-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaDeploying FLAREs to Visualize Functional Outcomes of Host—Pathogen Encounters
article has not abstract
Published in the journal: . PLoS Pathog 11(7): e32767. doi:10.1371/journal.ppat.1004912
Category: Pearls
doi: https://doi.org/10.1371/journal.ppat.1004912Summary
article has not abstract
Functional Microbial Reporters in Host—Pathogen Studies
One of the most exciting features of fluorescent probes is their rapid evolution from simple tracers to functional indicators in diverse research fields [1]. In mammalian cells, fluorescence has been coupled to synaptic transmission [2,3], neuronal differentiation [4], apoptotic cell death [5], fate mapping in different immune cells [6,7], and functional changes in cell physiology, for example, the induction of cytokines [8]. In microbial cells, fluorescence has long been utilized as a tracer for pathogenic microbes, revealing microbial localization, residence, and spread in host tissues [9]. However, measuring functional outcomes during individual encounters with host immune cells remains challenging.
In this Pearl, we describe fluorescence-based approaches, which we term “functional microbial reporters,” designed to relay changes in microbial physiology that occur in host cell and tissue environments. This class of microbial reporters typically harnesses fluorescence emission at two wavelengths. One of these signals functions as an invariant tracer of microbial cells and is generally unaffected by hostile conditions that are encountered during interactions with host cells, such as reactive oxygen species or acidified compartments. The other signal varies according to a change in microbial physiology or residence in the host. This second signal may act as a ratiometric indicator (i.e., a shift in the excitation or emission spectrum) to reflect a continuous variable such as the microbial growth rate. Alternatively, the second signal may act as an on-off indicator (i.e., extinction in the emission spectrum) to reflect a binary variable such as microbial viability. Three specific types of functional microbial reporters will be discussed, including reporters that distinguish live and killed fungal cells [10], indicate the mode of host cell entry by an intracellular parasite [11], and measure bacterial growth rates in host tissues [12–14].
Examples of Functional Microbial Reporters
The mold Aspergillus fumigatus is a major cause of infectious mortality in patients with acute leukemia and in hematopoietic and lung transplant recipients. Humans inhale many infectious conidia (non-dividing vegetative spores) daily, and a central function of the respiratory immune system is to prevent conidial germination into tissue-invasive hyphae. The molecular and cellular events that underlie conidial uptake and killing by leukocytes in the lung can be characterized using fluorescent Aspergillus reporter (FLARE) conidia [10]. FLARE conidia are coupled to the dye Alexa Fluor 633 (AF633), which acts as an invariant signal independent of viability, and express DsRed as an on-off indicator of fungal viability (Fig 1A). Thus, live FLARE conidia emit AF633 and DsRed fluorescence and are readily visualized within leukocytes. FLARE conidia that have been taken up and killed by leukocytes only emit AF633 fluorescence since DsRed is extinguished in the phagolysosome [15], temporally coincident with conidial killing. In FLARE-infected mice, lung leukocytes can be analyzed on the basis of conidial uptake and viability using flow cytometry and imaging cytometry (Fig 1A), in addition to microscopy-based techniques.
