-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaIncreased and Imbalanced dNTP Pools Symmetrically Promote Both Leading and Lagging Strand Replication Infidelity
The building blocks of DNA, dNTPs, are vital to life, and thus their production is carefully controlled within each cell. Under certain conditions, such as cancer, infection, or drugs, the overall dNTP level or dNTP balance can change. Using yeast genetics we manipulated the dNTP pool balance in unicellular baker's yeast and analysed the effects upon fidelity of DNA replication. We also disrupted mismatch repair, an internal safety system that corrects replication errors and is mutated in many cancers. By sequencing DNA from yeast cells with these alterations we gain insights into the mechanisms of mutation formation that contribute to genome instability. We find that the leading and lagging strand replication fidelity is affected similarly by the dNTP pool imbalance and that the mismatch repair machinery corrects replication errors driven by a dNTP pool imbalance with highly variable efficiencies.
Published in the journal: . PLoS Genet 10(12): e32767. doi:10.1371/journal.pgen.1004846
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1004846Summary
The building blocks of DNA, dNTPs, are vital to life, and thus their production is carefully controlled within each cell. Under certain conditions, such as cancer, infection, or drugs, the overall dNTP level or dNTP balance can change. Using yeast genetics we manipulated the dNTP pool balance in unicellular baker's yeast and analysed the effects upon fidelity of DNA replication. We also disrupted mismatch repair, an internal safety system that corrects replication errors and is mutated in many cancers. By sequencing DNA from yeast cells with these alterations we gain insights into the mechanisms of mutation formation that contribute to genome instability. We find that the leading and lagging strand replication fidelity is affected similarly by the dNTP pool imbalance and that the mismatch repair machinery corrects replication errors driven by a dNTP pool imbalance with highly variable efficiencies.
Introduction
The integrity of an organism's genome is vital to its continued survival, whether unicellular microbe or complex large mammal [1]. Therefore, there are highly conserved mechanisms involved in regulating and protecting genetic material both during and post DNA replication. One of the first safety systems for DNA replication is the stringent control of dNTP synthesis by ribonucleotide reductase (RNR), which maintains concentrations of the individual dNTPs at different levels [1], [2]. RNR catalyses the rate-limiting step in the production of all four dNTPs for the synthesis of nuclear and mitochondrial DNA [3], [4]. In yeast, RNR is a multi-subunit complex comprised of a large subunit, which exist as a homodimer of Rnr1 proteins or a heterodimer of Rnr1/Rnr3 proteins, and a small subunit comprised of Rnr2/Rnr4 proteins. The large subunits contain allosteric specificity sites that modulate enzyme activity and control the balance of the four dNTPs by influencing the specific ribonucleoside diphosphate reduction reaction within the catalytic sites [5]. A highly conserved loop of 13 amino acid residues (Loop 2) connects the allosteric specificity and catalytic sites and is crucial for the correct allosteric regulation of the enzyme [6], [7].
The DNA polymerase selectivity, proofreading and mismatch repair are subsequent safety systems that determine the fidelity of DNA replication. The DNA polymerase selectivity ensures insertion of the correct nucleotide during DNA synthesis. Although the major replicative polymerases alpha (Pol α), delta (Pol δ), and epsilon (Pol ε) are high fidelity enzymes, their accuracy is dependent upon the supply of dNTPs [8]. The second mechanism is proofreading in which errors are removed from primer termini during replication by a 3′–5′ exonuclease activity. Errors that escape proofreading can still be repaired post-replication, through the mismatch repair system (MMR) (reviewed in [9]). The major components of MMR are the homologs of the bacterial MutS proteins, a heterodimer of either Msh2-Msh6 or Msh2-Msh3 that recognise and bind to the mismatch. Msh2-Msh6 is mainly responsible for repairing single base-base mismatches, short insertions and deletions (indels) and small loops, whereas Msh2-Msh3 is involved in larger loop repair. Therefore, Msh2 is essential for MMR [10] and loss of this activity elevates mutation rates [11]. Mutation or loss of Msh2 in humans is associated with microsatellite instability and hereditary nonpolyposis colorectal cancer (HNPCC) [12] and gall bladder cancer [13].
The current model of the eukaryotic replication fork involves DNA polymerase complexes with very different subunit composition, enzymatic properties and fidelity. The leading strand is synthesized primarily by Pol ε, while the lagging strand is synthesized primarily by Pol α and Pol δ [14], [15]. Here we asked whether an imbalanced dNTP pool can force the leading and lagging strand polymerases to produce different errors. It is possible to answer this question by using a gene that is located close to an origin of replication and switching the leading and lagging strand synthesis. We previously created a panel of yeast strains with defined dNTP pool imbalances. The imbalances, in which none of dNTP levels was below normal, did not activate the genome integrity checkpoint and were highly mutagenic despite the availability of functional proofreading and MMR [16]. Utilizing a strain with elevated dTTP and dCTP concentrations and normal dATP and dGTP concentrations, we previously determined the rate and specificity of replication errors generated at the CAN1 locus [17]. As the CAN1 reporter gene is located close to a replication origin, by reversing the orientation of CAN1 and thereby switching the leading and lagging strand synthesis at this locus, we can analyse potential mutational strand bias. To determine the efficiency of DNA mismatch repair in the presence of this dNTP pool imbalance, we also created an rnr1-Y285A mutant strain that was MMR deficient. Our data demonstrate that the mutational potential of this dNTP pool imbalance overpowers the intrinsic differences in error specificity of the leading and lagging strand polymerases and reveals that MMR works with highly variable efficiency.
Results
dNTP pools of the rnr1-Y285A and msh2Δ strains
To examine potential differences in mutational specificity between the major replicative polymerases in the presence of the imbalanced dNTP pools, we reversed the orientation of the CAN1 gene (CAN1-OR2). To investigate the effect of this dNTP pool imbalance in the absence of MMR, we deleted MSH2 in the rnr1-Y285A CAN-OR1 strain. The msh2Δ single mutant strain had normal dNTP pools (Fig. 1). The dNTP pools in the double mutant had the same imbalance as in the single rnr1-Y285A [17], with approximately 26 - and 14-fold higher concentrations of dCTP and dTTP, respectively, compared to wild type (wt) whilst the concentration of dATP was about double and dGTP was normal (Fig. 1).
