-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaTranscriptional and Linkage Analyses Identify Loci that Mediate the Differential Macrophage Response to Inflammatory Stimuli and Infection
Macrophages provide a first line of defense against invading pathogens and play an important role in the initiation and resolution of immune responses. When in contact with pathogens or immune factors, such as cytokines, macrophages assume activation states that range between pro-inflammatory (classical activation) and anti-inflammatory (alternative activation). Even though it is known that macrophages from different individuals are biased towards one of the various activation states, the genetic factors that define individual differences in macrophage activation are not fully understood. Additionally, although macrophages are important in infectious disease pathogenesis, how individual differences in macrophage activation contribute to individual differences in susceptibility to infectious disease is just emerging. We used macrophages from genetically segregating mice to show that discrete transcriptional programs, which are modulated by specific genomic regions, modulate differences in macrophage activation. Murine macrophages differences in controlling Toxoplasma growth mapped to chromosome 3, proximal to the Guanylate binding protein (Gbp) locus that is known to modulate the murine macrophage response to Toxoplasma. Using a shRNA-mediated knockdown approach, we show that the DEAD box polypeptide 1 (Ddx1) modulates nitric oxide production in macrophages stimulated with interferon gamma and tumor necrosis factor. These findings are a step towards the identification of genes that regulate macrophage phenotypes and disease outcome.
Published in the journal: . PLoS Genet 11(10): e32767. doi:10.1371/journal.pgen.1005619
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1005619Summary
Macrophages provide a first line of defense against invading pathogens and play an important role in the initiation and resolution of immune responses. When in contact with pathogens or immune factors, such as cytokines, macrophages assume activation states that range between pro-inflammatory (classical activation) and anti-inflammatory (alternative activation). Even though it is known that macrophages from different individuals are biased towards one of the various activation states, the genetic factors that define individual differences in macrophage activation are not fully understood. Additionally, although macrophages are important in infectious disease pathogenesis, how individual differences in macrophage activation contribute to individual differences in susceptibility to infectious disease is just emerging. We used macrophages from genetically segregating mice to show that discrete transcriptional programs, which are modulated by specific genomic regions, modulate differences in macrophage activation. Murine macrophages differences in controlling Toxoplasma growth mapped to chromosome 3, proximal to the Guanylate binding protein (Gbp) locus that is known to modulate the murine macrophage response to Toxoplasma. Using a shRNA-mediated knockdown approach, we show that the DEAD box polypeptide 1 (Ddx1) modulates nitric oxide production in macrophages stimulated with interferon gamma and tumor necrosis factor. These findings are a step towards the identification of genes that regulate macrophage phenotypes and disease outcome.
Introduction
At the cellular level, innate immune cells, such as macrophages, are central to the development and prevention of infectious diseases. On engagement of surface signaling receptors or pattern recognition receptors (PRRs) such as toll-like receptors (TLRs), RIG-I-like receptors (RLRs) and the cytosolic NOD-like receptors (NLRs), by immune factors such as cytokines or conserved microbial products, macrophages can assume different activation states. The most extreme of these states are the classical (M1, M(IFNG)) and the alternative (M2, M(IL-4)) states, separated by several intermediate activation states [1–3] (We are following the recently described macrophage activation phenotype nomenclature [1]). Ultimately, macrophage activation results in pathogen clearance by downstream antimicrobial effector mechanisms, such as inflammasome activation, or activation of adaptive immune responses [4–6]. Although the outcome of macrophage activation is dependent on the stimulus engaged by the PRRs, emerging empirical data, from both human and mouse studies, indicate that the macrophage genetic background also plays a significant role.
The initiation of immune responses by macrophages can occur in the presence of pro-inflammatory cytokines such as interferon gamma (IFNG), while anti-inflammatory cytokines such as interleukin (IL)-4, and IL-13, prime macrophages for the resolution of immune responses and tissue repair [7–9]. This macrophage ability to initiate and resolve immune responses, while important in regulating immunopathology, can be exploited by pathogens to evade macrophage-associated immunity [10]. Indeed, to disseminate in their hosts most pathogens circumvent macrophage-mediated microbicidal mechanisms by modulating macrophage signaling pathways and activation phenotypes [11–16]. In addition to destroying pathogens, activated macrophages are important mediators in several inflammatory pathologies, including atherosclerosis, diabetes and cancer [17]. Studies in mice have linked several macrophage - or immune-related genes, such as Nramp1/Slc11a1, Icsbp1/Irf8, Csfgm, and Nos2, with the development of several infectious diseases, including salmonellosis, toxoplasmosis, and leishmaniasis [18–20]. Although the compendium of macrophage - or immune-related genes that modulate infectious disease pathogenesis is broad, the role of individual differences in macrophage activation phenotypes in determining individual differences in susceptibility to infectious disease is just emerging [21–25]. Furthermore, the genetic basis for individual differences in macrophage activation phenotypes has not been identified. Macrophage activation is likely modulated by complex gene and metabolite networks that cannot be defined one gene at a time, thus the difficulty in unraveling the genetics of macrophage activation. This hypothesis is reinforced by results from genetic perturbation experiments that have revealed multiple genes that individually modulate macrophage activation phenotypes, including IRF8, PPARG and AKT2 [26, 27]. Empirical data show that macrophages display distinct transcriptional programs in response to infectious and inflammatory stimuli [22] and that this macrophage response differs between genetically segregating individuals [22–25]. Our hypothesis is that inter-individual differences in susceptibility to infectious disease are partly due to genetic differences in the macrophage response to pathogens.
Quantitative trait locus (QTL) analyses have been used to elucidate the complex genetic basis for many traits in humans and model organisms [28, 29]. However, the region spanned by individual QTL is often large and encompasses multiple genes, making the transition from QTL to individual genes influencing disease (quantitative trait gene, QTG) difficult. It has been shown that differences in the abundance of certain transcripts can explain phenotypic variations between individuals [30, 31]. Forward genetics approaches that combine traditional QTL mapping with expression quantitative trait mapping (eQTL; in which case transcript abundance is the quantitative trait) [32] are increasingly being used to successfully transition from QTL to QTG [33–35]. Traditional QTL analysis will identify the genomic regions affecting trait variation, while eQTL analysis can help in understanding which genes, pathways, and biological processes are also under the influence of a given QTL. By examining the relationship between transcript location, the location of the eQTL and the pleiotropic effects of the eQTL, tools of systems biology such as network and functional analysis can be used to further delineate the complex genetic interactions modulating complex traits and reconstruct genetic pathways that underlie such traits [33–35].
In this study, we used the AXB/BXA recombinant inbred (RI) mice to investigate the relationship between the macrophage genotype and their response to inflammatory stimuli or infection with Toxoplasma, an obligate intracellular protozoan parasite. The AXB/BXA mice are derived from an initial reciprocal cross of AJ (A) and C57BL/6J (B) mice followed by multiple rounds (≥20) of inbreeding resulting in a stable mosaic of blocks of the parental alleles in their genomes [36]. These mice have been used to investigate the development and susceptibility to a variety of infectious and inflammatory diseases [19, 37–40]. Importantly, the parental strains, AJ and C57BL/6J, differ at key loci that regulate immune responses, including the complement 5 a (C5a) [41] and interleukin 3 receptor alpha (Il3ra) [25, 42]. These mice also exhibit differences in the amount of IL-10 and tumor necrosis factor (TNF) produced in response to bacterial infection [43, 44]. Furthermore, the parental AJ and C57BL/6J vary in susceptibility to a variety of pathogens, including Staphylococcus aureus, Toxoplasma gondii and Trypanosoma cruzi [38, 44–47]. By linking QTL analyses of defined macrophage phenotypes and macrophage transcriptional profiles, captured by high-throughput RNA-sequencing, we have identified many loci that affect the macrophage response to inflammatory stimuli and infection. These loci could provide the foundations for further studies in identifying the genetic basis for the differences in susceptibility to inflammation and infection in these mice. As an example, we report that differences in nitric oxide (NO) production, in AJ and C57BL/6J macrophages is due to differences in the expression of the RNA helicase Ddx1.
Results
AJ and C57BL/6J BMDM differ in their response to infectious and inflammatory stimuli
Although correlations between genetic variations in immune-related genes and the response to infectious and inflammatory stimuli have been reported in AJ and C57BL/6J (B6) [19, 43, 44], the role of specific immune cells in these phenotypic differences are mostly equivocal. Therefore, we stimulated AJ and B6 bone marrow-derived macrophages (BMDM) with interferon gamma (IFNG) and tumor necrosis alpha (TNF) (IFNG+TNF) or interleukin 4 (IL4). IFNG+TNF induces the classical (M(IFNG)) while IL-4 induces the alternative (M(IL-4)) macrophage activation phenotypes. Additionally, to mimic activation by bacteria and pathogen-associated molecular patterns (PAMPs), we stimulated the macrophages with lipopolysaccharide (LPS) (a component of gram-negative bacteria), or CpG (a synthetic oligonucleotide), respectively. A summary of the stimulation regimen and the corresponding phenotypes measured is shown in S1 Table. Next, we captured the macrophage response to the individual stimulus by measuring M(IFNG) and M(IL-4) markers. For M(IFNG) markers, we measured the amount of nitric oxide (NO) and IL-12, while for M(IL-4) markers, we quantified the amount of urea (a by-product of Arginase I enzyme activity), IL-10 and chemokine (C-C motif) ligand 22 (CCL22) (Fig 1). Finally, because: 1) IFNG is indispensable in the resistance to Toxoplasma gondii [48], 2) Toxoplasma is a master regulator of macrophage signaling pathways [14], and 3) AJ and B6 mice segregate for susceptibility to Toxoplasma [38, 49], we infected non-stimulated or IFNG+TNF-stimulated BMDM with a strain of Toxoplasma engineered to express firefly luciferase [50] and assessed parasite growth by measuring luciferase activity, which is often used to approximate Toxoplasma burden in in vitro or in vivo infection models [49–52]. We observed high amounts of NO and CCL22 in AJ BMDM, while the B6 BMDM produced higher amounts of IL-12, IL-10 and urea (Fig 1A–1E). Despite producing high amounts of M(IL-4) markers (urea and IL-10), the B6 BMDM also produced high amounts of an M(IFNG) marker (IL-12) relative to AJ BMDM. Similarly, the high amount of NO, an M(IFNG) marker, produced by AJ BMDM was accompanied by high amounts of CCL22, an M(IL-4) marker. This dual expression of M(IFNG) and M(IL-4) markers is perhaps indicative of complex molecular modulation of macrophage activation or the heterogeneity of macrophage activation phenotypes. Consistent with the known resistance of AJ mice to Toxoplasma relative to B6 mice [38, 49], and the divergent BMDM activation phenotypes, Toxoplasma growth in the IFNG+TNF-stimulated AJ BMDM was significantly reduced compared to its growth in B6 BMDM (Fig 1F). Thus, the variable susceptibility to Toxoplasma in AJ versus B6 observed in vivo [38, 49] can be recapitulated in vitro using AJ and B6 BMDM. As such, the AJ and B6 BMDM can be used to gain insight into the molecular mechanisms that underlie the differences in AJ versus B6 susceptibility to Toxoplasma. Together, the difference in macrophage response to IFNG+TNF stimulation, as evidenced by NO (a key toxostatic effector [53, 54]), and the differences in parasite growth in IFNG+TNF-stimulated BMDM, posit that the variable susceptibility to Toxoplasma between AJ and B6 mice is due to innate differences in the macrophage response to the parasite and/or IFNG+TNF.
Fig. 1. BMDM from AJ and B6 mice differ in their response to various stimuli. 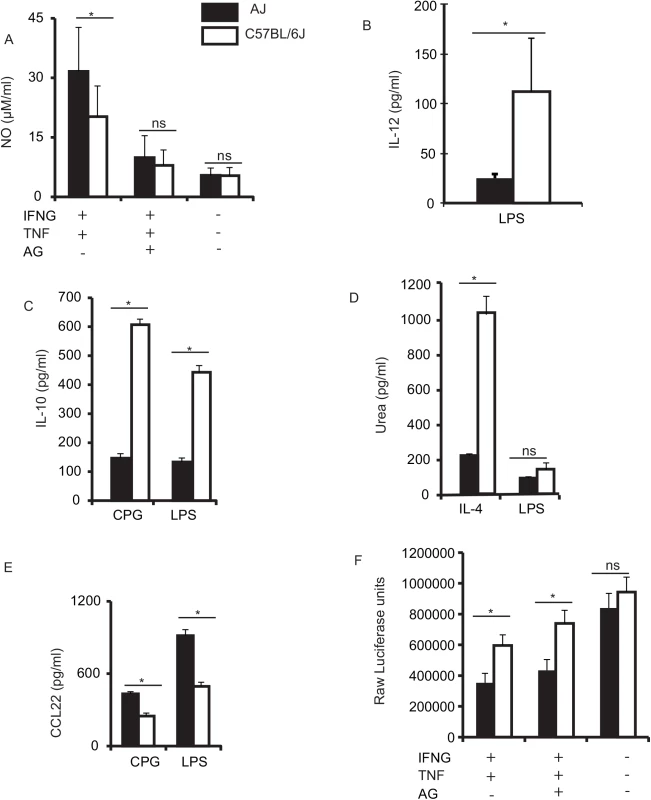
A) Stimulation of macrophages for ~ 18 hrs with IFNG+TNF resulted in high production of NO by AJ macrophages (black bars), while B) stimulation with LPS led to higher production of IL-12 in C57BL/6J (B6) macrophages (white bars). C) Stimulation of murine macrophages with LPS or CPG resulted in significantly higher production of IL-10 in B6 macrophages compared to the AJ macrophages. D) B6 macrophages stimulated with IL-4 produced significantly more urea, compared to AJ macrophages, while LPS stimulation induced low amounts of urea in both AJ and B6 BMDM. E) AJ macrophages stimulated with CPG or LPS produced significantly higher amounts of the chemokine CCL22 compared to B6 macrophages. F) The IFNG+TNF stimulated macrophages have increased toxoplasmacidal activity, which is slightly reduced in the presence of aminoguanidine (AG), an inducible nitric oxide synthase inhibitor. Where there was no detectable amount of cytokine/chemokine measured, including in all control (non-stimulated) BMDM, we do not include cytokine/chemokine data in the figures. Three independent replicates; Mean (SD) * p < 0.05 (Student’s t-test). The genetic basis for differential macrophage response to infectious and inflammatory stimuli
The differences in response to infectious and inflammatory stimuli in AJ and B6 mice have a genetic component [19, 38, 43]. Therefore, having established differences in parasite growth and activation phenotypes in AJ and B6 BMDM, we sought to establish the genetic basis for the differences in macrophage activation and toxoplasmacidal activity between AJ and B6 using the AXB/BXA mice. We stimulated BMDM obtained from 26 age-matched female AXB/BXA RI mice with IFNG+TNF, IL-4, CpG, or LPS and measured the amount of NO, urea, IL-12, IL-10, and CCL22 produced. We also infected IFNG+TNF stimulated BMDM with Toxoplasma that express luciferase and measured relative parasite growth by measuring luciferase activity. These phenotypes exhibited a continuous distribution in the RI mice, which is characteristic of quantitative traits (S1A–S1F Fig).
