-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaThe Endosymbiotic Bacterium Selectively Kills Male Hosts by Targeting the Masculinizing Gene
Pathogens are known to manipulate the physiology, behavior, and reproduction of their hosts for their own benefit. The endosymbiotic bacterium Wolbachia is known to manipulate the sex of its host's progeny. Male-killing is one of the phenotypes that Wolbachia induces, but the mechanism of how Wolbachia induces sex-specific death is unknown. Here we found a marked down-regulation of Masc, a lepidopteran-specific zinc finger protein gene, in embryos that are produced by Wolbachia-infected Ostrinia moths. We also observed that dosage compensation fails in Wolbachia-infected Ostrinia embryos. The findings of this study and our previous study using a lepidopteran model insect Bombyx mori indicate that Wolbachia has evolved to hijack the Masc-dependent, lepidopteran insect-specific sex determination system by capturing an unknown factor during Wolbachia-host coevolution.
Published in the journal: . PLoS Pathog 11(7): e32767. doi:10.1371/journal.ppat.1005048
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1005048Summary
Pathogens are known to manipulate the physiology, behavior, and reproduction of their hosts for their own benefit. The endosymbiotic bacterium Wolbachia is known to manipulate the sex of its host's progeny. Male-killing is one of the phenotypes that Wolbachia induces, but the mechanism of how Wolbachia induces sex-specific death is unknown. Here we found a marked down-regulation of Masc, a lepidopteran-specific zinc finger protein gene, in embryos that are produced by Wolbachia-infected Ostrinia moths. We also observed that dosage compensation fails in Wolbachia-infected Ostrinia embryos. The findings of this study and our previous study using a lepidopteran model insect Bombyx mori indicate that Wolbachia has evolved to hijack the Masc-dependent, lepidopteran insect-specific sex determination system by capturing an unknown factor during Wolbachia-host coevolution.
Introduction
Wolbachia is a genus in Rickettsiales, a diverse order of intracellular bacteria. Recent meta-sequencing analysis shows that over 65% of insect species possess Wolbachia [1], indicating that Wolbachia is the most widespread and common intracellular bacterium that infects insects. Wolbachia is a well-known example of a parasite that alters host reproduction to facilitate its own propagation. Wolbachia-induced phenotypes include parthenogenesis, feminization, cytoplasmic incompatibility, and male-killing, each of which is supposed to be adaptive for Wolbachia by enhancing the production of infected females [2].
Wolbachia-induced male-killing has been reported in three insect orders, Coleoptera [3], Diptera [4], and Lepidoptera [5], in which male-killing occurs mainly during embryogenesis. Recent advances in Wolbachia-induced male-killing have been made mainly from studies using Ostrinia moths [5–7]. Ostrinia species (corn borer) have a WZ/ZZ sex chromosome system and all of the progeny of Wolbachia-infected mother moths have a W chromosome, indicating the existence of male-killing [7]. The male-type splice variant of doublesex (dsx), a gene that acts at the downstream end of the sex differentiation cascade [8–9], is not detected in Ostrinia late embryos that originate from Wolbachia-infected mothers [7], suggesting that Wolbachia’s sexual manipulation is presumably established at an early embryonic stage. However, the molecular mechanism(s) by which Wolbachia manipulates sex and male lethality in Ostrinia moths has remained elusive.
The sex determination cascade in lepidopteran insects has been studied mainly using silkworm Bombyx mori as a model insect [10–11]. In B. mori, females have ZW sex chromosomes and males have two Z chromosomes. B. mori femaleness is strongly determined by the presence of the W chromosome irrespective of the Z chromosome number, suggesting that there is a dominant feminizing gene (Fem) on the W chromosome [12–13]. In 2014, we discovered that Fem is a precursor of a single W chromosome-derived PIWI-interacting RNA (piRNA) [14]. We also identified the target gene of Fem-derived piRNA (Fem piRNA), which is located on the Z chromosome. Depletion of this Z-linked gene, Masculinizer (Masc), in male embryos leads to the production of the female-type splicing of B. mori dsx, indicating that the product of Masc is a masculinizing factor. These results revealed that sex of B. mori is determined by the Fem piRNA-Masc cascade [14].
Further experiments showed that silencing of Masc in B. mori embryos results in male-specific lethality [14]. Deep sequencing (RNA-seq) of Masc mRNA-depleted embryos revealed that the Masc protein is required for the repression of global transcription from the Z chromosome in male embryos, and that a failure of this dosage compensation causes male-specific embryonic death [14]. Bioinformatic analysis also revealed that most of the Z-linked genes are dosage compensated by 72 hours post-oviposition (hpo) [15]. These results indicate that Masc-mediated control of dosage compensation at an early embryonic stage is essential for male development of B. mori.
In this study, we attempted to explore the mechanism by which Wolbachia accomplishes sexual manipulation using Wolbachia-infected Ostrinia moths as a model. We previously showed that artificial depletion of Masc mRNA in B. mori early embryos results in male-specific embryonic lethality [14]. This phenomenon likely mimics the way that Wolbachia induces male-killing in Ostrinia [7]. In the light of these facts, we first focused on the expression of Ostrinia homologue of Masc in Wolbachia-infected embryos. Transcriptome analysis by RNA-seq revealed that Wolbachia-induced male-killing is established by a failure of dosage compensation through Masc mRNA depletion. Furthermore, injection of Masc cRNA into Wolbachia-infected Ostrinia embryos prevented males from being killed at embryonic stages. This is the first study to identify the Wolbachia’s target that is utilized for sexual manipulation.
Results and Discussion
Characterization of a male-killing Wolbachia found in Ostrinia moths
We collected over 80 Ostrinia moths in the field and found 3 female moths that were infected with Wolbachia (Fig 1A). One of these moths produced only female progeny. By sex pheromone analysis, this female was found to be O. furnacalis. Subsequently, when the females were mated with other Wolbachia-free O. furnacalis males only female progeny were produced. There was no deviation in this pattern of female only progeny through 5 generations (in total 302 females and no males judged by external morphology at the adult stage) (Fig 1B).
