-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaFRA2A Is a CGG Repeat Expansion Associated with Silencing of
Some human genetic diseases are caused by dynamic mutations, or expansions of a short repeated sequence in the genome that can be unstably passed on from generation to generation. A subset of these dynamic mutations known as fragile sites can be seen as a break or gap on the chromosome when cells are cultured under specific conditions. To date eight folate-sensitive fragile sites (FSFS) have been characterized, and all are due to CGG-repeat expansions within the 5′ UTR or promoter region of the respective gene. When the repeat expands in size, it becomes hypermethylated and the adjacent gene or genes are transcriptionally silenced. For at least four of the eight known fragile sites this silencing of the associated gene(s) lead to intellectual disability syndromes such as fragile X. In this work we describe molecular characterization of an autosomal FSFS called FRA2A on chromosome 2. As the molecular cause of FRA2A, we identify an expansion of a CGG repeat which subsequently results in silencing of the neighbouring gene AFF3. This gene is one of the four autosomal paralogss of the AFF2/FMR2 gene which, when mutated, is the cause of the FRAXE syndrome. We find that FRA2A expression is associated with highly variable developmental anomalies in the three FRA2A families studied.
Published in the journal: . PLoS Genet 10(4): e32767. doi:10.1371/journal.pgen.1004242
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1004242Summary
Some human genetic diseases are caused by dynamic mutations, or expansions of a short repeated sequence in the genome that can be unstably passed on from generation to generation. A subset of these dynamic mutations known as fragile sites can be seen as a break or gap on the chromosome when cells are cultured under specific conditions. To date eight folate-sensitive fragile sites (FSFS) have been characterized, and all are due to CGG-repeat expansions within the 5′ UTR or promoter region of the respective gene. When the repeat expands in size, it becomes hypermethylated and the adjacent gene or genes are transcriptionally silenced. For at least four of the eight known fragile sites this silencing of the associated gene(s) lead to intellectual disability syndromes such as fragile X. In this work we describe molecular characterization of an autosomal FSFS called FRA2A on chromosome 2. As the molecular cause of FRA2A, we identify an expansion of a CGG repeat which subsequently results in silencing of the neighbouring gene AFF3. This gene is one of the four autosomal paralogss of the AFF2/FMR2 gene which, when mutated, is the cause of the FRAXE syndrome. We find that FRA2A expression is associated with highly variable developmental anomalies in the three FRA2A families studied.
Introduction
Dynamic mutations are heritable unstable expansions of short, genomic repeat sequences. Various pathogenic mechanisms have been associated with dynamic mutations [1], [2] and at least 40 neurological, neurodegenerative and neuromuscular disorders are known to be caused by these types of mutations [3], [4]. Expansions of these unstable sequences may occur in promoters, coding regions, introns and 3′ and 5′ untranslated regions (UTR) of genes [5], [6], [7]. Known and putative disease mechanisms include aberrant splicing [8], loss or gain of function of the encoded protein [9], [10], the expanded repeat itself [11] or its RNA transcript [12], [13] and Repeat Associated Non-ATG translation (RAN translation) [14], [15]. The size threshold at which a repeat becomes unstable and/or pathogenic varies widely, from the expansion of only a few trinucleotide repeats in e.g. ARX-associated infantile epileptic encephalopathy (MIM 308350) to over a thousand repeats in e.g. DMPK-associated myotonic dystrophy (MIM 160900), FXN-associated Friedreich ataxia (MIM 229300) and FMR1-associated fragile X syndrome (MIM 300624) [16], [17], [18].
Fragile sites represent a specific subset of dynamic mutations that are visible as gaps or breaks on metaphase chromosomes from cells cultured under specific conditions. Fragile sites are categorised by the nature of the inducing culture condition and the population frequency of the mutation [19]. FRAXA is a rare, folate sensitive fragile site (FSFS) associated with a trinucleotide repeat (CGG) expansion mutation in the 5′ UTR of the FMR1 gene resulting in fragile X syndrome, the most common inherited intellectual disability syndrome [20]. Twenty-six other FSFS have been reported cytogenetically but only eight of these have been molecularly characterized: FRAXA [20], FRAXE [21], FRAXF [22], FRA16A [23], FRA11B [24], FRA10A [25], FRA12A [26] and FRA11A [27]. To date, all characterized FSFS are due to a CCG/CGG trinucleotide repeat expansion. The expanded repeat and any adjacent CpG island become hypermethylated and transcriptionally silenced at a locus-specific repeat size-threshold [28]. At least four of the eight characterized rare, folate sensitive fragile sites are associated with a neurodevelopmental disorder. The relevance of folate sensitive fragile sites to intellectual disability (ID) is strengthened by five independent population studies that have all shown that autosomal folate sensitive fragile sites are overrepresented in people with ID compared to control groups without ID, with a prevalence of 1.2% and 0.27% respectively [reviewed in 29]. It thus seems likely that as yet uncharacterized CGG/CCG repeat expansions may be associated with neurodevelopmental problems.
An autosomal FSFS on chromosomal band 2q11.2-q12 has been previously described [30], [31], [32]. We studied three families with FRA2A-expression and a wide spectrum of neurodevelopmental and other anomalies. We identified expansion of an intronic CGG repeat leading to hypermethylation of at least one promoter of the AFF3 gene in all FRA2A carriers and we hypothesise that the associated transcriptional silencing of AFF3 in the brain may be responsible for some of the developmental features observed in FRA2A carriers.
Results
FRA2A is due to expansion of a polymorphic CGG repeat within AFF3
Using the simple repeat track on UCSC genome browser (GRCh37/hg19) we identified three candidate CGG repeats in the FRA2A containing region (2q11-12). One of these repeats is located within the LAF4/AFF3 gene (chr2 : 100721261–100721286; hg19), an autosomal homolog of the FRAXE-associated FMR2/AFF2 gene. In order to determine whether this CGG-repeat is expanded in FRA2A we used metaphase FISH analysis on a FRA2A-expressing individual (Figure 1; AII.3) with the BAC clone RP11-549H5 (chr2 : 100,588,792–100,759,365; hg19) encompassing the repeat. The FISH signal spanned the FRA2A fragile site (Figure 2A). Consistent with this the FISH signals from probes RP11-436F6 (AC010736) and RP11-506F3 (AC074387) were centromeric and telomeric to FRA2A, respectively. To establish co-location of the CGG repeat with the fragile site, long-range PCR-generated probes L10K (chr2 : 100721983–100733233; hg19) and L18K (chr2 : 100700447–100718834; hg19) were targeted to the genomic regions on either side of the (CGG)n repeat. These probes did indeed flank the fragile site, locating FRA2A to a 3.1 kb interval within the AFF3 gene (Figure 2B). The second red (L18K) FISH signal observed at 2q13 (Figure 2B) is the result of two copies of a 24 kb low copy repeat flanking the NPHP1 gene (Figure 2C) (chr2 : 110520380–110538822 and chr2 : 111347822–111366260; hg19). A copy of this sequence is also located adjacent to the CGG-repeat within the AFF3 gene in the region covered by L18K.
Fig. 1. Description of FRA2A Families A–C. 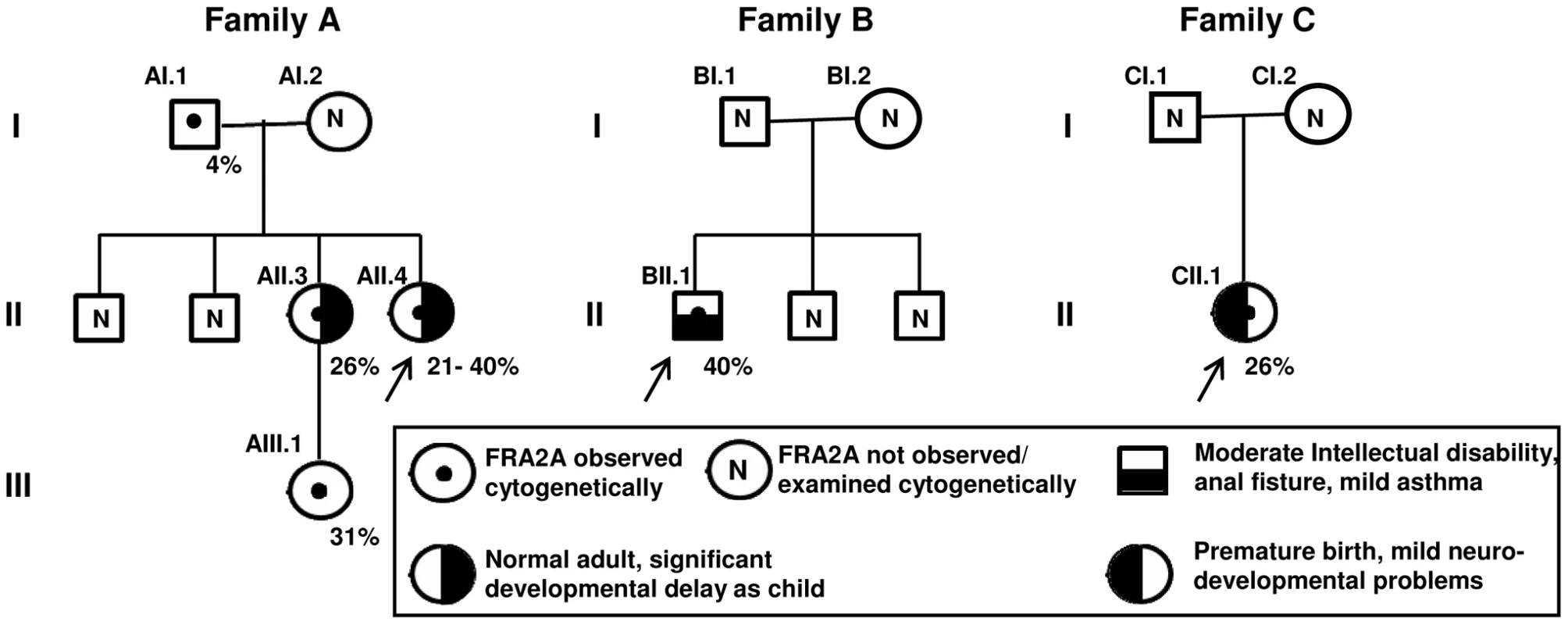
Females are represented by circles, males by squares. The percentage of cells showing FRA2A expression are indicated on the bottom right-hand side of the symbol. As discussed in the text the 4% FRA2A expression seen in individual AI.1 represents a false positive. The case number used to indicate the FRA2A carriers in the text is given on the top left-hand side of the symbol. N within a symbol indicated individuals expression of the fragile site was not examined or observed. The associated phenotypes in individuals AII.3, AII.4, BII.1 and CII.1 are detailed in the boxed symbol key. The proband in each family is arrowed. Fig. 2. FISH analysis of the FRA2A fragile site. 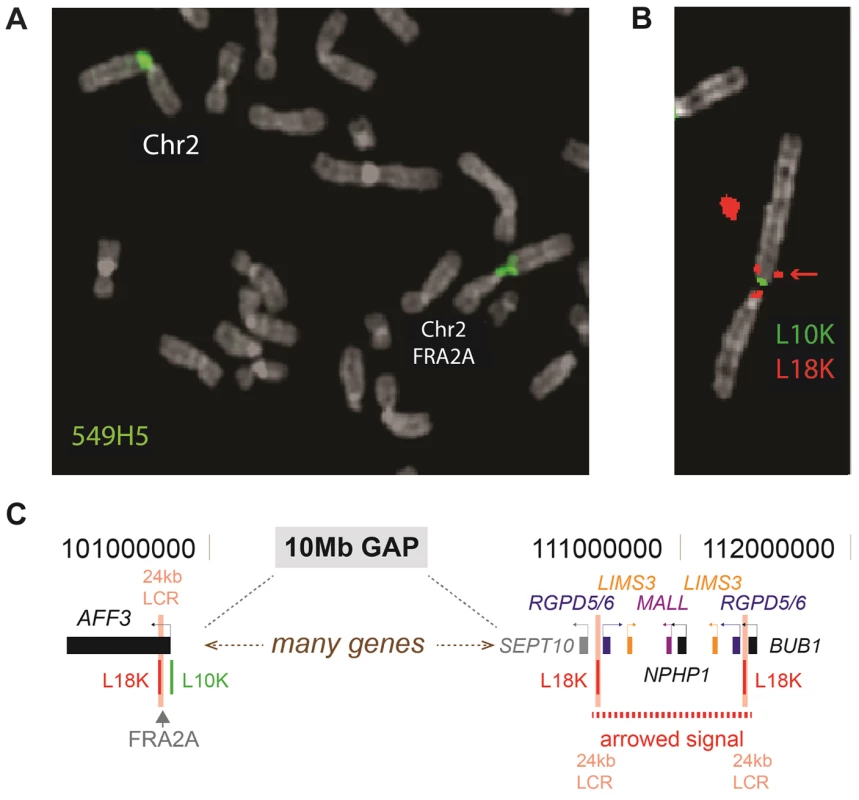
A. The BAC-clone 549H5 (labelled green) spans the fragile site FRA2A. B. FISH analysis using the 10 kb L10K (labelled green; chr2: 100721983–100733233; hg19) and 18 kb L18K (labelled red; chr2: 100700447–100718834; hg19) PCR-generated probes, targeted to map either side of FRA2A. The additional telomeric FISH signal on the red channel (red arrow chr2: 110520380–110538822 and chr2:111347822–111366260; hg19) is the result of a 24 kb low copy repeat (24 kb LCR pink text) encompassing L18K. C. A schematic representation of the position of the LCR. PCR-based amplification and sequencing of the AFF3 CGG repeat in 200 control chromosomes revealed it to be highly polymorphic with a length ranging from 3 to 20 copies (Figure 3). The most frequent CGG allele contains eight repeats (as does the genomic reference sequence; chr2 : 100721261–100721286; hg19). To exclude the possibility that apparently homozygous control individuals are in fact heterozygous for the detected allele in combination with a large expansion that is not detected by this protocol, we amplified these samples with gene specific repeat primed PCR (Asuragen Inc., Austin Texas, USA). This protocol enables us to detect expansions up to 300–500 repeats. However, no expanded repeats were detected in control chromosomes, and the genotype distribution agreed with Hardy-Weinberg equilibrium (P>0.05).
