-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaThe Polyproline Site in Hinge 2 Influences the Functional Capacity of Truncated Dystrophins
Mutations in dystrophin can lead to Duchenne muscular dystrophy or the more mild form of the disease, Becker muscular dystrophy. The hinge 3 region in the rod domain of dystrophin is particularly prone to deletion mutations. In-frame deletions of hinge 3 are predicted to lead to BMD, however the severity of disease can vary considerably. Here we performed extensive structure-function analyses of truncated dystrophins with modified hinges and spectrin-like repeats in mdx mice. We found that the polyproline site in hinge 2 profoundly influences the functional capacity of a microdystrophinΔR4-R23/ΔCT with a large deletion in the hinge 3 region. Inclusion of polyproline in microdystrophinΔR4-R23/ΔCT led to small myofibers (12% smaller than wild-type), Achilles myotendinous disruption, ringed fibers, and aberrant neuromuscular junctions in the mdx gastrocnemius muscles. Replacing hinge 2 of microdystrophinΔR4-R23/ΔCT with hinge 3 significantly improved the functional capacity to prevent muscle degeneration, increase muscle fiber area, and maintain the junctions. We conclude that the rigid α-helical structure of the polyproline site significantly impairs the functional capacity of truncated dystrophins to maintain appropriate connections between the cytoskeleton and extracellular matrix.
Published in the journal: . PLoS Genet 6(5): e32767. doi:10.1371/journal.pgen.1000958
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1000958Summary
Mutations in dystrophin can lead to Duchenne muscular dystrophy or the more mild form of the disease, Becker muscular dystrophy. The hinge 3 region in the rod domain of dystrophin is particularly prone to deletion mutations. In-frame deletions of hinge 3 are predicted to lead to BMD, however the severity of disease can vary considerably. Here we performed extensive structure-function analyses of truncated dystrophins with modified hinges and spectrin-like repeats in mdx mice. We found that the polyproline site in hinge 2 profoundly influences the functional capacity of a microdystrophinΔR4-R23/ΔCT with a large deletion in the hinge 3 region. Inclusion of polyproline in microdystrophinΔR4-R23/ΔCT led to small myofibers (12% smaller than wild-type), Achilles myotendinous disruption, ringed fibers, and aberrant neuromuscular junctions in the mdx gastrocnemius muscles. Replacing hinge 2 of microdystrophinΔR4-R23/ΔCT with hinge 3 significantly improved the functional capacity to prevent muscle degeneration, increase muscle fiber area, and maintain the junctions. We conclude that the rigid α-helical structure of the polyproline site significantly impairs the functional capacity of truncated dystrophins to maintain appropriate connections between the cytoskeleton and extracellular matrix.
Introduction
Duchenne muscular dystrophy (DMD) is a lethal X-linked recessive disease caused by mutations in the 2.2 MB dystrophin gene [1]–[3]. In skeletal muscle, dystrophin provides a flexible connection between the cytoskeleton and the dystrophin-glycoprotein complex at the sarcolemma, myotendinous junction (MTJ) and neuromuscular junction (NMJ) [4]–[6]. Mutations that affect the mechanical integrity of this molecular scaffold render muscles more susceptible to contraction-induced injury leading to cycles of necrosis and regeneration [3].
As a general rule, most frame-shift mutations in dystrophin lead to DMD whereas internal truncations (in-frame deletions) lead to a milder form of the disease called Becker muscular dystrophy (BMD) [7]–[14]. The severity of BMD can also vary depending on whether a critical region of dystrophin is deleted and the amount of dystrophin being expressed [7]–[14]. Dystrophin consists of a N-terminal actin-binding domain, a large central rod domain, a cysteine rich region and a C-terminal domain (Figure 1A) [15], [16]. The central rod domain contains 24 spectrin-like repeats, 4 hinges and a second actin-binding domain [15]–[20]. The locus encoding the N-terminal actin-binding domain and the region near hinge 3 of dystrophin are more susceptible to deletion mutations [7]–[13]. In-frame deletions of the central rod domain typically lead to a mild BMD [8]–[13]. However, in-frame deletions at the “hot spot” near hinge 3 can lead to more variable phenotypes [8]–[13], [21].
Fig. 1. The hinge domains of dystrophin influence muscle maturation and maintenance. 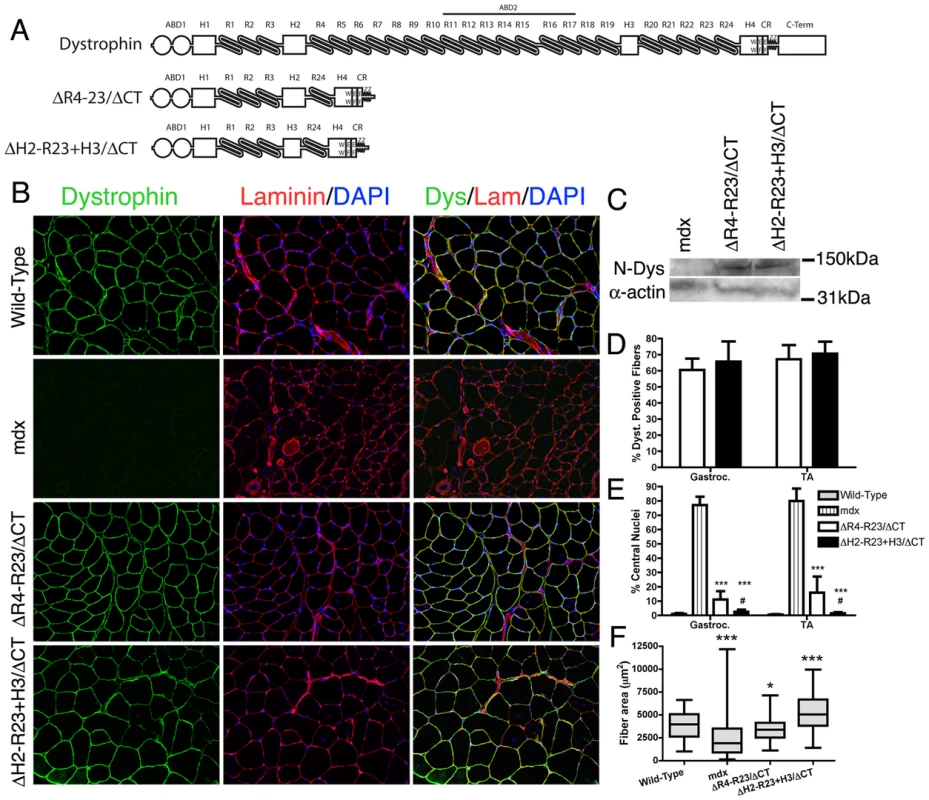
(A) The molecular structure of truncated dystrophins (reviewed in: [15]). ABD1 at the N-terminus is composed of two calponin homology domains denoted by the two circles. The central rod domain contains 24 spectrin-like repeats (R1-24), 4 hinge domains, a 20 amino acid insertion between spectrin-like repeats 15 and 16, and a central actin-binding domain (ABD2). A cluster of basic repeats forms ABD2 that bind to actin through an electrostatic interaction [20]. The hinge domains vary in that hinge 2 contains a polyproline site and hinge 4 contains a WW motif that is required for binding to β-dystroglycan [19], [68]. The cysteine-rich region contains two EF hands and a ZZ domain that is also required for binding to β-dystroglycan. The microdystrophins used in this study are shown below the full-length dystrophin. MicrodystrophinΔR4-R23/ΔCT lacks a large portion of the central rod domain between spectrin-like repeats 4 and 23 and also lacks the C-terminal domain (ΔR4-R23/ΔCT) [24]. Note that microdystrophinΔR4-R23/ΔCT and microdystrophinΔH2-R23+H3/ΔCT differ by a single hinge domain. (B) Transverse sections of gastrocnemius muscles from wild-type, mdx and mdx mice expressing either microdystrophinΔR4-R23/ΔCT or microdystrophinΔH2-R23+H3/ΔCT. Scale bar = 100 µm. (C) Expression of truncated dystrophins in treated mdx gastrocnemius muscles was similar. The western blots were performed using frozen muscles extracted from OCT. All lanes were loaded equally as shown by α-sarcomeric actin. (D) Shown is the mean +/− S.D. percentage of dystrophin-positive muscle fibers. TA is tibialis anterior. (E) Shown is the mean +/− S.D. of the percentage of muscle fibers containing centrally-located nuclei. MicrodystrophinΔH2-R23+H3/ΔCT with hinge 3 was more effective at preventing muscle degeneration in mdx mice compared to microdystrophinΔR4-R23/ΔCT with hinge 2. The treated muscles show the mean +/− SD for dystrophin-positive fibers only. ***P<0.001 compared to untreated mdx mice. #P<0.05 compared to microdystrophinΔR4-R23/ΔCT treated mdx muscles. (F) Hinge regions of dystrophin influence muscle fiber cross-sectional area. Shown is the mean +/− distribution (25th and 75th percentile (box) in addition to the farthest (whiskers) area of muscle fibers. *P<0.05 and ***P<0.001 compared to wild-type. The role of dystrophin in vivo has been largely defined by the structure-function relationship of truncated dystrophins in humans and mice [8]–[13], [22]–[24]. Rational design of dystrophin mini-genes has been highly effective in preventing and reversing functional abnormalities of dystrophic muscles [22]–[29]. In particular, we previously developed a microdystrophin (ΔR4-R23/ΔCT; defined as those with 4 or fewer spectrin-like repeats [24]) that accommodates the limited cloning capacity of recombinant adeno-associated viral vectors (rAAV) [24]. Intravenous injection of rAAV vectors pseudotyped with serotype 6 capsid (rAAV6) expressing microdystrophinΔR4-R23/ΔCT can prevent and reverse most aspects of dystrophic pathology in mdx muscles [24], [28], [30]–[35]. MicrodystrophinΔR4-R23/ΔCT also significantly protects muscles from contraction-induced injury [24], [28], [30]–[35].
