-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaGenome-Wide Analysis of DNA Methylation Dynamics during Early Human Development
DNA methylation reprogramming after fertilization is critical for normal mammalian development. Early embryos are sensitive to environmental stresses and a number of reports have pointed out the increased risk of DNA methylation errors associated with assisted reproduction technologies. Therefore, it is very important to understand normal DNA methylation patterns during early human development. Recent reduced representation bisulfite sequencing studies reported partial methylomes of human gametes and early embryos. To provide a more comprehensive view of DNA methylation dynamics during early human development, we report on whole genome bisulfite sequencing of human gametes and blastocysts. We show that the paternal genome is globally demethylated in blastocysts whereas the maternal genome is demethylated to a much lesser extent. We also reveal unique regulation of imprinted differentially methylated regions, gene bodies and repeat sequences during early human development. Our high-resolution methylome maps are essential to understand epigenetic reprogramming by human oocytes and will aid in the preimplantation epigenetic diagnosis of human embryos.
Published in the journal: . PLoS Genet 10(12): e32767. doi:10.1371/journal.pgen.1004868
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1004868Summary
DNA methylation reprogramming after fertilization is critical for normal mammalian development. Early embryos are sensitive to environmental stresses and a number of reports have pointed out the increased risk of DNA methylation errors associated with assisted reproduction technologies. Therefore, it is very important to understand normal DNA methylation patterns during early human development. Recent reduced representation bisulfite sequencing studies reported partial methylomes of human gametes and early embryos. To provide a more comprehensive view of DNA methylation dynamics during early human development, we report on whole genome bisulfite sequencing of human gametes and blastocysts. We show that the paternal genome is globally demethylated in blastocysts whereas the maternal genome is demethylated to a much lesser extent. We also reveal unique regulation of imprinted differentially methylated regions, gene bodies and repeat sequences during early human development. Our high-resolution methylome maps are essential to understand epigenetic reprogramming by human oocytes and will aid in the preimplantation epigenetic diagnosis of human embryos.
Introduction
In mammals, DNA methylation is essential for normal development and plays critical roles in repression of transposable elements, maintaining genome stability, genomic imprinting and X-chromosome inactivation. DNA methylation patterns are relatively stable in somatic cells but genome-wide reprogramming of DNA methylation occurs in primordial germ cells and preimplantation embryos [1]–[3]. During mouse preimplantation development, the maternal genome is passively demethylated in a replication-dependent manner while some oocyte-specific methylated regions maintain maternal allele-specific methylation at the blastocyst stage [4], [5]. In contrast, the paternal genome is actively and rapidly demethylated through the oxidation of 5-methylcytosine (5mC) to 5-hydroxymethylcytosine (5hmC) by ten-eleven translocation-3 [6]. In spite of the global demethylation, imprinted differentially methylated regions (DMRs) and some transposable elements (e.g. intracisternal A-particles (IAPs)) are specifically protected from demethylation [1].
During human preimplantation development, the paternal genome is reported to be actively demethylated as in the mouse [7], [8], but the regulatory mechanism and the genome-wide DNA methylation patterns in early embryos are not well understood. Recently, two studies employed reduced representation bisulfite sequencing (RRBS) of human gametes and early embryos to characterize the human methylome very early in development [7], [9]. According to these studies, the paternal genome is rapidly and globally demethylated after fertilization whereas demethylation of the maternal genome is more limited and some oocyte-specific methylated regions maintain monoallelic methylation during preimplantation development, similar to the mouse genome. RRBS is known to cover 5–10% of genomic CpGs, favoring those contained within CpG islands (CGIs) and promoter regions. To obtain an unbiased and more complete representation of the methylome during early human development, we performed whole genome bisulfite sequencing (WGBS) of human gametes and blastocysts that covered>70% of genomic CpGs. We found human-specific regulation of DNA methylation in various regions including oocyte-methylated CGIs, gene bodies and tandem repeat-containing regions.
Results
WGBS of human gametes and blastocysts
We performed WGBS of human oocytes, sperm, blastocysts and neonatal blood cells. For ethical reasons, we used only surplus germinal vesicle (GV) or metaphase I (MI) oocytes and blastocysts obtained from female patients undergoing in vitro fertilization (IVF) treatment. Sperm and blood cells were collected from healthy donors (see Materials and Methods for details). WGBS libraries were constructed using the amplification-free post-bisulfite adaptor tagging (PBAT) method [10] for all samples except the oocytes, which required PCR-amplification (PCR cycles = 10) to increase the read depth (Table 1). For each cell type, 87–96% of genomic CpGs were covered by at least one read, which was comparable to the reported methylome maps of mouse gametes [5], [11], [12] and human sperm [13]. We also compared two oocyte PBAT libraries prepared with and without PCR-amplification (Oocyte(+PCR) and Oocyte(−PCR)) (S1A Figure, S1B Figure, and Table 1). The methylation levels of individual CpGs were highly correlated (r = 0.83) between these two libraries. Furthermore, the average methylation levels were very similar: Oocyte(+PCR) at 53.1% versus Oocyte(−PCR) at 54.8%. These data demonstrate that our PCR-amplification protocol did not lead to significant bias in our data sets. Non-CpG methylation was observed in human oocytes, especially at CpA sites (mean = 5.6%), with a positive correlation between CpG and non-CpG methylation (S1C Figure, S1D Figure). Non-CpG methylation was not a significant feature of sperm or blastocysts (<1%). In the following analyses, only CpGs covered with ≥3 reads were considered for oocytes and those covered with ≥5 reads were considered for the other samples.
Tab. 1. Summary of whole genome bisulfite sequencing. 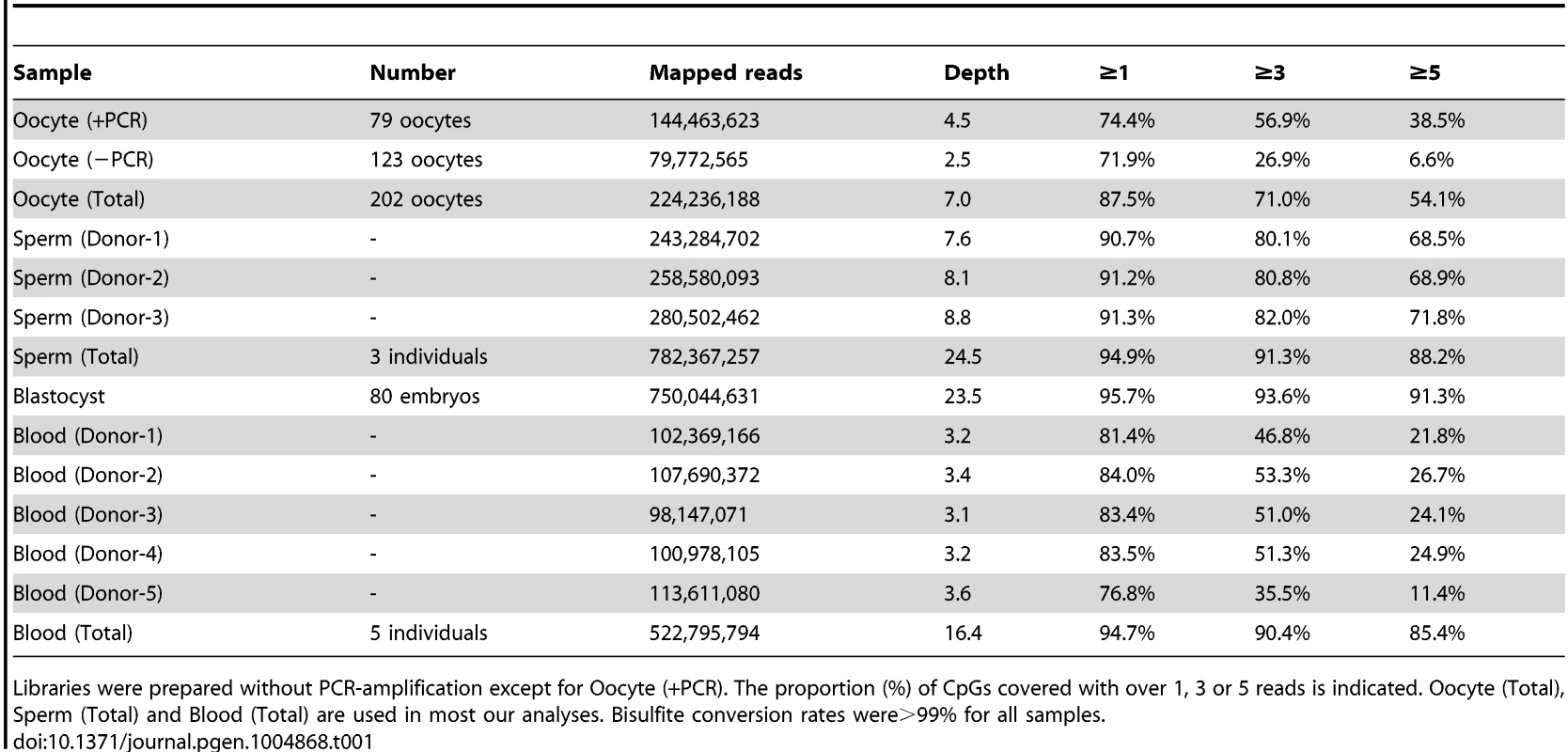
Libraries were prepared without PCR-amplification except for Oocyte (+PCR). The proportion (%) of CpGs covered with over 1, 3 or 5 reads is indicated. Oocyte (Total), Sperm (Total) and Blood (Total) are used in most our analyses. Bisulfite conversion rates were>99% for all samples. We confirmed that three imprinted DMRs and two pluripotency genes frequently observed to be abnormal in poor quality oocytes or embryos [14], [15] were normally methylated in our WGBS data (S1E Figure). We also compared our WGBS data with recently reported RRBS data of human oocytes, blastocysts and inner cell mass (ICM) and WGBS data of ICM [7], [9]. Our data substantially increased the coverage of genomic CpGs compared with the reported data (S1F Figure, S1G Figure). The methylation levels of CGIs showed high correlations (r = 0.96) between our WGBS data and the reported RRBS data (oocyte: S1H Figure, blastocyst: S1I Figure), validating the WGBS data.
