-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaTranscription-Associated R-Loop Formation across the Human CGG-Repeat Region
Expansion of a CGG-repeat element within the human FMR1 gene is responsible for multiple human diseases, including fragile X syndrome and fragile X-associated tremor/ataxia syndrome (FXTAS). These diseases occur in separate ranges of repeat length and are characterized by profoundly different molecular mechanisms. Fragile X syndrome results from FMR1 gene silencing, whereas FXTAS is associated with an increase in transcription and toxicity of the CGG-repeat-containing mRNA. This study introduces a previously unknown molecular feature of the FMR1 locus, namely the co-transcriptional formation of three-stranded R-loop structures upon re-annealing of the nascent FMR1 transcript to the template DNA strand. R-loops are involved in the normal function of human CpG island promoters in that they contribute to protecting these sequences from DNA methylation. However, excessive R-loop formation can lead to activation of the DNA damage response and result in genomic instability. We used antibody recognition and chemical single-stranded DNA footprinting to show that R-loops form at the FMR1 locus with increasing frequency and greater structural complexity as the CGG-repeat length increases. This discovery provides a missing piece of both the complex FMR1 molecular puzzle and the diseases resulting from CGG-repeat expansion.
Published in the journal: . PLoS Genet 10(4): e32767. doi:10.1371/journal.pgen.1004294
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1004294Summary
Expansion of a CGG-repeat element within the human FMR1 gene is responsible for multiple human diseases, including fragile X syndrome and fragile X-associated tremor/ataxia syndrome (FXTAS). These diseases occur in separate ranges of repeat length and are characterized by profoundly different molecular mechanisms. Fragile X syndrome results from FMR1 gene silencing, whereas FXTAS is associated with an increase in transcription and toxicity of the CGG-repeat-containing mRNA. This study introduces a previously unknown molecular feature of the FMR1 locus, namely the co-transcriptional formation of three-stranded R-loop structures upon re-annealing of the nascent FMR1 transcript to the template DNA strand. R-loops are involved in the normal function of human CpG island promoters in that they contribute to protecting these sequences from DNA methylation. However, excessive R-loop formation can lead to activation of the DNA damage response and result in genomic instability. We used antibody recognition and chemical single-stranded DNA footprinting to show that R-loops form at the FMR1 locus with increasing frequency and greater structural complexity as the CGG-repeat length increases. This discovery provides a missing piece of both the complex FMR1 molecular puzzle and the diseases resulting from CGG-repeat expansion.
Introduction
The human fragile X mental retardation 1 gene (FMR1; HGNC:3775) contains a (CGG)n trinucleotide repeat that is responsible for a family of heritable disorders affecting both early neurodevelopment (fragile X syndrome; FXS) and late-onset neurodegeneration (fragile X-associated tremor/ataxia syndrome; FXTAS) [1]–[4]. The repeat element is located in the 5′ untranslated region (5′UTR) of the gene, and is thus transcribed into mRNA but not translated into the amino acid sequence of the gene product, the FMR1 protein (FMRP).
Alleles in the ∼55–200 CGG-repeat range are historically referred to as “premutation” alleles in reference to increased instability and the tendency in maternal transmission to expand into the “full mutation” range of FXS (>200 CGG repeats) [3], [5], [6]. Premutation alleles are also variably associated with several clinical phenotypes; in addition to FXTAS, these phenotypes include primary ovarian insufficiency (FXPOI) [7] and neurodevelopmental involvement [8], [9]. Contrary to the gene silencing observed in FXS alleles, premutation alleles are associated with increased transcriptional activity. Indeed, FMR1 mRNA levels are positively correlated with size of the repeat expansion in the premutation range [10]. The molecular pathogenesis of the premutation disorders is generally considered to be a toxic RNA gain-of-function resulting from the expanded CGG-repeat region in the mRNA, but a definitive mechanism for the RNA involvement has not yet emerged [1], [11]–[15].
Stable RNA:DNA hybrids can form upon transcription of cytosine-rich template sequences because a guanine-rich RNA:cytosine-rich DNA heteroduplex is thermodynamically more stable than the corresponding DNA:DNA duplex [16], [17]. Recent work has revealed that such structures form throughout the human genome, particularly at CpG island promoters [18], [19]. Additionally, in vitro transcription experiments showed that CGG trinucleotide repeats alone are able to form R-loops [20].
R-loops at CpG island promoters serve a natural and important role in protecting CpG-rich regions from acquiring DNA methylation and becoming epigenetically silenced [18]. In addition, R-loop formation at the 3′ end of numerous human genes is thought to permit efficient transcription termination [19], [21]. However, R-loop formation has also been linked to genomic instability in numerous systems [22]–[24] and is thought to trigger recombination at class-switch regions [25], [26]. Recent results suggest that defects in mRNA processing can result in an R-loop-dependent activation of the DNA damage response, and to the accumulation of γH2AX, a histone variant associated with the repair of DNA breaks [27], [28]. R-loops at the Prader-Willi syndrome Snord116 locus are responsible for chromatin decondensation and for regulating the transcription of nearby imprinted genes [29]. Thus, it appears that R-loop formation in the genome is a widespread, dynamic process that is sensitive to perturbation, and has both physiological roles and potential “toxic” consequences through activation of the DNA damage response.
Herein we present evidence for R-loop formation at the endogenous human FMR1 locus, and explore the impact of CGG-repeat expansion and transcription induction on the extent of FMR1 R-loop formation.
Results
FMR1 5′UTR Sequence Composition Predicts R-Loop Formation
We examined the sequence of the human FMR1 5′UTR to identify important features for R-loop formation, including proximity to transcription start sites (TSSs), GC skew, and G-clusters [18], [30]. We calculated the GC content (GC%) together with the density in CpG dinucleotides (CpG observed/expected ratio; CpG O/E) and GC skew (G−C/G+C) across the 5′ end of the FMR1 gene (hg19 chrX:146,992,969–146,994,458; shown here for CGG = 100) (Figure 1). The multiple FMR1 TSSs are located upstream of the CGG repeats and constitute the upstream boundary of the UTR [31]–[33], as depicted in Figure 1. Overall, the promoter and 5′UTR are exceptionally GC-rich, with GC% peaking at 100% through the repeats, and staying above 60% through the entire UTR (Figure 1). Part of this region also shows an elevated frequency of CpG dinucleotides and can be classified as a CpG island (CGI). The FMR1 CGI overlaps with the promoter sequences and the 5′UTR through the CGG repeats (Figure 1), and therefore belongs to a large class of promoter CGIs [18]. It is notable that CGG-repeat expansions characteristic of FXTAS and FXS directly stretch out the 3′ boundary of the CGI promoter element, as defined by its high GC content and CpG density [34].