While the microdystrophinΔR4-R23 transgene provides a clear benefit to dystrophic muscles [24], more detailed analyses have revealed a potentially serious abnormality in some muscle groups. The microdystrophinΔR4-R23/mdx transgenic mice have chronic Achilles myotendinous strain injury, which leads to the formation of ringed fibers and fragmentation of the neuromuscular junctions [33], [36]. In the present study we examined whether the domain composition or the small size of microdystrophinΔR4-R23/ΔCT led to this myopathy in mdx mice. We found that the hinge regions of microdystrophin, rather than its small size can profoundly influence skeletal muscle maintenance, maturation and structure.
Results
Dystrophin hinge domains influence the maintenance and maturation of skeletal muscles
We initially screened several truncated dystrophins and found that inclusion of hinge 2, but not hinge 3 could lead to the structural abnormalities we observed in some muscles of the microdystrophinΔR4-R23 transgenic mice (Text S1; Figures S1, S2, S3). We subsequently compared the efficacy of two microdystrophins that differ only in their inclusion of hinge 2 (microdystrophinΔR4-R23/ΔCT) or hinge 3 (microdystrophinΔH2-R23+H3/ΔCT) (Figure 1A) to examine whether the hinge composition of microdystrophin could influence various aspects of muscle disease.
We administered a sub-optimal dose of 2×1012 vector genomes of a rAAV6 pseudotyped vector expressing either microdystrophinΔR4-R23/ΔCT or microdystrophinΔH2-R23+H3/ΔCT intravenously into 2 week-old mdx4cv mice. We used a sub-optimal dose of rAAV6-microdystrophins so that we could examine whether changing the hinge domain increased or decreased the functional capacity of microdystrophin. Six months after treatment, both microdystrophins were expressed in a similar percentage of gastrocnemius and tibialis anterior (TA) muscle fibers (ranging from approximately 61% to 71%; P = 0.238 when comparing between the microdystrophins; Figure 1B and 1D). Western blots confirmed similar expression levels of truncated dystrophins in treated gastrocnemius muscles (Figure 1C). Both microdystrophins restored dystrophin-associated proteins to the sarcolemma except for nNOS (Text S1; Figure S4). MicrodystrophinΔR4-R23/ΔCT containing hinge 2 significantly prevented muscle degeneration (∼11% central nuclei for treated muscles verse ∼78% for untreated mdx muscles; P<0.001), and limited the fiber area of skeletal muscles (12% smaller than wild-type; P<0.05; Figure 1E), consistent with previous studies [24], [32], [33]. MicrodystrophinΔH2-R23+H3/ΔCT containing hinge 3 was significantly better able to prevent muscle degeneration (1–2% central nuclei; P<0.05 compared to microdystrophinΔR4-R23/ΔCT), and surprisingly increased average muscle fiber cross sectional area (34% larger than wild-type; P<0.001; Figure 1E). Thus, replacing hinge 2 of microdystrophinΔR4-R23/ΔCT with hinge 3 significantly improved its capacity to prevent muscle degeneration and promote skeletal muscle maturation.
Dystrophin hinge domains influence myotendinous junction injury and formation of ringed fibers
The tendon extends deep folds into wild-type skeletal muscles to minimize membrane stress under shear (Figure 2)[37]. Most of the folds in the mdx junctions did not extend as far into the gastrocnemius muscles (Figure 2). rAAV6-microdystrophinΔR4-R23/ΔCT severely disrupted the Achilles myotendinous junctions in mdx mice. Many of the junctional folds were missing and myofibril degeneration was evident (Figure 2). Approximately 17% of the adjoining mdx gastrocnemius muscles had ringed fibers. In contrast, rAAV6 - microdystrophinΔH2-R23+H3/ΔCT with hinge 3 retained the normal architecture of the Achilles myotendinous junction and we found no ringed fibers in the adjoining gastrocnemius muscles (Figure 2). Thus, the hinge domains influenced whether microdystrophin was capable of maintaining the myotendinous junction and myofibril structure in mdx gastrocnemius muscles.
Fig. 2. The hinge domains of dystrophin influence myotendinous junction structure and ringed fiber formation. 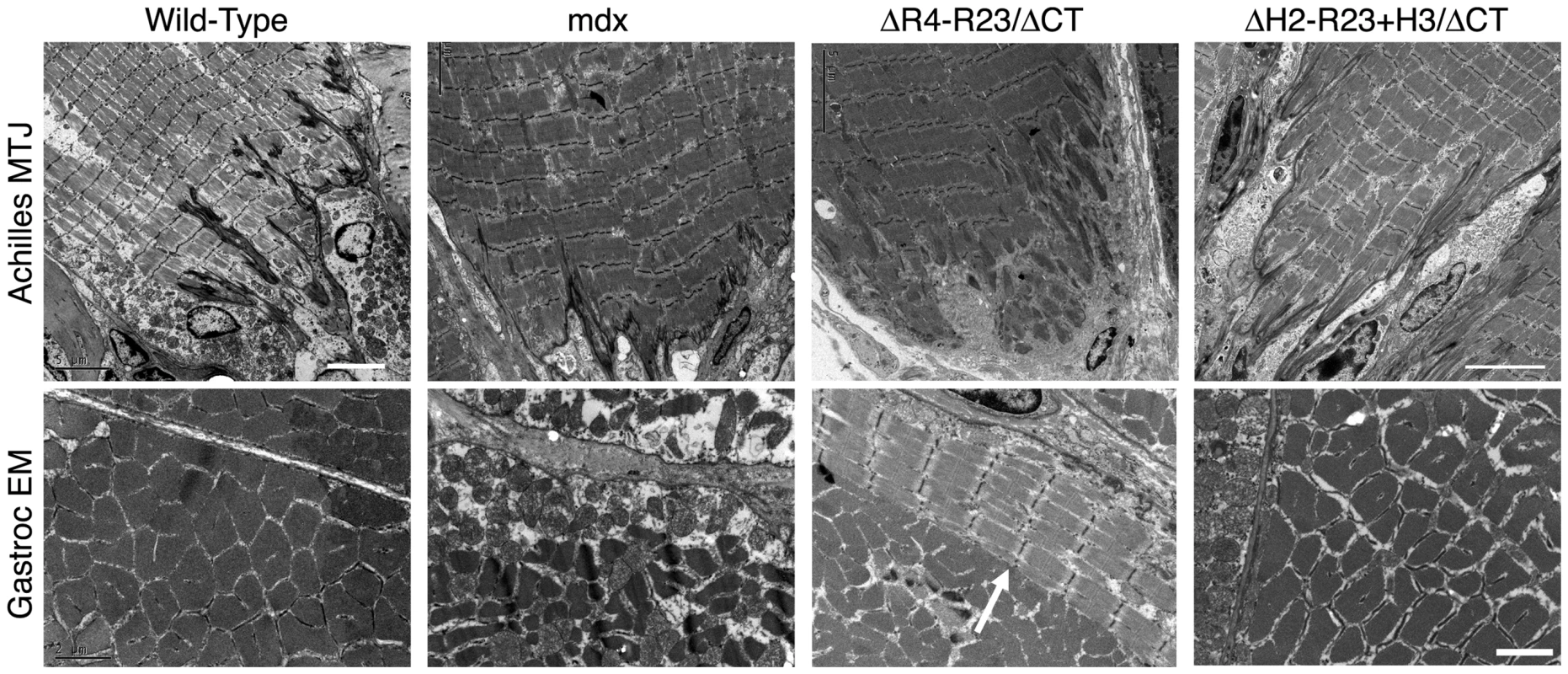
Shown are electron microscopy images of longitudinal sections of the Achilles myotendinous junctions in addition to transverse sections of gastrocnemius muscles. Note that expression of microdystrophin disrupted the myotendinous junctions and led to formation of ringed fibers (arrow). Scale bar = 5 µm for myotendinous junctions and 2 µm for transverse sections of gastrocnemius muscles. Dystrophin hinge domains influence neuromuscular synapse structure
We also examined neuromuscular synapses in mdx mice treated with rAAV6-microdystrophins. Most neuromuscular synapses in wild-type mice (∼97%) form a continuous tertiary structure as shown by staining whole muscle fibers with α-bungarotoxin (Figure 3A). Neuromuscular synapses in mdx mice begin to fragment temporally coincident with muscle degeneration [38]. Approximately 89% of neuromuscular synapses were fragmented in the gastrocnemius muscles of mdx mice (Figure 3B). We had previously shown that the neuromuscular synapses in transgenic microdystrophinΔR4-R23/mdx gastrocnemius muscles fragmented temporally coincident with the formation of ringed fibers [36]. In the present study we found that rAAV6 - microdystrophinΔR4-R23/ΔCT containing hinge 2 maintained continuous synapses in only 46% of the mdx gastrocnemius muscles (Figure 3A and 3B). In contrast, approximately 84% of synapses were continuous in mdx gastrocnemius muscles treated with rAAV6-microdystrophinΔH2-R23+H3/ΔCT containing hinge 3 (Figure 3A and 3B).