Fig. 1. dNTP pools of the strains with the imbalanced dNTP pools. 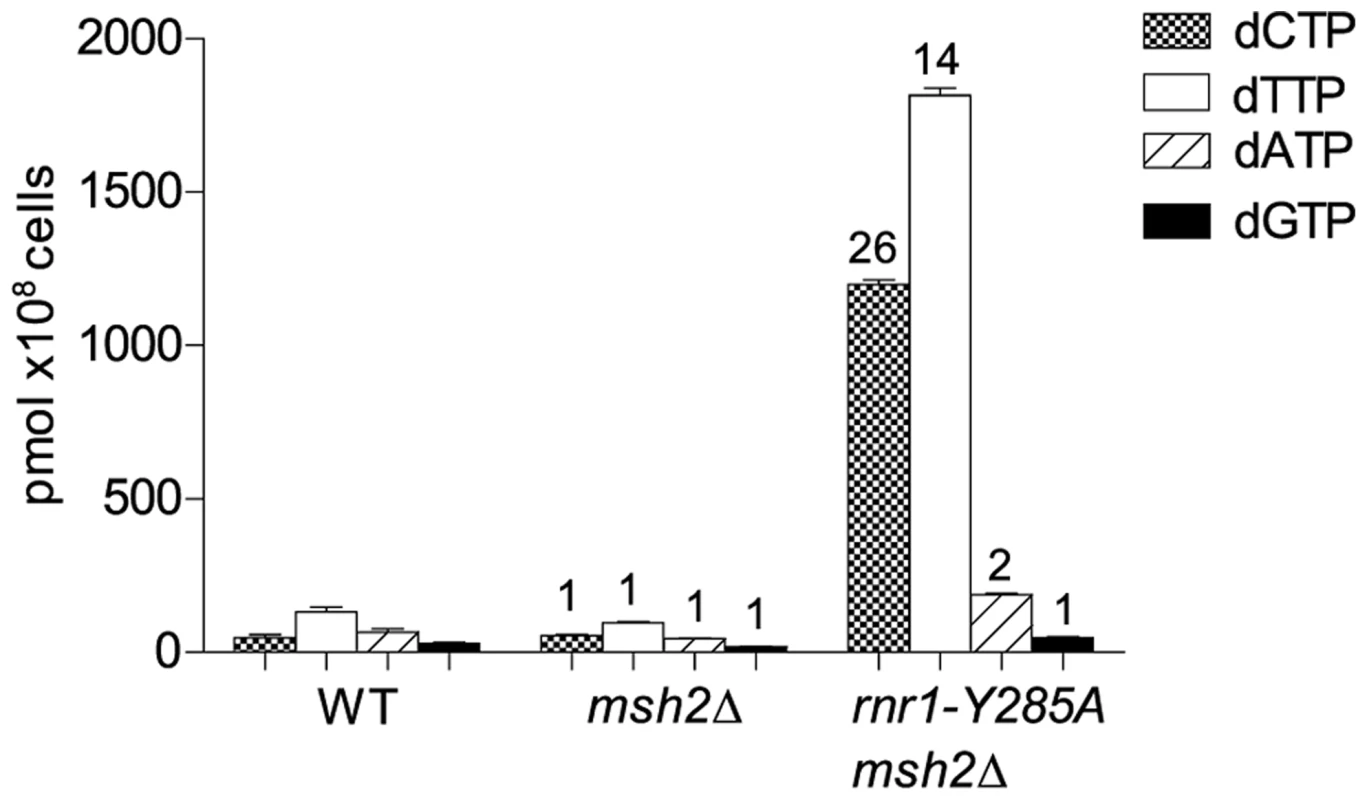
Numbers above columns show the factor increase over wt. Error bars show Standard Error of Mean. CAN1 spontaneous mutation rates and types
The spontaneous CAN1 mutation rate in the rnr1-Y285A CAN1-OR2 was 13-fold higher than wt (Table 1), which was similar to the CAN1-OR1 previously published OR1 [17]. The msh2Δ mutant had a 15-fold higher mutation rate compared to wt, however, the rnr1-Y285A msh2Δ strain mutation rate was over 500-fold greater than that of wt and over 30-fold either of the relative single mutants. Indels were the major mutation type observed in all four mutant strains whereas it was single base substitutions in wt (Fig. 2). The rnr1-Y285A, with CAN1 in both orientations, and msh2Δ strains had an average increase in the indel rate of more than 60-fold the wt strain (0.5×107 for wt versus 33×107 for OR1 [17], 37× for OR2, and 42×107 for msh2Δ). However, the double mutant indel rate was increased the most at more than 2000-fold over wt. In addition to single base indels, base substitutions were also significantly increased in the mutants to over 8-fold higher in the single mutants and 350-fold higher in rnr1-Y285A msh2Δ compared to wt. Complex mutations, defined as mutations involving insertions or deletions of multiple bases, were much more common in the double mutant, occurring at a rate over 30 times higher than that in wild type.
Fig. 2. Mutations rates by class. 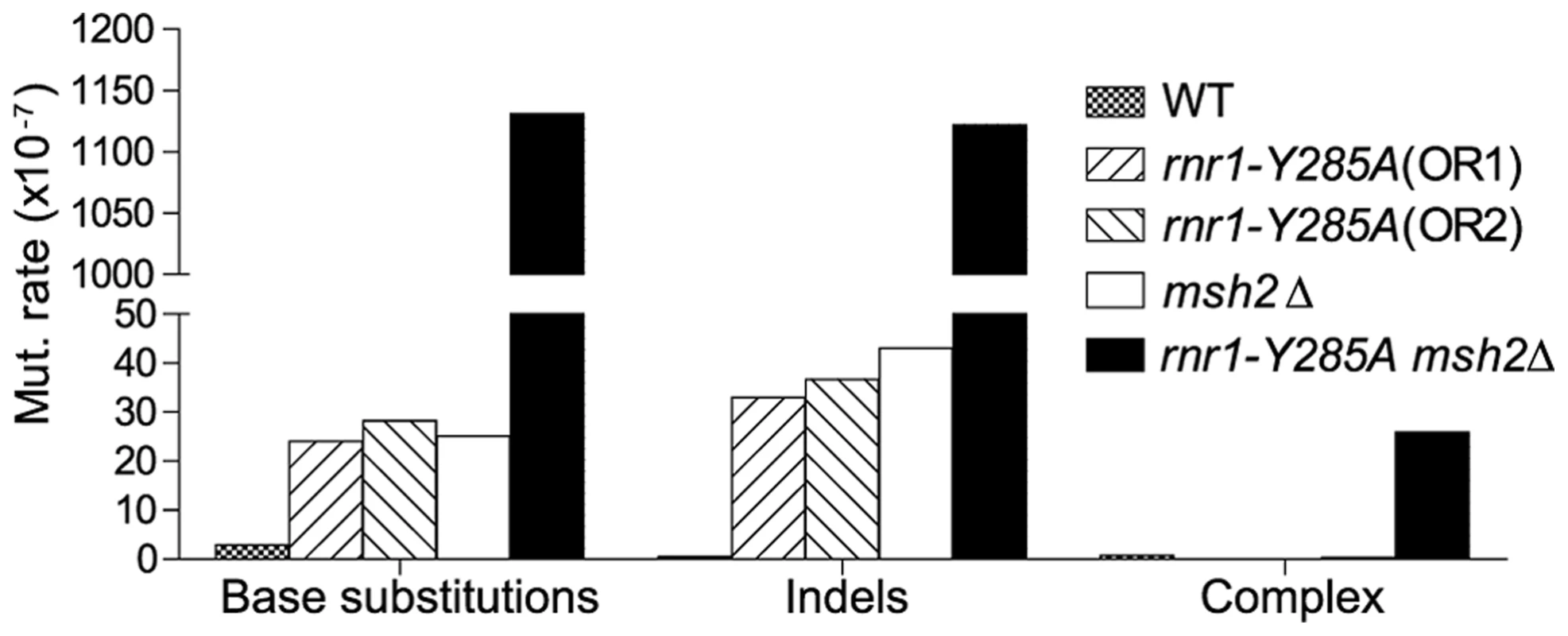
Tab. 1. CAN1 mutation rates and observed events.a 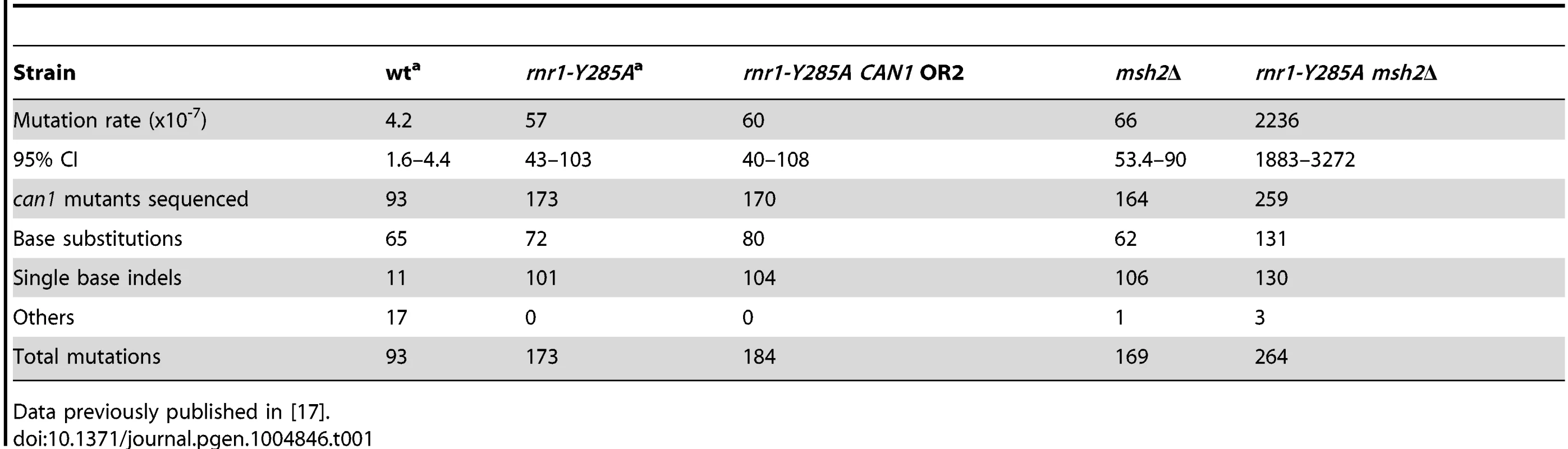
Data previously published in [17]. Mutation hotspots
Replication of the CAN1 gene originates from ARS507 and travels through the gene towards the telomere [18]–[20] (Fig. 3A). Therefore, in rnr1-Y285A CAN1-OR1 the leading strand polymerase, presumed to be Pol ε [21], uses the non-coding strand as the template while the coding strand is the template for lagging strand replication primarily by Pol δ or Pol α [14]. In CAN1-OR2, Pol