-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaA Phenomic Scan of the Norfolk Island Genetic Isolate Identifies a Major Pleiotropic Effect Locus Associated with Metabolic and Renal Disorder Markers
While many large genetic association studies have identified genes playing a role in complex disorders, there is still concern over the amount of missing genetic heritability. With this in mind, we have used a data reduction approach alongside pedigree-based association to obtain highly heritable components which explain 'hidden' variance of multiphenotypes within a large pedigree from the Norfolk Island genetic isolate. The most heritable of these components involved 7 traits reflecting metabolic and renal functionality, association of which locates to an intergenic region on chromosome 1p22.2. By integrating gene expression information, we identified enrichment of a purine metabolism pathway, further strengthening the metabolic nature of the observed association. Adding additional support to our approach, we show association of the tagging SNP (rs1396315) in an independent US population. The findings presented here are of particular interest as they implicate pleiotropic effect loci and newly associated biological pathways underlying cardiovascular disease risk.
Published in the journal: . PLoS Genet 11(10): e32767. doi:10.1371/journal.pgen.1005593
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1005593Summary
While many large genetic association studies have identified genes playing a role in complex disorders, there is still concern over the amount of missing genetic heritability. With this in mind, we have used a data reduction approach alongside pedigree-based association to obtain highly heritable components which explain 'hidden' variance of multiphenotypes within a large pedigree from the Norfolk Island genetic isolate. The most heritable of these components involved 7 traits reflecting metabolic and renal functionality, association of which locates to an intergenic region on chromosome 1p22.2. By integrating gene expression information, we identified enrichment of a purine metabolism pathway, further strengthening the metabolic nature of the observed association. Adding additional support to our approach, we show association of the tagging SNP (rs1396315) in an independent US population. The findings presented here are of particular interest as they implicate pleiotropic effect loci and newly associated biological pathways underlying cardiovascular disease risk.
Introduction
Cardiovascular diseases (CVD) are highly prevalent conditions and are the leading cause of morbidity and mortality in many developed nations [1]. In general, CVDs are comorbid conditions that arise from several major underlying risk factor traits, e.g. excess adiposity, hypertension and dyslipidemia. These component traits, and therefore risk of CVD, are influenced to varying degrees by inherited genetic factors. Many genome-wide association studies (GWASs) have been conducted in recent years and have identified common genetic variants associated with all major risk factor traits for CVD.
It is well known that risk factor traits for CVD tend to be correlated in populations suggesting the presence of underlying genetic variants that affect multiple different phenotypes, i.e. exert pleiotropic effects. GWAS analysis aimed at multiple CVD phenotypes simultaneously may reveal such genetic loci, which could remain undetected using univariate (single) phenotype analysis methods. For example, a large-scale population-based GWAS of >25,000 Americans revealed previously unrecognised variants within APOC1, BRAP and PLCG1 that may confer pleiotropic effects on composite traits associated with CVD [2]. More recently, a GWAS meta-analysis of 85,500 subjects identified 25 variants with pleiotropic associations to metabolic syndrome traits [3]. These results support a multiple phenotype (“phenomics” based approach) for mapping pleiotropic effect genes for CVD.
GWAS analysis of isolated populations have also been successfully utilised to map genes for CVD traits [4–6]. Features such as founder effect, reduced genetic and environmental diversity, as well as the availability of very large multi-generational pedigrees can offer advantages over general population GWAS approaches [4]. For example, pedigree information can allow accurate estimates of heritability to be calculated that can help prioritise genetically influenced phenotypes for GWAS analysis. Moreover, founder effect leading to reduced genetic heterogeneity in isolated populations can amplify genetic effects and thus improve power to detect susceptibility loci [7,8].
