-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaCell Type-Specific Functions of Genes Revealed by Novel Adipocyte and Hepatocyte Circadian Clock Models
Various aspects of our daily rhythms in physiology and behavior such as the sleep-wake cycle are regulated by endogenous circadian clocks that are present in nearly every cell. It is generally accepted that these oscillators share a similar biochemical negative feedback mechanism, consisting of transcriptional activators and repressors. In this study, we developed cell-autonomous, metabolically relevant clock models in mouse hepatocytes and adipocytes. Each clock model has an integrated luciferase reporter that allows for kinetic luminescence recording with an inexpensive microplate reader and thus is feasible for most laboratories. These models are amenable to high throughput screening of small molecules or genomic entities for impacts on cell-autonomous clocks relevant to metabolism. We validated these new models by RNA interference via lentivirus-mediated knockdown of known clock genes. As expected, we found that many core clock components have similar functions across cell types. To our surprise, however, we also uncovered previously under-appreciated cell type-specific functions of core clock genes, particularly Per1, Per2, and Per3. Because the circadian system is integrated with, and influenced by, the local physiology that is under its control, our studies provide important implications for future studies into cell type-specific mechanisms of various circadian systems.
Published in the journal: . PLoS Genet 10(4): e32767. doi:10.1371/journal.pgen.1004244
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1004244Summary
Various aspects of our daily rhythms in physiology and behavior such as the sleep-wake cycle are regulated by endogenous circadian clocks that are present in nearly every cell. It is generally accepted that these oscillators share a similar biochemical negative feedback mechanism, consisting of transcriptional activators and repressors. In this study, we developed cell-autonomous, metabolically relevant clock models in mouse hepatocytes and adipocytes. Each clock model has an integrated luciferase reporter that allows for kinetic luminescence recording with an inexpensive microplate reader and thus is feasible for most laboratories. These models are amenable to high throughput screening of small molecules or genomic entities for impacts on cell-autonomous clocks relevant to metabolism. We validated these new models by RNA interference via lentivirus-mediated knockdown of known clock genes. As expected, we found that many core clock components have similar functions across cell types. To our surprise, however, we also uncovered previously under-appreciated cell type-specific functions of core clock genes, particularly Per1, Per2, and Per3. Because the circadian system is integrated with, and influenced by, the local physiology that is under its control, our studies provide important implications for future studies into cell type-specific mechanisms of various circadian systems.
Introduction
In mammals, many aspects of daily behavior and physiology such as the sleep-wake cycle, body temperature, and liver metabolism are regulated by endogenous circadian clocks [1], [2]. The circadian time-keeping system is a hierarchical, multi-oscillator network, with the hypothalamic suprachiasmatic nucleus (SCN) acting as a central pacemaker at the top of the hierarchy. The SCN integrates external time cues and, through complex signaling cascades, synchronizes and coordinates extra-SCN oscillators in the brain and in peripheral clocks throughout the body, culminating in overt, coherent circadian rhythms at the organismal level [3], [4]. This time-keeping system is critical for normal physiology and behavior, and its disruption can lead to sleep disorders, metabolic syndrome, premature aging, and cancer (reviewed in [2], [5]).
Virtually all cells in our body have circadian oscillators [6]–[8]. Despite tissue-specific physiological differences, these oscillators share a highly conserved molecular mechanism – a negative feedback loop. This consists of transcriptional activators BMAL1 and CLOCK, which bind to E-box enhancers and activate the transcription of the Per and Cry families of repressors. These repressors then feed back to inhibit BMAL1/CLOCK activity and their own expression [9]. Each molecular component in the core clock loop is represented by multiple paralogs (Bmal1, Bmal2; Clock, Npas2; Per1, Per2, Per3; Cry1, Cry2), which provides the potential for functional redundancy and cell type specificity. In addition, post-translational modifications play critical roles in clock function. For example, the ubiquitin ligases FBXL3 and FBXL21 regulate period length and amplitude through ubiquitin-mediated degradation of CRY proteins and regulation of REV-ERBα activity [10]–[15].
This core clock loop integrates with other transcriptional systems such as the ROR/REV-ERB (via RORE) and DBP/E4BP4 (via D-box) accessory loops [16]. In the RORE loop, retinoic acid receptor-related orphan nuclear receptors (RORA, RORB, and RORC) act as activators, and REV-ERBs (REV-ERBα known as NR1D1 and REV-ERBβ known as NR1D2; referred to hereafter as NR1D1 and NR1D2) act as repressors to regulate rhythmic Bmal1 expression via the RORE cis-element in the Bmal1 promoter [17]–[19]. Similarly, DBP/TEF/HLF and E4BP4 serve as activators and repressors, respectively, to regulate D-box-mediated transcription of genes such as Per3 [16], [19]. These interlocking loops mediated by E-box, RORE, and D-box cis-elements form a complex clock network. These loops act individually or in combination to give rise to distinct waves of gene transcription [16], [20]. For example, while Nr1d1, Bmal1, and Per3 transcription are each mediated primarily by a single cis-element (i.e., primarily E-box, RORE, and D-box, respectively), many other clock genes (e.g., Cry1) are regulated via a combinatorial mechanism involving multiple circadian elements [21].
Cell-based models were instrumental in the identification and characterization of clock gene function in mammals [22], [23]. These studies relied on immortalized cell lines that display circadian rhythms of gene expression in a cell-autonomous manner (i.e., without systemic cues). We and others have used fibroblasts derived from clock component mutant mice expressing a clock gene reporter [17], [21], [24]. Cellular clock models for comprehensive genetic analysis, however, have so far been limited to 3T3 mouse fibroblasts and U2OS human osteosarcoma cells [22], [25], [26]. In the U2OS model, knockdowns of all clock components have been evaluated for impact on period length and amplitude [27]. In mice, 3T3 fibroblasts [22], [28] and more recently MMH-D3 hepatocytes [29] have been introduced as cellular clock models; however, unlike the U2OS model, these models haven't been fully characterized genetically.
An implicit assumption in all these studies is that the clock works the same way in all cell and tissue types, such that gene function determined in one cell or tissue type applies to all cells, regardless of local physiological inputs to the clock. However, while 3T3 cells may be an appropriate model of the fibroblast clock, it is likely not an appropriate model for other cells. Recent studies point to bidirectional interactions between circadian clocks and other cellular and physiological processes. Thus, the circadian system is integrated with, and influenced by, the local physiology. Of particular interest is the reciprocal interaction between clock function and metabolism [5], [30]–[33]. However, as yet, there aren't any characterized cellular models appropriate for the study of clock control of metabolism. Therefore, to reveal cell type-specific molecular, cellular, and physiological mechanisms of circadian clocks, new cell-autonomous, physiologically relevant peripheral clock models are needed.
To facilitate cell type-specific genetic characterization, we explored mouse cell lines relevant to major metabolic functions, focusing on 3T3-L1 adipocytes and MMH-D3 hepatocytes. The mouse 3T3-L1 adipocyte cell line reflects adipose tissue function and has been pivotal in advancing the understanding of basic cellular mechanisms associated with diabetes, obesity, and related disorders in thousands of studies (e.g., [34], [35]). In recent years, mouse MMH-D3 hepatocytes have become a prominent model reflecting hepatocyte function in the liver [36], [37]. Both cell lines have been shown to exhibit rhythmic expression of clock genes and other genes that are involved in and modulated by local physiology [29], [38], [39].
In this study, we used luciferase reporters of clock gene expression and established three high amplitude cell-autonomous clock models: 3T3 fibroblasts, 3T3-L1 adipocytes, and MMH-D3 hepatocytes, with 3T3 fibroblasts as a reference model. These reporter cells displayed persistent, high amplitude rhythms, which allowed longitudinal recording of clock gene rhythms with high temporal resolution using an inexpensive off-the-shelf microplate reader. For genetic perturbations, we developed a pipeline to produce high-quality lentiviral shRNAs to knock down any gene of interest, and validated these cellular models with shRNAs against a selected panel of known clock genes. We show that knockdown of many clock genes resulted in expected phenotypes in all tested cell lines. Unexpectedly, however, we also observed cell type-specific knockdown phenotypes, particularly within the Per gene family. This study has important implications for the tissue-specific mechanisms of circadian clocks.
Results and Discussion
Development of New Cell-Autonomous Clock Models
As an initial effort to develop new cellular clock models pertinent to metabolism, we screened cell lines for robust rhythms and chose 3T3-L1 adipocytes and MMH-D3 hepatocytes. We introduced a lentiviral reporter harboring the rapidly degradable firefly luciferase (dLuc) gene under the control of either mouse Per2 or Bmal1 gene promoters into cells [23]. Whereas the 3T3 reporter cells were directly used in bioluminescence recording, 3T3-L1 and MMH-D3 cells were first differentiated into mature adipocytes and hepatocytes, respectively, prior to recording. These cells displayed persistent bioluminescence rhythms in 35 mm culture dishes monitored in a LumiCycle luminometer (Figure 1A). In each cell line, Per2-dLuc and Bmal1-dLuc reporters displayed anti-phasic rhythms of bioluminescence, consistent with the function of E-box - and RORE-containing promoters in regulating distinct and opposite phases of gene expression.
