-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaGene Transposition Causing Natural Variation for Growth in
A major challenge in biology is to identify molecular polymorphisms responsible for variation in complex traits of evolutionary and agricultural interest. Using the advantages of Arabidopsis thaliana as a model species, we sought to identify new genes and genetic mechanisms underlying natural variation for shoot growth using quantitative genetic strategies. More quantitative trait loci (QTL) still need be resolved to draw a general picture as to how and where in the pathways adaptation is shaping natural variation and the type of molecular variation involved. Phenotypic variation for shoot growth in the Bur-0 × Col-0 recombinant inbred line set was decomposed into several QTLs. Nearly-isogenic lines generated from the residual heterozygosity segregating among lines revealed an even more complex picture, with major variation controlled by opposite linked loci and masked by the segregation bias due to the defective phenotype of SG3 (Shoot Growth-3), as well as epistasis with SG3i (SG3-interactor). Using principally a fine-mapping strategy, we have identified the underlying gene causing phenotypic variation at SG3: At4g30720 codes for a new chloroplast-located protein essential to ensure a correct electron flow through the photosynthetic chain and, hence, photosynthesis efficiency and normal growth. The SG3/SG3i interaction is the result of a structural polymorphism originating from the duplication of the gene followed by divergent paralogue's loss between parental accessions. Species-wide, our results illustrate the very dynamic rate of duplication/transposition, even over short periods of time, resulting in several divergent—but still functional—combinations of alleles fixed in different backgrounds. In predominantly selfing species like Arabidopsis, this variation remains hidden in wild populations but is potentially revealed when divergent individuals outcross. This work highlights the need for improved tools and algorithms to resolve structural variation polymorphisms using high-throughput sequencing, because it remains challenging to distinguish allelic from paralogous variation at this scale.
Published in the journal: . PLoS Genet 6(5): e32767. doi:10.1371/journal.pgen.1000945
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1000945Summary
A major challenge in biology is to identify molecular polymorphisms responsible for variation in complex traits of evolutionary and agricultural interest. Using the advantages of Arabidopsis thaliana as a model species, we sought to identify new genes and genetic mechanisms underlying natural variation for shoot growth using quantitative genetic strategies. More quantitative trait loci (QTL) still need be resolved to draw a general picture as to how and where in the pathways adaptation is shaping natural variation and the type of molecular variation involved. Phenotypic variation for shoot growth in the Bur-0 × Col-0 recombinant inbred line set was decomposed into several QTLs. Nearly-isogenic lines generated from the residual heterozygosity segregating among lines revealed an even more complex picture, with major variation controlled by opposite linked loci and masked by the segregation bias due to the defective phenotype of SG3 (Shoot Growth-3), as well as epistasis with SG3i (SG3-interactor). Using principally a fine-mapping strategy, we have identified the underlying gene causing phenotypic variation at SG3: At4g30720 codes for a new chloroplast-located protein essential to ensure a correct electron flow through the photosynthetic chain and, hence, photosynthesis efficiency and normal growth. The SG3/SG3i interaction is the result of a structural polymorphism originating from the duplication of the gene followed by divergent paralogue's loss between parental accessions. Species-wide, our results illustrate the very dynamic rate of duplication/transposition, even over short periods of time, resulting in several divergent—but still functional—combinations of alleles fixed in different backgrounds. In predominantly selfing species like Arabidopsis, this variation remains hidden in wild populations but is potentially revealed when divergent individuals outcross. This work highlights the need for improved tools and algorithms to resolve structural variation polymorphisms using high-throughput sequencing, because it remains challenging to distinguish allelic from paralogous variation at this scale.
Introduction
Natural phenotypic variation observed among different genotypes (accessions, varieties, populations, etc) is partly explained by alterations of the genetic material. Identifying the molecular basis of phenotypic variation provides candidates to test how and where in the pathway adaptation is shaping natural variation [1]. Of particular interest is the type of sequence variation from which intra-specific diversity originates. In Arabidopsis for example, there is reason to suspect that along with single nucleotide polymorphisms and short indels, structural variants in the genome as well may be an important source of natural variation [2]–[5]. Structural submicroscopic variants (smaller than those recognized microscopically) are defined as genomic alterations (insertions, deletions, inversions, duplications and transpositions) involving segments of DNA ranging from the kb to the Mb scale [6]. They can occur in genomes after large segmental duplications and subsequent gene loss, or as the result of unequal or illegitimate recombination (tandem duplications/insertions, deletions/gene loss) [7], DNA segment inversions [8] or transposable elements activity (dispersed gene duplication) [9].
Little is known about the prevalence of this phenomenon in plants and its phenotypic consequences, but it was recently found to be widespread in yeast [10] and in humans [11] where large structural variants (>50 kb) are confirmed to affect ∼4,000 genomic loci among healthy individuals [12]. Two individuals are also estimated to differ at approximately 1,000 copy-number variations (CNVs) alone [13]. In humans, structural variation may have a more significant impact on phenotypic variation than SNPs and they were implicated in gene expression variation, female fertility, susceptibility to HIV infection, systemic autoimmunity, genomic disorders… [13], [14].
Structural variation may also contribute to postzygotic isolation through the production of genetically deficient hybrids [15], as recently demonstrated among Arabidopsis strains in a study describing how the transposition of the functional copy of an essential gene (balanced structural polymorphism) results in recessive embryonic lethality in an intraspecific cross [16]. Complex structural variation at the GS-Elong locus, including tandem MAM-gene duplication, gene loss, gene conversion and CNV was shown to cause natural variation for insect herbivore resistance [17]. Another example of natural variation for disease resistance in Arabidopsis was explained by the reciprocal loss of R-gene paralogues located in an ancient segmental duplication, which resulted in certain combinations lacking either functional copy [18].
Dispersed kind of structural variation will result in epistasis in intraspecific crosses (if the functional structural variation has phenotypic consequences) and could therefore be detected in mapping populations. Although experimental mapping populations of the type of recombinant inbred lines (RILs) allow detection of genetic interactions between loci, the number of RILs observed as well as possible segregation distortions caused by lethality or reduced fitness of particular genotypes may hamper the power to detect interacting QTL [19]–[21]. In this context, the residual heterozygosity existing in some RIL sets is a plus, since deleterious genetic combinations can be restored and studied from their ‘latent’ heterozygous states.
Currently, not much interest has been manifested for the detection and consequences of structural polymorphisms in plants, probably because it is even less convenient to detect and complement than ‘simple’ coding sequence changes in a gene, for example. Although the global impact of structural variation is unknown, it might have dramatic consequences on phenotypic diversity [22]. Unfortunately, array-based re-sequencing projects are limited to this respect as they can only easily detect deletions relative to the reference sequence [5]. For A. thaliana, the short sequence reads produced by deep-sequencing on Illumina proved to support, in addition to SNPs, the detection of short to medium-size indels, and the discovery of new sequences (absent from the reference genome) but not their sequence context [2]. Therefore, nothing is obvious about the frequency of structural variation and its association with phenotype as no ideal methodologies yet allow direct detection of structural variants in plants. Here we give an example of a functional structural polymorphism resulting from the divergent evolution of duplicate genes among A. thaliana accessions.
Results/Discussion
We have used genome-wide molecular quantitative genetics to investigate natural genetic variation for shoot growth as a complex trait. Since the parental accessions were showing phenotypic differences with regard to shoot growth in our conditions, a subset of 164 Bur-0 × Col-0 Recombinant Inbred Lines optimized for QTL mapping [23] was grown and phenotyped in vitro in standard conditions in order to map loci affecting early stage shoot growth. Transgressive segregation of the shoot phenotypes observed among RILs (Figure S1A) indicates that the genetic potential for the study of shoot growth exists in this set. Indeed, four significant QTLs with LOD scores greater than 2.5 were mapped in this cross (Figure S1B).