ε now copies the coding strand and Pol δ/Pol α copy the non-coding strand (see Fig. 3A). An example is given in Fig. 3B, for the single base substitution at 648 bp. During leading strand synthesis in OR1, no mistake is made when copying template G due to high concentration of dCTP. However, during lagging strand replication dTTP is inserted opposite template C, as dTTP is at a much higher concentration than the dGTP required for correct pairing. As the succeeding incoming nucleotides are also at an increased concentration, rapid extension then follows stabilizing the C: dT mismatch. In the next round of replication, a C to A mutation arises. When the gene is reversed in rnr1-Y285A CAN1-OR2, Pol ε now copies the template C with low fidelity by misinserting dTTP, which ultimately results in a C to A mutation, and Pol δ/Pol α replication is error-free.
Fig. 3. Strand Assignment Model. 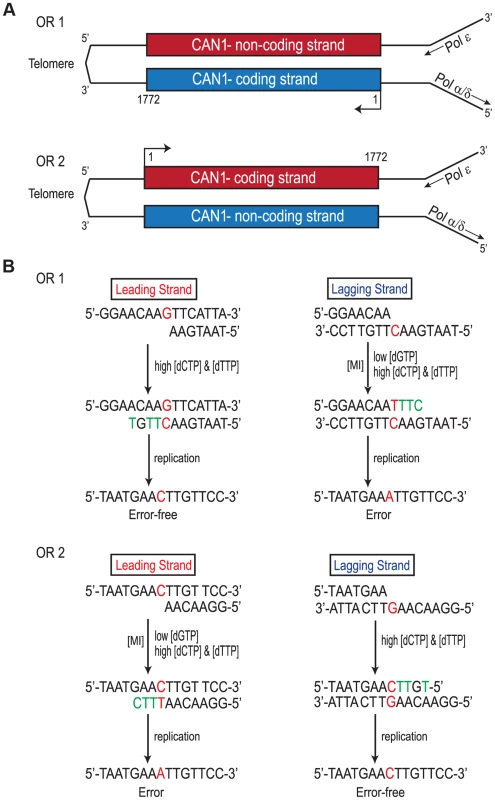
A. Cartoon representation of </emph>CAN1 orientation in rnr1-Y285A strains. B. Model showing strand assignment in the two rnr1-Y285A mutants (OR1 and OR2) using the hotspot at site 648 bp as an example. MI = Misinsertion. Red characters represent the mutational event and green characters represent bases where the dNTP is at an excessively high concentration. Hotspots, mutation sites where the rates were ≥10-fold greater than wt, were assigned to have occurred during leading or lagging strand synthesis by the nature of the mutation observed and dNTP pool imbalance. Simplified mutational spectra illustrating the hotspots in the CAN1 gene for each strain are shown in Fig. 4 with the full spectra in Figure S1-S3. The hotspot mutation rate was calculated by the equation [(frequency/total no. of samples) x CAN1 mutation rate]. The majority of hotspots in rnr1-Y285A CAN1-OR1 and OR2 show no leading – lagging strand bias and have similar mutation rates in both strains (Fig. 5). However, the major hotspot at position 425–427 bp was only seen in OR2 and had a mutation rate of 48×10−8 which was 15-fold higher than in OR1.