Fig. 1. Design, applications, and potential improvements for a functional microbial reporter of viability. 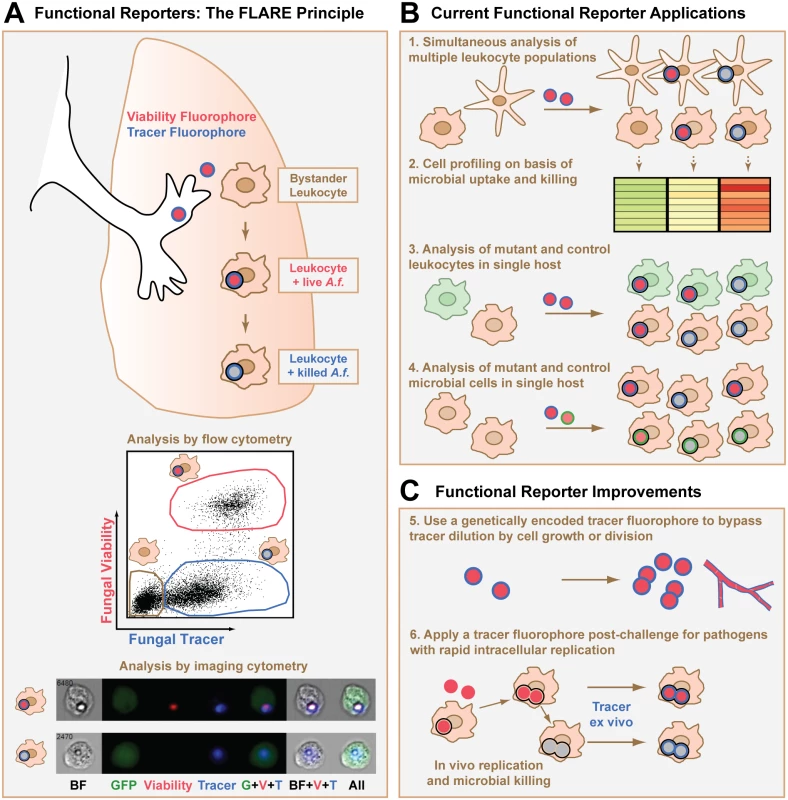
(A) Fluorescent Aspergillus reporter (FLARE) conidia represent an example of a functional microbial reporter. Live FLARE conidia emit two fluorescent signals: (i) a viability fluorophore DsRed (shown in red) that is extinguished at the time of conidial death, and (ii) a tracer fluorophore Alexa Fluor 633 (shown in blue) that persists after conidial death. A black ring indicates uptake of the conidium into the phagolysosome of a leukocyte. After experimental infection, flow cytometry can be used to distinguish bystander leukocytes (tan gate in flow cytometry plot) and fungus-engaged leukocytes that contain either live (red gate) or killed conidia (blue gate). Imaging cytometry can also be used to distinguish these different leukocyte groups. The imaging cytometry example shows green fluorescent protein (GFP)+ inflammatory monocytes that contain a live (top row) or killed (bottom row) FLARE conidium (image adapted from [20]). A.f., Aspergillus fumigatus conidium; BF, brightfield image. (B) Applications of a functional microbial reporter include (1) simultaneous analysis and (2) cell profiling of multiple leukocyte subsets on the basis of a microbial functional reporter readout, (3) simultaneous and focused analysis of microbial encounters with mutant and control leukocytes in a host with mixed chimerism, and (4) parallel analysis of the phenotype of mutant and control functional microbial reporters during cellular interactions with the immune system by using distinct tracer fluorophores for each microbial reporter. (C) Strategies to improve current functional microbial reporters include (5) creating functional microbial reporters in which the tracer and variant indicators are both genetically encoded so that the functional reporter is transmitted to progeny or (6) using microbial reporters that consist only of a variant indicator to infect the host and then staining with dyes or antibodies against the microbes for ex vivo quantification after the experimental conditions are complete. Toxoplasma gondii is an obligate intracellular parasite that infects one-third of humans worldwide and causes disease primarily in immune compromised individuals. Parasites that actively invade host cells multiply within specialized parasitophorous vacuoles (PVs). However, parasites that have been engulfed by host phagocytes are contained within acidified phagolysosomes. Two functional microbial reporters have been used to distinguish active invasion from phagocytic uptake. In the first approach, non-replicating parasites express pH-insensitive mCherry and are loaded with Cell Trace Violet, a pH-sensitive dye that dims under low pH conditions [11]. Following murine challenge, host cells that contain parasites within PVs emit both fluorophores, while host cells that phagocytose parasites retain mCherry and lose Cell Trace Violet fluorescence. In the second approach, transgenic red fluorescent protein (RFP)+ parasites inject bacteriophage Cre recombinase into the host cell cytosol upon active invasion, but not upon phagocytic uptake [16]. Cre then induces GFP expression in cells from Cre-regulated GFP reporter mice, thus marking actively invaded host cells. These functional microbial reporters have been used to study differences in the induction of parasite-specific cytokines [17,18] and effector T cell responses [11] based on the mode of parasite entry.
