-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaEstablishment of a Developmental Compartment Requires Interactions between Three Synergistic -regulatory Modules
The separation of cell populations into distinct functional units is essential for both vertebrate and invertebrate animal development. A classical paradigm for this phenomenon is the establishment of developmental compartments during Drosophila wing development. These compartments depend on the restricted expression of two selector genes, engrailed in the posterior compartment and apterous (ap) in the dorsal compartment. Yet, despite the central role these genes and their restricted expression patterns play in Drosophila development, we still do not understand how these patterns are established or maintained. Here, by dissecting the regulatory sequences required for ap expression, we solve this problem for this critical selector gene. We used a combination of experimental approaches to identify and functionally characterize the cis-regulatory modules (CRMs) that regulate ap expression during Drosophila wing development. For these analyses we implement a novel technique allowing us to study the function of these CRMs in vivo, at the native ap locus. We found three ap CRMs crucial for wing development: the Early (apE) and the D/V (apDV) enhancers and the ap PRE (apP). Only when all three regulatory elements are combined is a uniform and complete ap expression domain generated. In summary, our results indicate that ap is regulated in time and space by a three-step mechanism that generates a lineage compartment by integrating input from separate CRMs for the initiation, refinement and maintenance of its expression.
Published in the journal: . PLoS Genet 11(10): e32767. doi:10.1371/journal.pgen.1005376
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1005376Summary
The separation of cell populations into distinct functional units is essential for both vertebrate and invertebrate animal development. A classical paradigm for this phenomenon is the establishment of developmental compartments during Drosophila wing development. These compartments depend on the restricted expression of two selector genes, engrailed in the posterior compartment and apterous (ap) in the dorsal compartment. Yet, despite the central role these genes and their restricted expression patterns play in Drosophila development, we still do not understand how these patterns are established or maintained. Here, by dissecting the regulatory sequences required for ap expression, we solve this problem for this critical selector gene. We used a combination of experimental approaches to identify and functionally characterize the cis-regulatory modules (CRMs) that regulate ap expression during Drosophila wing development. For these analyses we implement a novel technique allowing us to study the function of these CRMs in vivo, at the native ap locus. We found three ap CRMs crucial for wing development: the Early (apE) and the D/V (apDV) enhancers and the ap PRE (apP). Only when all three regulatory elements are combined is a uniform and complete ap expression domain generated. In summary, our results indicate that ap is regulated in time and space by a three-step mechanism that generates a lineage compartment by integrating input from separate CRMs for the initiation, refinement and maintenance of its expression.
Introduction
Animal development requires the segregation of cell populations using both lineage and non-lineage boundaries. These cell boundaries act as signaling centers that organize the growth and patterning of specific tissues (reviewed in [1]). A paradigmatic example is the subdivision of the Drosophila wing disc into anterior-posterior (A/P) and dorsal-ventral (D/V) compartments. At the compartment boundaries, ligands encoded by decapentaplegic (dpp) and wingless (wg) are secreted and activate signaling pathways that orchestrate wing development [2–8]. The generation of the A/P and D/V compartments is directed by specific transcription factors, the selector genes engrailed (en) and apterous (ap), respectively, that define the identity of the cells using a binary code (on or off) [9–15]. Once the compartmental fates have been assigned, the cells in which en and ap are expressed as well as their descendants maintain that “determined” state. Unlike the A/P wing division, which is established during embryonic development, the D/V boundary is defined in the wing disc during the second larval stage by the expression of ap [16]. ap encodes a LIM-type homeodomain transcription factor and its activity depends on the formation of a complex with the LIM-domain binding protein Chip [17–19]. Since ap function is crucial to initiate the signaling center at the D/V boundary [20,21], ap null mutants completely lack the wing [16].
Due to its key role in wing disc development, ap function has been studied extensively. However, the transcriptional regulation of ap is poorly understood. How a sharp border of ap-expressing and non-expressing cells is generated de novo during the growth phase of the imaginal disc, and how the expression of ap is maintained and restricted to the dorsal compartment are critical unanswered aspects of wing development.
The spatial and temporal regulation of gene expression is mediated by the binding of transcription factors to discrete DNA sequences named cis-regulatory modules (CRMs). CRMs can be located up to hundreds of kilobases away from their target promoters. Synergistic interactions between CRMs may be required to faithfully regulate gene transcription (reviewed in [22]). Several CRMs have been identified controlling ap expression in different tissues, such as in muscle progenitors and in the embryonic nervous system [23,24]. A wing disc specific enhancer, named apC, has been reported to drive expression in the dorsal wing disc [24]. However, it has been demonstrated that this element is not sufficient for proper ap regulation in the wing [25]. ap expression is initially activated in future dorsal cells by the Drosophila Epidermal Growth Factor Receptor (EGFR) pathway through the secreted neuregulin-like signaling protein Vein (Vn) [26,27]. However, it is still unknown how ap expression is regulated after this initial EGFR-mediated activation. This is particularly critical in a highly proliferating tissue such as the wing imaginal disc.
The maintenance of selector gene expression domains through multiple rounds of cell divisions partially depends on the activity of the Polycomb and Trithorax group gene products (PcG and TrxG). These proteins either repress (PcG) or activate (TrxG) the expression of their target genes through cis-regulatory sequences called Polycomb Response Elements (PREs) (reviewed in [28,29]). It has been suggested that ap expression is repressed by PcG protein complexes in ventral wing disc cells [30].
In this study, we have analyzed the regulation of ap at the endogenous locus and identified three ap CRMs crucial for wing development: the Early (apE) and the D/V (apDV) enhancers and the ap PRE (apP). Importantly, we analyzed these CRMs in the endogenous locus using a novel in situ rescue system. We find that only when the three regulatory elements are combined, a uniform and complete ap expression domain is observed. Our results indicate that ap is regulated by a three-step mechanism that generates a lineage compartment through the integration of input from separate CRMs for the initiation, refinement and maintenance of its expression.
Results
Genetic characterization of the apterous promoter region
ap is expressed in multiple tissues during embryonic and larval stages. Four different transcripts starting from three different promoters have been annotated which give rise to three unique polypeptides (FlyBase). We have generated a series of deletions to identify which ap non-coding sequences are required for ap expression in the wing imaginal disc (Fig 1A–1C; see Materials and Methods for information about each allele). Unless otherwise stated, hemizygous phenotypes resulting from these deletions were analyzed over apDG3, a large deletion removing the bulk of the ap locus [25], and were classified as amorphs or hypomorphs depending on their severity. ap amorphs were defined by the absence of wing tissue in discs and adults.
Fig. 1. Deletion analysis at the ap locus. 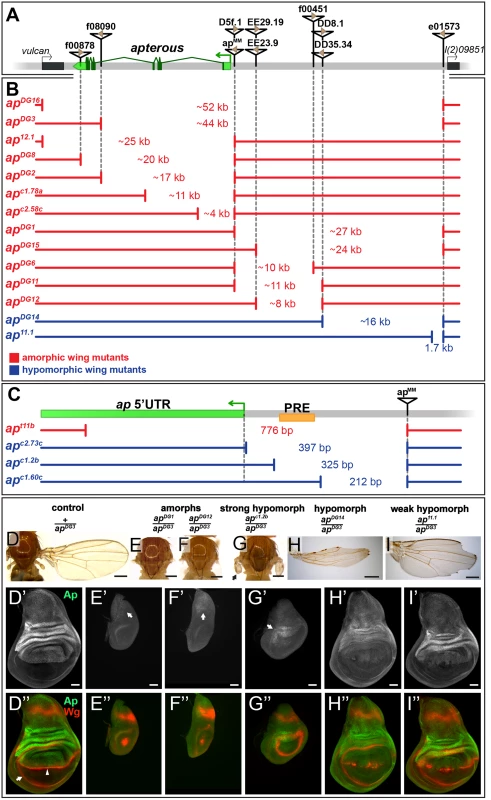
(A) Overview of the ap gene locus. ap transcript ap-RA is indicated in green and the arrow at the 5’ end demarcates its TSS. The flanking genes (indicated by black boxes) are vulcan on the proximal and l(2)09851 on the distal side. Relevant transposable elements used for the generation of deletions by flp-mediated recombination are displayed as black triangles with FRT sites within them as brown triangles. FRT orientation is indicated as defined by [82]. (B and C) Deletions at the ap locus. The number in between the break points indicates the approximate length of the deletion. Phenotypically, the deletions can be divided into amorphic (in red) or hypomorphic (in blue) wing alleles when hemizygous over apDG3. Please note the different scales of the maps depicted in B and C. (B) Deletions that affect the coding sequence all lead to a no wing phenotype (apDG16, apDG3, ap12.1, apDG8, apDG2, apc1.78a, and apc2.58c). Deletions in the upstream noncoding region between apMM and l(2)09851 either lead to amorphic (apDG1, apDG15, apDG6, apDG11, and apDG12) or hypomorphic wing phenotypes (apDG14 and ap11.1). (C) Blow up of the ap promoter region specific for transcripts ap-RA and ap-RC. The PRE core is depicted by a yellow box. apt11b, a deletion which removes the TSS as well as the PRE core, results in a no wing phenotype. The two deletions apc2.73c and apc1.2b leave the TSS intact but both remove the PRE core and both yield strong hypomorphic wing phenotypes. The weak hypomorphic allele apc1.60c leaves TSS and PRE core untouched. (D-I) Wings and 3rd instar wing discs of representative ap wing mutants stained for Wg (red) and Ap (green). (D) Wing and notum of a hemizygous +/apDG3 fly. Almost 100% of the wings look normal [25] (D’) Ap staining in the wing disc demarcates the dorsal compartment. (D”) Wg staining: the inner ring outlines the wing pouch (white arrow) and the stripe traversing it corresponds to the D/V compartment boundary (white arrowhead). (E and F) All wing tissue is lost in amophic wing mutants (apDG1/apDG3 and apDG12/apDG3). (E’ and F’) Only weak Ap staining is detectable in the notum (white arrow). (E” and F”) The wing pouch is completely lost and the inner Wg ring is reduced to a dot. (G) In strong hypomorphic conditions (apc1.2b/apDG3), only small wing and haltere stumps form (black arrow). (G’) Low Ap protein levels are detected (white arrow) mainly in the hinge region. (G”) The size of the wing pouch is drastically reduced and no Wg stripe along the D/V boundary is visible. (H) Hypomorphic mutants (apDG14/apDG3) developed considerably more wing tissue with no or little wing margin or hinge. (H’) Compared to control discs, weaker Ap staining is observed in the pouch region. (H”) The size of the wing pouch is comparable to wild type while the D/V Wg stripe is disrupted. (I) In weak hypomorphic mutants (ap11.1/apDG3) notching of the wing blade is prominent. (I’) Compared to control discs, ap expression is mostly compromised in the pouch region. (I”) Pouch size is similar to wild type and the Wg D/V stripe is locally disrupted. All scale bars are 50 μm. The shortest deletion in our collection with an ap null phenotype is apt11b (Fig 1C and S1D Fig). It specifically deletes the transcription start site (TSS) of transcripts ap-RA and ap-RC. Our in silico analysis indicates that this TSS is not controlled by a TATA-box promoter, but rather contains an Initiator (Inr) and a Downstream Promoter Element (DPE) (for review see [31]). In addition, apt11b removes a PRE located around 100 bp upstream of the ap-RA/ap-RC TSS. This PRE was defined by several chromatin immunoprecipitation studies with various anti-PcG antibodies [30,32–34]. The putative PRE core as defined by Oktaba et al [30] is indicated in Fig 1C. Two other small deletions with the same distal break point as apt11b were isolated: apc2.73c and apc1.2b. They leave the TSS, Inr and DPE intact, but remove the PRE core. In hemizygous flies, small wing stumps are often formed (Fig 1G and S1E Fig). In addition the wing pouches of 3rd instar wing discs are larger than in amorphic mutants (compare Fig 1G” with 1F”). Small amounts of Ap can only be detected in the presumptive hinge and notum (arrows in Fig 1G’ and S1E Fig). The Wg stripe along the compartment boundary is absent (Fig 1G” and S1E Fig). Hence, apc2.73c and apc1.2b behave as strong hypomorphic alleles. A dramatic improvement of the adult wing phenotype is observed for deletion apc1.60c which is a mere 113 bp shorter than apc1.2b (Fig 1C and S1F Fig). Note that it keeps the TSS, Inr, DPE as well as the PRE core in place. A weak phenotype becomes apparent in hemizygous condition: similar to other weak ap loss-of-function alleles, most wing margins have notches. Unexpectedly, this phenotype is brought about by partial ectopic ap expression in the ventral pouch compartment which correlates with gaps in the Wg stripe along the compartment boundary (S1F Fig).
In summary, these observations provide strong genetic evidence for an important contribution of the ap PRE to wing development. In addition, a region defined by apc1.60c appears to act as an auxiliary module, which helps to confine the established Ap pattern to the dorsal compartment.
Deletions affecting the intergenic spacer separating ap and l(2)09851 identify two regions important for ap function
As a next step, we generated alleles that retain an intact PRE/promoter region, but remove upstream non-coding regions of ap. In the mutant apDG1, 27 kb of the upstream non-coding region are deleted (Fig 1B) [35]. Hemizygous flies of this genotype can be considered as amorphic mutants, since no wing tissue was formed despite weak residual ap expression in the notum (Fig 1E’). Removing proximal upstream regions (apDG6, apDG11, and apDG12) also resulted in amorphic phenotypes (Fig 1B and 1F). These deletions remove the previously identified ap wing enhancer apC [24,25]. The distal part of the interval defined by apDG1 was deleted in apDG14. Hemizygous flies form wing stumps of almost normal length but wing margin formation is severely impaired (Fig 1H). In the corresponding wing discs, ap expression in the wing pouch is reduced and the Wg stripe along the D/V boundary is critically perturbed (Fig 1H’ and 1H”). The large size of the apDG14 deficiency precludes a precise localization of an enhancer element within the ~16 kb interval. However, a few small deletions extending only proximal to e01573 allowed us to narrow down its approximate distal end: one of them, ap11.1, deletes 1654 bp (Fig 1B; see also Materials and Methods). It can be maintained as a homozygous stock and wings look wild-type. Its weak hypomorphic nature is revealed in hemizygous ap11.1 flies: all wings have notches along the margin (Fig 1I). Their origin can be traced to gaps in the Wg stripe along the D/V compartment boundary due to reduced Ap levels in the pouch (Fig 1I’ and 1I”).
