-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaGlobal Analysis of Photosynthesis Transcriptional Regulatory Networks
Photosynthetic organisms are among the most abundant life forms on earth. Their unique ability to harvest solar energy and use it to fix atmospheric carbon dioxide is at the foundation of the global food chain. This paper reports the first comprehensive analysis of networks that control expression of photosynthesis genes using Rhodobacter sphaeroides, a microbe that has been studied for decades as a model of solar energy capture and other aspects of the photosynthetic lifestyle. We find a previously unappreciated complexity in the level of control of photosynthetic genes, while identifying new links between photosynthesis and central processes like iron availability. This organism is an ancestor of modern day plants, so our data can inform studies in other photosynthetic organisms and improve our ability to harness solar energy for food and industrial processes.
Published in the journal: . PLoS Genet 10(12): e32767. doi:10.1371/journal.pgen.1004837
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1004837Summary
Photosynthetic organisms are among the most abundant life forms on earth. Their unique ability to harvest solar energy and use it to fix atmospheric carbon dioxide is at the foundation of the global food chain. This paper reports the first comprehensive analysis of networks that control expression of photosynthesis genes using Rhodobacter sphaeroides, a microbe that has been studied for decades as a model of solar energy capture and other aspects of the photosynthetic lifestyle. We find a previously unappreciated complexity in the level of control of photosynthetic genes, while identifying new links between photosynthesis and central processes like iron availability. This organism is an ancestor of modern day plants, so our data can inform studies in other photosynthetic organisms and improve our ability to harness solar energy for food and industrial processes.
Introduction
Photosynthetic organisms are central to life on the planet. Their ability to harness solar energy and fix atmospheric carbon dioxide makes them integral parts of most ecosystems. Furthermore, many photosynthetic microbes, either naturally or via modifications, are capable of producing a variety of valuable commodities such as grain for food, hydrocarbons, hydrogen gas and valuable chemicals [1]–[4]. These properties will likely make them important in efforts to develop more sustainable societies. We are interested in obtaining new knowledge about the transcriptional networks of photosynthetic cells that underlie these important activities.
Anoxygenic photosynthetic bacteria have and continue to provide significant insight into the networks that govern photosynthetic activities because of their ease of growth, genetic tractability, and prior knowledge about solar energy capture and other aspects of this lifestyle [3], [5]. The advent of genomic approaches has allowed development of metabolic and transcriptional regulatory network (TRN) models for bacterial photosynthesis, the latter of which has led to predictions about regulatory networks in photosynthetic cells that extend beyond prior knowledge [6]–[10]. Thus, there is likely still much more to be learned about photosynthesis through testing the predictions of metabolic and TRN models in well-studied photosynthetic organisms.
To obtain this new knowledge, we analyze Rhodobacter sphaeroides, the best studied member of the purple non-sulfur bacteria – a group of photosynthetic microbes displaying great metabolic versatility and having significant biotechnological potential [1], [7], [11]–[18]. R. sphaeroides is capable of growing by aerobic respiration, anaerobic respiration and anaerobic anoxygenic photosynthesis. Prior analysis indicates that transitions between aerobic respiratory and anaerobic photosynthetic growth is achieved, in part, via a TRN involving 3 global transcription factors (TFs) – PrrA, FnrL and PpsR – that act to activate or repress relevant operons depending on the presence of oxygen or other signals. For instance, PrrA (the response regulator of the PrrAB two component system) and FnrL (the R. sphaeroides homolog of FNR) directly activate transcription of photosynthesis related genes at low oxygen tensions [9], [19]–[25]. On the other hand, PpsR represses the expression of photosynthesis related genes at high oxygen tensions [8], [26], [27]. In addition to these TFs, a small non-coding RNA, PcrZ has recently been implicated in the regulation of photosynthesis gene expression in R. sphaeroides [28]. While there is considerable information on how these regulators impact some photosynthesis genes, global information on their targets and how they act together to impact this lifestyle is lacking. Furthermore, a large-scale reconstruction of the R. sphaeroides TRN [10], which combined comparative genomics analysis with global gene expression data, predicted that two previously uncharacterized TFs, CrpK and RSP_2888 (hereafter referred to as modulator of photopigment genes, MppG), were involved in controlling the transcription of a number of operons that encode key functions involved in photosynthesis in R. sphaeroides, suggesting that the photosynthetic TRN of this organism is more complex than previously thought.
In this work, we use a combination of genetic, genomic and physiological analysis to dissect the roles of 4 TFs known or predicted to be involved in the regulation of the photosynthetic lifestyle of R. sphaeroides. The regulons of the previously characterized TFs, PrrA and FnrL, were refined and extended, while those of CrpK and MppG were characterized for the first time. Our analysis confirmed many predictions of the large-scale R. sphaeroides TRN, revealed the existence of significant overlap in direct targets for these TFs, as well as the high degree of combinational regulation of key operons. We also identified how components in this photosynthetic TRN provide robustness and fine-tuned expression of target genes. Overall, this study provides a large amount of new insight into the photosynthetic TRN of R. sphaeroides that is likely to be conserved in other related photosynthetic bacteria.
Results
Genome-wide analysis of known regulators of photosynthesis in R. sphaeroides
Based on previous analysis in R. sphaeroides and related purple non-sulfur bacteria, FnrL, PrrA and PpsR have been identified as key regulators of the photosynthetic lifestyle [8], [9], [20], [24], [27], [29]. We have previously characterized the genome-wide binding sites of PpsR via chromatin immunoprecipitation followed by sequencing (ChIP-seq) and gene expression analysis [10]. This analysis identified a total of 15 PpsR target operons, of which 13 had photosynthesis related functions. Here, we analyze the regulons of FnrL and PrrA using both ChIP-seq and global gene expression analysis.
FnrL—a global regulator of anaerobic growth in R. sphaeroides
FnrL is an iron-sulfur cluster-containing Crp-family TF that has been reported to be essential for both photosynthetic and anaerobic respiratory growth in R. sphaeroides [9], [25]. ChIP-chip analysis has previously been used to map genome-wide FnrL binding sites in vivo, identifying targets that indicate the direct involvement of this protein in a host of processes including those required for photosynthetic and anaerobic respiratory growth [24]. However, a large-scale reconstruction of R. sphaeroides TRN predicted that the FnrL regulon is significantly larger than previous analyses suggested. Thus, we re-examined the FnrL regulon using new and higher resolution complementary genomic approaches.
Analyzing the FnrL regulon using ChIP-seq
We determined the genome-wide FnrL binding sites using ChIP-seq with wild type (WT) cells grown under anoxygenic photosynthetic conditions. We reproducibly identified a total of 62 FnrL binding sites across 3 independent ChIP-seq experiments, corresponding to 52 known or predicted operons (S1 Table). These included several sites immediately upstream of genes involved in bacteriochlorophyll synthesis (bchEJGP), early steps in tetrapyrrole biosynthesis (hemN, hemZ and hemA), as well as genes that regulate anaerobic respiration using dimethyl sulfoxide (DMSO) as a terminal electron acceptor (dorS) (Fig. 1A, S1 Table). FnrL binding sites were also found upstream of genes encoding functions for iron transporter (feoABC) and iron sulfur cluster assembly (RSP_1949). When we compared this set of ChIP-seq identified FnrL binding sites to data from ChIP-chip analysis [24], 24 of the 27 FnrL binding sites identified previously were also detected in our analysis (S1 Table). The three previously identified FnrL binding sites not identified in our ChIP-seq analysis do not appear to contain a significant FnrL motif and likely represent false binding events. Furthermore, we found an additional 38 FnrL binding sites in the ChIP-seq dataset, implicating this TF as a direct regulator of a wide variety of new functions, ranging from protein synthesis and substrate transport to polyphosphate dependent phosphorylation and nitrogen metabolism (S1 Table, see Discussion). The larger number of sites identified in the ChIP-seq analysis compared to the previous ChIP-chip analysis for FnrL could be a reflection of the higher sensitivity and improved resolution obtainable with ChIP-seq analysis [30]. For instance, the higher signal to noise ratio of ChIP-seq could potentially allow for identification of relatively weak binding sites, which may be difficult to identify by ChIP-chip. Consistent with this, only 1 of the 24 ChIP-chip identified sites had lower than a 20-fold enrichment in the ChIP-seq dataset (S1 Table). On the other hand, some of these differences could also be the result of differences in peak calling algorithms and thresholds used to identify significant binding sites in the individual datasets.
Fig. 1. Analysis of the FnrL regulon in R. sphaeroides. 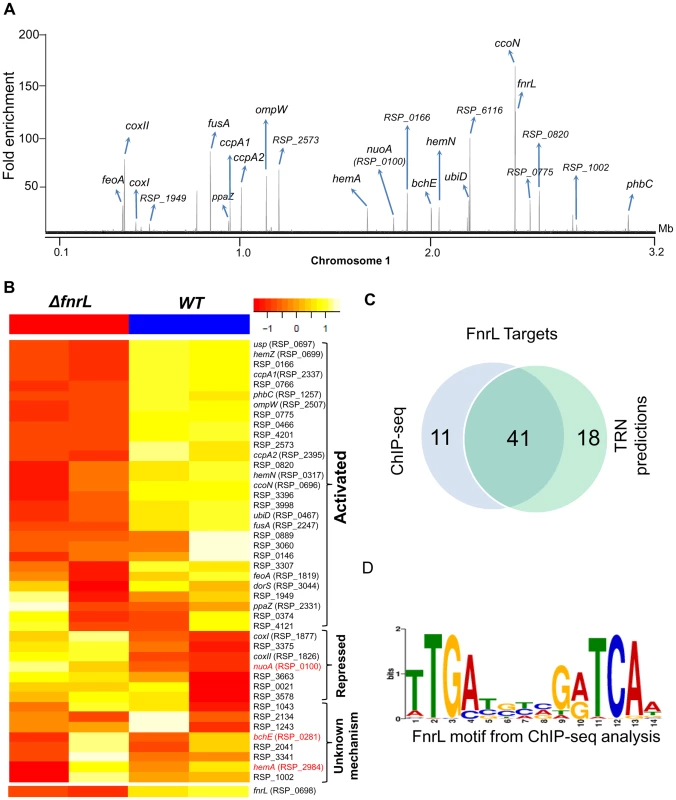
(A) A total of 62 FnrL binding sites were identified by ChIP-seq across the R. sphaeroides genome. Binding sites across chromosome 1 are highlighted. MochiView [90] was used for visualization of binding profile. (B) Heat map depicts the differentially expressed FnrL target operons between wild type (WT) and ΔfnrL cells grown on acetate-based media. For brevity only the first members of the target operons are presented. The relative expression of nuoA, bchE and hemA (highlighted in red), which are known to be positively regulated by FnrL, are either not differentially expressed or differentially expressed in the opposite direction. (C) Venn diagram depicting the overlap between our FnrL ChIP-seq analysis and prediction from the large-scale reconstruction of R. sphaeroides transcriptional network. (D) Position weight matrix logo generated for FnrL using targets identified by ChIP-seq. In addition to the 49 FnrL binding sites that were identified upstream of operons, 13 binding sites were outside upstream regulatory regions of any annotated genes. These sites could represent non-functional sites or binding sites in the upstream regulatory regions of other unannotated genomic elements. For instance, 2 of these 13 additional sites were in putative promoter regions of recently identified sRNA in R. sphaeroides - RSs0019 and RSs2461 [31]. Thus, it is conceivable that these other unassigned binding sites are located in the upstream regulatory regions of other as of yet unidentified genomic elements. It is also worth noting that 41 of 59 (69.5%) FnrL target operons predicted in the large-scale TRN reconstruction [10] were verified via this ChIP-seq analysis (Fig. 1C), including 17 operons that were novel predictions in that TRN inference study (S1 Table).
To independently assess the functional role of FnrL in the regulation of target genes identified in our ChIP-seq analysis, we conducted microarray analysis to compare the gene expression of WT cells to a ΔfnrL deletion strain [9], [25] during growth on acetate as the sole carbon source, a condition we found that allows photosynthetic growth of both WT and ΔfnrL strains (S1 Figure). Consistent with FnrL being a global regulator, a total of 303 genes were differentially expressed between the 2 strains (cutoff – 1.5 fold change (FC), p<0.01), with 166 and 137 genes showing increased and decreased transcript abundance, respectively, between WT and ΔfnrL cells (S2 Table). Of the 48 operons in which we found FnrL binding via ChIP-seq (and for which probes exist on the R. sphaeroides gene chip), 24 were differentially expressed between WT and ΔfnrL cells (Fig. 1B), indicating that at least under the conditions assayed, the expression of these target genes is significantly influenced by FnrL. While the change in expression of some of the FnrL targets did not meet the significance cut-off used for this analysis (S2 Table), changes in their expression in response to the loss of FnrL was sufficient to allow a tentative assignment of their control by FnrL (S1 Table, Fig. 1B). The direct and potential indirect influence of FnrL on global gene expression are further detailed in the Discussion. It should also be noted that transcripts for hemA and nuoA-N (RSP_0100-12) did not show a significant difference in levels between WT and ΔfnrL, while transcripts encoding bchEJGP were elevated in ΔfnrL cells, despite the proposed positive role of FnrL in transcription of these operons [24]. These results suggest that transcription of some FnrL target operons might be under control of other TFs, wherein loss of FnrL is partially or fully compensated for by the activity of other transcriptional regulators in the ΔfnrL strain. Indeed, hemA transcription is known to also be directly activated by the response regulator, PrrA under anaerobic conditions [32], [33].
PrrA—A global regulator of photosynthetic growth in R. sphaeroides
PrrA is the response regulator of the two component PrrAB system that has previously been proposed to be a major global TF in R. sphaeroides and related purple non-sulfur bacteria [20]. PrrA is essential for photosynthetic growth in R. sphaeroides and direct control of photosynthesis-related operons by PrrA has been shown via the use of in vitro experiments [22], [32]. To obtain a better understanding of the functional role of PrrA, we assessed PrrA activity using genome-wide gene expression data and ChIP-seq.
