-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaSyd/JIP3 and JNK Signaling Are Required for Myonuclear Positioning and Muscle Function
A common pathology found in numerous cases of muscle diseases, including congenital myopathies and muscular dystrophies, is aberrantly located nuclei within individual multinucleated muscle cells. However, whether or not mispositioned myonuclei are a cause or consequence of muscle disease states is currently debated. Here, we take advantage of the model organism, Drosophila melanogaster, which shares the conserved myofiber found in mammalian systems, to identify Syd as a novel regulator of myonuclear positioning. We show that Syd is responsible for mediating the activities of Kinesin and Dynein, two motor proteins that exert forces to pull myonuclei into place. Moreover, we demonstrate that Syd-dependent myonuclear positioning also requires intracellular signaling from the JNK MAPK cascade to direct when and how myonuclei are moved into proper position. This work thus identifies developmental cues that direct proper muscle morphogenesis, suggesting that cases of muscle disease may result from a failure to achieve initial spacing of myonuclei. Supporting this notion, we find that loss of Syd impairs muscle function, but resupplying Syd restores proper myonuclear spacing and muscle function. These findings are particularly important as mispositioned myonuclei gain traction as a potential contributing factor in cases of muscle disease.
Published in the journal: . PLoS Genet 10(12): e32767. doi:10.1371/journal.pgen.1004880
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1004880Summary
A common pathology found in numerous cases of muscle diseases, including congenital myopathies and muscular dystrophies, is aberrantly located nuclei within individual multinucleated muscle cells. However, whether or not mispositioned myonuclei are a cause or consequence of muscle disease states is currently debated. Here, we take advantage of the model organism, Drosophila melanogaster, which shares the conserved myofiber found in mammalian systems, to identify Syd as a novel regulator of myonuclear positioning. We show that Syd is responsible for mediating the activities of Kinesin and Dynein, two motor proteins that exert forces to pull myonuclei into place. Moreover, we demonstrate that Syd-dependent myonuclear positioning also requires intracellular signaling from the JNK MAPK cascade to direct when and how myonuclei are moved into proper position. This work thus identifies developmental cues that direct proper muscle morphogenesis, suggesting that cases of muscle disease may result from a failure to achieve initial spacing of myonuclei. Supporting this notion, we find that loss of Syd impairs muscle function, but resupplying Syd restores proper myonuclear spacing and muscle function. These findings are particularly important as mispositioned myonuclei gain traction as a potential contributing factor in cases of muscle disease.
Introduction
The intracellular location of the multiple nuclei within muscle cells has recently gained traction as a potential contributing factor to muscle disease. Improper myonuclear position strongly correlates with muscle disease [1], [2] and muscle weakness [3]–[5]; yet, the mechanisms of myonuclear movement and positioning have only recently begun to emerge.
Recent work in Drosophila melanogaster has identified myonuclear positioning as a microtubule-dependent process [4], [6], requiring both Kinesin-1 (Kinesin) and cytoplasmic Dynein (Dynein), the plus - and minus-end directed microtubule motor proteins, respectively [4], [5], [7]. Specifically, two spatially distinct Kinesin - and Dynein-dependent processes position myonuclei [5], [7]. In the first, Kinesin and Dynein exert forces directly on the nucleus: Kinesin extends the front of the myonucleus in the direction of travel, and Dynein is necessary for the retraction of the trailing edge of the myonucleus to complete a translocation step [7]. In the other, Kinesin transports Dynein to the cell cortex near the ends of the muscles where Dynein then pulls microtubule minus-ends and the attached myonuclei into place [5], [7]. Disruption of either pathway leads to mispositioned myonuclei [5], [7], but how Kinesin and Dynein are both used in two spatially segregated mechanisms, and whether their actions in these two different locations are connected or interdependent, is not known.
In many cellular contexts, adaptor proteins specify a variety of motor protein functions. Adaptors either recruit or restrict motors to particular cellular locations [8]–[11], mediate specific motor-cargo interactions [12]-[20], ensure proper temporal and spatial activation of motor function [11], [21]–[23], or enhance motor processivity [24]–[26]. Additionally, adaptor proteins that influence motor function respond to distinct stimuli. Some adaptor proteins regulate motors in response to physical changes of the cell brought on by mechanical strain [27], while others respond to environmental cues via the induction of signaling cascades [13], [28]–[30]. Thus, adaptors are ideal candidates to direct the activities of Kinesin and Dynein during myonuclear positioning.
The JNK signaling cascade is one of three classical mitogen-activated protein kinase (MAPK) cascades conserved across eukaryotes [31]–[37]. Each cascade consists of a MAPK kinase kinase → MAPK kinase → MAPK signaling module that impacts various cellular functions in response to specific stimuli. One function of the JNK MAPK signaling cascade is to phosphorylate JNK-interacting proteins (JIPs), a class of adaptor proteins that regulate Kinesin and Dynein activity in neurons [13], [38]–[41]. While all JIPs interact with both Kinesin and Dynein [14], [21], [38], [39], each adaptor has unique motor-binding domains [13], [38], [42], [43]. JIP1/2 proteins contain a shared motor-binding domain, while JIP3/4 family members have separate Kinesin - and Dynein-binding domains that facilitate binding to both motors simultaneously; thus, different JIPs coordinate motor functions via distinct mechanisms [13], [21], [38], [41]–[44]. Given these features, the JIP proteins could regulate the two Kinesin - and Dynein-dependent pathways governing myonuclear positioning.
To address this possibility, we examined the role of the JIP3 ortholog, Sunday Driver (Syd), during Drosophila muscle morphogenesis. We find that Syd responds to the activation of the JNK signaling cascade to regulate myonuclear positioning by specifically promoting Kinesin-dependent localization of Dynein to the muscle ends. Furthermore, we propose that Kinesin and Dynein are both initially perinuclear and that Syd specifies a subset of Kinesin to relocate Dynein to the muscle cell cortex to initiate cortical pulling of myonuclei. Finally, we demonstrate that disrupted JNK signaling or loss of Syd decreases muscle function, indicating that proper regulation of the JNK signaling cascade and the JIP adaptor proteins is not only necessary for intracellular organization but also critical for muscle function.
Results
Sunday Driver is expressed in muscle tissue
We hypothesized that the adaptor protein, Sunday Driver (Syd), may regulate one, or both, of the spatially distinct Kinesin - and Dynein-dependent pathways that impact myonuclear positioning from 1) the nucleus, and 2) the muscle end [5], [7]. In neurons, Syd physically interacts with both Kinesin and Dynein to coordinate motor activity and promote axonal transport (Fig. 1A) [39], [40], [45]. Because Syd has only been studied in the central nervous system (CNS), we immunostained for Syd and quantified the immunofluorescence intensity of the signal specifically in the Lateral Transverse (LT) muscles (Fig. 1B). Importantly, we compared Syd intensity to that of Tropomyosin, which did not significantly vary between genotypes (S1A-B Figs.) and therefore served as an internal immunostaining control. Intensities were plotted as a function of position (Fig. 1C), and both the peak intensity value (Fig. 1D) and the area under the curve (Fig. 1E), indicating the maximum and total fluorescence, respectively, were used as measures of Syd protein levels.
Fig. 1. Syd is expressed in muscle tissue. 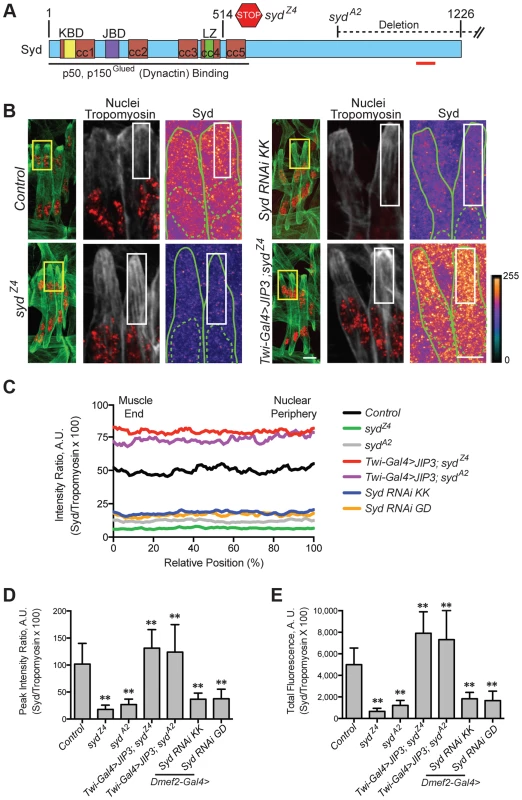
A) Diagram of Syd protein based on previous work and physical interaction data [39], [40], [45]. Amino acid number indicated above at distinct points. Yellow, Khc Binding Domain (KBD); purple, JNK Binding Domain (JBD); green, Leucine Zipper (LZ), mediates Klc binding; red, coiled coil (cc) domains 1–5. N-terminus mediates Dynactin binding (LZ for mammalian JIP3 [21]). syd alleles are noted [45]. Red line indicates the conserved region of Syd/JIP3 recognized by the C-terminal Syd antibody (S1A Fig.). B) (Left) Immunofluorescence projection images of the LT muscles in one hemisegment of stage 16 Drosophila embryos in the indicated genotypes. Green, Tropomyosin/muscles; red, dsRed/nuclei. Yellow boxes denote regions of higher magnification to the right. Scale bar, 10 µm. (Middle, Right) Higher magnification views used for analysis. Grayscale, Tropomyosin; red, nuclei (middle). Syd shown as a heatmap (right) to highlight regions of accumulation. Scale of relative intensities shown at lower right. Solid green lines outline the muscles as noted by Tropomyosin staining. Dotted green lines highlight the nuclei as noted by dsRed staining. White boxes denote regions used for analyses in C-E. Scale bar, 5 µm. C) Intensity profile of Syd immunofluorescence relative to Tropomyosin immunofluorescence plotted as a function of position normalized as a percentage of the total distance from the muscle end (left, 0%) to the nearest nucleus (right, 100%). D) Average peak intensity values for Syd immunofluorescence. E) Average total Syd immunofluorescence, determined by calculating the area under the curves in C. For each genotype in B-E, two LT muscles were measured in each of three hemisegments from ten embryos from at least three independent experiments. All error bars represent standard deviation. **, p<0.01 compared to controls (Student's t-test and ANOVA assessment). A.U., arbitrary units. These analyses demonstrated that Syd is highly expressed in the cytoplasm of muscle cells with no discernable regions of distinct accumulation (Fig. 1B–E, S1A–B Figs.), similar to observations of Syd in other tissues [39], [45]. This signal was lost in two different syd mutants (sydA2 and sydZ4) and in embryos that were depleted of Syd via GAL4/UAS-mediated [46] expression of Syd-RNAi specifically in the muscles (Fig. 1A–E, S1A–B Fig.). These analyses indicate that Syd is expressed in muscle tissue at the correct time and place to influence myonuclear positioning.
Syd is required for myonuclear positioning
To determine whether myonuclear positioning was disrupted in syd mutants, we measured the distance between the myonuclei and the ends of the muscles at embryonic stage 16 (16 h After Egg Laying, AEL) as previously described [5], [7]. At this developmental stage, the myonuclei in controls reside in two groups near the dorsal and ventral ends of the muscles. However, in both sydZ4 and sydA2 mutants, the myonuclei were located significantly further from the muscle ends relative to controls (Fig. 2A,C), suggesting that Syd is required for proper myonuclear positioning.