Fig. 1. Characterization of a male-killing Wolbachia found in adult Ostrinia. 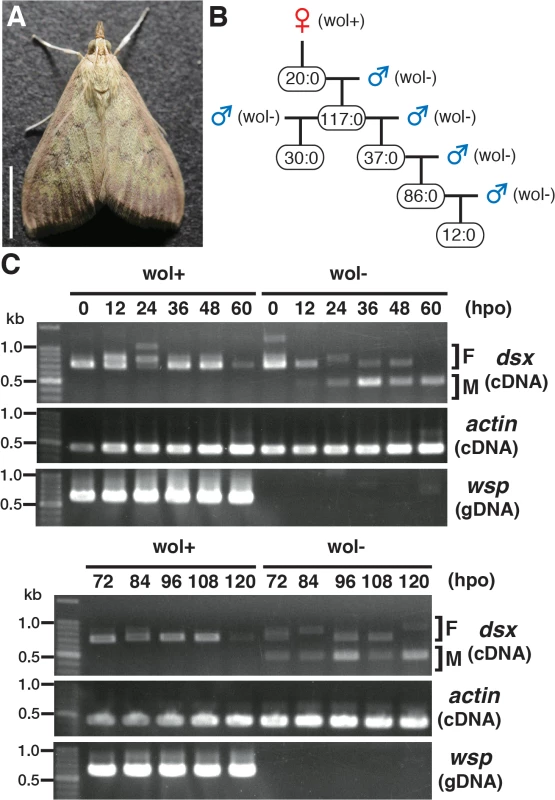
(A) Wolbachia-infected female moth of Ostrinia. Bar, 5 mm. (B) Brood sex ratios in a Wolbachia-infected matriline through 5 generations. The female:male ratio of each mating is shown. (C) Splicing patterns of Ostrinia dsx during embryonic development (0–120 hpo) of embryos that were infected with Wolbachia (wol+) or uninfected (wol-). Total RNA was prepared from Ostrinia embryos (25–50 embryos at each time point) and subjected to RT-PCR for dsx and actin. The F and M indicate female- and male-type splicing of Ostrinia dsx, respectively. actin was used as an internal control. Wolbachia infection was verified by PCR using wsp-specific primers. Note that in other experiments (S1E Fig), the female-type splicing variants were clearly detected when the same RNA pool prepared from Wolbachia-uninfected embryos at 60 hpo was used, although that was not observed in this case. We established the quantitative PCR (qPCR)-based molecular sexing method for O. furnacalis (S1A Fig), and verified that Wolbachia-infected moths were all female (S1B Fig). Using this method, we found that Wolbachia-infected embryos (just prior to hatching) contained both female and male individuals (S1C Fig). However, the hatched larvae were all female (S1C Fig), indicating that this Wolbachia induces male-specific embryonic lethality. In addition, when Wolbachia was eliminated from infected individuals by tetracycline treatment and Wolbachia-eliminated female moths were mated with other Wolbachia-free males, only male progeny were produced (S1D Fig). Taken together with these results, we concluded that this Wolbachia strain induces male-killing in O. furnacalis, which is similar to the phenotype observed in Wolbachia-infected O. scapulalis [7].
Sugimoto and Ishikawa [7] reported that the male-type splice variants of Ostrinia dsx [6] is not expressed in all 5-day-old O. scapulalis embryos (just prior to hatching) that are infected with a male-killing Wolbachia. In order to determine the precise developmental stage at which Wolbachia starts sexual manipulation in Ostrinia, we examined the splicing patterns of dsx in Wolbachia-infected Ostrinia embryos starting immediately after oviposition. Both male - and female-type variants of dsx were observed in uninfected embryos (Fig 1C). The male-type variant in uninfected embryos was detected from 12 hpo, indicating that the sex determination signal is transmitted prior to 12 hpo in Ostrinia. In contrast, the male-type dsx variant was not detected in Wolbachia-infected embryos throughout embryogenesis (Fig 1C). These results indicate that Wolbachia manipulates the sex of Ostrinia from the beginning of the sex determination period.
Marked down-regulation of Masc in Wolbachia-infected Ostrinia embryos
In order to investigate what occurs during Wolbachia-induced sexual manipulation, we performed RNA-seq experiments using RNAs prepared from Wolbachia-infected and-uninfected embryos (of both sexes) at 0, 12, 24, 36, and 48 hpo. We recently showed that Masc protein encodes a lepidopteran-specific CCCH-tandem zinc finger protein, which is required for both masculinization and dosage compensation in B. mori [14]. RNA interference-mediated knockdown experiments also show that depletion of Masc mRNA in B. mori early embryos results in male-specific death via a failure of dosage compensation [14]. In order to test whether this male-specific death in B. mori is related to the Wolbachia-induced male killing in Ostrinia, we first identified and characterized the Masc homolog of Ostrinia. Using RNA-seq data, we identified contigs that potentially encode a protein with significant homology to Masc proteins (Fig 2A and S2 Fig). Ostrinia Masc was composed of 583 amino acid residues and showed 28.1% identity to B. mori Masc (S2 Fig). Phylogenetic analysis based on the amino acid sequences of zinc finger domains revealed that Ostrinia Masc was closely related to that of Chilo suppressalis (Fig 2A).
Fig. 2. Identification and characterization of Ostrinia Masc protein. 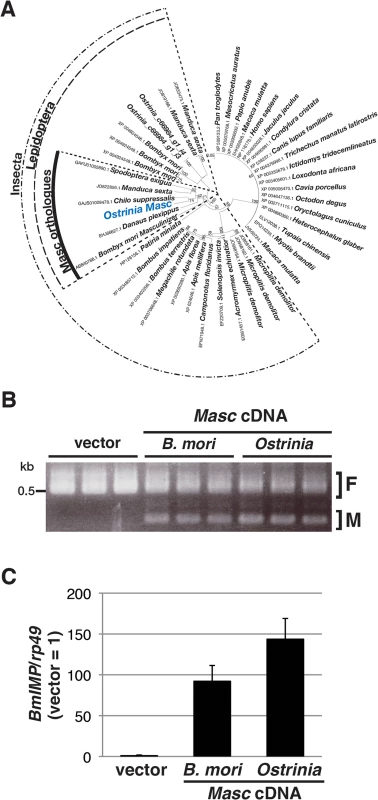
(A) Phylogenetic analysis of Ostrinia Masc protein. The neighbor-joining tree was generated using the amino acid sequences corresponding to the zinc finger domains from 43 proteins showing homology to B. mori Masc protein. The numbers on the internal branches represent the support value following bootstrap analysis (1,000 replicates). (B) Effect of Ostrinia Masc cDNA transfection on the Bmdsx splicing in BmN4 cells. The splicing patterns of Bmdsx in BmN4 cells transfected with Ostrinia or B. mori Masc expression vectors or empty vector were examined by RT–PCR. The F and M indicate female- and male-type splicing of Bmdsx, respectively. (C) Effect of Ostrinia Masc cDNA transfection on BmIMP expression. BmIMP expression was examined by RT-qPCR. rp49 was used as a normalization control. Data shown are means + standard deviation of triplicates. The silkworm ovarian cell line BmN4 expresses the female-type Bmdsx variant only, whereas transfection of B. mori Masc cDNA results in the production of the male-type splice variant [14]. Using this system, we examined whether Ostrinia Masc protein also has a masculinizing activity. As shown in Fig 2B, we observed the expression of the male-type variant of Bmdsx in BmN4 cells when transfected with Ostrinia Masc cDNA as well as B. mori Masc cDNA. In addition, we examined the expression of B. mori insulin-like growth factor II mRNA-binding protein (BmIMP), a factor that is involved in the male-specific splicing of Bmdsx [16], in Ostrinia Masc cDNA-transfected cells. We found that either B. mori or Ostrinia Masc cDNA markedly enhanced BmIMP expression (Fig 2C). These results strongly suggest that Ostrinia Masc encodes a masculinizing protein and may be required for masculinization in Ostrinia sex determination pathway.