Two conserved regions harbor essential wing enhancer elements
To further characterize the intergenic spacer between ap and l(2)09851, we engineered and validated a system which allowed us to investigate the role of given DNA stretches at the ap locus [25]. Briefly, we deleted the 27 kb (apDG1) upstream region of ap, and replaced it by an attP site juxtaposing the promoter/PRE (apattPΔEnh; Fig 2B’). In this amorphic situation, we were able to bring back sub-fragments of the previously deleted regulatory regions by ΦC31-integrase mediated insertion and test their ability to rescue wing development. Again, all the newly generated alleles were tested in hemizygous condition. According to sequence conservation and histone H3 lysine 4 trimethylation (H3K4me3) patterns, which have been reported to correlate with active promoters and enhancers [36], we have divided the upstream non-coding region of ap into 5 blocks (C1–5, Fig 2A). Combining all 5 conserved blocks and reintroducing them into apattPΔEnh (apC12345), fully rescued wing formation (Fig 2B) as well as the Ap and Wg pattern in wing imaginal discs (compare Figs 2E with 1D). Deleting the conserved blocks that showed no H3K4me3 mark (C1 and C4), had no consequence on wing phenotype (Fig 2B). Next, we deleted conserved regions with a methylation mark. Deleting C3 had no influence on wing formation (apC25, Fig 2B). In contrast, upon removal of C5, wing development was critically disturbed (apC2, Fig 2B). Long wing stumps with defective wing margin and hinge were formed that resembled the hypomorphic apDG14 mutant, which completely lacks the conserved C5 block (compare Figs 2B and 1H). In flies containing only C5, no wing tissue was formed (apC5).
Fig. 2. Analysis of the ap wing enhancer region. 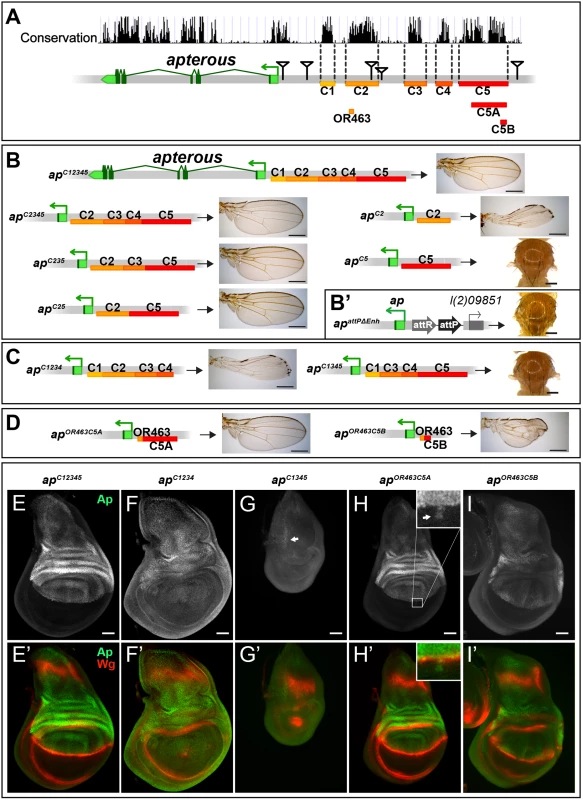
(A) Conservation of the ap locus (data from UCSC genome browser) and subdivision of the 27 kb intergenic region between ap and l(2)09851 into 5 conserved blocks (C1–C5) is shown. OR463, C5A and C5B are subfragments of C2 and C5, respectively. Black triangles mark the locations of the transposon used for the generation of the deletions. (B) Six different constructs consisting of variable combinations of conserved blocks and the corresponding hemizygous wing phenotypes are depicted. When all 5 conserved regions are present (apC12345), a normal sized and patterned wing develops. Gradual removal of C1 (apC2345), C4 (apC235), and C3 (apC25) has no effect on wing morphology. Removing C5 from apC25 results in hypomorphic wings (apC2). C5 alone (apC5) is an amophic allele, as no wings are formed. (B’) apattPΔEnh: the docking site of the in situ rescue system for the evaluation of DNA fragments originating from the 27 kb intergenic spacer is shown. An attP site located ~400 bp upstream of the ap TSS juxtaposes the promoter/PRE region. As in apDG1, the 27 kb intergenic region is deleted. (C) Removing C2 or C5 in the context of apC12345 (apC1234 and apC1345) leads to the same phenotype as each element alone (apC2 or apC5, respectively). (D) Enhancer bashing of C2 and C5 regions. OR463 and C5A in combination are the shortest fragments that still result in a normal wing (apOR463C5A). C5B, a sub-fragment of C5A, in combination with OR463 does not fully rescue wing formation (apOR463C5B). Wing size is reduced, but all margin structures are formed. (E-I) Third instar wing discs of different genotypes stained for Ap (green) and Wg (red). (E-E’) apC12345: Ap and Wg pattern is indistinguishable from wild type. (F-F’) apC1234: a significant reduction of Ap levels in the wing pouch is observed. The Wg stripe along the D/V border is almost completely lost. (G-G’) apC1345: scattered cells with little Ap protein are detectable in the notum (see arrow). Wing pouch is reduced to a small dot of wg expression. (H-H’) apOR463C5A: Ap and Wg patterns are similar to wild type. Ap protein can sometimes be detected in some cells of the ventral part of the disc (arrow in inset). (I-I’) apOR463C5B: although protein levels are reduced, the Ap pattern is close to wild type. Nevertheless, the appearance of the Wg stripe along the D/V border is not as smooth as in wild type. All scale bars are 50 μm. We then tested whether the C2 and C5 fragments were also necessary when all the other conserved elements were present. Removing C5 only (apC1234) had the same effect as maintaining C2 alone, since long wing stumps with little margin and hinge were formed (compare Fig 2C with 2B). Wing discs of this genotype showed drastically reduced ap expression in the pouch region and in most cases lost the Wg stripe along the D/V boundary (Fig 2F). As expected, when only C2 was removed (apC1345, see Fig 2C), no wing tissue was formed, Ap protein was only weakly detected in the notum and the Wg pattern was equivalent to amophic ap wing mutants (Fig 2G).
We also investigated a possible role of the positions of C2 and C5 relative to each other in apC52 and apC15342 flies. Both alleles yield wild-type wings in hemizygous flies, indicating that their order on the chromosome is not important (S2A Fig).
Next, we aimed at defining the minimal CRMs which were able to direct wing development. We have recently found that shorter sub-fragments of C2 retain its wing disc specific activity [25]. The combination of a 463 bp fragment of C2 (OR463) and 3.8 kb of C5 (C5A) in apOR463C5A fully rescued wing development (Fig 2D and 2H). Replacing C5A by C5B, a 600 bp subfragment of C5A, indicated that it lacks certain regulatory input (Fig 2D). The expression of ap in apOR463C5B wing discs was restricted to the dorsal compartment, but reduced compared to apC12345 (compare Fig 2I with 2E). Nevertheless, apart from small disruptions at the D/V boundary, wg expression appeared almost normal (Fig 2I’).
Finally, to investigate whether additional wing-specific CRMs reside within the intronic sequences, we replaced the coding sequences with an ap cDNA lacking most intronic sequences (apcDNAint2.3). This allele produces normal wings (S2B and S2C Fig). Thus, we conclude that no essential wing CRMs are present in the intronic regions of ap. In agreement with this notion, fragments taken from intronic sequences (see below and S2D Fig) failed to drive reporter gene expression in the wing disc. Note that the cDNA used for the construction of apcDNAint2.3 corresponds to the ap-RA/ap-RC transcripts.
Combining the results from the two complementary in vivo approaches (deletion analysis and the in situ rescue system), we have defined three distinct regions which are absolutely required for the correct ap expression in the wing disc: a region next to the ap TSS which contains a PRE and two enhancers with distinct regulatory input located in homology blocks C2 and C5.
Identification of ap cis-regulatory modules active in the wing imaginal disc
In parallel to the ap deletion and in situ rescue strategies, we performed an unbiased search for ap CRMs active in the wing imaginal disc. Using the Fly Light database [37] and self-made constructs (see Materials and Methods), we screened the ap genomic region for DNA fragments that activate the Gal4 gene in an ap-like expression pattern (Fig 3A). We found that 4 of the 17 lines tested partially recapitulated ap-like expression pattern in third instar wing imaginal disc (S3 Fig). Interestingly, lines 1 and 2 were active in a similar pattern in the wing pouch and hinge but were not active in the notum, while lines 7 and 8 showed identical expression pattern in the notum and hinge with low levels in the dorsal wing pouch (S3 Fig). Subsequently, we cloned the overlapping sequences between lines 1–2 and 7–8 in reporter constructs and compared their activity with ap expression as well as with each other during wing imaginal disc development (Fig 3B–3F; see Materials and Methods). apE (Early), the first element to be activated in early to mid-second instar imaginal discs, drove expression in all ap-expressing cells (Fig 3B and 3D). The other element, named apDV (Dorso-Ventral), was activated a few hours later in dorsal cells close to the D/V boundary (Fig 3C and 3E). As the wing imaginal disc developed, the activity of apE became mainly restricted to the notum and hinge with low expression remaining in the wing pouch (Fig 3B). In contrast, apDV was always restricted to dorsal wing pouch cells close to the D/V boundary, with some cells expressing the reporter in the dorsal wing hinge (Fig 3C).
Fig. 3. Activity patterns of apE and apDV enhancers. 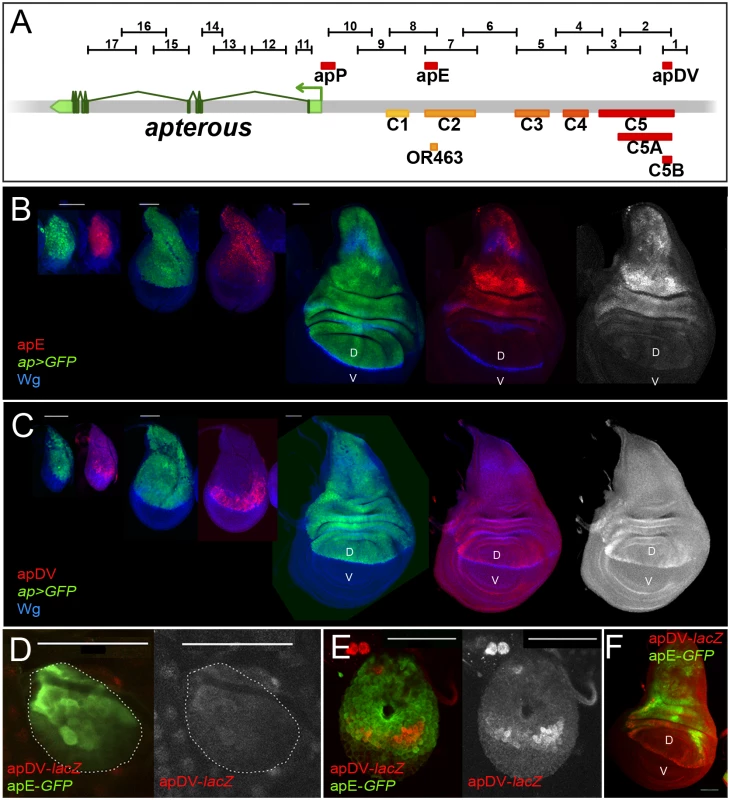
(A) Schematic representation of the ap genomic region is shown as a grey bar. ap transcript ap-RA is shown in green. In the upper part of the panel, horizontal bars represent the DNA elements for which Gal4 drivers were generated by the Janelia Farm consortium except for line 6 (see Materials and Methods) (http://flweb.janelia.org/cgi-bin/flew.cgi). Red bars represent regulatory elements apP, apE and apDV. At the bottom of the panel, 8 fragments tested with the ap in situ rescue system are indicated. (B-C) Pairs of wing imaginal discs isolated from second, early and late third instar larvae are shown (from left to right). They were stained for GFP (ap-Gal4>UAS-GFP, green) and Wg (blue) or apE-lacZ (red) in (B) or apDV-lacZ (red) in (C). Note that ap>GFP represents the complete ap pattern to which those of apE-lacZ and apDV-lacZ are compared. (B) In early discs, apE is active in all the cells that express ap. Later, its activity is restricted to a subset of ap-expressing cells mainly in the notum and hinge region. Expression in the wing pouch is very low. (C) apDV is active in dorsal-distal cells in early discs. Later, its activity is restricted to dorsal wing pouch cells close to the D/V boundary. (D-F) Early second (D), mid-second (E) and third instar imaginal discs (F) stained for apDV-lacZ (red) and apE-GFP (green) are shown. (D) apE is activated earlier than apDV in proximal wing disc cells. (E) apDV is activated in dorsal-distal cells that already have apE activity. (F) In third instar imaginal discs, apE and apDV occupy complementary territories. apDV is restricted to dorsal wing cells close to the D/V border. apE remains mainly active in the hinge and notum. All scale bars are 50 μm. D, dorsal and V, ventral. In line with our previous results, apE and apDV are located within the C2 and C5 regulatory fragments identified with the in situ rescue system, respectively, and overlap with OR463 and C5B (Fig 3A). Moreover, in a reporter gene construct, C2 and C5 reproduced the same expression pattern described for apE and apDV, respectively (S3A Fig).