Fig. 2. Syd is required to position myonuclei. 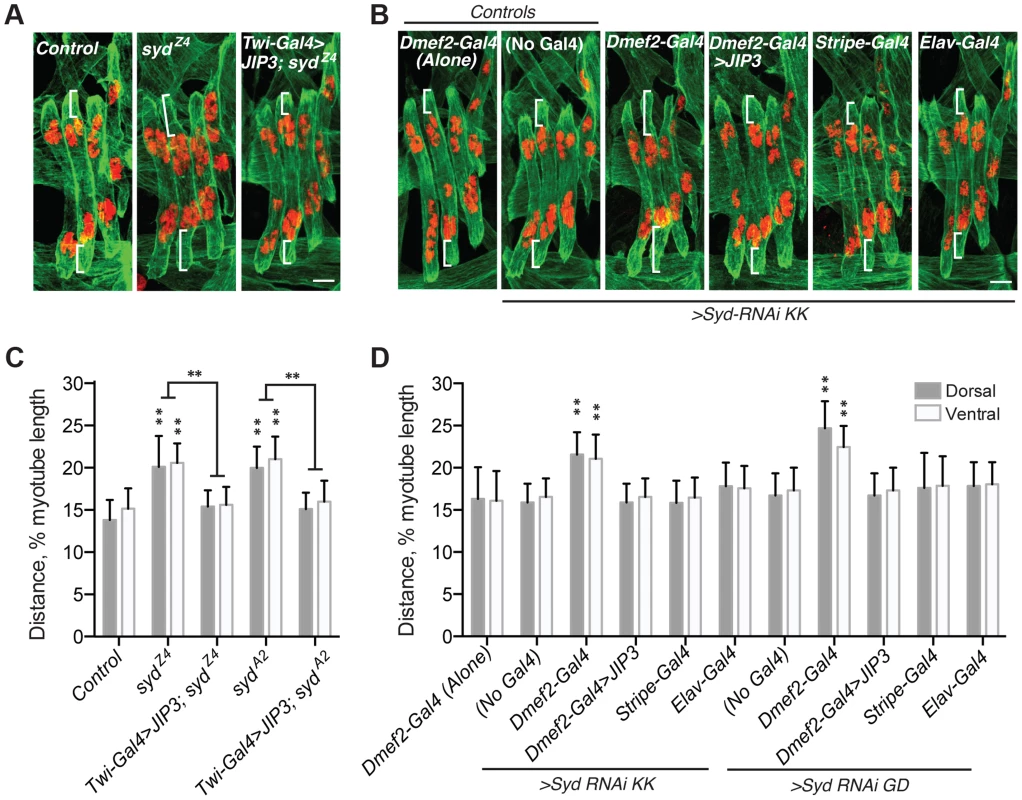
A–B) Immunofluorescence projection images of the LT muscles in one hemisegment of stage 16 Drosophila embryos of the indicated genotypes. Green, Tropomyosin/muscles; red, dsRed/nuclei. White brackets indicate the distance from the dorsal and ventral muscle ends and the nearest nucleus used for quantification of myonuclear position in C–D. Scale Bar, 10 µm. A) syd mutants B) Syd-RNAi expressed under the control of the indicated GAL4 driver. C–D) Histograms indicating the shortest distance between the indicated LT muscle end (Dorsal, grey; Ventral, white) and the nearest nucleus normalized for muscle length for the genotypes indicated in A–B. For each genotype in A–D, all four LT muscles were measured in each of three hemisegments from ten embryos from at least three independent experiments. All error bars represent standard deviation. **, p<0.01 compared to controls, ‘Dmef2-Gal4 Alone,’ and ‘No Gal4,’ unless otherwise noted by brackets (Student's t-test and ANOVA assessment). To confirm that the role of Syd in myonuclear positioning is muscle autonomous, we assessed myonuclear position when Syd-RNAi was expressed in the muscles (Dmef2-Gal4), tendons (Stripe-Gal4), or CNS (Elav-Gal4). Expression of Syd-RNAi specifically in the muscles phenocopied syd mutants, while RNAi-mediated depletion of Syd in either the tendons or the CNS had no effect on myonuclear position (Fig. 2B,D). These data indicate that Syd impacts myonuclear position in a muscle autonomous manner.
Finally, we rescued syd-related myonuclear positioning defects with GAL4/UAS-mediated expression of JIP3 (also known as mSyd2 and JSAP1). JIP3 is the mammalian ortholog of Syd, bearing 69% similarity and 42% identity to Drosophila Syd. Expressing JIP3 in the mesoderm using twist-Gal4 (Fig. 1A–E, S1A–B Fig. and S2A Fig.) rescued myonuclear positioning in syd mutants (Fig. 2A,C). Additionally, because mammalian JIP3 and Drosophila Syd do not align in the regions targeted by Syd-RNAi (S2B–E Fig.), we co-expressed JIP3 and Syd-RNAi in the muscles and rescued the effects of RNAi-mediated Syd depletion on myonuclear position (Fig. 2B,D). Collectively, these data emphasize the conservation of this protein across species and confirm that Syd is required in muscle tissue for proper positioning of myonuclei.
Syd regulates Kinesin - and Dynein-dependent myonuclear positioning
Myonuclear positioning defects in syd mutants are similar to those in both Kinesin heavy chain (Khc) and Dynein heavy chain (Dhc64C) homozygous mutants [4], [5], [7]. Given that Syd physically interacts with Kinesin and Dynein [39], [40], [45], we tested whether Syd genetically interacts with Kinesin and/or Dynein during myonuclear positioning by examining doubly heterozygous embryos. Importantly, the myonuclei are properly positioned in embryos heterozygous for Khc8 (null), Dhc64C4-19 (null), and either syd allele (S3A Fig.) [7]. However, double heterozygotes of Khc8/+; syd/+ and Dhc64C4-19,+/+,syd exhibited mispositioned myonuclei (Fig. 3A–C), suggesting that Syd regulates myonuclear positioning in a Kinesin - and Dynein-dependent manner. Reciprocal crosses to control for maternal loading effects produced identical results (S3B–C Fig.). These data suggest that Kinesin, Dynein, and Syd work in a common pathway to position myonuclei.
Fig. 3. syd genetically interacts with factors required for myonuclear positioning. 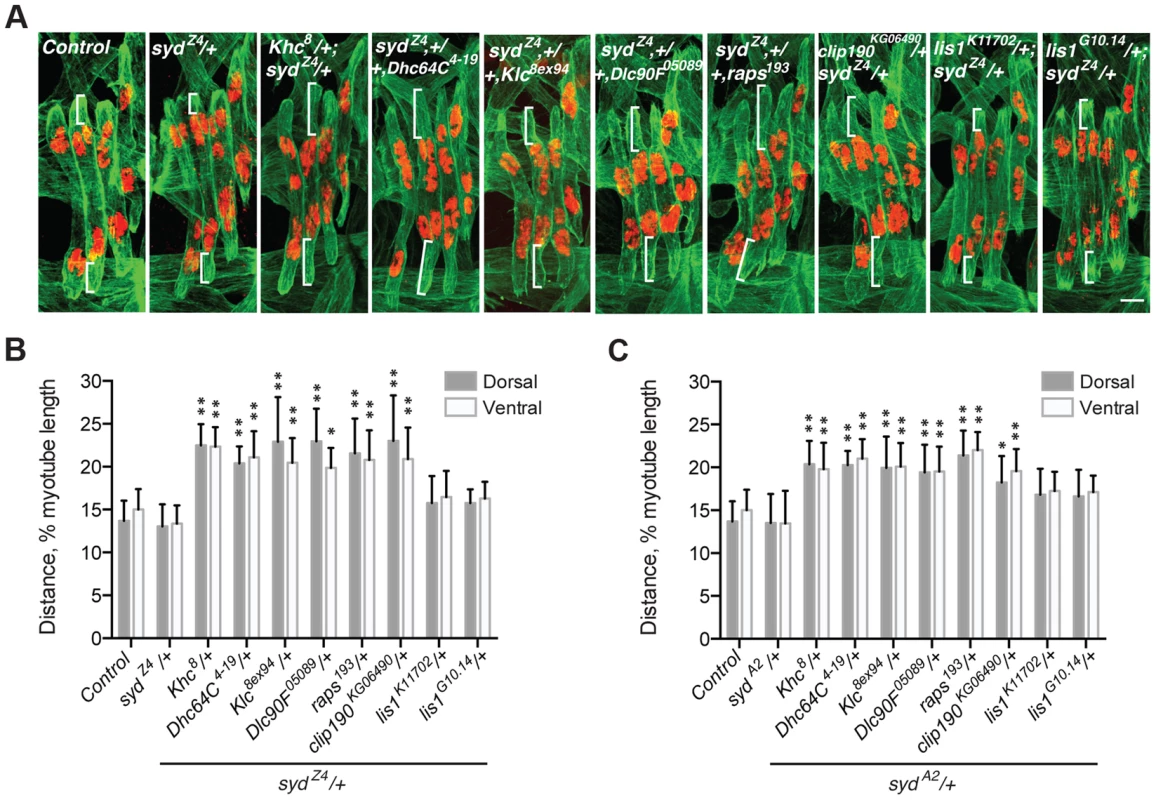
A) Immunofluorescence projection images of the LT muscles in one hemisegment of stage 16 Drosophila embryos of the indicated genotypes in which the syd allele was maternally provided. Green, Tropomyosin/muscles; red, dsRed/nuclei. Scale Bar, 10 µm. B) Histogram indicating the shortest distance between the indicated LT muscle end (Dorsal, grey; Ventral, white) and the nearest nucleus normalized for muscle length for the sydZ4 genetic interactions in A. C) Identical experiment as in A–B using the sydA2 allele in genetic interactions. For each genotype in A–C, all four LT muscles were measured in each of three hemisegments from ten embryos from at least three independent experiments. All error bars represent standard deviation. *, p<0.05; **, p<0.01 compared to wild-type and heterozygous controls (Student's t-test and ANOVA assessment). Adaptors often regulate motors by modulating motor localization. We therefore examined Kinesin (S4A–D Fig.) and Dynein (Fig. 4A–D) localization in control and syd mutant embryos, focusing on Kinesin concentrated near the nucleus and Dynein accumulation at the muscle ends, as we previously identified these localization patterns as necessary for proper myonuclear positioning [5], [7]. The intensity of Kinesin or Dynein immunofluorescence was measured relative to that of Tropomyosin, which remained constant between genotypes (S1C–D Fig and S4E–F Fig.), and values were plotted as a function of position. The location of peak intensity indicated where the motor was enriched, while the amplitude of peak intensity and the integrated total fluorescence approximated protein levels. The location and levels of Kinesin immunofluorescence were similar in both controls and syd mutants (S4A–F Fig.), suggesting that Kinesin is properly localized near the myonuclei in syd mutants. In contrast, Dynein was reduced at the ends of the muscles in syd mutants compared to controls (Fig. 4A). This reduction was not due to a decrease in the amount of Dynein present (Fig. 4C,D), but rather, the distribution of Dynein was shifted towards the nucleus in syd mutants (Fig. 4B). Consistent with previous reports [7], Dynein was similarly mislocalized in Khc8 mutants (Fig. 4A–D). Together, these data suggest that Syd does not affect Kinesin localization but is required for Kinesin-dependent localization of Dynein to the muscle ends in Drosophila.
