-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaPRL1, an RNA-Binding Protein, Positively Regulates the Accumulation of miRNAs and siRNAs in Arabidopsis
PRL1, a conserved WD-40 protein, is required for plant development and immune responses. However, its functional mechanisms are not well understood. Here, we show the positive impact of PRL1 on the accumulation of miRNAs and siRNAs, which are key regulators of plant growth and immunity. PRL1 interacts with multiple DCLs (the processors of miRNAs and siRNAs) and is required for their optimal activities, suggesting that PRL1 acts as a general factor to facilitate the production of miRNAs and siRNAs. In addition, PRL1 is an RNA-binding protein, binds pri-miRNAs in vivo and positively influences the levels of pri-miRNAs levels without affecting the promoter activities of genes encoding pri-miRNAs. These results suggest that PRL1 may also stabilize pri-miRNAs. We further show that RPL1 and its interactor CDC5 (a DNA-binding protein) synergistically regulate pri-miRNA levels, resulting in enhanced effects on miRNA accumulation, although they function together as a complex to facilitate DCL1 activity.
Published in the journal: . PLoS Genet 10(12): e32767. doi:10.1371/journal.pgen.1004841
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1004841Summary
PRL1, a conserved WD-40 protein, is required for plant development and immune responses. However, its functional mechanisms are not well understood. Here, we show the positive impact of PRL1 on the accumulation of miRNAs and siRNAs, which are key regulators of plant growth and immunity. PRL1 interacts with multiple DCLs (the processors of miRNAs and siRNAs) and is required for their optimal activities, suggesting that PRL1 acts as a general factor to facilitate the production of miRNAs and siRNAs. In addition, PRL1 is an RNA-binding protein, binds pri-miRNAs in vivo and positively influences the levels of pri-miRNAs levels without affecting the promoter activities of genes encoding pri-miRNAs. These results suggest that PRL1 may also stabilize pri-miRNAs. We further show that RPL1 and its interactor CDC5 (a DNA-binding protein) synergistically regulate pri-miRNA levels, resulting in enhanced effects on miRNA accumulation, although they function together as a complex to facilitate DCL1 activity.
Introduction
In plants and animals, microRNAs (miRNAs), ∼20–25 nucleotides (nt) in size, regulate gene expression in various biological processes such as development and metabolism [1]–[3]. They are produced as duplexes through precise excision from imperfect fold-back primary transcripts (pri-miRNAs) [1]–[3]. In the miRNA duplex, the miRNA strand is loaded into ARGONAUTE (AGO) proteins to repress the expression of target genes containing its complementary sequences while the other strand (passenger strand; miRNA*) is degraded [1]–[3]. Plants and animals also use small interfering RNAs (siRNAs) to repress gene expression. siRNAs are chemically identical to miRNAs [2]. However they are produced from long double stranded RNAs. The two major classes of plant siRNAs are siRNAs derived from repeated DNAs (ra-siRNAs) and trans-acting siRNAs (ta-siRNAs) [4], [5].
In plants, most pri-miRNAs are transcribed by DNA-dependent RNA polymerase II (Pol II) from endogenous miRNA encoding genes (MIR) [1], [2]. The mediator complex and Negative on TATA less2 (NOT2; a transcription factor) regulate the transcription of MIR [6], [7]. After generation, pri-miRNAs are proposed to be stabilized by DAWDLE (DDL), an RNA binding protein [8]. Pri-miRNAs are then processed to stem-loop precursors (pre-miRNAs) and finally to the miRNA/miRNA* duplex by Dicer-LIKE 1 (DCL1; an RNAse III enzyme) in the nucleus in plants [9], [10]. The C2H2 zinc-finger protein SERRATE (SE) and the RNA binding proteins HYPONASTIC LEAVES 1 (HYL1) and TOUGH (TGH) form a complex with DCL1 to ensure efficient and accurate process of pri-miRNAs [9], [11]–[17]. To ensure its proper function, HYL1 needs to be dephosphorylated during pri-miRNA processing [18]. Several other proteins including DDL, Cap-Binding Protein 20 (CBP20), CBP80, RACK1 and NOT2 are associated with the DCL1 complex to facilitate miRNA maturation [7], [8], [19]–[21]. NOT2 and MODIFIER OF SNC1, 2 (MOS2; an RNA binding protein) have been shown to guide the correct localization of the DCL1 complex [7], [22]. SICKLE (SIC; a proline rich protein) is shown to regulate the accumulation of some miRNAs [23]. Besides protein factors, the structure of pri-miRNAs plays essential roles in regulating DCL1 activity [24]–[27]. For instance, an imperfectly paired lower stem of ∼15 bp below the miRNA:miRNA* duplex is crucial for the initial pri-miRNA cleavage [25]–[27].
We previously showed that Cell Division Cycle 5 (CDC5), a DNA-binding protein, positively regulates miRNA biogenesis in Arabidopsis [28]. CDC5 interacts with Pol II and MIR promoters [28]. Lack of CDC5 reduces the occupancy of Pol II at MIR promoters and pri-miRNA levels, suggesting that CDC5 is a positive transcription factor of MIR [28]. Besides acting as a transcription factor, CDC5 functions as a co-factor of the DCL1 complex to participate pri-miRNA processing [28]. CDC5 is a component of the conserved MOS4-associated complex (MAC). MAC was first identified as a suppressor of snc1, which carries a gain-of-function mutation in the SNC gene and show constitutive resistance to a wide spectrum of pathogens [29]. Loss-of-function mutations in the MAC complex reduce plant immunity to bacterial infections and cause multiple developmental defects such as reduced fertility and delayed growth [29]. The counterparts of MAC in yeast and Human associate with spliceosome and function in splicing [29]. Other components of MAC include MOS4 (a coil-coil domain containing protein), PRL1 (a WD-40 protein), MAC3A and MAC3B (two functionally redundant U-box E3 ubiquitin ligases). Among these proteins, PRL1 and MOS4 have been shown to interact with CDC5 directly [29].
In this study, we show that PRL1 but not MOS4 plays important roles in the accumulation of miRNAs and siRNAs. Lack of PRL1 in prl1 reduces miRNA accumulation and pri-miRNA processing efficiency. In addition, PRL1 interacts with the DCL1 complex, suggesting it may function as co-factor of DCL1 to promote miRNA maturation. Pri-miRNA levels are reduced in prl1 relative to wild-type plants. However, MIR promoter activity is not affected by prl1, despite of the association of PRL1 with Pol II. Based on the facts that PRL1 is an RNA-binding protein and binds pri-miRNAs in vivo, we propose that PRL1 may stabilize pri-miRNAs. Furthermore, the levels of both miRNAs and pri-miRNAs are further reduced in cdc5 prl1 relative to either cdc5 or prl1. However, CDC5 and PRL1 do not show additive effects on the processing of pri-miRNAs. These results suggest that CDC5 and PRL1 may synergistically influence pri-miRNAs levels and act together as a complex to promote miRNA maturation. PRL1 also interacts with DCL3 and DCL4, which produces siRNAs, and is required for their optimal activities, suggesting that PRL1 may be a general accessory factor for the production of small RNAs.
Results
The accumulation of miRNAs and siRNAs is reduced in prl1-2
Given the role of CDC5 in miRNA biogenesis, it is possible that other components of the MAC complex may also be required for miRNA accumulation. Therefore, we examined the effect of the mutants mac3b (SALK_050811), mos4 (SALK_0090851C) and prl1-2 on miRNA abundance using Northern blot. We also included snc1 (SALK_047058C) in the analysis since SNC1 is related to the MAC complex and snc1 causes development defects. These mutants are likely null since the transcripts of corresponding genes could not be detected by RT-PCR (Figure S1A). Like in cdc5-1, the abundance of all four tested miRNAs (miR167, miR171, miR172 and miR173) was decreased in prl1-2 compared to Col (wild-type control). In contrast, miRNA levels in mos4, mac3b and snc1 were comparable with those in Col (Fig. 1A). We examined the accumulation of additional miRNAs in prl1-2 and found that all these miRNAs were reduced in abundance in prl1-2 relative to Col (Fig. 1B). In addition, expression a wild-type copy of PRL1 fused with a YFP tag under the control of its native promoter (pPRL1::PRL1-YFP) fully recovered miRNA levels in prl1-2 (Fig. 1B). These results demonstrated that PRL1 but not MOS4 and MAC3b is required for miRNA accumulation. We next analyzed the transcript levels of several miRNA targets (ARF3, CUC1, MYB33, MYB65 and PHV) in prl1-2 and Col by quantitative RT-PCR (qRT-PCR) in order to test the effect of prl1-2 on miRNA function. The transcription levels of these targets were slightly increased in prl1-2 relative to Col (Figure. S1B). The PRL1 transgene fully recovered miRNA function in prl1 (Figure S1B). We also asked if PRL1 has a role in siRNA biogenesis. The levels of nine examined siRNAs (three ta-siRNAs and six ra-siRNAs) were reduced compared to those in Col (Fig. 1B and 1C), which was complemented by the expression of pPRL1::PRL1-YFP. These results revealed that like CDC5, PRL1 positively regulates the levels of miRNAs and siRNAs in Arabidopsis.