Salmonella bacteria are a major cause of diarrheal illness and enteric fever. The emergence of antibiotic resistance by Salmonella species represents a public health threat. Two recently developed functional microbial reporters harness fluorescent protein dilution during cell division [14] or differences in the maturation rate of a DsRed variant called TIMER [12,19] to measure bacterial growth rates in antibiotic-treated mice. The latter reporter, termed TIMERbac-Salmonella, undergoes shifts in fluorescence over time from green to green/orange due to different kinetics of maturation and fluorescence resonance energy transfer between green and orange fluorophores [12]. These studies reveal that the majority of antibiotic survivors emerge from moderately growing, partially tolerant Salmonella [12], though non-replicating Salmonella contribute to the survivor pool as well [13,14].
Current Applications of Functional Microbial Reporters
Functional microbial reporters enable a number of experimental readouts at the level of individual microbial cell—host cell encounters. First, different host leukocyte subsets can be analyzed in parallel during encounters with a functional microbial reporter (Fig 1B, point 1). This application identified lung-infiltrating inflammatory monocytes as effector cells that kill FLARE conidia directly during respiratory A. fumigatus challenge [20].
Second, host leukocytes can be profiled at the transcriptional or proteomic level based on a specific readout, such as microbial uptake and killing (Fig 1B, point 2), parasite mode of entry, or intracellular bacterial replication rate. Using this application, T. gondii entry into mononuclear phagocytes was found to be dispensable for protective interleukin-12 p40 release [18], while parasite phagocytosis, but not active invasion, induced protective type I interferon responses in mammalian hosts [17].
Third, functional microbial reporters provide a means to compare gene-deficient and control leukocytes side-by-side with regard to a defined microbial readout (Fig 1B, point 3). This application is particularly informative in mixed bone marrow chimeric hosts that contain gene-deficient and control hematopoietic cells that are distinguished by congenic markers. In this experimental setup, the impact of a genetic alteration in host cells on microbial functional outcomes is revealed in a leukocyte-intrinsic manner, independent of secondary effects on microbial burden and tissue inflammation that often arise in the analysis of globally gene-deficient mice.
Fourth, to compare wild-type and mutant microbial strains in a single host using a mixed infection model, it is possible to engineer functional microbial reporters with fluorescence signatures that differentiate between the two strains while using the same functional readout (Fig 1B, point 4). For example, mutant and wild-type fungal cells can be distinguished by incorporating a different tracer fluorophore, such as Brilliant Violet 421 [21], instead of AF633 in FLARE conidia while using the same viability fluorophore DsRed.
Limitations and Possible Improvements of Functional Microbial Reporters
A key limitation of functional microbial reporters is the unwanted dilution of fluorophores that are not endogenously expressed. For instance, FLARE conidia have been an ideal model system for studying early innate antifungal immunity pathways that operate prior to the formation of fungal hyphae. However, the FLARE principle does not lend itself to the study of hyphae or other rapidly dividing microbes, since the exogenously applied tracer fluorophore is diluted as microbial cells germinate or divide. This limitation is apparent with the Toxoplasma mCherry-Cell Trace Violet reporter as well, since the pH-sensitive Cell Trace Violet dye is loaded into transgenic parasites that are replication-incompetent to avoid dilution of the pH indicator dye [11].