Fig. 4. Simplified CAN1 mutation spectra showing hotpots (where mutation rate is greater than 10-fold that in wt) for rnr1-Y285A CAN1-OR1 (n = 173), rnr1-Y285A CAN1-OR2 (n = 170), rnr1-Y285A msh2Δ (n = 259), and msh2Δ (n = 164) where n = number of individual colonies sequenced. 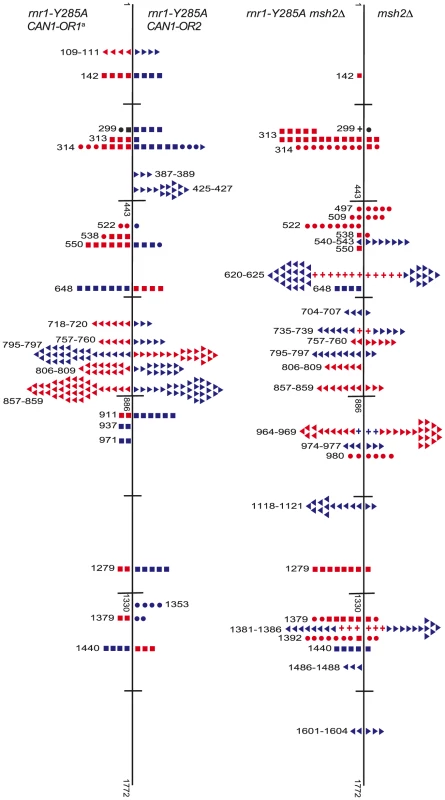
Symbols indicate the following: plus - additions, triangles - deletions, squares - transversions, circles - transitions, red - occur during leading strand synthesis, blue - occur during lagging strand synthesis, black - mutation cannot be assigned to a strand. a Reanalysed from the dataset published in [17]. Fig. 5. Comparison of CAN1 mutation rates at hotspots (predominant mutation at site) in rnr1-Y285A strains with natural (OR1) and reversed (OR2) orientation of the CAN1 gene. 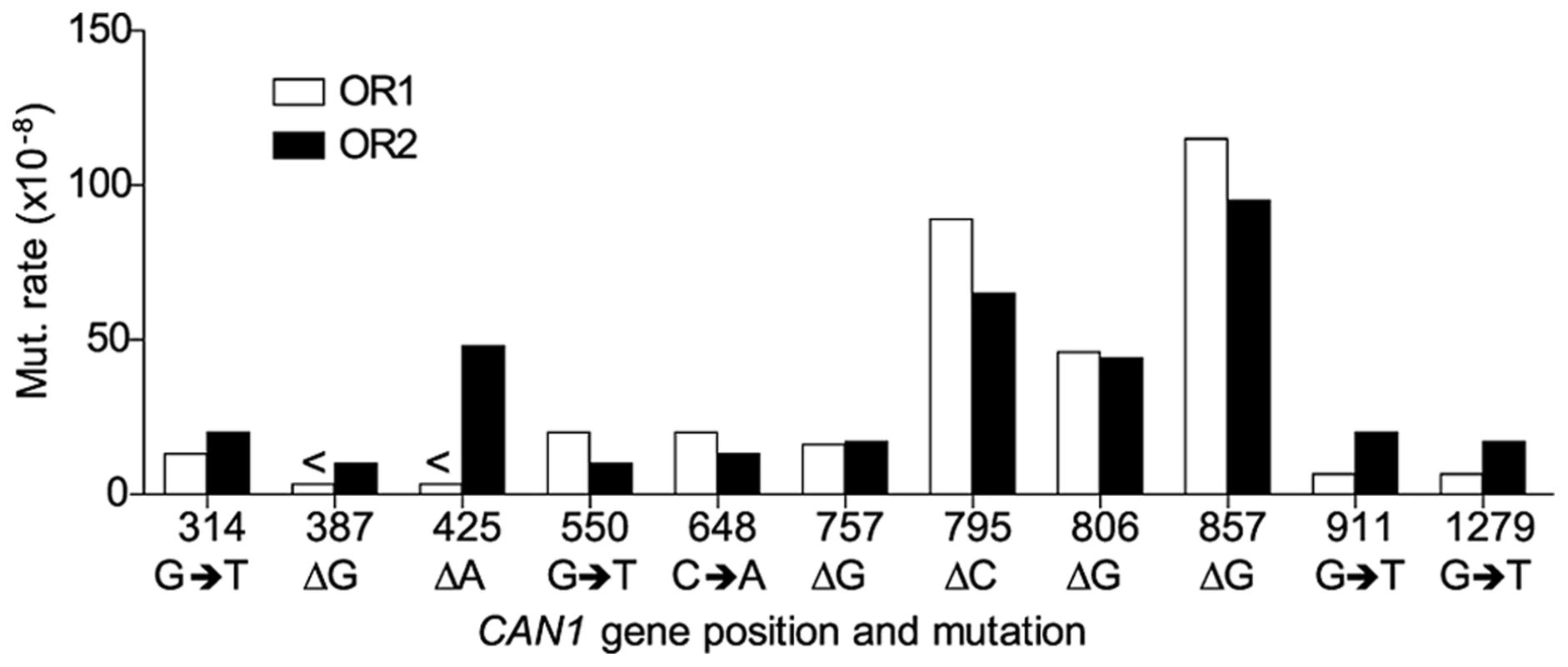
<Denotes that no events were detected. The rnr1-Y285A and msh2Δ strains had several shared major hotspots. Three single base deletions occurred in G: C homopolymeric runs at 757–760 bp, 795–797 bp, and 857–859 bp and two single base substitutions at 313 bp and 1379 bp (Fig. 4 and Table S1). Whilst the double mutant shared these five hotspots with both single mutants, there was a more than 100-fold increase in rates for base substitutions at 313 bp and 13791 bp (Fig. 6A and Table S1). The major hotspots in rnr1-Y285A msh2Δ were those seen in msh2Δ at 1118–1121 bp, 1392 bp, and especially the dominant deletion hotspots at 620–625 bp, 964–969 bp, and 1381–1386 bp. The site-specific mutation rates in the double mutant ranged from 4 - to 800-fold the single mutants.