Redefining the set of PrrA target genes
Gene expression profiling experiments comparing mRNA abundance in a ΔprrA strain to WT cells were previously conducted under anaerobic respiratory conditions using DMSO as a terminal electron acceptor – a condition which permits growth of the photosynthesis deficient ΔprrA strain and under which PrrA is active in WT cells. This analysis showed that over 1000 genes were differentially expressed in the absence of PrrA [20]. However, a large percentage of these genes encoded functions related to protein synthesis and cell growth [20], suggesting that these might also reflect differences in growth rates between the 2 strains, possibly resulting from unlinked mutations in the ΔprrA strain. Consistent with this, we found that the ΔprrA strain used in the previous analysis (PrrA2) grew significantly faster than our WT strain under anaerobic respiratory conditions (S2A Figure). In contrast, an independently constructed markerless ΔprrA strain made for this study (PrrA3) grew similarly to WT under anaerobic respiratory conditions (S2A Figure) and showed similar pigmentation phenotypes to the original ΔprrA mutant. Consequently, we reassessed differences in gene expression between WT and ΔprrA using the PrrA3 mutant strain. We found a total of 255 genes were differentially expressed between WT and ΔprrA (2 FC, p<0.01) (S3 Table), significantly less than the 1058 previously reported at a similar cut off [20]. In addition, this set of 255 differentially expressed genes did not include any protein synthesis genes. We believe this set of differentially expressed genes, which are essentially a subset of those previously identified [20], provide a better picture of potential PrrA target genes.
Consistent with previous knowledge on PrrA, the 255 differentially expressed genes that we identified were enriched for genes known or predicted to be involved in photosynthetic processes (Table 1). In addition to photosynthesis related functions, other GO terms significantly enriched for differentially expressed genes in this dataset include categories such as the TCA cycle, electron transport chain and iron binding (Table 1). Overall, of the 255 differentially expressed genes (corresponding to 182 operons), mRNA levels from 148 were increased in the presence of PrrA, while 107 were decreased, supporting previous suggestions that PrrA functions as both a transcriptional activator and repressor [20].
Tab. 1. GO functional categories significantly enriched for genes regulated by PrrA. 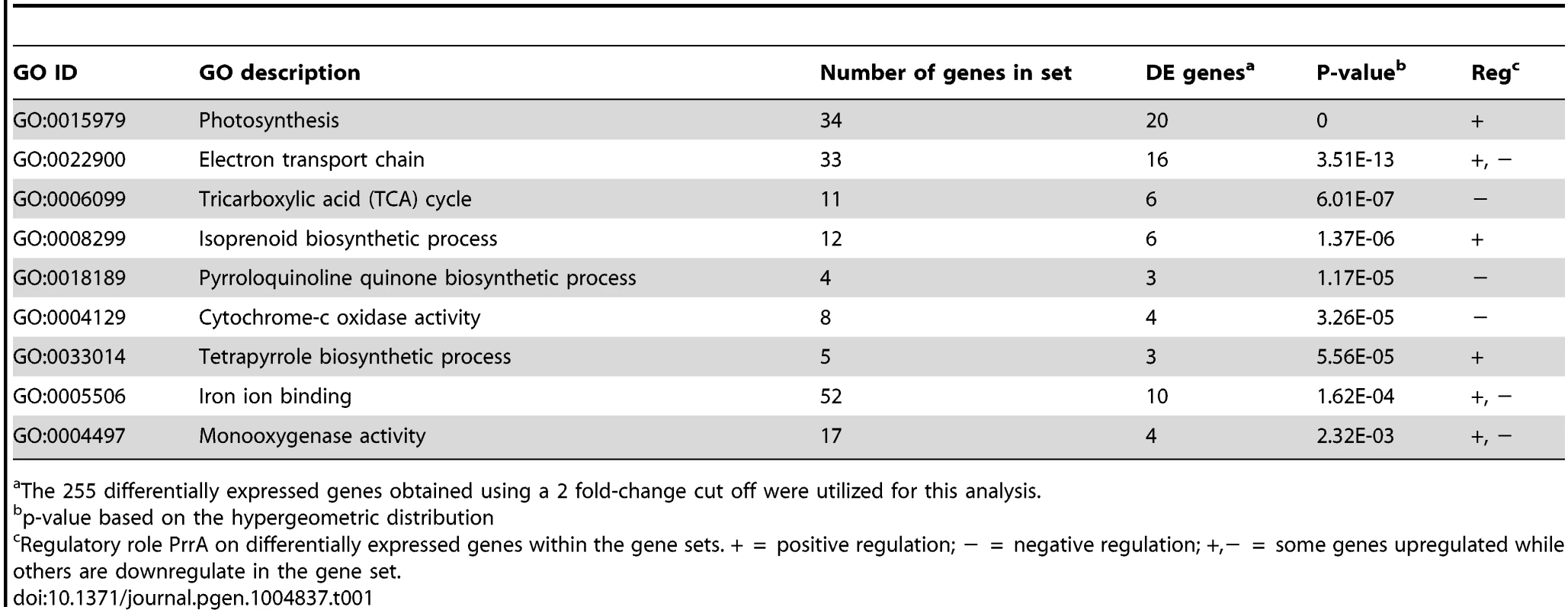
The 255 differentially expressed genes obtained using a 2 fold-change cut off were utilized for this analysis. PrrA regulates transcription from only a subset of its binding sites
To determine which differentially expressed genes are directly regulated by PrrA, we conducted ChIP-seq analysis on exponentially growing cells using a 3X myc-tagged PrrA protein that complements the photosynthetic growth defect of PrrA3 (S2B Figure). We observed significant enrichment for PrrA at ∼140 sites across the R. sphaeroides genome (Fig. 2A). Analysis of the sequences under all of these peaks did not reveal any strong consensus sequence shared by a significant number of these sites. Thus, to help determine the transcriptionally regulated direct targets of PrrA, only operons with both a significant peak and which were differentially expressed in PrrA3, were considered as candidate direct targets of this TF. A total of 34 operons met these criteria, including 18 photosynthesis related operons (Table 2, Figs. 2B and 2C). In addition to photosynthesis-related genes to which PrrA had previously been linked, these analyses indicate that PrrA is also a direct regulator of electron transport (regulating operons encoding fbcFBC, fbcQ-soxDA and RSP_0820 (cytochrome B561)), tetrapyrrole synthesis (hemA, hemC and hemE) and terpenoid backbone biosynthesis (dxr). Our data predict that all but one of these operons are positively regulated by PrrA, since RNA levels from these genes were lower in PrrA3 (Fig. 2B, Table 2). Other enriched sites in the genome not included in this set of transcriptionally regulated direct PrrA targets are provided in S4 Table.
Fig. 2. Analysis of the PrrA regulon in R. sphaeroides. 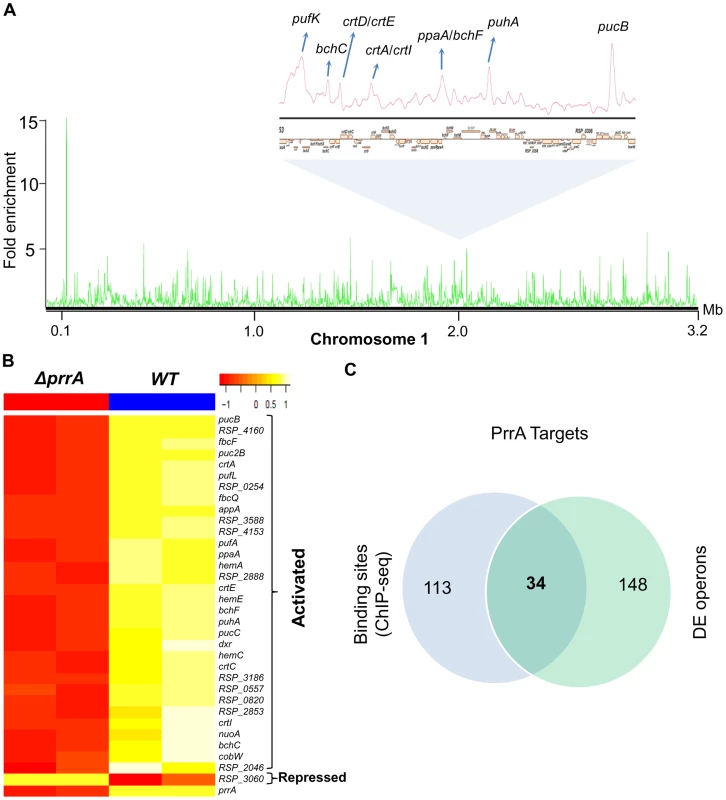
(A) PrrA binding sites across chromosome 1. Binding sites within the photosynthetic gene cluster are enlarged. (B) Heat map depicting the PrrA targets genes from a pair-wise comparison of transcript levels from WT and ΔprrA cells grown under anaerobic respiratory conditions. (C) Venn diagram showing the overlap between identified ChIP-seq binding sites for PrrA and differentially expressed operons from microarray analysis. It should be noted that some binding sites are located between two divergently transcribed operons, which were both differentially expressed. In such cases, both operons were considered as direct PrrA targets. Tab. 2. PrrA target genes identified by ChIP-seq and gene expression analysis of R. sphaeroides cells. 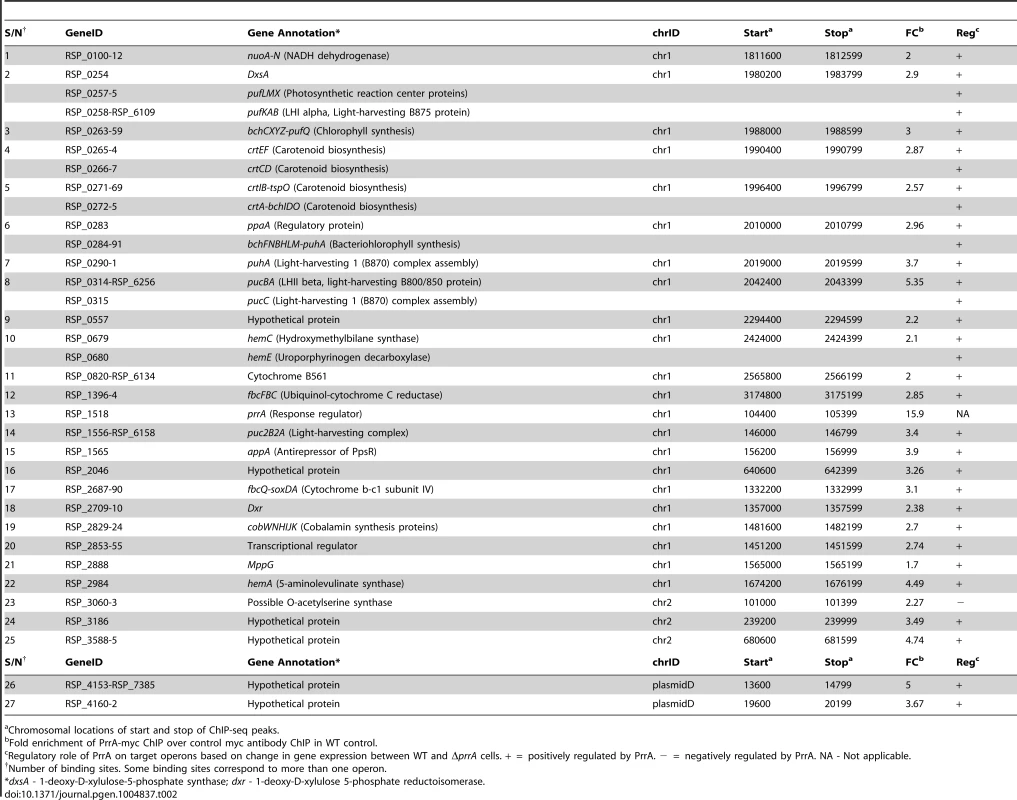
Chromosomal locations of start and stop of ChIP-seq peaks. Analysis of newly predicted regulators of photosynthesis in R. sphaeroides
A recent large-scale reconstruction of the TRN of R. sphaeroides predicted that there are additional regulators of photosynthesis. Among the highest scoring TFs that fell into this category were: (i) CrpK (RSP_2572), a Crp/Fnr-family regulator, and (ii) MppG, a BadM/Rrf2-family protein. Using a combination of physiological, genetic and genomic analysis, we investigated the contributions made by these proteins to regulation of photosynthesis in R. sphaeroides.
CrpK—A member of the Crp/FnrL family that controls many photosynthesis genes
CrpK is a Crp/Fnr-family TF, which based on Pfam analysis [34], shares similar cyclic nucleotide-binding and Crp-like helix-turn-helix domains as FnrL. However, unlike FnrL, CrpK does not possess the 4 cysteine residues at its N-terminus required for coordination with iron-sulfur clusters, suggesting CrpK might not directly sense oxygen. Nevertheless, ectopic expression of CrpK in an ΔfnrL mutant from an IPTG-inducible plasmid restores photosynthetic growth on succinate (S3A Figure), indicating CrpK might directly regulate a similar set of genes as FnrL. However, a markerless crpK deletion mutant is capable of photosynthetic growth on succinate (S3B Figure), indicating that FnrL and CrpK might also have distinct targets. In addition, CrpK transcript levels are ∼2 to 3 fold higher in photosynthetic cells relative to cells grown under aerobic or anaerobic respiratory conditions (indicating it may have a physiological role linked to photosynthesis), but the levels of CrpK-specific transcripts are lower than those coding for FnrL under all growth conditions that have so far been tested by global expression analysis.
The CrpK and FnrL regulons are overlapping but distinct
We conducted ChIP-seq analysis using a 3X myc tagged variant of CrpK, which was able to restore photosynthetic growth to a ΔfnrL deletion strain (ΔfnrL+pIND5-crpK-3Xmyc) (S3A Figure) confirming its functionality (the observed difference in growth rate between the strains complemented with a tagged or untagged CrpK protein may reflect alterations in activity or abundance of the individual proteins). We identified a total of 38 binding sites for CrpK in the R. sphaeroides genome (Table 3, Fig. 3A). Consistent with its predicted involvement in regulation of photosynthetic genes, CrpK was found to bind to the upstream regulatory regions of bchEJGP and hemA, both known to be involved in the biosynthesis of photopigments or their tetrapyrrole precursors [35]–[37]. CrpK binding sites were also found upstream of genes encoding iron transporters (feoABC, ccmD) and iron-sulfur cluster binding proteins (rdxBHIS). In addition, 23 (60.5%) of the identified CrpK sites were also identified as FnrL target sites (Table 3, S1 Table, S4 Figure), possibly providing an explanation for the ability of CrpK to at least partially compensate for the loss of FnrL (S3A Figure). The remaining 15 CrpK binding sites were not occupied by FnrL under the conditions we tested (Table 3). On the other hand, FnrL was found bound to 39 sites that were not recognized by CrpK. These observations of TF occupancy for a subset of these overlapping and distinct sites were also verified via independent ChIP-qPCR analysis (S3D Figure), but it should be noted that many of the “CrpK unique sites” (i.e., those not also bound by FnrL) are relatively low enrichment sites in ChIP-seq assays (S4 Figure). However, this set of “CrpK unique sites” all possessed a similar shared motif to other identified CrpK sites, so we consider it likely that these are actually direct targets for control by this TF. The ChIP-seq peaks for CrpK and FnrL at sites bound by both TFs were centered at the same location for both TFs (S4 Figure) and consequently the predicted binding motifs for both TFs bear strong DNA sequence similarity at both shared and unique sites (Fig. 3B). This observation is consistent with general motif type recognized by Crp/Fnr-family TFs and the relatively high degree of amino acid sequence similarity in the predicted DNA binding motifs of CrpK and FnrL [24]. However, subtle differences between the motifs that are assembled by analysis of the “FnrL and CrpK unique sites” could be discerned, which might allow for future computational or experimental discrimination between target sites for each TF (Fig. 3B).