Fig. 4. Syd is required for Dynein localization. 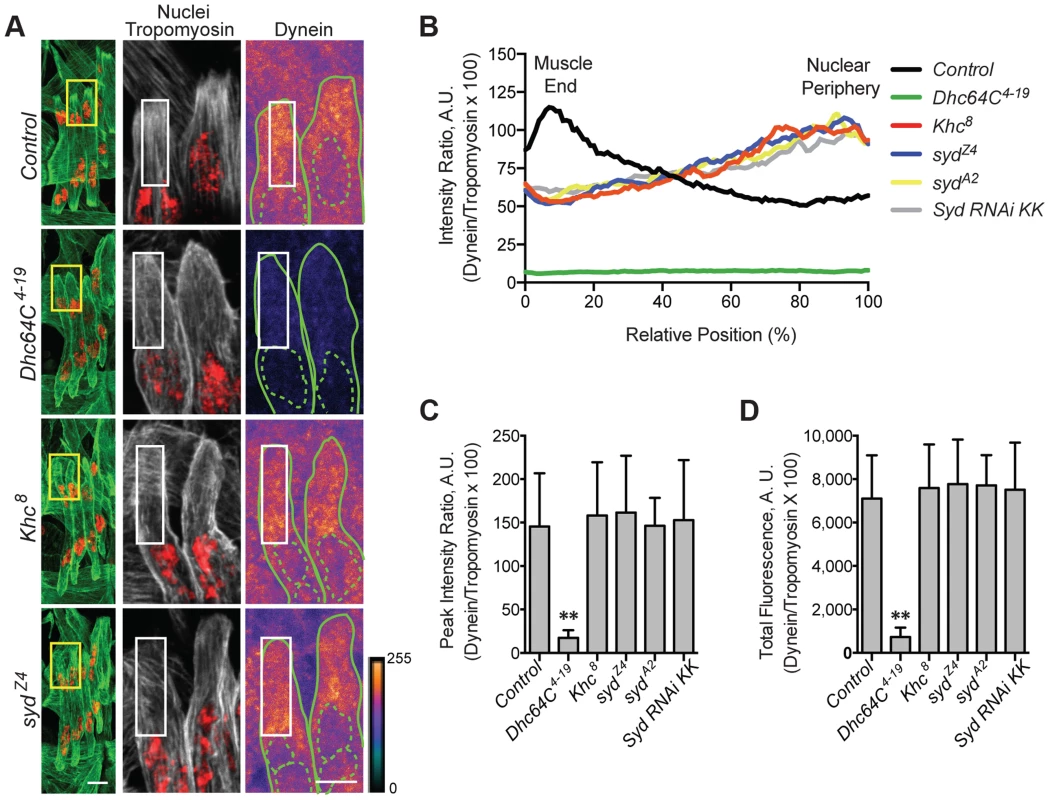
A) (Left) Immunofluorescence projection images of the LT muscles in one hemisegment of stage 16 Drosophila embryos for the indicated genotypes. Green, Tropomyosin/muscles; red, dsRed/nuclei. Yellow boxes denote regions of higher magnification to the right used for analysis. Scale bar, 10 µm. (Middle, Right) Grayscale, Tropomyosin/muscles; red, dsRed/nuclei (middle); Heatmap, Dynein (right). Intensity scale, lower right. Solid and dotted green lines note the perimeter of the muscles and nuclei, respectively. White boxes denote regions used for analyses in B–D. Scale bar, 5 µm. B) Intensity profile of Dynein/Tropomyosin immunofluorescence plotted as a function of normalized position (muscle end, left, 0%; nucleus, right, 100%). C) Average peak intensity for Dynein immunofluorescence. D) Average total Dynein immunofluorescence. For each genotype in A–D, two LT muscles were measured in each of three hemisegments from ten embryos from at least three independent experiments. All error bars represent standard deviation. **, p<0.01 compared to controls (Student's t-test and ANOVA assessment). A.U., arbitrary units. Importantly, inefficient transport of Dynein is not due to defects in the microtubule network, as gross microtubule organization is comparable to controls in both Khc8 [7] and syd mutants (S5A–C Fig.). Therefore, we hypothesized that, as a result of its ability to bind to Kinesin and Dynein [39], [40], [45], Syd mediates an association between the two motors to facilitate Kinesin-dependent localization of Dynein to the cell cortex at the muscle ends to promote proper myonuclear positioning.
This hypothesis implies that the intracellular distribution of Syd would be disrupted in Khc8 mutants but unaffected in Dhc64C4–19 mutants. Indeed, Syd was aberrantly enriched near the nucleus in Khc8 mutants, but Syd was found throughout the cytoplasm in controls and Dhc64C4–19 mutants (Fig. 5A–D, S1E–F Fig.). These data argue that the localization of Syd is Kinesin-dependent, but Dynein-independent.
Fig. 5. Kinesin is required for Syd localization. 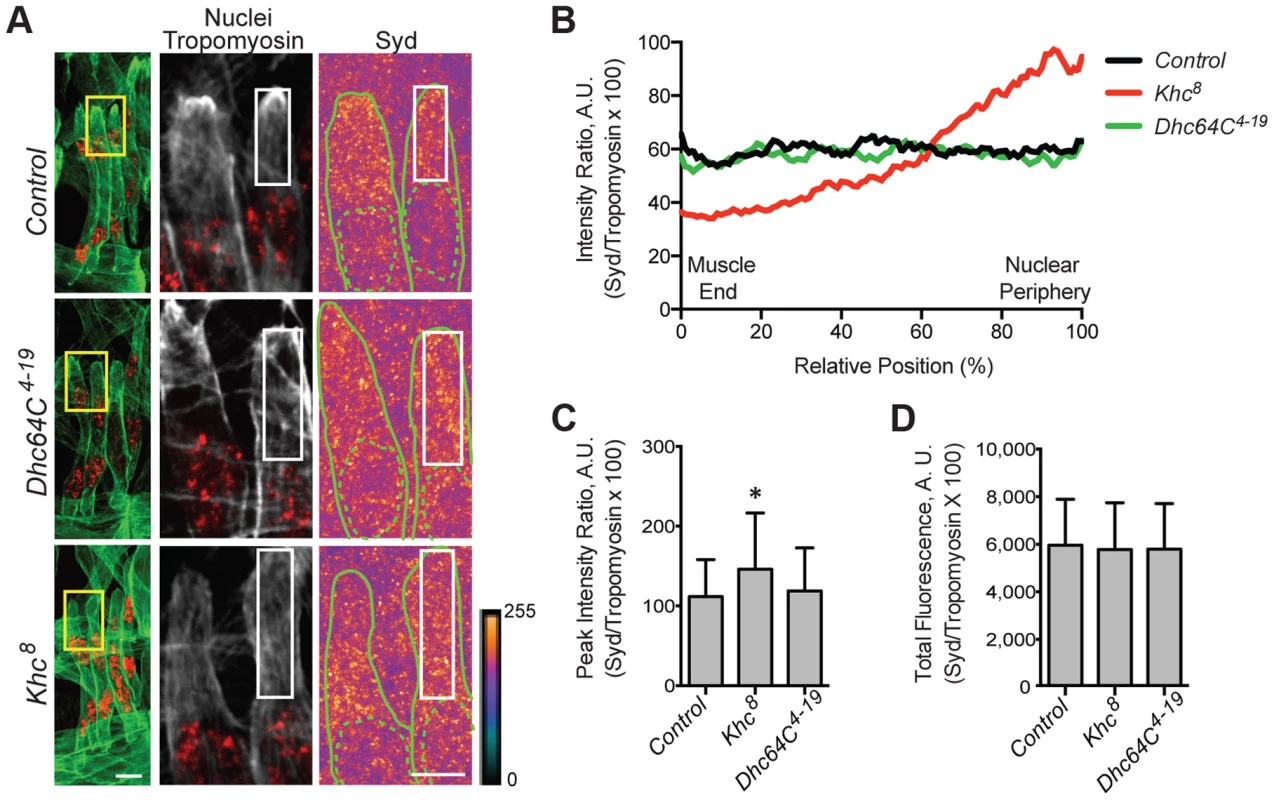
A) (Left) Immunofluorescence projection images of the LT muscles in one hemisegment of stage 16 Drosophila embryos for the indicated genotypes. Green, Tropomyosin/muscles; red, dsRed/nuclei. Yellow boxes denote regions of higher magnification to the right used for analysis. Scale bar, 10 µm. (Middle, Right) Grayscale, Tropomyosin/muscles; red, dsRed/nuclei (middle); Heatmap, Syd (right). Intensity scale, lower right. Solid and dotted green lines note the perimeter of the muscles and nuclei, respectively. White boxes denote regions used for analyses in B–D. Scale bar, 5 µm. B) Intensity profile of Syd/Tropomyosin immunofluorescence plotted as a function of normalized position (muscle end, left, 0%; nucleus, right, 100%). C) Average peak intensity of Syd immunofluorescence. D) Average total Syd immunofluorescence. For each genotype in A–D, two LT muscles were measured in each of three hemisegments from ten embryos from at least three independent experiments. All error bars represent standard deviation. **, p<0.01 compared to controls (Student's t-test and ANOVA assessment). A.U., arbitrary units. Syd affects cortical pulling of myonuclei
These data are consistent with a role for Syd in the cortical pulling mechanism of myonuclear positioning in which Kinesin transports Dynein to the muscle ends, and cortically-anchored Dynein pulls microtubules and the attached nuclei towards the muscle ends [5], [7]. To determine whether Syd functions in this pathway, we tested whether Syd functionally interacts with factors shown to be required for this pathway: Dynein light chain (Dlc90F), Raps, and CLIP-190. We also examined Kinesin light chain (Klc), a regulator of the Kinesin motor. Myonuclear position was unaffected in Klc8ex94, Dlc90F05089, raps193, or clip190KG06490 single heterozygous embryos (S3A Fig.). However, embryos doubly heterozygous for syd and each of the aforementioned alleles exhibited significantly mispositioned myonuclei (Fig. 3A–C; S3B–C Fig.), suggesting that Syd functionally interacts with each factor in the cortical pathway. Syd did not interact with two different alleles of lis1, a regulator of Dynein that does not impact myonuclear positioning in Drosophila [5], highlighting the specificity of the observed genetic effects (Fig. 3A–C; S3B–C Fig.). Together, these data suggest that Syd works with Khc, Klc, Dhc64C, Dlc90F, Raps (Pins), and CLIP-190 in a common pathway to position myonuclei. Moreover, these factors impact cortical pulling of myonuclei [5], [7], suggesting that Syd also affects this process.
A role for Syd in one pathway does not exclude its participation in parallel pathways that work towards the same goal. Kinesin and Dynein also exert forces directly on the front and back of myonuclei, respectively, to promote myonuclear movement [7]. This leads to dynamic nuclear shape changes in which myonuclei transition between spherical and elongated nuclear outlines during translocation. Using previously described time-lapse imaging techniques [7], we found that syd mutants exhibited myonuclear shape changes similar to controls, and that the myonuclei in syd mutants maintained the correct leading edge during translocation (S6A–D Fig.). These data indicate that Syd does not impact the activities of Kinesin and Dynein that influence nuclear dynamics to promote myonuclear positioning [7].
JNK signaling is required for myonuclear positioning
Together with previously documented physical interaction data [39], [40], [45], these new findings collectively suggest that Syd specifically mediates Kinesin-dependent localization of Dynein to the muscle ends to promote cortical pulling of myonuclei [5], [7]. Syd contains a JNK-binding domain (JBD; Fig. 1A, purple) [40], which mediates binding to JNK/p-JNK [39] and facilitates JNK-dependent phosphorylation of Syd/JIP3 [47]. Furthermore, Syd and related JIPs respond to JNK signaling in neurons to promote cargo binding and influence Kinesin-dependent axonal transport [38], [39], [41], [47], [48]. Thus, we hypothesized that Syd could respond to induction of the JNK signaling cascade to impact myonuclear positioning in muscle tissue.
In Drosophila, the JNK signaling cascade is composed of Tak1 (MAPKKK), Hep (MAPKK), and Bsk (MAPK), the Drosophila ortholog of mammalian JNK [49]. We used the GAL4/UAS system to deplete Tak1, Hep, or Bsk specifically in the muscles, and in each case, the myonuclei were significantly mispositioned further from the muscle ends (S3D Fig.), mimicking syd, Khc, and Dhc64C mutants. Furthermore, muscle-specific expression of dominant negative Bsk (Bsk-DN), which cannot be phosphorylated [50], similarly affected myonuclear position (Fig. 6A,B), indicating that loss of JNK signaling disrupts myonuclear positioning.