It should be noted that none of the single ap-CRMs identified, apDV or apE, nor the combination of them, apDV-lacZ+apE-GFP, was able to completely reproduce the endogenous ap expression pattern, suggesting that additional elements are necessary (Fig 3F and see below).
The EGFR pathway transiently regulates the apE element
The initial ap expression in the wing disc is activated by the EGFR signalling pathway at early stages of wing development (from early to mid-second instar), while its later expression is EGFR-independent [26,27]. Since the apE element was active in the entire ap expression domain in early wing discs, we tested whether this CRM is regulated by the EGFR pathway. Clones of cells expressing a dominant-negative form of the pathway effector Raf (RafDN) generated early in larval development (24–48 hrs after egg laying, AEL) were unable to activate apE (Fig 4A), while no effect was observed in clones generated later (72–96h AEL, Fig 4B). The same temporal EGFR-dependency of apE was found when the pathway was reduced in the entire wing disc using a temperature-sensitive EGFR allele (S4 Fig).
Fig. 4. apE is regulated by the EGFR pathway. 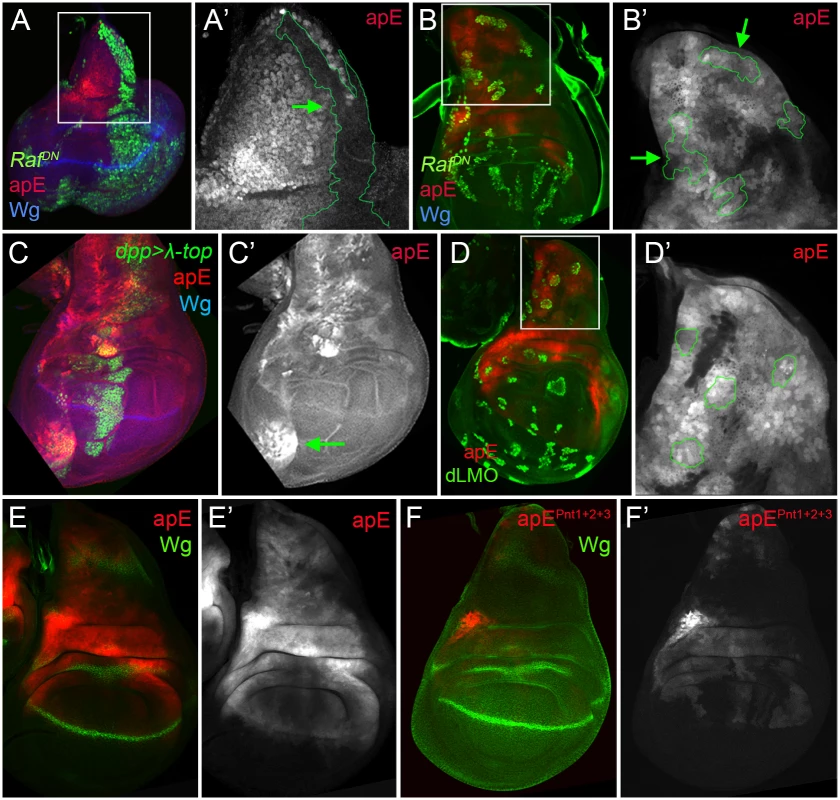
(A-B) Third instar wing imaginal disc with clones expressing a dominant negative version of Raf (RafDN) induced at different time points of larval development are marked by GFP (green). Discs were also stained for Wg (blue) and apE-lacZ activity (red). (A) Early induced RafDN clones (24–48hrs after egg laying, AEL) are unable to activate apE. (A´) Close-up of the disc in (A) with the clone outlined in green (green arrow). Note that apE is not activated within the clone. (B) Late induced RafDN clones (72–96hrs AEL) have no effect on apE activity. (B´) Close-up of the disc in (B) with clones outlined in green (green arrows). (C) dpp-Gal4; UAS-GFP, UAS-EGFRλtop4.2 (λ-top, green) wing disc stained for apE-lacZ (red) and Wg (blue). Ectopic activation of the EGFR pathway induces apE in ventral pleural cells. (C´) Single channel for apE. Green arrow points to ectopic apE activity. (D) Gain of function clones of the Ap activity repressor dLMO (green) has no effect on apE activity (red). (D´) Close-up of the disc in (D) with clones outlined in green. (E-F) Wing imaginal discs stained for Wg (green) and apE (E, red) and apEpnt1+2+3 (F, red) activity. Note that apE activity is strongly reduced after mutating the three identified Pnt binding sites (apEpnt1+2+3). All constructs have been inserted in the same genomic location. Images were obtained keeping the confocal settings constant. Consistent with the low levels of apE activity in dorsal wing pouch cells, misexpression of a constitutive active version of the EGFR receptor (EGFRλtop4.2) by dpp-Gal4 activated apE-lacZ expression in cells of the ventral pleura, while wing pouch cells were resilient to activate it (Fig 4C). To rule out a potential auto-regulatory input on the apE element, gain of function clones of the Ap activity repressor dLMO were made [38]. However, dLMO expression had no effect on apE activity (Fig 4D). Taken together, these results suggest that apE is activated by the EGFR pathway and that other factors regulate its expression afterwards.
To understand how the EGFR pathway regulates apE activity, we searched for putative binding sites of the ETS transcription factor Pnt [39]. Two highly conserved and one less conserved sites were identified. When all these sites were mutated simultaneously, the activity of apE was strongly reduced (compare Fig 4E with 4F). Altogether, these results suggest that apE is initially activated by the EGFR pathway and that this activation requires Pnt function.
Ap and Vg/Sd regulate the apDV CRM
While the apE element was activated in all ap-expressing cells in early second instar wing discs, the apDV element was induced only later and restricted to a subset of apE-positive cells (the wing pouch cells). Therefore, we tested whether the Ap protein itself is needed in an autoregulatory fashion for the restricted activity of the apDV element in the dorsal compartment. dLMO expressing clones cell-autonomously repressed the apDV element, while forced expression of ap in the ventral compartment cells ectopically activated it (Fig 5A and 5B). This suggests that Ap restricts the dorsal activity of apDV. Although ap is expressed in all dorsal wing disc cells, apDV is only active in dorsal wing cells close to the D/V boundary, which suggests additional input into this element. Therefore, we tested whether apDV activity is controlled by wg or vestigial (vg) [40], two key genes required for wing development. Downregulation or ectopic activation of the Wg pathway did not significantly affect apDV-lacZ expression (S4 Fig). However, knockdown of vg in the dpp domain eliminated apDV-lacZ expression (Fig 5C). Additionally, ectopic expression of vg strongly activated apDV in the dorsal compartment (Fig 5D). Remarkably, while apDV is not activated in the leg disc, forced expression of vg in this disc induced its activity in the distal domain of the leg, where a ring of endogenous ap expression has been described (Fig 5E and 5F)[16,41].
Fig. 5. apDV is regulated by Ap and Sd/Vg. 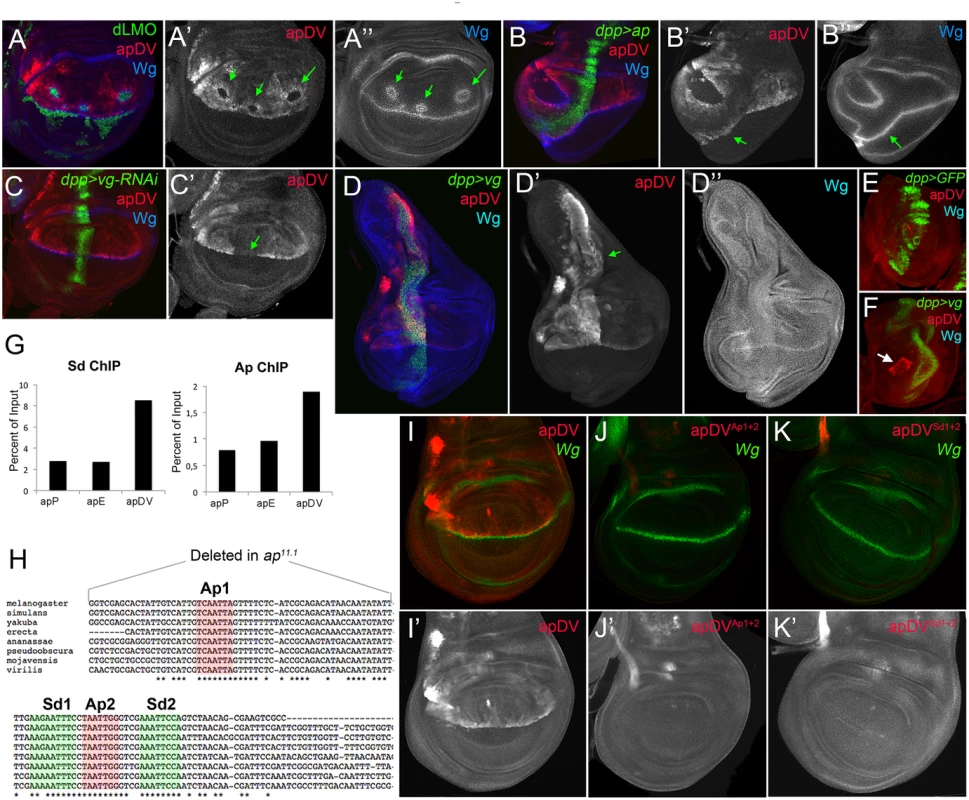
(A) In dLMO expressing clones (green), apDV-lacZ activity (red) is repressed. Wg (blue) is non-autonomously activated in cells surrounding the clones. Single channels are displayed for apDV-lacZ (A’) and Wg (A”). Green arrows point to dLMO expressing clones. (B) dpp-Gal4; UAS-GFP, UAS-ap (green): upon ectopic Ap induction by dpp-Gal4, apDV-lacZ (red) and Wg (blue) are induced in ventral compartment cells. Single channels are displayed for apDV-lacZ (B’) and Wg (B”). Green arrow points to ectopic apDV and wg expression. (C) dpp-Gal4; UAS-GFP, UAS-vg-RNAi: RNAi-induced knockdown of Vg in dpp domain (green). Wg is in blue. Note that apDV-lacZ expression (red) is strongly downregulated in the central part of the pouch. (C’) Single channel display of apDV-lacZ. Green arrow points to discontinuity in the apDV pattern. (D) dpp-Gal4; UAS-GFP, UAS-vg: ectopic vg expression induces apDV (red) along the dpp domain (green), but only in the dorsal compartment. Note that wg is not induced upon the ectopic expression of vg. Single channels are displayed for apDV-lacZ (D’; green arrow points to ectopic apDV-lacZ) and Wg (D”). (E-F) dpp-Gal4; UAS-GFP (E) and dpp-Gal4; UAS-GFP, UAS-vg leg discs (F): dpp-Gal4; UAS-GFP (green), apDV-lacZ (red) and Wg (blue) patterns are shown. Ectopic vg expression induces apDV activity in the distal domain of the leg disc (white arrow in F). (G) ChIP experiments with anti-Sd and anti-Ap antibodies. Quantifications of apP, apE and apDV DNA in immunoprecipitates demonstrate that Sd and Ap are preferentially bound to the apDV regulatory region. Representative enrichment values are shown for a single experiment that was conducted in triplicate. (H) DNA sequences of various Drosophilae species surrounding the identified Ap (red shade) and Sd (green shade) binding sites are shown. Note that Ap1 site is deleted in ap11.1 flies. (I-K) Wing imaginal discs stained for Wg (green) and apDV (I, red), apDVAp1+2 (J, red) and apDVSd1+2 (K, red) activity. Mutation of the Ap sites (J) or Sd sites (K) results in loss of apDV activity. (I’-K’) Single channel pictures are depicted for each apE wild type and mutant condition. All constructs have been inserted in the same genomic location and images were obtained keeping the confocal settings constant. As a next step, we tested whether Ap and Scalloped proteins (Sd), the transcriptional companion of Vg, directly bind to the apDV CRM. Using Ap and Sd chromatin immunoprecipitation (ChIP), we found that Ap and Sd were significantly enriched at the apDV regulatory region in comparison to apE or apP (Fig 5G). Moreover, we identified two conserved consensus-binding sites for Sd as well as for Ap in the apDV region. Mutation of these sites completely eliminated apDV activity (Fig 5H–5K). Intriguingly, loss of one of these Ap binding sites likely contributes to the wing defects seen in the ap11.1 mutant described previously (see Fig 1I).
Taken together, these results suggest that Ap and Vg/Sd directly regulate apDV in the wing pouch, with an Ap autoregulatory input restricting its activity to the dorsal compartment.
Synergistic effect of apE and apDV with the ap promoter directs ap expression in the wing disc
We have identified two ap CRMs (apE and apDV) that, when combined in a reporter assay, partially recapitulated ap expression in the wing disc (see Fig 3F), suggesting that other CRMs are needed for full expression. Since PRE-containing sequences are necessary for correct ap expression and proper wing development (Fig 1C), we tested if a region around the ap TSS including the PRE, named apPRE (apP), had an impact on the activity of the distal ap CRMs (Fig 3A). On its own, the apP drove weak expression in the wing disc in a pattern not related to the characteristic ap expression (Fig 6A). When placed together in a reporter construct with either apDV or apE (resulting in apDV+P-lacZ or apE+P-lacZ), the activity of the resulting reporter gene construct was the sum of both elements and did not reproduce faithfully the ap expression pattern (Fig 6B and 6C). Interestingly, when the three CRMs were placed together, the expression of the apDV+E+P-lacZ in third instar wing discs was more accurate than the expression of the previous CRMs combinations or the apDV+E-lacZ and more precisely reproduced the expression pattern of ap (compare Fig 6D with 6E).