Fig. 3. Analysis of the CrpK regulon in R. sphaeroides. 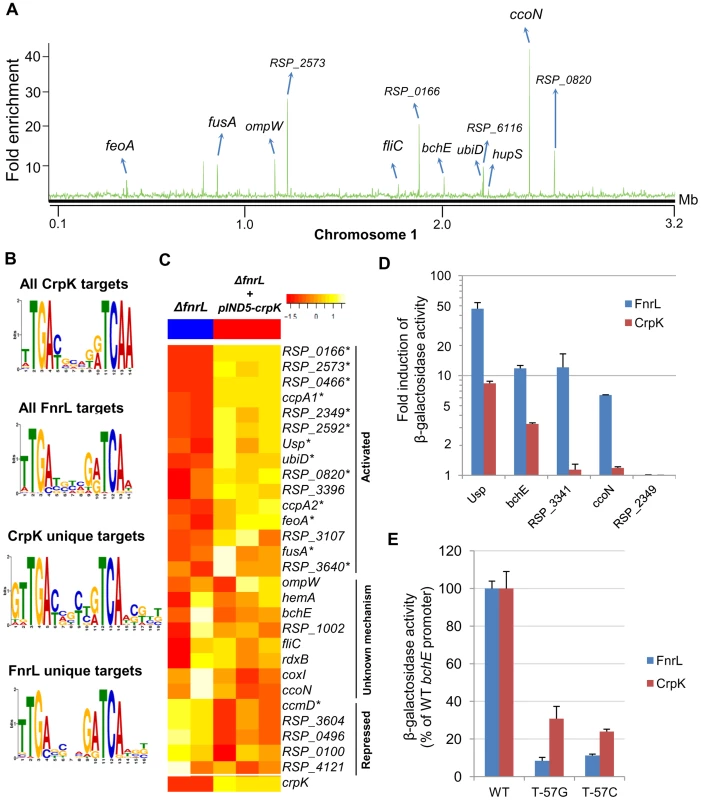
(A) CrpK binding sites across chromosome 1. (B) Comparison of motifs generated for all CrpK and FnrL ChIP-seq identified targets, as well as those for targets exclusive to the CrpK and FnrL ChIP-seq datasets. (C) Pair-wise comparison of the global transcript level data between a ΔfnrL strain and a ΔfnrL strain over-expressing CrpK (ΔfnrL+pIND5-crpk) for all ChIP-seq identified CrpK target operons. Only the first members of the operons are shown. Significantly differentially expressed genes are indicated with *. (D) Fold increase in β-galactosidase activity observed after inducing either CrpK or FnrL synthesis using promoter-lacZ fusions of the bchE, RSP_0697 (Usp), ccoN, RSP_3341 and RSP_2349 promoters, integrated into the chromosome of a ΔfnrL-ΔcrpK reporter strain. Fold inductions represent fold change over the measured basal β-galactosidase activity prior to induction of CrpK or FnrL. (E) Percentage of β-galactosidase activity at mutant bchE promoters relative to the WT promoter. The average growth rate of cultures with FnrL induced was 3.4±0.2 hrs, while that of cultures with CrpK induced was 3.1±0.2 hrs. The error bars in (D) and (E) represent standard error of mean of 3 independent biological replicates. Tab. 3. CrpK binding sites across the R. sphaeroides genome identified by ChIP-seq. 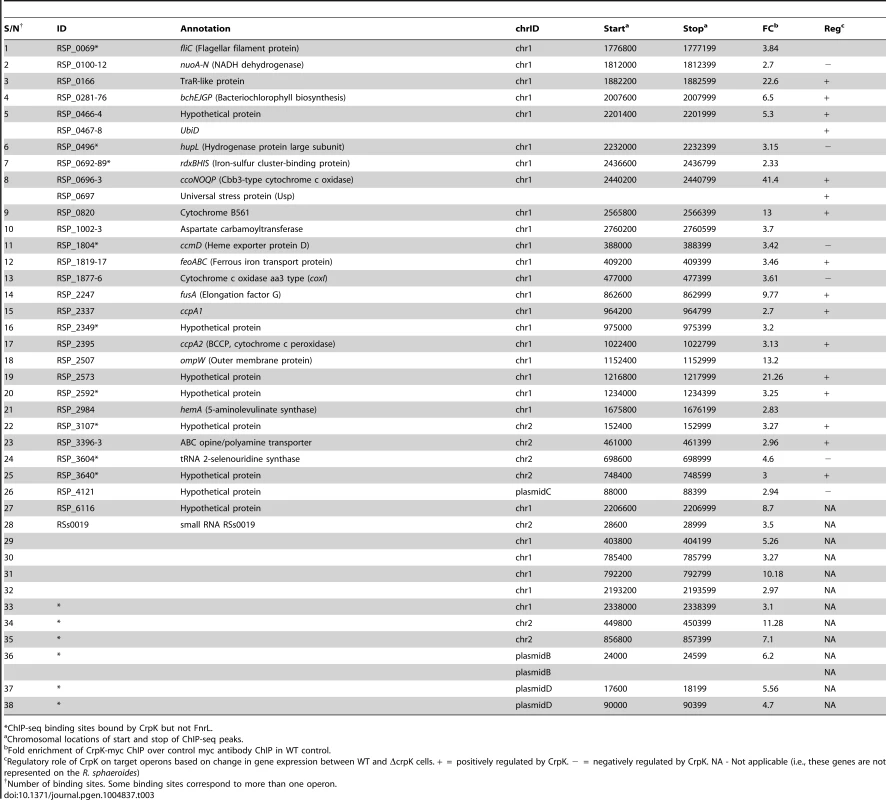
*ChIP-seq binding sites bound by CrpK but not FnrL. CrpK controls expression of its predicted target genes
To independently test the role of CrpK on its predicted target genes, we conducted microarray analysis comparing the expression of a ΔfnrL strain to a ΔfnrL strain expressing an untagged crpK gene from an IPTG-inducible plasmid (ΔfnrL+pIND5-crpK) (S3A and S3C Figures), to bypass the effect of FnrL on these target operons (a ΔfnrLΔcrpK strain was not viable under any anaerobic or photosynthetic conditions that we tested). Of the 28 CrpK target operons identified by ChIP-seq (and for which probes exist on the R. sphaeroides gene chip), 14 of these were differentially expressed (using a 1.5 FC, p<0.01) under these conditions (Fig. 3C, Table 3), predicting that CrpK controls expression from these target promoters in this reporter strain. Transcripts from 13 of these differentially expressed operons were increased including those encoding RSP_0166 (a TraR-like protein), UbiD, RPS_0697 (universal stress protein, Usp) and iron transporter FeoABC, whereas 1 operon was down regulated in the absence of CrpK (Fig. 3C). Just as in the case of FnrL, regulatory control at some of the CrpK target operons such as hemA, nuoA-N and bchEJGP might be obscured by reprogramming of the transcriptional network in the ΔfnrL strain (see Discussion). Thus, the number of functional CrpK targets is likely larger. In general, the predicted direction of regulation by CrpK in all of the differentially expressed operons (activation or repression) were the same as observed with FnrL (Table 3, Fig. 3C).
Given the gene expression patterns observed above, we analyzed expression of promoter-lacZ fusions, using a few CrpK and FnrL target genes, in ΔfnrLΔcrpK double mutant reporter strains, as an additional test of the ability of these proteins to control activity of candidate promoters. Consistent with the predictions of our genome-wide analysis, we observed that CrpK was able to increase β-galactosidase activity from the bchE, universal stress protein (RSP_0697) and ccoN promoters, similar to FnrL though albeit with lower β-galactosidase activity (Fig. 3D). This result also indicates a positive regulatory role for both FnrL and CrpK have on bchE, a fact which was not possible to infer from our global gene expression data. No significant β-galactosidase activity was observed with the RSP_3341 promoter upon ectopic expression of CrpK (1.1 fold induction), whereas significant β-galactosidase activity was obtained when FnrL synthesis was induced. These data are also consistent with the ChIP-seq data, which indicates that FnrL, but not CrpK, binds to the RSP_3341 promoter (S4 Figure). For the one unique CrpK site tested in this analysis, RSP_2349, we did not observe any increase in β-galactosidase activity from this promoter when either CrpK or FnrL was ectopically expressed, suggesting that this CrpK binding site might not be functional under the growth conditions tested. Nevertheless, the ability of CrpK to bind the upstream regulatory regions of several FnrL target genes such as bchEJGP and hemA, as well as control their expression, provides a direct explanation for the ability of increased CrpK expression to restore photosynthetic growth to an FnrL mutant of R. sphaeorides.
CrpK and FnrL share similar DNA binding determinants
To test the above prediction that CrpK and FnrL can recognize related DNA sequences, we made 2 separate base substitutions in the predicted FnrL/CrpK consensus sequence of the bchE promoter, substituting thymine of the TCAA (at position −57 relative to the start codon) with either a guanine or cytosine. When these base substitutions were introduced into the bchE promoter fused to a promoterless lacZ gene, we found that they caused a significant decrease in β-galactosidase activity relative to the WT promoter when either FnrL or CrpK synthesis was induced (Fig. 3E). An ∼90% reduction in β-galactosidase activity was observed with both promoter mutations when FnrL synthesis was induced, while ∼70 and 80% decreases were observed with the individual guanine and cytosine mutations, respectively, when CrpK synthesis was induced. These data indicate that both TFs recognize similar sequences, consistent with predictions from the motif finding analysis and the fact that they belong to the same TF family.
MppG—A newly identified modulator of photopigment biosynthesis
Another TF predicted by the large-scale TRN to play a role in the control of R. sphaeroides photosynthesis genes is the BadM/Rrf2 family TF, MppG. mppG transcript levels are increased under photosynthetic conditions in WT cells and mppG is predicted to be a direct target of PrrA (Table 2, Fig. 2B). Consistent with this, mppG transcript levels are more than 5 fold higher in WT cells relative to ΔprrA, being the most differentially expressed TF in that dataset (S3 Table). To test the role of MppG in regulation of photosynthesis, we conducted a combination of physiological, gene expression and protein-DNA binding assays for this TF.
MppG negatively regulates photopigment synthesis
To assess the physiological role of MppG, we constructed and analyzed the properties of a mppG deletion mutant (ΔMppG). Furthermore, ΔMppG was complemented with mppG from an IPTG-inducible plasmid (ΔMppG+pIND5-mppG). The WT and ΔMppG strains both exhibited similar growth rates (Fig. 4A), while the complemented strains also grew at similar rates up to 10 µM IPTG, beyond which photosynthetic growth, but not aerobic growth, was severely negatively impacted (Fig. 4A, S5A Figure). This suggested a role for MppG in one or more aspects of photosynthesis.
Fig. 4. Physiological and genomic analysis of MppG regulation. 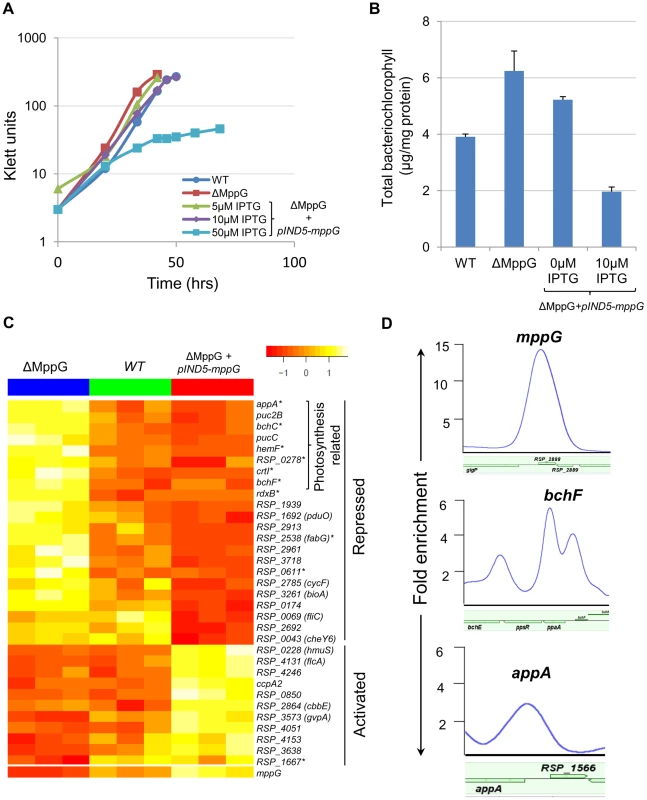
(A) Growth of WT, ΔMppG and ΔMppG+pIND5-mppG with increasing IPTG concentrations under photosynthetic conditions. (B) Amounts of bacteriochlorophyll produced in WT, ΔMppG and ΔMppG+pIND5-mppG. (C) Expression profiles of genes differentially expressed in response to the loss of MppG (ΔMppG) or over-expression of MppG (ΔMppG+pIND5-mppG) strains. Genes differentially expressed in the ΔMppG only are indicated with an asterisk (*). (D) ChIP-seq binding profile of MppG at the mppG, bchF and appA promoters. It appeared that MppG had some impact on photopigment biosynthesis as there was a reduction of colony pigmentation when this protein was expressed from an IPTG-inducible plasmid. To test this hypothesis, we assessed the total amount of bacteriochlorophyll in cells containing or lacking MppG. These experiments showed that the ΔMppG mutant strain produces >50% more bacteriochlorophyll than its WT parent (Fig. 4B). Furthermore, ectopic expression of MppG lowered the amount of cellular bacteriochlorophyll (a 2-fold decrease from WT levels at 10 uM IPTG), providing additional support for the role of MppG as a regulator of photopigment synthesis (Fig. 4B). These data also indicate that MppG functions to negatively modulate photopigment synthesis.
