-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaDimerization-Induced Allosteric Changes of the Oxyanion-Hole Loop Activate the Pseudorabies Virus Assemblin pUL26N, a Herpesvirus Serine Protease
Herpesviruses encode a unique serine protease, which is essential for herpesvirus capsid maturation and is therefore an interesting target for drug development. In solution, this protease exists in an equilibrium of an inactive monomeric and an active dimeric form. All currently available crystal structures of herpesvirus proteases represent complexes, particularly dimers. Here we show the first three-dimensional structure of the native monomeric form in addition to the native and the chemically inactivated dimeric form of the protease derived from the porcine herpesvirus pseudorabies virus. Comparison of the monomeric and dimeric form allows predictions on the structural changes that occur during dimerization and shed light onto the process of protease activation. These new crystal structures provide a rational base to develop drugs preventing dimerization and therefore impeding herpesvirus capsid maturation. Furthermore, it is likely that this mechanism is conserved throughout the herpesviruses.
Published in the journal: . PLoS Pathog 11(7): e32767. doi:10.1371/journal.ppat.1005045
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1005045Summary
Herpesviruses encode a unique serine protease, which is essential for herpesvirus capsid maturation and is therefore an interesting target for drug development. In solution, this protease exists in an equilibrium of an inactive monomeric and an active dimeric form. All currently available crystal structures of herpesvirus proteases represent complexes, particularly dimers. Here we show the first three-dimensional structure of the native monomeric form in addition to the native and the chemically inactivated dimeric form of the protease derived from the porcine herpesvirus pseudorabies virus. Comparison of the monomeric and dimeric form allows predictions on the structural changes that occur during dimerization and shed light onto the process of protease activation. These new crystal structures provide a rational base to develop drugs preventing dimerization and therefore impeding herpesvirus capsid maturation. Furthermore, it is likely that this mechanism is conserved throughout the herpesviruses.
Introduction
The family Herpesviridae is divided into the three subfamilies alpha-, beta - and gammaherpesviruses. These infectious agents cause a variety of diseases in many different hosts including humans. Pseudorabies virus (PrV) is a neurotropic porcine alphaherpesvirus [1] and the causative agent of Aujeszky's disease. The pig is the only susceptible species that can survive a PrV infection depending on the age of the animal and virulence of the virus, while most other mammals die within a few days. Only higher primates including humans and equids are resistant to infection. Due to its broad host range PrV has become an important model system to study herpesvirus biology in cell culture and in the natural host. PrV genome organization and protein content exhibit significant homology to that of the human herpes simplex virus type 1 (HSV-1) [2,3], which is among the best-studied herpesviruses.
Capsid assembly of HSV-1 has been intensively analyzed. However, since herpesvirus capsid proteins are well conserved, it is very likely that the process leading to mature, DNA-filled nucleocapsids is also similar. The proteolytic activity of the serine protease is essential for this process [4]. In HSV-1 and PrV, this protease is encoded by the UL26 gene [5], which is the longest open reading frame in a family of in-frame overlapping genes [5–8]. UL26 overlaps in frame with UL26.5 [3]. UL26 and UL26.5 possess identical 3'-termini, which encode a scaffold protein while the unique 5'-terminus of UL26 contains the protease domain. There are at least two target sites for the protease in the full-length UL26 protein (pUL26) [8,9]. Autoproteolytic activity at the release site (R-site) results in release of the N-terminal protease domain (pUL26N, also called VP24 or generic: assemblin) and the C-terminal part containing the scaffold protein (pUL26C, also called VP21 or generic: assembly protein) [10]. Due to the presence of a linker region pUL26C is 21 amino-acid residues longer than pUL26.5 (Fig 1). Near the C-terminus of the scaffold protein is the maturational site (M-site) where pUL26.5 and pUL26C are cleaved [10]. The scaffold protein binds to the major capsid protein pUL19 (VP5) and directs it to the nucleus [11–17]. During capsid assembly, the scaffold protein forms a scaffold core with the major capsid protein bound to the C-termini of pUL26/pUL26C/pUL26.5 [17]. When the capsid is fully assembled the scaffold is cleaved at its M-site releasing the ring-like scaffold structure from the capsid, which is then expelled during DNA packaging. In contrast, the protease remains in the nucleocapsid [18]. Without protease activity, the scaffold remains in the capsid resulting in capsids without viral DNA designated as B-capsids. Upon activation of the protease, the B-capsids mature and subsequent steps of viral replication occur as shown with a temperature-sensitive HSV-1 pUL26 mutant [19].
Fig. 1. Cleavage sites and products of PrV pUL26 and pUL26.5. 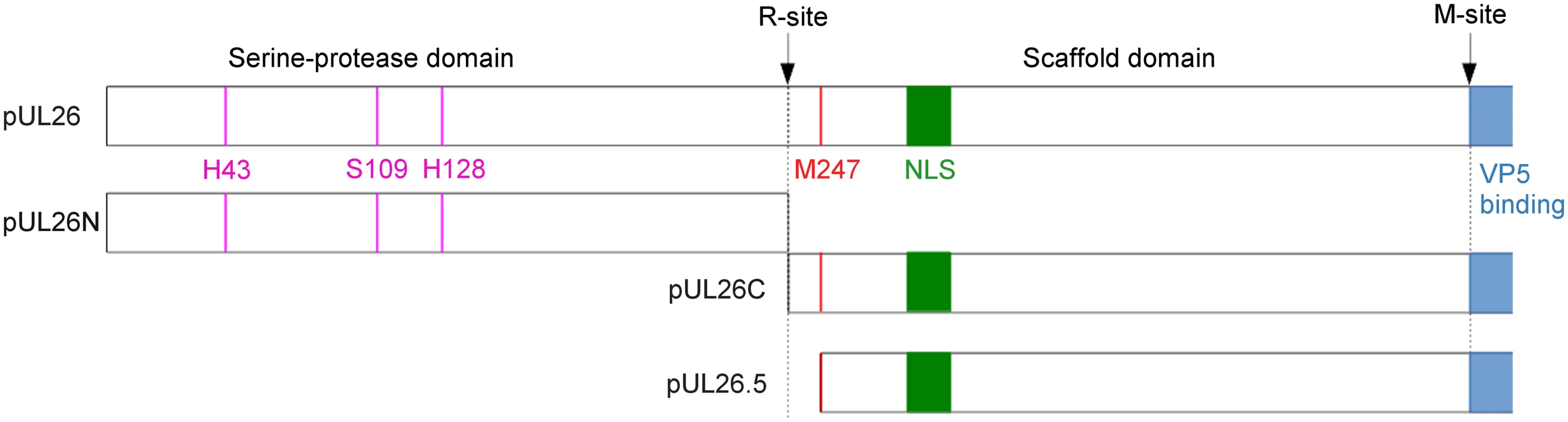
Schematic overview of the PrV UL26/26.5 gene products and their processing. Catalytic residues are shown in pink, the position of the nuclear localization sequence (NLS) is shown in green, the predicted major capsid protein (VP5) binding-region is shown in blue and the start of pUL26.5 is shown in red. Cleavage of PrV pUL26 at the R-site releases protease pUL26N from the scaffolding protein pUL26C, and cleavage of pUL26C/pUL26.5 at the putative M-site removes C-terminally 37 amino-acid residues thereby freeing them from interaction with the major capsid protein. The PrV R- and M-sites are most likely YLQA-T(226) and presumably VQA-S(488), respectively, as predicted from sequence alignments with the protease homologs of other herpesviruses [20,21]. The function of pUL26.5 can be taken over by pUL26C but with reduced efficiency as well as loss of an apparent core structure in the resulting capsids [22]. Proteases of several herpesviruses, such as human cytomegalovirus (HCMV) and Kaposi's sarcoma-associated herpesvirus (KSHV), have one or more internal cleavage sites to regulate activity or promote destabilization of the protease [23–25]. For proteases of PrV, HSV-1 and herpes simplex virus type 2 (HSV-2) no internal cleavage sites have been reported.
Herpesvirus maturational proteases exist in a monomer-dimer equilibrium. Dimers are weakly associated with dissociation constants (KD) in the micromolar range [26] and are active, while monomers are almost inactive [27]. It was shown that dimerization of the HCMV assemblin is dependent on protein concentration. The fraction of dimeric protease at 0.2 μM was demonstrated to be ~0.3 increasing to ~0.7 at 4.5 μM [27]. Additionally, dimerization is favored by high concentrations of kosmotropic compounds like glycerol [26,27].
Activity of the herpesvirus protease has to be strictly regulated. The enzyme is expressed as full-length pUL26 in the cytosol of infected cells where its concentration is low and thus the inactive, monomeric form is predominant [28]. The major capsid protein is bound by pUL26.5 and translocated to the nucleus via the nuclear localization sequence within pUL26.5 [29]. Since pUL26 encompasses pUL26.5, it also contains this nuclear localization sequence. Therefore, autoproteolytic activity of the protease in the cytosol would prevent its localization to the nucleus. When capsid assembly is completed, the protease has to become active to release the scaffold protein from the capsid and to allow DNA packaging. In the capsids, the concentration of protease is much higher than in the cytosol, thus promoting dimerization [28]. Additionally, it was proposed that the capsid environment itself might enhance proteolytic activity of the protease [30].
Several structures of homologous assemblins of other herpesviruses have been published, revealing the overall fold, active site and biological assembly [31–38]. The sequence identities of these structures to the PrV assemblin range from 60% (alphaherpesviruses) to 30% (beta - and gammaherpesviruses) [39,40]. All assemblins consist of 6–9 α-helices surrounding a β-barrel formed by two β-sheets. The catalytic triad is unique among the serine proteases and consists of one serine and two histidine residues. The active site is solvent accessible and distal to the dimer interface. Nevertheless, dimerization drastically influences the activity [27]. There is evidence, that upon dimerization the oxyanion hole is formed by structural changes of a loop containing two strictly conserved arginine residues [41,42].
Currently, crystal structures are available for native dimeric and covalently inhibited dimeric assemblins. Recently, three structures of the truncated KSHV assemblin in complex with helical-peptide mimetics were published [43,44]. These compounds bind to the dimerization area and disrupt the dimerization interface of full-length KSHV assemblin [45]. Here, we report crystal structure analyses of the active dimer, the diisopropyl phosphate-inhibited dimer as well as the non-inhibited monomer of pUL26N from PrV (224 amino-acid residues). The latter is the first-ever structure of an assemblin in its native monomeric state. Comparison of the monomeric and dimeric forms provides insight into the regulation of protease activity by dimerization and a structural basis for rational design of therapeutic substances that trap the protein in the inactive monomeric state.
Additional details and information about herpesvirus proteases and its involvement in capsid maturation are reviewed elsewhere [21,46–52].
Results and Discussion
Native and inhibited dimeric pUL26N
Overall fold and dimer interface
The tertiary structure of pUL26N is a β-barrel consisting of a four-stranded antiparallel and a three-stranded mixed β-sheet (Fig 2) surrounded by eight α-helices. The quaternary structure is a homodimer related by a two-fold non-crystallographic axis with an interface area of 1,273 Å2 (native dimer) and 1,258 Å2 (inhibited dimer) comparable to all previously published structures (~1,300 Å2, determined by PDBePISA [53]). The native and inhibited dimeric structures of pUL26N are nearly identical with an r.m.s. deviation of 0.39 Å (437 aligned Cα atoms, secondary-structure matching [54]) over the whole dimer (Fig 2). The covalently bound diisopropyl phosphate replaces a chloride ion at the active site serine of the native structure. B-factors of the Cα atoms are low throughout the β-barrel and dimer interface and are increased at the far side of the dimer interface (S1 Fig). Overall, B-factors in the inhibited dimeric structure are lower than in the native form (Table 1). This is most likely due to stabilization by inhibitor binding. The overall fold of dimeric pUL26N is similar to homologous structures [31–37]. A superposition of structures of representative assemblins from all herpesvirus subfamilies is shown in S2 Fig. The r.m.s. deviations range from 1.32 Å (pdb entry 1vzv, 410 aligned Cα atoms) to 2.58 Å (pdb entry 1cmv, 334 aligned Cα atoms) over the whole dimers.