Fig. 6. JNK signaling is required for Syd-mediated myonuclear positioning. 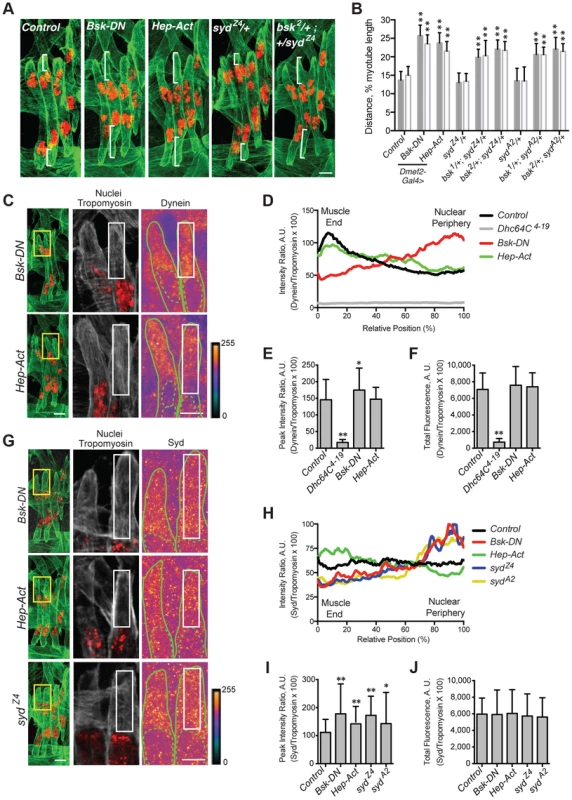
A–B) Identical assay as in Fig. 2. A) Stage 16 embryos of the indicated genotypes. Green, Tropomyosin/muscles; red, dsRed/nuclei. Scale Bar, 10 µm. B) Histogram indicating the shortest (normalized) distance of white brackets in A. C–F) Identical assay as in Fig. 4. C) (Left) Green, Tropomyosin; red, nuclei. Scale bar, 10 µm. (Middle, Right) Grayscale, Tropomyosin; red, nuclei; Heatmap, Dynein. Scale bar, 5 µm. Compare to control in Fig. 4A. D) Intensity profile of Dynein/Tropomyosin immunofluorescence plotted as a function of normalized position. E) Average peak intensity for Dynein immunofluorescence. F) Average total Dynein immunofluorescence. G–J) Identical assay as in Fig. 5. G) (Left) Green, Tropomyosin; red, nuclei. Scale bar, 10 µm. (Middle, Right) Grayscale, Tropomyosin; red, nuclei; Heatmap, Syd. Scale bar, 5 µm. Compare to control in Fig. 5A. H) Intensity profile of Syd/Tropomyosin immunofluorescence plotted as a function of normalized position. I) Average peak intensity for Syd immunofluorescence. J) Average total Syd immunofluorescence. For each genotype/experiment, all four LT muscles in A–B and at least two LT muscles in C-J were measured in each of three hemisegments from ten embryos from at least three independent experiments. All error bars represent standard deviation. *, p<0.05; **, p<0.01 compared to controls (Student's t-test and ANOVA assessment). A.U. arbitrary units. Interestingly, myonuclear position was also sensitive to the levels of JNK signaling. Overactivation of the cascade with muscle-specific expression of Hep-Act, a constitutively active phosphomimetic form of Hep [50], similarly resulted in mispositioned myonuclei (Fig. 6A,B), indicating that disrupted regulation of JNK signaling negatively impacts myonuclear positioning.
We next tested whether JNK signaling requires Syd to position myonuclei. bsk/+; syd/+ double heterozygotes had mispositioned myonuclei similar to syd homozygous mutants and embryos with disrupted JNK signaling (Fig. 6A,B; S3A Fig.), suggesting that Syd and Bsk (JNK) work in a common pathway to position myonuclei. Furthermore, embryos expressing Bsk-DN resembled both Khc8 and syd mutants, with both Dynein (Fig. 6C–F, S1G–H Fig.) and Syd (Fig. 6G-J, S1I–J Fig.) aberrantly enriched near the nucleus. These data argue that JNK signaling is required for the cortical pulling pathway of myonuclear positioning.
The effects of overactive JNK signaling were more intriguing. In embryos expressing Hep-Act, Dynein was often properly located at the muscle ends, resembling controls; however, in many instances, Dynein was found more diffusely throughout the cytoplasm (Fig. 6C–F, S1G–H Fig.). When averaged together, these data generated an intermediate distribution curve (Fig. 6D, green). In contrast, Syd consistently becomes aberrantly enriched at the muscle ends in embryos expressing Hep-Act (Fig. 6G–J, S1I–J Fig.). Collectively, these data support that JNK signaling is indeed required to promote Kinesin - and Syd-mediated localization of Dynein to the muscle ends. These data also suggest that improper regulation of JNK signaling affects the maintenance of both Dynein and Syd at the muscle ends.
The C-terminus of Syd is specifically required for Kinesin/Syd-dependent transport of Dynein to the muscle ends
Syd likely transduces JNK signaling through its JBD to influence Kinesin-dependent transport of Dynein. However, the syd alleles result in premature coding truncations that produce N-terminal fragments of Syd (Fig. 1A) [45]. Thus, the JBD as well as the Kinesin - and Dynein-binding domains are present in syd mutants; yet, Dynein still fails to localize to the muscles ends, suggesting that the C-terminus of Syd is critical for motor transport. Indeed, immunostaining with an N-terminal Syd antibody demonstrated that truncated Syd was enriched near the nucleus in syd mutants despite proper JNK signaling in these genetic backgrounds (Fig. 6G–J, S1I–J Fig.). These data demonstrate that, although no known domains have been identified in the C-terminus of Syd, the C-terminus is required for Kinesin - and JNK signaling-dependent transport of Syd, and therefore Dynein, to the muscle end.
Syd and JNK signaling are required for muscle function
We next investigated whether Syd and/or JNK signaling was necessary for muscle function by quantifying stage L3 larval locomotion using previously described tracking techniques [4], [5], [51]. However, consistent with previous reports [45], only 14–17% of syd mutants hatch into larvae (Fig. 7A), and the survivors die soon after hatching, preventing assessments of L3 larval velocity. In contrast, while muscle-specific depletion of Syd using RNAi also leads to decreased viability, 30% of embryos hatch and survive to adulthood, and this lethality can be fully rescued with muscle-specific expression of JIP3 (Fig. 7B). Similarly, while genetic mutations in JNK signaling components leads to embryonic lethality [49], muscle-specific expression of Bsk-DN or Hep-Act does not impair viability at larval stages of development (Fig. 7C). Thus, GAL4/UAS-mediated tissue-specific loss and gain of Syd and JNK signaling was used to assess larval locomotion.
Fig. 7. Syd impacts muscle function. 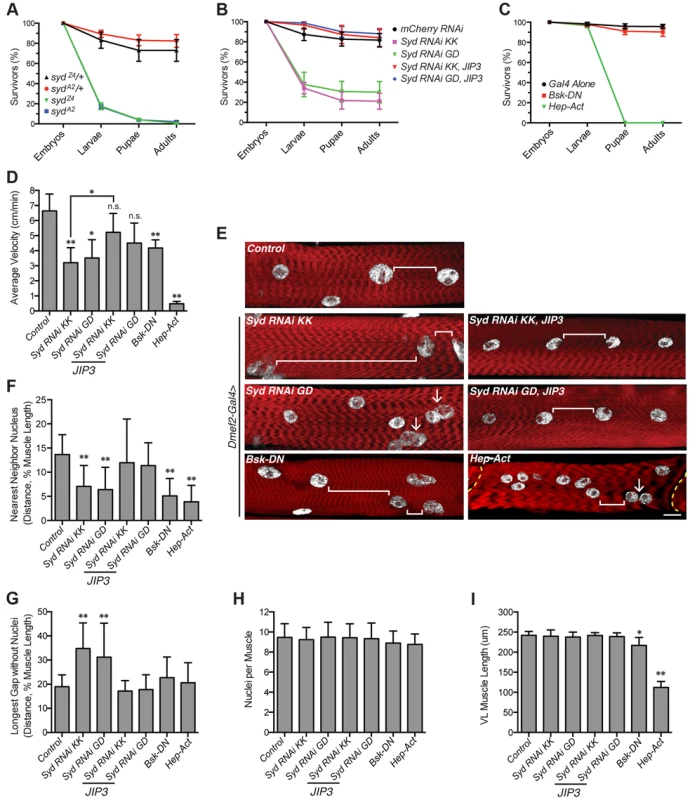
A) Viability of syd mutants. B) Viability of embryos expressing Syd-RNAi in the muscle tissue driven by Dmef2-Gal4. C) Viability of embryos with disrupted JNK signaling in muscle tissue via Dmef2-Gal4 expression. D) Average velocity of Drosophila larvae as they crawled towards a stimulus. E) Immunofluorescence projection images of VL muscles in dissected L3 larvae that were previously used in locomotion assays. Red, Phalloidin/sarcomeres; white, Hoescht/nuclei. White brackets highlight internuclear distance. Arrows denote clumped myonuclei. Yellow dashed line, segment border. Scale bar, 20 µm. F) Average distance between each myonucleus and its nearest neighbor normalized for muscle length. G) Average length of the longest region of muscle tissue devoid of nuclei normalized for muscle length. H) Average number of nuclei per VL muscle. I) Average muscle length in the anterior-posterior axis. For each genotype, at least 300 embryos were assessed in A–C and at least 30 larvae were tracked in D from at least three independent experiments each. For each genotype in E–I, three VL muscles (1, 2, and 4) in six hemisegments from five larvae from at least three independent experiments were measured/counted. All error bars represent standard deviation. *, p<0.05; **, p<0.01 compared to controls unless otherwise noted by brackets (Student's t-test and ANOVA assessment). n.s., not significant. We found that larvae lacking Syd in the muscles crawled significantly slower than controls, but expression of JIP3 in the muscles rescued these locomotion defects (Fig. 7D). Additionally, although overactivation of the cascade was more deleterious, any disruption in JNK signaling negatively affected locomotive ability (Fig. 7D). Importantly, no changes were detected in the muscle innervation sites or the number of boutons at the neuromuscular junctions (S7A–C Fig), both of which, if altered, would be evidence of impaired synaptic transmission [52], [53]. Together, these data indicate that larvae lacking Syd or proper JNK signaling in the muscles exhibit decreased locomotion due to muscle-specific defects independent of communication between the muscles and the CNS.
Myonuclei are mispositioned in larval muscles
We next examined myonuclear position in the Ventral Longitudinal (VL) muscles by two methods: 1) the Nearest Neighbor analysis described previously [4] to quantify the degree of clustering amongst adjacent myonuclei, and 2) a Longest Gap analysis to measure the greatest longitudinal span of the muscle devoid of nuclei. Importantly, both of these measures were normalized to muscle size to account for variation between genotypes (Fig. 7I). These analyses demonstrated that muscles depleted of Syd exhibit greater degrees of myonuclear clustering and longer spans of muscle tissue devoid of myonuclei compared to controls (Fig. 7E–G). Again, expression of JIP3 in these backgrounds restores all values to control levels (Fig. 7E–G). Importantly, the number of nuclei per VL muscle remains constant across genotypes (Fig. 7H), indicating that clustered nuclei and large muscle regions lacking nuclei are not due to gains or losses of myonuclei.