Confirming the chromosome 4 locus
In this work, we are now focusing on allelic variation in the genomic region underlying the QTL predicted between 14 and 15 Mb on chromosome 4. Confirmation of the phenotypic effect related to this locus was performed using specific NILs differing only for a small genomic region spanning a few cM around the QTL. NILs for this QTL were obtained by producing Heterogeneous Inbred Families (HIFs) which are easily generated taking advantage of the residual heterozygosity still segregating in F6 RILs [24], [25]. Initially, four candidate RILs (# 067, 081, 212 and 332), heterozygous only around the chromosome 4 locus, were used. In all HIFs except HIF332, the comparison of plants that were fixed for each parental allele (Col or Bur) at the QTL region revealed a highly significant difference in shoot growth (P<0.001). The HIFs[Bur] had marked phenotypic features (Figure 1) strongly reducing shoot size, as estimated either at a young stage in vitro (−25%) or at a later stage on soil (−70%), and chlorophyll content (−40% in greenhouse conditions). Then, bolting in plants carrying the Bur allele was delayed by approximately two weeks, plants were less robust with shorter inflorescence stems and yielded significantly fewer seeds than the HIFs[Col] plants, though flowers were normally developed and fully fertile. We named this QTL SG3 (Shoot Growth-3). Yet, the observed allelic effect was surprising: the Bur allele at the QTL was negatively affecting shoot size, while the QTL mapping had predicted that an opposite effect was segregating with this region (Figure S1). Moreover, HIF332 was not segregating for SG3 although its heterozygous region seemed to fully cover the QTL locus (and other positive HIFs' heterozygous regions), suggesting that the QTL is most probably involved in an epistatic interaction. The analysis of several (12) other independent RILs genotypically segregating at the locus revealed that SG3 was in complete interaction with a region at the top of chromosome 4, which we called SG3i (SG3-interactor; Figure 1). The phenotypic segregation of SG3 is conditioned by the presence of a Col allele at SG3i. Hence, with only two RILs (among our 164 subset) fixed for the combination of alleles giving rise to the defective growth phenotype, we could not have mapped this locus with the RILs and SG3 is a distinct locus than mapped initially in this region. Indeed, further analysis of HIFs genotypically segregating for the bottom of chromosome 4 region, but that do not segregate for SG3 (because harbouring a Bur allele at SG3i), showed that the locus mapped in the QTL analysis is real but distinct from SG3 and of much smaller–opposite–effect (HIF332; Figure S2). It is very likely that plants with the Col allele at SG3i and the Bur allele at SG3 (SG3i[Col]/SG3[Bur] combination) were unintentionally counter-selected during the single-seed descent process used for RIL fixation (despite the care to avoid any obvious bias) because of their retarded and small stature early after germination.
Fig. 1. Confirmation of the phenotypic effect of SG3 and its interaction with SG3i. 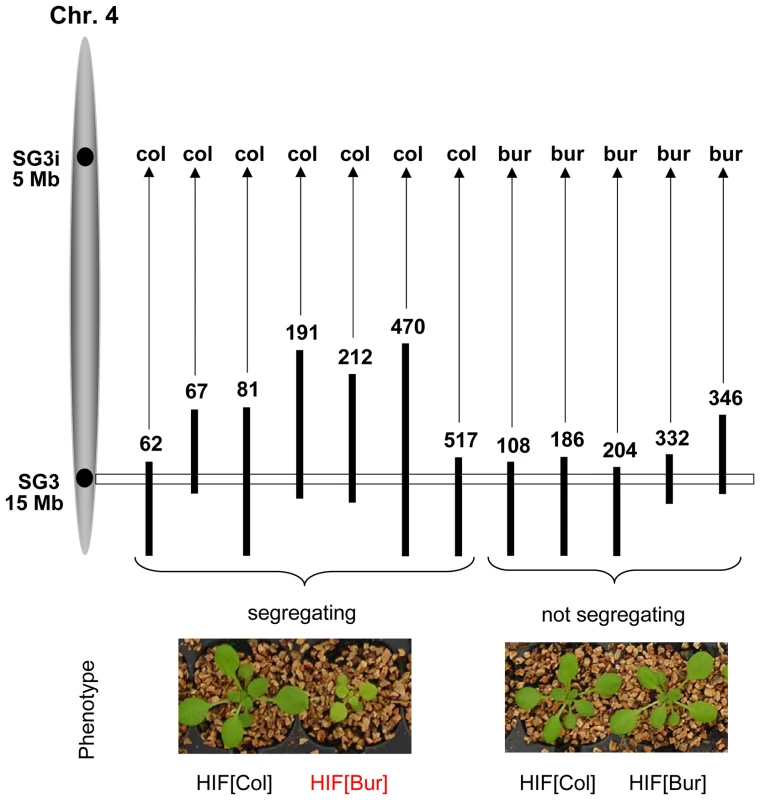
Black vertical bars represent heterozygous regions on chromosome 4 (represented in grey on the left) from different RILs used to generate HIFs and to check for the segregation of the defective growth/pale green phenotype. Photos of the plants were taken in the greenhouse three weeks after germination and show typical phenotypic segregation of individual HIFs. The phenotypic segregation of SG3 is conditioned by the presence of the Col allele at another locus: SG3i. The SG3i[Col]/SG3[Bur] combination causes a pale green pigmentation and defective growth. Map-based cloning of SG3
We used HIF212 to further perform the fine-mapping of SG3 by recurrent (genotypic) selection and (phenotypic) analysis of recombinants within the target interval (rHIF, see Materials and Methods), as described earlier [26]. A first set of nine recombinants were identified in the initial heterozygous region of ∼5 Mb, when fixing HIF212. Each recombinant was tested for the segregation of the phenotype and the region of interest was fast reduced to ∼1 Mb. A following screen of 600 plants resulted in the isolation of 35 recombination events in the remaining interval. Recombinants with homozygous Bur-genotype at the QTL were easily phenotyped during the selection process (the Col allele is dominant at SG3) and seven other interesting recombinants were analysed by progeny-testing. Reliable phenotyping results allowed us to further narrow down the candidate interval to ∼100 kb. Finally, a last screen of 5,000 plants, descending from one positive rHIF, provided 34 additional recombinants in the 100 kb interval. The analysis of 12 interesting recombinants delineated the region of interest to a 9 kb-interval (Figure 2), precisely 9,325 bp between bordering polymorphisms internal to the recombination events. Based on the most informative recombinants (rHIF212.60 and rHIF212.77), an ‘advanced rHIF cross’ (arHIF; see Materials and Methods and [26]) was designed in order to obtain a line (arHIF212.97) that was segregating solely for the final candidate interval. This confirmed the presence of SG3 within the 9 kb-interval when comparing arHIF lines fixed for each allele (arHIF[Col] vs arHIF[Bur]). Three predicted genes (At4g30720, At4g30730 and At4g30740) are present in this interval.
Fig. 2. Fine-mapping of SG3. 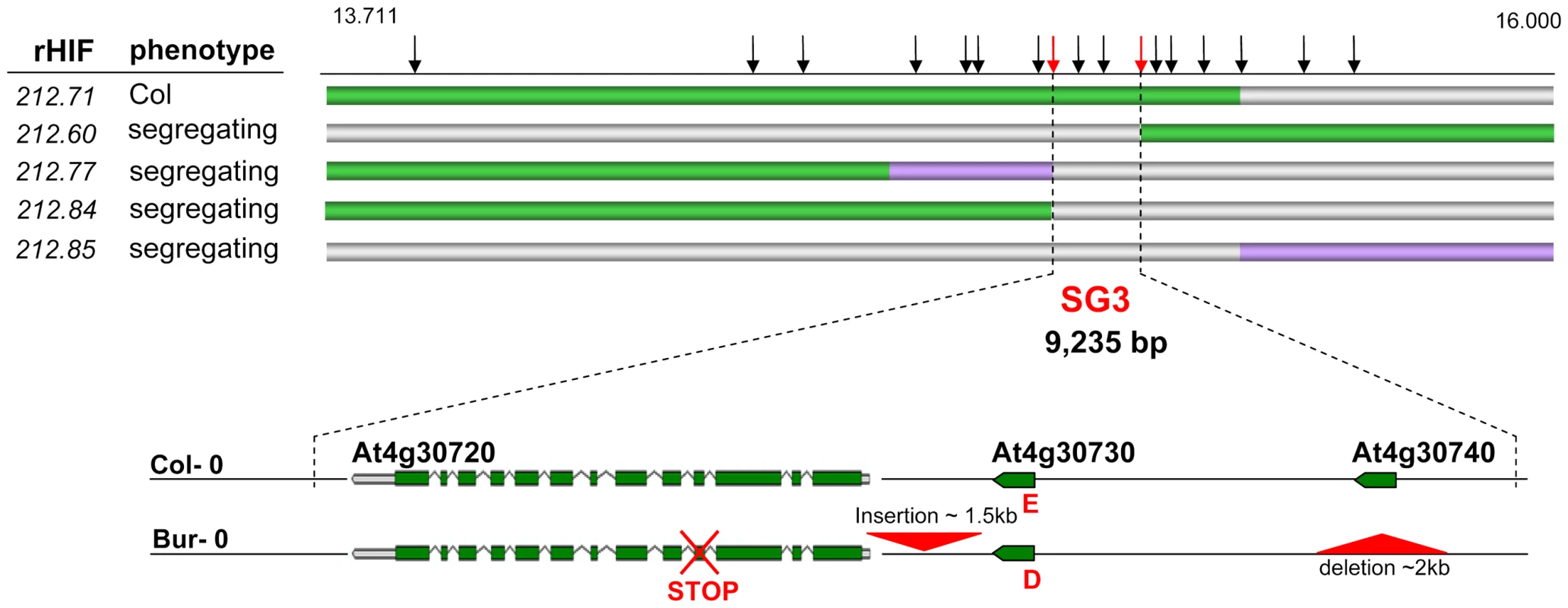
The genotype of the most informative recombinants (rHIFs) used to delineate the final 9 kb-candidate interval are represented with horizontal bars (green for Col allele, purple for Bur allele, grey for heterozygous). Vertical black arrows above represent frequently used markers for genotyping between 13.711 and 16.000 Mb on chromosome 4. Red arrows mark the most informative recombination breakpoints. Below, genes in the final candidate interval are illustrated for both parental accessions. Major polymorphisms are marked in red, including an E/D amino-acid change in At4g30730 and a premature STOP codon in At4g30720 in Bur-0. The 9 kb-interval was sequenced in both parental accessions to identify putative functional polymorphisms. The presence of “heterozygous” (double) peaks in the sequence chromatograms when amplifying from Bur-0 DNA with primers within At4g30720, was taken as predictive of a duplication of (at least part of) the candidate region. SNP information was isolated from the “heterozygous” peaks, converted into CAPS markers and the duplication was mapped thanks to the original RIL set genetic map [23] between two markers at physical positions 5,629 and 6,923 kb, i.e. in the region of SG3i.
