-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaAnalysis of the Genome, Transcriptome and Secretome Provides Insight into Its Pioneer Colonization Strategies of Wood
The wood decay fungus Phlebiopsis gigantea degrades all components of plant cell walls and is uniquely able to rapidly colonize freshly exposed conifer sapwood. However, mechanisms underlying its conversion of lignocellulose and resinous extractives have not been explored. We report here analyses of the genetic repertoire, transcriptome and secretome of P. gigantea. Numerous highly expressed hydrolases, together with lytic polysaccharide monooxygenases were implicated in P. gigantea's attack on cellulose, and an array of ligninolytic peroxidases and auxiliary enzymes were also identified. Comparisons of woody substrates with and without extractives revealed differentially expressed genes predicted to be involved in the transformation of resin. These expression patterns are likely key to the pioneer colonization of conifers by P. gigantea.
Published in the journal: . PLoS Genet 10(12): e32767. doi:10.1371/journal.pgen.1004759
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1004759Summary
The wood decay fungus Phlebiopsis gigantea degrades all components of plant cell walls and is uniquely able to rapidly colonize freshly exposed conifer sapwood. However, mechanisms underlying its conversion of lignocellulose and resinous extractives have not been explored. We report here analyses of the genetic repertoire, transcriptome and secretome of P. gigantea. Numerous highly expressed hydrolases, together with lytic polysaccharide monooxygenases were implicated in P. gigantea's attack on cellulose, and an array of ligninolytic peroxidases and auxiliary enzymes were also identified. Comparisons of woody substrates with and without extractives revealed differentially expressed genes predicted to be involved in the transformation of resin. These expression patterns are likely key to the pioneer colonization of conifers by P. gigantea.
Introduction
The most abundant source of terrestrial carbon is plant biomass, composed primarily of cellulose, hemicellulose, and lignin. Numerous microbes utilize cellulose and hemicellulose, but a much smaller group of filamentous fungi has the capacity to degrade lignin, the most recalcitrant component of plant cell walls. Uniquely, such ‘white-rot’ fungi efficiently depolymerize lignin to access cell wall carbohydrates for carbon and energy sources. As such, white-rot fungi play a key role in the carbon cycle.
White-rot basidiomycetes may differ in their substrate preference and morphological patterns of decay (for review see [1], [2]). The majority of lignin-degrading fungi, including Phanerochaete chrysosporium and Ceriporiopsis subvermispora, are unable to colonize freshly cut wood unless inhibitory compounds (extractives) are removed or transformed [2]–[5]. A few basidiomycetes, including Phlebiopsis gigantea, are pioneer colonizers of softwood because they tolerate and utilize resinous extractives (e.g., resin acids, triglycerides, long chain fatty acids, see Figure 1) which cause pitch deposits in paper pulp manufacturing [6]. It is this unusual capability that also led to the development of P. gigantea as a biocontrol agent against subsequent colonization of cut stumps by the root rot pathogen Heterobasidium annosum sensu lato (now considered several species) [7], [8] and of harvested wood by blue stain fungi [9], [10]. It seems likely that when applied to freshly cut wood, P. gigantea is able to rapidly metabolize accessible extractives and hemicellulose. As the hyphae continue to invade tracheids and ray parenchyma cells, the more recalcitrant cell wall polymers (cellulose, lignin; Figure 1) are eroded. Little is known of how some white-rot fungi degrade conifer extractives [11], [12] or interact with other fungi such as H. annosum [13].
Fig. 1. Schematic representations of lignocellulose components in cell walls of pine wood. 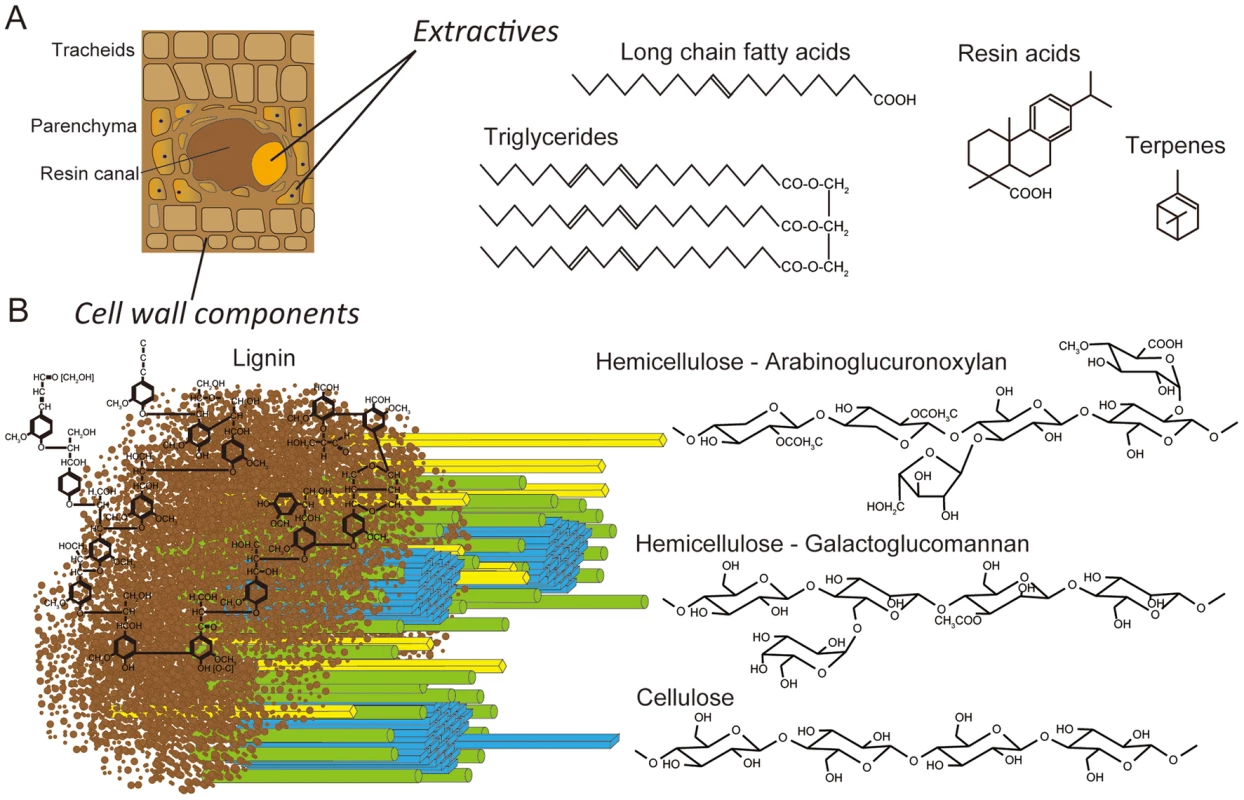
Panel A: The extractives (long chain fatty acids, triglycerides, resin acids and terpenes) are found primarily in the resin ducts, but damage to pine wood causes the release of these compounds across wounded areas. Panel B: In tracheid cell walls, the amorphous, phenylpropanoid polymer lignin (brown) form a matrix around the more structured carbohydrate polymers, hemicellulose (yellow and green) and cellulose (blue). White-rot fungi degrade major cell wall polymers through concerted action of hydrolytic and oxidative enzymes (reviewed in [14], [15]). Cellulose is attacked by a combination of exo-cellobiohydrolases and endoglucanases assigned to glycoside hydrolase families GH5, GH6, GH7 and possibly GH9, GH12, GH44 and GH45 [16], [17]. In addition to these hydrolases, recent evidence strongly supports the involvement of lytic polysaccharide monooxygenases (LPMOs) in cellulose degradation [18]–[20]. Lignin degradation is catalyzed by an array of oxidative enzymes, especially lignin peroxidase (LiP), manganese peroxidase (MnP) and versatile peroxidase (VP) belonging to class II of the plant-fungal-prokaryotic peroxidase superfamily. Recent genome investigations reveal that all efficient lignin degraders possess some combination of these class II ligninolytic peroxidases [21], [22]. In P. gigantea, four MnP sequences were previously identified [23].
In addition to peroxidases, laccases have been implicated in lignin degradation [24]–[26]. To date, multiple laccase isozymes and/or the corresponding genes have been characterized from most white-rot fungi except P. chrysosporium, an efficient lignocellulose degrader that lacks such enzymes [27]–[29]. The mechanism(s) by which laccases might degrade lignin remain unclear as the enzyme lacks sufficient oxidation potential to cleave non-phenolic linkages within the polymer. Interestingly, laccase activity has not been reported in P. gigantea.
Additional ‘auxiliary activities’ [30] commonly ascribed to ligninolytic systems include extracellular enzymes capable of generating H2O2. These enzymes may be physiologically coupled to peroxidases. Among them, aryl-alcohol oxidase (AAO), methanol oxidase (MOX), pyranose 2-oxidase (P2O), and copper radical oxidases (such as glyoxal oxidase, GLX) have been extensively studied. With the exception of P2O [31], none of these activities have been reported in P. gigantea cultures. In short, the repertoire of extracellular enzymes produced by P. gigantea is largely unknown, and its mechanism(s) for cell wall degradation remain unexplored.
Beyond extracellular systems, the complete degradation of lignin requires many intracellular enzymes for the complete mineralization of monomers to CO2 and H2O. Examples of enzymes that have been characterized from P. chrysosporium include cytochromes P450 (CYPs) [32]–[34], glutathione transferases [35], and aryl alcohol dehydrogenase (AAD) [36]. The role of such enzymes in P. gigantea, if any, is unknown.
Herein, we report analysis of the P. gigantea draft genome. Gene annotation, transcriptome analyses and secretome profiles identified numerous genes involved in lignocellulose degradation and in the metabolism of conifer extractives.
Results
Genome assembly and annotation
Following an assessment of wood decay properties (Figure 2), P. gigantea single basidiospore strain 5–6 was selected for sequencing using Illumina reads assembled with AllPathsLG. Genome size was estimated to be approximately 30 Mbp (Text S1), somewhat lower than closely related members of the ‘Phlebia clade’ [23], [37] such as C. subvermispora (39 Mbp) and P. chrysosporium (35 Mbp) [22], [27]. Aided by 17,915 mapped EST clusters, the JGI annotation pipeline predicted 11,891 genes. Proteins were assigned to 6412, 5615, 6932 and 2253 KOG categories, GO terms, pfam domains and EC numbers, respectively. Significant synteny with P. chrysosporium was observed (Figure S1). Detailed information on the assembly and annotations is available via the JGI portal MycoCosm [38].