The effects of disrupted JNK signaling on myonuclear position were more interesting. Both loss and gain of JNK signaling led to significant defects in myonuclear position by Nearest Neighbor assessment (Fig. 7F); however, concomitant defects in muscle length in these backgrounds were also evident (Fig. 7I), indicating that JNK signaling is required for multiple aspects of muscle development. We previously observed a similar developmental relationship between myonuclear positioning and muscle length/growth [5], highlighting that these two processes are closely co-regulated. Interestingly though, we do not observe similar defects in muscle length in larvae lacking Syd, suggesting that Syd functions downstream of JNK signaling to specifically impact myonuclear positioning. Given the well-characterized role of Syd as a JNK interacting protein (JIP) [39]–[41], [54], these data suggest that, while JNK signaling is required for multiple aspects of muscle development, Syd may be a key link that facilitates JNK signaling-mediated regulation of myonuclear position.
Discussion
We have used the Drosophila musculature to elucidate the cellular mechanisms and signaling pathways that impact myonuclear position in vivo. The stereotypic distribution of evenly spaced myonuclei is disrupted in embryos and larvae mutant for Sunday Driver (Syd), an adaptor protein known to regulate Kinesin and Dynein activity in neurons [39], [40], [45]. Here, we show that Syd is expressed and required in muscle tissue to regulate motor activity during myonuclear positioning. Moreover, while Kinesin and Dynein are known to influence myonuclear position via two spatially distinct mechanisms [5], [7], we demonstrate that Syd specifically regulates these motors in the context of cortical pulling at the muscle end without affecting motor activity at the nuclear surface. Syd is a member of the JNK-interacting protein (JIP) family, and we demonstrate that JNK signaling is required for Kinesin - and Syd-dependent localization of Dynein to the muscle ends, which facilitates Dynein-based cortical pulling of myonuclei (Fig. 8). Moreover, in the absence of Syd, both Kinesin and Dynein accumulate near the nucleus, where the motors are known to influence nuclear shape changes/dynamics to promote myonuclear translocation. Syd has no impact on nuclear dynamics; thus, we propose that, via instructive JNK signaling, Syd specifies and activates a population of Kinesin at the nucleus to relocate Dynein to the muscle ends to initiate cortical pulling of myonuclei (Fig. 8, numbers 2 and 7). Finally, we show that loss of Syd and disruption of JNK signaling impairs locomotive ability, indicating that Syd-dependent myonuclear positioning is critical for muscle function.
Fig. 8. Model of Syd-dependent myonuclear positioning. 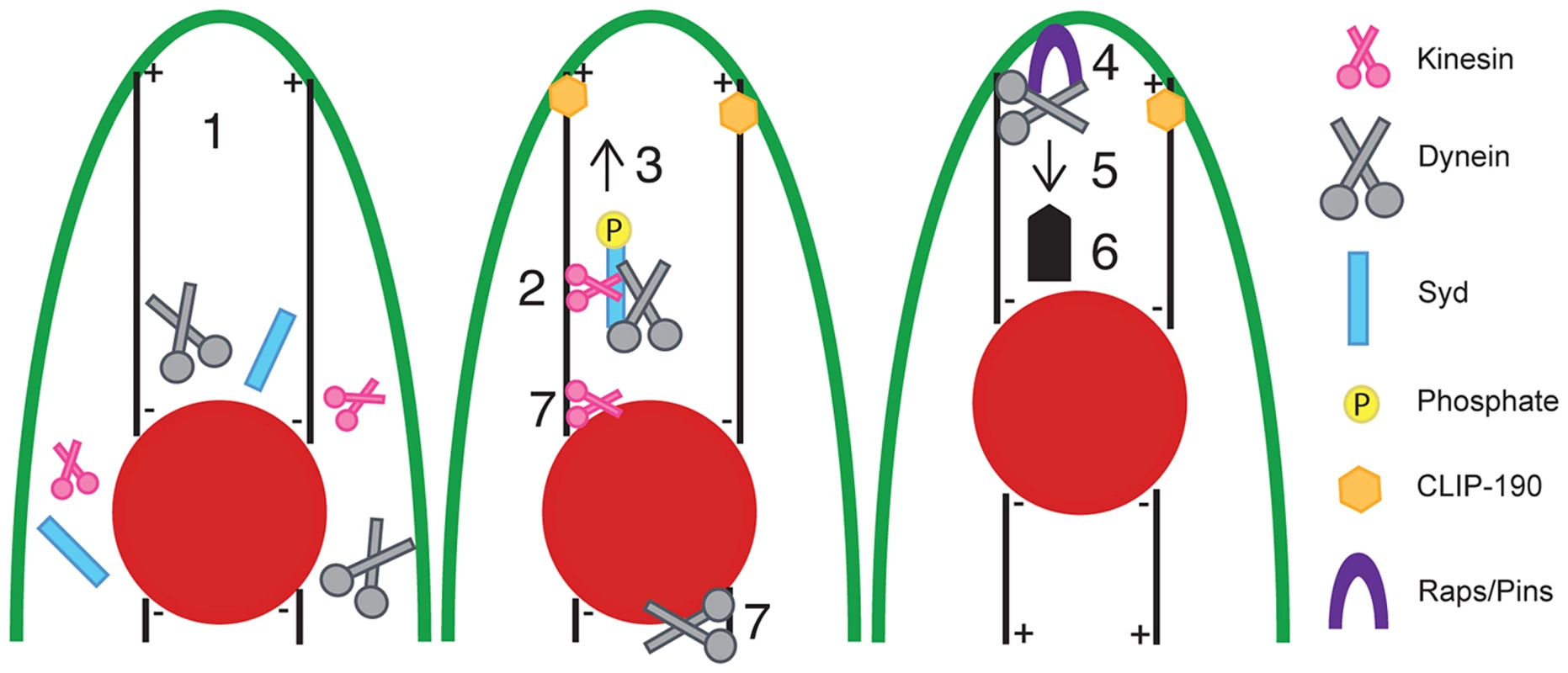
Syd works in the cortical pulling pathway of myonuclear positioning, which requires cellular transport between the myonuclei (red) and the end of the muscle (green outline). (1) Kinesin, Dynein, and Syd are cytoplasmic proteins. Syd mediates (2) complex formation between Kinesin and Dynein and (3) promotes Kinesin-dependent transport of Dynein to the muscle ends in a JNK signaling-dependent manner. Microtubules contact the cell cortex in a CLIP-190-dependent manner [5]. At the muscle end (4) Raps/Pins anchors Dynein to the cell cortex [5]. The Dynein motor becomes active (5, thin arrow) and the net result (6, thick arrow) of cortically stabilized Dynein activity ultimately pulls the myonuclei into proper position. Syd may also work to initially organize Kinesin and Dynein near the nucleus (7) to specify a population of Kinesin to relocate Dynein out to the muscle ends (2) while the remaining Kinesin and Dynein exert forces on the nucleus to promote nuclear dynamics (7). In this manner, both mechanisms of myonuclear positioning work together to move myonuclei into proper position. The finding that JNK signaling is required for this process is novel and significant. Prior to this study, work to elucidate the process of myonuclear positioning focused on identifying factors required for the physical mechanics of moving myonuclei through the cell. However, the present work newly identifies intracellular signals that regulate these activities. Specifically, we show that JNK signaling is required for Kinesin - and Syd-dependent localization of Dynein to the muscle ends. What remains unclear is whether JNK-mediated phosphorylation of Syd [39], [47] induces/permits Dynein binding to Kinesin or if a trimolecular Kinesin-Syd-Dynein complex forms prior to JNK signaling-dependent activation of Kinesin motor activity, which, in turn, transports Dynein to the muscle ends and leads to cortical pulling and proper positioning of myonuclei.
Regardless, our data are consistent with Syd being necessary to relay this instructive JNK signal. Specifically, the C-terminus of Syd is critical for this transport, as Kinesin, Dynein, and truncated Syd are all found near the nucleus in syd mutants, which express N-terminal fragments of Syd. This finding was initially unexpected because the minimal domains of Syd required for binding to Kinesin, Dynein, and JNK are all located in the N-terminus (Fig. 1A) [40]. Furthermore, the N-terminus of Syd is sufficient to support Kinesin-based transport in vitro [40], [41]. However, the C-terminus of Syd is more highly conserved than the well-annotated N-terminus [45], and biochemical analysis of JIP3 demonstrates that upstream components of the JNK MAPK signaling pathway bind to C-terminal regions of JIP3 [47], [54]. Consistent with these data, we show that mutants lacking the C-terminus of Syd fail to promote proper transport of Dynein to the muscle ends despite normal JNK signaling in these backgrounds. This argues that both the N - and C-termini of Syd are necessary to relay instructive cues from the JNK signaling cascade, which leads to proper myonuclear positioning.
Interestingly, while JNK signaling is indeed necessary for myonuclear positioning, we also demonstrate that constitutively active JNK signaling impairs this process. Despite the inconsistent phenotype, overall decreased amounts of Dynein are found at the muscle ends when Hep-Act is expressed in the muscles. This suggests that overactive JNK signaling either mildly impairs Dynein transport, or alternatively, prevents the maintenance of Dynein at the muscle end. We favor the latter interpretation based on the observation that increased levels of Syd are found at the muscle end when JNK signaling is overactivated. This suggests that efficient transport, and likely excess transport, indeed occurs in embryos expressing Hep-Act. This activity would transport Dynein to the muscle ends; thus, overactive JNK signaling likely inhibits the maintenance of Dynein at this location.
It is not clear how this inhibition could occur; however, one possibility is that failure to dephosphorylate Syd may impair the ability of Dynein to associate with ARF6-GTP, a membrane-bound protein that binds to JIP3/4 proteins [21]. ARF6-GTP and Klc compete for binding to JIP3, and phosphorylated JIPs preferentially bind and activate Kinesin [38]. However, JIP3–ARF6-GTP interactions enhance binding between ARF6-GTP and Dynactin, a well-known Dynein-interacting protein [21]. Thus, in our model, once the Kinesin-Syd-Dynein complex reaches the cell cortex, perhaps Syd must lose the JNK signal and become dephosphorylated to dissociate from Kinesin and bind to ARF6-GTP. This may facilitate hand-off of Dynein to ARF6-GTP, which would aid in securing Dynein to the muscle end.
Although the details of this final step remain unclear, our model (Fig. 8) likely represents a common mechanism by which Syd and other JIP3 orthologs act. Analogous to the proposed manner in which Syd selects certain Kinesin complexes to initiate transport, the C. elegans ortholog of Syd/JIP3, UNC-16, was found to exhibit similar gatekeeper characteristics by designating specific complexes to initiate axonal transport in neurons [55]. Furthermore, once specified, UNC-16 also mediates Kinesin-dependent transport of Dynein to the ends of nerve processes [14], [48]. Similarly, mammalian JIP proteins mediate Kinesin-driven transport of many large cargoes, including Dynein [21],[38],[41]. Finally, Syd, UNC-16, and JIP3 directly bind to Kinesin heavy and light chains, and all three orthologs bind to p50 and p150Glued, components of the Dynactin complex, a well-known regulator of Dynein [14], [21], [22], [39], [40]. Together with our data, these observations collectively suggest that this mechanism of JIP3-mediated motor coordination (Fig. 8) is well conserved across species and tissues.
These findings are interesting given that Kinesin and Dynein can directly interact in vitro [56]; however, adaptors such as Syd and other JIP proteins are likely required to mediate motor protein interactions to direct specific motor functions in vivo. Consistent with this notion, structural differences between the JIP1/2 subfamily [13], [42] and the JIP3/4 subfamily of proteins [13], [21], [43], [54] impact how each JIP binds to Kinesin and Dynein and influences motor activity [14], [21], [38], [41]. Although these different JIPs can cooperate towards a single goal [57], they often have unique roles and cannot fully compensate for loss of another in vivo [20], [22], [30], [58], [59].