Fig. 3. The hinge domains of dystrophin influence the structure of neuromuscular synapses. 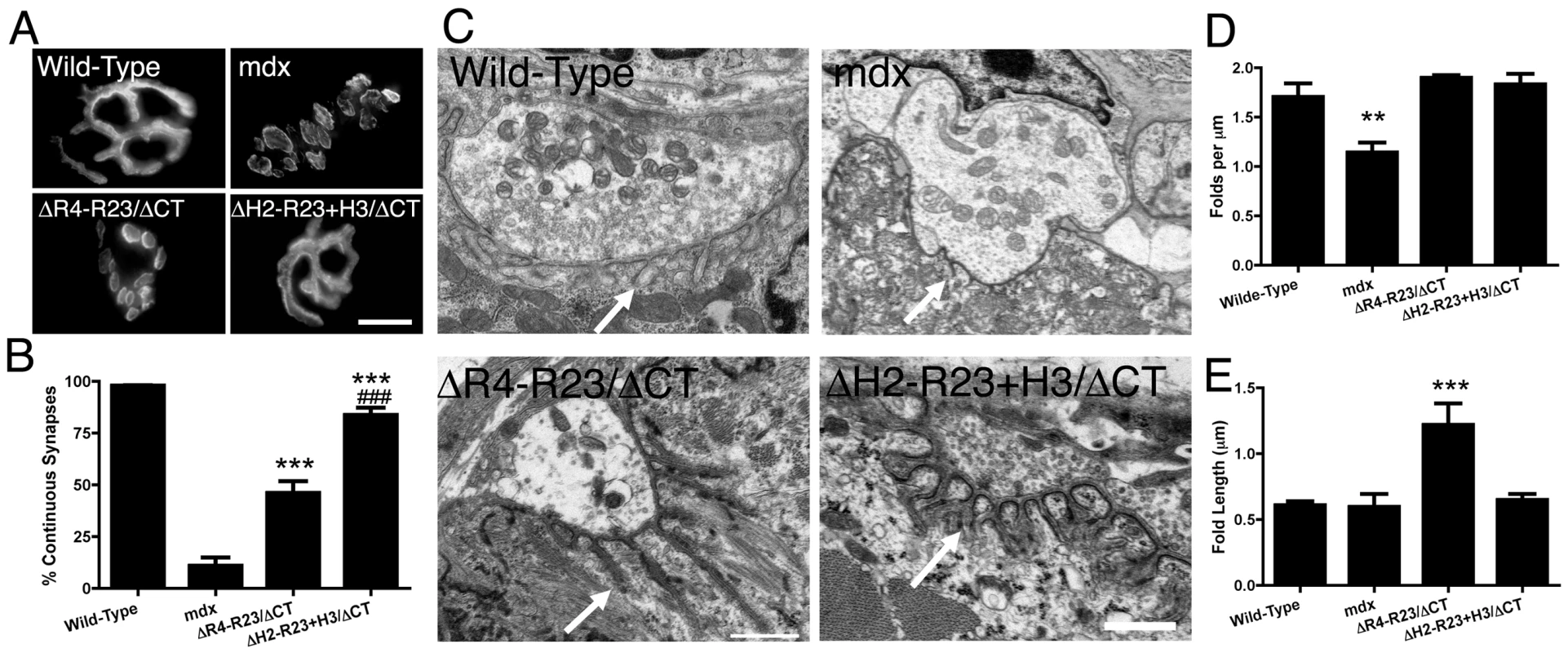
(A) Topographic view of AChR clusters stained with α-bungarotoxin. Scale bar = 10 µm. (B) Graph shows mean +/− SD percentage of continuous synapses. Significant difference to mdx ***P<0.001. Significant difference to microdystrophinΔR4-R23/ΔCT/mdx mice ###P<0.001. (C) Ultrastructure of neuromuscular synapses from wild-type, mdx and mdx gastrocnemius muscles expressing microdystrophinΔR4-R23/ΔCT and microdystrophinΔH2-R23/ΔCT+H3. Note that synaptic folding is reduced in mdx mice and increased in rAAV6-microdystrophinΔR4-R23/ΔCT treated muscles (arrows). Scale bar = 0.5 µm. (D) The graph shows the mean +/− SD number of folds per µm of postsynaptic membrane juxtaposed to the presynaptic cleft. Significant difference compared to wild-type **P<0.01. (E) The mean +/− SD depth of the folds was significantly increased in mdx muscles treated with rAAV6-microdystrophinΔR4-R23/ΔCT (***P<0.001). Neuromuscular synapses also contain folds in the postsynaptic membrane that align directly adjacent to vesicle release sites (active zones) in the pre-synaptic nerve terminal (arrows; Figure 3C). The number of synaptic folds in mdx mice was significantly reduced compared to wild-type (P<0.01; Figure 3C and 3D) as previously described [4], [39]. The number of folds was restored in microdystrophinΔR4-R23/ΔCT and microdystrophinΔH2-R23+H3/ΔCT treated muscles (Figure 3C and 3D). The synaptic folds extended significantly further into microdystrophinΔR4-R23/ΔCT treated mdx muscles compared to wild-type muscles (P<0.001; Figure 3C and 3E), as previously described in transgenic microdystrophinΔR4-R23/mdx mice [36]. In contrast, the number and length of synaptic folds in microdystrophinΔH2-R23+H3/ΔCT treated mdx muscles was similar to wild-type (Figure 3C–3E). Thus, microdystrophinΔH2-R23+H3/ΔCT containing hinge 3 can maintain the structure of neuromuscular junctions in mdx muscles.
Mechanical properties of muscles expressing microdystrophins with either hinge 2 or hinge 3
Contraction-induced injury can initiate muscle degeneration in mdx mice [40]. Skeletal muscles from mdx mice have a lower force producing capacity than wild-type muscles and are more susceptible to contraction-induced injury (Figure 4). We found that sub-optimal doses of both rAAV6-microdystrophinΔR4-R23/ΔCT and rAAV6 - microdystrophinΔH2-R23+H3/ΔCT maintained the peak force producing capacity of mdx gastrocnemius and tibialis anterior muscles (Figure 4A). Both microdystrophins also significantly improved the specific force (force per cross sectional area of muscle) production in mdx muscles (P<0.05; Figure 4B). The specific force was not restored to wild-type partly because the sub-optimal dose of rAAV6-microdystrophin did not prevent the pseudo hypertrophy normally found in mdx muscles (P = 0.454 when comparing the muscle mass between mdx and treated mdx muscles; one-way ANOVA). Each microdystrophin significantly protected the treated limb muscles from contraction-induced injury (P<0.001; Figure 4C and 4D). However, we found no significant difference between the peak force, specific force or protection from contraction-induced injury when comparing between the two microdystrophins with either hinge 2 or hinge 3.