Fig. 1. PRL1 is required for the accumulation of miRNAs. 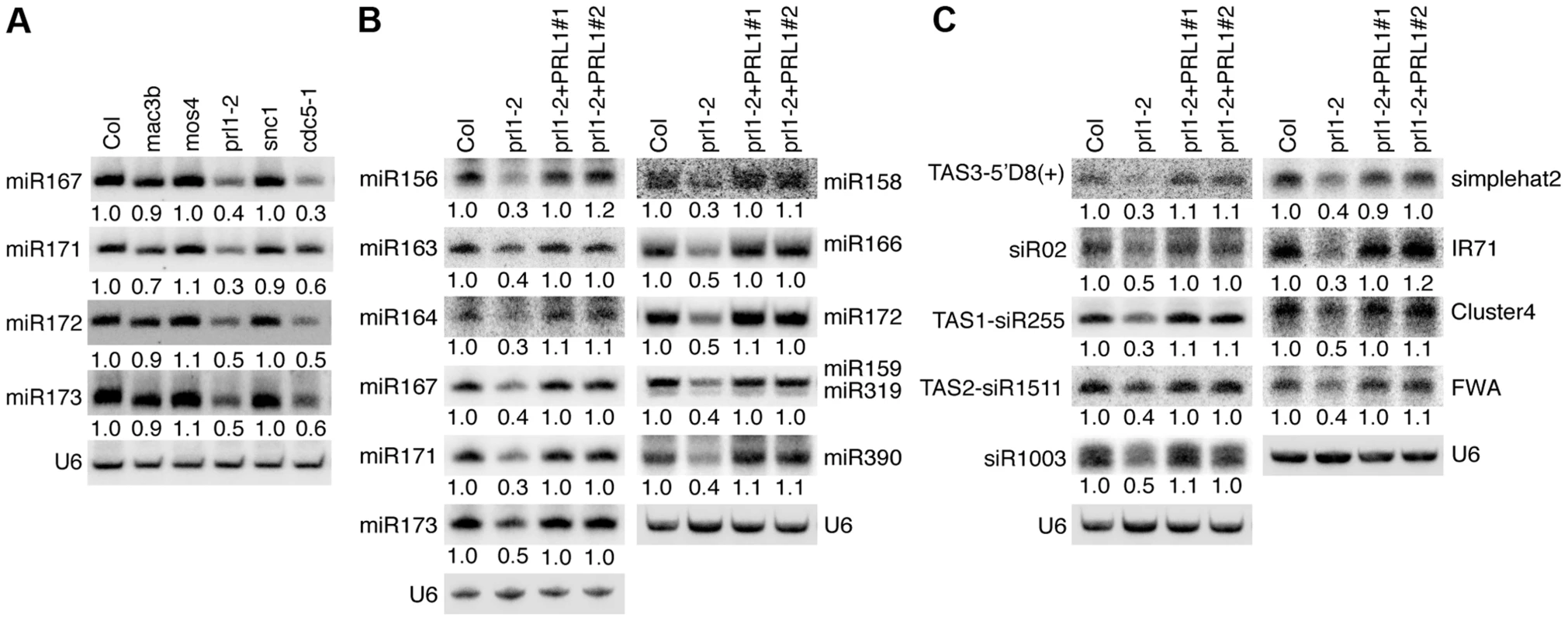
(A) The effect of various MAC components on the abundance of miRNAs (B) The levels of miRNAs are reduced in prl1-2. (C) The levels of siRNAs are reduced in prl1-2. Col: wild-type control. For miR159/319: upper band, miR159; lower band, miR319. Northern blot was used to detect small RNAs and the radioactive signals were quantified with ImageQuant (V5.2). To determine the amount of miRNAs/siRNAs in various mutants relative to that in Col, the radioactive signals of miRNAs/siRNAs were normalized to U6 RNA and compared with that in Col (set as 1). The numbers indicate the average value of three repeats (P<0.05). PRL1 associates with Pol II and DCL1
The PRL1-CDC5 interaction suggests that similar to CDC5, PRL1 may act as a co-factor of Pol II and DCL1 to regulate miRNA accumulation. To test these two possibilities, we first examined the PRL1-Pol II association using co-immunoprecipitation (co-IP) assay. In this experiment, anti-YFP and anti-RPB2 that detects the second largest subunit of Pol II (RPB2) [6] were used to capture the PRL1-YFP and Pol II complex, respectively, from the protein extracts of prl1-2 complementation line expressing the pPRL1::PRL1-YFP transgene. After IP, PRL1-YFP was detected in the Pol II precipitates whereas RPB2 existed in the PRL1-YFP complex (Fig. 2A and 2B). In contrast, no interaction was detected in the control reactions (Fig. 2A and 2B), demonstrating the PRL1-Pol II association.
Fig. 2. PRL1 associates with the Pol II and DCL1 complexes. 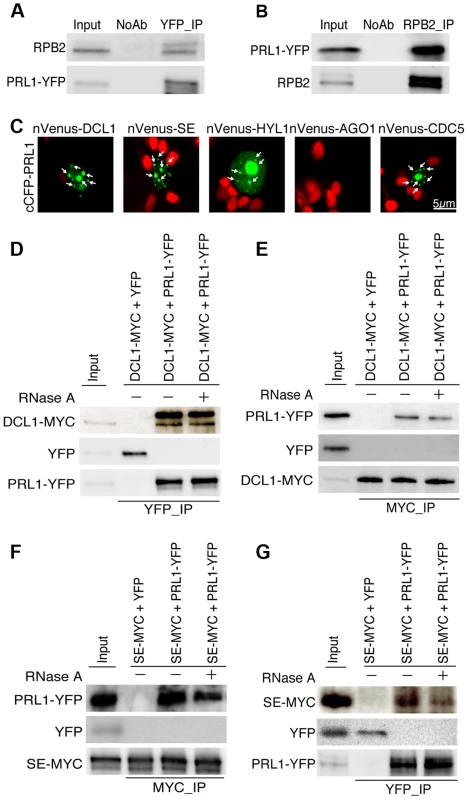
(A) and (B) Co-immunoprecipitation (Co-IP) between PRL1 and Pol II. Protein extracts from transgenic plants containing PRL1-YFP were incubated with Anti-YFP or anti-RPB2 antibodies to precipitate PRL1-YFP or Pol II. PRL1-YFP and RBP2 were detected with western blot and labeled on the left side of the picture. Ten percent of input proteins were used for IP and one percent of input proteins were used for Co-IP. (C) BiFC analysis of PRL1 with DCL1, HYL1, SE, AGO1 and CDC5. Paired cCFP- and nVenus-fusion proteins were co-infiltrated into N. benthamiana leaves. The BiFC signal (Yellow fluorescence) was detected at 48 h after infiltration by confocal microscopy, assigned as green color and marked with arrow. 30 nuclei were examined for each pair and an image is shown. Red: auto fluorescence of chlorophyll. (D) and (E) Co-immunoprecipitation between PRL1 and DCL1. (F) and (G) Co-immunoprecipitation between PRL1 and SE. PRL1-YFP or YFP were co-expressed with DCL1-MYC and SE-MYC in N. benthamiana, respectively. Anti-YFP and anti-MYC (MBL) antibodies were used to detect YFP- and MYC-fused proteins, respectively. The protein pairs in the protein extracts were indicated on the on tope of the picture and proteins detected by western blot were indicated on the left side of the picture. Ten percent of input proteins were used for IP and one percent of inputs proteins were used for Co-IP. We next tested the association of PRL1 with the components of DCL1 complex using a bimolecular fluorescence complementation (BiFC). In the BiFC assay, transient co-expression of PRL1 fused with C-terminal fragment of cyan fluorescent protein (cCFP) with DCL1, SE, HYL1 or CDC5 fused with the N-terminal fragment of Venus (nVenus) produced yellow fluorescence signals (Fig. 2C), suggesting that PRL1 might associate with the DCL1 complex. To verify this result, we tested co-IP of PRL1 with DCL1 and SE. After PRL1-YFP or YFP was transiently co-expressed with DCL1-MYC and SE-MYC fusion proteins in N. benthamiana, respectively, IPs were performed with anti-YFP or anti-MYC antibodies. Western blots detected PRL1-YFP in the DCL1-MYC and SE-MYC complexes and DCL1-MYC/SE-MYC in the PRL1-YFP precipitates, suggesting that PRL1-YFP and DCL1/SE reciprocally pulled down each other (Fig. 2D, 2E, 2F and 2G). As a control, YFP did not show interaction with either DCL1 or SE. These results demonstrated the association of PRL1 with DCL1 and SE.