Fig. 2. Wood decay characteristics. 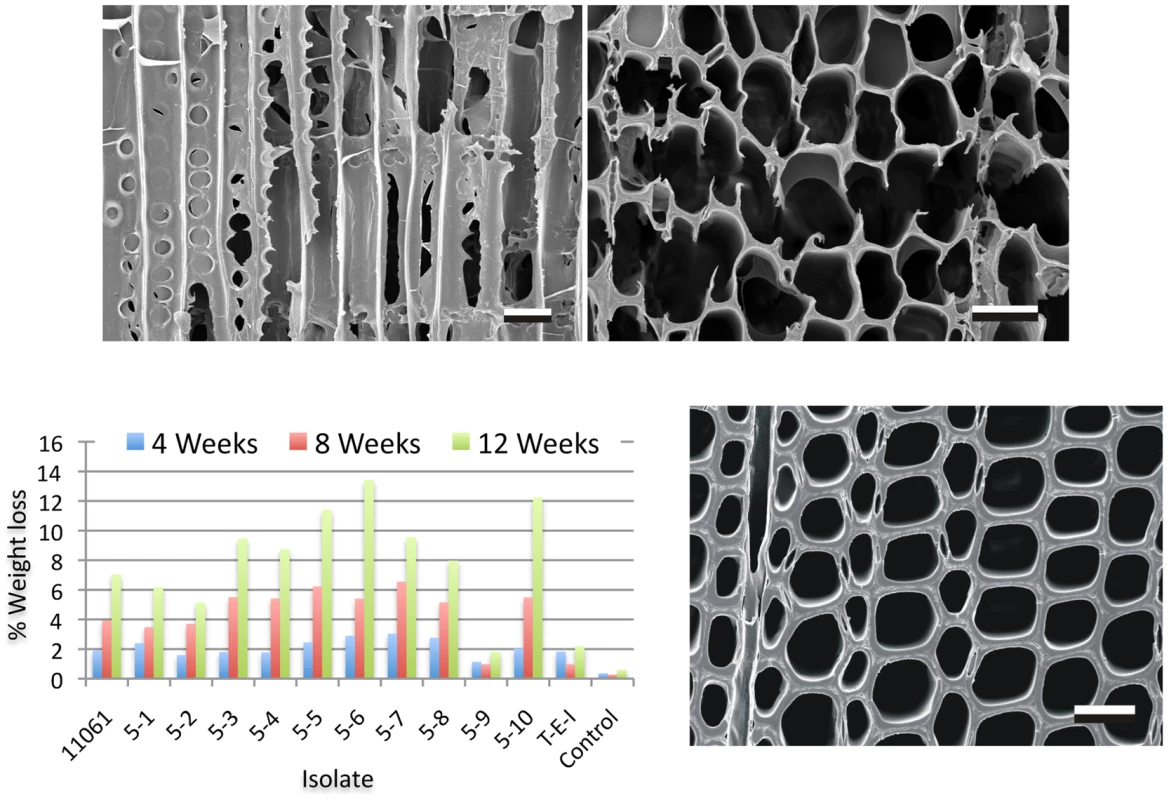
Comparative weight loss of parental strain 11061 and single basidiospore derivatives on colonized loblolly pine wood (Pinus taeda) wood wafers were determined after 4, 8 and 12 weeks incubation (bottom left panel) as described in Methods. Single basidiospore strain 5–6 also aggressively decayed birch and spruce (Text S1) and was selected for sequencing. Upper panels show scanning electron microscopy [68] of radial (left) and transverse (right) sections of pine wood tracheids that were substantially eroded or completely degraded by P. gigantea strain 5–6 by week twelve. Transverse section of sound wood (bottom photo) provides comparison. (Bar = 40 µm). Gene families
Principal component analysis (PCA), based on 73 and 12 families of carbohydrate active enzymes (CAZys, [16]) and auxiliary activities (AAs), [30]), respectively, clustered P. gigantea with other efficient lignin degraders ([39], Figures 3A and S2). Gene numbers were extracted from 21 fungal genomes and excluded genes encoding putative GMC oxidases such as methanol oxidase, alcohol oxidase and glucose oxidase (Dataset S1). Highest contribution of PC1 (50% of variance separating white-rot and brown-rot fungi) and PC2 (13.0% of variance)) values were those genes associated with degradation of plant cell wall polysaccharides and lignin, respectively (Text S1). Hierarchical clustering analysis with this dataset also categorized P. gigantea into a clade of white-rot fungi that included the polypore P. chrysosporium. The precise number and distribution of P. gigantea genes likely involved in lignocellulose degradation were similar, but not identical, to other polypores such as P. chrysosporium and C. subvermispora (Figure 4). Like P. chrysosporium and Phanerochaete flavido-alba, P. gigantea had no laccase sensu stricto genes. Interestingly, while both P. gigantea and the white-rot Russulales H. annosum are adapted to colonization of conifers, significant numbers of laccase sensu stricto genes were only observed in H. annosum (Figure 4). This important conifer pathogen also lacked GLX, LiP and representatives of GH5 subfamiles 15 and 31.
Fig. 3. Comparative analysis of gene repertoires associated with degradation of plant cell wall polymers and extractives in 21 fungal genomes. 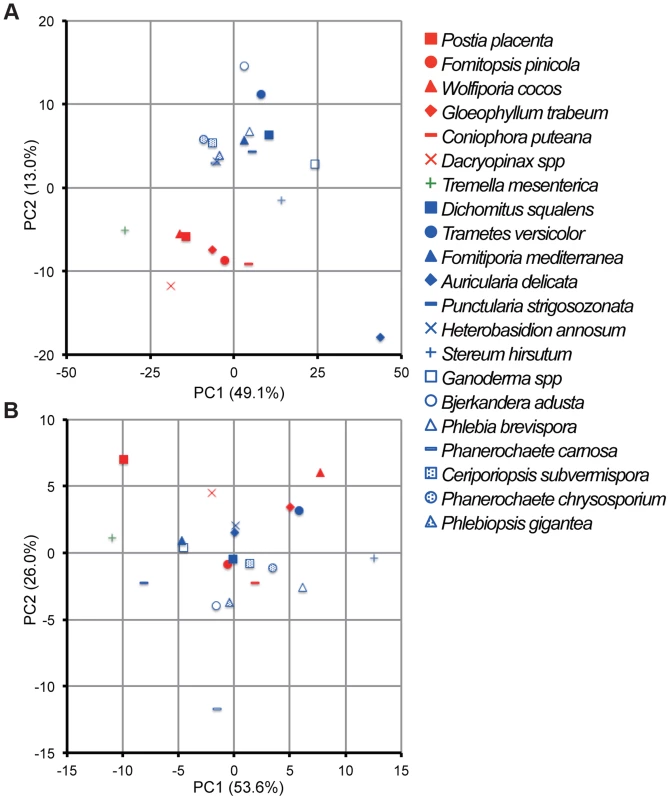
(A) Principal component analysis (PCA) of 21 fungi using 73 CAZy and 12 AA families (Dataset S1). GMC oxidoreductases methanol oxidase, glucose oxidase and aryl alcohol oxidase were excluded because confident functional assignments could not be made and/or their inclusion did not contribute to separation of white- and brown-rot species. (B) PCA of 21 fungi using genes encoding 14 enzymes involved in lipid metabolism (KEGG reference pathway 00071, Dataset S1). There is no significant segregation of white-rot and brown-rot fungi although P. gigantea was positioned in the third quadrant with B. adusta and P. carnosa. Symbols for white rot and brown rot fungi appear in blue and red, respectively. Tremella mesenterica is a mycoparasite. For raw data and contributions of the top 20 families see Dataset S1, Text S1 and Figures S2 and S3. Fig. 4. Number of genes identified in white rot fungi P. gigantea (Phlgi), P. chrysosporium (Phach)[27], C. subvermispora (Cersu)[22], and H. annosum (Hetan)[75], and the brown rot fungus P. placenta (Pospl)[45]. ![Number of genes identified in white rot fungi <i>P. gigantea</i> (Phlgi), <i>P. chrysosporium</i> (Phach)<em class="ref">[27]</em>, <i>C. subvermispora</i> (Cersu)<em class="ref">[22]</em>, and <i>H. annosum</i> (Hetan)<em class="ref">[75]</em>, and the brown rot fungus <i>P. placenta</i> (Pospl)<em class="ref">[45]</em>.](https://www.prolekarniky.cz/media/cache/resolve/media_object_image_small/media/image/c9a5b11469f551b2523f1e90354dbde5.png)
CROs were distinguished as previously described [76]. Lytic polysaccharide monooxygenases were formerly classified as GH61 within the CAZy system (http://www.cazy.org/; [16]). Glycoside hydrolase family GH5 was subdivided as described [77] (Figure S22). With regard to hemicellulose degradation, the genomes of conifer-adapted P. gigantea and H. annosum revealed increased numbers of genes involved in pectin degradation such as GH28 polygalacturonase, CE8 pectin methylesterase and CE12 rhamnogalacturonan acetylesterase (Figure 4). The major hemicellulose of conifer is galactoglucomannan ([40], Figure 1) but, in the case of mannan degradation, no significant increase in genes encoding GH2 β-mannosidase, GH5_7 endo-mannanase and GH27 α-galactosidase was observed relative to other wood decay fungi (Figure 4). Similarly, no significant differences in the number of genes involved in arabinoglucuronoxylan hydrolysis were identified, except for two transcriptionally convergent GH11 genes present in P. gigantea (Text S1). Encoding putative endo-1,4-β-xylanases, wood decay fungi typically harbor one or no GH11 genes. Auricularia delicata is another exception with three of these endoxylanases. Also unusual among white-rot fungi, none of the P. gigantea protein models were assigned to GH95 (Dataset S1). This family includes 1,2-α-fucosidases that hydrolyze the α-Fuc-1,2-Gal linkages in plant xyloglucans.
The P. gigantea genome includes representatives for all the peroxidase families reported in basidiomycetes, including LiP, MnP, heme-thiolate peroxidases, and dye-decolorizing type peroxidases (DyP), with the only exception of VP (Text S1; Figures S8–S13). MnP gene expansion is similar to that found in the C. subvermispora and H. annosum genomes. Beyond class II peroxidases and multicopper oxidases (MCOs), genes encoding auxiliary enzymes involved in ligninolysis were also found such as GMC oxidoreductases (Figures S14–S19; Table S5) and copper radical oxidases (CRO, Figure 4; Table S4). Among the latter group, GLX is coupled to P. chrysosporium LiPs via extracellular H2O2 generation [41]. Consistent with this physiological connection, the P. gigantea genome features both GLX - and LiP-encoding genes. GMC genes encoding putative AAO, MOX and glucose oxidase (GOX) may also be involved in H2O2 production by oxidation of low molecular weight aliphatic and aromatic alcohols. The P2O gene (protein model Phlgi1_130349) lies immediately adjacent to a putative pyranosone dehydratase (Phlgi1_16096) gene. This arrangement is conserved in several wood decay fungi and, in addition to peroxide generation, suggests a route for conversion of glucose to the pyrone antibiotic, cortalcerone [42], [43]. Genes encoding AAD, members of the zinc-type alcohol dehydrogenase superfamily [44], are also abundant in P. gigantea. Relatively few genes were predicted to encode CYPs which are generally considered important in the intracellular metabolism of lignin derivatives and related aromatic compounds (Figure S19; Dataset S2).
The repertoire of P. gigantea genes contrasts sharply with that of brown-rot polypores, such as Postia placenta [45], which lack ligninolytic class II peroxidases, cellobiohydrolases (GH6, GH7), and endoglucanases fused to cellulose binding modules [21], [46] (Figure 4). Unlike P. gigantea and other white-rot fungi, brown-rot fungi often lack genes encoding cellobiose dehydrogenase (CDH) and have relatively few lytic polysaccharide monooxygenase genes (LPMOs). Formerly classified as GH61 ‘hydrolases’, the LPMOs are now known to be copper-dependent monooxygenases [18]–[20] capable of enhancing cellulose attack by CDH and cellobiohydrolase (CBH) [47], [48]. With the exception of Gloeophyllum trabeum, genes encoding GH74 enzymes have not been found in brown-rot fungi. Two such xyloglucanase genes were identified in P. gigantea (Text S1).
In contrast to analysis of genes involved in lignocellulose degradation (Figure 3A), white-rot and brown-rot fungi were not clearly separated by principal component analysis of 14 enzymes involved in lipid metabolism (Figures 3B and S3). However, P. gigantea was grouped near B. adusta and P. carnosa. These associations seem in line with the preferential colonization of softwood substrates by P. carnosa [49] and with the efficient degradation of conifer extractives by B. adusta culture supernatants [50].The highest contribution to PC1 (26.0% variance) and PC2 (6.8% variance) were aldehyde dehydrogenase and long chain fatty acid CoA ligase, respectively (Figures 3A and S3, Text S1). Also potentially involved in intracellular lipid metabolism, CYP52 and CYP505 clans of cytochrome P450s are associated with degradation of fatty acids and alkanes. Relative to other white-rot fungi, P. gigantea had a slightly greater number of CYP52-encoding genes whereas CYP505 gene numbers were similar (Figure 4; Dataset S1; Figures S31, S32; Tables S13–S15).