MppG is a transcriptional repressor of photopigment and other photosynthetic genes
Given the observed negative role of MppG on photopigment synthesis, we conducted global gene expression analysis on the WT, ΔMppG and ΔMppG +pIND5-mppG strains under photosynthetic conditions. Consistent with the above observations, a variety of photosynthesis related genes were significantly differentially expressed between these strains. A total of 17 genes other than MppG were differentially expressed (cut-off −1.5 fold, p<0.05) between the WT and ΔMppG (Fig. 4C, S5 Table), with 12 of these genes having photosynthesis related functions. Most of these differentially expressed genes include genes from 7 operons (appA, rdxBHIS, bchCXYZ, hemF, bchF, crtIB, RSP_0278). Furthermore, transcript levels from all but one of the differentially expressed genes were down regulated in the presence of MppG, indicating that it functions as a transcriptional repressor. This predicted negative regulatory function for MppG is consistent with the decreased photopigment phenotype seen when this TF is over-expressed. Given that MppG transcript levels are increased under photosynthetic growth conditions relative to aerobic conditions in WT cells, its function is likely to fine-tune photopigment synthesis, similar to the predicted role for the sRNA, PcrZ [28].
By comparing the global gene expression profile of the ΔMppG strain with that of cells ectopically expressing this TF, we found a total of 36 genes differentially expressed (1.5 FC, p<0.05), including 14 of the 17 genes that were differentially expressed between WT and ΔMppG (S6 Table). The other 22 genes that were differentially expressed when MppG was ectopically expressed included additional photosynthesis related genes like those encoding light-harvesting proteins (pucC and puc2B), as well as genes involved in functions ranging from iron and heme transport to fatty acid biosynthesis (Fig. 4C, S6 Table). Twenty six of these 36 differentially expressed genes were also down regulated when MppG was ectopically expressed, consistent with this TF functioning as a repressor of photosynthesis and other functions.
MppG directly represses transcription of genes encoding photosynthesis related proteins
To further test the predicted direct role of MppG in regulation of photosynthesis, we performed 2 independent ChIP-seq analyses using a ΔMppG strain containing a 3X myc-tagged variant of MppG that complements the phenotype of the parent strain (ΔMppG+pIND5-mppG-3Xmyc) (S5B Figure). This ChIP-seq analysis identified a total of 52 MppG binding sites across the genome. We found that most of the genes downstream of these binding sites were not differentially expressed in the presence or absence of MppG in any of global gene expression datasets. In addition, we were unable to identify any conserved DNA motif shared by a significant number of these target sites. Thus, to identify potential direct targets of MppG, only operons that were both differentially expressed in either of our global gene expression datasets and had a significant ChIP-seq peak were considered. When these criteria were applied, we identified 9 potential MppG target operons: bchCXYZ, bchFNBH, rdxBHIS, appA, RSP_1257-4, RSP_2692, mppG, RSP_2961 and RSP_3718 (Table 4, Fig. 4D). Transcript levels from all of these operons were lower in the presence of MppG and they represent the most high confidence direct MppG targets in our dataset. The 43 other sites which showed significant enrichment for MppG are provided in S7 Table.
Tab. 4. MppG target genes identified by ChIP-seq and gene expression analysis. 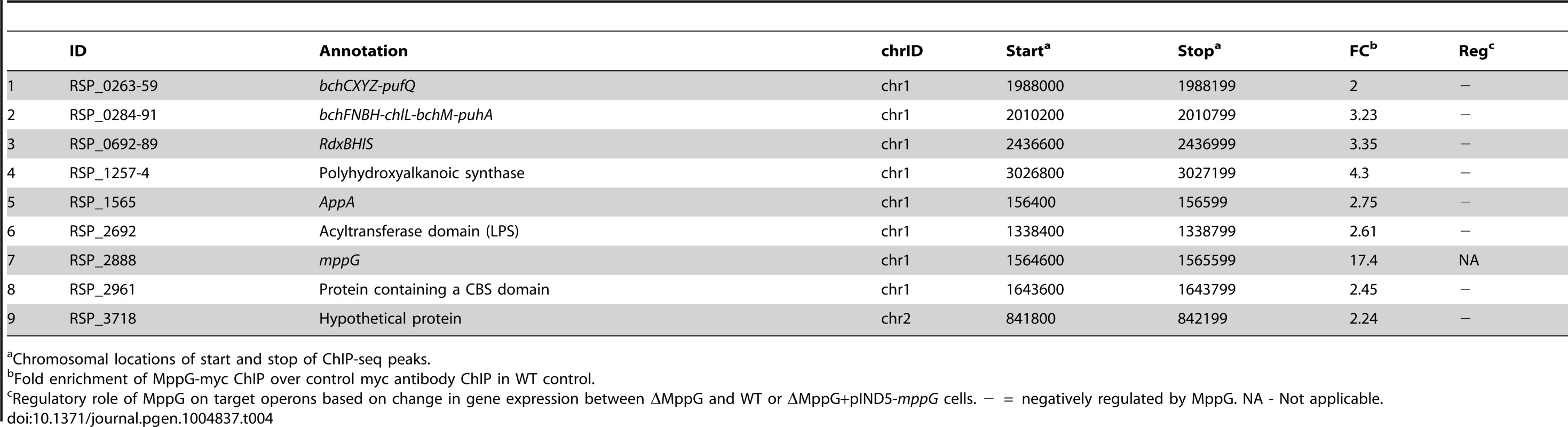
Chromosomal locations of start and stop of ChIP-seq peaks. Discussion
Our analyses have provided new information on the TRN controlling bacterial photosynthesis in R. sphaeroides. We confirmed the predicted involvement of two previously uncharacterized TFs, CrpK and MppG, in the regulation of photosynthesis related genes. We also extended the regulons of PrrA and FnrL, which had previously been implicated in regulation of the photosynthetic lifestyle of R. sphaeroides. Our analyses, combined with previous analyses of PrrA, FnrL and PpsR, illustrate the depth, complexity and robustness of the photosynthetic TRN. They also highlight significant combinatorial regulation of target genes, cross-talk between regulators and redundancy in the use of TFs within this network (Fig. 5), features that are likely to be broadly seen in the TRN of other biological systems.
Fig. 5. Photosynthetic gene regulatory network. 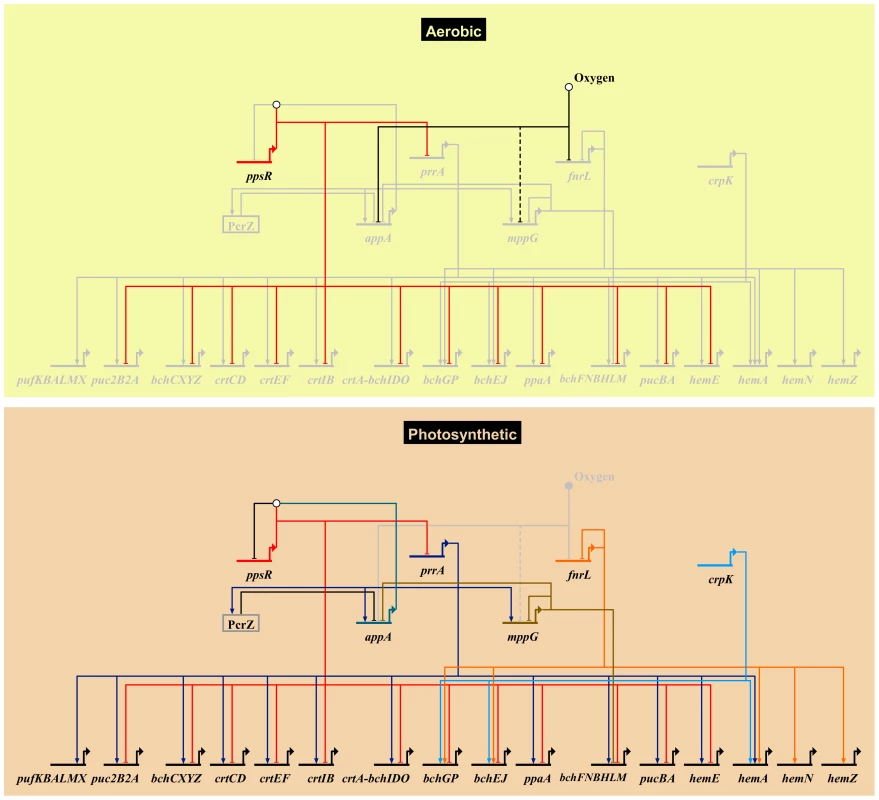
An overview of the R. sphaeroides photosynthetic gene regulatory network, showing known transcriptional regulators and their photosynthesis-related direct target genes. Grey nodes and edges indicate inactive genes and interactions. The top panel (Aerobic) depicts the regulatory control mechanism of photosynthesis genes under aerobic respiratory conditions in the dark. Under this condition the anti-repressor protein AppA, which has been proposed to be sensitive to oxygen and light, is inactivated. This allows its cognate repressor, PpsR, to downregulate the expression of several photosynthesis related genes including the gene encoding the transcription factor PrrA. This results in the inhibition of photosynthesis by preventing the production of photopigments. Molecular oxygen also inhibits the activity of the transcription factors FnrL and potentially MppG (the uncertain nature of effect is indicated by a dashed edge). Under anoxygenic photosynthetic conditions (bottom panel), AppA becomes active and directly interacts with PpsR(depicted by the white circle above ppsR) inhibiting its activity. In addition, the activators of photosynthesis, PrrA, FnrL and CrpK, become active under these conditions and induce the expression of photosynthetic genes. Under these conditions, the photopigment gene repressors MppG and the sRNA PcrZ are also active, negatively modulating photopigment gene expression. The expression of appA and the activity of its gene product is dependent on regulatory inputs from PrrA, MppG, PcrZ and oxygen. Biotapestry was used for network visualization [91]. New insights into regulation by PrrA and FnrL
Previous analysis of photosynthetic gene expression in R. sphaeroides had established the importance of 3 global TFs, PpsR, PrrA and FnrL, in the regulation of this lifestyle [8], [9], [19], [20], [24], [26], [27]. Our global analysis of these previously identified members of this TRN extends prior knowledge by comprehensively identifying the direct targets for two of these proteins, PrrA and FnrL. The work in this paper also, complements parallel genome-wide analysis of a global repressor of photosynthesis, PpsR [10].
For example, our analysis indicated the total number of genes, directly or indirectly, controlled by R. sphaeroides PrrA is ∼4 times smaller than previously reported, providing a picture of the PrrA regulon that is not influenced by apparent growth-rate differences between wild type and the mutant used previously [20]. In addition, our data verified the major direct role played by this TF in photosynthesis gene expression, significantly expanding previous analyses that reported the ability of PrrA to bind DNA in vitro at a handful of sites [32], [38], [39] Our data show that PrrA, controls expression of genes required for light energy capture, as well as a number of operons encoding proteins involved in electron transport both directly (e.g., fbcFBC complex, cytochrome B561 and cycA [40]) and indirectly (S3 Table).
Although we identified several new PrrA direct targets, we were unable to identify a strong consensus binding motif for this TF. While PrrA, and its analog in Rhodobacter capsulatus RegA, have been proposed to bind a degenerate GCG inverted repeat with a varying length spacer region, previous analyses have suggested that both DNA curvature and sequence specificity might contribute to target site recognition [23], [32]. These potential features, together with the GC-rich nature of the R. sphaeroides genome and the Fis-like nature of the PrrA DNA binding domain [23], possibly made it difficult to identify a shared motif among target genes from our analysis. Thus, despite identifying members of the PrrA regulon, it is not always possible to predict the consensus binding site for a given TF.
A large-scale reconstruction of the R. sphaeroides TRN predicted that the regulon of a second mater regulator, FnrL, was larger than previously described [24]. Our studies verified several of these predictions, significantly extending the size and function of genes in the FnrL regulon to include nitrogen regulatory proteins, iron sulfur assembly proteins, ABC transporters, additional TFs and recently identified sRNAs, all of which significantly increase the scope of genes and functions that are controlled by FnrL (see below). This illustrates how the predictions of a large-scale TRN can provide new insights into the functions that are regulated by even a previously well-studied TF.
Redundancy of the photosynthetic TRN
One of the previously uncharacterized TFs we tested for a role in the photosynthetic lifestyle was CrpK. Genome-wide analysis of CrpK targets revealed an overlapping but distinct regulon to that of FnrL, providing an explanation for the ability of CrpK to rescue the photosynthesis defect of an FnrL deletion strain.
While bacterial TRNs are often tightly controlled, they also need to be robust to allow cells to adapt to potentially deleterious changes to these networks. Given the central role of FnrL in regulating photosynthesis and a large number of anaerobic processes, the redundancy observed between this TF and the CrpK regulon might function to provide robustness to the R. sphaeroides photosynthetic TRN. Alternatively, CrpK might have a broader function under different conditions from FnrL. For instance, FnrL contains an O2-sensitive iron sulfur cluster that controls it DNA binding activity [24], so the absence of such a metal center in CrpK might allow this protein to function under conditions that would inactivate FnrL, facilitating photosynthesis or other metabolic functions during microaerophilic or semi-aerobic growth in nature. Interestingly, while FnrL binds upstream of dorS (encoding the histidine kinase of the DorSR two-component system involved in regulation of anaerobic respiratory growth on DMSO [41]), CrpK binding was not observed at this promoter (S4 Figure). This suggests that CrpK's ability to functionally replace FnrL might not extend to FnrL's role in regulation of anaerobic respiration.