PRL1 positively influences pri-miRNA levels without affecting MIR promoter activity
The interaction of PRL1 with Pol II suggests that PRL1 may positively regulate MIR transcription. If so, lack of PRL1 will impair MIR transcription, resulting in reduced levels of pri-miRNAs. To test this, we compared the pri-miRNA levels in prl1-2 with those in Col using qRT-PCR. In fact, the levels of all seven examined pri-miRNAs in prl1-2 were less than those in Col (Fig. 3A), which were recovered in the complementation line of prl1-2 (Fig. 3A). To test whether the reduction of pri-miRNA levels is due to impaired MIR promoter activity, we introduced the prl1-2 mutation into a Col transgenic line containing a single cope of GUS transgene driven by MIR167a promoter (pMIR167a::GUS), which was previously used to test the function of the mediator complex in regulating MIR transcription [6]. However, GUS staining and qRT-PCR analysis showed a similar GUS expression level in PRL1+ (PRL1/PRL1 or PRL1/prl1 genotype) and prl1-2 containing the pMIR167a::GUS transgene (Fig. 3B and 3C). This result demonstrated that PRL1 does not affect MIR promoter activity. Consistent with this notion, prl1 did not show obvious effect on MIR172b promoter activity (Figure S2A).
Fig. 3. PRL1 positively influences pri-miRNA levels. 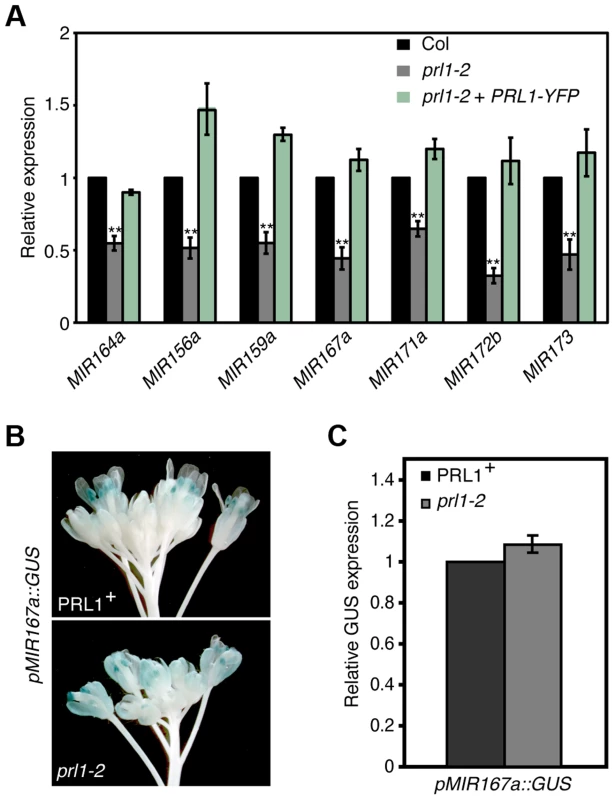
(A) The abundance of pri-miRNAs in inflorescences of prl1-2 and Col. The transcript levels of pri-miRNAs in prl1-2 were determined by quantitative RT-PCR (qRT-PCR), normalized to that of UBQUITIN5 (UBQ5) and compared with those in Col. Value of Col was set to 1. Error bars indicate standard deviation of three technical replications. **: P<0.01. (B) GUS expression in PRL1+ and prl1-2 harboring pMIR167a::GUS. PRL1+: PRL1/PRL1 or PRL1/prl1-2. Twenty plants containing GUS were stained for each genotype and an image was shown. (C) Relative GUS mRNA levels in PRL1+ and prl1-2 harboring pMIR167a::GUS. GUS transcript levels were determined by qRT-PCR and normalized to UBQ5. Value of PRL1+ was set to 1. Standard deviation of three technical replications was shown as error bars. P = 0.11 (t-test). We next evaluated the effect of prl1-2 on the half-lives of pri-miRNAs using cordycepin as a transcriptional inhibitor [30]. Two-week old plants were transferred to medium containing cordycepin. After incubation was stopped at various time points, we measured the levels of pri-miR164a and pri-miR167a using qRT-PCR. The results showed that the degradation rate of pri-miR164a and pri-miR167a in prl1-2 is similar to that in Col (Figure S2B).
PRL1 functions in miRNA maturation
We next asked whether PRL1 has a role in processing of miRNA precursors through an in vitro processing assay [13], [31] since it is associated with DCL1 and SE. In this experiment, a portion of pri-miR162b that contains the stem-loop of miR162b with 6-nt arms at each end (MIR162b; Fig. 4A) and pre-miR162b (Fig. 4B) were first produced through in vitro transcription in the presence of [α-32P] UTP. [32P]-labeled MIR162b and pre-miR162b were then processed in the protein extracts of young flower buds of prl1-2 or Col. The production of miR162b from both MIR162b and pre-miR162b in prl1-2 at various time points was less than that in Col (Fig. 4C and 4D). The processing of MIR162b and pre-miRNA162b was recovered in the PRL1 complementation line (Figure S3) The levels of miR162 produced from MIR162b and pre-miRNA162b in prl1 at 80 min were ∼40% of those produced in Col (Fig. 4E and 4F). These results suggested that PRL1 might have a role in promoting miRNA maturation.
Fig. 4. PRL1 is required for miRNA maturation in vitro. 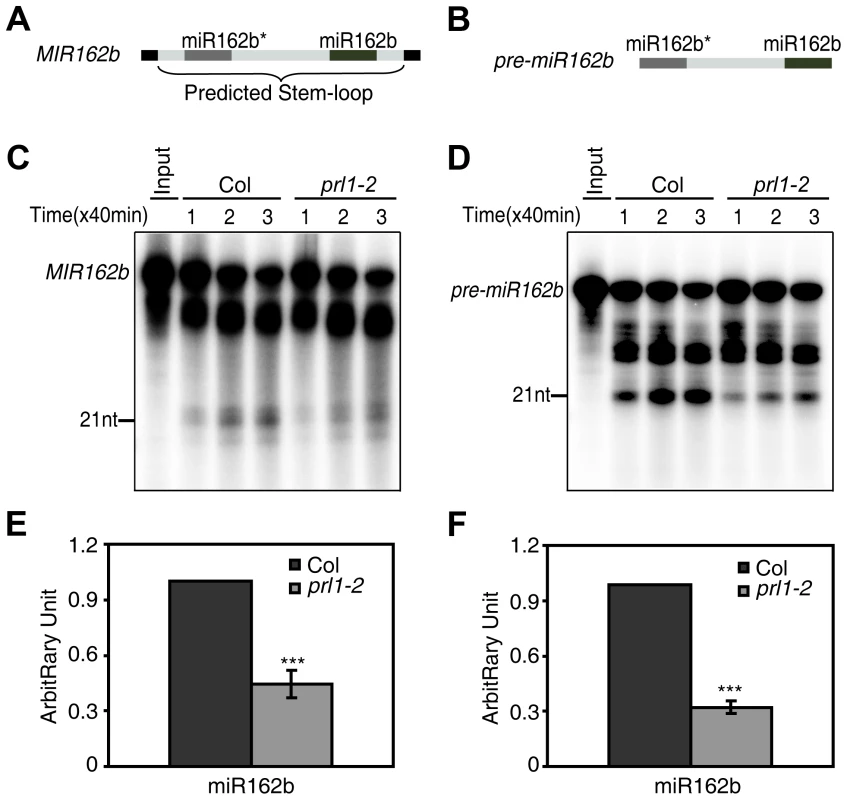
(A) and (B) A schematic diagram of the MIR162b (A) and pre-miR162b (B) used in vitro processing assay. (C) and (D) The amount of miR162b produced from MIR162b and pre-miR162b were reduced in prl1-2. Proteins were isolated from inflorescences of prl1-2 and Col and incubated with MIR162b or pre-miR162b. The reactions were stopped at various time points as indicated in the picture. (E) and (F) Quantification of miR162b production in prl1-2 compared to that in Col. Quantification analysis was performed at 80 min. The radioactive signal of miR162 were normalized to input and compared with that of Col. The amount of miR162 produced in Col was set as 1. The value represents mean of three repeats (*** P<0.001; t-test). The role of PRL1 in siRNA biognesis
We next asked the role of PRL1 in siRNA biogenesis, as prl1-2 reduces the accumulation of siRNAs. By analog, we examined the interaction of PRL1 with DCL3 and DCL4 and the effect of prl1-2 on dsRNA processing. To test the PRL1-DCL3/DCL4 interaction, we expressed a recombined PRL1 fused with a maltose-binding protein at its N-terminus (MBP-PRL1) and MBP in E.coli. The protein extracts containing MBP-PRL1 or MBP were mixed with protein extracts containing DCL3-YFP or DCL4-YFP, which were transiently expressed in N. benthamiana. Then the DCL3-YFP or DCL4-YFP complex was IPed with anti-YFP antibodies. MBP-PRL1, but not MBP, was co-IPed with DCL3-YFP and DCL4-YFP (Fig. 5A). In addition, YFP did not interact with MBP or MBP-PRL1. These results demonstrated that PRL1 interacts with DCL3 and DCL4 (Fig. 5A).