Fig. 4. Microdystrophins significantly improve the mechanical properties of mdx hind limb muscles. 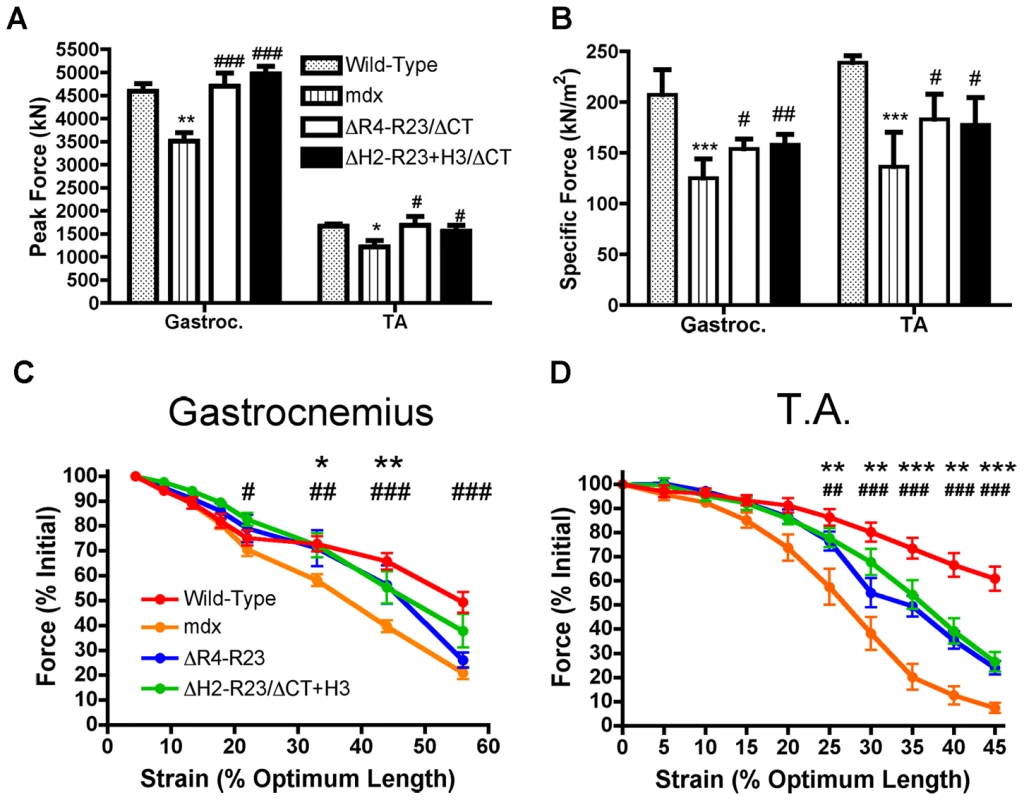
(A) Graph shows the mean +/− S.D. peak force. *P<0.05 and **P<0.01 compared to wild-type. #P<0.05 and ###P<0.001 compared to mdx. (B) Graph shows the mean +/− S.D. specific force. ***P<0.001 compared to wild-type. #P<0.05 and ##P<0.01 compared to mdx. The contractile performance of (C) gastrocnemius muscles and (D) tibialis anterior muscles immediately prior to increasing length changes during maximal force production. Bars represent the mean +/− S.D. percentage of the initial optimal muscle contraction. rAAV6-microdystrophinΔR4-R23/ΔCT treated muscles were significantly (*P<0.05; **P<0.01; ***P<0.001) protected from contraction-induced injury, as were rAAV6-microdystrophinΔH2-R23/ΔCT+H3 (#P<0.05; ##P<0.01; ###P<0.001) treated muscles when compared to mdx mice. Polyproline in hinge 2 influences the pathology of skeletal muscle fibers
Together, our results suggested that the structural abnormalities observed in some treated mdx muscles could be traced to the presence of hinge 2 within the microdystrophin. We next examined the molecular composition of the hinges to define what was unique about hinge 2. The hinges in dystrophin are defined as such because of the higher concentration of proline residues, which function to limit the continuation of the α-helical coiled-coils of the spectrin-like repeats through the entire length of the dystrophin rod domain [19]. Both hinge 2 and hinge 3 have six proline residues and the lengths of these hinges are similar [19]. We hypothesized that the placement of the prolines most likely results in their different functions [5], [19]. Hinge 2 has 5 consecutive proline residues (polyproline; Figure 5A) whereas the proline residues in hinge 3 are more evenly distributed throughout the hinge [19]. Polyproline residues are thought to have their own defined rigid helical structure [41], [42], and this could affect the functional capacity of microdystrophinΔR4-R23/ΔCT.
Fig. 5. MicrodystrophinΔR4-R23/ΔCT did not cause structural abnormalities when the polyproline site was deleted from hinge 2. 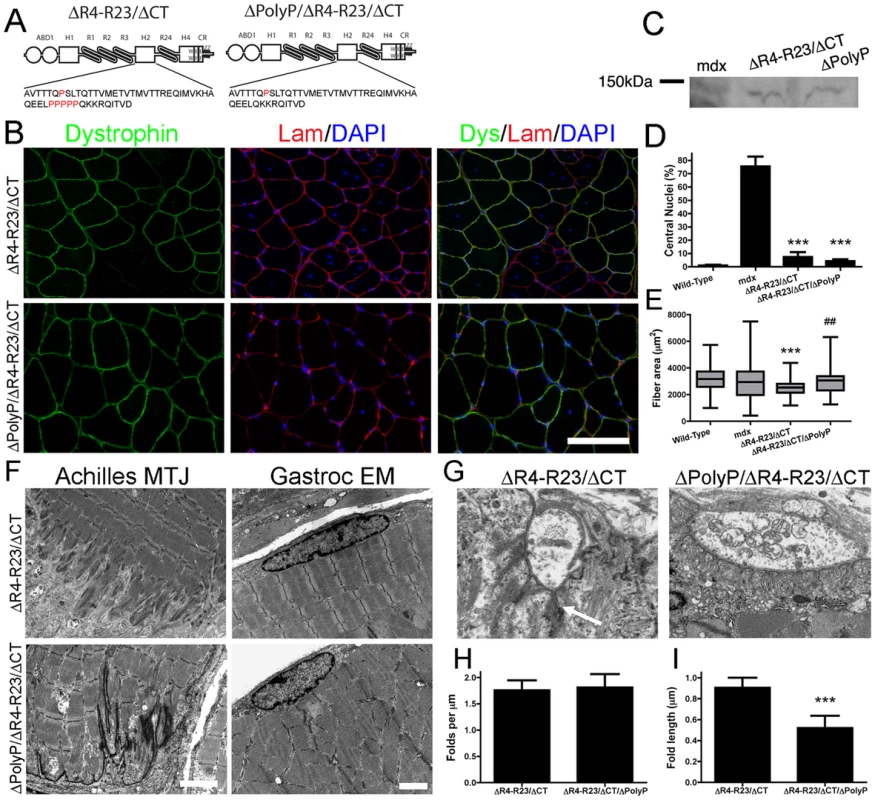
(A) The molecular structure of microdystrophins. Below each structure is the amino acid sequence of hinge 2. Note that proline residues are highlighted in red and that microdystrophinΔPolyP/ΔR4-R23/ΔCT lacks the polyproline site. (B) Transverse sections of gastrocnemius muscles from mdx mice expressing microdystrophinΔR4-R23/ΔCT (top panel) or microdystrophinΔPolyP/ΔR4-R23/ΔCT (lower panel). Scale bar = 100 µm. (C) Expression of truncated dystrophins from treated gastrocnemius muscles. The western blots were performed using tissue sections frozen in OCT. (D) Shown is the mean +/− S.D. percentage of myofibers with centrally-located nuclei. The treated muscles show the mean +/− SD for dystrophin positive fibers only. ***P<0.001 compared to untreated mdx mice. (E) The polyproline region influenced muscle fiber cross sectional area. Shown is the mean +/− distribution (25th and 75th percentile (box) in addition to the farthest (whiskers) of muscle fibers. ***P<0.001 compared to wild-type. ##P<0.01 compared to microdystrophinΔR4-R23/ΔCT. (F) MicrodystrophinΔR4-R23/ΔCT expression led to a disruption of the myotendinous junctions and ringed fiber formation (arrow; top panels) in mdx gastrocnemius muscles, but not when the polyproline site was deleted (lower panels). Scale bars = 2 µm. (G) Electron microscopy images of neuromuscular junctions (NMJ) in mdx muscles treated with rAAV6-microdystrophinΔR4-R23/ΔCT or rAAV6- microdystrophinΔPolyP/ΔR4-R23/ΔCT. Scale bar = 1 µm. (H) The graph shows the mean +/− SD number of folds per mm of postsynaptic membrane juxtaposed to the presynaptic cleft. (I) Deletion of the polyproline site from microdystrophinΔR4-R23/ΔCT restored the normal mean +/− SD length of synaptic folds (***P<0.001 compared to microdystrophinΔR4-R23/ΔCT). To test this hypothesis we compared muscles expressing the original microdystrophinΔR4-R23/ΔCT with a newly developed microdystrophinΔpolyP/ΔR4-R23/ΔCT that lacks the polyproline site in hinge 2 (Figure 5A). We delivered 6×1010 vg of each microdystrophin into mdx gastrocnemius muscles at 2 days of age and examined the mice 7 weeks after treatment. Both microdystrophins were expressed in a similar percentage of muscle fibers (Figure 5B; 59–68%), and were expressed at similar levels (Figure 5C). Each microdystrophin significantly reduced muscle fiber degeneration (Figure 5D). As expected, the original microdystrophinΔR4-R23/ΔCT limited muscle fiber cross-sectional area (Figure 5E), was associated with disrupted myotendinous junctions (Figure 5F), led to the formation of ringed fibers (Figure 5F), and perturbed neuromuscular junctions (Figure 5G–5I). In contrast, the mdx muscles treated with microdystrophinΔpolyP/ΔR4-R23/ΔCT did not show any abnormalities in muscle fiber maturation or structure (Figure 5). Thus, the presence of this polyproline site in hinge 2 of microdystrophinΔR4-R23/ΔCT prevented the appropriate integration of muscles into the nerve-tendon environment.