Fig. 1. Fibroblasts, adipocytes, and hepatocytes display bioluminescence rhythms. 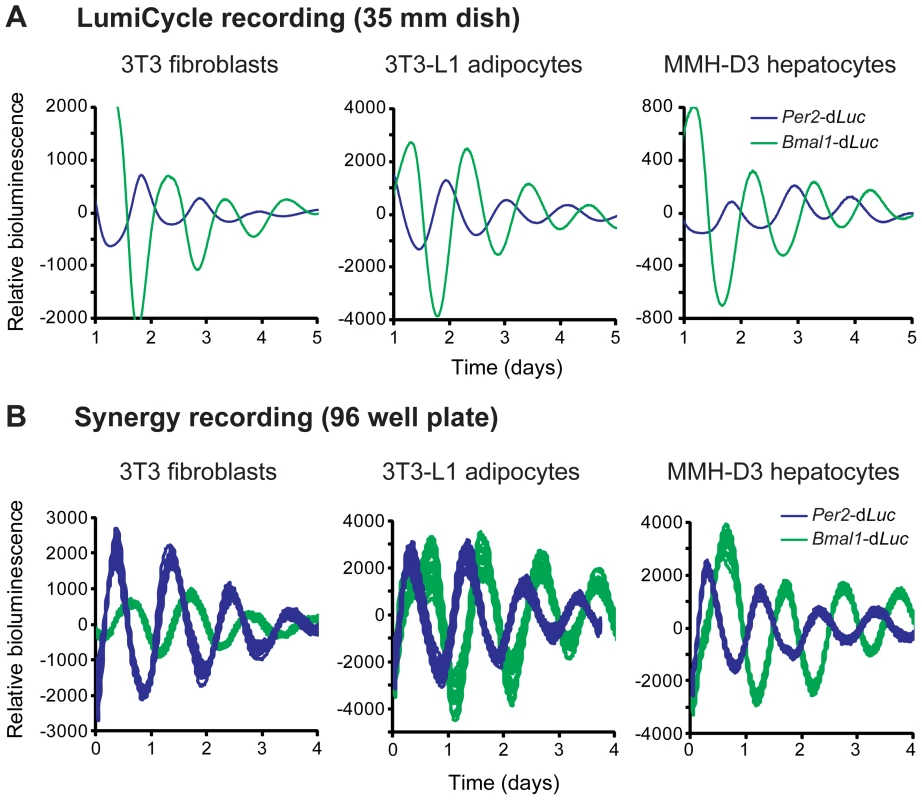
(A) Representative bioluminescence rhythms of reporter cells recorded in a LumiCycle luminometer on 35 mm dishes. Reporter cells were generated via lentiviral infection of either Per2-dLuc or Bmal1-dLuc luciferase reporter, and infected cell populations were recorded in a LumiCycle. Baseline-subtracted bioluminescence data of both reporter lines are plotted together to show the expected, approximately anti-phasic reporter expression for each cell type. (B) Representative bioluminescence rhythms of homogenous clonal cell lines recorded in a Synergy microplate reader on 96 well plates. Baseline-subtracted bioluminescence data of selected clonal lines representing both reporter types are plotted together to show anti-phasic reporter expression for each cell type. High reproducibility is illustrated by showing overlapping traces from 24 of the 96 wells for each reporter. The period lengths are highly consistent (mean ± SD, n = 24 for each line): 3T3 Per2-dLuc, 25.62 hr±0.21; 3T3 Bmal1-dLuc, 26.72 hr±0.31; 3T3-L1 Per2-dLuc, 24.60 hr±0.32; 3T3-L1 Bmal1-dLuc, 25.01 hr±0.19; MMH-D3 Per2-dLuc, 24.49 hr±0.18; MMH-D3 Bmal1-dLuc, 25.33 hr±0.13. Next, we adapted the LumiCycle reporter assay to high-throughput screening (HTS) formats on 96 well plates. For this, we performed single cell cloning and selected clonal cell lines that expressed high levels of bioluminescence. These reporter lines displayed persistent rhythms under optimized growth conditions when monitored on a microplate reader (Synergy 2 SL) with highly consistent period lengths (Figure 1B). These highly reproducible rhythms seen in 96 well plates were similar to those in the LumiCycle, a lower throughput but much more expensive recorder. Therefore, these lines represent a tangible advantage to many labs interested in exploring circadian biology in these metabolically relevant cell lines.
Generation of Lentiviral shRNAs for Gene Knockdown
For genetic perturbations, we developed a pipeline to produce high-quality, validated lentiviral shRNA vectors to knock down any mouse gene. We chose lentiviral shRNAs over transfected siRNAs because lentivirus-mediated delivery mediates potent transduction and stable integration in both dividing and non-dividing cells of various types in vitro and in vivo, thus circumventing low transfection efficiency for certain cells. We designed 6 target oligonucleotide sequences for each gene and cloned the shRNA expression cassette into the lentiviral pLL3.7GW Gateway vector, in which shRNA expression is under the control of the mouse U6 promoter, as we reported previously (Figure S1) [17]. Infectious lentiviral particles were produced in 293T cells using standard procedures and used to infect reporter cells. Infection efficiency was estimated by observing GFP co-expressed from a separate expression cassette under control of the CMV promoter (Figure S1).
We used this pipeline to generate a panel of shRNA constructs targeting the following selected 13 clock genes: Bmal1, Bmal2, Clock, Npas2 (core loop activators); Per1, Per2, Per3, Cry1, Cry2 (core loop repressors); Fbxl3 (core loop post-translational modifier); Nr1d1, Nr1d2 (RORE repressors); and E4bp4 (D-box repressor). Because of the more prominent roles of repressors in clock function, we chose to examine Nr1d1, Nr1d2, and E4bp4, the RORE and D-box negative factors, rather than the corresponding activators [17], [40], [41].
We tested shRNA knockdown (KD) efficiency for the 13 clock genes. Co-transfection of shRNA with Flag-tagged cDNA in 293T cells followed by Western blot analysis showed efficient KD of each gene at the protein level by at least two shRNAs (Figure S2). To check the KD efficiency for endogenous gene expression, mRNA levels of targeted clock genes were also measured using qPCR. For each gene, at least two shRNAs were effective in knocking down gene expression, a requirement to filter out off-target effects of shRNAs [42]. The versatility and efficiency of lentiviral shRNA allowed us to study all 13 known clock genes in all three cell type-specific clock models in parallel, which allows direct phenotypic comparison.
Ubiquitous Functions of Bmal1, Clock, Cry1, Cry2, and Fbxl3
Knockdown of Bmal1, Clock, Cry1, Cry2, and Fbxl3 in all three cellular models resulted in expected phenotypes similar to those in LumiCycle assays using 35 mm dishes and consistent with previous knockout and knockdown studies using human and mouse cellular models [17], [27], [31], [43]–[45]. Specifically, KD of Bmal1 or Clock results in rapid damping or arrhythmicity (Figure 2A and Tables 1, S1, S2, S3); Cry1 KD leads to low amplitude or rapid damping depending on KD efficiency, whereas Cry2 KD lengthens period and increases rhythm amplitude (Figure 2B). The phenotypic defects correlate with KD efficiency of the endogenous genes by the individual shRNAs as determined by qPCR analysis. Taken together, our data demonstrate that Bmal1, Clock, Cry1, and Cry2 play similar roles in the clock mechanism across tested cell types, which provides validation for the three cellular models.