We next compared the level of Masc mRNA in Wolbachia-infected and-uninfected embryos by mapping RNA-seq tags onto the Ostrinia Masc coding sequence. We found a significant decrease in Masc mRNA in Wolbachia-infected embryos prior to 12 hpo (Fig 3A). To elucidate this reduction in greater detail, we performed reverse transcription-qPCR (RT-qPCR) using total RNA isolated from Wolbachia-infected and uninfected embryos at 0, 6, 12, and 18 hpo. In uninfected embryos, Masc expression peaked at 6 hpo and decreased rapidly. In contrast, Masc expression in Wolbachia-infected embryos declined by 6 hpo, and remained at a low level compared with that observed in uninfected embryos (Fig 3B). These results clearly showed that Wolbachia infection markedly reduces Masc mRNA level during embryogenesis of Ostrinia. Together with transfection results (Fig 2B and 2C) and our previous results showing that knock down of Masc mRNA results in the production of the female-type dsx in male embryos of B. mori [14], we conclude that the lack of the male-type dsx in Wolbachia-infected Ostrinia embryos (Fig 1C) was caused by down-regulation of Masc mRNA (Fig 3A and 3B).
Fig. 3. A marked decrease in Masc mRNA was observed in Wolbachia-infected embryonic Ostrinia. 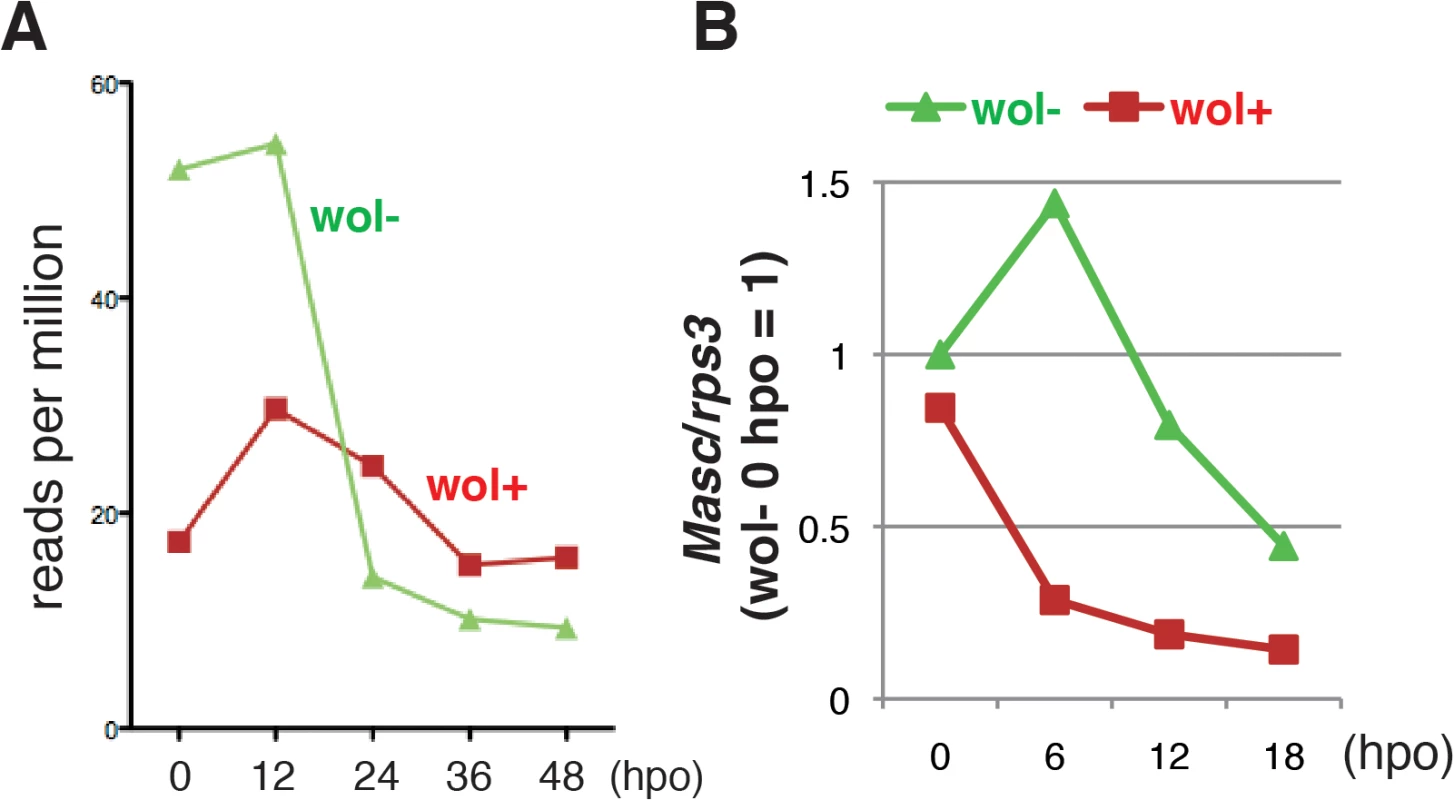
(A) Number of Ostrinia Masc coding sequence-derived tags found in RNA-seq library generated from Wolbachia-infected (wol+) and-uninfected (wol-) embryos at 0, 12, 24, 36, and 48 hpo. (B) Expression profile of Ostrinia Masc mRNA during early embryogenesis (0, 6, 12, and 18 hpo). Total RNA was prepared from embryos (25–50 embryos at each time point) that were wol+ or wol-. Rps3 was used as a normalization control for RT-qPCR. Failure of dosage compensation in Wolbachia-infected Ostrinia embryos
Considering our recent finding that depletion of Masc mRNA in early embryos of B. mori results in male-specific embryonic lethality due to a failure of dosage compensation [14], we hypothesized that a decrease in Masc mRNA in Wolbachia-infected Ostrinia embryos also affects dosage compensation, presumably resulting in a male-specific embryonic death. To test this hypothesis, we examined dosage compensation effects in Wolbachia-infected Ostrinia embryos using RNA-seq data. As reported previously [14], Z-linked transcripts are expressed at higher levels in Masc mRNA depleted B. mori embryos than in control embryos (Fig 4A, left panel). A similar transcriptional bias in putative Z-linked genes in Wolbachia-infected Ostrinia embryos at 48 hpo was also found (Fig 4A, right panel). A failure of dosage compensation of Z-linked genes was detected from 24 hpo and continued to 48 hpo (Fig 4B and S3 Fig). These results strongly support our hypothesis that Wolbachia infection leads to abnormally enhanced expression of Z-linked genes in male embryos via Masc mRNA down-regulation, resulting in a male-killing phenotype.