A potential solution to this limitation is to rely exclusively on fluorophores that are encoded in DNA, so that the fluorescent signals are self-replenishing as microbes divide or germinate (Fig 1C, point 5). This strategy was central to the generation of the Salmonella reporters of bacterial growth rate already discussed [12,14]. In the case of FLARE conidia, the use of a genetically encoded tracer fluorophore that does not quench after fungal death may improve the utility of these conidia and extend the FLARE principle to a broader range of pathogens, beyond spore-forming molds. For the Toxoplasma functional reporter, the use of genetically encoded pH-sensitive fluorescent proteins [2,22] anchored to the exterior parasite surface could conceivably replace the Cell Trace Violet dye and extend the functional reporter to replication-competent Toxoplasma strains. The feasibility of these genetic manipulations depends on the ease with which the target microbe can be mutated. Since genetically encoded fluorophores can be sensitive to environmental conditions, for example, ambient oxygen concentrations in host tissues [12], it is imperative to define the readouts of these functional microbial reporters under in vitro and in vivo conditions.
An alternative to genetically encoded tracer fluorophores is to use dyes or monoclonal antibodies to stain for all microbes, live or dead, in collected host tissues after the experimental conditions are complete (Fig 1C, point 6). For example, calcofluor white, a fluorescent dye for chitin found in fungal cell walls, could be used to reveal and quantify all fungal cells in permeabilized single cell suspensions from infected organs. Such an approach may extend the FLARE principle of coupling fluorescence to fungal viability to pathogenic fungi, like Cryptococcus neoformans or Histoplasma capsulatum, that grow as rapidly dividing yeast cells.
The kinetics of fluorophore degradation may limit the utility of functional reporters. In particular, if a viability indicator does not quench in a timely manner coincident with microbial death, significant outcomes may be overlooked. In experiments with recombinant Escherichia coli, DsRed degradation within Dictiostylium discoideum phagosomes occurred with a half-life of approximately 45 minutes [15]. Consistent with these results, the fluorescence signal in GFP-expressing Plasmodium yoelii-infected red blood cells was degraded with a half-life of 30–60 minutes in murine macrophage and dendritic cell phagosomes in vitro [23]. The mechanism by which DsRed and other genetically encoded fluorophores are quenched and degraded within mammalian phagosomes remains poorly defined. Therefore, the strict correlation between loss of fluorescence in sorted leukocyte populations and loss of microbial viability, as measured by colony-forming unit analysis, is critical to the success of the FLARE approach.
Future Prospects of Functional Microbial Reporters
Although this Pearl highlights a small number of functional microbial reporters, the vast library of fluorescent protein indicators to monitor ion and metabolite concentrations, enzymatic activities, and the induction of microbial virulence factors suggests that this class of probes will expand rapidly at the host—microbe research interface. Coupling fluorescence to microbial properties that facilitate survival or immune evasion in host environments, such as dimorphism in specific fungi, antigenic hypervariability, or latency [24], will enable more precise understanding of the host—microbe dynamic in native tissue contexts. The examples presented herein collectively illustrate that functional microbial reporters facilitate molecular, cellular, and pharmacologic investigation into fundamental events that guide the outcomes of individual encounters between host and microbial constituents in vivo. Continued development of new and improved functional microbial reporters will be an important approach to improving our ability to decipher host responses to many clinically important pathogens.
Zdroje
1. Tsien RY (2010) Fluorescence readouts of biochemistry in live cells and organisms. In: Weissleder R, Ross BD, Rehemtulla A, Gambhir SS, editors. Molecular Imaging: Principles and Practice. Shelton, CT: People's Medical Publishing House—USA. pp. 808–828.