Fig. 2. Knockdowns of Bmal1, Clock, Cry1, Cry2, and Fbxl3 lead to cell type-ubiquitous circadian phenotypes. 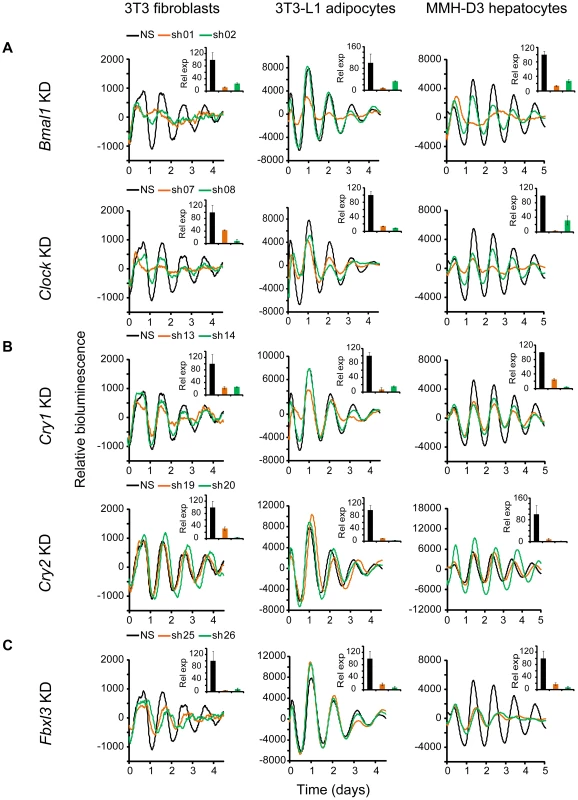
Bioluminescence expression patterns upon KD of Bmal1 or Clock (A), Cry1 or Cry2 (B), and Fbxl3 (C) in all three cell types. For clock phenotyping, both reporters were used for each cell line and phenotypes were independent of the reporter used. For phenotyping, we selected 3T3 cells expressing the Bmal1-dLuc reporter, and 3T3-L1 and MMH-D3 cells expressing the Per2-dLuc reporter. Cells were infected with specific lentiviral shRNAs as indicated. Real-time bioluminescence expression was recorded by Synergy microplate reader as in Figure 1. Out of the 6 shRNAs tested, two validated shRNAs (orange and green) are shown. NS, non-specific shRNA as control (black). While KD of Bmal1 or Clock resulted in low amplitude, rapid damping or arrhythmicity, Fbxl3 KD led to long period and low amplitude in 3T3, long period in 3T3-L1, and low amplitude and rapid damping in MMH-D3 cells. Cry1 KD caused rapid damping or low amplitude, and Cry2 KD lengthened period and increased rhythm amplitude. Bioluminescence data are representative of four independent experiments for 3T3 and 3T3-L1 cells, and three independent experiments for MMH-D3 cells. Knockdown of endogenous mRNA expression in non-synchronized cells was determined by qPCR (insert). Values for each gene are expressed as percentage of gene expression in NS control cells. qPCR data are mean ± SD (two samples/wells from one experiment). Tab. 1. Summary of knockdown phenotypes. 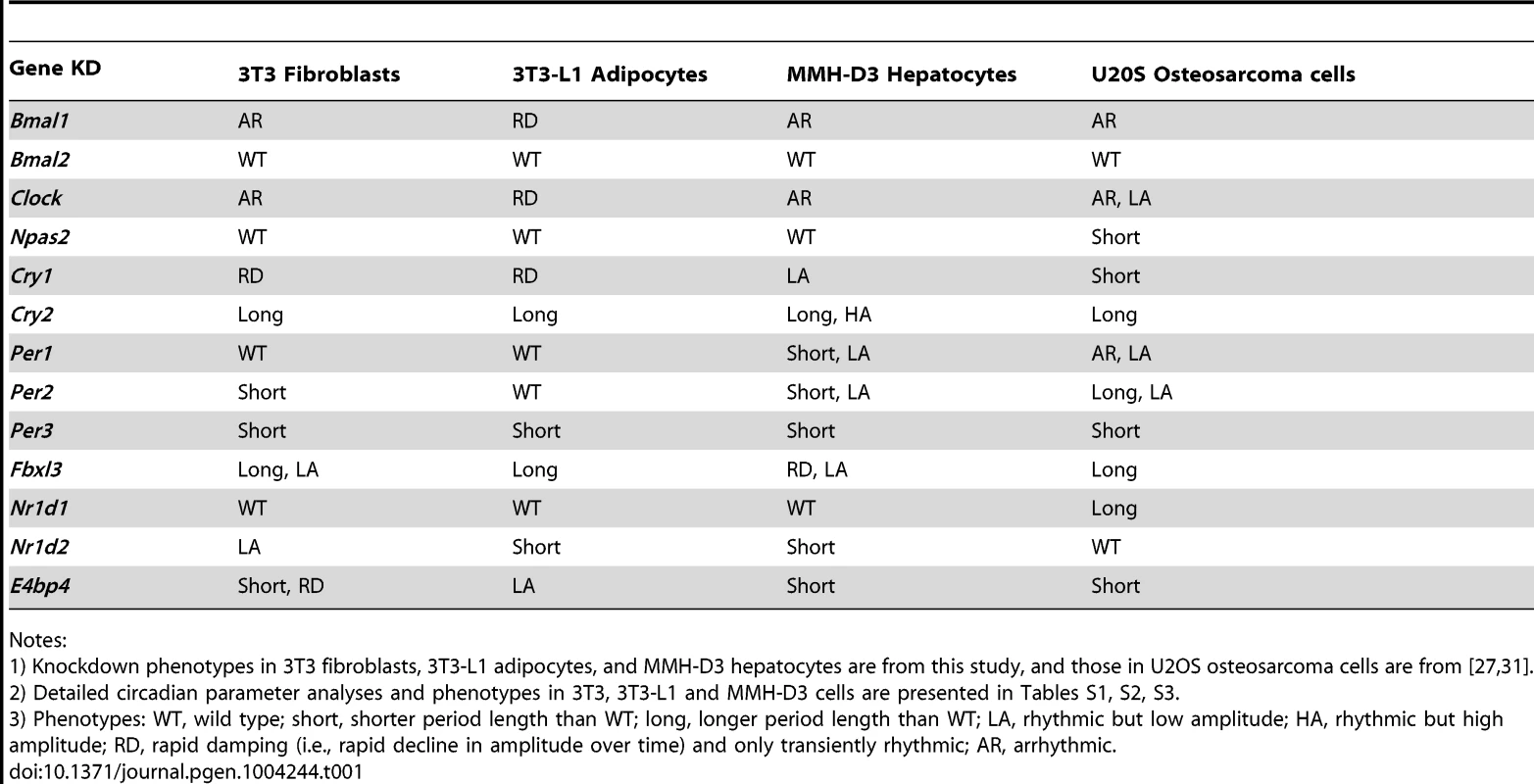
Notes: Knockdown of Bmal2 and Npas2 did not exhibit any obvious circadian phenotypes in any of the three cellular models (Figure S3), even though their expression was knocked down to levels similar to those for Bmal1 or Clock (Figure 2A and Table 1). These results are consistent with absence of observable circadian phenotypes of liver and lung tissues from Npas2−/− mice [46], though these mice do have deficits in circadian behavior and sleep homeostasis [47]. Despite potential functional redundancy between Bmal1 and Bmal2 [45], [48], and between Clock and Npas2 [46], our data suggest that Bmal2 and Npas2 are not necessary for the clock to operate in these cells.
We show that Fbxl3 KD caused long period and low amplitude in 3T3, long period in 3T3-L1, and low amplitude and rapid damping in MMH-D3 cells (Figure 2C). This cell-autonomous phenotype is much more extreme compared to the relatively modest period-lengthening phenotypes seen at the SCN tissue and behavioral levels [10]–[12], or in human U2OS cells [27], [44]. Notably, although KD of Fbxl3 or Cry2 both produced long periods, Cry2 down-regulation increased rhythm amplitude, whereas Fbxl3 silencing resulted in low amplitude, consistent with its dual role in ubiquitin-mediated degradation of CRY proteins and in regulation of NR1D1-mediated transcriptional suppression [10]–[13].
Nr1d1, Nr1d2, and E4bp4 Knockdown Caused Cell Type-Dependent Clock Phenotypes
Nr1d1 and Nr1d2 play overlapping but essential functions in regulating RORE-mediated transcription, and knockdown of either gene results in low amplitude rhythms, and in some cases, short period [17], [27]. We examined the effects of Nr1d1 or Nr1d2 KD on clock function in our cellular clock models. Knockdown of Nr1d1 resulted in largely normal rhythms in these cells (Figure 3A and Table 1), indicating potential overlapping functions of Nr1d2 [17]. This is different from the period lengthening produced by siRNA knockdown in U2OS cells [27], [31] or the period shortening and greater variability seen in behavioral rhythms of Nr1d1−/− mice [18], [49]. Nr1d2 KD, on the other hand, resulted in period shortening in MMH-D3 and 3T3-L1 cells but low amplitude in 3T3 cells (Figure 3A and Table 1). The Nr1d2 impact on overall rhythms was at least similar to, if not substantially stronger than, for Nr1d1. This is consistent with the reported redundant functions of Nr1d1 and Nr1d2 [17], but contrasts with previous studies showing that NR1D2 deficiency has no observable rhythm phenotype in U2OS cells [27] or in mice at the behavioral level [18]. Thus, Nr1d1 and Nr1d2 play different roles in clock function depending on tissue or cell type, and Nr1d2 may be more important than previously recognized.