P. gigantea also diverges from other Agaricomycetes with respect to genes encoding proteins that are more distantly connected to lignocellulose degradation, including hydrophobins (Figures S33 and S34; Tables S17–S19), transporters (Table S20) and non laccase MCOs (Figure S20). Detailed analyses are provided for CAZys (Tables S7–S10; Figures S22–S30; Dataset S1), peroxidases (Figures S8–S13), auxiliary proteins, cytochrome P450s (Figures S31–S32; Table S13–S15), potential regulatory genes (Figures S4–S7; Tables S3, S11–S12) and genes involved in secondary metabolite synthesis (Table S16).
Differential gene expression of P. gigantea in response to substrate
Transcript levels were determined in cultures in which the sole carbon source was glucose (Glc), freshly harvested loblolly pine wood (Pinus taeda; LP) extracted with acetone (ELP), or freshly harvested but not extracted loblolly pine wood (NELP) (Text S1). GC-MS analysis [51] identified the major extract components as resin acids (46%), triglycerides (13%) and fatty acids (11%) (Text S1; Figure S35; Table S21).
Excluding genes with relatively low transcript levels (RPKM values <10) in LP-containing media, transcripts of 187 genes were increased>2-fold (p<0.05) in NELP or ELP relative to Glc. Of those Glc-derived transcripts with RPKM values>10, 146 genes had higher transcripts in Glc relative to NELP or ELP (Figure 5; Dataset S2).
Fig. 5. P. gigantea transcriptome. 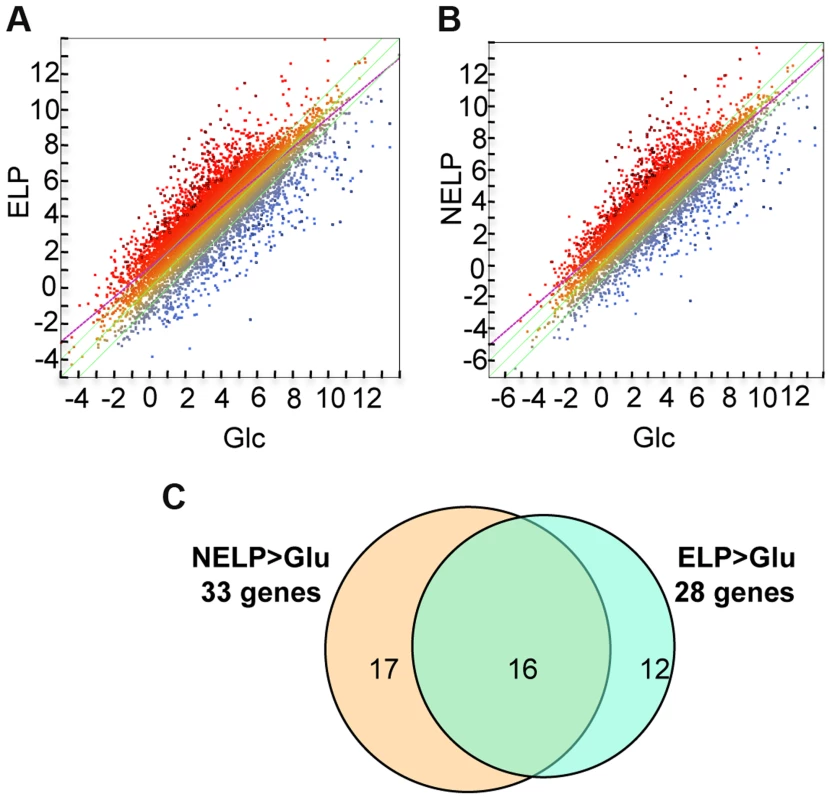
Scatterplots show the distribution of RNA-seq RPKM values (log2) for 11,376 P. gigantea genes when grown on basal salts containing A, acetone-extracted loblolly pine wood (ELP) or B, non-extracted loblolly pine wood (NELP) relative to glucose (Glc). Plot lines define 2-fold borders and best fit regression. Darkened points represent 164 (A) and 145 (B) transcripts accumulating>4-fold at p<0.01. Venn diagram (C) illustrates genes with RPKM signals>10 and upregulated>4-fold in NELP or ELP relative to Glc. Mass spectrometry (nanoLC-MS/MS) identified extracellular peptides corresponding to a total of 319 gene products in NELP and ELP cultures (Dataset S2). Most proteins were observed in both NELP and ELP culture filtrates, which contained 294 and 268 proteins, respectively. Approximate protein abundance, expressed as the exponentially modified protein abundance index (emPAI) [52], varied substantially within samples. As expected, gene products with predicted secretion signals and high transcript levels were often detected. Other detected proteins (e.g. MOX model Phlgi1_120749; [53]) may be loosely associated with cell walls and/or secreted via ‘non-classical’ mechanisms ([54]; www.cbs.dtu.dk/services/SecretomeP). Still other peptides correspond to true intracellular proteins released by cell lysis, e.g. ribosomal proteins (Dataset S2).
Glycoside hydrolase gene expression was heavily influenced by media composition with transcripts corresponding to 76 genes increasing>2-fold in NELP - or ELP-containing media relative to glucose medium (Figure 6). Some of these genes were highly expressed with RPKM values well over 100. For example, transcript and peptide levels matching GH7 cellobiohydrolase (CBH1; model Phlgi1_34136) were among the ten most highly expressed genes (Table 1). Indicative of a complete cellulolytic system, this CBH1 was accompanied by upregulated transcripts and extracellular proteins corresponding to another CBH1 (Phlgi1_13298), a GH6 family member CBH2 (Phlgi1_17701) and GH5_5 β-1,4 endoglucanases (EGs; Phlgi1_86144, Phlgi1_84111), all of which feature a family 1 carbohydrate binding module (CBM1). Also highly expressed were putative β-glucosidases (Phlgi1_127564, Phlgi1_18210) and a GH12 (Phlgi1_34479). Other glycoside hydrolases likely involved in degradation of cell wall hemicelluloses include GH5_7 endomannanases (Phlgi1_97727, Phlgi1_110296), a GH74 xyloglucanase (Phlgi1_98770), a GH27 α-galactosidase (Phlgi1_72848) and a GH10 endoxylanase (Phlgi1_85016).
Fig. 6. Number and expression of genes likely involved in lignocellulose degradation. 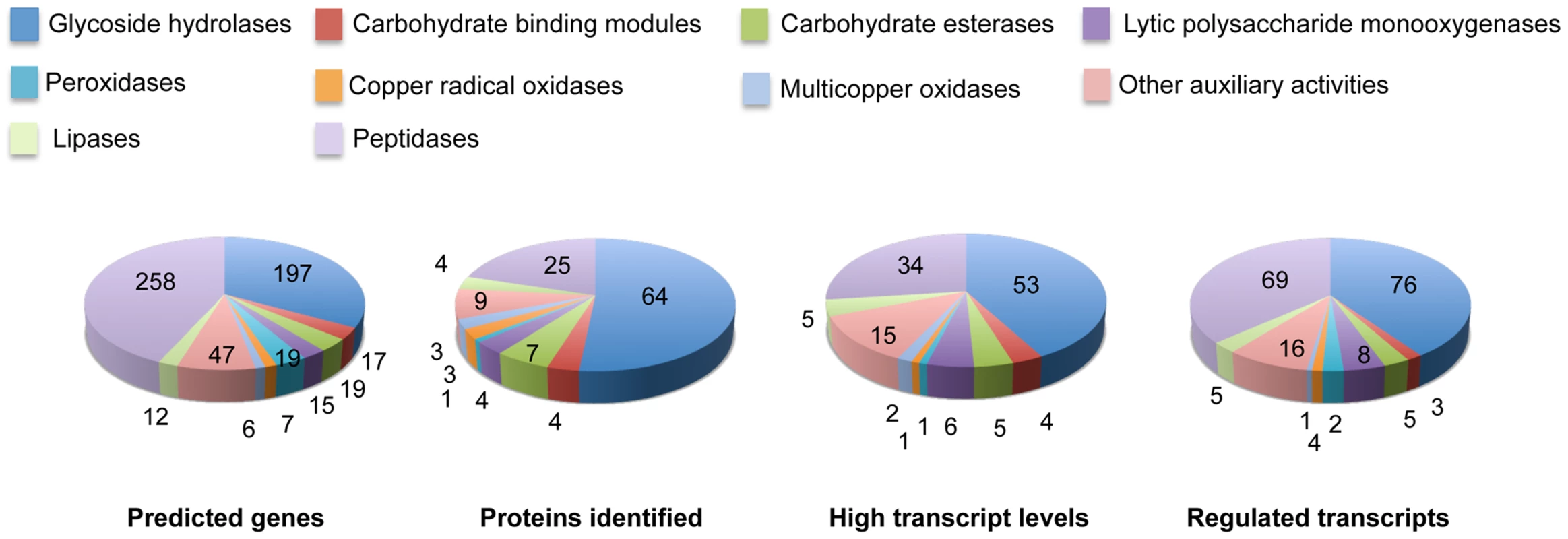
The number of genes encoding mass spectrometry-identified proteins was limited to those matching≥2 unique peptides after 5–9 days growth in media containing NELP or ELP. RPKM values>100 for RNA derived from these cultures were arbitrarily selected as the threshold for high transcript levels. Genes designated as ‘regulated’ showed significant accumulation (p<0.05;>2-fold) in NELP or ELP relative to glucose containing media. Methods and complete data are presented in Text S1 and Dataset S2. Tab. 1. Differentially regulated genes in media containing non-extracted loblolly pine wood (NELP), solvent extracted loblolly pine wood (ELP), or glucose (Glc) as sole carbon source. 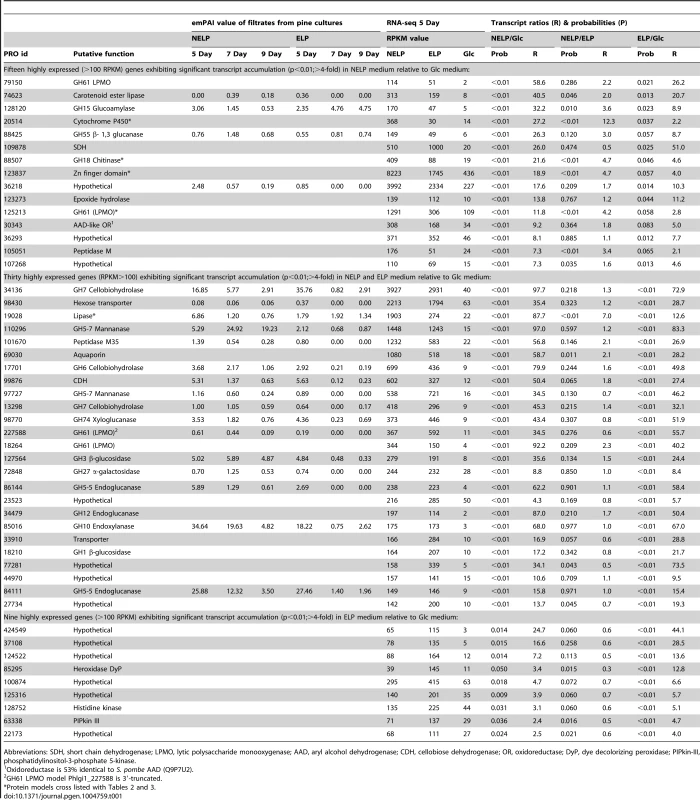
Abbreviations: SDH, short chain dehydrogenase; LPMO, lytic polysaccharide monooxygenase; AAD, aryl alcohol dehydrogenase; CDH, cellobiose dehydrogenase; OR, oxidoreductase; DyP, dye decolorizing peroxidase; PIPkin-III, phosphatidylinositol-3-phosphate 5-kinase. Expression of oxidative enzymes implicated in lignocellulose degradation was also influenced by growth on LP-media (NELP or ELP) relative to Glc-containing media. Transcripts corresponding to five LPMO-encoding genes showed significant regulation (P<0.01) in LP-medium, and three LPMO proteins were detected (Phlgi1_227588, Phlgi1_227560, Phlgi1_37310). An AAD-like oxidoreductase (Phlgi1_30343), possibly involved in the transformation of lignin metabolites, was also upregulated. However, we did not observe high expression of class II peroxidases under the conditions tested (Dataset S2). On the other hand, a DyP (Phlgi1_85295) was significantly upregulated in certain LP-containing media (Table 1). The importance of these peroxidases is further supported by the high protein levels of another DyP, Phlgi1_122124. Specifically, the latter protein showed emPAI values>17 after 5 days growth on LP media and, relative to Glc medium, its transcript ratios were>5-fold higher (p<0.04) (Dataset S2). High DyP gene expression has been observed in white-rot fungi Trametes versicolor and Dichomitus squalens [21], but no genes for these proteins are present in P. chrysosporium and C. subvermispora (Figure 4). The P. gigantea DyP (Phlgi1_122124) was also abundant in media containing microcrystalline cellulose (Avicel) as the sole carbon source (Dataset S2).