Global changes of DNA methylation during early human development
Similar to findings for the mouse, human oocytes showed an intermediate methylation level of CpGs and blastocysts were globally hypomethylated (Fig. 1A). To further characterize global DNA methylation changes, we used a system of sliding windows of 20 CpGs with a step size change of 10 CpGs. Windows were classified as increasing (or decreasing) if the methylation levels increased (or decreased) by>20% and the changes were statistically significant (Benjamini-Hochberg (BH) corrected P<0.05). We found that 57% and 83% of windows showed decreased methylation levels in blastocysts compared with oocytes and sperm, respectively (Fig. 1B). In contrast,>90% of windows showed increased methylation in ES or blood cells compared with blastocysts (Fig. 1B). To explore the differences in demethylation dynamics between parental genomes, we focused on windows hypermethylated in one gamete and hypomethylated in the other. In this study, we defined regions that were ≥80% methylated as hypermethylated and those that were ≤20% methylated as hypomethylated. Windows hypermethylated in sperm and hypomethylated in oocytes (sperm-specific methylated windows) were abundant in intergenic regions. In contrast, oocyte-specific ones showed a relatively uniform distribution (S2A Figure). In blastocysts, oocyte-specific methylated windows showed intermediate methylation levels (median = 35.1%), in contrast to the nearly complete demethylation of sperm-specific ones (Fig. 1C). Almost all windows hypomethylated in both gametes remained hypomethylated and very few windows (0.04%) were hypermethylated in blastocysts, suggesting that genome-wide de novo methylation occurred after implantation (Fig. 1C and S2B Figure). Consistently, the methylation patterns of oocytes and blastocysts were very similar to each other (Figs. 1E, F), suggesting that the global methylation pattern of the maternal genome, but not the paternal genome, was inherited by blastocysts.
Fig. 1. Global changes of DNA methylation during early human development. 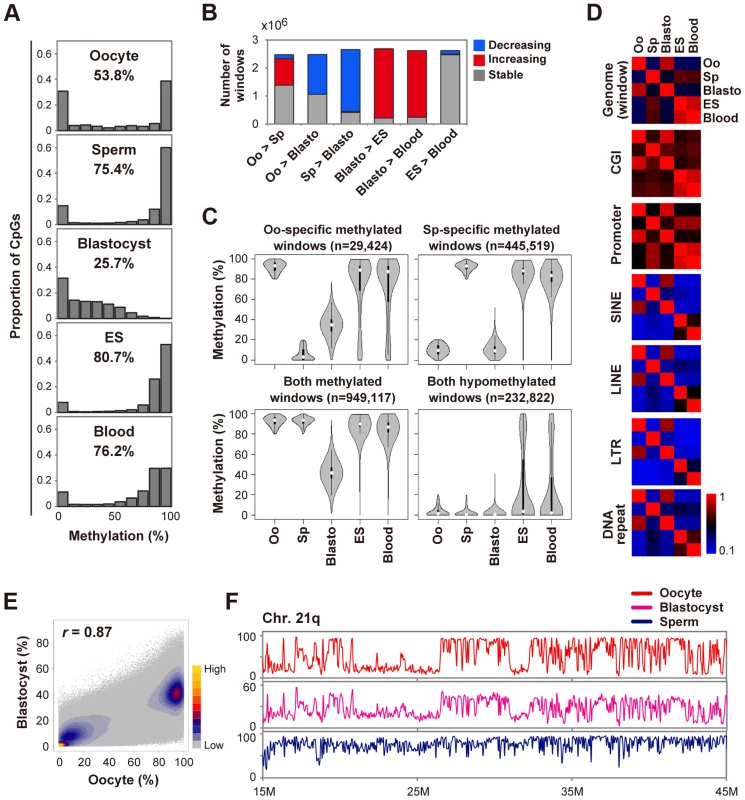
A, Distribution of methylation levels of individual CpGs. The mean methylation levels of CpGs are also indicated. We included human H9 ES cells (GEO accession number: GSM706059) for comparison. B, Detection of dynamic methylation changes using a sliding window (window size = 20 CpGs, step size = 10 CpGs). Windows were classified as increasing (or decreasing) if the methylation levels increased (or decreased) by>20% and the changes were significant (BH-corrected P<0.05). The other windows were classified as stable. Oo: Oocyte; Sp: Sperm; Blasto: Blastocyst. C, Violin plots of mean methylation levels of windows hypermethylated (≥80%) or hypomethylated (≤20%) in one or both gametes. Oo-specific (Sp-specific) methylated windows are defined as windows hypermethylated in oocytes (sperm) and hypomethylated in sperm (oocytes). Thin and thick lines are box plots and white dots indicate the median. D, Heatmaps of Pearson correlation coefficients. Correlation coefficients were calculated based on the mean methylation levels of individual windows, CGIs, promoters and repeat copies. Correlation coefficients are color-coded as shown. E, A density scatterplot of mean methylation levels of the sliding windows. The Pearson correlation coefficient between oocytes and blastocysts was high (r = 0.87). The density is color-coded as indicated. F, Methylation levels across the long arm of chromosome 21 (smoothed using 50 kb non-overlapping windows). Similar methylation patterns were observed for oocytes and blastocysts whereas the methylation levels of blastocysts were low (note that the vertical maximum scale is 60% for blastocysts). Next, we examined specific genomic features: CGIs, promoters and transposable elements. CGIs and promoters hypermethylated in sperm remained methylated in ES and blood cells. On the other hand, oocyte-specific methylated CGIs showed variable methylation levels and oocyte-specific methylated promoters were preferentially demethylated in ES and blood cells (S3A Figure and S3B Figure). In addition, the promoter methylation patterns of sperm, but not of oocytes, showed high correlations with those of ES and blood cells (r>0.8, Fig. 1D). These data highlighted the unique promoter methylation profile of oocytes. Short interspersed nuclear elements (SINEs), long interspersed nuclear elements (LINEs), long terminal repeats (LTRs) and DNA repeats were essentially highly methylated in ES and blood cells, whereas 20–30% and 3–8% of repeat copies were hypomethylated in oocytes and sperm, respectively (S2B Figure). These transposable elements were demethylated similarly to other genomic regions in blastocysts (S3 Figure).