Fig. 3. Knockdowns of Nr1d1, Nr1d2, and E4bp4 lead to cell type-specific circadian phenotypes. 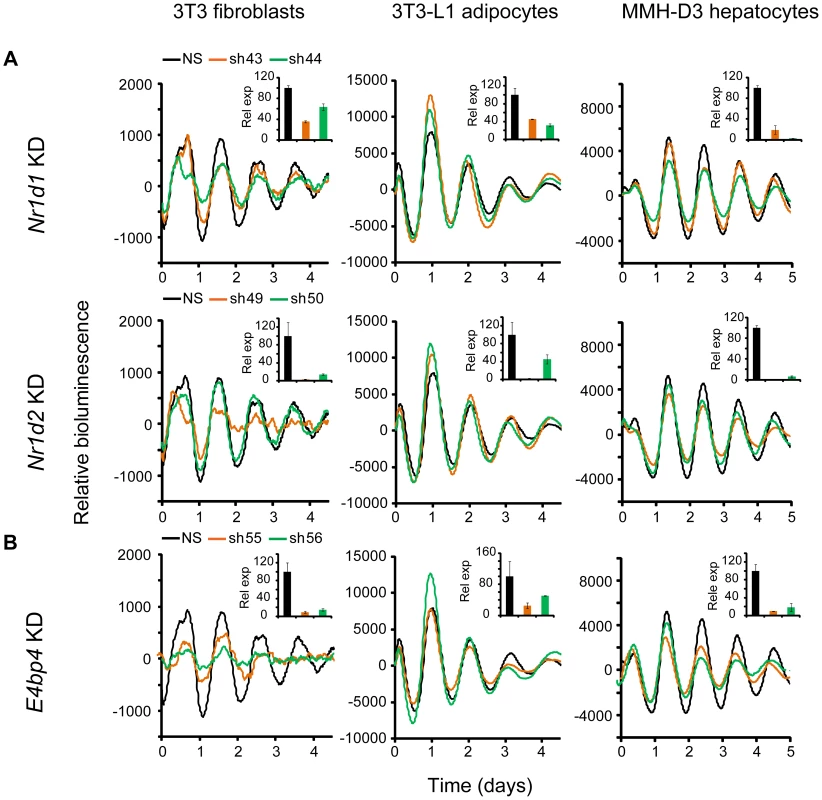
Bioluminescence expression patterns upon KD of Nr1d1 and Nr1d2 (A) and E4bp4 (B) in all three cell types. See Figure 2 for details. Note that, compared to the more prominent role of Nr1d1 in clock function previously found in U2OS cells or mouse behavioral rhythms [18], [27], [49], Nr1d2 plays a more prominent role in all three cells. E4bp4 KD caused period length and/or amplitude phenotypes depending cell type. Despite the widely accepted role of E4BP4 as the repressor of D-box-mediated transcription, definitive genetic evidence of clock function has been lacking. We show here that E4bp4 KD resulted in short period and rapid damping in 3T3 fibroblasts, low amplitude in 3T3-L1 cells, and short period in MMH-D3 cells (Figure 3B and Table 1). These data are in line with recent studies suggesting a prominent role of E4bp4 in regulating the phase of Cry1 transcription in mouse embryonic fibroblasts [21] and period length in Rat-1 fibroblasts [41]. Studies of E4BP4's function in the clock mechanism using E4bp4−/− mice are therefore needed to validate our findings in cellular clock models.
Cell Type-Specific Clock Functions of Per1, Per2, and Per3
The shRNA constructs against Per1, Per2, and Per3 down-regulated mRNA and protein expression (Figures S2 and 4). However, unlike Cry1 and Cry2 KDs, knockdown of the Per genes in our clock models resulted in cell type-specific clock phenotypes (Figure 4). First, compared to the dramatic circadian defects observed in peripheral tissue explants and fibroblasts of Per1−/− mice, or upon siRNA-mediated Per1 KD in U2OS cells [31], [43], [50], Per1 KD in our clock models had milder effects on clock function. Interestingly, these less dramatic phenotypes are cell type-specific: WT phenotype in 3T3 and 3T3-L1 cells, but significantly shorter period and low amplitude in MMH-D3 cells (Figures 4A and 4D; Table 1).
Fig. 4. shRNA-mediated knockdowns of Per1, Per2 and Per3 lead to cell type-specific circadian phenotypes. 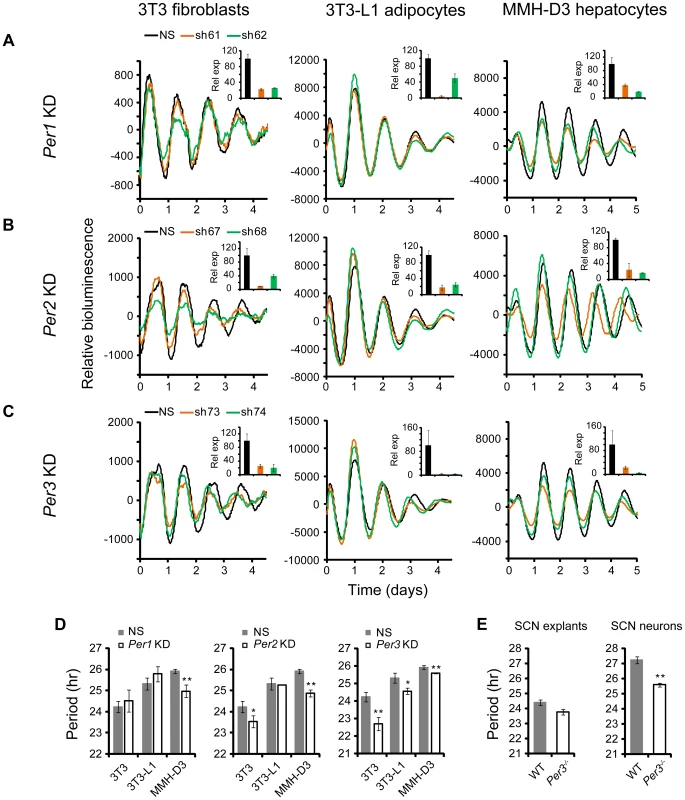
Bioluminescence expression patterns upon KD of Per1 (A), Per2 (B), and Per3 (C) in all three cell types. See Figure 2 for details. Whereas Per3 KD led to short periods in all three cell types, Per1 and Per2 KDs caused different clock phenotypes depending on cell type. (D) Summary of period length phenotypes. Data are mean ± SD (n = 4 independent experiments for 3T3 cells; n = 3 samples/wells of one experiment for 3T3-L1 and MMH-D3 cells). NS, non-specific shRNA. Compared to NS controls, significant difference in period length was detected in the following KDs: NS vs. Per1 KD in MMH-D3, t-test, p<0.001; NS vs. Per2 KD in 3T3, t-test, p = 0.013; NS vs. Per2 KD in MMH-D3, t-test, p<0.001; NS vs. Per3 KD in 3T3, t-test, p<0.001; NS vs. Per3 KD in 3T3-L1, t-test, p = 0.003; NS vs. Per3 KD in MMH-D3, t-test, p<0.001). *p<0.01; ** p<0.001. (E) Per3 deletion led to short period length defects in SCN explants (left) and even stronger defects in dissociated SCN neurons (right). Per3−/− SCN explants show a slightly shorter period than WT (mean ± SEM: WT, 24.4 hr±0.17, n = 5; Per3−/−, 23.78 hr±0.18, n = 5). The mean period of rhythms in Per3−/− neurons was substantially shorter than in WT cells (mean ± SEM: WT, 27.23 hr±0.24, n = 106; Per3−/−, 25.58 hr±0.12, n = 157; t-test, p<10E-10; ** p<0.001). Similarly, Per2 KD did not cause arrhythmicity in any of the three cellular clock models: only a modest reduction of amplitude in 3T3-L1 cells, but significantly shorter period in 3T3 and short period and low amplitude in MMH-D3 cells (Figures 4B and 4D; Table 1). The amplitude reduction in MMH-D3 cells was evident in both the subtracted data and raw data (compare Figure 4 with Figure S5). Although it is possible that the more modest phenotypes may be due to incomplete silencing, the knockdown levels were comparable to those of Bmal1, Clock, and Fbxl3 (Figures 2 and 4B); and as in the case of Per1 KD, the different phenotypes resulted from similar Per2 KD efficiency in different cells. This is unexpected given its essential role in circadian rhythms of mice at the behavioral level and in cultured fibroblasts and U2OS cells [27], [43], [51]–[53]. Interestingly, however, our finding of non-essential but cell type-specific role of Per2 is in line with a recent report showing that Per2−/− SCN explants displayed persistent rhythms with short periods, whereas Per2−/− pituitary explant rhythms were normal and lung explants displayed slightly long periods [54]. Thus, even though genetic knockout and knockdown (incomplete silencing) may cause variations in phenotypes, the loss-of-function phenotypes of Per1 and Per2 are largely consistent and are cell or tissue type specific.
While Per2 appears to be more important than Per1 for normal clock function in mice [52], [55], deletion of Per3 has only subtle effects on the SCN clock and is often not considered part of the core clock mechanism [43], [54], [56]. However, we show here that KD of Per3 in all three models produced significantly shorter periods than in control cells (Figures 4C and 4D; Table 1). These results are consistent with recent reports showing that tissue explants of Per3−/− mice, including liver, lung, and pituitary, also displayed short periods [43], [54]. In addition, knockdowns of Per1, Per2, and Per3 showed similar phenotypes in cells expressing a different reporter (Figures 4 and S4), confirming that the knockdown effects are reporter independent. Furthermore, the Per1 and Per3 knockdown effects are largely consistent with data of liver explants from Per1 and Per3 knockouts [43], [57], [58].
Per3 Plays an Important Role in the SCN Clock
The prominent role of Per3 in peripheral clock function led us to examine its function more carefully in both intact SCN explants and dissociated SCN neurons derived from Per3−/−:mPer2Luc mice. We detected persistent mPer2Luc rhythms in Per3−/− SCN explants with a slightly shorter period than WT (Figure 4E). This is consistent with the original study of Per3−/− mice, where a slightly shorter period of behavioral rhythms was reported [56]. We then dissociated Per3−/− SCN neurons and examined mPer2Luc bioluminescence from dispersed neurons at the single-cell level, as we have done previously for other genotypes [43]. We found that dissociated WT and Per3−/− neurons generally exhibited persistent rhythms with high amplitude. However, the mean period of rhythms in Per3−/− neurons was substantially shorter than in WT cells (Figure 4E). The weaker circadian defect at the SCN tissue level than in cell-autonomous preparations (either our cellular models or dissociated SCN neurons) is consistent with the principle that the SCN network confers robustness against genetic perturbations [43], [59], such that Per3 plays a less prominent role in the intact SCN due to compensation by the SCN network. Based on these results, we conclude that Per3 plays an important role in the SCN cellular clock as well as in peripheral oscillators and thus represents a bona fide clock component.