The Norfolk Island Health Study (NIHS) is a long running investigation of the genetics of CVD traits in the isolated population of Norfolk Island—situated off the Australian East Coast [9]. The Norfolk Island population was founded by a small group of European “Bounty” mutineers and their Polynesian wives in the late 1700s and now forms a 6000-member pedigree spanning 11 generations [10–12]. The Norfolk Island population exhibits features such as genetic founder effect and admixture [13], and has high rates of heritable CVD traits such as hypertension and obesity [9,14]. Recently, we completed pedigree-based GWASs of 37 CVD-related traits in the Norfolk pedigree and identified a number of loci to be associated with individual phenotypes (under review). The aim of this new study was to perform a multi-phenotype (or phenomics-based) GWAS in the genetic isolate of Norfolk Island in an effort to identify novel pleiotropic effect loci influencing CVD related traits.
Results
This study focused on a well-characterised core pedigree from the Norfolk Island genetic isolate [14]. This core-pedigree is comprised of 330 individuals and has been measured for 37 quantitative traits as part of the NIHS to assess the genetics of CVD. The study involved principal component analysis (PCA) of all 37 traits to assess inter-correlation, and subsequent heritability (h2) analysis to estimate genetic variance of resultant components. This was followed by pedigree-based GWAS of heritable components using SNPs to identify major pleiotropic effect loci for CVD risk phenotypes in this population.
Principle Component Analysis (PCA) of CVD traits
An ‘unsupervised’ PCA of 37 CVD related traits was conducted (trait summaries listed in S1 Table). The PCA extracted 13 composite phenotypes which together explained >75% of the total variance (Table 1). Trait loading scores for each of these components is available in S2 Table. Heritability values for all 13 components, as well as a total combined component were calculated. Nine components showed nominal significance with heritability estimates ranging from 0.22–0.55 (P<0.05). After correction for multiple testing was applied only Component 3 and Component 9 remained significant (P<0.0038). Component 3 was the most heritable (h2 = 0.55) and was loaded with 7 measures related to CVD—% body fat, waist-to-hip ratio, systolic and diastolic blood pressure, creatinine, blood urea nitrogen, and uric acid. The measures most strongly correlated with Component 3 were blood urea nitrogen (0.76), creatinine (0.74) and uric acid (0.61) which are all kidney function markers suggesting that the Component 3 phenotype may represent renal dysfunction. The relationship between all extracted components is displayed in Fig 1. Overlap between components is suggestive of co-morbidity among traits as well as the possibility of genetic commonalities. The clinical relevance of the Component 3 score was explored by comparison with the formal Framingham CVD score [15] and showed a positive correlation (r = 0.4, P = 1.2x10-8).
Tab. 1. PCA and heritability analysis of 37 quantitative traits. 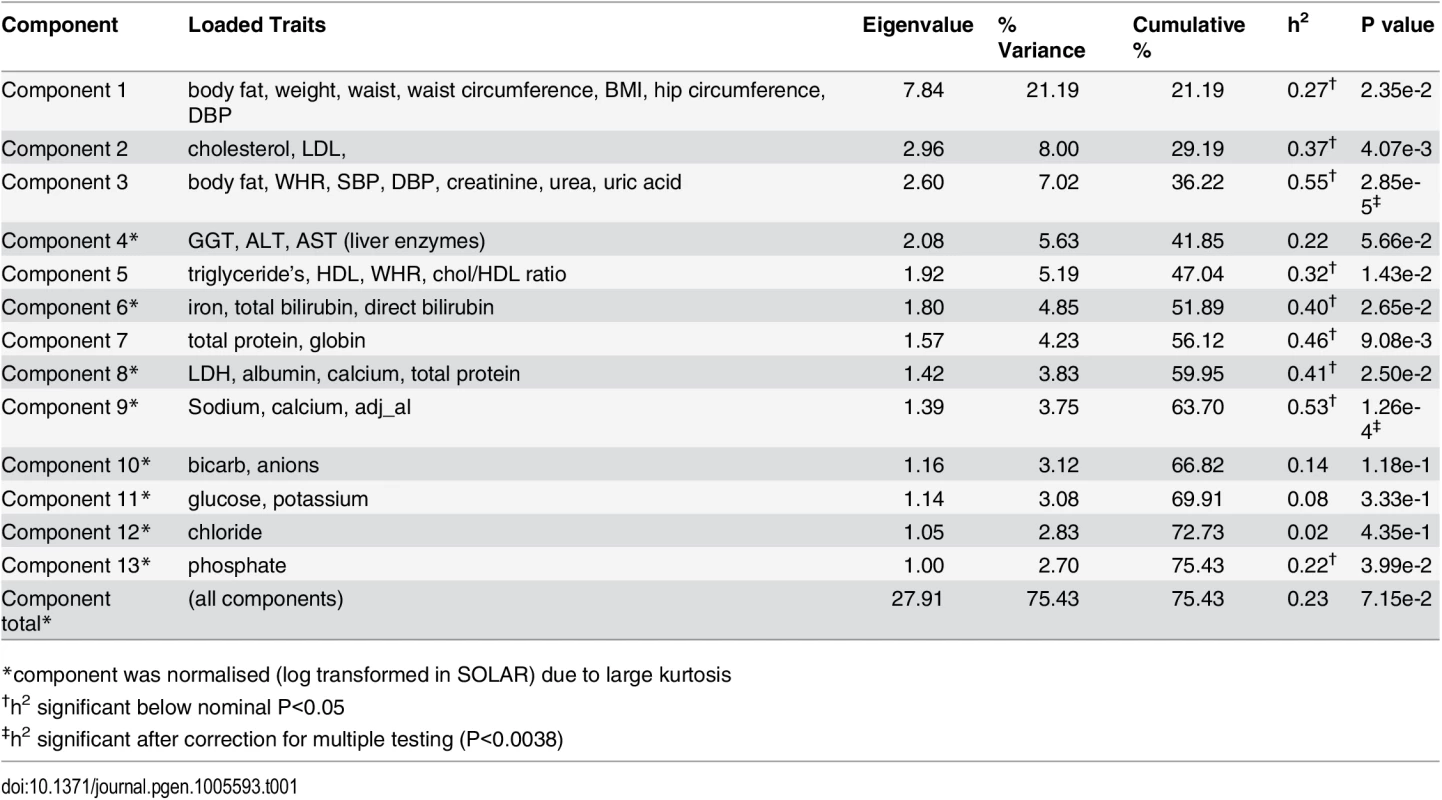
*component was normalised (log transformed in SOLAR) due to large kurtosis Fig. 1. Venn diagram showing the inter-correlation and heritabilities of components extracted from the PCA of 37 quantitative traits. 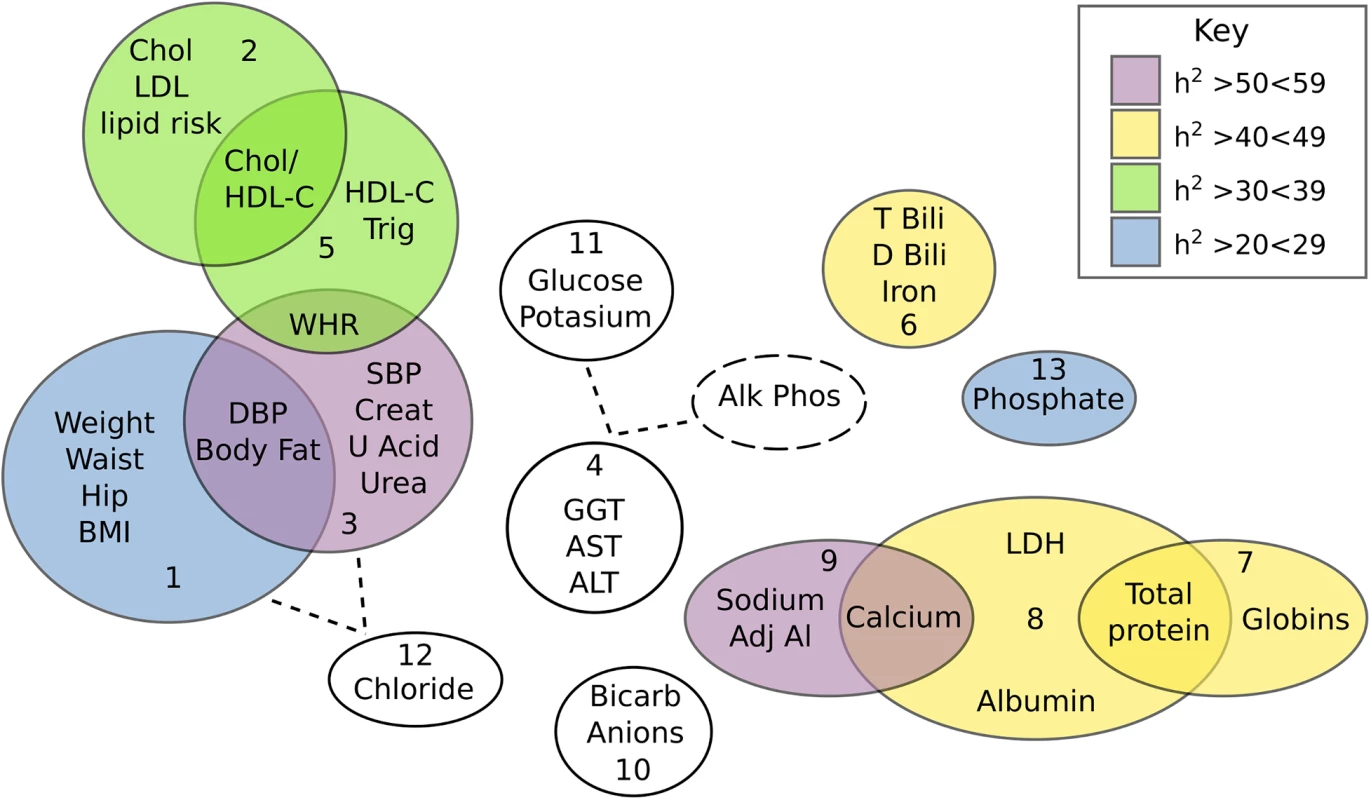
Relationships between components were inferred either by variable overlap, or loadings on a component greater than 0.35. Heritability of each component is colour coded (as per key) with the non-significant traits being coloured white. GWAS of principle components
Following the PCA, we performed GWAS analysis for the 9 nominally significantly (p<0.05) heritable components shown in Table 1. We decided to test all heritable components regardless of multiple testing to explore the potential biological relevance of association hits. Of the 9 components only 2 showed association peaks with clusters of SNPs that exceeded the genome-wide statistical significance threshold (P = 1.8x10-7). Fig 2A shows that Component 6—loaded with iron and bilirubin measures—yielded the strongest association peak on chr 2q37.1 (P = 1x10-11). Analysis of the individual component traits revealed that this peak was entirely associated with total and direct bilirubin, and maps to the well-known bilirubin metabolising gene—UGT1A1 [16–18], Fig 2B.
Fig. 2. Manhattan plot of Chromosome 2 showing Component 6 associations. 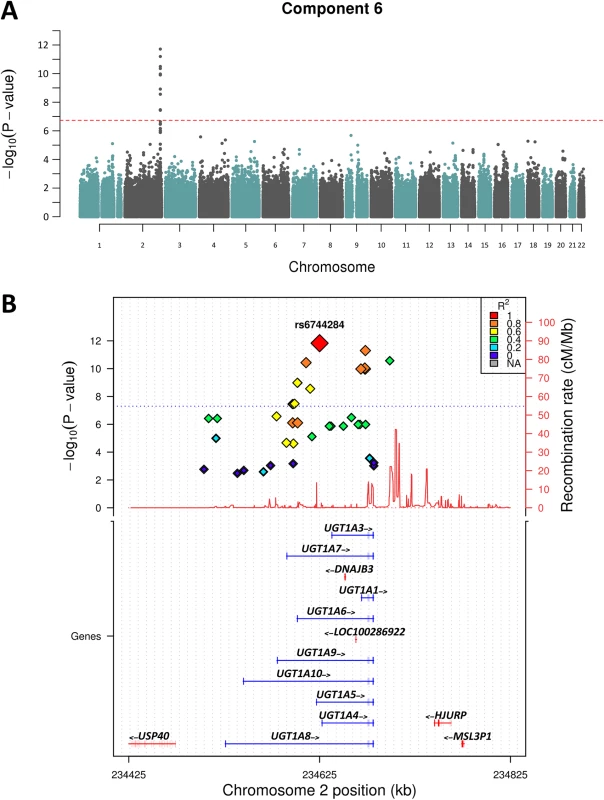
A) shows the genome-wide associations statistics with a distinct peak on chromosome 2 for bilirubin, B) refines the location of the peak to chr2q37.1, a region containing the UDP-glucuronosyltransferase gene family well known to metabolise bilirubin (UCSC track demonstrates the numerous isoforms). The blue dotted line indicates genome-wide significance level. The other component trait to yield a statistically significant association peak was the most heritable component, Component 3. Fig 3 shows that the GWAS of Component 3 yielded three associated SNPs located at chr 1p22.2 (min P = 1.3x10-9 for rs1396315), as well as a supportive peak below the threshold of study-wide significance. GWAS of each of the 7 traits loaded into Component 3 individually did not show any significant peak at this locus (S1 Fig). Therefore, unlike Component 6 (explained by bilirubin alone) the Component 3 hit indicated a potential pleotropic effect locus that was further investigated.