Fig. 4. A failure of dosage compensation in Wolbachia-infected embryos. 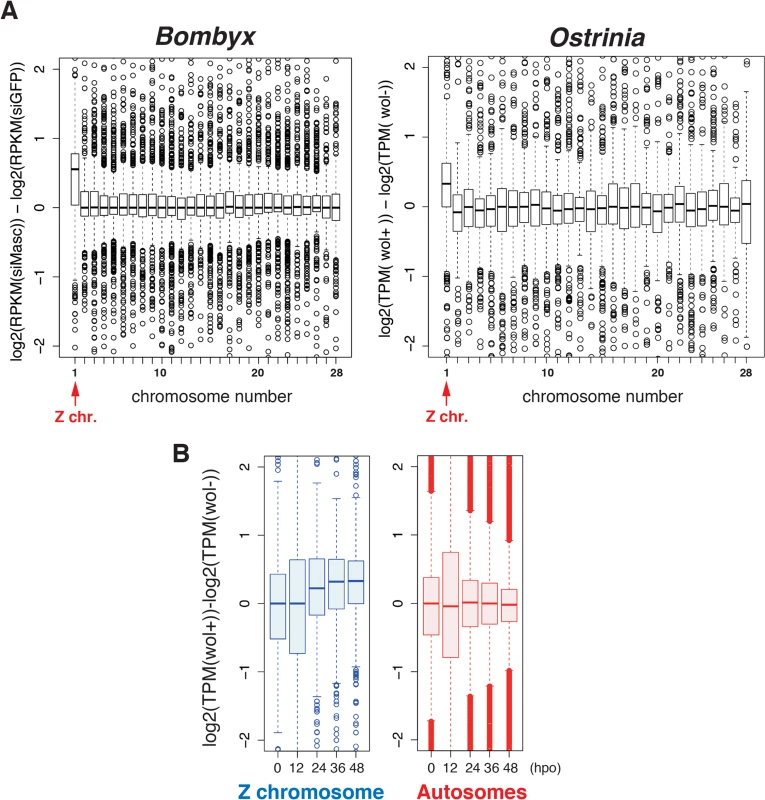
(A) Chromosomal distribution of differentially expressed transcripts in B. mori and Ostrinia embryos. The B. mori RNA-seq data of Masc RNAi experiments (GFP and Masc RNAi embryos of mixed sex, 72 h post-injection) were used as a control experiment (left panel). The Ostrinia RNA-seq data (Wolbachia-infected and uninfected embryos of both sexes at 48 hpo were used to draw the panel on the right. The chromosome number for each Ostrinia transcript-derived contig was assigned using B. mori gene models. The data are shown by box-and-whisker diagrams. The boxes represent the median and 25–75 percentile ranges of the expression ratios. (B) Time-course and Z chromosome-biased distribution of differentially expressed transcripts in Wolbachia-infected and uninfected Ostrinia embryos. The data are separately shown in Z chromosome (blue)- and autosome (red)-linked genes. The data are shown by box-and-whisker diagrams. The boxes represent the median and 25–75 percentile ranges of the expression ratios. Rescue of male progeny by injection of Masc cRNA into Wolbachia-infected Ostrinia embryos
To obtain direct evidence that down-regulation of Masc mRNA in Wolbachia-infected Ostrinia embryos results in the male-killing phenotype, we performed rescue experiments by injecting in vitro synthesized capped, poly(A)-tailed Masc cRNA into Wolbachia-infected embryos. As shown in Fig 5, the hatched larvae injected with control (GFP) cRNA were all female, whereas both male and female larvae were observed when injected with Masc cRNA. This clearly indicates that introduction of Masc cRNA into Wolbachia-infected embryos can rescue male embryos. Together with the results of transcriptome data, we conclude that a decrease in Masc mRNA in Wolbachia-infected embryos causes a male-killing phenotype via a failure of dosage compensation.
Fig. 5. Injection of Masc cRNA rescued the male-specific death of Wolbachia-infected embryonic Ostrinia. 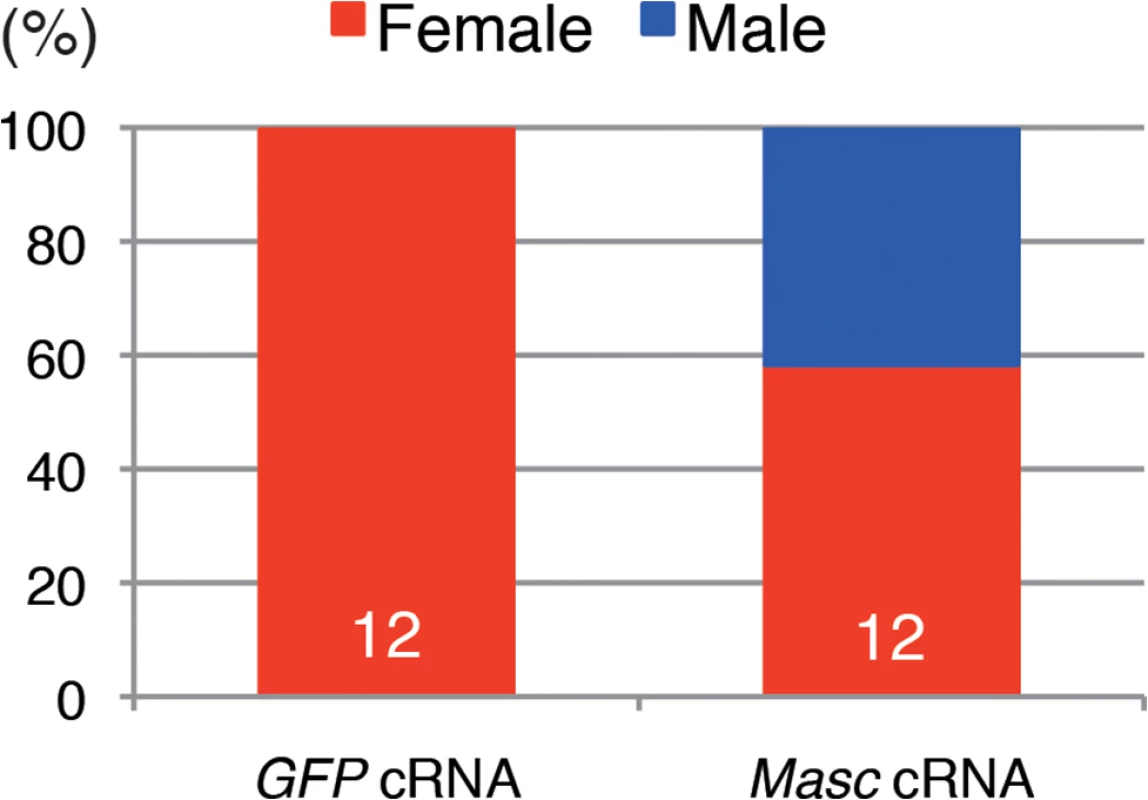
Capped, poly(A)-tailed Masc cRNA was synthesized and injected into Wolbachia-infected embryos immediately after oviposition. The hatched larvae were collected and molecularly sexed. The number indicates the sample size of each group. Conclusion and perspective
In conclusion, our study answered the question of how Wolbachia manipulates sex ratios in moths: in Ostrinia, Wolbachia targets Masc, a masculinizing gene that was originally characterized in B. mori, to establish male-killing (Fig 6). In Drosophila, the Sex lethal protein functions as a master switch for sex determination, and also controls dosage compensation by inhibiting translation of male-specific lethal 2 (msl-2) [17]. Veneti et al. reported that a male-killing Spiroplasma targets the dosage compensation complex, including msl-2, to kill male D. melanogaster [18]. Our current results demonstrate that a similar event occurs in Wolbachia-infected lepidopteran insects; Wolbachia infection leads to male-killing by down-regulating Masc (Fig 3A and 3B), which is an essential factor controlling both sex determination and dosage compensation pathways in lepidopteran insects [14]. This analogy comes from the fact that the sex determination cascade is often tightly associated with the control of dosage compensation in insects.