Due to the stable and unique combination of blocks of parental alleles in their genomes, the AXB/BXA RI mice are particularly suited for quantitative trait locus (QTL) mapping [55]. Therefore, we used the AXB/BXA genetic map (containing 934 informative genetic markers [56]) in a genome-wide scan in R/qtl [57] to identify the genomic regions that modulate differences in AJ and B6 BMDM phenotypes. To correct for multiple testing on the 934 genetic markers, we performed 1000 permutation tests on the individuals relative to their phenotypes [58] to obtain adjusted p-values for each QTL (Table 1). Parasite growth in the IFNG+TNF-stimulated BMDM mapped to chromosome 3 (147.7 Mb), proximal to the Guanylate binding protein (Gbp) locus (142.6 Mb) that is known to modulate the murine macrophage response to Toxoplasma [59–62]. Even though AJ and B6 BMDM display distinct polarization states following IFNG+TNF or IL-4 stimulation, except for IL-12 and CCL22, we did not observe statistically significant QTL peaks for any of the activation phenotypes. Instead we observed a suggestive QTL for NO on chromosome 12 (Table 1) and a second QTL for NO on chromosome 4 (S2A Fig). The additive QTL for NO also mapped to chromosome 4 (S2B Fig). Similar to observations in the parental BMDM, the AJ allele at the chromosome 12 QTL was associated with high amounts of NO (S2C Fig). However, the AJ allele at the chromosome 4 QTL was associated with low amounts of NO (S2D Fig). Although mapping in cis to Arg1 on chromosome 10, the QTL for the amount of urea did not reach statistical significance. Nevertheless, the allele effect at the suggestive urea QTL was consistent with the parental allele effect on urea.
Tab. 1. Linkage analysis of AJ and C57BL/6J phenotypic differences using the recombinant inbred AXB/BXA mice. 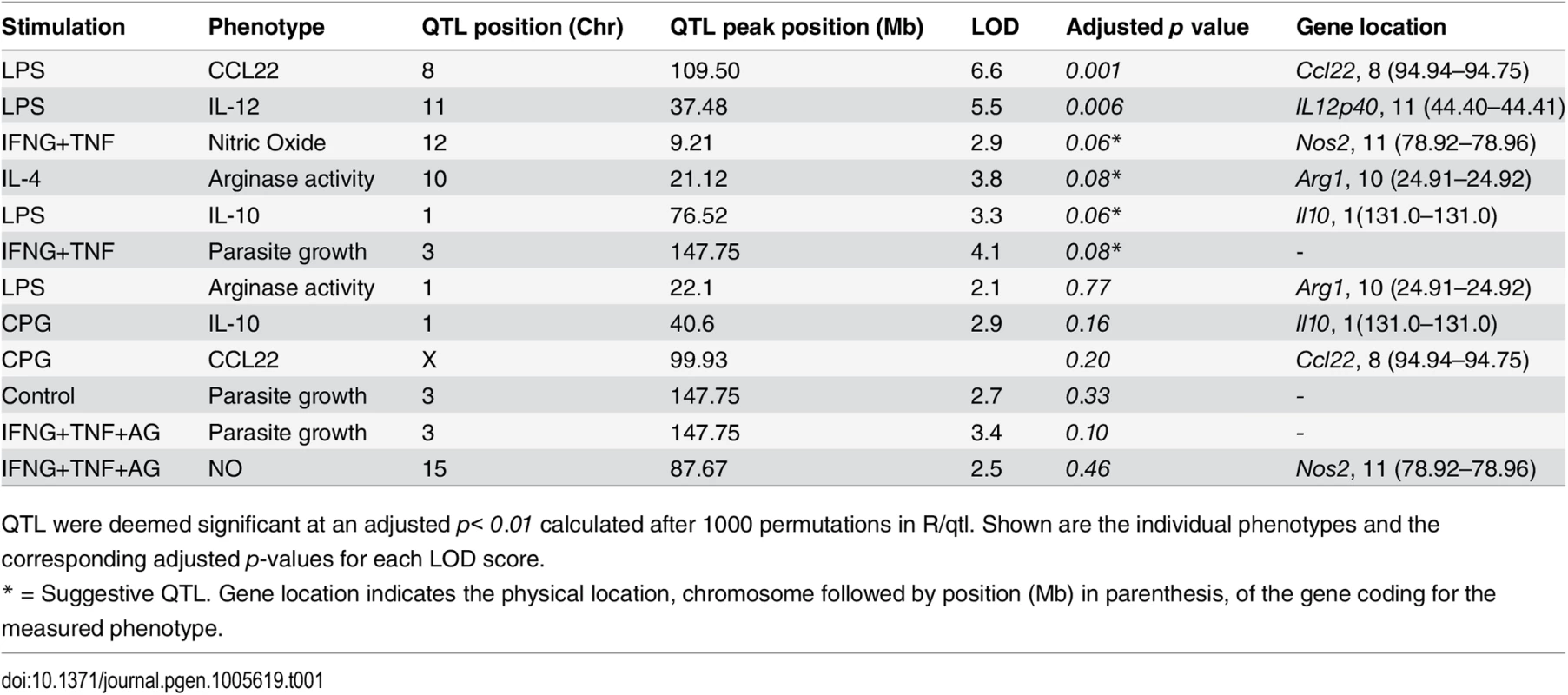
QTL were deemed significant at an adjusted p< 0.01 calculated after 1000 permutations in R/qtl. Shown are the individual phenotypes and the corresponding adjusted p-values for each LOD score. Finally, we selected the top 2 QTL for each phenotype (where there was marginal difference between the LOD scores for the second and third largest QTL, we picked both) and grouped the BMDM based on the genotypes at each QTL. We then estimated the QTL inheritance pattern by comparing the values for the corresponding phenotype amongst the genotypes using a one-way ANOVA with Tukey’s-Post-test (StatPlus, AnalystSoft Inc) [63] (Table 2).
Tab. 2. QTL inheritance and allele effects on different traits in AXB/BXA BMDM. 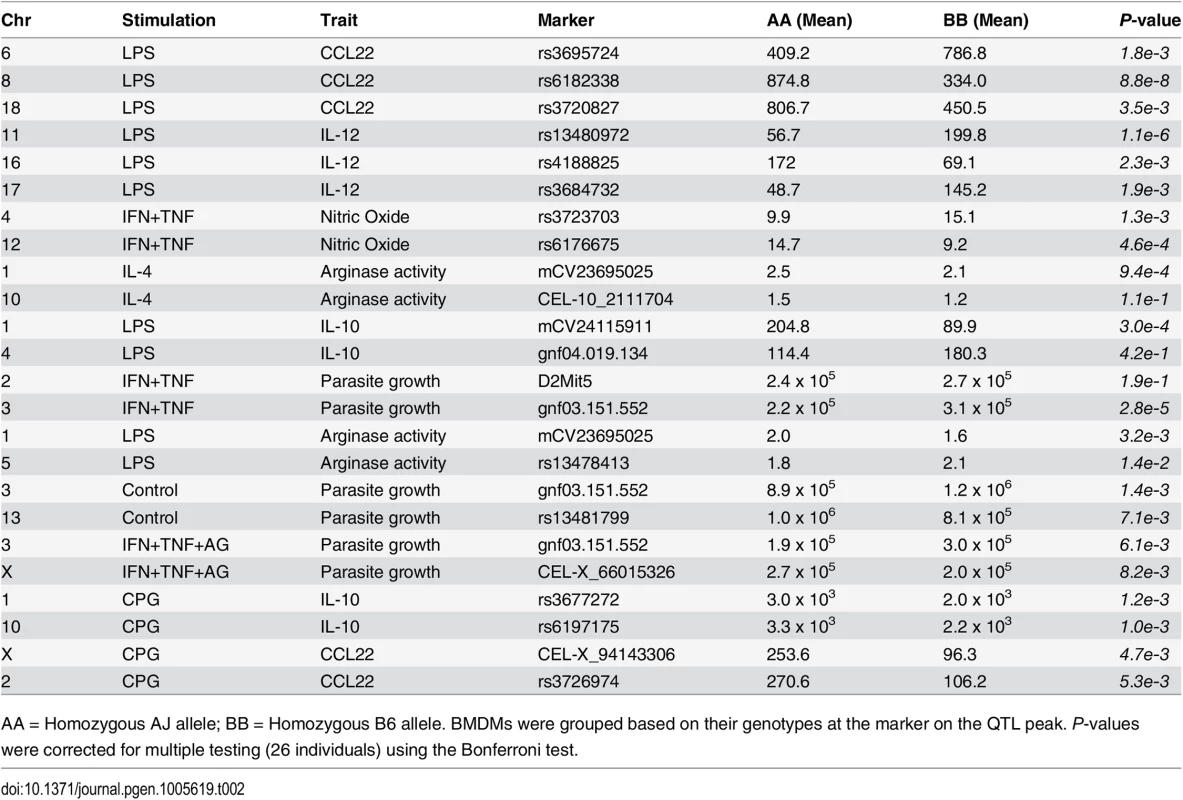
AA = Homozygous AJ allele; BB = Homozygous B6 allele. BMDMs were grouped based on their genotypes at the marker on the QTL peak. P-values were corrected for multiple testing (26 individuals) using the Bonferroni test. Divergent genomic loci modulate macrophage transcriptional response to infectious and inflammatory stimuli
Discrete transcriptional programs modulate the response of immune cells, including macrophages, to stimuli such as pathogens and immune factors [24, 31, 33]. As such transcriptional profiles can be used to gain insight into the intricate and incipient molecular networks that modulate complex traits, such as macrophage activation [24, 34, 64, 65]. Consequently, we investigated whether specific transcriptional programs modulate the differential activation of AJ and B6 BMDM. To do this, we performed high throughput RNA-sequencing (RNA-seq) on BMDM obtained from the same 26 age - and sex-matched AXB/BXA mice described above and their progenitors (28 samples in total), before (resting, controls) and after infection with Toxoplasma or stimulation with IFNG+TNF, or CpG. For each sample we generated at least 100 million paired-end reads, except for the IFNG+TNF-stimulated BMDM samples that were sequenced on a single end. Although all the samples were sequenced once, due to the unique recombination of the parental alleles and homozygosity at each of the informative genetic markers (934), there are at least 4 replicates (the minimum number of mice having the same allele at each marker) for each marker. Thereafter, we processed the RNA-seq data as previously described [25]. Briefly, we aligned the RNA-seq reads to the mouse reference genome (NCBI build GRcm38, downloaded from Illumina iGenomes; https://support.illumina.com/sequencing/sequencing_software/igenome.html) using TopHat [66]. To avoid read alignment bias due to sequence polymorphisms between AJ and B6 genomes, we made a synthetic reference genome in which all the polymorphic nucleotides between AJ and B6 were converted to a neutral nucleotide [67]. However, and consistent with a recent report [68], allele bias did not significantly affect read alignment to the genome. On average, about 70% of reads in each sample uniquely mapped to the synthetic genome, which was about 1% less than the number of reads uniquely aligned to the iGenome. Henceforth, unless otherwise stated, all RNA-seq data presented herein were processed using the synthetic genome. Transcript abundance was estimated using Cufflinks [69] and reported as fragment per kilobase exon per million reads (FPKM).
Next, using the FPKM values from each of the 26 RI BMDM and the corresponding AXB/BXA genetic map, we mapped the genomic loci that modulate gene expression (expression QTL, eQTL) [32] using R/qtl [57]. As previously described [25], we performed 1000 permutations to correct for multiple testing across the 934 informative genetic markers in the AXB/BXA cross. Next, we used a false discovery rate (FDR) ≤ 10%, calculated in the qvalue package [70], to correct for multiple testing on the transcripts and to nominate significant eQTL. Finally, to allow for meaningful comparisons, we only included in the downstream analyses, for each stimulation condition, eQTL for transcripts with an average FPKM ≥ 5 across the 26 RI BMDM. The linkage analyses and the subsequent filtering steps were performed separately in resting, IFNG+TNF-stimulated, CpG-stimulated, and Toxoplasma-infected BMDM and identified, 131, 367, 688 and 1008 significant eQTL, respectively, dispersed throughout the genome (Fig 2A and 2B and S1A–S1D Dataset). Thus, consistent with previous studies [24, 71], the genetic background influences macrophage gene expression profile. Importantly the different stimuli induced transcriptional programs that were modulated by distinct eQTL hotspots, which can be exploited to unravel the complex molecular networks that regulate macrophage activation states.
