-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaDNA Glycosylases Involved in Base Excision Repair May Be Associated with Cancer Risk in and Mutation Carriers
Women harboring a germ-line mutation in the BRCA1 or BRCA2 genes have a high lifetime risk to develop breast and/or ovarian cancer. However, not all carriers develop cancer and high variability exists regarding age of onset of the disease and type of tumor. One of the causes of this variability lies in other genetic factors that modulate the phenotype, the so-called modifier genes. Identification of these genes might have important implications for risk assessment and decision making regarding prevention of the disease. Given that BRCA1 and BRCA2 participate in the repair of DNA double strand breaks, here we have investigated whether variations, Single Nucleotide Polymorphisms (SNPs), in genes participating in other DNA repair pathway may be associated with cancer risk in BRCA carriers. We have selected the Base Excision Repair pathway because BRCA defective cells are extremely sensitive to the inhibition of one of its components, PARP1. Thanks to a large international collaborative effort, we have been able to identify at least two SNPs that are associated with increased cancer risk in BRCA1 and BRCA2 mutation carriers respectively. These findings could have implications not only for risk assessment, but also for treatment of BRCA1/2 mutation carriers with PARP inhibitors.
Published in the journal: . PLoS Genet 10(4): e32767. doi:10.1371/journal.pgen.1004256
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1004256Summary
Women harboring a germ-line mutation in the BRCA1 or BRCA2 genes have a high lifetime risk to develop breast and/or ovarian cancer. However, not all carriers develop cancer and high variability exists regarding age of onset of the disease and type of tumor. One of the causes of this variability lies in other genetic factors that modulate the phenotype, the so-called modifier genes. Identification of these genes might have important implications for risk assessment and decision making regarding prevention of the disease. Given that BRCA1 and BRCA2 participate in the repair of DNA double strand breaks, here we have investigated whether variations, Single Nucleotide Polymorphisms (SNPs), in genes participating in other DNA repair pathway may be associated with cancer risk in BRCA carriers. We have selected the Base Excision Repair pathway because BRCA defective cells are extremely sensitive to the inhibition of one of its components, PARP1. Thanks to a large international collaborative effort, we have been able to identify at least two SNPs that are associated with increased cancer risk in BRCA1 and BRCA2 mutation carriers respectively. These findings could have implications not only for risk assessment, but also for treatment of BRCA1/2 mutation carriers with PARP inhibitors.
Introduction
Carrying an inherited mutation in the BRCA1 or BRCA2 gene increases a woman's lifetime risk of developing breast, ovarian and other cancers. The estimated cumulative risk of developing breast cancer by the age of 70 in BRCA1 and BRCA2 mutation carriers varies between 43% to 88%; similarly, between 11% to 59% of mutation carriers will develop ovarian cancer by the age of 70 [1]–[3]. These considerable differences in disease manifestation suggest the existence of other genetic or environmental factors that modify the risk of cancer development. The Consortium of Investigators of Modifiers of BRCA1 and BRCA2 (CIMBA), was established in 2006 [4] and with more than 40,000 mutation carriers currently provides the largest sample size for reliable evaluation of even modest associations between single-nucleotide polymorphisms (SNPs) and cancer risk. CIMBA studies have so far demonstrated that more than 25 SNPs are associated with the risk of developing breast or ovarian cancer for BRCA1 or BRCA2 carriers. These were identified through genome-wide association studies (GWAS) of breast or ovarian cancer in the general population or through BRCA1- and BRCA2-specific GWAS [5]–[8]. Cells harboring mutations in BRCA1 or BRCA2 show impaired homologous recombination (HR) [9]–[11] and are thus critically dependent on other members of the DNA repair machinery such as poly ADP ribose polymerase (PARP1) involved in the Base Excision Repair (BER) pathway. The BER pathway is crucial for the replacement of aberrant bases generated by different causes [12]. A deficiency in BER can give rise to a further accumulation of double-strand DNA breaks which, in the presence of a defective BRCA1 or BRCA2 background, could persist and lead to cell cycle arrest or cell death; this makes BRCA-deficient cells extremely sensitive to PARP inhibitors, as previously demonstrated [13]. We hypothesize that SNPs in PARP1 and other members of BER may be associated with cancer risk in BRCA1 and BRCA2 mutation carriers. SNPs in XRCC1, one of the main components of BER, have been recently evaluated within the CIMBA consortium [14], however a comprehensive study has not yet been performed of either XRCC1 or the other genes participating in BER.
In the present study, we used a tagging SNP approach to evaluate whether the common genetic variation in the genes involved in the BER pathway could be associated with cancer risk in a large series of BRCA1/2 mutation carriers using a two-stage approach. The first stage involved an analysis of 144 tag SNPs in 1,787 Spanish and Italian BRCA1/2 mutation carriers. In stage II, the 36 SNPs showing the strongest evidence of association in stage I, were evaluated in a further 23,463 CIMBA mutation carriers included in the Collaborative Oncological Gene-environment Study (COGS) and genotyped using the iCOGS custom genotyping array.