Fig. 5. The role of PRL1 in siRNA biogenesis. 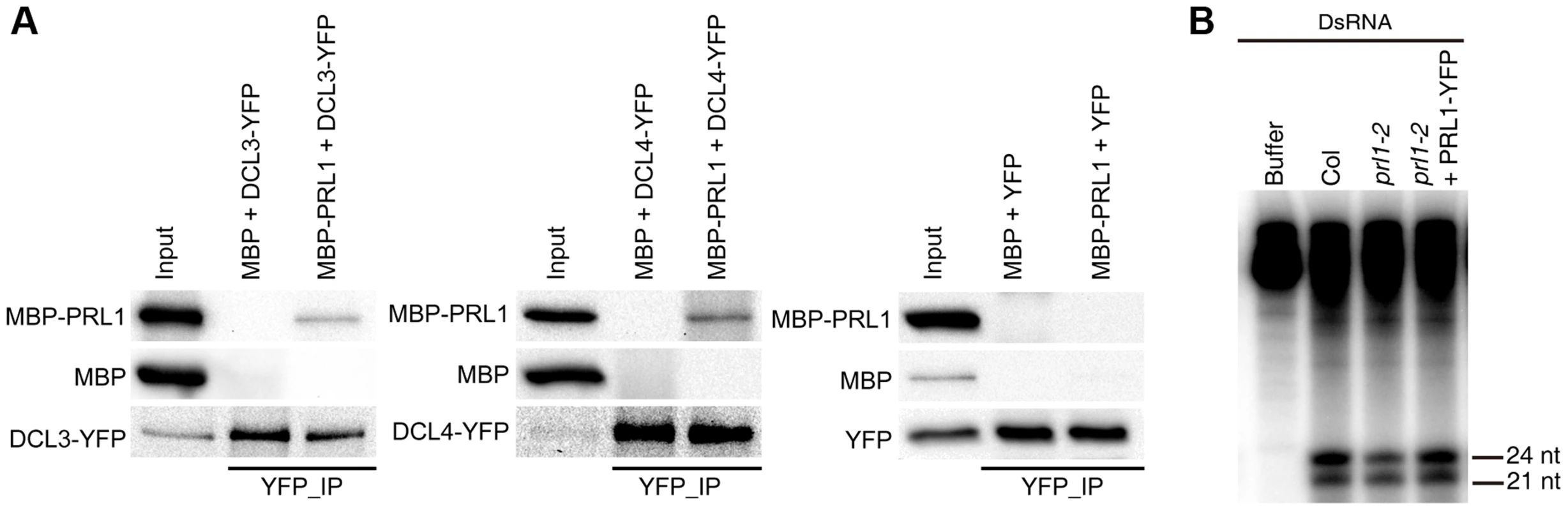
(A) PRL1 interacts with DCL3 and DCL4. Co-IP was performed to detect the interaction of PRL1 with DCL3 or DCL4. MBP and MBP-PRL1 fused protein were expressed in E.coli. YFP, DCL3-YFP and DCL4-YFP were expressed in N. benthamiana leaves. Anti-YFP was used for IP. For loading, ten percent and one percent of input proteins were used for IP and Co-IP, respectively. (B) prl1-2 impairs siRNA production from double-stranded RNAs (dsRNAs). Protein extracts isolated from inflorescences of Col, prl1-2 and prl1-2 containing a PRL1-YFP transgene were incubated dsRNAs for 120 min. dsRNAs were synthesized through in vitro transcription of a DNA fragment (5′ portion of UBQ5 gene, ∼460 bp) under the presence of [α-32P] UTP. To test the effect of prl1 on dsRNA processing, we generated ∼460 bp dsRNAs through in vitro transcription of a DNA fragment (5′ portion of UBIQUITIN 5) containing the T7 promoter at the end of each strand under the presence [α-32P] UTP. The radioactive labeled dsRNA then incubated with prl1 or Col protein extracts. The production of both 21 nt and 24 nt small RNAs was impaired in prl1 compared with that in Col and that in the complementation line (Fig. 5B). This result indicated that multiple DCL activities are impaired by prl1-2, because DCL3 is responsible for the production of 24 nt small RNAs and DCL1/DCL4 is involved in the production of 21 nt small RNAs from dsRNAs.
PRL1 and CDC5 synergistically regulate miRNA accumulation
CDC5 and PRL1 have been shown to directly interact with each other. Both CDC5 and PRL1 interact with DCL1 and positively regulate miRNA processing. These results raise a possibility that CDC5 and PRL1 may act as a complex to regulate DCL1 activity. In addition, CDC5 regulates MIR promoter activity while PRL1 does not. These suggest that PRL1 and CDC5 might act additionally in miRNA pathway. To test these two possibilities, we constructed a cdc5-1 prl1-2 double mutant by crossing prl1-2 into cdc5-1 and compared miRNA levels in cdc5-1 prl1-2 with those in cdc5-1 and prl1-2, respectively. The cdc5-1 prl1-2 double mutant displayed more severe developmental defects than either cdc5-1 or prl1-2, suggesting that PRL1 and CDC5 function additionally in regulating development (Fig. 6A). Northern blot analyses showed that the levels of several examined miRNAs in cdc5-1 prl1-2 were lower than those in either prl1-2 or cdc5-1 (Fig. 6B), indicating that PRL1 and CDC5 function synergistically in miRNA pathway.