Fig. 6. Mismatch repair efficiency. 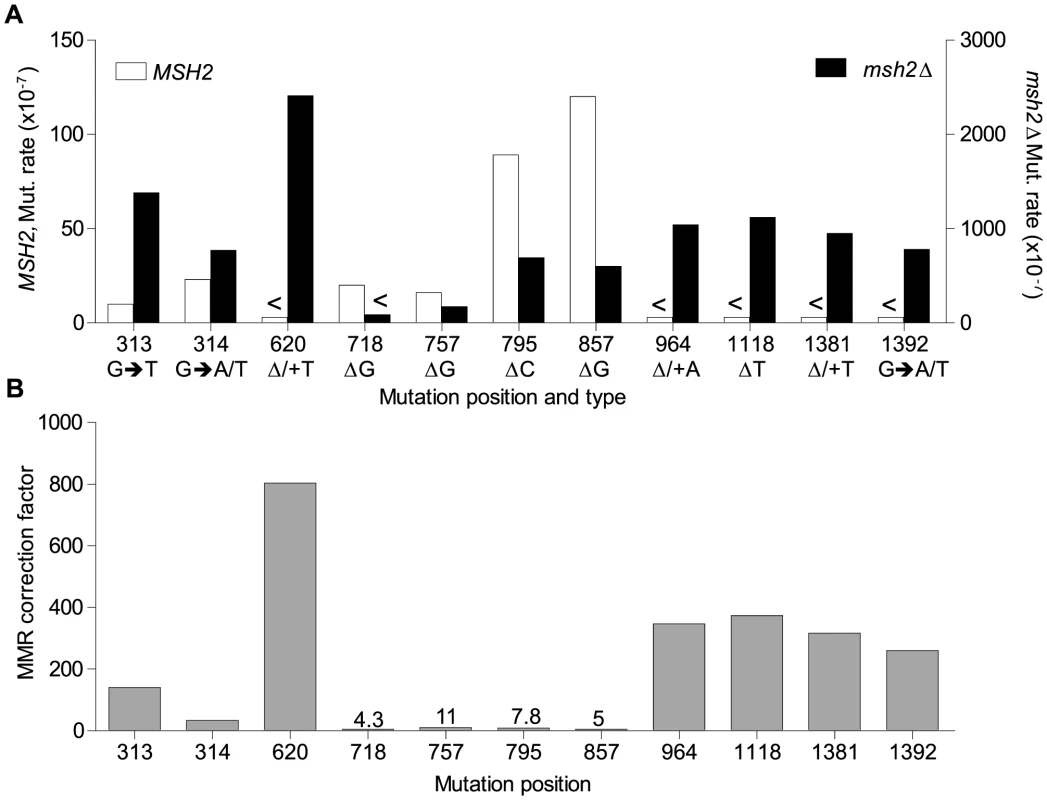
A. Comparison of site mutation rates at major hotspots in rnr1-Y285A with (left axis) and without MMR (right axis). B. MMR correction factor at these sites in the presence of an rnr1-Y285A dNTP pool imbalance. <Denotes no events observed. MMR efficiency
Analysis of the mutation spectra in the rnr1-Y285A and rnr1-Y285A msh2Δ strains showed that MMR had different efficiency at distinct mismatches. The ratio of mutation rates in the msh2Δ and MMR-proficient strains gave site-specific MMR correction efficiency (Fig. 6B and Table S1). The five hotspots (314, 718, 757, 795 and 857 bp), which include those with the highest mutation rates in the single rnr1-Y285A mutant, were the same sites that MMR was the least efficient at repairing errors. The correction factors were less than 30 and only around 10 in four of these sites (i.e., around 10% of errors at these sites will remain uncorrected by MMR). The majority of sites with the highest mutation rates in the double mutant (620, 964, 1118, 1381, and 1392 bp) were those that MMR was best at repairing, namely at T: A mononucleotide repeats. Loss of MSH2 increased the mutation rate at these sites by 260 - to 800-fold.
Discussion
Increased dCTP and dTTP drive different polymerases to make similar errors
Despite the inherent differences in complexity of continuous (leading strand) and non-continuous (lagging strand) synthesis, the increased dCTP and dTTP drive the same kind of mutations at identical sequences regardless of the replicative DNA polymerase. Most of the mutations occurred at a G: C base pair in which the cytosine served as the template for synthesis and was often flanked by a 5′-A or a tract of purines as exemplified in Fig. 3. With the concentration of dGTP being the lowest and dCTP and dTTP the highest, the deletion of a G: C base pair in a mononucleotide repeat is stabilized by the rapid incorporation of the next incoming nucleotide (dTTP opposite the template A), as described in detail in our previous report [17]. This dNTP imbalance and sequence context also explains the G: C to T: A base substitutions where dTTP is misinserted opposite template C and mismatch extension proceeds with the rapid incorporation of the pyrimidines opposite the flanking tract of purines. Thus, the mismatch remains at the expense of polymerase proofreading.
However, an exception was found at 425 bp (where there is a hotspot found only when the CAN1 gene is reversed to OR2). Although similar sequences (AT runs) show no variation in mutation rates between orientations it appears that polymerase δ or α could be making a mistake at this point but not Pol ε. There were also several minor hotspots that suggest polymerase specificity (positions 538, 937, and 971 were unique to OR1 whereas 387 and 1353 were seen only in OR2) which could be indicative of the differences in polymerase efficiency in certain sequence contexts. Whole genome sequencing may give insight into other sites and contexts that affect polymerase specificity and establish the patterns of mutations arising in the presence of this dNTP pool imbalance.
Given that the concentration of dATP was also lower than dCTP and dTTP, T: A to G: C transversions could also be expected in the base substitution hotspots where dCTP is misinserted opposite template T during replication. The nucleotide ratio of dCTP: dATP increased from ∼1∶1 in the wt strain to ∼6∶1 in the rnr1-Y285A strains. However, the increase in the nucleotide ratio of dTTP: dGTP was larger, from ∼4∶1 in the wt strain to ∼38∶1 in the rnr1-Y285A strains, which may explain the prevalence of the G: C to T: A transversions. Furthermore, the lack of T: A to G: C transversions may be due to the intrinsic difference in the rates at which the two errors are generated. Recent genome-wide studies in S. cerevisiae have reported that G: C to T: A transversions were observed at a higher rate than T: A to G: C in strains with normal dNTP pools [22], [23]. The three major replicative polymerases were more prone to generate G: C to T: A transversions but very rarely generated T: A to G: C transversions [23]. In addition, tumours with somatic mutations in the exonuclease domain of Pol ε have a higher prevalence of C to A mutations [24]–[28].
MMR repairs replication errors driven by a dNTP pool imbalance with highly variable efficiencies
MMR efficiency was dependent upon the site and mismatch generated from the dNTP pool imbalance. The increase in indels in the msh2Δ strains was not surprising as MMR is known to be highly active at repairing mistakes at mononucleotide repeats [29], [30]. The indels were almost entirely unique to sequences with ≥3 mononucleotide repeats in the double mutant (99.2%, 127 of 128) compared to 91% in the msh2Δ mutant and most frequently occurred in A: T runs. This can be predicted as A-T mononucleotide repeats are often the site of indels in MMR deficient strains [31] and are by far the most common in the CAN1 gene sequence (Figure S4). Indeed, it appears that the relationship between mutation rate and mononucleotide repeat length is exponential as others have found across the whole yeast genome [22].
The MMR correction factor for all indels in the rnr1-Y285A background was 32, which means that on average, 31 of 32 indels are corrected by MMR (compare Fig. 2 rates). Nevertheless, this is ∼3-fold lower than that in the wt RNR background suggesting that some indels driven by this dNTP pool imbalance escape MMR. In addition, there were several major indel hotspots, mainly at G: C base pairs in mononucleotide repeats, with correction factors of 10 compared to the indels at A: T repeats which ranged from 200 to 800. This is a huge variation in the vital post-replication repair machinery that supports the notion of MMR efficiency being dependent on the dNTP pool imbalance, sequence context, and identity of the mismatch.
There are several possibilities as to why MMR is not efficient at these sites in the rnr1-Y285A strain. First, there could be a saturation of MMR due to the volume of errors induced by the pool imbalance that are not corrected by proofreading [32]. Consider the hotspots at 795 and 857 bp which dominated the spectra in rnr1-Y285A. The correction factor for the wt RNR strain was more than 2 - and 5-fold higher than for the rnr1-Y285A mutant at the 795 and 857 hotspots, respectively (S1 Table). Therefore, MMR was more accurate at repairing deletions at these sites in the wt RNR strain with normal dNTP pools. Second, MMR itself may require a natural dNTP pool balance in order to correctly repair mistakes. If MMR complexes recognise the mismatches generated but recruit an error-prone or even high fidelity polymerase, the imbalanced dNTP concentration may result in the same mismatch; thus, the mutation is maintained. Finally, some mismatches may not be subjected to MMR if they are damaged or generated outside of DNA replication [33]–[37].
