-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaANK, a Host Cytoplasmic Receptor for the Cell-to-Cell Movement Protein, Facilitates Intercellular Transport through Plasmodesmata
Plasmodesma (PD) is a channel structure that spans the cell wall and provides symplastic connection between adjacent cells. Various macromolecules are known to be transported through PD in a highly regulated manner, and plant viruses utilize their movement proteins (MPs) to gate the PD to spread cell-to-cell. The mechanism by which MP modifies PD to enable intercelluar traffic remains obscure, due to the lack of knowledge about the host factors that mediate the process. Here, we describe the functional interaction between Tobacco mosaic virus (TMV) MP and a plant factor, an ankyrin repeat containing protein (ANK), during the viral cell-to-cell movement. We utilized a reverse genetics approach to gain insight into the possible involvement of ANK in viral movement. To this end, ANK overexpressor and suppressor lines were generated, and the movement of MP was tested. MP movement was facilitated in the ANK-overexpressing plants, and reduced in the ANK-suppressing plants, demonstrating that ANK is a host factor that facilitates MP cell-to-cell movement. Also, the TMV local infection was largely delayed in the ANK-suppressing lines, while enhanced in the ANK-overexpressing lines, showing that ANK is crucially involved in the infection process. Importantly, MP interacted with ANK at PD. Finally, simultaneous expression of MP and ANK markedly decreased the PD levels of callose, β-1,3-glucan, which is known to act as a molecular sphincter for PD. Thus, the MP-ANK interaction results in the downregulation of callose and increased cell-to-cell movement of the viral protein. These findings suggest that ANK represents a host cellular receptor exploited by MP to aid viral movement by gating PD through relaxation of their callose sphincters.
Published in the journal: . PLoS Pathog 6(11): e32767. doi:10.1371/journal.ppat.1001201
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1001201Summary
Plasmodesma (PD) is a channel structure that spans the cell wall and provides symplastic connection between adjacent cells. Various macromolecules are known to be transported through PD in a highly regulated manner, and plant viruses utilize their movement proteins (MPs) to gate the PD to spread cell-to-cell. The mechanism by which MP modifies PD to enable intercelluar traffic remains obscure, due to the lack of knowledge about the host factors that mediate the process. Here, we describe the functional interaction between Tobacco mosaic virus (TMV) MP and a plant factor, an ankyrin repeat containing protein (ANK), during the viral cell-to-cell movement. We utilized a reverse genetics approach to gain insight into the possible involvement of ANK in viral movement. To this end, ANK overexpressor and suppressor lines were generated, and the movement of MP was tested. MP movement was facilitated in the ANK-overexpressing plants, and reduced in the ANK-suppressing plants, demonstrating that ANK is a host factor that facilitates MP cell-to-cell movement. Also, the TMV local infection was largely delayed in the ANK-suppressing lines, while enhanced in the ANK-overexpressing lines, showing that ANK is crucially involved in the infection process. Importantly, MP interacted with ANK at PD. Finally, simultaneous expression of MP and ANK markedly decreased the PD levels of callose, β-1,3-glucan, which is known to act as a molecular sphincter for PD. Thus, the MP-ANK interaction results in the downregulation of callose and increased cell-to-cell movement of the viral protein. These findings suggest that ANK represents a host cellular receptor exploited by MP to aid viral movement by gating PD through relaxation of their callose sphincters.
Introduction
Plasmodesma (PD) is a cell-wall spanning channel that interconnects plant cells and is unique for plants. This structure is characterized by a neck-like constriction at its each end, is lined by the plasma membrane, and is traversed by a strand of appressed endoplasmic reticulum, providing endomembrane and cytoplasmic connections to adjacent cells. PD mediate direct macromolecular exchange between the connected cells, and this transport is highly regulated (for review, see [1], [2], [3]).
PD are also utilized by plant viruses for infection. After initial inoculation and replication in the infected cell, plant viruses spread to the neighboring cells and throughout the entire plant. For this transport, viruses utilize their cell-to-cell movement proteins, MPs [3], [4], [5], [6], [7], [8], [9]. The traffic of viral MPs represents a paradigm and a model system for macromolecular transport through PD. For example, Tobacco mosaic virus (TMV) MP, the archetype of many viral MPs, presumably associates with the viral genomic RNA to form a movement ribonucleocomplex [10], targets this complex to PD [11], [12], [13], [14], and increases the PD size exclusion limit to translocate the movement complex through the PD channel [15], [16].
To date, two cellular factors, actin filaments and callose have been implicated in control of transport through PD [17], [18], [19], [20], [21], [22], [23], [24]. A recent study suggested that the Cucumber mosaic virus and TMV MP may sever the actin filaments to aid their cell-to-cell transport [25], and callose, which accumulates at the neck region of PD [18], represents a molecular sphincter that restricts cell-to-cell transport of macromolecules [18], [19], [20], [21], [22], [23], [24]. The level of callose in the cell wall is primarily determined by a balance between the enzymatic activities of callose synthases and β-1,3-glucanases [26], [27], [28]. Callose deposits presumably affect transport through PD, because their elevated accumulation delays local and systemic movement of different viruses [21], [22], [23], [24]. Thus, it would make a biological sense if plant viruses had evolved a mechanism to regulate, via viral MPs and as yet unknown host factor(s), callose deposits to allow their own movement through PD. So far, however, the existence of such a mechanism has not been demonstrated.
To date, several and very diverse host proteins have been shown to bind viral MPs. For example, cytoskeletal elements, calreticulin, pectin methylesterases, and DnaJ chaperones have all been shown to interact with TMV MP [11], [29], [30], [31], [32], [33], [34]. Yet, none of these factors had any effects on callose deposits at PD. The role of one class of host proteins, ankyrin repeat-containing proteins (ANKs) — also known as TIP1-3, and initially reported to bind MP of the Potato virus X (PVX) in vitro [35] — in viral movement has received no attention since their discovery. Here, we investigated the potential involvement of ANK in regulation of mobility of TMV MP through PD. Our data indicate that ANK interacts with MP and promotes reduction in PD callose deposits and subsequent MP cell-to-cell transport.
Results
ANK promotes MP cell-to-cell movement
To evaluate TMV spread within infected plant tissues, it is useful to employ a plant host that does not develop necrosis upon infection with this virus. Thus, we chose the cultivar of tobacco (Nicotiana tabacum cv. Turk), which lacks the N-gene and, therefore, does not produce a hypersensitive cell death response to TMV [22], [36], [37]. Because the ANK homolog of this cultivar has not been isolated, we cloned its cDNA and compared the predicted amino acid sequence with several known ANK homologs from tobacco and Arabidopsis. Figure 1 shows that the ANK sequence is well conserved in these plants, with the highest degree of identity found in the C-terminal ankyrin repeats (underlined in blue), which represent the protein domains typically involved in protein-protein interactions [38], [39]. N-termini of ANK proteins also carry a loosely-defined PEST domain (underlined in red) [35], [40], [41], which often, but not always, serves as signal for proteolysis [42], [43], [44].
Fig. 1. Amino acid sequence alignment of ANK proteins from different tobacco cultivars and Arabidopsis. 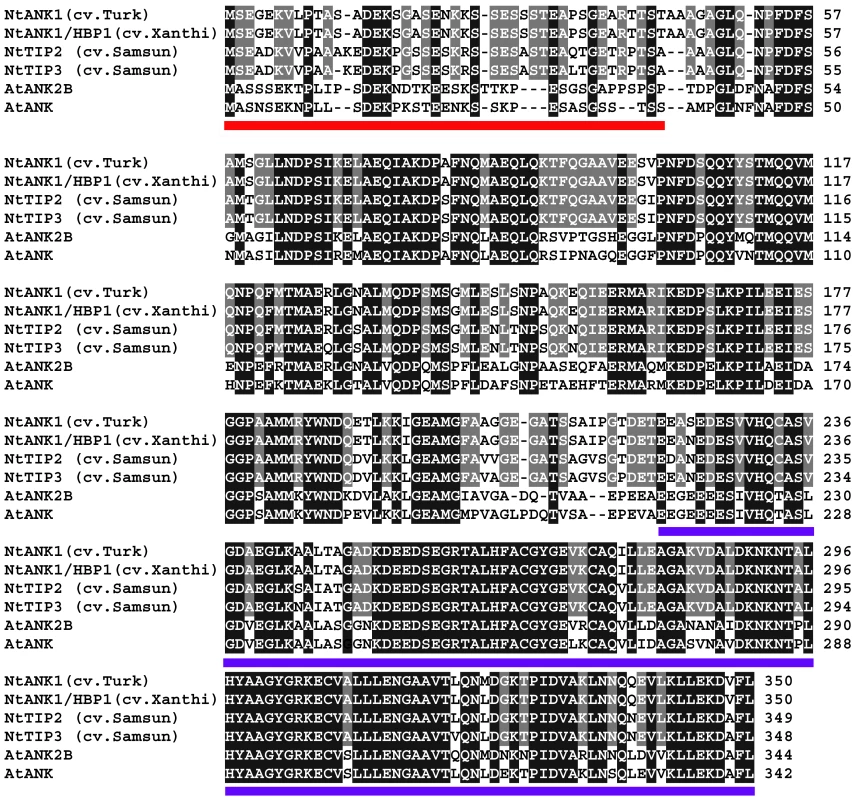
The ANK from N. tabacum cv. Turk (accession number GU320195) was aligned using the T-Coffee program (http://www.ebi.ac.uk/Tools/t-coffee/index.html) with its closest homologs from N. tabacum cv. Xanthi (NtANK1/HBP1, accession number AAK18619/AAN63819), N. tabacum cv. Samsun (NtTIP2 and NtTIP3, accession numbers AAO91861.1 and AAO91862.1, respectively), and Arabidopsis thaliana (AtAKR2B and AtAKR2, accession numbers NP_179331 and NP_849498, respectively). Note that NtTIP1 from N. tabacum cv. Samsun is identical to NtANK1/HBP1 [35]. Amino acid residues identical between tobacco ANKs are highlighted in gray and between all ANK homologs are highlighted in black. The PEST and ankyrin repeat-containing domains are underlined in red and blue, respectively. We then utilized reverse genetics to gain the first insight into the possible involvement of ANK in viral movement. We generated transgenic tobacco plants with RNAi-based suppression of the ANK gene. For specific suppression, we targeted the sequences of ANK that encode the N-terminal part of the protein, rather than its more conserved C-terminal ankyrin motifs (Figure S1). Based on quantitative real time PCR (qPCR) analysis of several independently-transformed lines, we identified severe and moderate suppressors, in which the ANK gene expression levels were reduced to 5% and 15–40%, respectively, of the wild-type expression level (Figure S2A). The severe suppressors also produced a markedly chlorotic phenotype (line RNAi ANK3, Figure S2B), consistent with the known involvement of ANK homologs in chloroplast biogenesis [45]. The moderate suppressors, however, appeared healthy and did not develop any detectible morphological or developmental phenotypes (line RNAi ANK1, Figure S2B). Two such transgenic lines, RNAi ANK1 and RNAi ANK2, were selected for further analyses.