2. Miesenbock G, De Angelis DA, Rothman JE (1998) Visualizing secretion and synaptic transmission with pH-sensitive green fluorescent proteins. Nature 394 : 192–195. 9671304
3. Akerboom J, Carreras Calderon N, Tian L, Wabnig S, Prigge M, et al. (2013) Genetically encoded calcium indicators for multi-color neural activity imaging and combination with optogenetics. Front Mol Neurosci 6 : 2. doi: 10.3389/fnmol.2013.00002 23459413
4. Kanki H, Shimabukuro MK, Miyawaki A, Okano H (2010) "Color Timer" mice: visualization of neuronal differentiation with fluorescent proteins. Mol Brain 3 : 5. doi: 10.1186/1756-6606-3-5 20205849
5. Nicholls SB, Chu J, Abbruzzese G, Tremblay KD, Hardy JA (2011) Mechanism of a genetically encoded dark-to-bright reporter for caspase activity. J Biol Chem 286 : 24977–24986. doi: 10.1074/jbc.M111.221648 21558267
6. Schraml BU, van Blijswijk J, Zelenay S, Whitney PG, Filby A, et al. (2013) Genetic Tracing via DNGR-1 Expression History Defines Dendritic Cells as a Hematopoietic Lineage. Cell 154 : 843–858. doi: 10.1016/j.cell.2013.07.014 23953115
7. Yona S, Kim KW, Wolf Y, Mildner A, Varol D, et al. (2013) Fate mapping reveals origins and dynamics of monocytes and tissue macrophages under homeostasis. Immunity 38 : 79–91. doi: 10.1016/j.immuni.2012.12.001 23273845
8. Croxford AL, Buch T (2011) Cytokine reporter mice in immunological research: perspectives and lessons learned. Immunology 132 : 1–8. doi: 10.1111/j.1365-2567.2010.03372.x 21070235
9. Coombes JL, Robey EA (2010) Dynamic imaging of host-pathogen interactions in vivo. Nat Rev Immunol 10 : 353–364. doi: 10.1038/nri2746 20395980
10. Jhingran A, Mar KB, Kumasaka DK, Knoblaugh SE, Ngo LY, et al. (2012) Tracing conidial fate and measuring host cell antifungal activity using a reporter of microbial viability in the lung. Cell Rep 2 : 1762–1773. doi: 10.1016/j.celrep.2012.10.026 23200858
11. Dupont CD, Christian DA, Selleck EM, Pepper M, Leney-Greene M, et al. (2014) Parasite fate and involvement of infected cells in the induction of CD4+ and CD8+ T cell responses to Toxoplasma gondii. PLoS Pathog 10: e1004047. doi: 10.1371/journal.ppat.1004047 24722202
12. Claudi B, Sprote P, Chirkova A, Personnic N, Zankl J, et al. (2014) Phenotypic variation of Salmonella in host tissues delays eradication by antimicrobial chemotherapy. Cell 158 : 722–733. doi: 10.1016/j.cell.2014.06.045 25126781
13. Helaine S, Cheverton AM, Watson KG, Faure LM, Matthews SA, et al. (2014) Internalization of Salmonella by macrophages induces formation of nonreplicating persisters. Science 343 : 204–208. doi: 10.1126/science.1244705 24408438
14. Helaine S, Thompson JA, Watson KG, Liu M, Boyle C, et al. (2010) Dynamics of intracellular bacterial replication at the single cell level. Proc Natl Acad Sci U S A 107 : 3746–3751. doi: 10.1073/pnas.1000041107 20133586
15. Maselli A, Laevsky G, Knecht DA (2002) Kinetics of binding, uptake and degradation of live fluorescent (DsRed) bacteria by Dictyostelium discoideum. Microbiology 148 : 413–420. 11832505
16. Koshy AA, Fouts AE, Lodoen MB, Alkan O, Blau HM, et al. (2010) Toxoplasma secreting Cre recombinase for analysis of host-parasite interactions. Nat Methods 7 : 307–309. doi: 10.1038/nmeth.1438 20208532
17. Han SJ, Melichar HJ, Coombes JL, Chan SW, Koshy AA, et al. (2014) Internalization and TLR-dependent type I interferon production by monocytes in response to Toxoplasma gondii. Immunol Cell Biol 92 : 872–881. doi: 10.1038/icb.2014.70 25155465
18. Christian DA, Koshy AA, Reuter MA, Betts MR, Boothroyd JC, et al. (2014) Use of transgenic parasites and host reporters to dissect events that promote interleukin-12 production during toxoplasmosis. Infect Immun 82 : 4056–4067. doi: 10.1128/IAI.01643-14 25024368
19. Terskikh A, Fradkov A, Ermakova G, Zaraisky A, Tan P, et al. (2000) "Fluorescent timer": protein that changes color with time. Science 290 : 1585–1588. 11090358
20. Espinosa V, Jhingran A, Dutta O, Kasahara S, Donnelly R, et al. (2014) Inflammatory monocytes orchestrate innate antifungal immunity in the lung. PLoS Pathog 10: e1003940. doi: 10.1371/journal.ppat.1003940 24586155
21. Shepardson KM, Jhingran A, Caffrey A, Obar JJ, Suratt BT, et al. (2014) Myeloid derived hypoxia inducible factor 1-alpha is required for protection against pulmonary Aspergillus fumigatus infection. PLoS Pathog 10: e1004378. doi: 10.1371/journal.ppat.1004378 25255025
22. Bencina M (2013) Illumination of the spatial order of intracellular pH by genetically encoded pH-sensitive sensors. Sensors (Basel) 13 : 16736–16758.