Polymorphisms at SG3
Some major polymorphisms were identified in the SG3 candidate region when comparing Bur-0 to Col-0 (Figure 2). A ∼2 kb region including At4g30740 is absent from the Bur-0 accession. Conversely, a large (1,130 bp) insertion is present in Bur-0, 180 bp upstream of At4g30720 (it includes LTR and ADN/MuDR transposon traces and a putative target site duplication TGATG/TGATG). Also, a 1 bp-deletion in exon four of At4g30720 results in a frame shift, predicting a premature stop codon which terminates the ORF after 5 amino acids. Finally, one non-synonymous SNP is present in the coding sequence of At4g30730, changing glutamic acid (E)#66 into aspartic acid (D).
Identification of the causal gene
The predicted At4g30720 is encoding a 707aa-putative oxidoreductase/electron carrier detected in the chloroplast stroma [27]. It has a predicted FAD-dependent oxidoreductase domain (IPR006076), adrenodoxin reductase domain (IPR000759) and N-terminal domain (www.arabidopsis.org; www.ncbi.nlm.nih.gov). Both At4g30730 and At4g30740 putatively encode very small proteins of unknown functions, if not pseudogenes (both are partially matching neighbouring genes, respectively At4g30750 and At4g30710, and they are not supported by ESTs and expression data).
T-DNA insertions in At4g30730 (SALK_049026-promoter) and At4g30740 (SALK_057859-3′UTR) had wild-type phenotypes for shoot growth. Instead, a homozygous mutant line (SALK_059716) with a T-DNA insertion in the fourth exon of At4g30720 in a Col-0 background, phenotypically resembled plants with SG3i[Col]/SG3[Bur] allelic combination (Figure 3). In addition, RT-PCR assays using primers annealing to the exon sequences flanking the T-DNA insertion site failed to amplify any full-length transcript, indicating that this mutant is very likely a knock-out.
Fig. 3. Quantitative complementation of an At4g30720 T–DNA allele. 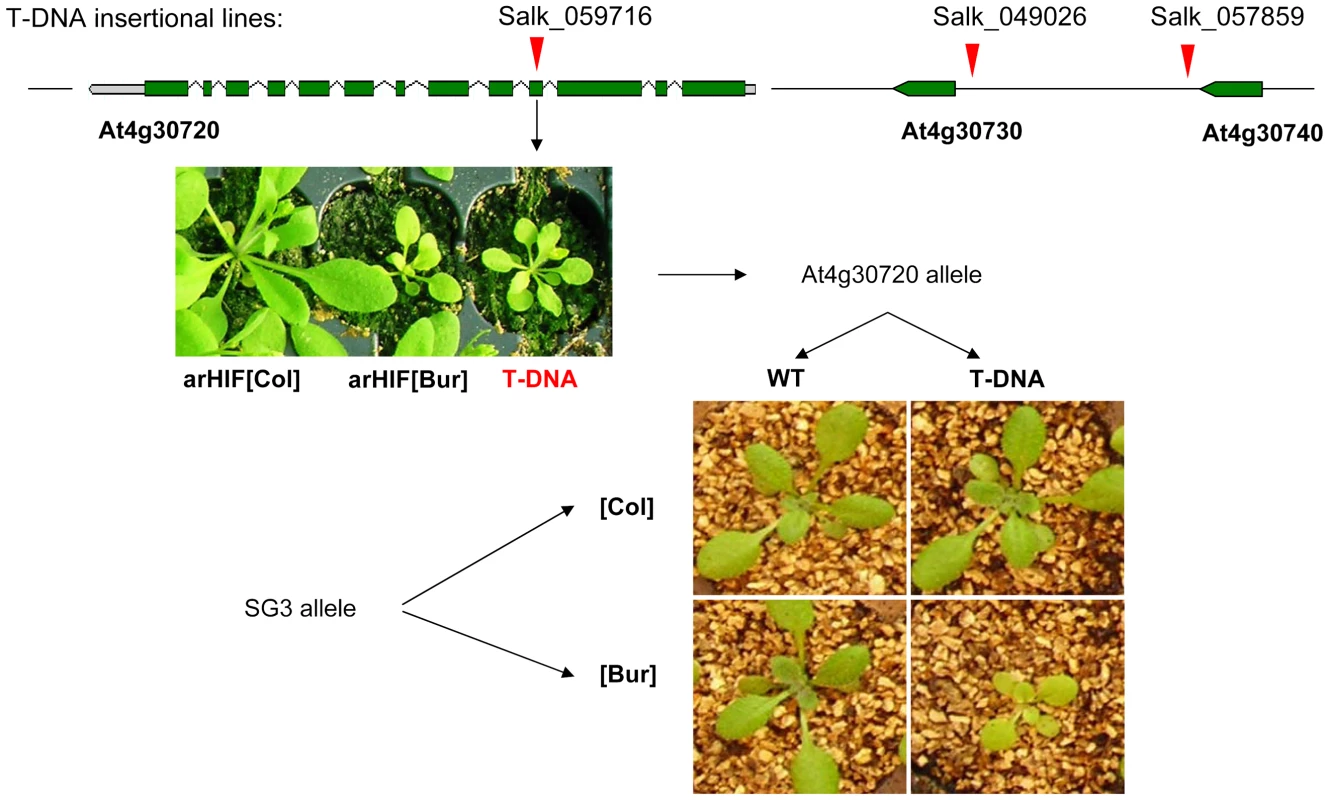
T-DNA insertion points are marked by red arrows. SALK_049026 hits the promoter of At4g30730 and SALK_057859 hits the 3′UTR of At4g30740 and have no phenotypic consequences in the Col-0 background. The phenotype of the SALK-059716 T-DNA homozygous mutant (an insertion in the fourth exon of At4g30720) is shown below and strikingly resembles the arHIF[Bur] phenotype. The complementation test combined the QTL alleles ([Col] or [Bur]) with either a mutant (T-DNA) or a wild type (WT, i.e. Col) allele at the candidate gene. Typical phenotypes resulting from the 4 combinations are shown and revealed that the SG3[Bur]-allele fails to complement the mutant phenotype. Quantitative complementation tests showed that the Col allele at SG3 was able to rescue the mutant allele phenotype, while, in an identical F1 genetic background, the Bur allele failed to complement the T-DNA allele (Figure 3). This indicates that the phenotype observed in arHIF[Bur] is probably due to a defect in At4g30720 and that (in the absence of any observed expression-level differences) the 1 bp-deletion found in At4g30720[Bur] is likely to be the causal polymorphism.