The more prominent role of Per3 in peripheral oscillators is in line with several recent studies showing that disruption of Per3 resulted in internal phase misalignment or desynchrony, or aberrant metabolic and sleep phenotypes [58], [60]–[62], all pointing to the role of Per3 in coherence of circadian organization. Thus, our findings suggest that tissue-specific function or dysfunction of clock genes in peripheral tissues can be an important contributing factor to human diseases, even when the behavioral effect of gene knockout is subtle. In this context, it is interesting to note that human polymorphisms in PER3 are associated with sleep and metabolic disorders [63], [64].
Taken together, our study expands our knowledge of the distinct functions of known clock genes across tissues. In particular, Per3 plays a more important role in both SCN and non-SCN cells than previously appreciated, and Per1 and Per2 appear to have different roles in different cell types. Results from this study are broadly consistent with previous findings from loss-of-function studies and collectively point to the previously under-appreciated cell type specificity of Per gene function in circadian physiology (Figure S6).
Composite Knockdown of the Per Genes in MMH-D3 Hepatocytes
Compared to other cell-autonomous models, the short period length in MMH-D3 cells after knockdown of each of the Per genes is unique (Figure S6 and Table 1), and therefore we sought to perform single and composite knockdowns for further phenotyping using the LumiCycle assay. Consistent with the Synergy assay, Per1, Per2, and Per3 single gene KD each gave short period phenotypes, about 2 hrs shorter than the control cells (Figures 5A and 5B; Table S4). Composite Per1/Per2 double KD and Per1/Per2/Per3 triple KD caused complete arrhythmicity (Figure 5A), indicating the prominent roles of Per1 and Per2 in the hepatocyte clock. Interestingly, Per1/Per3 and Per2/Per3 double KDs did not cause any further period shortening over single Per gene KDs (Figures 5A and 5B).
Fig. 5. shRNA-mediated single and composite knockdown effects of Per1, Per2 and Per3 in MMH-D3 hepatocytes. 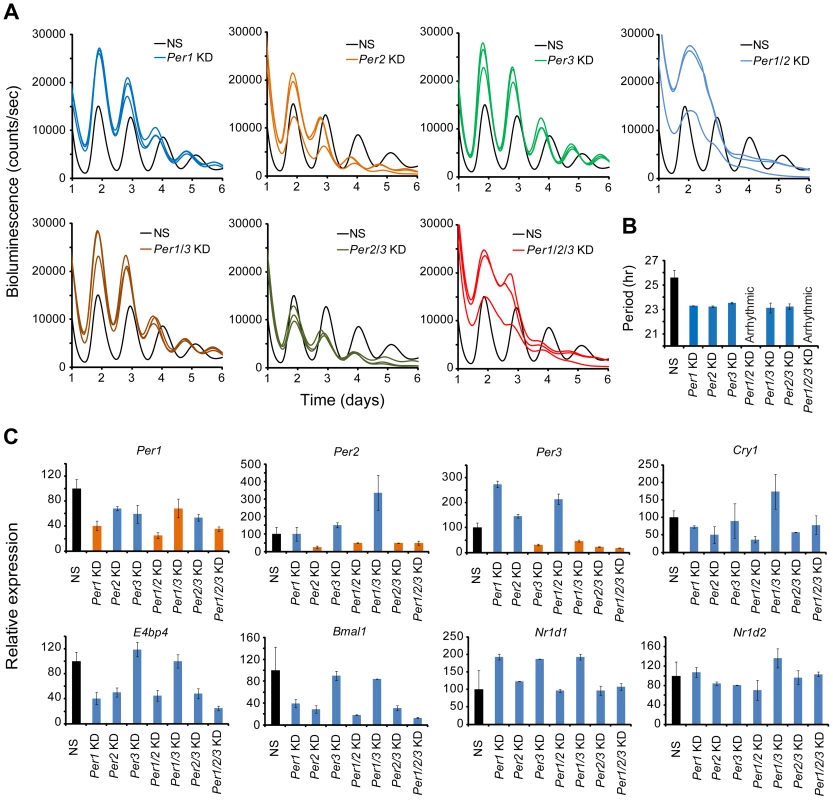
(A) Representative bioluminescence expression patterns recorded on LumiCycle upon knockdowns of Per1, Per2, Per3 (single KD); Per1/Per2, Per1/Per3, Per2/Per3 (double KD); and Per1/Per2/Per3 (triple KD) in MMH-D3 hepatocytes. sh62, sh67 and sh74 were used to knock down Per1, Per2 and Per3, respectively. All single KDs led to short periods in all three cell types, consistent with Synergy assays. Per1/Per2 double and Per1/Per2/Per3 triple KDs caused arrhythmicity. All other double composite KDs caused short period phenotype. (B) Summary of period length phenotypes. Data are mean ± SD (n = 3 independent samples/wells from one experiment). NS, non-specific shRNA. Compared to NS controls, significant difference in period length was detected (NS vs. Per1 KD, t-test, p<0.001; NS vs. Per2 KD, t-test, p<0.001; NS vs. Per3 KD, t-test, p<0.001; NS vs. Per1/Per3 KD, t-test, p<0.001; NS vs. Per2/Per3 KD, t-test, p<0.001). (C) qPCR analysis of clock gene expression upon Per KD in non-synchronized MMH-D3 cells. Values for each gene are expressed as percentage of gene expression in NS control cells. Data represent two samples/wells from one experiment. As an initial effort to probe the network features of the hepatocyte clock, we examined effects of Per KD on the expression of endogenous clock genes by qPCR. Per1, Per2, and Per3 were each knocked down in both single and composite KDs (Figure 5C). Compared to the U2OS model, paralog compensation among the Per genes in MMH-D3 cells is more pervasive. In our MMH-D3 model, both Per1 KD and Per2 KD upregulated Per3, and Per3 KD upregulated Per2. While composite Per1/Per2 KD upregulated Per3 and Per1/Per3 KD greatly upregulated Per2, Per2/Per3 KD did not increase Per1 expression. Interestingly, Per1 and/or Per2 KD had milder effects on the expression of E-box-containing genes (e.g., Nr1d1 and Nr1d2) than on RORE-containing genes such as Bmal1, E4bp4, and Cry1 (Figure 5C), in line with the notion that the PERs can directly and indirectly affect Bmal1 transcription [65], [66]. Overall, the network interactions in MMH-D3 cells appear to differ from those in the U2OS model in which PER1 plays a more dominant role than PER2 and PER3 [27], and is expected to differ from those in 3T3 and 3T3-L1 models. Thus, extensive investigation into the network features of these cellular models will require additional experiments and is warranted in future studies, as we have done with the U2OS model [27].
Concluding Remarks
Cell type-specific function of clock genes may result from their differential tissue expression and activity (i.e., expression levels, ratio of repressors to activators, rhythmicity, and relative amplitudes), compensatory mechanisms, alternative splice variants, and post-translational modifications (PMTs), all of which can be rendered cell type specific by local physiology. Recent studies have suggested a role for stoichiometric balance among clock proteins in circadian clock robustness and periodicity, and call for mechanistic studies in a tissue specific manner [67]–[69]. In the context of the liver and adipose tissue function, it is plausible that the basic core clock mechanism incorporates cell type-specific factors and forms distinctive functional networks to regulate (and in turn be regulated by) different local physiologies. It is interesting to note, as PTMs of clock factors (e.g., phosphorylation, ubiquitination, ADP-ribosylation, acetylation, and O-GlcNAcylation) represent critical regulatory mechanisms [33], [70], tissue-specific cellular functions and metabolic states that affect the PTMs would provide important inputs to adjust local circadian clocks, and vice versa. Thus, cell type specific clock gene function starts to make sense when local physiology is considered as inputs to the clock. This challenge surely provides an opportunity for deeper insights into mechanism of tissue specific clocks, akin to the recent realization that cyclin-dependent kinase networks in the cell cycle control program are tissue specific [71].
In summary, we established three new mouse cellular clock models: fibroblasts, adipocytes, and hepatocytes. These cellular clock models offer experimental tractability, efficiency, and versatility, which are more difficult or impossible to apply to traditional tissue or animal models. Of note, in contrast to previous cellular clock models, the new clock models are amenable to high throughput experiments with inexpensive off-the-shelf recording systems, making these lines especially suitable for screening small molecules or genomic entities for impacts on cell autonomous clocks relevant to metabolism. We validated these models by developing and testing an shRNA panel of selected known clock genes. Results from this study and others point to the previously under-appreciated cell type specificity of clock gene function in circadian physiology (Figure S6). The prevalence of tissue-specific clock gene function will have important implications for future studies of clock factors that affect local clock function. It is our hope that our findings in this study, along with the new cellular clock models, approaches, and tools developed here, can be applied to a greater variety of cell types in future studies, to reveal the full range of tissue-specific clock properties underlying local circadian biology.
Materials and Methods
Animals
Per3−/− mice were obtained from David Weaver at the University of Massachusetts. Knockout mice were bred with mPer2Luc reporter mice to obtain homozygous knockouts harboring the mPer2Luc reporter. Wheel-running assays were performed and analyzed as described previously [8]. All animal studies were conducted in accordance with the regulations of the Committees on Animal Care and Use at University of Memphis and UCSD.