Fig. 3. Manhattan plot for Component 3 showing an association peak on chr 1p22.2. 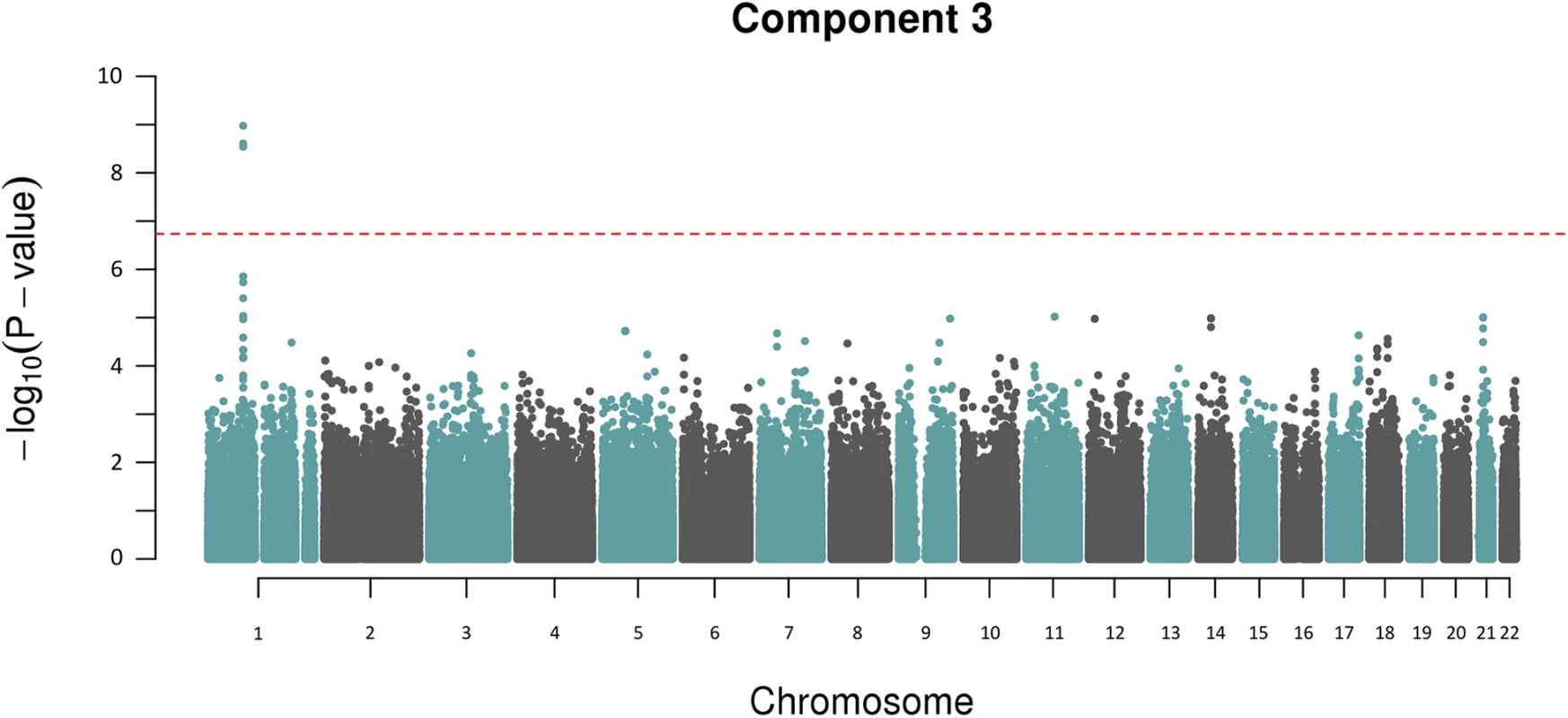
The red dotted line indicates the study-wide Meff significance threshold. To determine if there were any other additional independently associated variants in this region we performed an association analysis conditioning on rs1396315. This revealed no further associations indicating that only a single locus and resultant haplotype for Component 3 is present in this region. Linkage disequilibrium (LD) analysis of the SNPs spanning the Component 3 peak on chr 1p22.2 illustrated that rs1396315 tagged the associated haplotype and could be used as the reference SNP for further analysis (Fig 4). The allele frequency of the rarer allele (C) in the Norfolk pedigree was 0.24, which is consistent with the frequency in European populations [19]. Genotype association analysis of rs1396315 against the Component 3 score showed that the rare allele (C) was associated with elevated Component 3 score in an additive fashion. Locus-specific heritability estimates showed that this haplotype explained ~11% of the variation in the Component 3 score, indicating this is a major effect locus in the Norfolk Island population. Association analysis of rs1396315 against the individual phenotypes comprising Component 3 indicated that the strongest association was with blood urea nitrogen (P = 0.00006), followed by creatinine (P = 0.003) and uric acid (P = 0.005). The other traits loaded into Component 3 were not associated with rs1396315 when analysed independently (P>0.1). This indicates that the association of this SNP with Component 3 is primarily due to blood urea nitrogen, creatinine and uric acid—potential metabolic and renal functional markers.
Fig. 4. LocusTrack plot showing detailed annotation information of the associated chr1p22.2 region. 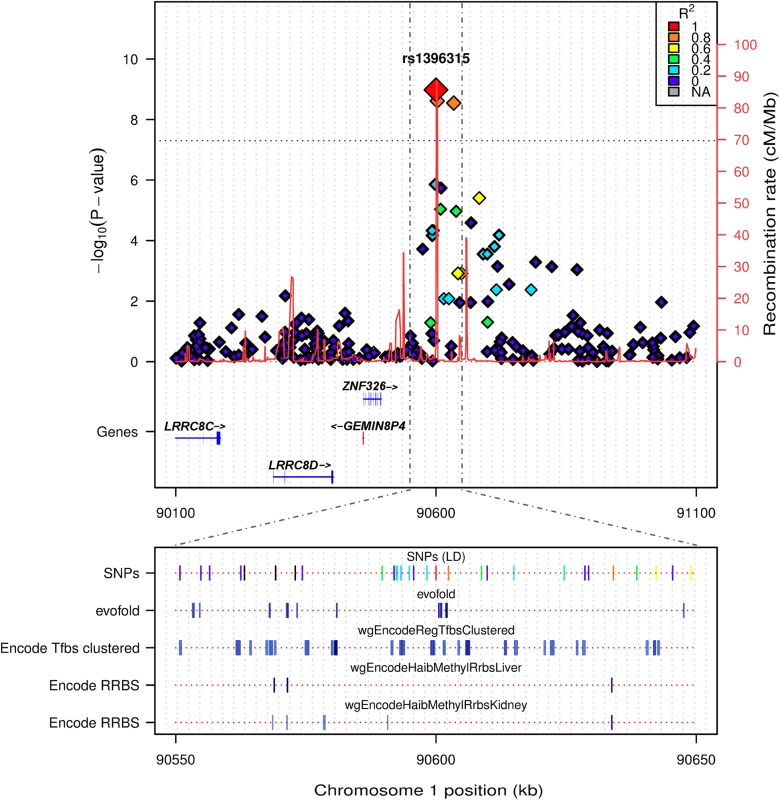
The significance level of association is given on the left y axis (-log10(p-value)), while genomic recombination is displayed on the right y axis. Pairwise LD between the tagging SNP and each other SNP is indicated by colour. In the bottom panel several relevant tracks of UCSC data are provided, including: evofold RNA loop prediction; ENCODE transcription factor binding sites from ChIP-SEQ, and tissue specific RRBS methylation information from liver and kidney (both relevant tissue types for the identified phenotype). The region directly surrounding the association peak on chr 1p22.2 did not reveal any known genes, with the closest, ZNF326, being ~325kb upstream (Fig 4). This led us to perform a detailed annotation survey of the region surrounding the peak on chr 1p22.2 using LocusTrack [20]. LocusTrack allows the incorporation of association statistics, SNP LD information, as well as several relevant UCSC annotation tracks, namely: evofold (bioinformatic prediction of RNA loop folding); ENCODE ChIP-SEQ transcription factor binding sites, and tissue specific methylation for liver and kidney (phenotypically relevant tissues) derived from ENCODE Reduced Representation Bisulfite Sequencing (RRBS). Fig 4 shows the chr 1p22.2 region of interest and all the relevant surrounding annotation information. It is interesting to note that while there may not be any genes directly annotated in the region, it does appear that there are distinct areas of potential control, i.e. transcription factor binding sites and tissue specific methylation, as indicated by the multiple UCSC tracks. Another annotated track of interest was that of the QTLs mapped from the rat genome database (RGB) to the human genome. There are 4 QTL specifically mapped to the area of interest at 1p22.2, all related to metabolic and renal dysfunction: Serum renin concentration QTL 2 (RGD:1300114); Non-insulin dependent diabetes mellitus QTL 15 and 20 (RGD:71115, RGD:70204), and Renal disease susceptibility QTL 4 (RGD:619619). Additional annotation is available for the chr 1p22.2 region in the form of a UCSC plot (S2 Fig), which contains the RGD QTL loci, and region specific information on DNase hypersensitivity clusters, as well as listing the individual transcription factor binding sites identified by ENCODE ChIP-SEQ. Finally, it is interesting to note that the association peak appears to lie on top of a region of increased genomic recombination (Fig 4).
Replication analysis
To investigate replication of the association signal seen for rs1396315 with Component 3 we utilised; a) an outgroup of the Norfolk Island cohort (n = 375) who are genetically unrelated (independent) to the core pedigree and for whom comparable genotype and phenotype data was available (S3 Table), and b) a completely independent cohort of 738 individuals from a general United States of America (USA) population (the CHDWB cohort) [21]. This cohort had measurements available for 6 out of the 7 traits loaded into Component 3 (uric acid was not available) as well as genotype data for rs1396315.