Results
Breast cancer association
In stage I, 144 selected Tag SNPs covering the 18 selected BER genes were genotyped in 968 BRCA1 and 819 BRCA2 mutation carriers from five CIMBA centres (Spanish National Cancer ResearchCentre (CNIO), Hospital Clínico San Carlos (HCSC), Catalan Institute of Oncology (ICO), Demokritos and Milan Breast Cancer Study Group (MBCSG). Of those, 50 were excluded because of low call-rates, minor allele frequency (MAF)<0.05, evidence of deviation from Hardy Weinberg Equilibrium (p-value<10−3) or monomorphism. Associations with breast cancer risk were assessed for 94 SNPs, as summarized in Table S1. The 36 SNPs that showed evidence of association at p≤0.05 were selected for analysis in stage II. Of the 36 SNPs successfully genotyped in the whole CIMBA series comprising 15,252 BRCA1 and 8211 BRCA2 mutation carriers, consistent evidence of association with breast cancer risk (p-trend<0.05) was observed for six SNPs (Table 1). The strongest evidence of association was observed for rs1466785 in the NEIL2 gene (HR: 1.09, 95% CI (1.03–1.16), p = 2.7×10−3) for association with breast cancer risk in BRCA2 mutation carriers. We had observed a consistent association in stage I in BRCA2 mutation carriers (HR: 1.25, p = 0.06). The SNP was primarily associated with ER-negative breast cancer (HR: 1.20, 95%CI (1.06–1.37), p = 4×10−3), although the difference in HRs for ER-positive and ER-negative disease was not statistically significant. The evidence of association in Stage II was somewhat stronger when considering the genotype-specific models, with the dominant being the best fitting (HR: 1.20 95% CI: 1.09–1.37, p = 1×10−4). The associations remained significant and the estimated effect sizes remained consistent with the overall analysis when the data were reanalyzed excluding samples used in stage I of the study (data not shown). Imputation using the 1000 genomes data showed that there were several SNPs in strong linkage disequilibrium (LD) with rs1466785 showing more significant associations (p<10−3) (Figure 1).
Fig. 1. p-values of association (−log10 scale) with breast cancer risk in BRCA2 carriers for genotyped and imputed SNPs in the NEIL2 gene. 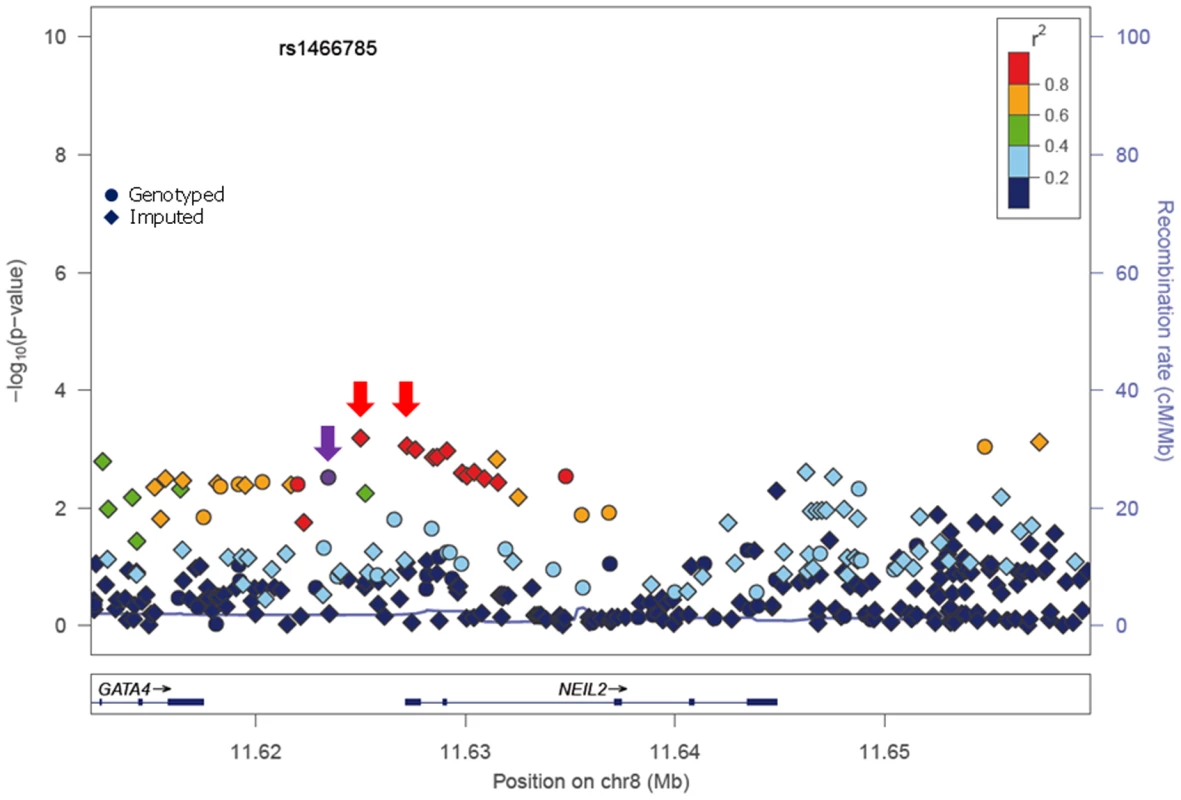
SNP rs1466785 is indicated with a purple arrow and the best causal imputed SNPs, rs804276 and rs804271 are indicated with a red arrow. Colors represent the pariwise r2. Plot generated with LocusZoom [42] (http://csg.sph.umich.edu/locuszoom/). Tab. 1. Associations with breast and ovarian cancer risk for SNPs observed at p-trend<0.05 in stage II of the experiment. 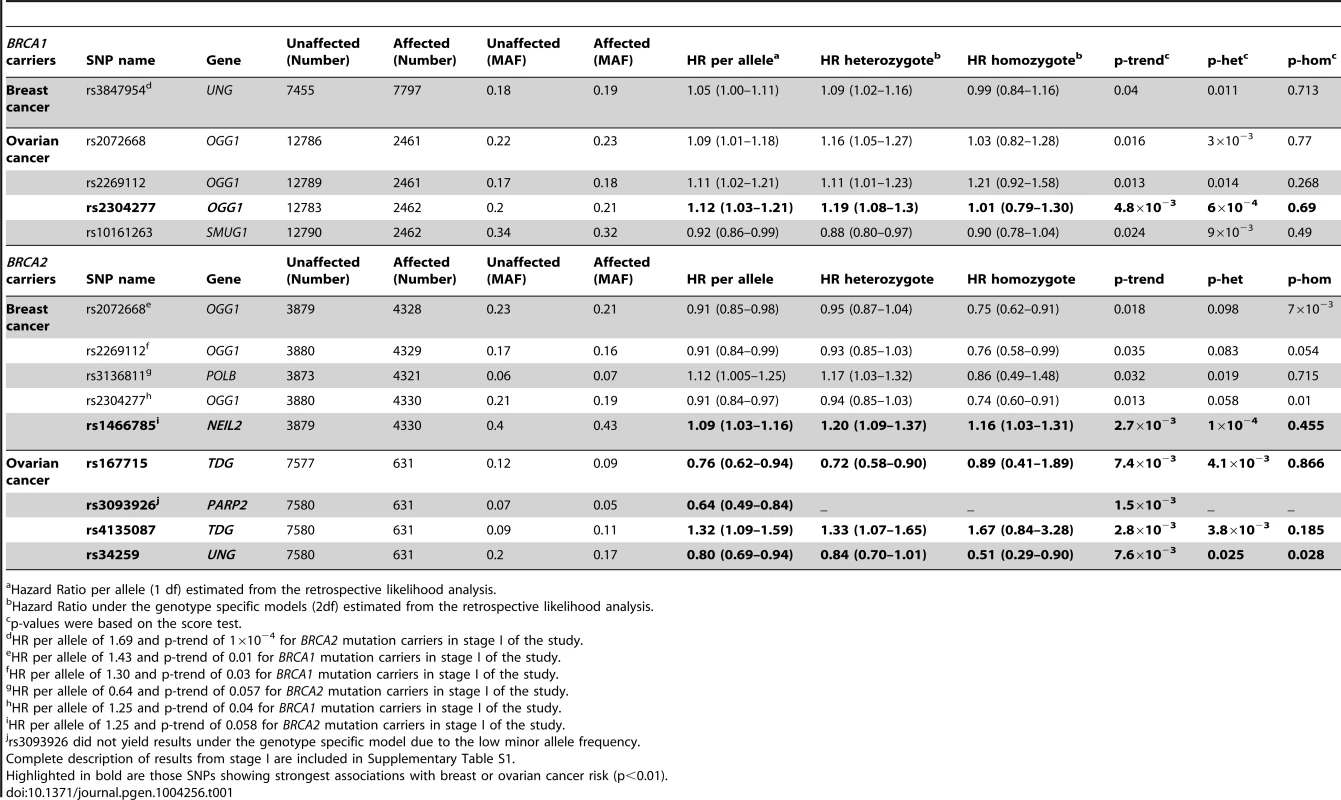
Hazard Ratio per allele (1 df) estimated from the retrospective likelihood analysis. Ovarian cancer association