Discussion
Most gene therapy strategies for DMD require the generation of highly functional truncated dystrophins. rAAV is an efficient and safe vector for systemically delivering truncated dystrophins to striated muscles to prevent muscle degeneration in animal models of DMD ([28]; reviewed in [43]). We had previously generated a microdystrophinΔR4-R23 that was highly capable of mitigating muscle degeneration and improving the mechanical function of mdx skeletal muscles [24], [28]. However, the microdystrophinΔR4-R23 transgene leads to chronic strain injury at the Achilles myotendinous junction [33]. This led to the formation of ringed fibers that function to protect skeletal muscles from contraction-induced injury, even better than wild-type mice [33]. The formation of the rings led to fragmentation of the neuromuscular junctions [36]. Other effects of the transgene included smaller muscle fibers [24], and increased length of synaptic folds [36]. Here we found that each of these phenotypic changes was recapitulated in mdx gastrocnemius muscles treated with rAAV6-microdystrophinΔR4-R23/ΔCT. A screen of several newly developed dystrophin mini-genes revealed that the hinge 2 region influenced the functional capacity of microdystrophinΔR4-R23/ΔCT. Replacing hinge 2 with hinge 3 led to several advantages such as better protection of skeletal muscles (only 1–2% central nuclei 6 months post treatment), larger muscle fibers and normal junctions. Deleting the polyproline site from hinge 2 of microdystrophinΔR4-R23/ΔCT also prevented these structural abnormalities.
Mechanical properties of skeletal muscles expressing microdystrophins
MicrodystrophinΔH2-R23+H3/ΔCT with hinge 3 significantly increased peak force, specific force and protected muscles from contraction-induced injury. However, the morphological improvements of microdystrophinΔH2-R23+H3/ΔCT treated muscles did not translate into a functional improvement compared to microdystrophinΔR4-R23/ΔCT treated muscles. This could result from the molecular and cellular responses to myotendinous strain injury that help protect the rAAV6-microdystrophinΔR4-R23/ΔCT treated muscles from contraction-induced injury [33]. Another possibility is that the presence of some dystrophin negative fibers masked any functional difference between the two proteins. The inclusion of hinge 2 in microdystrophin limited muscle fiber area whereas the inclusion of hinge 3 increased muscle fiber area (Figure 1). Larger muscle fibers in microdystrophinΔH2-R23+H3/ΔCT treated mice could have two distinct advantages: They could replace some of the muscle mass lost in advanced stages of disease and they could be better protected from contraction-induced injury [44]. However, the sub-optimal dose of either rAAV6-microdystrophin did not prevent the pseudo hypertrophy in mdx mice and no mechanical advantages could be discerned when comparing treatments. Saturating levels of rAAV6-microdystrophins or transgenic mice will most likely be required to detect minor differences in the mechanical properties of muscles expressing various truncated dystrophins.
How does polyproline influence the functional capacity of truncated dystrophins?
Our most effective truncated dystrophins developed for gene therapy have been designed to maximize functional interactions between specific spectrin-like repeats and hinge domains. This design has been influenced by genetic studies in mice and man as well as biophysical studies in vitro on the structure, folding and physical properties of both individual and tandemly expressed spectrin-like repeats and hinge domains [24], [45]–[52]. Individual spectrin-like repeats are not all interchangeable, and ones adjacent to hinges have distinct properties from those flanked by other spectrin-like repeats [21], [24], [47], [51], [52]. Also, spectrin-like repeats rarely function as isolated units [15], [24], [50]–[53]. Instead, they appear to fold into nested domains interrupted by various insertions (hinges) that disrupt the uniformity and rigidity of the spectrin-like repeat rod domain [24], [45]–[48], [53]–[55]. These interruptions appear important for the elastic and flexible structure that dystrophin requires in its role as a force transducer and shock absorber in muscle [56]–[59]. Our studies suggest that the most functional truncations of dystrophin retain a central hinge domain that is flanked by spectrin-like repeats found adjacent to a hinge in the wild-type dystrophin [24]. Disruption of this linkage could influence protein folding, stability and function leading to the variable phenotypes in patients associated with deletions at or near hinge 3, which is encoded on exons 50–51 [21].
Individual spectrin-like repeats are composed of 3 helical domains connected by non-helical linkers, which fold into a triple helical coiled coil structure (Figure 5A; [45], [47]). The linker regions between discreet repeats are also typically short and relatively unstructured to allow a smooth connection between the third helix of a preceding repeat and the first helix of the next repeat (Figure 6A). However, hinge domains interrupt the nested nature of adjacent spectrin-like repeats and allow more flexibility in the rod domain (Figure 6B). This degree of flexibility appears to be significantly different when hinge 2 or hinge 3 is present. While both hinges contain 6 prolines, which act to disrupt alpha helical structures, in hinge 3 they are dispersed whereas 5 of the 6 prolines in hinge 2 are clustered together (Figure 5A, Figure 6C and 6D; [10]). Polyproline residues form a rigid α-helix [41], [42], much like a molecular ruler [60]. We suggest that the location of this polyproline sequence within a highly truncated rod domain induces a severe structural disruption that can affect the ability of dystrophin to form a mechanically flexible connection between F-actin and β-dystroglycan. Spectrin-like repeats 1-3 have been shown to associate with the sarcolemmal membrane, while the WW domain in hinge 4 forms a critical portion of the β-dystroglycan binding domain [45], [61]. A rigid rod domain induced by polyproline in hinge 2 may directly impair the ability of microdystrophin to form a flexible interaction with either or both of these structures (Figure 6C). In contrast, when hinge 2 is present in full-length dystrophin, a significantly greater number of spectrin-like repeats are present between the hinge and the β-dystroglycan binding domain, allowing greater flexibility in the overall structure.
Fig. 6. Predicted nested structure of specific dystrophin spectrin-like repeats relevant to microdystrophins and how they interact with hinge domains. 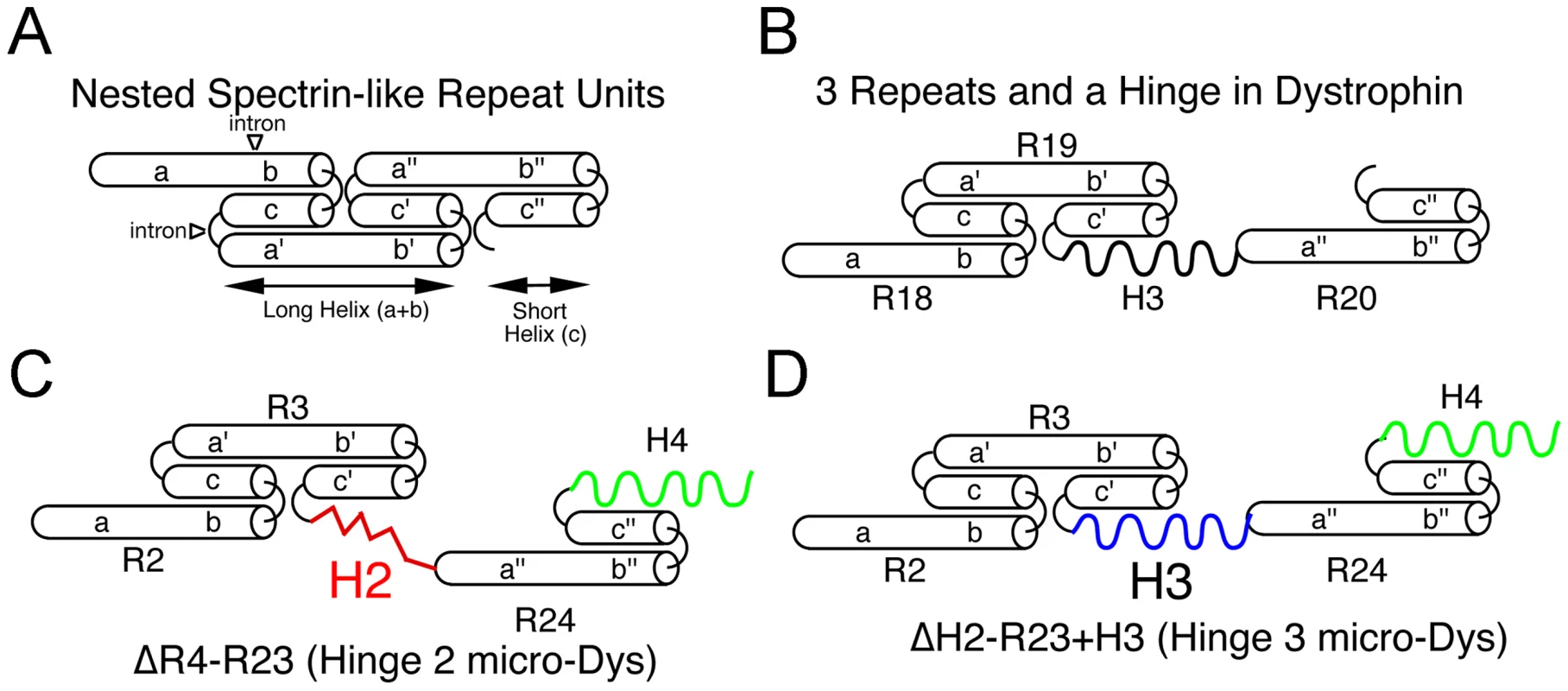
(A) Predicted structure of 3 nested repeats [15]. Each repeat is composed of 3 helical domains (a, b, c) connected by non-helical linkers. The triple helical coiled-coil repeat structure is formed by helices b and c of the preceding repeat interfacing with the N-terminal helix a' from the following repeat. The helices a and b fold together into a long repeat, while helix c folds into the short repeat. (B) In dystrophin, the repeat domains are interrupted by hinge 2 (between repeats 3 and 4) and by hinge 3 (Repeats19 and 20). Shown is the predicted structure of the repeat 19-hinge 3-repeat 20 domain in full-length dystrophin. (C) The hinge 2-microdystrophin induces an unusual structural alteration that disrupts the normal connection between adjacent repeats. (D) In contrast, use of hinge 3 generates a similar structure as in (B) except that hinge 3 joins to repeats 3 and 24 rather than repeats 19 and 20. It is difficult to predict the function of the polyproline site from patients with in frame deletions of exon 17 (hinge 2) of dystrophin. The described deletions (Leiden Muscular Dystrophy Pages) usually encompass larger regions of dystrophin than the polyproline site and it is not clear how these deletions affect protein stability. Our finding that hinge 3 microdystrophin can prevent muscle degeneration suggests that the polyproline site is not a necessary component of dystrophin, similar to previous reports on longer forms of truncated dystrophins [24], [62].