Due to the difficulties of finding an independent replication cohort with the exact same set of 37 traits with which to replicate the PCA and generate Component 3 scores we had to explore other options for replication. We decided to use a linear modelling approach to attempt to predict individual Component 3 scores using only the 7 most heavily loaded traits for that component from the discovery phase. The linear model as generated in the original Norfolk Island discovery cohort is (all equations are as implemented in base R):
The values from this model showed a very strong correlation with the original Component 3 scores and provided excellent predictive value (R2 0.92, p < 2.16x10-16). The model (fit1) was used to predict Component 3 scores in the n = 375 unrelated Norfolk Island outgroup using the 7 traits of interest. The regression equation required slight modification for the US replication cohort because uric acid measures were missing. To do this we tested the effects of dropping uric acid on the overall linear model and observed a minimal reduction in predictive value (R2 0.91, p < 2.16x10-16), suggesting that the updated model should still yield accurate Component 3 score prediction using only 6 trait measures. Predicted Component 3 scores were then incorporated into their respective replication data sets for association analyses to be performed.
Table 2 shows the results of the replication analysis. Association testing of the predicted Component 3 score showed a direction specific trend toward association between rs1396315 and Component 3 in the unrelated Norfolk Island outgroup (P = 0.048). Furthermore, the US replication cohort showed a significant association in the same direction as both the Norfolk Island discovery and outgroup cohorts (p = 1.13x10-5, Table 2). Given the tight familial clustering and known founder effects of the discovery cohort it is not surprising that the effect size for both replication cohorts drops substantially (>50%). Clearly, this effect size reduction has impacted on the statistical significance of the associations. Nevertheless, these replication findings provide compelling evidence for a robust association between rs1396315 and Component 3 and suggest a sample size of at least 400 unrelateds are required to detect the effect as statistically significant (P<0.05).
Tab. 2. Genotypic Association Analysis between rs1396315 and Component 3 in an unrelated Outgroup from Norfolk Island and an independent replication cohort. 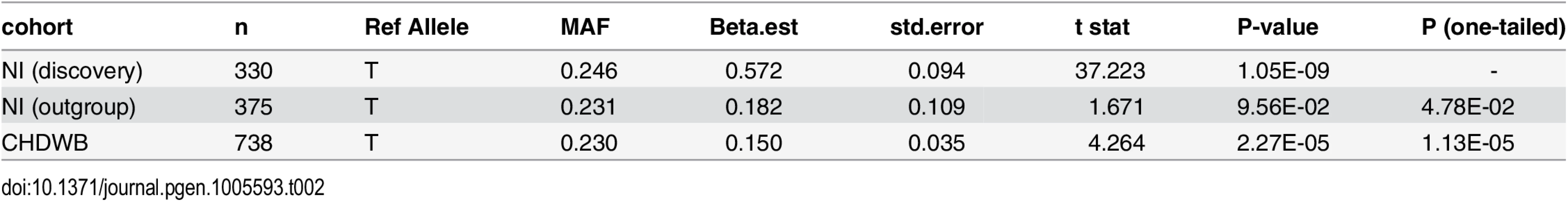
Gene expression and pathways analysis
Due to the lack of functional genes in close proximity to the identified region on chromosome 1p22.2 we explored the potential of trans (distal) associations with the rs1396315 and available gene expression data. We interrogated 1712 genetically heritable expression transcripts previously identified in the Norfolk Island population and measured in the same individuals [22]. Analysis revealed 55 significantly associated (P<0.05) transcripts, with the majority of transcripts showing positive association with this SNP (S3 Fig). When annotated, 35/55 transcripts were assigned to well documented genes of known function, with the remaining 20 residing in regions of the genome that are less well annotated. Gene set enrichment analysis of these 35 genes identified 5 functional pathways exhibiting significant enrichment (Fig 5). The most significantly enriched pathway was found to be that of purine metabolism (P = 0.0015), with 3 genes showing enrichment; PDE6D, GART, and NME2. This finding is interesting as previous work in Norfolk Island reported on a set of eQTL associated genes implicated with CVD-risk traits which also resided with the purine metabolism pathway [22].
Fig. 5. Significantly enriched pathways identified from KEGG enrichment analysis. 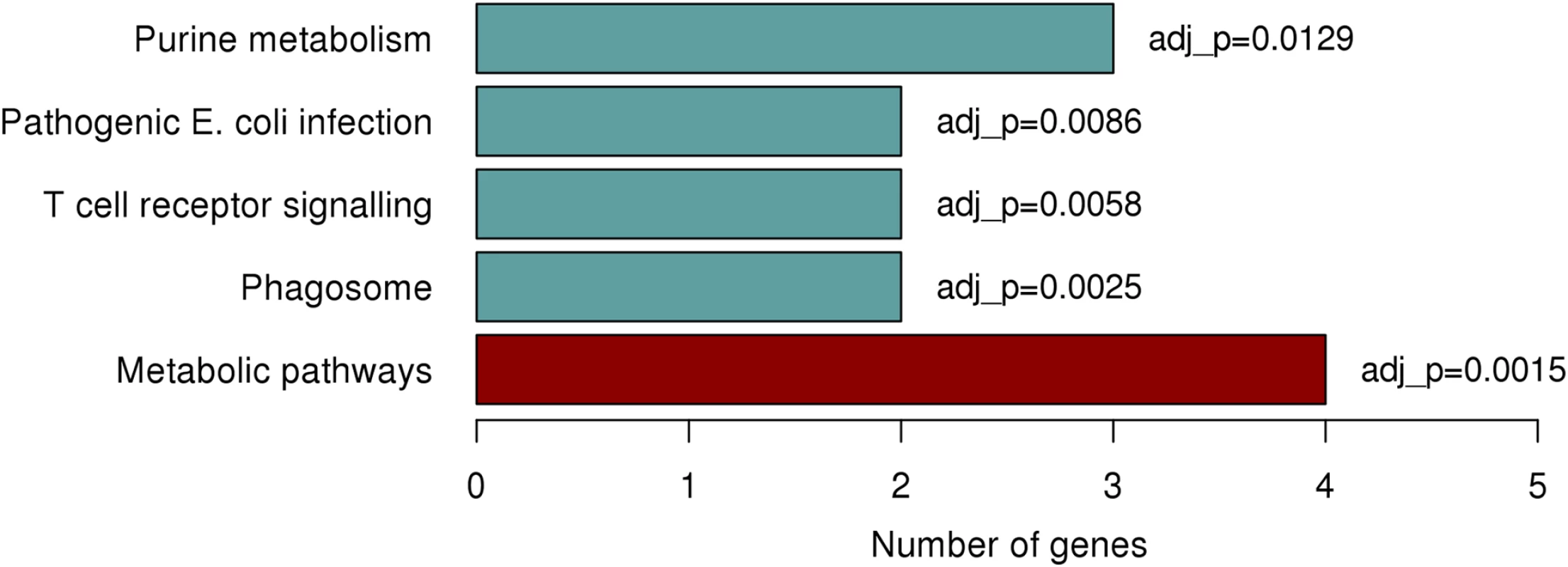
Discussion
Many heritable CVD risk traits tend to be correlated suggesting the presence of underlying genes acting pleiotropically to influence multiple traits. GWASs aimed at composite CVD traits may reveal such genes, which would remain undetected using conventional single phenotype analyses. Analysis of large pedigrees from isolated populations can offer advantages for mapping disease genes including the ability to estimate heritability for disease prioritisation and the possible presence of larger effect loci. Here we conducted a study to search for pleiotropic effect loci associated with CVD risk in the well characterized genetic isolate of Norfolk Island. We performed PCA and heritability testing and identified a heritable component trait strongly loaded with indices of metabolism and kidney function. We subsequently performed pedigree-based GWAS to identify a risk haplotype on chromosome 1p22.2 that explains 11% of the variance in this composite trait. This represents a very large effect exerted by a single locus and may reflect the fundamental importance of pleiotropic genes on this phenotype in this genetic isolate. Because the composite phenotype was not able to be standardised for comparison in external populations and given the unique nature of the Norfolk Island pedigree we could not perform conventional replication analysis, instead using a proxy prediction based approach. Replication analysis in an independent outgroup from Norfolk Island supported the association of rs1396315 and Component 3 (P = 0.048). We also investigated trait/rs1396315 association in a completely independent US cohort and observed significant association between the predicted Component 3 score and rs1396315 (P = 0.000011). Additionally, the clinical relevance of the Component 3 score was explored by comparison with the well-known Framingham CVD risk score [23] which showed a positive correlation indicating that the T allele of rs1396315 is associated with increased risk of CVD in the Norfolk population.
A number of GWAS studies have reported association of loci at chromosome 1p22.2 with CVD risk traits. Zabaneth et al., reported association of SNPs in this region to metabolic syndrome (including obesity) in 2554 Indian men [24]. Beck et al., reported association to liver enzyme indices in a population of 133,653 Europeans and Asians [25]. Hasstedt et al., reported association of chromosome 1p22.2 locus to coronary artery disease in 984 whites [26]. Additionally, several GWAS have identified SNPs within 1p22.2 associated with both obesity related traits [27] and liver enzyme levels (gamma-glutamyl transferase) [28]. Interestingly in another GWAS the same intergenic region of 1p22.2 was associated with atherosclerosis and coronary artery calcification [29].