Fig. 2. Superposition of dimeric pUL26N from PrV with and without bound inhibitor. 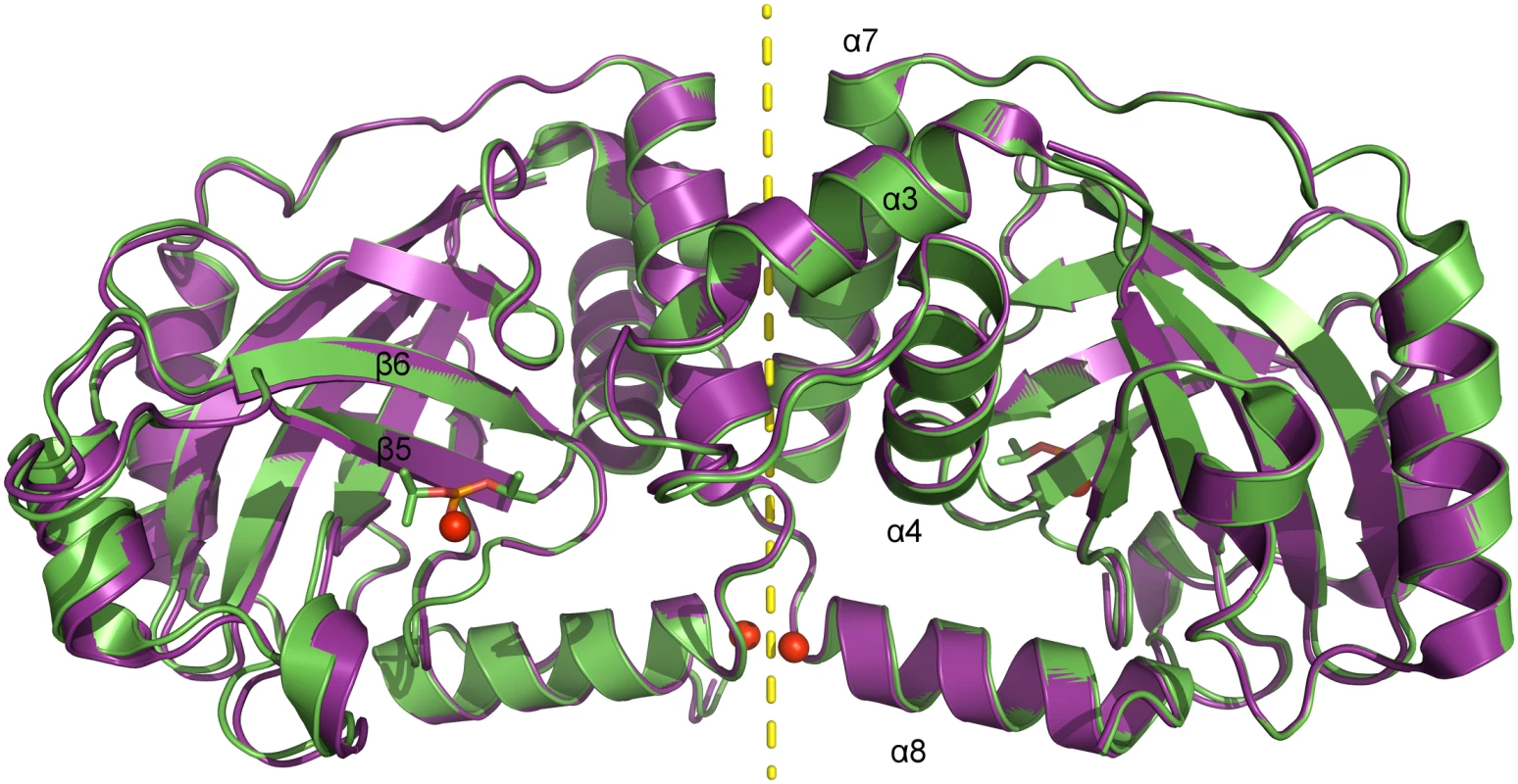
Superposition of native (pdb entry 4v07, shown in purple) and diisopropyl fluorophosphate-inhibited (pdb entry 4v08, shown in green) dimeric pUL26N from PrV. Diisopropyl phosphate is shown as stick model and chloride ions as spheres. The two-fold axis of the dimer is shown as a yellow dashed line. Tab. 1. Summary of data collection and refinement statistics. 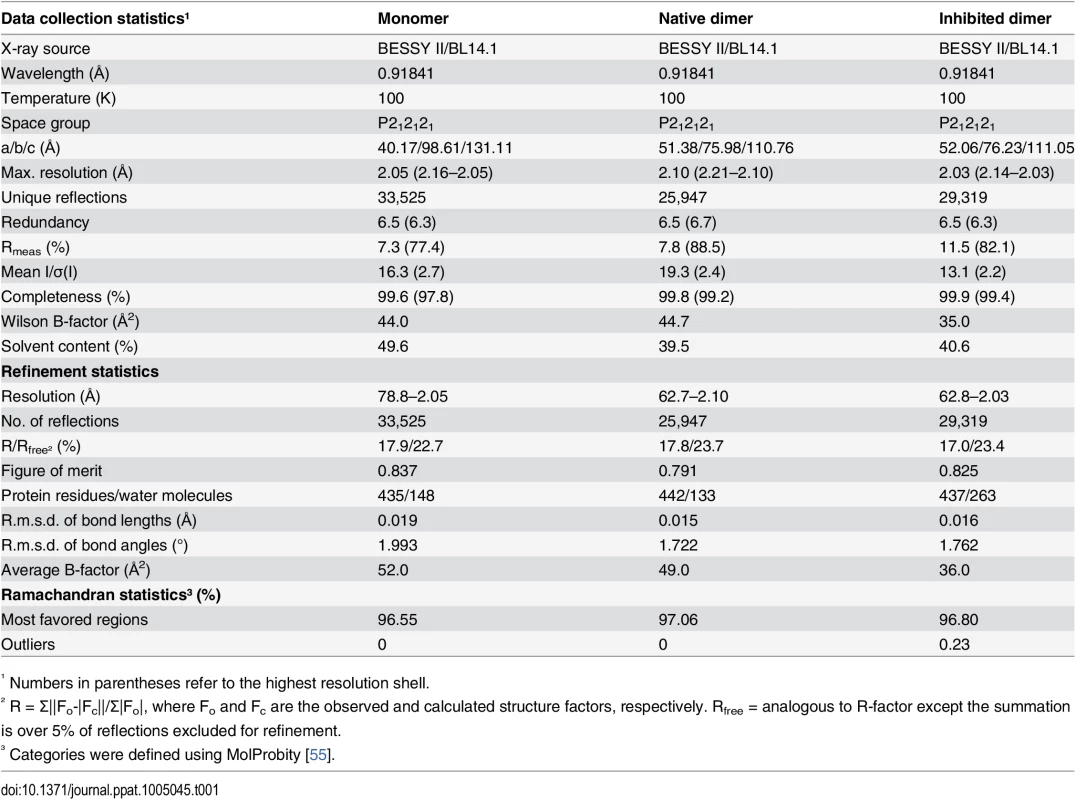
¹ Numbers in parentheses refer to the highest resolution shell. The dimer interface is formed by helices α3, α4 and α7 establishing a hydrophobic patch towards the adjacent monomer like the corresponding α-helices in the structures of assemblins from varicella zoster virus (VZV, pdb entry 1vzv) and HSV-2 (pdb entry 1at3). As already described before, dimer interfaces of alphaherpesvirus assemblins are slightly different from those of beta - and gammaherpesvirus assemblins. In dimers of beta - and gammaherpesvirus assemblins, the helices corresponding to helix α7 of dimeric PrV assemblin are nearly parallel, whereas in alphaherpesvirus assemblins they form an angle of 30–40° [35]. A difference to all other published structures is the loop α3-α4 in PrV pUL26N that forms one α-helical turn in the other structures, except for HSV-2 assemblin where this part is not modeled. Moreover, the N-termini of helices α8 of PrV pUL26N point towards each other with the positive dipole moment compensated by one chloride ion per monomer. The presence of such chloride ions has not been observed in previous structures. Probably, the solvent interaction is sufficient to compensate for the positive dipole moment in dimers of beta - and gammaherpesvirus assemblins.
Active site and oxyanion hole
The active site serine is solvent accessible and the catalytic residues are part of β-strands β5 (Ser109) and β6 (His128) and the β2-β3 loop (His43), like in earlier structures [31–37]. In our inhibitor complex, covalent binding of diisopropyl fluorophosphate has formed a phosphate ester with the active-site serine in a manner also observed in crystal structures of homologous assemblins [34]. In the native structure, a chloride ion occupies the oxyanion hole, which is formed by the β6-β7 loop (residues 133–142, further referred to as oxyanion-hole loop, OHL, Fig 3). Two consecutive arginine residues in this loop (Arg136 and Arg137) are strictly conserved throughout all herpesvirus maturational proteases (S3 Fig). The backbone N-H of Arg136 provides the first hydrogen-bond donor of the oxyanion hole. A water molecule as second hydrogen-bond donor supports anion stabilization, as does the positive local environment established by these two arginine residues. For HCMV assemblin it was shown that this water molecule plays a role in catalysis [56]. It is kept in place by a second water molecule that is positioned by hydrogen bonds of the peptide backbone oxygen of Leu10 (loop β1-α1) and the peptide N-H of Leu110 (β5). Both water molecules are present in the dimeric structures of assemblins from PrV (this report), HSV-2 (pdb entry 1at3), KSHV (pdb entries 1fl1, and 2pbk) and HCMV (pdb entries 1cmv, 1wpo, 1id4, 1iec, 1ied, 1ief, and 1ieg). The positions of these water molecules seem to be conserved in all active assemblins. Our findings for the composition of the oxyanion hole are consistent with those reported for HSV-2 [34].
Fig. 3. Active site and oxyanion hole loop of inhibited dimeric pUL26N from PrV. 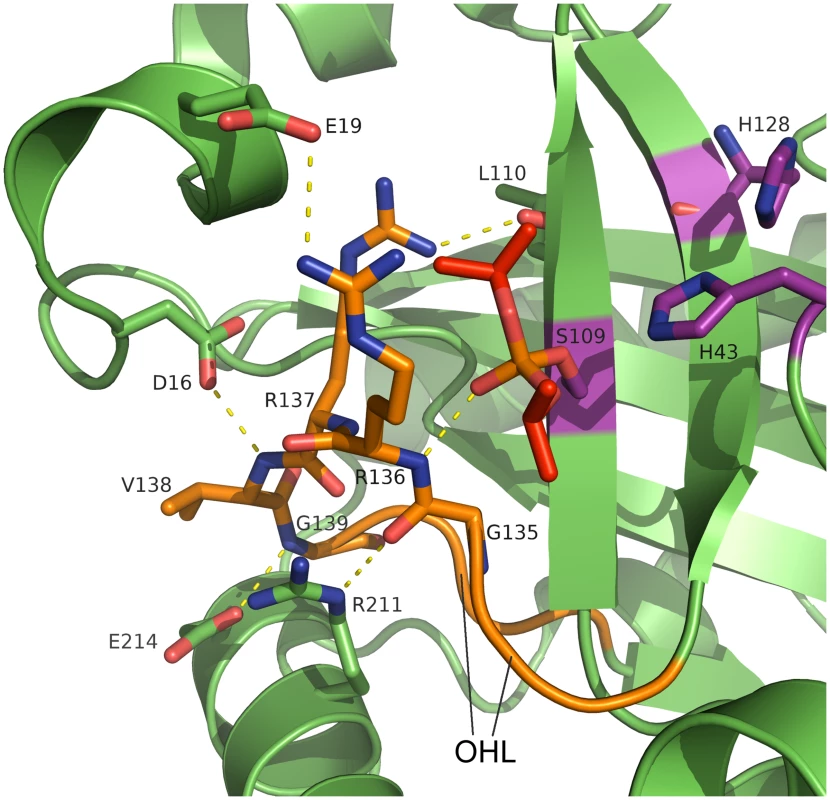
A detailed view of the active site and the hydrogen-bond network of the oxyanion hole is shown. The oxyanion-hole loop is colored orange, the covalently bound diisopropyl phosphate is colored red and the catalytic triad is colored purple. Dashed lines illustrate parts of the hydrogen bond network (distances in the range of 2.7–3.4 Å). The strictly conserved Arg136 forms the oxyanion hole with its peptide backbone N-H. Diisopropyl phosphate mimics the transition state of the natural substrate. An extensive network of hydrogen bonds stabilizes the OHL (S4 Fig). This network includes conserved water molecules and parts of the α1 region, α8 and β5. There are five strictly conserved residues in the OHL and each of these is involved in the network of hydrogen bonds that positions Arg136 and stabilizes the oxyanion hole.