Fig. 6. apP mediates ap expression maintenance and depends on Trx input (A-E) Third instar wing imaginal discs stained with α-βGal antibody to visualize lacZ activity. 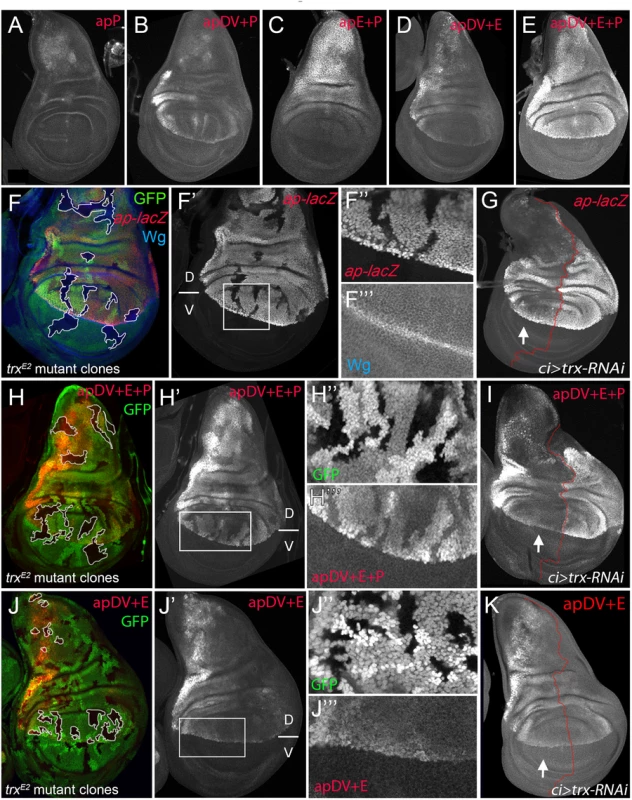
(A) apP activity is weak and not related to the endogenous ap expression pattern. (B) apDV+P activity is the sum of both elements. (C) The combination of apE+P leads to stronger and more homogeneous lacZ expression in the notum and hinge. Note that expression levels remain low in the dorsal wing pouch. (D) apDV+E activity is the sum of apDV and apE and does not reproduce the complete ap expression pattern. (E) Only the combination of apDV+E+P reproduces the endogenous ap pattern. All constructs were inserted in the same genomic location. (F, H and J) trxE2 mutant clones were generated 48–72hrs AEL and are marked by the absence of GFP (in each disc, several of them are outlined in white). Discs were stained for Wg (blue) and ap-lacZ (red, F), apDV+E+P-lacZ (red, H) and apDV+E-lacZ (red, J). (F’,H’,I’) single channel image of lacZ staining. (G, I, and K) ci-Gal4; UAS-trx-RNAi: RNAi-induced knockdown of Trx-activity in the anterior wing disc compartment. Imaginal disc were stained for β-Gal protein. White arrow points to anterior wing compartment. (G) ap-lacZ: enhancer trap aprK568, (I) apDV+E+P-lacZ and (K) apDV+E-lacZ. (F, F’) trxE2 mutant clones show downregulation of ap expression. (F” and F”’) Close-up of ap-lacZ and wg expression shown in F’. (G) Knockdown of Trx in the anterior compartment downregulates ap-lacZ expression. Note that the reduction of ap-lacZ is stronger in the notum and the wing pouch close to the hinge. (H and H’) trxE2 mutant clones show downregulation of apDV+E+P-lacZ expression. (H” and H”’) Close-up of GFP and apDV+E+P activity in H’. (I) Knockdown of Trx in the anterior compartment (arrow) downregulates apDV+E+P-lacZ expression. As ap-LacZ in (G), apDV+E+P activity is reduced in a spatial dependent manner. (J and J’) trxE2 mutant clones show no effect on apDV+E-lacZ expression. (J” and J”’) Close-up of GFP and apDV+E activity in J’. (K) Reducing Trx in the anterior compartment has no effect on apDV+E-lacZ expression. D, dorsal and V, ventral. Therefore, we tested whether these ap CRM combinations placed next to ap cDNA were sufficient to rescue wing development in an ap mutant background. As expected, apE+P-apcDNA was partially able to rescue wing growth, but completely lacks the D/V margin, whereas an apDV+P-apcDNA transgene, that lacks the apE enhancer, did not rescue wing formation (S5 Fig). Interestingly, the apDV+E+P-apcDNA transgene rescued wing formation in ap mutants. Although the rescue was not fully wild type, a clear wing margin was observed in wing discs and adult wings (S5 Fig).
In summary, we have identified three ap CRMs that, only when combined, can accurately reproduce the endogenous ap expression pattern in the wing imaginal disc.
Trx maintains robust ap expression via the apP element
It has been proposed that PcG proteins repress ap expression in ventral wing disc cells and that sequences around the ap TSS could function as a PRE [30]. However, the role of TrxG proteins in the control of ap expression in wing imaginal discs has not been tested previously. Therefore, we generated trxE2 mutant clones and studied ap expression with a lacZ-enhancer trap inserted immediately 5’ to the ap TSS (aprK568). We found that cells devoid of trx function show reduced ap-lacZ expression (Fig 6F). To analyze this result in more detail, we reduced trx mRNA levels in the anterior wing disc compartment (ci-Gal4>trx-RNAi) and compared the levels of ap-lacZ expression with the posterior control compartment (Fig 6G). Consistent with trx mutant clones, ap-lacZ expression was strongly reduced in the anterior compartment, although the reduction was more prominent in the notum and in the dorsal wing pouch close to the hinge.
To genetically confirm that the ap PRE (apP) functions as a Trithorax response element (TRE), we eliminated or downregulated Trx activity and analyzed the expression of the apDV+E+P-lacZ reporter construct. In trxE2 mutant clones, apDV+E+P-lacZ levels were strongly reduced, as it was the case for ap-lacZ (Fig 6H). In contrast, the same construct without the ap promoter, apDV+E-lacZ, was not altered in these trx mutant clones (Fig 6J). Accordingly, reducing the levels of Trx in the anterior compartment cells (ci>trx-RNAi) did not affect expression of apDV+E-lacZ (compare Fig 6K with 6I). Interestingly, the expression pattern of apDV+E+P-lacZ was strongly reduced upon Trx downregulation and resembled the pattern of wild type apDV+E-lacZ (the same construct without the apP, compare Fig 6G and 6I with 6K).
Altogether, our results suggest that the ap promoter region behaves as a PRE/TRE providing the information required to maintain ap expression.
Direct and continuous contact of the apDV and apE CRMs together with the apP element for ap maintenance
Classical transvection experiments usually deal with chromosomes harboring genes lacking either a functional promoter region or a functional enhancer. For combinations of members of the two groups, intragenic complementation can be observed, i.e. the corresponding phenotype is much less severe than seen in allelic combinations involving only one or the other group [42,43]; reviewed in [44]. We have previously reported that transvection is at work at ap [35]. For example, apDG12/apDG1 flies have no wings because both alleles delete wing enhancer apE. The same phenotype is observed in apt11b/apDG8 flies because both alleles remove the promoter region as well as the 5’ end of ap. In contrast, the wing phenotype of apt11b/apDG1 flies is much improved (S7B Fig). Models for transvection posit that the apE and apDV enhancers on chromosome apt11b can activate the transcription machinery of the functional ap gene on chromosome apDG1. However, the apt11b/apDG1 wings are consistently less well formed than those obtained from apDG3/+ flies (see S7 Fig). These observations suggest that the apP region on the one hand and the two enhancers on the other interact more efficiently if they are located in cis.
In our study, we have shown that the two ap wing enhancers are clearly separable units: (1) they lie ~10 kb apart and (2) the activity of apE is essential for auto-regulatory activation of apDV. From these premises, one would not a priori expect that the two enhancers must be in cis for full function. However, several allelic combinations containing only one or the other enhancer element (apE or apDV) generated discs and adult wings with defects at the D/V boundary: similar results were obtained for genotypes apC1234/apC1345, apDG14/apDG12, apC2/apDG12 or when a su(Hw) insulator element was inserted between apDV and apE in apf00451/apDG3 animals (Fig 7A and S7 Fig). Our transvection studies suggest that all three CRMs need to be in cis to fully rescue wing development.
Fig. 7. Evidence for genetic and physical interaction between apDV, apE and apP. 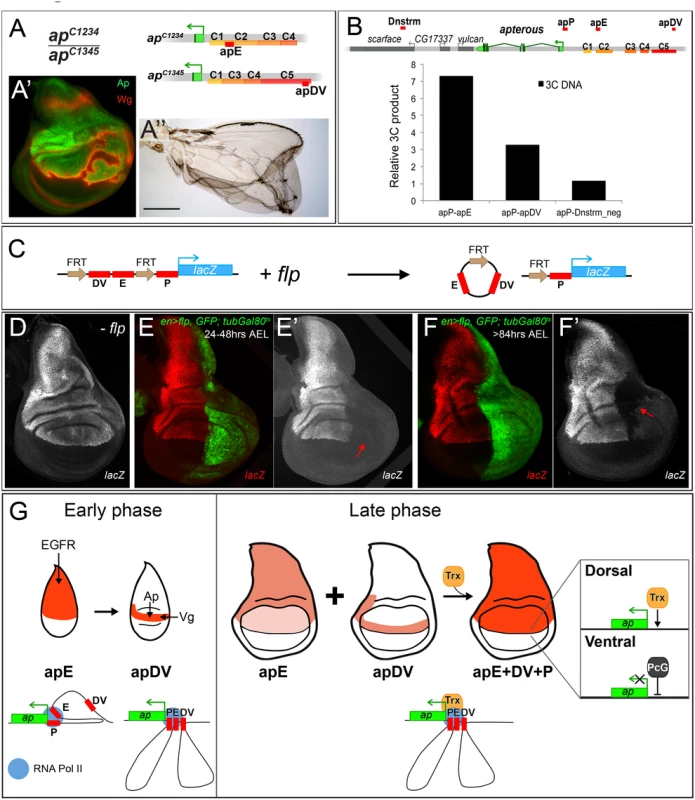
(A) At the top of the panel, the genetic constitution of apC1234/apC1345 flies is shown. Note that apE and apDV are present in trans and that apP (not indicated) is present on both chromosomes. (A’) Ap (green) in the wing disc is uneven leading to derepression of wg in cells with no Ap (red). (A”) Wings of apC1234/apC1345 flies frequently show wing patterning defects and outgrowths. (B) At the top of the panel, a schematic representation of the ap genomic locus and several flanking genes is shown. C1–C5 indicates the conserved homology regions. Red bars above the chromosome represent the different regions tested for direct interaction with apP using the 3C technique. A region downstream of ap, named Dnstrm, was used as a negative control. The diagram in the lower part of the panel summarizes the 3C data. In whole third instar larvae, apE and apDV elements are more frequently close to apP than a control DNA element located downstream (Dnstrm) the ap genomic locus. (C) Diagram of the FRT-apDV+E-FRT+P-lacZ reporter gene is depicted. Upon flp induction, the apDV+E cassette is deleted. The lacZ reporter remains under the control of apP only. (D-F) Expression of the FRT-apDV+E-FRT+P-lacZ reporter gene in third instar wing imaginal discs in the absence of flp (D) or after flp-induction at different times of larval development (E-F). Controlled flp-induction in the posterior pouch compartment (red arrow) was achieved and monitored in an en-Gal4; UAS-flp; UAS-GFP, tubGal80ts background. Wing discs were stained for lacZ (red) and GFP (green). (D) lacZ pattern without flp-induction resembles wild type ap expression. (E) flp-induction 24–48hrs AEL: deletion of the apDV+E cassette results in loss of lacZ expression in the posterior compartment. (F) flp-induction >84hrs AEL: Loss of lacZ expression in the posterior compartment after late flp-induction demonstrates the continuing requirement of apE and apDV. (G) ap cis-regulatory model for the establishment of Dorso-Ventral identity in the wing imaginal disc. During the early phase of ap activation, the EGFR pathway triggers ap expression via the apE CRM that directly interacts with apP. A few hours later, apDV is activated in dorsal cells close to the D/V boundary. It is activated by Vg/Sd in the future wing cells but its activity is restricted dorsally by Ap itself. apDV is also recruited to apP. In the late phase, apP maintains ap expression through Trx input in dorsal cells. Persistent ap transcription is required for the generation of a dorsal lineage compartment. It is dependent on the permanent presence of the apE and apDV enhancers and their continuing interaction with apP. PcG proteins repress ap in ventral compartment cells. To better understand how the synergy between the three regulatory elements is achieved, we used chromosome conformation capture (3C) [45], which allowed us to test in vivo whether there is direct physical contact between the apE and apDV CRMs with the apP. Indeed, as seen in Fig 7B, we found that in whole third instar larvae, apP preferentially contacted the apE and apDV elements, and did so more frequently when compared to sequences outside the ap genomic locus. This suggests that the distal apE and apDV regions are in close physical proximity to apP in vivo.
Next, we tested whether apDV and apE CRMs are required either continuously or only transiently to direct ap expression during wing disc development. To distinguish between these two possibilities, we generated an apDV+E+P-lacZ construct, in which the apDV+E is flanked by FRT sequences (FRT-apDV+E-FRT+P-lacZ, Fig 7C). This allowed us to remove the apDV+E cassette at different time points of wing development using Flp-mediated recombination [46,47] (see Materials and Methods). Deletion of apDV+E early in development in the posterior compartment completely abolished reporter expression compared to anterior control cells (compare Fig 7D and 7E). Deletion of the apDV+E at later stages also strongly decreased reporter gene expression (Fig 7F).
In summary, these experiments suggest that there is a direct contact between the apE and apDV with the ap promoter and that these three elements need to be in cis throughout wing disc development to confer optimal ap expression.