Our examination of the genomic landscape around the chromosome 1p22.2 region showed this to be a relatively gene-poor region with no obvious candidate genes for CVD risk; the nearest genes are ZNF326 (zinc finger protein) and BARHL2 (BarH-like homeobox 2) (Fig 4). We are unaware of any literature indicating an association between CVD and related traits with these genes, however a recent review implicates the role of zinc finger proteins in adipogenesis [30], suggesting that this could be a potential target gene to follow up in further studies. Despite the lack of gene annotation it is quite plausible that the presence of non-protein coding elements, eg. epigenetic marks, trans-acting eQTLs, or long-non-coding RNAs within this region may be influencing genes and/or other regulatory factors at a distance.
Following this line of thought, we investigated available annotation, including that from several pertinent UCSC tracks: evofold RNA loop prediction; ENCODE transcription factor binding sites from ChIP-SEQ, and tissue specific RRBS methylation information from liver and kidney (both relevant tissue types for the identified phenotype). This feature based annotation revealed important structures and control regions within, or in close proximity to, the association peak. This includes tissue specific epigenetic markers from kidney and liver, as well as several transcription factor binding sites (S2 Fig). While none of the transcription factor binding sites related to the trans-eQTLs explored in our analyses, it is interesting to note that there are several, such as FOXA1, that are directly associated with metabolism in tissues such as the liver. FOXA1 is of particular interest as its family members have been shown to play an important role in the differentiation of the pancreas and liver and regulate metabolism in mice [31]. Additionally, several transcription factor sites annotated directly under the association peak are known pioneer factors, meaning they can bind condensed chromatin and influence transcription and potentially even methylation. We also noted that there were 4 annotated QTL loci that have been mapped from the rat genome to the human genome (S2 Fig). While these QTL are coarsely mapped to the human genome from rat data, it is of particular interest to note that they are all metabolically related, and 2 are directly related to the function of the kidney. All these observations provide additional support for the association of an important pleiotropic site, associated with metabolism and kidney function, at 1p22.2
To investigate potential trans-acting mechanisms we conducted an association analysis between the top ranking Component 3 SNPs and heritable gene expression transcripts measured in the same individuals [22]. Pathways analysis of genes encoding transcripts showing significant association with SNPs revealed that the purine metabolism pathway was the most significantly enriched. This is consistent with our previous study which identified enrichment of NME1 and PAPPS1 in the purine pathway in the Norfolk Island cohort [22]. In the current study transcripts for GART, NME2 and PDE6D were found to be associated with rs1396315. Whereas NME1 encodes for the A isoform, NME2 encodes for the B isoform of nucleoside diphosphate kinase (NDPK) which maintains the balance of nucleosides and high energy nucleotides (ATP/GTP) via the reversible transfer of phosphate. Interestingly, GART plays a central role in the de novo synthesis of nucleotides where it catalyses three of the ten steps involved in the conversion of phosphoribosyl pyrophosphate into inosine monophosphate [32]. Genetic variation in both GART and NME2 would have the potential to have fundamentally important effects on nucleotide metabolism and homeostasis.
Modifications in the purine metabolic pathway are linked with a number of diseases including kidney-related diseases such as hyperuricaemia and gout [33]. Combined with our previous data there is evidence of a shift in the regulation of purine synthesis and metabolism in the Norfolk Island cohort that may be linked with the development of renal dysfunction and associated CVDs. Variation in PDE6D was also identified in relation to the purine pathway. PDE6D encodes phospohdiesterase (PDE) 6δ subunit of the enzyme PDE6 that specifically hydrolyzes cyclic GMP [34]. PDE6 is localised in rod or cone membranes of the mammalian retina focussing research on its involvement in visual dysfunction [35]. These findings may provide insight into the high incidence of ocular diseases, including glaucoma, in the Norfolk Island population [36]. Interestingly there is also data indicating that GART also plays a role in ocular development [37].
Overall, our findings provide convincing evidence for a significant pleiotropic effect locus at chromosome 1p22.2 associated with a composite trait representing metabolic and/or renal dysfunction. This study illustrates the value of the phenomics approach in large pedigrees and genetic isolates for identifying underlying genetic regulators of inter-correlated disease traits that would go undetected using conventional single phenotype approaches. Further studies are now warranted to better understand the importance of the associations in the broader population and to more comprehensively interrogate the functional relevance of this locus in terms of renal pathology and CVD risk more generally.
Materials and Methods
Sample/cohort collection, pedigree information and ethics
The Norfolk Island Health Study (NIHS) is well established [9,12,14,22,38]. The Norfolk Island pedigree structure was previously outlined in Bellis et al., 2008 [39], and has recently been updated Macgregor et al., 2010 [12]. The updated pedigree structure includes ~5700 Norfolk Island individuals, spanning 11 generations and ~200 years. In this study we focused on a reduced ‘core’ pedigree, meaning that individuals; a) were genetically related to the original founders, and b) had phenotype (S1 Table) and genotype information available. The total number of core pedigree members examined was 330 (excluding individuals under the age of 18) and spans 4 generations at its greatest depth. The Norfolk Island 'outgroup' population used as a validation cohort consisted of 375 individuals. These individuals are all unrelated to the core-pedigree members as inferred by lack of presence in the pedigree, or a genetic kinship of 0. A summary of the available phenotype data for these 375 individuals is available (S3 Table). All individuals gave written informed consent. Ethical approval was granted prior to the commencement of the study by the Griffith University Human Research Ethics Committee (ethical approval no: 1300000485). Ethics approval and management of the NIHS has since been transferred to Queensland University of Technology.
The US CHDWB replication cohort
The replication cohort in Atlanta is the Center for Health Discovery and Well Being (CHDWB) cohort of the Emory-Georgia Tech Predictive Health Institute [21]. Whole genome genotypes were imputed from either Illumina OmniQuad or Illumina Core+Exome genotyping arrays using Impute2 against the 1000 Genomes. Over 200 trait measures are available for the cohort, at baseline and between 3 and 5 subsequent visits over a 4 year period (n = 738). Participants are all employees of Emory University between the ages of 25 and 75 contacted at random and representing a broad cross-section of the geography of the city of Atlanta and socioeconomic diversity. Summarised clinical data for the seven major traits is available (S4 Table). Written informed consent for participation in genetic research was provided under the auspices of the Institutional Review Boards of Georgia Tech and Emory University.
Multiple phenotype analysis
Baseline statistics for the core-pedigree (discovery cohort) were calculated for 37 CVD-related measures including all the biochemical measures as well as body size and composition traits (S1 Table). Phenotypic baseline statistics were calculated in SPSS 18.0. All 37 traits were then subjected to Principle Component Analysis (PCA). Briefly, the PCA method is a multivariate reduction method that is commonly used to identify latent components in highly dimensional datasets. When applied to a set of observed variables, PCA will extract a smaller number of artificial variables (principle components) which should account for the majority of the total variance originally observed. Thus one of the major benefits of PCA is that once an underlying set of components has been identified the data can be compressed with very little loss of information. These component scores can then be retained and utilised in further analyses. The experimental method utilised in this study is termed unsupervised in the sense that the PCA was run across all the available CVD trait data, as opposed to selecting a subset of core CVD related traits (such as BMI, weight, lipids, blood pressure, and insulin). All previous published studies have utilised a more ‘supervised’ (selective) approach in this sense, and we hypothesise that this may impact on the outcome of underlying CVD component structures. PCA was conducted on 37 biochemical and anthropometric measures related to CVD. Components with eigenvalues greater than 1 were retained; the number of significant variables loading on a component was reduced through the implementation of orthogonal rotation (varimax). All PCA trait loadings for each component are available (S2 Table).
Heritability analysis
The genetic analysis program SOLAR [40] was utilised to calculate heritability estimates for all 37 traits as well as the 13 defined principle components. The extended Norfolk pedigree was used by SOLAR to estimate h2 (heritability). Both the individual and component traits were screened for covariant effects of age|sex interactions. All traits which SOLAR calculated as having a high kurtosis were log transformed before calculating heritability estimates.
Genome-wide genotyping
EDTA anticoagulated venous blood samples were collected from all participants. Genomic DNA was extracted from blood buffy coats using standard phenol-chloroform procedures (Qiagen). Genome-wide genotyping was carried out using the Illumina Human610-Quad v1.0 beadchip. Raw data from Illumina idat files was SNP genotyped in R using the CRLMM package [41]. Genotype data then underwent QC routines using PLINK [42]. Briefly, SNP analysis was restricted to autosomal SNPs with minor allele frequency >0.01, call rate >0.95 and Hardy-Weinberg equilibrium testing p-value >10−5. After quality control, 590,603 SNPs were used for association analyses. Genotype data was then exported from PLINK and imported into the CRAN package GenABEL [43] for analysis.