Fig. 3. Allele frequencies of the FRA2A associated CGG repeat in a population of 100 control individuals. 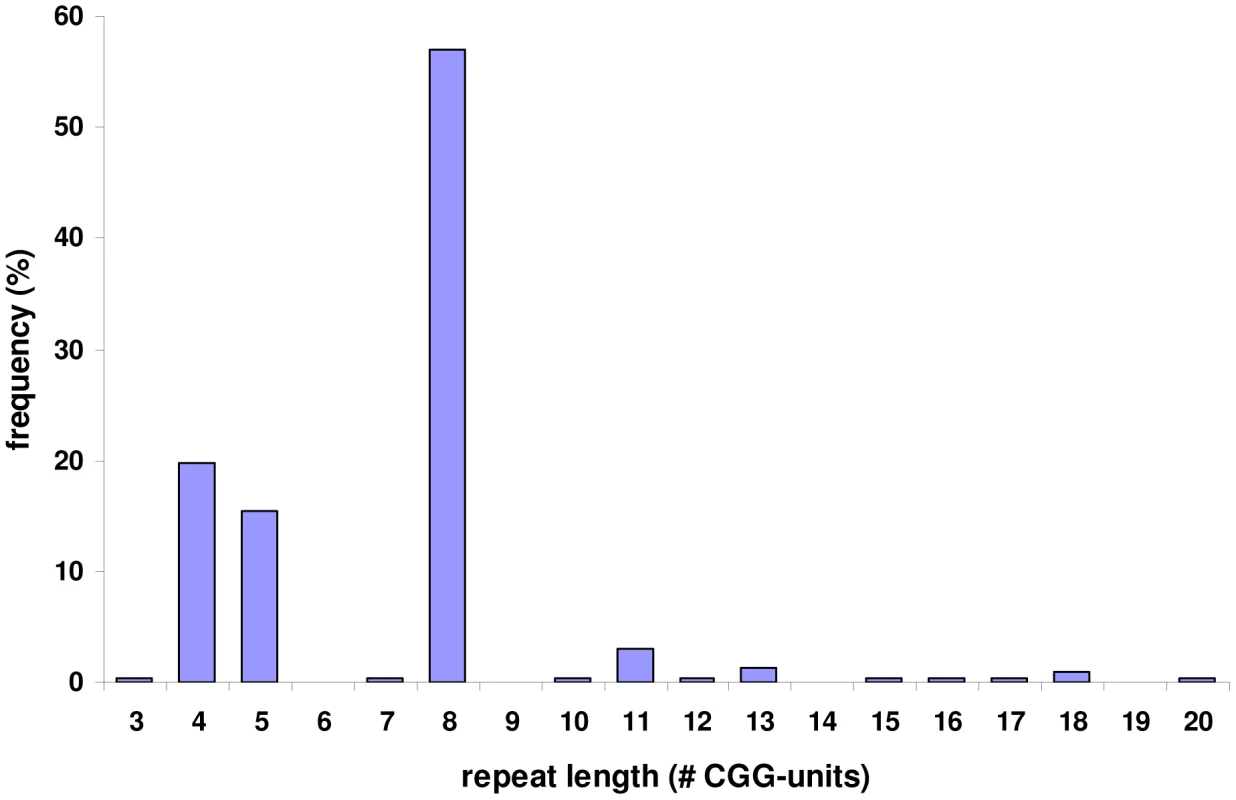
PCR amplification of the repeat and subsequent sequencing in 200 control chromosomes revealed that it is highly polymorphic with a length ranging from 3 to 20 copies. The most commonly found allele contains eight repeated units. PCR amplification of the repeat in the FRA2A-expressing individuals from AI.1, AII.3, AII.4, AIII.1, BII.1 and CII.1 (Figure 1) showed a single CGG allele in the normal size range. An additional smaller fragment was detected in subject AII.4. Sequence analysis of the smaller PCR product showed a 134-bp deletion encompassing the entire CGG repeat as well as some flanking sequences (Figure S1). This deletion was not detected in 800 control chromosomes. To visualize the repeat expansion in the FRA2A-positive individuals, we performed Southern blotting on HindIII digested genomic DNA of all available members of the three families (AI.1, AII.3, AII.4, AIII.1, BI.2, BII.1, CI.1, CI.2 and CII.1) and control samples. A 4.4 kb fragment was observed in all cases and controls. In five affected FRA2A-positive individuals we detected additional large fragments or smears compatible with the presence of an expanded allele (Figure 4). Two FRA2A-negative parents of FRA2A-positive individuals (BI.2 and CI.2) showed additional larger fragments indicative of repeat expansion. No evidence of an expanded fragment was observed in the control samples or in FRA2A-negative individual CI.1. Interestingly, individual AI.1 who had been reported as showing a low level of FRA2A-positive cells showed no fragments suggestive of an expansion mutation. A Southern blot of the same samples after NcoI digestion gave very similar results (data not shown).
Fig. 4. Southern blot analysis of the AFF3 CGG repeat in all available members of the three families and two unrelated control individuals (C). 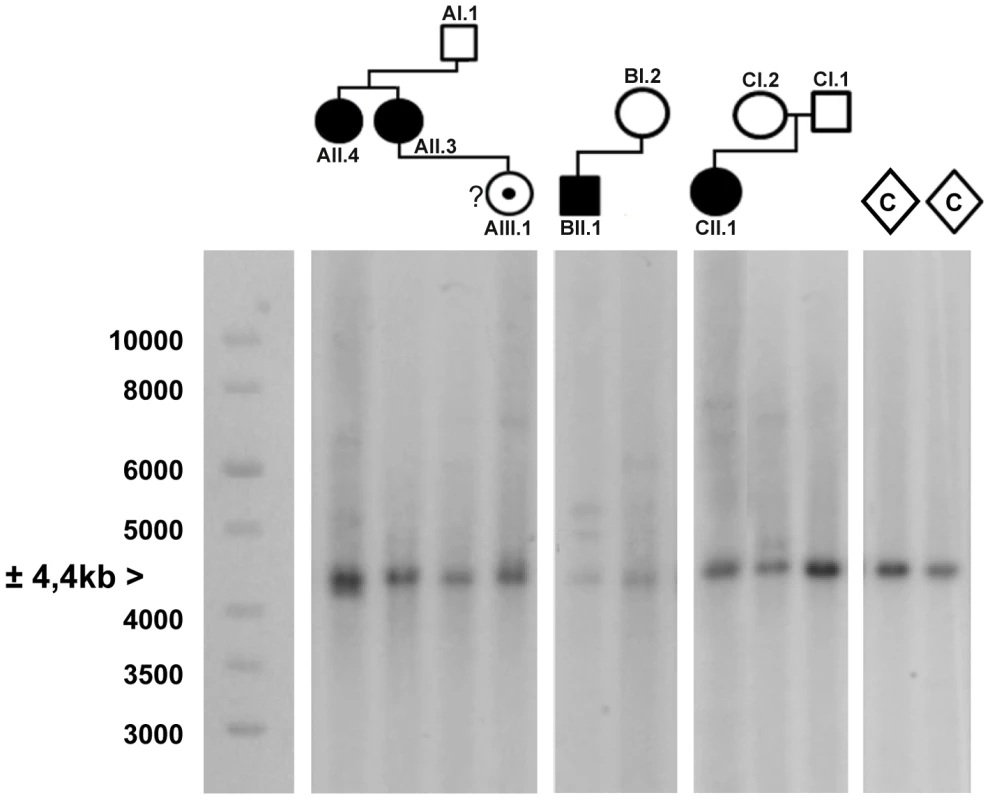
DNA restriction fragments obtained from blood samples of all available members of family A (lanes 1–4), B (lanes 5–6) and C (lanes 7–9) where blotted with a specific 32P-labeled probe (chr2:100088460–100089451; hg19) after digestion with HindIII. Lanes 10 and 11 contain DNA restriction fragments from two unrelated control samples (represented as diamonds) after digestion with HindIII. In addition to a normal size allele fragment of 4.4 kb, individuals CII.1, CI.2, BII.1, BI.2, AII.4, AII.3 and AIII.1 show additional fragments of a larger size, indicating repeat expansion. These expanded fragments were not present in the controls and individuals CI.1 and AI.1. A 1 kb length marker is presented at the left of the figure. A gene specific repeat-primed PCR assay was used for accurate sizing of the repeat expansion mutations. The mothers in family B and C both showed a slightly expanded allele (±120 and 106 repeat units, respectively) in addition to an allele in the normal size range (15 and 17 repeat units, respectively) while their offspring show one allele of 8 units compatible with paternal inheritance and one allele with a large expansion of more than 300 units (Figure S2). This strongly suggests that the expanded allele in both families was inherited from the mother (Table 1). In family A, the apparently FRA2A-positive individual AI.1 showed no evidence of an expanded allele on either Southern blot analysis or on repeat primed PCR. This apparent discrepancy could be resolved genetically using the microsatellite markers (D2S2209/AFMA246XE9 and D2S2311/AFMB355ZG1, Figure S3). The FRA2A-positive daughters of AI.1, AII.3 and AII.4, were shown to have inherited a different non-expanded allele (8 CGG units) from him, while they share a common allele with the mother (AI.2). Their FRA2A-positive grand-daughter, AIII.1 also inherited this maternal allele. The expansion was therefore probably inherited from AI.2, with the 4% FRA2A expression in AI.1 representing a false-positive cytogenetic result. Unfortunately DNA was not available from AI.2 to determine if she also carried an intermediately expanded allele.