Materials and Methods
Yeast strains
The CAN1 gene was replaced with URA3 from the pUG72 plasmid [38] (primers “CAN1 Del Ura3” in Materials and Methods S1) in the RNR mutated strain (rnr1-Y285A) previously described [16]. PCR amplified WT CAN1 in reversed orientation (primers “CAN1 orientation” in Materials and Methods S1) was then transformed into the can1:: URA3 strain to give rnr1-Y285A CAN1 OR2. 5-FOA selection allowed the elimination of any can1:: URA3 cells and the CAN1 reverse orientation was confirmed by sequencing (“can1ori scr” primers in Materials and Methods S1).
An MMR deficient strain was created by deleting MSH2 in the AC402 wt (to give msh2Δ) using the pAG32 plasmid and transfection technique [39] and the primers msh2_hphMX4, shown in Materials and Methods S1. The deletion was confirmed using primers flanking MSH2. This strain was then crossed with rnr1-Y285A [16], sporulated and dissected spores selected on Hygromycin and –Trp plates for the double mutant rnr1-Y285A msh2Δ.
Culture conditions and Canavanine resistance assay
All culturing was carried out at 30°C in YPAD (1% yeast extract, 2% bacto-peptone, 20 mg/l adenine, 2% agar for plates) liquid cultures in a shaking incubator at 160rpm. The Canavanine resistance assay was used to calculate mutation rates as previously described [17], [40], [41]. The Canr colonies were picked and the CAN1 gene amplified and sequenced (MWG Eurofins) using published primers [17] to produce the mutation spectra.
dNTP pool measurements
dNTP pools were measured in asynchronous cultures as described previously [16] with minor changes as described in [42]. Briefly, cells were harvested by filtration at a density of 0.4×107 to 0.5×107 cells/ml and NTPs and dNTPs were extracted in trichloroacetic acid and MgCl2 followed by a Freon-trioctylamine mix. dNTPs were separated using boronate columns (Affigel 601, Biorad) and analysed by HPLC on a LaChrom Elite UV detector (Hitachi) with a Partisphere SAX column (Hichrom, UK).
Supporting Information
Zdroje
1. KunzBA, KohalmiSE, KunkelTA, MathewsCK, McIntoshEM, et al. (1994) International Commission for Protection Against Environmental Mutagens and Carcinogens. Deoxyribonucleoside triphosphate levels: a critical factor in the maintenance of genetic stability. Mutation research 318 : 1–64.
2. Mathews CK (2014) Deoxyribonucleotides as genetic and metabolic regulators. FASEB J. E-pub ahead of print.
3. ReichardP (1988) Interactions between deoxyribonucleotide and DNA synthesis. Annual review of biochemistry 57 : 349–374.
4. ThelanderL (2007) Ribonucleotide reductase and mitochondrial DNA synthesis. Nature genetics 39 : 703–704.
5. ThelanderL, ReichardP (1979) Reduction of ribonucleotides. Annual review of biochemistry 48 : 133–158.
6. HoferA, CronaM, LoganDT, SjobergBM (2012) DNA building blocks: keeping control of manufacture. Critical reviews in biochemistry and molecular biology 47 : 50–63.
7. XuH, FaberC, UchikiT, FairmanJW, RaccaJ, et al. (2006) Structures of eukaryotic ribonucleotide reductase I provide insights into dNTP regulation. Proceedings of the National Academy of Sciences of the United States of America 103 : 4022–4027.
8. YaoNY, SchroederJW, YurievaO, SimmonsLA, O'DonnellME (2013) Cost of rNTP/dNTP pool imbalance at the replication fork. Proceedings of the National Academy of Sciences 110 : 12942–12947.
9. KunkelTA, ErieDA (2005) DNA mismatch repair. Annual review of biochemistry 74 : 681–710.
10. HarfeBD, Jinks-RobertsonS (2000) DNA mismatch repair and genetic instability. Annual review of genetics 34 : 359–399.
11. JohnsonRE, KovvaliGK, PrakashL, PrakashS (1996) Requirement of the Yeast MSH3 and MSH6 Genes for MSH2-dependent Genomic Stability. Journal of Biological Chemistry 271 : 7285–7288.
12. PeltomakiP, VasenHF (1997) Mutations predisposing to hereditary nonpolyposis colorectal cancer: Database and results of a collaborative study. The International Collaborative Group on Hereditary Nonpolyposis Colorectal Cancer. Gastroenterology 113 : 1146–1158.
13. SrivastavaK, SrivastavaA, MittalB (2010) Polymorphisms in ERCC2, MSH2, and OGG1 DNA repair genes and gallbladder cancer risk in a population of Northern India. Cancer 116 : 3160–3169.
14. Nick McElhinnySA, GordeninDA, StithCM, BurgersPM, KunkelTA (2008) Division of labor at the eukaryotic replication fork. Molecular Cell 30 : 137–144.
15. BurgersPMJ (2009) Polymerase Dynamics at the Eukaryotic DNA Replication Fork. Journal of Biological Chemistry 284 : 4041–4045.
16. KumarD, VibergJ, NilssonAK, ChabesA (2010) Highly mutagenic and severely imbalanced dNTP pools can escape detection by the S-phase checkpoint. Nucleic Acids Research 38 : 3975–3983.
17. KumarD, AbdulovicAL, VibergJ, NilssonAK, KunkelTA, et al. (2011) Mechanisms of mutagenesis in vivo due to imbalanced dNTP pools. Nucleic Acids Research 39 : 1360–1371.
18. RaghuramanMK, WinzelerEA, CollingwoodD, HuntS, WodickaL, et al. (2001) Replication dynamics of the yeast genome. Science 294 : 115–121.
19. YabukiN, TerashimaH, KitadaK (2002) Mapping of early firing origins on a replication profile of budding yeast. Genes to cells: devoted to molecular & cellular mechanisms 7 : 781–789.
20. KimH-M, NarayananV, MieczkowskiPA, PetesTD, KrasilnikovaMM, et al. (2008) Chromosome fragility at GAA tracts in yeast depends on repeat orientation and requires mismatch repair. EMBO J 27 : 2896–2906.
21. PursellZF, IsozI, LundstromEB, JohanssonE, KunkelTA (2007) Yeast DNA polymerase epsilon participates in leading-strand DNA replication. Science 317 : 127–130.