Fig. 1. Sequence analysis of the FMR1 promoter reveals signatures of R-loop formation. 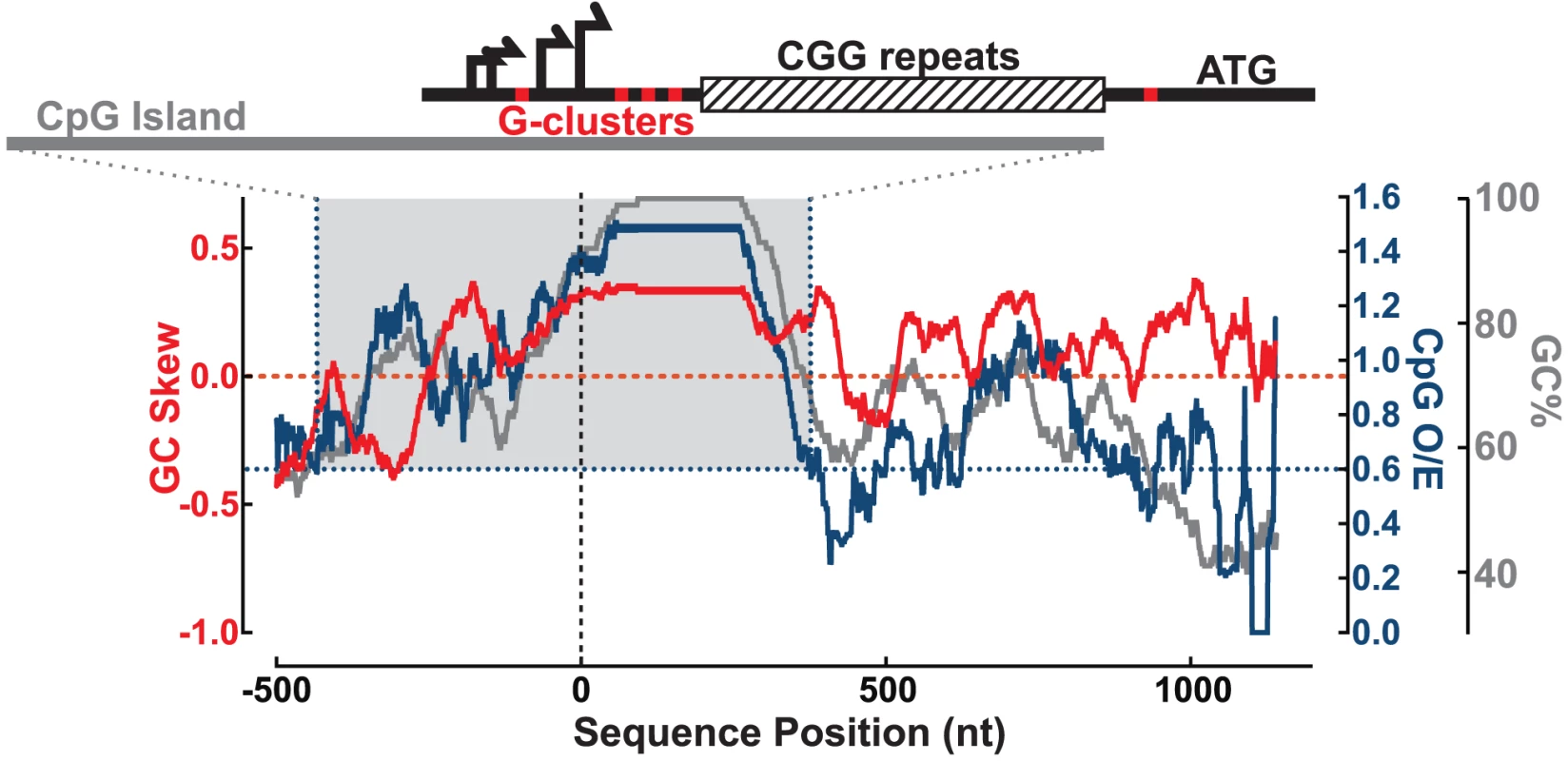
GC skew (red, left y-axis), CpG observed/expected ratio (CpG O/E; navy, right y-axis), and GC% (gray, right y-axis) calculated over a sliding 100 nt window from −500 to +1200 nt around the downstream-most known transcription start site (vertical dotted line). Gray-shaded box highlights CGI defined by CpG O/E>0.6 (navy dotted lines) and GC%>50% for at least 200 nt. Schematic at the top shows the FMR1 5′UTR with multiple transcription start sites (black arrows), G-clusters (red ticks), and CGG repeats (striped box), all overlapping the CGI (gray bar) for scale to the graph below. In addition to elevated GC% and CpG O/E, the FMR1 CGI is also characterized by elevated GC skew downstream of the TSSs and through the CGG repeats (Figure 1). As noted for CGI promoters and other regions in the human genome, GC skew is highly predictive of R-loop formation [18], [19]. As with GC skew, G-clusters (≥4 Gs in a row) act as nucleation points for RNA:DNA hybridization [30]. Five such clusters are found in the 5′UTR, as indicated by red ticks on the schematic in Figure 1, including one that is included/excluded in the transcript depending on TSS choice. In total, these features predict R-loop formation at FMR1 following transcription.
DNA:RNA Immunoprecipitation Indicates the Formation of Genomic FMR1 R-Loops
We used DNA:RNA immunoprecipitation (DRIP) to directly test the existence of R-loops at the endogenous FMR1 locus in human genomic DNA, and compared the relative abundance of R-loops across the range of transcribed CGG-repeat expansion alleles. The S9.6 antibody recognizes RNA:DNA hybrids without any known sequence preference or sensitivity to DNA methylation ([18], [21], [35]; unpublished data). After DRIP, we calculated the fold enrichment of FMR1 relative to input genomic DNA, and to a non-R-loop-forming genomic locus (ZNF554; HGNC:26629) using qPCR, where DRIP enrichment is not expected.
In genomic DNA from cultured human male dermal fibroblasts, we observed a 2.1 - to 13.9-fold enrichment for FMR1 across the range of CGG-repeat alleles tested (Figure 2A). Although there was substantial inter-subject variation in fold enrichment, both in control and premutation groups, the premutation group as a whole demonstrated greater enrichment (mean 9.0, SD 3.9, range 2.9–13.4) than the control group (mean 4.2, SD 2.4, range 1.6–8.6) (P = 0.0008; linear mixed-effects model, see: Material and Methods). By contrast, a positive control for a strong R-loop-forming locus, MYADM (HGNC:7544), showed consistently high enrichment (25 - to 50-fold), which was not influenced by FMR1 CGG-repeat size (Figure S1). As expected for R-loop formation, treatment with purified recombinant human RNases H1 and H2 eliminated DRIP pulldown. Hence, enrichment for FMR1 in 3 different fibroblast lines went from a mean of 6.36±2.31 (SEM, n = 4) to 1.59±0.219 (SEM, n = 4) upon RNase H treatment (Figure 2B), a significant reduction (unpaired t-test on log-transformed enrichment values, P = 0.0125). Likewise, elimination of DRIP enrichment following RNase H treatment was also observed at the positive MYADM locus (unpaired t-test on log-transformed enrichment values, P = 0.0002) (Figure 2B).
Fig. 2. R-loop pull-down in human dermal fibroblasts confirms R-loop formation in the genome. 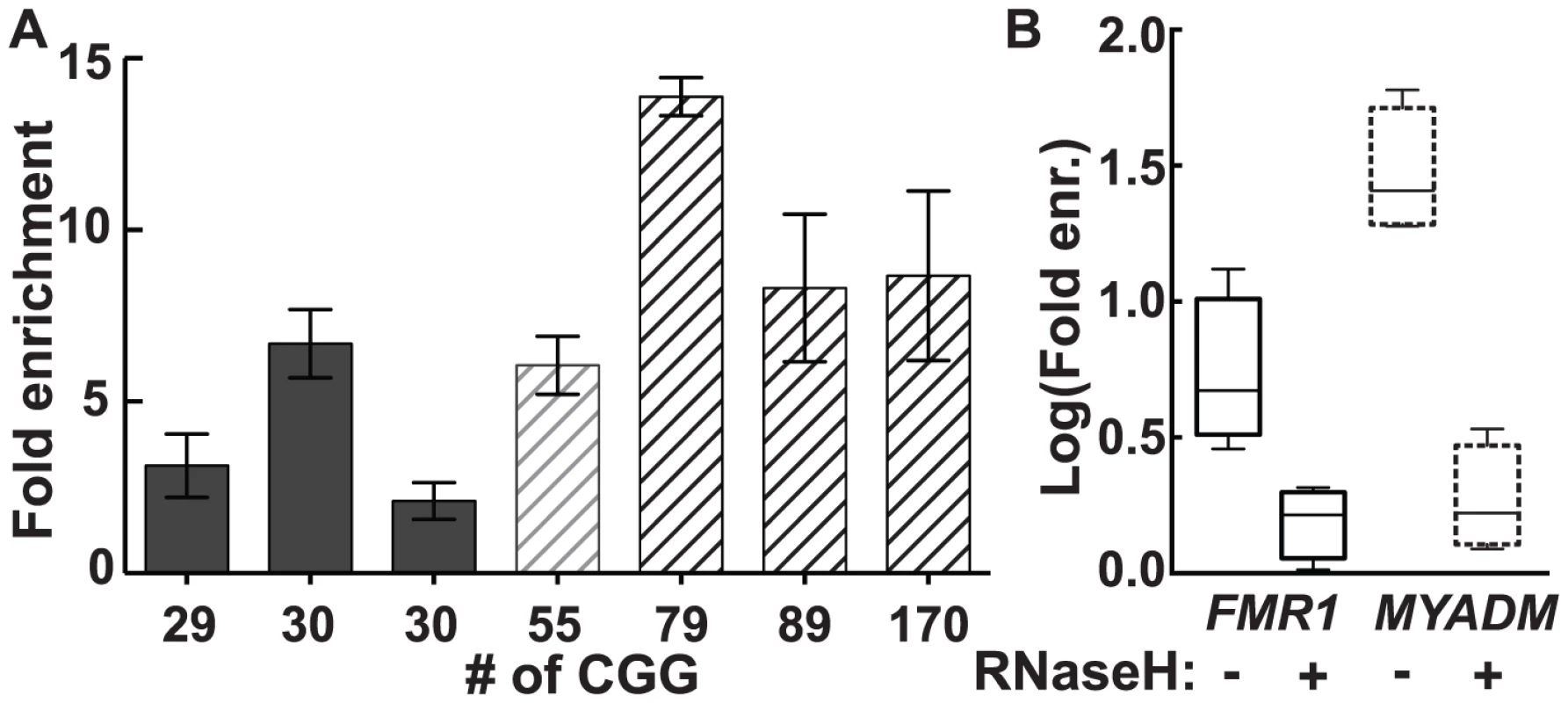
(A) Fold enrichment for FMR1 in dermal fibroblast cells cultured from seven individuals using a monoclonal antibody specific to hybrids. Enrichment is relative to input and a non-R-loop-forming genomic reference locus. (B) Treatment with recombinant RNases H1 and H2 (RNase H) eliminates enrichment seen for FMR1 (solid lines) and MYADM (broken lines). DOX-Induced Transcription and Expanded CGG Repeats Result in Enhanced FMR1 R-Loop Formation
We used a doxycycline (DOX)-inducible episomal system in SK-N-MC neuroepithelioma cells [36] to investigate the relationship between the frequency of transcription initiation and R-loop formation. The TRE-Tight promoter allows for precise control of transcription through an FMR1 5′UTR sequence harboring either a 95 or 30 CGG-repeat element, or a non-FMR1 linker sequence ([36]; Figure 3A). All three constructs include EGFP cDNA, which was used as a target for qPCR to avoid amplification from endogenous sequences. Treatment with DOX at 10 ng/mL and 100 ng/mL resulted in a clear induction of transcription with equal expression levels for all three constructs, relative to the no-DOX baseline (Figure 3B).