Fig. 2. The transcriptional response in BMDM is regulated by stimulation-specific trans-loci. 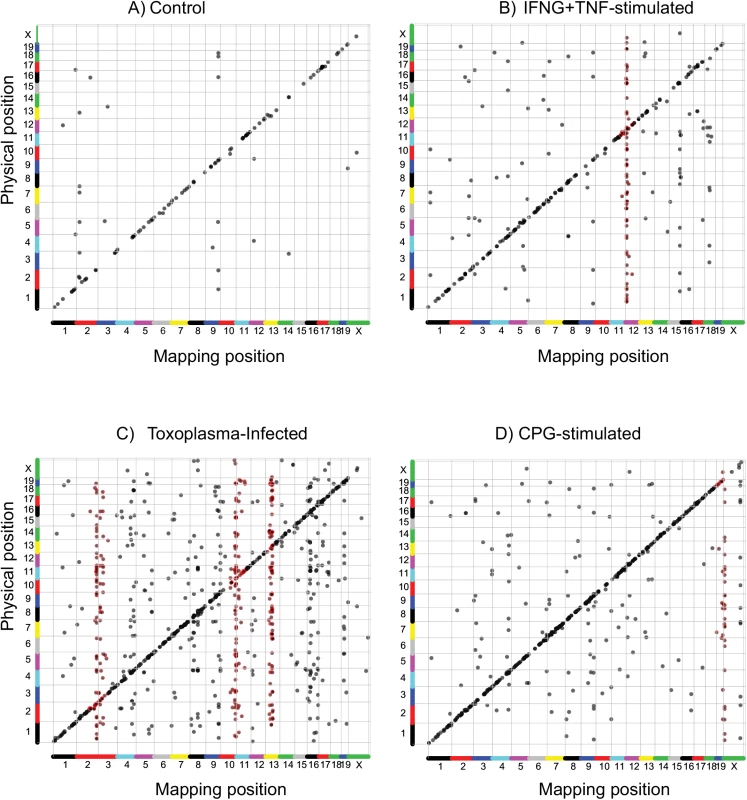
Expression quantitative trait loci (eQTL) in the RI mice were mapped in A) non-stimulated, B) IFNG+TNF-stimulated, C) Toxoplasma-infected, and D) CpG-Stimulated BMDM. Each dot represents a single eQTL (transcript). Significant eQTL located ≤ 10 Mb from the start of the physical location of the corresponding gene were designated as cis mapping (diagonal lines). All other eQTL were designated as trans-mapping (vertical lines). eQTL significance was calculated after 1000 permutations and reported at genome-wide thresholds corresponding to FDR ≤ 10%. Red spots identify genes mapping to a trans-eQTL hotpot (trans-band). QTL hotspots are enriched for biological processes and putative regulators that modulate the expression of multiple genes
Variable gene expression can be due to sequence or structural variations close to (cis) or further removed from (trans) the gene itself, such as polymorphism in the promoter regions or at a distal transcription factor, respectively. Consequently, relative to the physical location of the corresponding gene, an eQTL can be categorized as either cis or trans. Thus, we designated eQTL that co-localized within a 10 Mb genomic window with the corresponding gene as cis and all other eQTL as trans [72]. Except for the Toxoplasma-infected BMDM (407 cis vs. 601 trans), most of the eQTL were located in cis in: resting (99 cis vs. 32 trans), IFNG+TNF-stimulated (194 cis vs. 173 trans), and CpG-stimulated (482 cis vs. 206 trans) BMDM (S1A–S1D Dataset).
Suppose a common locus was to modulate the expression of multiple genes in trans, then in linkage analysis, we should expect the eQTL for these genes to co-localize in the vicinity of the common locus forming a trans-eQTL hotspot (trans-band). Indeed, similar to previous studies [23, 24, 73], and indicative of a common variant regulating the expression of multiple genes in trans, we detected trans-eQTL hotspots, within a 10 Mb window, in all the samples (Fig 2A and 2B and S1A–S1D Dataset). Because it is possible for eQTL to co-localize by chance alone, we used Bonferroni-corrected p-values and Poisson distribution to compute the number of trans-eQTL that can co-localize in a 10 Mb genomic window by chance. Using these cutoffs, we identified 2, 3, 5, and 15 trans-eQTL hotspots in the resting, IFNG+TNF-stimulated, CpG-stimulated, and Toxoplasma-infected BMDM, respectively (S1A–S1D Dataset).
Previously, it was reported that eQTL that localize close to the physical location of the relevant gene (cis-eQTL) are a consequence of single nucleotide polymorphisms (SNPs) [72] or larger structural variants (SV), such as insertions and deletions. Thus, we investigated the nucleotide sequence in a 2000 bp window upstream and downstream of the transcription start site (TSS) of all significant eQTL. Consistent with these reports, we found that most cis-eQTL were associated with genes reported to have structural or sequence variations within 2000 bp upstream or downstream of their TSS [74, 75]. That is, in control BMDM we found 89 out of 99 (Hypergeometric test P≤2.3e-7); in the IFNG+TNF-stimulated BMDM we found 156 out of 196 (Hypergeometric test P≤3.3e-10); in the CpG-stimulated BMDM we found 422 out of 482 (Hypergeometric test P≤3.8e-23); and in the Toxoplasma-infected BMDM we found 369 out of 407 (Hypergeometric test P≤1.6e-49) cis-eQTL with polymorphisms within 2000bp upstream or downstream of TSS. These included genes with known immunological functions such as Gbp1, Gbp2, Irak4 and Srebf1.
As indicated above, trans-eQTL hotspots can be due to transcriptional regulation of several genes by a common genetic variant [76], such as a polymorphic or differentially expressed transcription factor, enhancer or repressor. Alternatively, a trans-band can be a result of a differentially expressed or polymorphic signaling protein, such as a cell surface receptor, that could lead to differential activation of transcription factors and the genes downstream of these transcription factors. Consequently, the eQTL localizing at the trans-eQTL hotspot are likely to be enriched for binding sites for a common transcription factor(s) physically located at the trans-eQTL-hotspot, biological function, or signaling pathway. Therefore, we used gene ontology (GO) [77] and rVISTA [78] to functionally characterize and search for transcription factor binding site enrichment in each of the eQTL in the different trans-hotspots (Table 3). We found that the three largest trans-bands in the Toxoplasma infected BMDM mapped to loci containing genes with either known or putative roles in macrophage inflammatory or metabolic processes, which are important host responses against intracellular pathogens. For instance the trans-band on chromosome 13 (117.1–127.8 Mb) overlapped docking protein 3 (Dok3), chemokine (C-X-C motif) ligand 14 (Cxcl14), and AU RNA binding protein/enoyl-coenzyme A hydratase (Auh), which are known to regulate various aspects of cellular inflammatory and metabolic processes [79–81]. Indeed, this trans-band was enriched for, among others, “natural killer cell mediated immunity”. Similarly, the trans-band in the IFNG+TNF-stimulated BMDM on chromosome 15 (89.6–96.6 Mb), which was enriched for “regulation of leukocyte mediated immunity”, overlapped the interleukin-1 receptor-associated kinase 4 (Irak4), known to regulate the immune response to a variety of infectious and inflammatory stimuli [18, 82, 83]. Furthermore, Irak4 contains genetic insertions and deletions in AJ relative to the reference B6 mouse strain [84]. Thus, the trans-bands are functionally enriched in biologically relevant processes and can potentially reveal novel regulators and insight in the complex gene interactions that modulate macrophage response to exogenous stimuli.
Tab. 3. Functional enrichments in the large <i>trans</i>-eQTL hotspots in AXB/BXA macrophages following IFNG+TNF-stimulation, CpG-stimulation, or <i>Toxoplasma</i>-infection. 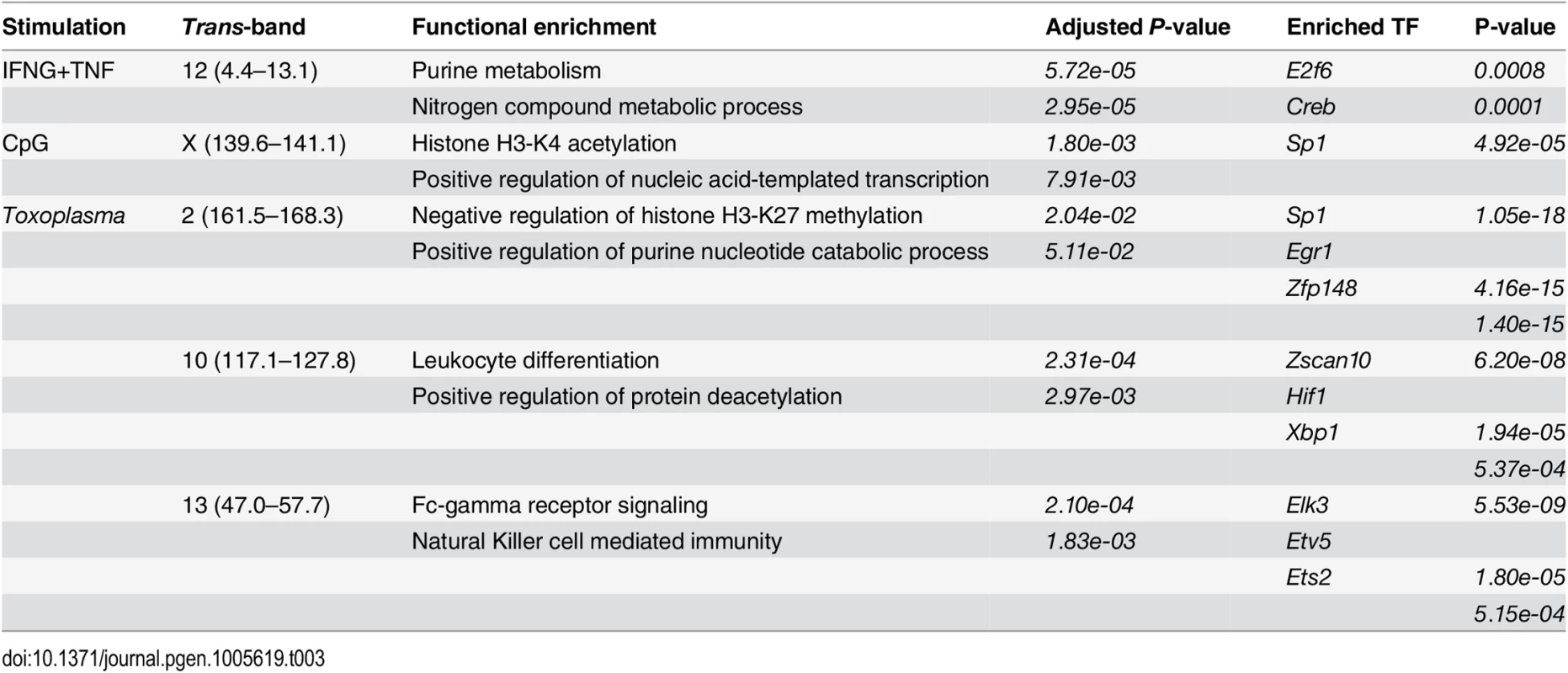
To identify putative regulators for the trans-bands, which can be variable transcription factors or signal transducers, we identified genes that were expressed (average FPKM ≥5), were physically located within 10 Mb on either side of the trans-eQTL hotspot, and were differentially expressed or had non-synonymous (NS) polymorphisms in AJ versus B6 mice. As an example, the chromosome 15 trans-band (between 79.7–99.7 Mb) contained 81 expressed coding and non-coding genes, 5 of which contained non-synonymous SNPs and 8 exhibited differential expression that mapped in cis. Of these genes, we considered Plxnb2, Irak4, and Apobec3 to be good candidates since they are known to be involved in immune response pathways [82, 85, 86], similar to the functional enrichment observed for the chromosome 15 trans-band. Additionally, these genes are polymorphic in AJ compared to B6. Due to insertions and deletions [87] and its function as an immune signaling adaptor, we considered Irak4 to be a strong candidate regulator for the chromosome 15 trans-band. Hence, we used shRNA to knockdown Irak4 in the IFNG+TNF-stimulated macrophages. Enrichment analysis on the perturbed genes following Irak4 knockdown revealed an overrepresentation (p = 0.005) of several chromosome 15 trans-eQTL (Table 4). shRNA-knockdown of the other putative candidate genes identified in this study did not perturbed most of the chromosome 15 trans-band eQTL (S1E Dataset), indicative of a specific effect of Irak4 knockdown on this trans-band.