Stability of imprinted DMRs and oocyte-specific methylated CGIs
Germline DMRs (gDMRs) frequently serve as imprinting control regions [16] and we were interested in how many gDMRs exist in the human genome. Among the 67 known imprinted DMRs [17], 46 DMRs were classified as gDMRs according to the following definition: DMRs hypermethylated in one gamete and hypomethylated in the other (Fig. 2A, B and S1 Table). Of these, 15 reportedly placenta-specific DMRs were lost in blood cells (Fig. 2A, C). The other 31 gDMRs showed intermediate methylation levels in blood cells, but about one-third of these gDMRs were not maintained in ES cells (H9 ES cells: Fig. 2A, H1 and HUES6 ES cells: S4A Figure), indicating the instability of gDMRs in human ES cells. Importantly, oocyte-specific methylated autosomal CGIs showed methylation levels very similar (median = 37.5%) to gDMRs (median = 39.2%) in human blastocysts (Fig. 2D). We confirmed monoallelic methylation of four autosomal CGIs in human blastocysts by using conventional bisulfite sequencing (Fig. 2E and S4B Figure). We also analyzed two X-linked CGIs hypermethylated in oocytes and found that these CGIs showed high methylation levels in male blastocysts (the X chromosome of male blastocysts is derived from oocytes) and monoallelic methylation in female blastocysts (Fig. 2F). Consistently, X-linked CGIs with oocyte-specific methylation showed higher methylation levels than autosomal ones in blastocysts (the WGBS data were derived from a pool of blastocysts) (Fig. 2D). A similar tendency was also observed in the sliding window-based analyses (S2C Figure). These data suggested that a substantial number of oocyte-specific methylated CGIs may maintain maternal allele-specific methylation in human blastocysts. In contrast, most oocyte-specific methylated CGIs were significantly demethylated compared with gDMRs in mouse blastocysts (Fig. 2D).
Fig. 2. Establishment and maintenance of imprinted DMRs. 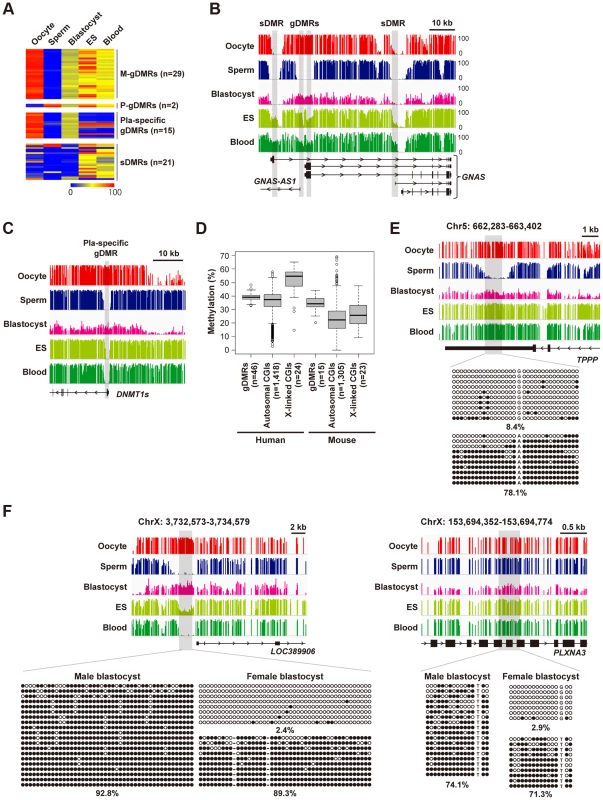
A, A heatmap of mean methylation levels of imprinted DMRs. We classified the 67 known human imprinted DMRs [17], and found that 44 were maternal germline DMRs (M-gDMRs), 2 were paternal germline DMRs (P-gDMRs) and 21 were secondary DMRs (sDMRs). 15 M-gDMRs are reported to be maintained only in the placenta and shown as “Pla-specific gDMRs”. gDMRs other than placenta-specific ones showed 35–65% methylation levels in blood cells but the intermediate methylation levels were not well maintained in ES cells (11/31 showed>75% methylation). Methylation levels are color coded as indicated. The raw data are shown in S1 Table. B, Methylation patterns at the human GNAS locus. The vertical axis indicates the methylation level (%). In this locus, there were two gDMRs and two sDMRs. All DMRs overlap promoter regions. C, Methylation patterns at the human DNMT1 locus. The promoter region of the somatic isoform of DNMT1 (DNMT1s) is known to show maternal allele-specific methylation in the placenta [45]. The DNMT1 DMR was hypomethylated in both ES and blood cells, suggesting placenta-specific protection of the maternal allele from demethylation. D, Box plots of mean methylation levels of gDMRs and oocyte-specific methylated CGIs in blastocysts. Boxes represent lower and upper quartiles and horizontal lines indicate the median. Whiskers extend to the most extreme data points within 1.5 times the interquartile range from the boxes. The open circles indicate the data points outside the whiskers. Methylation levels of mouse gDMRs and oocyte-specific methylated CGIs [5] are shown for comparison. E, Methylation patterns of an oocyte-specific methylated CGI. A single blastocyst was used for the analysis. Black and white circles indicate methylated and unmethylated residues, respectively. The percentages of methylated CpG sites are indicated. F, Bisulfite sequencing analyses of X-linked CGIs hypermethylated in oocytes. A single blastocyst was used for each bisulfite sequencing analysis. A bimodal gene body methylation pattern associated with transcription in human oocytes
In mouse oocytes, gene-body methylation levels are reported to positively correlate with the transcription levels [5]. In human oocytes, a positive correlation between gene-body methylation and transcription levels was also observed. Interestingly, there was an expression-level boundary at around log2(RPKM) = −5 (RPKM: reads per kilobase per million) (Fig. 3A). Genes with log2(RPKM)>−5 and <−5 may be transcriptionally active and inactive genes, respectively (Fig. 3B). We analyzed previously reported mouse methylome and transcriptome data and found that a bimodal distribution of gene body methylation was also observed while there was a boundary at around log2(RPKM) = 0 (Fig. 3A). It is unclear whether the difference between the human and mouse expression-level boundaries reflects experimental or functional differences. We found that 971 genes showed differential gene body methylation between human and mouse oocytes (Fig. 3C and S2 Table). Gene ontology (GO) analysis revealed an abundance of genes encoding cell adhesion molecules with human-specific gene body hypermethylation (Fig. 3D), which could have important roles during human oogenesis. In mouse oocytes, Dnmt3l and Zfp57 are highly expressed and essential for DNA methylation regulation [18], [19] whereas human DNMT3L is undetectable in oocytes [20]. Here we found that the gene body regions of DNMT3L and ZFP57 were hypomethylated in human oocytes and neither gene was expressed (Figs. 3E, F), implying that DNMT3L and ZFP57 might not be essential for regulation of DNA methylation in human oocytes.