22. SereroA, JubinC, LoeilletS, Legoix-NéP, NicolasAG (2014) Mutational landscape of yeast mutator strains. Proceedings of the National Academy of Sciences 111 : 1897–1902 doi:10.1073/pnas.1314423111
23. Lujan SA, Clausen AR, Clark AB, MacAlpine HK, MacAlpine DM, et al. (2014) Heterogeneous polymerase fidelity and mismatch repair bias genome variation and composition. Genome Res. doi:10.1101/gr.178335.114
24. KaneDP, ShcherbakovaPV (2014) A common cancer-associated DNA polymerase epsilon mutation causes an exceptionally strong mutator phenotype, indicating fidelity defects distinct from loss of proofreading. Cancer Research 74 : 1895–1901.
25. Shinbrot E, Henninger EE, Weinhold N, Covington KR, Goksenin AY, et al. (2014) Exonuclease mutations In DNA Polymerase Epsilon reveal replication strand specific mutation patterns and human origins of replication. Genome Research. doi:10.1101/gr.174789.114
26. AlexandrovLB, Nik-ZainalS, WedgeDC, AparicioSA, BehjatiS, et al. (2013) Signatures of mutational processes in human cancer. Nature 500 : 415–421.
27. MuznyDM, BainbridgeMN, ChangK, DinhHH, DrummondJA, et al. (2012) Comprehensive molecular characterization of human colon and rectal cancer. Nature 487 : 330–337.
28. GetzG, GabrielSB, CibulskisK, LanderE, SivachenkoA, et al. (2013) Integrated genomic characterization of endometrial carcinoma. Nature 497 : 67–73.
29. LujanSA, WilliamsJS, PursellZF, Abdulovic-CuiAA, ClarkAB, et al. (2012) Mismatch Repair Balances Leading and Lagging Strand DNA Replication Fidelity. PLoS Genet 8: e1003016.
30. GraggH, HarfeBD, Jinks-RobertsonS (2002) Base composition of mononucleotide runs affects DNA polymerase slippage and removal of frameshift intermediates by mismatch repair in Saccharomyces cerevisiae. Mol Cell Biol 22 : 8756–8762.
31. LehnerK, MudrakSV, MinesingerBK, Jinks-RobertsonS (2012) Frameshift mutagenesis: the roles of primer-template misalignment and the nonhomologous end-joining pathway in Saccharomyces cerevisiae. Genetics 190 : 501–510.
32. SchaaperRM, RadmanM (1989) The extreme mutator effect of Escherichia coli mutD5 results from saturation of mismatch repair by excessive DNA replication errors. The EMBO journal 8 : 3511–3516.
33. KramerB, KramerW, FritzHJ (1984) Different base/base mismatches are corrected with different efficiencies by the methyl-directed DNA mismatch-repair system of E. coli. Cell 38 : 879–887.
34. DohetC, WagnerR, RadmanM (1985) Repair of defined single base-pair mismatches in Escherichia coli. Proceedings of the National Academy of Sciences of the United States of America 82 : 503–505.
35. SchaaperRM, DunnRL (1991) Spontaneous mutation in the Escherichia coli lacI gene. Genetics 129 : 317–326.
36. SchaaperRM (1993) Base selection, proofreading, and mismatch repair during DNA replication in Escherichia coli. The Journal of biological chemistry 268 : 23762–23765.
37. Nick McElhinnySA, KisslingGE, KunkelTA (2010) Differential correction of lagging-strand replication errors made by DNA polymerases {alpha} and {delta}. Proceedings of the National Academy of Sciences of the United States of America 107 : 21070–21075.
38. GueldenerU, HeinischJ, KoehlerGJ, VossD, HegemannJH (2002) A second set of loxP marker cassettes for Cre-mediated multiple gene knockouts in budding yeast. Nucleic Acids Research 30: e23.
39. GoldsteinAL, McCuskerJH (1999) Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast 15 : 1541–1553.
40. DrakeJW (1991) A constant rate of spontaneous mutation in DNA-based microbes. Proceedings of the National Academy of Sciences of the United States of America 88 : 7160–7164.
41. LeaDE, CoulsonCA (1949) The distribution of the numbers of mutants in bacterial populations. Journal of genetics 49 : 264–285.
42. HochNC, ChenES, BucklandR, WangSC, FazioA, et al. (2013) Molecular basis of the essential s phase function of the rad53 checkpoint kinase. Molecular and cellular biology 33 : 3202–3213.
Štítky
Genetika Reprodukční medicína
Článek Large-scale Metabolomic Profiling Identifies Novel Biomarkers for Incident Coronary Heart DiseaseČlánek Notch Signaling Mediates the Age-Associated Decrease in Adhesion of Germline Stem Cells to the NicheČlánek Phosphorylation of Mitochondrial Polyubiquitin by PINK1 Promotes Parkin Mitochondrial TetheringČlánek Natural Variation Is Associated With Genome-Wide Methylation Changes and Temperature SeasonalityČlánek Overlapping and Non-overlapping Functions of Condensins I and II in Neural Stem Cell DivisionsČlánek Unisexual Reproduction Drives Meiotic Recombination and Phenotypic and Karyotypic Plasticity inČlánek Tetraspanin (TSP-17) Protects Dopaminergic Neurons against 6-OHDA-Induced Neurodegeneration inČlánek ABA-Mediated ROS in Mitochondria Regulate Root Meristem Activity by Controlling Expression inČlánek Mutations in Global Regulators Lead to Metabolic Selection during Adaptation to Complex EnvironmentsČlánek The Evolution of Sex Ratio Distorter Suppression Affects a 25 cM Genomic Region in the Butterfly
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2014 Číslo 12
-
Všechny články tohoto čísla
- Stratification by Smoking Status Reveals an Association of Genotype with Body Mass Index in Never Smokers
- Genome Wide Meta-analysis Highlights the Role of Genetic Variation in in the Regulation of Circulating Serum Chemerin
- Occupancy of Mitochondrial Single-Stranded DNA Binding Protein Supports the Strand Displacement Mode of DNA Replication
- Distinct Genealogies for Plasmids and Chromosome
- Large-scale Metabolomic Profiling Identifies Novel Biomarkers for Incident Coronary Heart Disease
- Non-coding RNAs Prevent the Binding of the MSL-complex to Heterochromatic Regions
- Plasmid Flux in ST131 Sublineages, Analyzed by Plasmid Constellation Network (PLACNET), a New Method for Plasmid Reconstruction from Whole Genome Sequences
- Epigenome-Guided Analysis of the Transcriptome of Plaque Macrophages during Atherosclerosis Regression Reveals Activation of the Wnt Signaling Pathway
- The Inventiveness of Nature: An Interview with Werner Arber
- Mediation Analysis Demonstrates That -eQTLs Are Often Explained by -Mediation: A Genome-Wide Analysis among 1,800 South Asians
- Generation of Antigenic Diversity in by Structured Rearrangement of Genes During Mitosis
- A Massively Parallel Pipeline to Clone DNA Variants and Examine Molecular Phenotypes of Human Disease Mutations
- Genetic Analysis of the Cardiac Methylome at Single Nucleotide Resolution in a Model of Human Cardiovascular Disease
- Genetic Analysis of Circadian Responses to Low Frequency Electromagnetic Fields in