In Drosophila, Syd is the only ortholog of the JIP3/4 family of proteins. Similarly, there is only one ortholog of the JIP1/2 family of proteins, Aplip1. Thus, in the context of myonuclear positioning, it is tempting to speculate that while Syd modulates motor activity to promote cortical pulling of myonuclei, perhaps Aplip1 affects the ability of the motors to regulate nuclear dynamics. Furthermore, perhaps JNK signaling is the necessary switch that shifts motor function from one mechanism to the other. Indeed, work in Drosophila neurons shows that activation of the JNK signaling cascade disrupts the association between Aplip1 (JIP1) and Kinesin [30], and here we show that JNK signaling is required for coordinated Kinesin-Syd (JIP3) function in the cortical pulling pathway of myonuclear positioning.
Regarding the biological relevance of Syd-dependent mechanisms, we demonstrate that Syd and proper regulation of JNK signaling are critical for muscle function. RNAi-mediated loss of Syd specifically in the muscles leads to decreased larval velocity, and similar locomotive dysfunction is observed in larvae with muscle-specific disruptions in JNK signaling. Consistent with previous reports [4], [5], we show that these locomotive defects are not due to impaired communication with the CNS, but rather correlate with mispositioned myonuclei. These analyses further revealed that muscle-specific disruptions of the JNK signaling cascade additionally impair muscle length/growth, which can also affect locomotion [5]. These findings indicate that JNK signaling is, not surprisingly, necessary for multiple aspects of muscle development. However, that similar concomitant defects are not observed in larvae lacking Syd argues that the role of Syd is more specific to mechanisms of myonuclear positioning. Finally, muscle-specific expression of mammalian JIP3 simultaneously restored myonuclear spacing and rescued larval crawling defects in larvae lacking Syd. These data highlight the high degree of conservation across species, emphasize the muscle autonomous role of Syd, and reiterate the strong correlation between mispositioned myonuclei and decreased muscle output observed previously [4]–[6]. In sum, we have identified that JNK signaling and the motor adaptor, Syd, are required for influencing specific functions of Kinesin and Dynein that lead to proper myonuclear positioning and muscle function, which has significant implications for muscle cell organization, development, and disease.
Materials and Methods
Drosophila genetics
All stocks were grown under standard conditions. Stocks used: apterousME-NLS::dsRed [60], Df(3L)sydA2 and sydZ4 [45], Khc8 [61], Dhc64C4−19 [62], Klc8ex94 [63], Dlc90F05089 [64], lis1K11702 [65], lis1G10.14 [66], raps193 [67], twist-Gal4 [68], Dmef2-Gal4 [69], Stripe-Gal4 (gift from T. Volk), Elav-Gal4 (gift from E. Lai), UAS-GFP-JIP3 (this study), bsk1 and bsk2 [49], UAS-Bsk-DN and UAS-Hep-Act [50]. Bloomington Drosophila Stock Center: clip190KG06490 (14493), UAS-Tak1-RNAi (33404, 35180), UAS-Hep-RNAi (28710, 35210), UAS-Bsk-RNAi (31323, 32977, 35594, 36643), UAS-mCherry-RNAi (35785). Vienna Drosophila RNAi Center: UAS-Syd-RNAi KK109225 (v101459), UAS-Syd-RNAi GD12383 (v35346). Mutants were balanced and identified using CTG (CyO, Twi-Gal4, UAS-2xeGFP) and TTG (TM3, Twi-Gal4, UAS-2xeGFP) [70].
Transgenics
Mouse JIP3 construct: pEGFP-C1/mSyd2 (gift from L. Goldstein) [45].
For Drosophila rescue experiments, mSyd2 (later re-annotated as JIP3) was amplified from the pEGFP-C1/mSyd2 construct using the following primers and cloned into the pUAST vector containing GFP: mSyd2 Forward: 5′-CACCGAATTCATGGAGATCCAGATGGACGAGGGA-3′; mSyd2 Reverse: 5′-GAATTCCTCAGGGGTGTAGGACACCTGCCA-3′.
All constructs were sequenced and verified, and pUAST-GFP/mSyd2 DNA was injected into Drosophila embryos using transposable-element-based insertion methods (Genetic Services, Inc.) to generate flies carrying UAS-GFP-JIP3 on either chromosome II or III that were used in experiments.
Immunohistochemistry
Whole mount embryo staining was performed as described [60]. For Dynein localization measurements, embryos were fixed with 10% formalin diluted 1∶1 in heptane for 20 minutes, then rinsed three times in PBS containing 0.3% Triton X-100, then fixed with 4% EM-grade paraformaldehyde in PBS diluted 1∶1 in heptane for 20 minutes. In all cases, embryos were devitellinized by vortexing in a 1∶1::methanol:heptane solution. Larvae were dissected in ice cold HL3.1 as previously described [71] and fixed with 10% Formalin (Sigma, HT501128-4L). Dynein antibody incubations were performed in PBS supplemented with 0.2% BSA and 0.15% Triton X-100. All other antibody incubations were performed in PBS supplemented with 0.1% BSA and 0.3% Triton X-100. Embryos and larvae were mounted in ProLong Gold (Invitrogen) for fluorescent immunostainings. Antibodies were preabsorbed (PA) as described [72] where noted and used at the indicated final dilutions: rabbit anti-dsRed (1∶400, Clontech, 632496), rat anti-Tropomyosin (PA, 1∶500, Abcam, ab50567), mouse anti-GFP (PA, 1∶200, Clontech, 632381), mouse anti-Dhc (1∶50, Developmental Studies Hybridoma Bank), mouse anti-Tubulin (1∶500, Sigma T9026), rabbit anti-Khc (1∶200, Cytoskeleton Inc., AKIN01), mouse anti-Discs large (1∶200, Developmental Studies Hybridoma Bank), rabbit anti-N-terminal-Syd (SN1) and rabbit anti-C-terminal-Syd (7704) (1∶300) (gifts from V. Cavalli and L. Goldstein) [45]. Alexa Fluor 488-, Alexa Fluor 555-, and Alexa Fluor 647-conjugated fluorescent secondary antibodies (1∶200), Alexa Fluor 546-conjugated Phalloidin (1∶100), and Hoechst-33342 (1 µg/1ml) were used for fluorescent stains (Invitrogen). Fluorescence projection images were acquired on a Leica SP5 laser scanning confocal microscope equipped with a 63× 1.4 NA HCX PL Apochromat oil objective and LAS AF 2.2 software unless noted otherwise. Maximum intensity projections of confocal Z-stacks were rendered using Volocity 6.1.1 Visualization software (Improvision). All resulting 2D projection images were cropped using Adobe Photoshop CS6.
Nuclear position and muscle length measurements
Analysis was performed as described [5].
Kinesin, Dynein, and Syd localization measurements
High magnification projection images of a set 2 µm depth-size were acquired as described [5], ensuring that the entire Z-stack was acquired from completely within the muscle fiber. Any projection images with a slice including the muscle cell membrane or any area outside the bounds of the muscle were discarded to eliminate variation in background fluorescence. Immunofluorescence intensity was assessed across defined regions using ImageJ (NIH). In all cases, boxed regions were unbiasedly selected in the Tropomyosin/nuclear projection image and transferred to identical regions in other channels. For Kinesin, a box of set dimensions was used. For Dynein and Syd, a box of set width and varying length was used to measure immunofluorescence between the end of the muscle and the nearest nucleus. Immunofluorescence intensity of Kinesin, Syd, or Dynein, was compared to Tropomyosin intensity, and the ratios were multiplied by 100. For Kinesin, average ratios were then plotted as a function of raw position. For Dynein and Syd, the total distance was normalized to 100 to account for myonuclear positioning defects, and average ratios were plotted as a function of normalized position (from muscle end to the nearest nucleus). The area under the curves (total fluorescence) were calculated in Microsoft Excel. The greatest pixel intensity denoted the peak fluorescence intensity.
Microtubule organization measurements
Analysis was performed as previously described [5] using ImageJ software (NIH) and projection images acquired at a 3X optical zoom on a Zeiss LSM510 laser-scanning confocal microscope equipped with a 63×1.4 NA HCX PL Apochromat oil DIC objective and ZEN 2009 software.
Time-lapse imaging and nuclear dynamics analysis
Experiments and analyses were performed as described [7].
Larval behavior, dissections, and analysis
Larval speed was assessed as previously described [5]. Tracked larvae were dissected, stained, and analyzed as described above. Using projection images, internuclear distance was assessed in muscles VL1, VL2, and VL4 using ImageJ (NIH) to measure the distance between each nucleus and the nearest neighboring nucleus. The greatest span of muscle devoid of nuclei was measured in the same manner. All distances were normalized to muscle length (measured from projection images using ImageJ). The number of nuclei per muscle, number of NMJs per muscle, and number of boutons per NMJ were quantified manually using a Leica SP5 laser-scanning confocal microscope with a 20×0.7 NA HCX PL Apochromat oil objective to view the muscles.
Viability assays
Embryos were collected at 25°C on yeasted apple juice agar plates using overnight lays. Stage 15–16 embryos lacking the balancer (where applicable) were staged by development of the gut and hand-selected for analysis. Embryos were counted, transferred to a lightly yeasted apple juice agar plate, and raised at 22°C overnight. The following day, L1 larvae were counted and transferred to vials of standard fly food at 22°C. Eight to twelve days later, the number of pupal cases and adults present in the vials were quantified. All values were normalized to 100%.
Alignments
Protein and nucleotide alignments were performed using Clustal Omega and ClustalW2, respectively (EMBL-EBI).
Statistics
All data sets were analyzed similarly. Error bars represent standard deviations calculated in Excel. Traditional pairwise comparisons using the Student's t-test were used to compare experimental values to control values. Additionally, Analysis of Variance (ANOVA) was used to compare the same data in the context of an entire experiment (three or more values per comparison), taking into account variations in multiple means and standard deviations amongst genotypes within a given experiment. With one-way ANOVA, multiple groups of data were compared simultaneously to determine significance (for example, comparing homozygous mutants to both wild-type and heterozygous controls). Values reaching a threshold of statistical significance in both the Student's t-test and the more stringent ANOVA assessment were considered significant and noted with asterisks in the relevant graphs. All analyses were performed using Prism 6.0.
Supporting Information
Zdroje
1. RomeroNB (2010) Centronuclear myopathies: a widening concept. Neuromuscul Disord 20 : 223–228.
2. JeannetPY, BassezG, EymardB, LaforetP, UrtizbereaJA, et al. (2004) Clinical and histologic findings in autosomal centronuclear myopathy. Neurology 62 : 1484–1490.
3. PuckelwartzMJ, KesslerE, ZhangY, HodzicD, RandlesKN, et al. (2009) Disruption of nesprin-1 produces an Emery Dreifuss muscular dystrophy-like phenotype in mice. Hum Mol Genet 18 : 607–620.
4. MetzgerT, GacheV, XuM, CadotB, FolkerES, et al. (2012) MAP and kinesin-dependent nuclear positioning is required for skeletal muscle function. Nature 484 : 120–124.
5. FolkerES, SchulmanVK, BayliesMK (2012) Muscle length and myonuclear position are independently regulated by distinct Dynein pathways. Development 139 : 3827–3837.
6. Elhanany-TamirH, YuYV, ShnayderM, JainA, WelteM, et al. (2012) Organelle positioning in muscles requires cooperation between two KASH proteins and microtubules. J Cell Biol 198 : 833–846.
7. FolkerES, SchulmanVK, BayliesMK (2014) Translocating myonuclei have distinct leading and lagging edges that require kinesin and dynein. Development 141 : 355–366.
8. GottaM, DongY, PetersonYK, LanierSM, AhringerJ (2003) Asymmetrically distributed C. elegans homologs of AGS3/PINS control spindle position in the early embryo. Curr Biol 13 : 1029–1037.