Fig. 6. A proposed model for Wolbachia-induced male-killing in Ostrinia. 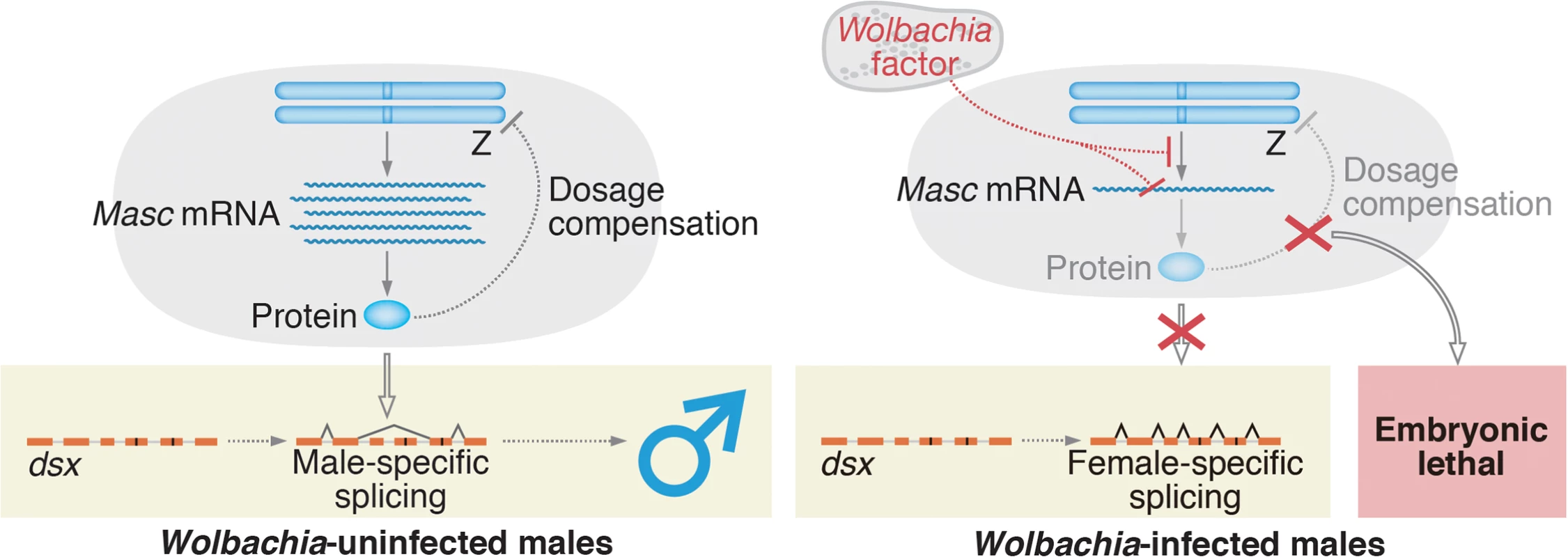
In uninfected Ostrinia (left panel), Masc protein is expressed and utilized for dosage compensation and masculinization in male development. By contrast, in Wolbachia-infected Ostrinia (right panel), Wolbachia reduces Masc mRNA level in early embryos in an unknown manner. Down-regulation of Masc mRNA inhibits a masculinizing pathway, resulting in loss of the male-type variants of dsx. Simultaneously, dosage compensation fails to establish and male-specific embryonic lethality occurs. In B. mori, femaleness is determined by Fem piRNA-mediated, highly tuned post-transcriptional regulation of Masc mRNA [14, 19]. Our findings suggest that Wolbachia has captured an unknown factor through evolution and succeeded in mimicking this sex determination system to execute the male-specific death. Our future goal is to identify a Wolbachia factor that decreases Masc mRNA post-transcriptionally or that directly inhibits Masc transcription (Fig 6).
Materials and Methods
Insects
Moths were collected at Matsudo (35.8° N, 139.9° E) and Nishi-Tokyo (35.4° N, 139.3° E), Japan, in early summer, 2014. GC-MS analysis of the pheromone gland extracts of the moths used in this study showed the presence of (E)-12 - and (Z)-12-tetradecenyl acetates (E12-14:OAc and Z12-14:OAc). The relative abundance of the two components was 1 : 1.6 (E12-14:OAc and Z12-14:OAc) in Wolbachia-infected and 1 : 2.7 in Wolbachia-uninfected moths, indicating that they were O. furnacalis (Lepidoptera: Crambidae). Wolbachia-infected strain was maintained by crossing with Wolbachia-free O. furnacalis male moths. Ostrinia larvae were reared on an artificial diet (Insecta LF, Nosan Corp.) at 23°C under a photoperiod of 16 L and 8 D. Tetracycline treatment was performed as described previously [7].
Molecular sexing
Molecular sexing of Ostrinia moths and embryos was performed by qPCR using two Z-linked genes triose phosphate isomerase (Tpi) and kettin as described by Kern et al. [20]. The autosomal gene EF-1α was used for normalization. Primers used for qPCR are listed below:
Tpi_F: 5'-ACGGAGGATCGGTTACTGGAGC-3'
Tpi_R: 5'-CGATGTCAACGAACTCTGGCTTGA-3'
kettin_F: 5'-AGGACTCTGGACGCATGGCT-3'
kettin_R: 5'-TGCAAGGCTATCAACAGGGCA-3'
EF1a_F: 5'-TTGCCACACAGCCCACATTG-3'
EF1a_R: 5'-TTGACAATGGCGGCATCACC-3'
RNA-seq analysis
Total RNA and genomic DNA were prepared simultaneously from Ostrinia embryos (25–50 embryos at each time point) using TRIzol reagent (Invitrogen) according to the manufacturer’s protocol. Libraries for RNA-seq were generated from 0, 12, 24, 36, 48 hpo embryos using the TruSeq RNA Sample Preparation kit (Illumina). The cDNAs were analyzed using the Illumina HiSeq 2500 platform with 100-bp paired-end reads according to the manufacturer's protocol [21]. De novo assembly of RNA-seq data from 10 data sets was performed as described previously [14]. Ostrinia Masc was identified from assembled contigs by BLAST using the B. mori Masc amino acid sequence as a query.