Fig. 6. PRL1 and CDC5 synergistically regulate miRNA accumulation. 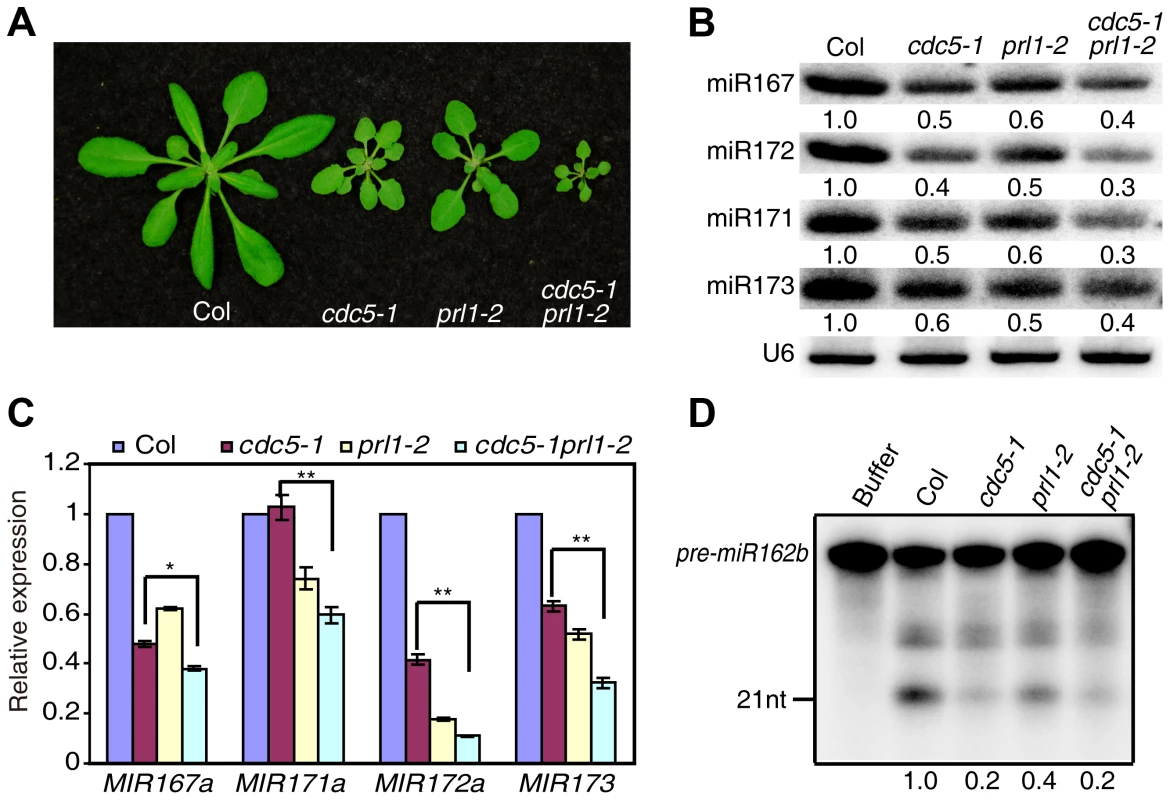
(A) Morphological phenotypes of Col, cdc5-1, prl1-2 and cdc5-1 prl1-2. (B) The abundance of miRNAs is lower in cdc5-1 prl1-2 than that in cdc5-1 or prl1-2. Small RNAs were detected by Northern Blot. To determine the amount of miRNAs, radioactive signals of miRNAs were normalized to U6 RNA. The number represents the relative abundance compared to Col (set as 1) quantified by three repeats (P<0.05). (C) The abundance of pri-miRNAs is reduced in cdc5-1 prl1-2. The levels of pri-miRNAs in various mutants were determined by qRT-PCR, normalized to UBQUITIN5 (UBQ5) and compared with those of Col (set as 1). Standard deviation of three technical replications was shown as error bars. **: P<0.01. (D) miR162b production from pre-miR162b in Col, cdc5-1 prl1-2, cdc5-1 and prl1-2. The reaction was stopped at 120 min. The radioactive signals of miR162b were normalized to input. The number represents the relative production in various genotypes compared to Col (set as 1) quantified by three repeats (P<0.05). There are at least two possible explanations for the further reduction of miRNA levels in cdc5-1 pr1l-2 relative to either cdc5-1 or prl1-2 based on the fact that both PRL1 and CDC5 positively regulate pri-miRNA levels and miRNA maturation. One is that pri-miRNA levels might be further reduced in cdc5-1 prl1-2. The other is that the processing efficiency of miRNA precursors might be lower than either cdc5-1 or prl1-2. To test these two possibilities, we first determined the pri-miRNA levels in cdc5-1 prl1-2, cdc5-1 and prl1-2 through qRT-PCR. The levels of several pri-miRNAs were decreased in cdc5-1 prl1-2 when compared with those in either cdc5-1 or prl1-2 (Fig. 6C), demonstrating that CDC5 and PRL1 indeed act synergistically in regulating pri-miRNA levels. Next, we evaluated the in vitro processing of pre-miR162b in cdc5-1 prl1-2. The amount of miR162b produced in cdc5-1 prl1-2 was similar to that in cdc5-1 and slightly lower than that in prl1-2 (Fig. 6D), suggesting that PRL1 and CDC5 may not act additionally in promoting miRNA maturation.
PRL1 binds pri-miRNAs in vitro and in vivo
Given the role of PRL1 in RNA metabolism, it is reasonable to speculate that PRL1 might have an RNA-binding activity. We therefore performed an in vitro RNA pull-down assay to test this possibility. In this assay, recombinant PRL1 fused with a maltose-binding protein at its N-terminus (MBP-PRL1) and MBP were expressed in E.coli and purified with amylose resin (Fig. 7A). MBP-PRL1 and MBP were then incubated with [32P]-labeled MIR162b or pre-miR162b, respectively. MBP-PRL1 but not MBP bound MIR162b and pre-miR162b after incubation. In addition, when excess amount of unlabeled MIR162b or pre-miR162b was added in the reaction, radioactive labeled MIR162b or pre-miR162b could not be retained in the MBP-PRL1 complex. These results suggested that PRL1 binds RNAs in vitro. We next tested the binding ability of MBP-PRL1 to dsRNA, ssRNA and DNA using the in vitro RNA pull-down assay described above. MBP-PRL1 was able to bind a ∼100-nt RNA corresponding to a portion of the 5′ end of the UBIQUITIN 5 (UBQ5) [13], but not an in vitro synthesized ∼50 bp DNA fragment [32] and a dsRNA generated through in vitro transcription vitro transcription of a DNA fragment (5′ portion of UBQ5, ∼460 bp) containing the T7 promoter at end of each strand [13] (Fig. 7B).
Fig. 7. PRL1 binds pri-miRNAs in vitro and in vivo. 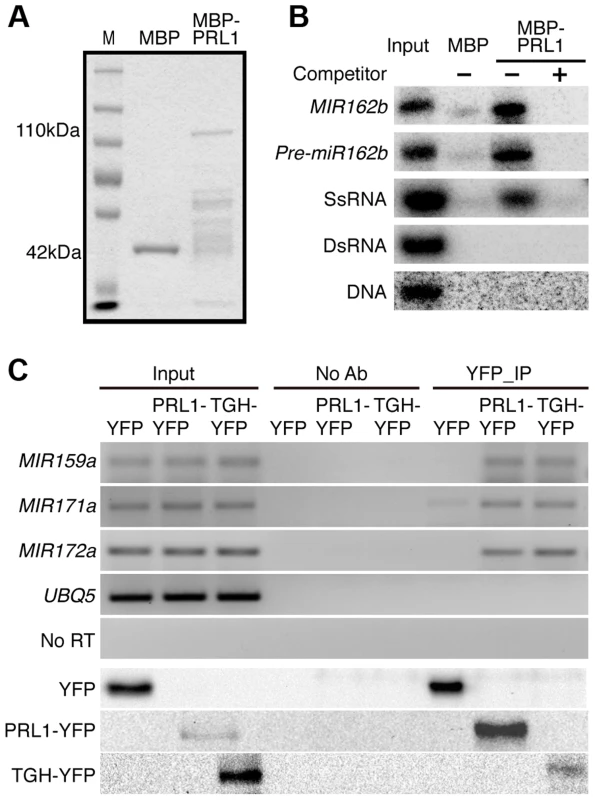
(A) Proteins used for in vitro RNA binding assay. Purified MBP and MBP-PRL1 proteins were resolved on SDS-page gel and stained by Coomassie Blue. The lower bands in MBP-PRL1 line are degraded MBP-PRL1 since they can be detected by anti-MBP antibody. M: marker. (B) PRL1 binds MIR162b, pre-miR162b and ssRNA in vitro. MIR162b, pre-miR162b and ssRNA were produced by in vitro transcription. SsRNA: single stranded-RNA; DsRNA: double stranded-RNA. (C) PRL1 binds pri-miRNAs in vivo. NoAb: No antibody control. Transgenic plants harboring pPRL1::PRL1-YFP, pTGH: TGH-YFP or YFP transgene were used for RIP assay with anti-YFP anti-bodies. Ten percent immunoprecipitates and two percent input proteins were analyzed by western blot. Five percent RNAs were used as input RNA. Next, we performed an RNA immunoprecipitation (RIP) assay to test whether PRL1 binds pri-miRNAs in vivo [13]. Seedlings of the prl1-2 complementation line expressing pPRL1::PRL1-YFP transgene and the control plants harboring YFP were used for RIP. After PRL1-YFP or YFP complex were precipitated with anti-YFP antibody, pri-miRNAs were detected with RT-PCR. Several tested pri-miRNAs (pri-miR159a, pri-miR167a, pri-miR171 and pri-miR172a) existed in the PRL1-YFP complex but not in the YFP complex and no Anti-body (NoAb) controls (Fig. 7C). These results suggested that PRL1 associates with pri-miRNAs in vivo.
Discussion
In this study, we identify PRL1, a WD-40 protein, as an important regulator of miRNA accumulation. Several evidences including reduced accumulation of pri-miRNAs and miRNAs in prl1, PRL1-DCL1 interaction and PRL1-pri-miRNA association demonstrate that PRL1 positively impacts miRNA biogenesis. It has been suggested that PRL1 influences plant immunity and development through its impacts on RNA processing [29], [33]. Given the essential roles of miRNAs in plant immunity and development, it is possible that reduced miRNA levels in prl1 may partially contribute to the observed phenotypes.
PRL1 likely has a role in promoting miRNA maturation, as lack of PRL1 reduces processing of MIR162b and pre-miR162b. PRL1 interacts with the DCL1 complex and does not positively regulate the transcription of genes involved in miRNA biogenesis (Figure S4), suggesting that PRL1 may act as a co-factor to regulate DCL1 activity. CDC5, a direct interactor of PRL1 also regulates the DCL1 activity through its interaction with the helicase and dsRNA binding domains of DCL1. The effect of PRL1 on pri-miRNA processing appears to be weaker than that of CDC5. The processing efficiency of MIR162b and pre-miR162b in cdc5-1 prl1-2 is similar to that in cdc5-1 and slightly lower than that in prl1-2. This result suggests that PRL1 and CDC5 may act together as a complex to regulate DCL1 activity. Furthermore, gel filtration analysis suggests that PRL1 may not affect DCL1-CDC5 association (Figure S5). Thus, it is possible that PRL1 may act as accessory factor to facilitate CDC5 function.