Fig. 3. A bimodal gene body methylation pattern associated with transcription in human oocytes. 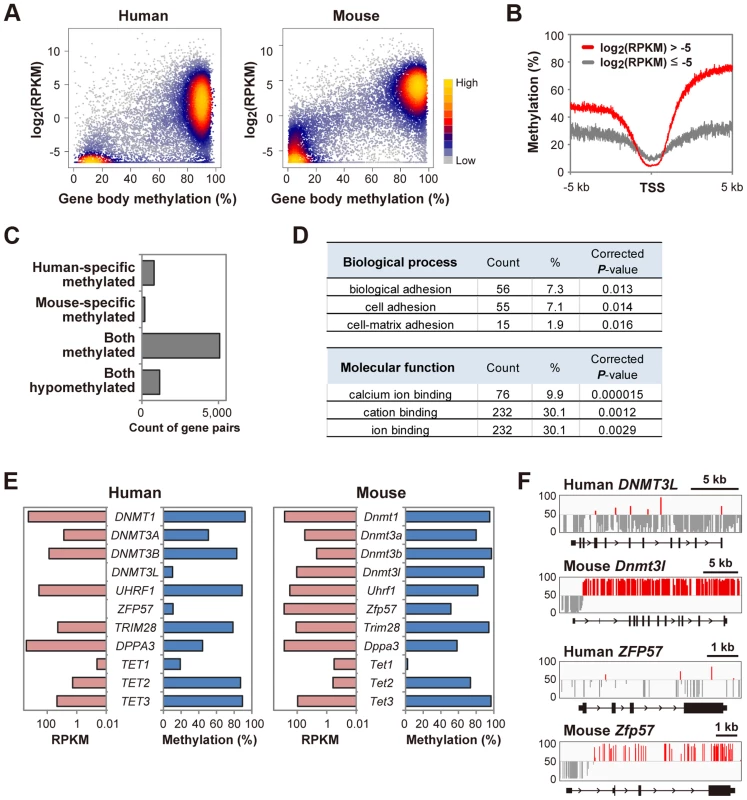
A, A density scatterplot of gene body methylation levels and transcription levels [43] in human oocytes. The data of mouse oocytes [5], [11] are also shown for comparison. Only genes longer than 5 kb were analyzed. For genes with RPKM less than 0.01, RPKM was set as 0.01. The density is color-coded as indicated. B, Mean methylation levels within 5 kb of transcription start sites (TSS) in human oocytes. Genes (>5 kb) were classified into two groups (log2(RPKM)>−5 and ≤−5). Methylation levels were smoothed using 5 bp non-overlapping sliding windows. C, Conservation of gene body methylation levels between human and mouse oocytes. 783 and 188 genes showed human-specific and mouse-specific gene body hypermethylation, respectively. 5076 and 1151 genes were hypermethylated and hypomethylated in both types of oocytes, respectively. The raw data are shown in S2 Table. D, GO analysis of 783 genes with human-specific gene body hypermethylation. The top three GO terms (biological process and molecular function) are indicated with gene counts, the proportion (%) and BH-corrected P-values. No GO term was enriched in genes with mouse-specific gene body hypermethylation. E, Gene body methylation levels and transcription levels of DNA methylation regulators in human and mouse oocytes. DNMT3L and ZFP57 showed gene body hypomethylation and were not expressed (RPKM<0.01) in human oocytes. DNMT3B (RPKM = 76.0) showed 10-fold higher expression than DNMT3A (RPKM = 7.6) in human oocytes. In contrast, Dnmt3b (RPKM = 4.9) showed ∼6-fold lower expression than Dnmt3a (RPKM = 30.6) in mouse oocytes. F, Methylation patterns at human DNMT3L and ZFP57 loci and mouse Dnmt3l and Zfp57 loci. The vertical line indicates the methylation level (%) and the baseline is set at 50% to highlight unmethylated CpGs. CpGs with>50% and <50% methylation are shown in red and grey, respectively. Unique regulations of tandem repeat-containing regions
As described above, global methylation changes of SINEs, LINEs, LTRs and DNA repeats were very similar to other genomic regions in early human embryos (S3 Figure). We further analyzed mean methylation levels of CpGs in various classes of these transposable elements (Fig. 4A, see also S3 Table for details). These repeat classes showed similar methylation changes: ∼60% methylated in oocytes, ∼80% methylated in sperm, ES and blood cells and ∼30% methylated in blastocysts. These data suggested that SINEs, LINEs, LTRs and DNA repeats were essentially not resistant to genome-wide demethylation after fertilization. Mouse IAPs are known to be protected from demethylation during preimplantation development [5], [21]. To identify transposable elements specifically protected from demethylation during human preimplantation development, we screened repeat copies overlapping windows showing>70% methylation in blastocysts (0.3% of all windows) (S4 Table). We found that SINE-VNTR-Alu (SVA) subfamilies, especially SVA_A, frequently overlapped the>70% methylated windows (Fig. 4B). SVA_A also showed the highest methylation level in blastocysts (59.2%) whereas the other repeat sequences were <50% methylated (Fig. 4A and S3 Table). SVA is a hominid-specific repeat family that remains active in the human genome [22]. Similar to mouse LTRs [5], methylation levels of CpGs within SVAs are positively correlated with CpG density in human oocytes and blastocysts (Fig. 4C and S5 Figure). LTR12 subfamilies, which are LTRs of HERV9, also tended to overlap the>70% methylated windows (Fig. 4B). Interestingly, both SVA and LTR12 subfamilies contain CpG-rich variable number tandem repeats (VNTRs) [22], [23]. We also noticed that whereas the MER34C2 consensus sequence does not contain VNTRs, MER34C2 copies overlapping the>70% methylated windows were all tandemly repeated in a single genomic locus (Fig. 4D). VNTRs were also found in the two paternal gDMRs (Fig. 4E). VNTRs were not a common feature of the maternal gDMRs, but a significantly higher proportion of the maternal gDMRs did contain VNTRs as compared with all CGIs (gDMRs: 11/44, CGIs: 1763/27718, chi-square P = 4.1×10−7). Therefore, we focused on CGIs hypermethylated in both gametes and found that CGIs containing VNTRs were preferentially protected from demethylation in blastocysts (Fig. 4F). A comparison between VNTRs of>70% and <50% methylated CGIs in blastocysts revealed that VNTRs with more repeats tended to be protected from demethylation, whereas no sequence motif was found (Fig. 4G). These data suggested that VNTRs might underlie silencing of specific transposable elements and the protection of paternal gDMRs.
Fig. 4. Unique regulation of tandem repeat-containing regions. 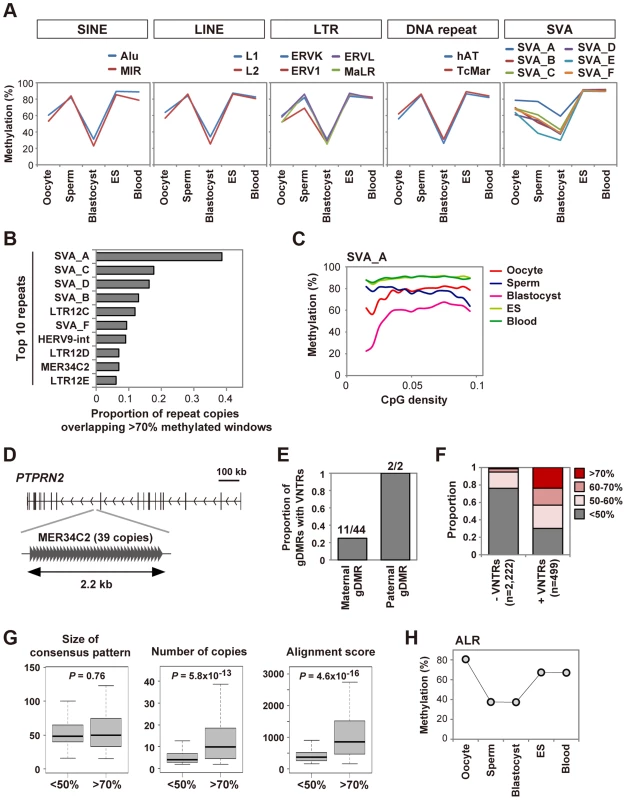
A, DNA methylation dynamics of transposable elements. Mean methylation levels of CpGs in various classes of SINEs, LINEs, LTRs and DNA repeats and SVA subfamilies are shown. SVA_A showed an especially high methylation level in blastocysts (59.2%). B, Proportions of repeat copies overlapping>70% methylated windows in human blastocysts. We analyzed only SINEs, LINEs, LTRs, DNA repeats, SVAs and satellites with>100 copies in the human genome. The top ten repeat names with the highest proportions are shown. The raw data are shown in S4 Table. C, Relationships between methylation levels and CpG densities. Mean methylation levels of CpGs in SVA_A are plotted against CpG densities. D, MER34C2 copies overlapping>70% methylated windows in human blastocysts. 39 MER34C2 copies are all tandemly repeated within the PTPRN2 gene locus. E, Proportions of maternal and paternal gDMRs containing VNTRs. Counts of gDMRs with VNTRs and total gDMRs are indicated. F, Proportions of mean methylation levels of CGIs with and without VNTRs in human blastocysts. Only autosomal CGIs hypermethylated in both gametes were analyzed. 118 of 499 CGIs with VNTRs and 31 of 2,222 CGIs without VNTRs showed>70% methylation (P = 0, chi-square test). G, Characteristics of VNTRs highly methylated in blastocysts. Using Tandem Repeats Finder [41], the size of the consensus pattern, the number of tandemly aligned copies and the alignment score were compared between VNTRs of <50% methylated CGIs and>70% methylated CGIs shown in (F). The alignment score calculated by Tandem Repeat Finder reflects the degree of similarity between repeat copies. When several VNTRs were found in a CGI, the VNTR with the highest alignment score was analyzed. Boxes represent lower and upper quartiles and horizontal lines indicate the median. Whiskers extend to the most extreme data points within 1.5 times the interquartile range from the boxes. The Mann-Whitney U test was used to calculate P-values. No sequence motif was found among the consensus patterns of the>70% methylated CGIs using DREME [42]. H, Mean methylation levels of CpGs in ALR. Oocytes showed the highest methylation level (80.6%). We also found that alpha satellite (ALR), which is a tandemly repeated DNA family found in centromeric and pericentromeric regions [24], was hypermethylated in human oocytes (80.6%) (Fig. 4H). Interestingly, DNMT3B was highly expressed in human oocytes (Fig. 3E), and DNMT3B is reported to interact with centromere protein CENP-C and contribute to DNA methylation of ALR [25]. Thus, it is possible that DNMT3B is involved in DNA methylation of ALR in human oocytes.