First, to confirm the specificity of the RNAi-based ANK suppression, we examined the expression levels of two genes, ASPARTATE AMINOTRANSFERASE (AATF) and MAGNESIUM PROTOPORPHYRIN IX (MgPP), which are unrelated to ANK, but contain a short, 17-bp nucleotide sequence that is also found in the ANK sequence used for the RNAi suppression (Figure S1). No differences were detected in the expression levels of AATF and MgPP in both RNAi ANK lines and in the wild type plants (Table S1), indicating that the ANK suppression in our RNAi ANK lines was specific.
We then used RNAi ANK1 and RNAi ANK2 to examine the effect of the reduction in the endogenous ANK expression on MP cell-to-cell movement. To this end, MP was tagged with YFP and the encoding constructs introduced into the wild-type and RNAi transgenic lines by microbombardment. The MP-YFP movement was determined by confocal microscopy two days after bombardment. Figure 2A shows that, in the wild-type plants, approximately 60% of the signal was distributed between 2 to 3 cells. These observations were consistent with the known rate of the MP movement in plant tissues [46], [47], and typical for this tobacco cultivar. In contrast, 70–75% of the signal remained confined to a single cell in RNAi ANK1 and RNAi ANK2 lines, indicating substantial decrease in the MP movement. Statistical evaluation of these data by the unpaired two-tailed Student's t-test confirmed that the MP movement capacities in the RNAi ANK1 and RNAi ANK2 plants were similar to each other, but different from those in the wild-type plants (Figure 2A). MP was found associated with PD in a punctate pattern characteristic for PD localization in both RNAi ANK1 (Figure 2B, D–F) and in RNAi ANK2 plants (not shown), as in wild-type (Figure 2C) [46], [48], [49], [50], [51]. Importantly, the MP colocalized with a PD marker, the PD callose binding protein (PDCB) [52] (Figure 2D–F), further confirming the targeting of the viral protein to PD in RNAi ANK background. MP localization to PD was also confirmed in both wild-type and RNAi ANK backgrounds at different time periods after bombardment (Figure S3A, B, D, E); furthermore, the expression levels of MP-YFP in these plants were essentially the same, indicating that the alteration of the ANK amount in the cell does not affect the accumulation of MP (Figure S3G). Thus, the ANK suppression most likely affected MP translocation through PD, rather than its PD targeting or level of expression.
Fig. 2. Reduced cell-to-cell movement of MP-YFP in RNAi ANK1 and RNAi ANK2 plants and colocalization of MP-YFP and PDCB-mCherry. 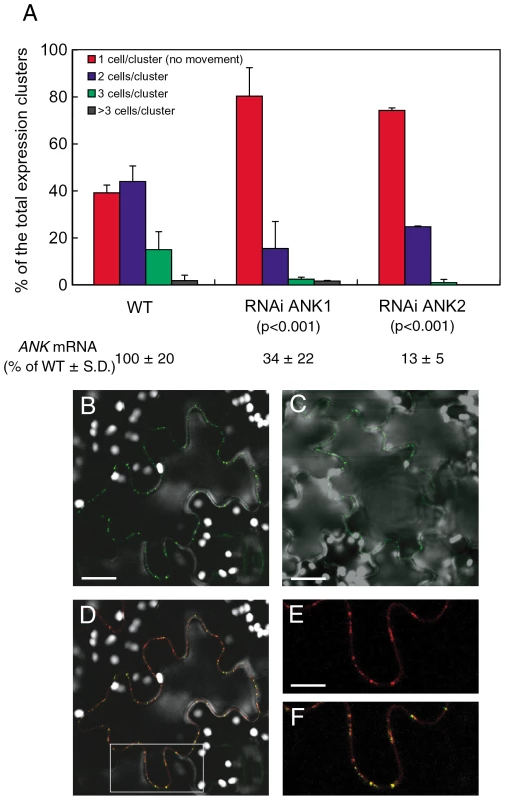
(A) Quantification of MP-YFP movement and ANK transcript levels. Standard deviations (S. D.) and the unpaired two tailed Student's t-test P-values for statistical significance of difference between the RNAi ANK1, RNAi ANK2, and wild-type (WT) plants are indicated. (B–F) Colocalization of MP-YFP and PDCB-mCherry to PD in RNAi ANK1 plants. Panels B, C represent YFP signal and show PD localization of MP-YFP in RNAi ANK1 and wild-type plants, respectively. Bars = 50 µm. Panel D represents merged YFP and mCherry signals and shows colocalization of MP-YFP and PDCB-mCherry in the same cell as in panel B. The area outlined by a white rectangle in panel D is presented at a larger magnification in panels E, F, showing PD localization of PDCB-mCherry and PD colocalization of PDCB-mCherry and MP-YFP, respectively. Bars = 25 µm. Note that MP/PDCB colocalization is not complete because MP preferentially localizes only to secondary PD [13]. YFP signal is in green, mCherry signal is in red, overlapping YFP/mCherry signal is in yellow, and plastid autofluorescence is in white. All images are single confocal sections. MP is known to associate with the host cell ER [53], [54], [55], [56], [57], [58]. Thus, we examined whether ANK is also involved in cell-to-cell transport of ER-associated plant proteins. Specifically, we tested the cell-to-cell diffusion of two of such proteins, the Arabidopsis calnexin (CNX) and calmodulin-regulated Ca2+-ATPase ER membrane protein (ACA2), which move between cells presumably due to membrane diffusion through the PD-spanning ER [53]. Table 1 shows no statistically significant effects of ANK suppression on this cell-to-cell diffusion of YFP-tagged CNX and ACA2 as well as free YFP that diffuses through cytoplasm. Thus, the effect of ANK on the MP movement is specific.
Tab. 1. Cell-to-cell diffusion of CNX and ACA2 in wild-type and RNAi ANK1 and RNAi ANK2 plants. 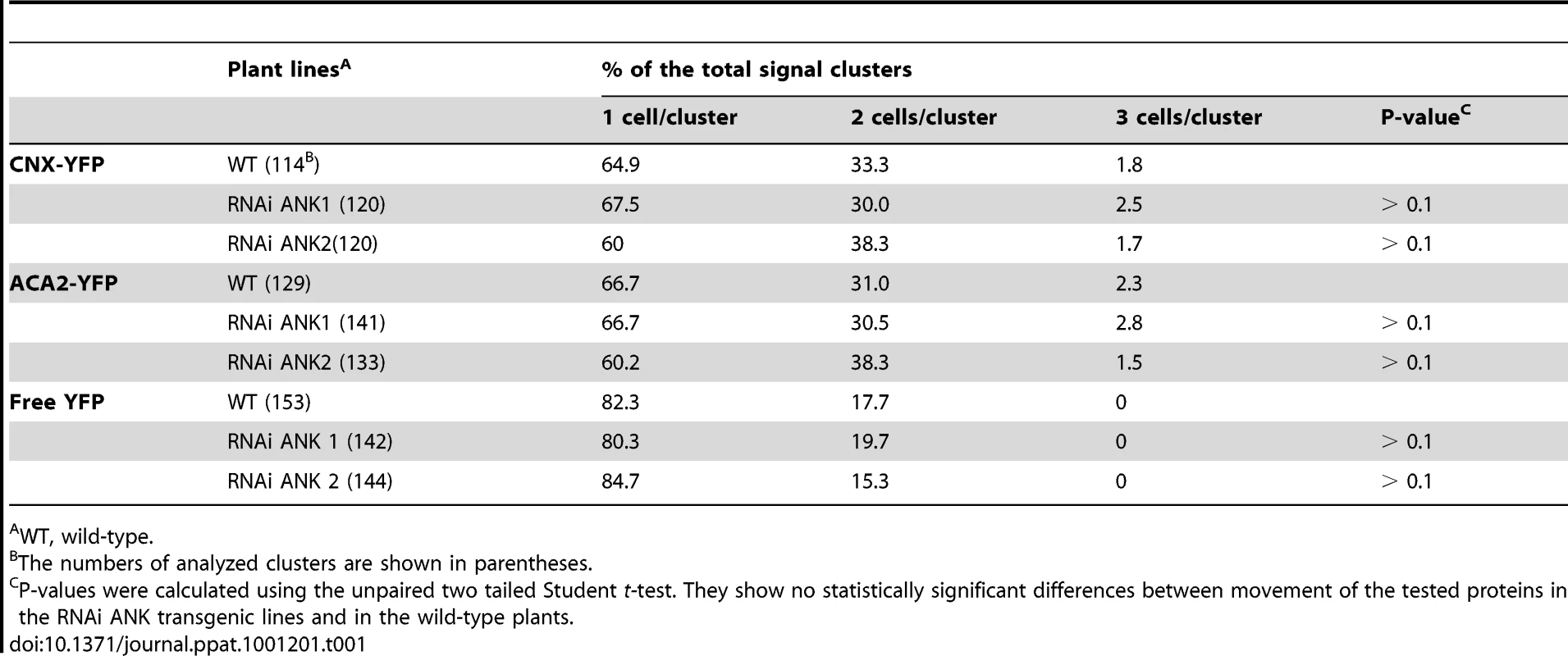
WT, wild-type. That the lack of ANK negatively affects MP movement is suggestive of ANK's involvement in this transport process. To support this notion further, however, it is useful also to demonstrate that overproduction of ANK enhances movement. To this end, we generated another series of transgenic lines, this time, expressing ANK under the control of a constitutive promoter. We then selected two of these lines, ANK1 and ANK2, which exhibited moderate levels of the ANK transcript overexpression (1.5–2 fold, see Figure 3). We could not measure directly the levels of the ANK protein in these lines due to the lack of a specific anti-ANK antibody. Instead, we produced transgenic lines overexpressing ANK tagged with a short (1 kDa) StrepII epitope [59], [60] and demonstrated close correlation between the levels of the ANK-StrepII mRNA and the ANK protein (Figure S4), suggesting that the increased levels of ANK transcription lead to the increased accumulation of ANK protein.