Fig. 3. Effect of transcription and repeat length on FMR1 R-loop formation. 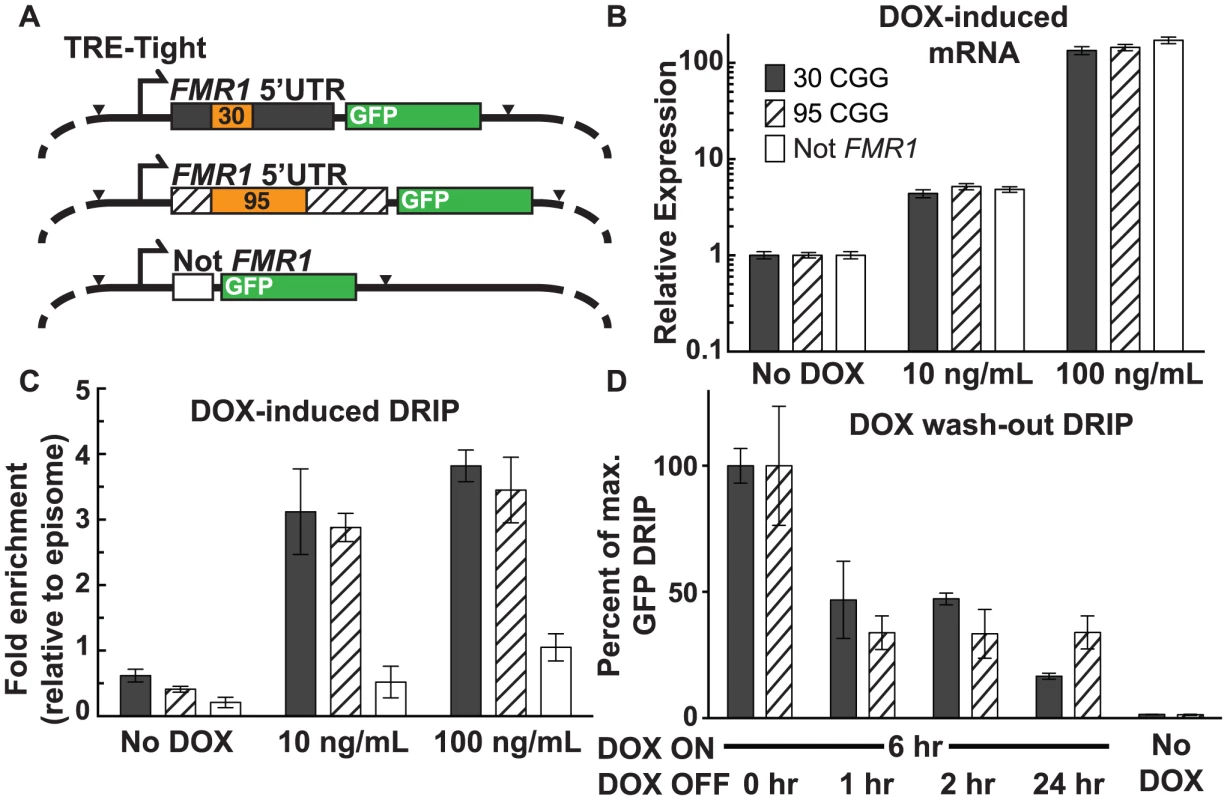
(A) Schematic of DOX-ON constructs with short or expanded FMR1 CGG repeats or non-FMR1 sequence, each with GFP reporter tags. Black arrowheads mark sites of restriction enzyme cleavage prior to DRIP, with EcoRI cutting at the start of the FMR1 5′UTR and XbaI cutting at the end of EGFP. (B) mRNA expression relative to non-induced cells for each construct. Error bars: SEM from 2 biological replicates. (C) DRIP fold enrichment of GFP fragment relative to the episome backbone. Error bars: SEM from 3 biological replicates. (D) DRIP percentage of input normalized to peak recovery (6 hours DOX ON) of GFP fragment at 0, 1, 2, and 24 hours post DOX washout, and No-DOX treatment. Error bars: SEM from 3 biological replicates. Using DRIP-qPCR, we observed increased R-loop formation through the FMR1 5′UTR, mirroring the transcriptional response to DOX induction (Figure 3C; Figure S2A). Fold enrichment for the 30 CGG-repeat allele increased from 0.62±0.096 (n = 3) without DOX, to 3.1±0.65 (n = 3) at 10 ng/mL DOX, and 3.8±0.24 (n = 3) at 100 ng/mL DOX. The 95 CGG-repeat allele increased from 0.41±0.044 (SEM, n = 3) without DOX, to 2.9±0.22 (n = 3) at 10 ng/mL DOX, and 3.45±0.50 (n = 3) at 100 ng/mL DOX. By contrast, the non-FMR1 control locus showed little to no increase upon induction (Fig. 3C). We note that the episome backbone also showed modestly increased pull-down efficiency with increasing expression (Figure S2B), which could result from R-loop formation around the EGFP poly(A) sequence. Indeed, R-loops have the propensity to form broad peaks around poly(A)-dependent termination regions ([19], [22]; F.C and L.S., unpublished observations). Given that R-loops inhibit the activity of restriction enzymes, this inhibition could prevent the cleavage required for separating the GFP restriction fragment from the background fragment and lead to apparent DOX-inducible R-loop formation over the episomal backbone. To account for this background, R-loop formation at the target GFP locus, as measured by DRIP-qPCR, was calculated relative to R-loop formation over the episome backbone and normalized to non-induced baseline (Figure 3C).
To assess the stability of R-loops once formed, we induced transcription for 6 hours with 100 ng/mL DOX, after which DOX was removed from the media and R-loop presence was measured by DRIP 1, 2, and 24 hours following the washout. Recovery of the 30-repeat allele decreased to 46.8%±15.3% (n = 2) of maximum after a 1-hour washout. It persisted at 47.2%±2.3% (n = 3) after a 2-hour washout, and dropped to 16.6%±1.2% (n = 3) after 24 hours (Figure 3D). Recovery of the 95-repeat allele decreased to 33.8%±6.7% (n = 2) of maximum after 1 hour, then remained essentially unchanged to 24 hours (33.9%±6.6%; n = 3) (Figure 3D). These data show that R-loop formation through the FMR1 5′UTR depends on active transcription initiation and that R-loops are dynamic structures, which are progressively formed and resolved.
Mapping the ssDNA Structure of the FMR1 Genomic R-Loop
We used non-denaturing sodium bisulfite treatment to map the extent of the displaced single-stranded DNA (ssDNA) constituting the FMR1 R-loop in human male fibroblast genomic DNA. Sodium bisulfite deaminates unmethylated cytosines, but only with high efficiency in ssDNA. When applied in a non-denaturing manner, it can therefore be used as an efficient probe for R-loop formation and has been used extensively to footprint R-loop structures at single-nucleotide resolution [18], [26]. As expected from DRIP data, non-denaturing bisulfite footprinting revealed extensive single-strandedness through the 5′UTR across the range of transcribed FMR1 alleles (CGG = 29, 30, 55, 79) (Figure 4). R-loop structures began at the first G-cluster downstream of the TSSs and continued through the repeat region for all four alleles examined. However, unlike any other region analyzed to date, stretches of unconverted cytosines indicative of double-stranded DNA were found inside the repeats. Furthermore, the extent of unconverted DNA was much greater for expanded alleles with higher CGG-repeat sizes (Figure 4). For normal CGG-repeat sizes, patterns of non-conversion were short and symmetrical and were centered on an AGG-repeat interruption not shown in the figure. The single converted CpG dinucleotide in the center of the unconverted CGG track of both 29 - and 30-repeat samples was located immediately adjacent to the AGG polymorphism. Such a pattern would be expected if a short hairpin formed within the repeat region, with the stem of the structure being double-stranded and protected from conversion while a short loop is exposed. For longer CGG repeats, the footprinting data suggests that a large region of ssDNA exists upstream and downstream of the CGG repeat, but that most of the repeat region itself is in fact protected from conversion, save a few scattered points of conversion (Figure 4). Note that, for a certain number of molecules in the 30-, 55-, and 79-CGG samples, R-loops seemed to initiate at a G-cluster downstream of the repeats. Overall, these data show that R-loop formation at FMR1 can initiate from different G-cluster seeding points, both upstream and downstream of the CGGs, and that R-loop formation through expanded CGG repeats may result in hairpin formation or other structural conformations.