Discussion
This work reports the genome-wide DNA methylation patterns of human gametes and blastocysts at single-base resolution. Our WGBS data of oocytes and blastocysts substantially increase the coverage of genomic CpGs adding to the reported RRBS data of oocytes and blastocysts and WGBS data of ICM [7], [9]. We confirmed that the paternal genome was globally demethylated as previously reported. However, the oocyte-specific methylated regions maintained intermediate methylation levels in human blastocysts (median = 35.1%). Consistently, the methylation patterns of oocytes and blastocysts were very similar to each other, suggesting that the global methylation pattern of the maternal genome was inherited by blastocysts. Furthermore, oocyte-specific methylated CGIs showed methylation levels very similar (median = 37.5%) to gDMRs (median = 39.2%). These data appear not to support replication-dependent global demethylation of the maternal genome during human early development, because oocyte-specific methylated regions should show ≤25% methylation after one replication-dependent global demethylation event. In mouse blastocysts, most oocyte-specific methylated CGIs were significantly demethylated compared with gDMRs, which may reflect the passive demethylation of the maternal genome [1], [2]. These data strongly suggest that the maternal genome is demethylated to a much lesser extent in human blastocysts than in mouse blastocysts.
We classified known imprinted DMRs [17] and discovered that there were at least 46 gDMRs in the human genome including 15 specific to the placenta. Our data suggested that a substantial number of oocyte-specific methylated CGIs may also maintain mono-allelic methylation in human blastocysts whereas they were essentially lost through hypermethylation or hypomethyaltion in blood cells. It is suggested that a significant portion of gene transcripts show mono-allelic expression in human 8-cell embryos and morulae [26], and the oocyte-specific methylated CGIs could regulate mono-allelic expression of some genes in human preimplantation embryos. In the mouse genome, ∼25 well defined gDMRs have been identified and only the Gpr1 DMR is reported to be placenta-specific [27], [28]. The demethylation resistance of oocyte-specific methylated CGIs during early human development may, in part, explain the increased number of placenta-specific gDMRs in the human genome. Interestingly, we found that ZFP57 was not expressed in human oocytes. Because replication-dependent global demethylation of the maternal genome is not likely to occur during human preimplantation development, we speculate that the protection of gDMRs by ZFP57 may be dispensable in human oocytes. These data contribute to our understanding of the regulatory mechanism of human-specific genomic imprinting.
Both human and mouse oocytes showed bimodal gene body methylation patterns associated with transcription. While it is unclear whether transcription is the only determinant, transcription may be an important determinant of the oocyte methylomes. In mammals, DNMT3A and DNMT3B are de novo DNA methyltransferases whereas DNMT3L acts in a recruiting role. In mouse oocytes, Dnmt3a and Dnmt3l are essential for de novo DNA methylation, whereas Dnmt3b is poorly expressed and essentially dispensable [11], [29]. In contrast, in human oocytes DNMT3B showed ∼10-fold higher expression than DNMT3A, and DNMT3L was not expressed, suggesting that DNMT3B may be the critical de novo DNA methyltransferase during human oocyte growth. Interestingly, centromeric satellite repeats were highly methylated in human oocytes. These regions are known to be hypomethylated in mouse oocytes [30]. Human DNMT3B is reported to interact with centromere protein CENP-C and contribute to DNA methylation of centromeric satellite repeats [25]. Similarly, centromeric satellite repeats are demethylated in Dnmt3b mutant mice [31]. Therefore, the differential expression pattern of DNMT3B could explain this human-specific hypermethylation of centromeric satellite repeats in oocytes.
It is suggested that evolutionarily young SINEs and LINEs are demethylated to a milder extent than older ones during human preimplantation development [7]. We found that SVAs and some LTRs containing CpG-rich VNTRs were much more preferentially protected from demethylation than SINEs and LINEs in human blastocysts. Paternal gDMRs also contained VNTRs and many VNTR-containing CGIs remained highly methylated in human blastocysts. Therefore, VNTRs might underlie the protection of paternal gDMRs and specific transposable elements from demethylation. The maintenance of DNA methylation of SVAs may be especially important because SVAs are currently active in the human genome and are involved in various human diseases [22], [32]. While the underlying mechanism of the protection of VNTR-containing regions is currently unknown, it is noteworthy that VNTRs are related to RNA-directed DNA methylation in plants [33]. Many transposable elements including SVAs are expressed in human early embryos [7], [9] and it is interesting to speculate that RNA might be involved in the demethylation resistance of VNTR-containing regions.
Overall, this work highlights both conserved and species-specific regulation of DNA methylation during early mammalian development. Our WGBS data of human gametes and blastocysts not only provide information to support our understanding of normal human developmental processes but also will be useful in interpreting studies on assisted reproductive technologies (ARTs). ARTs in humans are associated with an increased risk of imprinting disorders [34], [35], and our data will aid in the safety evaluation of ARTs and the preimplantation epigenetic diagnosis of human embryos.
Materials and Methods
Sample collection
Human oocytes, sperm, blastocysts and umbilical cord blood cells were obtained with signed informed consent of the donors or the couples, and the approval of the Ethics Committee of Tohoku University School of Medicine (Research license 2013-1-57), associated hospitals, the Japan Society of Obstetrics and Gynecology and the Ministry of Education, Culture, Sports, Science and Technology (Japan). Altogether, 202 surplus oocytes and 80 surplus blastocysts were obtained from female patients (ages 26–43) undergoing IVF treatment. The patients were healthy women with no habitual drug use and no particular past or familial disease history. We collected morphologically normal GV and MI oocytes from preovulatory follicles by intravaginal ultrasound-guided follicular aspiration after controlled ovarian hyperstimulation. To remove cumulus cells and the zona pellucida, oocytes were treated with hyaluronidase solution (JX Nippon Oil & Energy Corporation, Tokyo, Japan) and Tyrode's solution-Acidified (JX Nippon Oil & Energy Corporation) according to the manufacturer's instructions. Blastocysts were obtained by culturing early cleavage-stage embryos in Global Medium (LifeGlobal, Guilford, CT) overlaid with mineral oil. We used morphologically normal expanding or expanded blastocysts. The number of ICM cells is similar to, or a little lower than, that of trophectoderm (TE) cells in blastocysts at this stage [36]. Because ICM and TE cells show similar methylation levels [7], [9] and the available embryos in this study were limited, we performed WGBS using whole blastocysts. Ejaculated sperm samples with normal volume, counting and rates of mortality were collected. Only motile sperm cells isolated by the swim-up method [37] were used.
Construction and sequencing of PBAT libraries
Oocytes and blastocysts were incubated in a lysis solution (0.1% SDS, 1 mg/ml proteinase K, 50 ng/µl carrier RNA (QIAGEN, Valencia, CA)) for 60 min at 37°C and then 15 min at 98°C. Genomic DNA was purified with phenol/chloroform extraction and ethanol precipitation. Sperm genomic DNA was prepared as described [38]. Genomic DNA of cord blood cells was purified with phenol/chloroform extraction and ethanol precipitation. Isolated genomic DNA was spiked with 5% (for oocytes and blastocysts) or 0.5% (for sperm and cord blood cells) unmethylated lambda DNA (Promega, Madison, WI). Bisulfite treatment was performed using the MethylCode Bisulfite Conversion Kit (Invitrogen, Carlsbad, CA).
PBAT libraries were prepared as previously described [10]. Briefly, the first-strand DNA was synthesized with the Klenow fragment (3′-5′ exo-) (NEB, Beverly, MA) using BioPEA2N4 (5′-biotin-ACA CTC TTT CCC TAC ACG ACG CTC TTC CGA TCT NNN N-3′). The biotinylated first-strand DNA was captured using Dynabeads M-280 Streptavidin (Invitrogen). The second-strand DNA was synthesized with the Klenow fragment (3′-5′ exo-) using PE-reverse-N4 (5′-CAA GCA GAA GAC GGC ATA CGA GAT NNN N-3′). After removing the first-strand DNA, the second strand was double stranded with Phusion Hot Start II High-Fidelity DNA Polymerase (Finnzymes, Woburn, MA) using Primer-3 (5′-AAT GAT ACG GCG ACC ACC GAG ATC TAC ACT CTT TCC CTA CAC GAC GCT CTT CCG ATC T-3′). For an oocyte PBAT library, PCR-amplification was performed with KAPA HiFi HotStart Uracil+ ReadyMix (2×) (Kapa Biosystems, Woburn, MA) using primers, (5′-CAA GCA GAA GAC GGC ATA CGA GAT-3′) and (5′ - AAT GAT ACG GCG ACC ACC GAG ATC T-3′). The following program was used for the PCR-amplification: 10 cycles of 98°C for 15 sec, 65°C for 30 sec and 72°C for 30 sec. Concentrations of the PBAT libraries were measured by quantitative PCR (qPCR) using the Kapa Library Quantification Kit (Kapa Biosystems).