Fig. 3. Enhanced cell-to-cell movement of MP-YFP in ANK1 and ANK2 plants. 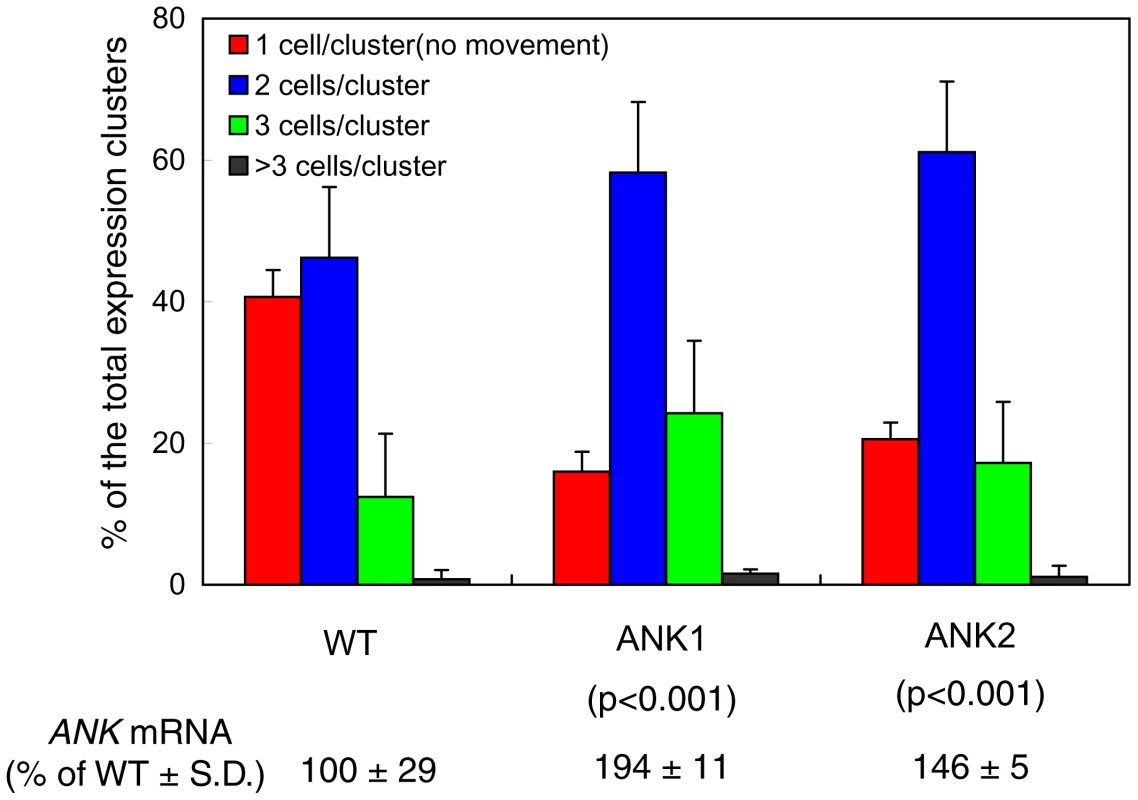
Quantification of MP-YFP movement and ANK transcript levels is shown with standard deviations (S. D.) and the unpaired two tailed Student's t-test P-values for statistical significance of difference between the ANK1 or ANK2 and wild-type (WT) plants. When the ANK1 and ANK2 plants were examined for their ability to support cell-to-cell movement of MP-YFP, the movement was enhanced in both lines. Specifically, whereas 58% of the signal moved in the wild-type plants, the extent of movement in the ANK transgenic plants reached a total of 84–86% (Figure 3). In addition, the extent of the movement was also slightly, but consistently enhanced, with 7% of the signal found in three cell-clusters in the wild-type tissues, and in 11–17% in the ANK1 and ANK2 plants. The unpaired two-tailed Student's t-test confirmed the statistical significance of the differences in cell-to-cell movement of MP between the wild-type and the ANK transgenic plants (Figure 3). As shown in Figure S3, the PD localization pattern or the accumulation level of MP were not affected in these plants. Collectively, these data indicate that the ANK may represent a plant factor that facilitates the MP cell-to-cell movement.
ANK is required for TMV infection
Biologically, suppression of MP mobility should lead to delayed infection, while increase of the MP trafficking through PD is expected to enhance the viral spread. To examine this possibility, we inoculated the ANK overexpressor and suppressor lines as well as the wild type plants with a recombinant virus that carries an autofluorescent tag DsRed (TMV-DsRed) in the place of its coat protein. The local movement of this virus was detected by the appearance and spread of the DsRed signal. Figure 4 shows that whereas the viral transport became visible in all plants approximately at the same time after inoculation (panels A, B), the subsequent viral spread occurred faster in the ANK1 and ANK2 lines than in the wild type plants (panels A, C, E). In contrast, in RNAi ANK1 and RNAi ANK2 lines, the virus moved significantly slower than in wild type plants (Figure 4A, C, D). Importantly, the viral movement in the ANK suppressor lines was delayed, but not arrested, and it reached the wild-type levels late in the infection process (Figure 4F, G). It is important to assertain whether the positive effect of ANK overexpression and negative effect of ANK suppression on the viral movement are not due to increased or decreased replication of the virus in the ANK overexpressor or suppressor lines, respectively. To this end, we produced protoplasts from ANK1, ANK2, RNAi ANK, RNAi ANK2, and wild type plants and infected them with TMV-DsRed. Figure 4H shows that no significant differences were detected in the replication levels of the virus between any of the tested plant lines, suporting the idea that the effect of ANK on the spread of the virus was due to its bona fide effect on the cell-to-cell movement capacity of the viral MP.
Fig. 4. Local movement of TMV-DsRed is facilitated in ANK1 and ANK2 plants and delayed in RNAi ANK1 and RNAi ANK2 plants. 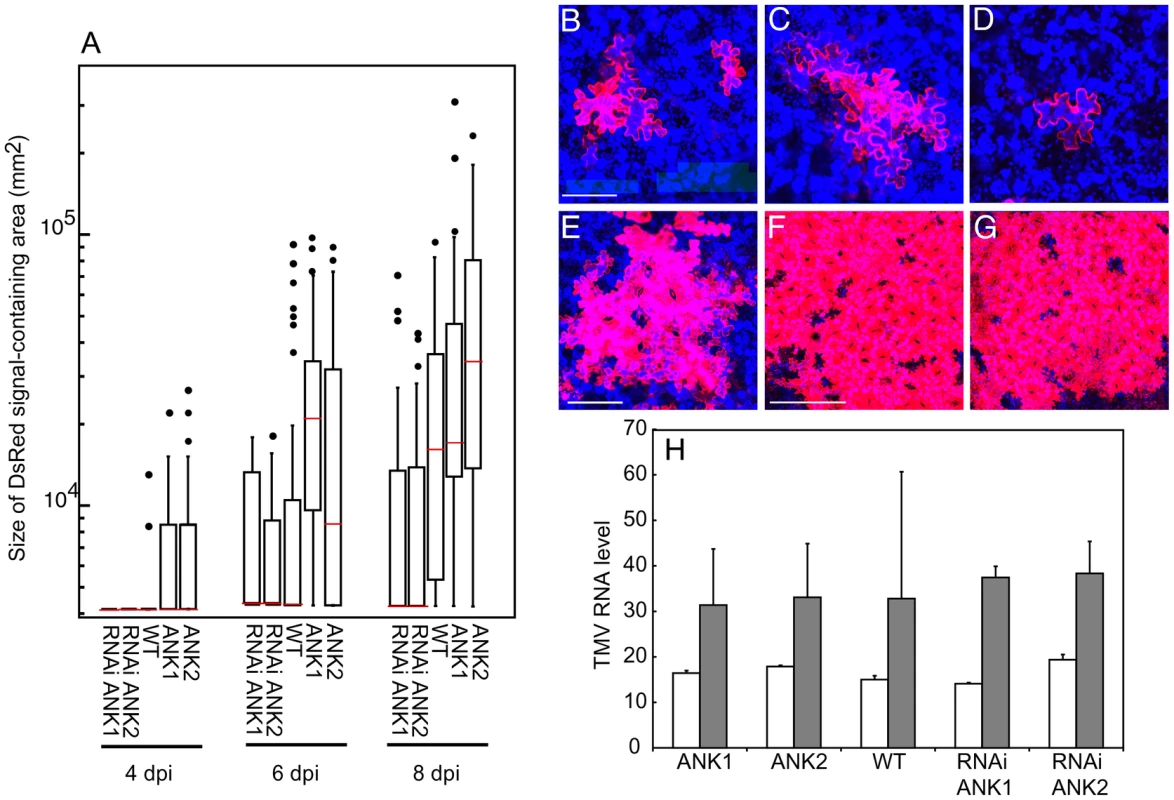
(A) Quantification of TMV-DsRed local movement in the indicated plant lines based on the size of DsRed signal-containing areas at 4, 6, and 8 days post inoculation (dpi). The box-and-whisker plots show the statistical distribution of the data, with horizontal red lines across the boxes representing the median size of the DsRed expression foci, the ends of boxes indicating the quartile results, and the circles representing outliers. Statistical significance of differences between each transgenic line and the wild-type (WT) plants was analyzed by ANOVA and calculated as P<0.0001. (B–G) TMV-DsRed signal in WT plants at 4 and 6 (panels B and C, respectively), RNAi ANK1 plant at 6 dpi (panel D), ANK2 plant at 6 dpi (panel E), WT plant at 10 dpi (panel F) and RNAi ANK1 plant at 15 dpi (panel G). Bars = 100, 200, 400 µm for panels B–D, E, F, respectively. DsRed signal is in red. All images are single confocal sections. (H) Quantification of TMV-DsRed replication efficiency based on the levels of TMV RNA in protoplasts prepared from the indicated plant lines at 16 and 40 h post inoculation (hpi) (open and shaded bars, respectively). The shown values were normalized to the amounts of TUBLIN transcript in the same samples. MP and ANK directly interact at PD
To gain insight into molecular mechanism of the ANK function in MP movement, we first examined the subcellular localizations of both proteins. When MP tagged with YFP and ANK tagged with CFP were transiently coexpressed in tobacco leaves following agroinfiltration, MP displayed the typical punctate pattern of PD localization (Figure 5A, B), whereas ANK was largely cytoplasmic with characteristic transvacuolar strands (Figure 5C, D), consistent with the known cytoplasmic localization of its homologs [41], [45]. Thus, the majority of the ANK population does not colocalize with MP. Furthermore, the presence of ANK did not detectibly alter the MP localization. Indeed, the MP localization pattern in the coexpressing cells (indicated with single asterisks) was essentially identical to that in the neighboring cells (indicated with double asterisks, Figure 5A–D). Taken together with the data in Figure 2 (panels B–F) and Figure S3 (panels A–F), these observations indicate that ANK does not affect PD targeting of MP.