Fig. 4. Non-denaturing bisulfite footprinting of the displaced DNA strand of the FMR1 R-loop. 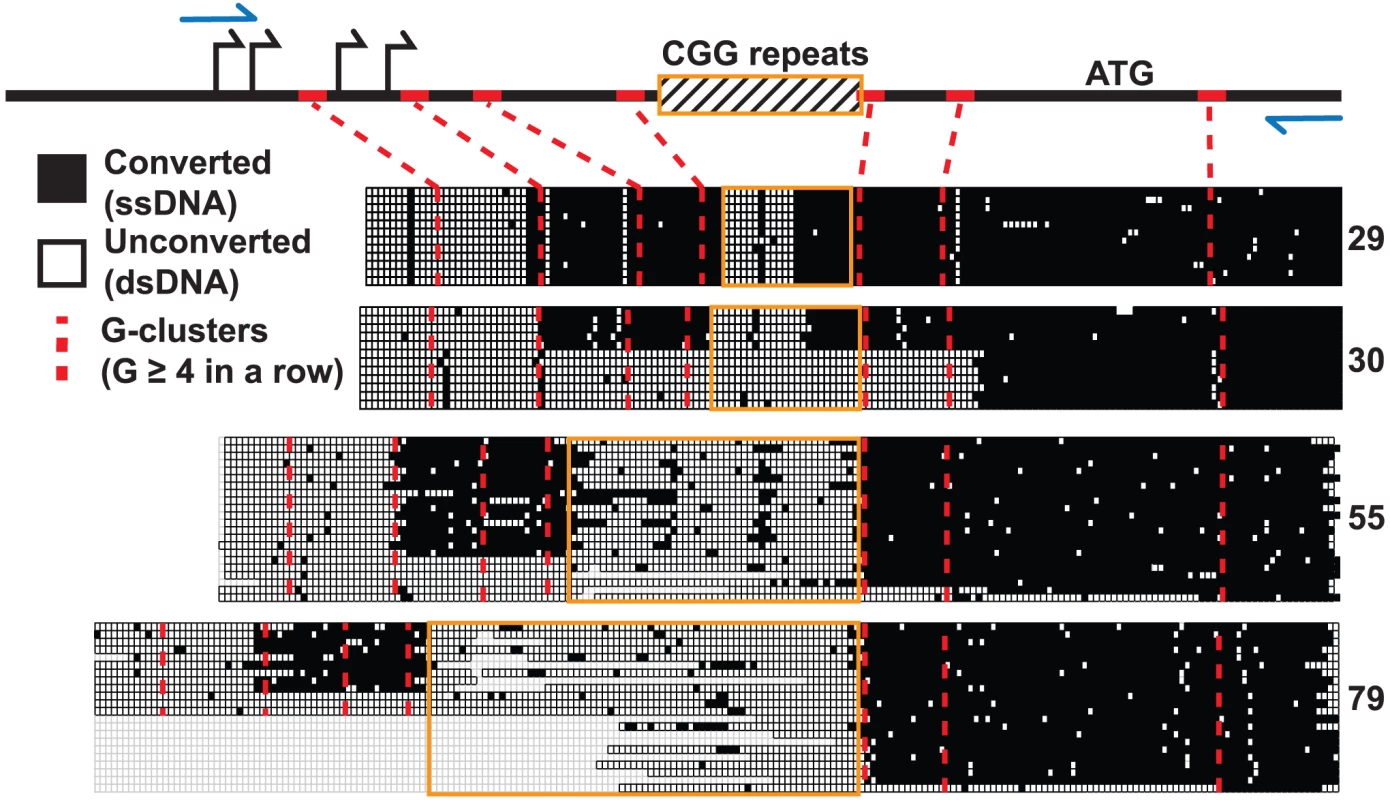
Each row represents an individual sequence clone, grouped together for each allele size, from cultured human dermal fibroblasts. Each column is a cytosine position, with filled boxes representing converted, single-stranded DNA and open boxes representing unconverted, double-stranded DNA. Empty boxes represent sequence gaps from bacterial deletion or loss of clean sequencing signal. Schematic diagram at the top represents the FMR1 5′UTR with marked TSSs (black arrows), translation start (ATG), CGG repeats (striped box with orange border), PCR primers (blue arrows), and G-clusters (red ticks; red dotted lines). Discussion
At the DNA sequence level, R-loop formation is best predicted by the combination of GC content and GC skew, which measures the density and strand asymmetry in the distribution of guanines and cytosines, and correlates with the stability of RNA:DNA hybrids. In comparison to other R-loop forming regions of the genome, the normal (unexpanded) FMR1 promoter matches “Class II” CGI promoters [18], [19]. This category is typical of skewed promoters on the X-chromosome and associates with marginally weaker GC skew [19]. As far as GC sequence composition is concerned, the human FMR1 CGI is at the extreme end of the spectrum in the genome. A GC content of greater than ∼65% is generally viewed as “high GC,” and only 22 CGIs are listed at >80% in the human reference genome [37]. Our analysis shows that GC% at the FMR1 promoter/5′UTR region peaks at 100% across the CGG-repeat, saturating this sequence characteristic. All three metrics of CGI composition and predictors of R-loop formation (GC%, CpG O/E, GC skew) peak at the repeat region. Importantly, CGG-repeat expansions associated with FXTAS and FXS will push FMR1 into the category of stronger skewed promoters by increasing the lengths of the GC content, GC skew, and CpG tracks. CGG expansions are also likely to increase R-loop formation efficiency in two additional ways. First, expanded (premutation) alleles trigger higher transcriptional rates [10], which should favor the frequency of co-transcriptional R-loops. Second, expanded alleles are characterized by a shift in the usage of transcription initiation sites to upstream sites [31], [32]. This shift is expected to allow the inclusion of additional G-clusters, which are de facto R-loop initiation points, in the transcript. The CGG-repeat expansions that associate with FXTAS and FXS are therefore unique in that they strongly enhance the propensity of the FMR1 CGI to form co-transcriptional R-loops.
Here, we provide direct experimental evidence that R-loops form at the endogenous genomic FMR1 locus, which was first detected by the ability of the S9.6 anti-RNA:DNA hybrid antibody to specifically immunoprecipitate the FMR1 locus. As expected, enrichment was lost following enzymatic resolution of the RNA:DNA hybrids using purified RNases H (Figure 2). These experiments are consistent with the notion that repeat expansions cause a corresponding increase in R-loops at the FMR1 locus. Indeed, an upward trend in the FMR1 S9.6-pulldown efficiency was observed for individuals with increasing CGG-repeat sizes (Figure 2A; Figure S3), despite the variation that exists between individuals within both control and premutation allele classes. Based on three control CGG subjects (2–3 independent replications per subject; n = 8 experiments) and four premutation subjects (2–4 independent replications per subject; n = 10 experiments), the fold enrichment in premutation (mean 9.0, SD 3.9, range 2.9–13.4) was significantly greater than for controls (mean 4.2, SD 2.4, range 1.6–8.6) (P = 0.0008; linear mixed-effects model; see: Materials and Methods).
Our DOX-inducible episomal FMR1 system provides a more controlled isogenic platform to directly parse out the impact of repeat expansion and transcription frequency on R-loop formation. As expected, R-loop formation increased in direct response to increasing transcription (Figure 3C). CGG-repeat length at equivalent transcription levels, however, appeared to have little effect on R-loop frequency. Removing DOX resulted in a corresponding decrease in R-loops in the episomal FMR1 5′UTR (Figure 3D), demonstrating the plasticity of R-loop formation at a given locus, with formation driven by active transcription and dissolution catalyzed by native enzymes such as RNases H, RNA:DNA helicases, or DNA topoisomerases [21], [38].
R-loops at CGI promoters were recently implicated in mediating protection against DNA methylation and epigenetic silencing [18]. R-loops at FMR1 likely contribute to the same function for alleles in the normal and premutation ranges, in which the 5′UTR remains essentially unmethylated. Additionally, R-loop formation, by fostering a more open chromatin environment [29], is possibly responsible for the increased transcription resulting from repeat expansion in FMR1, although there is no direct evidence for this suggestion. The FMR1 CGI is unique among CGIs in that it undergoes hypermethylation and silencing for full mutation alleles (≥200 CGG repeats), which suggests that, above a certain genetically-encoded threshold, the protection force operating at FMR1 may be overcome by an as-yet-undefined silencing mechanism (either at the DNA or histone level). This transition between protection and silencing regimes could in fact be mediated by the unusual nature of R-loops formed through expanded CGG repeats. We provide evidence here that the non-template strand of FMR1 R-loops presents stretches that are refractory to bisulfite footprinting (Figure 4), indicative of possible hairpin-like structures, which is consistent with the well-documented propensity of trinucleotide CGG repeats to fold into higher-order structures [39], [40]. Such structural characteristics distinguish FMR1 from other non-repetitive R-loop-forming CGI promoters [18], [19] and even from repetitive R-loop-forming sequences such as class-switch regions [26], [41].