The side chain of Arg137 forms hydrogen bonds to the peptide backbone of Leu20 (α1) and Leu110 (β5), which are also conserved in assemblins (S3 Fig). The OH-group of the conserved Thr140 forms a hydrogen bond to the backbone oxygen of Arg137 whereas the backbone oxygen of Thr140 accepts a hydrogen bond from the backbone N-H of Ala108 (β5). Inspection of other assemblin structures showed that these hydrogen bonds are present in all dimeric structures. In the PrV assemblin, the peptide oxygen of Val138 is connected to the peptide N-H of Leu12 via hydrogen bonds mediated by a conserved water molecule (found in HSV-2 and KSHV assemblins). Most likely this water molecule and hydrogen bonding pattern are present in all herpesvirus maturational proteases.
Additionally, the OHL of PrV pUL26N is maintained by hydrogen bonds of the side chains of Asp16 (α1), Glu214 and Arg211 (both from α8) with the peptide backbone of Val138, Gly139 and Gly135, respectively. These hydrogen bonds are also present in HSV-2 assemblin with Glu214 replaced by Gln. Identical residues are present in the VZV assemblin, but the side chains of Glu and Asp point in different directions and do not form the corresponding hydrogen bonds. This ambiguity may result from the limited resolution of that structural model (pdb entry 1vzv with a resolution of 3 Å, no structure factors were deposited). Most likely, the loop is stabilized comparable to HSV-2 and PrV assemblins.
Monomeric pUL26N
Diffraction datasets of dimeric pUL26N were collected from needle-shaped crystals and phasing by molecular replacement was successful either by using a monomer or the complete dimer as search models. For datasets derived from morphologically different, plate-shaped crystals, molecular replacement was only successful when using a monomer as a search model. In the resulting structure, two chains are present in the asymmetric unit. These chains and their adjacent symmetry mates are not in a proper position to form the known dimer. PDBePISA [53] suggests one assembly. This putative dimer has no local dyad, which is unusual for biologically relevant dimers [57,58]. The interface area (978 Å2) is much smaller than average for homodimers of this molecular weight (~1,500 Å2) [57]. Furthermore, the helices forming the interface have B-factors (60–100 Å2) above average (52 Å2). Thus, the suggested dimeric interaction is a mere crystal contact and the crystal structure actually corresponds to monomeric pUL26N.
The region corresponding to helix α1 in dimeric PrV pUL26N is disordered in the monomeric form. For ease of comparison, in this report numbering of the helices in the monomeric protease is adapted to that of the dimeric protease. The β-barrel and distal side of the dimerization area of chain A align very well with chain B of the asymmetric unit. The OHL and helices α3, α7 and α8 of the dimerization area, on the other hand, differ in chain A and B (Fig 4). The OHL has two alternative conformations in chain A with equal occupancies. One conformation correlates to the OHL of chain B and the other one is shifted towards α8 due to crystal contacts. Compared to chain B the helices α3 and α7 of chain A are bent and helix α8 is slightly tilted. Accordingly, the overall r.m.s. deviation of chain A and B is 1.03 Å (207 aligned Cα atoms). Furthermore, there is no electron density observed for residues 16–19 of chain A (β1-α2 loop) and very weak electron density for residues 14–18 and 194–196 of chain B (β1-α2 loop and the short α7-α8 loop, respectively).
Fig. 4. B-factor coloring of superposed chains of the asymmetric unit of monomeric pUL26N from PrV. 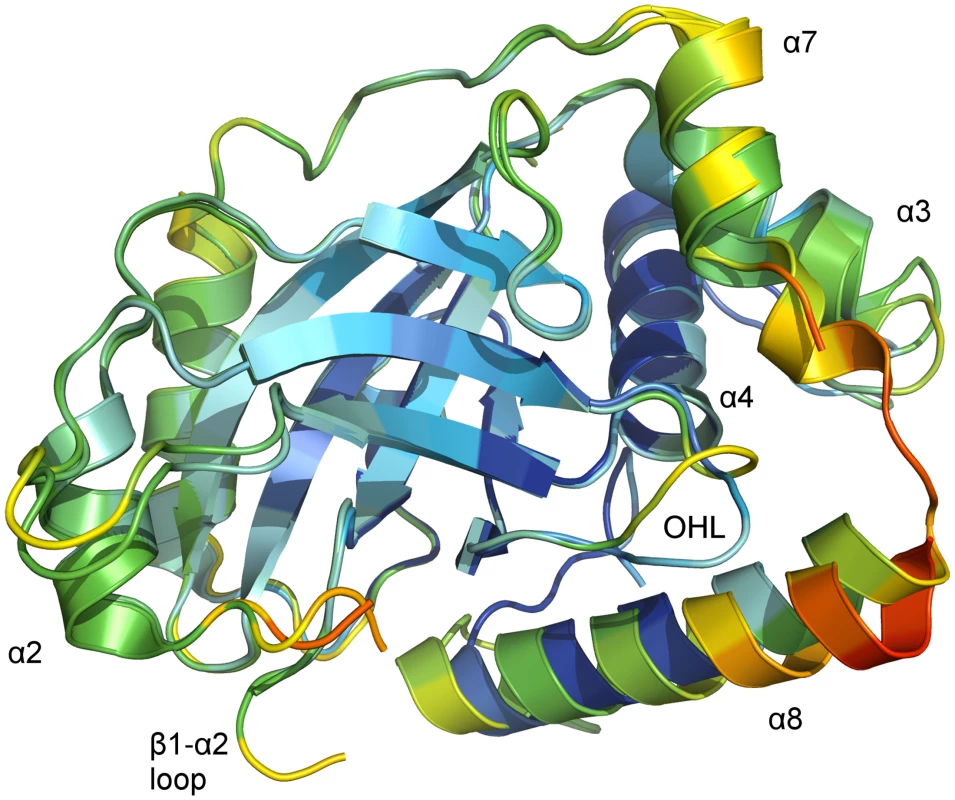
Superposition of chain A and chain B of monomeric PrV pUL26N (pdb entry 4v0t) with Cα atom B-factor coloring. B-factors below 40 Å2 are colored deep blue. B-factors above 100 Ų are colored orange or red. The OHL and the dimerization helices that differ in both chains are labeled. The β-barrel and the far side of the dimerization area show low to moderate B-factors. High B-factors are observed for the α7-α8 loop and the partial disordered loop β1-α2. The core of the monomer is rigid as indicated by low B-factors of Cα atoms in the β-barrel, helix α4 and most of helix α8 of chain A (Fig 4). In contrast, the periphery of both monomers is flexible as evidenced by increased B-factors of Cα atoms at the distal side of the dimerization area, the OHL, and the dimerization helices α3 and α7 as well as in helix α8 of chain B. B-factors around 100 Å2 are also observed at the N-termini of helices α8 in both chains.
Taken together, these observations show that the dimerization area and the parts necessary for formation of the oxyanion hole in dimeric pUL26N are flexible and not strictly ordered in monomeric pUL26N.
Comparison of monomeric and dimeric pUL26N from PrV
The tertiary structures of monomeric and dimeric PrV pUL26N are partially similar. The r.m.s. deviations of inhibited dimeric pUL26N chain B with chain A and chain B of monomeric protease are 0.93 Å (192 aligned Cα atoms) and 1.01 Å (190 aligned Cα atoms), respectively. The β-barrel, the distal side of the dimer interface, and helix α4 of the dimer interface are almost identical in monomeric and dimeric pUL26N (Fig 5). The r.m.s. deviations of these parts of inhibited dimeric pUL26N chain B with chain A and chain B of monomeric protease are 0.54 Å (151 aligned Cα atoms) and 0.63 Å (149 aligned Cα atoms), respectively. In contrast, significant differences are observed for the OHL, N - and C-termini of helices α3, α7 and α8 of the dimer interface, and the α7-α8 loop (Fig 5).
Fig. 5. Superposition of monomers of monomeric and dimeric PrV assemblin. 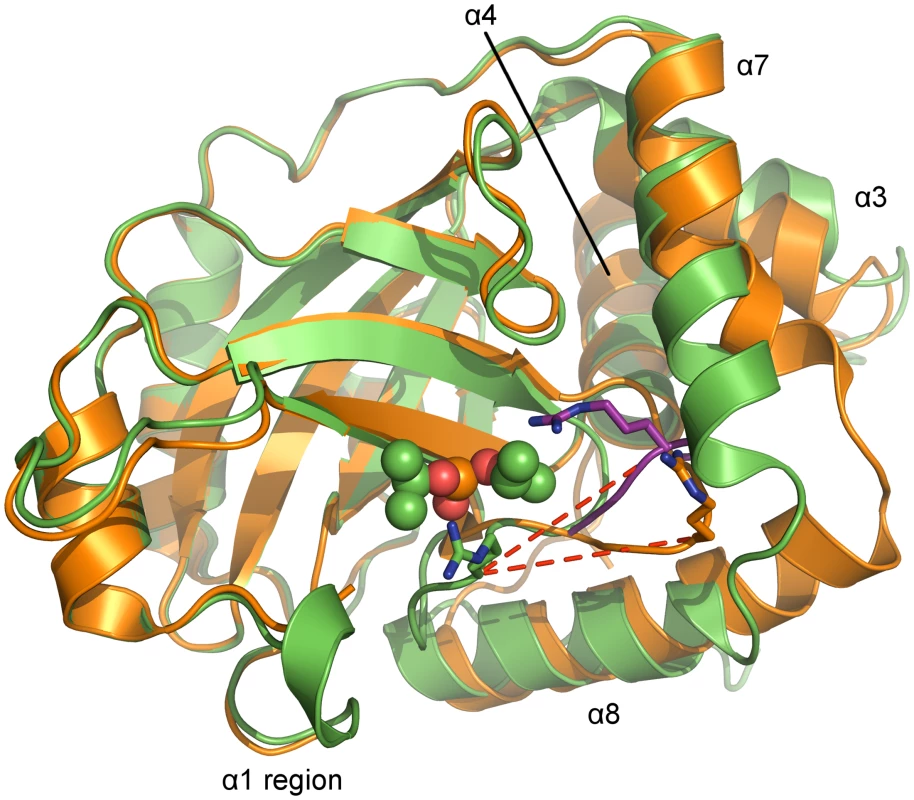
Superposition of chain A of monomeric (shown in orange, alternative conformation of the oxyanion-hole loop colored purple) and chain B of inhibited dimeric (shown in green) pUL26N from PrV. Arg136 (stick model) of the oxyanion-hole loop moves ~11 Å (red dashed lines) to form the oxyanion hole. The helices, which undergo significant changes upon dimerization, are labeled. Diisopropyl phosphate is represented as space-filling model. In the monomer, the N-terminus of helix α8 is one and a half turn longer, but the C-terminus of helix α7 is one turn shorter. Thus, the α7-α8 loop conformation is shifted towards α8 upon dimer formation. In dimeric pUL26N helices α3 and α7 form a hydrophobic cleft that is occupied by helix α7 of the second monomer within the dimer. This cleft is closed in the monomer by bending the C-termini of these helices towards each other (Fig 5).