Tab. 4. Genes from the chromosome 15 <i>trans</i>-band in IFNG+TNF-stimulated BMDM that were perturbed by at least 2 fold after <i>Irak4</i> knockdown and IFNG+TNF stimulation of murine BMDM. 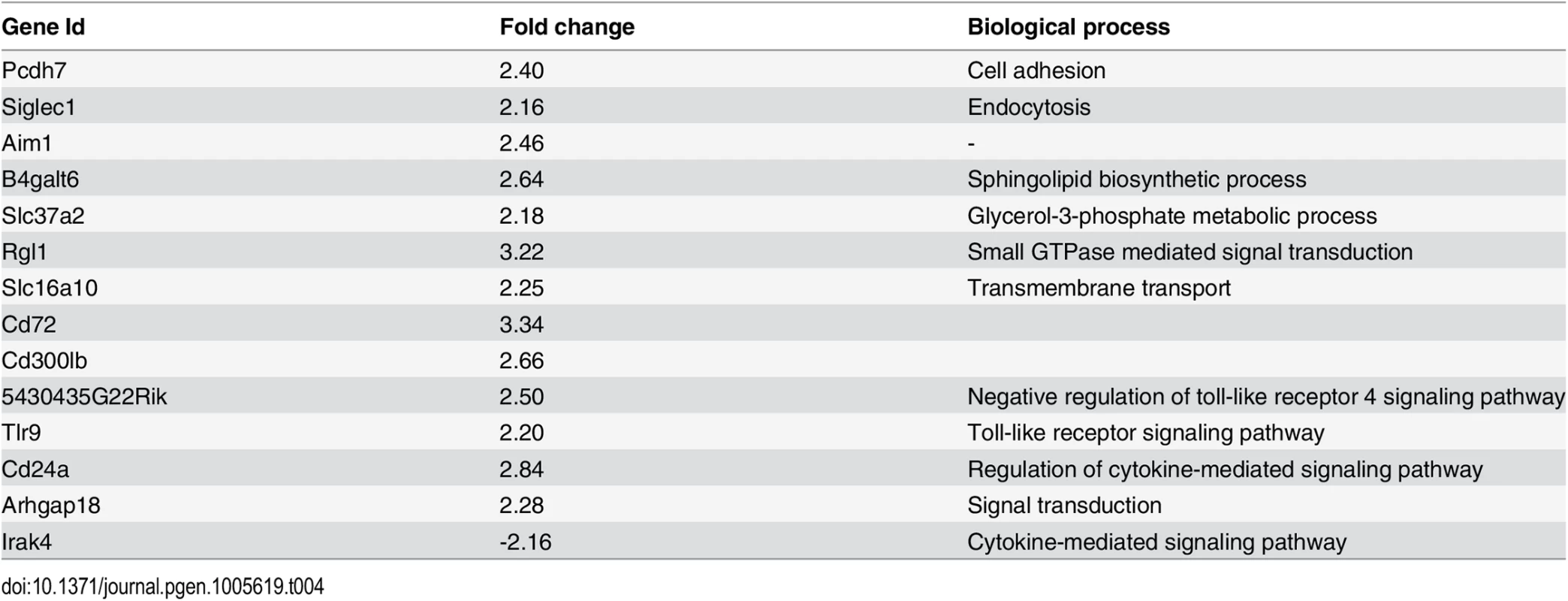
Leveraging eQTL analysis to identify candidate genes that modulate macrophage response to exogenous stimuli
Transcriptional networks capture the connectivity between genes modulating complex phenotypes and may provide a means to unravel the molecular mechanisms underlying complex traits. Because cis genetic variants account only for the phenotypes related to the gene they modulate, we reasoned that they are not good prototypes to illustrate how transcriptional, linkage and network analyses can be leveraged to systematically elucidate the genetic basis of a complex trait. Therefore, we used trans genetic variants, which potentially modulate multiple phenotypes, and followed a step-wise procedure [34] to identify the relationship between transcript levels, QTL, and BMDM phenotypes. First, to gain insight into the transcriptional architecture that modulate macrophage response to stimuli, we constructed gene co-expression network modules for each macrophage stimulation condition using the topological overlap matrix (TOM) in the weighed gene co-expression network analysis (WGCNA) program [88]. Next, using the eigenvalues for each module, we made correlations between each module and macrophage phenotypes. As proof of principle, we used this approach to nominate candidate genes that modulate the amount of NO produced in IFNG+TNF-stimulated BMDM. Because the amount of NO varied in the BMDM after IFNG+TNF, we correlated the co-expression modules with the amount of NO produced in the IFNG+TNF-stimulated BMDM (S3 Fig). Subsequently, we identified 4 modules (identified as white, light yellow, blue, and tan) that showed significant (P ≤ 0.01) correlation with NO levels (S3 Fig). It is important to note that, apart from being arbitrary identifiers for each module, the color code used to name each module reveals no further information. Each module, however, contains co-expressed genes (i.e. genes that exhibit transcriptional correlation across the 26 IFNG+TNF-stimulated BMDM). Of the 4 modules, the “white” module showed the greatest association with NO, hence we used it to illustrate our approach. Because the module-trait relationship is based on the correlation of the module eigenvalue with the amount of NO, not all the genes in the module will show significant association with the trait (amount of NO produced in the BMDM). Therefore, we further filtered the genes in the “white” module based on their individual relationship with NO (gene significance) (S1F Dataset). Expectedly, the eQTL for most of the genes in the module were localized on chromosome 12 and chromosome 4, the locations for NO QTL. To identify the potential regulator for cellular amounts of NO, we reasoned that if two genetic traits are both modulated by the same genetic variant, then the QTL for the two traits will co-localize at the common genetic variant [34]. Therefore, to further narrow down the significant genes and to identify the transcriptional network that likely modulate genetic differences in cellular amounts of NO produced after IFNG+TNF-stimulation, we searched for genes with eQTL that overlapped with the NO QTL on chromosome 12 (4.4–13.1 Mb) and found 11 eQTL (S1F Dataset). These eQTL were functionally enriched for “oxidoreductase activity” (p = 8.322e-04) and “nitrogen compound metabolism” (p = 1.345e-03) in gene ontology.
Because the common regulator for both NO levels and the chromosome 12 trans-band can be a polymorphic or differentially expressed transcription factor or signaling receptor physically located at the chromosome 12 trans-band locus, we narrowed the putative regulators by searching for cis-eQTL or expressed polymorphic genes at the chromosome 12 locus. Ddx1, a transcriptional regulator of cell cycle, maps in cis at this locus, and was considered a putative candidate for NO. Additionally, we used the Transcriptional Regulation Inference from Genetics of Gene Expression (Trigger) program [89] to establish the causal relationship between NO and the genes on chromosome 12, including Ddx1. Trigger utilizes randomized genetic backgrounds and phenotypes to test for causality between phenotypes that are linked to the same locus e.g. Ddx1 and NO. Of the chromosome 12 genes, only Ddx1 exhibited a minimum p-value (≤0.05) for causal relationship. As expected, the reciprocal analysis did not show NO or any of the chromosome 12 trans-eQTL as causal for Ddx1 differential expression. Indeed, shRNA-mediated knockdown of Ddx1 in immortalized B6 macrophages resulted in an increase in the amount of NO produced after IFNG+TNF-stimulation (Fig 3). Similar to NO, Ddx1 transcript abundance in IFNG+TNF-stimulated macrophages mapped to two loci on chromosome 4 and 12. However, Ddx1 transcript abundance exhibited an inverse relationship with the amount of macrophage NO at both loci (S4A–S4E Fig). Thus, we concluded that the expression of Ddx1, which is higher in B6 compared to AJ BMDM, inhibited NO production in the B6 macrophages.
Fig. 3. shRNA- mediated Ddx1 knockdown in C57BL/B6J immortalized macrophages relieves NO inhibition. 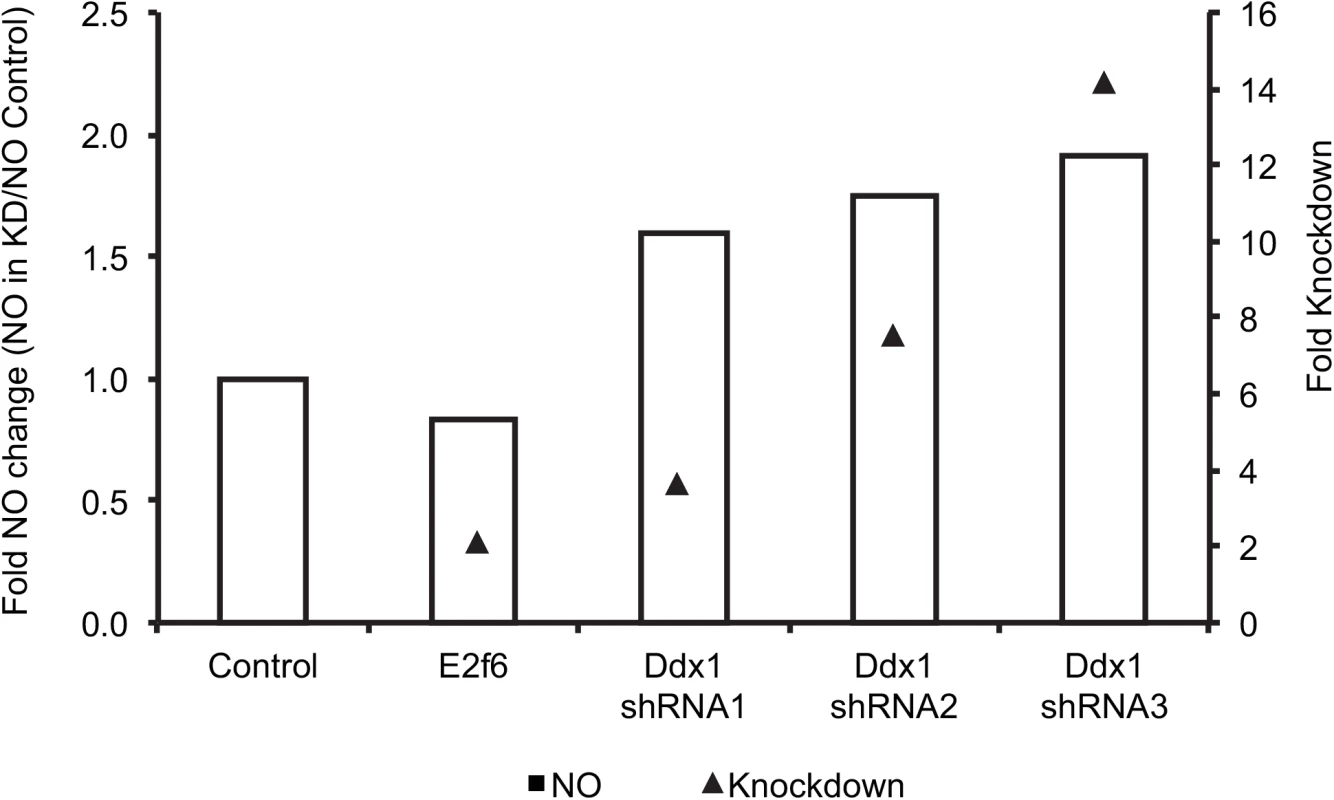
Fold knockdown of Ddx1 and fold change in NO levels are relative to the Ddx1 expression and the amount of NO in cells transduced with control shRNA (LacZ), respectively. Knocking down E2f6, the other candidate gene at this locus, did not affect the amount of NO produced. Shown are values of NO (μM) fold change obtained from two independent experiments using three different shRNA constructs. The knockdown level is indicated by the black triangles. Fold knockdown was calculated using the 2deltadelta Ct method. The shRNA transductions and NO measurements were done in 3 independent replicates. Due to the important role macrophages play in the pathology of various intracellular pathogens [90], we investigated whether there were overlaps between our trans-eQTL hotspots and other disease QTL in the AXB/BXA RI mice available in webQTL [91] and found several (S1A–S1D Dataset). For example the QTL for Listeria monocytogenes proliferation, which is attributed to macrophage inflammatory response [92], and susceptibility to hepatitis virus [93] overlapped the trans-eQTL hotspots on chromosome 15 and 7, respectively, in the IFNG+TNF-stimulated macrophages. Considering that IFNG+TNF is important in the pathogenesis of Listeria and Hepatitis infections [94–96], it is likely that these trans-bands harbor genes that modulate the outcome of infection with these and other pathogens.
Discussion
The activation of macrophages, in response to stimuli such as microbial components or immune factors, into the broadly defined classical, M(IFNG), and alternative, M(IL-4), phenotypes, determines whether they initiate or resolve immune responses. As gatekeepers against invading pathogens, the macrophage activation phenotype is essential in determining the persistence or resolution of infection. Thus, the hypothesis is that for infection to persist, the pathogen has to “trick” host macrophages to assume the “wrong” activation state and that variation in susceptibility to infection between hosts is due to genetic differences in macrophage response to the pathogen. Indeed empirical evidence shows that many pathogens can alter the macrophage activation to a phenotype that is favorable for its replication and persistence. For example, virulent strains of the intracellular parasite Toxoplasma gondii, which is vulnerable to M(IFNG) macrophages, induces the M(IL-4) phenotype in macrophages [12, 14, 97]. Similar observations have been made in leishmaniasis, in which the extent of macrophage modulation is dependent on the host [98]. On the other hand, macrophages from genetically segregating hosts have been shown to exhibit differential activation states, as captured by transcriptional and cytokine profiles [21, 71, 99–101]. Despite these documented pathogen-macrophage interplays and the variable inter-host macrophage activation phenotypes, the genetic basis for individual differences in macrophage polarization is just emerging. We describe differential activation of macrophages from the genetically segregating AJ and C57BL/6J (B6) mouse strains, which we have linked to the variable response to the obligate intracellular parasite, Toxoplasma gondii. Our in vitro model, which obviates the interference by other immune cells and involves naïve bone-marrow derived macrophages as opposed to elicited peritoneal macrophages, indicates that when stimulated with equal amounts of cytokines, AJ-derived macrophages produce a stronger M(IFNG) phenotype relative to B6 (as measured by NO). Additionally, we used a panel of genetically diverse recombinant inbred (RI) mice, derived from AJ and B6 mice, to investigate the specific loci responsible for this variable macrophage activation and the associated phenotypes.