Our non-denaturing bisulfite footprinting data also show that regions of ssDNA often appear to be located downstream of the CGG repeats themselves, particularly for larger repeats (Figure 4). This pattern is unusual in that GC skew should favor R-loop initiation within the repeats. It is possible that hairpin formation on the displaced G-rich strand may cause collapse of the R-loop structure by imposing torsional stress on the RNA:DNA hybrid (Figure 5). Such hairpin-mediated interconversion between a “regular” R-loop and a “collapsed” R-loop would explain the patterns observed and would be compatible with the unique sequence characteristics of the region. Interestingly, the structural complexities observed at FMR1 may have relevance to the transition from an active to a silenced state characteristic of fully expanded repeats. For instance, DNMT1, the most powerful DNA methyltransferase in human cell extracts, recognizes structured/hairpin DNA as a substrate for methylation [42]. This recognition could seed DNA methylation inside the repeats before spreading up - and down-stream over the rest of the UTR/promoter. Alternatively, collapse of the RNA:DNA hybrid inside the CGG repeats could potentially disrupt the protective effect of R-loops against DNA methylation, which has been observed at other CGI promoters [18]. Previous studies of FMR1 hypermethylation have not mapped high-resolution methylation patterns inside the repeats themselves [43], [44], and thus would easily overlook this repeat-centric model.
Fig. 5. Model of proposed CGG-repeat effects on the FMR1 R-loop. 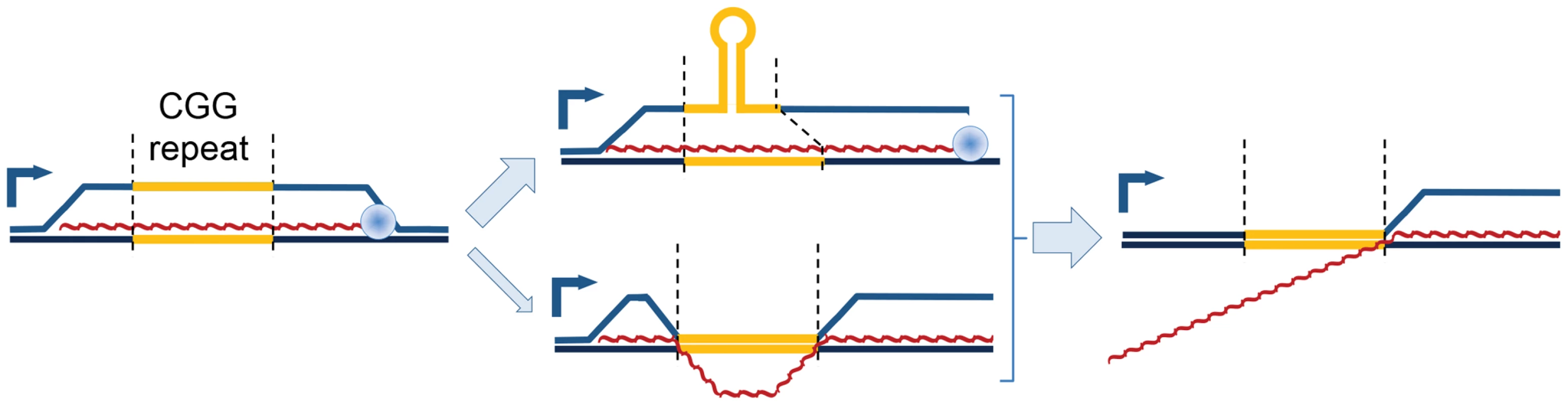
R-loops that span the FMR1 CGG-repeat region (yellow) during transcription could adopt a hairpin structure within the displaced CGG-repeat strand, thus protecting the CGG-repeat region from bisulfite conversion while leaving both 5′ and 3′ flanking regions exposed; the CGG-repeat is known to form such structures readily in vitro [61]. An alternative structure, although less energetically feasible, would involve maintenance of R-loops flanking the CGG-repeat element, which has collapsed into a dsDNA structure again. Loss of the upstream R-loop region would explain the absence of bisulfite conversion in ∼25–50% of molecules (Figure 4). Red, nascent RNA transcript; 90° arrow, start of transcription; blue sphere, Pol II. In addition to changes in secondary structure, repeat expansion and increased transcription could result in R-loop-driven activation of the DNA damage response and genomic instability [22], [27], [28], [45]–[49]. R-loops in the inappropriate context or timing result in DNA breaks, as indicated by recruitment of γH2AX. In this regard, we have previously reported activation of the double-stranded-break repair pathway in this same episomal system, but only in highly transcribed expanded CGG repeats [36]. Even though elevated exogenous expression likely exaggerates this effect in the model system, γH2AX is observed in the characteristic intranuclear protein inclusions of post-mortem neurons in FXTAS patients [36]. In addition to potential involvement in FXTAS pathology, R-loop formation and/or DNA damage are responsible for genomic instability generally [22], [48], and at FMR1 specifically [50], [51], including the eponymous fragile site [52]. In addition to toxicity and instability, DNA damage has been linked to aberrant DNA methylation [53]; increased R-loop formation and/or increased damage of a full mutation CGG R-loop could overcome the protective features of 5′UTR CGI R-loops and trigger the methylation and silencing characteristic of FXS.
We introduce here a previously unrecognized molecular feature of the FMR1 gene that is influenced by expansion of the CGG-repeat element. R-loop formation is a normal and important feature of the FMR1 promoter, but expansion of the CGG repeats, and the associated increase in transcription, results in increased formation of longer R-loops that are more prone to folding into complex secondary structures, which could trigger instability and hypermethylation associated with FMR1-repeat expansion. This discovery provides a novel area of inquiry for understanding the aberrant cellular responses to CGG-repeat expansion at FMR1, and at transcribed trinucleotide-repeat loci throughout the genome.
Materials and Methods
Cell Culture
Human dermal fibroblasts were originally cultured from skin biopsies acquired under an IRB-approved protocol, as previously described [54]. Cells were grown at 37°C, 5% CO2 in a 50∶50 mix of RPMI-1640, supplemented with 1× Amphotericin B (JR Scientific, Woodland, CA), 1× Penicillin-Streptomycin-Glutamine (Life Technologies, Carlsbad, CA), 1× MEM Non-Essential Amino Acids Solution (Life Technologies), and 10% Fetal Bovine Serum (JR Scientific) and AmnioMAX C100 media (Life Technologies). Fibroblasts were harvested at 80% confluency to avoid decreased transcription associated with contact inhibition.
SK-N-MC-rtTA cell lines harboring expanded CGG-repeat episomes were created, as previously described [36]. These cells were grown in DMEM (Life Technologies) +10% Tet-system-approved fetal bovine serum (Clontech, Mountain View, CA) and 1× Penicillin-streptomycin (Life Technologies) at 37°C, 5% CO2. DOX media was prepared from 1 mg/mL stock doxycycline hyclate (Sigma-Aldrich, St. Louis, MO) dissolved in sterile water. For the DOX washout, DOX media was aspirated and cells were washed once with DPBS (Life Technologies) before adding DOX-free media.
Harvesting Nucleic Acids for DRIP
Adherent cells were trypsinized (0.25% trypsin; Life Technologies) for fibroblasts and 0.05% trypsin (JR Scientific) for SK-N-MC cells for 4 minutes at 37°C before quenching with an equal volume of media and pelleting at low speed (200 RCF). Cell pellets were washed with DPBS (Life Technologies) and divided for DRIP or RNA harvests. Cell pellets for RNA harvest were lysed in RLT buffer (Qiagen, Hilden, Germany) and frozen at −80°C before processing at a later date according to the RNeasy kit (Qiagen). Cell pellets for DRIP were resuspended in 4 mL of 10 mM Tris-HCl, 10 mM EDTA, 100 mM NaCl pH 8, lysed with 0.5% SDS, and digested with 400 units of Proteinase K (Thermo Fisher Scientific, Waltham, MA) at 37°C overnight. Cell lysates were then extracted once with 1 volume of equilibrated phenol pH 8 (USB, Cleveland, OH) and twice with 1 volume of chloroform (Sigma-Aldrich). DNA was precipitated with 1 volume of isopropanol and 300 mM sodium acetate, and was swirled out of solution with a glass shepherd's hook. The DNA pellet was washed twice by rinsing the hook with 400 µL of 70% ethanol, and was rehydrated in 10 mM Tris-HCl pH 8.
DRIP
Harvested nucleic acids (∼50 µg) were digested using a restriction enzyme cocktail (20 units each of EcoRI, HindIII, BsrGI, XbaI) (New England Biolabs, Ipswich, MA; NEB) overnight at 37°C in 1× NEBuffer 2. Digests were cleaned by phenol and chloroform extraction followed by precipitation in isopropanol. The resulting fragmented DNA was pelleted at full speed (16,100× g) at 4°C and washed twice with 70% ethanol. Air-dried pellets were rehydrated in 10 mM Tris-HCl pH 7.5, 1 mM EDTA (TE).