In order to assess predicted cases of redundancy in a TRN or overlapping target genes of TFs, it is important to identify promoter elements that may allow discrimination between binding sites. For instance, although the predicted consensus motifs derived from the FnrL and CrpK binding sites were similar, the observation that both proteins can recognize unique, as well as overlapping sites, indicates there must be some subtle but functionally significant differences in DNA recognition by these TFs. Closer inspection of these DNA sequence motifs, suggest there may be a greater tolerance by FnrL for deviations from the TGA N6 TCA consensus, while bases within the spacer region or outside the core target site might play a role. Under the anaerobic photosynthetic growth conditions typically used in the lab, the CrpK transcript is present at a significantly lower level than that of FnrL. While it is possible that both proteins might compete for some shared binding sites under laboratory conditions, our analysis of a few shared or unique promoters suggested this was likely not a major factor in our experiments (S3E Figure). However, reproducibly greater FnrL enrichment was observed at the ccoN/RSP_0697 promoter in the absence of CrpK (S3D Figure), so the possibility that multiple TFs can compete for binding at selected sites cannot be ruled out without additional genetic and biochemical studies. This illustrates the need to couple predictions of TRN function with experimental studies.
Incoherent feed forward regulation of the photosynthesis TRN
The second photosynthesis-related TF characterized for the first time in this study was MppG. Our data showed that MppG functions as a direct transcriptional repressor of photopigment biosynthesis operons, including bchCXYZ and bchFNBHM, with high cellular levels of this protein inhibiting photosynthetic growth. In addition, transcripts from several other operons that encode photosynthesis-related functions were indirectly repressed by MppG. Our data predict that much of this indirect regulation of photosynthesis function is achieved through the direct regulation of the gene that encodes the anti-repressor, AppA, by MppG. Reduced cellular levels of AppA caused by the presence of MppG, would in principle cause accumulation of free PpsR under photosynthetic conditions, which would lead to repression of the photosynthesis-related genes that are PpsR targets (Fig. 5). Given that mppG transcript levels are significantly elevated during photosynthetic growth, its function in repressing photopigment synthesis would appear to be counterintuitive, similar to the observation for the sRNA, PcrZ [28]. Since no significant difference in photosynthetic growth was observed between WT and ΔMppG cells, the additional pigment produced in the ΔMppG mutant strain did provide increased fitness, potentially equating to a waste of cellular resources in the production of this extra pigment. In addition, the presence of excess photopigment could be a source of metabolic stress, especially since they can result in production of reactive oxygen species if light is present under microaerophilic conditions in the lab or in nature [42]. Thus, MppG may function as a negative modulator of pigment synthesis to ensure the optimal expression and tight coordination between expression of photopigment biosynthetic pathway genes and those for other components of the photosynthetic apparatus.
Newly-identified links between photosynthetic and iron homeostasis gene regulatory networks
In addition to its role in photosynthesis, MppG also regulates, either directly or indirectly, a variety genes encoding iron/heme dependent proteins (AppA, RdxBHIS, BchX, BchL, RSP_2785) and iron/heme transporters (RSP_2913, HmuS). MppG shares a high degree of amino acid sequence similarity to RirA, which was previously shown to regulate iron-responsive genes in Rhizobium leguminosarum [43], [44]. Thus, in addition to its role in regulation of photopigment synthesis, MppG appears to have a previously unidentified role in maintaining iron homeostasis during photosynthetic growth in R. sphaeroides. Furthermore, like RirA, MppG possesses a set of cysteine residues in its C-terminal region, which could coordinate an iron-sulfur cluster or some other metal, potentially allowing it to directly sense signals such as oxygen or metal availability.
Our data also provide new evidence that both FnrL and CrpK directly regulate genes encoding iron-dependent, iron transport and iron-sulfur biogenesis proteins, as well as several proteins involved in tetrapyrrole biosynthesis. In addition, we showed that FnrL directly activates expression of another RirA-like protein, RSP_3341, which has also been shown to directly regulate other iron dependent genes in R. sphaeroides [10]. Thus, we have provided new evidence that the TRNs and TFs controlling photosynthesis and iron homeostasis are tightly linked in R. sphaeroides. This link is likely, at least in part, due to the anaerobic anoxygenic mode of photosynthesis in R. sphaeroides, the sensitivity of Fe-S clusters to oxygen, and the involvement of a variety of iron-dependent proteins in light energy capture or other aspects of photosynthesis.
Cross-talk between TFs that regulate photosynthetic gene expression
For complex TRNs to function effectively, the components of the network often need to communicate with one another, and this is the case with the photosynthetic TRN. For example, our previous analysis of the PpsR regulon identified a PpsR binding site upstream of prrA, in an intra-operonic promoter shown to be occupied by σ70 under photosynthetic conditions [24]. If this PpsR binding site upstream of prrA is functional, it would provide an additional, previously unrecognized, mechanism to prevent aerobic expression of photosynthetic genes. We also found that PrrA directly activates both AppA and MppG, which in turn represses AppA, forming an incoherent feed-forward loop to control photosynthetic genes (Fig. 5) similar to the situation proposed for PcrZ [28]. Thus, our data provide new support for the previous hypotheses that the control of appA transcription serves as a major point of integration of regulatory signals, integrating opposing regulatory inputs from PrrA, MppG, PcrZ, oxygen and possibly other as of yet unidentified factors. Type 1 incoherent feedforward loops can enable significant acceleration in the expression of a target gene in response to a signal compared to simple activation [45]. Thus we predict that this network architecture likely results in a rapid response of cells to small environmental perturbations and allows optimal expression of photosynthetic genes under anaerobic conditions.
R. sphaeroides photosynthetic TRN as a multi-faceted network
Bacteria and other cells use a myriad of TRNs to respond to different types of stimuli, with these TRNs varying in depth and complexity [46], [47]. While the regulation of some cellular processes can be largely controlled by a TRN involving just one TF (for instance, LexA regulation of DNA repair in some bacteria [48]), other cellular processes involve the coordinated activities of multiple globally and locally acting TFs (e.g., the regulation of amino acid metabolism by ArgR, Lrp and TrpR in E. coli [49]). Generally, cellular processes which result in significant physiological or morphological changes (such as sporulation in Bacillus [50]–[53]) or are central to cell survival (such as central metabolism [54] or chemotaxis [55]) require highly interconnected TRNs involving multiple TFs and sensory components. The TRN network controlling photosynthesis in R. sphaeroides is multi-faceted, involving the activities of at least 5 TFs (including 4 global regulators) and one sRNA (Figs. 5, 6).
Fig. 6. Cellular processes regulated by photosynthesis global regulators. 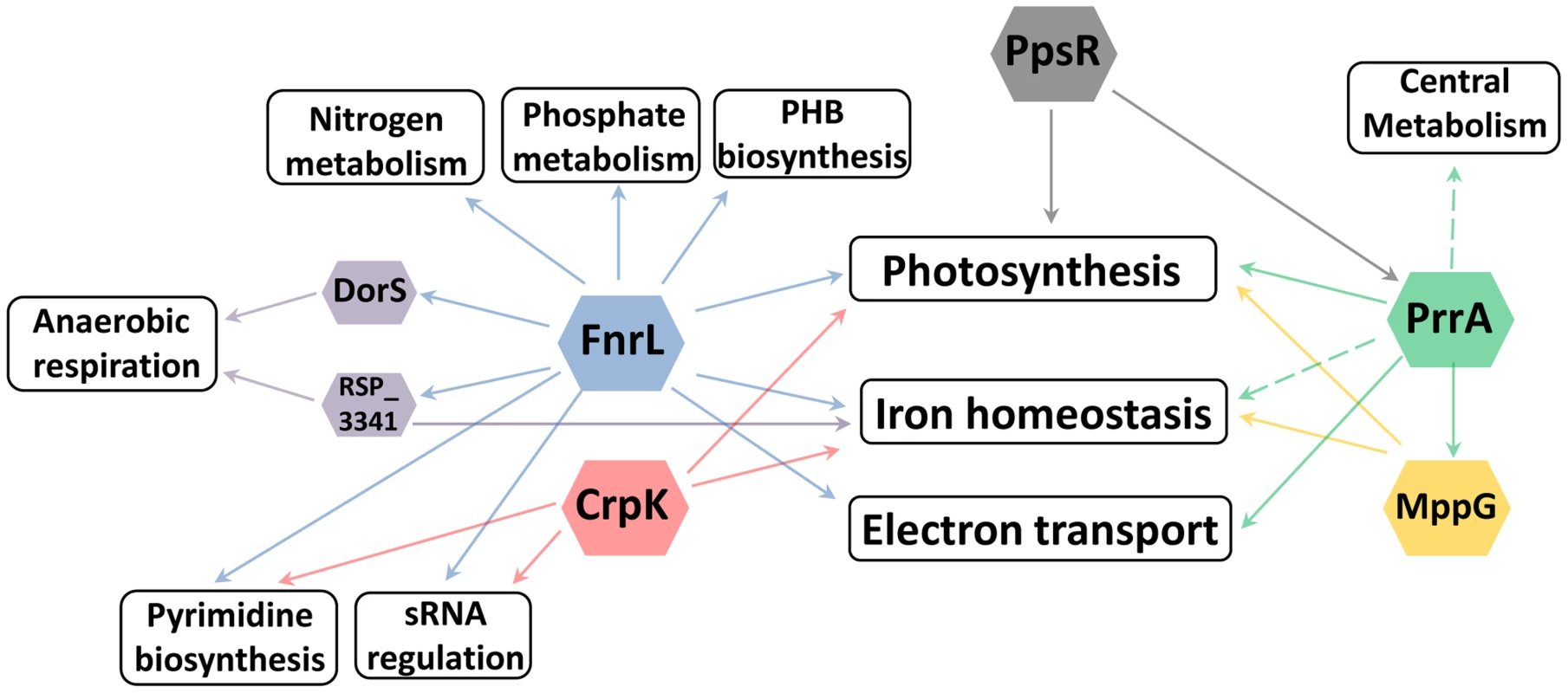
Overview of the different cellular processes coordinated by the TFs regulating photosynthesis in R. sphaeroides. It is not surprising to find that a photosynthetic TRN has these multiple elements given the number and types of functions that need to be coordinated to harvest and conserve the energy in sunlight. In addition, the transition to the photosynthetic lifestyle of R. sphaeroides involves profound changes gene expression [56], resulting in major physiological changes that culminate in the accumulation of photopigments and formation of specialized intracytoplasmic membrane structures to house the photosynthetic apparatus [57]. Achieving such seamless transitions requires the use of a multi-faceted TRN, parts of which have been described in this work.
Some aspects of the TRN controlling photosynthesis in R. sphaeroides show similarities to mechanisms employed in other photosynthetic cells, suggesting that design principles learned from this system will be applicable to other organisms. For instance, in the cyanobacterium Synechocystis sp., regulation of photosynthetic gene expression has been shown to be under the control of bacterial two-component systems, which are proposed to sense the redox state of the cell and coordinate the expression of the two photosystems (PSI and PSII) to maintain redox poise [58]–[60](a process referred to as photosystem stoichiometry adjustment [61]). In this organism, loss of the response regulator RppA or its cognate sensor kinase RppB, has been shown to result in dysfunctional regulation of PSI and PSII in response to changes in the redox state of the plastoquinone pool [58]. In addition, the sensor kinase Hik2 has also been proposed to sense redox signals [60] and activate its cognate response regulator Rre1 in cyanobacteria [62]. Homologs of Rre1 have been shown to bind to and regulate the expression of photosynthesis related genes in other organisms [60]. Hik2 has also been shown to interact with RppA, potentially controlling its activities in regulation of photosystem stoichiometry [62]. These characteristics could make these systems functionally analogous to the PrrAB system in R. sphaeroides, which has also been proposed to regulate photosynthetic gene expression in response to redox state of the ubiquinone pool [63]. A third cyanobacterial sensor kinase Hik33 is proposed to function as global regulator integrating multiple inputs from different environmental stress conditions including cold and nutrient stress, and high light intensities [60], [64]. Its cognate response regulator Rre26 has been shown to bind and/or regulate the expression of specific photosynthesis related genes [65], [66]. Thus, through the use of two-component systems sensing the cell's redox state, cyanobacteria are able to control expression of their photosynthetic apparatus.
Cyanobacteria are the ancestors of chloroplasts found in modern day algae and higher plants [61]. While the TRNs controlling photosynthetic gene expression are not as well characterized in plants, the nuclear-encoded homolog of Hik2, CSK (a chloroplast sensor kinase which has lost the conserved histidine residue that serves as the site of phosphorylation in bacterial sensor kinases), has been shown to be required for the normal expression of PSI and PSII in response to changes in the redox state of the chloroplast plastoquinone pool in Arabidopsis thaliana [61], [67]. While no cognate response regulator has been identified for CSK, it has been shown to directly interact with the chloroplast-encoded sigma factor SIG1 and PTK (plastid transcription kinase). CSK is proposed to phosphorylate SIG1 when the plastoquinone pool is oxidized resulting in transcriptional repression of PSI genes, while permitting expression of PSII genes [67]. Thus, aspects of the regulatory mechanisms that control transcription of photosynthetic genes appear to be conserved between purple bacteria, cyanobacteria and photosynthetic eukaryotes.
One link between cellular redox state and gene expression is achieved through the two-component PrrAB system in R. sphaeroides. However, in addition to this TF, our studies show that R. sphaeroides employs at least 4 other TFs to control photosynthesis. The requirement for these additional TFs is likely, in part, due to the anaerobic nature of photosynthesis in R. sphaeroides, requiring its photosynthetic TRN to incorporate additional systems that sense oxygen tensions and modulate gene expression to minimize production of reactive oxygen species that can damage cellular components [42]. Thus, the R. sphaeroides photosynthetic TRN likely integrates the signals of cellular redox state, oxygen tension and possibly light intensity [68] to achieve optimal expression of photosynthesis genes. It should be noted that in general, the TRNs controlling photosynthesis in cyanobacteria and higher plants, are not as well characterized as that of R. sphaeroides and thus the complexity of these systems are probably yet to be fully appreciated.