Due to lack of power we did not perform analysis of associations with ovarian cancer in stage I. However, we performed this analysis for the 36 SNPs tested in stage II. Although they had been selected based on their evidence of association with breast cancer risk, under the initial hypothesis they are also plausible modifiers of ovarian cancer risk for BRCA1 and BRCA2 mutation carriers. We found four SNPs associated with ovarian cancer risk with a p-trend<0.01 in BRCA1 or BRCA2 mutation carriers (Table 1). The strongest association was found for rs2304277 in OGG1 in BRCA1 mutation carriers (HR: 1.12, 95%CI: 1.03–1.21, p = 4.8×10−3). The association was somewhat stronger under the dominant model (HR: 1.19, 95%CI: 1.08–1.3, p = 6×10−4). Although three other SNPs were found to be associated with ovarian cancer risk in BRCA2 mutation carriers (p-trend<10−3), these results were based on a relatively small number of ovarian cancer cases. Imputed data did not show any SNPs with substantially more significant associations with ovarian cancer risk except for rs3093926 in PARP2, associated with ovarian cancer risk in BRCA2 mutation carriers for which there was a SNP, rs61995542, with a stronger association (HR: 0.67, p = 4.6×10−4) (Figure S1).
Discussion
Based on the interaction of synthetic lethality that has been described between PARP1 and both BRCA1 and BRCA2, we hypothesize that this and other genes involved in the BER pathway could potentially be associated with cancer risk in BRCA1/2 mutation carriers. Several studies have recently investigated the association of some of the BER genes with breast cancer, however, no definitive conclusions can be drawn, given that some publications suggest that SNPs in these genes can be associated with breast cancer risk with marginal p-values while others rule out a major role of these genes in the disease [15]–[21]. There is only one study from the CIMBA consortium which has evaluated the role of three of the most studied SNPs in the XRCC1 gene, c.-77C>T (rs3213245) p.Arg280His (rs25489) and p.Gln399Arg (rs25487), ruling out associations of these variants with cancer risk in BRCA1 and BRCA2 mutation carriers [14]. However, a comprehensive analysis of neither XRCC1 nor the other genes involved in the pathway in the context of BRCA mutation carriers has been performed. In the present study we have assessed the common genetic variation of 18 genes participating in BER by using a two stage strategy.
Eleven SNPs showed evidence of association with breast and/or ovarian cancer at p<0.05 in stage II of the experiment (Table 1). Of those, six showed a p-trend value<0.01 and were therefore considered the best candidates for further evaluation. Only one of those six, rs1466785 in the NEIL2 gene (endonuclease VIII-like 2) showed an association with breast cancer risk while the other five, rs2304277 in OGG1 (8-guanine DNA glycosylase), rs167715 and rs4135087 in TDG (thymine-DNA glycosylase), rs3093926 in PARP2 (Poly(ADP-ribose) polymerase 2) and rs34259 in UNG (uracil-DNA glycosylase) were associated with ovarian cancer risk.