23. Bettiol E, Van de Hoef DL, Carapau D, Rodriguez A (2010) Efficient phagosomal maturation and degradation of Plasmodium-infected erythrocytes by dendritic cells and macrophages. Parasite Immunol 32 : 389–398. doi: 10.1111/j.1365-3024.2010.01198.x 20500669
24. Finlay BB, McFadden G (2006) Anti-immunology: evasion of the host immune system by bacterial and viral pathogens. Cell 124 : 767–782. 16497587
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek Transmission Properties of Human PrP 102L Prions Challenge the Relevance of Mouse Models of GSSČlánek Decline of FoxP3+ Regulatory CD4 T Cells in Peripheral Blood of Children Heavily Exposed to MalariaČlánek IFNγ and IL-12 Restrict Th2 Responses during Helminth/ Co-Infection and Promote IFNγ from Th2 CellsČlánek Exploiting Fungal Virulence-Regulating Transcription Factors As Novel Antifungal Drug Targets
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2015 Číslo 7- Jak souvisí postcovidový syndrom s poškozením mozku?
- Měli bychom postcovidový syndrom léčit antidepresivy?
- Farmakovigilanční studie perorálních antivirotik indikovaných v léčbě COVID-19
- 10 bodů k očkování proti COVID-19: stanovisko České společnosti alergologie a klinické imunologie ČLS JEP
-
Všechny články tohoto čísla
- Basic Prion Science “Spreads” Insight
- Research Driven by Curiosity: The Journey from Basic Molecular Biology and Virology to Studies of Human Pathogenic Coronaviruses
- Cross Kingdom Activators of Five Classes of Bacterial Effectors
- Vaccination Drives Changes in Metabolic and Virulence Profiles of
- Expression of the Blood-Group-Related Gene Alters Susceptibility to Infection
- Transmission Properties of Human PrP 102L Prions Challenge the Relevance of Mouse Models of GSS
- Latent KSHV Infected Endothelial Cells Are Glutamine Addicted and Require Glutaminolysis for Survival
- The DSF Family of Cell–Cell Signals: An Expanding Class of Bacterial Virulence Regulators
- Intraperitoneal Infection of Wild-Type Mice with Synthetically Generated Mammalian Prion
- Vpr Promotes Macrophage-Dependent HIV-1 Infection of CD4 T Lymphocytes
- An In-Depth Comparison of Latency-Reversing Agent Combinations in Various and HIV-1 Latency Models Identified Bryostatin-1+JQ1 and Ingenol-B+JQ1 to Potently Reactivate Viral Gene Expression
- α-Macroglobulin Can Crosslink Multiple Erythrocyte Membrane Protein 1 (PfEMP1) Molecules and May Facilitate Adhesion of Parasitized Erythrocytes
- Should Symbionts Be Nice or Selfish? Antiviral Effects of Wolbachia Are Costly but Reproductive Parasitism Is Not
- A Unique Human Norovirus Lineage with a Distinct HBGA Binding Interface
- The Broad Neutralizing Antibody Responses after HIV-1 Superinfection Are Not Dominated by Antibodies Directed to Epitopes Common in Single Infection
- Rapidly Evolving Genes Are Key Players in Host Specialization and Virulence of the Fungal Wheat Pathogen ()
- MiR-21 in Extracellular Vesicles Leads to Neurotoxicity via TLR7 Signaling in SIV Neurological Disease
- Age-Dependent Cell Trafficking Defects in Draining Lymph Nodes Impair Adaptive Immunity and Control of West Nile Virus Infection