PBAT libraries were sequenced on the HiSeq 2000 or HiSeq 2500 platform (Illumina, CA, USA) with 100-bp single-end reads using the TruSeq SR Cluster Kit v3-cBot-HS and the TruSeq SBS Kit v3-HS (Illumina).
Mapping and methylation analysis
Sequenced reads were processed using the Illumina standard base-calling pipeline (v1.8.2) and the first 4 bases were trimmed to remove random primer sequences. The resulting reads were aligned to the reference genome (UCSC hg19) using Bismark [39] (v.0.9.0) with default parameters. For the oocyte library prepared with PCR-amplification, identical reads were treated as a single read to remove PCR duplicates. The methylation level of each cytosine was calculated using the Bismark methylation extractor. For CpG sites, reads from both strands were combined to calculate the methylation levels. Except for S1 Figure, methylation levels of CpGs covered with ≥3 reads were analyzed for oocytes and those of CpGs covered with ≥5 reads were analyzed for the other samples. Bisulfite conversion rates were estimated using reads that uniquely aligned to the lambda phage genome and were>99% for all samples. In this study, mC and hmC were indistinguishable because bisulfite sequencing cannot differentiate hmC from mC.
We also included available RRBS data of human oocytes, ICM and blastocysts [7], [9] and WGBS data of human ICM [7], ES cells (H1, H9 and HUES6) and mouse oocytes [11]. Processed methylation data were downloaded from NCBI GEO (http://www.ncbi.nlm.nih.gov/geo) for ICM (Accession number: GSE49828 and GSE51239), blastocysts (Accession number: GSE51239), H1 (Accession number: GSM429321), H9 (Accession number: GSM706059) and HUES6 ES cells (Accession number: GSM1173778). The RRBS data from biological replicates were combined. For mouse oocytes [11], the raw reads were mapped to the reference genome (UCSC mm9) and analyzed as described above (only CpGs covered with ≥5 reads were used).
Annotations of genomic regions
Annotations of Refseq genes, CGIs and repeat sequences were downloaded from the UCSC Genome Browser. Refseq genes shorter than 300 bp (encoding microRNAs or small nucleolar RNAs in most cases) were excluded from our analyses. Promoters were defined as regions 1 kb upstream and downstream from transcription start sites of Refseq transcripts. For calculation of the mean methylation levels, we analyzed only CGIs and promoters containing ≥10 CpGs with sufficient coverage for calculation of the methylation levels. Similarly, we considered only repeat copies containing ≥5 CpGs for calculation of the mean methylation levels of repeat copies. The gene bodies were defined as transcribed regions of Refseq transcripts except for promoters. When several Refseq transcripts were assigned to a Refseq gene, the transcribed regions were merged into a single gene body. Regions and names of the 67 imprinted DMRs were defined as previously reported [17].
The CpG density was defined for each CpG site as the density of CpGs within 100 bp upstream and downstream regions (the number of CpGs was divided by 200). Gene ontology analyses were performed using the Database for Annotation, Visualization and Integrated Discovery (DAVID) [40]. The list of human and mouse homologs including HomoloGene IDs was downloaded from Mouse Genome Informatics (MGI, http://www.informatics.jax.org/). VNTRs were identified using Tandem Repeats Finder [41] (alignment parameters = 2, 5, 7; minimum alignment score = 150; maximum period size = 500). Sequence motifs among VNTRs were searched using DREME [42] (the consensus patterns of VNTRs of>70% and <50% methylated CGIs in Fig. 4G were used as positive and negative sequences, respectively).
Transcriptome analysis
Transcriptome data of human and mouse oocytes were previously reported [5], [43]. The raw reads from biological replicates were combined and analyzed using Avadis NGS software with default parameters (version 1.5, Strand Scientific Intelligence).
Graphical presentation
Methylation levels of CpGs were visualized using Integrative Genomics Viewer (IGV) software (http://www.broadinstitute.org/igv/). Heatmaps and scatter plots were generated using the heatmap.2 function of the gplots package and the heatscatter function of the LSD package in R (http://www.R-project.org/), respectively. Violin plots were generated using the vioplot package (http://neoscientists.org/~plex/).
Sliding window-based analysis of methylation changes
We used a sliding window of 20 CpGs with a step size of 10 CpGs (the mean length was ∼2 kb) for consideration of the successful identification of imprinted DMRs using sliding windows of 10 CpGs [44] and 25 CpGs [17]. We considered only windows containing ≥10 CpGs with sufficient coverage for calculation of the methylation levels (84% of windows were covered in all samples shown in Fig. 1). Windows were classified as increasing (or decreasing) if the methylation levels increased (or decreased) by>20% and the changes were statistically significant according to Student's t-test with BH correction (P<0.05).
Bisulfite sequencing
DNA samples were treated with sodium bisulfite using an EZ DNA Methylation Kit (Zymo Research, Orange, CA) and PCR-amplified using TaKaRa EpiTaq™ HS (Takara Bio, Shiga, Japan). The PCR products were cloned into the pGEM-T Easy vector (Promega) and individual clones were sequenced. The following primers were used: chr5 : 662,283–663,402: (5′-GGG GTT AAG ATG GGA GTT ATG A-3′) and (5′-TAA ACA ACC CAA TCC CCA CA-3′), chr12 : 20,704,525-20,706,004: (5′-GGG AGG AGG AGG AGT AGT AGG A-3′) and (5′-CCC ACT AAA AAC AAA ATC AAT ACC-3′), chr15 : 89,952,271-89,953,061: (5′-GAT TTT TGT TAA TGA TTG GGT AGG A-3′) and (5′-CCC CAC AAT ATC TAC CCT CAT A-3′), chr21 : 32,716,044-32,716,485: (5′-AGA AGT TAA GGG GGA AAG ATG A-3′) and (5′-TTC ACA AAT TAC ACC CAC TAC CTC-3′′), chrX: 3,732,573-3,734,579: (5′-TTA ATG GGG TAA AGG GGT TAG A-3′) and (5′-ACC AAA TAA ACC CCA CCC AAA C-3′), chrX: 153,694,352-153,694,774: (5′-GTG GGG TTT AAG GAA GGA GGT A-3′) and (5′-CAA TCA CCC ACA CAC AAC TCC-3′). The sex of blastocysts was determined by PCR amplification of the male-specific SRY locus using bisulfite-converted DNA with the following primers: Forward: (5′ -TGA AAT TAA ATA TAA GAA AGT GAG GGT TG - 3′) and Reverse: (5′ -CCA CAC ACT CAA AAA TAA AAC ACC A - 3′).
Accession number
All sequencing data are deposited in the Japanese Genotype-phenotype Archive under the accession number JGAS00000000006.
Supporting Information
Zdroje
1. MesserschmidtDM, KnowlesBB, SolterD (2014) DNA methylation dynamics during epigenetic reprogramming in the germline and preimplantation embryos. Genes Dev 28 : 812–828.
2. SmithZD, MeissnerA (2013) DNA methylation: roles in mammalian development. Nat Rev Genet 14 : 204–220.
3. SaitouM, KagiwadaS, KurimotoK (2012) Epigenetic reprogramming in mouse pre-implantation development and primordial germ cells. Development 139 : 15–31.
4. SmallwoodSA, TomizawaS, KruegerF, RufN, CarliN, et al. (2011) Dynamic CpG island methylation landscape in oocytes and preimplantation embryos. Nat Genet 43 : 811–814.
5. KobayashiH, SakuraiT, ImaiM, TakahashiN, FukudaA, et al. (2012) Contribution of intragenic DNA methylation in mouse gametic DNA methylomes to establish oocyte-specific heritable marks. PLoS Genet 8: e1002440.
6. KohliRM, ZhangY (2013) TET enzymes, TDG and the dynamics of DNA demethylation. Nature 502 : 472–479.
7. GuoH, ZhuP, YanL, LiR, HuB, et al. (2014) The DNA methylation landscape of human early embryos. Nature 511 : 606–610.
8. BeaujeanN, HartshorneG, CavillaJ, TaylorJ, GardnerJ, et al. (2004) Non-conservation of mammalian preimplantation methylation dynamics. Curr Biol 14: R266–267.
9. SmithZD, ChanMM, HummKC, KarnikR, MekhoubadS, et al. (2014) DNA methylation dynamics of the human preimplantation embryo. Nature 511 : 611–615.