The minor allele of NEIL2-rs1466785 was associated with increased breast cancer risk in BRCA2 mutation carriers; moreover, when considering the genotype-specific risks observed that the best fitting model was the dominant one. NEIL2 is one of the oxidized base-specific DNA glycosylases that participate in the initial steps of BER and specifically removes oxidized bases from transcribing genes [22]. By imputing using the 1000 genome data we found six correlated SNPs in strong LD with rs1466785 (r2>0.8), located closer or inside the gene and showing slightly stronger and more significant associations with the disease and therefore being better candidate causal variants. From those, we considered rs804276 and rs804271 as the best candidates given that they showed the most significant associations (p = 6×10−4 and p = 8×10−4 respectively) and there were available epidemiological or functional data supporting their putative role in cancer. SNP rs804276 has been associated with disease recurrence in patients with bladder cancer treated with Bacillus Calmette-Guérin (BCG) (HR: 2.71, 95%CI (1.75–4.20), p = 9×10−6) [23]. SNP rs804271 is located in a positive regulatory region in the promoter of the gene, between two potential cis - binding sites for reactive oxygen species responsive transcription factors in which sequence variation has been proven to alter the transcriptional response to oxidative stress [24]. Moreover, this SNP has been proposed to partly explain the inter-individual variability observed in NEIL2 expression levels in the general population and has been proposed as a potential risk modifier of disease susceptibility [25].
Several studies have been published showing associations between SNPs in NEIL2 and lung or oropharyngeal cancer risk [26], [27] but to our knowledge, no association with breast cancer risk has been reported. We hypothesize that the potential association observed in the present study could be explained by the interaction between NEIL2 and BRCA2, each of them causing a deficiency in the BER and HR DNA repair pathways, respectively. This would explain why the breast cancer risk modification due to rs1466785 would only be detected in the context of BRCA2 mutation carriers and not in the general population.
The strongest evidence of association found in BRCA1 carriers was between rs2304277 in the OGG1 gene and ovarian cancer risk. The association was more significant when considering the dominant model. OGG1 removes 8-oxodeoxyguanosine which is generated by oxidative stress and is highly mutagenic, and it has been suggested that SNPs in the gene could be associated with cancer risk [28]–[31]. This is an interesting result, given that to date only one SNP, rs4691139 in the 4q35.3 region, also identified through the iCOGS effort, has been found to modify ovarian cancer risk specifically in BRCA1 carriers [32]. SNP rs2304277 is located in the 3′UTR (untranslated region) of the gene and is probably not the causal variant, however, in this case imputations through the 1000 Genome did not show better results for a more plausible causal SNP.
We have identified four SNPs associated with ovarian cancer risk in BRCA2 mutation carriers, rs167715 and rs4135087 in the TDG gene, rs34259 in the UNG gene and rs3093926 in PARP2. However, these last results should be interpreted with caution given that the number of BRCA2 carriers affected with ovarian cancer is four-fold lower than for BRCA1 carriers and the statistical power was therefore more limited, increasing the possibility of false-positives. In the case of PARP2, imputed data showed a lower p-value of association (4×10−4) for another SNP, rs61995542, that had a slightly higher MAF than rs3093926 (0.074 vs. 0.067) (Figure S1). However, it must still be interpreted with caution due to small number of ovarian cancer cases in the BRCA2 group.
It is worth noting that, four of the five genes for which strongest evidence of association was observed, are all DNA glycosylases participating in the initiation of BER by removing damaged or mismatched bases. Apart from the already mentioned NEIL2 and OGG1, TDG initiates repair of G/T and G/U mismatches commonly associated with CpG islands, while UNG removes uracil in DNA resulting from deamination of cytosine or replicative incorporation of dUMP. We have not found strong associations with SNPs in genes involved in any other parts of the pathway, such as strand incision, trimming of ends, gap filling or ligation. It has been suggested that at least in the case of uracil repair, base removal is the major rate-limiting step of BER [33]. This is consistent with our findings, suggesting that SNPs causing impairment in the function of these specific DNA glycosylases could give rise to accumulation of single strand breaks and subsequently DNA double strand breaks that, in the HR defective context of BRCA1/2 mutation carriers would increase breast and ovarian cancer risk.
The fact that the SNPs tested are located in genes participating in the same DNA repair pathway as PARP1, make them especially interesting, not only as risk modifiers but also because they could have an impact on patients' response to treatment with PARP inhibitors. BRCA1/2 mutation carriers harboring a potential modifier SNP in DNA glycosylases could be even more sensitive to PARPi due to a constitutional slight impairment of the BER activity. This is a hypothesis that should be confirmed in further studies. The design of this study in two stages, the hypothesis-based approach adopted to select genes, and that it is based on the largest possible series of BRCA1 and BRCA2 carriers available nowadays, mean that the results obtained are quite solid However, the study still has some limitations such as the possible existence of residual confounding due to environmental risk factors for which we did not have information.
In summary, we have identified at least two SNPs, rs1466785 and rs2304277, in the DNA glycolylases NEIL2 and OGG1, potentially associated with increased breast and ovarian cancer risks in BRCA2 and BRCA1 mutation carriers, respectively. Our results suggest that glycosylases involved in the first steps of the BER pathway may be cancer risk modifiers in BRCA1/2 mutation carriers and should be more comprehensively studied. If confirmed, these findings could have implications not only for risk assessment, but also for treatment of BRCA1/2 mutation carriers with PARP inhibitors.