- Decline of FoxP3+ Regulatory CD4 T Cells in Peripheral Blood of Children Heavily Exposed to Malaria
- Dimerization-Induced Allosteric Changes of the Oxyanion-Hole Loop Activate the Pseudorabies Virus Assemblin pUL26N, a Herpesvirus Serine Protease
- Macrophages Subvert Adaptive Immunity to Urinary Tract Infection
- Mycolactone-Dependent Depletion of Endothelial Cell Thrombomodulin Is Strongly Associated with Fibrin Deposition in Buruli Ulcer Lesions
- Activation of TLR2 and TLR6 by Dengue NS1 Protein and Its Implications in the Immunopathogenesis of Dengue Virus Infection
- K-bZIP Mediated SUMO-2/3 Specific Modification on the KSHV Genome Negatively Regulates Lytic Gene Expression and Viral Reactivation
- Phosphoproteomic Analysis of KSHV-Infected Cells Reveals Roles of ORF45-Activated RSK during Lytic Replication
- CR3 and Dectin-1 Collaborate in Macrophage Cytokine Response through Association on Lipid Rafts and Activation of Syk-JNK-AP-1 Pathway
- IFNγ and IL-12 Restrict Th2 Responses during Helminth/ Co-Infection and Promote IFNγ from Th2 Cells
- THY-1 Cell Surface Antigen (CD90) Has an Important Role in the Initial Stage of Human Cytomegalovirus Infection
- Human Enterovirus Nonstructural Protein 2C Functions as Both an RNA Helicase and ATP-Independent RNA Chaperone
- IL-27 Signaling Is Crucial for Survival of Mice Infected with African Trypanosomes via Preventing Lethal Effects of CD4 T Cells and IFN-γ
- Synergistic Reactivation of Latent HIV Expression by Ingenol-3-Angelate, PEP005, Targeted NF-kB Signaling in Combination with JQ1 Induced p-TEFb Activation
- Vpu Exploits the Cross-Talk between BST2 and the ILT7 Receptor to Suppress Anti-HIV-1 Responses by Plasmacytoid Dendritic Cells
- Herpesvirus Genome Recognition Induced Acetylation of Nuclear IFI16 Is Essential for Its Cytoplasmic Translocation, Inflammasome and IFN-β Responses
- A Comprehensive Analysis of Replicating Merkel Cell Polyomavirus Genomes Delineates the Viral Transcription Program and Suggests a Role for mcv-miR-M1 in Episomal Persistence
- Analysis of the SUMO2 Proteome during HSV-1 Infection
- Capacity of Broadly Neutralizing Antibodies to Inhibit HIV-1 Cell-Cell Transmission Is Strain- and Epitope-Dependent
- A Novel Antiviral Target Structure Involved in the RNA Binding, Dimerization, and Nuclear Export Functions of the Influenza A Virus Nucleoprotein
- Deploying FLAREs to Visualize Functional Outcomes of Host—Pathogen Encounters
- Mosquitoes Reset Malaria Parasites
- The Lung Microbiome: New Principles for Respiratory Bacteriology in Health and Disease
- Extracellular Virions: The Advance Guard of Poxvirus Infections
- Risks of Antibiotic Exposures Early in Life on the Developing Microbiome
- RNA Virus Reassortment: An Evolutionary Mechanism for Host Jumps and Immune Evasion
- Exploiting Fungal Virulence-Regulating Transcription Factors As Novel Antifungal Drug Targets
- N-acetylglucosamine Regulates Virulence Properties in Microbial Pathogens
- Periodontal Diseases: Bug Induced, Host Promoted
- Mechanisms of Host Behavioral Change in Rodent Association
- The Endosymbiotic Bacterium Selectively Kills Male Hosts by Targeting the Masculinizing Gene