10. MiuraF, EnomotoY, DairikiR, ItoT (2012) Amplification-free whole-genome bisulfite sequencing by post-bisulfite adaptor tagging. Nucleic Acids Res 40: e136.
11. ShiraneK, TohH, KobayashiH, MiuraF, ChibaH, et al. (2013) Mouse oocyte methylomes at base resolution reveal genome-wide accumulation of non-CpG methylation and role of DNA methyltransferases. PLoS Genet 9: e1003439.
12. WangL, ZhangJ, DuanJ, GaoX, ZhuW, et al. (2014) Programming and inheritance of parental DNA methylomes in mammals. Cell 157 : 979–991.
13. MolaroA, HodgesE, FangF, SongQ, McCombieWR, et al. (2011) Sperm methylation profiles reveal features of epigenetic inheritance and evolution in primates. Cell 146 : 1029–1041.
14. Al-KhtibM, BlachereT, GuerinJF, LefevreA (2012) Methylation profile of the promoters of Nanog and Oct4 in ICSI human embryos. Hum Reprod 27 : 2948–2954.
15. DenommeMM, MannMR (2012) Genomic imprints as a model for the analysis of epigenetic stability during assisted reproductive technologies. Reproduction 144 : 393–409.
16. Ferguson-SmithAC (2011) Genomic imprinting: the emergence of an epigenetic paradigm. Nat Rev Genet 12 : 565–575.
17. CourtF, TayamaC, RomanelliV, Martin-TrujilloA, Iglesias-PlatasI, et al. (2014) Genome-wide parent-of-origin DNA methylation analysis reveals the intricacies of human imprinting and suggests a germline methylation-independent mechanism of establishment. Genome Res 24 : 554–569.
18. LiX, ItoM, ZhouF, YoungsonN, ZuoX, et al. (2008) A maternal-zygotic effect gene, Zfp57, maintains both maternal and paternal imprints. Dev Cell 15 : 547–557.
19. Bourc'hisD, XuGL, LinCS, BollmanB, BestorTH (2001) Dnmt3L and the establishment of maternal genomic imprints. Science 294 : 2536–2539.
20. HuntrissJ, HinkinsM, OliverB, HarrisSE, BeazleyJC, et al. (2004) Expression of mRNAs for DNA methyltransferases and methyl-CpG-binding proteins in the human female germ line, preimplantation embryos, and embryonic stem cells. Mol Reprod Dev 67 : 323–336.
21. SmithZD, ChanMM, MikkelsenTS, GuH, GnirkeA, et al. (2012) A unique regulatory phase of DNA methylation in the early mammalian embryo. Nature 484 : 339–344.
22. WangH, XingJ, GroverD, HedgesDJ, HanK, et al. (2005) SVA elements: a hominid-specific retroposon family. J Mol Biol 354 : 994–1007.
23. LaniaL, Di CristofanoA, StrazzulloM, PengueG, MajelloB, et al. (1992) Structural and functional organization of the human endogenous retroviral ERV9 sequences. Virology 191 : 464–468.
24. SchuelerMG, SullivanBA (2006) Structural and functional dynamics of human centromeric chromatin. Annu Rev Genomics Hum Genet 7 : 301–313.
25. GopalakrishnanS, SullivanBA, TrazziS, Della ValleG, RobertsonKD (2009) DNMT3B interacts with constitutive centromere protein CENP-C to modulate DNA methylation and the histone code at centromeric regions. Hum Mol Genet 18 : 3178–3193.
26. XueZ, HuangK, CaiC, CaiL, JiangCY, et al. (2013) Genetic programs in human and mouse early embryos revealed by single-cell RNA sequencing. Nature 500 : 593–597.
27. KobayashiH, YanagisawaE, SakashitaA, SugawaraN, KumakuraS, et al. (2013) Epigenetic and transcriptional features of the novel human imprinted lncRNA GPR1AS suggest it is a functional ortholog to mouse Zdbf2linc. Epigenetics 8 : 635–645.
28. ProudhonC, DuffieR, AjjanS, CowleyM, IranzoJ, et al. (2012) Protection against de novo methylation is instrumental in maintaining parent-of-origin methylation inherited from the gametes. Mol Cell 47 : 909–920.
29. HirasawaR, ChibaH, KanedaM, TajimaS, LiE, et al. (2008) Maternal and zygotic Dnmt1 are necessary and sufficient for the maintenance of DNA methylation imprints during preimplantation development. Genes Dev 22 : 1607–1616.
30. YamagataK, YamazakiT, MikiH, OgonukiN, InoueK, et al. (2007) Centromeric DNA hypomethylation as an epigenetic signature discriminates between germ and somatic cell lineages. Dev Biol 312 : 419–426.
31. UedaY, OkanoM, WilliamsC, ChenT, GeorgopoulosK, et al. (2006) Roles for Dnmt3b in mammalian development: a mouse model for the ICF syndrome. Development 133 : 1183–1192.
32. HancksDC, KazazianHHJr (2010) SVA retrotransposons: Evolution and genetic instability. Semin Cancer Biol 20 : 234–245.
33. ChandlerVL (2010) Paramutation's properties and puzzles. Science 330 : 628–629.
34. van MontfoortAP, HanssenLL, de SutterP, VivilleS, GeraedtsJP, et al. (2012) Assisted reproduction treatment and epigenetic inheritance. Hum Reprod Update 18 : 171–197.
35. HiuraH, OkaeH, MiyauchiN, SatoF, SatoA, et al. (2012) Characterization of DNA methylation errors in patients with imprinting disorders conceived by assisted reproduction technologies. Hum Reprod 27 : 2541–2548.
36. HardyK, HandysideAH, WinstonRM (1989) The human blastocyst: cell number, death and allocation during late preimplantation development in vitro. Development 107 : 597–604.
37. UshijimaC, KumasakoY, KihailePE, HirotsuruK, UtsunomiyaT (2000) Analysis of chromosomal abnormalities in human spermatozoa using multi-colour fluorescence in-situ hybridization. Hum Reprod 15 : 1107–1111.
38. BahnakBR, WuQY, CoulombelL, DrouetL, Kerbiriou-NabiasD, et al. (1988) A simple and efficient method for isolating high molecular weight DNA from mammalian sperm. Nucleic Acids Res 16 : 1208.
39. KruegerF, AndrewsSR (2011) Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics 27 : 1571–1572.
40. Huang daW, ShermanBT, LempickiRA (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4 : 44–57.
41. BensonG (1999) Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res 27 : 573–580.
42. BaileyTL (2011) DREME: motif discovery in transcription factor ChIP-seq data. Bioinformatics 27 : 1653–1659.
43. YanL, YangM, GuoH, YangL, WuJ, et al. (2013) Single-cell RNA-Seq profiling of human preimplantation embryos and embryonic stem cells. Nat Struct Mol Biol 20 : 1131–1139.
44. FangF, HodgesE, MolaroA, DeanM, HannonGJ, et al. (2012) Genomic landscape of human allele-specific DNA methylation. Proc Natl Acad Sci U S A 109 : 7332–7337.
45. DasR, LeeYK, StrogantsevR, JinS, LimYC, et al. (2013) DNMT1 and AIM1 Imprinting in human placenta revealed through a genome-wide screen for allele-specific DNA methylation. BMC Genomics 14 : 685.