Because extensive synteny conservation is observed among several lepidopteran insects including Ostrinia [22–24], we identified putative corresponding chromosomes for Ostrinia RNA-seq derived contigs by BLAST using 13,789 B. mori gene models (putative protein-coding genes whose chromosomal locations are identified). Transcript abundance in each contig was quantified as described previously [14–15]. RNA-seq data of B. mori Masc RNAi experiments (GFP and Masc RNAi embryos of each sex, 72 h post-injection, 4 data sets) [14] were used as a control data set.
RT-PCR
Total RNA and genomic DNA were prepared from Ostrinia embryos (25–50 embryos at each time point) using TRIzol reagent (Invitrogen) according to the manufacturer’s protocol. Total RNA was subjected to reverse transcription using avian myeloblastosis virus (AMV) reverse transcriptase with an oligo-dT primer (TaKaRa). PCR was carried out with KOD FX-neo DNA polymerase (TOYOBO). Sex-specific splicing of Ostrinia dsx by RT-PCR and Wolbachia detection by wsp PCR were performed with primers reported previously [6]. RT-qPCR analyses were performed using a KAPATM SYBR FAST qPCR kit (Kapa Biosystems) and specific primers. The expression levels of rps3 were used to normalize transcript levels. Primers used for RT-qPCR are listed below:
OstriniaMasc_F: 5'-TTTGCCGCATTCATTCGCAG-3'
OstriniaMasc_R: 5'-TGGTTTTGGTGCAAGCAATTCG-3'
OstriniaRPS3_F: 5'-TGGCCACCAGAACTCAAAGC-3'
OstriniaRPS3_R: 5'-GAAACGCTTCTGGACTACGGA-3'
Transfection of Ostrinia Masc cDNA in BmN4 cells
Ostrinia Masc cDNA was cloned into the pIZ/V5-His vector (Invitrogen). Transfection experiments were performed as described previously [14]. In brief, BmN4 cells were transfected with plasmid DNAs using X-tremeGENE HP (Roche). Cells were collected at 72 h after transfection, and RT-PCR for Bmdsx and RT-qPCR for BmIMP were performed. B. mori Masc was used as a positive control for this experiment. BmIMP mRNA level was normalized to that of rp49. Primers used for RT-qPCR are listed below:
Bmdsx_F: 5'-AACCATGCCACCACTGATACCAAC-3'
Bmdsx_R: 5'-GCACAACGAATACTGCTGCAATCG-3'
rp49_F: 5'-CCCAACATTGGTTACGGTTC-3'
rp49_R: 5'-GCTCTTTCCACGATCAGCTT-3'
BmIMP_F: 5'-ATGCGGGAAGAAGGTTTTATG-3'
BmIMP_R: 5'-TCATCCCGCCTCAGACGATTG-3'
cRNA injection
The DNA template for cRNA synthesis was amplified by PCR using the pIZ/V5-His vector containing Ostrinia Masc (described above) or GFP (control) cDNA. Primers used for PCR are listed below:
pIZ-F-T7 : 5'-TAATACGACTCACTATAGGGAGACAGTTGAACAGCATCTGTTC-3'
pIZ-R: 5'-GACAATACAAACTAAGATTTAGTCAG-3'
Capped, poly(A)-tailed cRNA was synthesized using mMESSAGE mMACHINE T7 Ultra Kit (Ambion). cRNA injection was performed as described previously [14] with some modifications. We injected 1–2 nl of Masc or GFP cRNA solution (1 μg/μl in 100 mM potassium acetate, 2 mM magnesium acetate, 30 mM HEPES-KOH; pH7.4) into Wolbachia-infected Ostrinia embryos within 4 h after oviposition. The hatched larvae were collected and molecularly sexed by qPCR.
Phylogenetic analysis
Phylogenetic analysis of Ostrinia Masc protein was performed as described previously [14]. The amino acid sequences of proteins in the NCBI database and those deduced form the RNA-seq data obtained in this study with significant homology (E-value of < 1 × 10−9) to residues 51–122 of Masc were identified using the BLAST program. A neighbor-joining tree was constructed using 43 sequences [including 3 Ostrinia sequences (Ostrinia Masc, c66984_g1_i4, and c66984_g1_i3)] and the reliability of the tree was tested by bootstrap analysis with 1000 replications.
Sequence deposition
The nucleotide sequence of Ostrinia Masc has been submitted to the DDBJ/EMBL/GenBank data bank under the accession number LC028928. Deep sequencing data obtained in this study are available under the accession number DRA003038 (DDBJ).
Supporting Information
Zdroje
1. Hilgenboecker K, Hammerstein P, Schlattmann P, Telschow A, Werren JH. How many species are infected with Wolbachia?—a statistical analysis of current data. FEMS Microbiol Lett. 2008; 281 : 215–220. doi: 10.1111/j.1574-6968.2008.01110.x 18312577
2. Werren JH, Baldo L, Clark ME. Wolbachia: master manipulators of invertebrate biology. Nat Rev Microbiol. 2008;6 : 741–751. doi: 10.1038/nrmicro1969 18794912
3. Fialho RF, Stevens L. Male-killing Wolbachia in a flour beetle. Proc R Soc Lond B. 2000;267 : 1469–1473.
4. Dyer KA, Jaenike J. Evolutionarily stable infection by a male-killing endosymbiont in Drosophila innubila: molecular evidence from the host and parasite genomes. Genetics 2004;168 : 1443–1455. 15579697
5. Kageyama D, Traut W. Opposite sex-specific effects of Wolbachia and interference with the sex determination of its host Ostrinia scapulalis. Proc Biol Sci. 2004;271 : 251–258. 15058435
6. Sugimoto TN, Fujii T, Kayukawa T, Sakamoto H, Ishikawa Y. Expression of a doublesex homologue is altered in sexual mosaics of Ostrinia scapulalis moths infected with Wolbachia. Insect Biochem Mol Biol. 2010;40 : 847–854. doi: 10.1016/j.ibmb.2010.08.004 20728536
7. Sugimoto TN, Ishikawa Y. A male-killing Wolbachia carries a feminizing factor and is associated with degradation of the sex-determining system of its host. Biol Lett. 2012;8 : 412–415. doi: 10.1098/rsbl.2011.1114 22219393
8. Suzuki MG, Funaguma S, Kanda T, Tamura T, Shimada T. Analysis of the biological functions of a doublesex homologue in Bombyx mori. Dev Genes Evol. 2003;213 : 345–354. 12733073
9. Suzuki MG, Funaguma S, Kanda T, Tamura T, Shimada T. Role of the male BmDSX protein in the sexual differentiation of Bombyx mori. Evol Dev. 2005;7 : 58–68. 15642090
10. Traut W, Sahara K, Marec F. Sex chromosomes and sex determination in Lepidoptera. Sex Dev. 2007;1 : 332–346. doi: 10.1159/000111765 18391545
11. Fujii T, Shimada T. Sex determination in the silkworm, Bombyx mori: a female determinant on the W chromosome and the sex-determining gene cascade. Semin Cell Dev Biol. 2007;18 : 379–788. 17446095
12. Hasimoto H. The role of the W-chromosome in the sex determination of Bombyx mori [in Japanese]. Jpn J Genet. 1933;8 : 245–247.