In mice, nitric oxide (NO) production and L-Arginine metabolism are often used to define the M(IFNG) and M(IL-4) macrophage activation phenotypes, respectively [100, 102]. While the cellular levels of these factors vary between genetically divergent individuals and are known to contribute to the differential response to infection [101, 103, 104], the genetic loci that predispose macrophages to either the M(IFNG) or M(IL-4) phenotypes are not known. The general assumption is that a single locus will determine whether individual macrophages assume the M(IFNG) or M(IL-4) state. Results from the current study contradict this assumption and suggest that macrophage activation, as measured by levels of NO, IL-12, CCL22, IL-10 and Arginase I activity, is modulated by several loci, some with antagonistic effects. While the limited number of mice available from AXB/BXA RI line may partly explain the lack of statistically significant QTL for most of the macrophage phenotypes in this study, we submit that the genetic factors that modulate macrophage polarization form complex interaction networks that are not linked to a single locus. This conclusion is supported by the identification of two loci with contrasting effect on NO levels, the high levels of IL-12 and urea in B6 BMDM, and the high levels of NO and CCL22 in AJ BMDM. Furthermore, our observation of significant QTL peaks for IL-12 and CCL22 portends that the complex regulation of other phenotypes, such as NO, rather than the few number of mice, maybe the reason for the insignificant QTL peaks. Apart from the individual macrophage activation phenotypes, we did not observe the convergence of all the M(IFNG) (IL12, NO) at a single locus and the M(IL-4) phenotypes (IL10, CCL22 and urea) at another locus, instead each phenotype localized to a unique locus, again reinforcing the complex molecular circuits that modulate the M(IFNG) and M(IL-4) macrophage states. Although, the B6 mice carry a dominant negative mutation in the Slc7a2 gene, which is involved in L-Arginine transport and is postulated to contribute to the differential metabolism of L-Arginine in B6 relative to other mouse strains such as BALB/c [105], the QTL for L-Arginine metabolism did not localize to chromosome 8, which is the physical location of Slc7a2. Instead, the QTL for urea (a measure of Arginase activity) mapped to chromosome 10, proximal to Arg1, while the QTL for NO mapped to chromosome 12, respectively. It is plausible that both the transport and metabolism of L-Arginine contribute to the difference in urea and NO production in AJ and B6, hence the lack of significant QTL for these traits.
Modulation of host cellular signaling and transcriptional pathways by Toxoplasma is known to aid in immune evasion by the parasite and is achieved via the secretion of polymorphic effector proteins localized in the rhoptry and dense granule organelles [37, 106–108]. Specifically, the secretion of the polymorphic dense granule protein (GRA15) by the avirulent type II, and rhoptry kinase (ROP16) by the virulent type I Toxoplasma strains, is known to elicit classical and alternative macrophage activation, respectively [12]. Additionally, the secretion of these two Toxoplasma effector proteins is known to modulate intestinal pathology in the susceptible B6 mice [37]. However, to the best of our knowledge, the potential role of macrophages in Toxoplasma-induced intestinal pathology has not been shown. Furthermore, even though AJ and B6 are known to diverge in their response to in vivo Toxoplasma infection phenotypes, there has been no study showing that this variable response is due to a differential response of their macrophages to either IFNG or to the parasite itself. Together, the current study and our previous work [49], provide compelling evidence that macrophages may play an important role in Toxoplasma pathology. We postulate that alternative macrophage activation by Toxoplasma [12], and the differential AJ and B6 macrophage response to Toxoplasma and IFNG+TNF, provide the intersection of host-parasite interaction that harbors candidate genes mediating murine toxoplasmosis. Toxoplasma virulence appears to be related to its ability to skew macrophages towards alternative activation, which is abetted in susceptible animals, such as B6 mice.
In conclusion, our findings provide an extensive genetic analysis of the macrophage signaling processes in response to exogenous stimuli. Because activation of macrophages by IFNG and/or TNF confers resistance to a wide range of intracellular pathogens and human diseases, and because susceptibility loci for some of these phenotypes overlap, it is expected that this study will provide a framework to help identify candidate genes that mediate some of these disease phenotypes.
Material and Methods
Ethics statement
All animal experiments were performed in strict accordance with National Institutes of Health Guide for the Care and Use of Laboratory Animals and the Animal Welfare Act. The Massachusetts Institute of Technology Committee on Animal Care (assurance number A 3125–01) approved all protocols. All mice were maintained in specific pathogen-free conditions and euthanasia was performed in controlled CO2 chamber as approved by the MIT Animal Care Committee.
Primary bone marrow derived macrophages and parasites
Bone marrow-derived macrophages (BMDM) were obtained from 6–8 weeks old AJ, C57BL/6J and 26 female AXB/BXA recombinant inbred mice (Jackson Laboratories). Marrow cells were obtained from each mouse by flushing the femur and tibia with cold phosphate buffered saline (PBS; GIBCO-Invitrogen). The cells were then centrifuged at 500 x g for 5 minutes at 4°C and re-suspended in 4 ml red cell lysis buffer (Sigma) and incubated on ice for 5 minutes. Next, the cells were passed through a 70 μm cell strainer (BD Biosciences) and centrifuged at 500 x g for 5 minutes at 4 °C. The cells from each mouse were subsequently grown on four 10 cm non-tissue culture petri dishes (Corning) in Dulbecco's modified Eagle's medium (DMEM; GIBCO-Invitrogen) supplemented with 10% heat-inactivated fetal bovine serum (FBS; HyClone), 2 mM L-glutamine, 1 mM sodium pyruvate, 1X MEM nonessential amino acids, and 50 μg/ml each of penicillin and streptomycin. To differentiate the cells into macrophages, the DMEM was conditioned with 20% L929 cell supernatant (containing GM-CSF (40 ng/ml), hereafter 20% L929). After incubating the cells at 37°C and 5% CO2 for 3 days, the non-adherent cells were pipetted into 50 ml tubes and centrifuged at 500 x g for 5 minutes at 4°C and seeded in new 10 cm petri dishes. Simultaneously, the old 10 cm petri dishes were topped up with fresh media supplemented with 20% L929 i.e. in the end, for each mouse there were 8 petri dishes. After further incubation at 37°C and 5% CO2 for 4 days, the BMDM were harvested and stored in liquid nitrogen (5 million cells/ aliquot). The BMDM yield for each mouse strain ranged between 60–150 million cells, with the B6 mouse consistently producing more cells. This protocol has previously been shown to yield pure (>99%) macrophages [12]. A Pru (type II) Toxoplasma gondii strain engineered to express firefly luciferase and GFP (Pru ΔHXGPRT A7) [109], maintained in the laboratory by serial passage on Human Foreskin Fibroblasts (HFF), was used for all infections.
Immortalized macrophages
To immortalize macrophages, we used J2 recombinant retrovirus [110] produced from ψCREJ2 cells (a generous gift from John MacMicking, Yale University School of Medicine). The J2-expressing cells were grown to confluency in DMEM medium supplemented with 10% FBS (D10). The medium containing retroviral particles was collected and passed through 0.45 μM low protein-binding filters (Millipore). In parallel, 5 x 106 primary BMDM from AJ and C57BL/6J, obtained as described above, were thawed and grown for 2 days in DMEM medium supplemented with 10% FBS and 20% L929. After 2 days, the medium was replaced with the filtered J2-retrovirus-containing medium supplemented with 50% L929. After 24 hrs, the media was replaced with fresh D10 medium supplemented with 25% L929. Media was subsequently changed after every 24hrs with concomitant reduction in L929 until 10% L929 concentration was reached. Immortalized cells were harvested and stored in liquid nitrogen until use.
In vitro measurements
Unless otherwise stated, a total of 104 /well immortalized BMDM (iBMDM) or 105 /well primary BMDM were used in all the in vitro assays. Unless stated otherwise, before stimulation or infection, the primary BMDM or iBMDM were seeded overnight in D10 supplemented with 20% L929. Cellular nitric oxide levels were measured using the Griess reagent procedure on supernatant from non-stimulated or stimulated cells. Arginase activity was measured by quantifying the amount of urea as previously described [111]. Briefly, L-arginine was added to cell lysates and incubated at 37°C. After 1 hour, 175μl of an acid mixture containing sulfuric acid/phosphoric acid/water (H2SO4/H3PO4/H2O) in a 1 : 3:7 ratio, was added to each well to stop the enzymatic reaction. Urea was quantified calorimetrically at 540 nm after adding 1.25 μl of 1-phenyl-1,2-propanedione-2-oxime (ISPF) and heating at 95°C for 30–60 minutes. This procedure abrogates interference from other metabolites generated, such as L-citrulline [111]. IL10, and IL12 were measured on the relevant cell supernatants using ELISA kits as previously described [12]. For parasite growth assay, cells were either left unstimulated (control) or stimulated with recombinant mouse IFNG (100 ng/ml, Peprotech) and TNF (100 ng/ml, AbD serotec) for ~18 hr. The supernatant was removed for nitric oxide assay and replaced with D10 containing Toxoplasma at an MOI ~1. The parasites were allowed to infect and replicate for 24 hrs before luciferase activity was measured using a luciferase assay kit (Promega) according to the manufacturer recommendations.
RNA sequencing
Primary BMDM were plated (3 x106) overnight before stimulation or infection. For the stimulated samples, IFNG (100 ng/ml) and TNF (100 ng/ml) were added to each well for 18 hrs, while for the infected samples, a type II strain of Toxoplasma (Pru) was added to the confluent BMDM at an MOI of 1.3 for 8 hrs. Total RNA (Qiagen RNeasy Plus kit) was then isolated from the non-stimulated and non-infected cells (controls plated overnight), stimulated, and infected cells and the integrity, size, and concentration of RNA checked (Agilent 2100 Bioanalyser). The mRNA was then purified by polyA-tail enrichment (Dynabeads mRNA Purification Kit; Invitrogen), fragmented into 200–400 base-pairs, and reverse transcribed into cDNA before Illumina sequencing adapters were added to each end. Libraries were barcoded, multiplexed into 4 samples per sequencing lane in the Illumina HiSeq 2000, and sequenced from both ends resulting in 40 bp reads after discarding the barcodes. Our preliminary RNA-seq experiments with infected BMDM have shown that with 4 samples per lane, we still obtain enough read coverage for reliable gene expression analysis.
Gene expression measurements
Reads were initially mapped to the mouse genome (mm9) and the Toxoplasma (ME49 v8.2) genome using Bowtie (2.0.2) [112] and Tophat (v2.0.4) [66]. We then estimated gene expression levels in cufflinks (v2.0.0) [113] using the Illumina iGenomes refseq genome annotation (NCBI build 37.2) with the multi-read, compatible-hits corrections and upper quantile normalization options enabled. Because the reference genome to which we mapped the RNA-seq reads is based on the C57BL/6J genomic sequence, and due to the known polymorphisms between the AJ and C57BL/6J, we suspected that biases introduced at the read mapping stage might affect our expression results. To mitigate this potential bias towards the reference allele, we created a copy of the mouse genome in which all the known single nucleotide polymorphisms (SNPs) between AJ and B6, as annotated by the Wellcome Trust Sanger Institute sequencing (ftp://ftp-mouse.sanger.ac.uk/current_snps/), were converted to a third (neutral) nucleotide that is different from both the reference and AJ allele [67]. However, this did not substantially change the average proportion of uniquely mapped reads or expression profiles of individual genes in all the samples. In the end we used the mapping data generated from the synthetic genome to quantify gene expression levels.
Quantitative Trait Locus (QTL) mapping
To map QTL, we used 934 AXB/BXA genetic informative markers obtained from http://www.genenetwork.org. For all the in vitro measurements and gene expression linkage analysis, a genome-wide scan was performed using R/qtl [57]. Significance of QTL logarithm-of-odds (LOD) scores was assessed using 1000 permutations of the phenotype data [114] and the corresponding p-values reported. For the cellular phenotypes, QTL significance was reported at a genome-wide threshold corresponding to p < 0.05. However, for eQTL mapping, we further corrected for multiple testing on the multiple transcripts by using the p-values to estimate false discovery rate (FDR) in the qvalue package [115] and reported significant eQTL at FDR ≤ 10%. To identify cis- and trans - eQTL, we computed the distance from the position of the eQTL and the start of the physical location of the corresponding gene and designated any eQTL located <10 Mb from the corresponding gene as cis, otherwise trans eQTL. The procedure used to determine trans-bands has previously been described [25].
Validation of candidate regulators
We used shRNA to probe for functional or regulatory significance of some of the candidates identified in our analysis. To do this, we used C57BL/6J immortalized bone marrow-derived macrophages, described above. One day after plating, we added shRNA constructs containing a puromycin resistance marker (RNAi Platform, Broad Institute) in the presence of 8 μg/ml polybrene, followed by centrifugation at 800 x g for 2 hrs at 37°C. At the end of the spinfection, the cells were incubated for an additional 24 hrs at 37°C in 5% CO2. The cells were then grown in fresh cell culture medium for an additional 24 hrs before adding 4 μg/ml puromycin. Transcript knockdown was measured by quantitative reverse transcriptase polymerase chain reaction (qRT-PCR) using the KAPA SYBR FAST Universal 2X qPCR Master Mix (KAPA Biosystems) on a LightCycler qPCR instrument (Roche). Fold knockdown was measured using the 2 delta-delta method [116] relative to LacZ-shRNA transduced cells. The puromycin-selected cells were either left stimulated or stimulated with IFNG+TNF, as described above, and the cell supernatant collected for nitric oxide assay.
Data access
The microarray data is available at the NCBI Gene Expression Omnibus archive under accession number GSE47046.