9. HuangSH, DuanS, SunT, WangJ, ZhaoL, et al. (2011) JIP3 mediates TrkB axonal anterograde transport and enhances BDNF signaling by directly bridging TrkB with kinesin-1. J Neurosci 31 : 10602–10614.
10. SetouM, SeogDH, TanakaY, KanaiY, TakeiY, et al. (2002) Glutamate-receptor-interacting protein GRIP1 directly steers kinesin to dendrites. Nature 417 : 83–87.
11. SheemanB, CarvalhoP, SagotI, GeiserJ, KhoD, et al. (2003) Determinants of S. cerevisiae dynein localization and activation: implications for the mechanism of spindle positioning. Curr Biol 13 : 364–372.
12. AbeN, Almenar-QueraltA, LilloC, ShenZ, LozachJ, et al. (2009) Sunday driver interacts with two distinct classes of axonal organelles. J Biol Chem 284 : 34628–34639.
13. VerheyKJ, MeyerD, DeehanR, BlenisJ, SchnappBJ, et al. (2001) Cargo of kinesin identified as JIP scaffolding proteins and associated signaling molecules. J Cell Biol 152 : 959–970.
14. ArimotoM, KoushikaSP, ChoudharyBC, LiC, MatsumotoK, et al. (2011) The Caenorhabditis elegans JIP3 protein UNC-16 functions as an adaptor to link kinesin-1 with cytoplasmic dynein. J Neurosci 31 : 2216–2224.
15. DrerupCM, NechiporukAV (2013) JNK-interacting protein 3 mediates the retrograde transport of activated c-Jun N-terminal kinase and lysosomes. PLoS Genet 9: e1003303.
16. KamalA, StokinGB, YangZ, XiaCH, GoldsteinLS (2000) Axonal transport of amyloid precursor protein is mediated by direct binding to the kinesin light chain subunit of kinesin-I. Neuron 28 : 449–459.
17. MatsudaS, MatsudaY, D'AdamioL (2003) Amyloid beta protein precursor (AbetaPP), but not AbetaPP-like protein 2, is bridged to the kinesin light chain by the scaffold protein JNK-interacting protein 1. J Biol Chem 278 : 38601–38606.
18. McKenneyRJ, WeilSJ, SchererJ, ValleeRB (2011) Mutually exclusive cytoplasmic dynein regulation by NudE-Lis1 and dynactin. J Biol Chem 286 : 39615–39622.
19. Rosa-FerreiraC, MunroS (2011) Arl8 and SKIP act together to link lysosomes to kinesin-1. Dev Cell 21 : 1171–1178.
20. StockingerW, BrandesC, FaschingD, HermannM, GotthardtM, et al. (2000) The reelin receptor ApoER2 recruits JNK-interacting proteins-1 and -2. J Biol Chem 275 : 25625–25632.
21. MontagnacG, SibaritaJB, LouberyS, DavietL, RomaoM, et al. (2009) ARF6 Interacts with JIP4 to control a motor switch mechanism regulating endosome traffic in cytokinesis. Curr Biol 19 : 184–195.
22. SakamotoR, ByrdDT, BrownHM, HisamotoN, MatsumotoK, et al. (2005) The Caenorhabditis elegans UNC-14 RUN domain protein binds to the kinesin-1 and UNC-16 complex and regulates synaptic vesicle localization. Mol Biol Cell 16 : 483–496.
23. VerheyKJ, LizotteDL, AbramsonT, BarenboimL, SchnappBJ, et al. (1998) Light chain-dependent regulation of Kinesin's interaction with microtubules. J Cell Biol 143 : 1053–1066.
24. HsuCC, MoncaleanoJD, WagnerOI (2011) Sub-cellular distribution of UNC-104(KIF1A) upon binding to adaptors as UNC-16(JIP3), DNC-1(DCTN1/Glued) and SYD-2(Liprin-alpha) in C. elegans neurons. Neuroscience 176 : 39–52.
25. GillSR, SchroerTA, SzilakI, SteuerER, SheetzMP, et al. (1991) Dynactin, a conserved, ubiquitously expressed component of an activator of vesicle motility mediated by cytoplasmic dynein. J Cell Biol 115 : 1639–1650.
26. Waterman-StorerCM, KarkiSB, KuznetsovSA, TabbJS, WeissDG, et al. (1997) The interaction between cytoplasmic dynein and dynactin is required for fast axonal transport. Proc Natl Acad Sci U S A 94 : 12180–12185.
27. TanoueM, YamagaM, IdeJ, TakagiK (1996) Acute stretching of peripheral nerves inhibits retrograde axonal transport. J Hand Surg Br 21 : 358–363.
28. MiaoT, WuD, WheelerA, WangP, ZhangY, et al. (2011) Two cytokine signaling molecules co-operate to promote axonal transport and growth. Exp Neurol 228 : 165–172.
29. NikitinaLS, DorofeevaNA, KirillovaOD, KorotkovAA, GlazovaM, et al. (2014) Role of the ERK signaling pathway in regulating vasopressin secretion in dehydrated rats. Biotech Histochem 89 : 199–208.
30. HoriuchiD, CollinsCA, BhatP, BarkusRV, DiantonioA, et al. (2007) Control of a kinesin-cargo linkage mechanism by JNK pathway kinases. Curr Biol 17 : 1313–1317.
31. CanoE, MahadevanLC (1995) Parallel signal processing among mammalian MAPKs. Trends Biochem Sci 20 : 117–122.
32. GalloKA, JohnsonGL (2002) Mixed-lineage kinase control of JNK and p38 MAPK pathways. Nat Rev Mol Cell Biol 3 : 663–672.
33. HerskowitzI (1995) MAP kinase pathways in yeast: for mating and more. Cell 80 : 187–197.
34. Rios-BarreraLD, Riesgo-EscovarJR (2013) Regulating cell morphogenesis: the Drosophila Jun N-terminal kinase pathway. Genesis 51 : 147–162.
35. RodriguezMC, PetersenM, MundyJ (2010) Mitogen-activated protein kinase signaling in plants. Annu Rev Plant Biol 61 : 621–649.
36. WaskiewiczAJ, CooperJA (1995) Mitogen and stress response pathways: MAP kinase cascades and phosphatase regulation in mammals and yeast. Curr Opin Cell Biol 7 : 798–805.
37. GomezAR, Lopez-VareaA, MolnarC, de la Calle-MustienesE, Ruiz-GomezM, et al. (2005) Conserved cross-interactions in Drosophila and Xenopus between Ras/MAPK signaling and the dual-specificity phosphatase MKP3. Dev Dyn 232 : 695–708.
38. FuMM, HolzbaurEL (2013) JIP1 regulates the directionality of APP axonal transport by coordinating kinesin and dynein motors. J Cell Biol 202 : 495–508.
39. CavalliV, KujalaP, KlumpermanJ, GoldsteinLS (2005) Sunday Driver links axonal transport to damage signaling. J Cell Biol 168 : 775–787.
40. SunF, ZhuC, DixitR, CavalliV (2011) Sunday Driver/JIP3 binds kinesin heavy chain directly and enhances its motility. EMBO J 30 : 3416–3429.
41. SunT, YuN, ZhaiLK, LiN, ZhangC, et al. (2013) c-Jun NH2-terminal kinase (JNK)-interacting protein-3 (JIP3) regulates neuronal axon elongation in a kinesin - and JNK-dependent manner. J Biol Chem 288 : 14531–14543.
42. YasudaJ, WhitmarshAJ, CavanaghJ, SharmaM, DavisRJ (1999) The JIP group of mitogen-activated protein kinase scaffold proteins. Mol Cell Biol 19 : 7245–7254.
43. KelkarN, StandenCL, DavisRJ (2005) Role of the JIP4 scaffold protein in the regulation of mitogen-activated protein kinase signaling pathways. Mol Cell Biol 25 : 2733–2743.
44. IsabetT, MontagnacG, RegazzoniK, RaynalB, El KhadaliF, et al. (2009) The structural basis of Arf effector specificity: the crystal structure of ARF6 in a complex with JIP4. EMBO J 28 : 2835–2845.
45. BowmanAB, KamalA, RitchingsBW, PhilpAV, McGrailM, et al. (2000) Kinesin-dependent axonal transport is mediated by the sunday driver (SYD) protein. Cell 103 : 583–594.
46. BrandAH, PerrimonN (1993) Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118 : 401–415.
47. KelkarN, GuptaS, DickensM, DavisRJ (2000) Interaction of a mitogen-activated protein kinase signaling module with the neuronal protein JIP3. Mol Cell Biol 20 : 1030–1043.
48. ByrdDT, KawasakiM, WalcoffM, HisamotoN, MatsumotoK, et al. (2001) UNC-16, a JNK-signaling scaffold protein, regulates vesicle transport in C. elegans. Neuron 32 : 787–800.
49. SlussHK, HanZ, BarrettT, GoberdhanDC, WilsonC, et al. (1996) A JNK signal transduction pathway that mediates morphogenesis and an immune response in Drosophila. Genes Dev 10 : 2745–2758.
50. WeberU, ParicioN, MlodzikM (2000) Jun mediates Frizzled-induced R3/R4 cell fate distinction and planar polarity determination in the Drosophila eye. Development 127 : 3619–3629.
51. LouisM, PiccinottiS, VosshallLB (2008) High-resolution measurement of odor-driven behavior in Drosophila larvae. J Vis Exp.
52. HurdDD, SaxtonWM (1996) Kinesin mutations cause motor neuron disease phenotypes by disrupting fast axonal transport in Drosophila. Genetics 144 : 1075–1085.
53. GhoM, McDonaldK, GanetzkyB, SaxtonWM (1992) Effects of kinesin mutations on neuronal functions. Science 258 : 313–316.
54. ItoM, YoshiokaK, AkechiM, YamashitaS, TakamatsuN, et al. (1999) JSAP1, a novel jun N-terminal protein kinase (JNK)-binding protein that functions as a Scaffold factor in the JNK signaling pathway. Mol Cell Biol 19 : 7539–7548.
55. EdwardsSL, YuSC, HooverCM, PhillipsBC, RichmondJE, et al. (2013) An organelle gatekeeper function for Caenorhabditis elegans UNC-16 (JIP3) at the axon initial segment. Genetics 194 : 143–161.
56. LigonLA, TokitoM, FinklesteinJM, GrossmanFE, HolzbaurEL (2004) A direct interaction between cytoplasmic dynein and kinesin I may coordinate motor activity. J Biol Chem 279 : 19201–19208.
57. HammondJW, GriffinK, JihGT, StuckeyJ, VerheyKJ (2008) Co-operative versus independent transport of different cargoes by Kinesin-1. Traffic 9 : 725–741.
58. BlasiusTL, CaiD, JihGT, ToretCP, VerheyKJ (2007) Two binding partners cooperate to activate the molecular motor Kinesin-1. J Cell Biol 176 : 11–17.
59. HaHY, ChoIH, LeeKW, LeeKW, SongJY, et al. (2005) The axon guidance defect of the telencephalic commissures of the JSAP1-deficient brain was partially rescued by the transgenic expression of JIP1. Dev Biol 277 : 184–199.
60. RichardsonBE, BeckettK, NowakSJ, BayliesMK (2007) SCAR/WAVE and Arp2/3 are crucial for cytoskeletal remodeling at the site of myoblast fusion. Development 134 : 4357–4367.
61. BrendzaKM, RoseDJ, GilbertSP, SaxtonWM (1999) Lethal kinesin mutations reveal amino acids important for ATPase activation and structural coupling. J Biol Chem 274 : 31506–31514.
62. GepnerJ, LiM, LudmannS, KortasC, BoylanK, et al. (1996) Cytoplasmic dynein function is essential in Drosophila melanogaster. Genetics 142 : 865–878.