13. Tajima Y. Studies on chromosome aberrations in the silkworm. II. Translocation involving second and W-chromosomes [in Japanese]. Bull Seric Exp Stn. 1944;12 : 109–181.
14. Kiuchi T, Koga H, Kawamoto M, Shoji K, Sakai H, Arai Y, et al. A single female-specific piRNA is the primary determiner of sex in the silkworm. Nature 2014;509 : 633–636. doi: 10.1038/nature13315 24828047
15. Kawamoto M, Koga H, Kiuchi T, Shoji K, Sugano S, Shimada T, et al. Sexually biased transcripts at early embryonic stages of the silkworm depend on the sex chromosome constitution. Gene 2015;560 : 50–56. doi: 10.1016/j.gene.2015.01.036 25615878
16. Suzuki MG, Imanishi S, Dohmae N, Asanuma M, Matsumoto S. Identification of a male-specific RNA binding protein that regulates sex-specific splicing of Bmdsx by increasing RNA binding activity of BmPSI. Mol Cell Biol. 2010;30 : 5776–5786. doi: 10.1128/MCB.00444-10 20956562
17. Penalva LO, Sa´nchez L. RNA binding protein sex-lethal (Sxl) and control of Drosophila sex determination and dosage compensation. Microbiol Mol Biol Rev. 2003;67 : 343–359. 12966139
18. Veneti Z, Bentley JK, Koana T, Braig HR, Hurst GD. A functional dosage compensation complex required for male killing in Drosophila. 2005;307 : 1461–1463.
19. Katsuma S, Kawamoto M, Kiuchi T. Guardian small RNAs and sex determination. RNA Biol. 2014;11 : 1238–1242. doi: 10.1080/15476286.2014.996060 25588029
20. Kern P, Cook JM, Kageyama D, Riegler M. Double trouble: combined action of meiotic drive and Wolbachia feminization in Eurema butterflies. Biol. Lett. 2015;11 : 20150095. doi: 10.1098/rsbl.2015.0095 25948567
21. Sato Y, Yoshizato T, Shiraishi Y, Maekawa S, Okuno Y, Kamura T, et al. Integrated molecular analysis of clear-cell renal cell carcinoma. Nature Genet. 2013;45 : 860–867. doi: 10.1038/ng.2699 23797736
22. d'Alençon E, Sezutsu H, Legeai F, Permal E, Bernard-Samain S, Gimenez S, et al. Extensive synteny conservation of holocentric chromosomes in Lepidoptera despite high rates of local genome rearrangements. Proc Natl Acad Sci U S A. 2010;107 : 7680–7685. doi: 10.1073/pnas.0910413107 20388903
23. Sahara K, Yoshido A, Shibata F, Fujikawa-Kojima N, Okabe T, Tanaka-Okuyama M, et al. FISH identification of Helicoverpa armigera and Mamestra brassicae chromosomes by BAC and fosmid probes. Insect Biochem Mol Biol. 2013;43 : 644–653. doi: 10.1016/j.ibmb.2013.04.003 23628856
24. Kroemer JA, Coates BS, Nusawardani T, Rider SD Jr, Fraser LM, Hellmich RL. A rearrangement of the Z chromosome topology influences the sex-linked gene display in the European corn borer, Ostrinia nubilalis. Mol Genet Genomics. 2011;286 : 37–56. doi: 10.1007/s00438-011-0624-1 21573787
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek Transmission Properties of Human PrP 102L Prions Challenge the Relevance of Mouse Models of GSSČlánek Decline of FoxP3+ Regulatory CD4 T Cells in Peripheral Blood of Children Heavily Exposed to MalariaČlánek IFNγ and IL-12 Restrict Th2 Responses during Helminth/ Co-Infection and Promote IFNγ from Th2 CellsČlánek Exploiting Fungal Virulence-Regulating Transcription Factors As Novel Antifungal Drug Targets
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2015 Číslo 7- Jak souvisí postcovidový syndrom s poškozením mozku?
- Měli bychom postcovidový syndrom léčit antidepresivy?
- Farmakovigilanční studie perorálních antivirotik indikovaných v léčbě COVID-19
- 10 bodů k očkování proti COVID-19: stanovisko České společnosti alergologie a klinické imunologie ČLS JEP
-
Všechny články tohoto čísla
- Basic Prion Science “Spreads” Insight
- Research Driven by Curiosity: The Journey from Basic Molecular Biology and Virology to Studies of Human Pathogenic Coronaviruses
- Cross Kingdom Activators of Five Classes of Bacterial Effectors
- Vaccination Drives Changes in Metabolic and Virulence Profiles of
- Expression of the Blood-Group-Related Gene Alters Susceptibility to Infection
- Transmission Properties of Human PrP 102L Prions Challenge the Relevance of Mouse Models of GSS
- Latent KSHV Infected Endothelial Cells Are Glutamine Addicted and Require Glutaminolysis for Survival
- The DSF Family of Cell–Cell Signals: An Expanding Class of Bacterial Virulence Regulators
- Intraperitoneal Infection of Wild-Type Mice with Synthetically Generated Mammalian Prion
- Vpr Promotes Macrophage-Dependent HIV-1 Infection of CD4 T Lymphocytes
- An In-Depth Comparison of Latency-Reversing Agent Combinations in Various and HIV-1 Latency Models Identified Bryostatin-1+JQ1 and Ingenol-B+JQ1 to Potently Reactivate Viral Gene Expression
- α-Macroglobulin Can Crosslink Multiple Erythrocyte Membrane Protein 1 (PfEMP1) Molecules and May Facilitate Adhesion of Parasitized Erythrocytes
- Should Symbionts Be Nice or Selfish? Antiviral Effects of Wolbachia Are Costly but Reproductive Parasitism Is Not
- A Unique Human Norovirus Lineage with a Distinct HBGA Binding Interface
- The Broad Neutralizing Antibody Responses after HIV-1 Superinfection Are Not Dominated by Antibodies Directed to Epitopes Common in Single Infection
- Rapidly Evolving Genes Are Key Players in Host Specialization and Virulence of the Fungal Wheat Pathogen ()
- MiR-21 in Extracellular Vesicles Leads to Neurotoxicity via TLR7 Signaling in SIV Neurological Disease
- Age-Dependent Cell Trafficking Defects in Draining Lymph Nodes Impair Adaptive Immunity and Control of West Nile Virus Infection
- Decline of FoxP3+ Regulatory CD4 T Cells in Peripheral Blood of Children Heavily Exposed to Malaria
- Dimerization-Induced Allosteric Changes of the Oxyanion-Hole Loop Activate the Pseudorabies Virus Assemblin pUL26N, a Herpesvirus Serine Protease
- Macrophages Subvert Adaptive Immunity to Urinary Tract Infection
- Mycolactone-Dependent Depletion of Endothelial Cell Thrombomodulin Is Strongly Associated with Fibrin Deposition in Buruli Ulcer Lesions