Supporting Information
Zdroje
1. Murray PJ, Allen JE, Biswas SK, Fisher EA, Gilroy DW, Goerdt S, et al. Macrophage activation and polarization: nomenclature and experimental guidelines. Immunity. 2014;41(1):14–20. doi: 10.1016/j.immuni.2014.06.008 25035950
2. Biswas SK, Mantovani A. Macrophage plasticity and interaction with lymphocyte subsets: cancer as a paradigm. Nat Immunol. 2010;11(10):889–96. doi: 10.1038/ni.1937 20856220
3. Martinez FO, Gordon S. The M1 and M2 paradigm of macrophage activation: time for reassessment. F1000prime reports. 2014;6 : 13. doi: 10.12703/P6-13 24669294
4. Auffray C, Sieweke MH, Geissmann F. Blood monocytes: development, heterogeneity, and relationship with dendritic cells. Annual review of immunology. 2009;27 : 669–92. Epub 2009/01/10. doi: 10.1146/annurev.immunol.021908.132557 19132917
5. MacMicking JD. Recognizing macrophage activation and host defense. Cell Host & Microbe. 2009;5(5):405–7. Epub 2009/05/21.
6. Cirelli KM, Gorfu G, Hassan MA, Printz M, Crown D, Leppla SH, et al. Inflammasome sensor NLRP1 controls rat macrophage susceptibility to Toxoplasma gondii. PLoS Pathog. 2014;10(3):e1003927. doi: 10.1371/journal.ppat.1003927 24626226
7. Gordon S, Martinez FO. Alternative activation of macrophages: mechanism and functions. Immunity. 2010;32(5):593–604. Epub 2010/06/01. doi: 10.1016/j.immuni.2010.05.007 20510870
8. Gordon S, Taylor PR. Monocyte and macrophage heterogeneity. Nature reviews Immunology. 2005;5(12):953–64. Epub 2005/12/03. 16322748
9. Martinez FO, Helming L, Gordon S. Alternative activation of macrophages: an immunologic functional perspective. Annual review of immunology. 2009;27 : 451–83. Epub 2008/12/25. doi: 10.1146/annurev.immunol.021908.132532 19105661
10. Price JV, Vance RE. The macrophage paradox. Immunity. 2014;41(5):685–93. doi: 10.1016/j.immuni.2014.10.015 25517611
11. Benoit M, Desnues B, Mege JL. Macrophage polarization in bacterial infections. J Immunol. 2008;181(6):3733–9. 18768823
12. Jensen KDC, Wang Y, Wojno EDT, Shastri AJ, Hu K, Cornel L, et al. Toxoplasma Polymorphic Effectors Determine Macrophage Polarization and Intestinal Inflammation. Cell Host & Microbe. 2011;9(6):472–83.
13. Mege JL, Mehraj V, Capo C. Macrophage polarization and bacterial infections. Current opinion in infectious diseases. 2011;24(3):230–4. doi: 10.1097/QCO.0b013e328344b73e 21311324
14. Melo MB, Jensen KD, Saeij JP. Toxoplasma gondii effectors are master regulators of the inflammatory response. Trends in parasitology. 2011;27(11):487–95. Epub 2011/09/07. doi: 10.1016/j.pt.2011.08.001 21893432
15. Melo MB, Nguyen QP, Cordeiro C, Hassan MA, Yang N, McKell R, et al. Transcriptional Analysis of Murine Macrophages Infected with Different Toxoplasma Strains Identifies Novel Regulation of Host Signaling Pathways. PLoS Pathog. 2013
16. Thi EP, Lambertz U, Reiner NE. Sleeping with the enemy: how intracellular pathogens cope with a macrophage lifestyle. PLoS Pathog. 2012;8(3):e1002551. doi: 10.1371/journal.ppat.1002551 22457616
17. Mills C. M1 and m2 macrophages: oracles of health and disease. Critical reviews in immunology. 2012;32(6):463–88. Epub 2013/02/23. 23428224
18. Bela SR, Dutra MS, Mui E, Montpetit A, Oliveira FS, Oliveira SC, et al. Impaired innate immunity in mice deficient in interleukin-1 receptor-associated kinase 4 leads to defective type 1 T cell responses, B cell expansion, and enhanced susceptibility to infection with Toxoplasma gondii. Infection and immunity. 2012;80(12):4298–308. Epub 2012/10/03. doi: 10.1128/IAI.00328-12 23027530
19. Marquis JF, Gros P. Genetic analysis of resistance to infections in mice: A/J meets C57BL/6J. Current topics in microbiology and immunology. 2008;321 : 27–57. 18727486
20. Witola WH, Mui E, Hargrave A, Liu S, Hypolite M, Montpetit A, et al. NALP1 influences susceptibility to human congenital toxoplasmosis, proinflammatory cytokine response, and fate of Toxoplasma gondii-infected monocytic cells. Infect Immun. 2011;79(2):756–66. doi: 10.1128/IAI.00898-10 21098108
21. Kim S, Becker J, Bechheim M, Kaiser V, Noursadeghi M, Fricker N, et al. Characterizing the genetic basis of innate immune response in TLR4-activated human monocytes. Nature communications. 2014;5 : 5236. doi: 10.1038/ncomms6236 25327457
22. Xue J, Schmidt SV, Sander J, Draffehn A, Krebs W, Quester I, et al. Transcriptome-based network analysis reveals a spectrum model of human macrophage activation. Immunity. 2014;40(2):274–88. doi: 10.1016/j.immuni.2014.01.006 24530056
23. Barreiro LB, Tailleux L, Pai AA, Gicquel B, Marioni JC, Gilad Y. Deciphering the genetic architecture of variation in the immune response to Mycobacterium tuberculosis infection. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(4):1204–9. Epub 2012/01/12. doi: 10.1073/pnas.1115761109 22233810
24. Orozco LD, Bennett BJ, Farber CR, Ghazalpour A, Pan C, Che N, et al. Unraveling Inflammatory Responses using Systems Genetics and Gene-Environment Interactions in Macrophages. Cell. 2012;151(3):658–70. Epub 2012/10/30. doi: 10.1016/j.cell.2012.08.043 23101632
25. Hassan MA, Butty V, Jensen KD, Saeij JP. The genetic basis for individual differences in mRNA splicing and APOBEC1 editing activity in murine macrophages. Genome Res. 2014;24(3):377–89. doi: 10.1101/gr.166033.113 24249727
26. Chawla A. Control of macrophage activation and function by PPARs. Circulation research. 2010;106(10):1559–69. doi: 10.1161/CIRCRESAHA.110.216523 20508200
27. Date D, Das R, Narla G, Simon DI, Jain MK, Mahabeleshwar GH. Kruppel-like transcription factor 6 regulates inflammatory macrophage polarization. J Biol Chem. 2014;289(15):10318–29. doi: 10.1074/jbc.M113.526749 24385430
28. Almasy L, Blangero J. Human QTL linkage mapping. Genetica. 2009;136(2):333–40. doi: 10.1007/s10709-008-9305-3 18668207
29. McPeek MS. From mouse to human: fine mapping of quantitative trait loci in a model organism. Proc Natl Acad Sci U S A. 2000;97(23):12389–90. 11050190
30. Majewski J, Pastinen T. The study of eQTL variations by RNA-seq: from SNPs to phenotypes. Trends in genetics: TIG. 2011;27(2):72–9. Epub 2010/12/03. doi: 10.1016/j.tig.2010.10.006 21122937
31. Goring HH, Curran JE, Johnson MP, Dyer TD, Charlesworth J, Cole SA, et al. Discovery of expression QTLs using large-scale transcriptional profiling in human lymphocytes. Nature genetics. 2007;39(10):1208–16. 17873875
32. Jansen RC, Nap JP. Genetical genomics: the added value from segregation. Trends in genetics: TIG. 2001;17(7):388–91. Epub 2001/06/22. 11418218
33. Lee MN, Ye C, Villani AC, Raj T, Li W, Eisenhaure TM, et al. Common genetic variants modulate pathogen-sensing responses in human dendritic cells. Science. 2014;343(6175):1246980. doi: 10.1126/science.1246980 24604203
34. Schadt EE, Lamb J, Yang X, Zhu J, Edwards S, Guhathakurta D, et al. An integrative genomics approach to infer causal associations between gene expression and disease. Nature genetics. 2005;37(7):710–7. Epub 2005/06/21. 15965475
35. Tu Z, Keller MP, Zhang C, Rabaglia ME, Greenawalt DM, Yang X, et al. Integrative analysis of a cross-Loci regulation network identifies App as a gene regulating insulin secretion from pancreatic islets. Plos Genet. 2012;8(12):e1003107. Epub 2012/12/14. doi: 10.1371/journal.pgen.1003107 23236292
36. Nesbitt MN, Skamene E. Recombinant inbred mouse strains derived from A/J and C57BL/6J: a tool for the study of genetic mechanisms in host resistance to infection and malignancy. Journal of leukocyte biology. 1984;36(3):357–64. 6592283
37. Jensen KD, Hu K, Whitmarsh RJ, Hassan MA, Julien L, Lu D, et al. Toxoplasma rhoptry kinase ROP16 promotes host resistance to oral infection and intestinal inflammation only in the context of the dense granule protein GRA15. Infection and immunity. 2013. Epub 2013/04/03.
38. McLeod R, Skamene E, Brown CR, Eisenhauer PB, Mack DG. Genetic regulation of early survival and cyst number after peroral Toxoplasma gondii infection of A x B/B x A recombinant inbred and B10 congenic mice. Journal of immunology. 1989;143(9):3031–4. Epub 1989/11/01.
39. Rong J, Zhang W, Wang X, Fan H, Lu C, Yao H. Identification of candidate susceptibility and resistance genes of mice infected with Streptococcus suis type 2. PLoS One. 2012;7(2):e32150. doi: 10.1371/journal.pone.0032150 22384161
40. Tuite A, Elias M, Picard S, Mullick A, Gros P. Genetic control of susceptibility to Candida albicans in susceptible A/J and resistant C57BL/6J mice. Genes and immunity. 2005;6(8):672–82. 16079897
41. Mullick A, Elias M, Picard S, Bourget L, Jovcevski O, Gauthier S, et al. Dysregulated inflammatory response to Candida albicans in a C5-deficient mouse strain. Infect Immun. 2004;72(10):5868–76. 15385488
42. Ichihara M, Hara T, Takagi M, Cho LC, Gorman DM, Miyajima A. Impaired interleukin-3 (IL-3) response of the A/J mouse is caused by a branch point deletion in the IL-3 receptor alpha subunit gene. The EMBO journal. 1995;14(5):939–50. 7889941
43. Fulton WB, Reeves RH, Takeya M, De Maio A. A quantitative trait loci analysis to map genes involved in lipopolysaccharide-induced inflammatory response: identification of macrophage scavenger receptor 1 as a candidate gene. J Immunol. 2006;176(6):3767–73. 16517746
44. Dominguez-Punaro Mde L, Segura M, Radzioch D, Rivest S, Gottschalk M. Comparison of the susceptibilities of C57BL/6 and A/J mouse strains to Streptococcus suis serotype 2 infection. Infect Immun. 2008;76(9):3901–10. doi: 10.1128/IAI.00350-08 18573893
45. Ahn SH, Deshmukh H, Johnson N, Cowell LG, Rude TH, Scott WK, et al. Two genes on A/J chromosome 18 are associated with susceptibility to Staphylococcus aureus infection by combined microarray and QTL analyses. PLoS pathogens. 2010;6(9):e1001088. Epub 2010/09/09. doi: 10.1371/journal.ppat.1001088 20824097
46. Parekh PI, Petro AE, Tiller JM, Feinglos MN, Surwit RS. Reversal of diet-induced obesity and diabetes in C57BL/6J mice. Metabolism: clinical and experimental. 1998;47(9):1089–96. Epub 1998/09/29.
47. Silva GK, Cunha LD, Horta CV, Silva ALN, Gutierrez FRS, Silva JS, et al. A Parent-of-Origin Effect Determines the Susceptibility of a Non-Informative F1 Population to Trypanosoma cruzi Infection In Vivo. PLoS One. 2013;8(2).
48. Takacs AC, Swierzy IJ, Luder CG. Interferon-gamma restricts Toxoplasma gondii development in murine skeletal muscle cells via nitric oxide production and immunity-related GTPases. PloS one. 2012;7(9):e45440. Epub 2012/10/02. 23024821
49. Jensen KD, Hu K, Whitmarsh RJ, Hassan MA, Julien L, Lu D, et al. Toxoplasma gondii rhoptry 16 kinase promotes host resistance to oral infection and intestinal inflammation only in the context of the dense granule protein GRA15. Infect Immun. 2013;81(6):2156–67. Epub 2013/04/03. doi: 10.1128/IAI.01185-12 23545295
50. Kim SK, Karasov A, Boothroyd JC. Bradyzoite-specific surface antigen SRS9 plays a role in maintaining Toxoplasma gondii persistence in the brain and in host control of parasite replication in the intestine. Infect Immun. 2007;75(4):1626–34. 17261600
51. Rosowski EE, Lu D, Julien L, Rodda L, Gaiser RA, Jensen KD, et al. Strain-specific activation of the NF-kappaB pathway by GRA15, a novel Toxoplasma gondii dense granule protein. The Journal of experimental medicine. 2011;208(1):195–212. Epub 2011/01/05. doi: 10.1084/jem.20100717 21199955
52. Saeij JP, Boyle JP, Grigg ME, Arrizabalaga G, Boothroyd JC. Bioluminescence imaging of Toxoplasma gondii infection in living mice reveals dramatic differences between strains. Infect Immun. 2005;73(2):695–702. 15664907
53. Hunter CA, Sibley LD. Modulation of innate immunity by Toxoplasma gondii virulence effectors. Nature reviews Microbiology. 2012;10(11):766–78. Epub 2012/10/17. doi: 10.1038/nrmicro2858 23070557
54. Woods S, Schroeder J, McGachy HA, Plevin R, Roberts CW, Alexander J. MAP kinase phosphatase-2 plays a key role in the control of infection with Toxoplasma gondii by modulating iNOS and arginase-1 activities in mice. PLoS Pathog. 2013;9(8):e1003535. doi: 10.1371/journal.ppat.1003535 23966857
55. Boyle AE, Gill K. Sensitivity of AXB/BXA recombinant inbred lines of mice to the locomotor activating effects of cocaine: a quantitative trait loci analysis. Pharmacogenetics. 2001;11(3):255–64. 11337941
56. Sampson SB, Higgins DC, Elliot RW, Taylor BA, Lueders KK, Koza RA, et al. An edited linkage map for the AXB and BXA recombinant inbred mouse strains. Mammalian genome: official journal of the International Mammalian Genome Society. 1998;9(9):688–94. Epub 1998/08/26.