Cell and Tissue Culture
All cell culture media were from HyClone. 3T3 (also known as NIH 3T3) and 3T3-L1 cells were cultured in regular medium in which DMEM was supplemented with 10% FBS and 1× penicillin-streptomycin-glutamine (PSG). For 3T3-L1 differentiation, pre-adipocytes were first grown to confluence (Day 0). On Day 2, cells were fed with induction medium (regular medium with 1 µM dexamethasone, 0.5 mM isobutylmethylxanthine, and 2 µg/ml insulin). On Day 4, cells were changed to maintenance medium (regular medium containing 2 µg/ml insulin). From day 6 onward, cells were grown in regular medium until use. For bioluminescence recording, 3T3 and 3T3-L1 cells were grown in 25 mM HEPES-buffered regular medium (pH 7.4) containing 1 nM forskolin and 1 mM luciferin. Fully differentiated 3T3-L1 cells were used in all experiments.
MMH-D3 cells were grown in regular medium in which RPMI medium was supplemented with 10% FBS, 1× PSG, 10 µg/ml insulin, 55 ng/ml epidermal growth factor (EGF), and 16 ng/ml insulin like growth factor-II (IGF-II). For differentiation, pre-hepatocytes were first grown to 100% confluence. Two days later, cells were replaced with differentiation medium (regular medium with 2% DMSO). Medium change was repeated every 48 hours for 6–8 days for cells to be fully differentiated for use. Circadian rhythms of differentiated cells were synchronized with 200 nM dexamethasone followed by bioluminescence recording in 25 mM HEPES-buffered serum-free explant medium (pH 7.4) containing B-27 and 1 mM luciferin, as we have done previously [43]. Fully differentiated MMH-D3 cells were used in all experiments.
SCN explants and dissociated neuronal cells were prepared and cultured as previously described [43]. Bioluminescence recording of explants, single cell-imaging of individual SCN neurons, and respective data analysis were performed as previously described [43].
Generation of Reporter Cell Lines
Lentiviral luciferase reporters of the Per2 or Bmal1 promoter were described previously [17], [43]. Reporter cells and clonal lines were generated as previously described [23]. Briefly, reporter viral particles of high titer (>108 viral particles/ml) were obtained by ultracentrifugation and used to infect 3T3, 3T3-L1, and MMH-D3 cells. Clonal cell lines of homogenous cell populations were obtained by single cell sorting and cloning in 96 well plates. We then selected the clones that expressed high levels of luciferase and exhibited circadian properties comparable to infected parental cell populations. These brighter cells were used in high-throughput assays on 96 well plates. Knockdown phenotypes were confirmed to be independent of the reporter, either Per2-dLuc or Bmal1-dLuc, and thus phenotypic differences across cell types are unlikely due to different reporter insertion sites.
Construction of Lentiviral shRNA Vectors and Viral Preparation
We used an optimized shRNA design algorithm adapted from Aza-Blanc et al. [72] for target sequence prediction. This adapted algorithm selects for optimal target sequence for knockdown, and against homologous sequences to minimize off-target effects. We selected 6 target sequences for each gene as listed in Table S5. Each shRNA construct contained a sense and an antisense target sequence of 19 nucleotides (nts) in length, separated by 9 nts for a hairpin loop, and flanked by TTTG at 5′ and GATC at 3′ ends for cohesive end cloning. All oligos (55 nts) were synthesized by Integrated DNA Technologies (IDT). The annealed oligonucleotides were first cloned into the BbsI and SpeI sites of a pGWL-si2/U6 vector, in which the shRNA expression cassette is driven by an RNA polymerase III - based mouse U6 promoter. Subsequently, the U6-shRNA cassette was cloned into the lentiviral pLL3.7GW vector (modified from pLL3.7) [17], [73] in a Gateway LR Clonase reaction (Life Technologies), according to manufacturer's instructions.
Viral particles were prepared using standard methods in 293T cells on 12-well plates as previously described [23], [74]. Culture medium containing viral particles (∼106 viral particles/ml) were collected and used for subsequent infection of reporter cells. Transfection and infection efficiency were estimated by observing GFP co-expressed from a CMV promoter. To produce high titer viruses, crude viral particles were concentrated through ultracentrifugation and appropriate titers were used, as described previously [74]. This pipeline allowed us to generate a panel of shRNA constructs against all known clock factors for genetic perturbation and phenotyping.
Bioluminescence Recording and Data Analysis
We used a LumiCycle luminometer (version 2.31, Actimetrics) for bioluminescence recording of cells grown on 35 mm culture dishes, as described elsewhere [17], [23], [43]. The LumiCycle Analysis program version 2.53 (Actimetrics) was used to determine circadian parameters. Briefly, raw data were fitted to a linear baseline, and the baseline-subtracted data were fitted to a sine wave (damped), from which period length and goodness of fit and damping constant were determined. For samples that showed persistent rhythms, goodness-of-fit of >80% was usually achieved. Due to high transient luminescence upon medium change, the first cycle was usually excluded from rhythm analysis. Damping rate = 1/damping constant. For amplitude analysis, raw data from day 3 to day 5 were fitted to a linear baseline, and the baseline-subtracted (polynomial number = 1) data were fitted to a sine wave, from which the amplitude was determined.
We used a Synergy 2 SL microplate reader (Bio Tek) for bioluminescence recording of cells grown on 96 well plates, as previously described [23]. Synergy data were analyzed with the MultiCycle Analysis program (Actimetrics), in which bioluminescence data were baseline-subtracted and fit to a damped sine wave to determine period length, goodness of fit, and amplitude, as with LumiCycle Analysis. Due to the various reporter expression levels and for direct comparison of different rhythms, baseline subtracted data were plotted. Because there is no damping rate output function in the MultiCycle Analysis, we used a curve fitting program of “CellulaRhythm” to determine damping rate from Synergy data as previously developed [75].
Western Blot Analysis
For testing shRNA knockdown efficiency targeting each clock gene, the cDNA was cloned into a p3XFlag-CMV-14 vector. Flag-tagged cDNA was co-transfected with the indicated shRNA in 3T3 or 293T cells. Forty eight hours post-transfection, cells were lysed in RIPA buffer containing complete protease (Roche) and phosphatase inhibitors (Sigma). Protein expression was determined by Western blot analysis using an anti-Flag monoclonal antibody (Sigma). For all Western assays, PVDF membrane was used in protein transfer, and SuperSignal West Pico substrate (Thermo Scientific) was used for chemiluminescent detection.
Quantitative PCR (qPCR) Analysis
For qPCR analysis, parallel infection experiments were performed as with bioluminescence recording. Cells were harvested prior to medium change and were therefore unsynchronized. Total RNAs were prepared using the RNeasy 96 kit (Qiagen), as previously described [31]. Reverse transcription was performed using a high-capacity RNA to cDNA kit (Applied Biosystems), and qPCR was performed using SYBR Green PCR master mix (Thermo Scientific) on an iCycler thermal cycler (BioRad). The primers used in qPCR analysis are listed in Table S6. Transcript levels for each gene were normalized to Gapdh and values were expressed as percentage of expression in NS control cells, as previously described [17].
Supporting Information
Zdroje
1. ReppertSM, WeaverDR (2002) Coordination of circadian timing in mammals. Nature 418 : 935–941.
2. HastingsMH, ReddyAB, MaywoodES (2003) A clockwork web: circadian timing in brain and periphery, in health and disease. Nat Rev Neurosci 4 : 649–661.
3. LiuAC, LewisWG, KaySA (2007) Mammalian circadian signaling networks and therapeutic targets. Nat Chem Biol 3 : 630–639.
4. MohawkJA, GreenCB, TakahashiJS (2012) Central and peripheral circadian clocks in mammals. Annu Rev Neurosci 35 : 445–462.
5. GreenCB, TakahashiJS, BassJ (2008) The meter of metabolism. Cell 134 : 728–742.
6. NagoshiE, SainiC, BauerC, LarocheT, NaefF, et al. (2004) Circadian gene expression in individual fibroblasts: cell-autonomous and self-sustained oscillators pass time to daughter cells. Cell 119 : 693–705.
7. WelshDK, YooSH, LiuAC, TakahashiJS, KaySA (2004) Bioluminescence imaging of individual fibroblasts reveals persistent, independently phased circadian rhythms of clock gene expression. Curr Biol 14 : 2289–2295.
8. YooSH, YamazakiS, LowreyPL, ShimomuraK, KoCH, et al. (2004) PERIOD2::LUCIFERASE real-time reporting of circadian dynamics reveals persistent circadian oscillations in mouse peripheral tissues. Proc Natl Acad Sci U S A 101 : 5339–5346.
9. YoungMW, KaySA (2001) Time zones: a comparative genetics of circadian clocks. Nat Rev Genet 2 : 702–715.
10. BusinoL, BassermannF, MaiolicaA, LeeC, NolanPM, et al. (2007) SCFFbxl3 controls the oscillation of the circadian clock by directing the degradation of cryptochrome proteins. Science 316 : 900–904.
11. SiepkaSM, YooSH, ParkJ, SongW, KumarV, et al. (2007) Circadian mutant Overtime reveals F-box protein FBXL3 regulation of cryptochrome and period gene expression. Cell 129 : 1011–1023.