Tab. 1. Sizing the repeat in all available family members with gene specific repeat primed PCR. 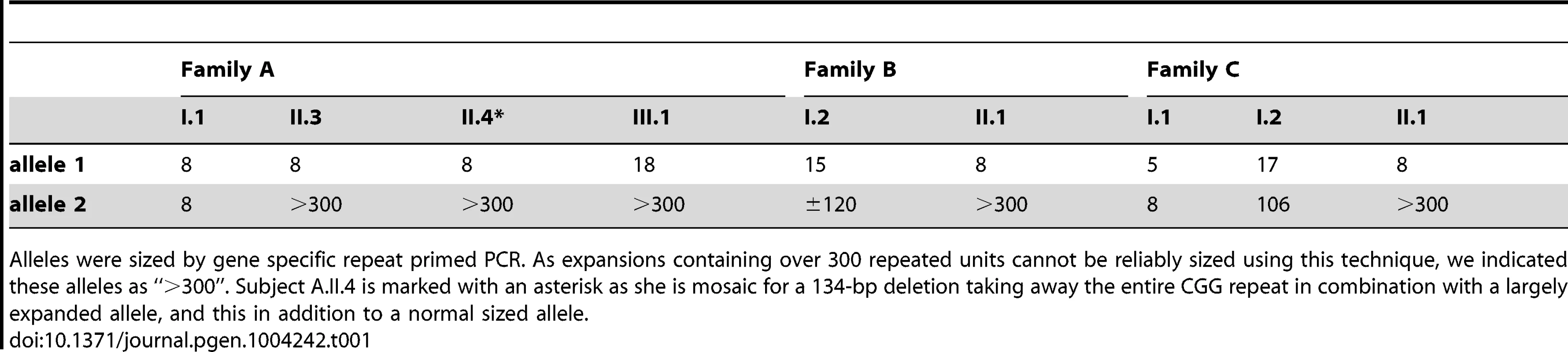
Alleles were sized by gene specific repeat primed PCR. As expansions containing over 300 repeated units cannot be reliably sized using this technique, we indicated these alleles as “>300”. Subject A.II.4 is marked with an asterisk as she is mosaic for a 134-bp deletion taking away the entire CGG repeat in combination with a largely expanded allele, and this in addition to a normal sized allele. Characterization of the two major AFF3 transcriptional start sites (TSS)
The RefSeq AFF3 gene model consists of 23 coding exons and two 5′ non-coding exons together spanning 558 kb genomic DNA [33]. Here, the 5′ non-coding exons are named exons 1 and 2 with the first coding exon called exon 3. An AFF3-specific cDNA probe encompassing the final 5 exons was used for northern blot analysis. A major transcript of approximately 8 kb, corresponding to the predicted size was detected in several tissues, including brain, placenta and lung (data not shown).
To determine the precise location of the AFF3 transcriptional start sites (TSSs) in relation to the expanded repeat we used Cap Analysis of Gene Expression (CAGE) data from the FANTOM4 consortium. FANTOM4 produces sequence tags from the 5′ end of mRNAs from many different tissue sources and species and maps these to the reference genome [34]. Mapped CAGE tags reveal the sites of transcription initiation at single nucleotide resolution and provides a semi-quantitative measure of steady state mRNA levels for those transcripts using a tags per million (TPM) metric. The TPM scores for three different human tissue groups (brain, immune and other tissues) are plotted in Figure 5B. AFF3 is transcribed in telomeric to centromeric orientation and the x axis of these graphs represent the hg19 genomic coordinates. The location of the 25 annotated exons of the RefSeq model of the AFF3 gene (NM_001025108) is represented above the graphs using the same genomic coordinates. To assess the transcriptional activity of AFF3 during early brain development we mapped transcriptome sequencing reads of mRNA (RNA-seq) from three different human fetal brain samples to the regions surrounding the TSS identified by CAGE (Figure 5C).
Fig. 5. AFF3 protein-domain and gene structure. 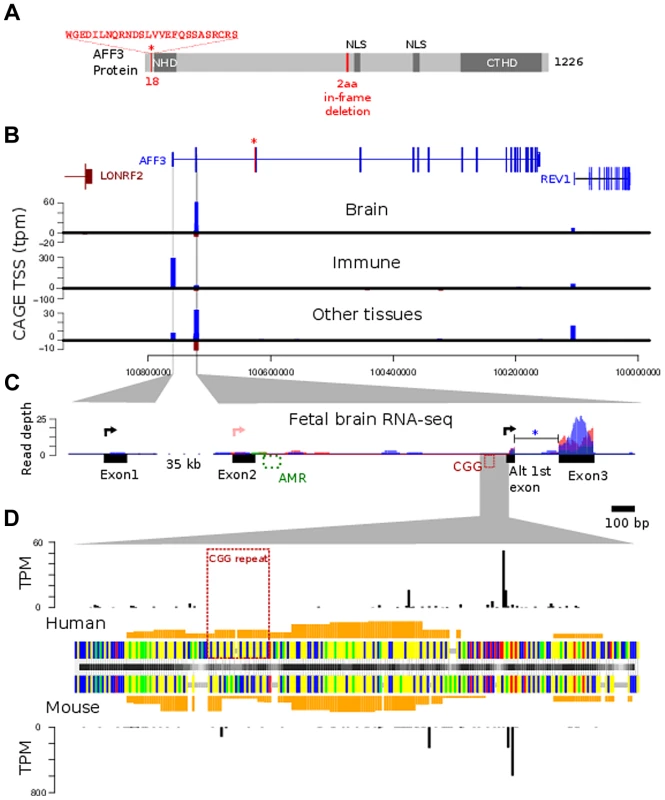
A. diagrammatic representation of the AFF3 protein, showing the N-terminal homology domain (NHD), C-terminal homology domain (CTHD) and two predicted nuclear localisation signals. The 18 amino acids encoded by the alternatively spliced exon 4 are represented as an “insertion” at position 18 of isoform 1. The 6-bp in-frame deletion we identified in exon 14 of subjects AI.1 and AII.4, removing two amino acids (position 619 and 621 respectively) is indicated in red. This deletion was predicted to be benign. B. Genomic structure of AFF3 with the alternately used spliced exon 4 shown in red with asterisk. AFF3 exons 2, 3 and the alternate first exon are not separately resolvable at this resolution but shown in detail in panel C. Transcription left to right is shown in blue, right to left in brown. CAGE tag defined transcription start sites are shown aligned with annotated gene structure (blue forward strand transcription, brown reverse strand transcription shown with negative counts). Y-axis values show average tags per million (TPM) from CAGE libraries from the indicated tissue groups. C. Finer details of the AFF3 TSS regions are shown along with histograms of human fetal brain RNA-seq read coverage; three replicates are coloured separately. Splicing of the intron between the alternate first exon and exon 3 (blue asterisk) was supported by 9 independent RNA-seq reads and found in all three replicates. The CGG repeat and abnormally methylated region (AMR) are shown in red and green respectively, The major transcription start sites are shown as black arrowed lines. There is no supporting evidence for the RefSeq TSS represented by the pink arrowed line in our data. D. Alignment of human CGG repeat region and associated TSS with the orthologous mouse region. Nucleotides are color coded (A = green, G = yellow, C = blue, T = red, alignment gaps are grey). Orange histograms show the predicted G-quadruplex forming potential of the human and mouse sequences. Outer histograms show CAGE tag 5′ ends at single nucleotide resolution in both human (top) and mouse (bottom). TPM counts shown are the average from brain derived CAGE libraries in each species and represent the precise location of transcription initiations. CAGE tag sequencing demonstrates two robust TSSs. The most 5′ TSS corresponds to the 5′ end of exon 1 at position chr2 : 100759169 (GRCh37/hg19). This TSS is highly expressed in immune tissues with a mean of 300 tags per million shown by the blue bar in the middle graph (Figure 5B). There is no obvious transcription of exon 1 on RNA-seq from human fetal brain (Figure 5C, left-hand side). A second robust TSS was identified in CAGE data from brain and other tissues which mapped within intron 2 as shown by the blue bars in the top and bottom graphs in Figure 5B. The highest levels are seen in the brain (mean of 60 tags per million). An expanded representation of this region is shown in the right-hand side of Figure 5C. This shows no evidence of transcription of exon 2 but strong expression in exon 3. This also shows evidence of an alternative exon 1 immediately 3′ to the TSS (Figure 5C, right hand side, black arrow). The TSS lies immediately downstream of the CGG repeat (Figure 5D) suggesting this expansion prone repeat is located within the core of an alternate AFF3 promoter.
The location and tissue specific activity of the AFF3 promoters are similar in mouse Aff3
FANTOM4 CAGE data also encompasses a range of mouse tissues. From this we can demonstrate that both the exon 1 TSS and intron 2 TSS are evolutionarily conserved and functional TSSs. Although the CGG repeat itself is not conserved, a region of low compositional complexity flanked by highly constrained non-coding sequences is a conserved feature of the intron 2 TSS promoter (Figure 5).
Whole mount in situ hybridization (WISH) using riboprobes targeted to the 3′UTR of Aff3 was used to determine the developmental expression pattern in mouse embryos at 9.5, 10.5, 11.5 and 12.5 days post coitus (dpc). This has shown site and stage specific expression of Aff3. The most striking areas of expression are in the somites, the upper limb buds, the diencephalon/prosomere I and the fusing primary palate (Figure 6).