57. Broman KW, Wu H, Sen S, Churchill GA. R/qtl: QTL mapping in experimental crosses. Bioinformatics. 2003;19(7):889–90. Epub 2003/05/02. 12724300
58. Burrage LC, Baskin-Hill AE, Sinasac DS, Singer JB, Croniger CM, Kirby A, et al. Genetic resistance to diet-induced obesity in chromosome substitution strains of mice. Mammalian Genome. 2010;21(3–4):115–29. doi: 10.1007/s00335-010-9247-9 20127486
59. Selleck EM, Fentress SJ, Beatty WL, Degrandi D, Pfeffer K, Virgin HW, et al. Guanylate-binding Protein 1 (Gbp1) Contributes to Cell-autonomous Immunity against Toxoplasma gondii. PLoS pathogens. 2013;9(4):e1003320. doi: 10.1371/journal.ppat.1003320 23633952
60. Virreira Winter S, Niedelman W, Jensen KD, Rosowski EE, Julien L, Spooner E, et al. Determinants of GBP recruitment to Toxoplasma gondii vacuoles and the parasitic factors that control it. PloS one. 2011;6(9):e24434. Epub 2011/09/21. doi: 10.1371/journal.pone.0024434 21931713; PubMed Central PMCID: PMC3169597.
61. Degrandi D, Kravets E, Konermann C, Beuter-Gunia C, Klumpers V, Lahme S, et al. Murine guanylate binding protein 2 (mGBP2) controls Toxoplasma gondii replication. Proc Natl Acad Sci U S A. 2013;110(1):294–9. doi: 10.1073/pnas.1205635110 23248289
62. Yamamoto M, Okuyama M, Ma JS, Kimura T, Kamiyama N, Saiga H, et al. A cluster of interferon-gamma-inducible p65 GTPases plays a critical role in host defense against Toxoplasma gondii. Immunity. 2012;37(2):302–13. Epub 2012/07/17. 22795875
63. Burrage LC, Baskin-Hill AE, Sinasac DS, Singer JB, Croniger CM, Kirby A, et al. Genetic resistance to diet-induced obesity in chromosome substitution strains of mice. Mamm Genome. 2010;21(3–4):115–29. doi: 10.1007/s00335-010-9247-9 20127486
64. Schadt EE. Novel integrative genomics strategies to identify genes for complex traits. Animal Genetics. 2006;37 : 18–23. 16886998
65. Raza S, Barnett MW, Barnett-Itzhaki Z, Amit I, Hume DA, Freeman TC. Analysis of the transcriptional networks underpinning the activation of murine macrophages by inflammatory mediators. Journal of leukocyte biology. 2014;96(2):167–83. doi: 10.1189/jlb.6HI0313-169R 24721704
66. Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25(9):1105–11. doi: 10.1093/bioinformatics/btp120 19289445
67. Degner JF, Marioni JC, Pai AA, Pickrell JK, Nkadori E, Gilad Y, et al. Effect of read-mapping biases on detecting allele-specific expression from RNA-sequencing data. Bioinformatics. 2009;25(24):3207–12. Epub 2009/10/08. doi: 10.1093/bioinformatics/btp579 19808877
68. Panousis NI, Gutierrez-Arcelus M, Dermitzakis ET, Lappalainen T. Allelic mapping bias in RNA-sequencing is not a major confounder in eQTL studies. Genome biology. 2014;15(9):467. doi: 10.1186/s13059-014-0467-2 25239376
69. Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nature protocols. 2012;7(3):562–78. Epub 2012/03/03. doi: 10.1038/nprot.2012.016 22383036
70. Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci U S A. 2003;100(16):9440–5. Epub 2003/07/29. 12883005
71. Fairfax BP, Humburg P, Makino S, Naranbhai V, Wong D, Lau E, et al. Innate immune activity conditions the effect of regulatory variants upon monocyte gene expression. Science. 2014;343(6175):1246949. doi: 10.1126/science.1246949 24604202
72. Doss S, Schadt EE, Drake TA, Lusis AJ. Cis-acting expression quantitative trait loci in mice. Genome Res. 2005;15(5):681–91. Epub 2005/04/20. 15837804
73. Ghazalpour A, Doss S, Kang H, Farber C, Wen PZ, Brozell A, et al. High-resolution mapping of gene expression using association in an outbred mouse stock. Plos Genet. 2008;4(8):e1000149. Epub 2008/08/09. doi: 10.1371/journal.pgen.1000149 18688273; PubMed Central PMCID: PMC2483929.
74. Keane TM, Goodstadt L, Danecek P, White MA, Wong K, Yalcin B, et al. Mouse genomic variation and its effect on phenotypes and gene regulation. Nature. 2011;477(7364):289–94. Epub 2011/09/17. doi: 10.1038/nature10413 21921910
75. Maddatu TP, Grubb SC, Bult CJ, Bogue MA. Mouse Phenome Database (MPD). Nucleic acids research. 2012;40(Database issue):D887–94. Epub 2011/11/22. doi: 10.1093/nar/gkr1061 22102583
76. Wu C, Delano DL, Mitro N, Su SV, Janes J, McClurg P, et al. Gene set enrichment in eQTL data identifies novel annotations and pathway regulators. Plos Genet. 2008;4(5):e1000070. Epub 2008/05/10. doi: 10.1371/journal.pgen.1000070 18464898
77. Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nature genetics. 2000;25(1):25–9. 10802651
78. Dubchak I, Munoz M, Poliakov A, Salomonis N, Minovitsky S, Bodmer R, et al. Whole-Genome rVISTA: a tool to determine enrichment of transcription factor binding sites in gene promoters from transcriptomic data. Bioinformatics. 2013;29(16):2059–61. doi: 10.1093/bioinformatics/btt318 23736530
79. Ozawa S, Kato Y, Kubota E, Hata R. BRAK/CXCL14 expression in oral carcinoma cells completely suppresses tumor cell xenografts in SCID mouse. Biomedical research. 2009;30(5):315–8. 19887729
80. Leite TC, Coelho RG, Da Silva D, Coelho WS, Marinho-Carvalho MM, Sola-Penna M. Lactate downregulates the glycolytic enzymes hexokinase and phosphofructokinase in diverse tissues from mice. FEBS letters. 2011;585(1):92–8. doi: 10.1016/j.febslet.2010.11.009 21074528
81. Nakagawa J, Waldner H, Meyer-Monard S, Hofsteenge J, Jeno P, Moroni C. AUH, a gene encoding an AU-specific RNA binding protein with intrinsic enoyl-CoA hydratase activity. Proc Natl Acad Sci U S A. 1995;92(6):2051–5. 7892223
82. Pennini ME, Perkins DJ, Salazar AM, Lipsky M, Vogel SN. Complete Dependence on IRAK4 Kinase Activity in TLR2, but Not TLR4, Signaling Pathways Underlies Decreased Cytokine Production and Increased Susceptibility to Streptococcus pneumoniae Infection in IRAK4 Kinase-Inactive Mice. Journal of immunology. 2013;190(1):307–16. Epub 2012/12/05.
83. Xiong Y, Medvedev AE. Induction of endotoxin tolerance in vivo inhibits activation of IRAK4 and increases negative regulators IRAK-M, SHIP-1, and A20. Journal of leukocyte biology. 2011;90(6):1141–8. doi: 10.1189/jlb.0611273 21934070
84. Yalcin B, Wong K, Agam A, Goodson M, Keane TM, Gan XC, et al. Sequence-based characterization of structural variation in the mouse genome. Nature. 2011;477(7364):326–9. doi: 10.1038/nature10432 21921916
85. Holl EK, Roney KE, Allen IC, Steinbach E, Arthur JC, Buntzman A, et al. Plexin-B2 and Plexin-D1 in dendritic cells: expression and IL-12/IL-23p40 production. PloS one. 2012;7(8):e43333. Epub 2012/08/24. doi: 10.1371/journal.pone.0043333 22916243
86. Mehta HV, Jones PH, Weiss JP, Okeoma CM. IFN-alpha and lipopolysaccharide upregulate APOBEC3 mRNA through different signaling pathways. Journal of immunology. 2012;189(8):4088–103. Epub 2012/09/14.
87. Yalcin B, Wong K, Agam A, Goodson M, Keane TM, Gan X, et al. Sequence-based characterization of structural variation in the mouse genome. Nature. 2011;477(7364):326–9. Epub 2011/09/17. doi: 10.1038/nature10432 21921916
88. Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC bioinformatics. 2008;9 : 559. doi: 10.1186/1471-2105-9-559 19114008
89. Chen LS, Emmert-Streib F, Storey JD. Harnessing naturally randomized transcription to infer regulatory relationships among genes. Genome biology. 2007;8(10):R219. 17931418
90. Jankovic D, Liu Z, Gause WC. Th1 - and Th2-cell commitment during infectious disease: asymmetry in divergent pathways. Trends in immunology. 2001;22(8):450–7. Epub 2001/07/28. 11473835
91. Wang J, Williams RW, Manly KF. WebQTL: web-based complex trait analysis. Neuroinformatics. 2003;1(4):299–308. Epub 2004/03/27. 15043217
92. Stevenson MM, Gervais F, Skamene E. Natural-Resistance to Listeriosis—Role of Host Inflammatory Responsiveness. Clinical and Investigative Medicine-Medecine Clinique Et Experimentale. 1984;7(4):297–301. 6442652
93. Dindzans VJ, Skamene E, Levy GA. Susceptibility Resistance to Mouse Hepatitis-Virus Strain-3 and Macrophage Procoagulant Activity Are Genetically Linked and Controlled by 2 Non-H-2-Linked Genes. Journal of immunology. 1986;137(7):2355–60.
94. Yin J, Ferguson TA. Identification of an IFN-gamma-producing neutrophil early in the response to Listeria monocytogenes. J Immunol. 2009;182(11):7069–73. doi: 10.4049/jimmunol.0802410 19454704
95. Hennig BJ, Fielding K, Broxholme J, Diatta M, Mendy M, Moore C, et al. Host genetic factors and vaccine-induced immunity to hepatitis B virus infection. PLoS One. 2008;3(3):e1898. doi: 10.1371/journal.pone.0001898 18365030
96. Xu H, Li H, Cao D, Wu Y, Chen Y. Tumor necrosis factor alpha (TNF-alpha) receptor-I is required for TNF-alpha-mediated fulminant virus hepatitis caused by murine hepatitis virus strain-3 infection. Immunology letters. 2014;158(1–2):25–32. doi: 10.1016/j.imlet.2013.11.008 24286726
97. Butcher BA, Fox BA, Rommereim LM, Kim SG, Maurer KJ, Yarovinsky F, et al. Toxoplasma gondii rhoptry kinase ROP16 activates STAT3 and STAT6 resulting in cytokine inhibition and arginase-1-dependent growth control. PLoS Pathog. 2011;7(9):e1002236. doi: 10.1371/journal.ppat.1002236 21931552
98. Wanderley JL, Moreira ME, Benjamin A, Bonomo AC, Barcinski MA. Mimicry of apoptotic cells by exposing phosphatidylserine participates in the establishment of amastigotes of Leishmania (L) amazonensis in mammalian hosts. J Immunol. 2006;176(3):1834–9. 16424214
99. Zhao ZJ, Zhang J, Wei J, Li Z, Wang T, Yi SQ, et al. Lower expression of inducible nitric oxide synthase and higher expression of arginase in rat alveolar macrophages are linked to their susceptibility to Toxoplasma gondii infection. PLoS One. 2013;8(5):e63650. doi: 10.1371/journal.pone.0063650 23691079
100. Mills CD, Kincaid K, Alt JM, Heilman MJ, Hill AM. M-1/M-2 macrophages and the Th1/Th2 paradigm. J Immunol. 2000;164(12):6166–73. 10843666
101. Li Z, Zhao ZJ, Zhu XQ, Ren QS, Nie FF, Gao JM, et al. Differences in iNOS and arginase expression and activity in the macrophages of rats are responsible for the resistance against T. gondii infection. PloS one. 2012;7(4):e35834. Epub 2012/05/05. doi: 10.1371/journal.pone.0035834 22558235
102. Mills CD. Molecular basis of "suppressor" macrophages. Arginine metabolism via the nitric oxide synthetase pathway. J Immunol. 1991;146(8):2719–23. 1707918
103. MacMicking J, Xie QW, Nathan C. Nitric oxide and macrophage function. Annual review of immunology. 1997;15 : 323–50. 9143691
104. Karupiah G, Xie QW, Buller RM, Nathan C, Duarte C, MacMicking JD. Inhibition of viral replication by interferon-gamma-induced nitric oxide synthase. Science. 1993;261(5127):1445–8. 7690156
105. Sans-Fons MG, Yeramian A, Pereira-Lopes S, Santamaria-Babi LF, Modolell M, Lloberas J, et al. Arginine Transport Is Impaired in C57Bl/6 Mouse Macrophages as a Result of a Deletion in the Promoter of Slc7a2 (CAT2), and Susceptibility to Leishmania Infection Is Reduced. J Infect Dis. 2013;207(11):1684–93. doi: 10.1093/infdis/jit084 23460752
106. Fentress SJ, Behnke MS, Dunay IR, Mashayekhi M, Rommereim LM, Fox BA, et al. Phosphorylation of immunity-related GTPases by a Toxoplasma gondii-secreted kinase promotes macrophage survival and virulence. Cell Host & Microbe. 2010;8(6):484–95. Epub 2010/12/15.