Discussion
The selector gene ap encodes for a transcription factor that confers dorsal identity in the wing imaginal disc. A precise border of ap-expressing and non-expressing cells is absolutely necessary for wing growth and pattern formation. Although the role of ap as a dorsal selector gene has been extensively studied, how its specific spatial expression pattern is brought about during wing development has remained unclear. In this work, we have used complementary strategies to identify and molecularly characterize the endogenous CRMs that regulate ap expression during wing development.
ap cis-regulatory logic for Dorso-Ventral identity in the wing imaginal disc
Our genetic and cis-regulatory analysis provides information about the logic of ap expression during wing development. We propose that ap expression is controlled by at least three CRMs that act in combination (Fig 7G). The first element, apE is the earliest to be activated in proximal wing disc cells via the EGFR pathway; its expression subsequently weakens in the wing pouch. Deletion of this early enhancer (e.g. apDG12 or apC1345) completely abolishes wing formation. The asymmetry of ap expression to the proximal domain of the wing disc is probably due to the localized activation of the EGFR pathway by its ligand Vn and a distal repression by Wg signaling [26,48–50]. We have genetically and molecularly confirmed the initial activation of the apE by the EGFR pathway; however, other inputs are required for the continuous activation of this CRM in later wing discs.
A few hours after apE activation, a second CRM, apDV, is activated in a subset of apE positive cells. In contrast to apE, apDV is restricted to the dorsal-distal domain of the wing pouch by direct positive inputs from Ap and Vg/Sd (Fig 7G). The direct Ap autoregulatory input defines the time window when the apDV element is activated; apDV can only be active after the induction of Ap by the early enhancer (apE). It has been shown that Ap induces vg expression by triggering Notch signaling at the D/V boundary [20,21,48,51,52]. Thus, the (direct) input of Vg/Sd on apDV can be regarded as an indirect positive autoregulation, which delimits the spatial domain where apDV can be actived. Consequently, the interface of Ap and Vg expression defines the region of apDV activity via positive autoregulation.
The third ap CRM is the ap PRE/TRE region (apP), that, when deleted, leads to a strong hypomorphic wing phenotype (apc1.2b). The apP requires Trx input and maintains ap expression when placed in cis with the apDV and apE CRMs (Fig 7G). Only the combination of the three CRMs faithfully reproduces ap expression in the wing disc. Moreover, our regulatory in locus deletion and in situ rescue analysis provide strong functional relevance for these CRMs.
Ultimately, this cascade of ap CRMs provides a mechanism to initiate, refine and maintain ap expression during wing imaginal disc development, in which the later CRMs depend on the activity of the early ones (Fig 7G). A similar mechanism has been described for Distal-less (Dll) regulation in the leg primordia where separate CRMs trigger and maintain Dll expression in part by an autoregulatory mechanism [46,53].
It has been proposed that positive autoregulation may help to maintain the epigenetic memory of differentiation [54]. In the case of ap, we demonstrate that autoregulation works in conjunction with a PRE/TRE system; this might make the system very robust and refractory to perturbations.
The role of ap promoter in maintenance
ChIP experiments have shown that many developmentally important genes are associated with a promoter proximal PRE as found at ap [30]. The role of such a PRE has been studied at the engrailed (en) locus. It has been demonstrated that in imaginal discs, the promoter as well as the promoter proximal PRE are important for the long-range action of en enhancers [55,56]. These authors propose that this PRE brings chromatin together, allowing both positive and negative regulatory interactions between distantly located DNA fragments.
Our results indicate that sequences around the transcription start of ap (apP) may serve a similar function. First, this element, when placed in cis with the ap CRMs (apE and apDV), maintains the ap expression pattern and keeps reporter gene expression off in cells where low or no activity of apDV and apE has been observed. Second, in the absence of trx, the expression of ap and apDV+E+P-lacZ is strongly reduced. All these data suggest that sequences within the apP integrate Trx input, thereby maintaining ap expression in a highly proliferative tissue such as the wing disc. Interestingly, trx mutant clones were not round and did not show ectopic wg activation (Fig 6F), which is a hallmark of ap loss-of-function clones. This suggests that in trx mutant clones enough Ap protein is still present to maintain wg expression off. However, we found derepression of the ventral-specific integrin αPS2 in trx mutant clones in the wing pouch as previously described for ap mutant clones [14] (S6 Fig).
It has been suggested that TrxG proteins could act passively antagonizing PcG silencing, rather than playing an active role as co-activators of gene transcription [57,58]. For example, Ubx expression in the leg and haltere does not require Trx in the absence of Polycomb repression [59]. We tested these possibilities and generated trx mutant clones that were also mutant for the PcG member Sex combs on midlegs (Scm). Dorsally-located Scm- trx- double mutant clones still downregulate ap-lacZ expression while ventral-induced ones are unable to derepress ap-lacZ as we observed for Scm- single mutant clones (S6 Fig). Therefore, our results, in addition with previous findings by Oktaba et al [30], suggest that TrxG maintains ap expression in dorsal cells, while ap expression is repressed in the ventral compartment by PcG proteins. Moreover, it has been shown that the sequences around the ap transcription start, including the PRE, are occupied by PcG complexes PRC1 and PRC2, as well as Trx [30,60].
Direct and continuous interactions between the apE and apDV with the apP
Enhancers-promoter interactions initiate transcription but their dynamics during development have remained poorly understood. Our chromosome conformation capture (3C) experiment provides evidence for the direct interaction between the ap CRMs apE and apDV with the maintenance element encoded by the apP. Beyond this, we also find that these elements cooperate continuously during wing development. Our flip-out experiments, in which we removed the apDV and apE CRMs at different time points, suggest that these elements need to be present continuously to ensure correct ap expression. Additionally, flies carrying apE only on one chromosome and apDV only on the homologue were unable to fully rescue wing development suggesting that these CRMs need to be in cis. It is conceivable that in cis configuration of the three ap CRMs facilitates and stabilizes enhancer-promoter looping. It could also help to rapidly establish relevant chromatin contacts after each cell division. These results are in accordance with previous observations, in which constant interactions between ap enhancers and promoter during embryogenesis have been described [61]. Our results extend these observations to the wing disc, a highly proliferative tissue, where the expression of the trans-factors that regulate the activity of the apE and apDV is very dynamic. This raises the question on how this contact is re-assembled over many cell generations. It is possible that some epigenetic modifications are laid down in the activated apE and apDV CRMs, which are then inherited during cell divisions to ensure contact with apP. Studies of the chromatin status of these elements will be required to fully understand this process.
Developmental transcriptional regulation during tissue growth
A key question in developmental biology is how transcriptional regulation is coupled to tissue growth to precisely regulate gene expression in a spatio-temporal manner. For example, during Drosophila leg development, initial activation of the ventral appendage gene Dll by high levels of Wg and Dpp initiates a cascade of cross-regulation between Dll and Dachshund (Dac) and positive feedback loops that patterns the proximo-distal axis [46,62]. Other mechanisms to expand gene expression patterns depend on memory modules such as PREs, as it is the case for the Hox genes or other developmental genes like hh [63–65]. To direct wing formation, expression of ap in the highly proliferative tissue of the wing disc must be precisely induced to generate and maintain the D/V border. Our in-depth analyses at the ap locus provide a functional and molecular explanation of how expression of this dorsal selector gene is initiated, refined at the D/V border, and maintained during wing disc development. We propose that this three-step mechanism may be common for developmental patterning genes to make the developmental program robust to perturbations.
Materials and Methods
Stocks used in this study
Flies were grown on standard cornmeal agar. ap-lacZ (P{PZ}aprK568), ap-Gal4, apUGO35, trxE2, ScmD1, trxE2 ScmD1 (gift from Jürg Müller)[59], EGFRtsa, UAS-EGFRλtop4.2, UAS-RafDn UAS-armS10, UAS-TCFDN, UAS-vg, UAS-dLMO, UAS-ap, dpp-Gal4; UAS-GFP, ci-Gal4; UAS-GFP, tubGal80ts. act5C>stop>lacZ; UAS-flp, P{hsFLP}12, y1 w*, TM3, ryRK Sb1 Ser1 P{Δ2–3}99B, P{EPgy2}l(2)09851EY06365, al1 b1 c1 sp1, y1 w67c23; nocSco / CyO, P{Crew}DH1, y1 w*; Mi{y[+mDint2] = MIC}MI00964, y1 w*; Mi{y[+mDint2] = MIC}MI02330/SM6a as well as all the Janelia Farm Gal4 drivers were obtained from the Bloomington Drosophila Stock Center except as indicated. These are described in the Fly light data base (http://flweb.janelia.org/cgi-bin/flew.cgi): 1-GMR_39E04, 2-GMR_42A06, 3-GMR_42D11, 4-GMR_41B09, 5 - GMR_41E03, 7-GMR_42B11, 8-GMR_41D11, 9-GMR_41D03, 10-GMR_40H04, 11-GMR_39B07, 12-GMR_40A08, 13-GMR_39G10, 14-GMR_ 39C09, 15-GMR_40A07, 16-GMR_41A02, 17-GMR_41C10. For the lineage analyses of the Janelia lines we used the act5C>stop>lacZ; UAS-flp [47]. UAS-vg-RNAi, UAS-sd-RNAi and UAS-trx-RNAi are available at the Vienna Drosophila Resource Center (VDRC). RNAi knock-down experiments were performed in a UAS-Dcr–2 background. en-Gal4; UAS-flp, UAS-GFP was a gift from Laura Johnston. PBac{RB}e01573, apf08090 (PBac{WH}f08090),apf00878 (PBac{WH}f08090) and apf00451 (PBac{WH}f00451) were purchased from the Exelixis stock collection at Harvard Medical School. y w M{vas-int.Dm}zh-2A, a stock producing ФC31-integrase under the control of the vasa promoter, and docking site M{3xP3-RFP.attP}zh-86Fb were obtained from Johannes Bischof [66]. The GFP knock-in allele apGFP is described in Caussinus et al, 2012 [67]. apMM and apMM-Mcp have been described previously [35]. They contain a P-element insertion ~400 bp upstream of the ap TSS. The ΦC31-integrase platforms apattPΔCDS and apattPΔEnh used for the in situ rescue system are described in detail in Caussinus et al, 2012 and Bieli et al, 2015, respectively [25,67]. The generation of all deficiencies shown in Fig 1A and 1B is described below.
Adult wings were dissected and mounted in Hoyer’s and baked at 58°Celsius for a few hours. Pictures were taken with a Nikon Microphot-FXA microscope with a Sony NEX-5RK digital camera.
The notums of adult flies were photographed with a Leica M125 binocular equipped with a Leica DFC420C camera.
Generation of deletions
Df(2R)apDG1 is described in Gohl et al, 2008, where it is called apDG [35].
Df(2R)apDG3, Df(2R)apDG8 and Df(2R)apDG11 are described in Bieli et al, 2015 [25].
Df(2R)ap12.1, al b was obtained in an attempt to isolate male-recombination events to the right of P-element insertion apMM-Mcp. Molecular characterization identified the proximal breakpoint in a P-element insertion hot spot at the 5’ end of the vulcan gene (Genome release R6 FB2015_01 : 2R:5702133). It also verified the integrity of apMM-Mcp at its original insertion site. This deletion is referred to as Df(2R)ap12.1-Mcp, al b. In order to delete the Mcp element located between 2 loxP sites on apMM-Mcp, Df(2R)ap12.1-Mcp, al b was treated with Cre recombinase [68] and Df(2R)ap12.1, al b was obtained. Homozygous flies of this genotype make it to the pharate adult stage. Dissected individuals have neither wings nor halteres. In this study, Df(2R)ap12.1, al b is referred to as ap12.1. Note that it is associated with a FRT site left within apMM.
The following 6 deletions were created by flp-mediated recombination [69] between 2 FRT sites located in trans to each other in 2 different transposons (below, their names are indicated in parenthesis; their positions within the ap locus is depicted in Fig 1A):
Df(2R)apDG16, al b (ap12.1; e01573). Referred to in the text as apDG16. This chromosome is deficient for vulcan and ap. Homozygous apDG16 flies are pharate adult lethal. Dissected individuals have neither wings nor halteres.
Df(2R)apDG2 (f08090; apMM-Mcp). Referred to in the text as apDG2. Note that Mcp is lost upon flp-mediated recombination and that this deletion is associated with an array of Su(Hw) binding sites originating from f08090.
Df(2R)apDG15 (apEE23.9; e01573). Referred to in the text as apDG15. apEE23.9 (as well as apEE29.19) is a ΦC31-integrase mediated insertion of a plasmid containing mini-white, FRT and mini-yellow in docking site MI00964 [70].
Df(2R)apDG6, al (apD5f.1; f00451). Referred to in the text as apDG6. apD5f.1 is a ΦC31-integrase mediated insertion of a plasmid containing mini-white, FRT and mini-yellow in docking site apc1.4b [25]. Note that this deletion is associated with an array of Su(Hw) binding sites originating from f00451. apDG6 flies have neither wings nor halteres. These phenotypes are not modified in a su(Hw)- background.
Df(2R)apDG12 (apEE29.19; apDD8.1). Referred to in the text as apDG12. apDD8.1 (as well as apDD35.34) is a ΦC31-integrase mediated insertion of a plasmid containing mini-white, FRT and mini-yellow in docking site MI02330 [70].
Df(2R)apDG14 (apDD35.34; e01573). Referred to in the text as apDG14.
Df(2R)apc2.73c: this short deletion was obtained by direct gene conversion [71,72]. A detailed account on our experimental approach is given in Bieli et al, 2015 [25]. apc2.73c was obtained according to the exact same procedure as apc1.4b, except that the left homology arm on the gene conversion template plasmid was only 502 bp long, leading to a 397 bp deletion just proximal to apMM. Our gene conversion approach also introduced a cassette consisting of a GFP reporter driven by a minimal hsp70 promoter flanked by two inverted attP sites for Recombination Mediated Cassette Exchange (RMCE) [73].
The following five deletions were obtained by imprecise excision of insert apMM during the generation of gene conversion events apc1.4b and apc2.73c. In all five cases, the deletion extends only to the left of apMM.
