polymerase (3Dpol). Divalent cation modulation of primer, template and nucleotide selection. J Biol Chem 274 37060 37069
40. IuchiS
2001 Three classes of C2H2 zinc finger proteins. Cell Mol Life Sci 58 625 635
41. Gomis-RuthXF
2009 Catalytic domain architecture of metzincin metalloproteases. J Biol Chem 284 15353 15357
42. YapTL
XuT
ChenY-L
MaletH
EgloffM-P
2007 Crystal Structure of the Dengue Virus RNA-Dependent RNA Polymerase Catalytic Domain at 1.85-Angstrom Resolution. J Virol 81 4753 4765
43. WinekCL
BuehlerEV
1966 Intravenous toxicity of zinc pyridinethione and several zinc salts. Toxicol Appl Pharmacol 9 296 273
44. SnyderDR
de JesusCP
TowfighiJ
JacobyRO
WedigJH
1979 Neurological, microscopic and enzyme-histochemical assessment of zinc pyrithione toxicity. Food Cosmet Toxicol 17 651 660
45. MagdaD
LecaneP
WangZ
HuW
ThiemannP
2008 Synthesis and anticancer properties of water-soluble zinc ionophores. Cancer Res 68 5318 5325
46. SnijderEJ
van der MeerY
Zevenhoven-DobbeJ
OnderwaterJJM
van der MeulenJ
2006 Ultrastructure and Origin of Membrane Vesicles Associated with the Severe Acute Respiratory Syndrome Coronavirus Replication Complex. J Virol 80 5927 5940
47. StudierF
2005 Protein production by auto-induction in high density shaking cultures. Protein Expr Purif 41 207 234
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek Tick Histamine Release Factor Is Critical for Engorgement and Transmission of the Lyme Disease AgentČlánek TGF-b2 Induction Regulates Invasiveness of -Transformed Leukocytes and Disease SusceptibilityČlánek The Origin of Intraspecific Variation of Virulence in an Eukaryotic Immune Suppressive ParasiteČlánek Glycosylation Focuses Sequence Variation in the Influenza A Virus H1 Hemagglutinin Globular Domain
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2010 Číslo 11- Jak souvisí postcovidový syndrom s poškozením mozku?
- Měli bychom postcovidový syndrom léčit antidepresivy?
- Farmakovigilanční studie perorálních antivirotik indikovaných v léčbě COVID-19
- 10 bodů k očkování proti COVID-19: stanovisko České společnosti alergologie a klinické imunologie ČLS JEP
-
Všechny články tohoto čísla
- Patients with Discordant Responses to Antiretroviral Therapy Have Impaired Killing of HIV-Infected T Cells
- A Molecular Mechanism for Eflornithine Resistance in African Trypanosomes
- Tyrosine Sulfation of the Amino Terminus of PSGL-1 Is Critical for Enterovirus 71 Infection
- Autoimmunity as a Predisposition for Infectious Diseases
- The Subtilisin-Like Protease AprV2 Is Required for Virulence and Uses a Novel Disulphide-Tethered Exosite to Bind Substrates
- Structural Analysis of HIV-1 Maturation Using Cryo-Electron Tomography
- Tick Histamine Release Factor Is Critical for Engorgement and Transmission of the Lyme Disease Agent
- Interferon-Inducible CXC Chemokines Directly Contribute to Host Defense against Inhalational Anthrax in a Murine Model of Infection
- TGF-b2 Induction Regulates Invasiveness of -Transformed Leukocytes and Disease Susceptibility
- The Origin of Intraspecific Variation of Virulence in an Eukaryotic Immune Suppressive Parasite
- CO Acts as a Signalling Molecule in Populations of the Fungal Pathogen
- SV2 Mediates Entry of Tetanus Neurotoxin into Central Neurons
- MAP Kinase Phosphatase-2 Plays a Critical Role in Response to Infection by
- Glycosylation Focuses Sequence Variation in the Influenza A Virus H1 Hemagglutinin Globular Domain
- Potentiation of Epithelial Innate Host Responses by Intercellular Communication
- Fcγ Receptor I Alpha Chain (CD64) Expression in Macrophages Is Critical for the Onset of Meningitis by K1
- ANK, a Host Cytoplasmic Receptor for the Cell-to-Cell Movement Protein, Facilitates Intercellular Transport through Plasmodesmata
- Analysis of the Initiating Events in HIV-1 Particle Assembly and Genome Packaging
- Evolution of Linked Avirulence Effectors in Is Affected by Genomic Environment and Exposure to Resistance Genes in Host Plants
- Structural Basis of HIV-1 Neutralization by Affinity Matured Fabs Directed against the Internal Trimeric Coiled-Coil of gp41
- Hepatitis C Virus (HCV) Evades NKG2D-Dependent NK Cell Responses through NS5A-Mediated Imbalance of Inflammatory Cytokines
- Host Cell Invasion and Virulence Mediated by Ssa1
- Global Gene Expression in Urine from Women with Urinary Tract Infection
- Should the Human Microbiome Be Considered When Developing Vaccines?
- HapX Positively and Negatively Regulates the Transcriptional Response to Iron Deprivation in
- Enhancing Oral Vaccine Potency by Targeting Intestinal M Cells
- Herpes Simplex Virus Reorganizes the Cellular DNA Repair and Protein Quality Control Machinery
- The Female Lower Genital Tract Is a Privileged Compartment with IL-10 Producing Dendritic Cells and Poor Th1 Immunity following Infection
- Zn Inhibits Coronavirus and Arterivirus RNA Polymerase Activity and Zinc Ionophores Block the Replication of These Viruses in Cell Culture
- Cryo Electron Tomography of Native HIV-1 Budding Sites
- Crystal Structure and Size-Dependent Neutralization Properties of HK20, a Human Monoclonal Antibody Binding to the Highly Conserved Heptad Repeat 1 of gp41
- Modelling the Evolution and Spread of HIV Immune Escape Mutants
- The Arabidopsis Resistance-Like Gene Is Activated by Mutations in and Contributes to Resistance to the Bacterial Effector AvrRps4
- Platelet-Activating Factor Receptor Plays a Role in Lung Injury and Death Caused by Influenza A in Mice
- Genetic and Structural Basis for Selection of a Ubiquitous T Cell Receptor Deployed in Epstein-Barr Virus Infection
- Ubiquitin-Regulated Nuclear-Cytoplasmic Trafficking of the Nipah Virus Matrix Protein Is Important for Viral Budding
- Pneumolysin Activates the NLRP3 Inflammasome and Promotes Proinflammatory Cytokines Independently of TLR4
- Immune Evasion by : Differential Targeting of Dendritic Cell Subpopulations
- Survival in Selective Sand Fly Vector Requires a Specific -Encoded Lipophosphoglycan Galactosylation Pattern
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Zn Inhibits Coronavirus and Arterivirus RNA Polymerase Activity and Zinc Ionophores Block the Replication of These Viruses in Cell Culture
- The Female Lower Genital Tract Is a Privileged Compartment with IL-10 Producing Dendritic Cells and Poor Th1 Immunity following Infection
- Crystal Structure and Size-Dependent Neutralization Properties of HK20, a Human Monoclonal Antibody Binding to the Highly Conserved Heptad Repeat 1 of gp41
- The Arabidopsis Resistance-Like Gene Is Activated by Mutations in and Contributes to Resistance to the Bacterial Effector AvrRps4
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