12. GodinhoSI, MaywoodES, ShawL, TucciV, BarnardAR, et al. (2007) The after-hours mutant reveals a role for Fbxl3 in determining mammalian circadian period. Science 316 : 897–900.
13. ShiG, XingL, LiuZ, QuZ, WuX, et al. (2013) Dual roles of FBXL3 in the mammalian circadian feedback loops are important for period determination and robustness of the clock. Proc Natl Acad Sci U S A 110 : 4750–4755.
14. YooSH, MohawkJA, SiepkaSM, ShanY, HuhSK, et al. (2013) Competing E3 ubiquitin ligases govern circadian periodicity by degradation of CRY in nucleus and cytoplasm. Cell 152 : 1091–1105.
15. HiranoA, YumimotoK, TsunematsuR, MatsumotoM, OyamaM, et al. (2013) FBXL21 regulates oscillation of the circadian clock through ubiquitination and stabilization of cryptochromes. Cell 152 : 1106–1118.
16. UedaHR, HayashiS, ChenW, SanoM, MachidaM, et al. (2005) System-level identification of transcriptional circuits underlying mammalian circadian clocks. Nat Genet 37 : 187–192.
17. LiuAC, TranHG, ZhangEE, PriestAA, WelshDK, et al. (2008) Redundant function of REV-ERBalpha and beta and non-essential role for Bmal1 cycling in transcriptional regulation of intracellular circadian rhythms. PLoS Genet 4: e1000023.
18. ChoH, ZhaoX, HatoriM, YuRT, BarishGD, et al. (2012) Regulation of circadian behaviour and metabolism by REV-ERB-alpha and REV-ERB-beta. Nature 485 : 123–127.
19. KoikeN, YooSH, HuangHC, KumarV, LeeC, et al. (2012) Transcriptional architecture and chromatin landscape of the core circadian clock in mammals. Science 338 : 349–354.
20. DeBruyneJP, HogeneschJB (2011) A CRY in the Night. Dev Cell 20 : 144–145.
21. Ukai-TadenumaM, YamadaRG, XuH, RippergerJA, LiuAC, et al. (2011) Delay in feedback repression by cryptochrome 1 is required for circadian clock function. Cell 144 : 268–281.
22. HogeneschJB, UedaHR (2011) Understanding systems-level properties: timely stories from the study of clocks. Nat Rev Genet 12 : 407–416.
23. RamanathanC, KhanSK, KathaleND, XuH, LiuAC (2012) Monitoring cell-autonomous circadian clock rhythms of gene expression using luciferase bioluminescence reporters. J Vis Exp e4234 doi 4210.37914234.:4210.3791/4234.
24. KhanSK, XuH, Ukai-TadenumaM, BurtonB, WangY, et al. (2012) Identification of a novel cryptochrome differentiating domain required for feedback repression in circadian clock function. J Biol Chem 287 : 25917–25926.
25. HogeneschJB, HerzogED (2011) Intracellular and intercellular processes determine robustness of the circadian clock. FEBS Lett 585 : 1427–1434.
26. UkaiH, UedaHR (2010) Systems biology of mammalian circadian clocks. Annu Rev Physiol 72 : 579–603.
27. BaggsJE, PriceTS, DiTacchioL, PandaS, FitzgeraldGA, et al. (2009) Network features of the mammalian circadian clock. PLoS Biol 7: e52.
28. SatoTK, YamadaRG, UkaiH, BaggsJE, MiragliaLJ, et al. (2006) Feedback repression is required for mammalian circadian clock function. Nat Genet 38 : 312–319.
29. AtwoodA, DeCondeR, WangSS, MocklerTC, SabirJS, et al. (2011) Cell-autonomous circadian clock of hepatocytes drives rhythms in transcription and polyamine synthesis. Proc Natl Acad Sci U S A 108 : 18560–18565.
30. PandaS, AntochMP, MillerBH, SuAI, SchookAB, et al. (2002) Coordinated transcription of key pathways in the mouse by the circadian clock. Cell 109 : 307–320.
31. ZhangEE, LiuAC, HirotaT, MiragliaLJ, WelchG, et al. (2009) A genome-wide RNAi screen for modifiers of the circadian clock in human cells. Cell 139 : 199–210.
32. Eckel-MahanK, Sassone-CorsiP (2009) Metabolism control by the circadian clock and vice versa. Nat Struct Mol Biol 16 : 462–467.
33. AsherG, SchiblerU (2011) Crosstalk between components of circadian and metabolic cycles in mammals. Cell Metab 13 : 125–137.
34. GreenH, KehindeO (1975) An established preadipose cell line and its differentiation in culture. II. Factors affecting the adipose conversion. Cell 5 : 19–27.
35. KallenCB, LazarMA (1996) Antidiabetic thiazolidinediones inhibit leptin (ob) gene expression in 3T3-L1 adipocytes. Proc Natl Acad Sci U S A 93 : 5793–5796.
36. AmiconeL, SpagnoliFM, SpathG, GiordanoS, TommasiniC, et al. (1997) Transgenic expression in the liver of truncated Met blocks apoptosis and permits immortalization of hepatocytes. EMBO J 16 : 495–503.
37. FeigelstockDA, ThompsonP, KaplanGG (2005) Growth of hepatitis A virus in a mouse liver cell line. J Virol 79 : 2950–2955.
38. OtwayDT, FrostG, JohnstonJD (2009) Circadian rhythmicity in murine pre-adipocyte and adipocyte cells. Chronobiol Int 26 : 1340–1354.
39. AoyagiT, ShimbaS, TezukaM (2005) Characteristics of Circadian Gene Expression in Mice White Adipose Tissue and 3T3-L1 Adipocytes. Journal of Health Science 51 : 21–32.
40. GachonF, OlelaFF, SchaadO, DescombesP, SchiblerU (2006) The circadian PAR-domain basic leucine zipper transcription factors DBP, TEF, and HLF modulate basal and inducible xenobiotic detoxification. Cell Metab 4 : 25–36.
41. YamajukuD, ShibataY, KitazawaM, KatakuraT, UrataH, et al. (2011) Cellular DBP and E4BP4 proteins are critical for determining the period length of the circadian oscillator. FEBS Lett 585 : 2217–2222.
42. EcheverriCJ, BeachyPA, BaumB, BoutrosM, BuchholzF, et al. (2006) Minimizing the risk of reporting false positives in large-scale RNAi screens. Nat Methods 3 : 777–779.
43. LiuAC, WelshDK, KoCH, TranHG, ZhangEE, et al. (2007) Intercellular coupling confers robustness against mutations in the SCN circadian clock network. Cell 129 : 605–616.
44. MaierB, WendtS, VanselowJT, WallachT, ReischlS, et al. (2009) A large-scale functional RNAi screen reveals a role for CK2 in the mammalian circadian clock. Genes Dev 23 : 708–718.
45. KoCH, YamadaYR, WelshDK, BuhrED, LiuAC, et al. (2010) Emergence of noise-induced oscillations in the central circadian pacemaker. PLoS Biol 8: e1000513.
46. DeBruyneJP, WeaverDR, ReppertSM (2007) Peripheral circadian oscillators require CLOCK. Curr Biol 17: R538–539.
47. DudleyCA, Erbel-SielerC, EstillSJ, ReickM, FrankenP, et al. (2003) Altered patterns of sleep and behavioral adaptability in NPAS2-deficient mice. Science 301 : 379–383.
48. ShiS, HidaA, McGuinnessOP, WassermanDH, YamazakiS, et al. (2010) Circadian clock gene Bmal1 is not essential; functional replacement with its paralog, Bmal2. Curr Biol 20 : 316–321.
49. PreitnerN, DamiolaF, Lopez-MolinaL, ZakanyJ, DubouleD, et al. (2002) The orphan nuclear receptor REV-ERBalpha controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell 110 : 251–260.
50. PendergastJS, FridayRC, YamazakiS (2009) Endogenous rhythms in Period1 mutant suprachiasmatic nuclei in vitro do not represent circadian behavior. J Neurosci 29 : 14681–14686.
51. BaeK, JinX, MaywoodES, HastingsMH, ReppertSM, et al. (2001) Differential functions of mPer1, mPer2, and mPer3 in the SCN circadian clock. Neuron 30 : 525–536.
52. ZhengB, AlbrechtU, KaasikK, SageM, LuW, et al. (2001) Nonredundant roles of the mPer1 and mPer2 genes in the mammalian circadian clock. Cell 105 : 683–694.
53. BrownSA, Fleury-OlelaF, NagoshiE, HauserC, JugeC, et al. (2005) The period length of fibroblast circadian gene expression varies widely among human individuals. PLoS Biol 3: e338.
54. PendergastJS, FridayRC, YamazakiS (2010) Distinct functions of Period2 and Period3 in the mouse circadian system revealed by in vitro analysis. PLoS One 5: e8552.
55. ChenR, SchirmerA, LeeY, LeeH, KumarV, et al. (2009) Rhythmic PER abundance defines a critical nodal point for negative feedback within the circadian clock mechanism. Mol Cell 36 : 417–430.
56. ShearmanLP, JinX, LeeC, ReppertSM, WeaverDR (2000) Targeted disruption of the mPer3 gene: subtle effects on circadian clock function. Mol Cell Biol 20 : 6269–6275.