Fig. 6. Site and stage specific expression of Aff3 in mouse embryos. 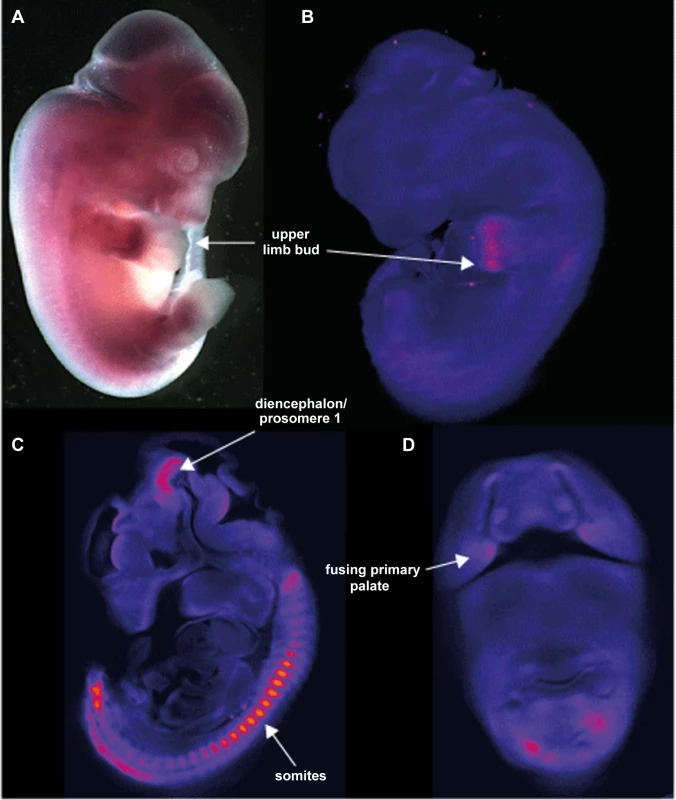
A. photomicrograph of a lateral view of a 12.5Aff3. The only area of strong expression at this stage is in the hand plate. B. show a false coloured but unthresholded image of the brightfield optical projection tomography (OPT) scan of a 11.5 dpc embryo with digital sagittal and coronal sections from the same embryo shown in C and D. Strong expression is seen in the somites, upper limb bud, the fusing primary palate and the diencephalon and prosomere1 regions of the developing brain. AFF3 promoter regions are hypermethylated in individuals with the FRA2A CGG-repeat expansion
In all rare, folate-sensitive sites characterised to date, CGG repeat expansions are associated with hypermethylation of the surrounding CpG island. Bisulfite sequencing indicated hypermethylation of the CpG island in all five affected FRA2A carriers AII.3, AII.4, AIII.1, BII.1 and CII.1, while in healthy control individuals this region was not methylated (figure S4). In order to quantify the methylation level, we subsequently subjected all samples to pyrosequencing. This technique allows accurate quantification of methylation across individual CpG sites [35]. A methylation frequency of 50% would be consistent with complete methylation of the expanded allele as all affected individuals in this study are heterozygous. We analyzed a short region of genomic DNA (chr2 : 100721843–100721885; hg19) adjacent to the CGG repeat in all available family members, containing four analysable CpG dinucleotides (Figure S1 and Table 2). Methylation percentages are congruous across the 4 CpG-sites in each individual (p-values ranging from 0.417 to 1.000) and are consistent with hypermethylation of the CGG repeat region in individuals carrying an expanded allele. There is some suggestion that the methylation frequency may be increasing upon generational transmission.
Tab. 2. Pyrosequencing reliably quantified the methylation of four consecutive CpG dinucleotides per individual (chr2:100721843–100721885; hg19). 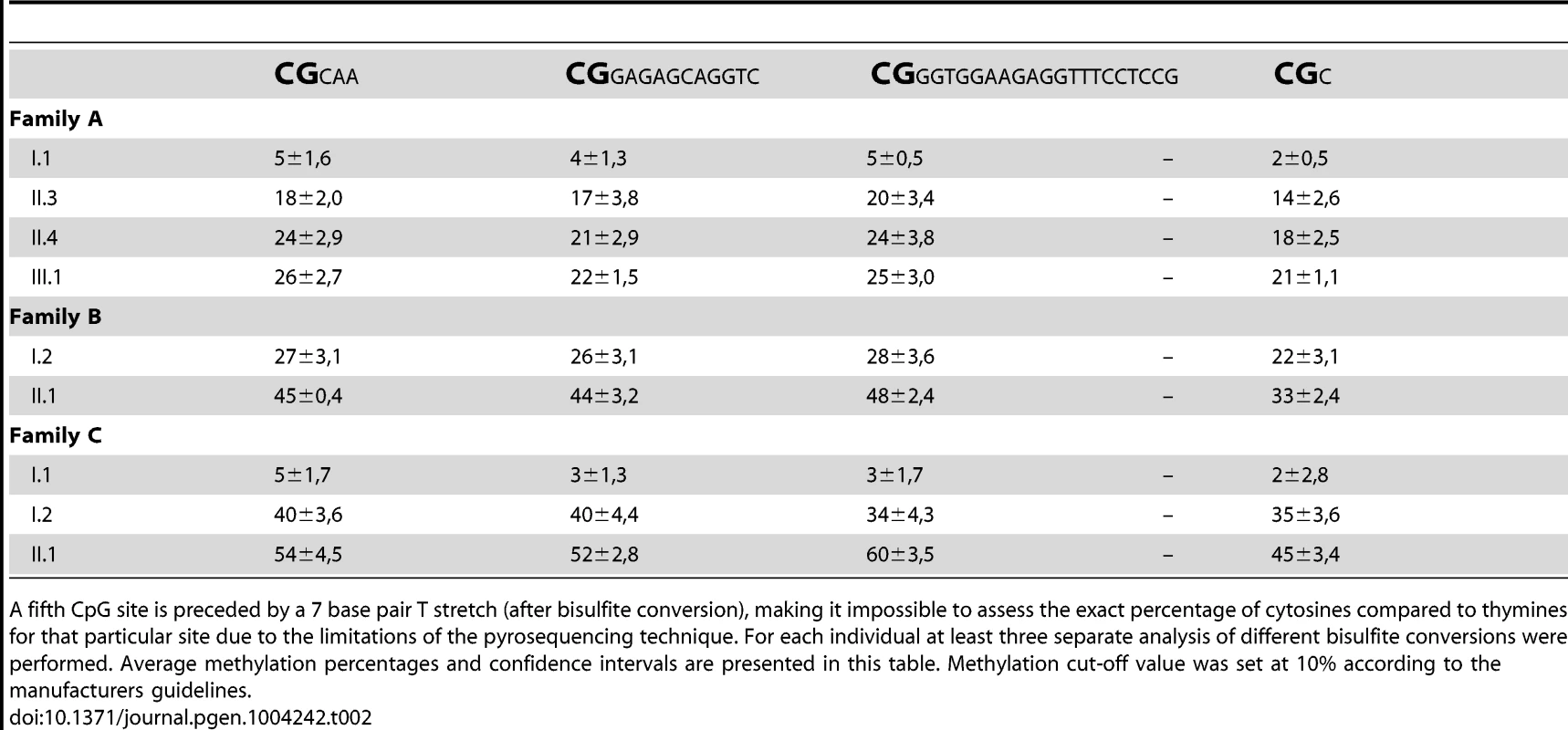
A fifth CpG site is preceded by a 7 base pair T stretch (after bisulfite conversion), making it impossible to assess the exact percentage of cytosines compared to thymines for that particular site due to the limitations of the pyrosequencing technique. For each individual at least three separate analysis of different bisulfite conversions were performed. Average methylation percentages and confidence intervals are presented in this table. Methylation cut-off value was set at 10% according to the manufacturers guidelines. To exclude a non-specific effect of increasing age on the methylation of this region, we pyrosequenced 72 individuals from 24 unrelated two-generation families. The ages within this control group varied between 0 and 11 years for the children and between 23 and 53 years for the parents, which is comparable to the age distribution in our FRA2A families at the time of DNA collection. No methylation above the threshold was detected in any control individual (data not shown). In the FRA2A-carriers the methylation level for each of the four CpG sites differed significantly from the level determined in this control population (p-values≤0.001 for CpG site 1,2 and 4 and p<0.004 for CpG site 3), suggesting that expansion of the repeat is associated with hypermethylation of the region surrounding the repeat.
In AII.4, mosaic for a CGG-repeat expansion and a deletion, the promoter region on the expanded allele was hypermethylated as determined by bisulfite sequencing. The allele with the 134-bp deletion was not methylated, as determined by Southern blotting after double digestion with BamHI and NotI (data not shown).
Monoallelic expression of the AFF3 gene in FRA2A carriers
The level of AFF3 expression in lymphoblastoid cell lines is too low to be reliably measured by RT-qPCR. To determine if FRA2A results in transcriptional silencing of AFF3 in cis, we utilized single nucleotide polymorphisms (SNP) mapping within the open reading frame. Two such SNPs were found to be heterozygous in the genomic DNA of affected FRA2A carrier BII.1. Rs4851214 maps to exon 14 and heterozygous individuals display both an T and a C peak (c.1499T/C) on Sanger sequencing, while rs13427251 maps to exon 25 and heterozygotes for this SNP show both an A and a T peak (c.5475A/T). Sequencing cDNA from BII.1, revealed only a C peak at rs4851214 (Figure 7) and only the A allele peak was seen for rs13427251. These results are consistent with monoallelic expression of AFF3 in this individual. Genomic DNA from BII.1's mother (BI.2) is homozygous for the rs13427251 T allele (g.5475T/T) indicating that it is the maternal T allele carrying the CGG expansion that is silenced in BII.1. cDNA of BI.2 showed a heterozygous signal for rs4851214 (c.1499T/C) indicating that both AFF3 alleles are transcribed in the mother despite partial methylation of her expanded allele.
Fig. 7. Analysis of rs4851214 which maps to coding exon 14 of AFF3 using paired genomic DNA and cDNA templates from the unaffected carrier mother BI.2 and the affected carrier son BII.1 from family B. 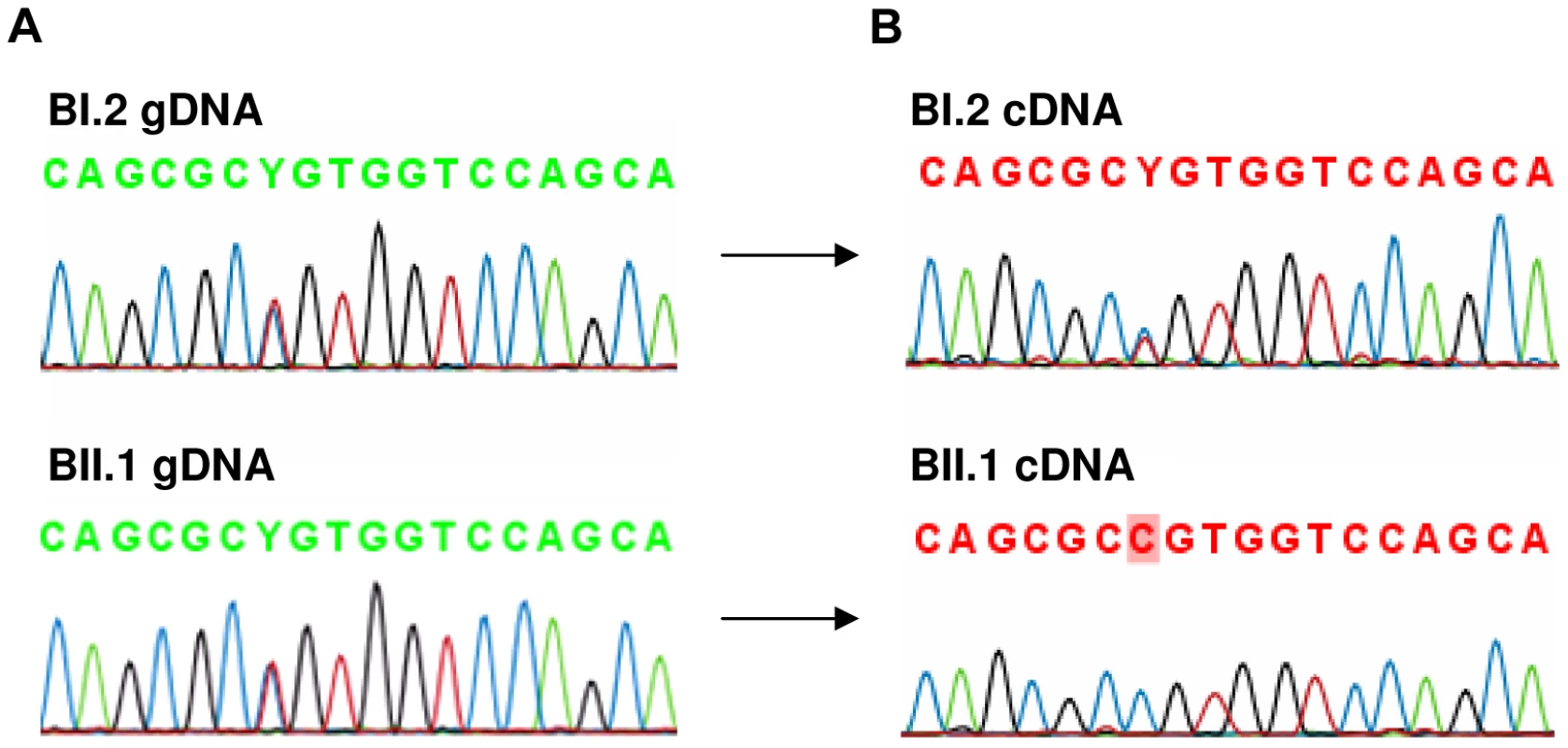
A: Both BI.2 and BII.1 are heterozygous for SNP rs4851214 in genomic DNA as T and C peaks are visible at this site. B: Analysis of cDNA from subject BII.1, shows that only the C allele could be detected in this patient, indicating monoallelic expression, while in cDNA of subject BI.2 the heterozygous T/C signal was observed, indicating transcription of both alleles. Analysis of clinical phenotypes associated with FRA2A
Six FRA2A carriers were initially included in this study (Figure 1, Table 3), four in Family A and one each in Families B and C. Individual AIII.1 is currently too young to make any conclusion about cognitive development. Individual A1.1 has no discernible affected phenotype and he also has the lowest expression of the fragile site. The molecular analysis presented above strongly suggests that the 4% apparent expression of this case represents a false positive and so was excluded from the clinical analysis. Two FRA2A carriers AII.3 and AII.4 are adults; both had global delay in their neurocognitive development to a level that merited genetic investigations during childhood and their long-term placement in special educational facilities. However, as adults both of these individuals are functioning at a normal level and are in full time employment. This raises the possibility that they had a true delay in development rather than a fixed disability. Something similar is observed in FRAXE patients as most adult FRAXE males adapt to live a normal life. Individual CII.1 was born prematurely and had significant respiratory distress, which confounds the unambiguous interpretation of the cause of her mild developmental delay. BII.1 has the most significant delay, currently without a plausible non-genetic explanation. Thus all four of the characterized true FRA2A carriers did have significant delay in their motor and language development.