To identify enzymes involved in tolerance to and/or degradation of extractives, comparisons were made of gene expression in ground loblolly pine wood that had been extensively extracted with acetone (ELP) versus non-extracted loblolly pine wood (NELP) (Figure 7A). In general, this treatment had little impact on gene expression. For example, glycoside hydrolase transcript and protein patterns showed only minor differences (Figure 8). Nevertheless, transcripts corresponding to 22 genes showed significantly increased levels (>4-fold; p<0.01) in NELP relative to ELP (Figure 7B; Table 2). Of particular interest were genes potentially involved in metabolism of resin acids (e.g. CYPs; [55]), in altering the accessibility of cell wall components (e.g., an endoxylanase), and in regulating gene expression (e.g. proteins containing putative Zn finger domains or HMG-Box transcription factors). Integration of transcript profiles of genes involved in intracellular lipid and oxalate metabolism, together with TCA and glyoxylate cycles, strongly supports a central role for β-oxidation in triglyceride and terpenoid transformation by P. gigantea (Figure 9).
Fig. 7. P. gigantea transcriptome. 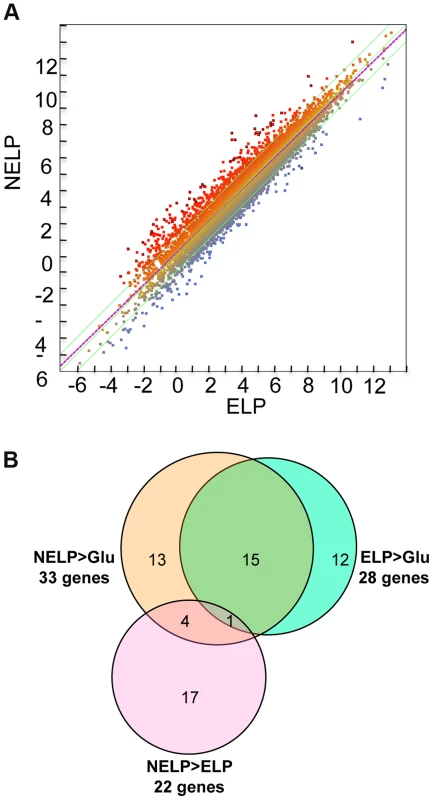
Scatterplot (A) shows the distribution of RNA-seq RPKM values (log2) for 11,376 P. gigantea genes when grown on basal salts containing acetone-extracted loblolly pine wood (ELP) or non-extracted loblolly pine wood (NELP). Lines define 2-fold borders and best fit regression. Darkened points represent 44 transcripts accumulating>4-fold at p<0.01. Venn diagram (B) illustrates genes with RPKM signals>10 and upregulated>4-fold in NELP relative to ELP. Twenty-two genes showed significant transcript accumulation in NELP relative to ELP suggesting potential response to resin and pitch content. Under these stringent thresholds (p<0.01;>4-fold), only one gene, a MCO model Phlgi1_129839, showed significant transcript accumulation in ELP relative to NELP. Additional detail appears in Tables 1-3. Detailed methods and complete data are presented in Text S1 and Dataset S2. Fig. 8. Glycoside hydrolase encoding genes show similar patterns of expression in media containing freshly ground and non-extracted loblolly pine wood (NELP) relative to the same substrate but extracted with acetone (ELP) to remove pitch and resins. 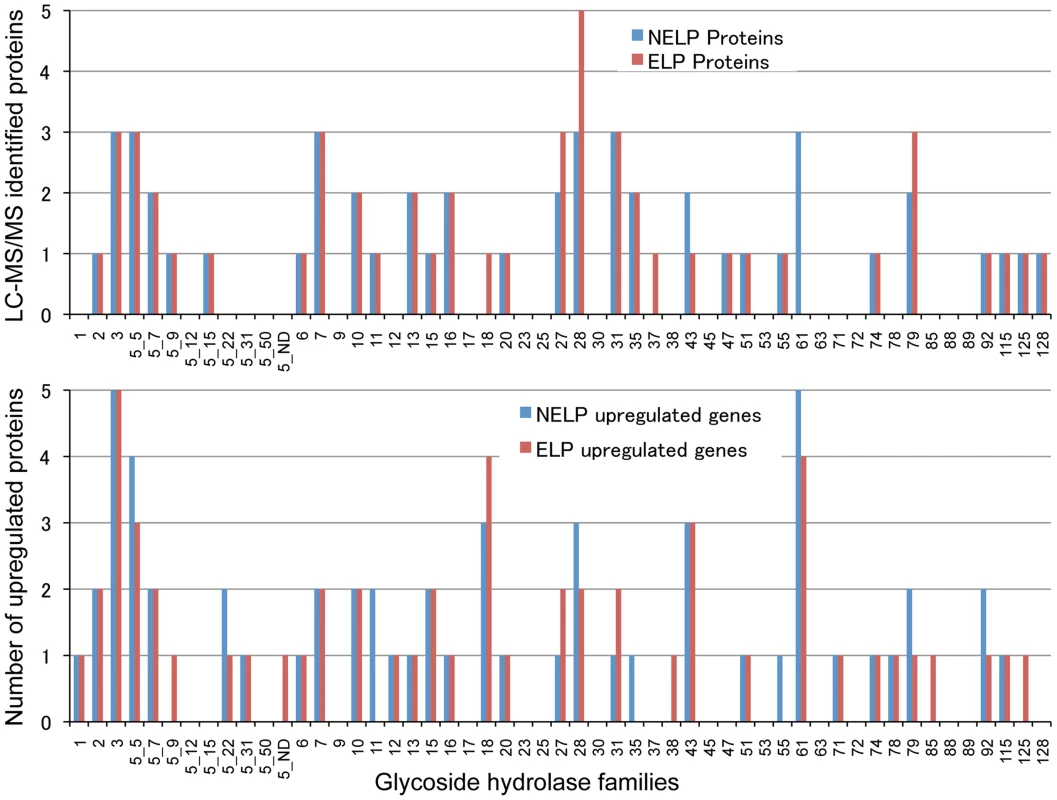
Proteins (upper panel) and transcripts (lower panel) were identified by LC-MS/MS and RNA-seq, respectively. Protein identification was limited to those with>2 unique peptides after five days incubation. Transcript upregulation was limited to significant accumulation (p<0.05;>2-fold) on NELP or ELP relative to glucose-containing medium. Secretome and transcriptome experimental details and complete data are presented in Text S1 and Dataset S2. Fig. 9. Glyoxalate shunt and proposed relationship to lipid oxidation when P. gigantea is cultivated on wood-containing media (ELP or NELP) relative to Glc medium. 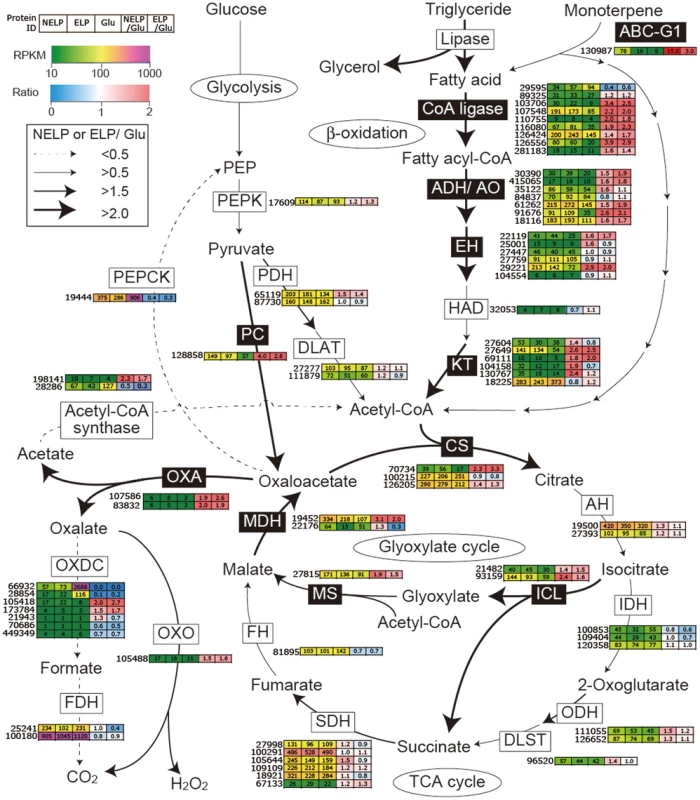
Enzymes encoded by upregulated genes are black highlighted and associated with thickened arrows. Abbreviations: ABC-G1, ABC transporter associated with monoterpene tolerance; ADH/AO, Acyl-CoA dehydrogenase/oxidase; AH, Aconitate hydratase; CoA ligase, long fatty acid-CoA ligase; DLAT, Dihydrolipoyllysine-residue acetyltransferase; DLST, Dihydrolipoyllysine-residue succinyltransferase; EH, Enoyl-CoA hydratase; FDH, Formate dehydrogenase; FH, Fumarate hydratase; KT, Ketothiolase (acetyl-CoA C-acyltransferase); HAD, 3-Hydroxyacyl-CoA dehydrogenase; ICL, Isocitrate lyase; IDH, Isocitrate dehydrogenase; MDH, Malate dehydrogenase; MS, Malate synthase; ODH, Oxoglutarate dehydrogenase; OXA, Oxaloacetase; OXDC, Oxalate decarboxylase; OXO, Oxalate oxidase; PC, Pyruvate carboxylase; PDH, Pyruvate dehydrogenase; PEP, Phosphoenolpyruvate; PEPCK, Phosphoenolpyruvate carboxykinase; PEPK, Phosphoenolpyruvate kinase; SDH, succinate dehydrogenase. See Dataset S2 for detailed gene expression data. Tab. 2. Transcripts accumulating>4-fold in non-extracted loblolly pine wood (NELP) relative to extracted loblolly pine wood (ELP).1 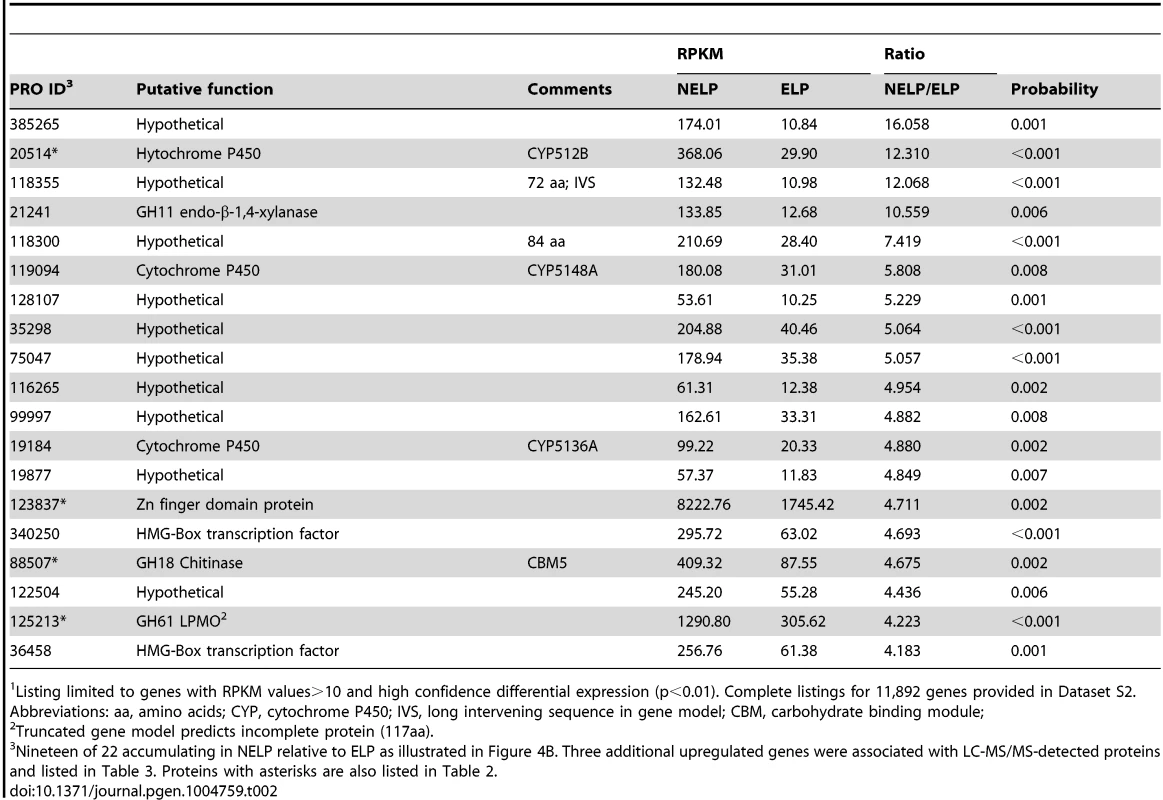
Listing limited to genes with RPKM values>10 and high confidence differential expression (p<0.01). Complete listings for 11,892 genes provided in Dataset S2. Abbreviations: aa, amino acids; CYP, cytochrome P450; IVS, long intervening sequence in gene model; CBM, carbohydrate binding module; Relaxing the transcript fold-change threshold (>2-fold; p<0.01) and focusing on mass spectrometry-identified proteins revealed 14 additional genes potentially involved in metabolism and/or tolerance to loblolly pine wood extractives (Table 3).Among these extract-induced genes, lipases Phlgi1_19028 and Phlgi1_36659 likely hydrolyze the significant levels of triglycerides. The substrate specificity of aldehyde dehydrogenases such as Phlgi1_115040 is difficult to assess based on sequence, although several have been implicated in the degradation of pine wood resins by bacteria [56]. Secretome patterns in media containing microcrystalline cellulose (Avicel) as sole carbon source generally supported the importance of the same proteins in the metabolism of pine wood extractives (Table 3, Dataset S2). Specifically, lipases Phlgi1_19028 and Phlgi1_36659 and aldehyde dehydrogenase Phlg1_115040 were more abundant in loblolly pine wood and in Avicel media relative to the same media without extractives. The role of peroxiredoxin (Phlgi1_95619) and glutathione S-transferase (Phlgi1_113065) are less clear, but transformations involving H2O2 reduction and glutathione conjugation are possible. A single MCO (Phlgi1_129839) and its corresponding transcripts, were observed to be upregulated in ELP relative to NELP. Although lacking the L2 signature common to laccases [57], the MCO4 protein may have iron oxidase activity provided that an imperfectly aligned Glu residue serves in catalysis (Text S1; Figures S20 and S21; Table S6).