Df(2R)apc1.78a: 12 bp are left between the break points, 8 of them can be identified as belonging to the end of the P-element 3’ foot. Referred to in the text as apc1.78a.
Df(2R)apc2.58c: the most 3’ ~1.6 kb of apMM are left at the break point, including the wing enhancer of the yellow gene. Referred to in the text as apc2.58c.
Df(2R)apt11b: the terminal 17 bp of the P-element 3’-foot are left between the breakpoints. Referred to in the text as apt11b.
Df(2R)apc1.2b: the intact apMM insert is left at the break point. Referred to in the text as apc1.2b.
Df(2R)apc1.60c, sp: the intact apMM insert is left at the break point. This small deletion can be maintained as a homozygous stock and most wings look wild-type. Referred to in the text as apc1.60c.
Finally, Df(2R)ap11.1, c sp was obtained by transposase treatment of EY06365, a P-element inserted in the 5’ end of l(2)09851, the gene immediately distal to ap (see Fig 1A). In an attempt to isolate deletions extending proximal to EY06365, dysgenic males of the genotype y w; al apDG3{w+} + + + / + + EY06365{y+ w+} c sp; TM3, Sb, Δ2–3 / + were crossed with y w; al b c sp / SM6, al sp females. Progeny was screened for candidates with no eye colour, y+ body colour and carrying the c and sp markers. 2 of the candidate chromosomes (isolation numbers 11.1 and 34.1) gave rise to notched wings in trans to apDG3, a phenotype reminiscent of weak ap alleles. SM6 balanced stocks were established. Homozygous flies readily hatch and show no or only very weak wing phenotypes. Molecular characterization of EY06365 and the 2 candidates detected in all three a ~400 bp LTR of the springer retrotransposon at position 2R:5751931 (Genome release R6 FB2015_01). EY06365/apDG3 flies have normal wings indicating that the LTR doesn’t have phenotypic consequences. Furthermore, remarkably similar rearrangements could be detected in candidates 11.1 and 34.1: EY06365 has relocated into exactly the same site in the hybrid piggyBac present on apDG3 (obtained by flp-mediated recombination between FRTs in f08090 and e01573) in between mini-white and FRT. On the proximal side of the relocated EY element and next to the 3’ P-element foot, 11.1 contains ~100 bp of DNA originating from the 5’ end of CR44953, while 34.1 contains ~200 bp of DNA originating from the rosy locus. These insertions of heterologous DNA normally found on chromosome arm 3R are abutted by a ~1.7 kb deletion that extends to the left into the apterous region, 11.1 removing 8 bp more than 34.1. The two rearrangements are referred to as ap11.1 and ap34.1. Apart from these, two other very similar rearrangements associated with smaller deficiencies were isolated. Their names are ap72.2 and ap62.3. Their distal break point is the same as for ap11.1 and ap34.1 but they are smaller: 657 bp and 480 bp are missing, respectively. In both cases, hemizygous flies have normal wings, implying that the different position of their proximal deletion break is responsible for the wing phenotype observed for ap11.1 and ap34.1. These observations map the distal end of the ap regulatory domain to a 1 kb interval between the proximal ends of deficiencies ap11.1 and ap72.2.
Generation of α-Ap antibody
DNA corresponding to amino acids 312 to 469 of ap cDNA clone HL02012 (DGRC, Indiana University) was amplified by PCR and cloned into pET22b(+) bacterial expression vector (Novagen) via NcoI and NotI sites. This fragment contains the Ap homeodomain (apHD), which is shared by all different Ap isoforms. The pelB leader sequence of pET22b vector was subsequently removed via mutagenesis PCR [74], resulting in the final expression plasmid pETapHD. BL21(DE3) bacteria (NEB) were transformed with pETapHD, grown to OD600nm 0.6. T7 polymerase was induced with 0.1 mM IPTG. The protein was produced overnight at 18°C. Bacterial cells were lysed using a French press, then the lysate was loaded on a HisTrap HP column (GE Healthcare Life Sciences). apHD was purified with an ÄKTA HPLC machine. 3 mg of pure apHD were sent to Perbio Sciences Switzerland, where two rabbits were immunized. After 80 days, the serum of one positive rabbit was used to perform affinity purification of polyclonal antibody pool (final concentration: 0.67 mg/ml). For imaginal disc staining, the antibody is used at a dilution of 1 : 1000–2000.
Cloning of in situ rescue constructs
First, fragments C1 (size: 1.6 kb), C2 (3.6 kb), C3 (2.5 kb), C4 (1.6 kb), C5 (5.3 kb), C5A (3.8 kb), C5B (600 bp) and OR463 (463 bp) were amplified by PCR from clone BACR45O18 (Berkeley Drosophila Genome Project). The PCR primers had AvrII or XmaI sites overhangs, respectively (see S1 Table for Primer sequences). PCR-fragments were cut with AvrII and XmaI and subcloned into pBS KSII(+) vector, in which the XbaI site had previously been mutated into an AvrII site. Primers containing the XmaI site additionally had a SpeI site. AvrII and SpeI produce compatible sticky ends, which –when ligated - cannot be cut again by any of these enzymes. To combine the different fragments in the desired order, the following strategy was used: one fragment was cut out with AvrII and XmaI, and cloned into another pBSKSII subclone, that had a different fragment, via SpeI and XmaI sites. In the new subclone two different fragments were combined, which could be cut out again via AvrII and XmaI sites and cloned into another SpeI/XmaI cut plasmid. Subsequently, the combined fragments were cut out with AvrII/XmaI and cloned into AvrII/AgeI cut pEnh-Reentry plasmids, resulting in the final pEnh-Reentry constructs. Detailed description of the pEnh-Reentry plasmid can be found elsewhere [25]. Transgenic flies were obtained by injecting these plasmids (300ng/μl final concentration) into y w M{vas-int.Dm}zh-2A; apattPΔEnh/CyO embryos and stocks were established according to standard genetic practice [75].
Cloning of ap coding sequence in situ rescue constructs
ap cDNA was amplified from clone HL02012, the ap promoter region was PCRed from BAC clone BACR45O18 (Berkeley Drosophila Genome Project). The two fragments where combined by fusion PCR, and subcloned into pCR-XL-TOPO (Invitrogen). The ap promoter-cDNA fusion fragment was cloned into pCDS-Reentry vector [67] via NotI and AscI sites, to produce plasmid pCDS-Reentry-apcDNA. The pCDS-Reentry-apcDNAint2.3 construct, which contains the intron 2 and 3 of ap at the correct position, was synthesized by Genewiz, Inc. Transgenic flies were obtained by injecting these plasmids (300ng/μl final concentration) into y w M{vas-int.Dm}zh-2A; apattPΔCDS/CyO embryos and stocks were established according to standard genetic practice.
Generation of lacZ reporter and rescue transgenic lines
To generate C1–C5 and int2.3 reporter constructs, DNA from ap locus was amplified by PCR from y1 w67c23 genomic DNA with primers containing restriction enzyme sites as overhangs, and subsequently cloned into plasmid pAttBLaZ [76] sing the respective enzymes (See S1 Table for primers and restriction enzymes). apE, apDV and apP were cloned into two reporter genes vectors, attB-hs43-nuc-lacZ [62] and attB-pHPdesteGFP [77]. The putative Pnt, Ap and Sd binding sites were identified on the basis of a bioinformatics analysis combining data from the JASPAR CORE Insecta database (http://jaspar.genereg.net/) and the Target Explorer tool [78].
Mutagenesis of the putative Pnt, Sd and Ap binding sites was performed using the QuikChange Site-Directed Mutagenesis Kit (Stratagene). See S1 Table for sequence of all primers used in this study. All the reporter constructs were inserted and analysed at the same landing attP site. The reporter FRT-apDV+E-FRT-P-lacZ was generated cloning PCR FRT sequences flanking the apDV and apE elements with the apP following the last FRT. To delete the apDV and apE casette at different time points of development we drove flp in the posterior compartment by crossing FRT-apDV+E-FRT+P-lacZ containing flies to en-Gal4, UAS-flp, UAS-GFP; tubGal80ts. Larvae were kept at 17°C to keep Gal4 off. At the desired time of development, the fly vials were shifted to 29°C for flp induction.
ap rescue experiments were done replacing the lacZ reporter gene of the attB-hs43-nuc-lacZ with the ap cDNA using EcoRI and KpnI in the different ap CRMs combinations. All ap rescue transgenes were inserted in the same attP site (86Fb).
trx and Scm mutant clonal analysis
Loss-of-function clones were generated by heat shocking the larvae for 1 hour at 37°C. The following genotypes were used:
y w hs FLP122; FRT 82B ubiGFP/ FRT 82B trxE2
y w hs FLP122; FRT 82B ubiGFP/ FRT 82B ScmD1
y w hs FLP122; FRT 82B ubiGFP/ FRT 82B trxE2 ScmD1
Immunostaining
Imaginal discs were prepared and stained using standard procedures. The primary antibodies used were: rabbit and mouse anti-β-Gal (1 : 1000, Cappel and Promega), mouse anti-Wg (1 : 50, Developmental Studies Hybridoma Bank), rat-αPS2 (1 : 5, gift from Martín Bermudo) and rabbit anti-Ap (1 : 1000, this study)
Chromatin immunoprecipitation experiments
Third instar larvae were dissected and wing imaginal discs were collected in PBS on ice. Discs were fixed with 1.8% formaldehyde. Chromatin preparation and immunoprecipitation were performed as described [79]. For Ap ChIPs, 1.5 μg anti-Ap (dN–20, Santa Cruz Biotechnologies) was used for each immunoprecipitation, and specificity was tested by parallel “mock” immunoprecipitations carried out with normal goat IgG (Santa Cruz Biotechnologies). ChIP enrichment values were normalized relative to “mock” enrichment values to control for any signal that could be attributed to highly accessible chromatin [80]. Three real-time PCR amplicons surrounding the apP (chr2R, 1614425–1614545; coordinates based on dm3 build of Drosophila genome), apE (chr2R, 1622079–1622182), or apDV (chr2R, 1639774–1639867) elements were used to quantify immunoprecipitated chromatin. For Sd ChIP, maximum enrichment signals from Sd ChIP-chip data [79] for the corresponding apP, apE, and apDV regions were normalized to the same “mock” enrichment values used in the Ap ChIP experiments. Importantly, the Sd peak at apDV was called as statistically significant in the previously published genome-wide ChIP data [79].
Chromosome conformation capture (3C)
Chromosome conformation capture (3C) was performed as described in Webber et al, 2013 [81] with slight modification. Approximately 200 early third instar larvae were homogenized at room temperature in a crosslinking solution (1.8% formaldehyde, 50 mM HEPES, 1 mM EDTA, 0.5 mM EGTA, 100 mM NaCl). Total crosslinking time was limited to 20 minutes and followed by a 5-minute quench with glycine (0.125 M Glycine, 1xPBS, 0.01% Triton). Crude, fixed homogenate was then washed twice with PBS with 1% Triton, washed twice with a HEPES buffer (10 mM HEPES pH 7.6, 10 mM EDTA, 0.5 mM EGTA, 0.25% Triton), then Dounce homogenized in Buffer A (15 mM HEPES at pH 7.6, 10 mM KCl, 5 mM MgCl2, 0.1 mM EDTA, 0.5 mM EGTA, 350 mM sucrose, 1 mM DTT). After a brief centrifugation (400g for 1 minute) to remove cuticle and large debris, homogenate was centrifuged for 15 min at 10,000 rpm. Nuclei were resuspended in 100 μl of 1.2X DpnII Buffer with BSA (New England BioLabs), and then passed through a 27G syringe needle 10 times. 1.5 μl of 20% SDS was added to the nuclei-containing solution, which was then incubated for 30 min at 37°C, followed by 10 minutes at 65°C, addition of 10 μl 20% Triton X–100, and then incubation for 1 hour at 37°C. 100 units of DpnII were then added to the nuclei-containing solution, followed by overnight incubation at 37°C. The digestion reaction was stopped by adding 16 μl 10% SDS and incubating at 65°C for 10 minutes. From this point on, 3C was carried out as described in [81]. Ligation products were analyzed by qPCR (primer sequences available upon request). The amount of 3C amplicon product was normalized relative to an amplicon in the ap promoter that does not span a DpnII site and gives a measure of the total DNA in the reaction.