Fig. 5. Subcellular localization of MP and ANK and interaction between MP and ANK in planta and in vitro. 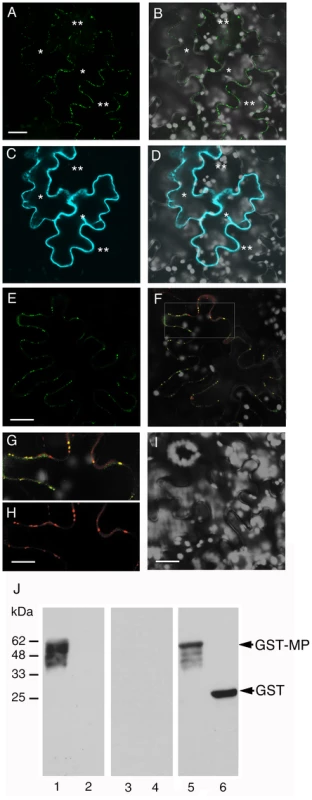
(A, B) PD localization of MP-YFP coexpressed with CFP-ANK. (C, D) Cytoplasmic localization of CFP-ANK coexpressed with MP-YFP. Single asterisks indicate initially expressing cells, and double asterisks indicate cells to which MP-YFP has moved. Panels A, C represent YFP and CFP signals, respectively, and panels B, D represent merged images of YFP (in green) or CFP (in blue) and plastid autofluorescence (in white) signals. Images are projections of three confocal sections. Bars = 20 µm. (E–H) BiFC assay for the MP-ANK interaction and colocalization of MP-cYFP/nYFP-ANK complexes and PDCB-mCherry in planta. Panel E represents YFP signal and shows interaction between MP-cYFP and nYFP-ANK and PD localization of the interacting proteins. Bar = 20 µm. Panel F represents merged YFP and mCherry signals and shows colocalization of MP-cYFP/nYFP-ANK and PDCB-mCherry in the same cell as in panel E. The area outlined by a white rectangle in panel F is presented at a larger magnification in panels G, H, showing PD colocalization of MP-cYFP/nYFP-ANK and PD localization of PDCB-mCherry, respectively. Bar = 10 µm. (I) MP-cYFP does not interact with nYFP-NADK3 in the BiFC assay. Bar = 20 µm. YFP BiFC signal is in green, mCherry signal is in red, overlapping YFP/mCherry signal is in yellow, and plastid autofluorescence is in white. Images are single confocal sections. (J) Renatured gel blot overlay assay for the MP-ANK interaction in vitro. Lane 1, GST-MP + ANK-StrepII; lane 2, GST + ANK-StrepII; lane 3, GST-MP + NADK3-StrepII; lane 4, GST + NADK3-StrepII. Lanes 1–4 were probed with the anti-StrepII antibody. Lane 5, GST-MP + ANK-StrepII; lane 6, GST + ANK-StrepII. Lanes 5, 6 were probed with the anti-GST antibody. Arrows indicate the positions of GST-MP and unfused GST. The numbers on the left indicate molecular mass standards in thousands of Daltons. Although the expression pattern of ANK is different from that of MP, MP is known to possesses cytoplasmic domain [56], [58], providing physical basis for potential interaction between a proportion of the ANK population and MP. Thus, we tested whether ANK can directly recognize and bind MP in vivo. To this end, we utilized the bimolecular fluorescence complementation (BiFC) assay in planta [61]. To date, BiFC represents one of the best assays for protein-protein interactions and subcellular localization of the interacting proteins within living cells. In this approach, proteins are tagged with N-terminal (nYFP) and C-terminal (cYFP) halves of YFP, neither of which fluoresces on its own. Interaction of the tagged proteins results in reconstruction of the YFP signal [62]. In addition, to avoid potentially non-specific effects on protein overexpression on the interaction [63], MP-cYFP was expressed from a relatively weak nopaline synthase promoter [64]. Under these conditions, coexpression of MP-cYFP and nYFP-ANK in tobacco leaf epidermal cells resulted in appearance of strong YFP signal, indicating protein interaction (Figure 5E–G). Importantly, this YFP signal faithfully colocalized with the PDCB-mCherry (Figure 5F–H), clearly demonstrating that the MP-ANK interaction occurs at PD.
The specificity of the MP-ANK interaction was verified in negative control experiments, for which we chose two unrelated cytoplasmic proteins similar in size to ANK (37 kDa), the Arabidopsis cytoplasmic NADH kinase (NADK3, 36 kDa) [65], and a fragment of the bacterial ß-glucuronidase (GUS, 37 kDa). Neither NADK3 (Figure 5I) nor the GUS fragment (not shown) promoted reconstruction of the YFP signal when they were tagged with nYFP and coexpressed with MP-cYFP. Thus, the in vivo interaction between MP and ANK at PD was specific.
This BiFC data were confirmed by an independent approach, using a renatured gel blot overlay assay for protein interaction [33]. Figure 5J shows that ANK tagged with StrepII specifically interacted with the membrane-immobilized recombinant MP tagged with glutathione S-transferase (GST) (lane 1), whereas no such binding was observed to the immobilized unfused GST (lane 2). Binding of MP to ANK was specific because it did not occurred between MP and NADK3 (Figure 5J, lanes 3, 4) or between MP and CNX or ACA2 (not shown). Thus, MP can specifically recognize and bind ANK both in vivo and in vitro.
ANK, in concert with MP, down-regulate callose deposits at PD
Arabidopsis ANK2 is involved in ROS scavenging through its interaction with ascorbate peroxidase [40], and ROS may, in turn, affect PD transport [66], [67]. Thus, we tested whether expression of ANK alone or together with the MP alters the ROS content of plant tissues. Figure 6A shows histochemical staining for ROS revealed no detectible differences in ROS content between the control, mock-transformed tissues and those expressing ANK or ANK and the MP. Also, the ROS levels in transgenic plants, both ANK overexpressor and suppressor lines, were not significantly different from wild-type lines (Figure S5). Most likely, therefore, ANK facilitates MP movement through the mechanism independent from ROS.
Fig. 6. Effects of coexpression of ANK and MP on the content of ROS and callose in plant tissues. 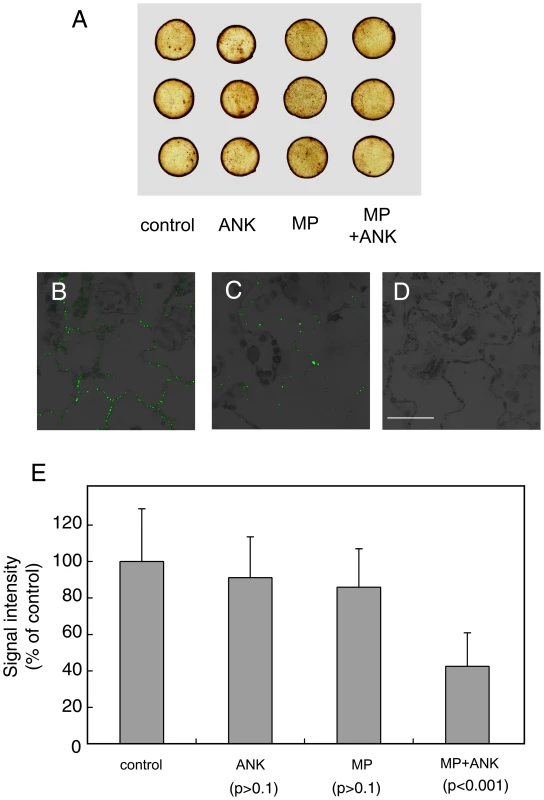
(A) Histochemical detection of ROS. (B) Immunodetection of callose deposits at PD in control, mock-transformed plant tissues. (C) Immunodetection of callose deposits at PD following coexpression of ANK and MP. (D) Immunostaining in the absence of the primary, anti-callose antobody in control tissue. Panels B–D represent merged images of the specific signal from callose staining (green) and tissue autofluorescence signal (black and grey). Bar = 50 µm. (E) Quantification of callose deposits at PD with indicated standard deviations and the unpaired two tailed Student's t-test P-values for statistical significance of difference between plants coexpressing ANK and MP and plants expressing separately either ANK or MP or the control plants. Another possibility is that ANK is involved in regulation of callose sphincters of PD. Callose deposits are known to affect viral movement through PD [20], [21], [22], [23], [24], presumably functioning as a sphincter which physically restricts PD mediated macromolecular trafficking. Thus, ANK may facilitate MP movement by reducing the PD callose deposits. To test this idea, we transiently expressed ANK and MP separately or together with each other in tobacco leaves. The amounts of callose in the expressing tissues were assayed by immunostaining using anti-callose antibody with previously demonstrated specificity [68], [69], followed by quantitative confocal microscopy.
Figure 6 shows that this technique readily visualizes callose deposits in a typical distribution pattern specific for PD with virtually no background signal (panels B and C), and no signal at all in the absence of the primary, anti-callose antibody (panel D). Quantification of the callose-specific signal (Figure 6E) revealed that expression of ANK alone or MP alone resulted in virtually no reduction in callose levels as compared to control tissues. However, when both ANK and MP were coexpressed in the same tissues, the callose-specific signal decreased substantially (Figure 6C, E), to as low as 37% of the control. These results were statistically significant as demonstrated by the unpaired two tailed Student's t-test (Figure 6E). Thus, ANK most likely potentiates MP movement through PD by reducing PD callose deposits, and this down-regulation of the callose content requires the presence of both ANK and MP.
Discussion
The molecular mechanism by which MP gates PD for viral trafficking remains obscure even two decades after the discovery of this activity of MP [15]. This is mainly due to the lack of knowledge about the host factor(s) with which MP interacts during the PD gating process. Once such factor, actin, is suggested by a recent study showing that several viral MPs may sever actin filaments to relax PD during their cell-to-cell movement [25]. Here, we characterized another cellular factor, a tobacco cytoplasmic protein ANK, which likely facilitates MP transport through PD. Our data demonstrate direct interaction between ANK and MP in vivo and in vitro and indicate that this interaction results in reduction of callose deposits at PD and enhanced cell-to-cell movement of MP. Conversely, ANK deficit results in reduced movement of MP and slower spread of viral infection. Mechanistically, ANK promotes increase in PD transport by reducing callose deposits at PD and, by implication, relaxing the callose sphincter around the PD channel.
There are two interesting aspects to this activity of ANK. First, it binds and affects PD transport of MP, which is known to target specifically to PD and gate and traverse them [13], [15], [16]. On the other hand, it does not bind to or affect PD transport of proteins, such as CNX, ACA2 and free YFP, that do not specifically localize to PD and move through them presumably by simple diffusion which does not involve PD gating [53]. Thus, ANK most likely is involved in active PD gating, but not in mere endomembrane-mediated diffusion through PD. Second, ANK cannot elicit its effects on PD transport alone, but does so in concert with MP. Thus, MP does not simply use the existing PD transport pathway, but actively interacts with the host components and participates in the gating activity. MP, thought to associate with endomembranes [54], [55], [57], [70], interacts with ANK most likely through its cytoplasmic domains [56], [58]. Because MP specifically targets to PD [13], it may help direct ANK, a predominantly cytoplasmic protein, to PD.
ANK is a multifunctional protein, the roles of which in different plant species are just beginning to emerge. In Arabidopsis, there are two highly conserved ANK proteins, both of which are involved in chloroplast biogenesis by binding to its outer envelope membrane proteins and delivering them to their destination [45]. The Arabidopsis AKR2 also participates in ROS scavenging via interaction with ascorbate peroxidase [40]. Also, both Arabidopsis and tobacco ANKs are involved in disease resistance against Pseudomonas syringe [40], [41], and ANK/HBP1/TIP1 from the N-gene containing N. tabacum cv. Xanthi in hypersensitive cell death response to TMV [41]. ANK/HBP1/TIP1 and two ANKs from N. tabacum cv. Samsun, i.e., TIP2 and TIP3, may bind MP of PVX; these interactions, however, were shown only in the yeast two-hybrid system, and potential involvement of these ANKs in potexviral movement has not been explored [35]. Finally, also in the yeast two-hybrid system, ANK/HBP1/TIP1, TIP2 and TIP3 interact with vacuolar β-1,3-glucanases [35], [71]; because these ANKs and β-1,3-glucanases reside in different subcellular compartments, this interaction, presumably, is not biological [72]. Furthermore, all known β-1,3-glucanases that might degrade callose are predicted to be either vacuolar or secreted [28], [73], [74], [75], [76], [77], [78], [79], [80] and are not known to possess cytoplasmic domains. The predicted subcellular localization suggests that it is physically impossible for these enzymes to interact with cytoplasmic ANK. One possibility is that ANK may be crucially involved during the targeting of β-1,3-glucanases containing ER-derived vesicles by MP to PD, as proposed recently [3], [53], by directly binding to MP. Another possible target for the ANK activity is cellular callose synthases. Callose is presumed to have a rapid turnover [81], and active callose synthases are likely critical to maintain the PD callose deposits. These enzymes usually contain several subunits, many of which have not been sufficiently characterized; however, at least some of them have been predicted to represent transmembrane proteins with cytoplasmic domains [82], [83], [84]. This topology of callose synthases would allow interactions with ANK and/or ANK - MP complexes. Regardless of its potential downstream targets in the molecular pathway of PD gating, ANK most likely represents a cellular factor that recognizes MP and acts synergistically with it to gate PD and mediate MP transport through these channels.