We adapted the previously described DRIP protocol [18]. Six to eight µg of digested nucleic acids were diluted in 450 µL of TE, and 10 µL was reserved as input for qPCR. Fifty-two µL of 10× IP buffer was added for a final buffer concentration of 10 mM sodium phosphate, 140 mM sodium chloride, 0.05% Triton X-100, and 20 µL of S9.6 antibody (1 mg/ml; prepared from ascites, as previously described [18]). The samples were incubated with the antibody at 4°C for 2 hours. This incubation and all wash steps were performed on a rotisserie mixer. Forty µL of Protein A/G Agarose slurry (Pierce, Rockford, IL) was washed twice with 800 µL of 1× IP buffer for 5 minutes at room temperature. After adding agarose slurry to each sample, they were incubated for 2 hours at 4°C. Each DRIP was then washed three times with 700 µL 1× IP buffer for 10 minutes per wash at room temperature. After the final wash, the agarose slurry was resuspended in 250 µL of 1× IP buffer and incubated with 60 units of Proteinase K for 30 minutes at 50°C. Digested DRIP samples were then cleaned with phenol/chloroform extraction and isopropanol precipitation. Air-dried DRIP pellets were resuspended in 80 µL of 10 mM Tris-HCl pH 8.
We used 12 µL reactions with Sensi-FAST Lo-Rox 2× qPCR mix (Bioline, London, UK) to assay for genomic loci: FMR1 (200 nM each) (F: TTGCCCCTTAGTTCCCTGAG; R:TCTTCCATCAGTGCAGACCA), MYADM (300 nM each) (F: CGTAGGTGCCCTAGTTGGAG; R: TCCATTCTCATTCCCAAACC), and ZNF554 (300 nM each) (F: CGGGGAAAAGCCCTATAAAT; R: TCCACATTCACTGCATTCGT). For the episomal DRIP experiments, we assayed for EGFP (F: TCAAGATCCGCCACAACATC; R:TTCTCGTTGGGGTCTTTGCT) and the pCEP4 backbone (F:ATCCCCATCCCTACCGTCCA; R:CCCCATCCTCCGAACCATCC) using 5 µL of 1∶500 diluted template or 5 µL undiluted DRIP output (from 80 µL total). Reactions were incubated with the following program on a Viia 7 System (Life Technologies): 50°C 2 minutes, 95°C 10 minutes, 40 cycles of 95°C 15 seconds, 64°C 1 minute, followed by a melt curve: 95°C 15 seconds, 60°C 1 minute, 0.05°C/second to 95°C 15 seconds. For each DRIP sample, 5 µL of the output and 5 µL of diluted input (1∶100) were assayed in triplicate. Fold enrichment for a given locus (i.e., FMR1 or EGFP) was calculated using the comparative Ct method [55], relative first to input and then to the appropriate reference (i.e., ZNF554 or pCEP4 backbone).
Comparison of fold enrichment between premutation (4 subjects, 2–4 independent replications per subject; n = 10 experiments) and control (3 subjects, 2–3 independent replications per subject; n = 8 experiments) subjects (Figure 2) was based on a linear mixed-effects model to account for correlation between repeated measurements on the same subjects. The analysis was done using SAS version 9.3.
Non-denaturing Sodium Bisulfite Mapping
Harvested nucleic acids (4–10 µg) were digested with HindIII (20 units, ∼5 hours at 37°C; NEB) and then treated with the sodium bisulfite conversion mix from the EZ-DNA Methylation Kit (Zymo Research, Irvine, CA) overnight at 37°C. Bisulfite-treated DNA was then desulphonated and cleaned according to kit protocol and was eluted in 10 µL 10 mM Tris-HCl pH 8.
Bisulfite-treated DNA was amplified using a method adapted for CGG-repeat amplification [56]. One to two µL of bisulfite-treated DNA was amplified in a 30 µL reaction with 0.5 mM dNTPs, 2.25 M betaine (Sigma), 333 nM of each primer in 1× buffer and 0.2 µL of enzyme mix from the Expand Long Template Kit (Roche, Basel, Switzerland). Enzyme and buffer were added after 8 minutes at 98°C, followed by an additional 2 minutes at 98°C, then 10 cycles at 97°C for 35 seconds, 64°C for 35 seconds, 68°C for 4 minutes, 25 cycles at 97°C for 35 seconds, 64°C for 35 seconds, 68°C for 4 minutes, plus a 20-second increment for each cycle, and a final extension at 68°C for 10 minutes. In order to successfully and cleanly amplify through the bisulfite-converted CGG repeats, we used two rounds of amplification with a nested primer set (first round: F:GAGGGAACAGCGTTGATCACGTG R: CACTTAACACCAATTTCAACCCTTCCCACC; second round: F: GGAACAGCGTTGATCACGTGACGTGGTTTC R: CTTCCCTCCCAACAACATCCCACCAAAC).
PCR-amplified DNA was sub-cloned using the Qiagen PCR Cloning Kit. Chemically competent E. coli Top10 cells (Life Technologies) were transformed by heat-shock with ligated plasmid, and were grown overnight at 37°C on LB agar plates with 100 mg/ml ampicillin selection. Picked colonies were grown in 4 mL LB broth with 100 mg/ml ampicillin at 30°C with 150 rpm shaking overnight; plasmid DNA was extracted using the Qiagen plasmid miniprep kit.
Plasmid DNA PCR clones were sequenced (Davis Sequencing, Davis, CA) with M13R or SP6 primers, depending on orientation of the PCR insert. Clean sequence clones were then aligned to an unconverted reference sequence with Clustal W2 [57] to score cytosine conversion events.
The full-length cDNA for human RNASEH1 (ATCC, Manassas, VA) was PCR-amplified, excluding the first 26 amino acids of the protein corresponding to the mitochondrial localization signal [58]. The amplified fragment was recloned in frame in a modified pMAL vector [59] to generate an MBP-RNASEH1 fusion protein. Protein expression was induced for 2 hours at 37°C in E. coli Rosetta cells grown in exponential phase in Terrific Broth. Cells were harvested and lysed with a microfluidizer in amylose buffer (10% glycerol, 25 mM Tris-HCl pH 7.5, 250 mM NaCl, 1 mM DTT, 0.5 mM EDTA supplemented with complete protease inhibitor cocktail; Roche), and the lysate was spun for 1 hour at 30,000× g. The supernatant was then applied to a 20 ml amylose column (NEB) equilibrated in 1× amylose buffer, after which the column was washed with 10 column volumes of binding buffer. The MBP-RNASEH1 protein was eluted in batch in binding buffer supplemented with 20 mM maltose. The protein was then dialyzed against Q buffer (20 mM Tris-HCl pH 7.5, 50 mM NaCl, 0.1 mM β-mercaptoethanol) overnight at 4°C and was applied to a pre-equilibrated 5 ml FastFlow Q column (GE Healthcare, Little Chalfont, UK) using an Akta FPLC system. The protein mostly flowed through. The flow-through was then re-applied to a Mono-Q column (GE Heathcare) to separate the protein from any contaminating nucleic-acid species. The flow-through was again collected, concentrated, and dialyzed against storage buffer (20 mM Tris-HCl pH 7.5, 50 mM NaCl, 1 mM DTT, 0.1 mM EDTA, 20% glycerol), then aliquoted and snap frozen in liquid nitrogen before storage at −80°C. The concentration of the preparation was calculated to be 17.7 µM (1.3 mg/ml) using an extinction coefficient of 112,710 M−1cm−1. The pMAR22 expression vector for the heterotrimeric RNASEH2 complex was a kind gift from Dr. Reijns; the complex was purified essentially as described [60]. The protein was stored as described for RNASEH1. The concentration of the preparation was calculated to be 15 µM (1.34 mg/ml) using an extinction coefficient of 81,050 M−1cm−1. Both preparations were ∼98% pure, as judged from Coomassie-stained SDS-PAGE gels, and gave expected sizes of either one single band for MBP-RNASEH1 or three equimolar bands for the RNASEH2 complex. Both preparations were devoid of detectable endo - or exonuclease activity after incubating 1 µl of undiluted protein with double-stranded circular or linear DNA substrates for 4 hours at 37°C (data not shown). Both preparations were highly active even under 10,000-fold diluted concentrations against artificial R-loop substrates prepared by in vitro transcription (data not shown).
Supporting Information
Zdroje
1. HagermanP (2013) Fragile X-associated tremor/ataxia syndrome (FXTAS): pathology and mechanisms. Acta Neuropathol 126 : 1–19.
2. AmiriK, HagermanRJ, HagermanPJ (2008) Fragile X-associated tremor/ataxia syndrome: an aging face of the fragile X gene. Arch Neurol 65 : 19–25.
3. FuYH, KuhlDP, PizzutiA, PierettiM, SutcliffeJS, et al. (1991) Variation of the CGG repeat at the fragile X site results in genetic instability: resolution of the Sherman paradox. Cell 67 : 1047–1058.