Coordinated regulation of other pathways by global transcriptional regulators of photosynthesis
Historically, TFs were often identified while studying individual metabolic or developmental pathways. Our results illustrate how the ability to globally predict and analyze the roles of TFs provides insight into previously unknown roles and connections between these proteins. For example, in characterizing the regulons of global regulators involved in the transcriptional control of photosynthesis in R. sphaeroides: PrrA, FnrL and CrpK, we found that each of these TFs directly or indirectly regulates a broad range of other cellular processes. Of the total number of operons directly regulated by these TFs, only 18 (∼53%), 5 (∼6%) and 3 (∼10%) (for PrrA, FnrL, and CrpK respectively), correspond to operons directly involved in photosynthesis, suggesting that these TFs may coordinate the control of photosynthesis with other processes that are beneficial or even required for the photosynthetic lifestyle. In addition to the previously noted example of iron homeostasis, FnrL is predicted to directly regulate the expression of the nitrogen regulatory proteins GlnB and GlnK (homologs of nitrogen regulatory protein P-II), which modulate the synthesis and activity of glutamine synthetase, implicating FnrL in the regulation of nitrogen metabolism [69], [70]. In addition, both FnrL and CrpK are predicted to directly regulate the expression of aspartate carbamoyltransferase (RSP_1002), which catalyzes the first step in the pyrimidine biosynthetic pathway. Furthermore, FnrL and CrpK also both directly regulate key enzymes in the electron transport chain including NuoA-N (RSP_0100-12) and CcoNOQP (Cbb3-type cytochrome c oxidase) (Table 3, S1 Table). On the other hand, PrrA is predicted to directly control the expression genes encoding a variety of electron transport chain enzymes including nuoA-N (RSP_0100-12), fbcFBC, fbcQ and cytochrome B561 (Table 2).
In addition to these metabolic functions, PrrA and FnrL are also predicted to directly regulate the expression of a number of other transcriptional regulators. For instance, PrrA directly activates both MppG and RSP_2854 (a TetR family TF) in addition to the anti-repressor of PpsR, AppA. On the other hand, FnrL is predicted to directly regulate 4 other TFs: DorS (a direct regulator of DMSO reductase), RSP_3341 (a direct regulator of nitrate reductase), RSP_4201 (an ArsR family TF) and RSP_1243 (a LacI family TF), as well as 2 proposed small RNAs (RSs0019 and RSs2461). While the cellular roles of some of these putative transcriptional regulators are unknown, the expression of their target genes is likely to be indirectly affected by FnrL and PrrA under appropriate conditions. Consistent with this, over 85% of the 303 differentially expressed genes between WT and ΔFnrL cells did not correspond to direct targets of FnrL. Instead this set of indirectly regulated genes included known direct target of DorS and RSP_3341, a variety iron transport and iron-dependent genes, genes involved in electron transport (such as quinol oxidases), nitrogen metabolism, and several photosynthetic genes. The set of indirectly controlled PrrA genes included those encoding several enzymes involved in central carbon metabolism (TCA cycle) and electron transport. These data predict that, through direct and indirect mechanisms, FnrL and PrrA serve to integrate and coordinate of the processes of photosynthesis, central metabolism, nitrogen metabolism, anaerobic respiration, electron transport, pyrimidine biosynthesis, PHB biosynthesis, phosphate metabolism and iron homeostasis during anaerobiosis (Fig. 6). These findings provide insight into previously unrecognized processes controlled by these TFs that could potentially be conserved by homologues of these TFs in other organisms.
In E. coli, the sRNA FnrS is directly regulated by FNR [71] and functions to repress several target RNAs under anaerobic conditions [72]. The two sRNAs (RSs0019 and RSs2461), which we found to be direct targets of FnrL, are yet to be functionally characterized [31] and do not share sequence similarities to FnrS. Additionally, the regulatory influence of FnrL on these target sRNAs could not be established in this study, as they are not represented on the R. sphaeroides Affymetrix gene chip used in our analyses. Nevertheless, one might also expect that the indirect targets of FnrL captured in our global expression analysis also includes downstream targets of these regulatory elements.
Interestingly, only about 50% of the genes predicted to be directly regulated by FnrL and CrpK were observed to be differentially expressed when the expression profile of the cognate deletion mutants were compared to that of wild type cells. Similar observations have previously been reported for another Crp family protein, FNR, in E. coli [71]. Thus, these observations could reflect the requirement of as yet uncharacterized TFs, which could function as condition-dependent co-activators, for controlling the expression of their target genes [71]. Alternatively, these observation could be the result of condition-dependent repression of specific operons by alternative TFs that obscured the regulatory influence of FnrL and CrpK on their target promoters under the specific conditions used for our global gene expression assays. Our analyses have also shown that several of the photosynthetic operons are under the control of multiple TFs, raising the possibility that the regulatory effect of each of these TFs, could potentially be compensated for by others at some of these operons, as shown for FnrL and CrpK. If this is the case, it could equip the cells with increased robustness in the expression of specific operons, obscuring the regulatory influence of individual TFs. This feature of the TRN would better enable cells to adapt to potentially deleterious changes.
Several of the genomic locations that were enriched for PrrA and MppG binding also did not show significant changes in gene expression under the conditions we tested and thus were not considered further as direct targets of these TFs (see Results). In addition, we observed that the TF enrichment at target sites was much lower for PrrA and MppG, than at FnrL and CrpK sites. These observations could reflect fundamental differences in the binding properties of these TFs. For instance, the predicted NMR structure of the PrrA DNA binding domain indicates that it is a Fis-like protein [23]. Previous studies in E. coli have shown that Fis, a nucleoid associated protein, binds to DNA in both a sequence specific and non-specific manner [73], [74] and that only about a fifth of bound sites are differentially expressed upon deletion of Fis [73]. If PrrA exhibits similar properties, it could account for the large number of bindings sites observed in our ChIP-seq data that do not correspond to genes that are differentially expressed in a PrrA-dependent manner.
Concluding remarks
Using a combination of genetic, genomic and physiological approaches, guided in large part by computational predictions from a large-scale reconstruction of the R. sphaeroides TRN, we obtained a significant amount of new knowledge about regulation of photosynthesis in R. sphaeroides. Our analyses highlight the important role computational predictions can play in guiding biological discovery, as novel components of the photosynthetic TRN, not previously identified using traditional approaches, were identified computationally, with those predictions serving as the basis for this work. We expect that predictions from this large-scale TRN will continue to provide new insights into other aspects of R. sphaeroides diverse metabolic and energetic lifestyles, including those involved in production of high-value commodities such as biofuel precursors. In addition, given the ancestral relationship of R. sphaeroides to plants and other oxygenic phototrophs, we predict that knowledge of this photosynthetic TRN will help inform parallel or future studies in other photosynthetic organisms. Integration of the available large-scale network models of metabolism and transcriptional regulation for R. sphaeroides, will broaden the predictive capabilities of these models and further guide future experimental efforts.
Materials and Methods
Bacterial strains and growth conditions
R. sphaeroides 2.4.1 was used as the parental (wild type) strain and all mutants were constructed in this background except the ΔfnrLΔcrpK double deletion strain which was constructed in an existing ΔfnrL mutant background [9] (S8 Table). E. coli DH5α was used as a plasmid host, and E. coli S17-1 was used to conjugate DNA into R. sphaeroides. R. sphaeroides cultures were incubated at 30°C in Sistrom's minimal medium (SMM) [75]. Anaerobic cultures were started from cells obtained from exponentially growing aerobic cultures. When required, the media was supplemented with 100 µM IPTG, 25 µg/mL kanamycin, 25 µg/mL spectinomycin, or 1 µg/mL tetracycline. E. coli cells were grown in Luria Bertani medium at 37°C, supplemented with 50 µg/mL kanamycin, 25 µg/mL spectinomycin, or 20 µg/mL tetracycline where needed [11].
Quantification of bacteriochlorophyll
Photosynthetic pigments were quantified in R. sphaeroides strains grown photosynthetically in screw cap tubes at a light intensity of ∼10 W/m2 as previously described [76]. Briefly, 5 mL of culture was centrifuged and supernatant discarded. Cells were resuspended in 100 µL of water, transferred to 15 mL glass corex centrifuge tubes held in centrifuge adaptors and covered with rubber stoppers to prevent exposure to light. 4.9 mL of a 7∶2 mixture of acetone and methanol was added to the cell suspension and vortexed thoroughly in the dark. Samples were centrifuged for 10 minutes at 10000 g. Absorbance of the supernatant was measured at 775 nm and total bacteriochlorophyll was determined as follows: Abs775 * total volume of sample (5 mL) * (bacteriochlorophyll molecular weight (914 g/mol)/bacteriochlorophyll millimolar extinction coefficient (75 mM−1 cm−1)). Total bacteriochlorophyll in each sample was normalized to total protein content of samples determined using the Lowry assay [77].
Construction of mutants and expression plasmids
All mutants constructed for this study contained in-frame markerless deletions, which were constructed as previously described [11], [78]. Briefly, regions spanning ∼1500 bp upstream and downstream of the target gene were amplified using sequence-specific primers containing restriction sites for EcoRI, XbaI or HindIII. These fragments were digested with the appropriate restriction enzymes and ligated into pK18mobsacB [79], which had been digested with EcoRI and HindIII, by three-way ligation to generate the various gene deletion constructs, which were confirmed by sequencing. The pK18mobsacB-based plasmids were separately mobilized from E. coli S17-1 into relevant R. sphaeroides strains. Cells in which the plasmid had successfully integrated into the genome via homologous recombination were identified by selection on SMM plates supplemented with kanamycin. These cells were then grown overnight in SMM without kanamycin [11]. Cells that had lost the sacB gene via a second recombination event were identified by growth on SMM plates supplemented with 10% sucrose [11]. Gene deletions were confirmed by PCR and sequencing with specific primers (S8 Table).
To construct plasmids for the ectopic expression of 3x Myc tagged proteins, we modified pIND5 [80] to include 3 copies of a codon optimized Myc tag (EQKLISEEDL – GAGCAGAAGCTGATCTCGGAGGAGGACCTG) within the plasmid's multiple cloning site. New multiple cloning sites were added to allowing tagging of proteins either C-terminally (NdeI-PstI-NcoI) or N-terminally (BamHI-SalI-BglII-HindIII). Individual expression plasmids were made by amplifying the target genes from the genome using sequence specific primers (S8 Table) containing restriction sites for NdeI and BglII, HindIII or BamHI for cloning into pIND5 [11] and NdeI/NcoI or BamHI/HindIII for cloning into pIND5-3xMyc. These DNA fragments were digested with the appropriate enzymes and cloned into pIND5 or pIND5-3xMyc digested with the same enzymes. These plasmids were conjugated from E. coli S17-1 into the relevant R. sphaeroides strains. Cells which harbor the desired plasmid were identified by selection on SMM plates supplemented with kanamycin [11].
Construction of lacZ reporter promoter fusions and β-galactosidase assays
To assay the activity of FnrL and CrpK in vivo, β-galactosidase assays were conducted, as previously described [78], [81], in ΔfnrLΔcrpK deletion strains containing different promoter-lacZ fusions integrated into the genome. To construct these reporter strains, ∼200–300 bp regions upstream of putative target genes (RSP_0281 (bchE), RSP_0696 (ccoN), RSP_0697 (usp), RSP_2346 and RSP_3341), were amplified from genomic DNA using specific primers having NcoI and XbaI restriction sites at their ends (S8 Table). The amplified DNA fragments, as well as a pSUP202 suicide vector containing a promoterless lacZ gene [78], were digested with NcoI-XbaI. DNA fragments containing the upstream regulatory sequences were cloned into pSUP202. These promoter-lacZ fusion plasmids were then individually conjugated into the ΔfnrLΔcrpK strain, generating single copy promoter-lacZ fusions integrated in the genome after selecting for the plasmid-encoded tetracycline resistance activity. The fnrL and crpK genes cloned into pIND5 were conjugated into individual reporter strains and cells harboring the reporter construct and the ectopic expression plasmid were identified by selection with tetracycline and kanamycin. These strains were grown aerobically by shaking 10 mL of culture in 125 mL conical flasks until exponential phase, then were treated with 100 µM IPTG for 3 hrs to increase expression of the indicated TF before measuring β-galactosidase activity as previously described [81].
To assess the contribution of specific bases to FnrL and CrpK activity, β-galactosidase assays were conducted in ΔfnrLΔcrpK double deletion strains containing reporter gene fusions of the RSP_0281 (bchE) upstream regulatory region with individual point mutations (see Results). These reporter strains were constructed as described above, with individual point mutations being generated by overlap extension PCR. β-galactosidase assays were conducted as described above [11], [78], [81].
RNA extraction, qRT-PCR and microarray analyses
RNA was isolated from exponential phase cultures of R. sphaeroides strains that were grown photosynthetically in 16 mL screw cap tubes or 500 ml cultures in roux bottles with bubbling (95% N2, 5% CO2) [11],[78]. RNA isolation and subsequent cDNA synthesis, labeling and hybridization to R. sphaeroides GeneChip microarrays (Affymetrix, Santa Clara, CA) were performed as previously described [82]. Microarray datasets were normalized by Robust Multichip Average (RMA) to log2 scale with background adjustment and quantile normalization [83]. Statistical analysis of normalized data to identify differentially expressed genes was done using the limma package [84]. Correction for multiple testing was done using Benjamini-Hochberg correction [85]. All analyses were conducted in the R statistical programming environment (http://www.R-project.org).
Chromatin immunoprecipitation analysis (ChIP-qPCR and ChIP-seq analysis)
R. sphaeroides cells were grown photosynthetically in 500 ml cultures (see above). For FnrL studies, 3 independent ChIP-seq experiments were conducted for WT cells grown photosynthetically with succinate (2 replicates) or acetate as sole carbon source. For tagged TFs, plasmids expressing the tagged variant of the gene from an IPTG inducible promoter, were cloned into the appropriate mutants (S8 Table). Protein expression was induced with IPTG concentrations (MppG (5 µM), PrrA (10 µM) and CrpK (10 µM)), which restored WT-like growth or pigmentation phenotypes. Cells were harvested at mid-exponential phase and chromatin immunoprecipitation was conducted as previously described [86], using polyclonal antibody against FnrL [24] or against the Myc epitope tag (ab9132, Abcam plc) for all other TFs analyzed. Immunoprecipitated DNA samples were PCR-amplified, gel purified (size selection ∼200 bp) and sequenced at the UW Biotechnoloy Center using the HiSeq 2000 sequencing system (Illumina, Inc). The initial 100 bp sequence tags were trimmed to 70 bp, to remove less reliable DNA sequences, and mapped to the R. sphaeroides strain 2.4.1 genome (ftp://ftp.ncbi.nih.gov/genomes/Bacteria/Rhodobacter_sphaeroides_2_4_1_uid57653/) using SOAP version 2.21 [87], allowing a maximum of 3 mismatches and no gaps. Peaks that represent potential TF binding sites were identified using MOSAiCS [88] at a false discovery rate of 0.05. The MOSAiCS analysis was conducted as a two-sample analysis, with control ChIP-seq data generated from ΔfnrL grown on acetate (for FnrL analysis), myc antibody ChIP in WT cells (for myc-tagged proteins) or input DNA. Only peaks that were called as significant using both input DNA and an appropriate ChIP control were considered as true peaks. Motifs were identified from within peak regions using MEME [89].
