-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaGenome-Wide Copy Number Variation in Epilepsy: Novel Susceptibility Loci in Idiopathic Generalized and Focal Epilepsies
Epilepsy is one of the most common neurological disorders in humans with a prevalence of 1% and a lifetime incidence of 3%. Several genes have been identified in rare autosomal dominant and severe sporadic forms of epilepsy, but the genetic cause is unknown in the vast majority of cases. Copy number variants (CNVs) are known to play an important role in the genetic etiology of many neurodevelopmental disorders, including intellectual disability (ID), autism, and schizophrenia. Genome-wide studies of copy number variation in epilepsy have not been performed. We have applied whole-genome oligonucleotide array comparative genomic hybridization to a cohort of 517 individuals with various idiopathic, non-lesional epilepsies. We detected one or more rare genic CNVs in 8.9% of affected individuals that are not present in 2,493 controls; five individuals had two rare CNVs. We identified CNVs in genes previously implicated in other neurodevelopmental disorders, including two deletions in AUTS2 and one deletion in CNTNAP2. Therefore, our findings indicate that rare CNVs are likely to contribute to a broad range of generalized and focal epilepsies. In addition, we find that 2.9% of patients carry deletions at 15q11.2, 15q13.3, or 16p13.11, genomic hotspots previously associated with ID, autism, or schizophrenia. In summary, our findings suggest common etiological factors for seemingly diverse diseases such as ID, autism, schizophrenia, and epilepsy.
Published in the journal: . PLoS Genet 6(5): e32767. doi:10.1371/journal.pgen.1000962
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1000962Summary
Epilepsy is one of the most common neurological disorders in humans with a prevalence of 1% and a lifetime incidence of 3%. Several genes have been identified in rare autosomal dominant and severe sporadic forms of epilepsy, but the genetic cause is unknown in the vast majority of cases. Copy number variants (CNVs) are known to play an important role in the genetic etiology of many neurodevelopmental disorders, including intellectual disability (ID), autism, and schizophrenia. Genome-wide studies of copy number variation in epilepsy have not been performed. We have applied whole-genome oligonucleotide array comparative genomic hybridization to a cohort of 517 individuals with various idiopathic, non-lesional epilepsies. We detected one or more rare genic CNVs in 8.9% of affected individuals that are not present in 2,493 controls; five individuals had two rare CNVs. We identified CNVs in genes previously implicated in other neurodevelopmental disorders, including two deletions in AUTS2 and one deletion in CNTNAP2. Therefore, our findings indicate that rare CNVs are likely to contribute to a broad range of generalized and focal epilepsies. In addition, we find that 2.9% of patients carry deletions at 15q11.2, 15q13.3, or 16p13.11, genomic hotspots previously associated with ID, autism, or schizophrenia. In summary, our findings suggest common etiological factors for seemingly diverse diseases such as ID, autism, schizophrenia, and epilepsy.
Introduction
Epilepsy is one of the most common neurological disorders in humans with a prevalence of ∼1% and a lifetime incidence of up to 3% [1]. The epilepsies present with a broad range of clinical features, and over 50 distinct epilepsy syndromes are now recognized. Particularly in a pediatric setting, a broad range of different epilepsy syndromes can be distinguished. Seizure disorders can roughly be divided into idiopathic or symptomatic epilepsies. While symptomatic epilepsies are due to an identifiable cause such as metabolic disorders, brain trauma or intracranial tumors, idiopathic seizure disorders occur in the absence of identifiable causal factors and are thought to have a strong genetic contribution.
Although it has long been observed that the idiopathic epilepsies have a genetic component, the genetic etiology of only a small fraction of cases can be determined. The role of copy number variants (CNVs) in intellectual disability (ID) [2]–[8], autism [9]–[14] and schizophrenia [15]–[19] has been extensively investigated. It has become increasingly clear that, collectively, rare variants contribute significantly to the etiology of these common diseases–following the rare variant common disease hypothesis. We hypothesize this can be extended to other neurological disorders and that rare CNVs significantly contribute to the genetic etiology of epilepsy.
Recently, in a study targeted to six genomic regions, recurrent microdeletions on chromosome 15q13.3, 16p13.11 and 15q11.2 were identified as important genetic factors predisposing to idiopathic generalized epilepsy (IGE) [20]–[22]. Here, we carry out whole-genome array comparative genomic hybridization (CGH) in a cohort of 517 individuals with mixed types of idiopathic epilepsy in order to discover novel copy number changes associated with epilepsy. We find recurrent microdeletions of 15q13.3, 16p13.11 and 15q11.2 each in ∼1% of affected individuals, confirming previous studies [20]–[22]. In addition to recurrent rearrangements at rearrangement-prone regions, we show that, overall, 8.9% of affected individuals have one or more rare copy number changes involving at least one gene.
Results
We performed genome-wide array CGH to detect copy number changes in 517 patients with mixed types of epilepsy. Of these, 399 have idiopathic generalized epilepsy (IGE), 50 have benign epilepsy with centrotemporal spikes (BECTS) and 68 have other types of idiopathic seizure disorders (Table 1). We used a custom microarray with high-density targeted coverage of 107 regions of the genome flanked by large, highly homologous duplications, termed rearrangement hotspots [23]. In addition, probes were evenly spaced throughout the remainder of the genome with average probe spacing of ∼38 kb. Overall, we find that 46 probands (8.9%) carry one or more rare CNVs not previously reported in the 2493 unrelated controls [24]. The rare CNVs detected in our cohort range in size from 13 kb to 15.9 Mb (average 1.2 Mb; median 600 kb), and the majority (69%) are deletion events.