Characterizing SG3i
A Bur-0 BAC genomic library was used to sequence the duplicated copy of At4g30720 at SG3i. Although a non-synonymous SNP is causing an amino-acid change compared to At4g30720[Col], converting alanine #43 into threonine, it seems that this copy must be encoding a functional protein and explains why the Bur-0 accession itself does not have a small, pale green phenotype. Our sequencing results on the BAC identify a 10 kb-region (including At4g30720' paralogue) at SG3i clearly corresponding to the SG3 region surrounding At4g30720. Efforts have been made but we did not manage to obtain any good sequence (corresponding to any known stretch of DNA that would correspond to our reference sequence, Col-0) when sequencing from the 10 kb duplicated region toward the outside. Direct sequencing of the BAC was then impossible due to double priming which suggests the presence of repetitive DNA around the duplicated region. In addition, attempts to amplify specific loci (especially genes neighbouring the 10 kb duplicated region from SG3, like At4g30710, At4g30750, …) from the SG3i BAC gave no results, so we consider likely that the paralogues arose from a small-scale dispersed duplication event.
Genetic mechanism
Overall, our results support the following: At4g30720 is responsible for the observed phenotype; two copies are present in the Bur-0 genome, one of which is not functional (at SG3); only one (functional) copy is present in the Col-0 genome (at SG3). Thus, plants that are homozygous for the Col allele at the SG3i locus and at the same time homozygous for the Bur allele at SG3, have no functional copy of At4g30720, resulting in the defective growth phenotype (Figure 4). This theory is supported by the mutant line SALK_059716 which represents an equivalent combination of alleles and by the observation of the segregation of the phenotype in other independent crosses (see below). We suggest that the transposition of the functional copy of the gene in Bur-0 (compared to Col-0) is the result of divergent evolution of the paralogues after an ancestral (dispersed) single gene duplication. This type of structural variation could have been previously underestimated as it has no phenotypic impact on the parental accessions themselves but seems to evolve quickly and could therefore represent a major source of intraspecific diversity and constraints [16].
Fig. 4. Species-wide patterns of diversity for the SG3/SG3i duplication of At4g30720. 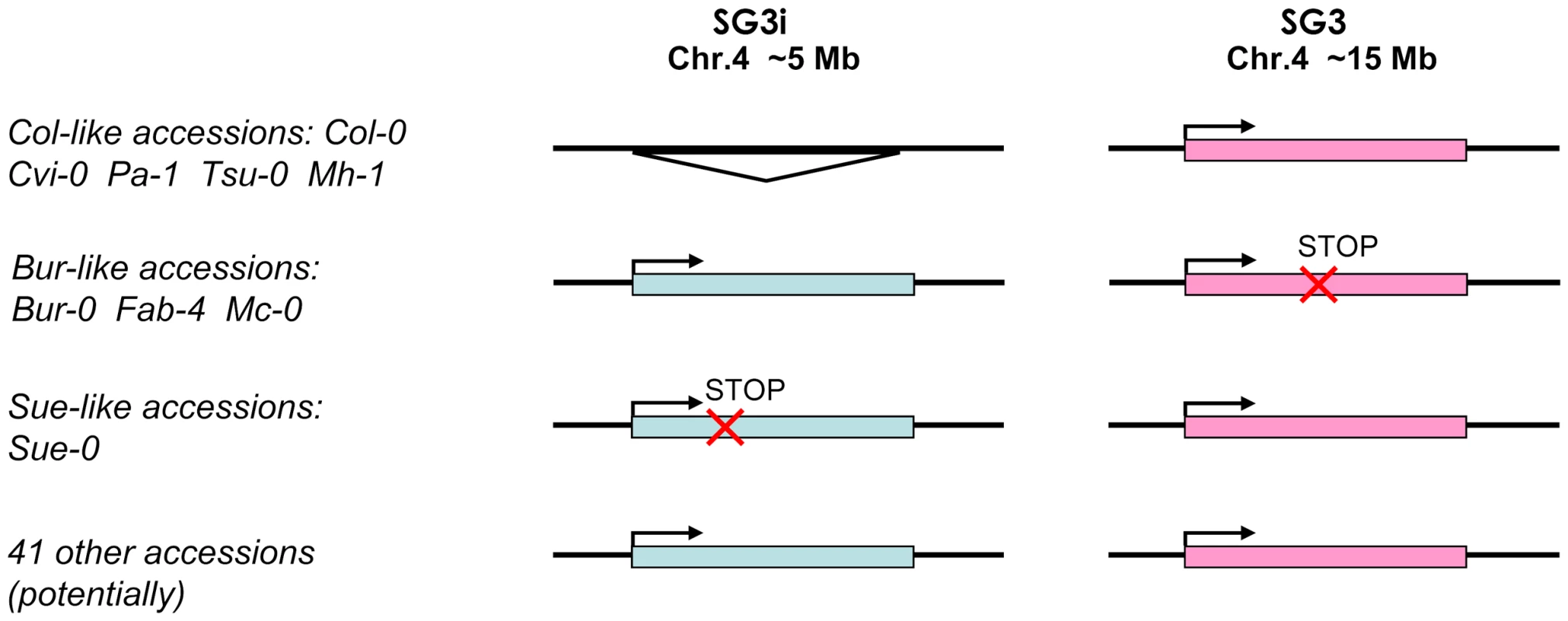
Accessions are grouped according to the divergence of the duplicate gene pair schematized on the right. A triangle indicates the complete absence of the paralogue. The ‘stop’ mark indicates the presence of a premature stop codon. The majority of accessions fall in the last group where no major functional polymorphism has been detected for both copies. Species-wide diversity
We sequenced ∼3,400 bp of At4g30720 from 52 different accessions (essentially our core-collection of 48 [28]; Table S1) to investigate the species-wide patterns of diversity for this duplication event. The SG3i amplicons were distinguished from SG3 by a specific 7 bp-indel located 125 bp upstream of the start codon. The vast majority of accessions analysed bear two copies, while only 5 have a single copy, like Col-0. Most of the two copies-accessions seem to have both copies functional (no obvious functional polymorphisms in the coding sequence). Bur-0, Mc-0 and Fab-4 share the same polymorphism resulting in a premature stop codon in At4g30720, while Sue-0 presents a distinct 1 bp-indel causing a premature stop codon in the copy present at SG3i (Figure 4). RT-PCR analyses were performed for a subset of 26 accessions. In all two copies-accessions tested, transcripts were detected from both paralogues, with no exceptions.
In addition, crosses between Bur-like and Col-like accessions were designed in order to independently validate the candidate genetic mechanism and the functional group in which accessions belong (Figure 4). The F2 progeny of a cross between the single copy-accession Cvi-0 and Bur-0 is clearly segregating for the small, pale green phenotype in accordance with segregation of alleles at SG3/SG3i loci, indicating that Cvi-0 has a functional copy of At4g30720 at SG3 despite bearing a haplotype much closer to Bur-0 than to Col-0 at this locus (see below). Also, the F2 progeny of reciprocal crosses between Col-0 and either Mc-0 or Fab-4 (both Bur-like accessions) confirmed that a similar allelic combination is present in these Bur-like accessions. As expected, segregation of the phenotype was also observed in the reciprocal crosses between Sue-0 and Bur-0, in agreement with our sequencing results. In each case the causality link between SG3/SG3i allelic combination and the phenotype was confirmed by genotyping at these two loci. As negative controls, no phenotype was observed in the Sue-0 × Col-0, Cvi-0 × Col-0 reciprocal crosses' progeny, as well as in crosses between Bur-0 and other Bur-like accessions. The clear link observed between the segregation of diverse functional alleles at SG3/SG3i and the phenotype in distinct unrelated genetic backgrounds is another strong confirmation of the identity of the QTL.
Phylogenetic analysis, using A. lyrata as outgroup, essentially clustered the two paralogues in distinct haplotype groups. The SG3 cluster, contains most of the SG3 copies except for those from Bur-0, Fab-4, Mc-0, Sue-0 and Cvi-0 which branch together as a divergent group with a distinct haplotype (Figure 5). Overall, each of the three clusters have distinctive polymorphisms and we see no obvious genealogy between the two main clusters and either the minor cluster or A. lyrata (Table S2). It seems that the minor cluster evolved, in some way, away from the rest of the SG3 copies. Ectopic recombination and gene conversion (the extents of which remain to be shown in plants) could also participate in generating new haplotypes during duplicated gene evolution [29]. Very likely, the At4g30720 alleles found in the minor cluster derive from a common ancestor. It is nevertheless surprising to see the very divergent path followed by the five accessions bearing this haplotype, with at least one belonging to each of the first three functional groups depicted on Figure 4. The Bur-0, Fab-4 and Mc-0 SG3 copies lost their function (premature stop codon) probably very recently; while the Cvi-0 and Sue-0 copies maintained undifferentiated functions (as confirmed in the crosses) and are found associated with, respectively, a deleted and a non-functional paralogue at SG3i. Hybridization between accessions bearing different alleles could explain the diversity of allelic combinations detected at these unlinked loci.