Tab. 3. Clinical findings in FRA2A carriers. 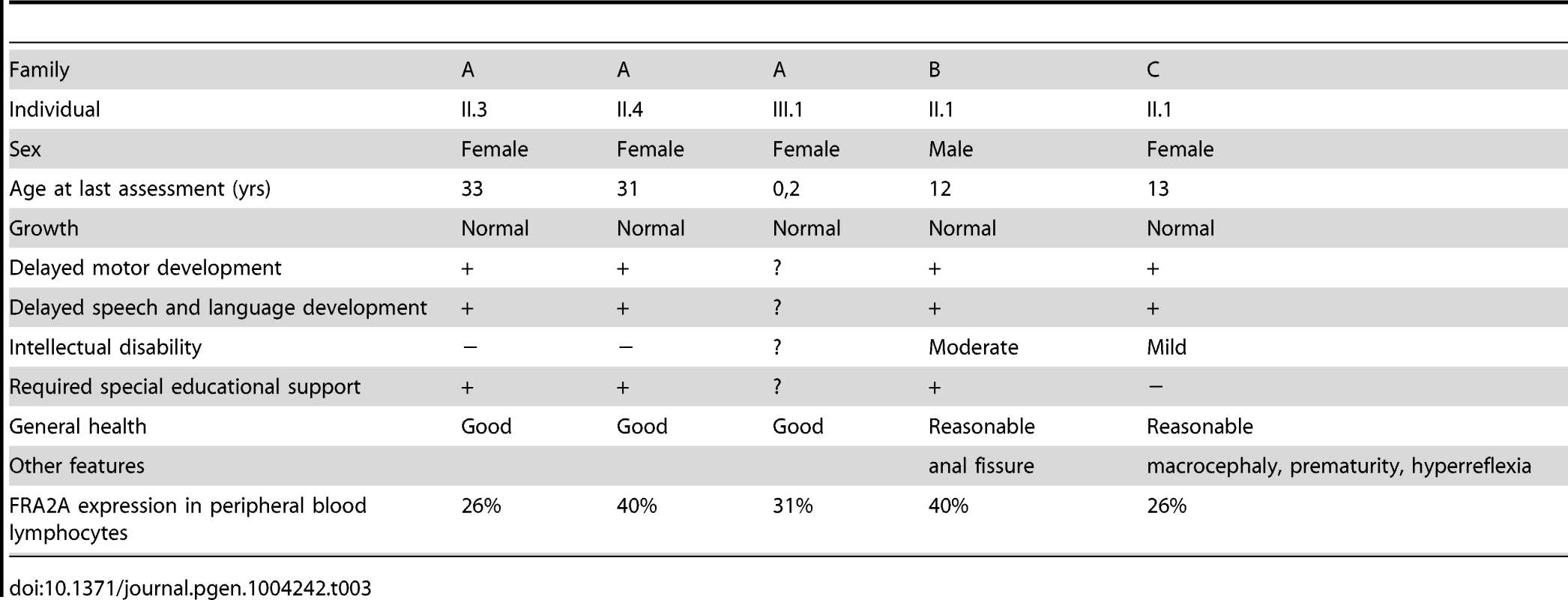
To determine whether the FRA2A carriers with neurodevelopmental anomalies had additional mutations in the protein coding region of the AFF3 gene, mutation analysis of all coding exons was performed. No sequence abnormalities were detected in any of the affected FRA2A carriers, except in subject AII.4, in which a 6-bp deletion was identified in exon 14, removing two amino acids: Threonine and Alanine (position 619 and 621 respectively) (Figure 5A). Both amino acids are found in a region, enriched with proline, serine and glutamic acid residues and located between the transactivation domain and the nuclear localisation signal (NLS). According to different prediction software (mutationtaster, mutation assessor, Indelz), the deletion is benign. Moreover, this 6-bp in-frame deletion was also present in the unaffected father (AI.1).
Discussion
We provide compelling evidence that the molecular basis of the FSFS FRA2A is a CGG repeat expansion in an alternative promoter which is active in the brain and is located in the intron immediately 5′ to the first coding exon of the major AFF3 transcript. The FRA2A-associated repeat is polymorphic in the general population. Repeat primed PCR showed all individuals with an expansion of over 300 repeat units expressed FRA2A in more than 20% of their cells. The expansion was associated with hypermethylation of a CpG island adjacent to the alternative promoter and, in at least one case, resulted in transcriptional silencing of AFF3. These results are consistent with the epigenetic effects that have been described in other FSFS. Within each of the three families studied here higher levels of methylation correlate well with neurodevelopmental delay, higher repeat size and silencing of AFF3. However, there are striking disparities in the absolute levels of methylation observed between the families. For example individuals AII.3 and AII.4 both have >300 repeats and had evidence of neurodevelopmental delay during childhood but have lower levels of methylation than BI.2 (∼120 repeats, biallelic expression of AFF3) and C1.2 (106 repeats) neither of whom showed evidence of developmental delay. A likely explanation for this is that the assay used here was performed on peripheral blood-derived cells whereas the phenotype in which we are interested is developmental and neural. Many developmental loci appear to show tissue specific differences in DNA methylation [36]. In this regard the ability to model brain development using cerebral organoids from patient-derived pluripotent cells [37] may enable more interpretable transcriptional and epigenomic analyses of the consequences of CGG-repeat expansion on AFF3.
Nonetheless, all individuals for whom a significant methylation frequency was measured, show an expanded allele in the pre - or full mutation range. Repeat sizes of >300 do correlate with neurodevelopmental effects and expression of the fragile site in a significant percentage of cells. Carriers of an expanded allele in the premutation range are phenotypically normal but may show lower levels of expression of the fragile site.
In one individual (AII.4) a mosaic deletion of 134-bp removed the entire CGG repeat and the CpG island on the deleted allele remains unmethylated (data not shown). A similar combination of a full mutation with an expanded repeat and a deletion encompassing the repeat has been reported in several fragile X syndrome patients and recently also in a myotonic dystrophy type 1 case [38], [39]. In the fragile X syndrome, the phenotype of deletion cases is often indistinguishable from that of carriers of an expanded repeat, a reported exception being an unaffected individual where the deletion is the major allele present, and the transcription and translation start sites are preserved [40].
AFF3 belongs to a family of nuclear transcription activating factors including AFF1/AF4, AFF2/FMR2 and AFF4/AF5q31 [33], [41], [42]. These proteins form super elongating complexes (SEC) with active P-TEFb (positive transcription elongation factor) and AF9/ENL. SECs regulate the induction and expression of different subsets of genes. AFF3 is the closest paralog of AFF2, and is 36% identical on the amino acid sequence level. They share functional domains including the N-terminal Homology Domain (NHD), the C-terminal Homology Domain (CTHD) involved in intranuclear localization and binding of G-quadruplex RNA structures, and the ALF domain that promotes protein degradation through the proteasome pathway and the transactivation domain (TAD). Intriguingly, the highly conserved intron 2 TSS sequences and to a lesser extent the CGG repeat itself, are predicted to have a strong propensity to form G-quadruplex structures (Figure 5D, orange bars) with the most downstream of these being present in the 5′ UTR of the produced transcript. Given that AFF3 is known to bind G-quadruplexes, there is the potential for AFF3 autoregulation at this promoter. Both AFF2 and AFF3 localize to nuclear speckles and modulate splicing efficiency [43]. The expression pattern of murine Aff3 overlaps to a considerable extent with that of murine Aff2 [43], [44].
FRAXE is associated with loss of expression of AFF2 through dynamic repeat expansion of a CGG repeat in the 5′ UTR. FRAXE causes an X-linked non-syndromic intellectual disability [45]. AFF2 may play an important role in learning, memory, and language-learning processes [46]. Moreover, rare missense variations in the highly evolutionary conserved sites of the AFF2 gene might be associated with autism spectrum disorder [47]. The Aff2 knockout mouse model shows specific cognitive deficits, including an impaired conditioned fear memory over longer periods and enhanced long-term potentiation in the hippocampus [48], [49]. Aff3 expression is upregulated in cortical neurons during the initial steps of cortical differentiation and is downregulated in postnatal cortex, indicating its involvement in brain development [44]. We have shown that Aff3 shows strong regional expression in the developing mouse brain.
AFF3 is thus a reasonable candidate for the neurodevelopmental features seen in FRA2A carriers in our families. Our data predict that FRA2A carriers are haploinsufficient for AFF3, at least in a subset of tissues. A confident assignment of causality to the association of AFF3-associated repeat expansion mutations with neurodevelopmental anomalies is confounded by the rarity of the fragile site and the strong bias in clinical ascertainment. It is, however, intriguing that delay in motor and language development appears to be a common feature in the individuals presented here and this may represent a true delay in development rather than a fixed disability. AFF3 deficiency may then be involved in the speed of skill acquisition without impairing the developmental capacity.