107. Braun L, Brenier-Pinchart MP, Yogavel M, Curt-Varesano A, Curt-Bertini RL, Hussain T, et al. A Toxoplasma dense granule protein, GRA24, modulates the early immune response to infection by promoting a direct and sustained host p38 MAPK activation. J Exp Med. 2013;210(10):2071–86. doi: 10.1084/jem.20130103 24043761
108. Bougdour A, Durandau E, Brenier-Pinchart MP, Ortet P, Barakat M, Kieffer S, et al. Host cell subversion by Toxoplasma GRA16, an exported dense granule protein that targets the host cell nucleus and alters gene expression. Cell Host Microbe. 2013;13(4):489–500. doi: 10.1016/j.chom.2013.03.002 23601110
109. Kim S-K, Karasov A, Boothroyd JC. Bradyzoite-Specific Surface Antigen SRS9 Plays a Role in Maintaining Toxoplasma gondii Persistence in the Brain and in Host Control of Parasite Replication in the Intestine. Infection & Immunity. 2007;75(4):1626–34.
110. Cox GW, Mathieson BJ, Gandino L, Blasi E, Radzioch D, Varesio L. Heterogeneity of hematopoietic cells immortalized by v-myc/v-raf recombinant retrovirus infection of bone marrow or fetal liver. Journal of the National Cancer Institute. 1989;81(19):1492–6. 2778838
111. Corraliza IM, Campo ML, Soler G, Modolell M. Determination of arginase activity in macrophages: a micromethod. J Immunol Methods. 1994;174(1–2):231–5. 8083527
112. Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10(3):R25. Epub 2009/03/06. doi: 10.1186/gb-2009-10-3-r25 19261174
113. Trapnell C, Hendrickson DG, Sauvageau M, Goff L, Rinn JL, Pachter L. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nature Biotechnology. 2012. Epub 2012/12/12.
114. Churchill GA, Doerge RW. Empirical threshold values for quantitative trait mapping. Genetics. 1994;138(3):963–71. Epub 1994/11/01. 7851788
115. Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci U S A. 2003;100(16):9440–5. 12883005
116. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–8. 11846609
Štítky
Genetika Reprodukční medicína
Článek Evidence of Selection against Complex Mitotic-Origin Aneuploidy during Preimplantation DevelopmentČlánek A Novel Route Controlling Begomovirus Resistance by the Messenger RNA Surveillance Factor PelotaČlánek A Follicle Rupture Assay Reveals an Essential Role for Follicular Adrenergic Signaling in OvulationČlánek Canonical Poly(A) Polymerase Activity Promotes the Decay of a Wide Variety of Mammalian Nuclear RNAsČlánek FANCI Regulates Recruitment of the FA Core Complex at Sites of DNA Damage Independently of FANCD2Článek Hsp90-Associated Immunophilin Homolog Cpr7 Is Required for the Mitotic Stability of [URE3] Prion inČlánek The Dedicated Chaperone Acl4 Escorts Ribosomal Protein Rpl4 to Its Nuclear Pre-60S Assembly SiteČlánek Chromatin-Remodelling Complex NURF Is Essential for Differentiation of Adult Melanocyte Stem CellsČlánek A Systems Approach Identifies Essential FOXO3 Functions at Key Steps of Terminal ErythropoiesisČlánek Integration of Posttranscriptional Gene Networks into Metabolic Adaptation and Biofilm Maturation inČlánek Lateral and End-On Kinetochore Attachments Are Coordinated to Achieve Bi-orientation in OocytesČlánek MET18 Connects the Cytosolic Iron-Sulfur Cluster Assembly Pathway to Active DNA Demethylation in
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2015 Číslo 10
-
Všechny články tohoto čísla
- Gene-Regulatory Logic to Induce and Maintain a Developmental Compartment
- A Decad(e) of Reasons to Contribute to a PLOS Community-Run Journal
- DNA Methylation Landscapes of Human Fetal Development
- Single Strand Annealing Plays a Major Role in RecA-Independent Recombination between Repeated Sequences in the Radioresistant Bacterium
- Evidence of Selection against Complex Mitotic-Origin Aneuploidy during Preimplantation Development
- Transcriptional Derepression Uncovers Cryptic Higher-Order Genetic Interactions
- Silencing of X-Linked MicroRNAs by Meiotic Sex Chromosome Inactivation
- Virus Satellites Drive Viral Evolution and Ecology
- A Novel Route Controlling Begomovirus Resistance by the Messenger RNA Surveillance Factor Pelota
- Sequence to Medical Phenotypes: A Framework for Interpretation of Human Whole Genome DNA Sequence Data
- Your Data to Explore: An Interview with Anne Wojcicki
- Modulation of Ambient Temperature-Dependent Flowering in by Natural Variation of
- The Ciliopathy Protein CC2D2A Associates with NINL and Functions in RAB8-MICAL3-Regulated Vesicle Trafficking
- PPP2R5C Couples Hepatic Glucose and Lipid Homeostasis
- DCA1 Acts as a Transcriptional Co-activator of DST and Contributes to Drought and Salt Tolerance in Rice
- Intermediate Levels of CodY Activity Are Required for Derepression of the Branched-Chain Amino Acid Permease, BraB
- "Missing" G x E Variation Controls Flowering Time in
- The Rise and Fall of an Evolutionary Innovation: Contrasting Strategies of Venom Evolution in Ancient and Young Animals
- Type IV Collagen Controls the Axogenesis of Cerebellar Granule Cells by Regulating Basement Membrane Integrity in Zebrafish
- Loss of a Conserved tRNA Anticodon Modification Perturbs Plant Immunity
- Genome-Wide Association Analysis of Adaptation Using Environmentally Predicted Traits
- Oriented Cell Division in the . Embryo Is Coordinated by G-Protein Signaling Dependent on the Adhesion GPCR LAT-1
- Disproportionate Contributions of Select Genomic Compartments and Cell Types to Genetic Risk for Coronary Artery Disease
- A Follicle Rupture Assay Reveals an Essential Role for Follicular Adrenergic Signaling in Ovulation
- The RNAPII-CTD Maintains Genome Integrity through Inhibition of Retrotransposon Gene Expression and Transposition
- Canonical Poly(A) Polymerase Activity Promotes the Decay of a Wide Variety of Mammalian Nuclear RNAs
- Allelic Variation of Cytochrome P450s Drives Resistance to Bednet Insecticides in a Major Malaria Vector
- SCARN a Novel Class of SCAR Protein That Is Required for Root-Hair Infection during Legume Nodulation
- IBR5 Modulates Temperature-Dependent, R Protein CHS3-Mediated Defense Responses in
- NINL and DZANK1 Co-function in Vesicle Transport and Are Essential for Photoreceptor Development in Zebrafish
- Decay-Initiating Endoribonucleolytic Cleavage by RNase Y Is Kept under Tight Control via Sequence Preference and Sub-cellular Localisation
- Large-Scale Analysis of Kinase Signaling in Yeast Pseudohyphal Development Identifies Regulation of Ribonucleoprotein Granules
- FANCI Regulates Recruitment of the FA Core Complex at Sites of DNA Damage Independently of FANCD2
- LINE-1 Mediated Insertion into (Protein of Centriole 1 A) Causes Growth Insufficiency and Male Infertility in Mice
- Hsp90-Associated Immunophilin Homolog Cpr7 Is Required for the Mitotic Stability of [URE3] Prion in
- Genome-Scale Mapping of σ Reveals Widespread, Conserved Intragenic Binding
- Uncovering Hidden Layers of Cell Cycle Regulation through Integrative Multi-omic Analysis
- Functional Diversification of Motor Neuron-specific Enhancers during Evolution
- The GTP- and Phospholipid-Binding Protein TTD14 Regulates Trafficking of the TRPL Ion Channel in Photoreceptor Cells
- The Gyc76C Receptor Guanylyl Cyclase and the Foraging cGMP-Dependent Kinase Regulate Extracellular Matrix Organization and BMP Signaling in the Developing Wing of
- The Ty1 Retrotransposon Restriction Factor p22 Targets Gag
- Functional Impact and Evolution of a Novel Human Polymorphic Inversion That Disrupts a Gene and Creates a Fusion Transcript
- The Dedicated Chaperone Acl4 Escorts Ribosomal Protein Rpl4 to Its Nuclear Pre-60S Assembly Site
- The Influence of Age and Sex on Genetic Associations with Adult Body Size and Shape: A Large-Scale Genome-Wide Interaction Study
- Parent-of-Origin Effects of the Gene on Adiposity in Young Adults
- Chromatin-Remodelling Complex NURF Is Essential for Differentiation of Adult Melanocyte Stem Cells
- Retinoic Acid Receptors Control Spermatogonia Cell-Fate and Induce Expression of the SALL4A Transcription Factor
- A Systems Approach Identifies Essential FOXO3 Functions at Key Steps of Terminal Erythropoiesis
- Protein O-Glucosyltransferase 1 (POGLUT1) Promotes Mouse Gastrulation through Modification of the Apical Polarity Protein CRUMBS2
- KIF7 Controls the Proliferation of Cells of the Respiratory Airway through Distinct Microtubule Dependent Mechanisms
- Integration of Posttranscriptional Gene Networks into Metabolic Adaptation and Biofilm Maturation in
- Lateral and End-On Kinetochore Attachments Are Coordinated to Achieve Bi-orientation in Oocytes
- Protein Homeostasis Imposes a Barrier on Functional Integration of Horizontally Transferred Genes in Bacteria
- A New Method for Detecting Associations with Rare Copy-Number Variants
- Histone H2AFX Links Meiotic Chromosome Asynapsis to Prophase I Oocyte Loss in Mammals
- The Genomic Aftermath of Hybridization in the Opportunistic Pathogen
- A Role for the Chaperone Complex BAG3-HSPB8 in Actin Dynamics, Spindle Orientation and Proper Chromosome Segregation during Mitosis
- Establishment of a Developmental Compartment Requires Interactions between Three Synergistic -regulatory Modules
- Regulation of Spore Formation by the SpoIIQ and SpoIIIA Proteins
- Association of the Long Non-coding RNA Steroid Receptor RNA Activator (SRA) with TrxG and PRC2 Complexes
- Alkaline Ceramidase 3 Deficiency Results in Purkinje Cell Degeneration and Cerebellar Ataxia Due to Dyshomeostasis of Sphingolipids in the Brain
- ACLY and ACC1 Regulate Hypoxia-Induced Apoptosis by Modulating ETV4 via α-ketoglutarate
- Quantitative Differences in Nuclear β-catenin and TCF Pattern Embryonic Cells in .
- HENMT1 and piRNA Stability Are Required for Adult Male Germ Cell Transposon Repression and to Define the Spermatogenic Program in the Mouse
- Axon Regeneration Is Regulated by Ets–C/EBP Transcription Complexes Generated by Activation of the cAMP/Ca Signaling Pathways
- A Phenomic Scan of the Norfolk Island Genetic Isolate Identifies a Major Pleiotropic Effect Locus Associated with Metabolic and Renal Disorder Markers
- The Roles of CDF2 in Transcriptional and Posttranscriptional Regulation of Primary MicroRNAs
- A Genetic Cascade of Modulates Nucleolar Size and rRNA Pool in
- Inter-population Differences in Retrogene Loss and Expression in Humans
- Cationic Peptides Facilitate Iron-induced Mutagenesis in Bacteria
- EP4 Receptor–Associated Protein in Macrophages Ameliorates Colitis and Colitis-Associated Tumorigenesis
- Fungal Infection Induces Sex-Specific Transcriptional Changes and Alters Sexual Dimorphism in the Dioecious Plant
- FLCN and AMPK Confer Resistance to Hyperosmotic Stress via Remodeling of Glycogen Stores
- MET18 Connects the Cytosolic Iron-Sulfur Cluster Assembly Pathway to Active DNA Demethylation in
- Sex Bias and Maternal Contribution to Gene Expression Divergence in Blastoderm Embryos
- Transcriptional and Linkage Analyses Identify Loci that Mediate the Differential Macrophage Response to Inflammatory Stimuli and Infection
- Mre11 and Blm-Dependent Formation of ALT-Like Telomeres in Ku-Deficient
- Genome Wide Identification of SARS-CoV Susceptibility Loci Using the Collaborative Cross
- Identification of a Single Strand Origin of Replication in the Integrative and Conjugative Element ICE of
- The Type VI Secretion TssEFGK-VgrG Phage-Like Baseplate Is Recruited to the TssJLM Membrane Complex via Multiple Contacts and Serves As Assembly Platform for Tail Tube/Sheath Polymerization
- The Dynamic Genome and Transcriptome of the Human Fungal Pathogen and Close Relative
- Secondary Structure across the Bacterial Transcriptome Reveals Versatile Roles in mRNA Regulation and Function
- ROS-Induced JNK and p38 Signaling Is Required for Unpaired Cytokine Activation during Regeneration
- Pelle Modulates dFoxO-Mediated Cell Death in
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Single Strand Annealing Plays a Major Role in RecA-Independent Recombination between Repeated Sequences in the Radioresistant Bacterium
- The Rise and Fall of an Evolutionary Innovation: Contrasting Strategies of Venom Evolution in Ancient and Young Animals
- Genome Wide Identification of SARS-CoV Susceptibility Loci Using the Collaborative Cross
- DCA1 Acts as a Transcriptional Co-activator of DST and Contributes to Drought and Salt Tolerance in Rice
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