Fig. 5. Phylogenetic analysis of the paralogues. 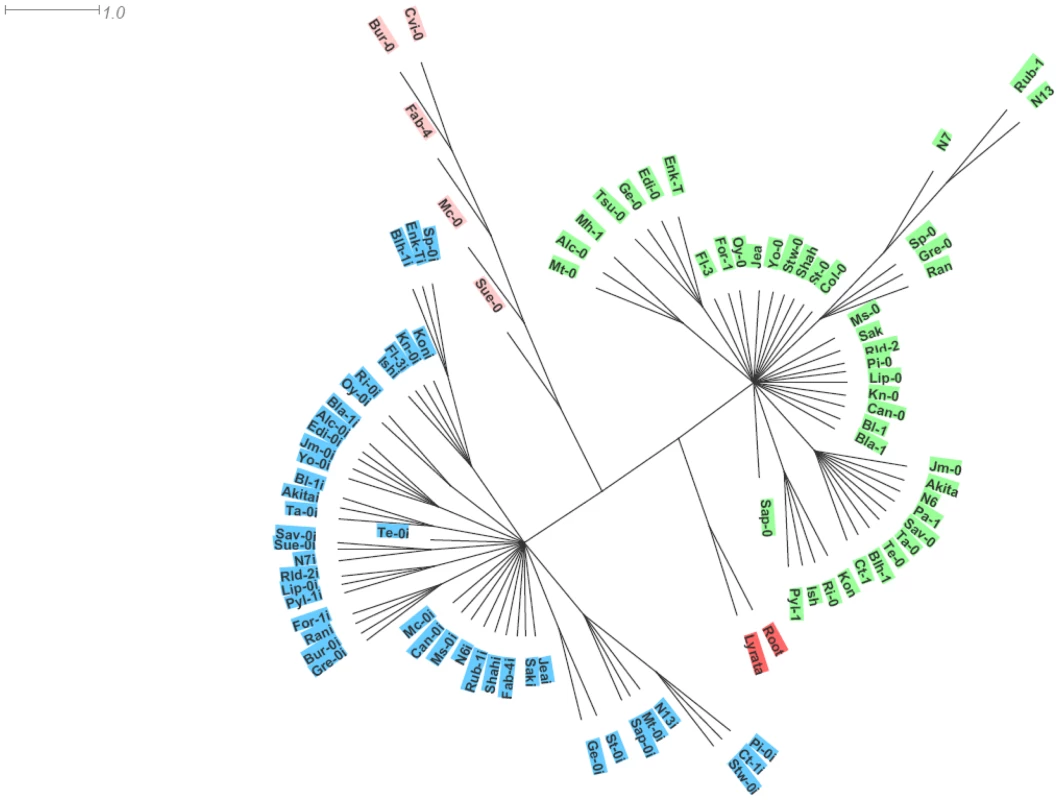
A rooted haplotype network is shown, generated from the genomic locus sequences and using A. lyrata as outgroup. To distinguish SG3i copies, the names of accessions are followed by an ‘I’ for this paralogue. SG3i copies all cluster together and form a first haplotype group (highlighted in blue). A second cluster (highlighted in green) groups most of the SG3 copies, with the exception of Bur-0, Fab-4, Mc-0, Sue-0 and Cvi-0 which branch together as a third cluster (highlighted in pink). As found previously [16], we saw no clear correlation between our haplotype network and the population structure described on a very similar accession sample [30].
Molecular population genetics
A recent duplication
In a similar fashion as described by Moore and Purugganan [31], i.e. based on Ks–the number of silent substitutions per site between duplicate gene pairs–and a molecular clock calibrated with an A. thaliana/A. lyrata divergence date 5.2 million years ago (mya) [32], we estimate the date of duplication of our paralogues to be about 0.25+/−0.035 mya. We conclude that the two copies of At4g30720 at SG3 and SG3i arose from a duplication event that occurred late after the divergence of A. thaliana from its close relative A. lyrata and before the wide recent colonization event/population expansion [33].
It was not possible to clearly infer the progenitor locus from the haplotype network generated from At4g30720 and its paralogue's sequences rooted with A. lyrata (Table S2). However, conserved synteny between the region of SG3 in A. thaliana and the region of the unique copy of this gene in A. lyrata suggests that SG3 is the progenitor locus. As described above, it seems that this duplicate gene pair would then be the result of a small-scale duplication of ∼10 kb.
Tests of selection
According to our estimates, total sites nucleotide diversity, θπ, varied between 0.0009 and 0.0019 for the paralogues at SG3 and SG3i respectively, consistently lower than the nucleotide diversity observed for the average Arabidopsis nuclear gene (0.0071; [34], [35]). Nevertheless, the copies at SG3i are overall more polymorphic than the ones at SG3, indicating that they may have evolved more rapidly (Table S3). To test if the reduced level of nucleotide polymorphism observed in both copies is associated with an hypothetical directional selection event, we performed a multilocus HKA test [36]: we compared sequence polymorphism and divergence at SG3 to four reference loci previously described as neutral (COI1, EDR1, NDR1 and TGA2; [37]). The results showed no significant difference (Chi2 = 4.715; P = 0.3178), indicating that our genes have levels of variation that do not differ from those of neutral reference loci. For SG3, both Tajima's D (D = −2.0506) and Fu and Li statistics D* (D* = −3.1489) indicate an excess of rare polymorphisms (singletons), significantly deviating from expectations based on the neutral-equilibrium model of molecular evolution [38], [39]. But this excess of singletons, also described at the genome scale [35], could simply reflect the inbreeding nature of the plant or a rapid post-Pleistocene expansion [40]–[42].
Toward the role of At4g30720
Phenotypically, at the vegetative stage, SG3[Bur] plants are always pale green, with fewer leaves and rosettes about 70% smaller than the SG3[Col] plants (Figure 6A). SG3[Bur] plants contain less chlorophyll (a and b) and the chlorophyll a/chlorophyll b ratio is significantly modified (Table S4).
Fig. 6. SG3 affects the photosynthetic electron transfer rate with consequences on shoot growth. 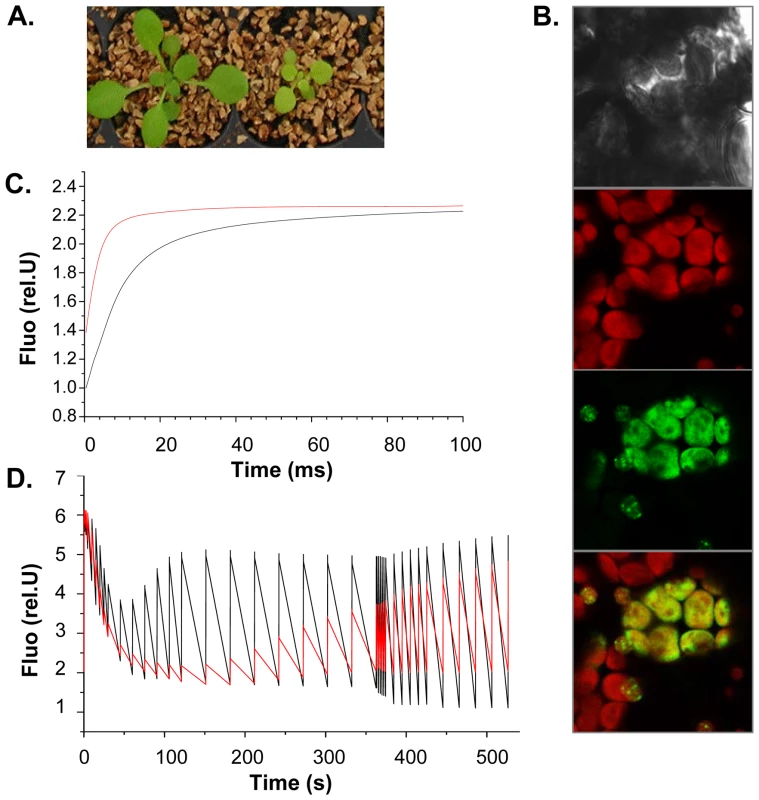
(A) Severe consequences of SG3 on pigmentation and growth observed three weeks after sowing in arHIF[Bur] (bottom) compared to arHIF[Col] (top). (B) Transient expression in Arabidopsis cotyledons of the At4g30720-GFP fused target peptide showing the localization of the protein into the chloroplast, as shown by colocalization with the chloroplast autofluorescence in red. (C) Light-induced fluorescence changes in the presence of DCMU (inhibitor of PSII) suggesting that the overall PSII + PSI content is likely to be decreased in arHIF[Bur] (red) compared to arHIF[Col] (black). (D) Differences in fluorescence observed between arHIF[Col] and arHIF[Bur] under continuous illumination. A strongly quenched Fmax' and its delayed recovery suggest a severely delayed activation of the Benson-Calvin cycle and a decrease in the electron flow through the photosynthetic chain in arHIF[Bur]. Transient expression assays of the GFP-fused (N-terminal) target peptide in Arabidopsis cotyledons confirmed the exclusive localization of the protein into the chloroplast (Figure 6B).