- HIV Reactivation from Latency after Treatment Interruption Occurs on Average Every 5-8 Days—Implications for HIV Remission
- Ubiquilin 1 Promotes IFN-γ-Induced Xenophagy of
- Transfer of Immunity from Mother to Offspring Is Mediated via Egg-Yolk Protein Vitellogenin
- Suppression of Long-Lived Humoral Immunity Following Infection
- The Role of VP1 Amino Acid Residue 145 of Enterovirus 71 in Viral Fitness and Pathogenesis in a Cynomolgus Monkey Model
- Utilizing Chemical Genomics to Identify Cytochrome as a Novel Drug Target for Chagas Disease
- The Emerging Role for RNA Polymerase II in Regulating Virulence Gene Expression in Malaria Parasites
- Turning Up the Heat: Inflammasome Activation by Fungal Pathogens
- On and Under the Skin: Emerging Basidiomycetous Yeast Infections Caused by Species
- EhVps32 Is a Vacuole-Associated Protein Involved in Pinocytosis and Phagocytosis of
- Characterization of a Prefusion-Specific Antibody That Recognizes a Quaternary, Cleavage-Dependent Epitope on the RSV Fusion Glycoprotein
- The Serine Protease EspC from Enteropathogenic Regulates Pore Formation and Cytotoxicity Mediated by the Type III Secretion System
- Existing Infection Facilitates Establishment and Density of Malaria Parasites in Their Mosquito Vector
- Evaluating Human T-Cell Therapy of Cytomegalovirus Organ Disease in HLA-Transgenic Mice
- Neuronal Interferon Signaling Is Required for Protection against Herpes Simplex Virus Replication and Pathogenesis
- Epstein-Barr Virus Proteins EBNA3A and EBNA3C Together Induce Expression of the Oncogenic MicroRNA Cluster miR-221/miR-222 and Ablate Expression of Its Target p57
- Colonization of the Mouse Gastrointestinal Tract Is Modulated by Wall Teichoic Acid, Capsule, and Surface Proteins
- Virulence of Group A Streptococci Is Enhanced by Human Complement Inhibitors
- Identification of Caspase Cleavage Sites in KSHV Latency-Associated Nuclear Antigen and Their Effects on Caspase-Related Host Defense Responses
- Calprotectin Increases the Activity of the SaeRS Two Component System and Murine Mortality during Infections
- Type VI Secretion System Transports Zn to Combat Multiple Stresses and Host Immunity
- Lv4 Is a Capsid-Specific Antiviral Activity in Human Blood Cells That Restricts Viruses of the SIV/SIV/HIV-2 Lineage Prior to Integration
- Phenylbutyrate Is Bacteriostatic against and Regulates the Macrophage Response to Infection, Synergistically with 25-Hydroxy-Vitamin D₃
- An Internally Translated MAVS Variant Exposes Its Amino-terminal TRAF-Binding Motifs to Deregulate Interferon Induction
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- RNA Virus Reassortment: An Evolutionary Mechanism for Host Jumps and Immune Evasion
- Activation of TLR2 and TLR6 by Dengue NS1 Protein and Its Implications in the Immunopathogenesis of Dengue Virus Infection
- N-acetylglucosamine Regulates Virulence Properties in Microbial Pathogens
- Characterization of a Prefusion-Specific Antibody That Recognizes a Quaternary, Cleavage-Dependent Epitope on the RSV Fusion Glycoprotein
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