Tab. 3. Genes encoding LC-MS/MS detected proteins and exhibiting>2-fold regulation in comparisons of NELP and ELP cultures.1 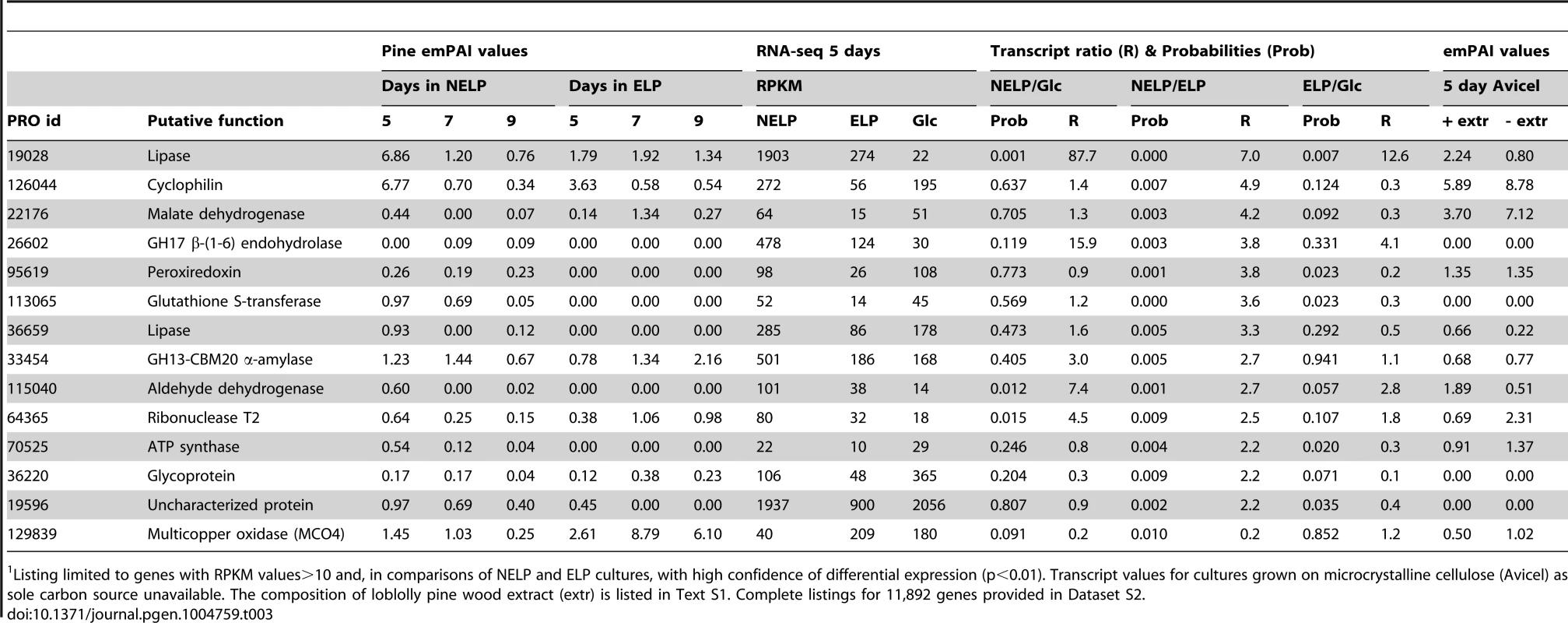
Listing limited to genes with RPKM values>10 and, in comparisons of NELP and ELP cultures, with high confidence of differential expression (p<0.01). Transcript values for cultures grown on microcrystalline cellulose (Avicel) as sole carbon source unavailable. The composition of loblolly pine wood extract (extr) is listed in Text S1. Complete listings for 11,892 genes provided in Dataset S2. Discussion
The distinctive repertoire and regulation of P. gigantea genes underlie a unique and efficient system for degrading all components of conifer sapwood. Transcriptome and proteome analyses demonstrate an active system of hydrolases and LPMOs involved in the complete deconstruction of cellulose and hemicellulose. The overall enzymatic strategy is therefore similar to most cellulolytic microbes, but unlike closely related brown-rot decay Agaricomycetes such as P. placenta.
With regard to ligninolysis, key genes were identified including LiPs, MnPs, CROs and GMC oxidoreductases. As in P. chrysosporium, the presence of both LiP - and GLX-encoding genes is consistent with a close physiological connection involving peroxide generation [41]. We also annotated non-class II peroxidases HTPs and DyPs some of which have been implicated in metabolism of lignin derivatives [58], [59]. The distribution and expression of DyP-encoding genes are notable; with no genes present in P. chrysosporium and C. subvermispora but several highly expressed genes in T. versicolor, D. squalens [21] and P. gigantea (Table 2). Physiological roles of DyP are likely diverse, but oxidation of lignin-related aromatic compounds has been demonstrated [59].
In addition to lignin, oxidative mechanisms likely play a central role in P. gigantea cellulose attack. Of 15 LPMO-encoding genes, transcripts of six genes were regulated (>2-fold; p<0.01) and peptides corresponding to three were unambiguously identified in NELP - or ELP-containing media. Our inability to detect any LPMO proteins in Avicel media (Dataset S2) suggests induction by substrates other than cellulose [60]. Beyond this, the CDH gene was highly expressed (transcripts and protein) in LP media. The observed coordinate expression of CDH and LPMO may reflect oxidative ‘boosting’ as recently demonstrated [19], [20], [47], [61]. However, we did not detect elevated transcripts or peptides corresponding to the two P. gigantea aldose 1-epimerase genes even though these were previously observed in culture filtrates of various white-rot fungi [21], [62], including Bjerkandera adusta, Ganoderma sp, and Phlebia brevispora [17]. Thus, it seems unlikely that enzymatic conversion of oligosaccharides to their β-anomers is necessary for efficient CDH catalysis.
Softwood hemicellulose composition typically includes 15-20% galactoglucomannan while hardwoods contain 15–30% glucuronoxylan [40]. Consistent with an adaption to conifer hemicellulose, GH5_7 β-mannanases were highly expressed in both NELP and ELP cultures, together with a GH27 α-galactosidase (Table 1). GH11 endoxylanase and CE carbohydrate esterase peptides were also detected in the pine wood-containing media, but not in Avicel cultures (Dataset S2). In aggregate, these results demonstrate P. gigantea adaptation to conifer hemicellulose degradation.
P. gigantea's gene expression patterns reveal multiple strategies for overcoming the challenging composition of resinous sapwood. Tolerance to monoterpenes may be mediated in part by a putative ABC efflux transporter (Phlbi1_130987, Figure 9). Of the 51 ABC proteins of P. gigantea, this protein is most closely related to the GcABC-G1 gene of the ascomycete Grosmannia clavigera, a pathogen of Pinus contorta [63]. The GcABC-G1 gene is upregulated in response to various terpenes and appears to be a key element against the host defenses. Consistent with a similar function, our analysis showed the P. gigantea homolog to be upregulated>4.9-fold (p = 0.02) in NELP relative to ELP media (Dataset S2). Other transcripts accumulating in NELP-derived mycelia included three CYPs (Table 2) potentially involved in the hydroxylation of diterpenoids and related resin acids [55]. Differential regulation also implicates glutathione S-transferase, aldehyde dehydrogenase and peroxiredoxin in the transformation and detoxification of extractives (Table 2). Three putative transcription regulators were similarly regulated (Table 3). Aldehyde dehydrogenase - and AAD-encoding genes, some of which are upregulated in P. gigantea LP cultures relative to Glc cultures (Tables 1), are induced by aromatic compounds in P. chrysosporium [64], [65].
Predicted to degrade triglycerides, a total of nine lipase-encoding genes were identified in the P. gigantea genome and four of these were upregulated>2-fold (p<0.01) in LP media compared to Glc medium (Dataset S2). Two lipases displayed similar patterns of transcript and protein upregulation on NELP relative to ELP (Table 3), and the pine wood extractive also induced accumulation of these lipases in Avicel media (Table 3). Further metabolism of triglycerides is uncertain, although a putative glycerol uptake facilitator (Phlbi1_99331) was highly expressed (RPKM value of 2532) and significantly (p<0.02) upregulated (2.1-fold) in NELP compared to ELP (Dataset S2). Fatty acids activation and β-oxidation can be inferred by the expression of fatty acid CoA ligase (Phlgi1_107548, Phlgi1_126556, Phlgi1_89325), β-ketothiolase (Phlgi1_27649, Phlgi1_130767), and fatty acid desaturase (Phlgi1_100083, Phlgi1_115799). Upregulation of a mitochondrial malate dehydrogenase (Phlgi1_22176, Table 3), together with relatively high transcript levels of other TCA cycle components (citrate synthases Phlgi1_126205, Phlgi1_100215; 2-oxoglutarate dehydrogenase, Phlgi1_126652) may complete fatty acid oxidation. In this connection, we also observed high expression of isocitrate lyase (Phlgi1_21482, Phlgi1_93159) and malate synthase (Phlgi1_27815), which partially explain oxalate accumulation [66] and strongly support an active glyoxylate shunt [45], [67] (Figure 9). Upregulation of glycoside hydrolases, transcription factors, cyclophilins, ATP synthase and ribonuclease may also reflect broad shifts in metabolism or reduced accessibility of the unextracted substrate (Tables 2 and 3).