- The Dissection of Meiotic Chromosome Movement in Mice Using an Electroporation Technique
- Altered Chromatin Occupancy of Master Regulators Underlies Evolutionary Divergence in the Transcriptional Landscape of Erythroid Differentiation
- Syd/JIP3 and JNK Signaling Are Required for Myonuclear Positioning and Muscle Function
- Notch Signaling Mediates the Age-Associated Decrease in Adhesion of Germline Stem Cells to the Niche
- Mutation of Leads to Blurred Tonotopic Organization of Central Auditory Circuits in Mice
- The IKAROS Interaction with a Complex Including Chromatin Remodeling and Transcription Elongation Activities Is Required for Hematopoiesis
- RAN-Binding Protein 9 is Involved in Alternative Splicing and is Critical for Male Germ Cell Development and Male Fertility
- Enhanced Longevity by Ibuprofen, Conserved in Multiple Species, Occurs in Yeast through Inhibition of Tryptophan Import
- Phosphorylation of Mitochondrial Polyubiquitin by PINK1 Promotes Parkin Mitochondrial Tethering
- Recurrent Loss of Specific Introns during Angiosperm Evolution
- Natural Variation Is Associated With Genome-Wide Methylation Changes and Temperature Seasonality
- SEEDSTICK is a Master Regulator of Development and Metabolism in the Arabidopsis Seed Coat
- Overlapping and Non-overlapping Functions of Condensins I and II in Neural Stem Cell Divisions
- Unisexual Reproduction Drives Meiotic Recombination and Phenotypic and Karyotypic Plasticity in
- Tetraspanin (TSP-17) Protects Dopaminergic Neurons against 6-OHDA-Induced Neurodegeneration in
- ABA-Mediated ROS in Mitochondria Regulate Root Meristem Activity by Controlling Expression in
- Mutations in Global Regulators Lead to Metabolic Selection during Adaptation to Complex Environments
- Global Analysis of Photosynthesis Transcriptional Regulatory Networks
- Mucolipin Co-deficiency Causes Accelerated Endolysosomal Vacuolation of Enterocytes and Failure-to-Thrive from Birth to Weaning
- Controlling Pre-leukemic Thymocyte Self-Renewal
- How Malaria Parasites Avoid Running Out of Ammo
- Echoes of the Past: Hereditarianism and
- Deep Reads: Strands in the History of Molecular Genetics
- Keep on Laying Eggs Mama, RNAi My Reproductive Aging Blues Away
- Analysis of a Plant Complex Resistance Gene Locus Underlying Immune-Related Hybrid Incompatibility and Its Occurrence in Nature
- Epistatic Adaptive Evolution of Human Color Vision
- Increased and Imbalanced dNTP Pools Symmetrically Promote Both Leading and Lagging Strand Replication Infidelity
- Genetic Basis of Haloperidol Resistance in Is Complex and Dose Dependent
- Genome-Wide Analysis of DNA Methylation Dynamics during Early Human Development
- Interaction between Conjugative and Retrotransposable Elements in Horizontal Gene Transfer
- The Evolution of Sex Ratio Distorter Suppression Affects a 25 cM Genomic Region in the Butterfly
- is Required for Adult Maintenance of Dopaminergic Neurons in the Ventral Substantia Nigra
- PRL1, an RNA-Binding Protein, Positively Regulates the Accumulation of miRNAs and siRNAs in Arabidopsis
- Genetic Control of Contagious Asexuality in the Pea Aphid
- Early Mesozoic Coexistence of Amniotes and Hepadnaviridae
- Local and Systemic Regulation of Plant Root System Architecture and Symbiotic Nodulation by a Receptor-Like Kinase
- Gene Pathways That Delay Reproductive Senescence
- The Evolution of Fungal Metabolic Pathways
- Maf1 Is a Novel Target of PTEN and PI3K Signaling That Negatively Regulates Oncogenesis and Lipid Metabolism
- Formation of Linear Amplicons with Inverted Duplications in Requires the MRE11 Nuclease
- Identification of Rare Causal Variants in Sequence-Based Studies: Methods and Applications to , a Gene Involved in Cohen Syndrome and Autism
- Rrp12 and the Exportin Crm1 Participate in Late Assembly Events in the Nucleolus during 40S Ribosomal Subunit Biogenesis
- The Mutations in the ATP-Binding Groove of the Rad3/XPD Helicase Lead to -Cockayne Syndrome-Like Phenotypes
- Topoisomerase I Plays a Critical Role in Suppressing Genome Instability at a Highly Transcribed G-Quadruplex-Forming Sequence
- A Cbx8-Containing Polycomb Complex Facilitates the Transition to Gene Activation during ES Cell Differentiation
- Transcriptional Frameshifting Rescues Type VI Secretion by the Production of Two Length Variants from the Prematurely Interrupted Gene
- Association Mapping across Numerous Traits Reveals Patterns of Functional Variation in Maize
- Genome-Wide Analysis of -Regulated and Phased Small RNAs Underscores the Importance of the ta-siRNA Pathway to Maize Development
- Dissemination of Cephalosporin Resistance Genes between Strains from Farm Animals and Humans by Specific Plasmid Lineages
- The Tau Tubulin Kinases TTBK1/2 Promote Accumulation of Pathological TDP-43
- Germline Signals Deploy NHR-49 to Modulate Fatty-Acid β-Oxidation and Desaturation in Somatic Tissues of
- Microevolution of in Macrophages Restores Filamentation in a Nonfilamentous Mutant
- Vangl2-Regulated Polarisation of Second Heart Field-Derived Cells Is Required for Outflow Tract Lengthening during Cardiac Development
- Chondrocytes Transdifferentiate into Osteoblasts in Endochondral Bone during Development, Postnatal Growth and Fracture Healing in Mice
- A ABC Transporter Regulates Lifespan
- RA and FGF Signalling Are Required in the Zebrafish Otic Vesicle to Pattern and Maintain Ventral Otic Identities
- , and Reprogram Thymocytes into Self-Renewing Cells
- The miR9863 Family Regulates Distinct Alleles in Barley to Attenuate NLR Receptor-Triggered Disease Resistance and Cell-Death Signaling
- Detection of Pleiotropy through a Phenome-Wide Association Study (PheWAS) of Epidemiologic Data as Part of the Environmental Architecture for Genes Linked to Environment (EAGLE) Study
- Extensive Copy-Number Variation of Young Genes across Stickleback Populations
- The and Genetic Modules Interact to Regulate Ciliogenesis and Ciliary Microtubule Patterning in
- Analysis of the Genome, Transcriptome and Secretome Provides Insight into Its Pioneer Colonization Strategies of Wood
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Tetraspanin (TSP-17) Protects Dopaminergic Neurons against 6-OHDA-Induced Neurodegeneration in
- Maf1 Is a Novel Target of PTEN and PI3K Signaling That Negatively Regulates Oncogenesis and Lipid Metabolism
- The IKAROS Interaction with a Complex Including Chromatin Remodeling and Transcription Elongation Activities Is Required for Hematopoiesis
- Echoes of the Past: Hereditarianism and
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