Tab. 1. Phenotypes of probands evaluated by array CGH. 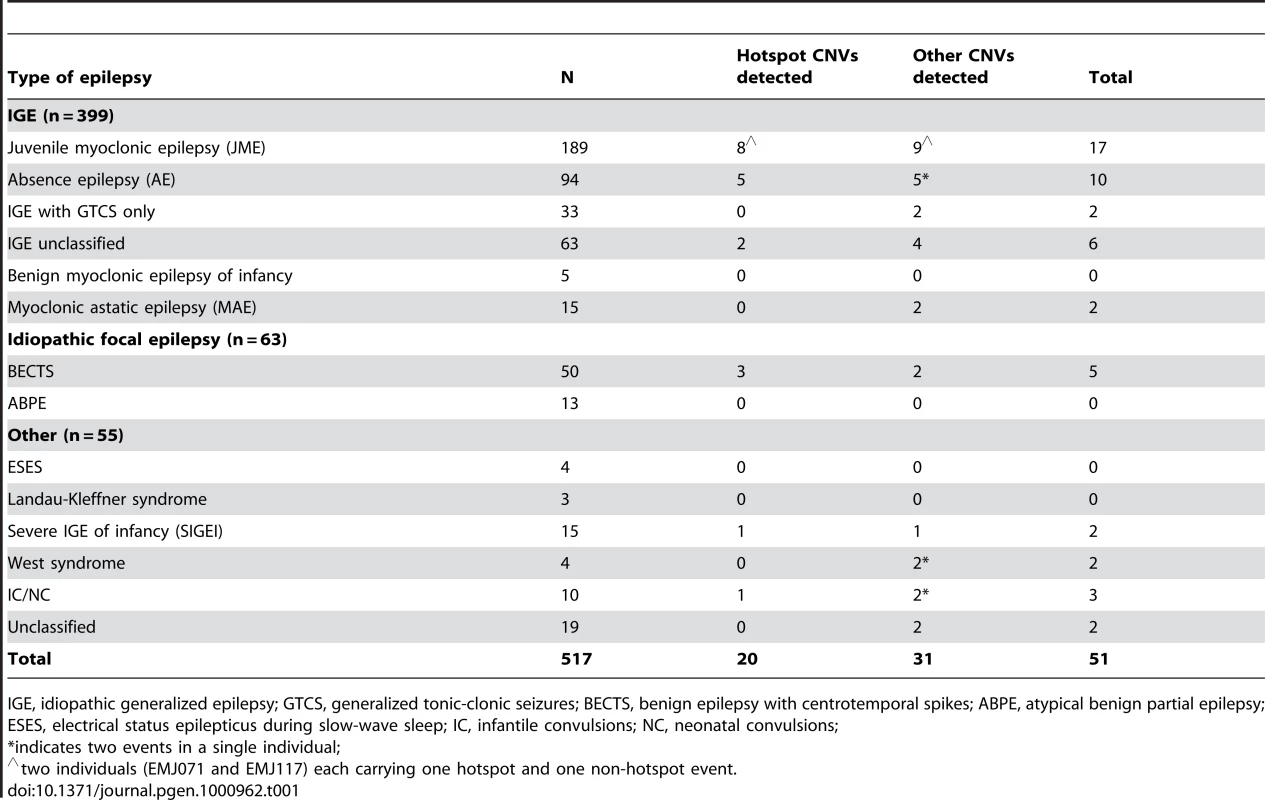
IGE, idiopathic generalized epilepsy; GTCS, generalized tonic-clonic seizures; BECTS, benign epilepsy with centrotemporal spikes; ABPE, atypical benign partial epilepsy; ESES, electrical status epilepticus during slow-wave sleep; IC, infantile convulsions; NC, neonatal convulsions; Rearrangements at genomic hotspots
We first evaluated rearrangement hotspots for copy number changes. We found 20 probands (3.9%) with copy number changes at known rearrangement hotspots including 15q13.3 deletions (n = 5), 16p13.11 deletions (n = 5), 15q11.2 BP1–BP2 deletions (n = 5), 1q21.1 deletions (n = 2), a 16p12.1 deletion (n = 1), a 16p11.2 duplication (n = 1) and a more distal 16p11.2 deletion (n = 1) (Table 2, Figure 1). We also identified four individuals with duplications of 15q11.2 BP1–BP2; because duplications of this region are frequent in the general population, we classified these duplications as polymorphic events. These results confirm our previous studies and emphasize the importance of deletions of 15q13.3, 16p13.11 and 15q11.2 BP1–BP2 as frequent genetic susceptibility factors in epilepsy [20]–[22]. All three regions have also been associated with ID, autism and/or schizophrenia [15], [17], [25]–[32], as have deletions at 1q21.1 [33], [34], two distinct regions of 16p11.2 [10], [14], [35]–[37] and 16p12 [38], which were also detected in our cohort. Deletions of 16p13.11 (5/517 vs 0/2493 controls, p = 0.00014, Fisher's exact test), 15q13.3 (5/517 vs 0/2493, p = 0.00014) and 15q11.2 (5/517 vs. 4/2493, p = 0.010) are significantly enriched in our epilepsy cohort and together account for 2.9% of cases.