Methods
ANK cDNA
Total RNA was extracted from the N. tabacum cv. Turk plants by Tri-Reagent (Ambion), and its cDNA was synthesized using the RevertAid First Strand cDNA Synthesis Kit (Fermentas) with the oligo-dT primer. The full-length ANK cDNA was obtained by PCR using the primer pair 5′GCGAATTCTATGTCTGAGGGAGAGAAAGTTTTGCC3′/5′TCCCCCGGGTCACAGAAACACATCTTTCTCGAGCAG3′ and the total cDNA as template.
Vectors for generation of transgenic plants and expression of untagged ANK and MP
For pANK, the PCR-amplified ANK cDNA was inserted into the EcoRI-SmaI sites of pSAT4A-35SP-MCS-35ST [85]. For pRNAi-ANK, the ACTIN intron 11 sequence from pSK-int [86] was first inserted into the HindIII-EcoRI sites of pSAT4A-35SP-MCS-35ST, resulting in pRNAi. To suppress the ANK gene specifically, we designed RNAi constructs that target the unique ANK cDNA sequences located between positions 1 and 406. These sequences of ANK were obtained from pANK by digesting it with BamHI-HindIII and BamHI-EcoRI and cloned into the BglII-HindIII and EcoRI-BamHI sites, respectively, of pRNAi. The resulting pRNAi-ANK construct carried the inverted repeat of the first 406 base pairs of the ANK cDNA, flanked by the ACTIN intron 11 sequences.
The entire expression cassettes from pRNAi-ANK and pANK were then cloned as I-SceI fragments into the binary vector pRCS2-nptII, which contains a kanamycin resistance selection marker [87], resulting in pRCS-RNAi-ANK and pRCS-ANK, respectively.
For pTMV MP, the MP coding sequence was PCR-amplified using the primer pair 5′AAGAATTCATGGCTCTAGTTGTTAAAGGAAAAGTG3′/5′TGCATCCCGGGTTAAAACGAATCCGATTCGGCGACAGT3′ and pTMV004 [88] as template. The amplified product was digested with the EcoRI and SmaI and inserted into the same sites of pSAT4A-nosP-MCS-nosT, which was constructed by transferring the nosP-MCS-nosT expression cassette as an AgeI-NotI fragment of pSAT2A-nosP-MCS-nosT [85] into the same sites of pSAT4-EGFP-C1 [87], replacing the EGFP-C1 expression cassette. Then, the expression cassette from pTMV MP was cloned as an I-SceI fragment into pRCS2-nptII, resulting in pRCS-TMV MP.
Vectors for movement assay, subcellular localization, and BiFC
To construct pCFP-ANK and pnYFP-ANK, the PCR-amplified ANK cDNA was digested with EcoRI and SmaI, and the resulting fragment cloned into the same sites of pSAT4-ECFP-C1 and pSAT4-nEYFP-C1 [61], respectively. pSAT4-ECFP-C1 was made by transferring the ECFP-C1 expression cassette as an AgeI-NotI fragment of pSAT6-ECFP-C1 [87] into the same sites of pSAT4-EGFP-C1 [87], replacing the EGFP-C1 expression cassette. Then, the expression cassettes from pCFP-ANK and pnYFP-ANK were cloned as I-SceI fragments into pRCS2-nptII, resulting in pRCS-CFP-ANK and pRCS-nYFP-ANK.
For pTMV MP-YFP and pTMV MP-cYFP, the TMV MP-encoding sequence was first PCR-amplified using the primer pair 5′AAGAATTCATGGCTCTAGTTGTTAAAGGAAAAGTG3′/5′GGCCGGTACCGAAACGAATCCGATTCGGCGACAGT3′ and pTMV004 as template. The amplified fragment was digested with EcoRI and KpnI, inserted into the EcoRI-XbaI sites of pSAT4A-nosP-MCS-nosT by double-ligation with the YFP and cYFP coding sequences excised with KpnI-XbaI from pSAT4-EYFP-N1 and pSAT4-cEYFP-N1 [61], respectively. pSAT4-EYFP-N1 was made by transferring the EYFP-N1 expression cassette as an AgeI-NotI fragment of pSAT6-EYFP-N1 [87] into the same sites of pSAT4-EGFP-C1 [87], replacing the EGFP-C1 expression cassette. Then, the expression cassettes from pTMV MP-YFP and pTMV MP-cYFP were cloned as I-SceI fragments into pRCS2-nptII, resulting in pRCS-TMV MP-YFP and pRCS-TMV MP-cYFP.
For pnYFP-NADK3, NADK3 cDNA sequence was obtained by PCR using primers 5′GCGAATTCATGGCGATTAGGAAGCTTTTGCTTCTTTTG3′/5′CATCCCGGGCTAGTACCTTGATCTGATCTGAG3′ and Arabidopsis cDNA library [89] as a template. The PCR-amplified NADK3 cDNA was digested with EcoRI and SmaI, and the resulting fragment was cloned into the same sites of pSAT4-nEYFP-C1 [61].
Then, the expression cassette from pnYFP-NADK3 was cloned as an I-SceI fragment into pRCS2-nptII, resulting in pRCS-nYFP-NADK3.
For pCNX-YFP and pACA2-YFP, the CNX and ACA2 cDNAs were amplified from the Arabidopsis cDNA library [89] using the respective sets of primers, 5′GCGAATTCTATGAGACAACGGCAACTATTTTCCG3′/5′GGCCGGTACCATTATCACGTCTCGGTTGCCTTTTGC3′, and 5′GCGAATTCTATGGAGAGTTACCTAAACGAGAAT3′/5′GGCCGGTACCAACGGGAATCGTCTTCAGTCCAGCG3′. The amplified products were digested with EcoRI and KpnI, and cloned into the same sites of pSAT4A-YFP-N1.
The PDCB-mCherry expression construct was kindly provided by Dr. Maule [52].
Vectors for renatured gel blot overlay assay
The StrepII coding sequence [59] was included in the reverse primer of the primer pair 5′ GCGAATTCTATGTCTGAGGGAGAGAAAGTTTTGCC 3′/5′GCGGCCGCTTATTTTTCAAACTGCGGATGGCTCCAGGTACCCAGAAACACATCTTTCTCGAGCAG3′ and pANK as template. The StrepII coding sequence was also included in the reverse primer of the primer pair 5′GCGAATTCATGGCGATTAGGAAGCTTTTGCTTCTTTTG3′/5′GCGGCCGCTTATTTTTCAAACTGCGGATGGCTCCAGTACCTTGATCTGATCTGAGA3′ and used to amplify the NADK3 cDNA from the pnYFP-NADK3. The amplified products were digested with EcoRI and EagI and inserted into the same sites of pET28c(+) (Novagen), producing pET28-ANK-StrepII and pET28-NADK3-StrepII, respectively.
To produce GST-MP, the MP coding sequence was excised from pTMV MP by digesting it with EcoRI and SmaI inserted into the same sites of pGEX-5X-1 (GE Healthcare Life Sciences), producing pGEX-TMV MP.
All PCR reactions were performed using a high-fidelity proofreading Pfu Turbo DNA polymerase (Stratagene), and their products were verified by DNA sequencing using an Applied Biosystems 3730 Genetic Analyzer.
Generation of transgenic plants with suppressed or enhanced ANK expression
pRCS-RNAi-ANK and pRCS-ANK were introduced into the disarmed Agrobacterium strain EHA105, which was then used to transform N. tabacum cv. Turk as described [90]. The resulting transgenic plants were selected on kanamycin-containing media, and maintained according to our standard protocol [22]. The transgenic plants were vegetatively propagated, and the suppression or overexpression of ANK in the ANK and RNAi ANK lines, respectively, was monitored by reverse transcription (RT) followed by quantitative (q) PCR. Control, wild-type tobacco plants were grown as described [22]. All plants were transferred to soil and grown in an environment-controlled chamber at 22–24°C under long day conditions of 16 h white light (70–80 µmol photons m-2 s-1) and 8 h dark. All experiments utilized 5–6-week-old plants with 6–8 leaves.
RT-PCR and qPCR
Total RNA was extracted from tissue samples using Tri-Reagent (Ambion), treated with RNase-free DNase I (Fermentas), and 0.5-µg samples of the resulting preparations were reverse-transcribed using the RevertAid First Strand cDNA Synthesis Kit (Fermentas), according to the manufacturer's instructions. RT reaction products were amplified by qPCR in a 7300 Real-Time PCR System (Applied Biosystems) with Maxima™ SYBR Green qPCR Master Mix (Fermentas) using primer pairs specific for ANK (5′AGGCTGCACTAACTGCTGGT3′/5′TTACAGCGGCTCCATTCTCT3′), MP (5′AAAGATTTCAGTTCAAGGTCGTTCC 3′/5′TCCGTCTCTCACGTTTGTAATCTTC3′), ACTIN (5′TCACTGAAGCACCTCTTAACC3′/5′CAGCTTCCATTCCAATCATTG3′), and TUBLIN(5′TACACAGGGGAAGGAATGG/CTCGAAACCAACGCTTATC3′).
Relative abundance of the ANK or MP mRNA-specific products was normalized to the amount of the product specific for ACTIN and TUBLIN, respectively, which represented an internal control of a constitutively expressed gene. The absence of potential residual genomic DNA contamination was confirmed by control qPCR reactions performed without RT.
Agroinfiltration and microbombardment
For agroinfiltration, binary vectors were introduced into the Agrobacterium EHA105 strain as described [91], grown overnight at 28°C, and infiltrated into intact N. tabacum leaves as described [92], [93]. Microbombardment experiments were performed as described [46]. Briefly, 100 µg of DNA preparations was adsorbed onto 10 mg of 1-µm gold particles, which were then microbombarded into the lower leaf epidermis at a pressure of 80–110 psi, using a portable Helios gene gun system (Model PDS-1000/He, Bio-Rad). All experiments were repeated at least three times.