Materials and Methods
Subjects
Eligible subjects were female carriers of deleterious mutations in BRCA1 or BRCA2 aged 18 years or older [6]. A total of 55 collaborating CIMBA studies contributed genotypes for the study. Numbers of samples included from each are provided in Table S2. A total of 1,787 mutation carriers (968 with mutations in BRCA1 and 819 with mutations in BRCA2) from the CNIO, HCSC, ICO, Demokritos and MBCSG were genotyped in the first stage of the study. Stage II included 23,463 CIMBA samples (15,252 with mutations in BRCA1 and 8,211 with mutations in BRCA2). All carriers participated in clinical and/or research studies at the host institution under IRB-approved protocols.
Methods stage I
Selection and genotyping of SNPs
Eighteen genes (UNG, SMUG1, MBD4, TDG, OGG1, MUTYH, NTHL1, MPG, NEIL1, NEIL2, APEX1, APEX2, LIG3, XRCC1, PNKP, POLB, PARP1 and PARP2) involved in the BER pathway were selected, based on the information available at http://www.cgal.icnet.uk/DNA_Repair_Genes.html as at the 31st December, 2009. Tag SNPs for the selected genes were defined using Haploview v.4.0 (http://www.broad.mit.edu/mpg/haploview) with an r2 threshold of 0.8 and a minimum minor allele frequency of 0.05. In addition, SNPs with potentially functional effects already described in the literature were selected. A final number of 144 SNPs was included in an oligonucleotide pool assay for genotyping using the Illumina Veracode technology (Illumina Inc., San Diego, CA). Three hundred nanograms of DNA from each sample were genotyped using the GoldenGate Genotyping Assay with Veracode technology according to the published Illumina protocol. Genotype clustering and calling were carried out using the GenomeStudio software. SNPs with a call rate <0.95 were excluded from further analysis. Duplicate samples and CEPH trios (Coriell Cell Repository, Camden, NJ) were genotyped across the plates. SNPs showing Mendelian allele-transmission errors or showing discordant genotypes across duplicates were excluded.
Statistical analysis
To test for departure from Hardy-Weinberg equilibrium (HWE), a single individual was randomly selected from each family and Pearson's X2 Test (1df) was applied to genotypes from this set of individuals. The association of the SNPs with breast cancer risk was assessed by estimating hazard ratios (HR) and their corresponding 95% confidence intervals (CI) using weighted multivariable Cox proportional hazards regression with robust estimates of variance [34]. For each mutation carrier, we modeled the time to diagnosis of breast cancer from birth, censoring at the first of the following events: bilateral prophylactic mastectomy, breast cancer diagnosis, ovarian cancer diagnosis, death or date last know to be alive. Subjects were considered affected if their age at censoring corresponded to their age at diagnosis of breast cancer and unaffected otherwise. Weights were assigned separately for carriers of mutations in BRCA1 and BRCA2, by age and affection status, so that the weighted observed incidences in the sample agreed with established estimates for mutation carriers [1]; [34].
We considered log-additive and co-dominant genetic models and tested for departure from HR = 1 by applying a Wald test based on the log-HR estimate and its standard error. Additional independent variables included in all analyses were year of study, centre and country. All statistical analyses were carried out using Stata: Release 10 (StataCorp. 2007. Stata Statistical Software: Release 10.0. College Station, TX: Stata Corporation LP). Robust estimates of variance were calculated using the cluster subcommand, applied to an identifier variable unique to each family.
Methods stage II
iCOGS SNP array
Stage II of the experiment was performed as part of the iCOGS genotyping experiment. The iCOGS custom array was designed in collaboration between the Breast Cancer Association Consortium (BCAC), the Ovarian Cancer Association Consortium (OCAC), the Prostate Cancer Association Group to Investigate Cancer Associated in the Genome (PRACTICAL) and CIMBA. The final design comprised 211,155 successfully manufactured SNPs of which approximately 17.5% had been proposed by CIMBA. A total of 43 SNPs were nominated for inclusion on iCOGS based on statistical evidence of association in stage I of the present study (p≤0.05). Of these, 36 were successfully manufactured and genotyped in CIMBA mutation carriers.
iCOGS genotyping and quality control
Genotyping was performed at Mayo Clinic and the McGill University and Génome Québec Innovation Centre (Montreal, Canada). Genotypes were called using Illumina's GenCall algorithm. Sample and quality control process have been described in detail elsewhere [32], [35]. After the quality control process a total of 23,463 carriers were genotyped for the 36 selected SNPs.
Statistical analysis
Both breast and ovarian cancer associations were evaluated in stage II. Censoring for breast cancer followed the same approach as in stage I. Censoring for ovarian cancer risk occurred at risk-reducing salpingo-oophorectomy or last follow-up.
The genotype-disease associations were evaluated within a survival analysis framework, by modelling the retrospective likelihood of the observed genotypes conditional on the disease phenotypes [9], [34], [36], [37]. The associations between genotype and breast or ovarian cancer risk were assessed using the 1 d.f. score test statistic based on this retrospective likelihood. To allow for the non-independence among related individuals, we accounted for the correlation between the genotypes by estimating the kinship coefficient for each pair of individuals using the available genomic data [34], [38], [39]. These analyses were performed in R using the GenABELlibraries and custom-written functions in FORTRAN and Python.
To estimate the magnitude of the associations (HRs), the effect of each SNP was modeled either as a per-allele HR (multiplicative model) or as genotype-specific HRs, and was estimated on the log-scale by maximizing the retrospective likelihood. The retrospective likelihood was fitted using the pedigree-analysis software MENDEL. The variances of the parameter estimates were obtained by robust variance estimation based on reported family membership. All analyses were stratified by country of residence and based on calendar-year and cohort-specific breast cancer incidence rates for mutation carriers. Countries with small number of mutation carriers were combined with neighbouring countries to ensure sufficiently large numbers within each stratum. USA and Canada were further stratified by reported Ashkenazi Jewish (AJ) ancestry.