4. ChonchaiyaW, SchneiderA, HagermanRJ (2009) Fragile X: a family of disorders. Adv Pediatr 56 : 165–186.
5. OberleI, RousseauF, HeitzD, KretzC, DevysD, et al. (1991) Instability of a 550-base pair DNA segment and abnormal methylation in fragile X syndrome. Science 252 : 1097–1102.
6. EichlerEE, HoldenJJ, PopovichBW, ReissAL, SnowK, et al. (1994) Length of uninterrupted CGG repeats determines instability in the FMR1 gene. Nat Genet 8 : 88–94.
7. SullivanSD, WeltC, ShermanS (2011) FMR1 and the continuum of primary ovarian insufficiency. Semin Reprod Med 29 : 299–307.
8. RendaMM, VoigtRG, Babovic-VuksanovicD, HighsmithWE, VinsonSS, et al. (2012) Neurodevelopmental disabilities in children with intermediate and premutation range fragile X cytosine-guanine-guanine expansions. J Child Neurol 29 : 326–30.
9. HagermanR, AuJ, HagermanP (2011) FMR1 premutation and full mutation molecular mechanisms related to autism. J Neurodev Disord 3 : 211–224.
10. TassoneF, HagermanRJ, TaylorAK, GaneLW, GodfreyTE, et al. (2000) Elevated levels of FMR1 mRNA in carrier males: a new mechanism of involvement in the fragile-X syndrome. Am J Hum Genet 66 : 6–15.
11. Garcia-ArocenaD, HagermanPJ (2010) Advances in understanding the molecular basis of FXTAS. Hum Mol Genet 19: R83–89.
12. Ross-IntaC, Omanska-KlusekA, WongS, BarrowC, Garcia-ArocenaD, et al. (2010) Evidence of mitochondrial dysfunction in fragile X-associated tremor/ataxia syndrome. Biochem J 429 : 545–552.
13. SellierC, RauF, LiuY, TassoneF, HukemaRK, et al. (2010) Sam68 sequestration and partial loss of function are associated with splicing alterations in FXTAS patients. EMBO J 29 : 1248–1261.
14. SellierC, FreyermuthF, TabetR, TranT, HeF, et al. (2013) Sequestration of DROSHA and DGCR8 by expanded CGG RNA repeats alters microRNA processing in fragile X-associated tremor/ataxia syndrome. Cell Rep 3 : 869–880.
15. ToddPK, OhSY, KransA, HeF, SellierC, et al. (2013) CGG repeat-associated translation mediates neurodegeneration in fragile X tremor ataxia syndrome. Neuron 78 : 440–455.
16. RoyD, YuK, LieberMR (2008) Mechanism of R-loop formation at immunoglobulin class switch sequences. Mol Cell Biol 28 : 50–60.
17. MasukataH, TomizawaJ (1990) A mechanism of formation of a persistent hybrid between elongating RNA and template DNA. Cell 62 : 331–338.
18. GinnoPA, LottPL, ChristensenHC, KorfI, ChedinF (2012) R-loop formation is a distinctive characteristic of unmethylated human CpG island promoters. Mol Cell 45 : 814–825.
19. GinnoPA, LimYW, LottPL, KorfI, ChedinF (2013) GC skew at the 5′ and 3′ ends of human genes links R-loop formation to epigenetic regulation and transcription termination. Genome Res 23 : 1590–1600.
20. ReddyK, TamM, BowaterRP, BarberM, TomlinsonM, et al. (2011) Determinants of R-loop formation at convergent bidirectionally transcribed trinucleotide repeats. Nucleic Acids Res 39 : 1749–1762.
21. Skourti-StathakiK, ProudfootNJ, GromakN (2011) Human senataxin resolves RNA/DNA hybrids formed at transcriptional pause sites to promote Xrn2-dependent termination. Mol Cell 42 : 794–805.
22. McIvorEI, PolakU, NapieralaM (2010) New insights into repeat instability: role of RNA*DNA hybrids. RNA Biol 7 : 551–558.
23. AguileraA, Garcia-MuseT (2012) R loops: from transcription byproducts to threats to genome stability. Mol Cell 46 : 115–124.
24. HelmrichA, BallarinoM, NudlerE, ToraL (2013) Transcription-replication encounters, consequences and genomic instability. Nat Struct Mol Biol 20 : 412–418.
25. TsaiAG, LieberMR (2010) Mechanisms of chromosomal rearrangement in the human genome. BMC Genomics 11 Suppl 1: S1.
26. YuK, ChedinF, HsiehCL, WilsonTE, LieberMR (2003) R-loops at immunoglobulin class switch regions in the chromosomes of stimulated B cells. Nat Immunol 4 : 442–451.
27. PaulsenRD, SoniDV, WollmanR, HahnAT, YeeMC, et al. (2009) A genome-wide siRNA screen reveals diverse cellular processes and pathways that mediate genome stability. Mol Cell 35 : 228–239.
28. SordetO, NakamuraAJ, RedonCE, PommierY (2010) DNA double-strand breaks and ATM activation by transcription-blocking DNA lesions. Cell Cycle 9 : 274–278.
29. PowellWT, CoulsonRL, GonzalesML, CraryFK, WongSS, et al. (2013) R-loop formation at Snord116 mediates topotecan inhibition of Ube3a-antisense and allele-specific chromatin decondensation. Proc Natl Acad Sci U S A 110 : 13938–13943.
30. RoyD, LieberMR (2009) G clustering is important for the initiation of transcription-induced R-loops in vitro, whereas high G density without clustering is sufficient thereafter. Mol Cell Biol 29 : 3124–3133.
31. TassoneF, De RubeisS, CarosiC, La FataG, SerpaG, et al. (2011) Differential usage of transcriptional start sites and polyadenylation sites in FMR1 premutation alleles. Nucleic Acids Res 39 : 6172–6185.
32. BeilinaA, TassoneF, SchwartzPH, SahotaP, HagermanPJ (2004) Redistribution of transcription start sites within the FMR1 promoter region with expansion of the downstream CGG-repeat element. Hum Mol Genet 13 : 543–549.
33. StrausbergRL, FeingoldEA, GrouseLH, DergeJG, KlausnerRD, et al. (2002) Generation and initial analysis of more than 15,000 full-length human and mouse cDNA sequences. Proc Natl Acad Sci U S A 99 : 16899–16903.
34. Gardiner-GardenM, FrommerM (1987) CpG islands in vertebrate genomes. J Mol Biol 196 : 261–282.
35. BoguslawskiSJ, SmithDE, MichalakMA, MickelsonKE, YehleCO, et al. (1986) Characterization of monoclonal antibody to DNA.RNA and its application to immunodetection of hybrids. J Immunol Methods 89 : 123–130.
36. HoemG, RaskeCR, Garcia-ArocenaD, TassoneF, SanchezE, et al. (2011) CGG-repeat length threshold for FMR1 RNA pathogenesis in a cellular model for FXTAS. Hum Mol Genet 20 : 2161–2170.
37. LanderES, LintonLM, BirrenB, NusbaumC, ZodyMC, et al. (2001) Initial sequencing and analysis of the human genome. Nature 409 : 860–921.
38. CerritelliSM, CrouchRJ (2009) Ribonuclease H: the enzymes in eukaryotes. FEBS J 276 : 1494–1505.
39. ChenX, MariappanSV, CatastiP, RatliffR, MoyzisRK, et al. (1995) Hairpins are formed by the single DNA strands of the fragile X triplet repeats: structure and biological implications. Proc Natl Acad Sci U S A 92 : 5199–5203.
40. GacyAM, GoellnerG, JuranicN, MacuraS, McMurrayCT (1995) Trinucleotide repeats that expand in human disease form hairpin structures in vitro. Cell 81 : 533–540.
41. HuangFT, YuK, HsiehCL, LieberMR (2006) Downstream boundary of chromosomal R-loops at murine switch regions: implications for the mechanism of class switch recombination. Proc Natl Acad Sci U S A 103 : 5030–5035.
42. SmithSS, LaayounA, LingemanRG, BakerDJ, RileyJ (1994) Hypermethylation of telomere-like foldbacks at codon 12 of the human c-Ha-ras gene and the trinucleotide repeat of the FMR-1 gene of fragile X. J Mol Biol 243 : 143–151.
43. NaumannA, HochsteinN, WeberS, FanningE, DoerflerW (2009) A distinct DNA-methylation boundary in the 5′-upstream sequence of the FMR1 promoter binds nuclear proteins and is lost in fragile X syndrome. Am J Hum Genet 85 : 606–616.
44. StogerR, GenereuxDP, HagermanRJ, HagermanPJ, TassoneF, et al. (2011) Testing the FMR1 promoter for mosaicism in DNA methylation among CpG sites, strands, and cells in FMR1-expressing males with fragile X syndrome. PLoS One 6: e23648.
45. HuertasP, AguileraA (2003) Cotranscriptionally formed DNA:RNA hybrids mediate transcription elongation impairment and transcription-associated recombination. Mol Cell 12 : 711–721.