Genome-wide association analysis, LD testing and genotype association
A pedigree based GWAS analysis of the 9 heritable components was batched using custom R scripts and the package GenABEL, using the polygenic model with age and sex interactions, as well as genetic structure (the top 2 principal components of the complete SNP set as calculated by KING [44]). Specifically, the polygenic model function implemented in GenABEL is capable of estimating the narrow sense heritability of a trait (h2). The polygenic model takes into account the fact that potentially thousands of genetic variants contribute to a trait phenotype. The function implemented in GenABEL maximises the likelihood of the data under the polygenic model with covariates and reports twice negative maximum likelihood estimates and the inverse of the variance-covariance matrix at the point of maximum likelihood. GenABEL also estimates residuals of the trait and the inverse of the variance-covariance matrix for further use in association analysis with the mmscore function. The polygenic model within GenABEL implements variance components defined to account for linked major gene effects, background polygenic effects, and environmental effects. Age and sex were included as covariates, as well as genetic structure which was assessed by principal components analysis using the KING [44] program. The top two components were chosen as covariates because we found that these explained the majority of the variance in the outcomes being tested and inclusion of additional, less informative, components only served to reduce the parsimony of the models. All heritable transcripts were then treated as phenotypes and batched GWAS were run. The mmscore function as implemented in GenABEL was used. This function represents a mixed model approximation analysis for association between a trait and genetic polymorphism, and is specifically designed for association testing in samples of related individuals. This allows for per SNP association testing using a polygenic (mixed) model approach [43]. The study-wide significance was set based on Meff adjustment (P = 1.84x10−7). Haplotype/LD testing was conducted in Haploview 4.2 [45]. Exploration of the region surrounding the peak on chr 1p22.2 and integration of LD information and UCSC track data was performed using LocusTrack [20] within R.
Replication analysis
Ideally replication would be conducted on the exact same set of 37 clinical traits, and involve generating component scores using the original coefficients. However, it is extremely difficult to obtain a separate population with the exact same phenotype information, making this option not possible. We ultimately have performed a linear model approach, using the 7 most highly loaded Component 3 traits to predict the score in our replication cohorts, the unrelated Norfolk Island outgroup (n = 375), and the US CHDWB replication cohort (n = 738). Details of the modelling are given in results, but overall the model was constructed from the discovery cohort (NI core-pedigree, n = 330) using the lm (linear models) base R function:
Predictions were then made using the predict function in base R as follows:
Association testing of Component 3 (predicted) and rs1396315 in the replication cohorts was performed using linear modelling, with the predicted component score as the outcome and the genotype as the independent variable. The genotype was coded in an additive fashion: 0 (CC); 1 (CT), 2 (TT).
eQTL association analysis
The generation and list of eQTLs in the Norfolk Island cohort has previously described in detail [22]. Blood from the same n = 330 Norfolk Island core-pedigree samples was collected and stored at -20°C in PAXgene tubes (Qiagen, Valencia, CA). PAXgene Blood miRNA Kits kits (Qiagen) were used to extract total RNA according to the manufacturers’ instructions and RNA was assessed for quality using the Bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA). A total of 250ng RNA was amplified and labelled using the Illumina TotalPrep-96 RNA Amplification Kit (Life Technologies, Grand Island, NY), according to the manufacturers’ instructions. Expression profiling was performed using the Illumina HumanHT-12 v4.0 Expression BeadChip Kit (Illumina, San Diego, CA) using 750ng of amplified RNA and following the Whole-Genome Gene Expression Direct Hybridization Assay Guide. Array images were scanned on the Illumina iScan and analyzed initially using the Gene Expression Module of GenomeStudio (V2011.1). Background subtraction was applied and missing bead types were imputed using GenomeStudio. Based on the number of expressed probes (at “detection p-values” < = 0.05), mean raw expression values across probes, and correlations (across probes) between samples, all samples provided high quality data, except for one sample that was of questionable quality and that was removed. Significantly expressed probes were then determined at a false discovery rate (FDR) of 5%, based on p-values generated in a binomial test on the counts of samples in which a probe generated “detection p-values” < = 0.05 (success) and >0.05 (failure). Subsequently, the raw expression levels of probes detecting significant expression were shifted by a constant amount so that the minimum observed value of any probe in any sample was 1.0, followed by log2 transformation and quantile normalization. A series of custom filters were designed to identify cis/trans eQTL’s; presence of multiple adjacent SNPs in a peak (within +/-20Kb), a quick parse SNP/CHR location filter, a chromosome quadrants filter, and a graphical filter (modified Manhattan Plots with kern smoothing to facilitate peak identification) [22]. All eQTLs were defined by the 'tagging' SNP, the SNP with showed the most significant association with the given transcript. A set of n = 2200 SNPs representing all those within eQTL peaks which surpassed the Meff adjusted threshold (1.84x10-7) were extracted and used as a basis for an eQTL-centric association analysis. This analysis involved Component 3 phenotype being run in a GWAS-based approach in GenABEL [43] using the extracted SNP set. A short list of traits was obtained by using a relaxed significance threshold 1.0x10-2.
Supporting Information
Zdroje
1. Laslett LJ, Alagona P, Clark BA, Drozda JP, Saldivar F, Wilson SR, et al. The worldwide environment of cardiovascular disease: prevalence, diagnosis, therapy, and policy issues: a report from the American College of Cardiology. J Am Coll Cardiol. 2012;60: S1–49. doi: 10.1016/j.jacc.2012.11.002 23257320
2. Avery CL, He Q, North KE, Ambite JL, Boerwinkle E, Fornage M, et al. A Phenomics-Based Strategy Identifies Loci on APOC1, BRAP, and PLCG1 Associated with Metabolic Syndrome Phenotype Domains. PLoS Genet. Public Library of Science; 2011;7: e1002322.
3. Kraja AT, Chasman DI, North KE, Reiner AP, Yanek LR, Kilpeläinen TO, et al. Pleiotropic genes for metabolic syndrome and inflammation. Mol Genet Metab. 2014;112 : 317–338. doi: 10.1016/j.ymgme.2014.04.007 24981077
4. Lowe JK, Maller JB, Pe’er I, Neale BM, Salit J, Kenny EE, et al. Genome-Wide Association Studies in an Isolated Founder Population from the Pacific Island of Kosrae. PLoS Genet. Public Library of Science; 2009;5: e1000365.
5. Sanna S, Li B, Mulas A, Sidore C, Kang HM, Jackson AU, et al. Fine mapping of five loci associated with low-density lipoprotein cholesterol detects variants that double the explained heritability. Gibson G, editor. PLoS Genet. Public Library of Science; 2011;7: e1002198.
6. Traglia M, Sala C, Masciullo C, Cverhova V, Lori F, Pistis G, et al. Heritability and demographic analyses in the large isolated population of Val Borbera suggest advantages in mapping complex traits genes. PLoS One. 2009;4: e7554. doi: 10.1371/journal.pone.0007554 19847309
7. Shifman S, Darvasi A. The value of isolated populations. Nat Genet. 2001;28 : 309–310. <Go to ISI>://000170174800006 11479587
8. Heutink P, Oostra BA. Gene finding in genetically isolated populations. Hum Mol Genet. 2002;11 : 2507–2515. Available: <Go to ISI>://000178515900019 12351587
9. Bellis C, Hughes RM, Begley KN, Quinlan S, Lea RA, Heath SC, et al. Phenotypical characterisation of the isolated Norfolk Island population focusing on epidemiological indicators of cardiovascular disease. Hum Hered. 2006;60 : 211–219.
10. Hoare M. Norfolk Island: A Revised and Enlarged History 1774–1998. Rockhampton: Central Queensland University Press; 1999.
11. Edgecombe J. Norfolk Island—South Pacific: Island of History and Many Delights. Thornleigh: Australia: J.M. Edgecombe; 1999.
12. Macgregor S, Bellis C, Lea RA, Cox H, Dyer T, Blangero J, et al. Legacy of mutiny on the Bounty: founder effect and admixture on Norfolk Island. Eur J Hum Genet. 2010;18 : 67–72. doi: 10.1038/ejhg.2009.111 19584896
13. McEvoy BP, Zhao ZZ, Macgregor S, Bellis C, Lea RA, Cox H, et al. European and Polynesian admixture in the Norfolk Island population. Heredity (Edinb). 2010; 229–234.
14. Cox HC, Bellis C, Lea RA, Quinlan S, Hughes R, Dyer T, et al. Principal Component and Linkage Analysis of Cardiovascular Risk Traits in the Norfolk Isolate. Hum Hered. 2009;68 : 55–64. doi: 10.1159/000210449 19339786
15. Lotufo PA. Framingham score for cardiovascular diseases. Medicina (B Aires). 2008;87 : 232–237.
16. Lin J-P, Cupples LA, Wilson PWF, Heard-Costa N, O’Donnell CJ. Evidence for a gene influencing serum bilirubin on chromosome 2q telomere: a genomewide scan in the Framingham study. Am J Hum Genet. 2003;72 : 1029–34. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=1180333&tool=pmcentrez&rendertype=abstract 12618960
17. Cox AJ, Ng MC-Y, Xu J, Langefeld CD, Koch KL, Dawson PA, et al. Association of SNPs in the UGT1A gene cluster with total bilirubin and mortality in the Diabetes Heart Study. Atherosclerosis. 2013;229 : 155–60. doi: 10.1016/j.atherosclerosis.2013.04.008 23642732
18. Johnson AD, Kavousi M, Smith A V, Chen M-H, Dehghan A, Aspelund T, et al. Genome-wide association meta-analysis for total serum bilirubin levels. Hum Mol Genet. 2009;18 : 2700–10. doi: 10.1093/hmg/ddp202 19414484
19. Frazer KA, Ballinger DG, Cox DR, Hinds DA, Stuve LL, Gibbs RA, et al. A second generation human haplotype map of over 3.1 million SNPs. Nature. Nature Publishing Group; 2007;449 : 851–61.