In the monomeric pUL26N, the OHL is positioned near the N-terminus of α8. Thus, the activation of pUL26N relies on dimerization-induced allosteric changes, which shift the OHL towards the active site (Fig 6A). The top of the OHL (Arg136Cα) moves approximately 11 Å. In the dimer, the N-terminus of α8 is unwound, so some residues which point towards the protein core in the monomeric pUL26N are buried by the adjacent monomer of the dimer (Fig 6B). However, the side chain positions of the catalytic triad are unchanged as suspected by Batra et al. [41].
Fig. 6. Active site and dimer interface helices of superposed monomers of monomeric and dimeric PrV assemblin. 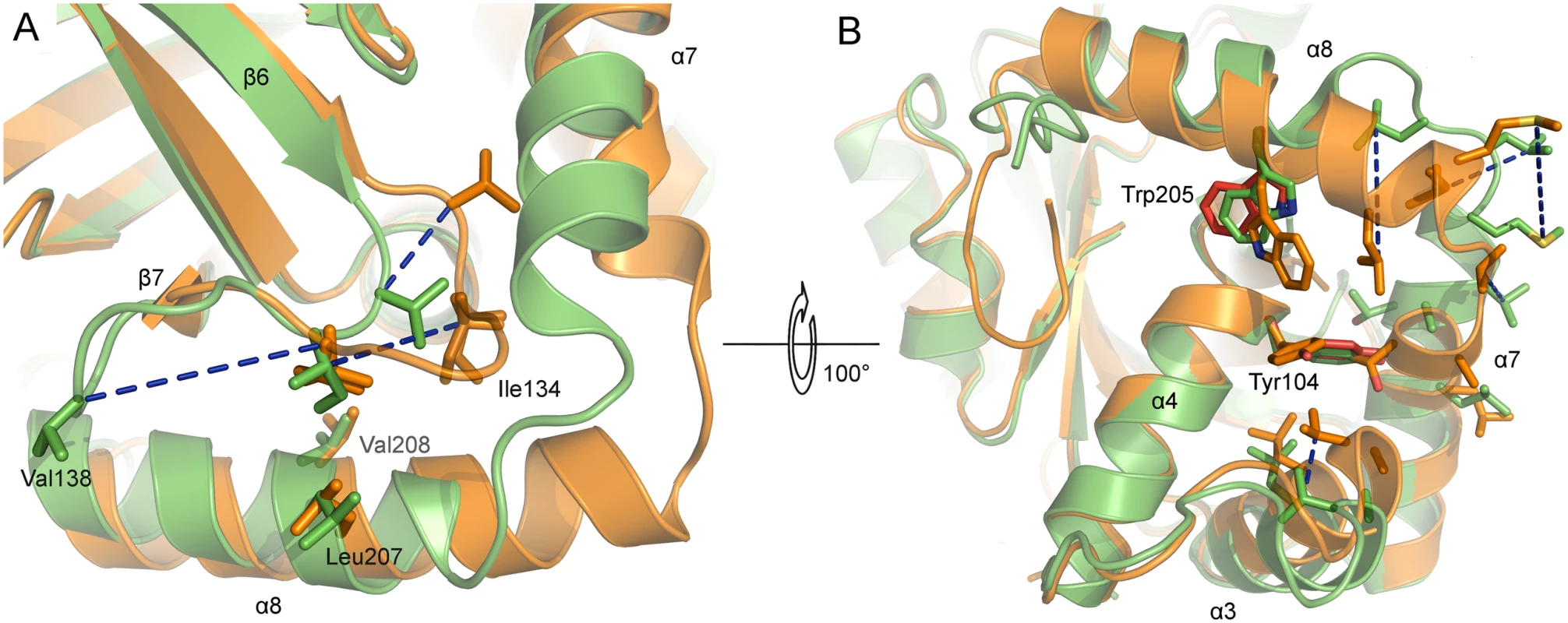
Superposed A-chains of monomeric (shown in orange) and inhibited dimeric (shown in green) PrV protease. Blue dashed lines connect corresponding residues of both structures. (A) Hydrophobic residues of the oxyanion-hole loop (OHL, only one conformation shown). (B) Hydrophobic residues of α3, α7 and α8, which undergo changes, are shown. The OHL is omitted for clarity. Close to the OHL and the dimerization area the residues Tyr104 (α4) and Trp205 (α8) (both colored red) have different side-chain rotamers in chain A and B, due to crystal contacts of chain A indicating the not established dimer interface. In monomeric protease, the helices α3 and α7 move towards each other. After dimer formation, this space is occupied by α7 of the second monomer (not shown). Hydrophobic residues of helices of the dimerization area in monomeric protease are clustering together to be less exposed to solvent. In the dimeric form, these residues are enabling dimerization. Comparison of the active and inactive conformations of the OHL reveals a key location in the structure that is occupied by alternative hydrophobic residues. In dimeric PrV pUL26N, Ile134 is present at this position, whereas in the monomeric form Val138 takes its place (Fig 6A). The hydrophobic character of the Ile134 position is highly conserved in all sequences and three-dimensional structures of assemblins (S3 Fig). The hydrophobic character of Val138 is type-conserved in alphaherpesvirus sequences. Both residues are kept in place by hydrophobic interactions with two side chains of helix α8 (Leu207, Val208) or the corresponding helices in related structures. The hydrophobic character of these residues is conserved in herpesvirus proteases, although Thr and long, partially aliphatic side chains appear to be tolerated (e.g. Thr in Epstein-Barr virus (EBV) assemblin, or Lys in KSHV assemblin, S3 Fig).
Another consequence of the different conformation of the OHL in the monomeric form is that the Asp16-containing region and α1 are disordered. This loop, the OHL and major parts of the dimer interface area are flexible in monomeric pUL26N as indicated by partially disordered segments and high B-factors. Thus, the crystallographically observed increase of order upon dimerization of PrV assemblin is in line with the disorder-to-order mechanism of dimerization previously proposed for KSHV assemblin [28,43].
Impact of a C-terminal deletion
In our crystallization assays we used a construct of PrV pUL26N that was shortened by one C-terminal amino-acid residue (Ala225) compared to the in vivo form. The resulting overall fold, conformation of the OHL, and position and orientation of the residues of the catalytic triad are identical to those in previously determined dimeric full-length assemblin structures of related herpesviruses, strongly suggesting that the C-terminal deletion does not affect the activity or structure of the protease. Moreover, native and inhibited dimers of PrV pUL26N crystallized isomorphously and the diisopropyl phosphate-ester of the active-site serine is quantitatively observed in the inhibited structure.
The inhibitor diisopropyl fluorophosphate reacts specifically with the active-site serine, but hydrolyzes in aqueous solutions with a half-life of one hour at pH 7.5 and even faster at higher pH [59]. After incubation of PrV pUL26N with a sevenfold excess of diisopropyl fluorophosphate (5 mM) in a buffer at pH 7.5 at room temperature for one hour, the protein was crystallized at pH 8. Based on these constraints quantitative inhibition of PrV pUL26N (minimum 90% in the crystal) prior to inhibitor hydrolysis can be calculated to occur at a second order reaction rate of at least 0.09 s−1 M−1. This is consistent with the known reaction rate of full-length HSV-1 assemblin with diisopropyl fluorophosphate of 1 s−1 M−1 at higher pH (pH 8.0) and higher temperature (30°C) [60]. Thus, the catalytic site of PrV pUL26N remains fully reactive with respect to this substrate-like inhibitor. Furthermore, earlier work has shown that proteolytic activity with respect to substrates containing the M-site is not significantly reduced by 3 - or 8-residue C-terminal truncations [61]. C-terminally extended assemblins on the other hand do show a considerable decrease in activity [37,61,62], presumably because such an extension sterically interferes with the proper positioning of helix α8, which is required to establish the correct conformation of the OHL. The absence of Ala225, however, does not interfere in any way with helix α8, the conformation of the active site, and the OHL. Indeed, in the crystal structure of VZV assemblin the C-termini are disordered [35], further confirming that the C-terminal region does not have a substantial impact on the overall protein fold.
Monomer-dimer equilibrium
Structural properties and self-association behavior of PrV pUL26N in solution were characterized by small-angle X-ray scattering (SAXS). Data were recorded at protein concentrations between 0.5 mg/ml and 10 mg/ml. Normalized SAXS intensities vary with protein concentration, suggesting the presence of more than one oligomerization state. The experimental SAXS curves can be accurately described assuming a monomer-dimer equilibrium and using form factors for the individual states derived from the monomeric and dimeric crystallographic models (a representative example is shown in S5 Fig). Monomer volume fractions resulting from curve fitting with the program OLIGOMER [63] are shown in S5 Fig and S1 Table. At the highest protein concentration investigated (10 mg/ml, which approximately corresponds to the starting concentration in our crystallization experiments), a considerable volume fraction (30%) of pUL26N is present in the monomeric form. This observation is consistent with the fact that crystals of the monomer could be obtained under these conditions. In contrast, inclusion of 0.2 M MgCl2 in the buffer used for SAXS measurements markedly shifts the monomer-dimer equilibrium, resulting in nearly complete dimerization at a protein concentration of 10 mg/ml. This result may explain why crystallization of the dimeric form of pUL26N required the presence of MgCl2. Particle shape under conditions that result in virtually complete dimerization according to the analysis with OLIGOMER (i.e. 10 mg/ml protein in the presence of MgCl2) was also determined independently by means of an ab initio approach. The size and oblong shape of the final bead model obtained with DAMMIN [64] are in good agreement with the crystallographic structure of the pUL26N dimer (S5 Fig), further confirming the nature of the concentration-dependent self-association that is observed here. The dissociation constants with and without MgCl2 can be estimated to 200 μM and 50 μM, respectively (S5 Fig). Higher oligomers than dimers are not observed.
Monomeric PrV pUL26N compared to truncated KSHV assemblin in complex with helical-peptide mimetics
Three structures of KSHV assemblin (KA) with helical-peptide mimetics (HPMs) have been published to date with pdb entries 3njq, 4p2t, and 4p3h [43,44]. These HPMs disrupt dimer formation in full-length KA as shown by size-exclusion chromatography [45]. In solution, both C-terminal helices of the monomeric KA are unfolded according to NMR - and CD-spectroscopic data [28,43]. Therefore, 34 C-terminal residues of KA were truncated for crystallization of these HPM complexes. One of these truncated helices is the major dimer-interface helix, so this truncated KA is obligate monomeric. Consequently, the models of these HPM complexes were stated as monomeric [43,44]. Inspection of these models led us to the conclusion, that the HPMs function as an artificial dimer interface for this truncated KA. PDBePISA suggests dimers or higher association states for HPM complexes (buried surface area of ~1,200 Å2 per monomer). The monomer is termed as unstable in solution, because the hydrophobic HPMs would be heavily solvent exposed. Compared to the native dimeric structure of full-length KA, one monomer is rotated approximately 80° around an axis roughly perpendicular to the dimer interface in the artificial, inactive HPM complexes of truncated KA (S6 Fig).