Beyond genetic regulation, certain constitutively expressed genes are also likely involved in the degradation of all plant cell wall components, including complex resins and triglycerides. For example MOX (Phlgi1_120749) is among the most abundant transcripts in both NELP and ELP (Dataset S2), suggesting an important role in H2O2 production associated with lignin demethylation [53]. Extracellular peroxide generation is key to peroxidase activity, and MOX fulfills this role along with CRO, AAO, and P2O. Along these lines, we also observed high extracellular protein levels of DyP (Phlgi1_122124) under all culture conditions.
Most problematic, many P. gigantea genes and proteins exhibited little or no homology to NCBI NR or Swiss-Prot entries. Some of these ‘hypothetical’ or ‘uncharacterized’ proteins are undoubtedly important, particularly those that are highly expressed, regulated and/or secreted. For example, of 92 genes upregulated (>2-fold; p<0.01) in NELP relative to ELP, 51 were designated as hypothetical (Table 2; Dataset S2). Three of these featured predicted secretion signals and peptides were detected in one case. In the absence of biochemical characterization and/or genetic evidence, assigning function to these genes represents a major undertaking. Nevertheless, high throughput transcript and secretome profiling substantially filtered the number of potential targets from a genome-wide estimate of 4744 ‘hypothetical’ genes to the more manageable numbers reported here. More broadly, the results advance understanding of the early and exclusive colonization of coniferous wood by P. gigantea and also provide a framework for developing effective wood protection strategies, improving biocontrol agents and identifying useful enzymes [6], [9], [10].
Methods
Wood colonization assays
Wood wafers (1 cm by 1 cm by 2 mm) were cut from the sapwood of aspen (Populus tremuloides), pine (P. taeda) and spruce (Picea glauca) and sterilized by autoclaving. Following inoculation by contact with mycelium growing on malt extract agar (15 g malt extract [Difco, Detroit, MI] and 15 g agar per liter of water) in Petri dishes, colonized wafers were harvested 30, 60 and 90 days. Noninoculated wood wafers placed on the same media in Petri dishes served as controls. Wafers were removed 30, 60 or 90 days later, weighed and percent weight loss was determined. Additional wafers were removed at the same time period, immediately frozen to −20°C and prepared for scanning electron microscopy as previously described [68].
Sequencing and annotation
The genome was sequenced using Illumina and annotated using the JGI Annotation Pipeline [69]. Assembly and annotations are available from JGI portal MycoCosm [38] and deposited to DDBJ/EMBL/GenBank under accession AZAG00000000. The version described in this paper is version AZAG01000000. The completeness of the P. gigantea genome was assessed by finding 99.1% of CEGMA proteins conserved across sequenced genomes of eukaryotes [70](Text S1; Tables S1, S2).
RNA-seq
Mycelium was derived from triplicate cultures of 250 ml basal salts containing: i. 1.25 g freshly-harvested, ground (1mm mesh) loblolly pine wood that had been ‘spiked’ with acetone and thoroughly dried (NELP); or ii. the same material following extended acetone extraction in a Soxhlet apparatus and drying (ELP). The composition of the extract (Text S1) was determined by GC-MS [51]. Duplicate cultures of basal salts medium with glucose as sole carbon source served as a reference. After 5 days incubation, total RNA was purified from frozen mycelium as described [22], [71]. Multiplexed libraries were constructed and sequenced on an Illumina HiSeq2000. DNAStar Inc (Madison, WI) modules SeqNGen and Qseq were used for mapping reads and statistical analysis. Transcriptome data was deposited to the NCBI Gene Expression Omnibus (GEO) database and assigned accession GSE53112 (Reviewer access via http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=ilovmswixtajjez&acc=GSE53112). Experimental details are provided in Text S1 and all transcriptome analyses are summarized in Dataset S2.
Secretome analysis
With minor modification, NanoLC-MS/MS analysis identified extracellular proteins in culture filtrates as described [22], [72]. For each of the two woody substrates (e.g NELP and ELP), cultures were harvested after 5, 7 and 9 days. Mass spectrometric protein identifications were accepted if they could be established at greater than 95.0% probability within 0.9% False Discovery Rate and contained at least two identified peptides. Protein probabilities were assigned by the Protein Prophet algorithm [73]. To verify the effects of pine wood extractives in a well-defined substrate, media containing microcrystalline cellulose (Avicel) were also employed [22], [45], [74]. Filtrates from these cultures, with or without addition of loblolly pine wood acetone extract, were collected after 5 days and analyzed. Approximate protein abundance in each of the cultures was expressed as the number of unique peptide and the exponentially modified protein abundance index (emPAI) value [52] (See Text S1 for detailed methods).
Supporting Information
Zdroje
1. BlanchetteR (1991) Delignification by wood-decay fungi. Ann Rev Phytopath 29 : 381–398.
2. Eriksson K-EL, Blanchette RA, Ander P (1990) Microbial and enzymatic degradation of wood and wood components; Timell TE, editor. Berlin: Springer-Verlag.
3. Rayner ADM, Boddy L (1988) Fungal decomposition of wood: its biology and ecology. Chichester: John Wiley and Sons.
4. Kaarik AA (1974) Decomposition of wood. In: Dickinson CH, Pugh GJF, editors.Biology of plant litter composition.New York: Academic Press. pp.129–174.
5. ShigoAL (1967) Succession of organisms in discoloration and decay wood. Int Rev For Res 2 : 237–299.
6. GutierrezA, Del RioJC, MartinezAT (2009) Microbial and enzymatic control of pitch in the pulp and paper industry. Appl Microbiol Biotechnol 82 : 1005–1018.
7. Garbelotto M, Guglielmo F, Mascheretti S, Croucher PJ, Gonthier P (2013) Population genetic analyses provide insights on the introduction pathway and spread patterns of the North American forest pathogen Heterobasidion irregulare in Italy. Mol Ecol 10.1111/mec.12452.
8. RishbethJ (1963) Stump protection against Fomes annosus. Annals of Applied Biology 52 : 63–77.
9. BehrendtCJ, BlanchetteRA (1997) Biological Processing of Pine Logs for Pulp and Paper Production with Phlebiopsis gigantea. Appl Environ Microbiol 63 : 1995–2000.
10. BehrendtCJ, BlanchetteRA (2001) Biological control of blue stain in pulpwood: mechanisms of control used by Phlebiopsis gigantea. Holzforschung 55 : 238–245.
11. FischerK, AkhtarM, BlanchetteRA, BurnesTA, MessnerK, et al. (1994) Reduction in resin content in wood chips during experimental biological pulping processes. Holzforschung 48 : 285–290.
12. Martinez-InigoMJ, ImmerzeelP, GutierrezA, del RioJC, Sierra-AlvarezR (1999) Biodegradability of extractives in sapwood and heartwood from Scots Pine by sapstain and white rot fungi. Holzforschung 53 : 247–252.
13. AdomasA, EklundM, JohanssonM, AsiegbuFO (2006) Identification and analysis of differentially expressed cDNAs during nonself-competitive interaction between Phlebiopsis gigantea and Heterobasidion parviporum. FEMS Microbiol Ecol 57 : 26–39.
14. Kersten P, Cullen D (2013) Recent advances on the genomics of litter - and soil-inhabiting Agaricomycetes. Genomics of Soil - and Plant-Associated Fungi: Springer. pp. 311–332.
15. BaldrianP, ValaskovaV (2008) Degradation of cellulose by basidiomycetous fungi. FEMS Microbiol Rev 32 : 501–521.
16. CantarelBL, CoutinhoPM, RancurelC, BernardT, LombardV, et al. (2009) The Carbohydrate-Active EnZymes database (CAZy): an expert resource for Glycogenomics. Nucleic Acids Res 37: D233–238.
17. HoriC, GaskellJ, IgarashiK, SamejimaM, HibbettD, et al. (2013) Genome-wide analysis of polysaccharide degrading enzymes in eleven white - and brown-rot polyporales provides insight into mechanisms of wood decay. Mycologia 105 : 1412–1427.
18. BeyM, ZhouS, PoidevinL, HenrissatB, CoutinhoPM, et al. (2013) Cello-oligosaccharide oxidation reveals differences between two lytic polysaccharide monooxygenases (family GH61) from Podospora anserina. Appl Environ Microbiol 79 : 488–496.
19. QuinlanRJ, SweeneyMD, Lo LeggioL, OttenH, PoulsenJC, et al. (2011) Insights into the oxidative degradation of cellulose by a copper metalloenzyme that exploits biomass components. Proc Natl Acad Sci U S A 108 : 15079–15084.
20. WesterengB, IshidaT, Vaaje-KolstadG, WuM, EijsinkVG, et al. (2011) The putative endoglucanase PcGH61D from Phanerochaete chrysosporium is a metal-dependent oxidative enzyme that cleaves cellulose. PLoS ONE 6: e27807.
21. FloudasD, BinderM, RileyR, BarryK, BlanchetteRA, et al. (2012) The Paleozoic origin of enzymatic lignin decomposition reconstructed from 31 fungal genomes. Science 336 : 1715–1719.
22. Fernandez-FueyoE, Ruiz-DuenasFJ, FerreiraP, FloudasD, HibbettDS, et al. (2012) Comparative genomics of Ceriporiopsis subvermispora and Phanerochaete chrysosporium provide insight into selective ligninolysis. Proc Natl Acad Sci U S A 109 : 5458–5463.
23. MorgensternI, KlopmanS, HibbettDS (2008) Molecular evolution and diversity of lignin degrading heme peroxidases in the Agaricomycetes. J Mol Evol 66 : 243–257.
24. KawaiS, AsukaiM, OhyaN, OkitaK, ItoT, et al. (1999) Degradation of non-phenolic beta-O-4 susbstructure and of polymeric lignin model compounds by laccase of Coriolus versicolor in the presence of 1-hydroxybenzotriazole. FEMS Microbiol Lett 170 : 51–57.
25. BourbonnaisR, PaiceMG, FreiermuthB, BodieE, BornemanS (1997) Reactivities of various mediators and laccases with kraft pulp and lignin model compounds. Appl Environ Microbiol 63 : 4627–4632.
26. EggertC, TempU, ErikssonK (1997) Laccase is essential for lignin degradation by the white-rot fungus Pycnoporus cinnabarinus. FEBS Lett 407 : 89–92.
27. MartinezD, LarrondoLF, PutnamN, Sollewijn GelpkeMD, HuangK, et al. (2004) Genome sequence of the lignocellulose degrading fungus Phanerochaete chrysosporium strain RP78. Nat Biotechnol 22 : 695–700.
28. LarrondoL, SalasL, MeloF, VicunaR, CullenD (2003) A novel extracellular multicopper oxidase from Phanerochaete chrysosporium with ferroxidase activity. Appl Environ Microbiol 69 : 6257–6263.
29. HoeggerPJ, KilaruS, JamesTY, ThackerJR, KüesU (2006) Phylogenetic comparison and classification of laccase and related multicopper oxidase protein sequences. FEBS J 273 : 2308–2326.
30. LevasseurA, DrulaE, LombardV, CoutinhoPM, HenrissatB (2013) Expansion of the enzymatic repertoire of the CAZy database to integrate auxiliary redox enzymes. Biotechnol Biofuels 6 : 41.