A de novo microdeletion of 500 kb on chromosome 2q11.2 removing only AFF3 [50] has been reported in a girl with a severe multisystem disorder consisting of a mesomelic skeletal dysplasia (fibular agenesis, abnormal and triangular tibiae, short neck), urogenital tract malformations, delayed psychomotor development and recurrent apnoea leading to respiratory arrest at the age of 4 months. This clinical presentation is clearly very different to those associated with FRA2A but would be consistent with the expression pattern we demonstrate in mouse embryos. The clinical differences may be explained by the fact that the methylation of the repeat and thus the inactivation of the AFF3 gene presumably takes place several weeks after fertilization, so that development during the first weeks is not affected [51]. It is also plausible that the transcriptional silencing associated with FRA2A may by tissue specific given that the alternative promoter that is immediately adjacent to the expansion mutation shows evolutionarily-conserved tissue-specific activity, and appears to be the main driver of AFF3/Aff3 transcription in the brain in humans and mouse.
Both rare and common fragile sites often co-localize with evolutionary breakpoints as was postulated previously by Ruiz-Herrera et al. [52], [53]. We have shown through FISH and BLAST-analysis that the region close to the AFF3 repeat is indeed involved in a chromosomal rearrangement including a duplication and inversion of a 24 kb sequence from 2q13 to 2q11.2 followed by an ancestral head-to–head chromosomal fusion that led at 2q13 that led to the formation of human chromosome 2. The 2q11.2 breakpoint of this rearrangement falls within base pairs of the repeat and is also present in other primates. The 2q13 region also co-localizes with FRA2B, an as yet to be characterized rare fragile site of the same type.
In conclusion, we report a CGG repeat expansion mutation as the molecular cause of the fragile site FRA2A. FRA2A expression is associated with methylation of an AFF3 promoter and apparent transcriptional silencing of AFF3. It is currently difficult to unequivocally link FRA2A to a specific neurodevelopmental phenotype but it is plausible that haploinsufficiency for AFF3 in the developing brain is related to a true developmental delay and possibly mild intellectual disability.
Materials and Methods
Ethics statement
The ethics committees of the participating study centers approved the study protocol and all participants gave their written informed consent. The study was in accordance with the principles of the current version of the Declaration of Helsinki. The fetal brain tissue was collected with informed written consent and ethical approval by Southampton and South West Hants LREC. The fetal tissue was obtained following surgical termination of pregnancy and staged according to the Carnegie Classification [54], [55].
Family description: Clinical diagnosis and chromosome analysis
Family A
The proband (AII.4, Figure 1) was originally investigated at the age of 7.5-years for mild to moderate learning disability and enuresis. She was born at term following an uneventful pregnancy. There were no neonatal problems but her early motor and language development was reported to be slow. Her general health was good and her weight was on the 25th centile for her age. She attended a school for children with special educational needs. When last seen at the age of 20-years she was healthy and was working as a checkout operative in a high-street store and had no evidence of a significant functional neurocognitive deficit. Her elder sister (AII.3) had been investigated several years earlier for moderate global learning disability and had attended the same a school for children with special educational needs. Again she displayed no evidence of significant cognitive impairment when seen at the age of 26-years. Indeed she was very much valued in the workplace for remembering numerical codes for almost the entire stock inventory. AII.3 has a healthy daughter (AIII.1), born after an uneventful pregnancy. Evaluation of AIII.1 at the age of 3 weeks revealed no phenotypic abnormality. No intellectual problems were apparent in any other relatives of the three-generation family tree. DNA analysis of the FRAXA locus was normal and permission to use the remaining peripheral blood DNA for research purposes was obtained from patient AII.4 (Figure 1). An Epstein-Barr-transformed lymphoblastoid cell line was established at ECACC (Cambridge, UK) from a peripheral blood sample of proband AII.4. The index patient showed FRA2A expression in 40% of the examined cells. At the age of 20-years, chromosome analysis was repeated and confirmed FRA2A expression, this time in 21% of the examined cells. Subjects AII.3 and AIII.1 expressed the fragile site FRA2A in respectively 26% and 31% of the examined cells. The unaffected father (subject AI.1) showed FRA2A expression in 4% of his cells, whereas in two unaffected siblings and the mother no indications were found for FRA2A expression (Figure 1).
Family B
The proband (II.1, Figure 1) was born at 39 weeks after an uneventful pregnancy. He was the third child of a non-consanguineous Caucasian couple. There were no problems in the neonatal period. At 8 weeks of age, an anal fistula was diagnosed and required three surgical procedures between 5 and 8 months for cure. He developed mild asthma at 10 months of age. Food allergies were demonstrated at 12 months of age and were managed by dietary modification. His height and head circumference were within the normal range, but his development was slow. He had mild dysmorphic features, including telecanthus, slightly short palpebral fissures, smooth philtrum, thin upper lip and a small mouth. Psychological assessment at the age of 12 using the WISC-IV showed a full-scale IQ in the range 40–52 (<0.1 centile), within the moderate range of intellectual disability. The Vineland Adaptive Behaviour Scales 11 gave an Adaptive Behaviour Composite in the range 55–67, below the first centile and in the low range. His parents and two elder brothers had normal intelligence and were not dysmorphic. Routine chromosomes, subtelomere FISH of chromosomes, molecular testing for fragile X syndrome and urinary amino acids/organic acids/mucopolysaccharides gave normal results. The proband expressed the fragile site FRA2A in 40% of the examined cells. Fragile site expression was not examined in the other family members.
Family C
The proband (II.1, see Figure 1) was born at 33 weeks after an uneventful pregnancy. She had respiratory distress syndrome and was treated with surfactant and ventilated for five days. There were no significant complications in the neonatal period. Her early development was delayed and she suffered from intermittent asthma and required ventilator tubes for serious otitis media. At three years of age, she was seen because of speech delay – her expressive language was estimated to be at the 2 year level while receptive language and general development were assessed at the 2.5–3.0 year level. She was noted to be macrocephalic, hypotonic and hyperreflexic. Height was normal. At 13 years she is in an age appropriate class in high school and undertakes all subjects, except mathematics, with her peers, but struggles to keep up. She has poor concentration, is easily distractible and does not like changes in routine or to immediate expectations. She is doing well socially. The following investigations gave normal results: CT brain scan, urine amino acids and organic acids, chromosomes, creatine kinase and thyroid function tests. The proband showed FRA2A expression in 26% of the examined cells. Fragile site expression was not examined in the other family members.
Fluorescent in situ hybridisation (FISH) mapping
Peripheral blood lymphocyte-derived metaphase chromosome preparations from individual AII.3 were obtained using standard methods. An AFF3-containing BAC-clone from the RPCI library, RP11-549H5 (AC092667), and clones mapping centromeric (RP11-436F6, AC010736) and telomeric (RP11-506F3, AC074387) to AFF3 were obtained from the BACPAC Resource Center (Oakland, California, USA). Long Range-PCR (LR-PCR) was used to generate probes of 10 kb and 18 kb situated respectively immediately 5′ and 3′ to the promoter region of AFF3. The following primer pairs were used: L10K (forward 5′-TGCAGGAATGAATGAAGGGCAAGCAA-3′ and reverse 5′-TGGCCTCTGGGTGTCGACTTCAAACT-3′) and L18K (forward 5′-ACAGTTTGGCTTGACCGGGAGGGTTT-3′ and reverse 5′ - TCAAAAATGTTCCCTTGCCCACAGTGC-3′). LR-PCR was performed using the Expand Long Template PCR System (Roche, Basel, Switzerland) according to the manufacturer's instructions. The amount of BAC DNA used per reaction was 5–10 ng. All probes were labelled with digoxigenin-11-dUTP or biotin-16-dUTP (Roche, Indianapolis, IN) by nick translation. DNA hybridisation and antibody detection were carried out as described previously [56]. At least five metaphases were analysed for each hybridisation, using a Zeiss Axioplan 2 fluorescence microscope equipped with a triple band-pass filter (#83000 for DAPI, FITC and Texas Red; Chroma Technology, Brattleboro, VT). Images were collected using a cooled CCD camera (Princeton Instruments Pentamax, Roper Scientific, Trenton, NJ) and analysed using IPLab software (Scanalytics, Vienna, VA).
PCR amplification and hybridization of the FRA2A-associated CGG repeat
PCR amplification of the normal FRA2A CGG repeat was performed with the aid of 2.5× PCR Enhancer solution (Invitrogen, Carlsbad, CA, USA) using a forward primer P1 (5′-GGCCGTAAAAGCCACGAGAGAGGG-3′) and a reverse primer P2 (5′-CTTGCGCGCAGGCACACTCAAGAG-3′) derived from the sequences flanking the repeat. PCR products were sequenced and subsequently analysed by use of an ABI Prism 3130 Genetic Analyzer (Applied Biosystems, Foster City, CA, USA).
A Southern blot was created by digesting 10 µg of DNA, extracted from blood or Epstein-Barr-virus transformed cells, with the restriction enzymes HindIII and NcoI (Fermentas GmbH, St. Leon-Rot, Germany) in separate reactions. The digested DNA was then separated by electrophoresis on a 0.7% agarose gel. No ethidium bromide was added during this electrophoresis step to avoid product-related smearing on the gel that would cause overestimation of the mosaicism of repeat sizes [57]. After subsequent denaturation and neutralisation the DNA fragments were transferred to Hybond N+ membranes. Hybridisation was performed at 65°C using a specific 32P-labeled 992 bp PCR probe (forward primer 5′ - AGCCTTTGTTCCTGGGAATGCTGTCTCAAT -3′ and reverse primer 5′ - GGAAAGGCAGGTGATCAGCTAGAAGGGTG -3′).
Repeat primed PCR was performed to interrogate the number of CGG repeats in the AFF3 gene with Asuragens CGG Repeat Primed PCR system designed for detection of Fragile X expanded alleles. Triplet repeat primed PCR (TP-PCR) uses a locus-specific forward primer that flanks the repeat. The reverse PCR primer of the primer pair is designed to hybridise within the CGG repeat region as it contains a (GCC)5 tail. This generates amplicons of various sizes as the reverse primers bind to multiple locations during TP-PCR. As the number of CGG repeats increases, a characteristic ladder profile appears on the fluorescence electropherogram enabling the rapid and inexpensive identification of expanded repeats that may have been missed using current PCR methods. Samples were PCR-amplified by preparing a master mix containing 11.45 µl of GC-rich AMP buffer, 0.25 µl of FAM-labelled AFF3 forward primer (5′-GGCCGTAAAAGCCACGAGAGAGGG-3′), 0.25 µl of AFF3 reverse primer (5′-CTTGCGCGCAGGCACACTCAAGAG-3′), 0.5 µl of CGG primer (5′ - TACGCATCCCAGTTTGAGACGGCCGCCGCCGCCGCC-3′), 0.5 µl of nuclease-free water, and 0.05 µl of GC-rich polymerase mix from Asuragen (Austin, TX). 2 µl of the DNA sample, typically at 20 ng/µl, was added before transferring the plate to a thermal cycler (9700, Applied Biosystems, Foster City, CA). Samples were amplified with an initial heat denaturation step of 95°C for 5 minutes, followed by 10 cycles of 97°C for 35 seconds, 62°C for 35 seconds, and 68°C for 4 minutes, and then 20 cycles of 97°C for 35 seconds, 62°C for 35 seconds, and 68°C for 4 minutes with a 20 second autoextension at each cycle. The final extension step was 72°C for 10 minutes. After PCR, 2 µl of the PCR product was added to a mix with 11 µl HDF and 2 µl Rox 1000 standard. After a brief denaturation step, samples were analysed using the ABI Prism 3130 Genetic Analyzer.