PRL1 also positively regulates the pri-miRNA levels since prl1 reduces the accumulation of pri-miRNAs. We previously showed that CDC5 interacts with Pol II and positively regulate MIR transcription [28]. Since PRL1 associates with Pol II as well, it is possible that PRL1 acts as a component of the CDC5 complex to regulate MIR promoter activity. However this seems not to be the case, as loss-of-function of PRL1 does not affect the GUS levels driven by the MIR167a promoter. Consistent with this notion, the levels of pri-miRNAs are further reduced in cdc5-1 prl1-2 compared with cdc5-1 or prl1-2. Given the fact that PRL1 binds pri-miRNAs in vitro and vivo, we propose that PRL1 may stabilize pri-miRNAs. Indeed, the fact that the half-life of pri-miR164a and pri-miR167a in prl1 is similar to that in Col suggests that the degradation of pri-miRNAs may be increased in prl1, because less efficient processing may lead to increased abundance of pri-miRNAs in prl1. However, we cannot rule out the possibility that PRL1 acts in MIR transcription after initiation, as it associates with Pol II.
In summary, we reveal that PRL1 positively regulates miRNA levels through its impacts on pri-miRNA levels and processing. PRL1 functions additively with its interactor CDC5 as miRNA abundance is lower in cdc5-1 prl1-2 than in cdc5-1 or prl1-2. The synergistic effect of CDC5 and PRL1 on miRNA levels can be explained by their different roles in controlling pri-miRNA levels rather than their function in promoting miRNA maturation. Besides CDC5 and PRL1, the core components MAC complex includes MOS4, MAC3A and MAC3B [29]. We show that MOS4 and MAC3b have no impact on miRNA levels. However, whether MAC3B has a role in miRNA accumulation needs to be further explored since it acts redundantly with MAC3A [29]. The MAC complex appears to have a role in siRNA biogenesis. Both CDC5 and PRL1 promote the accumulation of siRNA [28] while MOS4 is required for the accumulation of ra-siRNAs [34]. How does MAC participate in siRNA biogenesis? We have showed both PRL1 and CDC5 interact with the DCL1 complex and regulate its activity. By analogy, it is possible that the MAC complex associates with the DCL3 complex to regulate its activity. In fact, DCL3 interacts with PRL1. prl1 also reduces the abundance of ta-siRNAs, whose production requires DCL4 and DCL1-dependent miRNAs. Since PRL1 interacts with DCL4 and is required for the accumulation of DCL1-dependent miRNAs, it may promote ta-siRNA production through facilitating DCL4 function and miRNA production. The MAC complex is an evolutionarily conserved complex [29]. As many aspects of small RNA pathway are conserved, it is tempting to propose that the counterparts of MAC play some roles in small RNA pathways in other organisms.
Materials and Methods
Plant materials
The mac3b (SALK_050811), mos4 (SALK_0090851C), prl1-2 (Salk_008466), snc1 (SALK_047058C) and cdc5-1 (SAIL_207_F03) mutants were ordered from Arabidopsis Biological Resources Center (ABRC). All of them are in the Columbia-0 genetic background. Transgenic line containing a single copy of pMIR167a::GUS was crossed to prl1-2. In the F2 population, PRL1/PRL1, PRL1/prl1-2 and prl1-2/prl1-2 harboring pMIR167a::GUS were identified through PCR genotyping for prl1-2 and GUS.
Plasmid construction
PRL1 genomic DNA was amplified from Col genome and cloned to pMDC204 binary vector to generate pPRL1::PRL1-YFP construct. The construct was transformed to prl1-2. The full-length PRL1 cDNA was amplified by RT-PCR and ligated to pMAL-c5x (NEB) to generate MBP-PRL1. To generate the cCFP-PRL1 fusion vector, the PRL1 cDNA was first cloned into the pSAT4-cCFP-C vector [35]. The DNA fragment containing cCFP-PRL1 was released by I-SecI restriction enzyme and subsequently cloned into the pPZP-RCS2-ocs-bar vector. All the primers are listed in Table S1.
Co-IP assay
In the PRL1-PoII co-IP experiment, proteins were extracted from the transgenic plants harboring the pPRL1::PRL1-YFP transgene and incubated with anti-YFP (Clontech) or anti-RPB2 antibodies coupled to protein G agarose beads (Clontech) for 4 h at 4°C. After the beads were washed five times with protein extraction buffer, proteins were resolved by SDS/PAGE. Anti-YFP and anti-RPB2 antibodies were then used to detect PRL1-YFP and RPB2, respectively. To test the interaction of PRL1 with components of the DCL1 complex, PRL1-YFP (YFP) was co-expressed with DCL1-MYC or SE-MYC in N. benthamiana. Protein extracts were then incubated with anti-YFP or anti-MYC antibodies coupled to protein G agarose beads. Anti-YFP and anti-MYC (MBL) antibodies were used to detect PRL1-YFP/YFP and DCL1-MYC/SE-MYC, respectively.
Dicer activity assay
In vitro dicer activity assay was performed according to Qi et al and Ren et al [13], [31]. MIR162b and pre-miR162b RNAs were produced by in vitro transcription under the presence of [α-32P] UTP. In the dicer activity assay, protein extractions were incubated with [32P] labeled MIR162b or pre-miR162b in reaction buffer containing 100 mM NaCl, 1 mM ATP, 0.2 mM GTP, 1.2 mM MgCl2, 25 mM creatine phosphate, 30 µg/ml creatine kinase and 4 U RNase inhibitor at 25°C. After the reactions were stopped at 40, 80 or 120 mins, respectively, RNAs were extracted and resolved on PAGE gel. Radioactive signals were detected with a PhosphorImager and quantified by ImageQuant version 5.2.
BiFC assay
Paired cCFP and nVenus constructs were co-infiltrated in N. benthamiana leaves for 40 h. YFP signals were then detected with a confocal microscopy (Fluoview 500 workstation; Olympus) at 488 nm with a narrow barrier (505–525 nm, BA505–525; Olympus).
RNA analysis
Northern blot was used to detect small RNA abundance as described [13]. qRT - PCR was performed to detect the levels of pri-miRNAs, transcripts of miRNA targets and GUS using cDNA templates reverse transcribed by the SuperScript III (Invitrogen) and oligo dT18 primer. qRT-PCR was run on an iCycler apparatus (Bio-Rad). RNA pull-down were performed according to Ren et al [13]. MBP and MBP-PRL1 were expressed in E.coli. MIR162b, pre-miR162b, dsRNAs and ssRNA were produced by in vitro transcription with T7 RNA polymerase at the presence [α-32P] UTP whereas DNA was synthesized at IDT and labeled with T4 PNK at the presence [γ-32P] ATP. [32P]-labeled probes are incubated with amylose resin beads combined MBP or MBP-PRL1 at 4°C for 1 hour. After 4 times wash with washing buffer, DNA or RNA are extracted and resolved on PAGE gel. Radioactive signals were detected with a PhosphorImager and quantified by ImageQuant version 5.2. RIP was performed according to [13], [36]. Seedlings of transgenic plants harboring the pPRL1::PRL1-YFP transgene or YFP were used to examine the RNA binding activity of PRL1 in vivo. All the primers are listed in Table S1.
RNA half-life assay was performed according to Lidder et al [30]. Two-week-old Col and prl1-2 seedlings were transferred to flask with incubation buffer (1/2 MS medium), respectively. After 30 min incubation, 3′-deoxyadenosine (Cordycepin, Sigma) was added to final concentration of 0.6 mM (time 0). Seedlings were collected at various time points (0, 15, 30, 60, 90, 120 and 240 min). qRT - PCR then was performed to detect the transcript levels of pri-miRNAs and DDL. For quantification, the transcript levels of pri-miRNAs and DDL at various time points were normalized to that of eIF4a, respectively. Value of time 0 was set to 1. Error bars indicate standard deviation of three technical replications. Three biological repeats were performed and similar results were obtained.
Gel filtration analysis
The gel filtration was performed on an HPLC system and a HiPrep 16/60 Sephacryl S-300 HR column (GE Healthcare) at a rate of 0.5 ml/min, and 0.5 ml fractions were collected every minute. Fractions were separated by 8% SDS–PAGE and analyzed by Western blotting using antibodies recognizing CDC or YFP. The protein standards (Bio-Rad, http://www.bio-rad.com/) were used to calibrate the column contain five size standards.