Flanigan et. al., 2009 has proposed that approximately 62% of all DMD patients could be treated with oligonucleotides that skip exons 45–55 (from spectrin-like repeat 18–22)[14]. This would create a truncated dystrophin that contains hinge 2 but not hinge 3, similar to, but much larger than our microdystrophinΔR4-R23 transgene. It will therefore be of interest to determine whether the polyproline site in hinge 2 can influence the functional capacity of larger, truncated dystrophins. It will also be of interest to examine whether the polyproline site affects the functional capacity of truncated utrophin constructs that are designed for gene therapy of DMD [63], [64].
Materials and Methods
Mice and ethics statement
We utilized C57Bl/10 wild-type mice and mdx4cv mice. All experiments are in accordance with the institution of animal care and use committee (IACUC) of the University Of Washington.
Generation of constructs
The expression vector CMV-ΔR4-R23/ΔCT which uses the cytomegalovirus immediate early promoter and enhancer to drive expression of a microdystrophin cDNA was generated as previously described [32]. We generated the ΔH2-R24/ΔCT, ΔR2-R23+R18-H3/ΔCT, ΔH2-R23+H3/ΔCT and ΔPolyP/ΔR4-R23/ΔCT constructs using recombination PCR with CMV-ΔR4-R23/ΔCT as the template [65]. The primers used to generate ΔH2-R24/ΔCT, ΔR2-R23+R18-H3/ΔCT, ΔH2-R23+H3/ΔCT and ΔPolyP/ΔR4-R23/ΔCT are found in Table S1. The resulting expression vectors were sequenced and co-transfected with the pDGM6 packaging plasmid into HEK293 cells to generate recombinant AAV vectors comprising serotype 6 capsids that were harvested, purified, and quantitated as described previously [29]. The resulting titer was determined by comparison to previously known concentrations of rAAV6-CMV-lacZ and ΔR4-R23/ΔCT by Southern analyses with a probe to the CMV promoter. The rAAV6-microdystrophins were delivered intravenously by tail vein injection at two weeks of age or directly into the mdx gastrocnemius muscles at 2 days of age while the mice were anaesthetized.
Gross muscle morphology and morphometry
Gross muscle morphology was analyzed as previously described [24], [32]. Primary antibodies included the N-terminus of dystrophin (1∶800; [23]), utrophin A (1∶300; gift from Stanley Froehner, University of Washington), mouse monoclonal anti-α-dystrobrevin (Transduction laboratories; 1∶200), rabbit polyclonal anti-Syn17 (α-syntrophin; 1∶200; [66]), rabbit polyclonal anti-nNOS (Alexis; 1∶200). Secondary antibodies included Alexa 488, Alexa 594 rabbit polyclonal or Alexa 488 mouse monoclonal secondary antibodies (Molecular Probes; 1∶800). The sections were mounted in anti-fade mounting media containing DAPI (Vector Labs). Fluorescent sections were imaged using a Nikon eclipse E1000 fluorescent microscope (Nikon; NY) and captured using a DeltaVision fluorescence microscope. Muscle fiber areas were quantified using Image J (NIH).
Immunoblotting
For immunoblots, n = 4 gastrocnemius muscles from mdx mice and mdx mice treated with rAAV6-microdystrophinΔR4-R23/ΔCT or rAAV6-microdystrophinΔH2-R23+H3/ΔCT were thawed from OCT blocks and placed into extract buffer (50 mM Tris-HCl, 150 mM NaCl, 0.2% sodium dodecyl sulfate, 10% glycerol, 24 mM Na Deoxycholate, 1% NP40, 47.6 mM Na Fluoride, 200 mM Na orthovanadate, Roche). Protein concentrations were determined by Coomassie Plus Bradford Assay (Peirce). Equal amounts of protein (15 mg) were resolved on a 4–12% SDS polyacrylamide gel. The blots were incubated in rabbit polyclonal antibodies to dystrophin (1∶500; kind gift from James Ervasti, University of Minnesota) and mouse monoclonal antibodies to α-sarcomeric actin (1∶500; SIGMA).
We also performed immunoblots on frozen tissue sections from n = 4 gastrocnemius muscles treated with rAAV6-microdystrophinΔR4-R23/ΔCT and microdystrophinΔPolyP/ΔR4-R23/ΔCT as previously described [67], with minor modifications. Briefly, we cut twenty-five 20 µm sections and diluted the sections into 200 µl lysis buffer (4% SDS, 25 mM Tris pH 8.8, 40% glycerol, 0.5 M phenylmethylsulfonyl fluoride, 100 mM dithiothreitol and bromophenol blue). Samples were briefly sonicated (10 sec at 4°C), heated to 95°C for 5 minutes, centrifuged for 5 minutes at 13,200×g and electrophoresed on a 4–12% SDS-polyacrylamide gel. The blots were incubated in primary rabbit polyclonal antibody against the N-terminus of dystrophin (1∶500; kind gift from James Ervasti, University of Minnesota). All blots were developed with ECL Plus (Pierce) and scanned with the Storm 860 imaging system (Amersham Biosciences).
Electron microscopy
Electron microscopy was performed as previously described [33]. The junctional fold number and lengths were measured from n = 4 mice at 6 months of age using Image J (NIH) and compared using Students t-test (Prism). The counts represent the fold numbers and lengths from all fibers (dystrophin positive and negative).
Quantitation of ringed fibers
We quantitated the number of ringed myofibers in EM images and thick (1 µm) toluidine blue sections from at least 4 animals per group. At least 300 muscle fibers from n = 4 gastrocnemius muscles were examined from wild-type, mdx4cv and mdx4cv mice expressing the various microdystrophins.
Quantification of neuromuscular synapses
Neuromuscular synapses were analyzed in whole mount immunofluorescence stained muscles and quantitated as previously described [36]. The acetylcholine receptor clusters were stained with TRITC conjugated α-bungarotoxin (αBTX; 1∶800; Molecular Probes). Synapses were classified as continuous if they presented with 3 or less continuous regions of AChR clustering and discontinuous if they presented with more than 3 regions of AChR clustering. More than 50 synapses were analyzed from treated and untreated gastrocnemius skeletal muscle fibers from n = 4 mice. The counts in treated muscles include both dystrophin positive and negative fibers. We compared the proportion of continuous synapses using a Students t-test.
Muscle physiology
Muscle physiology was performed as previously described for tibialis anterior [29] and gastrocnemius [33] muscles. We examined six-month-old wild-type, mdx, and mdx mice treated with rAAV6-microdystrophinΔR4-R23/ΔCT or rAAV6-microdystrophinΔH2-R23+H3/ΔCT (n = 5).