Cell-to-cell movement assay
Leaves with the length of 18 cm, excluding petiole, from 4–5 week-old plants were selected for all experiments. Constructs expressing YFP-tagged tested proteins, i.e., pTMV MP-YFP, pCNX-YFP and pACA2-YFP, were microbombarded into the lower epidermis at the equivalent locations on each leaf. When movement of two proteins was compared, their expression constructs were introduced at the symmetrical locations relatively to the mid-rib of the leaf. At least 120 YFP-expressing clusters in each microbombarded tissue were observed under a Zeiss LSM 5 Pascal confocal microscope. After 24 h, all expression clusters were represented by single cells and considered to indicate the absence of movement. After 48 h, the number of fluorescent cells in each expression cluster varied due to cell-to-cell movement. At this time, the number of cells in each expression cluster was recorded, followed by statistical evaluation of resulting data by the unpaired two-tailed Student's t-test. Differences between sets of measurements with p-values less than 0.001, corresponding to the statistical probability of greater than 99.9%, were considered statistically significant.
TMV infection assay
pTRBO-DsRed (kindly provided by Dr. Jens Tilsner, University of Edinburgh) was agroinfiltrated into 18-cm, excluding petiole, leaves from 4–5 week-old plants. Four to 14 days after inoculation, at least 40 DsRed-expressing clusters in each infiltrated tissue were analyzed by confocal microscopy, followed by statistical evaluation of the DsRed-expressing surface area measurements by the unpaired two-tailed Student's t-test. Differences between sets of measurements with P-values <0.001, corresponding to the statistical probability of greater than 99.9%, were considered statistically significant.
Protoplast preparation and TMV RNA replication assay
Protoplasts were prepared from the 3–8-cm, excluding petiole, leaves from 4–5 week old plants. The leaves were sterilized in 50% bleach with 0.1% SDS for 5 min, followed by 5 rinses in distilled water and overnight incubation at room temperature in the protoplasting enzyme mixture (40 mg/ml cellulase Onozuka R-10 (Phytotechnology Laboratories) and 1.5 mg/ml macerozyme (MP Biomedicals) in 500 mM D-sorbitol, 1 mM CaCl2, 5 mM MES, pH 5.5). The protoplasts were collected by centrifugation at 600 x g for 5 min, resuspended in the MMC buffer (0.7 M mannitol, 10 mM CaCl2, 5 mM MES, pH 5.8) at 2×107 cells/ml. For transformation with the viral infectious clone, 5 ml of 30% PEG 8000 in the MMC buffer and 4 µg of the pTRBO-DsRed DNA were added to 2 ml of protoplast suspension and mixed gently. After 15-min incubation at room temperature in the dark, 40 ml of the culture medium (3% sucrose, 500 mM D-mannitol, MS salts, 5 mM MES, pH 5.7) was added to the protoplast suspension. The incubation was continued for an additional 45 min, after which the protoplasts were collected by centrifugation, resuspended in 35 ml of the culture medium, and incubated further to allow replication of the virus. Protoplast samples (5 ml) were harvested 16, 28, 40, 52, and 64 h after transformation, and their total RNA was extracted by the TRIzol (Invitrogen) method according to the manufacturer's instructions.
Subcellular localization and BiFC
For subcellular localization, pTMV MP-YFP and pCFP-ANK were expressed in tobacco leaves following microbombardment, and the subcellular fluorescence pattern was analyzed by confocal microscopy. For BiFC, pRCS-nYFP-ANK and pRCS-TMV MP-cYFP, or pRCS-nYFP-ANK and pRCS-nYFP-NADK3 binary constructs were expressed in tobacco leaves following agroinfiltration, and BiFC was detected by confocal microscopy as described [61]. For the combination of pRCS-nYFP-ANK and pRCS-TMV MP-cYFP, we used Agrobacterium cells at OD600 = 0.0015. For negative controls, bacterial cultures at OD600 of up to 0.6 were used to confirm the absence of reconstructed YFP signal even at very high inocula.
Renatured gel blot overlay assay
Recombinant GST-MP, unfused GST, ANK-StrepII, and NADK3-StrepII were produced as described [94] in BL21(DE3) Escherichia coli strain (Novagen) from the pGEX-TMV MP, pGEX-5X-1, pET28-ANK-StrepII and pET28-NADK3-StrepII vectors. The identity of these proteins was confirmed by western blot analysis using anti-GST (Santa Cruz) and anti-StrepII (Genscript) polyclonal antibodies, and the amounts of these proteins in the total bacterial extracts were estimated by scanning densitometry of the corresponding bands on SDS-polyacrylamide gels stained with Coomassie Brilliant Blue R-250, using the known amounts of BSA as standard. Protein extracts containing 1 µg of GST-MP or unfused GST were resolved on 15% SDS-polyacrylamide gels, followed by eletrotransfer to a nitrocellulose membrane. The membrane-immobilized proteins were incubated with 0.5 µg/ml of ANK-StrepII or NADK3-StrepII and processed as described [32]. Binding of the tested proteins to the immobilized proteins was detected by probing the membranes with anti-StrepII rabbit polyclonal antibody (Genscript), followed by anti-rabbit IgG+M secondary antibody conjugated to HRP.
Detection of ROS
Three days after agroinfiltration with binary constructs expressing the tested proteins, i.e., pRCS-ANK and/or pRCS-TMV MP, leaf disks from the agroinfiltrated areas were excised and transferred to the 10 mM MES (pH 5.6) containing SIGMAFAST 3,3′-diaminobenzidine (DAB) (Sigma-Aldrich). DAB was vacuum-infiltrated into the tissues for 20 min, followed by incubation of the leaf disks in the DAB staining solution for 4 h in the light and 16 h in the dark. After four brief washes in double-distilled water, the stained leaf disks were transferred to 95% ethanol and heated at 55°C to remove chlorophyll. The tissue was rehydrated, and imaged using an EPSON Perfection 4490 photo scanner.
Whole-mount callose staining
Three days after agroinfiltration with pRCS-ANK and/or pRCS-TMV MP, tissue samples (<2×5 mm) from the agroinfiltrated areas were fixed by vacuum infiltration with 4% paraformaldehyde in the PIPES buffer (100 mM PIPES, 5 mM EGTA, 2 mM MgCl2, pH 6.9) followed by overnight incubation at 25°C with gentle agitation. The samples were then washed twice in double-distilled water and depleted of chlorophyll by incubation for 2 h at 25°C with gentle agitation in a graded series of ethanol (25, 50, 75, and 95%). For permeabilization, the dehydrated samples were air-dried, transferred to the PIPES buffer, and freeze-shattered as described [95]. Finally, the samples were transferred to a microscope slide, blocked in 1% BSA in PBS, and reacted with 1/200 dilution of anti-callose mouse monoclonal antibody (Biosupplies, Parkville, Australia) in 1% BSA in PBS, followed by the Alexa-488-conjugated anti-mouse IgG+M secondary antibody (Jackson ImmunoResearch, 1/200 dilution in 1% BSA in PBS). After three 10-min washes in PBS, the immunostained samples were mounted on microscope slide, using BioMount (Electron Microscopy Sciences), with the abaxial side of the leaf facing up and observed under a confocal microscope. To avoid measuring callose induced by the wounding made by the initial step of the sample preparation, the section in the middle of the tissue piece was selected for the observation. For each experiment, the confocal microscopy analysis included five samples immunostained independently. The total signal intensity in a 318×318 µm-area was measured using the fluorescence intensity quantification function of the LSM 5 Pascal software for least five fields per sample or a total 25 fields per experiment. To measure background signal intensities, mock-transformed samples, i.e., tissues infiltrated with Agrobacterium carrying empty expression vector, were immunostained in the absence of the primary antibody. These background signal values were subtracted from the signal values obtained from the samples, the average signal intensities with standard error for the each experiment were calculated and evaluated statistically by the unpaired two-tailed Student's t-test as described for analyses of the cell-to-cell movement data.
Experimental conditions for the supplemental figures
All the procedures for the experiments documented in Figures S1, S2, S3, S4 and S5 and Table S1 are presented in File S1(Supplemental Methods).
Accession numbers
The GenBank accession number for the ANK sequence reported in this paper is GU320195. The accession numbers for the ANK homologs are: NtANK1/HBP1 from N. tabacum cv. Xanthi; AAK18619/AAN63819, NtTIP2 and NtTIP3 from N. tabacum cv. Samsun; AAO91861.1 and AAO91862.1, AtAKR2B and AtAKR2 from Arabidopsis thaliana; NP_179331 and NP_849498. The accession numbers for the two genes that contains 17-bp identical to the sequence found in ANK are: N. tabacum AATF; AB126259.1, and MgPP; AF014052.1.
Supporting Information
Zdroje
1. OparkaKJ
2005 Plasmodesmata. Oxford Blackwell Publishing 311
2. MauleAJ
2008 Plasmodesmata: structure, function and biogenesis. Curr Opin Plant Biol 11 680 686
3. EpelBL
2009 Plant viruses spread by diffusion on ER-associated movement-protein-rafts through plasmodesmata gated by viral induced host beta-1,3-glucanases. Semin Cell Dev Biol 20 1074 1081
4. RheeY
TzfiraT
ChenMH
WaigmannE
CitovskyV
2000 Cell-to-cell movement of tobacco mosaic virus: enigmas and explanations. Mol Plant Pathol 1 33 39
5. WaigmannE
UekiS
TrutnyevaK
CitovskyV
2004 The ins and outs of non-destructive cell-to-cell and systemic movement of plant viruses. Crit Rev Plant Sci 23 195 250
6. LazarowitzSG
BeachyRN
1999 Viral movement proteins as probes for intracellular and intercellular trafficking in plants. Plant Cell 11 535 548
7. LucasWJ
2006 Plant viral movement proteins: agents for cell-to-cell trafficking of viral genomes. Virology 344 169 184
8. BeachyRN
HeinleinM
2000 Role of P30 in replication and spread of TMV. Traffic 1 540 544
9. BoevinkP
OparkaKJ
2005 Virus-host interactions during movement processes. Plant Physiol 138 1815 1821
10. CitovskyV
KnorrD
SchusterG
ZambryskiPC
1990 The P30 movement protein of tobacco mosaic virus is a single-strand nucleic acid binding protein. Cell 60 637 647
11. HeinleinM
EpelBL
PadgettHS
BeachyRN
1995 Interaction of tobamovirus movement proteins with the plant cytoskeleton. Science 270 1983 1985
12. TomeniusK
ClaphamD
MeshiT
1987 Localization by immunogold cytochemistry of the virus coded 30 K protein in plasmodesmata of leaves infected with tobacco mosaic virus. Virology 160 363 371
13. DingB
HaudenshieldJS
HullRJ
WolfS
BeachyRN
1992 Secondary plasmodesmata are specific sites of localization of the tobacco mosaic virus movement protein in transgenic tobacco plants. Plant Cell 4 915 928
14. MeshiT
WatanabeY
SaitoT
SugimotoA
MaedaT
1987 Function of the 30 kd protein of tobacco mosaic virus: involvement in cell-to-cell movement and dispensability for replication. EMBO J 6 2557 2563
15. WolfS
DeomCM
BeachyRN
LucasWJ
1989 Movement protein of tobacco mosaic virus modifies plasmodesmatal size exclusion limit. Science 246 377 379
16. WaigmannE
LucasWJ
CitovskyV
ZambryskiPC
1994 Direct functional assay for tobacco mosaic virus cell-to-cell movement protein and identification of a domain involved in increasing plasmodesmal permeability. Proc Natl Acad Sci USA 91 1433 1437
17. DingB
KwonMO
1996 Evidence that actin filaments are involved in controlling the permeability of plasmodesmata in tobacco mesophyll. Plant J 10 157 164
18. NorthcoteDH
DaveyR
LayJ
1989 Use of antisera to localize callose, xylan and arabinogalactan in the cell-plate, primary and secondary cell walls of plant cells. Planta 178 353 366
19. BothaCE
CrossRH
2000 Towards reconciliation of structure with function in plasmodesmata - who is the gatekeeper? Micron 31 713 721
20. BucherGL
TarinaC
HeinleinM
Di SerioF
MeinsFJr
2001 Local expression of enzymatically active class I beta-1,3-glucanase enhances symptoms of TMV infection in tobacco. Plant J 28 361 369
21. IglesiasVA
MeinsFJr
2000 Movement of plant viruses is delayed in a beta-1,3-glucanase-deficient mutant showing a reduced plasmodesmatal size exclusion limit and enhanced callose deposition. Plant J 21 157 166
22. UekiS
CitovskyV
2002 Cadmium ion-induced glycine-rich protein inhibits systemic movement of a tobamovirus. Nat Cell Biol 4 478 485
23. BeffaRS
HoferRM
ThomasM
MeinsFJr
1996 Decreased susceptibility to virus disease of beta-1,3-glucanase-deficient plants generated by antisense transformation. Plant Cell 8 1001 1011
24. BeffaR
MeinsFJr
1996 Pathogenesis-related functions of plant beta-1,3-glucanases investigated by antisense transformation - a review. Gene 179 97 103
25. SuS
LiuZ
ChenC
ZhangY
WangX
2010 Cucumber mosaic virus movement protein severs actin filaments to increase the plasmodesmal size exclusion limit in tobacco. Plant Cell 22 1373 1387