Supporting Information
Zdroje
1. BartelDP (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116 : 281–297.
2. ChenX (2005) MicroRNA biogenesis and function in plants. FEBS Lett 579 : 5923–5931.
3. VoinnetO (2009) Origin, biogenesis, and activity of plant microRNAs. Cell 136 : 669–687.
4. BrodersenP, VoinnetO (2006) The diversity of RNA silencing pathways in plants. Trends Genet 22 : 268–280.
5. HerrAJ, BaulcombeDC (2004) RNA silencing pathways in plants. Cold Spring Harb Symp Quant Biol 69 : 363–370.
6. KimYJ, ZhengB, YuY, WonSY, MoB, et al. (2011) The role of Mediator in small and long noncoding RNA production in Arabidopsis thaliana. EMBO J 30 : 814–822.
7. WangL, SongX, GuL, LiX, CaoS, et al. (2013) NOT2 proteins promote polymerase II-dependent transcription and interact with multiple MicroRNA biogenesis factors in Arabidopsis. Plant Cell 25 : 715–727.
8. YuB, BiL, ZhengB, JiL, ChevalierD, et al. (2008) The FHA domain proteins DAWDLE in Arabidopsis and SNIP1 in humans act in small RNA biogenesis. Proc Natl Acad Sci U S A 105 : 10073–10078.
9. SongL, HanMH, LesickaJ, FedoroffN (2007) Arabidopsis primary microRNA processing proteins HYL1 and DCL1 define a nuclear body distinct from the Cajal body. Proc Natl Acad Sci U S A 104 : 5437–5442.
10. ParkW, LiJ, SongR, MessingJ, ChenX (2002) CARPEL FACTORY, a Dicer homolog, and HEN1, a novel protein, act in microRNA metabolism in Arabidopsis thaliana. Curr Biol 12 : 1484–1495.
11. VazquezF, GasciolliV, CreteP, VaucheretH (2004) The nuclear dsRNA binding protein HYL1 is required for microRNA accumulation and plant development, but not posttranscriptional transgene silencing. Curr Biol 14 : 346–351.
12. YangL, LiuZ, LuF, DongA, HuangH (2006) SERRATE is a novel nuclear regulator in primary microRNA processing in Arabidopsis. Plant J 47 : 841–850.
13. RenG, XieM, DouY, ZhangS, ZhangC, et al. (2012) Regulation of miRNA abundance by RNA binding protein TOUGH in Arabidopsis. Proc Natl Acad Sci U S A 109 : 12817–12821.
14. RenG, YuB (2012) Critical roles of RNA-binding proteins in miRNA biogenesis in Arabidopsis. RNA Biol 9 : 1424–1428.
15. DongZ, HanMH, FedoroffN (2008) The RNA-binding proteins HYL1 and SE promote accurate in vitro processing of pri-miRNA by DCL1. Proc Natl Acad Sci U S A 105 : 9970–9975.
16. FangY, SpectorDL (2007) Identification of nuclear dicing bodies containing proteins for microRNA biogenesis in living Arabidopsis plants. Curr Biol 17 : 818–823.
17. FujiokaY, UtsumiM, OhbaY, WatanabeY (2007) Location of a possible miRNA processing site in SmD3/SmB nuclear bodies in Arabidopsis. Plant Cell Physiol 48 : 1243–1253.
18. ManavellaPA, HagmannJ, OttF, LaubingerS, FranzM, et al. (2012) Fast-forward genetics identifies plant CPL phosphatases as regulators of miRNA processing factor HYL1. Cell 151 : 859–870.
19. SpethC, WillingEM, RauschS, SchneebergerK, LaubingerS (2013) RACK1 scaffold proteins influence miRNA abundance in Arabidopsis. Plant J 76 : 433–445.
20. LaubingerS, SachsenbergT, ZellerG, BuschW, LohmannJU, et al. (2008) Dual roles of the nuclear cap-binding complex and SERRATE in pre-mRNA splicing and microRNA processing in Arabidopsis thaliana. Proc Natl Acad Sci U S A 105 : 8795–8800.
21. GregoryBD, O'MalleyRC, ListerR, UrichMA, Tonti-FilippiniJ, et al. (2008) A link between RNA metabolism and silencing affecting Arabidopsis development. Dev Cell 14 : 854–866.
22. WuX, ShiY, LiJ, XuL, FangY, et al. (2013) A role for the RNA-binding protein MOS2 in microRNA maturation in Arabidopsis. Cell Res 23 : 645–657.
23. ZhanX, WangB, LiH, LiuR, KaliaRK, et al. (2012) Arabidopsis proline-rich protein important for development and abiotic stress tolerance is involved in microRNA biogenesis. Proc Natl Acad Sci U S A 109 : 18198–18203.
24. ZhuH, ZhouY, Castillo-GonzalezC, LuA, GeC, et al. (2013) Bidirectional processing of pri-miRNAs with branched terminal loops by Arabidopsis Dicer-like1. Nat Struct Mol Biol 20 : 1106–1115.
25. MateosJL, BolognaNG, ChorosteckiU, PalatnikJF (2010) Identification of microRNA processing determinants by random mutagenesis of Arabidopsis MIR172a precursor. Curr Biol 20 : 49–54.
26. SongL, AxtellMJ, FedoroffNV (2010) RNA secondary structural determinants of miRNA precursor processing in Arabidopsis. Curr Biol 20 : 37–41.
27. WernerS, WollmannH, SchneebergerK, WeigelD (2010) Structure determinants for accurate processing of miR172a in Arabidopsis thaliana. Curr Biol 20 : 42–48.
28. ZhangS, XieM, RenG, YuB (2013) CDC5, a DNA binding protein, positively regulates posttranscriptional processing and/or transcription of primary microRNA transcripts. Proc Natl Acad Sci U S A 110 : 17588–17593.
29. PalmaK, ZhaoQ, ChengYT, BiD, MonaghanJ, et al. (2007) Regulation of plant innate immunity by three proteins in a complex conserved across the plant and animal kingdoms. Genes Dev 21 : 1484–1493.
30. LidderP, GutierrezRA, SalomePA, McClungCR, GreenPJ (2005) Circadian control of messenger RNA stability. Association with a sequence-specific messenger RNA decay pathway. Plant Physiol 138 : 2374–2385.
31. QiY, DenliAM, HannonGJ (2005) Biochemical specialization within Arabidopsis RNA silencing pathways. Mol Cell 19 : 421–428.
32. XieM, RenG, ZhangC, YuB (2012) The DNA - and RNA-binding protein FACTOR of DNA METHYLATION 1 requires XH domain-mediated complex formation for its function in RNA-directed DNA methylation. Plant J 72 : 491–500.
33. XuF, XuS, WiermerM, ZhangY, LiX (2012) The cyclin L homolog MOS12 and the MOS4-associated complex are required for the proper splicing of plant resistance genes. Plant J 70 : 916–928.
34. ZhangCJ, ZhouJX, LiuJ, MaZY, ZhangSW, et al. (2013) The splicing machinery promotes RNA-directed DNA methylation and transcriptional silencing in Arabidopsis. EMBO J 32 : 1128–1140.
35. LeeLY, FangMJ, KuangLY, GelvinSB (2008) Vectors for multi-color bimolecular fluorescence complementation to investigate protein-protein interactions in living plant cells. Plant Methods 4 : 24.
36. WierzbickiAT, HaagJR, PikaardCS (2008) Noncoding transcription by RNA polymerase Pol IVb/Pol V mediates transcriptional silencing of overlapping and adjacent genes. Cell 135 : 635–648.