57. YagitaK, TamaniniF, van Der HorstGT, OkamuraH (2001) Molecular mechanisms of the biological clock in cultured fibroblasts. Science 292 : 278–281.
58. PendergastJS, NiswenderKD, YamazakiS (2012) Tissue-specific function of Period3 in circadian rhythmicity. PLoS One 7: e30254.
59. EvansJA, PanH, LiuAC, WelshDK (2012) Cry1−/ − circadian rhythmicity depends on SCN intercellular coupling. J Biol Rhythms 27 : 443–452.
60. HasanS, van der VeenDR, Winsky-SommererR, DijkDJ, ArcherSN (2011) Altered sleep and behavioral activity phenotypes in PER3-deficient mice. Am J Physiol Regul Integr Comp Physiol 301: R1821–1830.
61. DallmannR, WeaverDR (2010) Altered body mass regulation in male mPeriod mutant mice on high-fat diet. Chronobiol Int 27 : 1317–1328.
62. CostaMJ, SoAY, KaasikK, KruegerKC, PillsburyML, et al. (2011) Circadian rhythm gene period 3 is an inhibitor of the adipocyte cell fate. J Biol Chem 286 : 9063–9070.
63. BarnardAR, NolanPM (2008) When clocks go bad: neurobehavioural consequences of disrupted circadian timing. PLoS Genet 4: e1000040.
64. DijkDJ, ArcherSN (2009) PERIOD3, circadian phenotypes, and sleep homeostasis. Sleep Med Rev 14 : 151–160.
65. ShearmanLP, SriramS, WeaverDR, MaywoodES, ChavesI, et al. (2000) Interacting molecular loops in the mammalian circadian clock. Science 288 : 1013–1019.
66. SchmutzI, RippergerJA, Baeriswyl-AebischerS, AlbrechtU (2010) The mammalian clock component PERIOD2 coordinates circadian output by interaction with nuclear receptors. Genes Dev 24 : 345–357.
67. KimJK, ForgerDB (2012) A mechanism for robust circadian timekeeping via stoichiometric balance. Mol Syst Biol 8 : 630.
68. LeeY, ChenR, LeeHM, LeeC (2011) Stoichiometric relationship among clock proteins determines robustness of circadian rhythms. J Biol Chem 286 : 7033–7042.
69. YeR, SelbyCP, OzturkN, AnnayevY, SancarA (2011) Biochemical analysis of the canonical model for the mammalian circadian clock. J Biol Chem 286 : 25891–25902.
70. HartGW (2013) How sugar tunes your clock. Cell Metab 17 : 155–156.
71. PaganoM, JacksonPK (2004) Wagging the dogma; tissue-specific cell cycle control in the mouse embryo. Cell 118 : 535–538.
72. Aza-BlancP, CooperCL, WagnerK, BatalovS, DeverauxQL, et al. (2003) Identification of modulators of TRAIL-induced apoptosis via RNAi-based phenotypic screening. Mol Cell 12 : 627–637.
73. RubinsonDA, DillonCP, KwiatkowskiAV, SieversC, YangL, et al. (2003) A lentivirus-based system to functionally silence genes in primary mammalian cells, stem cells and transgenic mice by RNA interference. Nat Genet 33 : 401–406.
74. TiscorniaG, SingerO, VermaIM (2006) Production and purification of lentiviral vectors. Nat Protoc 1 : 241–245.
75. HirotaT, LewisWG, LiuAC, LeeJW, SchultzPG, et al. (2008) A chemical biology approach reveals period shortening of the mammalian circadian clock by specific inhibition of GSK-3beta. Proc Natl Acad Sci U S A 105 : 20746–20751.
Štítky
Genetika Reprodukční medicína
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2014 Číslo 4
-
Všechny články tohoto čísla
- The Challenges of Mitochondrial Replacement
- Concocting Cholinergy
- Genome-Wide Diet-Gene Interaction Analyses for Risk of Colorectal Cancer
- Statistical Power to Detect Genetic (Co)Variance of Complex Traits Using SNP Data in Unrelated Samples
- Mouse Pulmonary Adenoma Susceptibility 1 Locus Is an Expression QTL Modulating -4A
- Transcription-Associated R-Loop Formation across the Human CGG-Repeat Region
- Epigenetic Regulation by Heritable RNA
- Protein Quantitative Trait Loci Identify Novel Candidates Modulating Cellular Response to Chemotherapy
- Genome-Wide Profiling of Yeast DNA:RNA Hybrid Prone Sites with DRIP-Chip
- The Mechanism of Gene Targeting in Human Somatic Cells
- A LINE-1 Insertion in DLX6 Is Responsible for Cleft Palate and Mandibular Abnormalities in a Canine Model of Pierre Robin Sequence
- Interaction between Two Timing MicroRNAs Controls Trichome Distribution in
- DNA Glycosylases Involved in Base Excision Repair May Be Associated with Cancer Risk in and Mutation Carriers
- The Myc-Mondo/Mad Complexes Integrate Diverse Longevity Signals
- Evolutionarily Diverged Regulation of X-chromosomal Genes as a Primal Event in Mouse Reproductive Isolation
- Mutations in Conserved Residues of the microRNA Argonaute ALG-1 Identify Separable Functions in ALG-1 miRISC Loading and Target Repression
- Genetic Predisposition to In Situ and Invasive Lobular Carcinoma of the Breast
- Isl1 Directly Controls a Cholinergic Neuronal Identity in the Developing Forebrain and Spinal Cord by Forming Cell Type-Specific Complexes
- A Synthetic Community Approach Reveals Plant Genotypes Affecting the Phyllosphere Microbiota
- The Sequence-Specific Transcription Factor c-Jun Targets Cockayne Syndrome Protein B to Regulate Transcription and Chromatin Structure
- Determining the Control Circuitry of Redox Metabolism at the Genome-Scale
- DNA Repair Pathway Selection Caused by Defects in , , and Telomere Addition Generates Specific Chromosomal Rearrangement Signatures
- Methylome Diversification through Changes in DNA Methyltransferase Sequence Specificity
- Folliculin Regulates Ampk-Dependent Autophagy and Metabolic Stress Survival
- Fine Mapping of Dominant -Linked Incompatibility Alleles in Hybrids
- Unexpected Role of the Steroid-Deficiency Protein Ecdysoneless in Pre-mRNA Splicing
- Three Groups of Transposable Elements with Contrasting Copy Number Dynamics and Host Responses in the Maize ( ssp. ) Genome
- Sox5 Functions as a Fate Switch in Medaka Pigment Cell Development
- Synergistic Interactions between the Molecular and Neuronal Circadian Networks Drive Robust Behavioral Circadian Rhythms in
- Chromatin Landscapes of Retroviral and Transposon Integration Profiles
- Widespread Use of Non-productive Alternative Splice Sites in
- Ras GTPase-Like Protein MglA, a Controller of Bacterial Social-Motility in Myxobacteria, Has Evolved to Control Bacterial Predation by
- Cell Type-Specific Functions of Genes Revealed by Novel Adipocyte and Hepatocyte Circadian Clock Models
- Phenotype Ontologies and Cross-Species Analysis for Translational Research
- Embryogenesis Scales Uniformly across Temperature in Developmentally Diverse Species
- In Pursuit of the Gene: An Interview with James Schwartz
- Molecular Mechanisms of Hypoxic Responses via Unique Roles of Ras1, Cdc24 and Ptp3 in a Human Fungal Pathogen
- Analysis of the Genome and Transcriptome of var. Reveals Complex RNA Expression and Microevolution Leading to Virulence Attenuation
- Genotypic and Functional Impact of HIV-1 Adaptation to Its Host Population during the North American Epidemic
- RNA Editome in Rhesus Macaque Shaped by Purifying Selection
- Proper Actin Ring Formation and Septum Constriction Requires Coordinated Regulation of SIN and MOR Pathways through the Germinal Centre Kinase MST-1
- Interplay of the Serine/Threonine-Kinase StkP and the Paralogs DivIVA and GpsB in Pneumococcal Cell Elongation and Division
- A Quality Control Mechanism Coordinates Meiotic Prophase Events to Promote Crossover Assurance
- CNNM2 Mutations Cause Impaired Brain Development and Seizures in Patients with Hypomagnesemia
- The RNA-Binding Protein QKI Suppresses Cancer-Associated Aberrant Splicing
- Uncoupling Transcription from Covalent Histone Modification
- Rad51–Rad52 Mediated Maintenance of Centromeric Chromatin in
- FRA2A Is a CGG Repeat Expansion Associated with Silencing of
- A General Approach for Haplotype Phasing across the Full Spectrum of Relatedness
- A Novel Highly Divergent Protein Family Identified from a Viviparous Insect by RNA-seq Analysis: A Potential Target for Tsetse Fly-Specific Abortifacients
- A Central Role for in Regulation of Islet Function in Man
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- The Sequence-Specific Transcription Factor c-Jun Targets Cockayne Syndrome Protein B to Regulate Transcription and Chromatin Structure
- The Mechanism of Gene Targeting in Human Somatic Cells
- Genetic Predisposition to In Situ and Invasive Lobular Carcinoma of the Breast
- Widespread Use of Non-productive Alternative Splice Sites in
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