26. KaussH
1996 Callose synthesis.
SmallwoodM
KnoxJP
BowlesDJ
Membranes: Specialized Functions in Plants Oxford BIOS Scientific Publishers 77 92
27. KaussH
1985 Callose biosynthesis as a Ca2+-regulated process and possible relations to the induction of other metabolic changes. J Cell Sci Suppl 2 89 103
28. LevyA
ErlangerM
RosenthalM
EpelBL
2007 A plasmodesmata-associated beta-1,3-glucanase in Arabidopsis. Plant J 49 669 682
29. McLeanBG
ZupanJ
ZambryskiPC
1995 Tobacco mosaic virus movement protein associates with the cytoskeleton in tobacco cells. Plant Cell 7 2101 2114
30. ShimizuT
YoshiiA
SakuraiK
HamadaK
YamajiY
2009 Identification of a novel tobacco DnaJ-like protein that interacts with the movement protein of tobacco mosaic virus. Arch Virol 154 959 967
31. von BargenS
SalchertK
PaapeM
PiechullaB
KellmannJ
2001 Interactions between the tomato spotted wilt virus movement protein and plant proteins showing homologies to myosin, kinesin, and DnaJ-like chaperons. Plant Physiol Biochem 39 1083 1093
32. ChenMH
TianGW
GafniY
CitovskyV
2005 Effects of calreticulin on viral cell-to-cell movement. Plant Physiol 138 1866 1876
33. ChenMH
ShengJ
HindG
HandaA
CitovskyV
2000 Interaction between the tobacco mosaic virus movement protein and host cell pectin methylesterases is required for viral cell-to-cell movement. EMBO J 19 913 920
34. DorokhovYL
MakinenK
FrolovaOY
MeritsA
SaarinenJ
1999 A novel function for a ubiquitous plant enzyme pectin methylesterase: the host-cell receptor for the tobacco mosaic virus movement protein. FEBS Lett 461 223 228
35. FridborgI
GraingerJ
PageA
ColemanM
FindlayK
2003 TIP, a novel host factor linking callose degradation with the cell-to-cell movement of Potato virus X. Mol Plant-Microbe Interact 16 132 140
36. GhoshroyS
FreedmanK
LarteyR
CitovskyV
1998 Inhibition of plant viral systemic infection by non-toxic concentrations of cadmium. Plant J 13 591 602
37. CitovskyV
GhoshroyS
TsuiF
KlessigDF
1998 Non-toxic concentrations of cadmium inhibit tobamoviral systemic movement by a salicylic acid-independent mechanism. Plant J 16 13 20
38. MosaviLK
CammettTJ
DesrosiersDC
PengZY
2004 The ankyrin repeat as molecular architecture for protein recognition. Protein Sci 13 1435 1448
39. BjörklundAK
EkmanD
ElofssonA
2006 Expansion of protein domain repeats. PLoS Comput Biol 2 e114
40. YanJ
WangJ
ZhangH
2002 An ankyrin repeat-containing protein plays a role in both disease resistance and antioxidation metabolism. Plant J 29 193 202
41. KuhlmannM
HorvayK
StrathmannA
HeinekampT
FischerU
2003 The alpha-helical D1 domain of the tobacco bZIP transcription factor BZI-1 interacts with the ankyrin-repeat protein ANK1 and is important for BZI-1 function, both in auxin signaling and pathogen response. J Biol Chem 278 8786 8794
42. SpencerML
TheodosiouM
NoonanDJ
2004 NPDC-1, a novel regulator of neuronal proliferation, is degraded by the ubiquitin/proteasome system through a PEST degradation motif. J Biol Chem 279 37069 37078
43. ShumwaySD
MakiM
MiyamotoS
1999 The PEST domain of IkappaBalpha is necessary and sufficient for in vitro degradation by mu-calpain. J Biol Chem 274 30874 30881
44. RechsteinerM
RogersSW
1996 PEST sequences and regulation by proteolysis. Trends Biochem Sci 21 267 271
45. BaeW
LeeYJ
KimDH
LeeJ
KimS
2008 AKR2A-mediated import of chloroplast outer membrane proteins is essential for chloroplast biogenesis. Nat Cell Biol 10 220 227
46. UekiS
LacroixB
KrichevskyA
LazarowitzSG
CitovskyV
2009 Functional transient genetic transformation of Arabidopsis leaves by biolistic bombardment. Nat Protoc 4 71 77
47. LewisJD
LazarowitzSG
2010 Arabidopsis synaptotagmin SYTA regulates endocytosis and virus movement protein cell-to-cell transport. Proc Nat Acad Sci USA 9 2491 2496
48. BoykoV
van Der LaakJ
FerralliJ
SuslovaE
KwonMO
2000 Cellular targets of functional and dysfunctional mutants of tobacco mosaic virus movement protein fused to green fluorescent protein. J Virol 74 11339 11346
49. OparkaKJ
PriorDAM
Santa-CruzS
PadgettHS
BeachyRN
1997 Gating of epidermal plasmodesmata is restricted to the leading edge of expanding infection sites of tobacco mosaic virus (TMV). Plant J 12 781 789
50. CrawfordKM
ZambryskiPC
2001 Non-targeted and targeted protein movement through plasmodesmata in leaves in different developmental and physiological states. Plant Physiol 125 1802 1812
51. KotlizkyG
KatzA
van der LaakJ
BoykoV
LapidotM
2001 A dysfunctional movement protein of Tobacco mosaic virus interferes with targeting of wild-type movement protein to microtubules. Mol Plant-Microbe Interact 14 895 904
52. SimpsonC
ThomasCL
FindlayK
BayerE
MauleAJ
2009 An Arabidopsis GPI-anchor plasmodesmal neck protein with callose binding activity and potential to regulate cell-to-cell trafficking. Plant Cell 21 581 594
53. Guenoune-GelbartD
ElbaumM
SagiG
LevyA
EpelBL
2008 Tobacco mosaic virus (TMV) replicase and movement protein function synergistically in facilitating TMV spread by lateral diffusion in the plasmodesmal desmotubule of Nicotiana benthamiana. Mol Plant-Microbe Interact 21 335 345
54. HeinleinM
PadgettHS
GensJS
PickardBG
CasperSJ
1998 Changing patterns of localization of the tobacco mosaic virus movement protein and replicase to the endoplasmic reticulum and microtubules during infection. Plant Cell 10 1107 1120
55. ReichelC
BeachyRN
1999 The role of the ER and cytoskeleton in plant viral trafficking. Trends Plant Sci 4 458 462
56. BrillLM
NunnRS
KahnTW
YeagerM
BeachyRN
2000 Recombinant tobacco mosaic virus movement protein is an RNA-binding, alpha-helical membrane protein. Proc Natl Acad Sci USA 97 7112 7117
57. MasP
BeachyRN
1999 Replication of tobacco mosaic virus on endoplasmic reticulum and role of the cytoskeleton and virus movement protein in intracellular distribution of viral RNA. J Cell Biol 147 945 958
58. FujikiM
KawakamiS
KimRW
BeachyRN
2006 Domains of tobacco mosaic virus movement protein essential for its membrane association. J Gen Virol 87 2699 2707
59. VossS
SkerraA
1997 Mutagenesis of a flexible loop in streptavidin leads to higher affinity for the Strep-tag II peptide and improved performance in recombinant protein purification. Protein Eng 10 975 982
60. WitteCP
NöelLD
GielbertJ
ParkerJE
RomeisT
2004 Rapid one-step protein purification from plant material using the eight-amino acid StrepII epitope. Plant Mol Biol 55 135 147
61. CitovskyV
LeeLY
VyasS
GlickE
ChenMH
2006 Subcellular localization of interacting proteins by bimolecular fluorescence complementation in planta. J Mol Biol 362 1120 1131
62. HuCD
ChinenovY
KerppolaTK
2002 Visualization of interactions among bZIP and Rel family proteins in living cells using bimolecular fluorescence complementation. Mol Cell 9 789 798
63. ZamyatninAAJr
SolovyevAG
BozhkovPV
ValkonenJPT
MorozovSY
2006 Assessment of the integral membrane protein topology in living cells. Plant J 46 145 154
64. AnG
CostaMA
HaSB
1990 Nopaline synthase promoter is wound inducible and auxin inducible. Plant Cell 2 225 233
65. ChaiMF
WeiPC
ChenQJ
AnR
ChenJ
2006 NADK3, a novel cytoplasmic source of NADPH, is required under conditions of oxidative stress and modulates abscisic acid responses in Arabidopsis. Plant J 47 665 674
66. Benitez-AlfonsoY
CiliaM
San RomanA
ThomasCL
MauleAJ
2009 Control of Arabidopsis meristem development by thioredoxin-dependent regulation of intercellular transport. Proc Natl Acad Sci USA 106 3615 3620
67. StonebloomS
Burch-SmithT
KimI
MeinkeD
MindrinosM
2009 Loss of the plant DEAD-box protein ISE1 leads to defective mitochondria and increased cell-to-cell transport via plasmodesmata. Proc Nat Acad Sci USA 106 17229 17234
68. SivaguruM
FujiwaraT
SamajJ
BaluškaF
YangZ
2000 Aluminum-induced 1-->3-beta-D-glucan inhibits cell-to-cell trafficking of molecules through plasmodesmata. A new mechanism of aluminum toxicity in plants. Plant Physiol 124 991 1006
69. UekiS
CitovskyV
2005 Identification of an interactor of cadmium ion-induced glycine-rich protein involved in regulation of callose levels in plant vasculature. Proc Natl Acad Sci USA 102 12089 12094
70. ReichelC
BeachyRN
1998 Tobacco mosaic virus infection induces severe morphological changes of the endoplasmic reticulum. Proc Natl Acad Sci USA 95 11169 11174
71. WirdnamC
MotoyamaA
Arn-BouldoiresE
van EedenS
IglesiasA
2004 Altered expression of an ankyrin-repeat protein results in leaf abnormalities, necrotic lesions, and the elaboration of a systemic signal. Plant Mol Biol 56 717 730
72. LevyA
Guenoune-GelbartD
EpelBL
2007 beta-1,3-Glucanases: plasmodesmal gate keepers for intercellular communication. Plant Signal Behav 2 404 407
73. SuenDF
WuSS
ChangHC
DhuggaKS
HuangAH
2003 Cell wall reactive proteins in the coat and wall of maize pollen: potential role in pollen tube growth on the stigma and through the style. J Biol Chem 278 43672 43681
74. DelpG
PalvaET