Imputation
Genotypes were imputed separately for BRCA1 and BRCA2 mutation carriers using the v3 April 2012 release (Genomes Project et al., 2012) as reference panel. To improve computation efficiency we used a two-step procedure which involved pre-phasing in the first step and imputation of the phased data in the second. Pre-phasing was carried out using the SHAPEIT software [40]. The IMPUTE version 2 software was used for the subsequent imputation [41]. SNPs were excluded from the association analysis if their imputation accuracy was r2<0.3 or MAF<0.005 in any of the data sets. For the final analysis we only took in account those SNPs with an imputation accuracy r2>0.7, MAF>0.01 and being located in the region comprised within 15 kilo bases (kb) downstream and upstream the gene where the genotyped SNP showing an association was located (Table 1). Associations between imputed genotypes and breast cancer risk were evaluated using a version of the score test as described above but with the posterior genotype probabilities replacing the genotypes.
Supporting Information
Zdroje
1. AntoniouA, PharoahPD, NarodS, RischHA, EyfjordJE, et al. (2003) Average risks of breast and ovarian cancer associated with BRCA1 or BRCA2 mutations detected in case Series unselected for family history: a combined analysis of 22 studies. Am J Hum Genet 72 : 1117–1130.
2. ChenS, IversenES, FriebelT, FinkelsteinD, WeberBL, et al. (2006) Characterization of BRCA1 and BRCA2 mutations in a large United States sample. J Clin Oncol 24 : 863–871.
3. MilneRL, OsorioA, CajalTR, VegaA, LlortG, et al. (2008) The average cumulative risks of breast and ovarian cancer for carriers of mutations in BRCA1 and BRCA2 attending genetic counseling units in Spain. Clin Cancer Res 14 : 2861–2869.
4. Chenevix-TrenchG, MilneRL, AntoniouAC, CouchFJ, EastonDF, et al. (2007) An international initiative to identify genetic modifiers of cancer risk in BRCA1 and BRCA2 mutation carriers: the Consortium of Investigators of Modifiers of BRCA1 and BRCA2 (CIMBA). Breast Cancer Res 9 : 104.
5. AntoniouAC, KuchenbaeckerKB, SoucyP, BeesleyJ, ChenX, et al. (2012) Common variants at 12p11, 12q24, 9p21, 9q31.2 and in ZNF365 are associated with breast cancer risk for BRCA1 and/or BRCA2 mutation carriers. Breast Cancer Res 14: R33.
6. AntoniouAC, SpurdleAB, SinilnikovaOM, HealeyS, PooleyKA, et al. (2008) Common breast cancer-predisposition alleles are associated with breast cancer risk in BRCA1 and BRCA2 mutation carriers. Am J Hum Genet 82 : 937–948.
7. AntoniouAC, SinilnikovaOM, McGuffogL, HealeyS, NevanlinnaH, et al. (2009) Common variants in LSP1, 2q35 and 8q24 and breast cancer risk for BRCA1 and BRCA2 mutation carriers. Hum Mol Genet 18 : 4442–4456.
8. AntoniouAC, WangX, FredericksenZS, McGuffogL, TarrellR, et al. (2010) A locus on 19p13 modifies risk of breast cancer in BRCA1 mutation carriers and is associated with hormone receptor-negative breast cancer in the general population. Nat Genet 42 : 885–892.
9. AntoniouAC, SinilnikovaOM, SimardJ, LeoneM, DumontM, et al. (2007) RAD51 135G→C modifies breast cancer risk among BRCA2 mutation carriers: results from a combined analysis of 19 studies. Am J Hum Genet 81 : 1186–1200.
10. MoynahanME, ChiuJW, KollerBH, JasinM (1999) Brca1 controls homology-directed DNA repair. Mol Cell 4 : 511–518.
11. PatelKJ, YuVP, LeeH, CorcoranA, ThistlethwaiteFC, et al. (1998) Involvement of Brca2 in DNA repair. Mol Cell 1 : 347–357.
12. XuG, HerzigM, RotreklV, WalterCA (2008) Base excision repair, aging and health span. Mech Ageing Dev 129 : 366–382.
13. FarmerH, McCabeN, LordCJ, TuttAN, JohnsonDA, et al. (2005) Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature 434 : 917–921.
14. OsorioA, MilneRL, AlonsoR, PitaG, PeterlongoP, et al. (2011) Evaluation of the XRCC1 gene as a phenotypic modifier in BRCA1/2 mutation carriers. Results from the consortium of investigators of modifiers of BRCA1/BRCA2. Br J Cancer 104 : 1356–1361.
15. ZhangY, NewcombPA, EganKM, Titus-ErnstoffL, ChanockS, et al. (2006) Genetic polymorphisms in base-excision repair pathway genes and risk of breast cancer. Cancer Epidemiol Biomarkers Prev 15 : 353–358.
16. ZipprichA, KussO, RogowskiS, KleberG, LottererE, et al. (2010) Incorporating indocyanin green clearance into the Model for End Stage Liver Disease (MELD-ICG) improves prognostic accuracy in intermediate to advanced cirrhosis. Gut 59 : 963–968.
17. PopandaO, SeiboldP, NikolovI, OakesCC, BurwinkelB, et al. (2013) Germline variants of base excision repair genes and breast cancer: A polymorphism in DNA polymerase gamma modifies gene expression and breast cancer risk. Int J Cancer 132 : 55–62.