31. BastianS, RekowskiMJ, WitteK, Heckmann-PohlDM, GiffhornF (2005) Engineering of pyranose 2-oxidase from Peniophora gigantea towards improved thermostability and catalytic efficiency. Appl Microbiol Biotechnol 67 : 654–663.
32. YadavJS, SoellnerMB, LoperJC, MishraPK (2003) Tandem cytochrome P450 monooxygenase genes and splice variants in the white rot fungus Phanerochaete chrysosporium: cloning, sequence analysis, and regulation of differential expression. Fungal Genet Biol 38 : 10–21.
33. SyedK, PorolloA, LamYW, GrimmettPE, YadavJS (2013) CYP63A2, a catalytically versatile fungal P450 monooxygenase capable of oxidizing higher-molecular-weight polycyclic aromatic hydrocarbons, alkylphenols, and alkanes. Appl Environ Microbiol 79 : 2692–2702.
34. SyedK, YadavJS (2012) P450 monooxygenases (P450ome) of the model white rot fungus Phanerochaete chrysosporium. Crit Rev Microbiol 38 : 339–363.
35. DowdCA, BuckleyCM, SheehanD (1997) Glutathione S-transferases from the white-rot fungus, Phanerochaete chrysosporium. Biochem J 324 (Pt 1): 243–248.
36. ReiserJ, MuheimA, HardeggerM, FrankG, FiechterA (1994) Aryl-alcohol dehydrogenase from the white-rot fungus Phanerochaete chrysosporium: gene cloning, sequence analysis, expression and purification of recombinant protein. J Biol Chem 269 : 28152–28159.
37. HibbettDS, DonoghueMJ (2001) Analysis of character correlations among wood decay mechanisms, mating systems, and substrate ranges in homobasidiomycetes. Syst Biol 50 : 215–242.
38. GrigorievIV, NordbergH, ShabalovI, AertsA, CantorM, et al. (2012) The genome portal of the Department of Energy Joint Genome Institute. Nucleic Acids Res 40: D26–32.
39. RileyR, SalamovAA, BrownDW, NagyLG, FloudasD, et al. (2014) Extensive sampling of basidiomycete genomes demonstrates inadequacy of the white-rot/brown-rot paradigm for wood decay fungi. Proc Natl Acad Sci U S A 111 : 9923–9928.
40. Sjöström E (1993) Wood Chemistry. Fundamentals and Applications. San Diego: Academic Press. 293 p.
41. KerstenPJ (1990) Glyoxal oxidase of Phanerochaete chrysosporium: Its characterization and activation by lignin peroxidase. Proc Natl Acad Sci USA 87 : 2936–2940.
42. de KokerTH, MozuchMD, CullenD, GaskellJ, KerstenPJ (2004) Pyranose 2-oxidase from Phanerochaete chrysosporium: isolation from solid substrate, protein purification, and characterization of gene structure and regulation. Appl Environ Microbiol 70 : 5794–5800.
43. GiffhornF (2000) Fungal pyranose oxidases: occurrence, properties and biotechnical applications in carbohydrate chemistry. Appl Microbiol Biotechnol 54 : 727–740.
44. SunHW, PlappBV (1992) Progressive sequence alignment and molecular evolution of the Zn-containing alcohol dehydrogenase family. J Mol Evol 34 : 522–535.
45. MartinezD, ChallacombeJ, MorgensternI, HibbettD, SchmollM, et al. (2009) Genome, transcriptome, and secretome analysis of wood decay fungus Postia placenta supports unique mechanisms of lignocellulose conversion. Proc Natl Acad Sci U S A 106 : 1954–1959.
46. EastwoodDC, FloudasD, BinderM, MajcherczykA, SchneiderP, et al. (2011) The plant cell wall-decomposing machinery underlies the functional diversity of forest fungi. Science 333 : 762–765.
47. LangstonJA, ShaghasiT, AbbateE, XuF, VlasenkoE, et al. (2011) Oxidoreductive cellulose depolymerization by the enzymes cellobiose dehydrogenase and glycoside hydrolase 61. Appl Environ Microbiol 77 : 7007–7015.
48. PhillipsCM, BeesonWT, CateJH, MarlettaMA (2011) Cellobiose dehydrogenase and a copper-dependent polysaccharide monooxygenase potentiate cellulose degradation by Neurospora crassa. ACS Chem Biol 6 : 1399–1406.
49. MacdonaldJ, DoeringM, CanamT, GongY, GuttmanDS, et al. (2011) Transcriptomic responses of the softwood-degrading white-rot fungus Phanerochaete carnosa during growth on coniferous and deciduous wood. Appl Environ Microbiol 77 : 3211–3218.
50. DoradoJ, ClaassenFW, van BeekTA, LenonG, WijnbergJB, et al. (2000) Elimination and detoxification of softwood extractives by white-rot fungi. J Biotechnol 80 : 231–240.
51. GutierrezA, del RioJC, Gonzalez-VilaFJ, MartinF (1998) Analysis of lipophilic extractives from wood and pitch deposits by solid-phase extraction and gas chromatography. J Chromatogr 823 : 449–455.
52. IshihamaY, OdaY, TabataT, SatoT, NagasuT, et al. (2005) Exponentially modified protein abundance index (emPAI) for estimation of absolute protein amount in proteomics by the number of sequenced peptides per protein. Mol Cell Proteomics 4 : 1265–1272.
53. DanielG, VolcJ, FilonovaL, PlihalO, KubatovaE, et al. (2007) Characteristics of Gloeophyllum trabeum alcohol oxidase, an extracellular source of H2O2 in brown rot decay of wood. Appl Environ Microbiol 73 : 6241–6253.
54. BendtsenJD, JensenLJ, BlomN, Von HeijneG, BrunakS (2004) Feature-based prediction of non-classical and leaderless protein secretion. Protein engineering, design & selection: PEDS 17 : 349–356.
55. van BeekTA, ClaassenFW, DoradoJ, GodejohannM, Sierra-AlvarezR, et al. (2007) Fungal biotransformation products of dehydroabietic acid. J Nat Prod 70 : 154–159.
56. AdamsAS, AylwardFO, AdamsSM, ErbilginN, AukemaBH, et al. (2013) Mountain pine beetles colonizing historical and naive host trees are associated with a bacterial community highly enriched in genes contributing to terpene metabolism. Appl Environ Microbiol 79 : 3468–3475.
57. KumarSV, PhalePS, DuraniS, WangikarPP (2003) Combined sequence and structure analysis of the fungal laccase family. Biotechnol Bioeng 83 : 386–394.
58. HofrichterM, UllrichR, PecynaMJ, LiersC, LundellT (2010) New and classic families of secreted fungal heme peroxidases. Appl Microbiol Biotechnol 87 : 871–897.
59. LiersC, PecynaMJ, KellnerH, WorrichA, ZornH, et al. (2013) Substrate oxidation by dye-decolorizing peroxidases (DyPs) from wood - and litter-degrading agaricomycetes compared to other fungal and plant heme-peroxidases. Appl Microbiol Biotechnol 97 : 5839–5849.
60. Agger JW, Isaksen T, Varnai A, Vidal-Melgosa S, Willats WG, et al. (2014) Discovery of LPMO activity on hemicelluloses shows the importance of oxidative processes in plant cell wall degradation. Proc Natl Acad Sci U S A 10.1073/pnas.1323629111.
61. Vaaje-KolstadG, WesterengB, HornSJ, LiuZ, ZhaiH, et al. (2010) An oxidative enzyme boosting the enzymatic conversion of recalcitrant polysaccharides. Science 330 : 219–222.
62. Vanden WymelenbergA, MingesP, SabatG, MartinezD, AertsA, et al. (2006) Computational analysis of the Phanerochaete chrysosporium v2.0 genome database and mass spectrometry identification of peptides in ligninolytic cultures reveals complex mixtures of secreted proteins. Fungal Genet Biol 43 : 343–356.
63. WangY, LimL, DiGuistiniS, RobertsonG, BohlmannJ, et al. (2013) A specialized ABC efflux transporter GcABC-G1 confers monoterpene resistance to Grosmannia clavigera, a bark beetle-associated fungal pathogen of pine trees. New Phytol 197 : 886–898.
64. MatsuzakiF, ShimizuM, WariishiH (2008) Proteomic and metabolomic analyses of the white-rot fungus Phanerochaete chrysosporium exposed to exogenous benzoic acid. J Proteome Res 7 : 2342–2350.
65. ShimizuM, YudaN, NakamuraT, TanakaH, WariishiH (2005) Metabolic regulation at the tricarboxylic acid and glyoxylate cycles of the lignin-degrading basidiomycete Phanerochaete chrysosporium against exogenous addition of vanillin. Proteomics 5 : 3919–3931.
66. AnnesiT, CurcioG, D'AmicoL, MottaE (2005) Biological control of Heterobasidion annosum on Pinus pinea by Phlebiopsis gigantea. Forest Pathology 35 : 127–134.
67. MunirE, YoonJJ, TokimatsuT, HattoriT, ShimadaM (2001) A physiological role for oxalic acid biosynthesis in the wood-rotting basidiomycete Fomitopsis palustris. Proc Natl Acad Sci USA 98 : 11126–11130.
68. BlanchetteRA, HeldBW, ArenzBE, JurgensJA, BaltesNJ, et al. (2010) An Antarctic hot spot for fungi at Shackleton's historic hut on Cape Royds. Microb Ecol 60 : 29–38.
69. Grigoriev IV, Martinez DA, Salamov AA (2006) Fungal genomic annotation. In: Arora DK, Berka RA, Singh GB, editors.Applied Mycology and Biotechnology.Amsterdam: Elsevier. pp. 123–142.
70. ParraG, BradnamK, KorfI (2007) CEGMA: a pipeline to accurately annotate core genes in eukaryotic genomes. Bioinformatics 23 : 1061–1067.
71. Vanden WymelenbergA, GaskellJ, MozuchMD, SabatG, RalphJ, et al. (2010) Comparative transcriptome and secretome analysis of wood decay fungi Postia placenta and Phanerochaete chrysosporium. Appl Environ Microbiol 76 : 3599–3610.
72. RyuJS, SharyS, HoutmanCJ, PaniskoEA, KorripallyP, et al. (2011) Proteomic and functional analysis of the cellulase system expressed by Postia placenta during brown rot of solid wood. Appl Environ Microbiol 77 : 7933–7941.
73. NesvizhskiiAI, KellerA, KolkerE, AebersoldR (2003) A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem 75 : 4646–4658.
74. Vanden WymelenbergA, GaskellJ, MozuchMD, KerstenP, SabatG, et al. (2009) Transcriptome and secretome analysis of Phanerochaete chrysosporium reveal complex patterns of gene expression. Appl Environ Microbiol 75 : 4058–4068.
75. OlsonA, AertsA, AsiegbuF, BelbahriL, BouzidO, et al. (2012) Insight into trade-off between wood decay and parasitism from the genome of a fungal forest pathogen. New Phytol 194 : 1001–1013.
76. Vanden WymelenbergA, SabatG, MozuchMD, KerstenP, CullenD, et al. (2006) Structure, organization, and transcriptional regulation of a family of copper radical oxidase genes in the lignin-degrading basidiomycete Phanerochaete chrysosporium. Appl Environ Microbiol 72 : 4871–4877.
77. AspeborgH, CoutinhoPM, WangY, BrumerH3rd, HenrissatB (2012) Evolution, substrate specificity and subfamily classification of glycoside hydrolase family 5 (GH5). BMC Evol Biol 12 : 186.
78. RzhetskyA, NeiM (1992) Statistical properties of the ordinary least-squares, generalized least-squares, and minimum-evolution methods of phylogenetic inference. J Mol Evol 35 : 367–375.
79. Zuckerkandl J, Pauling L (1965) Evolutionary divergence and convergence in proteins. In: Bryson V, Vogel HJ, editors.Evolving genes and proteins. New York: Academic Press.