Chlorophyll fluorescence studies were undertaken to find evidence of the physiological role of At4g30720. Light-induced absorption changes in the absence/presence of DCMU and hydroxylamine (PSII inhibitors) at 520 nm measured 100 µs after a single turnover flash was employed to quantify the PSII/PSI stoichiometry in arHIFs. The PSII/PSI ratio seems not to be altered in the arHIF[Bur] compared to the arHIF[Col]. However, the arHIF[Bur] showed a lower Fmax/F0 ratio suggesting a higher F0 and, hence, a significant amount of PSII antenna not being connected to the PSII photochemical trap (Figure 6C). Together with the unaltered PSII/PSI stoichiometry, these data suggest that the overall PSII + PSI content is likely to be lower in arHIF[Bur].
In line with this hypothesis, the fluorescence changes observed during an illumination of a few minutes show that the line with a Bur allele at SG3 is marked by strongly quenched Fmax' and that the recovery of this quenching is severely delayed (Figure 6D). This may reflect the delayed activation of the Benson-Calvin cycle which would stem from the decrease in the electron flow through the photosynthetic chain.
We tend to think that it is not a specific effect on one of the photosynthetic complexes because the PSII/PSI stoichiometry is not affected. Rather, by one way or another, the main consequence on the photosynthetic chain is a decrease in the number of complexes involved in electron transfer, resulting in an overall impaired CO2 assimilation. The resulting defect in carbon metabolism could justify the delay in growth observed in the arHIF[Bur] and T-DNA mutant.
General conclusion
Our work highlights the very dynamic rate of evolution of duplicate genes in Arabidopsis where multiple divergent–but still functional–combinations of alleles can be fixed in different backgrounds over a limited period of time. Even genes essential for fundamental processes, like photosynthesis here, can be affected. In the present work, just as in previously described examples from Arabidopsis [16], [18], the structural variation has its origin in a duplication event followed by paralogue's extinction, a loss that can occur early in the process of duplicate gene evolution [31]. Due to the numerous large-scale segmental duplications and dispersed small-scale gene duplications, we can expect a high prevalence of this phenomenon in plants, potentially significantly impacting and constraining phenotypic variation generated at the intraspecific level.
The real extent of structural variation remains to be evaluated and sequencing projects at the species level are willing to consider this [22]. However, to enable a more comprehensive detection of structural variation contributing to intraspecific genetic diversity, significantly longer reads and paired-end sequencing–at the least–are necessary [11], [43] as well as new algorithms to analyse this data [44]–[46]. This should reveal at least CNV polymorphisms, but an extreme case of structural variation is balanced gene transposition which remains challenging to solve because it is then difficult to distinguish allelic from paralogous variation [10]. In Arabidopsis species-wide sequencing studies, one should expect to commonly face new DNA sequences, for which we have no reference and/or no idea of the insertion context, as it is clear that most Arabidopsis accessions have genome sizes 5 to 10% larger than the reference Col-0 genome [3]. A true de novo assembly of high-quality complete Arabidopsis genomes should elucidate all types of polymorphisms that can dictate natural variation.
Materials and Methods
Plant material
A subset of 164 Bur-0 × Col-0 RILs (http://dbsgap.versailles.inra.fr/vnat/; [23]) optimized for QTL mapping was grown in vitro on standard media and phenotyped to map QTLs affecting early-stage shoot growth (see below). RILs 067, 081, 212, 332 that still segregated only for a limited region around 15 Mb on chromosome 4 were used to generate HIFs [24], which enabled the comparison of lines containing either of the parental alleles at the locus of interest in an otherwise identical background. The progeny of RIL212 was genotypically screened to find recombinants used in the fine-mapping process (see below). All (22) lines in the complete RIL population that were still heterozygous around 15 Mb were analysed by progeny-testing to identify and roughly map an interactor controlling SG3 phenotypic segregation. T-DNA insertion lines at At4g30720 (SALK_059716), At4g30730 (SALK_049026) and At4g30740 (SALK_057859) were ordered from NASC and grown under greenhouse conditions. The 52 accessions used to explore species-wide diversity at SG3/SG3i are listed and described in Table S1. They represent most of the core-collection of 48 accessions from the Versailles Center for Biological Resources ([28]; http://dbsgap.versailles.inra.fr/vnat/), plus a few more references and specifically selected accessions.
Shoot measurements
Seeds were surface-sterilized by soaking 10 minutes in 70% EtOH, 0.1% TritonX-100, followed by one wash with 95% EtOH for another 10 minutes. Under sterile conditions, the seeds were suspended in a 0.1% agar solution and stratified in the dark at 4°C for 4 days. Then, seeds were sown on square Petri dishes (120 mm) containing classical Arabidopsis media [47], with 9 RILs per plate and 9 seeds representing each RIL. Plants were grown for 11 days in a culture room (21°C, 16 hours light/8 hours dark cycle) where plates were rotated daily. At 10 DAG (days after germination) plantlets were carefully flattened onto the surface of the media and scanned on a flatbed scanner. Projected shoot area was measured with Optimas 6.5 as an estimate of shoot growth.
QTL mapping
To find genetic loci that affect trait variation, average shoot areas were used as quantitative values to carry out a simple interval mapping using the WebQTL tool (www.genenetwork.org). A 1000 permutations-test was used to estimate a significance threshold at 2.5 LOD. Scanning of the genome with 2 loci mapped simultaneously including their interaction (pair-scan) was performed to search for complex epistasis between pairs of loci that could explain the trait variation.
Fine-mapping
Phenotyping for the confirmation of the QTL segregation during the fine-mapping process was performed as described above, except that seeds were not surface-sterilized and plants were grown in the greenhouse on soil. For each RIL, 48 plants were grown and genotyped to isolate individuals fixed for the parental alleles in the interval (HIF) as well as possible recombinants within the heterozygous region (rHIF). For the chosen HIF (HIF212), more recombinants were searched in two successive screens of respectively 600 and 5,000 plants. Genotyping during screens involved microsatellite or indel markers to identify recombination events within the candidate region. Once recombinants had been identified, CAPS markers and direct sequencing were used to refine and localize recombination breakpoints to smaller intervals when needed. Interesting (informative) rHIFs were then tested for the segregation of the defective growth/pale green phenotype by progeny-testing.
Advanced rHIF line 212.97 that segregate solely for the candidate region (and, hence, for the phenotype) was obtained from crosses between two different rHIFs lines with adequate genotypes (rHIFs recombined immediately to the north or immediately to the south of the SG3 final interval and with adequate genotype elsewhere), following a strategy initiated earlier [21] and as described by Loudet et al. [26].
Allelic complementation
F1 plants used for the quantitative complementation assay were generated by reciprocally crossing heterozygous arHIF212.97 to the heterozygous T-DNA insertion line, SALK_059716. F1 plants were phenotyped and genotyped to detect which allelic combination at SG3 was restoring the wild-type phenotype or not.
Sequencing At4g30720 paralogues
Bur-0 SG3 and SG3i copies were sequenced using a BAC library (Amplicon Express). Corresponding BACs were selected by PCR using primers for IND414975 amplifying a fragment of ∼1,300 bp at SG3 and respectively ∼100 bp at SG3i. BAC DNA was then extracted with the NucleoBond BAC 100 kit (Macherey-Nagel) and sent for sequencing.
For the sequencing of the 52 accessions, a specific primer (INDEL'75R) based on an indel polymorphism was used to specifically amplify a 4.5 kb-fragment covering the SG3i copy of the gene. The obtained PCR product was used as template to further PCR amplify and sequence five overlapping fragments. We inferred the SG3 sequence from an unspecific sequencing reaction by manually subtracting the SG3i polymorphisms (according to the specific SG3i sequence above), assuming none of our accessions had residual heterozygosity at these loci (which is extremely likely after the numerous SSD cycles they have been through). Primers used for sequencing are listed in Table S5. A. lyrata sequence was obtained from the Joint Genome Institute sequencing project data (http://genomeportal.jgi-psf.org/Araly1/Araly1.home.html).