Methylation analysis of the AFF3 promoter
The methylation status of the AFF3 associated CGG repeat and the surrounding region, was analysed by bisulfite sequencing. Genomic DNA collected from lymphoblastoid cell lines and saliva (subject AIII.1) was bisulfite-treated using the EpiTect Bisulfite Kit (Qiagen, Venlo, Netherlands) according to manufacturer's guidelines. Bisulfite treatment converts all non-methylated cytosines into thymines, while methylated cytosines remain unchanged. Primers specific for the methylated bisulfite converted DNA (forward 5′ - GGTGAGAAATAAAAAGAAAGGAG -3′ and reverse 5′ - CCTCAACAACCCTAAAATACC -3′) were designed. After PCR amplification, the CGG surrounding area (chr2 : 100721494–100721911; hg19) was sequenced using an ABI Prism 3130 DNA sequencer. Moreover we have performed pyrosequencing using the AFF3_002 PyroMark CpG assay according to manufacturer's instructions (Qiagen, Venlo, Netherlands) and analysed the results on a PyroMark Q24. Methylation cut-off value was set at 10%.
Gene-expression analysis
The expression pattern of AFF3 in human tissues was studied using a multiple-tissue Northern blot (FirstChoice Northern Human Blot I, Ambion, Austin, TX, USA). The specific AFF3 probe was a 655-bp PCR product [forward primer 5′-TATCGAGTGTGGAAATGCAA-3′ and reverse primer 5′-TGAGGTCCCTATGACAGGTG-3′] and radiolabelled by the addition of 32P-dATP and 32P-dCTP (MP, Irvine, California, USA). Hybridisation was performed according to the manufacturer's instructions.
Total RNA-sequencing data from the Illumina Human Body Map 2.0 project (GSE30611) was obtained from the NCBI Gene Expression Omnibus. Data from brain (ERR030882, female, 77y), ovary (ERR030874, female, 47yj) and lymph node (ERR030878, female, 86y) was downloaded in the form of 2×50 bp reads and imported into CLC genomics workbench v6.01. Transcriptomics analysis was performed within CLC Genomics using the human reference genome version hg19. Default settings were used, apart from a smaller expected insert size of 50 bp. Additionally total RNA was isolated from the human fetal brain tissues (FB1 54 gestational days (GD), FB2 47 GD, FB3 59 GD) according to the Trizol (Invitrogen) protocol. The preparation of amplicon libraries and RNA-Seq analysis were performed following standard Illumina TruSeq protocols and reads of length 50 bp were produced on the Illumina GAIIx platform. The fetal tissue was obtained following surgical termination of pregnancy and staged according to the Carnegie Classification [54], [55]. CAGE tag data was obtained from the FANTOM4 consortium as both pre-defined CAGE tag clusters (http://fantom.gsc.riken.jp/download/Tables/human/CAGE/promoters/tag_clusters/ and http://fantom.gsc.riken.jp/4/download/Tables/mouse/CAGE/promoters/tag_clusters/) and as genome aligned individual tags (http://fantom.gsc.riken.jp/4/download//Tables/human/CAGE/mapping/). Coordinates were converted from the hg18 reference genome assembly to hg19 using LiftOver (http://genome.ucsc.edu/cgi-bin/hgLiftOver). Statistical analysis was performed R (http://www.R-project.org/; version 3.0.0).
The AFF3 transcript NM_002285 (http://www.ncbi.nlm.nih.gov/) was searched for cSNP's. The cSNP's in FRA2A patients were tested by PCR followed by sequencing at the genomic and at the cDNA level. RNA was isolated from Epstein-Barr-transformed lymphoblastoid cells using Trizol (Invitrogen, Carlsbad, CA, USA) and converted to cDNA using Superscript III reverse transcriptase (Invitrogen, Carlsbad, CA, USA). The following primer sets were used: SNP1 (rs4851214) in exon 14 (forward primer 5′ - AGTGATGAAGAGGAGAATGAACA -3′ and reverse primer 5′ - ATAGGAGGCTTGTGGGGATTA -3′) and SNP2 (rs13427251) in exon 25 (forward primer 5′-GTGTGTCTGGTATGTTTACAC-3′ and reverse primer 5′-GGATCAGCATCTAGTCTAAG-3′). Sequencing products were analysed on an ABI 3130 Prism automatic sequencer.
The Aff3 riboprobe for whole mount in situ hybridisation to mouse embryos was generated by in vitro transcription from a PCR template amplified from the Aff3 3′UTR using mouse genomic DNA as a template. T3 (for sense probe) and T7 (for antisense probe) binding sites were added to the forward (5′-AATTAACCCTCACTAAAGGCTCTCCAACCGGATCCAGAAT-3′) and reverse (5′-TAATACGACTCACTATAGGAGCCCATGGCACCTCTCT-3′) primers. The WISH protocol and OPT scanning was performed exactly as previously described [58].
Mutation analysis of the AFF3 gene and marker analysis
All coding exons of the AFF3 gene were PCR amplified at the genomic level using standard protocols for all patients and relatives to exclude the presence of any other disease-causing mutation. PCR products were enzymatically purified and sequenced. Sequences were analysed with an ABI Prism 3130 DNA sequencer.
For the marker analysis, genomic DNA was isolated from peripheral blood from all available family members using standard procedures. Highly polymorphic microsatellite markers, D2S2209 and D2S2311, were selected from the Marshfield genetic map in the proximity of the repeat. These markers are both dinucleotides with an average heterozygosity of 71%. Analysis was performed by a Go Taq DNA polymerase mediated PCR, with fluorescently labeled primers. Fragment analysis of amplified products was performed using an ABI PRISM 3130 XL Genetic Analyzer (Applied Biosystems). Allele identification was done with Gene mapper v3.7 software (Applied Biosystems).
Supporting Information
Zdroje
1. WojciechowskaM, KrzyzosiakWJ (2011) CAG repeat RNA as an auxiliary toxic agent in polyglutamine disorders. RNA Biol 8 : 565–571.
2. Costa LimaMA, PimentelMM (2004) Dynamic mutation and human disorders: the spinocerebellar ataxias. Int J Mol Med 13 : 3.
3. RudnickiDD, MargolisRL, PearsonCE, KrzyzosiakWJ (2012) Diced triplets expose neurons to RISC. Plos Genet 8: e1002545.
4. PearsonCE, Nichol EdamuraK, ClearyJD (2005) Repeat instability: mechanisms of dynamic mutations. Nat Rev Genet 6 : 729–742.
5. KumariD, UsdinK (2009) Chromatin remodeling in the noncoding repeat expansion diseases. J Biol Chem 284 : 5.
6. McMurrayC (2010) Mechanisms of trinucleotide repeat instability during human development. Nat Rev Genet 11 : 786–799.
7. Dudek RW (2006) High-yield cell and molecular biology. Philadelphia; Baltimore; New York: Lippincott Williams & Wilkins.
8. LicatalosiDD, DarnellRB (2006) Splicing regulation in neurologic disease. Neuron 52 : 93–101.
9. PierettiM, ZhangFP, FuYH, WarrenST, OostraBA, et al. (1991) Absence of expression of the FMR-1 gene in fragile X syndrome. Cell 66 : 817–822.
10. TrottierY, LutzY, StevaninG, ImbertG, DevysD, et al. (1996) Polyglutamine expansion as a pathological epitope in Huntington's disease and four dominant cerebellar ataxias. Nature 378 : 403–406.
11. LinYaW, JH (2011) Transcription-induced DNA toxicity at trinucleotide repeats. Cell Cycle 10 : 611–618.
12. RichardsRI (2001) Fragile and unstable chromosomes in cancer: causes and consequences. Trends Genet 17 : 339–345.
13. RichardsRI (2001) Dynamic mutations: a decade of unstable expanded repeats in human genetic disease. Hum Mol Genet 10 : 2187–2194.
14. ZuT, GibbensB, DotyNS, Gomes-PereiraM, HuguetA, et al. (2011) Non-ATG-initiated translation directed by microsatellite expansions. Proc Natl Acad Sci U S A 108 : 260–265.
15. PearsonCE (2011) Repeat associated non-ATG translation initiation: one DNA, two transcripts, seven reading frames, potentially nine toxic entities!. PLoS Genet 7: e1002018.
16. BenítezJ (1999) Clinical and genetic implications of dynamic mutations in neuropediatric practice. Rev Neurol 28 : 4.
17. La SpadaAR, TaylorJP (2010) Repeat expansion disease: progress and puzzles in disease pathogenesis. Nat Rev Genet 11 : 13.
18. KatoM (2006) A new paradigm for West syndrome based on molecular and cell biology. Epilepsy Res 70 Suppl 1: S87–95.
19. DebackerK, KooyRF (2007) Fragile sites and human disease. Hum Mol Genet 16: R150–158.
20. VerkerkAJMH, PierettiM, SutcliffeJS, FuY-H, KuhlDPA, et al. (1991) Identification of a gene (FMR-1) containing a CGG repeat coincident with a breakpoint cluster region exhibiting length variation in fragile X syndrome. Cell 65 : 905–914.
21. KnightSJL, FlanneryAV, HirstMC, CampbellL, ChristodoulouZ, et al. (1993) Trinucleotide repeat amplification and hypermethylation of a CpG island in FRAXE mental retardation. Cell 74 : 127–134.
22. ParrishJE, OostraBA, VerkerkAJMH, RichardsCS, ReynoldsJ, et al. (1994) Isolation of a GCC repeat showing expansion in FRAXF, a fragile site distal to FRAXA and FRAXE. Nat Genet 8 : 229–235.
23. NancarrowJK, KremerE, HolmanK, EyreH, DoggettNA, et al. (1994) Implications of FRA16A structure for the mechanism of chromosomal fragile site genesis. Science 264 : 1938–1941.
24. JonesC, PennyL, MattinaT, YuS, BakerE, et al. (1995) Association of a chromosome deletion syndrome with a fragile site within the proto-oncogene CBL2. Nature 376 : 145–149.
25. SarafidouT, KahlC, Martinez-GarayI, MangelsdorfM, GeskS, et al. (2004) Folate-sensitive fragile site FRA10A is due to an expansion of a CGG repeat in a novel gene, FRA10AC1, encoding a nuclear protein. Genomics 84 : 69–81.
26. WinnepenninckxB, DebackerK, RamsayJ, SmeetsD, SmitsA, et al. (2007) CGG repeat expansion in the DIP2B gene is associated with the fragile site FRA12A on chromosome 12q13.1. Am J Hum Genet 80 : 221–231.
27. DebackerK, WinnepenninckxB, LongmanC, ColganJ, TolmieJ, et al. (2007) The molecular basis of the folate-sensitive fragile site FRA11A at 11q13. Cytogenet Genome Res 119 : 9–14.
28. PierettiM, ZhangF, FuY-H, WarrenST, OostraBA, et al. (1991) Absence of expression of the FMR-1 gene in fragile X syndrome. Cell 66 : 817–822.
29. Kooy RF (2009) Fragile sites and human disease. Encyclopedia of Life Sciences. Chichester, UK: John Wiley & Sons, Ltd. pp. DOI:10.1002/9780470015902.a9780470021457.
30. TukunA, RendaY, TopcuM, TuncaliT, BokesoyI (2000) Mental retardation with rare fragile site expressed at 2q11. Brain Dev 22 : 498–500.
31. MurthyDS, TeebiAS, SundareshanTS, al-AwadiSA (1990) Familial fragile secondary constriction on chromosome 2 (2q11) with unusual features and psychomotor retadation. Indian Pediatr 57 : 257–260.