80. SaitouN, NeiM (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4 : 406–425.
81. TamuraK, DudleyJ, NeiM, KumarS (2007) MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol 24 : 1596–1599.
82. GuillenF, EvansCS (1994) Anisaldehyde and veratraldehyde acting as redox cycling agents for H2O2 production by Pleurotus eryngii. Appl Environ Microbiol 60 : 2811–2817.
83. FerreiraP, Ruiz-DuenasFJ, MartinezMJ, van BerkelWJ, MartinezAT (2006) Site-directed mutagenesis of selected residues at the active site of aryl-alcohol oxidase, an H2O2-producing ligninolytic enzyme. FEBS J 273 : 4878–4888.
84. CavenerDR (1992) GMC oxidoreductases. A newly defined family of homologous proteins with diverse catalytic activities. J Mol Biol 223 : 811–814.
85. ThompsonJD, HigginsDG, GibsonTJ (1994) ClustalW improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22 : 2552–2556.
86. HallbergBM, HenrikssonG, PetterssonG, VasellaA, DivneC (2003) Mechanism of the reductive half-reaction in cellobiose dehydrogenase. J Biol Chem 278 : 7160–7166.
87. LetteraV, PiscitelliA, LeoG, BiroloL, PezzellaC, et al. (2010) Identification of a new member of Pleurotus ostreatus laccase family from mature fruiting body. Fungal Biol 114 : 724–730.
88. KilaruS, HoeggerPJ, KüesU (2006) The laccase multi-gene family in Coprinopsis cinerea has seventeen different members that divide into two distinct subfamilies. Curr Genet 50 : 45–60.
89. RühlM, MajcherczykA, KuesU (2013) Lcc1 and Lcc5 are the main laccases secreted in liquid cultures of Coprinopsis cinerea strains. Antonie van Leeuwenhoek 103 : 1029–1039.
90. KüesU, RühlM (2011) Multiple multi-copper oxidase gene families in basidiomycetes - what for? Curr Genomics 12 : 72–94.
91. AltschulSF, MaddenTL, SchafferAA, ZhangJ, ZhangZ, et al. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25 : 3389–3402.
92. TamuraK, PetersonD, PetersonN, StecherG, NeiM, et al. (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28 : 2731–2739.
Štítky
Genetika Reprodukční medicína
Článek Large-scale Metabolomic Profiling Identifies Novel Biomarkers for Incident Coronary Heart DiseaseČlánek Notch Signaling Mediates the Age-Associated Decrease in Adhesion of Germline Stem Cells to the NicheČlánek Phosphorylation of Mitochondrial Polyubiquitin by PINK1 Promotes Parkin Mitochondrial TetheringČlánek Natural Variation Is Associated With Genome-Wide Methylation Changes and Temperature SeasonalityČlánek Overlapping and Non-overlapping Functions of Condensins I and II in Neural Stem Cell DivisionsČlánek Unisexual Reproduction Drives Meiotic Recombination and Phenotypic and Karyotypic Plasticity inČlánek Tetraspanin (TSP-17) Protects Dopaminergic Neurons against 6-OHDA-Induced Neurodegeneration inČlánek ABA-Mediated ROS in Mitochondria Regulate Root Meristem Activity by Controlling Expression inČlánek Mutations in Global Regulators Lead to Metabolic Selection during Adaptation to Complex Environments
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2014 Číslo 12
-
Všechny články tohoto čísla
- Stratification by Smoking Status Reveals an Association of Genotype with Body Mass Index in Never Smokers
- Genome Wide Meta-analysis Highlights the Role of Genetic Variation in in the Regulation of Circulating Serum Chemerin
- Occupancy of Mitochondrial Single-Stranded DNA Binding Protein Supports the Strand Displacement Mode of DNA Replication
- Distinct Genealogies for Plasmids and Chromosome
- Large-scale Metabolomic Profiling Identifies Novel Biomarkers for Incident Coronary Heart Disease
- Non-coding RNAs Prevent the Binding of the MSL-complex to Heterochromatic Regions
- Plasmid Flux in ST131 Sublineages, Analyzed by Plasmid Constellation Network (PLACNET), a New Method for Plasmid Reconstruction from Whole Genome Sequences
- Epigenome-Guided Analysis of the Transcriptome of Plaque Macrophages during Atherosclerosis Regression Reveals Activation of the Wnt Signaling Pathway
- The Inventiveness of Nature: An Interview with Werner Arber
- Mediation Analysis Demonstrates That -eQTLs Are Often Explained by -Mediation: A Genome-Wide Analysis among 1,800 South Asians
- Generation of Antigenic Diversity in by Structured Rearrangement of Genes During Mitosis
- A Massively Parallel Pipeline to Clone DNA Variants and Examine Molecular Phenotypes of Human Disease Mutations
- Genetic Analysis of the Cardiac Methylome at Single Nucleotide Resolution in a Model of Human Cardiovascular Disease
- Genetic Analysis of Circadian Responses to Low Frequency Electromagnetic Fields in
- The Dissection of Meiotic Chromosome Movement in Mice Using an Electroporation Technique
- Altered Chromatin Occupancy of Master Regulators Underlies Evolutionary Divergence in the Transcriptional Landscape of Erythroid Differentiation
- Syd/JIP3 and JNK Signaling Are Required for Myonuclear Positioning and Muscle Function
- Notch Signaling Mediates the Age-Associated Decrease in Adhesion of Germline Stem Cells to the Niche
- Mutation of Leads to Blurred Tonotopic Organization of Central Auditory Circuits in Mice
- The IKAROS Interaction with a Complex Including Chromatin Remodeling and Transcription Elongation Activities Is Required for Hematopoiesis
- RAN-Binding Protein 9 is Involved in Alternative Splicing and is Critical for Male Germ Cell Development and Male Fertility
- Enhanced Longevity by Ibuprofen, Conserved in Multiple Species, Occurs in Yeast through Inhibition of Tryptophan Import
- Phosphorylation of Mitochondrial Polyubiquitin by PINK1 Promotes Parkin Mitochondrial Tethering
- Recurrent Loss of Specific Introns during Angiosperm Evolution
- Natural Variation Is Associated With Genome-Wide Methylation Changes and Temperature Seasonality
- SEEDSTICK is a Master Regulator of Development and Metabolism in the Arabidopsis Seed Coat
- Overlapping and Non-overlapping Functions of Condensins I and II in Neural Stem Cell Divisions
- Unisexual Reproduction Drives Meiotic Recombination and Phenotypic and Karyotypic Plasticity in
- Tetraspanin (TSP-17) Protects Dopaminergic Neurons against 6-OHDA-Induced Neurodegeneration in
- ABA-Mediated ROS in Mitochondria Regulate Root Meristem Activity by Controlling Expression in
- Mutations in Global Regulators Lead to Metabolic Selection during Adaptation to Complex Environments
- Global Analysis of Photosynthesis Transcriptional Regulatory Networks
- Mucolipin Co-deficiency Causes Accelerated Endolysosomal Vacuolation of Enterocytes and Failure-to-Thrive from Birth to Weaning
- Controlling Pre-leukemic Thymocyte Self-Renewal
- How Malaria Parasites Avoid Running Out of Ammo
- Echoes of the Past: Hereditarianism and
- Deep Reads: Strands in the History of Molecular Genetics
- Keep on Laying Eggs Mama, RNAi My Reproductive Aging Blues Away
- Analysis of a Plant Complex Resistance Gene Locus Underlying Immune-Related Hybrid Incompatibility and Its Occurrence in Nature
- Epistatic Adaptive Evolution of Human Color Vision
- Increased and Imbalanced dNTP Pools Symmetrically Promote Both Leading and Lagging Strand Replication Infidelity
- Genetic Basis of Haloperidol Resistance in Is Complex and Dose Dependent
- Genome-Wide Analysis of DNA Methylation Dynamics during Early Human Development
- Interaction between Conjugative and Retrotransposable Elements in Horizontal Gene Transfer
- The Evolution of Sex Ratio Distorter Suppression Affects a 25 cM Genomic Region in the Butterfly
- is Required for Adult Maintenance of Dopaminergic Neurons in the Ventral Substantia Nigra
- PRL1, an RNA-Binding Protein, Positively Regulates the Accumulation of miRNAs and siRNAs in Arabidopsis
- Genetic Control of Contagious Asexuality in the Pea Aphid
- Early Mesozoic Coexistence of Amniotes and Hepadnaviridae
- Local and Systemic Regulation of Plant Root System Architecture and Symbiotic Nodulation by a Receptor-Like Kinase
- Gene Pathways That Delay Reproductive Senescence
- The Evolution of Fungal Metabolic Pathways
- Maf1 Is a Novel Target of PTEN and PI3K Signaling That Negatively Regulates Oncogenesis and Lipid Metabolism
- Formation of Linear Amplicons with Inverted Duplications in Requires the MRE11 Nuclease
- Identification of Rare Causal Variants in Sequence-Based Studies: Methods and Applications to , a Gene Involved in Cohen Syndrome and Autism
- Rrp12 and the Exportin Crm1 Participate in Late Assembly Events in the Nucleolus during 40S Ribosomal Subunit Biogenesis
- The Mutations in the ATP-Binding Groove of the Rad3/XPD Helicase Lead to -Cockayne Syndrome-Like Phenotypes
- Topoisomerase I Plays a Critical Role in Suppressing Genome Instability at a Highly Transcribed G-Quadruplex-Forming Sequence
- A Cbx8-Containing Polycomb Complex Facilitates the Transition to Gene Activation during ES Cell Differentiation
- Transcriptional Frameshifting Rescues Type VI Secretion by the Production of Two Length Variants from the Prematurely Interrupted Gene
- Association Mapping across Numerous Traits Reveals Patterns of Functional Variation in Maize
- Genome-Wide Analysis of -Regulated and Phased Small RNAs Underscores the Importance of the ta-siRNA Pathway to Maize Development
- Dissemination of Cephalosporin Resistance Genes between Strains from Farm Animals and Humans by Specific Plasmid Lineages
- The Tau Tubulin Kinases TTBK1/2 Promote Accumulation of Pathological TDP-43
- Germline Signals Deploy NHR-49 to Modulate Fatty-Acid β-Oxidation and Desaturation in Somatic Tissues of
- Microevolution of in Macrophages Restores Filamentation in a Nonfilamentous Mutant
- Vangl2-Regulated Polarisation of Second Heart Field-Derived Cells Is Required for Outflow Tract Lengthening during Cardiac Development
- Chondrocytes Transdifferentiate into Osteoblasts in Endochondral Bone during Development, Postnatal Growth and Fracture Healing in Mice
- A ABC Transporter Regulates Lifespan
- RA and FGF Signalling Are Required in the Zebrafish Otic Vesicle to Pattern and Maintain Ventral Otic Identities
- , and Reprogram Thymocytes into Self-Renewing Cells
- The miR9863 Family Regulates Distinct Alleles in Barley to Attenuate NLR Receptor-Triggered Disease Resistance and Cell-Death Signaling
- Detection of Pleiotropy through a Phenome-Wide Association Study (PheWAS) of Epidemiologic Data as Part of the Environmental Architecture for Genes Linked to Environment (EAGLE) Study
- Extensive Copy-Number Variation of Young Genes across Stickleback Populations
- The and Genetic Modules Interact to Regulate Ciliogenesis and Ciliary Microtubule Patterning in
- Analysis of the Genome, Transcriptome and Secretome Provides Insight into Its Pioneer Colonization Strategies of Wood
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Tetraspanin (TSP-17) Protects Dopaminergic Neurons against 6-OHDA-Induced Neurodegeneration in
- Maf1 Is a Novel Target of PTEN and PI3K Signaling That Negatively Regulates Oncogenesis and Lipid Metabolism
- The IKAROS Interaction with a Complex Including Chromatin Remodeling and Transcription Elongation Activities Is Required for Hematopoiesis
- Echoes of the Past: Hereditarianism and
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