Accession numbers
All microarray and ChIP-seq datasets generated for this study have been deposited in GEO under the accession GSE58717.
Supporting Information
Zdroje
1. AtsumiS, HigashideW, LiaoJC (2009) Direct photosynthetic recycling of carbon dioxide to isobutyraldehyde. Nat Biotechnol 27 : 1177–1180.
2. GronenbergLS, MarcheschiRJ, LiaoJC (2013) Next generation biofuel engineering in prokaryotes. Curr Opin Chem Biol 17 : 462–471.
3. Hunter CN, Daldal F., Thurnauer, M C. and Beatty, J T. (2009) The purple phototrophic bacteria: Springer.
4. Peralta-YahyaPP, ZhangF, del CardayreSB, KeaslingJD (2012) Microbial engineering for the production of advanced biofuels. Nature 488 : 320–328.
5. Blankenship RE, Madigan MT, Bauer CE (1995) Anoxygenic photosynthetic bacteria; Blankenship R E, editor.1330 p.
6. GomelskyL, MoskvinOV, StenzelRA, JonesDF, DonohueTJ, et al. (2008) Hierarchical regulation of photosynthesis gene expression by the oxygen-responsive PrrBA and AppA-PpsR systems of Rhodobacter sphaeroides. J Bacteriol 190 : 8106–8114.
7. MackenzieC, ErasoJM, ChoudharyM, RohJH, ZengX, et al. (2007) Postgenomic adventures with Rhodobacter sphaeroides. Annu Rev Microbiol 61 : 283–307.
8. Zeilstra-RyallsJ, GomelskyM, ErasoJM, YeliseevA, O'GaraJ, et al. (1998) Control of photosystem formation in Rhodobacter sphaeroides. J Bacteriol 180 : 2801–2809.
9. Zeilstra-RyallsJH, KaplanS (1995) Aerobic and anaerobic regulation in Rhodobacter sphaeroides 2.4.1: the role of the fnrL gene. J Bacteriol 177 : 6422–6431.
10. Imam S (2014) Integrated modeling of metabolism and transcriptional regulation in Rhodobacter sphaeroides. Madison: University of Wisconsin-Madison. Available: http://gradworks.umi.com/36/18/3618539.html p.
11. ImamS, NogueraDR, DonohueTJ (2013) Global insights into energetic and metabolic networks in Rhodobacter sphaeroides. BMC Syst Biol 7 : 89.
12. ImamS, YilmazS, SohmenU, GorzalskiAS, ReedJL, et al. (2011) iRsp1095: a genome-scale reconstruction of the Rhodobacter sphaeroides metabolic network. BMC Syst Biol 5 : 116.
13. KhatipovE, MiyakeM, MiyakeJ, YAsada (1999) Polyhydroxybutyrate accumulation and hydrogen evolution by Rhodobacter sphaeroides as a function of nitrogen availability. Biohydrogen III: 157–161.
14. KienNB, KongIS, LeeMG, KimJK (2010) Coenzyme Q10 production in a 150-l reactor by a mutant strain of Rhodobacter sphaeroides. J Ind Microbiol Biotechnol 37 : 521–529.
15. KonturWS, ZiegelhofferEC, SperoMA, ImamS, NogueraDR, et al. (2011) Pathways involved in reductant distribution during photobiological H2 production by Rhodobacter sphaeroides. Appl Environ Microbiol 77 : 7425–7429.
16. Tabita FR (1995) The biochemistry and metabolic regulation of carbon metabolism and CO2-fixation in purple bacteria. In: Blankenship RE, Madigan MT, Bauer CE, editors. Anoxygenic photosynthetic bacteria. The Netherlands: Kluwer Academic Publishers.pp.885–914.
17. WahlundTM, ConwayT, TabitaFR (1996) Bioconversion of CO2 to ethanol and other compounds. American Chemical Society Division of Fuel Chemistry 3 : 1403–1405.
18. Yilmaz LS, Kontur WS, Sanders AP, Sohmen U, Donohue TJ, et al. (2010) Electron partitioning during light - and nutrient-powered hydrogen production by Rhodobacter sphaeroides. Bioenerg Res Volume: 55–66.
19. ErasoJM, KaplanS (1994) prrA, a putative response regulator involved in oxygen regulation of photosynthesis gene expression in Rhodobacter sphaeroides. J Bacteriol 176 : 32–43.
20. ErasoJM, RohJH, ZengX, CallisterSJ, LiptonMS, et al. (2008) Role of the global transcriptional regulator PrrA in Rhodobacter sphaeroides 2.4.1: combined transcriptome and proteome analysis. J Bacteriol 190 : 4831–4848.
21. ErasoJM, KaplanS (1995) Oxygen-insensitive synthesis of the photosynthetic membranes of Rhodobacter sphaeroides: a mutant histidine kinase. J Bacteriol 177 : 2695–2706.
22. DangelAW, TabitaFR (2009) Protein-protein interactions between CbbR and RegA (PrrA), transcriptional regulators of the cbb operons of Rhodobacter sphaeroides. Mol Microbiol 71 : 717–729.
23. LaguriC, Phillips-JonesMK, WilliamsonMP (2003) Solution structure and DNA binding of the effector domain from the global regulator PrrA (RegA) from Rhodobacter sphaeroides: insights into DNA binding specificity. Nucleic Acids Res 31 : 6778–6787.
24. DufourYS, KileyPJ, DonohueTJ (2010) Reconstruction of the core and extended regulons of global transcription factors. PLoS Genet 6: e1001027.
25. Zeilstra-RyallsJH, KaplanS (1998) Role of the fnrL gene in photosystem gene expression and photosynthetic growth of Rhodobacter sphaeroides 2.4.1. J Bacteriol 180 : 1496–1503.
26. BruscellaP, ErasoJM, RohJH, KaplanS (2008) The use of chromatin immunoprecipitation to define PpsR binding activity in Rhodobacter sphaeroides 2.4.1. J Bacteriol 190 : 6817–6828.
27. GomelskyM, KaplanS (1995) Genetic evidence that PpsR from Rhodobacter sphaeroides 2.4.1 functions as a repressor of puc and bchF expression. J Bacteriol 177 : 1634–1637.
28. MankNN, BerghoffBA, HermannsYN, KlugG (2012) Regulation of bacterial photosynthesis genes by the small noncoding RNA PcrZ. Proc Natl Acad Sci U S A 109 : 16306–16311.
29. GomelskyM, KaplanS (1995) appA, a novel gene encoding a trans-acting factor involved in the regulation of photosynthesis gene expression in Rhodobacter sphaeroides 2.4.1. J Bacteriol 177 : 4609–4618.
30. ParkDM, AkhtarMS, AnsariAZ, LandickR, KileyPJ (2013) The bacterial response regulator ArcA uses a diverse binding site architecture to regulate carbon oxidation globally. PLoS Genet 9: e1003839.
31. BerghoffBA, GlaeserJ, SharmaCM, VogelJ, KlugG (2009) Photooxidative stress-induced and abundant small RNAs in Rhodobacter sphaeroides. Mol Microbiol 74 : 1497–1512.
32. Ranson-OlsonB, JonesDF, DonohueTJ, Zeilstra-RyallsJH (2006) In vitro and in vivo analysis of the role of PrrA in Rhodobacter sphaeroides 2.4.1 hemA gene expression. J Bacteriol 188 : 3208–3218.
33. WillettJ, SmartJL, BauerCE (2007) RegA control of bacteriochlorophyll and carotenoid synthesis in Rhodobacter capsulatus. J Bacteriol 189 : 7765–7773.
34. BatemanA, CoinL, DurbinR, FinnRD, HollichV, et al. (2004) The Pfam protein families database. Nucleic Acids Res 32: D138–141.
35. AddleseeHA, FiedorL, HunterCN (2000) Physical mapping of bchG, orf427, and orf177 in the photosynthesis gene cluster of Rhodobacter sphaeroides: functional assignment of the bacteriochlorophyll synthetase gene. J Bacteriol 182 : 3175–3182.
36. AddleseeHA, HunterCN (1999) Physical mapping and functional assignment of the geranylgeranyl-bacteriochlorophyll reductase gene, bchP, of Rhodobacter sphaeroides. J Bacteriol 181 : 7248–7255.
37. OhJI, ErasoJM, KaplanS (2000) Interacting regulatory circuits involved in orderly control of photosynthesis gene expression in Rhodobacter sphaeroides 2.4.1. J Bacteriol 182 : 3081–3087.
38. ErasoJM, KaplanS (2009) Regulation of gene expression by PrrA in Rhodobacter sphaeroides 2.4.1: role of polyamines and DNA topology. J Bacteriol 191 : 4341–4352.
39. ErasoJM, KaplanS (2009) Half-Site DNA sequence and spacing length contributions to PrrA binding to PrrA site 2 of RSP3361 in Rhodobacter sphaeroides 2.4.1. J Bacteriol 191 : 4353–4364.
40. ComolliJC, CarlAJ, HallC, DonohueT (2002) Transcriptional activation of the Rhodobacter sphaeroides cytochrome c(2) gene P2 promoter by the response regulator PrrA. J Bacteriol 184 : 390–399.
41. MounceyNJ, KaplanS (1998) Cascade regulation of dimethyl sulfoxide reductase (dor) gene expression in the facultative phototroph Rhodobacter sphaeroides 2.4.1T. J Bacteriol 180 : 2924–2930.
42. ZiegelhofferEC, DonohueTJ (2009) Bacterial responses to photo-oxidative stress. Nat Rev Microbiol 7 : 856–863.
43. ToddJD, WexlerM, SawersG, YeomanKH, PoolePS, et al. (2002) RirA, an iron-responsive regulator in the symbiotic bacterium Rhizobium leguminosarum. Microbiology 148 : 4059–4071.
44. YeomanKH, CursonAR, ToddJD, SawersG, JohnstonAW (2004) Evidence that the Rhizobium regulatory protein RirA binds to cis-acting iron-responsive operators (IROs) at promoters of some Fe-regulated genes. Microbiology 150 : 4065–4074.
45. AlonU (2007) Network motifs: theory and experimental approaches. Nat Rev Genet 8 : 450–461.
46. BallezaE, Lopez-BojorquezLN, Martinez-AntonioA, Resendis-AntonioO, Lozada-ChavezI, et al. (2009) Regulation by transcription factors in bacteria: beyond description. FEMS Microbiol Rev 33 : 133–151.
47. BalajiS, BabuMM, AravindL (2007) Interplay between network structures, regulatory modes and sensing mechanisms of transcription factors in the transcriptional regulatory network of E. coli. J Mol Biol 372 : 1108–1122.
48. ButalaM, Zgur-BertokD, BusbySJ (2009) The bacterial LexA transcriptional repressor. Cell Mol Life Sci 66 : 82–93.
49. ChoBK, FederowiczS, ParkYS, ZenglerK, PalssonBO (2011) Deciphering the transcriptional regulatory logic of amino acid metabolism. Nat Chem Biol 8 : 65–71.
50. KroosL (2007) The Bacillus and Myxococcus developmental networks and their transcriptional regulators. Annu Rev Genet 41 : 13–39.
51. ZhengLB, LosickR (1990) Cascade regulation of spore coat gene expression in Bacillus subtilis. J Mol Biol 212 : 645–660.
52. Freyre-GonzalezJA, Manjarrez-CasasAM, MerinoE, Martinez-NunezM, Perez-RuedaE, et al. (2013) Lessons from the modular organization of the transcriptional regulatory network of Bacillus subtilis. BMC Syst Biol 7 : 127.
53. EichenbergerP, FujitaM, JensenST, ConlonEM, RudnerDZ, et al. (2004) The program of gene transcription for a single differentiating cell type during sporulation in Bacillus subtilis. PLoS Biol 2: e328.
54. ChubukovV, GerosaL, KochanowskiK, SauerU (2014) Coordination of microbial metabolism. Nat Rev Microbiol 12 : 327–340.
55. PorterSL, WadhamsGH, ArmitageJP (2011) Signal processing in complex chemotaxis pathways. Nat Rev Microbiol 9 : 153–165.
56. AraiH, RohJH, KaplanS (2008) Transcriptome dynamics during the transition from anaerobic photosynthesis to aerobic respiration in Rhodobacter sphaeroides 2.4.1. J Bacteriol 190 : 286–299.
57. KileyPJ, KaplanS (1988) Molecular genetics of photosynthetic membrane biosynthesis in Rhodobacter sphaeroides. Microbiol Rev 52 : 50–69.
58. LiH, ShermanLA (2000) A redox-responsive regulator of photosynthesis gene expression in the cyanobacterium Synechocystis sp. Strain PCC 6803. J Bacteriol 182 : 4268–4277.
59. AlfonsoM, PerewoskaI, KirilovskyD (2000) Redox control of psbA gene expression in the cyanobacterium Synechocystis PCC 6803. Involvement of the cytochrome b(6)/f complex. Plant Physiol 122 : 505–516.
60. PuthiyaveetilS, AllenJF (2009) Chloroplast two-component systems: evolution of the link between photosynthesis and gene expression. Proc Biol Sci 276 : 2133–2145.
61. PuthiyaveetilS, KavanaghTA, CainP, SullivanJA, NewellCA, et al. (2008) The ancestral symbiont sensor kinase CSK links photosynthesis with gene expression in chloroplasts. Proc Natl Acad Sci U S A 105 : 10061–10066.
62. SatoS, ShimodaY, MurakiA, KoharaM, NakamuraY, et al. (2007) A large-scale protein protein interaction analysis in Synechocystis sp. PCC6803. DNA Res 14 : 207–216.
63. SwemLR, GongX, YuCA, BauerCE (2006) Identification of a ubiquinone-binding site that affects autophosphorylation of the sensor kinase RegB. J Biol Chem 281 : 6768–6775.
64. MikamiK, KanesakiY, SuzukiI, MurataN (2002) The histidine kinase Hik33 perceives osmotic stress and cold stress in Synechocystis sp PCC 6803. Mol Microbiol 46 : 905–915.
65. AshbyMK, MullineauxCW (1999) Cyanobacterial ycf27 gene products regulate energy transfer from phycobilisomes to photosystems I and II. FEMS Microbiol Lett 181 : 253–260.