Supporting Information
Zdroje
1. Irvine KD, Rauskolb C (2001) Boundaries in development: formation and function. Annu Rev Cell Dev Biol 17 : 189–214. 11687488
2. Garcia-Bellido A, Ripoll P, Morata G (1976) Developmental compartmentalization in the dorsal mesothoracic disc of Drosophila. Dev Biol 48 : 132–147. 1245256
3. Diaz-Benjumea FJ, Cohen SM (1995) Serrate signals through Notch to establish a Wingless-dependent organizer at the dorsal/ventral compartment boundary of the Drosophila wing. Development 121 : 4215–4225. 8575321
4. Basler K, Struhl G (1994) Compartment boundaries and the control of Drosophila limb pattern by hedgehog protein. Nature 368 : 208–214. 8145818
5. Nellen D, Burke R, Struhl G, Basler K (1996) Direct and long-range action of a DPP morphogen gradient. Cell 85 : 357–368. 8616891
6. Zecca M, Basler K, Struhl G (1996) Direct and long-range action of a wingless morphogen gradient. Cell 87 : 833–844. 8945511
7. Lecuit T, Brook WJ, Ng M, Calleja M, Sun H, et al. (1996) Two distinct mechanisms for long-range patterning by Decapentaplegic in the Drosophila wing. Nature 381 : 387–393. 8632795
8. Neumann CJ, Cohen SM (1997) Long-range action of Wingless organizes the dorsal-ventral axis of the Drosophila wing. Development 124 : 871–880. 9043068
9. Morata G, Lawrence PA (1975) Control of compartment development by the engrailed gene in Drosophila. Nature 255 : 614–617. 1134551
10. Lawrence PA, Struhl G (1982) Further studies of the engrailed phenotype in Drosophila. EMBO J 1 : 827–833. 6152896
11. Tabata T, Schwartz C, Gustavson E, Ali Z, Kornberg TB (1995) Creating a Drosophila wing de novo, the role of engrailed, and the compartment border hypothesis. Development 121 : 3359–3369. 7588069
12. Zecca M, Basler K, Struhl G (1995) Sequential organizing activities of engrailed, hedgehog and decapentaplegic in the Drosophila wing. Development 121 : 2265–2278. 7671794
13. Lawrence PA, Morata G (1976) Compartments in the wing of Drosophila: a study of the engrailed gene. Dev Biol 50 : 321–337. 1278589
14. Blair SS, Brower DL, Thomas JB, Zavortink M (1994) The role of apterous in the control of dorsoventral compartmentalization and PS integrin gene expression in the developing wing of Drosophila. Development 120 : 1805–1815. 7924988
15. Diaz-Benjumea FJ, Cohen SM (1993) Interaction between dorsal and ventral cells in the imaginal disc directs wing development in Drosophila. Cell 75 : 741–752. 8242746
16. Cohen B, McGuffin ME, Pfeifle C, Segal D, Cohen SM (1992) apterous, a gene required for imaginal disc development in Drosophila encodes a member of the LIM family of developmental regulatory proteins. Genes Dev 6 : 715–729. 1349545
17. Fernandez-Funez P, Lu CH, Rincon-Limas DE, Garcia-Bellido A, Botas J (1998) The relative expression amounts of apterous and its co-factor dLdb/Chip are critical for dorso-ventral compartmentalization in the Drosophila wing. EMBO J 17 : 6846–6853. 9843490
18. Milan M, Cohen SM (1999) Regulation of LIM homeodomain activity in vivo: a tetramer of dLDB and apterous confers activity and capacity for regulation by dLMO. Mol Cell 4 : 267–273. 10488342
19. van Meyel DJ, O'Keefe DD, Jurata LW, Thor S, Gill GN, et al. (1999) Chip and apterous physically interact to form a functional complex during Drosophila development. Mol Cell 4 : 259–265. 10488341
20. Kim J, Sebring A, Esch JJ, Kraus ME, Vorwerk K, et al. (1996) Integration of positional signals and regulation of wing formation and identity by Drosophila vestigial gene. Nature 382 : 133–138. 8700202
21. Rulifson EJ, Blair SS (1995) Notch regulates wingless expression and is not required for reception of the paracrine wingless signal during wing margin neurogenesis in Drosophila. Development 121 : 2813–2824. 7555709
22. Spitz F, Furlong EE (2012) Transcription factors: from enhancer binding to developmental control. Nat Rev Genet 13 : 613–626. doi: 10.1038/nrg3207 22868264
23. Capovilla M, Kambris Z, Botas J (2001) Direct regulation of the muscle-identity gene apterous by a Hox protein in the somatic mesoderm. Development 128 : 1221–1230. 11262224
24. Lundgren SE, Callahan CA, Thor S, Thomas JB (1995) Control of neuronal pathway selection by the Drosophila LIM homeodomain gene apterous. Development 121 : 1769–1773. 7600992
25. Bieli D, Kanca O, Gohl D, Denes A, Schedl P, et al. (2015) The Drosophila melanogaster Mutants apblot and apXasta Affect an Essential apterous Wing Enhancer. G3 5 : 1129–1143. doi: 10.1534/g3.115.017707 25840432
26. Wang SH, Simcox A, Campbell G (2000) Dual role for Drosophila epidermal growth factor receptor signaling in early wing disc development. Genes Dev 14 : 2271–2276. 10995384
27. Zecca M, Struhl G (2002) Control of growth and patterning of the Drosophila wing imaginal disc by EGFR-mediated signaling. Development 129 : 1369–1376. 11880346
28. Kassis JA, Brown JL (2013) Polycomb group response elements in Drosophila and vertebrates. Adv Genet 81 : 83–118. doi: 10.1016/B978-0-12-407677-8.00003-8 23419717
29. Kennison JA (1995) The Polycomb and trithorax group proteins of Drosophila: trans-regulators of homeotic gene function. Annu Rev Genet 29 : 289–303. 8825476
30. Oktaba K, Gutierrez L, Gagneur J, Girardot C, Sengupta AK, et al. (2008) Dynamic regulation by polycomb group protein complexes controls pattern formation and the cell cycle in Drosophila. Dev Cell 15 : 877–889. doi: 10.1016/j.devcel.2008.10.005 18993116
31. Butler JE, Kadonaga JT (2002) The RNA polymerase II core promoter: a key component in the regulation of gene expression. Genes Dev 16 : 2583–2592. 12381658
32. Negre N, Hennetin J, Sun LV, Lavrov S, Bellis M, et al. (2006) Chromosomal distribution of PcG proteins during Drosophila development. PLoS Biol 4: e170. 16613483
33. Tolhuis B, de Wit E, Muijrers I, Teunissen H, Talhout W, et al. (2006) Genome-wide profiling of PRC1 and PRC2 Polycomb chromatin binding in Drosophila melanogaster. Nat Genet 38 : 694–699. 16628213
34. Schwartz YB, Kahn TG, Nix DA, Li XY, Bourgon R, et al. (2006) Genome-wide analysis of Polycomb targets in Drosophila melanogaster. Nat Genet 38 : 700–705. 16732288
35. Gohl D, Muller M, Pirrotta V, Affolter M, Schedl P (2008) Enhancer blocking and transvection at the Drosophila apterous locus. Genetics 178 : 127–143. doi: 10.1534/genetics.107.077768 18202363
36. Pekowska A, Benoukraf T, Zacarias-Cabeza J, Belhocine M, Koch F, et al. (2011) H3K4 tri-methylation provides an epigenetic signature of active enhancers. EMBO J 30 : 4198–4210. doi: 10.1038/emboj.2011.295 21847099
37. Jory A, Estella C, Giorgianni MW, Slattery M, Laverty TR, et al. (2012) A survey of 6,300 genomic fragments for cis-regulatory activity in the imaginal discs of Drosophila melanogaster. Cell Rep 2 : 1014–1024. doi: 10.1016/j.celrep.2012.09.010 23063361
38. Milan M, Diaz-Benjumea FJ, Cohen SM (1998) Beadex encodes an LMO protein that regulates Apterous LIM-homeodomain activity in Drosophila wing development: a model for LMO oncogene function. Genes Dev 12 : 2912–2920. 9744867
39. Rebay I (2002) Keeping the receptor tyrosine kinase signaling pathway in check: lessons from Drosophila. Dev Biol 251 : 1–17. 12413894
40. Halder G, Polaczyk P, Kraus ME, Hudson A, Kim J, et al. (1998) The Vestigial and Scalloped proteins act together to directly regulate wing-specific gene expression in Drosophila. Genes Dev 12 : 3900–3909. 9869643
41. Pueyo JI, Galindo MI, Bishop SA, Couso JP (2000) Proximal-distal leg development in Drosophila requires the apterous gene and the Lim1 homologue dlim1. Development 127 : 5391–5402. 11076760
42. Morris JR, Petrov DA, Lee AM, Wu CT (2004) Enhancer choice in cis and in trans in Drosophila melanogaster: role of the promoter. Genetics 167 : 1739–1747. 15342512
43. Morris JR, Chen J, Filandrinos ST, Dunn RC, Fisk R, et al. (1999) An analysis of transvection at the yellow locus of Drosophila melanogaster. Genetics 151 : 633–651. 9927457
44. Duncan IW (2002) Transvection effects in Drosophila. Annu Rev Genet 36 : 521–556. 12429702
45. Dekker J, Rippe K, Dekker M, Kleckner N (2002) Capturing chromosome conformation. Science 295 : 1306–1311. 11847345
46. Estella C, McKay DJ, Mann RS (2008) Molecular integration of wingless, decapentaplegic, and autoregulatory inputs into Distalless during Drosophila leg development. Dev Cell 14 : 86–96. doi: 10.1016/j.devcel.2007.11.002 18194655
47. Struhl G, Basler K (1993) Organizing activity of wingless protein in Drosophila. Cell 72 : 527–540. 8440019
48. Williams JA, Paddock SW, Carroll SB (1993) Pattern formation in a secondary field: a hierarchy of regulatory genes subdivides the developing Drosophila wing disc into discrete subregions. Development 117 : 571–584. 8330528
49. Paul L, Wang SH, Manivannan SN, Bonanno L, Lewis S, et al. (2013) Dpp-induced Egfr signaling triggers postembryonic wing development in Drosophila. Proc Natl Acad Sci U S A 110 : 5058–5063. doi: 10.1073/pnas.1217538110 23479629
50. Simcox AA, Grumbling G, Schnepp B, Bennington-Mathias C, Hersperger E, et al. (1996) Molecular, phenotypic, and expression analysis of vein, a gene required for growth of the Drosophila wing disc. Dev Biol 177 : 475–489. 8806825
51. de Celis JF, Bray S (1997) Feed-back mechanisms affecting Notch activation at the dorsoventral boundary in the Drosophila wing. Development 124 : 3241–3251. 9310319
52. Klein T, Arias AM (1998) Different spatial and temporal interactions between Notch, wingless, and vestigial specify proximal and distal pattern elements of the wing in Drosophila. Dev Biol 194 : 196–212. 9501029
53. McKay DJ, Estella C, Mann RS (2009) The origins of the Drosophila leg revealed by the cis-regulatory architecture of the Distalless gene. Development 136 : 61–71. doi: 10.1242/dev.029975 19036798
54. Crews ST, Pearson JC (2009) Transcriptional autoregulation in development. Curr Biol 19: R241–246. doi: 10.1016/j.cub.2009.01.015 19321138
55. DeVido SK, Kwon D, Brown JL, Kassis JA (2008) The role of Polycomb-group response elements in regulation of engrailed transcription in Drosophila. Development 135 : 669–676. doi: 10.1242/dev.014779 18199580
56. Kwon D, Mucci D, Langlais KK, Americo JL, DeVido SK, et al. (2009) Enhancer-promoter communication at the Drosophila engrailed locus. Development 136 : 3067–3075. doi: 10.1242/dev.036426 19675130
57. Milne TA, Briggs SD, Brock HW, Martin ME, Gibbs D, et al. (2002) MLL targets SET domain methyltransferase activity to Hox gene promoters. Mol Cell 10 : 1107–1117. 12453418
58. Poux S, Horard B, Sigrist CJ, Pirrotta V (2002) The Drosophila trithorax protein is a coactivator required to prevent re-establishment of polycomb silencing. Development 129 : 2483–2493. 11973279
59. Klymenko T, Muller J (2004) The histone methyltransferases Trithorax and Ash1 prevent transcriptional silencing by Polycomb group proteins. EMBO Rep 5 : 373–377. 15031712
60. Schuettengruber B, Ganapathi M, Leblanc B, Portoso M, Jaschek R, et al. (2009) Functional anatomy of polycomb and trithorax chromatin landscapes in Drosophila embryos. PLoS Biol 7: e13. doi: 10.1371/journal.pbio.1000013 19143474
61. Ghavi-Helm Y, Klein FA, Pakozdi T, Ciglar L, Noordermeer D, et al. (2014) Enhancer loops appear stable during development and are associated with paused polymerase. Nature 512 : 96–100. doi: 10.1038/nature13417 25043061
62. Giorgianni MW, Mann RS (2011) Establishment of medial fates along the proximodistal axis of the Drosophila leg through direct activation of dachshund by Distalless. Dev Cell 20 : 455–468. doi: 10.1016/j.devcel.2011.03.017 21497759
63. Perez L, Barrio L, Cano D, Fiuza UM, Muzzopappa M, et al. (2011) Enhancer-PRE communication contributes to the expansion of gene expression domains in proliferating primordia. Development 138 : 3125–3134. doi: 10.1242/dev.065599 21715425
64. Bejarano F, Milan M (2009) Genetic and epigenetic mechanisms regulating hedgehog expression in the Drosophila wing. Dev Biol 327 : 508–515. doi: 10.1016/j.ydbio.2009.01.006 19210960
65. Steffen PA, Ringrose L (2014) What are memories made of? How Polycomb and Trithorax proteins mediate epigenetic memory. Nat Rev Mol Cell Biol 15 : 340–356. doi: 10.1038/nrm3789 24755934
66. Bischof J, Maeda RK, Hediger M, Karch F, Basler K (2007) An optimized transgenesis system for Drosophila using germ-line-specific phiC31 integrases. Proc Natl Acad Sci U S A 104 : 3312–3317. 17360644
67. Caussinus E, Kanca O, Affolter M (2012) Fluorescent fusion protein knockout mediated by anti-GFP nanobody. Nat Struct Mol Biol 19 : 117–121.