32. AnnerénG, GustavsonKH (1981) A fragile secondary constriction on chromosome 2 in five patients with different clinical features. Hereditas 95 : 63–67.
33. MaC, StaudtL (1996) LAF-4 encodes a lymphoid nuclear protein with transactivation potential that is homologous to AF-4, the gene fused to MLL in t(4;11) leukemias. Blood 87 : 734–745.
34. KodziusR, KojimaM, NishiyoriH, NakamuraM, FukudaS, et al. (2006) CAGE: cap analysis of gene expression. Nat Methods 3 : 211–222.
35. TostJ, GutIG (2007) DNA methylation analysis by pyrosequencing. Nat Protocols 2 : 2265–2275.
36. IllingworthR, KerrA, DesousaD, JorgensenH, EllisP, et al. (2008) A novel CpG island set identifies tissue-specific methylation at developmental gene loci. PLoS Biol 6: e22.
37. LancasterMA, RennerM, MartinCA, WenzelD, BicknellLS, et al. (2013) Cerebral organoids model human brain development and microcephaly. Nature 501 : 373–379.
38. de GraaffE, RouillardP, WillemsPJ, SmitsAPT, RousseauF, et al. (1995) Hotspot for deletions in the CGG repeat region of FMR1 in fragile X patients. Hum Mol Genet 4 : 45–49.
39. AxfordMM, Lopez-CastelA, NakamoriM, ThorntonCA, PearsonCE (2011) Replacement of the myotonic dystrophy type 1 CTG repeat with ‘non-CTG repeat’ insertions in specific tissues. J Med Genet 48 : 438–443.
40. GronskovK, HjalgrimH, BjeragerMO, Brondum-NielsenK (1997) Deletion of all CGG repeats plus flanking sequences in FMR1 does not abolish gene expression. Am J Hum Genet 61 : 961–967.
41. TakiT, KanoH, TaniwakiM, SakoM, YanagisawaM, et al. (1999) AF5q31, a newly identified AF4-related gene, is fused to MLL in infant acute lymphoblastic leukemia with ins(5;11) (q31;q13q23). Proc Natl Acad Sci USA 96 : 14535–14540.
42. LiaoX, MaC, TraskB, MassaH, GilbertD, et al. (1996) LAF4 maps to mouse chromosome 1 and human chromosome 2q11.2-q12. Mamm Genome 7 : 467–468.
43. MelkoM, DouguetD, BensaidM, ZongaroS, VerheggenC, et al. (2011) Functional characterization of the AFF (AF4/FMR2) family of RNA-binding proteins: insights into the molecular pathology of FRAXE intellectual disability. Hum Mol Genet 20 : 1873–1885.
44. BritanovaO, LukyanovS, GrussP, TarabykinV (2002) The mouse Laf4 gene: Exon/intron organization, cDNA sequence, alternative splicing, and expression during central nervous system development. Genomics 80 : 31–37.
45. GéczJ, OostraBA, HockeyA, CarbonellP, TurnerG, et al. (1997) FMR2 expression in families with FRAXE mental retardation. Hum Mol Genet 6 : 435–441.
46. ChakrabartiL, BristulfJ, FossGS, DaviesKE (1998) Expression of the murine homologue of FMR2 in mouse brain and during development. Hum Mol Genet 7 : 441–448.
47. MondalK, RamachandranD, PatelVC, HagenKR, BoseP, et al. (2012) Excess variants in AFF2 detected by massively parallel sequencing of males with autism spectrum disorder. Hum Mol Genet 21 : 4356–4364.
48. GuY, NelsonDL (2003) FMR2 function: insight from a mouse knockout model. Cytogenet Genome Res 100 : 129–139.
49. DaviesKE, OliverPL, JonesEL, JeansA, ClarkJ, et al. (2003) Functional studies of Af4 in the robotic mouse. Am J Hum Genet 73 Suppl: A86.
50. Steichen-GersdorfE, GassnerI, Superti-FurgaA, UllmannR, StrickerS, et al. (2008) Triangular tibia with fibular aplasia associated with a microdeletion on 2q11.2 encompassing LAF4. Clin Genet 74 : 560–565.
51. WillemsenR, BontekoeCJ, SeverijnenLA, OostraBA (2002) Timing of the absence of FMR1 expression in full mutation chorionic villi. Hum Genet 110 : 601–605.
52. Ruiz-HerreraA, GarciaF, GiulottoE, AttoliniC, EgozcueJ, et al. (2005) Evolutionary breakpoints are co-localized with fragile sites and intrachromosomal telomeric sequences in primates. Cytogenet Genome Res 108 : 234–247.
53. Ruiz-HerreraA, CastresanaJ, RobinsonTJ (2006) Is mammalian chromosomal evolution driven by regions of genome fragility? Genome Biol 7: R115.
54. O'RahillyR, MullerF (1987) Developmental stages in human embryos: revised and new measurements. Cells Tissues Organs 192 : 73–84.
55. Bullen P, Wilson D, editors(1997) The Carnegie Staging of Human Embryos: a Practical Guide. Oxford: Bios Scientific Publishers. 27–50 p.
56. ChongSS, PackSD, RoschkeAV, TanigamiA, CarrozzoR, et al. (1997) A revision of the lissencephaly and Miller-Dieker syndrome critical regions in chromosome 17p13.3. Hum Mol Genet 6 : 147–155.
57. NolinSL, BrownWT, GlicksmanA, HouckGEJr, GarganoAD, et al. (2003) Expansion of the fragile X CGG repeat in females with premutation or intermediate alleles. Am J Hum Genet 72 : 454–464.
58. RaingerJ, BenganiH, CampbellL, AndersonE, SokhiK, et al. (2012) Miller (Genee-Wiedemann) syndrome represents a clinically and biochemically distinct subgroup of postaxial acrofacial dysostosis associated with partial deficiency of DHODH. Hum Mol Genet 21 : 3969–3983.
Štítky
Genetika Reprodukční medicína
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2014 Číslo 4
-
Všechny články tohoto čísla
- The Challenges of Mitochondrial Replacement
- Concocting Cholinergy
- Genome-Wide Diet-Gene Interaction Analyses for Risk of Colorectal Cancer
- Statistical Power to Detect Genetic (Co)Variance of Complex Traits Using SNP Data in Unrelated Samples
- Mouse Pulmonary Adenoma Susceptibility 1 Locus Is an Expression QTL Modulating -4A
- Transcription-Associated R-Loop Formation across the Human CGG-Repeat Region
- Epigenetic Regulation by Heritable RNA
- Protein Quantitative Trait Loci Identify Novel Candidates Modulating Cellular Response to Chemotherapy
- Genome-Wide Profiling of Yeast DNA:RNA Hybrid Prone Sites with DRIP-Chip
- The Mechanism of Gene Targeting in Human Somatic Cells
- A LINE-1 Insertion in DLX6 Is Responsible for Cleft Palate and Mandibular Abnormalities in a Canine Model of Pierre Robin Sequence
- Interaction between Two Timing MicroRNAs Controls Trichome Distribution in
- DNA Glycosylases Involved in Base Excision Repair May Be Associated with Cancer Risk in and Mutation Carriers
- The Myc-Mondo/Mad Complexes Integrate Diverse Longevity Signals
- Evolutionarily Diverged Regulation of X-chromosomal Genes as a Primal Event in Mouse Reproductive Isolation
- Mutations in Conserved Residues of the microRNA Argonaute ALG-1 Identify Separable Functions in ALG-1 miRISC Loading and Target Repression
- Genetic Predisposition to In Situ and Invasive Lobular Carcinoma of the Breast
- Isl1 Directly Controls a Cholinergic Neuronal Identity in the Developing Forebrain and Spinal Cord by Forming Cell Type-Specific Complexes
- A Synthetic Community Approach Reveals Plant Genotypes Affecting the Phyllosphere Microbiota
- The Sequence-Specific Transcription Factor c-Jun Targets Cockayne Syndrome Protein B to Regulate Transcription and Chromatin Structure
- Determining the Control Circuitry of Redox Metabolism at the Genome-Scale
- DNA Repair Pathway Selection Caused by Defects in , , and Telomere Addition Generates Specific Chromosomal Rearrangement Signatures
- Methylome Diversification through Changes in DNA Methyltransferase Sequence Specificity
- Folliculin Regulates Ampk-Dependent Autophagy and Metabolic Stress Survival
- Fine Mapping of Dominant -Linked Incompatibility Alleles in Hybrids
- Unexpected Role of the Steroid-Deficiency Protein Ecdysoneless in Pre-mRNA Splicing
- Three Groups of Transposable Elements with Contrasting Copy Number Dynamics and Host Responses in the Maize ( ssp. ) Genome
- Sox5 Functions as a Fate Switch in Medaka Pigment Cell Development
- Synergistic Interactions between the Molecular and Neuronal Circadian Networks Drive Robust Behavioral Circadian Rhythms in
- Chromatin Landscapes of Retroviral and Transposon Integration Profiles
- Widespread Use of Non-productive Alternative Splice Sites in
- Ras GTPase-Like Protein MglA, a Controller of Bacterial Social-Motility in Myxobacteria, Has Evolved to Control Bacterial Predation by
- Cell Type-Specific Functions of Genes Revealed by Novel Adipocyte and Hepatocyte Circadian Clock Models
- Phenotype Ontologies and Cross-Species Analysis for Translational Research
- Embryogenesis Scales Uniformly across Temperature in Developmentally Diverse Species
- In Pursuit of the Gene: An Interview with James Schwartz
- Molecular Mechanisms of Hypoxic Responses via Unique Roles of Ras1, Cdc24 and Ptp3 in a Human Fungal Pathogen
- Analysis of the Genome and Transcriptome of var. Reveals Complex RNA Expression and Microevolution Leading to Virulence Attenuation
- Genotypic and Functional Impact of HIV-1 Adaptation to Its Host Population during the North American Epidemic
- RNA Editome in Rhesus Macaque Shaped by Purifying Selection
- Proper Actin Ring Formation and Septum Constriction Requires Coordinated Regulation of SIN and MOR Pathways through the Germinal Centre Kinase MST-1
- Interplay of the Serine/Threonine-Kinase StkP and the Paralogs DivIVA and GpsB in Pneumococcal Cell Elongation and Division
- A Quality Control Mechanism Coordinates Meiotic Prophase Events to Promote Crossover Assurance
- CNNM2 Mutations Cause Impaired Brain Development and Seizures in Patients with Hypomagnesemia
- The RNA-Binding Protein QKI Suppresses Cancer-Associated Aberrant Splicing
- Uncoupling Transcription from Covalent Histone Modification
- Rad51–Rad52 Mediated Maintenance of Centromeric Chromatin in
- FRA2A Is a CGG Repeat Expansion Associated with Silencing of
- A General Approach for Haplotype Phasing across the Full Spectrum of Relatedness
- A Novel Highly Divergent Protein Family Identified from a Viviparous Insect by RNA-seq Analysis: A Potential Target for Tsetse Fly-Specific Abortifacients
- A Central Role for in Regulation of Islet Function in Man
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- The Sequence-Specific Transcription Factor c-Jun Targets Cockayne Syndrome Protein B to Regulate Transcription and Chromatin Structure
- The Mechanism of Gene Targeting in Human Somatic Cells
- Genetic Predisposition to In Situ and Invasive Lobular Carcinoma of the Breast
- Widespread Use of Non-productive Alternative Splice Sites in
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