20. Cuellar-Partida G, Renteria ME, MacGregor S. LocusTrack: Integrated visualization of GWAS results and genomic annotation. Source Code Biol Med. 2015;10 : 1. doi: 10.1186/s13029-015-0032-8 25750659
21. Patel CJ, Sivadas A, Tabassum R, Preeprem T, Zhao J, Arafat D, et al. Whole genome sequencing in support of wellness and health maintenance. Genome Med. 2013;5 : 58. 23806097
22. Benton MC, Lea RA, Macartney-Coxson D, Carless MA, Göring HH, Bellis C, et al. Mapping eQTLs in the Norfolk Island Genetic Isolate Identifies Candidate Genes for CVD Risk Traits. Am J Hum Genet. Elsevier; 2013;93 : 1087–99.
23. D’Agostino RB, Vasan RS, Pencina MJ, Wolf PA, Cobain M, Massaro JM, et al. General cardiovascular risk profile for use in primary care: the Framingham Heart Study. Circulation. 2008;117 : 743–53. doi: 10.1161/CIRCULATIONAHA.107.699579 18212285
24. Zabaneh D, Balding DJ. A Genome-Wide Association Study of the Metabolic Syndrome in Indian Asian Men. PLoS One. Public Library of Science; 2010;5: e11961.
25. Beck SR, Brown WM, Williams AH, Pierce J, Rich SS, Langefeld CD. Age-stratified QTL genome scan analyses for anthropometric measures. BMC Genet. 2003;4 Suppl 1: S31. 14975099
26. Hasstedt SJ, Hanis CL, Elbein SC. Univariate and bivariate linkage analysis identifies pleiotropic loci underlying lipid levels and type 2 diabetes risk. Ann Hum Genet. 2010;74 : 308–15. doi: 10.1111/j.1469-1809.2010.00589.x 20597901
27. Comuzzie AG, Cole SA, Laston SL, Voruganti VS, Haack K, Gibbs RA, et al. Novel genetic loci identified for the pathophysiology of childhood obesity in the Hispanic population. PLoS One. 2012;7: e51954. doi: 10.1371/journal.pone.0051954 23251661
28. Chambers JC, Zhang W, Sehmi J, Li X, Wass MN, Van der Harst P, et al. Genome-wide association study identifies loci influencing concentrations of liver enzymes in plasma. Nat Genet. 2011;43 : 1131–8. doi: 10.1038/ng.970 22001757
29. O’Donnell CJ, Cupples LA, D’Agostino RB, Fox CS, Hoffmann U, Hwang S-J, et al. Genome-wide association study for subclinical atherosclerosis in major arterial territories in the NHLBI’s Framingham Heart Study. BMC Med Genet. 2007;8 Suppl 1: S4. 17903303
30. Wei S, Zhang L, Zhou X, Du M, Jiang Z, Hausman GJ, et al. Emerging roles of zinc finger proteins in regulating adipogenesis. Cell Mol Life Sci. 2013;
31. Kaestner KH. The hepatocyte nuclear factor 3 (HNF3 or FOXA) family in metabolism. Trends Endocrinol Metab. 2000;11 : 281–5. http://www.ncbi.nlm.nih.gov/pubmed/10920385 10920385
32. Welin M, Grossmann JG, Flodin S, Nyman T, Stenmark P, Trésaugues L, et al. Structural studies of tri-functional human GART. Nucleic Acids Res. 2010;38 : 7308–19. doi: 10.1093/nar/gkq595 20631005
33. Nyhan WL. Disorders of purine and pyrimidine metabolism. Mol Genet Metab. 2005;86 : 25–33. 16176880
34. Li N, Florio SK, Pettenati MJ, Rao PN, Beavo JA, Baehr W. Characterization of human and mouse rod cGMP phosphodiesterase delta subunit (PDE6D) and chromosomal localization of the human gene. Genomics. 1998;49 : 76–82. 9570951
35. Tsang SH, Tsui I, Chou CL, Zernant J, Haamer E, Iranmanesh R, et al. A novel mutation and phenotypes in phosphodiesterase 6 deficiency. Am J Ophthalmol. 2008;146 : 780–8. doi: 10.1016/j.ajo.2008.06.017 18723146
36. Sherwin JC, Kearns LS, Hewitt AW, Ma Y, Kelly J, Griffiths LR, et al. Prevalence of Chronic Ocular Diseases in a Genetic Isolate: The Norfolk Island Eye Study (NIES). Ophthalmic Epidemiol. 2011;18 : 61–71. 21401413
37. Ng A, Uribe RA, Yieh L, Nuckels R, Gross JM. Zebrafish mutations in gart and paics identify crucial roles for de novo purine synthesis in vertebrate pigmentation and ocular development. Development. 2009;136 : 2601–11. doi: 10.1242/dev.038315 19570845
38. Bellis C, Cox HC, Dyer TD, Charlesworth JC, Begley KN, Quinlan S, et al. Linkage mapping of CVD risk traits in the isolated Norfolk Island population. Hum Genet. 2008;124 : 543–552. doi: 10.1007/s00439-008-0580-y 18975005
39. Bellis C, Cox HC, Ovcaric M, Begley KN, Lea RA, Quinlan S, et al. Linkage disequilibrium analysis in the genetically isolated Norfolk Island population. Heredity (Edinb). 2008;100 : 366–373.
40. Almasy L, Blangero J. Multipoint quantitative-trait linkage analysis in general pedigrees. Am J Hum Genet. 1998;62 : 1198–1211. <Go to ISI>://000073487000025 9545414
41. Scharpf RB, Irizarry RA, Ritchie ME, Carvalho B, Ruczinski I. Using the R Package crlmm for Genotyping and Copy Number Estimation. J Stat Softw. 2011;40 : 1–32. <Go to ISI>://WOS:000290086900001 22523482
42. Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, et al. PLINK: A Tool Set for Whole-Genome Association and Population-Based Linkage Analyses. Am J Hum Genet. University of Chicago Press; 2007;81 : 559–575. http://linkinghub.elsevier.com/retrieve/pii/S0002929707613524
43. Aulchenko YS, Ripke S, Isaacs A, Van Duijn CM. GenABEL: an R library for genorne-wide association analysis. Bioinformatics. 2007;23 : 1294–1296. 17384015
44. Manichaikul A, Mychaleckyj JC, Rich SS, Daly K, Sale M, Chen WM. Robust relationship inference in genome-wide association studies. Bioinformatics. 2010;26 : 2867–2873. doi: 10.1093/bioinformatics/btq559 20926424
45. Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21 : 263–265. 15297300
Štítky
Genetika Reprodukční medicína
Článek Evidence of Selection against Complex Mitotic-Origin Aneuploidy during Preimplantation DevelopmentČlánek A Novel Route Controlling Begomovirus Resistance by the Messenger RNA Surveillance Factor PelotaČlánek A Follicle Rupture Assay Reveals an Essential Role for Follicular Adrenergic Signaling in OvulationČlánek Canonical Poly(A) Polymerase Activity Promotes the Decay of a Wide Variety of Mammalian Nuclear RNAsČlánek FANCI Regulates Recruitment of the FA Core Complex at Sites of DNA Damage Independently of FANCD2Článek Hsp90-Associated Immunophilin Homolog Cpr7 Is Required for the Mitotic Stability of [URE3] Prion inČlánek The Dedicated Chaperone Acl4 Escorts Ribosomal Protein Rpl4 to Its Nuclear Pre-60S Assembly SiteČlánek Chromatin-Remodelling Complex NURF Is Essential for Differentiation of Adult Melanocyte Stem CellsČlánek A Systems Approach Identifies Essential FOXO3 Functions at Key Steps of Terminal ErythropoiesisČlánek Integration of Posttranscriptional Gene Networks into Metabolic Adaptation and Biofilm Maturation inČlánek Lateral and End-On Kinetochore Attachments Are Coordinated to Achieve Bi-orientation in OocytesČlánek MET18 Connects the Cytosolic Iron-Sulfur Cluster Assembly Pathway to Active DNA Demethylation in
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2015 Číslo 10
-
Všechny články tohoto čísla
- Gene-Regulatory Logic to Induce and Maintain a Developmental Compartment
- A Decad(e) of Reasons to Contribute to a PLOS Community-Run Journal
- DNA Methylation Landscapes of Human Fetal Development
- Single Strand Annealing Plays a Major Role in RecA-Independent Recombination between Repeated Sequences in the Radioresistant Bacterium
- Evidence of Selection against Complex Mitotic-Origin Aneuploidy during Preimplantation Development
- Transcriptional Derepression Uncovers Cryptic Higher-Order Genetic Interactions
- Silencing of X-Linked MicroRNAs by Meiotic Sex Chromosome Inactivation
- Virus Satellites Drive Viral Evolution and Ecology
- A Novel Route Controlling Begomovirus Resistance by the Messenger RNA Surveillance Factor Pelota
- Sequence to Medical Phenotypes: A Framework for Interpretation of Human Whole Genome DNA Sequence Data
- Your Data to Explore: An Interview with Anne Wojcicki
- Modulation of Ambient Temperature-Dependent Flowering in by Natural Variation of
- The Ciliopathy Protein CC2D2A Associates with NINL and Functions in RAB8-MICAL3-Regulated Vesicle Trafficking
- PPP2R5C Couples Hepatic Glucose and Lipid Homeostasis