At4g30720 expression studies
At4g30720 expression in the transgenic plants was analyzed by RT-PCR. RNA was extracted with the RNeasy Plant Mini Kit (Qiagen) and the reverse transcription performed with the RevertAid First Strand cDNA Synthesis Kit (Fermentas) to yield single-strand cDNA. Transcription was tested with the following primers, forward primer 5′-CGTTTTCAACACCGCTAGAAC-3′ and reverse primer 5′ - TGGTTTGTTTGCTGCTCTTG-3′, flanking the T-DNA insertion site. The adenine phosphoribosyl-transferase gene was used as control in the RT-PCR reaction.
Accessions were grown in vitro for 2 weeks then shoots were frozen and ground in liquid nitrogen. RNA was extracted and reverse-transcribed as above. PCR was carried out with the following primers, forward primer 5′ - TGGTTTGTTTGCTGCTCTTG -3′ and reverse primer 5′ - AACAACACGAGAATCCTCTACCA -3′, complementary to both SG3 and SG3i copies in all accessions. Amplicons generated (410 bp) were digested with BclI (Fermentas). Due to a SNP in the sequence shared by all SG3i copies among accessions tested, SG3i amplicons are digested while SG3 amplicons remain undigested, allowing to distinguish each paralogue's expression.
Molecular population genetics data analysis
All accessions' sequences were aligned using CodonCode Aligner v3.01 and manually checked for all polymorphisms. The haplotype network was generated using Splitstree4 V4.10 on all sequenced accessions [48]. Population genetics analyses were generated on a subset of 41 accessions with both SG3 and SG3i functional copies, and A. lyrata ortholog was used as the outgroup in the analyses. Estimates of nucleotide diversity θπ, nucleotide divergence (Ks), Tajima's D and Fu et Li D* were performed using DnaSP5 [49]. The multilocus HKA test was performed on silent sites using a program kindly provided by J. Hey (Rutgers University, Piscataway, NJ).
Chlorophyll fluorescence
The fluorescence changes were measured as described previously [50]. The fluorescence changes were induced by a continuous green light (100 µE.m2.s−1) allowing the measurement of the fluorescence yield of the dark-adapted sample (F0) and the fluorescence yield reached under quasi steady-state conditions (Fstat). The maximum fluorescence yield was measured 50 µs after applying an intense (5,000 µE.m2.s−1) light-pulse of 200 ms duration.
Supporting Information
Zdroje
1. Alonso-BlancoC
AartsMG
BentsinkL
KeurentjesJJ
ReymondM
2009 What has natural variation taught us about plant development, physiology, and adaptation? Plant Cell 21 1877 1896
2. OssowskiS
SchneebergerK
ClarkRM
LanzC
WarthmannN
2008 Sequencing of natural strains of Arabidopsis thaliana with short reads. Genome Res 18 2024 2033
3. SchmuthsH
MeisterA
HorresR
BachmannK
2004 Genome size variation among accessions of Arabidopsis thaliana. Ann Bot 93 317 321
4. ZiolkowskiPA
BlancG
SadowskiJ
2003 Structural divergence of chromosomal segments that arose from successive duplication events in the Arabidopsis genome. Nucleic Acids Res 31 1339 1350
5. ClarkRM
SchweikertG
ToomajianC
OssowskiS
ZellerG
2007 Common sequence polymorphisms shaping genetic diversity in Arabidopsis thaliana. Science 317 338 342
6. FeukL
CarsonAR
SchererSW
2006 Structural variation in the human genome. Nat Rev Genet 7 85 97
7. AchazG
CoissacE
ViariA
NetterP
2000 Analysis of intrachromosomal duplications in yeast Saccharomyces cerevisiae: a possible model for their origin. Mol Biol Evol 17 1268 1275
8. FranszPF
ArmstrongS
de JongJH
ParnellLD
van DrunenC
2000 Integrated cytogenetic map of chromosome arm 4S of A. thaliana: structural organization of heterochromatic knob and centromere region. Cell 100 367 376
9. HughesAL
FriedmanR
EkolluV
RoseJR
2003 Non-random association of transposable elements with duplicated genomic blocks in Arabidopsis thaliana. Mol Phylogenet Evol 29 410 416
10. FaddahDA
GankoEW
McCoachC
PickrellJK
HanlonSE
2009 Systematic identification of balanced transposition polymorphisms in Saccharomyces cerevisiae. PLoS Genet 5 e1000502 doi:10.1371/journal.pgen.1000502
11. KorbelJO
UrbanAE
AffourtitJP
GodwinB
GrubertF
2007 Paired-end mapping reveals extensive structural variation in the human genome. Science 318 420 426
12. IafrateAJ
FeukL
RiveraMN
ListewnikML
DonahoePK
2004 Detection of large-scale variation in the human genome. Nat Genet 36 949 951
13. ConradDF
PintoD
RedonR
FeukL
GokcumenO
2009 Origins and functional impact of copy number variation in the human genome. Nature 7 7
14. BuchananJA
SchererSW
2008 Contemplating effects of genomic structural variation. Genet Med 10 639 647
15. LynchM
ConeryJS
2003 The evolutionary demography of duplicate genes. J Struct Funct Genomics 3 35 44
16. BikardD
PatelD
Le MettéC
GiorgiV
CamilleriC
2009 Divergent evolution of duplicate genes leads to genetic incompatibilities within A. thaliana. Science 323 623 626
17. KroymannJ
DonnerhackeS
SchnabelrauchD
Mitchell-OldsT
2003 Evolutionary dynamics of an Arabidopsis insect resistance quantitative trait locus. Proc Natl Acad Sci USA 100 14587 14592
18. StaalJ
KaliffM
BohmanS
DixeliusC
2006 Transgressive segregation reveals two Arabidopsis TIR-NB-LRR resistance genes effective against Leptosphaeria maculans, causal agent of blackleg disease. Plant J 46 218 230
19. KeurentjesJJ
BentsinkL
Alonso-BlancoC
HanhartCJ
Blankestijn-De VriesH
2007 Development of a near-isogenic line population of Arabidopsis thaliana and comparison of mapping power with a recombinant inbred line population. Genetics 175 891 905
20. MelchingerAE
PiephoHP
UtzHF
MuminovicJ
WegenastT
2007 Genetic basis of heterosis for growth-related traits in Arabidopsis investigated by testcross progenies of near-isogenic lines reveals a significant role of epistasis. Genetics 177 1827 1837
21. KroymannJ
Mitchell-OldsT
2005 Epistasis and balanced polymorphism influencing complex trait variation. Nature 435 95 98
22. WeigelD
MottR
2009 The 1001 genomes project for Arabidopsis thaliana. Genome Biol 10 107
23. SimonM
LoudetO
DurandS
BérardA
BrunelD
2008 QTL mapping in five new large RIL populations of Arabidopsis thaliana genotyped with consensus SNP markers. Genetics 178 2253 2264
24. LoudetO
GaudonV
TrubuilA
Daniel-VedeleF
2005 Quantitative trait loci controlling root growth and architecture in Arabidopsis thaliana confirmed by heterogeneous inbred family. Theor Appl Genet 110 742 753
25. TuinstraMR
EjetaG
GoldsbroughPB
1997 Heterogeneous inbred family (HIF) analysis: a method for developing near-isogenic lines that differ at quantitative trait loci. Theor Appl Genet 95 1005 1011
26. LoudetO
MichaelTP
BurgerBT
Le MettéC
MocklerTC
2008 A zinc knuckle protein that negatively controls morning-specific growth in Arabidopsis thaliana. Proc Natl Acad Sci USA 105 17193 17198
27. ZybailovB
RutschowH
FrisoG
RudellaA
EmanuelssonO
2008 Sorting signals, N-terminal modifications and abundance of the chloroplast proteome. PLoS ONE 3 e1994 doi:10.1371/journal.pone.0001994
28. McKhannHI
CamilleriC
BerardA
BataillonT
DavidJL
2004 Nested core collections maximizing genetic diversity in Arabidopsis thaliana. Plant J 38 193 202
29. KelleherES
MarkowTA
2009 Duplication, selection and gene conversion in a Drosophila mojavensis female reproductive protein family. Genetics 181 1451 1465
30. OstrowskiMF
DavidJ
SantoniS
McKhannH
ReboudX
2006 Evidence for a large-scale population structure among accessions of Arabidopsis thaliana: possible causes and consequences for the distribution of linkage disequilibrium. Mol Ecol 15 1507 1517
31. MooreRC
PuruggananMD
2003 The early stages of duplicate gene evolution. Proc Natl Acad Sci USA 100 15682 15687
32. KochMA
HauboldB
Mitchell-OldsT
2000 Comparative evolutionary analysis of chalcone synthase and alcohol dehydrogenase loci in Arabidopsis, Arabis, and related genera (Brassicaceae). Mol Biol Evol 17 1483 1498