Fig. 1. Deletions and duplications at genomic rearrangement hotspots in 20 probands. 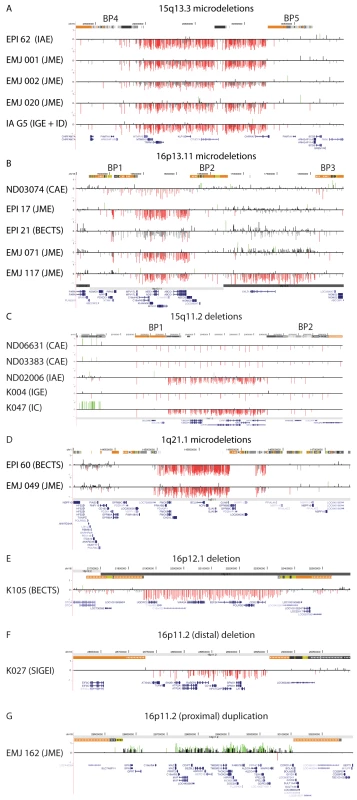
Array CGH results are depicted for (A) 15q13.3, chr15: 28.0–31.0 Mb, (B) 16p13.11, chr16: 14.5–18.5 Mb, (C) 15q11.2, chr15: 20.0–20.9 Mb, (D) 1q21.1, chr1: 144.0–147.5 Mb, (E) 16p12.1, chr16: 21.6–22.6 Mb, (F) 16p11.2, chr16:28.6–29.1 Mb, and (G) 16p11.2, chr16: 29.0–30.3 Mb. For each individual, deviations of probe log2 ratios from 0 are depicted by gray and black lines. Those exceeding a threshold of 1.5 s.d. from the mean probe ratio are colored green and red to represent relative gains and losses, respectively. Segmental duplications of increasing similarity (90–98%, 98–99%, and >99%) are represented by gray, yellow, and orange bars, respectively. RefSeq genes are depicted in blue. Tab. 2. Rare copy number variants in 517 patients with epilepsy. 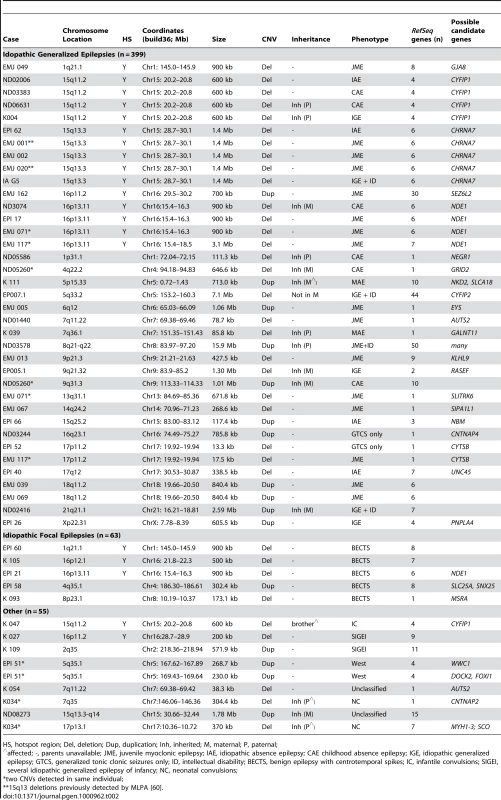
HS, hotspot region; Del, deletion; Dup, duplication; Inh, inherited; M, maternal; P, paternal; Rare or unique deletions involving potential candidate genes
We next focused on non-hotspot CNVs that overlap one or more genes and are not present in the control cohort of 2493 individuals [24]. We identified 28 individuals with at least one rare gene-containing deletion or duplication, and five individuals each carry two rare CNVs (Table 2). Fifteen of the events we detected involve a single gene. Two genes were altered in two patients each: AUTS2 deletions were identified in one proband with juvenile myoclonic epilepsy (JME) and one proband with unclassified non-lesional epilepsy with features of atypical benign partial epilepsy (ABPE) [39]. Deletions involving CTYSB (SPECC1) were identified in two probands with IGE. All other single-gene CNVs were seen only once. Seventeen events involved multiple genes, one of which was observed in two different individuals with JME (duplication of 18q11, Table 2).
Individuals with multiple rare CNVs
We found five individuals with two rare CNVs (Figure 2). Two patients with JME and a deletion of 16p13.11 (EMJ071 and EMJ117) each have a second rare deletion. EMJ071 has a large deletion on chromosome 13 that removes the SLITRK6 gene, a member of the SLITRK gene family involved in controlling neurite outgrowth; individual EMJ117 also has a deletion involving the CTYSB gene. Case ND05260 (childhood absence epilepsy, CAE) carries a 647-kb deletion within the GRID2 gene, which encodes a glutamate receptor expressed in the cerebellum, and a 1-Mb duplication of 9q31. Though both are maternally inherited, neither has been reported in controls. Case EPI 51 (idiopathic West syndrome) has two apparently independent duplications of chromosome 5q35, each containing several genes. Finally, we identified one proband with neonatal convulsions (NC) carrying a deletion within the CNTNAP2 gene that spans exons 2–4 as well as a 370-kb deletion of 17p13 involving 7 genes.
Fig. 2. Two rare CNVs in five probands. 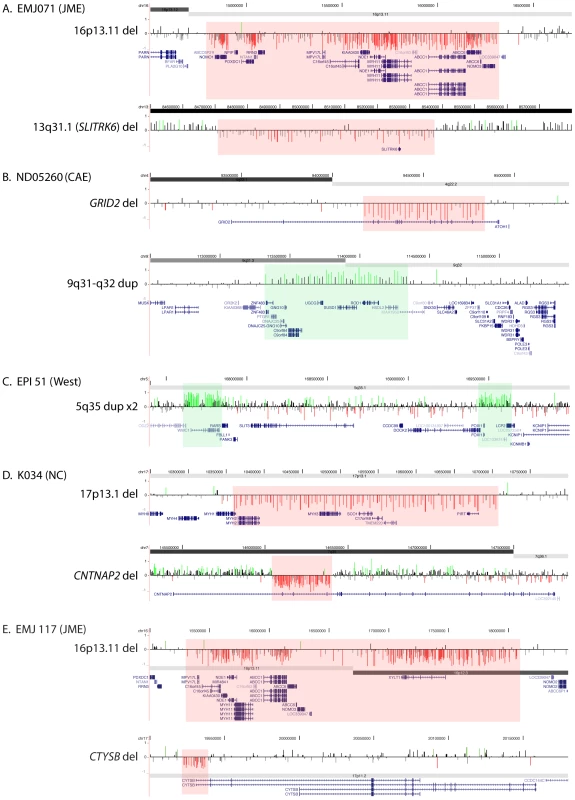
Array CGH results are shown for the for two rare CNVs detected in probands EMJ071 (A), ND05260 (B), EPI51 (C), K034 (D), and EMJ117 (E). Array CGH results are depicted as in Figure 1; segmental duplications are not shown in this figure. DNA from one of more family members was available for analysis in 14 cases. Inheritance, if determined, is shown in Table 2. In twelve cases, we determined that one or both CNVs in the proband were inherited; in three cases the transmitting parent is also affected. In one case (EP007.1), the CNV was not found in the mother, but the father was unavailable. In another case (K047), parents were unavailable, but a brother was found to carry the same CNV suggesting one of the parents carries the same CNV.