Although artificial, the HPM-interface underlines the importance of hydrophobic interfaces as suitable drug targets. The used HPMs were reported to bind to assemblin dimer interfaces of all herpesvirus classes. The IC50-values for these HPMs against the representative alphaherpesvirus assemblin (HSV-2 assemblin) were significantly higher than those against HCMV, EBV and KSHV assemblins, indicating much weaker binding to HSV-2 assemblin [44]. The C-terminal helices of alphaherpesvirus assemblins are likely ordered due to the hydrophobic key position being occupied by alternative conserved hydrophobic residues of the OHL. Thus, the HPMs have to compete against the C-terminal helices for binding which explains the observed higher IC50-values. Additionally, the most important residues for dimerization in KA, the so-called “hot spot” residues [65], are Trp109 (α4) [43] and maybe Phe76 (α3). These correspond to Tyr (α4) and Leu (α3), respectively, in PrV, VZV, HSV-1 and HSV-2 assemblins (S3 Fig). Neither Leu nor Tyr are considered as “hot spot” residues [65] and thus, binding of these HPMs is likely weakened. Screening for suitable mimetics is necessary to achieve specific and efficient inactivation of alphaherpesvirus proteases.
As mentioned above the HPMs force an artificial assembly of truncated KA in contrast to our native monomeric PrV assemblin. Therefore, a detailed comparison of monomeric PrV assemblin and HPM complexes of truncated KA is discussed in the supplement (S1 Text and S7–S10 Figs).
Inspection of a putative cation binding-site
A putative cation was found in both dimeric structures from PrV. A distorted octahedral arrangement of the coordinated water molecules with mean distances of 2.1 Å strongly suggests the presence of a cation rather than water. Since the crystallization buffers contain MgCl2, it is highly probable that these cations are Mg2+ ions. For verification, divalent cations with higher electron density were tested in the crystallization procedure. Mn2+ was able to substitute for Mg2+ but the resulting crystals diffracted considerably less well. The best dataset that we managed to collect at a wavelength near the Mn absorption edge had a resolution of 3 Å, but no significant anomalous signal was detected and electron density at the putative Mg2+/Mn2+ position was too weak to unambiguously confirm the presence of a Mn2+ ion. Presumably, the presence of putative Mg2+ ions in the structures is a direct result of the crystallization conditions (containing 400 mM MgCl2) and does not reflect functional Mg2+ binding by the native protein since the metal ions compensate negative charges from two adjacent dimers, rather than within one dimer (S11 Fig). Thus, Mg2+ ions or divalent cations with related properties are presumably necessary for the observed crystal packing of dimeric pUL26N from PrV.
Presumable temperature-sensitive pUL26 phenotypes
For HSV-1 protease a temperature-sensitive (ts) phenotype was described by mutating Y30F and A48V [66]. Mutating the corresponding residues in PrV protease (Y13F/A30V), however, did not result in the desired phenotype. Hence, these mutations may not sufficiently destabilize PrV pUL26N. In HSV-1 protease, it is highly probable that the result of these mutations is a destabilization of the region around helix α1. This helix and surrounding loops stabilize the OHL in the active conformation. In the PrV assemblin Arg24 is located at the N-terminus of α2. This residue stabilizes the α1 region, because its side chain forms a hydrogen bond to the peptide oxygen of Asp59 (Fig 7). This hydrogen bond is missing in HSV-1 assemblin, since the corresponding residue is Pro42. Consequently, we propose a Y13F/R24P/A30V mutant for achieving the desired ts phenotype in PrV assemblin. If the additional R24P exchange completely inactivates the protease, an R24K variant could also be taken into account. The slightly shorter side chain of Lys may cause a varied hydrogen-bond network and a weaker hydrogen bond to the peptide oxygen of Asp59.
Fig. 7. Superposition of inhibited dimeric pUL26N from PrV and HSV-2. 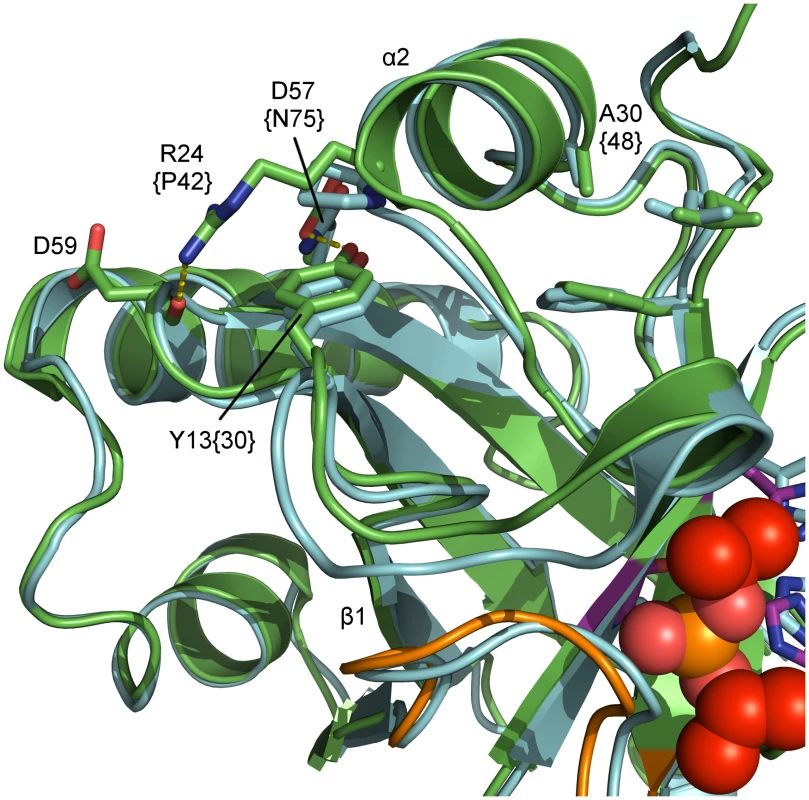
Detailed view of relevant residues for the proposed temperature-sensitive (ts) variant of pUL26N from PrV. Shown are a superposition of inhibited dimeric PrV pUL26N (colored green) and HSV-2 (pdb entry 1at3, colored cyan). Residues for the temperature-sensitive phenotype of HSV-2 assemblin (identifier in braces), the proposed residues for ts mutant of PrV assemblin and interaction partners are labeled. The oxyanion-hole loop (OHL) is shown in orange, diisopropyl phosphate in red and the catalytic triad in purple. The polypeptide around residues 15–21, including helix α1, is likely disordered at the non-permissive temperature. This results in destabilization of the OHL and reduced activity. Conclusions
During capsid assembly, pUL26 accumulates in the nascent capsids because of its C-terminal scaffold-protein part. Thus, the local high concentration of the protease is promoting dimerization and autoproteolysis occurs to release the scaffold from capsids for DNA packaging. In comparison with its dimeric structure, the monomer of pUL26N reveals changes at the dimerization area, in line with allosteric changes of a loop forming the oxyanion hole. As previously anticipated, the core of the protease including the positions and orientation of the active-site residues remains unchanged, but the oxyanion hole is disrupted in the monomeric form [41,42]. The monomeric structure presented here is not truncated and does not contain any inhibitors. Thus, it constitutes the first reliable model for native monomeric structures of other assemblins.
Dimerization induces the following allosteric events: helix α7 of a monomer interleaves between helices α3 and α7 of a second monomer moving these helices farther apart. At the same time, helix α7 is elongated by one turn at its C-terminus to the cost of one N-terminal turn of helix α8. This allosteric process forces the OHL to shift to the vicinity of the active-site serine and builds a far-reaching network of hydrogen bonds with side chains of helix α8 and the polypeptide of strand β5. Furthermore, residues 13–20 of the β1-α2 loop become ordered and form helix α1 in the dimer by getting involved in that network of hydrogen bonds. In this position, the OHL forms the oxyanion hole and activity of the protease is established. The extent of disorder at the dimerization area will vary in assemblins of different herpesviruses, but the general disorder-to-order mechanism of dimerization [28,43] will very likely hold for all assemblins.
The molecular structure of dimeric pUL26N will help to engineer a temperature-sensitive phenotype of the PrV protease. A temperature-sensitive variant will be a powerful tool to observe subsequent steps of viral replication in a synchronous wave [19]. This will provide valuable data on kinetics for cleavage, packaging of the DNA, nuclear egress and intracellular trafficking of the virions.
The structure of monomeric PrV assemblin is the paradigm for monomeric states of other assemblins, primarily from alphaherpesviruses. Detailed knowledge of this structure, conformational changes and sequence specific contacts upon dimerization are a rational basis for the development of drugs binding to the dimerization area and, thus, trapping the inactive monomeric state [45]. Inhibition of dimerization suppresses protease activity and therefore prevents the assembly of fully functional virus capsids.
Small-angle X-ray scattering (SAXS) of PrV assemblin in solution revealed that dimerization increases with protein concentration and in the presence of MgCl2. Dissociation constants in the micromolar range are comparable to those observed for other assemblins [26,27]. Divalent cations like Mg2+ or Mn2+ are required for crystallization of the dimeric PrV assemblin, because these cations support crystal packing by compensating negative charges of neighboring dimers. Since MgCl2 shifts the condition of equilibrium towards dimeric pUL26N, the concentration of the monomeric form is likely below the critical nucleation concentration resulting in crystals of the dimeric form only. Accordingly, crystallization of the monomeric assemblin requires absence of divalent cations. The monomeric fraction of ~0.3 is sufficient for nucleation and the monomer-dimer equilibrium provides a steady supply of monomeric pUL26N. The monomeric form is increasingly favored since crystallization of monomeric pUL26N decreases the concentration of pUL26N in solution. Similar cases with a minor monomeric fraction crystallizing from a monomer-dimer equilibrium were reported earlier [67].
Materials and Methods
Expression and purification
Full-length pUL26 cleaves itself at two positions, and therefore expression of the full-length protein leads to an inhomogeneous product that is unlikely to crystallize. Accordingly, cleavage was prevented by cloning a stop codon behind Gln224. The resulting coding sequence contains the protease fraction of pUL26 (pUL26N) only.
N-terminally (His)6-tagged pUL26N was expressed using a pET28a+ vector in E.coli strain BL-21 (DE3). Cells were grown in LB medium to an OD600 of 0.5–0.8 at 37°C and then induced by addition of isopropyl β-D-1-thiogalactopyranoside to a final concentration of 1 mM. Cells were lysed by sonication. The protein was purified by performing immobilized metal-ion affinity chromatography using a Poros MC 20 column loaded with Ni2+ ions (0.5 M NaCl, 50 mM Tris/HCl pH 7.5, 5% glycerol, eluted with a gradient of 0–250 mM imidazole). The protein was checked for its purity by SDS-PAGE and then concentrated to ~20 mg/ml. Aliquots were stored at −80°C. The purified protein was not tested for enzymatic activity.
Crystallization and cryo-protection
Crystals were obtained using the hanging-drop vapor diffusion method at 22°C. First crystals grew in drops containing 1 μl pUL26N concentrate and 1 μl precipitant solution (0.1 M Hepes pH 7.5, 20% PEG 8,000) within several days. The quality and size of the crystals could be increased by optimization of the composition of the precipitant solution and the ratio of protein solution to precipitant solution. The morphology of the crystals changed from plate-shaped to needle-shaped when MgCl2 was used as an additive in the crystallization procedure. Plate-shaped crystals turned out to be monomeric pUL26N, whereas the dimer formed needle-shaped crystals. For crystallization with inhibitor, the concentrated protein solution was incubated with a final concentration of 5 mM diisopropyl fluorophosphate for 1 hour prior to crystallization.
Best crystals of native dimeric, inhibited dimeric and monomeric pUL26N grew in drops containing 2 μl pUL26N concentrate and 1 μl precipitant solution. Precipitant solution for monomeric pUL26N consisted of 0.1 M Tris/HCl pH 8, 8% PEG 8,000 and crystals grew within one week. Precipitant solutions for native dimeric and inhibited pUL26N consisted of 0.1 M Tris/HCl pH 8, 14% PEG 8,000, 0.4 M MgCl2 and 0.1 M Tris/HCl pH 8, 20% PEG 8,000, 0.2 M MgCl2, respectively.