63. GindhartJGJr, DesaiCJ, BeushausenS, ZinnK, GoldsteinLS (1998) Kinesin light chains are essential for axonal transport in Drosophila. J Cell Biol 141 : 443–454.
64. CaggeseC, MoschettiR, RagoneG, BarsantiP, CaizziR (2001) dtctex-1, the Drosophila melanogaster homolog of a putative murine t-complex distorter encoding a dynein light chain, is required for production of functional sperm. Mol Genet Genomics 265 : 436–444.
65. LeiY, WarriorR (2000) The Drosophila Lissencephaly1 (DLis1) gene is required for nuclear migration. Dev Biol 226 : 57–72.
66. LiuZ, XieT, StewardR (1999) Lis1, the Drosophila homolog of a human lissencephaly disease gene, is required for germline cell division and oocyte differentiation. Development 126 : 4477–4488.
67. ParmentierML, WoodsD, GreigS, PhanPG, RadovicA, et al. (2000) Rapsynoid/partner of inscuteable controls asymmetric division of larval neuroblasts in Drosophila. J Neurosci 20: RC84.
68. BayliesMK, BateM (1996) twist: a myogenic switch in Drosophila. Science 272 : 1481–1484.
69. HalfonMS, CarmenaA, GisselbrechtS, SackersonCM, JimenezF, et al. (2000) Ras pathway specificity is determined by the integration of multiple signal-activated and tissue-restricted transcription factors. Cell 103 : 63–74.
70. HalfonMS, GisselbrechtS, LuJ, EstradaB, KeshishianH, et al. (2002) New fluorescent protein reporters for use with the Drosophila Gal4 expression system and for vital detection of balancer chromosomes. Genesis 34 : 135–138.
71. BrentJR, WernerKM, McCabeBD (2009) Drosophila larval NMJ dissection. J Vis Exp.
72. SchulmanVK, FolkerES, BayliesMK (2013) A method for reversible drug delivery to internal tissues of Drosophila embryos. Fly (Austin) 7 : 193–203.
Štítky
Genetika Reprodukční medicína
Článek Large-scale Metabolomic Profiling Identifies Novel Biomarkers for Incident Coronary Heart DiseaseČlánek Notch Signaling Mediates the Age-Associated Decrease in Adhesion of Germline Stem Cells to the NicheČlánek Phosphorylation of Mitochondrial Polyubiquitin by PINK1 Promotes Parkin Mitochondrial TetheringČlánek Natural Variation Is Associated With Genome-Wide Methylation Changes and Temperature SeasonalityČlánek Overlapping and Non-overlapping Functions of Condensins I and II in Neural Stem Cell DivisionsČlánek Unisexual Reproduction Drives Meiotic Recombination and Phenotypic and Karyotypic Plasticity inČlánek Tetraspanin (TSP-17) Protects Dopaminergic Neurons against 6-OHDA-Induced Neurodegeneration inČlánek ABA-Mediated ROS in Mitochondria Regulate Root Meristem Activity by Controlling Expression inČlánek Mutations in Global Regulators Lead to Metabolic Selection during Adaptation to Complex EnvironmentsČlánek The Evolution of Sex Ratio Distorter Suppression Affects a 25 cM Genomic Region in the Butterfly
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2014 Číslo 12
-
Všechny články tohoto čísla
- Stratification by Smoking Status Reveals an Association of Genotype with Body Mass Index in Never Smokers
- Genome Wide Meta-analysis Highlights the Role of Genetic Variation in in the Regulation of Circulating Serum Chemerin
- Occupancy of Mitochondrial Single-Stranded DNA Binding Protein Supports the Strand Displacement Mode of DNA Replication
- Distinct Genealogies for Plasmids and Chromosome
- Large-scale Metabolomic Profiling Identifies Novel Biomarkers for Incident Coronary Heart Disease
- Non-coding RNAs Prevent the Binding of the MSL-complex to Heterochromatic Regions
- Plasmid Flux in ST131 Sublineages, Analyzed by Plasmid Constellation Network (PLACNET), a New Method for Plasmid Reconstruction from Whole Genome Sequences
- Epigenome-Guided Analysis of the Transcriptome of Plaque Macrophages during Atherosclerosis Regression Reveals Activation of the Wnt Signaling Pathway
- The Inventiveness of Nature: An Interview with Werner Arber
- Mediation Analysis Demonstrates That -eQTLs Are Often Explained by -Mediation: A Genome-Wide Analysis among 1,800 South Asians
- Generation of Antigenic Diversity in by Structured Rearrangement of Genes During Mitosis
- A Massively Parallel Pipeline to Clone DNA Variants and Examine Molecular Phenotypes of Human Disease Mutations
- Genetic Analysis of the Cardiac Methylome at Single Nucleotide Resolution in a Model of Human Cardiovascular Disease
- Genetic Analysis of Circadian Responses to Low Frequency Electromagnetic Fields in
- The Dissection of Meiotic Chromosome Movement in Mice Using an Electroporation Technique
- Altered Chromatin Occupancy of Master Regulators Underlies Evolutionary Divergence in the Transcriptional Landscape of Erythroid Differentiation
- Syd/JIP3 and JNK Signaling Are Required for Myonuclear Positioning and Muscle Function
- Notch Signaling Mediates the Age-Associated Decrease in Adhesion of Germline Stem Cells to the Niche
- Mutation of Leads to Blurred Tonotopic Organization of Central Auditory Circuits in Mice
- The IKAROS Interaction with a Complex Including Chromatin Remodeling and Transcription Elongation Activities Is Required for Hematopoiesis
- RAN-Binding Protein 9 is Involved in Alternative Splicing and is Critical for Male Germ Cell Development and Male Fertility
- Enhanced Longevity by Ibuprofen, Conserved in Multiple Species, Occurs in Yeast through Inhibition of Tryptophan Import
- Phosphorylation of Mitochondrial Polyubiquitin by PINK1 Promotes Parkin Mitochondrial Tethering
- Recurrent Loss of Specific Introns during Angiosperm Evolution
- Natural Variation Is Associated With Genome-Wide Methylation Changes and Temperature Seasonality
- SEEDSTICK is a Master Regulator of Development and Metabolism in the Arabidopsis Seed Coat
- Overlapping and Non-overlapping Functions of Condensins I and II in Neural Stem Cell Divisions
- Unisexual Reproduction Drives Meiotic Recombination and Phenotypic and Karyotypic Plasticity in
- Tetraspanin (TSP-17) Protects Dopaminergic Neurons against 6-OHDA-Induced Neurodegeneration in
- ABA-Mediated ROS in Mitochondria Regulate Root Meristem Activity by Controlling Expression in
- Mutations in Global Regulators Lead to Metabolic Selection during Adaptation to Complex Environments
- Global Analysis of Photosynthesis Transcriptional Regulatory Networks
- Mucolipin Co-deficiency Causes Accelerated Endolysosomal Vacuolation of Enterocytes and Failure-to-Thrive from Birth to Weaning
- Controlling Pre-leukemic Thymocyte Self-Renewal
- How Malaria Parasites Avoid Running Out of Ammo
- Echoes of the Past: Hereditarianism and
- Deep Reads: Strands in the History of Molecular Genetics
- Keep on Laying Eggs Mama, RNAi My Reproductive Aging Blues Away
- Analysis of a Plant Complex Resistance Gene Locus Underlying Immune-Related Hybrid Incompatibility and Its Occurrence in Nature
- Epistatic Adaptive Evolution of Human Color Vision
- Increased and Imbalanced dNTP Pools Symmetrically Promote Both Leading and Lagging Strand Replication Infidelity
- Genetic Basis of Haloperidol Resistance in Is Complex and Dose Dependent
- Genome-Wide Analysis of DNA Methylation Dynamics during Early Human Development
- Interaction between Conjugative and Retrotransposable Elements in Horizontal Gene Transfer
- The Evolution of Sex Ratio Distorter Suppression Affects a 25 cM Genomic Region in the Butterfly
- is Required for Adult Maintenance of Dopaminergic Neurons in the Ventral Substantia Nigra
- PRL1, an RNA-Binding Protein, Positively Regulates the Accumulation of miRNAs and siRNAs in Arabidopsis
- Genetic Control of Contagious Asexuality in the Pea Aphid
- Early Mesozoic Coexistence of Amniotes and Hepadnaviridae
- Local and Systemic Regulation of Plant Root System Architecture and Symbiotic Nodulation by a Receptor-Like Kinase
- Gene Pathways That Delay Reproductive Senescence
- The Evolution of Fungal Metabolic Pathways
- Maf1 Is a Novel Target of PTEN and PI3K Signaling That Negatively Regulates Oncogenesis and Lipid Metabolism
- Formation of Linear Amplicons with Inverted Duplications in Requires the MRE11 Nuclease
- Identification of Rare Causal Variants in Sequence-Based Studies: Methods and Applications to , a Gene Involved in Cohen Syndrome and Autism
- Rrp12 and the Exportin Crm1 Participate in Late Assembly Events in the Nucleolus during 40S Ribosomal Subunit Biogenesis
- The Mutations in the ATP-Binding Groove of the Rad3/XPD Helicase Lead to -Cockayne Syndrome-Like Phenotypes
- Topoisomerase I Plays a Critical Role in Suppressing Genome Instability at a Highly Transcribed G-Quadruplex-Forming Sequence
- A Cbx8-Containing Polycomb Complex Facilitates the Transition to Gene Activation during ES Cell Differentiation
- Transcriptional Frameshifting Rescues Type VI Secretion by the Production of Two Length Variants from the Prematurely Interrupted Gene
- Association Mapping across Numerous Traits Reveals Patterns of Functional Variation in Maize
- Genome-Wide Analysis of -Regulated and Phased Small RNAs Underscores the Importance of the ta-siRNA Pathway to Maize Development
- Dissemination of Cephalosporin Resistance Genes between Strains from Farm Animals and Humans by Specific Plasmid Lineages
- The Tau Tubulin Kinases TTBK1/2 Promote Accumulation of Pathological TDP-43
- Germline Signals Deploy NHR-49 to Modulate Fatty-Acid β-Oxidation and Desaturation in Somatic Tissues of
- Microevolution of in Macrophages Restores Filamentation in a Nonfilamentous Mutant
- Vangl2-Regulated Polarisation of Second Heart Field-Derived Cells Is Required for Outflow Tract Lengthening during Cardiac Development
- Chondrocytes Transdifferentiate into Osteoblasts in Endochondral Bone during Development, Postnatal Growth and Fracture Healing in Mice
- A ABC Transporter Regulates Lifespan
- RA and FGF Signalling Are Required in the Zebrafish Otic Vesicle to Pattern and Maintain Ventral Otic Identities
- , and Reprogram Thymocytes into Self-Renewing Cells
- The miR9863 Family Regulates Distinct Alleles in Barley to Attenuate NLR Receptor-Triggered Disease Resistance and Cell-Death Signaling
- Detection of Pleiotropy through a Phenome-Wide Association Study (PheWAS) of Epidemiologic Data as Part of the Environmental Architecture for Genes Linked to Environment (EAGLE) Study
- Extensive Copy-Number Variation of Young Genes across Stickleback Populations
- The and Genetic Modules Interact to Regulate Ciliogenesis and Ciliary Microtubule Patterning in
- Analysis of the Genome, Transcriptome and Secretome Provides Insight into Its Pioneer Colonization Strategies of Wood
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Tetraspanin (TSP-17) Protects Dopaminergic Neurons against 6-OHDA-Induced Neurodegeneration in
- Maf1 Is a Novel Target of PTEN and PI3K Signaling That Negatively Regulates Oncogenesis and Lipid Metabolism
- The IKAROS Interaction with a Complex Including Chromatin Remodeling and Transcription Elongation Activities Is Required for Hematopoiesis
- Echoes of the Past: Hereditarianism and
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