18. RobertsMR, ShieldsPG, AmbrosoneCB, NieJ, MarianC, et al. (2011) Single-nucleotide polymorphisms in DNA repair genes and association with breast cancer risk in the web study. Carcinogenesis 32 : 1223–1230.
19. SangrajrangS, SchmezerP, BurkholderI, WaasP, BoffettaP, et al. (2008) Polymorphisms in three base excision repair genes and breast cancer risk in Thai women. Breast Cancer Res Treat 111 : 279–288.
20. Ming-ShieanH, YuJC, WangHW, ChenST, HsiungCN, et al. (2010) Synergistic effects of polymorphisms in DNA repair genes and endogenous estrogen exposure on female breast cancer risk. Ann Surg Oncol 17 : 760–771.
21. ZipprichJ, TerryMB, Brandt-RaufP, FreyerGA, LiaoY, et al. (2010) XRCC1 polymorphisms and breast cancer risk from the New York Site of the Breast Cancer Family Registry: A family-based case-control study. J Carcinog 9 : 4.
22. BanerjeeD, MandalSM, DasA, HegdeML, DasS, et al. (2011) Preferential repair of oxidized base damage in the transcribed genes of mammalian cells. J Biol Chem 286 : 6006–6016.
23. WeiH, KamatA, ChenM, KeHL, ChangDW, et al. (2012) Association of polymorphisms in oxidative stress genes with clinical outcomes for bladder cancer treated with Bacillus Calmette-Guerin. PLoS One 7: e38533.
24. KinslowCJ, El-ZeinRA, RondelliCM, HillCE, WickliffeJK, et al. (2010) Regulatory regions responsive to oxidative stress in the promoter of the human DNA glycosylase gene NEIL2. Mutagenesis 25 : 171–177.
25. KinslowCJ, El-ZeinRA, HillCE, WickliffeJK, Abdel-RahmanSZ (2008) Single nucleotide polymorphisms 5′ upstream the coding region of the NEIL2 gene influence gene transcription levels and alter levels of genetic damage. Genes Chromosomes Cancer 47 : 923–932.
26. DeyS, MaitiAK, HegdeML, HegdePM, BoldoghI, et al. (2012) Increased risk of lung cancer associated with a functionally impaired polymorphic variant of the human DNA glycosylase NEIL2. DNA Repair (Amst) 11 : 570–578.
27. ZhaiX, ZhaoH, LiuZ, WangLE, El-NaggarAK, et al. (2008) Functional variants of the NEIL1 and NEIL2 genes and risk and progression of squamous cell carcinoma of the oral cavity and oropharynx. Clin Cancer Res 14 : 4345–4352.
28. ArcandSL, ProvencherD, Mes-MassonAM, ToninPN (2005) OGG1 Cys326 variant, allelic imbalance of chromosome band 3p25.3 and TP53 mutations in ovarian cancer. Int J Oncol 27 : 1315–1320.
29. RossnerPJr, TerryMB, GammonMD, ZhangFF, TeitelbaumSL, et al. (2006) OGG1 polymorphisms and breast cancer risk. Cancer Epidemiol Biomarkers Prev 15 : 811–815.
30. WeiW, HeXF, QinJB, SuJ, LiSX, et al. (2012) Association between the OGG1 Ser326Cys and APEX1 Asp148Glu polymorphisms and lung cancer risk: a meta-analysis. Mol Biol Rep 39 : 11249–11262.
31. XieY, YangH, MillerJH, ShihDM, HicksGG, et al. (2008) Cells deficient in oxidative DNA damage repair genes Myh and Ogg1 are sensitive to oxidants with increased G2/M arrest and multinucleation. Carcinogenesis 29 : 722–728.
32. CouchFJ, WangX, McGuffogL, LeeA, OlswoldC, et al. (2013) Genome-Wide Association Study in BRCA1 Mutation Carriers Identifies Novel Loci Associated with Breast and Ovarian Cancer Risk. PLoS Genet 9: e1003212.
33. VisnesT, AkbariM, HagenL, SlupphaugG, KrokanHE (2008) The rate of base excision repair of uracil is controlled by the initiating glycosylase. DNA Repair (Amst) 7 : 1869–1881.
34. AntoniouAC, GoldgarDE, AndrieuN, Chang-ClaudeJ, BrohetR, et al. (2005) A weighted cohort approach for analysing factors modifying disease risks in carriers of high-risk susceptibility genes. Genet Epidemiol 29 : 1–11.
35. GaudetMM, KuchenbaeckerKB, VijaiJ, KleinRJ, KirchhoffT, et al. (2013) Identification of a BRCA2-Specific Modifier Locus at 6p24 Related to Breast Cancer Risk. PLoS Genet 9: e1003173.
36. BarnesDR, LeeA, EastonDF, AntoniouAC (2012) Evaluation of association methods for analysing modifiers of disease risk in carriers of high-risk mutations. Genet Epidemiol 36 : 274–291.
37. BarnesDR, AntoniouAC (2012) Unravelling modifiers of breast and ovarian cancer risk for BRCA1 and BRCA2 mutation carriers: update on genetic modifiers. J Intern Med 271 : 331–343.
38. AminN, van DuijnCM, AulchenkoYS (2007) A genomic background based method for association analysis in related individuals. PLoS One 2: e1274.
39. LeuteneggerAL, PrumB, GeninE, VernyC, LemainqueA, et al. (2003) Estimation of the inbreeding coefficient through use of genomic data. Am J Hum Genet 73 : 516–523.
40. DelaneauO, ZaguryJF (2012) Haplotype inference. Methods Mol Biol 888 : 177–196.
41. HowieBN, DonnellyP, MarchiniJ (2009) A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet 5: e1000529.
42. PruimRJ, WelchRP, SannaS, TeslovichTM, ChinesPS, et al. (2010) LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics 26 : 2336–2337.