Discussion
In this study, we performed whole-genome array CGH in a series of 517 individuals with a presenting diagnosis of idiopathic epilepsy in order to discover novel copy number changes associated with epilepsy. While our previous studies were targeted to specific genomic regions in probands with IGE [21], [22], here we present data from whole-genome analysis on probands with IGE and extend our analysis to other idiopathic epilepsy syndromes. In total, we identified 46 individuals (8.9%) with 51 rearrangements that may be pathogenic as they were not found in controls or were significantly enriched in our epilepsy cohort.
Hotspot rearrangements
Rearrangements at several genomic hotspots have been associated with a range of neurocognitive disorders. In our cohort of 517 probands with epilepsy, we find deletions at 15q13.3, 16p13.11 and 15q11.2 in 2.9% of our cases. Interestingly, all of the deletions of 15q13.3 (n = 5) and 4/5 deletions at 16p13.11 and 15q11.2 were in probands with IGE, accounting for 3.3% of the patients with IGE in our cohort confirming our previous findings. While it is possible that deletions of 15q13.3 are also predisposing to non-IGE epilepsy syndromes, we did not find this to be the case in our series (n = 118). Additional large cohorts of patients with focal epilepsy or epileptic encephalopathy will be required to determine whether these deletions also play a significant role in other subtypes of epilepsy.
Deletions of 16p13.11 have previously been associated with intellectual disability +/ − congenital anomalies in one study [26]. Three of four probands with 16p13.11 deletions in that series had epilepsy; two further fetal cases had brain abnormalities. The findings in this cohort and one previous study of IGE [20] suggest that deletions of 16p13.11 are more frequent in epilepsy (0.5–1% of cases) than in other phenotypes including ID and autism [26], [27], [32], and may be as frequent as 15q13.3 deletions in individuals with IGE. Deletions and duplications of this region have also been reported in schizophrenia, though the associations have not been statistically significant [16], [29].
Deletions of 15q13.3, detected in five individuals with IGE in our series, have been associated with a wide range of phenotypes including ID, autism, epilepsy and schizophrenia [15], [17], [20]–[22], [25], [28], [30], [31], [40]. The gene within the 15q13.3 region that is most likely responsible for the epilepsy phenotype is CHRNA7, a subunit of the nicotinic acetylcholine receptor. At least two small studies have failed to identify causal point mutations in the CHRNA7 gene in autosomal dominant nocturnal frontal lobe epilepsy [41] and JME [42], but additional studies should be performed to further evaluate affected individuals for mutations. A recent publication identifying atypical rearrangements with exclusive deletions of CHRNA7 further emphasizes the importance of CHRNA7 as the main candidate gene in this region [43].
Compared to the above structural genomic variants, copy number variation at 15q11.2 between breakpoints BP1 and BP2 of the Prader-Willi and Angelman syndrome region is more common in the general population with the BP1–BP2 deletion present in 0.2% of unaffected individuals. Despite this, deletions between BP1 and BP2 have now been reported as enriched in patients with schizophrenia [16], [17], ID [27] and epilepsy [20]. Furthermore, there is evidence that patients with Prader-Willi or Angelman syndrome who have deletions including BP1–BP2 are more severely affected [44]–[46]. In this study, we also find enrichment of deletions at this locus in affected individuals. Together, these studies suggest that deletion of the 15q11.2 BP1–BP2 region confers susceptibility to a wide range of neuropsychiatric conditions, albeit with incomplete penetrance.
Two patients in our series, one each with JME and BECTS, have deletions of 1q21.1, which have been previously associated with a wide range of phenotypes, including intellectually disability and developmental delay [33], [34], schizophrenia [15], [17], [18], congenital heart disease [47], [48] and cataracts [34], [49]. In two large studies of patients who present primarily with cognitive or developmental delay, 5/42 (11.9%) patients also had seizures [33], [34]; 1 of 10 patients with schizophrenia and a 1q21.1 deletion also had epilepsy [15]. Identifying 1q21.1 microdeletions in patients with idiopathic generalized and idiopathic focal epilepsies suggests that variation at this locus predisposes to a broad range of seizure disorders crossing traditional diagnostic boundaries.
In addition, we identified one patient (EMJ162) with JME and a duplication of 16p11.2 (chr16 : 29.5–30.2 Mb), which has been associated with autism, developmental delay and schizophrenia [10]–[12], [14], [27], [35], [37]. Finally, we identified one individual with severe idiopathic generalized epilepsy of infancy (SIGEI) (K027) with a more distal deletion of 16p11.2 (chr16 : 27.7–28.9 Mb), recently associated with severe early-onset obesity and ID [36], and one patient with BECTS (K105) and a deletion of 16p12.1 (chr16 : 20.2–20.8 Mb), also associated with ID and other neurodevelopmental defects [38]. Thus, our data adds to the phenotypic spectrum associated with rearrangements at several genomic hotspot regions. In particular, we identify hotspot deletions in two patients with BECTS. Gene identification in BECTS, despite representing the most common focal epilepsy syndrome of childhood, has been elusive so far. Here, we suggest that some recurrent hotspot deletions might predispose to both idiopathic generalized and focal epilepsies.
Non-hotspot rearrangements
We detected 18 deletions and 16 duplications that are not associated with rearrangement hotspots. Fifteen events involve a single gene; of these, 12 are deletions. Although all of the CNVs reported here are not found in our control set of 2493 individuals, it is possible that some are rare but benign CNVs. However, many of the CNVs we identified contain one of more plausible candidate genes for epilepsy (Table 2).
We identified a deletion of exons 2–4 in the CNTNAP2 gene in a proband with neonatal seizures. CNTNAP2 has been identified as a candidate gene for autism [50]–[52], and heterozygous deletions involving the gene were reported in three patients with schizophrenia and autism [53]. The deletion is predicted to cause an in-frame deletion of 153 amino acids in the resulting protein. The same patient has a 370-kb deletion of 17p13 that deletes seven genes and has not been seen in our control cohort. We also identified a patient with a duplication encompassing a related gene, CNTNAP4. Finally, two individuals in our cohort have overlapping deletions within AUTS2. This gene is disrupted by de novo balanced translocations in three unrelated individuals with mental retardation [54] and a pair of twins with autism and mental retardation [55], suggesting a role for AUTS2 in normal cognitive development. The two deletions we detected are intragenic and overlapping.
CNVs in epilepsy subtypes
Previous studies of CNVs in epilepsy have focused on probands with IGE. It is known from studies of families with autosomal dominant epilepsy that a wide range of seizure types can be caused by the same single-gene mutation. For example, Dravet syndrome, a severe early-onset disorder associated with poor cognitive outcome, and the milder generalized epilepsy with febrile seizures plus (GEFS+) syndrome are both caused by mutations in the SCN1A gene [56]–[58]. Therefore, we included probands with common idiopathic focal epilepsies and non-lesional, idiopathic epilepsies. Some of our probands were diagnosed with specific epilepsy syndromes, including myoclonic astatic epilepsy (Doose Syndrome), atypical benign partial epilepsy [39], Landau-Kleffner syndrome, idiopathic West syndrome, severe idiopathic generalized epilepsy of infancy [59] and benign neonatal or infantile seizures. These particular epilepsy syndromes are usually associated with normal MRI results. We find that 6.6% of probands with IGE and 7.9% of those with idiopathic focal epilepsy harbor rare CNVs that may underlie their epilepsy phenotype. Notably, 12.7% of patients with other, often more severe forms of epilepsy in our series carry one or more rare CNVs. In our series, the vast majority of patients with deletions of 15q13.3, 16p13.11 and 15q11.2 BP1–BP2 were in the IGE cohort, accounting for 3.3% of cases. In the non-IGE patients, a deletion of 15q11.2 was found in a single patient with infantile seizures and a deletion of 16p13.11 was found in one patient with BECTS, suggesting that deletions at these three genomic hotspots confer greater risk for IGE than other types of epilepsy.
In summary, we find that 46/517 probands (8.9%) with various forms of idiopathic epilepsy carry one or more rare CNVs that may predispose to seizures, a frequency similar to that in studies of patients who present with other neurocognitive phenotypes, including ID, autism and schizophrenia. Furthermore, we identified CNVs involving genes and genomic regions previously identified in patients with the neurocognitive phenotypes listed above, suggesting common genetic etiological factors for these disorders. Our data suggest that rare CNVs are important in many subtypes of idiopathic epilepsies, including idiopathic generalized and idiopathic focal epilepsies as well as specific idiopathic, non-lesional epilepsy syndromes. The genomic regions and genes identified in this study are potential novel candidate genes for epilepsy.
Materials and Methods
Ethics statement
Patients were collected at five centers after appropriate human subjects approval and informed consent at each site.
Patient cohorts
Patients were collected at five centers: (1) 140 probands with a primary diagnosis of JME, CAE, absence epilepsy, IGE or idiopathic epilepsy were selected from the NINDS repository (http://ccr.coriell.org/ninds); (2) 160 patients are probands with a primary diagnosis of JME from Switzerland. Patients from cohorts (1) and (2) were previously analyzed using MLPA for the CHRNA7 gene [60], and two probands (EMJ001 and EMJ020) were determined to have 15q13.3 microdeletions by that method; they were not previously analyzed for any other copy number changes. (3) 186 German patients came from two cohorts: 76 patients from a population-based cohort from Northern Germany (POPGEN cohort) and 110 patients with childhood-onset epilepsy collected at the University of Kiel. Finally, 41 patients with various idiopathic generalized epilepsies collected at (4) the University of Iowa and (5) at Washington University, St. Louis. DNA from the NINDS repository was derived from cell lines; DNA from all other cohorts was directly from blood. Patients were diagnosed according to the widely used 1989 ILAE classification [61]. In addition, several pediatric patients were diagnosed with specific syndromes not yet recognized in the ILAE classification (Table 1). Patients with non-lesional, idiopathic epilepsies in which diagnostic criteria of the recent ILAE classification for particular epilepsy syndromes were not met were labeled as “unclassified”.
Array comparative genomic hybridization (CGH)
Array CGH was performed using either custom or commercially available oligonucleotide arrays containing 135,000 isothermal probes (Roche NimbleGen, Inc.). Customized arrays (459 samples) were designed with higher density probe coverage in known rearrangement hotspot regions (average probe spacing 2.5 kb) with lower density whole-genome backbone coverage (average probe spacing 38 kb). A subset of samples (n = 62) was analyzed using a commercially available whole-genome array (Roche NimbleGen 12×135 k whole-genome tiling array) with average probe spacing throughout the genome of 21 kb.
Data analysis
Data were analyzed according to manufacturer's instructions using NimbleScan software to generate normalized log2 fluorescence intensity ratios. Then, for each sample, normalized log intensity ratios are transformed into z-scores using the chromosome-specific mean and standard deviation. Z-scores are subsequently used to classify probes as “increased”, “normal” and “decreased” copy-number using a three-state Hidden Markov Model (HMM). The HMM was implemented using HMMSeg [62], which assumes Gaussian emission probabilities. The “increased” and “decreased” states are defined to have the same standard deviation as the “normal” state but with mean z-score two standard deviations above and below the mean, respectively. Probe-by-probe HMM state assignments are merged into segments according to the following criteria: consecutive probes of the same state less than 50 kb apart are merged, and if two segments of the same state are separated by an intervening sequence of ≤5 probes and ≤10 kb, both segments and intervening sequence are called as a single variant. CNV calls are filtered to eliminate (i) events containing <5 probes, (ii) CNVs with >50% overlap in a series of 2493 control individuals [24] and (iii) events that had no overlap with RefSeq genes. In addition, when comparing CNV calls to control CNVs, we eliminated calls for which there was insufficient probe coverage (<5 probes) in the control data to identify the same or similar CNV. Filtered copy number changes are also visually inspected in a genome browser.
Zdroje
1. HauserWA
AnnegersJF
RoccaWA
1996 Descriptive epidemiology of epilepsy: contributions of population-based studies from Rochester, Minnesota. Mayo Clin Proc 71 576 586
2. de VriesBB
PfundtR
LeisinkM
KoolenDA
VissersLE
2005 Diagnostic genome profiling in mental retardation. Am J Hum Genet 77 606 616
3. FriedmanJM
BarossA
DelaneyAD
AllyA
ArbourL
2006 Oligonucleotide microarray analysis of genomic imbalance in children with mental retardation. Am J Hum Genet 79 500 513
4. KoolenDA
VissersLE
PfundtR
de LeeuwN
KnightSJ
2006 A new chromosome 17q21.31 microdeletion syndrome associated with a common inversion polymorphism. Nat Genet 38 999 1001
5. SagooG
ButterworthA
SandersonS
Shaw-SmithC
HigginsJ
2009 Array CGH in patients with learning disability (mental retardation) and congenital anomalies: updated systematic review and meta-analysis of 19 studies and 13,926 subjects. Genet Med 11 139 146
6. ShafferLG
KashorkCD
SalekiR
RoremE
SundinK
2006 Targeted genomic microarray analysis for identification of chromosome abnormalities in 1500 consecutive clinical cases. J Pediatr 149 98 102
7. SharpAJ
HansenS
SelzerRR
ChengZ
ReganR
2006 Discovery of previously unidentified genomic disorders from the duplication architecture of the human genome. Nat Genet 38 1038 1042
8. Shaw-SmithC
PittmanAM
WillattL
MartinH
RickmanL
2006 Microdeletion encompassing MAPT at chromosome 17q21.3 is associated with developmental delay and learning disability. Nat Genet 38 1032 1037
9. ChristianSL
BruneCW
SudiJ
KumarRA
LiuS
2008 Novel submicroscopic chromosomal abnormalities detected in autism spectrum disorder. Biol Psychiatry 63 1111 1117
10. KumarRA
KaraMohamedS
SudiJ
ConradDF
BruneC
2008 Recurrent 16p11.2 microdeletions in autism. Hum Mol Genet 17 628 638
11. MarshallCR
NoorA
VincentJB
LionelAC
FeukL
2008 Structural variation of chromosomes in autism spectrum disorder. Am J Hum Genet 82 477 488
12. SebatJ
LakshmiB
MalhotraD
TrogeJ
Lese-MartinC
2007 Strong association of de novo copy number mutations with autism. Science 316 445 449
13. SzatmariP
PatersonAD
ZwaigenbaumL
RobertsW
BrianJ
2007 Mapping autism risk loci using genetic linkage and chromosomal rearrangements. Nat Genet 39 319 328
14. WeissLA
ShenY
KornJM
ArkingDE
MillerDT
2008 Association between microdeletion and microduplication at 16p11.2 and autism. N Engl J Med 358 667 675
15. International Schizophrenia Consortium 2008 Rare chromosomal deletions and duplications increase risk of schizophrenia. Nature 455 237 241
16. KirovG
GrozevaD
NortonN
IvanovD
MantripragadaKK
2009 Support for the involvement of large cnvs in the pathogenesis of schizophrenia. Hum Mol Genet 18 1497 1503
17. StefanssonH
RujescuD
CichonS
PietilainenOP
IngasonA
2008 Large recurrent microdeletions associated with schizophrenia. Nature 455 232 236
18. WalshT
McClellanJM
McCarthySE
AddingtonAM
PierceSB
2008 Rare structural variants disrupt multiple genes in neurodevelopmental pathways in schizophrenia. Science 320 539 543
19. XuB
RoosJL
LevyS
van RensburgEJ
GogosJA
2008 Strong association of de novo copy number mutations with sporadic schizophrenia. Nat Genet 40 880 885
20. de KovelCG
TrucksH
HelbigI
MeffordHC
BakerC
2009 Recurrent microdeletions at 15q11.2 and 16p13.11 predispose to idiopathic generalized epilepsies. Brain
21. DibbensLM
MullenS
HelbigI
MeffordHC
BaylyMA
2009 Familial and sporadic 15q13.3 microdeletions in idiopathic generalized epilepsy: precedent for disorders with complex inheritance. Hum Mol Genet 18 3626 3631
22. HelbigI
MeffordHC
SharpAJ
GuipponiM
FicheraM
2009 15q13.3 microdeletions increase risk of idiopathic generalized epilepsy. Nat Genet 41 160 162
23. BaileyJA
GuZ
ClarkRA
ReinertK
SamonteRV
2002 Recent segmental duplications in the human genome. Science 297 1003 1007
24. ItsaraA
CooperGM
BakerC
GirirajanS
LiJ
2009 Population analysis of large copy number variants and hotspots of human genetic disease. Am J Hum Genet 84 148 161
25. Ben-ShacharS
LanpherB
GermanJR
QasaymehM
PotockiL
2009 Microdeletion 15q13.3: a locus with incomplete penetrance for autism, mental retardation, and psychiatric disorders. J Med Genet 46 382 388
26. HannesFD
SharpAJ
MeffordHC
de RavelT
RuivenkampCA
2009 Recurrent reciprocal deletions and duplications of 16p13.11: the deletion is a risk factor for MR/MCA while the duplication may be a rare benign variant. J Med Genet 46 223 232
27. MeffordHC
CooperGM
ZerrT
SmithJD
BakerC
2009 A method for rapid, targeted CNV genotyping identifies rare variants associated with neurocognitive disease. Genome Res 19 1579 1585
28. MillerDT
ShenY
WeissLA
KornJ
AnselmI
2008 Microdeletion/duplication at 15q13.2q13.3 among individuals with features of autism and other neuropsychiatirc disorders. J Med Genet 46 242 248
29. NeedAC
GeD
WealeME
MaiaJ
FengS
2009 A genome-wide investigation of SNPs and CNVs in schizophrenia. PLoS Genet 5 e1000373 doi:10.1371/journal.pgen.1000373
30. PagnamentaAT
WingK
AkhaES
KnightSJ
BolteS
2009 A 15q13.3 microdeletion segregating with autism. Eur J Hum Genet 17 687 692
31. SharpAJ
MeffordHC
LiK
BakerC
SkinnerC
2008 A recurrent 15q13.3 microdeletion syndrome associated with mental retardation and seizures. Nat Genet 40 322 328
32. UllmannR
TurnerG
KirchhoffM
ChenW
TongeB
2007 Array CGH identifies reciprocal 16p13.1 duplications and deletions that predispose to autism and/or mental retardation. Hum Mutat 28 674 682
33. Brunetti-PierriN
BergJS
ScagliaF
BelmontJ
BacinoCA
2008 Recurrent reciprocal 1q21.1 deletions and duplications associated with microcephaly or macrocephaly and developmental and behavioral abnormalities. Nat Genet 40 1466 1471
34. MeffordHC
SharpAJ
BakerC
ItsaraA
JiangZ
2008 Recurrent rearrangements of chromosome 1q21.1 and variable pediatric phenotypes. N Engl J Med 359 1685 1699
35. BijlsmaEK
GijsbersAC
Schuurs-HoeijmakersJH
van HaeringenA
Fransen van de PutteDE
2009 Extending the phenotype of recurrent rearrangements of 16p11.2: Deletions in mentally retarded patients without autism and in normal individuals. Eur J Med Genet 52 77 87
36. BochukovaEG
HuangN
KeoghJ
HenningE
PurmannC
2010 Large, rare chromosomal deletions associated with severe early-onset obesity. Nature 463 666 670
37. McCarthySE
MakarovV
KirovG
AddingtonAM
McClellanJ
2009 Microduplications of 16p11.2 are associated with schizophrenia. Nat Genet 41 1223 1227
38. GirirajanS
RosenfeldJA
CooperGM
AntonacciF
SiswaraP
2010 A recurrent 16p12.1 microdeletion supports a two-hit model for severe developmental delay. Nat Genet 42 203 209
39. DooseH
HahnA
NeubauerBA
PistohlJ
StephaniU
2001 Atypical “benign” partial epilepsy of childhood or pseudo-lennox syndrome. Part II: family study. Neuropediatrics 32 9 13
40. van BonBW
MeffordHC
MentenB
KoolenDA
SharpAJ
2009 Further delineation of the 15q13 microdeletion and duplication syndromes: a clinical spectrum varying from non-pathogenic to a severe outcome. J Med Genet 46 511 523
41. BonatiMT
CombiR
AsseltaR
DugaS
MalcovatiM
2002 Exclusion of linkage of nine neuronal nicotinic acetylcholine receptor subunit genes expressed in brain in autosomal dominant nocturnal frontal lobe epilepsy in four unrelated families. J Neurol 249 967 974
42. TaskeNL
WilliamsonMP
MakoffA
BateL
CurtisD
2002 Evaluation of the positional candidate gene CHRNA7 at the juvenile myoclonic epilepsy locus (EJM2) on chromosome 15q13-14. Epilepsy Res 49 157 172
43. ShinawiM
SchaafCP
BhattSS
XiaZ
PatelA
2009 A small recurrent deletion within 15q13.3 is associated with a range of neurodevelopmental phenotypes. Nat Genet 41 1269 1271
44. ButlerMG
BittelDC
KibiryevaN
TalebizadehZ
ThompsonT
2004 Behavioral differences among subjects with Prader-Willi syndrome and type I or type II deletion and maternal disomy. Pediatrics 113 565 573
45. HartleySL
MacleanWEJr
ButlerMG
ZarconeJ
ThompsonT
2005 Maladaptive behaviors and risk factors among the genetic subtypes of Prader-Willi syndrome. Am J Med Genet A 136 140 145
46. SahooT
PetersSU
MadduriNS
GlazeDG
GermanJR
2006 Microarray based comparative genomic hybridization testing in deletion bearing patients with Angelman syndrome: genotype-phenotype correlations. J Med Genet 43 512 516
47. ChristiansenJ
DyckJD
ElyasBG
LilleyM
BamforthJS
2004 Chromosome 1q21.1 contiguous gene deletion is associated with congenital heart disease. Circ Res 94 1429 1435
48. GreenwaySC
PereiraAC
LinJC
DePalmaSR
IsraelSJ
2009 De novo copy number variants identify new genes and loci in isolated sporadic tetralogy of Fallot. Nat Genet 41 931 935
49. RedonR
IshikawaS
FitchKR
FeukL
PerryGH
2006 Global variation in copy number in the human genome. Nature 444 444 454
50. AlarconM
AbrahamsBS
StoneJL
DuvallJA
PerederiyJV
2008 Linkage, association, and gene-expression analyses identify CNTNAP2 as an autism-susceptibility gene. Am J Hum Genet 82 150 159
51. ArkingDE
CutlerDJ
BruneCW
TeslovichTM
WestK
2008 A common genetic variant in the neurexin superfamily member CNTNAP2 increases familial risk of autism. Am J Hum Genet 82 160 164
52. BakkalogluB
O'RoakBJ
LouviA
GuptaAR
AbelsonJF
2008 Molecular cytogenetic analysis and resequencing of contactin associated protein-like 2 in autism spectrum disorders. Am J Hum Genet 82 165 173
53. FriedmanJI
VrijenhoekT
MarkxS
JanssenIM
van der VlietWA
2008 CNTNAP2 gene dosage variation is associated with schizophrenia and epilepsy. Mol Psychiatry 13 261 266
54. KalscheuerVM
FitzPatrickD
TommerupN
BuggeM
NiebuhrE
2007 Mutations in autism susceptibility candidate 2 (AUTS2) in patients with mental retardation. Hum Genet 121 501 509
55. SultanaR
YuCE
YuJ
MunsonJ
ChenD
2002 Identification of a novel gene on chromosome 7q11.2 interrupted by a translocation breakpoint in a pair of autistic twins. Genomics 80 129 134
56. ClaesL
CeulemansB
AudenaertD
SmetsK
LofgrenA
2003 De novo SCN1A mutations are a major cause of severe myoclonic epilepsy of infancy. Hum Mutat 21 615 621
57. EscaygA
MacDonaldBT
MeislerMH
BaulacS
HuberfeldG
2000 Mutations of SCN1A, encoding a neuronal sodium channel, in two families with GEFS+2. Nat Genet 24 343 345
58. FujiwaraT
2006 Clinical spectrum of mutations in SCN1A gene: severe myoclonic epilepsy in infancy and related epilepsies. Epilepsy Res 70 Suppl 1 S223 230
59. DooseH
LunauH
CastiglioneE
WaltzS
1998 Severe idiopathic generalized epilepsy of infancy with generalized tonic-clonic seizures. Neuropediatrics 29 229 238
60. HelbigI
MeffordHC
SharpAJ
GuipponiM
FicheraM
2009 15q13.3 microdeletions increase risk of idiopathic generalized epilepsy. Nat Genet 41 160 162
61. ILAE 1989 Proposal for revised classification of epilepsies and epileptic syndromes. Commission on Classification and Terminology of the International League Against Epilepsy. Epilepsia 30 389 399
62. DayN
HemmaplardhA
ThurmanRE
StamatoyannopoulosJA
NobleWS
2007 Unsupervised segmentation of continuous genomic data. Bioinformatics 23 1424 1426
Štítky
Genetika Reprodukční medicína
Článek Epistasis of Transcriptomes Reveals Synergism between Transcriptional Activators Hnf1α and Hnf4αČlánek SMA-10/LRIG Is a Conserved Transmembrane Protein that Enhances Bone Morphogenetic Protein SignalingČlánek B1 SOX Coordinate Cell Specification with Patterning and Morphogenesis in the Early Zebrafish EmbryoČlánek Bulk Segregant Analysis by High-Throughput Sequencing Reveals a Novel Xylose Utilization Gene fromČlánek Abundant Quantitative Trait Loci Exist for DNA Methylation and Gene Expression in Human Brain
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2010 Číslo 5
-
Všechny články tohoto čísla
- Digital Quantification of Human Eye Color Highlights Genetic Association of Three New Loci
- Common Genetic Variants near the Brittle Cornea Syndrome Locus Influence the Blinding Disease Risk Factor Central Corneal Thickness
- Age- and Temperature-Dependent Somatic Mutation Accumulation in
- All About Mitochondrial Eve: An Interview with Rebecca Cann
- Aging and Chronic Sun Exposure Cause Distinct Epigenetic Changes in Human Skin
- Transposed Genes in Arabidopsis Are Often Associated with Flanking Repeats
- A Survey of Genomic Traces Reveals a Common Sequencing Error, RNA Editing, and DNA Editing
- Gene Transposition Causing Natural Variation for Growth in
- The Polyproline Site in Hinge 2 Influences the Functional Capacity of Truncated Dystrophins
- Epistasis of Transcriptomes Reveals Synergism between Transcriptional Activators Hnf1α and Hnf4α
- Integration of Light Signals by the Retinoblastoma Pathway in the Control of S Phase Entry in the Picophytoplanktonic Cell
- The Proprotein Convertase Encoded by () Is Required in Corpora Cardiaca Endocrine Cells Producing the Glucose Regulatory Hormone AKH
- Sgs1 and Exo1 Redundantly Inhibit Break-Induced Replication and Telomere Addition at Broken Chromosome Ends
- A MATE-Family Efflux Pump Rescues the 8-Oxoguanine-Repair-Deficient Mutator Phenotype and Protects Against HO Killing
- The Relationship among Gene Expression, the Evolution of Gene Dosage, and the Rate of Protein Evolution
- SMA-10/LRIG Is a Conserved Transmembrane Protein that Enhances Bone Morphogenetic Protein Signaling
- The Nuclear Receptor DHR3 Modulates dS6 Kinase–Dependent Growth in
- Involvement of Global Genome Repair, Transcription Coupled Repair, and Chromatin Remodeling in UV DNA Damage Response Changes during Development
- B1 SOX Coordinate Cell Specification with Patterning and Morphogenesis in the Early Zebrafish Embryo
- Linkage and Association Mapping of Flowering Time in Nature
- Bulk Segregant Analysis by High-Throughput Sequencing Reveals a Novel Xylose Utilization Gene from
- Abundant Quantitative Trait Loci Exist for DNA Methylation and Gene Expression in Human Brain
- Ablation of Whirlin Long Isoform Disrupts the USH2 Protein Complex and Causes Vision and Hearing Loss
- Characterization of Oxidative Guanine Damage and Repair in Mammalian Telomeres
- DNA Adenine Methylation Is Required to Replicate Both Chromosomes Once per Cell Cycle
- Genome-Wide Copy Number Variation in Epilepsy: Novel Susceptibility Loci in Idiopathic Generalized and Focal Epilepsies
- FACT Prevents the Accumulation of Free Histones Evicted from Transcribed Chromatin and a Subsequent Cell Cycle Delay in G1
- GC-Biased Evolution Near Human Accelerated Regions
- Liver and Adipose Expression Associated SNPs Are Enriched for Association to Type 2 Diabetes
- Myeloid Cell-Restricted Insulin Receptor Deficiency Protects Against Obesity-Induced Inflammation and Systemic Insulin Resistance
- The Mating Type Locus () and Sexual Reproduction of : Insights into the Evolution of Sex and Sex-Determining Chromosomal Regions in Fungi
- B-Cyclin/CDKs Regulate Mitotic Spindle Assembly by Phosphorylating Kinesins-5 in Budding Yeast
- Post-Replication Repair Suppresses Duplication-Mediated Genome Instability
- Genome Sequence of the Plant Growth Promoting Endophytic Bacterium sp. 638
- Transcription Factors Mat2 and Znf2 Operate Cellular Circuits Orchestrating Opposite- and Same-Sex Mating in
- The Use of Orthologous Sequences to Predict the Impact of Amino Acid Substitutions on Protein Function
- Mutation in Archain 1, a Subunit of COPI Coatomer Complex, Causes Diluted Coat Color and Purkinje Cell Degeneration
- Shelterin-Like Proteins and Yku Inhibit Nucleolytic Processing of Telomeres
- Affecting Function Causes a Dilated Heart in Adult
- Manipulation of Behavioral Decline in with the Rag GTPase
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Common Genetic Variants near the Brittle Cornea Syndrome Locus Influence the Blinding Disease Risk Factor Central Corneal Thickness
- All About Mitochondrial Eve: An Interview with Rebecca Cann
- The Relationship among Gene Expression, the Evolution of Gene Dosage, and the Rate of Protein Evolution
- SMA-10/LRIG Is a Conserved Transmembrane Protein that Enhances Bone Morphogenetic Protein Signaling
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