Supporting Information
Zdroje
1. HoffmanEP
BrownRHJr
KunkelLM
1987 Dystrophin: the protein product of the Duchenne muscular dystrophy locus. Cell 51 919 928
2. KoenigM
HoffmanEP
BertelsonCJ
MonacoAP
FeenerC
1987 Complete cloning of the Duchenne muscular dystrophy (DMD) cDNA and preliminary genomic organization of the DMD gene in normal and affected individuals. Cell 50 509 517
3. EmeryAE
Muntoni
2003 Duchenne Muscular Dystrophy., 3rd edn. Oxford University Press, Oxford
4. BanksGB
FuhrerC
AdamsME
FroehnerSC
2003 The postsynaptic submembrane machinery at the neuromuscular junction: requirement for rapsyn and the utrophin/dystrophin-associated complex. J Neurocytol 32 709 726
5. BhasinN
LawR
LiaoG
SaferD
EllmerJ
2005 Molecular extensibility of mini-dystrophins and a dystrophin rod construct. J Mol Biol 352 795 806
6. ErvastiJM
2007 Dystrophin, its interactions with other proteins, and implications for muscular dystrophy. Biochim Biophys Acta 1772 108 117
7. ChamberlainJS
GibbsRA
RanierJE
NguyenPN
CaskeyCT
1988 Deletion screening of the Duchenne muscular dystrophy locus via multiplex DNA amplification. Nucleic Acids Res 16 11141 11156
8. BaumbachLL
ChamberlainJS
WardPA
FarwellNJ
CaskeyCT
1989 Molecular and clinical correlations of deletions leading to Duchenne and Becker muscular dystrophies. Neurology 39 465 474
9. GillardEF
ChamberlainJS
MurphyEG
DuffCL
SmithB
1989 Molecular and phenotypic analysis of patients with deletions within the deletion-rich region of the Duchenne muscular dystrophy (DMD) gene. Am J Hum Genet 45 507 520
10. KoenigM
BeggsAH
MoyerM
ScherpfS
HeindrichK
1989 The molecular basis for Duchenne versus Becker muscular dystrophy: correlation of severity with type of deletion. Am J Hum Genet 45 498 506
11. WerneckLC
ScolaRH
MaegawaGH
WerneckMC
2001 Comparative analysis of PCR-deletion detection and immunohistochemistry in Brazilian Duchenne and Becker muscular dystrophy patients. Am J Med Genet 103 115 120
12. Den DunnenJT
GrootscholtenPM
BakkerE
BlondenLA
GinjaarHB
1989 Topography of the Duchenne muscular dystrophy (DMD) gene: FIGE and cDNA analysis of 194 cases reveals 115 deletions and 13 duplications. Am J Hum Genet 45 835 847
13. Coral-VazquezR
ArenasD
CisnerosB
PenalozaL
KofmanS
1993 Analysis of dystrophin gene deletions in patients from the Mexican population with Duchenne/Becker muscular dystrophy. Arch Med Res 24 1 6
14. FlaniganKM
DunnDM
von NiederhausernA
SoltanzadehP
GappmaierE
2009 Mutational spectrum of DMD mutations in dystrophinopathy patients: application of modern diagnostic techniques to a large cohort. Hum Mutat 30 1657 1666
15. AbmayrS
ChamberlainJ
2006 The structure and function of dystrophin.
WinderSJ
Molecular Mechanisms of Muscular Dystrophies Georgetown Landes Biosciences 14 34
16. BanksGB
ChamberlainJS
2008 The value of mammalian models for duchenne muscular dystrophy in developing therapeutic strategies. Curr Top Dev Biol 84 431 453
17. KoenigM
MonacoAP
KunkelLM
1988 The complete sequence of dystrophin predicts a rod-shaped cytoskeletal protein. Cell 53 219 226
18. DavisonMD
BaronMD
CritchleyDR
WoottonJC
1989 Structural analysis of homologous repeated domains in alpha-actinin and spectrin. Int J Biol Macromol 11 81 90
19. KoenigM
KunkelLM
1990 Detailed analysis of the repeat domain of dystrophin reveals four potential hinge segments that may confer flexibility. J Biol Chem 265 4560 4566
20. AmannKJ
RenleyBA
ErvastiJM
1998 A cluster of basic repeats in the dystrophin rod domain binds F-actin through an electrostatic interaction. J Biol Chem 273 28419 28423
21. CarsanaA
FrissoG
TremolaterraMR
LanzilloR
VitaleDF
2005 Analysis of dystrophin gene deletions indicates that the hinge III region of the protein correlates with disease severity. Ann Hum Genet 69 253 259
22. PhelpsSF
HauserMA
ColeNM
RafaelJA
HinkleRT
1995 Expression of full-length and truncated dystrophin mini-genes in transgenic mdx mice. Hum Mol Genet 4 1251 1258
23. RafaelJA
CoxGA
CorradoK
JungD
CampbellKP
1996 Forced expression of dystrophin deletion constructs reveals structure-function correlations. J Cell Biol 134 93 102
24. HarperSQ
HauserMA
DelloRussoC
DuanD
CrawfordRW
2002 Modular flexibility of dystrophin: implications for gene therapy of Duchenne muscular dystrophy. Nat Med 8 253 261
25. CrawfordGE
FaulknerJA
CrosbieRH
CampbellKP
FroehnerSC
2000 Assembly of the dystrophin-associated protein complex does not require the dystrophin COOH-terminal domain. J Cell Biol 150 1399 1410
26. WangB
LiJ
XiaoX
2000 Adeno-associated virus vector carrying human minidystrophin genes effectively ameliorates muscular dystrophy in mdx mouse model. Proc Natl Acad Sci U S A 97 13714 13719
27. YuasaK
MiyagoeY
YamamotoK
NabeshimaY
DicksonG
1998 Effective restoration of dystrophin-associated proteins in vivo by adenovirus-mediated transfer of truncated dystrophin cDNAs. FEBS Lett 425 329 336
28. GregorevicP
BlankinshipMJ
AllenJM
CrawfordRW
MeuseL
2004 Systemic delivery of genes to striated muscles using adeno-associated viral vectors. Nat Med 10 828 834
29. GregorevicP
AllenJM
MinamiE
BlankinshipMJ
HaraguchiM
2006 rAAV6-microdystrophin preserves muscle function and extends lifespan in severely dystrophic mice. Nat Med 12 787 789
30. YoshimuraM
SakamotoM
IkemotoM
MochizukiY
YuasaK
2004 AAV vector-mediated microdystrophin expression in a relatively small percentage of mdx myofibers improved the mdx phenotype. Mol Ther 10 821 828
31. YueY
LiuM
DuanD
2006 C-terminal-truncated microdystrophin recruits dystrobrevin and syntrophin to the dystrophin-associated glycoprotein complex and reduces muscular dystrophy in symptomatic utrophin/dystrophin double-knockout mice. Mol Ther 14 79 87
32. BanksGB
GregorevicP
AllenJM
FinnEE
ChamberlainJS
2007 Functional capacity of dystrophins carrying deletions in the N-terminal actin-binding domain. Hum Mol Genet 16(17) 2105 2113
33. BanksGB
CombsAC
ChamberlainJR
ChamberlainJS
2008 Molecular and cellular adaptations to chronic myotendinous strain injury in mdx mice expressing a truncated dystrophin. Hum Mol Genet (17)24 3975 3986
34. FosterH
SharpPS
AthanasopoulosT
TrolletC
GrahamIR
2008 Codon and mRNA Sequence Optimization of Microdystrophin Transgenes Improves Expression and Physiological Outcome in Dystrophic mdx Mice Following AAV2/8 Gene Transfer. Mol Ther
35. GregorevicP
BlankinshipMJ
AllenJM
ChamberlainJS
2008 Systemic microdystrophin gene delivery improves skeletal muscle structure and function in old dystrophic mdx mice. Mol Ther 16 657 664
36. BanksGB
ChamberlainJS
FroehnerSC
2009 Truncated dystrophins can influence neuromuscular synapse structure. Mol Cell Neurosci 40 433 441
37. TidballJG
1991 Force transmission across muscle cell membranes. J Biomech 24 Suppl 1 43 52
38. LyonsPR
SlaterCR
1991 Structure and function of the neuromuscular junction in young adult mdx mice. J Neurocytol 20 969 981
39. GradyRM
TengH
NicholMC
CunninghamJC
WilkinsonRS
1997 Skeletal and cardiac myopathies in mice lacking utrophin and dystrophin: a model for Duchenne muscular dystrophy. Cell 90 729 738
40. LynchGS
2004 Role of contraction-induced injury in the mechanisms of muscle damage in muscular dystrophy. Clin Exp Pharmacol Physiol 31 557 561
41. KayBK
WilliamsonMP
SudolM
2000 The importance of being proline: the interaction of proline-rich motifs in signaling proteins with their cognate domains. Faseb J 14 231 241
42. RathA
DavidsonAR
DeberCM
2005 The structure of “unstructured” regions in peptides and proteins: role of the polyproline II helix in protein folding and recognition. Biopolymers 80 179 185
43. JudgeLM
ChamberlainJS
2005 Gene therapy for Duchenne muscular dystrophy: AAV leads the way. Acta Myol 24 184 193
44. GehrigSM
KoopmanR
NaimT
TjoakarfaC
LynchGS
2009 Making Fast-Twitch Dystrophic Muscles Bigger Protects Them from Contraction Injury and Attenuates the Dystrophic Pathology. Am J Pathol
45. CrossRA
StewartM
Kendrick-JonesJ
1990 Structural predictions for the central domain of dystrophin. FEBS Lett 262 87 92
46. YanY
WinogradE
VielA
CroninT
HarrisonSC
1993 Crystal structure of the repetitive segments of spectrin. Science 262 2027 2030
47. KahanaE
MarshPJ
HenryAJ
WayM
GratzerWB
1994 conformation and phasing of dystrophin structural repeats. J Mol Biol 235 1271 1277
48. CalvertR
KahanaE
GratzerWB
1996 Stability of the dystrophin rod domain fold: evidence for nested repeating units. Biophys J 71 1605 1610
49. PascualJ
CastresanaJ
SarasteM
1997 Evolution of the spectrin repeat. Bioessays 19 811 817
50. BroderickMJ
WinderSJ
2002 Towards a complete atomic structure of spectrin family proteins. J Struct Biol 137 184 193
51. HarperSQ
CrawfordRW
DelloRussoC
ChamberlainJS
2002 Spectrin-like repeats from dystrophin and alpha-actinin-2 are not functionally interchangeable. Hum Mol Genet 11 1807 1815
52. SaadatL
PittmanL
MenhartN
2006 Structural cooperativity in spectrin type repeats motifs of dystrophin. Biochim Biophys Acta 1764 943 954
53. WinderSJ
GibsonTJ
Kendrick-JonesJ
1995 Dystrophin and utrophin: the missing links! FEBS Lett 369 27 33
54. KahanaE
GratzerWB
1995 Minimum folding unit of dystrophin rod domain. Biochemistry 34 8110 8114
55. PascualJ
PfuhlM
WaltherD
SarasteM
NilgesM
1997 Solution structure of the spectrin repeat: a left-handed antiparallel triple-helical coiled-coil. J Mol Biol 273 740 751
56. ErvastiJM
CampbellKP
1993 A role for the dystrophin-glycoprotein complex as a transmembrane linker between laminin and actin. J Cell Biol 122 809 823
57. RybakovaIN
PatelJR
ErvastiJM
2000 The dystrophin complex forms a mechanically strong link between the sarcolemma and costameric actin. J Cell Biol 150 1209 1214