66. KappellAD, van WaasbergenLG (2007) The response regulator RpaB binds the high light regulatory 1 sequence upstream of the high-light-inducible hliB gene from the cyanobacterium Synechocystis PCC 6803. Arch Microbiol 187 : 337–342.
67. PuthiyaveetilS, IbrahimIM, JelicicB, TomasicA, FulgosiH, et al. (2010) Transcriptional control of photosynthesis genes: the evolutionarily conserved regulatory mechanism in plastid genome function. Genome Biol Evol 2 : 888–896.
68. YinL, DragneaV, FeldmanG, HammadLA, KartyJA, et al. (2013) Redox and light control the heme-sensing activity of AppA. MBio 4: e00563–00513.
69. EnglemanEG, FrancisSH (1978) Cascade control of E. coli glutamine synthetase. II. Metabolite regulation of the enzymes in the cascade. Arch Biochem Biophys 191 : 602–612.
70. van HeeswijkWC, RabenbergM, WesterhoffHV, KahnD (1993) The genes of the glutamine synthetase adenylylation cascade are not regulated by nitrogen in Escherichia coli. Mol Microbiol 9 : 443–457.
71. MyersKS, YanH, OngIM, ChungD, LiangK, et al. (2013) Genome-scale analysis of Escherichia coli FNR reveals complex features of transcription factor binding. PLoS Genet 9: e1003565.
72. DurandS, StorzG (2010) Reprogramming of anaerobic metabolism by the FnrS small RNA. Mol Microbiol 75 : 1215–1231.
73. ChoBK, KnightEM, BarrettCL, PalssonBO (2008) Genome-wide analysis of Fis binding in Escherichia coli indicates a causative role for A-/AT-tracts. Genome Res 18 : 900–910.
74. BetermierM, GalasDJ, ChandlerM (1994) Interaction of Fis protein with DNA: bending and specificity of binding. Biochimie 76 : 958–967.
75. SistromWR (1960) A requirement for sodium in the growth of Rhodopseudomonas spheroides. J Gen Microbiol 22 : 778–785.
76. Cohen-BazireG, SistromWR, StanierRY (1957) Kinetic studies of pigment synthesis by non-sulfur purple bacteria. J Cell Physiol 49 : 25–68.
77. LowryOH, RosebroughNJ, FarrAL, RandallRJ (1951) Protein measurement with the Folin phenol reagent. J Biol Chem 193 : 265–275.
78. DufourYS, ImamS, KooBM, GreenHA, DonohueTJ (2012) Convergence of the transcriptional responses to heat shock and singlet oxygen stresses. PLoS Genet 8: e1002929.
79. SchaferA, TauchA, JagerW, KalinowskiJ, ThierbachG, et al. (1994) Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145 : 69–73.
80. IndAC, PorterSL, BrownMT, BylesED, de BeyerJA, et al. (2009) Inducible-expression plasmid for Rhodobacter sphaeroides and Paracoccus denitrificans. Appl Environ Microbiol 75 : 6613–6615.
81. SchilkeBA, DonohueTJ (1995) ChrR positively regulates transcription of the Rhodobacter sphaeroides cytochrome c2 gene. J Bacteriol 177 : 1929–1937.
82. TavanoCL, PodevelsAM, DonohueTJ (2005) Identification of genes required for recycling reducing power during photosynthetic growth. J Bacteriol 187 : 5249–5258.
83. BolstadBM, IrizarryRA, AstrandM, SpeedTP (2003) A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 19 : 185–193.
84. Smyth G (2004) Applications in genetics and molecular biology 3: Berkeley Electronic Press.
85. BenjaminiY, HochbergY (1995) Controlling the false discovery rate - a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society Series B-Methodological 57 : 289–300.
86. DufourYS, LandickR, DonohueTJ (2008) Organization and evolution of the biological response to singlet oxygen stress. J Mol Biol 383 : 713–730.
87. LiR, YuC, LiY, LamTW, YiuSM, et al. (2009) SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics 25 : 1966–1967.
88. KuanPF, ChungD, PanG, ThomsonJA, StewartR, et al. (2011) A statistical framework for the analysis of ChIP-seq data. Journal of the American Statistical Association 106 : 891–903.
89. BaileyTL, BodenM, BuskeFA, FrithM, GrantCE, et al. (2009) MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res 37: W202–208.
90. HomannOR, JohnsonAD (2010) MochiView: versatile software for genome browsing and DNA motif analysis. BMC Biol 8 : 49.
91. LongabaughWJ, DavidsonEH, BolouriH (2009) Visualization, documentation, analysis, and communication of large-scale gene regulatory networks. Biochim Biophys Acta 1789 : 363–374.
Štítky
Genetika Reprodukční medicína
Článek Large-scale Metabolomic Profiling Identifies Novel Biomarkers for Incident Coronary Heart DiseaseČlánek Notch Signaling Mediates the Age-Associated Decrease in Adhesion of Germline Stem Cells to the NicheČlánek Phosphorylation of Mitochondrial Polyubiquitin by PINK1 Promotes Parkin Mitochondrial TetheringČlánek Natural Variation Is Associated With Genome-Wide Methylation Changes and Temperature SeasonalityČlánek Overlapping and Non-overlapping Functions of Condensins I and II in Neural Stem Cell DivisionsČlánek Unisexual Reproduction Drives Meiotic Recombination and Phenotypic and Karyotypic Plasticity inČlánek Tetraspanin (TSP-17) Protects Dopaminergic Neurons against 6-OHDA-Induced Neurodegeneration inČlánek ABA-Mediated ROS in Mitochondria Regulate Root Meristem Activity by Controlling Expression inČlánek Mutations in Global Regulators Lead to Metabolic Selection during Adaptation to Complex EnvironmentsČlánek The Evolution of Sex Ratio Distorter Suppression Affects a 25 cM Genomic Region in the Butterfly
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2014 Číslo 12
-
Všechny články tohoto čísla
- Stratification by Smoking Status Reveals an Association of Genotype with Body Mass Index in Never Smokers
- Genome Wide Meta-analysis Highlights the Role of Genetic Variation in in the Regulation of Circulating Serum Chemerin
- Occupancy of Mitochondrial Single-Stranded DNA Binding Protein Supports the Strand Displacement Mode of DNA Replication
- Distinct Genealogies for Plasmids and Chromosome
- Large-scale Metabolomic Profiling Identifies Novel Biomarkers for Incident Coronary Heart Disease
- Non-coding RNAs Prevent the Binding of the MSL-complex to Heterochromatic Regions
- Plasmid Flux in ST131 Sublineages, Analyzed by Plasmid Constellation Network (PLACNET), a New Method for Plasmid Reconstruction from Whole Genome Sequences
- Epigenome-Guided Analysis of the Transcriptome of Plaque Macrophages during Atherosclerosis Regression Reveals Activation of the Wnt Signaling Pathway
- The Inventiveness of Nature: An Interview with Werner Arber
- Mediation Analysis Demonstrates That -eQTLs Are Often Explained by -Mediation: A Genome-Wide Analysis among 1,800 South Asians
- Generation of Antigenic Diversity in by Structured Rearrangement of Genes During Mitosis
- A Massively Parallel Pipeline to Clone DNA Variants and Examine Molecular Phenotypes of Human Disease Mutations
- Genetic Analysis of the Cardiac Methylome at Single Nucleotide Resolution in a Model of Human Cardiovascular Disease
- Genetic Analysis of Circadian Responses to Low Frequency Electromagnetic Fields in
- The Dissection of Meiotic Chromosome Movement in Mice Using an Electroporation Technique
- Altered Chromatin Occupancy of Master Regulators Underlies Evolutionary Divergence in the Transcriptional Landscape of Erythroid Differentiation
- Syd/JIP3 and JNK Signaling Are Required for Myonuclear Positioning and Muscle Function
- Notch Signaling Mediates the Age-Associated Decrease in Adhesion of Germline Stem Cells to the Niche
- Mutation of Leads to Blurred Tonotopic Organization of Central Auditory Circuits in Mice
- The IKAROS Interaction with a Complex Including Chromatin Remodeling and Transcription Elongation Activities Is Required for Hematopoiesis
- RAN-Binding Protein 9 is Involved in Alternative Splicing and is Critical for Male Germ Cell Development and Male Fertility
- Enhanced Longevity by Ibuprofen, Conserved in Multiple Species, Occurs in Yeast through Inhibition of Tryptophan Import
- Phosphorylation of Mitochondrial Polyubiquitin by PINK1 Promotes Parkin Mitochondrial Tethering
- Recurrent Loss of Specific Introns during Angiosperm Evolution
- Natural Variation Is Associated With Genome-Wide Methylation Changes and Temperature Seasonality
- SEEDSTICK is a Master Regulator of Development and Metabolism in the Arabidopsis Seed Coat
- Overlapping and Non-overlapping Functions of Condensins I and II in Neural Stem Cell Divisions
- Unisexual Reproduction Drives Meiotic Recombination and Phenotypic and Karyotypic Plasticity in
- Tetraspanin (TSP-17) Protects Dopaminergic Neurons against 6-OHDA-Induced Neurodegeneration in
- ABA-Mediated ROS in Mitochondria Regulate Root Meristem Activity by Controlling Expression in
- Mutations in Global Regulators Lead to Metabolic Selection during Adaptation to Complex Environments
- Global Analysis of Photosynthesis Transcriptional Regulatory Networks
- Mucolipin Co-deficiency Causes Accelerated Endolysosomal Vacuolation of Enterocytes and Failure-to-Thrive from Birth to Weaning
- Controlling Pre-leukemic Thymocyte Self-Renewal
- How Malaria Parasites Avoid Running Out of Ammo
- Echoes of the Past: Hereditarianism and
- Deep Reads: Strands in the History of Molecular Genetics
- Keep on Laying Eggs Mama, RNAi My Reproductive Aging Blues Away
- Analysis of a Plant Complex Resistance Gene Locus Underlying Immune-Related Hybrid Incompatibility and Its Occurrence in Nature
- Epistatic Adaptive Evolution of Human Color Vision
- Increased and Imbalanced dNTP Pools Symmetrically Promote Both Leading and Lagging Strand Replication Infidelity
- Genetic Basis of Haloperidol Resistance in Is Complex and Dose Dependent
- Genome-Wide Analysis of DNA Methylation Dynamics during Early Human Development
- Interaction between Conjugative and Retrotransposable Elements in Horizontal Gene Transfer
- The Evolution of Sex Ratio Distorter Suppression Affects a 25 cM Genomic Region in the Butterfly
- is Required for Adult Maintenance of Dopaminergic Neurons in the Ventral Substantia Nigra
- PRL1, an RNA-Binding Protein, Positively Regulates the Accumulation of miRNAs and siRNAs in Arabidopsis
- Genetic Control of Contagious Asexuality in the Pea Aphid
- Early Mesozoic Coexistence of Amniotes and Hepadnaviridae
- Local and Systemic Regulation of Plant Root System Architecture and Symbiotic Nodulation by a Receptor-Like Kinase
- Gene Pathways That Delay Reproductive Senescence
- The Evolution of Fungal Metabolic Pathways
- Maf1 Is a Novel Target of PTEN and PI3K Signaling That Negatively Regulates Oncogenesis and Lipid Metabolism
- Formation of Linear Amplicons with Inverted Duplications in Requires the MRE11 Nuclease
- Identification of Rare Causal Variants in Sequence-Based Studies: Methods and Applications to , a Gene Involved in Cohen Syndrome and Autism
- Rrp12 and the Exportin Crm1 Participate in Late Assembly Events in the Nucleolus during 40S Ribosomal Subunit Biogenesis
- The Mutations in the ATP-Binding Groove of the Rad3/XPD Helicase Lead to -Cockayne Syndrome-Like Phenotypes
- Topoisomerase I Plays a Critical Role in Suppressing Genome Instability at a Highly Transcribed G-Quadruplex-Forming Sequence
- A Cbx8-Containing Polycomb Complex Facilitates the Transition to Gene Activation during ES Cell Differentiation
- Transcriptional Frameshifting Rescues Type VI Secretion by the Production of Two Length Variants from the Prematurely Interrupted Gene
- Association Mapping across Numerous Traits Reveals Patterns of Functional Variation in Maize
- Genome-Wide Analysis of -Regulated and Phased Small RNAs Underscores the Importance of the ta-siRNA Pathway to Maize Development
- Dissemination of Cephalosporin Resistance Genes between Strains from Farm Animals and Humans by Specific Plasmid Lineages
- The Tau Tubulin Kinases TTBK1/2 Promote Accumulation of Pathological TDP-43
- Germline Signals Deploy NHR-49 to Modulate Fatty-Acid β-Oxidation and Desaturation in Somatic Tissues of
- Microevolution of in Macrophages Restores Filamentation in a Nonfilamentous Mutant
- Vangl2-Regulated Polarisation of Second Heart Field-Derived Cells Is Required for Outflow Tract Lengthening during Cardiac Development
- Chondrocytes Transdifferentiate into Osteoblasts in Endochondral Bone during Development, Postnatal Growth and Fracture Healing in Mice
- A ABC Transporter Regulates Lifespan
- RA and FGF Signalling Are Required in the Zebrafish Otic Vesicle to Pattern and Maintain Ventral Otic Identities
- , and Reprogram Thymocytes into Self-Renewing Cells
- The miR9863 Family Regulates Distinct Alleles in Barley to Attenuate NLR Receptor-Triggered Disease Resistance and Cell-Death Signaling
- Detection of Pleiotropy through a Phenome-Wide Association Study (PheWAS) of Epidemiologic Data as Part of the Environmental Architecture for Genes Linked to Environment (EAGLE) Study
- Extensive Copy-Number Variation of Young Genes across Stickleback Populations
- The and Genetic Modules Interact to Regulate Ciliogenesis and Ciliary Microtubule Patterning in
- Analysis of the Genome, Transcriptome and Secretome Provides Insight into Its Pioneer Colonization Strategies of Wood
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Tetraspanin (TSP-17) Protects Dopaminergic Neurons against 6-OHDA-Induced Neurodegeneration in
- Maf1 Is a Novel Target of PTEN and PI3K Signaling That Negatively Regulates Oncogenesis and Lipid Metabolism
- The IKAROS Interaction with a Complex Including Chromatin Remodeling and Transcription Elongation Activities Is Required for Hematopoiesis
- Echoes of the Past: Hereditarianism and
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