All crystals were cryo-protected by soaking for 15 s in drops of precipitant solution with increasing amounts of PEG 400. Final concentrations of PEG 400 were 25% (monomer), 18% (native dimer) and 17% (inhibited dimer). Cryo-protected crystals were flash-frozen in liquid nitrogen.
X-ray diffraction, phasing and model refinement
Datasets were measured at 100 K with a PILATUS-6M detector at beamline BL14.1, operated by the Helmholtz-Zentrum Berlin (HZB) at the BESSY II electron storage ring (Berlin-Adlershof, Germany) [68]. 1,800 images were collected at X-ray wavelength 0.91841 Å with an exposure time of 0.5 s and an oscillation range of 0.1°.
All datasets were processed using XDS and Aimless [69–72]. Further details on data collection and processing are listed in Table 1. For determination of the Matthews coefficient and solvent content of the unit cell the CCP4 Program suite (version 6.4.0) was used [73–76].
The structures of monomeric and native dimeric pUL26N were solved via molecular replacement using Phaser [77]. The starting model for native dimeric pUL26N was the A-chain of pdb entry 1at3 (the homologous protein from HSV-2) edited with Chainsaw [78,79]. The A-chain of native dimeric pUL26N (pdb entry 4v07) was used as the starting model for monomeric pUL26N. Since both dimeric forms were isomorphous, the inhibited structure could be solved by refining the native structure against the dataset of the inhibited form. Cycles of model building and refinement were carried out using Coot (version 0.7.1) and Refmac5 (version 5.8.0073), respectively [80–88]. Further details on refinement are listed in Table 1. The oligomerization states were confirmed by PDBePISA [53]. All figures representing structural models were prepared using PyMOL version 1.7.1.3 [89].
The diffraction data and refined models of monomeric, native dimeric and inhibited dimeric pUL26N were deposited with the Protein Data Bank under entry codes 4v0t, 4v07, and 4v08, respectively. A preliminary dataset of the monomeric pUL26N at 2.5 Å resolution was deposited with entry code 4cx8.
Small-angle X-ray scattering
SAXS data were recorded at beamline P12 of the EMBL outstation at PETRA III, DESY, Hamburg [90], using a PILATUS 2M pixel detector, a sample-to-detector distance of 3.1 m and a wavelength of 1.24 Å. Solutions contained 0.5 M NaCl, 50 mM Tris pH 7.5, 0.25 M imidazole, 5% glycerol, 50 mM urea and pUL26N as indicated. In all experiments the sample temperature was 283 K. Measurements covered the momentum transfer range 0.008 < s < 0.47 Å-1 (s = 4π sin(θ) / λ, where 2θ is the scattering angle and λ is the X-ray wavelength). To monitor radiation damage, 20 successive 50 ms exposures of protein solutions were compared, revealing no significant change. The data were normalized to the intensity of the transmitted beam and radially averaged. Scattering of the buffer was subtracted and the difference curves were scaled to unity protein concentration (1 mg/ml). For further data analysis, version 2.6.0 of the ATSAS package was used [91]. Form factors were generated from the monomeric and dimeric crystallographic models by means of the program FFMAKER. For subsequent curve fitting, the program OLIGOMER [63] was used. An ab initio model corresponding to the highest protein concentration in the presence of MgCl2 was generated using the programs DAMMIF [92], DAMAVER [93] and DAMMIN [64] via the PRIMUS interface [63], in "slow" mode and without imposing particle symmetry.
The scattering data, structural models and curve fittings of dimeric PrV pUL26N were deposited with the small-angle scattering biological data bank (SASBDB) with entry code SASDA58 [94].
Accession numbers/ID numbers of genes and proteins
PrV UL26: Gene ID 2952508
HSV-1 UL26: Gene ID 2703453
PrV UL26.5: Gene ID 2952525
HSV-1 UL26.5: Gene ID 2703454
PrV pUL26: Q83417 in UniProtKB, S21.001 in MEROPS [95]
PrV pUL26N: Amino-acid residues 1–224 of Q83417 in UniProtKB, S21.001 in MEROPS [95]
PrV pUL26C: Amino-acid residues 226–524 of Q83417 in UniProtKB
PrV pUL26.5: Q83418 in UniProtKB
KA: Q2HRB6 in UniProtKB, S21.006 in MEROPS [95]
Supporting Information
Zdroje
1. Card JP, Rinaman L, Schwaber JS, Miselis RR, Whealy ME, Robbins AK, et al. (1990) Neurotropic properties of pseudorabies virus: uptake and transneuronal passage in the rat central nervous system. J Neurosci 10 : 1974–1994. 2162388
2. Ben-Porat T, Veach RA, Ihara S (1983) Localization of the regions of homology between the genomes of herpes simplex virus, type 1, and pseudorabies virus. Virology 127 : 194–204. 6305015
3. Klupp BG, Hengartner CJ, Mettenleiter TC, Enquist LW (2004) Complete, annotated sequence of the pseudorabies virus genome. J Virol 78 : 424–440. 14671123
4. Preston VG, Coates JA, Rixon FJ (1983) Identification and characterization of a herpes simplex virus gene product required for encapsidation of virus DNA. J Virol 45 : 1056–1064. 6300447
5. Liu FY, Roizman B (1991) The herpes simplex virus 1 gene encoding a protease also contains within its coding domain the gene encoding the more abundant substrate. J Virol 65 : 5149–5156. 1654435
6. Liu FY, Roizman B (1991) The promoter, transcriptional unit, and coding sequence of herpes simplex virus 1 family 35 proteins are contained within and in frame with the UL26 open reading frame. J Virol 65 : 206–212. 1845885
7. Welch AR, McNally LM, Gibson W (1991) Cytomegalovirus assembly protein nested gene family: four 3'-coterminal transcripts encode four in-frame, overlapping proteins. J Virol 65 : 4091–4100. 1649317
8. Welch AR, Woods AS, McNally LM, Cotter RJ, Gibson W (1991) A herpesvirus maturational proteinase, assemblin: identification of its gene, putative active site domain, and cleavage site. Proc Natl Acad Sci USA 88 : 10792–10796. 1961747
9. Stevens JT, Mapelli C, Tsao J, Hail M, O'Boyle II D, et al. (1994) In vitro proteolytic activity and active-site identification of the human cytomegalovirus protease. Eur J Biochem 226 : 361–367. 8001553
10. DiIanni CL, Drier DA, Deckman IC, McCann PJ, Liu FY, et al. (1993) Identification of the herpes simplex virus-1 protease cleavage sites by direct sequence analysis of autoproteolytic cleavage products. J Biol Chem 268 : 2048–2051. 8380586
11. Beaudet-Miller M, Zhang R, Durkin J, Gibson W, Kwong AD, et al. (1996) Virus-specific interaction between the human cytomegalovirus major capsid protein and the C terminus of the assembly protein precursor. J Virol 70 : 8081–8088. 8892933
12. Desai P, Person S (1996) Molecular Interactions between the HSV-1 Capsid Proteins as Measured by the Yeast Two-Hybrid System. Virology 220 : 516–521. 8661404
13. Hong Z, Beaudet-Miller M, Durkin J, Zhang R, Kwong AD (1996) Identification of a minimal hydrophobic domain in the herpes simplex virus type 1 scaffolding protein which is required for interaction with the major capsid protein. J Virol 70 : 533–540. 8523566
14. Nicholson P, Addison C, Cross A, Kennard J, Preston VG, et al. (1994) Localization of the herpes simplex virus type 1 major capsid protein VP5 to the cell nucleus requires the abundant scaffolding protein VP22a. J Gen Virol 75 : 1091–1099. 8176370
15. Pelletier A, Dô F, Brisebois JJ, Lagacé L, Cordingley MG (1997) Self-association of herpes simplex virus type 1 ICP35 is via coiled-coil interactions and promotes stable interaction with the major capsid protein. J Virol 71 : 5197–5208. 9188587
16. Thomsen DR, Newcomb WW, Brown JC, Homa FL (1995) Assembly of the herpes simplex virus capsid: requirement for the carboxyl-terminal twenty-five amino acids of the proteins encoded by the UL26 and UL26. 5 genes. J Virol 69 : 3690–3703. 7745718
17. Wood LJ, Baxter MK, Plafker SM, Gibson W (1997) Human cytomegalovirus capsid assembly protein precursor (pUL80.5) interacts with itself and with the major capsid protein (pUL86) through two different domains. J Virol 71 : 179–190. 8985337
18. Sheaffer AK, Newcomb WW, Brown JC, Gao M, Weller SK, Tenney DJ (2000) Evidence for controlled incorporation of herpes simplex virus type 1 UL26 protease into capsids. J Virol 74 : 6838–6848. 10888623
19. Church GA, Wilson DW (1997) Study of herpes simplex virus maturation during a synchronous wave of assembly. J Virol 71 : 3603–3612. 9094633
20. Dezélée S, Bras F, Vende P, Simonet B, Nguyen X, et al. (1996) The BamHI fragment 9 of pseudorabies virus contains genes homologous to the UL24, UL25, UL26, and UL 26.5 genes of herpes simplex virus type 1. Virus Res 42 : 27–39. 8806172
21. Gibson W, Welch AR, Hall MRT (1994) Assemblin, a herpes virus serine maturational proteinase and new molecular target for antivirals. Perspect Drug Discov 2 : 413–426.
22. Thomsen DR, Roof LL, Homa FL (1994) Assembly of herpes simplex virus (HSV) intermediate capsids in insect cells infected with recombinant baculoviruses expressing HSV capsid proteins. J Virol 68 : 2442–2457. 8139029
23. Pray TR, Nomura AM, Pennington MW, Craik CS (1999) Auto-inactivation by cleavage within the dimer interface of Kaposi’s sarcoma-associated herpesvirus protease. J Mol Biol 289 : 197–203. 10366498
24. Loveland AN, Chan C-K, Brignole EJ, Gibson W (2005) Cleavage of human cytomegalovirus protease pUL80a at internal and cryptic sites is not essential but enhances infectivity. J Virol 79 : 12961–12968. 16188998
25. Holwerda BC, Wittwer AJ, Duffin KL, Smith C, Toth MV, et al. (1994) Activity of two-chain recombinant human cytomegalovirus protease. J Biol Chem 269 : 25911–25915. 7929296
26. Waxman L, Darke PL (2000) The herpesvirus proteases as targets for antiviral chemotherapy. Antivir Chem Chemother 11 : 1–22. 10693650
27. Darke PL, Cole JL, Waxman L, Hall DL, Sardana MK, et al. (1996) Active human cytomegalovirus protease is a dimer. J Biol Chem 271 : 7445–7449. 8631772
28. Nomura AM, Marnett AB, Shimba N, Dotsch V, Craik CS (2005) Induced structure of a helical switch as a mechanism to regulate enzymatic activity. Nat Struct Mol Biol 12 : 1019–1020. 16244665
29. Plafker SM, Gibson W (1998) Cytomegalovirus assembly protein precursor and proteinase precursor contain two nuclear localization signals that mediate their own nuclear translocation and that of the major capsid protein. J Virol 72 : 7722–7732. 9733808
30. Robertson BJ, McCann PJ, Matusick-Kumar L, Newcomb WW, Brown JC, et al. (1996) Separate functional domains of the herpes simplex virus type 1 protease: evidence for cleavage inside capsids. J Virol 70 : 4317–4328. 8676454
31. Tong L, Qian C, Massariol M-J, Bonneau PR, Cordingley MG, et.al (1996) A new serine-protease fold revealed by the crystal structure of human cytomegalovirus protease. Nature 383 : 272–275. 8805706
32. Qiu X, Culp JS, DiLella AG, Hellmig B, Hoog SS, et al. (1996) Unique fold and active site in cytomegalovirus protease. Nature 383 : 275–279. 8805707
33. Shieh H-S, Kurumbail RG, Stevens AM, Stegeman RA, Sturman EJ, et al. (1996) Three-dimensional structure of human cytomegalovirus protease. Nature 383 : 279–282. 8805708
34. Hoog SS, Smith WW, Qiu X, Janson CA, Hellmig B, et al. (1997) Active site cavity of herpesvirus proteases revealed by the crystal structure of herpes simplex virus protease/inhibitor complex. Biochemistry 36 : 14023–14029. 9369473
35. Qiu X, Janson CA, Culp JS, Richardson SB, Debouck C, et al. (1997) Crystal structure of varicella-zoster virus protease. Proc Natl Acad Sci U S A 94 : 2874–2879. 9096314
36. Reiling KK, Pray TR, Craik CS, Stroud RM (2000) Functional consequences of the Kaposi's sarcoma-associated herpesvirus protease structure: regulation of activity and dimerization by conserved structural elements. Biochemistry 39 : 12796–12803. 11041844
37. Buisson M, Hernandez J-F, Lascoux D, Schoehn G, Forest E, et al. (2002) The crystal structure of the Epstein-Barr virus protease shows rearrangement of the processed C terminus. J Mol Biol 324 : 89–103. 12421561
38. Lazic A, Goetz DH, Nomura AM, Marnett AB, Craik CS (2007) Substrate modulation of enzyme activity in the herpesvirus protease family. J Mol Biol 373 : 913–923. 17870089
39. Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, et al. (2011) Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7 : 539. doi: 10.1038/msb.2011.75 21988835
40. Goujon M, McWilliam H, Li W, Valentin F, Squizzato S, et al. (2010) A new bioinformatics analysis tools framework at EMBL-EBI. Nucleic Acids Res 38: W695–W699. doi: 10.1093/nar/gkq313 20439314
41. Batra R, Khayat R, Tong L (2001) Molecular mechanism for dimerization to regulate the catalytic activity of human cytomegalovirus protease. Nat Struct Mol Biol 8 : 810–817.