Štítky
Genetika Reprodukční medicína
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2014 Číslo 4
-
Všechny články tohoto čísla
- The Challenges of Mitochondrial Replacement
- Concocting Cholinergy
- Genome-Wide Diet-Gene Interaction Analyses for Risk of Colorectal Cancer
- Statistical Power to Detect Genetic (Co)Variance of Complex Traits Using SNP Data in Unrelated Samples
- Mouse Pulmonary Adenoma Susceptibility 1 Locus Is an Expression QTL Modulating -4A
- Transcription-Associated R-Loop Formation across the Human CGG-Repeat Region
- Epigenetic Regulation by Heritable RNA
- Protein Quantitative Trait Loci Identify Novel Candidates Modulating Cellular Response to Chemotherapy
- Genome-Wide Profiling of Yeast DNA:RNA Hybrid Prone Sites with DRIP-Chip
- The Mechanism of Gene Targeting in Human Somatic Cells
- A LINE-1 Insertion in DLX6 Is Responsible for Cleft Palate and Mandibular Abnormalities in a Canine Model of Pierre Robin Sequence
- Interaction between Two Timing MicroRNAs Controls Trichome Distribution in
- DNA Glycosylases Involved in Base Excision Repair May Be Associated with Cancer Risk in and Mutation Carriers
- The Myc-Mondo/Mad Complexes Integrate Diverse Longevity Signals
- Evolutionarily Diverged Regulation of X-chromosomal Genes as a Primal Event in Mouse Reproductive Isolation
- Mutations in Conserved Residues of the microRNA Argonaute ALG-1 Identify Separable Functions in ALG-1 miRISC Loading and Target Repression
- Genetic Predisposition to In Situ and Invasive Lobular Carcinoma of the Breast
- Isl1 Directly Controls a Cholinergic Neuronal Identity in the Developing Forebrain and Spinal Cord by Forming Cell Type-Specific Complexes
- A Synthetic Community Approach Reveals Plant Genotypes Affecting the Phyllosphere Microbiota
- The Sequence-Specific Transcription Factor c-Jun Targets Cockayne Syndrome Protein B to Regulate Transcription and Chromatin Structure
- Determining the Control Circuitry of Redox Metabolism at the Genome-Scale
- DNA Repair Pathway Selection Caused by Defects in , , and Telomere Addition Generates Specific Chromosomal Rearrangement Signatures
- Methylome Diversification through Changes in DNA Methyltransferase Sequence Specificity
- Folliculin Regulates Ampk-Dependent Autophagy and Metabolic Stress Survival
- Fine Mapping of Dominant -Linked Incompatibility Alleles in Hybrids
- Unexpected Role of the Steroid-Deficiency Protein Ecdysoneless in Pre-mRNA Splicing
- Three Groups of Transposable Elements with Contrasting Copy Number Dynamics and Host Responses in the Maize ( ssp. ) Genome
- Sox5 Functions as a Fate Switch in Medaka Pigment Cell Development
- Synergistic Interactions between the Molecular and Neuronal Circadian Networks Drive Robust Behavioral Circadian Rhythms in
- Chromatin Landscapes of Retroviral and Transposon Integration Profiles
- Widespread Use of Non-productive Alternative Splice Sites in
- Ras GTPase-Like Protein MglA, a Controller of Bacterial Social-Motility in Myxobacteria, Has Evolved to Control Bacterial Predation by
- Cell Type-Specific Functions of Genes Revealed by Novel Adipocyte and Hepatocyte Circadian Clock Models
- Phenotype Ontologies and Cross-Species Analysis for Translational Research
- Embryogenesis Scales Uniformly across Temperature in Developmentally Diverse Species
- In Pursuit of the Gene: An Interview with James Schwartz
- Molecular Mechanisms of Hypoxic Responses via Unique Roles of Ras1, Cdc24 and Ptp3 in a Human Fungal Pathogen
- Analysis of the Genome and Transcriptome of var. Reveals Complex RNA Expression and Microevolution Leading to Virulence Attenuation
- Genotypic and Functional Impact of HIV-1 Adaptation to Its Host Population during the North American Epidemic
- RNA Editome in Rhesus Macaque Shaped by Purifying Selection
- Proper Actin Ring Formation and Septum Constriction Requires Coordinated Regulation of SIN and MOR Pathways through the Germinal Centre Kinase MST-1
- Interplay of the Serine/Threonine-Kinase StkP and the Paralogs DivIVA and GpsB in Pneumococcal Cell Elongation and Division
- A Quality Control Mechanism Coordinates Meiotic Prophase Events to Promote Crossover Assurance
- CNNM2 Mutations Cause Impaired Brain Development and Seizures in Patients with Hypomagnesemia
- The RNA-Binding Protein QKI Suppresses Cancer-Associated Aberrant Splicing
- Uncoupling Transcription from Covalent Histone Modification
- Rad51–Rad52 Mediated Maintenance of Centromeric Chromatin in
- FRA2A Is a CGG Repeat Expansion Associated with Silencing of
- A General Approach for Haplotype Phasing across the Full Spectrum of Relatedness
- A Novel Highly Divergent Protein Family Identified from a Viviparous Insect by RNA-seq Analysis: A Potential Target for Tsetse Fly-Specific Abortifacients
- A Central Role for in Regulation of Islet Function in Man
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- The Sequence-Specific Transcription Factor c-Jun Targets Cockayne Syndrome Protein B to Regulate Transcription and Chromatin Structure
- The Mechanism of Gene Targeting in Human Somatic Cells
- Genetic Predisposition to In Situ and Invasive Lobular Carcinoma of the Breast
- Widespread Use of Non-productive Alternative Splice Sites in
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