1999 A novel flower-specific Arabidopsis gene related to both pathogen-induced and developmentally regulated plant beta-1,3-glucanase genes. Plant Mol Biol 39 565 575
75. BolJF
LinthorstHJM
CornelissenBJC
1990 Plant pathogenesis-related proteins induced by virus infection. Annu Rev Phytopathol 28 113 138
76. NishikawaM
SuzukiK
YoshidaK
1990 Structural and functional stability of IncP plasmids during stepwise transmission by trans-kingdom mating: promiscuous conjugation of Escherichia coli and Saccharomyces cerevisiae. Jpn J Genet 65 323 334
77. HuG
RijkenbergFH
1998 Subcellular localization of beta-1,3-glucanase in Puccinia recondita f.sp. tritici-infected wheat leaves. Planta 204 324 334
78. CastresanaC
de CarvalhoF
GheysenG
HabetsM
InzéD
1990 Tissue-specific and pathogen-induced regulation of a Nicotiana plumbaginifolia beta-1,3-glucanase gene. Plant Cell 2 1131 1143
79. RinnePLH
KaikurantaPM
van der SchootC
2001 The shoot apical meristem restores its symplasmic organization during chilling-induced release from dormancy. Plant J 26 249 264
80. RinnePLH
van der SchootC
2003 Plasmodesmata at the crossroads between development, dormancy, and defense. Can J Bot 81 1182 1197
81. MeierH
BuchsL
BuchalaAJ
HomewoodT
1981 (1-->3) - {lower case beta}-D-glucan (callose) is a probable intermediate in biosynthesis of cellulose fibers. Nature 289 821 822
82. RichmondTA
SomervilleCR
2001 Integrative approaches to determining Csl function. Plant Mol Biol 47 131 143
83. DhuggaKS
2001 Building the wall: genes and enzyme complexes for polysaccharide synthases. Curr Opin Plant Biol 4 488 493
84. VermaDP
HongZ
2001 Plant callose synthase complexes. Plant Mol Biol 47 693 701
85. ChungSM
FrankmanEL
TzfiraT
2005 A versatile vector system for multiple gene expression in plants. Trends Plant Sci 10 357 361
86. GuoHS
FeiJF
XieQ
ChuaNH
2003 A chemical-regulated inducible RNAi system in plants. Plant J 34 383 392
87. TzfiraT
TianGW
LacroixB
VyasS
LiJ
2005 pSAT vectors: a modular series of plasmids for fluorescent protein tagging and expression of multiple genes in plants. Plant Mol Biol 57 503 516
88. ShivprasadS
PogueGP
LewandowskiDJ
HidalgoJ
DonsonJ
1999 Heterologous sequences greatly affect foreign gene expression in tobacco mosaic virus-based vectors. Virology 255 312 323
89. TzfiraT
VaidyaM
CitovskyV
2001 VIP1, an Arabidopsis protein that interacts with Agrobacterium VirE2, is involved in VirE2 nuclear import and Agrobacterium infectivity. EMBO J 20 3596 3607
90. HorschRB
FryJE
HoffmanNL
EichholtzDA
RogersSG
1985 A simple and general method for transferring genes into plants. Science 227 1229 1231
91. TzfiraT
JensenCS
WangxiaW
ZukerA
AltmanA
1997 Transgenic Populus: a step-by-step protocol for its Agrobacterium-mediated transformation. Plant Mol Biol Rep 15 219 235
92. KapilaJ
De RyckeR
Van MontaguM
AngenonG
1997 An Agrobacterium-mediated transient gene expression system for intact leaves. Plant Sci 122 101 108
93. WroblewskiT
TomczakA
MichelmoreR
2005 Optimization of Agrobacterium-mediated transient assays of gene expression in lettuce, tomato and Arabidopsis. Plant Biotech J 3 259 273
94. LacroixB
LoyterA
CitovskyV
2008 Association of the Agrobacterium T-DNA-protein complex with plant nucleosomes. Proc Natl Acad Sci USA 105 15429 15434
95. WrightKM
WoodNT
RobertsAG
ChapmanS
BoevinkP
2007 Targeting of TMV movement protein to plasmodesmata requires the actin/ER network; evidence from FRAP. Traffic 8 21 31
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek Tick Histamine Release Factor Is Critical for Engorgement and Transmission of the Lyme Disease AgentČlánek TGF-b2 Induction Regulates Invasiveness of -Transformed Leukocytes and Disease SusceptibilityČlánek The Origin of Intraspecific Variation of Virulence in an Eukaryotic Immune Suppressive ParasiteČlánek Glycosylation Focuses Sequence Variation in the Influenza A Virus H1 Hemagglutinin Globular Domain
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2010 Číslo 11- Jak souvisí postcovidový syndrom s poškozením mozku?
- Měli bychom postcovidový syndrom léčit antidepresivy?
- Farmakovigilanční studie perorálních antivirotik indikovaných v léčbě COVID-19
- 10 bodů k očkování proti COVID-19: stanovisko České společnosti alergologie a klinické imunologie ČLS JEP
-
Všechny články tohoto čísla
- Patients with Discordant Responses to Antiretroviral Therapy Have Impaired Killing of HIV-Infected T Cells
- A Molecular Mechanism for Eflornithine Resistance in African Trypanosomes
- Tyrosine Sulfation of the Amino Terminus of PSGL-1 Is Critical for Enterovirus 71 Infection
- Autoimmunity as a Predisposition for Infectious Diseases
- The Subtilisin-Like Protease AprV2 Is Required for Virulence and Uses a Novel Disulphide-Tethered Exosite to Bind Substrates
- Structural Analysis of HIV-1 Maturation Using Cryo-Electron Tomography
- Tick Histamine Release Factor Is Critical for Engorgement and Transmission of the Lyme Disease Agent
- Interferon-Inducible CXC Chemokines Directly Contribute to Host Defense against Inhalational Anthrax in a Murine Model of Infection
- TGF-b2 Induction Regulates Invasiveness of -Transformed Leukocytes and Disease Susceptibility
- The Origin of Intraspecific Variation of Virulence in an Eukaryotic Immune Suppressive Parasite
- CO Acts as a Signalling Molecule in Populations of the Fungal Pathogen
- SV2 Mediates Entry of Tetanus Neurotoxin into Central Neurons
- MAP Kinase Phosphatase-2 Plays a Critical Role in Response to Infection by
- Glycosylation Focuses Sequence Variation in the Influenza A Virus H1 Hemagglutinin Globular Domain
- Potentiation of Epithelial Innate Host Responses by Intercellular Communication
- Fcγ Receptor I Alpha Chain (CD64) Expression in Macrophages Is Critical for the Onset of Meningitis by K1
- ANK, a Host Cytoplasmic Receptor for the Cell-to-Cell Movement Protein, Facilitates Intercellular Transport through Plasmodesmata
- Analysis of the Initiating Events in HIV-1 Particle Assembly and Genome Packaging
- Evolution of Linked Avirulence Effectors in Is Affected by Genomic Environment and Exposure to Resistance Genes in Host Plants
- Structural Basis of HIV-1 Neutralization by Affinity Matured Fabs Directed against the Internal Trimeric Coiled-Coil of gp41
- Hepatitis C Virus (HCV) Evades NKG2D-Dependent NK Cell Responses through NS5A-Mediated Imbalance of Inflammatory Cytokines
- Host Cell Invasion and Virulence Mediated by Ssa1
- Global Gene Expression in Urine from Women with Urinary Tract Infection
- Should the Human Microbiome Be Considered When Developing Vaccines?
- HapX Positively and Negatively Regulates the Transcriptional Response to Iron Deprivation in
- Enhancing Oral Vaccine Potency by Targeting Intestinal M Cells
- Herpes Simplex Virus Reorganizes the Cellular DNA Repair and Protein Quality Control Machinery
- The Female Lower Genital Tract Is a Privileged Compartment with IL-10 Producing Dendritic Cells and Poor Th1 Immunity following Infection
- Zn Inhibits Coronavirus and Arterivirus RNA Polymerase Activity and Zinc Ionophores Block the Replication of These Viruses in Cell Culture
- Cryo Electron Tomography of Native HIV-1 Budding Sites
- Crystal Structure and Size-Dependent Neutralization Properties of HK20, a Human Monoclonal Antibody Binding to the Highly Conserved Heptad Repeat 1 of gp41
- Modelling the Evolution and Spread of HIV Immune Escape Mutants
- The Arabidopsis Resistance-Like Gene Is Activated by Mutations in and Contributes to Resistance to the Bacterial Effector AvrRps4
- Platelet-Activating Factor Receptor Plays a Role in Lung Injury and Death Caused by Influenza A in Mice
- Genetic and Structural Basis for Selection of a Ubiquitous T Cell Receptor Deployed in Epstein-Barr Virus Infection
- Ubiquitin-Regulated Nuclear-Cytoplasmic Trafficking of the Nipah Virus Matrix Protein Is Important for Viral Budding
- Pneumolysin Activates the NLRP3 Inflammasome and Promotes Proinflammatory Cytokines Independently of TLR4
- Immune Evasion by : Differential Targeting of Dendritic Cell Subpopulations
- Survival in Selective Sand Fly Vector Requires a Specific -Encoded Lipophosphoglycan Galactosylation Pattern
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Zn Inhibits Coronavirus and Arterivirus RNA Polymerase Activity and Zinc Ionophores Block the Replication of These Viruses in Cell Culture
- The Female Lower Genital Tract Is a Privileged Compartment with IL-10 Producing Dendritic Cells and Poor Th1 Immunity following Infection
- Crystal Structure and Size-Dependent Neutralization Properties of HK20, a Human Monoclonal Antibody Binding to the Highly Conserved Heptad Repeat 1 of gp41
- The Arabidopsis Resistance-Like Gene Is Activated by Mutations in and Contributes to Resistance to the Bacterial Effector AvrRps4
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