42. Liang P, Brun KA, Feild JA, O'Donnell K, Doyle ML, et al. (1998) Site-Directed Mutagenesis Probing the Catalytic Role of Arginines 165 and 166 of Human Cytomegalovirus Protease. Biochemistry 37 : 5923–5929. 9558326
43. Lee GM, Shahian T, Baharuddin A, Gable JE, Craik CS (2011) Enzyme inhibition by allosteric capture of an inactive conformation. J Mol Biol 411 : 999–1016. doi: 10.1016/j.jmb.2011.06.032 21723875
44. Gable JE, Lee GM, Jaishankar P, Hearn BR, Waddling CA, et al. (2014) Broad-spectrum allosteric inhibition of herpesvirus proteases. Biochemistry 53 : 4648–4660. doi: 10.1021/bi5003234 24977643
45. Shahian T, Lee GM, Lazic A, Arnold LA, Velusamy P, et al. (2009) Inhibition of a viral enzyme by a small-molecule dimer disruptor. Nat Chem Biol 5 : 640–646. doi: 10.1038/nchembio.192 19633659
46. Conway JF, Homa FL Nucleocapsid structure, assembly and DNA packaging of Herpes simplex virus. In: Weller SK, editors. Alphaherpesviruses: Molecular Virology. Norfolk, UK: Caister Academic Press; 2011. pp. 175–193.
47. Gibson W. Chapter 783—Cytomegalovirus assemblin and precursor. In: Rawlings ND, Salvesen GS, editors. Handbook of proteolytic enzymes. London: Academic Press, Elsevier; 2013. pp. 3540–3545.
48. Shahian T. Craik CS. Chapter 784—Kaposi's Sarcoma Virus Assemblin (Herpesvirus 8-type Assemblin). In: Rawlings ND, Salvesen GS, editors. Handbook of proteolytic enzymes. London: Academic Press, Elsevier; 2013. pp. 3545–3550.
49. Jupp R, Mills J, Tigue NJ, Kay J. Chapter 785—Human Herpesvirus Type 6 Assemblin. In: Rawlings ND, Salvesen GS, editors. Handbook of proteolytic enzymes. London: Academic Press, Elsevier; 2013. pp. 3550–3552.
50. Jupp R, Ritchie A, Robinson M, Broadhurst A, Mills J. Chapter 786—Epstein-Barr Virus Assemblin. In: Rawlings ND, Salvesen GS, editors. Handbook of proteolytic enzymes. London: Academic Press, Elsevier; 2013. pp. 3552–3554.
51. McMillan D, Jupp R, Mills J, Kay J. Chapter 787—Varicella-Zoster Virus Assemblin. In: Rawlings ND, Salvesen GS, editors. Handbook of proteolytic enzymes. London: Academic Press, Elsevier; 2013. pp. 3555–3556.
52. Steven AC, Spear PG. Herpesvirus capsid assembly and envelopment. In: Chiu W, Burnett RM, Garcea RL, editors. Structural Biology of Viruses. New York, NY: Oxford University Press; 1997. pp. 312–351.
53. Krissinel E, Henrick K (2007) Inference of macromolecular assemblies from crystalline state. J Mol Biol 372 : 774–797. 17681537
54. Krissinel E, Henrick K (2004) Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr D Biol Crystallogr 60 : 2256–2268. 15572779
55. Chen VB, Arendall WB, Headd JJ, Keedy DA, Immormino RM, et al. (2009) MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr 66 (Pt 1): 12–21. doi: 10.1107/S0907444909042073 20057044
56. Khayat R, Batra R, Massariol M, Lagacé L, Tong L (2001) Investigating the role of histidine 157 in the catalytic activity of human cytomegalovirus protease. Biochemistry 40 : 6344–6351. 11371196
57. Jones S, Thornton JM (1996) Principles of protein-protein interactions. Proc Natl Acad Sci U S A 93 : 13–20. 8552589
58. Goodsell DS, Olson AJ (2000) Structural symmetry and protein function. Review. Annu Rev Biophys Biomol Struct 29 : 105–153. 10940245
59. Diisopropyl fluorophosphate in the product catalogue from Sigma-Aldrich: http://www.sigmaaldrich.com/catalog/product/sigma/d0879
60. DiIanni CL, Stevens JT, Bolgar M, O'Boyle DR, Weinheimer SP, et al. (1994) Identification of the serine residue at the active site of the herpes simplex virus type 1 protease. J Biol Chem 269 : 12672–12676. 8175677
61. Baum EZ, Bebernitz GA, Hulmes JD, Muzithras VP, Jones TR, et al. (1993) Expression and analysis of the human cytomegalovirus UL80-encoded protease: identification of autoproteolytic sites. J Virol 67 : 497–506. 8380089
62. Smith MC, Giordano J, Cook JA, Wakulchik M, Villarreal EC, et al. (1994) Purification and kinetic characterization of human cytomegalovirus assemblin. Meth Enzymol 244 : 412–423. 7845223
63. Konarev PV, Volkov VV, Sokolova AV, Koch MHJ, Svergun DI (2003) PRIMUS: a Windows PC-based system for small-angle scattering data analysis. J Appl Cryst 36 : 1277–1282.
64. Svergun DI (1996) Restoring low resolution structure of biological macromolecules from solution scattering using simulated annealing. Biophys J 76 : 2879–2886.
65. Ma B, Nussinov R (2007) Trp/Met/Phe hot spots in protein-protein interactions: potential targets in drug design. Curr Top Med Chem 10 : 999–1005.
66. Register RB, Shafer JA (1996) A facile system for construction of HSV-1 variants: site directed mutation of the UL26 protease gene in HSV-1. J Virol Methods 57 : 181–193. 8801230
67. Palm GJ, Zdanov A, Gaitanaris GA, Stauber R, Pavlakis GN, et al. (1997) The structural basis for spectral variations in green fluorescent protein. Nat Struct Mol Biol 4 : 361–365.
68. Mueller U, Darowski N, Fuchs MR, Förster R, Hellmig M, Paithankar KS, et al. (2012) Facilities for Macromolecular Crystallography at the Helmholtz-Zentrum Berlin. J Synch Rad 19 : 442–449.
69. Kabsch W (2010) XDS. Acta Crystallogr D Biol Crystallogr 66 (Pt 2): 125–132. doi: 10.1107/S0907444909047337 20124692
70. Evans PR (2011) An introduction to data reduction: space-group determination, scaling and intensity statistics. Acta Crystallogr D Biol Crystallogr 67 (Pt 4): 282–292. doi: 10.1107/S090744491003982X 21460446
71. Evans PR, Murshudov GN (2013) How good are my data and what is the resolution?. Acta Crystallogr D Biol Crystallogr 69 (Pt 7): 1204–1214. doi: 10.1107/S0907444913000061 23793146
72. Evans P (2005) Scaling and assessment of data quality. Acta Crystallogr D Biol Crystallogr 62 (Pt 1): 72–82. 16369096
73. Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, et al. (2011) Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr 67 (Pt 4): 235–242. doi: 10.1107/S0907444910045749 21460441
74. Potterton EA, Briggs PJ, Turkenburg MG, Dodson EJ (2003) A graphical user interface to the CCP4 program suite. Acta Crystallogr D Biol Crystallogr 59 (Pt 7): 1131–1137. 12832755
75. Kantardjieff KA, Rupp B (2003) Matthews coefficient probabilities: improved estimates for unit cell contents of proteins, DNA, and protein-nucleic acid complex crystals. Protein Sci 12 : 1865–1871. 12930986
76. Matthews BW (1968) Solvent content of protein crystals. J Mol Biol 33 : 491–497. 5700707
77. McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, et al. (2007) Phaser crystallographic software. J Appl Crystallogr 40 : 658–674. 19461840
78. Stein N (2008) CHAINSAW: a program for mutating pdb files used as templates in molecular replacement. J Appl Crystallogr 41 : 641–643.
79. Schwarzenbacher R, Godzik A, Grzechnik SK, Jaroszewski L (2004) The importance of alignment accuracy for molecular replacement. Acta Crystallogr D Biol Crystallogr 60 (Pt 7): 1229–1236. 15213384
80. Emsley P, Lohkamp B, Scott W, Cowtan KD (2010) Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66 (Pt 4): 486–501. doi: 10.1107/S0907444910007493 20383002
81. Winn MD, Murshudov GN, Papiz MZ (2003) Macromolecular TLS Refinement in REFMAC at Moderate Resolutions. Methods Enzymol 374 : 300–321. 14696379
82. Skubák P, Murshudov GN, Pannu NS (2004) Direct incorporation of experimental phase information in model refinement. Acta Crystallogr D Biol Crystallogr 60 (Pt 12 Pt 1): 2196–2201. 15572772
83. Murshudov GN, Vagin AA, Lebedev A, Wilson KS, Dodson EJ (1999) Efficient anisotropic refinement of macromolecular structures using FFT. Acta Crystallogr D Biol Crystallogr 55 (Pt 1): 247–255. 10089417
84. Steiner RA, Lebedev AA, Murshudov GN (2003) Fisher's information in maximum-likelihood macromolecular crystallographic refinement. Acta Crystallogr D Biol Crystallogr 59 (Pt 12): 2114–2124. 14646069
85. Pannu NS, Murshudov GN, Dodson EJ, Read RJ (1998) Incorporation of prior phase information strengthens maximum-likelihood structure refinement. Acta Crystallogr D Biol Crystallogr 54 (Pt 6 Pt 2): 1285–1294. 10089505
86. Murshudov GN, Vagin AA, Dodson EJ (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr 53 (Pt 3): 240–255. 15299926
87. Vagin AA, Steiner RA, Lebedev AA, Potterton L, McNicholas S, et al. (2004) REFMAC5 dictionary: organization of prior chemical knowledge and guidelines for its use. Acta Crystallogr D Biol Crystallogr 60 (Pt 12 Pt 1): 2184–2195. 15572771
88. Winn MD, Isupov MN, Murshudov GN (2001) Use of TLS parameters to model anisotropic displacements in macromolecular refinement. Acta Crystallogr D Biol Crystallogr 57 (Pt 1): 122–133. 11134934
89. Schrödinger, LLC (2014) The PyMOL Molecular Graphics System, Version 1.7.1.3.
90. Blanchet CE, Spilotros A, Schwemmer F, Graewert MA, Kikhney A (2015) Versatile sample environments and automation for biological solution X-ray scattering experiments at the P12 beamline (PETRA III, DESY). J Appl Cryst 48 : 431–443.