Štítky
Genetika Reprodukční medicína
Článek Large-scale Metabolomic Profiling Identifies Novel Biomarkers for Incident Coronary Heart DiseaseČlánek Notch Signaling Mediates the Age-Associated Decrease in Adhesion of Germline Stem Cells to the NicheČlánek Phosphorylation of Mitochondrial Polyubiquitin by PINK1 Promotes Parkin Mitochondrial TetheringČlánek Natural Variation Is Associated With Genome-Wide Methylation Changes and Temperature SeasonalityČlánek Overlapping and Non-overlapping Functions of Condensins I and II in Neural Stem Cell DivisionsČlánek Unisexual Reproduction Drives Meiotic Recombination and Phenotypic and Karyotypic Plasticity inČlánek Tetraspanin (TSP-17) Protects Dopaminergic Neurons against 6-OHDA-Induced Neurodegeneration inČlánek ABA-Mediated ROS in Mitochondria Regulate Root Meristem Activity by Controlling Expression inČlánek Mutations in Global Regulators Lead to Metabolic Selection during Adaptation to Complex EnvironmentsČlánek The Evolution of Sex Ratio Distorter Suppression Affects a 25 cM Genomic Region in the Butterfly
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2014 Číslo 12
-
Všechny články tohoto čísla
- Stratification by Smoking Status Reveals an Association of Genotype with Body Mass Index in Never Smokers
- Genome Wide Meta-analysis Highlights the Role of Genetic Variation in in the Regulation of Circulating Serum Chemerin
- Occupancy of Mitochondrial Single-Stranded DNA Binding Protein Supports the Strand Displacement Mode of DNA Replication
- Distinct Genealogies for Plasmids and Chromosome
- Large-scale Metabolomic Profiling Identifies Novel Biomarkers for Incident Coronary Heart Disease
- Non-coding RNAs Prevent the Binding of the MSL-complex to Heterochromatic Regions
- Plasmid Flux in ST131 Sublineages, Analyzed by Plasmid Constellation Network (PLACNET), a New Method for Plasmid Reconstruction from Whole Genome Sequences
- Epigenome-Guided Analysis of the Transcriptome of Plaque Macrophages during Atherosclerosis Regression Reveals Activation of the Wnt Signaling Pathway
- The Inventiveness of Nature: An Interview with Werner Arber
- Mediation Analysis Demonstrates That -eQTLs Are Often Explained by -Mediation: A Genome-Wide Analysis among 1,800 South Asians
- Generation of Antigenic Diversity in by Structured Rearrangement of Genes During Mitosis
- A Massively Parallel Pipeline to Clone DNA Variants and Examine Molecular Phenotypes of Human Disease Mutations
- Genetic Analysis of the Cardiac Methylome at Single Nucleotide Resolution in a Model of Human Cardiovascular Disease
- Genetic Analysis of Circadian Responses to Low Frequency Electromagnetic Fields in
- The Dissection of Meiotic Chromosome Movement in Mice Using an Electroporation Technique
- Altered Chromatin Occupancy of Master Regulators Underlies Evolutionary Divergence in the Transcriptional Landscape of Erythroid Differentiation
- Syd/JIP3 and JNK Signaling Are Required for Myonuclear Positioning and Muscle Function
- Notch Signaling Mediates the Age-Associated Decrease in Adhesion of Germline Stem Cells to the Niche
- Mutation of Leads to Blurred Tonotopic Organization of Central Auditory Circuits in Mice
- The IKAROS Interaction with a Complex Including Chromatin Remodeling and Transcription Elongation Activities Is Required for Hematopoiesis
- RAN-Binding Protein 9 is Involved in Alternative Splicing and is Critical for Male Germ Cell Development and Male Fertility
- Enhanced Longevity by Ibuprofen, Conserved in Multiple Species, Occurs in Yeast through Inhibition of Tryptophan Import
- Phosphorylation of Mitochondrial Polyubiquitin by PINK1 Promotes Parkin Mitochondrial Tethering
- Recurrent Loss of Specific Introns during Angiosperm Evolution
- Natural Variation Is Associated With Genome-Wide Methylation Changes and Temperature Seasonality
- SEEDSTICK is a Master Regulator of Development and Metabolism in the Arabidopsis Seed Coat
- Overlapping and Non-overlapping Functions of Condensins I and II in Neural Stem Cell Divisions
- Unisexual Reproduction Drives Meiotic Recombination and Phenotypic and Karyotypic Plasticity in
- Tetraspanin (TSP-17) Protects Dopaminergic Neurons against 6-OHDA-Induced Neurodegeneration in
- ABA-Mediated ROS in Mitochondria Regulate Root Meristem Activity by Controlling Expression in
- Mutations in Global Regulators Lead to Metabolic Selection during Adaptation to Complex Environments
- Global Analysis of Photosynthesis Transcriptional Regulatory Networks
- Mucolipin Co-deficiency Causes Accelerated Endolysosomal Vacuolation of Enterocytes and Failure-to-Thrive from Birth to Weaning
- Controlling Pre-leukemic Thymocyte Self-Renewal
- How Malaria Parasites Avoid Running Out of Ammo
- Echoes of the Past: Hereditarianism and
- Deep Reads: Strands in the History of Molecular Genetics
- Keep on Laying Eggs Mama, RNAi My Reproductive Aging Blues Away
- Analysis of a Plant Complex Resistance Gene Locus Underlying Immune-Related Hybrid Incompatibility and Its Occurrence in Nature
- Epistatic Adaptive Evolution of Human Color Vision
- Increased and Imbalanced dNTP Pools Symmetrically Promote Both Leading and Lagging Strand Replication Infidelity
- Genetic Basis of Haloperidol Resistance in Is Complex and Dose Dependent
- Genome-Wide Analysis of DNA Methylation Dynamics during Early Human Development
- Interaction between Conjugative and Retrotransposable Elements in Horizontal Gene Transfer
- The Evolution of Sex Ratio Distorter Suppression Affects a 25 cM Genomic Region in the Butterfly
- is Required for Adult Maintenance of Dopaminergic Neurons in the Ventral Substantia Nigra
- PRL1, an RNA-Binding Protein, Positively Regulates the Accumulation of miRNAs and siRNAs in Arabidopsis
- Genetic Control of Contagious Asexuality in the Pea Aphid
- Early Mesozoic Coexistence of Amniotes and Hepadnaviridae
- Local and Systemic Regulation of Plant Root System Architecture and Symbiotic Nodulation by a Receptor-Like Kinase
- Gene Pathways That Delay Reproductive Senescence
- The Evolution of Fungal Metabolic Pathways
- Maf1 Is a Novel Target of PTEN and PI3K Signaling That Negatively Regulates Oncogenesis and Lipid Metabolism
- Formation of Linear Amplicons with Inverted Duplications in Requires the MRE11 Nuclease
- Identification of Rare Causal Variants in Sequence-Based Studies: Methods and Applications to , a Gene Involved in Cohen Syndrome and Autism
- Rrp12 and the Exportin Crm1 Participate in Late Assembly Events in the Nucleolus during 40S Ribosomal Subunit Biogenesis
- The Mutations in the ATP-Binding Groove of the Rad3/XPD Helicase Lead to -Cockayne Syndrome-Like Phenotypes
- Topoisomerase I Plays a Critical Role in Suppressing Genome Instability at a Highly Transcribed G-Quadruplex-Forming Sequence
- A Cbx8-Containing Polycomb Complex Facilitates the Transition to Gene Activation during ES Cell Differentiation
- Transcriptional Frameshifting Rescues Type VI Secretion by the Production of Two Length Variants from the Prematurely Interrupted Gene
- Association Mapping across Numerous Traits Reveals Patterns of Functional Variation in Maize
- Genome-Wide Analysis of -Regulated and Phased Small RNAs Underscores the Importance of the ta-siRNA Pathway to Maize Development
- Dissemination of Cephalosporin Resistance Genes between Strains from Farm Animals and Humans by Specific Plasmid Lineages
- The Tau Tubulin Kinases TTBK1/2 Promote Accumulation of Pathological TDP-43
- Germline Signals Deploy NHR-49 to Modulate Fatty-Acid β-Oxidation and Desaturation in Somatic Tissues of
- Microevolution of in Macrophages Restores Filamentation in a Nonfilamentous Mutant
- Vangl2-Regulated Polarisation of Second Heart Field-Derived Cells Is Required for Outflow Tract Lengthening during Cardiac Development
- Chondrocytes Transdifferentiate into Osteoblasts in Endochondral Bone during Development, Postnatal Growth and Fracture Healing in Mice
- A ABC Transporter Regulates Lifespan
- RA and FGF Signalling Are Required in the Zebrafish Otic Vesicle to Pattern and Maintain Ventral Otic Identities
- , and Reprogram Thymocytes into Self-Renewing Cells
- The miR9863 Family Regulates Distinct Alleles in Barley to Attenuate NLR Receptor-Triggered Disease Resistance and Cell-Death Signaling
- Detection of Pleiotropy through a Phenome-Wide Association Study (PheWAS) of Epidemiologic Data as Part of the Environmental Architecture for Genes Linked to Environment (EAGLE) Study
- Extensive Copy-Number Variation of Young Genes across Stickleback Populations
- The and Genetic Modules Interact to Regulate Ciliogenesis and Ciliary Microtubule Patterning in
- Analysis of the Genome, Transcriptome and Secretome Provides Insight into Its Pioneer Colonization Strategies of Wood
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Tetraspanin (TSP-17) Protects Dopaminergic Neurons against 6-OHDA-Induced Neurodegeneration in
- Maf1 Is a Novel Target of PTEN and PI3K Signaling That Negatively Regulates Oncogenesis and Lipid Metabolism
- The IKAROS Interaction with a Complex Including Chromatin Remodeling and Transcription Elongation Activities Is Required for Hematopoiesis
- Echoes of the Past: Hereditarianism and
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