- Activation of TLR2 and TLR6 by Dengue NS1 Protein and Its Implications in the Immunopathogenesis of Dengue Virus Infection
- K-bZIP Mediated SUMO-2/3 Specific Modification on the KSHV Genome Negatively Regulates Lytic Gene Expression and Viral Reactivation
- Phosphoproteomic Analysis of KSHV-Infected Cells Reveals Roles of ORF45-Activated RSK during Lytic Replication
- CR3 and Dectin-1 Collaborate in Macrophage Cytokine Response through Association on Lipid Rafts and Activation of Syk-JNK-AP-1 Pathway
- IFNγ and IL-12 Restrict Th2 Responses during Helminth/ Co-Infection and Promote IFNγ from Th2 Cells
- THY-1 Cell Surface Antigen (CD90) Has an Important Role in the Initial Stage of Human Cytomegalovirus Infection
- Human Enterovirus Nonstructural Protein 2C Functions as Both an RNA Helicase and ATP-Independent RNA Chaperone
- IL-27 Signaling Is Crucial for Survival of Mice Infected with African Trypanosomes via Preventing Lethal Effects of CD4 T Cells and IFN-γ
- Synergistic Reactivation of Latent HIV Expression by Ingenol-3-Angelate, PEP005, Targeted NF-kB Signaling in Combination with JQ1 Induced p-TEFb Activation
- Vpu Exploits the Cross-Talk between BST2 and the ILT7 Receptor to Suppress Anti-HIV-1 Responses by Plasmacytoid Dendritic Cells
- Herpesvirus Genome Recognition Induced Acetylation of Nuclear IFI16 Is Essential for Its Cytoplasmic Translocation, Inflammasome and IFN-β Responses
- A Comprehensive Analysis of Replicating Merkel Cell Polyomavirus Genomes Delineates the Viral Transcription Program and Suggests a Role for mcv-miR-M1 in Episomal Persistence
- Analysis of the SUMO2 Proteome during HSV-1 Infection
- Capacity of Broadly Neutralizing Antibodies to Inhibit HIV-1 Cell-Cell Transmission Is Strain- and Epitope-Dependent
- A Novel Antiviral Target Structure Involved in the RNA Binding, Dimerization, and Nuclear Export Functions of the Influenza A Virus Nucleoprotein
- Deploying FLAREs to Visualize Functional Outcomes of Host—Pathogen Encounters
- Mosquitoes Reset Malaria Parasites
- The Lung Microbiome: New Principles for Respiratory Bacteriology in Health and Disease
- Extracellular Virions: The Advance Guard of Poxvirus Infections
- Risks of Antibiotic Exposures Early in Life on the Developing Microbiome
- RNA Virus Reassortment: An Evolutionary Mechanism for Host Jumps and Immune Evasion
- Exploiting Fungal Virulence-Regulating Transcription Factors As Novel Antifungal Drug Targets
- N-acetylglucosamine Regulates Virulence Properties in Microbial Pathogens
- Periodontal Diseases: Bug Induced, Host Promoted
- Mechanisms of Host Behavioral Change in Rodent Association
- The Endosymbiotic Bacterium Selectively Kills Male Hosts by Targeting the Masculinizing Gene
- HIV Reactivation from Latency after Treatment Interruption Occurs on Average Every 5-8 Days—Implications for HIV Remission
- Ubiquilin 1 Promotes IFN-γ-Induced Xenophagy of
- Transfer of Immunity from Mother to Offspring Is Mediated via Egg-Yolk Protein Vitellogenin
- Suppression of Long-Lived Humoral Immunity Following Infection
- The Role of VP1 Amino Acid Residue 145 of Enterovirus 71 in Viral Fitness and Pathogenesis in a Cynomolgus Monkey Model
- Utilizing Chemical Genomics to Identify Cytochrome as a Novel Drug Target for Chagas Disease
- The Emerging Role for RNA Polymerase II in Regulating Virulence Gene Expression in Malaria Parasites
- Turning Up the Heat: Inflammasome Activation by Fungal Pathogens
- On and Under the Skin: Emerging Basidiomycetous Yeast Infections Caused by Species
- EhVps32 Is a Vacuole-Associated Protein Involved in Pinocytosis and Phagocytosis of
- Characterization of a Prefusion-Specific Antibody That Recognizes a Quaternary, Cleavage-Dependent Epitope on the RSV Fusion Glycoprotein
- The Serine Protease EspC from Enteropathogenic Regulates Pore Formation and Cytotoxicity Mediated by the Type III Secretion System
- Existing Infection Facilitates Establishment and Density of Malaria Parasites in Their Mosquito Vector
- Evaluating Human T-Cell Therapy of Cytomegalovirus Organ Disease in HLA-Transgenic Mice
- Neuronal Interferon Signaling Is Required for Protection against Herpes Simplex Virus Replication and Pathogenesis
- Epstein-Barr Virus Proteins EBNA3A and EBNA3C Together Induce Expression of the Oncogenic MicroRNA Cluster miR-221/miR-222 and Ablate Expression of Its Target p57
- Colonization of the Mouse Gastrointestinal Tract Is Modulated by Wall Teichoic Acid, Capsule, and Surface Proteins
- Virulence of Group A Streptococci Is Enhanced by Human Complement Inhibitors
- Identification of Caspase Cleavage Sites in KSHV Latency-Associated Nuclear Antigen and Their Effects on Caspase-Related Host Defense Responses
- Calprotectin Increases the Activity of the SaeRS Two Component System and Murine Mortality during Infections
- Type VI Secretion System Transports Zn to Combat Multiple Stresses and Host Immunity
- Lv4 Is a Capsid-Specific Antiviral Activity in Human Blood Cells That Restricts Viruses of the SIV/SIV/HIV-2 Lineage Prior to Integration
- Phenylbutyrate Is Bacteriostatic against and Regulates the Macrophage Response to Infection, Synergistically with 25-Hydroxy-Vitamin D₃
- An Internally Translated MAVS Variant Exposes Its Amino-terminal TRAF-Binding Motifs to Deregulate Interferon Induction
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- RNA Virus Reassortment: An Evolutionary Mechanism for Host Jumps and Immune Evasion
- Activation of TLR2 and TLR6 by Dengue NS1 Protein and Its Implications in the Immunopathogenesis of Dengue Virus Infection
- N-acetylglucosamine Regulates Virulence Properties in Microbial Pathogens
- Characterization of a Prefusion-Specific Antibody That Recognizes a Quaternary, Cleavage-Dependent Epitope on the RSV Fusion Glycoprotein
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