33. SharbelTF
HauboldB
Mitchell-OldsT
2000 Genetic isolation by distance in Arabidopsis thaliana: biogeography and post-glacial colonization of Europe. Mol Ecol 9 2109 2118
34. SchmidKJ
Ramos-OnsinsS
Ringys-BecksteinH
WeisshaarB
Mitchell-OldsT
2005 A multilocus sequence survey in Arabidopsis thaliana reveals a genome-wide departure from a neutral model of DNA sequence polymorphism. Genetics 169 1601 1615
35. NordborgM
HuTT
IshinoY
JhaveriJ
ToomajianC
2005 The pattern of polymorphism in Arabidopsis thaliana. PLoS Biol 3 e196 doi:10.1371/journal.pbio.0030196
36. HudsonRR
KreitmanM
AguadeM
1987 A test of neutral molecular evolution based on nucleotide data. Genetics 116 153 159
37. CaldwellKS
MichelmoreRW
2009 Arabidopsis thaliana genes encoding defense signaling and recognition proteins exhibit contrasting evolutionary dynamics. Genetics 181 671 684
38. TajimaF
1989 Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123 585 595
39. FuYX
LiWH
1993 Statistical tests of neutrality of mutations. Genetics 133 693 709
40. PuruggananMD
SuddithJI
1999 Molecular population genetics of floral homeotic loci. Departures from the equilibrium-neutral model at the APETALA3 and PISTILLATA genes of Arabidopsis thaliana. Genetics 151 839 848
41. KuittinenH
AguadeM
2000 Nucleotide variation at the CHALCONE ISOMERASE locus in Arabidopsis thaliana. Genetics 155 863 872
42. AguadeM
2001 Nucleotide sequence variation at two genes of the phenylpropanoid pathway, the FAH1 and F3H genes, in Arabidopsis thaliana. Mol Biol Evol 18 1 9
43. ListerR
GregoryBD
EckerJR
2009 Next is now: new technologies for sequencing of genomes, transcriptomes, and beyond. Curr Opin Plant Biol 12 107 118
44. AlkanC
KiddJM
Marques-BonetT
AksayG
AntonacciF
2009 Personalized copy number and segmental duplication maps using next-generation sequencing. Nat Genet 41 1061 1067
45. HormozdiariF
AlkanC
EichlerEE
SahinalpSC
2009 Combinatorial algorithms for structural variation detection in high-throughput sequenced genomes. Genome Res 19 1270 1278
46. SchneebergerK
HagmannJ
OssowskiS
WarthmannN
GesingS
2009 Simultaneous alignment of short reads against multiple genomes. Genome Biol 10 R98
47. EstelleMA
SomervilleC
1987 Auxin-resistant mutants of Arabidopsis thaliana with an altered morphology. Mol Gen Genet 206 200 206
48. HusonDH
BryantD
2006 Application of phylogenetic networks in evolutionary studies. Mol Biol Evol 23 254 267
49. LibradoP
RozasJ
2009 DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25 1451 1452
50. RappaportF
BealD
JoliotA
JoliotP
2007 On the advantages of using green light to study fluorescence yield changes in leaves. Biochim Biophys Acta 1767 56 65
Štítky
Genetika Reprodukční medicína
Článek Epistasis of Transcriptomes Reveals Synergism between Transcriptional Activators Hnf1α and Hnf4αČlánek SMA-10/LRIG Is a Conserved Transmembrane Protein that Enhances Bone Morphogenetic Protein SignalingČlánek B1 SOX Coordinate Cell Specification with Patterning and Morphogenesis in the Early Zebrafish EmbryoČlánek Bulk Segregant Analysis by High-Throughput Sequencing Reveals a Novel Xylose Utilization Gene fromČlánek Abundant Quantitative Trait Loci Exist for DNA Methylation and Gene Expression in Human Brain
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2010 Číslo 5
-
Všechny články tohoto čísla
- Digital Quantification of Human Eye Color Highlights Genetic Association of Three New Loci
- Common Genetic Variants near the Brittle Cornea Syndrome Locus Influence the Blinding Disease Risk Factor Central Corneal Thickness
- Age- and Temperature-Dependent Somatic Mutation Accumulation in
- All About Mitochondrial Eve: An Interview with Rebecca Cann
- Aging and Chronic Sun Exposure Cause Distinct Epigenetic Changes in Human Skin
- Transposed Genes in Arabidopsis Are Often Associated with Flanking Repeats
- A Survey of Genomic Traces Reveals a Common Sequencing Error, RNA Editing, and DNA Editing
- Gene Transposition Causing Natural Variation for Growth in
- The Polyproline Site in Hinge 2 Influences the Functional Capacity of Truncated Dystrophins
- Epistasis of Transcriptomes Reveals Synergism between Transcriptional Activators Hnf1α and Hnf4α
- Integration of Light Signals by the Retinoblastoma Pathway in the Control of S Phase Entry in the Picophytoplanktonic Cell
- The Proprotein Convertase Encoded by () Is Required in Corpora Cardiaca Endocrine Cells Producing the Glucose Regulatory Hormone AKH
- Sgs1 and Exo1 Redundantly Inhibit Break-Induced Replication and Telomere Addition at Broken Chromosome Ends
- A MATE-Family Efflux Pump Rescues the 8-Oxoguanine-Repair-Deficient Mutator Phenotype and Protects Against HO Killing
- The Relationship among Gene Expression, the Evolution of Gene Dosage, and the Rate of Protein Evolution
- SMA-10/LRIG Is a Conserved Transmembrane Protein that Enhances Bone Morphogenetic Protein Signaling
- The Nuclear Receptor DHR3 Modulates dS6 Kinase–Dependent Growth in
- Involvement of Global Genome Repair, Transcription Coupled Repair, and Chromatin Remodeling in UV DNA Damage Response Changes during Development
- B1 SOX Coordinate Cell Specification with Patterning and Morphogenesis in the Early Zebrafish Embryo
- Linkage and Association Mapping of Flowering Time in Nature
- Bulk Segregant Analysis by High-Throughput Sequencing Reveals a Novel Xylose Utilization Gene from
- Abundant Quantitative Trait Loci Exist for DNA Methylation and Gene Expression in Human Brain
- Ablation of Whirlin Long Isoform Disrupts the USH2 Protein Complex and Causes Vision and Hearing Loss
- Characterization of Oxidative Guanine Damage and Repair in Mammalian Telomeres
- DNA Adenine Methylation Is Required to Replicate Both Chromosomes Once per Cell Cycle
- Genome-Wide Copy Number Variation in Epilepsy: Novel Susceptibility Loci in Idiopathic Generalized and Focal Epilepsies
- FACT Prevents the Accumulation of Free Histones Evicted from Transcribed Chromatin and a Subsequent Cell Cycle Delay in G1
- GC-Biased Evolution Near Human Accelerated Regions
- Liver and Adipose Expression Associated SNPs Are Enriched for Association to Type 2 Diabetes
- Myeloid Cell-Restricted Insulin Receptor Deficiency Protects Against Obesity-Induced Inflammation and Systemic Insulin Resistance
- The Mating Type Locus () and Sexual Reproduction of : Insights into the Evolution of Sex and Sex-Determining Chromosomal Regions in Fungi
- B-Cyclin/CDKs Regulate Mitotic Spindle Assembly by Phosphorylating Kinesins-5 in Budding Yeast
- Post-Replication Repair Suppresses Duplication-Mediated Genome Instability
- Genome Sequence of the Plant Growth Promoting Endophytic Bacterium sp. 638
- Transcription Factors Mat2 and Znf2 Operate Cellular Circuits Orchestrating Opposite- and Same-Sex Mating in
- The Use of Orthologous Sequences to Predict the Impact of Amino Acid Substitutions on Protein Function
- Mutation in Archain 1, a Subunit of COPI Coatomer Complex, Causes Diluted Coat Color and Purkinje Cell Degeneration
- Shelterin-Like Proteins and Yku Inhibit Nucleolytic Processing of Telomeres
- Affecting Function Causes a Dilated Heart in Adult
- Manipulation of Behavioral Decline in with the Rag GTPase
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Common Genetic Variants near the Brittle Cornea Syndrome Locus Influence the Blinding Disease Risk Factor Central Corneal Thickness
- All About Mitochondrial Eve: An Interview with Rebecca Cann
- The Relationship among Gene Expression, the Evolution of Gene Dosage, and the Rate of Protein Evolution
- SMA-10/LRIG Is a Conserved Transmembrane Protein that Enhances Bone Morphogenetic Protein Signaling
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