46. SordetO, RedonCE, Guirouilh-BarbatJ, SmithS, SolierS, et al. (2009) Ataxia telangiectasia mutated activation by transcription - and topoisomerase I-induced DNA double-strand breaks. EMBO Rep 10 : 887–893.
47. MischoHE, Gomez-GonzalezB, GrzechnikP, RondonAG, WeiW, et al. (2011) Yeast Sen1 helicase protects the genome from transcription-associated instability. Mol Cell 41 : 21–32.
48. HelmrichA, BallarinoM, ToraL (2011) Collisions between replication and transcription complexes cause common fragile site instability at the longest human genes. Mol Cell 44 : 966–977.
49. StirlingPC, ChanYA, MinakerSW, AristizabalMJ, BarrettI, et al. (2012) R-loop-mediated genome instability in mRNA cleavage and polyadenylation mutants. Genes Dev 26 : 163–175.
50. EntezamA, UsdinK (2008) ATR protects the genome against CGG.CCG-repeat expansion in Fragile X premutation mice. Nucleic Acids Res 36 : 1050–1056.
51. EntezamA, UsdinK (2009) ATM and ATR protect the genome against two different types of tandem repeat instability in Fragile X premutation mice. Nucleic Acids Res 37 : 6371–6377.
52. KumariD, SommaV, NakamuraAJ, BonnerWM, D'AmbrosioE, et al. (2009) The role of DNA damage response pathways in chromosome fragility in Fragile X syndrome. Nucleic Acids Res 37 : 4385–4392.
53. CuozzoC, PorcelliniA, AngrisanoT, MoranoA, LeeB, et al. (2007) DNA damage, homology-directed repair, and DNA methylation. PLoS Genet 3: e110.
54. Garcia-ArocenaD, YangJE, BrouwerJR, TassoneF, IwahashiC, et al. (2010) Fibroblast phenotype in male carriers of FMR1 premutation alleles. Hum Mol Genet 19 : 299–312.
55. SchmittgenTD, LivakKJ (2008) Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc 3 : 1101–1108.
56. SalutoA, BrussinoA, TassoneF, ArduinoC, CagnoliC, et al. (2005) An enhanced polymerase chain reaction assay to detect pre - and full mutation alleles of the fragile X mental retardation 1 gene. J Mol Diagn 7 : 605–612.
57. LarkinMA, BlackshieldsG, BrownNP, ChennaR, McGettiganPA, et al. (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23 : 2947–2948.
58. CerritelliSM, FrolovaEG, FengC, GrinbergA, LovePE, et al. (2003) Failure to produce mitochondrial DNA results in embryonic lethality in Rnaseh1 null mice. Mol Cell 11 : 807–815.
59. MoarefiAH, ChedinF (2011) ICF syndrome mutations cause a broad spectrum of biochemical defects in DNMT3B-mediated de novo DNA methylation. J Mol Biol 409 : 758–772.
60. ReijnsMA, BubeckD, GibsonLC, GrahamSC, BaillieGS, et al. (2011) The structure of the human RNase H2 complex defines key interaction interfaces relevant to enzyme function and human disease. J Biol Chem 286 : 10530–10539.
61. ZumwaltM, LudwigA, HagermanPJ, DieckmannT (2007) Secondary structure and dynamics of the r(CGG) repeat in the mRNA of the fragile X mental retardation 1 (FMR1) gene. RNA Biol 4 : 93–100.
Štítky
Genetika Reprodukční medicína
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2014 Číslo 4
-
Všechny články tohoto čísla
- The Challenges of Mitochondrial Replacement
- Concocting Cholinergy
- Genome-Wide Diet-Gene Interaction Analyses for Risk of Colorectal Cancer
- Statistical Power to Detect Genetic (Co)Variance of Complex Traits Using SNP Data in Unrelated Samples
- Mouse Pulmonary Adenoma Susceptibility 1 Locus Is an Expression QTL Modulating -4A
- Transcription-Associated R-Loop Formation across the Human CGG-Repeat Region
- Epigenetic Regulation by Heritable RNA
- Protein Quantitative Trait Loci Identify Novel Candidates Modulating Cellular Response to Chemotherapy
- Genome-Wide Profiling of Yeast DNA:RNA Hybrid Prone Sites with DRIP-Chip
- The Mechanism of Gene Targeting in Human Somatic Cells
- A LINE-1 Insertion in DLX6 Is Responsible for Cleft Palate and Mandibular Abnormalities in a Canine Model of Pierre Robin Sequence
- Interaction between Two Timing MicroRNAs Controls Trichome Distribution in
- DNA Glycosylases Involved in Base Excision Repair May Be Associated with Cancer Risk in and Mutation Carriers
- The Myc-Mondo/Mad Complexes Integrate Diverse Longevity Signals
- Evolutionarily Diverged Regulation of X-chromosomal Genes as a Primal Event in Mouse Reproductive Isolation
- Mutations in Conserved Residues of the microRNA Argonaute ALG-1 Identify Separable Functions in ALG-1 miRISC Loading and Target Repression
- Genetic Predisposition to In Situ and Invasive Lobular Carcinoma of the Breast
- Isl1 Directly Controls a Cholinergic Neuronal Identity in the Developing Forebrain and Spinal Cord by Forming Cell Type-Specific Complexes
- A Synthetic Community Approach Reveals Plant Genotypes Affecting the Phyllosphere Microbiota
- The Sequence-Specific Transcription Factor c-Jun Targets Cockayne Syndrome Protein B to Regulate Transcription and Chromatin Structure
- Determining the Control Circuitry of Redox Metabolism at the Genome-Scale
- DNA Repair Pathway Selection Caused by Defects in , , and Telomere Addition Generates Specific Chromosomal Rearrangement Signatures
- Methylome Diversification through Changes in DNA Methyltransferase Sequence Specificity
- Folliculin Regulates Ampk-Dependent Autophagy and Metabolic Stress Survival
- Fine Mapping of Dominant -Linked Incompatibility Alleles in Hybrids
- Unexpected Role of the Steroid-Deficiency Protein Ecdysoneless in Pre-mRNA Splicing
- Three Groups of Transposable Elements with Contrasting Copy Number Dynamics and Host Responses in the Maize ( ssp. ) Genome
- Sox5 Functions as a Fate Switch in Medaka Pigment Cell Development
- Synergistic Interactions between the Molecular and Neuronal Circadian Networks Drive Robust Behavioral Circadian Rhythms in
- Chromatin Landscapes of Retroviral and Transposon Integration Profiles
- Widespread Use of Non-productive Alternative Splice Sites in
- Ras GTPase-Like Protein MglA, a Controller of Bacterial Social-Motility in Myxobacteria, Has Evolved to Control Bacterial Predation by
- Cell Type-Specific Functions of Genes Revealed by Novel Adipocyte and Hepatocyte Circadian Clock Models
- Phenotype Ontologies and Cross-Species Analysis for Translational Research
- Embryogenesis Scales Uniformly across Temperature in Developmentally Diverse Species
- In Pursuit of the Gene: An Interview with James Schwartz
- Molecular Mechanisms of Hypoxic Responses via Unique Roles of Ras1, Cdc24 and Ptp3 in a Human Fungal Pathogen
- Analysis of the Genome and Transcriptome of var. Reveals Complex RNA Expression and Microevolution Leading to Virulence Attenuation
- Genotypic and Functional Impact of HIV-1 Adaptation to Its Host Population during the North American Epidemic
- RNA Editome in Rhesus Macaque Shaped by Purifying Selection
- Proper Actin Ring Formation and Septum Constriction Requires Coordinated Regulation of SIN and MOR Pathways through the Germinal Centre Kinase MST-1
- Interplay of the Serine/Threonine-Kinase StkP and the Paralogs DivIVA and GpsB in Pneumococcal Cell Elongation and Division
- A Quality Control Mechanism Coordinates Meiotic Prophase Events to Promote Crossover Assurance
- CNNM2 Mutations Cause Impaired Brain Development and Seizures in Patients with Hypomagnesemia
- The RNA-Binding Protein QKI Suppresses Cancer-Associated Aberrant Splicing
- Uncoupling Transcription from Covalent Histone Modification
- Rad51–Rad52 Mediated Maintenance of Centromeric Chromatin in
- FRA2A Is a CGG Repeat Expansion Associated with Silencing of
- A General Approach for Haplotype Phasing across the Full Spectrum of Relatedness
- A Novel Highly Divergent Protein Family Identified from a Viviparous Insect by RNA-seq Analysis: A Potential Target for Tsetse Fly-Specific Abortifacients
- A Central Role for in Regulation of Islet Function in Man
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- The Sequence-Specific Transcription Factor c-Jun Targets Cockayne Syndrome Protein B to Regulate Transcription and Chromatin Structure
- The Mechanism of Gene Targeting in Human Somatic Cells
- Genetic Predisposition to In Situ and Invasive Lobular Carcinoma of the Breast
- Widespread Use of Non-productive Alternative Splice Sites in
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