68. Siegal ML, Hartl DL (1996) Transgene Coplacement and high efficiency site-specific recombination with the Cre/loxP system in Drosophila. Genetics 144 : 715–726. 8889532
69. Golic KG, Golic MM (1996) Engineering the Drosophila genome: chromosome rearrangements by design. Genetics 144 : 1693–1711. 8978056
70. Venken KJ, Schulze KL, Haelterman NA, Pan H, He Y, et al. (2011) MiMIC: a highly versatile transposon insertion resource for engineering Drosophila melanogaster genes. Nat Methods 8 : 737–743. 21985007
71. Sipos L, Kozma G, Molnar E, Bender W (2007) In situ dissection of a Polycomb response element in Drosophila melanogaster. Proc Natl Acad Sci U S A 104 : 12416–12421. 17640916
72. Gloor GB, Nassif NA, Johnson-Schlitz DM, Preston CR, Engels WR (1991) Targeted gene replacement in Drosophila via P element-induced gap repair. Science 253 : 1110–1117. 1653452
73. Bateman JR, Lee AM, Wu CT (2006) Site-specific transformation of Drosophila via phiC31 integrase-mediated cassette exchange. Genetics 173 : 769–777. 16547094
74. Makarova O, Kamberov E, Margolis B (2000) Generation of deletion and point mutations with one primer in a single cloning step. Biotechniques 29 : 970–972. 11084856
75. Greenspan R (1997) Fly pushing: The theory and practice of Drosophila genetics.
76. Weiss A, Charbonnier E, Ellertsdottir E, Tsirigos A, Wolf C, et al. (2010) A conserved activation element in BMP signaling during Drosophila development. Nat Struct Mol Biol 17 : 69–76. doi: 10.1038/nsmb.1715 20010841
77. Boy AL, Zhai Z, Habring-Muller A, Kussler-Schneider Y, Kaspar P, et al. (2010) Vectors for efficient and high-throughput construction of fluorescent drosophila reporters using the PhiC31 site-specific integration system. Genesis 48 : 452–456. doi: 10.1002/dvg.20637 20506180
78. Sosinsky A, Bonin CP, Mann RS, Honig B (2003) Target Explorer: An automated tool for the identification of new target genes for a specified set of transcription factors. Nucleic Acids Res 31 : 3589–3592. 12824372
79. Slattery M, Voutev R, Ma L, Negre N, White KP, et al. (2013) Divergent transcriptional regulatory logic at the intersection of tissue growth and developmental patterning. PLoS Genet 9: e1003753. doi: 10.1371/journal.pgen.1003753 24039600
80. Teytelman L, Thurtle DM, Rine J, van Oudenaarden A (2013) Highly expressed loci are vulnerable to misleading ChIP localization of multiple unrelated proteins. Proc Natl Acad Sci U S A 110 : 18602–18607. doi: 10.1073/pnas.1316064110 24173036
81. Webber JL, Zhang J, Mitchell-Dick A, Rebay I (2013) 3D chromatin interactions organize Yan chromatin occupancy and repression at the even-skipped locus. Genes Dev 27 : 2293–2298.
82. Thibault ST, Singer MA, Miyazaki WY, Milash B, Dompe NA, et al. (2004) A complementary transposon tool kit for Drosophila melanogaster using P and piggyBac. Nat Genet 36 : 283–287. 14981521
83. Moabbi AM, Agarwal N, El Kaderi B, Ansari A (2012) Role for gene looping in intron-mediated enhancement of transcription. Proc Natl Acad Sci U S A 109 : 8505–8510. doi: 10.1073/pnas.1112400109 22586116
Štítky
Genetika Reprodukční medicína
Článek Evidence of Selection against Complex Mitotic-Origin Aneuploidy during Preimplantation DevelopmentČlánek A Novel Route Controlling Begomovirus Resistance by the Messenger RNA Surveillance Factor PelotaČlánek A Follicle Rupture Assay Reveals an Essential Role for Follicular Adrenergic Signaling in OvulationČlánek Canonical Poly(A) Polymerase Activity Promotes the Decay of a Wide Variety of Mammalian Nuclear RNAsČlánek FANCI Regulates Recruitment of the FA Core Complex at Sites of DNA Damage Independently of FANCD2Článek Hsp90-Associated Immunophilin Homolog Cpr7 Is Required for the Mitotic Stability of [URE3] Prion inČlánek The Dedicated Chaperone Acl4 Escorts Ribosomal Protein Rpl4 to Its Nuclear Pre-60S Assembly SiteČlánek Chromatin-Remodelling Complex NURF Is Essential for Differentiation of Adult Melanocyte Stem CellsČlánek A Systems Approach Identifies Essential FOXO3 Functions at Key Steps of Terminal ErythropoiesisČlánek Integration of Posttranscriptional Gene Networks into Metabolic Adaptation and Biofilm Maturation inČlánek Lateral and End-On Kinetochore Attachments Are Coordinated to Achieve Bi-orientation in OocytesČlánek MET18 Connects the Cytosolic Iron-Sulfur Cluster Assembly Pathway to Active DNA Demethylation in
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2015 Číslo 10
-
Všechny články tohoto čísla
- Gene-Regulatory Logic to Induce and Maintain a Developmental Compartment
- A Decad(e) of Reasons to Contribute to a PLOS Community-Run Journal
- DNA Methylation Landscapes of Human Fetal Development
- Single Strand Annealing Plays a Major Role in RecA-Independent Recombination between Repeated Sequences in the Radioresistant Bacterium
- Evidence of Selection against Complex Mitotic-Origin Aneuploidy during Preimplantation Development
- Transcriptional Derepression Uncovers Cryptic Higher-Order Genetic Interactions
- Silencing of X-Linked MicroRNAs by Meiotic Sex Chromosome Inactivation
- Virus Satellites Drive Viral Evolution and Ecology
- A Novel Route Controlling Begomovirus Resistance by the Messenger RNA Surveillance Factor Pelota
- Sequence to Medical Phenotypes: A Framework for Interpretation of Human Whole Genome DNA Sequence Data
- Your Data to Explore: An Interview with Anne Wojcicki
- Modulation of Ambient Temperature-Dependent Flowering in by Natural Variation of
- The Ciliopathy Protein CC2D2A Associates with NINL and Functions in RAB8-MICAL3-Regulated Vesicle Trafficking
- PPP2R5C Couples Hepatic Glucose and Lipid Homeostasis
- DCA1 Acts as a Transcriptional Co-activator of DST and Contributes to Drought and Salt Tolerance in Rice
- Intermediate Levels of CodY Activity Are Required for Derepression of the Branched-Chain Amino Acid Permease, BraB
- "Missing" G x E Variation Controls Flowering Time in
- The Rise and Fall of an Evolutionary Innovation: Contrasting Strategies of Venom Evolution in Ancient and Young Animals
- Type IV Collagen Controls the Axogenesis of Cerebellar Granule Cells by Regulating Basement Membrane Integrity in Zebrafish
- Loss of a Conserved tRNA Anticodon Modification Perturbs Plant Immunity
- Genome-Wide Association Analysis of Adaptation Using Environmentally Predicted Traits
- Oriented Cell Division in the . Embryo Is Coordinated by G-Protein Signaling Dependent on the Adhesion GPCR LAT-1
- Disproportionate Contributions of Select Genomic Compartments and Cell Types to Genetic Risk for Coronary Artery Disease
- A Follicle Rupture Assay Reveals an Essential Role for Follicular Adrenergic Signaling in Ovulation
- The RNAPII-CTD Maintains Genome Integrity through Inhibition of Retrotransposon Gene Expression and Transposition
- Canonical Poly(A) Polymerase Activity Promotes the Decay of a Wide Variety of Mammalian Nuclear RNAs
- Allelic Variation of Cytochrome P450s Drives Resistance to Bednet Insecticides in a Major Malaria Vector
- SCARN a Novel Class of SCAR Protein That Is Required for Root-Hair Infection during Legume Nodulation
- IBR5 Modulates Temperature-Dependent, R Protein CHS3-Mediated Defense Responses in
- NINL and DZANK1 Co-function in Vesicle Transport and Are Essential for Photoreceptor Development in Zebrafish
- Decay-Initiating Endoribonucleolytic Cleavage by RNase Y Is Kept under Tight Control via Sequence Preference and Sub-cellular Localisation
- Large-Scale Analysis of Kinase Signaling in Yeast Pseudohyphal Development Identifies Regulation of Ribonucleoprotein Granules
- FANCI Regulates Recruitment of the FA Core Complex at Sites of DNA Damage Independently of FANCD2
- LINE-1 Mediated Insertion into (Protein of Centriole 1 A) Causes Growth Insufficiency and Male Infertility in Mice
- Hsp90-Associated Immunophilin Homolog Cpr7 Is Required for the Mitotic Stability of [URE3] Prion in
- Genome-Scale Mapping of σ Reveals Widespread, Conserved Intragenic Binding
- Uncovering Hidden Layers of Cell Cycle Regulation through Integrative Multi-omic Analysis
- Functional Diversification of Motor Neuron-specific Enhancers during Evolution
- The GTP- and Phospholipid-Binding Protein TTD14 Regulates Trafficking of the TRPL Ion Channel in Photoreceptor Cells
- The Gyc76C Receptor Guanylyl Cyclase and the Foraging cGMP-Dependent Kinase Regulate Extracellular Matrix Organization and BMP Signaling in the Developing Wing of
- The Ty1 Retrotransposon Restriction Factor p22 Targets Gag
- Functional Impact and Evolution of a Novel Human Polymorphic Inversion That Disrupts a Gene and Creates a Fusion Transcript
- The Dedicated Chaperone Acl4 Escorts Ribosomal Protein Rpl4 to Its Nuclear Pre-60S Assembly Site
- The Influence of Age and Sex on Genetic Associations with Adult Body Size and Shape: A Large-Scale Genome-Wide Interaction Study
- Parent-of-Origin Effects of the Gene on Adiposity in Young Adults
- Chromatin-Remodelling Complex NURF Is Essential for Differentiation of Adult Melanocyte Stem Cells
- Retinoic Acid Receptors Control Spermatogonia Cell-Fate and Induce Expression of the SALL4A Transcription Factor
- A Systems Approach Identifies Essential FOXO3 Functions at Key Steps of Terminal Erythropoiesis
- Protein O-Glucosyltransferase 1 (POGLUT1) Promotes Mouse Gastrulation through Modification of the Apical Polarity Protein CRUMBS2
- KIF7 Controls the Proliferation of Cells of the Respiratory Airway through Distinct Microtubule Dependent Mechanisms
- Integration of Posttranscriptional Gene Networks into Metabolic Adaptation and Biofilm Maturation in
- Lateral and End-On Kinetochore Attachments Are Coordinated to Achieve Bi-orientation in Oocytes
- Protein Homeostasis Imposes a Barrier on Functional Integration of Horizontally Transferred Genes in Bacteria
- A New Method for Detecting Associations with Rare Copy-Number Variants
- Histone H2AFX Links Meiotic Chromosome Asynapsis to Prophase I Oocyte Loss in Mammals
- The Genomic Aftermath of Hybridization in the Opportunistic Pathogen
- A Role for the Chaperone Complex BAG3-HSPB8 in Actin Dynamics, Spindle Orientation and Proper Chromosome Segregation during Mitosis
- Establishment of a Developmental Compartment Requires Interactions between Three Synergistic -regulatory Modules
- Regulation of Spore Formation by the SpoIIQ and SpoIIIA Proteins
- Association of the Long Non-coding RNA Steroid Receptor RNA Activator (SRA) with TrxG and PRC2 Complexes
- Alkaline Ceramidase 3 Deficiency Results in Purkinje Cell Degeneration and Cerebellar Ataxia Due to Dyshomeostasis of Sphingolipids in the Brain
- ACLY and ACC1 Regulate Hypoxia-Induced Apoptosis by Modulating ETV4 via α-ketoglutarate
- Quantitative Differences in Nuclear β-catenin and TCF Pattern Embryonic Cells in .
- HENMT1 and piRNA Stability Are Required for Adult Male Germ Cell Transposon Repression and to Define the Spermatogenic Program in the Mouse
- Axon Regeneration Is Regulated by Ets–C/EBP Transcription Complexes Generated by Activation of the cAMP/Ca Signaling Pathways
- A Phenomic Scan of the Norfolk Island Genetic Isolate Identifies a Major Pleiotropic Effect Locus Associated with Metabolic and Renal Disorder Markers
- The Roles of CDF2 in Transcriptional and Posttranscriptional Regulation of Primary MicroRNAs
- A Genetic Cascade of Modulates Nucleolar Size and rRNA Pool in
- Inter-population Differences in Retrogene Loss and Expression in Humans
- Cationic Peptides Facilitate Iron-induced Mutagenesis in Bacteria
- EP4 Receptor–Associated Protein in Macrophages Ameliorates Colitis and Colitis-Associated Tumorigenesis
- Fungal Infection Induces Sex-Specific Transcriptional Changes and Alters Sexual Dimorphism in the Dioecious Plant
- FLCN and AMPK Confer Resistance to Hyperosmotic Stress via Remodeling of Glycogen Stores
- MET18 Connects the Cytosolic Iron-Sulfur Cluster Assembly Pathway to Active DNA Demethylation in
- Sex Bias and Maternal Contribution to Gene Expression Divergence in Blastoderm Embryos
- Transcriptional and Linkage Analyses Identify Loci that Mediate the Differential Macrophage Response to Inflammatory Stimuli and Infection
- Mre11 and Blm-Dependent Formation of ALT-Like Telomeres in Ku-Deficient
- Genome Wide Identification of SARS-CoV Susceptibility Loci Using the Collaborative Cross
- Identification of a Single Strand Origin of Replication in the Integrative and Conjugative Element ICE of
- The Type VI Secretion TssEFGK-VgrG Phage-Like Baseplate Is Recruited to the TssJLM Membrane Complex via Multiple Contacts and Serves As Assembly Platform for Tail Tube/Sheath Polymerization
- The Dynamic Genome and Transcriptome of the Human Fungal Pathogen and Close Relative
- Secondary Structure across the Bacterial Transcriptome Reveals Versatile Roles in mRNA Regulation and Function
- ROS-Induced JNK and p38 Signaling Is Required for Unpaired Cytokine Activation during Regeneration
- Pelle Modulates dFoxO-Mediated Cell Death in
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Single Strand Annealing Plays a Major Role in RecA-Independent Recombination between Repeated Sequences in the Radioresistant Bacterium
- The Rise and Fall of an Evolutionary Innovation: Contrasting Strategies of Venom Evolution in Ancient and Young Animals
- Genome Wide Identification of SARS-CoV Susceptibility Loci Using the Collaborative Cross
- DCA1 Acts as a Transcriptional Co-activator of DST and Contributes to Drought and Salt Tolerance in Rice
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