- DCA1 Acts as a Transcriptional Co-activator of DST and Contributes to Drought and Salt Tolerance in Rice
- Intermediate Levels of CodY Activity Are Required for Derepression of the Branched-Chain Amino Acid Permease, BraB
- "Missing" G x E Variation Controls Flowering Time in
- The Rise and Fall of an Evolutionary Innovation: Contrasting Strategies of Venom Evolution in Ancient and Young Animals
- Type IV Collagen Controls the Axogenesis of Cerebellar Granule Cells by Regulating Basement Membrane Integrity in Zebrafish
- Loss of a Conserved tRNA Anticodon Modification Perturbs Plant Immunity
- Genome-Wide Association Analysis of Adaptation Using Environmentally Predicted Traits
- Oriented Cell Division in the . Embryo Is Coordinated by G-Protein Signaling Dependent on the Adhesion GPCR LAT-1
- Disproportionate Contributions of Select Genomic Compartments and Cell Types to Genetic Risk for Coronary Artery Disease
- A Follicle Rupture Assay Reveals an Essential Role for Follicular Adrenergic Signaling in Ovulation
- The RNAPII-CTD Maintains Genome Integrity through Inhibition of Retrotransposon Gene Expression and Transposition
- Canonical Poly(A) Polymerase Activity Promotes the Decay of a Wide Variety of Mammalian Nuclear RNAs
- Allelic Variation of Cytochrome P450s Drives Resistance to Bednet Insecticides in a Major Malaria Vector
- SCARN a Novel Class of SCAR Protein That Is Required for Root-Hair Infection during Legume Nodulation
- IBR5 Modulates Temperature-Dependent, R Protein CHS3-Mediated Defense Responses in
- NINL and DZANK1 Co-function in Vesicle Transport and Are Essential for Photoreceptor Development in Zebrafish
- Decay-Initiating Endoribonucleolytic Cleavage by RNase Y Is Kept under Tight Control via Sequence Preference and Sub-cellular Localisation
- Large-Scale Analysis of Kinase Signaling in Yeast Pseudohyphal Development Identifies Regulation of Ribonucleoprotein Granules
- FANCI Regulates Recruitment of the FA Core Complex at Sites of DNA Damage Independently of FANCD2
- LINE-1 Mediated Insertion into (Protein of Centriole 1 A) Causes Growth Insufficiency and Male Infertility in Mice
- Hsp90-Associated Immunophilin Homolog Cpr7 Is Required for the Mitotic Stability of [URE3] Prion in
- Genome-Scale Mapping of σ Reveals Widespread, Conserved Intragenic Binding
- Uncovering Hidden Layers of Cell Cycle Regulation through Integrative Multi-omic Analysis
- Functional Diversification of Motor Neuron-specific Enhancers during Evolution
- The GTP- and Phospholipid-Binding Protein TTD14 Regulates Trafficking of the TRPL Ion Channel in Photoreceptor Cells
- The Gyc76C Receptor Guanylyl Cyclase and the Foraging cGMP-Dependent Kinase Regulate Extracellular Matrix Organization and BMP Signaling in the Developing Wing of
- The Ty1 Retrotransposon Restriction Factor p22 Targets Gag
- Functional Impact and Evolution of a Novel Human Polymorphic Inversion That Disrupts a Gene and Creates a Fusion Transcript
- The Dedicated Chaperone Acl4 Escorts Ribosomal Protein Rpl4 to Its Nuclear Pre-60S Assembly Site
- The Influence of Age and Sex on Genetic Associations with Adult Body Size and Shape: A Large-Scale Genome-Wide Interaction Study
- Parent-of-Origin Effects of the Gene on Adiposity in Young Adults
- Chromatin-Remodelling Complex NURF Is Essential for Differentiation of Adult Melanocyte Stem Cells
- Retinoic Acid Receptors Control Spermatogonia Cell-Fate and Induce Expression of the SALL4A Transcription Factor
- A Systems Approach Identifies Essential FOXO3 Functions at Key Steps of Terminal Erythropoiesis
- Protein O-Glucosyltransferase 1 (POGLUT1) Promotes Mouse Gastrulation through Modification of the Apical Polarity Protein CRUMBS2
- KIF7 Controls the Proliferation of Cells of the Respiratory Airway through Distinct Microtubule Dependent Mechanisms
- Integration of Posttranscriptional Gene Networks into Metabolic Adaptation and Biofilm Maturation in
- Lateral and End-On Kinetochore Attachments Are Coordinated to Achieve Bi-orientation in Oocytes
- Protein Homeostasis Imposes a Barrier on Functional Integration of Horizontally Transferred Genes in Bacteria
- A New Method for Detecting Associations with Rare Copy-Number Variants
- Histone H2AFX Links Meiotic Chromosome Asynapsis to Prophase I Oocyte Loss in Mammals
- The Genomic Aftermath of Hybridization in the Opportunistic Pathogen
- A Role for the Chaperone Complex BAG3-HSPB8 in Actin Dynamics, Spindle Orientation and Proper Chromosome Segregation during Mitosis
- Establishment of a Developmental Compartment Requires Interactions between Three Synergistic -regulatory Modules
- Regulation of Spore Formation by the SpoIIQ and SpoIIIA Proteins
- Association of the Long Non-coding RNA Steroid Receptor RNA Activator (SRA) with TrxG and PRC2 Complexes
- Alkaline Ceramidase 3 Deficiency Results in Purkinje Cell Degeneration and Cerebellar Ataxia Due to Dyshomeostasis of Sphingolipids in the Brain
- ACLY and ACC1 Regulate Hypoxia-Induced Apoptosis by Modulating ETV4 via α-ketoglutarate
- Quantitative Differences in Nuclear β-catenin and TCF Pattern Embryonic Cells in .
- HENMT1 and piRNA Stability Are Required for Adult Male Germ Cell Transposon Repression and to Define the Spermatogenic Program in the Mouse
- Axon Regeneration Is Regulated by Ets–C/EBP Transcription Complexes Generated by Activation of the cAMP/Ca Signaling Pathways
- A Phenomic Scan of the Norfolk Island Genetic Isolate Identifies a Major Pleiotropic Effect Locus Associated with Metabolic and Renal Disorder Markers
- The Roles of CDF2 in Transcriptional and Posttranscriptional Regulation of Primary MicroRNAs
- A Genetic Cascade of Modulates Nucleolar Size and rRNA Pool in
- Inter-population Differences in Retrogene Loss and Expression in Humans
- Cationic Peptides Facilitate Iron-induced Mutagenesis in Bacteria
- EP4 Receptor–Associated Protein in Macrophages Ameliorates Colitis and Colitis-Associated Tumorigenesis
- Fungal Infection Induces Sex-Specific Transcriptional Changes and Alters Sexual Dimorphism in the Dioecious Plant
- FLCN and AMPK Confer Resistance to Hyperosmotic Stress via Remodeling of Glycogen Stores
- MET18 Connects the Cytosolic Iron-Sulfur Cluster Assembly Pathway to Active DNA Demethylation in
- Sex Bias and Maternal Contribution to Gene Expression Divergence in Blastoderm Embryos
- Transcriptional and Linkage Analyses Identify Loci that Mediate the Differential Macrophage Response to Inflammatory Stimuli and Infection
- Mre11 and Blm-Dependent Formation of ALT-Like Telomeres in Ku-Deficient
- Genome Wide Identification of SARS-CoV Susceptibility Loci Using the Collaborative Cross
- Identification of a Single Strand Origin of Replication in the Integrative and Conjugative Element ICE of
- The Type VI Secretion TssEFGK-VgrG Phage-Like Baseplate Is Recruited to the TssJLM Membrane Complex via Multiple Contacts and Serves As Assembly Platform for Tail Tube/Sheath Polymerization
- The Dynamic Genome and Transcriptome of the Human Fungal Pathogen and Close Relative
- Secondary Structure across the Bacterial Transcriptome Reveals Versatile Roles in mRNA Regulation and Function
- ROS-Induced JNK and p38 Signaling Is Required for Unpaired Cytokine Activation during Regeneration
- Pelle Modulates dFoxO-Mediated Cell Death in
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Single Strand Annealing Plays a Major Role in RecA-Independent Recombination between Repeated Sequences in the Radioresistant Bacterium
- The Rise and Fall of an Evolutionary Innovation: Contrasting Strategies of Venom Evolution in Ancient and Young Animals
- Genome Wide Identification of SARS-CoV Susceptibility Loci Using the Collaborative Cross
- DCA1 Acts as a Transcriptional Co-activator of DST and Contributes to Drought and Salt Tolerance in Rice
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání






