-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaEpistasis of Transcriptomes Reveals Synergism between Transcriptional Activators Hnf1α and Hnf4α
The transcription of individual genes is determined by combinatorial interactions between DNA–binding transcription factors. The current challenge is to understand how such combinatorial interactions regulate broad genetic programs that underlie cellular functions and disease. The transcription factors Hnf1α and Hnf4α control pancreatic islet β-cell function and growth, and mutations in their genes cause closely related forms of diabetes. We have now exploited genetic epistasis to examine how Hnf1α and Hnf4α functionally interact in pancreatic islets. Expression profiling in islets from either Hnf1a+/− or pancreas-specific Hnf4a mutant mice showed that the two transcription factors regulate a strikingly similar set of genes. We integrated expression and genomic binding studies and show that the shared transcriptional phenotype of these two mutant models is linked to common direct targets, rather than to known effects of Hnf1α on Hnf4a gene transcription. Epistasis analysis with transcriptomes of single - and double-mutant islets revealed that Hnf1α and Hnf4α regulate common targets synergistically. Hnf1α binding in Hnf4a-deficient islets was decreased in selected targets, but remained unaltered in others, thus suggesting that the mechanisms for synergistic regulation are gene-specific. These findings provide an in vivo strategy to study combinatorial gene regulation and reveal how Hnf1α and Hnf4α control a common islet-cell regulatory program that is defective in human monogenic diabetes.
Published in the journal: . PLoS Genet 6(5): e32767. doi:10.1371/journal.pgen.1000970
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1000970Summary
The transcription of individual genes is determined by combinatorial interactions between DNA–binding transcription factors. The current challenge is to understand how such combinatorial interactions regulate broad genetic programs that underlie cellular functions and disease. The transcription factors Hnf1α and Hnf4α control pancreatic islet β-cell function and growth, and mutations in their genes cause closely related forms of diabetes. We have now exploited genetic epistasis to examine how Hnf1α and Hnf4α functionally interact in pancreatic islets. Expression profiling in islets from either Hnf1a+/− or pancreas-specific Hnf4a mutant mice showed that the two transcription factors regulate a strikingly similar set of genes. We integrated expression and genomic binding studies and show that the shared transcriptional phenotype of these two mutant models is linked to common direct targets, rather than to known effects of Hnf1α on Hnf4a gene transcription. Epistasis analysis with transcriptomes of single - and double-mutant islets revealed that Hnf1α and Hnf4α regulate common targets synergistically. Hnf1α binding in Hnf4a-deficient islets was decreased in selected targets, but remained unaltered in others, thus suggesting that the mechanisms for synergistic regulation are gene-specific. These findings provide an in vivo strategy to study combinatorial gene regulation and reveal how Hnf1α and Hnf4α control a common islet-cell regulatory program that is defective in human monogenic diabetes.
Introduction
In all eukaryotic organisms a limited number of DNA binding transcriptional regulators determine a much greater number of genetic programs. This is made possible by a code whereby unique combinations of regulators define cellular fates or functions. The combinatorial nature of transcriptional regulation has been demonstrated in countless studies that have dissected individual gene regulatory regions [1]–[4]. A major underlying principle is that DNA-binding transcriptional activators often function synergistically due to cooperativity in binding or recruitment of regulatory complexes [1], [3]. Other common functional interactions include redundancy or antagonism between different factors binding to the same regulatory region [1]–[4].
A true understanding of transcriptional programs will require the dissection of transcription factor interactions in global cellular contexts, rather than in single genes. In recent years, the function of several mammalian transcription factors has been examined by profiling gene expression in genetically perturbed cells [5]. Such studies provide a broad inventory of genes that are dependent on selected transcription factors, but they do not in themselves reveal how different factors interact functionally. Other studies have determined the genomic binding sites of single or multiple transcription factors [6], [7]. However, knowing that a regulator binds to a gene does not clarify if the binding event leads to positive, negative, or no regulation. Numerous studies, in fact, suggest that a major fraction of transcription factor binding events might be functionally dispensable [8]–[13]. Similarly, when more than one factor binds to the same gene, several functional interactions are possible. New approaches are therefore necessary to understand how transcriptional regulators engage in the combinatorial interactions that regulate cellular programs.
The genetics of human diabetes provides a paradigm to study transcriptional programs in pancreatic β-cells [14]–[17]. Heterozygous mutations in several genes encoding DNA binding transcription factors cause autosomal dominant diabetes, or Maturity Onset Diabetes of the Young (MODY) [14]–[16], [18]. Mutations in HNF1A and HNF4A (encoding for hepatocyte nuclear factor 1α and 4α) are responsible for the most common form of monogenic diabetes [14], [15]. Despite transient differences in newborns, the diabetic phenotype in HNF1A and HNF4A patients shares many features, including similar disease progression curves, insulin secretory responses, and sensitivity to hypoglycemic drugs [18]. Human genetics therefore suggests that HNF1A and HNF4A may be involved in a common regulatory network in β-cells.
One simple explanation for the shared HNF1A and HNF4A-deficient phenotype is that Hnf1α regulates the transcription of the Hnf4a pancreas-specific promoter[19]–[21]. However, several lines of evidence point to additional regulatory interactions. For example, a large-scale binding study found that many Hnf1α-bound genes are also bound by Hnf4α [6]. Other studies have shown that Hnf1α physically interacts in vitro and in vivo with the Hnf4α AF2 domain [22]–[24]. Such interactions have been linked to observations that overexpression of Hnf1α inhibits Hnf4α-regulation of targets, and overexpression of Hnf4α inhibits Hnf1α function [23]–[25]. Other studies showed that Hnf4α can increase Hnf1α function in synthetic promoters that only contain an Hnf1α binding site [22], or in promoters containing binding sites for both factors [2], [26], [27]. Because so far most functional studies have employed overexpression systems in cultured non-β cell lines, the true functional consequences of Hnf1α/Hnf4α interactions in islet-cells remain unclear.
We have now developed a strategy to study the integrated function of Hnf1α and Hnf4α in pancreatic islet cells. We profiled gene expression in genetic models with weak phenotypes and show that Hnf1α and Hnf4α regulate a remarkably similar set of genes. Using binding studies and epistasis analysis of transcriptome phenotypes, we demonstrate that the common function of Hnf1α and Hnf4α in pancreatic islet cells is in part due to global synergistic interactions between the two factors at common direct targets. The results provide an approach to decipher transcriptional networks in mammalian cells, and reveal novel insight into a common regulatory program that underlies human monogenic diabetes.
Results
Hnf4a-deficient pancreatic islets exhibit an impaired transcriptional program
The goal of this study was to understand the integrated transcriptional function of Hnf1α and Hnf4α in pancreatic islets. To study Hnf4α function, we generated pancreas-specific Hnf4a knock-out mice (Hnf4apKO), and confirmed that this targeted deletion caused a marked reduction of Hnf4a gene mRNA in islet-cells (Figure 1A).
Fig. 1. Hnf1a and Hnf4a expression in mutant models. 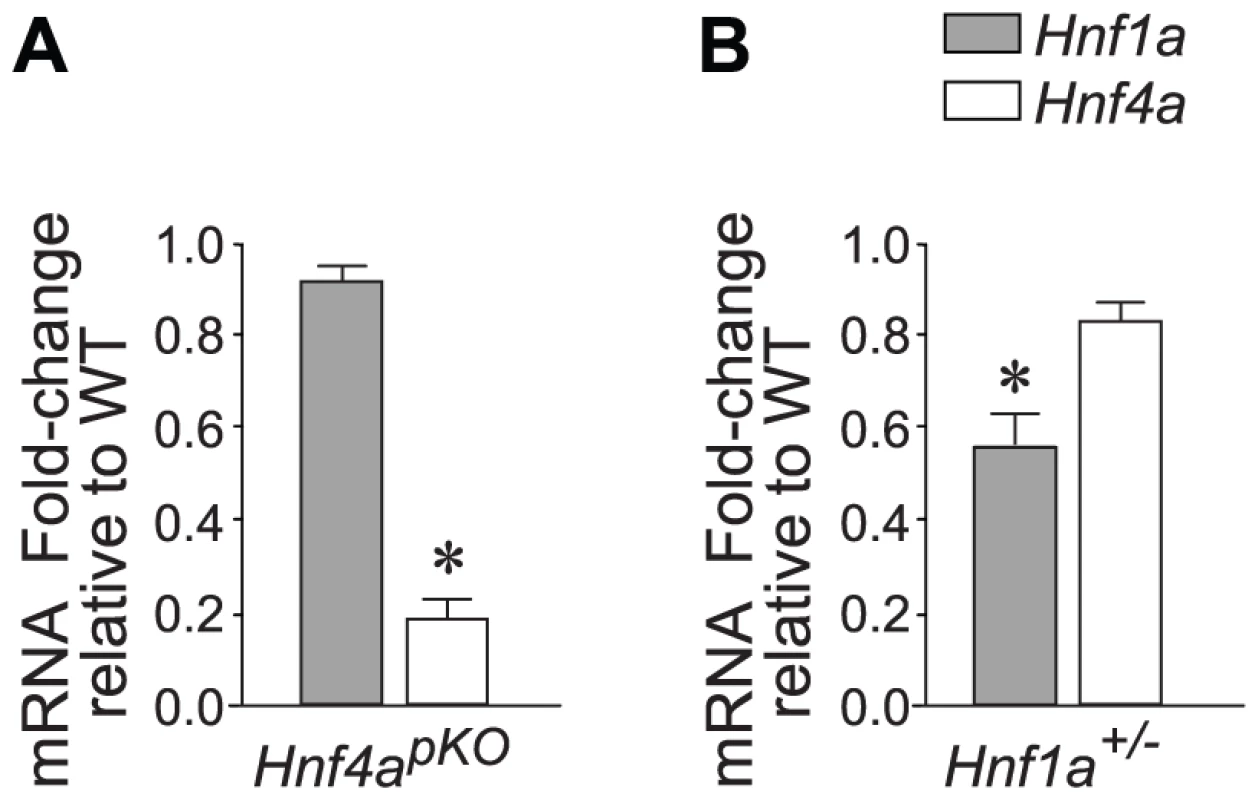
(A,B) Expression of Hnf1a and Hnf4a mRNA in islets from (A) Hnf4apKO and (B) Hnf1a+/− male mice. Results were normalized to Hprt mRNA and are expressed relative to littermate wild-type controls. * P<0.05. In keeping with previous studies of mice with β-cell specific ablation of Hnf4a (Hnf4abetaKO) [28]–[30], Hnf4apKO mice developed a mild complex phenotype, with very subtle glucose intolerance and a slightly reduced fasting glycemia (Figure S1).
Despite this mild metabolic phenotype, Hnf4apKO mice showed a clear islet transcriptional phenotype (Table S1). Downregulated genes encoded for varied cellular roles, including the metabolism of steroids, glucose, and amino acids (Table S1 and Table S2). Others encoded for regulators of signal transduction and cell growth, consistent with a previous report in Hnf4abetaKO islets [31], or were in keeping with the proposed role of Hnf4α in epithelial differentiation [32] (Table S1 and Table S2). Upregulated genes included genes known to form part of the epithelial mesenchymal transition process (Table S2). Overall, the functional classes that were perturbed in Hnf4a-deficient islets were remarkably similar to those reported in Hnf1a-/- islets[11].
Hnf1a haploinsufficient mice reveal Hnf1a-dependent transcription in islets
To study Hnf1α transcriptional function, we used Hnf1a+/− mice. As opposed to mice with homozygous Hnf1a mutations, which develop diabetes, this model has no documented in vitro or in vivo metabolic disturbances (Figure S2)[33]–[36]. Also in keeping with previous studies, Hnf1a+/− islets exhibit only marginal downregulation of Hnf4α (∼70–90% of normal values) (Figure 1B)[36]. Hnf1a+/− mice thus lack two elements that are thought to exert an indirect impact on islet gene expression in homozygous Hnf1a mutant islets. Array analysis revealed a transcriptional phenotype in Hnf1a+/− islets, with 196 non-redundant genes downregulated >1.5-fold in Hnf1a+/− islets at a nominal P value<0.01. We validated this dataset with gene-specific assays in 20 genes from independent Hnf1a+/− mice (Figure S4A). Furthermore, genes that were bound by Hnf1α and downregulated in homozygous Hnf1a mutant islets were significantly downregulated in Hnf1a+/− islets (Figure S4B). Thus, expression profiling in Hnf1a+/− islets provides a tool to assess the transcriptional function of Hnf1α in this tissue.
Hnf4a - and Hnf1a-deficient islets share a common transcriptional signature
We next compared expression changes in Hnf1a+/− and Hnf4apKO islets. This revealed a striking correlation between the two models (r = 0.57, P = 10−6) (Figure 2A–2C). Gene set enrichment analysis (GSEA) showed that genes that were significantly downregulated in Hnf4apKO islets were downregulated in Hnf1a+/− islets (Figure 2D, P<0.001). Conversely, genes downregulated in Hnf1a+/− islets were downregulated in Hnf4apKO islets (Figure 2E, P<0.001). Not surprisingly, gene expression changes were consistently lower in the Hnf1a haploinsufficient islets compared with islets with biallelic inactivation of Hnf4a (Figure 2A–2E). We confirmed the correlation with gene-specific assays (Figure 2B), and with an independent comparison of Hnf1a+/− versus Hnf4apKO mice of 16 rather than 8 weeks of age (not shown, and Table S1).
Fig. 2. Hnf1α and Hnf4α regulate a common set of genes. 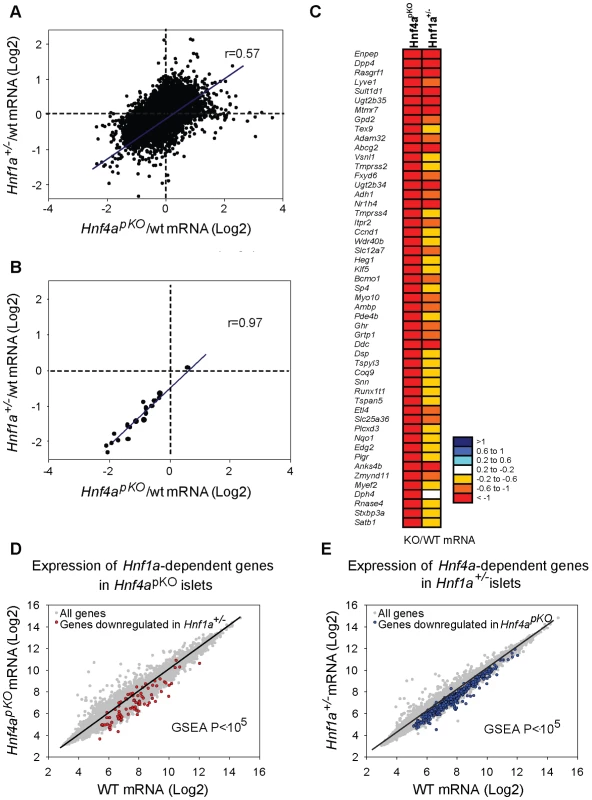
(A) Correlation of mutant/wild-type Log2 gene expression ratios in Hnf4apKO versus Hnf1a+/− islets. (B) Validation of 22 genes using gene-specific qPCR. (C) Heatmap of expression ratios in Hnf4apKO and Hnf1a+/− islets for the 50 most downregulated genes in Hnf4apKO islets. (D) Expression of Hnf1a-dependent genes in Hnf4apKO islets. Grey dots represent average expression values of genes in Hnf4apKO versus control islets. We superimposed red dots to show the subset of genes downregulated in Hnf1a+/− islets. (E) Expression of Hnf4a-dependent genes in Hnf1a+/− islets. Grey dots are expression values of all genes, superimposed blue dots are the subset of genes downregulated in Hnf4apKO islets. These common gene expression changes were unexpected, because Hnf1a expression in Hnf4apKO islets was unperturbed (Figure 1A) (as previously shown for Hnf4abetaKO mice[28]–[30]) and Hnf4a expression was only marginally reduced in Hnf1a+/− islets (Figure 1B, and [36]). In conclusion, the analysis of models that minimize the impact of indirect perturbations showed that Hnf1α and Hnf4α regulate a common set of genes in pancreatic islets.
Hnf1α targets are similarly impaired in Hnf4a - and Hnf1a-deficient islets
Previous studies provide two possible mechanisms whereby Hnf1a+/− versus Hnf4apKO islets could exhibit a similar transcriptional phenotype (Figure 3A). One is that Hnf1α and Hnf4α frequently bind the same genes in human liver and islets [6]. We confirmed this finding using mouse liver binding datasets reported elsewhere [8], [11] (Figure 3B), after estimating that ∼75% of Hnf4α-bound genes in islets may also bound in liver (Figure. S8). However, co-occupancy does not per se explain the similar gene expression changes in the two mutant models, because for most genes bound by Hnf1α or Hnf4α, gene expression is not altered in the respective knock-out tissues [8], [11]. An alternate explanation for the similar transcriptional phenotypes is that Hnf1α regulates Hnf4a gene transcription in islets (Figure 3A) [11], [19]–[21]. In the current study we used heterozygous Hnf1a mutant islets because Hnf4a was not significantly altered (unlike homozygous Hnf1a mutants). However, it remained possible that a subtle decrease in Hnf4a expression in Hnf1a+/− islets caused the common transcriptional phenotype (which would thus result from a perturbation of Hnf4α in both models). We predicted that if this were true we should observe impaired expression of direct targets of Hnf4α in the two models, whereas the direct targets of the upstream factor of this hierarchy, Hnf1α, should be impaired only in Hnf1a-deficient islets (Figure 3A).
Fig. 3. Expression of Hnf1α targets is impaired in Hnf4a-deficient islets. 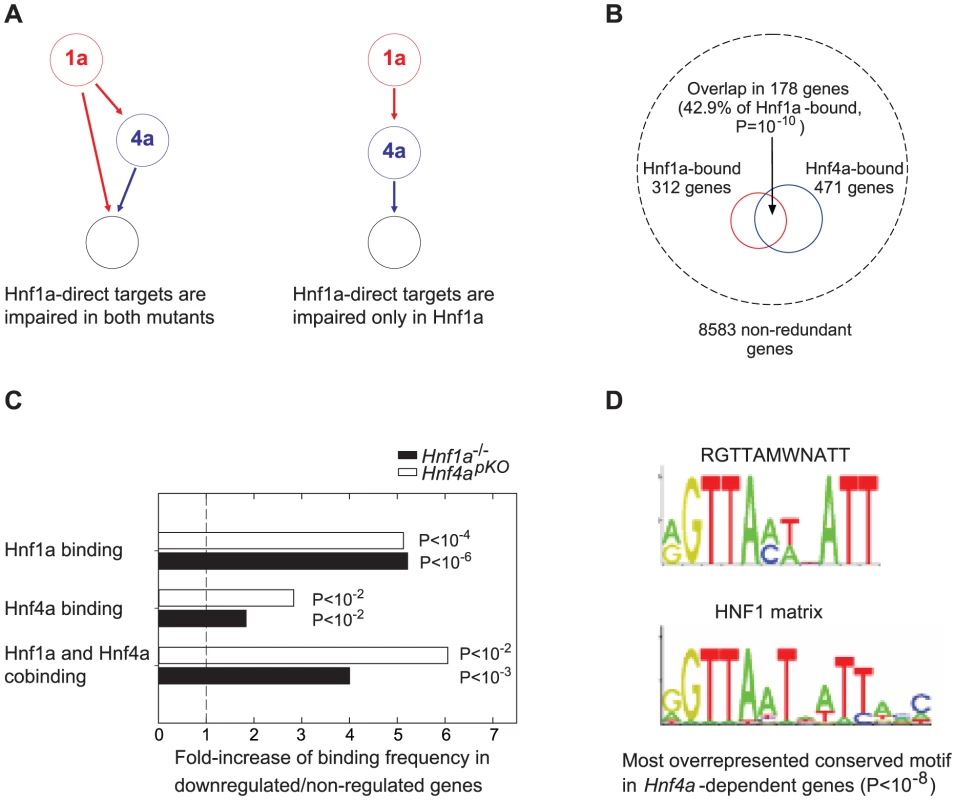
(A) Alternate models of Hnf1α and Hnf4α network structures that could potentially underlie the similar transcriptomes in Hnf4apKO and Hnf1a+/− islets, and expected functional perturbation of Hnf1α bound genes in each case. (B) The analysis of previously reported [8], [11] mouse liver binding datasets showed that Hnf1α and Hnf4α preferentially bind the same genes, as reported in human islets and liver[6]. Hypergeometric distributions were tested to calculate significance values. (C) Hnf1α, Hnf4α and Hnf1α/Hnf4α binding were enriched in genes that were significantly downregulated 2-fold in Hnf4apKO and Hnf1a-/- islets. Hypergeometric distributions were tested to calculate significance values. (D) Most significant over-represented evolutionary conserved sequence element in 10 Kb surrounding transcription start sites of genes that were downregulated in Hnf4apKO islets. The canonical HNF1 matrix is shown below. Motifs matching Hnf4α, or Hnf1α and Hnf4α binding sequences were also overrepresented in genes downregulated in Hnf4apKO and Hnf1a-/- islets, respectively (not shown). We thus tested if transcription of Hnf1α-bound genes was impaired in mice deficient for either factor. Hnf1α binding frequency was increased 5-fold among genes that were downregulated in homozygous Hnf1a mutant islets (Figure 3C). Remarkably, Hnf1α binding frequency was also increased 5-fold in promoters of genes that were downregulated in Hnf4a-deficient islets (Figure 3C). We also found that Hnf1α bound genes that were downregulated in Hnf1a-/- islets were downregulated in Hnf4a-deficient islets (Figure S5). In silico studies confirmed these findings, as the most overrepresented conserved motif in genes downregulated in Hnf4apKO islets was identical to the canonical HNF1 binding sequence (Figure 3D). Thus, Hnf1α targets are impaired in Hnf4a deficient islets to a similar extent as in Hnf1a deficient islets.
Genes that were downregulated in both Hnf4a - and Hnf1a-deficient islets also showed significantly enriched Hnf4α binding and a 4 to 6-fold higher co-occupancy rate than non-regulated genes (Figure 3C). Collectively, these results argue that the shared transcriptome in our models is not due to the known Hnf1a-Hnf4a transcriptional hierarchy, and instead support that it is linked to the regulation of common target genes.
Hnf1α and Hnf4α function is interdependent in pancreatic islets
The above findings were consistent with an interdependent function of Hnf1α and Hnf4α, or alternatively with the regulation of common genes in a mechanistically independent manner. To discriminate among these possibilities, we compared expression profiles in single mutant (Hnf1a+/− and Hnf4apKO) versus double mutant (Hnf1a+/− Hnf4apKO) islets (Figure 4A). We predicted that if Hnf1α and Hnf4α act independently in any gene that is downregulated in both single mutant islets, the expression ratio in double mutant (Hnf1a+/− Hnf4apKO) islets should reflect the product of the two single mutant expression ratios. By contrast, if the two factors regulate a gene in an interdependent manner, the expression in double mutant islets should differ from this expectation (Figure 4A).
Fig. 4. Epistasis reveals functional synergism between Hnf1α and Hnf4α. 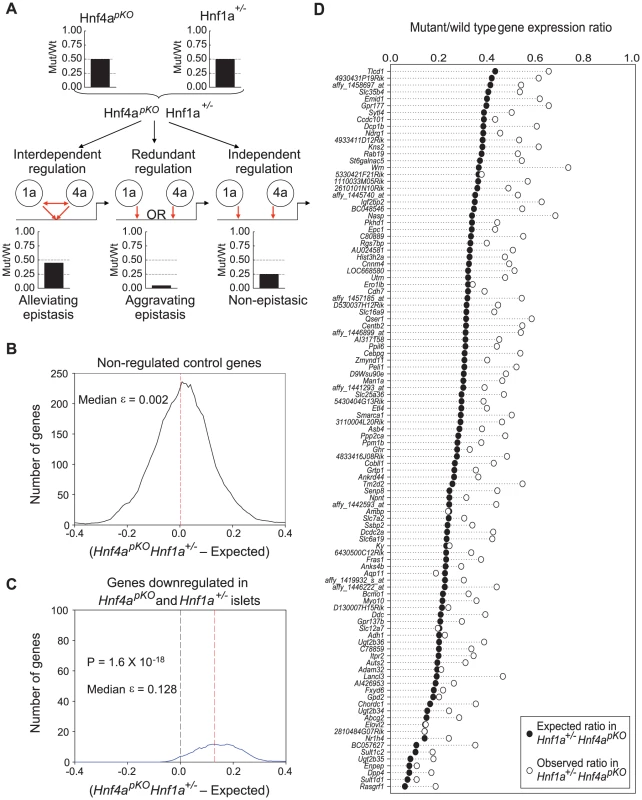
(A) Schematic representation of the genetic approach used to test functional interactions between Hnf1a and Hnf4a in a hypothetical gene that is downregulated 50% of wild type values in both Hnf4apKO and Hnf1a+/− islets. (B,C) Distribution of ε values (see results for explanation) for control genes (B), or for all genes that were downregulated in both single mutant mice (C). (D) Observed gene expression ratios in Hnf4apKO Hnf1a+/− islets (white circles) and expected changes in a non-epistatic model (black circles) for each gene that was significantly downregulated in both single mutant mice. For each gene that was downregulated in both Hnf1a+/− and Hnf4apKO single mutant islets we calculated an epistasis ε value that measures the deviation from expectation. An ε value > 0 indicates that the expression ratio in Hnf1a+/− Hnf4apKO islets is higher (less perturbed) than expected from the independent effects of the two single mutant values. As shown in Figure 4C, the distribution of ε values was unambiguously greater than 0. Figure 4D further illustrates this concept, showing that the perturbation of individual genes in Hnf1a+/− Hnf4apKO islets was epistatic (P = 10−35). Similar findings were confirmed in independent mice using qPCR (quantitative PCR) rather than oligonucleotide chips (Figure S6).
The high co-occupancy rate of Hnf1α and Hnf4α also raises the question whether these two factors might exert redundant functions at some targets. Because selecting genes that are downregulated in the single mutant islets can represent a bias against redundancy, we also performed this analysis in all genes that were downregulated >3 fold in double Hnf1a+/− Hnf4apKO mutant islets. Most such genes were markedly downregulated in the single mutants, and again had ε values exceeding 0 (Figure S7), thus showing that although Hnf1α and Hnf4α often bind to the same genes, their function is not redundant in islets. In keeping with these findings, the mild glucose intolerance phenotype observed in Hnf4apKO mice was not further impaired in Hnf1a+/− Hnf4apKO mice (Figure S3). In summary, these results indicate that Hnf1α and Hnf4α bind to similar targets and act through interdependent regulatory mechanisms in islets, thus leading to a common transcriptional phenotype in Hnf1a+/− and Hnf4apKO islets.
Gene-specific mechanisms for interdependent activation
To assess the mechanisms underlying the interdependent function of Hnf1α and Hnf4α in common target genes, we examined whether their binding is interdependent. Because Hnf1α expression is unaltered in Hnf4a-deficient mouse islets, we were able to use this model to study whether Hnf4α is required for Hnf1α binding to common targets. We selected eight genes that we have previously shown are bound by Hnf1α and are functionally dependent on Hnf1α in islets [11](Figure 5A). All of them were also directly bound by Hnf4α in wild type islets (Figure 5C), and were markedly downregulated in Hnf4a-deficient islets (Figure 5B). Thus, all 8 selected genes were co-occupied and were functionally dependent on the two factors in islets. We next examined Hnf1α binding to these sites in Hnf4a-deficient islets. We found that in 5 of 8 genes, Hnf1α binding was maintained in islets that lack Hnf4α (Figure 5D). In two other Hnf4a-dependent genes, Hnf1α binding was significantly reduced in Hnf4a-deficient islets, and was completely abrogated in one case (Figure 5D). The difference between genes concerning Hnf1α binding in Hnf4a-deficient islets could not be linked to differences in the affinity of Hnf1α and Hnf4α binding sites. Thus, the synergistic activation of common targets by Hnf1α and Hnf4α can reflect gene-specific interdependent mechanisms at both binding and post-binding levels.
Fig. 5. Gene-specific mechanisms for functional synergism. 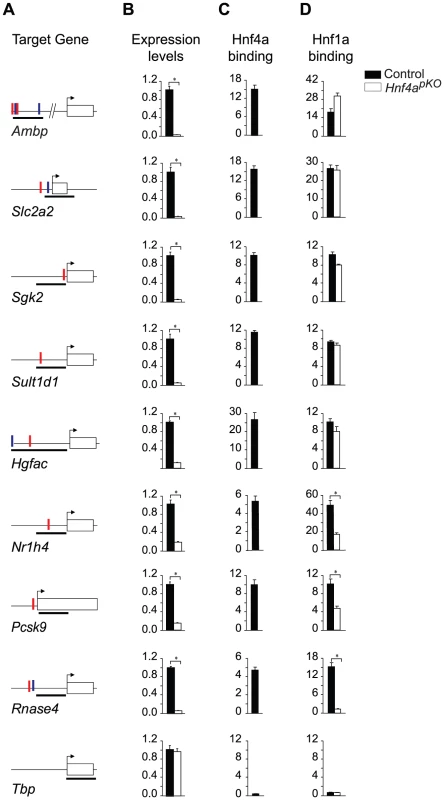
We tested Hnf1α binding in Hnf4a-deficient islets in 8 genes that are bound by Hnf1α and Hnf4α in wild type islets and are downregulated in Hnf1a and Hnf4a-deficient islets. Hnf1α binding in Hnf4a-deficient islets was unaltered in 5/8 genes examined, was partially reduced in two genes, and was abrogated in one gene. (A) Schematic representations of PCR products (black thick lines) used for Hnf1α and Hnf4α ChIPs, and high affinity HNF1 (red vertical lines) and HNF4 (blue vertical lines) binding sequences. (B) Gene expression in wild type and Hnf4a-deficient islets assayed by quantitative PCR. Results are normalized by expression levels of Actb mRNA, and are shown as fold-changes relative to wild type islets. (C,D) Hnf1α and Hnf4α binding in wild type (black bars) and Hnf4a-deficient (white bars) islets. Results are expressed as fold over Actb negative control regions. Tbp is shown as an independent negative control for both ChIP and gene expression studies. * P<0.05. Discussion
Epistasis reveals functional interdependence of Hnf1α and Hnf4α in islets
We have addressed how transcription factors establish functional interactions in an in vivo context. To achieve this, we studied epistatic relationships of transcriptional phenotypes. Our approach follows recent studies that used epistasis of transcriptomes to study functional interactions between regulators of protein kinase A in Dictyostelium, Mediator subunits in yeast, and most recently to unravel yeast transcription factor networks [37]–[39]. Our use of epistasis is analogous to classic studies that studied synergism or redundancy between transcription factors by comparing cells transfected with reporter minigenes along with single versus multiple transcription factors[1]. In the reverse approach we employed, we studied the transcriptome of mice with single and double transcription factor mutations. This allowed us to study combinatorial function in vivo, in endogenous genes of primary mammalian cells. It also enabled a global analysis, rather than studying specific gene targets that do not necessarily reflect a predominant regulatory strategy. This approach complements studies that compare the genomic location of different transcription factors without assessing their functional interactions. Our analysis thus confirmed previous observations that Hnf1α and Hnf4α bind to common targets, and suggests that these two factors function as obligate interdependent regulators in pancreatic islets. We thus demonstrate a role for epistasis to unravel the function of transcription factor networks in mammalian cells.
Value of weak genetic phenotypes to study regulatory networks
Previous studies showed that homozygous Hnf1a mutant mice exhibit full blown diabetes, in contrast to β-cell and pancreas-specific Hnf4a mutations which only result in glucose intolerance [21], [28], [29], [31], [34], [40]. In light of these differences it was somewhat unexpected that Hnf1α and Hnf4α regulate similar islet genes. We observed this coregulation using heterozygous Hnf1a and pancreas-specific Hnf4a mutations, which do not have common metabolic disturbances that could confound the comparison. They are also selective models: four studies have now shown that Hnf4a-deficient islets express normal Hnf1α levels (this study, and [28]–[30]), and, in contrast to Hnf1a-/- mice, Hnf4α levels are only minimally altered in Hnf1a+/− islets (this study, and [36]). Plausibly, differences in the phenotype of Hnf1a and Hnf4a-deficient models reported so far are due to the use of different types of genetic inactivation systems. More generally, we believe that weak genetic perturbations can be of great interest in studying transcription factor function, because although they only cause mild target expression changes, they are also less likely to disrupt downstream regulatory networks, thus limiting the magnitude of indirect effects.
Hnf1α and Hnf4α regulate common targets
Most genes bound by Hnf4α or Hnf1α are not affected by mutations of these two factors[11], [19]. It was thus necessary to integrate binding studies with genetic perturbation models to understand the functional interactions between these transcriptional regulators.
We observed that functional Hnf1α targets were similarly perturbed in Hnf4a-deficient and Hnf1a-deficient islets. This suggests that Hnf4α regulates Hnf1α function, and discards that epistasis was simply due to the known transcriptional hierarchy in which Hnf1a is upstream of Hnf4a. Together with the high Hnf1α/Hnf4α co-occupancy rate reported here and previously in human liver and islets[6], these findings suggest that epistasis between Hnf1a and Hnf4a is at least in part due to interdependent interactions at common direct targets. We thus propose a model for the integrated function of Hnf1α and Hnf4α in islets whereby Hnf1α regulates Hnf4a transcription [19], [20], and furthermore both factors act as interdependent transcriptional partners in islet-cell targets (Figure 6). Importantly, this network model is not based solely on binding studies, but integrates information on the combinatorial functional interactions between these factors.
Fig. 6. Model of the Hnf1α/Hnf4α regulatory network in pancreatic islets. 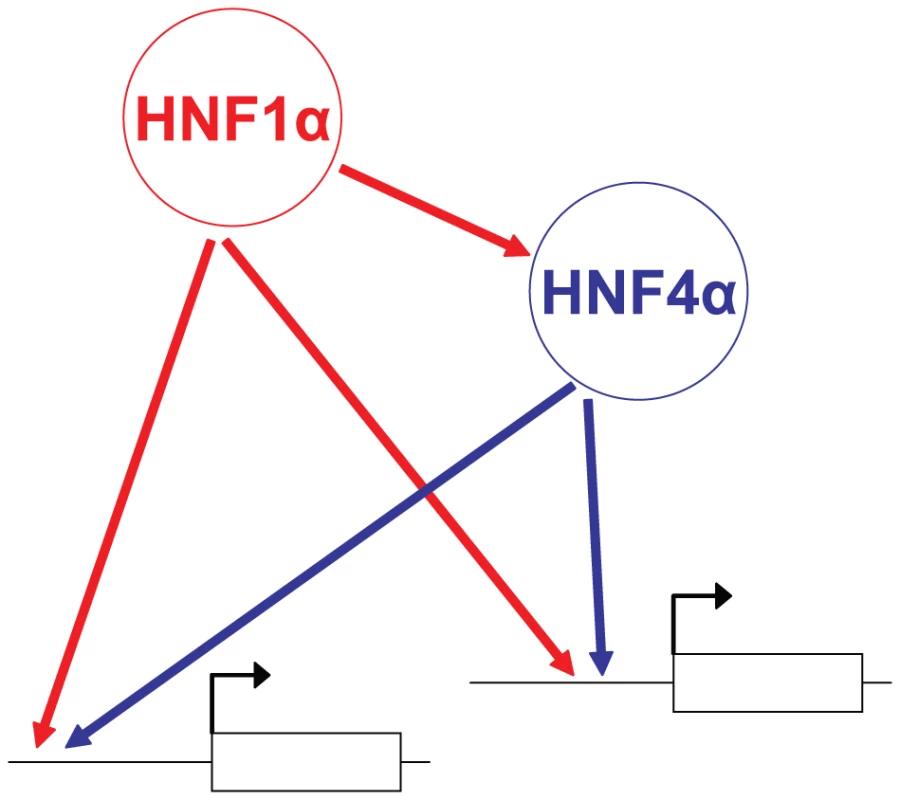
In islets, Hnf1α controls Hnf4a gene transcription, while both Hnf1α and Hnf4α activate common targets synergistically. Mechanisms underlying interdependent function
Several reports have demonstrated protein-protein interactions between Hnf1α and Hnf4α [22]–[24]. Such interactions can lead to functional inhibition [23], [24]. We show that although inhibitory consequences may be prevalent in other tissues or may occur in a small subset of genes, Hnf1α and Hnf4α largely activate genes synergistically in pancreatic islets. Earlier gene-specific studies have shown cooperative binding of Hnf1α and Hnf4α [2], [26], [27], while Hnf4α has been shown to co-activate a gene that is only directly bound by Hnf1α[22]. Our in vivo data showed that binding was interdependent in a subset of targets, but also showed that in many targets Hnf4a-deficiency does not entail decreased binding of Hnf1α. In the latter genes Hnf4α is likely required for post-binding functions of Hnf1α, such as the recruitment of co-regulatory complexes required for chromatin remodelling and/or assembly of the preinitiation complex. The mechanisms underlying functional synergism between Hnf1α and Hnf4α therefore appear to vary across target genes.
Implications for monogenic diabetes
Our study predicts a common islet transcriptome defect in the pathophysiology of HNF1A and HNF4A diabetes. This is consistent with the clinically indistinguishable diabetic phenotype of adult HNF1A and HNF4A patients[18], [41]. An exception to this notion is that HNF4A mutations cause transient in utero and neonatal hyperinsulinism, which later evolves to decreased insulin secretion, whereas HNF1A mutations develop the latter phenotype without early hyperinsulinism [30]. This may result if Hnf4α has Hnf1α-independent functions during prenatal and neonatal developmental stages.
The interdependent function of Hnf1α and Hnf4α is also relevant to our understanding of how haploinsufficiency of HNF1A and HNF4A leads to β-cell dysfunction and diabetes. We previously proposed that HNF1A or HNF4A haploinsufficiency could lead to the disruption of a Hnf1α/Hnf4α positive cross-regulatory network in β-cells[42]. Our current model depicted in Figure 6 provides new elements to assess the consequences of haploinsufficiency for the complex Hnf1α/Hnf4α network. Synergistic Hnf1α/Hnf4α-dependent activation is expected to result in steeper activator-response curves, and thus greater vulnerability to decreased gene dosage. This may be more pronounced in islet-cells, where Hnf1α and Hnf4α concentrations are much lower than in liver and other tissues that are not clinically afflicted in MODY[11], [43]. Haploinsufficiency of either HNF1A or HNF4A may in this manner disrupt the function of both HNF1α and HNF4α and the common transcriptional program, which include essential genes for the proper function of pancreatic islets. In conclusion, these studies provide an approach to understand the in vivo function of a regulatory network, and increase our understanding of the mechanisms underlying monogenic diabetes.
Materials and Methods
Mouse models and isolation of cells
Hnf4aLoxP mice were obtained from The Jackson Laboratory[44], Hnf1a+/− mice were provided by Frank Gonzalez (NCI)[34]. Pdx1Cre transgenic mice were provided by Pedro Herrera (U. Geneva) [45]. All studies were performed according to procedures approved by the institutional animal care and use committee. Animals were maintained on C57B/l6 backgrounds and genotyped as described [40]. For Hnf4a studies, Hnf4aLoxP littermates lacking Pdx1Cre and Pdx1Cre mice lacking Hnf4aLoxP alleles were used as controls unless stated otherwise. Pancreatic islets from 2 - to 4-month old mice were isolated as described [40]. Islets were cultured for 48 hr at 37°C, 5% CO2 in RPMI (Invitrogen) containing 11mM glucose supplemented with 10% FCS.
Glucose tolerance
Animals were fasted overnight and injected glucose intraperitoneally (2 gm/Kg). Glucose was measured from the tail vein at 0,15,30,60 and 120 min. Fasted plasma insulin was measured by ELISA (Mercodia).
Gene expression analysis
RNA from purified islets was isolated with Trizol (Invitrogen) and tested with an Agilent 2100 Bioanalyzer to ascertain RNA integrity. The reduction of Hnf4a mRNA in Hnf4apKO mice varied between 5–50% of wild type islets, most likely due the inherent variability of Cre-based recombination. We thus assessed Hnf4a mRNA by semiquantitative PCR [46] and used samples with >80% reduction for further analysis.
For each array replicate, RNAs from 2–4 male mice were pooled, and 50 ng was used in two cycles of cDNA synthesis for hybridization of Affymetrix 430 2.0 arrays. For epistasis experiments we used 8 week-old male mice. We separately compared 16 week-old male Hnf4aLoxP and control mice. Three arrays (a total of 8–12 mice) were analyzed per genotype, normalized with RMA, and analyzed with the LIMMA package to identify downregulated genes using a multiple test adjusted P value <0.05. To select genes downregulated in Hnf1a+/− islets we used a nominal (unadjusted) P<0.01 threshold, and validated this set with gene-specific assays. Gene-specific expression was assessed either with Taqman Low Density Arrays (Applied Biosystems) using unpooled islet RNA samples from 2–3 additional mice per genotype, or by qPCR using SybrGreen detection system as described [11]. Gene expression datasets are available in ArrayExpress (http://www.ebi.ac.uk) (Accession number: E-MEXP-1729).
Chromatin immunoprecipitation (ChIP) assays
Approximately 2000 purified islets from Hnf4aLoxP Pdx1Cre and littermates lacking the Pdx1Cre transgene were used for ChIP assays essentially as described [11], [19]. For each genotype we processed islets from two independent pools of ∼10 mice separately, we measured in duplicate the enrichment of immunoprecipitated DNA relative to input DNA, and corrected for the same values obtained in Actb as a negative control gene.
Computational sequence analysis
We used oPOSSUM [47], which computes a Fisher exact test to measure over-representation of sequence elements in a gene set relative to a background comprising all genes. We analyzed evolutionary conserved sequences 5 Kb upstream and 5 Kb downstream of transcription start sites of all downregulated genes (M<−0.6), and searched for conserved sequence elements as described in [48]. The empirical recommendations to identify binding sites oPOSSUM are a Z-score>10 and a Fisher P value<0.01. Overrepresented motifs were tested against the JASPAR CORE database of binding site profiles.
Epistasis and statistical analyses
Our analysis of epistasis of transcriptome phenotypes is based on previous large-scale studies of epistasis among yeast mutants regulating cell growth [49], [50]. We selected 105 genes that were downregulated (M<−0.6) in both Hnf1a+/− and Hnf4apKO islets, and compared gene expression changes with that of double mutant islets. For each gene we calculated an ε epistasis value that measures the deviation of the observed mutant/wild type expression ratio (R) in Hnf1a+/− Hnf4apKO double mutant islets from the expected ratio based on the product of the two single mutant values (ε = R Hnf1a+/− Hnf4apKO - [R Hnf1a+/− x R Hnf4apKO]). We performed a similar analysis for a control set of genes that showed no regulation in the two single mutant islets (M-1.1 to 1.1, p>0.2,). We calculated statistical significance with Student's t test, comparing the experimental and control ε value distributions, or with a paired Student's t test, comparing Hnf1a+/− Hnf4apKO R values to expected [R Hnf1a+/− x R Hnf4apKO] values in each gene.
Enrichment of functional annotations was examined with GSEAv2.0 (http://www.broad.mit.edu/gsea/) using gene sets as the permutation type and 1000 permutations, and with DAVID (http://david.abcc.ncifcrf.gov/). Statistical significance in binding comparisons was calculated with two-sided Fisher's exact test, or by testing the Hypergeometric distribution.
Supporting Information
Zdroje
1. CareyM
SmaleST
2000 Transcriptional regulation in eukaryotes concepts, strategies, and techniques. Cold Spring Harbor, NY Cold Spring Harbor Laboratory Press
2. CostaRH
GraysonDR
DarnellJEJr
1989 Multiple hepatocyte-enriched nuclear factors function in the regulation of transthyretin and alpha 1-antitrypsin genes. Mol Cell Biol 9 1415 1425
3. DuW
ThanosD
ManiatisT
1993 Mechanisms of transcriptional synergism between distinct virus-inducible enhancer elements. Cell 74 887 898
4. YuhCH
BolouriH
DavidsonEH
1998 Genomic cis-regulatory logic: experimental and computational analysis of a sea urchin gene. Science 279 1896 1902
5. HortonJD
ShahNA
WarringtonJA
AndersonNN
ParkSW
2003 Combined analysis of oligonucleotide microarray data from transgenic and knockout mice identifies direct SREBP target genes. Proc Natl Acad Sci U S A 100 12027 12032
6. OdomDT
ZizlspergerN
GordonDB
BellGW
RinaldiNJ
2004 Control of pancreas and liver gene expression by HNF transcription factors. Science 303 1378 1381
7. BlaisA
TsikitisM
Acosta-AlvearD
SharanR
KlugerY
2005 An initial blueprint for myogenic differentiation. Genes Dev 19 553 569
8. BojSF
ServitjaJM
MartinD
RiosM
TalianidisI
2009 The functional targets of the monogenic diabetes transcription factors HNF1{alpha} and HNF4{alpha} are highly conserved between mice and humans. Diabetes
9. LiXY
MacArthurS
BourgonR
NixD
PollardDA
2008 Transcription factors bind thousands of active and inactive regions in the Drosophila blastoderm. PLoS Biol 6 e27 doi:10.1371/journal.pbio.0060027
10. LohYH
WuQ
ChewJL
VegaVB
ZhangW
2006 The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat Genet 38 431 440
11. ServitjaJM
PignatelliM
MaestroMA
CardaldaC
BojSF
2009 Hnf1{alpha} (MODY3) controls tissue-specific transcriptional programs and exerts opposed effects on cell growth in pancreatic islets and liver. Mol Cell Biol
12. YangA
ZhuZ
KapranovP
McKeonF
ChurchGM
2006 Relationships between p63 binding, DNA sequence, transcription activity, and biological function in human cells. Mol Cell 24 593 602
13. PhucLP
FriedmanJR
SchugJ
BrestelliJE
ParkerJB
2005 Glucocorticoid receptor-dependent gene regulatory networks. PLoS Genet 1 e16 doi:10.1371/journal.pgen.0010016
14. YamagataK
FurutaH
OdaN
KaisakiPJ
MenzelS
1996 Mutations In the hepatocyte nuclear factor-4 alpha gene in maturity-onset diabetes of the young (MODY1). Nature 384 458 460
15. YamagataK
OdaN
KaisakiPJ
MenzelS
FurutaH
1996 Mutations in the hepatocyte nuclear factor-1 alpha gene in maturity-onset diabetes of the young (MODY3). Nature 384 455 458
16. StoffersDA
FerrerJ
ClarkeWL
HabenerJF
1997 Early-onset type-II diabetes mellitus (MODY4) linked to IPF1. Nat Genet 17 138 139
17. ServitjaJM
FerrerJ
2004 Transcriptional networks controlling pancreatic development and beta cell function. Diabetologia 47 597 613
18. MurphyR
EllardS
HattersleyAT
2008 Clinical implications of a molecular genetic classification of monogenic beta-cell diabetes. Nat Clin Pract Endocrinol Metab 4 200 213
19. BojSF
ParrizasM
MaestroMA
FerrerJ
2001 A transcription factor regulatory circuit in differentiated pancreatic cells. Proceedings of the National Academy of Sciences of the United States of America 98 14481 14486
20. ThomasH
JaschkowitzK
BulmanM
FraylingTM
MitchellSM
2001 A distant upstream promoter of the HNF-4alpha gene connects the transcription factors involved in maturity-onset diabetes of the young. Hum Mol Genet 10 2089 2097
21. ShihDQ
ScreenanS
MunozKN
PhilipsonL
PontoglioM
2001 Loss of HNF-1 alpha function in mice leads to abnormal expression of genes involved in pancreatic islet development and metabolism. Diabetes 50 2472 2480
22. EeckhouteJ
FormstecherP
LaineB
2004 Hepatocyte nuclear factor 4alpha enhances the hepatocyte nuclear factor 1alpha-mediated activation of transcription. Nucleic Acids Res 32 2586 2593
23. KtistakiE
TalianidisI
1997 Modulation of hepatic gene expression by hepatocyte nuclear factor 1. Science 277 109 112
24. RowleyCW
StalochLJ
DivineJK
McCaulSP
SimonTC
2006 Mechanisms of mutual functional interactions between HNF-4alpha and HNF-1alpha revealed by mutations that cause maturity onset diabetes of the young. Am J Physiol Gastrointest Liver Physiol 290 G466 G475
25. KyrmiziI
HatzisP
KatrakiliN
TroncheF
GonzalezFJ
2006 Plasticity and expanding complexity of the hepatic transcription factor network during liver development. Genes Dev 20 2293 2305
26. HuC
PerlmutterDH
1999 Regulation of alpha1-antitrypsin gene expression in human intestinal epithelial cell line caco-2 by HNF-1alpha and HNF-4. Am J Physiol 276 G1181 G1194
27. OzekiT
TakahashiY
KumeT
NakayamaK
YokoiT
2001 Co-operative regulation of the transcription of human dihydrodiol dehydrogenase (DD)4/aldo-keto reductase (AKR)1C4 gene by hepatocyte nuclear factor (HNF)-4alpha/gamma and HNF-1alpha. Biochem J 355 537 544
28. GuptaRK
VatamaniukMZ
LeeCS
FlaschenRC
FulmerJT
2005 The MODY1 gene HNF-4alpha regulates selected genes involved in insulin secretion. J Clin Invest 115 1006 1015
29. MiuraA
YamagataK
KakeiM
HatakeyamaH
TakahashiN
2006 Hepatocyte nuclear factor-4alpha is essential for glucose-stimulated insulin secretion by pancreatic beta-cells. J Biol Chem 281 5246 5257
30. PearsonER
BojSF
SteeleAM
BarrettT
StalsK
2007 Macrosomia and hyperinsulinaemic hypoglycaemia in patients with heterozygous mutations in the HNF4A gene. PLoS Med 4 e118 doi:10.1371/journal.pmed.0040118
31. GuptaRK
GaoN
GorskiRK
WhiteP
HardyOT
2007 Expansion of adult beta-cell mass in response to increased metabolic demand is dependent on HNF-4alpha. Genes Dev 21 756 769
32. ParvizF
MatulloC
GarrisonWD
SavatskiL
AdamsonJW
2003 Hepatocyte nuclear factor 4alpha controls the development of a hepatic epithelium and liver morphogenesis. Nat Genet 34 292 296
33. DukesID
SreenanS
RoeMW
LevisettiM
ZhouYP
1998 Defective pancreatic beta-cell glycolytic signaling in hepatocyte nuclear factor-1alpha-deficient mice. J Biol Chem 273 24457 24464
34. LeeYH
SauerB
GonzalezFJ
1998 Laron dwarfism and non-insulin-dependent diabetes mellitus in the Hnf - 1alpha knockout mouse. Mol Cell Biol 18 3059 3068
35. PontoglioM
SreenanS
RoeM
PughW
OstregaD
1998 Defective insulin secretion in hepatocyte nuclear factor 1 alpha-deficient mice. Journal of Clinical Investigation 101 2215 2222
36. ShihDQ
HeimesaatM
KuwajimaS
SteinR
WrightCV
2002 Profound defects in pancreatic beta -cell function in mice with combined heterozygous mutations in Pdx-1, Hnf-1alpha, and Hnf-3beta. Proc Natl Acad Sci U S A 99 3818 3823
37. van dePJ
KettelarijN
van BakelH
KockelkornTT
van LeenenD
2005 Mediator expression profiling epistasis reveals a signal transduction pathway with antagonistic submodules and highly specific downstream targets. Mol Cell 19 511 522
38. Van DriesscheN
DemsarJ
BoothEO
HillP
JuvanP
2005 Epistasis analysis with global transcriptional phenotypes. Nat Genet 37 471 477
39. CapaldiAP
KaplanT
LiuY
HabibN
RegevA
2008 Structure and function of a transcriptional network activated by the MAPK Hog1. Nat Genet 40 1300 1306
40. ParrizasM
MaestroMA
BojSF
PaniaguaA
CasamitjanaR
2001 Hepatic nuclear factor 1-alpha directs nucleosomal hyperacetylation to its tissue-specific transcriptional targets. Mol Cell Biol 21 3234 3243
41. BellGI
PolonskyKS
2001 Diabetes mellitus and genetically programmed defects in beta-cell function. Nature 414 788 791
42. FerrerJ
2002 A genetic switch in pancreatic beta-cells - Implications for differentiation and haploinsufficiency. Diabetes 51 2355 2362
43. IharaA
YamagataK
NammoT
MiuraA
YuanM
2005 Functional characterization of the HNF4alpha isoform (HNF4alpha8) expressed in pancreatic beta-cells. Biochem Biophys Res Commun 329 984 990
44. HayhurstGP
LeeYH
LambertG
WardJM
GonzalezFJ
2001 Hepatocyte nuclear factor 4alpha (nuclear receptor 2A1) is essential for maintenance of hepatic gene expression and lipid homeostasis. Mol Cell Biol 21 1393 1403
45. HerreraPL
2000 Adult insulin - and glucagon-producing cells differentiate from two independent cell lineages. Development 127 2317 2322
46. LucoRF
MaestroMA
SadoniN
ZinkD
FerrerJ
2008 Targeted deficiency of the transcriptional activator Hnf1alpha alters subnuclear positioning of its genomic targets. PLoS Genet 4 e1000079 doi:10.1371/journal.pgen.1000079
47. Ho SuiSJ
FultonDL
ArenillasDJ
KwonAT
WassermanWW
2007 oPOSSUM: integrated tools for analysis of regulatory motif over-representation. Nucleic Acids Res 35 W245 W252
48. XieX
LuJ
KulbokasEJ
GolubTR
MoothaV
2005 Systematic discovery of regulatory motifs in human promoters and 3′ UTRs by comparison of several mammals. Nature 434 338 345
49. St OngeRP
ManiR
OhJ
ProctorM
FungE
2007 Systematic pathway analysis using high-resolution fitness profiling of combinatorial gene deletions. Nat Genet 39 199 206
50. JasnosL
KoronaR
2007 Epistatic buffering of fitness loss in yeast double deletion strains. Nat Genet 39 550 554
Štítky
Genetika Reprodukční medicína
Článek SMA-10/LRIG Is a Conserved Transmembrane Protein that Enhances Bone Morphogenetic Protein SignalingČlánek B1 SOX Coordinate Cell Specification with Patterning and Morphogenesis in the Early Zebrafish EmbryoČlánek Bulk Segregant Analysis by High-Throughput Sequencing Reveals a Novel Xylose Utilization Gene fromČlánek Abundant Quantitative Trait Loci Exist for DNA Methylation and Gene Expression in Human Brain
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2010 Číslo 5
-
Všechny články tohoto čísla
- Digital Quantification of Human Eye Color Highlights Genetic Association of Three New Loci
- Common Genetic Variants near the Brittle Cornea Syndrome Locus Influence the Blinding Disease Risk Factor Central Corneal Thickness
- Age- and Temperature-Dependent Somatic Mutation Accumulation in
- All About Mitochondrial Eve: An Interview with Rebecca Cann
- Aging and Chronic Sun Exposure Cause Distinct Epigenetic Changes in Human Skin
- Transposed Genes in Arabidopsis Are Often Associated with Flanking Repeats
- A Survey of Genomic Traces Reveals a Common Sequencing Error, RNA Editing, and DNA Editing
- Gene Transposition Causing Natural Variation for Growth in
- The Polyproline Site in Hinge 2 Influences the Functional Capacity of Truncated Dystrophins
- Epistasis of Transcriptomes Reveals Synergism between Transcriptional Activators Hnf1α and Hnf4α
- Integration of Light Signals by the Retinoblastoma Pathway in the Control of S Phase Entry in the Picophytoplanktonic Cell
- The Proprotein Convertase Encoded by () Is Required in Corpora Cardiaca Endocrine Cells Producing the Glucose Regulatory Hormone AKH
- Sgs1 and Exo1 Redundantly Inhibit Break-Induced Replication and Telomere Addition at Broken Chromosome Ends
- A MATE-Family Efflux Pump Rescues the 8-Oxoguanine-Repair-Deficient Mutator Phenotype and Protects Against HO Killing
- The Relationship among Gene Expression, the Evolution of Gene Dosage, and the Rate of Protein Evolution
- SMA-10/LRIG Is a Conserved Transmembrane Protein that Enhances Bone Morphogenetic Protein Signaling
- The Nuclear Receptor DHR3 Modulates dS6 Kinase–Dependent Growth in
- Involvement of Global Genome Repair, Transcription Coupled Repair, and Chromatin Remodeling in UV DNA Damage Response Changes during Development
- B1 SOX Coordinate Cell Specification with Patterning and Morphogenesis in the Early Zebrafish Embryo
- Linkage and Association Mapping of Flowering Time in Nature
- Bulk Segregant Analysis by High-Throughput Sequencing Reveals a Novel Xylose Utilization Gene from
- Abundant Quantitative Trait Loci Exist for DNA Methylation and Gene Expression in Human Brain
- Ablation of Whirlin Long Isoform Disrupts the USH2 Protein Complex and Causes Vision and Hearing Loss
- Characterization of Oxidative Guanine Damage and Repair in Mammalian Telomeres
- DNA Adenine Methylation Is Required to Replicate Both Chromosomes Once per Cell Cycle
- Genome-Wide Copy Number Variation in Epilepsy: Novel Susceptibility Loci in Idiopathic Generalized and Focal Epilepsies
- FACT Prevents the Accumulation of Free Histones Evicted from Transcribed Chromatin and a Subsequent Cell Cycle Delay in G1
- GC-Biased Evolution Near Human Accelerated Regions
- Liver and Adipose Expression Associated SNPs Are Enriched for Association to Type 2 Diabetes
- Myeloid Cell-Restricted Insulin Receptor Deficiency Protects Against Obesity-Induced Inflammation and Systemic Insulin Resistance
- The Mating Type Locus () and Sexual Reproduction of : Insights into the Evolution of Sex and Sex-Determining Chromosomal Regions in Fungi
- B-Cyclin/CDKs Regulate Mitotic Spindle Assembly by Phosphorylating Kinesins-5 in Budding Yeast
- Post-Replication Repair Suppresses Duplication-Mediated Genome Instability
- Genome Sequence of the Plant Growth Promoting Endophytic Bacterium sp. 638
- Transcription Factors Mat2 and Znf2 Operate Cellular Circuits Orchestrating Opposite- and Same-Sex Mating in
- The Use of Orthologous Sequences to Predict the Impact of Amino Acid Substitutions on Protein Function
- Mutation in Archain 1, a Subunit of COPI Coatomer Complex, Causes Diluted Coat Color and Purkinje Cell Degeneration
- Shelterin-Like Proteins and Yku Inhibit Nucleolytic Processing of Telomeres
- Affecting Function Causes a Dilated Heart in Adult
- Manipulation of Behavioral Decline in with the Rag GTPase
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Common Genetic Variants near the Brittle Cornea Syndrome Locus Influence the Blinding Disease Risk Factor Central Corneal Thickness
- All About Mitochondrial Eve: An Interview with Rebecca Cann
- The Relationship among Gene Expression, the Evolution of Gene Dosage, and the Rate of Protein Evolution
- SMA-10/LRIG Is a Conserved Transmembrane Protein that Enhances Bone Morphogenetic Protein Signaling
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