91. Petoukhov MV, Franke D, Shkumatov A, Tria G, Kikhney AG, et al. (2012) New developments in the ATSAS program package for small-angle scattering data analysis. J Appl Cryst 45 : 342–350.
92. Franke D, Svergun DI (2009) DAMMIF, a program for rapid ab-initio shape determination in small-angle scattering. J Appl Cryst 42 : 342–346.
93. Volkov VV, Svergun DI (2003) Uniqueness of ab initio shape determination in small-angle scattering. J Appl Cryst 36 : 860–864.
94. Valentini E, Kikhney AG, Previtaly G, Jeffries CM, Svergun DI (2015) SASBDB, a repository for biological small-angle scattering data. Nucl Acids Res 43: D357–D363. doi: 10.1093/nar/gku1047 25352555
95. Rawlings ND, Waller M, Barrett AJ, Bateman A (2014) MEROPS: the database of proteolytic enzymes, their substrates and inhibitors. Nucl Acids Res 42: D503–D509. doi: 10.1093/nar/gkt953 24157837
96. Bond CS, Schüttelkopf AW (2009) ALINE: a WYSIWYG protein-sequence alignment editor for publication-quality alignments. Acta Crystallogr D Biol Crystallogr 65 : 510–512. doi: 10.1107/S0907444909007835 19390156
97. Kozin MB, Svergun DI (2001) Automated matching of high - and low-resolution structural models. J Appl Cryst 34 : 33–41.
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek Transmission Properties of Human PrP 102L Prions Challenge the Relevance of Mouse Models of GSSČlánek Decline of FoxP3+ Regulatory CD4 T Cells in Peripheral Blood of Children Heavily Exposed to MalariaČlánek IFNγ and IL-12 Restrict Th2 Responses during Helminth/ Co-Infection and Promote IFNγ from Th2 CellsČlánek Exploiting Fungal Virulence-Regulating Transcription Factors As Novel Antifungal Drug Targets
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2015 Číslo 7- Jak souvisí postcovidový syndrom s poškozením mozku?
- Měli bychom postcovidový syndrom léčit antidepresivy?
- Farmakovigilanční studie perorálních antivirotik indikovaných v léčbě COVID-19
- 10 bodů k očkování proti COVID-19: stanovisko České společnosti alergologie a klinické imunologie ČLS JEP
-
Všechny články tohoto čísla
- Basic Prion Science “Spreads” Insight
- Research Driven by Curiosity: The Journey from Basic Molecular Biology and Virology to Studies of Human Pathogenic Coronaviruses
- Cross Kingdom Activators of Five Classes of Bacterial Effectors
- Vaccination Drives Changes in Metabolic and Virulence Profiles of
- Expression of the Blood-Group-Related Gene Alters Susceptibility to Infection
- Transmission Properties of Human PrP 102L Prions Challenge the Relevance of Mouse Models of GSS
- Latent KSHV Infected Endothelial Cells Are Glutamine Addicted and Require Glutaminolysis for Survival
- The DSF Family of Cell–Cell Signals: An Expanding Class of Bacterial Virulence Regulators
- Intraperitoneal Infection of Wild-Type Mice with Synthetically Generated Mammalian Prion
- Vpr Promotes Macrophage-Dependent HIV-1 Infection of CD4 T Lymphocytes
- An In-Depth Comparison of Latency-Reversing Agent Combinations in Various and HIV-1 Latency Models Identified Bryostatin-1+JQ1 and Ingenol-B+JQ1 to Potently Reactivate Viral Gene Expression
- α-Macroglobulin Can Crosslink Multiple Erythrocyte Membrane Protein 1 (PfEMP1) Molecules and May Facilitate Adhesion of Parasitized Erythrocytes
- Should Symbionts Be Nice or Selfish? Antiviral Effects of Wolbachia Are Costly but Reproductive Parasitism Is Not
- A Unique Human Norovirus Lineage with a Distinct HBGA Binding Interface
- The Broad Neutralizing Antibody Responses after HIV-1 Superinfection Are Not Dominated by Antibodies Directed to Epitopes Common in Single Infection
- Rapidly Evolving Genes Are Key Players in Host Specialization and Virulence of the Fungal Wheat Pathogen ()
- MiR-21 in Extracellular Vesicles Leads to Neurotoxicity via TLR7 Signaling in SIV Neurological Disease
- Age-Dependent Cell Trafficking Defects in Draining Lymph Nodes Impair Adaptive Immunity and Control of West Nile Virus Infection
- Decline of FoxP3+ Regulatory CD4 T Cells in Peripheral Blood of Children Heavily Exposed to Malaria
- Dimerization-Induced Allosteric Changes of the Oxyanion-Hole Loop Activate the Pseudorabies Virus Assemblin pUL26N, a Herpesvirus Serine Protease
- Macrophages Subvert Adaptive Immunity to Urinary Tract Infection
- Mycolactone-Dependent Depletion of Endothelial Cell Thrombomodulin Is Strongly Associated with Fibrin Deposition in Buruli Ulcer Lesions
- Activation of TLR2 and TLR6 by Dengue NS1 Protein and Its Implications in the Immunopathogenesis of Dengue Virus Infection
- K-bZIP Mediated SUMO-2/3 Specific Modification on the KSHV Genome Negatively Regulates Lytic Gene Expression and Viral Reactivation
- Phosphoproteomic Analysis of KSHV-Infected Cells Reveals Roles of ORF45-Activated RSK during Lytic Replication
- CR3 and Dectin-1 Collaborate in Macrophage Cytokine Response through Association on Lipid Rafts and Activation of Syk-JNK-AP-1 Pathway
- IFNγ and IL-12 Restrict Th2 Responses during Helminth/ Co-Infection and Promote IFNγ from Th2 Cells
- THY-1 Cell Surface Antigen (CD90) Has an Important Role in the Initial Stage of Human Cytomegalovirus Infection
- Human Enterovirus Nonstructural Protein 2C Functions as Both an RNA Helicase and ATP-Independent RNA Chaperone
- IL-27 Signaling Is Crucial for Survival of Mice Infected with African Trypanosomes via Preventing Lethal Effects of CD4 T Cells and IFN-γ
- Synergistic Reactivation of Latent HIV Expression by Ingenol-3-Angelate, PEP005, Targeted NF-kB Signaling in Combination with JQ1 Induced p-TEFb Activation
- Vpu Exploits the Cross-Talk between BST2 and the ILT7 Receptor to Suppress Anti-HIV-1 Responses by Plasmacytoid Dendritic Cells
- Herpesvirus Genome Recognition Induced Acetylation of Nuclear IFI16 Is Essential for Its Cytoplasmic Translocation, Inflammasome and IFN-β Responses
- A Comprehensive Analysis of Replicating Merkel Cell Polyomavirus Genomes Delineates the Viral Transcription Program and Suggests a Role for mcv-miR-M1 in Episomal Persistence
- Analysis of the SUMO2 Proteome during HSV-1 Infection
- Capacity of Broadly Neutralizing Antibodies to Inhibit HIV-1 Cell-Cell Transmission Is Strain- and Epitope-Dependent
- A Novel Antiviral Target Structure Involved in the RNA Binding, Dimerization, and Nuclear Export Functions of the Influenza A Virus Nucleoprotein
- Deploying FLAREs to Visualize Functional Outcomes of Host—Pathogen Encounters
- Mosquitoes Reset Malaria Parasites
- The Lung Microbiome: New Principles for Respiratory Bacteriology in Health and Disease
- Extracellular Virions: The Advance Guard of Poxvirus Infections
- Risks of Antibiotic Exposures Early in Life on the Developing Microbiome
- RNA Virus Reassortment: An Evolutionary Mechanism for Host Jumps and Immune Evasion
- Exploiting Fungal Virulence-Regulating Transcription Factors As Novel Antifungal Drug Targets
- N-acetylglucosamine Regulates Virulence Properties in Microbial Pathogens
- Periodontal Diseases: Bug Induced, Host Promoted
- Mechanisms of Host Behavioral Change in Rodent Association
- The Endosymbiotic Bacterium Selectively Kills Male Hosts by Targeting the Masculinizing Gene
- HIV Reactivation from Latency after Treatment Interruption Occurs on Average Every 5-8 Days—Implications for HIV Remission
- Ubiquilin 1 Promotes IFN-γ-Induced Xenophagy of
- Transfer of Immunity from Mother to Offspring Is Mediated via Egg-Yolk Protein Vitellogenin
- Suppression of Long-Lived Humoral Immunity Following Infection
- The Role of VP1 Amino Acid Residue 145 of Enterovirus 71 in Viral Fitness and Pathogenesis in a Cynomolgus Monkey Model
- Utilizing Chemical Genomics to Identify Cytochrome as a Novel Drug Target for Chagas Disease
- The Emerging Role for RNA Polymerase II in Regulating Virulence Gene Expression in Malaria Parasites
- Turning Up the Heat: Inflammasome Activation by Fungal Pathogens
- On and Under the Skin: Emerging Basidiomycetous Yeast Infections Caused by Species
- EhVps32 Is a Vacuole-Associated Protein Involved in Pinocytosis and Phagocytosis of
- Characterization of a Prefusion-Specific Antibody That Recognizes a Quaternary, Cleavage-Dependent Epitope on the RSV Fusion Glycoprotein
- The Serine Protease EspC from Enteropathogenic Regulates Pore Formation and Cytotoxicity Mediated by the Type III Secretion System
- Existing Infection Facilitates Establishment and Density of Malaria Parasites in Their Mosquito Vector
- Evaluating Human T-Cell Therapy of Cytomegalovirus Organ Disease in HLA-Transgenic Mice
- Neuronal Interferon Signaling Is Required for Protection against Herpes Simplex Virus Replication and Pathogenesis
- Epstein-Barr Virus Proteins EBNA3A and EBNA3C Together Induce Expression of the Oncogenic MicroRNA Cluster miR-221/miR-222 and Ablate Expression of Its Target p57
- Colonization of the Mouse Gastrointestinal Tract Is Modulated by Wall Teichoic Acid, Capsule, and Surface Proteins
- Virulence of Group A Streptococci Is Enhanced by Human Complement Inhibitors
- Identification of Caspase Cleavage Sites in KSHV Latency-Associated Nuclear Antigen and Their Effects on Caspase-Related Host Defense Responses
- Calprotectin Increases the Activity of the SaeRS Two Component System and Murine Mortality during Infections
- Type VI Secretion System Transports Zn to Combat Multiple Stresses and Host Immunity
- Lv4 Is a Capsid-Specific Antiviral Activity in Human Blood Cells That Restricts Viruses of the SIV/SIV/HIV-2 Lineage Prior to Integration
- Phenylbutyrate Is Bacteriostatic against and Regulates the Macrophage Response to Infection, Synergistically with 25-Hydroxy-Vitamin D₃
- An Internally Translated MAVS Variant Exposes Its Amino-terminal TRAF-Binding Motifs to Deregulate Interferon Induction
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- RNA Virus Reassortment: An Evolutionary Mechanism for Host Jumps and Immune Evasion
- Activation of TLR2 and TLR6 by Dengue NS1 Protein and Its Implications in the Immunopathogenesis of Dengue Virus Infection
- N-acetylglucosamine Regulates Virulence Properties in Microbial Pathogens
- Characterization of a Prefusion-Specific Antibody That Recognizes a Quaternary, Cleavage-Dependent Epitope on the RSV Fusion Glycoprotein
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