58. ErvastiJM
2003 Costameres: the Achilles' heel of Herculean muscle. J Biol Chem 278 13591 13594
59. OzawaE
2006 The functional biology of dystrophin: structural components and the pathogenesis of Duchenne muscular dystrophy.
ChamberlainJS
RandoTA
Duchenne Muscular Dystrophy: Advances in Therapeutics New York Taylor and Francis 21 54
60. MoradiM
BabinV
RolandC
DardenTA
SaguiC
2009 Conformations and free energy landscapes of polyproline peptides. Proc Natl Acad Sci U S A
61. Ishikawa-SakuraiM
YoshidaM
ImamuraM
DaviesKE
OzawaE
2004 ZZ domain is essentially required for the physiological binding of dystrophin and utrophin to beta-dystroglycan. Hum Mol Genet 13 693 702
62. LaiY
ThomasGD
YueY
YangHT
LiD
2009 Dystrophins carrying spectrin-like repeats 16 and 17 anchor nNOS to the sarcolemma and enhance exercise performance in a mouse model of muscular dystrophy. J Clin Invest 119 624 635
63. OdomGL
GregorevicP
AllenJM
FinnE
ChamberlainJS
2008 Microutrophin delivery through rAAV6 increases lifespan and improves muscle function in dystrophic dystrophin/utrophin-deficient mice. Mol Ther 16 1539 1545
64. SonnemannKJ
Heun-JohnsonH
TurnerAJ
BaltgalvisKA
LoweDA
2009 Functional substitution by TAT-utrophin in dystrophin-deficient mice. PLoS Med 6 e1000083 doi:10.1371/journal.pmed.1000083
65. ChamberlainJ
2004 PCR-mediated mutagenesis. In: Nature Encyclopedia of Life Sciences. Nature Publishing Group, London
66. PetersMF
AdamsME
FroehnerSC
1997 Differential association of syntrophin pairs with the dystrophin complex. J Cell Biol 138 81 93
67. CooperST
LoHP
NorthKN
2003 Single section Western blot: improving the molecular diagnosis of the muscular dystrophies. Neurology 61 93 97
68. ErvastiJM
OhlendieckK
KahlSD
GaverMG
CampbellKP
1990 Deficiency of a glycoprotein component of the dystrophin complex in dystrophic muscle. Nature 345 315 319
Štítky
Genetika Reprodukční medicína
Článek Epistasis of Transcriptomes Reveals Synergism between Transcriptional Activators Hnf1α and Hnf4αČlánek SMA-10/LRIG Is a Conserved Transmembrane Protein that Enhances Bone Morphogenetic Protein SignalingČlánek B1 SOX Coordinate Cell Specification with Patterning and Morphogenesis in the Early Zebrafish EmbryoČlánek Bulk Segregant Analysis by High-Throughput Sequencing Reveals a Novel Xylose Utilization Gene fromČlánek Abundant Quantitative Trait Loci Exist for DNA Methylation and Gene Expression in Human Brain
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2010 Číslo 5
-
Všechny články tohoto čísla
- Digital Quantification of Human Eye Color Highlights Genetic Association of Three New Loci
- Common Genetic Variants near the Brittle Cornea Syndrome Locus Influence the Blinding Disease Risk Factor Central Corneal Thickness
- Age- and Temperature-Dependent Somatic Mutation Accumulation in
- All About Mitochondrial Eve: An Interview with Rebecca Cann
- Aging and Chronic Sun Exposure Cause Distinct Epigenetic Changes in Human Skin
- Transposed Genes in Arabidopsis Are Often Associated with Flanking Repeats
- A Survey of Genomic Traces Reveals a Common Sequencing Error, RNA Editing, and DNA Editing
- Gene Transposition Causing Natural Variation for Growth in
- The Polyproline Site in Hinge 2 Influences the Functional Capacity of Truncated Dystrophins
- Epistasis of Transcriptomes Reveals Synergism between Transcriptional Activators Hnf1α and Hnf4α
- Integration of Light Signals by the Retinoblastoma Pathway in the Control of S Phase Entry in the Picophytoplanktonic Cell
- The Proprotein Convertase Encoded by () Is Required in Corpora Cardiaca Endocrine Cells Producing the Glucose Regulatory Hormone AKH
- Sgs1 and Exo1 Redundantly Inhibit Break-Induced Replication and Telomere Addition at Broken Chromosome Ends
- A MATE-Family Efflux Pump Rescues the 8-Oxoguanine-Repair-Deficient Mutator Phenotype and Protects Against HO Killing
- The Relationship among Gene Expression, the Evolution of Gene Dosage, and the Rate of Protein Evolution
- SMA-10/LRIG Is a Conserved Transmembrane Protein that Enhances Bone Morphogenetic Protein Signaling
- The Nuclear Receptor DHR3 Modulates dS6 Kinase–Dependent Growth in
- Involvement of Global Genome Repair, Transcription Coupled Repair, and Chromatin Remodeling in UV DNA Damage Response Changes during Development
- B1 SOX Coordinate Cell Specification with Patterning and Morphogenesis in the Early Zebrafish Embryo
- Linkage and Association Mapping of Flowering Time in Nature
- Bulk Segregant Analysis by High-Throughput Sequencing Reveals a Novel Xylose Utilization Gene from
- Abundant Quantitative Trait Loci Exist for DNA Methylation and Gene Expression in Human Brain
- Ablation of Whirlin Long Isoform Disrupts the USH2 Protein Complex and Causes Vision and Hearing Loss
- Characterization of Oxidative Guanine Damage and Repair in Mammalian Telomeres
- DNA Adenine Methylation Is Required to Replicate Both Chromosomes Once per Cell Cycle
- Genome-Wide Copy Number Variation in Epilepsy: Novel Susceptibility Loci in Idiopathic Generalized and Focal Epilepsies
- FACT Prevents the Accumulation of Free Histones Evicted from Transcribed Chromatin and a Subsequent Cell Cycle Delay in G1
- GC-Biased Evolution Near Human Accelerated Regions
- Liver and Adipose Expression Associated SNPs Are Enriched for Association to Type 2 Diabetes
- Myeloid Cell-Restricted Insulin Receptor Deficiency Protects Against Obesity-Induced Inflammation and Systemic Insulin Resistance
- The Mating Type Locus () and Sexual Reproduction of : Insights into the Evolution of Sex and Sex-Determining Chromosomal Regions in Fungi
- B-Cyclin/CDKs Regulate Mitotic Spindle Assembly by Phosphorylating Kinesins-5 in Budding Yeast
- Post-Replication Repair Suppresses Duplication-Mediated Genome Instability
- Genome Sequence of the Plant Growth Promoting Endophytic Bacterium sp. 638
- Transcription Factors Mat2 and Znf2 Operate Cellular Circuits Orchestrating Opposite- and Same-Sex Mating in
- The Use of Orthologous Sequences to Predict the Impact of Amino Acid Substitutions on Protein Function
- Mutation in Archain 1, a Subunit of COPI Coatomer Complex, Causes Diluted Coat Color and Purkinje Cell Degeneration
- Shelterin-Like Proteins and Yku Inhibit Nucleolytic Processing of Telomeres
- Affecting Function Causes a Dilated Heart in Adult
- Manipulation of Behavioral Decline in with the Rag GTPase
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Common Genetic Variants near the Brittle Cornea Syndrome Locus Influence the Blinding Disease Risk Factor Central Corneal Thickness
- All About Mitochondrial Eve: An Interview with Rebecca Cann
- The Relationship among Gene Expression, the Evolution of Gene Dosage, and the Rate of Protein Evolution
- SMA-10/LRIG Is a Conserved Transmembrane Protein that Enhances Bone Morphogenetic Protein Signaling
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