Štítky
Genetika Reprodukční medicína
Článek Large-scale Metabolomic Profiling Identifies Novel Biomarkers for Incident Coronary Heart DiseaseČlánek Notch Signaling Mediates the Age-Associated Decrease in Adhesion of Germline Stem Cells to the NicheČlánek Phosphorylation of Mitochondrial Polyubiquitin by PINK1 Promotes Parkin Mitochondrial TetheringČlánek Natural Variation Is Associated With Genome-Wide Methylation Changes and Temperature SeasonalityČlánek Overlapping and Non-overlapping Functions of Condensins I and II in Neural Stem Cell DivisionsČlánek Unisexual Reproduction Drives Meiotic Recombination and Phenotypic and Karyotypic Plasticity inČlánek Tetraspanin (TSP-17) Protects Dopaminergic Neurons against 6-OHDA-Induced Neurodegeneration inČlánek ABA-Mediated ROS in Mitochondria Regulate Root Meristem Activity by Controlling Expression inČlánek Mutations in Global Regulators Lead to Metabolic Selection during Adaptation to Complex EnvironmentsČlánek The Evolution of Sex Ratio Distorter Suppression Affects a 25 cM Genomic Region in the Butterfly
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2014 Číslo 12
-
Všechny články tohoto čísla
- Stratification by Smoking Status Reveals an Association of Genotype with Body Mass Index in Never Smokers
- Genome Wide Meta-analysis Highlights the Role of Genetic Variation in in the Regulation of Circulating Serum Chemerin
- Occupancy of Mitochondrial Single-Stranded DNA Binding Protein Supports the Strand Displacement Mode of DNA Replication
- Distinct Genealogies for Plasmids and Chromosome
- Large-scale Metabolomic Profiling Identifies Novel Biomarkers for Incident Coronary Heart Disease
- Non-coding RNAs Prevent the Binding of the MSL-complex to Heterochromatic Regions
- Plasmid Flux in ST131 Sublineages, Analyzed by Plasmid Constellation Network (PLACNET), a New Method for Plasmid Reconstruction from Whole Genome Sequences
- Epigenome-Guided Analysis of the Transcriptome of Plaque Macrophages during Atherosclerosis Regression Reveals Activation of the Wnt Signaling Pathway
- The Inventiveness of Nature: An Interview with Werner Arber
- Mediation Analysis Demonstrates That -eQTLs Are Often Explained by -Mediation: A Genome-Wide Analysis among 1,800 South Asians
- Generation of Antigenic Diversity in by Structured Rearrangement of Genes During Mitosis
- A Massively Parallel Pipeline to Clone DNA Variants and Examine Molecular Phenotypes of Human Disease Mutations
- Genetic Analysis of the Cardiac Methylome at Single Nucleotide Resolution in a Model of Human Cardiovascular Disease
- Genetic Analysis of Circadian Responses to Low Frequency Electromagnetic Fields in
- The Dissection of Meiotic Chromosome Movement in Mice Using an Electroporation Technique
- Altered Chromatin Occupancy of Master Regulators Underlies Evolutionary Divergence in the Transcriptional Landscape of Erythroid Differentiation
- Syd/JIP3 and JNK Signaling Are Required for Myonuclear Positioning and Muscle Function
- Notch Signaling Mediates the Age-Associated Decrease in Adhesion of Germline Stem Cells to the Niche
- Mutation of Leads to Blurred Tonotopic Organization of Central Auditory Circuits in Mice
- The IKAROS Interaction with a Complex Including Chromatin Remodeling and Transcription Elongation Activities Is Required for Hematopoiesis
- RAN-Binding Protein 9 is Involved in Alternative Splicing and is Critical for Male Germ Cell Development and Male Fertility
- Enhanced Longevity by Ibuprofen, Conserved in Multiple Species, Occurs in Yeast through Inhibition of Tryptophan Import
- Phosphorylation of Mitochondrial Polyubiquitin by PINK1 Promotes Parkin Mitochondrial Tethering
- Recurrent Loss of Specific Introns during Angiosperm Evolution
- Natural Variation Is Associated With Genome-Wide Methylation Changes and Temperature Seasonality
- SEEDSTICK is a Master Regulator of Development and Metabolism in the Arabidopsis Seed Coat
- Overlapping and Non-overlapping Functions of Condensins I and II in Neural Stem Cell Divisions
- Unisexual Reproduction Drives Meiotic Recombination and Phenotypic and Karyotypic Plasticity in
- Tetraspanin (TSP-17) Protects Dopaminergic Neurons against 6-OHDA-Induced Neurodegeneration in
- ABA-Mediated ROS in Mitochondria Regulate Root Meristem Activity by Controlling Expression in
- Mutations in Global Regulators Lead to Metabolic Selection during Adaptation to Complex Environments
- Global Analysis of Photosynthesis Transcriptional Regulatory Networks
- Mucolipin Co-deficiency Causes Accelerated Endolysosomal Vacuolation of Enterocytes and Failure-to-Thrive from Birth to Weaning
- Controlling Pre-leukemic Thymocyte Self-Renewal
- How Malaria Parasites Avoid Running Out of Ammo
- Echoes of the Past: Hereditarianism and
- Deep Reads: Strands in the History of Molecular Genetics
- Keep on Laying Eggs Mama, RNAi My Reproductive Aging Blues Away
- Analysis of a Plant Complex Resistance Gene Locus Underlying Immune-Related Hybrid Incompatibility and Its Occurrence in Nature
- Epistatic Adaptive Evolution of Human Color Vision
- Increased and Imbalanced dNTP Pools Symmetrically Promote Both Leading and Lagging Strand Replication Infidelity
- Genetic Basis of Haloperidol Resistance in Is Complex and Dose Dependent
- Genome-Wide Analysis of DNA Methylation Dynamics during Early Human Development
- Interaction between Conjugative and Retrotransposable Elements in Horizontal Gene Transfer
- The Evolution of Sex Ratio Distorter Suppression Affects a 25 cM Genomic Region in the Butterfly
- is Required for Adult Maintenance of Dopaminergic Neurons in the Ventral Substantia Nigra
- PRL1, an RNA-Binding Protein, Positively Regulates the Accumulation of miRNAs and siRNAs in Arabidopsis
- Genetic Control of Contagious Asexuality in the Pea Aphid
- Early Mesozoic Coexistence of Amniotes and Hepadnaviridae
- Local and Systemic Regulation of Plant Root System Architecture and Symbiotic Nodulation by a Receptor-Like Kinase
- Gene Pathways That Delay Reproductive Senescence
- The Evolution of Fungal Metabolic Pathways
- Maf1 Is a Novel Target of PTEN and PI3K Signaling That Negatively Regulates Oncogenesis and Lipid Metabolism
- Formation of Linear Amplicons with Inverted Duplications in Requires the MRE11 Nuclease
- Identification of Rare Causal Variants in Sequence-Based Studies: Methods and Applications to , a Gene Involved in Cohen Syndrome and Autism
- Rrp12 and the Exportin Crm1 Participate in Late Assembly Events in the Nucleolus during 40S Ribosomal Subunit Biogenesis
- The Mutations in the ATP-Binding Groove of the Rad3/XPD Helicase Lead to -Cockayne Syndrome-Like Phenotypes
- Topoisomerase I Plays a Critical Role in Suppressing Genome Instability at a Highly Transcribed G-Quadruplex-Forming Sequence
- A Cbx8-Containing Polycomb Complex Facilitates the Transition to Gene Activation during ES Cell Differentiation
- Transcriptional Frameshifting Rescues Type VI Secretion by the Production of Two Length Variants from the Prematurely Interrupted Gene
- Association Mapping across Numerous Traits Reveals Patterns of Functional Variation in Maize
- Genome-Wide Analysis of -Regulated and Phased Small RNAs Underscores the Importance of the ta-siRNA Pathway to Maize Development
- Dissemination of Cephalosporin Resistance Genes between Strains from Farm Animals and Humans by Specific Plasmid Lineages
- The Tau Tubulin Kinases TTBK1/2 Promote Accumulation of Pathological TDP-43
- Germline Signals Deploy NHR-49 to Modulate Fatty-Acid β-Oxidation and Desaturation in Somatic Tissues of
- Microevolution of in Macrophages Restores Filamentation in a Nonfilamentous Mutant
- Vangl2-Regulated Polarisation of Second Heart Field-Derived Cells Is Required for Outflow Tract Lengthening during Cardiac Development
- Chondrocytes Transdifferentiate into Osteoblasts in Endochondral Bone during Development, Postnatal Growth and Fracture Healing in Mice
- A ABC Transporter Regulates Lifespan
- RA and FGF Signalling Are Required in the Zebrafish Otic Vesicle to Pattern and Maintain Ventral Otic Identities
- , and Reprogram Thymocytes into Self-Renewing Cells
- The miR9863 Family Regulates Distinct Alleles in Barley to Attenuate NLR Receptor-Triggered Disease Resistance and Cell-Death Signaling
- Detection of Pleiotropy through a Phenome-Wide Association Study (PheWAS) of Epidemiologic Data as Part of the Environmental Architecture for Genes Linked to Environment (EAGLE) Study
- Extensive Copy-Number Variation of Young Genes across Stickleback Populations
- The and Genetic Modules Interact to Regulate Ciliogenesis and Ciliary Microtubule Patterning in
- Analysis of the Genome, Transcriptome and Secretome Provides Insight into Its Pioneer Colonization Strategies of Wood
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Tetraspanin (TSP-17) Protects Dopaminergic Neurons against 6-OHDA-Induced Neurodegeneration in
- Maf1 Is a Novel Target of PTEN and PI3K Signaling That Negatively Regulates Oncogenesis and Lipid Metabolism
- The IKAROS Interaction with a Complex Including Chromatin Remodeling and Transcription Elongation Activities Is Required for Hematopoiesis
- Echoes of the Past: Hereditarianism and
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



